
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Trop. Dis. , 20 April 2023
Sec. Antimicrobial Resistance
Volume 4 - 2023 | https://doi.org/10.3389/fitd.2023.1152422
Salmonella Enteritidis is the most reported non-typhoidal Salmonella serovar and is implicated in both gastroenteritis and invasive non-typhoidal disease. Whole-genome sequence comparison of Salmonella Enteritidis isolates from across the world identified four definitive clades: Outlier, Global Epidemic, East African and West African. Three of these clades were implicated in epidemics: the Global Epidemic clade was linked to poultry-associated gastroenteritis, while the two African clades were related to invasive disease. Despite the recognition of different Salmonella Enteritidis clades, the distribution and epidemiology of these clades across Africa is poorly understood. In our proof-of-concept study, we classified 618 Salmonella Enteritidis isolates originating from four South African provinces over a period of two years (2012 – 2013) into clades using a clade-typing real-time PCR assay. Associations between clades and province of collection, antimicrobial resistance and HIV status were explored using statistical analysis. Majority of the South African isolates were classified within the Outlier clade (61.00%), with fewer classified within the Global Epidemic clade (38.83%) and only one isolate classified within the West African Clade (0.16%). Of note, were the isolates within the Global clade, which were strongly associated with ciprofloxacin resistance (15.42%, OR: 7.45, CI: 3.526 – 15.751) and invasive disease (58.33%, OR: 1.57, CI: 1.13 – 2.17) in humans. The increase in poultry consumption and importation in South Africa has placed the country at risk of a Salmonella Enteritidis epidemic. Thus, there is a necessity for routine monitoring of S. Enteritidis along the farm-to-fork continuum to implement preventative measures.
Sub-Saharan African (sSA) countries are under pressure to increase the production of affordable protein-rich foods often in a nascent regulatory environment with limited veterinary or environmental surveillance capabilities (1). Poultry has long been valued as an affordable source of animal protein in sSA. Previously poultry was predominantly produced by subsistence farmers, however intensive farming is rapidly emerging (2). A global epidemic of Salmonella enterica serotype Enteritidis (S. Enteritidis) began early in the 1980s, in association with consumption of contaminated poultry meat and eggs, which often results in S. Enteritidis infection in humans (3). That S. Enteritidis proliferates alongside the industrial production of poultry is well described, but its simultaneous expansion in many parts of the world remains a mystery, although this has recently been linked to S. Enteritidis dissemination via centralized sourcing and international trade of breeding stocks (4). The lack of biosafety measures in poultry production has resulted in the NTS serovar, S. Enteritidis becoming the most reported foodborne pathogen in sSA (5). Generally, S. Enteritidis infections are associated with outbreaks of gastroenteritis in Europe and the United States (6). However, S. Enteritidis infections in sSA are additionally associated with life threatening bloodstream infections, known as invasive non-typhoidal Salmonella disease (hereafter named iNTS disease) (7). Indeed, in 2017, cases of iNTS disease in sSA were estimated to account for 79% of the global burden (8).
The distinct clinical presentation of Salmonella infections in sSA has been linked both to a high prevalence of immunosuppressive conditions (i.e., HIV) and to Salmonella Enteritidis clades that are phyletically distinct from those associated with enterocolitis in the United States and Europe (9). A 2016 study investigated the diversity of S. Enteritidis in sSA and identified two geographically distinct groups of S. Enteritidis strains circulating in sSA, namely the West African and the Central/Eastern African clade (hereafter the “East African clade”) in addition to the poultry associated lineage, the Global Epidemic clade (9). These African clades are also strongly associated with multidrug resistance (resistance to 3 or more antimicrobial classes), resulting in longer hospital stays and reliance on more expensive, intravenous antibiotics that complicate treatments in the resource-constrained health systems in countries of sSA (9).
Despite the recognition of distinct S. Enteritidis clades and the severity of iNTS disease, the distribution of these clades across sSA remains poorly understood (9). In part, the scarcity of S. Enteritidis clade data is due to the lack of a robust, reproducible molecular typing system. South Africa is an example of a middle-income country, with a well-developed poultry industry and public health surveillance systems, but also with a high prevalence of HIV. Here, we have leveraged this strength to classify 618 S. Enteritidis isolates submitted to the National Institute of Communicable Diseases Centre for Enteric Diseases in South Africa using a multiplex clade typing real-time PCR assay and discuss the epidemiological significance of our findings.
Since 2003, approximately 50 public and private clinical laboratories across all provinces of South Africa have been participating in the GERMS-SA active laboratory-based surveillance program (https://www.nicd.ac.za/internal-publications/germs-sa/). Pathogens of public health concern (AIDS-related opportunistic infections, epidemic-prone diseases, vaccine- preventable diseases, nosocomial infections) are submitted to the National Institute for Communicable Diseases (NICD) for confirmation and further characterisation. Non-typhoidal Salmonella, which includes Salmonella Enteritidis, is classified as an epidemic-prone disease. Approximately 83.1% of the South African population have access to public healthcare and are therefore under surveillance by the GERMS-SA laboratory network.
Clinical metadata (age, gender, and HIV status of the patient) and microbiological metadata (invasive/non-invasive, year and province of collection and antimicrobial susceptibility) were captured in the GERMS-SA database for each S. Enteritidis isolate. For this study we used the captured metadata to select 618 S. Enteritidis isolates from the frozen GERMS-SA repository that accurately represent the invasive (isolated from blood and cerebral spinal fluid) and non-invasive (isolated from stool) Salmonella Enteritidis isolates referred to the NICD by Gauteng, KwaZulu-Natal, Mpumalanga, and Western Cape laboratories for the years 2012 and 2013.
From 2012 to 2014 there was a significant rise in S. Enteritidis outbreaks in South African poultry (10) and this surge in cases is consistent with the behaviour of this pathogen in other settings around the world (3, 4, 11). During this period (2012 and 2013), there was also a surge in the number of S. Enteritidis reported in humans, surpassing all other Salmonella serovars collected during these two years. Laboratories from Gauteng, KwaZulu-Natal, Mpumalanga, and the Western Cape contributed the majority (94.6%, 1 866/1 973) of all Salmonella Enteritidis isolates received in 2012 and 2013. Northern Cape, North West, Limpopo, Eastern Cape and Free State provinces collectively contributed only a small proportion of S. Enteritidis isolates (5.4%, 107/1 973) in 2012 and 2013, and were therefore excluded from the study.
All S. Enteritidis isolates were stored at -80°C in Cryogenic Tubes (ThermoFisher Scientific, California, USA) containing 1 mL Tryptic Soy broth (1 L distilled water, 17 g casein, 5 g sodium chloride, 3 g soytone, 2.5 g dextrose, 2.5 g dipotassium phosphate, adjusted to pH 7.3) in the GERMS- SA repository at the NICD (Johannesburg, South Africa).
For this study, we retrieved the Cryogenic Tubes with the 618 selected S. Enteritidis isolates and thawed the tubes at room temperature. The thawed culture was then streaked onto 5% blood agar (Diagnostic Media Products, Johannesburg, South Africa) using a 10 μl plastic classic Microloop (Medical Wire and Equipment Co. Ltd, Wiltshire, UK). We then incubated the agar plates in an IN 750 incubator (Memmert, Schwabach, Germany) set at 37°C for a minimum of 18 hours. Single colonies were picked off each agar plate using a 5 μl plastic classic Microloop (Medical Wire and Equipment Co. Ltd, Wiltshire, UK) and placed in 400 μl of 10 X TE buffer (800 mL distilled water, 2.92 g Tris, 15.76 g EDTA (pH 8)) in a 1.2 mL tube (Eppendorf, Hamburg, Germany). The solution was thoroughly mixed using a Vortex-Genie 2 (Lasec SA (Pty) Ltd., Midrand, South Africa) and boiled in a Dry Bath Plus heating block (Lasec SA (Pty) Ltd., Midrand, South Africa) set at 95°C for 25 minutes. The boiled cell solutions were then centrifuged using the Centrifuge 5415R (Eppendorf, Hamburg, Germany) at 22 673 g and 20 μl of the supernatant was transferred into 80 μl of 10 X TE buffer. We determined the DNA concentrations fluorometrically using the Qubit 2.0 Fluorometer (ThermoFisher Scientific, California, USA). A minimum DNA concentration of 10 ng/μL was accepted for use in the clade typing real-time PCR assay.
We previously established a multiplexed clade typing real-time PCR assay that can group a S. Enteritidis isolate within a particular clade (12). Approximately 2μL of crude DNA from each of the 618 S. Enteritidis isolates was used in the clade typing real-time PCR assay according to the method described in 12, without any modifications. As previously disclosed, the multiplexed clade typing real-time PCR assay is limited to confirmed S. Enteritidis isolates.
Hypothesising that these clades were associated with different clinical and microbiological characteristics, once the bacteria had been typed, the clinical and phenotypic information associated with each isolate was used to undertake a case-to-case analysis (Global Epidemic clade isolates compared with Outlier clade isolates) to determine the association between clade and particular characteristics (i.e., invasive diseases and antimicrobial resistance). Simple bivariate logistic models were preformed to obtain odds ratios with 95% confidence intervals using Stata version 14 (StataCorp LD, Texas, USA).
Ethical clearance for all laboratory-based surveillance and research (approved 12 November 2018) was obtained from the University of Witwaterstrand, Johannesburg, South Africa (Wits protocol no. M140159) by the Centre for Enteric Diseases, NICD. The sensitive data used within this study (i.e., HIV status) was anonymised through the removal of patient names.
A total of 618 confirmed Salmonella Enteritidis isolates were selected from the GERMS-SA 2012 and 2013 archive. From the accompanying patient data, we determined that the cohort consisted of similar proportions of male (329/618, 53.24%) and female (284/618, 45.95%) patients. The age range of these patients spanned 0-92 years, with the highest proportion of S. Enteritidis cases in children under the age of five years (27.99%, 173/618) (Table 1). HIV test result data was available for 228 (228/618, 36.89%) of the patients. A total of 165 (165/228, 72.37%) patients were HIV positive and 63 (63/228, 27.63%) were HIV negative (Table 1). The outstanding HIV status data for the remaining 390 patients were regarded as unknown.
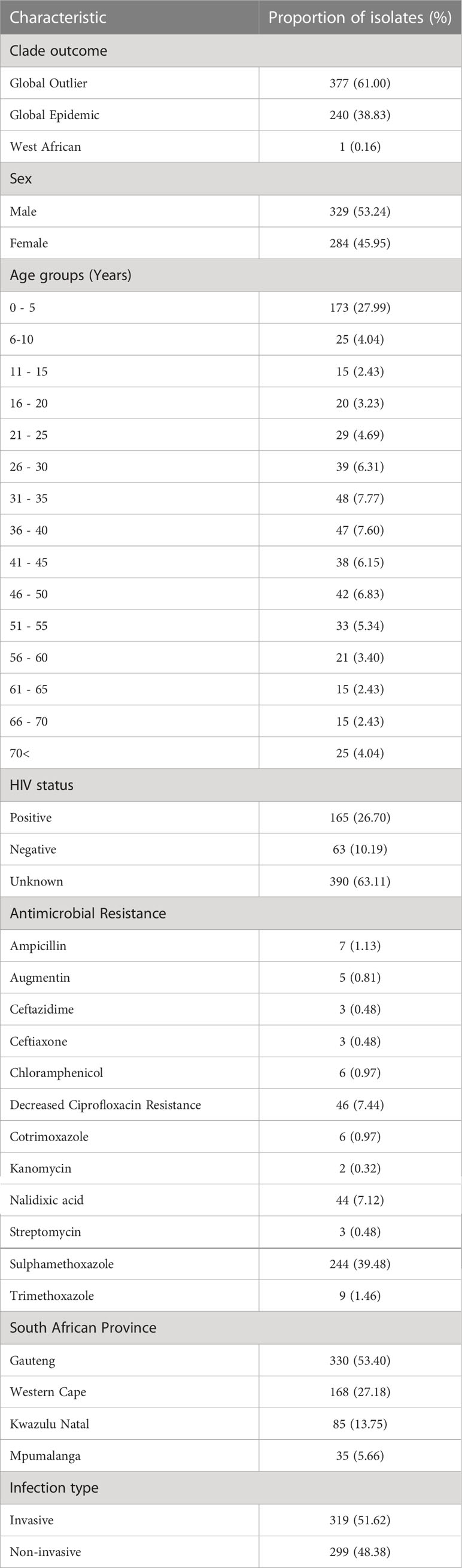
Table 1 Clinical metadata associated with and microbiological characteristics of the 618 selected Salmonella Enteritidis isolates.
The corresponding microbiological information for the S. Enteritidis isolates showed that approximately half of the isolates (319/618, 51.6%) were associated with invasive disease. A large proportion of the isolates were collected from the Gauteng province (330/618, 53.4%), while a smaller proportion were collected from the Western Cape (168/618, 27.18%), KwaZulu-Natal (85/618, 13.8%) and Mpumalanga (35/618, 5.7%). Interestingly, the greatest proportion of invasive isolates were from KwaZulu-Natal (63/85, 74.12%, OR: 2.60, CI: 1.528 – 4.421), while the Western Cape contributed the smallest proportion of invasive isolates (63/168, 37.50%, OR: 0.54, CI: 0.372 – 0.796).
Using the clade typing real-time PCR assay (12), the S. Enteritidis isolates were successfully typed according to clade. The isolates were typed within the Global Epidemic Clade (240/618, 38.9%), the paraphyletic outlier cluster to the Global Epidemic clade (Global Outlier clade) (377/618, 61.0%), and the West African clade (1/618, 0.2%) (Figure 1), however no East African Clade isolates were identified (9). Our study found a significant association between Global Epidemic clade isolates and decreased ciprofloxacin susceptibility (DCS) (15.42%, OR: 7.45, CI: 3.526 – 15.751) (Table 2). A further significant association was observed between the Global Epidemic clade and iNTS disease (58.33%, OR: 1.57, CI: 1.13 – 2.17) (Table 2).
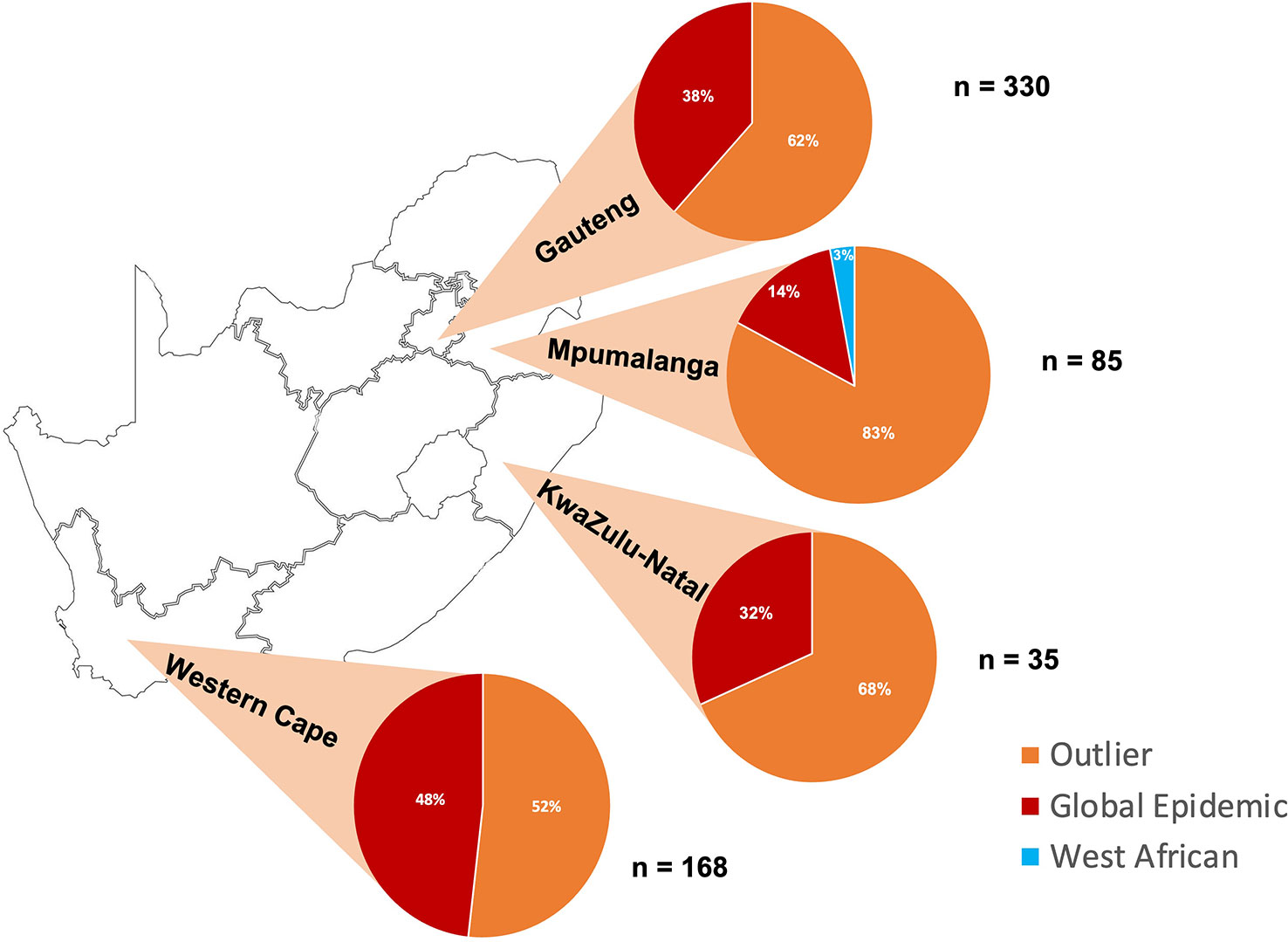
Figure 1 Proportion (%) of Salmonella Enteritidis isolates from four South African provinces (Gauteng, Mpumalanga, KwaZulu-Natal and Western Cape) classified within the Outlier, Global Epidemic and West African clade.
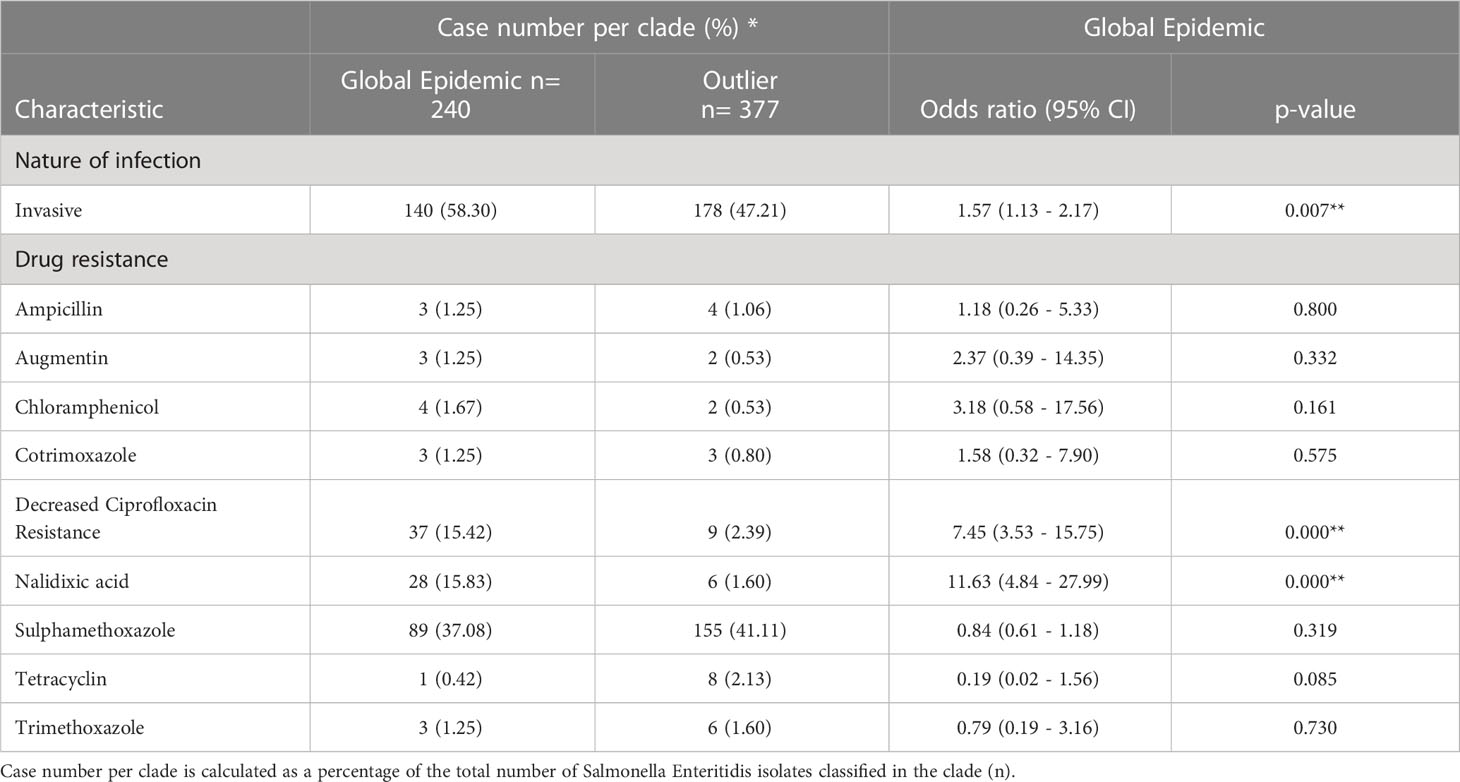
Table 2 Case-to-case (Global Epidemic clade compared with Outlier clade) statistical analysis of the association between Salmonella Enteritidis clades and characteristics (nature of infection and drug resistance).
Here we report our findings after typing 618 South African S. Enteritidis isolates according to previously defined clades (9). We found that the predominant clade was the paraphyletic outlier cluster to the Global Epidemic clade (Global Outlier clade), followed by the Global Epidemic clade and the West African clade. Our study showed that the Global clade isolates in South Africa are associated with invasive disease and decreased ciprofloxacin susceptibility (DCS).
The Global Epidemic clade is thought to have originated in Europe and spread to the US and Brazil through the commercial exchange of poultry between these countries (4). The genetic homogeneity of the clade suggests rapid clonal expansion in each country (9). South Africa’s increased poultry consumption from 2003 and subsequent increase in the import of poultry after 2010, was the most likely source of Global Epidemic clade introduction and spread within the country (4). The Department of Agriculture, Forestry and Fisheries has indeed reported an increase in S. Enteritidis outbreaks in poultry in the subsequent years, with less than 5 cases reported in 2011, 14 cases reported in 2012 and more than 23 cases reported in 2013 (10).
The complexity of South African poultry distribution as well as differences in food safety practices between rural and urban settings presents a network of transmission opportunities both nationally and regionally (13). Gauteng, KwaZulu-Natal and Western Cape are the major areas commercial production and trade of poultry in South Africa (14). This corresponds with the high proportion of the Global Epidemic clade that is present in Gauteng, KwaZulu-Natal and Western Cape (Figure 1), assuming the Global Epidemic clade has been introduced through international trade. The smaller proportion of the Global Epidemic clade in Mpumalanga (Figure 1) may be due to a heavier reliance on small scale local farms. There is a major risk of S. Enteritidis transmission throughout poultry stocks in the nascent industry across the region, with the potential for further outbreaks S. Enteritidis in settings with poor disease surveillance and a high prevalence of immunosuppressive disease.
We found that the Global Epidemic clade was associated with invasive disease in South Africa, which contrasts with genomic data from the European and US epidemics that revealed no association between Global Epidemic clade isolates and iNTS disease (9). However, this is likely due to the much higher prevalence of immunosuppressive disease (importantly, HIV) in sSA, which predisposes individuals to iNTS disease (8). In line with this, we found that the greatest proportion of invasive S. Enteritidis isolates were collected from KwaZulu-Natal, which is also the province with the highest HIV-infected population in South Africa (15).
Since 2010, South Africa has reported an increase in DCS, with approximately 1% of all S. Enteritidis surveillance isolates exhibiting DCS (16). The increase in DCS amongst S. Enteritidis isolates is thought to have been facilitated by the clonal expansion and spread of the Global Epidemic clade within South Africa (17). This picture is however mirrored in other African settings (18). In cases of DCS, patients are generally treated with longer courses of alternative intravenous antimicrobials, increasing the cost of treatment, posing a major problem to low-income countries where healthcare resources in general and reserve antibiotics are limited.
The S. Enteritidis isolates used in our study were collected using a passive laboratory-based surveillance program. These programs are limited by the health-seeking behaviour of an ill person, prevailing specimen collection practices, diagnostic practices at laboratories, and voluntary submission of isolates (19). In particular, the difference in stool collection practices when investigating adults versus children suffering from diarrhoea in healthcare facilities affects the number of confirmed NTS gastroenteritis cases reported (20). Furthermore, stool collection is more likely for patients seen in private and tertiary hospitals, which have the available resources. We acknowledge that the invasive and non-invasive NTS surveillance isolates received by the NICD do not represent the entire exposed population and therefore the results from this study could not be used to infer risk factors or incident rates. All these limitations point to the true burden being higher rather than spuriously lower than we have highlighted and serve to emphasise the case for improved surveillance.
Food safety remains a concern in South Africa for both the public health and poultry sectors. Cases and outbreaks of disease caused by S. Enteritidis continue to be reported throughout the country for both human and poultry infections (15, 21). S. Enteritidis outbreaks present a significant healthcare burden to a Middle-Income Country, such as South Africa. An integrated One Health surveillance approach to S. Enteritidis clades present in the environment, livestock, and health sector, is necessary to provide cohesive data for targeted public health action to mitigate Salmonella transmission, acknowledging that the clades are phenotypically distinct pathogens with distinct distribution or reservoirs. This will facilitate a better understanding of the routes of transmission and will assist with the public health response within South Africa as well as limit the exportation of the disease to surrounding sSA countries with rapidly developing poultry sectors. It is our hope that an increase in the surveillance S. Enteritidis clades in South Africa and abroad will help enable public health authorities to prevent future outbreaks and provide safer poultry for all.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
SG was responsible for the conception and design of the study (with help from AS and NF), laboratory work and wrote the first draft of the manuscript. SG and NR carried out the statistical analysis of the epidemiological data. NF revised, edited and wrote sections of the manuscript. NF, AS, and JT supervised SG throughout the study. All authors contributed to the article and approved the submitted version.
This work was supported by the German Federal Ministry of Education and Research (BMBF grant number: 81203616).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. OECD/Food and Agriculture Organization of the United Nations. OECD-FAO agricultural outlook 2022-2031. In: “Executive summary,”. Paris: OECD Publishing (2022).
2. Aworh OC. Food safety issues in fresh produce supply chain with particular reference to sub-Saharan Africa. Food Control (2021) 123. doi: 10.1016/J.FOODCONT.2020.107737
3. Ward LR, Threlfall J, Smith HR, O’Brien SJ. Salmonella enteritidis epidemic. Sci (1979) (2000) 287:1753–6.
4. Li S, He Y, Mann DA, Deng X. Global spread of salmonella enteritidis via centralized sourcing and international trade of poultry breeding stocks. Nat Commun 2021 12:1 (2021) 12:1–12. doi: 10.1038/s41467-021-25319-7
5. Thomas KM, de Glanville WA, Barker GC, Benschop J, Buza JJ, Cleaveland S, et al. Prevalence of campylobacter and salmonella in African food animals and meat: a systematic review and meta-analysis. Int J Food Microbiol (2020) 315. doi: 10.1016/J.IJFOODMICRO.2019.108382
6. Tack DM, Marder EP, Griffin PM, Cieslak PR, Dunn J, Hurd S, et al. Preliminary incidence and trends of infections with pathogens transmitted commonly through food — foodborne diseases active surveillance network, 10 U.S. sites 2015–2018. Morbidity Mortality Weekly Rep (2019) 68:369. doi: 10.15585/MMWR.MM6816A2
7. Gordon MA, Feasey NA, Nyirenda TS, Graham SM. Nontyphoid salmonella disease. Hunter’s Trop Med Emerging Infect Dis (2020) 5:500–6. doi: 10.1016/B978-0-323-55512-8.00049-1
8. Stanaway JD, Parisi A, Sarkar K, Blacker BF, Reiner RC, Hay SI, et al. The global burden of non-typhoidal salmonella invasive disease: a systematic analysis for the global burden of disease study 2017. Lancet Infect Dis (2019) 19:418–9. doi: 10.1016/S1473-3099(19)30418-9
9. Feasey NA, Hadfield J, Keddy KH, Dallman TJ, Jacobs J, Deng X, et al. Distinct salmonella enteritidis lineages associated with enterocolitis in high-income settings and invasive disease in low-income settings. Nat Genet (2016) 48:1211–7. doi: 10.1038/ng.3644
10. Magwedere K, Rauff D, De Klerk G, Keddy KH, Dziva F. Incidence of nontyphoidal salmonella in food-producing animals, animal feed, and the associated environment in south Africa 2012–2014. Clin Infect Dis (2015) 61:S283–9. doi: 10.1093/CID/CIV663
11. Silva E, Duarte A. Salmonella enteritidis in poultry: retrospective in Brazil. Rev Bras Cienc Avic (2002) 4:85–100. doi: 10.1590/S1516-635X2002000200001
12. Gallichan S, Perez-Sepulveda BM, Feasey NA, Hinton JCD, Thomas J, Smith AM. Multiplex PCR assay for clade typing of salmonella enterica serovar enteritidis. Microbiol Spectr (2022). doi: 10.1128/SPECTRUM.03182-22
13. Hawkes C, Ruel M. The links between agriculture and health: an intersectoral opportunity to improve the health and livelihoods of the poor. Bull World Health Organ (2006) 84:984–90. doi: 10.2471/blt.05.025650
14. South African Poultry Association. 2020 industry profile (2020). Available at: https://www.sapoultry.co.za/wp-content/uploads/2022/03/SAPA-INDUSTRY-PROFILE-2020.pdf (Accessed January 16, 2023).
15. National Institute for Communicable Diseases. GERMS-SA: Annual surveillance review (2020). Available at: https://www.nicd.ac.za/wp-content/uploads/2022/02/2020-GERMS-SA-Annual-Review.pdf (Accessed January 4, 2023).
16. Crowther-Gibson P, Govender N, Lewis D, Bamford C, Brink A, von Gottenberg A, et al. Human infections and antibiotic resistance. South Afr Med J (2011) 101:567–78.
17. Govender N. Molecular epidemiology and mechanism of resistance of invasive QuinoloneResistant south African isolates of salmonella enterica. (2009) Johannesburg, South Africa. pp. 2004–6.
18. Aldrich C, Hartman H, Feasey N, Chattaway MA, Dekker D, Al-Emran HM, et al. Emergence of phylogenetically diverse and fluoroquinolone resistant salmonella enteritidis as a cause of invasive nontyphoidal salmonella disease in Ghana. PLoS Negl Trop Dis (2019) 13:e0007485. doi: 10.1371/journal.pntd.0007485
19. Crump JA, Youssef FG, Luby SP, Wasfy MO, Rangel JM, Taalat M, et al. Estimating the incidence of typhoid fever and other febrile illnesses in developing countries. Emerg Infect Dis (2003) 9:539–544. doi: 10.3201/eid0905.020428
20. World Health Organization: Food and Agricultural Organization of the United Nation. Risk Assessments of Salmonella in Eggs and Broiler Chickens (2022). Second. Rome, Italy: WHO Library Cataloguing-in-Publication Data.
Keywords: non-typhoidal Salmonella, molecular epidemiology, food safety, antimicrobial resistance, sub-Sahara African countries
Citation: Gallichan S, Ramalwa N, Thomas J, Feasey N and Smith AM (2023) Salmonella Enteritidis clades in South Africa: why we should be paying more attention. Front. Trop. Dis 4:1152422. doi: 10.3389/fitd.2023.1152422
Received: 27 January 2023; Accepted: 28 March 2023;
Published: 20 April 2023.
Edited by:
Alex Owusu-Ofori, Kwame Nkrumah University of Science and Technology, GhanaReviewed by:
Joshua Mbanga, National University of Science and Technology, ZimbabweCopyright © 2023 Gallichan, Ramalwa, Thomas, Feasey and Smith. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sarah Gallichan, c2FyYWhhZ2FsbGljaGFuQGdtYWlsLmNvbQ==; Anthony M. Smith, YW50aG9ueXNAbmljZC5hYy56YQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.