
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Synth. Biol. , 06 May 2024
Sec. Genetic Systems and Circuit Design
Volume 2 - 2024 | https://doi.org/10.3389/fsybi.2024.1397533
Cell-free synthetic biology has gained increasing popularity owing to its ability to elucidate biological functions in isolation from intricate cellular environments and the capacity to build increasingly complex biochemical systems in vitro. But cell-free transcription—translation systems are often only available in small volumes which has been a major challenge in the field. Microfluidic technologies helped address this challenge by enabling miniaturization and implementation of robust multi-step workflows. In this review, we highlight microfluidic technologies which have been applied to cell-free applications and discuss various ways in which they have advanced the boundaries of cell-free synthetic biology.
Described as the manipulation of fluids on the microliter to femtoliter scale, microfluidics enables processing and handling minuscule amounts of fluid samples (Gravesen et al., 1993; Beebe et al., 2002; Todd and Quake, 2005; Whitesides, 2006). Miniaturization of fluidics can eliminate non-linearities in flow behaviour and also introduces new phenomena not observed at the macroscale (Howard et al., 2004; Li, 2004; Di Carlo, 2009; Seemann et al., 2011). Several multi-step protocols have been implemented in so-called “lab-on-a-chip” and “micro total analysis” microfluidic devices (Dittrich et al., 2006; Abgrall and Gue, 2007; Maerkl and Quake, 2007). Design and fabrication of such microfluidic devices most commonly employs a combination of UV lithography and soft lithography (Whitesides et al., 2001). The use of polydimethylsiloxane (PDMS) for soft lithography of microfluidic devices served as a major improvement, facilitating rapid prototyping and design-build-test cycles (Xia and Whitesides, 1998). By enabling parallelization, minimal reagent consumption, faster sample processing, and the ability to carry out complex multi-step protocols, microfluidics has played an important role in the development of cell-free synthetic biology.
Originating from the discovery of fermentation with yeast extract by Eduard Buchner in 1897 (Jaenicke, 2007), cell-free systems (CFS) have recently seen increasing use in building complex synthetic gene networks and understanding biological pathways in vitro. Working with biomolecular components outside of cells allows researchers to elucidate their function often more precisely and quantitatively than is possible within the complex cellular environment. CFS are also increasingly used for applications including biosensing (Tinafar et al., 2022), metabolic engineering (Dudley et al., 2015), and product synthesis (Kumar Dondapati et al., 2020). More recently, researchers are beginning to tackle the question whether a living system could be built from the bottom-up using non-living components and CFS are playing a major role in this endeavor (Maerkl, 2023). However, cell-free systems often require extensive component optimization or the large-scale analysis of a particular biochemical system. Cost of reagents and economic feasibility of scaling up reactions are important factors to consider in using CFS, and can often prevent tapping the full potential of CFS in benchtop settings.
The field of microfluidics has become a major driving force in pushing the boundaries of cell-free synthetic biology. Performing cell-free reactions in microfluidic devices enables many applications and uses of CFS that would often be impossible to achieve using standard benchtop methods and pipetting. For example, the development of microfluidic chemostats enabled the implementation of continuous cell-free transcription - translation reactions running under steady state conditions, where it is crucial to maintain the influx of fresh reagents and components while diluting out the products and waste, with the smallest volumes possible (Niederholtmeyer et al., 2013; Karzbrun et al., 2014). Application of microfluidics in cell-free setups enables high-throughput characterization of biomolecular-components and allows multistep long-term studies (Niederholtmeyer et al., 2015; Chang et al., 2018; Swank et al., 2019; Swank and Maerkl, 2021) with minimal reagent consumption in an automated fashion. A growing number of examples can be found in literature showcasing microfluidics enabling long-term, continuous implementation of cell-free systems (Niederholtmeyer et al., 2013; Karzbrun et al., 2014; Niederholtmeyer et al., 2015; Swank et al., 2019; Lavickova et al., 2020; Laohakunakorn et al., 2021; Swank and Maerkl, 2021; Lavickova et al., 2022), generation and manipulation of artificial cells and organelles (Cho and Lu, 2020; Lavickova et al., 2020), cell-free biosensors (Pardee et al., 2014; Pardee et al., 2016; Amalfitano et al., 2021), high-throughput studies of complex biological systems (Chang et al., 2018; Swank et al., 2019; Laohakunakorn et al., 2021; Swank and Maerkl, 2021), and high-throughput directed evolution studies (Fallah-Araghi et al., 2012; Dodevski et al., 2015; Holstein et al., 2021). Many of the drawbacks and challenges in cell-free studies discussed above could be addressed with microfluidic setups, and only a few reviews of microfluidic cell-free systems have been published (Damiati et al., 2018; Ayoubi-Joshaghani et al., 2020).
In this review we provide a comprehensive overview and discuss recent technological developments in microfluidics that contributed to advances in cell-free synthetic biology. We begin with brief descriptions of cell-free synthetic biology and the physical phenomena in microfluidics relevant to cell-free systems. This is followed by discussions on single-phase systems, focusing on passive paper-based devices, that operate via autonomous microfluidic flow and recent applications in CFS. Next, we discuss active single phase as well as multiphase microfluidic systems actuated via peripheral devices, enabling high-throughput multistep cell-free reactions and describe how microfluidics has pushed the envelope in research towards creating artificial cells. We also cover digital microfluidic devices and discuss their potential for cell-free applications. Finally, we discuss the potential of microfluidic technologies and challenges to be addressed to further improve them for enabling cell-free synthetic biology.
The term “synthetic biology” is used to describe efforts to design or redesign life on a molecular level (Benner and Sismour, 2005; Shankar and Van Oudenaarden, 2009; Slusarczyk et al., 2012; Church et al., 2014; Smanski et al., 2016). It involves development of complex systems from DNA, proteins, and other biomolecules, which constitute the building blocks of biology (Heinemann and Panke, 2006; Purnick and Weiss, 2009). The major motivations behind synthetic biology lie in understanding the complex mechanisms behind natural biology and building synthetic systems that could perform novel, useful functions (Purnick and Weiss, 2009; Nandagopal and Elowitz, 2011; Jeffrey et al., 2014). The former often involves characterizing synthetic constructs consisting of components involved in a biological process of interest, thus allowing studies of their interactions in a robust and isolated manner (Andrianantoandro et al., 2006). The foundation for synthetic biology was laid in 1970s with fundamental studies on DNA sequencing, molecular cloning, and polymerase chain reactions (PCR) (Cameron et al., 2014). The construction of the first synthetic gene circuit elements—a genetic toggle switch (Gardner et al., 2000) and a genetic oscillator (Elowitz and Leibler, 2000), in 2000, were the cornerstones of synthetic biology. This development led to design and engineering of genetic circuits with feedback loops and logic gates, analogous to transistor logic circuits (Hasty et al., 2001a; Hasty et al., 2001b; Hasty et al., 2002; Guido et al., 2006). Since then, the field has grown by leaps and bounds, with many examples of complex gene regulatory networks and reverse-engineered cellular networks found in literature and commercial products (Slusarczyk et al., 2012; Sedlmayer et al., 2018; Xie and Fussenegger, 2018). The timeline of synthetic biology and key developments over the last 2 decades have been intricately covered in excellent reviews by Cameron et al., 2014 and Meng and Ellis, 2020. Several commercial examples of synthetic biology can now be found in applications in cancer therapeutics, drugs and fertilizers (Khalil and Collins, 2010; Higashikuni et al., 2017; Voigt, 2020).
However, cells are extremely complex environments and many challenges relating to system variability and standardization arise from engineering synthetic networks in living cells (Bruggeman and Westerhoff, 2007; Cardinale and Arkin, 2012). This has resulted in an increased focus on using “cell-free” systems for studies and applications (Eric Hodgman and Jewett, 2012; Perez et al., 2016; Yuan, 2017; Laohakunakorn et al., 2020) Cell-free systems can be created by extracting the cellular machinery from cultured cells, followed by reconstituting them with energy components, additional enzymes and co-factors to run in vitro transcription and translation (IVTT) (Ashty and Jewett, 2018; Jiang et al., 2018; Koch et al., 2018). This process of isolation and reconstitution allows precise characterization of processes and interactions of biomolecules devoid of influence from secondary processes, including cell growth (Lee and Kim, 2013; Smith et al., 2014; Morgado et al., 2018), motility (Ni et al., 2020), quorum sensing (Melissa and Bassler, 2001; Keller and Surette, 2006) or interactions with extracellular environment (Bissell and Barcellos-Hoff, 1987). Researchers have shown robust cell-free protein synthesis (CFPS) from different cell lysates (Katzen et al., 2005; Kim et al., 2006; Gregorio et al., 2019). Lysate-based systems have well-established production protocols (Garamella et al., 2016). While they have proven to be powerful systems in synthetic biology, lysate-based systems remain highly complex and undefined. An alternative approach is protein expression using recombinant elements (PURE), where all necessary proteins are individually purified prior to reconstituting them in a mixture (Shimizu et al., 2001). Cell-free systems based on lysate or PURE are commercially available in the form of ready-to-use kits. Aside from commercial protein expression kits, there are also several cheaper, self-made alternatives for CFS. Lavickova and Maerkl, 2019 recently introduced a robust and low-cost method for producing the PURE system. They showed protein synthesis capacity similar to the PURExpress commercial kits at 6% cost per microliter, thus providing a cheaper alternative and broadening access to cell-free systems.
Cell-free applications range from biosensor design (Soltani et al., 2018; Zhang et al., 2020), protein synthesis (Niederholtmeyer et al., 2013; Chang et al., 2018; Lavickova et al., 2020; Laohakunakorn et al., 2021), rapid prototyping of gene regulatory components via rapid design-build-test cycles (Maerkl and Quake, 2007; Niederholtmeyer et al., 2015; Swank et al., 2019) to building complex gene circuits (Silverman et al., 2020; Garenne et al., 2021). One grand challenge that has emerged in cell-free research is the bottom-up development of a synthetic cell (Maerkl, 2023). Efforts towards this challenge include creation of DNA replication (Sakatani et al., 2018; Van Nies et al., 2018; Ueno et al., 2021; Grasemann et al., 2023), tRNA replication (Hibi et al., 2020), energy and metabolism (Berhanu et al., 2019; Miller et al., 2020), vesicle structures and membrane synthesis (Deshpande et al., 2016; Bhattacharya et al., 2019; Blanken et al., 2020), and communication systems (Li et al., 2019; Buddingh et al., 2020). We recently demonstrated continuous self-replication of essential proteins in the PURE system (Lavickova et al., 2020). Microfluidic devices have proven to be absolutely essential in many of these developments and a great variety of research over the last decade focused on utilizing microfluidics to tackle challenges in cell-free systems. There is a growing body of research on compartmentalization of cell-free reactions in microdroplets (Petra et al., 2005; Holstein et al., 2021) and hydrogels (Park et al., 2009; Cui et al., 2020), construction of artificial vesicles to imitate cellular membranes (Arriaga et al., 2014; Deshpande et al., 2016), co-encapsulation of liposomes (Stucki et al., 2021a; Nuti et al., 2022) and coacervates (Pir Cakmak et al., 2019; Abbas et al., 2022) to imitate intracellular organelles and establishing cellular processes such as cell growth (Scott et al., 2016; Bhattacharya et al., 2019), division (Deshpande et al., 2018; Steinkühler et al., 2020; De Franceschi et al., 2024), fusion (Gong et al., 2008; Caschera et al., 2011) and transcription-translation (TX-TL) (Vincent and Libchaber, 2004; Vincent et al., 2005) within these artificial cell constructs. There have also been applications of microfluidic devices for high-throughput characterization of binding affinities (Maerkl and Quake, 2007; Swank et al., 2019) and microfluidic chemostats enabling continuous steady-state cell-free reactions (Niederholtmeyer et al., 2013; Karzbrun et al., 2014; Lavickova et al., 2020; Laohakunakorn et al., 2021; Swank and Maerkl, 2021).
Microfluidics involves the interplay of various physical phenomena which have to be optimized for particular applications. A few key dimensionless numbers representing the relative influences of these phenomena help us understand the fluid parameter space and are often considered paramount for optimizing a given microfluidic system (Todd and Quake, 2005; Whitesides, 2006; Convery and Gadegaard, 2019). Microfluidics often deals with flows having a low Reynold’s Number (Re) (Eq. 1) (Brody et al., 1996), which is the ratio of inertial to viscous forces in the system:
where ρ is the density of fluid, η gives the dynamic viscosity, v is the flow velocity and L is the characteristic width of the flow path. At low values of Re (Edward, 1977), several non-linear fluidic effects become negligible, resulting in linear and predictable flows (Karimi et al., 2015). This streamlines applications of microfluidics as fluid flows are often governed by linear characteristics. Typical values of Re range from 10–6 to 10. This implies inertial fluid effects are mostly negligible and other dimensionless numbers, such as the Péclet number and the capillary number, are of much greater importance. The Péclet number (Pe) is the ratio between convective and diffusive forces (Eq. 2), and an important parameter to estimate the extent of mixing in a microfluidic system:
where D is the diffusion constant of the molecule of interest. While random eddies and turbulent flows are the major driving factors for mixing within fluids on the macroscale, diffusion is the major driving force for mixing in microfluidics (Knight et al., 1998). A molecule in a microfluidic system with low Pe would have greater mixing efficiency, as diffusion would dominate convection. This is crucial to take into consideration in setups with large biomolecules, such as long DNA molecules, as they have very low diffusion constants (Arosio et al., 2016). Herringbone structures can be introduced into microchannels to introduce chaotic streamlines and facilitate mixing via chaotic advection (Williams et al., 2008), similar to turbulent mixing in macroscale fluidics. Therefore, greater surface area and chaotic advection are the main techniques to maximize mixing within microfluidic reactors (Suh and Kang, 2010). Another important dimensionless parameter to consider is the capillary number (Ca) (Eq. 3), especially in the case of multiphase microfluidics:
where γ is the interfacial tension between two immiscible fluids in flow. Multiphase systems involve the flow of immiscible fluids, with surface tension present at their interface. The interplay of viscous forces and the interfacial surface tension can lead to formation of emulsion droplets in such flow systems. Ca gives the ratio between the viscous stresses and the interfacial surface stresses and its optimization is crucial for production of monodisperse emulsions. At sufficiently low values of Ca (
Microfluidic devices based on autonomous fluid flow (capillary flow) have come to prominence in diagnostics and point-of-care (POC) applications primarily because of their ultra-low cost and simplicity (Gervais and Delamarche, 2009; Curtis et al., 2012; Hua and Steckl, 2018; Pandey et al., 2018; Hassan and Zhang, 2020). These devices utilize capillary forces to shunt liquids along pre-defined pathways in an autonomous fashion (Tan et al., 2021). Classic examples of this are lateral flow assay (LFA) devices (Hemmig et al., 2019; Kasetsirikul et al., 2020; Liu et al., 2021), widely used in pregnancy self-test kits and COVID-19 self-test kits (Zhou et al., 2021). Following the landmark publication of a patterned paper microfluidic device from the Whitesides group in 2007 (Martinez et al., 2007), the field of capillary and paper microfluidics has boomed over the last decade and a half. Today, the potential to generate flows without requiring external pumps or controls, work with microliter-sized samples, low fabrication cost and ease of use have made capillary microfluidic devices popular options for cell-free POC and diagnostic applications, with commercialized products serving as good examples (Pardee et al., 2014; Slomovic et al., 2015; Zhang et al., 2020; Tinafar et al., 2022). Capillary forces are governed by factors such as liquid surface tension, surface geometry, and material properties of the solid surface (Olanrewaju et al., 2018). Key manipulations such as valving, pumping and limited control on flow rates can be mimicked in capillary devices via an interplay of capillary effects and surface modification (Wang et al., 2021). Diagnostic platforms often utilize multistep protocols, which in turn require careful design of fluidic circuits to isolate reactions and reagents. Recognizing this, there have been several key developments in design of fluidic circuits with “capillaric” elements (Safavieh and Juncker, 2013) in combination with magnetic, electronic and optical techniques over the last decade (Novo et al., 2014; Liu et al., 2022).
Paper-based microfluidic devices (µPADs) constitute a large subset of capillary microfluidic devices and are fabricated by using variations of cellulose paper or chromatography paper as flow substrates (Carrell et al., 2019; Soum et al., 2019; Qin et al., 2021). Here fluids flow through the paper substrate instead of fabricated microchannels. The desired flow path is defined by patterning a hydrophobic coating, such as SU-8, on the substrate. Use of paper as a substrate has led to technological developments of various valve designs to mimic functions of multilayer soft valves in PDMS microfluidic devices (Cate et al., 2015; Fu and Downs, 2017). A good example is the swelling hydrogel valve introduced by Toley et al. (Toley et al., 2015). The valve is made of a hydrogel column connected to an actuation channel on one end and impermeably to channel A at the other end. When actuation fluid reaches the hydrogel, it swells to push channel A upwards until it establishes contact with channel B, thus closing the fluidic circuit. They showed the potential for temporal and volumetric control using this valve, and successfully performed a multistep ELISA on the device.
µPADs show great potential as cell-free biosensors due to their low cost, ease of use and implementation, long term stability of surface dried reagents and rapid processing (Li et al., 2012; Noviana et al., 2021; Yuan, 2022). The Collins group at MIT has developed robust µPADs for executing cell-free biosensing (Pardee et al., 2014; Pardee et al., 2016; Takahashi et al., 2018) and recently applied them for detection and quantification of RNA sequences in complex samples (Karlikow et al., 2022). The integrated setup for their device is shown in Figure 1A. They achieved clinically relevant detection limits for Zika and chikungunya viruses in field tests at costs close to two orders of magnitude lower compared to standard clinical tests. Their workflow (Figure 1B) integrates a section for nucleic acid sequence based amplification (NASBA) upstream of a cell-free RNA detection assay. NASBA enabled amplification of the RNA of interest to improve the detection limit of the device and bring it closer to clinically relevant levels (Deiman et al., 2002). They also integrated an RNA-specific toehold switch sensor and CRISPR-Cas9-mediated selection to eliminate the effect of contaminants from samples and distinguish between variant strains of viral RNA. Similarly, Ma et al. used a PURExpress kit to detect target novoviral RNA on a µPAD and reached a 5000-fold improvement in the detection limit of their assay by using a synbody-based concentration step (Ma et al., 2018).
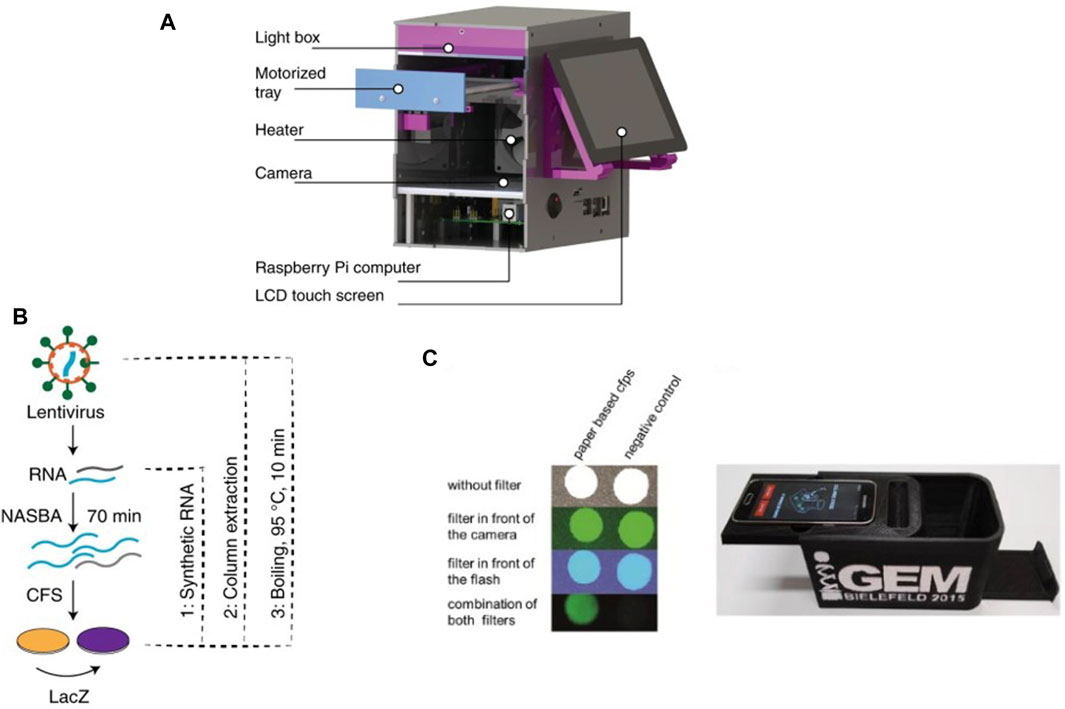
Figure 1. Recent examples of cell-free µPADs: (A) 3D rendered image of the integrated CFS-based paper device used by Karlikow et al. for field testing for the Zika virus (Karlikow et al., 2022). (B) Workflow developed for detecting viral RNA where RNA was first amplified by NASBA followed by detection via a cell-free reaction on paper. (C) Design of the integrated paper biosensor from Gräwe et al., 2019.
The development of µPADs was pushed by the need for rapid POC diagnostic kits during the COVID-19 global pandemic. Several concepts for disposable and low-cost µPADs, including some based on CFS, were developed for POC applications during the pandemic, and covered by excellent reviews (Bhalla et al., 2020; Morales-Narváez and Dincer, 2020; Pinheiro et al., 2021). Hunt et al. developed a workflow for such a µPAD to detect SARS-CoV-2 RNA sequences from saliva samples (Porter Hunt et al., 2022). They freeze-dried CFPS reagents on a hybrid cellulose-chromatography paper and housed it in a polymer cassette for long-term storage. When rehydrated with saliva samples, the device returned a bioluminescent readout in roughly 7 minutes at an estimated cost of less than 0.50 USD per device. Another growing field where paper-based cell-free biosensors have been applied is environmental water testing (Kung et al., 2019; Zheng et al., 2021; Zhang Daohong et al., 2022). Duyen et al. utilized the device concept introduced earlier by Pardee et al. for colorimetric detection of antibiotics inhibiting bacterial protein synthesis (Tran et al., 2017). They used µPADs with freeze-dried reagents for IVTT and were able to detect and quantify aparomomycin in spiked river water samples, thus showing the potential of this concept in tracking antibiotic contamination in water bodies. Similarly, Zhang et al. developed a µPAD for detecting presence of heavy metal ions such as Pb2+ and Hg2+ in water via in vitro transcription (IVT) (Zhang Yongkang et al., 2022). They employed allosteric transcription factors (aTFs) sensitive to the presence of these ions to regulate transcription in the IVT system. Pb2+ and Hg2+ would bind to the aTFs and allow them to dissociate from DNA templates, following which transcription could be carried out in the device. Gräwe et al. also developed a CFS-based paper biosensor for detection of heavy metals using aTFs (Figure 1C) (Gräwe et al., 2019). They further modified it by adding a repressor protein which was inactivated by γ -hydroxybutyrate, a recreational drug (Busardo and Jones, 2015), thus enabling detection of γ -hydroxybutyrate via the biosensor.
With commercial products and devices already in the market, we expect rapid growth and development to continue in µPADs for the foreseeable future (Lisowski and Zarzycki, 2013; Sackmann et al., 2014). The major drawback with these devices is usually limitations in carrying out multistep or complex protocols, thus only permitting the implementation of relatively simple assays and biosensing applications (Sharma et al., 2015; Morbioli et al., 2017; Yamada et al., 2017). Limited integration with modules such as electrodes or magnetics for example, can restrict their functionality to single-step assays. In addition, there are very few options for valving or metering of flows in µPADs, in comparison to active microfluidics, discussed in the next section. Owing to multiple choices for substrates, there is also an emphasis on choosing the right substrate keeping in mind the biochemistry of a desired reaction. Improvements can also be made in the quality of paper substrate after long-term storage, which is vital for commercialization. The advent of 3D printing in recent years could greatly benefit paper-substrate based 3D devices by enabling sample flow in both lateral and vertical directions, thereby introducing another avenue for multistep assays. We expect recent technological advances and workflow designs to enable the design and adaptation of more complex and multistep µPADs, with the potential for PoC and diagnostic applications in the future (Gong and Sinton, 2017; Kim et al., 2020).
Active microfluidics refers to the subset of microfluidic systems that utilize external pressure sources for inducing fluid flows and precisely executing complex protocols such as dosing (Beate Bußmann et al., 2021), mixing (Green et al., 2007), multiplexing (Todd et al., 2002), and separation (Sajeesh and Kumar Sen, 2014). Active microfluidic systems often employ peripheral control systems such as external solenoid valves in tandem with imaging and electronics. While this increases the difficulty of setting up and running active microfluidic systems (Sharma et al., 2015; Suea-Ngam et al., 2019; Yang et al., 2022), they offer the possibility of performing complex fluidic manipulations, which would not be possible in passive microfluidics. As a result, active microfluidic systems are most commonly employed in cutting edge research in academia and applications in industry.
Building complex fluidic circuits in microfluidic devices took off with several designs for microvalves reported in the early 2000s (Oh and Ahn, 2006). These included a variety of rotary (Hasegawa et al., 2003) and membrane (Hosokawa and Maeda, 2000) microvalves. Many designs for PDMS membrane microvalves integrated with PDMS and silicon devices were developed with the aim of manipulating microfluidic flows. Popular examples include monolithic design for membrane valves in glass devices introduced by Grover et al., 2003 and an in-plane microvalve design by Go and Shoji, 2004. But perhaps the most influential, and now the most commonly implemented, design was described in 2000 (Unger et al., 2000), when a protocol for multilayer PDMS soft lithography to generate microfluidic valves on-chip was published. Fabricated by alignment and bonding of two PDMS layers, these valves can be pneumatically actuated to fully close or open microfluidic channels. Sequential activation of multiple such valves in series in a microchannel could create a peristaltic micropump to push fluids.
Todd et al., 2002 showed the potential of these valves in building microfluidic circuits by achieving large-scale microfluidic integration, analogous to electronic integrated devices. They demonstrated a microfluidic multiplexer based on binary trees which allowed them independent control of 1000 pL-volume chambers based on actuation of microvalves via 20 control channels, which were in turn actuated by solenoid valves. They also developed a device for microfluidic loading of multiple reagents, mixing and selective recovery in 256 independent chambers, isolated by microvalves. The ability to reliably mass-integrate thousands of valves and pneumatically actuate them directly on-chip led to rapid development of lab-on-a-chip platforms for executing multistep laboratory processes in PDMS microchips. One of the earliest examples of this phenomenon was the microfluidic formulator and metering device, utilizing microvalves for its functions, introduced by Hansen et al. (Hansen et al., 2004). Their device allowed rapid formulation of complex mixtures from various components to screen over 4000 solubility conditions for a protein using minimal volumes. They also employed peristaltic pumps using microvalves to push liquids through the device or for continuous mixing within reactors. Balagaddé et al., 2005 adapted this microformulator to design a microchemostat more than 2 orders of magnitude smaller in volume than the previously smallest chemostat design. They used it for long-term culture (over hundreds of hours) of bacteria via manipulation of microvalves, which showed the potential of this valving technique in building complex lab-on-a-chip devices.
With the aim of implementing complex biological systems such as genetic regulation and synthetic oscillators in vitro, Niederholtmeyer et al., 2013 adapted and repurposed the microformulator introduced by Hansen et al., 2004 for performing continuous cell-free transcription-translation under steady state conditions. Their modified device consisted of 8 independent 33-nL reactors, each independently addressable with different reagents and programmable dilution fractions (Figure 2A). The reactors were initialized with all required components for starting a CFPS reaction, followed by dilutions in discrete time-steps to add in fresh components of the IVTT master mix, simultaneously removing equivalent reaction volumes from the bioreactors (Figure 2B). A set of valves was used as a peristaltic pump to facilitate mixing of components inside the bioreactors. They achieved continuous steady state protein expression (Figure 2C) in these bioreactors for up to 30 h and showcased the ability of this device to implement and run the first in vitro genetic oscillator network (Figure 2D). They further showed the potential of this microfluidic device in rapid prototyping of genetic networks (Niederholtmeyer et al., 2015). They first emulated the repressilator network introduced by Elowitz and Leibler in 2000 (Elowitz and Leibler, 2000) and then went a step further by building five-node repressilator oscillating networks and highlighted the dependence of oscillation periods and amplitudes on dilution period and component concentrations. Four-node repressilator networks were also tested showing toggle-switch behaviour as predicted. They successfully transplanted and tested the above designed genetic networks in E. coli using a mother machine microfluidic device. This work provided a proof-of-concept for rapid forward engineering of genetic circuits in CFS. de Maddalena et al., 2016 also used the microchemostat to study effects of adding transcription elongation factors to a PURE mix. They showed greatly enhanced expression in PURE by using the elongation factors and studied regulation of natural and synthetic E. coli promoters in steady-state conditions. Laohakunakorn et al. modified the microchemostat design by introducing hardcoded dilution fractions and showed its application in long term CFPS (Laohakunakorn et al., 2021).
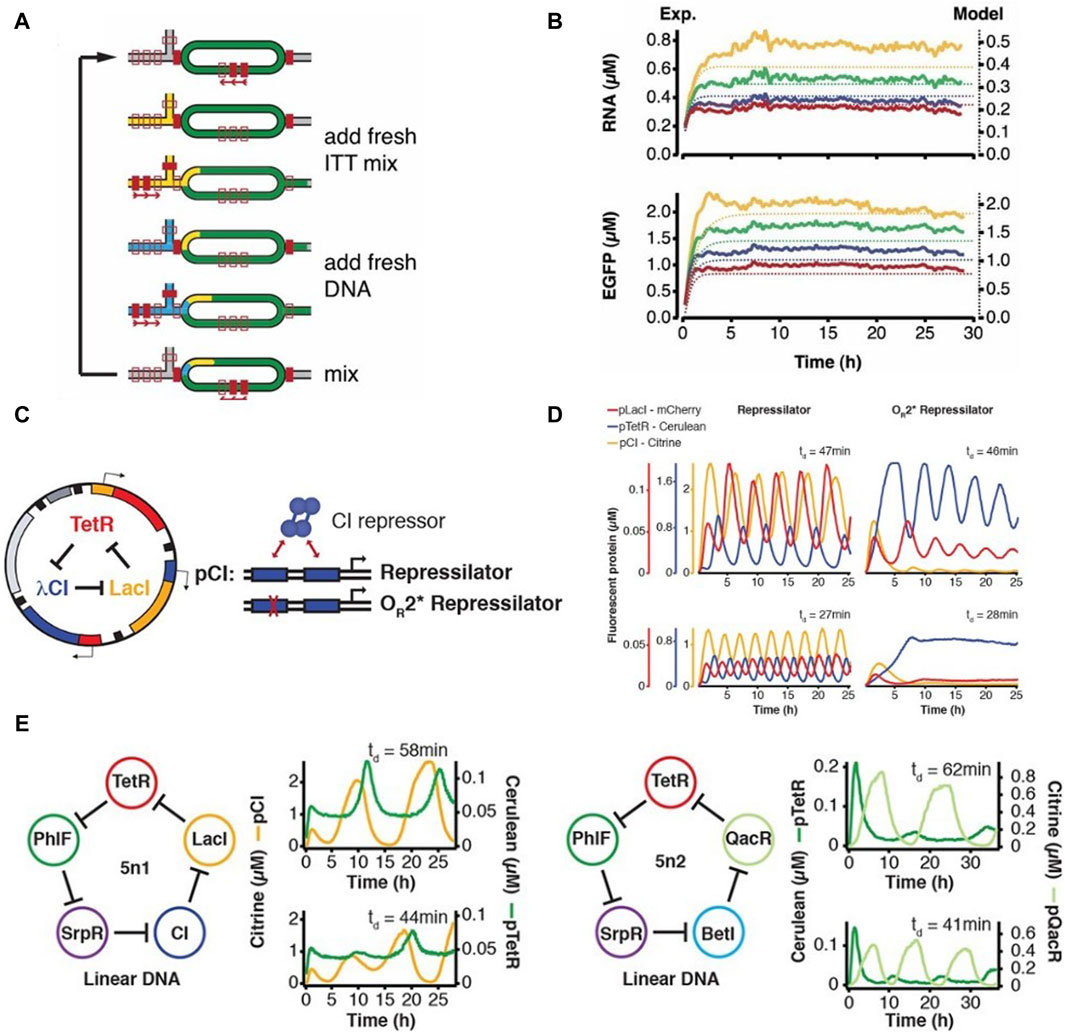
Figure 2. The programmable microfluidic chemostat for continuous CFPS introduced by Niederholtmeyer et al., 2013 (A) Schematic and functional description of the microchemostat device. (B) Steady-state CFPS achieved within these microchemostats. (C) The three-node repressilator circuit designed by Elowitz and Leibler (Elowitz and Leibler, 2000) (D) Microfluidic implementation of the repressilator circuit by Niederholtmeyer et al. showing variations in oscillatory behavior dependent on dilution times in the microchemostat (Niederholtmeyer et al., 2015) (E) Design and microfluidic implementation of the first five-node repressilator circuits showing oscillatory behaviours as theoretically predicted.
Working towards the goal of a fully self-regenerating CFS, Lavickova et al., 2020 used this device to demonstrate the possibility of the PURE system to self-regenerate several essential PURE proteins over extended periods of time (Figure 3A). They started out with a complete homemade PURE system along with DNA templates coding for PURE proteins as well as a template coding for a fluorescent reporter protein. This was followed by switching to a PURE system lacking these particular PURE proteins. They observed that the system was able to self-sustain by regenerating the proteins from their respective DNA templates Figure 3B. They successfully self-regenerated up to 7 aminoacyl tRNA synthetases (AARSs), thus achieving a partially self-regenerating synthetic cell. They theorized self-regeneration of all 36 non-ribosomal PURE proteins would require either a more efficient PURE system and/or a lower dilution rate. But simply decreasing the dilution rate led to concomitant decreases in synthesis rate due to limited availability of small molecule building blocks thus requiring the need for a dialysis setup in their microfluidic device. Following this, Lavickova et al., 2022 improved the microchemostat by adding semipermeable hydrogel membranes for continuous dialysis of small biomolecules. Each microreactor in the chip was connected to its respective feeding chamber via semipermeable membranes (Figure 3C), and the contents of the feeding chamber could be replenished independently of the reactor itself. The membrane was designed to allow diffusion of energy components from the feeding chamber into the reactor while preventing DNA and proteins from diffusing out of the reactors. An anti-evaporation layer was assembled on top of the fluidic layer to prevent evaporation in the microreactors. They performed batch cell-free protein synthesis reactions with continuous dialysis on-chip and observed a threefold increase in enhanced Green Fluorescent Protein (eGFP) production compared to the batch reaction without replenishment of the feeding chamber. Following this, they successfully carried out a cell-free reaction under steady-state conditions enabling lower dilution rates compared to older chemostat models, without negatively affecting protein synthesis rates by continuously replenishing the feeding chamber, thus proving the potential of this device to push the limits of self-regeneration in CFS.
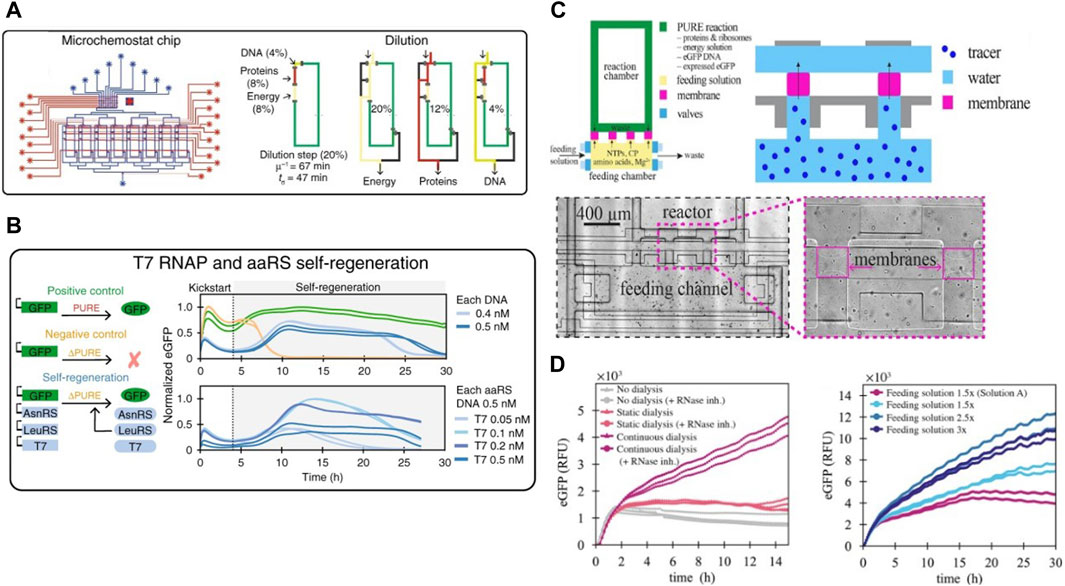
Figure 3. The second-generation cell-free microchemostat and third-generation dialysis microchemostat: (A) Schematic design of the second generation microchemostat microfluidic chip consisting of 8 microchemostats using hard-coded dilution fractions used by Lavickova et al., 2020 for achieving a partially self-regenerating synthetic cell. Their protocol for continuous dilution and replenishment in each chemostat is also shown (B) Successful self-regeneration of T7 RNA Polymerase and several AARSs in the cell-free system. (C) Schematic design of a third generation microchemostat augmented with a dialysis chamber for sustained self-regeneration in CFS and close-up view of fabricated dialysis membranes under the microscope (Lavickova et al., 2022). (D) Protein synthesis for over 30 h on the dialysis chemostat enabled by continuous replenishment of the feeding chamber. Amount of protein synthesized within the reactors increased with increase of concentration of the feeding solution.
Sluijs et al. adapted the microchemostat developed by Niederholtmeyer et al. to characterize performance of a library of repressors and promoters in steady-state CFS (van der Linden et al., 2019; van Sluijs et al., 2022). They implemented automated design-characterize-test workflows by executing incoherent feed-forward loops in chemostats (Figure 4A). This allowed them to characterize their chosen six repressors and obtain their parameter sets for simulation models. These parameter sets were used to accurately predict the behaviour of two genetic networks, thus proving the potential of their workflow in generating rapid design-build-test-learn cycles for study of complex genetic networks. Aufinger et al. used microchemostats to elucidate complex genetic oscillation dynamics and feed-forward loops in CFS (Aufinger et al., 2022). They demonstrated how cycling periods could be set externally in an oscillator by modulating concentrations of one of its components at discrete points. Periodic forcing also led to introduction of period doubling, shown in Figure 4B, and even quadrupling bifurcation within the system. With the help of simulations, they showed these non-linear effects could be studied in cell-free systems to elucidate complex chaotic dynamics.
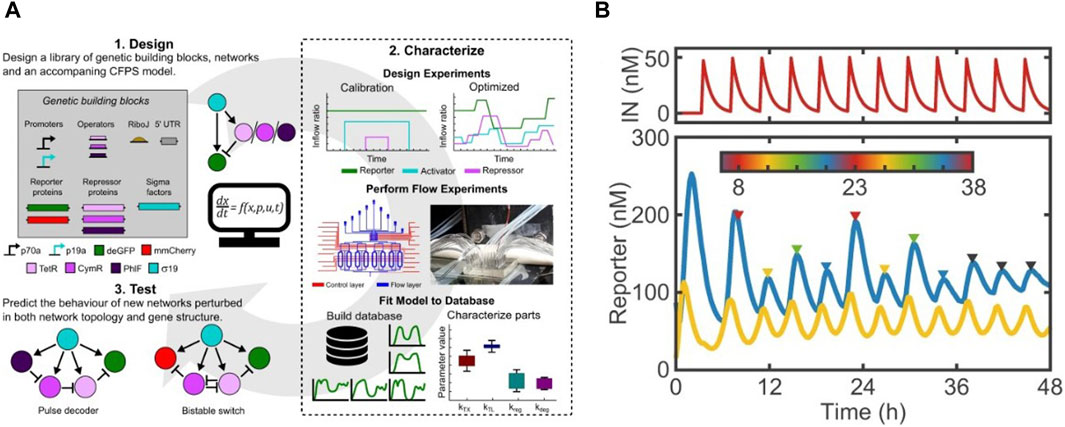
Figure 4. Examples of cell-free studies enabled by microchemostats (Niederholtmeyer et al., 2013): (A) The design-characterize-test workflow designed by Sluijs et al. for characterizing a library of repressors and promotors (van Sluijs et al., 2022). (B) Period doubling observed by Aufinger et al. upon introducing cycling periods in steady state chemostats (Aufinger et al., 2022).
Aside from enabling long-term studies in CFS, the high-throughput usually associated with microfluidics, combined with microvalves, has driven developments in high-throughput cell-free studies. In one of the earliest examples, Maerkl et al. introduced a microfluidic device to measure binding affinities of transcription factors (TFs) in high-throughput (Maerkl and Quake, 2007). Using a network of microfluidic valves, the device contained 2400 isolated microfluidic compartments aligned above a spotted DNA array, thus allowing for 2400 independent binding affinity measurements per experiment. They formulated a multistep protocol to carry out surface modification and patterning to immobilize the transcription factor being studied (Figure 5A). A novel mechanism based on mechanically induced trapping of molecular interactions (MITOMI) was invented to enable precise quantification of low-affinity or highly transient molecular interactions. They used MITOMI to generate quantitative binding energy landscapes of multiple TFs. Blackburn et al. adapted the MITOMI device for characterizing binding affinities of over 400 synthetic Zinc Finger (ZF) TFs synthesized on-chip (Blackburn et al., 2016). They showed that ZF-TF binding affinity could be tuned independent of specificity. Swank et al. repurposed this MITOMI device to synthesize ZF TFs on-chip in 768 independent cell-free reactions and to assess their capacity to repress a large library of synthetic target promoters (Swank et al., 2019) (Figure 5B). This system was used to study effects of binding affinities, binding site positions and cooperative interactions between TFs and promoters on transcriptional regulation. The high-throughput microfluidic devices allowed them to comprehensively characterize a complex TF-promoter system, which in turn allowed them to engineer novel higher order systems creating OR, AND, NOT and NAND logic gates as well as combinations thereof (Figure 5C).
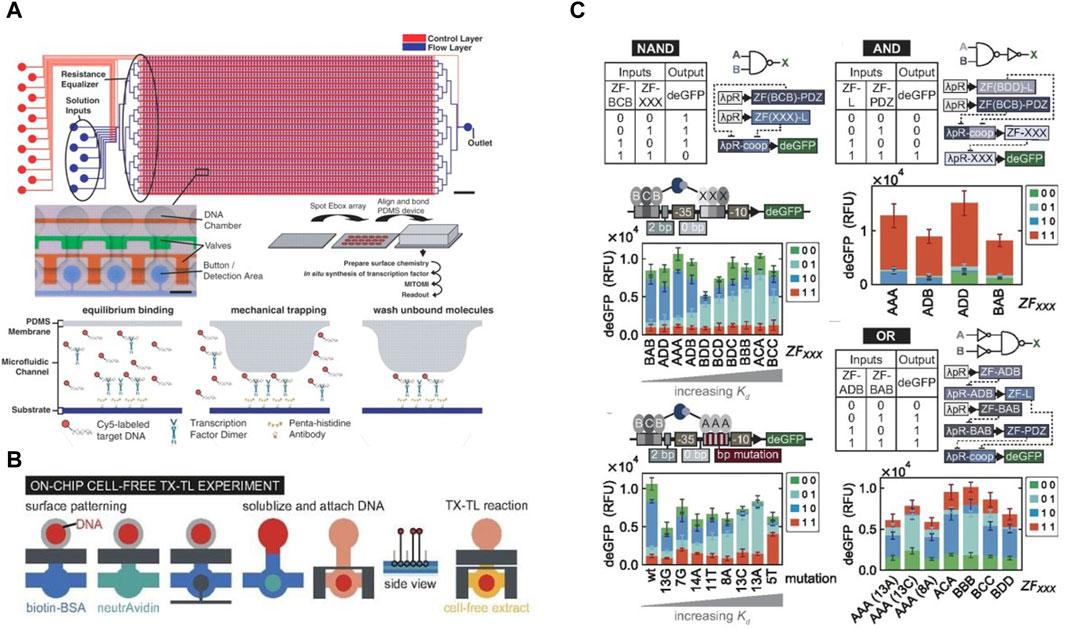
Figure 5. The MITOMI device designed for high-throughput analysis of protein binding affinities: (A) Schematics of the MITOMI device designed by Maerkl et al. for measuring binding energy landscapes, along with an illustration of mechanical trapping of molecular interactions (Maerkl and Quake, 2007). (B) Workflow of the MITOMI device used by Swank et al. for high-throughput cell-free synthetic biology. The protocol for surface functionalization and carrying out cell-free tx-tl reactions on-chip is shown (Swank et al., 2019). (C) Illustration of the genetic AND, NAND, and OR logic gates developed and characterized using the MITOMI chip.
Another device based on MITOMI for high-throughput cell-free studies was designed by Swank et al., where they added a connecting channel to each chamber to achieve steady-state CFPS in 280 independent chambers (Swank and Maerkl, 2021) following the design of the continuous-flow chemostat developed by Karzbrun et al., 2014 described in detail in the next section. The connecting channel removed products from the chambers and brought in fresh reagents via diffusion. This in effect created a high-throughput device containing 280 independently addressable chemostats (Figure 6A). A pulse width modulation microfluidic module (Woodruff and Maerkl, 2018) was included as well to allow fully automated and programmable real-time generation of reaction conditions. They successfully characterized dynamic gene repression and toggle switch circuits within these chemostats (Figure 6B).
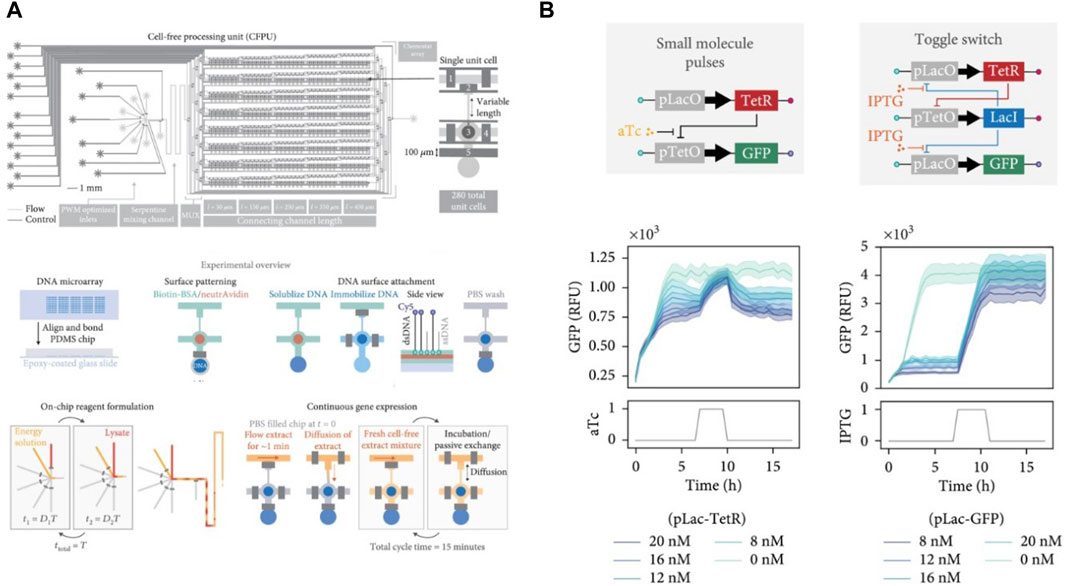
Figure 6. Design and applications of the high-throughput microchemostat designed by Swank et al. (Swank and Maerkl, 2021): (A) Workflow of the high-throughput chemostat showing the device schematic and multistep protocol for on-chip experiments to run 280 independent steady-state reactions in parallel. (B) Implementation and characterization of transcriptional repression (left) and a genetic toggle switch (right) in the high-throughput microchemostat.
Buxboim et al., 2007 introduced a microfluidic approach based on DNA immobilization and subsequent protein trapping on-chip for building synthetic gene networks. They designed a molecule called “Daisy,” a polyethylene glycol (PEG) with a silane group at one end and an amine group at the other end, protected by a Nvoc moiety sensitive to UV exposure. This Daisy molecule formed a self-assembled monolayer on the silicon dioxide surface of a wafer and could covalently couple with a variety of biomolecules upon UV exposure, due to de-protection of the amine end. This was used to generate microtraps for tagged proteins on the Daisy-coated regions. Unlike the approach developed by the Maerkl et al. which allows surface patterning of thousands of different molecules using a microarrayer (Maerkl and Quake, 2007), only one DNA template or template mixture could be immobilized with this approach. They demonstrated a cascading gene circuit by immobilization of gene constructs in alternating stripes. Karzbrun et al., 2014 took this technology further to develop and implement genetic oscillatory circuits, enabling oscillating protein expression patterns and expression gradients, similar to what was previously demonstrated by Niederholtmeyer et al., 2013. E. coli lysate based cell-free extract was supplied via a main flow channel and allowed to diffuse via narrower channels into small circular dead-end compartments containing immobilized DNA. Proteins synthesized within these compartments could also diffuse out of the chamber into the main channel, thus creating a linear concentration gradient along the connecting channel. They showed formation of positive feedback and negative feedback circuits, and were able to implement a genetic oscillator. By connecting compartments in series via additional capillaries, they implemented activator-repressor networks by separating components between the compartments, resulting in a spatio-temporal variation in gene expression.
Tayar et al., 2017 improved this device design to integrate as many as 80 DNA compartments for on-chip cell-free expression (Figure 7A), although compartments could only be programmed with the same DNA templates. They designed and successfully implemented a non-linear genetic oscillator by expressing proteins for sequestering the activator as well as degrading the repressor protein. They further coupled compartments acting as oscillators via thin capillaries running in parallel to the main flow channel, enabling protein transfer between compartments via diffusion. Autonomous synchronization of coupled oscillators operating at different frequencies was observed (Figure 7B), with the oscillator at lower frequency always entraining the faster one owing to the inherent negative feedback in the network. Their device enabled communication and synchronization across networks along with temporal variation. Efrat et al., 2018 showed the potential for integration with other technologies by fabricating gold electrodes close to the immobilized DNA in the compartments. This was used to generate a localized electric field gradient which trapped biomolecules away from the DNA and inhibited CFPS. Turning off the electrodes again led to homogenous distribution of the components in the compartments and thus restoring eGFP expression. Contrary to using repressors and activators for gene circuit regulation, this device provided an electrical mechanism for spatio-temporal control of cell-free reactions.
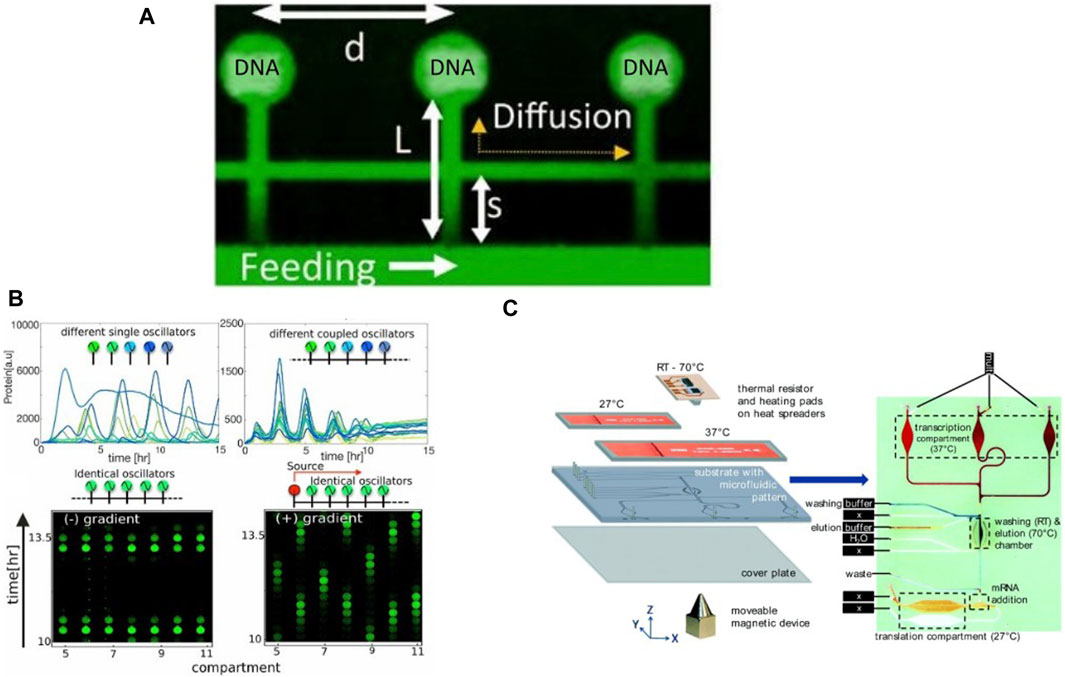
Figure 7. Examples of valve-less microfluidic devices developed for continuous cell-free expression: (A) The microfluidic device used by Tayar et al., 2017 containing a main flow channel with CFPS components diffusing to compartments containing immobilized DNA brushes (B) GFP expression in compartments acting as single or coupled oscillators under different microfluidic configurations, forming synchronized gene networks (Tayar et al., 2017). (C) Visualization of the integrated microfluidic TRITT device designed by Georgi et al., 2016 where transcription and translation are separated by microfluidic compartments.
Levy et al., 2020 theorized that the high density of biomolecules observed in the DNA brushes immobilized on-chip could be leveraged to facilitate assembly of ribosomal subunits post-synthesis from their templates in the DNA brushes. Thus, they used the concept of immobilized DNA compartments to show autonomous synthesis and assembly of ribosomal subunits on-chip and verified that high density of biomolecules within the DNA brushes was the driving factor for assembly of ribosomal subunits. Levy et al., 2021 also demonstrated higher concentration of ribosomes within immobilized DNA brushes, recording up to 17-fold increased concentration at peak activity in comparison to bulk concentrations. They were able to decouple transcription from translation, similar to spatial separation of transcription and translation in eukaryotic cells, by pre-immobilizing ribosomes close to DNA brushes and supplying cell-extract solution void of ribosomes.
Georgi et al., 2016 introduced another microfluidic protocol for CFPS, termed Transcription—mRNA Immobilization and Transfer—Translation (TRITT). Transcription and translation was conducted in separate compartments on a microfluidic chip by transfer of transcribed mRNA to a translation chamber (Figure 7C). Magnetic particles were used to immobilize mRNA generated by transcription, which was followed by a washing and elution step to separate the mRNA from the transcription mix and the particles. The eluted mRNA was then transferred by flow to another chamber for translation. The particles could be moved back and forth via magnets for subsequent cycles of transcription. The device was integrated with thermal sensors and heating pads to enable a fully automated protocol for CFPS. Another microfluidic continuous flow device was designed by Siuti et al., 2011, containing porous 40 μm wide and 15 μm tall on-chip porous reaction containers for CFPS. Fabricated using a combination of UV microlithography and electron beam lithography, each reactor contained pores which allowed for diffusion of small molecules required for cell-free reactions while trapping the template DNA, synthesized RNA, and protein components of the cell-free reaction within. The reactors were filled with components required for cell-free synthesis and sealed with a PDMS membrane, following which a solution containing small molecule precursors and secondary energy sources were continuously flowed through the chip to sustain the cell-free reaction in each reactor for up to 25 h.
A drawback with the devices discussed in this section lies in the relatively large volumes required to run cell-free reactions. Living cells show stochasticity in gene expression alongside various other sources ranging from microenvironment and epigenetic variation (Stewart et al., 2005) to cell growth (Cookson et al., 2010) and partitioning of components during cell division (Rosenfeld et al., 2005). Gene expression in low-volume cell-free systems could permit the study of gene expression stochasticity in isolation from the other noise sources described above. To address this challenge, Karig et al., 2013 introduced a microwell array system to conduct cell-free expression in biologically relevant volumes down to 20 fL. They produced these wells in PDMS devices and sealed them with a glass coverslip to isolate cell-free reactions in each well. Significant variation was observed in protein expression across wells in each experiment, which they hypothesized might be due to spontaneous crowding of cell-free components upon compartmentalization in small volumes (Luigi Luisi et al., 2010). They observed noise in each well to be of much higher magnitude than expected due to Poisson distribution, and theorized this may be due to translational bursting, where each mRNA is transcribed multiple times to obtain proteins. This study provides an interesting step towards modelling noise in cell-like systems by separating the intrinsic noise from gene expression from extrinsic sources of noise.
Active single-phase microfluidic devices also suffer from being inherently limited in throughput by design. Adding reaction chambers or increasing reaction throughput in all of these devices requires additional microvalves, which only increases operational complexity. Adding reaction chambers also takes up additional space on-chip, which might not always be a viable solution when scaling-up. Due to this, throughputs in single-phase devices is often limited to the thousands, while multiphase systems, explained in the next section, often offer millions in throughput per experiment. Thus, they might not be the best option in massively high-throughput studies such as directed evolution. Furthermore, scaling down to reach biologically relevant dimensions often leads to increasingly complex and expensive fabrication strategies, which in turn might restrict accessibility of such technologies. Therefore, it is imperative to keep these restrictions in mind when choosing active single-phase microfluidic systems for cell-free applications.
Despite the restrictions mentioned above, it is clear from examples in this section that active single-phase microfluidics has and will continue to greatly contribute to the advancement of cell-free synthetic biology. We highlighted key microfluidic technologies and advances that enabled long-term and high-throughput cell-free studies by implementation of microfluidic valves or continuous flow systems. It can sometimes be challenging to implement these setups owing to their reliance on peripheral control systems, but several simple DIY protocols describing all necessary hardware and software elements are available (Brower et al., 2018; White and Streets, 2018). Along with this, the ability to develop complex, high-throughput fluid handling protocols with minimal use of reagents makes single-phase microfluidics an attractive option for cell-free synthetic biology research and applications.
While single-phase microfluidic devices have been implemented for several applications in cell-free synthetic biology they do not emulate living cells in one key aspect: compartmentalization by a phospholipid bilayer. Implementing life-like compartmentalization of cell-free reactions in vesicles would be an important step towards realization of a synthetic cell (Maerkl, 2023), which cannot be realized in single-phase systems. This challenge can be addressed by multiphase microfluidic systems, which utilize flow of two or more fluid phases and are commonly employed in high-throughput production of single or multiphase emulsions. The earliest examples for emulsion generation were using bulk emulsification, which involved stirring an aqueous phase into an oil phase to generate large populations of water-in-oil (W/O) emulsions (Sjöblom, 2006). This enabled high-throughput directed evolution studies, which rely on scanning large libraries of DNA or enzymes over multiple rounds of evolution to select the most efficient mutants (Tawfik and Griffiths, 1998; Oliver et al., 2006). The need for generating uniform compartments at high-throughput led to development of microfluidic devices for on-chip generation and manipulation of monodisperse emulsions (Todd et al., 2001; Teh et al., 2008; Shang et al., 2017). Emulsions of water droplets in oil (W/O droplets) can be formed in microfluidic devices by breakup of an aqueous stream at a T-junction or a flow-focusing junction (Todd and Jon, 2013; Zhu and Wang, 2017). The ability to create monodisperse volumes of fluid in high-throughput makes droplet microfluidic systems very attractive for several applications in high-throughput screening assays including mass spectrometry (Wang et al., 2015), sorting (Xi et al., 2017), and directed evolution (Stucki et al., 2021b). Droplets of volumes in the range of nanoliters to femtoliters can be generated and analyzed at a throughput of several kilohertz (kHz) (Todd and Jon, 2013; Trantidou et al., 2018). The generation of discrete, deformable volumes in a flow system enables new possibilities such as reversible droplet trapping (Niu et al., 2011; Saint-Sardos et al., 2020), high-throughput sorting and building droplet interface bilayer (DIB) networks (Elani et al., 2012). Fluorescence-assisted droplet sorting (FADS) (Tabuchi and Yokobayashi, 2022), droplet fusion, droplet splitting (Zeng et al., 2011) and droplet trapping (Huebner et al., 2009) are some of the most popular examples of fluidic manipulations possible in droplet microfluidic systems (Theberge et al., 2010).
Droplet microfluidics involves study and optimization of a diverse set of parameters including, but not limited to, surfactant choice and concentration (Baret, 2012), dimensionless numbers such as capillary number, and choice of carrier fluid (Sohrabi et al., 2020). For example, fluorinated oils can carry atmospheric gases, are immiscible with water and don’t attract hydrophobic biomolecules (Wagner et al., 2016), thus utilizing them as the carrier phase enables microdroplets to function as biochemical reactors for cell-free applications. Another factor to consider is the type and concentration of surfactants used, as they stabilize droplets in the carrier phase, which prevents uncontrolled droplet fusion and cross-talk between droplets in long-term studies (Debon et al., 2015). Furthermore, similar to fluidic networks developed in single-phase systems, transport of microdroplets can be manipulated to establish robust microdroplet networks in both active and passive formats (Schindler and Ajdari, 2008; Biral and Zanella, 2013; Medina et al., 2019; Grimmer and Wille, 2020). Geometric parameters can be varied to establish precise droplet flow paths, and generate temporally oscillating flow networks (Cybulski et al., 2019). Complex flow dynamics relevant to droplet microfluidics and their applications are covered in detail in an excellent review by Baroud et al., 2010. Microdroplets flow in the form of plugs in a majority of droplet microfluidic applications. While flowing, recirculating flow fields are formed inside these plugs that facilitate continuous mixing of components (Fowler et al., 2002). Mixing inside droplets can be further enhanced by adding turns to the flow channel, which leads to chaotic advection of components inside droplets (Bringer et al., 2004). This possibility of mixing components in droplets post-formation permits droplet generation from co-flow of aqueous streams, which can be used to segregate components that cannot be pre-mixed, as is often the case in CFS. Microvalves can be used to add another layer of control to microdroplet devices by introducing an active control element (Chang et al., 2018). Integration of microelectrodes into droplet-based devices adds yet another control component, by enabling dielectrophoretic (Singh and Aubry, 2007) manipulation of droplets and their contents, commonly employed in active droplet sorting (Ahn et al., 2006). Droplet fusion enables introduction of components into a microdroplet (Mazutis et al., 2009; Abate et al., 2010), thus allowing for precise control over reaction kinetics. Droplets can also be arbitrarily split using Y-junctions (Ma et al., 2021) or T-junctions (Hoang et al., 2013) with the ratio of splitting dependent on flow resistance in downstream branches. This can be applied in combination with electrophoresis (Saucedo-Espinosa and Dittrich, 2020) or dielectrophoresis (Huang et al., 2021) to up-concentrate biomolecules in droplets prior to analysis or further processing. The advantages arising from the microfluidic phenomena exclusive to droplet microfluidics coupled with high-throughput generation and analysis makes single emulsion systems attractive for cell-free studies, especially in directed evolution and cell-free assays utilizing IVTT.
Tawfik and Griffiths showcased the first example of CFPS in microdroplets (Tawfik and Griffiths, 1998). They generated droplets in bulk with a mean diameter of 2.6 µm by stirring an aqueous IVTT mix into oil containing surfactants. They demonstrated cell-free expression of several proteins from template DNA encapsulated within droplets, thus providing the first proof-of-concept for compartmentalization of IVTT reactions. Following this, several examples of high-throughput cell-free studies enabled by microdroplets generated via bulk emulsification were published (Pietrini and Luisi, 2004; Swartz et al., 2004; Vincent and Libchaber, 2004; Oliver et al., 2006; Rothe et al., 2006; Stapleton and Swartz, 2010). In 2005, Dittrich et al. showcased CFPS in W/O droplets generated on-chip (Petra et al., 2005), where they expressed GFP in droplets containing IVTT mix. Hori et al. further developed this concept by encapsulating different combinations of repressor and inducer concentrations to map their effect on protein synthesis (Hori et al., 2017). They generated picoliter-sized droplets at rates of over 30 kHz and barcoded them using dyes with final concentrations corresponding to the combinations of inputs encapsulated in each droplet. An incoherent feed-forward loop (Haseltine and Arnold, 2007) was designed in droplets to study the effect of a transcriptional activator (AraC) and a repressor (TetR) on deGFP expression. Wang et al. reported successful mammalian cell-free expression in droplets using a HeLa-based lysate and incorporated a nucleic acid probe to simultaneously track transcription and translation in droplets (Wang et al., 2018).
Droplet microfluidic devices for executing complex multistep processes have been developed by several groups. As an early example demonstrating the potential of multistep droplet microfluidics, Fallah-Araghi et al., 2012 conducted cell-free directed evolution in droplets. They isolated single copies of genes in droplets and performed PCR amplification in them. Elevated temperatures led to unwanted fusion of some droplets during PCR due to the effect of temperature on surfactants and surface tension. Thus, a size sorting step was employed to collect a monodisperse population of droplets. This was followed by one-to-one fusion with droplets containing IVTT mix and fluorogenic substrate, followed by incubation at 37°C for 2 hours. Finally, the droplets with fluorescent signal were collected by coalescing them into an aqueous stream. Recently, Holstein et al., 2021 developed a robust protocol for cell-free directed evolution in droplets generated on-chip. They encapsulated DNA template coding for the enzyme Savinase along with a master mix to perform rolling circle amplification (RCA) in 14 pL-sized droplets. This avoided challenges with elevated temperatures in PCR reported by Fallah-Araghi et al., 2012. Following this, PURE mix and fluorogenic substrate were sequentially pico-injected for IVTT and fluorescence detection of Savinase. Finally, droplets were sorted on-chip to collect the drop population with high fluorescence and DNA was recovered from them for further rounds of evolution (Figure 8A). This droplet microfluidic protocol allowed for screening of over a million droplets and 106 variants per evolution cycle. Multistep protocols can also be executed by encapsulation of smaller compartments such as liposomes followed by their controlled disintegration. Gan et al. described a integrated protocol using droplet microfluidics to achieve rapid and efficient screening of gene circuit regulatory components (Gan et al., 2022) (Figure 8B). Their protocol combines generation of droplets containing CFPS mix, droplet sorting and next-generation sequencing to form a robust workflow, with an additional step of encapsulation in double emulsions for increased compatibility with standard flow cytometry.
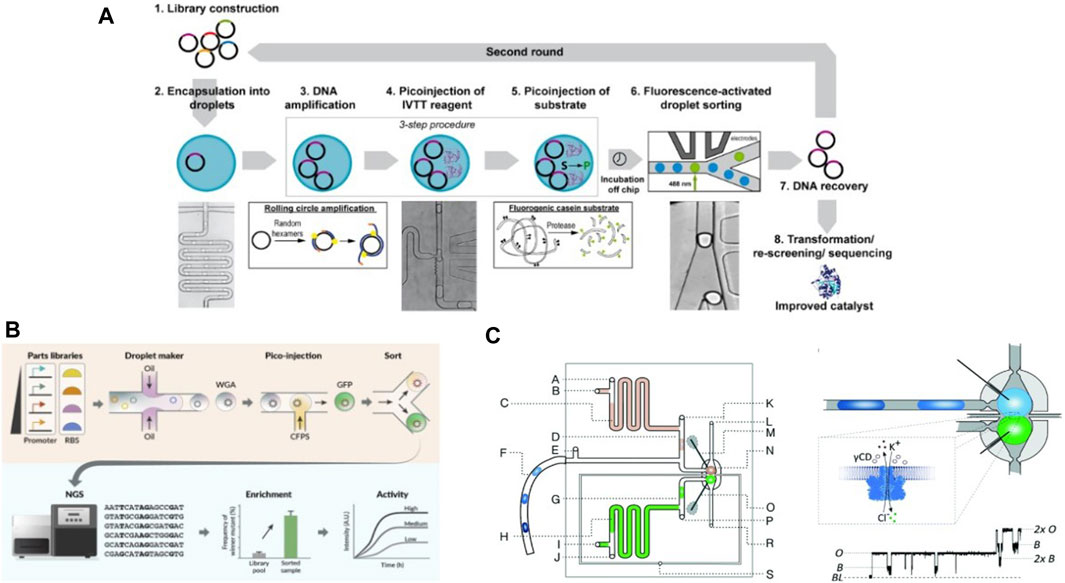
Figure 8. Examples of droplet microfluidic devices and protocols developed within the context of cell-free applications: (A) Workflow for multiple rounds of cell-free directed evolution in droplets designed by Holstein et al., 2021 (B) Droplet microfluidics protocol designed by Gan et al., 2022 for rapid iteration and testing of gene circuit components (C) Device schematic for generating droplet bilayer network by Czekalska et al., 2015 with electrodes incorporated for measuring current across the bilayer due to exchange of ions and biomolecules.
Aside from droplet generation via microfluidic devices, large arrays of microdroplets can be generated on substrates via hydrophilic-hydrophobic patterned surfaces. Zhang et al., 2019 developed a protocol for high-throughput screening via CFPS in microarrayed femtoliter droplets. They etched an array of more than a million droplet wells per cm2 in CYTOP, a hydrophobic perfluoropolymer, The bottom of each well is hydrophilic glass. A PURE mixture containing template DNA and CFPS components was first flown over the array to fill all wells followed by oil to flush the aqueous solution from the CYTOP surface. The hydrophilic glass surface retained CFPS solution within the wells. Finally, a sealing oil was introduced to eliminate droplet evaporation and to stabilize the emulsions. They showed CFPS of various fluorescent proteins using this protocol and demonstrated CFPS using cell lysates as well. They were able to use a glass micropipette to manually recover DNA from individual droplets following CFPS, enabling high-throughput directed evolution of enzymes. Mutant DNA coding for the E. coli enzyme ALP was used for tx-tl in over half a million droplets per experiment. Droplets showing high enzyme activity were sorted and DNA recovered from them was used for the next round of CFPS to confirm improved enzyme activity.
Another area of interest in droplet microfluidics relevant to cell-free systems is enabling inter-droplet communication via the droplet interface. When two droplets stabilized by single layers of phospholipids are brought into contact, it results in the formation of a phospholipid bilayer at the droplet interface (Funakoshi et al., 2006; Holden et al., 2007; Stanley et al., 2010). Transport of ions and small molecules occurs via simple diffusion across this bilayer while inclusion of protein pores within the bilayer allows transport of biomolecules and larger biocomponents. This strategy can be used to produce cascading cell-free reactions in 2D, or even 3D, droplet networks (Stanley et al., 2010; Friddin et al., 2016; Xavier et al., 2016; Trantidou et al., 2017; Trantidou et al., 2018; Strutt et al., 2022). Syeda et al., 2008 also demonstrated CFPS of membrane proteins inside droplets in DIB network, which were incorporated into the bilayer after synthesis. However, they observed a decrease in stability of the bilayer in the presence of IVTT components. Friddin et al. studied this bilayer destabilization effect in detail and observed that addition of lipid vesicles to the ITT mix stabilized the bilayer, thus paving the way for long-term cell-free studies in DIB setups in the future (Friddin et al., 2013). Along with the possibility of electrical measurements on membrane proteins incorporated into a bilayer (Czekalska et al., 2015) (Figure 8C), DIB setups show great promise for enabling studies on bilayer membrane proteins and movement of biomolecules across bilayers and tissue-like systems to enable material exchange and intercellular communication studies.
The strategy of building 2D and 3D droplet networks connected via DIBs was further developed to generate 3D-printed “synthetic tissue” like structures by Villar et al., 2013 They formed 3D networks of picoliter-sized lipid droplets, with a single lipid bilayer present at all droplet interfaces in the material. Booth et al., 2016 demonstrated application of such a 3D-printed synthetic tissue in visualizing intercellular electrical and biomolecular communication, resembling a functional mimic of neuronal transmission. They designed a DNA expressing GFP linked to α-hemolysin, a membrane pore protein, downstream of a promoter sequence activated by light. Upon bringing two droplets to form a DIB, one containing this DNA in a PURE system and one droplet lacking this DNA, they observed localization of expressed GFP-α-hemolysin at the DIB only upon exposure to UV (Figure 9A). They verified an electrical signal travelled between the 2 droplets only post-expression and localization of α-hemolysin at the DIB. They printed a synthetic tissue patterned with droplets containing light-activated DNA in a PURE system and droplets missing this DNA to show propagation of electric signals along patterned light-activated droplets pathways, marked by cell-free expression of mVenus proteins within these droplets.
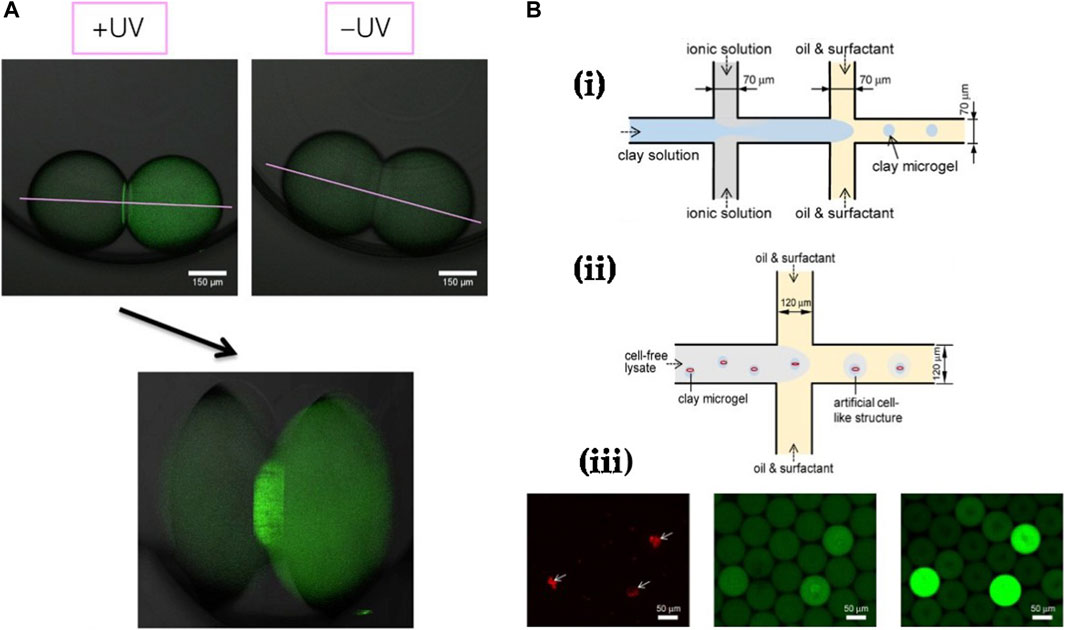
Figure 9. Further examples of cell-free studies enabled by droplet microfluidics: (A) Cell-free expression of membrane proteins in droplets linked via a DIB, performed by Booth et al., 2016. Expression was only observed under UV exposure and localization of expressed GFP-α-hemolysin is clearly visible at the droplet bilayer interface. (B) CFPS enabled via clay-DNA hydrogels in droplets by Jiao et al. (Yi et al., 2018) showing (i) The protocol for fabricating clay hydrogels, (ii) Protocol for encapsulating fabricated hydrogels together with cell-free solution in droplets and (iii) Fluorescent timelapse images of clay hydrogels encapsulated inside droplets and confirmation of cell-free expression only in droplets containing the DNA-clay hydrogels.
Droplet microfluidics has also been employed in high-throughput production of gel beads for CFS. Protein expression rates can be enhanced by executing CFPS in a clay matrix, shown by Jiao et al., where they mixed ionic and clay aqueous solutions prior to crossflow with an oil phase in a microfluidic device to spontaneously generate clay microgels (Yi et al., 2018) (Figure 9B). Plasmids of interest could be directly integrated into these microgels due to their inherent hydrophilicity and favourable electrostatic interactions with the clay matrix. Following this, they introduced E. coli cell-free lysate solution to plasmid-integrated microgels containing magnetic nanoparticles and showed robust eGFP synthesis over multiple rounds of lysate replenishment. Finally, to imitate cell-like structures, they encapsulated clay microgels in microdroplets containing lysate solution by using a droplet microfluidic generator and achieved eGFP expression within the droplets. Using a similar protocol, Aufinger et al. demonstrated localized transcription and translation reactions in agarose microbeads co-encapsulated in droplets (Aufinger and Friedrich, 2018). Each droplet contained two agarose microbeads, one with covalent immobilization of template DNA and another with immobilized DNA sequences corresponding to transcribed mRNA. They observed cell-free expression only within droplets containing both beads.
While droplets show great potential as compartments for cell-free reactions, there are some challenges which need to be addressed for their utilization in the field. Studies have shown significant differences in IVTT rates in droplet versus bulk reactions, especially at sub-nanoliter droplet volumes. Hansen et al. performed an extensive study discussing and characterizing stochastic elements and possible sources of noise in picoliter-sized droplets (Hansen et al., 2016). They found that introduction of crowding agents inside droplets acted as a significant noise source, and hypothesized the reason to be heterogenous expression, localization and translation of mRNA inside these small droplets. Macromolecular crowding also has significant effects on transcription and translation rates in CFPS. Studies have shown crowding leading to increases in transcription rates but decreases in translation rates in cell-free reactions (Xumeng et al., 2011; Sokolova et al., 2013). Another significant noise source is the stochastic variation of CFPS components in droplet populations (Reginald Beer et al., 2007; Courtois et al., 2008; Reginald Beer et al., 2008) and low copy number DNA in small droplets (Shahrezaei and Swain, 2008). Aside from these factors, it is also important to verify functionality of droplets as isolated compartments. Studies have shown that leakage of biomolecules between droplets or into the carrier phase can occur based on the choice of surfactant and fluids (Debon et al., 2015). This can severely compromise studies by allowing cross-talk between compartments, but this issue can be mitigated by choosing appropriate combinations of fluid phases and appropriate surfactants. Finally, evaporation can also be a significant factor in long-term studies with microdroplets in PDMS-based devices as PDMS is permeable to most gases (Shim et al., 2007). These factors must always be taken into consideration when implementing CFS in droplet microfluidics.
Looking towards the future, we imagine droplet microfluidic systems will continue to play an important role in cell-free studies owing to their robustness, simpler experimental setups in comparison to single-phase systems and high throughput. As shown in few examples in this section, droplets also have great potential to act as isolated environments for studying intercellular communication of multiple artificial cells or vesicles encapsulated within them. Combined with recent studies on DIBs and potential for incorporation of microvalves to build complex droplet systems for enabling multistep cell-free protocols, we see several future directions for adaptation of droplet microfluidics in cell-free studies.
Recent developments in microfluidic techniques and bench techniques have enabled robust generation of water-in-oil-in-water (w/o/w) double emulsions (DEs) (Wang et al., 2014; Vladisavljević et al., 2017). In 2004, Okushima et al., 2004 were the first to generate DEs in microfluidic chips by utilizing two T-junctions in series to generate w/o/w emulsions. This was followed by research published by Shah et al., 2008, where they utilized glass microcapillary devices to generate monodisperse multiphase emulsions with high-throughput and showed encapsulation of emulsions within emulsions. Since then, the field of DEs has come a long way, with focus shifting from their use as simple reaction compartments to generating complex liposomes and artificial cell bodies (Carugo et al., 2016; Martino and deMello, 2016; Has and P Sunthar, 2020). Monodisperse double emulsions are typically generated in microfluidic devices by using two flow-focusing junctions in series (Arriaga et al., 2015; Vladisavljević et al., 2017). Reagents and oil phase can be adapted to generate vesicles or liposomes in high-throughput, where the oil phase dissociates from the oil shell of a double emulsion leaving behind an aqueous phase compartmentalized by a lipid bilayer. DEs have several benefits over single emulsions, such as increased stability during storage and incubation, no volume loss due to evaporation and ease of integration with existing systems that primarily operate with aqueous media. Thus, DEs can serve as excellent compartments for biochemical reactions as well as a great starting point for generation of liposomes and artificial cells. DEs can also be used to generate polymeric capsules with finely tuned pore sizes (Kim et al., 2015; Lee et al., 2016; Mytnyk et al., 2017; Gao and Chen, 2019; Michielin and Maerkl, 2022).
These advantages have seen implementation of double emulsions as compartments in several cell-free studies. For example, Nuti et al. encapsulated artificial vesicles in DEs to screen for activity of anti-microbial peptides (AMPs) (Nuti et al., 2022). Since AMPs are known to disrupt bacterial membranes and also show slight toxicity towards mammalian cell membranes (Magana et al., 2020), CFPS is a good alternative for their production and testing. The authors first encapsulated self-quenching fluorescent dyes in bacteria-like and mammalian-like large unilamellar vesicles (LUVs) off-chip. This was followed by co-encapsulation of these LUVs with DNA templates coding for AMPs and a CFPS mix in double emulsions followed by trapping for long-term incubation and downstream fluorescence-assisted droplet sorting (FADS) (Figure 10A). Production of AMPs (meucin-25 and Cathelicidin-BF) in DEs caused rupture of bacteria-like LUVs but not mammalian-like LUVs. This was tracked by increases in fluorescence in DEs from leakage of dyes previously encapsulated in the bacteria-like LUVs, thus proving specific membranolytic activity of the AMPs. Similarly, Stucki et al., 2021a used addition of sodium dodecyl sulfate (SDS) as a trigger for solubilization of LUVs encapsulated in DEs, thus establishing a robust platform for carrying out multistep CFPS protocols by using DEs and vesicles as compartments. Chang et al., 2018 were able to generate real-time IVTT formulations containing different concentrations of lysate extract, template DNA, and energy solutions on-chip using pulse-width modulation (Woodruff and Maerkl, 2018). Following mixing facilitated via herringbone structures, the reaction mix was encapsulated into double emulsions by using a DE generator on-chip. They reported successful GFP expression with varying concentrations of DNA encapsulated in the DEs, thus providing a proof of concept for running massively parallelized reactions with minimal volumes by formulating the reaction mix on-chip.
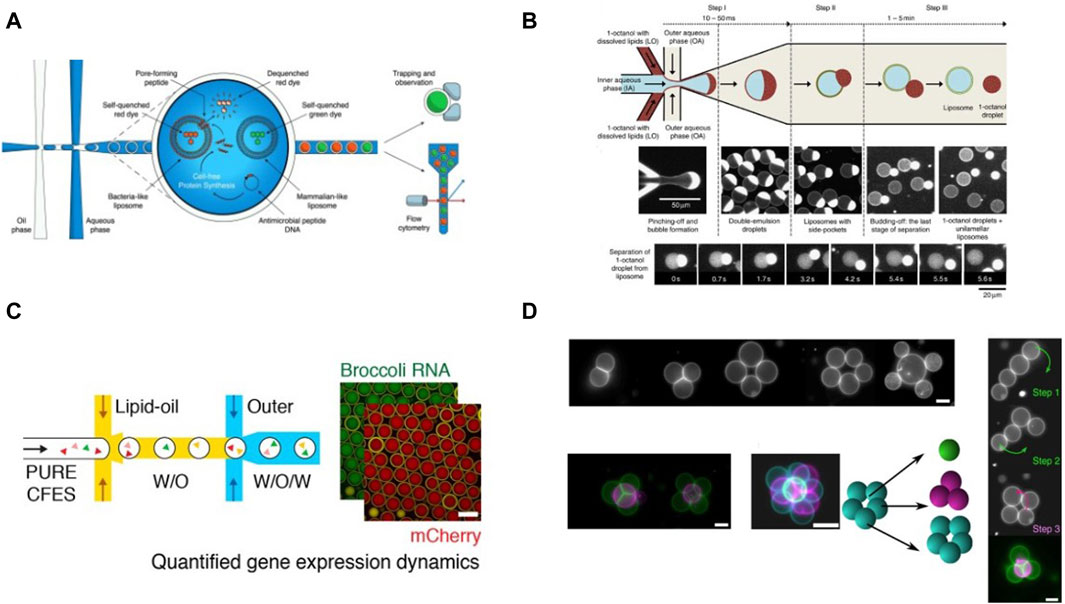
Figure 10. Examples of cell-free studies enabled by microfluidic manipulation of DEs and artificial vesicles: (A) Workflow for screening activity of AMPs using vesicles encapsulated in DEs, designed by Nuti et al., 2022 (B) Octanol assisted liposome generation on-chip described by Deshpande et al., 2016 (C) Workflow for CFPS in liposomes generated via microfluidics. (Gonzales et al., 2022). (D) Generation of 2D and 3D vesicle networks and fusion of vesicles enabled by optical tweezers, demonstrated by Bolognesi et al., 2018.
Polymeric capsules with controlled permeabilities, generated via double-emulsion microfluidic systems, have been used as artificial cell models to study population behaviours such as quorum sensing (Niederholtmeyer et al., 2018) and intercellular communication (Michielin and Maerkl, 2022). Michielin and Maerkl demonstrated encapsulation of biomolecules in semi-permeable polymeric capsules which were fabricated from DEs (Michielin and Maerkl, 2022). They formed these polymeric capsules by UV polymerization of poly (ethylene glycol) diacrylate (PEG-DA 258) in the oil shell. The size of pores in the capsule could be controlled by altering the percentage of polymer to solvent in the oil phase. This enabled size-based permeation of biomolecules across the polymeric shells. They employed these capsules to perform cascading DNA strand displacement (DSD) reactions (Simmel et al., 2019), where DNA strands were first immobilized in capsules. Following addition of input ssDNA and subsequent DSD, a Cy5 fluorophore was unquenched along with the simultaneous release of a signal strand. The strand diffused out of these shells into a second population of polymeric capsules functionalized with a DSD gate corresponding to this signal strand. This released a second signal strand, unquenching a Cy3 fluorophore. The delayed onset of Cy3 fluorescence following Cy5 fluorescence confirmed the successful execution of the cascading reaction. Niederholtmeyer et al. used polymeric capsules to implement communication and quorum sensing in a population of artificial cells, similar to collective behaviours of cells in tissues and biofilms (Niederholtmeyer et al., 2018). They used T3 RNA polymerase, expressed in a cell mimic, for diffusive signaling to drive expression in another population of reporter cell mimics. The expression in reporter cell mimics was found to only switch on when activator cell mimic concentration was above a threshold value, thereby establishing quorum sensing in this system. Li et al., 2022 showed selective compartmentalization of components required for CFPS by engineering permeability of hydrogel compartments produced from DEs. The permeability of the compartments was controlled by changing pore size by tuning the gel-to-crosslinker ratio. The authors used these compartments to conduct CFPS where transcription was isolated within the compartments and translation occurred in the external environment. This was achieved by co-encapsulation of plasmids and RNA polymerase within compartments and providing energy components and proteins required for translation in the external solution; mRNA transcribed within compartments subsequently diffused into the external solution for protein translation.
One of the grand challenges in cell-free synthetic biology and biochemistry is the bottom-up construction of an artificial cell (Schwille, 2011; Laohakunakorn et al., 2020; Maerkl, 2023). This implies creation of an in vitro system that mimics a cell’s structure including its complex lipid bilayer membrane (Yeagle, 2016) and possibly even intracellular vesicles (Van Niel et al., 2018). Thus, generation of liposomes and vesicles for enabling CFPS within them is an important aspect of cell-free synthetic biology (Jia et al., 2017). Several protocols have been designed for generation of liposomes or DEs with ultra-thin oil shells functioning as bilayer membrane (Göpfrich et al., 2018; Ugrinic et al., 2019; Ai et al., 2020). One of the more robust techniques, referenced in several recent research papers on artificial vesicles, was introduced by Deshpande et al., where they generate monodisperse liposomes by using octanol as the oil phase (Figure 10B) in a microfluidic device designed for generating DEs with ultra-thin oil shells (Deshpande et al., 2016).
With the advent of robust techniques for forming artificial vesicles, interest has grown in enabling cell-free reactions within them post-generation. Gonzales et al., 2022 demonstrated a robust protocol for CFPS in liposomes fabricated on-chip using PURExpress (Figure 10C) and observed differences in expression rates inside liposomes compared to bulk. They attributed this difference to the limited diffusion of ions and materials across lipid bilayer membranes, an effect previously reported by Noireaux and Libchaber (Vincent and Libchaber, 2004). Noireaux and Libchaber used PURExpress CFPS solution containing DNA template coding for a reporter protein as the inner aqueous phase, and prepared a feeding mix to use as the outer aqueous phase to sustain CFPS inside liposomes. As expected, composition of the outer aqueous phase, despite being of the same osmolarity as the inner aqueous phase, had a significant influence on protein expression dynamics inside liposomes. Dilution of feeding mix led to decreased protein expression within liposomes, thus proving that diffusion of biomolecules across the lipid bilayer sustains CFPS inside liposomes. To resolve limited diffusion of biomolecules from feeding mix into liposomes, Noireaux and Libchaber added a membrane pore protein, α-hemolysin, to selectively allow diffusion of small molecules across the bilayer. They successfully demonstrated sustained CFPS using E. coli extract inside liposomes for up to 4 days. Soga et al., 2020 introduced a technique for generation of femtoliter-sized liposomes and executed cell-free expression within them, thereby moving one step closer to cell-like functional structures. They used a microarray of femtoliter sized wells to first create W/O emulsions, followed by bulk emulsion transfer to obtain liposomes of 0.6–5.3 µm in diameter. Successful integration of α - hemolysin pores was also achieved to allow material transfer with the outer aqueous phase. They verified biocompatibility of their liposomes by executing cell-free synthesis using PURE components after appropriate optimization of phospholipid composition used in bilayer formation, and highlighted this platform’s potential in studying gene expression stochasticity in CFSs.
Vesicles can also be used to build signalling pathways in artificial cells by manipulating their membrane composition and placing them in close proximity to allow transfer of biomolecules. Such setups are also used for carrying out cascading biochemical reactions using vesicles as reactors. Similarly, cascading reactions can also be carried out in droplets stabilized by phospholipids. Bolognesi et al., 2018 developed a vesicle network by using optical tweezers for bringing vesicles into contact, which allowed transfer of molecules between them. They employed α-hemolysin in bilayer membranes for selective permeation between vesicles and added a pore blocking protein in the outer phase to prevent leakage of molecules from vesicles to the outer phase. They demonstrated fusion of vesicles by using gold nanoparticles on the outer layer of the membrane to rapidly heat and disrupt the membrane between two vesicles, which was followed by them fusing in a few minutes. They expressed GFP using a PURExpress mix and template DNA split between three vesicles, which were later fused together, thus bringing all the components together to perform CFPS (Figure 10D).
While double emulsions can function as model compartments for reactions and have also pushed the boundaries of synthetic cell studies, their usage is still hindered by several limitations. Injecting components into double emulsions and vesicles post-formation is a challenge due to the presence of an oil shell or a lipid bilayer. Material exchange can only occur in these cases by diffusion across the oil shell or lipid bilayer (Needham, 2017). This prohibits more direct ways of forming a cascading reaction, as all larger-sized components required have to be encapsulated in the DEs or artificial cells right from the formation step. The solutions currently popular for this involve integration of various membrane proteins expressed and folded in CFS inside liposomes, thus allowing for material exchange with the environment (Kalmbach et al., 2007; Long et al., 2012; Focke et al., 2016). Another way to address this challenge in capsules has been reported by Li et al., where they fabricated charge-selective shells for controlled uptake and release of fluorescein as a model for small molecules (Chuen-Ru and Lutz-Bueno, 2023). Incorporating DNA nanopores for selective protein and biomolecular transport could be another solution (Diederichs et al., 2019; Luo et al., 2023). However, it remains to be seen whether such techniques can be adapted for cell-free applications. The composition of lipids used to generate vesicles also demands highly specific tuning based on the desired application. A recent study by Miwa et al. highlighted the importance of optimizing the bilayer lipid composition for cell-free expression in vesicles, where they reported higher transcription and translation rates within vesicles containing positively charged lipids in comparison to neutrally or negatively charged lipids (Miwa et al., 2024). Studies have reported leakage of small biomolecules and reaction components across emulsion drops via simple diffusion across their interface when using certain surfactants, thus highlighting a need for selecting appropriate surfactants to eliminate cross-contamination (Etienne et al., 2018). Many studies have also reported challenges with bio-compatibility when using the PURE system or other CFS in artificial vesicles due to high protein concentration (Syeda et al., 2008). With the goal of constructing a fully self-regenerating artificial cell, which must be resolved in order to build IVTT-enabling artificial cells in the future. Further investigation is also required to increase stability of artificially constructed vesicles (Sedighi et al., 2019), as they are sensitive to bilayer composition (Needham and Nunn, 1990), solvent used (Webb et al., 2019), changes in pH (Sułkowski et al., 2005) and other environmental factors.
Addressing these limitations would drive future applications of microfluidics in building artificial cells and vesicle systems. With proof-of-concept studies established on vesicle networks and communication models (Pereira De Souza et al., 2017), one can imagine how such setups would enable the study of complex intercellular communication models and signal propagation. The ability to precisely control flows in microfluidic devices could also enable chemotaxis studies (Jin et al., 2017) in artificial cell populations in the future. Resolving challenges in biocompatibility arising from high protein concentrations within vesicles would enable robust studies of IVTT systems in artificial cells and bring us closer to building a living cell from its biological components.
Translation of multistep parallelized laboratory processes into the microscale involves development of precise manipulation techniques for individual microliter-sized volumes. These techniques are often associated with valve-based microfluidics and digital microfluidics (DMF). DMF broadly covers the range of integrated microfluidic systems for manipulation of droplets using microelectrode arrays (Choi et al., 2012; Jebrail et al., 2012; Gach et al., 2017; Kothamachu et al., 2020). Digital microfluidic systems allow for various precise manipulations such as generation, fusion, mixing and splitting of discrete droplets on an open surface by applying a defined series of electric potentials (Cho et al., 2003; Aaron, 2008). The versatile setup in these systems also enables integration with thermal, optic or most commonly, magnetic actuation systems. Altogether, these systems allow researchers to replicate multistep procedures such as precise metering of reagents, formation of drops with different reagent concentrations, purification and sequencing of DNA, and multistep proteomics. They also provide advantages associated with microfluidics such as minimal reagent consumption and ability to multiplex reactions. DMF devices often offer the potential for automation to a high degree and do not require micropumps or microtubing like conventional microfluidic setups.
DMF devices can be differentiated into single-plate or parallel-plate devices. In single-plate devices, a droplet is manipulated on a surface that has an electrode array underneath consisting of both ground and actuation electrodes. In parallel-plate devices, droplets are sandwiched between two surfaces, where one plate has an electrode array for actuation and the opposite plate acts as the ground electrode. The electrode array is passivated with an insulating dielectric layer to generate the field and charge gradients required for droplet actuation (Lee et al., 2002; Lu et al., 2008; Nelson and Kim, 2012). The static contact angle between the droplet and the surface changes with the square of the potential difference applied across them. Using the electrode array, differential contact angles can be created across the droplet, which results in capillary pressure driving the droplet along the desired path, a phenomenon called electrowetting (Berge, 1993; Pollack et al., 2002). The surfaces that come in contact with the droplet have a hydrophobic coating on them to facilitate smooth droplet manipulation in the device. Furthermore, in order to prevent adsorption of biomolecules to the surface, oils are often used as a thin filler layer (Srinivasan et al., 2004). The ability to precisely execute multistep processes in an automated fashion, generate multiple droplets of submicroliter volumes, integrate with other techniques and prevent biofouling make DMF devices a potentially attractive option for cell-free systems although the number of publications so far lags behind the other microfluidic device types. Currently DMF devices are used in DNA-based applications, immunoassays, and diagnostic applications (Pollack et al., 2011; Shen et al., 2014; Samiei et al., 2016; Sergio, 2016).
A great example for the potential applications of a DMF device in biological studies is the OpenDrop microfluidic chip developed by Alistar and Gaudenz (Alistar and Gaudenz, 2017) (Figure 11A). Their biochip allows for merging, dispensing, splitting and mixing of droplets on an electrode array, along with the possibility of moving droplets in any direction on the array using their software. This resulted in a versatile microfluidic chip with possibilities in tissue printing, synthetic biology, etc. This OpenDrop device was later adapted for CFPS by Liu et al., 2020 to demonstrate cell-free glucose detection and protein screening (Figure 11B). They measured the effect of Mg2+ concentration on cell-free protein production, thus showing that CFPS reactions could be carried out in DMF devices.
Figure 11. Applications of DMF devices in enabling robust, multi-step CFPS protocols: (A) The various droplet manipulations possible in the OpenDrop platform: droplet generation from reservoirs, splitting, merging and movement on the electrode array (Alistar and Gaudenz, 2017). (B) Workflow for enabling CFPS in a DMF device proposed by Liu et al., 2020.
Nuclera recently introduced an eProtein Discovery platform for rapid CFPS enabled by digital microfluidics (Nuclera, 2023). Their platform allows for screening of proteins from 24 gene constructs, in 8 cell-free mixes, resulting in 192 independent screens per run. The platform allows for precise control of 80 nL droplets. Modules for fluorescence analysis, magnetic-bead-based protein purification and thermal control are integrated into the DMF device with pre-loaded protocols for automation. As a proof-of-concept, they successfully expressed and purified functional VEGF, a protein difficult to express in E. coli (Lee et al., 2011). They claim production of micrograms of purified proteins in 48 h using the platform with less than 2 hours of hands-on time, along with integrated characterization of purified proteins. We imagine integrated DMF platforms such as the one from Nuclera, as well as the Tierra Protein Platform (Tierra biosciences, 2023), might mature into useful research tools for robust protein synthesis in cell-free systems.
Despite their advantages, one of the biggest challenges in DMF devices remains their complex fabrication. There is currently a lack of commercial instrumentation for mass-production of DMF devices. However, the field of digital microfluidics still holds promise due to the versatility and possibility of integration with other techniques. With future improvements in technology, automation, and instrument miniaturization, we imagine DMF devices could be useful for developing rapid, portable solutions for multistep cell-free applications.
Focused on applications in cell-free systems, numerous device concepts and proof of principles for both active and passive flow systems have been covered in this review. As interest grows in development of cell-free systems to characterize and build complex biochemical systems, microfluidics will continue to enable studies in this domain owing to its capacity for high-throughput and execution of complex multistep protocols, useful in executing genetic circuits and studying protein binding interactions, for example, With cell-free reaction components often being expensive to acquire, microfluidic setups offer a great advantage by drastically reducing the required reaction volumes. With continuously evolving microfluidic technologies and further developments in microfluidic integration of cell-free workflows, we believe microfluidics will continue to play a substantial role in driving the development and applications of cell-free synthetic biology in academia and industry.
However, as is the case with most technologies, utilization of microfluidics comes with its own challenges, many of which have been covered in the sections above. There are many interesting engineering challenges still being addressed in the implementation of microfluidics in cell-free systems (Gulati et al., 2009; Damiati et al., 2018). The increase of surface area to volume ratio in the micro scale in turn increases influence of factors such as evaporation, particle and fluid diffusion rates, and thus must be taken into consideration while designing a micro-bioreactor. Furthermore, choice of substrate and surface treatment also play a crucial role in optimization of these setups.
Enabling IVTT reactions in cell-like constructs is a stepping stone in the bottom-up construction of complex synthetic biological systems and artificial cells (Schwille, 2011; Cho and Lu, 2020; Laohakunakorn et al., 2020; Maerkl, 2023). There is great potential in developing systems for communication via material transfer between compartmentalized cell-free reactions, which would require complex and robust microfluidic protocols. Robust cell-like division (Deshpande et al., 2018) or vesicles-in-vesicle multi-compartmentalization (Haller et al., 2018; Weiss et al., 2018) in artificial cells is another field where microfluidic systems could be beneficial. The possibility of complete automation, combined with active feedback loops and rapid data processing, will inevitably lead to more advanced microfluidic devices over the next decade. With future developments in microfluidic technologies and adaptation of already existing technologies for cell-free studies, we imagine a more synergistic future in these two fields. This would involve development of robust microfluidic technologies specifically for addressing prominent questions in cell-free synthetic biology. Cell-free systems will continue to greatly benefit from the advantages offered by microfluidics in terms of automation, small volume requirements, rapid prototyping and reproducible flow environments, thus bringing us ever closer to fully integrated lab-on-chip devices for enabling new discoveries and inventions in cell-free synthetic biology.
AB: Conceptualization, Writing–original draft, Writing–review and editing. SM: Conceptualization, Supervision, Validation, Writing–review and editing, Funding acquisition, Writing–original draft.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by a Swiss National Science Foundation grant (Grant No. 214843).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aaron, R. W. (2008). Chemistry. Putting electrowetting to work. Science 322 (5901), 539–540. doi:10.1126/science.1165719
Abate, A. R., Hung, T., Mary, P., Agresti, J. J., and Weitz, D. A. (2010). High-throughput injection with microfluidics using picoinjectors. Proc. Natl. Acad. Sci. 107 (45), 19163–19166. doi:10.1073/pnas.1006888107
Abbas, M., Law, J. O., Grellscheid, S. N., Huck, W. T. S., and Spruijt, E. (2022). Peptide-based coacervate-core vesicles with semipermeable membranes. Adv. Mater. 34 (34), 2202913. doi:10.1002/adma.202202913
Abgrall, P., and Gue, A. M. (2007). Lab-on-chip technologies: making a microfluidic network and coupling it into a complete microsystem—a review. J. Micromechanics Microengineering 17 (5), R15–R49. doi:10.1088/0960-1317/17/5/r01
Ahn, K., Kerbage, C., Hunt, T. P., Westervelt, R. M., Link, D. R., and Weitz, D. A. (2006). Dielectrophoretic manipulation of drops for high-speed microfluidic sorting devices. Appl. Phys. Lett. 88 (2), 024104. doi:10.1063/1.2164911
Ai, Y., Xie, R., Xiong, J., and Liang, Q. (2020). Microfluidics for biosynthesizing: from droplets and vesicles to artificial cells. Small 16 (9), 1903940. doi:10.1002/smll.201903940
Alistar, M., and Gaudenz, U. (2017). OpenDrop: an integrated do-it-yourself platform for personal use of biochips. Bioengineering 4 (2), 45. doi:10.3390/bioengineering4020045
Amalfitano, E., Karlikow, M., Norouzi, M., Jaenes, K., Cicek, S., Masum, F., et al. (2021). A glucose meter interface for point-of-care gene circuit-based diagnostics. Nat. Commun. 12 (1), 724. doi:10.1038/s41467-020-20639-6
Andrianantoandro, E., Basu, S., Karig, D. K., and Weiss, R. (2006). Synthetic biology: new engineering rules for an emerging discipline. Mol. Syst. Biol. 2 (1), 2006–0028. doi:10.1038/msb4100073
Arosio, P., Müller, T., Rajah, L., Yates, E. V., Aprile, F. A., Zhang, Y., et al. (2016). Microfluidic diffusion analysis of the sizes and interactions of proteins under native solution conditions. ACS Nano 10 (1), 333–341. doi:10.1021/acsnano.5b04713
Arriaga, L. R., Amstad, E., and Weitz, D. A. (2015). Scalable single-step microfluidic production of single-core double emulsions with ultra-thin shells. Lab a Chip 15 (16), 3335–3340. doi:10.1039/c5lc00631g
Arriaga, L. R., Datta, S. S., Kim, S.-H., Amstad, E., Kodger, T. E., Monroy, F., et al. (2014). Ultrathin shell double emulsion templated giant unilamellar lipid vesicles with controlled microdomain formation. Small 10 (5), 950–956. doi:10.1002/smll.201301904
Ashty, S. K., and Jewett, M. C. (2018). Cell-free synthetic biology for pathway prototyping. Methods Enzym. 608, 31–57. doi:10.1016/bs.mie.2018.04.029
Aufinger, L., Brenner, J., and Simmel, F. C. (2022). Complex dynamics in a synchronized cell-free genetic clock. Nat. Commun. 13 (1), 2852. doi:10.1038/s41467-022-30478-2
Aufinger, L., and Friedrich, C. S. (2018). Artificial gel-based organelles for spatial organization of cell-free gene expression reactions. Angew. Chem. Int. Ed. 57 (52), 17245–17248. doi:10.1002/anie.201809374
Ayoubi-Joshaghani, M. H., Dianat-Moghadam, H., Seidi, K., Jahanban-Esfahalan, A., Zare, P., and Jahanban-Esfahlan, R. (2020). Cell-free protein synthesis: the transition from batch reactions to minimal cells and microfluidic devices. Biotechnol. Bioeng. 117 (4), 1204–1229. doi:10.1002/bit.27248
Balagaddé, F. K., You, L., Hansen, C. L., Arnold, F. H., and Quake, S. R. (2005). Long-term monitoring of bacteria undergoing programmed population control in a microchemostat. Science 309 (5731), 137–140. doi:10.1126/science.1109173
Baret, J.-C. (2012). Surfactants in droplet-based microfluidics. Lab a Chip 12 (3), 422–433. doi:10.1039/c1lc20582j
Baroud, C. N., Gallaire, F., and Dangla, R. (2010). Dynamics of microfluidic droplets. Lab a Chip 10 (16), 2032–2045. doi:10.1039/c001191f
Beate Bußmann, A., Grünerbel, L. M., Patricia Durasiewicz, C., Alexander Thalhofer, T., Wille, A., and Richter, M. (2021). Microdosing for drug delivery application — a review. Sensors Actuators A Phys. 330, 112820. doi:10.1016/j.sna.2021.112820
Beebe, D. J., Mensing, G. A., and Walker, G. M. (2002). Physics and applications of microfluidics in biology. Annu. Rev. Biomed. Eng. 4 (1), 261–286. doi:10.1146/annurev.bioeng.4.112601.125916
Benner, S. A., and Sismour, A. M. (2005). Synthetic biology. Nat. Rev. Genet. 6 (7), 533–543. doi:10.1038/nrg1637
Berge, B. (1993). Electrocapillarity and wetting of insulator films by water. Comptes Rendus De. L Acad. Des. Sci. Ser. Ii 317 (2), 157–163.
Berhanu, S., Ueda, T., and Kuruma, Y. (2019). Artificial photosynthetic cell producing energy for protein synthesis. Nat. Commun. 10 (1), 1325. doi:10.1038/s41467-019-09147-4
Bhalla, N., Pan, Y., Yang, Z., and Payam, A. F. (2020). Opportunities and challenges for biosensors and nanoscale analytical tools for pandemics: COVID-19. ACS Nano 14 (7), 7783–7807. doi:10.1021/acsnano.0c04421
Bhattacharya, A., Brea, R. J., Niederholtmeyer, H., and Devaraj, N. K. (2019). A minimal biochemical route towards de novo formation of synthetic phospholipid membranes. Nat. Commun. 10 (1), 300. doi:10.1038/s41467-018-08174-x
Biral, A., and Zanella, A. (2013). Introducing purely hydrodynamic networking functionalities into microfluidic systems. Nano Commun. Netw. 4 (4), 205–215. doi:10.1016/j.nancom.2013.09.001
Bissell, M. J., and Barcellos-Hoff, M. H. (1987). The influence of extracellular matrix on gene expression: is structure the message? J. Cell Sci. 1987 (8), 327–343. doi:10.1242/jcs.1987.supplement_8.18
Blackburn, M. C., Petrova, E., Correia, B. E., and Maerkl, S. J. (2016). Integrating gene synthesis and microfluidic protein analysis for rapid protein engineering. Nucleic Acids Res. 44 (7), e68. doi:10.1093/nar/gkv1497
Blanken, D., Foschepoth, D., Serrão, A. C., and Danelon, C. (2020). Genetically controlled membrane synthesis in liposomes. Nat. Commun. 11 (1), 4317. doi:10.1038/s41467-020-17863-5
Bolognesi, G., Friddin, M. S., Salehi-Reyhani, A., Barlow, N. E., Brooks, N. J., Ces, O., et al. (2018). Sculpting and fusing biomimetic vesicle networks using optical tweezers. Nat. Commun. 9 (1), 1882. doi:10.1038/s41467-018-04282-w
Booth, M. J., Restrepo Schild, V., Graham, A. D., Olof, S. N., and Bayley, H. (2016). Light-activated communication in synthetic tissues. Sci. Adv. 2 (4), e1600056. doi:10.1126/sciadv.1600056
Bringer, M. R., Gerdts, C. J., Song, H., Tice, J. D., and Ismagilov, R. F. (2004). Microfluidic systems for chemical kinetics that rely on chaotic mixing in droplets. Philos. Trans. A Math. Phys. Eng. Sci. 362 (1818), 1087–1104. doi:10.1098/rsta.2003.1364
Brody, J. P., Yager, P., Goldstein, R. E., and Austin, R. H. (1996). Biotechnology at low Reynolds numbers. Biophysical J. 71 (6), 3430–3441. doi:10.1016/S0006-3495(96)79538-3
Brower, K., Puccinelli, R. R., Markin, C. J., Tyler, C. S., Longwell, S. A., Cruz, B., et al. (2018). An open-source, programmable pneumatic setup for operation and automated control of single-and multi-layer microfluidic devices. HardwareX 3, 117–134. doi:10.1016/j.ohx.2017.10.001
Bruggeman, F. J., and Westerhoff, H. V. (2007). The nature of systems biology. Trends Microbiol. 15 (1), 45–50. doi:10.1016/j.tim.2006.11.003
Buddingh, B. C., Elzinga, J., and van Hest, J. C. M. (2020). Intercellular communication between artificial cells by allosteric amplification of a molecular signal. Nat. Commun. 11 (1), 1652. doi:10.1038/s41467-020-15482-8
Busardo, F. P., and Jones, A. W. (2015). GHB pharmacology and toxicology: acute intoxication, concentrations in blood and urine in forensic cases and treatment of the withdrawal syndrome. Curr. Neuropharmacol. 13 (1), 47–70. doi:10.2174/1570159X13666141210215423
Buxboim, A., Bar-Dagan, M., Frydman, V., Zbaida, D., Morpurgo, M., and Bar-Ziv, R. (2007). A single-step photolithographic interface for cell-free gene expression and active biochips. Small 3 (3), 500–510. doi:10.1002/smll.200600489
Cameron, D. E., Bashor, C. J., and Collins, J. J. (2014). A brief history of synthetic biology. Nat. Rev. Microbiol. 12 (5), 381–390. doi:10.1038/nrmicro3239
Cardinale, S., and Arkin, A. P. (2012). Contextualizing context for synthetic biology–identifying causes of failure of synthetic biological systems. Biotechnol. J. 7 (7), 856–866. doi:10.1002/biot.201200085
Carrell, C., Kava, A., Nguyen, M., Menger, R., Munshi, Z., Call, Z., et al. (2019). Beyond the lateral flow assay: a review of paper-based microfluidics. Microelectron. Eng. 206, 45–54. doi:10.1016/j.mee.2018.12.002
Carugo, D., Bottaro, E., Owen, J., Stride, E., and Nastruzzi, C. (2016). Liposome production by microfluidics: potential and limiting factors. Sci. Rep. 6 (1), 25876–25915. doi:10.1038/srep25876
Caschera, F., Sunami, T., Matsuura, T., Suzuki, H., Hanczyc, M. M., and Yomo, T. (2011). Programmed vesicle fusion triggers gene expression. Langmuir 27 (21), 13082–13090. doi:10.1021/la202648h
Cate, D. M., Adkins, J. A., Mettakoonpitak, J., and Henry, C. S. (2015). Recent developments in paper-based microfluidic devices. Anal. Chem. 87 (1), 19–41. doi:10.1021/ac503968p
Chang, J.-C., Swank, Z., Keiser, O., Maerkl, S. J., and Amstad, E. (2018). Microfluidic device for real-time formulation of reagents and their subsequent encapsulation into double emulsions. Sci. Rep. 8 (1), 8143. doi:10.1038/s41598-018-26542-x
Cho, E., and Lu, Y. (2020). Compartmentalizing cell-free systems: toward creating life-like artificial cells and beyond. ACS Synth. Biol. 9 (11), 2881–2901. doi:10.1021/acssynbio.0c00433
Cho, S. K., Moon, H., and Kim, C.-J. (2003). Creating, transporting, cutting, and merging liquid droplets by electrowetting-based actuation for digital microfluidic circuits. J. Microelectromechanical Syst. 12 (1), 70–80. doi:10.1109/jmems.2002.807467
Choi, K., Ng, A. H. C., Fobel, R., and Wheeler, A. R. (2012). Digital microfluidics. Annu. Rev. Anal. Chem. 5, 413–440. doi:10.1146/annurev-anchem-062011-143028
Chuen-Ru, L., and Lutz-Bueno, V. (2023). Charge-selectively permeable self-healing microcapsules. Adv. Mater. Interfaces 10 (35), 2300531. doi:10.1002/admi.202300531
Church, G. M., Elowitz, M. B., Smolke, C. D., Voigt, C. A., and Weiss, R. (2014). Realizing the potential of synthetic biology. Nat. Rev. Mol. Cell Biol. 15 (4), 289–294. doi:10.1038/nrm3767
Convery, N., and Gadegaard, N. (2019). 30 years of microfluidics. Micro Nano Eng. 2, 76–91. doi:10.1016/j.mne.2019.01.003
Cookson, N. A., Cookson, S. W., Tsimring, L. S., and Hasty, J. (2010). Cell cycle-dependent variations in protein concentration. Nucleic Acids Res. 38 (8), 2676–2681. doi:10.1093/nar/gkp1069
Courtois, F., Olguin, L. F., Whyte, G., Bratton, D., Huck, W. T. S., Abell, C., et al. (2008). An integrated device for monitoring time-dependent in vitro expression from single genes in picolitre droplets. ChemBioChem 9 (3), 439–446. doi:10.1002/cbic.200700536
Cui, J., Wu, D., Sun, Q., Yang, X., Wang, D., Zhuang, M., et al. (2020). A PEGDA/DNA hybrid hydrogel for cell-free protein synthesis. Front. Chem. 8, 28. doi:10.3389/fchem.2020.00028
Curtis, D. C., Vincent, L., and Sia, S. K. (2012). Commercialization of microfluidic point-of-care diagnostic devices. Lab a Chip 12 (12), 2118–2134. doi:10.1039/c2lc21204h
Cybulski, O., Garstecki, P., and Grzybowski, B. A. (2019). Oscillating droplet trains in microfluidic networks and their suppression in blood flow. Nat. Phys. 15 (7), 706–713. doi:10.1038/s41567-019-0486-8
Czekalska, M. A., Kaminski, T. S., Jakiela, S., Sapra, K. T., Bayley, H., and Garstecki, P. (2015). A droplet microfluidic system for sequential generation of lipid bilayers and transmembrane electrical recordings. Lab a Chip 15 (2), 541–548. doi:10.1039/c4lc00985a
Damiati, S., Mhanna, R., Kodzius, R., and Ehmoser, E.-K. (2018). Cell-free approaches in synthetic biology utilizing microfluidics. Genes 9 (3), 144. doi:10.3390/genes9030144
Debon, A. P., Wootton, R. C. R., and Elvira, K. S. (2015). Droplet confinement and leakage: causes, underlying effects, and amelioration strategies. Biomicrofluidics 9 (2), 024119. doi:10.1063/1.4917343
De Franceschi, N., Barth, R., Meindlhumer, S., Fragasso, A., and Dekker, C. (2024). Dynamin A as a one-component division machinery for synthetic cells. Nat. Nanotechnol. 19 (1), 70–76. doi:10.1038/s41565-023-01510-3
Deiman, B., van Aarle, P., and Sillekens, P. (2002). Characteristics and applications of nucleic acid sequence-based amplification (NASBA). Mol. Biotechnol. 20 (2), 163–179. doi:10.1385/MB:20:2:163
de Maddalena, L. L., Niederholtmeyer, H., Turtola, M., Swank, Z. N., Belogurov, G. A., and Maerkl, S. J. (2016). Grea and GreB enhance expression of Escherichia coli RNA polymerase promoters in a reconstituted transcription–translation system. ACS Synth. Biol. 5 (9), 929–935. doi:10.1021/acssynbio.6b00017
Deshpande, S., Caspi, Y., Meijering, A. E. C., and Dekker, C. (2016). Octanol-assisted liposome assembly on chip. Nat. Commun. 7 (1), 10447. doi:10.1038/ncomms10447
Deshpande, S., Spoelstra, W. K., Van Doorn, M., Jacob, K., and Dekker, C. (2018). Mechanical division of cell-sized liposomes. ACS Nano 12 (3), 2560–2568. doi:10.1021/acsnano.7b08411
Diederichs, T., Pugh, G., Dorey, A., Xing, Y., Burns, J. R., Nguyen, Q. H., et al. (2019). Synthetic protein-conductive membrane nanopores built with DNA. Nat. Commun. 10 (1), 5018. doi:10.1038/s41467-019-12639-y
Dittrich, P. S., Tachikawa, K., and Manz, A. (2006). Micro total analysis systems. latest advancements and trends. Anal. Chem. 78 (12), 3887–3908. doi:10.1021/ac0605602
Dodevski, I., George, C. M., and Sarkar, C. A. (2015). Conceptual and methodological advances in cell-free directed evolution. Curr. Opin. Struct. Biol. 33, 1–7. doi:10.1016/j.sbi.2015.04.008
Dudley, Q. M., Karim, A. S., and Jewett, M. C. (2015). Cell-free metabolic engineering: biomanufacturing beyond the cell. Biotechnol. J. 10 (1), 69–82. doi:10.1002/biot.201400330
Efrat, Y., Tayar, A. M., Daube, S. S., Levy, M., and Bar-Ziv, R. H. (2018). Electric-field manipulation of a compartmentalized cell-free gene expression reaction. ACS Synth. Biol. 7 (8), 1829–1833. doi:10.1021/acssynbio.8b00160
Elani, Y., Demello, A. J., Niu, X., and Ces, O. (2012). Novel technologies for the formation of 2-D and 3-D droplet interface bilayer networks. Lab a Chip 12 (18), 3514–3520. doi:10.1039/c2lc40287d
Elowitz, M. B., and Leibler, S. (2000). A synthetic oscillatory network of transcriptional regulators. Nature 403 (6767), 335–338. doi:10.1038/35002125
Eric Hodgman, C., and Jewett, M. C. (2012). Cell-free synthetic biology: thinking outside the cell. Metab. Eng. 14 (3), 261–269. doi:10.1016/j.ymben.2011.09.002
Etienne, G., Vian, A., Biočanin, M., Deplancke, B., and Amstad, E. (2018). Cross-talk between emulsion drops: how are hydrophilic reagents transported across oil phases? Lab a Chip 18 (24), 3903–3912. doi:10.1039/c8lc01000e
Fallah-Araghi, A., Baret, J.-C., Ryckelynck, M., and Griffiths, A. D. (2012). A completely in vitro ultrahigh-throughput droplet-based microfluidic screening system for protein engineering and directed evolution. Lab a Chip 12 (5), 882–891. doi:10.1039/c2lc21035e
Focke, P. J., Hein, C., Hoffmann, B., Matulef, K., Bernhard, F., Dotsch, V., et al. (2016). Combining in vitro folding with cell free protein synthesis for membrane protein expression. Biochemistry 55 (30), 4212–4219. doi:10.1021/acs.biochem.6b00488
Fowler, J., Moon, H., and Kim, C.-J. (2002). “Enhancement of mixing by droplet-based microfluidics,” in Technical digest. MEMS 2002 IEEE international conference. Fifteenth IEEE international conference on micro electro mechanical systems (cat. No. 02CH37266) (IEEE), 97–100.
Friddin, M. S., Bolognesi, G., Elani, Y., Brooks, N. J., Law, R. V., Seddon, J. M., et al. (2016). Optically assembled droplet interface bilayer (OptiDIB) networks from cell-sized microdroplets. Soft Matter 12 (37), 7731–7734. doi:10.1039/c6sm01357k
Friddin, M. S., Morgan, H., and de Planque, M. R. R. (2013). Cell-free protein expression systems in microdroplets: stabilization of interdroplet bilayers. Biomicrofluidics 7 (1), 014108. doi:10.1063/1.4791651
Fu, E., and Downs, C. (2017). Progress in the development and integration of fluid flow control tools in paper microfluidics. Lab a Chip 17 (4), 614–628. doi:10.1039/c6lc01451h
Funakoshi, K., Suzuki, H., and Takeuchi, S. (2006). Lipid bilayer formation by contacting monolayers in a microfluidic device for membrane protein analysis. Anal. Chem. 78 (24), 8169–8174. doi:10.1021/ac0613479
Gach, P. C., Iwai, K., Kim, P. W., Hillson, N. J., and Singh, A. K. (2017). Droplet microfluidics for synthetic biology. Lab a Chip 17 (20), 3388–3400. doi:10.1039/c7lc00576h
Gan, R., Cabezas, M. D., Pan, M., Zhang, H., Hu, G., Clark, L. G., et al. (2022). High-throughput regulatory part prototyping and analysis by cell-free protein synthesis and droplet microfluidics. ACS Synth. Biol. 11 (6), 2108–2120. doi:10.1021/acssynbio.2c00050
Gao, W., and Chen, Y. (2019). Microencapsulation of solid cores to prepare double emulsion droplets by microfluidics. Int. J. Heat Mass Transf. 135, 158–163. doi:10.1016/j.ijheatmasstransfer.2019.01.136
Garamella, J., Marshall, R., Rustad, M., and Vincent, N. (2016). The all E. coli TX-TL toolbox 2.0: a platform for cell-free synthetic biology. ACS Synth. Biol. 5 (4), 344–355. doi:10.1021/acssynbio.5b00296
Gardner, T. S., Cantor, C. R., and Collins, J. J. (2000). Construction of a genetic toggle switch in Escherichia coli. Nature 403 (6767), 339–342. doi:10.1038/35002131
Garenne, D., Haines, M. C., Romantseva, E. F., Paul, F., Strychalski, E. A., and Vincent, N. (2021). Cell-free gene expression. Nat. Rev. Methods Prim. 1 (1), 49. doi:10.1038/s43586-021-00046-x
Garstecki, P., Fuerstman, M. J., Stone, H. A., and Whitesides, G. M. (2006). Formation of droplets and bubbles in a microfluidic T-junction—scaling and mechanism of break-up. Lab a Chip 6 (3), 437–446. doi:10.1039/b510841a
Georgi, V., Georgi, L., Blechert, M., Bergmeister, M., Zwanzig, M., Wüstenhagen, D. A., et al. (2016). On-chip automation of cell-free protein synthesis: new opportunities due to a novel reaction mode. Lab a Chip 16 (2), 269–281. doi:10.1039/c5lc00700c
Gervais, L., and Delamarche, E. (2009). Toward one-step point-of-care immunodiagnostics using capillary-driven microfluidics and PDMS substrates. Lab a Chip 9 (23), 3330–3337. doi:10.1039/b906523g
Go, J. S., and Shoji, S. (2004). A disposable, dead volume-free and leak-free in-plane PDMS microvalve. Sensors Actuators A Phys. 114 (2-3), 438–444. doi:10.1016/j.sna.2003.12.028
Gong, M. M., and Sinton, D. (2017). Turning the page: advancing paper-based microfluidics for broad diagnostic application. Chem. Rev. 117 (12), 8447–8480. doi:10.1021/acs.chemrev.7b00024
Gong, Y., Ma, M., Luo, Y., and Bong, D. (2008). Functional determinants of a synthetic vesicle fusion system. J. Am. Chem. Soc. 130 (19), 6196–6205. doi:10.1021/ja711184u
Gonzales, D. T., Yandrapalli, N., Robinson, T., Zechner, C., and Tang, T. Y. D. (2022). Cell-free gene expression dynamics in synthetic cell populations. ACS Synth. Biol. 11 (1), 205–215. doi:10.1021/acssynbio.1c00376
Göpfrich, K., Platzman, I., and Spatz, J. P. (2018). Mastering complexity: towards bottom-up construction of multifunctional eukaryotic synthetic cells. Trends Biotechnol. 36 (9), 938–951. doi:10.1016/j.tibtech.2018.03.008
Grasemann, L., Thiel Pizarro, P., and Maerkl, S. J. (2023). C2CAplus: a one-pot isothermal circle-to-circle DNA amplification system. ACS Synth. Biol. 12 (10), 3137–3142. doi:10.1021/acssynbio.3c00390
Gravesen, P., Branebjerg, J., and Jensen, O. S. (1993). Microfluidics - a review. J. Micromechanics Microengineering 3 (4), 168–182. doi:10.1088/0960-1317/3/4/002
Gräwe, A., Dreyer, A., Vornholt, T., Barteczko, U., Buchholz, L., Drews, G., et al. (2019). A paper-based, cell-free biosensor system for the detection of heavy metals and date rape drugs. PLOS ONE 14 (3), e0210940. doi:10.1371/journal.pone.0210940
Green, J., Holdø, A., and Khan, A. (2007). A review of passive and active mixing systems in microfluidic devices. Int. J. Multiphysics 1 (1), 1–32. doi:10.1260/175095407780130544
Gregorio, N. E., Levine, M. Z., and Oza, J. P. (2019). A user’s guide to cell-free protein synthesis. Methods Protoc. 2 (1), 24. doi:10.3390/mps2010024
Grover, W. H., Skelley, A. M., Liu, C. N., Lagally, E. T., and Mathies, R. A. (2003). Monolithic membrane valves and diaphragm pumps for practical large-scale integration into glass microfluidic devices. Sensors Actuators B Chem. 89 (3), 315–323. doi:10.1016/s0925-4005(02)00468-9
Guido, N. J., Wang, X., Adalsteinsson, D., McMillen, D., Hasty, J., Cantor, C. R., et al. (2006). A bottom-up approach to gene regulation. Nature 439 (7078), 856–860. doi:10.1038/nature04473
Gulati, S., Vincent, R., Niu, X., Chappell, J., Kitney, R. I., Edel, J. B., et al. (2009). Opportunities for microfluidic technologies in synthetic biology. J. R. Soc. Interface 6, S493–S506. doi:10.1098/rsif.2009.0083.focus
Haller, B., Göpfrich, K., Schröter, M., Janiesch, J.-W., Platzman, I., and Spatz, J. P. (2018). Charge-controlled microfluidic formation of lipid-based single-and multicompartment systems. Lab a Chip 18 (17), 2665–2674. doi:10.1039/c8lc00582f
Hansen, C. L., Sommer, M. O. A., and Quake, S. R. (2004). Systematic investigation of protein phase behavior with a microfluidic formulator. Proc. Natl. Acad. Sci. 101 (40), 14431–14436. doi:10.1073/pnas.0405847101
Hansen, M. M. K., Meijer, L. H. H., Spruijt, E., Maas, R. J. M., Ventosa Rosquelles, M., Groen, J., et al. (2016). Macromolecular crowding creates heterogeneous environments of gene expression in picolitre droplets. Nat. Nanotechnol. 11 (2), 191–197. doi:10.1038/nnano.2015.243
Has and P Sunthar, C. (2020). A comprehensive review on recent preparation techniques of liposomes. J. Liposome Res. 30 (4), 336–365. doi:10.1080/08982104.2019.1668010
Hasegawa, T., Hasegawa, T., Ikuta, K. I. K., Nakashima, K. N. K., and Hasegawa, T. (2003). “10-way micro switching valve chip with rotary mechanism for multi-directional flow control,” in The seventh international symposium on micro total analysis system, 215–218.
Haseltine, E. L., and Arnold, F. H. (2007). Synthetic gene circuits: design with directed evolution. Annu. Rev. Biophys. Biomol. Struct. 36, 1–19. doi:10.1146/annurev.biophys.36.040306.132600
Hassan, S.-ul, and Zhang, X. (2020). Design and fabrication of capillary-driven flow device for point-of-care diagnostics. Biosensors 10 (4), 39. doi:10.3390/bios10040039
Hasty, J., Isaacs, F., Dolnik, M., McMillen, D., and Collins, J. J. (2001b). Designer gene networks: towards fundamental cellular control. Chaos Interdiscip. J. Nonlinear Sci. 11 (1), 207–220. doi:10.1063/1.1345702
Hasty, J., McMillen, D., and Collins, J. J. (2002). Engineered gene circuits. Nature 420 (6912), 224–230. doi:10.1038/nature01257
Hasty, J., McMillen, D., Isaacs, F., and Collins, J. J. (2001a). Computational studies of gene regulatory networks: in numero molecular biology. Nat. Rev. Genet. 2 (4), 268–279. doi:10.1038/35066056
Heinemann, M., and Panke, S. (2006). Synthetic biology — putting engineering into biology. Bioinformatics 22 (22), 2790–2799. doi:10.1093/bioinformatics/btl469
Hemmig, E., Temiz, Y., Gökçe, O., Lovchik, R. D., and Delamarche, E. (2019). Transposing lateral flow immunoassays to capillary-driven microfluidics using self-coalescence modules and capillary-assembled receptor carriers. Anal. Chem. 92 (1), 940–946. doi:10.1021/acs.analchem.9b03792
Hibi, K., Amikura, K., Sugiura, N., Masuda, K., Ohno, S., Yokogawa, T., et al. (2020). Reconstituted cell-free protein synthesis using in vitro transcribed tRNAs. Commun. Biol. 3 (1), 350. doi:10.1038/s42003-020-1074-2
Higashikuni, Y., Chen, W. C. W., and Lu, T. K. (2017). Advancing therapeutic applications of synthetic gene circuits. Curr. Opin. Biotechnol. 47, 133–141. doi:10.1016/j.copbio.2017.06.011
Hoang, D. A., Portela, L. M., Kleijn, C. R., Kreutzer, M. T., and Steijn, V. V. (2013). Dynamics of droplet breakup in a T-junction. J. Fluid Mech. 717, R4. doi:10.1017/jfm.2013.18
Holden, M. A., Needham, D., and Bayley, H. (2007). Functional bionetworks from nanoliter water droplets. J. Am. Chem. Soc. 129 (27), 8650–8655. doi:10.1021/ja072292a
Holstein, J. M., Gylstorff, C., and Hollfelder, F. (2021). Cell-free directed evolution of a protease in microdroplets at ultrahigh throughput. ACS Synth. Biol. 10 (2), 252–257. doi:10.1021/acssynbio.0c00538
Hori, Y., Kantak, C., Murray, R. M., and Abate, A. R. (2017). Cell-free extract based optimization of biomolecular circuits with droplet microfluidics. Lab a Chip 17 (18), 3037–3042. doi:10.1039/c7lc00552k
Hosokawa, K., and Maeda, R. (2000). A pneumatically-actuated three-way microvalve fabricated with polydimethylsiloxane using the membrane transfer technique. J. Micromechanics Microengineering 10 (3), 415–420. doi:10.1088/0960-1317/10/3/317
Howard, A. S., Stroock, A. D., and Ajdari, A. (2004). Engineering flows in small devices: microfluidics toward a lab-on-a-chip. Annu. Rev. Fluid Mech. 36, 381–411. doi:10.1146/annurev.fluid.36.050802.122124
Hua, Li, and Steckl, A. J. (2018). Paper microfluidics for point-of-care blood-based analysis and diagnostics. Anal. Chem. 91 (1), 352–371. doi:10.1021/acs.analchem.8b03636
Huang, C., Zhang, H., Han, S.-I., and Han, A. (2021). Cell washing and solution exchange in droplet microfluidic systems. Anal. Chem. 93 (24), 8622–8630. doi:10.1021/acs.analchem.1c01558
Huebner, A., Bratton, D., Whyte, G., Yang, M., Demello, A. J., Abell, C., et al. (2009). Static microdroplet arrays: a microfluidic device for droplet trapping, incubation and release for enzymatic and cell-based assays. Lab a Chip 9 (5), 692–698. doi:10.1039/b813709a
Jaenicke, L. (2007). Centenary of the award of a Nobel Prize to Eduard Buchner, the father of biochemistry in a test tube and thus of experimental molecular bioscience. Angew. Chem. Int. Ed. 46 (36), 6776–6782. doi:10.1002/anie.200700390
Jebrail, M. J., Bartsch, M. S., and Patel, K. D. (2012). Digital microfluidics: a versatile tool for applications in chemistry, biology and medicine. Lab a Chip 12 (14), 2452–2463. doi:10.1039/c2lc40318h
Jeffrey, C. W., Collins, J. J., Keasling, J. D., and Silver, P. A. (2014). Integrating biological redesign: where synthetic biology came from and where it needs to go. Cell 157 (1), 151–161. doi:10.1016/j.cell.2014.02.039
Jia, H., Heymann, M., Bernhard, F., Schwille, P., and Kai, L. (2017). Cell-free protein synthesis in micro compartments: building a minimal cell from biobricks. New Biotechnol. 39, 199–205. doi:10.1016/j.nbt.2017.06.014
Jiang, L., Zhao, J., Lian, J., and Xu, Z. (2018). Cell-free protein synthesis enabled rapid prototyping for metabolic engineering and synthetic biology. Synthetic Syst. Biotechnol. 3 (2), 90–96. doi:10.1016/j.synbio.2018.02.003
Jin, C., Krüger, C., and Maass, C. C. (2017). Chemotaxis and autochemotaxis of self-propelling droplet swimmers. Proc. Natl. Acad. Sci. 114 (20), 5089–5094. doi:10.1073/pnas.1619783114
Kalmbach, R., Chizhov, I., Schumacher, M. C., Friedrich, T., Bamberg, E., and Engelhard, M. (2007). Functional cell-free synthesis of a seven helix membrane protein: in situ insertion of bacteriorhodopsin into liposomes. J. Mol. Biol. 371 (3), 639–648. doi:10.1016/j.jmb.2007.05.087
Karig, D. K., Jung, S.-Y., Srijanto, B., Collier, C. P., and Simpson, M. L. (2013). Probing cell-free gene expression noise in femtoliter volumes. ACS Synth. Biol. 2 (9), 497–505. doi:10.1021/sb400028c
Karimi, A., Karig, D., Kumar, A., and Ardekani, A. M. (2015). Interplay of physical mechanisms and biofilm processes: review of microfluidic methods. Lab a Chip 15 (1), 23–42. doi:10.1039/c4lc01095g
Karlikow, M., Guo, Y., Cicek, S., Krokovsky, L., Homme, P., Xiong, Y., et al. (2022). Field validation of the performance of paper-based tests for the detection of the Zika and chikungunya viruses in serum samples. Nat. Biomed. Eng. 6 (3), 246–256. doi:10.1038/s41551-022-00850-0
Karzbrun, E., Tayar, A. M., Vincent, N., and Bar-Ziv, R. H. (2014). Synthetic biology. Programmable on-chip DNA compartments as artificial cells. Science 345 (6198), 829–832. doi:10.1126/science.1255550
Kasetsirikul, S., Shiddiky, M. J. A., and Nguyen, N.-T. (2020). Challenges and perspectives in the development of paper-based lateral flow assays. Microfluid. Nanofluidics 24, 17–18. doi:10.1007/s10404-020-2321-z
Katzen, F., Chang, G., and Kudlicki, W. (2005). The past, present and future of cell-free protein synthesis. Trends Biotechnol. 23 (3), 150–156. doi:10.1016/j.tibtech.2005.01.003
Keller, L., and Surette, M. G. (2006). Communication in bacteria: an ecological and evolutionary perspective. Nat. Rev. Microbiol. 4 (4), 249–258. doi:10.1038/nrmicro1383
Khalil, A. S., and Collins, J. J. (2010). Synthetic biology: applications come of age. Nat. Rev. Genet. 11 (5), 367–379. doi:10.1038/nrg2775
Kim, B., Jeon, T. Y., Oh, Y.-K., and Kim, S.-H. (2015). Microfluidic production of semipermeable microcapsules by polymerization-induced phase separation. Langmuir 31 (22), 6027–6034. doi:10.1021/acs.langmuir.5b01129
Kim, T. H., Hahn, Y.Ki, and Kim, M. S. (2020). Recent advances of fluid manipulation technologies in microfluidic paper-based analytical devices (μPADs) toward multi-step assays. Micromachines 11 (3), 269. doi:10.3390/mi11030269
Kim, T.-W., Keum, J.-W., Oh, S., Choi, C.-Y., Park, C.-G., and Kim, D.-M. (2006). Simple procedures for the construction of a robust and cost-effective cell-free protein synthesis system. J. Biotechnol. 126 (4), 554–561. doi:10.1016/j.jbiotec.2006.05.014
Knight, J. B., Vishwanath, A., Brody, J. P., and Austin, R. H. (1998). Hydrodynamic focusing on a silicon chip: mixing nanoliters in microseconds. Phys. Rev. Lett. 80 (17), 3863–3866. doi:10.1103/physrevlett.80.3863
Koch, M., Faulon, J.-L., and Borkowski, O. (2018). Models for cell-free synthetic biology: make prototyping easier, better, and faster. Front. Bioeng. Biotechnol. 6, 182. doi:10.3389/fbioe.2018.00182
Kothamachu, V. B., Zaini, S., and Muffatto, F. (2020). Role of digital microfluidics in enabling access to laboratory automation and making biology programmable. SLAS Technol. 25 (5), 411–426. doi:10.1177/2472630320931794
Kumar Dondapati, S., Stech, M., Zemella, A., and Kubick, S. (2020). Cell-free protein synthesis: a promising option for future drug development. BioDrugs 34 (3), 327–348. doi:10.1007/s40259-020-00417-y
Kung, C.-T., Hou, C.-Y., Wang, Y.-N., and Fu, L.-M. (2019). Microfluidic paper-based analytical devices for environmental analysis of soil, air, ecology and river water. Sensors Actuators B Chem. 301, 126855. doi:10.1016/j.snb.2019.126855
Laohakunakorn, N., Grasemann, L., Lavickova, B., Michielin, G., Shahein, A., Swank, Z., et al. (2020). Bottom-up construction of complex biomolecular systems with cell-free synthetic biology. Front. Bioeng. Biotechnol. 8, 213. doi:10.3389/fbioe.2020.00213
Laohakunakorn, N., Lavickova, B., Swank, Z., Laurent, J., and Maerkl, S. J. (2021). Steady-state cell-free gene expression with microfluidic chemostats. Synthetic Gene Circuits Methods Protoc. 2229, 189–203. doi:10.1007/978-1-0716-1032-9_9
Lavickova, B., Grasemann, L., and Maerkl, S. J. (2022). Improved cell-free transcription–translation reactions in microfluidic chemostats augmented with hydrogel membranes for continuous small molecule dialysis. ACS Synth. Biol. 11 (12), 4134–4141. doi:10.1021/acssynbio.2c00453
Lavickova, B., Laohakunakorn, N., and Maerkl, S. J. (2020). A partially self-regenerating synthetic cell. Nat. Commun. 11 (1), 6340. doi:10.1038/s41467-020-20180-6
Lavickova, B., and Maerkl, S. J. (2019). A simple, robust, and low-cost method to produce the PURE cell-free system. ACS Synth. Biol. 8 (2), 455–462. doi:10.1021/acssynbio.8b00427
Lee, I.-L., Li, P.-S., Yu, W.-L., and Shen, H.-H. (2011). Prokaryotic expression, refolding, and purification of functional human vascular endothelial growth factor isoform 165: purification procedures and refolding conditions revisited. Protein Expr. Purif. 76 (1), 54–58. doi:10.1016/j.pep.2010.08.014
Lee, J., Moon, H., Fowler, J., Schoellhammer, T., and Kim, C.-J. (2002). Electrowetting and electrowetting-on-dielectric for microscale liquid handling. Sensors Actuators A Phys. 95 (2-3), 259–268. doi:10.1016/s0924-4247(01)00734-8
Lee, K.-H., and Kim, D.-M. (2013). Applications of cell-free protein synthesis in synthetic biology: interfacing bio-machinery with synthetic environments. Biotechnol. J. 8 (11), 1292–1300. doi:10.1002/biot.201200385
Lee, T. Y., Choi, T. M., Shim, T. S., Frijns, R. A. M., and Kim, S.-H. (2016). Microfluidic production of multiple emulsions and functional microcapsules. Lab a Chip 16 (18), 3415–3440. doi:10.1039/c6lc00809g
Levy, M., Falkovich, R., Daube, S. S., and Bar-Ziv, R. H. (2020). Autonomous synthesis and assembly of a ribosomal subunit on a chip. Sci. Adv. 6 (16), eaaz6020. doi:10.1126/sciadv.aaz6020
Levy, M., Falkovich, R., Vonshak, O., Bracha, D., Tayar, A. M., Shimizu, Y., et al. (2021). Boundary-free ribosome compartmentalization by gene expression on a surface. ACS Synth. Biol. 10 (3), 609–619. doi:10.1021/acssynbio.0c00613
Li, L., Zhang, R., Chen, L., Tian, X., Li, T., Pu, B., et al. (2022). Permeability-engineered compartmentalization enables in vitro reconstitution of sustained synthetic biology systems. Adv. Sci. 9 (34), 2203652. doi:10.1002/advs.202203652
Li, S., Wang, X., Mu, W., and Han, X. (2019). Chemical signal communication between two protoorganelles in a lipid-based artificial cell. Anal. Chem. 91 (10), 6859–6864. doi:10.1021/acs.analchem.9b01128
Li, Xu, Ballerini, D. R., and Shen, W. (2012). A perspective on paper-based microfluidics: current status and future trends. Biomicrofluidics 6 (1), 011301–1130113. doi:10.1063/1.3687398
Lisowski, P., and Zarzycki, P. K. (2013). Microfluidic paper-based analytical devices (μPADs) and micro total analysis systems (μTAS): development, applications and future trends. Chromatographia 76 (19), 1201–1214. doi:10.1007/s10337-013-2413-y
Liu, D., Wang, Y., Li, X., Li, M., Wu, Q., Song, Y., et al. (2022). Integrated microfluidic devices for in vitro diagnostics at point of care. Aggregate 3 (5), e184. doi:10.1002/agt2.184
Liu, D., Yang, Z., Zhang, L., Wei, M., and Lu, Y. (2020). Cell-free biology using remote-controlled digital microfluidics for individual droplet control. RSC Adv. 10 (45), 26972–26981. doi:10.1039/d0ra04588h
Liu, Y., Zhan, L., Qin, Z., Sackrison, J., and Bischof, J. C. (2021). Ultrasensitive and highly specific lateral flow assays for point-of-care diagnosis. ACS Nano 15 (3), 3593–3611. doi:10.1021/acsnano.0c10035
Long, A. R., O’Brien, C. C., and Alder, N. N. (2012). The cell-free integration of a polytopic mitochondrial membrane protein into liposomes occurs cotranslationally and in a lipid-dependent manner. PLOS ONE 7, 463322–e46411. doi:10.1371/journal.pone.0046332
Lu, H.-W., Bottausci, F., Fowler, J. D., Bertozzi, A. L., Meinhart, C., and Kim, C. J. C. J. (2008). A study of EWOD-driven droplets by PIV investigation. Lab a Chip 8 (3), 456–461. doi:10.1039/b717141b
Luigi Luisi, P., Allegretti, M., Pereira de Souza, T., Steiniger, F., Fahr, A., and Stano, P. (2010). Spontaneous protein crowding in liposomes: a new vista for the origin of cellular metabolism. ChemBioChem 11 (14), 1989–1992. doi:10.1002/cbic.201000381
Luo, L., Manda, S., Park, Y., Demir, B., Sanchez, J., Anantram, M. P., et al. (2023). DNA nanopores as artificial membrane channels for bioprotonics. Nat. Commun. 14 (1), 5364. doi:10.1038/s41467-023-40870-1
Ma, D., Liang, D., Zhu, C., Fu, T., Ma, Y., Yuan, X., et al. (2021). The breakup dynamics and mechanism of viscous droplets in Y-shaped microchannels. Chem. Eng. Sci. 231, 116300. doi:10.1016/j.ces.2020.116300
Ma, D., Shen, L., Wu, K., Diehnelt, C. W., and Green, A. A. (2018). Low-cost detection of norovirus using paper-based cell-free systems and synbody-based viral enrichment. Synth. Biol. 3 (1), ysy018. doi:10.1093/synbio/ysy018
Maerkl, S. J. (2023). On biochemical constructors and synthetic cells. Interface Focus 13 (5), 20230014. doi:10.1098/rsfs.2023.0014
Maerkl, S. J., and Quake, S. R. (2007). A systems approach to measuring the binding energy landscapes of transcription factors. Science 315 (5809), 233–237. doi:10.1126/science.1131007
Magana, M., Pushpanathan, M., Santos, A. L., Leanse, L., Fernandez, M., Ioannidis, A., et al. (2020). The value of antimicrobial peptides in the age of resistance. Lancet Infect. Dis. 20 (9), e216–e230. doi:10.1016/S1473-3099(20)30327-3
Martinez, A. W., Phillips, S. T., Butte, M. J., and Whitesides, G. M. (2007). Patterned paper as a platform for inexpensive, low-volume, portable bioassays. Angew. Chem. 119 (8), 1340–1342. doi:10.1002/ange.200603817
Martino, C., and deMello, A. J. (2016). Droplet-based microfluidics for artificial cell generation: a brief review. Interface Focus 6 (4), 20160011. doi:10.1098/rsfs.2016.0011
Mazutis, L., Baret, J.-C., and Griffiths, A. D. (2009). A fast and efficient microfluidic system for highly selective one-to-one droplet fusion. Lab a Chip 9 (18), 2665–2672. doi:10.1039/b903608c
Medina, H., Werner, H., Grimmer, A., Wille, R., and Springer, A. (2019). Passive droplet control in microfluidic networks: a survey and new perspectives on their practical realization. Nano Commun. Netw. 19, 33–46. doi:10.1016/j.nancom.2018.10.002
Melissa, B. M., and Bassler, B. L. (2001). Quorum sensing in bacteria. Annu. Rev. Microbiol. 55 (1), 165–199. doi:10.1146/annurev.micro.55.1.165
Meng, F., and Ellis, T. (2020). The second decade of synthetic biology: 2010–2020. Nat. Commun. 11 (1), 5174. doi:10.1038/s41467-020-19092-2
Michielin, G., and Maerkl, S. J. (2022). Direct encapsulation of biomolecules in semi-permeable microcapsules produced with double-emulsions. Sci. Rep. 12 (1), 21391. doi:10.1038/s41598-022-25895-8
Miller, T. E., Beneyton, T., Schwander, T., Diehl, C., Girault, M., McLean, R., et al. (2020). Light-powered CO2 fixation in a chloroplast mimic with natural and synthetic parts. Science 368 (6491), 649–654. doi:10.1126/science.aaz6802
Miwa, A., Wakamori, M., Ariyoshi, T., Okada, Y., Shirouzu, M., Umehara, T., et al. (2024). Efficiency of transcription and translation of cell-free protein synthesis systems in cell-sized lipid vesicles with changing lipid composition determined by fluorescence measurements. Sci. Rep. 14 (1), 2852. doi:10.1038/s41598-024-53135-8
Morales-Narváez, E., and Dincer, C. (2020). The impact of biosensing in a pandemic outbreak: COVID-19. Biosens. Bioelectron. 163, 112274. doi:10.1016/j.bios.2020.112274
Morbioli, G. G., Mazzu-Nascimento, T., Stockton, A. M., and Carrilho, E. (2017). Technical aspects and challenges of colorimetric detection with microfluidic paper-based analytical devices (μPADs) - a review. Anal. Chim. Acta 970, 1–22. doi:10.1016/j.aca.2017.03.037
Morgado, G., Gerngross, D., Roberts, T. M., and Panke, S. (2018). Synthetic biology for cell-free biosynthesis: fundamentals of designing novel in vitro multi-enzyme reaction networks. Synth. Biol. – Metab. Eng. 162, 117–146. doi:10.1007/10_2016_13
Mytnyk, S., Ziemecka, I., Olive, A. G. L., van der Meer, J. W. M., Totlani, K. A., Sander, O., et al. (2017). Microcapsules with a permeable hydrogel shell and an aqueous core continuously produced in a 3D microdevice by all-aqueous microfluidics. RSC Adv. 7 (19), 11331–11337. doi:10.1039/c7ra00452d
Nandagopal, N., and Elowitz, M. B. (2011). Synthetic biology: integrated gene circuits. Science 333 (6047), 1244–1248. doi:10.1126/science.1207084
Needham, D. (2017). “Cohesion and permeability of lipid bilayer vesicles,” in Permeability and stability of lipid bilayers (CRC Press), 49–76.
Needham, D., and Nunn, R. S. (1990). Elastic deformation and failure of lipid bilayer membranes containing cholesterol. Biophysical J. 58 (4), 997–1009. doi:10.1016/S0006-3495(90)82444-9
Nelson, W. C., and Kim, C. J. (2012). Droplet actuation by electrowetting-on-dielectric (EWOD): a review. J. Adhesion Sci. Technol. 26 (12-17), 1747–1771. doi:10.1163/156856111x599562
Ni, B., Colin, R., Link, H., Endres, R. G., and Sourjik, V. (2020). Growth-rate dependent resource investment in bacterial motile behavior quantitatively follows potential benefit of chemotaxis. Proc. Natl. Acad. Sci. 117 (1), 595–601. doi:10.1073/pnas.1910849117
Niederholtmeyer, H., Chaggan, C., and Devaraj, N. K. (2018). Communication and quorum sensing in non-living mimics of eukaryotic cells. Nat. Commun. 9 (1), 5027. doi:10.1038/s41467-018-07473-7
Niederholtmeyer, H., Stepanova, V., and Maerkl, S. J. (2013). Implementation of cell-free biological networks at steady state. Proc. Natl. Acad. Sci. 110 (40), 15985–15990. doi:10.1073/pnas.1311166110
Niederholtmeyer, H., Sun, Z. Z., Hori, Y., Yeung, E., Verpoorte, A., Murray, R. M., et al. (2015). Rapid cell-free forward engineering of novel genetic ring oscillators. eLife 4, e09771. doi:10.7554/eLife.09771
Niu, X., Gielen, F., Edel, J. B., and Demello, A. J. (2011). A microdroplet dilutor for high-throughput screening. Nat. Chem. 3 (6), 437–442. doi:10.1038/nchem.1046
Noviana, E., Ozer, T., Carrell, C. S., Link, J. S., McMahon, C., Jang, I., et al. (2021). Microfluidic paper-based analytical devices: from design to applications. Chem. Rev. 121 (19), 11835–11885. doi:10.1021/acs.chemrev.0c01335
Novo, P., Chu, V., and Conde, J. P. (2014). Integrated optical detection of autonomous capillary microfluidic immunoassays: a hand-held point-of-care prototype. Biosens. Bioelectron. 57, 284–291. doi:10.1016/j.bios.2014.02.009
Nuclera (2023). Nuclera. Available at: https://www.nuclera.com/ (Accessed October 26, 2023).
Nuti, N., Rottmann, P., Stucki, A., Koch, P., Panke, S., and Dittrich, P. S. (2022). A multiplexed cell-free assay to screen for antimicrobial peptides in double emulsion droplets. Angew. Chem. Int. Ed. 61 (13), e202114632. doi:10.1002/anie.202114632
Oh, K. W., and Ahn, C. H. (2006). A review of microvalves. J. Micromechanics Microengineering 16 (5), R13–R39. doi:10.1088/0960-1317/16/5/r01
Okushima, S., Nisisako, T., Torii, T., and Higuchi, T. (2004). Controlled production of monodisperse double emulsions by two-step droplet breakup in microfluidic devices. Langmuir 20 (23), 9905–9908. doi:10.1021/la0480336
Olanrewaju, A., Beaugrand, M., Mohamed, Y., and Juncker, D. (2018). Capillary microfluidics in microchannels: from microfluidic networks to capillaric circuits. Lab a Chip 18 (16), 2323–2347. doi:10.1039/c8lc00458g
Oliver, J. M., Bernath, K., Agresti, J. J., Amitai, G., Kelly, B. T., Mastrobattista, E., et al. (2006). Directed evolution by in vitro compartmentalization. Nat. Methods 3 (7), 561–570. doi:10.1038/nmeth897
Pandey, C. M., Augustine, S., Kumar, S., Kumar, S., Nara, S., Srivastava, S., et al. (2018). Microfluidics based point-of-care diagnostics. Biotechnol. J. 13 (1), 1700047. doi:10.1002/biot.201700047
Pardee, K., Green, A. A., Ferrante, T., Cameron, D. E., DaleyKeyser, A., Yin, P., et al. (2014). Paper-based synthetic gene networks. Cell 159 (4), 940–954. doi:10.1016/j.cell.2014.10.004
Pardee, K., Green, A. A., Takahashi, M. K., Braff, D., Lambert, G., Lee, J. W., et al. (2016). Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell 165 (5), 1255–1266. doi:10.1016/j.cell.2016.04.059
Park, N., Ho Um, S., Funabashi, H., Xu, J., and Luo, D. (2009). A cell-free protein-producing gel. Nat. Mater. 8 (5), 432–437. doi:10.1038/nmat2419
Pereira De Souza, T., Bossa, G. V., Stano, P., Steiniger, F., May, S., Luisi, P. L., et al. (2017). Vesicle aggregates as a model for primitive cellular assemblies. Phys. Chem. Chem. Phys. 19 (30), 20082–20092. doi:10.1039/c7cp03751a
Perez, J. G., Stark, J. C., and Jewett, M. C. (2016). Cell-free synthetic biology: engineering beyond the cell. Cold Spring Harb. Perspect. Biol. 8 (12), a023853. doi:10.1101/cshperspect.a023853
Petra, S. D., Jahnz, M., and Schwille, P. (2005). A new embedded process for compartmentalized cell-free protein expression and on-line detection in microfluidic devices. ChemBioChem 6 (5), 811–814. doi:10.1002/cbic.200400321
Pietrini, A. V., and Luisi, P. L. (2004). Cell-free protein synthesis through solubilisate exchange in water/oil emulsion compartments. ChemBioChem 5 (8), 1055–1062. doi:10.1002/cbic.200400014
Pinheiro, T., Cardoso, A. R., Sousa, C. E. A., Marques, A. C., Tavares, A. P. M., Matos, A. M., et al. (2021). Paper-based biosensors for COVID-19: a review of innovative tools for controlling the pandemic. ACS Omega 6 (44), 29268–29290. doi:10.1021/acsomega.1c04012
Pir Cakmak, F., Grigas, A. T., and Keating, C. D. (2019). Lipid vesicle-coated complex coacervates. Langmuir 35 (24), 7830–7840. doi:10.1021/acs.langmuir.9b00213
Pollack, M. G., Pamula, V. K., Srinivasan, V., and Eckhardt, A. E. (2011). Applications of electrowetting-based digital microfluidics in clinical diagnostics. Expert Rev. Mol. Diagnostics 11 (4), 393–407. doi:10.1586/erm.11.22
Pollack, M. G., Shenderov, A. D., and Fair, R. B. (2002). Electrowetting-based actuation of droplets for integrated microfluidics. Lab a Chip 2 (2), 96–101. doi:10.1039/b110474h
Porter Hunt, J., Zhao, E. L., Free, T. J., Soltani, M., Warr, C. A., Benedict, A. B., et al. (2022). Towards detection of SARS-CoV-2 RNA in human saliva: a paper-based cell-free toehold switch biosensor with a visual bioluminescent output. New Biotechnol. 66, 53–60. doi:10.1016/j.nbt.2021.09.002
Purnick, P. E. M., and Weiss, R. (2009). The second wave of synthetic biology: from modules to systems. Nat. Rev. Mol. Cell Biol. 10 (6), 410–422. doi:10.1038/nrm2698
Qin, X., Liu, J., Zhang, Z., Li, J., Yuan, L., Zhang, Z., et al. (2021). Microfluidic paper-based chips in rapid detection: current status, challenges, and perspectives. TrAC Trends Anal. Chem. 143, 116371. doi:10.1016/j.trac.2021.116371
Reginald Beer, N., Hindson, B. J., Wheeler, E. K., Hall, S. B., Rose, K. A., Kennedy, I. M., et al. (2007). On-chip, real-time, single-copy polymerase chain reaction in picoliter droplets. Anal. Chem. 79 (22), 8471–8475. doi:10.1021/ac701809w
Reginald Beer, N., Wheeler, E. K., Lee-Houghton, L., Watkins, N., Nasarabadi, S., Hebert, N., et al. (2008). On-chip single-copy real-time reverse-transcription pcr in isolated picoliter droplets. Anal. Chem. 80 (6), 1854–1858. doi:10.1021/ac800048k
Rosenfeld, N., Young, J. W., Alon, U., Swain, P. S., and Elowitz, M. B. (2005). Gene regulation at the single-cell level. Science 307 (5717), 1962–1965. doi:10.1126/science.1106914
Rothe, A., Surjadi, R. N., and Power, B. E. (2006). Novel proteins in emulsions using in vitro compartmentalization. Trends Biotechnol. 24 (12), 587–592. doi:10.1016/j.tibtech.2006.10.007
Sackmann, E. K., Fulton, A. L., and Beebe, D. J. (2014). The present and future role of microfluidics in biomedical research. Nature 507 (7491), 181–189. doi:10.1038/nature13118
Safavieh, R., and Juncker, D. (2013). Capillarics: pre-programmed, self-powered microfluidic circuits built from capillary elements. Lab a Chip 13 (21), 4180–4189. doi:10.1039/c3lc50691f
Saint-Sardos, A., Sart, S., Lippera, K., Brient-Litzler, E., Michelin, S., Amselem, G., et al. (2020). High-throughput measurements of intra-cellular and secreted cytokine from single spheroids using anchored microfluidic droplets. Small 16 (49), 2002303. doi:10.1002/smll.202002303
Sajeesh, P., and Kumar Sen, A. (2014). Particle separation and sorting in microfluidic devices: a review. Microfluid. Nanofluidics 17, 1–52. doi:10.1007/s10404-013-1291-9
Sakatani, Y., Yomo, T., and Ichihashi, N. (2018). Self-replication of circular DNA by a self-encoded DNA polymerase through rolling-circle replication and recombination. Sci. Rep. 8 (1), 13089. doi:10.1038/s41598-018-31585-1
Samiei, E., Tabrizian, M., and Hoorfar, M. (2016). A review of digital microfluidics as portable platforms for lab-on a-chip applications. Lab a Chip 16 (13), 2376–2396. doi:10.1039/c6lc00387g
Saucedo-Espinosa, M. A., and Dittrich, P. S. (2020). In-droplet electrophoretic separation and enrichment of biomolecules. Anal. Chem. 92 (12), 8414–8421. doi:10.1021/acs.analchem.0c01044
Schindler, M., and Ajdari, A. (2008). Droplet traffic in microfluidic networks: a simple model for understanding and designing. Phys. Rev. Lett. 100 (4), 044501. doi:10.1103/PhysRevLett.100.044501
Schwille, P. (2011). Bottom-up synthetic biology: engineering in a tinkerer’s world. Science 333 (6047), 1252–1254. doi:10.1126/science.1211701
Scott, A., Noga, M. J., de Graaf, P., Westerlaken, I., Yildirim, E., and Danelon, C. (2016). Cell-free phospholipid biosynthesis by gene-encoded enzymes reconstituted in liposomes. PLOS ONE 11 (10), e0163058. doi:10.1371/journal.pone.0163058
Sedighi, M., Sieber, S., Rahimi, F., Shahbazi, M.-A., Rezayan, A. H., Huwyler, J., et al. (2019). Rapid optimization of liposome characteristics using a combined microfluidics and design-of-experiment approach. Drug Deliv. Transl. Res. 9, 404–413. doi:10.1007/s13346-018-0587-4
Sedlmayer, F., Aubel, D., and Fussenegger, M. (2018). Synthetic gene circuits for the detection, elimination and prevention of disease. Nat. Biomed. Eng. 2 (6), 399–415. doi:10.1038/s41551-018-0215-0
Seemann, R., Brinkmann, M., Pfohl, T., and Herminghaus, S. (2011). Droplet based microfluidics. Rep. Prog. Phys. 75 (1), 016601. doi:10.1088/0034-4885/75/1/016601
Sergio, L. S. F. (2016). Perspectives on digital microfluidics. Sensors Actuators A Phys. 250, 15–28. doi:10.1016/j.sna.2016.08.007
Shah, R. K., Cheung Shum, Ho, Rowat, A. C., Lee, D., Agresti, J. J., Utada, A. S., et al. (2008). Designer emulsions using microfluidics. Mater. Today 11 (4), 18–27. doi:10.1016/s1369-7021(08)70053-1
Shahrezaei, V., and Swain, P. S. (2008). The stochastic nature of biochemical networks. Curr. Opin. Biotechnol. 19 (4), 369–374. doi:10.1016/j.copbio.2008.06.011
Shang, L., Cheng, Y., and Zhao, Y. (2017). Emerging droplet microfluidics. Chem. Rev. 117 (12), 7964–8040. doi:10.1021/acs.chemrev.6b00848
Shankar, M., and Van Oudenaarden, A. (2009). Synthetic biology: understanding biological design from synthetic circuits. Nat. Rev. Genet. 10 (12), 859–871. doi:10.1038/nrg2697
Sharma, S., Zapatero-Rodríguez, J., Estrela, P., and Kennedy, R. O. (2015). Point-of-care diagnostics in low resource settings: present status and future role of microfluidics. Biosensors 5 (3), 577–601. doi:10.3390/bios5030577
Shen, H.-H., Fan, S.-K., Kim, C.-J., and Yao, D.-J. (2014). EWOD microfluidic systems for biomedical applications. Microfluid. Nanofluidics 16, 965–987. doi:10.1007/s10404-014-1386-y
Shim, J.-U., Cristobal, G., Link, D. R., Thorsen, T., Jia, Y., Piattelli, K., et al. (2007). Control and measurement of the phase behavior of aqueous solutions using microfluidics. J. Am. Chem. Soc. 129 (28), 8825–8835. doi:10.1021/ja071820f
Shimizu, Y., Inoue, A., Tomari, Y., Suzuki, T., Yokogawa, T., Nishikawa, K., et al. (2001). Cell-free translation reconstituted with purified components. Nat. Biotechnol. 19 (8), 751–755. doi:10.1038/90802
Silverman, A. D., Karim, A. S., and Jewett, M. C. (2020). Cell-free gene expression: an expanded repertoire of applications. Nat. Rev. Genet. 21 (3), 151–170. doi:10.1038/s41576-019-0186-3
Simmel, F. C., Yurke, B., and Singh, H. R. (2019). Principles and applications of nucleic acid strand displacement reactions. Chem. Rev. 119 (10), 6326–6369. doi:10.1021/acs.chemrev.8b00580
Singh, P., and Aubry, N. (2007). Transport and deformation of droplets in a microdevice using dielectrophoresis. Electrophoresis 28 (4), 644–657. doi:10.1002/elps.200600549
Siuti, P., Retterer, S. T., and Doktycz, M. J. (2011). Continuous protein production in nanoporous, picolitre volume containers. Lab a Chip 11 (20), 3523–3529. doi:10.1039/c1lc20462a
Sjöblom, J. (2006). Emulsions and emulsion stability, 45. NY, United States: Taylor & Francis New York.
Slomovic, S., Pardee, K., and Collins, J. J. (2015). Synthetic biology devices for in vitro and in vivo diagnostics. Proc. Natl. Acad. Sci. 112 (47), 14429–14435. doi:10.1073/pnas.1508521112
Slusarczyk, A. L., Lin, A., and Weiss, R. (2012). Foundations for the design and implementation of synthetic genetic circuits. Nat. Rev. Genet. 13 (6), 406–420. doi:10.1038/nrg3227
Smanski, M. J., Zhou, H., Claesen, J., Shen, B., Fischbach, M. A., and Voigt, C. A. (2016). Synthetic biology to access and expand nature’s chemical diversity. Nat. Rev. Microbiol. 14 (3), 135–149. doi:10.1038/nrmicro.2015.24
Smith, M. T., Wilding, K. M., Hunt, J. M., Bennett, A. M., and Bundy, B. C. (2014). The emerging age of cell-free synthetic biology. FEBS Lett. 588 (17), 2755–2761. doi:10.1016/j.febslet.2014.05.062
Soga, N., Ota, A., Nakajima, K., Watanabe, R., Ueno, H., and Noji, H. (2020). Monodisperse liposomes with femtoliter volume enable quantitative digital bioassays of membrane transporters and cell-free gene expression. ACS Nano 14 (9), 11700–11711. doi:10.1021/acsnano.0c04354
Sohrabi, S., Moraveji, M. K., and Keshavarz Moraveji, M. (2020). Droplet microfluidics: fundamentals and its advanced applications. RSC Adv. 10 (46), 27560–27574. doi:10.1039/d0ra04566g
Sokolova, E., Spruijt, E., Hansen, M. M. K., Dubuc, E., Groen, J., Chokkalingam, V., et al. (2013). Enhanced transcription rates in membrane-free protocells formed by coacervation of cell lysate. Proc. Natl. Acad. Sci. 110 (29), 11692–11697. doi:10.1073/pnas.1222321110
Soltani, M., Brady, R. D., Ford, H., Nelson, J. A. D., and Bundy, B. C. (2018). Reengineering cell-free protein synthesis as a biosensor: biosensing with transcription, translation, and protein-folding. Biochem. Eng. J. 138, 165–171. doi:10.1016/j.bej.2018.06.014
Soum, V., Park, S., Brilian, A. I., Kwon, O.-S., and Shin, K. (2019). Programmable paper-based microfluidic devices for biomarker detections. Micromachines 10 (8), 516. doi:10.3390/mi10080516
Srinivasan, V., Pamula, V. K., and Fair, R. B. (2004). An integrated digital microfluidic lab-on-a-chip for clinical diagnostics on human physiological fluids. Lab a Chip 4 (4), 310–315. doi:10.1039/b403341h
Stanley, C. E., Elvira, K. S., Niu, X. Z., Gee, A. D., Ces, O., Edel, J. B., et al. (2010). A microfluidic approach for high-throughput droplet interface bilayer (DIB) formation. Chem. Commun. 46 (10), 1620–1622. doi:10.1039/b924897h
Stapleton, J. A., and Swartz, J. R. (2010). Development of an in vitro compartmentalization screen for high-throughput directed evolution of [FeFe] hydrogenases. PLOS ONE 5 (12), e15275. doi:10.1371/journal.pone.0015275
Steinkühler, J., Knorr, R. L., Zhao, Z., Bhatia, T., Bartelt, S. M., Wegner, S., et al. (2020). Controlled division of cell-sized vesicles by low densities of membrane-bound proteins. Nat. Commun. 11 (1), 905. doi:10.1038/s41467-020-14696-0
Stewart, E. J., Madden, R., Paul, G., and Taddei, F. (2005). Aging and death in an organism that reproduces by morphologically symmetric division. PLoS Biol. 3 (2), e45. doi:10.1371/journal.pbio.0030045
Strutt, R., Sheffield, F., Barlow, N. E., Flemming, A. J., Harling, J. D., Law, R. V., et al. (2022). UV-DIB: label-free permeability determination using droplet interface bilayers. Lab a Chip 22 (5), 972–985. doi:10.1039/d1lc01155c
Stucki, A., Jusková, P., Nuti, N., Schmitt, S., and Dittrich, P. S. (2021a). Synchronized reagent delivery in double emulsions for triggering chemical reactions and gene expression. Small Methods 5 (8), 2100331. doi:10.1002/smtd.202100331
Stucki, A., Vallapurackal, J., Ward, T. R., and Dittrich, P. S. (2021b). Droplet microfluidics and directed evolution of enzymes: an intertwined journey. Angew. Chem. Int. Ed. 60 (46), 24368–24387. doi:10.1002/anie.202016154
Suea-Ngam, A., Howes, P. D., Srisa-Art, M., and DeMello, A. J. (2019). Droplet microfluidics: from proof-of-concept to real-world utility? Chem. Commun. 55 (67), 9895–9903. doi:10.1039/c9cc04750f
Suh, Y. K., and Kang, S. (2010). A review on mixing in microfluidics. Micromachines 1 (3), 82–111. doi:10.3390/mi1030082
Sułkowski, W. W., Pentak, D., Nowak, K., and Sułkowska, A. (2005). The influence of temperature, cholesterol content and pH on liposome stability. J. Mol. Struct. 744, 737–747. doi:10.1016/j.molstruc.2004.11.075
Swank, Z., Laohakunakorn, N., and Maerkl, S. J. (2019). Cell-free gene-regulatory network engineering with synthetic transcription factors. Proc. Natl. Acad. Sci. 116 (13), 5892–5901. doi:10.1073/pnas.1816591116
Swank, Z., and Maerkl, S. J. (2021). CFPU: a cell-free processing unit for high-throughput, automated in vitro circuit characterization in steady-state conditions. BioDesign Res. 2021, 2021. doi:10.34133/2021/2968181
Swartz, J. R., Jewett, M. C., and Woodrow, K. A. (2004). Cell-free protein synthesis with prokaryotic combined transcription-translation. Recomb. Gene Expr. Rev. Protoc. 267, 169–182. doi:10.1385/1-59259-774-2:169
Syeda, R., Holden, M. A., Hwang, W. L., and Bayley, H. (2008). Screening blockers against a potassium channel with a droplet interface bilayer array. J. Am. Chem. Soc. 130 (46), 15543–15548. doi:10.1021/ja804968g
Tabuchi, T., and Yokobayashi, Y. (2022). High-throughput screening of cell-free riboswitches by fluorescence-activated droplet sorting. Nucleic Acids Res. 50 (6), 3535–3550. doi:10.1093/nar/gkac152
Takahashi, M. K., Tan, X., Dy, A. J., Braff, D., Akana, R. T., Furuta, Y., et al. (2018). A low-cost paper-based synthetic biology platform for analyzing gut microbiota and host biomarkers. Nat. Commun. 9 (1), 3347. doi:10.1038/s41467-018-05864-4
Tan, W., Powles, E., Zhang, L., and Shen, W. (2021). Go with the capillary flow. Simple thread-based microfluidics. Sensors Actuators B Chem. 334, 129670. doi:10.1016/j.snb.2021.129670
Tawfik, D. S., and Griffiths, A. D. (1998). Man-made cell-like compartments for molecular evolution. Nat. Biotechnol. 16 (7), 652–656. doi:10.1038/nbt0798-652
Tayar, A. M., Karzbrun, E., Vincent, N., and Bar-Ziv, R. H. (2017). Synchrony and pattern formation of coupled genetic oscillators on a chip of artificial cells. Proc. Natl. Acad. Sci. 114 (44), 11609–11614. doi:10.1073/pnas.1710620114
Teh, S.-Y., Lin, R., Hung, L.-H., and Lee, A. P. (2008). Droplet microfluidics. Lab a Chip 8 (2), 198–220. doi:10.1039/b715524g
Theberge, A. B., Courtois, F., Schaerli, Y., Fischlechner, M., Abell, C., Hollfelder, F., et al. (2010). Microdroplets in microfluidics: an evolving platform for discoveries in chemistry and biology. Angew. Chem. Int. Ed. 49 (34), 5846–5868. doi:10.1002/anie.200906653
Tierra biosciences (2023). Tierra biosciences. Available at: https://www.tierrabiosciences.com/ (Accessed October 26, 2023).
Tinafar, A., Zhou, Y., Hong, F., Swingle, K. L., Tang, A. A., Green, A. A., et al. (2022). Cell-free biosensors: synthetic biology without borders. Handb. Cell Biosens., 243–281. doi:10.1007/978-3-030-23217-7_130
Todd, M. S., and Quake, S. R. (2005). Microfluidics: fluid physics at the nanoliter scale. Rev. Mod. Phys. 77 (3), 977–1026. doi:10.1103/revmodphys.77.977
Todd, P. L., and Jon, F. E. (2013). A review of the theory, methods and recent applications of high-throughput single-cell droplet microfluidics. J. Phys. D Appl. Phys. 46 (11), 114005. doi:10.1088/0022-3727/46/11/114005
Todd, T., Maerkl, S. J., and Quake, S. R. (2002). Microfluidic large-scale integration. Science 298 (5593), 580–584. doi:10.1126/science.1076996
Todd, T., Roberts, R. W., Arnold, F. H., and Quake, S. R. (2001). Dynamic pattern formation in a vesicle-generating microfluidic device. Phys. Rev. Lett. 86 (18), 4163–4166. doi:10.1103/PhysRevLett.86.4163
Toley, B. J., Wang, J. A., Gupta, M., Buser, J. R., Lafleur, L. K., Lutz, B. R., et al. (2015). A versatile valving toolkit for automating fluidic operations in paper microfluidic devices. Lab a Chip 15 (6), 1432–1444. doi:10.1039/c4lc01155d
Tran, T.My D., Matsuura, H., Ujiie, K., Muraoka, M., Harada, K., and Hirata, K. (2017). Paper-based colorimetric biosensor for antibiotics inhibiting bacterial protein synthesis. J. Biosci. Bioeng. 123 (1), 96–100. doi:10.1016/j.jbiosc.2016.07.015
Trantidou, T., Friddin, M., Elani, Y., Brooks, N. J., Law, R. V., Seddon, J. M., et al. (2017). Engineering compartmentalized biomimetic micro-and nanocontainers. ACS Nano 11 (7), 6549–6565. doi:10.1021/acsnano.7b03245
Trantidou, T., Friddin, M. S., Salehi-Reyhani, A., Ces, O., and Elani, Y. (2018). Droplet microfluidics for the construction of compartmentalised model membranes. Lab a Chip 18 (17), 2488–2509. doi:10.1039/c8lc00028j
Ueno, H., Sawada, H., Soga, N., Sano, M., Nara, S., Tabata, K. V., et al. (2021). Amplification of over 100 kbp DNA from single template molecules in femtoliter droplets. ACS Synth. Biol. 10 (9), 2179–2186. doi:10.1021/acssynbio.0c00584
Ugrinic, M., deMello, A., and Tang, T.-Y. D. (2019). Microfluidic tools for bottom-up synthetic cellularity. Chem 5 (7), 1727–1742. doi:10.1016/j.chempr.2019.03.012
Unger, M. A., Chou, H.-P., Todd, T., Scherer, A., and Quake, S. R. (2000). Monolithic microfabricated valves and pumps by multilayer soft lithography. Science 288 (5463), 113–116. doi:10.1126/science.288.5463.113
van der Linden, A. J., Yelleswarapu, M., Pieters, P. A., Swank, Z., Huck, W. T. S., Maerkl, S. J., et al. (2019). A multilayer microfluidic platform for the conduction of prolonged cell-free gene expression. JoVE J. Vis. Exp. 152, e59655. doi:10.3791/59655
Van Niel, G., d’Angelo, G., and Raposo, G. (2018). Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 19 (4), 213–228. doi:10.1038/nrm.2017.125
Van Nies, P., Westerlaken, I., Blanken, D., Salas, M., Mencía, M., and Danelon, C. (2018). Self-replication of DNA by its encoded proteins in liposome-based synthetic cells. Nat. Commun. 9 (1), 1583. doi:10.1038/s41467-018-03926-1
van Sluijs, B., Maas, R. J. M., van der Linden, A. J., Greef, T. F. A. de, and Huck, W. T. S. (2022). A microfluidic optimal experimental design platform for forward design of cell-free genetic networks. Nat. Commun. 13 (1), 3626. doi:10.1038/s41467-022-31306-3
Villar, G., Alexander, D. G., and Bayley, H. (2013). A tissue-like printed material. Science 340 (6128), 48–52. doi:10.1126/science.1229495
Vincent, N., Bar-Ziv, R., Godefroy, J., Hanna, S., and Libchaber, A. (2005). Toward an artificial cell based on gene expression in vesicles. Phys. Biol. 2 (3), P1–P8. doi:10.1088/1478-3975/2/3/P01
Vincent, N., and Libchaber, A. (2004). A vesicle bioreactor as a step toward an artificial cell assembly. Proc. Natl. Acad. Sci. 101 (51), 17669–17674. doi:10.1073/pnas.0408236101
Vladisavljević, G. T., Nuumani, R. A., and Ali Nabavi, S. (2017). Microfluidic production of multiple emulsions. Micromachines 8 (3), 75. doi:10.3390/mi8030075
Voigt, C. A. (2020). Synthetic biology 2020–2030: six commercially-available products that are changing our world. Nat. Commun. 11 (1), 6379. doi:10.1038/s41467-020-20122-2
Wagner, O., Thiele, J., Weinhart, M., Mazutis, L., Weitz, D. A., Huck, W. T. S., et al. (2016). Biocompatible fluorinated polyglycerols for droplet microfluidics as an alternative to PEG-based copolymer surfactants. Lab a Chip 16 (1), 65–69. doi:10.1039/c5lc00823a
Wang, S., Majumder, S., Emery, N. J., and Liu, A. P. (2018). Simultaneous monitoring of transcription and translation in mammalian cell-free expression in bulk and in cell-sized droplets. Synth. Biol. 3 (1), ysy005. doi:10.1093/synbio/ysy005
Wang, S., Zhang, X., Ma, C., Yan, S., Inglis, D., and Feng, S. (2021). A review of capillary pressure control valves in microfluidics. Biosensors 11 (10), 405. doi:10.3390/bios11100405
Wang, W., Zhang, M.-J., and Chu, L.-Y. (2014). Microfluidic approach for encapsulation via double emulsions. Curr. Opin. Pharmacol. 18, 35–41. doi:10.1016/j.coph.2014.08.003
Wang, X., Yi, L., Mukhitov, N., Schrell, A. M., Dhumpa, R., and Roper, M. G. (2015). Microfluidics-to-mass spectrometry: a review of coupling methods and applications. J. Chromatogr. A 1382, 98–116. doi:10.1016/j.chroma.2014.10.039
Webb, C., Khadke, S., Schmidt, S. T., Roces, C. B., Forbes, N., Berrie, G., et al. (2019). The impact of solvent selection: strategies to guide the manufacturing of liposomes using microfluidics. Pharmaceutics 11 (12), 653. doi:10.3390/pharmaceutics11120653
Weiss, M., Frohnmayer, J. P., Benk, L. T., Haller, B., Janiesch, J.-W., Heitkamp, T., et al. (2018). Sequential bottom-up assembly of mechanically stabilized synthetic cells by microfluidics. Nat. Mater. 17 (1), 89–96. doi:10.1038/nmat5005
White, J. A., and Streets, A. M. (2018). Controller for microfluidic large-scale integration. HardwareX 3, 135–145. doi:10.1016/j.ohx.2017.10.002
Whitesides, G. M. (2006). The origins and the future of microfluidics. Nature 442 (7101), 368–373. doi:10.1038/nature05058
Whitesides, G. M., Ostuni, E., Takayama, S., Jiang, X., and Ingber, D. E. (2001). Soft lithography in biology and biochemistry. Annu. Rev. Biomed. Eng. 3 (1), 335–373. doi:10.1146/annurev.bioeng.3.1.335
Williams, M. S., Longmuir, K. J., and Paul, Y. (2008). A practical guide to the staggered herringbone mixer. Lab a Chip 8 (7), 1121–1129. doi:10.1039/b802562b
Woodruff, K., and Maerkl, S. J. (2018). Microfluidic module for real-time generation of complex multimolecule temporal concentration profiles. Anal. Chem. 90 (1), 696–701. doi:10.1021/acs.analchem.7b04099
Xavier, C. I., Solvas, X. C. I., Edel, J. B., Law, R. V., and Ces, O. (2016). Microfluidic generation of encapsulated droplet interface bilayer networks (multisomes) and their use as cell-like reactors. Chem. Commun. 52 (35), 5961–5964. doi:10.1039/c6cc01434h
Xi, H.-D., Zheng, H., Guo, W., Gañán-Calvo, A. M., Ai, Y., Chia-Wen, T., et al. (2017). Active droplet sorting in microfluidics: a review. Lab a Chip 17 (5), 751–771. doi:10.1039/c6lc01435f
Xia, Y., and Whitesides, G. M. (1998). Soft lithography. Annu. Rev. Mater. Sci. 28 (1), 153–184. doi:10.1146/annurev.matsci.28.1.153
Xie, M., and Fussenegger, M. (2018). Designing cell function: assembly of synthetic gene circuits for cell biology applications. Nat. Rev. Mol. Cell Biol. 19 (8), 507–525. doi:10.1038/s41580-018-0024-z
Xumeng, G., Luo, D., and Xu, J. (2011). Cell-free protein expression under macromolecular crowding conditions. PLOS ONE 6 (12), e28707. doi:10.1371/journal.pone.0028707
Yamada, K., Shibata, H., Suzuki, K., and Citterio, D. (2017). Toward practical application of paper-based microfluidics for medical diagnostics: state-of-the-art and challenges. Lab a Chip 17 (7), 1206–1249. doi:10.1039/c6lc01577h
Yang, S.-M., Lv, S., Zhang, W., and Cui, Y. (2022). Microfluidic point-of-care (POC) devices in early diagnosis: a review of opportunities and challenges. Sensors 22 (4), 1620. doi:10.3390/s22041620
Yi, J., Yang, L., Luo, D., Huck, W. T. S., and Yang, D. (2018). Microfluidic-assisted fabrication of clay microgels for cell-free protein synthesis. ACS Appl. Mater. Interfaces 10 (35), 29308–29313. doi:10.1021/acsami.8b09324
Yuan, Lu (2017). Cell-free synthetic biology: engineering in an open world. Synthetic Syst. Biotechnol. 2 (1), 23–27. doi:10.1016/j.synbio.2017.02.003
Yuan, Lu (2022). Textile-embedded cell-free biosensors. Nat. Biomed. Eng. 6 (3), 225–226. doi:10.1038/s41551-022-00869-3
Zeng, S., Liu, X., Xie, H., and Lin, B. (2011). Basic technologies for droplet microfluidics. Microfluid. Technol. Appl. 304, 69–90. doi:10.1007/128_2011_149
Zhang, D., Li, C., Ji, D., and Wang, Y. (2022a). Paper-based microfluidic sensors for onsite environmental detection: a critical review. Crit. Rev. Anal. Chem. 52 (6), 1432–1449. doi:10.1080/10408347.2021.1886900
Zhang, L., Guo, W., and Lu, Y. (2020). Advances in cell-free biosensors: principle, mechanism, and applications. Biotechnol. J. 15 (9), 2000187. doi:10.1002/biot.202000187
Zhang, Y., Minagawa, Y., Kizoe, H., Miyazaki, K., Iino, R., Ueno, H., et al. (2019). Accurate high-throughput screening based on digital protein synthesis in a massively parallel femtoliter droplet array. Sci. Adv. 5 (8), eaav8185. doi:10.1126/sciadv.aav8185
Zhang, Y., Zhao, C., Bi, H., Zhang, X., Xue, B., Li, C., et al. (2022b). A cell-free paper-based biosensor dependent on allosteric transcription factors (aTFs) for on-site detection of harmful metals Hg2+ and Pb2+ in water. J. Hazard. Mater. 438, 129499. doi:10.1016/j.jhazmat.2022.129499
Zheng, W., Wang, K., Xu, H., Zheng, C., Cao, B., Qi, Q., et al. (2021). Strategies for the detection of target analytes using microfluidic paper-based analytical devices. Anal. Bioanal. Chem. 413, 2429–2445. doi:10.1007/s00216-021-03213-x
Zhou, Y., Wu, Y., Ding, L., Huang, X., and Xiong, Y. (2021). Point-of-care COVID-19 diagnostics powered by lateral flow assay. TrAC Trends Anal. Chem. 145, 116452. doi:10.1016/j.trac.2021.116452
Keywords: microfluidics, PDMS, synthetic biology, molecular engineering, cell-free synthetic biology, artificial cells
Citation: Baranwal AK and Maerkl SJ (2024) A comprehensive review of Microfluidic approaches in cell-free synthetic biology. Front. Synth. Biol. 2:1397533. doi: 10.3389/fsybi.2024.1397533
Received: 08 March 2024; Accepted: 12 April 2024;
Published: 06 May 2024.
Edited by:
Corentin Briat, ETH Zürich, SwitzerlandReviewed by:
Richard Murray, California Institute of Technology, United StatesCopyright © 2024 Baranwal and Maerkl. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sebastian J. Maerkl, c2ViYXN0aWFuLm1hZXJrbEBlcGZsLmNo
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.