
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Transplant., 27 November 2023
Sec. Transplantation Immunology
Volume 2 - 2023 | https://doi.org/10.3389/frtra.2023.1283275
This article is part of the Research TopicRising Stars: Transplantation Immunology 2022View all 3 articles
Regulatory T cells are fundamental for the induction and maintenance of immune homeostasis, with their dysfunction resulting in uncontrolled immune responses and tissue destruction predisposing to autoimmunity, transplant rejection and several inflammatory and metabolic disorders. Recent discoveries have demonstrated that metabolic processes and mitochondrial function are critical for the appropriate functioning of these cells in health, with their metabolic adaptation, influenced by microenvironmental factors, seen in several pathological processes. Upon activation regulatory T cells rearrange their oxidation-reduction (redox) system, which in turn supports their metabolic reprogramming, adding a layer of complexity to our understanding of cellular metabolism. Here we review the literature surrounding redox homeostasis and metabolism of regulatory T cells to highlight new mechanistic insights of these interlinked pathways in immune regulation.
Oxidative stress, teamed with heightened levels of inflammation and metabolic disturbances, underlies the pathology of many conditions, including cardiovascular diseases, cancer, autoimmune and neurodegenerative disorders (1–4). This phenomenon is the result of either an overproduction of reactive oxygen species (ROS) and/or an imbalance between production of oxidants and antioxidants defenses (5). However, redox states have an important role in immunity and T cell function. It is well known that moderate levels of ROS are essential as pivotal second messengers for T cell receptor signaling and activation and participate in chemotaxis and antigen cross-presentation (6–8), whilst elevated levels of ROS have a detrimental effect on immune regulation (9, 10), causing damage of proteins, DNA and carbohydrates.
Under inflammatory conditions, regulatory T cells (Tregs), a subset of immune cells, play an important role at curbing excessive immune activation and maintaining immune homeostasis (11). With the growing appreciation of the relationship between metabolism and immunity, we now realise that the phenotypic stability, suppressive function and survival of Tregs are tightly determined by specific metabolic requirements and programs, with their metabolic adaptation seen in various pathological conditions. Previous studies, mainly based in murine models, have described the engagement of different metabolic pathways by Tregs to exert their function. Unlike effector T cells (Teff) that use aerobic glycolysis to meet their bioenergetic demands under steady state conditions, Tregs largely rely on mitochondrial metabolism through a selective dependency on fatty acid oxidation to sustain the tricarboxylic acid (TCA) cycle and oxidative phosphorylation (OXPHOS) reactions (12, 13). Likewise, Tregs show a distinct metabolic profile during proliferation, migration, and suppressive functions. Following stimulation, Treg proliferation is dependent on an oscillatory switch of glycolysis dictated by increased activity of the PI3K/Akt and mTOR pathways at the early phase (14). However, in contrast to Teff, Tregs also use an array of substrates from the fatty acid oxidation and amino acid metabolism to fuel the TCA cycle. During migration, Tregs engage in glycolysis like most migratory cells (15). Although glycolysis promotes Treg growth and motility, the immunosuppressive function and stability of Tregs is compromised. To exert their suppressive functions, Tregs require a stable FOXP3 expression which in turn controls the induction of OXPHOS activity and downregulation of glycolysis via AMPK signaling (16). Of note, in autoimmunity and various other inflammatory diseases, oxidative stress can disrupt the mitochondrial network integrity, compromise OXPHOS, deplete cellular ATP, and alter cellular metabolic pathways (17). In addition, the systemic and tissue microenvironmental changes seen in many disorders, including deficiencies in metabolites and nutrients as well as changes in oxygen levels can result in alterations in Treg function and stability via their metabolic reprogramming. In the absence of oxygen, the hypoxia-inducible factors (HIFs) orchestrate the transcription of an array of target genes that regulate metabolic pathways to adapt cells to low-oxygen environments by promoting glycolysis and suppressing mitochondrial respiration (18, 19). In line with the latter notion, studies have shown that hypoxia can impair Treg differentiation by promoting aerobic glycolysis by stabilizing one of the HIF isoforms (HIF1α) (20). Moreover, Yamamoto et al. provided further supporting evidence, showing that stabilization of both isoforms HIF1α and HIF2α in adult mice, through silencing of the prolyl hydroxylase domain 2 enzyme (PHD2), results in an impaired ability of Tregs to suppress either skin allograft rejection or in vitro responder T cell proliferation (21). However, despite the extensive literature to date, the molecular mechanisms that define Treg cell fate and plasticity and the interlinked processes of metabolism and redox homeostasis are still not well described in health and the changes seen in disease. This is of paramount importance given the fundamental role of impaired Treg function in an array of pathological conditions (22–25), with an increased Treg number and function reported in several mouse and human cancers (26–29), representing a major barrier to anti-tumour immunity. To gain an understanding of the integration of these processes the next sections review the key antioxidant and nutrient sensing pathways that determine Treg redox homeostasis and function and the impact of microenvironmental metabolic perturbations and nutrient availability.
Tregs predominantly use fatty acid oxidation that generates acetyl-CoA to fuel the TCA cycle. In a series of enzymatic reactions, the TCA cycle generates the reducing equivalents NADH and FADH2 which are required to transfer electrons to the mitochondrial respiratory chain. The energy released from these oxidation-reduction (redox) reactions, along with the creation of a proton gradient, results in the synthesis of ATP by OXPHOS reactions. During this process, 0.2%–2% of the electrons leak out of the transfer and interact with O2 to produce ROS, such as superoxide or hydrogen peroxide (30, 31). Although most mitochondrial ROS results from OXPHOS, another three reactions during the TCA cycle generate ROS: pyruvate to acetyl-CoA (32, 33), α-ketoglutarate to succinyl-CoA (34, 35) and fumarate to succinate (36). Despite this, mitochondrial ROS levels can also be increased when there is disruption to the electron transport chain, resulting several pathological processes such as ageing, autoimmunity (17) and neurodegeneration (37).
Aside from the mitochondria, NADPH oxidases (NOXs) are also a source of ROS, with NOX2 described as a major ROS-generating enzyme in T cells (38). The negative impact of ROS in Tregs has been highlighted by studies reporting that NOX2 deficient Tregs exhibit higher suppressive capacity than wild-type Tregs both in vitro and in vivo (39, 40). This, therefore, highlights that the balance between production and consumption of ROS is an important factor in the development and function of Tregs. In line with this, and in the setting of chronic liver disease, recent work from our group has shown that Treg function and viability is negatively impacted by oxidative stress and increased intracellular ROS (41). Mechanistic insights revealed this to be secondary to an imbalance in redox homeostasis with an increased NOX-2 activity and dysregulated endogenous antioxidant signaling pathway.
The transcription factor NF (erythroid-derived 2)-like 2 (NRF2) is a basic leucine zipper transcription factor that serves as a master regulator of antioxidant defense against the cytotoxic effects of oxidative stress by regulating an array of detoxifying and antioxidant genes. Under quiescent conditions, NRF2 is retained and degraded in the proteasome by Kelch like ECH-associated protein 1 (Keap1) in concert with the E3 ubiquitin ligase Cullin 3 (Cul3) (42). Oxidative stress induces conformational changes in Keap1 that allow NRF2 release and translocation into the nucleus where it promotes the expression of phase II detoxifying enzymes and antioxidant proteins (43). Of note, NRF2 also targets the expression of several components of the glutathione (GSH) and thioredoxin (TRX)-1 antioxidant systems (44). Activated T cells, maintain the concentrations of intracellular ROS by using GSH (45) which triggers cytosolic ROS depletion either in a direct way or through enzymatic catalysis mediated by glutathione peroxidases (GPX). Cellular GSH content is largely determined by de novo synthesis, a process mediated by two ATP-dependent ligases, glutamate-cysteine ligase (GCL) and glutathione synthase (GS), as well as the availability of its constituent amino acids, cysteine, glycine and glutamate (46). TRX1 is also critical for Treg's resistance to oxidative stress. This enzyme scavenges reactive oxygen species and regulates other enzymes metabolizing H2O2 (47). Moreover, TRX1 maintains sustained surface expression of thiols as the first line of defense against ROS and is sensitive to proinflammatory stimuli, mainly tumor necrosis factor-α, in a nuclear factor-κB-dependent fashion, which likely boosts Treg survival under inflammatory conditions (48).
Despite their role in the neutralization of excess ROS, the cellular antioxidant mechanisms can modulate Treg metabolic rewiring via engagement of the mammalian target of rapamycin (mTOR) pathway.
mTOR is a conserved serine/threonine kinase that senses and integrates diverse immune receptor signaling pathways and environmental cues to coordinately regulate metabolic programs that decipher T cell differentiation, proliferation, and function (49). mTOR is a component of two distinct multi-protein complexes termed mTOR complex 1 (mTORC1) and 2 (mTORC2) which share the catalytic subunit mTOR and the associated proteins Deptor and mLST8. PRAS40 and Raptor proteins are unique components of mTORC1, while mTORC2 is characterized by the expression of mSIN1 and Rictor (50). The upstream and downstream components of mTOR signaling differ in each complex. mTORC1 activity is mainly dependent on the small G protein RHEB (Ras homologue enriched in brain) which in turn is modulated by the tuberous sclerosis 1 (TSC1)–TSC2 complex (51, 52). This complex integrates positive and negative upstream signals transduced from the PI3/AKT and AMPK (AMP-activated protein kinase) pathways (53). An array of stimuli induces the PI3K/AKT pathway including TCR stimulation (54, 55) and co-stimulation signals mediated by CD28 or OX40 (56, 57), toll-like receptor signals (58, 59), cytokines (60, 61), growth factors and hormones (e.g., leptin and bioactive lipid sphingosine-1-phosphate-S1P) (14, 62). Besides PI3K/AKT dependent mechanisms, essential and non-essential amino acids can also activate mTORC1 via RAG GTPases (63). Moreover, other nutrients such as glucose and lipids can influence mTORC1 activation, however the mechanism for such regulation remains to be elucidated. During energy deprivation, AMPK downregulates mTORC1 directly by phosphorylation of Raptor and indirectly by phosphorylation of TSC2 (64, 65). Compared to mTORC1, regulation of mTORC2 is far less well understood. Studies in non-T cell lineages have postulated that its activity is modulated by PI3K signaling in response to growth factors and insulin which lastly mediates mTORC2-ribosome binding (66, 67). Moreover, other signals independent of PI3K enhance mTORC2 activity such as amino acid, glucose and glutamine starvation (68–70.) Following activation, mTORC1 drives c-MYC and HIF1α signaling via multiple mechanisms involving S6K1 and 4EBP1 to ensure induction of aerobic glycolysis and glutaminolysis necessary for conventional T cell activation and differentiation (71, 72). Meanwhile, mTORC2 activates AGC kinases, including Akt, SGK1 and PKCα, to promote cell survival, cell cycle progression and anabolism (73–75).
Observations regarding mTOR activity in Tregs have been complex and at times seemingly controversial. First, several studies revealed that the efficiency of Treg generation both in vivo and in vitro could be markedly enhanced in the presence of rapamycin (76–80). This result was further supported by genetic deletion of components of the mTOR signaling pathway (81–83). In addition, it was found that constitutive activation of the PI3K/Akt/mTOR network antagonized FoxP3 induction along with other Treg-signature transcripts (82, 84), which are essential in the generation and sustenance of the Treg lineage (85–87). Additionally, induced-Treg cell generation requires a metabolic switch to fatty acid oxidation via AMPK, thereby inhibiting mTORC1 signaling (88). In line with these results, studies demonstrated that negative regulation upstream of mTOR in Tregs is required for the maintenance of their suppressive function, thus indicating that the glycolytic metabolism induced by mTOR is dispensable for Treg cell function (28, 89–91). Despite these data, it seems paradoxical that other studies revealed that mTOR activity was increased in Tregs compared to Teff at steady state and promotes Treg proliferation (14, 92). According to these findings, Zeng et al. showed that Raptor/mTORC1 signaling coordinates cholesterol and lipid metabolic programs to support Treg proliferation, upregulation of the suppressive molecules CTLA-4 and ICOS, and functional fitness in part through inhibiting mTORC2 (92). Likewise, effector Tregs were characterized by increased mTOR activity, glycolysis, and effector molecule upregulation after robust TCR stimulation and in presence of a suppressive microenvironment (TGF-β) (93). Finally, Chapman et al. showed that in vivo inhibition of mTOR disrupts Treg suppressive function and leads to uncontrolled conventional T cell activation, especially associated with an excess of Th2 at mucosal tissues (94). Mechanistically, mTOR regulates the expression of the transcription factors IRF4 and GATA3 that are essential to activate the machinery involved in Treg-dependent suppression (95). Moreover, mTOR promotes mitochondrial fitness most likely via Raptor-mTORC1 as Raptor-deficient Tregs were not able to evade T helper responses in a colitis mouse model while Rictor-deficient Tregs showed normal mitochondrial activity and in vitro suppressive capacity (92). mTORC2 signaling by CD28 stimulation is required for Treg migration and involves the induction of the enzyme glucokinase (GCK) which is essential to maintain the glycolytic flux (96). These observations supported a key role for Raptor-mTORC1 pathway in establishing the fate and function of activated Tregs. However, the local environmental signals that may orchestrate mTOR tuning and hence the Treg function in health and the changes seen in various pathologies remain to be deciphered.
The antioxidant mechanisms GSH, NRF2 and TRX1 can regulate mTOR activity by different mechanisms (Figure 1). NRF2 directly modulates mTOR transcription by binding to the mTOR promoter (97). Although it has also been reported that NRF2 can upregulate mTOR activity indirectly by increasing the expression of RagD, a protein known as an activator of mTOR or by modulating activity of components of the PI3K/Akt pathway (63, 98). In line with these observations, Klemm et al. showed an increased phosphorylation of mTOR and of the mTOR target p-S6 in Tregs of Foxp3cre mice with constitutive NRF2 activation. Their results suggest that enhanced mTOR signaling led to elevated glucose uptake that impacts Treg long term survival and lineage stability with reduced FoxP3 expression and immunosuppressive function (99), supporting the role of mTORC1 as a negative regulator of Treg cell function. A previous study also showed that absence of NRF2 in donor T cells enhanced persistence of Tregs and reduced systemic inflammation in graft-vs.-host disease mice (100). Additionally, Noel et al. demonstrated that T cell-specific activation of NRF2 increases the number of Tregs and improves Treg-mediated suppression of inflammation in an ischemia reperfusion-induced acute kidney injury mouse model (101).
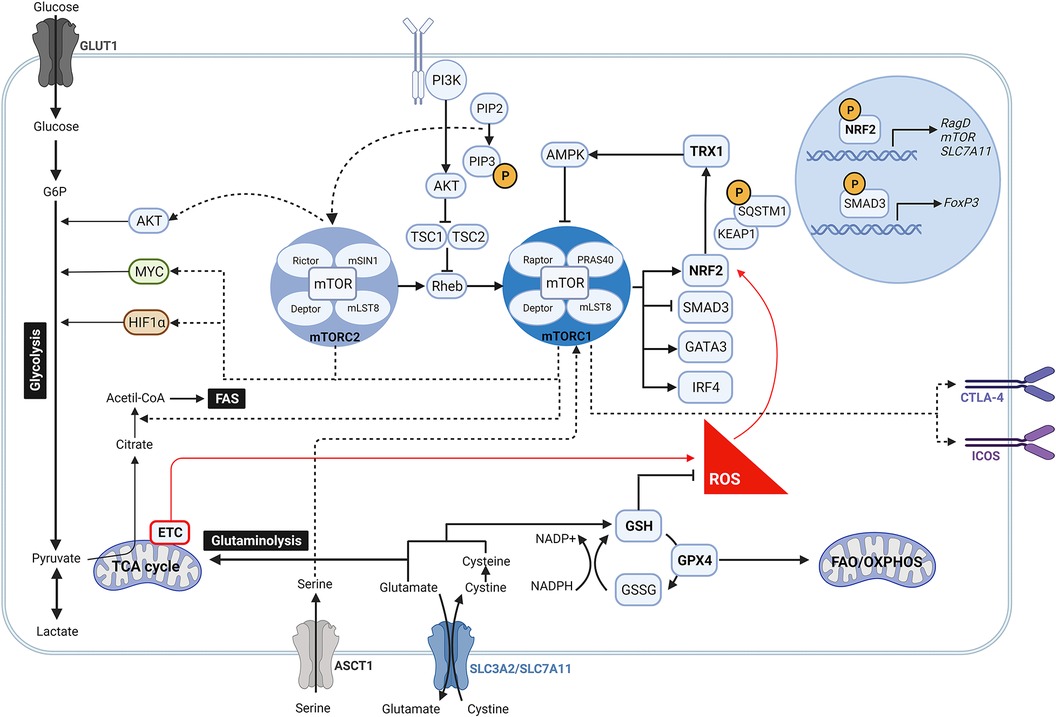
Figure 1. Crosstalk between mTOR pathway and anti-oxidant signalling in regulatory T cells. During oxidative stress, high levels of ROS induce the activity of NRF2 by disrupting its binding to KEAP1. Once NRF2 is released, it is translocated into the nucleus where it promotes the expression of mTOR, RagD and components of the PI3K/AKT pathway, which contribute to activate the mTOR complex signalling pathway. Likewise, increased mTORC1 activity may be link to an intracellular accumulation of serine due to an upregulated expression of its transporter ASCT1 by NRF2. Following activation, mTORC1 and mTORC2 are involved in many different signalling pathways. While mTORC2 modulates AKT, mTORC1 drives MYC and HIF1α downstream signalling so as to ensure aerobic glycolysis. mTORC1 reciprocally prompts NRF2 activity by phosphorylation of SQSTM1, inhibits the transcription of FoxP3 by SMAD3, and regulates the expression of the transcription factors IRF4 and GATA3. Moreover, mTORC1 is linked to the upregulation of CTLA-4 and ICOS, and to the induction of fatty acid biosynthesis programs. At the same time, NRF2 targets the expression of the glutamate/cystine transporter SLC7A11 that supports the GSH system and glutaminolysis. GSH boosts the removal of ROS which in turn reduce the activation of NRF2 and antagonize mTOR induction. Along this mechanism, NRF2 can also fine-tune mTOR activity via TRX1 that releases AMPK.
More recent data have outlined the role of NRF2 along with other antioxidant mechanisms in controlling mTOR activity and Treg function. To diminish mTOR activity, GSH scavenges ROS to control NRF2 activation which decreases the amino acid transporter ASCT1 and therefore serine uptake (102). In line with this, intracellular accumulation of serine has been shown to result in Treg proliferation at the cost of FoxP3 expression and suppressive function (102). As GSH controls NRF2 activation, which in turn releases TRX1 that inhibits AMPK (103), it appears evident that Tregs also use GSH to indirectly modulate mTOR signaling (Figure 1). The antagonistic effect of TRX1 in controlling mTOR activation is further supported by Chakraborty et al., who did not report an increase in the quantity and suppressive quality of induced Tregs generated by using a recombinant Trx (104).
As the cellular antioxidant mechanisms control mTOR activity, mTOR can reciprocally regulate them. In this context, mTORC1 enables NRF2 release from its principal negative regulator KEAP1 by phosphorylation of sequestosome 1 (SQSTM1), thus inducing NRF2 stabilization and translocation into the nucleus (105–108), where it drives cystine/glutamate antiporter solute carrier (SLC)7A11 expression. This transporter together with SLC3A2 (or CD98) are responsible for maintaining the cysteine intracellular pool, which in addition to protein synthesis, is essential to produce GSH and boost redox balance (109–111). In an elegant study Procaccini et al. described this mechanism as a key determinant of Treg proliferative capacity in patients with relapsing-remitting multiple sclerosis (RRMS) (112). In their study they showed a decreased expression of SLC7A11 by patient-derived Tregs, linked to a dysregulated NRF2 signaling, which was rescued upon treatment of these patients with dimethyl fumarate (DMF), small molecule inducer of NRF2 licensed for the treatment of RRMS (112). These findings highlight NRF2 as an indispensable checkpoint to support redox homeostasis and determine Treg cell expansion (12).
The antioxidant mechanisms can directly modulate Treg cell outcome regardless of mTOR activity. Upon activation, Tregs markedly reprogram lipid synthesis and fatty acid oxidation in support of their expansion and suppressive function (88, 113). In fact, several studies have suggested that Tregs rely on fatty acid synthesis and oxidation to survive and proliferate in the hostile tumor environment (114, 115). However, this mechanism potentially enhances generation of toxic lipid peroxides and subsequent ferroptosis, that can impair cellular redox homeostasis. One of the members of the antioxidant GPX family, GPX4, directly reduces membrane phospholipid hydroperoxides and oxidizes lipoproteins by using GSH as a cofactor to maintain redox balance under active lipid metabolism (116–118). Along these lines, a recent study demonstrated that inducing ferroptosis of intra-tumoral Treg cells by GPX4 deficiency disrupted mitochondrial fitness and enhances interleukin (IL)-1β production, which potentiates Th17 responses (119).
Iron homeostasis is tightly linked to lipid metabolism and ROS production. In short, excess iron ions (Fe2+) convert the hydrogen peroxide (H2O2), produced by mitochondrial respiration, into the toxic free hydroxyl radical (OH) through Fenton/Haber-Weiss reactions, which in turn need electrons from lipids resulting in an accelerated lipid peroxidation and ferroptosis (120). The phosphoinositide-dependent kinase 1 (PDK1) has been shown to play a key role in maintaining iron-dependent ROS levels and the suppressive function of Tregs. A recent study described that PDK1 inhibits MEK-Erk signaling and thus expression of the transferrin receptor protein 1 (CD71) and iron uptake (121). Apart from iron metabolism, PDK1 regulates multiple signaling pathways including the canonical NF-kB pathway and Akt/mTOR pathways (122). Indeed, the previous study showed that PDK1-deficient Tregs exhibit downregulation of the mTORC1 pathway and an altered mitochondrial metabolism which likely explains the impaired Treg function. Moreover, Hyunju Oh et al. reported that PDK1 controls Treg signature gene expression by regulating the canonical NF-κB pathway and that Treg-specific deletion of PDK1 in mice results in a severe systemic inflammation, accompanied by reduced number and suppressive function of Tregs (123).
Besides the antioxidant mechanisms, different metabolic pathways may also play a key role in the resistance of Tregs to oxidative stress in certain environments. As already outlined in previous sections, Tregs are less dependent on glycolysis and largely use OXPHOS to fulfill their bioenergetic demands (12, 13). In the setting of metabolic stress, as seen in solid tumors, the alterations in the availability of nutrients, such as glucose, glutamine, and tryptophan and the enrichment in lactic acid and kynurenines (124), have an impact on the tumour immune compartment. In this regard, extensive literature reports the negative impact of this metabolic environment on the cytotoxicity of effector T cell responses, which in-essence are dependent on glycolysis (125–127). In contrast, intra-tumoral Treg's metabolic reprogramming in this environment is evident by the use of alternative metabolites, such as lactic acid, to support their survival and function (128). Indeed, FoxP3 has been shown to favor oxidation of L-lactate to pyruvate while quenching glycolysis by downregulation of Myc (114). In addition to their role as substrates to produce ATP, lactate and pyruvate have also been described to have potent roles in resistance against oxidative stress. In this regard, in their study, Tauffenberger et al. show that oxidative metabolism of L-lactate, and to a lesser extent pyruvate, boosts mitochondrial activity and promotes a moderate increase in ROS production that is sufficient to induce NRF2 activity, PI3k/AKT and mTOR signaling and thereby support cell survival (129). However, this mechanism has been described in an in vitro model, and it remains to be elucidated whether lactate activates detoxifying mechanisms under oxidative stress conditions in vivo.
The molecular pathways described above provide multiple opportunities to finely tune ROS homeostasis and metabolic programs in Tregs by targeting signaling cascades, enzymes and metabolites. In so doing, we anticipate it will be possible to either boost Treg function (to promote tolerance in autoimmunity or transplantation) or inhibit it (in cancer or chronic infections).
For instance, by specifically targeting a ROS-producing enzyme, Trevelin et al. observed that the depletion of Nox2 within Tregs in murine heart transplant recipients promoted the engraftment of allogeneic grafts. Mechanistically, Nox2-deficient Tregs exhibited notable changes: (1) increased expression of chemokine receptors CCR4 and CCR8, promoting their migration toward the allograft; (2) heightened capacity to suppress the proliferation of CD8+ T effector cells and the production of interferon-gamma (IFN-γ); and (3) improved survival (40). The understanding of the metabolic pathways underpinning the heightened functionality of Nox2-deficient Tregs, however, remains to be elucidated.
mTOR is another target influencing Treg metabolism that has been explored for therapeutic purposes, although its downstream effect on Treg function appear to be highly context dependent and far from being well understood. Thus, several studies have reported that mTOR inhibition, using Rapamycin or Everolimus (EVR), facilitates the selective ex vivo expansion of Tregs to generate cell therapy products for adoptive transfer into transplant recipients or autoimmune patients (130–132). To uncover the underlying mechanisms of mTOR inhibition in this context, Gedaly and colleagues conducted a metabolic characterization of autologous Tregs from renal transplant patients during the initial phase of their ex vivo expansion in the presence of Rapamycin or EVR (133). During the first 5 days of expansion, both compounds reduced the glycolytic rates and promoted an OXPHOS energy profile in Tregs, although the EVR-treated Tregs showed a reduced OXPHOS activity that was associated with diminished mTORC2 signaling and slow initial expansion rates. The distinct bioenergetic profiles and the relative reliance on OXPHOS and glycolysis during the expansion of Tregs and Tconv cells contributed to the selective expansion of Tregs. Despite such a study, several questions remain to be addressed, such as the metabolic characteristics and redox state of Tregs during expansion and the impact of the microenvironment on the infused cells. This information would lend itself to optimization of the current protocols/process to ensure the expansion of a product that is stable/ functional and resilient to the stressors of the microenvironment, once injected.
Of note, the difficulties of standardizing Treg manufacture protocols, together with the known efficacy advantages of conferring antigen-specificity to Treg products (134), have prompted the use of the chimeric antigen receptor (CAR)-engineered technology. Human leukocyte antigen (HLA)-A2-targeted CAR-Tregs have been shown to efficiently prevent graft-vs.-host disease and skin graft rejection (135–138), and are currently in clinical trials in kidney and liver transplantation. There is limited information, however, concerning their metabolic requirements, and whether their function, lineage stability, proliferative potential, and survival can be further optimized by modifying metabolic programs.
Examples of how this could be achieved are from lessons learnt from the CAR-T oncology field, where genetic modifications and extracellular metabolite supplementation strategies have demonstrated the ability to influence mitochondrial metabolism, extend the longevity of CAR-T cells, and counteract their exhaustion. For instance, incorporating the co-stimulatory endodomain 4-1BB into CAR-T molecules has been shown to promote the upregulation of the peroxisome proliferator-activated receptor (PPAR)-γ coactivator (PGC)-1α via p38-mitogen-activated protein kinases (MAPK) pathway. This leads to mitochondrial fusion and biogenesis, significantly boosting T cell respiratory capacity (139). Such CAR-T configurations have proven clinically effective, enhancing antitumor immunity and long-term T cell survival in leukemia patients (140, 141). Notably, a recent publication has underscored the benefits of including PGC-1α in the CAR structure (142). Alternatively, Ligtenberg et al. described a different strategy involving the co-expression of catalase with the CAR construct (CAR-CAT) in adoptive T cells. This approach neutralizes H2O2 and bolsters cellular antioxidative capacity, thereby sustaining CAR-T viability and antitumor function under oxidative stress (143). The pre-conditioning employed to facilitate the engraftment of CAR-T cells, which results in increased levels of IL-2, IL-7, IL-15, and IL-21 in the circulation, offers additional opportunities to influence metabolism. IL-2 promotes glycolysis through the Akt-mTOR pathway, and differentiation of CD8+ T cells into an effector memory phenotype (144); IL-7 enhances memory CD8+ T cell formation by inducing the expression of glycerol channels and increasing triglyceride synthesis (145); IL-15 increases the spare respiratory capacity and oxidative metabolism rate by enhancing mitochondrial biogenesis and CPT1a (Carnitine Palmitoyl transferase 1a) expression, which results in a stem cell memory T cell phenotype (146); IL-21 modulates the induction of central memory T cells and acquisition of the exhaustion phenotype, increasing antitumor efficacy in an fatty acid oxidation-dependent manner (147). Furthermore, pre-treatment of CAR-T cells with compounds that modulate the PI3K-AKT-mTOR pathway, such as PI3K inhibitors, Rapamycin, AMPK activators like Metformin or MAPK inhibitors, have demonstrated the potential to enhance the fate and potency of CAR-T cells. This approach helps preserve memory-like characteristics by overall promotion of fatty acid oxidation and mitochondrial biogenesis (148–152). Finally, the use of mitochondria-targeted antioxidants offers a novel strategy to counteract cytotoxic T cell exhaustion. Antioxidant treatment effectively reduces the levels of ROS generated by chronically stimulated T cells and mitigates the decline in mitochondrial fitness (153).
In addition to these mechanisms, therapeutically shaping the local microenvironment to mitigate oxidative stress and foster redox homeostasis offers additional opportunities to protect/boost Tregs. In this context, the gut microbiota appears to be key. Thus, various metabolites produced by bacteria have been linked to immune homeostasis and systemic inflammation (154, 155). Some of them have a strong effect on Tregs: microbial short-chain fatty acids and tryptophan promote the suppressive function of Tregs (156, 157). On the other hand, certain bacteria possess the ability to generate and counteract reactive oxygen species (ROS), thereby regulating the host's redox state (158–160). Currently, innovative therapies are in development to modulate the gut microbiota, particularly in gastrointestinal diseases characterized by excessive ROS production, such as inflammatory bowel disease and colorectal cancer (161, 162). Further studies are needed to understand whether these approaches hold promise in supporting redox homeostasis and metabolic fitness of Tregs.
The mechanisms underlying the complex and interlink networks between redox homeostasis and metabolism as determinants of Treg fate are ill defined. In this review, we summarize the signaling and metabolic pathways that are known to be fine-tuned by oxidative stress in Tregs with special attention to the antioxidant mechanisms that coordinate many of the signaling pathways that contribute to metabolic rewiring, ultimately leading to Treg survival and function. In this regard, Tregs from healthy subjects express and secrete high levels of antioxidants, specially TRX1, indicating a higher endogenous antioxidant capacity than Tconv (121, 122). However, it is still controversial whether Tregs exhibit greater resistance to oxidative stress-induced cell death, particularly in the context of an oxidant/antioxidant imbalance (122, 123). Indeed, further insight is needed into the precise modulatory switches that may delineate Treg fate under steady state and the effect of oxidative stress and various pathological processes. It is also important to highlight the main challenges involved in the study of these interlinked networks. This includes the complexity of environmental cues: factors such as hypoxia, nutrient deprivation and the presence of a vast array of extracellular metabolites, all affecting intracellular regulatory mechanisms. Additionally, the heterogeneity of Tregs, activation states and interactions with other immune cells, will inevitably be governed by these mechanistic processes. Therefore, the future will see an integration of not only systems-based approaches, but computational and mathematical modeling of these cellular networks. The application of these approaches will advance our understanding of these complex networks and interactions as determinants of Treg function, inevitably leading to therapeutic targets in several conditions governed by aberrant immune responses.
EP: Conceptualization and Writing – original draft. AS-F: Writing – review & editing. NS: Conceptualization, Writing – original draft and Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article.
NS is recipient of a Wellcome Trust Fellowship (211113/A/18/Z). AS-F and EP were supported by the CRUK HUNTER (Hepatocellular Carcinoma Expediter Network). We additionally acknowledge the support of the Research Councils United Kingdom (RCUK) and the National Institute for Health Research (NIHR) Biomedical Research Centre based at Guy’s and St Thomas' NHS Foundation Trust and King’s College London.
AS-F is a founder of Quell Therapeutics Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Peoples JN, Saraf A, Ghazal N, Pham TT, Kwong JQ. Mitochondrial dysfunction and oxidative stress in heart disease. Exp Mol Med. (2019) 51:1–13. doi: 10.1038/s12276-019-0355-7
2. Lin T, Michael Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nat Rev. (2006) 443:787–95. doi: 10.1038/nature05292
3. Hayes JD, Dinkova-Kostova AT, Tew KD. Oxidative stress in cancer. Cancer Cell. (2020) 38:167–97. doi: 10.1016/j.ccell.2020.06.001
4. Di Dalmazi G, Hirshberg J, Lyle D, Freij JB, Caturegli P. Reactive oxygen species in organ-specific autoimmunity. Autoimmun Highlights. (2016) 7:1–11. doi: 10.1007/s13317-016-0083-0
5. Liebert MA, Jones DP. Redefining oxidative stress. Antioxid Redox Signal. (2006) 8:1865–79. doi: 10.1089/ars.2006.8.1865
6. Padgett LE, Tse HM. NADPH Oxidase–derived superoxide provides a third signal for CD4 T cell effector responses. J Immunol. (2016) 197:1733–42. doi: 10.4049/jimmunol.1502581
7. Hattori H, Subramanian KK, Sakai J, Jia Y, Li Y, Porter TF, et al. Small-molecule screen identifies reactive oxygen species as key regulators of neutrophil chemotaxis. Proc Natl Acad Sci U S A. (2010) 107:3546–51. doi: 10.1073/pnas.0914351107
8. Oberkampf M, Guillerey C, Mouriès J, Rosenbaum P, Fayolle C, Bobard A, et al. Mitochondrial reactive oxygen species regulate the induction of CD8+ T cells by plasmacytoid dendritic cells. Nat Commun. (2018) 9:1–14. doi: 10.1038/s41467-018-04686-8
9. Hildeman DA, Mitchell T, Teague TK, Henson P, Day BJ, Kappler J, et al. Reactive oxygen Species regulate activation-induced T cell apoptosis. Immunity. (1999) 10:735–44. doi: 10.1016/S1074-7613(00)80072-2
10. Cemerski S, van Meerwijk JPM, Romagnoli P. Oxidative-stress-induced T lymphocyte hyporesponsiveness is caused by structural modification rather than proteasomal degradation of crucial TCR signaling molecules. Eur J Immunol. (2003) 33:2178–85. doi: 10.1002/eji.200323898
11. Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol. (2012) 30:531–64. doi: 10.1146/annurev.immunol.25.022106.141623
12. Almeida L, Lochner M, Berod L, Sparwasser T. Metabolic pathways in T cell activation and lineage differentiation. Semin Immunol. (2016) 28:514–24. doi: 10.1016/j.smim.2016.10.009
13. Pearce EL, Poffenberger MC, Chang CH, Jones RG. Fueling immunity: insights into metabolism and lymphocyte function. Science (80-.). (2013) 342:1242454.1–11. doi: 10.1126/science.1242454
14. Procaccini C, De Rosa V, Galgani M, Abanni L, Calì G, Porcellini A, et al. An oscillatory switch in mTOR kinase activity sets regulatory T cell responsiveness. Immunity. (2010) 33:929–41. doi: 10.1016/j.immuni.2010.11.024
15. Kempkes RWM, Joosten I, Koenen HJPM, He X. Metabolic pathways involved in regulatory T cell functionality. Front Immunol. (2019) 10:2839.1–13. doi: 10.3389/fimmu.2019.02839
16. Gerriets VA, Kishton RJ, Johnson MO, Cohen S, Siska PJ, Nichols AG, et al. Foxp3 and toll-like receptor signaling balance treg cell anabolic metabolism for suppression. Nat Immunol. (2016) 17:1459–66. doi: 10.1038/ni.3577
17. Alissafi T, Kalafati L, Lazari M, Filia A, Kloukina I, Manifava M, et al. Mitochondrial oxidative damage underlies regulatory T cell defects in autoimmunity. Cell Metab. (2020) 32:591–604. doi: 10.1016/j.cmet.2020.07.001
18. Schofield CJ, Ratcliffe PJ. Oxygen sensing by HIF hydroxylases. Nat Rev Mol Cell Biol. (2004) 5:343–54. doi: 10.1038/nrm1366
19. Kaelin WG, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. (2008) 30:393–402. doi: 10.1016/j.molcel.2008.04.009
20. Nutsch K, Hsieh C. When T cells run out of breath: the HIF-1α story. Cell. (2011) 146:673–4. doi: 10.1016/j.cell.2011.08.018
21. Yamamoto A, Hester J, Macklin PS, Kawai K, Uchiyama M, Biggs D, et al. Systemic silencing of Phd2 causes reversible immune regulatory dysfunction. J Clin Invest. (2019) 129:3640–56. doi: 10.1172/JCI124099
22. Wigren M, Björkbacka H, Andersson L, Ljungcrantz I, Fredrikson GN, Persson M, et al. Low levels of circulating CD4 + FoxP3+ T cells are associated with an increased risk for development of myocardial infarction but not for stroke. Arterioscler Thromb Vasc Biol. (2012) 32:2000–7. doi: 10.1161/ATVBAHA.112.251579
23. Rohm I, Atiskova Y, Drobnik S, Fritzenwanger M, Kretzschmar D, Pistulli R, et al. Decreased regulatory T cells in vulnerable atherosclerotic lesions: imbalance between pro- and anti-inflammatory cells in atherosclerosis. Mediators Inflamm. (2015) 2015:364710.1–13. doi: 10.1155/2015/364710
24. Valencia X, Yarboro C, Illei G, Lipsky PE. Deficient CD4+CD25 high T regulatory cell function in patients with active systemic lupus erythematosus. J Immunol. (2007) 178:2579–88. doi: 10.4049/jimmunol.178.4.2579
25. Bending D, Pesenacker AM, Ursu S, Wu Q, Lom H, Thirugnanabalan B, et al. Hypomethylation at the regulatory T cell–specific demethylated region in CD25 hi T cells is decoupled from FOXP3 expression at the inflamed site in childhood arthritis. J Immunol. (2014) 193:2699–708. doi: 10.4049/jimmunol.1400599
26. Curiel TJ, Coukos G, Zou L, Alvarez X, Cheng P, Mottram P, et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat. Med. (2004) 10:942–9. doi: 10.1038/nm1093
27. Roychoudhuri R, Eil RL, Restifo NP. The interplay of effector and regulatory T cells in cancer. Curr Opin Immunol. (2015) 33:101–11. doi: 10.1016/j.coi.2015.02.003
28. Delgoffe GM, Woo SR, Turnis ME, Gravano DM, Guy C, Overacre AE, et al. Stability and function of regulatory T cells is maintained by a neuropilin-1-semaphorin-4a axis. Nature. (2013) 501:252–6. doi: 10.1038/nature12428
29. Saito T, Nishikawa H, Wada H, Nagano Y, Sugiyama D, Atarashi K, et al. Two FOXP3+CD4+T cell subpopulations distinctly control the prognosis of colorectal cancers. Nat Med. (2016) 22:679–84. doi: 10.1038/nm.4086
30. Turrens JF. Mitochondrial formation of reactive oxygen species. J Physiol. (2003) 552:335–44. doi: 10.1113/jphysiol.2003.049478
31. Cadenas E, Davies KJA. Mitochondrial free radical generation, oxidative stress, and aging. Free Radic Biol Med. (2000) 29:222–30. doi: 10.1016/S0891-5849(00)00317-8
32. Woolbright BL, Rajendran G, Harris RA, Taylor JA. Metabolic flexibility in cancer: targeting the pyruvate dehydrogenase kinase:pyruvate dehydrogenase axis. Mol Cancer Ther. (2019) 18:1673–81. doi: 10.1158/1535-7163.MCT-19-0079
33. O’Brien M, Chalker J, Slade L, Gardiner D, Mailloux RJ. Protein S-glutathionylation alters superoxide/hydrogen peroxide emission from pyruvate dehydrogenase complex. Free Radic Biol Med. (2017) 106:302–14. doi: 10.1016/j.freeradbiomed.2017.02.046
34. Starkov AA, Fiskum G, Chinopoulos C, Lorenzo BJ, Browne SE, Patel MS, et al. Mitochondrial α-ketoglutarate dehydrogenase complex generates reactive oxygen species. J Neurosci. (2004) 24:7779–88. doi: 10.1523/JNEUROSCI.1899-04.2004
35. Tretter L, Adam-Vizi V. Generation of reactive oxygen species in the reaction catalyzed by α-ketoglutarate dehydrogenase. J Neurosci. (2004) 24:7771–8. doi: 10.1523/JNEUROSCI.1842-04.2004
36. Zhang Y, Zhang M, Zhu W, Yu J, Wang Q, Zhang J, et al. Succinate accumulation induces mitochondrial reactive oxygen species generation and promotes status epilepticus in the kainic acid rat model. Redox Biol. (2020) 28:101365. doi: 10.1016/j.redox.2019.101365
37. Sun N, Joule RJ, Finkel T. The mitochondrial basis of aging nuo. Mol Cell. (2016) 315–316:654–66. doi: 10.1016/j.molcel.2016.01.028
38. Jackson SH, Devadas S, Kwon J, Pinto LA, Williams MS. T cells express a phagocyte-type NADPH oxidase that is activated after T cell receptor stimulation. Nat Immunol. (2004) 5:818–27. doi: 10.1038/ni1096
39. Emmerson A, Trevelin SC, Mongue-Din H, Becker PD, Ortiz C, Smyth LA, et al. Nox2 in regulatory T cells promotes angiotensin II–induced cardiovascular remodeling. J Clin Invest. (2018) 128:3088–101. doi: 10.1172/JCI97490
40. Trevelin SC, Zampetaki A, Sawyer G, Ivetic A, Brewer AC, Smyth LA, et al. Nox2-deficient tregs improve heart transplant outcomes via their increased graft recruitment and enhanced potency. JCI Insight. (2021) 6:e149301.1–11. doi: 10.1172/jci.insight.149301
41. Vaikunthanathan T, Landmann E, Correa DM, Romano M, Trevelin SC, Peng Q, et al. Dysregulated anti-oxidant signalling and compromised mitochondrial integrity negatively influence regulatory T cell function and viability in liver disease. eBioMedicine. (2023) In Press.37657135
42. Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. (2003) 23:8137–51. doi: 10.1128/MCB.23.22.8137-8151.2003
43. Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. (2007) 47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046
44. Hayes JD, Dinkova-Kostova AT. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem Sci. (2014) 39:199–218. doi: 10.1016/j.tibs.2014.02.002
45. Mak TW, Grusdat M, Duncan GS, Dostert C, Nonnenmacher Y, Cox M, et al. Glutathione primes T cell metabolism for inflammation. Immunity. (2017) 46:675–89. doi: 10.1016/j.immuni.2017.03.019
46. Lu SC. Regulation of glutathione synthesis. Mol Aspects Med. (2009) 30:42–59. doi: 10.1016/j.mam.2008.05.005
47. Holmgren A, Lu J. Thioredoxin and thioredoxin reductase: current research with special reference to human disease. Biochem Biophys Res Commun. (2010) 396:120–4. doi: 10.1016/j.bbrc.2010.03.083
48. Mougiakakos D, Johansson CC, Jitschin R, Böttcher M, Kiessling R. Increased thioredoxin-1 production in human naturally occurring regulatory T cells confers enhanced tolerance to oxidative stress. Blood. (2011) 117:857–61. doi: 10.1182/blood-2010-09-307041
49. Chi H. Regulation and function of mTOR signalling in T cell fate decisions. Nat Rev Immunol. (2012) 12:325–38. doi: 10.1038/nri3198
50. Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. (2009) 10:307–18. doi: 10.1038/nrm2672
51. Manning BD, Cantley LC. Rheb fills a GAP between TSC and TOR. TRENDS Biochem Sci. (2003) 28:573–6. doi: 10.1016/j.tibs.2003.09.003
52. Kwiatkowski DJ, Manning BD. Tuberous sclerosis: a GAP at the crossroads of multiple signaling pathways. Hum. Mol. Genet. (2005) 14:R251–8. doi: 10.1093/hmg/ddi260
53. Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. (2011) 12:21–35. doi: 10.1038/nrm3025
54. Turner MS, Kane LP, Morel PA. Dominant role of antigen dose in CD4+Foxp3+regulatory T cell induction and expansion. J Immunol. (2009) 183:4895–903. doi: 10.4049/jimmunol.0901459
55. Katzman SD, O'Gorman WE, Villarino AV, Gallo E, Friedman RS, Krummel MF, et al. Duration of antigen receptor signaling determines T-cell tolerance or activation. Proc Natl Acad Sci U S A. (2010) 107:18085–90. doi: 10.1073/pnas.1010560107
56. Colombetti S, Basso V, Mueller DL, Mondino A. Prolonged TCR/CD28 engagement drives IL-2-independent T cell clonal expansion through signaling mediated by the mammalian target of rapamycin. J Immunol. (2006) 176:2730–8. doi: 10.4049/jimmunol.176.5.2730
57. So T, Choi H, Croft M. OX40 Complexes with phosphoinositide 3-kinase and protein kinase B (PKB) to augment TCR-dependent PKB signaling. J Immunol. (2011) 186:3547–55. doi: 10.4049/jimmunol.1003156
58. Geng D, Zheng L, Srivastava R, Asprodites N, Velasco-Gonzalez C, Davila E. When toll-like receptor and T-cell receptor signals collide: a mechanism for enhanced CD8 T-cell effector function. Blood. (2010) 116:3494–504. doi: 10.1182/blood-2010-02-268169
59. Gerriets VA, Kishton RJ, Johnson MO, Cohen S, Siska PJ, Nichols AG, et al. Foxp3 and toll-like receptor signaling balance T reg cell anabolic metabolism for suppression. Nat Immunol. (2016) 17:1459–66. doi: 10.1038/ni.3577
60. Wofford JA, Wieman HL, Jacobs SR, Zhao Y, Rathmell JC. IL-7 promotes Glut1 trafficking and glucose uptake via STAT5-mediated activation of akt to support T-cell survival. Blood. (2008) 111:2101–11. doi: 10.1182/blood-2007-06-096297
61. Rao RR, Li Q, Odunsi K, Shrikant PA. The mTOR kinase determines effector versus memory CD8+ T cell fate by regulating the expression of transcription factors T-bet and eomesodermin. Immunity. (2010) 32:67–78. doi: 10.1016/j.immuni.2009.10.010
62. Liu G, Burns S, Huang G, Boyd K, Proia RL, Flavell RA, et al. The receptor S1P1 overrides regulatory T cell-mediated immune suppression through akt-mTOR. Nat. Immunol. (2009) 10:769–77. doi: 10.1038/ni.1743
63. Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, et al. The rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science (80-.). (2008) 320:1496–501. doi: 10.1126/science.1157535
64. Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Vasquez DS, Turk BE, et al. AMPK Phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. (2008) 30:214–26. doi: 10.1016/j.molcel.2008.03.003
65. Inoki K, Zhu T, Guan K-L. TSC2 Mediates cellular energy response to control cell growth and survival. Cell. (2003) 115:577–90. doi: 10.1016/S0092-8674(03)00929-2
66. Gan X, Wang J, Su B, Wu D. Evidence for direct activation of mTORC2 kinase activity by phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. (2011) 286:10998–1002. doi: 10.1074/jbc.M110.195016
67. Zinzalla V, Stracka D, Oppliger W, Hall MN. Activation of mTORC2 by association with the ribosome. Cell. (2011) 144:757–68. doi: 10.1016/j.cell.2011.02.014
68. Moloughney JG, Kim PK, Vega-Cotto NM, Wu C, Zhang S, Adlam M, et al. mTORC2 responds to glutamine catabolite levels to modulate the hexosamine biosynthesis enzyme GFAT1. Mol Cell. (2016) 63:811–26. doi: 10.1016/j.molcel.2016.07.015
69. Byun JK, Choi YK, Kim JH, Jeong JY, Jeon HJ, Kim MK, et al. A positive feedback loop between Sestrin2 and mTORC2 is required for the survival of glutamine-depleted lung cancer cells. Cell Rep. (2017) 20:586–99. doi: 10.1016/j.celrep.2017.06.066
70. Tao R, Xiong X, Liangpunsakul S, Dong XC. Sestrin 3 protein enhances hepatic insulin sensitivity by direct activation of the mTORC2-akt signaling. Diabetes. (2015) 64:1211–23. doi: 10.2337/db14-0539
71. Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, et al. The transcription factor myc controls metabolic reprogramming upon T lymphocyte activation ruoning. Immunity. (2011) 35:871–82. doi: 10.1016/j.immuni.2011.09.021
72. Doedens AL, Phan AT, Stradner MH, Fujimoto JK, Nguyen JV, Yang E, et al. Hypoxia-inducible factors enhance the effector responses of CD8+T cells to persistent antigen. Nat Immunol. (2013) 14:1173–82. doi: 10.1038/ni.2714
73. Facchinetti V, Ouyang W, Wei H, Soto N, Lazorchak A, Gould C, et al. The mammalian target of rapamycin complex 2 controls folding and stability of akt and protein kinase C. EMBO J. (2008) 27:1932–43. doi: 10.1038/emboj.2008.120
74. García-Martínez JM, Alessi DR. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1). Biochem J. (2008) 416:375–85. doi: 10.1042/BJ20081668
75. Ikenoue T, Inoki K, Yang Q, Zhou X, Guan KL. Essential function of TORC2 in PKC and akt turn motif phosphorylation, maturation and signalling. EMBO J. (2008) 27:1919–31. doi: 10.1038/emboj.2008.119
76. Battaglia M, Stabilini A, Migliavacca B, Horejs-Hoeck J, Kaupper T, Roncarolo MG. Rapamycin promotes expansion of functional CD4+CD25+FOXP3+regulatory T cells of both healthy subjects and type 1 diabetic patients. J Immunol. (2006) 177:8338–47. doi: 10.4049/jimmunol.177.12.8338
77. Valmori D, Tosello V, Souleimanian NE, Godefroy E, Scotto L, Wang Y, et al. Rapamycin-Mediated enrichment of T cells with regulatory activity in stimulated CD4+T cell cultures is not due to the selective expansion of naturally occurring regulatory T cells but to the induction of regulatory functions in conventional CD4+T cel. J Immunol. (2006) 177:944–9. doi: 10.4049/jimmunol.177.2.944
78. Turnquist HR, Raimondi G, Zahorchak AF, Fischer RT, Wang Z, Thomson AW. Rapamycin-Conditioned dendritic cells are poor stimulators of allogeneic CD4+T cells, but enrich for antigen-specific Foxp3+T regulatory cells and promote organ transplant tolerance. J Immunol. (2007) 178:7018–31. doi: 10.4049/jimmunol.178.11.7018
79. Strauss L, Czystowska M, Szajnik M, Mandapathil M, Whiteside TL. Differential responses of human regulatory T cells (treg) and effector T cells to rapamycin. PLoS One. (2009) 4:e5994. doi: 10.1371/journal.pone.0005994
80. Zeiser R, Leveson-Gower DB, Zambricki EA, Kambham N, Beilhack A, Loh J, et al. Differential impact of mammalian target of rapamycin inhibition on CD4 +CD25 + Foxp3 + regulatory T cells compared with conventional CD4+ T cells. Blood. (2008) 111:453–62. doi: 10.1182/blood-2007-06-094482
81. Delgoffe GM, Kole TP, Zheng Y, Zarek PE, Matthews KL, Xiao B, et al. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. (2009) 30:832–44. doi: 10.1016/j.immuni.2009.04.014
82. Haxhinasto S, Mathis D, Benoist C. The AKT-mTOR axis regulates de novo differentiation of CD4 +Foxp3 + cells. J Exp Med. (2008) 205:565–74. doi: 10.1084/jem.20071477
83. Chang X, Lazorchak AS, Liu D, Su B. Sin1 regulates treg-cell development but is not required for T-cell growth and proliferation. Eur J Immunol. (2012) 42:1639–47. doi: 10.1002/eji.201142066
84. Sauer S, Bruno L, Hertweck A, Finlay D, Leleu M, Spivakov M, et al. T cell receptor signaling controls Foxp3 expression via PI3K, akt, and mTOR. Proc Natl Acad Sci U S A. (2008) 105:7797–802. doi: 10.1073/pnas.0800928105
85. Gavin MA, Rasmussen JP, Fontenot JD, Vasta V, Manganiello VC, Beavo JA, et al. Foxp3-dependent programme of regulatory T-cell differentiation. Nature. (2007) 445:771–5. doi: 10.1038/nature05543
86. Lin W, Haribhai D, Relland LM, Truong N, Carlson MR, Williams CB, et al. Regulatory T cell development in the absence of functional Foxp3. Nat Immunol. (2007) 8:359–68. doi: 10.1038/ni1445
87. Hill JA, Feuerer M, Tash K, Haxhinasto S, Perez J, Melamed R, et al. Foxp3 transcription-factor-dependent and -independent regulation of the regulatory T cell transcriptional signature. Immunity. (2007) 27:786–800. doi: 10.1016/j.immuni.2007.09.010
88. Wilson MS, Metink-Kane MM. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol. (2011) 186:3299–303. doi: 10.4049/jimmunol.1003613
89. Shrestha S, Yang K, Guy C, Vogel P, Neale G, Program BS. Regulatory T cells require the phosphatase PTEN to restrain type 1 and follicular helper T-cell responses. Nat Immunol. (2015) 16:178–87. doi: 10.1038/ni.3076
90. Apostolidis SA, Rodríguez-Rodríguez N, Suárez-Fueyo A, Dioufa N, Ozcan E, Crispín JC, et al. Phosphatase PP2A is requisite for the function of regulatory T cells. Nat Immunol. (2016) 17:556–64. doi: 10.1038/ni.3390
91. Wei J, Long L, Yang K, Guy C, Shrestha S, Chen Z, et al. Autophagy enforces functional integrity of regulatory T cells by coupling environmental cues and metabolic homeostasis. Nat Immunol. (2016) 17:277–85. doi: 10.1038/ni.3365
92. Zeng H, Yang K, Cloer C, Neale G, Vogel P, Chi H. mTORC1 couples immune signals and metabolic programming to establish T reg-cell function. Nature. (2013) 499:485–90. doi: 10.1038/nature12297
93. Sun I-H, Oh M-H, Zhao L, Patel CH, Arwood ML, Xu W, et al. mTOR Complex 1 signaling regulates the generation and function of central and effector Foxp3+regulatory T cells. J Immunol. (2018) 201:481–92. doi: 10.4049/jimmunol.1701477
94. Chapman NM, Zeng H, Nguyen TLM, Wang Y, Vogel P, Dhungana Y, et al. mTOR coordinates transcriptional programs and mitochondrial metabolism of activated treg subsets to protect tissue homeostasis. Nat Commun. (2018) 9:2095.1–15. doi: 10.1038/s41467-018-04392-5
95. Yu F, Sharma S, Edwards J, Feigenbaum L, Zhu J. Dynamic expression of T-bet and GATA3 by regulatory T cells maintains immune tolerance. Nat Immunol. (2015) 16:1–23. doi: 10.1038/ni.3053
96. Kishore M, Cheung KCP, Fu H, Bonacina F, Wang G, Coe D, et al. Regulatory T cell migration is dependent on glucokinase-mediated glycolysis. Immunity. (2017) 47:875–889.e10. doi: 10.1016/j.immuni.2017.10.017
97. Bendavit G, Aboulkassim T, Hilmi K, Shah S, Batist G. Nrf2 transcription factor can directly regulate mTOR: linking cytoprotective gene expression to a major metabolic regulator that generates redox activity. J Biol Chem. (2016) 291:25476–88. doi: 10.1074/jbc.M116.760249
98. Shin JM, Kim MY, Sohn KC, Jung SY, Lee HE, Lim JW, Kim S, Lee YH, et al. Nrf2 negatively regulates melanogenesis by modulating PI3K/akt signaling. PLoS One. (2014) 9:1–7. doi: 10.1371/journal.pone.0096035
99. Klemm P, Rajendiran A, Fragoulis A, Wruck C, Schippers A, Wagner N, et al. Nrf2 expression driven by Foxp3 specific deletion of Keap1 results in loss of immune tolerance in mice. Eur J Immunol. (2020) 50:515–24. doi: 10.1002/eji.201948285
100. Tsai JJ, Velardi E, Shono Y, Argyropoulos KV, Holland AM, Smith OM, et al. Nrf2 regulates CD41 T cell–induced acute graft-versus-host disease in mice. Blood. (2018) 132:2763–74. doi: 10.1182/blood-2017-10-812941
101. Noel S, Martina MN, Bandapalle S, Racusen LC, Potteti HR, Hamad ARA, et al. T lymphocyte-specific activation of Nrf2 protects from AKI. J Am Soc Nephrol. (2015) 26:2989–3000. doi: 10.1681/ASN.2014100978
102. Kurniawan H, Franchina DG, Guerra L, Bonetti L, Soriano- L , Grusdat M, et al. Glutathione restricts serine metabolism to preserve regulatory T cell function. Cell Metab. (2020) 31:920–36. doi: 10.1016/j.cmet.2020.03.004
103. Shao D, Oka SI, Liu T, Zhai P, Ago T, Sciarretta S, et al. A redox-dependent mechanism for regulation of AMPK activation by thioredoxin1 during energy starvation. Cell Metab. (2014) 19:232–45. doi: 10.1016/j.cmet.2013.12.013
104. Chakraborty P, Chatterjee S, Kesarwani P, Thyagarajan K, Iamsawat S, Dalheim A, et al. Thioredoxin-1 improves the immunometabolic phenotype of antitumor T cells. J Biol Chem. (2019) 294:9198–212. doi: 10.1074/jbc.RA118.006753
105. Ichimura Y, Waguri S, Sou YS, Kageyama S, Hasegawa J, Ishimura R, et al. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol Cell. (2013) 51:618–31. doi: 10.1016/j.molcel.2013.08.003
106. Jain A, Lamark T, Sjøttem E, Larsen KB, Awuh JA, Øvervatn A, et al. P62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem. (2010) 285:22576–91. doi: 10.1074/jbc.M110.118976
107. Katsuragi Y, Ichimura Y, Komatsu M. Regulation of the Keap1–Nrf2 pathway by p62/SQSTM1. Curr Opin Toxicol. (2016) 1:54–61. doi: 10.1016/j.cotox.2016.09.005
108. Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. (2010) 12:213–23. doi: 10.1038/ncb2021
109. Ishii T, Mann GE. Redox status in mammalian cells and stem cells during culture in vitro: critical roles of Nrf2 and cystine transporter activity in themaintenance of redox balance. Redox Biol. (2014) 2:786–94. doi: 10.1016/j.redox.2014.04.008
110. Sha LK, Sha W, Kuchler L, Daiber A, Giegerich AK, Weigert A, et al. Loss of Nrf2 in bone marrow-derived macrophages impairs antigen-driven CD8+ T cell function by limiting GSH and cys availability. Free Radic Biol Med. (2015) 83:77–88. doi: 10.1016/j.freeradbiomed.2015.02.004
111. Ye P, Mimura J, Okada T, Sato H, Liu T, Maruyama A, et al. Nrf2- and ATF4-dependent upregulation of xCT modulates the sensitivity of T24 bladder carcinoma cells to proteasome inhibition. Mol Cell Biol. (2014) 34:3421–34. doi: 10.1128/MCB.00221-14
112. Procaccini C, Garavelli S, Carbone F, Di Silvestre D, La Rocca C, Greco D, et al. Signals of pseudo-starvation unveil the amino acid transporter SLC7A11 as key determinant in the control of treg cell proliferative potential. Immunity. (2021) 54:1543–1560.e6. doi: 10.1016/j.immuni.2021.04.014
113. Field CS, Baixauli F, Kyle RL, Puleston DJ, Cameron AM, Sanin DE, et al. Mitochondrial integrity regulated by lipid metabolism is a cell-intrinsic checkpoint for treg suppressive function. Cell Metab. (2020) 31:422–437.e5. doi: 10.1016/j.cmet.2019.11.021
114. Angelin A, Gil-de-Gómez L, Dahiya S, Jiao J, Guo L, Levine MH, et al. Foxp3 reprograms T cell metabolism to function in low-glucose, high-lactate environments. Cell Metab. (2017) 25:1282–1293.e7. doi: 10.1016/j.cmet.2016.12.018
115. Pacella I, Procaccini C, Focaccetti C, Miacci S, Timperi E, Faicchia D, et al. Fatty acid metabolism complements glycolysis in th selective regulatory t cell expansion during tumor growth. Proc Natl Acad Sci U S A. (2018) 115:E6546–55. doi: 10.1073/pnas.1720113115
116. Thomas JP, Maiorino M, Ursini F, Girotti AW. Protective action of phospholipid hydroperoxide glutathione peroxidase against membrane-damaging lipid peroxidation. In situ reduction of phospholipid and cholesterol hydroperoxides. J Biol Chem. (1990) 265:454–61. doi: 10.1016/S0021-9258(19)40252-4
117. Sattler W, Maiorino M, Stocker R. Reduction of HDL- and LDL-associated cholesterylester and phospholipid hydroperoxides by phospholipid hydroperoxide glutathione peroxidase and ebselen (PZ 51). Arch Biochem Biophys. (1994) 309(2):214–21. doi: 10.1006/abbi.1994.1105
118. Seiler A, Schneider M, Förster H, Roth S, Wirth EK, Culmsee C, et al. Glutathione peroxidase 4 senses and translates oxidative stress into 12/15-lipoxygenase dependent- and AIF-mediated cell death. Cell Metab. (2008) 8:237–48. doi: 10.1016/j.cmet.2008.07.005
119. Xu C, Sun S, Johnson T, Qi R, Zhang S, Zhang J, et al. The glutathione peroxidase Gpx4 prevents lipid peroxidation and ferroptosis to sustain treg cell activation and suppression of antitumor immunity. Cell Rep. (2021) 35:109235. doi: 10.1016/j.celrep.2021.109235
120. Matsushita M, Freigang S, Schneider C, Conrad M, Bornkamm GW, Kopf M. T cell lipid peroxidation induces ferroptosis and prevents immunity to infection. J Exp Med. (2015) 212:555–68. doi: 10.1084/jem.20140857
121. Feng P, Yang Q, Luo L, Sun Y, Lv W, Wan S, et al. The kinase PDK1 regulates regulatory T cell survival via controlling redox homeostasis. Theranostics. (2021) 11:9503–18. doi: 10.7150/thno.63992
122. Finlay DK, Rosenzweig E, Sinclair LV, Carmen FC, Hukelmann JL, Rolf J, et al. PDK1 Regulation of mTOR and hypoxia-inducible factor 1 integrate metabolism and migration of CD8+ T cells. J Exp Med. (2012) 209:2441–53. doi: 10.1084/jem.20112607
123. Oh H, Zhao J, Grinberg-Bleyer Y, Postler TS, Wang P, Park S-G, et al. PDK1 Is required for maintenance of CD4+Foxp3+regulatory T cell function. J Immunol. (2021) 206:1776–83. doi: 10.4049/jimmunol.2000051
124. Singer K, Gottfried E, Kreutz M, Mackensen A. Suppression of T-cell responses by tumor metabolites. Cancer Immunol Immunother. (2011) 60:425–31. doi: 10.1007/s00262-010-0967-1
125. Chang CH, Curtis JD, Maggi LB, Faubert B, Villarino AV, O'Sullivan D, et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. (2013) 153:1239. doi: 10.1016/j.cell.2013.05.016
126. Ho PC, Bihuniak JD, MacIntyre AN, Staron M, Liu X, Amezquita R, et al. Phosphoenolpyruvate is a metabolic checkpoint of anti-tumor T cell responses. Cell. (2015) 162:1217–28. doi: 10.1016/j.cell.2015.08.012
127. Macintyre AN, Gerriets VA, Nichols AG, Michalek RD, Rudolph MC, Deoliveira D, et al. The glucose transporter Glut1 is selectively essential for CD4 T cell activation and effector function. Cell Metab. (2014) 20:61–72. doi: 10.1016/j.cmet.2014.05.004
128. Watson MLJ, Vignali PDA, Mullett SJ, Overacre-Delgoffe AE, Peralta RM, Grebinoski S, et al. Metabolic support of tumour-infiltrating regulatory T cells by lactic acid. Nature. (2021) 591:645–51. doi: 10.1038/s41586-020-03045-2
129. Tauffenberger A, Fiumelli H, Almustafa S, Magistretti PJ. Lactate and pyruvate promote oxidative stress resistance through hormetic ROS signaling. Cell Death Dis. (2019) 10:653.1–16. doi: 10.1038/s41419-019-1877-6
130. Battaglia M, Stabilini A, Roncarolo MG. Rapamycin selectively expands CD4 + CD25 + FoxP3+regulatory T cells. Blood. (2005) 105:4743–8. doi: 10.1182/blood-2004-10-3932
131. Strauss L, Whiteside TL, Knights A, Bergmann C, Knuth A, Zippelius A. Selective survival of naturally occurring human CD4 + CD25 + Foxp3+ regulatory T cells cultured with rapamycin. J Immunol. (2007) 178:320–9. doi: 10.4049/jimmunol.178.1.320
132. Fraser H, Safinia N, Grageda N, Thirkell S, Lowe K, Fry LJ, et al. A rapamycin-based GMP-compatible process for the isolation and expansion of regulatory T cells for clinical trials. Mol Ther Methods Clin Dev. (2018) 8:198–209. doi: 10.1016/j.omtm.2018.01.006
133. Gedaly R, De Stefano F, Turcios L, Hill M, Hidalgo G, Mitov MI, et al. Mtor inhibitor everolimus in regulatory t cell expansion for clinical application in transplantation. Transplantation. (2019) 103:705–15. doi: 10.1097/TP.0000000000002495
134. Trzonkowski P, Bacchetta R, Battaglia M, Berglund D, Bohnenkamp HR, Ten Brinke A, et al. Hurdles in therapy with regulatory T cells. Sci Transl Med. (2015) 7:1–11. doi: 10.1126/scitranslmed.aaa7721
135. Boardman DA, Philippeos C, Fruhwirth GO, Ibrahim MAA, Hannen RF, Cooper D, et al. Expression of a chimeric antigen receptor specific for donor HLA class I enhances the potency of human regulatory T cells in preventing human skin transplant rejection. Am J Transplant. (2017) 17:931–43. doi: 10.1111/ajt.14185
136. Noyan F, Zimmermann K, Hardtke-Wolenski M, Knoefel A, Schulde E, Geffers R, et al. Prevention of allograft rejection by use of regulatory T cells with an MHC-specific chimeric antigen receptor. Am J Transplant. (2017) 17:917–30. doi: 10.1111/ajt.14175
137. Pierini A, Iliopoulou BP, Peiris H, Pérez-Cruz M, Baker J, Hsu K, et al. T cells expressing chimeric antigen receptor promote immune tolerance. JCI Insight. (2017) 2:e92865.1–17. doi: 10.1172/jci.insight.92865
138. Dawson NAJ, Rosado-Sánchez I, Novakovsky GE, Fung VCW, Huang Q, McIver E, et al. Functional effects of chimeric antigen receptor co-receptor signaling domains in human regulatory T cells. Sci Transl Med. (2020) 12:eaaz3866.1–16. doi: 10.1126/scitranslmed.aaz3866
139. Menk AV, Scharping NE, Rivadeneira DB, Calderon MJ, Watson MJ, Dunstane D, et al. 4-1BB Costimulation induces T cell mitochondrial function and biogenesis enabling cancer immunotherapeutic responses. J Exp Med. (2018) 215:1091–100. doi: 10.1084/jem.20171068
140. Brentjens RJ, Rivière I, Park JH, Davila ML, Wang X, Stefanski J, et al. Safety and persistence of adoptively transferred autologous CD19-targeted T cells in patients with relapsed or chemotherapy refractory B-cell leukemias. Blood. (2011) 118:4817–28. doi: 10.1182/blood-2011-04-348540
141. Porter DL, Hwang WT, Frey NV, Lacey SF, Shaw PA, Loren AW, et al. Chimeric antigen receptor T cells persist and induce sustained remissions in relapsed refractory chronic lymphocytic leukemia. Sci Transl Med. (2015) 7:1–13. doi: 10.1126/scitranslmed.aac5415
142. Lontos K, Wang Y, Joshi SK, Frisch AT, Watson MJ, Kumar A, et al. Metabolic reprogramming via an engineered PGC-1α improves human chimeric antigen receptor T-cell therapy against solid tumors. J Immunother Cancer. (2023) 11:1–10. doi: 10.1136/jitc-2022-006522
143. Ligtenberg MA, Mougiakakos D, Mukhopadhyay M, Witt K, Lladser A, Chmielewski M, et al. Coexpressed catalase protects chimeric antigen receptor–redirected T cells as well as bystander cells from oxidative stress–induced loss of antitumor activity. J Immunol. (2016) 196:759–66. doi: 10.4049/jimmunol.1401710
144. Liao W, Lin JX, Leonard WJ. Interleukin-2 at the crossroads of effector responses, tolerance, and immunotherapy. Immunity. (2013) 38:13–25. doi: 10.1016/j.immuni.2013.01.004
145. Cui G, Staron MM, Gray SM, Ho PC, Amezquita RA, Wu J, et al. IL-7-induced glycerol transport and TAG synthesis promotes memory CD8+ T cell longevity. Cell. (2015) 161:750–61. doi: 10.1016/j.cell.2015.03.021
146. Alizadeh D, Wong RA, Yang X, Wang D, Pecoraro JR, Kuo C, et al. IL15 Enhances CAR-T-cell antitumor activity by reducing mTORC1 activity and preserving their stem cell memory phenotype. Cancer Immunol Res. (2019) 7:759–72. doi: 10.1158/2326-6066.CIR-18-0466
147. Loschinski R, Böttcher M, Stoll A, Bruns H, Mackensen A, Mougiakakos D. IL-21 modulates memory and exhaustion phenotype of T-cells in a fatty acid oxidation-dependent manner. Oncotarget. (2018) 9:13125–38. doi: 10.18632/oncotarget.24442
148. Dwyer CJ, Arhontoulis DC, Rangel Rivera GO, Knochelmann HM, Smith AS, Wyatt MM, et al. Ex vivo blockade of PI3K gamma or delta signaling enhances the antitumor potency of adoptively transferred CD8+ T cells. Eur J Immunol. (2020) 50:1386–99. doi: 10.1002/eji.201948455
149. Urak R, Walter M, Lim L, Wong CLW, Budde LE, Thomas S, et al. Ex vivo akt inhibition promotes the generation of potent CD19CAR T cells for adoptive immunotherapy. J Immunother Cancer. (2017) 5:1–13. doi: 10.1186/s40425-017-0227-4
150. Mousset CM, Hobo W, Ji Y, Fredrix H, De Giorgi V, Allison RD, et al. Ex vivo AKT-inhibition facilitates generation of polyfunctional stem cell memory-like CD8+ T cells for adoptive immunotherapy. Oncoimmunology. (2018) 7:1–14. doi: 10.1080/2162402X.2018.1488565
151. Verma V, Jafarzadeh N, Boi S, Kundu S, Jiang Z, Fan Y, et al. MEK Inhibition reprograms CD8+ T lymphocytes into memory stem cells with potent antitumor effects. Nat Immunol. (2021) 22:53–66. doi: 10.1038/s41590-020-00818-9
152. Crompton JG, Sukumar M, Roychoudhuri R, Clever D, Gros A, Eil RL, et al. Akt inhibition enhances expansion of potent tumor-specific lymphocytes with memory cell characteristics. Cancer Res. (2015) 75:296–305. doi: 10.1158/0008-5472.CAN-14-2277
153. Yu YR, Imrichova H, Wang H, Chao T, Xiao Z, Gao M, et al. Disturbed mitochondrial dynamics in CD8+ TILs reinforce T cell exhaustion. Nat Immunol. (2020) 21:1540–51. doi: 10.1038/s41590-020-0793-3
154. Sharon G, Garg N, Debelius J, Knight R, Dorrestein PC, Mazmanian SK. Specialized metabolites from the microbiome in health and disease. Cell Metab. (2014) 20:719–30. doi: 10.1016/j.cmet.2014.10.016
155. Thorburn AN, Macia L, Mackay CR. Diet, metabolites, and ‘western-lifestyle’ inflammatory diseases. Immunity. (2014) 40:833–42. doi: 10.1016/j.immuni.2014.05.014
156. Arpaia N, Campbell C, Fan X, Dikiy S, Van Der Veeken J, Deroos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. (2013) 504:451–5. doi: 10.1038/nature12726
157. Roager HM, Licht TR. Microbial tryptophan catabolites in health and disease. Nat Commun. (2018) 9:1–10. doi: 10.1038/s41467-018-05470-4
158. Prete R, Garcia-Gonzalez N, Di Mattia CD, Corsetti A, Battista N. Food-borne lactiplantibacillus plantarum protect normal intestinal cells against inflammation by modulating reactive oxygen species and IL-23/IL-17 axis. Sci Rep. (2020) 10:1–11. doi: 10.1038/s41598-019-56847-4
159. Wang Y, Wu Y, Wang Y, Xu H, Mei X, Yu D, et al. Antioxidant properties of probiotic bacteria. Nutrients. (2017) 9:521.1–15. doi: 10.3390/nu9050521
160. Uchiyama J, Akiyama M, Hase K, Kumagai Y, Kim YG. Gut microbiota reinforce host antioxidant capacity via the generation of reactive sulfur species. Cell Rep. (2022) 38:110479. doi: 10.1016/j.celrep.2022.110479
161. Eeckhaut V, Machiels K, Perrier C, Romero C, Maes S, Flahou B, et al. Butyricicoccus pullicaecorum in inflammatory bowel disease. Gut. (2013) 62:1745–52. doi: 10.1136/gutjnl-2012-303611
Keywords: regulatory T cells, reactive oxygen species, anti-oxidants, NRF2, mTOR
Citation: Perpiñán E, Sanchez-Fueyo A and Safinia N (2023) Immunoregulation: the interplay between metabolism and redox homeostasis. Front. Transplant. 2:1283275. doi: 10.3389/frtra.2023.1283275
Received: 25 August 2023; Accepted: 13 November 2023;
Published: 27 November 2023.
Edited by:
Fadi Issa, University of Oxford, United KingdomReviewed by:
Masateru Uchiyama, Teikyo University, Japan© 2023 Perpiñán, Sanchez-Fueyo and Safinia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: N. Safinia bmlsb3VmYXIuMS5zYWZpbmlhQGtjbC5hYy51aw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.