- 1Menzies School of Health Research, Charles Darwin University, Darwin, NT, Australia
- 2National Centre for Epidemiology and Population Health, Australia National University, Canberra, ACT, Australia
Despite a high use of antibiotics and a significant burden of infectious disease, ongoing monitoring and reporting of antimicrobial resistant pathogens in rural and regional Australia is insufficient. Many geographically isolated regions of Australia have limited infrastructure, resources and fall outside of surveillance reach, limiting health services’ ability to provide an early warning signal and appropriate response. To monitor trends in the development of antimicrobial resistance (AMR), identify high-risk populations and to evaluate effectiveness of control and prevention in rural and regional Australia, a subnational surveillance system termed HOTspots was developed. To promote the best use of public health resources through the development of effective and efficient surveillance systems, we evaluated HOTspots and its prototype surveillance platform for data quality, acceptability, representativeness, and timeliness. We used the Centers for Disease Prevention and Control (CDC) guidelines for evaluating public health surveillance systems and assessed the four attributes using a descriptive analysis of quantitative data and a thematic analysis of qualitative data. We report that the HOTspots surveillance system and its prototype platform effectively captures and represents AMR data across Northern Australia. The descriptive analysis of HOTspots data demonstrated some variation in data completeness but that data validity and representativeness were high. Thematic analysis of interview transcripts found that the system was acceptable, with almost all study participants identifying timeliness, online accessibility, and community representativeness as drivers for adoption of the system, and that the system provided timely data. The evaluation also identified areas for improvement and made recommendations to the HOTspots surveillance system and its associated prototype platform.
1 Introduction
Surveillance of antimicrobial resistance (AMR) is fundamental to contain the spread of AMR by generating evidence for local, national global guidance and actions (1). The purpose of AMR surveillance includes (a) gathering and disseminating local evidence for empiric treatment and clinical decision making; (b) to assess the effect of antimicrobial stewardship program and infection control strategies; (c) to determine the burden of disease; and (d) to track temporal and geographic trends for outbreak detection (1, 2). Surveillance can be comprehensive for the entire population at risk, or sentinel reaching a limited catchment area. The frequency of surveillance is either continuous, episodic or periodic using routinely collected data (passive) or active collection of data which would otherwise be unavailable (2, 3). Passive AMR surveillance using routine pathology data of clinical isolates from sentinel sites is the most commonly used approach (4). A key assumption is that sentinel surveillance is representative of the population at risk as closely as possible.
Despite a high overall health status of many Australians, rural and regional areas within this country continue to experience comparatively poor health outcomes (5). Northern Australia is a large geographical region (comprising half of the Australian landmass) which is home to only 5.2% of the total population (1.3 million), and 30% of the total Aboriginal and Torres Strait Islander population (6). Despite the sparse population, it is a region of high AMR prevalence (7) combined with a complex interplay of socio-economic factors that are likely contributing to the disease burden in this setting (8). National progress to track and respond to AMR systematically in regional and rural Australia are insufficient (7, 8) and are not timely (9).
To achieve comprehensive AMR surveillance in Northern Australia and timely data provision to those who need it, a subnational surveillance system was developed using an innovative web-based platform (10). Here we report an evaluation of the AMR surveillance system, known as “HOTspots”, and its prototype platform (11). This evaluation focuses on the end-users of AMR surveillance data who work across health disciplines in rural and regional Australia. We described the newly developed and implemented HOTspots surveillance system (termed HOTspots) and, using the CDC framework we assessed the utility and acceptability of the system. Recommendations were made to improve the system and its contribution to AMR prevention and control activities in Northern Australia.
2 Methods
2.1 Evaluation Approach
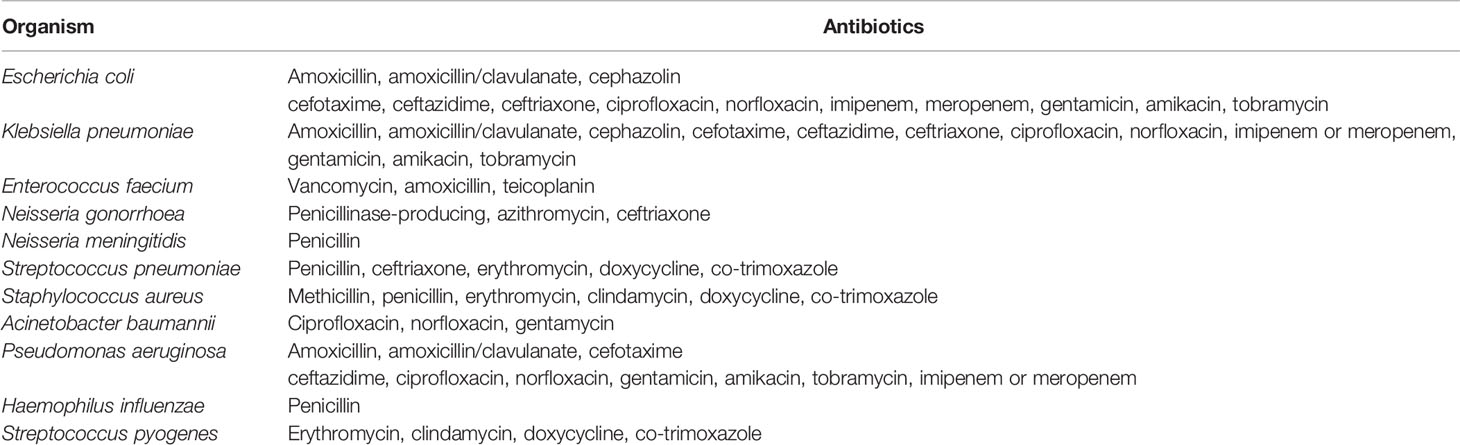
The Centers for Disease Prevention and Control (CDC) guidelines for evaluating public health surveillance systems were used as a framework to assess the performance of HOTspots (12). First, we describe the surveillance system in terms of the purpose and operation of the system. Next, we assessed the CDC surveillance system attributes of data quality, acceptability, representativeness, and timeliness. These attributes were chosen as they reflected the main aims and objectives of the HOTspots surveillance system. Definitions of the attributes and the criteria used to assess these are outlined in Table 1.
2.2 Data Collection and Analysis
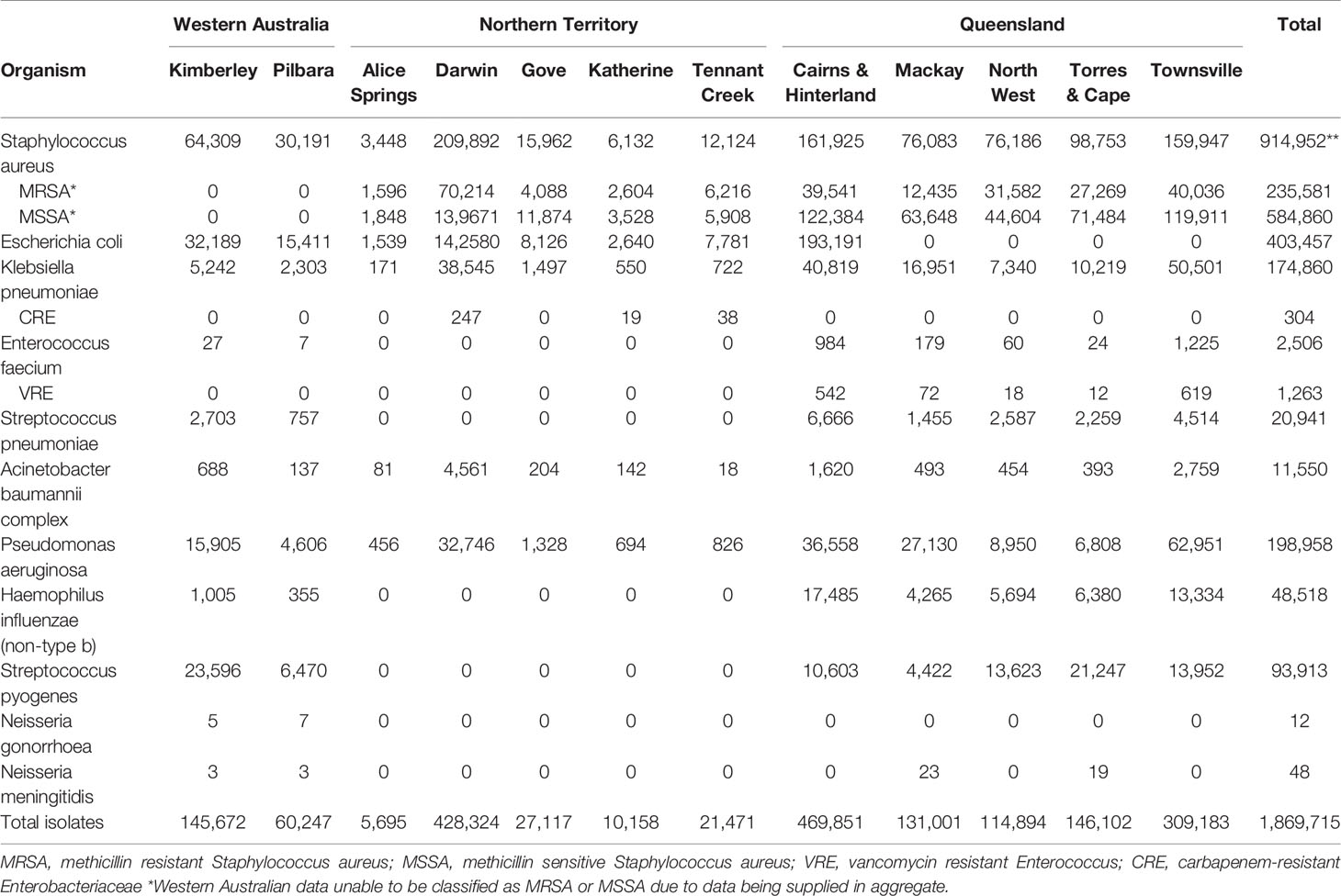
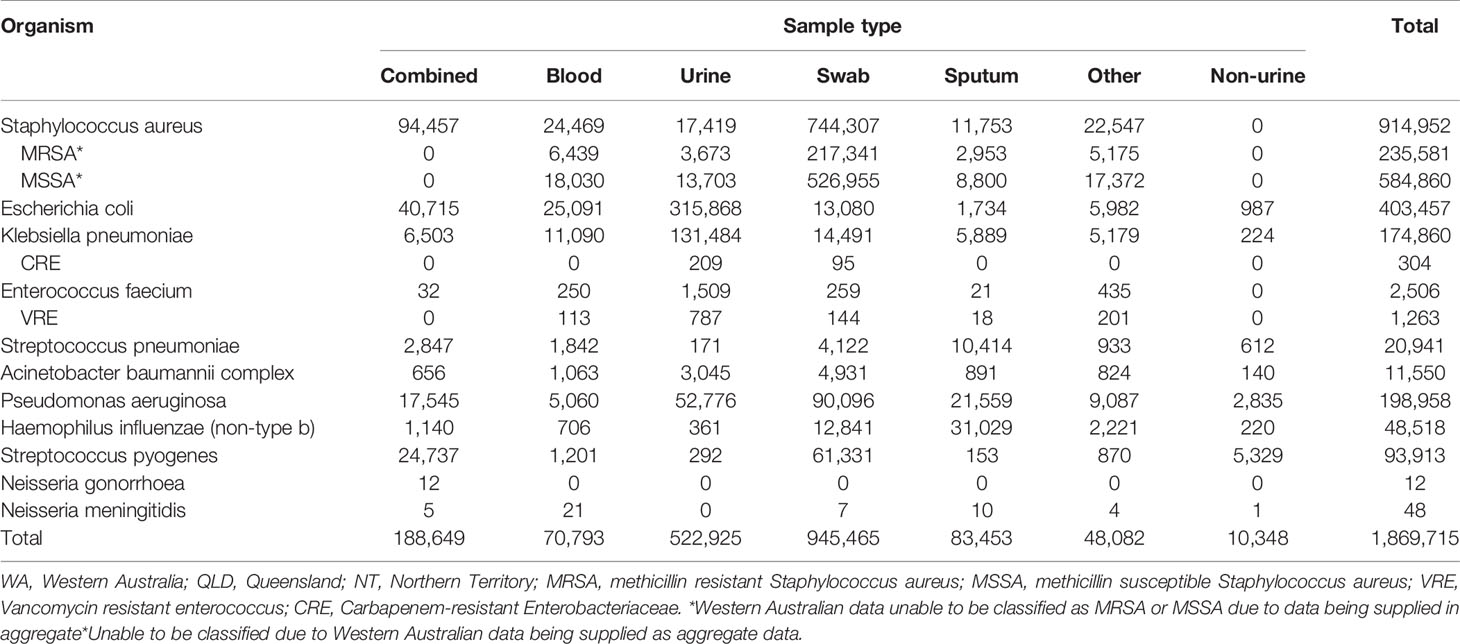
Quantitative and qualitative data were collected and analysed in the assessment of the system attributes. To assess the CDC surveillance system attribute data quality (4.2.1), we measured the completeness and validity of HOTspots data. Data were extracted from HOTspots in September 2019 for all organisms and antibiotics under surveillance (Table 2). Completeness was assessed by calculating the number and proportion of isolates by organism, year of sample collection, sample type, and geographical location (Tables 3, 4).
To measure data validity, we compared data fields collected by HOTspots against World Health Organisation (WHO) criteria for AMR surveillance systems and conducted a sub-analysis of the system’s sensitivity (Table 6). The agreement was calculated between the proportion of HOTspots Staphylococcus aureus (S. aureus) blood isolates collected in the Northern Territory in 2017 and those reported in the 2017 Northern Territory hospital antibiogram (13). Agreement was calculated using the below formula, where M denotes percent agreement, R denotes the reference susceptibility and A denotes the susceptibility (14):
The sub-analysis was limited to the antibiotics methicillin, sulfamethoxazole and trimethoprim (SXT), and clindamycin due to their use as first and second-line therapy in Australia (15). In an email from infectious disease physician Dr Nicholas Douglas (July 2020), it was clarified that in the NT hospital antibiogram, methicillin-resistant S. aureus (MRSA) is divided into health care-associated MRSA (equivalent to multi-resistant MRSA) and non-multi-resistant MRSA. These were combined for the purpose of a comparison with HOTspots MRSA, which includes both multi-resistant and non-multi-resistant MRSA. Data were analysed using Stata/IC version 15.1 and Microsoft Excel.
The representativeness (4.2.2) of the system was determined through a quantitative analysis of AMR occurrence over time and geographical location (Tables 3, 5). We also described the representativeness of the population by setting (community or hospital) and by pathology service provider coverage across three jurisdictions that are captured by HOTspots.
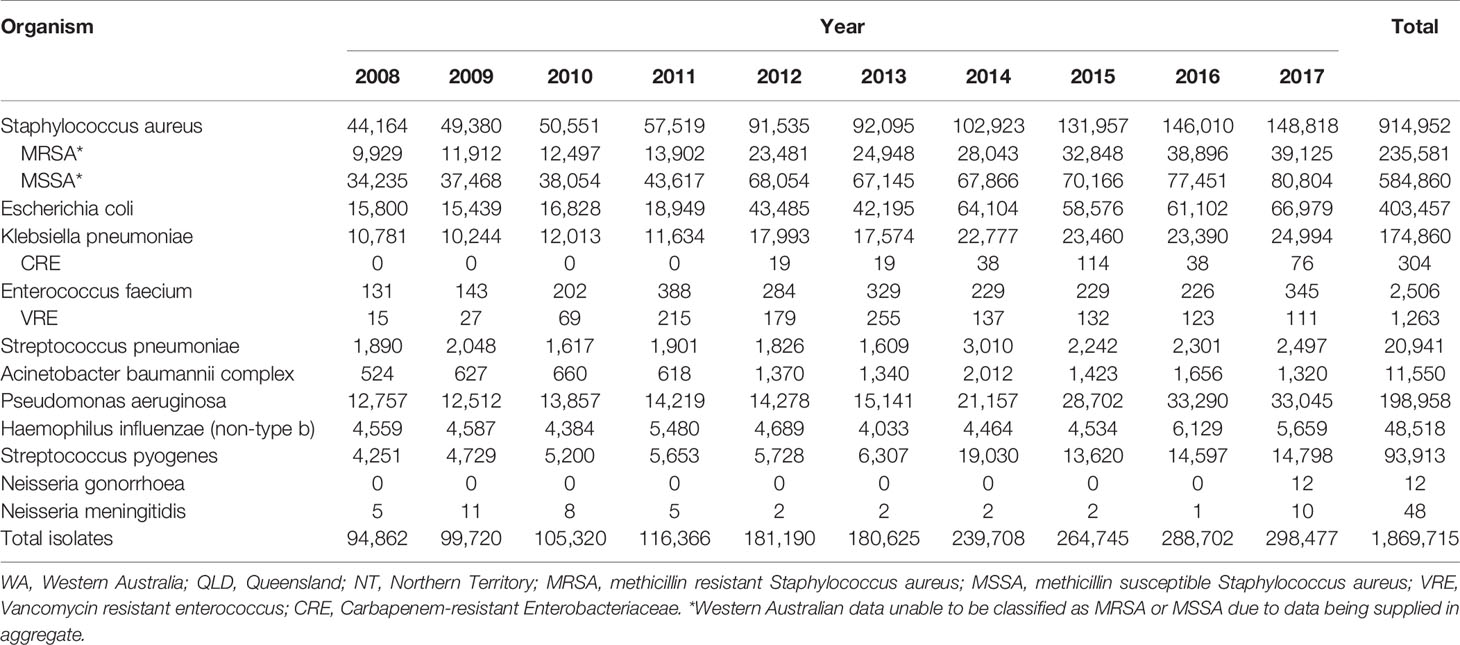
Table 5 HOTspots isolates by microorganism, jurisdiction and year of collection, northern Australia, 2008-2017.
Qualitative data were used to assess the CDC surveillance system attributes acceptability (4.2.3) and timeliness (4.2.4). These data were collected through semi-structured interviews with a defined group of AMR surveillance data end-users, including: antibiotic prescribers (doctors) and drug administrators (nurses, Aboriginal Health Practitioners, pharmacists); scientists and microbiologists; those involved in communicable disease surveillance and control (epidemiologists, public health nurses, infectious disease and public health physicians); policymakers; and content experts of treatment guideline development groups.
For practicality, a combination of convenience and purposive sampling was used to recruit study participants. Participants were provided with a participant information sheet and a consent form prior to the interview. Interviews were conducted either in person or via telephone, took on average thirty minutes to complete, and were audio-recorded and transcribed verbatim. Responses were manually coded against the CDC framework’s surveillance system attributes. Twenty-three end-users were invited to participate in the evaluation and nineteen agreed to participate (response rate of 83%). Eleven one-on-one interviews and two focus group discussions were conducted.
2.3 Ethics Statement
Ethics approval was provided by the Human Research Ethics Committee of the Northern Territory Department of Health and Menzies School of Health Research (2019–3425) and the Australian National University Human Research Ethics Committee (2017/909). Research governance authorisation and site-specific authorisation was obtained from Top End Health Services.
3 Results
3.1 Description of the Purpose and Operation of the HOTspots System
HOTspots aims to improve population-level surveillance of, and response to, AMR in Northern Australia through the timely dissemination of local antibiotic susceptibility test (AST) data. At the time of the evaluation, three major pathology providers (Territory Pathology, Queensland Pathology, PathWest) that service primary and tertiary health care services across Northern Australia contributed AST data to HOTspots for the period between 2008-2017 (10).
Participating pathologies provided data using two widely used international susceptibility methods, Clinical and Laboratory Standards Institute (CLSI) and European Committee on Antimicrobial Susceptibility Testing (EUCAST). Susceptibility results were provided as interpreted (Susceptible, Intermediate and Resistant) or minimum inhibitory concentration values for each organism. Resistant and intermediate results were combined as “resistant” for the purpose of phenotypic analysis. Microbiological data provided from the participating service providers include year of test, location of sample collection, sample type (blood, urine, or swab), organism isolated and susceptibility to a list of pre-specified antibiotics, entered as a line listing of individual de-duplicated isolates.
A digital interactive platform, HOTspots reports temporal and spatial trends for eleven AMR organisms of public health significance (Table 2) (10).
3.2 Surveillance System Attributes
3.2.1 Data Quality
The CDC guidelines define data quality as the completeness (proportion of missing values) and validity (comparison between the data and the metadata) of the data recorded in the surveillance system (12). In total, there were 1,869,715 isolates collated for the period 2008-2017, with 63% of isolates from Queensland (QLD), 26% of isolates from Northern Territory (NT), and 11% of isolates from Western Australia (WA). The analysis of AMR data by organism, year of sample collection, sample type, and geographical location demonstrated that completeness was high for all, except sample type (Tables 3–5). Sample type could not be determined for 13% of all samples, due to these pathology datasets being supplied in an aggregated format for the sample variable (Table 4).
Validity was assessed by comparing data collected by HOTspots against WHO criteria for AMR surveillance systems (16). HOTspots met WHO’s core criteria of an isolate-level database that collates relevant microbiological and demographic data, though demographic data were limited to only geographic location or ‘place of specimen collection’. Due to the limited diagnostics and microbiological capacity in remote settings of Northern Australia, patients who are acutely unwell in remote primary health care, are commonly transferred to major tertiary hospitals (often in metropolitan areas) where their specimen samples are processed and infections are treated. Therefore place of specimen collection may not be an accurate proxy for geolocation and should be supplemented with postcode of residence. Additional demographic data such as age and sex were requested and planned for future surveillance activities. Data validity was also collected using sensitivity analysis against a gold standard estimate for that setting. We compared estimates of MRSA from HOTspots to MRSA rates from a major tertiary health care centre, identified in a hospital antibiogram. We found strong agreement, with 100% for MSSA and 92.4% for MRSA between HOTspots data and gold standard for S. aureus blood isolates (Table 6).
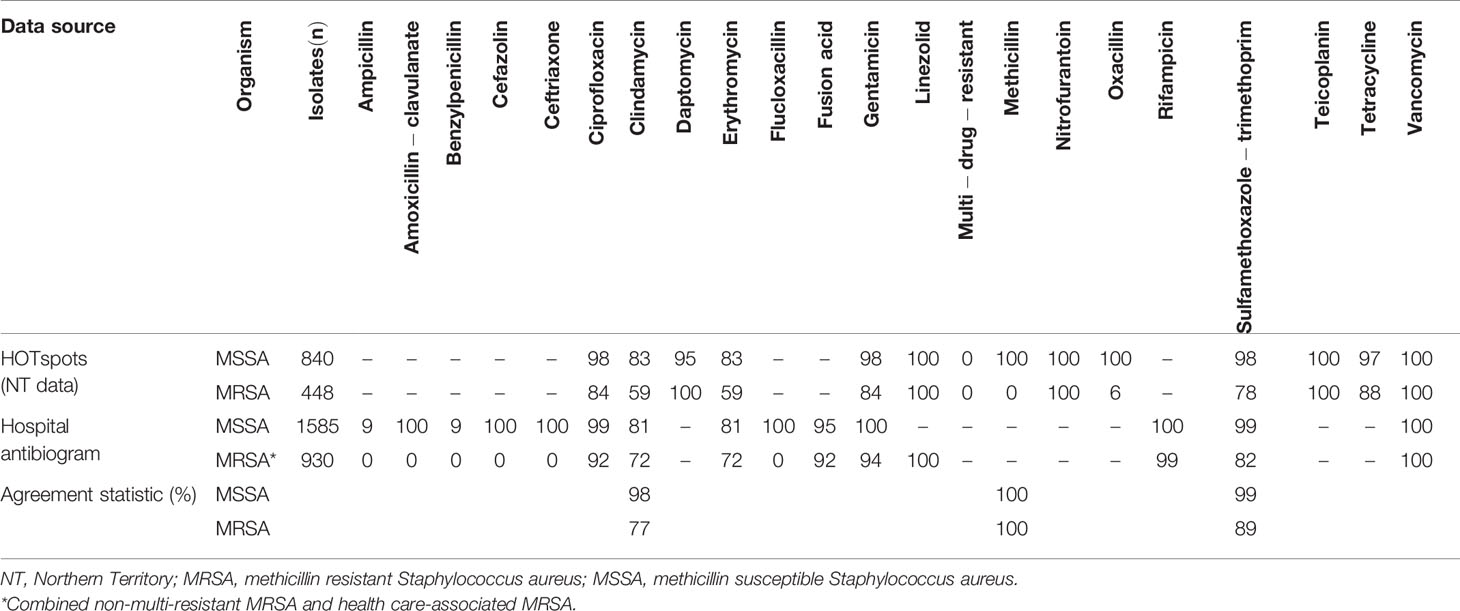
Table 6 Antibiogram showing the percent susceptible of HOTspots and hospital antibiogram S. aureus blood isolates collected in 2017.
3.2.2 Representativeness
Representativeness was defined as a system’s accuracy in describing the occurrence of the event over time and its distribution in the population by person and place (12). At the time of the evaluation, HOTspots did not yet include age or sex data. Furthermore, information on Indigeneity is not included in pathology request forms and is therefore not readily available through laboratory data (17). Given these limitations in assessing the representativeness of HOTspots, we assessed the occurrence of AMR over time (Table 5) and by geographical location (Table 3).
Data were reported for each year of specimen collection for all eleven of HOTspots organisms except for Neisseria gonorrhoea, which for an unknown reason only had data reported for 2017, and for Neisseria meningitidis between 2012-2016 (Table 5). The reason for the reduction in the number of N. meningitidis cases from QLD during this period is unclear but may be related to testing or a delay in notifications.
By region, there were variations in the representativeness of HOTspots data (Table 3). One jurisdiction in Northern Australia (QLD) did not supply data for all regions for Escherichia coli isolates and N. meningitidis isolates. There were no data provided for Enterococcus faecium, Streptococcus pneumoniae, Haemophilus influenzae (non-type B), Streptococcus pyogenes, N. gonorrhoea, or N. meningitidis for the NT. Our evaluation found that it is unclear as to the reason for missing data for certain organisms but it may be related to testing and diagnostic capacity.
In addition to this quantitative analysis, we also described the representativeness of the population by setting (primary or tertiary) and by pathology service provider coverage across HOTspots’ three jurisdictions. As a laboratory-based system, the representativeness of HOTspots is dependent on the participation of laboratories in Northern Australia and the population and geographical areas they service. To our knowledge, the pathology service providers operating in Northern Australia include Territory Pathology, Pathology Queensland, Sullivan Nicolaides Pathology, Western Diagnostics, PathWest, Australian Clinical Laboratories and Queensland Medical Laboratory Pathology.
At the time of evaluation, HOTspots received data from three of these pathology service providers (Territory Pathology, Pathology Queensland, and PathWest) and has since received AMR data from an additional provider, Western Diagnostics. These pathology providers service a mixture of private and public health care services across primary and tertiary health care throughout Northern Australia (Table 7). We determined that HOTspots covers most public hospital and community settings as far north as Kununurra in WA (18), all four public hospitals in NT’s Top End (19), all public hospitals and some community settings in QLD and (with the recent addition of data from Western Diagnostics) will cover almost all community settings across WA and NT (20).
3.2.3 Acceptability
Acceptability of the surveillance system was defined as the willingness of persons and organisations to participate in the surveillance system (12) and was informed by participant interviews. HOTspots is a voluntary, laboratory-based system, and therefore relies on the participation of laboratories. To influence local treatment guidelines, policies, and the response to AMR more broadly, HOTspots requires the participation of antibiotic prescribers, drug administrators, policymakers, guideline contributors and public health professionals.
For laboratory staff, the simple nature of the HOTspots system and minimal additional workload in data extraction and collation was discussed in participant interviews as a facilitator for acceptability. While the availability of direct testing (e.g. from Vitek) feed data and provision of minimum inhibitory concentrations is becoming increasingly possible within the participating laboratories, integration with larger AMR surveillance systems can be complex and resource intensive for both laboratories and the governing body of the surveillance system.
Participants identified timeliness, online accessibility, community representativeness, and potential outputs (e.g. community antibiograms) of the data as drivers for adoption of the system. For example, community-based clinicians interviewed as part of this evaluation discussed having limited access to susceptibility data to inform practice. This may be due to a delay in individual pathology results caused by remoteness as well as the limited availability of population susceptibility data in the community setting. In remote settings, pathology is only sent on certain days of the week when there are planes. Therefore, clinicians who practice within remote and regional settings perceived value in accessing AMR data from the HOTspots surveillance system. On the other hand, public health professionals (including epidemiologists, public health nurses, and infectious diseases and public health physicians) who participated in two focus group discussions did not perceive much value in accessing HOTspots surveillance data as they did not consider community-associated AMR infections to be within their organisational scope, means or responsibility. However, where there was alignment with a jurisdiction’s notifiable diseases register, these public health professionals expressed a greater interest in using data from the HOTspots surveillance system.
Three main barriers were identified by interview participants, which impede acceptability of the HOTspots surveillance system. These include (1) additional time taken to access online portal as a prescribing reference (2); ability to support clinical decision-making; and (3) accuracy of the data. The first barrier was discussed by clinicians who are often time-poor and therefore rely on treatment guidelines (i.e. Australian Therapeutic Guidelines) to determine treatment options. For some prescribers, accessing another online portal during an already time-limited consultation was not ideal.
Secondly, pharmacists, guideline contributors, and policymakers, reported a need for antibiograms or thresholds levels for switching antibiotic treatment to facilitate decision-making. For example, the process of writing the infectious diseases sections in treatment guidelines requires constant checking against local antibiogram data. Guideline contributors identified that having more in-depth information (than what is currently readily available) would be helpful for this process and to understand at what threshold guidelines should be changed to recommend alternative antibiotics. Public health physicians working in communicable disease prevention and control, identified the lack of molecular surveillance data for organisms such as N. gonorrhoea, which relies on genotypic AST for samples collected in remote geographical areas, as an impediment to HOTspots supporting decision-making.
The last barrier to acceptability and uptake of the surveillance system, was data accuracy and deviation from clinical guidelines and protocols. Guidelines are greatly relied on in regional and remote settings that are staffed mainly by remote area nurses and Aboriginal health practitioners. Clinicians working in primary health care raised concerns that access to AMR trend data that differs to treatment guidelines may lead to confusion of how best to manage patients and lead to clinical variation.
3.2.4 Timeliness
Timeliness is defined as the speed between steps in the surveillance system, or in the case of HOTspots, between the production of data (i.e., the point at which AST is performed and results become available) to the point at which this information is made available to end-users. Interview participants were asked how frequently data should be updated through the HOTspots web-based platform. There was consensus among the participants interviewed that four to twelve-monthly data updates would be an ideal frequency of updates. Currently, hospital antibiograms are typically generated on an annual basis (21).
3.3 Evaluation Recommendations
The three following recommendations were made in response to the evaluation findings:
3.3.1 Recommendation 1: Review and Standardise Data Collection
The completeness and validity of HOTspots data is high but could be improved with the addition of some core data, as defined by the WHO criteria for AMR surveillance systems, and by data standardisation. Defining data specifications in a data dictionary, in partnership with participating laboratories, would be a simple method to achieve a basic level of standardised data provision. In doing so, data completeness would be improved (e.g. sample type was not able to be disaggregated for Western Australia). The addition of core data fields such as age, sex and residential postcode, and linkages with clinical or administrative datasets to obtain ethnicity data, would not only improve the validity of the data, but would enable a more comprehensive analysis of the representativeness of the data collected by HOTspots against population data from the three jurisdictions.
3.3.2 Recommendation 2: Support End User Decision-Making
The evaluation identified that a barrier to the acceptability and use of the data in the surveillance system was for end users to have decision-making support. Apart from the addition of demographic and genotypic data, enhanced functionality such as the ability to generate antibiograms (summary tables) and explicit recommendations for antibiotic threshold levels to support switch therapies would further support clinical decision-making. Concurrent to the implementation of these additional features, HOTspots program managers should work with guideline contributors to align existing therapeutic guidelines with the most up-to-date data.
3.3.3 Recommendation 3: Update HOTspots Data at Least Annually
One of the main strengths of HOTspots is delivery of timely data through the HOTspots platform. The evaluation found that users preferred quarterly or six-monthly data updates. At a minimum, surveillance data should be updated annually to inform AMR trends in high-prevalence settings.
4 Discussion
We report an evaluation of an AMR surveillance system and its prototype web-based platform in a high-prevalence region of Northern Australia. The evaluation found that HOTspots surveillance provides an acceptable, stable and high-quality system suitable to the resource-constrained context of this region. A unique feature of this surveillance system was inclusion of data from the primary health care setting. While Australia continues to invest in surveillance of AMR and in particular to improve the geographical and population coverage, many rural populations remain outside of surveillance reach (22). Surveillance of AMR across both primary and tertiary health care setting is particularly important, given the urgent need to improve antimicrobial stewardship strategies in the remote regions of central and Northern Australia (23).
In addition to representativeness, early detection, timely and appropriate response are essential attributes of a functioning surveillance system, given the significant health burden of AMR and infectious disease which continues to increase in this region (10) and in neighbouring low and middle-income countries (24). Our analysis of the sensitivity of HOTspots demonstrates its ability to accurately detect disease, and the speed at which HOTspots is able to make these data available to end users is an advantage over more traditional communicable surveillance systems that typically require a data access request. The delay in access to and dissemination of data from these systems means that they are not as well suited to early detection and public health response.
However, recommendations for the improvement of the HOTspots surveillance system and its prototype web-based platform were made and these are currently being used to inform the system and the development of a new platform (25). Recommendations to improve data quality included strengthening data collection and management, such as developing a data dictionary and a data request proforma and collecting other demographic data, specifically age, sex and Indigeneity. Age and sex data were requested from pathology service providers and though they were initially not provided due to privacy concerns these data have since been supplied and are incorporated in the new platform. Unfortunately, Indigeneity data are limited as this is not routinely collected on pathology forms (17), however data linkage with clinical or administrative datasets may be an option in the future. In addition, the evaluation found that ‘place of specimen collection’ may not be an accurate proxy for geolocation for pathology providers who service patients in remote locations and it should be supplemented with postcode of residence. Furthermore, HOTspots is a passive system reporting data on phenotypic AMR isolated from blood, urine and swab specimens. Including these data would also facilitate a more detailed analysis of the representativeness of the surveillance system.
To improve the acceptability of the system, it was recommended that HOTspots program managers continue to engage with end users to ensure surveillance data are available in such a way that they can be readily utilised in decision-making. Engagement with end users also provides the opportunity to address end user concerns, such as data misinterpretation by non-medical prescribers and potential contraindications with clinical guidelines. For example, an antibiogram function has since been added to the HOTspots platform (25) and continued collaboration with clinical guideline developers will reduce variation in empiric treatment of patients at the point of care.
A limitation of this evaluation is that while the interviews highlight important considerations in the uptake and utilisation of HOTspots, these views may not be representative of end-users across Northern Australia. This is due to both the convenience method of sampling and because of the limited stakeholder participation in the northern regions of WA and QLD; seventeen of the nineteen interviewees were from the NT. Unfortunately, it was not possible to interview more end-users from QLD and WA due to the complexity of obtaining ethics approval in multiple jurisdictions and the time and resource constraints on the evaluation. Despite these limitations, the participants interviewed presented broad perspectives from stakeholders in the NT and the challenges identified in this study are likely to be similar across Northern Australia.
Strengths of the evaluation are that, as the first evaluation of HOTspots and one of only a handful of evaluations of AMR surveillance systems, this evaluation provides important baseline data for future evaluations and improvements. The evaluation also highlights how an innovative and collaborative approach to AMR surveillance can bring much needed data directly to end users in a high-prevalence, resource-poor setting.
5 Conclusion
HOTspots effectively collects, analyses and interprets AMR data from rural and remote settings of Northern Australia. The evaluation found that it provides evidence for action directly to end-users. Specifically, trends developing in AMR data over time and geographical region, which can be used to update treatment guidelines, guide screening and diagnostic testing, inform pharmaceutical and infection control policies, and identify geographic areas or at-risk population.
The strength of HOTspots is the large geographical coverage of previously unsurveyed regions and inclusion of primary health care data. By providing timely and region-specific evidence, the surveillance system can assess the populations-at-risk, evaluate effectiveness of control measures, support health planning and empower local decision-makers to contain AMR effectively. The geospatial digital platform and timely access to local AMR data, has the capacity to provide an early warning and almost real-time updates of AMR by regions to facilitate cross-jurisdictional communication and data informatics needed for a response. The evaluation findings and associated recommendations were implemented in 2020 and an updated HOTspots surveillance platform has been developed.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by Human Research Ethics Committee of the Northern Territory Department of Health and Menzies School of Health Research (2019–3425) and the Australian National University Human Research Ethics Committee (2017/909). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
LG contributed to the study design, data collection and analysis, and writing of the article. TW conceptualised the project, coordinated the development of the online platform, and contributed to the study design, writing, and revising of the article. All authors contributed to the article and approved the submitted version.
Funding
The project is funded by the Australian National Health and Medical Research Council (NHMRC)-funded ‘Improving Health Outcomes in the Tropical North: A multidisciplinary collaboration (HOT NORTH)’, GNT1131932.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
This project was undertaken as part of the Master of Applied Epidemiology (MAE) program at Australian National University. The authors would like to thank all those who participated in the study as interviewees for their time and valuable insights.
References
1. Bielicki J, de Kraker M, Gandra S, Gastmeier P, Gilchrist G, Gikas A, et al. Surveillance for Control of Antimicrobial Resistance. Lancet Infect Dis (2018) 18(3):e99–e106. doi: 10.1016/S1473-3099(17)30485-1
2. World Health Organization. Surveillance Standards for Antimicrobial Resistance. Switzerland. Geneva: World Health Organization (2002). Contract No.: WHO/CDS/CSR/DRS/2001.5.
3. Mather AE, Reeve R, Mellor DJ, Matthews L, Reid-Smith RJ, Dutil L, et al. Detection of Rare Antimicrobial Resistance Profiles by Active and Passive Surveillance Approaches. PloS One (2016) 11(7):e0158515. doi: 10.1371/journal.pone.0158515
4. Ashley EA, Recht J, Chua A, Dance D, Dhorda M, Thomas NV, et al. An Inventory of Supranational Antimicrobial Resistance Surveillance Networks Involving Low- and Middle-Income Countries Since 2000. J Antimicrob Chemother (2018) 73(7):1737–49. doi: 10.1093/jac/dky026
6. Australian Government Department of Industry Science Energy and Resources. Our North, Our Future: White Paper on Developing Northern Australia. Canberra: ACT: Australian Government Department of Industry, Science, Energy and Resources (2015).
7. Cameron JK, Hall L, Tong SYC, Paterson DL, Halton K. Incidence of Community Onset MRSA in Australia: Least Reported Where it is Most Prevalent. Antimicrobial Resistance Infect Control (2019) 8(1):1–9. doi: 10.1186/s13756-019-0485-7
8. Bowen AC, Daveson K, Anderson L, Tong SYC. An Urgent Need for Antimicrobial Stewardship in Indigenous Rural and Remote Primary Health Care. Med J Aust (2019) 211(1):9–11. doi: 10.5694/mja2.50216
9. Australian Commission on Safety and Quality in Health Care (ACSQHC). AURA 2019: Third Australian Report on Antimicrobial Use and Resistance in Human Health. Sydney, NSW: Australian Commission on Safety and Quality in Health Care (2019).
10. Wozniak TM, Cuningham W, Buchanan S, Coulter S, Baird RW, Nimmo GR, et al. Geospatial Epidemiology of Staphylococcus Aureus in a Tropical Setting: An Enabling Digital Surveillance Platform. Sci Rep (2020) 10(1):1–10. doi: 10.1038/s41598-020-69312-4
11. Menzies School of Health Research. HOTspots Darwin, Northern Territory (2019). Available at: https://amrhotspots.com.au.
12. German RR, Lee ML, Horan JM, Milstein RL, Pertowski CA, Waller MN, et al. Updated Guidelines for Evaluating Public Health Surveillance Systems: Recommendations From the Guidelines Working Group. Morbidity Mortality Weekly Report: Recommendations Rep (2001) 50(RR-13):i–35.
14. Macfadden D, Fisman D, Andre J, Ara Y, Bogoch I, Daneman N, et al. A Platform for Monitoring Regional Antimicrobial Resistance, Using Online Data Sources: ResistanceOpen. J Infect Dis (2016) 1(suppl_4):S393–8. doi: 10.1093/infdis/jiw343
15. Therapeutic Guidlines: Antibiotic. Boils and Carbuncles (2019). Available at: https://tgldcdp.tg.org.au/viewTopic?topicfile=boils-carbuncles#toc_d1e62.
16. The WHO Advisory Group on Integrated Surveillance of Antimicrobial Resistance (AGISAR). Integrated Surveillance of Antimicrobial Resistance: Guidance From a WHO Advisory Group. Geneva, Switzerland: World Health Organisation (2013).
17. Australian Institute of Health and Welfare (AIHW). The Inclusion of Indigenous Status on Pathology Request Forms. Canberra, ACT: AIHW (2013). Contract No.: Cat. no. IHW 103.
18. Government of Western Australia. PathWest Collection Centres. Available at: https://pathwest.health.wa.gov.au/Patients/Pages/Collection-Centre-Locations.aspx.
19. Government of Northern Territory. Pathology. Available at: https://nt.gov.au/wellbeing/hospitals-health-services/general-hospital-services/pathology.
20. Government of Queensland. Collection Centres. Available at: https://www.health.qld.gov.au/healthsupport/businesses/pathology-queensland/patients/collection-centres.
21. Australian Commission on Safety and Quality in Health Care (ACSQHC). Specification for a Hospital Cumulative Antibiogram Sydney: ACSQHC (2013). Available at: https://www.safetyandquality.gov.au/sites/default/files/migrated/A-Specification-for-Hospital-Cumulative-Antibiograms-December-2013.pdf.
22. Wozniak TM, Smith-Vaughan H, Andrews R. Convergence of Surveillance Blind Spots With Antimicrobial Resistance Hotspots. Aust New Z J Public Health (2021). doi: 10.1111/1753-6405.13165
23. Davidson L, Knight J, Bowen AC. Skin Infections in Australian Aboriginal Children: A Narrative Review. Med J Aust (2020) 212(5):231–7. doi: 10.5694/mja2.50361
24. Yam ELY, Hsu LY, Yap EP, Yeo TW, Lee V, Schlundt J, et al. Antimicrobial Resistance in the Asia Pacific Region: A Meeting Report. Antimicrob Resist Infect Control (2019) 8:202. doi: 10.1186/s13756-019-0654-8
25. Menzies School of Health Research. HOTspots Darwin, Northern Territory (2021). Available at: https://anti-microbe-res.shinyapps.io/hotspots/.
Keywords: surveillance, public health, antibiotic resistance, evaluation, digital health
Citation: Goddard L and Wozniak TM (2021) Antimicrobial Resistance Surveillance to Support Decision-Making in a High-Prevalence Region: An Evaluation. Front. Trop. Dis 2:772491. doi: 10.3389/fitd.2021.772491
Received: 08 September 2021; Accepted: 11 October 2021;
Published: 26 October 2021.
Edited by:
Daniel Gyamfi Amoako, National Institute for Communicable Diseases (NICD), South AfricaReviewed by:
Mohamed Hamid, King Khalid University, Saudi ArabiaLucas Naam-kayagre Amenga-Etego, University of Ghana, Ghana
Copyright © 2021 Goddard and Wozniak. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Teresa M. Wozniak, dGVyZXNhLndvem5pYWtAbWVuemllcy5lZHUuYXU=
 Laura Goddard
Laura Goddard Teresa M. Wozniak
Teresa M. Wozniak