
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Trop. Dis. , 10 August 2021
Sec. Vector Biology
Volume 2 - 2021 | https://doi.org/10.3389/fitd.2021.708817
This article is part of the Research Topic Highlights in Vector Biology View all 8 articles
Understanding the mosquito antiviral response could reveal target pathways or genes of interest that could form the basis of new disease control applications. However, there is a paucity of data in the current literature in understanding antiviral response during the replication period. To illuminate the gene expression patterns in the replication stage, we collected gene expression data at 2.5 days after Dengue-2 virus (DENV-2) infection. We sequenced the whole transcriptome of the midgut tissue and compared gene expression levels between the control and virus-infected group. We identified 31 differentially expressed genes. Based on their function, we identified that those genes fell into two major functional categories - (1) nucleic acid/protein process and (2) immunity/oxidative stress response. Our study has identified candidate genes that can be followed up for gene overexpression/inhibition experiments to examine if the perturbed gene interaction may impact the mosquito’s immune response against DENV. This is an important step to understanding how mosquitoes eliminate the virus and provides an important foundation for further research in developing novel dengue control strategies.
Dengue virus (DENV) is a mosquito-borne virus that has greatly impacted human health and is a global issue in tropical and sub-tropical areas (1). The dengue disease burden worldwide is estimated to range from 100 to 400 million infections each year (2), and its occurrence closely mirrors that of its primary vector, Aedes aegypti. According to the World Health Organization, the largest number of dengue cases reported in 2019 were 3.1 million cases in the American region and more than 1 million cases around southeast Asia. Dengue virus is described as having four serotypes, DENV-1, DENV-2, DENV-3, and DENV-4, all of which cause dengue fever. In some individuals, dengue fever can progress to severe dengue with life threatening complications (3, 4).
The time required for a virus to replicate and become transmissible in the mosquito is called the extrinsic incubation period (EIP) (5). To become transmissible, the virus must pass through several major barriers in the mosquito: the midgut infection barrier (MIB), the midgut escape barrier (MEB), the salivary gland infection barrier (SGIB), and the salivary gland escape barrier (SGEB) (6). After a female Ae. aegypti ingests virus particles contained in a blood meal, the virus binds to receptors on the midgut-cell surface and enters the cell through endocytosis (7). The initiation of infection and replication in the midgut during the first two days is called the eclipse period, in which the virus titer decreases on day 1 and then continues to decline until day 2 post-ingestion (8). At 2 days post-infection, around 30% of midgut epithelial cells are infected (9). After 3 days post-infection, the virus can spread laterally to infect neighboring midgut epithelial cells (9). Infected midgut cells often become degenerated and cell-cell adhesion is often disrupted which may be the path for viral escape from the midgut epithelium (6). The duration of the EIP varies depending on the genotypes of both the mosquito and the virus, as well as on environmental conditions (10).
These processes rely on complex chains of virus-host interactions, many of which are still poorly characterized. The virus must evade the host immune system and co-opt host machinery to complete its replication cycle. There are a wide range of different host immune processes that can prevent the virus from using resources or disturbing critical processes essential for cell function. This includes passive processes and processes triggered by recognition of pathogen-associated molecules, which have been shown to limit pathogen infection and transmission (11). The RNA interference (RNAi) pathway, which limits viral replication by targeting viral RNAs for degradation has been found to play a major role in the antiviral response (12–14). Three other innate immunity pathways involving pattern recognition and signal transduction have an identified antiviral role: the Toll (15, 16), IMD (17, 18), and JAK/STAT (19, 20) pathways. Activation of these pathways leads to the induction of antimicrobial peptides and other immune effectors. A further pathway, apoptosis, the process of programmed cell death, is also crucial to limiting viral titer (21). Infection with DENV, Zika, and chikungunya viruses often induces the expression of other immune genes, including cysteine-rich venom proteins, clip-domain serine proteases, C-type lectins, and serine proteases, with the effect varying between mosquito populations (22–25).
In addition to those canonical immune pathways, substantial numbers of other mosquito genes unrelated to immunity often display altered transcription levels in response to virus infection, and some might impact virus replication in mosquitoes (26). For example, transcript levels of genes related to the cytoskeleton (15, 27, 28), heat-shock response (27, 29), detoxification (e.g., cytochrome p450s) (27, 30), cell proliferation (30, 31), DNA replication, and fatty acid biosynthesis (30, 32, 33) exhibited altered expression patterns during arboviral infection and some non-immune-related genes have a demonstrated impact on virus infection (31, 34, 35).
The early phase of the viral replication cycle is critical, as it shapes the course of infection and ultimately determines whether the mosquito becomes competent to transmit the virus (36). Most transcriptomic assays in mosquitoes that examined arbovirus infection were conducted within 48 hours of infection (27, 30, 32, 34) or 7-10 days post-DENV infection (16, 17, 19, 37). However, the time between 2 and 3 days after virus infection may be a crucial time point corresponding to the end of an infection bottleneck before viral titers spike and the virus particles escape the midgut epithelium (8, 9, 36). Mosquito host-virus interactions at this stage are still not well defined, so examining transcript differences associated with infection at this key time point could improve our overall understanding of DENV infection in mosquitoes, and produce candidate genes that might be used to develop for new strategies aimed at preventing arbovirus transmission.
Here we investigate transcriptomic changes in mosquito midguts after 60 hours of DENV infection, which corresponds to the end of the eclipse phase when virus replication rates and DENV titers begin to increase (36). Through RNA-Seq analysis, we evaluate the impact of DENV infection in the Ae. aegypti Vero Beach strain to identify differentially expressed genes associated with the virus replication cycle. A total of 31 genes had transcripts that showed differential expression following DENV infection. Our results provide important insight into DENV replication and indicate differential regulation of genes in two major functional categories - (1) nucleic acid/protein process and (2) immunity/oxidative stress response, during this stage of infection.
Aedes aegypti Vero strain F20 was used in this study. The strain was originally collected in Vero Beach, Florida in 2015. Mosquitoes were reared at 28°C and 60-80% relative humidity in a climate-controlled room with a light: dark cycle of 14:10 hours. Upon hatching, larvae were separated into pans (35cm x 20cm x 6cm) with approximately 200 larvae per pan and fed a mixture of 0.5% brewer’s yeast and 0.5% liver powder (MP Biomedicals, Solon, OH, USA). Adults were fed with 20% sucrose solution-soaked cotton rolls (Carolina Absorbent Cotton, Charlotte, NC, USA). Mosquito eggs were collected to maintain the colony as described in (38), and female mosquitoes were blood-fed from chickens following standard approved protocols (IACUC protocol 201807682).
Four- to five- day old adult female mosquitoes were transferred to 16 oz cardboard cartons (WebstaurantStore, Lancaster, PA, USA) one day before infection and placed in an incubator at 28°C and 60-80% relative humidity. The sucrose solution-soaked cotton rolls were removed and only water-soaked cotton rolls provided. In the virus-infected group, DENV-2 strain New Guinea C (GenBank Accession # KM204118) was incubated at a multiplicity of infection of 0.01 viruses per cell in Vero cells (African green monkey kidney cell line) for 5 days. Supernatant from the virus culture was mixed with defibrinated bovine blood (Hemostat, Dixon, CA, USA) and fed to female mosquitoes with an artificial feeding apparatus (Hemotek, Lancashire, United Kingdom) with membranes made from sausage casings. In the control group, bovine blood was mixed with M199 culture media (Corning, Manassas, VA, USA) from uninfected Vero cells 5 days after splitting following the same procedure as the infected group. Three fully blood-engorged mosquitoes were collected at 1-hour post-feeding to quantify the ingested DENV titer. After the removal of non-blood fed mosquitoes across both feeding group, the remaining mosquitoes were placed into cartons and maintained in the incubator at 28°C and 60-80% relative humidity. 20% sucrose solution-soaked cotton rolls were provided and changed every day. At 60 hours post-infection, 180 mosquitoes from both treatment and control groups were immobilized on ice. Midguts were dissected in sterile saline solution and immediately transferred to an eppendorf tube on dry ice. All the tissue samples were stored at -80°C.
A total of 90 midguts from treatment and control groups, separately, were divided in triplicate sample each contained with 30 midguts. RNA from a total of 6 samples was extracted using the standard Trizol protocol (38, 39). The three biological replications of control and three infected group samples were sent to the Novogene Bioinformatics Technology Co. (Durham, NC, USA) for library generation and Illumina HiSeq6000 paired-end 150bp sequencing.
All RNA samples passed quality control for RNA purity, integrity and potential contamination before library construction, and mRNA was enriched using oligo dT beads. The enriched mRNA was fragmented and cDNA synthesized using random hexamers. After cDNA was synthesized, a custom second-strand synthesis Illumina buffer was added with dNTPs, RNase H, and Escherichia coli polymerase I to generate the second strand through nick-translation. The cDNA library was finalized after the application of terminal repair, poly-A tailing, sequencing adapter ligation, size selection, and PCR enrichment. The cDNA libraries were quantified and placed into Illumina sequencer to generate the sequencing reads (Novogene Bioinformatics Technology Co., Durham, NC, USA).
The data from Illumina sequencing were transformed and recorded in a FASTQ file. Quality control indicated the clean reads were more than 98% and the error rate was below 0.03%. The HISAT2 version 2.1.0-beta was used to map the filtered sequenced reads to Aedes aegypti LVP_AGWG AaegL5 chromosome and transcripts reference files available from VectorBase (https://vectorbase.org/) (40). Gene expression level was measured as the Fragments Per Kilobase of transcript sequence per Millions of base pairs sequenced (FPKM) values. HTSeq software version v0.6.1 was used to calculate FPKM.
To analyze the difference of FPKM between control and infected groups, T-tests were used to determine if average expression levels between the control and infected groups were equal. We then calculated the false discovery rate (FDR) with the Benjamini-Hochberg procedure (41) implemented in Python SciPy version 1.5.4 (https://scipy.org/), with a FDR under 0.2 considered as a significant difference. This method has been routinely used over the classic correction methods such as Bonferroni (42) or Sidak (43) in high-throughput science allowing discovery of novel genes or genic regions with relatively small sample sizes (44, 45).
To identify the function of gene, several approaches were applied. Gene IDs were annotated using VectorBase (https://vectorbase.org/) and Uniprot (https://www.uniprot.org/) to obtain gene names and full-length sequences. Gene ontology (GO) terms were also used to estimate gene function, as well as homology with putative orthologs from other mosquito species. Sequences were then examined using BLASTx (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to match identities and characterize conserved domains. Information on key differentially expressed genes was also collected and predicted through literature review.
To assess the impact of DENV infection on Ae. aegypti immune pathways, we compiled a list of 352 genes that fall into 23 gene families or 4 functional groups, all implicated in the innate immune response of dipteran insects to bacterial, viral, or parasitic infections. This list was derived from the ImmunoDB database (http://cegg.unige.ch/Insecta/immunodb), which categorizes loci into families based on homology among Ae. aegypti, Anopheles gambiae, and Drosophila melanogaster (46). Gene accession numbers and annotations were updated using the Ae. aegypti genome (AaegL5.3) on VectorBase (www.vectorbase.org). Accession numbers of previously identified genes were searched, and their functional roles were confirmed by at least one of three sources: protein domain information, orthology/paralogy with a gene(s) of similar function, or gene ontology (GO) terms. In addition, VectorBase was searched using the name of each gene family with results filtered by the organism (Ae. aegypti, genome release AaegL 5.3). The names of immunity genes and gene families in our list are in accordance with annotations from ImmunoDB. The final list was intended to err on the side of inclusivity and may include genes without a confirmed role in innate immunity but are closely related to loci (i.e., belong to the same gene family) with empirical support for a role in some aspect of the immune response.
RNA from the same samples used to generate the transcriptome was used to validate expression profiles of a subset of genes determined to have differential expression due to DENV infection via RNA-Seq. Gene expression was quantified using a Bio-Rad CFX96™ Real-Time PCR system and the iTaq Universal SYBR Green One-step kit (Bio-Rad, Hercules, CA, USA), with specific primer sets designed for each gene (Table S3). Four candidate genes were selected from both upregulated and downregulated expression groups and from the two major functional categories we identified (Tables 1, 2). The PCR products from specific primer sets were sequenced to confirm the targets were correct. Ae. aegypti ribosomal protein S7 gene (GenBank Accession # AY380336) was picked as a control for standardizing gene expression level (39). The qRT-PCR conditions were one cycle of 50°C for 10 min, one cycle of 95°C for 1 min, then 40 cycles of 95°C for 10 s, and 60°C for 30 s, followed by melt curve analysis. The 2[-Delta Delta C(T)] method was applied to analyze the relative changes in gene expression from real-time quantitative PCR results (47). Gene expression data were analyzed by the Wilcoxon Method from JMP Pro (www.jmp.com), to comparing group means, calculate the p-value and determine the presence of significant differences between treatment groups.
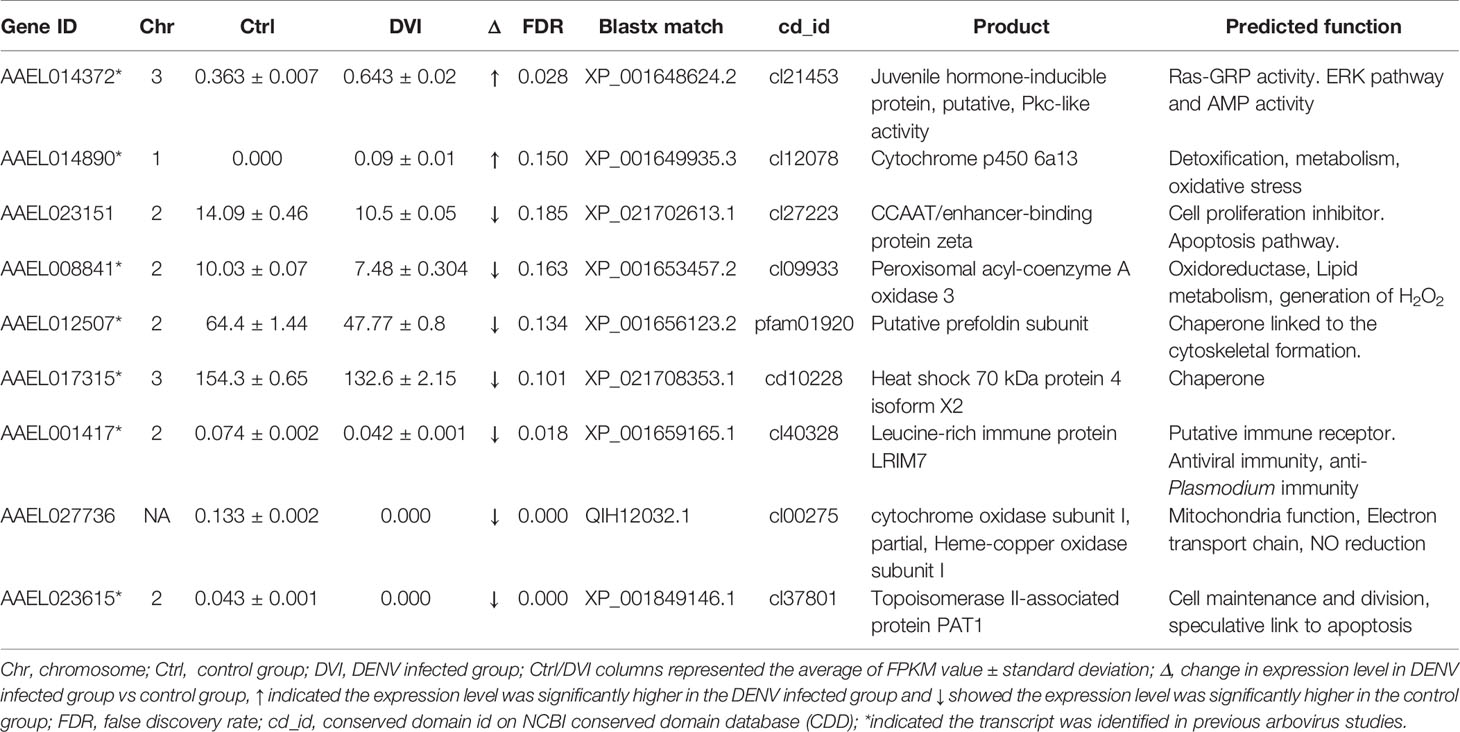
Table 1 The immunity/oxidative stress response-related genes with differences between the control and DENV-2 infected midgut.
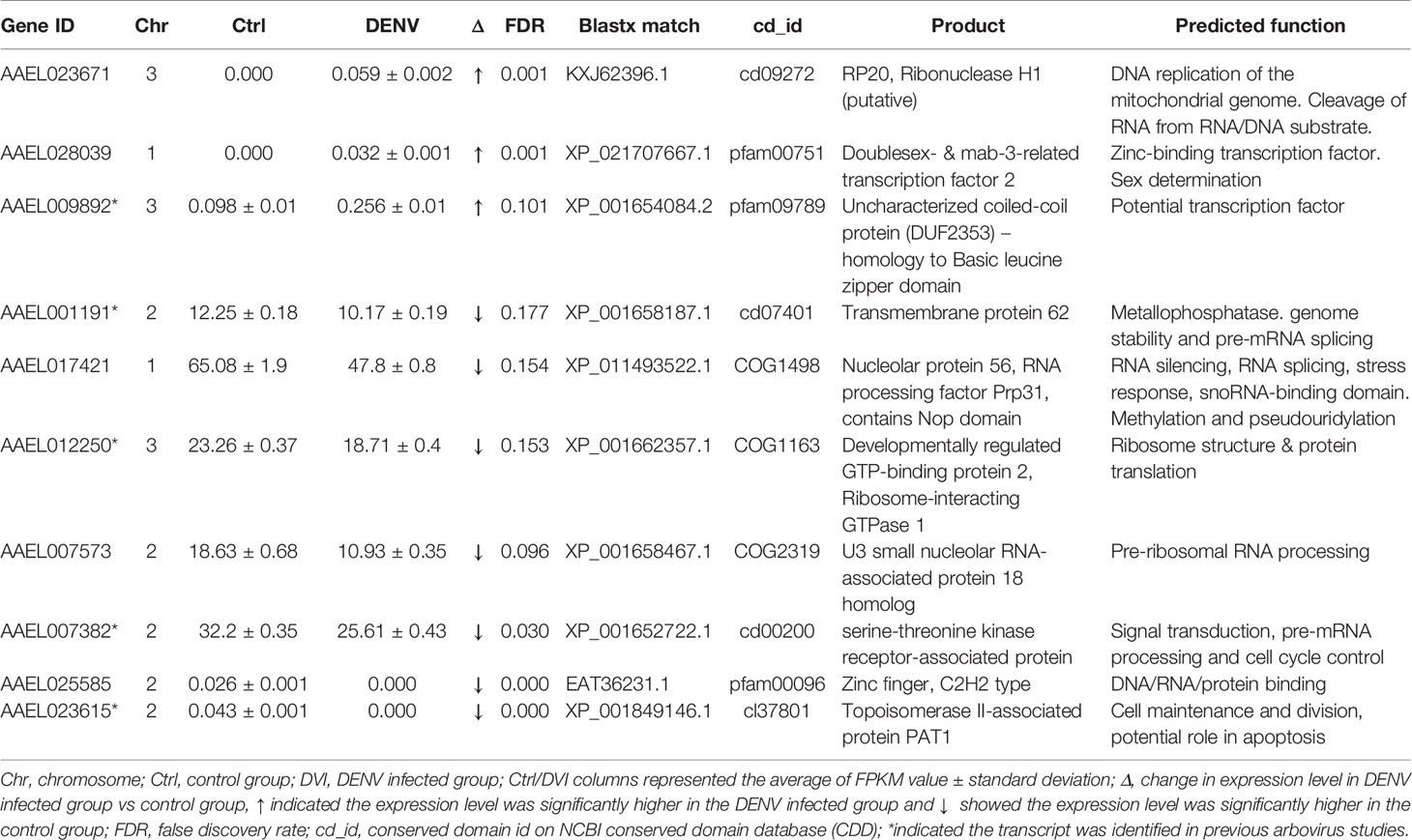
Table 2 The nucleic acid/protein process-related genes with differences between the control and DENV-2 infected midgut.
To determine virus genome equivalents in blood and freshly fed mosquitoes from the infection study, DENV-2 specific primers (38) were used with the iTaq Universal SYBR Green One-step kit and the Bio-Rad CFX96™ Real-Time PCR (Bio-Rad, Hercules, CA, USA). The qRT-PCR conditions were as described above. Viral genome equivalents were calculated based on a standard curve and recorded as log (base 10) plaque-forming unit equivalents per milliliter (log PFUe/ml). The standard curves for DENV genome equivalents were described previously (38). The blood mixture contained 9.3 log PFUe/ml of DENV-2. The freshly fed mosquito contained 5.78 ± 0.07 log PFUe/ml of DENV-2. The virus titer at 60 hours after infection in the midgut was 6.25 ± 0.74 log PFUe/ml of DENV-2.
An average of 48.4 million reads was generated per sample, of which 90% were clean reads. The average sequencing error rate was 0.03 [Qphred=-10log10(e)] and the average GC content was 46.86%. The mapping percentage (total number of reads that could be mapped to the reference genome) was 87.7%, and more than 90% of mapped reads corresponded to known exon regions. An average of 9.25% reads were mapped to multiple locations and 78.42% reads were uniquely mapped. The full dataset of FKPM values and the FDR rate for each of 15,173 genes are provided as supplemental Table S1.
We identified 31 genes that were significantly differentially expressed (DEGs) between the control and infected groups based on a false discovery rate of 0.2 (Tables 1–3). These included 8 genes located on Chromosome 1, 15 genes on Chromosome 2, and 7 genes on Chromosome 3 (Figure 1). Eleven genes were upregulated, and 20 genes were downregulated after 60 hours of virus infection in the midgut.
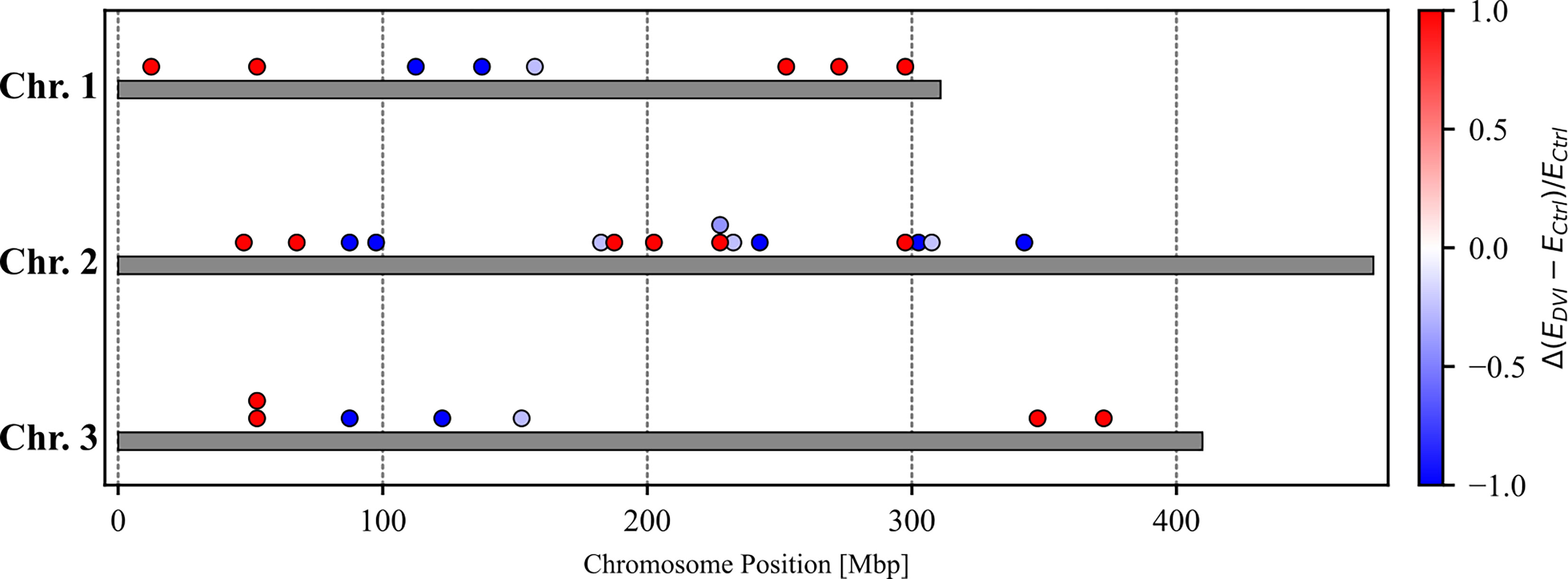
Figure 1 The chromosomal location of differentially expressed genes with relative expression differences [=(Edvi-E ctrl)/Ectrl where Ectrl denotes FPKM value in the control group and Edvi denote FPKM value in DENV-infected group]. Red dots indicate genes where the DENV-infected group had higher gene expression level. Blue dots indicate genes where the control group had higher gene expression levels.
Based on data from Vectorbase, only 7 of the DEGs had an annotated function, with 24 described as having an ‘unspecified product’ based on AaegL5 annotation. Through BLASTx analysis we identified conserved domains in a further 13 DEGs, making a total of 20 annotated genes. Based on their predicted functions from Vectorbase and BLASTx analysis, 9 DEGs were grouped as having a role in immunity or oxidative stress response (Table 1). A further 10 DEGs had a putative role relating to nucleic acid or protein process (Table 2). One gene, AAEL023615-RA, was associated with both immunity and nucleic acid processes. The 13 DEGs not linked to these functions are recorded in Table 3.
The 352 genes known to be involved in the dipteran immune response covered 23 families including Toll, IMD, JAK/STAT, and RNAi pathways. The list was split into 4 functional categories, defined as (i) pathogen/parasite recognition, (ii) signal modulation, (iii) signal transduction, and (iv) effector, oxidation-reduction, and reactive oxygen species. However, when the expression level differences between control and DENV-2 infected mosquitoes at 2.5 days post-infection in the data from this study were examined, 315 genes had FPKM data but none of the canonical immune genes showed expression level differences with a FDR below 0.2 (Table S2).
Four genes were validated with qRT-PCR and supported the RNA-Seq results with the same pattern of expression (Figure 2). The relative transcript expression fold change of AAEL014890 was 2.15 ± 0.96 (p = 0.02), AAEL028039 was 3.1 ± 1.68 (p = 0.02), AAEL008841 was 0.87 ± 0.15 (p = 0.38) and AAEL001417 was 1.04 ± 0.41 (p = 0.77). Although two transcripts did not have statistically significant differences, the relative expression patterns were the same as the RNA-Seq FPKM results.
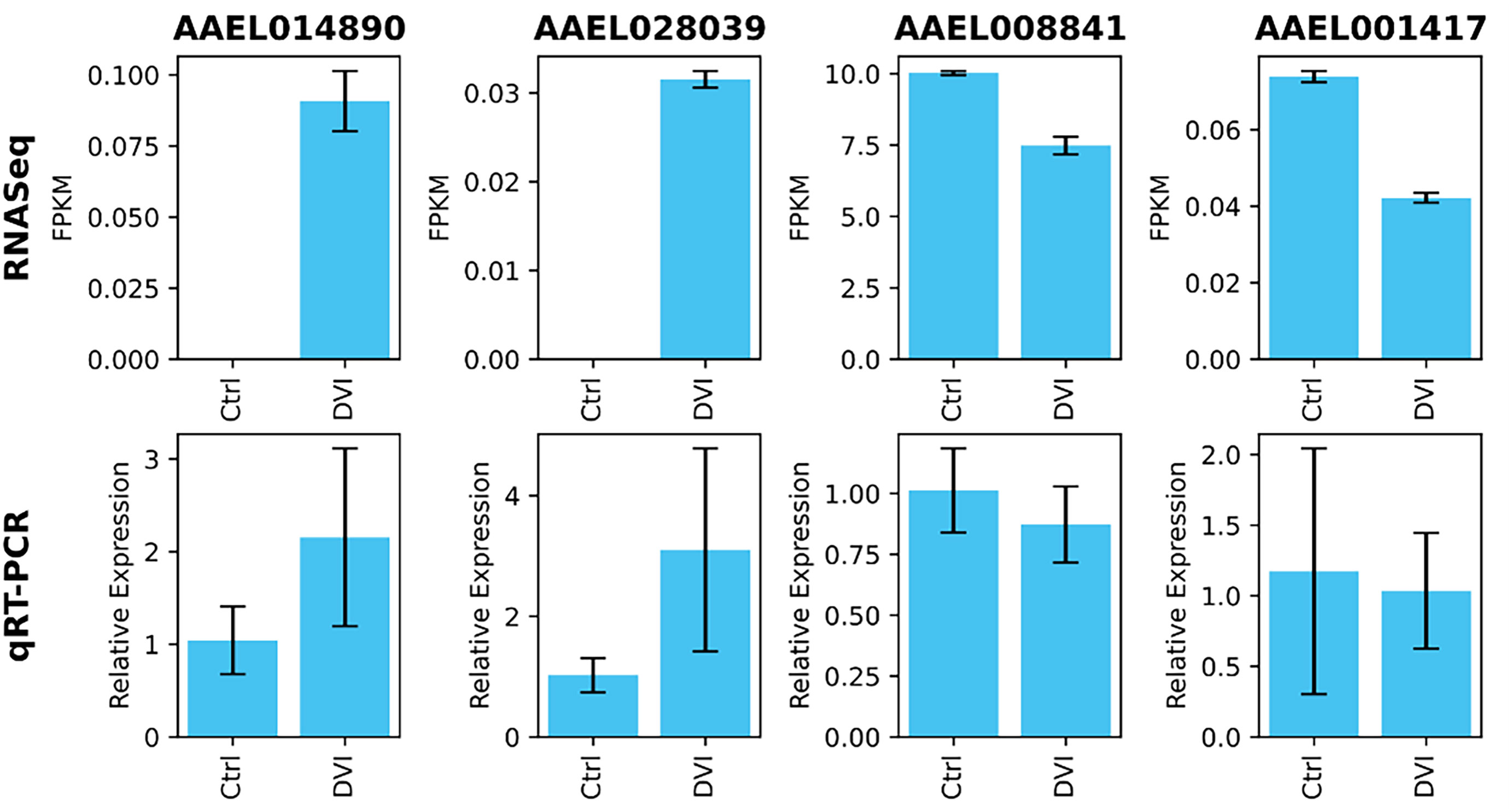
Figure 2 Validation of expression levels on selected genes using qRT-PCR. The differences in relative expression level between the control (Ctrl) and DENV infected (DVI) groups showed a patterns consistent with the RNA-Seq FPKM data.
We investigated the transcriptional response in midgut tissue of Ae. aegypti at 2.5 days post-infection with DENV-2. Although DENV is detectable in mosquito midgut epithelial cells at 2 days post-infection, the subsequent 24 hours are crucial for the virus to replicate and infect surrounding cells. This makes 2.5 days post-infection a critical timepoint for studying viral replication. We identified 31 transcripts with significantly altered expression levels following DENV infection, and based on their putative functions, grouped the majority as being linked either to immunity/oxidative stress response or to nucleic acid/protein processes. This finding provides new information on the nature of mosquito-virus interactions during the replication and assembly stages of the DENV replication cycle, and offers 7 new target genes with their potential function that could interfere with the virus infection process in a mosquito host.
Canonical immune pathways and genes are generally thought to play a crucial role in a mosquito’s ability to restrict viral infection, with many canonical immune transcripts displaying altered activity after infection (16, 30). Several immune pathways such as the Toll (16), JAK/STAT (19) and RNAi pathways (12) have a demonstrated ability to limit DENV infection in the mosquito. However, in our dataset there were no significant differences in the expression of 352 canonical immune genes (Table S2), which suggests there might be a distinct, uncharacterized aspect of the immune response and host-virus interactions at this time point and in the midgut tissue we investigated.
Based on previous midgut transcriptome studies, canonical immune genes were generally not identified as significantly associated with the DENV replication cycle, although there were a few exceptions. The Inhibitor of apoptosis protein-1 (AAEL009074) and SCF ubiquitin ligase Rbx1 component (AAEL004691) were increased in the susceptible population after 48 hours of infection (48). The cecropin gene (AAEL017211), an antimicrobial peptide, was downregulated after 24 hours of infection (27). Several immunity-related genes including serine protease inhibitors (AAEL003182 and AAEL002704), Clip domain serine protease (AAEL002124 and AAEL001098), C-type lectin (AAEL005641), and fibrinogen-related protein genes (AAEL006704) were differentially expressed after 4 days of infection in the midgut (27). However, the canonical immune gene activity described in these studies was noted to occur either in the early infection stage or late replication phase (36) but not during the early replication phase. These observations in conjunction with our results suggest that distinct patterns of gene expression might underlie host-virus interactions during different phases of the DENV replication cycle. Moreover, gene expression likely varies among different Ae. aegypti populations if they have different genetic backgrounds. A previous study showed genome-wide divergence of Ae. aegypti populations (49).
Based on conserved domains, we identified 9 other DEGs with a putative role in immunity or redox-associated processes, which can be immunomodulatory in Ae. aegypti (50–52). Two DEGs, AAEL014372-RA and AAEL014890-RA, both linked to oxidative stress and immunity, were upregulated after infection (Table 1). AAEL014372-RA is a juvenile hormone-inducible protein, that was thought to regulate the expression of many other genes in D. melanogaster (53). The conserved protein domain cl21453 from AAEL014372-RA belongs to the PKc_like superfamily which was mainly composed of the catalytic domains of serine/threonine-specific and tyrosine-specific protein kinases. The Ras/ERK signaling pathway is included in the PKc_like superfamily and was shown to play a role in resistance of Aedes mosquitoes to DENV infection (52). AAEL014890-RA is a known Cytochrome p450 gene associated with metabolic detoxification and insecticide resistance (54). Cytochrome p450 genes as monooxygenases were upregulated in the refractory Ae. aegypti in response to DENV infection (30). Although cytochrome p450s are associated with viral infection-induced inflammation and oxidative stress in mammals (51), the role in virus infection in mosquitoes is still unclear.
Most of the downregulated DEGs related to oxidative stress and immunity were associated with cell death and survival, which are key processes associated with virus infection (55, 56). Seven DEGs, AAEL023151-RA, AAEL008841-RA, AAEL012507-RA, AAEL017315-RA, AAEL001417-RA, AAEL023615-RA and AAEL027736-RA were downregulated in response to DENV infection (Table 1). Based on BLASTx results, AAEL023151-RA is a CCAAT/enhancer-binding protein zeta, which has been implicated in growth arrest and the control of apoptosis in mammals (57). However, its role in mosquitoes still needs further investigation. AAEL008841-RA is acyl-coenzyme A oxidase which produces H2O2 and is involved in lipid metabolism (58). AAEL012507-RA has the conserved protein domain pfam01920, indicating it is likely a prefoldin. Prefoldin is a co-chaperone protein that transfers unfolded polypeptides to the chaperonin containing tailless complex polypeptide (59). On the other hand, AAEL017315-RA has the conserved protein domain cd10228 that suggests it is a 70 -kDa heat shock protein 4 (Hsp70) which is expressed under stress and is an important component of the chaperone system (60). Together, both AAEL012507-RA and AAEL017315-RA might be involved in chaperone activity where the chaperone function of Hsp70 protects the cell from stress-induced apoptosis (61). AAEL001417-RA is a leucine-rich immune protein (LRIM) which is activated as a complement-like defense response (62). Most of the studies of LRIMs have focused on their ability to act against malaria parasites in Anopheles mosquitoes (63). However, increased levels of LRIM16 transcripts were observed in Mayaro Virus-infected Aedes mosquitoes (64), although their exact role in arboviral infection remains to be determined. The presence of a conserved protein domain cl37801 in AAEL023615-RA could indicate a function similar to that of a topoisomerase II-associated protein (PAT1), which has conserved roles in the 5→3 mRNA decay pathway in both Drosophila and humans (65). Furthermore, PAT1 might affect cell survival and regulation of viral gene expression (66). AAEL027736-RA is predicted as cytochrome oxidase subunit I which is one of the proteins encoded from mitochondria DNA (67). The cl00275 domain AAEL027736-RA contained in the Heme-copper oxidase superfamily (68) which is involved in the reduction of oxygen.
Based on the potential functions of oxidative stress and immunity, we propose that DENV induces stress in midgut cells after entry. Altered mitochondrial function leads to changes in oxidative stress and oxidative homeostasis. Mass production of viral protein induces the unfolded protein response (69). Although the genes did not have a direct link to mosquito immunity, the higher levels of reactive oxygen species and unfolded protein could stimulate apoptosis (70, 71). Further experiments must be done to establish the connections between the genes we identified in the midgut and apoptosis and, moreover, their potential to moderate virus infection.
We identified 9 DEGs associated with nucleic acid/protein processes (Table 2). Three DEGs, AAEL023671-RA, AAEL028039-RA, and AAEL009892-RA, were upregulated after 2.5 days post-infection and were associated with DNA binding and DNA replication. Cd09272 is the conserved domain of AAEL023671-RA and belongs to the family of Ribonuclease H (RNase H). RNase H cleaves the RNA of RNA/DNA hybrids and is critical during DNA replication (72). AAEL028039-RA maintains the conserved protein domain pfam00751 which is a DNA binding domain (73) and has been shown to dimerize and bind palindromic DNA (74). The AAEL009892-RA conserved protein domain pfam09789 identifies as an uncharacterized coiled-coil protein with homology to the basic leucine zipper domain, a large group of transcription factors (75).
There were a further 7 DEGs related to the nucleic acid/protein process that were downregulated (Table 2). Most of these genes were associated with mRNA and rRNA processing. The mRNA splicing machinery was shown to be hijacked by the DENV NS5 protein to improve viral replication (76). On the other hand, DENV NS1 protein interacted with ribosomal proteins associated with DENV translation and replication (77). The possible involvement of DENV proteins might be a reason we identified the genes related to mRNA and rRNA processing. AAEL001191-RA has the conserved protein domain similar to that of transmembrane protein 62, which is a member of the metallophosphatase (MPP) superfamily. MPPs have diverse functions including Mre11/SbcD-like exonucleases which are critical to genome stability (78) and Dbr1-like RNA lariat debranching enzymes which are key factors of pre-mRNA splicing (79). AAEL017421-RA is orthologous to nucleolar protein 56. Although a specific function has not been identified (80), other nucleolar proteins are important for the maintenance of heterochromatin and ribosomal RNA (81). Moreover, AAEL017421-RA has a conserved domain, COG1498, that indicates similarity to RNA processing factor Prp31. The Nop domain in RNA processing factor Prp31 is required for ribosome biogenesis (82) and pre-mRNA splicing (83). The conserved protein domain COG1163 in AAEL012250-RA suggests ribosome interacting GTPase 1 which is important to ribosomal structure and biogenesis (84). AAEL007573 is the U3 small nucleolar RNA-associated protein 18 homolog which functions in the maturation of the small subunit rRNA (85). AAEL025585-RA contains a conserved protein domain pfam00096 and is the structure of the zinc finger that recognizes and binds DNA (86). AAEL007382-RA has the conserved protein domain cd00200 indicates WD40 which involved in many biological processes such as signal transduction, pre-mRNA processing, and cell cycle control (87, 88).
Since viruses utilize host transcription and translation machinery to replicate (76, 77), the genes we identified in Table 2 could be linked to viral RNA translation and replication. Furthermore, the virus might subvert the host cell cycle or nucleic acid processes to keep the cell in a state where viral replication is promoted (89). However, more studies are needed to understand the DENV replication process and to investigate interactions between the DEGs and virus.
We reported 13 DEGs were not listed in either immunity/oxidative stress response or to nucleic acid/protein processes (Table 3). Twelve DEGs were identified as non-coding RNAs (ncRNA), specifically long non-coding RNAs (lncRNA) (90). Expression of cellular lncRNAs may be altered in response to viral replication and induced antiviral response (91). A previous study has shown that DENV-2 infection in Ae. aegypti increased the abundance of a number of long ncRNAs, which might be involved in the response to viral infection (92).
To determine if any of the 31 DEGs had a previously discovered change in expression due to arboviral infection, we compared our data with previous reports describing transcriptomic response to arboviral infection in Ae. aegypti (22, 93). Four DEGs were previously described as having transcript level differences under DENV or Zika virus infection (AAEL001417-RA, AAEL023122-RA, AAEL028030-RA and AAEL023254-RA). Moreover, if we expand the database to a virus-injected transcriptomic response study (34), a total of 15 of our DEGs were reported to have expression levels altered in response to infection with at least one of DENV, Zika virus, Yellow fever virus, or West Nile virus. However, 16 DEGs from our studies were not reported in previous studies. For the previously reported 15 DEGs, 9 were affected by DENV at 1 day post infection (dpi), 2dpi or 7dpi (AAEL014372-RA, AAEL008841-RA, AAEL001417-RA, AAEL023615-RA, AAEL009892-RA, AAEL023122-RA, AAEL023254-RA, AAEL025431-RA and AAEL007382-RA) and 6 DEGs (AAEL014890-RA, AAEL012507-RA, AAEL001191-RA, AAEL012250-RA, AAEL017315-RA and AAEL028030-RA) were only expressed differentially following infection with other viruses. Elucidation of potential genes involved in the replication cycle revealed altered expression of 5 DEGs at 1dpi or 2dpi after DENV infection (AAEL023615-RA, AAEL023122-RA, AAEL023254-RA, AAEL025431-RA, and AAEL007382-RA).
The AAEL023615-RA (PAT1) and AAEL007382-RA was described above and is crucial for cell survival (66, 88). AAEL025431-RA has the conserved domain Josephin, which might regulate cell motility, and endocytosis (94). The Josephin domain-containing protein is also involved in de-ubiquitination (95). AAEL023122-RA and AAEL023254-RA are potential lncRNAs that could participate in antiviral responses (92). The fact that the five DEGs exhibited differential expression across multiple studies of DENV or ZIKV infection in mosquitoes could indicate that they play important roles during arboviral infection, making them important candidates for future study.
A quantitative trait locus (QTL) is a chromosomal region responsible for variation of a quantitative trait and several QTLs that control Ae. aegypti susceptibility to virus infection and dissemination have been identified (96–98). Across the 31 DEGs we identified, there were 8 genes located on Chromosome 1, 15 genes on Chromosome 2, 7 genes on Chromosome 3, and 1 that was not assigned (Figure 1). To date, six DENV-2 midgut infection barrier (MIB) QTLs and two DENV-2 midguts escape barrier (MEB) QTLs have been identified across all three chromosomes (36, 99). However, none of the 31 genes we discovered overlapped with the predicted QTL locations. On the other hand, Aubry et al. detected five highly significant QTLs and four QTLs with lower statistical support Chromosome 2 that were associated with Zika virus infection status in Ae. aegypti (100). Three of our DEGs, AAEL022994-RA, AAEL012507-RA, and AAEL008841-RA, fell in the highly significant QTLs regions. Additionally, three further DEGs, AAEL023696-RA, AAEL027382-RA, and AAEL007573-RA, were located within the less significant QTLs regions. These findings suggest that Zika virus-associated QTLs might also be involved in the response to DENV infection.
A greater number of differentially expressed genes may have been identified as significant if sample sizes had been larger. For example, if the same expression pattern held but we had 6 control and treatment groups, the number of genes below P-value of 0.05 after Sidak correction would have been 121. The number of genes greater than 95% power using false discovery rate 0.2 would have been 1,480. Nevertheless, the 31 DEGs we have identified here are those that demonstrated the most extensive response to DENV infection.
Our data revealed key time-specific differences in gene expression when compared to previous transcriptomic studies of DENV-infected midguts (27, 48). Given differences in gene expression between carcass and midgut samples after DENV infection (27), our findings highlight the value of time- and tissue-specific transcriptional studies to investigate viral infection in mosquitoes. Further research in this area will likely be crucial to improving understanding of the virus infection process. For example, DENV-2 dissemination to the abdominal fat body was detected in 35% of mosquitoes between 2 and 3 days post-infection (9). The fat body-mediated antiviral response of mosquitoes (101) and transcriptomes also changed after blood feeding (102). On the other hand, expanding the sampling time points for transcriptional response during the virus replication process could be applied with dual RNA-seq and provide a different view of pathogen- host interactions, moreover, the impact from the endogenous viral elements (103). All in all, our results provide a missing piece of mosquito’s early response against DENV and could offer potential targets for developing novel arbovirus control strategies.
The original contributions presented in the study are publicly available. This data can be found here in NCBI under accession number PRJNA729510.
The animal study was reviewed and approved by Institutional Animal Care and Use Committee University of Florida.
Conceived and designed the experiments, CTS and T-YC. Performed the experiments, CTS and T-YC. Analyzed the data, CTS, T-YC, YL and EPC. Wrote the paper, CTS, T-YC, YL, XW, DM and EPC. All authors contributed to the article and approved the submitted version.
This work is supported by the USDA National Institute of Food and Agriculture, Hatch project 1025565, UF/IFAS Florida Medical Entomology Laboratory fellowship to T-YC, and NSF CAMTech IUCRC Phase II grant (AWD05009_MOD0030).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Dr. Travis C. Collier (University of Florida – Florida Medical Entomology Laboratory) for providing python script to generate gene location figure. We thank Dr. Bryony Bonning (University of Florida – Department of Entomology & Nematology) and Dr. Ana Romero-Weaver (University of Florida – Florida Medical Entomology Laboratory) for reviewing the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fitd.2021.708817/full#supplementary-material
Supplementary Table 1 | Gene expression data.
Supplementary Table 2 | Immune-related gene expression data.
Supplementary Table 3 | Primer sequences for gene validation.
2. Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, et al. The Global Distribution and Burden of Dengue. Nature (2013) 496:504–7. doi: 10.1038/nature12060
3. Rigau-Pérez JG, Clark GG, Gubler DJ, Reiter P, Sanders EJ, Vance Vorndam A. Dengue and Dengue Haemorrhagic Fever. Lancet (1998) 352:971–7. doi: 10.1016/S0140-6736(97)12483-7
4. Simmons CP, Farrar JJ, van Vinh Chau N, Wills B. Dengue. N Engl J Med (2012) 366:1423–32. doi: 10.1056/NEJMra1110265
5. Black WC, Bennett KE, Gorrochótegui-Escalante N, Barillas-Mury CV, Fernández-Salas I, de Lourdes Muñoz M, et al. Flavivirus Susceptibility in Aedes Aegypti. Arch Med Res (2002) 33:379–88. doi: 10.1016/S0188-4409(02)00373-9
6. Franz AWE, Kantor AM, Passarelli AL, Clem RJ. Tissue Barriers to Arbovirus Infection in Mosquitoes. Viruses (2015) 7:3741–67. doi: 10.3390/v7072795
7. Hidari KI, Suzuki T. Dengue Virus Receptor. Trop Med Health (2011) 39:37–43. doi: 10.2149/tmh.2011-S03
8. Richardson J, Molina-Cruz A, Salazar MI, Black W4. Quantitative Analysis of Dengue-2 Virus RNA During the Extrinsic Incubation Period in Individual Aedes Aegypti. Am J Trop Med Hyg (2006) 74:132–41. doi: 10.4269/ajtmh.2006.74.132
9. Salazar MI, Richardson JH, Sánchez-Vargas I, Olson KE, Beaty BJ. Dengue Virus Type 2: Replication and Tropisms in Orally Infected Aedes Aegypti Mosquitoes. BMC Microbiol (2007) 7:9. doi: 10.1186/1471-2180-7-9
10. Souza-Neto JA, Powell JR, Bonizzoni M. Aedes Aegypti Vector Competence Studies: Areview. Infect Genet Evol (2019) 67:191–209. doi: 10.1016/j.meegid.2018.11.009
11. Tikhe CV, Dimopoulos G. Mosquito Antiviral Immune Pathways. Dev Comp Immunol (2021) 116:103964. doi: 10.1016/j.dci.2020.103964
12. Sánchez-Vargas I, Scott JC, Poole-Smith BK, Franz AWE, Barbosa-Solomieu V, Wilusz J, et al. Dengue Virus Type 2 Infections of Aedes Aegypti are Modulated by the Mosquito’s RNA Interference Pathway. PloS Pathog (2009) 5:e1000299–e1000299. doi: 10.1371/journal.ppat.1000299
13. McFarlane M, Arias-Goeta C, Martin E, O’Hara Z, Lulla A, Mousson L, et al. Characterization of Aedes Aegypti Innate-Immune Pathways That Limit Chikungunya Virus Replication. PloS Negl Trop Dis (2014) 8:e2994. doi: 10.1371/journal.pntd.0002994
14. Feng X, Zhou S, Wang J, Hu W. MicroRNA Profiles and Functions in Mosquitoes. PloS Negl Trop Dis (2018) 12:e0006463–e0006463. doi: 10.1371/journal.pntd.0006463
15. Sanders HR, Foy BD, Evans AM, Ross LS, Beaty BJ, Olson KE, et al. Sindbis Virus Induces Transport Processes and Alters Expression of Innate Immunity Pathway Genes in the Midgut of the Disease Vector, Aedes Aegypti. Insect Biochem Mol Biol (2005) 35:1293–307. doi: 10.1016/j.ibmb.2005.07.006
16. Xi Z, Ramirez JL, Dimopoulos G. The Aedes Aegypti Toll Pathway Controls Dengue Virus Infection. PloS Pathog (2008) 4:e1000098–e1000098. doi: 10.1371/journal.ppat.1000098
17. Sim S, Jupatanakul N, Ramirez JL, Kang S, Romero-Vivas CM, Mohammed H, et al. Transcriptomic Profiling of Diverse Aedes Aegypti Strains Reveals Increased Basal-Level Immune Activation in Dengue Virus-Refractory Populations and Identifies Novel Virus-Vector Molecular Interactions. PloS Negl Trop Dis (2013) 7:e2295–5. doi: 10.1371/journal.pntd.0002295
18. Barletta ABF, Nascimento-Silva MCL, Talyuli OAC, Oliveira JHM, Pereira LOR, Oliveira PL, et al. Microbiota Activates IMD Pathway and Limits Sindbis Infection in Aedes Aegypti. Parasit Vectors (2017) 10:103–3. doi: 10.1186/s13071-017-2040-9
19. Souza-Neto JA, Sim S, Dimopoulos G. An Evolutionary Conserved Function of the JAK-STAT Pathway in Anti-Dengue Defense. Proc Natl Acad Sci USA (2009) 106:17841–6. doi: 10.1073/pnas.0905006106
20. Jupatanakul N, Sim S, Angleró-Rodríguez YI, Souza-Neto J, Das S, Poti KE, et al. Engineered Aedes Aegypti JAK/STAT Pathway-Mediated Immunity to Dengue Virus. PloS Negl Trop Dis (2017) 11:e0005187–e0005187. doi: 10.1371/journal.pntd.0005187
21. Ocampo CB, Caicedo PA, Jaramillo G, Bedoya RU, Baron O, Serrato IM, et al. Differential Expression of Apoptosis Related Genes in Selected Strains of Aedes Aegypti With Different Susceptibilities to Dengue Virus. PloS One (2013) 8:10. doi: 10.1371/journal.pone.0061187
22. Etebari K, Hegde S, Saldaña MA, Widen SG, Wood TG, Asgari S, et al. Global Transcriptome Analysis of Aedes Aegypti Mosquitoes in Response to Zika Virus Infection. mSphere (2017) 2:e00456–17. doi: 10.1128/mSphere.00456-17
23. Zhao L, Alto BW, Shin D, Yu F. The Effect of Permethrin Resistance on Aedes Aegypti Transcriptome Following Ingestion of Zika Virus Infected Blood. Viruses (2018) 10:470. doi: 10.3390/v10090470
24. Caicedo PA, Serrato IM, Sim S, Dimopoulos G, Coatsworth H, Lowenberger C, et al. Immune Response-Related Genes Associated to Blocking Midgut Dengue Virus Infection in Aedes Aegypti Strains That Differ in Susceptibility. Insect Sci (2019) 26:635–48. doi: 10.1111/1744-7917.12573
25. Shin D, Kang S, Smartt CT. Profiling Transcripts of Vector Competence Between Two Different Aedes Aegypti Populations in Florida. Viruses (2020) 12:823. doi: 10.3390/v12080823
26. Sigle LT, McGraw EA. Expanding the Canon: Non-Classical Mosquito Genes at the Interface of Arboviral Infection. Insect Biochem Mol Biol (2019) 109:72–80. doi: 10.1016/j.ibmb.2019.04.004
27. Bonizzoni M, Dunn WA, Campbell CL, Olson KE, Marinotti O, James AA. Complex Modulation of the Aedes Aegypti Transcriptome in Response to Dengue Virus Infection. PloS One (2012) 7:e50512. doi: 10.1371/journal.pone.0050512
28. Shrinet J, Srivastava P, Sunil S. Transcriptome Analysis of Aedes Aegypti in Response to Mono-Infections and Co-Infections of Dengue Virus-2 and Chikungunya Virus. Interdiscip Approaches Study Flavivirus (2017) 492:617–23. doi: 10.1016/j.bbrc.2017.01.162
29. Angleró-Rodríguez YI, MacLeod HJ, Kang S, Carlson JS, Jupatanakul N, Dimopoulos G. Aedes Aegypti Molecular Responses to Zika Virus: Modulation of Infection by the Toll and Jak/Stat Immune Pathways and Virus Host Factors. Front Microbiol (2017) 8:2050. doi: 10.3389/fmicb.2017.02050
30. Behura SK, Gomez-Machorro C, Harker BW, deBruyn B, Lovin DD, Hemme RR, et al. Global Cross-Talk of Genes of the Mosquito Aedes Aegypti in Response to Dengue Virus Infection. PloS Negl Trop Dis (2011) 5:e1385–5. doi: 10.1371/journal.pntd.0001385
31. Serrato-Salas J, Hernández-Martínez S, Martínez-Barnetche J, Condé R, Alvarado-Delgado A, Zumaya-Estrada F, et al. De Novo DNA Synthesis in Aedes Aegypti Midgut Cells as a Complementary Strategy to Limit Dengue Viral Replication. Front Microbiol (2018) 9:801. doi: 10.3389/fmicb.2018.00801
32. Behura SK, Gomez-Machorro C, deBruyn B, Lovin DD, Harker BW, Romero-Severson J, et al. Influence of Mosquito Genotype on Transcriptional Response to Dengue Virus Infection. Funct Integr Genomics (2014) 14:581–9. doi: 10.1007/s10142-014-0376-1
33. Kang DS, Barron MS, Lovin DD, Cunningham JM, Eng MW, Chadee DD, et al. A Transcriptomic Survey of the Impact of Environmental Stress on Response to Dengue Virus in the Mosquito, Aedes Aegypti. PloS Negl Trop Dis (2018) 12:e0006568. doi: 10.1371/journal.pntd.0006568
34. Colpitts TM, Cox J, Vanlandingham DL, Feitosa FM, Cheng G, Kurscheid S, et al. Alterations in the Aedes Aegypti Transcriptome During Infection With West Nile, Dengue and Yellow Fever Viruses. PloS Pathog (2011) 7:e1002189–e1002189. doi: 10.1371/journal.ppat.1002189
35. Taguwa S, Maringer K, Li X, Bernal-Rubio D, Rauch JN, Gestwicki JE, et al. Defining Hsp70 Subnetworks in Dengue Virus Replication Reveals Key Vulnerability in Flavivirus Infection. Cell (2015) 163:1108–23. doi: 10.1016/j.cell.2015.10.046
36. Severson DW, Behura SK. Genome Investigations of Vector Competence in Aedes Aegypti to Inform Novel Arbovirus Disease Control Approaches. Insects (2016) 7:58. doi: 10.3390/insects7040058
37. Sim S, Ramirez JL, Dimopoulos G. Dengue Virus Infection of the Aedes Aegypti Salivary Gland and Chemosensory Apparatus Induces Genes That Modulate Infection and Blood-Feeding Behavior. PloS Pathog (2012) 8:e1002631–e1002631. doi: 10.1371/journal.ppat.1002631
38. Smartt CT, Shin D, Alto BW. Dengue Serotype-Specific Immune Response in Aedes Aegypti and Aedes Albopictus. Mem Inst Oswaldo Cruz (2017) 112:829–37. doi: 10.1590/0074-02760170182
39. Chen T-Y, Smartt CT, Shin D. Permethrin Resistance in Aedes Aegypti Affects Aspects of Vectorial Capacity. Insects (2021) 12:71. doi: 10.3390/insects12010071
40. Matthews BJ, Dudchenko O, Kingan SB, Koren S, Antoshechkin I, Crawford JE, et al. Improved Reference Genome of Aedes Aegypti Informs Arbovirus Vector Control. Nature (2018) 563:501–7. doi: 10.1038/s41586-018-0692-z
41. Benjamini Y, Yekutieli D. The Control of the False Discovery Rate in Multiple Testing Under Dependency. Ann Stat (2001) 29:1165–88. doi: 10.1214/aos/1013699998
42. Dunn OJ. Multiple Comparisons Among Means. J Am Stat Assoc (1961) 56:52–64. doi: 10.2307/2282330
43. Šidák Z. Rectangular Confidence Regions for the Means of Multivariate Normal Distributions. J Am Stat Assoc (1967) 62:626–33. doi: 10.1080/01621459.1967.10482935
44. Storey JD, Tibshirani R. Statistical Significance for Genomewide Studies. Proc Natl Acad Sci (2003) 100:9440. doi: 10.1073/pnas.1530509100
45. Lee Y, Collier TC, Sanford MR, Marsden CD, Fofana A, Cornel AJ, et al. Chromosome Inversions, Genomic Differentiation and Speciation in the African Malaria Mosquito Anopheles Gambiae. PloS One (2013) 8:e57887. doi: 10.1371/journal.pone.0057887
46. Waterhouse RM, Kriventseva EV, Meister S, Xi Z, Alvarez KS, Bartholomay LC, et al. Evolutionary Dynamics of Immune-Related Genes and Pathways in Disease-Vector Mosquitoes. Science (2007) 316:1738. doi: 10.1126/science.1139862
47. Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2–ΔΔCT Method. Methods (2001) 25:402–8. doi: 10.1006/meth.2001.1262
48. Baron OL, Ursic-Bedoya RJ, Lowenberger CA, Ocampo CB. Differential Gene Expression From Midguts of Refractory and Susceptible Lines of the Mosquito, Aedes Aegypti, Infected With Dengue-2 Virus. J Insect Sci (2010) 10:23. doi: 10.1673/031.010.4101
49. Lee Y, Schmidt H, Collier TC, Conner WR, Hanemaaijer MJ, Slatkin M, et al. Genome-Wide Divergence Among Invasive Populations of Aedes Aegypti in California. BMC Genomics (2019) 20:204. doi: 10.1186/s12864-019-5586-4
50. Bottino-Rojas V, Talyuli OAC, Carrara L, Martins AJ, James AA, Oliveira PL, et al. The Redox-Sensing Gene Nrf2 Affects Intestinal Homeostasis, Insecticide Resistance, and Zika Virus Susceptibility in the Mosquito Aedes aegypti. J Biol Chem (2018) 293:9053–63. doi: 10.1074/jbc.RA117.001589
51. Stavropoulou E, Pircalabioru GG, Bezirtzoglou E. The Role of Cytochromes P450 in Infection. Front Immunol (2018) 9:89. doi: 10.3389/fimmu.2018.00089
52. Liu W-Q, Chen S-Q, Bai H-Q, Wei Q-M, Zhang S-N, Chen C, et al. The Ras/ERK Signaling Pathway Couples Antimicrobial Peptides to Mediate Resistance to Dengue Virus in Aedes Mosquitoes. PloS Negl Trop Dis (2020) 14:e0008660. doi: 10.1371/journal.pntd.0008660
53. Dubrovsky EB, Dubrovskaya VA, Bilderback AL, Berger EM. The Isolation of Two Juvenile Hormone-Inducible Genes in Drosophila Melanogaster. Dev Biol (2000) 224:486–95. doi: 10.1006/dbio.2000.9800
54. Faucon F, Dusfour I, Gaude T, Navratil V, Boyer F, Chandre F, et al. Identifying Genomic Changes Associated With Insecticide Resistance in the Dengue Mosquito Aedes Aegypti by Deep Targeted Sequencing. Genome Res (2015) 25:1347–59. doi: 10.1101/gr.189225.115
55. Ghosh Roy S, Sadigh B, Datan E, Lockshin RA, Zakeri Z. Regulation of Cell Survival and Death During Flavivirus Infections. World J Biol Chem (2014) 5:93–105. doi: 10.4331/wjbc.v5.i2.93
56. Datan E, Roy SG, Germain G, Zali N, McLean JE, Golshan G, et al. Dengue-Induced Autophagy, Virus Replication and Protection From Cell Death Require ER Stress (PERK) Pathway Activation. Cell Death Dis (2016) 7:e2127–7. doi: 10.1038/cddis.2015.409
57. Ramji DP, Foka P. CCAAT/enhancer-Binding Proteins: Structure, Function and Regulation. Biochem J (2002) 365:561–75. doi: 10.1042/BJ20020508
58. Inestrosa NC, Bronfman M, Leighton F. Detection of Peroxisomal Fatty Acyl-Coenzyme A Oxidase Activity. Biochem J (1979) 182:779–88. doi: 10.1042/bj1820779
59. Kubota H, Hynes G, Carne A, Ashworth A, Willison K. Identification of Six Tcp-1-Related Genes Encoding Divergent Subunits of the TCP-1-Containing Chaperonin. Curr Biol (1994) 4:89–99. doi: 10.1016/S0960-9822(94)00024-2
60. Kiang JG, Tsokos GC. Heat Shock Protein 70 kDa: Molecular Biology, Biochemistry, and Physiology. Pharmacol Ther (1998) 80:183–201. doi: 10.1016/S0163-7258(98)00028-X
61. Mosser DD, Caron AW, Bourget L, Meriin AB, Sherman MY, Morimoto RI, et al. The Chaperone Function of Hsp70 Is Required for Protection Against Stress-Induced Apoptosis. Mol Cell Biol (2000) 20:7146. doi: 10.1128/MCB.20.19.7146-7159.2000
62. Waterhouse RM, Povelones M, Christophides GK. Sequence-Structure-Function Relations of the Mosquito Leucine-Rich Repeat Immune Proteins. BMC Genomics (2010) 11:531–1. doi: 10.1186/1471-2164-11-531
63. Clayton AM, Dong Y, Dimopoulos G. The Anopheles Innate Immune System in the Defense Against Malaria Infection. J Innate Immun (2014) 6:169–81. doi: 10.1159/000353602
64. Diop F, Alout H, Diagne CT, Bengue M, Baronti C, Hamel R, et al. Differential Susceptibility and Innate Immune Response of Aedes Aegypti and Aedes Albopictus to the Haitian Strain of the Mayaro Virus. Viruses (2019) 11:924. doi: 10.3390/v11100924
65. Marnef A, Standart N. Pat1 Proteins: A Life in Translation, Translation Repression and mRNA Decay. Biochem Soc Trans (2010) 38:1602–7. doi: 10.1042/BST0381602
66. Standart N, Marnef A. Pat1 Proteins: Regulating mRNAs From Birth to Death? Biomol Concepts (2012) 3:295–306. doi: 10.1515/bmc-2012-0005
67. Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, et al. The Whole Structure of the 13-Subunit Oxidized Cytochrome C Oxidase at 2.8 Å. Science (1996) 272:1136. doi: 10.1126/science.272.5265.1136
68. Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, et al. CDD/SPARCLE: The Conserved Domain Database in 2020. Nucleic Acids Res (2020) 48:D265–8. doi: 10.1093/nar/gkz991
69. Zhang L, Wang A. Virus-Induced ER Stress and the Unfolded Protein Response. Front Plant Sci (2012) 3:293. doi: 10.3389/fpls.2012.00293
70. Fribley A, Zhang K, Kaufman RJ. Regulation of Apoptosis by the Unfolded Protein Response. Methods Mol Biol Clifton NJ (2009) 559:191–204. doi: 10.1007/978-1-60327-017-5_14
71. Redza-Dutordoir M, Averill-Bates DA. Activation of Apoptosis Signalling Pathways by Reactive Oxygen Species. Biochim Biophys Acta BBA - Mol Cell Res (2016) 1863:2977–92. doi: 10.1016/j.bbamcr.2016.09.012
72. Cerritelli SM, Crouch RJ. Ribonuclease H: The Enzymes in Eukaryotes. FEBS J (2009) 276:1494–505. doi: 10.1111/j.1742-4658.2009.06908.x
73. Erdman SE, Burtis KC. The Drosophila Doublesex Proteins Share a Novel Zinc Finger Related DNA Binding Domain. EMBO J (1993) 12:527–35. doi: 10.1002/j.1460-2075.1993.tb05684.x
74. Erdman SE, Chen HJ, Burtis KC. Functional and Genetic Characterization of the Oligomerization and DNA Binding Properties of the Drosophila Doublesex Proteins. Genetics (1996) 144:1639–52. doi: 10.1093/genetics/144.4.1639
75. Vinson CR, Sigler PB, McKnight SL. Scissors-Grip Model for DNA Recognition by a Family of Leucine Zipper Proteins. Science (1989) 246:911–6. doi: 10.1126/science.2683088
76. De Maio FA, Risso G, Iglesias NG, Shah P, Pozzi B, Gebhard LG, et al. The Dengue Virus NS5 Protein Intrudes in the Cellular Spliceosome and Modulates Splicing. PloS Pathog (2016) 12:e1005841. doi: 10.1371/journal.ppat.1005841
77. Cervantes-Salazar M, Angel-Ambrocio AH, Soto-Acosta R, Bautista-Carbajal P, Hurtado-Monzon AM, Alcaraz-Estrada SL, et al. Dengue Virus NS1 Protein Interacts With the Ribosomal Protein RPL18: This Interaction Is Required for Viral Translation and Replication in Huh-7 Cells. Virology (2015) 484:113–26. doi: 10.1016/j.virol.2015.05.017
78. Wendel BM, Cole JM, Courcelle CT, Courcelle J. SbcC-SbcD and ExoI Process Convergent Forks to Complete Chromosome Replication. Proc Natl Acad Sci USA (2018) 115:349–54. doi: 10.1073/pnas.1715960114
79. Clark NE, Katolik A, Roberts KM, Taylor AB, Holloway SP, Schuermann JP, et al. Metal Dependence and Branched RNA Cocrystal Structures of the RNA Lariat Debranching Enzyme Dbr1. Proc Natl Acad Sci (2016) 113:14727. doi: 10.1073/pnas.1612729114
80. Campbell CL, Dickson LB, Lozano-Fuentes S, Juneja P, Jiggins FM, Black WC,4. Alternative Patterns of Sex Chromosome Differentiation in Aedes Aegypti (L). BMC Genomics (2017) 18:943–3. doi: 10.1186/s12864-017-4348-4
81. Marinho J, Martins T, Neto M, Casares F, Pereira PS. The Nucleolar Protein Viriato/Nol12 Is Required for the Growth and Differentiation Progression Activities of the Dpp Pathway During Drosophila Eye Development. Dev Biol (2013) 377:154–65. doi: 10.1016/j.ydbio.2013.02.003
82. Gautier T, Bergès T, Tollervey D, Hurt E. Nucleolar KKE/D Repeat Proteins Nop56p and Nop58p Interact With Nop1p and are Required for Ribosome Biogenesis. Mol Cell Biol (1997) 17:7088–98. doi: 10.1128/mcb.17.12.7088
83. Weidenhammer EM, Singh M, Ruiz-Noriega M, Woolford JL Jr. The PRP31 Gene Encodes a Novel Protein Required for pre-mRNA Splicing in Saccharomyces Cerevisiae. Nucleic Acids Res (1996) 24:1164–70. doi: 10.1093/nar/24.6.1164
84. Karbstein K. Role of GTPases in Ribosome Assembly. Biopolymers (2007) 87:1–11. doi: 10.1002/bip.20762
85. Hughes JMX. Functional Base-Pairing Interaction Between Highly Conserved Elements of U3 Small Nucleolar RNA and the Small Ribosomal Subunit RNA. J Mol Biol (1996) 259:645–54. doi: 10.1006/jmbi.1996.0346
86. Böhm S, Frishman D, Mewes HW. Variations of the C2H2 Zinc Finger Motif in the Yeast Genome and Classification of Yeast Zinc Finger Proteins. Nucleic Acids Res (1997) 25:2464–9. doi: 10.1093/nar/25.12.2464
87. Xu C, Min J. Structure and Function of WD40 Domain Proteins. Protein Cell (2011) 2:202–14. doi: 10.1007/s13238-011-1018-1
88. Zhang C, Zhang F. The Multifunctions of WD40 Proteins in Genome Integrity and Cell Cycle Progression. J Genomics (2015) 3:40–50. doi: 10.7150/jgen.11015
89. Fan Y, Sanyal S, Bruzzone R. Breaking Bad: How Viruses Subvert the Cell Cycle. Front Cell Infect Microbiol (2018) 8:396. doi: 10.3389/fcimb.2018.00396
90. Clark MB, Mattick JS. Long Noncoding RNAs in Cell Biology. Semin Cell Dev Biol (2011) 22:366–76. doi: 10.1016/j.semcdb.2011.01.001
91. Fortes P, Morris KV. Long Noncoding RNAs in Viral Infections. Virus Res (2016) 212:1–11. doi: 10.1016/j.virusres.2015.10.002
92. Etebari K, Asad S, Zhang G, Asgari S. Identification of Aedes Aegypti Long Intergenic non-Coding RNAs and Their Association With Wolbachia and Dengue Virus Infection. PloS Negl Trop Dis (2016) 10:e0005069. doi: 10.1371/journal.pntd.0005069
93. Raquin V, Merkling SH, Gausson V, Moltini-Conclois I, Frangeul L, Varet H, et al. Individual Co-Variation Between Viral RNA Load and Gene Expression Reveals Novel Host Factors During Early Dengue Virus Infection of the Aedes Aegypti Midgut. PloS Negl Trop Dis (2017) 11:e0006152. doi: 10.1371/journal.pntd.0006152
94. Seki T, Gong L, Williams AJ, Sakai N, Todi SV, Paulson HL. JosD1, a Membrane-Targeted Deubiquitinating Enzyme, Is Activated by Ubiquitination and Regulates Membrane Dynamics, Cell Motility, and Endocytosis. J Biol Chem (2013) 288:17145–55. doi: 10.1074/jbc.M113.463406
95. Tzvetkov N, Breuer P. Josephin Domain-Containing Proteins From a Variety of Species Are Active De-Ubiquitination Enzymes. Biol Chem (2007) 388:973–8. doi: 10.1515/BC.2007.107
96. Bosio CF, Fulton RE, Salasek ML, Beaty BJ, Black WC,4. Quantitative Trait Loci That Control Vector Competence for Dengue-2 Virus in the Mosquito Aedes Aegypti. Genetics (2000) 156:687–98. doi: 10.1093/genetics/156.2.687
97. Gomez-Machorro C, Bennett KE, Del Lourdes Munoz M, Black WC IV. Quantitative Trait Loci Affecting Dengue Midgut Infection Barriers in an Advanced Intercross Line of Aedes Aegypti. Insect Mol Biol (2004) 13:637–48. doi: 10.1111/j.0962-1075.2004.00522.x
98. Bennett KE, Flick D, Fleming KH, Jochim R, Beaty BJ, Black WC,4. Quantitative Trait Loci That Control Dengue-2 Virus Dissemination in the Mosquito Aedes Aegypti. Genetics (2005) 170:185–94. doi: 10.1534/genetics.104.035634
99. Timoshevskiy VA, Severson DW, Debruyn BS, Black WC, Sharakhov IV, Sharakhova MV. An Integrated Linkage, Chromosome, and Genome Map for the Yellow Fever Mosquito Aedes Aegypti. PloS Negl Trop Dis (2013) 7:e2052–2. doi: 10.1371/journal.pntd.0002052
100. Aubry F, Dabo S, Manet C, Filipović I, Rose NH, Miot EF, et al. Enhanced Zika Virus Susceptibility of Globally Invasive Aedes Aegypti Populations. Science (2020) 370:991. doi: 10.1126/science.abd3663
101. Lee W-S, Webster JA, Madzokere ET, Stephenson EB, Herrero LJ. Mosquito Antiviral Defense Mechanisms: A Delicate Balance Between Innate Immunity and Persistent Viral Infection. Parasites Vectors (2019) 12:165. doi: 10.1186/s13071-019-3433-8
102. Price DP, Nagarajan V, Churbanov A, Houde P, Milligan B, Drake LL, et al. The Fat Body Transcriptomes of the Yellow Fever Mosquito Aedes Aegypti, Pre- and Post-Blood Meal. PloS One (2011) 6:e22573. doi: 10.1371/journal.pone.0022573
Keywords: RNA-Seq, mosquito, immune response, gene expression, dengue virus
Citation: Chen T-Y, Lee Y, Wang X, Mathias D, Caragata EP and Smartt CT (2021) Profiling Transcriptional Response of Dengue-2 Virus Infection in Midgut Tissue of Aedes aegypti. Front. Trop. Dis 2:708817. doi: 10.3389/fitd.2021.708817
Received: 12 May 2021; Accepted: 26 July 2021;
Published: 10 August 2021.
Edited by:
Hassan Vatandoost, Tehran University of Medical Sciences, IranReviewed by:
Paulo Eduardo Martins Ribolla, São Paulo State University, BrazilCopyright © 2021 Chen, Lee, Wang, Mathias, Caragata and Smartt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chelsea T. Smartt, Y3RzbWFydEB1ZmwuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.