- Department of Cell and Developmental Biology, School of Biological Sciences, University of California, San Diego, San Diego, CA, United States
Genetic-based technologies are emerging as promising tools to support vector population control. Vectors of human malaria and dengue have been the main focus of these development efforts, but in recent years these technologies have become more flexible and adaptable and may therefore have more wide-ranging applications. Culex quinquefasciatus, for example, is the primary vector of avian malaria in Hawaii and other tropical islands. Avian malaria has led to the extinction of numerous native bird species and many native bird species continue to be threatened as climate change is expanding the range of this mosquito. Genetic-based technologies would be ideal to support avian malaria control as they would offer alternatives to interventions that are difficult to implement in natural areas, such as larval source reduction, and limit the need for chemical insecticides, which can harm beneficial species in these natural areas. This mosquito is also an important vector of human diseases, such as West Nile and Saint Louis encephalitis viruses, so genetic-based control efforts for this species could also have a direct impact on human health. This commentary will discuss the current state of development and future needs for genetic-based technologies in lesser studied, but important disease vectors, such as C. quinquefasciatus, and make comparisons to technologies available in more studied vectors. While most current genetic control focuses on human disease, we will address the impact that these technologies could have on both disease and conservation focused vector control efforts and what is needed to prepare these technologies for evaluation in the field. The versatility of genetic-based technologies may result in the development of many important tools to control a variety of vectors that impact human, animal, and ecosystem health.
Introduction
Billions of people are at risk of contracting vector-borne diseases. Approximately 3.4 billion people residing in 92 countries are presently at risk of malaria infection alone and about a third of those are at high risk with a greater than 1 and 1000 chance of contracting malaria each year (https://www.who.int/data/gho/data/themes/malaria, September 23, 2021). Recent estimates of the costs of malaria prevention and treatment efforts exceed 4 billion annually (1), but this does not account for the economic impact on individuals to treat and prevent malaria as well as time away from work, school and household activities for the infected and their caretakers (2). Research and development of new vector-borne disease prevention technologies have resulted in the creation of a variety of novel vector control tools. Many of these emerging technologies are specific to malaria or dengue vectors or so broad that they negatively impact non-target species, but some, particularly genetic-based engineered technologies, have the potential to be adaptable to potentially any species without negatively impacting non-target species.
In addition to the impact of vector-borne disease on human health, invasive insect species cause extensive agricultural and environmental damage. They are one of the biggest threats to forest ecology in the United States, with potentially high impact species established every 2-3 years (3). The economic loss of these introductions is billions of dollars annually (4, 5). Despite the expansion of genetic tools and their applications in many model organisms, most disease vectors and invasive species have limited genetic tools to address basic biological questions and even fewer tools necessary to develop genetic control technologies. Numerous insect pests could benefit from the recent expansion of genetic-based tools, which have accelerated research in other fields. These tools include targeted nucleases like CRISPR, advanced sequencing methodologies, and improved genomics analysis, but again most currently lack even the basic technologies to develop these tools, such as insect husbandry or transgenesis techniques for these species. More investment into tools and other resources to advance the state of genetic and genomic technologies in understudied disease vectors and invasive species could foster advancements in genetic control capabilities that could have an incredible impact on human health and the global economy.
Overview of genetic control strategies
Genetic control strategies have focused on two main approaches to reduce the negative impacts of a species. One approach termed population modification, or population replacement, aims to modify the target population with desired traits. In the mosquito field, for example, many groups have developed mosquitoes that are incapable of transmitting one or more deadly human pathogens (6–13). If these disease-resistant mosquitoes are robust enough to stably function in diverse natural pathogen populations (14) and then released into the environment such that they achieve a high enough frequency in the population, then the population could have reduced or eliminated disease transmission capacity [reviewed in (14–16)]. Conceivably, other desired traits such as insecticide susceptibility, temperature sensitivity, or other characteristics to control the size or spread of a target population could be generated, given that tools are available to identify and build transgenes with these effects. For conservation applications, however, the desired trait, such as disease resistance, can be spread into a protected or beneficial population to prevent the spread of disease negatively impacting that population. While a population modification approach may be appropriate in some contexts (17), modifying protected or endangered populations, which would likely be the goal of conservation efforts, may be problematic. There are not only logistical and technical obstacles to modifying and propagating most protected species but modifying a sensitive species already at low population numbers is inherently risky (17). There are some applications, such as developing insecticide or pathogen-resistant honey bees (18), that are still being researched, but this approach will not be the focus of this manuscript.
The alternative approach, known as population suppression, is ideal for many applications, including conservation, particularly those that aim to eradicate problematic invasive species. Population suppression aims to reduce the target population by biasing the inheritance of gene modifications that decrease fertility or survival. As these modifications increase in frequency in a population, the population size subsequently decreases. Local elimination or global eradication of a target species may even be possible, but this is technology specific and dependent on the efficacy, stability, and spread of the suppression technologies and their release scheme. These technologies can be used to reduce or prevent the spread of invasive species that negatively impact the environment and human, plant, or animal health.
There are many strategies for genetically controlling a target population, either through suppression of the species or by spreading genes of interest through the population. Some traditional suppression methods, such as the sterile insect technique (SIT), were developed prior to modern genome engineering and use ionizing radiation to cause heritable germline chromosomal breaks and, consequently, sterilization (19–21) (Figure 1). These chromosomal aberrations, however, are random, imprecise, and complete sterilization cannot always be achieved. There are also typically radiation associated fitness impacts, which significantly reduce field competitiveness of released animals, thereby limiting the effectiveness and increasing effort required to suppress populations. While SIT has successfully controlled many insects (19, 20, 22–25), particularly agricultural pests, SIT has shown limited success in wide-scale management of mosquito populations. SIT requires inundative releases of the sterile individuals to ensure frequent matings with the target wild individuals. There are also large fitness costs associated with irradiating pupae (26–29). Often, SIT simply cannot be cost-effectively scaled to support the suppression and halt the spread of many invasive species due to the high numbers of individuals required for release and the frequency of releases. Even minimal occurrences of incomplete sterilization and radiation associated fitness costs can further increase the number of individuals needed for release. There are also many insect pests where the detrimental effects of releasing large numbers of pest species would preclude frequent releases. This may include insects that are voracious eaters or inhabit already stressed or sensitive environments where the frequent influx of high numbers of the target species could cause additional issues. Certainly, irradiation based SIT is a proven technology for controlling multiple insect agricultural pests, but its many disadvantages make this approach difficult to implement for all insects.
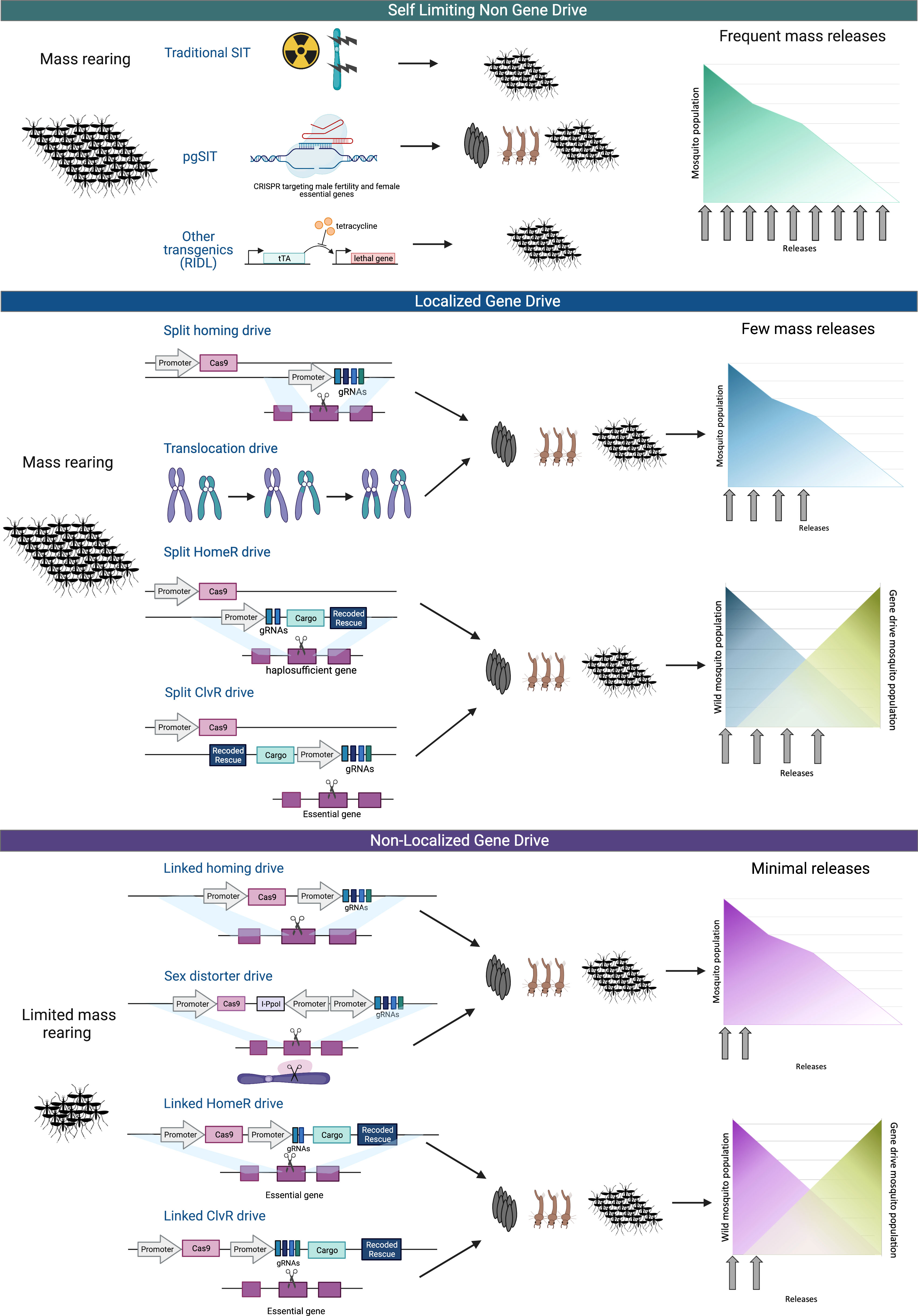
Figure 1 Select genetic and gene drive population suppression and modification technologies. Genetic control technologies can generally be divided into three categories based on the capability of the drive to spread beyond the target site; self-limiting non-gene drive (top), localized gene drive (middle), and non-localized gene drive (bottom). (Top) The self-limiting non-gene drives are population suppression technologies that reduce populations through the release of large numbers of sterile male. Traditional sterilization techniques, such as sterile insect technique (SIT) use radiation to cause DNA damage that sterilizes the insects. pgSIT utilizes CRISPR to target female essential and male fertility genes to kill female offspring and sterilize the male offspring. Release of Insects carrying a Dominant Lethal (RIDL) suppresses population by relying on tetracycline repressible dominant lethal phenotype. Female-specific RIDL (fsRIDL) uses the same method as RIDL, but with female specific lethality which can allow for greater effect by males passing on lethal genes to offspring. (Middle) Localized gene drives are designed to be locally and sometimes temporally confinable and require multiple releases to suppress a population. These technologies are not expected to spread far beyond the release site. The split homing drive consists of Cas9 and gRNA transgenes that are unlinked, so when breeding, the inherited transgenes do not necessarily concur and therefore the drive is limited to only offspring that inherit both drive components. Translocation drives create chromosomal rearrangements or inversions to bias inheritance of the translocation homozygote, in a phenomenon called underdominance. Translocation heterozygotes are inviable due to their inheritance of an unbalanced set of chromosomes. Additionally, there are homing drives that are combined with a Toxin/Antidote system by disrupting a necessary gene via Cas9 (toxin), requiring a healthy, Cas9-immune variant of the gene(antidote). This is accomplished by providing the Cas9-immune gene at the site of the target gene through a process called Home-and-Replace (HomeR) or by supplying the antidote gene at another site in the genome through an approach called Cleave-and-Rescue (ClvR). These approaches act as a selection mechanism for these gene drives to propagate themselves, selecting for the inheritance of the Cas9-immune gene (antidote). This can lead to a robust gene drive with a high frequency of inheritance, ideal for non-localized drives. To make these drive systems localized for better control, only half of the gene elements necessary for a gene drive are driven in a super-Mendelian inheritance manner. In this depiction, Cas9 is the separated element and the antidote gene, and the gRNAs are inherited in a super-Mendelian manner. This leads to a limited gene drive that will only propagate if the Cas9 element is co-inherited, causing the drive to be incapable of spreading indefinitely. (Bottom) Non-localized gene drives are designed to link the primary components needed for super Mendelian inheritance, so the entire drive is inherited and, therefore, rapidly spread into a population and beyond the initial release area. While in practice, this system does not often result in complete inheritance due to resistance mutations, in principle, these drives could spread without limit. Sex distorter drives utilize Cas9 to drive the disruption of necessary sex chromosomes or essential sex chromosome-linked genes. Linked HomeR and ClvR drives are as previously described, but the drive elements are now all linked. These systems are more robust than a simpler homing gene drive as they are built to address escape mutations and additionally allow for this system to be leveraged as a replacement or extinction drive. Created with BioRender.com.
Precision guided SIT (pgSIT) is a genetic based suppression technology that also produces sterile males but uses CRISPR genome editing to precisely target female essential genes and genes associated with male fertility during development (Figure 1). CRISPR genome editing is a proven technology in a variety of species and the ease with which these systems can be programmed to specifically target endogenous genes, together makes a CRISPR-based approach to generating sterile males advantageous for many applications. Furthermore, this programming flexibility also makes it possible to engineer these systems to target earlier stages of development. The pgSIT systems built to date (30–32) simultaneously target female essential and male fertility genes during development. Eggs, or any other life stage, can therefore be released to maximize any life stage-specific release benefits (e.g. density dependence) and eliminate the need to manually handle and release the fragile adults, which can also reduce fitness. Most other male sterility and female killing technologies developed to date, including conventional SIT, act in later developmental stages and require the release of adults, making them increasingly difficult to distribute and scale. Many insect species also have some degree of embryonic dormancy (diapause or quiescence) that would allow long-term storage and stocking of pgSIT eggs for some species (e.g. Aedes species), which would be propitious for scaling and distribution.
Other genetic suppression technologies, including Release of Insects carrying a Dominant Lethal (RIDL) (Figure 1) and female-specific flightless (fsRIDL), use a dominant lethal transactivator and operon system with a tetracycline repressor (Tet-off). In the absence of tetracycline in the RIDL system, the transactivator binds the operon system to generate a toxin expressed in early developmental stages (33). In fsRIDL, this toxin is female flight-specific, generating females that die shortly after emergence due to their inability to fly (34, 35). The RIDL technology field assessments are ongoing but have been shown to successfully suppress vector populations in the field (36–39) and are being developed for other invasive species that impact agricultural commodities, such as the diamondback moth (40) and the Mediterranean fruit fly (41). While these technologies are promising for other conservation applications, the technical requirements to develop a Tet-off system may be prohibitive for some species as compared to the more flexible CRISPR based systems. In addition, the use of the broad-spectrum antibiotic tetracycline can alter the insect microbiome and cause potential fitness issues that may be unsuitable for some applications.
In each of the above examples, the genetic suppression systems require frequent inundative releases of large numbers of individuals to achieve suppression. For many species, scaling and releasing large numbers of individuals may be impractical, so for many years, there has been a focus on the development of gene drive technologies to rapidly spread desired traits into populations (42–45). When linked to genetic cargo, gene drives bias the inheritance of the cargo at a higher frequency than expected by Mendelian inheritance. This “selfish” transmission facilitates the rapid spread of the gene drive and any linked cargo to a high frequency in a population and, therefore, can accomplish population suppression or modification with limited releases. There are many different categories of gene drives (reviewed here (46–48) and elsewhere) that differ by mechanism, performance, and predicted behavior in populations.
Some notable gene drive systems developed to date include underdominance systems, which spread into a population by conferring a disadvantage to heterozygotes and an advantage to homozygotes (49–54). Translocation drives are a type of stable underdominance drive and can be engineered to generate chromosomal rearrangements unique to the drive population. When scaled and released above a critical threshold into a wild population, individuals heterozygous for the translocation drive are inviable due to their inheritance of an unbalanced set of chromosomes. Over time, this advantage conferred by the drive homozygotes supports the gradual increase of the drive in the population. Additional underdominance drives have been developed and reviewed elsewhere (46, 47). Given that these drives only facilitate local population modification or replacement, they may prove valuable to modify local populations with beneficial traits, such as disease resistance, without imposing the risk of invasive spread.
There have been multiple toxin antidote gene drives developed to date. Inspired by a system found in nature (55), the maternal-effect dominant embryonic arrest (Medea) drive was the first example of a reverse engineered maternal toxin and zygotic antidote drive system (56–58). The Medea drive biases the drive inheritance by killing offspring that fail to inherit the antidote, which is linked to the toxin. The Medea drive has only been designed for population replacement to date but could be configured for population suppression (47, 57). Medea drives may be difficult to develop in many species, however, as they require the identification of maternal and zygotic promoters and the identification or design of appropriate toxins, which will likely be difficult for some species. Cleave and Rescue (ClvR), simplifies the development of toxin antidote systems by utilizing CRISPR. This CRISPR-based toxin antidote drive, is designed to cleave an essential gene (toxin) in trans but also encodes a cleavage-resistant copy of the target gene (antidote) (59). Other related CRISPR-based toxin antidote systems, such as toxin-antidote dominant embryo (TADE) and toxin-antidote dominant sperm (TADS) drives, target haplosufficient essential genes, however, they can conceivably be designed for population suppression (60), but this remains to be demonstrated.
Homing endonuclease gene (HEG) drives and X-shredders are two primary approaches that have been taken to generate population suppression gene drives. X-shredders are engineered to express an endonuclease on the Y chromosome and to target genes specific to the X-chromosome resulting in male-only progeny (61). X-shredders are only useful in species with genetically distinct sex chromosomes, so this drive system is not suitable for many insects, including grasshoppers, cockroaches, and some mosquitoes, which do not have a Y chromosome (62–65). As the name suggests, HEGs utilize a genome integrated endonuclease to cleave and copy itself into a corresponding locus on the homologous chromosome, thereby converting a drive heterozygote to a homozygote. When this conversion occurs in the germline, the drive can be inherited by subsequent generations at a rate higher than expected by Mendelian inheritance. Homing endonuclease drives were first described as a means to support population control nearly two decades ago (66). Only a few years later, the first demonstration of HEGs in Drosophila melanogaster used the I-SceI and I-OnuI nucleases to achieve only modest drive inheritance rates (67, 68). The zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) were then used to generate nuclease homing-based drives systems in D. melanogaster, but again, the homing efficiencies were only modest (69). Around the same time, multiple CRISPR homing-based drives CHDs were developed with high, sometimes remarkably >99% conversion rates (70–73). These high conversion rates, ease of programming precise genome modifications with CRISPR, and the capacity for CRISPR to function in a variety of species have resulted in CHDs becoming the predominant gene drive system in development today. There are, however, issues with CHDs that may limit their long term stability in the field. There are multiple DNA repair pathways that can either result in the accurate copying of the gene drive into the programmed location in the genome, or they can result in mutations at the target site. Some of these mutations will be resistant to cleavage by the gene drives, and as these cleavage-resistant mutations accumulate in the population, the drive becomes ineffective in that subset of the population (74, 75). Other mutations can occur from loss of the recipient chromosome, recombination events, and partial inheritance of the drive element. Additionally, cleavage associated with the maternal deposition of the Cas9 endonuclease into the embryo can result in the generation of resistant alleles (76–78). Multiple design considerations can be used to mitigate these adverse events, which will be discussed throughout this review.
One approach to prevent the development of drive resistance is to utilize a system that disadvantages the accumulation of resistance alleles and selects for the inheritance of drive susceptible copies after DNA repair. The innovative Home-and-Rescue (HomeR) drive shares the same toxin-antidote principle as Medea, and ClvR drives while utilizing the copying mechanism of CHDs. HomeR drives rely on locus-specific targeted repair that will result in an embryonic toxic effect if the repair fails (79, 80). This drive requires a haplosufficient gene, which is viable when one copy of the gene is functional or is supplied in trans, but is lethal if both copies are non-functional. The HomeR drive also includes a recoded rescue of the haplosufficient gene, which contains a drive susceptible allele that can rescue both gene and drive function. By selecting this target gene mutations tend to be non-functional, leading to embryonic lethality if the rescue gene is not inherited. Silent escape mutants, those that alter the nucleotide but not the amino acid sequence, may still be generated with this system but were not predicted to impact fitness and therefore are expected to spread into the population. Cas9 primarily causes insertions and deletions rather than base changes, which more likely result in amino acid changes, but unlikely repair outcomes could occur, and there may be naturally occurring drive-resistant alleles in the population. To address these risks, gRNAs can also be multiplexed to avoid escape mutant resistance by requiring multiple mutations for each gRNA target (81, 82). This approach can avoid both the development and accumulation of inframe functional resistant mutants resulting from CRISPR editing and the accumulation of naturally occurring escape alleles that likely exist in wild populations.
There are multiple technologies that can support the development of these drives, but most have yet to be adapted to understudied insect pests or vectors. Algorithms are available that can provide general estimates of CRISPR repair outcomes and frameshift rates for the selected CRISPR targets. These tools have yet to be optimized for analyzing repair outcomes in insect species but can provide insight into expected DNA repair outcomes (83, 84). With growing interest in HomeR, ClvR, and CHD based drive systems for insect population control, this CRISPR prediction software may be further developed to be species-specific. Recent work has shown differences in repair template preference between more traditional mammalian models and mosquitoes, suggesting that further study of CRISPR activity in mosquitoes or other insects of interest could reveal different repair preferences (85). The plethora of genetic control tools and methodologies allow for the selection of systems specific to the application, environment, and desired rate of spread and outcome of the system (e.g. population reduction, local elimination, or global eradication). In many cases, it would be prudent to prevent drives from spreading beyond the target site, while other target sites and species may require a robust system requiring fewer releases.
Genetic technologies for vector control and invasive insects
Mosquito genetic transformation started in the 1990s using transposable elements and transposases to integrate transgenes into the genome. Due to their impact on human health, genetic transformation technologies were developed for many disease vectors, including Aedes aegypti(86; 87), the primary yellow fever, dengue, Zika and chikungunya vector; Anopheles gambiae (88), a primary malaria vector in Africa; Culex quinquefasciatus(89), a primary West Nile virus, lymphatic filariasis, and avian malaria vector; Anopheles stephensi (90; 91), a primary malaria vector in Asia; Aedes albopictus (92), a global vector of chikungunya virus, dengue virus and dirofilariasis; Anopheles albimanus (93), a key new world malaria vector, and Anopheles sinensis (94), a malaria and lymphatic filariasis vector in Asia. There are many other vectors, particularly vectors of neglected tropical diseases (NTDs), or which are secondary vectors of malaria, in which there are no genetic tools to generate transgenic lines. Additional investment into genetic tools for these species would improve research capabilities and foster the generation of genetic control tools for these species.
Limited effort has been put into developing genetic tools for invasive species. Invasive species have profoundly impacted the environment, ecology, and human health or food security. The red imported ant, Solenopsis invicta (95), social wasps, Vespula spp. (96) and the gypsy moth, Lymantria dispar (97), are just a few insects that have had a profound negative impact on agriculture, wildlife or forest health when introduced to new areas. Invasive insect species are one of the biggest threats to forest health in the United States, for example, with potentially high impact invasive species established every two to three years (3). The economic loss of these introductions is billions of dollars annually (4, 5). Invasive species introductions require an integrated pest management (IPM) approach to limit the spread or eradicate invasive populations; however, current technologies are few, often expensive and inefficient (98). There has been a lot of recent interest in developing genetic tools to support IPM approaches to control invasive species (99–103); nonetheless, there are limited genetic technologies currently available for even basic functional genetic studies in many invasive insects.
Culex quinquefasciatus, a disease vector and threat to bird conservation
C. quinquefasciatus is an ideal target for genetic suppression technologies. It is a member of a complex of mosquitoes known as the Culex pipiens complex and is one mosquito species that poses a substantial threat to both human health and conservation. C. quinquefasciatus is a vector of both viral and parasitic pathogens of humans and wildlife and is a nuisance biter. C. quinquefasciatus is also invasive in many sensitive island habitats where it transmits avian malaria, Plasmodium relictum, and Avipoxvirus (APV), which has led to the extinction of unique island bird species (104–108), and threatens many more, such as Hawaiian honeycreepers, some of the most threatened bird species in the world (109). Even in the absence of disease transmission, this species has a negative impact on the tourism industry due to its nuisance biting. (110) Removing this species would, therefore, result in the elimination of multiple pathogens, improve outcomes for endangered species, and provide economic benefits to communities.
Culex quinquefasciatus has spread worldwide with human travel and has a global tropical and subtropical distribution (111). It is an opportunistic feeder of avian and mammalian hosts, making it a bridge vector for arboviruses, such as West Nile virus (WNV) (112). Bridge vectors are the species that transition pathogens from their wild, enzootic cycle to humans. When introduced into naive avian populations in the United States during the late 1990s, WNV also led to the death of thousands of wild birds (113). WNV may also play a role in increased mortality amongst conservation flagship species, such as raptors (114, 115) and other endangered birds (116, 117). Culex quinquefasciatus is also a vector of other arboviruses, such as Saint Louis encephalitis virus (SLEV), which is an arbovirus that is maintained in an enzootic cycle between birds and mosquitoes, including C. quinquefasciatus. This virus is endemic to the Americas, but there have been more human cases since the recent introductions into the western United States (118, 119) This mosquito is also considered to be a secondary vector of Japanese encephalitis virus (JEV) throughout Southeast Asia, but its vector competence for JEV (120–123), opportunistic feeding behaviors and urban distribution make it a high risk for becoming an enzootic vector (123) and urban bridge vector of JEV (124) throughout its distribution.
Insecticide treatment of passenger and commercial vessels is the main tool to prevent this species’ further spread. Once established, the predominant methods to control C. quinquefasciatus are through larval source reduction and insecticide application, but in areas with high usage of insecticides, C. quinquefasciatus can be highly insecticide resistant (125). Larval source reduction is also difficult since they prefer diverse, nutrient-rich larval habitats ranging from artificial containers to man-made water bodies, such as ponds and drainage areas. There is a need to develop more tools to prevent the spread of C. quinquefasciatus and to control C. quinquefasciatus where it has been established and causes negative impacts on human and wildlife health. CRISPR-based mutagenesis was demonstrated in C. quinquefasciatus (126–129). In the earliest study, CRISPR was used in C. quinquefasciatus to generate frameshift mutations to disrupt the function of an insecticide resistance gene. The more recent CRISPR mutagenesis studies targeted genes with observable markers that could be useful in generating additional genetic tools (127, 128). More recently, CRISPR homology directed repair (HDR) based transgenesis was also demonstrated, making it possible to site-specifically engineer this mosquito (129). Transgenesis with conventional transposable elements and transposase has been achieved in the past (89, 130) but has proven difficult overall. These new CRISPR based mutagenesis and transgenesis technologies make it more likely that more comprehensive genetic research and genetic control tools can be accomplished in the future.
Essential technologies for genetic engineering and gene drives
There are many technical obstacles to developing genetic technologies for understudied vectors and insects of interest to conservation programs, particularly control technologies (Table 1). First and foremost, the target species must be amenable to laboratory colonization and ideally have shorter generation times (days or weeks as opposed to many months) to expedite the creation and expansion of genetic lines and streamline mass rearing for large-scale production. Target species would also benefit from having an easily sourceable, effective, and affordable diet, ensuring the downstream products are scalable. Straightforward sexing methods are also ideal for accelerating genetic tool development and are essential for many control technologies that will rely on the release of non-biting males. In species that are difficult to sex sort, for example, species without easily and rapidly screenable sexual dimorphisms in pre-mating stages, then other sex sorting techniques may need to be engineered into the species, such as drug inducible (164) or fluorescent marker (165) based methods for sex sorting. Many other biological considerations may also preclude a potential target species from being a suitable candidate for genetic control, including reproductive capacity, egg storage, and handling and shipping conditions, among others. There are numerous understudied species, however, that may have characteristics that may warrant investment into improving their genetic toolset for future research and control efforts.
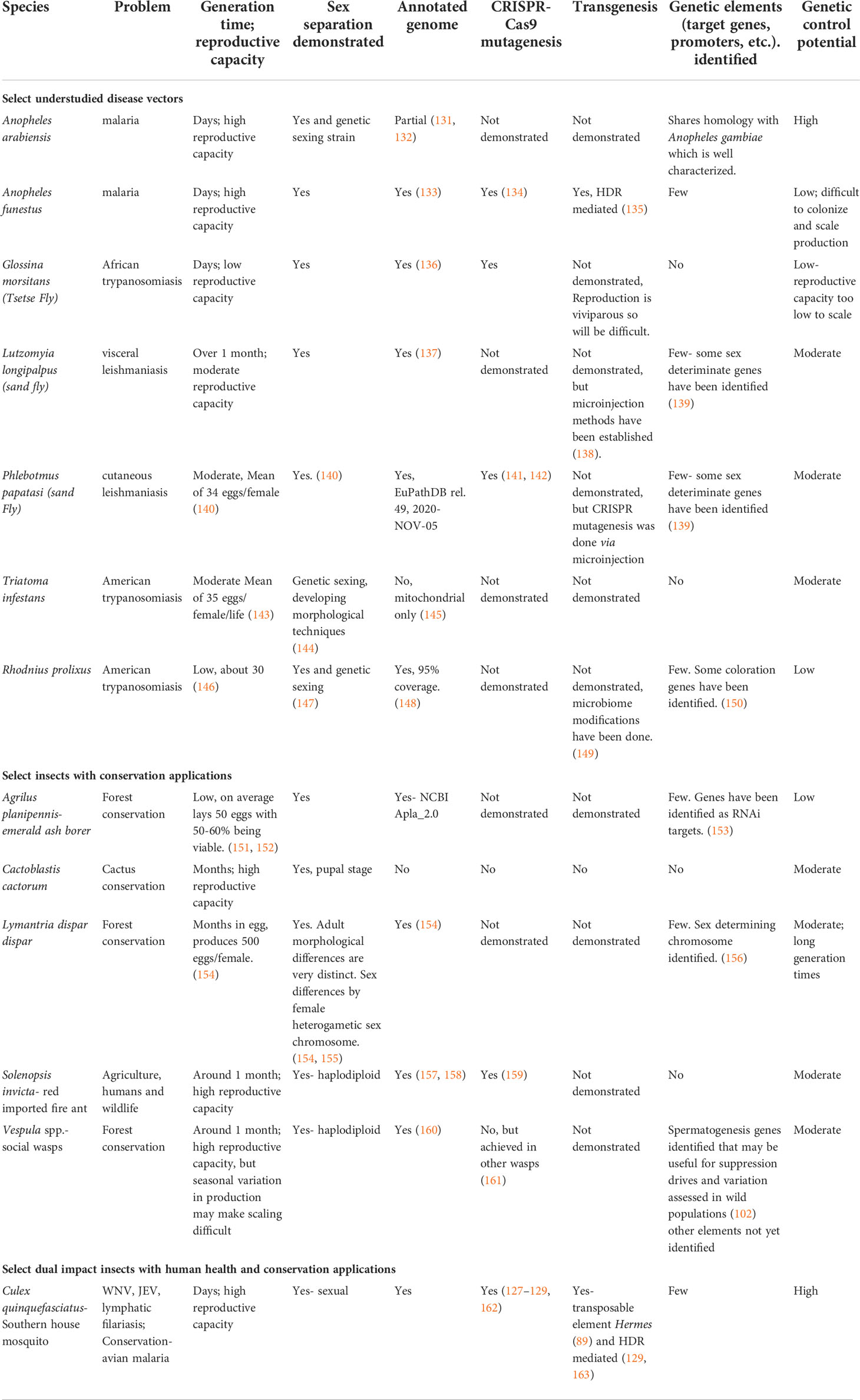
Table 1 State of genetic capabilities in selected colonizable, but genetically understudied insects of importance to human health and/or conservation.
If these biological conditions are reasonably met, then a sequenced genome, preferably one assembled and annotated, is required to identify target genes and genetic elements, such as promoters and other regulatory sequences useful for the expression and regulation of transgenes. For understudied species closely related to well-studied species, such as the case with the Anopheles gambiae complex where the molecular genetics in all but Anopheles gambiae is understudied, some genetic elements and target genes may be orthologous and similar enough to function adequately between species. Genomic information is scarce though for many invasive species or species of interest to conservation biologists (166).
Certainly, other considerations will facilitate the development of genetic technologies in understudied species. Characterization of genes that also encode easily observable characteristics, which can be used to rapidly evaluate genome engineering tools, and be designed as gene drive insertion targets allowing rapid evaluation and tracking of gene drives. When serving as markers, disrupting these phenotypic genes would ideally have minimal fitness costs (167). Without these types of phenotypic markers, genetic engineering efforts will be more difficult. Characterizing promoters that can be used for the tissue or stage-specific expression of fluorescent markers necessary for tracking transgene inheritance would also be useful. Promoters that drive the expression of fluorescent proteins before the adult stage, for instance, facilitate downstream genetic crosses, and promoters that drive distinct tissue-specific fluorescent protein expression allow the simultaneous tracking of multiple linked elements. Flexible options for marker expression can also reduce fitness costs associated with marker insertion and expression.
The considerations discussed so far are needed for basic genetic engineering, but the genetic population suppression technologies, such as pgSIT and RIDL, and self-propagating gene drive technologies have additional considerations. Population suppression technologies have multiple attributes that must be considered when designing a drive. First, it is important to consider targeting highly conserved regions of essential or fertility genes. If a mutation occurs at a highly conserved site, it will likely destroy the gene function. This design consideration is particularly important if drive cleavage resistant mutations arise at the cut sites as cleavage resistant mutants will be eliminated from the population if the essential gene is nonfunctional. Knowledge of genomics, population genetics, and population structure of the target wild population can also facilitate the selection of conserved targets. With the high fitness costs of suppression drives, any natural population variation that is resistant to the drive would have an advantage and therefore increase to a high frequency in the population. Better insight into the population genetics and structure can also be used to support efforts to model the impact of the drive on a wild population, a necessity for assessing drive performance and a precursor to planning any field release. The gene drive can also be designed to target multiple unique sequences in one or more target genes (66, 75, 81, 168, 169), so if a functional cleavage resistant mutation arises at one target site, then the drive will persist as the other target sites can compensate.
For the generation of CRISPR homing suppression drives, many additional elements are needed to support the drive, such as identifying functional Polymerase III U6 promoters or other promoters to drive the gRNA production. Some of these may function across many taxa, but further studies are needed to validate this in vivo(170). Flanking ribozymes or tRNA sequences have also been used to process gRNAs in other species and could be utilized to time gRNA expression or co-expression from the same transcript as the Cas9 (171–173). These designs may allow gRNAs to be expressed from any promoter, not just type III RNA polymerase III promoters. This added flexibility may better regulate CRISPR activity by limiting the window in which both the Cas9 and the gRNA are expressed; however, they are more complicated to design, which is an obstacle to their development, particularly in the many understudied species of interest to conservation. The selection of the Cas9 promoter also significantly impacts drive performance. Cas9 promoters with minimal somatic expression and appropriate timing can reduce the parental deposition of Cas9, which can lower drive fitness and contribute to the generation of drive cut site resistance. Germline specific promoters, however, are often difficult to identify even in well-characterized model organisms. Temporal control of Cas9 may be further refined by putting the gRNA under any promoter that shares some expression time with the Cas9 promoter. Alternatively, endogenous genes with appropriately timed expression can be used to drive the expression of Cas9 (174). Even with this knowledge, considerable resources will need to be expended to generate the genomic data needed to support the rational development of gene drives in many species.
Considerations for using genetic control tools for conservation
Genetic tools have many attributes that make them beneficial to conservation programs. Many conventional methods of pest control use insecticides, traps, or other methods that are not species specific. These methods often kill unintended species targets while trying to control the invasive target population. Due to their broad effects, pesticide use, for example, impacts many trophic levels and ecosystems and is a threat to biodiversity worldwide (175–178) Genetic tools can be designed to have high species specificity so that they will have minimal impact on unintended species.
Spatial confinability is essential for genetic control tools for conservation applications. Species that are invasive and cause negative impacts in one area may be beneficial in another area or within their native range. Gene drives with the ability to spread globally would, therefore, be inappropriate for controlling many invasive species. Gene drives where the active drive components are unlinked (split drives or daisy drives) (47, 99) or only target multiple mutations unique to the target population (99) would prevent the global spread of gene drives and therefore secure biodiversity outside of areas where the target species is undesirable. Additional considerations are required for eradicating a native species with a negative impact on human interests or eradicating a long-established invasive species (179). In these cases, environmental impact assessments may be needed to determine if eradication will have downstream effects on predator or prey species. The potential impact of gene drive technology on native species should be considered in the development of these technologies. Criteria for reducing hazards to endangered species and humans should be considered when planning field trials of any genetic and gene drive suppression systems (180, 181).
Another approach to spatially and temporally confining gene drives is to make their non-Mendelian inheritance conditional. These conditionally functional drives can be dependent on environmental conditions (e.g. temperature) or another natural or synthetic factor (e.g. small molecule activators or inhibitors). Small molecule gene drive activators or inhibitors have been developed in multiple species. The small molecule dependent gene drive was a self-limiting drive in Drosophila designed to excise Cas9 via a site-specific recombinase, Rippase, under the control of an RU486 inducible promoter (182). Small molecule gene drive activators have also been developed (183); however, it is still unclear if these have utility outside of the laboratory for many insect species. The function of these systems is drug dose-dependent, and the expression and shutoff of these systems are heavily reliant on the fidelity of the expression of the main components of these systems. The drug dose required for these current systems to function will likely be higher than feasible in natural environments, and most promoters have leaky basal expression, resulting in incomplete activation or deactivation of the drive. Novel inducible agents should also be explored for gene drive control due to the potential off-target effects of the RU486 and trimethoprim chemicals used to date. These gene drive systems are regulated at the DNA and protein level, but could also be regulated at the mRNA level, as seen in some examples of gene therapy (184). This approach uses an expression cassette that contains a self-cleaving ribozyme that translates protein when the ribozyme is disrupted by a morpholino, an oligomer that can modify gene expression. In a gene therapy context, it is possible to deliver morpholinos to the patient directly, but leveraging this technology in a gene drive system will require alternate designs and delivery mechanisms. Again, however, developing small molecule inducible or repressible drives is complicated and beyond the current genetic capabilities available for most invasive species. Substantial effort would be required to generate tools to develop these technologies in most species.
To limit their spread, gene drive organisms can also be designed to function in response to environmental factors. A recent example of this approach is a heat-sensitive ClvR suppression drive developed in Drosophila melanogaster that functions in response to seasonal changes in temperature. This ClvR system was designed to target an essential gene, dribble, and encodes a recoded cleavage resistant rescue gene that contains a temperature sensitive intein. At low temperatures, the intein splices itself out of the recoded gene and maintains gene function (185). At high temperatures, however, the intein is not spliced out of the recoded gene and cannot rescue the gene function, thereby killing all individuals that inherit the drive. If this temperature sensitive ClvR system persists for multiple generations in the population at low temperatures, it can therefore drive itself into the population at a high frequency facilitating rapid population suppression upon an increase in temperature. This system can limit the drive spread to regions that do not reach lethal or sterilizing temperatures or to seasons with lower temperatures. Another proof of concept drive system developed in D. melanogaster is designed to replace insecticide resistant alleles with a susceptible wildtype allele (186). This drive could be used to restore insecticide susceptibility to target populations, and by simply applying insecticides to the environment, the drive could be removed from a population. Certainly, these are innovative approaches to spatially or temporally confining a gene drive but are currently beyond the technical capabilities of many organisms of interest to conservation. These examples do, nonetheless, demonstrate the future technologies that may be available for conservation and NTD applications with further investment into genetic and genome technologies in these fields.
Ethical and regulatory considerations for deploying gene drive
Genetic suppression and gene drive technologies are promising technologies with the potential to address many problems in conservation and other fields. Current approaches to controlling invasive species have many unintended off-target effects on human health and the environment. Many of these conventional approaches are also becoming increasingly ineffective at controlling target species. These genetic engineering approaches have the potential to effectively address NTDs and conservation issues by targeting the species of interest directly, which avoids the side effects of pesticides on non-target species. As with most novel technologies, genetic and gene drive technologies are not without risk, and there is a potential for inadvertent and unexpected outcomes. To maximize the benefits of genetic and gene drive technologies while minimizing potential harm, principles have been set to guide future work in the conservation field (187). In brief, the key principles outlined are to ensure tools used for conservation applications 1) have public and expert approval, 2) state and international governance, 3) a broad consideration of ethical and moral systems, and 4) developers of the technology must self regulate the development of these technologies. While this list is succinct, these principles are complex and their execution more so. Developing and maintaining a dialogue with the communities affected by these technologies is paramount, but how this is achieved will vary by region, country, and community. Focusing on more localized and confinable technologies rather than non-localizations systems, particularly early in the development of these technologies, may help improve and maintain public trust throughout this process. In addition to public support, it is important to engage experts in many fields, including developers, social scientists, basic researchers, regulators, and experts in public health, conservation, and the local community, who can provide insights into the technology, community, and potential risks. While a consensus may not be made among all relevant experts, some guiding principles on the use of genetic and gene drive technologies in the laboratory and the field should be considered. The literature is abundant with discussions of these principles (187–193). There is also a need for governance to approve the use of these technologies and when required, this governance needs to be able to negotiate the use of this technology on an international level. Genetic and gene drive technologies are currently regulated at the national level, but as many of these technologies have the potential to spread beyond their release area, this may preclude the use of some of these technologies in target species with distributions spanning multiple countries. Several multinational entities, such as the World Health Organization and the African Union, have recognized that these technologies will require international cooperation and consent and have begun to discuss the use of these technologies and these issues (194, 195). With further development of gene drive technology and the expansion of these technologies to NTD and conservation targets, this technology will likely require additional international governance. Finally, the development of these technologies is centered in laboratories and institutions throughout the world by experts in the field of genetics and gene drive, but there is a need to better communicate this science and risks to the public and other experts. While other institutions will assist with these important duties, we must keep these goals in mind as stewards of genetic suppression technology.
There are also multiple practical examples of genetic technologies that have been studied in the field. Within the United States, the non-gene drive suppression technologies developed by Oxitec and MosquitoMate have required approvals from multiple regulatory agencies, which evaluate the safety of the technology and deployment plan. Oxitec was the first to market, and therefore faced regulatory jurisdictional conflicts prior to its assignment to the Environmental Protection Agency (196). Brazil has had multiple field trials with self-limiting systems and will soon deploy these technologies on a country-wide scale (37, 197, 198). Brazil also has had multiple regulatory, health, and biosafety agencies take part in determining the safety of mosquito deployment (199). As these technologies become more commonly used, there will likely be more standardization in the regulatory processes and more regulations to guide the development of safe and effective genetic control systems. To date, however, only non-gene drive mosquitoes have been studied in the field. Due to their ability to spread beyond their release point and persist in the environment, the regulatory requirements for gene drive technologies is much more uncertain.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
Funding
This work was supported by funding from an NIH award (R01AI151004), a Pacific Islands Fish and Wildlife Office ward (F21AS00155), and an Environmental Protection Agency award (84020401) awarded to O.S.A. The views, opinions, and/or findings expressed are those of the authors and should not be interpreted as representing the official views or policies of the U.S. government.
Conflict of interest
OA is a founder of Agragene, Inc. and Synvect, Inc. with equity interest. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Haakenstad A, Harle AC, Tsakalos G, Micah AE, Tao T, Anjomshoa M, et al. Tracking spending on malaria by source in 106 countries 2000–16: an economic modelling study. Lancet Infect Dis (2019) 19:703–16. doi: 10.1016/s1473-3099(19)30165-3
2. Institute of Medicine (US) Committee on the Economics of Antimalarial Drugs, Arrow KJ, Panosian C, Gelband H. The human and economic burden of malaria. US: National Academies Press (2004).
3. Lovett GM, Weiss M, Liebhold AM, Holmes TP, Leung B, Lambert KF, et al. Nonnative forest insects and pathogens in the united states: Impacts and policy options. Ecol Appl (2016) 26:1437–55. doi: 10.1890/15-1176
4. Pimentel D, Lach L, Zuniga R, Morrison D. Environmental and economic costs of nonindigenous species in the united states. BioScience (2000) 50:53. doi: 10.1641/0006-3568(2000)050[0053:eaecon]2.3.co;2
5. Aukema JE, Leung B, Kovacs K, Chivers C, Britton KO, Englin J, et al. Economic impacts of non-native forest insects in the continental United States. PloS One (2011) 6:e24587. doi: 10.1371/journal.pone.0024587
6. Franz AWE, Sanchez-Vargas I, Adelman ZN, Blair CD, Beaty BJ, James AA, et al. Engineering RNA interference-based resistance to dengue virus type 2 in genetically modified aedes aegypti. Proc Natl Acad Sci U S A (2006) 103:4198–203. doi: 10.1073/pnas.0600479103
7. Franz AWE, Sanchez-Vargas I, Raban RR, Black WC, James AA, Olson KE. Fitness impact and stability of a transgene conferring resistance to dengue-2 virus following introgression into a genetically diverse aedes aegypti strain. PloS Negl Trop Dis (2014) 8:e2833. doi: 10.1371/journal.pntd.0002833
8. Mishra P, Furey C, Balaraman V, Fraser MJ. Antiviral hammerhead ribozymes are effective for developing transgenic suppression of chikungunya virus in aedes aegypti mosquitoes. Viruses (2016) 8:169. doi: 10.3390/v8060163
9. Yen P-S, James A, Li J-C, Chen C-H, Failloux A-B. Synthetic miRNAs induce dual arboviral-resistance phenotypes in the vector mosquito aedes aegypti. Commun Biol (2018) 1:11. doi: 10.1038/s42003-017-0011-5
10. Buchman A, Gamez S, Li M, Antoshechkin I, Li H-H, Wang H-W, et al. Engineered resistance to zika virus in transgenic expressing a polycistronic cluster of synthetic small RNAs. Proc Natl Acad Sci U S A (2019) 116:3656–61. doi: 10.1073/pnas.1810771116
11. Buchman A, Gamez S, Li M, Antoshechkin I, Li H-H, Wang H-W, et al. Broad dengue neutralization in mosquitoes expressing an engineered antibody. PloS Pathog (2020) 16:e1008103. doi: 10.1371/journal.ppat.1008103
12. Williams AE, Sanchez-Vargas I, Reid WR, Lin J, Franz AWE, Olson KE. The antiviral small-interfering RNA pathway induces zika virus resistance in transgenic aedes aegypti. Viruses (2020) 12:1231. doi: 10.3390/v12111231
13. Liu W-L, Hsu C-W, Chan S-P, Yen P-S, Su MP, Li J-C, et al. Transgenic refractory aedes aegypti lines are resistant to multiple serotypes of dengue virus. Sci Rep (2021) 11:23865. doi: 10.1038/s41598-021-03229-4
14. Marshall JM, Raban RR, Kandul NP, Edula JR, León TM, Akbari OS. Winning the tug-of-War between effector gene design and pathogen evolution in vector population replacement strategies. Front Genet (2019) 10:1072. doi: 10.3389/fgene.2019.01072
15. Carballar-Lejarazú R, James AA. Population modification of anopheline species to control malaria transmission. Pathog Glob Health (2017) 111:424–35. doi: 10.1080/20477724.2018.1427192
16. Reid WR, Olson KE, Franz AWE. Current effector and gene-drive developments to engineer arbovirus-resistant aedes aegypti (Diptera: Culicidae) for a sustainable population replacement strategy in the field. J Med Entomol (2021) 58:1987–96. doi: 10.1093/jme/tjab030
17. Rode NO, Estoup A, Bourguet D, Courtier-Orgogozo V, Débarre F. Population management using gene drive: Molecular design, models of spread dynamics and assessment of ecological risks. Conserv Genet (2019) 20:671–90. doi: 10.1007/s10592-019-01165-5
18. Grozinger CM, Robinson GE. The power and promise of applying genomics to honey bee health. Curr Opin Insect Sci (2015) 10:124–32. doi: 10.1016/j.cois.2015.03.007
19. Knipling EF. Possibilities of insect control or eradication through the use of sexually sterile Males1. J Econ Entomol (1955) 48:459–62. doi: 10.1093/jee/48.4.459
20. Knipling EF. Screwworm eradication: Concepts and research leading to the sterile-male method. (USA: Smithsonian Institution Washington, DC) (1959).
21. Bakri A, Mehta K, Lance DR. Sterilizing insects with ionizing radiation. In: Dyck VA., and Hendrichs J, Robinson AS. (eds.) Sterile insect technique: Principles and practice in area-wide integrated pest management (2005). p. 355–98. (Dordrecht, Netherlands:Springer)
22. Politzar H, Cuisance D. An integrated campaign against riverine tsetse, glossina palpalis gambiensis and glossina tachinoides by trapping, and the release of sterile males. Int J Trop Insect Sci (1984) 5:439–42. doi: 10.1017/S1742758400008791
23. Takken W, Oladunmade MA, Dengwat L. The eradication of glossina palpalis palpalis using traps, insecticide impregnated targets and the sterile insect technique in central Nigeria. Bull Entomol Res (1986) 76:275–86. doi: 10.1017/S0007485300014747
24. Msangi AR, Kiwia N, Malele II, Mramba F, Saleh KM, Mussa WA, et al.(Ed.) (2000). Success in Zanzibar: Eradication of tsetse. (Malaysia:Penerbit Universiti Sains Malaysia)
25. Curtis CF. Review of previous applications of genetics to vector control. (2006), 33–43. InKnols B.G.J., Louis C.. (Eds.) Bridging laboratory and field research for genetic control of disease vectors. Proceedings volume of the joint WHO/TDR, NIH/NIAID, IAEA and Frontis workshop on bridging laboratory and field research for genetic control of disease vectors Nairobi, Kenya:Springer/Frontis, Volume 11:pp 225
26. Smittle BJ, Mount GA, Das M, Rajapaksa N. Apholate and gamma irradiation compared as sterilants for culex pipiens quinquefasciatus say (Diptera: Culicidae). Mosq News (1968) 28:201–4.
27. Andreasen MH, Curtis CF. Optimal life stage for radiation sterilization of anopheles males and their fitness for release. Med Vet Entomol (2005) 19:238–44. doi: 10.1111/j.1365-2915.2005.00565.x
28. Maïga H, Damiens D, Niang A, Sawadogo SP, Fatherhaman O, Lees RS, et al. Mating competitiveness of sterile male anopheles coluzzii in large cages. Malar J (2014) 13:460. doi: 10.1186/1475-2875-13-460
29. Yamada H, Vreysen MJB, Gilles JRL, Munhenga G, Damiens DD. The effects of genetic manipulation, dieldrin treatment and irradiation on the mating competitiveness of male anopheles arabiensis in field cages. Malar J (2014) 13:318. doi: 10.1186/1475-2875-13-318
30. Kandul NP, Liu J, Sanchez C HM, Wu SL, Marshall JM, Akbari OS. Transforming insect population control with precision guided sterile males with demonstration in flies. Nat Commun (2019) 10:84. doi: 10.1038/s41467-018-07964-7
31. Kandul NP, Liu J, Akbari OS. Temperature-inducible precision-guided sterile insect technique. CRISPR J (2021) 4:822–35. doi: 10.1089/crispr.2021.0077
32. Li M, Yang T, Bui M, Gamez S, Wise T, Kandul NP, et al. Suppressing mosquito populations with precision guided sterile males. Nat Commun. (2021) 12:5374. doi: 10.1038/s41467-021-25421-w
33. Thomas DD, Donnelly CA, Wood RJ, Alphey LS. Insect population control using a dominant, repressible, lethal genetic system. Science (2000) 287:2474–6. doi: 10.1126/science.287.5462.2474
34. Fu G, Lees RS, Nimmo D, Aw D, Jin L, Gray P, et al. Female-specific flightless phenotype for mosquito control. Proc Natl Acad Sci U S A (2010) 107:4550–4. doi: 10.1073/pnas.1000251107
35. Labbé GMC, Scaife S, Morgan SA, Curtis ZH, Alphey L. Female-specific flightless (fsRIDL) phenotype for control of aedes albopictus. PloS Negl Trop Dis (2012) 6:e1724. doi: 10.1371/journal.pntd.0001724
36. Harris AF, Nimmo D, McKemey AR, Kelly N, Scaife S, Donnelly CA, et al. Field performance of engineered male mosquitoes. Nat Biotechnol (2011) 29:1034–7. doi: 10.1038/nbt.2019
37. Carvalho DO, McKemey AR, Garziera L, Lacroix R, Donnelly CA, Alphey L, et al. Suppression of a field population of aedes aegypti in Brazil by sustained release of transgenic Male mosquitoes. PloS Negl Trop Dis (2015) 9:e0003864. doi: 10.1371/journal.pntd.0003864
38. Gorman K, Young J, Pineda L, Márquez R, Sosa N, Bernal D, et al. Short-term suppression of aedes aegypti using genetic control does not facilitate aedes albopictus. Pest Manage Sci (2016) 72:618–28. doi: 10.1002/ps.4151
39. Waltz E. First genetically modified mosquitoes released in the united states. Nature (2021) 593:175–6. doi: 10.1038/d41586-021-01186-6
40. Martins S, Naish N, Walker AS, Morrison NI, Scaife S, Fu G, et al. Germline transformation of the diamondback moth, plutella xylostella l., using the piggyBac transposable element. Insect Mol Biol (2012) 21:414–21. doi: 10.1111/j.1365-2583.2012.01146.x
41. Leftwich PT, Koukidou M, Rempoulakis P, Gong H-F, Zacharopoulou A, Fu G, et al. Genetic elimination of field-cage populations of Mediterranean fruit flies. Proc Biol Sci (2014) 281:20141372. doi: 10.1098/rspb.2014.1372
42. Sandler L, Novitski E. Meiotic drive as an evolutionary force. Am Nat (1957) 91:105–10. doi: 10.1086/281969
43. Hamiton WD. Extraordinary sex ratios. A sex-ratio theory for sex linkage and inbreeding has new implications in cytogenetics and entomology. Science (1967) 156:477–88. doi: 10.1126/science.156.3774.477
44. Curtis CF. Possible use of translocations to fix desirable genes in insect pest populations. Nature (1968) 218:368–9. doi: 10.1038/218368a0
45. International Atomic Energy Agency. (1969). Sterile-male Technique for Eradication or Control of Harmful Insects, Panel Proceedings Series - International Atomic Energy Agency No. (Vienna:IAEA)
46. Champer J, Buchman A, Akbari OS. Cheating evolution: Engineering gene drives to manipulate the fate of wild populations. Nat Rev Genet (2016) 17:146–59. doi: 10.1038/nrg.2015.34
47. Raban RR, Marshall JM, Akbari OS. Progress towards engineering gene drives for population control. J Exp Biol (2020) 223:jeb208181. doi: 10.1242/jeb.208181
48. Wang G-H, Gamez S, Raban RR, Marshall JM, Alphey L, Li M, et al. Combating mosquito-borne diseases using genetic control technologies. Nat Commun (2021) 12:4388. doi: 10.1038/s41467-021-24654-z
49. Akbari OS, Matzen KD, Marshall JM, Huang H, Ward CM, Hay BA. A synthetic gene drive system for local, reversible modification and suppression of insect populations. Curr Biol (2013) 23:671–7. doi: 10.1016/j.cub.2013.02.059
50. Buchman AB, Ivy T, Marshall JM, Akbari OS, Hay BA. Engineered reciprocal chromosome translocations drive high threshold, reversible population replacement in drosophila. ACS Synth Biol (2018) 7:1359–70. doi: 10.1021/acssynbio.7b00451
51. Buchman A, Shriner I, Yang T, Liu J, Antoshechkin I, Marshall JM, et al. Engineered reproductively isolated species drive reversible population replacement. Nat Commun (2021) 12:3281. doi: 10.1038/s41467-021-23531-z
52. Gantz VM, Akbari OS. Gene editing technologies and applications for insects. Curr Opin Insect Sci (2018) 28:66–72. doi: 10.1016/j.cois.2018.05.006
53. Maselko M, Feltman N, Upadhyay A, Hayward A, Das S, Myslicki N, et al. Engineering multiple species-like genetic incompatibilities in insects. Nat Commun (2020) 11:4468. doi: 10.1038/s41467-020-18348-1
54. Webster SH, Vella MR, Scott MJ. Development and testing of a novel killer–rescue self-limiting gene drive system in drosophila melanogaster. Proc R Soc B: Biol Sci (2020) 287:20192994. doi: 10.1098/rspb.2019.2994
55. Beeman RW, Friesen KS. Properties and natural occurrence of maternal-effect selfish genes (“Medea” factors) in the red flour beetle, tribolium castaneum. Heredity (1999) 82:529–34. doi: 10.1038/sj.hdy.6885150
56. Chen C-H, Huang H, Ward CM, Su JT, Schaeffer LV, Guo M, et al. A synthetic maternal-effect selfish genetic element drives population replacement in drosophila. Science (2007) 316:597–600. doi: 10.1126/science.1138595
57. Akbari OS, Chen C-H, Marshall JM, Huang H, Antoshechkin I, Hay BA. Novel synthetic medea selfish genetic elements drive population replacement in drosophila; a theoretical exploration of medea-dependent population suppression. ACS Synth Biol (2014) 3:915–28. doi: 10.1021/sb300079h
58. Buchman A, Marshall JM, Ostrovski D, Yang T, Akbari OS. Synthetically engineered medea gene drive system in the worldwide crop pest drosophila suzukii. Proc Natl Acad Sci U S A (2018) 115:4725–30. doi: 10.1073/pnas.1713139115
59. Oberhofer G, Ivy T, Hay BA. Cleave and rescue, a novel selfish genetic element and general strategy for gene drive. Proc Natl Acad Sci U S A (2019) 116:6250–9. doi: 10.1073/pnas.1816928116
60. Champer J, Kim IK, Champer SE, Clark AG, Messer PW. Performance analysis of novel toxin-antidote CRISPR gene drive systems. BMC Biol (2020) 18:27. doi: 10.1186/s12915-020-0761-2
61. Windbichler N, Papathanos PA, Crisanti A. Targeting the X chromosome during spermatogenesis induces y chromosome transmission ratio distortion and early dominant embryo lethality in anopheles gambiae. PloS Genet (2008) 4:e1000291. doi: 10.1371/journal.pgen.1000291
62. Palacios-Gimenez OM, Castillo ER, Martí DA, Cabral-de-Mello DC. Tracking the evolution of sex chromosome systems in melanoplinae grasshoppers through chromosomal mapping of repetitive DNA sequences. BMC Evol Biol (2013) 13:167. doi: 10.1186/1471-2148-13-167
63. Hall AB, Basu S, Jiang X, Qi Y, Timoshevskiy VA, Biedler JK, et al. A male-determining factor in the mosquito Aedes aegypti. Science (2015) 348:1268–70. doi: 10.1126/science.aaa2850
64. Turner J, Krishna R, Van’t Hof AE, Sutton ER, Matzen K, Darby AC. The sequence of a male-specific genome region containing the sex determination switch in aedes aegypti. Parasitol Vectors (2018) 11:549. doi: 10.1186/s13071-018-3090-3
65. Meisel RP, Delclos PJ, Wexler JR. The X chromosome of the German cockroach, blattella germanica, is homologous to a fly X chromosome despite 400 million years divergence. BMC Biol (2019) 17:100. doi: 10.1186/s12915-019-0721-x
66. Burt A. Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc Biol Sci (2003) 270:921–8. doi: 10.1098/rspb.2002.2319
67. Chan Y-S, Naujoks DA, Huen DS, Russell S. Insect population control by homing endonuclease-based gene drive: An evaluation in drosophila melanogaster. Genetics (2011) 188:33–44. doi: 10.1534/genetics.111.127506
68. Chan Y-S, Huen DS, Glauert R, Whiteway E, Russell S. Optimising homing endonuclease gene drive performance in a semi-refractory species: The drosophila melanogaster experience. PloS One (2013) 8:e54130. doi: 10.1371/journal.pone.0054130
69. Simoni A, Siniscalchi C, Chan YS, Huen DS, Russell S, Windbichler N, et al. Development of synthetic selfish elements based on modular nucleases in drosophila melanogaster. Nucleic Acids Res (2015) 43:2991–1. doi: 10.1093/nar/gkv117
70. DiCarlo JE, Chavez A, Dietz SL, Esvelt KM, Church GM. Safeguarding CRISPR-Cas9 gene drives in yeast. Nat Biotechnol (2015) 33:1250–5. doi: 10.1038/nbt.3412
71. Gantz VM, Bier E. The mutagenic chain reaction: A method for converting heterozygous to homozygous mutations. Science (2015) 348:442–4. doi: 10.1126/science.aaa5945
72. Gantz VM, Jasinskiene N, Tatarenkova O, Fazekas A, Macias VM, Bier E, et al. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito anopheles stephensi. Proc Natl Acad Sci U S A (2015) 112:E6736–43. doi: 10.1073/pnas.1521077112
73. Hammond A, Galizi R, Kyrou K, Simoni A, Siniscalchi C, Katsanos D, et al. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector anopheles gambiae. Nat Biotechnol (2016) 34:78–83. doi: 10.1038/nbt.3439
74. Hammond AM, Kyrou K, Bruttini M, North A, Galizi R, Karlsson X, et al. The creation and selection of mutations resistant to a gene drive over multiple generations in the malaria mosquito. PloS Genet (2017) 13:e1007039. doi: 10.1371/journal.pgen.1007039
75. Marshall JM, Buchman A, Sánchez C HM, Akbari OS. Overcoming evolved resistance to population-suppressing homing-based gene drives. Sci Rep (2017) 7:3776. doi: 10.1038/s41598-017-02744-7
76. Lin C-C, Potter CJ. Non-mendelian dominant maternal effects caused by CRISPR/Cas9 transgenic components in drosophila melanogaster. G3 (2016) 6:3685–91. doi: 10.1534/g3.116.034884
77. Champer J, Chung J, Lee YL, Liu C, Yang E, Wen Z, et al. Molecular safeguarding of CRISPR gene drive experiments. eLife (2019) 8:e41439. doi: 10.7554/elife.41439
78. Kandul NP, Liu J, Buchman A, Gantz VM, Bier E, Akbari OS. Assessment of a split homing based gene drive for efficient knockout of multiple genes. G3 (2020) 10:827–37. doi: 10.1534/g3.119.400985
79. Kandul NP, Liu J, Bennett JB, Marshall JM, Akbari OS. A confinable home-and-rescue gene drive for population modification. Elife (2021) 10:e65939. doi: 10.7554/eLife.65939
80. Terradas G, Buchman AB, Bennett JB, Shriner I, Marshall JM, Akbari OS, et al. Inherently confinable split-drive systems in drosophila. Nat Commun (2021) 12:1480. doi: 10.1038/s41467-021-21771-7
81. Champer SE, Oh SY, Liu C, Wen Z, Clark AG, Messer PW, et al. Computational and experimental performance of CRISPR homing gene drive strategies with multiplexed gRNAs. Sci Adv (2020) 6:eaaz0525. doi: 10.1126/sciadv.aaz0525
82. Yang E, Metzloff M, Langmüller AM, Xu X, Clark AG, Messer PW, et al. A homing suppression gene drive with multiplexed gRNAs maintains high drive conversion efficiency and avoids functional resistance alleles. G3 (2022) 12:jkac081. doi: 10.1093/g3journal/jkac081
83. Shen MW, Arbab M, Hsu JY, Worstell D, Culbertson SJ, Krabbe O, et al. Author correction: Predictable and precise template-free CRISPR editing of pathogenic variants. Nature (2019) 567:E1–2. doi: 10.1038/s41586-019-0938-4
84. Liu X, Wang S, Ai D. Predicting CRISPR/Cas9 repair outcomes by attention-based deep learning framework. Cells (2022) 11:1847. doi: 10.3390/cells11111847
85. Ang JXD, Nevard K, Ireland R, Purusothaman D-K, Verkuijl SAN, Shackleford L, et al. Considerations for homology-based DNA repair in mosquitoes: Impact of sequence heterology and donor template source. PloS Genet (2022) 18:e1010060. doi: 10.1371/journal.pgen.1010060
86. Coates CJ, Jasinskiene N, Miyashiro L, James AA. Mariner transposition and transformation of the yellow fever mosquito, aedes aegypti. Proc Natl Acad Sci U S A (1998) 95:3748–51. doi: 10.1073/pnas.95.7.3748
87. Jasinskiene N, Coates CJ, Benedict MQ, Cornel AJ, Rafferty CS, James AA, et al. Stable transformation of the yellow fever mosquito, aedes aegypti, with the Hermes element from the housefly. Proc Natl Acad Sci U S A (1998) 95:3743–7. doi: 10.1073/pnas.95.7.3743
88. Grossman GL, Rafferty CS, Clayton JR, Stevens TK, Mukabayire O, Benedict MQ. Germline transformation of the malaria vector, anopheles gambiae, with the piggyBac transposable element. Insect Mol Biol (2001) 10:597–604. doi: 10.1046/j.0962-1075.2001.00299.x
89. Allen ML, O’Brochta DA, Atkinson PW, Levesque CS. Stable, germ-line transformation of Culex quinquefasciatus (Diptera: Culicidae). J Med Entomol (2001) 38:701–10. doi: 10.1603/0022-2585-38.5.701
90. Catteruccia F, Nolan T, Loukeris TG, Blass C, Savakis C, Kafatos FC, et al. Stable germline transformation of the malaria mosquito anopheles stephensi. Nature (2000) 405:959–62. doi: 10.1038/35016096
91. Nolan T, Bower TM, Brown AE, Crisanti A, Catteruccia F. piggyBac-mediated germline transformation of the malaria mosquito anopheles stephensi using the red fluorescent protein dsRED as a selectable marker. J Biol Chem (2002) 277:8759–62. doi: 10.1074/jbc.C100766200
92. Labbé GMC, Nimmo DD, Alphey L. Piggybac- and PhiC31-mediated genetic transformation of the Asian tiger mosquito, aedes albopictus (Skuse). PloS Negl Trop Dis (2010) 4:e788. doi: 10.1371/journal.pntd.0000788
93. Perera OP, Harrell RA II, Handler AM. Germ-line transformation of the south American malaria vector, anopheles albimanus, with a piggyBac/EGFP transposon vector is routine and highly efficient. Insect Mol Biol (2002) 11:291–7. doi: 10.1046/j.1365-2583.2002.00336.x
94. Liu J-G, Qiao L, Zhang J-J, Chen B, He Z-B. piggyBac-mediated germline transformation of the malaria mosquito anopheles sinensis (Diptera: Culicidae). Insect Sci (2021) 28:1202–6. doi: 10.1111/1744-7917.12836
95. Pimentel D, Zuniga R, Morrison D. Update on the environmental and economic costs associated with alien-invasive species in the united states. Ecol Econ (2005) 52:273–88. doi: 10.1016/j.ecolecon.2004.10.002
96. MacIntyre P, Hellstrom J. An evaluation of the costs of pest wasps (Vespula species) in. Int Pest Control (2015) 57:162–3.
97. Campbell FT, Schlarbaum SE. Fading forests III. In: North American trees and the threat of exotic pests report (2014). (Virginia, USA:The Nature Conservancy, Arlington).
98. Teem JL, Alphey L, Descamps S, Edgington MP, Edwards O, Gemmell N, et al. Genetic biocontrol for invasive species. Front Bioeng Biotechnol (2020) 8:452. doi: 10.3389/fbioe.2020.00452
99. Esvelt KM, Gemmell NJ. Conservation demands safe gene drive. PloS Biol (2017) 15:e2003850. doi: 10.1371/journal.pbio.2003850
100. Leitschuh CM, Kanavy D, Backus GA, Valdez RX, Serr M, Pitts EA, et al. Developing gene drive technologies to eradicate invasive rodents from islands. J Responsible Innovation (2018) 5:S121–38. doi: 10.1080/23299460.2017.1365232
101. Godwin J, Serr M, Barnhill-Dilling SK, Blondel DV, Brown PR, Campbell K, et al. Rodent gene drives for conservation: Opportunities and data needs. Proc Biol Sci (2019) 286:20191606. doi: 10.1098/rspb.2019.1606
102. Lester PJ, Bulgarella M, Baty JW, Dearden PK, Guhlin J, Kean JM. The potential for a CRISPR gene drive to eradicate or suppress globally invasive social wasps. Sci Rep (2020) 10:12398. doi: 10.1038/s41598-020-69259-6
103. Simberloff D. Maintenance management and eradication of established aquatic invaders. Hydrobiologia (2021) 848:2399–420. doi: 10.1007/s10750-020-04352-5
104. Warner RE. The role of introduced diseases in the extinction of the endemic Hawaiian avifauna. Condor (1968) 70:101–20. doi: 10.2307/1365954
105. van Riper C, van Riper SG, Lee Goff M, Laird M. The epizootiology and ecological significance of malaria in Hawaiian land birds. Ecol Monogr (1986) 56:327–44. doi: 10.2307/1942550
106. Atkinson CT, Woods KL, Dusek RJ, Sileo LS, Iko WM. Wildlife disease and conservation in Hawaii: pathogenicity of avian malaria (Plasmodium relictum) in experimentally infected iiwi (Vestiaria coccinea). Parasitology (1995) 111 Suppl:S59–69. doi: 10.1017/S003118200007582X
107. Atkinson CT, Dusek RJ, Woods KL, Iko WM. Pathogenicity of avian malaria in experimentally-infected Hawaii amakihi. J Wildl Dis (2000) 36:197–204. doi: 10.7589/0090-3558-36.2.197
108. Lapointe DA, Atkinson CT, Samuel MD. Ecology and conservation biology of avian malaria. Ann N Y Acad Sci (2012) 1249:211–26. doi: 10.1111/j.1749-6632.2011.06431.x
109. Buchanan GM, Donald PF, Butchart SHM. Identifying priority areas for conservation: A global assessment for forest-dependent birds. PloS One (2011) 6:e29080. doi: 10.1371/journal.pone.0029080
110. Negi C, Verma P. Review on Culex quinquefasciatus: Southern house mosquito. Int J Life-Sci Sci Res (2018) 4:1563–66. doi: 10.21276/ijlssr.2018.4.1.9
111. Alaniz AJ, Carvajal MA, Bacigalupo A, Cattan PE. Global spatial assessment of aedes aegypti and Culex quinquefasciatus: A scenario of zika virus exposure. Epidemiol Infect (2019) 147:e52. doi: 10.1017/S0950268818003102
112. Farajollahi A, Fonseca DM, Kramer LD, Marm Kilpatrick A. “Bird biting” mosquitoes and human disease: A review of the role of culex pipiens complex mosquitoes in epidemiology. Infect Genet Evol (2011) 11:1577–85. doi: 10.1016/j.meegid.2011.08.013
113. Gamino V, Höfle U. Pathology and tissue tropism of natural West Nile virus infection in birds: A review. Vet Res (2013) 44:39. doi: 10.1186/1297-9716-44-39
114. Saito EK, Sileo L, Green DE, Meteyer CU, McLaughlin GS, Converse KA, et al. Raptor mortality due to West Nile virus in the united states 2002. J Wildl Dis (2007) 43:206–13. doi: 10.7589/0090-3558-43.2.206
115. Vidaña B, Busquets N, Napp S, Pérez-Ramírez E, Jiménez-Clavero MÁ, Johnson N. The role of birds of prey in West Nile virus epidemiology. Vaccines (Basel) (2020) 8:550. doi: 10.3390/vaccines8030550
116. Höfle U, Blanco JM, Crespo E, Naranjo V, Jiménez-Clavero MA, Sanchez A, et al. West Nile Virus in the endangered Spanish imperial eagle. Vet Microbiol (2008) 129:171–8. doi: 10.1016/j.vetmic.2007.11.006
117. LaPointe DA, Hofmeister EK, Atkinson CT, Porter RE, Dusek RJ. Experimental infection of hawai`i `amakihi (hemignathus virens) with west nile virus and competence of a co-occurring vector, Culex quinquefasciatus: Potential impacts on endemic hawaiian avifauna. J Wildlife Dis (2009) 45:257–71. doi: 10.7589/0090-3558-45.2.257
118. White GS, Symmes K, Sun P, Fang Y, Garcia S, Steiner C, et al. Reemergence of st. Louis encephalitis virus, California 2015. Emerg Infect Dis (2016) 22:2185–8. doi: 10.3201/eid2212.160805
119. Swetnam DM, Stuart JB, Young K, Maharaj PD, Fang Y, Garcia S, et al. Movement of st. Louis encephalitis virus in the Western united states 2014- 2018. PloS Negl Trop Dis (2020) 14:e0008343.
120. Van Den Hurk AF, Nisbet DJ, Hall RA, Kay BH, Mackenzie JS, Ritchie SA. Vector competence of Australian mosquitoes (Diptera: Culicidae) for Japanese encephalitis virus. J Med Entomol (2003) 40:82–90. doi: 10.1603/0022-2585-40.1.82
121. Turell MJ, Mores CN, Dohm DJ, Lee W-J, Kim H-C, Klein TA. Laboratory transmission of Japanese encephalitis, West Nile, and getah viruses by mosquitoes (Diptera: Culicidae) collected near camp greaves, gyeonggi province, republic of Korea 2003. J Med Entomol (2006) 43:1076–81. doi: 10.1093/jmedent/43.5.1076
122. Huang Y-JS, Harbin JN, Hettenbach SM, Maki E, Cohnstaedt LW, Barrett ADT, et al. Susceptibility of a north American Culex quinquefasciatus to Japanese encephalitis virus. Vector Borne Zoonotic Dis (2015) 15:709–11. doi: 10.1089/vbz.2015.1821
123. Huang Y-JS, Hettenbach SM, Park SL, Higgs S, Barrett ADT, Hsu W-W, et al. Differential infectivities among different Japanese encephalitis virus genotypes in Culex quinquefasciatus mosquitoes. PloS Negl Trop Dis (2016) 10:e0005038. doi: 10.1371/journal.pntd.0005038
124. Lindahl J, Chirico J, Boqvist S, Thu HTV, Magnusson U. Occurrence of Japanese encephalitis virus mosquito vectors in relation to urban pig holdings. Am J Trop Med Hyg (2012) 87:1076–82. doi: 10.4269/ajtmh.2012.12-0315
125. Talipouo A, Mavridis K, Nchoutpouen E, Djiappi-Tchamen B, Fotakis EA, Kopya E, et al. High insecticide resistance mediated by different mechanisms in Culex quinquefasciatus populations from the city of yaoundé, Cameroon. Sci Rep (2021) 11:7322. doi: 10.1038/s41598-021-86850-7
126. Itokawa K, Komagata O, Kasai S, Ogawa K, Tomita T. Testing the causality between CYP9M10 and pyrethroid resistance using the TALEN and CRISPR/Cas9 technologies. Sci Rep (2016) 6:24652. doi: 10.1038/srep24652
127. Anderson ME, Mavica J, Shackleford L, Flis I, Fochler S, Basu S, et al. CRISPR/Cas9 gene editing in the West Nile virus vector, Culex quinquefasciatus say. PloS One (2019) 14:e0224857. doi: 10.1371/journal.pone.0224857
128. Li M, Li T, Liu N, Raban RR, Wang X, Akbari OS. Methods for the generation of heritable germline mutations in the disease vector Culex quinquefasciatus using clustered regularly interspaced short palindrome repeats-associated protein 9. Insect Mol Biol (2020) 29:214–20. doi: 10.1111/imb.12626
129. Feng X, Del Amo VL, Mameli E, Lee M, Bishop AL, Perrimon N, et al. Optimized CRISPR tools and site-directed transgenesis towards gene drive development in Culex quinquefasciatus mosquitoes. Nat Commun (2021) 12:2960. doi: 10.1038/s41467-021-23239-0
130. Allen ML, Christensen BM. Flight muscle-specific expression of act88F: GFP in transgenic Culex quinquefasciatus say (Diptera: Culicidae). Parasitol Int (2004) 53:307–14. doi: 10.1016/j.parint.2004.04.002
131. Zamyatin A, Avdeyev P, Liang J, Sharma A, Chen C, Lukyanchikova V, et al. Chromosome-level genome assemblies of the malaria vectors Anopheles coluzzii and Anopheles arabiensis. Gigascience (2021) 10:giab017 doi: 10.1093/gigascience/giab01
132. Neafsey DE, Waterhouse RM, Abai MR, Aganezov SS, Alekseyev MA, Allen JE, et al. Mosquito genomics. highly evolvable malaria vectors: the genomes of 16 anopheles mosquitoes. Science (2015) 347:1258522. doi: 10.1126/science.1258522
133. Ghurye J, Koren S, Small ST, Redmond S, Howell P, Phillippy AM, et al. A chromosome-scale assembly of the major African malaria vector anopheles funestus. Gigascience (2019) 8:giz063. doi: 10.1093/gigascience/giz063
134. Li M, Akbari OS, White BJ. Highly efficient site-specific mutagenesis in malaria mosquitoes using CRISPR. G3 Genes|Genomes|Genetics (2018) 8:653–8. doi: 10.1534/g3.117.1134
135. Quinn C, Anthousi A, Wondji C, Nolan T. CRISPR-mediated knock-in of transgenes into the malaria vector anopheles funestus. G3 (2021) 11:jkab201. doi: 10.1093/g3journal/jkab201
136. International Glossina Genome Initiative. Genome sequence of the tsetse fly (Glossina morsitans): Vector of African trypanosomiasis. Science (2014) 344:380–6. doi: 10.1126/science.1249656
137. Yang Z, Wu Y. Improved annotation of lutzomyia longipalpis genome using bioinformatics analysis. PeerJ (2019) 7:e7862. doi: 10.7717/peerj.7862
138. Jeffries CL, Rogers ME, Walker T. Establishment of a method for lutzomyia longipalpis sand fly embryo microinjection: The first step towards potential novel control strategies for leishmaniasis. Wellcome Open Res (2018) 3:55. doi: 10.12688/wellcomeopenres.14555.1
139. Petrella V, Aceto S, Colonna V, Saccone G, Sanges R, Polanska N, et al. Identification of sex determination genes and their evolution in phlebotominae sand flies (Diptera, nematocera). BMC Genomics (2019) 20:522. doi: 10.1186/s12864-019-5898-4
140. Lawyer P, Killick-Kendrick M, Rowland T, Rowton E, Volf P. Laboratory colonization and mass rearing of phlebotomine sand flies (Diptera, psychodidae). Parasite (2017) 165:24–42. doi: 10.1051/parasite/2017041
141. Louradour I, Ghosh K, Inbar E, Sacks DL. CRISPR/Cas9 mutagenesis in phlebotomus papatasi: The immune deficiency pathway impacts vector competence for leishmania major. MBio (2019) 10:e01941–19. doi: 10.1128/mBio.01941-19
142. Louradour I, Ghosh K, Inbar E, Sacks DL, Aluvihare C, Harrell RA 2nd. Sand fly (Phlebotomus papatasi) embryo microinjection for CRISPR/Cas9 mutagenesis. J Vis Exp (2020):165. doi: 10.3791/61924
143. Forlani L, Pedrini N, Girotti JR, Mijailovsky SJ, Cardozo RM, Gentile AG, et al. Biological control of the chagas disease vector triatoma infestans with the entomopathogenic fungus beauveria bassiana combined with an aggregation cue: Field, laboratory and mathematical modeling assessment. PloS Negl Trop Dis (2015) 9:e0003778. doi: 10.1371/journal.pntd.0003778
144. Vilaseca C, Méndez MA, Pinto CF, Lemic D, Benítez HA. Unraveling the morphological variation of triatoma infestans in the peridomestic habitats of chuquisaca Bolivia: A geometric morphometric approach. Insects (2021) 12:185. doi: 10.3390/insects12020185
145. Pita S, Panzera F, Vela J, Mora P, Palomeque T, Lorite P. Complete mitochondrial genome of triatoma infestans (Hemiptera, reduviidae, triatominae), main vector of chagas disease. Infect Genet Evol (2017) 54:158–63. doi: 10.1016/j.meegid.2017.06.003
146. Chiang RG, Chiang JA. Reproductive physiology in the blood feeding insect, rhodnius prolixus, from copulation to the control of egg production. J Insect Physiol (2017) 97:27–37. doi: 10.1016/j.jinsphys.2016.06.001
147. Koerich LB, Dupim EG, Faria LL, Dias FA, Dias AF, Trindade GS, et al. First report of y-linked genes in the kissing bug rhodnius prolixus. BMC Genomics (2016) 17:100. doi: 10.1186/s12864-016-2425-8
148. Mesquita RD, Vionette-Amaral RJ, Lowenberger C, Rivera-Pomar R, Monteiro FA, Minx P, et al. Genome of rhodnius prolixus, an insect vector of chagas disease, reveals unique adaptations to hematophagy and parasite infection. Proc Natl Acad Sci U S A (2015) 112:14936–41. doi: 10.1073/pnas.1506226112
149. Durvasula RV, Gumbs A, Panackal A, Kruglov O, Taneja J, Kang AS, et al. Expression of a functional antibody fragment in the gut of rhodnius prolixus via transgenic bacterial symbiont rhodococcus rhodnii. Med Vet Entomol (1999) 13:115–9. doi: 10.1046/j.1365-2915.1999.00175.x
150. Berni M, Bressan D, Simão Y, Julio A, Oliveira PL, Pane A, et al. Pigmentation loci as markers for genome editing in the chagas disease vector rhodnius prolixus. bioRxiv (2020) 06793. doi: 10.1101/2020.04.29.067934
151. Rutledge CE, Keena MA. Mating frequency and fecundity in the emerald ash borer agrilus planipennis (Coleoptera: Buprestidae). Ann Entomol Soc Am (2012) 105:66–72. doi: 10.1038/nature.2016.19804
152. Chamorro ML, Jendek E, Haack RA, Petrice TR, Woodley NE, Grebennikov VV, et al. Illustrated guide to the emerald ash borer agrilus planipennis fairmaire and related species (Coleoptera, buprestidae) (Sofia, Bulgaria:Pensoft) (2015).
153. Rodrigues TB, Rieske LK, J Duan J, Mogilicherla K, Palli SR. Development of RNAi method for screening candidate genes to control emerald ash borer, agrilus planipennis. Sci Rep (2017) 7:7379. doi: 10.1038/s41598-017-07605-x
154. Doane CC, McManus ML. The gypsy moth. In: Research toward integrated pest management., (Washington D.C., USA:U.S. Department of Agriculture) (1981).
155. Sahara K, Yoshido A, Traut W. Sex chromosome evolution in moths and butterflies. Chromosome Res (2012) 20:83–94. doi: 10.1007/s10577-011-9262-z
156. Zhang J, Cong Q, Rex EA, Hallwachs W, Janzen DH, Grishin NV, et al. Gypsy moth genome provides insights into flight capability and virus–host interactions. Proc Natl Acad Sci (2019) 116:1669–78. doi: 10.1073/pnas.1818283116
157. Wang J, Ross KG, Keller L. Genome-wide expression patterns and the genetic architecture of a fundamental social trait. PloS Genet (2008) 4:e1000127. doi: 10.1371/journal.pgen.1000127
158. Wurm Y, Wang J, Riba-Grognuz O, Corona M, Nygaard S, Hunt BG, et al. The genome of the fire ant solenopsis invicta. Proc Natl Acad Sci U S A (2011) 108:5679–84. doi: 10.1073/pnas.1009690108
159. Chiu Y-K, Hsu J-C, Chang T, Huang Y-C, Wang J. Mutagenesis mediated by CRISPR/Cas9 in the red imported fire ant, solenopsis invicta. Insectes Soc (2020) 67:317–26. doi: 10.1007/s00040-020-00755-8
160. Harrop TWR, Guhlin J, McLaughlin GM, Permina E, Stockwell P, Gilligan J, et al. High-quality assemblies for three invasive social wasps from the vespula genus. G3 (2020) 10:3479–88. doi: 10.1534/g3.120.401579
161. Li M, Bui M, Akbari OS. Embryo microinjection and transplantation technique for nasonia vitripennis genome manipulation. J Vis Exp (2017) 130:56990. doi: 10.3791/56990
162. Bui M, Li M, Raban RR, Liu N, Akbari OS. Embryo microinjection techniques for efficient site-specific mutagenesis in Culex quinquefasciatus. J Visualized Experiments (2020):159. doi: 10.3791/61375
163. Purusothaman D-K, Shackleford L, Anderson MAE, Harvey-Samuel T, Alphey L. CRISPR/Cas-9 mediated knock-in by homology dependent repair in the West Nile virus vector Culex quinquefasciatus say. Sci Rep (2021) 11:14964. doi: 10.1038/s41598-021-94065-z
164. Kandul NP, Liu J, Hsu AD, Hay BA, Akbari OS. A drug-inducible sex-separation technique for insects. Nat Commun (2020) 11:2106. doi: 10.1038/s41467-020-16020-2
165. Condon KC, Condon GC, Dafa’alla TH, Fu G, Phillips CE, Jin L, et al. Genetic sexing through the use of y-linked transgenes. Insect Biochem Mol Biol (2007) 37:1168–76. doi: 10.1016/j.ibmb.2007.07.006
166. Novak BJ, Fraser D, Maloney TH. Transforming ocean conservation: Applying the genetic rescue toolkit. Genes (2020) 11:209. doi: 10.3390/genes11020209
167. Pham TB, Phong CH, Bennett JB, Hwang K, Jasinskiene N, Parker K, et al. Experimental population modification of the malaria vector mosquito, anopheles stephensi. PloS Genet (2019) 15:e1008440. doi: 10.1371/journal.pgen.1008440
168. Deredec A, Godfray HCJ, Burt A. Requirements for effective malaria control with homing endonuclease genes. Proc Natl Acad Sci U S A (2011) 108:E874–80. doi: 10.1073/pnas.1110717108
169. Esvelt KM, Smidler AL, Catteruccia F, Church GM. Concerning RNA-guided gene drives for the alteration of wild populations. eLife (2014) 3:e03401. doi: 10.7554/elife.03401
170. Anderson MAE, Purcell J, Verkuijl SAN, Norman VC, Leftwich PT, Harvey-Samuel T, et al. Expanding the CRISPR toolbox in culicine mosquitoes: In vitro validation of pol III promoters. ACS Synth Biol (2020) 9:678–81. doi: 10.1021/acssynbio.9b00436
171. Gao Y, Zhao Y. Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. J Integr Plant Biol (2014) 56:343–9. doi: 10.1111/jipb.12152
172. Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol (2014) 32:569–76. doi: 10.1038/nbt.2908
173. Xie K, Minkenberg B, Yang Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci U S A (2015) 112:3570–5. doi: 10.1073/pnas.1420294112
174. Hoermann A, Tapanelli S, Capriotti P, Del Corsano G, Masters EKG, Habtewold T, et al. Author response: Converting endogenous genes of the malaria mosquito into simple non-autonomous gene drives for population replacement. (2021). doi: 10.7554/elife.58791.sa2
175. Iyaniwura TT. Non-target and environmental hazards of pesticides. Rev Environ Health (1991) 9:161–76. doi: 10.1515/REVEH.1991.9.3.161
176. Geiger F, Bengtsson J, Berendse F, Weisser WW, Emmerson M, Morales MB, et al. Persistent negative effects of pesticides on biodiversity and biological control potential on European farmland. Basic Appl Ecol (2010) 11:97–105. doi: 10.1016/j.baae.2009.12.001
177. Beketov MA, Kefford BJ, Schäfer RB, Liess M. Pesticides reduce regional biodiversity of stream invertebrates. Proc Natl Acad Sci U S A (2013) 110:11039–43. doi: 10.1073/pnas.1305618110
178. Zaller JG, Brühl CA. Editorial: Non-target effects of pesticides on organisms inhabiting agroecosystems. Front Environ Sci Eng China (2019) 7:75. doi: 10.3389/fenvs.2019.00075
179. Medina RF. Gene drives and the management of agricultural pests. J Responsible Innovation (2018) 5:S255–62. doi: 10.1080/23299460.2017.1407913
180. Brown DM, Alphey LS, McKemey A, Beech C, James AA. Criteria for identifying and evaluating candidate sites for open-field trials of genetically engineered mosquitoes. Vector Borne Zoonotic Dis (2014) 14:291–9. doi: 10.1089/vbz.2013.1364
181. Rašić G, Lobo NF, Jeffrey Gutiérrez EH, Sánchez C HM, Marshall JM. Monitoring needs for gene drive mosquito projects: Lessons from vector control field trials and invasive species. Front Genet (2021) 12:780327.
182. Chae D, Lee J, Lee N, Park K, Moon SJ, Kim HH. Chemical controllable gene drive in drosophila. ACS Synth Biol (2020) 9:2362–77. doi: 10.1021/acssynbio.0c00117
183. López Del Amo V, Leger BS, Cox KJ, Gill S, Bishop AL, Scanlon GD, et al. Small-molecule control of super-mendelian inheritance in gene drives. Cell Rep (2020) 31:107841. doi: 10.1016/j.celrep.2020.107841
184. Zhong G, Wang H, He W, Li Y, Mou H, Tickner ZJ, et al. A reversible RNA on-switch that controls gene expression of AAV-delivered therapeutics in vivo. Nat Biotechnol (2020) 38:169–75. doi: 10.1038/s41587-019-0357-y
185. Oberhofer G, Ivy T, Hay BA. Gene drive that results in addiction to a temperature-sensitive version of an essential gene triggers population collapse in drosophila. Proc Natl Acad Sci U S A (2021) 118:e2107413118. doi: 10.1073/pnas.2107413118
186. Kaduskar B, Kushwah RBS, Auradkar A, Guichard A, Li M, Bennett JB, et al. Reversing insecticide resistance with allelic-drive in drosophila melanogaster. Nat Commun (2022) 13:291. doi: 10.1038/s41467-021-27654-1
187. Hartley S, Taitingfong R, Fidelman P. The principles driving gene drives for conservation. Environ Sci Policy (2022) 135:36–45. doi: 10.1016/j.envsci.2022.04.021
188. Akbari OS, Bellen HJ, Bier E, Bullock SL, Burt A, Church GM, et al. BIOSAFETY. safeguarding gene drive experiments in the laboratory. Science (2015) 349:927–9. doi: 10.1126/science.aac7932
189. Mitchell HJ, Bartsch D. Regulation of GM organisms for invasive species control. Front Bioeng Biotechnol (2019) 7:454. doi: 10.3389/fbioe.2019.00454
190. Long KC, Alphey L, Annas GJ, Bloss CS, Campbell KJ, Champer J, et al. Core commitments for field trials of gene drive organisms. Science (2020) 370:1417–9. doi: 10.1126/science.abd1908
191. American Committee of Medical Entomology, American Society of Tropical Medicine and Hygiene. Containment practices for arthropods modified with engineered transgenes capable of gene drive addendum 1 to the arthropod containment guidelines, version 3.2. Vector Borne Zoonotic Dis (2022) 22:3–17.
192. Millett P, Alexanian T, Palmer MJ, Evans SW, Kuiken T, Oye K. iGEM and gene drives: A case study for governance. Health Secur (2022) 20:26–34. doi: 10.1089/hs.2021.0157
193. Warmbrod KL, Kobokovich AL, West R, Gronvall GK, Montague M. The need for a tiered registry for US gene drive governance. Health Secur (2022) 20:43–9. doi: 10.1089/hs.2021.0139
194. Health Organization W. Guidance framework for testing genetically modified mosquitoes . Available at: https://apps.who.int/iris/bitstream/handle/10665/341370/9789240025233-eng.pdf?sequence=1 (Accessed June 27, 2022).
195. Union A, Others. Gene drives for malaria control and elimination in Africa. (2018). (Gauteng, South Africa).
196. Schairer CE, Najera J, James AA, Akbari OS, Bloss CS. Oxitec and MosquitoMate in the united states: Lessons for the future of gene drive mosquito control. Pathog Glob Health (2021) 115:365–76. doi: 10.1080/20477724.2021.1919378
197. Evans BR, Kotsakiozi P, Costa-da-Silva AL, Ioshino RS, Garziera L, Pedrosa MC, et al. Transgenic aedes aegypti mosquitoes transfer genes into a natural population. Sci Rep (2019) 9:13047. doi: 10.1038/s41598-019-49660-6
198. Servick K. Brazil Will release billions of lab-grown mosquitoes to combat infectious disease. will it work? (2021). 2016.
Keywords: conservation, vector-borne disease, Culex quinquefasciatus, gene drive, precision guided sterile insect technique (pgSIT), CRISPR-Cas9, invasive species, genetic control
Citation: Raban R, Gendron WAC and Akbari OS (2022) A perspective on the expansion of the genetic technologies to support the control of neglected vector-borne diseases and conservation. Front. Trop. Dis. 3:999273. doi: 10.3389/fitd.2022.999273
Received: 20 July 2022; Accepted: 17 August 2022;
Published: 14 October 2022.
Edited by:
Janet Midega, Wellcome Trust (WT), United KingdomReviewed by:
M. Renee Bellinger, United States Geological Survey (USGS), United StatesKeun Chae, Texas A&M University, United States
Copyright © 2022 Raban, Gendron and Akbari. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Omar S. Akbari, b2FrYmFyaUB1Y3NkLmVkdQ==
 Robyn Raban
Robyn Raban William A. C. Gendron
William A. C. Gendron Omar S. Akbari
Omar S. Akbari