
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Trop. Dis. , 13 October 2022
Sec. Neglected Tropical Diseases
Volume 3 - 2022 | https://doi.org/10.3389/fitd.2022.1017900
This article is part of the Research Topic Novel Therapeutics for Parasitic NTDs View all 5 articles
 Marina Nick1
Marina Nick1 Frederick A. Partridge1
Frederick A. Partridge1 Ruth Forman2
Ruth Forman2 Carole J. R. Bataille3,4
Carole J. R. Bataille3,4 Kathryn J. Else2
Kathryn J. Else2 Angela J. Russell3,4
Angela J. Russell3,4 David B. Sattelle1*
David B. Sattelle1*The 3 major classes of soil transmitted helminths (whipworm, hookworm and Ascaris) affect 1.5 billion people worldwide mostly in poor countries, where they have adverse effects on child development, nutrition, and the work capacity of adults. Although there are drugs effective on Ascaris, notably the benzimidazoles, those same drugs show poor efficacy particularly against whipworm (Trichuris trichiura) and to a certain extent hookworm. Parasitic nematodes also infect farm livestock and companion animals. Resistance to currently deployed human and veterinary anthelmintic drugs is a growing problem. Therefore, new chemical anthelmintic lead compounds are urgently needed. One of the fastest routes to a novel therapeutic lead is to screen libraries of drugs which are either already approved for human use or have already been part of clinical trials. We have pursued this approach to anthelmintic lead discovery using an invertebrate automated phenotyping platform (INVAPP) for screening chemicals and the well-established nematode genetic model organism Caenorhabditis elegans. The 400 compound Medicines for Malaria Venture (MMV) Pandemic Response Box library was screened with each compound tested initially at 1.0x10-4 M. We identified 6 compounds (MMV1593515 (vorapaxar), MMV102270 (diphyllin), MMV1581032 (ABX464), MMV1580796 (rubitecan), MMV1580505 and MMV1593531) active in both an L1-L4 growth/motility assay and in an L4 motility assay. For vorapaxar, an EC50 of 5.7x10-7 M was observed, a value comparable to those of some commercial anthelmintics. Although not a parasite, the ease with which high-throughput screens can be pursued on the free-living nematode C. elegans makes this a useful approach to identify chemical leads and complements the often lower-throughput experiments on parasitic nematode models.
The major human gastrointestinal tract parasites collectively known as soil transmitted helminths (STHs) are the whipworm (Trichuris trichiura), the roundworm (Ascaris lumbricoides), and hookworms (Necator americanus and Ancylostoma duodenale). Over one billion people are estimated to be infected with at least one STH (1). STH infection is a significant but neglected cause of morbidity (2, 3). According to Hotez and Kamath (2) more than 50 million school-aged children and 7 million child-bearing-age women in Sub-Saharan Africa are infected with one or more STH (2). Due to the remoteness and inaccessibility of the worst-affected places, the worldwide burden of STHs has most likely been underestimated (4).
Currently, preventive chemotherapy, based on single-dose, mass drug administration (MDA), is used to treat STHs in endemic regions, with the goal of minimising morbidity in pre-school and school-aged children with moderate to heavy infections. Such treatments, which use the benzimidazole drugs albendazole and mebendazole remain effective against ascariasis but less so against hookworm and whipworm. (3–7). Therefore, treatment for the STHs is an example of an unmet clinical need that necessitates the development of novel therapeutics. Recent experience from the animal health field showed that resistance to the anthelmintic monepantel (8) appeared within two years of its introduction (9). Therefore, current MDA programmes may lead to similar selection for resistance, and the efficacy of, for example, benzimidazole drugs against T. trichiura has fallen in recent years (10, 11).
Despite the long history of anthelmintic medication development, the majority of currently used anthelmintics were discovered by phenotypic screening of chemical candidates. Medicines that act on ligand-gated ion channels (12) and drugs that target the cytoskeletal protein tubulin are among the products of this method (13). In the search for new anthelmintic drug candidates, several laboratories have developed high-throughput screening methods (14). In our laboratory an invertebrate automated phenotyping platform (INVAPP) and the algorithm Paragon (15) have been developed. This system has been used to identify two novel chemical classes of anthelmintics, the 2, 4-dihydrobenz[e][1,4]oxazepin-2(3H)-ones (11, 16) and the diaminothieno[3,2-d]pyrimidines (17). We have therefore adopted the INVAPP/Paragon system to explore a new chemical library.
C. elegans is a nematode genetic model organism (18) that is frequently employed in anthelmintic discovery research (8, 14, 19, 20). The Pandemic Response Box is a chemical library supplied by Medicines for Malaria Venture (MMV) and the Drugs for Neglected Diseases Initiative (DNDi). It includes 400 drug-like compounds that are either already on the market or in various stages of research for uses other than as anthelmintics. The library was selected by experts from Academia and Industry with the aim of pursuing an Open Source approach to discovering new chemical leads that could impact on neglected, pandemic-scale diseases with unmet clinical needs (21). The library contains 201 antibacterial compounds, 153 antivirals, and 46 antifungals. Compounds of interest from this library may therefore offer a fast-track route into clinical trials in the search for new anthelmintics. We have used INVAPP to assess the activities of all 400 drugs on C. elegans L1-L4 stages using a growth/motility assay and on the L4 stage utilising a motility assay in order to understand the potential of library compounds for re-purposing as candidate anthelmintic drug leads.
The Pandemic Response Box library was supplied by the Medicines for Malaria Venture. All stock compounds were supplied as 10-2 M stocks in DMSO.
The C. elegans wild type N2 strain was maintained at 20°C on nematode growth medium (NGM) agar seeded with the Escherichia coli strain OP50. To prepare worm populations for screening, first a mixed population was obtained. 5 to 7 NGM plates were seeded with 250 μl of the E. coli strain OP50 and incubated at 37°C for 3 to 5 days to form a lawn. A 2.5cm square section of the agar plate rich in worms was transferred to each NGM plate and maintained at 20°C for 5 days.
Such C. elegans cultures contain worms of different ages. To eliminate variation caused by age differences, worms need to be synchronised. To obtain a synchronised L1 population we used filtration. Each NGM plate was washed with 50 mL of S-basal medium into a falcon tube and centrifuged at 3000 x g at 20°C for 4 min. The pellet was then re-washed (3x) and re-centrifuged, using the same procedure, to clear any remaining bacteria. Worms were then filtered (100 μm filter) to remove any adult and late-stage larvae. Finally, they were passed through a 40 μm sieve (3x) to obtain a synchronous L1 larval population.
To obtain a synchronised L4 larval population, a population enriched in L4s was first obtained by incubating the NGM plates, prepared as described, for between 8-10 days after transferring C. elegans. Plates with the highest number of L4 larvae were selected and washed with S-basal medium. Worms were then filtered (100 μm filter) to capture any L4 stage larvae and remove earlier stages. The filter was rinsed with S-basal media into a falcon tube and centrifuged at 3000 x g at 20°C for 4 min. The pellet was then re-washed (2x) using the same procedure to remove any residual bacteria.
The INVAPP/Paragon system used in these experiments has already been described in detail (15, 22). Two hundred frame movies were captured at 25 frames s-1 for 8 s using μManager (23). Movies were analysed using MATLAB scripts (available at https://github.com/fpartridge/invapp-paragon) and the variance determined through time for each pixel. The distribution of these pixel variances was then considered, and pixels whose variance was above the threshold (those greater than one standard deviation away from the mean variance) were considered ‘motile’. Motile pixels within each well were counted to obtain a movement score.
Screening experiments were conducted in 96-well plate liquid cultures. Synchronised L1s were diluted to approximately 15-25 worms per 50 μl in S complete buffer with 1% w/v HB101 E. coli. Assay plates were prepared with 99 μL of L1 suspension and 1 μL of each compound per well. As a control 16 wells in each plate were prepared with 1 µL DMSO solution (1% v/v final concentration). Plates were incubated at 25°C and motility was recorded using the INVAPP/Paragon system 3 days later. By this time control worms developed to L4 or adult stage. Movies were recorded and the median growth/motility score measured using INVAPP as described above. This assay measures both growth and motility in a combined score (15). A schematic of this assay is shown in Figure 1A.
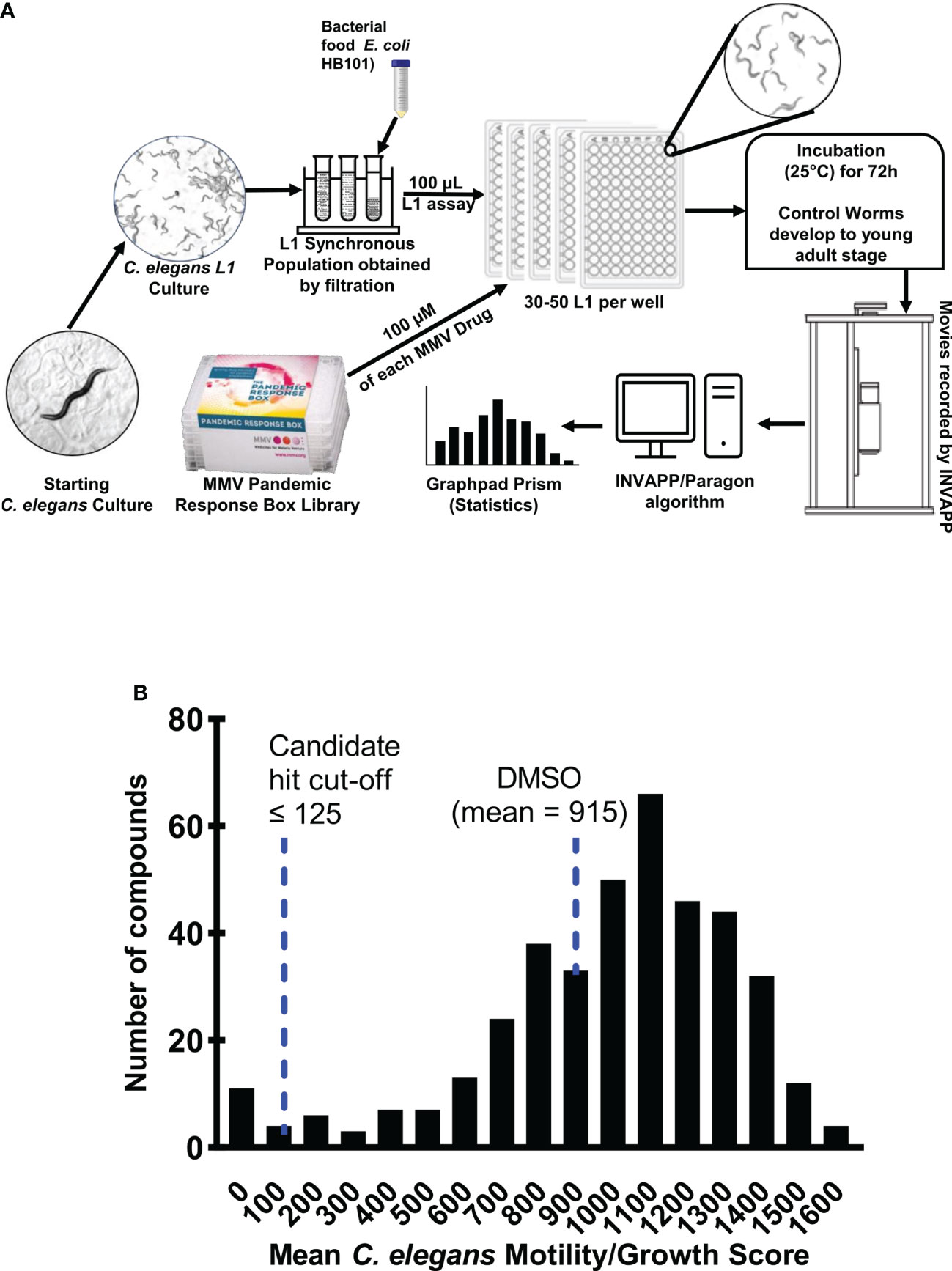
Figure 1 (A) Cartoon illustration of preparation and screening of C. elegans growth/motility assay. By the time that the movies are recorded, surviving C. elegans worms are expected to have developed to the (larger) L4 or adult stages. Therefore, their movement recorded by INVAPP/Paragon is a Growth/Motility score. (B) Frequency distribution of the mean C. elegans growth/motility scores for each of the Pandemic Response Box compounds tested at 1.0x10-4 M concentration, along with the DMSO-only control. Each bin is a range of scores from 50 below the axis value to 50 above the axis value. For example, the 200 bin counts the number of compounds scoring between 150 and 250. The screen was performed on three separate occasions (n = 3). The mean of the three C. elegans growth/motility scores were plotted. A cut-off score for candidate hits of 125 was selected.
Synchronised L4 were diluted to approximately 15-25 worms per 50 μL in S basal buffer. No bacterial food is used in this assay. Assay plates were prepared with 99 μL of L4 suspension and 1 μL of each compound per well. As a control 16 wells in each plate were prepared with 1 µL DMSO solution (1% v/v final concentration). Plates were incubated at 25°C before the motility of C. elegans was recorded using the INVAPP/Paragon system after 24 h. Due to the absence of food, worms do not grow or develop in this assay, which therefore exclusively measures changes in motility and is recorded as a motility score. A schematic of this assay is shown in Figure 3A.
The 400 drugs of the MMV Pandemic Response Box library were screened at 1.0x10-4 M on wild type C. elegans in a growth/motility assay primary screen (L1 to L4 or adult development). 1% v/v DMSO was used as a negative control. Three sets of identical assay plates were prepared for each experiment and the entire screen was repeated on 3 different days (n=3).
The 18 potential candidate lead compounds identified in the primary screen of the MMV Pandemic Response Box library were re-tested on C. elegans in the growth/motility assay, at 7.5x10-5 M (0.75% v/v DMSO) and 5.0x10-5 M (0.5% v/v DMSO). In addition, C. elegans L4 animals were screened at 1.0x10-4 M in the motility assay. In all cases, screens were undertaken on three separate occasions (n=3), each time with 4 assay repeats. Levamisole at the same concentration was the positive control and 1% v/v DMSO the negative control.
The concentration-response relationship for selected compounds was determined by testing activity in the C. elegans growth/motility assay. Compounds were tested at each of 12 concentrations from 5.0x10-5 M to 2.0x10-8 M (10 to 12 replicates tested on three occasions so n=3). EC50 values were estimated by fitting curves using a four-parameter log-logistic function in Graphpad Prism 9.3.
Primarily the Pandemic Response Box library was screened using the C. elegans growth/motility assay. The 400 compounds were screened at 1.0x10-4 M and the growth/motility score recorded (15). A histogram showing the distribution of the mean C. elegans growth/motility scores for each compound, along with the DMSO-only control is shown in Figure 1B.
The primary screen was used to prioritise the most active compounds for confirmatory rescreening. We chose the 14 compounds with the lowest mean growth/motility score (3.5% of the library) for rescreening (each compound had a mean growth/motility score below 125). In addition, we selected a further 4 compounds for rescreening with scores close to this cut-off value. The full data for all three repeats of the primary screen using the C. elegans growth/motility assay at 1.0x10-4 M are presented in the Table S1.
To confirm the activity of the lead compounds, we conducted a secondary screen. The 18 candidates were re-tested in the same C. elegans growth/motility assay at two lower concentrations (7.5x10- 5 M and 5.0x10-5 M). The results for the 7.5x10-5 M secondary screen are shown in Figure 2A. A one-way ANOVA showed there was a significant effect of compound treatment (P ≤ 0.0001). The effectiveness of each compound compared to the DMSO-only control was determined using Dunnett’s multiple comparison test. All 18 candidate lead compounds significantly reduced the C. elegans growth/motility score at 7.5x10-5 M (Figure 2A, Table S3).
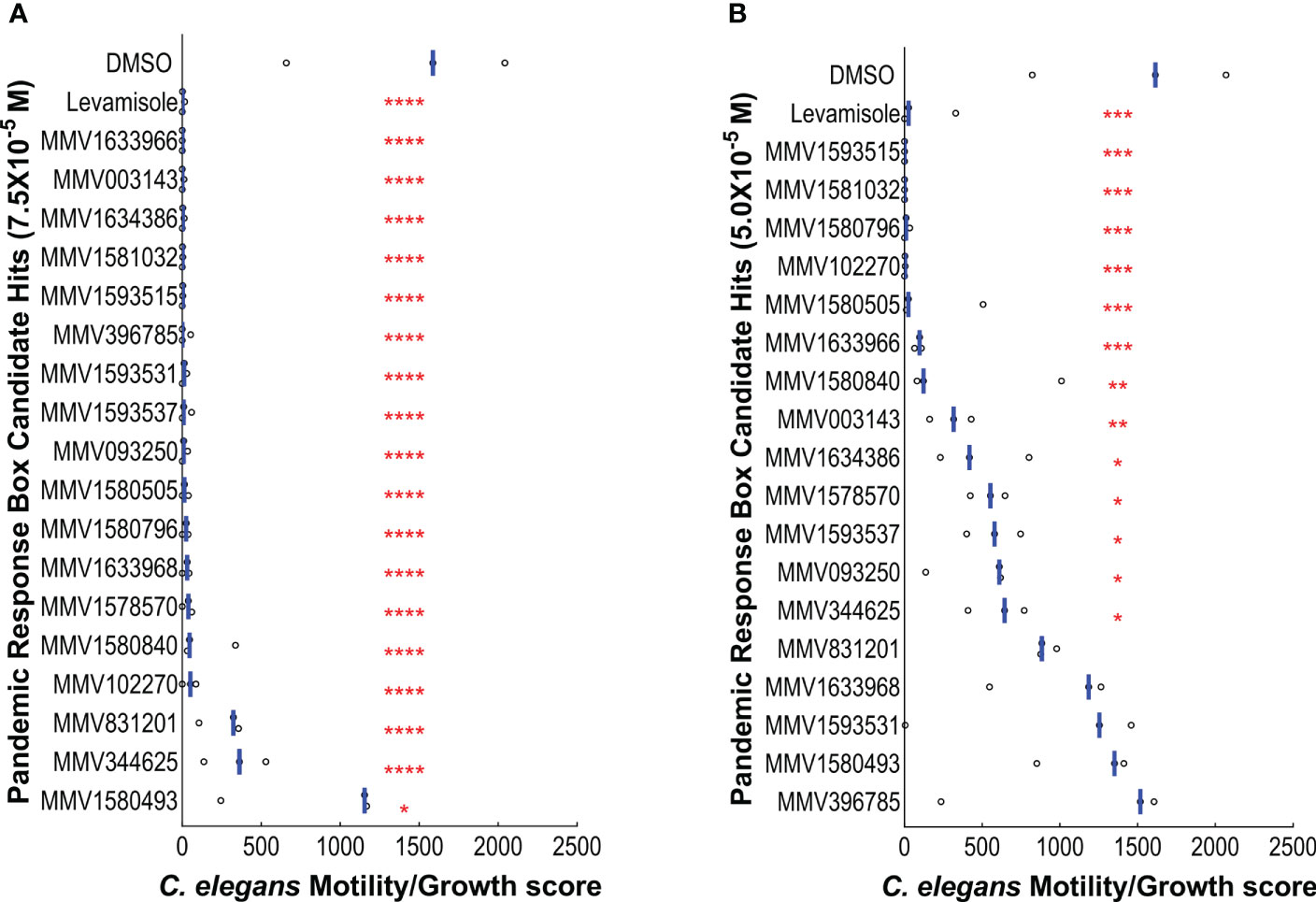
Figure 2 Retesting the 18 lead candidates as well as levamisole and DMSO as controls in the C. elegans growth/motility assay at (A) 7.5x10-5 M and (B) 5.0x10-5 M. Each point shows one well. The blue bar indicates the mean movement index for each treatment. The screen was performed with four replicates and on three occasions (n = 3). A one-way ANOVA for the 7.5x10-5 M concentration dataset showed there was a significant effect of compound treatment P < 0.0001. A post-hoc Dunnett’s test compared each compound to the DMSO-only control (P values*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001).
The results of the C. elegans growth/motility screen at 5.0x10-5 M are shown in Figure 2B. A one-way ANOVA showed when there was a significant effect of compound treatment (P ≤ 0.0001). The effectiveness of each compound was determined using Dunnett’s multiple comparison test with reference to the DMSO-only control. 13 Pandemic Response Box compounds significantly reduced the C. elegans growth/motility score at 5.0x10-5 M, as indicated by red asterisks in Figure 2B. Six compounds were highly effective (P values ≤ 0.0001). Data for the secondary screen of the C. elegans growth/motility assay at 5.0x10-5 M are presented in Table S2.
Many existing anthelmintics act by reducing motility, for example by acting on ion channels that function in the nervous system and/or at neuromuscular junctions (24–26). We therefore wanted to determine which of the candidate lead compounds impair motility. The 18 candidate lead compounds were tested on C. elegans L4 stage worms in a pure motility assay at 1.0x10-4 M. This allowed us to verify whether, in addition to showing activity in the C. elegans growth/motility assay over 72 h, the candidate hit compounds were also effective in an assay using C. elegans L4 stage worms without bacterial food, over 24 h. The method is shown in Figure 3A and the results are presented in Figure 3B. A one-way ANOVA for this dataset showed there was a significant effect of compound treatment (P ≤ 0.0001). The effectiveness of each compound was then determined using Dunnett’s multiple comparison test compared to the DMSO-only control. In addition to the positive control drug levamisole, 6 compounds that significantly reduced the C. elegans L4 motility score were identified and indicated with red asterisks (P values: * P ≤ 0.05, ** P ≤ 0.01). Data for the C. elegans L4 motility assay screen is presented in the Table S4.
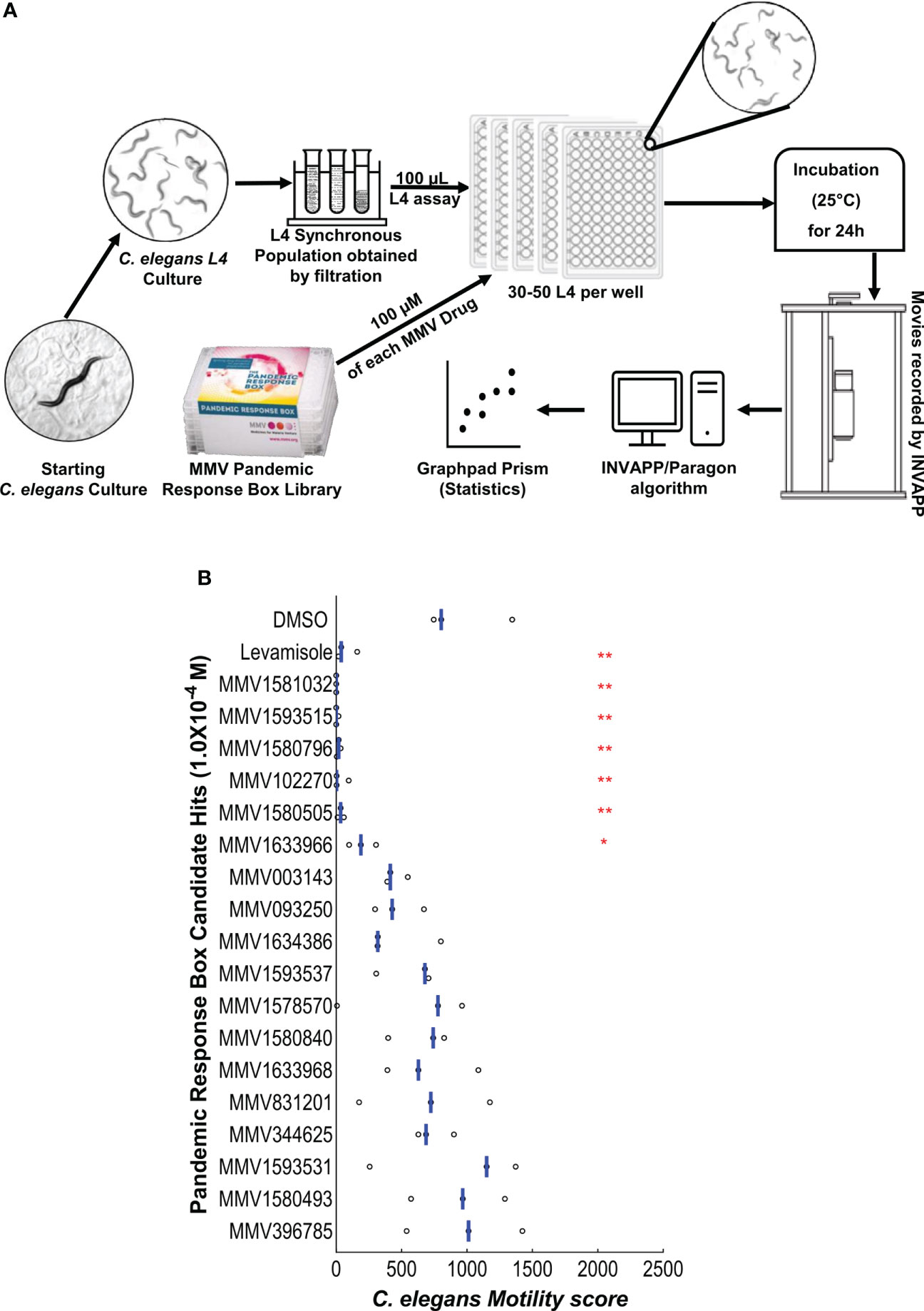
Figure 3 (A) Cartoon illustration of preparation and screening C. elegans motility assay. When the movies are recorded, surviving C. elegans worms have not developed further due to the lack of bacterial food. Therefore, their movement recorded by INVAPP/Paragon is a pure motility score. (B) Retesting the candidates in the C. elegans L4 motility assay at 1.0x10-4 M. Each point shows one well. The blue bar indicates the mean movement index for each treatment. The screen was performed with four replicates and on three occasions (n = 3). A one-way ANOVA for the 1.0x10-4 M concentration dataset showed there was a significant effect of compound treatment (P ≤0.0001). A post-hoc Dunnett’s test compared each compound to the DMSO-only control (P values: *P ≤ 0.05, **P ≤ 0.01).
Thus, six of the hit compounds from the Pandemic Response Box library were significantly active on both the C. elegans growth/motility assay at 1.0x10-4 M, 5.0 x 10- 5 M and 7.5x10-5 M as well as the L4 motility assay at 1.0x10-4 M. These compounds were MMV1593515 (vorapaxar), MMV102270 (diphyllin), MMV1581032 (ABX464), MMV1580796 (rubitecan), MMV1580505 and MMV1593531. Four are previously described antivirals and two have antibacterial activity (21). Vorapaxar is a human approved drug while diphyllin, ABX464 and rubitecan are in various human clinical trials (Table 1).
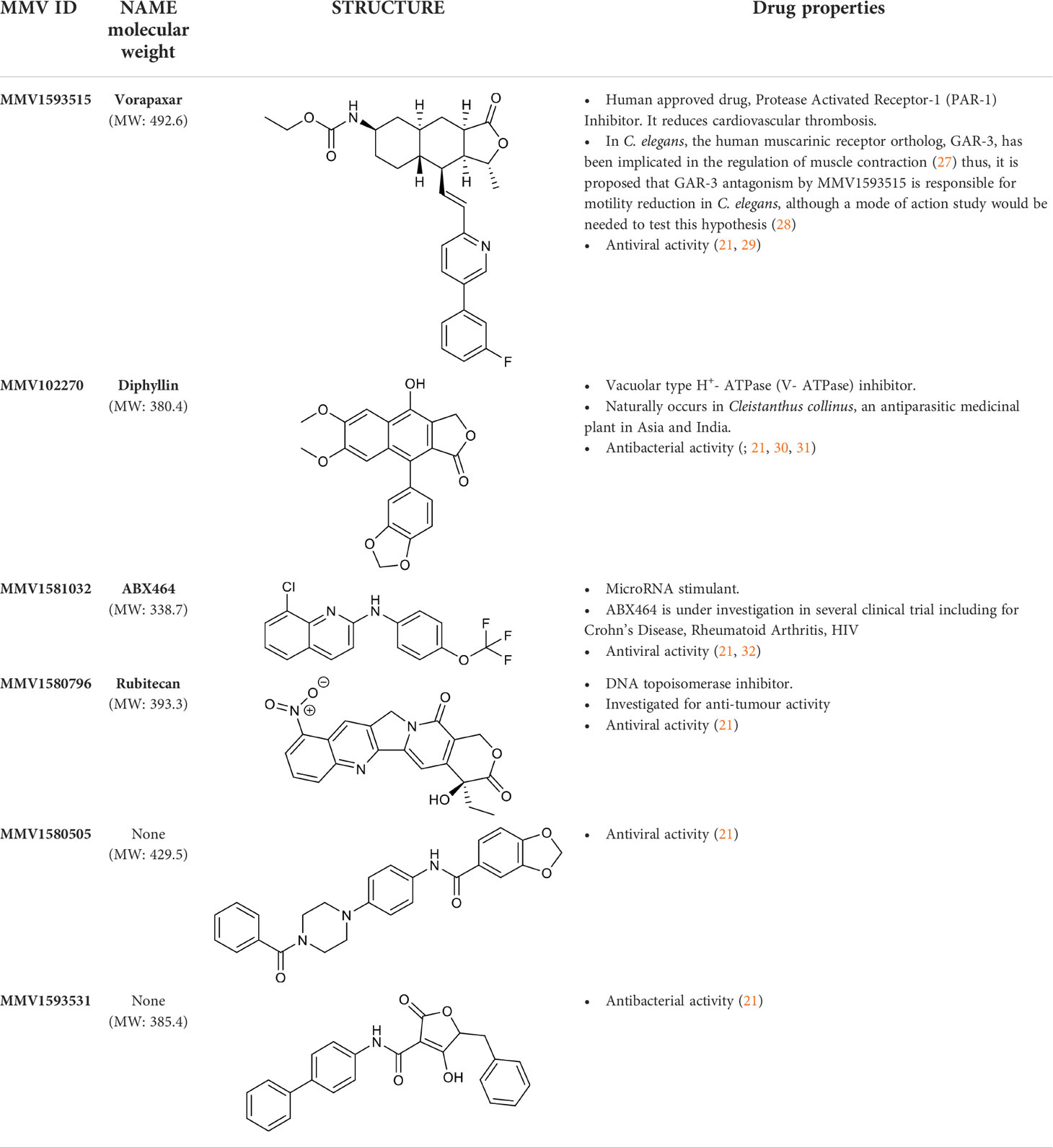
Table 1 List of Pandemic Response Box hit compounds from the C. elegans growth/motility screen at 1.0x10- 4 M, 5.0x10-5 M and 7.5x10-5 M and C. elegans L4 motility screen at 1.0x10-4 M.
We wanted to ensure that the anthelmintic compounds we identified showed concentration-dependent activity and to estimate their relative potency to inform future work. We used the efficacy of the best candidate lead compounds in the secondary screens to prioritize which to investigate. Of the six compounds significantly reducing L1 growth/motility at both 5.0x10-5 M and 7.5x10-5 M, as well as significantly reducing L4 motility, four (ABX464, diphyllin, rubitecan and vorapaxar) were readily available as solid material. Concentration-response curves for these four compounds are shown in Figure 4. It was encouraging to see EC50 values in the low micromolar – ABX464 (EC50 = 2.3x10-6 M), diphyllin (EC50 = 3.9x10-6 M) and rubitecan (EC50 = 1.2x10-5 M) and even sub-micromolar range – vorapaxar (EC50 = 5.7x10-7 M).
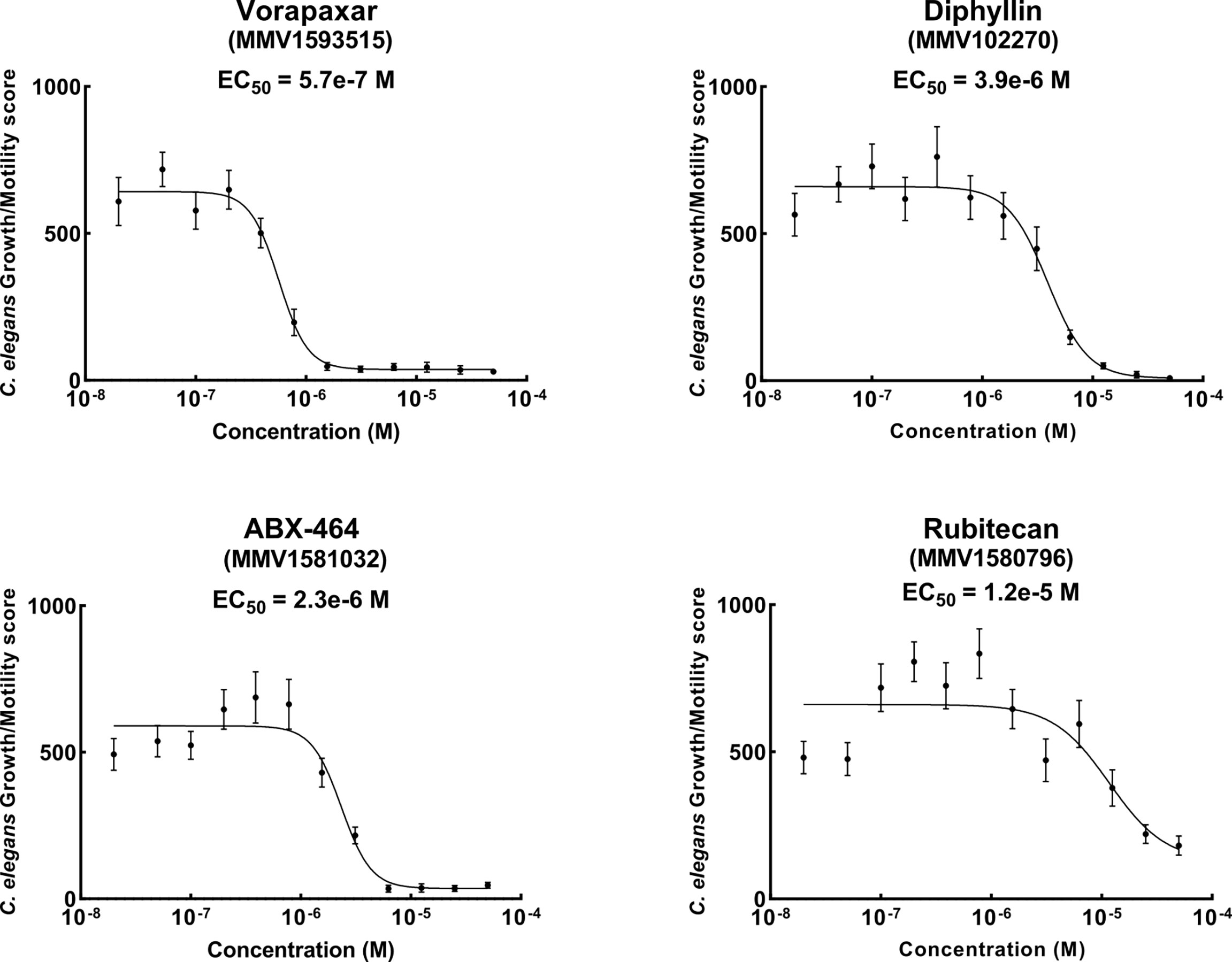
Figure 4 EC50 values are determined from concentration-response curves for the 4 compounds consistently showing activity in the growth/motility assay. Each point shows the mean and standard error (error bars) for each concentration tested. Compounds were tested between 10 to 12 replicates and 12 concentrations ranging from 5.0x10-5 M to 2.0x10-8 M (tested on three occasions so n = 3). EC50 curves were fitted using a four-parameter log-logistic function in Graphpad Prism 9.3.
In mammalian studies, MMV1593515 (vorapaxar) the human approved drug is a protease activated receptor-1 (PAR-1) inhibitor (33, 34), MMV102270 (diphyllin) is a vacuolar type H+- ATPase (V- ATPase) inhibitor (35), MMV1581032 (ABX464) is a microRNA stimulant (36) and MMV1580796 (rubitecan) is a DNA topoisomerase inhibitor (37). Thus the 4 candidate compounds of interest have diverse mechanisms of action but all showed anthelmintic activity. MMV1593515 (vorapaxar), MMV1581032 (ABX464) and MMV1580796 (rubitecan) are antivirals and MMV102270 (diphyllin) is an antibacterial drug. Their mechanisms of action in nematodes remain to be determined.
The 400 compound MMV Pandemic Response Box library has proved useful in identifying from library screens compounds of interest for re-purposing as antiparasitics for helminth control, and the control of schistosome and protozoan parasites. A recent study reported a screen of the MMV Pandemic Response Box against C. elegans young adults in a motility screening assay. This assay is similar to our L4 motility assay that is reported in Figure 3. They also screened against Haemonchus contortus exsheathed third-stage larvae (xL3s) in motility and development screening assays (28).
Shanley et al. identified two compounds, MMV1581032 (ABX464) and MMV1593515 (vorapaxar), that inhibited C. elegans motility. These compounds are among our four most active compounds, and we confirm their relatively low EC50 values. In addition, Shanley et al. (28) showed that MMV1581032 (ABX464), as well as another drug, MMV1593539, have anthelmintic activity against H. contortus. As in their studies, we also found that MMV1593539 is not active against C. elegans. The accord between our findings and those of Shanley et al. (28) confirms the utility of our INVAPP screening approach.
However, in this study we also identify an additional 16 active anthelmintic compounds. This difference is likely due to our use of a C. elegans assay that measures growth/motility as worms develop from the L1 stage. This might reflect either a greater sensitivity of the L1 stages, or the choice of a higher initial screening concentration. Differential sensitivity of different life stages of nematodes and the importance of this in the choice of screening assays in order to avoid false negatives has been well described elsewhere (1). There are likely more biological processes (and hence drug targets) that could be targeted by anthelmintic assays in our growth/motility assay compared to a pure motility assay, which may account for the greater diversity of compounds detected with anthelmintic properties. The known inactivity of the approved anthelmintic drug mebendazole on L4 motility in C. elegans, compared to its high activity in the L1 growth/motility assay (15) demonstrates that viable anthelmintics will be missed by pure motility screens. Our study demonstrates the utility of INVAPP/Paragon and is complementary to that of Shanley et al. (28).
MMV1581032 (ABX464) is a first-in-class, clinical-stage, oral small molecule immunomodulator (32). ABX464 is reported to bind to the RNA cap-binding complex, which modulates both viral and cellular RNA biogenesis (36). ABX464 was originally developed for its antiviral potential but it was redeployed for chronic inflammatory diseases due to potent anti-inflammatory effects in preclinical testing. It has shown safety and tolerability in clinical trials, which makes it a good candidate for repurposing as an anthelmintic.
Diphyllin occurs naturally in Cleistanthus collinus, which is known as an antiparasitic medicinal plant in Asia and India (38). Diphyllin proved deadly for the promastigote and amastigote stages of the protozoan parasite Leishmania infantum (39), while the structurally related compound justicidin β suppressed the growth of the parasites causing sleeping sickness Trypanosoma brucei rhodesiense and T. cruzi (40).
The MMV Pandemic Response Box library has also been screened against Schistosoma mansoni with the aim of discovering an effective drug for the neglected tropical disease schistosomiasis (41). The study identified 26 compounds active against newly transformed schistosomula, of which 17 were active against adult S. mansoni. Three compounds with anti-schistosomal activity (MMV396785 [alexidine], MMV1634386 [oteseconazole] and MMV1578570) showed activity against C. elegans growth/motility in this study. All three of these compounds showed efficacy against adult S. mansoni at 10 µM but not at 1 µM. Of particular interest alexidine was one of only two compounds which was lethal to adult S. mansoni at 10 µM but was not tested in vivo. In addition, oteseconazole showed good efficacy in vitro but did not alter worm burdens in vivo when dosed by a single oral gavage of 200 mg/kg 7 weeks post infection. These compounds therefore have potential for development as a broad-spectrum anthelmintics.
Some of our confirmed active compounds have also shown activity against other infectious diseases. MMV1593537 has antifungal activity against Cryptococcus neoformans, Cryptococcus deuterogattii, and the emerging global threat Candida auris (42). MMV1578570, MMV396785 (alexidine), MMV1634386 and MMV1580796 (rubitecan) are active against the pathogenic amoebae Balamuthia mandrillaris, Naegleria fowleri, and Acanthamoeba castellanii (43). Rubitecan is one of our four most active anthelmintic compounds. It prevents DNA from unwinding during replication via DNA topoisomerase 1, therefore interfering with tumour growth (44). Rubitecan is a derivative of a compound extracted from the Camptotheca acuminata tree with potent antitumor and antiviral properties.
This study identified 18 candidate lead compounds which impair nematode growth or motility, and the four most active include one drug (vorapaxar) with a sub-micromolar EC50, comparable to some current commercial anthelmintic drugs. It is of interest that comparisons between the growth/motility assay and the pure motility assay show that some candidate leads could be missed in assays monitoring only motility. Vorapaxar emerged as a lead compound of particular interest as it is approved for human use. Some vorapaxar analogues have been reported (45, 46) and in future it would be interesting to explore these compounds to see how the activity of such closely related structures compare with that of vorapaxar itself. The original targets of these re-purposed drugs are not necessarily those responsible for their anthelmintic activity. Important future work to identify their nematode molecular targets will be facilitated by studies on C. elegans, via genetic screens (47, 48). Another line of future research of interest would be to explore whether newly identified candidate lead compounds such as vorapaxar can circumvent benzimidazole resistance in C. elegans lines expressing resistance mutations in tubulin genes (49, 50). Finally, where that is possible, it will also be useful to explore on parasitic worms the actions of chemical leads derived from model organism studies of the type described here.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
MN, FP, DS, KE, AR contributed to conception and design of the study. MN performed the experiments, and the statistical analysis was performed by MN and FP. MN, FP, and DS wrote the first draft of the manuscript. All authors MN, FP, DS, CB, AR, RF and KE contributed to manuscript draft revision, read, and approved the submitted version.
DS and FP were supported by Medical Research Council grant MR/N024842/1 and a UCL Therapeutic Innovation Networks Pilot Data Scheme award supported by funding from MRC UCL Confidence in Concept (CiC6) 2017 MC_PC_17180.
The authors would like to thank Medicines for Malaria Venture (MMV) for supplying the Pandemic Response Box.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fitd.2022.1017900/full#supplementary-material
Supplementary Table 1 | Result of Primary screen of MMV Pandemic Response Box library at 1.0x10-4 M using growth/motility assay and DMSO-only as negative control. Table S1 contains data for three separate screen batches undertaken at three separate occasions each with 3 x identical replicates for each compound. INVAPP score indicates the rate of movement for each well of 96 well plates containing growth/motility assay. The table also contains the position of each compound at the experiments. Median rate for each replicate also included in the table with MMV name of each drug, Trival name, Disease area and Smile structure of each compound. The cut off to identify active compounds also presented.
Supplementary Table 2 | Result of Secondary screen of the 18 hit compounds identified in the primary screen of the MMV Pandemic Response Box library. They were re-tested on C. elegans L1s growth/motility assay at 5.0x10-5 M and (0.5% v/v DMSO) as negative control. Screens were undertaken at 3 separate occasions with 4 replicates each (n=3) with similar concentration of levamisole as a positive control. INVAPP score indicates the rate of movement for each well of 96 well plates containing growth/motility assay. Table also contains the position of each compound at the experiments, median rate for each replicate with MMV name of each compound.
Supplementary Table 3 | Result of Secondary screen of the 18 hit compounds identified in the primary screen of the MMV Pandemic Response Box library. They were re-tested on C. elegans L1s growth/motility assay at 7.5x10-5 M and (0.75% v/v DMSO) as negative control. Screens were undertaken at 3 separate occasions with 4 replicates each (n=3) with similar concentration of levamisole as a positive control. INVAPP score indicates the rate of movement for each well of 96 well plates containing growth/motility assay. Table also contains the position of each compound at the experiments, median rate for each replicate with MMV name of each compound.
Supplementary Table 4 | Result of Secondary screen of the 18 hit compounds identified in the primary screen of the MMV Pandemic Response Box library. They were re-tested on C. elegans L4 motility assay at 1.0x10-4 M and DMSO-only as negative control. Screens were undertaken at 3 separate occasions with 4 replicates each (n=3) with similar concentration of levamisole as a positive control. INVAPP score indicates the rate of movement for each well of 96 well plates containing motility assay. Table also contains the position of each compound at the experiments, median rate for each replicate with MMV name of each compound.
1. Elfawal MA, Savinov SN, Aroian RV. Drug screening for discovery of broad-spectrum agents for soil-transmitted nematodes. Sci Rep (2019) 9:12347. doi: 10.1038/s41598-019-48720-1
2. Hotez PJ, Kamath A. Neglected tropical diseases in Sub-Saharan Africa: Review of their prevalence, distribution, and disease burden. PloS Negl Trop Dis (2009) 3:e412. doi: 10.1371/journal.pntd.0000412
3. Else KJ, Keiser J, Holland CV, Grencis RK, Sattelle DB, Fujiwara RT, et al. Whipworm and roundworm infections. Nat Rev Dis Primer (2020) 6:44. doi: 10.1038/s41572-020-0171-3
4. Moser W, Schindler C, Keiser J. Drug combinations against soil-transmitted helminth infections. Adv Parasitol (2019) 103:91–115. doi: 10.1016/bs.apar.2018.08.002
5. Speich B, Ali SM, Ame SM, Bogoch II, Alles R, Huwyler J, et al. Efficacy and safety of albendazole plus ivermectin, albendazole plus mebendazole, albendazole plus oxantel pamoate, and mebendazole alone against trichuris trichiura and concomitant soil-transmitted helminth infections: a four-arm, randomised controlled trial. Lancet Infect Dis (2015) 15:277–84. doi: 10.1016/S1473-3099(14)71050-3
6. Moser W, Schindler C, Keiser J. Efficacy of recommended drugs against soil transmitted helminths: systematic review and network meta-analysis. BMJ (2017) 358:j4307. doi: 10.1136/bmj.j4307
7. World Health Organization. Soil-transmitted helminthiases 2019 data (2019) Available at: https://www.who.int/data/gho/data/themes/topics/soil-transmitted-helminthiases. (Accessed December 2, 2021).
8. Kaminsky R, Ducray P, Jung M, Clover R, Rufener L, Bouvier J, et al. A new class of anthelmintics effective against drug-resistant nematodes. Nature (2008) 452:176–80. doi: 10.1038/nature06722
9. Bartley DJ, Devin L, Nath M, Morrison AA. Selection and characterisation of monepantel resistance in teladorsagia circumcincta isolates. Int J Parasitol Drugs Drug Resist (2015) 5:69–76. doi: 10.1016/j.ijpddr.2015.05.001
10. Adegnika AA, Lötsch F, Mba RMO, Ramharter M. Update on treatment and resistance of human trichuriasis. Curr Trop Med Rep (2015) 2:218–23. doi: 10.1007/s40475-015-0061-z
11. Partridge FA, Bataille CJR, Forman R, Marriott AE, Forde-Thomas J, Häberli C, et al. Structural requirements for dihydrobenzoxazepinone anthelmintics: Actions against medically important and model parasites: Trichuris muris, brugia malayi, heligmosomoides polygyrus, and schistosoma mansoni. ACS Infect Dis (2021) 7:1260–74. doi: 10.1021/acsinfecdis.1c00025
12. Lees K, Sluder A, Shannan N, Hammerland L, Sattelle D. “Ligand-gated ion channels as targets for anthelmintic drugs: Past, current, and future perspectives,”. In: Caffrey CR, editor. Parasitic helminths. Weinheim, Germany: Wiley-VCH Verlag GmbH & Co. KGaA (2012). doi: 10.1002/9783527652969.ch1
13. Sharma S, Abuzar S. The benzimidazole anthelmintics–chemistry and biological activity. Prog Drug Res Fortschr Arzneimittelforschung Progres Rech. Pharm (1983) 27:85–161. doi: 10.1007/978-3-0348-7115-0_3
14. Partridge FA, Forman R, Bataille CJR, Wynne GM, Nick M, Russell AJ, et al. Anthelmintic drug discovery: Target identification, screening methods and the role of open science. Beilstein J Org Chem (2020) 16:1203–24. doi: 10.3762/bjoc.16.105
15. Partridge FA, Brown AE, Buckingham SD, Willis NJ, Wynne GM, Forman R, et al. An automated high-throughput system for phenotypic screening of chemical libraries on c. elegans and parasitic nematodes. Int J Parasitol Drugs Drug Resist (2018) 8:8–21. doi: 10.1016/j.ijpddr.2017.11.004
16. Partridge FA, Murphy EA, Willis NJ, Bataille CJR, Forman R, Heyer-Chauhan N, et al. Dihydrobenz[e][1,4]oxazepin-2(3H)-ones, a new anthelmintic chemotype immobilising whipworm and reducing infectivity in vivo. PloS Negl Trop Dis (2017) 11:e0005359. doi: 10.1371/journal.pntd.0005359
17. Partridge FA, Forman R, Willis NJ, Bataille CJR, Murphy EA, Brown AE, et al. 2,4-Diaminothieno[3,2-d]pyrimidines, a new class of anthelmintic with activity against adult and egg stages of whipworm. PloS Negl Trop Dis (2018) 12:e0006487. doi: 10.1371/journal.pntd.0006487
18. Brenner S. The genetics of caenorhabditis elegans. Genetics (1974) 77:71–94. doi: 10.1093/genetics/77.1.71
19. Holden-Dye L, Joyner M, O’Connor V, Walker RJ. Nicotinic acetylcholine receptors: a comparison of the nAChRs of Caenorhabditis elegans and parasitic nematodes. Parasitol Int (2013) 62:606–15. doi: 10.1016/j.parint.2013.03.004
20. Burns AR, Luciani GM, Musso G, Bagg R, Yeo M, Zhang Y, et al. Caenorhabditis elegans is a useful model for anthelmintic discovery. Nat Commun (2015) 6:7485. doi: 10.1038/ncomms8485
21. Samby K, Besson D, Dutta A, Patra B, Doy A, Glossop P, et al. The pandemic response Box─Accelerating drug discovery efforts after disease outbreaks. ACS Infect Dis (2022) 8:713–20. doi: 10.1021/acsinfecdis.1c00527
22. Buckingham SD, Partridge FA, Poulton BC, Miller BS, McKendry RA, Lycett GJ, et al. Automated phenotyping of mosquito larvae enables high-throughput screening for novel larvicides and offers potential for smartphone-based detection of larval insecticide resistance. PloS Negl Trop Dis (2021) 15:e0008639. doi: 10.1371/journal.pntd.0008639
23. Edelstein AD, Tsuchida MA, Amodaj N, Pinkard H, Vale RD, Stuurman N. Advanced methods of microscope control using μManager software. J Biol Methods (2014) 1:e10. doi: 10.14440/jbm.2014.36
24. Raymond V, Sattelle DB. Novel animal-health drug targets from ligand-gated chloride channels. Nat Rev Drug Discov (2002) 1:427–36. doi: 10.1038/nrd821
25. Geary TG, Sakanari JA, Caffrey CR. Anthelmintic drug discovery: Into the future. J Parasitol (2015) 101:125–33. doi: 10.1645/14-703.1
26. Holden-Dye L, Walker RJ. Anthelmintic drugs and nematicides: studies in Caenorhabditis elegans (2018). Available at: http://www.wormbook.org. WormBook (Accessed June 1, 2022).
27. Steger KA, Avery L. The GAR-3 muscarinic receptor cooperates with calcium signals to regulate muscle contraction in the caenorhabditis elegans pharynx. Genetics (2004) 167:633–43. doi: 10.1534/genetics.103.020230
28. Shanley HT, Taki AC, Byrne JJ, Jabbar A, Wells TNC, Samby K, et al. A high-throughput phenotypic screen of the ‘Pandemic response box’ identifies a quinoline derivative with significant anthelmintic activity. Pharmaceuticals (2022) 15:257. doi: 10.3390/ph15020257
29. Abosheasha MA, El-Gowily AH, Elfiky AA. Potential antiviral properties of antiplatelet agents against SARS-CoV-2 infection: an in silico perspective. J Thromb Thrombolysis (2022) 53:273–81. doi: 10.1007/s11239-021-02558-5
30. Thamburaj S, Ramaraj E, Sethupathy S, Kamalanathan C, Raji A, Rajasekharan SK. Antibacterial and antibiofilm activities of diphyllin against fish pathogens. Microb Pathog (2020) 145:104232. doi: 10.1016/j.micpath.2020.104232
31. Plescia CB, Lindstrom AR, Quintero MV, Keiser P, Anantpadma M, Davey R, et al. Evaluation of phenol-substituted diphyllin derivatives as selective antagonists for Ebola virus entry. ACS Infect Dis (2022) 8:942–57. doi: 10.1021/acsinfecdis.1c00474
32. Vautrin A, Manchon L, Garcel A, Campos N, Lapasset L, Laaref AM, et al. Both anti-inflammatory and antiviral properties of novel drug candidate ABX464 are mediated by modulation of RNA splicing. Sci Rep (2019) 9:792. doi: 10.1038/s41598-018-37813-y
33. Heuberger DM, Schuepbach RA. Protease-activated receptors (PARs): mechanisms of action and potential therapeutic modulators in PAR-driven inflammatory diseases. Thromb J (2019) 17:4. doi: 10.1186/s12959-019-0194-8
34. Zhang H, Pan D, Wu X, Su W, Tang X, Zhao D, et al. Platelet protease activated receptor 1 Is involved in the hemostatic effect of 20(S)-protopanaxadiol by regulating calcium signaling. Front Pharmacol (2020) 11:549150. doi: 10.3389/fphar.2020.549150
35. Chen H, Liu P, Zhang T, Gao Y, Zhang Y, Shen X, et al. Effects of diphyllin as a novel V-ATPase inhibitor on TE-1 and ECA-109 cells. Oncol Rep (2018) 39:921–8. doi: 10.3892/or.2018.6191
36. Tazi J, Begon-Pescia C, Campos N, Apolit C, Garcel A, Scherrer D. Specific and selective induction of miR-124 in immune cells by the quinoline ABX464: a transformative therapy for inflammatory diseases. Drug Discov Today (2021) 26:1030–9. doi: 10.1016/j.drudis.2020.12.019
37. Burris HA, Rivkin S, Reynolds R, Harris J, Wax A, Gerstein H, et al. Phase II trial of oral rubitecan in previously treated pancreatic cancer patients. Oncol. (2005) 10:183–90. doi: 10.1634/theoncologist.10-3-183
38. Suman T, Elangomathavan R, Kasipandi M, Chakkaravarthi K, Tamilvendan D, Parimelazhagan T. Diphyllin: An effective anticandidal agent isolated from Cleistanthus collinus leaf extract. Egypt. J Basic Appl Sci (2018) 5:130–7. doi: 10.1016/j.ejbas.2018.05.004
39. Di Giorgio C, Delmas F, Akhmedjnova V, Ollivier E, Bessonova I, Riad E, Timon-David P. In vitro antileishmanial activity of diphyllin isolated from Haplophyllum bucharicum. Planta Med. (2005) 71:366–369. doi: 10.1055/s-2005-864106
40. Gertsch J, Tobler RT, Brun R, Sticher O, Heilmann J. Antifungal, antiprotozoal, cytotoxic and piscicidal properties of justicidin b and a new arylnaphthalide lignan from Phyllanthus piscatorum. Planta Med (2003) 69:420–4. doi: 10.1055/s-2003-39706
41. Biendl S, Häberli C, Keiser J. Discovery of novel antischistosomal scaffolds from the open access pandemic response box. Expert Rev Anti Infect Ther (2022) 20:621–9. doi: 10.1080/14787210.2022.1990042
42. de Oliveira HC, Castelli RF, Reis FCG, Samby K, Nosanchuk JD, Alves LR, et al. Screening of the pandemic response box reveals an association between antifungal effects of MMV1593537 and the cell wall of cryptococcus neoformans, cryptococcus deuterogattii, and candida auris. Microbiol Spectr. (2022) 10: e0060122. doi: 10.1128/spectrum.00601-22
43. Rice CA, Troth EV, Russell AC, Kyle DE. Discovery of anti-amoebic inhibitors from screening the MMV pandemic response box on balamuthia mandrillaris, naegleria fowleri, and acanthamoeba castellanii. Pathogens (2020) 9:476. doi: 10.3390/pathogens9060476
44. Lian X, Dong J, Zhang J, Teng Y, Lin Q, Fu Y, et al. Soluplus(®) based 9-nitrocamptothecin solid dispersion for peroral administration: preparation, characterization, in vitro and in vivo evaluation. Int J Pharm (2014) 477:399–407. doi: 10.1016/j.ijpharm.2014.10.055
45. Knight E, Robinson E, Smoktunowicz N, Chambers RC, Aliev AE, Inglis GG, et al. Synthesis of novel and potent vorapaxar analogues. Org Biomol Chem (2016) 14:3264–74. doi: 10.1039/c5ob02541a
46. Fan M, Han M, Xia Y, Zhang Y, Chu Y, Bai G, et al. Design and synthesis of potent PAR-1 antagonists based on vorapaxar. Bioorg. Med Chem Lett (2020) 30:127046. doi: 10.1016/j.bmcl.2020.127046
47. Jorgensen EM, Mango SE. The art and design of genetic screens: Caenorhabditis elegans. Nat Rev Genet (2002) 3:356–69. doi: 10.1038/nrg794
48. Jones AK, Buckingham SD, Sattelle DB. Chemistry-to-gene screens in Caenorhabditis elegans. Nat Rev Drug Discov (2005) 4:321–30. doi: 10.1038/nrd1692
49. Dilks CM, Koury EJ, Buchanan CM, Andersen EC. Newly identified parasitic nematode beta-tubulin alleles confer resistance to benzimidazoles. Int J Parasitol Drugs Drug Resist (2021) 17:168–75. doi: 10.1016/j.ijpddr.2021.09.006
Keywords: Medicines for Malaria Venture (MMV) Pandemic Response Box, phenotypic chemical library screen, anthelmintic drug candidates, drug re-purposing, invertebrate automated phenotyping platform (INVAPP), Caenorhabditis elegans
Citation: Nick M, Partridge FA, Forman R, Bataille CJR, Else KJ, Russell AJ and Sattelle DB (2022) Screening the Medicines for Malaria Venture (MMV) Pandemic Response Box chemical library on Caenorhabditis elegans identifies re-profiled candidate anthelmintic drug leads. Front. Trop. Dis. 3:1017900. doi: 10.3389/fitd.2022.1017900
Received: 12 August 2022; Accepted: 09 September 2022;
Published: 13 October 2022.
Edited by:
Kenneth Pfarr, University Hospital Bonn, GermanyReviewed by:
Ivan Scandale, Drugs for Neglected Diseases initiative, SwitzerlandCopyright © 2022 Nick, Partridge, Forman, Bataille, Else, Russell and Sattelle. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David B. Sattelle, ZC5zYXR0ZWxsZUB1Y2wuYWMudWs=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.