
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Trop. Dis. , 17 February 2022
Sec. Antimicrobial Resistance
Volume 2 - 2021 | https://doi.org/10.3389/fitd.2021.810863
This article is part of the Research Topic Insights in Antimicrobial Resistance: 2021 View all 5 articles
 Sirjana Bhandari1†
Sirjana Bhandari1† Sanjib Adhikari1†
Sanjib Adhikari1† Deepa Karki1
Deepa Karki1 Arun Bahadur Chand2
Arun Bahadur Chand2 Sanjeep Sapkota3
Sanjeep Sapkota3 Binod Dhungel1
Binod Dhungel1 Megha Raj Banjara1
Megha Raj Banjara1 Prakash Joshi4
Prakash Joshi4 Binod Lekhak1
Binod Lekhak1 Komal Raj Rijal1*
Komal Raj Rijal1*Efflux-pump system and biofilm formation are two important mechanisms Pseudomonas aeruginosa deploys to escape the effects of antibiotics. The current study was undertaken from September 2019 to March 2020 at a tertiary-care hospital in Kathmandu in order to ascertain the burden of P. aeruginosa in clinical specimens, examine their biofilm-forming ability and determine their antibiotic susceptibility pattern along with the possession of two efflux-pump genes-mexA and mexB. Altogether 2820 clinical specimens were collected aseptically from the patients attending the hospital and processed according to standard microbiological procedures. Identification of P. aeruginosa was done by Gram stain microscopy and an array of biochemical tests. All the P. aeruginosa isolates were subjected to in vitro antibiotic susceptibility testing and their biofilm-forming ability was also examined. Presence of mexA and mexB efflux-pump genes was analyzed by Polymerase Chain Reaction (PCR) using specific primers. Out of 603 culture positive isolates, 31 (5.14%) were found to be P. aeruginosa, of which 55% were multi-drug resistant (MDR). Out of 13 commonly used antibiotics tested by Kirby-Bauer disc diffusion method, greatest resistance was shown against piperacillin-tazobactam 15 (48.4%) and ceftazidime 15 (48.4%), and least against meropenem 6 (19.4%) and ofloxacin 5 (16.2%). Of all 17 MDR isolates subjected to biofilm detection, strong biofilm formation was exhibited by 11 (65%) and 14 (82%) isolates with microtiter plate method and tube method respectively. Out of 17 isolates tested, 12 (70.6%) isolates possessed mexA and mexB genes indicating the presence of active efflux-pump system. Higher number of the isolates recovered from sputum 7 (58.3%) and pus 5 (41.7%) possessed mexA/mexB genes while the genes were not detected at all in the isolates recovered from the urine (p<0.05). This study assessed no significant association between biofilm production and multi-drug resistance (p>0.05). Adoption of stern measures by the concerned authorities to curb the incidence of multi-drug resistant and biofilm-forming isolates is recommended to prevent their dissemination in the hospital settings.
Pseudomonas aeruginosa is extensively spread opportunistic human pathogen associated with numerous acute and chronic human infections, such as respiratory and urinary tract infection, pneumonia and bacteremia (1). It can thrive in a broad range of different natural and artificial environments including medical surfaces, due to its suitability and its strong intrinsic antibiotic resistance (2, 3). P. aeruginosa is the sixth most common nosocomial pathogen that causes health care associated infections from 2011 to 2014 and the second most common pathogen in ventilator-associated pneumonia in hospitals of USA (4).
In recent years, increasing antibiotic resistance is becoming a global problem, and drug resistance in P. aeruginosa has become a serious concern due to its potential to cause numerous nosocomial and chronic infections (2). It is naturally resistant to a wide range of antimicrobial drugs and it may also evolve resistance to other antibiotic classes by mutating or acquiring foreign resistance genes (5). Besides, several resistance mechanisms include efflux-mediated, beta-lactamase production, biofilm production, pori-related resistance and target site modification (6–8). Efflux pumps are membrane proteins that are involved in the export of noxious substances from the bacterial cell into the external environment. According to their composition, the number of transmembrane spanning regions, energy sources and substrates, bacterial efflux pumps is classified into six families: ATP-binding cassette (ABC) superfamily, the small multidrug resistance (SMR) family, the major facilitator superfamily (MFS), the resistance-nodulation-division (RND) superfamily, the drug metabolite transporter (DMT) superfamily and the multidrug and toxic compound extrusion (MATE) family (9, 10). P. aeruginosa has a significant number of efflux pumps, including four potent RND-type multidrug resistance efflux pumps (Mex) that remove harmful chemicals from the periplasm and cytoplasm (6). In particular, the MexAB-oprM efflux pump system was reported to contribute in conferring inherent resistance to chloramphenicol, β-lactams, quinolones, macrolides novobiocin and tetracyclines (11). Overproduction of such types of efflux systems plays a significant role in development of multidrug-resistant (MDR) bacteria (12).
P. aeruginosa adherence to host cells via type IV pili, along with the secretion of extracellular polysaccharide mediated by polysaccharide synthesis locus (psl), enhances biofilm development and conceals the pathogen from the immune system of the host (13, 14). The biofilm formation is widely known for promoting antimicrobial resistance, and its presence during the infection process has been regarded as a limiting factor for treatment effectiveness (15).
In developing countries like Nepal, antibiotic resistances are usually detected merely by phenotypic methods (16). In fact, interpretations by such methods are not sufficient and uncommon resistant patterns are often complicated by false-positive or false-negative results which may lead to inappropriate antibiotic prescriptions and treatment failure. Furthermore, a phenotypic test does not allow detailed molecular identification of antimicrobial resistance which is needed for epidemiological and infection control purposes. The molecular methods can also aid in quick, accurate and sensitive determination of resistance status among such pathogens. Additionally, drug-resistant isolates are becoming more common in Nepal’s hospitals and clinics (16–20). The data available in the country for identifying resistant patterns is insufficient to define the antimicrobial resistance trend. Therefore, this study attempts to investigate the presence of two efflux-pump genes in clinical isolates of P. aeruginosa besides phenotypic detection of antibiotic resistance and biofilm production.
This was a hospital-based cross-sectional study conducted in Kist Medical College and Teaching Hospital, Lalitpur, Nepal from September 2019 to March 2020. A total of 2820 clinical specimens (pus, urine, sputum, urethral swab, CSF and pleural fluid) were collected during the study period. The samples were collected in sterile, leak-proof, clearly-labelled containers and processed immediately in the Microbiology laboratory of the hospital.
The clinical specimens were cultivated on Blood agar, CLED Agar (Cystine-Lactose-Electrolyte-Deficient Agar), Nutrient agar, MacConkey agar and brain heart infusion broth as well as bile broth (HiMedia, Mumbai, India)) depending on requirements and the isolated colonies were identified following standard microbiological procedures that involved studying the colonial morphology, Gram staining and various biochemical tests (21).
Screening of biofilm producing P. aeruginosa was done using tube method and tissue culture plate method (TCP). The method standardised by Mathur et al. (22) was followed to develop biofilms using tube method. In brief, a loopful of the overnight bacterial culture was inoculated in glass tube containing 10 mL of trypticase soy broth with 1% glucose. The inoculated tubes were then incubated at 37°C for 24 hours. After incubation, the tubes were emptied, washed with phosphate buffer saline and left to dry. Crystal violet (0.1%) was used to stain the dried tubes for 15 minutes. Excess stain was removed by washing the tubes with deionized water. The tubes were then dried in inverted position and examined for biofilm production. Formation of a stained ring at the air-liquid interface indicated a negative result for biofilm creation, whereas the presence of a visible film along the bottom and wall of the tube indicated a positive result for biofilm production.
Tissue culture plate method was executed following protocols described by Mathur et al. (22) in 96 flat-bottom wells. Briefly, a loopful of freshly cultured isolates was inoculated in 10 ml of trypticase soy broth with 1% glucose and kept in the incubator at 37°C for 24 hours. Bacterial suspension was further diluted (1:100) with fresh medium. Separate wells of a sterile polystyrene tissue culture plate, composed of 96 flat bottom wells, were filled by 200 µL of the prepared bacterial suspension and incubated at 37°C for 24 hours. The plate was gently tapped to remove the content of the wells followed by washing with 200 µL of phosphate buffer saline for four times. Sodium acetate (2%) was added to the wells and kept for 30 minutes to fix the biofilms formed by bacteria attached to the wells. Staining of the fixed biofilms was conducted using crystal violet (0.1%). After 30 minutes, the wells were thoroughly washed by deionized water. After drying, a micro-ELISA reader (at 570 nm wave length) was used to measure the optical densities (OD) of stained bacterial biofilms. The experiment was carried out in triplicate and three times, after which the data were averaged and the standard deviation was determined. To account for background absorbance, OD measurements from the sterile medium, fixative, and dye were averaged and subtracted from all test results. The mean OD value obtained from the medium control well was subtracted from all of the test OD readings.
The antibiotic-susceptibility testing was performed on Muller Hinton agar (MHA) by the modified Kirby-Bauer disc diffusion method following Clinical and Laboratory Standard guidelines (CLSI) 2018 (23). The antibiotic disks used in this study were procured from HiMedia Laboratories, Mumbai, India and included amikacin (30 µg), levofloxacin (5 µg), cefepime (50 µg), ceftazidime (30 µg), meropenem (10 µg), ciprofloxacin (5 µg), imipenem (10 µg), tobramycin (10 µg), piperacillin (100 µg), piperacillin-tazobactam (100/10 µg), gentamicin (10 µg), ofloxacin (5 µg) and aztreonam (30 µg). A suspension of bacteria adjusted to the 0.5 McFarland standards was swabbed using sterile swabs on MHA plates, and antibiotic discs were placed on top. The plates were incubated for 24 h at 37°C. The diameter of the zone of inhibition for each antibiotic was measured and interpreted as recommended by CLSI guidelines (23). Strains were considered multidrug-resistant (MDR) if they were resistant to at least one agent in three or more antimicrobial categories (24).
For the amplification of mexA and mexB gene, chromosomal DNA was extracted from each P. aeruginosa isolates following chromosomal DNA extraction method (25, 26). Briefly, the bacterial cells were lysed with 0.5 mg/mL lysozyme in the presence of 1/10 volume of 10% sodium dodecyl sulfate (SDS) at high pH and the lysate was then neutralized. Subsequent deproteinization with 1:1 phenol: chloroform, chromosomal DNA was precipitated with ethanol by spinning at high speed and extracted DNA was stored at -20°C. The primers used to detect the mexA and mexB gene have been enlisted in Table 1.
The polymerase chain reaction targeting mexAB gene was done in 25 μL reaction mixture consisting of 21 μL of 1X Qiagen master mix, 0.5 μL each of forward and reverse primers and 3 μL of extracted DNA template. Amplification was performed following conditions for mexAB gene which comprised of initial heating at 94°C for 3 min, then 32 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 45 s and extension at 72°C for 1 min and final extension at 72°C for 7 min. The PCR amplification products and 100bp DNA ladder were fractionated by electrophoresis through 1.5% (for 280bp) and 1% (for 503bp) agarose gel visualized by staining with ethidium bromide (1μg/mL).
The efficiency of laboratory equipments such as an incubator, autoclave, hot air oven, and refrigerator was frequently evaluated. The expiry date and storage conditions of reagents, media, and antibiotic discs were thoroughly examined. Likewise, the quality of each batch of culture and biochemical media was assured by incubating a randomly sampled medium at 37°C for 24 h. A reference strain of Pseudomonas aeruginosa ATCC 27853 was used to maintain the quality control of AST. Similarly, the thickness and pH of MHA were maintained at 4 mm and 7.2-7.4 respectively.
All the data obtained were entered into SPSS and analyzed using SPSS (Version 22.0). Chi square test was performed to draw association between the related variables and a value of p ≤ 0.05 was considered significant wherever applicable.
A total of 2820 clinical specimen were collected and processed for microbial analysis of which 603 (21.4%) were culture positive. The highest percentage of microbial growth was reported in urine 313 (51.9%) followed by sputum 117 (28.4%) and body fluids 119 (19.7%). Bacterial growth was not reported in pleural fluid and CSF (Figure 1).
Out of 603 culture positive isolates, 31 isolates were reported as P. aeruginosa. The prevalence of P. aeruginosa among culture positive isolates was 5.14% (Figure 2).
Out of 2820 clinical samples processed, 603 were culture positive, among which E. coli (49.6%) was the most frequently isolated bacteria followed by K. pneumoniae (14.6%), Acinetobacter spp. (9.8%), and Staphylococcus aureus (8.4%). The most common bacteria isolated from urine, sputum and body fluids were E. coli (78.9%), K. pneumoniae (30.9%) and Staphylococcus aureus (38.7%) respectively. Highest incidence of P. aeruginosa was observed in sputum 14 (45.16%) samples (Table 2).
The antibiotic susceptibility pattern of P. aeruginosa isolates revealed that ofloxacin, meropenem and amikacin were the most effective antibiotics in vitro with sensitivities of 83.8%, 80.6% and 74.2% respectively. In contrast, piperacillin-tazobactam and ceftazidime were the least effective antibiotics inhibiting the growth of 48.4% isolates. The antibiotic susceptibility pattern of P. aeruginosa isolates is depicted in Table 3. Out of 31 P. aeruginosa isolates subjected to AST, 17 (54.84%) were identified to be MDR (Figure 3).
All seventeen MDR isolates were selected and tested for biofilm production by both tube culture method and microtiter plate method. Out of 17 isolates of P. aeruginosa subjected to biofilm detection, 11 (65%) showed strong biofilm with microtiter plate method while 14 (82%) showed strong biofilm production with tube method. There was no significant association between two methods for biofilm formation (p>0.05). Among thirteen antibiotics tested, resistance to piperacillin, cefepime, aztreonam, imipenem and meropenem showed significant association with strong biofilm production (p<0.05) (Table 4).
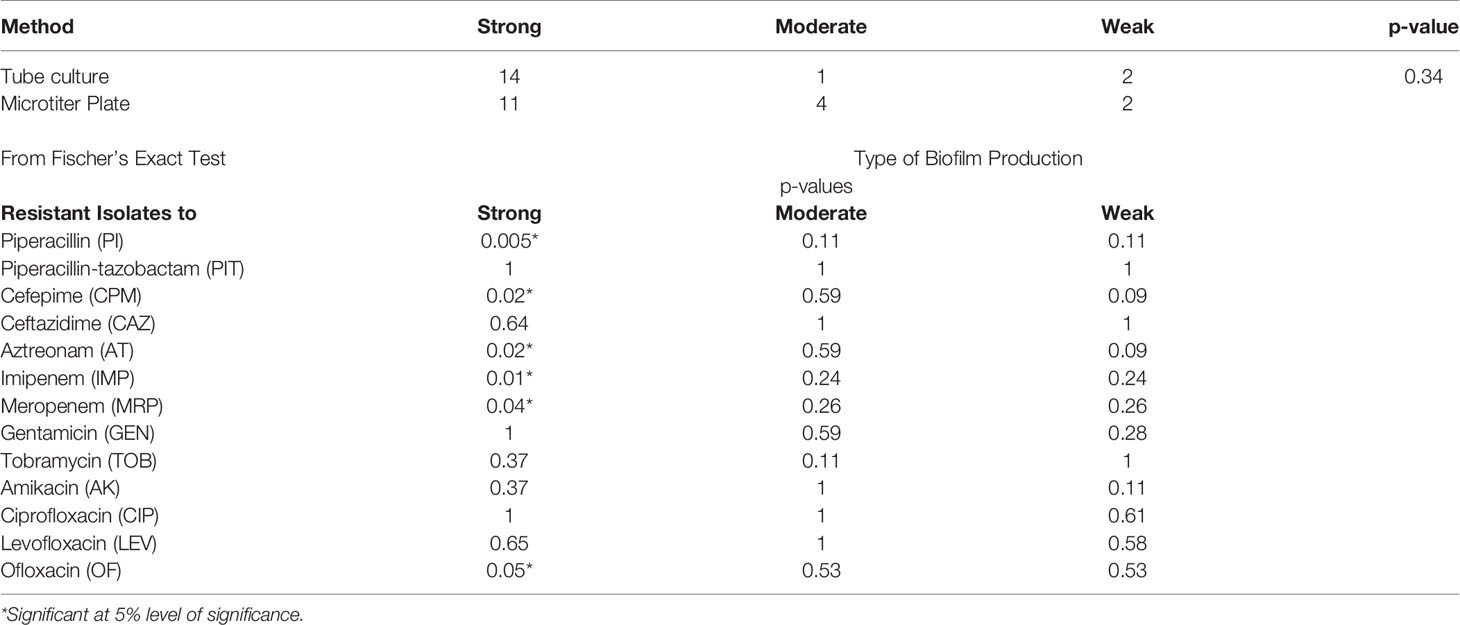
Table 4 Biofilm production and its association with antibiotic resistance in Pseudomonas aeruginosa (n=17).
Among thirty-one isolates of P. aeruginosa, only the MDR isolates (17) were subjected to PCR amplification for the detection of mexA and mexB genes. Among them, twelve (71%) isolates were found to possess both mexA and mexB genes. The PCR amplified product size of mexA and mexB genes were 503 bp and 280 bp respectively (Figures 4A, B).
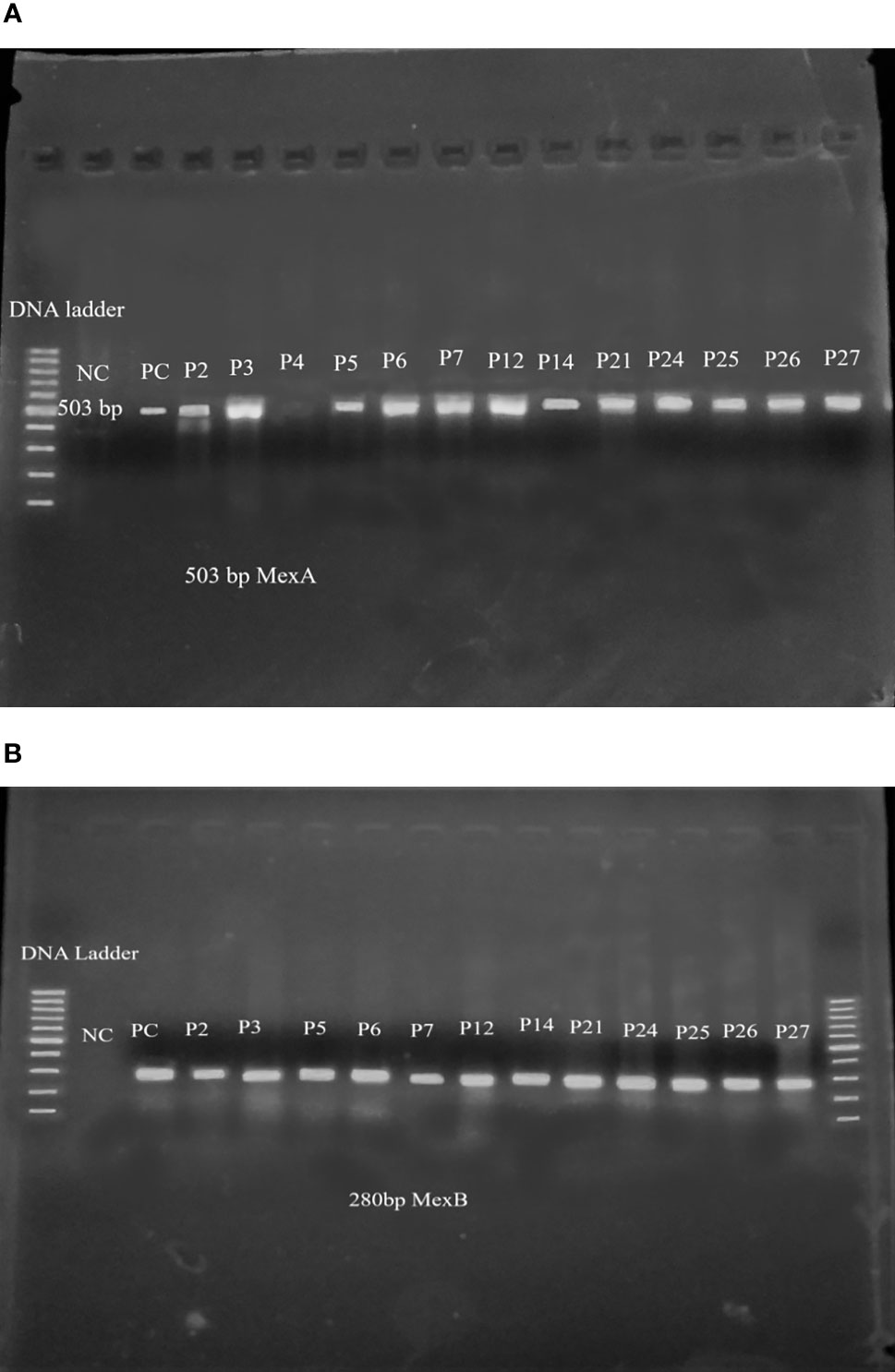
Figure 4 (A) mexA gene amplified product after PCR in agarose gel electrophoresis. Lane 1-100bp DNA ladder, lane 2- negative control, lane 3- positive control, lane4-P2, lane5-P3, lane6-P4, lane7-P5, lane8-P6, lane9-P7, lane10- P12, lane11-P14, lane12- P21, lane13-P24, lane14- P25, lane15- P26, lane16- P27. (B) mexB gene amplified product after PCR in agarose gel electrophoresis. Lane 1-100bp DNA ladder, lane 2- negative control, lane 3- positive control, lane 4-P2, lane 5-P3, lane 6-P5, lane7-P6, lane8-P7, lane9-P12, lane10- P14, lane11-P21, lane12- P24, lane13-P25, lane14- P26, lane15- P27, lane16- 100bp DNA ladder.
A substantial number of P. aeruginosa isolates recovered from sputum 7 (58.3%) and body fluids 5 (41.7%) possessed both mexA and mexB genes (p<0.05). The occurrence of mexA and mexB genes was found more in the isolates that were resistant to levofloxacin (p>0.05). Majority of the MDR isolates possessed both mexA and mexB genes (p>0.05). Moreover, the prevalence of the mexA and mexB gene was found more in the isolates that were resistant to beta-lactam antibiotics (p>0.05) (Table 5).
P. aeruginosa is regarded as a diverse opportunistic human pathogen, with final infection comprising attachment, colonization, local invasion and spread as a systemic disease (27, 28). This study documented its growth in 21.4% of total clinical specimens. Different studies conducted in Nepal reported its growth rate of 13-58% in clinical specimens (29–31). The prevalence of P. aeruginosa in this study was 5.14% which is in tune with the earlier studies done in Nepal (32, 33); however, the prevalence rate is lower than the previous studies done in Nepal (34), India (35) and Egypt (36). Growth of bacteria in different clinical specimens differs with the hospitals, number of samples, type of patients, and time duration of the study.
In this study, E. coli was the most predominant Gram-negative bacterial isolate whereas S. aureus was the predominant Gram-positive bacterial isolate. This finding was in line with other studies reported from different hospitals/laboratory settings in Nepal (37–40). Observation of higher number of Gram-negative bacterial isolates in clinical specimens in different studies conducted in Nepal indicates Gram-negative bacteria are emerging as important health-care associated problems (41, 42). The highest incidence of E. coli and S. aureus may be due to factors such as their ubiquity in hospital environments and propensity to form biofilm on numerous uninhabitable surfaces.
This study reported ofloxacin and meropenem as the choice of drug for P. aeruginosa infection whereas ceftazidime was the least effective drug. Similar to this finding, several studies conducted in Nepal reported ceftazidime as the least effective drug (29, 43, 44). The presence of naturally occurring inducible cephalosporinase in P. aeruginosa imparts a low-level resistance to cephalosporins and aminopenicillins (45). Prevalence of MDR among P. aeruginosa isolates was 55% which was higher than the prevalence rate reported in other studies (29, 42). The widespread use of antibiotics in humans and animals, as well as inadequate infection control and increased mobility of people might have all contributed to the fast spread of multidrug-resistant bacteria.
The pathogenesis of the majority of chronic P. aeruginosa infections is mainly due to the capacity of this organism to form biofilm that imparts resistance to many antibiotics. All MDR P. aeruginosa isolates showed strong to weak biofilm formation in the current study. This finding echoes with the study done in Nigeria (46); however, a significant lower incidence (33.7%) of biofilm forming P. aeruginosa was reported in the study done in India (47). Biofilms embedded bacteria are difficult to eradicate by conventional antibiotic therapy as bacterial cells living inside biofilms are more tolerant towards antibiotics than their planktonic counterparts (48). P. aeruginosa resistant to piperacillin, cefepime, aztreonam, imipenem and meropenem were reported as the strong biofilm formation. Biofilm producing P. aeruginosa exhibit increased resistance to several antibiotics including tetracycline, chloramphenicol, quinolones, β-lactams and these resistance patterns could be closely related to drug profiles that are actively effluxed by the MexAB-OprM pump (49).
In the current study, all seventeen MDR isolates were selected for PCR amplification, twelve isolates (70.58%) showed the presence of both mexA and mexB genes. A similar study done in Suez Canal University Hospital, Egypt reported both mexA and mexB genes in 58.8% Pseudomonas strains which is in commensurate with our finding (50). On the other hand, Abbas et al. reported mexA and mexB genes in 100% isolates (25). RND efflux pumps (MexAB–OprM) are transport proteins in P. aeruginosa that may play a role in the extrusion of all types of therapeutically relevant antimicrobial drugs from the cell into the external environment (51). This study reported an insignificant association between the presence of mexA and mexB genes with fluoroquinolone and beta-lactams. The result of this study is in contrast with that of the result of a work by Dupont et al. who concluded that MexAB-OprM is responsible for efflux of beta-lactams and quinolones (52). Apart from the MexAB-OprM efflux pump system, three additional efflux pump systems (MexCD-OprJ, MexEF-OprN, and MexXY-OprM) also contribute to antibiotic resistance. Biofilm and efflux pumps are not the only variables that contribute to an organism’s antibiotic resistance, but many other factors also play a part in creating an organism drug resistant.
This study reported 21.4% bacterial growth in various clinical specimens at a tertiary-care hospital of which 5.1% were Pseudomonas aeruginosa and more than half of them were multi-drug resistant. All the isolates tested showed biofilm-forming capacity with majority being strong producers. The efflux-pump genes mexA and mexB were detected in 70.58% of the isolates tested. It’s high time the concerned authorities employ serious and robust measures to prevent the dissemination of MDR isolates with biofilm-forming potential within and outside the hospital environments.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
The studies involving human participants were reviewed and approved by Institutional Review Committee (IRC) of Institute of Science and Technology (IOST), Tribhuvan University, Nepal. The patients/participants provided their written informed consent to participate in this study.
Conceiving and study design: SB, KR, BL, and MR. Lab work and data collection: SB and DK. Supervision: BL, SA, AC, PJ, and KR. Data curation: SB, BD, MR, and KR. Data analysis: SS, BD, SA, and KR. Drafting the manuscript: SS, SA, BD, and KR. Review, editing and finalizing the manuscript: SA, BD, BL, and KR. All authors have read and approved to the published version of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to express our sincere thanks to all the staffs of KIST Medical College, Gwarko, Lalitpur, Nepal for their support and coordination to carry out this study. We are very much thankful to the laboratory staffs, Central Department of Microbiology, Kirtipur, Kathmandu for support. Above all, we are grateful to all the patients for their involvement in this study.
1. Gellatly SL, Hancock RE. Pseudomonas Aeruginosa: New Insights Into Pathogenesis and Host Defenses. Pathog Dis (2013) 67:159–73. doi: 10.1111/2049-632X.12033
2. Moradali MF, Ghods S, Rehm BH. Pseudomonas Aeruginosa Lifestyle: A Paradigm for Adaptation, Survival, and Persistence. Front Cell Infect Microbiol (2017) 7:39. doi: 10.3389/fcimb.2017.00039
3. Redfern J, Enright MC. Further Understanding of Pseudomonas Aeruginosa’s Ability to Horizontally Acquire Virulence: Possible Intervention Strategies. Expert Rev Anti-Infective Ther (2020) 18:539–49. doi: 10.1080/14787210.2020.1751610
4. Weiner-Lastinger LM, Abner S, Benin AL, Edwards JR, Kallen AJ, Karlsson M, et al. Antimicrobial-Resistant Pathogens Associated With Pediatric Healthcare-Associated Infections: Summary of Data Reported to the National Healthcare Safety Network, 2015–2017. Infect Control Hosp Epidemiol (2020) 41:19–30. doi: 10.1017/ice.2019.297
5. Strateva T, Yordanov D. Pseudomonas Aeruginosa–a Phenomenon of Bacterial Resistance. J Med Microbiol (2009) 58:1133–48. doi: 10.1099/jmm.0.009142-0
6. Santajit S, Indrawattana N. Mechanisms of Antimicrobial Resistance in ESKAPE Pathogens. BioMed Res Int (2016) 2016:2475067. doi: 10.1155/2016/2475067
7. Kriengkauykiat J, Porter E, Lomovskaya O, Wong-Beringer A. Use of an Efflux Pump Inhibitor to Determine the Prevalence of Efflux Pump-Mediated Fluoroquinolone Resistance and Multidrug Resistance in Pseudomonas Aeruginosa. Antimicrob Agents Chemother (2005) 49:565–70. doi: 10.1128/AAC.49.2.565-570.2005
8. Lima J, Alves LR, Jacomé P, Bezerra JP, Maciel MAV, Morais M. Biofilm Production by Clinical Isolates of Pseudomonas Aeruginosa and Structural Changes in LasR Protein of Isolates non Biofilm-Producing. Braz J Infect Dis (2018) 22:129–36. doi: 10.1016/j.bjid.2018.03.003
9. Sun J, Deng Z, Yan A. Bacterial Multidrug Efflux Pumps: Mechanisms, Physiology and Pharmacological Exploitations. Biochem Biophys Res Commun (2014) 453:254–67. doi: 10.1016/j.bbrc.2014.05.090
10. Piddock LJ. Clinically Relevant Chromosomally Encoded Multidrug Resistance Efflux Pumps in Bacteria. Clin Microbiol Rev (2006) 19:382–402. doi: 10.1128/CMR.19.2.382-402.2006
11. Horna G, López M, Guerra H, Saénz Y, Ruiz J. Interplay Between MexAB-OprM and MexEF-OprN in Clinical Isolates of Pseudomonas Aeruginosa. Sci Rep (2018) 8:1–11. doi: 10.1038/s41598-018-34694-z
12. Schweizer HP. Efflux as a Mechanism of Resistance to Antimicrobials in Pseudomonas Aeruginosa and Related Bacteria: Unanswered Questions. Genet Mol Res (2003) 2:48–62.
13. Burrows LL. Pseudomonas Aeruginosa Twitching Motility: Type IV Pili in Action. Annu Rev Microbiol (2012) 66:493–520. doi: 10.1146/annurev-micro-092611-150055
14. Hayashi N, Nishizawa H, Kitao S, Deguchi S, Nakamura T, Fujimoto A, et al. Pseudomonas Aeruginosa Injects Type III Effector ExoS Into Epithelial Cells Through the Function of Type IV Pili. FEBS Lett (2015) 589:890–6. doi: 10.1016/j.febslet.2015.02.031
15. Worlitzsch D, Tarran R, Ulrich M, Schwab U, Cekici A, Meyer KC, et al. Effects of Reduced Mucus Oxygen Concentration in Airway Pseudomonas Infections of Cystic Fibrosis Patients. J Clin Invest (2002) 109:317–25. doi: 10.1172/JCI0213870
16. Rijal KR, Banjara MR, Dhungel B, Kafle S, Gautam K, Ghimire B, et al. Use of Antimicrobials and Antimicrobial Resistance in Nepal: A Nationwide Survey. Sci Rep (2021) 11:11554. doi: 10.1038/s41598-021-90812-4
17. Aryal SC, Upreti MK, Sah AK, Ansari M, Nepal K, Dhungel B, et al. Plasmid-Mediated AmpC Beta-Lactamase CITM and DHAM Genes Among Gram-Negative Clinical Isolates. Infect Drug Resist (2020) 13:4249–61. doi: 10.2147/IDR.S284751
18. Raut S, Rijal KR, Khatiwada S, Karna S, Khanal R, Adhikari J, et al. Trend and Characteristics of Acinetobacter Baumannii Infections in Patients Attending Universal College of Medical Sciences, Bhairahawa, Western Nepal: A Longitudinal Study of 2018. Infect Drug Resist (2020) 13:1631–41. doi: 10.2147/IDR.S257851
19. Muktan B, Shrestha UT, Dhungel B, Mishra BC, Shrestha N, Adhikari N, et al. Plasmid Mediated Colistin Resistant Mcr-1 and Co-Existence of OXA-48 Among Escherichia Coli From Clinical and Poultry Isolates: First Report From Nepal. Gut Pathog (2020) 12:1–9. doi: 10.1186/s13099-020-00382-5
20. Gurung S, Kafle S, Dhungel B, Adhikari N, Thapa Shrestha U, Adhikari B, et al. Detection of OXA-48 Gene in Carbapenem-Resistant Escherichia Coli and Klebsiella Pneumoniae From Urine Samples. Infect Drug Resist (2020) 13:2311–21. doi: 10.2147/IDR.S259967
21. Forbes B, Sahm D, Weissfeld A. Scott’s, Diagnostic Microbiology, Mosby Elsevier. St. Louis MO USA (2007).
22. Mathur T, Singhal S, Khan S, Upadhyay D, Fatma T, Rattan A. Detection of Biofilm Formation Among the Clinical Isolates of Staphylococci: An Evaluation of Three Different Screening Methods. Indian J Med Microbiol (2006) 24:25–9. doi: 10.1016/S0255-0857(21)02466-X
23. Clinical and Laboratory Standards Institute. Performance of Standards for Antimicrobial Susceptibility Testing, Vol. M100. Wayne, Pennsylvania (2018).
24. Magiorakos A-P, Srinivasan A, Carey R, Carmeli Y, Falagas M, Giske C, et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin Microbiol Infect (2012) 18:268–81. doi: 10.1111/j.1469-0691.2011.03570.x
25. Abbas HA, El-Ganiny AM, Kamel HA. Phenotypic and Genotypic Detection of Antibiotic Resistance of Pseudomonas Aeruginosa Isolated From Urinary Tract Infections. Afr Health Sci (2018) 18:11–21. doi: 10.4314/ahs.v18i1.3
26. Al-Grawi IGA, khdier Al-Absali A, Kareem NH, Belal SA-L. Occurrence of MexAB-OprM Efflux Pump Operon on Septicemic Pseudomonas Aeruginosa Chromosome. Iraqi Postgraduate Med J (2012) 11:97–102.
27. Mesaros N, Nordmann P, Plésiat P, Roussel-Delvallez M, Van Eldere J, Glupczynski Y, et al. Pseudomonas Aeruginosa: Resistance and Therapeutic Options at the Turn of the New Millennium. Clin Microbiol Infect (2007) 13:560–78. doi: 10.1111/j.1469-0691.2007.01681.x
28. Liew SM, Rajasekaram G, Puthucheary SA, Chua KH. Antimicrobial Susceptibility and Virulence Genes of Clinical and Environmental Isolates of Pseudomonas Aeruginosa. PeerJ (2019) 7:e6217. doi: 10.7717/peerj.6217
29. Pokharel K, Dawadi BR, Bhatt CP, Gupte S. Prevalence of Pseudomonas Aeruginosa and its Antibiotic Sensitivity Pattern. J Nepal Health Res Council (2019) 17:109–13. doi: 10.33314/jnhrc.v17i01.1877
30. Thapa E, Thapa R, Singh A, Pokharel BM, Devkota U. Antibiotic Resistance Pattern of Bacterial Isolates From Post-Tracheostomised Patients Attending Tertiary Care Hospital in Nepal. J Institute Sci Technol (2016) 21:129–32. doi: 10.3126/jist.v21i1.16064
31. Thakur P, Ghimire P, Rijal KR, Singh GK. Antimicrobial Resistance Pattern of Escherichia Coli Isolated From Urine Samples in Patients Visiting Tertiary Health Care Centre in Eastern Nepal. Sunsari Tech Coll J (2012) 1(1):22–6. doi: 10.3126/stcj.v1i1.8657
32. Adhikari S, Khadka S, Sapkota S, Rana JC, Khanal S, Neupane A, et al. Prevalence and Antibiograms of Uropathogens From the Suspected Cases of Urinary Tract Infections in Bharatpur Hospital, Nepal. J Coll Med Sciences Nepal (2019) 15:260–6. doi: 10.3126/jcmsn.v15i4.20856
33. Gurung R, Adhikari S, Adhikari N, Sapkota S, Rana JC, Dhungel B, et al. Efficacy of Urine Dipstick Test in Diagnosing Urinary Tract Infection and Detection of the blaCTX-M Gene Among ESBL-Producing Escherichia Coli. Diseases (2021) 9:59. doi: 10.3390/diseases9030059
34. Sales A, Fathi R, Mobaiyen H, Bonab F, Kondlaji K. Molecular Study of the Prevalence of CTX-M1, CTX-M2, CTXM3 in Pseudomanas aeruginosa Isolated From Clinical Samples in Tabriz Town, Iran. Electronic J Biol (2017) 13:253–9.
35. Ranjan KP, Ranjan N, Bansal SK, Arora D. Prevalence of Pseudomonas Aeruginosa in Post-Operative Wound Infection in a Referral Hospital in Haryana, India. J Lab Physicians (2010) 2:074–7. doi: 10.4103/0974-2727.72153
36. Gad GF, El-Domany RA, Zaki S, Ashour HM. Characterization of Pseudomonas Aeruginosa Isolated From Clinical and Environmental Samples in Minia, Egypt: Prevalence, Antibiogram and Resistance Mechanisms. J Antimicrob Chemother (2007) 60:1010–7. doi: 10.1093/jac/dkm348
37. Guragain N, Pradhan A, Dhungel B, Banjara MR, Rijal KR, Ghimire P. Extended Spectrum β-Lactamase Producing Gram Negative Bacterial Isolates From Urine of Patients Visiting Everest Hospital, Kathmandu, Nepal. TUJM (2019) 6(1):26–31. doi: 10.3126/tujm.v6i0.26575
38. Upadhyaya G Bhattarai A, Rijal KR, Ghimire P, Upadhyaya B. Urinary Tract Infections in Kidney Transplant Patients of Kathmandu Valley. Int J Microbiol Res Rev (2013) 3(1):1–6.
39. Karki D, Dhungel B, Bhandari S, Kunwar A, Joshi PR, Shrestha B, et al. Antibiotic Resistance and Detection of Plasmid Mediated Colistin Resistance Mcr-1 Gene Among Escherichia Coli and Klebsiella Pneumoniae Isolated From Clinical Samples. Gut Pathog (2021) 13(1):45. doi: 10.1186/s13099-021-00441-5
40. Dhungel S, Rijal KR, Yadav B, Dhungel B, Adhikari N, Shrestha UT, et al. Methicillin-Resistant Staphylococcus Aureus (MRSA): Prevalence, Antimicrobial Susceptibility Pattern, and Detection of mecA Gene Among Cardiac Patients From a Tertiary Care Heart Center in Kathmandu, Nepal. Infect Dis (Auckl) (2021) 14:11786337211037355. doi: 10.1177/11786337211037355
41. Koirala S, Khadka S, Sapkota S, Sharma S, Khanal S, Thapa A, et al. Prevalence of CTX-M b-Lactamases Producing Multidrug Resistant Escherichia Coli and Klebsiella Pneumoniae Among Patients Attending Bir Hospital, Nepal. BioMed Res Int (2021) 2021:9958294. doi: 10.1155/2021/9958294
42. Sah RSP, Dhungel B, Yadav BK, Adhikari N, Thapa Shrestha U, Lekhak B, et al. Detection of TEM and CTX-M Genes in Escherichia Coli Isolated From Clinical Specimens at Tertiary Care Heart Hospital, Kathmandu, Nepal. Diseases (2021) 9:15. doi: 10.3390/diseases9010015
43. Bhandari S, Banjara MR, Lekhak B, Bhatta DR, Regmi SR. Multi-Drug and Pan-Drug Resistant Pseudomonas Aeruginosa: A Challenge in Post-Antibiotic Era. Nepal J Sci Technol (2012) 13:197–202. doi: 10.3126/njst.v13i2.7736
44. Dahal RH, Chaudhary DK. Microbial Infections and Antimicrobial Resistance in Nepal: Current Trends and Recommendations. Open Microbiol J (2018) 12:230. doi: 10.2174/1874285801812010230
45. Chen H, Yuan M, Livermore D. Mechanisms of Resistance to β-Lactam Antibiotics Amongst Pseudomonas Aeruginosa Isolates Collected in the UK in 1993. J Med Microbiol (1995) 43:300–9. doi: 10.1099/00222615-43-4-300
46. Ugwuanyi FC, Ajayi A, Ojo DA, Adeleye AI, Smith SI. Evaluation of Efflux Pump Activity and Biofilm Formation in Multidrug Resistant Clinical Isolates of Pseudomonas Aeruginosa Isolated From a Federal Medical Center in Nigeria. Ann Clin Microbiol Antimicrob (2021) 20:1–7. doi: 10.1186/s12941-021-00417-y
47. Gurung J, Khyriem AB, Banik A, Lyngdoh WV, Choudhury B, Bhattacharyya P. Association of Biofilm Production With Multidrug Resistance Among Clinical Isolates of Acinetobacter Baumannii and Pseudomonas Aeruginosa From Intensive Care Unit. Indian J Crit Care Medicine: Peer-Reviewed Off Publ Indian Soc Crit Care Med (2013) 17:214. doi: 10.4103/0972-5229.118416
48. Furiga A, Lajoie B, El Hage S, Baziard G, Roques C. Impairment of Pseudomonas Aeruginosa Biofilm Resistance to Antibiotics by Combining the Drugs With a New Quorum-Sensing Inhibitor. Antimicrob Agents Chemother (2015) 60:1676–86. doi: 10.1128/AAC.02533-15
49. Soto SM. Role of Efflux Pumps in the Antibiotic Resistance of Bacteria Embedded in a Biofilm. Virulence (2013) 4:223–9. doi: 10.4161/viru.23724
50. Kishk RM, Abdalla MO, Hashish AA, Nemr NA, El Nahhas N, Alkahtani S, et al. Efflux MexAB-Mediated Resistance in P. Aeruginosa Isolated From Patients With Healthcare Associated Infections. Pathogens (2020) 9:471. doi: 10.3390/pathogens9060471
51. Pan YP, Xu YH, Wang ZX, Fang YP, Shen Jl. Overexpression of MexAB-OprM Efflux Pump in Carbapenem-Resistant Pseudomonas Aeruginosa. Arch Microbiol (2016) 198:565–71. doi: 10.1007/s00203-016-1215-7
Keywords: mexA and mexB gene, MDR, efflux pump, PCR, biofilm
Citation: Bhandari S, Adhikari S, Karki D, Chand AB, Sapkota S, Dhungel B, Banjara MR, Joshi P, Lekhak B and Rijal KR (2022) Antibiotic Resistance, Biofilm Formation and Detection of mexA/mexB Efflux-Pump Genes Among Clinical Isolates of Pseudomonas aeruginosa in a Tertiary Care Hospital, Nepal. Front. Trop. Dis 2:810863. doi: 10.3389/fitd.2021.810863
Received: 08 November 2021; Accepted: 30 December 2021;
Published: 17 February 2022.
Edited by:
Dennis Nurjadi, Heidelberg University Hospital, GermanyReviewed by:
Luria Leslie Founou, Centre of Expertise and Biological Diagnostic of Cameroon (CEDBCAM), CameroonCopyright © 2022 Bhandari, Adhikari, Karki, Chand, Sapkota, Dhungel, Banjara, Joshi, Lekhak and Rijal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Komal Raj Rijal, a29tYWwucmlqYWxAY2RtaS50dS5lZHUubnA=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.