- 1The W. Harry Feinstone Department of Molecular Microbiology and Immunology, The Johns Hopkins Malaria Research Institute, Johns Hopkins Bloomberg School of Public Health, Baltimore, MD, United States
- 2Department of Biological Sciences, Purdue University, West Lafayette, IN, United States
- 3Tropical Diseases Research Centre, Ndola, Zambia
- 4Macha Research Trust, Choma, Zambia
- 5Florida Medical Entomology Laboratory, Department of Entomology and Nematology, Institute of Food and Agricultural Sciences, University of Florida, Vero Beach, FL, United States
Residual vector populations that do not come in contact with the most frequently utilized indoor-directed interventions present major challenges to global malaria eradication. Many of these residual populations are mosquito species about which little is known. As part of a study to assess the threat of outdoor exposure to malaria mosquitoes within the Southern and Central Africa International Centers of Excellence for Malaria Research, foraging female anophelines were collected outside households in Nchelenge District, northern Zambia. These anophelines proved to be more diverse than had previously been reported in the area. In order to further characterize the anopheline species, sequencing and phylogenetic approaches were utilized. Anopheline mosquitoes were collected from outdoor light traps, morphologically identified, and sent to Johns Hopkins Bloomberg School of Public Health for sequencing. Sanger sequencing from 115 field-derived samples yielded mitochondrial COI sequences, which were aligned with a homologous 488 bp gene segment from known anophelines (n = 140) retrieved from NCBI. Nuclear ITS2 sequences (n = 57) for at least one individual from each unique COI clade were generated and compared against NCBI’s nucleotide BLAST database to provide additional evidence for taxonomical identity and structure. Molecular and morphological data were combined for assignment of species or higher taxonomy. Twelve phylogenetic groups were characterized from the COI and ITS2 sequence data, including the primary vector species Anopheles funestus s.s. and An. gambiae s.s. An unexpectedly large proportion of the field collections were identified as An. coustani and An. sp. 6. Six phylogenetic groups remain unidentified to species-level. Outdoor collections of anopheline mosquitoes in areas frequented by people in Nchelenge, northern Zambia, proved to be extremely diverse. Morphological misidentification and underrepresentation of some anopheline species in sequence databases confound efforts to confirm identity of potential malaria vector species. The large number of unidentified anophelines could compromise the malaria vector surveillance and malaria control efforts not only in northern Zambia but other places where surveillance and control are focused on indoor-foraging and resting anophelines. Therefore, it is critical to continue development of methodologies that allow better identification of these populations and revisiting and cleaning current genomic databases.
Introduction
Human malaria is transmitted by species of mosquitoes in the genus Anopheles. There are approximately 450 recognized species of anopheline mosquitoes worldwide, which are placed into six main subgenera: Anopheles, Cellia, Kerteszia, Lophopodomyia, Nyssorhynchus, and Stethomyia. The largest of these subgenera by far are Anopheles (183 species) and Cellia (224 species) (1). Cellia has an old-world distribution, Anopheles is cosmopolitan, and the remaining subgenera are neotropical in distribution. Despite this diversity, there are fewer than 50 species within the entire Anopheles genus that are classically considered important to maintaining human malaria transmission (2).
Subgenera of anophelines can be further divided into smaller taxonomic units, including Sections, Series, Groups, and species complexes. Members of species complexes are morphologically difficult to distinguish, so a combination of morphological, behavioral/ecological, and molecular approaches must be used to identify species (3). Species-level identification is important because even at the level of highly-related species within a complex, behavior, ecology, vector competence and susceptibility to vector control can be highly variable (4–11). Members of the An. funestus sensu lato (s.l.) group, for example, differ in terms of host preference, foraging behavior, insecticide resistance, and ecological niche (12–14). For well-studied systems like the Anopheles gambiae complex, it has been repeatedly shown the importance of good genetic markers for any association studies investigating insecticide resistance (9, 15) or vector competence (11, 16).
In pre-elimination settings where the principal malaria vectors have been largely reduced, persistent malaria transmission has been frustratingly difficult to control. In these malaria endemic regions, vector surveillance has been challenging partly due to limited resources for molecular and genetic species verification for understudied, undescribed or morphologically cryptic anophelines. To date, most phylogenetic studies have focused on classically-recognized malaria vectors (17). Although morphology remains the primary and most cost-effective method for anopheline identification, newly-discovered and largely undescribed anopheline species are increasingly recognized as important to malaria transmission (18–23), and the scarcity of genetic data for development of molecular diagnostics for a wider range of anopheline species significantly hinder rapid confirmation of potential vectors of malaria transmission (24).
When diverse specimens from field collections remain unidentified using routine morphological and polymerase chain reaction (PCR)-based methods, sequence comparison to National Center for Biotechnology Information (NCBI) non-redundant nucleotide database using the Basic Local Alignment Search Tool (BLAST) may be used (25). This method has limited power for identifying understudied vector species because of the paucity of well-documented reference sequences for neglected anopheline species. For example, Anopheles gambiae has over 300,000 sequences in the NCBI Nucleotide database. Other well studied members of the Anopheles gambiae complex and An. funestus have just over 1000 entries in the NCBI Nucleotide database. In contrast, understudied species have at most 200 entries and most of them have under 80 entries (Figure 1). Due to recent changes some species designations (3) these could be misidentified. Unverified specimens, due to discrepancies in feature annotations, could also be excluded from a BLAST search. As this is much more common for understudied species, a BLAST search may have limited utility as a method of species identification, depending on genomic target.
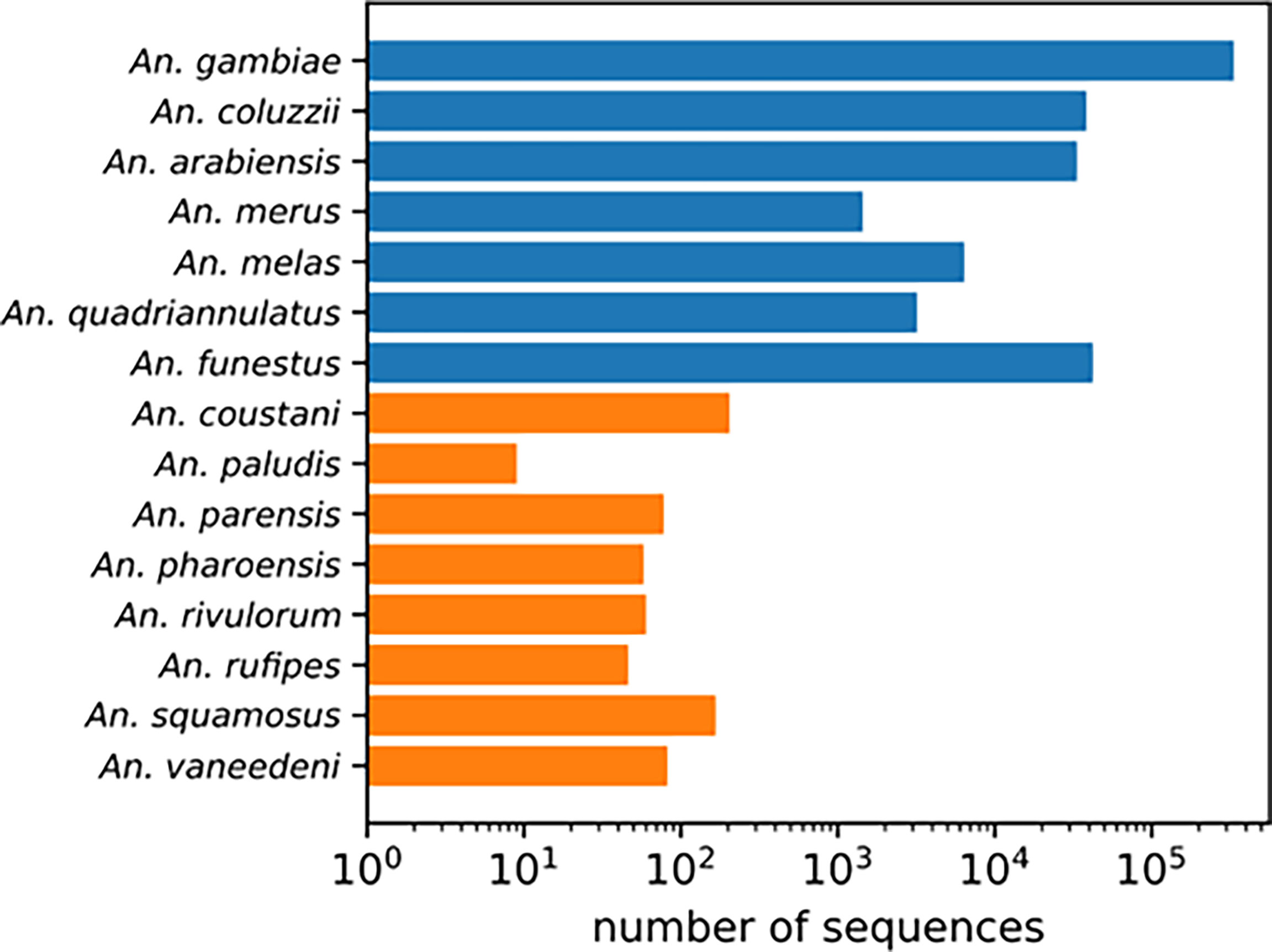
Figure 1 NCBI Nucleotide database search results for each species name. Well-studied An. gambiae complex sibling species and An. funestus s.s. in blue. Understudied species in orange.
The mitochondrial cytochrome oxidase c subunit I (COI) gene is often targeted for species-level identification as a so-called ‘barcode’ for many taxa, including many vector insects and anophelines (26–29). The COI gene is targeted because efforts to catalogue global species diversity using the Barcode of Life (BOL) have led to many sequences being available for this locus (30), and the balance of sequence conservation and polymorphism in COI allow for comparison at the level of closely related species, such as within the complicated species complexes common to anophelines. However, reliance on mitochondrial loci for species relatedness could be problematic for mosquitoes because researchers have demonstrated the lack of species-specific markers in the An. gambiae complex (31) or different gene genealogies between mitochondrial and nuclear markers in other mosquito species such as Ae. aegypti (32). For this reason, the internal transcribed spacer 2 (ITS2) ribosomal region could be used an alternative target in the nuclear genome for species identification. In the Anopheles gambiae complex, ITS2 experiences a higher rate of divergence than COI and has better discriminating power for phylogenetic relationships at the level of species complexes (31, 33). The use of these complementary targets also allows for a fuller picture of phylogenetic relationships, with COI representing mitochondrial/maternal inheritance and ITS2 reflecting nuclear inheritance (24).
A few recent studies have combined morphological data and sequencing of COI and ITS2 to illuminate previously-unrecognized diversity of anophelines in southern and east Africa (20, 34, 35). In eastern Zambia, molecular examination found a large number of species and species groups which had been misidentified solely using morphological identification (34). Malaria parasites were detected in many of these specimens. In numerous instances, these anophelines did not have a DNA sequence match in publicly available databases (34). This was not only observed in Zambia. Studies from western Kenya also demonstrated high species diversity of indoor and outdoor collections of anophelines, with infected specimens found in anopheline species with no corresponding published molecular sequences (20). From the outdoor collections, one species, designated An. sp. 1, dominated collections with an infection rate similar to that seen for An. funestus (20). The presence of such cryptic but significant malaria vectors escaping typical malaria entomological surveillance seriously compromises the assessment and control of malaria.
Indoor-based collections have been conducted extensively in Nchelenge district, northern Zambia. From these collections, An. funestus s.s. is considered the primary vector both as a result of a high malaria parasite infection rate and its occurrence in substantially higher numbers than other anophelines. Anopheles gambiae is a secondary contributor to transmission in this region (36–38). Anopheles coustani and An. leesoni, which are not traditionally considered malaria vector species in Zambia, are found in small numbers indoors, and only a handful of other anopheline species have been identified in the area (36, 37, 39). To date, collections have been almost exclusively focused indoors and indoor-focused interventions, such as widespread use of long-lasting insecticide treated nets (LLINs) and indoor residual spraying (IRS), have had minimal impact in reducing malaria in Nchelenge (40–42).
Based on these facts, we hypothesized that there could be unrecognized vectors contributing to malaria transmission that are missed by solely relying on indoor surveillance and that are not targeted by indoor-directed vector control efforts. In order to capture other anophelines, we utilized Centers for Disease Control light traps (CDC LTs) in outdoor locations in Nchelenge District. We sequenced COI and ITS2 segments of collected specimens and searched the NCBI Nucleotide database to find matching species. Here, we report our findings on species diversity, species identification success rates, and implications on future research, surveillance, and control of malaria vectors in Zambia and Africa.
Materials and Methods
Sample Collection
Anopheles mosquitoes were collected from outdoor locations in Nchelenge District in northern Zambia. Nchelenge shares a border with the Democratic Republic of the Congo that bisects Lake Mweru. It is a marshy region lying ~800 meters above sea level. There are three seasons: a rainy season from November to May, a cool dry season from May to August, and a hot dry season from August to November. Anopheles funestus and An. gambiae are considered the primary vectors in the area, although An. funestus contributes more to transmission in Nchelenge due to its much greater abundance and infection rates (37, 39). The An. funestus population peaks during the dry season when An. gambiae numbers are at their lowest (37). Sample collections for this study were conducted in August 2016 during the dry season, through which high malaria transmission is maintained. CDC LTs were set in a total of thirteen households in two different locations in Nchelenge, either within a kilometer from the lake or in a village more than 7 kilometers inland (Figure 2). Traps were set outside overnight adjacent to areas where people gathered in the evening, close to outdoor washing areas/latrines, and near animal pens. Traps were rotated through households for nine nights. Due to samples lost during shipping, the collections here represent collections from a total of 74 trap nights.
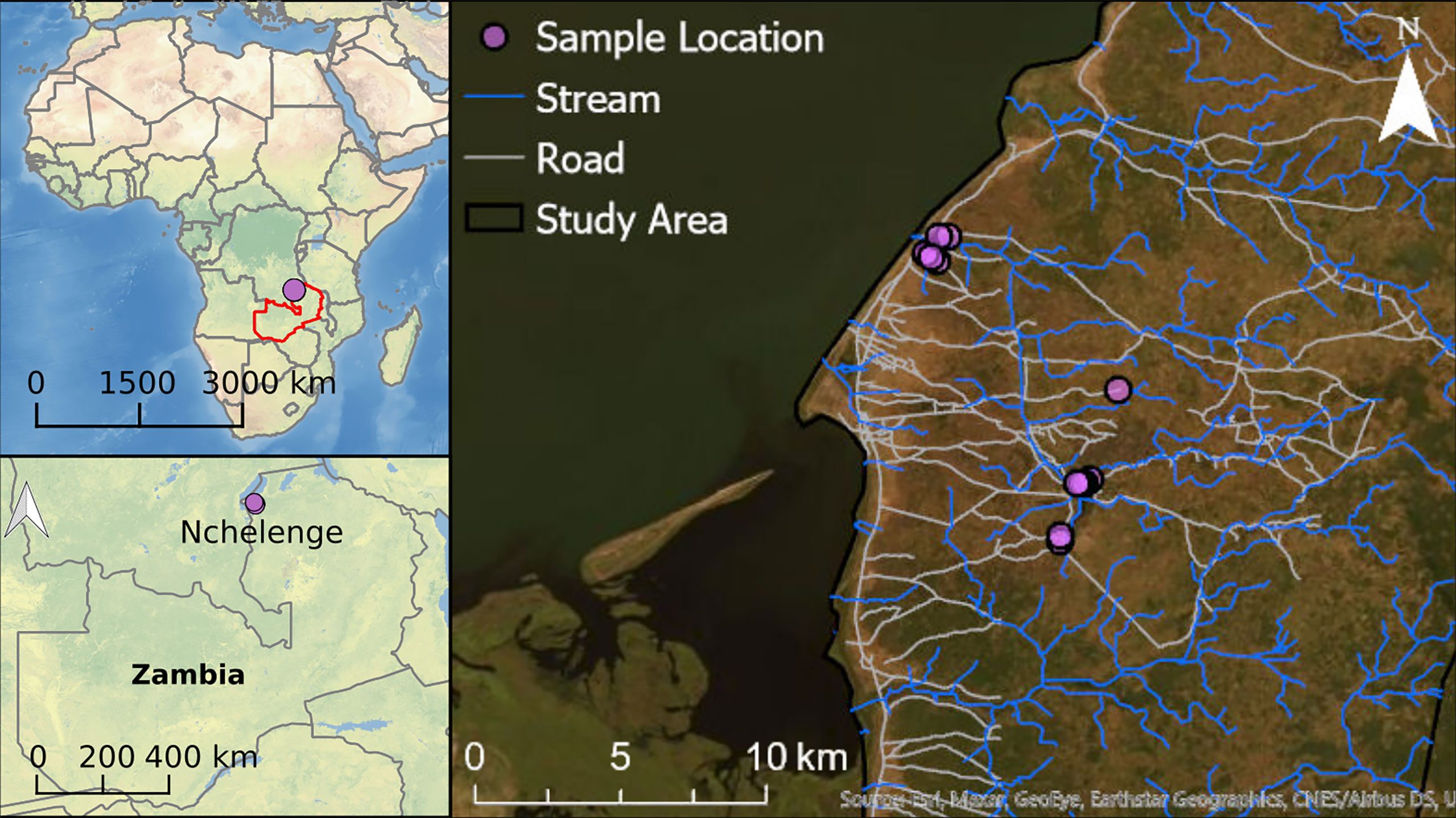
Figure 2 Collection site map. Each purple dot represents a household in Nchelenge. The public domain map, CleanTopo2 (43) was used for the base map on left two panels. World imagery from ArcGIS® software by Esri was used for the base map for the right panel.
DNA Extraction and Species Identification
Mosquitoes were morphologically identified to species using standard keys by trained personnel (12, 13). Anopheline mosquitoes were placed individually into labelled 0.6 mL microcentrifuge tubes containing silica gel desiccant and cotton wool and stored at room temperature. Abdomens and heads/thoraces were split and placed into two separate tubes. DNA was extracted from abdomen tissue of each mosquito using a modified salt extraction protocol as previously described (36, 39, 44).
For anopheline molecular species identification, a series of PCR assays was conducted and products visualized by electrophoresis on 2% agarose gels. For specimens morphologically identified as members of the An. funestus or An. gambiae complex, respective species-complex-specific PCR diagnostics were run (45, 46). For specimens that did not amplify following species-complex-specific PCR diagnostics for An. funestus or An. gambiae species groups or for which morphological identification indicated they were not of these two major vector groups, a more general differential PCR based on the ITS2 region of rDNA was used (26, 35, 47). For those samples that either did not amplify using the ITS2 assay, or that gave an ambiguous fragment size, a COI-based BOL (cytochrome oxidase I - Barcode of Life) PCR protocol was used to amplify a ~500 bp long fragment followed by Sanger sequencing (34). When samples failed to amplify, pellets from the abdomen DNA extraction that had been saved and stored at -20°C were re-extracted using the Qiagen DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany), and the product was again subjected to the COI PCR assays.
Sequencing and Phylogenetic Analysis
Samples that amplified following the COI-based BOL PCR were purified using the QIAquick PCR Purification Kit (Qiagen, Hilden, Germany). COI BOL amplicons were sequenced at the Johns Hopkins Medical Institutions (JHMI) Synthesis and Sequencing Facility using the LCO1490 (5’-GGT CAA CAA ATC ATA AAG ATA TTG G-3’) and HCO2198 (5’-TAA ACT TCA GGG TGA CCA AAA AAT CA-3’) primers described by Hebert et al. (26). Forward and reverse sequences were trimmed to remove ends with low quality and then high-quality trimmed sequences were aligned to generate a single consensus sequences using Geneious v11.1.5 (48). Corresponding sequences of the COI from known anopheline species, as well as several taxonomically unassigned species (e.g. “An. sp. 1”), were downloaded from the NCBI database (n = 140) to represent a spectrum of taxa in the genus Anopheles as well as several sequences from sister genera for analysis as outgroups (Supplementary Table S1). All sequences (n = 256) were trimmed to a final length of 488 bp and aligned in Geneious v11.1.5 (48). Identical sequences were collapsed to a single unique sequence for analysis using FaBox, resulting in a final 196 unique sequences (49). Phylogenetic trees were built using the Jukes-Cantor Genetic Distance method (50) and the Neighbor-Joining tree build method (51) implemented in Geneious (48). Two hundred replicates were used to calculate bootstrap values. Nodes with low support (lower than 75% support threshold) were collapsed into polytomies, and an Ae. aegypti sequence or most distant Anopheles were used as outgroups.
To validate results from COI BOL sequencing, representative specimens of each phylogenetic group from the COI tree (Figure 3A) were sequenced using the internal transcribed spacer region (ITS2) in the nuclear genome. ITS2 amplicons were purified and sent for sequencing using the forward and reverse ITS2A (5’- TGT GAA CTG CAG GAC ACA T -3’) and ITS2B (5’- TAT GCT TAA ATT CAG GGG GT -3’) primers (46). Individual forward and reverse sequences were trimmed to remove low quality ends and then the resulting trimmed high-quality sequences were aligned in a pairwise fashion to generate single consensus sequences. In the few cases where either the forward or reverse sequence of a samples failed, the single high-quality trimmed read was used instead for further analyses. Final ITS2 sequences were compared using BLASTN against the NCBI non-redundant nucleotide database. Hits with a high percentage of query coverage (>70%), a high percentage sequence identity (>80%), and/or E-value < 1x10-21 were considered good hits.
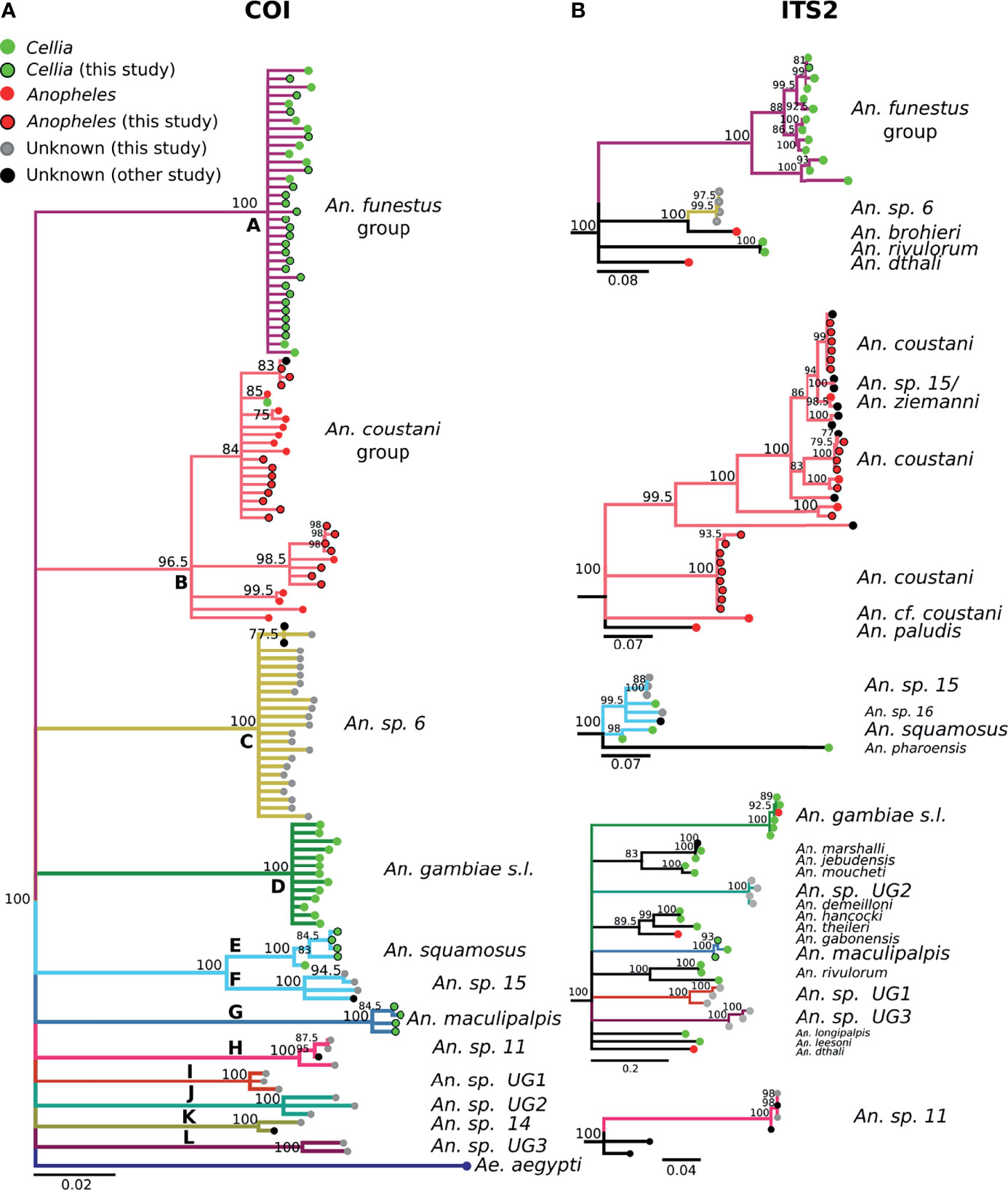
Figure 3 (A) Phylogenetic tree based on COI sequences. (B) Tree based on ITS2 sequences. Trees include both well-defined sequences from this study (green or red node termini outlined in black) and from NCBI (green or red node termini without black outline), as well as sequences without a proper species identification (gray from this study and black from NCBI).
Annotation and Data Availability
COI sequences generated in this study are available in GenBank with the following accession numbers: MK016543-MK016657. ITS2 sequences accession numbers are: MK592014-MK592096.
Map Generation
Maps of anopheline species composition at geolocated study households were created with QGIS version x QGIS version 3.18 (https://www.qgis.org/). CleanTOPO2 (43) World imagery from ArcGIS® software by Esri was used for the base map.
Results
A total of 790 female anophelines were molecularly processed for this study, generating 115 representative COI BOL sequences from morphological groups to assist with species verification. For 43/790 (5.4%) samples, which repeatedly failed to amplify for any PCR attempted, genetic data could not be generated, and thus were excluded from subsequent analyses. Of those 43 samples, 25 (58.1%) were morphologically identified as An. funestus, 7 (16.3%) were An. coustani, 7 (16.3%) were inconclusive, 3 (7.0%) were An. squamosus/cydippis, and 1 (2.3%) was An. gambiae. The majority of the species-validated specimens (as determined through a combination of morphological and molecular analyses) were An. funestus s.s. (644/747, 86.2%), with only a few An. gambiae s.s. (14/747, 1.9%), and an unexpectedly high diversity of additional species (89/747, 11.9%, Table 1).
Phylogenetic Analyses
The 488 bp multi-alignment for the COI BOL included 132 unique haplotypes and 98 ITS2 sequences were used to construct phylogenetic trees (Figure 3). Anopheles funestus morphological identifications were confirmed by molecular and sequence analysis and these samples fell in a single well-supported clade with NCBI sequences from An. funestus group (Figure 3, clade A). Two sequenced An. gambiae samples fell into a well-supported clade with sequences representing the An. gambiae complex (data not shown). The phylogenetic tree revealed approximately 10 additional clades of sequenced study specimens outside of An. funestus and An. gambiae (Table 1 and Figure 3).
Some species groups and complexes clustered into well-supported clades using COI, which is consistent with widespread use of COI as a good discriminator at the approximate level of species groups (26, 30, 31). However, some sequences were clustered in exclusive clades without any recognized species included in them (clades H-L in Figure 3). When possible, we assigned species identifiers from corresponding closely-matched and previously identified species. A total of 12 clades were identified from our collection based on the COI tree (Figure 3A). There were 3 clades, namely I, J, and L, with no match in NCBI. We named these clades as Unknown Group 1 (UG1), UG2, and UG3, respectively. ITS2 sequences were too divergent to construct a single phylogenetic tree with a common root (Figure 3B). We summarized the relationship between morphological identification and sequence-based identification in Table 1.
Species groups An. funestus, An. coustani, and An. gambiae grouped together independently based on ITS2 sequences (Figure 3B). An. sp. 6 sample were distantly related to An. funestus group, An. brohieri, and An. rivulorum (Figure 3B). The An. coustani group formed multiple clades with other study samples recognized as An. sp. 15 or An. ziemanni (Figure 3B), suggesting further studies are needed to resolve members of this species complex. An. squamosus and An. sp. 15/16 formed distinct but closely related clades (Figures 3A, B). The COI sequence similarity with An. sp. 15/16 was over 98% and ITS2 sequence similarity with An. sp. 15/16 over 92% (Table 1). The COI tree was not able to resolve the species relationship with clades including An. sp. 11 and 14 as well as the UG1-3 groups (Figure 3A), while ITS2 sequences revealed potentially distant relationships (ITS2 sequence similarity between 70-80%) to An. demeilloni, An. marshalli, and An. hancocki (Table 1 and Figure 3B).
Subgenus Cellia
Multiple species in the An. funestus group, An. parensis, An. vaneedeni, An. longipalpis, and An. funestus s.s., formed a monophyletic clade based on COI sequences (Figure 4A). ITS2 sequences were better at resolving different member species within the An. funestus group and all our samples fall within An. funestus s.s. (Figure 3B). Sequence similarity of our An. funestus s.s. sample was 85-87% with An. longipalpis, 85-86% with An. vaneedeni, and 72-75% with An. parensis.
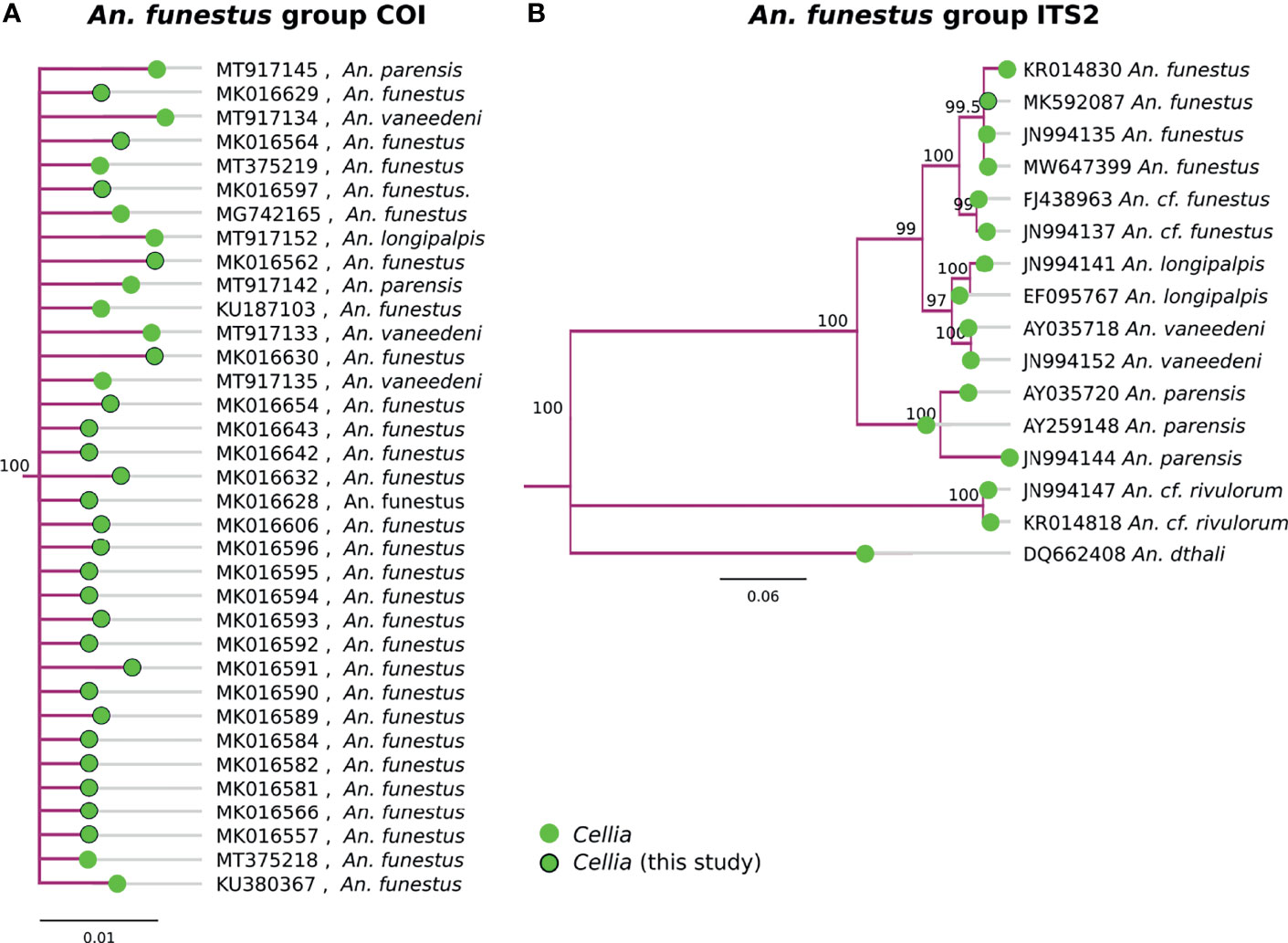
Figure 4 (A) Phylogenetic tree based on COI sequences of An. funestus group samples. (B) Tree based on ITS2 sequences. Trees include both well-defined sequences from this study (green node termini outlined in black) and from NCBI (green node termini without black outline).
A group of 4 specimens clustered with An. squamosus COI sequences with high support (Figure 3A, clade E). This clade formed a group with 3 additional specimens that more closely matched a NCBI sequence for “An. sp. 15,” (clade F in Figure 3A) but remained monophyletically clustered with sequence similarity around 94-95% between clades E and F. This is well outside the typical conspecific COI sequence similarity of >98%. All 7 samples were morphologically identified as An. squamosus and corresponding ITS2 sequences returned BLAST matches to An. squamosus. Given the high relatedness of the An. squamous and An. sp. 15 COI and ITS2 groupings, it is likely that these belong to the same subgenus, Cellia. Variation in ITS2 sequences within An. squamosus (82-99% sequence similarities within An. squamosus ITS sequences), suggest that there could be higher variation in genetic background within an An. squamosus group that could include An. sp. 15 and An. sp. 16 (Figure 5). Fixed chromosome inversion arrangements have also been identified in An. squamosus (personal communication with Maureen Coetzee, University of the Witwatersrand), which supports the likelihood of An. squamosus existing as an incompletely described species complex.
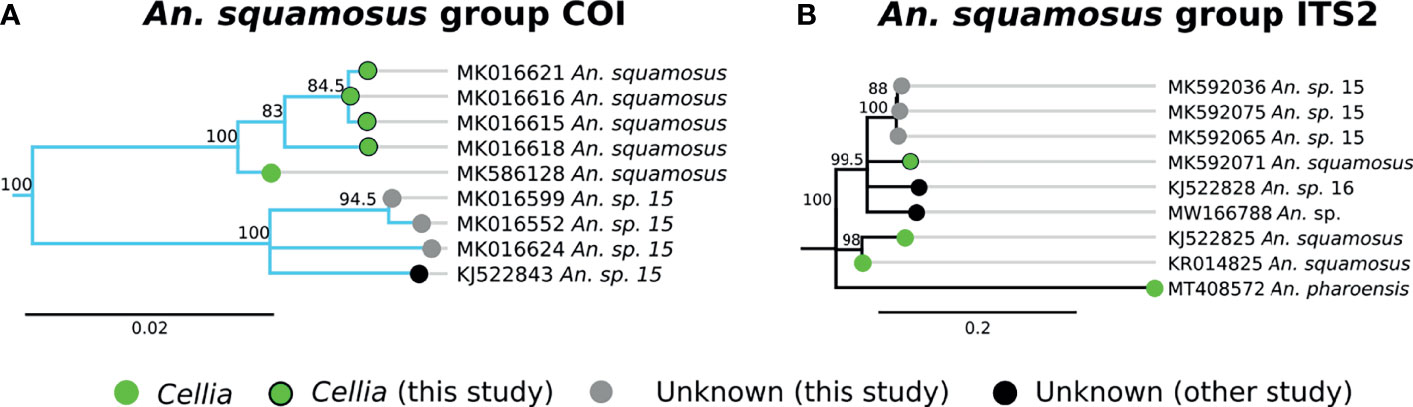
Figure 5 (A) Phylogenetic tree based on COI sequences of An. squamosus and An. sp. 15 group samples. (B) Tree based on ITS2 sequences. Trees include both well-defined sequences from this study (green node termini outlined in black) and from NCBI (green node termini without black outline) as well as sequences without a proper species identification (gray from this study and black from NCBI).
Subgenus Anopheles
Two distinct clades were detected within the coustani group based on COI sequences (Figure 6). The sequence similarity between the two COI clades (labeled B1 and B2 in Figure 6A) were 96%, which is outside of within-species similarity (>98%) observed in this study. B1 clade includes An. coustani, a chromosomal form (cf) of An. coustani, An. paludis, An. tenebrosus, and An. rufipes. As there was only one An. rufipes sequence, a species in the Cellia subgenus group (52), inclusion in clade B1 could be due to misclassification. However, if the pattern were repeated with An. rufipes samples from other regions, the subgenus designation may need to be revisited. The majority of samples similar to our An. coustani group samples belong to the Anopheles subgenus. Clade B2 includes one An. paludis sample sequenced by another group as well as 7 unique sequences generated from our Zambian samples (Figure 6A).
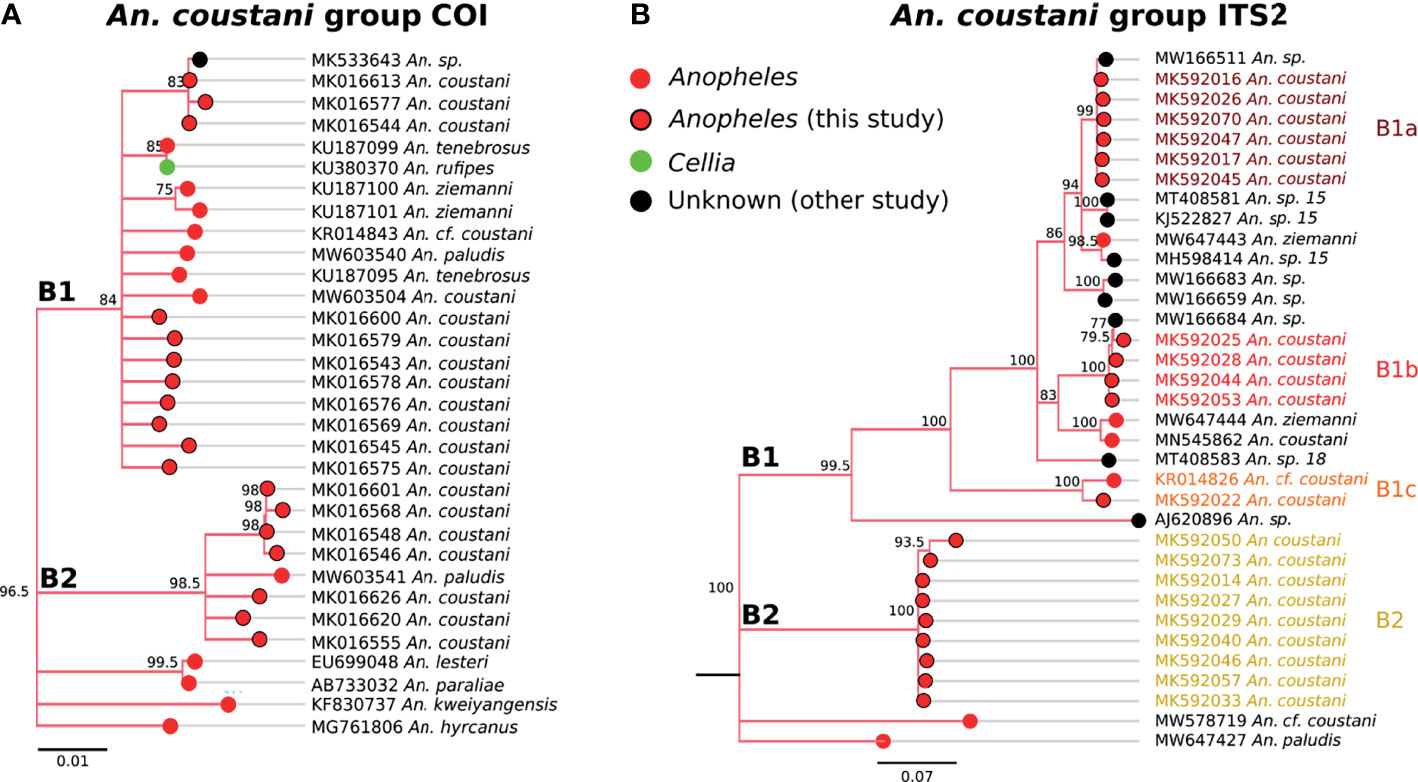
Figure 6 (A) Phylogenetic tree based on COI sequences of An. coustani group samples. (B) Tree based on ITS2 sequences. Trees include both well-defined sequences from this study (red node termini outlined in black) and from NCBI (green or red node termini without black outline), as well as sequences without a proper species identification (black from NCBI).
Genetic relationships among species within clade B1 appear to be complicated and warrant further genetic studies for clarification. Our An. coustani samples roughly divided into four groups with one belonging to clade B2 that is distantly related to clade B1 (~58% ITS2 sequence similarity with clade B1). Two closely related ITS2 clades (namely B1a and B1b with 84-85% sequence similarity) and one more distantly related ITS2 clade (B1c with 70% sequence similarity to B1a and B1b) were detected within the B1 clade. Other clades that were not represented in our Zambian samples including An. sp. 15 and 18, also grouped with clade B1.
Other Groups
The second-most abundant group collected in the study (31/747, 4.1%) fell within a single highly-supported clade C (Figure 3A). Due to the inclusion in this clade of the sequence of “An. sp. 6” from NCBI, these samples have been classified as An. sp. 6. The An. sp. 6 from other studies (MT375225 and KJ522834 in Figure 7A) were from Kenya. The morphological identifications for An. sp. 6 specimens were inconsistent and varied: 21/31 (67.7%) were identified as An. funestus s.s., 7/31 (22.6%) were identified as An. gambiae, and 3/31 (9.68%) were morphologically unclassified. Due to this variability across morphologically disparate groups, the morphology may be best defined as indeterminant. ITS2 sequence similarity between An. sp. 6 and An. theileri was ~76% (Figure 7B).
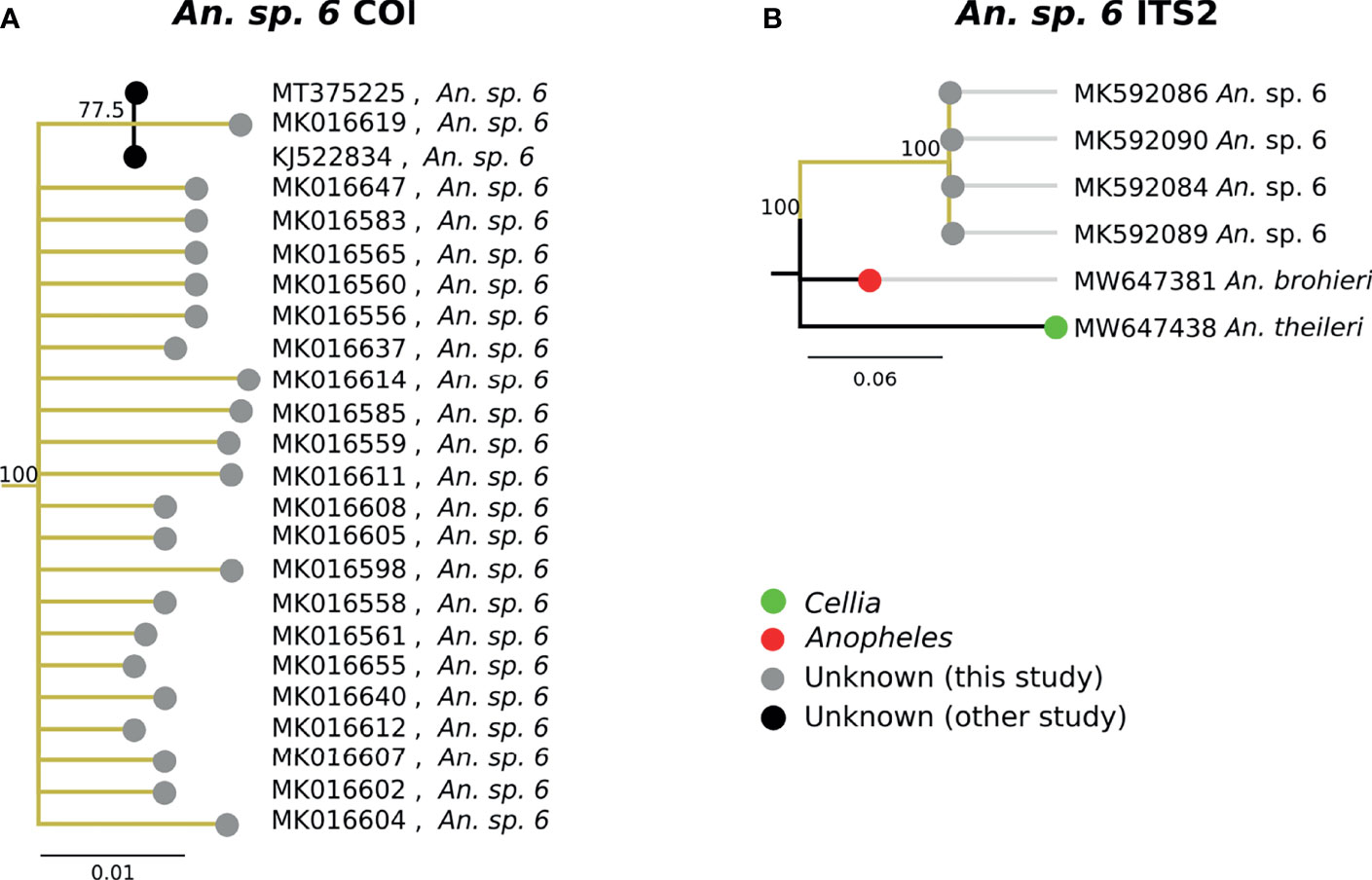
Figure 7 (A) Phylogenetic tree based on COI sequences of An. sp. 6 samples. (B) Tree based on ITS2 sequences. Trees include both well-defined sequences from NCBI (green or red node termini without black outline) as well as sequences without a proper species identification (gray from this study and black from NCBI).
Three specimens clustered tightly with An. sp. 11 based on COI and ITS2 sequences, and were therefore classified as such (Figure 8). All three were morphologically identified as An. squamosus, though neither COI nor ITS2 sequences matched An. squamosus. There was no named species that were similar to the An. sp. 11 clade. The closest ITS2 sequence matches were from unknown Anopheles species from Zambia (53) with 67-68% sequence similarity (Figure 8).
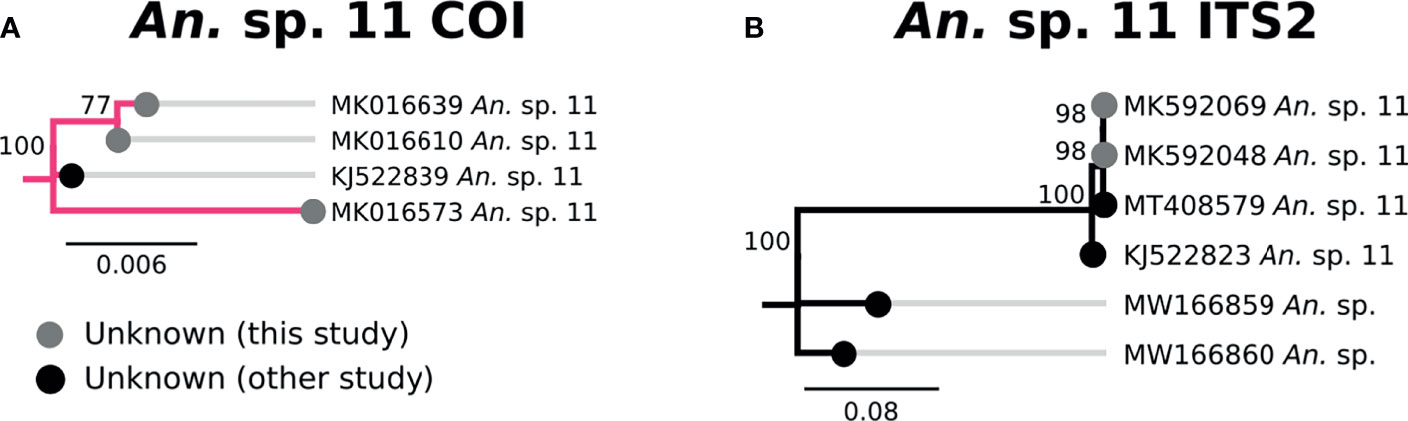
Figure 8 (A) Phylogenetic tree based on COI sequences of An. sp. 11 samples. (B) Tree based on ITS2 sequences. Trees include sequences without a proper species identification (gray from this study and black from NCBI).
The single An. sp. 14 specimen was morphologically identified as An. gambiae. The COI sequences form its own clade (Figures 1, 9) and we did not find any match with ITS2 sequences (Table 1). Consequently, its subgenus placement remains unclear. Three specimens fell into Unknown Group 1 (UG1), five into Unknown Group 2 (UG2), and five into Unknown Group 3 (UG3) (Figure 9). UG1 samples were morphologically identified as An. funestus (n = 2) and unidentified (n = 1). ITS2 sequences for this group did not match any existing NCBI data. UG2 and UG3 had varied morphological identifications. One member of UG2 was morphologically unidentified, 2/5 were identified as An. brunnipes, and 2/5 as An. rhodesiensis. Two members of UG3 were morphologically unidentified, 2/5 were identified as An. funestus, and 1/5 as An. tchekedii. Neither UG2 nor UG3 COI clustered with significant support with NCBI entries for An. funestus and An. rhodesiensis, and no COI sequences for An. brunnipes nor An. tchekedii were available. BLAST results for ITS2 sequences from both groups returned only poor matches to existing sequences in NCBI. Based on our blast results, we can exclude An. marshallii, An. jebudensis, An. moucheti, An. demeilloni, An. hancocki, An. theileri, An. rivulorum, An. longipalpis, An. leesoni, and An. dthali (Figure 9).
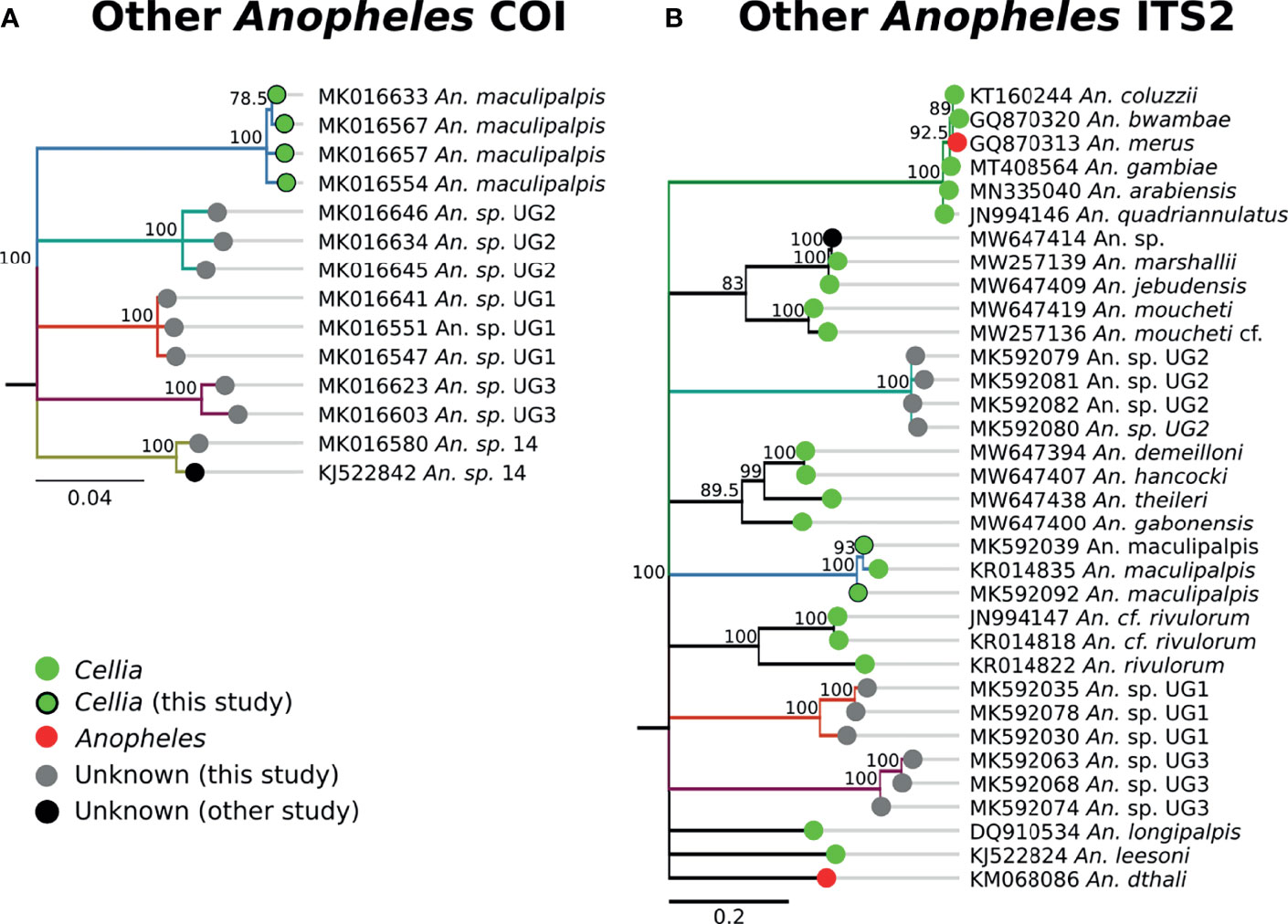
Figure 9 (A) Phylogenetic tree based on COI sequences of other Anopheles samples. (B) Tree based on ITS2 sequences. Trees include both well-defined sequences from this study (green node termini outlined in black) and from NCBI (green or red node termini without black outline), as well as sequences without a proper species identification (gray from this study and black from NCBI).
Discussion
We examined anopheline COI and ITS2 sequences of outdoor foraging mosquitoes to characterize understudied anopheline species and potential malaria vectors in Nchelenge District, Zambia. The collected anophelines included both easily-identified and commonly-recognized major malaria vector species, as well as many species for which species-level identification by morphology was not easily obtained. COI sequences were collected to attain phylogenetic placement of some of these unassigned specimens with well-characterized anopheline species. This allowed for positive identification of some specimens as known species and novel clades for which well-referenced genetic data are not yet available. Determining the species composition of outdoor foraging mosquitoes is critical for guiding malaria surveillance and appropriate interventions as understudied and potential vector species may comprise a large proportion of such collections (34, 37, 54–57).
Diversity
This collection from outdoor sampling represents a higher diversity of anopheline species than has previously been documented in Nchelenge District, despite extensive sampling in the region spanning almost two decades (38, 39, 58). These studies were focused on indoor collections and predominantly reported An. funestus s.s. and An. gambiae s.s. The unexpected diversity reported in this study may be explained by several non-mutually exclusive factors. Firstly, this is one of very few studies in the region in which mosquito collections were conducted outdoors, and multiple studies have documented higher species diversity of anophelines outdoors, especially next to livestock. Often routine malaria entomological surveillance is not conducted in such locations (59, 60). Secondly, several consecutive years of IRS combined with insecticide-treated nets (ITNs) targeting An. funestus and An. gambiae, the most abundant anophelines, may have reduced their populations and other species are now being revealed (42, 61). Finally, the lack of extensive sequencing and rigorous identification of unidentified and assumed non-vector specimens in previous collections may have overlooked existing species (20, 34, 54).
Similar to the An. gambiae complex (31), COI was not sufficient to delineate species with in the An. funestus group (Figure 4). ITS2 sequences, however, were informative in clustering the clades by known member species. All of our An. funestus samples from Nchelenge are An. funestus s.s. and no other funestus group species were detected in this collection (Figure 4).
An. squamosus and its sister species An. pharoensis have long been considered secondary vectors of Plasmodium to humans (13, 21). Findings from southern Zambia indicate that An. squamosus may also serve as a vector of malaria parasites in that region (21). Seven specimens in this study have been tentatively classified as An. squamosus based on morphological and molecular data. Three of them match closely (91% ITS2 similarity) with a sequence reported as An. sp. 15 (GenBank Accession: KJ522843), which we also tentatively classify as An. squamosus on the basis of both morphological identification and molecular analysis (20). An. squamosus in southern Zambia has been reported to be comprised of two COI clades, with a strong phylogenetic relationship to An. sp. 15 (62), lending support to the hypothesis that these are members of an undescribed species complex. Another sequence reported as An. sp. 16 (GenBank Accession: KJ522828) as well as an unknown Anopheles sample from Zambia (GenBank Accession: MW166788) are somewhat similar (86-90% ITS2 similarity) to An. squamosus ITS2 sequences. Given the close phylogenetic relationship between An. squamosus and An. sp. 15, further investigations on the role of these species as alternative vectors of malaria are warranted.
An. coustani and the closely-related species An. ziemanni and An. paludis have likewise increased in notoriety as potentially important malaria vectors in sub-Saharan Africa (55, 63). In some cases, these species have served as major vectors, ranging from Cameroon and the Central Africa Republic to Kenya and Madagascar (64–67). One reason these species have been overlooked for so long is that they have been recognized primarily as exophagic and zoophilic mosquitoes, and so were assumed not to be important in human malaria transmission. However, reports of high degrees of anthropophily in some regions or potentially in some cryptic populations indicate that their vectorial capacity may be much higher (68). The An. coustani specimens within this study fell into multiple clades within An. coustani group (Figure 6). One subgroup may be An. coustani s.s., while the other clades may represent either subpopulations or distinct but highly related species, perhaps within a single species complex. These data are more extensively examined in the context of additional samples from northern Zambia in Ciubotariu et al. (22).
The best match for the second most abundant group of anophelines collected in this study was an NCBI database sequence for “An. sp. 6” (GenBank Accession: KJ522834), which was identified as An. theileri group F by another group (20). However, its ITS2 sequences were too divergent from An. theileri (~76% ITS2 sequence similarity) to be considered as the An. theileri species group. The closest match was An. brohieri with 83% ITS2 sequence similarity. An. brohieri is in the Anopheles subgenus while An. theileri is in the Cellia subgenus. Available data are not sufficient to conclude that An. sp. 6 belongs to the Cellia or Anopheles subgenus.
An. maculipalpis, according to Gillies and De Meillon, is generally a low-abundance species found throughout savannah- and tropical-type environments in Africa and tends to be zoophilic and rest outdoors (69). It has never been implicated as a disease vector of any significant importance. Six samples from our study were identified as An. maculipalpis through molecular analysis, although only one of these six was morphologically identified as such. The others were not able to be morphologically identified (N=2) or identified as An. coustani (N=3).
Specimens which lack definitive species designations from this study (An. sp. 11, UG1-3) represent anopheline populations which remain unidentified. Proper identification of specimens such as these require not only additional field material for more accurate morphology, but corresponding genetic data from taxonomically-verified specimens. Further studies must be undertaken to properly document these populations to determine if they represent previously undocumented species or are simply species for which we lack genomic information.
Limitations
The relationships of the anopheline subgenera to one another remain unclear and somewhat contentious (1, 20, 70–74). Studies based on combinations of nuclear and mitochondrial DNA as well as amino acid sequence and morphological characters show that sections/series and even subgenera of anophelines are para- or polyphyletic, i.e., they do not have a single phylogenetic origin. This may be unsurprising, as the original taxonomic classification of anopheline mosquitoes was based largely on morphological characters. As closely-related anopheline species can be morphologically distinct, and distantly-related species remarkably similar, morphological classification may suffer from some degree of evolutionary inaccuracy.
There have been relatively few molecular phylogenetic studies of anopheline mosquitoes at a broad geographic scale. One study used full mitochondrial genomes to analyze the phylogenetics of Anophelinae below the genus level (1). Even with much more extensive sequence data than was used in this study, there was low support when using nucleotide data. We attempted to use corresponding amino acid translations for distantly related species, but it did not add discriminatory value to our analyses.
Although the COI BOL is among the most common targets used for phylogenetic analysis in this group of organisms, its utility is likely limited to comparing relatively closely related species as evident by a recent study of the An. gambiae complex (31) and An. funestus member species relationships illustrated in this study (Figure 4A). Reports have been mixed with regard to the useful phylogenetic signal in COI for comparing subgenera within Anopheles (35, 75). To more accurately place ambiguous groups from this study, alternative sequences or multiple targets would be helpful. For instance, ND5 from the mitochondrial genome, along with D2 from the nuclear genome, have been successfully used to resolve relationships at the subgenus level (75, 76). In addition, ITS2 is a very common locus that might be a useful addition and validation of COI-based phylogenetics, although it is difficult to align across taxa and therefore may lead to inaccurate phylogenetic reconstructions.
Morphological misidentification remains a problem, even for experienced investigators (Table 1), particularly when specimens are damaged as commonly occurs during trapping and processing of samples. Misidentification of anophelines for An. gambiae specimens in this small sample set was common with 39% (9/23) of specimens morphologically identified as An. gambiae being molecularly identified as something else. Comparatively, only 6.2% of morphologically identified An. funestus were misidentified. A high proportion of the remaining specimens were morphologically mis- or unidentified, which is likely due to inexperience with identification of relatively rarely observed species of anophelines, as well as damage to specimens in the field or during collection. More extensive documentation of species, including verified voucher specimens for comparison and genetic sequence from such specimens, would be of great benefit to malaria researchers and vector biologists.
Conclusions
By going beyond standard PCR assays for speciation of samples and conducting phylogenetic analysis, this study was able to show an unprecedented diversity of anophelines in Nchelenge District, northern Zambia. Several of these anophelines represent species known to be or emerging as important vectors for malaria transmission in other regions of Africa. At such low numbers in this collection, it is impossible to estimate their contribution to transmission in Nchelenge District and long-term studies of outdoor anophelines spanning larger parts of the district and region are required to further determine their role and distribution. Other specimens in this study remain unverified and represent either unnamed species or named species which have yet to be genetically characterized. Future taxonomic efforts are clearly needed to link anopheline morphology to genomic data.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by the Institutional Review Board at the Johns Hopkins Bloomberg School of Public Health (Baltimore, MD USA, IRB No: 00003467) the Ethics Review Committee of the Tropical Diseases Research Centre (Ndola, Zambia, Protocol No: TDRC/ERC/2010/14/11). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
CJ and DN were responsible for study design and concept. MM, JL, GC, DM, and JS carried out collections. MM, JS, LS, TM, CJ, and IC identified specimens. CJ, GC, YL, AM, KK, and DN analyzed and interpreted data. All authors contributed to the article and approved the submitted version.
Funding
Funding for this study was provided in part by the NIH as part of the International Centers of Excellence for Malaria Research (2U19AI089680), the USDA National Institute of Food and Agriculture (Hatch project 1025565), NIH T32 Training Grant (T32AI0074717) support to CJ and MG, as well as support to CJ, GC, JS, and DN by the Bloomberg Philanthropies and the Johns Hopkins Malaria Research Institute.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
This work would not be possible without the hard work and assistance of scientists and personnel working at International Centers of Excellence for Malaria Research site in Nchelenge District. We would like to thank community members in Nchelenge for their graciousness and patience during the course of this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fitd.2021.780664/full#supplementary-material
Supplementary Table S1 | Genbank accession numbers of COI and ITS2 sequences used in our analysis.
References
1. Foster PG, De Oliveira TMP, Bergo ES, Conn JE, Sant’ana DC, Nagaki SS, et al. Phylogeny of Anophelinae Using Mitochondrial Protein Coding Genes. R Soc Open Sci (2017) 4:170758. doi: 10.1098/rsos.170758
2. The publisher is CRC Press. Detailed info about this book can be found here: https://www.taylorfrancis.com/books/edit/10.1201/9780203756621/essential-malariology-david-warrell-herbert-gilles.
3. Coetzee M, Hunt RH, Wilkerson RC, Della Torre A, Coulibaly MB, Besansky NJ. Anopheles Coluzzii and Anopheles Amharicus, New Members of the Anopheles Gambiae Complex. Zootaxa (2013) 3619:246–74. doi: 10.11646/zootaxa.3619.3.2
4. Lee Y, Cornel AJ, Meneses CR, Fofana A, Andrianarivo AG, Mcabee RD, et al. Ecological and Genetic Relationships of the Forest-M Form Among Chromosomal and Molecular Forms of the Malaria Vector Anopheles Gambiae Sensu Stricto. Malar J (2009) 8:75. doi: 10.1186/1475-2875-8-75
5. Lee Y, Meneses CR, Fofana A, Lanzaro GC. Desiccation Resistance Among Subpopulations of Anopheles Gambiae s.s. From Selinkenyi, Mali. J Med Entomol (2009) 46:316–20. doi: 10.1603/033.046.0216
6. Sanford MR, Demirci B, Marsden CD, Lee Y, Cornel AJ, Lanzaro GC. Morphological Differentiation may Mediate Mate-Choice Between Incipient Species of Anopheles Gambiae s.s. PloS One (2011) 6:e27920. doi: 10.1371/journal.pone.0027920
7. Fryxell RT, Nieman CC, Fofana A, Lee Y, Traore SF, Cornel AJ, et al. Differential Plasmodium Falciparum Infection of Anopheles Gambiae s.s. Molecular and Chromosomal Forms in Mali. Malar J (2012) 11:133. doi: 10.1186/1475-2875-11-133
8. Sanford MR, Ramsay S, Cornel AJ, Marsden CD, Norris LC, Patchoke S, et al. A Preliminary Investigation of the Relationship Between Water Quality and Anopheles Gambiae Larval Habitats in Western Cameroon. Malar J (2013) 12:225. doi: 10.1186/1475-2875-12-225
9. Mitri C, Markianos K, Guelbeogo WM, Bischoff E, Gneme A, Eiglmeier K, et al. The Kdr-Bearing Haplotype and Susceptibility to Plasmodium Falciparum in Anopheles Gambiae: Genetic Correlation and Functional Testing. Malar J (2015) 14:391. doi: 10.1186/s12936-015-0924-8
10. Hanemaaijer MJ, Higgins H, Eralp I, Yamasaki Y, Becker N, Kirstein OD, et al. Introgression Between Anopheles Gambiae and Anopheles Coluzzii in Burkina Faso and its Associations With Kdr Resistance and Plasmodium Infection. Malar J (2019) 18:127. doi: 10.1186/s12936-019-2759-1
11. Lee Y, Souvannaseng L, Collier TC, Main BJ, Norris LC, Fofana A, et al. Evidence for Divergent Selection on Immune Genes Between the African Malaria Vectors. Anopheles coluzzii A. gambiae Insects (2020) 11(12):893-905. doi: 10.3390/insects11120893
12. Gillies MT, De Meillon B. The Anophelinae of Africa South of the Sahara (Ethiopian Zoogeographical Region). Johannesburg, South Africa: The South African Institute for Medical Research (1968).
13. Gillies MT, Coetzee M. A Supplement to the Anophelinae of Africa South of the Sahara. Johannesburg, South Africa: The South African Institute for Medical Research (1987).
14. Dia I, Guelbeogo MW, Ayala D. Advances and Perspectives in the Study of the Malaria Mosquito Anopheles funestus, Anopheles mosquitoes - New insights into malaria vectors, Sylvie Manguin, IntechOpen, doi: 10.5772/55389. Available from: https://www.intechopen.com/chapters/43973
15. Main BJ, Lee Y, Collier TC, Norris LC, Brisco K, Fofana A, et al. Complex Genome Evolution in Anopheles Coluzzii Associated With Increased Insecticide Usage in Mali. Mol Ecol (2015) 24:5145–57. doi: 10.1111/mec.13382
16. Riehle MM, Guelbeogo WM, Gneme A, Eiglmeier K, Holm I, Bischoff E, et al. A Cryptic Subgroup of Anopheles Gambiae is Highly Susceptible to Human Malaria Parasites. Science (2011) 331:596–8. doi: 10.1126/science.1196759
17. Harbach RE. The Classification of Genus Anopheles (Diptera: Culicidae): A Working Hypothesis of Phylogenetic Relationships. Bull Entomol. Res (2004) 94:537–53. doi: 10.1079/BER2004321
18. Carrara GC, Petrarca V, Niang M, Coluzzi M. Anopheles Pharoensis and Transmission of Plasmodium Falciparum in the Senegal River Delta, West Africa. Med Vet Entomol (1990) 4:421–4. doi: 10.1111/j.1365-2915.1990.tb00460.x
19. Awono-Ambene HP, Kengne P, Simard F, Antonio-Nkondjio C, Fontenille D. Description and Bionomics of Anopheles (Cellia) Ovengensis (Diptera: Culicidae), a New Malaria Vector Species of the Anopheles Nili Group From South Cameroon. J Med Entomol (2004) 41:561–8. doi: 10.1603/0022-2585-41.4.561
20. St Laurent B, Cooke M, Krishnankutty SM, Asih P, Mueller JD, Kahindi S, et al. Molecular Characterization Reveals Diverse and Unknown Malaria Vectors in the Western Kenyan Highlands. Am J Trop Med Hyg (2016) 94:327–35. doi: 10.4269/ajtmh.15-0562
21. Stevenson JC, Simubali L, Mbambara S, Musonda M, Mweetwa S, Mudenda T, et al. Detection of Plasmodium Falciparum Infection in Anopheles Squamosus (Diptera: Culicidae) in an Area Targeted for Malaria Elimination, Southern Zambia. J Med Entomol (2016) 53:1482–7. doi: 10.1093/jme/tjw091
22. Ciubotariu II, Jones CM, Kobayashi T, Bobanga T, Muleba M, Pringle JC, et al. Genetic Diversity of Anopheles Coustani (Diptera: Culicidae) in Malaria Transmission Foci in Southern and Central Africa. J Med Entomol (2020) 57:1782–92. doi: 10.1093/jme/tjaa132
23. Goupeyou-Youmsi J, Rakotondranaivo T, Puchot N, Peterson I, Girod R, Vigan-Womas I, et al. Differential Contribution of Anopheles Coustani and Anopheles Arabiensis to the Transmission of Plasmodium Falciparum and Plasmodium Vivax in Two Neighbouring Villages of Madagascar. Parasit Vectors (2020) 13:430. doi: 10.1186/s13071-020-04282-0
24. Yao H, Song J, Liu C, Luo K, Han J, Li Y, et al. Use of ITS2 Region as the Universal DNA Barcode for Plants and Animals. PloS One (2010) 5(10):e13102. doi: 10.1371/journal.pone.0013102
25. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic Local Alignment Search Tool. J Mol Biol (1990) 215:403–10. doi: 10.1016/S0022-2836(05)80360-2
26. Hebert PD, Cywinska A, Ball SL, Dewaard JR. Biological Identifications Through DNA Barcodes. Proc Biol Sci (2003) 270:313–21. doi: 10.1098/rspb.2002.2218
27. Cywinska A, Hunter FF, Hebert PD. Identifying Canadian Mosquito Species Through DNA Barcodes. Med Vet Entomol (2006) 20:413–24. doi: 10.1111/j.1365-2915.2006.00653.x
28. Chan A, Chiang LP, Hapuarachchi HC, Tan CH, Pang SC, Lee R, et al. DNA Barcoding: Complementing Morphological Identification of Mosquito Species in Singapore. Parasit Vectors (2014) 7:569. doi: 10.1186/s13071-014-0569-4
29. Batovska J, Blacket MJ, Brown K, Lynch SE. Molecular Identification of Mosquitoes (Diptera: Culicidae) in Southeastern Australia. Ecol Evol (2016) 6:3001–11. doi: 10.1002/ece3.2095
30. Ratnasingham S, Hebert PD. Bold: The Barcode of Life Data System (Http://Www.Barcodinglife.Org). Mol Ecol Notes (2007) 7:355–64. doi: 10.1111/j.1471-8286.2007.01678.x
31. Hanemaaijer MJ, Houston PD, Collier TC, Norris LC, Fofana A, Lanzaro GC, et al. Mitochondrial Genomes of Anopheles Arabiensis, An. Gambiae and An. Coluzzii Show No Clear Species Division. F1000Res (2018) 7:347. doi: 10.12688/f1000research.13807.1
32. Lee Y, Schmidt H, Collier TC, Conner WR, Hanemaaijer MJ, Slatkin M, et al. Genome-Wide Divergence Among Invasive Populations of Aedes Aegypti in California. BMC Genomics (2019) 20:204. doi: 10.1186/s12864-019-5586-4
33. Paskewitz SM, Wesson DM, Collins FH. The Internal Transcribed Spacers of Ribosomal DNA in Five Members of the Anopheles Gambiae Species Complex. Insect Mol Biol (1993) 2:247–57. doi: 10.1111/j.1365-2583.1994.tb00144.x
34. Lobo NF, Laurent BS, Sikaala CH, Hamainza B, Chanda J, Chinula D, et al. Unexpected Diversity of Anopheles Species in Eastern Zambia: Implications for Evaluating Vector Behavior and Interventions Using Molecular Tools. Sci Rep (2015) 5:17952. doi: 10.1038/srep17952
35. Norris LC, Norris DE. Phylogeny of Anopheline (Diptera: Culicidae) Species in Southern Africa, Based on Nuclear and Mitochondrial Genes. J Vector Ecol (2015) 40:16–27. doi: 10.1111/jvec.12128
36. Das S, Henning TC, Simubali L, Hamapumbu H, Nzira L, Mamini E, et al. Underestimation of Foraging Behaviour by Standard Field Methods in Malaria Vector Mosquitoes in Southern Africa. Malar J (2015) 14:12. doi: 10.1186/s12936-014-0527-9
37. Stevenson JC, Pinchoff J, Muleba M, Lupiya J, Chilusu H, Mwelwa I, et al. Spatio-Temporal Heterogeneity of Malaria Vectors in Northern Zambia: Implications for Vector Control. Parasit Vectors (2016) 9:510. doi: 10.1186/s13071-016-1786-9
38. Hast MA, Stevenson JC, Muleba M, Chaponda M, Kabuya JB, Mulenga M, et al. Risk Factors for Household Vector Abundance Using Indoor CDC Light Traps in a High Malaria Transmission Area of Northern Zambia. Am J Trop Med Hyg (2019) 101:126–36. doi: 10.4269/ajtmh.18-0875
39. Das S, Muleba M, Stevenson JC, Norris DE, Southern Africa International Centers of Excellence for Malaria Research T. Habitat Partitioning of Malaria Vectors in Nchelenge District, Zambia. Am J Trop Med Hyg (2016) 94:1234–44. doi: 10.4269/ajtmh.15-0735
40. Moss WJ, Norris DE, Mharakurwa S, Scott A, Mulenga M, Mason PR, et al. Challenges and Prospects for Malaria Elimination in the Southern Africa Region. Acta Trop (2012) 121:207–11. doi: 10.1016/j.actatropica.2011.06.019
41. Mukonka VM, Chanda E, Haque U, Kamuliwo M, Mushinge G, Chileshe J, et al. High Burden of Malaria Following Scale-Up of Control Interventions in Nchelenge District, Luapula Province, Zambia. Malar J (2014) 13:153. doi: 10.1186/1475-2875-13-153
42. Hast MA, Chaponda M, Muleba M, Kabuya JB, Lupiya J, Kobayashi T, et al. The Impact of 3 Years of Targeted Indoor Residual Spraying With Pirimiphos-Methyl on Malaria Parasite Prevalence in a High-Transmission Area of Northern Zambia. Am J Epidemiol (2019) 188:2120–30. doi: 10.1093/aje/kwz107
43. Patterson T. “CleanTOPO2: Edited SRTM30 Plus World Elevation Data”. (Online (Author Communicated That His Data is Published in Public Domain and Free to Use). (2008). Available at: https://gisuser.com/2006/02/introducing-cleantopo2-edited-srtm30-plus-world-elevation-data/
44. Kent RJ, Norris DE. Identification of Mammalian Blood Meals in Mosquitoes by a Multiplexed Polymerase Chain Reaction Targeting Cytochrome B. Am J Trop Med Hyg (2005) 73:336–42. doi: 10.4269/ajtmh.2005.73.336
45. Scott JA, Brogdon WG, Collins FH. Identification of Single Specimens of the Anopheles Gambiae Complex by the Polymerase Chain Reaction. Am J Trop Med Hyg (1993) 49:520–9. doi: 10.4269/ajtmh.1993.49.520
46. Koekemoer LL, Kamau L, Hunt RH, Coetzee M. A Cocktail Polymerase Chain Reaction Assay to Identify Members of the Anopheles Funestus (Diptera: Culicidae) Group. Am J Trop Med Hyg (2002) 66:804–11. doi: 10.4269/ajtmh.2002.66.804
47. Mohanty A, Swain S, Kar SK, Hazra RK. Analysis of the Phylogenetic Relationship of Anopheles Species, Subgenus Cellia (Diptera: Culicidae) and Using it to Define the Relationship of Morphologically Similar Species. Infect Genet Evol (2009) 9:1204–24. doi: 10.1016/j.meegid.2009.06.021
48. Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics (2012) 28:1647–9. doi: 10.1093/bioinformatics/bts199
49. Villesen P. FaBox: An Online Toolbox for Fasta Sequences. Mol Ecol Notes (2007) 7:965–8. doi: 10.1111/j.1471-8286.2007.01821.x
50. Jukes TH, Cantor CR. "Evolution of Protein Molecules". In: Munro HN, editor. Evolution of Protein Molecules. New York: Academic Press (1969). p. 21–132.
51. Saitou N, Nei M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol Biol Evol (1987) 4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454
52. Ajamma YU, Villinger J, Omondi D, Salifu D, Onchuru TO, Njoroge L, et al. Composition and Genetic Diversity of Mosquitoes (Diptera: Culicidae) on Islands and Mainland Shores of Kenya's Lakes Victoria and Baringo. J Med Entomol (2016) 53:1348–63. doi: 10.1093/jme/tjw102
53. Cross DE, Thomas C, Mckeown N, Siaziyu V, Healey A, Willis T, et al. Geographically Extensive Larval Surveys Reveal an Unexpected Scarcity of Primary Vector Mosquitoes in a Region of Persistent Malaria Transmission in Western Zambia. Parasit Vectors (2021) 14:91. doi: 10.1186/s13071-020-04540-1
54. Stevenson J, St Laurent B, Lobo NF, Cooke MK, Kahindi SC, Oriango RM, et al. Novel Vectors of Malaria Parasites in the Western Highlands of Kenya. Emerg Infect Dis (2012) 18:1547–9. doi: 10.3201/eid1809.120283
55. Tabue RN, Nem T, Atangana J, Bigoga JD, Patchoke S, Tchouine F, et al. Anopheles Ziemanni a Locally Important Malaria Vector in Ndop Health District, North West Region of Cameroon. Parasit Vectors (2014) 7:262. doi: 10.1186/1756-3305-7-262
56. Stevenson JC, Norris DE. Implicating Cryptic and Novel Anophelines as Malaria Vectors in Africa. Insects (2017) 8:1–18. doi: 10.3390/insects8010001
57. Tabue RN, Awono-Ambene P, Etang J, Atangana J, Toto JC, Patchoke S, et al. Role of Anopheles (Cellia) Rufipes (Goug) and Other Local Anophelines in Human Malaria Transmission in the Northern Savannah of Cameroon: A Cross-Sectional Survey. Parasit Vectors (2017) 10:22. doi: 10.1186/s13071-016-1933-3
58. Steketee RW, Sipilanyambe N, Chimumbwa J, Banda JJ, Mohamed A, Miller J, et al. National Malaria Control and Scaling Up for Impact: The Zambia Experience Through 2006. Am J Trop Med Hyg (2008) 79:45–52. doi: 10.4269/ajtmh.2008.79.45
59. Jumbam DT, Stevenson JC, Matoba J, Grieco JP, Ahern LN, Hamainza B, et al. Knowledge, Attitudes and Practices Assessment of Malaria Interventions in Rural Zambia. BMC Public Health (2020) 20:216. doi: 10.1186/s12889-020-8235-6
60. Sougoufara S, Ottih EC, Tripet F. The Need for New Vector Control Approaches Targeting Outdoor Biting Anopheline Malaria Vector Communities. Parasit Vectors (2020) 13:295. doi: 10.1186/s13071-020-04170-7
61. Larsen DA, Martin A, Pollard D, Nielsen CF, Hamainza B, Burns M, et al. Leveraging Risk Maps of Malaria Vector Abundance to Guide Control Efforts Reduces Malaria Incidence in Eastern Province, Zambia. Sci Rep (2020) 10:10307. doi: 10.1038/s41598-020-66968-w
62. Hoffman JE, Ciubotariu II, Simubali L, Mudenda T, Moss WJ, Carpi G, et al. Phylogenetic Complexity of Morphologically Identified Anopheles Squamosus in Southern Zambia. Insects (2021) 12(2):146-58. doi: 10.3390/insects12020146
63. Karch S, Mouchet J. Anopheles Paludis: Important Vector of Malaria in Zaire. Bull Soc Pathol Exot (1992) 85:388–9. doi: 10.3390/insects12020146
64. Antonio-Nkondjio C, Kerah CH, Simard F, Awono-Ambene P, Chouaibou M, Tchuinkam T, et al. Complexity of the Malaria Vectorial System in Cameroon: Contribution of Secondary Vectors to Malaria Transmission. J Med Entomol (2006) 43:1215–21. doi: 10.1093/jmedent/43.6.1215
65. Nepomichene TN, Tata E, Boyer S. Malaria Case in Madagascar, Probable Implication of a New Vector, Anopheles Coustani. Malar J (2015) 14:475. doi: 10.1186/s12936-015-1004-9
66. Ndiath MO, Eiglmeier K, Ole Sangba ML, Holm I, Kazanji M, Vernick KD. Composition and Genetics of Malaria Vector Populations in the Central African Republic. Malar J (2016) 15:387. doi: 10.1186/s12936-016-1431-2
67. Ogola E, Villinger J, Mabuka D, Omondi D, Orindi B, Mutunga J, et al. Composition of Anopheles Mosquitoes, Their Blood-Meal Hosts, and Plasmodium Falciparum Infection Rates in Three Islands With Disparate Bed Net Coverage in Lake Victoria, Kenya. Malar J (2017) 16:360. doi: 10.1186/s12936-017-2015-5
68. Fornadel CM, Norris LC, Franco V, Norris DE. Unexpected Anthropophily in the Potential Secondary Malaria Vectors Anopheles Coustani s.L. And Anopheles Squamosus in Macha, Zambia. Vector Borne Zoonotic Dis (2011) 11:1173–9. doi: 10.1089/vbz.2010.0082
69. Gillies M, De Meillon B. “The Anophelinae of Africa South of Sahara”. 2 ed. Johannesburg: The South African Institute for Medical Research (1968).
70. Foley DH, Bryan JH, Yeates D, Saul A. Evolution and Systematics of Anopheles: Insights From a Molecular Phylogeny of Australasian Mosquitoes. Mol Phylogenet Evol (1998) 9:262–75. doi: 10.1006/mpev.1997.0457
71. Krzywinski J, Wilkerson RC, Besansky NJ. Toward Understanding Anophelinae (Diptera, Culicidae) Phylogeny: Insights From Nuclear Single-Copy Genes and the Weight of Evidence. Syst Biol (2001) 50:540–56. doi: 10.1080/106351501750435095
72. Krzywinski J, Besansky NJ. Molecular Systematics of. Anopheles: subgenera to subpopulations Annu Rev Entomol (2003) 48:111–39. doi: 10.1146/annurev.ento.48.091801.112647
73. Manoukis NC, Powell JR, Toure MB, Sacko A, Edillo FE, Coulibaly MB, et al. A Test of the Chromosomal Theory of Ecotypic Speciation in Anopheles Gambiae. Proc Natl Acad Sci USA (2008) 105:2940–5. doi: 10.1073/pnas.0709806105
74. Freitas LA, Russo CA, Voloch CM, Mutaquiha OC, Marques LP, Schrago CG. Diversification of the Genus Anopheles and a Neotropical Clade From the Late Cretaceous. PloS One (2015) 10:e0134462. doi: 10.1371/journal.pone.0134462
75. Sallum M, Schultz TR, Foster PG, Aronstein K, Wirtz RA, Wilkerson RC. Phylogeny of Anophelinae (Diptera : Culicidae) Based on Nuclear Ribosomal and Mitochondrial DNA Sequences. Systematic Entomol (2002) 27:361–82. doi: 10.1046/j.1365-3113.2002.00182.x
Keywords: mosquito, phylogenetics, malaria, residual transmission, Anopheles, Zambia
Citation: Jones CM, Ciubotariu II, Muleba M, Lupiya J, Mbewe D, Simubali L, Mudenda T, Gebhardt ME, Carpi G, Malcolm AN, Kosinski KJ, Romero-Weaver AL, Stevenson JC, Lee Y and Norris DE (2021) Multiple Novel Clades of Anopheline Mosquitoes Caught Outdoors in Northern Zambia. Front. Trop. Dis 2:780664. doi: 10.3389/fitd.2021.780664
Received: 21 September 2021; Accepted: 15 November 2021;
Published: 09 December 2021.
Edited by:
Baldwyn Torto, International Centre of Insect Physiology and Ecology (ICIPE), KenyaReviewed by:
Marco Pombi, Sapienza University of Rome, ItalyEunice Owino, University of Nairobi, Kenya
Copyright © 2021 Jones, Ciubotariu, Muleba, Lupiya, Mbewe, Simubali, Mudenda, Gebhardt, Carpi, Malcolm, Kosinski, Romero-Weaver, Stevenson, Lee and Norris. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yoosook Lee, eW9vc29vay5sZWVAdWZsLmVkdQ==; Douglas E. Norris, ZG91Z2xhcy5ub3JyaXNAamh1LmVkdQ==
 Christine M. Jones
Christine M. Jones Ilinca I. Ciubotariu
Ilinca I. Ciubotariu Kyle J. Kosinski
Kyle J. Kosinski Ana L. Romero-Weaver
Ana L. Romero-Weaver Jennifer C. Stevenson
Jennifer C. Stevenson Yoosook Lee
Yoosook Lee Douglas E. Norris for the Southern Central Africa International Centers of Excellence for Malaria Research1*
Douglas E. Norris for the Southern Central Africa International Centers of Excellence for Malaria Research1*