
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Trop. Dis. , 11 February 2022
Sec. Vector Biology
Volume 3 - 2022 | https://doi.org/10.3389/fitd.2022.842523
This article is part of the Research Topic Insights in Vector Biology: 2021 View all 8 articles
Host association is among the most important factors that drive the transmission dynamics of mosquito-vectored pathogens. Competent vectors that feed exclusively upon non-competent hosts will not transmit pathogens, and highly competent hosts that are not bitten by competent vectors will not contribute to pathogen amplification. Therefore, characterizing the blood-feeding ecology of vector mosquitoes is critical to understanding how zoonotic pathogens amplify within ecosystems and spillover to humans and domesticated animals. In North America, mosquito species of the subgenus Culex are considered the most important vectors of zoonotic Flaviviruses, particularly West Nile virus (WNV), St. Louis encephalitis virus (SLEV), and western equine encephalitis virus. Many species of the Culex subgenus Culex are thought to feed predominantly upon birds and mammals, a behavior that facilitates the amplification and spillover of these zoonotic pathogens. Much of our understanding of the host associations of Culex vectors is based on research conducted in the 1960s and 1970s that used serological methods to infer host group(s). Here we reevaluate host associations of six Culex species from the southern US (Florida and Arizona) using DNA barcoding-based blood meal analysis. Our results demonstrate that reptiles, particularly lizards, constitute an important, and previously underappreciated, group of vertebrate hosts for several subgenus Culex mosquitoes. In Florida, >25% of Culex nigripalpus blood meals were derived from lizards (mainly Anolis spp.), and reptile host use generally increased from north to south with ~10%, ~25% and ~60% of Cx. nigripalpus blood meals derived from reptiles in northern, central, and southern Florida, respectively. In southern Arizona, lizards (mainly Sceloporus spp. and Urosaurus ornatus) constituted 40-45% of blood meals of Culex tarsalis, Culex thriambus, and Culex stigmatosoma. Other species of the subgenus Culex, including Culex quinquefasciatus, were not found to feed upon reptiles at the same sites, suggesting host association variation within Culex subgenus Culex. Whether or not lizards contribute to or dilute amplification of zoonotic Flaviviruses depends upon host competency of the lizard species bitten for WNV and SLEV. To date, very few studies have evaluated host competence of lizards for these viruses, so their roles in transmission cycles of zoonotic Flaviviruses remains obscure.
West Nile virus (WNV) and St. Louis encephalitis virus (SLEV) are pathogenic mosquito-borne zoonotic Flaviviruses that are primarily transmitted by subgenus Culex mosquitoes in North America. Passeriform (WNV) and columbiform birds (SLEV) are considered to be the major amplifying hosts of these zoonotic viruses (1). Several major vectors of WNV and SLEV, including Culex nigripalpus (2), Culex quinquefasciatus (3), and Culex tarsalis (4), feed heavily upon birds in the amplification phase of the virus’ transmission cycle (typically spring and early summer), then shift to feeding more heavily upon mammals in the epidemic/epizootic phase (late summer and fall). These seasonal shifts from avian to mammalian hosts by the vector mosquitoes, are thought to drive the amplification, as well as the spillover of these viruses to humans and other mammals (5). Therefore, the host associations of these Culex mosquitoes are critical determinants of WNV and SLEV transmission, and, by extension, the risk these viruses pose to public health in the US.
Our understanding of mosquito host associations has been heavily influenced by early (1960s – 1970s) large-scale studies using serology-based blood meal analysis. Working mainly in Florida and California, respectively, John D. Edman and Constantine H. Tempelis, analyzed tens of thousands of mosquito blood meals of numerous species to determine their vertebrate sources, typically identifying mosquito hosts to class or family. Their methodology for blood meal analyses relied largely on the precipitin test, in which blood from a vertebrate animal was injected into a laboratory animal, typically rabbit or game bird, to produce “anti-sera” that reacts when combined with blood from the original vertebrate animal and related species (6, 7). In this way, blood-engorged mosquito homogenates could be diluted and aliquoted into antisera from diverse vertebrate groups with serological reactions indicating an association. Importantly, the sensitivity of the assays was dependent upon the relatedness of organisms, such that cross reactivity was desired within members of a related taxonomic group (family or order, for example) but not between classes. These differences in antisera reactiveness have influenced our understanding of the host associations of important mosquito vectors, including the Culex vectors of WNV and SLEV.
Reptiles have been historically overlooked as vertebrate hosts for vector mosquitoes. In their 25-page review of mosquito host blood meal identification, Washino and Tempelis (8) mention reptiles just once, in reference to the difficulty obtaining “sufficient animal sera to prepare the necessary antibody … that will react with small reptiles, birds, or mammals”. The importance of this statement is reinforced by Edman (2, 9) and Tempelis et al. (4) in their papers on host feeding patterns of Culex spp. mosquitoes. In Florida and California, respectively, Edman (2) and Tempelis et al. (4), found that Cx. quinquefasciatus, Cx. nigripalpus and Cx. tarsalis feed almost exclusively from birds and mammals. Less than 1% of blood meals from any of these three major vectors of WNV and SLEV were determined to originate from reptiles or amphibians. Importantly, both Edman (2) and Tempelis et al. (4) used Iguana iguana (green iguana) as the source species for producing anti-sera (6, 7), a species that was not breeding on mainland USA at that time (10). In his ultimate paper on the blood-feeding patterns of Florida mosquitoes (9), Edman revealed that Iguana antisera “reacted poorly with the blood of local lizards”, presumably Anolis spp. lizards. Iguana iguana was likely chosen for their availability in the pet trade and large size, as the local lizards “were too small to obtain sufficient blood for antibody production”. It is therefore unclear to what extent major vectors of zoonotic Flaviviruses feed upon the blood of reptiles, and what implications this may have for the transmission of WNV and SLEV in the US.
Recent research in Lyme disease transmission dynamics has demonstrated that lizards can be consequential organisms for the transmission systems of arthropod-transmitted pathogens. Borrelia burgdorferi is a tick-borne pathogen that causes Lyme disease. In the US, Lyme disease is most prevalent in northern states, while rare in the southeastern states, despite the presence of Borrelia burgdorferi, and its tick vectors and rodent amplifying hosts. This pattern in the geographic prevalence of Lyme is linked, in part, to the host associations of tick vectors: in the southern US, ticks selectively attach to lizards, which are poor amplifiers of B. burgdorferi, leading to fewer bites on competent rodent amplifying hosts, fewer ticks infected with B. burgdorferi and thus, reduced incidence of human disease (11). Lizards could exert a similarly consequential effect within the transmission systems of mosquito-vectored pathogens, particularly WNV and SLEV, if the host associations of their primary vectors encompass lizards, and the lizard species that are fed upon are poor amplifiers of these viruses.
Here we re-investigate the host associations of Culex (Culex) spp. mosquitoes in two southern US states (Florida and Arizona) using polymerase chain reaction (PCR)- and sequencing-based blood meal analysis. We show that lizards are important hosts for several Culex spp., including major zoonotic Flavivirus vectors. We investigate the potential effects of location and season on host use and discuss how determining host competence of a few preferred lizard species could shed light on the potential suppressive effect of lizards on arbovirus transmission.
Female mosquitoes for these analyses were collected from numerous localities, using several methods, over a seven-year period (Table 1 and Supplemental Table 1). Our intent was not to intensively sample at any one location, but to gather samples from diverse settings, minimizing biases that occur due to method, habitat, or season. In Florida, mosquitoes were collected from five counties, spanning northern (Alachua, Levy), central (Indian River, Pinellas) and southern (Miami-Dade) portions of the state. In Arizona, mosquitoes were collected in the Sky Islands Region from the three southeasternmost contiguous counties (Cochise, Pima, Santa Cruz), in or near the Huachuca, Pajarito and Santa Rita Mountain Ranges. Mosquitoes were sampled primarily with a large-diameter aspirator (12), resting shelters (13), or carbon dioxide-baited light traps. In Florida, mosquitoes were generally sampled during all seasons, while in Arizona sampling only occurred during summer months (July and August), corresponding with the onset of the North American monsoon.
Collected mosquitoes were killed by exposure to low temperature, either by placing collection chambers containing mosquitoes into -20°C freezers for >10 minutes, or into dry ice-filled coolers. Samples were examined under a stereoscope and blood fed females were visually identified and separated from the sample. Blood fed female mosquitoes were identified morphologically with dichotomous keys (14, 15). To ensure correct identification of the morphologically similar Cx. stigmatosoma and Cx. thriambus, each specimen morphologically identified as these species was verified using molecular identification following the protocol described in Reeves et al. (16) for sequence-based mosquito identification using the DNA barcoding region of the cytochrome c oxidase subunit I gene (COI). All other Culex specimens were identified using morphological characters. After morphological identification of the mosquito species, each blood meal was individually preserved by smearing the contents of the mosquito abdomen onto the surface of a Whatman Flinders Technology Associates (FTA) Classic Card with a sterile pipette tip (17). Extent of digestion was visually estimated (18). The cards were stored at room temperature until molecular processing.
We used the Hot Sodium Hydroxide and Tris (HotSHOT) method (19) to extract DNA from the FTA Card-preserved blood meals, following the protocol described in Reeves et al. (18), with some adjustments to reagent volumes and incubation times. We removed two 1 mm (diameter) round punches from each blood meal using a hole-punch. Both punches were transferred to the same 0.2 ml tube. Thirty µl of lysis buffer were added to each tube, and the samples, with lysis buffer, were incubated in a thermocycler for three hours at 95°C followed by 4°C for five minutes. After incubation, 30 µl of neutralization buffer were added to each tube, and the tubes were stored at -20°C until PCR.
Extracted DNA was used as template in PCRs intended to amplify a fragment of the DNA barcoding region of the vertebrate COI gene. We applied a hierarchical approach to amplification of vertebrate templates using the Reeves et al. (18) primer combinations. Under this approach, extracted DNA from each blood meal was used in an initial PCR using the VertCOI_7194_F and Mod_RepCOI_R primer combination. The PCR products were electrophoresed on a 1.5% agarose gel and visualized under a transilluminator. Reaction products exhibiting an amplicon of the expected size were sent to Eurofins Genomics (Louisville, Kentucky) for sequencing using the Sanger method (20). In cases in which a DNA template failed to produce an amplicon of the expected size, the DNA template was used in a second PCR with the Mod_RepCOI_F and VertCOI_7216_R primer combination. Of these, templates from any failed reaction were used in a third reaction using the Mod_RepCOI_F and Mod_RepCOI_R primer combination. If a template failed to produce an amplicon in all three reactions, no further steps were taken, and the blood meal was not identified. All reactions were performed in a final volume of 20 µl consisting of 10 µl 2.0X Apex Taq RED Master Mix (Genesee Scientific Corp., San Diego, CA), 0.75 µl forward primer (10µM), 0.75 µl reverse primer (10µM), 7.5 µl ultra-pure water, and 1 µl extracted DNA. Thermal cycling conditions for all reactions consisted of 95°C for 3 minutes, followed by 40 cycles of 95°C for 40 seconds, 45°C for 30 seconds, and 72°C for 3 minutes, with a final extension step of 72°C for 7 minutes.
Resulting vertebrate COI sequence chromatograms were examined and edited for quality using the bioinformatic software Geneious Prime Version 11.0.3. Unambiguous stretches of sequences were submitted to the Barcode of Life Datasystem (BOLD) v. 4 Identification Engine for species-level identification by alignment to reference sequences (21). Sequences that were 98-100% similar to reference sequences were considered conspecific with the species from which the reference sequences were derived. In a few cases, host COI sequences were poor matches (<98% similar) to reference sequences. Of these, nearly all were matched (98-100%) to independently collected reference sequences (18) for Sylvilagus palustris (marsh rabbit) or Anolis carolinensis (green anole). For S. palustris, reference sequences are not yet available on BOLD, while A. carolinensis has high levels of intraspecific variation with some geographic structuring (22) and introgression via hybridization with introduced Anolis porcatus (Cuban green anole) from Cuba (23). Sequences that were 98-100% similar to independently collected S. palustris or A. carolinensis reference sequences were considered conspecific with the corresponding species. Odocoileus virginianus (white-tailed deer) and Odocoileus hemionus (mule deer) could not be distinguished molecularly, but all sequences derived from these hosts were assumed to be O. virginianus based on geographic distribution and the absence of O. hemionus occuring naturally in Florida.
We compiled lists of species of birds, mammals, reptiles, and amphibians recorded from Florida and Arizona to explore patterns of diversity and host use as it relates to availability of vertebrate hosts, the number of species utilized by Culex mosquitoes, and status (exotic, domesticated or native) from published sources (24–31). We used chi-square tests of independence to test for differences in the distributions of available hosts and blood meals between location and season. Rank abundance curves (32) were plotted to visualize the relative abundance of host species detected for Cx. nigripalpus, Cx. quinquefasciatus, Cx. thriambus, the three mosquito species for which more than 100 blood meals were successfully identified. Rank abundance curves were plotted using the Vegan (33) and BiodiversityR (34) packages in R.
A total of 1,166 blood-engorged female Culex (Culex) mosquitoes were analyzed by PCR, of which, 88.1% produced an identifiable sequence. The overall rate of PCR and sequencing success was high (85.3% on average), but varied by species and location, the lowest being Culex coronator from Indian River County (47.1%). Culex (Culex) spp. mosquitoes in Florida and Arizona were found to feed upon diverse species of birds, reptiles and mammals, but, somewhat surprisingly, not amphibians (Table 2). Across Culex (Culex) species, birds were a major host group in both Florida and Arizona (Table 2), constituting 69.2% and 44.2% of total identified blood meals, respectively. Mammals were also important hosts in both states (19.0 and 35.7%, in Florida and Arizona respectively) as were reptiles (11.8 and 20.1%, in Florida and Arizona respectively). The overall distribution of blood meals between the vertebrate classes differed significantly between Arizona and Florida (X2 = 109.96, df=2, P<0.001).
When vertebrates are examined according to ecological groupings, songbirds (order Passeriformes) and lizards (suborder Lacertilia) are notable groups that were important to our samples of Culex mosquitoes from both Florida and Arizona (Figure 1). In both states, songbirds outnumber all other vertebrate groups in terms of species available (species richness) and fraction of total blood meals (Figure 1). However, the number of blood meals per songbird species bitten is relatively low, a function of blood meals being spread over a large number of species. Lizards, while not the most species rich group of reptiles in either state (Figure 1), contributed relatively large fractions of total blood meals in Arizona (41.9%) and Florida (10.9%) across Culex (Culex) species. The numbers of blood meals per lizard species bitten were relatively high, a function of blood meals being focused on a relatively small number of species (Figure 1). Other vertebrate groups for which the number of blood meals per species bitten was relatively high included landfowl (mainly domestic chicken), carnivores (mainly raccoon), and marsupials (North American opossum).
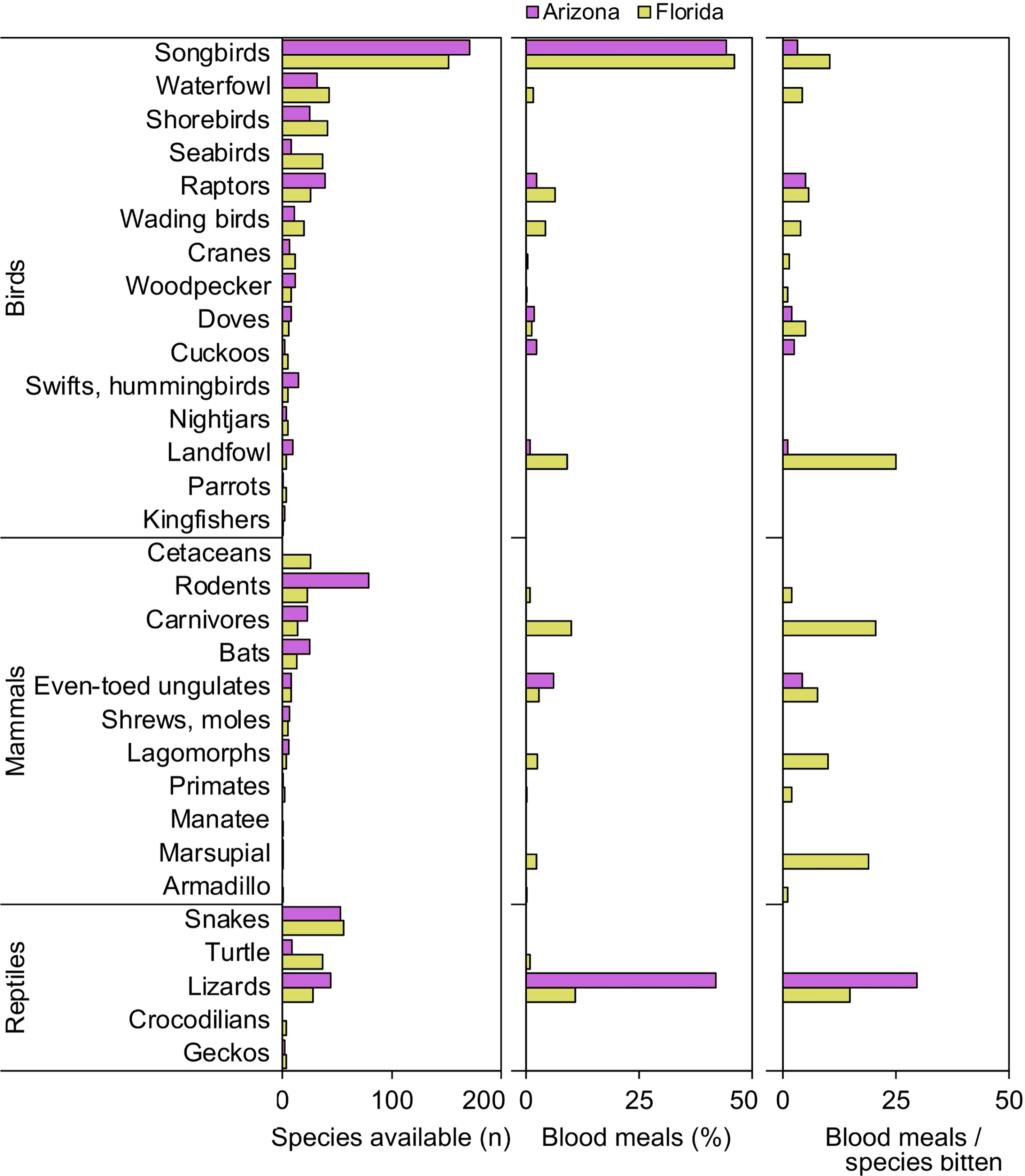
Figure 1 Availability and host use of vertebrate groups for Culex mosquitoes in Arizona and Florida, USA. Numbers of host species available in each ecological group were derived from published sources. Blood meal numbers were combined for 4-6 Culex (Culex) spp. in each state. Vertebrate host species were determined by PCR and Sanger sequencing of field-collected blood-engorged females. Data were combined from three counties (Arizona) and five counties (Florida).
In Florida, class-level host use varied between Culex species, location and, in some cases, season. Culex coronator and Culex interrogator fed exclusively upon birds and mammals (Table 2). Culex restuans blood meals were from birds, though sample size was small (n=2). Culex declarator, Cx. nigripalpus and Cx. quinquefasciatus fed upon birds, mammals, and reptiles to varying degrees (Table 2). Class-level host use of Cx. nigripalpus varied by season and location. The proportion of Cx. nigripalpus blood meals from reptiles generally increased from north to south in Florida (Figure 2), which corresponded with decreasing use of avian and mammalian hosts with decreasing latitude. Reptiles constituted just 9.0% (n=13/145) of total Cx. nigripalpus blood meals in northern Florida (Alachua County), for example, but 55.3% (n=52/94) in southern Florida (Miami-Dade County). In Alachua, Indian River and Miami-Dade Counties, Cx. nigripalpus fed predominantly on birds in winter and spring (Figure 3), but predominantly fed upon other host classes in other seasons. The distribution of Cx. nigripalpus blood meals between the vertebrate classes differed significantly between fall and winter in Alachua County (X2 = 7.92, df=2, P=0.019), in Indian River County and in Florida, across all three counties (X2 = 12.09, df=2, P=0.002), and between spring and summer (X2 = 14.25, df=2, P<0.001) and summer and fall (X2 = 17.34, df=2, P<0.001) in Miami-Dade County. Culex quinquefasciatus fed predominantly upon birds in Alachua, Indian River and Pinellas Counties, constituting 66.7 – 80% of all blood meals from these counties (Table 2). No seasonal shifts in host use were observed for Cx. quinquefasciatus however the distribution of class-level host use differed significantly between spring and summer (X2 = 10.57, df=2, P=0.005) and winter and spring (X2 = 8.73, df=2, P=0.013) in Alachua County (Figure 3). Relatively small sample sizes for some species in Florida (2 Cx. restuans, 11 Cx. declarator, 6 Cx. interrogator) likely affect the probability of detecting blood meals from other vertebrate classes and limit the ability to draw conclusions about the host associations of these species.
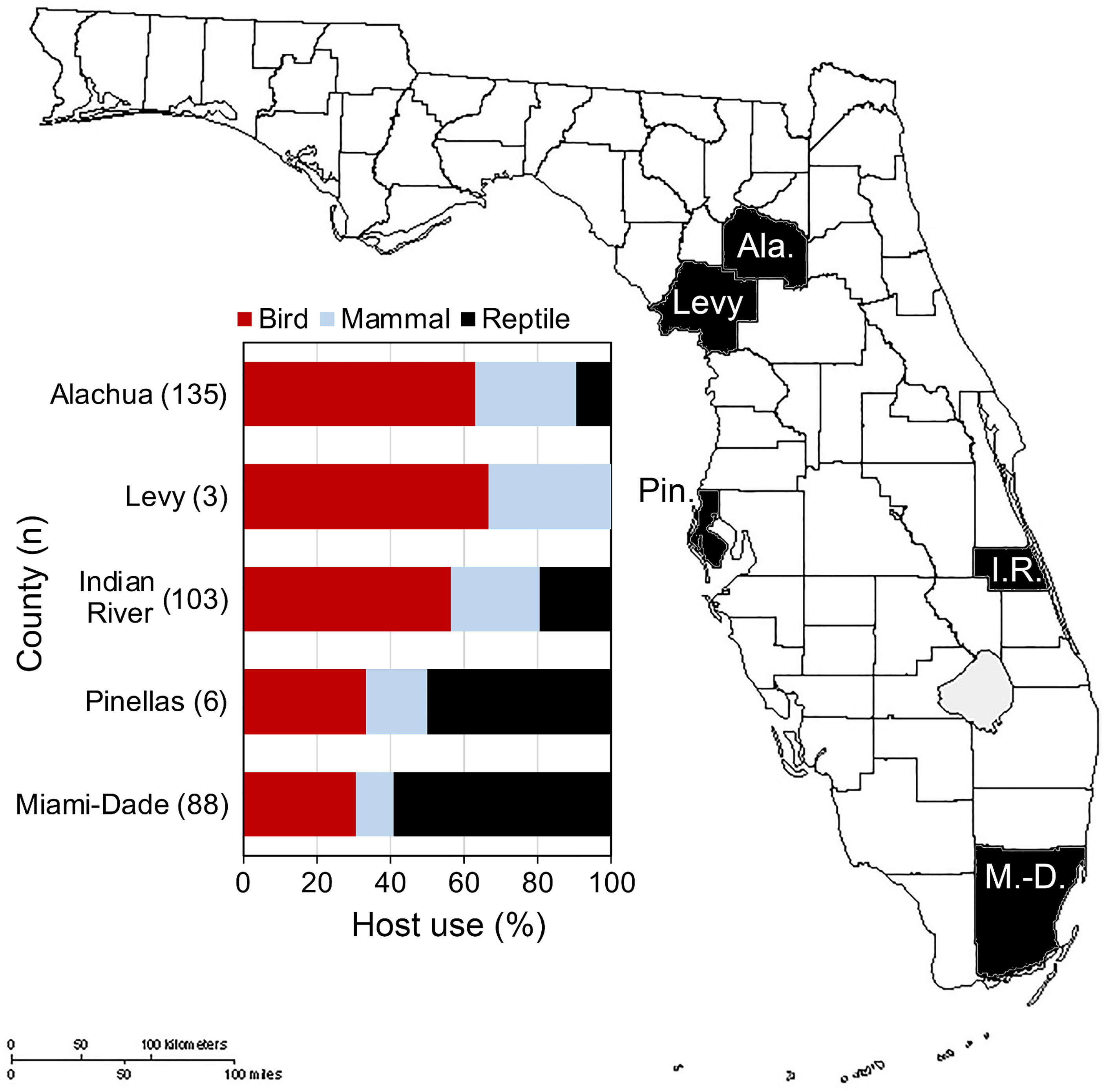
Figure 2 Class-level host use of Culex nigripalpus in Florida. Vertebrate host species were determined by PCR and Sanger sequencing of field-collected blood-engorged females from five counties.
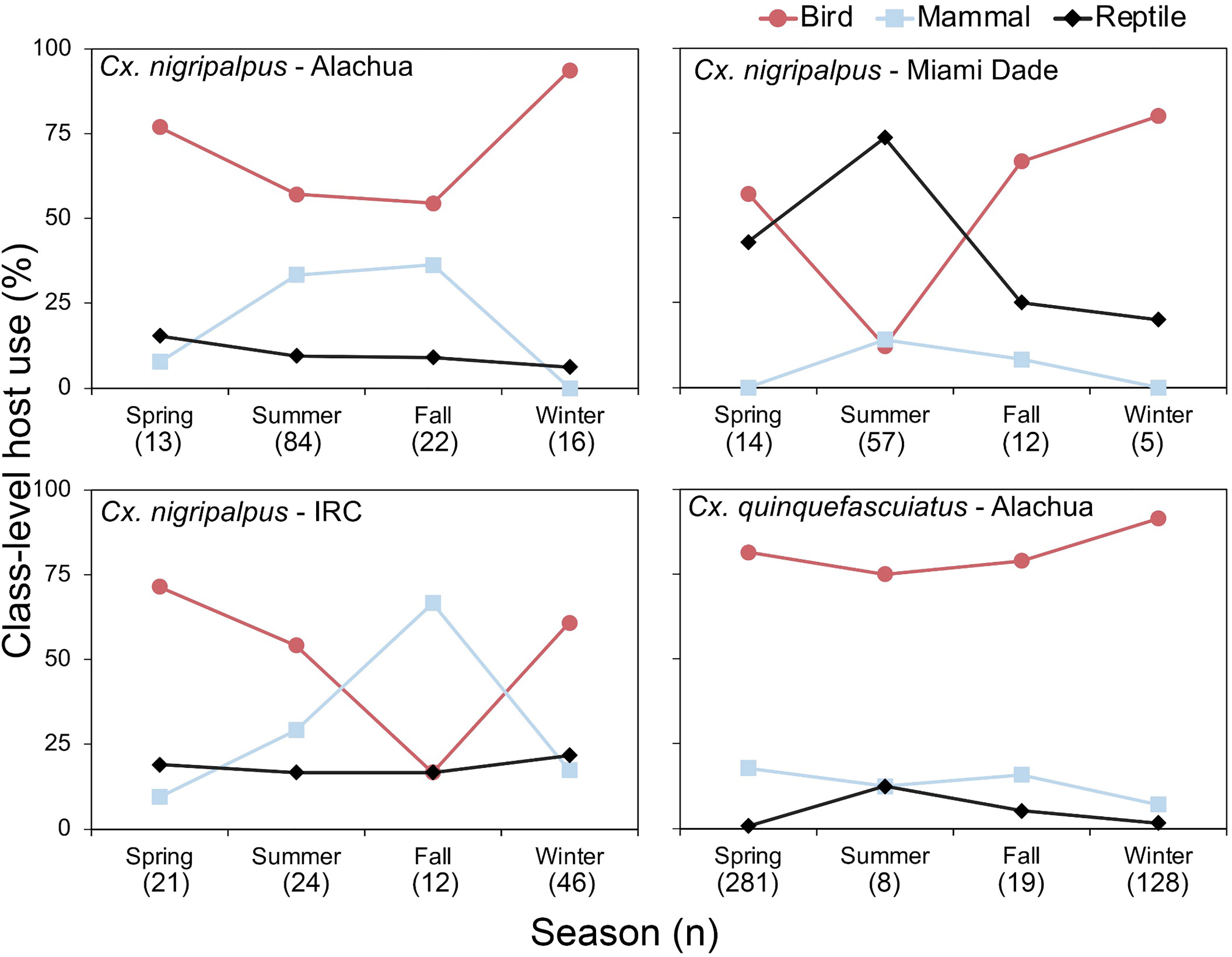
Figure 3 Seasonal patterns of class-level host use of zoonotic Flavivirus vector mosquitoes in Florida. Vertebrate host species were determined by PCR and Sanger sequencing of field-collected blood-engorged females of Culex nigripalpus and Culex quinquefasciatus. Season dates correspond to astronomical calendar.
In Arizona, class-level host use did not vary significantly between counties for mosquito species, with the exception of Cx. thriambus (X2 = 28.98, df=4, P<0.001), which took more total blood meals from reptiles in Cochise County (50.6%) and Pima County (46.0%) than in Santa Cruz County (14.5%). Culex thriambus took a substantial fraction of blood meals from birds in all three counties, ranging from 30.2 to 63.2% of total blood meals (Table 2), while mammals contributed <2% of blood meals. Culex tarsalis and Cx. stigmatosoma were only found to feed upon birds and reptiles (Table 2), although sample sizes for these species were relatively low (19 Cx. stigmatosoma, 10 Cx. tarsalis). Culex erythrothorax fed mainly upon mammals (12 of 13 identified blood meals), with a single avian blood meal detected (Table 1).
Rank abundance curves of the host species identified from Cx. nigripalpus, Cx. quinquefasciatus, and Cx. thriambus, the three species for which sample size of identified blood meals was >100, indicated that these mosquitoes have strong hosts associations: a small number of host species contributed a majority of blood meals while a large number of host species were bitten occasionally (Figure 4). For all three Culex species, a single host species dominated the blood meal set and was fed upon to a greater extent than all others (1.8 – 4.6 times more frequently than the second most frequently fed upon host species). For Cx. nigripalpus and Cx. thriambus, species of diurnal, tree-roosting lizards (Anolis sagrei, brown anole, and Sceloporus clarkii, Clark’s spiny lizard, respectively) were the most frequently detected hosts, constituting 1.8 – 2.3 times more blood meals than the second most frequently detected host (birds in both cases) for each mosquito species. The three most frequently fed upon hosts for Cx. nigripalpus were a lizard (Anolis sagrei, 60 detections), Gallus gallus (domestic chicken, 34 detections), and Procyon lotor (raccoon, 20 detections). For Cx. thriambus, lizard species were the first (Sceloporus clarkii, 56 detections) and third (Urosaurus ornatus, ornate tree lizard, 21 detections) most frequently detected species, while a songbird (Thryomanes bewickii, Bewick’s wren, 24 detections) was the second most frequently bitten host. Lizards were not among the most frequently bitten hosts for Cx. quinquefasciatus, and out of 445 identified blood meals, only three were derived from lizards (two from A. carolinensis, one from A. sagrei). Cardinalis cardinalis (northern cardinal), was by far the most frequently detected host species for Cx. quinquefasciatus (167 detections), followed by G. gallus and P. lotor (36 and 25 detections, respectively).
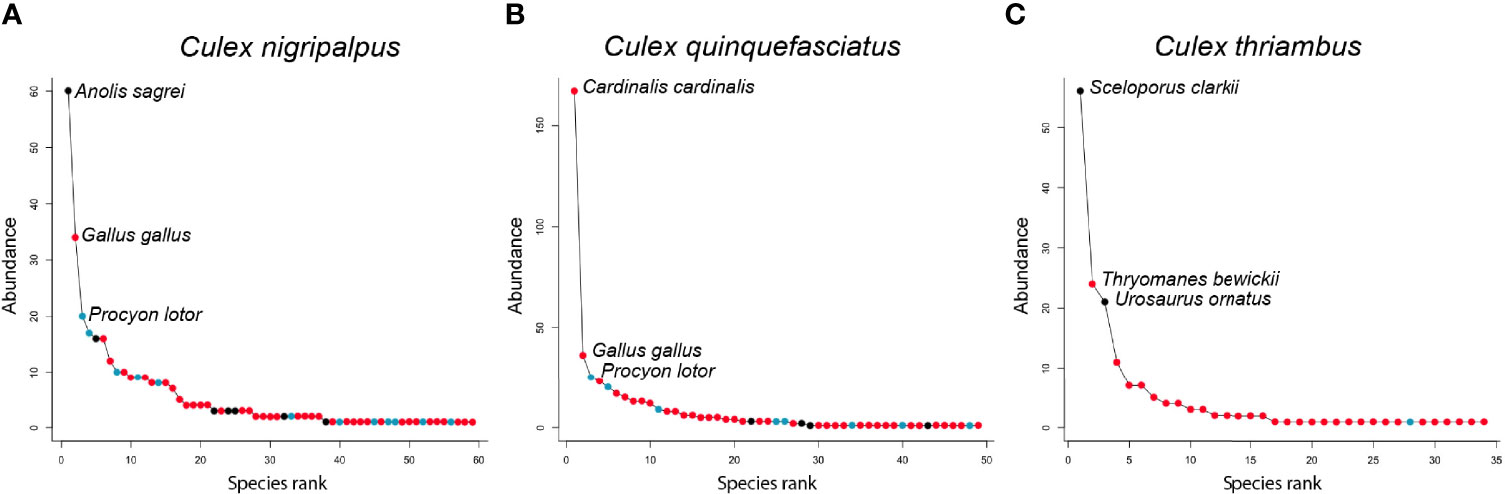
Figure 4 Rank abundance curves of host species detected by Culex nigripalpus (A), Culex quinquefasciatus (B) and Culex thriambus (C). Points on each curve indicate the abundance of a host species among the blood meal sample for each species, with coloring indicating vertebrate class of the host species: Aves (red), Mammalia (blue), Reptilia (black). Note that the y-axis scale differs between panels.
Exotic (nonnative and domesticated) animals contributed a substantial fraction of blood meals, particularly in Florida (Figure 5). Compared to the relative number of exotic species available, the fraction of blood meals from exotic birds and reptiles was greater than from the native species of their respective classes (Figure 5A). Exotic reptiles, in particular, constituted 75% of the total reptile blood meals in Florida, but make up 39% of available reptile species (Figure 5A; X2 = 37.91, df=1, P<0.001). Anolis sagrei was the most commonly bitten reptile, with 64.6% (n=62/96) of reptile blood meals arising from this nonnative lizard. Other nonnative reptiles bitten (three or less blood meals) include Anolis equestris (knight anole), I. iguana, Basiliscus vittatus (brown basilisk), Anolis cristatellus (Puerto Rican crested anole), and Python bivittatus (Burmese python). Domestic birds (mainly chickens) and domestic mammals (mainly house cat) contributed a relatively large proportion of blood meals in Florida, 15.4% and 26.9% of avian and mammalian blood meals, respectively. In contrast, domestic mammals (cow) contributed a relatively large percentage of blood meals derived from mammals (77.0%), whereas few blood meals were detected from exotic birds (0.9%) or reptiles (0%) in Arizona (Figure 5B).
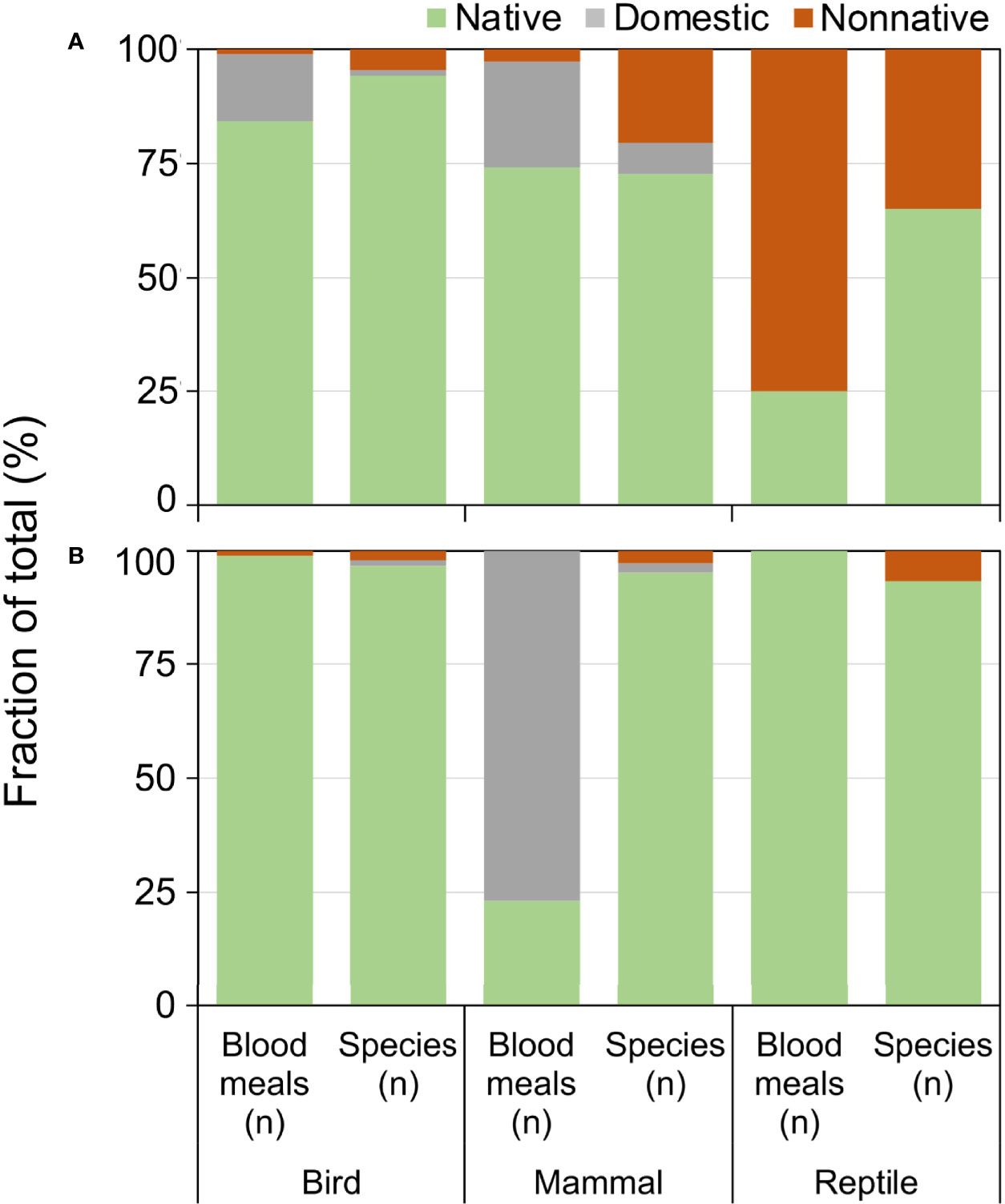
Figure 5 Relationship between availability and host use of exotic and native vertebrate classes for Culex (Culex) spp. mosquitoes collected in Florida (A) and Arizona (B). Numbers of host species available in each class were derived from published sources. Blood meal numbers were combined for 4-6 Culex (Culex) spp. in each state. Vertebrate host blood meal species were determined by PCR and Sanger sequencing of field-collected blood-engorged females. Data were combined from three counties (Arizona) and 5 counties (Florida).
The class-level patterns of host use of some Culex (Culex) spp. mosquitoes observed in this study are contradictory to findings of previous studies of their host associations. While birds and mammals are acknowledged as important host groups for well-studied vector species such as Cx. quinquefasciatus, Cx. nigripalpus and Cx. tarsalis, few studies have documented significant utilization of reptilian hosts by these same species (Table 2). The potential reasons for this disparity include inadequacy of assays (serological or molecular) to recognize or amplify reptile blood proteins or DNA targets, respectively (9), geographical variation in the US in reptile diversity and abundance, and/or shifts in host association due to recent introductions, establishments and geographic expansions of reptile species that colonized the US in recent decades. On average, studies that used PCR and sequencing-based blood meal analysis (including this study) found much higher reptile host use (20.0%) by Cx. nigripalpus than studies which employed serological methods (0.5%). This suggests that methodological limitations contribute to the disparity. Our own blood meal results (Figure 3) suggest that latitude and its inverse relationship with reptile diversity and abundance (35) may also contribute to the high level of reptile-biting observed in our own study, compared to most prior studies of subgenus Culex host use, which have largely focused on more northerly regions of the continental US. Interestingly, in Puerto Rico half (five of 10) of Cx. nigripalpus blood meals were from reptiles (36). Similarly, reptiles constituted a surprisingly large proportion of blood meals in Arizona (Table 1). Culex thriambus, for example, took 36.7% of 218 total blood meals from reptiles in southern Arizona. Previous studies of host use by this species found 96-100% of blood meals were derived from birds (37, 38) in two California counties. Similarly, Cx. tarsalis, an important arbovirus vector species, for which numerous blood meal studies have been published, took 40% (four of 10 blood meals) from reptiles in Arizona, while prior studies had reported between 1 and 5% of blood meals from reptiles (39–43). Fewer studies have been published on the host associations of Cx. stigmatosoma, but in California, McPhatter et al. (44) reported that 98.6%, 1.2% and 0.2% of 647 blood meals were derived from birds, mammals, and reptiles (Sceloporus occidentalis), respectively.
Our findings that lizards were one of the two most bitten host groups, in terms of percentage of total blood meals and blood meals per available species (Figure 1), indicates that lizards are important hosts for some vector mosquitoes, relative to other reptiles, and even most other vertebrate groups. The finding that the blood meals were focused on a relatively small number of lizard species indicates that a few species of lizard (A. sagrei in Florida, S. clarkii and U. ornatus in Arizona) have a disproportionately strong influence on host use patterns at locations where these species occur (Figure 1). For songbirds, however, blood meals were distributed across a large number of species (Figure 1), although C. cardinalis was an especially important host to Cx. quinquefasciatus (Figure 4). Therefore, determining host competence of songbirds in a region requires evaluating many species in laboratory experiments, sensu Komar et al. (45). For reptiles however, understanding the host competence of these three lizard species (A. sagrei, S. clarkii, U. ornatus) would provide valuable information on their role in amplifying or diluting arboviruses in nature and could help explain geographical patterns of zoonotic Flavivirus prevalence in North America, i.e., why incidence of WNV and SLEV have been historically lower in tropical regions than in temperate regions. As discussed above, Lyme disease prevalence is higher in northern states than in southern states, due in part to tick vectors selectively attaching to lizards in the southern US (11). Lizards are poor amplifiers of B. burgdorferi, leading to fewer bites on competent rodent amplifying hosts. If lizards are poor amplifiers of WNV and SLEV, they should have a similar suppressive impact on arbovirus transmission.
In Florida and Arizona, blood meals from lizards were derived from only a subset of the available lizard species in both states. In Florida (Figure 6), Culex mosquitoes fed from A. sagrei, A. carolinensis, A. equestris, A. cristatellus, B. vittatus, and I. iguana, while in Arizona, they fed only from S. clarkii, Sceloporus cowlesii (southwestern fence lizard), and U. ornatus. These lizards are all diurnal and, to varying extents, arboreal, associated with trees or vegetation (25, 27). Conversely, diurnal lizard species that are abundant but associated with the ground, rocks, or crevices were absent from our blood meal samples. In both states, there are abundant and relatively diverse terrestrial lizard communities that include Agama agama (African rock agama), Leiocephalus carinatus (northern curly-tailed lizard), Sceloporus undulatus (eastern fence lizard), Aspidoscelis sexlineata (six-lined racerunner) and various species of Eumeces skinks in Florida, and various species of Aspidoscelis whiptails, Phrynosoma horned lizards, and other Sceloporus species, among many others, in Arizona (25, 27). Similarly, blood meals derived from nocturnally active lizards (e.g., Heloderma suspectum, Coleonyx banded geckos, Hemidactylus house geckos) were not found. A possible explanation for the assemblage of lizard species detected in Culex blood meals, and the absence of ground-associated and nocturnal species, is that nocturnally active, host-seeking Culex females forage in trees or vegetation, where they encounter and feed from lizard species that roost on exposed branches or leaves, while they do not encounter those that rest underground or in crevices, or those that are active by night.
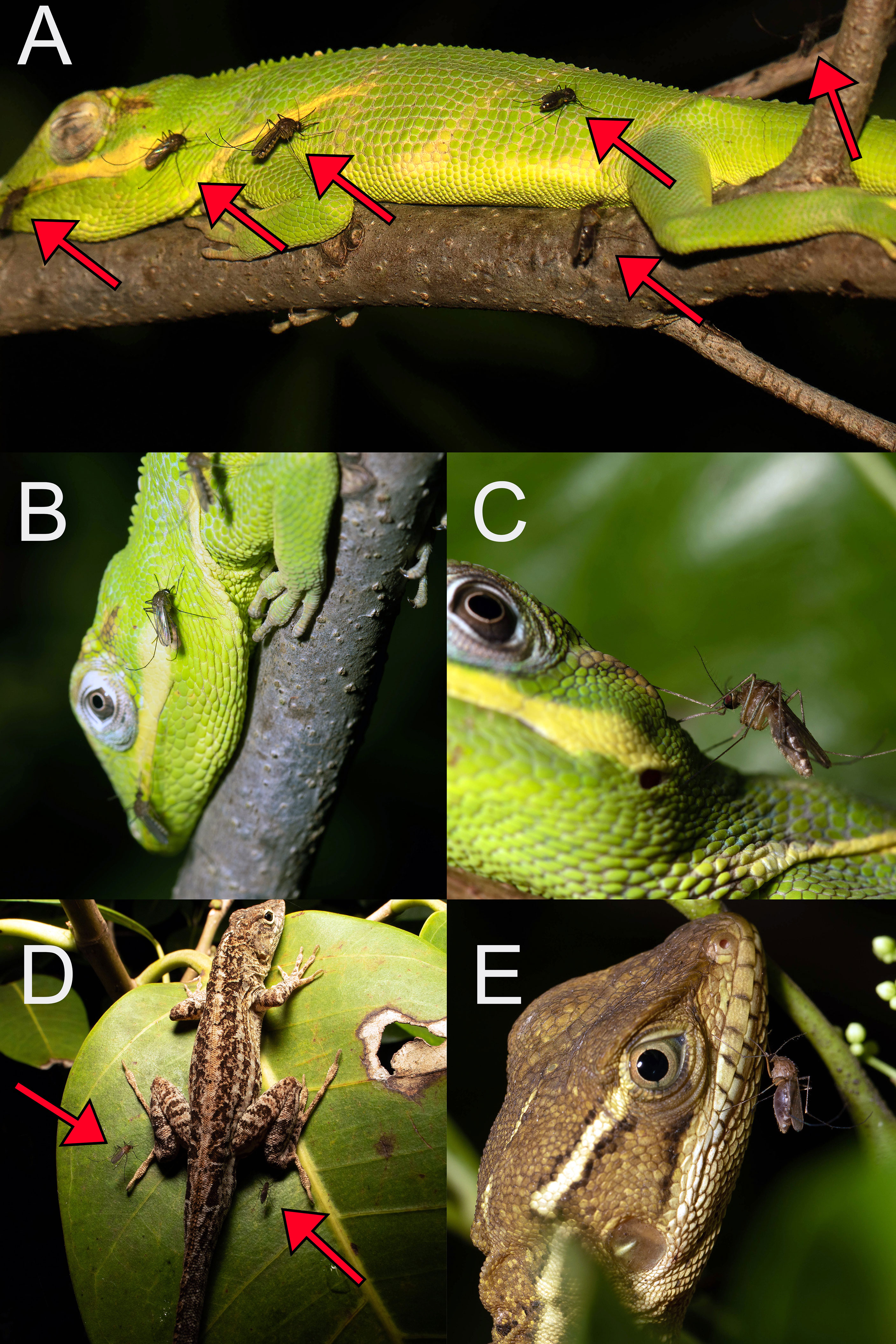
Figure 6 Adult female Culex nigripalpus mosquitoes blood feeding from non-native, diurnal lizards in Miami-Dade County, Florida, USA: Anolis equestris (A, B), 11 October 2015; Anolis equestris (C), 10 October 2015; Anolis sagrei (D), 9 July 2016; Basiliscus vittatus (E), 11 October 2015. Red arrows indicate position of Culex nigripalpus females on or near lizards.
Seasonal shifts in bird-to-mammal host use observed for Cx. nigripalpus in Indian River County, Florida (Figure 3) confirms findings of Edman and Taylor (46). Interestingly, this same vector species shifted seasonally between birds and reptiles in Miami-Dade County (Figure 3), suggesting that class-level host shifts are labile within a given species. Edman and Taylor (46) postulated that Cx. nigripalpus bird-to-mammal host shifts were driven by seasonal weather-driven changes in foraging behavior of the mosquitoes, i.e., that females forage in open fields in the late summer and fall, where they are more likely to encounter mammals. Our prior work with Culiseta melanura indicated that seasonal bird-to-reptile host shifts were due to weather-induced changes in host behavior, in that Anolis lizards were more likely to sleep on exposed surfaces such as vegetation or terminal branches of trees or shrubs when temperatures were higher (47), which is in agreement with the summertime peak of lizard biting in Miami-Dade County observed here (Figure 3). The county-level differences in class-level host use observed for several species is not unexpected, as several studies have reported these differences. For example, Thiemann et al. (42) found that avian host use by Cx. tarsalis varied from 51.5 to 96.0% between four California counties.
Our results demonstrate that introductions of vertebrates outside of their native distributions can affect the species-level host associations of mosquitoes, and may even affect the class-level host associations when a favored exotic species establishes in a new area. The disproportionately high fraction of blood meals derived from exotic vertebrates in Florida is reflective of the large number of invasive species in the state (48). The very large fraction of reptile blood meals from nonnative reptile species (75%) is due primarily to blood meals from A. sagrei. Anolis sagrei was the most important host in our sample of Cx. nigripalpus blood meals, and was fed upon nearly twice as often as the next most abundant host (Figure 4). This small lizard, native to Caribbean islands, has been in Florida since the 1880s, when they were first reported in the lower Florida Keys (49). By 2002, A. sagrei had been recorded from every Florida county, across a largely contiguous distribution (50) and has become one of the most abundant lizards in the state. Previous work, based on blood meals collected in the 1960s (7, 46) at a time when A. sagrei was still rare in Florida and had not yet become widespread and abundant in the state (25), forms the basis of our current understanding of the host associations of Cx. nigripalpus in the state. The timeline of invasion and dispersal of this preferred reptile species could explain the increase in reptilian host use by species of Culex (this study) and Culiseta (51) observed in Florida over the past 40-50 years. Not only has the colonization of Florida by A. sagrei provided mosquitoes with an omnipresent new vertebrate host, but the presence of this anole impacts the behavior of the native A. carolinensis, pushing them to higher perches within vegetation (52), which may affect their susceptibility to mosquito feeding.
The lizard species fed upon by Culex mosquitoes in Arizona were all native species. The relatively lower importance of exotic host species, overall, in Arizona is representative of the low numbers of invasive vertebrates in that state, compared to Florida. While the changing vertebrate host community of Florida with the colonization of the state by nonnative lizards may, in part, explain the incongruence of host associations between previous studies and this study, in Arizona, Culex host associations have not been thoroughly assessed. Blood meal data are available only for Cx. quinquefasciatus (n=143) and Cx. tarsalis (n=3) collected in residential and metropolitan areas around Phoenix, Maricopa Co. and Tucson, Pima Co. (53, 54). For Cx. quinquefasciatus, the results of these studies paralleled our results for Florida, with most blood meals derived from birds and, to a lesser extent, mammals, and less than 1% derived from lizards (U. ornatus), while all three Cx. tarsalis had fed from birds. Our sampling efforts for blood fed Culex in Arizona were exclusively focused on undeveloped areas of largely natural habitat in the Sky Islands Region, which, coupled with the paucity of information on Culex host associations in Arizona, may contribute to the novel finding that Cx. thriambus, Cx. stigmatosoma, and Cx. tarsalis feed from tree-associated lizards in these areas. Distribution and abundance patterns of diurnal and arboreal lizard species could further explain the high levels of lizard feeding we found across multiple sites in southern Arizona. Sceloporus clarkii has a limited distribution in the southwestern US (55), and U. ornatus, although more widespread, is largely absent from California (56), where much of the host association work has been focused for Cx. tarsalis, Cx. thriambus, and Cx. stigmatosoma (4, 37–40, 42, 44).
We found that host association varied among the Culex species we assessed, and not all Culex species fed from lizards to the same extent. For Cx. nigripalpus, Cx. stigmatosoma, Cx. tarsalis, and Cx. thriambus, lizards made up a large proportion (24–57%) of the identified blood meals, and diurnal/arboreal lizard species were the most important hosts for Cx. nigripalpus and Cx. thriambus. Lizard-derived blood meals were not found in Cx. interrogator, Cx. restuans, Cx. coronator, and Cx. erythrothorax, though for all, sample size was relatively small. While the former two species fed predominantly from birds, Cx. coronator and Cx. erythrothorax fed almost exclusively from mammals, findings that are in line with the results of previous studies (57, 58). Together, this suggests that Cx. coronator and Cx. erythrothorax are largely associated with mammalian hosts, with occasional use of avian hosts. For Cx. quinquefasciatus, an important Flavivirus vector in the US and elsewhere, our sample included 445 identified blood meals, derived almost entirely from birds, particularly C. cardinalis, and mammals. The majority of this sample (434/445 blood meals) was collected at the same site in Alachua Co., Florida where 13/139 (9%) Cx. nigripalpus blood meals were derived from lizards. At this site, only 3/434 (0.7%) identified Cx. quinquefasciatus blood meals were derived from lizards, suggesting that Cx. quinquefasciatus does not feed from lizards to the same extent as Cx. nigripalpus. This discrepancy could be due to innate preferences for certain host classes over others, to differences in foraging strategies or foraging times, or to differences in the importance of host-seeking cues between these species.
Recognizing the potential for nonnative vertebrate hosts to impact mosquito host associations and vector-borne disease transmission is important, as climate change is likely to increasingly facilitate establishments of nonnative species (59). Shift in vector distribution is typically considered the major implication of climate change for vector-borne disease [e.g., (60)]. However, as exotic mosquito vectors can change the transmission dynamics and public health risks of mosquito-vectored pathogens, so, too, can introductions of nonindigenous vertebrate hosts (61). Since Edman’s work on Culex host associations in the 1960s, Florida’s vertebrate community has transformed. Notably, there have been numerous establishments of nonindigenous lizard and other reptile species, to the extent that Florida ranks first in the world in number of nonnative reptile species (28). We found that in Florida many of these nonnative species, the diurnal and arboreal lizards, are important hosts for Cx. nigripalpus, but not for Cx. quinquefasciatus, both primary vectors of WNV and SLEV. In southern Arizona, an unexpectedly large proportion of Cx. thriambus, Cx. tarsalis, and Cx. stigmatosoma blood meals were derived from native lizard species, possibly due to the availability of these hosts in the ecosystems of our study sites. If lizards are found to be dilution hosts for WNV and SLEV, it may be possible to encourage or support populations of native diurnal/arboreal lizards to serve as dilution effect biological controls, particularly in metropolitan areas, with the intent of dampening circulation of these Flaviviruses among competent hosts.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
LER and NDB-C designed studies, performed fieldwork. LER performed molecular analysis. NDB-C and LER performed statistical analysis. Both authors drafted the manuscript and approved the final version.
This work was supported by State of Florida funds managed by the Florida Department of Agriculture and Consumer Services under the Applied Mosquito Research Program Contract Number 26452. This material is based on the work supported by the National Science Foundation Graduate Research Fellowship under grant 00107251 to LER.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors thank Candelaria Domingo for performing molecular processing of samples from Arizona. Aaron Chambers assisted with fieldwork and mosquito sampling in Arizona. Kristin Sloyer of FMEL contributed specimens from Pinellas County. The authors thank Tom Walker and the University of Florida, Natural Area Teaching Lab for support collecting mosquitoes in Alachua Co., Phillip Kaufman for laboratory space and support for molecular processing some mosquito specimens from Florida, Florida, PJ Walker and the National Park Service for granting permits and their assistance at Everglades National Park (permit numbers EVER-2015-SCI-0054 and EVER-2017-SCI-0011), and the Reeves Family, WH Reeves and FC Reeves, for allowing mosquitoes to be collected in their yard.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fitd.2022.842523/full#supplementary-material
2. Edman JD. Host-Feeding Patterns of Florida Mosquitoes: III. Culex (Culex) and Culex (Neoculex). J Med Entomol (1974) 11:95–104. doi: 10.1093/jmedent/11.1.95
3. Molaei G, Andreadis TG, Armstrong PM, Bueno R Jr., Dennett JA, Real SV, et al. Host Feeding Pattern of Culex quinquefasciatus (Diptera: Culicidae) and its Role in Transmission of West Nile Virus in Harris County, Texas. Am J Trop Med Hyg (2007) 77:73–81. doi: 10.4269/ajtmh.2007.77.73
4. Tempelis CH, Reeves WC, Bellamy RE, Lofy MF. A Three Year Study of the Feeding Habits of Culex tarsalis in Kern County, California. Am J Trop Med Hyg (1965) 14:170–77. doi: 10.4269/ajtmh.1965.14.170
5. Kilpatrick AM, Kramer LD, Jones MJ, Marra PP, Daszak P. West Nile Virus Epidemics in North America are Driven by Shifts in Mosquito Feeding Behavior. PloS Biol (2006) 4:e82. doi: 10.1371/journal.pbio.0040082
6. Tempelis CH, Lofy MF. A Modified Precipitin Method for Identification of Mosquito Blood-Meals. Am J Trop Med Hyg (1963) 12:825–31. doi: 10.4269/ajtmh.1963.12.825
7. Edman JD. Host-Feeding Patterns of Florida Mosquitoes I. Aedes, Anopheles, Coquillettidia, Mansonia and Psorophora. J Med Entomol (1971) 8:687–95. doi: 10.1093/jmedent/8.6.687
8. Washino RK, Tempelis CH. Mosquito Host Bloodmeal Identification: Methodology and Data Analysis. Annu Rev Entomol (1983) 28:179–201. doi: 10.1146/annurev.en.28.010183.001143
9. Edman JD. Host-Feeding Patterns of Florida Mosquitoes (Diptera: Culicidae) VI. Culex (Melanoconion). J Med Entomol (1979) 15:521–5. doi: 10.1093/jmedent/15.5-6.521
10. Krysko KL, Enge KM, Donlan EM, Seitz JC, Golden EA. Distribution, Natural History, and Impacts of the Introduced Green Iguana (Iguana iguana) in Florida. Iguana (2007) 3:2–17.
11. Ginsberg HS, Hickling GJ, Burke RL, Ogden NH, Beati L, LeBrun RA, et al. Why Lyme Disease is Common in the Northern US, But Rare in the South: The Roles of Host Choice, Host-Seeking Behavior, and Tick Density. PloS Biol (2021) 19:e3001066. doi: 10.1371/journal.pbio.3001066
12. Nasci RS. A Lightweight Battery-Powered Aspirator for Collecting Resting Mosquito in the Field. Mosquito News (1981) 41:808–11.
13. Burkett-Cadena ND, Hoyer I, Blosser E, Reeves LE. Human-Powered Pop-Up Resting Shelter for Sampling Cavity-Resting Mosquitoes. Acta Tropica (2019) 190:288–92. doi: 10.1016/j.actatropica.2018.12.002
14. Darsie RF, Morris CD. Keys to the Adult Females and Fourth Instar Larvae of the Mosquitoes of Florida (Diptera, Culicidae). Fort Myers: Florida Mosquito Control Association (2003).
15. Darsie RF, Ward RA. Identification and Geographical Distribution of the Mosquitoes of North America, North of Mexico. Gainesville: University Press of Florida (2005).
16. Reeves LE, Medina J, Miqueli E, Sloyer KE, Petrie W, Vasquez C, et al. Establishment of Aedes scapularis (Diptera: Culicidae) in Mainland Florida, With Notes on the Ochlerotatus Group in the United States. J Med Entomol (2021) 58:717–29. doi: 10.1093/jme/tjaa250
17. Reeves LE, Holderman CJ, Gillett-Kaufman JL, Kawahara AY, Kaufman PE. Maintenance of Host DNA Integrity in Field-Preserved Mosquito (Diptera: Culicidae) Blood Meals for Identification by DNA Barcoding. Parasites Vectors (2016) 9:503. doi: 10.1186/s13071-016-1791-z
18. Reeves LE, Gillett-Kaufman JL, Kawahara AY, Kaufman PE. Barcoding Blood Meals: New Vertebrate-Specific Primer Sets for Assigning Taxonomic Identities to Host DNA From Mosquito Blood Meals. PloS Negl Trop Dis (2018) 12:e0006767. doi: 10.1371/journal.pntd.0006767
19. Truett GE, Heeger P, Mynatt RL, Truett AA, Walker JA, Warman ML. Preparation of PCR Quality Mouse Genomic DNA With Sodium Hydroxide and Tris (HotSHOT). BioTechniques (2000) 29:52–4. doi: 10.2144/00291bm09
20. Sanger F, Nicklen S, Coulson AR. DNA Sequencing With Chain-Terminating Inhibitors. Proc Nat Acad Sci (1977) 74:5463–7. doi: 10.1073/pnas.74.12.5463
21. Ratnasingham S, Hebert PDN. BOLD: The Barcode of Life Datasystem. Mol Ecol Notes (2007) 7:355–64. (Http://Www.Barcodinglife.Org). doi: 10.1111/j.1471-8286.2007.01678.x
22. Tollis M, Boissinot S. Genetic Variation in the Green Anole Lizard (Anolis carolinensis) Reveals Island Refugia and a Fragmented Florida During the Quaternary. Genetica (2014) 142:59–72. doi: 10.1007/s10709-013-9754-1
23. Wegener JE, Pita-Aquina JN, Atutubo J, Moreno A, Kolbe JJ. Hybridization and Rapid Differentiation After Secondary Contact Between the Native Green Anole (Anolis carolinensis) and the Introduced Green Anole (Anolis Porcatus). Ecol Evol (2019) 9:4138–48. doi: 10.1002/ece3.5042
25. Bartlett RD, Bartlett PR. A Field Guide to Florida Reptiles and Amphibians. Houston: Gulf Publishing Company (1999).
27. Brennan TC, Holycross AT. A Field Guide to Amphibians and Reptiles in Arizona. Phoenix: Arizona Game and Fish Department (2006).
28. Krysko KL, Burgess JP, Rochford MR, Gillette C, Cueva D, Enge KM, et al. Verified non-Indigenous Amphibians and Reptiles in Florida From 1863 Through 2010: Outlining the Invasion Process and Identifying Invasion Pathways and Stages. Zootaxa (2011) 3028:1–64. doi: 10.11646/zootaxa.3028.1.1
29. Arizona Bird Committee. Checklist of the Birds of Arizona. Available at: http://abc.azfo.org/lists/state_list.html.
30. Florida Ornithological Society. Official Florida State Bird List, Available at: http://www.fosbirds.org/florida-bird-list.html June 22, 2021. Retrieved August 10, 2021.
31. Chesser RT, Billerman SM, Burns KJ, Cicero C, Dunn JL, Hernández-Baños BE, et al. Check-List of North American Birds (Online). Chicago: American Ornithological Society (2021). Available at: http://checklist.aou.org/taxa.
33. Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, et al. Vegan: Community Ecology Package. R Package Version 2.5-7. (2020).
34. Kindt R, Coe R. Tree Diversity Analysis. A Manual and Software for Common Statistical Methods for Ecological and Biodiversity Studies. Nairobi: World Agroforestry Centre (2005).
35. Qian H, Ricklefs RE. Global Concordance in Diversity Patterns of Vascular Plants and Terrestrial Vertebrates. Ecol Lett (2008) 11:547–53. doi: 10.1111/j.1461-0248.2008.01168.x
36. Barrera R, Amador M, Young G, Komar N. Mosquito (Diptera: Culicidae) Bloodmeal Sources During a Period of West Nile Virus Transmission in Puerto Rico. J Med Entomol (2011) 48:701–4. doi: 10.1603/ME10281
37. Tempelis CH, Reeves WC. Feeding Habits of One Anopheline and Three Culicine Mosquitoes by the Precipitin Test. J Med Entomol (1964) 1:148–51. doi: 10.1093/jmedent/1.2.148
38. Nelms BM, Thiemann TC, Bridges DN, Williams AE, Koschik ML, Ryan BM, et al. Bionomics and Vector Potential of Culex thriambus (Diptera: Culicidae) Mosquitoes in Lake County, California. J Med Entomol (2016) 53:473–81. doi: 10.1093/jme/tjw123
39. Tempelis CH, Washino RK. Host-Feeding Patterns of Culex tarsalis in the Sacramento Valley, California, With Notes on Other Species. J Med Entomol (1967) 4:315–8. doi: 10.1093/jmedent/4.3.315
40. Wekesa JW, Yuval B, Washino RK, De Vasquez AM. Blood Feeding Patterns of Anopheles freeborni and Culex tarsalis (Diptera: Culicidae): Effects of Habitat and Host Abundance. Bull Entomol Res (1997) 87:633–41. doi: 10.1017/S0007485300038736
41. Kent R, Juliusson L, Weissmann M, Evans S, Komar N. Seasonal Blood-Feeding Behavior of Culex tarsalis (Diptera: Culicidae) in Weld County, Colorado 2007. J Med Entomol (2009) 46:380–90. doi: 10.1603/033.046.0226
42. Thiemann TC, Wheeler SS, Barker CM, Reisen WK. Mosquito Host Selection Varies Seasonally With Host Availability and Mosquito Density. PloS Negl Trop Dis (2011) 5:e1452. doi: 10.1371/journal.pntd.0001452
43. Reisen WK, Lothrop HD, Thiemann T. Host Selection Patterns of Culex tarsalis (Diptera: Culicidae) at Wetlands Near the Salton Sea, Coachella Valley, California 1998–2002. J Med Entomol (2013) 50:1071–6. doi: 10.1603/ME13078
44. McPhatter LP, Su T, Williams G, Cheng M-L, Dhillon M, Gerry AC. Host-Feeding Patterns of Culex stigmatosoma (Diptera: Culicidae) in Southern California. J Med Entomol (2017) 54:1750–7. doi: 10.1093/jme/tjx154
45. Komar N, Langevin S, Hinten S, Nemeth N, Edwards E, Hettler D, et al. Experimental Infection of North American Birds With the New York 1999 Strain of West Nile Virus. Emerg Infect Dis (2003) 9:311. doi: 10.3201/eid0903.020628
46. Edman JD, Taylor DJ. Culex nigripalpus: Seasonal Shift in the Bird-Mammal Feeding Ratio in a Mosquito Vector of Human Encephalitis. Sci (1968) 161:67–8. doi: 10.1126/science.161.3836.67
47. Blosser EM, Lord CC, Stenn T, Acevedo C, Hassan HK, Reeves LE, et al. Environmental Drivers of Seasonal Patterns of Host Utilization by Culiseta melanura (Diptera: Culicidae) in Florida. J Med Entomol (2017) 54:1365–74. doi: 10.1093/jme/tjx140
48. Rodgers L, Bodle M, Black D, Laroche F, Pratt P, Boughton A. Status of Nonindigenous Species. South Florida Environ Rep (2021) 2012:1–35.
49. Garman S. On West Indian Reptiles in the Museum of Comparative Zoology, at Cambridge, Mass. Proc Am Philos Soc (1887) 24:278–86.
50. Campbell TS. The Introduced Brown Anole (Anolis sagrei) Occurs in Every County in Peninsular Florida. Herpetol Rev (2003) 34:173.
51. West RG, Mathias DK, Day JF, Acevedo C, Unnasch TR, Burkett-Cadena ND. Season Changes of Host Use by Culiseta melanura (Diptera: Culicidae) in Central Florida. J Med Entomol (2020) 57:1627–34. doi: 10.1093/jme/tjaa067
52. Kamath A, Stuart YE, Campbell TS. Behavioral Partitioning by the Native Lizard Anolis carolinensis in the Presence and Absence of the Invasive Anolis sagrei in Florida. Breviora (2013) 535:1–10. doi: 10.3099/MCZ8.1
53. Zinser M, Ramberg F, Willott E. Scientific Note Culex quinquefasciatus (Diptera: Culicidae) as a Potential West Nile Virus Vector in Tucson, Arizona: Blood Meal Analysis Indicates Feeding on Both Humans and Birds. J Insect Sci (2004) 4:20. doi: 10.1673/031.004.2001
54. Godsey MS, Burkhalter K, Young G, Delorey M, Smith K, Townsend J, et al. Entomologic Investigations During an Outbreak of West Nile Virus Disease in Maricopa County, Arizona 2010. Am J Trop Med Hyg (2012) 87:1125–31. doi: 10.4269/ajtmh.2012.11-0700
55. Rivera JA, Lawnig AM, Martins EP. Reconstructing Historical Shifts in Suitable Habitat of Sceloporus Lineages Using Phylogenetic Niche Modeling. J Biogeogr (2020) 47:2117–28. doi: 10.1111/jbi.13915
56. Haenel GJ. Phylogeography of the Tree Lizard, Urosaurus ornatus: Responses of Populations to Past Climate Change. Mol Ecol (2007) 16:4321–34. doi: 10.1111/j.1365-294X.2007.03515.x
57. Walton WE, Workman PD, Tempelis CH. Dispersal, Survivorship, and Host Selection of Culex erythrothorax (Diptera: Culicidae) Associated With a Constructed Wetland in Southern California. J Med Entomol (1999) 36:30–40. doi: 10.1093/jmedent/36.1.30
58. Mackay AJ, Kramer WL, Meece JK, Brumfield RT, Foil LD. Host Feeding Patterns of Culex Mosquitoes (Diptera: Culicidae) in East Baton Rouge Parish, Louisiana. J Med Entomol (2010) 47:238–48. doi: 10.1603/ME09168
59. Mainka SA, Howard GW. Climate Change and Invasive Species: Double Jeopardy. Integr Zool (2010) 5:102–11. doi: 10.1111/j.1749-4877.2010.00193.x
60. Mills JN, Gage KL, Khan AS. Potential Influence of Climate Change on Vector-Borne and Zoonotic Diseases: A Review and Proposed Research Plan. Environ Health Perspect (2010) 118:1507–14. doi: 10.1289/ehp.0901389
Keywords: mosquito, dilution effect, arbovirus, vectorial capacity, Anolis, blood meal, invasive species
Citation: Reeves LE and Burkett-Cadena ND (2022) Lizards Are Important Hosts for Zoonotic Flavivirus Vectors, Subgenus Culex, in the Southern USA. Front. Trop. Dis 3:842523. doi: 10.3389/fitd.2022.842523
Received: 23 December 2021; Accepted: 11 January 2022;
Published: 11 February 2022.
Edited by:
Cleber Galvão, Oswaldo Cruz Foundation (Fiocruz), BrazilReviewed by:
Shih Keng Loong, University of Malaya, MalaysiaCopyright © 2022 Reeves and Burkett-Cadena. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lawrence E. Reeves, bGVyZWV2ZXNAdWZsLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.