- 1Department of Biotechnology and Systems Biology, National Institute of Biology, Ljubljana, Slovenia
- 2Faculty of Chemistry and Chemical Technology, University of Ljubljana, Ljubljana, Slovenia
- 3Department of Environmental Toxicology, Eawag—Swiss Federal Institute of Aquatic Science and Technology, Duebendorf, Switzerland
The last decade has seen the adverse outcome pathways (AOP) framework become one of the most powerful tools in chemical risk assessment, but the development of new AOPs remains a slow and manually intensive process. Here, we present a faster approach for AOP generation, based on manually curated causal toxicological networks. As a case study, we took a recently published zebrafish developmental neurotoxicity network, which contains causally connected molecular events leading to neuropathologies, and developed two new adverse outcome pathways: Inhibition of Fyna (Src family tyrosine kinase A) leading to increased mortality via decreased eye size (AOP 399 on AOP-Wiki) and GSK3beta (Glycogen synthase kinase 3 beta) inactivation leading to increased mortality via defects in developing inner ear (AOP 410). The approach consists of an automatic separation of the toxicological network into candidate AOPs, filtering the AOPs according to available evidence and length as well as manual development of new AOPs and weight-of-evidence evaluation. The semiautomatic approach described here provides a new opportunity for fast and straightforward AOP development based on large network resources.
Introduction
A decade ago, adverse outcome pathways (AOPs) (Ankley et al., 2010) have been put forward as a tool for organizing toxicological knowledge across different levels of biological organization, from the initial interaction of chemicals with the biological system (MIE = molecular initiating event) (Allen et al., 2014) to the individual and population level effects relevant for environmental risk assessment (AO = adverse outcome). The main idea of AOPs is collecting basic knowledge about biological systems and their chemical perturbations, and organizing it in easy to understand sequences of causally connected biological events (KE = key events; KER = key event relationship, i.e., how one KE is connected to another) (Villeneuve et al., 2014b) which allows risk assessors and toxicologists to identify chemicals likely to cause environmental harm. Additionally, through identification of knowledge gaps, AOPs inform future research and the development of novel biological assays that allow more specific in vitro chemical testing and reduction of animal testing (Groh et al., 2015). The long-term goal of the AOP framework is to develop AOPs that cover the whole space of chemically-induced biological perturbations and a complete set of assays required for comprehensive chemical risk assessment (Zupanic et al., 2018).
From their conception, several tools have been developed for easier development and management of AOPs. The AOP-wiki and the AOP knowledge base (https://aopwiki.org/; AOP-KB: https://aopkb.oecd.org/index.html) form an online portal hosted by the OECD that serves as the central AOP hub (Groh et al., 2015; Fay et al., 2017). The accepted core principles (Villeneuve et al., 2014a) and a handbook for AOP development (OECD iLibrary, 2021) serve as a standard that enables the development of high-quality and structurally similar AOPs, with comparable weight-of-evidence (WoE) evaluations. On top, several helper tools for AOP development and visualization have been made available to the community (e.g., http://datasciburgoon.github.io/aopxplorer). The AOPs in the AOP-wiki have been useful resources for quite diverse toxicological studies, e.g., to find chemicals likely to activate the AOPs (Jeong et al., 2019), to evaluate the hazard associated with specific chemicals and chemical groups (Carvaillo et al., 2019; Negi et al., 2021), to develop assays for in vitro assessment of mixture toxicity (Pistollato et al., 2020), to develop a new tiered testing approach for thyroid hormone disruptors (Knapen et al., 2020), to find the mechanisms of nanomaterial toxicity (Murugadoss et al., 2021) and to develop quantitative AOPs, mathematical models that can be used directly in chemical risk assessment (Margiotta-Casaluci et al., 2016; Doering et al., 2018; Perkins et al., 2019; Burgoon et al., 2020; Lillicrap et al., 2020).
As of August 2021, more than 400 AOPs and 5017 KEs have been developed [a large increase from 219 AOPs of April 2018 (Pollesch et al., 2019)], covering a range of species from nematodes to humans. However, these still cover only a very small part of the biological perturbations caused by chemical exposure and also only a few taxonomic groups. AOP development remains relatively slow, because each AOP requires searching for scientific literature, its manual curation, the formatting of the acquired knowledge into a user-friendly AOP and performing a WoE assessment (Vinken, 2013). To come closer to the final goal of a complete AOP space, the development of new AOPs needs to be accelerated.
There have been some attempts to do this already. The United States Environmental Protection Agency (EPA) has recently developed the Adverse Outcome Pathway database (AOP-DB), which can help with the annotation of AOP pathways under development, by connecting the information present on the AOP-wiki with various public resources [e.g., the NCBI gene, STRING (Szklarczyk et al., 2021) and Comparative Toxicogenomics Database (CTD) (Pittman et al., 2018; Davis et al., 2020)]. Other studies have tried to integrate publicly available resources [e.g., (ToxCast (Dix et al., 2007), CTD, Reactome (Jassal et al., 2020)] to develop AOP-like networks, which can serve as starting point for computationally predicted AOPs (cpAOPs). Several studies have used association rule mining to generate computationally predicted AOPs, mostly at the molecular level (Bell et al., 2016; Oki et al., 2016). Doktorova et al. have further developed a filtering approach for refining the molecular AOP-like networks using gene expression data from TG-Gates database (Igarashi et al., 2015) and manual curation to arrive at a putative AOP-network ending in the AO non-genotoxic induced hepatocellular carcinoma (Doktorova et al., 2020). The AOP-helpFinder tool uses text mining to find potential connections between key events (Jornod et al., 2021), while the computational pipeline developed by Jin et al. uses chemical-specific toxicogenomic data, pathway enrichment analysis and biomarker selection to develop putative cpAOPs (Jin et al., 2021).
Here we present a new approach for development of AOPs, based on a thus far AOP-untapped toxicological resource—causal toxicological networks (CTN) (Boué et al., 2015). CTNs are computational networks, which describe causally connected, mostly molecular, cellular and tissue events. We hypothesize that CTNs are especially appropriate as a starting resource for AOP development, as they are highly curated and all the connections in such networks are annotated by evidence. Compared to de novo AOP development, starting from such a network resource should decrease the time needed to search and curate scientific literature, but, since the CTNs are made from connected nodes and relationships between them, also make it easier to format the AOP into KE and KERs. The described approach is semiautomatic. It allows for automatic generation of a large number of candidate AOPs from a CTN, while the WoE evaluation and formatting of the evidence remains a manual process, as described by (Becker et al., 2015).
As a case study, we developed two developmental neurotoxicity AOPs. The choice of developmental neurotoxicity was made, because AOPs related to developmental neurotoxicity are still underrepresented in the AOP-wiki (Bal-Price et al., 2015; Knapen et al., 2015; Pistollato et al., 2020) and because of the recent activity in incorporation of AOP-based developmental neurotoxicity in vitro assays into chemical risk assessment (Sachana et al., 2021). This also allowed us to use our recently developed zebrafish causal developmental neurotoxicity network as starting point (Li et al., 2021). The new AOPs can be found on the AOP-wiki under Ids 399 and 410 (https://aopwiki.org/aops/399 and https://aopwiki.org/aops/410). Here, we present the details of the taken approach, the analysis of the differences between the CTNs and AOPs and the consequences for using the former as a source for the latter. We also provide tools and guidance for the development of AOPs from large network resources for the toxicological community.
Methods
From the Causal Toxicological Network to AOP Candidates
The zebrafish causal developmental neurotoxicity network, which is the basis of our AOP development, has been developed in an earlier publication, therefore we here only summarize the most relevant properties of the network for this study, and refer the reader to original publication for details (Li et al., 2021). CTN network development usually starts from a known adverse outcome of interest. For the network used as starting point for AOP development here, these were known zebrafish developmental nervous system-related pathologies (megalencephaly, microcephaly, microphthalmos, seizures, neurogenic inflammation, hydrocephalus). Then evidence for events leading to the pathologies are found in the scientific literature, specifically the evidence needs to be present in a peer reviewed publication. The scientific literature is queried using search engines such as Google Scholar, using keywords representing the specific molecular events. The evidence needs to show causality, i.e., a performed upstream perturbation leading to a measured downstream perturbation. With literature curation, upstream nodes directly affecting the pathology are added, then further upstream nodes directly connected to these nodes and so on, until no more evidence (in the form of causal experimentally validated relationships) is found in the literature. From the time of the first publication of this causal developmental neurotoxicity network, the network has been further extended and the version used as starting point in this paper (NTOX_BEL; Supplementary Material) features 515 nodes (ranging from protein activations and gene ontology terms to pathologies), with 682 edges between them. It was built based on evidence gathered from 90 zebrafish specific scientific articles and each edge in the network is annotated by the evidence behind it in the form of a BEL statement (Slater, 2014) and the reference from which the evidence is taken.
The original toxicological network (NTOX_BEL, Supplementary Material) contained nodes (molecular events) connected by edges (causal relationships between the nodes) and was written in Biological Expression Language (BEL) formalism. From the AOP development point of view it contained several non-necessary nodes (e.g., mRNA expression or protein activation of the same gene). To make it easier to manipulate with standard network analysis tools, we first generated a simplified abstracted network (NTOX_ABSTRACTED, Supplementary Material), i.e., the BEL formalism was simplified by removing unnecessary details from the BEL node and edge definitions (Figures 1A,1B; Supplementary Table S1). This abstracted network was then imported into Cytoscape [v3.8.0 (Shannon et al., 2003)] and further reduced by removing all self-loops. Finally, all nodes not connected to pathologies were removed by first selecting all nodes that had no outgoing connections, and then deleting anything that was not a pathology (Figures 1B,C). This step had to be repeated several times, until all the nodes in the network were connected to a pathology.
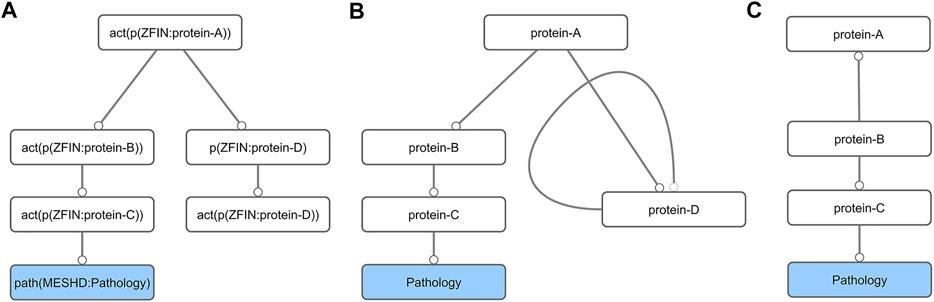
FIGURE 1. Schematic diagram of the network preparation procedure. First, the BEL network (A) is converted into its abstract form (B). This simplification results in the formation of self-loops (protein-D), which are removed together with all nodes that do not lead to a pathology node (C).
In addition to original annotations that the abstracted network inherited from the BEL formalism (e.g., references, experimental method), nodes were further manually curated and annotated. Each node in the network was first defined as a 1) coding or non-coding gene, the former further split into enzymes, transcription factors, receptors and transporters/channels, depending on the known or predicted function they perform (information taken from ZFIN and/or Gene Cards websites); 2) small molecule, which can be a metabolite (internal to the organism) or a chemical (externally added) (information taken from Chebi and/or KEGG databases; 3) biological process or a 4) pathology. All coding or non-coding genes were further annotated with gene identifiers (info taken from ZFIN, NCBI and/or ENSEMBL) and gene descriptions (ZFIN and UniProt). All nodes annotated as transporter/channel proteins, receptor proteins, enzymes and all targets of small molecules (metabolite or chemical) in the network were then marked as candidate MIE. These categories were chosen because they represent a vast majority of all MIEs currently in the AOP-wiki.
Next, all simple paths (defined as linear paths with every node present only once in a path) in the abstracted network starting at candidate MIEs and ending in nodes annotated as pathologies were found with the igraph package [v1.2.5 (Csardi and Nepusz, 2006)] in R [v4.0.2 (R Core Team, 2020)]. The resulting lists of simple paths were reformatted with a set of custom scripts (Supplementary Material) and form the basis for our choice of candidate AOPs to develop further.
From all candidate AOPs, we selected two that would be further developed into full feature AOPs, with additional condition that the new AOPs cannot be similar to any AOPs already present in the AOP-Wiki (on date: 10 Jan 2021) and that the new AOPs must feature more than two events above the cellular/tissues KE level. All the scripts for the network manipulations and instructions on how to use them can be found in the Supplementary material.
Adverse Outcome Pathway Development and Weight-Of-Evidence Analysis
Although evidence on the KERs was already provided in the original toxicological network, they were based on a few articles, which were focused mostly on the causality between molecular events in zebrafish (so, mostly on biological plausibility of the KERs). This by itself is not enough for AOP development or WoE evaluation. We have therefore performed an additional search for evidence in the literature, seeking additional empirical evidence on dose and time concordance between connected KEs, additional evidence on the essentiality of the KEs and evidence on taxonomic applicability of the KERs. This search was performed using the search engines Google Scholar and Web of Science, using keywords relating to the key events. Additionally, papers citing the evidence already present in the CTN and scientific papers cited in the CTN evidence were used in the search. Also, as the candidate AOPs obtained from the CTN ended in a neuropathology, which conforms nicely to an organ/tissue level KE, we needed to find additional evidence to link this KE to AOs at the individual and population level (as per AOP definition). For all KEs and KERs in the two AOPs, we searched for new evidence through keyword based searches in GoogleScholar/Pubmed, but also took advantage of information already available in the AOP-wiki. We did this by connecting the KEs obtained from the CTN to KEs and KERs that have already been developed on the AOP-Wiki by other researchers. Whenever we made such a connection, we analyzed the evidence already present on the AOP-Wiki and contacted the authors of the KEs to ask for permission to add additional evidence to their KEs and KERs and to connect our AOP under development to them. After collecting all evidence, we performed a WoE evaluation.
The AOPs were developed based on the OECD Users’ handbook for developing and assessing AOPs (OECD, 2014). According to OECD instructions, the level of biological organization, taxonomic, life stage and sex applicability of the KE, as well as how the KE can be measured, were described. The description of KERs was based on their biological plausibility. WoE assessment of the AOPs was done through application of the modified Bradford-Hill criteria for biological plausibility and empirical evidence of KERs and essentiality of KEs (Becker et al., 2015). The WoE assessments were conducted for the individual KERs and KEs of the AOP as well as for the overall AOP. The level of support for the KEs and KERs was classified as high, moderate or low, using the criteria outlined in the OECD Users’ handbook (OECD, 2014).
Results
From Causal Networks to Candidate Adverse Outcome Pathways
The first step of the network analysis was cleaning the original toxicological network (NTOX_BEL), i.e., adding and removing annotations and joining network nodes representing the same genes (NTOX_ABSTRACTED), then removing self-loops and unconnected components and finally removing all nodes that are not directly connected to pathologies. Table 1 shows how the size of the network changed with each manipulation step, going from 515 nodes and 682 edges in the original network to 125 nodes and 212 edges after the final step (Figure 2, Supplementary material)
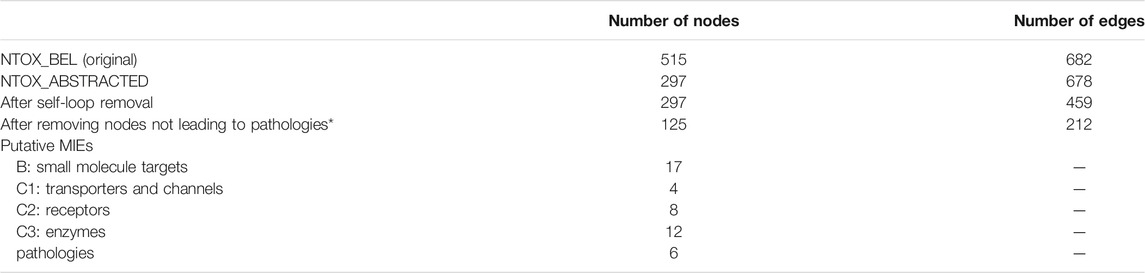
TABLE 1. Zebrafish causal developmental network reduction through the different network manipulation steps described in Materials and Methods.
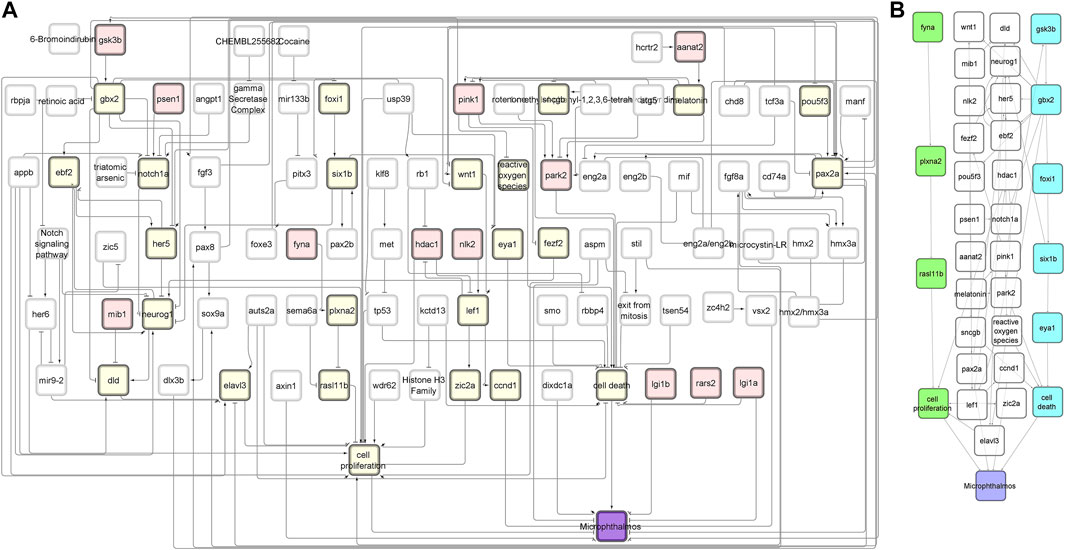
FIGURE 2. NTOX network. (A) The reduced NTOX network (125 nodes, 212 connections) where the start nodes are enzymes (red) and the final node is microphthalmos (purple). All other nodes that are part of the candidate AOPs are marked in yellow. (B) The subnetwork of the candidate AOPs that end with microphthalmos. The two candidate AOPs that were further developed into AOPs are marked in green (AOP 399) and blue (AOP 410).
The network was separated into individual simple paths from candidate MIE nodes to pathology nodes. In total, 391 simple paths were found to exist and be reachable between the candidate MIE and the pathologies (Table 2). We call these paths candidate AOP from here on, although they are not yet connected to an individual or population level AO. When connecting all simple paths from individual type of MIE to each pathology into subnetworks, the microphthalmos and microcephaly subnetworks were the largest, with more than 100 simple paths leading to each of them, none were found for megalencephaly, while only a couple of simple paths leading to the other four pathologies (Table 2). To reduce the number of simple paths further, we removed all those paths that had less than two molecular level KEs. We decided to do this, because the strength of the original toxicological network lies at the molecular level, therefore it makes more sense to use the toxicological network to develop AOPs with a strong molecular KE component. This reduced the total number of candidate AOPs to 329 (Table 2).
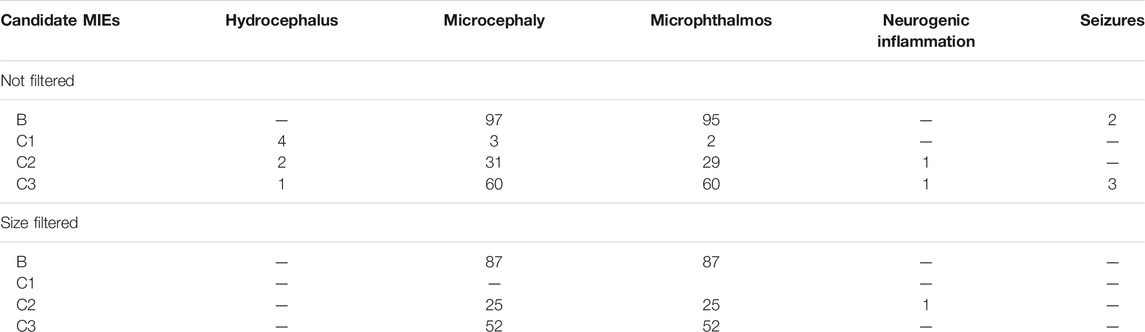
TABLE 2. The number of simple paths (candidate AOPs) found for each candidate MIE node type/pathology combination.
Adverse Outcome Pathway Development
Since even after network manipulations described in the previous section, there were still many candidate AOPs left with high-quality evidence, we were not able to come to a choice of best candidate AOPs based on the list alone. We therefore arbitrarily selected two candidate AOPs ending in microphthalmos (reduced eye size), which matched the scientific interest of the authors in the impairment of sensory organ development.
As has become custom in publications describing AOPs, the following AOP descriptions include the relevant chemical initiators of the AOPs, the KEs and KERs, the associated WoE for each, and the methods available for KE measurement (LaLone et al., 2017). As the focus of this paper is to evaluate WoE associated with AOPs generated from CTNs, biological plausibility of the KERs, essentiality of the KEs and empirical evidence as well as lack thereof are all discussed in detail.
Contrary to the authors of the earliest adverse outcome pathways in the AOP-Wiki, who had to describe all the KEs and KERs anew, some of the KEs and KERs of the new AOPs developed in this study were already described in the AOP-Wiki. We were thus able to take advantage of these descriptions, and only added new evidence when necessary to fit better with the zebrafish focus of our work. In this paper, for all KEs and KERs that are part of the new AOPs described here, but have been started by other researchers, we provide url links to the AOP-wiki. It is also worth mentioning that the AOP description in the following pages present a detailed, but still partly summarized description of all the information available on the AOP-Wiki. For a complete description, the reader is therefore invited to look up the AOPs 399 and 410 in whole on the AOP-Wiki.
Adverse Outcome Pathway Description
Inhibition of Fyna Leading to Increased Mortality via Decreased Eye Size (Microphthalmos) [AOP 399] https://aopwiki.org/aops/399
The first AOP to be developed was the »Inhibition of Fyna leading to increased mortality via decreased eye size (Microphthalmos)«, which can be found in the AOP-Wiki under AOP 399. The simple path that served as a basis for this AOP was found in the subnetwork containing all simple paths starting with enzymes and ending with microphthalmos (Figure 2A). The starting point of the development of AOP 399 was the MIE: inhibition of Fyna and the ending point the organ level KE: Decreased eye size. Since the original toxicological network did not feature any nodes interpretable as adverse outcomes at the individual or population level, we had to find additional evidence leading from KE: decreased eye size downstream to make a complete AOP. The final AOP 399 is visualized in Figure 3.
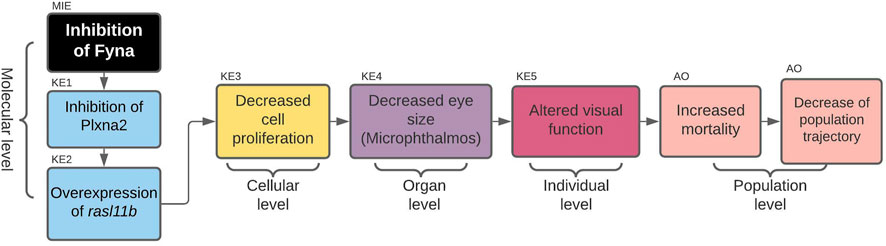
FIGURE 3. AOP 399. Inhibition of Fyna leads to increased mortality via decreased eye size (microphthalmos).
Chemical Initiators
Src family tyrosine kinase A (Fyna) is known to be inhibited by the herbal plant secondary metabolite rosmarinic acid (Jelić et al., 2007), by the microbial alkaloid staurosporine (Kinoshita et al., 2006) and saracatinib, a dual src family kinases (SFK) inhibitor, considered as a candidate drug for thyroid cancer and Alzheimer’s disease (Green et al., 2009). None of these chemical initiators is currently expected to be found in the environment in concentrations causing immediate concern. However, as FYN (the human ortholog of Fyna) is a potential target for medical treatments, drugs targeting FYN have the potential to become environmentally relevant pollutants.
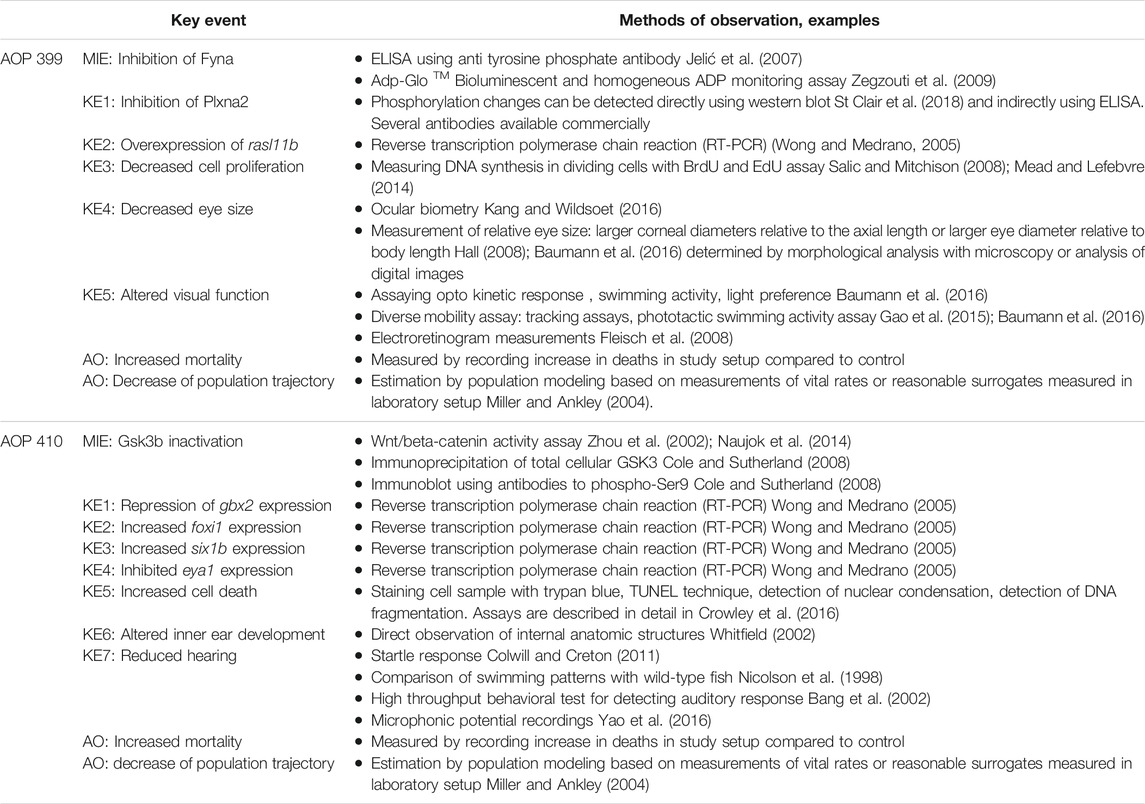
MIE: Inhibition of Fyna
Fyna is a member of non-receptor tyrosine kinases, a group of enzymes that, after activation, transmit signals from several surface receptors to target proteins by phosphorylating tyrosine residues (Sen and Johnson, 2011). In zebrafish, Fyna was shown to be involved in several neurodevelopmental processes, such as gastrulation, adherens junction maintenance and regulation of axon growth, and to be required for normal brain development (Sharma et al., 2005). The human ortholog of fyna (FYN) is implicated in Alzheimer’s disease and schizophrenia (Challa and Chatti, 2013).
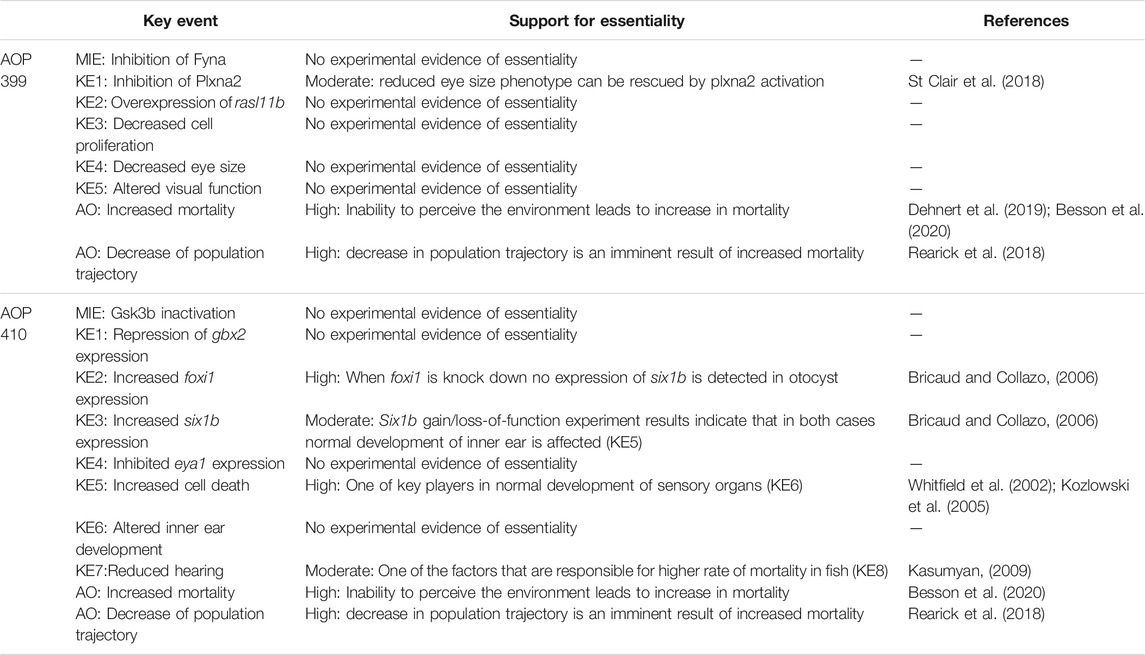
Essentiality—no experimental evidence found.
KER1: Inhibition of Fyna Leads to Inhibition of Plxna2
In zebrafish, mouse and human cell lines, Fyna was shown to constitutively associate with and phosphorylate the intracellular region of Plexin A1a (Plxna1) and Plxna2 (Sasaki et al., 2002; St Clair et al., 2018). To show that FYN induces PLXNA2 tyrosine phosphorylation, HEK293 cells were transfected with expression plasmids encoding PLXNA2 and either Fyn wild-type (WT) or a kinase dead (KD) point mutant. PLXNA2 showed prominent tyrosine phosphorylation when FYN WT was expressed and this phosphorylation was absent when FYN KD was expressed (St Clair et al., 2018). In zebrafish, it was found that Fyna-dependent Plxna2 phosphorylation is critical for zebrafish eye development in vivo. Antisense oligonucleotides targeted to plxna2 resulted in decreased proliferation of retinal precursor cells (Emerson et al., 2017).
Biological plausibility—moderate. Relationship evidence of phosphorylating activity of Fyna kinase and consequent changes in activity of Plxna2 is sufficiently researched and described for vertebrate models (Sasaki et al., 2002; Franco and Tamagnone, 2008; St. Clair et al., 2018). Fyna phosphorylation of intracellular domains of Plxna2 is crucial for translation of Plxna2 signals and normal development of eyes in zebrafish embryos (St. Clair et al., 2018). For better understanding of the relationship, data on the chemical inhibition of zebrafish Fyna kinase, its effect on phosphorylation of Plxna2 tyrosine residues, and further signaling is needed.
KE1: Inhibition of Plxna2
Plexins (Plxns) are receptors encoded by the members of the plexin gene family. They are the primary transducers of vertebrate Semaphorin (Sema) signals (Tamagnone et al., 1999). Semaphorins are members of a large gene family of secreted and membrane-anchored proteins, initially characterized as axon guidance factors (Nakamura et al., 2000). The receptors belonging to the Plexin family function as Semaphorin receptors (Neufeld and Kessler, 2008). Semas were initially discovered with respect to their role as repulsive guidance cues for migrating axons, although it is now appreciated that they have much broader roles in development. Semas and Plxns have tissue-specific expression patterns, and many Semas can signal through multiple Plxn family members (Luo et al., 1993). Plxna2 is involved in optic vesicle formation, and is predicted to localize to integral components of the plasma membrane and the Semaphorin receptor complex. It has been shown to be critical to zebrafish eye development (Ebert et al., 2014; St Clair et al., 2018).
Essentiality—moderate. Inhibition of Fyna kinase inhibits the function of plxna2 because of the lack of tyrosine residue phosphorylation in plxna2 and causes the reduction of zebrafish eye size. The same was also achieved by mutating the tyrosine residues of plxna2 to phenylalanine. However, the reduced eye size phenotype can be rescued by adding wild-type human PLXNA2 mRNA to the larva, indicating essentiality of plxna2 for this AOP (St. Clair et al., 2018).
KER2: Inhibition of Plxna2 Leads to Overexpression of rasl11b
RAS-like, family 11, member B (Rasl11b) is negatively regulated downstream of Sema6a/Plxna2 (semaphoring 6A) signaling and when overexpressed, decreases retinal precursor cells proliferation and eye size (Emerson et al., 2017). Rasl11b was found to be a target of Plxna2 regulation in a microarray study using zebrafish embryos deficient in either sema6a or plxna2. Rasl11b had a 2.18 log-fold change in expression (logFC) in sema6a morphants and a 1.58 logFC in plxna2 morphants. The microarray results were confirmed in independent experiments, using RT-PCR as readout. Further characterization rasl11b revealed its role in regulating retinal progenitor cell (RPC) proliferation.
Biological plausibility—low. Current knowledge of interactions between Plxna2 and Rasl11b are consistent in scientific literature, but there are one or more missing molecular links between the KE1 and KE2.
KE2: Overexpression of rasl11b
Rasl11b is a member of the small GTPase protein family with a high degree of similarity to RAS proteins (Stolle et al., 2007). Rasl11b is highly conserved among vertebrates, sharing on average 94% homology with its mammalian orthologs. In zebrafish, it has been shown to be involved in mesendoderm development (Pézeron et al., 2008). Ras proteins are otherwise known to be involved in the mitogen-activated protein kinase (MAPK) pathway (Emerson et al., 2017).
Essentiality—no experimental evidence found.
KER3: Overexpression of rasl11b Leads to Decreased Cell Proliferation
It is hypothesized that Rasl11b acts as a negative regulator of MAPK by outcompeting Ras for its effectors such as Raf, leading to decreased cell proliferation. In zebrafish, it has been shown that after overexpression of rasl11b, the G0/G1 phase cell cycle arrest is induced and the proliferation of retinal progenitor cells (RPCs) is inhibited in a dose-dependent manner (Emerson et al., 2017).
Biological plausibility—high. Impact of Rasl11b on overall proliferation is scientifically well supported, and the evidence from studies on zebrafish (Emerson et al., 2017) and human cancer cells (He et al., 2018) is consistent.
KE3: Decreased Cell Proliferation (https://aopwiki.org/events/1821)
Essentiality—no experimental evidence found.
KER4: Decreased cell proliferation leads to decreased eye sizeIn the same overexpression experiment described above, it was also determined that overexpression of rasl11b resulted in smaller eyes. It was confirmed through morpholino knockdown, that Sema6a/Plxna2 signaling regulates proliferation and cohesive migration of RPCs in developing optic vesicles in zebrafish (Emerson et al., 2017).
Biological plausibility—high. There is extensive evidence linking decreased proliferation of retinal precursor cells to decreased eye size in vertebrates. For the relationship support there are numerous studies of the effect of knockdown/knockout genes (rasl11b, rx3, vsx2) in vertebrates involved in the proliferation of retinal progenitor cells, which leads to reduced size of the eye or even absence of it (Burmeister et al., 1996; Kennedy et al., 2004; Graw, 2010; Zagozewski et al., 2014; Emerson et al., 2017). Also there are some studies of stressor (DEAB, citral) effects to support the relationship (Marsh-Armstrong et al., 1994; Le et al., 2012)
KE4: Decreased Eye Size (https://aopwiki.org/events/1878)
Essentiality—no experimental evidence found.
KER5: Decreased Eye Size Leads to Altered Visual Function (https://aopwiki.org/relationships/2377)
Biological plausibility - high. The impact of changes to the eye size on altered visual function has been well established and researched. Although the change in eye size is not the only indicator of impaired eye function, it is a good indicator of changes in eye development that can lead to impaired eye function.
KE5: Altered Visual Function (https://aopwiki.org/events/1643)
Essentiality—no experimental evidence found.
KER6: Altered Visual Function Leads to Increased Mortality (https://aopwiki.org/relationships/2375)
Biological plausibility—high. The link between impaired visual function and increased mortality is well accepted and proven. Evidence has been obtained in fish that have been exposed during development to various disruptors that affect eye development.
AO: Increased Mortality (https://aopwiki.org/events/351)
Essentiality—high. Mortality on individual level must increase for population trajectory to decrease.
Overall Assessment of the AOP 399
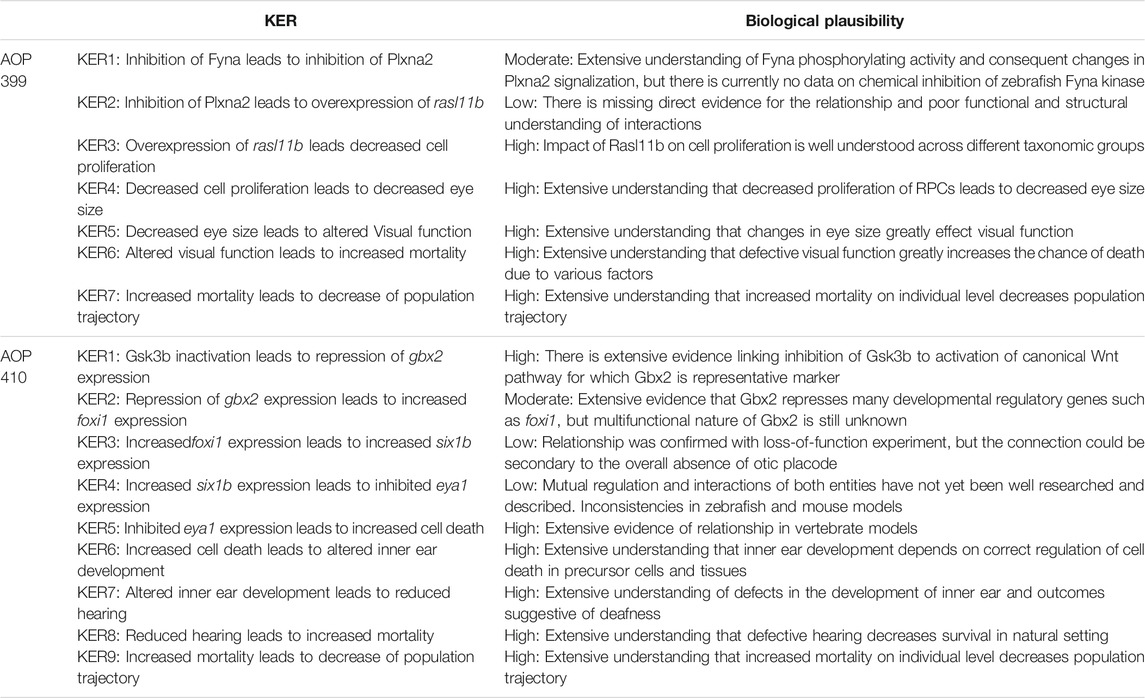
An overall assessment of this AOP shows that there is moderate biological plausibility to support a qualitative link between the Fyna kinase inhibition to the KE5 of altered visual function and high evidence linking KE5 to decreased population trajectory. Biological plausibility is considered moderate because there is ample evidence from gain- and loss- of function experiments and knock out animal models that support the relationships between key events and are consistent with current biological knowledge. A score of high in this respect would require further evidence for chemical inhibition or experimental downregulation of zebrafish Fyna kinase and direct or more extensive evidence linking Plxna2 inhibition to rasl11b overexpression. No empirical evidence in the form of dose response, time concordance and incidence concordance was found for the KERs of the AOP, and only little evidence for the essentiality of the KEs. The summarized information on KER biological plausibility, KE essentiality and KE observation methods of AOP 399 can be found in Tables 3–5.
Gsk3beta Inactivation Leads to Defects in Developing Inner Ear and Consequently to Increased Mortality [AOP 410] https://aopwiki.org/aops/410
The linear path from which we started to develop our second AOP (AOP 410) was located in the subnetwork that contained all linear paths that started with enzymes and led to microphthalmia (reduced eye size) (Figure 2). By comparing the above sentence to the title of the AOP, it is going to be immediately clear to the reader that the development of AOP 410 did not go as smoothly as for AOP 399 and we have ended up developing a different AOP than was initially planned.
This is because, during the review of the evidence for AOP 410, we encountered a problem: Evidence linking cell death to microphthalmos was inconsistent with evidence for upstream KERs. Namely, all additional evidence we have found for upstream KERs referred to perturbations during inner ear development (Solomon et al., 2003; Kozlowski et al., 2005; Bricaud and Collazo, 2006) while the KER linking cell death and microphthalmia in the original CTN referred to cell death in developing eye (Tsai et al., 2015; Schultz et al., 2018). Since the evidence did not relate to a common pathology, we reversed back to the full version of the toxicological network to see if any nodes have been lost while applying network filters. We found that the selected pathway upstream of cell death also led to node otic vesicle formation (Figure 4) which got removed during filtering, since it did not lead to any pathologies that was selected to be part of the original neurotoxicity network. As the evidence throughout the pathway was therefore more consistent for KE:altered inner ear development, we decided to switch our focus to the alternate AOP. The final developed AOP 410 is visualized in Figure 5.
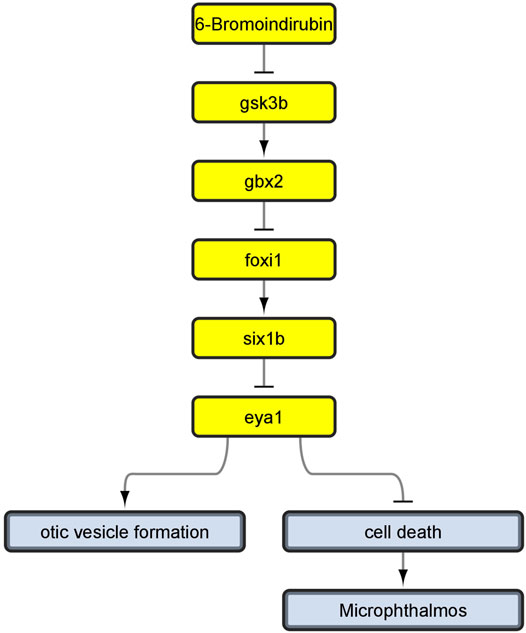
FIGURE 4. Linear path which we developed into AOP 410 and its connection to otic vesicle formation node before removing non-pathology-ends. Yellow nodes represent linear path upstream of cell death.
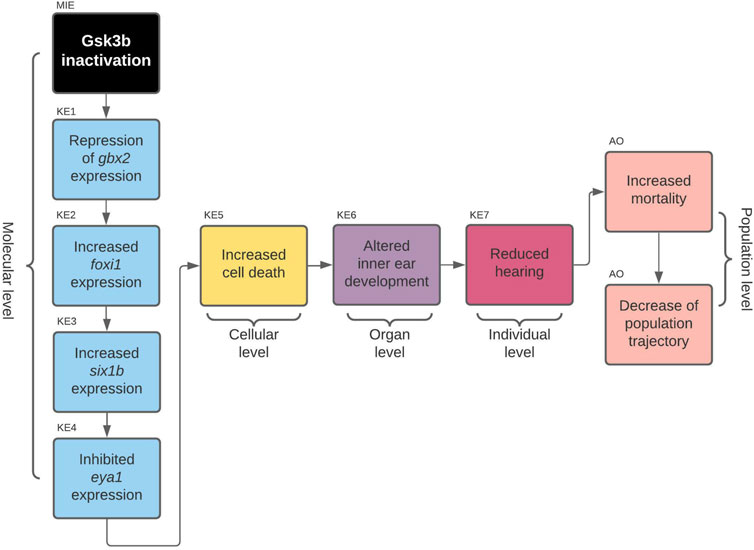
FIGURE 5. AOP 410. Gsk3beta inactivation leads to increased mortality via defects in development of the inner ear.
Chemical Initiators
Glycogen synthase kinase three beta (Gsk3b) has been shown to be inactivated by 6-bromoindirubin-3′-oxime (BIO), a selective GSK3 inhibitor that activates Wnt signaling, su5402, a growth factor receptor inhibitor, and retinoic acid, a metabolite involved in embryonic development. This regulation was shown in mouse (Sato et al., 2004) and zebrafish embryos (Wang et al., 2018). In the zebrafish experiments, chemical treatment with the chemical initiators has started between 14 and 17 hpf (hours post fertilization) and gastrulation brain homeobox 2 (gbx2) expression was examined at 18 hpf. The effects of the chemicals on gbx2 expression were dependent on the time of exposure, with later time points (17 hpf) being less effective. Therefore, the timing of the chemical insult seems to be crucial for the toxicity of the chemicals (Wang et al., 2018).
MIE: Gsk3b Inactivation
Gsk3b kinase is constitutively active in resting cells and undergoes a rapid and transient inhibition in response to a number of external signals. Gsk3b activity is regulated by site-specific phosphorylation. Phosphorylation of Ser9 is the most common and important regulatory mechanism (Grimes and Jope, 2001; Luo, 2012). Gsk3b regulates diverse developmental events in the immature brain, including neurogenesis, neuronal migration, differentiation and survival in vertebrates (Kim and Kimmel, 2000). Human ortholog(s) of this gene are implicated in Alzheimer’s disease (Grassilli et al., 2014). Among the signaling proteins regulated by GSK3b are many transcription factors, including activator protein-1 (AP-1), cyclic AMP response element binding protein (CREB), heat shock factor-1 (HSF-1), nuclear factor of activated T cells (NFAT), MYC Proto-Oncogene (MYC), b-catenin, CCAAT/enhancer binding protein (C/EBP), and nuclear factor kappa-light-chain-enhancer of activated B cells (NFkB) (Grimes and Jope, 2001)
Essentiality—no experimental evidence found.
KER1: Gsk3b Inactivation Leads to Repression of gbx2 Expression
Wnt signaling is implicated in anteroposterior (AP) axis patterning and midbrain specification in animals. Gbx2 is one of the representative AP markers and is downregulated in activation of Wnt signal pathway, under Gsk3b inhibition. In zebrafish embryos, selective GSK3 inbition with BIO that activates Wnt signaling caused reduced expression of gbx2 in the telencephalon (Wang et al., 2018), while in human ESC-derived cells GSKB inhibition with BIO was shown to downregulate GBX2 expression in a dose dependent manner (Kim et al., 2018). The role of Wnt signaling in gbx2 regulation was confirmed via another Wnt signaling inhibitor (LiCl) that resulted in similar gene expression patterns of GBX2 in ESC-derived cells (Kim et al., 2018).
Biological plausibility—high. Inhibition of Gsk3b leads to activation of canonical Wnt signaling pathway (Grassilli et al., 2014), which is confirmed in many independent studies. Wnt signaling is implicated in AP axis patterning and midbrain specification in several vertebrate species, including humans. Gbx2 is one of the representative AP markers and is downregulated in activation of the Wnt signal pathway (Gsk3b inhibition) (Kim et al., 2018; Wang et al., 2018).
KE1: Repression of gbx2 Expression
During vertebrate brain development, the Gbx2 is expressed expressed in the forebrain (Wang et al., 2018). The genes encoding the Gbx-type homeodomain transcription factors have been identified in a variety of vertebrates, and are primarily implicated in the regulation of various aspects of vertebrate brain development (Nakayama et al., 2017). Gbx2 is involved in cerebellum development, iridophore differentiation, telencephalon regionalization and the anterior hindbrain development, where its role seem to be conserved at least in zebrafish and mice (Burroughs-Garcia et al., 2011). A number of studies have shown that Gbx2 represses many developmental regulatory genes during midbrain-hindbrain boundary development, including forkhead box i1 (foxi1) (Simeone, 2000; Nakamura, 2001). Thus, Gbx2 may be a multifunctional transcriptional factor, although the mechanisms of the differential regulation of its activity during development are unknown (Nakayama et al., 2017).
Essentiality—no experimental evidence found.
KER2: Repression of gbx2 Expression Leads to Increased foxi1 Expression
The downregulation of foxi1 by Gbx2 in zebrafish embryos has been demonstrated in a transgenic fish line, using transient induction of gbx2 and microarray analysis, later confirmed also by qPCR (Nakayama et al., 2017).
Biological plausibility—moderate. KER2 is supported based on a number of studies showing that Gbx2 represses many developmental regulatory genes (foxi1) (Simeone, 2000; Nakamura, 2001). Multifunctional nature of Gbx2 is still unknown and Gbx2 regulation of foxi1 was measured indirectly with microarray analysis and later confirmed with qPCR. WISH (whole mount in situ hybridization) failed to confirm alterations of foxi1 after gbx2 expression but qPCR analysis showed immediate downregulation of foxi1 (Nakayama et al., 2017).
KE2: Increased foxi1 Expression
Foxi1 is a transcription factor, involved in several processes, including animal organ development, epidermal cell fate specification, neuron development and the inner ear development, where it is involved in the induction of the otic-epibranchial progenitor domain (Hans et al., 2013).
Essentiality—high. When foxi1 is knocked down, no expression of SIX homeobox 1b (six1b) is detected in the otocyst (Bricaud and Collazo, 2006). Therefore increase of foxi1 expression is essential for downstream events in AOP to occur.
KER3: Increased foxi1 Expression Leads to Increased six1b Expression
When foxi1 is knocked down, the ear anlagen is either entirely missing or greatly reduced (Solomon et al., 2003) and no expression of six1b is detectable (Bricaud and Collazo, 2006).
Biological plausibility—low. Foxi1 transcription factor regulates six1b and EYA transcriptional coactivator and phosphatase 1 (eya1) gene expression (Bricaud and Collazo, 2006; Hans et al., 2013). Both are responsible for zebrafish inner ear development (Solomon et al., 2003). Regulation of six1b by Foxi1 was confirmed with loss-of-function experiment, but connection could be secondary to the overall absence of the otic placode attributable to foxi1 loss-of-function (Bricaud and Collazo, 2006).
KE3: Increased six1b Expression
Six1b is predicted to have DNA-binding transcription factor activity, RNA polymerase II-specific and RNA polymerase II cis-regulatory region sequence-specific DNA binding activity. It is involved in several processes, including muscle organ development, nervous system development and regulation of skeletal muscle cell proliferation. Six1b is a Member of the Pax–Six1b–Eya–Dach (paired box–sine oculis homeobox–eyes absent–dachshund) gene regulatory network, involved in the development of numerous organs and tissues (Bessarab et al., 2004; Bricaud and Collazo, 2006). It has been proposed to play an important role in inner ear development. Six1b promotes hair cell fate and, conversely, inhibits neuronal fate by differentially affecting cell proliferation and cell death in these lineages. Gain/loss-of-function experiment results indicate that when six1b is overexpressed, not only are fewer neural progenitors formed, but many of these progenitors do not go on to differentiate into neurons (Bricaud and Collazo, 2006).
Essentiality—moderate. Six1b promotes hair cell fate and, conversely, inhibits neuronal fate by differentially affecting cell proliferation and cell death in these lineages. Six1b gain/loss-of-function experiment results indicate that in both cases normal development of inner ear is affected (KE5) (Bricaud and Collazo, 2006).
KER4: Increased six1b Expression Leads to Inhibited eya1 Expression
Eya1 and Six1b together with the Dach protein directly interact to form a functional transcription factor. In this complex, the DNA binding function is provided by the Six protein, while Eya mediates transcriptional activation and Dach proteins appear to function as cofactors (López-Ríos et al., 2003). Six1b gain-of-function experiment results showed that overexpression of six1b in zebrafish developing inner ear reduced the expression of eya1 (Bricaud and Collazo, 2006).
Biological plausibility—low. Although there is evidence to support the relationship, there is still too little known about interactions and regulation of both entities, as it is thought to act in a complex like transcription factor (López-Ríos et al., 2003). In addition, interactions between Six1b and Eya1 in zebrafish and mice are not conserved, according to several studies (Xu et al., 1999; Li et al., 2003; Zheng et al., 2003).
KE4: Inhibited eya1 Expression
Eya1 is predicted to have protein tyrosine phosphatase activity and is involved in adenohypophysis development, otic vesicle morphogenesis, and otolith development in both vertebrates and invertebrates. In zebrafish, the eya1 gene is widely expressed in placode-derived sensory organs during embryogenesis. Eya1 function appears to be primarily required for survival of sensory hair cells in the developing ear and lateral line neuromasts (Kozlowski et al., 2005).
Essentiality—no experimental evidence found.
KER5: Inhibited eya1 Expression Leads to Increased Cell Death
Zebrafish Eya1 has a role in development of the cristae, statoacoustic ganglia, and lateral line system. An eya1 disrupted zebrafish mutant (named dog-eared) features premature apoptosis in precursors to these structures. Because of the large number of apoptotic cells observed within the otic vesicle of the mutants, it has been proposed that Eya1 could act as a suppressor of apoptosis (Kozlowski et al., 2005). In mammals EYA1 dephosphorylates histone variant H2AX and thereby affects DNA repair and cell survival (Cook et al., 2009).
Biological plausibility—high. Eya1 role in regulating cell death within developing otic vesicles is well established in vertebrates (Xu et al., 1999; Cook et al., 2009). Zebrafish dog-eared mutants are one of the models for human deafness disorders. Dog-eared zebrafish ear phenotype shows sensory and non-sensory defects (Whitfield, 2002).
KE5: Increased Cell Death (https://aopwiki.org/events/1825)
Essentiality—high. While not the sole contributor to altered inner ear development, cell death is one of the key players in normal development of sensory organs (Whitfield, 2002; Kozlowski et al., 2005).
KER6: Increased Cell Death Leads to Altered Inner Ear Development
Increased levels of apoptosis occur in the migrating primordia of the posterior lateral line in the dog-eared zebrafish embryo mutants, resulting in smaller otic vesicles (Kozlowski et al., 2005). The lateral line placodes of fishes and amphibians also give rise to hair cells and supporting cells, which form small mechanosensory organs (neuromasts) distributed in lines along the body surface and involved in the detection of water movements. They also produce the sensory neurons innervating these receptor organs (Schlosser, 2014). After six1b or eya1 loss of function, the numbers of sensory receptors and neurons in the sense organs and ganglia derived from the olfactory, otic, lateral line, profundal/trigeminal, and epibranchial placodes are reduced, and only small, malformed sensory organs develop that are abnormally patterned and functionally deficient (Schlosser, 2014).
Biological plausibility—high. There is a number of consistent studies that support the relationship of increased cell death during the development and defect in the development of the inner ear (Whitfield et al., 1996; Kozlowski et al., 2005; Schlosser, 2014).
KE6: Altered Inner Ear Development
Zebrafish do not possess outer or middle ears, but have a fairly typical vertebrate inner ear, the normal development and anatomy of which has been extensively described (Haddon and Lewis, 1996; Bang et al., 2001). Although the zebrafish ear does not contain a specialized hearing organ—there is no equivalent of the mammalian cochlea—many features are conserved with other vertebrate species (Whitfield et al., 2002). The mature inner ear, found in all jawed vertebrates, has two functions: It serves as an auditory system, which detects sound waves, and as a vestibular system, which detects linear and angular accelerations, enabling the organism to maintain balance (Whitfield et al., 1996).
Essentiality—no experimental evidence found.
KER7: Altered Inner Ear Development Leads to Reduced Hearing
Mutations in several genes connected to development of inner ear affect morphology and patterning of the inner ear epithelium, including formation of the semicircular canals and development of sensory patches (maculae and cristae). Dog-eared mutants show abnormal development of semicircular canals and lack cristae within the ear (Kozlowski et al., 2005). Zebrafish mutant embryos with defects of the inner ear fail to balance correctly, and may swim on their sides, upside down, or in circles (Whitfield et al., 1996).
Biological plausibility—high. The inner ear is the vertebrate organ of hearing and balance (Whitfield, 2002). Several zebrafish mutants for studying development of inner ear like dog-eared or van gogh show defects in inner ear development and irregular swimming patterns (Whitfield et al., 1996).
KE7: Reduced Hearing
Hearing refers to the ability to perceive sound vibrations propagated as pressure changes through a medium such as air or water. Reduced hearing in the context of this key event can refer to reduction in the perceived volume of a sound relative to the amplitude of sound waves. Reduced hearing may also refer to a reduced range of frequencies that can be perceived. Zebrafish serves as a model organism for hearing and deafness. Zebrafish mutant embryos fail to balance correctly, and may swim on their sides, upside down, or in circles (Whitfield et al., 1996).
Essentiality—no experimental evidence found.
KER8: Reduced Hearing Leads to Increased Mortality
Although we are not aware of any studies directly looking into increased mortality as a results of reduced hearing ability in fish, it is known that hearing ability is very important for fish, with roles in everything from reproduction to swimming (Kasumyan, 2009). It is therefore very likely that a reduction in the vital sensory ability would negatively affect fish survival.
Biological plausibility—high. Impaired hearing can result in changes in ecologically relevant endpoints, such as predator avoidance and prey capture. Therefore, it can be assumed that an effect on hearing could reduce young of year survival. The relationship is well accepted and corresponds to the logic of nature.
AO: Increased Mortality (https://aopwiki.org/events/351) Overall Assessment of the AOP 410
An overall assessment of this AOP shows that there is low to moderate biological plausibility to suggest a qualitative link between the inactivation of Gsk3b to the KE4-cell death within developing inner ear and high evidence linking KE5 to increased mortality (AO). Biological plausibility is considered moderate because there is ample evidence from gain- and loss- of function experiments and knock out animal models that support the relationships between key events which are consistent with current biological knowledge, but there is mostly indirect evidence linking KEs on molecular level. KEs on molecular level have some uncertainties like foxi1 loss of function experiment resulting in no expression of six1b in otic placode (due to absence of otic placode) and inconsistencies across species (zebrafish, mouse). No empirical evidence in the form of dose response, time concordance and incidence concordance was found for the KERs of the AOP, and only little evidence for the essentiality of the KEs. The summarized information on KER biological plausibility, KE essentiality and KE observation methods of AOP 410 can be found in Tables 3–5.
Discussion
In this study we evaluated the suitability of causal toxicological networks (CTNs) for AOP development. We have developed a semi-automatic pipeline and scripts that take a CTN in BEL format, remove unneeded node and connection annotations and add new functional ones, then reduce the network to a size similar to an AOP network (Pollesch et al., 2019). We performed these operations on a recently developed zebrafish developmental neurotoxicity network as a case study (Li et al., 2021). The network was then separated into subnetworks, based on different starting points (candidate MIEs of different types) and endpoints (pathologies), with a size suitable for close visual inspection. Finally, we separated the subnetworks into a set of candidate AOPs related to zebrafish neurotoxicity. Both the pipeline and the set of candidate AOPs can be found in the Supplementary Material and will hopefully be used for further endeavors in neurotoxicity AOP development.
The second contribution of the study is the development of two new AOPs, one centered on reduced eye size and decrease visual function (AOP 399) and another on alterations in inner ear development and decreased hearing (AOP 410) of the zebrafish. Both newly developed AOPs are available in the AOP-Wiki and have added to the very few neurotoxicity AOPs already there. Neurotoxicity AOPs are especially difficult to develop, as the brain is an extremely complex organ, comprised of a variety of highly specialized neural and glial cell types with diverse cellular functions. This implies the existence of a potentially large number of AOPs and the great need to develop more as soon as possible (Bal-price et al., 2015). As far as the WoE analysis of AOPs 399 and 410, the biological plausibility for most KERs in both AOPs is high, with a few KERs where plausibility is moderate/low. The evidence for the essentiality of the KEs is mostly missing. This means that AOP 399 and AOP 410 require further improvements before regulatory use and that we should probably still refer to them as plausible AOPs. However, even if not immediately useful for regulation purposes, plausible AOPs lacking some evidence are useful for identification of missing knowledge and targeted design of new experiments that can fill the identified gaps (Perkins et al., 2015).
But, perhaps most importantly, while developing AOPs 399 and 410, we have also encountered some advantages and challenges that are specific to development of AOPs from CTNs. A very important advantage is that, because of the nature of the design/development of CTNs (Li et al., 2020), which is based on direct experimental evidence connecting the nodes in the network, the biological plausibility of the KERs of the derived AOPs is mostly high/moderate. Since biological plausibility is the most important characteristic of AOPs (OECD, 2014; Fay et al., 2017), this makes CTNs an excellent starting point for AOP development. On the other hand, because of the same reason, much less evidence can be found for KE essentiality and even less empirical evidence in the form of dose response, time concordance and incidence concordance, which are essential for qAOP development. Nevertheless, the AOPs developed from CTNs are consistent with the core principles of AOP development (Villeneuve et al., 2014a).
In our results section we describe all the details that came up when developing the AOP, which will hopefully be of use for future AOP development, either with the same zebrafish neurotoxicity network or with another CTN. As our described development of AOP 410 suggests, the network manipulations necessary to come to a smaller, easier to handle network that can be used as AOP starting points, can also cause some nodes and evidence to disappear. Therefore, although a CTN may make it easier to construct a new AOP, for some KE and KERs it is still necessary to add additional evidence and handle WoE assessment with utmost care. In our case, in the original network cell death and otic vesicle formation nodes were not causally connected, probably because not enough literature was added to the initial network or the computational pipeline did not detect causal dependence of entities in it. It is therefore worth noting that although a lot of information is stored in CTNs, they are by no means complete. When comparing the number of references in the CTN-based candidate AOPs that we started from to the number of references in the final AOPs 399 and 410, we can see that only ca. 20 % were already in the CTN and ca. 80 % were added through the search for additional evidence. However, the original 20% were a very good starting point, as many of the additional references were added based on searching for scientific studies cites or citing these original 20 %.
As we (the authors of this paper) have little experience in developing AOPs using other strategies, it is difficult to assess the practical advantages or disadvantages of the proposed approach, either with respect of time investment or expertise needed. It is in our opinion certain that developing sets of candidate adverse outcome pathways that are all backed by sound evidence can be done very quickly using the proposed methodology. From an experimental point of view, the candidate AOPs represent working hypotheses, which can help guide targeted experiments that add to our toxicological knowledge base. However, choosing among this candidate set, developing the AOP and performing the WoE will still require the same amount of time and expertise as with any other approach that we are aware of, as the evidence available in the CTN cannot be taken for granted, but needs to be reevaluated for each new AOP. It has been argued that AOP networks, with their increased complexity have advantages in representing the toxic effects of chemicals (Knapen et al., 2018; Villeneuve et al., 2018), so the question arises why not use CTNs to directly develop AOP networks, instead of AOPs, as we did here. However, AOP networks are defined as networks of individual AOPs, joined at shared KEs. The focus is therefore on KEs and KERs and the evidence supporting them. Once the weight of evidence analysis is performed, AOP networks can be easily put together from the AOP-Wiki, as demonstrated recently (Ankley et al., 2020).
When we started the development of the new AOPs, there were no AOPs on the AOP-wiki that would feature the KEs related to eye size and ear development. We were thus pleasantly surprised when soon afterwards the reduced eye size KE appeared and we could link the evidence we found to a collection of evidence already there (but found from different sources). Surprisingly, although we purposefully avoided developing AOPs that would already be present in the AOP-Wiki, for both developed AOPs we ended up connecting to other KE in other AOPs. For AOP 399 it was because AOP 364 (entitled Thyroperoxidase inhibition leading to altered visual function via decreased eye size) added the same KE only a couple of weeks before we did. For AOP 410 it was due to the lack of evidence for reduced eye size caused by gbx2 expression inhibition that we moved the development towards the inner ear and the upstream MIE of Gsk3b inactivation, which was also already present in the AOP 298 (entitled Chronic ROS leading to human treatment-resistant gastric cancer). With more AOPs being developed, we expect that more and more KEs and KERs will be shared among them and that the AOP world will quite soon become a large AOP network. This should in turn speed up the development of new AOPs.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
AZ designed the study and co-developed the AOPs. ZR annotated and analysed the networks and wrote all the scripts. VM annotated the networks and developed the AOPs. CB and RL provided the neurotoxicology expert knowledge and co-developed the AOPs. All authors have read and approved the manuscript.
Funding
Funding for this research was provided by the ARRS Research Program P4-0165 and the CRACK IT 2017 DART Challenge.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Lucia Vergauwen (University of Antwerp) for letting us link to the KE reduced eye size, which her team was the first to enter into AOP-Wiki, You Song (Norwegian Institute for Water Research) for letting us link to the KE cell death and Shihori Tanabe for letting us link to the KE GSK3beta inactivation. We would also like to thank the AOP-Wiki Administrator group for the help with getting the new AOPs into the correct format for the AOP-Wiki.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/ftox.2022.815754/full#supplementary-material
References
Allen, T. E. H., Goodman, J. M., Gutsell, S., and Russell, P. J. (2014). Defining Molecular Initiating Events in the Adverse Outcome Pathway Framework for Risk Assessment. Chem. Res. Toxicol. 27, 2100–2112. doi:10.1021/TX500345J
Ankley, G. T., Bennett, R. S., Erickson, R. J., Hoff, D. J., Hornung, M. W., Johnson, R. D., et al. (2010). Adverse Outcome Pathways: A Conceptual Framework to Support Ecotoxicology Research and Risk Assessment. Environ. Toxicol. Chem. 29, 730–741. doi:10.1002/ETC.34
Ankley, G. T., Blackwell, B. R., Cavallin, J. E., Doering, J. A., Feifarek, D. J., Jensen, K. M., et al. (2020). Adverse Outcome Pathway Network-Based Assessment of the Interactive Effects of an Androgen Receptor Agonist and an Aromatase Inhibitor on Fish Endocrine Function. Environ. Toxicol. Chem. 39, 913–922. doi:10.1002/etc.4668
Bal-Price, A., Crofton, K. M., Sachana, M., Shafer, T. J., Behl, M., Forsby, A., et al. (2015). Putative Adverse Outcome Pathways Relevant to Neurotoxicity. Crit. Rev. Toxicol. 45, 83–91. doi:10.3109/10408444.2014.981331
Bang, P. I., Sewell, W. F., and Malicki, J. J. (2001). Morphology and Cell Type Heterogeneities of the Inner Ear Epithelia in Adult and Juvenile Zebrafish (Danio rerio). J. Comp. Neurol. 438, 173–190. doi:10.1002/CNE.1308
Bang, P. I., Yelick, P. C., Malicki, J. J., and Sewell, W. F. (2002). High-throughput Behavioral Screening Method for Detecting Auditory Response Defects in Zebrafish. J. Neurosci. Methods 118, 177–187. doi:10.1016/S0165-0270(02)00118-8
Baumann, L., Ros, A., Rehberger, K., Neuhauss, S. C. F., and Segner, H. (2016). Thyroid Disruption in Zebrafish (Danio rerio) Larvae: Different Molecular Response Patterns lead to Impaired Eye Development and Visual Functions. Aquat. Toxicol. 172, 44–55. doi:10.1016/J.AQUATOX.2015.12.015
Becker, R. A., Ankley, G. T., Edwards, S. W., Kennedy, S. W., Linkov, I., Meek, B., et al. (2015). Increasing Scientific Confidence in Adverse Outcome Pathways: Application of Tailored Bradford-Hill Considerations for Evaluating Weight of Evidence. Regul. Toxicol. Pharmacol. 72, 514–537. doi:10.1016/J.YRTPH.2015.04.004
Bell, S. M., Angrish, M. M., Wood, C. E., and Edwards, S. W. (2016). Integrating Publicly Available Data to Generate Computationally Predicted Adverse Outcome Pathways for Fatty Liver. Toxicol. Sci. 150, 510–520. doi:10.1093/toxsci/kfw017
Bessarab, D. A., Chong, S.-W., and Korzh, V. (2004). Expression of Zebrafishsix1 during Sensory Organ Development and Myogenesis. Dev. Dyn. 230, 781–786. doi:10.1002/DVDY.20093
Besson, M., Feeney, W. E., Moniz, I., François, L., Brooker, R. M., Holzer, G., et al. (2020). Anthropogenic Stressors Impact Fish Sensory Development and Survival via Thyroid Disruption. Nat. Commun. 11, 1–10. doi:10.1038/s41467-020-17450-8
Boué, S., Talikka, M., Westra, J. W., Hayes, W., Di Fabio, A., Park, J., et al. (2015). Causal Biological Network Database: a Comprehensive Platform of Causal Biological Network Models Focused on the Pulmonary and Vascular Systems. Database 2015, 1–14. doi:10.1093/DATABASE/BAV030
Bricaud, O., and Collazo, A. (2006). The Transcription Factor Six1 Inhibits Neuronal and Promotes Hair Cell Fate in the Developing Zebrafish (Danio rerio) Inner Ear. J. Neurosci. 26, 10438–10451. doi:10.1523/JNEUROSCI.1025-06.2006
Burgoon, L. D., Angrish, M., Garcia‐Reyero, N., Pollesch, N., Zupanic, A., and Perkins, E. (2020). Predicting the Probability that a Chemical Causes Steatosis Using Adverse Outcome Pathway Bayesian Networks (AOPBNs). Risk Anal. 40, 512–523. doi:10.1111/risa.13423
Burmeister, M., Novak, J., Liang, M.-Y., Basu, S., Ploder, L., Hawes, N. L., et al. (1996). Ocular Retardation Mouse Caused by Chx10 Homeobox Null Allele: Impaired Retinal Progenitor Proliferation and Bipolar Cell Differentiation. Nat. Genet. 12, 376–384. doi:10.1038/ng0496-376
Burroughs-Garcia, J., Sittaramane, V., Chandrasekhar, A., and Waters, S. T. (2011). Evolutionarily Conserved Function of Gbx2 in Anterior Hindbrain Development. Dev. Dyn. 240, 828–838. doi:10.1002/DVDY.22589
Carvaillo, J.-C., Barouki, R., Coumoul, X., and Audouze, K. (2019). Linking Bisphenol S to Adverse Outcome Pathways Using a Combined Text Mining and Systems Biology Approach. Environ. Health Perspect. 127, 047005. doi:10.1289/EHP4200
Challa, A. K., and Chatti, K. (2013). Conservation and Early Expression of Zebrafish Tyrosine Kinases Support the Utility of Zebrafish as a Model for Tyrosine Kinase Biology. Zebrafish 10, 264–274. Available at: https://home.liebertpub.com/zeb 10. doi:10.1089/zeb.2012.0781
Cole, A. R., and Sutherland, C. (2008). Measuring GSK3 Expression and Activity in Cells. Methods Mol. Biol. 468, 45–65. doi:10.1007/978-1-59745-249-6_4
Colwill, R. M., and Creton, R. (2011). Imaging Escape and Avoidance Behavior in Zebrafish Larvae. Rev. Neurosci. 22, 63–73. doi:10.1515/RNS.2011.008
Cook, P. J., Ju, B. G., Telese, F., Wang, X., Glass, C. K., and Rosenfeld, M. G. (2009). Tyrosine Dephosphorylation of H2AX Modulates Apoptosis and Survival Decisions. Nature 458, 591–596. doi:10.1038/nature07849
Crowley, L. C., Marfell, B. J., Scott, A. P., Boughaba, J. A., Chojnowski, G., Christensen, M. E., et al. (2016). Dead Cert: Measuring Cell Death. Cold Spring Harb. Protoc. 2016, pdb.top070318–1072. doi:10.1101/pdb.top070318
Csardi, G., and Nepusz, T. (2006). The Igraph Software Package for Complex Network Research. Int. J. Complex Syst., 1695, 1–9.
Davis, A. P., Grondin, C. J., Johnson, R. J., Sciaky, D., Wiegers, J., Wiegers, T. C., et al. (2020). Comparative Toxicogenomics Database (CTD): Update 2021. Nucleic Acids Res. 49, D1138–D1143. doi:10.1093/nar/gkaa891
Dehnert, G. K., Karasov, W. H., and Wolman, M. A. (2019). 2,4-Dichlorophenoxyacetic Acid Containing Herbicide Impairs Essential Visually Guided Behaviors of Larval Fish. Aquat. Toxicol. 209, 1–12. doi:10.1016/J.AQUATOX.2019.01.015
Dix, D. J., Houck, K. A., Martin, M. T., Richard, A. M., Setzer, R. W., and Kavlock, R. J. (2007). The ToxCast Program for Prioritizing Toxicity Testing of Environmental Chemicals. Toxicol. Sci. 95, 5–12. doi:10.1093/TOXSCI/KFL103
Doering, J. A., Wiseman, S., Giesy, J. P., and Hecker, M. (2018). A Cross-Species Quantitative Adverse Outcome Pathway for Activation of the Aryl Hydrocarbon Receptor Leading to Early Life Stage Mortality in Birds and Fishes. Environ. Sci. Technol. 52, 7524–7533. doi:10.1021/acs.est.8b01438
Doktorova, T. Y., Oki, N. O., Mohorič, T., Exner, T. E., and Hardy, B. (2020). A Semi-automated Workflow for Adverse Outcome Pathway Hypothesis Generation: The Use Case of Non-genotoxic Induced Hepatocellular Carcinoma. Regul. Toxicol. Pharmacol. 114, 104652. doi:10.1016/j.yrtph.2020.104652
Ebert, A. M., Childs, S. J., Hehr, C. L., Cechmanek, P. B., and McFarlane, S. (2014). Sema6a and Plxna2 Mediate Spatially Regulated Repulsion within the Developing Eye to Promote Eye Vesicle Cohesion. Dev 141, 2473–2482. doi:10.1242/DEV.103499/VIDEO-2
Emerson, S. E., St. Clair, R. M., Waldron, A. L., Bruno, S. R., Duong, A., Driscoll, H. E., et al. (2017). Identification of Target Genes Downstream of semaphorin6A/PlexinA2 Signaling in Zebrafish. Dev. Dyn. 246, 539–549. doi:10.1002/DVDY.24512
Fay, K. A., Villeneuve, D. L., LaLone, C. A., Song, Y., Tollefsen, K. E., and Ankley, G. T. (2017). Practical Approaches to Adverse Outcome Pathway Development and Weight-Of-Evidence Evaluation as Illustrated by Ecotoxicological Case Studies. Environ. Toxicol. Chem. 36, 1429–1449. doi:10.1002/etc.3770
Fleisch, V. C., Jametti, T., and Neuhauss, S. C. F. (2008). Electroretinogram (ERG) Measurements in Larval Zebrafish. Cold Spring Harb. Protoc. 2008, pdb.prot4973. doi:10.1101/PDB.PROT4973
Franco, M., and Tamagnone, L. (2008). Tyrosine Phosphorylation in Semaphorin Signalling: Shifting into Overdrive. EMBO Rep. 9, 865–871. doi:10.1038/embor.2008.139
Gao, D., Wu, M., Wang, C., Wang, Y., and Zuo, Z. (2015). Chronic Exposure to Low Benzo[a]pyrene Level Causes Neurodegenerative Disease-like Syndromes in Zebrafish (Danio rerio). Aquat. Toxicol. 167, 200–208. doi:10.1016/j.aquatox.2015.08.013
Grassilli, E., Ianzano, L., Bonomo, S., Missaglia, C., Cerrito, M. G., Giovannoni, R., et al. (2014). GSK3A Is Redundant with GSK3B in Modulating Drug Resistance and Chemotherapy-Induced Necroptosis. PLoS One 9, e100947–8. doi:10.1371/journal.pone.0100947
Graw, J. (2010). Eye Development. Eye Development. Curr. Top. Dev. Biol. 90, 343–386. doi:10.1016/S0070-2153(10)90010-0
Green, T. P., Fennell, M., Whittaker, R., Curwen, J., Jacobs, V., Allen, J., et al. (2009). Preclinical Anticancer Activity of the Potent, Oral Src Inhibitor AZD0530. Mol. Oncol. 3, 248–261. doi:10.1016/J.MOLONC.2009.01.002
Grimes, C. A., and Jope, R. S. (2001). The Multifaceted Roles of Glycogen Synthase Kinase 3β in Cellular Signaling. Prog. Neurobiol. 65, 391–426. doi:10.1016/S0301-0082(01)00011-9
Groh, K. J., Carvalho, R. N., Chipman, J. K., Denslow, N. D., Halder, M., Murphy, C. A., et al. (2015). Development and Application of the Adverse Outcome Pathway Framework for Understanding and Predicting Chronic Toxicity: I. Challenges and Research Needs in Ecotoxicology. Chemosphere 120, 764–777. doi:10.1016/J.CHEMOSPHERE.2014.09.068
Haddon, C., and Lewis, J. (1996). Early Ear Development in the Embryo of the Zebrafish,Danio rerio. J. Comp. Neurol. 365, 113–128. doi:10.1002/(sici)1096-9861(19960129)365:1<113::aid-cne9>3.0.co;2-6
Hall, M. I. (2008). Comparative Analysis of the Size and Shape of the Lizard Eye. Zoology 111, 62–75. doi:10.1016/J.ZOOL.2007.04.003
Hans, S., Irmscher, A., and Brand, M. (2013). Zebrafish Foxi1 Provides a Neuronal Ground State during Inner Ear Induction Preceding the Dlx3b/4b-Regulated Sensory Lineage. Development 140, 1936–1945. doi:10.1242/DEV.087718
He, H., Dai, J., Zhuo, R., Zhao, J., Wang, H., Sun, F., et al. (2018). Study on the Mechanism behind lncRNA MEG3 Affecting clear Cell Renal Cell Carcinoma by Regulating miR‐7/RASL11B Signaling. J. Cel. Physiol. 233, 9503–9515. doi:10.1002/jcp.26849
Igarashi, Y., Nakatsu, N., Yamashita, T., Ono, A., Ohno, Y., Urushidani, T., et al. (2015). Open TG-GATEs: A Large-Scale Toxicogenomics Database. Nucleic Acids Res. 43, D921–D927. doi:10.1093/nar/gku955
Jassal, B., Matthews, L., Viteri, G., Gong, C., Lorente, P., Fabregat, A., et al. (2020). The Reactome Pathway Knowledgebase. Nucleic Acids Res. 48, D498–D503. doi:10.1093/NAR/GKZ1031
Jelić, D., Mildner, B., Koštrun, S., Nujić, K., Verbanac, D., Čulić, O., et al. (2007). Homology Modeling of Human Fyn Kinase Structure: Discovery of Rosmarinic Acid as a New Fyn Kinase Inhibitor and In Silico Study of its Possible Binding Modes. J. Med. Chem. 50, 1090–1100. doi:10.1021/JM0607202
Jeong, J., Garcia-Reyero, N., Burgoon, L., Perkins, E., Park, T., Kim, C., et al. (2019). Development of Adverse Outcome Pathway for PPARγ Antagonism Leading to Pulmonary Fibrosis and Chemical Selection for its Validation: ToxCast Database and a Deep Learning Artificial Neural Network Model-Based Approach. Chem. Res. Toxicol. 32, 1212–1222. doi:10.1021/acs.chemrestox.9b00040
Jin, Y., Feng, M., Ma, W., Wei, Y., Qi, G., Luo, J., et al. (2021). A Toxicity Pathway-Oriented Approach to Develop Adverse Outcome Pathway: AHR Activation as a Case Study. Environ. Pollut. 268, 115733. doi:10.1016/j.envpol.2020.115733
Jornod, F., Jaylet, T., Blaha, L., Sarigiannis, D., Tamisier, L., and Audouze, K. (2021). AOP-helpFinder Webserver: a Tool for Comprehensive Analysis of the Literature to Support Adverse Outcome Pathways Development. Bioinformatics 38, 1173–1175. doi:10.1093/BIOINFORMATICS/BTAB750
Kang, P., and Wildsoet, C. F. (2016). Acute and Short-Term Changes in Visual Function with Multifocal Soft Contact Lens Wear in Young Adults. Contact Lens Anterior Eye 39, 133–140. doi:10.1016/j.clae.2015.09.004
Kasumyan, A. O. (2009). Acoustic Signaling in Fish. J. Ichthyol. 49, 963–1020. doi:10.1134/S0032945209110010
Kennedy, B. N., Stearns, G. W., Smyth, V. A., Ramamurthy, V., Van Eeden, F., Ankoudinova, I., et al. (2004). Zebrafish Rx3 and Mab21l2 Are Required during Eye Morphogenesis. Dev. Biol. 270, 336–349. doi:10.1016/j.ydbio.2004.02.026
Kim, L., and Kimmel, A. R. (2000). GSK3, a Master Switch Regulating Cell-Fate Specification and Tumorigenesis. Curr. Opin. Genet. Dev. 10, 508–514. doi:10.1016/S0959-437X(00)00120-9
Kim, J. Y., Lee, J. S., Hwang, H. S., Lee, D. R., Park, C.-Y., Jung, S. J., et al. (2018). Wnt Signal Activation Induces Midbrain Specification through Direct Binding of the Beta-catenin/TCF4 Complex to the EN1 Promoter in Human Pluripotent Stem Cells. Exp. Mol. Med. 50, 1–13. doi:10.1038/s12276-018-0044-y
Kinoshita, T., Matsubara, M., Ishiguro, H., Okita, K., and Tada, T. (2006). Structure of Human Fyn Kinase Domain Complexed with Staurosporine. Biochem. Biophys. Res. Commun. 346, 840–844. doi:10.1016/J.BBRC.2006.05.212
Knapen, D., Vergauwen, L., Villeneuve, D. L., and Ankley, G. T. (2015). The Potential of AOP Networks for Reproductive and Developmental Toxicity Assay Development. Reprod. Toxicol. 56, 52–55. doi:10.1016/J.REPROTOX.2015.04.003
Knapen, D., Angrish, M. M., Fortin, M. C., Katsiadaki, I., Leonard, M., Margiotta-Casaluci, L., et al. (2018). Adverse Outcome Pathway Networks I: Development and Applications. Environ. Toxicol. Chem. 37, 1723–1733. doi:10.1002/etc.4125
Knapen, D., Stinckens, E., Cavallin, J. E., Ankley, G. T., Holbech, H., Villeneuve, D. L., et al. (2020). Toward an AOP Network-Based Tiered Testing Strategy for the Assessment of Thyroid Hormone Disruption. Environ. Sci. Technol. 54, 8491–8499. doi:10.1021/acs.est.9b07205
Kozlowski, D. J., Whitfield, T. T., Hukriede, N. A., Lam, W. K., and Weinberg, E. S. (2005). The Zebrafish Dog-Eared Mutation Disrupts Eya1, a Gene Required for Cell Survival and Differentiation in the Inner Ear and Lateral Line. Dev. Biol. 277, 27–41. doi:10.1016/J.YDBIO.2004.08.033
LaLone, C. A., Villeneuve, D. L., Wu-Smart, J., Milsk, R. Y., Sappington, K., Garber, K. V., et al. (2017). Weight of Evidence Evaluation of a Network of Adverse Outcome Pathways Linking Activation of the Nicotinic Acetylcholine Receptor in Honey Bees to colony Death. Sci. Total Environ. 584-585, 751–775. doi:10.1016/J.SCITOTENV.2017.01.113
Le, H.-G. T., Dowling, J. E., and Cameron, D. J. (2012). Early Retinoic Acid Deprivation in Developing Zebrafish Results in Microphthalmia. Vis. Neurosci. 29, 219–228. doi:10.1017/S0952523812000296
Li, X., Ohgi, K. A., Zhang, J., Krones, A., Bush, K. T., Glass, C. K., et al. (2003). Eya Protein Phosphatase Activity Regulates Six1-Dach-Eya Transcriptional Effects in Mammalian Organogenesis. Nature 426, 247–254. doi:10.1038/nature02083
Li, R., Zupanic, A., Talikka, M., Belcastro, V., Madan, S., Dörpinghaus, J., et al. (2020). Systems Toxicology Approach for Testing Chemical Cardiotoxicity in Larval Zebrafish. Chem. Res. Toxicol. 33, 2550–2564. doi:10.1021/acs.chemrestox.0c00095
Li, R. A., Talikka, M., Gubian, S., vom Berg, C., Martin, F., Peitsch, M. C., et al. (2021). Systems Toxicology Approach for Assessing Developmental Neurotoxicity in Larval Zebrafish. Front. Genet. 12, 993. doi:10.3389/FGENE.2021.652632
Lillicrap, A., Moe, S. J., Wolf, R., Connors, K. A., Rawlings, J. M., Landis, W. G., et al. (2020). Evaluation of a Bayesian Network for Strengthening the Weight of Evidence to Predict Acute Fish Toxicity from Fish Embryo Toxicity Data. Integr. Environ. Assess. Manag. 16, 452–460. doi:10.1002/ieam.4258
Lopez-Rios, J., Tessmar, K., Loosli, F., Wittbrodt, J., and Bovolenta, P. (2003). Six3 and Six6 Activity Is Modulated by Members of the Groucho Family. Development 130, 185–195. doi:10.1242/dev.00185
Luo, Y., Raible, D., and Raper, J. A. (1993). Collapsin: A Protein in Brain that Induces the Collapse and Paralysis of Neuronal Growth Cones. Cell 75, 217–227. doi:10.1016/0092-8674(93)80064-L
Luo, J. (2012). The Role of GSK3beta in the Development of the central Nervous System. Front. Biol. 7, 212–220. doi:10.1007/s11515-012-1222-2
Margiotta-Casaluci, L., Owen, S. F., Huerta, B., Rodríguez-Mozaz, S., Kugathas, S., Barceló, D., et al. (2016). Internal Exposure Dynamics Drive the Adverse Outcome Pathways of Synthetic Glucocorticoids in Fish. Sci. Rep. 6, 1–13. doi:10.1038/srep21978
Marsh-Armstrong, N., Mccaffery, P., Gilbert, W., Dowling, J. E., and Drager, U. C. (1994). Retinoic Acid Is Necessary for Development of the Ventral Retina in Zebrafish. Proc. Natl. Acad. Sci. 91, 7286–7290. doi:10.1073/pnas.91.15.7286
Mead, T. J., and Lefebvre, V. (2014). Proliferation Assays (BrdU and EdU) on Skeletal Tissue Sections. Methods Mol. Biol. 1130, 233–243. doi:10.1007/978-1-62703-989-510.1007/978-1-62703-989-5_17
Miller, D. H., and Ankley, G. T. (2004). Modeling Impacts on Populations: Fathead Minnow (Pimephales promelas) Exposure to the Endocrine Disruptor 17β-Trenbolone as a Case Study. Ecotoxicol. Environ. Saf. 59, 1–9. doi:10.1016/j.ecoenv.2004.05.005
Murugadoss, S., Vrček, I. V., Pem, B., Jagiello, K., Judzinska, B., Sosnowska, A., et al. (2021). A Strategy towards the Generation of Testable Adverse Outcome Pathways for Nanomaterials. ALTEX 38 (4), 580–594. doi:10.14573/altex.2102191
Nakamura, F., Kalb, R. G., and Strittmatter, S. M. (2000). Molecular Basis of Semaphorin-Mediated Axon Guidance. J. Neurobiol. 44, 219–229. doi:10.1002/1097-4695(200008)44:2<219::aid-neu11>3.0.co;2-w
Nakamura, H. (2001). Regionalization of the Optic Tectum: Combinations of Gene Expression that Define the Tectum. Trends Neurosci. 24, 32–39. doi:10.1016/S0166-2236(00)01676-3
Nakayama, Y., Inomata, C., Yuikawa, T., Tsuda, S., and Yamasu, K. (2017). Comprehensive Analysis of Target Genes in Zebrafish Embryos Reveals Gbx2 Involvement in Neurogenesis. Dev. Biol. 430, 237–248. doi:10.1016/J.YDBIO.2017.07.015
Naujok, O., Lentes, J., Diekmann, U., Davenport, C., and Lenzen, S. (2014). Cytotoxicity and Activation of the Wnt/beta-Catenin Pathway in Mouse Embryonic Stem Cells Treated with Four GSK3 Inhibitors. BMC Res. Notes 7, 1–8. doi:10.1186/1756-0500-7-273
Negi, C. K., Bajard, L., Kohoutek, J., and Blaha, L. (2021). An Adverse Outcome Pathway Based In Vitro Characterization of Novel Flame Retardants-Induced Hepatic Steatosis. Environ. Pollut. 289, 117855. doi:10.1016/J.ENVPOL.2021.117855
Neufeld, G., and Kessler, O. (2008). The Semaphorins: Versatile Regulators of Tumour Progression and Tumour Angiogenesis. Nat. Rev. Cancer 8, 632–645. doi:10.1038/nrc2404
Nicolson, T., Rüsch, A., Friedrich, R. W., Granato, M., Ruppersberg, J. P., and Nüsslein-Volhard, C. (1998). Genetic Analysis of Vertebrate Sensory Hair Cell Mechanosensation: The Zebrafish Circler Mutants. Neuron 20, 271–283. doi:10.1016/S0896-6273(00)80455-9
OECD (2016). Users’ Handbook Supplement to the Guidance Document for Developing and Assessing Adverse Outcome Pathways. OECD Series on Adverse Outcome Pathways, No. 1. Paris: OECD Publishing. Available at: https://www.oecd-ilibrary.org/environment/users-handbook-supplement-to-the-guidance-document-for-developing-and-assessing-adverse-outcome-pathways_5jlv1m9d1g32-en (Accessed August 24, 2021).
OECD (2014). Users’ Handbook Supplement to the Guidance Document for Developing and Assessing AOPs. OECD Ser. Test. Assess.
Oki, N. O., Nelms, M. D., Bell, S. M., Mortensen, H. M., and Edwards, S. W. (2016). Accelerating Adverse Outcome Pathway Development Using Publicly Available Data Sources. Curr. Envir Health Rpt 3, 53–63. doi:10.1007/s40572-016-0079-y
Pézeron, G., Lambert, G., Dickmeis, T., Strähle, U., Rosa, F. M., and Mourrain, P. (2008). Rasl11b Knock Down in Zebrafish Suppresses One-Eyed-Pinhead Mutant Phenotype. PLoS One 3, e1434. doi:10.1371/JOURNAL.PONE.0001434
Perkins, E. J., Antczak, P., Burgoon, L., Falciani, F., Garcia-Reyero, N., Gutsell, S., et al. (2015). Adverse Outcome Pathways for Regulatory Applications: Examination of Four Case Studies with Different Degrees of Completeness and Scientific Confidence. Toxicol. Sci. 148, 14–25. doi:10.1093/TOXSCI/KFV181
Perkins, E. J., Ashauer, R., Burgoon, L., Conolly, R., Landesmann, B., Mackay, C., et al. (2019). Building and Applying Quantitative Adverse Outcome Pathway Models for Chemical Hazard and Risk Assessment. Environ. Toxicol. Chem. 38, 1850–1865. doi:10.1002/etc.4505
Pistollato, F., De Gyves, E. M., Carpi, D., Bopp, S. K., Nunes, C., Worth, A., et al. (2020). Assessment of Developmental Neurotoxicity Induced by Chemical Mixtures Using an Adverse Outcome Pathway Concept. Environ. Health 19, 1–26. doi:10.1186/s12940-020-00578-x
Pittman, M. E., Edwards, S. W., Ives, C., and Mortensen, H. M. (2018). AOP-DB: A Database Resource for the Exploration of Adverse Outcome Pathways through Integrated Association Networks. Toxicol. Appl. Pharmacol. 343, 71–83. doi:10.1016/j.taap.2018.02.006
Pollesch, N. L., Villeneuve, D. L., O’Brien, J. M., and O’Brien, J. M. (2019). Extracting and Benchmarking Emerging Adverse Outcome Pathway Knowledge. Toxicol. Sci. 168, 349–364. doi:10.1093/toxsci/kfz006
R Core Team, A. (2020). R: A Language and Environment for Statistical Computing. Available at: https://www.r-project.org/. (Accessed from January 1, 2021).
Rearick, D. C., Ward, J., Venturelli, P., and Schoenfuss, H. (2018). Environmental Oestrogens Cause Predation-Induced Population Decline in a Freshwater Fish. R. Soc. Open Sci. 5, 181065. doi:10.1098/rsos.181065
Sachana, M., Willett, C., Pistollato, F., and Bal-Price, A. (2021). The Potential of Mechanistic Information Organised within the AOP Framework to Increase Regulatory Uptake of the Developmental Neurotoxicity (DNT) In Vitro Battery of Assays. Reprod. Toxicol. 103, 159–170. doi:10.1016/j.reprotox.2021.06.006
Salic, A., and Mitchison, T. J. (2008). A Chemical Method for Fast and Sensitive Detection of DNA Synthesis In Vivo. Proc. Natl. Acad. Sci. 105, 2415–2420. doi:10.1073/pnas.0712168105
Sasaki, Y., Cheng, C., Uchida, Y., Nakajima, O., Ohshima, T., Yagi, T., et al. (2002). Fyn and Cdk5 Mediate semaphorin-3A Signaling, Which Is Involved in Regulation of Dendrite Orientation in Cerebral Cortex. Neuron 35, 907–920. doi:10.1016/S0896-6273(02)00857-7
Sato, N., Meijer, L., Skaltsounis, L., Greengard, P., and Brivanlou, A. H. (2004). Maintenance of Pluripotency in Human and Mouse Embryonic Stem Cells through Activation of Wnt Signaling by a Pharmacological GSK-3-Specific Inhibitor. Nat. Med. 10, 55–63. doi:10.1038/NM979
Schlosser, G. (2014). Early Embryonic Specification of Vertebrate Cranial Placodes. Wires Dev. Biol. 3, 349–363. doi:10.1002/WDEV.142
Schultz, L. E., Haltom, J. A., Almeida, M. P., Wierson, W. A., Solin, S. L., Weiss, T. J., et al. (2018). Epigenetic Regulators Rbbp4 and Hdac1 Are Overexpressed in a Zebrafish Model of RB1 Embryonal Brain Tumor, and Are Required for Neural Progenitor Survival and Proliferation. DMM Dis. Model. Mech. 11, 1–15. doi:10.1242/dmm.034124
Sen, B., and Johnson, F. M. (2011). Regulation of Src Family Kinases in Human Cancers. J. Signal Transduction 2011, 1–14. doi:10.1155/2011/865819
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 13, 2498–2504. doi:10.1101/GR.1239303
Sharma, D., Holets, L., Zhang, X., and Kinsey, W. H. (2005). Role of Fyn Kinase in Signaling Associated with Epiboly during Zebrafish Development. Dev. Biol. 285, 462–476. doi:10.1016/J.YDBIO.2005.07.018
Simeone, A. (2000). Positioning the Isthmic Organizer: Where Otx2 and Gbx2 Meet. Trends Genet. 16, 237–240. doi:10.1016/S0168-9525(00)02000-X
Slater, T. (2014). Recent Advances in Modeling Languages for Pathway Maps and Computable Biological Networks. Drug Discov. Today 19, 193–198. doi:10.1016/J.DRUDIS.2013.12.011
Solomon, K. S., Kudoh, T., Dawid, I. B., and Fritz, A. (2003). Zebrafish Foxi1 Mediates Otic Placode Formation and Jaw Development. Development 130, 929–940. doi:10.1242/DEV.00308
St. Clair, R. M., Emerson, S. E., D'Elia, K. P., Weir, M. E., Schmoker, A. M., Ebert, A. M., et al. (2018). Fyn‐Dependent Phosphorylation of PlexinA1 and PlexinA2 at Conserved Tyrosines Is Essential for Zebrafish Eye Development. FEBS J. 285, 72–86. doi:10.1111/FEBS.14313
Stolle, K., Schnoor, M., Fuellen, G., Spitzer, M., Cullen, P., and Lorkowski, S. (2007). Cloning, Genomic Organization, and Tissue-specific Expression of the RASL11B Gene. Biochim. Biophys. Acta (Bba) - Gene Struct. Expr. 1769, 514–524. doi:10.1016/J.BBAEXP.2007.05.005
Szklarczyk, D., Gable, A. L., Nastou, K. C., Lyon, D., Kirsch, R., Pyysalo, S., et al. (2021). The STRING Database in 2021: Customizable Protein-Protein Networks, and Functional Characterization of User-Uploaded Gene/Measurement Sets. Nucleic Acids Res. 49, D605–D612. doi:10.1093/NAR/GKAA1074
Tamagnone, L., Artigiani, S., Chen, H., He, Z., Ming, G.-l., Song, H.-j., et al. (1999). Plexins Are a Large Family of Receptors for Transmembrane, Secreted, and GPI-Anchored Semaphorins in Vertebrates. Cell 99, 71–80. doi:10.1016/S0092-8674(00)80063-X
Tsai, M.-Y., Lu, Y.-F., Liu, Y.-H., Lien, H.-W., Huang, C.-J., Wu, J.-L., et al. (2015). Modulation Ofp53andmetexpression by Krüppel-like Factor 8 Regulates Zebrafish Cerebellar Development. Devel Neurobio. 75, 908–926. doi:10.1002/dneu.22258
Villeneuve, D. L., Crump, D., Garcia-Reyero, N., Hecker, M., Hutchinson, T. H., LaLone, C. A., et al. (2014a). Adverse Outcome Pathway (AOP) Development I: Strategies and Principles. Toxicol. Sci. 142, 312–320. doi:10.1093/TOXSCI/KFU199
Villeneuve, D. L., Crump, D., Garcia-Reyero, N., Hecker, M., Hutchinson, T. H., LaLone, C. A., et al. (2014b). Adverse Outcome Pathway Development II: Best Practices. Toxicol. Sci. 142, 321–330. doi:10.1093/TOXSCI/KFU200
Villeneuve, D. L., Angrish, M. M., Fortin, M. C., Katsiadaki, I., Leonard, M., Margiotta-Casaluci, L., et al. (2018). Adverse Outcome Pathway Networks II: Network Analytics. Environ. Toxicol. Chem. 37, 1734–1748. doi:10.1002/ETC.4124
Vinken, M. (2013). The Adverse Outcome Pathway Concept: A Pragmatic Tool in Toxicology. Toxicology 312, 158–165. doi:10.1016/J.TOX.2013.08.011
Wang, Z., Nakayama, Y., Tsuda, S., and Yamasu, K. (2018). The Role of Gastrulation Brain Homeobox 2 (Gbx2) in the Development of the Ventral Telencephalon in Zebrafish Embryos. Differentiation 99, 28–40. doi:10.1016/J.DIFF.2017.12.005
Whitfield, T. T., Granato, M., Van Eeden, F. J., Schach, U., Brand, M., Furutani-Seiki, M., et al. (1996). Mutations Affecting Development of the Zebrafish Inner Ear and Lateral Line. Development 123, 241–254. doi:10.1242/dev.123.1.241
Whitfield, T. T., Riley, B. B., Chiang, M.-Y., and Phillips, B. (2002). Development of the Zebrafish Inner Ear. Dev. Dyn. 223, 427–458. doi:10.1002/DVDY.10073
Whitfield, T. T. (2002). Zebrafish as a Model for Hearing and Deafness. J. Neurobiol. 53, 157–171. doi:10.1002/neu.10123
Wong, M. L., and Medrano, J. F. (2005). Real-time PCR for mRNA Quantitation. BioTechniques 39, 75–85. doi:10.2144/05391RV01
Xu, P.-X., Adams, J., Peters, H., Brown, M. C., Heaney, S., and Maas, R. (1999). Eya1-deficient Mice Lack Ears and Kidneys and Show Abnormal Apoptosis of Organ Primordia. Nat. Genet. 23, 113–117. doi:10.1038/12722
Yao, Q., Desmidt, A. A., Tekin, M., Liu, X., and Lu, Z. (2016). Hearing Assessment in Zebrafish during the First Week Postfertilization. Zebrafish 13, 79–86. doi:10.1089/zeb.2015.1166
Zagozewski, J. L., Zhang, Q., and Eisenstat, D. D. (2014). Genetic Regulation of Vertebrate Eye Development. Clin. Genet. 86, 453–460. doi:10.1111/cge.12493
Zegzouti, H., Zdanovskaia, M., Hsiao, K., and Goueli, S. A. (2009). ADP-Glo: A Bioluminescent and Homogeneous Adp Monitoring Assay for Kinases. ASSAY Drug Dev. Tech. 7, 560–572. doi:10.1089/adt.2009.0222
Zheng, W., Huang, L., Wei, Z.-B., Silvius, D., Tang, B., and Xu, P.-X. (2003). The Role ofSix1in Mammalian Auditory System Development. Development 130, 3989–4000. doi:10.1242/dev.00628
Zhou, L., An, N., Jiang, W., Haydon, R., Cheng, H., Zhou, Q., et al. (2002). Fluorescence-Based Functional Assay for Wnt/β-Catenin Signaling Activity. Biotechniques 33, 1126–1135. doi:10.2144/02335dd07
Keywords: systems toxicology, adverse outcome pathway, causal network, toxicological network, neurotoxicity
Citation: Ramšak Ž, Modic V, Li RA, vom Berg C and Zupanic A (2022) From Causal Networks to Adverse Outcome Pathways: A Developmental Neurotoxicity Case Study. Front. Toxicol. 4:815754. doi: 10.3389/ftox.2022.815754
Received: 15 November 2021; Accepted: 31 January 2022;
Published: 07 March 2022.
Edited by:
Daniel Villeneuve, United States Environmental Protection Agency, United StatesReviewed by:
Lucia Vergauwen, University of Antwerp, BelgiumJui-Hua Hsieh, National Toxicology Program Division (NIEHS), United States
Copyright © 2022 Ramšak, Modic, Li, vom Berg and Zupanic. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anze Zupanic, YW56ZS56dXBhbmljQG5pYi5zaQ==
 Živa Ramšak
Živa Ramšak Vid Modic
Vid Modic Roman A. Li
Roman A. Li Colette vom Berg
Colette vom Berg Anze Zupanic
Anze Zupanic