Abstract
Odor from preferred/non-preferred tsetse fly vertebrate hosts have been exploited in R&D of attractants/repellents of the fly for human and livestock protection. Odors from vertebrate hosts of Glossina austeni and Glossina pallidipes tsetse flies can facilitate formulation of novel attractants effective against G. austeni or improvement of existing attractant blends for G. pallidipes. We compared vertebrate blood meal sources of both fly species at Shimba Hills National Reserve, Kenya, to establish putative preferred host of either species, hence potential source of G. austeni or G. pallidipes specific odors. We trapped sympatric adult flies in 2021 and 2022 using NGU traps/sticky panels baited with 3-propylphenol, octenol, p-cresol, and acetone (POCA), collected their blood meals and characterized the meals using High Resolution Melting (HRM) vertebrate 16S rRNA- PCR (for host identification), and compared host profiles using GLM and Fisher’s exact tests. We collected 168 and 62 sympatric G. pallidipes and G. austeni with bloodmeal, respectively in 2021 and, 230 and 142 respectively in 2022. In 2021, we identified putative hosts of 65.48 and 69.35% of the G. pallidipes and G. austeni respectively and 82.61 and 80.28%, respectively in 2022. In 2021, we detected harnessed bushbuck, buffalo, common warthog and cattle putative host bloodmeals, and additionally bushpig and suni antelope bloodmeals in 2022. Putative vertebrate bloodmeal sources were significantly different by tsetse fly species (χ²(1, N=457) = 43.215, p < 0.001) and sampling year (χ²(1, N=457) = 8.044, p = 0.005). Frequency of common warthog bloodmeals was higher in G. pallidipes (65.79%) than in G. austeni (38.60%), and that of suni antelope and harnessed bushbuck putative bloodmeals higher in G. austeni (21.05-28.07%) than in G. pallidipes (6.84 - 17.37%) in 2022. There was an apparent change in putative feeding preference/host choices in both fly species between 2021 and 2022. Host bloodmeals in G. pallidipes or G. austeni predominantly from putative harnessed bushbuck, suni antelope or common warthog reveal that these vertebrates have potential odors that can be harnessed and formulated into appropriate attractants for respective species and integrated into routine control regiment for G. pallidipes and/or G. austeni.
Introduction
Human African Trypanosomiasis (HAT) and Animal African Trypanosomiasis (AAT) are among the most Neglected Tropical Diseases with devastating health and economic consequences in sub-Sahara Africa (1, 2). African trypanosomes that cause HAT and AAT are transmitted by different groups of tsetse species that have far-reaching impacts on human and animal health. With no effective vaccines against HAT and AAT, vector control remains the cornerstone of disease suppression and eradication. Bait technologies based on visual and olfactory responses to natural visual cues and synthetic blends of attractants, that mimic those of natural hosts, have successfully been applied in tsetse fly control (3). These technologies are environment friendly (4), and especially applicable for savanna species. Attractants include various phenolic derivatives (5, 6) of host-emitted and excreted metabolic products, carbon dioxide, acetone, and 1-octen-3-ol (7–12), which have been formulated into POCA (3-propylphenol, octenol, p-cresol and acetone) with enhanced attraction to and routinely used for control of Glossina pallidipes and most savannah species (10, 13). Natural responses in tsetse flies to this and/or other odors vary between species among conspecific populations and sexes, (11, 14–16). Relative avoidance of tsetse fly-refractory animals is mediated by specific repellent constituents or blends emitted by these animals. These compounds include guaiacol (methylphenols), δ-octalactone, methylketones (14, 17) and 2-methoxy-4-methylphenol (18). This has stimulated the development of ‘push-pull’ tactics that integrate attractant baits with use of controlled release of repellents or blends on preferred hosts to push target tsetse flies to the attractant bait. Compositions of odor profiles of attractive or refractory host animals, which can aid in the formulation of attractant and repellent blends effective on Glossina austeni (for which there are no known attractants or repellents), or new compounds, which can enhance the attraction of the current blend (19) or POCA (which are sub-optimally active compared to natural odor from attractive hosts) to G. pallidipes and other savannah tsetse species have not been established. To date, no effective attractants or repellents have been characterized for G. austeni tsetse fly and riverine tsetse species, such as Glossina fuscipes fuscipes.
In Kenya, at least two tsetse species occur in sympatry, which necessitates development of tsetse control tools that target the species in sympatry within tsetse habitats for significant disruption of trypanosome transmission. In Shimba Hills National Reserve along the coast of Kenya, G. pallidipes occur in sympatry with G. austeni (20). Glossina pallidipes is attracted mainly to Bovidae and warthog as revealed by analyses of host bloodmeals from field caught flies (21, 22). The results are consistent with previous studies on the attraction of body odor of buffalo and ox (17) and blends of constituents of fermented cattle urine to G. pallidipes (8, 10, 11, 23). Common warthog (Phacochoerus africanus), hippopotamus (Hippopotamus amphibious), African elephant (Loxodonta africana), giraffe (Giraffa camelopardis) and baboon (Papio spp.) have been identified as optional blood meal sources for G. pallidipes (24, 25), feeding on them when available. While G. pallidipes responds well to POCA, G. austeni does not (26), indicating potential differential host preferences between the two tsetse fly species. No effective attractant has been established against G. austeni and only single forensic bloodmeal assessment of limited laboratory curated specimens has associated G. austeni host preference to bushpig (Potamochoerus larvatus) (27). Identification of natural differential host preferences between the two sympatric species can help establish additional host candidates that can be exploited to formulate new or improved attractants against either species.
We therefore initiated this study to see if there are differences in feeding profiles of sympatric G. austeni and G. pallidipes during 2021 and 2022 in Shimba Hills National Reserve, as revealed by their bloodmeals and associated vertebrate hosts.
Materials and methods
Ethics statement
Clearance for this research in protected areas was provided by Kenya Wildlife Service (KWS), and Wildlife Research and Training Institute Research Authorization committee via Permits KWS/BRM/5001 and WRTI-0198-06-22.
Study location
We conducted our studies at Shimba Hills National Reserve (004° 15’ 26’’S, 039° 23’16’’E) (altitude 403 m) in Kwale County, Kenya where wild populations of G. pallidipes and G. austeni occur in sympatry. The reserve is an enclosed area of about 250 km2 surrounded by human habitats, where livestock herding is common. The reserve has several large mammal populations that include elephants, harnessed bushbuck, duikers, giraffes, leopards, monkeys, hartebeest, common warthogs, buffalos, suni antelope, sable antelopes and bush pigs. The reserve is rich in flora characterized by areas of coastal rain forest, dense semi-evergreen woodland, open woodland, savannah and open grasslands. The area typically experiences long and short rainy seasons from April to June, and October to November, respectively. Mean annual rainfall in the reserve ranges between 855 and 1682 mm. Maximum daily temperatures are highest in March and November, often reaching 30-31°C. June to July are the coolest months, with daily maximum temperatures of 26-27°C.
Study design and sample collection
Due to dense forest and other vegetation cover of our study area, we initiated our study by mapping for tsetse presence in accessible sites, within the reserve where G. austeni and G. pallidipes are sympatric. To achieve this, we identified blocks of accessible areas, randomly placed NGU cloth traps baited with POCA (28, 29) 1-5 km apart within the blocks and collected trapped tsetse flies 24 hrs post deployments between December 2020 and June 2021. Each trapped tsetse fly was identified using taxonomic keys (30). The data informed us on blocks with sympatric G. austeni and G. pallidipes populations. We subsequently selected these blocks for our definitive sampling from August 28-September 24, 2021, and June 24-July 9, 2022. For the definitive sampling, we placed NGU cloth traps baited with POCA and sticky panels (31) totaling to 146 around tree trunks in alternating pattern at intervals of at least 100 meters. While G. pallidipes were attracted by both the visual reflectance of the traps as well as the olfactory attractants deployed, G. austeni were attracted to the trap visually (32), since no effective attractants of G. austeni have been characterized and formulated. We deliberately incorporated sticky panels since our study targeted only fed flies that typically rest on cool and shaded areas and avoid baited NGU cloth traps which are biased towards sampling hungry host-seeking flies. We geo-referenced all our sampling sites. We deployed 24 traps and sticky panels within our four definitive sampling blocks (Figure 1B) and collected trapped flies at 1700 hrs each day to cover the morning and afternoon peaks, coincident with significant interaction between tsetse flies and their respective hosts (22). This duration covered the bimodal and unimodal tsetse peak activity periods for G. pallidipes and G. austeni respectively. We labelled the cages at collection, sedated the flies with chloroform, transferred them into 15 ml falcon tubes, preserved them in cool boxes containing icepacks and transported them to the laboratory. In the laboratory, we identified the flies to species level using taxonomic keys (30). Briefly, we distinguished G. pallidipes from G. austeni species using differences in 1) position of their median lobes between superior claspers, 2) adnominal colors and banding patterns and 3) color of the hind tarsi morphological features. Median lobes in G. austeni project out beyond general line of the superior claspers while those in G. pallidipes do not. Dorsal surface in G. austeni is sandy reddish-brown and not strongly banded on abdomen while in G. pallidipes the surface is dark, and the abdomen strongly banded. Dark color on hind tarsi in G. pallidipes is limited to last two tarsal segments, but not in G. austeni. We further distinguished the two species from Glossina brevipalpis sympatric with the two species in the study area (but not involved in our study) by presence of hairy fringe to the squamae (in G. brevipalpis). We visually established feeding status of the tsetse flies and confirmed by microscopic dissection and examination of their gut contents. We removed and either squashed the midgut contents on a partitioned and clearly labelled Whatman® filter paper no. 1 or placed them, individually, in 1.5ml of 70% ethanol in Eppendorf tubes. We prepared and documented the smears and tubes from samples collected in each site and trap/panel for tsetse species (30) to get a representative sample per species. We air dried the smears under shade, wrapped them with grease proof aluminum foil and stored the samples inside an airtight desiccator. We kept the samples dry using a self-indicating silica gel which we replaced as necessary until the samples were transported to the Kenya Wildlife Services (KWS) Forensic and Genetics Laboratory. We stored both smeared and ethanol preserved samples at 4°C.
Figure 1

Map of sampling sites for (G) pallidipes and (G) austeni tsetse flies in Shimba Hills National Reserve in Kwale County, Kenya. Tsetse flies were sampled in the reserve in two phases. First phase identified spatial locations/sites within reserve with (G) pallidipes and/or (G) austeni tsetse flies (A). Second phase focused on the locations/sites among those identified in the first phase where both tsetse fly species were sympatric (B).
Molecular analysis for host species identification
We cut out discs of about 1 cm diameter from blood spots on the sample holding filter papers, shredded them into finer pieces and separately placed them into 1.5ml centrifuge tubes for DNA extraction. Genomic DNA was extracted using extraction kits (Bioline, London, UK) in accordance with manufacturer’s instructions. We also extracted DNA from ethanol-preserved fly gut samples using DNeasy Blood and Tissue extraction kit (Qiagen, Germany) following manufacturer’s instructions. In both cases, we stored genomic DNA of all samples at -20°C until required for host blood-meal identification. For host blood-meal identification, we employed PCR coupled with high-resolution melting (HRM) analysis of vertebrate 16S rRNA gene product as previously described (33) using PCR-HRM thermal cycler (Qiagen, Germany). We targeted two (cytochrome b and 16S ribosomal RNA) vertebrate mitochondrial genes to select the most sensitive gene. We amplified 16S ribosomal RNA gene with Vert 16S (Forward primer 5’ -GAGAAGACCCTRTGGARCTT-3’ and Vert 16S Reverse primer 5’ -CGCTGTTATCCC TAGGGTA-3’) targeting approximately 200bp region. We used 15µl reaction volumes constituting 4µl Hot firepol 5X Evergreen (Master Mix), 0.5µl of 10 pmol vertebrate 16S rRNA gene forward and reverse primers (34), 8µl of PCR grade water and 2µl of template DNA. We set the touch-down PCR amplification conditions as follows: initial holding temperature at 95°C for 15 minutes followed by 10 cycles at 94°C for 30 seconds, 63.5°C for 30 seconds and 72°C for 45 seconds. We set another 25 cycles at 94°C for 30 seconds, 50.5°C for 30 seconds and 72°C for 45 seconds, and the final extension at 72°C for 10 minutes. We included both positive and negative controls (non-template) for assay quality assurance. We conducted PCR and HRM analyses using Rotor-Gene Q software version.2.1.0.9. We conducted HRM ramping from 75°C to 95°C, rising by 0.1°C each step with a wait of 2 seconds for each step afterwards as described by Nyamota et al. (35). We analyzed HRM profiles using Rotor gene 2.1.0.9 software, with normalized regions between 78°C and 92°C. We then purified amplicons representative of each unique HRM profile using ExoSAP-IT™ (Applied Biosystems) according to the manufacturer’s instructions. We submitted the amplicons to Inqaba Biotec™, South Africa, for unidirectional Sanger sequencing. We analyzed and aligned the sequences using Bioedit version 7.0.5.3 software (36), and confirmed the vertebrate species by sequence alignment and ≥99% homology via Basic Alignment Search Tool (BLAST) (37) against National Center for BioTechnology Information (NCBI) non-redundant (nr) nucleotide-nucleotide database (https://www.ncbi.nlm.nih.gov/nucleotide/) accessed on 15, 10, 2022.
Data analysis
The focus of our analysis was to determine differences in feeding preferences between G. pallidipes and G. austeni, based on counts of host identified in our 2021 or 2022 sampling periods.
We coincidentally therefore fitted a generalized linear model (GLM) with Poisson distribution incorporating negative binomial log link function to establish overall or specific association between tsetse fly species (G. pallidipes, G. austeni) or year of sampling (2021/2022) (independent variables) and frequency of bloodmeals from putative vertebrate hosts (dependent covariates). We dropped data that did not fit the model. In all analyses, we considered P value below 0.05 as significant. We conducted all analyses using IBM SPSS version 22.0 (IBM Corp, Armonk, New York, USA).
Results
Sympatric tsetse fly populations
We collected 11691 and 1843 G. pallidipes and G. austeni tsetse flies respectively in our initial assessment of spatial distribution of both species across the reserve between December 2020 and June 2021 (Figure 1A). We subsequently identified 24 sites where the fly species were sympatric (Figure 1B) and from where we collected 168 and 62 G. pallidipes and G. austeni flies with bloodmeals, respectively between August 28 and September 24, 2021. We successfully extracted DNA from bloodmeals of 110 G. pallidipes and 43 G. austeni, of this population. We collected additional 230 G. pallidipes and 142 G. austeni, sympatric flies from the same sites (Figure 1B) between June 24 and July 9, 2022. Out of these we successfully extracted DNA from bloodmeals of 190 and 114 of G. pallidipes and G. austeni respectively.
Putative vertebrate host identification
Our PCR-HRM analysis of bloodmeals identified putative sources (hosts) of bloodmeal for 87 G. pallidipes and 41 G. austeni in 2021, and 190 and 90 of the samples, respectively collected in 2022. Among these, the 2021 bloodmeal samples were putatively associated with harnessed bushbuck (Tragelaphus scriptus, 16S RNA GenBank accession JN632707), buffalo (Syncerus caffer, 16S RNA GenBank accession JQ235547), common warthog (Phacochoerus africanus) (16S RNA GenBank accession OK183892) and cattle (Bos Taurus, 16S RNA GenBank accession OK183899), (Figure S1A). The blood meal samples collected in 2022 were also associated with all species we identified in the samples we collected in 2021, and additionally included bushpig (Potamochoerus larvatus, 16S RNA GenBank accession GQ338948) and suni antelope (Neotragus moschatus, 16S RNA GenBank accession JN632669) (Figure S1B) for both tsetse fly species. Overall, profiles of putative vertebrate bloodmeal sources were significantly different by tsetse fly species (χ²(1, N=457) = 43.215, p < 0.001) and year of sampling (χ²(1, N=457) = 8.044, p = 0.005), with more vertebrates bloodmeals associated with G. pallidipes than G. austeni, and with higher frequencies in 2022 than in 2021 (Figure 2). We detected significantly more putative harnessed bushbuck bloodmeals in G. austeni than in G. pallidipes (χ²(1, N=457) = 4.516, p =0.034). Frequency of putative bloodmeals from buffalo was similar among tsetse fly species (χ²(1, N=457) = 2.250, p =0.134) and year of sampling (χ²(1, N=457) = 3.063, p =0.080). On the other hand, bloodmeals from putative common warthog were more frequent in 2022 than in 2021 (χ²(1, N=457) = 19.472, p < 0.001) and in G. pallidipes than in G. austeni (χ²(1, N=457) = 9.878, p =0.002). We only detected bloodmeals from putative bushpig (χ²(1, N=457) = 27.04, p < 0.001), or suni antelope (χ²(1, N=457) = 45.256, p < 0.001) hosts in 2022, with bloodmeals from putative bushpig significantly predominant in G. pallidipes than in G. austeni (χ²(1, N=457) = 4.000, p = 0.046), and those from putative suni antelope more predominant in G. austeni than in G. pallidipes (χ²(1, N=457) = 4.000, p = 0.046). Data on bloodmeal associated with putative bloodmeals from cattle did not fit the GLM model and were thus not considered for analysis.
Figure 2
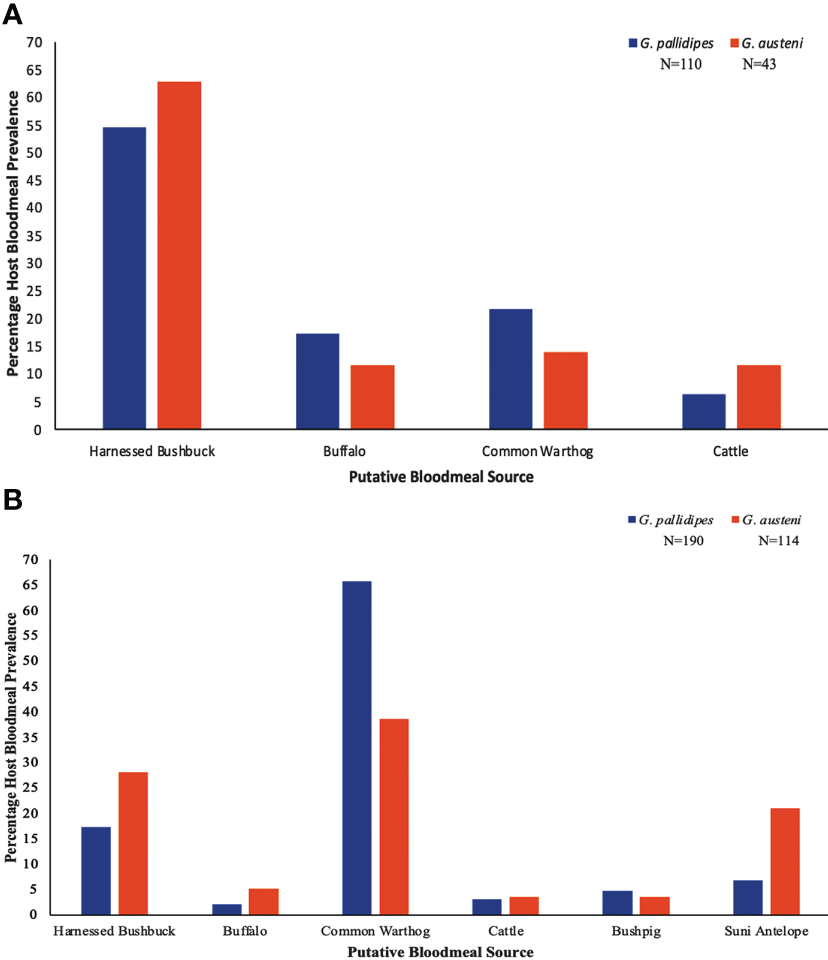
Proportions (%) of putative vertebrate hosts associated with (G) pallidipes and (G) austeni tsetse fly bloodmeals in 2021 (A) and 2022 (B) in Shimba Hills National Reserve in Kwale County, Kenya.
Discussion
The current study was focused on identifying vertebrate hosts associated with bloodmeals in sympatric G. pallidipes and G. austeni tsetse flies in Shimba Hills National Reserve, Kwale, Kenya in 2021 and 2022. We established that although bloodmeals in both tsetse fly species were derived from six vertebrate species (bushbuck, buffalo, common warthog, cattle, bushpig and suni antelope), in 2022 those from warthog predominated in G. pallidipes, and those from bushbuck and suni antelope predominated in G. austeni. These bloodmeal identities potentially reflect actual host choices/preferences in the two species and could have also been influenced by available vertebrate host sustaining the fly populations in the reserve. These findings are particularly interesting since previous studies presented preferential feeding predisposition in G. pallidipes and G. austeni for buffalo (38) and bushpig, respectively (30; 27). These findings could be attributed to adaptation of the flies to ecology (most abundant and available species) of the reserve, with the low frequency of bushpig likely due to differences in activity times between the fly and the host. Glossina austeni are most active during the day with U-shaped unimodal pattern during hottest part of the day (39). They spend the rest of the day and night resting on surfaces such as tree trunks and leaves. On the other hand, bushpigs are largely nocturnal (40) and hide in dense cover of thick forests during the day and tend to avoid open forests or savannas where G. austeni are most active. Thus, low frequency feeding on bushpig in our record may not be attributed to active avoidance of bushpig by the fly, but rather on unavailability of the bushpig hosts when the tsetse flies are most active. Nevertheless, the additional preferred hosts of G. pallidipes or G. austeni we identified provide additional sources of odor cues for formulation of attractants against the flies. This can however only be verified by isolation of the odors followed by extensive laboratory and field bioassays.
We coincidentally observed similar host profiles in bloodmeal patterns between G. pallidipes and G. austeni in samples collected in 2021, although the bloodmeal samples of fed tsetse flies were less than those we sampled and characterized (bloodmeal) in 2022. The underlying reason behind this phenomenon is not obvious but, it may be due to prevailing environmental conditions. In harsh conditions, plants typically defend themselves against herbivores by producing physical structures (trichomes and spines) or toxins and certain digestibility reducers (41) in addition to releasing volatile organic compounds (VOCs) (42) that help them adapt to the harsh weather/climatic conditions. Consequently, the tsetse bloodmeal hosts are likely to forage on flora whose phytochemical components could qualitatively alter the kind of the odor cues they release to the environment, with the likelihood of differences in odor cues emanating from the hosts during the wet and dry seasons. In addition, it may be due to a reduction in the availability of the preferred host, compelling the flies to source bloodmeals from non-preferred hosts, as previously observed with zebra and waterbuck (43, 44). This phenomenon further reveals plasticity in host preference in tsetse flies probably an adaptation in response to potential ecological changes. The higher feeding frequency in cattle in 2021 relative to 2022 in both tsetse fly species was probably due to a reduction in available alternative hosts. This can consequently increase the risk of trypanosome transmission to livestock and increased disease burden. The difference in host bloodmeal identities between 2021 and 2022 can also be attributed to the difference in sample sizes of the two periods.
In conclusion, preference of harnessed bushbuck and suni antelope by G. austeni, and common warthog by G. pallidipes suggest that these hosts can potentially provide odor cues inclined to either species. These cues can find application in integrated management of tsetse and trypanosomiasis. Both flies display host preference plasticity, probably an adaptation response to ecological changes, which sometimes enhances feeding of the tsetse flies on cattle. The bloodmeals also reveal potential reservoirs of trypanosomes that drive dynamics of local trypanosomiasis transmission in Shimba Hills, Kwale, Kenya.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
Clearance for this research in protected areas was provided by Kenya Wildlife Service (KWS), and Wildlife Research and Training Institute Research Authorization committee via Permits KWS/BRM/5001 and WRTI-0198-06-22.
Author contributions
KOO, TO and BB collected and analyzed the samples, drafted and reviewed the manuscript; MYO collected the samples, guided sample analysis and reviewed the manuscript; CMM generated figures and reviewed the manuscript; BO collected the samples and reviewed the manuscript; MOO, JM, AH and PO reviewed the manuscript; POM Conceived the experiments, wrote and reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by National Institutes of Health (NIH) grant No. R01 AI169503 to PM. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acknowledgments
We thank Mr. Paul Thande, Mr. Patrick Obore (Biotechnology Research Institute, Kenya Agricultural and Livestock Research Organization, Kikuyu, Kenya) and Mr. Ali Mwabilo for their technical field assistance in collection, storage and analysis of the samples. We also thank Mr. Elias Thuranira (National Agricultural Laboratories, Kenya Agricultural and Livestock Research Organization) for assistance with GLM data analysis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fitd.2023.1145993/full#supplementary-material
Supplementary Figure 1Distinct PCR-HRM profiles for putative vertebrate host species. The profiles show differentiation of bushbuck, cattle, buffalo and common warthog (A) and bushpig, common warthog, African Buffalo harnessed bushpig, cattle and suni antelope (B). The normalized HRM profiles are for 16S rRNA markers represented as percent fluorescence.
References
1
Hotez PJ Fenwick A Savioli L Molyneux DH . Rescuing the bottom billion through control of neglected tropical diseases. Lancet (2009) 373(9674):1570–5. doi: 10.1016/S0140-6736(09)60233-6
2
Brun R Blum J Chappuis F Burri C . Human african trypanosomiasis. Lancet (2010) 375(9709):148–59. doi: 10.1016/S0140-6736(09)60829-1
3
Mangwiro TN Torr SJ Cox JR Holloway MT . The efficacy of various pyrethroid insecticides for use on odour-baited targets to control tsetse. Med Vet Entomol (1999) 13(3):315–23. doi: 10.1046/j.1365-2915.1999.00165.x
4
Allsopp R . Options for vector control against trypanosomiasis in Africa. Trends Parasitol (2001) 17(1):15–9. doi: 10.1016/s1471-4922(00)01828-6
5
Den Otter CJ Van der Goes van Naters W . Responses of individual olfactory cells of tsetse flies (Glossina m. morsitans) to phenols from cattle urine. Physiol Entomol (1993) 18:43–9. doi: 10.1111/j.1365-3032.1993.tb00447.x
6
Saini RK Hassanali A Andoke J Ahuya P Ouma WP . Identification of major components of larviposition pheromone from larvae of tsetse flies Glossina morsitans morsitans Westwood and Glossina morsitans centralis machado. J Chem Ecol (1996) 22(7):1211–20. doi: 10.1007/BF02266961
7
Hall DR Beevor PS Cork A Nesbitt BF Vale GA . 1-Octen-3-ol. a potent olfactory stimulant and attractant for tsetse isolated from cattle odours. Int J Trop Insect Sci (1984) 5(5):335–9. doi: 10.1017/S1742758400008626
8
Hassanali A McDowell PG Owaga MLA Saini RK . Identification of tsetse attractants from excretory products of a wild host animal, Syncerus caffer. Int J Trop Insect Sci (1986) 7(1):5–9. doi: 10.1017/S1742758400003027
9
Vale GA Hargrove JW Cockbill GF Phelps RJ . Field trials of baits to control populations of Glossina morsitans morsitans westw. and G. pallidipes austen (Diptera: Glossinidae). Bull entomological Res (1986) 76(4):179–94. doi: 10.1017/S000748530001467X
10
Owaga ML Hassanali A McDowell PG . The role of 4-cresol and 3-n-propylphenol in the attraction of tsetse flies to buffalo urine. Int J Trop Insect Sci (1988) 9(1):95–100. doi: 10.1017/S1742758400010110
11
Vale GA Hall DR Gough AJE . The olfactory responses of tsetse flies, Glossina spp.(Diptera: Glossinidae), to phenols and urine in the field. Bull Entomological Res (1988) 78(2):293–300. doi: 10.1017/S0007485300013055
12
Bogner F . Response properties of CO2-sensitive receptors in tsetse flies (Diptera: Glossina palpalis). Physiol Entomol (1992) 17(1):19–24. doi: 10.1111/j.1365-3032.1992.tb00985.x
13
Rayaisse JB Tirados I Kaba D Dewhirst SY Logan JG Diarrassouba A et al . Prospects for the development of odour baits to control the tsetse flies Glossina tachinoides and g. palpalis sl. PloS Negl Trop Dis (2010) 4(3):e632. doi: 10.1371/journal.pntd.0000632
14
Gikonyo NK Hassanali A Njagi PG Saini RK . Responses of Glossina morsitans morsitans to blends of electroantennographically active compounds in the odors of its preferred (buffalo and ox) and nonpreferred (waterbuck) hosts. J Chem Ecol (2003) 29(10):2331–45. doi: 10.1023/a:1026230615877
15
Mireji PO Mabveni AM Dube BN Ogembo JG Matoka CM Mangwiro TNC . Field responses of tsetse flies (Glossinidae) and other diptera to oils in formulations of deltamethrin. Int J Trop Insect Sci (2003) 23(4):317–23. doi: 10.1017/S1742758400012388
16
Mwangi MT Gikonyo NK Ndiege IO . Repellent properties of delta-octalactone against the tsetse fly, Glossina morsitans morsitans. Int J Trop Insect Sci (2008) 8:1–4. doi: 10.1673/031.008.4301
17
Gikonyo NK Hassanali A Njagi PG Gitu PM Midiwo JO . Odor composition of preferred (buffalo and ox) and nonpreferred (waterbuck) hosts of some savanna tsetse flies. J Chem Ecol (2002) 28(5):969–81. doi: 10.1023/a:1015205716921
18
Saini RK Hassanali A . A 4-alkyl-substituted analogue of guaiacol shows greater repellency to savannah tsetse (Glossina spp.). J Chem Ecol (2007) 33(5):985–95. doi: 10.1007/s10886-007-9272-7
19
Wachira BM Kabaka JM Mireji PO Okoth SO Murilla GA Hassanali A . Blending studies with selected waterbuck odor constituents or analogues in the development of a potent repellent blend against savannah tsetse. Acta Tropica (2020) 211:105597. doi: 10.1016/j.actatropica.2020.105597
20
Ngari NN Gamba DO Olet PA Zhao W Paone M Cecchi G . Developing a national atlas to support the progressive control of tsetse-transmitted animal trypanosomosis in Kenya. Parasites Vectors (2020) 13(1):1–12. doi: 10.1186/s13071-020-04156-5
21
Weitz B . The feeding habits of glossina. Bull World Health Organ (1963) 28(5-6):711–29.
22
Okoth SO Kokwaro ED Kiragu JM Murila GA . Glossina pallidipes and host interactions: implications of host preference on transmission risk of Rhodesian sleeping sickness in Kenya. Trends App Sci Res (2007) 2(5):386–94.
23
Okech M Hassanali A . The origin of phenolic tsetse attractants from host urine: Studies on the pro-attractants and microbes involved. Int J Trop Insect Sci (1990) 11(3):363–8. doi: 10.1017/S1742758400012789
24
Auty H Cleaveland S Malele I Masoy J Lembo T Bessell P et al . Quantifying heterogeneity in host-vector contact: Tsetse (Glossina swynnertoni and g. pallidipes) host choice in Serengeti national park, Tanzania. PloS One (2016) 11(10):e0161291. doi: 10.1371/journal.pone.0161291
25
Ebhodaghe FI Okal MN Kalayou S Bastos AD Masiga DK . Tsetse bloodmeal analyses incriminate the common warthog phacochoerus africanus as an important cryptic host of animal trypanosomes in smallholder cattle farming communities in shimba hills, Kenya. Pathogens (2021) 10(11):1501. doi: 10.3390/pathogens10111501
26
Kappmeier K Nevill EM . Evaluation of conventional odour attractants for Glossina brevipalpis and Glossina austeni (Diptera: Glossinidae) in south Africa. Onderstepoort J Vet Res (1999) 66(4):307–16.
27
Clausen PH Adeyemi I Bauer B Breloeer M Salchow F Staak C . Host preferences of tsetse (Diptera: Glossinidae) based on bloodmeal identifications. Med veterinary entomology (1998) 12(2):169–80. doi: 10.1046/j.1365-2915.1998.00097.x
28
Dransfield RD Brightwell R Chaudhury MF Golder TK Tarimo SAR . The use of odour attractants for sampling Glossina pallidipes austen (Diptera: Glossinidae) at nguruman, Kenya. Bull Entomological Res (1986) 76(4):607–19. doi: 10.1017/S000748530001511X
29
Torr SJ Mangwiro TNC Hall DR . Responses of Glossina pallidipes (Diptera: Glossinidae) to synthetic repellents in the field. Bull Entomological Res (1996) 86(5):609–16. doi: 10.1016/j.actatropica.2015.02.017
30
Pollock ME . Description and keys for the identification of glossina species. Training Manual tsetse Control personnel (1982) 1:156–201.
31
Vreysen MJ Khamis IS van der Vloedt AM . Evaluation of sticky panels to monitor populations of Glossina austeni (Diptera: Glossinidae) on unguja island of Zanzibar. Bull entomological Res (1996) 86(3):289–96. doi: 10.1017/S0007485300052585
32
Kappmeier K Nevill EM . Evaluation of coloured targets for the attraction of Glossina brevipalpis and Glossina austeni (Diptera: Glossinidae) in south Africa. Onderstepoort J Vet Res (1999) 66(4):291–305.
33
Ouso DO Otiende MY Jeneby MM Oundo JW Bargul JL Miller SE et al . Three-gene PCR and high-resolution melting analysis for differentiating vertebrate species mitochondrial DNA for biodiversity research and complementing forensic surveillance. Sci Rep (2020) 10(1):1–13. doi: 10.1038/s41598-020-61600-3
34
Omondi D Masiga DK Ajamma YU Fielding BC Njoroge L Villinger J . Unraveling host-vector-arbovirus interactions by two-gene high resolution melting mosquito bloodmeal analysis in a Kenyan wildlife-livestock interface. PloS One (2015) 10(7):e0134375. doi: 10.1371/journal.pone.0134375
35
Nyamota R Owino V Murungi EK Villinger J Otiende M Masiga D et al . Broad diversity of simian immunodeficiency virus infecting chlorocebus species (African green monkey) and evidence of cross-species infection in papio anubis (olive baboon) in Kenya. J Med Primatology (2020) 49(4):165–78. doi: 10.1111/jmp.12461
36
Hall TA . BioEdit: A user-friendly biological sequence alignment Editor and analysis program for windows 95/98/NT. Nucleic Acids Symposium Ser (1999) 41:95–8.
37
Altschul SF Gish W Miller W Myers EW Lipman DJ . Basic local alignment search tool. J Mol Biol (1990) 215(3):403–10. doi: 10.1016/S0022-2836(05)80360-2
38
Moloo SK . The distribution of glossina species in Africa and their natural hosts. Int J Trop Insect Sci (1993) 14(4):511–27. doi: 10.1017/S1742758400014211
39
Kappmeier K . Diurnal activity patterns of Glossina brevipalpis and G austeni (Diptera: Glossinidae) in south Africa, with reference to season and meteorological factors. Onderstepoort J Vet Res (2000) 67(3):179–89.
40
Hoffman LC Cawthorn D . Species of meat animals | game and exotic animals. In: DikemanMDevineC, editors. Encyclopedia of meat sciences, 2nd ed.Cambridge, Massachusetts, USA:Academic Press (2014) p.345–356 doi: 10.1016/B978-0-12-384731-7.00081-7
41
Schoonhoven LM van Loon JJA Dicke M . Insect-plant biology. Oxford, UK: Oxford University Press (2005).
42
Turlings TCJ Tumlinson JH Lewis WJ . Exploitation of herbivore-induced plant odors by host-seeking parasitic wasps. Science (1990) 250:1251–3. doi: 10.1126/science.250.4985.1251
43
Negash M Girma M Seyoum E . Epizootiological importance of Glossina morsitans submorsitans (Diptera: Glossinidae)(Newstead) in the ghibe river valley, southwest Ethiopia. Acta tropica (2007) 102(2):100–5. doi: 10.1016/j.actatropica.2007.04.004
44
Bett B Irungu P Nyamwaro SO Murilla G Kitala P Gathuma J et al . Estimation of tsetse challenge and its relationship with trypanosomosis incidence in cattle kept under pastoral production systems in Kenya. Vet Parasitol (2008) 155(3-4):287–98. doi: 10.1016/j.vetpar.2008.05.028
Summary
Keywords
tsetse fly, bloodmeal identification, tsetse fly (Glossina spp.), G. pallidipes , G. austeni , host preference
Citation
Ogolla KO, Onyango T, Bwana BK, Otiende MY, Mang’era CM, Ochieng B, Omolo MO, Mugambi JM, Hassanali A, Omondi P and Mireji PO (2023) Bloodmeal host identities among sympatric Glossina austeni and Glossina pallidipes tsetse flies in Shimba Hills National Reserve, Kwale, Kenya. Front. Trop. Dis 4:1145993. doi: 10.3389/fitd.2023.1145993
Received
16 January 2023
Accepted
24 March 2023
Published
14 April 2023
Volume
4 - 2023
Edited by
Om P. Singh, National Institute of Malaria Research (ICMR), India
Reviewed by
Barbara K. Mable, University of Glasgow, United Kingdom; Inaki Tirados, Liverpool School of Tropical Medicine, United Kingdom
Updates
Copyright
© 2023 Ogolla, Onyango, Bwana, Otiende, Mang’era, Ochieng, Omolo, Mugambi, Hassanali, Omondi and Mireji.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paul O. Mireji, Mireji.paul@gmail.com
This article was submitted to Vector Biology, a section of the journal Frontiers in Tropical Diseases
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.