- 1Laboratory of Spatial Analysis in Health (LAES), Department of Epidemiology, School of Public Health, University of São Paulo (FSP/USP), São Paulo, SP, Brazil
- 2Section of Vector Borne Diseases, Pasteur Institute, State of São Paulo Health Secretariat, São Paulo, SP, Brazil
Introduction: Biomphalaria glabrata, B. tenagophila, and B. straminea occurrence are crucial for estimating the risk of infectious human schistosomiasis in the neotropics. How different geographic sample strategies influence snail genetic diversity estimations were here investigated for three Schistosoma mansoni hosts.
Methods: Mitochondrial gene sequences were employed for Cytochrome C Oxidase I (COI), ribosomal RNA (rRNA) 16S, and a dataset with concatenated gene sequences (COI+16S), resulting in an improved scientific hypothesis regarding the geographical distribution of snail species. This study compared the sequences of snails from the Middle Paranapanema (MP) hydrographic basin in a geographically restricted area (inner group) to snails widely distributed across a broad geographical range in São Paulo (outer group), Brazil from 1999 to 2017. DNA sequence polymorphisms and haplotype diversity were estimated using DNAsp software. Haplotype network trees were constructed using a network program. The geographical distribution of the haplotypes was mapped using QGIS. Haplotype variation and distribution were tested for population structure using analysis of molecular variance (AMOVA).
Results and discussion: The genetic diversity of B. glabrata, sampled from disconnected but geographically close freshwater collections, was partitioned into two sequence groups. The haplotype network showed that the diversity of B. straminea was more spatially partitioned than in B. tenagophila, which exhibited two population groups. The haplotype distribution pattern for B. tenagophila showed many unique and exclusive haplotypes for all three loci. AMOVA showed that genetic diversity could be high in species inhabiting small geographical areas, and a large river is not a local geographical barrier for snail migration. This study found that the survey dimensions and snail samplings influenced the genetic diversity results obtained by mitochondrial DNA molecular markers.
Introduction
Biomphalaria glabrata (Say, 1818) (Basommatophora: Planorbidae), B. straminea (Dunker, 1848), and B. tenagophila (Orbigny, 1835) freshwater snails act as intermediate hosts for Schistosoma mansoni Sambon 1907 (Strigeatida: Schistosomatidae), being the presence of these snails a key factor in the environmental management of these species that aims to establish snail control programs to prevent, control, and eliminate human schistosomiasis (1). Therefore, these snails are typically sampled from freshwater bodies on two geographic scales: a local scale to investigate hotspots for ongoing disease transmission and a regional scale to monitor the likelihood of future disease transmission. On both scales, numerous factors influence the occurrence and distribution range patterns of Biomphalaria (Preston, 1910) freshwater snails. Factors include the size and connectivity of the freshwater bodies (2), climatic and microclimate conditions (3, 4), large or small geographical areas (5), and land use for agriculture or urbanization. These factors act synergistically in colonizing freshwater habitats by Biomphalaria species, including non-native geographical areas.
Although animals have a low ability to move, Biomphalaria snails can disperse between different freshwater bodies with local, regional, or intercontinental ranges (6–8). According to genetic evidence, 14–33% of sampled snails in freshwater bodies in China have been identified as migrants from locations approximately 50 km away (7). The B. straminea invasion in Hong Kong in the 1970s and its subsequent spread to South China are good examples of the high potential for invasion of the host intermediate snail, increasing the risk of human schistosomiasis (8).
A significant change was recently observed in the geographic area occupied by B. tenagophila and B. straminea, as seen by studying previous surveys on freshwater collections located in the hydrographic basin of the Middle Paranapanema (MP) (9–12). Biomphalaria tenagophila suffered a marked reduction in previously occupied habitats. At the same time, B. straminea spread its geographical ranger across the MP region (13). In contrast, B. glabrata persisted in the same region as historically recorded.
Due to population growth, changes in land use, severe droughts, and other anthropogenic landscape changes, Southeast Brazil, particularly São Paulo (14), has experienced a plethora of disturbances that, in many cases, have led to local freshwater fragmentation and eutrophication (15). As a result, B. tenagophila and B. straminea, which often occur in São Paulo freshwater reservoirs, can now be found in contaminated and eutrophicated freshwater habitats, frequently close to urban areas (16, 17). The survival of the Biomphalaria species is boosted by a combination of their capacity to survive desiccation physiologically and self-fertilization (18–20).
The genetic diversity partitioning of different classes of molecular markers in S. mansoni snail hosts for different populations is potentially related to the mode of reproduction (especially inbreeding), population bottlenecks, and drift (21–28). Different populations include snails within the same or different water bodies (21, 29), different habitats (22–24), different regional scales (25, 26), and different countries (27, 28).
Investigating the genetic diversity of Biomphalaria in freshwater bodies in São Paulo led to some complex issues. An example includes whether the genetic variability characterized for the intermediate host species for S. mansoni at a few sites depicts the diversity inherent to these species or whether it is only a subset of the whole population diversity (13).
We conducted a genetic analysis of the three hosts of S. mansoni using two mitochondrial genes (Cytochrome C Oxidase I - COI and 16S ribosomal RNA - rRNA) and a dataset with concatenated gene sequences (COI+16S), owing to the robust results acquired using a couple of mitochondrial genes (30). In addition, we compared mtDNA sequences acquired from one watershed with a dataset of snail sequences sampled from eight water resource management units (UGRHI) in São Paulo’s hydrographic regions (31). This comparison was made to explore factors that affect genetic variation at local and broad geographic scales. Finally, we deepen our analysis to investigate whether a large river potentially acts as a geographical barrier to genetic diversity among populations of the same species.
Materials and methods
Study area and approaches for the obtention of DNA sequences and genetic analyses
This study used 306 COI, 181 rRNA 16S, and 144 COI+16S mitochondrial DNA sequences with a length of 546 bp, 344 bp and 894 bp nucleotides. These sequences were obtained from snails sampled from freshwater bodies in nine hydrographic basins in São Paulo (SP), Brazil, from 1999 to 2017. The nine hydrographic basins include: 1-”Alto Tietê” (AT), 2-Sorocaba-Middle Tietê (MT) and 3-Piracicaba-Capivari-Jundiaí (MT), 4-”Jacaré-Tietê” (JT), 5-”Paraíba do Sul” (PS), 6-”Ribeira do Iguape” valley (VR), 7-Northern Coast (LN), 8-”Pontal do Paranapanema” (PP), and 9- Middle Paranapanema (MP) (Figure 1; Table 1). In addition, the Sorocaba-Middle Tietê, and Piracicaba-Capivari-Jundiaí watershed data were combined due to a low number of sequences. All the freshwater bodies where the snails were surveyed are an extensive array of shallow and lentic freshwater bodies surrounded by urban and rural areas.
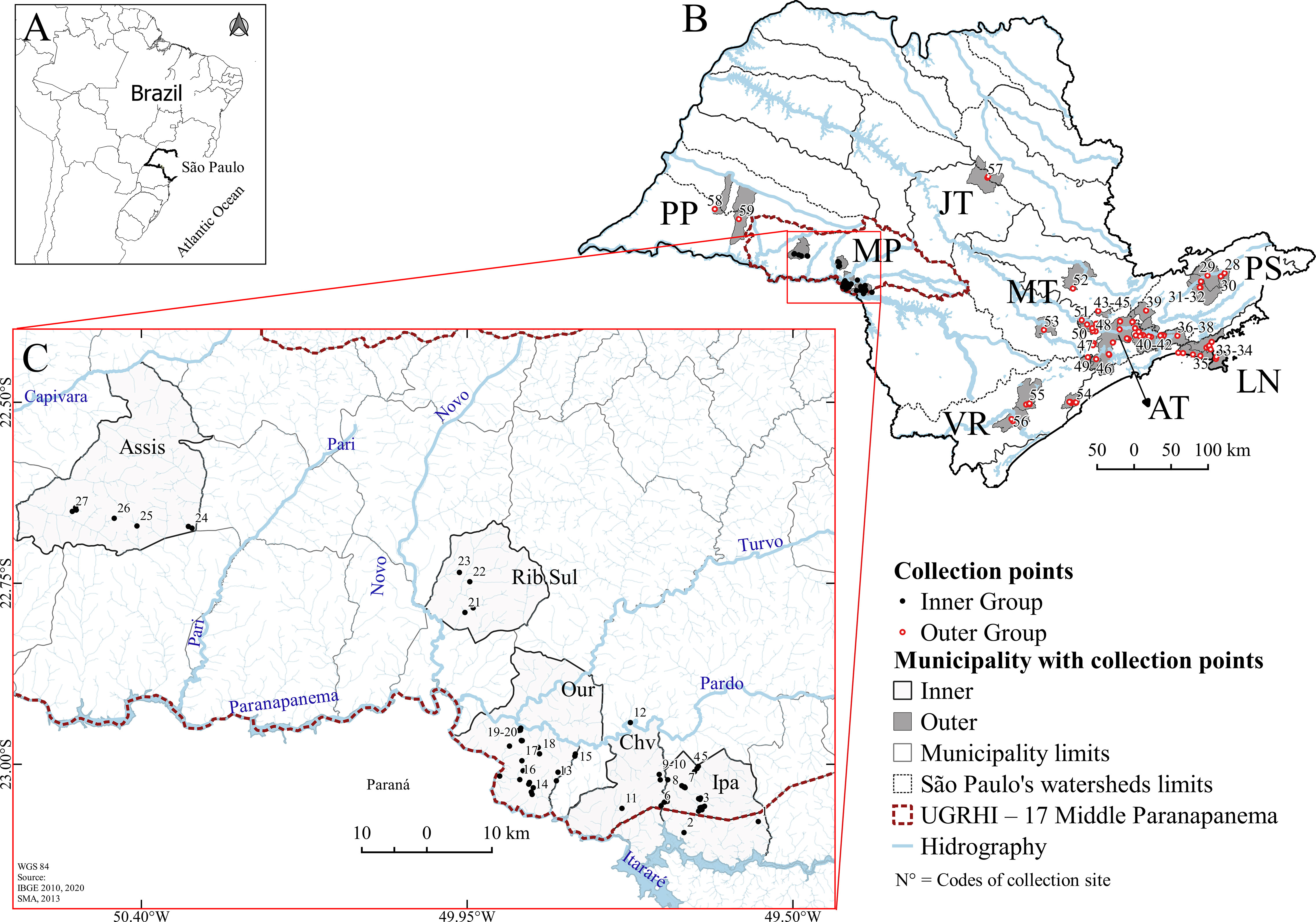
Figure 1 Collection points of Biomphalaria glabrata, B. tenagophila, and B. straminea. (A) Maps of South America, Brazil, the state of São Paulo; (B) Map showing the distribution of collection points in freshwater bodies in nine hydrographic basins in São Paulo; (C) Map of the Middle Paranapanema (MP) region, São Paulo, Brazil, from 1999 to 2018. (B) Hydrographic Basins: Outer Group – “Alto Tietê” (AT), Sorocaba-Middle Tietê and Piracicaba-Capivari-Jundiaí (MT), “Jacaré-Tietê” (JT), “Paraíba do Sul” (PS), “Ribeira do Iguape” valley (VR), Northern Coast (LN), “Pontal do Paranapanema” (PP) and Inner Group -Middle Paranapanema (MP); (C) Municipalities of: Assis (Assis), Ribeirão do Sul (Rib. Sul), Ourinhos (Our), Chavantes (Chv) and Ipaussu (Ipa), Water Resource Management Unit (UGRHI); Codes of the numbers of the water collection sites in Table 1. Source: 13, 26, 31, 34–38.
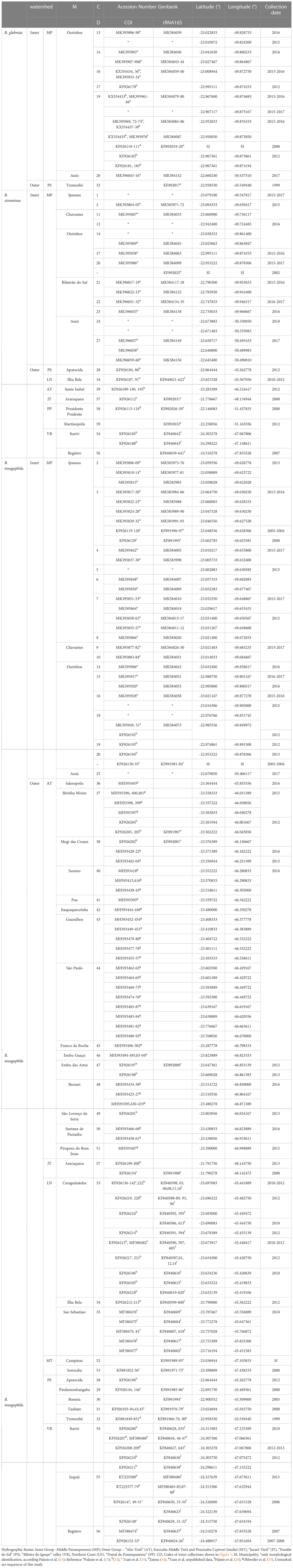
Table 1 GenBank accession numbers of nucleotide sequences for mitochondrial DNA COI and 16S rRNA and their respective geographic coordinates and references from snail species identified in the nine watersheds of the study, São Paulo, Brazil, from 1999 to 2018.
The methods for collecting snails, taxonomical identification based on the anatomical characters of reproductive systems, percent identity for each DNA Barcode sequences in NCBI reference library, and statistical tests DNA Barcode are described by Palasio et al. (13). Geographical coordinates of each collection site were provided by a Garmin eTrex GPS device (Garmin, Olathe, USA). All snails were identified simultaneously by morphology and DNA barcoding. In the case that DNA extraction was unsuccessful the morphology was used for identification (Table 1).
Amplification and sequencing of the mitochondrial genes used Folmer-LCO HCO primers (39) to amplify the mitochondrial COI region of ±600 bp, and the 16S ar/16S br primers (40) to amplify ±500 bp before aligned. Palasio et al. (32) and Tuan et al. (33) described the PCR reaction components and conditions. Alignments were performed using MAFFT version 7 (41) https://mafft.cbrc.jp/alignment/server/) under the Q-INS-I parameter. The final aligned sequences were edited and corrected using BioEdit version 7.2.5 (42, 43).
The COI and 16S sequences were concatenated using the SeaView 4 program (44). The 16S sequences obtained from snails sampled from MP were submitted to NCBI GenBank® (45) www.ncbi.nlm.nih.gov/genbank/) with accession numbers MK583964–MK584156. We also used COI and 16S sequences deposited in GenBank from Zanna (34), Tuan et al. (33), Palasio et al. (13, 26, 32), and Ohlweiler et al. (35) (Table 1).
The sequences were separated into the inner (MP watershed only) and outer group (AT+ MT+JT+PS+VR+LN+PP). Together, the inner and outer regions cover the most developed region in South America, with great changes in land use, including multiple hydroelectric power plants (46).
The inner group was marked by an intensive and extensive local sampling of B. glabrata, B. straminea, and B. tenagophila, along all the freshwater bodies located within the middle stretch of the Paranapanema river (MP, drainage area of ±16.749 Km2) (47). This basin is situated between -50.57325, -23.14839, -49.52913, and -22.44481 in São Paulo, Brazil. This area of MP is bathed mainly by the Paranapanema and Pardo rivers and their tributaries, the Turvo, Pari, Novo, and Capivara rivers (Figure 1) (47). The Paranapanema River flows westward from the headwaters in São Paulo into the Paraná River, defining the natural border between Brazil, Paraguay, and Argentina (48). These samples were for a scientific research program developed from 2015 to 2018 (13) and studies previously from 2002 to 2015 (32–34) (Table 1).
The outer group includes COI and 16S mtDNA sequences from studies previously published on snails distributed in freshwater bodies over eight different river basins: PS, AT, MT (two watersheds), JT, LN, VR, and PP, from 1999 to 2016 (26, 32–35) (Figure 1; Table 1). The outer drainage area encompasses <89.416 km2 (31) of suitable habitat areas for B. tenagophila, B. straminea, and B. glabrata (only one site).
The inner group counts B. glabrata (n = 33, 15, and 14 for COI, 16S, and COI+16S sequences, respectively), B. straminea (N = 18, 15, and 14), and B. tenagophila (N = 78, 51, and 50). The outer group contains B. glabrata (n = 1 for the 16S sequence), B. straminea (n = 16, 14, and 9 for COI, 16S, and COI+16S sequences, respectively), and B. tenagophila (n = 161, 85, and 57) (Table 2).
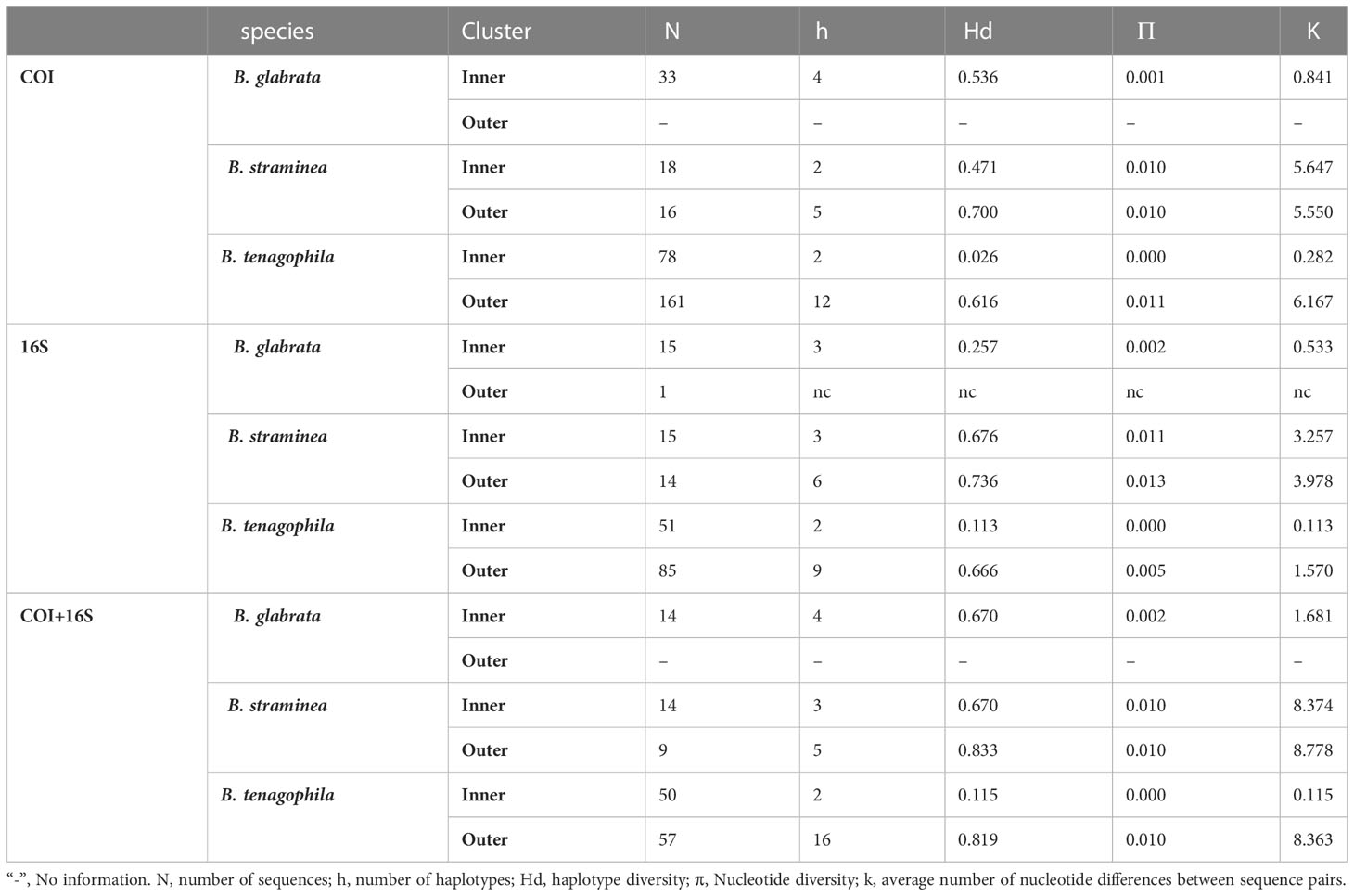
Table 2 Haplotype genetic diversity indices for Biomphalaria glabrata, B. straminea, and B. tenagophila calculated for the inner (Middle Paranapanema - MP) and outer (eight watersheds in São Paulo, Brazil) group in DNAsp, from 1999 to 2017.
DNA sequence polymorphism and haplotype diversity of 129 COI, 81 16S mitochondrial, and 78 COI+16S sequences were obtained for B. glabrata, B. tenagophila, and B. straminea of the inner group. Further, this was also conducted for the 177 COI, 100 16S, and 66 COI+16S sequences obtained for the three species sampled assigned to the outer group. Next, the results were analyzed in DNAsp version 6.12 (49) to provide estimated values of gene diversity related to mitochondrial haplotypes (h, Hd), nucleotide diversity (π), and the average number of nucleotide differences between sequence pairs (k).
Intraspecific haplotype networks and spatial distribution of haplotypes
The distribution of the COI, 16S, and COI+16S sequences into haplotypes for each species was inferred using DNAsp version 6.12 (49). The resulting nexus data files were used to infer haplotype network trees based on the median-joining algorithm (50) for COI, 16S, and COI+16S through Network program version 5.0.1.1 (Fluxus Technology Ltd, http://www.fluxus-engineering.com).
The mitochondrial haplotypes of COI+16S rRNA genes obtained from DNAsp (49) for the inner group sequences were associated with the geographic reference data (corresponding latitudes/longitudes) of the freshwater collections. In addition, the mitochondrial haplotypes for sequences for the outer group were associated with the centroid of watersheds. These associations were conducted to produce graph-type diagram maps from their attributes using the QGIS program version 3.16 (51). The cartographic materials were obtained from the Brazilian Institute of Geography and Statistics (IBGE), the Integrated System for Water Resources Management (SigRH), and the Secretary of Environment of the State of São Paulo (SMA) (31, 36–38).
AMOVA
The gene-population structuring hypothesis was tested on COI, 16S, and COI+16S sequence clusters of species with differentiated haplotypes through analysis of molecular variance (AMOVA) using Arlequin v.3.5.2.2 software (52). This analysis was run following 1023 permutations on the data to simulate the null hypothesis and was estimated using standard F-statistics. In addition, population structure hypotheses were tested for the species B. glabrata, B. straminea, and B. tenagophila at two geographic scales. This first geographic scale is in the inner group (MP region) from 2015 to 2017 with genetic differences between mitochondrial haplotypes of B. straminea obtained from snails collected from the right and left banks of the Pardo River. In addition, B. glabrata snails were obtained in disconnected and geographically close freshwater streams. The second geographic scale is in all the São Paulo regions with genetic differences between of B. straminea, and B. tenagophila from inner and outer group. The genetic differentiation among groups was considered low when the fixation index (Fst) < 0.05, moderate when Fst = 0.05–0.15, high when Fst = 0.15–0.25, and very high when Fst > 0.25 (53).
Results
Genetics and haplotype distribution analysis
We analyzed a length of 546 bp of the COI gene for 306 Biomphalaria snails (B. glabrata =33, B. straminea= 34, B. tenagophila=239), a 344 bp in length of 16S rRNA gene for 181 snails (B. glabrata= 16, B. straminea= 29 and B. tenagophila= 139), a dataset of 894 bp for concatenated COI+ 16S sequences for 181 snails (B. glabrata= 14, B. straminea= 23 and B. tenagophila= 107), sampled in freshwater bodies in São Paulo state, Brazil (Figure 1; Table 1). The number of sequences used for B. tenagophila is 7-10 higher than those obtained for B. glabrata and B. straminea, being this difference the result of the large distribution and abundance of B. tenagophila in the freshwater bodies in São Paulo.
A summary of the parameters for h, Hd, π, and K for the haplotype sequences used for statistics is shown in Table 2. The values were estimated for haplotype-based sequences according to whether they belonged to the inner or outer groups. For the nucleotide diversity (π) of sequences, the values were generally low, between 0 and 0.02, showing no differences in the patterns among all the genes and species analyzed. The patterns of genetic diversity based on the haplotype diversity (Hd) and the average number of nucleotide differences (K) seem to be a more realistic metric to indicate the potential population divergence for the three loci here analyzed.
The analysis of the genetic diversity (Table 2) for B. glabrata is limited to the inner group, an area of São Paulo that is considered a hot-spot for the presence of the species. Because only one sequence was available for B. glabrata from the inner group of sequences, the number of haplotypes (h=3-4) for B. glabrata reflects only the potential diversity of the entire population sampled in the whole study area. Nevertheless, the genetic diversity of B. glabrata was partitioned in two different streams (Christoni and Sobra) into the inner group of sequences: a substantial amount of this variability was found in the Christoni stream which seems isolated from Sobra stream population (Figures 2, 3).
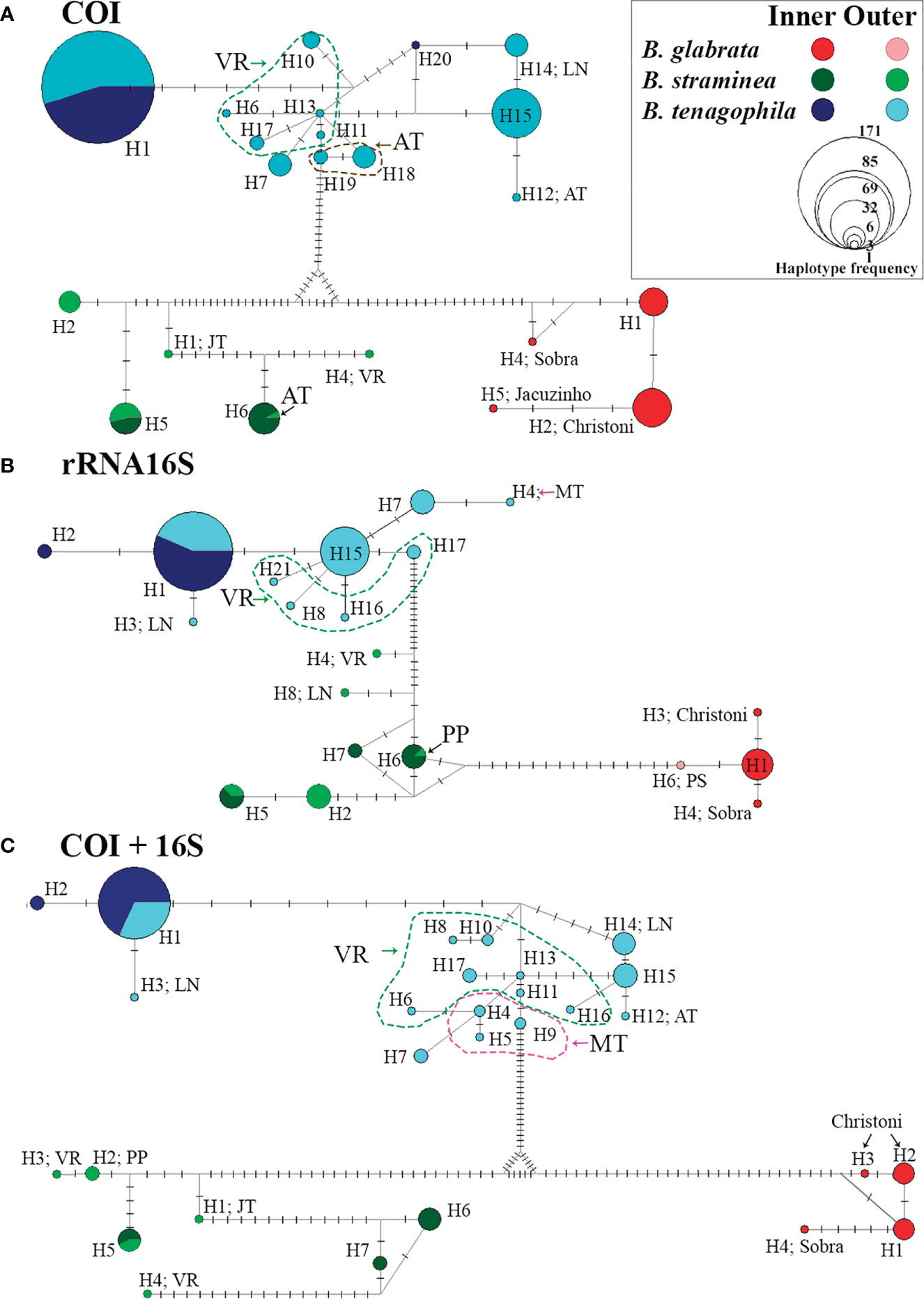
Figure 2 Haplotype network between the sequences: COI (A), 16S (B), and COI+16S (C) concatenated from the intermediate host species in the Middle Paranapanema (MP) region (inner) and the other eight watersheds (outer), São Paulo, Brazil, from 1999 to 2017. “Alto Tietê” (AT), Sorocaba- Middle Tietê and Piracicaba-Capivari-Jundiaí (MT), “Jacaré-Tietê” (JT), “Paraíba do Sul” (PS), “Ribeira do Iguape” valley (VR), Northern Coast (LN), “Pontal do Paranapanema” (PP).
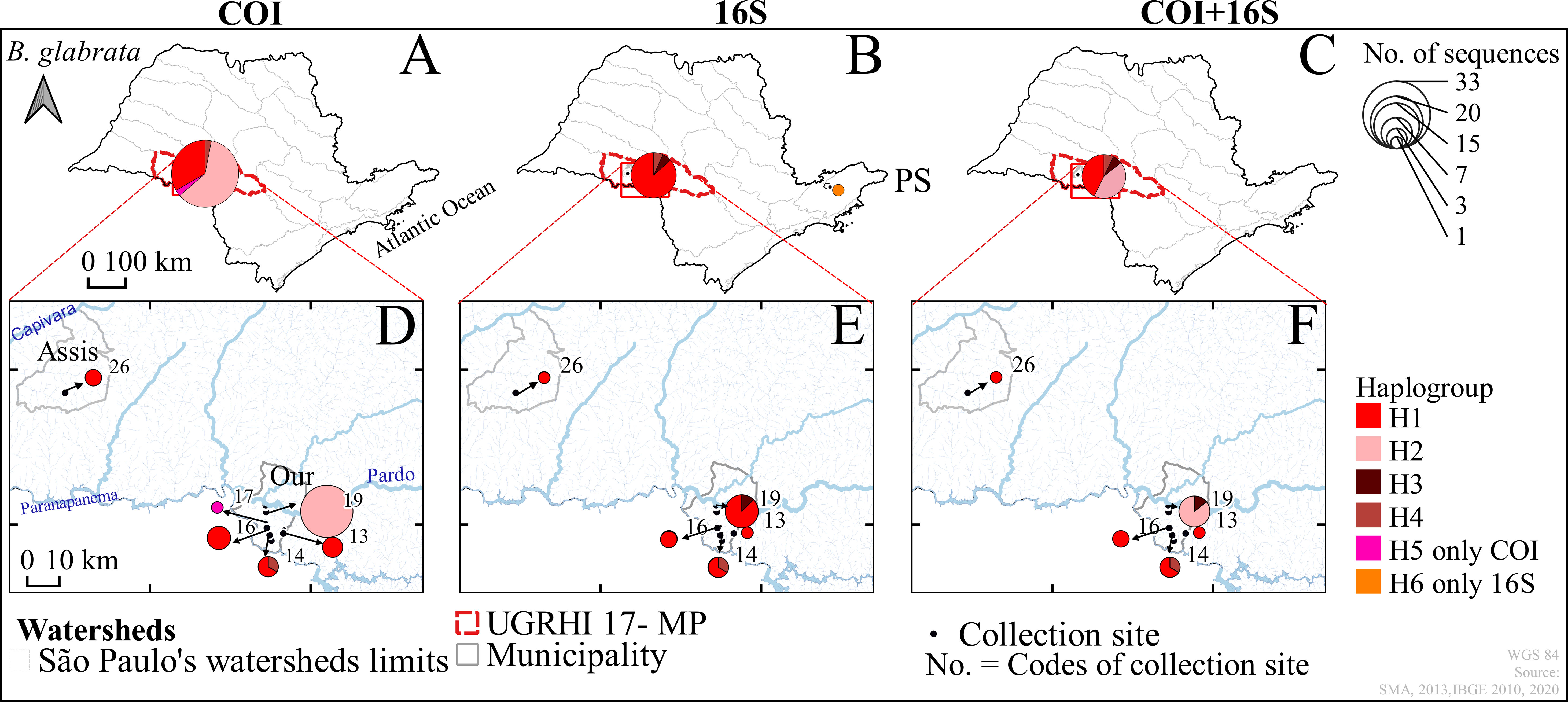
Figure 3 Haplotype distribution maps of the genes of Biomphalaria glabrata: COI, 16S, and COI+16S rRNA concatenated in the outer group (A–C) and the inner group (D–F), São Paulo, Brazil, from 1999 to 2017. Hydrographic Basins: Outer Group – “Paraíba do Sul” (PS) and Inner Group -Middle Paranapanema (MP); Municipalities of: Assis (Assis) and Ourinhos (Our); * Codes of the numbers of the water collection sites in Table 1. Code - Name water Collection: 13-Lageadinho, 14-Sobra, 16-Jacu, 17-Jacuzinho, 19-Christoni, 26-Fortuninha.
The haplotype diversity (hd) calculated for B. straminea show quantitative differences between inner and outer groups, with a high chance of obtaining samples from distinct haplotypes in the outer group. Notably, a wide distribution of B. straminea haplotypes from 5 to 7 hydrographic basins across the area was observed in the study. Another interesting result can be seen in the haplotype network which shows that for B. straminea, the diversity was more spatially partitioned than that for B. tenagophila, which exhibited two different population groups (Figures 2, 4).
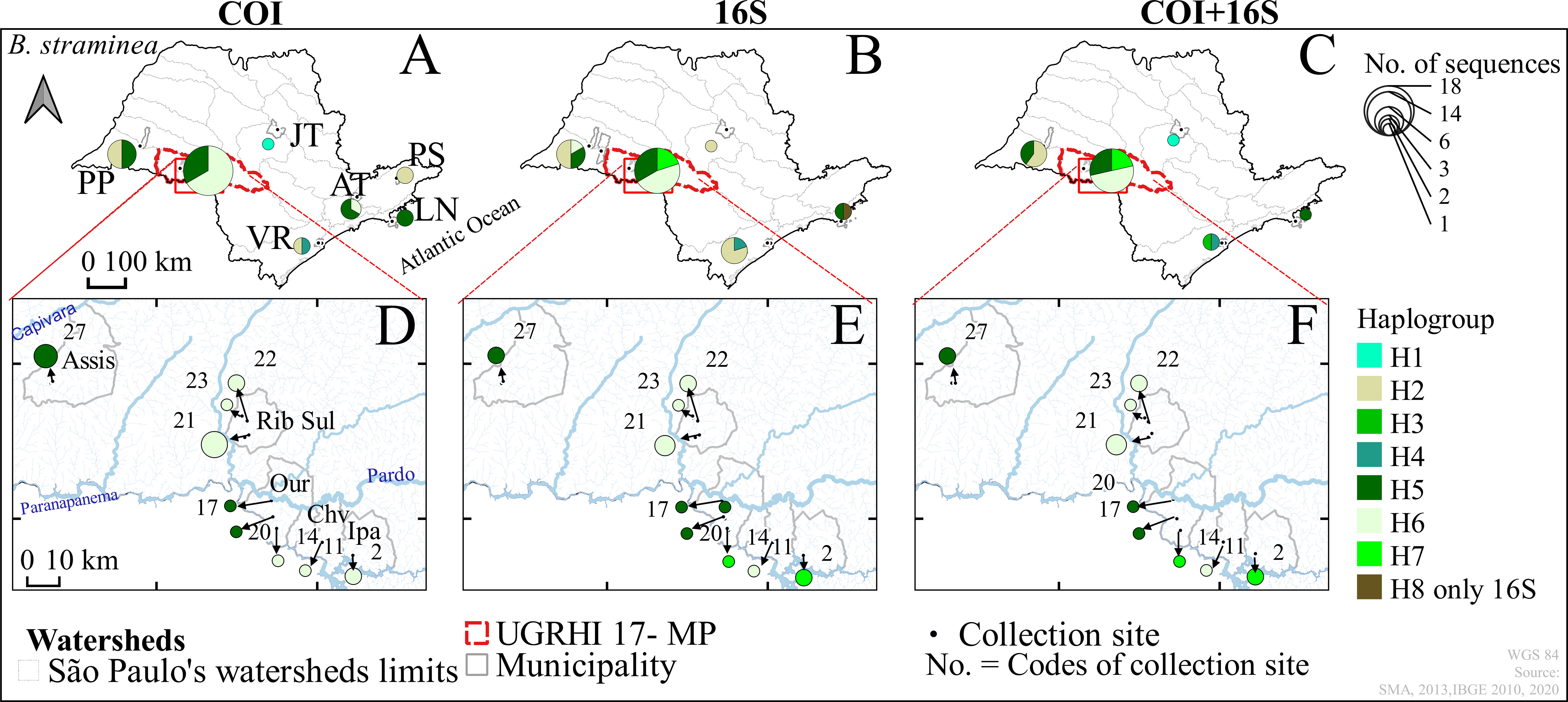
Figure 4 Haplotype distribution maps of the genes of Biomphalaria straminea: COI, 16S, and COI+16S rRNA concatenated in the outer group (A–C) and the inner group (D–F), São Paulo, Brazil, from 2002 to 2017. Hydrographic Basins: Outer Group – “Alto Tietê” (AT), “Jacaré-Tietê” (JT), “Paraíba do Sul” (PS), “Ribeira do Iguape” valley (VR), Northern Coast (LN), “Pontal do Paranapanema” (PP) and Inner Group -Middle Paranapanema (MP); Municipalities of: Assis (Assis), Ribeirão do Sul (Rib. Sul), Ourinhos (Our), Chavantes (Chv) and Ipaussu (Ipa); * Codes of the numbers of the water collection sites in Table 1. Code - Name water Collection: 2-Boa Vista, 11-Colossinho, 14-Sobra, 17-Jacuzinho, 20-Agua da Veada, 21-Ribeirão do Pinto, 22-Matão, 23-Lagoa, 27-Baixadão.
For B. tenagophila a remarkable loss of genetic diversity was observed in the inner group, as indicated by hd and K (Table 2). In general, the number of singletons was highest in the snails sampled from the outer group, a large area, encompassing water bodies across São Paulo. The geographical distribution of genetic variation for B. tenagophila was also widespread across all the hydrographic basins. The highest genetic diversity within B. tenagophila from the outer group was linked to the highest number of unique haplotypes, especially in the VR hydrographic basin (Figures 2, 5).
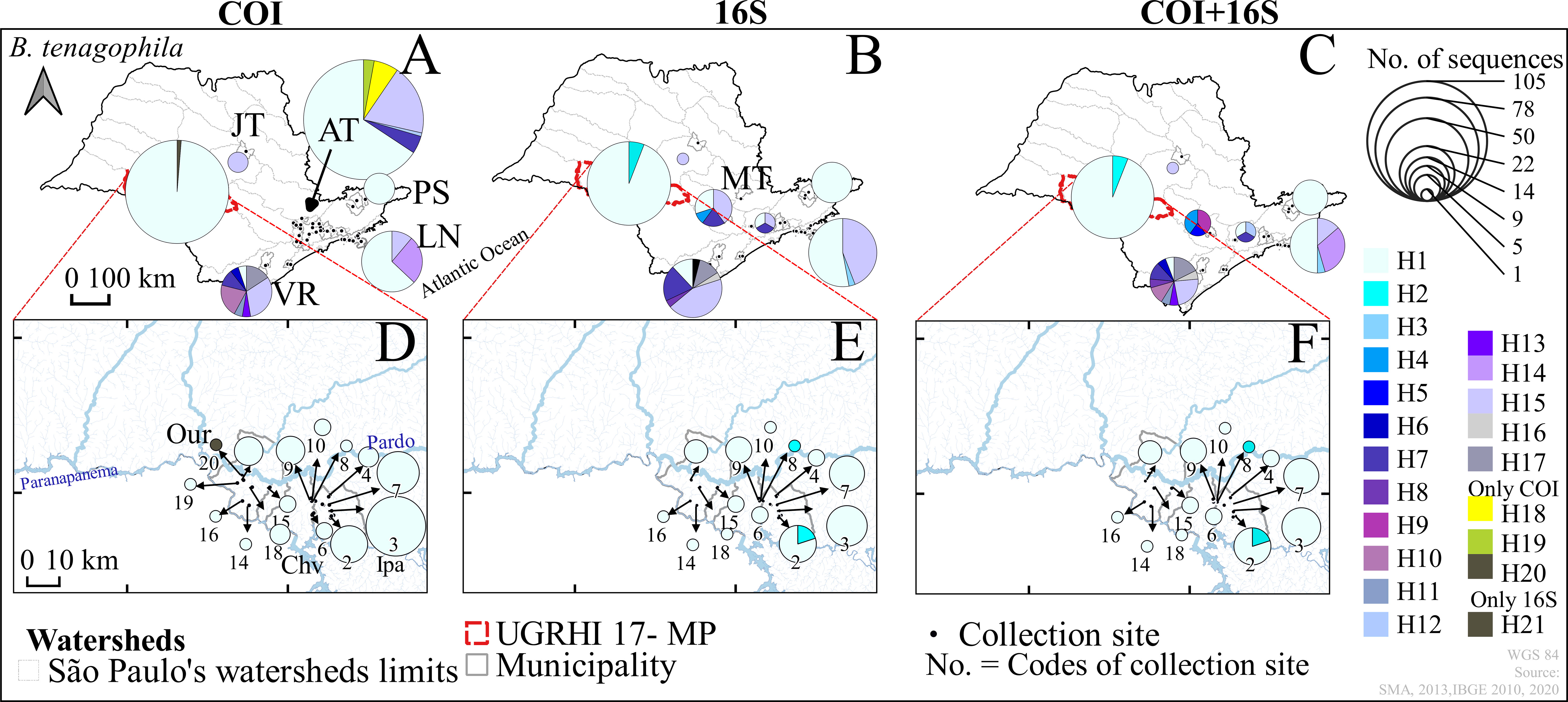
Figure 5 Haplotype distribution maps of the genes of Biomphalaria tenagophila COI, 16S, and COI+16S rRNA concatenated in the outer group (A–C) and the inner group (D–F), São Paulo, Brazil, from 1999 to 2017. Hydrographic Basins: Outer Group – “Alto Tietê” (AT), Sorocaba-Middle Tietê and Piracicaba-Capivari-Jundiaí (MT), “Jacaré-Tietê” (JT), “Paraíba do Sul” (PS), “Ribeira do Iguape” valley (VR), Northern Coast (LN) and Inner Group -Middle Paranapanema (MP); Municipalities of: Ourinhos (Our), Chavantes (Chv) and Ipaussu (Ipa); * Codes of the numbers of the water collection sites in Table 1. Code - Name water Collection: 2-Boa Vista, 3-São Luiz, 4-Mombuquinha, 6-Barranco Vermelho, 7-Ribeirão Grande, 8-Boa Vista (Divisa), 9-Santo Antonio, 10-Piranhas, 14-Sobra, 15-Barreirinha, 16-Jacu, 18-Furninhas, 19-Chumbiadinha, 20-Agua da Veada.
AMOVA
For all three mtDNA markers, the estimated genetic variation for B. straminea (72-82%) and B. tenagophila (70-81%) was higher within each group than among the groups. This difference shows that, for both species, no genetic structure was observed. Gene flow occurs between B. straminea snails (Fst=0.28) sampled from inner group (MP) despite a large geographical barrier, such as the Pardo River. The larger genetic variation among B. glabrata snails (72-93%) sampled from disconnected streams indicates a higher likelihood for the genetic variation observed in small and local populations rather than in different geographical regions. The values for Fst (0.67-0.92) also suggest a considerable level of differentiation among B. glabrata sampled in different freshwater in a local geographical scale (Table 3).
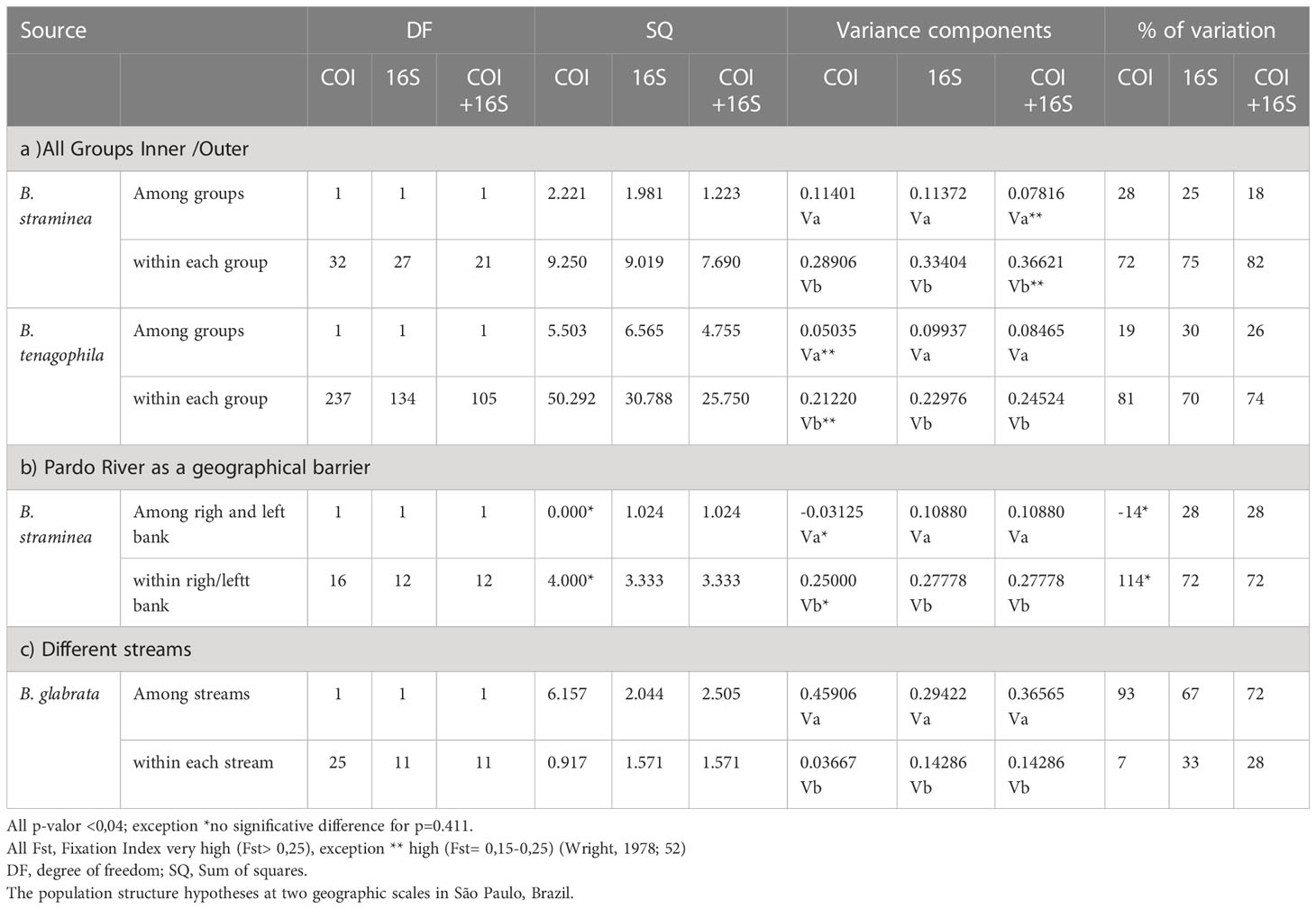
Table 3 Analysis of Molecular variance (AMOVA) of the frequency of the 16S and COI haplotypes separated and concatenated (COI+16S) among and within the groups of Biomphalaria glabrata, B. tenagophila, and B. straminea.
Discussion
Intraspecific genetic diversity has been little explored as an important metric in understanding the potential for snail expansion leading to the risk of spreading schistosomiasis. An important aspect of the current study is the extent to which the geographical scales of snail sampling influence the assessment of genetic diversity. The results of this study show that the genetic diversity varies within all three species at different spatial collection scales. This finding allows for speculation regarding the contemporary and recent past processes that have driven the population differences.
In Brazil, the main range of B. glabrata is found in Minas Gerais and Bahia freshwater bodies. However, focal sites, which are separated from the main range, can be found in Southeast Brazil, in bodies of water in São Paulo, Paraná, and Rio Grande do Sul (54). Any analysis of the species carried out in Southeast Brazil is, therefore, limited by the fact that this species only occurs in a very limited number of freshwater bodies, which are hydrologically disconnected from the main breeding sites of the species. The geographical range of B. glabrata in the MP river basin is a good example of this pattern; only four in Ourinhos and one in Assis municipality out of more than 100 streams are inhabited by this species (13).
The distribution of B. glabrata in fragmented sites across Southeast Brazil limits any analysis of the species due to snail numbers and limited geographic range. Therefore, further analysis must be based on sequences of the COI and 16S genes deposited in GenBank. Nevertheless, GenBank has a few sequences from specimens collected in Brazil, outside São Paulo. These include 16S (N = 26) or COI (N = 8) sequences (N° of GenBank 16S: JQ886405, JQ886408, AF449597–AF449599, AY030208, AY198084–AY198089, AY198091–AY198092, AY198095–AY198104, AY204640, and AY737280 and COI: AF199091– AF199092, AF199094– AF199096, and KF926107– KF926109). The low number of sequences is impressive, given the epidemiological relevance of this species for schistosomiasis transmission in Brazil and Africa. Furthermore, the genetic diversity indices calculated for B. glabrata sequences of the inner group for 16S (pi = 0.01, h = 13, Hd = 0.952, and k = 6.867) and COI (pi = 0.02, H = 4, Hd = 0.867, and k = 13.733) genes were about 10 times lower than in DNAsp from the GenBank database, which includes sequences from animals collected in Brazil. This result means that the MP population represents only a fraction of the potential whole genetic diversity of the species.
It is unknown why the species has not spread to adjacent sites even though the inner group was sampled from a high-risk area for flooding, overflow, and inundation (55). In consequence, if floods and inundations occur regularly such factors should not be seen as significant drivers for spreading the species. The limited genetic variability of B. glabrata in the inner group could explain the containment of the species to these few streams. It would be worth considering the adaptability of snails to specific ecological and environmental factors as a possible explanation for their containment in the MP region. The low genetic diversity associated with mitochondrial genes would also hypothetically be an important factor, since, snails can alter their oxidative systems to decrease their plasticity tolerance under stressors and environmental changes (56, 57).
Biomphalaria tenagophila sequences showed identical mitochondrial genotypes in the inner group. Compared to the outer group, the lower genetic diversity found in B. tenagophila in the inner group suggests that the genetic variation differences might result from differences in inbreeding rates. Also, the difference between self-fertilization could, in theory, protect clonal lineages from the negative impacts of population bottlenecks that affect this species due to environmental droughts.
We suggest, for B. tenagophila the possibility of two independent genetic lineages: a clonal lineage, which would result from the selection of animals with high potential for self-fecundation, and a lineage of animals in which self-fecundation is used more parsimoniously. The clonal lineage would have the advantage of adaptability to new environments and being able to be dispersed on a large scale, which relies on a comparison between B. tenagophila samples collected throughout the distribution area of the species.
In B. tenagophila, it may be possible to find significant genetic differentiation among animals, even in close and similar freshwater environments in the outer group, as exemplified by the haplotype in the VR basin. This variation mostly occurs because of the high frequency of unique haplotypes, a pattern that may result from genetic drift.
In B. straminea, the genetic variation in both groups was similar, possibly reflecting a more recent spatial colonization potentialized by the high ability of the species to colonize new habitats (13, 58). The expansion of the heat shock protein family in the genome of B. straminea (59) can improve the adaptability of the species to environmental stressors, causing B. straminea to spread to new environments across local and continental geographic scales.
The values for genetic variations and the gene flow between the sampled snails on opposite banks of the Pardo River show that the river does not represent a geographical barrier to the spread of the species. The colonization of new environments can be achieved through natural connections between collections of water by the passive displacement of snails through floods and overflows (60, 61) and by dispersal over a very large distance by the resident or migratory waterbirds (62).
In conclusion, this study presents the first genetic analysis of all three Neotropical schistosomiasis hosts based on the distribution patterns of different mitochondrial DNA sequence loci. The results show that the geographical scale of B. glabrata and B. tenagophila sampling may lead to discrepancies in the assessment of genetic diversity in both species. Additional studies must be conducted to achieve new insights into the biological differences between both species, which may be linked to adaptive constraints for their spreading. The same argument does not seem to apply to B. straminea because of their high ability to inhabit diverse habitats across a wide range of geographical scales.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article.
Ethics statement
This study has the approval by the School of Public Health Committee for Ethics in Research (COEP) of USP through the Plataforma Brasil system (Ministry of Health, National Health Council) under reference number 53559816.0.0000.5421.
Author contributions
RP: The manuscript was part of the Doctoral thesis of RP. The author contributed with field and molecular data collection, analysis and interpretation of molecular data, geospatial mapping and manuscript preparation adding intellectual content. FC-N: Contribution to Geospatial Data, Analysis and interpretation, manuscript preparation, adding intellectual content. RT: Concept, design and coordination of the research project funded by Health Secretariat of São Paulo State. Molecular and morphological data analysis and interpretation. Manuscript preparation. Critical revision, adding intellectual content. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and by the Fapesp [grant number 2021/10212-1] for the scholarship to RGSP, by Health Secretariat of São Paulo State for providing financial support for fieldwork activities and laboratory work analysis. Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) by research productivity grant awarded to FCN (PQ-1C, 304391/2022-0) and Fapesp [2020/01596-8]. All institution and research grant provides no fund for Open Access fees support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Sokolow SH, Wood CL, Jones IJ, Swartz SJ, Lopez M, Hsieh MH, et al. Global assessment of schistosomiasis control over the past century shows targeting the snail intermediate host works best. PloS Negl Trop Dis (2016) 10(7):e0004794. doi: 10.1371/journal.pntd.0004794
2. Lamy T, Gimenez O, Pointier JP, Jarne P, David P. Metapopulation dynamics of species with cryptic life stages. Am Nat (2013) 181(4):479–91. doi: 10.5061/dryad.s7v4b
3. Yang Y, Cheng W, Wu X, Huang S, Deng Z, Zeng X, et al. Prediction of the potential global distribution for Biomphalaria straminea. an intermediate host for Schistosoma mansoni. PloS Negl Trop Dis (2018) 12(5):e0006548. doi: 10.1371/journal.pntd.0006548
4. Rabone M, Wiethase JH, Allan F, Gouvras AN, Pennance T, Hamidou AA, et al. Freshwater snails of biomedical importance in the Niger river valley: evidence of temporal and spatial patterns in abundance. distribution and infection with Schistosoma spp. Parasites Vectors (2019) 12:498. doi: 10.1186/s13071-019-3745-8
5. Kock KN, Wolmarans CT, Bornman M. Distribution and habitats of Biomphalaria pfeifferi. snail intermediate host of Schistosoma mansoni. in south Africa. Waters SA (2004) 30(1):29–36. doi: 10520/EJC116133
6. Pointier JP, David P, Jarne P. Biological invasions: the case of planorbid snails. J Helminthology (2005) 79(3):249–56. doi: 10.1079/JOH2005292
7. Head JR, Chang H, Li Q, Hoover CM, Wilke T, Clewing C, et al. Genetic evidence of contemporary dispersal of the intermediate snail host of Schistosoma japonicum: movement of an NTD host is facilitated by land use and landscape connectivity. PloS Negl Trop Dis (2016) 10(12):e0005151. doi: 10.1371/journal.pntd.0005151
8. Lin D, Zeng X, Sanogo B, He P, Xiang S, Du S, et al. The potential risk of Schistosoma mansoni transmission by the invasive freshwater snail Biomphalaria straminea in south China. PloS Negl Trop Dis (2020) 14(6):e0008310. doi: 10.1371/journal.pntd.0008310
9. Piza JDT, Ramos ADS, Moraes LDC, Corrêa RDR, Takaku L, Pinto ADM. Carta planorbídica do estado de são paulo. In: Campanha de combate à esquistossomose. São Paulo, Secretaria de Estado da Saúde (1972). CACEsq, sd.
10. Teles HMS, Vaz JF. Distribuição de Biomphalaria glabrata (Say, 1818) (Pulmonata, planorbidae) no estado de são paulo, brasil. Rev Saúde Pública (1987) 21(6):508–12. doi: 10.1590/S0034-89101987000600007
11. Teles HMS. Distribuição geográfica das espécies dos caramujos transmissores de Schistosoma mansoni no estado de são paulo. Rev Soc Bras Med Trop (2005) 38(5):426–32. doi: 10.1590/S0037-86822005000500013
12. Tuan R. Diversidade e distribuição de espécies do gênero Biomphalaria em microrregiões localizadas no médio paranapanema, são paulo, brasil. Biota Neotrop (2009) 9(1):279–83. doi: 10.1590/S1676-06032009000100031
13. Palasio RGS, Rosa-Xavier IG, Chiaravalloti-Neto F, Tuan R. Diversity of Biomphalaria spp. freshwater snails and associated limnic mollusks in areas with schistosomiasis risk, using molecular and spatial analysis tools. Biota Neotrop (2019) 19(4):e20190746. doi: 10.1590/1676-0611-BN-2019-0746
14. Santarosa LV, Gastmans D, Gilmore TE, Boll J, Betancur SB, Gonçalves VFM. Baseflow and water resilience variability in two water management units in southeastern Brazil. Int J River Basin Manage (2022) 20:1–14. doi: 10.1080/15715124.2021.2002346
15. Santos DF, Silva JM, Becker V. Increasy eutrophication symptoms during a prolonged drought event in tropical semi-arid reservoirs, Brazil. RBRH (2021) 26:e39. doi: 10.1590/2318-0331.262120210097
16. Vendramini BM, Arruda EP. Freshwater mollusk species of itupararanga reservoir, são paulo, Brazil. Check List (2022) 18(3):629–708. doi: 10.15560/18.3.692
17. Paredes MG, Bianco KA, Menéndez-Helman RJ, Kristoff G. Aquatic contamination in lugano lake (Lugano lake ecological reserve, Buenos Aires, Argentina) cause negative effects on the reproduction and juvenile survival of the native gastropod. Biomphalaria straminea. Front Physiol (2022) 13:954868. doi: 10.3389/fphys.2022.954868
18. Tuan R, Simões LCG. Spermatogenesis and desiccation in Biomphalaria tenagophila (orbigny, 1835) (gastropoda, planorbidae). Rev Bras Genet (1989) 12(4):881–5.
19. Mavárez J, Amarista M, Pointier JP, Jarne P. Fine-scale population structure and dispersal in Biomphalaria glabrata, the intermediate snail host of Schistosoma mansoni, in Venezuela. Mol Ecol (2002) 11:879–89. doi: 10.1046/j.1365-294X.2002.01486.x
20. Tian-Bi YN, Jarne P, Konan JN, Utzinger J, N'goran EK. Contrasting the distribution of phenotypic and molecular variation in the freshwater snail Biomphalaria pfeifferi, the intermediate host of Schistosoma mansoni. Heredity (2013) 110(5):466–74. doi: 10.1038/hdy.2012.115
21. Charbonnel N, Angers B, Rasatavonjizay R, Bremond P, Jarne P. Evolutionary aspects of the metapopulation dynamics of Biomphalaria pfeifferi, the intermediate host of Schistosoma mansoni. J Evolutionary Biol (2002) 15(2):248–61. doi: 10.1046/j.1420-9101.2002.00381.x
22. Thiele EA, Corrêa-Oliveira G, Gazzinelli A, Minchella DJ. Elucidating the temporal and spatial dynamics of Biomphalaria glabrata genetic diversity in three Brazilian villages. Trop Med Int Health (2023) 18(10):1164–73. doi: 10.1111/tmi.12164
23. Mkize LS, Mukaratirwa S, Zishiri OT. Population genetic structure of the freshwater snail, Bulinus globosus (Gastropoda: planorbidae) from selected habitats of KwaZulu-natal, south Africa. Acta tropica (2016) 161:91–9. doi: 10.1016/j.actatropica.2016.06.001
24. Chibwana FD, Tumwebaze I, Mahulu A, Sands AF, Albrecht C. Assessing the diversity and distribution of potential intermediate hosts snails for urogenital schistosomiasis: Bulinus spp.(Gastropoda: planorbidae) of lake Victoria. Parasites Vectors (2020) 13(1):418. doi: 10.1186/s13071-020-04281-1
25. Standley CJ, Goodacre SL, Wade CM, Stothard JR. The population genetic structure of Biomphalaria choanomphala in lake Victoria, East Africa: implications for schistosomiasis transmission. Parasites Vectors (2014) 7(1):524. doi: 10.1186/s13071-014-0524-4
26. Palasio RGS, Zanotti-Magalhães EM, Tuan R. Genetic diversity of the freshwater snail Biomphalaria tenagophila (d’Orbigny, 1835) (Gastropoda: hygrophila: planorbidae) across two coastal areas of southeast Brazil. Folia Malacológica (2018) 26(4):221–9. doi: 10.12657/folmal.026.019
27. Mavárez J, Pointier JP, David P, Delay B, Jarne P. Genetic differentiation, dispersal and mating system in the schistosome-transmitting freshwater snail. Biomphalaria glabrata. Heredity (2002) 89(4):258–65. doi: 10.1038/sj.hdy.6800127
28. Mavárez J, Steiner C, Pointier JP, Jarne P. Evolutionary history and phylogeography of the schistosome-vector freshwater snail Biomphalaria glabrata based on nuclear and mitochondrial DNA sequences. Heredity (2002) 89(4):266–72. doi: 10.1038/sj.hdy.6800128
29. Webster JP, Davies CM, Hoffman JI, Ndamba J, Noble LR, Woolhouse MEJ. Population genetics of the schistosome intermediate host Biomphalaria pfeifferi in the Zimbabwean highveld: implications for co-evolutionary theory. Ann Trop Med Parasitol (2001) 95(2):203–14. doi: 10.1080/00034983.2001.11813630
30. Ghiselli F, Gomes-dos-Santos A, Adema CM, Lopes-Lima M, Sharbrough J, Boore JL. Molluscan mitochondrial genomes break the rules. Phil. Trans R Soc (2021) 376(1825):20200159. doi: 10.1098/rstb.2020.0159
31. Sistema Integrado de Gerenciamento dos Recursos Hídricos Divisão Hidrográfica – SigRH. As regiões hidrográficas do estado de são paulo (2023). Available at: https://sigrh.sp.gov.br/divisaohidrografica (Accessed Januery 04, 2023).
32. Palasio RGS, Guimarães MCA, Ohlweiler FP, Tuan R. Molecular and morphological identification of Biomphalaria species from the state of são paulo, Brazil. ZooKeys (2017) 668:11–32. doi: 10.3897/zookeys.668.10562
33. Tuan R, Ohweiller F, Palasio RGS, Zanna R, Guimaráes MCA. Chapter 15: pattern of genetic divergence of mitochondrial DNA sequences in biomphalaria tenagophila complex species under barcoding perspective. In: Rokni MB, editor. Schistosomiasis. Tehran, Iran: Intech (2012). p. 293–309. doi: 10.5772/25550
34. Zanna RLD. Diversidade genética de biomphalaria tenagophila (Orbigny, 1835) (Gastropoda: planorbidae) no estado de são paulo, brasil. Secretaria de Estado da Saúde de São Paulo. (2010).
35. Ohlweiler FP, Rossignol TDJ, Palasio RGS, Tuan R. Taxonomic diversity of Biomphalaria (Planorbidae) in são paulo state, Brazil. Biota Neotrop (2020) 20(2):e20200975. doi: 10.1590/1676-0611-BN-2018-0718
36. Secretaria de Meio Ambiente do Estado de São Paulo – SMA. Rede de drenagem do estado de são paulo obtida a partir da basedo gisat (cartas topográficas na escala 1:50.000) por processo automático - coordenadoria de planejamento ambiental, in: Meio ambiente, infraestrutura e logística (2013). Available at: https://www.infraestruturameioambiente.sp.gov.br/cpla/mapa-da-rede-de-drenagem-do-estado-de-sao-paulo/ (Accessed January 04, 2023).
37. Instituto Brasileiro de Geografia e Estatística - IBGE. Malha municipal 2020, in: Malhas digitais (2020). Available at: https://www.ibge.gov.br/geociencias/organizacao-do-territorio/malhas-territoriais/ (Accessed January 04, 2023).
38. Instituto Brasileiro de Geografia e Estatística – IBGE. Base contínua 250 mil, hidrografia. In: Mapas, interativos (2010). Available at: https://www.ibge.gov.br/geociencias/cartas-e-mapas/bases-cartograficas-continuas/.
39. Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA Primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol (1994) 3:294–9.
40. Palumbi S, Martin A, Romano S, McMillan WO, Stice L, Grabowski G. The simple fools guide to PCR. Honolulu: Depart. of Zoology, University of Hawaii (1991).
41. Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings Bioinf (2019) 20(4):1160–6. doi: 10.1093/bib/bbx108
42. Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucl Acids Symp Ser (1999) 41:95–8.
44. Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol (2010) 27(2):221–4. doi: 10.1093/molbev/msp259
45. Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Ostell J, Pruitt KD, et al. GenBank. Nucleic Acids Res (2018) 46(D1):D41–7. doi: 10.1093/nar/gkx1094
46. Souza-Dias V, Luz MP, Medero GM, Nascimento DTF. An overview of hydropower reservoirs in Brazil: current situation, future perspectives and impacts of climate change. Water (2018) 10(5):592. doi: 10.3390/w10050592
47. Comitê da Bacia Hidrográfica do Médio Paranapanema- CBH-MP. Relatório de situação dos recursos hídricos 2018 (2018). Available at: http://cbhmp.org/publicacoes/relatorios/ (Accessed January 04, 2023).
48. Brasil, Agência Nacional de Águas – ANA. Plano integrado de recursos hídricos da unidade de gestão de recursos hídricos paranapanema. (2016). p. 354.
49. Rozas J, Ferrer-Mata A, Sánchez-Del BJC, Guirao-Rico S, Librado P, Ramos-Onsins SE, et al. DnaSP 6: DNA sequence polymorphism analysis of Large datasets. Mol Biol Evol (Brasília, Brazil: Agencia Nacional das Águas) (2017) 34(2):3299–302. doi: 10.1093/molbev/msx248
50. Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol (1999) 16(1):37–48. doi: 10.1093/oxfordjournals.molbev.a026036
52. Excoffier L, Lischer HE. Arlequin suite verson 3.5: a new series of programs to perform population genetics analyses under Linux and windows. Mol Ecol Resour (Brazil: IBGE) (2010) 10(3):564–7. doi: 10.1111/j.1755-0998.2010.02847.x
53. Wright S. Evolution and the genetics of populations. variability within and among natural populations Chicago Vol. 4. University of Chicago (1978). p. 590.
54. Carvalho ODS, Mendonça CLFD, Marcelino JMDR, Passos LKJ, Fernandez MA, Leal RDS, et al. Distribuição geográfica dos hospedeiros intermediários do Schistosoma mansoni nos estados do Paraná, Minas Gerais, Bahia, Pernambuco e Rio Grande do Norte, 2012-2014. Epidemiologia e Serviços Saúde (2018) 27(3):e2017343. doi: 10.5123/S1679-49742018000300016
55. Guerra FC, Zacharias AA. Mapeamento das áreas de riscos hidrológicos e as políticas públicas de sustentabilidade: o caso de Ourinhos/SP. Rev Nacional Gerenciamento Cidades (2016) 4(26):75–94.
56. Coyne VE. The importance of ATP in the immune system of molluscs. Invertebrate Survival J (2011) 8(1):48–55.
57. Matoo OB, Neiman M. Bringing disciplines and people together to characterize the plastic and genetic responses of molluscs to environmental change. Integr Comp Biol (2021) 61(5):1689–98. doi: 10.1093/icb/icab186
58. Palasio RGS, Azevedo TS, Tuan R, Chiaravalloti-Neto F. Modelling the present and future distribution of Biomphalaria species along the watershed of the middle paranapanema region, são paulo, Brazil. Acta Tropica (2021) 214:105764. doi: 10.1016/j.actatropica.2020.105764
59. Nong W, Yu Y, Aase-Remedios ME, Xie Y, So WL, Li Y, et al. Genome of the ramshorn snail Biomphalaria straminea–an obligate intermediate host of schistosomiasis. GigaScience (2022) 11:1–13. doi: 10.1093/gigascience/giac012
60. Maciorowski G, Urbańska M, Gierszal H. An example of passive dispersal of land snails by birds – short note. Folia Malacologica (2012) 20(2):139–41. doi: 10.2478/v10125-012-0010-6
61. Van Leeuwen CHA, Huig N, van der Velde G, Van Alen TA, Wagemaker CAM, Sherman CDH, et al. How did this snail get here? several dispersal vectors inferred for an aquatic invasive species. Freshw Biol (2013) 58(1):88–99. doi: 10.1111/fwb.12041
Keywords: schistosomiasis, Biomphalaria, mtDNA, haplotype diversity, spatial analysis, molecular diversity, sampling strategy
Citation: Palasio RGS, Chiaravalloti-Neto F and Tuan R (2023) Distribution of genetic diversity of neotropical Biomphalaria (Preston 1910) (Basommatophora: Planorbidae) intermediate hosts for schistosomiasis in Southeast Brazil. Front. Trop. Dis 4:1143186. doi: 10.3389/fitd.2023.1143186
Received: 13 January 2023; Accepted: 10 April 2023;
Published: 16 May 2023.
Edited by:
Paul O. Mireji, Kenya Agricultural and Livestock Research Organization, KenyaReviewed by:
Kaio Cesar Chaboli Alevi, Sao Paulo State Universty, BrazilManuela Calado, New University of Lisbon, Portugal
Copyright © 2023 Palasio, Chiaravalloti-Neto and Tuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Raquel Gardini Sanches Palasio, cmFxdWVscGFsYXNpb0BhbHVtbmkudXNwLmJy
 Raquel Gardini Sanches Palasio
Raquel Gardini Sanches Palasio Francisco Chiaravalloti-Neto
Francisco Chiaravalloti-Neto Roseli Tuan
Roseli Tuan