- 1Laboratory of Malaria Immunology and Vaccinology (LMIV), National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), Bethesda, MD, United States
- 2Malaria Research and Training Center (MRTC)/University of Sciences, Techniques, and Technologies of Bamako (USTTB), Bamako, Mali
- 3Laboratory of Clinical Immunology and Microbiology (LCIM), National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), Bethesda, MD, United States
Malaria may affect the reliability of SARS-CoV-2 seroassay performance and limit understanding of SARS-CoV-2 epidemiology in malaria-endemic regions. We present our experience conducting SARS-CoV-2 serosurveillance in seasonal malaria-affected communities in Mali and discuss relevant literature regarding the effect of malaria on the performance of SARS-CoV-2 serodiagnostics, including approaches to minimize the effect of malaria-associated assay interference.
Introduction
There are many possible interfaces for COVID-19 and malaria in malaria-endemic regions, including pandemic-related program disruptions, shifts in clinical disease burden, and the effect of malaria on SARS-CoV-2 test performance (1, 2). In partnership with the Malian Ministry of Health, the Malaria Research and Training Center (MRTC) and the National Institutes of Health sought to assist in the public health response to COVID-19 in Mali. MRTC maintains clinical and research laboratory infrastructure in many regions of Mali to develop tools and strategies for malaria control and elimination. This infrastructure was leveraged to develop COVID-19 serosurveillance testing capacity, and forms part of a large in-country response to swiftly redirect existing research resources to address the pandemic (3). It is critical to ensure that serosurveillance tools provide reliable data for use in the local public health response. We discuss relevant literature regarding the effect of malaria on the performance of SARS-CoV-2 serodiagnostics, present our experience qualifying assay performance and conducting SARS-CoV-2 serosurveillance in seasonal malaria-affected communities in Mali, West Africa (4, 5), present new data regarding the effect of recent malaria infection on SARS-CoV-2 serostatus, and outline approaches to optimize assay performance in malaria-endemic regions.
Malaria and SARS-CoV-2 Serodiagnostics
For serological surveillance to be useful in the public health response to the COVID-19 pandemic, test selection and validation in the target population must be addressed. High rates of false-positivity have been described for multiple commercial SARS-CoV-2 serological assays in sub-Saharan Africa (6–11), and in other regions (12, 13), conceivably due to cross-reactivity with other coronaviruses or other endemic infections, including malaria. This is not a new phenomenon, for example high rates of false-positivity in HIV antibody tests have been previously described in Africa (14–16). Poor SARS-CoV-2 test specificity, particularly in the low or unknown prevalence setting, risks overestimation of community burden and may cause unnecessary harm, including diversion of limited public health resources and inaccurate estimations of population-level exposure or immunity. To ensure reliable serosurveillance data, the primary aim of assay qualification must be understanding the pattern and degree of background reactivity to SARS-CoV-2 antigens in order to optimize assay performance, while understanding the nature and causes of this reactivity an important secondary aim. As a result, we conducted extensive assay qualification to optimize test performance in Mali prior to conducting SARS-CoV-2 community serosurveillance (5). This included evaluating negative control samples collected prior to 2020 for background IgG reactivity to commonly tested SARS-CoV-2 antigens, other betacoronaviruses, and a panel of P. falciparum antigens, assessing in vitro SARS-CoV-2 pseudovirus neutralizing activity in negative controls, and evaluating the performance of several test configurations using local positive and negative controls.
While there has been minimal correlation between exposure to other human coronaviruses and SARS-CoV-2 seropositivity in pre-pandemic samples (5, 6, 11, 12, 17), malaria has been variably associated with poor assay performance (5, 7, 9, 17). In malaria-endemic regions including Mali, background antibody reactivity to SARS-CoV-2 antigens is common, increases with age group, and varies regionally (5, 17). Increased reactivity with age may suggest cumulative exposures may be responsible for background signal, and that assay performance may vary in different age groups. Background SARS-CoV-2 antigen reactivity may also vary seasonally, although year-round samples were not available for assessment in a Malian community with highly seasonal malaria transmission (Supplementary Figure 1). These observations implicate malaria as a potential confounder when considering SARS-CoV-2 seroassay performance.
Several mechanisms for malaria-associated SARS-CoV-2 antigen cross-reactivity have been proposed. SARS-CoV-2 nucleocapsid antibody reactivity by commercial assay has been associated with the presence of IgG antibodies to some Plasmodium sp. antigens, and several other neglected tropical diseases (9), although this is not consistent between SARS-CoV-2 assays. We did not demonstrate any substantial correlation between SARS-CoV-2 antigen reactivity using our reference ELISA and a panel of 11 P. falciparum antigens responsible for both short-lived and long-lived serological response following malaria infection (5, 18). A weak positive correlation has been reported between the same SARS-Cov-2 antigen constructs and AMA-1 ELISA reactivity in malaria-experienced Cambodian samples (12). While it remains possible that specific antibodies to Plasmodium sp. antigens could cross-react with SARS-CoV-2 antigens, the effect may not be substantial or predictable across assays. Non-specific polyclonal antibodies arising from acute malaria may also contribute to poor assay performance. Symptomatic and asymptomatic malaria have been associated with transient SARS-CoV-2 spike protein antibody reactivity, attributed to cross-reactive antibodies targeting N-linked glycans (17). Additionally, commercial spike protein and nucleocapsid based SARS-CoV-2 assay false-positivity has been associated with higher parasitemia malaria infections, although the presence of malaria alone versus no malaria did not reach statistical significance in this study (7). While acute malaria may affect SARS-CoV-2 assay performance, similar to cross-reactive Plasmodium sp. antibodies, this phenomenon may not be consistent across all groups and assays. As a result, there may not be a single distinct mechanism through which malaria induces SARS-CoV-2 assay reactivity. Irrespective of the stimulus, antibodies responsible for SARS-CoV-2 antigen background reactivity in malaria-endemic areas do not demonstrate functional activity in vitro (5, 7, 12, 17).
Optimizing Test Performance in Malaria-Endemic Areas
The concept of malaria-associated cross-reactivity presents several challenges to SARS-CoV-2 assay implementation, where performance must be balanced with cost and the timeliness of results. Specific collaboration with groups conducting intensive malaria studies may be warranted to obtain control samples and address the issue of malaria-associated assay interference. Ultimately any testing approach must be pragmatic and suited to local conditions and available resources. Approaches that may improve test performance include: comparison of different antigens to select constructs with lower background reactivity (5, 11, 13, 17), cutoff adjustment to set population-specific thresholds or relative to local background reactivity (5, 19), dual antigen testing (5, 19, 20), and avidity testing to eliminate low-affinity cross-reactive antibodies (9) (Table 1). Groups responsible for testing in malaria-endemic regions should consider determining background reactivity/assay false positivity rates in pre-pandemic samples and make compensatory adjustments if required before implementing high volume testing. This approach is outlined in Figure 1 and may include selection of one or several methods to optimize test specificity.
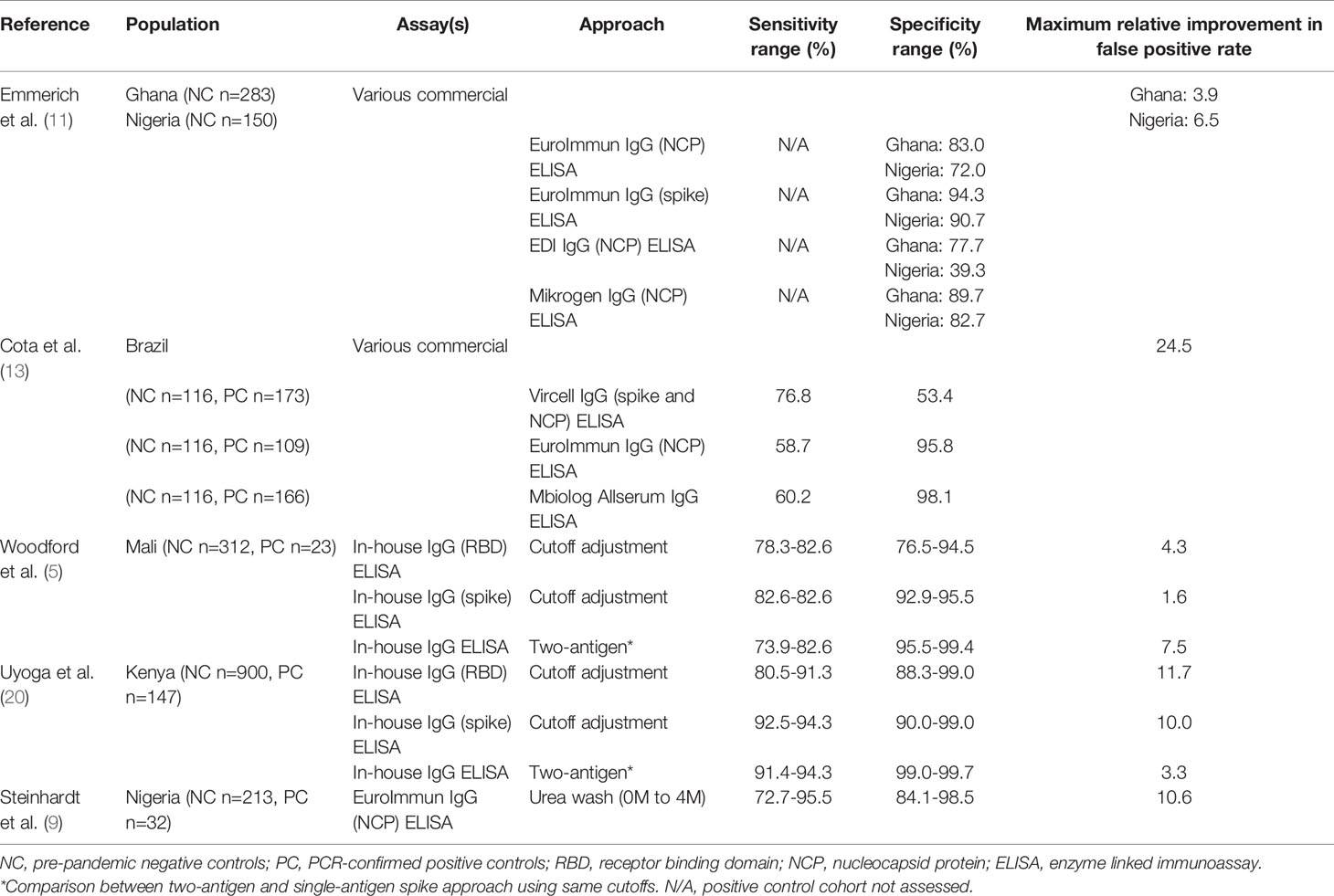
Table 1 Effect of several approaches to improve SARS-CoV-2 serology test performance in malaria-endemic regions.
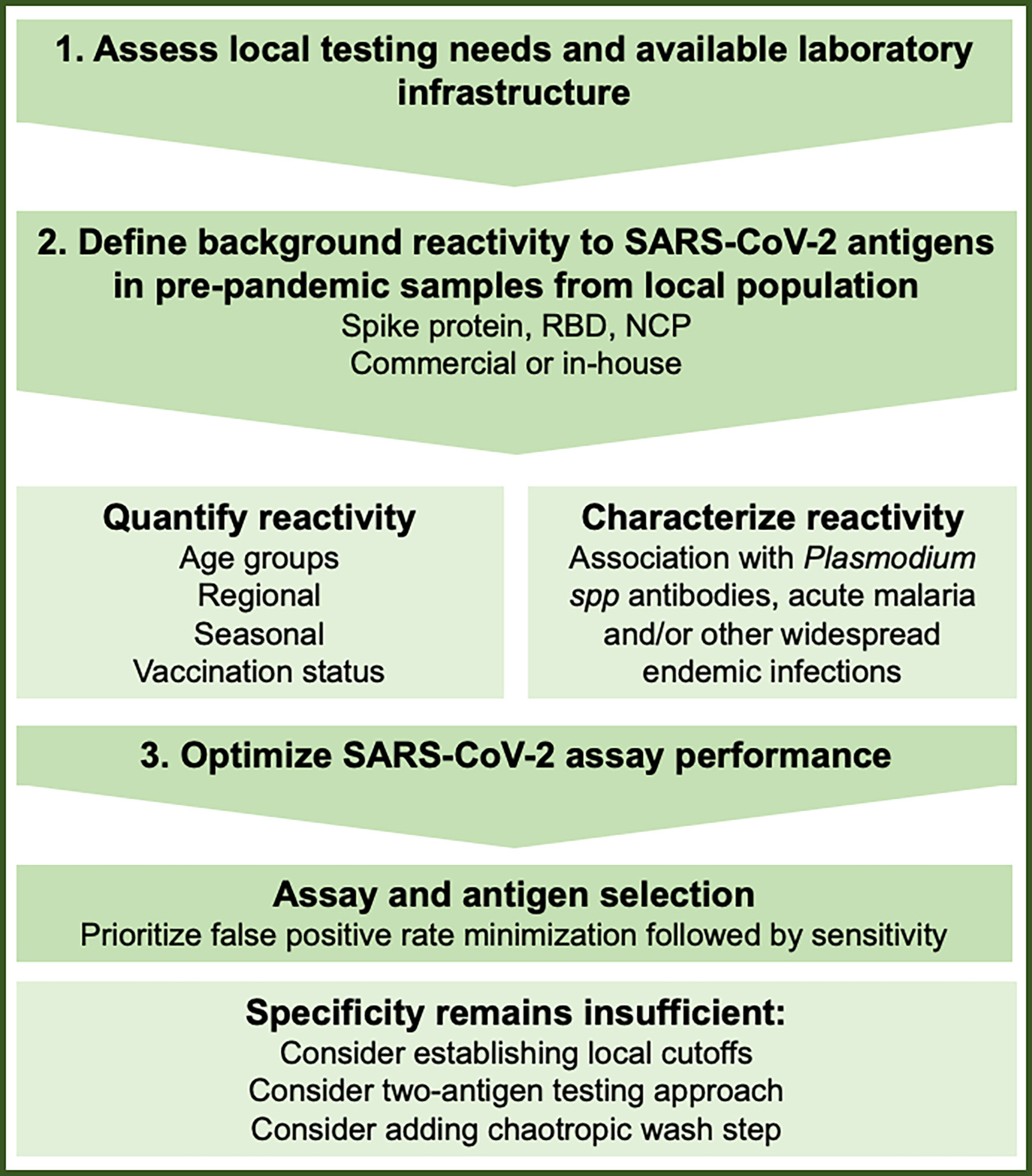
Figure 1 Considerations to understand and minimize the effect of SARS-CoV-2 serology background reactivity in malaria endemic regions.
In our study, a two-antigen ELISA assessing mammalian cell-expressed SARS-COV-2 spike protein and RBD antigens with population-specific cutoffs was selected based on the performance of this configuration in a Malian control population and the availability of in-country laboratory facilities and expertise (4, 5). Background ELISA reactivity in Malian pre-pandemic samples was most pronounced to SARS-CoV-2 nucleocapsid protein. While spike protein and RBD signals were comparatively lower, neither was sufficiently specific for a single-antigen approach without using prohibitively high assay cutoffs (5). Interestingly, there was minimal correlation between SARS-CoV-2 antigen reactivity in pre-pandemic samples (5), consolidating the concept that a two-antigen approach may be useful to minimize background signal.
To understand the practical effect of seasonal malaria on the performance of our assay, we analyzed our Malian serosurveillance data with respect to malaria transmission at a community and individual level. Overall, SARS-CoV-2 community seroprevalence was inversely proportional to seasonal malaria transmission intensity. Both before and after the 2020 malaria season, SARS-CoV-2 seroprevalence was consistently higher urban areas compared to rural regions with higher entomological inoculation rates (4) (Supplementary Text). In a population of 1037 individuals of all ages that were SARS-CoV-2 seronegative at the beginning of the 2020 malaria season, subjects underwent comprehensive malaria monitoring as part of an MRTC clinical trial supported by EDCTP (21) at the rural village of Donéguébougou. In this cohort, 33.3% (345/1037) experienced at least one episode of acute malaria between serosurvey visit 1 and visit 2, and 24.3% (252/1037) demonstrated SARS-CoV-2 two-antigen seroconversion during the malaria season. There was no difference in the rate of malaria diagnosis in the SARS-CoV-2 seropositive group compared to the seronegative group (34.9% (88/252) vs 32.7% (257/785), Fisher exact test p=0.54). In a multiple logistic regression model including age, an episode of acute malaria was not associated with SARS-CoV-2 seropositivity (OR 1.13, 95% CI: 0.83-1.53) (Figure 2A). In the subset of 345 individuals experiencing at least one acute malaria episode, both SARS-CoV-2 spike protein and RBD reactivity were not correlated with time since recent malaria diagnosis (Spearman r=-0.04, p=0.46 for spike protein and r=-0.04, p=0.52 for RBD respectively) (Figure 2B). Furthermore, in a multivariate analysis of these cases, time since most recent malaria diagnosis (OR 1.00, 95% CI: 0.99-1.01), and number of acute malaria episodes since visit 1 (OR 0.86, 95% CI: 0.51-1.40) were not associated with SARS-CoV-2 seropositivity at visit 2. Paired with the overall observation that SARS-CoV-2 seroprevalence was lowest in communities with the most intense malaria transmission (4) (Supplementary Text), the lack of association between recent acute malaria and SARS-CoV-2 serostatus suggests the two-antigen reference ELISA selected for community serosurveillance in Mali minimizes the possible interference of seasonal malaria on SARS-CoV-2 antibody testing.
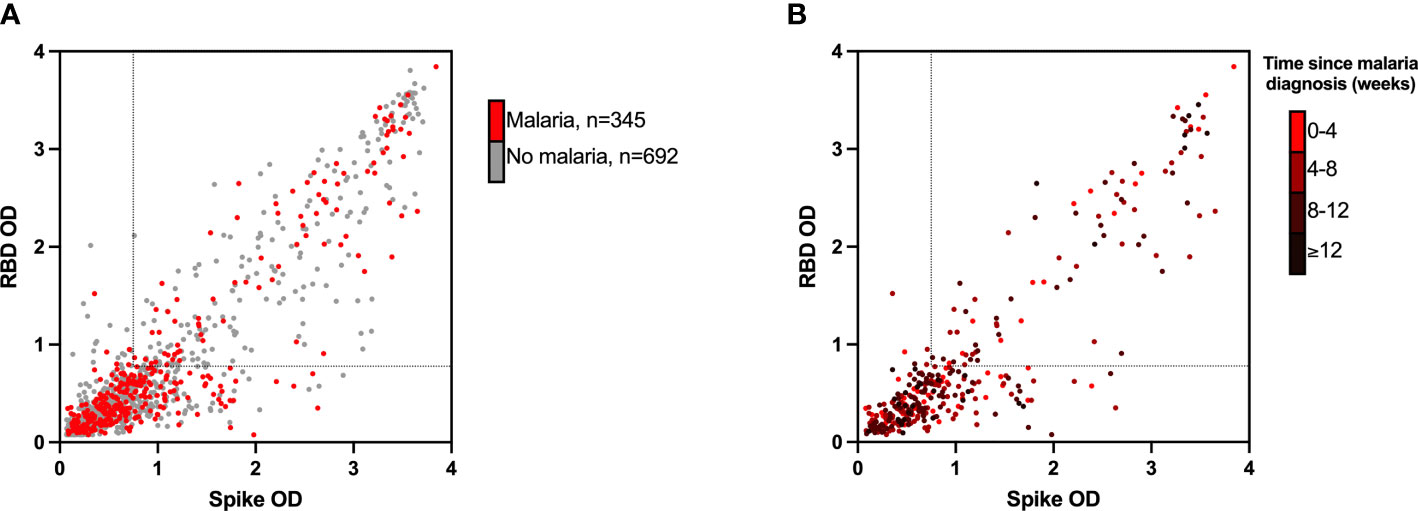
Figure 2 Assay absorbance values (OD) for RBD (y-axis) and spike protein (x-axis) at visit 2 (A) in all 1037 participants undergoing intensive malaria surveillance and (B) in 345 participants experiencing at least one acute malaria episode, stratified by time since time since malaria diagnosis. RBD, receptor binding domain; OD, optical density. Dotted lines represent assay cutoffs for SARS-CoV-2 seropositivity. In panel (B) darker data points represent a longer time since most recent malaria diagnosis (weeks).
Discussion
To understand interactions between COVID-19 and malaria it is critical that reliable SARS-CoV-2 surveillance is available in malaria-endemic regions. In many cases, these regions may be both disproportionately affected by uncertain assay performance and most in need of reliable serological tools due to limited access to gold-standard molecular diagnostics. Careful assay qualification is required prior to use, particularly where demographics and exposure histories differ significantly from the original assay validation population (5, 22). While it appears that a spike-based assay is likely to have better specificity compared to other antigens, we are unable to recommend a specific SARS-CoV-2 serological assay for use in malaria-endemic regions, as there are limited head-to-head studies, performance varies regionally, and there may be other local confounding factors that need to be addressed. Rather, a tailored approach is needed based on the target population, where one or several of the approaches presented above are likely to assist in test optimization. By using a two-antigen test, adjusting seropositivity thresholds, and reviewing the effect of intercurrent malaria on serostatus following implementation, we believe that we have minimized the effect of malaria on SARS-CoV-2 seroprevalence interpretation in Mali.
Reliable SARS-CoV-2 serosurveillance in communities with intense seasonal malaria transmission allows for a better understanding of the twin burdens of COVID-19 and malaria in Mali. We have identified high rates of seasonal malaria and COVID-19 infection in Mali, including many individuals with evidence of adjacent or concurrent infections. Despite rapidly increasing seroprevalence, we did not identify a large burden of COVID-19 attributable clinical disease in community serosurveillance (4). Combined malaria and SARS-CoV-2 surveillance allows for the informed allocation of resources to fight the emergent COVID-19 virus and the long-standing malaria epidemic. Understanding of the COVID-19 pandemic in Mali may also allow for the safe continuation of the malaria elimination agenda, and avoidance of excess malaria mortality due to program disruptions.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Deidentified data collected for this study may be made available to others after approval of and with a signed data access agreement.
Ethics Statement
The studies contributing to this manuscript were reviewed and approved by the USTTB FMOS/FAPH ethics committee and the NIH IRB. Written informed consent or assent was obtained from all study participants.
Author Contributions
JW collated and analyzed the data and drafted the manuscript. All authors contributed to the conception and design of the studies contributing data to the analysis. All authors contributed to manuscript revision, and approved the submitted version.
Funding
This project was funded by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health. Clinical trial NCT03917654 was conducted as part of the EDCTP2 program supported by the European Union (grant number RIA2018SV-2311: 2019-2024).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We gratefully acknowledge the support of the Malian COVID-19 Coordinator and Ministry of Health for permission to partner in developing serosurveillance capacity in Mali; Emily Higbee, Rathy Mohan, and Sara Healy (LMIV/NIAID/NIH) for providing access to NCT03917654 datasets; the EDCTP2 program for supporting ongoing MRTC malaria clinical trials and encouraging COVID-19 surveillance during the study period; and Kevin Tetteh and Chris Drakeley (London School of Hygiene and Tropical Medicine) for materials to perform P. falciparum multiplex suspension bead assay.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fitd.2021.781586/full#supplementary-material
References
1. Sherrard-Smith E, Hogan AB, Hamlet A, Watson OJ, Whittaker C, Winskill P, et al. The Potential Public Health Consequences of COVID-19 on Malaria in Africa. Nat Med (2020) 26(9):1411–6. doi: 10.1038/s41591-020-1025-y
2. Gutman JR, Lucchi NW, Cantey PT, Steinhardt LC, Samuels AM, Kamb ML, et al. Malaria and Parasitic Neglected Tropical Diseases: Potential Syndemics With COVID-19? Am J Trop Med Hyg (2020) 103(2):572–7. doi: 10.4269/ajtmh.20-0516
3. Doumbia S, Sow Y, Diakite M, Lau CY. Coordinating the Research Response to COVID-19: Mali’s Approach. Health Res Policy Syst (2020) 18(1):105. doi: 10.1186/s12961-020-00623-8
4. Sagara I, Woodford J, Kone M, Assadou MH, Katile A, Attaher O, et al. Rapidly Increasing SARS-CoV-2 Seroprevalence and Limited Clinical Disease in Three Malian Communities: A Prospective Cohort Study. Clin Infect Dis (2021) ciab589. doi: 10.1101/2021.04.26.21256016
5. Woodford J, Sagara I, Dicko A, Zeguime A, Doucoure M, Kwan J, et al. SARS-CoV-2 Seroassay Performance and Optimization in a Population With High Background Reactivity in Mali. J Infect Dis (2021) jiab579. doi: 10.1093/infdis/jiab498
6. Tso FY, Lidenge SJ, Pena PB, Clegg AA, Ngowi JR, Mwaiselage J, et al. High Prevalence of Pre-Existing Serological Cross-Reactivity Against Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) in Sub-Saharan Africa. Int J Infect Dis (2021) 102:577–83. doi: 10.1016/j.ijid.2020.10.104
7. Yadouleton A, Sander AL, Moreira-Soto A, Tchibozo C, Hounkanrin G, Badou Y, et al. Limited Specificity of Serologic Tests for SARS-CoV-2 Antibody Detection, Benin. Emerg Infect Dis (2021) 27(1):233–7. doi: 10.3201/eid2701.203281
8. Nkuba Ndaye A, Hoxha A, Madinga J, Marien J, Peeters M, Leendertz FH, et al. Challenges in Interpreting SARS-CoV-2 Serological Results in African Countries. Lancet Glob Health (2021) 9(5):e588–9. doi: 10.1016/S2214-109X(21)00060-7
9. Steinhardt LC, Ige F, Iriemenam NC, Greby SM, Hamada Y, Uwandu M, et al. Cross-Reactivity of Two SARS-CoV-2 Serological Assays in a Malaria-Endemic Setting. J Clin Microbiol, (2021) 59(7):e0051421. doi: 10.1128/JCM.00514-21
10. Fischer PU, Fischer K, Curtis KC, Huang Y, Fetcho N, Goss CW, et al. Evaluation of Commercial Rapid Lateral Flow Tests, Alone or in Combination, for SARS-CoV-2 Antibody Testing. Am J Trop Med Hyg (2021) 105(2):378–86. doi: 10.4269/ajtmh.20-1390
11. Emmerich P, Murawski C, Ehmen C, von Possel R, Pekarek N, Oestereich L, et al. Limited Specificity of Commercially Available SARS-CoV-2 IgG ELISAs in Serum Samples of African Origin. Trop Med Int Health (2021) 26(6):621–31. doi: 10.1101/2020.09.15.20159749
12. Manning J, Zaidi I, Lon C, Rosas LA, Park JK, Ponce A, et al. Pre-Pandemic SARS-CoV-2 Serological Cross-Reactivity in Rural Malaria-Experienced Cambodians. Emerg Infect Dis (2021). doi: 10.1101/2021.09.27.21264000
13. Cota G, Freire ML, de Souza CS, Pedras MJ, Saliba JW, Faria V, et al. Diagnostic Performance of Commercially Available COVID-19 Serology Tests in Brazil. Int J Infect Dis (2020) 101:382–90. doi: 10.1016/j.ijid.2020.10.008
14. Gasasira AF, Dorsey G, Kamya MR, Havlir D, Kiggundu M, Rosenthal PJ, et al. False-Positive Results of Enzyme Immunoassays for Human Immunodeficiency Virus in Patients With Uncomplicated Malaria. J Clin Microbiol (2006) 44(8):3021–4. doi: 10.1128/JCM.02207-05
15. Everett DB, Baisely KJ, McNerney R, Hambleton I, Chirwa T, Ross DA, et al. Association of Schistosomiasis With False-Positive HIV Test Results in an African Adolescent Population. J Clin Microbiol (2010) 48(5):1570–7. doi: 10.1128/JCM.02264-09
16. Lejon V, Ngoyi DM, Ilunga M, Beelaert G, Maes I, Buscher P, et al. Low Specificities of HIV Diagnostic Tests Caused by Trypanosoma Brucei Gambiense Sleeping Sickness. J Clin Microbiol (2010) 48(8):2836–9. doi: 10.1128/JCM.00456-10
17. Lapidus S, Liu F, Casanovas-Massana A, Dai Y, Huck JD, Lucas C, et al. Plasmodium Infection Induces Cross-Reactive Antibodies to Carbohydrate Epitopes on the SARS-CoV-2 Spike Protein. MedRxiv (2021) doi: 2021.05.10.21256855
18. Wu L, Hall T, Ssewanyana I, Oulton T, Patterson C, Vasileva H, et al. Optimisation and Standardisation of a Multiplex Immunoassay of Diverse Plasmodium Falciparum Antigens to Assess Changes in Malaria Transmission Using Sero-Epidemiology. Wellcome Open Res (2019) 4:26. doi: 10.12688/wellcomeopenres.14950.1
19. Uyoga S, Adetifa IMO, Karanja HK, Nyagwange J, Tuju J, Wanjiku P, et al. Seroprevalence of Anti-SARS-CoV-2 IgG Antibodies in Kenyan Blood Donors. Science (2021) 371(6524):79–82. doi: 10.1126/science.abe1916
20. Klumpp-Thomas C, Kalish H, Drew M, Hunsberger S, Snead K, Fay MP, et al. Standardization of ELISA Protocols for Serosurveys of the SARS-CoV-2 Pandemic Using Clinical and at-Home Blood Sampling. Nat Commun (2021) 12(1):113. doi: 10.1038/s41467-020-20383-x
21. Duffy PE. NCT03917654: Pfs230D1M-EPA/AS01 Vaccine, a Transmission Blocking Vaccine Against Plasmodium Falciparum, in an Age De-Escalation Trial of Children and a Family Compound Trial in Mali. (2019). Available at: https://clinicaltrials.gov/ct2/show/NCT03917654.
Keywords: malaria, COVID-19, SARS-CoV-2, serology, antibody, cross-reactivity
Citation: Woodford J, Sagara I, Kwan J, Zaidi I, Dicko A and Duffy PE (2021) Assessing and Minimizing the Effect of Malaria on SARS-CoV-2 Serodiagnostics. Front. Trop. Dis 2:781586. doi: 10.3389/fitd.2021.781586
Received: 22 September 2021; Accepted: 18 November 2021;
Published: 13 December 2021.
Edited by:
Herman Kosasih, Indonesia Research Partnership on Infectious Disease (INA-RESPOND), IndonesiaReviewed by:
Amy Bei, Yale University, United StatesKalpana Nathan, Stanford University, United States
Sukmawati Basuki, Airlangga University, Indonesia
Copyright © 2021 Woodford, Sagara, Kwan, Zaidi, Dicko and Duffy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John Woodford, am9obi53b29kZm9yZEBuaWguZ292
 John Woodford
John Woodford Issaka Sagara2
Issaka Sagara2 Jennifer Kwan
Jennifer Kwan Irfan Zaidi
Irfan Zaidi Patrick E. Duffy
Patrick E. Duffy