
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Trop. Dis., 29 January 2024
Sec. Major Tropical Diseases
Volume 5 - 2024 | https://doi.org/10.3389/fitd.2024.1287025
This article is part of the Research TopicChallenges For Diagnosis, Treatment And Elimination Of MalariaView all 11 articles
 Leon E. Hugo1
Leon E. Hugo1 Karla van Huyssteen2,3
Karla van Huyssteen2,3 Olamide Oloniniyi2,3
Olamide Oloniniyi2,3 Laura Donnelly2,3
Laura Donnelly2,3 Anna Conn2,3
Anna Conn2,3 Katharine A. Collins4
Katharine A. Collins4 Hayley Mitchell5
Hayley Mitchell5 James S. McCarthy4
James S. McCarthy4 Joanne Macdonald2,3*
Joanne Macdonald2,3*Vector surveillance of Plasmodium falciparum is critical for monitoring and reducing one of the most severe forms of malaria, which causes high morbidity and mortality in children under five and pregnant women. Here we developed a rapid and highly sensitive test for the detection of P. falciparum (Pf)-infected mosquitoes (Rapid Pf test), with high suitability for low-resource vector surveillance implementation. The Rapid Pf test had similar analytical sensitivity to laboratory-based tests, detecting down to 4 copies/μL of a 18S rRNA DNA standard. In addition, the Rapid Pf test could be completed in less than 30 minutes, and only required a liquid sample preparation reagent, pestle, tube, and 39°C heating block for operation, indicating amenability for low-resource implementation. Diagnostic testing was performed using Anopheles stephensi mosquitoes, either uninfected, or fed with P. falciparum gametocyte cultures. These P. falciparum fed mosquitoes were determined to have 79% infection prevalence based on parallel microscopy and qPCR testing on a subset of 19 mosquitoes. However, our Rapid Pf test determined a 90% positive test rate when testing individual infected mosquitoes (n=30), and did not detect 40 uninfected mosquitoes regardless of blood-fed status (n=40), suggesting the true prevalence of infection in the mosquitoes may have been higher than calculated by qPCR and microscopy. The Rapid Pf test was demonstrated to detect infection in individual mosquitoes (both fresh and frozen/thawed), as well as pools of 1 infected mosquito mixed with 19 known uninfected mosquitoes, and individual mosquitoes left in traps for up to 8 days. After testing on infected and uninfected mosquitoes (n=148) the Rapid Pf test was conservatively estimated to achieve 100% diagnostic sensitivity (95% confidence interval, CI: 91%-100%) and 97% diagnostic specificity (CI: 92%-99%) compared to the estimated prevalence from combined microscopy and qPCR results. These results indicate the Rapid Pf test could provide a highly effective tool for weekly surveillance of infected mosquitoes, to assist with P. falciparum monitoring and intervention studies.
Malaria is a mosquito-borne disease that causes significant morbidity and mortality, especially in sub-Saharan Africa (1, 2). Global malaria incidence decreased by 27% between 2000-2015, the result of measures funded by initiatives including Roll Back Malaria, 2000, the Global Fund, 2002, and the United States Presidents Initiative against Malaria, 2005 (3). Concerningly, global malaria incidence increased from 2020-2021, due in part to the disruption of malaria prevention and control strategies by the COVID-19 pandemic (2, 4). In 2021, there were an estimated 247 million cases of malaria, 619,000 deaths and with approximately 95% of these cases occurring in Africa (2).
The causative agents of malaria are protozoan parasites of the genus Plasmodium, which are transmitted by female Anopheles mosquitoes. Of the five species of protozoan parasites that cause human malaria, P. falciparum causes the most severe form of the disease with high morbidity and mortality, especially in children under five and pregnant women. Mosquitoes ingest gametocytes while blood feeding on an infected person. The sexual stage of the parasite life cycle is completed in the mosquito as the gametocytes develop through a series of intermediate stages into oocysts embedded in the walls of the mosquito stomach, or midgut. Sporozoites emerge 10-12 days later (5). Sporozoites migrate to the salivary gland of the mosquito and are transmitted to new host when the mosquito takes its next blood meal. Given the crucial role mosquitoes play in the transmission of malaria, vector control remains an important method in the quest to eradicate malaria (6). Indoor residual spraying (IRS), insecticide treated nets (ITNs), and larval source management are critical entomological tools to achieve this goal (7).
Monitoring the prevalence of Plasmodium infection in mosquito populations is a critical part of the management of malaria. Detection of Plasmodium prevalence in mosquitoes can be applied as a xenomonitoring tool for detecting the presence of human reservoirs of the parasite. Sensitive and non-invasive detection of reservoirs is becoming increasingly important as nations approach malaria elimination.
There are several methods for detection of Plasmodium parasites in mosquitoes. The traditional method and gold standard for detection is dissection and microscopic examination of mosquito midguts for the presence of oocysts (indicating infection of the mosquito) and salivary glands for the presence of sporozoites (indicating the potential for onward transmission of the parasite) (8). However, these methods are time consuming and require expert technical assistance. Circumsporozoite protein enzyme-linked immunosorbent assay (CS-ELISA) is another method which is used to detect circumsporozoite antigen, but there are issues with detection when parasites are in low abundance (9). A number of PCR assays have been developed for the detection of Plasmodium in mosquitoes. In particular, the nested PCR assay designed by Snounou et al. (10) is considered the gold standard among PCR assays for its sensitivity, however post PCR processing of samples is required and the assay requires hours to complete. Newer quantitative PCR (qPCR) assays provide sensitive detection and the ability to differentiate between the separate species of Plasmodium without any requirement for post PCR processing (11–14). These assays, however, require the use of expensive thermocyclers and are not suitable for use in resource poor settings. Several rapid diagnostic tests (RDTs) based on immuno-chromatography assays have been developed for the detection of human infection with Plasmodium, including tests targeting P. falciparum histidine rich protein 2 (HRP2) and Plasmodium lactate dehydrogenase (pLDH) (15–17). However, these tests have not been applied to test infections in mosquitoes and in some cases antigens may not be expressed during the mosquito life stages. A dipstick enzyme immunoassay based test was developed to detect sporozoite protein antigens from P. falciparum and two P. vivax strain infections in mosquitoes (18, 19). This was developed commercially as the VectorTest (formerly Vectest™) Malaria Antigen Panel (VecTOR Test Systems, Thousand Oaks, CA, USA). Rigorous testing demonstrated VecTest to have equivalent sensitivity with marginally lower sensitivity than CS-ELISA (20). However, sensitivity was substantially lower when compared to PCR (21). Novel diagnostic assays include isothermal nucleic acid-based tests that offer the sensitivity of PCR combined with portability and applicability to low-resource settings. These include tests based on Loop mediated isothermal amplification (LAMP) and Recombinase Polymerase Amplification (RPA) (22). These technologies have not been applied for the detection of Plasmodium in mosquitoes or applied in a limited capacity.
Here we report the development of a test for the detection of P. falciparum (Rapid Pf test) in mosquitoes that is suitable for low-resource implementation and offers equivalent sensitivity to PCR. The test uses a novel 10-minute sample preparation method that requires only tube, pestle, and a liquid reagent (23–28). Sensitive 10-minute isothermal amplification of the Plasmodium 18S rRNA gene is performed using recombinase polymerase amplification (RPA) followed by lateral flow detection, as described for other viruses and bacteria (23–34), thus requiring only a single temperature heating block for operation. In this study, we report analytical sensitivity of the test, and demonstrate detection of P. falciparum in experimentally infected Anopheles stephensi mosquitoes. Mosquito testing was performed both individually and in pools, using freshly-infected and frozen mosquitoes, as well as mosquitoes left in traps for up to 8 days in a simulated field environment.
An. stephensi (Sind-Kasur strain) were obtained from the University of Nijmegen, The Netherlands, and maintained in colony at the QIMR Berghofer Medical Research Institute insectary at 27°C ± 1, 70-80% relative humidity and 12:12 hour day:night light cycle. Adult mosquitoes were fed on 8% sucrose with para-aminobenzoic acid (PABA) (14). Female mosquitoes were collected at 3-4 days old, starved for 15 hours, and 100 mosquitoes were placed into paper cups with gauze lids. For P. falciparum-exposed mosquitoes, a mosquito blood-meal (0.22% mature gametocytemia) was prepared by mixing 650 µL of Plasmodium gametocyte infected red blood cells obtained from a P. falciparum (NF54 strain) gametocyte culture with 650 µL of AB blood serum. Mosquitoes were allowed to feed on the blood-meal maintained at 37°C via an artificial membrane feeding apparatus for 30 minutes. After feeding, mosquitoes that were not engorged were discarded and 110 remaining mosquitoes were incubated in an environmental growth chamber (Panasonic MLR-352H-PE, Panasonic, Japan) set at 28 ± 1°C, 75% relative humidity and 12:12 hr light cycle for 10 days.
After incubation, (1) 39 P. falciparum-exposed mosquitoes were dissected to observe oocysts in midguts, and of these, 19 were frozen and then tested by qPCR to determine the prevalence of Plasmodium infection; (2) Forty-eight P. falciparum-exposed mosquitoes were used to trial the RPA test: (i) 20 were tested immediately; (ii) 10 were frozen at –20°C for 7 days and then thawed before testing; (iii) 9 were used for testing pools of mosquitoes, and (iv) 9 were left in traps for 8 days in a chamber that simulated a tropical environment, as described in the next paragraph.
We tested the performance of the Rapid Pf test in a real-world scenario in which mosquitoes were tested only after a holding period in a mosquito trap under field conditions. We placed mosquito samples at 10 d after blood-feeding into gauze bags and placed the bags into a BG Sentinel mosquito trap (Biogents AG, Regensburg, Germany) positioned inside a second environmental growth chamber that was set to simulate tropical ambient conditions, including a daily cycling temperature profile varying between 23.5-31°C, 70% relative humidity and 12:12 hr day:night lighting.
De-Cifer P. falciparum RPA primers and probe (BioCifer Pty Ltd, Brisbane, QLD) were used in the Rapid Pf test, which target the 18S rRNA gene. A 157bp DNA sequence from P. falciparum 18S rRNA gene (accession number KJ170099.1) cloned into the pBIC-A plasmid, was used as the 18S rRNA DNA standard. The concentration (18S gene copies per ml) of the resuspended plasmid was determined using the dsDNA HS Assay Kit (Life Technologies, Singapore).
Mosquitoes were dissected 10 d after feeding, cold-anaesthetized and dissected to remove midguts into PBS. The midguts were then stained using 0.5% mercurochrome in PBS and examined to determine the presence and quantity of oocysts using a brightfield compound microscope.
Quantitative PCR was performed as described by Wang et al. (14) with some modifications. DNA was extracted from dissected midguts using the QIAGEN DNeasy blood and tissue DNA extraction kit (QIAGEN, Hilden, Germany). Samples were homogenized in 50 µl of QIAGEN ATL buffer using a micropestle and stored at -20°C. Samples were thawed, 130 µl of QIAGEN ATL buffer and 20 µL Proteinase K was added to each tube and samples were incubated at 56°C overnight. DNA was then extracted from samples according to the manufacturer’s instructions. Quantitative PCR was performed using the QIAGEN QuantiNova Probe qPCR kit. qPCR reactions consisted of 7.5 µl of QIAGEN QuantiNova 2x qPCR master mix, 1.5 µl of 1:1 dilution of DNA and QuantiNova yellow sample dilution buffer, 0.1 µM of each primer and 0.1 µM of a CY5-conjugated Taqman probe targeting a conserved region of the P. falciparum 18S gene (35) in a total volume of 15 µl. The qPCR assay was performed on a Corbett Rotorgene 6000 thermocycler (Corbett Research, Sydney, Australia) under the following conditions: 95°C for 2 min, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s.
P. falciparum fed individual whole mosquitoes were crushed with a disposable blue polypropylene tube pestle (Sigma-Aldrich, Castle Hill, NSW, AU), in 50 μL of TNA-Cifer Reagent (BioCifer Pty Ltd, Brisbane, QLD), and mosquito pools (1 P. falciparum-exposed mosquito with 19 uninfected mosquitoes, or 20 uninfected mosquitoes) were crushed in 200 μL TNA-Cifer Reagent. Samples were incubated for 10 minutes at room temperature, and then 10 μL was added to 40 μL of RNAse and DNAse-free water and 1 μL immediately used for isothermal amplification.
Sample (1 μL) was mixed with 8 μL recombinase polymerase amplification (RPA) mixture, followed by addition of 1 μL 140 mM Magnesium acetate to start the reactions, and incubation at 39°C for 10 mins. The final concentration of reactants in each tube was: 420 nM Forward and Reverse primers, 120 nM probe, 1x Rehydration buffer & pellet mixture, and 14 mM Magnesium acetate.
Immediately following incubation, 2 μL of the amplicons were transferred to the sample pad of a HybriDetect lateral flow strip (Milenia Biotec GmbH, Gießen, Germany), which detects DNA dual labelled with Biotin/fluorescein. Strips had been pre-prepared by the addition of 8 μL blocking buffer (0.4% casein, 0.1% Tween in PBS, pH 9) to the sample pads of the strips (32). Strips were subsequently placed into tubes with the sample pads immersed in 100 μL of borate buffer (100 mM H3BO3, 100 mM Na2B4O7, 1% BSA, 0.05% Tween 20, pH 8.8), and left to wick along the strips for 5 minutes before appearance of test bands was observed by eye and imaged.
Strips were imaged using the MultiDoc-It™ Digital Imaging System (Upland, CA, USA) or a flatbed scanner (Hewlett-Packard) and analyzed using ImageJ software (National Institutes of Health, MD, USA). Greyscale-converted images were used to determine band-intensity, by measuring the mean grey value (limit to threshold), using a fixed area measurement, and subtracting from the maximum threshold value. For each test band, an average of the neighboring white space of all LF strips in that experiment was subtracted from the band intensity to normalize the results. To define a sample as positive the test band values were standardized by subtracting a cutoff value (three standard deviations above the average of the negative control test band intensities). A standardized value of greater than 0 was a positive result.
Comparative analysis of tests was performed using a diagnostic sensitivity and specificity calculator with exact Clopper-Pearson 95% confidence intervals (36).
To develop a rapid and sensitive test for P. falciparum, we chose recombinase polymerase amplification (RPA) followed by lateral flow detection (LFD) (29–34), which enables sensitive and rapid amplification, but requires only a simple heating block for operation. Analytical sensitivity of our Pf RPA-LFD test (Figure 1) using a quantified DNA standard indicated strips producing clearly visible test lines at all concentrations tested (Figure 1A). Analytical sensitivity analysis, as determined both by eye (Figure 1A) and by image analysis of pixel density (Figures 1B-D), indicated a limit of detection of 4 copies/μL (the lowest concentration evaluated).
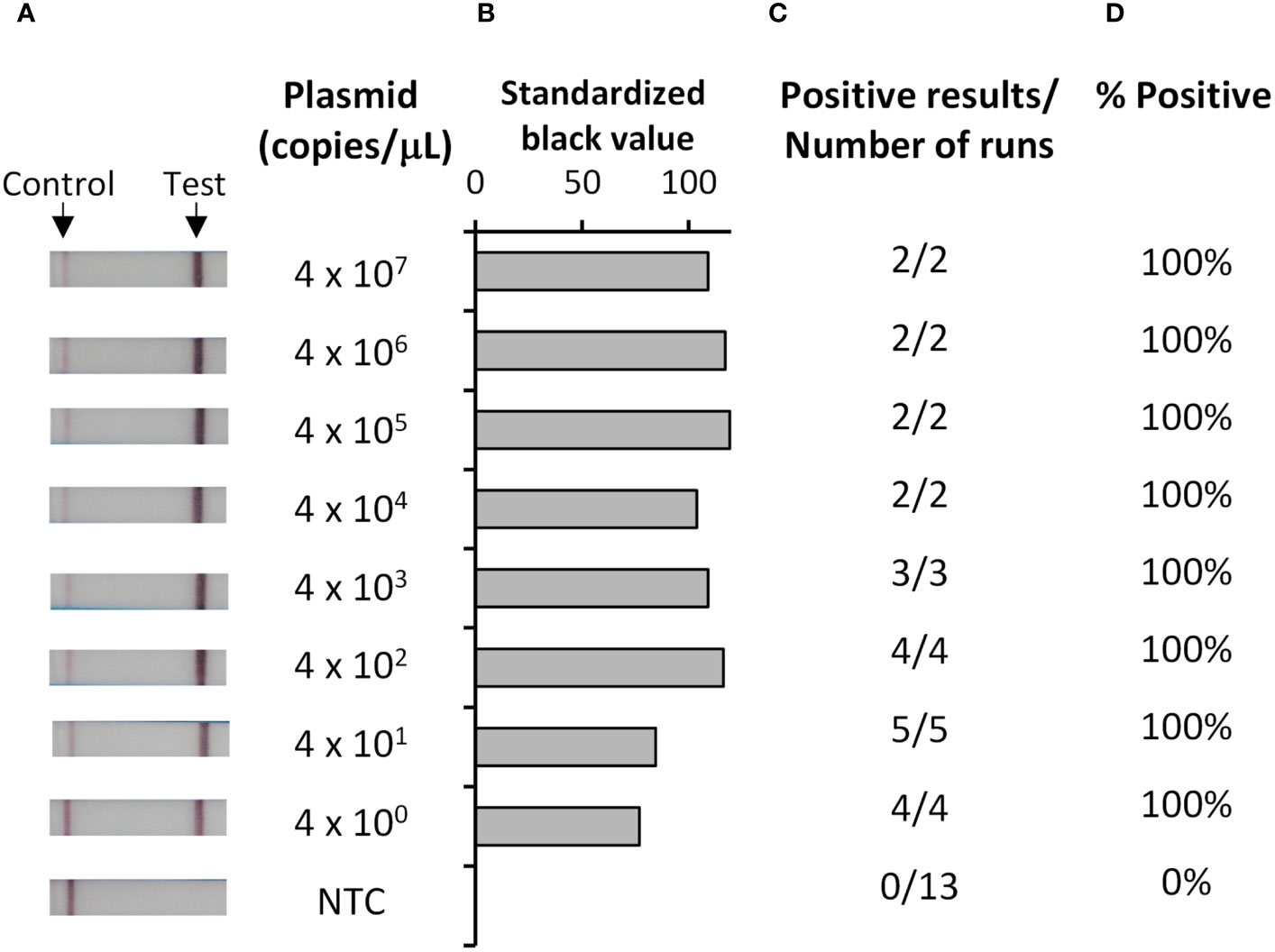
Figure 1 Analytical sensitivity of the Pf RPA-LFD test was 4 copies/µL. Ten-fold dilutions of plasmid containing a portion of the P. falciparum 18S rDNA target gene, or a water no template control (NTC), were tested with the Pf RPA-LFD test. Representative photographs of resultant lateral flow strips (A) show position of control and test lines alongside plasmid concentration (copies/µL) from which standardized black value (B) was determined by ImageJ analysis of test line. The number of times a positive result was obtained per total number of replicate experiments (C) was used to determine percentage positive tests (D).
To evaluate our developed test for detection of infected mosquitoes, we first had to determine the infection prevalence of P. falciparum in a batch of P. falciparum-exposed An. stephensi mosquitoes, which had fed on P. falciparum gametocyte cultures and harvested 10 days post blood-meal. Microscopy was performed on 39 An. stephensi mosquitoes fed on P. falciparum gametocyte cultures. A total of 24 mosquitoes were observed to have oocysts in their midgut (oocysts range 1 – 19, mean 4), indicating a microscopy positive prevalence rate of 62% (24/39; Table 1). While microscopy is considered the gold standard of detection of Plasmodium in mosquitoes, there is the possibility of failing to identify oocysts that have not stained well, or are not clearly visible, especially when present at low numbers. Therefore, DNA was extracted from the midguts of 19 of the mosquitoes already examined by microscopy and evaluated using qPCR, resulting in 14 mosquitoes testing positive by qPCR (Table 2) and indicating a qPCR-positive prevalence rate of 73% (14/19). Two additional microscopy negative mosquitoes were detected as positive by qPCR, and one qPCR negative result contained the largest oocyst number by microscopy, indicating this sample was also positive. By combining results, we determined four mosquitoes were negative by both tests, suggesting an estimated “true” infection prevalence of 79% (15 positives out of 19 mosquitoes; Table 2).
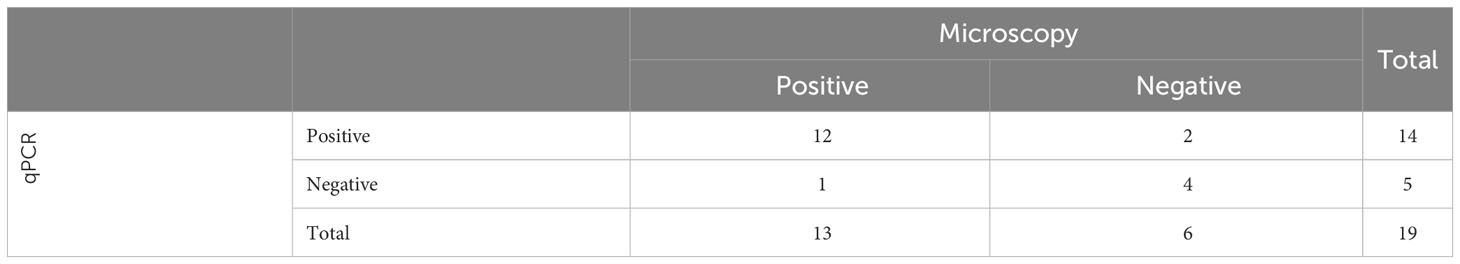
Table 2 Estimation of Infection prevalence for P. falciparum-exposed An. stephensi, determined by microscopy and qPCR on a subset of 19 mosquitoes.
To construct our low-resource Rapid Pf test for detection of infected mosquitoes, our Pf RPA-LFD was combined with a novel TNA-Cifer Reagent (BioCifer Pty Ltd, Brisbane, QLD), which enables low-resource sample preparation for molecular testing (23–28). Thirty P. falciparum-exposed mosquitoes, from the same batch assessed previously by qPCR and microscopy, were tested (20 tested fresh, and 10 frozen and subsequently thawed for testing; Figure 2). Of the P. falciparum-exposed mosquitoes, 27 were positive using the Rapid Pf test, indicating a 90% prevalence of P. falciparum in the blood-fed mosquitoes (Table 1). We also tested 40 known uninfected mosquitoes (20 fed on uninfected blood cultures, and 20 not blood-fed; Supplementary Figure S1), which produced a clear negative result for all uninfected mosquitoes tested, demonstrating 100% diagnostic specificity (95% Confidence interval, CI: 91% - 100%; n=40). These results indicated that the crushed blood-fed mosquito background did not affect test specificity, and that any observable band, regardless of intensity, was indicative of a mosquito that had been fed on P. falciparum. A comparison of microscopy, qPCR and our Rapid Pf test for determination of infection prevalence in the mosquitoes is shown in Table 1. Comparing our Rapid Pf test results to the “gold standard” combined microscopy and qPCR determined infection rate of 79%, our Rapid Pf test would be determined have a diagnostic sensitivity of 100% (95% confidence interval, CI: 86-100%) and 93% specificity (CI:82%-99%). However, as our testing showed improved sensitivity compared to the estimated prevalence from microscopy and qPCR, and did not have false positives when detecting known uninfected mosquitoes, our test is likely more accurate compared to the observed microscopy and qPCR results, with 100% diagnostic sensitivity (CI: 87% to 100%) and 100% diagnostic specificity (CI: 92% to 100%).
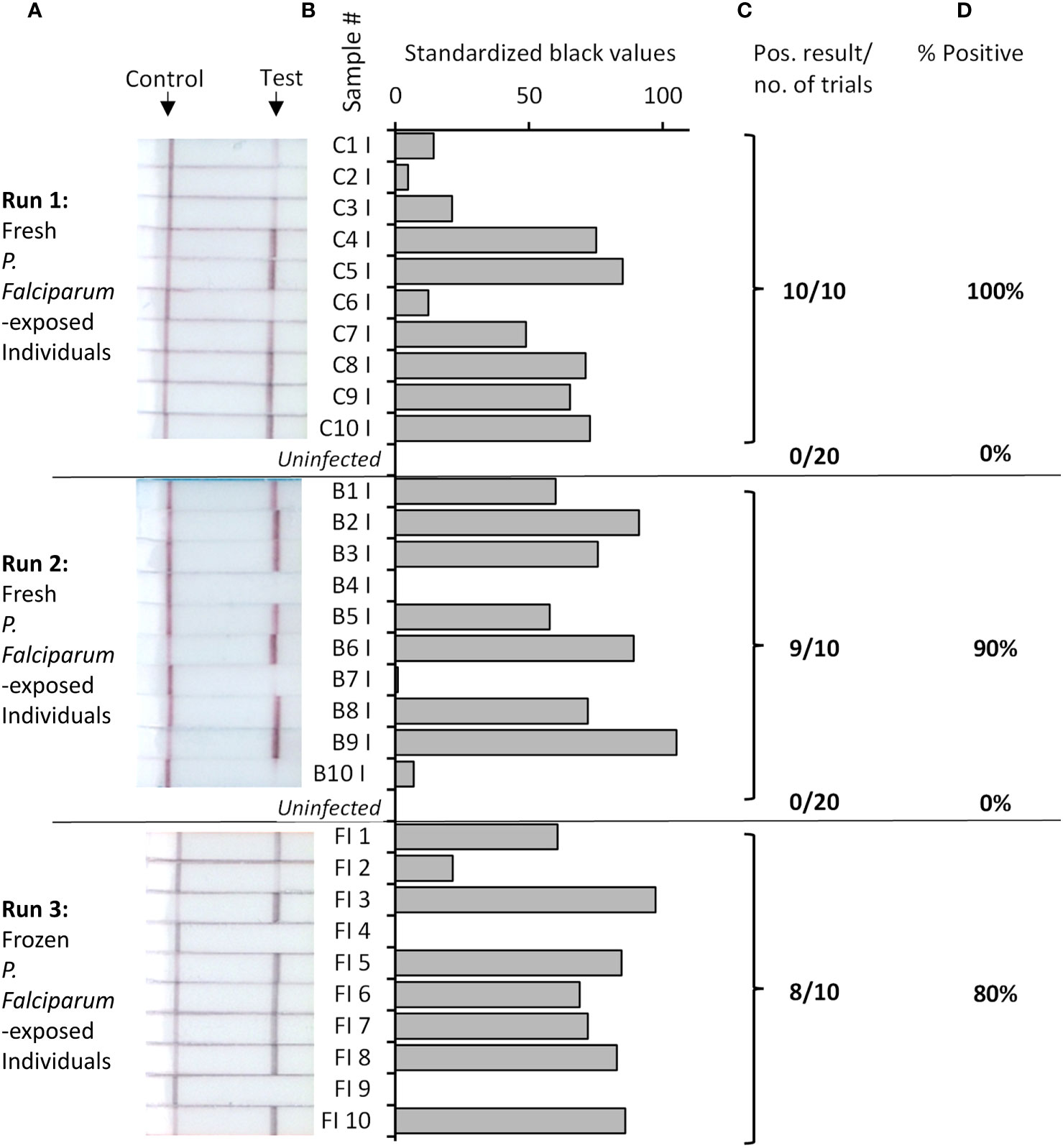
Figure 2 Rapid Pf test applied to detection of P. falciparum-exposed or uninfected An. stephensi mosquitoes. Fresh or frozen P. falciparum-exposed mosquitoes were tested in batches as indicated, alongside uninfected mosquitoes (pictures of all uninfected mosquitoes strips provided in Supplementary Figure S1). Photographs of resultant lateral flow strips (A) show position of control and test lines alongside mosquito sample groups from which standardized black value (B) was determined by ImageJ analysis of test line. The number of times a positive result was obtained per total number of samples tested (C) was used to determine percentage positive (D).
Parasite detection in wild caught mosquitoes is usually performed from pools of mosquitoes, due to the large number collected for examination in the field. We therefore tested if our Rapid Pf test could detect parasites when a single P. falciparum-exposed mosquito was pooled with 19 known uninfected mosquitoes. Because of the larger sample mass, pools were homogenized in 200 μL TNA-Cifer Reagent. Eight of 9 pools were determined to be positive (Figure 3, top panel), indicating an 89% positive rate, which was consistent with the individual mosquito testing estimates. All three pools containing only uninfected mosquitoes were negative, confirming the specificity of the Rapid Pf test. We also sought to determine if the rapid P. falciparum test could detect infected mosquitoes left in traps for up to a week, to simulate a weekly testing regime. Individual P. falciparum-exposed mosquitoes were left in traps for 8 days in a chamber that simulated a tropical environment. Six of 9 individual P. falciparum-exposed mosquitoes tested using the rapid P. falciparum test were positive after being held in these conditions (Figure 3, bottom panel). This 67% positive rate is lower than the positive rate obtained from individual and fresh pool testing, but within the range of the standard error, indicating the rapid P. falciparum test could be informative for weekly trap testing regimes. A summary of results from all mosquito trials and the resultant positive rate is provided in Table 3. Using the results from all Rapid Pf trials combined, and comparing our test results to the observed combined microscopy and qPCR determined infection rate of 79%, our test is conservatively calculated to have a diagnostic sensitivity of 100% (CI: 91-100%) and 97% specificity (CI:92%-99%).
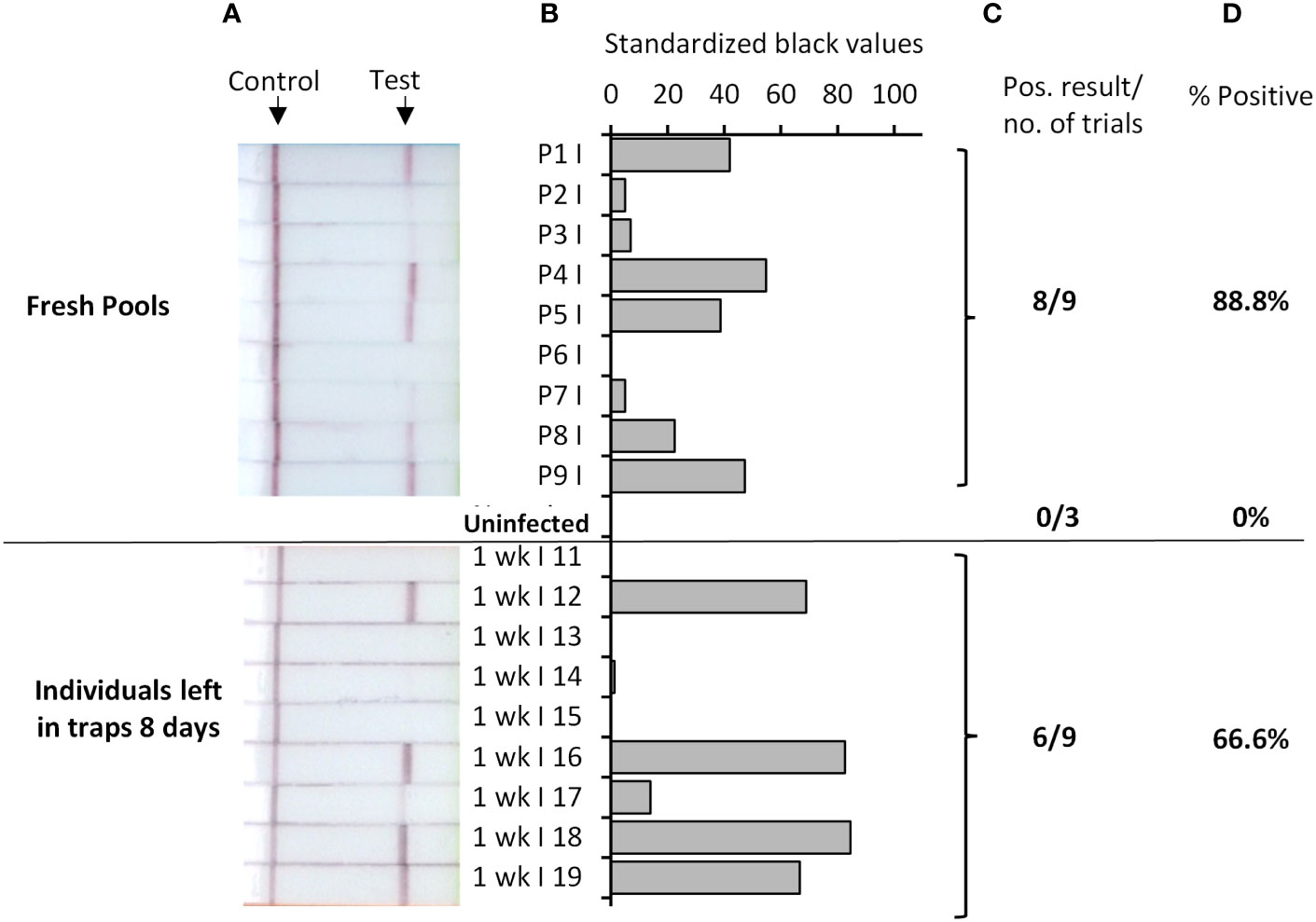
Figure 3 Rapid Pf test applied to detection of P. falciparum-exposed An. stephensi mosquitoes in pools (top), or when left in traps for up to 8 days (bottom). Top panel shows test results for P. falciparum-exposed pools that each contained one An. stephensi mosquito fed from P. falciparum gametocyte blood cultures, mixed with 19 known uninfected mosquitoes (or 20 known uninfected mosquitoes; Uninfected pools). Bottom panel shows results from testing individual P. falciparum-exposed An. stephensi mosquitoes (fed with P. falciparum infected blood cultures), that were subsequently left in environmental chambers simulating a tropical environment for 8 days, prior to testing. Photographs of resultant lateral flow strips (A) show position of control and test lines alongside mosquito sample groups from which standardized black value (B) was determined by ImageJ analysis of test line. The number of times a positive result was obtained per total number of samples tested (C) was used to determine percentage positive (D).
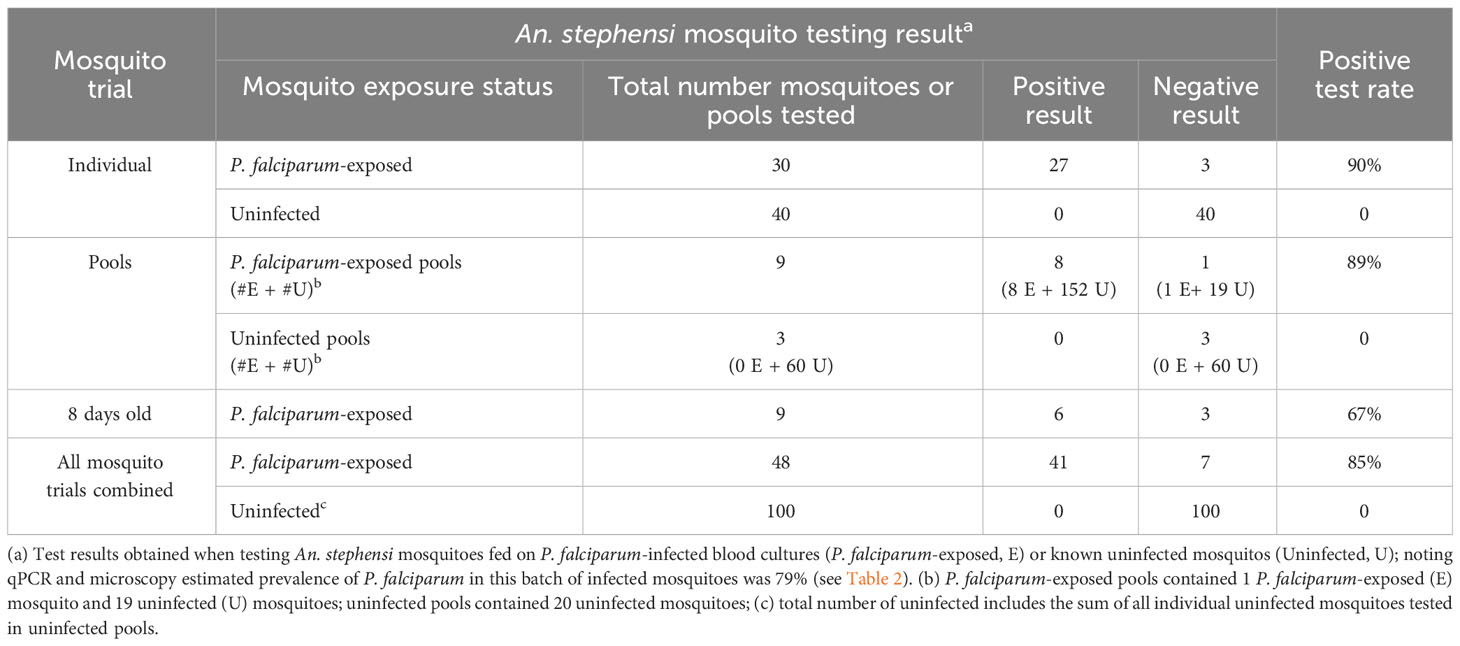
Table 3 Positive test rate of the Rapid Pf test when applied to the different trials performed in this study.
Elimination of malaria is an expressly stated goal in both the WHO Millennium Development Goals (MDG) and the current Sustainable Development Goals (SDG; Target 3.3) (37). To achieve these goals, improved vector surveillance and monitoring/evaluation of interventions aimed at vector control is critical. We sought to improve vector surveillance capabilities by developing a rapid test for detecting mosquitoes infected with P. falciparum, which causes the most severe form of malaria, causing high morbidity and mortality in children under five and pregnant women. Here we report a rapid P. falciparum test (Rapid Pf test) with specifications equivalent to laboratory-based tests, but with potential to be implemented in low-resource settings and be effective under the rigors of standard vector screening protocols, such as testing of mosquitoes collected in traps and left for up to one week, or processing large numbers of mosquito pools.
Our Rapid Pf test is a molecular test, yet suitable for implementation in low-resource settings. The test employs a novel TNA-Cifer Reagent that prepares the mosquito sample for molecular testing in as little as 10 minutes, using only a liquid reagent, tube, and pestle. This reagent has previously been demonstrated useful for detection of viruses (23–28), and here we report, for the first time, the utility of the reagent for detection of a parasitic infection. After sample preparation, a simple dilution step reduces inhibitory amounts of reagent and mosquito debris, allowing an isothermal amplification protocol that requires incubation for only 10 minutes using a simple 39˚ C heating block. Detection is performed using a lateral flow strip, such that the appearance of a test band indicates presence of P. falciparum. The simplicity of the test is exemplified when considering what equipment is not required for testing, including standard DNA extraction and testing equipment: centrifuges, silica membrane spin-columns, magnetic beads, and thermocycling machines. In addition, the entire test, from sample to result, can be performed in less than 30 minutes.
Testing of detection parameters in this study demonstrated excellent analytical sensitivity, down to 4 copies/μL of a P. falciparum 18S rRNA DNA standard. Diagnostic testing of P. falciparum-exposed or known uninfected individual mosquitoes indicated the Rapid Pf test detected more positives than would have been estimated by microscopy and qPCR, resulting in 100% diagnostic sensitivity (CI: 86%-100%), but lowering the diagnostic specificity (93%; CI: 82-99%). Further pool testing, however, continued to demonstrate excellent diagnostic specificity when testing known uninfected mosquitoes. Combining all testing results together (n=148), the diagnostic sensitivity was conservatively calculated to be 100% (CI: 91-100%), and diagnostic specificity was 97% (CI: 92%-99%). Testing mosquito pools demonstrated the Rapid Pf test could detect a single P. falciparum-exposed mosquito when mixed with 19 uninfected mosquitoes, giving the same diagnostic sensitivity and specificity of testing compared to individual mosquito testing. In addition, we demonstrated the Rapid Pf test could detect mosquitoes left in tropical humidity conditions for up to 8 days with similar positive rates to microscopy, qPCR, and fresh or frozen/thawed Rapid Pf test results. These results indicate our Rapid Pf test is highly suitable for low-resource implementation, and could enable effective weekly surveillance of P. falciparum prevalence in hot-spot areas.
The diagnostic specificity and sensitivity of our Rapid Pf test is class-leading for rapid assays detecting Plasmodium in mosquitoes. The only commercial rapid assay specifically designed for detection of Plasmodium in mosquitoes is the Vectortest Malaria Sporozoite Antigen Assay. A large multicenter trial found the assay had an overall diagnostic sensitivity of 92% and specificity of 98.1% when compared against the circumsporozoite (CS) ELISA assay (19). However, sensitivities dropped to below 18% when compared against PCR as the gold standard (21). Our experiments were designed to overcome inherent difficulties in determining the “true” positives for the purpose of determined the diagnostic sensitivity and specificity of our assay. We calculated “true” prevalence from a combination of positive determined by microscopy (which can underestimate prevalence) and qPCR (which may detect residual non-oocyst DNA and can be affected by intracellular inhibitors). Like qPCR and other molecular assays, our Rapid Pf test may also detect P. falciparum residual DNA from mosquito feeding but previous research indicates this is unlikely (14). However, our observation that a single midgut containing a large number of oocysts was qPCR negative was unexpected, which could be due to a sample processing error or PCR inhibition from mercurochrome. Regardless, testing with our Rapid Pf test showed higher positive rates compared to both microscopy and qPCR combined, and did not show false-positive results when testing uninfected mosquitoes, regardless of blood-fed status. These results highlight the difficulty in determining diagnostic sensitivity and specificity when a new test shows improved results compared to the gold standard, which could result in potentially true positives being labelled as false positives.
To the best of our knowledge, our Rapid Pf test is the first assay to apply RPA and LF to the detection of Plasmodium infection in mosquitoes. As an indirect means of comparison, the diagnostic specificity of our Rapid Pf test was equivalent to an 18S P. falciparum RPA and LF assay developed for clinical application (38). Specificity to P. falciparum was 100% among genomic DNA samples obtained from a range of prokaryotic and eukaryotic organisms and sensitivity was 100 fg of gDNA. Similarly, an 18S RPA-LF assay against Plasmodium knowlesi achieved 100% specificity and sensitivity of 10 parasites per µl (39). As with other RDTs targeting Plasmodium 18S, the Rapid Pf test will detect all P. falciparum life stages present in mosquitoes and will not be limited to the detection of sporozoites. Without specific quantification of sporozoites, it is not possible to directly calculate the entomological inoculation rate for specific epidemiological studies (9). However, targeting P. falciparum 18S as a ubiquitous marker of infection provides increased sensitivity (39). Detection of 18S provides a sensitive assay for the detection of parasite reservoirs and a high-throughput means of determining infectivity. Strong agreement between 18S and CS ELISA detections from the same mosquitoes provides a rationale for testing 18S as a high throughput proxy measure for transmission (40). Oocyst infection intensities from our experimentally infected mosquitos were lower than that observed from a study of Plasmodium infection intensities in wild-caught mosquitoes (41). The arithmetic mean number of oocysts for our infected mosquitoes was 4.72 oocysts per midgut, whereas the mean number of oocytes that developed in wild-collected mosquitoes ranged from 10.3 to 14.7. Thus, while additional studies testing more Anopheles spp., including mosquitoes naturally infected with P. falciparum, are required to confirm test diagnostic sensitivity, our results suggest that our test will also perform well with field-caught infected mosquitoes.
Limitations of this study include the inability to use the same panel of mosquitoes for both PCR and rapid testing, as to demonstrate the simplicity of the TNA-Cifer Reagent sample preparation, an entire mosquito was used; testing if TNA-Cifer Reagent prepared samples could be used for parallel PCR testing would also be valuable, as this could assist with standard diagnostic sensitivity testing. Testing more replicates and pools for mosquitoes stored for 1 week, as well as frozen mosquitoes, would also be valuable; while our study showed some drop in the number of positives, however, these differences were not statistically significant due to the sample size tested, and were still equivalent to the infection rate of our mosquito population when estimated by PCR and microscopy. In addition, analytical specificity should be confirmed by testing mosquitoes infected with other Plasmodium species and other pathogens. Further replicate studies (n=>20 at each DNA concentration) should be performed to confirm the true limit of detection as well as repeatability and reproducibility. In addition, a simple lateral flow strip reader would assist with low-resource interpretation of results. We also note that a low-resource test should also remain low cost, noting that cost is often country-dependent, and calculation of cost is outside the scope of this publication.
In conclusion, in this study we report a Rapid Pf test with similar analytical sensitivity to laboratory-based molecular testing; detecting down to 4 copies/µL of a 18S rRNA DNA standard. The Rapid Pf test was superior to microscopy and equivalent to qPCR, yet the entire Rapid Pf test could be completed in less than 30 minutes, and only required a liquid sample preparation reagent, pestle, tube, and 39°C heating block for operation, indicating suitability for low-resource implementation. Testing demonstrated excellent diagnostics sensitivity (100%, CI: 91-100%) and specificity (97%, CI: 92-99%) (n=148). The test successfully detected infection in individual mosquitoes both fresh and frozen/thawed, as well as pools of 1 P. falciparum-exposed mosquito mixed with 19 known uninfected mosquitoes. This, combined with detection of individual mosquitoes left in traps for up to 8 days, indicates our Rapid Pf test could provide a highly effective tool for weekly surveillance of P. falciparum levels in infected mosquitoes, to assist with elimination of malaria. Having demonstrated highly sensitive and robust detection of total P. falciparum life stages in mosquitoes, further developments will include detection of sporozoites to determine sporozoite rates.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
LH: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. Kv: Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. OO: Data curation, Formal analysis, Investigation, Validation, Visualization, Writing – original draft. LD: Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. AC: Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. KC: Investigation, Resources, Writing – review & editing. HM: Investigation, Resources, Writing – review & editing. JaM: Resources, Supervision, Writing – review & editing. JoM: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This project was funded by the Bill and Melinda Gates Foundation [OPP1140133]. Under the grant conditions of the Foundation, a Creative Commons Attribution 4.0 Generic License has already been assigned to the author Accepted Manuscript version that might arise from this submission. The project was also funded by the National Foundation of Medical Research Innovation and the NSW Department of Primary Industries. The Bill and Melinda Gates Foundation, the National Foundation of Medical Research Innovation, and the NSW Department of Primary Industries had no involvement in the study design, collection, analysis, interpretation of data, the writing of this article, or the decision to submit it for publication.
The authors are grateful to Matthew Adams, Greg Robinson, and Bridget Barber (Clinical Tropical Medicine Laboratory, QIMR-Berghofer Medical Research Institute) for assistance with mosquito infection and dissections, and guidance with P. falciparum qPCR. The authors are also grateful to Claire Wang (Queensland Paediatric Infectious Diseases (QPID) Research Group, University of Queensland) for Guidance with Plasmodium qPCR and provision of qPCR standards. The authors are also grateful to Nina Pollak (Centre for Bioinnovation, University of the Sunshine Coast) for assistance with supervision of students.
JoM is a co-founder, shareholder, and director of BioCifer Pty. Ltd., who has licensed the technology. Information in this paper has been included in an International Patent Application PCT/AU2022/051506 PCT filing date 14th Dec 2022; Applicants BioCifer Pty Ltd, University of the Sunshine Coast, DMTC Limited; Title: Sample preparation reagents; Inventor: JoM.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fitd.2024.1287025/full#supplementary-material
1. Phillips MA, Burrows JN, Manyando C, van Huijsduijnen RH, Van Voorhis WC, Wells TNC. Malaria. Nat Rev Dis Primers (2017) 3(1):17050. doi: 10.1038/nrdp.2017.50
2. World Malaria Report 2022. Geneva: World Health Organization (2022). Licence: CC BY-NC-SA 3.0 IGO.
3. Cohen JM, Kandula D, Smith DL, Le Menach A. How long is the last mile? Evaluating successful malaria elimination trajectories. Malaria J (2022) 21(1):330. doi: 10.1186/s12936-022-04368-3
4. González-Sanz M, Berzosa P, Norman FF. Updates on malaria epidemiology and prevention strategies. Curr Infect Dis Rep (2023) 25:131–9. doi: 10.1007/s11908-023-00805-9
5. Aly AS, Vaughan AM, Kappe SH. Malaria parasite development in the mosquito and infection of the mammalian host. Annu Rev Microbiol (2009) 63:195–221. doi: 10.1146/annurev.micro.091208.073403
6. Bhatt S, Weiss DJ, Cameron E, Bisanzio D, Mappin B, Dalrymple U, et al. The effect of malaria control on Plasmodium falciparum in Africa between 2000 and 2015. Nature (2015) 526(7572):207–11. doi: 10.1038/nature15535
7. World Malaria Report 2019. Geneva: World Health Organization (2019) p. 1–232. Licence: CC BY-NC-SA 3.0 IGO.
8. Timinao L, Vinit R, Katusele M, Koleala T, Nate E, Czeher C, et al. Infectivity of symptomatic malaria patients to Anopheles farauti colony mosquitoes in Papua New Guinea. Front Cell Infect Microbiol (2021) 11:771233. doi: 10.3389/fcimb.2021.771233
9. Wirtz RA, Zavala F, Charoenvit Y, Campbell GH, Burkot TR, Schneider I, et al. Comparative testing of monoclonal antibodies against Plasmodium falciparum sporozoites for ELISA development. Bull World Health Organ (1987) 65(1):39–45.
10. Snounou G, Viriyakosol S, Zhu XP, Jarra W, Pinheiro L, do Rosario VE, et al. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol Biochem Parasitol (1993) 61(2):315–20. doi: 10.1016/0166-6851(93)90077-b
11. Bass C, Nikou D, Blagborough AM, Vontas J, Sinden RE, Williamson MS, et al. PCR-based detection of Plasmodium in Anopheles mosquitoes: A comparison of a new high-throughput assay with existing methods. Malar J (2008) 7:177. doi: 10.1186/1475-2875-7-177
12. Kefi M, Mavridis K, Simoes ML, Dimopoulos G, Siden-Kiamos I, Vontas J. New rapid one-step PCR diagnostic assay for Plasmodium falciparum infective mosquitoes. Sci Rep (2018) 8(1):1462. doi: 10.1038/s41598-018-19780-6
13. Bass C, Williamson MS, Field LM. Development of a multiplex real-time PCR assay for identification of members of the Anopheles gambiae species complex. Acta Trop (2008) 107(1):50–3. doi: 10.1016/j.actatropica.2008.04.009
14. Wang CYT, McCarthy JS, Stone WJ, Bousema T, Collins KA, Bialasiewicz S. Assessing Plasmodium falciparum transmission in mosquito-feeding assays using quantitative PCR. Malaria J (2018) 17(1):249. doi: 10.1186/s12936-018-2382-6
15. Oyegoke OO, Maharaj L, Akoniyon OP, Kwoji I, Roux AT, Adewumi TS, et al. Malaria diagnostic methods with the elimination goal in view. Parasitol Res (2022) 121(7):1867–85. doi: 10.1007/s00436-022-07512-9
16. Mouatcho JC, Goldring JPD. Malaria rapid diagnostic tests: challenges and prospects. J Med Microbiol (2013) 62(Pt 10):1491–505. doi: 10.1099/jmm.0.052506-0
17. Cunningham J, Jones S, Gatton ML, Barnwell JW, Cheng Q, Chiodini PL, et al. A review of the WHO malaria rapid diagnostic test product testing programme (2008-2018): performance, procurement and policy. Malar J (2019) 18(1):387. doi: 10.1186/s12936-019-3028-z
18. Coleman RE, Barth JF, Turell MJ, Gordon SW, Sattabongkot J, Copeland R, et al. Development and evaluation of a dipstick assay for detection of Plasmodium falciparum and P. vivax sporozoites in mosquitoes (Diptera: Culicidae). J Med Entomol (2000) 37(4):581–7. doi: 10.1603/0022-2585-37.4.581
19. Ryan JR, Davé K, Collins KM, Hochberg L, Sattabongkot J, Coleman RE, et al. Extensive multiple test centre evaluation of the vectest malaria antigen panel assay. Med Vet Entomol (2002) 16(3):321–7. doi: 10.1046/j.1365-2915.2002.00368.x
20. Sattabongkot J, Kiattibut C, Kumpitak C, Ponlawat A, Ryan JR, Chan AST, et al. Evaluation of the VecTest malaria antigen panel assay for the detection of Plasmodium falciparum and P. vivax circumsporozoite protein in anopheline mosquitoes in Thailand. J Med Entomology (2004) 41(2):209–14. doi: 10.1603/0022-2585-41.2.209
21. Moreno M, Cano J, Nzambo S, Bobuakasi L, Buatiche JN, Ondo M, et al. Malaria panel assay versus PCR: detection of naturally infected Anopheles melat in a coastal village of Equatorial Guinea. Malar J (2004) 3:20. doi: 10.1186/1475-2875-3-20
22. Lalremruata A, Nguyen TT, McCall MBB, Mombo-Ngoma G, Agnandji ST, Adegnika AA, et al. Recombinase polymerase amplification and lateral flow assay for ultrasensitive detection of low-density Plasmodium falciparum infection from controlled human malaria infection studies and naturally acquired infections. J Clin Microbiol (2020) 58(5):e01879-19. doi: 10.1128/jcm.01879-19
23. Pollak NM, Olsson M, Ahmed M, Tan J, Lim G, Setoh YX, et al. Rapid diagnostic tests for the detection of the four dengue virus serotypes in clinically relevant matrices. Microbiol Spectr (2023) 11(1):e0279622. doi: 10.1128/spectrum.02796-22
24. Pollak NM, Marsh GA, Olsson M, McMillan D, Macdonald J. Rapid, sensitive, and specific, low-resource molecular detection of Hendra virus. One Health (2023) 16:100504. doi: 10.1016/j.onehlt.2023.100504
25. Pollak NM, Olsson M, Marsh GA, Macdonald J, McMillan D. Evaluation of three rapid low-resource molecular tests for Nipah virus. Front Microbiol (2022) 13:1101914. doi: 10.3389/fmicb.2022.1101914
26. Pollak NM, Fais O, Kristoffersen J, Phuthaworn C, Knibb W, Macdonald J. Rapid sample preparation and low-resource molecular detection of hepatopancreatic parvoviruses (Hpv) by recombinase polymerase amplification lateral flow detection assay in shrimps (Fenneropenaeus merguiensis). PloS One (2022) 17(11):e0276164. doi: 10.1371/journal.pone.0276164
27. Ahmed M, Pollak NM, Hugo LE, van den Hurk AF, Hobson-Peters J, Macdonald J. Rapid molecular assays for the detection of the four dengue viruses in infected mosquitoes. Gates Open Res (2022) 6:81. doi: 10.12688/gatesopenres.13534.2
28. Ahmed M, Nath NS, Hugo LE, Devine GJ, Macdonald J, Pollak NM. Rapid detection of kdr mutation F1534c in Aedes aegypti using recombinase polymerase amplification and lateral flow dipsticks. Pestic Biochem Physiol (2022) 187:105209. doi: 10.1016/j.pestbp.2022.105209
29. Daher RK, Stewart G, Boissinot M, Bergeron MG. Recombinase polymerase amplification for diagnostic applications. Clin Chem (2016) 62(7):947–58. doi: 10.1373/clinchem.2015.245829
30. James AS, Todd S, Pollak NM, Marsh GA, Macdonald J. Ebolavirus diagnosis made simple, comparable and faster than molecular detection methods: preparing for the future. Virol J (2018) 15(1):75. doi: 10.1186/s12985-018-0985-8
31. Escadafal C, Faye O, Sall AA, Faye O, Weidmann M, Strohmeier O, et al. Rapid molecular assays for the detection of yellow fever virus in low-resource settings. PLoS Negl Trop Dis (2014) 8(3):e2730. doi: 10.1371/journal.pntd.0002730
32. Rames EK, Macdonald J. Rapid assessment of viral water quality using a novel recombinase polymerase amplification test for human adenovirus. Appl Microbiol Biotechnol (2019) 103(19):8115–25. doi: 10.1007/s00253-019-10077-w
33. Li J, Macdonald J, von Stetten F. Review: A comprehensive summary of a decade development of the recombinase polymerase amplification. Analyst (2018) 144(1):31–67. doi: 10.1039/c8an01621f
34. Li J, Macdonald J. Advances in isothermal amplification: novel strategies inspired by biological processes. Biosens Bioelectron (2015) 64:196–211. doi: 10.1016/j.bios.2014.08.069
35. Rockett RJ, Tozer SJ, Peatey C, Bialasiewicz S, Whiley DM, Nissen MD, et al. A real-time, quantitative PCR method using hydrolysis probes for the monitoring of Plasmodium falciparum load in experimentally infected human volunteers. Malar J (2011) 10:48. doi: 10.1186/1475-2875-10-48
36. MedCalc Software Ltd. Diagnostic test evaluation calculator. Available at: https://www.medcalc.org/calc/diagnostic_test.php (Version 22.018; accessed January 22, 2024).
37. Sdg Target 3.3 End the Epidemics of Aids, Tuberculosis, Malaria and Neglected Tropical Diseases and Combat Hepatitis, Water-Borne Diseases and Other Communicable Diseases. World Health Organisation (2024). Available at: https://www.who.int/data/gho/data/themes/topics/sdg-target-3_3-communicable-diseases.
38. Kersting S, Rausch V, Bier FF, von Nickisch-Rosenegk M. Rapid detection of Plasmodium falciparum with isothermal recombinase polymerase amplification and lateral flow analysis. Malar J (2014) 13:99. doi: 10.1186/1475-2875-13-99
39. Lai MY, Ooi CH, Lau YL. Recombinase polymerase amplification combined with a lateral flow strip for the detection of Plasmodium knowlesi. Am J Trop Med Hyg (2018) 98(3):700–3. doi: 10.4269/ajtmh.17-0738
40. Stone WJR, Eldering M, van Gemert G-J, Lanke KHW, Grignard L, van de Vegte-Bolmer MG, et al. The relevance and applicability of oocyst prevalence as a read-out for mosquito feeding assays. Sci Rep (2013) 3(1):3418. doi: 10.1038/srep03418
Keywords: isothermal (DNA) amplification, rapid test, malaria - mosquito vectors, rapid sample preparation, nucleic acid lateral flow, Plasmodium falciparum
Citation: Hugo LE, van Huyssteen K, Oloniniyi O, Donnelly L, Conn A, Collins KA, Mitchell H, McCarthy JS and Macdonald J (2024) Rapid low-resource detection of Plasmodium falciparum in infected Anopheles mosquitoes. Front. Trop. Dis 5:1287025. doi: 10.3389/fitd.2024.1287025
Received: 01 September 2023; Accepted: 08 January 2024;
Published: 29 January 2024.
Edited by:
Tais Nobrega De Sousa, Oswaldo Cruz Foundation (Fiocruz), BrazilReviewed by:
Denise Anete Madureira Alvarenga, Fiocruz Minas, IRR, BrazilCopyright © 2024 Hugo, van Huyssteen, Oloniniyi, Donnelly, Conn, Collins, Mitchell, McCarthy and Macdonald. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joanne Macdonald, am1hY2RvbjFAdXNjLmVkdS5hdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.