
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Trop. Dis. , 21 June 2022
Sec. Neglected Tropical Diseases
Volume 3 - 2022 | https://doi.org/10.3389/fitd.2022.898814
This article is part of the Research Topic Evolution of Drug Resistance among Neglected Tropical Diseases in the Context of Preventive Chemotherapy and Intensified Disease Management View all 4 articles
The efficacy of benzimidazole anthelmintics can vary depending on the target parasite, with Ascaris nematodes being highly responsive, and whipworms being less responsive. Anthelmintic resistance has become widespread, particularly in strongyle nematodes such as Haemonchus contortus in ruminants, and resistance has recently been detected in hookworms of humans and dogs. Past work has shown that there are multiple β-tubulin isotypes in helminths, yet only a few of these contribute to benzimidazole interactions and resistance. The β-tubulin isotypes of ascarids and soil-transmitted helminths were identified by mining available genome data, and phylogenetic analysis showed that the ascarids share a similar repertoire of seven β-tubulin isotypes. Strongyles also have a consistent pattern of four β-tubulin isotypes. In contrast, the whipworms only have two isotypes, with one of these clustering more basally and distinct from any other group. Key β-tubulin isotypes selected based on previous studies were the focus of in silico molecular docking simulations to look at the interactions with benzimidazoles. These showed that all β-tubulins had similar interactions with benzimidazoles and maintained the key bond with residue E198 in all species, indicating similar mechanisms of action. However, the interaction was stronger and more consistent in the strongyles and whipworms than it was in the ascarids. Alteration of β-tubulin isotypes with the common resistance-associated mutations originally identified in H. contortus resulted in similar interaction modeling for all species. In conclusion, ascarids, strongyles, and whipworms all have their own unique repertoire of β-tubulins, which could explain why benzimidazole resistance and susceptibility varies between these groups of parasites. These data complement recent work that has highlighted the roles of essential residues in benzimidazole drug binding and shows that there is a separation between strongyle parasites that frequently develop resistance and ascarid parasites, which have been much less prone to developing resistance.
Due to the early emergence of benzimidazole (BZ) resistance in nematodes of veterinary importance, such as Haemonchus contortus, and its link with β-tubulins, there is growing interest in the diversity of β-tubulins in nematodes (1, 2). The model organism Caenorhabditis elegans, a free-living nematode, is the best source of information regarding the diversity of β-tubulins and the mechanisms of resistance. There are six β-tubulin genes (isotypes) within the genome of C. elegans and other helminths (1, 3–9). Benzimidazole resistance was specifically associated with one of the isotypes called ben-1 in C. elegans, and orthologues of this isotype have been associated with BZ resistance in strongyle helminths of veterinary importance, such as Cooperia oncophora, Haemonchus contortus, and Ostertagia ostertagia (1, 4). There are currently four known β-tubulin isotypes in H. contortus, and non-strongyle parasites such as Trichuris and Ascaris also have multiple β-tubulins, although in these helminths, the β-tubulin isotypes linked with BZ interactions are not orthologues of C. elegans ben-1 (1, 4).
Treatment of the ruminant nematode H. contortus has been most impacted by drug resistance and has subsequently been the focus of most research. Previous work has shown that of the four β-tubulins found in the H. contortus genome, only isotypes 1 and 2 are expressed in adults at high enough levels to be impacted by BZs, and isotype 1 has been the focus of research into drug resistance (1). A range of mutations in three amino acids have been shown to be associated with BZ resistance in several nematode species (10–17). A limited number of studies have used in silico methods to identify the exact mechanisms at play in β-tubulin–BZ interactions and the effects of resistance-associated mutations on these interactions (18, 19).
Recent works in Ascaris have provided the first in-depth investigations into the β-tubulins of Ascaris, their role in BZ interactions, and the potential consequences of resistance-associated mutations (7, 20). We now know that similar to strongyle parasites, Ascaris has several highly expressed isotypes capable of interacting with BZs, with isotype A being the most likely candidate (7). In silico work has highlighted the differences in drug binding between wild-type and mutated Ascaris β-tubulins, providing a basis for future work on parasite β-tubulins (20). There have also been multiple studies on Parascaris and Ascaridia, both members of the ascarid family of helminths, which have shown that resistance is emerging in these ascarids, though the mutations responsible are yet to be confirmed (9, 21, 22).
The efficacy of BZs against the soil-transmitted helminth (STH) Ascaris has remained generally high. However, this is not the case for other STHs such as hookworms and whipworms. Meta-analysis found that while average cure rates for Ascaris lumbricoides were between 88% and 95%, for hookworm, they were lower: between 15% and 72% depending on which BZ was used. Whipworm showed consistently lower susceptibility to BZ with average cure between 28% and 36% (23). More recent studies have continued to find lower susceptibility of whipworms to BZ, suggesting that whipworms may be innately less susceptible (24–26). The typical resistance-associated single-nucleotide polymorphisms (SNPs) (F167Y, E198A, and F200Y) have been found in both hookworms and whipworms, demonstrating that there has likely been a selective pressure by BZs leading to reduced treatment response (27–31).
While there have been investigations into BZ docking in a number of organisms, including parasitic nematodes and BZ-susceptible and refractory protozoan parasites and mammals, the recent work on Ascaris was the first to look into STHs of public health importance (18–20, 32). No work has been carried out on the remaining two groups of STH, the hookworms (Ancylostoma and Necator americanus) and whipworms (Trichiura), even though BZ resistance is a known problem in the control of STHs.
The aim of this work was to identify and compare the relationships between the β-tubulins of STHs and Ascaridomorpha parasites, herein referred to as ascarids, and use those isotypes most likely to interact with BZs to perform in silico drug docking simulations. We aimed to determine if all ascarids share the same grouping of β-tubulins as Ascaris and see how this compares to other groups of helminths. We also aimed to investigate whether there are any intrinsic differences in the binding capacity of ascarid, hookworm, and whipworm β-tubulins that might explain the varying efficacies of BZs against different nematodes.
The genomes of common STHs and all ascarid species with genomes accessible in Wormbase-Parasite were searched to identify β-tubulin isotypes (33, 34). A BLAST search of the genomes was performed using the A. lumbricoides isotype A peptide sequence as a reference (35). Genomes were also searched using the term “Tubulin beta” to ensure as many genes as possible could be found. The results of this were then filtered based on size and tubulin type to remove small fragment sequences and α-tubulins. The published β-tubulins of ascarids available from NCBI were also included (36). The nematode species included in this analysis were as follows: Ancylostoma caninum, Ancylostoma ceylanicum, Ancylostoma duodenale, Necator americanus, Trichuris trichiura, Trichuris suis, Parascaris equorum, Parascaris univalens, Anasakis simplex, Toxocara canis, and Ascaridia galli. Although Asc. galli does not have a genome available, the sequence for a β-tubulin was found in the BLAST search. A full list of β-tubulin genes identified can be seen in Supplementary Table 1. Sequences were aligned in BioEdit (version 7.0.5.3) (37). The full sequence alignment is shown in Supplementary Figure 1.
To compare the genetic structuring of the identified β-tubulins, phylogenetic analysis of the peptide sequences was performed in MEGA X (38). The four β-tubulins described in H. contortus and the six β-tubulins from C. elegans were included in the phylogenies as a comparison of a better studied organism, and the β-tubulin sequences described previously in Ascaris were included to show the full β-tubulin repertoire of ascarids (7, 20). The sequences were aligned using the MUSCLE algorithm, the best evolutionary model was found, and a maximum likelihood tree was constructed using the LG+G model with 1,000 bootstraps.
Molecular dynamics simulations were carried out on the β-tubulin isotype 1 proteins for the hookworms and whipworms, and isotype A proteins for the ascarids. Isotype A was chosen for this analysis as it is the isotype that is associated with the highest expression levels in ascarids and is therefore the most likely to be affected by BZs. For the other STHs, isotype 1 was chosen as this is the one that has previously been shown to cause resistance when mutated. To create 3D structures of the β-tubulin sequences, homology models were constructed in SWISS-MODEL using the PDB structure 6fkj as a reference (available at: https://swissmodel.expasy.org/ [Accessed February 22, 2021]) (39–41). All homology models were minimized using the YASARA energy minimization server (available at: http://www.yasara.org/minimizationserver.htm [Accessed October 6, 2021]) and quality checks of the structures were carried out using ProSA-web (available at: https://prosa.services.came.sbg.ac.at/prosa.php [Accessed October 6, 2021]), Verify3D, and PROCHECK (42–47). Verify3D and PROCHECK are part of the UCLA SAVES v6.0 server (available at: https://saves.mbi.ucla.edu/ [Accessed October 6, 2021]). Default settings were used for all servers.
Albendazole sulfoxide (ABZSO) was used to perform molecular dynamics simulation using Molecular Operating Environment (MOE) version 2020.01 (48). This drug was chosen as it is the active metabolite of albendazole, one of the most commonly administered BZs to treat helminths. Molecular dynamics simulations were carried out in MOE following the methods set out in Jones et al. (20). Briefly, β-tubulin structures were optimized using the Protonate3D command within MOE, the drug binding pockets were identified using the Site Finder algorithm, and ABZSO was docked into the pocket corresponding to the known binding region of BZs. The 10 best docking poses were returned based on the GBVI/WSA dG scoring method, and from this, the best binding pose was chosen for molecular dynamics simulations. Molecular dynamics simulations were carried out using the NPA algorithm and the Amber10:EHT forcefield. An equilibration step of 100 picoseconds (ps) at 300°K was run initially, followed by a further 500-ps run with a time step of 0.002 ps. Samples were taken for comparison every 50 ps and these structures were minimized to find drug binding affinity; the pose with the strongest affinity was taken as the final result.
The three canonical resistance-associated mutations (F167Y, E198A, and F200Y) were introduced into the Anc. duodenale and Tr. trichiura sequences by manually changing the sequence residues. Homology models were created and minimized and molecular dynamics simulations were carried out using the same methods as previously stated.
The initial genome search resulted in the identification of 107 β-tubulin sequences in 13 genomes, representing An. simplex, P. equorum, P. univalens, T. canis, Anc. caninum, Anc. ceylanicum, Anc. duodenale, N. americanus, Tr. trichiura, and Tr. suis. Additional sequences for Asc. galli and P. equorum were added from the NCBI database. Deduplication and removal of most incomplete sequences and α-tubulins left 64 β-tubulin sequences, which included the 14 Ascaris and four H. contortus reference sequences (Supplementary Table 1). Some incomplete sequences were included in the analysis as long as they were more than 200 amino acids long and included the region of the protein that contained the predicted binding pocket and resistance-associated SNP sites in the corresponding gene.
The phylogenetic analysis showed that the ascarid β-tubulins resembled the Ascaris β-tubulins, with representatives being present in each isotype cluster (Figure 1). Parascaris univalens was the only organism containing all seven β-tubulin isotypes. Toxocara canis lacked isotype C or D, and for An. simplex, which had one of the lowest qualities of genome assembly according to the BUSCO and CEGMA metrics, isotypes D, E, F, and G could not be identified. This may well be a result of the lower quality of the available genomes, rather than a lack of these isotypes being present in these species. There were several additional isotypes found only in T. canis that were very similar to isotype A. Of these, two had large insertions and so were disregarded, but one was of a similar size to all other β-tubulin isotypes and so was designated as isotype A.
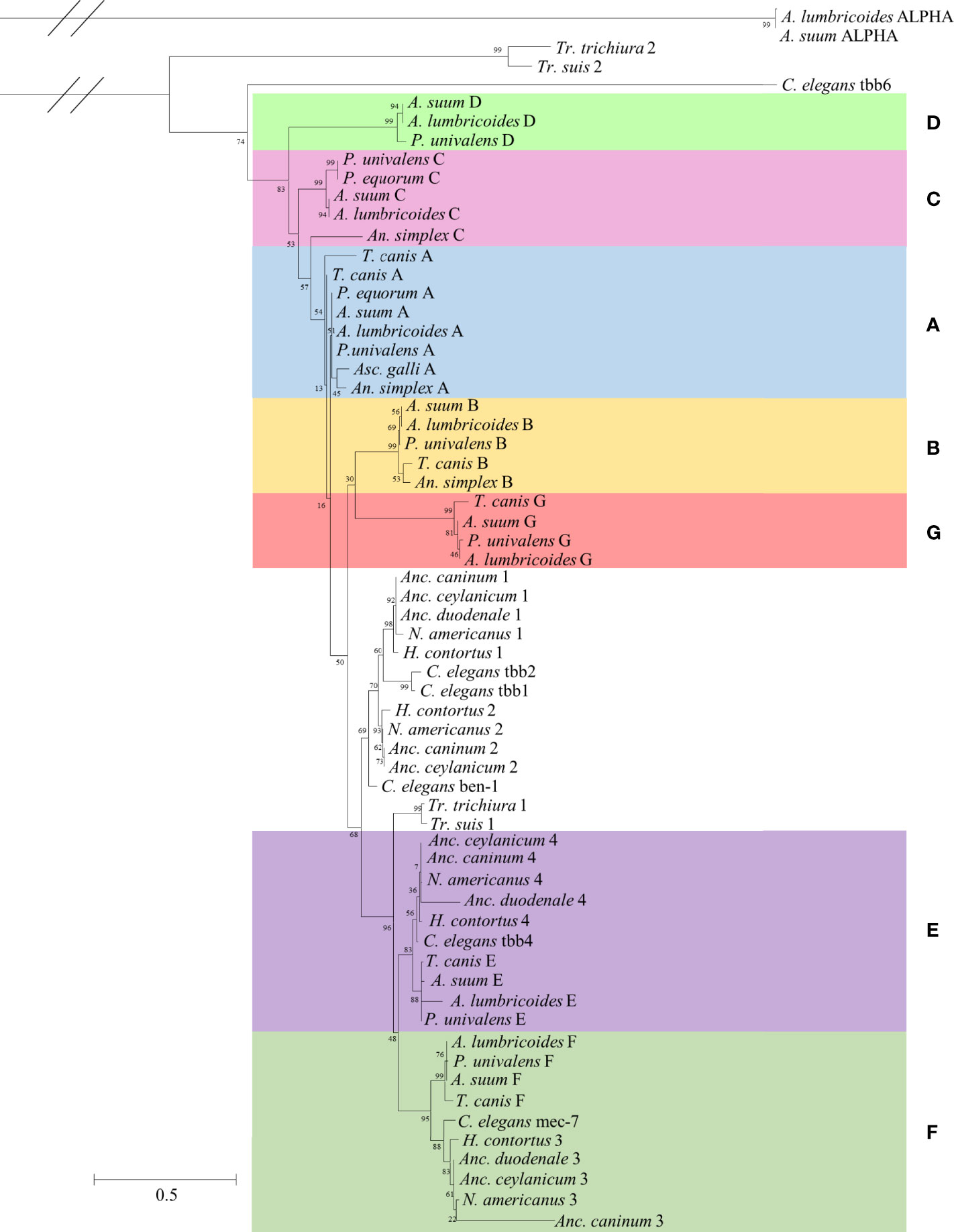
Figure 1 Maximum likelihood phylogeny showing β-tubulin isotypes from the model organism Caenorhabditis elegans, common soil-transmitted helminths, and other Ascaridomorpha species. The phylogeny was created using an LG+G model with 1,000 bootstraps. Ascaridomorpha isotypes were named based on Ascaris nomenclature and the strongyle and Trichuris isotypes were numbered based on established nomenclature. The Ascaridomorpha species follow a similar pattern of β-tubulin isotypes to Ascaris, organizing into seven groups [clusters (A–G) colored separately based on Ascaris isotype nomenclature]. In some cases, isotypes are not present for all species of Ascaridomorpha, likely due to the incompleteness of the available genomes. The strongyles (H. contortus and the hookworms) have their own β-tubulin profiles; however, these are closely related to the β-tubulins seen in the C. elegans. The strongyles contain four isotypes, two cluster distinctly from any Ascaridomorpha group (isotype 1 and 2), one clusters with group E (isotype 4), and one clusters with group F (isotype 3). The Trichuris have the most distinct β-tubulin profile, with only two isotypes found. One of these clusters basally to groups (E, F), and the other is distinct from any other β-tubulin isotype and forms its own separate group. Ascaris α-tubulins were used as the outgroup.
The non-ascarids (C. elegans, hookworms, H. contortus, and Trichuris) are more distantly related to Ascaris than the ascarids, and their β-tubulin repertoire reflected this. The strongyle genomes contained sequences that clustered with isotypes E and F, isotypes 4 and 3, respectively. However, within these isotype groupings, they formed their own clusters, with the corresponding C. elegans β-tubulins separate from the other ascarids. In the strongyles, isotypes 1 and 2 formed a distinct cluster separate from the ascarid isotypes. In C. elegans, isotypes 1 and 2 did not correspond to the strongyle β-tubulins, instead forming their own cluster closely related to isotype 1. The C. elegans ben-1 isotype was isolated basal to the strongyle isotypes 1 and 2 cluster. For Anc. duodenale, only isotype 1 was found, which may be due to the incompleteness of the genome (Figure 1). There were some nodes that showed relatively low bootstrap values.
The Trichuris genomes contained only two β-tubulin isotypes. Isotype 1 clustered basal to the β-tubulin E/4 and F/3 clade. The second isotype did not cluster with any other group and was shown to be the most basal of the β-tubulins analyzed (Figure 1). A phylogeny was also created omitting the divergent Trichuris isotype 2 sequences, which resulted in no major changes and so was discarded (data not shown).
Quality checks of the structures allow us to assess how realistic the homology models are compared to experimentally determined proteins by employing various tests. The quality checks of minimized homology models showed acceptable results with Z-scores within the expected range and verify3D scores over 80% (Supplementary Figures 2–17, Supplementary Tables 2 and 3). Most models showed that all residues within the proteins were in allowable conformations as PROCHECK found no residues with unfavorable torsion angles; however, the Trichuris isotype 1 proteins showed some outlier residues in the PROCHECK Ramachandran plots (Supplementary Tables 2 and 3).
Molecular dynamics results showed that there were many interactions between the β-tubulins and BZs shared across species. All models showed strong binding between ABZSO and residue E198, except for P. equorum, where only a relatively weak bond between BZ and E198 was seen (Figures 2 and 3, Table 1). Interactions with Q134, L253, and N256 were the most frequently seen after E198, with bonds seen in four, five, and four β-tubulins, respectively. Simulations for the ascarid isotype A structures showed similar results to each other and to the molecular dynamic simulations performed for Ascaris described previously (20) (Figure 2, Table 1). The results for the hookworm and whipworm species showed that the strong bond with E198 was seen in all species (Figure 3, Table 1). The results for the hookworms showed different preferences between species. Ancylostoma caninum and Anc. ceylanicum both shared the same binding profile, and Anc. duodenale and N. americanus both had different binding profiles with no overlap except for the E198 interaction that is shared with all nematodes (Figures 3A–D, Table 1). Due to a deletion at residue 177 in Trichuris isotype 1, E197, F199, and N255 correlate to E198, F200, and N256 in the other nematodes, respectively. In both whipworm species, there was binding to E197 and F199. There was an additional bond with N255 in Tr. trichiura (Figures 3E, F, Table 1). Binding affinities for each isotype and individual residue binding energies can be seen in Table 1.
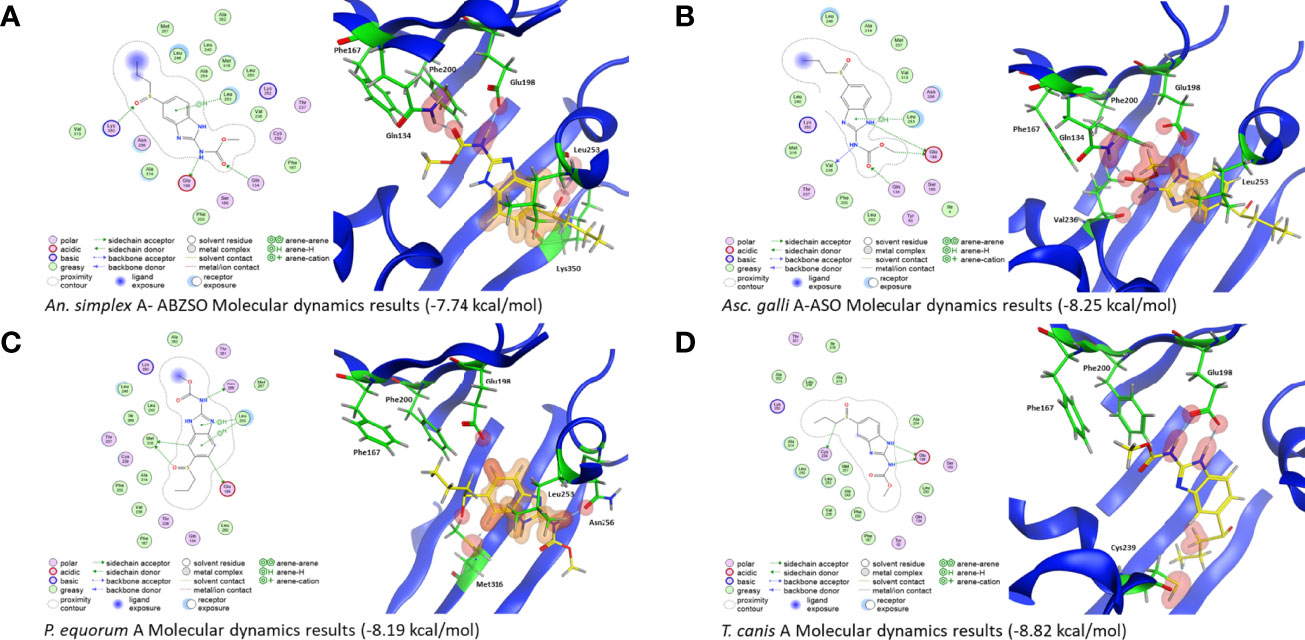
Figure 2 2D and 3D representations of the Ascaridomorpha isotype A and ABZSO molecular dynamics simulations. (A) Anisakis simplex model shows H-bonds between the drug and Q134, E198, and K350. An additional arene bond is also formed with L253. The overall affinity of the binding is −7.74 kcal/mol. (B) The Ascaridia galli model shows H-bonds between the drug and Q134, E198, and V236. An arene bond is also formed with L253. The overall affinity of the binding is −8.25 kcal/mol. (C) The Parascaris equorum model shows H-bonds between the drug and E198, N256, and M316. Arene bonds are formed with L253. The overall affinity of the binding is −8.19 kcal/mol. (D) The Toxocara canis isotype A model shows H-bonds between the drug and E198 and Cys239. The overall affinity of the binding is −8.82 kcal/mol.
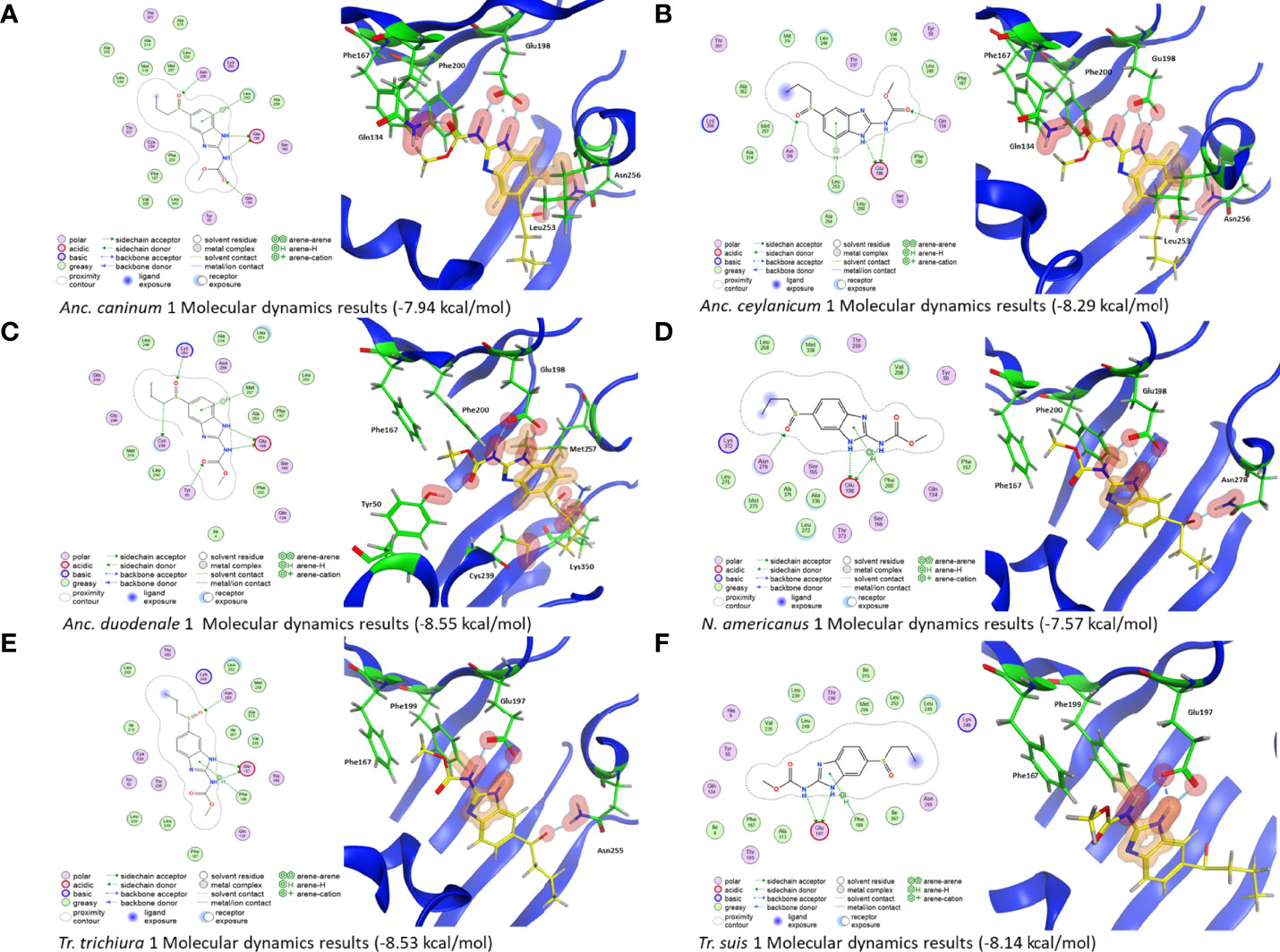
Figure 3 2D and 3D representations of the hookworm and whipworm isotype 1 models complexed with ABZSO from the molecular dynamics simulations. (A) The Ancylostoma caninum isotype 1 model shows H-bonds between the drug and Q134, E198, and N256. An arene bond is also formed with L253. The overall affinity of the binding is −7.94 kcal/mol. (B) The Ancylostoma ceylanicum isotype 1 model shows H-bonds between the drug and Q134, E198, and N256. An arene bond is also formed with L253. The overall affinity of the binding is −8.29 kcal/mol. (C) The Ancylostoma duodenale isotype 1 models H-bonds between the drug and Y50, E198, C239, and K350. An arene bond is also formed with M257. The overall affinity of the binding is −8.55 kcal/mol. (D) The Necator americanus isotype 1 model shows H-bonds between the drug and E198 and N278. An arene bond is also formed with F200. The overall affinity of the binding is −7.57 kcal/mol. In N. americanus 1, there is a 22-residue insertion at 228 that changes the residue numbers seen in all other species. In Trichuris isotype 1, there is a deletion at position 177 that changes the residue numbers seen in all other species. (E) The Trichuris suis model shows H-bonds between the drug and E197. An arene bond is also formed with F199. The overall affinity of the binding is −8.14 kcal/mol. (F) The Trichuris trichiura model shows H-bonds between the drug and E197 and Asn255. An arene bond is also formed with F199. The overall affinity of the binding is −8.53 kcal/mol.
Molecular dynamics with the Anc. duodenale F167Y model showed that the strong bond between BZ and E198 was still present. A self-binding interaction was observed between Q134 and F167Y, and a strong bond with K350 was also seen (Figure 4A, Table 2). In the Anc. duodenale E198A model, the bond between BZ and residue 198 was lost, and no bonds of similar strength were present (Figure 4B, Table 2). The Anc. duodenale F200Y model showed that the E198 bond was not present, and a self-binding interaction was seen between E198 and F200Y. The strong bond with K350 was again present, indicating that this residue may have an important role in drug stability in the binding pocket (Figure 4C, Table 2).
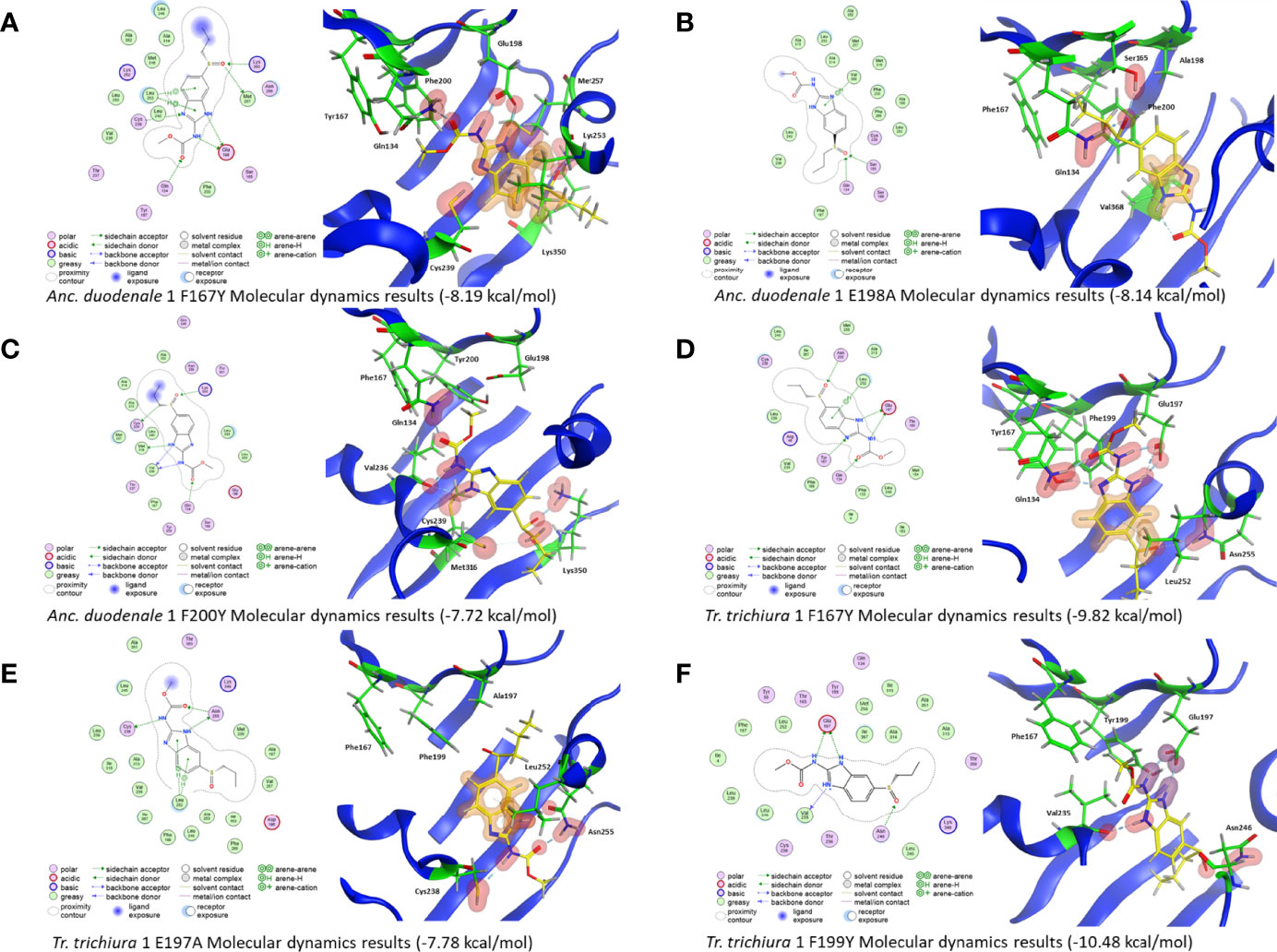
Figure 4 2D and 3D representations of the mutated Ancylostoma duodenale and Trichuris trichiura model. (A) The mutated Anc. duodenale F167Y isotype 1 shows H-bonds between the drug and Q134, E198, C239, M257, and K350. An arene bond is also formed with L253. The overall affinity of the binding is −8.19 kcal/mol. (B) The mutated Anc. duodenale E198A isotype 1 shows H-bonds between the drug and Q134 and S165. An arene bond is also formed with V368. The overall affinity of the binding is −8.14 kcal/mol. (C) The mutated Anc. duodenale F200Y isotype 1 shows H-bonds between the drug and Q134, V236, C239, M316, and K350. The overall affinity of the binding is −7.72 kcal/mol. (D) The mutated Tr. trichiura F167Y isotype 1 shows H-bonds between the drug and Q134, F167Y, E197, and N255. An arene bond is also formed with L252. The overall affinity of the binding is −9.82 kcal/mol. (E) The mutated Tr. trichiura E197A isotype 1 shows H-bonds between the drug and C238 and N255. An arene bond is also formed with L252. The overall affinity of the binding is −7.78 kcal/mol. (F) The mutated Tr. trichiura F199Y isotype 1 shows H-bonds between the drug and V235 and N255. An ionic hydrogen bond is also formed with E197. The overall affinity of the binding is −10.48 kcal/mol. The overall affinity of the binding is shown below each model.
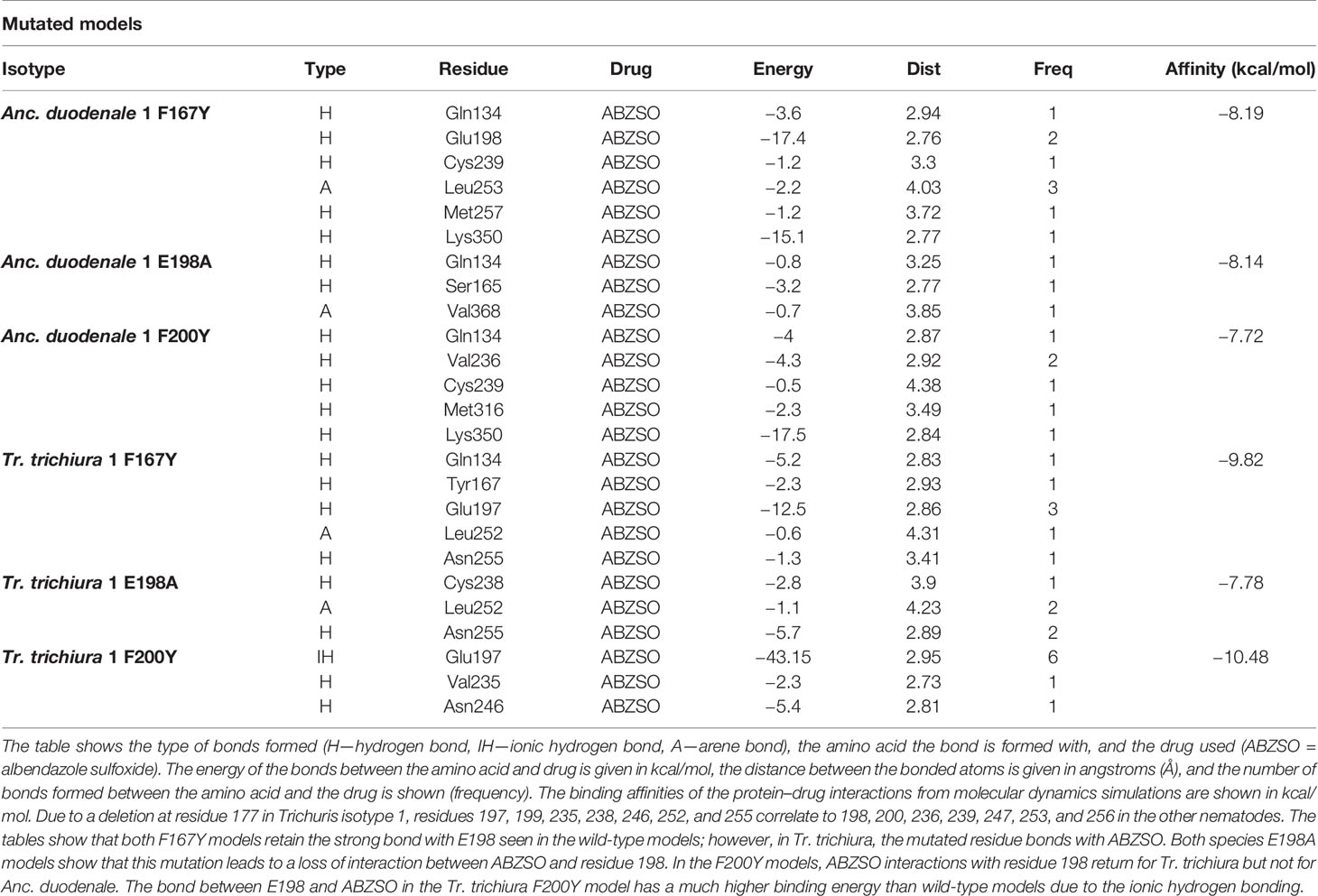
Table 2 Molecular dynamics results for mutated Ancylostoma duodenale and Trichuris trichiura isotype 1 models.
The results of molecular dynamics simulations for the Tr. trichiura F167Y model showed the presence of BZ binding with E197. The mutated F167Y residue also formed a bond with BZ (Figure 4D, Table 2). In the E197A model, the key interaction with residue 197 was not present (Figure 4E, Table 2). The residues that BZ did bind with meant that the drug was pulled away from the usual binding region and no interactions were made with residues 167, 197, and 199 (167, 198, and 200 in all other species). In the Tr. trichiura F199Y model, BZ binds with the key E197 residue. However, in this model, an ionic bond is formed rather than a hydrogen bond, which increases the binding energy (Figure 4F, Table 2). A self-binding interaction was also seen between E197 and F199Y in Tr. trichiura (Figure 4F).
This research has identified multiple β-tubulin isotypes in several parasites of human and veterinary importance. We have shown using phylogenetic analyses that the β-tubulin profiles are similar for the ascarids with seven groupings of β-tubulin isotypes. The strongyles also share similar β-tubulin profiles to each other using only four isotypes. The in silico work performed has shown that BZs appear to bind in the same way in all nematode β-tubulins investigated here and highlights the importance of residue 198 in the BZ mechanism of action. Due to the understudied nature of the organisms investigated here, in-depth genome assemblies are not available for all species. There were major disparities in the quality of the genomes used for this study, based on the quality metrics in Wormbase-Parasite and the number of tubulin genes that were recovered. For Asc. galli, a genome was not available, although a single sequence for the β-tubulin gene that had been previously studied in relation to BZ resistance was available (6). As Asc. galli is an important parasite of poultry, and with the recent report of BZ resistance being seen in a related species, Ascaridia dissimilis, it was determined that it would be important and of wider interest to include this single sequence in the study (22).
The clearest differences were seen in the Trichuris β-tubulins. Only two isotypes were identified: one that clusters basal to groups E and F of the other nematode β-tubulins, and another that clusters basally to the other helminth β-tubulins. This would suggest that Trichuris diverged from the other helminths prior to the β-tubulin duplications that gave rise to the β-tubulin families seen in the rest of the helminths. This fits with the taxonomy that shows that Trichuris are of a different class (Enoplea) to the strongyles and ascarids (Chromadorea). However, with this in mind, we should then expect the Trichuris isotype 1 to be basal to all Chromadorea β-tubulins, something that will require further investigation. In this study, we have used the current nomenclature for Trichuris β-tubulins of isotype 1 and 2. In the past, tubulins have been named based on the order in which they were discovered, or their expression level, and this has caused problems in the nomenclature, with the β-tubulin isotype 1 from different groups not being orthologues (1, 4, 8, 9). This problem has been addressed recently for Ascaris where the naming convention was changed from a numerical system to alphabetical (A–G) to clarify the difference in evolutionary history and roles of β-tubulins in these groups of parasites (7). The Trichuris β-tubulin isotypes 1 and 2 are also not orthologues of the strongyle β-tubulins, and the argument that the naming of these isotypes is misleading could be made. However, the reclassification of the Trichuris β-tubulin nomenclature is beyond the scope of this study and would require further examination of other species more closely related to Trichuris to put forward a new nomenclature that would best fit these species.
The full repertoire of β-tubulins characterized in Ascaris was also seen in P. univalens but none of the other ascarids. It is likely that this is the result of incomplete genome assemblies. This is evidenced by looking at the isotypes found in An. simplex and T. canis, which together represent all isotypes except isotype D. As Asc. galli is the only non-Ascaridoidea included in this group and a genome is not available, the only evidence to suggest that it would contain most, if not all the Ascaris isotypes is the fact that the strongyles, which are more distinct, also share some of these isotypes. Within the ascarids group, T. canis does show an extra isotype within group A. Without further investigation, it cannot be concluded if this is the result of recent duplication in this species or problems with genome assembly.
The phylogenies show that there are noticeable differences between the structuring of the ascarid and strongyle β-tubulins included in this study. There are some noticeable differences seen between the phylogeny presented here and phylogenies presented in recent studies of ascarid β-tubulins (7, 9, 49). While the general ordering of isotypes in our study is similar to that reported in previous studies, the exact clustering and evolutionary relationship is different. In our phylogeny, we see a continuous branching off of isotypes A, C, and D, with B and G, and E and F forming separate subsequent clusters. In previous work A, C, and D cluster together with B and G forming a separate cluster, and they together form a larger clade separate from E and F (7, 9, 49). Again, in previous work, the strongyle isotype 1 and 2 clusters were found to be at a basal position within the larger A, C, D, B, and G clade, whereas in our phylogeny, we see them form a separate clade (7, 9, 49). Within this clade, the relationship between the strongyles and C. elegans corresponds to previous work with C. elegans isotypes 1 and 2 being closely related to but isolated from strongyle isotype 1 and C. elegans ben-1 being basal to this clade. Trichuris isotype 1 is basal to isotypes E and F as was seen previously. However, in this former analysis, Tr. trichiura was also basal to all other nematode β-tubulins (7). Conversely, another investigation of nematode tubulins showed Tr. trichiura isotype 1 to sit within a clade with A. suum isotype E and C. elegans isotype 4 (4). Finally, the relationship between ascarid isotype F and strongyle isotype 3 and between ascarid isotype E and strongyle isotype 4 matches that seen previously with these isotypes forming their own clade (7, 9, 49). There are several possible causes for the differences seen between our phylogeny and those previously published such as the method used to construct the phylogeny, or the sequences used. Our study included more sequences than previous analyses have, including more divergent isotypes such as Trichuris isotype 2, C. elegans isotype 6, and Ascaris α-tubulin, which give us an insight into the history of β-tubulin evolution. The inclusion of these sequences may be the reason for the slightly different clustering of isotypes and the low bootstrap values seen for some nodes, although no major differences were seen when Trichuris isotype 2 was removed from analysis. The isotypes found in the hookworm genomes match the four β-tubulin isotypes that have been previously identified in H. contortus. Previous work has shown that of the four isotypes, only isotype 1 and isotype 2 are highly expressed in H. contortus (1). As most BZ resistance is linked to isotype 1 in strongyles, it was this isotype that was chosen to be the focus of further experiments.
In the molecular dynamics simulations performed in this study, most of the nematode β-tubulins show similar BZ binding results to the H. contortus and Ascaris models in previous studies (19, 20). A strong bond formation is seen between BZ and the E198 residue, which is key to BZ sensitivity. The exceptions to this were in P. equorum, which had a weaker bond with E198 compared to other helminths. While, so far, there has been a lack of evidence for BZ resistance in Ascaris, in Parascaris and Ascaridia, reports of resistance are beginning to be seen (9, 22, 50, 51). However, there is no molecular evidence so far of the canonical BZ resistance-associated SNPs in resistant populations of ascarids (9).
The hookworms have the same repertoire of β-tubulins as the more well-studied strongyles such as H. contortus. Most importantly, they share the two closely related isotypes associated with BZ interaction that are highly expressed (1). The theory of redundancy has been posited for β-tubulins in C. elegans, whereby the loss of function of either β-tubulin 1 or 2 rarely causes cellular defects whereas missense mutations lead to more severe effects (3, 52). Recent work has also proven that the canonical resistance-associated mutations cause resistance in gene-edited C. elegans β-tubulins yet have no effect on fitness in controls (53). It could be the case that as there are two proteins performing the same job in strongyles, this allows one to gain mutations that confer resistance but may reduce fitness, while the other protein can take on the needs of the cells counteracting this burden.
Most evidence of BZ resistance has come from strongyle parasites with mutations in isotypes 1 and 2 being the main cause (1, 10, 12, 13, 54–57). These two isotypes do not group closely with any ascarid β-tubulin isotype (Figure 1). It may be the close genetic relationship between these isotypes that provides the redundancy needed for mutations to develop. The ascarids have the most β-tubulin isotypes, yet BZs still seem to work efficiently against them. The greater divergences between the ascarid β-tubulin isotypes may make it less likely that they can take on the roles of isotype A and therefore the fitness cost of the resistance-associated SNPs would make them deleterious.
Trichuris are much more genetically divergent from both ascarids and strongyles, and hence have a very distinct repertoire of β-tubulins. Although there is little difference seen in terms of overall binding and affinity between Trichuris β-tubulins and BZs, the mechanisms may be different in these parasites. For example, it is possible that both β-tubulins are highly expressed, explaining why normal doses of BZs have been ineffective, yet higher doses or prolonged treatment increases the efficacy of BZs in Trichuris (25, 58).
Although there has been a documented decline in the efficacy of BZs against whipworms, there is still little evidence for the presence of the canonical resistance-associated SNPs (59). In several studies, these SNPs could not be found; in one study, only the F200Y SNP was found, and in another, all three SNPs were found in different populations (27, 60–64). In a population where all three SNPs were found, the proportion of the F167Y SNP was decreased after treatment (27). While there has been a slower development of resistance in hookworms compared to ruminant strongyles, reports are beginning to emerge with evidence of all three canonical resistance-associated SNPs in various hookworm species (27, 30, 65–68).
The mutated models again showed that residue E198 is likely to be a key player in BZ binding in β-tubulins. Similar to previous work on the strongyle parasite H. contortus, in Anc. duodenale, while the F167Y mutation does not directly affect the interaction of BZ, with the bond between BZ and E198 still being seen, it may have an indirect effect (19). We saw a self-binding interaction between the mutated F167Y residue and Q134. The interaction between these two residues has been previously predicted to affect the shape of the entry space of the binding pocket and therefore may affect BZ’s ability to enter the binding region, thereby leading to resistance (19). This same self-binding interaction was not seen in the Tr. trichiura 167Y model, nor was it seen in Ascaris β-tubulin models in a previous study (20). This, alongside the evidence of the F167Y SNP in BZ-susceptible populations of Ascaris and the findings of a reduction on the proportion of F167Y SNPs post-treatment in Tr. trichiura, could suggest that the entry blocking interaction between Q134 and F167Y may be a strongyle-specific trait (27).
In both the Anc. duodenale E198A and Tr. trichiura E197A models, we saw a complete lack of binding between the BZ and this key residue. There was also a lack of any bonds of comparable strength in these models, suggesting that the loss of this key residue leads to a less stable binding conformation of BZs. The biggest difference between Anc. duodenale and Tr. trichiura was seen with the F200Y and F199Y SNPs, respectively. In Anc. duodenale, we saw the loss of the key binding interaction with E198, and a self-binding interaction was seen between the mutated F200Y residue and E198. In Tr. trichiura, the binding between E197 and F199Y was also seen, although strong ionic bonds were also formed between BZ and E198. The interaction between the mutated F200Y residue and E198 was similarly seen in other parasites, and it was predicted that it could lead to the destabilization of BZ in the binding pocket and therefore cause resistance (19, 20).
The results presented here show that BZs act via the same mechanisms in all species tested and once again highlights E198 as a key residue for BZ binding. This analysis includes a wide range of helminths designated as STHs of human importance and closely related parasites of animals. The major difference between the groups of helminths is in the number of β-tubulin isotypes found within the genome rather than the diversity of these isotypes.
We can postulate from this that the differences seen in the development of resistance are not due to the structural changes in β-tubulin proteins themselves but may be the result of the differences in the β-tubulin isotype abundance and expression. However, it must be noted that the finding of the Q134–F167Y interaction in Anc. duodenale together with evidence of this F167Y SNP in susceptible Ascaris populations and its reduction in frequency after BZ treatment in Tr. trichiura may suggest that mutations in F167Y have more of an impact in strongyle parasite than other STHs.
There is still a need to be vigilant for the development of resistance in ascarid species and other nematodes of veterinary and medical importance. As we are beginning to see reports of treatment failure in ascarids and STHs, genetic investigation will aid in determining the underlying causes in this group of nematodes that have, until recently, been the only group of parasitic nematodes with no significant evidence of BZ resistance.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
BJ, MB, AV, and EL: conceptualization. BJ and MB: writing of original manuscript. BJ, MB, AV, and EL: Review and editing of manuscript. All authors contributed to the article and approved the submitted version.
This study was funded by the Kenneth Longhurst legacy PhD studentship Award from the University of Surrey.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fitd.2022.898814/full#supplementary-material
1. Saunders GI, Wasmuth JD, Beech R, Laing R, Hunt M, Naghra H, et al. Characterization and Comparative Analysis of the Complete Haemonchus Contortus β-Tubulin Gene Family and Implications for Benzimidazole Resistance in Strongylid Nematodes. Int J Parasitol (2013) 43(6):465–75. doi: 10.1016/j.ijpara.2012.12.011
2. Geary TG, Nulf SC, Favreau MA, Tang L, Prichard RK, Hatzenbuhler NT, et al. Three β-Tubulin cDNAs From the Parasitic Nematode Haemonchus Contortus. Mol Biochem Parasitol (1992) 50(2):295–306. doi: 10.1016/0166-6851(92)90227-B
4. Demeler J, Krüger N, Krücken J, von der Heyden VC, Ramünke S, Küttler U, et al. Phylogenetic Characterization of β-Tubulins and Development of Pyrosequencing Assays for Benzimidazole Resistance in Cattle Nematodes. PloS One (2013) 8(8):e70212. doi: 10.1371/journal.pone.0070212
5. Ryan LA, Hoey E, Trudgett A, Fairweather I, Fuchs M, Robinson MW, et al. Fasciola Hepatica Expresses Multiple α- and β-Tubulin Isotypes. Mol Biochem Parasitol (2008) 159(1):73–8. doi: 10.1016/j.molbiopara.2008.02.001
6. Tydén E, Engström A, Morrison DA, Höglund J. Sequencing of the β-Tubulin Genes in the Ascarid Nematodes Parascaris Equorum and Ascaridia Galli. Mol Biochem Parasitol (2013) 190(1):38–43. doi: 10.1016/j.molbiopara.2013.05.003
7. Roose S, Avramenko RW, Pollo SMJ, Wasmuth JD, Ame S, Ayana M, et al. Characterization of the β-Tubulin Gene Family in Ascaris Lumbricoides and Ascaris Suum and Its Implication for the Molecular Detection of Benzimidazole Resistance. PloS Negl Trop Dis (2021) 15(9):e0009777. doi: 10.1371/journal.pntd.0009777
8. Martis MM, Tarbiat B, Tydén E, Jansson DS, Höglund J. RNA-Seq De Novo Assembly and Differential Transcriptome Analysis of the Nematode Ascaridia Galli in Relation to In Vivo Exposure to Flubendazole. PloS One (2017) 12(11):e0185182. doi: 10.1371/journal.pone.0185182
9. Martin F, Halvarsson P, Delhomme N, Höglund J, Tydén E. Exploring the β-Tubulin Gene Family in a Benzimidazole-Resistant Parascaris Univalens Population. Int J Parasitol Drugs Drug Resist (2021) 17:84–91. doi: 10.1016/j.ijpddr.2021.08.004
10. Kwa MSG, Veenstra JG, Roos MH. Benzimidazole Resistance in Haemonchus Contortus Is Correlated With a Conserved Mutation at Amino Acid 200 in β-Tubulin Isotype 1. Mol Biochem Parasitol (1994) 63(2):299–303. doi: 10.1016/0166-6851(94)90066-3
11. Whittaker JH, Carlson SA, Jones DE, Brewer MT. Molecular Mechanisms for Anthelmintic Resistance in Strongyle Nematode Parasites of Veterinary Importance. J Vet Pharmacol Ther (2017) 40(2):105–15. doi: 10.1111/jvp.12330
12. Silvestre A, Cabaret J. Mutation in Position 167 of Isotype 1 β-Tubulin Gene of Trichostrongylid Nematodes: Role in Benzimidazole Resistance? Mol Biochem Parasitol (2002) 120(2):297–300. doi: 10.1016/S0166-6851(01)00455-8
13. Ghisi M, Kaminsky R, Mäser P. Phenotyping and Genotyping of Haemonchus Contortus Isolates Reveals a New Putative Candidate Mutation for Benzimidazole Resistance in Nematodes. Vet Parasitol (2006) 144(3–4):313–20. doi: 10.1016/j.vetpar.2006.10.003
14. Avramenko RW, Redman EM, Windeyer C, Gilleard JS. Assessing Anthelmintic Resistance Risk in the Post-Genomic Era: A Proof-of-Concept Study Assessing the Potential for Widespread Benzimidazole-Resistant Gastrointestinal Nematodes in North American Cattle and Bison. Parasitology (2020) 147(8):897–906. doi: 10.1017/S0031182020000426
15. Avramenko RW, Redman EM, Melville L, Bartley Y, Wit J, Queiroz C, et al. Deep Amplicon Sequencing as a Powerful New Tool to Screen for Sequence Polymorphisms Associated With Anthelmintic Resistance in Parasitic Nematode Populations. Int J Parasitol (2019) 49(1):13–26. doi: 10.1016/j.ijpara.2018.10.005
16. Redman E, Whitelaw F, Tait A, Burgess C, Bartley Y, Skuce PJ, et al. The Emergence of Resistance to the Benzimidazole Anthlemintics in Parasitic Nematodes of Livestock Is Characterised by Multiple Independent Hard and Soft Selective Sweeps. PloS Negl Trop Dis (2015) 9(2):1–24. doi: 10.1371/journal.pntd.0003494
17. Mohammedsalih KM, Krücken J, Khalafalla A, Bashar A, Juma F-R, Abakar A, et al. New Codon 198 β-Tubulin Polymorphisms in Highly Benzimidazole Resistant Haemonchus Contortus From Goats in Three Different States in Sudan. Parasit Vectors (2020) 13(1):1–15. doi: 10.1186/s13071-020-3978-6
18. Robinson MW, McFerran N, Trudgett A, Hoey L, Fairweather I. A Possible Model of Benzimidazole Binding to β-Tubulin Disclosed by Invoking an Inter-Domain Movement. J Mol Graph Model (2004) 23(3):275–84. doi: 10.1016/j.jmgm.2004.08.001
19. Aguayo-Ortiz R, Méndez-Lucio O, Romo-Mancillas A, Castillo R, Yépez-Mulia L, Medina-Franco JL, et al. Molecular Basis for Benzimidazole Resistance From a Novel β-Tubulin Binding Site Model. J Mol Graph Model (2013) 45:26–37. doi: 10.1016/j.jmgm.2013.07.008
20. Jones BP, van Vliet AHM, La Course EJ, Betson M. Identification of Key Interactions of Benzimidazole Resistance-Associated Amino Acid Mutations in Ascaris β-Tubulins by Molecular Docking Simulations. bioRxiv (2021) 2021:11.03.467184. doi: 10.1101/2021.11.03.467184
21. Tarbiat B, Jansson DS, Tydén E, Höglund J. Evaluation of Benzimidazole Resistance Status in Ascaridia Galli. Parasitology (2017) 144(10):1338–45. doi: 10.1017/S0031182017000531
22. Collins JB, Jordan B, Baldwin L, Hebron C, Paras K, Vidyashankar AN, et al. Resistance to Fenbendazole in Ascaridia Dissimilis, an Important Nematode Parasite of Turkeys. Poult Sci (2019) 98(11):5412–5. doi: 10.3382/ps/pez379
23. Keiser J, Utzinger J. Efficacy of Current Drugs Against Soil-Transmitted Helminth Infections. JAMA (2008) 299(16):1937–48. doi: 10.1001/jama.299.16.1937
24. Suteno E, Pasaribu AP, Husin N, Wijaya W, Pasaribu S. Efficacy of Albendazole and Albendazole-Mebendazole Against Trichuris Trichiura Infections. J Med Sci (2020) 8(B):1162–6. doi: 10.3889/oamjms.2020.5329
25. Adegnika AA, Lötsch F, Mba RMO, Ramharter M. Update on Treatment and Resistance of Human Trichuriasis. Curr Trop Med Rep (2015) 2(4):218–23. doi: 10.1007/s40475-015-0061-z
26. Mekonnen Z, Levecke B, Boulet G, Bogers JP, Vercruysse J. Efficacy of Different Albendazole and Mebendazole Regimens Against Heavy Intensity Trichuris Trichiura Infections in School Children, Jimma Town, Ethiopia. Pathog Glob Health (2013) 107(4):207–9. doi: 10.1179/2047773213Y.0000000092
27. Diawara A, Halpenny CM, Churcher TS, Mwandawiro C, Kihara J, Kaplan RM, et al. Association Between Response to Albendazole Treatment and β-Tubulin Genotype Frequencies in Soil-Transmitted Helminths. PloS Negl Trop Dis (2013) 7(5):e2247. doi: 10.1371/journal.pntd.0002247
28. Diawara A, Drake LJ, Suswillo RR, Kihara J, Bundy DAP, Scott ME, et al. Assays to Detect β-Tubulin Codon 200 Polymorphism in Trichuris Trichiura and Ascaris Lumbricoides. PloS Negl Trop Dis (2009) 3(3):e397. doi: 10.1371/journal.pntd.0000397
29. Diawara A, Schwenkenbecher JM, Kaplan RM, Prichard RK. Molecular and Biological Diagnostic Tests for Monitoring Benzimidazole Resistance in Human Soil-Transmitted Helminths. Am J Trop Med Hyg (2013) 88(6):1052–61. doi: 10.4269/ajtmh.12-0484
30. Furtado LFV, Bello ACP de P, dos Santos HA, Carvalho MRS, Rabelo ÉML. First Identification of the F200Y SNP in the β-Tubulin Gene Linked to Benzimidazole Resistance in Ancylostoma Caninum. Vet Parasitol (2014) 206(3–4):313–6. doi: 10.1016/j.vetpar.2014.10.021
31. Furtado LFV, de Paiva Bello ACP, Rabelo ÉML. Benzimidazole Resistance in Helminths: From Problem to Diagnosis. Acta Trop (2016) 162:95–102. doi: 10.1016/j.actatropica.2016.06.021
32. Aguayo-Ortiz R, Méndez-Lucio O, Medina-Franco JL, Castillo R, Yépez-Mulia L, Hernández-Luis F, et al. Towards the Identification of the Binding Site of Benzimidazoles to β-Tubulin of Trichinella Spiralis: Insights From Computational and Experimental Data. J Mol Graph Model (2013) 41:12–9. doi: 10.1016/j.jmgm.2013.01.007
33. Howe KL, Bolt BJ, Cain S, Chan J, Chen WJ, Davis P, et al. WormBase 2016: Expanding to Enable Helminth Genomic Research. Nucleic Acids Res (2015) 44(D1):D774–80. doi: 10.1093/nar/gkv1217
34. Howe KL, Bolt BJ, Shafie M, Kersey P, Berriman M. WormBase ParaSite – A Comprehensive Resource for Helminth Genomics. Mol Biochem Parasitol (2017) 215:2–10. doi: 10.1016/j.molbiopara.2016.11.005
35. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic Local Alignment Search Tool. J Mol Biol (1990) 215(3):403–10. doi: 10.1016/S0022-2836(05)80360-2
36. Sayers EW, Bolton EE, Brister JR, Canese K, Chan J, Comeau DC, et al. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res (2022) 50(D1):D20–6. doi: 10.1093/nar/gkab1112
37. Hall TA. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/Nt. Nucleic Acids Symp Ser (1999) 41:95–8.
38. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis Across Computing Platforms. Mol Biol Evol (2018) 35(6):1547. doi: 10.1093/molbev/msy096
39. Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res (2018) 46(W1):W296–303. doi: 10.1093/nar/gky427
40. Bienert S, Waterhouse A, De Beer TAP, Tauriello G, Studer G, Bordoli L, et al. The SWISS-MODEL Repository-New Features and Functionality. Nucleic Acids Res (2016) 45:313–9. doi: 10.1093/nar/gkw1132
41. Bueno O, Estévez Gallego J, Martins S, Prota AE, Gago F, Gómez-Sanjuan A, et al. High-Affinity Ligands of the Colchicine Domain in Tubulin Based on a Structure-Guided Design. Sci Rep (2018) 8(1):1–17. doi: 10.1038/s41598-018-22382-x
42. Wiederstein M, Sippl MJ. ProSA-Web: Interactive Web Service for the Recognition of Errors in Three-Dimensional Structures of Proteins. Nucleic Acids Res (2007) 35(SUPPL.2):407–10. doi: 10.1093/nar/gkm290
43. Sippl MJ. Recognition of Errors in Three-Dimensional Structures of Proteins. Proteins Struct Funct Genet (1993) 17(4):355–62. doi: 10.1002/prot.340170404
44. Luthy R, Bowie JU, Eisenberg D. Assessment of Protein Models With Three-Dimensional Profiles. Nature (1992) 356(6364):83–5. doi: 10.1038/356083a0
45. Bowie JU, Luthy R, Eisenberg D. A Method to Identify Protein Sequences That Fold Into a Known Three-Dimensional Stucture. Science (1991) 253(5016):164–70. doi: 10.1126/science.1853201
46. Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A Program to Check the Stereochemical Quality of Protein Structures. J Appl Crystallogr (1993) 26(2):283–91. doi: 10.1107/S0021889892009944
47. Krieger E, Joo K, Lee J, Lee J, Raman S, Thompson J, et al. Improving Physical Realism, Stereochemistry, and Side-Chain Accuracy in Homology Modeling: Four Approaches That Performed Well in CASP8. Proteins Struct Funct Bioinforma (2009) 77(SUPPL. 9):114–22. doi: 10.1002/prot.22570
48. ULC CCG. Molecular Operating Environment (MOE). Montreal, Canada: Chemical Computing Group ULC (2020).
49. Özben M, Von Samson-Himmelstjerna G, Freiin Von Streit MKB, Wilkes EJA, Hughes KJ, Krücken J. Absence of Polymorphisms in Codons 167, 198 and 200 of All Seven β-Tubulin Isotypes of Benzimidazole Susceptible and Resistant Parascaris Spp. Specimens From Australia. Pathog (2022) 11:490. doi: 10.3390/pathogens11050490
50. Armstrong SK, Woodgate RG, Gough S, Heller J, Sangster NC, Hughes KJ. The Efficacy of Ivermectin, Pyrantel and Fenbendazole Against Parascaris Equorum Infection in Foals on Farms in Australia. Vet Parasitol (2014) 205(3–4):575–80. doi: 10.1016/j.vetpar.2014.08.028
51. Alanazi AD, Mukbel RM, Alyousif MS, AlShehri ZS, Alanazi IO, Al-Mohammed HI. A Field Study on the Anthelmintic Resistance of Parascaris Spp. In Arab Foals in the Riyadh Region, Saudi Arabia. Vet Q (2017) 37(1):200–5. doi: 10.1080/01652176.2017.1334981
52. Ellis GC, Phillips JB, O’Rourke S, Lyczak R, Bowerman B. Maternally Expressed and Partially Redundant β-Tubulins in Caenorhabditis Elegans are Autoregulated. J Cell Sci (2004) 117(3):457–64. doi: 10.1242/jcs.00869
53. Dilks CM, Hahnel SR, Sheng Q, Long L, McGrath PT, Andersen EC. Quantitative Benzimidazole Resistance and Fitness Effects of Parasitic Nematode Beta-Tubulin Alleles. Int J Parasitol Drugs Drug Resist (2020) 14:28–36. doi: 10.1016/j.ijpddr.2020.08.003
54. Melville LA, Redman E, Morrison AA, Rebecca Chen PC, Avramenko R, Mitchell S, et al. Large Scale Screening for Benzimidazole Resistance Mutations in Nematodirus Battus, Using Both Pyrosequence Genotyping and Deep Amplicon Sequencing, Indicates the Early Emergence of Resistance on UK Sheep Farms. Int J Parasitol Drugs Drug Resist (2020) 12:68–76. doi: 10.1016/j.ijpddr.2020.03.001
55. Chaudhry U, Redman EM, Raman M, Gilleard JS. Genetic Evidence for the Spread of a Benzimidazole Resistance Mutation Across Southern India From a Single Origin in the Parasitic Nematode Haemonchus Contortus. Int J Parasitol (2015) 45(11):721–8. doi: 10.1016/j.ijpara.2015.04.007
56. Elard L, Humbert JF. Importance of the Mutation of Amino Acid 200 of the Isotype 1 β-Tubulin Gene in the Benzimidazole Resistance of the Small-Ruminant Parasite Teladorsagia Circumcincta. Parasitol Res (1999) 85(6):452–6. doi: 10.1007/s004360050577
57. Rufener L, Kaminsky R, Mäser P. In Vitro Selection of Haemonchus Contortus for Benzimidazole Resistance Reveals a Mutation at Amino Acid 198 of β-Tubulin. Mol Biochem Parasitol (2009) 168(1):120–2. doi: 10.1016/j.molbiopara.2009.07.002
58. Adegnika AA, Zinsou JF, Issifou S, Ateba-Ngoa U, Kassa RF, Feugap EN, et al. Randomized, Controlled, Assessor-Blind Clinical Trial to Assess the Efficacy of Single-Versus Repeated-Dose Albendazole to Treat Ascaris Lumbricoides, Trichuris Trichiura, and Hookworm Infection. Antimicrob Agents Chemother (2014) 58(5):2535–40. doi: 10.1128/AAC.01317-13
59. Moser W, Schindler C, Keiser J. Efficacy of Recommended Drugs Against Soil Transmitted Helminths: Systematic Review and Network Meta-Analysis. BMJ (2017) 358:4307. doi: 10.1136/bmj.j4307
60. Matamoros G, Rueda MM, Rodríguez C, Gabrie JA, Canales M, Fontecha G, et al. High Endemicity of Soil-Transmitted Helminths in a Population Frequently Exposed to Albendazole But No Evidence of Antiparasitic Resistance. Trop Med Infect Dis (2019) 4(2):73. doi: 10.3390/tropicalmed4020073
61. Bennett AB, Anderson TJC, Barker GC, Michael E, Bundy DAP. Sequence Variation in the Trichuris Trichiura Beta-Tubulin Locus: Implications for the Development of Benzimidazole Resistance. Int J Parasitol (2002) 32(12):1519–28. doi: 10.1016/S0020-7519(02)00155-8
62. Hansen TV, Thamsborg SM, Olsen A, Prichard RK, Nejsum P. Genetic Variations in the Beta-Tubulin Gene and the Internal Transcribed Spacer 2 Region of Trichuris Species From Man and Baboons. Parasit Vectors (2013) 6(1):1–9. doi: 10.1186/1756-3305-6-236
63. Hansen TVA, Nejsum P, Olsen A, Thamsborg SM. Genetic Variation in Codons 167, 198 and 200 of the Beta-Tubulin Gene in Whipworms (Trichuris Spp.) From a Range of Domestic Animals and Wildlife. Vet Parasitol (2013) 193(1–3):141–9. doi: 10.1016/j.vetpar.2012.12.003
64. Rashwan N, Scott M, Prichard R. Rapid Genotyping of β-Tubulin Polymorphisms in Trichuris Trichiura and Ascaris Lumbricoides. PloS Negl Trop Dis (2017) 11(1):e0005205. doi: 10.1371/journal.pntd.0005205
65. Orr AR, Quagraine JE, Suwondo P, George S, Harrison LM, Dornas FP, et al. Genetic Markers of Benzimidazole Resistance Among Human Hookworms (Necator Americanus) in Kintampo North Municipality, Ghana. Am J Trop Med Hyg (2019) 100(2):351. doi: 10.4269/ajtmh.18-0727
66. Zuccherato LW, Furtado LF, Medeiros C da S, Pinheiro C da S, Rabelo ÉM. PCR-RFLP Screening of Polymorphisms Associated With Benzimidazole Resistance in Necator Americanus and Ascaris Lumbricoides From Different Geographical Regions in Brazil. PloS Negl Trop Dis (2018) 12(9):1–13. doi: 10.1371/journal.pntd.0006766
67. Castro PDJ, Howell SB, Schaefer JJ, Avramenko RW, Gilleard JS, Kaplan R. m. Multiple Drug Resistance in the Canine Hookworm Ancylostoma Caninum: An Emerging Threat? Parasit Vectors (2019) 12(1):576. doi: 10.1186/s13071-019-3828-6
Keywords: nematode, benzimidazole, drug resistance, molecular simulations, soil-transmitted helminth
Citation: Jones BP, van Vliet AHM, LaCourse EJ and Betson M (2022) In Silico Docking of Nematode β-Tubulins With Benzimidazoles Points to Gene Expression and Orthologue Variation as Factors in Anthelmintic Resistance. Front. Trop. Dis 3:898814. doi: 10.3389/fitd.2022.898814
Received: 17 March 2022; Accepted: 09 May 2022;
Published: 21 June 2022.
Edited by:
Matthew Denwood, University of Copenhagen, DenmarkReviewed by:
John Stuart Gilleard, University of Calgary, CanadaCopyright © 2022 Jones, van Vliet, LaCourse and Betson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Martha Betson, bS5iZXRzb25Ac3VycmV5LmFjLnVr
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.