- 1Núcleo de Medicina Tropical, Universidade de Brasília, Brasília, Brazil
- 2Laboratório de Genética Humana, Instituto de Ciências Biológicas, Universidade de Brasília, Brasília, Brazil
- 3Programa de Pós-graduação em Biologia Animal, Instituto de Ciências Biológicas, Universidade de Brasília, Brasília, Brazil
- 4Campus Paranaíba, Universidade Federal de Mato Grosso do Sul, Paranaíba, Brazil
- 5Laboratório de Diagnóstico Molecular do Hospital Universitário de Brasília, Brasília, Brazil
- 6Instituto Nacional de Ciência e Tecnologia para Avaliação de Tecnologia em Saúde, Porto Alegre, Brazil
- 7Programa de Pós-graduação em Geografia, Departamento de Geografia, Universidade de Brasília, Brasília, Brazil
- 8Faculdade de Ceilândia, Universidade de Brasília, Brasília, Brazil
Background: Development and validation of point-of-care (POC) diagnostic tests with high accuracy is critical for underrepresented populations, allowing for wider access to diagnosis. Here, we evaluate the performance of the Panbio™ antigen-rapid test device (Ag-RTD) for SARS-CoV-2, our index test, having RT-qPCR as the reference standard.
Methods: This phase III validation study was conducted concomitantly with a primary health care center routine tending to a low-income Brazilian population. Eligibility criteria were residing at Cidade Estrutural and presenting flu-like/respiratory symptoms for 3-10 days.
Results: Among the 505 participants, 45.15% (228/505) tested positive for RT-qPCR and 54.85% (277/505) for the Ag-RTD. Overall sensitivity was 76.32% (CI95% 70.39-81.37) and specificity was 98.92% (96.02-99.82).
Conclusions: Our results show that the Panbio™ Ag-RTD does not meet the minimum performance requirements established by the World Health Organization (≥ 80% sensitivity and ≥ 97% specificity compared to a reference test in suspected COVID-19 cases). Thus, we do not recommend the implementation of Panbio™Ag-RTD as a single diagnostic tool in underrepresented and disadvantaged populations. Finally, we discuss a possible setting for the use of Panbio™Ag-RTD under combined sensitivity.
Introduction
The coronavirus disease-2019 (COVID-19) is caused by the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Host biological and socioeconomic factors, in addition to health conditions and other risk factors, are positively associated with the spread and unfavorable outcomes of COVID-19 (1–4). The current pandemic disproportionately affects historically underrepresented, disadvantaged groups, and exacerbates preexisting and current social inequalities (5).
In Brazil, populations from municipalities with higher socioeconomic vulnerability experienced higher death rates from COVID-19 and greater disease spread during the initial phase of the epidemic; moreover, socioeconomic inequalities appear to have had stronger effects on the epidemic rather than biological and other risk factors for COVID-19 (6). Publicly available state-level reports also revealed a strong positive correlation between mortality and socioeconomic vulnerability in a Brazilian pediatric population (7). Controlling the spread and reducing negative outcomes of COVID-19 require early and rapid detection for isolation of symptomatic patients and close contact tracing; nevertheless, health care and proper diagnostic tools are often unavailable for underrepresented populations.
Real-time reverse-transcription polymerase chain reaction (RT-qPCR) from nasopharyngeal (NP) samples is the reference standard test for the detection of SARS-CoV-2 (8). Specialized instruments, trained human resources, and equipped and safe laboratories are required to conduct the high-cost RT-qPCR essays, limiting testing availability and posing an obstacle to mass testing. Point-of-care (POC) SARS-CoV-2 antigen rapid test devices (Ag-RTDs), such as Panbio™ Ag-RTD, emerge as rapid, easy, and less expensive and laborious alternatives to diagnose ongoing SARS-CoV-2 infections in several settings.
The World Health Organization’s interim guidance from October 2021 advises the use of SARS-CoV-2 Ag-RTDs that meet a minimum performance of ≥80% sensitivity and ≥97% specificity compared to RT-qPCR (8). Sensitivity of the Panbio™ Ag-RTD evaluated at POC in symptomatic patients ranges from ~50 to 100%, whereas specificity ranges from ~80 to 100% (9–30). However, all but three studies with small samples and/or insufficient description of the sampled population (10, 17, 27) evaluated Panbio™ Ag-RTD in populations from high-income countries, and none of them included disadvantaged populations.
The case of the COVID-19 pandemic has raised concerns that the lack of representation of disadvantaged groups might bias investigation (31), as well as the perception of health and access to diagnosis and treatment. Thus, it is crucial to validate diagnostic tools for early detection of COVID-19 in underrepresented, disadvantaged groups and to account for patient characteristics such as race/ethnicity/ancestry, sex, age, and preexisting medical conditions. Here, we aimed to assess the accuracy of the Panbio™ Ag-RTD performed at POC at a primary health care center tending to an urban area inhabited by underrepresented groups in Brazil. Populations such as this, presenting flu-like/respiratory syndrome symptoms for 3 to 10 days, would benefit from opportune, accurate, fast, and cheaper diagnostic tools, as long as those tools meet WHO’s interim guidance from October 2021 for the adoption of rapid antigen detection tests.
Materials and methods
We conducted a prospective diagnostic accuracy study, type phase III (32), for the validation of a diagnostic test for the detection of SARS-CoV-2 in patients with flu-like/respiratory syndrome symptoms who sought medical assistance at a primary health center. This work was conducted in accordance with the Ethical Principles for Medical Research in Human Subjects (Declaration of Helsinki) and the Brazilian regulations on ethics in human research. Data and sample collection were approved by the Research Ethics Committee of the Faculty of Medicine at University of Brasília (CEP-FM/UnB, CAAE 39892420.7.1001.5558; CAAE 40557020.6.3001.5553) and Fundação de Ensino e Pesquisa em Ciências da Saúde (FEPECS/SES/DF, CAAE 40557020.6.3001.5553). All participants were volunteers and informed about research goals and confidentiality of data, and signed instruments of consent previously to data and sample collection. This study was conducted according to the updated Standards for Reporting Diagnostic Accuracy Studies (STARD) (33). A checklist of STARD essential items is presented in S1 Checklist.
Study population
Cidade Estrutural (RA XXV SCIA/Estrutural, DF, Brazil; Figure 1) is inhabited by a socioeconomically vulnerable population occupying a stretch of land bordered by highly degraded and preserved environments (Figure 1). It was founded as an irregular occupation area, which until 2017, housed an enormous untreated refuse disposal site known as “lixão da Estrutural’’ (Estrutural dump), where around 4,000 people worked scavenging waste (34). Today, the refuse disposal site has been turned into an official rubble receiving unit (URE, Unidade de Recebimento de Entulho), though different dumping sites often emerge in Estrutural. Furthermore, Estrutural borders the Brasília National Park (a 423.60 km2 conservation area), the Brasília National Forest, the Cabeceira do Valo river and its area of relevant ecological interest a.k.a ARIE (i.e., small extension area of sustainable use, protected by law, which houses rare specimens of fauna and flora). Even though Cidade Estrutural was founded during the 50’s, it is still expanding into surrounding areas, mostly nature preserves. Neighborhoods vary widely in urban infrastructure. On one extreme, Santa Luzia, the most socioeconomically vulnerable neighborhood, is situated on the border with the Brasilia National Park, making it susceptible to spillovers and outbreaks, as well as to political tension concerning its expansion. Streets are not paved, and there is no sanitation infrastructure. On the other hand, Setor Leste is a long-consolidated neighborhood and has a better infrastructure (Figure 1). Cidade Estrutural is inhabited by over 35,000 people (35) all tended to by a single public primary health care center, where our samples were collected. According to official data (35), sociodemographic data depicts a population with low income (R$ 573,00 or US$ 112,00 per capita), low education (6.4% are illiterate, 9.6% have completed only elementary school), and low Human Development Index (HDI, 0.616) in a Federative Unit (the Brazilian Federal District - DF) with a Gini coefficient of 0.553 (36). The resident population identifies mostly as black (15%) and mixed race (62%), and most of the remaining self-declared white. The age-sex pyramid is characteristic of developing populations (35).
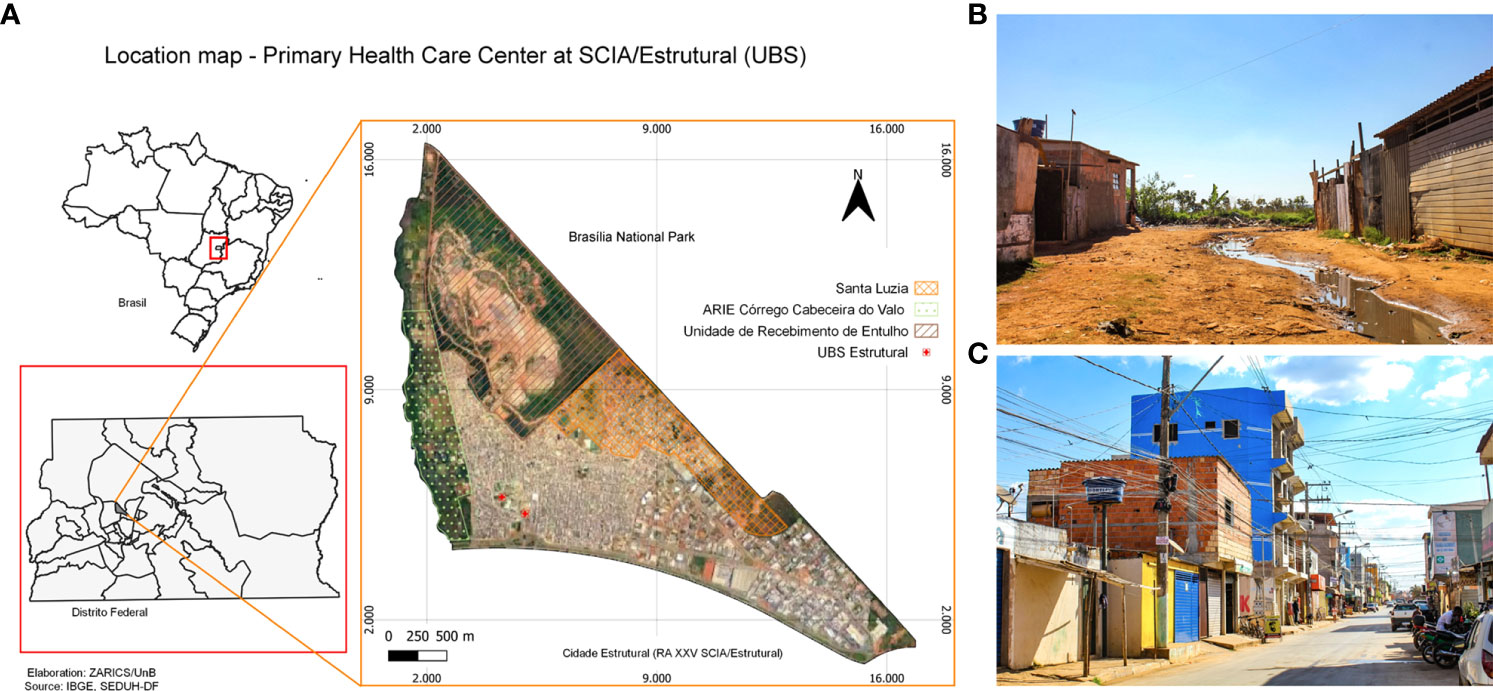
Figure 1 Characterization of Estrutural. (A) Location map of the primary health care center at Estrutural. The primary health care center (UBS) is pointed in red; Santa Luzia, Cabeceira do Valo river and its ecological conservation unit (Unidade de Conservação Área de Relevante Interesse Ecológico do Córrego Cabeceira do Valo (ARIE), Brasília National Park and the rubble receiving unit (URE - Unidade de Recebimento de Entulho) are also highlighted. (B) Santa Luzia, the most socioeconomically vulnerable neighborhood in Estrutural. The photograph depicts the border between Estrutural and the Brasília National Park. (C) A street in Setor Leste, a neighborhood with better urban infrastructure.
Sample size
Sample size was calculated as N= Z² [(p(1-p)]/(D²)], in which p was the frequency of the expected event and D was the semi-amplitude of a bi-caudal precision. According to the Abbott Panbio™ Ag-RTD insert, test sensitivity was 0.91 and specificity was 0.98. To calculate the sample size to assess sensitivity, we used 0.91 as the frequency of the expected event and a precision of 4%, resulting in a sample size of approximately 196 participants. To calculate the sample size to assess specificity, we used 0.98 as the frequency of the expected event and a precision of 4%, which resulted in a sample size of 188 participants. Therefore, our final sample size was at least 384 volunteers.
Clinical assessment and eligibility criteria
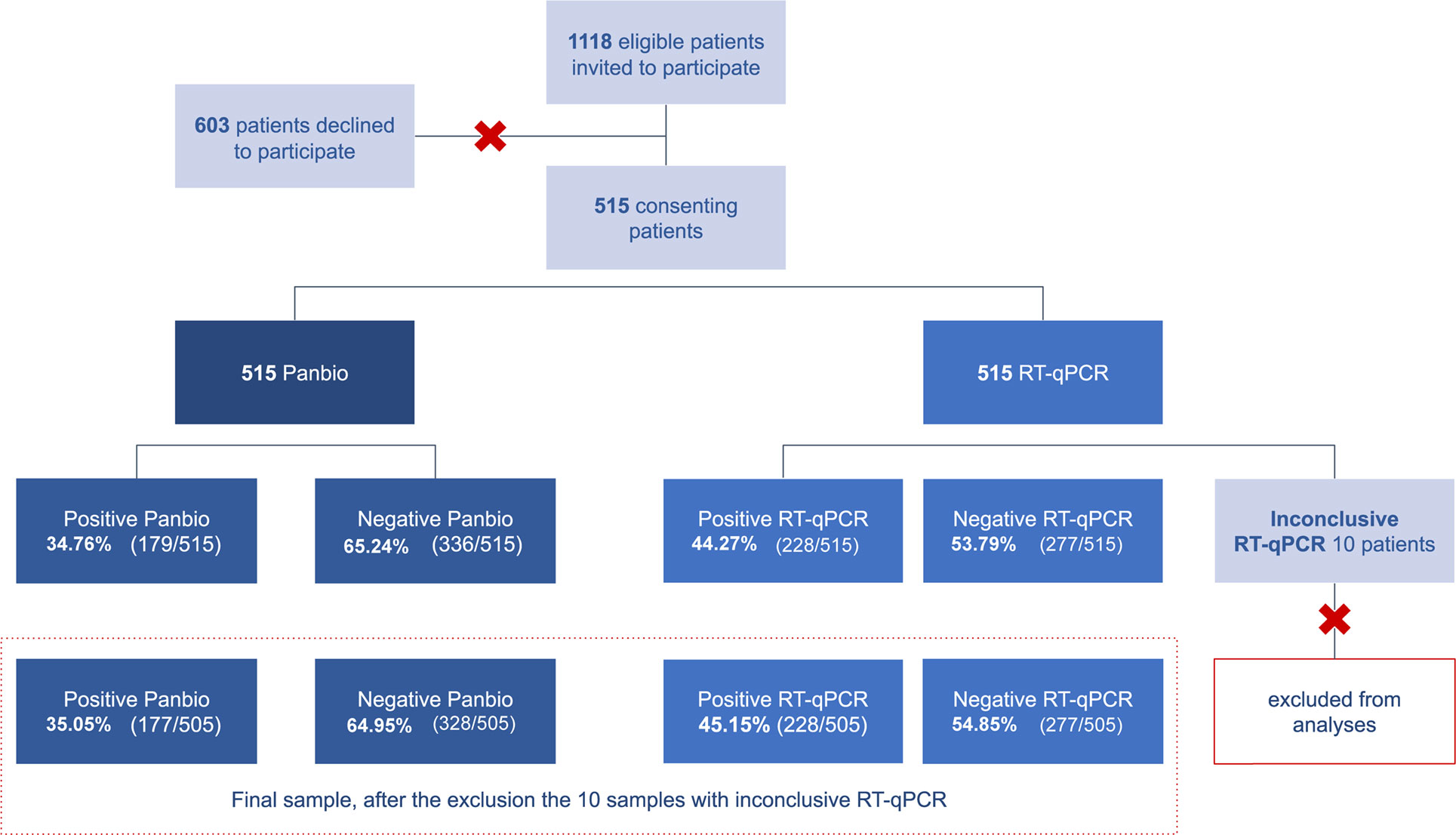
Eligibility criteria were: 1. residing in Cidade Estrutural; 2. presenting flu-like/respiratory syndrome symptoms for 3 to 10 days as defined by the Brazilian Ministry of Health (37); and 3. seeking assistance at the Cidade Estrutural primary health care center. All patients with flu-like/respiratory syndrome symptoms referred for medical care were invited to participate in the study (Figure S1). All participants were informed about research goals and confidentiality of data and signed instruments of consent previously to data and sample collection.
Data collection
In March 2021, our research team joined efforts with the Cidade Estrutural primary health care center staff and became responsible for the surveillance system for COVID-19. Primary data were collected by semi-structured interviews utilizing standardized, pre-tested questionnaires using the data capture software Research Electronic Data Capture (REDCap) (38) hosted at the University of Brasília. Beginning on March 16th, 515 patients agreed to participate in our validation study.
Sample collection for RT-qPCR reference standard test and case definition
Our trained staff collected NP swab specimens for diagnosis by RT-qPCR reference standard test from each participant at the Cidade Estrutural primary health care center. A single sterile swab was introduced in both nostrils until nasopharyngeal resistance was met, rotated 5 times, and stored in a viral transport medium (VTM - LaborClin, Brazil). Samples were transported at 4-6°C to the Laboratório de Diagnóstico Molecular (Laboratory of Molecular Diagnosis) at Hospital Universitário de Brasília (HUB), University of Brasília (UnB), where RT-qPCR assays were performed. All patients with flu-like symptoms plus SARS-CoV-2 RNA detected by RT-qPCR were considered COVID-19 cases, whereas patients with flu-like symptoms and undetected SARS-CoV-2 RNA were considered non-cases.
Execution of RT-qPCR reference standard test
RT-qPCR assays were performed by blinded, trained technical staff who had no prior knowledge of the clinical presentation of patients. SARS-CoV-2 RNA was isolated using the EXTRACTA 32 kit (MVXA-P016 FAST) in a Loccus automated extractor following the manufacturer’s instructions. SARS-CoV-2 was detected by the RT-qPCR Allplex™ 2019-nCoV Assay (Seegene Inc.) for the amplification of genes E, RdRP, N, as well as an internal control gene, according to the manufacturer’s protocol. RT-qPCR results were considered positive (SARS-CoV-2 RNA detected) when the internal control and at least two genes were amplified, negative (SARS-CoV-2 RNA not detected) when the internal control and none or only one gene was amplified, and inconclusive when the internal control did not amplify. Inconclusive RT-qPCR reactions were repeated once.
Over the months since the first sample collection, results of the ongoing analyses (RT-qPCR for diagnosis) were informed to the participants and to the public health services that attend to the community for follow-up and treatment, and for epidemiologic bulletins.
Sample collection and execution of the Panbio™ Antigen Rapid Test Device
Abbott’s Panbio™ Ag-RTD contains a swab to collect NP specimens, a buffer solution where the swab must be inserted immediately after the NP specimens collection, and an apparatus to run the test. This apparatus contains a membrane strip pre-coated with antibodies to detect the nucleo-capsid (N) protein of SARS-CoV-2 in human NP swab specimens. NP specimens were collected from both nostrils immediately after swab collection for RT-qPCR at POC (the primary health care center at Cidade Estrutural). Specimens were immediately inserted into the test buffer and then dripped on the spot indicated in the membrane strip as per the manufacturer’s instructions. A visible control line is required to validate the test result. A second line indicates a positive result, and its absence, a negative result. The absence of a control line defines an invalid test, which should be canceled, and the participant excluded from the study. All tests were performed according to the manufacturer’s protocol and each test was evaluated by two independent observers at the same time.
Statistical analysis
Test results and patient data were tabulated and analyzed using SPSS version 22 (39). Patients with missing data were excluded from the study. Data were plotted using RStudio version 1.1.463 (40) and the ggplot2 package (41). The overall sensitivity, specificity, and likelihood ratios of the Panbio™ Ag-RTD and their respective 95% confidence intervals (CI95%) were calculated in Microsoft Excel; we also calculated these values for the test regarding the self-reported symptoms and days from symptom onset. The kappa index was used to test for agreement between the results reported by two independent observers. For all statistical analyses, the first observer results were used.
Results
From March 16th to July 1st of 2021, 1118 patients with flu-like/respiratory syndrome symptoms sought medical care at the primary healthcare center at Cidade Estrutural and were invited to join our research. A total of 515 patients consented to participate and signed consent instruments, as described above. RT-qPCR results for 10 participants remained inconclusive even after repeating RNA extraction and RT-qPCR essay, and were therefore excluded from the accuracy analysis of the index test. Thus, the sample size for the accuracy analysis of the index test was 505 participants (Figure 2).
Sample description
Out of the 505 participants, 61.39% (310/505) were female and 79.60% (402/505) self-declared mixed-race or black. Age ranged from 5 to 73 years old (median age 33 and quartiles 24-42.25). A total of 43.56% (220/505) participants declared to have at least one comorbidity; hypertension was the most common self-reported comorbidity (15.25%, 77/505). Descriptive analysis of the sample characteristics according to RT-qPCR and Panbio™ Ag-RTD results are summarized in Table 1, and additional information on other self-reported coexisting conditions is shown in Table S1.
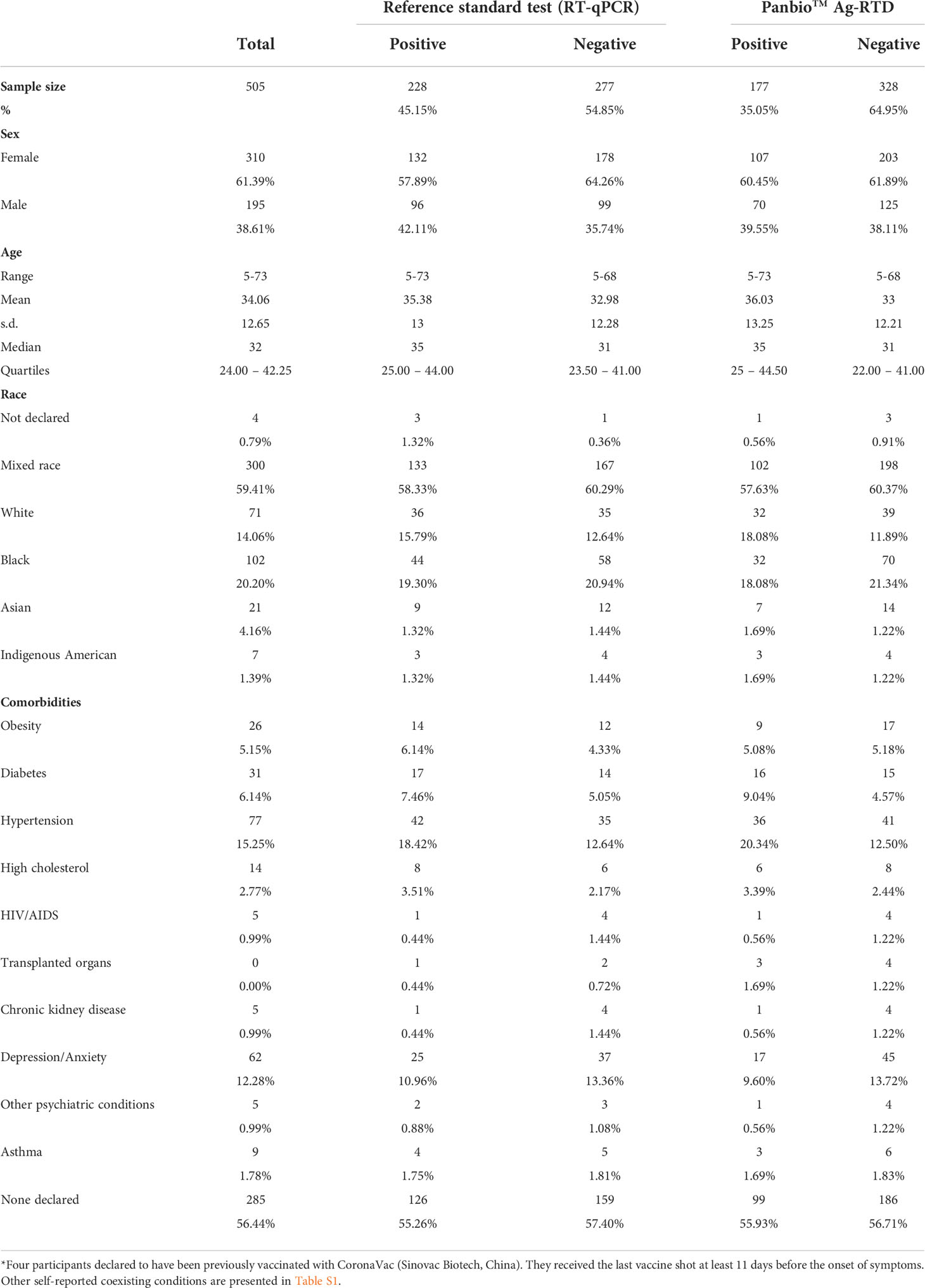
Table 1 Baseline characteristics of the 505 participants enrolled in Panbio™ antigen-rapid test devices (Ag-RTD) accuracy assessment.
Headache was the most reported symptom (75.64%, 382/505), followed by fever (67.33%, 340/505) and cough (66.93%, 338/505). Headache, cough, and fever were also the most commonly reported symptoms among the 228 participants that tested positive for RT-qPCR (72.37%, 165/228; 69.74%, 159/228; 68.42%, 156/228), as well as among participants with positive results for the Panbio™ Ag-RTD (headache: 71.19%, 126/177; cough: 70.62%, 125/177; fever: 67.80%, 120/177). Out of the 136 participants that reported ageusia, 63.23% (86/136) tested positive with RT-qPCR, and 53.68% (73/136) tested positive with the Ag-RTD. Similarly, anosmia was reported by 131 patients, 68.70% of which tested (90/131) positive with RT-qPCR and 57.25% (75/131) tested positive with Panbio™ Ag-RTD. Self-reported symptoms are summarized in Table 2, and additional information on other self-reported symptoms/signs/discomforts is presented in Table S1.
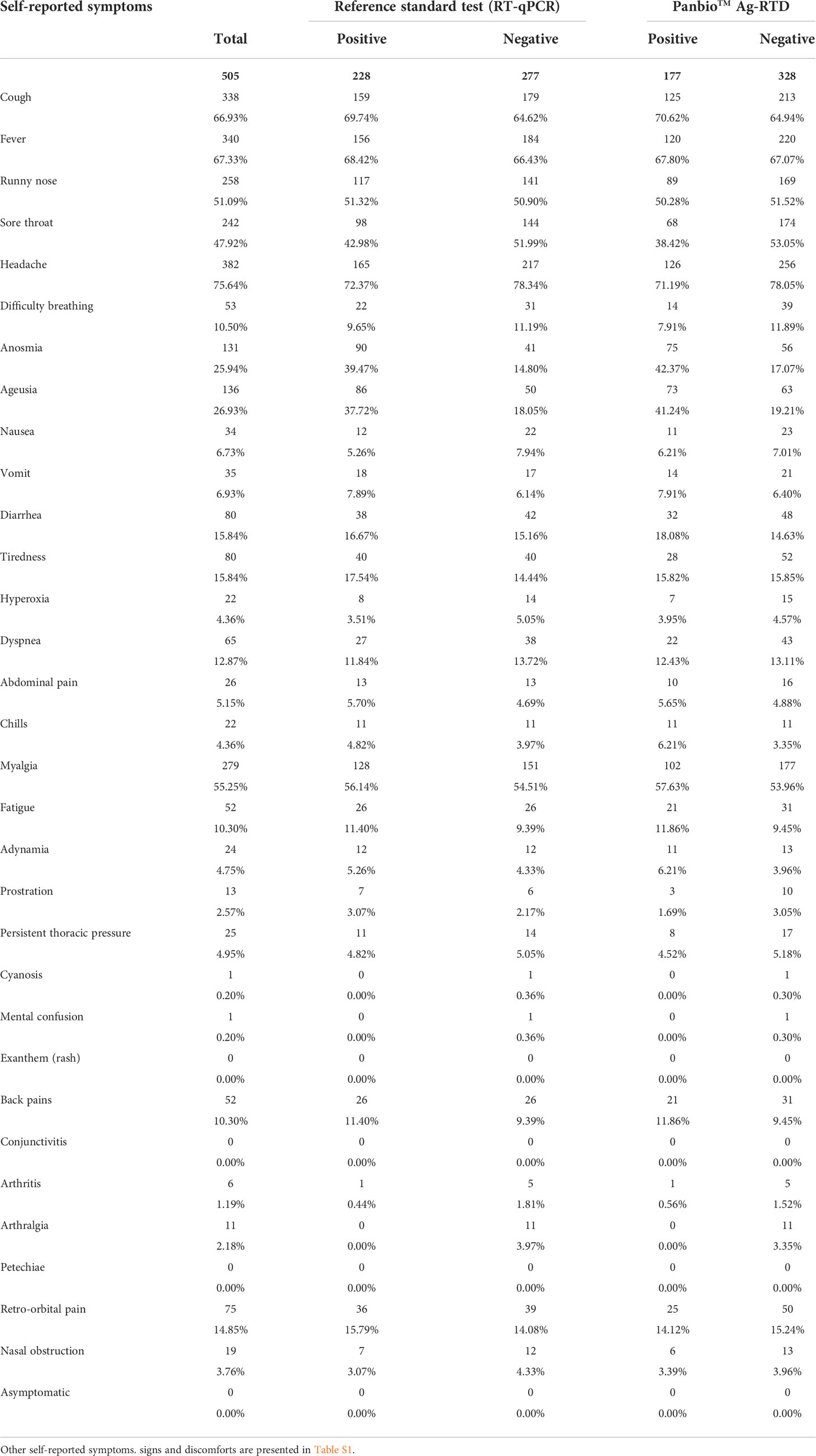
Table 2 Symptoms reported by the 505 participants enrolled in the Panbio™ antigen-rapid test devices (Ag-RTD) accuracy assessment.
Overall Panbio™ Ag-RTD accuracy
Considering our sample size of 505 participants, the Panbio™ Ag-RTD obtained 54.85% (277/505) positive results, whereas the reference standard test (RT-qPCR) obtained 45.15% (228/505). Accuracy assessment (Table 3) showed sensitivity of 76.32% (CI95% 70.39-81.37) and specificity of 98.92% (CI95% 96.02-99.82). Positive likelihood ratio was 70.46 (CI95% 22.81-217.64) and negative likelihood ratio was 0.24 (CI95% 0.19-0.30). There were no inconclusive results in the Panbio™ Ag-RTD. The raw concordance between results was 97.82% (494/505), and the kappa index for the Panbio™ Ag-RTD evaluating the agreement between results of two independent observers was 0.96.
Panbio™ Ag-RTD accuracy regarding symptoms and symptoms onset
The days from symptom onset to medical assistance at the primary health care center and, hence, the diagnosis, were similar between volunteers with positive RT-qPCR and Panbio™ Ag-RTD. Average days of self-reported symptoms for RT-qPCR positive participants was 5.47 days (SD 1.94), whereas for Panbio™ Ag-RTD it was 5.45 days (SD 1.93) (Figure S2). Sensitivity of Panbio™ Ag-RTD slightly increased in patients with ≤5 days from symptom onset to 77.17% (CI95% 69.13-83.60), whereas specificity virtually did not change 99.41% (CI95% 98.54-100) (see Table 3). In patients with >5 from symptom onset, sensitivity and specificity (75.25%, 66.01-82.64 and 98.13% 96.29-99.97, respectively) slightly decreased. We also found that the Panbio™ Ag-RTD sensitivity was improved to 81.55% (CI95% 72.98-87.86) in patients who declared having anosmia and/or ageusia; in these cases, specificity slightly decreased to 98.39% (CI95% 96.47-100).
Discussion
Our results show that the Panbio™ Ag-RTD has overall sensitivity and specificity of 76.32% (CI95% 70.39-81.37) and 98.92% (CI95% 98.02-99.82), respectively, compared to the reference standard RT-qPCR test for patients with mild to moderate flu-like/respiratory symptoms. The kappa index for agreement between results evaluated by two independent observers was 0.96, which is labeled as a strong agreement.
Other Panbio™ POC accuracy studies with symptomatic patients reported variable sensitivity and specificity values ranging from ~50-100% and ~80-100%, respectively (9–30). Although these are all phase III studies (32), such wide ranges are noteworthy. Possible explanations relate to the origin and collection of the samples used for RT-qPCR reference standard tests and index Ag-RTD. Some authors performed RT-qPCR as reference test from NP and/or oropharyngeal (OP) swab samples (11–13, 19, 23, 24, 30), whereas others used saliva or combined saliva and NP samples for the reference test or for the Panbio™ Ag-RTD (9, 16, 17). Akingba et al., 2021 (10) performed RT-qPCR using a single NP swab sample for both RT-qPCR and Panbio™ testing instead of collecting two NP swab samples from each nostril – one for RT-qPCR and another for Ag-RTD, as other authors chose to do (18, 22, 26, 28). Nsoga et al., 2021 (20) used OP swab samples to perform the Panbio™ Ag-RTD tests and NP samples for the RT-qPCR. Furthermore, other authors did not specify whether NP swab samples were collected from one or two nostrils (14, 15, 21, 25).
In the present study, we first collected a NP swab sample from both nostrils for diagnosis by RT-qPCR, and then collected a second one from each nostril for the Panbio™ Ag-RTD. By doing so, we expect to have established a more robust methodological approach for the accuracy study for two main reasons. First, the chosen workflow ensures the collection of a biological sample even in the eventuality of nasal obstruction due to anatomical variation (i.e., deviated septum), which might prevent a swab from reaching the nasopharynx. Second, the chosen method ensures that the two samples are collected exactly in the same way and excludes variation in sample collection as a factor for disagreement between RT-qPCR and Ag-RTD.
To our knowledge, this is the first Panbio™ Ag-RTD accuracy study carried out at POC in socioeconomically vulnerable, disadvantaged groups. All but three of the studies mentioned above were performed in populations from high-income countries (10, 17, 27). The study by Akingba and colleagues (2021) (10) does not fully describe the sampled population and given the high inequality rates in South Africa (Gini index 0.63) (42) it is not possible to ascertain whether high or low-income groups were included. The same problem was found in the other two studies in populations from São Paulo, Brazil (17, 27). Four out of six studies reporting Panbio™ Ag-RTD sensitivity >80% (12, 18, 20, 25) were carried out in European populations from Spain, Germany, or Switzerland. Although two studies found an overall sensitivity >80% in a population from Brazil (17, 27), they evaluated the Panbio™ accuracy in a very small sample (n=127 and 112, respectively). Moreover, the Matsuda et al., 2021 study (17) comprised patients from public and private hospitals – in addition to using saliva combined with NP swab samples to perform RT-qPCRs – whereas Faíco-Filho and colleagues (2021) (27) did not describe the population nor the hospital where samples were collected.
Unlike previously reported by Merino et al., 2021 and Bulilete et al., 2021 (18, 25), we did not see a great increase in sensitivity for Panbio™ Ag-RTD in patients with ≤ 5 days from symptom onset. Even though the relationship between viral load in COVID-19 (estimated by Cycle threshold [Ct] values) and days from symptom onset are not yet clear, a higher viral load is expected early in the course of the disease [e.g (18, 25)] – and, hence, a higher sensitivity of the test. However, it is important to stress that the precision of information like days from symptom onset in a population might affect the results of accuracy evaluation. Moreover, disadvantaged populations like those in our study, which in Brazil are mostly black/mixed race, are overall underrepresented in investigations on health, as well as on accuracy assessment of diagnostic tools like Panbio™ Ag-RTD. Such studies are worldwide centered on European and European-derived populations, which might introduce biases derived from ancestry and socioeconomic and sociodemographic characteristics. In spite of that, it should be noted that the presence of anosmia and ageusia, commonly reported in European COVID-19 patients (43), seems to improve the sensitivity of the Panbio™ Ag-RTD in our study population.
Development and validation of POC diagnostic tests with high accuracy would be pivotal for populations like that of Estrutural, Brazil, allowing for quick, efficient, and cheaper diagnosis, as well as mass testing. Tests such as the Panbio™ Ag-RTD could theoretically be used as a mass screening tool and aid a physician to identify and treat infected patients, given its high positive likelihood ratio (LR); nevertheless, its negative LR speaks against it. In scenarios in which patients have negative results in Panbio™ Ag-RTD, a second diagnostic tool (like the RT-LAMP test with 91% of sensitivity described by Jiang et al. (2020) (44), would increase sensitivity by 20%. Regardless, a combination of different diagnostic tools like this (i.e., combined sensitivity) is not feasible for most underrepresented and disadvantaged populations and should always be carefully evaluated before being implemented.
Caveats
It is important to note that our study included mild-to-moderate symptomatic patients; asymptomatic and severe cases were not assessed. However, Brazilian health care centers do not routinely test asymptomatic patients and collecting the two NP swabs from each nostril might not be possible in severe cases. Therefore, we tested the accuracy of the Panbio™ Ag-RTD in a real scenario of primary health care in a socioeconomically vulnerable area in Brazil.
During the collection of NP samples, we faced a methodological conundrum: could the amount of virus in the first swab be greater than the amount of virus in the second swab and, hence, affect the test sensitivity? We cannot answer this question without measuring the viral load in the swabs destined for RT-qPCR and Panbio™ Ag-RTD. Another possibility would be comparing our study and others that used a similar approach for sample collection. However, to our knowledge, the order of swab collection – and if it was collected from one or two nostrils – has not been addressed in any accuracy studies of the Panbio™ Ag-RTD, which limits our capacity to answer that question.
Our collection strategy might have diminished patient enrollment in the study because of the discomfort of NP swab collection. In spite of a large number of refusals, all patients who agreed to participate in our study i) reported symptoms of a very well-described flu-like syndrome, ii) had mild-to-moderate symptoms, and ii) all NP samples for both tests (reference and index) were collected at the same time – which, therefore, discards the possibility of introducing a selection bias based on prior knowledge of the patient’s condition of being infected or not infected by SARS-CoV-2. Therefore, we stand behind our choice for the reasons described above, as we followed a rigorous methodology for evaluating the test’s accuracy, ensuring biological material from all patients – mainly those who have nasal obstruction due to anatomical variation – was properly collected. In addition, samples collected for RT-qPCR were immediately stored in VTM and kept at 4-6°C until processed on the next day, guaranteeing sample integrity. Samples collected for Panbio™ Ag-RTD were immediately inserted into the test buffer and then dripped on the spot indicated in the membrane strip as per manufacturer’s instructions. Finally, our methodological approach for collection and processing ensures biological material from all patients was collected and processed exactly the same way.
Conclusions
Our results show that the Panbio™ Ag-RTD does not meet WHO’s interim guidance from October 2021 in our study population. Its sole use at POC could mislead patients with false negative results, who would in turn refrain from adopting isolation and other preventative recommendations. Hence, the apparent benefits from fast, cheap testing are outweighed by the possible harms precipitated by a false sense of security and health, and might hinder control of the spread of SARS-CoV-2 even further in populations where actual isolation and social distancing are nearly unattainable. Therefore, even though Panbio™ Ag-RTD might be useful in specific scenarios (as those recommended by WHO), we do not recommend its implementation as a single diagnostic tool in underrepresented and disadvantaged populations.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving human participants were reviewed and approved by the Research Ethics Committee of the Faculty of Medicine at University of Brasília (CEP-FM/UnB, CAAE 39892420.7.1001.5558; CAAE 40557020.6.3001.5553) and Fundação de Ensino e Pesquisa em Ciências da Saúde (FEPECS/SES/DF, CAAE 40557020.6.3001.5553). The procedures used in this study adhere to the tenets of the Ethical Principles for Medical Research in Human Subjects (Declaration of Helsinki) and the Brazilian regulations on ethics in human research. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author contributions
CG: Conceptualization and design of the study, Data acquisition and curation, Formal analysis and interpretation of data, Investigation, Methodology, Validation, Writing (Original draft, Review & Editing), Final approval. RB: Conceptualization and design of the study, Data acquisition and curation, Formal analysis and interpretation of data, Investigation, Methodology, Validation, Writing (Original draft, Review & Editing), Final approval. AT: Conceptualization and design of the study, Data acquisition and curation, Formal analysis and interpretation of data, Investigation, Methodology, Validation, Writing (Original draft, Review & Editing), Final approval. GR: Conceptualization and design of the study, Interpretation of the data, Validation, Resources, Funding acquisition, Writing (Review & Editing), Final approval. PP: Data acquisition, Writing (review & editing), Final approval. WR: Data acquisition, Resources, Funding acquisition, Writing (Review & Editing), Final approval. RH: Data acquisition, Resource, Funding acquisition, Writing (Review & Editing), Final approval. EN: Data acquisition, Resource, Funding acquisition, Writing (Review & Editing), Final approval. WA: Conceptualization and design of the study, Interpretation of data, Investigation, Methodology, Validation, Resource, Funding acquisition, Writing (Review & Editing), Final approval. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Ministério da Educação (MEC, http://portal.mec.gov.br /; grant number 23106.028855/2020-74) and Fundação de Amparo à Pesquisa do Distrito Federal (FAP-DF, http://www.fap.df.gov.br /; grant number 00193-00000495/2020-72). The funding sources were not involved in the study design, in the collection, analysis, and interpretation of the data, in the preparation of the manuscript, or in the decision to submit the manuscript to publication.
Acknowledgments
We thank the managers of the Estrutural primary health care center (Unidade Básica de Saúde da Estrutural, UBS) Rossana Michelli Ferreira de Pontes and Américo Yuiti Mori for supporting our field work, the technical staff of the Laboratory of Molecular Diagnosis (LDM) at Hospital Universitário de Brasília (HUB), and the Central Laboratory of Public Health (LACEN), Brasília, for providing technical support in executing RT-qPCR. We also thank the COVID-19 surveillance staff of the Estrutural UBS, DIRAPS Centro/Sul and the Distrito Federal Health Department (SES-DF), Brasília.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fitd.2022.929524/full#supplementary-material
Supplementary Figure 1 | Flowchart showing the route a patient seeking medical attention with flu-like/respiratory syndrome follows at the primary healthcare center where our study was conducted. The diagram shows the route from triage to enrollment in research (or decline) to diagnosis by RT-qPCR and Panbio™ Ag-RTD testing. *Flu-like/respiratory syndrome is characterized by the presence of at least two non-specific symptoms, such as coughing, sore throat, runny nose, anosmia, ageusia, diarrhea, abdominal pain, fever, chills, myalgia, fatigue, headache
Supplementary Figure 2 | Violin and boxplots showing results obtained from RT-qPCR and Panbio™ Ag-RTD, as well as agreement/disagreement of said tests over days from symptom onset. Dates were self-declared. Samples were collected from participants within a 3-10 days of symptoms interval
References
1. Clouston SAP, Natale G, Link BG. Socioeconomic inequalities in the spread of coronavirus-19 in the united states: A examination of the emergence of social inequalities. Soc Sci Med (2021) 268:113554. doi: 10.1016/j.socscimed.2020.113554
2. Gray DM, Anyane-Yeboa A, Balzora S, Issaka RB, May FP. COVID-19 and the other pandemic: Populations made vulnerable by systemic inequity. Nat Rev Gastroenterol Hepatol (2020) 17:520–2. doi: 10.1038/s41575-020-0330-8
3. Tavares FF, Betti G. The pandemic of poverty, vulnerability, and COVID-19: Evidence from a fuzzy multidimensional analysis of deprivations in Brazil. World Dev (2021) 139:105307. doi: 10.1016/j.worlddev.2020.105307
4. Williamson EJ, Walker AJ, Bhaskaran K, Bacon S, Bates C, Morton CE, et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature (2020) 584:430–6. doi: 10.1038/s41586-020-2521-4
5. Perry BL, Aronson B, Pescosolido BA. Pandemic precarity: COVID-19 is exposing and exacerbating inequalities in the American heartland. Proc Natl Acad Sci U S A (2021) 118:1–6. doi: 10.1073/pnas.2020685118
6. Rocha R, Atun R, Massuda A, Rache B, Spinola P, Nunes L, et al. Effect of socioeconomic inequalities and vulnerabilities on health-system preparedness and response to COVID-19 in Brazil: A comprehensive analysis. Lancet Glob Heal (2021) 9:e782–92. doi: 10.1016/S2214-109X(21)00081-4
7. Martins-filho PR, Quintans-júnior LJ, Araújo AADS, Sposato KB. Socio-economic inequalities and COVID-19 incidence and mortality in Brazilian children: A nationwide register-based study.Public Health (2020) 190:4–6. doi: 10.1016/j.puhe.2020.11.005
8. World Health Organization. Antigen-detection in the diagnosis of SARS-CoV-2 infection. interim guid (2021). Available at: https://www.who.int/publications/i/item/antigen-detection-in-the-diagnosis-of-sars-cov-2infection-using-rapid-immunoassays.
9. Agulló V, Fernández-González M, Ortiz de la Tabla V, Gonzalo-Jiménez N, García JA, Masiá M, et al. Evaluation of the rapid antigen test panbio COVID-19 in saliva and nasal swabs in a population-based point-of-care study. J Infect (2021) 82:186–230. doi: 10.1016/j.jinf.2020.12.007
10. Akingba OL, Sprong K, Marais G, Hardie DR. Field performance evaluation of the PanBio rapid SARS-CoV-2 antigen assay in an epidemic driven by the B.1.351 variant in the Eastern cape, south Africa. J Clin Virol Plus (2021) 1:100013. doi: 10.1016/j.jcvp.2021.100013
11. Kolwijck E, Brouwers-Boers M, Broertjes J, van Heeswijk K, Runderkamp N, Meijer A, et al. Validation and implementation of the panbio COVID-19 Ag rapid test for the diagnosis of SARS-CoV-2 infection in symptomatic hospital healthcare workers. Infect Prev Pract (2021) 3:100142. doi: 10.1016/j.infpip.2021.100142
12. Krüger LJ, Gaeddert M, Tobian F, Lainati F, Gottschalk C, Klein JAF, et al. The Abbott PanBio WHO emergency use listed, rapid, antigen-detecting point-of-care diagnostic test for SARS-CoV-2–evaluation of the accuracy and ease-of-use. PLoS One (2021) 16:1–11. doi: 10.1371/journal.pone.0247918
13. Landaas ET, Storm ML, Tollånes MC, Barlinn R, Kran A-MB, Bragstad K, et al. Diagnostic performance of a SARS-CoV-2 rapid antigen test in a large, Norwegian cohort. J Clin Virology (2021), 137:104789. doi: 10.1016/j.jcv.2021.104789
14. Lanser L, Bellmann-Weiler R, Öttl KW, Huber L, Griesmacher A, Theurl I, et al. Evaluating the clinical utility and sensitivity of SARS-CoV-2 antigen testing in relation to RT-PCR ct values. Infection (2021) 49:555–7. doi: 10.1007/s15010-020-01542-0
15. Linares M, Pérez-Tanoira R, Carrero A, Romanyk J, Pérez-García F, Gómez-Herruz P, et al. Panbio antigen rapid test is reliable to diagnose SARS-CoV-2 infection in the first 7 days after the onset of symptoms. J Clin Virol (2020) 133:3–6. doi: 10.1016/j.jcv.2020.104659
16. Masiá M, Fernández-González M, Sánchez M, Carvajal M, García JA, Gonzalo-Jiménez N, et al. Nasopharyngeal panbio COVID-19 antigen performed at point-of-Care has a high sensitivity in symptomatic and asymptomatic patients with higher risk for transmission and older age. Open Forum Infect Dis (2021) 8:1–9. doi: 10.1093/ofid/ofab059
17. Matsuda EM, de Campos IB, de Oliveira IP, Colpas DR, Carmo AM dos S, Brígido LF de M. Field evaluation of COVID-19 antigen tests versus RNA based detection: Potential lower sensitivity compensated by immediate results, technical simplicity, and low cost. J Med Virol (2021) 93:4405–10. doi: 10.1002/jmv.26985
18. Merino P, Guinea J, Muñoz-Gallego I, González-Donapetry P, Galán JC, Antona N, et al. Multicenter evaluation of the PanbioTM COVID-19 rapid antigen-detection test for the diagnosis of SARS-CoV-2 infection. Clin Microbiol Infect (2021) 27:758–61. doi: 10.1016/j.cmi.2021.02.001
19. Muhi S, Tayler N, Hoang T, Ballard SA, Graham M, Rojek A, et al. Multi-site assessment of rapid, point-of-care antigen testing for the diagnosis of SARS-CoV-2 infection in a low-prevalence setting: A validation and implementation study. The Lancet Regional Health - Western Pacific (2021) 9:100115. doi: 10.1016/j.lanwpc.2021.1001152666-6065/©
20. Nsoga MTN, Kronig I, Rodriguez FJP, Sattonnet-Roche P, Da Silva D, Helbling J, et al. Diagnostic accuracy of panbio rapid antigen tests on oropharyngeal swabs for detection of SARS-CoV-2. PLoS One (2021) 16:1–7. doi: 10.1371/journal.pone.0253321
21. Albert E, Torres I, Bueno F, Huntley D, Molla E, Fernández-Fuentes MÁ, et al. Field evaluation of a rapid antigen test (PanbioTM COVID-19 Ag rapid test device) for COVID-19 diagnosis in primary healthcare centres. Clin Microbiol Infect (2021) 27:472.e7–472.e10. doi: 10.1016/j.cmi.2020.11.004
22. Rubio JC, Costa Zamora M, Canals Aracil M, Pulgar Feio M, Mata Martínez A, Carrasco Munera A. Evaluación de la prueba diagnóstica de detección rápida de antígeno de covid-19 (Panbio covid rapid test) en atención primaria. Med Fam Semer (2021) 8:508–14. doi: 10.1016/j.semerg.2021.06.001
23. Stokes W, Berenger BM, Portnoy D, Scott B, Szelewicki J, Singh T, et al. Clinical performance of the Abbott panbio with nasopharyngeal, throat, and saliva swabs among symptomatic individuals with COVID-19. Eur J Clin Microbiol Infect Dis (2021) 40:1721–6. doi: 10.1007/S10096-021-04202-9
24. Bruins MJ, dos Santos CO, Spoelman-Lunsche M, Kromhout MI, van den B, Debast SB. Evaluation of the panbio. medRxiv (2021) 84. doi: 10.1101/2021.06.21.21259234
25. Bulilete O, Lorente P, Leiva A, Carandell E, Oliver A, Rojo E, et al. PanbioTM rapid antigen test for SARS-CoV-2 has acceptable accuracy in symptomatic patients in primary health care. J Infect (2021) 82:391–8. doi: 10.1016/j.jinf.2021.02.014
26. Carbonell-Sahuquillo S, Lázaro-Carreño MI, Camacho J, Barrés-Fernández A, Albert E, Torres I, et al. Evaluation of a rapid antigen detection test (PanbioTM COVID-19 Ag rapid test device) as a point-of-care diagnostic tool for COVID-19 in a pediatric emergency department. J Med Virol (2021) 12:6803–7. doi: 10.1002/jmv.27220
27. Faíco-Filho KS, Júnior FEF, Moreira LVL, Lins PRG, Justo AFO, Bellei N. Evaluation of the PanbioTM COVID-19 Ag rapid test at an emergency room in a hospital in São Paulo, Brazil. Braz J Infect Dis (2022) 26(2):102349. doi: 10.1101/2021.03.15.21253313
28. Fenollar F, Bouam A, Ballouche M, Fuster L, Prudent E, Colson P, et al. Evaluation of the panbio COVID-19 rapid antigen detection test device for the screening of patients with COVID-19. J Clin Microbiol (2021) 59:19–21. doi: 10.1128/JCM.02589-20
29. Ferté T, Ramel V, Cazanave C, Lafon ME, Bébéar C, Malvy D, et al. Accuracy of COVID-19 rapid antigenic tests compared to RT-PCR in a student population: The StudyCov study. J Clin Virol (2021) 141:5–8. doi: 10.1016/j.jcv.2021.104878
30. Gremmels H, Winkel BMF, Schuurman R, Rosingh A, Rigter NAM, Rodriguez O, et al. Real-life validation of the PanbioTM COVID-19 antigen rapid test (Abbott) in community-dwelling subjects with symptoms of potential SARS-CoV-2 infection. EClinicalMedicine (2021) 31:100677. doi: 10.1016/j.eclinm.2020.100677
31. Yudell M, Roberts D, DeSalle R, Tishkoff S. NIH Must confront the use of race in science. Science (2020) 369:1313–4. doi: 10.1126/SCIENCE.ABD4842
32. Leeflang MMG, Allerberger F. How to: evaluate a diagnostic test. Clin Microbiol Infect (2019) 25:54–9. doi: 10.1016/j.cmi.2018.06.011
33. Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig L, et al. STARD 2015: An updated list of essential items for reporting diagnostic accuracy studies. Clin Chem (2015) 61:1446–52. doi: 10.1373/clinchem.2015.246280
34. Sellera PEG, Moro MFSA, Albuquerque RDHE, Braga LI, De Souza MDS, Lima ASG, et al. The activation of socio-technical networks in the estrutural city/df brazil: Building a healthy and sustainable territory. Cienc e Saude Coletiva. (2019) 24:2185–91. doi: 10.1590/1413-81232018246.07982019
35. CODEPLAN. PDAD 2018, SCIA/Estrutural. Brazil: Secr Estado da Fazendo, Planejamento, Orçamento e Gestão do Dist Fed - SEFP (2019).
36. IBGE. (2018) Instituto Brasileiro de Geografia e Estatística. Síntese de indicadores sociais: Uma análise das condições de vida da população brasileira 2018 [Synthesis of social index: An analysis of the living standars of the Brazilian population 2018].
37. Guia de vigilância epidemiológica: emergência de saúde pública de importância nacional pela doença pelo coronavírus 2019 – covid-19 Vol. 86. Ministério da Saúde, Brazil: Secretaria de Vigilância em Saúde (2021). BRASIL. Ministério da Saúde. Secretaria de Vigilância.
38. Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)-a metadata-driven methodology and workflow process for providing translational research informatics support. J BioMed Inform (2009) 42:377–81. doi: 10.1016/j.jbi.2008.08.010
39. SPSS Statistics. IBM. Available at: https://www.ibm.com/products/spss-statistics?lnk=ushpv18ct7.
40. R Development Core Team. RStudio | open source & professional software for data science teams - RStudio. Available at: https://www.rstudio.com/.
41. Wickham H. ggplot2-elegant graphics for data analysis. Cham, Switz: Springer International Publishing (2016).
42. World Bank. South africa. gini index (2014). Available at: https://data.worldbank.org/indicator/SI.POV.GINI?locations=ZA.
43. Vaira LA, Salzano G, Deiana G, De Riu G. Anosmia and ageusia: Common findings in COVID-19 patients. Laryngoscope (2020) 130:1787. doi: 10.1002/lary.28692
44. Jiang M, Pan W, Arasthfer A, Fang W, Ling L, Fang H, et al. Development and validation of a rapid, single-step reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) system potentially to be used for reliable and high-throughput screening of COVID-19. Front Cell Infect Microbiol (2020) 10:331. doi: 10.3389/fcimb.2020.00331
Keywords: accuracy, antigen rapid detection test, socioeconomically vulnerable population, COVID-19, diagnostics
Citation: Gontijo CC, Brito RNd, Teixeira AIP, Romero GAS, Pedrette P, Ramalho WM, Noronha E, Haddad R and Araújo WNd (2022) Accuracy of point-of-care Panbio™ SARS-CoV-2 antigen-detection test in a socioeconomically vulnerable population in Brazil. Front. Trop. Dis 3:929524. doi: 10.3389/fitd.2022.929524
Received: 27 April 2022; Accepted: 05 August 2022;
Published: 13 September 2022.
Edited by:
Allassane Foungoye Ouattara, Université Nangui Abrogoua, Côte d’IvoireReviewed by:
Chitra Pattabiraman, Infectious Disease Research Foundation, IndiaPatricio Alejandro Vega-Mariño, Agency for the Regulation and Control of Biosafety and Quarantine for Galapagos (ABG), Ecuador
Copyright © 2022 Gontijo, Brito, Teixeira, Romero, Pedrette, Ramalho, Noronha, Haddad and Araújo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Raíssa Nogueira de Brito, cmFpc3Nhbm9ndWVpcmFicml0b0BnbWFpbC5jb20=; UmFpc3NhLkJyaXRvQHVnYS5lZHU=
†Present Address: Raíssa Nogueira de Brito, Department of Anthropology, University of Georgia, Athens, GA, United States
‡These authors have contributed equally to this work and share first authorship
 Carolina Carvalho Gontijo1,2,3‡
Carolina Carvalho Gontijo1,2,3‡ Raíssa Nogueira de Brito
Raíssa Nogueira de Brito Ana Izabel Passarella Teixeira
Ana Izabel Passarella Teixeira Gustavo Adolfo Sierra Romero
Gustavo Adolfo Sierra Romero Rodrigo Haddad
Rodrigo Haddad Wildo Navegantes de Araújo
Wildo Navegantes de Araújo