
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Trop. Dis., 26 July 2023
Sec. Emerging Tropical Diseases
Volume 4 - 2023 | https://doi.org/10.3389/fitd.2023.1221804
This article is part of the Research TopicThe Intersection of Covid and Tropical DiseasesView all 7 articles
 Kenneth B. Yeh1*
Kenneth B. Yeh1* Michael D. Powers2
Michael D. Powers2 Ami Patel1
Ami Patel1 Falgunee Parekh3
Falgunee Parekh3 Ashley S. Tseng4
Ashley S. Tseng4 Elaine Bradford1
Elaine Bradford1 Kyle Parker1
Kyle Parker1 Ricky Soong1
Ricky Soong1 Gene Olinger1
Gene Olinger1 Illich Mombo5
Illich Mombo5The landscape of in vitro diagnostic (IVD) devices encompasses a broad range of tests that have been used to detect and diagnose pathogens, especially tropical diseases, for decades. The COVID-19 pandemic exemplified the greater need for bringing IVDs from the laboratory directly to the consumer, and recent outbreaks such as mpox, Sudan ebolavirus, and Marburg virus further reinforce this need. The increased emergence of tropical disease outbreaks requires more agile development, higher performance, and mass production of IVD devices. Furthermore, lessons learned in previous device developments can sometimes be used to accelerate new disease diagnostic applications. As an example, we describe one case history of an earlier pan-orthopox viral assay that detected smallpox variola and vaccinia strains, and also discerned related strains including mpox. This work established the foundation for the molecular detection of orthopox viruses, which could be mobilized to address public health needs once an emergency declaration was made that opened the FDA pathway for issuing an emergency use authorization for the use of these assays. Thus, the utilization of knowledge from earlier investments was shown to enhance preparedness and readiness. Here in this retrospective, we elaborate on the processes that enable this approach, including multi-disciplinary and multisectoral collaborations to accomplish a holistic, one health world.
The detection, identification, and characterization of biological agents, including bacteria, fungi, viruses, and toxins, pose constant challenges. In November 2022, the World Health Organization (WHO) revised their list of priority pathogens termed as 'Disease X' to guide research investments for emerging, novel, and unknown threats. Effective biodetection—e.g., before symptoms of illness arise—has always been a race against time based on Koch’s postulates for microbial culture. PCR revolutionized molecular diagnostics, providing high sensitivity and specificity, faster results, and the foundation for genomic sequencing. Regarding the development of in vitro diagnostic (IVD) devices, a target product profile (TPP) guides the identification of the unmet clinical need and creates an effective roadmap from conception to market to keep the product development pipeline well defined (1). These IVD devices are ideally portable, easy to use, have easy-to-interpret results, and have limited logistical or storage requirements during the journey from manufacturer to end user. As the market for infectious disease IVD devices continues to expand, it highlights the need for a practical approach to development that considers a wide scope of factors, including detection technology, market assessments, and regulatory considerations (Figure 1). Together, these factors underscore the need for a clearly defined path from conceptualization to market.
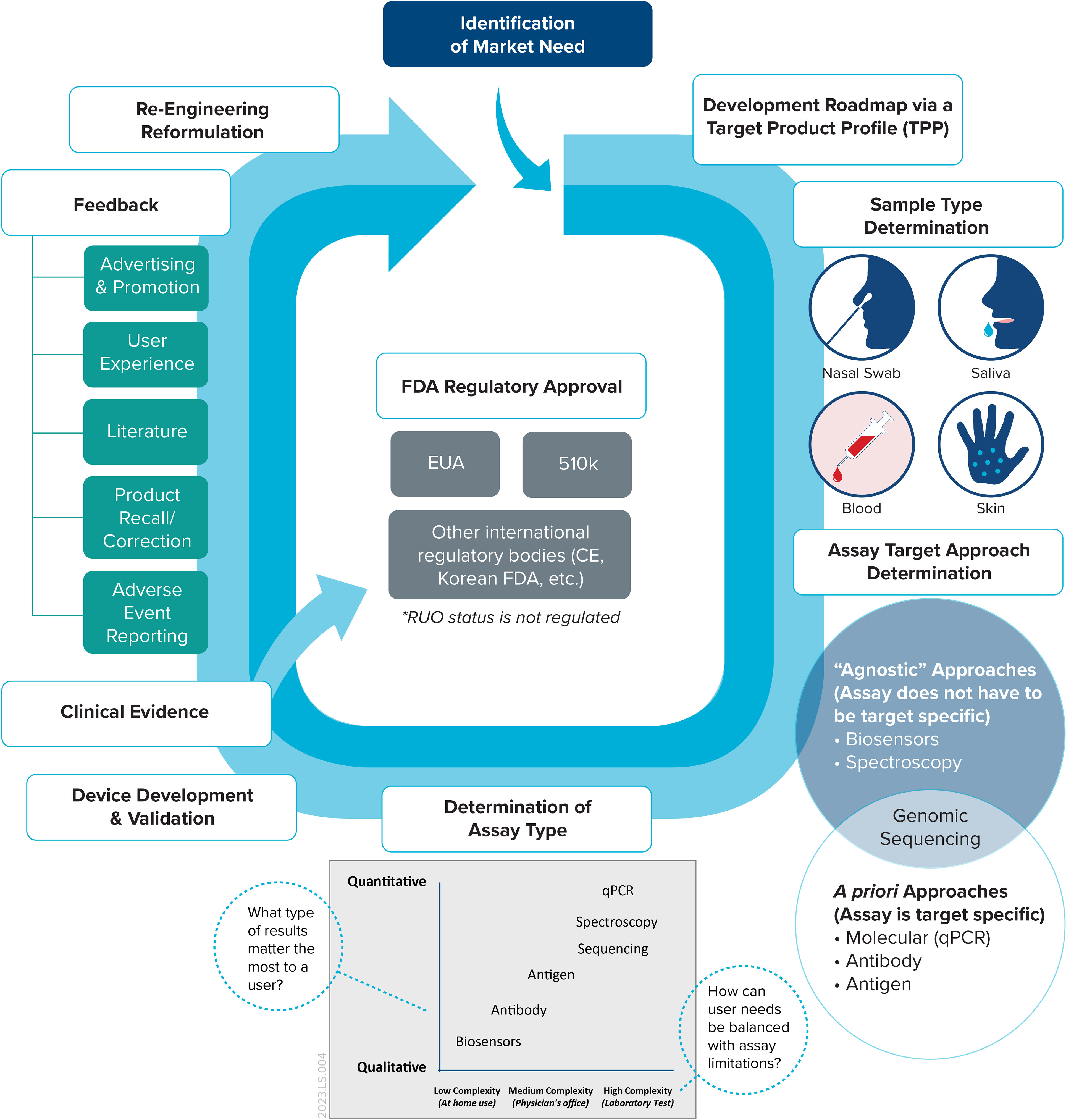
Figure 1 The IVD device lifecycle. This framework maps a typical lifecycle, which ideally begins with a TPP that outlines the overall scope of the device once a market need is identified.
Our earlier perspective examined the capabilities and activities of organizations performing research and development, testing and evaluation of clinical samples and diagnostics during COVID-19, highlighting the significance of operating high-containment biological laboratories (HCBL) and competing demands for human resources and infrastructure (2). University research labs are another example of organizations with skilled laboratory staff that adapted their high-throughput platforms to run real-time PCR assays to detect SARS-CoV-2 from patient samples (3). Here, we continued to examine the significance of biosafety and biosecurity, laboratory capabilities and capacities, and regulatory requirements in relation to mpox.
The clinical diagnostics market is highly competitive, especially for IVD devices, as demonstrated by the record number of Emergency Use Authorizations (EUAs) that the US Food and Drug Administration (FDA) issued for COVID-19. The FDA issued an EUA for mpox, which provided a regulatory pathway for medical diagnostics manufacturers to sell their tests to the consumer, and was a major market driver for IVD manufacturers. Furthermore, national government funding is often static, if not reactive, and investments do not always align with commercial market priorities, as in the case of the biodefense market that rapidly grew after 2001. For those fortunate first movers and fast followers, national government funding is extensive and offers opportunities to develop niche products in diagnostics medical countermeasures and vaccines for tropical infectious diseases (4). In the early 2000s, a qPCR clinical diagnostics instrument, which the US military fielded according to its portable and rugged design, became the first FDA-approved platform for the diagnosis of biological threats (5). Nevertheless, to understand the mpox diagnostics landscape we first focus on earlier work to detect the pan-orthopox virus that drove subsequent IVD developments in the family Poxviridae.
Poxviruses, which are in the family Poxviridae and ubiquitous in nature, are the largest known animal viruses and can be viewed by light microscopy. Orthopox strains, especially variola, remain potential biothreats given the state-sponsored programs that develop biological weapons and the waning vaccinated population after WHO declared smallpox eradicated on 8 May 1980. Potential biosecurity risks and implications remain as a result of the de novo synthesis of horsepox virus in relation to smallpox vaccine development (6). The 2022 mpox outbreak reinforces the reality that mpox is now the most important orthopox pathogen and was also not widely researched outside of Africa before 2022 (7). Only Clade I of mpox, variola, and members of the Capripoxvirus (CaPV) genus such as goatpox and sheeppox are on the US Department of Health and Human Services and US Department of Agriculture (USDA) Select Agents and Toxins list, which limits the ability to culture and propagate the virus in laboratories that are registered with the federal government and have access to BSL-3 facilities. The status of mpox as a Select Agent could significantly impact the ability of the research community to respond to an outbreak, or pandemic, on the scale of COVID-19. While CaPV are not considered tropical diseases, a world connected through global trade and challenged with climate change underscores the need for a comprehensive one health approach.
Mpox is caused by the MPXV virus of the genus Orthopoxvirus which includes variola (smallpox), cowpox, camelpox, and vaccinia viruses (8). Mpox was identified for the first time in 1958 in Denmark during an outbreak of vesicular disease observed in captive monkeys coming from Africa (9). Within 1 to 5 days after the onset of fever, the infected person develops a rash or skin lesions on the face, followed by dissemination to other body parts (10, 11). The first human case of mpox was a 9-month-old boy from Zaïre—now the Democratic Republic of Congo (DRC)—and was reported in 1970 (12). Genomic studies have phylogenetically divided MPXV into two subtypes, Clade I (Central African or Congo Basin) and Clade II (West African), with differences in epidemiology and clinical manifestation (13). The case fatality rate of Clade I is up to 10% whereas for Clade II, responsible for most outbreaks outside of Africa, the case fatality rate is about 1% (7, 14). Although the natural reservoir of the virus is not well defined, various African rodents are suspected to be the animal reservoirs.
As a result, mpox is transmitted zoonotically or via human-to-human contact. The zoonotic transmission of mpox occurs through scratches or bites from infected animals; during handling through direct contact with blood, body fluids and lesions; or by eating improperly cooked infected animals (10, 14). Human-to-human transmission also occurs through direct exposure to the respiratory droplets, skin, and body fluids of infected patients. The virus can also be transmitted during sexual contact, from pregnant woman to the fetus, and to newborns by close contact during birth (15, 16). Mpox became endemic in the DRC, eventually spreading to over 15 West and Central African countries.
During the 1970s, only a few sporadic cases were reported and remained limited to a few countries in Central and West Africa (17). During the 1980s, the number of cases increased nine-fold compared with the previous decade, with the active monitoring program implemented by the WHO discovering 338 cases and 33 fatalities between 1981 and 1986 in the DRC. Only 13 cases were reported from 1986 to 1992 and there were no cases reported until 1996 where an outbreak in the DRC resulted in 511 suspected cases, likely attributed to the decreased collective immunity of local populations after the cessation of smallpox vaccination (18). Between 1998 and 2019, continuous reporting occurred in the DRC annually with cumulative cases in Africa reaching about 30,000 (19). Until now, mpox was considered a neglected tropical disease due to the geographic proximity of outbreaks in the tropics.

Mpox remained an ignored global public health threat until 2003 when it gained international attention as a result of a US outbreak, the first outside of Africa (21). The outbreak comprised 47 recorded cases and was probably transmitted by infected prairie dogs that were in close contact with animals imported from Ghana (22). Since then, additional cases outside of Africa have been reported in the UK and Israel in 2018, and in Singapore in 2019 (23–25). The 2022 mpox outbreak, which is now the largest ever, spread to the Americas, Europe, Asia, and the Western Pacific and Eastern Mediterranean regions, and saw 98% of cases (n=85,059) reported in non-endemic countries with no history of local outbreaks. The global spread of mpox was due to human-to-human transmission and caused by Clade IIb, which has a low mortality rate due to perceived genetic differences from Clade I (26). A confirmed case of mpox transmission from a human to a domesticated dog also occurred, which emphasizes the risk of close skin-to-skin contact and suggests further infections via pets (27). As the 2022 mpox outbreak wanes, cases are still expected to flare up during summertime activities as the public health restrictions implemented owing to COVID-19 and mpox are lifted. The reality of mpox becoming endemic outside of Africa further reinforces the need to prioritize health systems, diagnostics, and vaccines in Africa (28). In fact, as the world recognized Smallpox Eradication Day on 8 May 2023 and marked the 43rd anniversary of the WHO declaration, we were reminded that public health surveillance and eradication initiatives analogous to what was accomplished for smallpox are needed to truly address mpox endemicity.
The US response to and mitigation of the mpox outbreak using testing and prevention was slowed due to health equity issues and a familiar pattern of bureaucracy, although related diagnostics, vaccines, and treatments existed for the orthopox virus (29, 30). One likely factor that slowed the development of mpox diagnostics, relative to SARS-COV-2 diagnostics, is the classification of mpox as a Select Agent, allowing only CDC- or USDA-registered laboratory spaces to work with the virus. Although the actual number of BSL-3 laboratories in the United States is currently unknown, the number which have CDC or USDA Select Agent registration is low, which could be perceived as a gap in preparedness for outbreak response. In Africa, mpox can be misdiagnosed due to the lack of adequate facilities and tools for differential diagnoses to rule out other diseases. For example, it is estimated that up to 50% of suspected cases in the DRC can be attributed to chickenpox (31, 32). Mpox cases continue to be overlooked or misdiagnosed as symptoms are similar to other diseases, such as chickenpox or other pox viruses, and physicians are unfamiliar with the disease (33, 34). Open-source tools such as a free phone app designed to distinguish mpox lesions from other skin lesions can also help those with a suspected infection seek professional healthcare (35). These examples emphasize the importance of a holistic approach to the diagnosis of mpox based on the “sum total of patient history, clinical presentation, and confirmatory laboratory testing.”
In the USA, despite the current infrastructure and preparedness built during the COVID-19 pandemic and existing pan-orthopox assays, the first commercial mpox diagnostics test took 3 months to develop and receive FDA EUA approval. (Figure 2). Organizations involved in R&D and national surveillance for MPVX offered vaccines to their laboratory scientists starting with ACAM2000 and then Jynneos as it became available, highlighting the continued need for a readily available stockpile of vaccines. The continued use of second-generation vaccines such as ACAM2000 speaks to the need for the development of safer, next-generation mRNA vaccines. The current PCR-based commercial mpox test only detects MPXV in swab samples from skin or other lesions. Commercial tests are not yet available for other sample matrices despite the ability to detect MPXV in urine, blood, and other human samples (36). As a part of readiness, applicable research data should be used to develop commercial tests that support higher sensitivity, greater accuracy, and faster turnaround times for earlier testing.
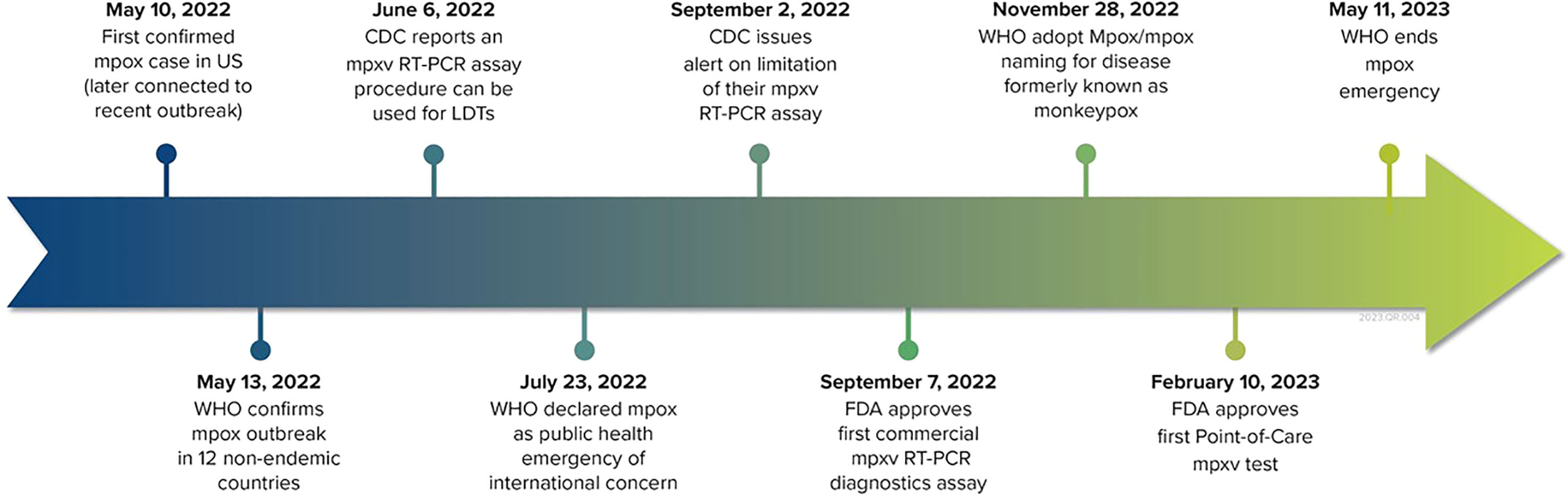
Figure 2 Timeline of the mpox outbreak and commercial diagnostics development. Since September 2022, the FDA has issued eight EUAs for IVD devices.
The 2022 mpox outbreak has solely focused on the community of men who have sex with men, which resulted in the common stereotype that mpox was only an issue in a certain, often stigmatized, high-risk population. This misperception of disease risk posed a barrier to an effective response to and management of the outbreak, rather than enabling targeted approaches. Given this outbreak was the first with multiple cases reported in non-endemic countries and focused on a specific high-risk population, further attention to sex and gender is needed to address various socioeconomic factors among populations in or low and middle and high-income countries (37). Ideally, robust surveillance systems will enable the identification of key risk factors and/or high-risk populations associated with infection and disease transmission while enhancing our ability to provide data-driven scientific information to the public. Fortunately, further outbreaks have not occurred in large population groups at college campuses and military bases following public awareness measures on practices for self-isolation and limiting skin-to-skin contact. The limited availability of early diagnostics and testing sample matrices besides those for skin lesions presents further challenges to at-risk groups, such as athletes engaged in contact sports (38). The US Government recently reported on the vital role of research in the mpox response and how important it is that this serves as a model for future outbreaks (39). Overall, the COVID-19 pandemic and 2022 mpox outbreak have emphasized the importance of R&D and pandemic readiness, which can be achieved by focusing on prevention (vaccine development), detection (diagnostics), treatment (therapeutics), surveillance (public health infrastructure), and biorisk management.
Many of the orthopox viruses are of veterinary, public health, and, consequently, economic concern, making this genus a priority for better screening assays that provide meaningful differentiation between genus members. For this reason, a trio of assays were developed: variola-specific, pan-orthopox, and vaccinia-specific assays. The terminal region of the genome allows for differentiation of the Orthopoxvirus genus (40, 41). Previous work led to the development of a variola-specific hybridization probe assay targeting the hemagglutinin (HA) gene, located in the terminal region of the genome (42). The gene region selected allows for the specific detection of variola major and minor, the causative agents of smallpox. The systemic infection manifested from variola major, a strictly human pathogen, has a fatality rate of 30%, while variola minor’s fatality rate is 1% in unvaccinated populations. These dramatic differences in fatality rates necessitate the importance of being able to distinguish variola major and minor.
Due to the limited access to variola DNA, oligonucleotides were used to generate an artificial template to test and optimize the probe design for this assay. After assay optimization was completed, a limited panel containing camelpox (Somalia), cowpox (Brighton), monkeypox (Zaire 1996), vaccinia (Copenhagen), rabbitpox (unknown), VAR-2 (Plasmid USAMRIID), and VAR-1 (Plasmid IT) was used for specificity testing. The only members of this panel detected were the two plasmid targets VAR-1 and VAR-2, generating strong evidence that this design is specific to variola. To further confirm cross-reactivity specificity a panel containing 50 bacterial and viral pathogens of medical significance was evaluated with all members of this panel testing negative. The panel included dengue, Enterovirus, influenza A, parainfluenza sub 1–3, Staphylococcus aureus, Salmonella typhi, Yersinia pestis, and Vibrio cholerae N16961.
The pan-orthopox assay was developed, using the same terminal region of the genome designed for the HA gene of vaccinia. Once the assay was optimized, the same orthopox panel described above was used for inclusivity testing. All members of the panel were detected with three distinct melting profiles generated. Since the vaccinia virus is used to vaccinate against smallpox, distinguishing adverse vaccine reactions from true smallpox is of value. To accomplish this, a vaccinia-specific assay designed for the surface antigen gene of vaccinia has been designed and tested. The location of the primers and probes leads to the exclusion or distinction, based on melting curve profiles, of all non-vaccinia virus targets.
The approach used for designing these three assays allows for the detection and meaningful differentiation of the Poxviridae family. This type of differentiation in testing allows for a triage approach where the result can inform the action to be taken. While the pan-orthopox assay is an older design, which does not differentiate camelpox, monkeypox, and VAR-2 plasmid (likely variola), there is the possibility of good differentiation among the Orthopoxvirus genus, similar to what has been observed in the nearest neighbor species. While the FDA EUA prioritized regulatory guidance for mpox IVD devices, the assay designs have various targets that are proprietary in nature and often undisclosed. For biosafety reasons stated earlier, limited access to viral material for testing and validating assays adds further time to design pipelines.
In the last five decades, incredible innovations and technological developments have shaped how modern clinical laboratories are equipped, staffed, and operated. Commercial and public health laboratories worldwide must continue to reinforce the infrastructure established before and during the COVID-19 pandemic to keep their laboratories ready for testing in any future epidemic or pandemic. The US has created a strong R&D readiness that combines diagnostic test platforms, whole genome sequencing consortiums such as SPHERES, FDA EUAs, and public health surveillance networks to address future pandemics. One strategy to bolster public health preparedness efforts is to use and reinforce the existing infrastructure mentioned above, especially HCBLs, biosafety risk assessments, instruments, expertise, personnel, and readiness responses. An integrated approach lays the groundwork for public health epidemiologists to identify and track disease cases, study disease transmission, and help prevent future outbreaks locally and globally.
In clinical diagnostics, there has been a general transition from stationary large-scale clinical laboratories to smaller point-of-care devices. Parallel to this trend, assay readiness should also include thoughtfully designed molecular assays, such as multiplexed molecular diagnostic assays for symptomatic near neighbors as described in our case history. In the future, the ability to combine affinity and molecular assays, especially for a point-of-care device, will enable IVDs that define susceptibility, early infection, and phases of disease by measuring host biomarkers and pathogens directly and simultaneously. Combined, this approach will enable informed clinical decisions for treatment and empower patients and their families in disease control and treatment. Eventually, it is hoped these assays will be cost-effective and even have prognostic capability.
As point-of-care and at-home testing become more common, investments in infectious disease surveillance systems that can capture the data from these IVD devices must be made in order to truly improve overall readiness to respond to future outbreaks. Systems that are agile and able to accommodate defined data standards while accounting for underreporting will be key. Supporting the accurate and timely reporting of cases is critical to effective outbreak response and curbing continued transmission. The data not only provide case counts, but help identify key risk factors associated with infection, disease transmission, and severity of disease. Understanding key factors, like age and geographic distribution, that can identify potential high-risk groups who may experience increased infection and complications is important during outbreaks to inform research and enable the development of targeted prevention, response, and treatment strategies. Thus, both clinical and laboratory diagnostic data are the basis of infectious disease surveillance systems and are inherently linked.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
KY, MP, and IM developed the concept for this work. AP, FP, AT, EB, KP, RS, and GO contributed their respective content. All authors contributed to the article and approved the submitted version.
This article was developed without any external funding. Special thanks to Drs. Barbara Price, Zvonko Orehovec, and Slavko Bovan for their help in finding a presentation from their CBMTS proceedings. Thank you Mr. Garrett Dalton for his expert graphic art. The authors thank the reviewers and editors for their critique.
KY, AP, EB, KP, RS, and GO are employed by MRIGlobal. FP was employed by the company EpiPointe, LLC.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. World Health Organization. High-priority target product profiles for new tuberculosis diagnostics: report of a consensus meeting (2014). Available at: https://apps.who.int/iris/bitstream/handle/10665/135617/WHO_HTM_TB_2014.18_eng.pdf?sequence=1 (Accessed December 21, 2022).
2. Yeh KB, Tabynov K, Parekh FK, Mombo I, Parker K, Tabynov K, et al. Significance of high-containment biological laboratories performing work during the COVID-19 pandemic: biosafety level-3 and -4 labs. Front Bioeng Biotechnol (2021) 9:720315. doi: 10.3389/fbioe.2021.720315
3. Ranoa DR, Holland RL, Alnaji FG, Green KJ, Wang L, Brooke CB, et al. Saliva-based molecular testing for SARS-CoV-2 that bypasses RNA extraction. biorxiv (2020) 18:2020–06. doi: 10.1101/2020.06.18.159434
4. Yeh KB, Du E, Olinger G, Boston D. Biotechnology and biodefense enterprise: an industry perspective on defence acquisition. Global Security: Health Sci Policy (2022) 7(1):37–43. doi: 10.1080/23779497.2022.2102527
5. Wilson SA. A novel approach for the development and acquisition of a diagnostic medical device from concept to fielding: the Joint Biological Agent Identification and Diagnostic System (JBAIDS). George Mason University (2006). https://www.proquest.com/openview/71b746761df02f6cd8ad4abadce5c815/1?pq-origsite=gscholar&cbl=18750&diss=y.
6. Noyce RS, Lederman S, Evans DH. Construction of an infectious horsepox virus vaccine from chemically synthesized DNA fragments. PloS One (2018) 13(1):e0188453. doi: 10.1371/journal.pone.0188453
7. Sklenovská N, Van Ranst M. Emergence of monkeypox as the most important orthopoxvirus infection in humans. Front Public Health (2018) 6:241. doi: 10.3389/fpubh.2018.00241
8. International Committee on Taxonomy of Viruses. (2023). Available at: https://ictv.global/taxonomy.
9. Magnus PV, Andersen EK, Petersen KB, Birch-Andersen A. A pox-like disease in cynomolgus monkeys. Acta Pathol Microbiol Scand (1959) 46(2):156–76. doi: 10.1111/j.1699-0463.1959.tb00328.x
10. Petersen E, Kantele A, Koopmans M, Asogun D, Yinka-Ogunleye A, Ihekweazu C, et al. Human monkeypox: epidemiologic and clinical characteristics, diagnosis, and prevention. Infect Dis Clin (2019) 33(4):1027–43. doi: 10.1016/j.idc.2019.03.001
11. Kumar N, Acharya A, Gendelman HE, Byrareddy SN. The 2022 outbreak and the pathobiology of the monkeypox virus. J Autoimmun (2022) 25:102855. doi: 10.1016/j.jaut.2022.102855
12. Marennikova SS, Šeluhina EM, Mal’Ceva NN, Čimiškjan KL, Macevič GR. Isolation and properties of the causal agent of a new variola-like disease (monkeypox) in man. Bull World Health Organ (1972) 46(5):599–611. https://apps.who.int/iris/handle/10665/263482.
13. Likos AM, Sammons SA, Olson VA, Frace AM, Li Y, Olsen-Rasmussen M, et al. A tale of two clades: monkeypox viruses. J Gen Virol (2005) 86(10):2661–72. doi: 10.1099/vir.0.81215-0
14. Gong Q, Wang C, Chuai X, Chiu S. Monkeypox virus: a re-emergent threat to humans. Virol Sin (2022). doi: 10.1016/j.virs.2022.07.006
15. Alakunle E, Moens U, Nchinda G, Okeke MI. Monkeypox virus in Nigeria: infection biology, epidemiology, and evolution. Viruses (2020) 12(11):1257. doi: 10.3390/v12111257
16. Hatmal MM, Al-Hatamleh MA, Olaimat AN, Ahmad S, Hasan H, Ahmad Suhaimi NA, et al. Comprehensive literature review of monkeypox. Emerg Microbes Infect (2022) 11(1):2600–31. doi: 10.1080/22221751.2022.2132882
17. Breman JG, Kalisa-Ruti, Steniowski MV, Zanotto E, Gromyko AI, Arita I. Human monkeypox 1970-1979 / by J.G. Bremen, Kalisa-Ruti, M.V. Steniowski, E. Zanotto, A.I. Gromyko and I. Arita. World Health Organization (1980). https://apps.who.int/iris/handle/10665/67095.
18. Hutin YJ, Williams RJ, Malfait P, Pebody R, Loparev VN, Ropp SL, et al. Outbreak of human monkeypox, Democratic Republic of Congo 1996 to 1997. Emerg Infect Dis (2001) 7(3):434. doi: 10.3201/eid0703.010311
19. Bunge EM, Hoet B, Chen L, Lienert F, Weidenthaler H, Baer LR, et al. The changing epidemiology of human monkeypox–a potential threat? a systematic review. PloS Negl Trop Dis (2022) 16(2):e0010141. doi: 10.1371/journal.pntd.0010141
20. Berthet N, Descorps-Declère S, Besombes C, Curaudeau M, Nkili Meyong AA, Selekon B, et al. Genomic history of human monkey pox infections in the Central African Republic between 2001 and 2018. Sci Rep (2021) 11(1):13085. doi: 10.1038/s41598-021-92315-8
21. Centers for Disease Control and Prevention (CDC). Update: multistate outbreak of monkeypox–Illinois, Indiana, Kansas, Missouri, Ohio, and Wisconsin. MMWR Morb Mortal Wkly Rep (2003) 52(24):561–4. https://www.cdc.gov/mmwr/preview/mmwrhtml/mm5226a5.htm.
22. Reynolds MG, Yorita KL, Kuehnert MJ, Davidson WB, Huhn GD, Holman RC, et al. Clinical manifestations of human monkeypox influenced by route of infection. J Infect Dis (2006) 194(6):773–80. doi: 10.1086/505880
23. Vaughan A, Aarons E, Astbury J, Brooks T, Chand M, Flegg P, et al. Human-to-human transmission of monkeypox virus, united kingdom, October 2018. Emerg Infect Dis (2020) 26(4):782. doi: 10.3201/eid2604.191164
24. Erez N, Achdout H, Milrot E, Schwartz Y, Wiener-Well Y, Paran N, et al. Diagnosis of imported monkeypox, Israel 2018. Emerg Infect Dis (2019) 25(5):980. doi: 10.3201/eid2505.190076
25. Yong SE, Ng OT, Ho ZJ, Mak TM, Marimuthu K, Vasoo S, et al. Imported monkeypox, Singapore. Emerg Infect Dis (2020) 26(8):1826. doi: 10.3201/eid2608.191387
26. Americo JL, Earl PL, Moss B. Virulence differences of mpox (monkeypox) virus clades I, IIa, and IIb.1 in a small animal model. PNAS (2023) 120(8):e2220415120. doi: 10.1073/pnas.22204151
27. Seang S, Burrel S, Todesco E, Leducq V, Monsel G, Le Pluart D, et al. Evidence of human-to-dog transmission of monkeypox virus. Lancet (2022) 400(10353):658–9. doi: 10.1016/S0140-6736(22)01487-8
28. Adetifa I, Muyembe JJ, Bausch DG, Heymann DL. Mpox neglect and the smallpox niche: a problem for Africa, a problem for the world. Lancet (2023) 401(10390):1822–4. doi: 10.1016/S0140-6736(23)00588-3
29. Centers for Disease Control and Prevention (CDC). (2023). Available at: https://www.cdc.gov/poxvirus/mpox/resources/toolkits/equity.html.
30. Scientific American. (2022). Available at: https://www.scientificamerican.com/article/monkeypox-explained-transmission-symptoms-vaccines-and-treatment/.
31. Jezek Z, Szczeniowski M, Paluku KM, Mutombo M, Grab B. Human monkeypox: confusion with chickenpox. Acta Tropica (1988) 45(4):297–307.
32. Meyer H, Perrichot M, Stemmler M, Emmerich P, Schmitz H, Varaine F, et al. Outbreaks of disease suspected of being due to human monkeypox virus infection in the democratic republic of Congo in 2001. J Clin Microbiol (2002) 40(8):2919–21. doi: 10.1128/JCM.40.8.2919-2921.2002
33. White House. MPox research gathering 03/31/2023 . Available at: https://www.hiv.gov/hiv-basics/staying-in-hiv-care/other-related-health-issues/monkeypox/mpox-research-gathering/.
34. Wieder-Feinsod A, Zilberman T, Erster O, Kolasko GW, Biber A, Gophen R, et al. Overlooked monkeypox cases among men having sex with men during the 2022 outbreak–a retrospective study. Int J Infect Dis (2023) 128:58–60. doi: 10.1016/j.ijid.2022.12.014
35. Stanford University. PoxApp . Available at: https://poxapp.stanford.edu (Accessed 23 June, 2023).
36. Palich R, Burrel S, Monsel G, Nouche A, Bleibtreu A, Seang S, et al. Viral loads in clinical samples of men with monkeypox virus infection: a French case series. Lancet Infect Dis (2023) 23(1):74–80. doi: 10.1016/S1473-3099(22)00586-2
37. Fusco D, Martínez-Pérez GZ, Remkes A, De Pascali AM, Ortalli M, Varani S, et al. A sex and gender perspective for neglected zoonotic diseases. Front Microbiol (2022) 13:1031683. doi: 10.3389/fmicb.2022.1031683
38. Smoliga JM. Mpox and monkeypox virus: special considerations for athletes in contact sports. Sports Med (2023) 27:1–3. doi: 10.1007/s40279-023-01812-5
39. White House. (2023). Available at: https://www.whitehouse.gov/ostp/news-updates/2023/04/28/the-vital-role-of-research-in-mpox-response-a-model-for-other-diseases-and-future-outbreaks/.
40. Espy MJ, Cockerill FR III, Meyer RF, Bowen MD, Poland GA, Hadfield TL, et al. Detection of smallpox virus DNA by LightCycler PCR. J Clin Microbiol (2002) 40(6):1985–8. doi: 10.1128/JCM.40.6.1985-1988.2002
41. Sofi Ibrahim M, Kulesh DA, Saleh SS, Damon IK, Esposito JJ, Schmaljohn AL, et al. Real-time PCR assay to detect smallpox virus. J Clin Microbiol (2003) 41(8):3835–9. doi: 10.1128/JCM.41.8.3835-3839.2003
Keywords: in vitro diagnostic (IVD), mpox, one health, Orthopoxvirus, research and development (R & D)
Citation: Yeh KB, Powers MD, Patel A, Parekh F, Tseng AS, Bradford E, Parker K, Soong R, Olinger G and Mombo I (2023) Retrospective on molecular assay design for detecting pan-orthopox viruses and prospective on mpox laboratory preparedness and readiness. Front. Trop. Dis 4:1221804. doi: 10.3389/fitd.2023.1221804
Received: 12 May 2023; Accepted: 27 June 2023;
Published: 26 July 2023.
Edited by:
Edwin Michael, University of South Florida, United StatesReviewed by:
Patrick K. Mukadi, Nagasaki University, JapanCopyright © 2023 Yeh, Powers, Patel, Parekh, Tseng, Bradford, Parker, Soong, Olinger and Mombo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kenneth B. Yeh, a3llaEBtcmlnbG9iYWwub3Jn
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.