- Department of Computer Science, St. Francis Xavier University, Antigonish, NS, Canada
Radiation’s harmful effects on biological organisms have long been studied through mainly evaluating pathological changes in cells, tissues, or organs. Recently, there have been more accessible gene expression datasets relating to radiation exposure studies. This provides an opportunity to analyze responses at the molecular level toward revealing phenotypic differences. Biomarkers in toxicogenomics have been suggested as indicators of radiation exposure and seem to react differently to various dosages of radiation. This study proposes a predictive gene signature specific to radiation exposure and can be used in automatically diagnosing the exposure dose. In searching for a reliable gene set that will correctly identify the exposure dose, consideration needs to be given to the size of the set. For this reason, we experimented with the number of genes used for training and testing. Gene set sizes of 28, 100, 200, 300, 400, 500, 600, 700, 800, 900 and 1,000 were tested to find the size that provided the best accuracy across three datasets. Models were then trained and tested using multiple datasets in various ways, including an external validation. The dissimilarities between these datasets provide an analogy to real-world conditions where data from multiple sources are likely to have variances in format, settings, time parameters, participants, processes, and machine tolerances, so a robust training dataset from many heterogeneous samples should provide better predictability. All three datasets showed positive results with the correct classification of the radiation exposure dose. The average accuracy of all three models was 88% for gene sets of both 400 and 1,000 genes. R400 provided the best results when testing the three datasets used in this study. A literature validation of top selected genes shows high relevance of perturbations to adverse effects reported during cancer radiotherapy.
Introduction
Radiation is all around us from both nature and human sources. Studies of radiation exposure over the past 75 years—including longitudinal studies of Hiroshima and Nagasaki survivors, nuclear accidents, radiology workers and scientists, space travel, and nuclear medicine—have produced lots of data (Lacombe et al., 2018). Such a diagnostic method could be used in testing a large number of people exposed during a natural or artificial disaster with mass radiation exposure in a timely manner. The current gold standard for ionizing radiation diagnosis is dicentric chromosome assay (DCA) but it takes time and expertise to complete each sample (Ostheim et al., 2022). This means that throughput is limited by the availability of capable clinicians. Samples will need to travel to the lab for manual examination and the results will need to be communicated to the triage center adding delay to time sensitive diagnosis and treatment. Determining an optimal dose predictive gene set could help develop a cost-effective, high-throughput testing system for use on an exposed population to allow faster triage according to dose (Broustas C. G. et al., 2017; Biolatti et al., 2021).
For the purposes of this study, concentration was given to looking for diagnostic biomarkers, which are characteristics that either indicate the presence of a disease or subtype of a disease. These characteristics, such as gene expression, indicate exposure to ionizing radiation (Ghandhi et al., 2015). Many biomarkers in toxicogenomics have been suggested and studied with the idea that these biomarkers can provide evidence of radiation exposure dose, duration, and class (Simon, 2011). Radiation biomarker studies have identified injuries due to exposure (Howe et al., 2021, p. 4). The number of genes identified in the literature review varied by study (Shuryak et al., 2020) (Panera et al., 2021). Although these studies are varied over a wide variety of circumstances and factors (such as species), some common biomarkers were found across studies which have shown to have a measurable response to radiation exposure. There has been research on identifying radiation sensitive genes (Broustas CG. et al., 2017). In this study, we looked to see if the number of genes used in a predictive signature affected the models’ accuracies. We also looked at which machine learning classifier performed the best across gene set sizes and datasets. Then we tested merging datasets so that the training samples were more heterogeneous and would simulate adding a variety of real-world samples to build a more robust diagnosing model. Lastly, we trained models using two different datasets and then tested the models on a third dataset’s samples.
Identifying affected genes and studying their reactions to exposure doses may provide the means to predict future health outcomes by connecting known diseases to specific genes and cellular pathways. Gene expressions biomarkers are affected by different toxins and aberrations appear quickly after exposure. Environmental factors such as chemicals can interact with genes and cause the gene’s production rate to change. Sometimes genes are up-regulated by chemicals and production is increased (or vice-versa) (Howe et al., 2021). These expression changes are the biomarkers used in this study to find identifiable signatures of sets of genes (Ghandhi et al., 2015). Since biomarkers should be able to be used to determine the dose of radiation exposure, a treatment can be prescribed which will be most effective when started as early as possible. Due to the fast rate of gene expression, diagnosis could be done before symptoms appear. One event can cause varying doses among its subjects, so a method is needed to quickly identify the dose of each person. Determination of dose can also influence treatment so fast diagnosis is important. A mass exposure event will need triaging so that appropriate treatments can be applied to individuals. Since expertise and lab space are needed for the current gold standard, a means to diagnose using a high-throughput method would help categorize victims so that they can receive the best treatment. Genomic assays can be taken during such events so that diagnoses can be conducted very quickly, and appropriate treatments applied to each person.
This paper proposes a predictive gene signature that is specific to radiation exposure and can be used in automatic diagnosing of the exposure dose. Having a robust training dataset from many heterogeneous samples should provide better predictability across a broad population of diverse people. Our study concentrated on samples exposed to X-ray, Beta, and Gamma ray types of radiation. To ensure that the most inclusive model is built which will be sensitive to gene responses across diverse populations, a wide range of samples from all demographics and geographical locations should be used for training. The datasets downloaded in this study were based on radiation and their effects on gene expressions and found on Gene Expression Omnibus (GEO) (www.ncbi.nlm.nih.gov/geo). Analysis of the combined responses of these genes to differing levels of radiation show promise of identifying predictive signatures of exposure. This paper found that a machine learning model trained on a signature set of genes’ expressions can provide accurate predictions of the dose of radiation exposure. A signature gene set was developed for this purpose and is called R400. A signature of a larger number of genes was also developed called R1000 which extends the R400 gene set to include 1,000 genes. The genes for each signature are listed in the Supplementary Tables S4, S5 respectively. Three datasets were used: GSE90909, GSE58613, and GSE65292. All three datasets showed positive results with correct classification of the dose of the radiation exposure. The accuracy of each model was 91%, 74%, and 100% respectively. The average accuracy of all three models was 88% for gene sets of both R400 and R1000 genes. All scripts and programs used in this study are available online on GitHub at https://github.com/Howaboutthis1/R400.
Materials and methods
GEO datasets
From Gene Expression Omnibus (GEO), the GSE90909, GSE58613, and GSE65292 datasets (see Table 1) were downloaded for the use of this study. A disease-gene correlation scheme was built and run on the GSE90909 dataset.
GSE90909[12288 genes, 51 samples (only x-ray samples were selected)] was built in a study which exposed blood samples to various doses of radiation ex vivo. There were 12 different healthy human donors (6 females and 6 males) and a total of 92 samples included in the dataset (Broustas CG. et al., 2017).
GSE58613[21225 genes; 53 samples (only healthy samples were selected)] contained samples from healthy humans and humans with various illnesses, aged between 21 and 66. Some samples were irradiated ex-vivo and some were taken after total body irradiation was performed. Blood samples were taken before and after patients underwent radiation treatment. Various exposure doses and lengths of time since irradiation were used in the original study (Lucas et al., 2014).
GSE65292[12073 genes; 35 samples] has gene expression data for human blood samples that were irradiated ex vivo with a variety of doses. The radiation was applied at two dose rates denoted as “acute” and “low” (Ghandhi et al., 2015).
The aim of considering several datasets is to see if predictions can be made to diagnose the exposure dose using a specific gene expression signature, we call R400. Moreover, recent studies are showing that certain genes are differentially responsive at various dosages. Lacombe et al. stated that combinations will be needed rather than individual biomarkers and that more studies are needed to overcome the differences in studies’ protocols and methods (Lacombe et al., 2018). The three datasets used in this study had limited demographic information available. Expanding and adding datasets to the training sets with samples from diverse populations should improve the efficacy across various demographics.
Proposed framework
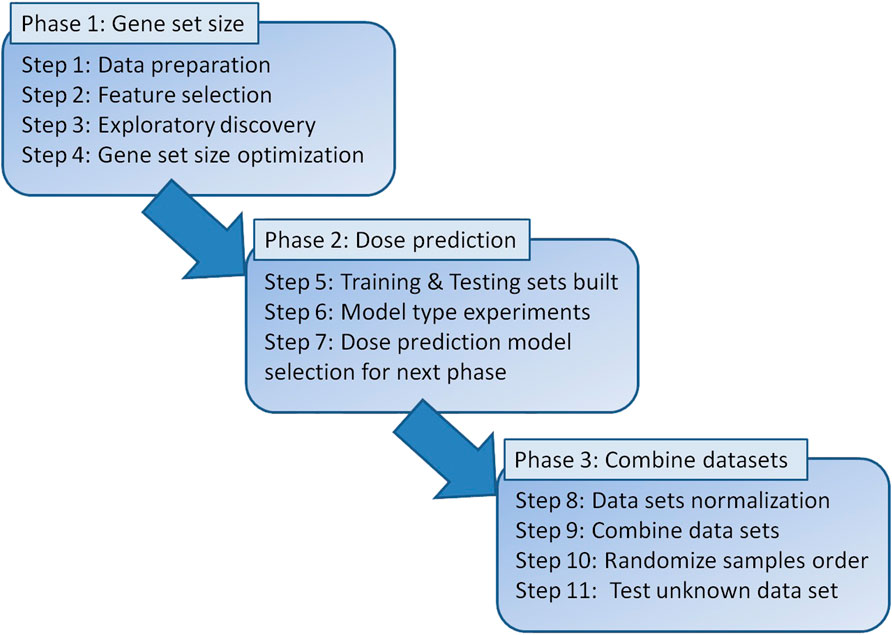
Figure 1 shows the workflow of the steps outlined in this paper. Phase 1 consisted of cleaning and preparing the datasets for feature selection which resulted in a correlation score for each gene. This allowed for various gene set sizes to be built from the top genes and tested throughout the study.
Phase 2 was training models with various gene set sizes and various classifier models using 5-fold cross validation. Testing accuracies were used to select the model with the best predictive capabilities for the external validation by combining datasets in Phase 3.
Phase 3 was external validation which was performed such that two datasets were processed as described earlier and merged into a training set. The third dataset was then used to test the model. GSE90909 was selected as the test set since its range of doses was within the range of doses in each of the other two datasets and its models showed accurate predictions during individual dataset testing. The raw data is prepared, and feature selection is done considering that each gene selected as a feature must be present in all three datasets. Next, the two datasets were merged, normalized, and used to train a classifier model. The third dataset, used for training, was then normalized, and tested on the classifier for dose prediction.
Raw data pre-processing
The raw datasets were imported into R using Bioconductor, Biobase, and GEOquery packages. When the gene expression samples were extracted, some of the data required for training and testing were not included in a machine-readable format. The doses of each sample had to be added so that the classifier could learn which dose was exposed to each sample. Gene assays sometimes use probe names rather than gene symbols to identify the expressions of each gene. Gene names were, therefore, mapped and extracted from each dataset’s metadata for readability during gene selection and test results. The datasets were imported from the Gene Expression Omnibus (GEO) repository in raw format and needed to be formatted so that dose, gene name, and sample id lined up among the datasets. The three databases were left as untouched as possible with the only change being made at this point was to match the dose measurements of centiGrays (cGy) to Grays (Gy) by dividing them by 100. This was done so that the classifier would evaluate the doses in each dataset on the same scale. The expression data was further prepared by replacing any NAs with averages of each of that gene’s expressions. The dose data was transformed to numeric values and added to each of the samples’ gene expressions for the machine learning training and testing steps.
GSE90909 contained samples which were exposed to x-ray or neutron radiation. The doses for this dataset ranged from 0 to 4 Gy. For radiation type consistency across datasets, only the x-ray and control samples were used providing 51 samples for testing. GSE58613 contained samples from healthy and total body irradiated (TBI) individuals which either received no treatment, Granulocyte colony stimulating factor (G-CSF), or lipopolysaccharide (LPS) treatments. The radiation source was Caesium-137 (Cs137) which emits beta and gamma rays. The doses in this dataset ranged from 0 to 600 cGy which were converted into 0–6 Gy to match the dose scale of the other two datasets. For consistency across datasets only the healthy, non-treated samples were used resulting in 26 samples for testing. All of the samples in GSE65292 were used. There are 35 samples with doses ranging from 0 to 4.45 Gy. Radiation was applied using a x-ray irradiator.
Gene ranking and selection
The three datasets chosen for this study have 12,288 genes (GSE90909), 22,277 genes (GSE58613), and 12,073 genes (GSE65292). Including large number of genes would limit applicability for high-throughput radiation exposure testing especially when many genes are non-reactive. In addition, including more genes (or features) can impact the performance of the machine learning (ML) algorithm (Soufan et al., 2015). Therefore, we reduced the number of genes using the Pearson’s correlation method which has shown improved performance in gene expression datasets (Soufan et al., 2015; Spainhour et al., 2019). Pearson’s correlation method was used to rank all of the genes by calculating a correlation score for each gene to allow the top dose-correlated genes to be sorted and selected. These scores were then used to select a set of top genes and then perform evaluation using machine learning models. As external verification, the genes discovered were compared to genes listed in current literature and the dose response was verified using fastBMD (www.fastbmd.ca), an online benchmark dose analysis service which accepts GEO datasets that conform to their standard file format (Ewald et al., 2021). Different gene set sizes were modeled to find the optimum size for best prediction accuracy. Choosing genes in GSE90909 with correlation scores above 0.80 defined 28 genes as candidates as predictive biomarkers. Other tested gene set sizes were 100, 200, 300, 400, 500, 600, 700, 800, 900 and 1,000 of the top correlated genes in each dataset.
Correlation analysis of gene to dose was used to sort all of the genes for feature selection. Only genes that were included in all three of the datasets used in this study were included. To determine the optimal gene set size, machine learning was run on Matlab’s classifier app using 15 of the built-in classifier types with 5-fold cross validation when training. This was done for gene set sizes of: 28, 100, 200, 300, 400, 500, 600, 700, 800, 900 and 1,000. Two datasets were used separately to determine the best gene set size across datasets using the validation accuracies. The ML model type with the best results was exported and used for testing unknown datasets on a model trained on a two-dataset training set. The Linear Discriminant classifier most often obtained the best results and was chosen as the model type for the 70/30 split experiments.
Machine learning for dose classification prediction
Before setting out to determine the optimal gene set size, the best model type needed to be found for model testing. Review of past toxicogenomics literature revealed 1,000 genes has been successfully used in feature selection for dose-response models like in T1000 and L1000 (Soufan et al., 2019). To find the model type to use going forward, we tested 15 machine learning model types on 11 gene set sizes. The machine learning model types are listed in the ranking table (Supplementary Table S3) in the supplemental files. Gene set sizes of 28, 100, 200, 300, 400, 500, 600, 700, 800, 900, and 1,000 were used for training and testing on each of the 15 model types using two different datasets separately. In total, 330 models were built. All of the training was done using 5-fold cross-validation and the training results are tabulated in the supplementary file (Supplementary Table S3). Matlab’s linear discriminant classifier (LDA) most often had the best training validation accuracies across gene set sizes.
Performance evaluation and experimental setup
The performance of each model was evaluated by looking at the correctly predicted percentage of testing samples. Accuracy = # of correct predictions/# of testing cases (such that # of correct predictions = number of predictions where the predicted label matches that actual dose). Models’ predictions either correctly matched actual doses or they did not. Correctly matched predictions were counted as True Positives (TP). There are no True Negatives (TN) predicted so for all accuracy calculations in this paper TN = 0.
Each model’s results were summarized on a multi-class confusion matrix which is a table consisting of two axes: one axis for the model’s predicted doses and one axis for the actual doses of the samples. The same dose classes are used and ordered on both axes, so that predicted and actual doses intersect diagonally. When a predicted dose is correctly classified as the actual dose, they meet on the corresponding cell of the confusion matrix so that the diagonal line of cells from the top, left corner to the bottom, right corner represents the correctly predicted samples.
The classification models use decision boundaries to best identify discrete dose classes. This allows calculations of prediction accuracies based upon the number of correctly (on the diagonal) and incorrectly (off the diagonal) predicted doses.
Evaluation 1: Internal testing setup
Each dataset’s samples were split into 70% training sets and a 30% testing sets. Then, 5-fold cross-validation was executed within each training set. Cross-validation was used to enhance the internal parameters of the ML models. The 30% partitions were tested on the models and the accuracies of each experiment are shown in Table 2. We have applied this testing setup on all three datasets separately and reported the results.
Evaluation 2: External testing setup
We have considered a different testing setup as an external validation of the model. Since the Linear Discriminant model provided the overall best results in this study (Supplementary Table S3), it was used to train a model on two datasets combined and reported the results on the third dataset. Two datasets were combined to provide the highest range coverage of doses (0–6 Gy). Then, the third dataset was used for testing only. It was chosen because all of its doses were within the range of doses of the combined dataset. We report performance scores using the separate test and consider this as an external validation of the model.
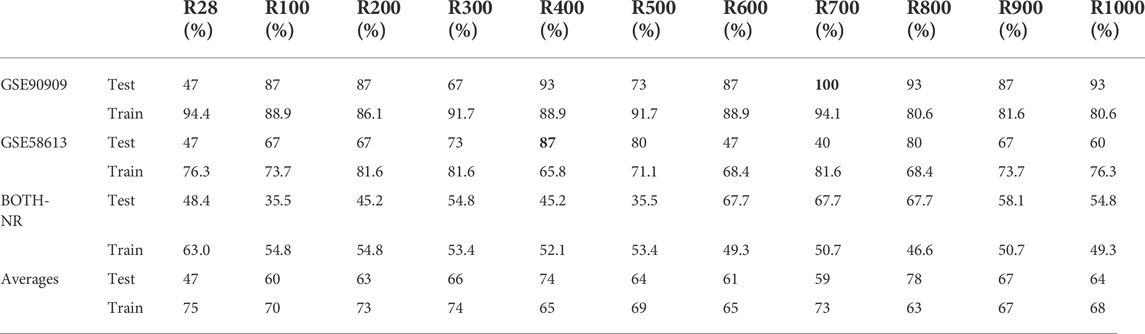
R400 and R1000 construction
R400 and R1000 were built by pulling the top 400 and 1,000 genes with the highest correlation scores that exist in all three datasets. Then the genes were ordered by correlation rank according to GSE90909’s order. GSE90909 was used as the test dataset on the models built using the other two datasets. The results in Table 2 show that GSE90909 had eight out the top ten testing accuracies. The best accuracy that was not from GSE90909 was for GSE58613 at 400 genes (87%) and GSE90909’s second best score was using 400 genes (93%). The overall accuracies of the models built using 400 genes outperformed the other gene set sizes. It should be noted that high accuracies were also noted at gene set sizes of 700 and 800 but individual dataset accuracies of these sets were not as high as at 400 genes (Table 2; Figure 2). One problem that may occur with a patient’s sample is that it may not contain all the genes found in R400 so a larger gene set, called R1000, was created. It contains R400 plus 600 more genes so that the top 400 genes found in the patient’s sample can be pulled from R1000 for testing.
Results
Dose-response prediction results
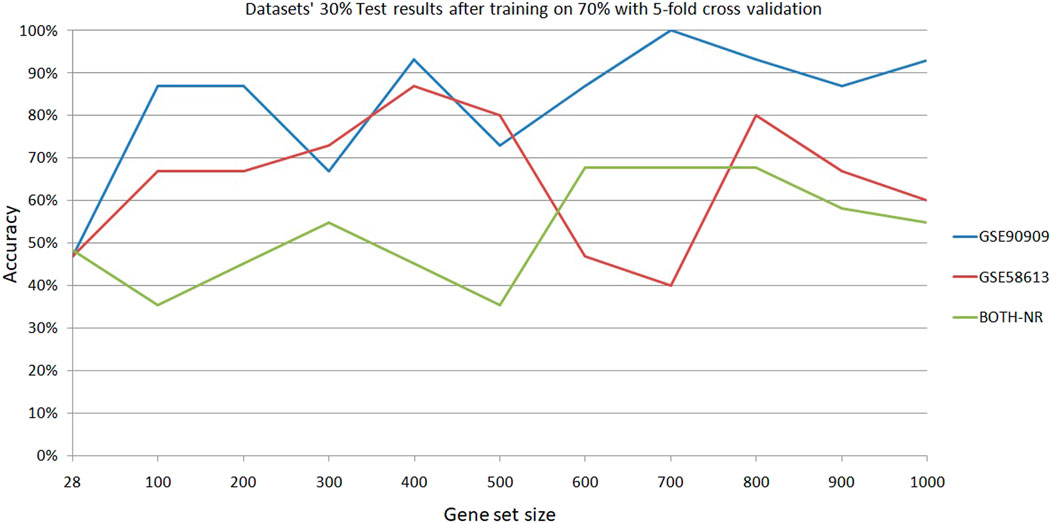
We have performed 330 experiments using 15 classifiers across 11 gene set sizes. For dataset 1, linear discriminant classifier (LDA) achieved the highest accuracy 9/11 times (82%). For dataset 2, LDA had the highest accuracy 4/11 times (36%). For each dataset, LDA most often had the highest accuracy. The average test results of the two datasets reached its maximum of 90% at 400 genes (Figure 2). For these reasons, gene sets sizes of 400 and 1,000 were decided upon for testing models built on multiple datasets and testing datasets which were not used in training. To confirm the preliminarily test results, Matlab’s linear discriminant classifier with 5-fold cross validation was used on all x-ray and control samples of GSE90909. All healthy, non-treatment and control samples from GSE58613 were used for the second model and all samples of GSE65292 were used for the third model. Two datasets were combined and partitioned at 30% test/70% train. The results show promise because there is a clear clustering near the correct predictions diagonal of the confusion matrix (Figure 3A). If tolerances are increased, the accuracy can be improved from 61.3% to 96.8% by catching neighbouring cells in the confusion matrix (Figure 3B). This is reasonable given that the two datasets have variances in their data.
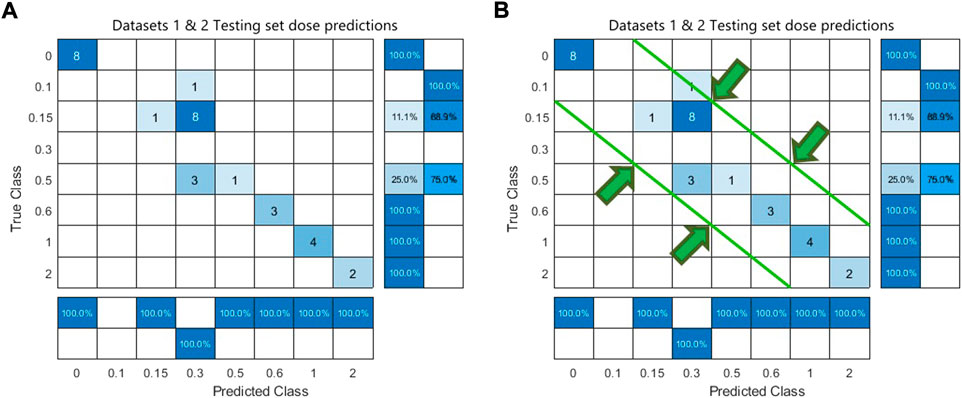
FIGURE 3. Confusion matrix of combined Datasets 1 & 2. Panel (A) shows a strict accuracy of 61.3% where TP are predictions that match true classes. Panel (B) allows a prediction that lands adjacent to the true class to be counted at a TP which brings the accuracy up to 96.8%. Widening the tolerance of allowed cells is reasonable due to datasets having different dose classes. i.e., Dataset 3 has no 0.3 class but it does have a 0.25 class.
Analysis of sizes of gene sets
The accuracies of the ML classifier predictions in this study indicate that a predictive gene signature for diagnosing the dose of exposure to a sample has merit and that the number of genes used in the gene signature affected the predictive accuracies of classifier models. There appears to be an optimal number of genes which increases the accuracy of predictions. Although there were variations in the results as the gene set size changed, they generally increased at 400 genes. This was seen during the classifier selecting stage and in the 70train/30test runs. Test accuracies at 400 genes were 93% for GSE90909, 87% for GSE58613 (Figure 2). As datasets became more heterogeneous by mixing two datasets, the accuracy of the 400 gene model was 48%. Accuracies dropped further when testing samples were from a dataset that was not included in the model training (Figure 4). The testing accuracies of the mixed dataset model had a narrow range of 20%–22% across gene set sizes. The training validation accuracies, however, crested above 80% in the curve from 200 genes (83.6%) to 400 genes (80.3%) and hit the maximum at 300 genes (85.2%) (Supplementary Table S2). The results found during this study support the proposal of an optimized gene signature based on 400 genes. We have named this set of genes R400.
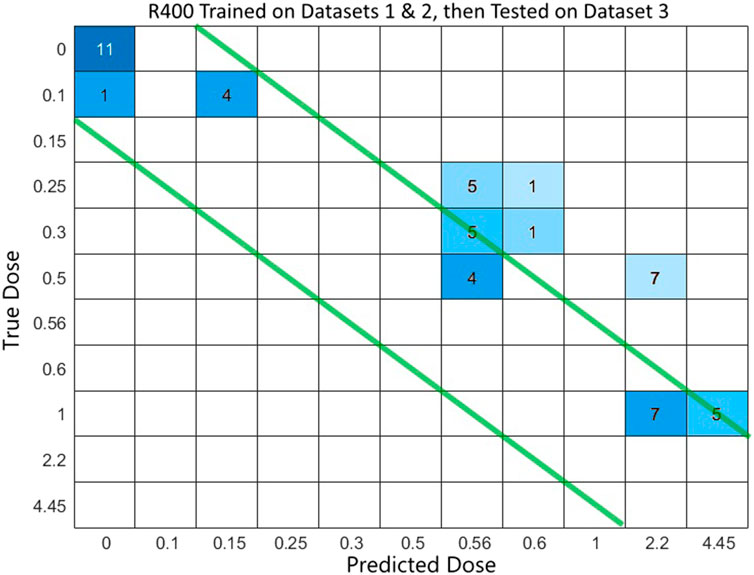
FIGURE 4. Confusion matrix of dose prediction from model trained on Datasets 1 & 2, then tested on Dataset 3.
Models were able to predict the dose of exposure after being trained on datasets. Although accuracies varied throughout the range of experiments performed in this study, there was an underlying trend of predictability in each of the models. Accuracies dropped as complexities were added to the experiments. In this study, we found that 400 genes provided the best overall results across GSE90909 and GSE58613. At 700 genes, GSE90909 (by itself) obtained the highest testing accuracy of 100% but GSE 58613 scored 40% which averages to 70%. At 400 genes, they scored 93% and 87% respectively which averages to 90% and is the best performance (Table 2).
Predicting dose from one dataset on a model built using multiple other datasets using Evaluation 2
For a diagnostic model to be useful, it must be able to diagnose a sample that is not from a known dataset, and it must be able to incorporate different datasets in its training. To test if such a model would achieve good results, a multiple-dataset model was constructed. This time, the two datasets GSE58613 and GSE65292 were normalized and combined and used to train machine learning classifiers in Matlab. The dataset GSE90909 was normalized and used for testing the model. All three datasets were normalized separately and then two datasets were combined.
The combined dataset of 1&2 was used as the training set for classification machine learning app of Matlab then recorded model validation results and exported model. The model was used to predict doses of samples from a 3rd dataset. Multiple gene set sizes were tested ranging from 28 to 1,000. When the test sets were tested on the models, the dose prediction can be improved if the correct detection diagonal is again widened to catch adjacent cells in the confusion matrix for nearest dose diagnosis (Figure 4). That will improve usability in the field when the training dataset may not have seen samples similar to the unknown samples that they are testing. The predictions appear to have a tangent line which seems to imply a bias or skewing of the results (Supplementary Figures S2–S4) which should be possible to account for so that predictive capabilities are maintained across a wide range of patients. This linear skewing/bias suggests a linear relationship with dose for this gene set so linear regression was done using Matlab.
Linear regression models were trained on the combined, normalized dataset of GSE58613 and GSE65292 which was used for the training of the ML classification models. Then the unknown samples from GSE90909 were tested on two models built using R400 and R1000. The Root Mean Square Error (RMSE) of the test results from R400 were slightly better than those from R1000 and the plots of predicted vs. actual doses seem to show a skew or bias between the datasets because the plotted data all falls in similar linear clumping pattern to the left of the correct prediction diagonal line (Supplementary Figure S4).
Discussion
The work in this paper is intended to see if a specific test or assay can be developed to diagnose the dose of ionizing radiation exposure of samples efficiently and accurately from various situations and locations. We looked for an optimized number of genes with the best predictability. Having an optimized gene set size would allow radiation-dose prediction specific assay equipment to be developed so that machine learning models can be used in the place of expert examinations for high-throughput diagnoses. We run a total of 2,475 experiments (i.e., 15models x 5folds x 11gene sets x 3datasets) with gene set sizes of 28, 100, 200, 300, 400, 500, 600, 700, 800, 900, and 1,000 across three datasets. For such models to be as accurate as possible, they will need to be trained using as wide of a population base as available to have the best real-world matching heterogeneity. For this study only genes that were included in all three datasets were considered candidates for the R400/R1000 signatures so that the machine learning models would not have any missing features’ values for training or testing. There appears to be an optimized size of the gene set used in machine learning for radiation exposure dose prediction. Preliminary reduction of a gene set size from 12,288 to 28 genes, selected for correlation scores above 0.80, resulted in improved prediction accuracy from 51.9% to 94.2%. The optimum number of genes seems to peak between 400 and 800 genes depending on the dataset used. Data across studies have variations in the experiments that can cause results from one study to have dissimilarities in datasets from a similar study. Things such as equipment differences, procedures, and donor population sampling can make similar biological reactions have differently recorded results compared to results from other studies. For this reason, a specific gene set used in multiple studies can provide differing results. This study found an optimal ordered gene set comprised of the 400 top selected genes which we call R400. Since some samples presented for testing may not contain all genes in R400 and because predictive accuracy also peaked at 800 genes (Figure 2), a gene set was created with 1,000 genes which we called R1000. As such, R400 is a subset of R1000; it is comprised of the top 400 available genes of R1000. We think these two gene sets are important because the accuracy peaked at a number close to 400 genes and again at close to 1,000 genes. The order of the gene set affected the model’s accuracy. This means that the accuracy of a model built on R400 will have best results if the order of the genes is maintained as is listed in the Supplementary Table S4. The same goes for R1000 which is provided as Supplementary Table S5.
The gene set size for this paper was initially chosen to be 28 by limiting inclusion to only those genes with a Pearson correlation score >0.80. Much of the literature found on radiation effects on specific genes listed two to six genes as being reactive to exposure. Other toxicogenomics studies have narrowed the number of genes to 1,000 and 1,500 (Subramanian et al., 2017). Earlier we have reported T1000 and illustrated better results were obtained with the gene set size of 1,000 (Soufan et al., 2019). Further, we explored grouping these genes and summarizing their expression into modules to support deciding if a chemical exposure was toxic or non-toxic (Ewald et al., 2021, 2020). Similarly, this study also found higher accuracies by including up to 1,000 genes (Figure 2; Table 2).
In this study, as the heterogeneousness of the samples increased, the predictions became less accurate. For example, when two datasets were merged to create a larger, more diverse dataset, accuracies dropped (Table 2). Even lower accuracies were seen when models were trained on one or more datasets and then samples from another dataset were used for testing. Even so, the predictions seemed to maintain a straight, diagonal pattern but were skewed from the left-down diagonal (Figure 4).
Machine learning was able to predict the dose of radiation exposure of samples across different datasets using the same gene set. Average accuracies for each gene set size peaked at 79% for R800 and 76% for R400 (Table 2). When a model was trained on one or two datasets, it was able to predict dose for samples from a dataset not used in the training. Accuracies fell when a new dataset was used on a model created from other datasets, but the results still showed a strong predictive capability when consideration is taken of the discrete nature of the classification model and the fact that the model may not have a specific class for the actual dose in a sample. To allow for this, tolerances can be increased by accepting predictions which are adjacent to the diagonal (Figure 3) and dose measures can be expanded so that the neighboring classifications can be combined when appropriate. For example, a dose of 0.56 Gy is the closest class available in the model to the actual dose of 0.50 Gy in the sample being tested. An allowance can be made here because of the predictability of diagnosis is still relevant and helpful at this point. When determining the accuracy of tests, the size of the differences between predicted and actual doses were considered.
The limitations of the predictive model that is proposed here comes from the training data it depends upon. Variations in the conditions and settings under which the samples were collected can have great influence on the accuracy of the model. Variations in populations and the limited number of samples available also limit the robustness of the model. If a sample is submitted for testing that is very dissimilar to the training samples, the accuracy of the prediction may suffer. To overcome this shortcoming, a wide range of samples should be used for training.
Another variance that seems to be possible is a skewing or bias effect shown in the results of datasets on models built from other datasets (Figure 4; Supplementary Figures S2–S4). This is an important point of consideration since a practical use of R400 would be to test samples with unknown exposure doses for patient diagnosis. A calibration method could be developed to allow correct predictions that fall on the skew line. A model trained on combined datasets does seem to have dose prediction capability on unknown samples. More datasets can be added to the model to increase its predictive comprehensiveness for samples with unknown exposure situations. The positive results of combining two datasets and testing on a third dataset imply a model that can be more broadly used by adding more samples from new datasets. This should make the mode more applicable to a wider population making it better able to test unknown samples from varied clinical scenarios.
Having a robust training dataset from many heterogeneous samples should provide better predictability across a broad population of diverse people. To ensure that the most inclusive model is built which will be sensitive and specific to gene responses across diverse populations, a wide range of samples from all demographics and geographical locations should be used for training. We set out to test the validity of using a training set built from multiple datasets to increase the predictability of samples from unknown sources. To do so, datasets had to be merged to simulate the on-going collection of training data for a robust, real-world model.
Literature validation of some selected genes
Radiation exposure is often encountered when treating patients with cancer (e.g., lung or breast cancer). While about half of the patients are tolerant to radiotherapy, others experience adverse effects (Borrego-Soto et al., 2015). Particles from ionizing radiation can transfer some of their high energy into atoms and release electrons that will affect the covalent bonds in DNA (i.e., cause common bonds in DNA to break) (Borrego-Soto et al., 2015). This can lead to apoptosis or even uncontrolled proliferation (Borrego-Soto et al., 2015). The in-place mechanisms consist of a cascade of biochemical events triggered by gene regulation in response to such radiation exposure events (Borrego-Soto et al., 2015). Cells have mechanisms to counteract such exposures, including mechanisms to fix single-stranded DNA breaks, double-stranded DNA breaks, frame shift mutations and even generation of ROS or reactive oxygenation species (Borrego-Soto et al., 2015). Figure 5 highlights how specifically radiation interacts with p53 and what possible pathways it is triggering using our top 5 selected genes.
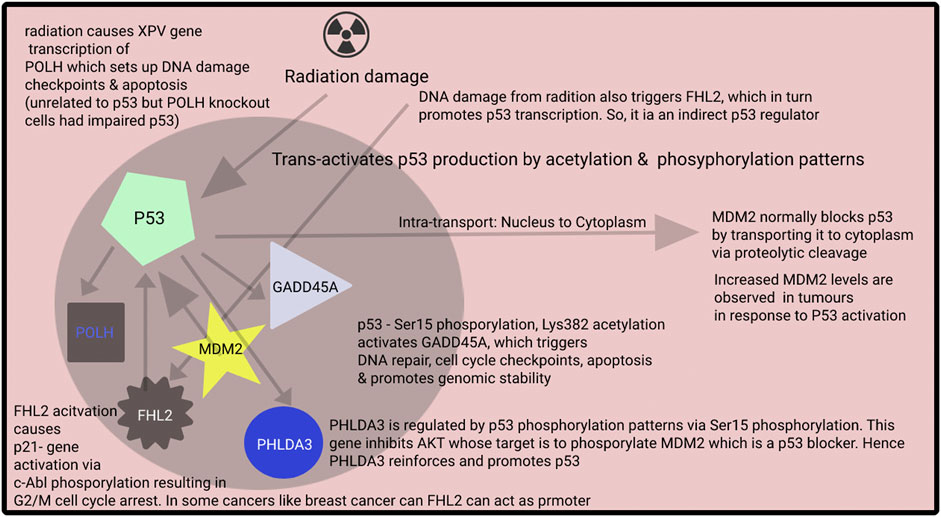
FIGURE 5. Although there are many pathways for cellular stress, usually cellular stress acts via phosphorylation of p53 tumor suppressor genes. P53 is referred to as the guardian of the genome as it is involved in various repair mechanisms. Based on the phosphorylation pattern of p53, various biochemical pathways for cell repair are activated. There are other pathways for, e.g., FHL2 pathway which also handle cellular stress and may have bivalent functions, i.e., act as tumor suppressor or also promoter.
One of the essential anti-cancer or cell repair mechanisms in our bodies is the p53 pathway. Typically, p53 produced is ubiquitylated by the cell’s MDM2 ligase and hydrolyzed 26S proteasome (Roy et al., 2022). However, radiation exposure or any metabolic stress causes the phosphorylation of p53 to various degrees at the N-terminus (He et al., 2019). These effects occur while undergoing acetylation, phosphorylation or sumoylation via cyclin-dependent kinases, checkpoint kinases, or Homeodomain-Interacting Protein Kinase-2 (Zhao and Malik, 2022). These phosphorylation patterns prevent MDM2 from binding and the ubiquitination of p53 (Koo et al., 2022). Based on the degree and the pattern phosphorylation of p53, a different pattern of down cascade biochemical events occurs.
Gamma radiation exposure is reported to affect death receptors based on the Fas gene, belonging to a family of death receptors called Tumor necrosis factor (Lhuillier et al., 2021). Activation of Fas receptors promotes apoptosis of cells. There are also other pathways involved in the extrinsic p53 pathway. The intrinsic p53 pathway, on the other hand, consists of the Bcl-2 class of proteins, which relays the release of Cytochrome-C via permeabilization (Zhang et al., 2021). During metabolic stress, pro-apoptotic proteins BAX, PUMA and NOXA are released, stimulating the release of Cytochrome-C from mitochondria (Chota et al., 2021). Apoptotic Protease-Activating Factor-1, Procaspase-9 and Cytochrome-C combine to form apoptosome complexes. The caspase-9 part of the apoptosome activates caspase3, caspase6 and caspase9, promoting the final demise of the aberrant cell (Avrutsky and Troy, 2021).
Radiation causes similar metabolic stress-related events, and our results indicate a significant similarity between radiation and other metabolic cascades in cancer-related events. We picked several top genes from the R400 signature to evaluate our findings and reported a literature review as supporting evidence.
The GADD45A gene (ranked 1 in our list) is expressed as a result of stressful growth conditions, including radiation and was also shown to have a marked increase in expression over 56% of patients, in stage 0 esophageal cancer (Ishiguro et al., 2016). The activation was independent but sometimes correlated with the activation of p53 tumor suppressor genes. Patients with increased expression of GADD45A showed higher survival rates (Ishiguro et al., 2016). Moreover, it was found that GADD45A overexpression by transfection of cancerous cells using Si-RNA resulted in the cells being stronger and caused fewer cells to become apoptotic when exposed to radiation (Zhang et al., 2011).
The MDM2 gene (ranked 2 in our list) belongs to a class of oncoproteins that is responsible for carrying p53 from the nucleus to the cytoplasm, where p53 is degraded by proteolytic enzymes (Mendoza et al., 2014, p. 2). It was discovered as a result of amplification of amplicons obtained from tumorigenic mouse cell line 3T3DM. It inhibits the p53 tumor suppressor gene, which is triggered during stressful growth conditions such as radiation and cancer. P53 is responsible for activating various repair mechanisms such as antiangiogenic genes, arresting cell growth cycle, apoptosis, DNA repair and autophagy. Essentially P53 triggers the transcriptional activation of MDM2, which in turn blocks p53 unless more p53 is released with additional or incremental stress on the cell (Mendoza et al., 2014). It is also reported that exposure of cells to UV or ionizing radiation was directly correlated to increased MDM2 expression (Perry, 2004, p. 2). The TNFSF4 gene (ranked 3 in our list) belongs to the TNF superfamily, also known as tumor necrosis factor (“TNFSF4 TNF superfamily member 4 [ Homo sapiens (human) ]”, National Library of Medicine, 2022), and was found to be abnormally expressed in breast carcinomas (Li K et al., 2021). As cancers become more and more aggressive, the immune system weakens. The reactivation of immune cells was found to be correlated with increased expression of the TNFSF4 gene (Li K et al., 2021). A dose-dependent increase in TNFSF4 was found in response to X-ray radiation (Li et al., 2014). However, its expression was also found to be associated with chemotherapeutic resistance of carcinoma cells and reduced apoptotic behavior of carcinomas when exposed to radiation along with drugs like cisplatin (Li Y et al., 2021).
The FHL2 (ranked 4 in our list) was reported to be extremely important for pancreatic cancer survival, and it was found that a decrease in its expression was correlated to low survival cells (Zienert et al., 2015). It was found to arrest the cell cycle of cancerous cells or cells exposed to radiation (Wang et al., 2021, p. 2). FHL2 was also found to be one of the genes activated during p53 tumor suppressor activation genes when cells are exposed to stressful conditions such as cancer or DNA damage as a result of radiation, although it is not directly related to p53 (Xu et al., 2014).
POLH (ranked 5 in our list) belongs to a family of DNA polymerases and was found to be upregulated by DNA damage as a result of exposure to ionizing radiation. It was also found that knocking out or downregulating the XPV gene causes mutations in cells and renders them sensitive to UV radiation and also causes issues as it impairs p53 activating during DNA damage and also increases the cancerous cell’s resistance to apoptosis (Liu and Chen, 2006). PHLDA3 (ranked 6 in our list) is yet another downstream product of p53 activation. It is one of the many genres p53 activates as a part of its DNA repair suite. It was found to be activated in many lung carcinomas, indicating a role as a protective mechanism against cancers. PHLDA3 was found to deactivate Akt, which helps the MDM2 protein block p53 in their autoregulatory feedback loop (Kawase et al., 2009). The inactivation of the PHLDA3 gene promotes pancreatic carcinoma development. It was found that the expression of the PHLDA3 gene was also related to exposure to UV or gamma radiation (Ohki et al., 2014). These findings confirm that changes in expression of several of our top-ranked genes can lead to changes in the cell as part of specific pathways given radiation exposure.
Conclusion
There seems to exist a specific geneset that will provide an optimal predictive capability in diagnosing a patient with exposure to an unknown dose of radiation. The size and order of these genes when used for model training are important for accurate and reliable results on test samples. R400 provided the best results when testing on the three datasets used in this study. Since other datasets or samples may not contain expressions from all of R400’s genes, R1000 was created and included here so that the best 400 genes available in the test data could be selected.
For widespread applicability, more datasets should be incorporated in model building because more samples can build a more robust model due to the wider population representation incorporated by using more donors. Although the datasets used in this study had limited donor variability, the combining of datasets was intended to test the viability of merging dissimilar datasets to increase the gene signature’s efficacy among diverse populations. Next efforts should be to find and incorporate radiation-exposure datasets that contain samples that would increase the variability of the training data across each of these characteristics. Time/duration of radiation exposure affects different genes differently. Having a robust dataset upon which to build the model is important to ensure its applicability to various scenarios. This includes the factor of time/duration because as time passes each individual gene in the signature will continue to react in its predictable fashion. This suggests that the R400/R1000 gene signature will have a “predictable” response over time, and by dose. In this paper, we did not eliminate or include any samples based on time/duration but instead allowed the inherent heterogeneity of the datasets to be used in building the models. Each of the datasets had their own timelines which we also felt contributed to the variation of time/duration in this study. We can say the genes showed responses by at least 24 h because that is the shortest amount of time of testing among the datasets used in this study. Since the R400/R1000 model evaluates the expressions of each gene, which each change in predictable ways, the model can learn to even predict the time/duration of samples given enough heterogeneity in the training data. This makes the R400/R1000 applicable to times ranging from earliest changes to latest changes. In this study, the model also correctly classified non-irradiated samples implying that the model can be used as soon as radiation exposure is suspected.
The results suggest a somewhat linear relationship between R400 and dose. This correlation is further supported by the results shown in the skewing of the predictions of a third dataset tested on a model trained on two other datasets.
Other papers found in the literature review often only listed three to twenty genes and some listed up to 1,000. Correlation results of this study looked at a gene set size selected by limiting the genes to only those with correlation scores above 0.80. This resulted in a gene set size of 28 for the first dataset. Testing revealed that more genes than that provided better predictive results which peaked around 400 and 800 genes. The top 400 genes found in all three datasets gave the best results and was called R400. A second gene set of the top 1,000 genes found in all three datasets were ordered and called R1000. Its main purpose is to provide a greater selection of genes to create a R400 set for any given sample which may not have all of the genes listed found in the R400 of this paper.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
FS and OS conceptualized the problem. FS was responsible for solution development and implementation. SV and OS were responsible for validating the new predictions. FS and OS reviewed the text and the evaluation of the work. OS supervised the study.
Funding
This research was supported by the Heaps Chair Endowment Fund at St. Francis Xavier University through Dr. H. Stanley & Doreen Alley Heaps Chairship.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsysb.2022.1022486/full#supplementary-material
References
Avrutsky, M. I., and Troy, C. M. (2021). Caspase-9: a multimodal therapeutic target with diverse cellular expression in human disease. Front. Pharmacol. 12, 701301. doi:10.3389/fphar.2021.701301
Biolatti, V., Negrin, L., Bellora, N., and Ibañez, I. L. (2021). High-throughput meta-analysis and validation of differentially expressed genes as potential biomarkers of ionizing radiation-response. Radiother. Oncol. 154, 21–28. doi:10.1016/j.radonc.2020.09.010
Borrego-Soto, G., Ortiz-López, R., and Rojas-Martínez, A. (2015). Ionizing radiation-induced DNA injury and damage detection in patients with breast cancer. Genet. Mol. Biol. 38, 420–432. doi:10.1590/S1415-475738420150019
Broustas, C. G., Xu, Y., Harken, A. D., Garty, G., and Amundson, S. A. (2017a). Comparison of gene expression response to neutron and x-ray irradiation using mouse blood. BMC genomics 18, 2–13. doi:10.1186/s12864-016-3436-1
Broustas, C. G., Xu, Y., Harken, A. D., Chowdhury, M., Guy, G., and Amundson, S. A. (2017b). Impact of neutron exposure on global gene expression in a human peripheral blood model. Radiat. Res. 187, 433–440. doi:10.1667/RR0005.1
Chota, A., George, B. P., and Abrahamse, H. (2021). Interactions of multidomain pro-apoptotic and anti-apoptotic proteins in cancer cell death. Oncotarget 12, 1615–1626. doi:10.18632/oncotarget.28031
Ewald, J. D., Soufan, O., Crump, D., Hecker, M., Xia, J., and Basu, N. (2020). EcoToxModules: custom gene sets to organize and analyze toxicogenomics data from ecological species. Environ. Sci. Technol. 54, 4376–4387. doi:10.1021/acs.est.9b06607
Ewald, J., Soufan, O., Xia, J., and Basu, N. (2021). FastBMD: an online tool for rapid benchmark dose–response analysis of transcriptomics data. Bioinformatics 37, 1035–1036. doi:10.1093/bioinformatics/btaa700
Ghandhi, S. A., Smilenov, L. B., Elliston, C. D., Chowdhury, M., and Amundson, S. A. (2015). Radiation dose-rate effects on gene expression for human biodosimetry. BMC Med. Genomics 8, 22–10. doi:10.1186/s12920-015-0097-x
He, F., Borcherds, W., Song, T., Wei, X., Das, M., Chen, L., et al. (2019). Interaction between p53 N terminus and core domain regulates specific and nonspecific DNA binding. Proc. Natl. Acad. Sci. U. S. A. 116, 8859–8868. doi:10.1073/pnas.1903077116
Howe, O., White, L., Cullen, D., O’Brien, G., Shields, L., Bryant, J., et al. (2021). A 4-gene signature of CDKN1, FDXR, SESN1 and PCNA radiation biomarkers for prediction of patient radiosensitivity. Int. J. Mol. Sci. 22, 10607. doi:10.3390/ijms221910607
Ishiguro, H., Kimura, M., Takahashi, H., Tanaka, T., Mizoguchi, K., and Takeyama, H. (2016). GADD45A expression is correlated with patient prognosis in esophageal cancer. Oncol. Lett. 11, 277–282. doi:10.3892/ol.2015.3882
Kawase, T., Ohki, R., Shibata, T., Tsutsumi, S., Kamimura, N., Inazawa, J., et al. (2009). PH domain-only protein PHLDA3 is a p53-regulated repressor of Akt. Cell 136, 535–550. doi:10.1016/j.cell.2008.12.002
Koo, N., Sharma, A. K., and Narayan, S. (2022). Therapeutics targeting p53-MDM2 interaction to induce cancer cell death. Int. J. Mol. Sci. 23, 5005. doi:10.3390/ijms23095005
Lacombe, J., Sima, C., Amundson, S. A., and Zenhausern, F. (2018). Candidate gene biodosimetry markers of exposure to external ionizing radiation in human blood: A systematic review. PLOS ONE 6 13, e0198851. doi:10.1371/journal.pone.0198851
Lhuillier, C., Rudqvist, N.-P., Yamazaki, T., Zhang, T., Charpentier, M., Galluzzi, L., et al. (2021). Radiotherapy-exposed CD8+ and CD4+ neoantigens enhance tumor control. J. Clin. Invest. 131, 138740. doi:10.1172/JCI138740
Li, K., Ma, L., Sun, Y., Li, X., Ren, H., Tang, S.-C., et al. (2021). The immunotherapy candidate TNFSF4 may help the induction of a promising immunological response in breast carcinomas. Sci. Rep. 11, 18587. doi:10.1038/s41598-021-98131-4
Li, S. E., Guo, F., Wang, P., Han, L., Guo, Y., Wang, X. A., et al. (2014). X-ray-induced expression changes of TNFSF4 gene in human peripheral blood. Biomed. Environ. Sci. 27, 729–732. doi:10.3967/bes2014.107
Li, Y., Chen, Y., Miao, L., Wang, Y., Yu, M., Yan, X., et al. (2021). Stress-induced upregulation of TNFSF4 in cancer-associated fibroblast facilitates chemoresistance of lung adenocarcinoma through inhibiting apoptosis of tumor cells. Cancer Lett. 497, 212–220. doi:10.1016/j.canlet.2020.10.032
Liu, G., and Chen, X. (2006). DNA polymerase η, the product of the xeroderma pigmentosum variant gene and a target of p53, modulates the DNA damage checkpoint and p53 activation. Mol. Cell. Biol. 26, 1398–1413. doi:10.1128/MCB.26.4.1398-1413.2006
Lucas, J., Dressman, H. K., Suchindran, S., Nakamura, M., Chao, N. J., Himburg, H., et al. (2014). A translatable predictor of human radiation exposure. PloS one 9, e107897. doi:10.1371/journal.pone.0107897
Mendoza, M., Mandani, G., and Momand, J. (2014). The MDM2 gene family. Biomol. Concepts 5, 9–19. doi:10.1515/bmc-2013-0027
National Library of Medicine (2022) TNFSF4 TNF superfamily member 4 [ Homo sapiens (human) ]. USA: National Library of Medicine US Gene ID: 7292 updated on 23-Jun-2022.
Ohki, R., Saito, K., Chen, Y., Kawase, T., Hiraoka, N., Saigawa, R., et al. (2014). PHLDA3 is a novel tumor suppressor of pancreatic neuroendocrine tumors. Proc. Natl. Acad. Sci. U. S. A. 111, E2404–E2413. doi:10.1073/pnas.1319962111
Ostheim, P., Amundson, S. A., Badie, C., Bazyka, D., Evans, A. C., Ghandhi, S. A., et al. (2022). Gene expression for biodosimetry and effect prediction purposes: promises, pitfalls and future directions – key session ConRad 2021. Int. J. Radiat. Biol. 98, 843–854. doi:10.1080/09553002.2021.1987571
Panera, N., Camisa, V., Brugaletta, R., Vinci, M. R., Santoro, A., Coscia, E., et al. (2021). Blood cell gene expression profiles: a narrative review of biomarkers and effects of low-dose ionizing radiation exposure. J. Health Soc. Sci. 6, 349. doi:10.19204/2021/bldc9
Perry, M. E. (2004). Mdm2 in the response to radiation. Mol. Cancer Res. 2, 9–19. doi:10.1158/1541-7786.9.2.1
Roy, P. K., Biswas, A., Deepak, K., and Mandal, M. (2022). An insight into the ubiquitin-proteasomal axis and related therapeutic approaches towards central nervous system malignancies. Biochimica Biophysica Acta (BBA)-Reviews Cancer 1877, 188734. doi:10.1016/j.bbcan.2022.188734
Shuryak, I., Ghandhi, S. A., Turner, H. C., Weber, W., Melo, D., Amundson, S. A., et al. (2020). Dose and dose-rate effects in a mouse model of internal exposure from 137Cs. Part 2: integration of gamma-H2AX and gene expression biomarkers for retrospective radiation biodosimetry. Radiat. Res. 196, 491–500. doi:10.1667/RADE-20-00042.1
Simon, R. (2011). Genomic biomarkers in predictive medicine. An interim analysis. EMBO Mol. Med. 3 (3), 429–435. doi:10.1002/emmm.201100153
Soufan, O., Ewald, J., Viau, C., Crump, D., Hecker, M., Basu, N., et al. (2019). T1000: a reduced gene set prioritized for toxicogenomic studies. PeerJ 13 7, e7975. doi:10.7717/peerj.7975
Soufan, O., Kleftogiannis, D., Kalnis, P., and Bajic, V. B. (2015). DWFS: a wrapper feature selection tool based on a parallel genetic algorithm. PloS one 10, e0117988. doi:10.1371/journal.pone.0117988
Spainhour, J. C., Lim, H. S., Yi, S. V., and Qiu, P. (2019). Correlation patterns between DNA methylation and gene expression in the Cancer Genome Atlas. Cancer Inf. 18, 1176935119828776. doi:10.1177/1176935119828776
Subramanian, A., Narayan, R., Corsello, S. M., Peck, D. D., Natoli, T. E., Lu, X., et al. (2017). A next generation connectivity map: L1000 platform and the first 1, 000, 000 profiles. Cell 12 171, 1437–1452. doi:10.1016/j.cell.2017.10.049
Wang, G., Niu, X., Liu, H., Dong, Q., Yao, Y., Wang, D., et al. (2021). c‐Abl kinase regulates cell proliferation and ionizing radiation‐induced G2/M arrest via phosphorylation of FHL2. FEBS Open bio 11, 1731–1738. doi:10.1002/2211-5463.13177
Xu, J., Zhou, J., Li, M.-S., Ng, C.-F., Ng, Y.-K., Lai, P. B.-S., et al. (2014). Transcriptional regulation of the tumor suppressor FHL2 by p53 in human kidney and liver cells. PloS one 9, e99359. doi:10.1371/journal.pone.0099359
Zhang, X., Qu, X., Wang, C., Zhou, C., Liu, G., Wei, F., et al. (2011). Over-expression of Gadd45a enhances radiotherapy efficacy in human Tca8113 cell line. Acta Pharmacol. Sin. 32, 253–258. doi:10.1038/aps.2010.208
Zhang, Y., Yapryntseva, M. A., Vdovin, A., Maximchik, P., Zhivotovsky, B., and Gogvadze, V. (2021). Modeling hypoxia facilitates cancer cell survival through downregulation of p53 expression. Chem. Biol. Interact. 345, 109553. doi:10.1016/j.cbi.2021.109553
Zhao, P., and Malik, S. (2022). The phosphorylation to acetylation/methylation cascade in transcriptional regulation: how kinases regulate transcriptional activities of DNA/histone-modifying enzymes. Cell Biosci. 12, 83–23. doi:10.1186/s13578-022-00821-7
Keywords: gene signature, dose prediction, p53 pathway, ionizing radiation, gene expression analysis, machine learning, point-of-care (POC) diagnosis
Citation: St. Peter F, Vadrev SM and Soufan O (2022) R400: A novel gene signature for dose prediction in radiation exposure studies in humans. Front. Syst. Biol. 2:1022486. doi: 10.3389/fsysb.2022.1022486
Received: 18 August 2022; Accepted: 12 October 2022;
Published: 26 October 2022.
Edited by:
Tarunendu Mapder, Bristol Myers Squibb, United StatesReviewed by:
Sohini Chakraborty, Grossman School of Medicine, New York University, United StatesSukanya Saha, National Institute of Environmental Health Sciences (NIH), United States
Copyright © 2022 St. Peter, Vadrev and Soufan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Othman Soufan, b3NvdWZhbkBzdGZ4LmNh
 Frederick St. Peter
Frederick St. Peter Srinivas Mukund Vadrev
Srinivas Mukund Vadrev Othman Soufan
Othman Soufan