
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Trop. Dis. , 02 November 2021
Sec. Vector Biology
Volume 2 - 2021 | https://doi.org/10.3389/fitd.2021.737909
This article is part of the Research Topic Parasite Interactions with Insect Hosts in Tropical Diseases View all 5 articles
The interaction between Rhodnius prolixus and Trypanosoma cruzi has huge medical importance because it responds to the transmission of Chagas disease, a neglected tropical disease that affects about eight million people worldwide. It is known that trypanosomatid pathogens depend on active lipid endocytosis from the insect host to meet growth and differentiation requirements. However, until now, knowledge on how the parasite affects the lipid physiology of individual insect organs was largely unknown. Herein, the biochemical and molecular dynamics of the triatomine R. prolixus lipid metabolism in response to T. cruzi acute infection were investigated. A qRT-PCR approach was used to determine the expression profile of 12 protein-coding genes involved in R. prolixus lipid physiology. In addition, microscopic and biochemical assays revealed the lipid droplet profile and the levels of the different identified lipid classes. Finally, spectrometry analyses were used to determine fatty acid and sterol composition and their modulation towards the infection. T. cruzi infection downregulated the transcript levels of protein-coding genes for lipid biosynthetic and degrading pathways in individual triatomine organs. On the other hand, upregulation of lipid receptor transcripts indicates an attempt to capture more lipids from hemolymphatic lipoproteins. Consequently, several lipid classes (such as monoacylglycerol, diacylglycerol, triacylglycerol, cholesteryl ester, phosphatidylcholine, and phosphatidylethanolamine) were involved in the response to the parasite challenge, although modulating only the insect fat body. T. cruzi never leaves the insect gut and yet it modulates non-infected tissues, suggesting that the association between the parasite and the vector organs is reached by cell signaling molecules. This hypothesis raises several intriguing issues to inspire future studies in the parasite-vector interaction field.
Insects are vectors of the most prevalent human diseases. About eight million people worldwide are estimated to be affected by Chagas disease in Latin America, where this neglected tropical disease remains a major social and health problem (1). Triatomine bugs, such as Rhodnius prolixus (Insecta: Hemiptera), are hematophagous insects and play a role in the transmission of Trypanosoma cruzi parasites, the causative agent of Chagas disease, during the ingestion of the blood meal. The blood meal represents a significant challenge for hematophagous insects since it is carried out at low frequencies, in huge amounts, rich in proteins, and relatively poor in lipids and carbohydrates (2). The ingestion of the blood meal also triggers important metabolic and physiological events such as oogenesis and molting, through gene expression regulation (2).
Regarding lipid metabolization, triacylglycerol lipases (as also Brummer lipases – Bmm) catalyze the initial lipolysis step, converting triacylglycerol (TG) to diacylglycerol (DG) and fatty acid (FA) for epithelial gut cell absorption (3). In the midgut, lipids, absorbed as FAs, are used to synthesize TG, DG, and phospholipids (PLs), which are then transferred to lipophorin (Lp), and directed to organs that require these lipids (4, 5). Lp is a hemolymphatic insect lipoprotein composed of two protein subunits: apolipophorin-I (ApoLp-I, ~ 240 KDa) and apolipophorin-II (ApoLp-II, ~ 80 KDa). Although mostly composed of DGs and PLs, Lp transports all lipid classes from sites of storage, absorption, or synthesis to utilization sites (6). Lipid capture is associated with Lp recognition by the Lp receptor (LpR) located on cell surfaces, which can internalize Lp particles through dependent (7–11) or independent (12) endocytosis mechanisms.
In addition to the blood-feeding, lipids can also be produced by non-lipid substrates through de novo FA synthesis. This metabolic pathway requires the consecutive action of two enzymes. First, acetyl-CoA carboxylase (ACC) carboxylates acetyl-CoA to produce malonyl-CoA (13). Next, fatty acid synthase I (FAS I) sequentially condensates several malonyl-CoA molecules with one acetyl-CoA primer to build up FA, generally ending in a C16 chain (14). The reaction catalyzed by malonyl-CoA decarboxylase (MCD) is also important for FA synthesis. This enzyme converts a malonyl-CoA to acetyl-CoA, decreasing substrate levels for the lipogenic process and inducing FA oxidation (15). Free FA can be successively processed to form TG, where the final and committed step is catalyzed by the diacylglycerol acyltransferase (DGAT) that forms TG from DG and FA (16). TG is stored in lipid droplets (LDs), neutral lipid-rich organelles surrounded by a PL monolayer, which are essential for animals that display low feeding frequency, like hematophagous insects. In addition to lipids, LDs also contain a high diversity of associated proteins, such as DGAT (16) and TG lipase (17).
An extremely important point concerning hematophagous insect lipid physiology is their interaction with parasites. After being ingested through blood-feeding, the parasite establishes infection in the insect gut, reproducing and differentiating. And, in the case of T. cruzi, never leaving the gut. Subsequent fecal transmission occurs in the following triatomine feeding (18). During the infection, T. cruzi depends on active lipid endocytosis from the host system to meet growth and differentiation requirements (19, 20). Cholesterol (CO) and cholesteryl ester (CE) derivatives are remarkably important, and not synthesized by trypanosomatids, being constantly acquired through the uptake of host lipoproteins (21, 22).
Noteworthy, both male and female hematophagous Hemiptera feed on blood, presenting the same potential as vector insects. However, male physiological processes are greatly neglected, and knowledge regarding their lipid metabolism and the molecular aspects of insect-parasite interaction are largely unknown. In the present study, the effects of acute T. cruzi infection on lipid metabolism modulation in adult male R. prolixus were evaluated. The findings indicate that the parasite downregulates lipid degradation and synthesis pathways in different triatomine organs. The transcription of lipid receptor proteins is upregulated as a countermeasure, probably as an attempt to capture more lipids from Lp. Consequently, several lipids are involved in response to T. cruzi infection, although modulating only insect fat bodies, non-infected tissues. Some effects on hemolymphatic phospholipids were also observed. Therefore, the success of R. prolixus and T. cruzi interaction appears to be reached by cell signaling molecules that communicate the parasite retained in the closed gut environment with other insect organs.
In this study, the Rhodnius prolixus adult males on the second feeding cycle, from a colony, were maintained at 28°C and 70–75% relative humidity at the Federal University of Rio de Janeiro, Brazil. Adults were fed at 21-day intervals on rabbit blood.
The animal study was reviewed and approved by Committee for Evaluation of Animal Use for Research at the Federal University of Rio de Janeiro, CAUAP-UFRJ, under the registry #IBQM067-05/16 and 24154319.5.0000.5257, and the NIH Guide for the Care and Use of Laboratory Animals (ISBN 0-309-05377-3). Animal facility technicians at the Leopoldo de Meis Medical Biochemistry Institute (UFRJ) performed all aspects related to rabbit husbandry under strict guidelines to ensure careful and consistent animal handling.
The epimastigotes forms of T. cruzi (y strain) were cultivated at 28°C in liver infusion tryptose (LIT) medium supplemented with 10% fetal bovine serum (FBS). Population growth was measured by direct cell counting in a hemocytometer. In all experiments, cells were used in the exponential phase of growth. To perform the infection through an artificial blood-feeding system (23), blood was taken from a central rabbit ear vein using heparin (1:50) as an anticoagulant. After centrifugation, the plasma was inactivated by incubation at 56°C for 1 h and blood cells were washed three times with PBS (PBS: 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4, pH 7.4). The inactivated plasma and washed cells were mixed and 1x107 parasites were added to the infected group. The non-infected group received the same blood meal without parasites.
To confirm R. prolixus infection with T. cruzi, gut compartments were dissected from three insects (from each group) and immediately placed on ice. DNA was extracted using the PowerSoil® DNA Isolation Kit (MoBio) following the manufacturer’s standard protocol. Subsequently, the DNA was used for T. cruzi kDNA detection by conventional PCR with Taq DNA polymerase (Invitrogen™) according to the manufacturer’s standard protocol. Primers used to amplify the kDNA were previously published (24). Rp18S was used as a reference gene. The primer nucleotide sequences are presented in Supplementary Table S1. The standard PCR conditions were also based on Guedes et al. (24) as follows: 50°C (2 min), 95°C (10 min), 40 cycles at 94°C (30 s), 58°C (30 s) and 72°C (1 min), followed by a final extension at 72°C (5 min). After PCR, electrophoresis using a 2% agarose gel (UBS, Cleveland, OH, USA) was carried out to determine the presence of the amplicons.
Based on the domains present in the first lipoprotein receptor (LpR) described in insects from Locusta migratoria (25) (CAA03855) we performed an HMMER search using FAT (26) against the R. prolixus protein dataset v 3.3 (VectorBase; http://www.vectorbase.org). The Pfam domains used in the search were: Low-density lipoprotein receptor domain class A (PF00057), Low-density lipoprotein receptor repeat class B (PF00058). As a result of this search, we identified RpLpR (RPRC011390) (27), and other five low-density lipoprotein receptors (LDLR) family members in R. prolixus. RpLDLR genes: RPRC000138, RPRC000060, RPRC000281, RPRC000270, and RPRC000551. Similarly, we used the Malonyl-CoA decarboxylase C-terminal domain (PF05292) from a human MCD sequence (NP_036345.2) to perform an HMMER search sing FAT (26) against the R. prolixus protein dataset v 3.3 (VectorBase; http://www.vectorbase.org). Only one protein was found in R. prolixus – RPRC009749.
Three days post-infection, the organs of adult males were dissected, washed in PBS, and immediately frozen on dry ice and stored at -70°C until use. The anterior midgut (amg), posterior midgut (pmg), hindgut (hg), fat body (fb), flight muscle (fm), and testis (ts) were isolated. A pool of 5 organs was collected in each experiment (N= 8). Total RNA was extracted using TRIzol Reagent (Thermo Fisher Scientific, Waltham, MA USA) according to the manufacturer’s standard protocol and quantified with NanoDrop Lite Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). After, 1 μg of total RNA was treated with 1U of DNase I (Invitrogen™, Carlsbad, CA, USA) for 30 min at 37°C in a final volume of 10uL to remove any potential DNA contamination. The reaction was stopped with incubation at 65°C for 10 min and the addition of 50 mM EDTA. The treated RNA was used as a template for complementary DNA (cDNA) synthesis using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems™) following the manufacturer’s standard protocol. The qPCR was performed in a StepOnePlus™ Real Time PCR System (Applied Biosystems™) using Power SYBR™ Green PCR Master Mix (Thermo Fisher Scientific). The qPCR was performed using the following parameters: 95°C for 10 min followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. The 2-ΔCT method (28) was used to calculate the relative abundance of the transcripts employing Rp18S as the reference gene (29). The primer nucleotide sequences used in this study are provided in the Supplementary Table S1.
Three days post-infection, adult males (pool of 5, N=8) had their hemolymph (hm) collected into a tube containing a few grains of phenylthiourea crystals, by cutting the first pairs of legs. A 0.15 M saline solution containing 1 mM EDTA and 1% protease inhibitor cocktail (Sigma-Aldrich®) was added to the tube. After centrifugation at 13.000 x g for 5 min, the supernatant was collected and stored at -70°C for subsequent analysis. Samples (1.5 µl of hemolymph/lane) were loaded onto an 8% (w/v) polyacrylamide gel. The electrophoresis was performed as previously described (30). After the electrophoresis run, the gel was stained for 30 min with Coomassie Brilliant Blue G-250 (Thermo Fisher Scientific) and destained for 24 h in 10% acetic acid.
Three days post-infection, the anterior midgut (amg), posterior midgut (pmg), hindgut (hg), fat body (fb), flight muscle (fm), and testis (ts) were isolated from adult males. A pool of 5 organs was collected in each experiment (N= 8). To analyze the lipid profile of individual R. prolixus organs, lipids were extracted following the protocol by Bligh and Dyer (31). Briefly, a methanol, chloroform, and water solution (2:1:0.8, v/v) was mixed with the samples, followed by intermittent agitation for 1h. After centrifugation (20 min at 3.000 x g), the supernatant was collected, and the pellet was subjected to a second lipid extraction for 1h. Both supernatants were pooled together and a 1:1 chloroform and water solution was added. The samples were then centrifuged (30 min at 3.000 x g) and the organic phase was collected and dried under a gentle nitrogen stream.
Extracted lipids were identified by one-dimensional TLC on Silica Gel 60 plates (Merck) for neutral lipid determination. Lipid separation was performed using a mixture of hexane, diethyl ether, and acetic acid (60:40:1 v/v) as solvent. Each lipid spot was identified by comparison with lipid standards run in parallel. A total of 15 μg of 1-oleoyl-rac-glycerol (MG), 1,3-diolein (DG), glycerol trioleate (GT), cholesterol (CO), cholesteryl palmitate (CE), and oleic acid (FA) were used as the lipid standards, purchased from Sigma-Aldrich®. The plates were stained with a charring reagent (10% CuSO4 w/v in 8% phosphoric acid v/v), revealed at 200°C (32), digitized, and subjected to a densitometry analysis using the TotalLab Quant v11 (TotalLab Ltd, Newcastle, United Kingdom).
Extracted lipids were also used in one-dimensional HPTLC on Silica Gel 60 plates (Merck) for phospholipid identification. Lipid separation was performed using a mixture of chloroform, acetone, methanol, acetic acid, and distilled water (20:7.5:6.5:6:4 v/v). Each lipid spot was identified by comparison with lipid standards run in parallel. A total of 15 μg of 3-sn-phosphatidic acid (PA), L-α-phosphatidylethanolamine (PE), L-α-phosphatidylcholine (PC), 3-sn-phosphatidyl-L-serine (PS), L-α-phosphatidylinositol (PI), L-α-phosphatidyl-DL-glycerol (PG), L-α-lysophosphatidylcholine (LPC), and sphingomyelin (SM) were used as the lipid standards, purchased from Sigma-Aldrich®. Lipids were visualized using a charring reagent (10% CuSO4 w/v in 8% phosphoric acid v/v), after heating at 200°C (32). Subsequently, digital images of chromatography plates were analyzed by densitometric determinations using the Image TotalLab Quant v11 (TotalLab Ltd, Newcastle, United Kingdom).
Lipids extracted from the fat bodies were subjected to mass spectrometry analyses for FA and sterol composition determinations. For the FA profile, lipids were prepared and analyzed according to Kluck et al., 2018 (33), whereas sterol composition was determined by generating saponified and non-saponified samples, as described by Pereira et al., 2011 (20).
The TG contents of fat bodies, flight muscle, and testes were evaluated using the enzymatic TG determination Triglicérides 120 kit (Doles Reagentes) according to the manufacturer’s recommendations. Gut compartments and hemolymph were not considered in this experiment, as the color of their homogenates is similar to the color generated by the kit enzymatic reaction.
To assess LD accumulation, R. prolixus fat bodies were isolated from infected and non-infected insects (15 of each group). The tissues were incubated with 0.001% Nile red (Molecular Probes™) and 10 μg/mL DAPI (Molecular Probes™) in 75% glycerol. After 15 min of incubation at room temperature in the dark, the fat bodies were washed in 100% glycerol and transferred to a glass slide with a drop of 100% glycerol. The slides were analyzed under a Leica TCS-SPE laser scanning confocal microscopy with the filter set for Nile red (Ex/Em: ~552/636 nm) and DAPI (Ex/Em: ~358/461 nm) fluorescence. Images were obtained from 40 to 60 optical sections (around 1.5 µm each) using a 20x objective, employing the “z-stack” function and an overlap of 350 µm. Figures are presented as a 3D reconstitution of such images, which were taken applying the same acquisition conditions between the control and infected groups.
For all experiments, values were submitted to Grubb´s test to detect outliers (34). Statistical comparisons were performed using Student’s t-test, with a 95% confidence interval (GraphPad Prism 7.0 software), to analyze infected insects against their respective control group. Differences were considered significant at p < 0.05. Data are presented as the means ± S.E.M of eight independent experiments (four samples were used for the GC-MS analysis).
The LDLR family has as a defining structural characteristic the presence of an ectodomain containing three types of protein modules: LDL receptor type A (LA), Epidermal Growth factor (EGF), and LDL receptor type B (LY – containing the YWTD motif) modules (35). HMMER searches with these domains yielded six genes – RPRC011390, RPRC000060, RPRC000281, RPRC000138, RPRC000551, and RPRC000270, which mainly differ in the number and combination of the three protein modules (Supplementary Figure S1). Among them, the protein encoded by the RPRC011390 gene exhibited 67% identity against the lipophorin receptor from Locusta migratoria (e-value 0.0, coverage 95%), presenting the unique set of sequence motifs of the C-terminal domains that characterize insect LpRs (36). Consequently, it was considered the true LpR homolog for R. prolixus. It is important to note that a 3’-RACE-PCR (manuscript under preparation) extended the RpLpR sequence from that deposited in VectorBase, enhancing our bioinformatics analysis. Based on the domain composition, we suggest that the other five proteins from the LDLR family comprise one LDL-related protein 1 (RpLrp1) – RPRC000138; two LDL-related proteins 2 (RpLrp2 or megalin) – RPRC000060 and RPRC000281; one vitellogenin receptor (RpVtgR) – RPRC000551.
The highest abundance of RpLpR (RPRC011390) mRNA was found in the midgut compartments, while the lowest level was detected in fat bodies. In non-infected insects, low abundances were also detected in the flight muscle, and testes (Figure 1A). A trend for increased LpR mRNA levels in all R. prolixus organs towards T. cruzi infection was observed (Figure 1A). T. cruzi upregulated RpLpR mRNA levels, which were only significantly different in the flight muscle (p = 0.0257) and testes (p = 0.0444).
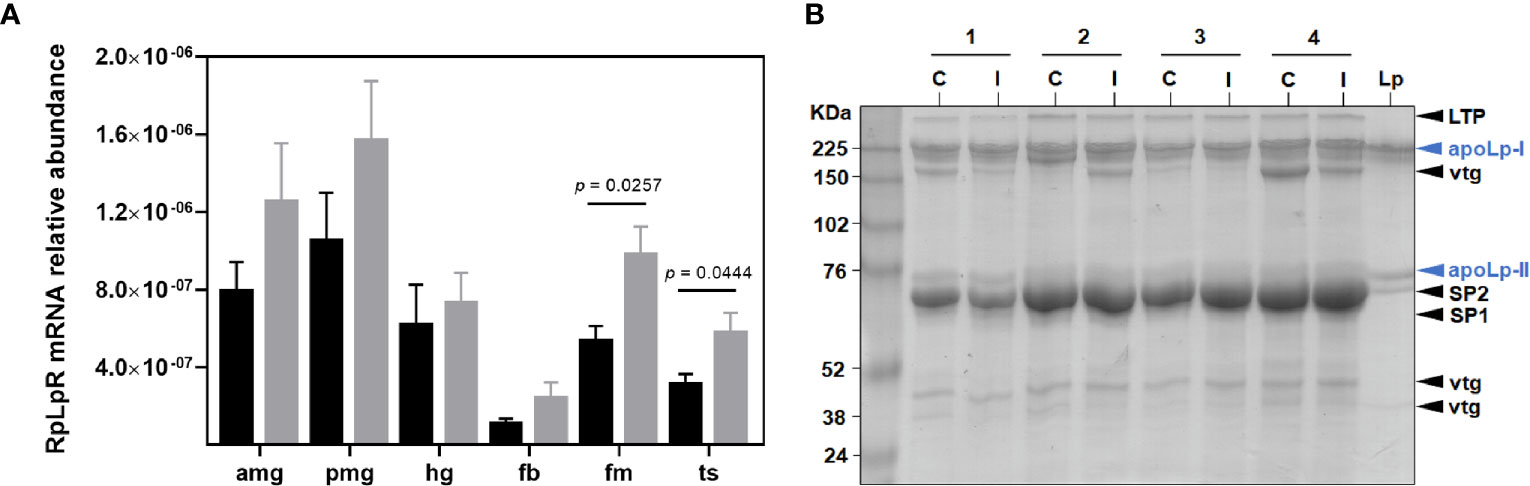
Figure 1 Lipid absorption and transport in R. prolixus males. (A) RpLpR transcript levels in individual triatomine organs. Analyses were performed using qRT-PCR and the 2−ΔCT method, with Rp18S as the reference gene. Data are displayed as means ± S.E.M. from eight replicates with t-test for parametric data comparing control (black columns) against T. cruzi infected (grey columns) insects. (B) Lp levels (apoLp-I and apoLp-II subunits) in the control (C) and infected (I) male insect hemolymph. The experiment was performed using four different samples (1 – 4) and a purified R. prolixus Lp as standard. Amg, anterior midgut; pmg, posterior midgut; hg, hindgut; fb, fat bodies; fm, flight muscle; ts, testes; LTP, lipid transfer particle; apoLp-I, apolipophorin I; vtg, vitellogenin; apoLp-II, apolipophorin II; SP1, stored protein 1 and SP2, stored protein 2.
Coomassie staining was used to detect the relative amount of Lp in male hemolymph. Two prominent Lp particle bands, corresponding to ApoLp-I (~ 240 KDa) and ApoLp-II (~ 80 KDa) subunits were detected. T. cruzi challenge did not alter Lp protein levels and, as expected, other proteins were also detectable in male circulation (Figure 1B).
As the exact function of the other LDLRs is still uncertain, the mRNA levels of the four LDLR family genes found in the R. prolixus genome were also evaluated. RpLrp1 (RPRC000138) transcripts were less abundant in the anterior midgut and flight muscle, with similar levels identified in the other investigated organs. T. cruzi infection did not modify the expression of this gene (Figure 2A). RpLrp2 (RPRC000060) mRNA levels were most abundant in the hindgut, flight muscle, and testes, with lower abundances detected in the other organs (Figure 2B). T. cruzi infection upregulated RpLrp2 (RPRC000060) mRNA expression in the hindgut (p = 0.0169), fat body (p = 0.0140), flight muscle (p = 0.0037) and testes (p = 0.0333), modifying the mRNA abundance profile due to the higher detection of this transcript in fat bodies (Figure 2B). The other RpLrp2 gene (RPRC000281) displayed a similar mRNA expression pattern (Figure 2C). T. cruzi infection, however, only upregulated mRNA abundance in fat bodies (p = 0.0361). Lastly, the highest transcript levels of RpVtgR (RPRC000551) were detected in the testes (Figure 2D). T. cruzi infection upregulated the anterior midgut RpVtgR (RPRC000551) mRNA expression (p = 0.0364).
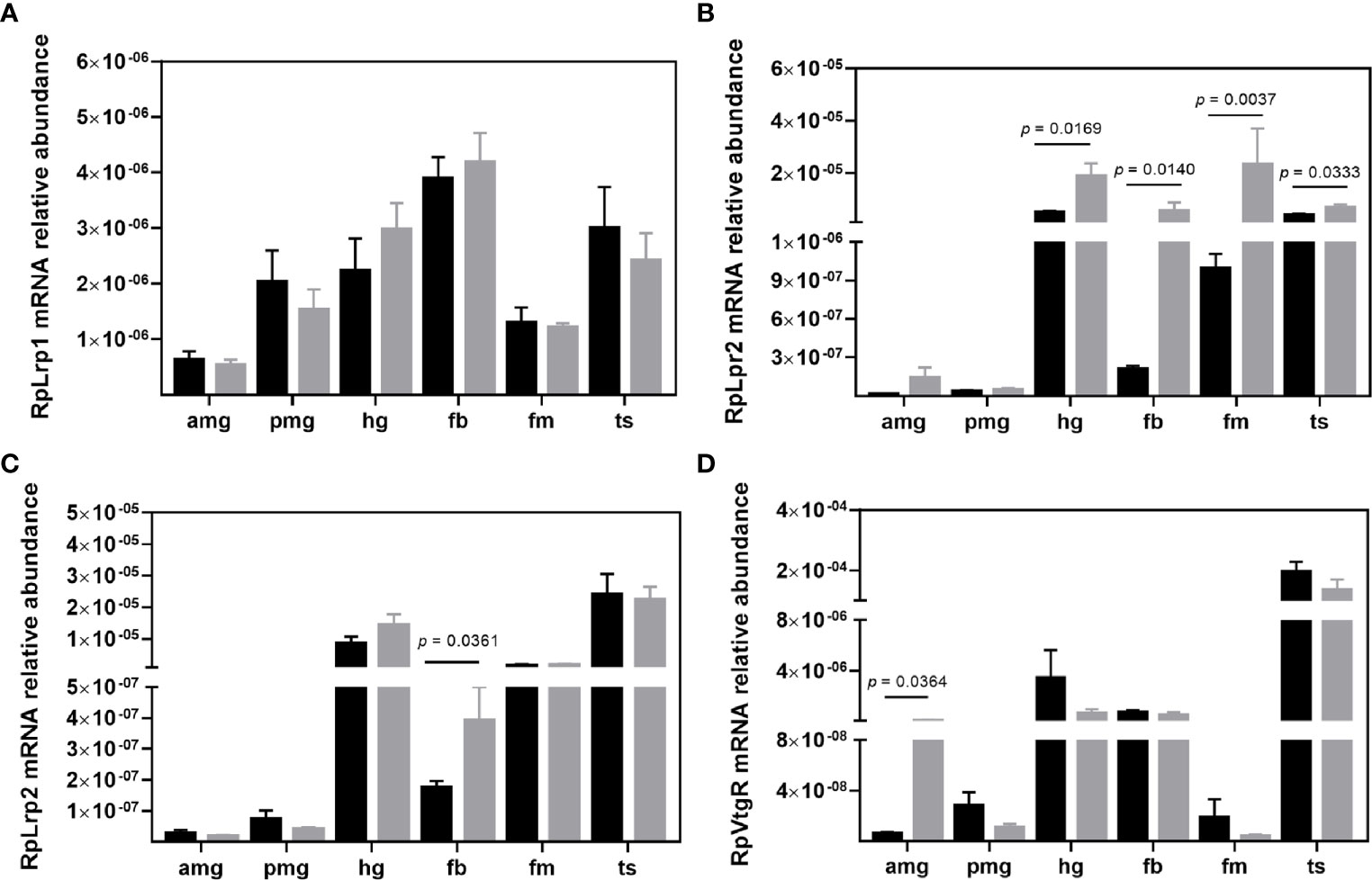
Figure 2 Transcript abundance of LDLR family members in R. prolixus males. mRNA levels of (A) RpLrp1 (RPRC000138), (B) RpLrp2 (RPRC000060), (C) RpLrp2 (RPRC000281), and (D) RpVtgR (RPRC000551) in individual triatomine organs. Analyses were performed using qRT-PCR and the 2−ΔCT method, with Rp18S as the reference gene. Data are displayed as means ± S.E.M. from eight replicates with a t-test for parametric data comparing control (black columns) against T. cruzi infected (grey columns) insects. Amg, anterior midgut; pmg, posterior midgut; hg, hindgut; fb, fat bodies; fm, flight muscle; and ts, testes.
To investigate the putative role of de novo FA synthesis during T. cruzi infection we evaluated the expression of RpACC (RPRC013987) (37, 38) in adult males. The transcript levels of the RpACC gene were most abundant in non-infected insect fat bodies, compared to other R. prolixus organs. Infected insects exhibited a reduced RpACC mRNA abundance in fat body (p = 0.0084), anterior midgut (p = 0.0296), and hindgut (p = 0.0313) (Figure 3A). The MCD enzyme catalyzes the ACC opposite reaction, carboxylate malonyl-CoA to acetyl-CoA, and its transcripts in R. prolixus males were mainly found in the flight muscle. T. cruzi infection did not modify the mRNA expression of this gene (Supplementary Figure S2).
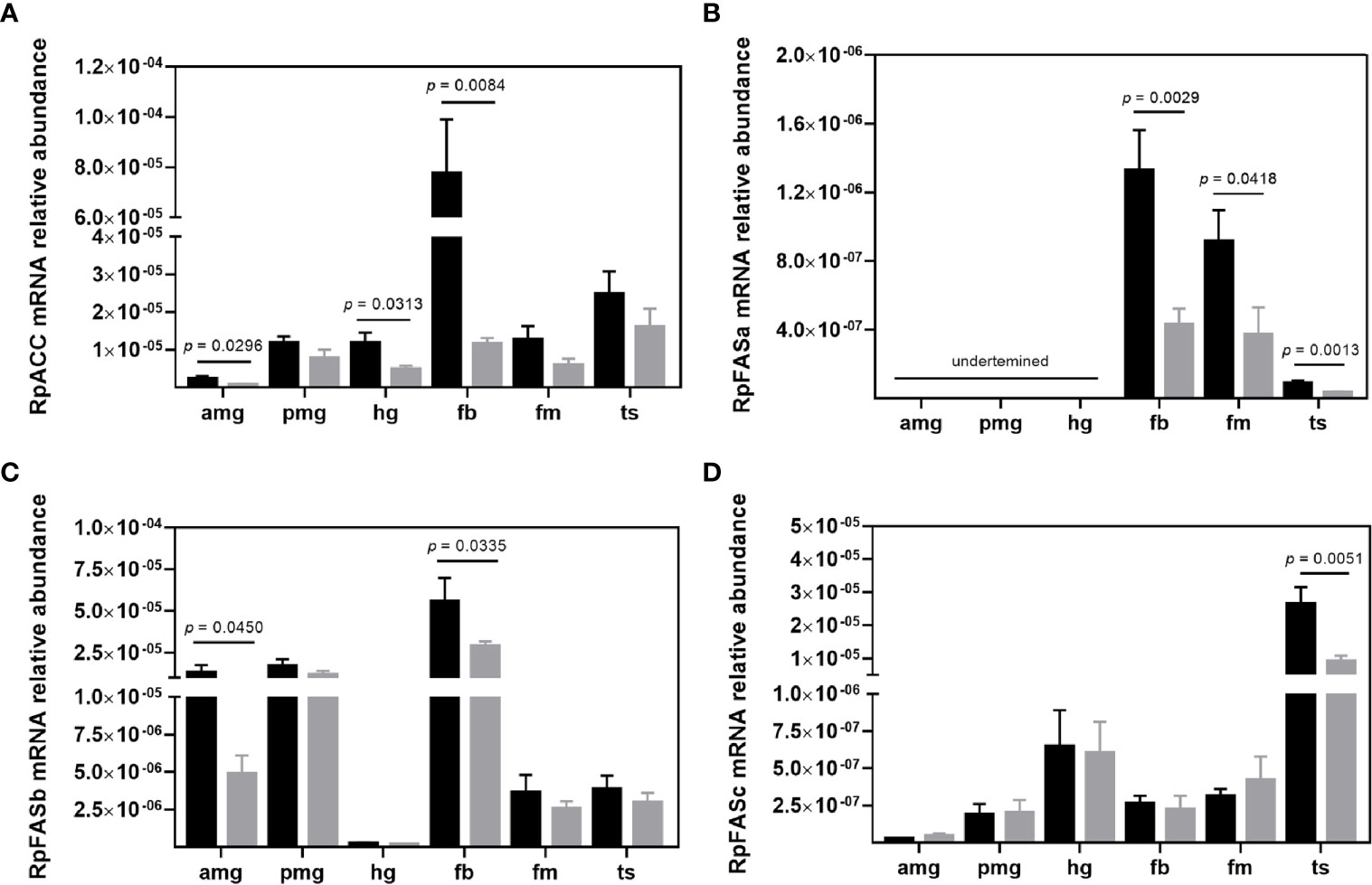
Figure 3 Transcript abundance of de novo fatty acid synthesis enzymes in R. prolixus males. mRNA levels of (A) RpACC, (B) RpFASa, (C) RpFASb, and (D) RpFASc in individual triatomine organs. Analyses were performed using qRT-PCR and the 2−ΔCT method, with Rp18S as the reference gene. Data are displayed as means ± S.E.M. from eight replicates with a t-test for parametric data comparing control (black columns) against T. cruzi infected (grey columns) insects. Amg, anterior midgut; pmg, posterior midgut; hg, hindgut; fb, fat bodies; fm, flight muscle; and ts, testes.
Fatty acid synthase I (FAS I) catalyzes the next step of FA synthesis, condensing malonyl-CoA molecules with one acetyl-CoA to build up FAs. Recent genomic analysis of triatomines lipid metabolism genes (39) reported three distinct type I FAS (FAS I) genes – RPRC000123, RPRC000269, and RPRC002909 (herein termed RpFASa, RpFASb, and RpFASc, respectively), which were all evaluated in the present study. RpFASa (RPRC000123) transcripts were not detected in any of the gut compartments. Its mRNA was mainly found in the fat body and flight muscle, and the infection with T. cruzi downregulated the transcript abundance in the fat body (p = 0.0029), flight muscle (p = 0.0418), and testes (p = 0.0013) (Figure 3B). The second FAS I gene – RpFASb (RPRC000269) was mainly transcripted in the fat body and midgut compartments (Figure 3C). Infected insects presented the same profile of mRNA abundance through the analyzed organs, with a reduction in the levels detected in the anterior midgut (p = 0.0450) and fat body (p = 0.0335) (Figure 3C). Transcripts of the third FAS I gene – RpFASc (RPRC002909) were predominantly found in the testes. The mRNA level profiles remained unaltered in infected insects, but the parasite-induced a reduction in RpFASc mRNA abundance in the testes (p = 0.0051) (Figure 3D).
Given the importance of lipid storage for hematophagous insect physiology, enzymes related to TG storage and degradation, as well as TG itself, were investigated. Although identified in all evaluated organs, the highest RpDGAT (RPRC003808) (39) transcript level was detected in flight muscle. T. cruzi infection induced a downregulation of this mRNA in the anterior midgut (p = 0.0324), hindgut (p = 0.0118), and fat body (p = 0.0165) (Figure 4A). The Bmm lipase (39) catalyzes the DGAT opposite reaction, and in R. prolixus males the highest abundance of its mRNA was found in the posterior midgut, hindgut, and fat body. Infected insects presented a reduction in RpBmm (RPRC002097) mRNA levels in the hindgut (p = 0.0362) and fat body (p = 0.0460) (Figure 4B).
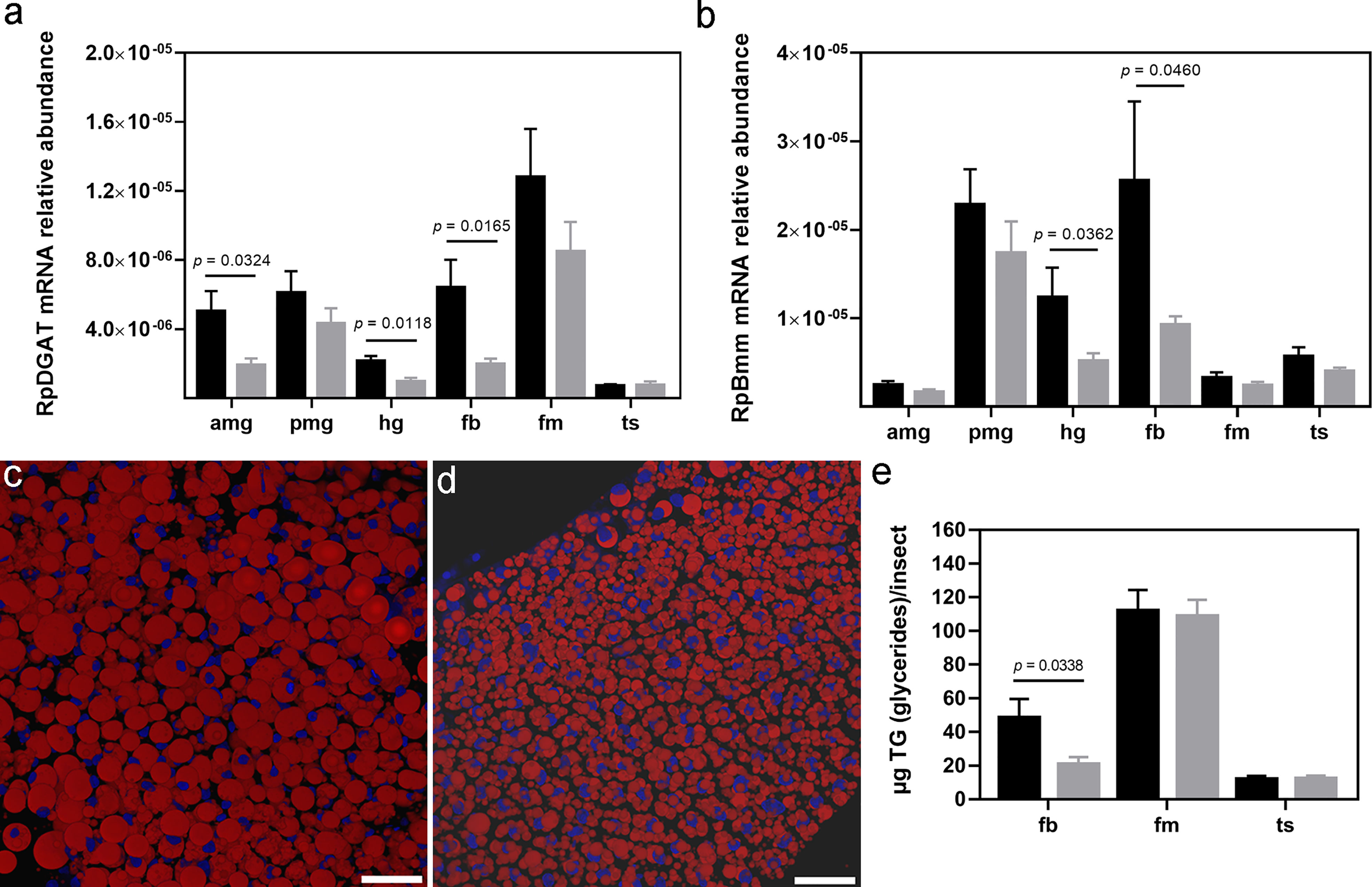
Figure 4 Lipid storage profile in R. prolixus males. (A) RpDGAT and (B) RpBmm mRNA levels in individual triatomine organs. Analyses were performed using qRT-PCR and the 2−ΔCT method, with Rp18S as the reference gene. Representative R. prolixus male fat bodies were stained with Nile red (red) and DAPI (blue) and imaged under a confocal microscope from (C) control and (D) infected insects. Images are a 3D reconstitution from optical sections of ~ 60 µm, bars = 50 µm. (E) TG quantification by colorimetric assay. Data are displayed as means ± S.E.M. from eight replicates with a t-test for parametric data comparing control (black columns) against T. cruzi infected (grey columns) insects. Amg, anterior midgut; pmg, posterior midgut; hg, hindgut; fb, fat bodies; fm, flight muscle; and ts, testes.
Regarding TG levels, the fat bodies of uninfected R. prolixus males mainly presented a single and large LD in the cytoplasm, delimiting cell nucleus size and shape (Figure 4C). Conversely, in T. cruzi infected males, the fat body cells exhibited numerous smaller LDs surrounding a large and rounded nucleus (Figure 4D). Colorimetric assays confirmed a reduction in TG content in the fat bodies of T. cruzi infected males (p = 0.0338). However, the infection did not alter flight muscle and testes TG content (Figure 4E).
To examine the lipid content of individual organs, lipids were extracted and separated by chromatography for neutral lipid (Supplementary Figure S3A) and phospholipid (Supplementary Figure S4A) identification. It is important to note that this chromatographic assay does not allow for efficient separation of 1,2-DG from CO. Consequently, the presented 1,2-DG level may contain some CO. T. cruzi infection modulated lipid levels only in triatomine fat bodies and hemolymph (Figure 5).
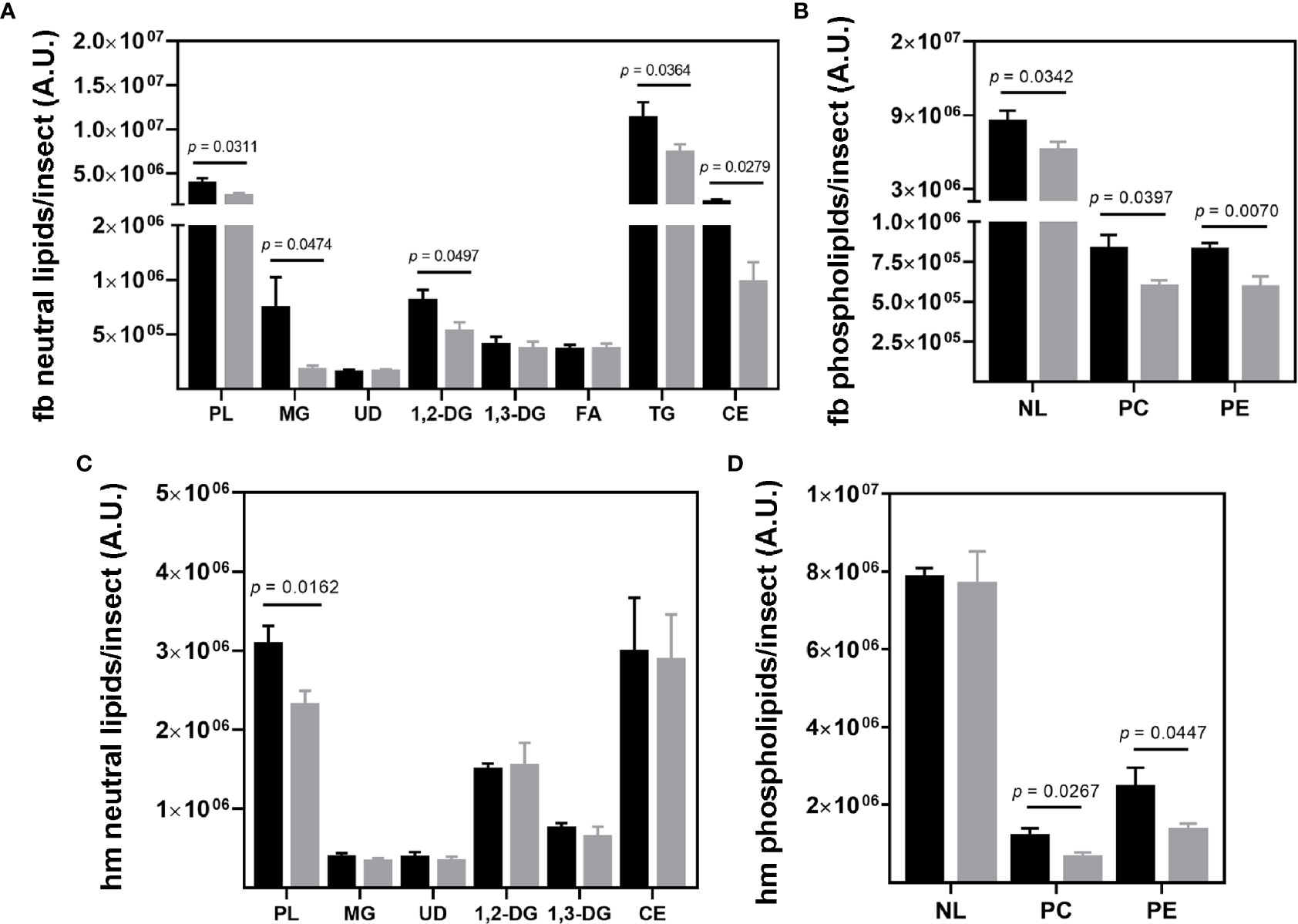
Figure 5 Lipid composition of R. prolixus male fat bodies and hemolymph. Levels of (A, C) neutral lipids and (B, D) phospholipids in the fat body (A, C), and hemolymph (B, D) of the triatomine. Lipids were extracted, separated, and identified by chromatography for subsequent densitometric analysis. Data are displayed as means ± S.E.M. expressed in arbitrary units (A. U.) from eight replicates with a t-test for parametric data comparing the lipid levels of control (black columns) against T. cruzi infected (grey columns) insects. Fb, fat bodies; hm, hemolymph; PL, phospholipids; MG, monoacylglycerol; UD, undeterminate lipid; 1;2-DG, 1;2-diacylglycerol (and cholesterol); 1,3-DG, 1,3-diacylglycerol; FA, fatty acid; TG, triacylglycerol; CE, cholesteryl ester; NL, neutral lipid; PC, phosphatidylcholine and PE, phosphatidylethanolamine.
TG and CE represented the most abundant neutral lipid classes in the fat body. A high level of PLs was also identified. Infected insects presented a reduction in MG (p = 0.0474), 1,2-DG/CO (p = 0.0497), TG (p = 0.0364), CE (p = 0.0279), and PL (p = 0.0311) levels (Figure 5A). The fat body phospholipids were composed of phosphatidylcholine (PC) and phosphatidylethanolamine (PE), both also downregulated by T. cruzi infection, at p = 0.0397 and 0.0070, respectively (Figure 5B). Other phospholipids, such as sphingomyelin, phosphatidylinositol, and phosphatidic acid, were also identified in the fat bodies, however at very low levels and frequency (in two of eight experiments). Thus, they were not considered representative in this study.
CE, 1,2-DG/CO, and PLs were the most abundant lipid classes in hemolymph, (Figure 5C). Only PLs were reduced (p = 0.0162) by T. cruzi infection, specifically PC (p = 0.0267) and PE (p = 0.0447) (Figures 5C, D). The neutral lipid and phospholipid composition of the other R. prolixus organs are presented in Supplementary Figure S3, S4, respectively.
To further investigate the effects of T. cruzi infection on the modulation of R. prolixus fat body metabolism, extracted lipids were subjected to a mass spectrometry analysis for FA and sterols identification. Several FA types were identified (Supplementary Figure S5), but at levels so low that they could not be quantified. The most abundant types were myristic (C14:0), pentadecanoic (C15:0), palmitic (C16:0), palmitoleic (C16:1), margaric (C17:0), stearic (C18:0) and elaidic (C18:1n9t) acids. The insect challenge with T. cruzi did not alter fat body FA composition or levels (Figure 6A). On the other hand, the sterol analysis revealed CO as the only sterol present in R. prolixus male fat body (Supplementary Figure S6). Additionally, T. cruzi infection did not modulate CO levels in this organ (Figure 6B).
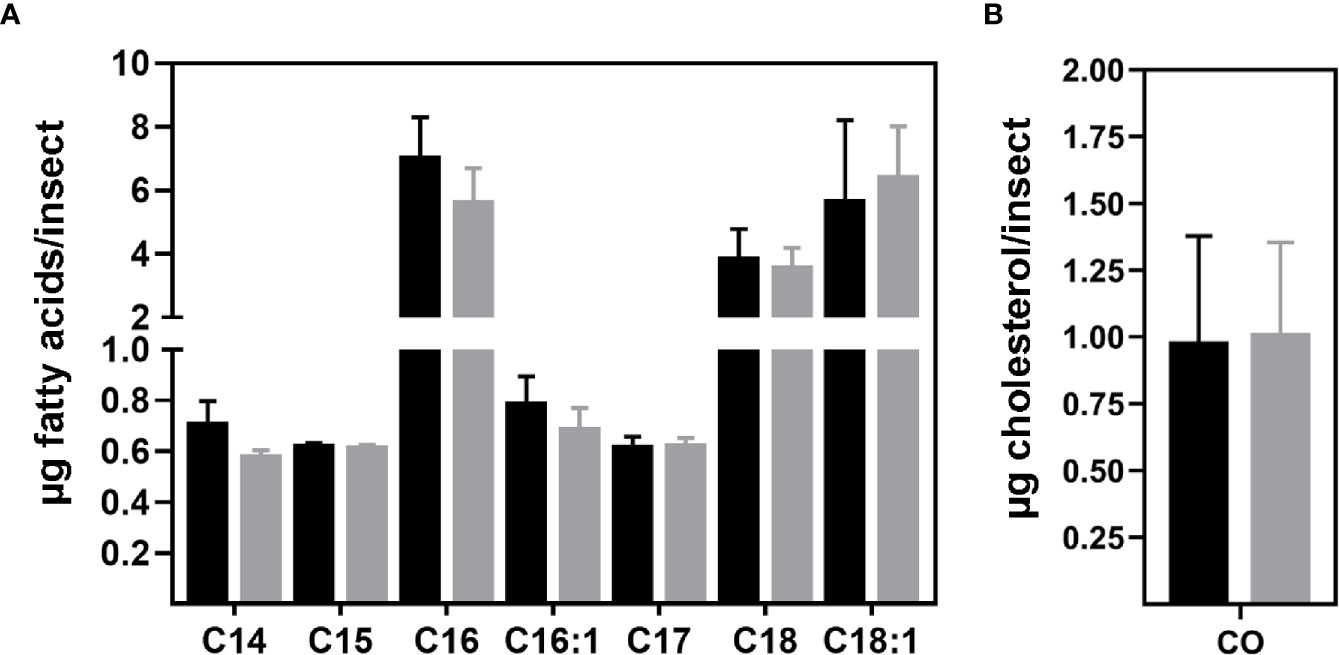
Figure 6 Fatty acid and sterol composition of R. prolixus male fat bodies. (A) FA and (B) sterol levels identified in triatomine fat body. After extraction, lipids were prepared and analyzed by gas chromatography coupled to mass spectrometry. Nonadecanoic acid (C19:0) and 7-dehydrocholesterol were used as internal standards, respectively. Data are displayed as the means ± S.E.M. from four replicates with a t-test for parametric data comparing the FA or sterol composition of control (black columns) against T. cruzi infected (grey columns) insects. C14:0 – myristic, C15:0 – pentadecanoic, C16:0 – palmitic, C16:1 – palmitoleic, C17:0 – margaric, C18:0 – stearic, C18:1n9t – elaidic acids and CO – cholesterol.
While a significant amount of studies regarding the lipid metabolism in R. prolixus females are available (4, 5, 37, 39–44), scarce work has been conducted on male metabolism. Additionally, how T. cruzi infection affects the lipid physiology of individual R. prolixus organs is insufficiently understood. In this context, several lipid metabolism parameters in adult male R. prolixus specimens were investigated herein, revealing that T. cruzi infection significantly alters several molecular and biochemical aspects of insect lipid physiology.
Although the R. prolixus genome contains five genes closely related to LDLR family members, the RPRC011390 gene is the only one similar to other described LpRs in insects (12, 36, 45). The tissue-specific expression pattern of this gene three days after feeding supports an Lp lipid loading route through the gut compartments to subsequent fat body storage. This supports previously observed Lp binding capacity in the midgut (42) and concomitant transference of phospholipids and neutral lipids from the posterior midgut to Lp (41) in R. prolixus females. The upregulation of RpLpR mRNA expression in the flight muscle and testes of T. cruzi infected insects may be a strategy to increase parasite dispersal through greater flight activity and reproduction, both sustained by additional lipid capitation. The infection response observed herein is similar to that of Plasmodium gallinaceum infected Aedes aegypti, where LpR expression is enhanced by the infection (46).
Since the LDLR protein family presents multi-ligand binding properties interacting with over 40 different molecules (47) and some insect LpRs are not essential for TG storage or mobilization in fat bodies (12), a potential role of the other R. prolixus LDLRs on Lp internalization cannot be disregarded. The tissue-specific mRNA expression pattern of these genes is different from that of the true RpLpR and also responsive to the parasite challenge, supporting possible participation in Lp uptake.
Despite the critical role Lp plays in lipid transport (6), the finding that Lp protein levels were unchanged in T. cruzi infection appears to conflict with the other results presented herein. The working hypothesis is that the parasite affects only the Lp lipid portion, which still requires further investigation. Supporting this finding, insect LpRs favor the endocytosis of delipidated Lp particles leading to downregulation when circulating Lp is fully lipidated (11), which occurs three days after R. prolixus feeding (41).
Concerning the lipid biosynthetic pathway, the first enzyme committed to the de novo FA synthesis, ACC, was mainly transcribed in the fat body. This may sustain lipid storage until the transference of dietary lipids from the midgut, which is highest around days two-three after feeding (5, 41, 42). The next enzyme in this route, FAS, was found in three copies in the R. prolixus genome. Interestingly, each was mostly transcribed at a specific site, ensuring the non-dietary lipid supply in all investigated insect organs. Intriguingly, RpACC and RpFAS were downregulated by T. cruzi infection.
As transcriptional factors, sterol regulatory-element binding proteins (SREBPs) are key lipogenic activators (48, 49). Among their control mechanisms, SREBPs can be inhibited by sirtuin (SIRT) deacetylation (48, 50, 51). An increase in the abundance of SIRT mRNAs in the hindgut and fat bodies of infected R. prolixus was observed in the infection system investigated herein (manuscript under preparation). Thus, T. cruzi may inhibit SREBP activity (through SIRT-dependent or independent manners) resulting in RpACC and RpFAS mRNA expression downregulation and subsequent decreased lipid levels and storage in the insect fat bodies.
RpDGAT transcription was also downregulated in the parasite challenge, which strongly corroborates with lower LDs, TG, CE, and PLs accumulation in the insect fat bodies. Considering that T. cruzi physiology is dependent on active lipid endocytic processes (19, 20) and that R. prolixus fat bodies are a rich source of lipids, it is not surprising that the parasite has evolved mechanisms to promote the mobilization of these stocks. In agreement, the most abundant lipids in T. cruzi storage – CE, TG, and PLs (20) were all downregulated in infected insects. Additionally, during the invertebrate host life cycle, the parasite also takes nutrients directly from vertebrate blood ingested by the insect. It is intriguing to note, however, that T. cruzi never leaves the R. prolixus gut lumen (18) and yet regulates lipid fat body metabolism. It has been shown in other insects that parasites can modulate a great diversity of cell signaling molecules to establish an association from the closed gut environment with the other insect organs leading to effects, such as immune response, change in behavior, modulation of host development (55–57). microRNAs, which are known to be significantly modulated by blood-feeding in vector arthropods (58, 59), may be responsible for this association between the parasite and the insect system.
Conversely, during the vertebrate host life cycle, intracellular T. cruzi forms induce the accumulation of LDs (60, 61). It is well established that a variety of intracellular pathogens, such as viruses and bacteria, leads to LD proliferation and increases in certain lipid levels (62–67). Indeed, a central immune response to infection triggers LD accumulation as a primary source of precursors for inflammatory mediators (such as prostaglandins, thromboxanes, leukotrienes, lipoxins, resolvins, eoxins, and others) biosynthesis (67). Neutral lipid and phospholipid downregulation in R. prolixus may be rerouting these molecules to form anti-immunity mediators, to allow T. cruzi growth and differentiation in the invertebrate host gut. It is important to note that a homeostatic mechanism that maintains stored TG levels in fat bodies unchanged even during a lipid-free diet period is present in other insect models (68). In addition, a well-known correlation between increased TG synthesis and LD formation and cell stress is known (69). T. cruzi thus appears to have evolved strategies to overcome triatomine homeostasis, stress, and immune system responses.
In addition to lower synthesis, lipid decreases may also be related to greater oxidation. TG recruitment undergoes the action of TG lipases to provide free FA (70). However, T. cruzi infection also decreased RpBmm transcription. It has been recently reported that induction of autophagy-dependent processing of LDs and TG (lipophagy) occurs in other infection systems to favor pathogen development (71). Lipophagy may answer for the modification identified in R. prolixus fat bodies without a concomitant increase in RpBmm expression or RpBmm transcription mat not correlated with enzyme activity.
Lipid importance in T. cruzi infections is evident when the parasite regulates insect lipid physiology in its favor. It is proposed that T. cruzi uptakes the lipid component of R. prolixus Lp, affecting dietary lipid storage. It is known that protozoan parasites such as T. rangeli and P. gallinaceum uptake their invertebrate host Lps as their main lipid source (72, 73), strongly supporting the results presented herein. Concomitantly, the parasite interferes with lipid degradation and synthesis pathways. As a countermeasure, the transcription of LpR and other proteins possibly related to lipid uptake is upregulated, probably as an attempt to capture more Lp. All lipid level modulations were identified in non-infected tissues. This suggests that T. cruzi displays a great diversity of cell signaling molecules to maintain its association from the closed gut environment with insect fat bodies. The parasite may modulate several other metabolic pathways that affect all insect organs in a different manner, which raises several intriguing issues for future studies in the parasite-vector interaction field.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
The animal study was reviewed and approved by Committee for Evaluation of Animal Use for Research at the Federal University of Rio de Janeiro, CAUAP-UFRJ, under the registry #IBQM067-05/16 and 24154319.5.0000.5257, and the NIH Guide for the Care and Use of Laboratory Animals (ISBN 0-309-05377-3). Animal facility technicians at the Leopoldo de Meis Medical Biochemistry Institute (UFRJ) performed all aspects related to rabbit husbandry under strict guidelines to ensure careful and consistent animal handling.
GA conceived the idea of study and designed the methodology. GS prepared all the experimental insect samples. GS and SC participated in data analysis and wrote sections of the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the Conselho Nacional de Desenvolvimento Cientifico e Tecnológico (CNPq), the Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), and the Instituto Nacional de Ciência e Tecnologia em Entomologia Molecular (INCT-EM).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to express our gratitude to Mileane Bush for her support with the GC-MS analysis and technical advice, and Yasmin Paule Gutierrez and Desenir Pedro for insect care.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fitd.2021.737909/full#supplementary-material
Supplementary Table S1 | Primer nucleotide sequences were used for the qRT-PCR analyses.
Supplementary Figure S1 | Modular structure of the LDLR family in R. prolixus. An analysis of the triatomine genome resulted in the identification of six genes encoding proteins belonging to the LDLR family. The RPRC011390 gene is the true insect LpR; RPRC000138 is an LDL-related protein 1 (Lrp1); RPRC000060 and RPRC000281 are LDL-related proteins 2 (Lrp2 or megalin); RPRC000551 is a vitellogenin receptor (VtgR); and, finally, RPRC000270 appears to be a gene related to lipoprotein intracellular transport. FN3 – fibronectin 3 and VPS10 – vacuolar protein sorting/targeting protein 10. Figure elaborated based on domain architecture acquired from the Smart Domain database (http://smart.embl-heidelberg.de/).
Supplementary Figure S2 | Transcript abundance of malonyl-CoA decarboxylase in R. prolixus males. RpMCD mRNA levels in individual triatomine organs. Analyses were performed using qRT-PCR and the 2−ΔCT method, with Rp18S as the reference gene. Data are displayed as means ± S.E.M. from eight replicates with a t-test for parametric data comparing control (black columns) against T. cruzi infected (grey columns) insects. Amg – anterior midgut, pmg – posterior midgut, hg – hindgut, fb – fat bodies, fm – flight muscle, and ts – testes.
Supplementary Figure S3 | Neutral lipid content in individual organs of R. prolixus males. Lipids were extracted, separated, and identified by chromatography for subsequent densitometric analysis. (A) Representative thin layer chromatography showing the profile of neutral lipids in the triatomine organs. Neutral lipid levels from the (B) anterior midgut, (C) posterior midgut, (D) hindgut, (E) flight muscle, and f testes. Data are displayed as means ± S.E.M. expressed in arbitrary units (A. U.) from eight replicates with a t-test for parametric data comparing the lipid levels of control (C/black columns) against T. cruzi infected (I/grey columns) insects. PL – phospholipids, MG – monoacylglycerol, UD – undeterminate lipid, 1,2-DG – 1,2-diacylglycerol (and cholesterol), 1,3-DG – 1,3-diacylglycerol, FA – fatty acid, TG - triacylglycerol, and CE – cholesteryl ester.
Supplementary Figure S4 | Phospholipid content in individual organs of R. prolixus males. Lipids were extracted, separated, and identified by chromatography for subsequent densitometric analysis. (A) Representative high-performance thin-layer chromatography showing the profile of phospholipids in the triatomine organs. Phospholipid levels from the (B) anterior midgut, (C) posterior midgut, (D) hindgut, (E) flight muscle, and f testes. Data are displayed as means ± S.E.M. expressed in arbitrary units (A. U.) from eight replicates with a t-test for parametric data comparing the lipid levels of control (C/black columns) against T. cruzi infected (I/grey columns) insects. NL – neutral lipids, LPC – lysophosphatidylcholine, SM – sphingomyelin, PG – phosphatidylglycerol, PI – phosphatidylinositol, PS – phosphatidylserine, PC – phosphatidylcholine, PE – phosphatidylethanolamine and PA – phosphatidic acid.
Supplementary Figure S5 | Chromatogram of the fatty acid composition of R. prolixus fat bodies. Analysis from control (A) and infected (B) insects. After extraction, lipids were prepared and evaluated by gas chromatography coupled to mass spectrometry. Nonadecanoic acid (C19:0) was used as an internal standard. The fatty acids identified were C12:0 - lauric, C14:0 – myristic, C15:0 – pentadecanoic, C16:1 – palmitoleic, C16:0 – palmitic, C17:0 – margaric, C18:2n6c – linoleic, C18:1n9c – oleic, C18:1n9t – elaidic, C18:0 – stearic, C19:0 – nonadecanoic (standard), C20:4n6 – arachidonic, C20:0 – arachidic, C21:0 – heneicosanoic, C22:1n9 – erucic and C24:0 – lignoceric.
Supplementary Figure S6 | Chromatogram of sterol composition of R. prolixus fat bodies. Analysis from control (A) and infected (B) insects. After extraction, lipids were prepared and evaluated by gas chromatography coupled to mass spectrometry. 7-Dehydrocholesterol was used as an internal standard. Only cholesterol was identified in this analysis.
1. Pérez-Molina JA, Molina I. Chagas Disease. Lancet (2018) 391(10115):82–94. doi: 10.1016/S0140-6736(17)31612-4
2. Gondim KC, Atella GC, Pontes EG, Majerowicz D. Lipid Metabolism in Insect Disease Vectors. Insect Biochem Mol Biol (2018) 101:108–23. doi: 10.1016/j.ibmb.2018.08.005
3. Canavoso LE, Jouni ZE, Karnas KJ, Pennington JE, Wells MA. Fat Metabolism in Insects. Annu Rev Nut (2001) 21:23–46. doi: 10.1146/annurev.nutr.21.1.23
4. Atella GC, Arruda MA, Masuda H, Gondim KC. Fatty Acid Incorporation by Rhodnius Prolixus Midgut. Arch Insect Biochem Physiol (2000) 43:99–107. doi: 10.1002/(SICI)1520-6327(200003)43:3<99::AID-ARCH1>3.0.CO;2-3
5. Grillo LA, Majerowicz D, Gondim KC. Lipid Metabolism in Rhodnius Prolixus (Hemiptera: Reduviidae): Role of a Midgut Triacylglycerol-Lipase. Insect Biochem Mol Biol (2007) 37:579–88. doi: 10.1016/j.ibmb.2007.03.002
6. Chino H, Kitazawa K. Diacylglycerol-Carrying Lipoprotein of Hemolymph of the Locust and Some Insects. J Lipid Res (1981) 22:1042–52. doi: 10.1016/S0022-2275(20)40661-3
7. Dantuma NP, Pijnenburg MAP, Diederen JHB, van der Horst DJ. Developmental Down-Regulation of Receptor-Mediated Endocytosis of an Insect Lipoprotein. J Lipid Res (1997) 38:254–65. doi: 10.1016/S0022-2275(20)37438-1
8. Van Hoof D, Rodenburg KW, van der Horst DJ. Insect Lipoprotein Follows a Transferrin-Like Recycling Pathway That Is Mediated by the Insect LDL Receptor Homologue. J Cell Sci (2002) 115:4001–12. doi: 10.1242/jcs.00113
9. Van Hoof D, Rodenburg KW, van der Horst DJ. Receptor-Mediated Endocytosis and Intracellular Trafficking of Lipoproteins and Transferrin in Insect Cells. Insect Biochem Mol Biol (2005) 35:117–28. doi: 10.1016/j.ibmb.2004.09.009
10. Roosendaal SD, Kerver J, Schipper M, Rodenburg KW, van der Horst DJ. The Complex of the Insect LDL Receptor Homolog, Lipophorin Receptor, LpR, and Its Lipoprotein Ligand Does Not Dissociate Under Endosomal Conditions. FEBS J (2008) 275:1751–66. doi: 10.1111/j.1742-4658.2008.06334.x
11. Roosendaal SD, Van Doorn JM, Valentijn KM, van der Horst DJ, Rodenburg KW. Delipidation of Insect Lipoprotein, Lipophorin, Affects Its Binding to the Lipophorin Receptor, LpR: Implications for the Role of LpR-Mediated Endocytosis. Insect Biochem Mol Biol (2009) 39:135–44. doi: 10.1016/j.ibmb.2008.10.013
12. Parra-Peralbo E, Culi J. Drosophila Lipophorin Receptors Mediate the Uptake of Neutral Lipids in Oocytes and Imaginal Disc Cells by an Endocytosis-Independent Mechanism. PloS Genet (2011) 7:e1001297. doi: 10.1371/journal.pgen.1001297
13. Barber MC, Price NT, Travers MT. Structure and Regulation of Acetyl-CoA Carboxylase Genes of Metazoa. Biochim Biophys Acta (2005) 1733:1–28. doi: 10.1016/j.bbalip.2004.12.001
14. Smith S, Tsai SC. The Type I Fatty Acid and Polyketide Synthases: A Tale of Two Megasynthases. Nat Prod Rep (2007) 24:1041–72. doi: 10.1039/b603600g
15. Laurent G, German NJ, Saha AK, de Boer VC, Davies M, Koves TR, et al. SIRT4 Coordinates the Balance Between Lipid Synthesis and Catabolism by Repressing Malonyl CoA Decarboxylase. Mol Cell (2013) 50:686–98. doi: 10.1016/j.molcel.2013.05.012
16. McFie PJ, Banman SL, Kary S, Stone SJ. Murine Diacylglycerol Acyltransferase-2 (DGAT2) Can Catalyze Triacylglycerol Synthesis and Promote Lipid Droplet Formation Independent of Its Localization to the Endoplasmic Reticulum. J Biol Chem (2011) 286:28235–46. doi: 10.1074/jbc.M111.256008
17. Grönke S, Mildner A, Fellert S, Tennagels N, Petry S, Müller G, et al. Brummer Lipase Is an Evolutionary Conserved Fat Storage Regulator in Drosophila. Cell Metab (2005) 1:323–30. doi: 10.1016/j.cmet.2005.04.003
18. Azambuja P, Garcia ES, Waniek PJ, Vieira CS, Figueiredo MB, Gonzalez MS, et al. Rhodnius Prolixus: From Physiology by Wigglesworth to Recent Studies of Immune System Modulation by Trypanosoma Cruzi and Trypanosoma Rangeli. J Insect Physiol (2017) 97:45–65. doi: 10.1016/j.jinsphys.2016.11.006
19. Pereira MG, Nakayasu ES, Sant’Anna C, De Cicco NNT, Atella GC, de Souza W, et al. Trypanosoma Cruzi Epimastigotes Are Able to Store and Mobilize High Amounts of Cholesterol in Reservosome Lipid Inclusions. PloS One (2011) 6:e22359. doi: 10.1371/journal.pone.0022359
20. Pereira MG, Visbal G, Costa TFR, Frases S, de Souza W, Atella GC, et al. Trypanosoma Cruzi Epimastigotes Store Cholesteryl Esters in Lipid Droplets After Cholesterol Endocytosis. Mol Biochem Parasitol (2018) 224:6–16. doi: 10.1016/j.molbiopara.2018.07.004
21. Soares MJ, De Souza W. Endocytosis of Gold-Labeled Proteins and LDL by Trypanosoma Cruzi. Parasitol Res (1991) 77:461–9. doi: 10.1007/BF00928410
22. de Souza W, Rodrigues JC. Sterol Biosynthesis Pathway as Target for Anti-Trypanosomatid Drugs. Interdiscip Perspect Infect Dis (2009) 2009:642502. doi: 10.1155/2009/642502
23. Garcia ES, Garcia MLM, Macarini JD, Ubatuba FB. Alimentação De Rhodnius Prolixus No Laboratório. Acad Bras Ciênc (1975) 47:537–45.
24. Guedes PMM, Gutierrez FRS, Maia FL, Milanezi CM, Silva GK, Pavanelli WR, et al. IL-17 Produced During Trypanosoma Cruzi Infection Plays a Central Role in Regulating Parasite-Induced Myocarditis. PloS Negl Trop Dis (2010) 4:e604. doi: 10.1371/journal.pntd.0000604
25. Van Hoof D, Rodenburg KW, van der Horst DJ. Lipophorin Receptor-Mediated Lipoprotein Endocytosis in Insect Fat Body Cells. J Lipid Res (2003) 44(8):1431–40. doi: 10.1194/jlr.M300022-JLR200
27. Silva-Oliveira G, De Paula IF, Medina JM, Alves-Bezerra M, Gondim KC. Insulin Receptor Deficiency Reduces Lipid Synthesis and Reproductive Function in the Insect Rhodnius Prolixus. Biochim Biophys Acta Mol Cell Biol Lipids (2021) 1866(2):158851. doi: 10.1016/j.bbalip.2020.158851
28. Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (2001) 25:402–8. doi: 10.1006/meth.2001.1262
29. Majerowicz D, Alves-Bezerra M, Logullo R, Fonseca-de-Souza AL, Meyer-Fernandes JR, Braz GRC, et al. Looking for Reference Genes for Real-Time Quantitative PCR Experiments in Rhodnius Prolixus (Hemiptera: Reduviidae). Insect Mol Biol (2011) 20:713–22. doi: 10.1111/j.1365-2583.2011.01101.x
30. Laemmli UK. Cleavage of Structural Proteins During the Assembly of the Head of Bacteriophage T4. Nature (1970) 227:680–5. doi: 10.1038/227680a0
31. Bligh EG, Dyer WJ. A Rapid Method of Total Lipid Extraction and Purification. Can J Biochem Physiol (1959) 37:911–7. doi: 10.1139/o59-099
32. Ruiz JI, Ochoa B. Quantification in the Subnanomolar Range of Phospholipids and Neutral Lipids by Monodimensional Thin-Layer Chromatography and Image Analysis. J Lipid Res (1997) 38:1482–9. doi: 10.1016/S0022-2275(20)37430-7
33. Kluck G, Regis KC, De Cicco NNT, Silva-Cardoso L, Pereira MG, Fampa P, et al. An Evaluation of Lipid Metabolism in the Insect Trypanosomatid Herpetomonas Muscarum Uncovers a Pathway for the Uptake of Extracellular Insect Lipoproteins. Parasitol Int (2018) 67:97–106. doi: 10.1016/j.parint.2017.10.013
34. Burns MJ, Nixon GJ, Foy CA, Harris N. Standardisation of Data From Real-Time Quantitative PCR Methods – Evaluation of Outliers and Comparison of Calibration Curves. BMC Biotechnol (2005) 5:31. doi: 10.1186/1472-6750-5-31
35. Schneider WJ, Nimpf J. LDL Receptor Relatives at the Crossroad of Endocytosis and Signaling. Cell Mol Life Sci (2003) 60:892–903. doi: 10.1007/s00018-003-2183-Z
36. Rodenburg KW, Smolenaars MM, Van Hoof D, van der Horst DJ. Sequence Analysis of the Non-Recurring C-Terminal Domains Shows That Insect Lipoprotein Receptors Constitute a Distinct Group of LDL Receptor Family Members. Insect Biochem Mol Biol (2006) 36:250–63. doi: 10.1016/j.ibmb.2006.01.003
37. Alves-Bezerra M, Gondim KC. Triacylglycerol Biosynthesis Occurs via the Glycerol-3-Phosphate Pathway in the Insect Rhodnius Prolixus. Biochim Biophys Acta (2012) 1821:1462–71. doi: 10.1016/j.bbalip.2012.08.002
38. Majerowicz D, Hannibal-Bach HK, Castro RSC, Bozaquel-Morais BL, Alves-Bezerra M, Grillo LAM, et al. The ACBP Gene Family in Rhodnius Prolixus: Expression, Characterization and Function of RpACBP-1. Insect Biochem Mol Biol (2016) 72:41–52. doi: 10.1016/j.ibmb.2016.03.002
39. Majerowicz D, Calderón-Fernández GM, Alves-Bezerra M, De Paula IF, Cardoso LS, Juárez MP, et al. Lipid Metabolism in Rhodnius Prolixus: Lessons From the Genome. Gene (2017) 5:27–44. doi: 10.1016/j.gene.2016.09.045
40. Gondim KC, Atella GC, Kawooya JK, Masuda H. Role of Phospholipids in the Lipophorin Particles of Rhodnius Prolixus. Arch Insect Biochem Physiol (1992) 20:303–14. doi: 10.1002/arch.940200406
41. Coelho HSL, Atella GC, Moreira MF, Gondim KC, Masuda H. Lipophorin Density Variation During Oogenesis in Rhodnius Prolixus. Arch Insect Biochem Physiol (1997) 35:301–13. doi: 10.1002/(SICI)1520-6327(199705)35:3<301::AID-ARCH4>3.0.CO;2-W
42. Grillo LAM, Pontes EG, Gondim KC. Lipophorin Interaction With the Midgut of Rhodnius Prolixus: Characterization and Changes in Binding Capacity. Insect Biochem Mol Biol (2003) 33:429–38. doi: 10.1016/S0965-1748(03)00007-9
43. Oliveira GA, Baptista DL, Guimarães-Motta H, Almeida IC, Masuda H, Atella GC. Flight-Oogenesis Syndrome in a Blood-Sucking Bug: Biochemical Aspects of Lipid Metabolism. Arch Insect Biochem Physiol (2006) 62:164–75. doi: 10.1002/arch.20132
44. Bittencourt-Cunha PR, Silva-Cardoso L, Oliveira GA, Silva JR, Silveira AB, Kluck GEG, et al. Perimicrovillar Membrane Assembly: The Fate of Phospholipids Synthesised by the Midgut of Rhodnius Prolixus. Mem Inst Oswaldo Cruz (2013) 108:494–500. doi: 10.1590/S0074-0276108042013016
45. Lu K, Chen X, Li Y, Li W, Zhou Q. Lipophorin Receptor Regulates Nilaparvata Lugens Fecundity by Promoting Lipid Accumulation and Vitellogenin Biosynthesis. Comp Biochem Physiol A Mol Integr Physiol (2018) 219–220:28–37. doi: 10.1016/j.cbpa.2018.02.008
46. Cheon HM, Shin SW, Bian G, Park JH, Raikhel AS. Regulation of Lipid Metabolism Genes, Lipid Carrier Protein Lipophorin, and Its Receptor During Immune Challenge in the Mosquito Aedes Aegypti. J Biol Chem (2006) 281:8426–35. doi: 10.1074/jbc.M510957200
47. May P, Woldt E, Matz RL, Boucher P. The LDL Receptor-Related Protein (LRP) Family: An Old Family of Proteins With New Physiological Functions. Ann Med (2007) 39:219–28. doi: 10.1080/07853890701214881
48. Ponugoti B, Kim DH, Xiao Z, Smith Z, Miao J, Zang M, et al. SIRT1 Deacetylates and Inhibits SREBP-1C Activity in Regulation of Hepatic Lipid Metabolism. J Biol Chem (2010) 285:33959–70. doi: 10.1074/jbc.M110.122978
49. Shimano H, Sato R. SREBP-Regulated Lipid Metabolism: Convergent Physiology – Divergent Pathophysiology. Nat Rev Endocrinol (2017) 13:710–30. doi: 10.1038/nrendo.2017.91
50. Rodgers JT, Puigserver P. Fasting-Dependent Glucose and Lipid Metabolic Response Through Hepatic Sirtuin 1. Proc Natl Acad Sci USA (2007) 104:12861–6. doi: 10.1073/pnas.0702509104
51. Walker AK, Yang F, Jiang K, Ji JY, Watts JL, Purushotham A, et al. Conserved Role of SIRT1 Orthologs in Fasting-Dependent Inhibition of the Lipid/Cholesterol Regulator SREBP. Genes Dev (2010) 24:1403–17. doi: 10.1101/gad.1901210
52. Wang Z, Ye X, Shi M, Li F, Wang Z, Zhou Y, et al. Parasitic Insect-Derived miRNAs Modulate Host Development. Nat Commun (2018) 9:2205. doi: 10.1038/s41467-018-04504-1
53. Libersat F, Delago A, Gal R. Manipulation of Host Behavior by Parasitic Insects and Insect Parasites. Annu Rev Entomol (2009) 54:189–207. doi: 10.1146/annurev.ento.54.110807.090556
54. Adamo S,A. Modulating the Modulators: Parasites, Neuromodulators and Host Behavioral Change. Brain Behav Evol (2002) 60:370–7. doi: 10.1159/000067790
55. Zhou J, Zhou Y, Cao J, Zhang H, Yu Y. Distinctive microRNA Profiles in the Salivary Glands of Haemaphysalis Longicornis Related to Tick Blood-Feeding. Exp Appl Acarol (2013) 59:339–49. doi: 10.1007/s10493-012-9604-3
56. Malik MI, Nawaz M, Hassan IA, Zhang H, Gong H, Cao J, et al. A microRNA Profile of Saliva and Role of miR-375 in Haemaphysalis Longicornis (Ixodida: Ixodidae). Parasit Vectors (2019) 12:68. doi: 10.1186/s13071-019-3318-x
57. Melo RCN, D’Avila HD, Fabrino DL, Almeida PE, Bozza PT. Macrophage Lipid Body Induction by Chagas Disease In Vivo: Putative Intracellular Domains for Eicosanoid Formation During Infection. Tissue Cell (2003) 35:59–67. doi: 10.1016/S0040-8166(02)00105-2
58. Melo RCN, Fabrino DL, Dias FF, Parreira GG. Lipid Bodies: Structural Markers of Inflammatory Macrophages in Innate Immunity. Inflamm Res (2006) 55:342–8. doi: 10.1007/s00011-006-5205-0
59. Cocchiaro JL, Kumar Y, Fischer ER, Hackstadt T, Valdivia RH. Cytoplasmic Lipid Droplets are Translocated Into the Lumen of the Chlamydia Trachomatis Parasitophorous Vacuole. Proc Natl Acad Sci USA (2008) 105:9379–84. doi: 10.1073/pnas.0712241105
60. Perera R, Riley C, Isaac G, Hopf-Jannasch AS, Moore RJ, Weitz KW, et al. Dengue Virus Infection Perturbs Lipid Homeostasis in Infected Mosquito Cells. PloS Pathog (2012) 8:e1002584. doi: 10.1371/journal.ppat.1002584
61. Saka HA, Valdivia R. Emerging Roles for Lipid Droplets in Immunity and Host-Pathogen Interactions. Annu Rev Cell Dev Biol (2012) 28:411–37. doi: 10.1146/annurev-cellbio-092910-153958
62. Rodrigues, Melo CFO, Oliveira DN, Lima EO, Guerreiro TM, Esteves CZ, et al. A Lipidomics Approach in the Characterization of Zika-Infected Mosquito Cells: Potential Targets for Breaking the Transmission Cycle. PloS One (2016) 11:e0164377. doi: 10.1371/journal.pone.0164377
63. Rodrigues Melo CFO, Delafiori J, Dabaja MZ, Oliveira DN, Guerreiro TM, Colombo TE, et al. The Role of Lipids in the Inception, Maintenance and Complications of Dengue Virus Infection. Sci Rep (2018) 8:11826. doi: 10.1038/s41598-018-30385-x
64. Bennett M, Gilroy DW. Lipid Mediators in Inflammation. Microbiol Spectr (2016) 4:6. doi: 10.1128/microbiolspec.MCHD-0035-2016
65. Palm W, Sampaio JL, Brankatschk M, Carvalho M, Mahmoud A, Shevchenko A, et al. Lipoproteins in Drosophila Melanogaster – Assembly, Function, and Influence on Tissue Lipid Composition. PloS Genet (2012) 8:e1002828. doi: 10.1371/journal.pgen.1002828
66. Gao Q, Goodman JM. The Lipid Droplet-a Well-Connected Organelle. Front Cell Dev Biol (2015) 3:49. doi: 10.3389/fcell.2015.00049
67. Haemmerle G, Zimmermann R, Zechner R. Letting Lipids Go: Hormone-Sensitive Lipase. Curr Opin Lipidol (2003) 14:289–97. doi: 10.1097/00041433-200306000-00009
68. Heaton NS, Randall G. Dengue Virus-Induced Autophagy Regulates Lipid Metabolism. Cell Host Microbe (2010) 8:422–32. doi: 10.1016/j.chom.2010.10.006
69. Folly E, Cunha e Silva NL, Lopes AH, Silva-Neto MA, Atella GC. Trypanosoma Rangeli Uptakes the Main Lipoprotein From the Hemolymph of Its Invertebrate Host. Biochem Biophys Res Commun (2003) 310:555–61. doi: 10.1016/j.bbrc.2003.09.038
70. Atella GC, Bittencourt-Cunha PR, Nunes RD, Shahabuddin M, Silva-Neto MA. The Major Insect Lipoprotein Is a Lipid Source to Mosquito Stages of Malaria Parasite. Acta Trop (2009) 109:159–62. doi: 10.1016/j.actatropica.2008.10.004
71. Mesquita RD, Vionette-Amaral RJ, Lowenberger C, Rivera-Pomar R, Monteiro FA, Minx P, et al. Genome of Rhodnius Prolixus, an Insect Vector of Chagas Disease, Reveals Unique Adaptations to Hematophagy and Parasite Infection. Proc Natl Acad Sci USA (2015) 112:14936–41. doi: 10.1073/pnas.1506226112
72. Ribeiro JMC, Genta FA, Sorgine MH, Logullo R, Mesquita RD, Paiva-Silva GO, et al. An Insight Into the Transcriptome of the Digestive Tract of the Bloodsucking Bug, Rhodnius Prolixus. PloS Negl Trop Dis (2014) 8:e2594. doi: 10.1371/journal.pntd.0002594
Keywords: lipid metabolism, Chagas disease, lipophorin receptor, Trypanosoma cruzi, Rhodnius prolixus, hematophagy
Citation: Sousa G, de Carvalho SS and Atella GC (2021) Trypanosoma cruzi Affects Rhodnius prolixus Lipid Metabolism During Acute Infection. Front. Trop. Dis 2:737909. doi: 10.3389/fitd.2021.737909
Received: 07 July 2021; Accepted: 06 October 2021;
Published: 02 November 2021.
Edited by:
Ana Beatriz Barletta Ferreira, National Institutes of Health Clinical Center (NIH), United StatesReviewed by:
Cecilia Stahl Vieira, Oswaldo Cruz Foundation (Fiocruz), BrazilCopyright © 2021 Sousa, de Carvalho and Atella. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Georgia Correa Atella, YXRlbGxhQGJpb3FtZWQudWZyai5icg==
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.