- Department of Radiation Oncology, School of Medicine, University of Missouri, Columbia, MO, United States
Tuberculosis (TB) is one of the preeminent causes of death among infectious diseases and remains a global threat to human health. Mycobacterium tuberculosis (Mtb) has coevolved with the human host and is an extremely successful pathogen by abusing the human system in different ways. Interestingly, Mtb can remain undetected in the human host for years as latent TB, so there is an urgent need to develop new therapies to combat Mtb. In the recent past, host-directed therapies have attracted the research community as a promising approach to combat TB, and thus novel host targets are of interest. In the present editorial, we have explored and suggested Notch signaling as a potential host target to develop a new therapeutic strategy against Mtb.
Introduction
Tuberculosis (TB) is a highly infectious disease caused by Mtb, carried in airborne particles. TB is one of the greatest causes of death in human history throughout the world. According to the Global Tuberculosis Report, 2022 by the World Health Organization, 10.6 million people fell ill and 1.6 million deaths estimated due to TB in the year 2021 (https://www.who.int/publications/i/item/9789240061729). Therefore, there is an urgent need for new methods, tools, or strategies for the prevention, diagnosis, and identification of novel disease biomarkers and host factors to fight the battle against TB.
Host-pathogen interaction is a crucial aspect in studying infectious diseases as pathogens co-evolve with the host and establish the disease pathogenesis by subverting host factors. Therefore, targeting the host factors using host-directed therapies is an efficient way to develop new treatment strategies against Mtb. Host-directed therapies to combat TB may include any compound, repurposed drug, or strategies targeting host signaling pathways that can boost host-defense mechanisms or modulate the inflammatory response, resulting in better clinical outcomes to combat Mtb (1, 2).
The Notch signaling pathway is one of the highly conserved signaling pathways that transmit signals via direct contact with adjacent cells in a very short range of cellular communications (3). There are four Notch receptors (Notch 1-4) in humans which are activated by binding with ligands (Jagged1, 2 and Delta-like ligands (Dll)1, 3, and 4) on the surface of adjacent cells (4). Notch is a transcriptional regulator originally expressed as a membrane-bound cell surface receptor. Following by binding to the ligands on the adjacent cells, Notch receptors are cleaved by a proteolytic process in their membrane-bound form and thus released their intracellular active fragment followed by nuclear translocation and regulation of gene transcription (5). Components of Notch signaling and ligands are constitutively expressed at different stages in the immune cells such as monocytes, lymphocytes, macrophages, and dendritic cells that further impact the development, effector functions, and many immunological mechanisms during the pro-inflammatory immune responses including activation of T cells and cytokine profile development (6–8). The distinct expression profile of Notch receptors in different cells of the respiratory tract is not clear, however, notch signaling is very well conserved during the development of the human respiratory system.
Interestingly, Notch is significantly involved in lung regeneration during both in health and disease conditions (9, 10). Active involvement and regulation of notch signaling in the lung regeneration and respiratory diseases suggested that Notch could be a promising target to develop host-directed therapy to fight against lung pathologies (11). In the present article, we have explored the role of Notch signaling during Mtb infection that suggested Notch as a potential host factor that could be targeted and developed for clinical therapeutics against TB.
Notch positively regulates TB progression
Recent evidence suggested Notch is a player important in TB pathogenesis via modulating the immune response dynamics during mycobacterial infections. An upregulated expression of Notch1 is observed in Mycobacterium bovis Bacille Calmette-Guérin (M. bovis BCG) infected macrophages, leading to the expression of suppressor of cytokine signaling (SOCS) that is a major negative regulator of cytokine signaling (12). Similarly, a recent case study of children with active tuberculosis, showed elevated mRNA levels of Notch1, Notch2, and Dll4 when compared to the control group (13). Increased levels of Notch ligands Dll1 and Dll4 are also reported in a serum-based case study of tuberculosis patients co-infected with HIV (14). Notch signaling is also reported in regulating apoptosis during M. bovis infection in macrophages. Results concluded that Notch1 inhibits the apoptosis that can be reversed by using a gamma-secretase inhibitor (GSI) which is known to inhibit Notch signaling and suggests an involvement of Notch upregulation in M. bovis infection (15). Next, a newly found non-coding circular RNA (circAGFG1) is reported to increase autophagy and reduced apoptosis via the miRNA-1257/Notch axis. Diminished levels of Notch2 are reported in infected macrophages from active TB patients upon treatment of sh-circAGFG1 or by mimicking miRNA1257 (target of miRNA1257) where miRNA-1257/Notch axis is suggested as a new therapeutic target (16). Additionally, a recent study on Chinese tuberculosis patients reported a positive correlation between missense mutation in Notch4 and increased susceptibility to tuberculosis (17). Another population-based case-control study confirmed that Mtb induces Notch4 expression in the macrophages during inflection and results in severe TB outcomes (18).
In correlation with these results, Notch inhibition by a GSI (DAPT) has also been reported to reverse the imbalance of Th1/Th2 cytokines in TB patients (19). Further, up-regulated expression of Notch ligand Dll4 is reported in the Mtb infected individuals that alter the immune response, evident by increased levels of IFN-γ and IL-17 cytokines released by T cells. Next, similar results are observed in the peripheral blood mononuclear cells (PBMCs) of active TB patients (20, 21). Also, an upregulated expression of Dll4 is observed in antigen-presenting cells (APCs) and bone marrow progenitor cells in mice infected with M. bovis BCG. Similarly, Mtb-infected patients are reported to have an increased number of Dll4-positive monocytes and concluded that Mtb infection regulates the expression of Dll4 that can modulate CD4+ T cell response (21). Results suggested that Notch ligands such as Dll4 could be used as potential biomarkers to identify the disease progression in TB and as a host-directed target to develop anti-TB therapeutics. Further, a Chinese herbal compound isoliquiritigenin, is reported to overcome the Mtb-induced inflammation by inhibiting Notch1/NF-κB signaling pathway in murine macrophages (22). Overall, experimental evidence suggested that Notch positively regulates TB progressions and could be a potential target for host-directed therapies.
Notch negatively regulates the Mtb-induced inflammatory immune response
Proinflammatory cytokines IL-6, IL-12, IFN-γ, and TNFα along with CD4+ T cells known to regulate the immune response during the Mtb infection (23). It has been reported that IL-6 knockout mice showed high mortality due to the failure of Mtb growth control during infection (24). Similarly, IL-12 deficient mice demonstrated to have less IFN-γ production and high incidences of Mtb infections (25, 26). Importantly, IL-12 is involved in the regulation of protective immunity against Mtb via the development of IFN-γ producing CD4+ Th1 cells (27). In a recent study, it has been shown that Mtb-induced expression of IL-6 and IL-12 is improved while an increased number of IFN-γ secreting Th1 cells is observed in a Notch4 deficient murine infection model which is further evident by reduced Mtb burden and lung damage (28). Another study reported that Notch interacts with TAK1 and TRAF6 while TAK1 forms a complex with TRAF6. TRAF6 activated TAK1 in a ubiquitination-dependent manner and further TAK1 is found involved in the TLR-induced production of proinflammatory cytokines (29, 30). Next, it has been confirmed that Notch4 inhibits the proinflammatory cytokines in the infected macrophage by restricting the phosphorylation and ubiquitination of TAK1. Finally, it is confirmed that Notch inhibition by DAPT is also able to abrogate the Mtb infection (28). Further, the anti-inflammatory effects of Andrographolide (a traditional Chinese herbal medicine) is reported in Mtb-infected macrophages via regulation of the Notch1/Akt/NF-κB axis (31). Additionally, patients with active TB are reported to have high expression of Notch ligand Dll4 when compared to healthy controls. Also, an upregulated expression of Notch1 is observed in patients having moderate or advanced lung injury (32). Interestingly, increased resistance to TB is demonstrated both in vitro and in vivo, using Notch deficient mice and by the treatment with the γ-secretase inhibitor (28). Another recent study by the same group reported Mtb induces Notch4 and jagged1 expression in macrophages of TB patients via TLR2/p38 and TLR2/ERK signaling, respectively. Conclusively, this study suggested that Notch4 negatively regulates the Mtb-induced inflammation and thus higher expression and gene polymorphism of Notch4 is positively associated with server TB outcomes (18). Notch4 induction is also reported in response to the hypoxia model of Erdman (a virulent Mtb strain) in rhesus macaques which it confirmed to stimulate undifferentiated cellular state with large necrosis lesions (33).
Overall, available studies suggested Notch signaling and its components essentially intervened with the severity level of Mtb infection via positive and negative regulation of the TB infection and Mtb-induced host inflammatory response, respectively. In light of the evidence, we suggested Notch signaling and its components as a potential biomarker for host-directed therapies and to monitor the disease progression and severity as well.
Discussion
Notch signaling and its components, especially Dll1 and Dll4, present in monocytes, macrophages, and dendritic cells played an important role during the pro-inflammatory immune responses and are involved in several immune mechanisms such as T cell and B cell development in both central and peripheral lymphoid organs (7, 34, 35). Interestingly, an increased expression level of Notch ligand Dll4 is found in the monocytes of active TB patients. Similar results observed in the Mtb-infected monocytes that suggested Mtb-induced expression of Notch ligands which is also positively correlated with the degree of lung injury (32). Notch signaling played an active role in the effector functions of T cells where Notch1 and Notch2 receptors are constitutively expressed in the different stages of lymphocyte development. NICD is the activated form of Notch signaling, that translocates into the nucleus after activation. In the nucleus, NICD interacts with the RBP-J and further induces the downstream expression of target genes such as Hes1, Hes5, Hey1, Hey2, and Dtx (36) (Figure 1). Next, Notch signaling regulates a feedback inhibition of TLR-induced cytokines via the downstream target genes, Hes1, and Hey1 (37). By using the feedback loop, Notch signaling negatively regulates the TLR-triggered immune response and thus the expression of pro-inflammatory cytokines (38). A higher expression of Hes1 and Dtx genes is observed in active TB patients in comparison to healthy individuals. This suggested systemic activation of Notch and its downstream signaling pathway during Mtb infection and thus Notch can be targeted to overcome the detrimental effects of TB (32). Further, an active transcriptional complex of Notch signaling is found to have a direct binding ability with the promoter regions of Rorc and IL-17 genes that can alter the activation of these genes. Interestingly, Notch ligand Dll4 showed to enhance the expression of both genes Rorc and IL-17 as they both are the direct transcriptional target of Notch signaling (39). Collectively, Notch and Dll4 expression in monocytes and T cells is confirmed to have a positive correlation with plasma levels of IFN-γ, IL-17A, and IL-2 (32, 40) (Figure 1). Additionally, TNF-α is known as an important cytokine during the Mtb infection that boosts the protection against TB by activation of monocytes and macrophages and new inflammatory cells at the site of infection (41). Enhanced production of TNF-α is demonstrated in mononuclear cell cultures upon blocking the Notch ligand Dll4 by using an anti-hDll4 antibody (32) (Figure 1). It seems there is an uncoupled functional relation between Notch and Dll4 as it has been shown that Notch inhibition altered the cytokine production during TB that are important for the activation of macrophages while Dll4 blocking improved the monocytes-mediated phagocytosis of Mtb. This again suggested the potential of Notch and Notch ligands as promising host targets for the development of anti-TB therapeutics.
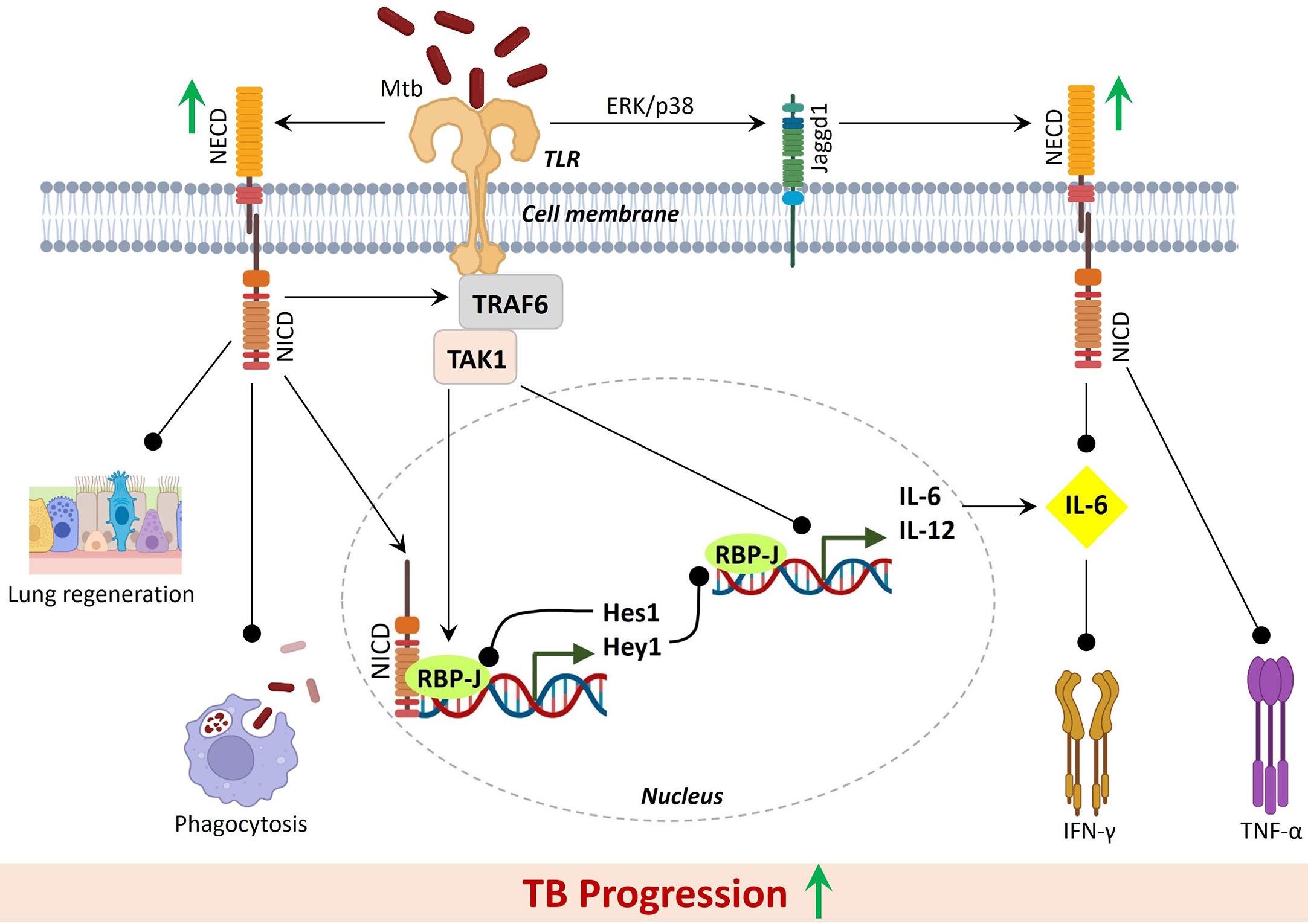
Figure 1 Active Notch signaling during Mtb infection negatively regulates inflammatory response against Mtb and results in severe TB progression. Black lines with round heads showed inhibition. Green upright thick arrows showed increased expression.
Overall, reports showed that Mtb infection promotes the activation of Notch signaling that subsequently altered the pro and anti-inflammatory response against TB while active TB patients reported to have higher expression of Notch and reduced levels of IL-6. This inverse correlation of Notch and IL-6 might result in divergent inflammatory states during diverse disease outcomes. In total, an altered immune response against Mtb infection positively assists the TB progression by further modulating the functional mechanisms of monocytes, such as phagocytosis and cytokine production via upregulation of the Notch signaling. Also, enhanced Notch expression in T cells and Dll4 expression in intermediate monocytes is directly associated with the severe form of TB and thus suggested as potential host-directed targets against Mtb. Notch activation largely depends on the activity of the γ-secretase complex, and thus GSIs are suggested as a promising therapeutic target for notch inhibition (42). GSIs are already under various clinical trials for different indications (43, 44). Till now several chemical classes of GSIs have been developed and most of them are competitive inhibitors of the catalytic activity of the presenilins (45). Recently, a novel potent GSI, GSI LY3039478 confirmed as the recommended phase II dose for advanced or metastatic cancer patients (NCT01695005) (46). Interestingly, one of the GSI, PF-03084014 has recently been in phase II clinical trials and found promising for desmoid tumors, known as aggressive fibromatosis (NCT01981551) (47). Receptor/ligand antibodies, GSIs, and Notch transcription complex inhibitors are available which could be tested to check the Notch inhibition and effects on Mtb infection (4, 48). Interestingly, DAPT (a GSI) showed to protect the mice from Mtb infection, evidenced by decreased pathological changes in the lungs, reduced bacterial load, and enhanced cytokine expression (28). Further, several GSIs have already undergone phase I/II clinical trials which can be further evaluated for their effects on immune response modulation during TB. Additionally, Notch signaling is essential for tissue regeneration of the various cell type in the lungs after injury (49). Excessive expression of Notch signaling may inhibit the regeneration of different cell types involved in lung airway functions and its tightly regulated inhibition is essential for normal regeneration, diverse cell differentiation, maintenance and restoration of lung airways for healing after injury (11, 50).
Conclusively, available studies and evidence suggested a relationship between the expression of Notch signaling components and the respective host immune response to Mtb infection. Further, evidence suggested that Notch signaling positively regulated the TB progression and negatively regulates the innate immunity response outside of its transcriptional activity. Thus, inhibition of Notch signaling might provide new hope for the development of therapeutics to fight against TB. Here we suggested further investigation of different GSIs including other Notch inhibitors to develop them as host-directed therapeutics against Mtb.
Conclusion
Overall, our analysis and proposed hypothesis suggested, Notch, signaling as one of the central regulators of Mtb-induced cellular immune response and inflammation during the disease progression of TB. As a high expression of Notch signaling and its ligands observed in active TB patients, a Notch-based-diagnostic assay could also be developed for the early estimation of disease severity and for developing a treatment strategy accordingly. We have also visualized the molecular interaction network of Notch with other host factors that represents Notch as a complicated pathway that is polygenic, multi-ligand, and post-translationally regulated (Figure 2). Notch has multiple different effects at the cell and tissue levels that have an important role in development, activation, differentiation, proliferation, and regeneration. Therefore, there is a high possibility of unwanted on-target and off-disease side effects upon pharmacologic targeting of Notch. However, evidence suggested Notch inhibition could be a promising strategy to reverse the deleterious effects of disease during Mtb infection and so, there is a possible provision of specific Notch-inhibitor drugs such as GSIs for a certain time during the disease progression. In summary, the present work avails the evidence and sheds light on Notch signaling as a potential host-target that can be considered in clinical trials for the development of treatment strategies against Mtb infection and management of TB.
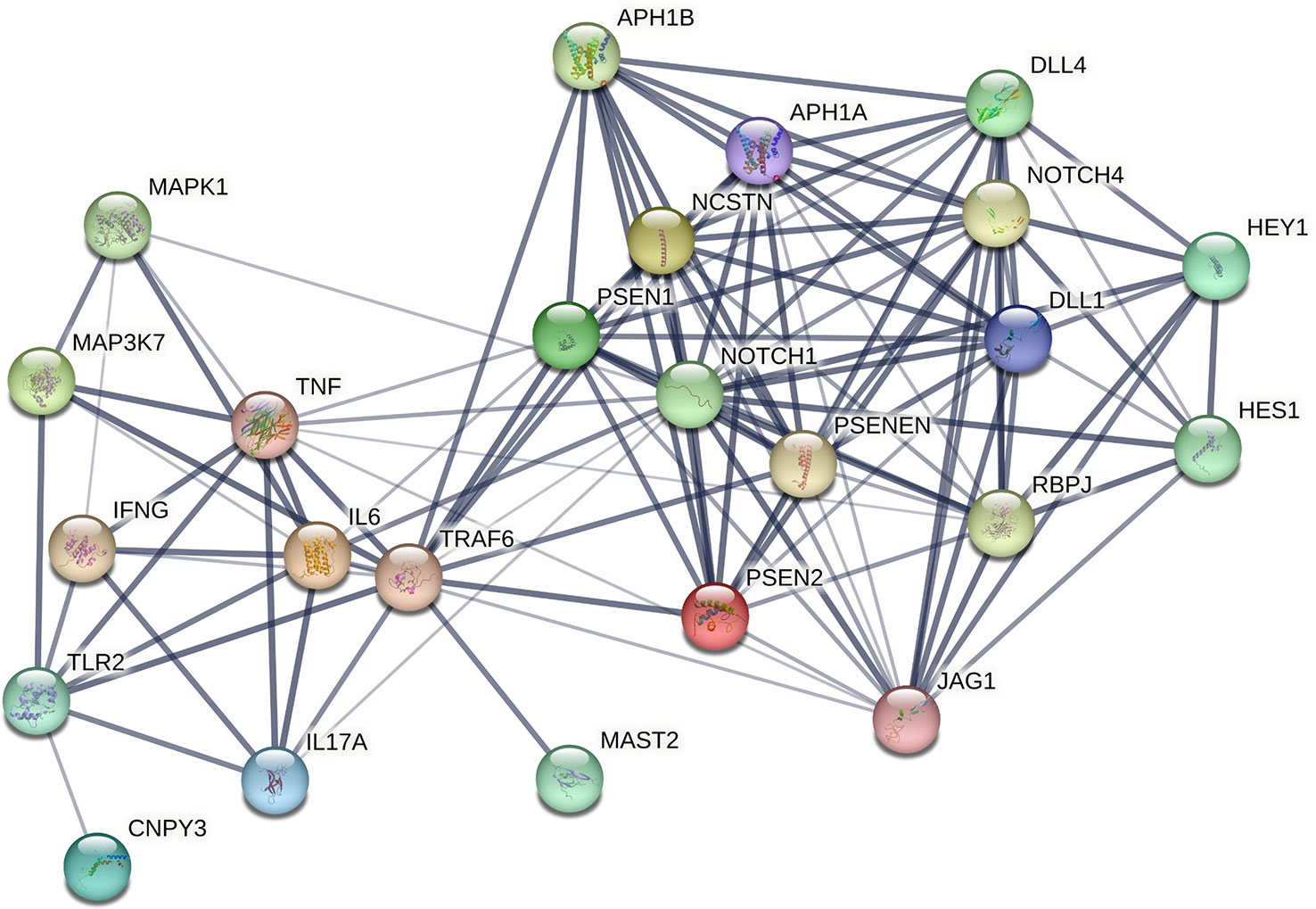
Figure 2 Physical protein-protein interaction network representing cross-talk of Notch with other host factors involved in TB progression. This network is generated using STRING v11.5: functional protein association networks. The thickness of the connecting edges showed the confidence level.
Limitation
There are limitations associated with the proposed hypothesis. First of all Notch signaling is involved in various development processes and so further, detailed studies are needed to confirm the regulation of Notch signaling during TB. Also, we suggested the use of Notch inhibitors which might promote severe inflammation thus, the time of treatment and other side effects should be properly explored using clinical trials before consideration as therapeutic drugs to treat TB.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Author contributions
PB wrote the manuscript, prepared the figures, conceived the sole idea, and led the project. PB prepared the protein-protein interaction network and proofread the manuscript. PB approved the submitted the final version.
Acknowledgments
The author acknowledges the Department of Radiation Oncology, School of Medicine, University of Missouri, Columbia, MO for providing the space and necessary facilities to write this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Baindara P. Host-directed therapies to combat tuberculosis and associated non-communicable diseases. Microb Pathog (2019) 130:156–68. doi: 10.1016/j.micpath.2019.03.003
2. Baindara P, Agrawal S, Franco OL. Host-directed therapies for malaria and tuberculosis: Common infection strategies and repurposed drugs. Expert Rev Anti Infect Ther (2022) 20:849–69. doi: 10.1080/14787210.2022.2044794
3. Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: Cell fate control and signal integration in development. Science (1999) 284:770–6. doi: 10.1126/science.284.5415.770
4. Aquila G, Kostina A, Vieceli Dalla Sega F, Shlyakhto E, Kostareva A, Marracino L, et al. The notch pathway: A novel therapeutic target for cardiovascular diseases? Expert Opin Ther Targets (2019) 23:695–710. doi: 10.1080/14728222.2019.1641198
5. Kopan R, Ilagan MXG. The canonical notch signaling pathway: Unfolding the activation mechanism. Cell (2009) 137:216–33. doi: 10.1016/j.cell.2009.03.045
6. Yamaguchi E, Chiba S, Kumano K, Kunisato A, Takahashi T, Takahashi T, et al. Expression of notch ligands, Jagged1, 2 and Delta1 in antigen presenting cells in mice. Immunol Lett (2002) 81:59–64. doi: 10.1016/S0165-2478(01)00326-1
7. Amsen D, Blander JM, Lee GR, Tanigaki K, Honjo T, Flavell RA. Instruction of distinct CD4 T helper cell fates by different notch ligands on antigen-presenting cells. Cell (2004) 117:515–26. doi: 10.1016/S0092-8674(04)00451-9
8. Radtke F, MacDonald HR, Tacchini-Cottier F. Regulation of innate and adaptive immunity by notch. Nat Rev Immunol (2013) 13:427–37. doi: 10.1038/nri3445
9. Tsao PN, Matsuoka C, Wei SC, Sato A, Sato S, Hasegawa K, et al. Epithelial notch signaling regulates lung alveolar morphogenesis and airway epithelial integrity. Proc Natl Acad Sci U S A (2016) 113:8242–7. doi: 10.1073/pnas.1511236113
10. Kiyokawa H, Morimoto M. Notch signaling in the mammalian respiratory system, specifically the trachea and lungs, in development, homeostasis, regeneration, and disease. Dev Growth Differ (2020) 62:67–79. doi: 10.1111/dgd.12628
11. Baindara P, Sarker B, Earhart AP, Mandal SM, Schrum AG. NOTCH signaling in COVID-19: A central hub controlling genes, proteins, and cells that mediate SARS-CoV-2 entry, the inflammatory response, and lung regeneration. Front Cell Infect Microbiol (2022) 4:928704. doi: 10.3389/fcimb.2022.928704
12. Narayana Y, Balaji KN. NOTCH1 up-regulation and signaling involved in mycobacterium bovis BCG-induced SOCS3 expression in macrophages. J Biol Chem (2008) 283:12501–11. doi: 10.1074/jbc.M709960200
13. Li QF, He XY, Xin T. Role of the notch signaling pathway in children with tuberculosis. Chin J Contemp Pediatr (2019) 21:1012–5. doi: 10.7499/j.issn.1008-8830.2019.10.011
14. Bermick JR, Lincoln PM, Allen RM, Kunkel SL, Schaller MA. Elevated notch ligands in serum are associated with HIV/TB coinfection. J Clin Tuberc Other Mycobact Dis (2021) 27:24. doi: 10.1016/j.jctube.2021.100258
15. Palaga T, Ratanabunyong S, Pattarakankul T, Sangphech N, Wongchana W, Hadae Y, et al. Notch signaling regulates expression of mcl-1 and apoptosis in PPD-treated macrophages. Cell Mol Immunol (2013) 10:444–52. doi: 10.1038/cmi.2013.22
16. Shi Q, Wang J, Yang Z, Liu Y. CircAGFG1modulates autophagy and apoptosis of macrophages infected by mycobacterium tuberculosis via the notch signaling pathway. Ann Transl Med (2020) 8:645–5. doi: 10.21037/atm.2020-20-3048
17. Zhang J, Jiao L, Bai H, Wu Q, Wu T, Liu T, et al. A Notch4 missense mutation is associated with susceptibility to tuberculosis in Chinese population. Infect Genet Evol (2020) 78:104145. doi: 10.1016/j.meegid.2019.104145
18. Fang W, Liu H, Qin L, Wang J, Huang X, Pan S, et al. Polymorphisms and gene expression of Notch4 in pulmonary tuberculosis. Front Immunol (2023) 14:1081483. doi: 10.3389/fimmu.2023.1081483
19. Li Q, Zhang H, Yu L, Wu C, Luo X, Sun H, et al. Down-regulation of notch signaling pathway reverses the Th1/Th2 imbalance in tuberculosis patients. Int Immunopharmacol (2018) 54:24–32. doi: 10.1016/j.intimp.2017.10.026
20. Ito T, Schaller M, Hogaboam CM, Standiford TJ, Sandor M, Lukacs NW, et al. TLR9 regulates the mycobacteria-elicited pulmonary granulomatous immune response in mice through DC-derived notch ligand delta-like 4. J Clin Invest (2009) 119:33–46. doi: 10.1172/JCI35647
21. Schaller MA, Allen RM, Kimura S, Day CL, Kunkel SL. Systemic expression of notch ligand delta-like 4 during mycobacterial infection alters the T cell immune response. Front Immunol (2016) 7:527. doi: 10.3389/fimmu.2016.00527
22. Sun J, Zhang Q, Yang G, Li Y, Fu Y, Zheng Y, et al. The licorice flavonoid isoliquiritigenin attenuates mycobacterium tuberculosis-induced inflammation through Notch1/NF-κB and MAPK signaling pathways. J Ethnopharmacol (2022) 294:115368. doi: 10.1016/j.jep.2022.115368
23. de Martino M, Lodi L, Galli L, Chiappini E. Immune response to mycobacterium tuberculosis: A narrative review. Front Pediatr (2019) 7:350. doi: 10.3389/fped.2019.00350
24. Martinez AN, Mehra S, Kaushal D. Role of interleukin 6 in innate immunity to mycobacterium tuberculosis infection. J Infect Dis (2013) 207:1253–61. doi: 10.1093/infdis/jit037
25. Cooper AM, Magram J, Ferrante J, Orme IM. Interleukin 12 (IL-12) is crucial to the development of protective immunity in mice intravenously infected with mycobacterium tuberculosis. J Exp Med (1997) 186:39–45. doi: 10.1084/jem.186.1.39
26. Cooper AM, Solache A, Khader SA. Interleukin-12 and tuberculosis: An old story revisited. Curr Opin Immunol (2007) 19:441–7. doi: 10.1016/j.coi.2007.07.004
27. Cooper AM, Khader SA. The role of cytokines in the initiation, expansion, and control of cellular immunity to tuberculosis. Immunol Rev (2008) 226:191–204. doi: 10.1111/j.1600-065X.2008.00702.x
28. Zheng R, Liu H, Zhou Y, Yan D, Chen J, Ma D, et al. Notch4 negatively regulates the inflammatory response to mycobacterium tuberculosis infection by inhibiting TAK1 activation. J Infect Dis (2018) 218:312–23. doi: 10.1093/infdis/jix636
29. Adhikari A, Xu M, Chen ZJ. Ubiquitin-mediated activation of TAK1 and IKK. Oncogene (2007) 26:3214–26. doi: 10.1038/sj.onc.1210413
30. Chen ZJ. Ubiquitination in signaling to and activation of IKK. Immunol Rev (2012) 246:95–106. doi: 10.1111/j.1600-065X.2012.01108.x
31. He W, Sun J, Zhang Q, Li Y, Fu Y, Zheng Y, et al. Andrographolide exerts anti-inflammatory effects in mycobacterium tuberculosis-infected macrophages by regulating the Notch1/Akt/NF-κB axis. J Leukoc Biol (2020) 108:1747–64. doi: 10.1002/JLB.3MA1119-584RRR
32. Castro RC, Zambuzi FA, Fontanari C, de Morais FR, Bollela VR, Kunkel SL, et al. NOTCH1 and DLL4 are involved in the human tuberculosis progression and immune response activation. Tuberculosis (2020) 124:101980. doi: 10.1016/j.tube.2020.101980
33. Bucşan AN, Veatch A, Singh DK, Akter S, Golden NA, Melanie K, et al. Response to hypoxia and the ensuing dysregulation of inflammation impacts mycobacterium tuberculosis pathogenicity. Am J Respir Crit Care Med (2022) 1:94–104. doi: 10.1164/rccm.202112-2747OC
34. Fung E, Tang SMT, Canner JP, Morishige K, Arboleda-Velasquez JF, Cardoso AA, et al. Delta-like 4 induces notch signaling in macrophages: Implications for inflammation. Circulation (2007) 115:2948–56. doi: 10.1161/CIRCULATIONAHA.106.675462
35. Shang Y, Smith S, Hu X. Role of notch signaling in regulating innate immunity and inflammation in health and disease. Protein Cell (2016) 7:159–74. doi: 10.1007/s13238-016-0250-0
36. Bray SJ. Notch signalling in context. Nat Rev Mol Cell Biol (2016) 17:722–35. doi: 10.1038/nrm.2016.94
37. Hu X, Chung AY, Wu I, Foldi J, Chen J, Ji JD, et al. Integrated regulation of toll-like receptor responses by notch and interferon-γ pathways. Immunity (2008) 29:691–703. doi: 10.1016/j.immuni.2008.08.016
38. Zhang Q, Wang C, Liu Z, Liu X, Han C, Cao X, et al. Notch signal suppresses toll-like receptor-triggered inflammatory responses in macrophages by inhibiting extracellular signal-regulated kinase 1/2-mediated nuclear factor κB activation. J Biol Chem (2012) 287:6208–17. doi: 10.1074/jbc.M111.310375
39. Mukherjee S, Schaller MA, Neupane R, Kunkel SL, Lukacs NW. Regulation of T cell activation by notch ligand, DLL4, promotes IL-17 production and rorc activation. J Immunol (2009) 182:7381–8. doi: 10.4049/jimmunol.0804322
40. Adler SH, Chiffoleau E, Xu L, Dalton NM, Burg JM, Wells AD, et al. Notch signaling augments T cell responsiveness by enhancing CD25 expression. J Immunol (2003) 171:2896–903. doi: 10.4049/jimmunol.171.6.2896
41. Lin PL, Plessner HL, Voitenok NN, Flynn JAL. Tumor necrosis factor and tuberculosis. J Investig Dermatol Symp Proc (2007) 12:22–5. doi: 10.1038/sj.jidsymp.5650027
42. Aster JC, Pear WS, Blacklow SC. Notch signaling in leukemia. Annu Rev Pathol Mech Dis (2008) 3:587–613. doi: 10.1146/annurev.pathmechdis.3.121806.154300
43. Espinoza I, Miele L. Notch inhibitors for cancer treatment. Pharmacol Ther (2013). doi: 10.1016/j.pharmthera.2013.02.003
44. Yuan X, Wu H, Xu H, Xiong H, Chu Q, Yu S, et al. Notch signaling: An emerging therapeutic target for cancer treatment. Cancer Lett (2015) 369:20–7. doi: 10.1016/j.canlet.2015.07.048
45. Golde TE, Koo EH, Felsenstein KM, Osborne BA, Miele L. γ-secretase inhibitors and modulators. Biochim Biophys Acta Biomembr (2013) 1828:2898–907. doi: 10.1016/j.bbamem.2013.06.005
46. Massard C, Azaro A, Soria JC, Lassen U, Le Tourneau C, Sarker D, et al. First-in-human study of LY3039478, an oral notch signaling inhibitor in advanced or metastatic cancer. Ann Oncol (2018) 29(9):1911–7. doi: 10.1093/annonc/mdy244
47. Kummar S, Coyne GOS, Do KT, Turkbey B, Meltzer PS, Polley E, et al. Clinical activity of the γ-secretase inhibitor PF-03084014 in adults with desmoid tumors (aggressive fibromatosis). J Clin Oncol (2017) 35(14):1561–9. doi: 10.1200/JCO.2016.71.1994
48. Moore G, Annett S, McClements L, Robson T. Top notch targeting strategies in cancer: A detailed overview of recent insights and current perspectives. Cells (2020) 9:1503. doi: 10.3390/cells9061503
49. Kotton DN, Morrisey EE. Lung regeneration: Mechanisms, applications and emerging stem cell populations. Nat Med (2014) 20:822–32. doi: 10.1038/nm.3642
Keywords: Mycobacterium tuberculosis, tuberculosis, host-targets, host-directed therapies, infection
Citation: Baindara P (2023) Notch signaling: A potential target for the development of host-directed therapies against tuberculosis. Front. Trop. Dis 4:1100526. doi: 10.3389/fitd.2023.1100526
Received: 16 November 2022; Accepted: 27 February 2023;
Published: 24 March 2023.
Edited by:
Selvakumar Subbian, Public Health Research Institute, Rutgers, The State University of New Jersey, United StatesReviewed by:
Armel Hervé Nwabo Kamdje, University of Garoua, CameroonRanjeet Kumar, Public Health Research Institute, Rutgers, The State University of New Jersey, United States
Copyright © 2023 Baindara. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Piyush Baindara, cGJhaW5kYXJhQGhlYWx0aC5taXNzb3VyaS5lZHU=
 Piyush Baindara
Piyush Baindara