- 1Institute of Chemistry and Biochemistry - Organic Chemistry, Freie Universität Berlin, Berlin, Germany
- 2Molecular Environmental Biotechnology, Helmholtz Centre for Environmental Research - UFZ, Leipzig, Germany
- 3Chair of Geobiotechnology, Technische Universität Berlin, Berlin, Germany
- 4Department of Chemistry, University of Manitoba, Winnipeg, MB, Canada
- 5Biocatalysis Group, Institute for Chemistry, Technische Universität Berlin, Berlin, Germany
Nature has scarcely evolved a biochemistry around fluorine. However, modern science has shown that fluorinated organic molecules are suitable building blocks for biopolymers, from peptides and proteins up to entire organisms. Here, we conducted adaptive laboratory evolution (ALE) experiments to introduce organofluorine into living microorganisms. By cultivating Escherichia coli with fluorinated indole analogs, we successfully evolved microbial cells capable of utilizing either 6-fluoroindole or 7-fluoroindole for growth. Our improved ALE protocols enabled us to overcome previous challenges and adaptation was achieved, enabling a former growth inhibiting unnatural molecule to become a substrate for the cell’s protein synthesis machinery to the extent that the entire proteome underwent Trp to F-Trp substitution. In the ALE experiments, we supplied fluoroindoles to Trp-auxotrophic E. coli bacteria, exerting strong selective pressure that led to microbial adaptation. Within the cells, these indoles were converted into the corresponding amino acids (6- and 7-fluorotryptophan) and globally incorporated into the proteome at tryptophan sites. This study is a first step and establishes a strong foundation for further exploration of the mechanisms underlying fluorine-based life and how a former antimetabolite can become a vital nutrient.
1 Introduction
Although fluorine is abundant in the earth’s crust, it is scarcely found in biomolecules. The reasons for this, including extremely high heat of hydration and redox potential as well as low nucleophilicity, are well discussed in the literature (Gribble, 2003; Merkel and Budisa, 2012; Gribble, 2015; O’Hagan and Deng, 2015). Only a few plants and microorganisms can produce organofluorine compounds (Harper and OHagan, 1994; Carvalho and Oliveira, 2017). One prominent example is the well-studied Streptomyces cattleya that biosynthesizes the amino acid 4-fluorothreonine (4FT) and at the same time produces toxic fluoroacetate (FA) (Sanada et al., 1986). In terms of chemistry, this is a challenging task as a C-H bond has to be replaced with a polar-reversed C-F bond. In fact, there is such an enzyme known as fluorinase, that enables C-F bond formation in biological systems (O’Hagan et al., 2002; Deng et al., 2004). The immense biotechnological potential of these biotransformations has led to extensive research in this field, which demonstrates the current high level of interest and research in this direction (Markakis et al., 2020; Eustáquio et al., 2010; Calero et al., 2020; Dall’Angelo et al., 2013).
Microbes, in particular bacteria, have an astonishing capacity to overcome environmental stress through adaptation, as is observed in resistant and extremophile populations (Gupta et al., 2014; Merino et al., 2019). For example, adaptation to pollutants impressively demonstrates that evolution can take place over relatively short time scales (Sangwan et al., 2014; Yoshida et al., 2016). One further aspect of high current relevance is microbe-mediated catabolism of organofluorine compounds, in particular poly- and perfluoroalkyl substances (PFASs), which represent an extremely serious environmental problem due to their persistence (Wang et al., 2017). The natural process of evolution can be realized under controlled laboratory conditions by means of adaptive laboratory evolution (ALE) in which a particular experimental selection pressure is applied (Dragosits and Mattanovich, 2013; Sandberg et al., 2019; Connolly et al., 2021; Mavrommati et al., 2022). Key examples of prior tryptophan-based ALE studies are: 1) the replacement of tryptophan by 4-fluorotryptophan (4FTrp) in Bacillus subtilis while using an external mutagen to accelerate the adaptation process, (Wong, 1983), which was later extended with the analogs 5-fluorotryptophan (5FTrp) and 6-fluorotryptophan (6FTrp) (Mat et al., 2010; Yu et al., 2014); 2) the adaptation of 4FTrp tolerant Escherichia coli (Bacher and Ellington, 2001); and 3) the proteome-wide substitution of Trp by L-ß-(thieno [3,2-b]pyrrolyl)alanine (Hoesl et al., 2015) as well as 4FTrp and 5FTrp (Agostini et al., 2021), in which the advantages of using indole precursors instead of amino acid analogs were demonstrated. The chemical synthesis of tryptophan analogs always bears the risk of contaminating traces of natural Trp, which narrows the selective pressure and might subvert the adaptation process, whereas indole and its derivatives are commercially available at extremely high purity levels. Our 2021 study on the adaptation of E. coli to 4-fluoroindole (4Fi) and 5-fluoroindole (5Fi) showed via multi-omics analyses that the two evolved populations were remarkably distinct with respect to the observed rearrangements in regulatory networks, membrane integrity, and quality control of protein folding (Agostini et al., 2021).
Here we report ALE experiments with 6-fluoroindole (6Fi) and 7-fluoroindole (7Fi), the latter of which to our knowledge has not been published in this context. We have further improved the protocols and experimental setup for the ALE procedure to obtain bacterial populations that are fully “addicted” (Tack et al., 2016) to these former antimetabolites and thus provide a reliable platform and methodologies for the experimental evolution of novel organisms with fluorine as a bioelement. It is applicable for any ALE experiment on microbial adaptation to non-canonical amino acids created by mimicking natural selection in the laboratory without external influences like mutagens.
2 Materials and methods
2.1 Adaptive laboratory evolution (ALE) experiments
For the adaptation of E. coli towards usage of 6- and 7-fluoroindole a serial transfer regime using batch cultures under aerobic conditions was applied. The cells were propagated in synthetic minimal medium (New Minimal Medium, NMM) (Budisa et al., 1995) which consisted of: 7.5 mM (NH4)2SO4, 8.5 mM NaCl, 22.5 mM KH2PO4, 50 mM K2HPO4, 1 mM MgSO4, 20 mM D-glucose, 1 mg/L Ca2+ (as CaCl2), 1 mg/L Fe2+ (as FeCl2), 10 mg/L biotin, 10 mg/mL thiamine and 10 ng/L trace elements (Cu2+, Zn2+, Mn2+, MoO42-). In addition, the medium was variably enriched with indole (2.5 µM–0 µM), fluoroindole (70 μM–30 µM) and amino acids (50 mg/L), and was dynamically adjusted during the adaptation process (Supplementary Tables S1–S4).
The cultivation of each experiment (adaptation to 6Fi, 7Fi, W, Ind) was executed in parallel using three biological replicates (assumed to be isogenic) of our metabolic prototype TUB00 (ΔtrpLEDC ΔtnaA), which is a derivative of E. coli MG1655. 10 mL culture were incubated in 100 mL-shaking flasks at 30°C, 180 rpm. When the cells reached their early stationary phase (usually after 50–65 h) fresh medium was inoculated to an OD600 of 0.02 with the parent culture. The adjustment of the medium composition (increasing the selective pressure by depleting the indole and cAA concentration) was carried out when the cells proliferated sufficiently at least for two consequential passages under the parent conditions or no further growth improvement could be detected. The transfer steps of this serial dilution approach are indicated as “passages” and samples of each passage were preserved as cryo-stocks (25% glycerol, −80°C) for further analysis.
Beforehand ancestral TUB00 cultures were supplemented with different concentrations of indole (0 μM–30 µM) and fluoroindole (5 μM–1,000 µM) to determine the starting conditions. 2.5 µM indole and 70 µM of 6Fi or 7Fi were found to allow the cells to grow to a sufficient optical density of OD600 0.7–1.0. Within the ALE setup, the concentration of fluoroindole was kept high; then first the concentration of indole was gradually reduced until complete depletion (ALE phase 1) and subsequently the amino acid supply was stepwise ceased (ALE phase 2). For the amino acid removal two different approaches OC (overall concentration) and MB (metabolic blocks) were applied (Figures 1, 2). At this stage the evolving lineages separated into either three (6TUB128-OC, 6TUB165-MB4, 6TUB165-MB3) or two (7TUB165-OC, 7TUB165-MB) independent strains. In case of the 7-fluoroindole ALEs the growth conditions had to be relaxed (reduction of 7Fi concentration and increasing of cAA supply) in order to retain sufficient cell growth.
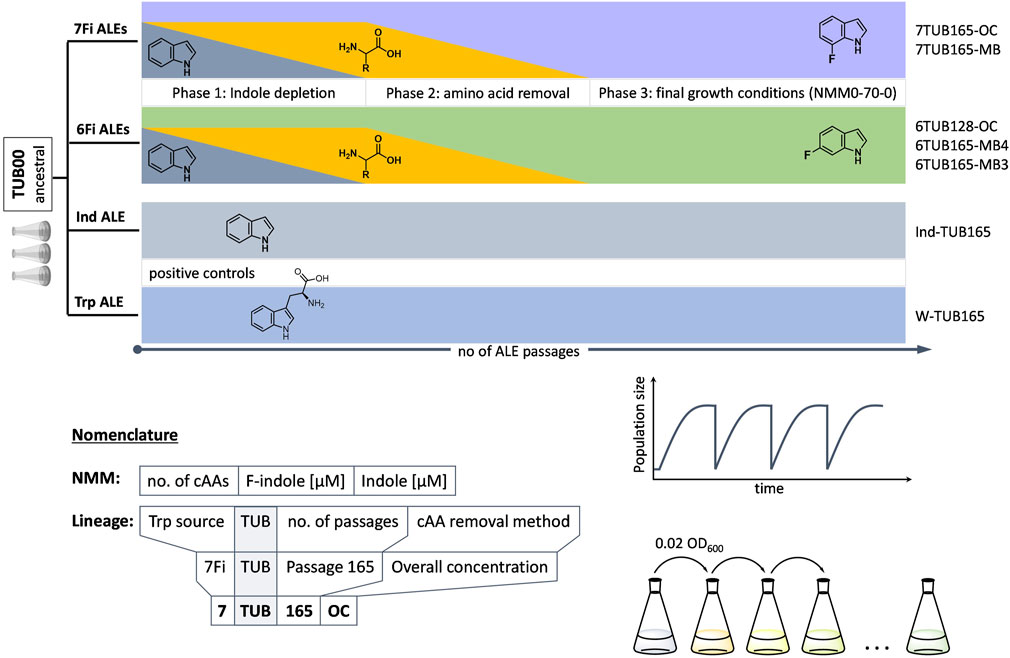
FIGURE 1. Conceptual ALE trajectories. Four parallel evolution experiments on populations/lineages over the course of a number of passages under different growth conditions were conducted: two positive controls (indole and Trp, lower grey bars) and adaptation to 6Fi (green middle cartoon) or 7Fi (purple upper cartoon). Molecular structures of indole, Trp, 6Fi and 7Fi are shown for the respective experiment. Increasing passage number (independent variable) is indicated by lowest black right arrow; lineage nomenclature follows Z-TUB-X-OC/MB pattern where “Z” indicates the Trp-source (Ind, Trp, 6Fi or 7Fi), “X” the passage number and “OC/MB” the cAA removal method; growth conditions were abbreviated in all cases as NMM([x]a-b-c), where x is the concentration of cAAs present in mg/L (only indicated when the concentration was below 50 mg/L); a is the number of different cAAs present; b is the concentration of 6Fi or 7Fi in µM; and c is the concentration of indole in µM. Phases 1 (indole depletion), 2 (amino acid depletion) and 3 (adaptation in presence of constant concentration of only 6Fi or 7Fi as supplement) are indicated. Phase 2 realized according to overall concentration (OC) or metabolic block (MB) strategies (see text for details).
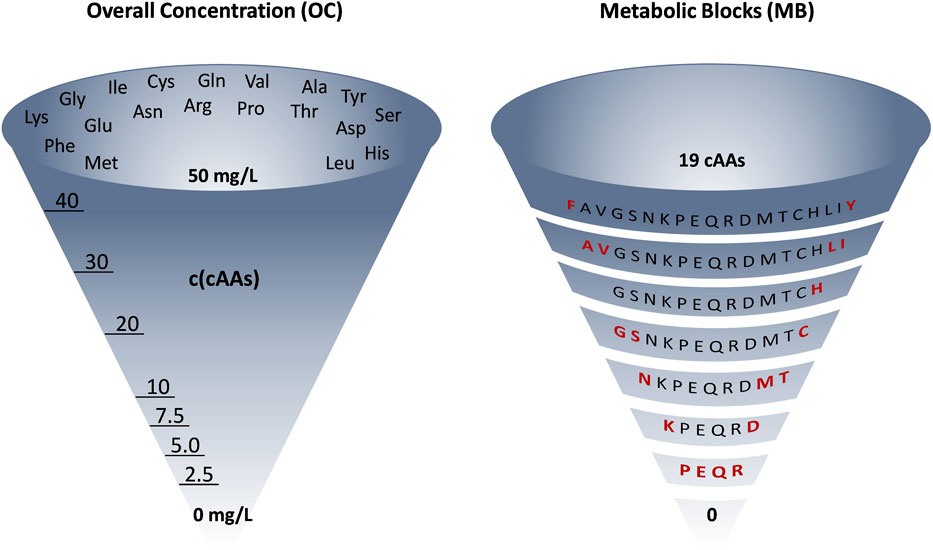
FIGURE 2. Graphical representation of the differences between OC and MB approaches for amino acid removal in ALE Phase 2, which is a general design concept of our ALE experiments. OC: mixture of 19 amino acids stepwise reduced in concentration from 50 to 0 mg/L. MB: 19 amino acids removed by identity (shown in red), as metabolic blocks or one by one (in case of the 7Fi ALE). This is a representative example (6TUB128-OC adaptation for the OC approach and 6TUB165-MB4 adaptation for the MB approach) but does not reflect the experimental details for all ALE experiments.
The same procedure as for the Fi-adaptation was followed with the positive controls, adapted to grow on 70 µM of either indole (Ind-TUBX) or tryptophan (W-TUBX). These cells were cultivated under the same growth regime for 165 passages, except for the amino acid removal process of ALE phase 2 (NMM19-0-70), which was omitted (Supplementary Figure S8).
2.2 Global Trp for F-Trp substitution (proteomics)
Final isolates of the adapted strains 6TUB128-OC, 6TUB165-MB4, 6TUB165-MB3, 7TUB165-OC, 7TUB165-MB and positive controls Ind-TUB165, W-TUB165 as well as TUB00 were grown in the respective NMM supplemented with either 70 µM indole, 6Fi or 7Fi. Exponential growing cells were harvested by centrifugation (18,000 g, 4°C, 20 min) and pellets were stored at −20°C. E. coli extracts were briefly run into an SDS-gel until the dye front had migrated about 5 mm into the gel and only the 3 mm behind the dye front were used for protein digestions. This step removes all positively charged molecules, small highly-mobile negatively charged ions, uncharged molecules and very large agglomerates that did not enter the gel. Following a protocol described by Kublik et. al. (Kublik et al., 2016), the gel slices were washed with ddH2O, and the proteins were reduced with dithiothreitol to break disulfide bridges. To protect the reduced cysteine residues, carbamidylation using iodoacetamide was performed. Subsequently, an overnight trypsin digestion was initiated to generate peptides. The resulting peptides were extracted from the gel pieces and desalted using C18-tips through zip-tipping. These extracted peptide samples were then analyzed by nLC-MS/MS on a nanoUPLC system (nanoAcquity, Waters) coupled to an Orbitrap Fusion mass spectrometer (Thermo Scientific), as previously described (Seidel et al., 2018). Protein identification was achieved using Proteome Discoverer (v2.4, Thermo Fisher Scientific) with the E. coli fasta genome database and SequestHT as the search engine, setting a false discovery rate threshold of 1% for peptide identification using the Target Decoy PSM Validator node. The abundance of proteins was quantified through label-free quantification based on intensity values in precursor scans using the Minora node in Proteome Discoverer. For the identification of peptides, carbamidomethylation of cysteine residues was taken as a fixed modification, oxidation of methionine residues and fluorination of tryptophan residues were taken as dynamic modifications. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD048225 (Perez-Riverol et al., 2022; Deutsch et al., 2023). Fluorination was calculated as the substitution of a hydrogen atom by fluorine giving a mass change of +17.991 Da (Supplementary Figure S9).
2.3 Growth behavior of the adapted strains
Isolates of the ancestral strain TUB00 and of the final isolates W-TUB165, Ind-TUB165, 6TUB128-OC, 6TUB165-MB4, 6TUB165-MB3, 7TUB165-OC and 7TUB165-MB were subjected to comprehensive growth analysis using different medium compositions along the adaptation course (growth curves see Figure 8, Supplementary Figure S11). These comprised LB (rich, undefined medium), NMM19-0-70 (Ind), NMM19-70-1 (6Fi or 7Fi, Ind), NMM19-70-0 (6Fi or 7Fi) and NMM0-70-0 (6Fi or 7Fi). The lineage nomenclature follows Z-TUB-X-OC/MB pattern where “Z” indicates the Trp-source (Ind, Trp, 6Fi or 7Fi), “X” the passage number and “OC/MB” the cAA removal method; and the growth conditions were abbreviated in all cases as NMM([x]a-b-c), where x is the concentration of cAAs present in mg/L (only indicated when the concentration was below 50 mg/L); a is the number of different cAAs present; b is the concentration of 6Fi or 7Fi in µM; and c is the concentration of indole in µM. Thereby, 6Fi adapted cells could be directly resuscitated in NMM0-70-0 (6Fi) from cryo stock, whereas 7Fi adapted cells had to be revived in nutrient rich LB medium, followed by two-times serial inoculation in their final adapted medium NMM0-70-0 (7Fi supplemented, in order to restore the intracellular Trp-free environment). Subsequently, the cells were washed twice with NMM0-0-0 to remove residual Ind, 6Fi or 7Fi, normalized to OD600 = 1, and then 200 µL of the respective medium composition were inoculated to OD600 = 0.02. The growth was monitored in 96-well plates (Greiner, flat bottom clear), sealed with breathable membrane (Breathe-Easy sealing membrane, Sigma Aldrich) at 30°C with continuous shaking using a microplate reader (Tecan reader Infinite M200) that measured the absorbance at 600 nm in 10 min intervals. The measurements were performed at least twice and in triplicates (biological replicates).
2.4 Viability assay (CCK-8)
The cell viability was determined using cell counting kit-8 (CCK-8; Sigma-Aldrich), which is based on dehydrogenase activity detection in viable cells. For the proliferation assay 190 µL of 5.0 × 1007 cells/mL suspension were seeded, added with 10 µL of the CCK-8 solution and incubated for 4 h at 30°C without shaking. Then, the absorbance at 450 nm was measured using microplate reader (Tecan reader Infinite M200), which is directly proportional to the number of proliferating cells. As background control medium without cells was used and the viability of TUB00 was set to 100% in comparison to the adapted strains. Measurements were performed in triplicates and repeated at least twice.
2.5 Susceptibility and tolerance of evolved strains to vancomycin
For testing the antibiotic susceptibility, the minimal inhibitory concentration (MIC) of vancomycin was determined (Figure 8, Supplementary Figure S12). Isolates from cryo stocks of the respective strains were resuscitated either in 2–3 mL LB or appropriate NMM, incubated at 30°C, 200 rpm and inoculated into NMM if necessary. Subsequently, cells were used to inoculate 3 mL NMM in culture tubes as follows: NMM0-0-70 (Ind or Trp) for TUB00, W-TUB165, Ind-TUB165 and NMM0-70-0 (6Fi or 7Fi) for adapted strains 6TUB128-OC, 6TUB165-MB4, 6TUB165-MB3 and 7TUB165-OC, 7TUB165-MB. For determination of the MIC a cell density corresponding to 0.5 McFarland standard (OD600 = 0.1) was used and cells were treated with serial dilutions of vancomycin (0, 25, 50, 100, 200, 400 μg/mL). Cell growth (incubation at 30°C, 200 rpm) was monitored by measuring the OD600 after 24 h and MIC was defined as the lowest concentration at which no increase in cell density could be observed (OD600 = 0.1). Measurements were performed using biological triplicates.
2.6 Expression of 6FTrp- and 7FTrp-substituted variants of EGFP and ECFP in ALE strains
Variants of the green fluorescent protein (GFP) were expressed in the final isolates 6TUB128-OC, 6TUB165-MB4, 6TUB165-MB3 and 7TUB165-OC, 7TUB165-MB, as well as TUB00. These variants are the C-terminal his-tagged enhanced green fluorescent protein (EGFP-H6) and the N-terminal his-tagged enhanced cyan fluorescent protein (H6-ECFP). The strains were rendered electro competent (by three times washing in 10% glycerol) and transformed with the respective expression plasmids pQE80L EGFP-H6 or pQE80L H6-ECFP (both endowed with ori ColEI, AmpR).
Inoculated from cryo stocks, precultures of transformed cells were incubated in NMM0-70-0 (6Fi or 7Fi), supplemented with 100 μg/mL ampicillin (Amp) and subsequently 150 mL expression culture (NMM0-70-0 (6Fi or 7Fi), 100 μg/mL Amp) were inoculated to an OD600 of 0.05. NMM0-0-70 (Ind) was used accordingly for the TUB00 control. The cultures were incubated at 30°C, 180 rpm until they reach exponential growth phase (OD600 0.4–0.7). Recombinant protein expression was induced by adding 0.5 mM isopropyl ß-D-1-thiogalactopyranoside (IPTG) and performed overnight at 30°C, 180 rpm.
Cells were harvested by centrifugation (20 min, 10,000 g, 4°C) and resuspended in B-PER™ (Bacterial Protein Extraction Reagent, Thermo Fisher Scientific) according to manufacturer’s instructions. The lysis mixture contained B-PER™, lysozyme, DNAse and was incubated 10–15 min at room temperature. After centrifugation of cell debris (5 min, 15,000 g, RT), the cell lysate was filtered (Rotilabo PVDF Filter, 0.45 µm pore diameter) and target proteins were purified by using affinity chromatography on Protino Ni-NTA column (1 mL Fast Flow, Machery-Nagel). The purification was carried out using a single-channel peristaltic pump P-1 (VWR) and sodium phosphate-based purification buffer (50 mM Na-P, 300 mM NaCl, two-step imidazole gradient: 20 mM and 40 mM, pH 7.8); followed by elution with 500 mM imidazole. EGFP or else ECFP containing fractions were identified and chromophore maturation was proved by detection of fluorescence by irradiation with a UV lamp at 365 nm. Respective elution fractions were collected, and imidazole was removed by dialysis (Microsep™ Advanced Centrifugal Devices, MWCO 3kDa, Pall Laboratory) against 50 mM Na-P, 100 mM NaCl (pH 7.8). Finally, protein identity and incorporation of 6- and 7-fluorotryptophan was confirmed by LC-ESI-Q-TOF mass spectrometry (Agilent 6530 Accurate-Mass Q-TOF). Protein samples were analyzed in a concentration of 0.1 mg/mL using the following HPLC parameters: linear gradient from 5% to 80% buffer A within 20 min (A: 0.1% formic acid in MQ-H2O, B: 0.1% formic acid in acetonitrile), flow rate 0.3 mL/min, injection volume 5 µL. For the MS spectrum a range of 27,000–29,000 amu was selected in the total ion current (TIC) plot and the maximum entropy deconvolution algorithm was applied. The MS spectra are shown in Figure 9 and Supplementary Figure S13, the mass values are shown in Supplementary Table S6 and the isolated yields in Supplementary Table S7.
3 Results and discussion
3.1 Design of adaptation experiments
We started the ALE experiments with the metabolic prototype TUB00 (E. coli MG1655 ΔtrpLEDC ΔtnaA) as ancestral strain, which possesses stringent Trp auxotrophy due to genomic deletions in the endogenous pathways of Trp biosynthesis and degradation (Supplementary Figure S4). However, the Trp processing enzymes tryptophan synthase (TrpS; trpBA) (Wilcox, 1974; Goss and Newill, 2006; Watkins-Dulaney et al., 2021) and tryptophanyl-tRNA synthetase (TrpRS; trpS) (Wong, 1983; Bacher and Ellington, 2001; Azim and Budisa, 2008) remained in the genome, exhibit a broad substrate promiscuity and enable TUB00 to produce FTrp from F-indole and to incorporate FTrp in the protein biosynthetic process (Supplementary Figure S5). Since the metabolic sources of indole (tnaA) and Trp (trpLEDC) are eliminated the ALE experiment requires bacteria to accept indole or a fluorinated indole analog and produce the corresponding amino acid via TrpS and L-serine to survive. Thus, depriving the cells of canonical indole while supplying them continually with the fluorinated indole analog exerts selective pressure and brings about the adaptation process.
A conceptual overview of the ALE experiments we performed is shown in Figure 1. Four parallel experiments were designed: 1) a positive “indole control”, 2) a positive “Trp control”, 3) adaptation to 6Fi and 4) adaptation to 7Fi. For a legend explaining the nomenclature for all conditions and lineages obtained please refer to Figure 1. The controls serve to determine fluorine-induced effects and general consequences of long-term cultivation, respectively. We opted to conduct the experiments by allowing liquid cultures to grow under a given set of conditions until they reached early stationary phase; once this occurred, a constant cell number corresponding to an OD600 value of 0.02 was used to inoculate the subsequent culture (defined as a “passage”). Unlike the alternative approach of “continuous cultivation”, this kind of serial transfer regime is characterized by alternating “feeding-and-starvation” regimes or “seasonal environments” (Vasi et al., 1994) (i.e., a population bottleneck), but benefits from its flexibility when adjustments to cultivation conditions are needed (Gresham and Dunham, 2014). Furthermore, we chose the serial transfer approach because this type of experimental management is closest to natural selection in that it mimics changing conditions in the real environment of natural populations and enabled cells to undergo all growth phases under each set of new conditions. Consistent inoculation of each new liquid culture with a normalized amount of cells ensures that even poorly growing cultures do not suffer an evolutionary disadvantage (e.g., our primary goal was not to select for rapidly growing populations) (LaCroix et al., 2017).
Three biological replicates of the starting point strain TUB00, which we assumed to be isogenic, were allowed to proliferate in synthetic minimal medium (New Minimal Medium, NMM) containing essential nutrients such as phosphate, ammonium, glucose, and vitamins. NMM was differentially supplemented as follows. Indole control populations grew in NMM with a constant concentration of 70 µM indole and high concentration of 19 of the 20 canonical amino acids (cAA); that is, all except Trp. Trp control populations grew in NMM with no indole but a constant concentration of 70 µM Trp and high concentration of the 19 cAAs (cultivation schemes shown in Supplementary Figure S8). Populations adapting to 6Fi and 7Fi grew in NMM variably enriched (see below) with indole, the 19 cAAs, and the respective fluorinated indole (see Supplementary Tables S1–S4 for medium compositions). To ensure the purity of the purchased 6Fi and 7Fi precursors, we determined the level of parent indole in batches by GC-MS and, indeed, indole was not detected within the limits of the method (see Supplementary Figures S1–S3).
The three phases of adaptation to 6Fi and 7Fi are represented in Figure 1. In phase 1, indole concentration is gradually, stepwise reduced to zero. In phase 2 the 19 cAAs are gradually, stepwise removed from the growth medium according to two distinct strategies (see paragraph below) to exclude possible contamination by traces of indole or tryptophan, which would disturb the adaptation process (Bacher and Ellington, 2001). Phase 3 represents the period of evolution in which only the fluorinated indole analog is present as NMM supplement. As noted above, the adaptation experiments designed here proceeded according to “passages” and it is important to note that the criteria that a population had to meet in order to be subjected to the subsequent growth condition were as follows: the population had to have reached early stationary phase under the given condition and proceeded through at least two full passages under that given condition; in some cases more than 2 passages were required to observe stable growth, or no further growth improvement was observed. This is why the number of days of growth does not correspond 1:1 with passage number. Only after these criteria were met were the conditions made more stringent to increase the evolutionary pressure being placed on the population.
Phase 2 was designed according to two different approaches: so-called “overall concentration (OC)” or “metabolic blocks (MB)”. OC and MB are visually represented in Figure 2. In the OC approach, the overall concentration of all 19 amino acids was equally reduced gradually and stepwise from 50 to 0 mg/L. The removal of amino acids in the MB approach was not based on their concentration, but rather on their origin in core metabolism (i.e., their metabolic identity). Canonical amino acids were also assigned to metabolic blocks considering factors such as their metabolic cost in terms of energy and precursor requirements (Akashi and Gojobori, 2002; Tolle et al., 2023). For example, the first block that was removed comprised the aromatic amino acids tyrosine and phenylalanine, both requiring laborious multistep biosynthesis and originating from the same starting metabolites (phosphoenolpyruvate and erythrose 4-phosphate). Whereas we and others successfully applied the MB approach in previous adaption experiments (Hoesl et al., 2015; Agostini et al., 2021), the OC approach presented here represents an innovation. We thought that the gradual reduction of all amino acids is more gentle to cells than the sudden elimination of whole metabolic blocks. This approach gently prompts cells to initiate their amino acid biosynthesis processes, avoiding abrupt ribosome arrest and mistranslation effects that result from a sudden depletion of amino acids (Wong et al., 2018).
3.2 Adaptive laboratory evolution experimental setup for 6-fluoroindole (6Fi)
Overall, Trp-auxotrophic E. coli tolerated 6Fi remarkably well (Figures 3–5). In phase 1, cells responded sensitively to the challenge of indole depletion, as evidenced by strongly fluctuating OD600 values. However, after the populations ceased to be dependent on indole, cell growth remained stable and at a high level.
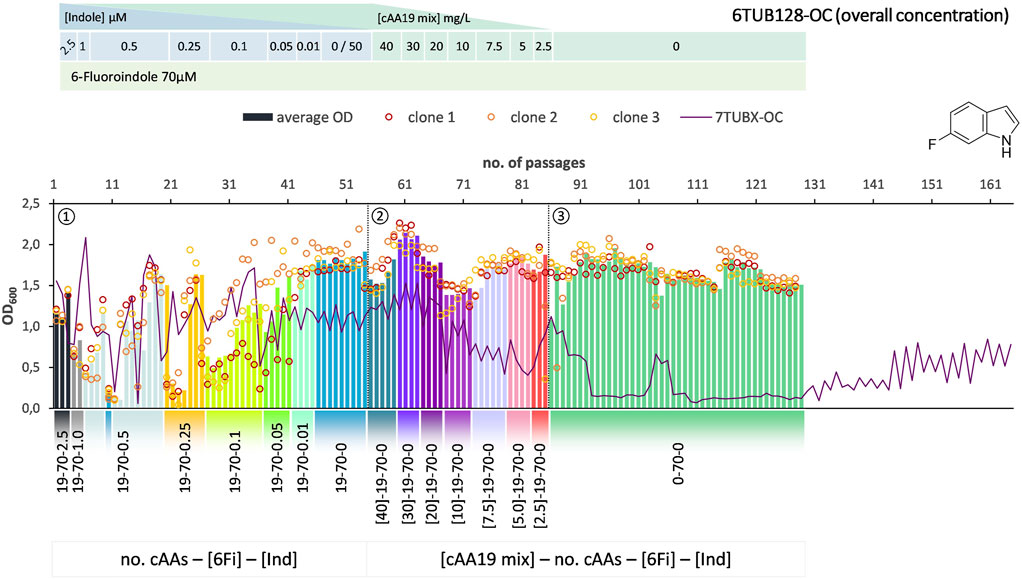
FIGURE 3. Cultivation scheme of the 6TUB128-OC lineage. Optical density (OD600) is plotted against the number of passages (reinoculation steps); average OD600 is shown as bars and individual clones are shown as circles. The corresponding 7TUBX-OC counterpart is depicted as purple line. ALE phases ①, ②, ③ and medium composition are illustrated; in the OC approach the concentration (mg/L) of the cAA19 mix is indicated in squared brackets [cAA conc.]-a-b-c.
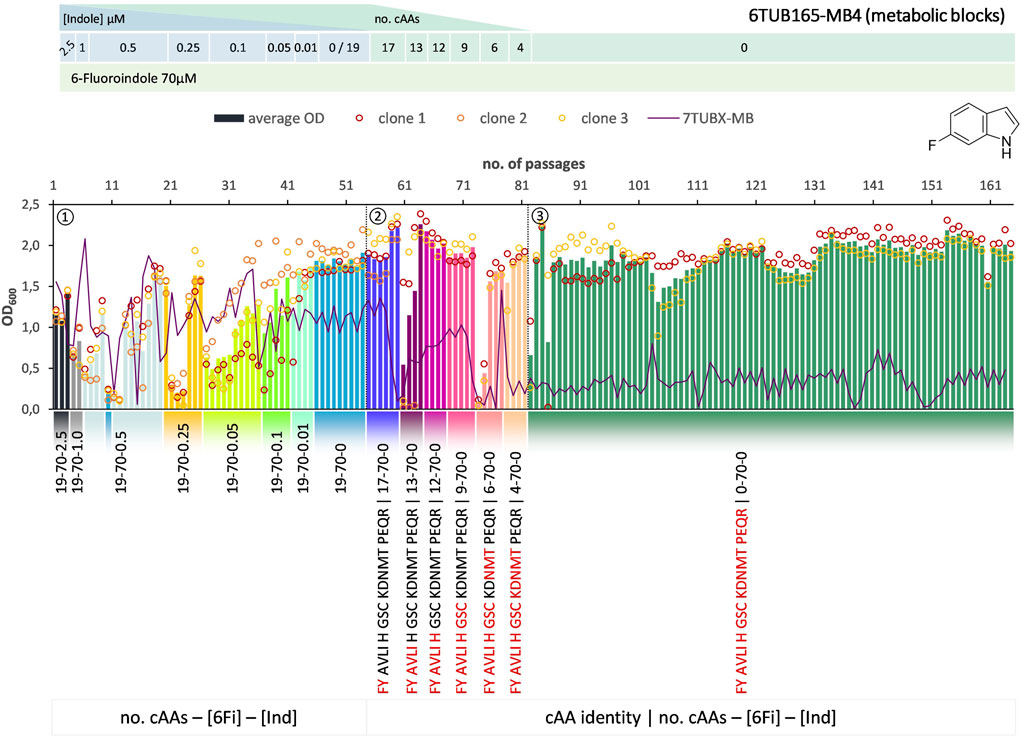
FIGURE 4. Cultivation scheme of the 6TUB165-MB4 lineage. Optical density (OD600) is plotted against the number of passages (reinoculation steps); average OD600 is shown as bars and individual clones are shown as circles. The corresponding 7TUBX-MB counterpart is depicted as purple line. ALE phases ①, ②, ③ and medium composition are illustrated; in the MB approach the amino acid composition is shown where red letters representing removed cAAs.
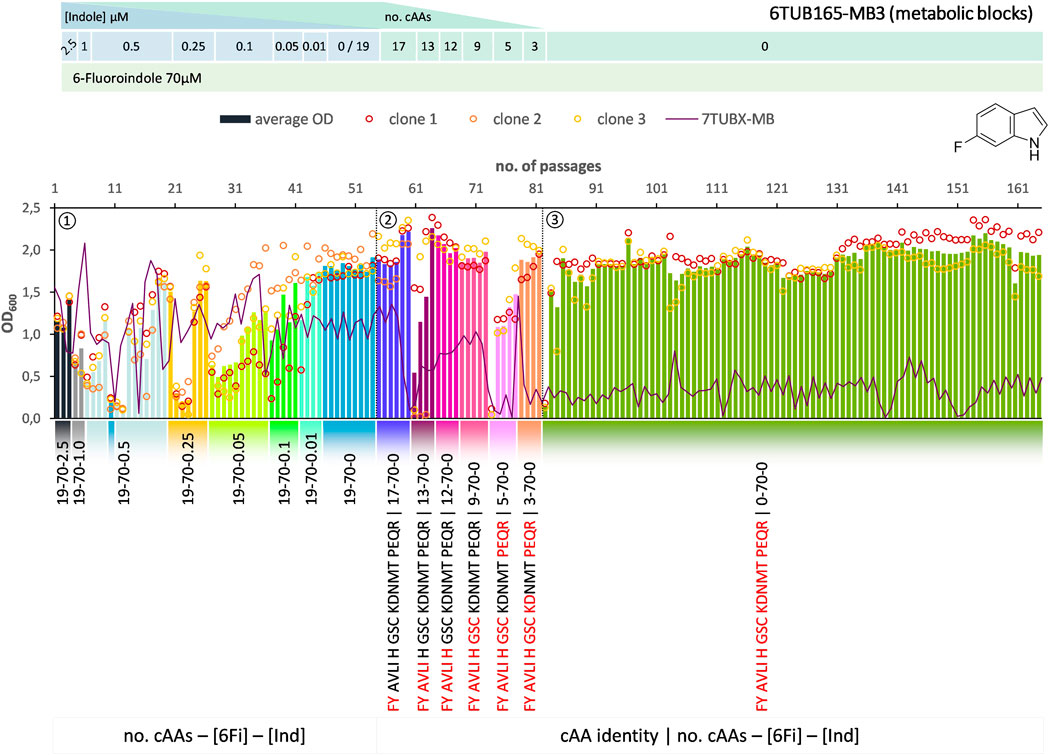
FIGURE 5. Cultivation scheme of the 6TUB165-MB3 lineage. Optical density (OD600) is plotted against the number of passages (reinoculation steps); average OD600 is shown as bars and individual clones are shown as circles. The corresponding 7TUBX-MB counterpart is depicted as purple line. ALE phases ①, ②, ③ and medium composition are illustrated; in the MB approach the amino acid composition is shown where red letters representing removed cAAs.
Surprisingly, the OC and MB approaches give approximately equal results in terms of adaptation speed and success. Adaptation took place in a similar time frame and the different lineages show equally good growth behavior. The mutual phase 1 lasted 53 passages, whereas the cells adapted via the OC approach took 31 passages in phase 2 and those for the MB approach (MB4 and MB3 cells) took 28 passages (Figures 3–5). When the amino acid supply in the MB-lineage was almost depleted (9 cAAs left, at passage 73) we tried the removal of different metabolic blocks, which yielded separate lineages 6TUB165-MB4 (Figure 4) and 6TUB165-MB3 (Figure 5). Although the cultivation schemes of the MB-lineages show large variations in cell densities upon removing the amino acid blocks “AVLI” and “NMT” (MB4) as well as “PEQR” (MB3), even removing different blocks was useful here, and the cells recovered very quickly.
Furthermore, both MB-lineages began to prefer 6Fi and to reject the non-fluorinated counterpart, because it was found that they grew better in 6Fi-supplemented NMM than in indole-supplemented NMM (Supplementary Figure S10). This behavior was not observed in clones of the 6TUBX-OC lineage; thus, its cultivation was terminated after 128 passages (in contrast to the 165 passages of the 6TUBX-MB lineages). In the course of a subpopulation screening (details described in the Supplementary Material), we selected five promising clones and started an additional ALE experiment with increased selection pressure by means of increased 6Fi concentration, from 70 µM to 85 µM to 100 µM, in order to provoke this phenotype. Ultimately, our goal was to create organisms that exhibit inherent adaptation to fluorine, aiming for cells that go beyond being merely facultative FTrp users. Unfortunately, this phenotype did not become dominant.
3.3 Adaptive laboratory evolution experimental setup for 7-fluoroindole (7Fi)
In contrast to 6Fi, tolerance towards 7Fi appears to be significantly more challenging as indicated by the low population sizes especially during the late adaptation process (Figures 6, 7). Already in phase 1, a high clonal variation in the acceptance to the growth conditions is observed; however, in sum similar high OD600 values were sustained as in the same phase of the 6Fi adaptation experiments, indicated by the 6TUBX-OC/MB trace (green line) in Figures 6, 7. In phase 2 cell density decreased continuously in both lineages (OC and MB). We hypothesize that this resulted from the low-grade toxicity of 7Fi that became increasingly difficult for cells to compensate while undergoing cAA starvation. In case of the MB lineage, we alternatively tried the removal of different amino acid as well as performed replay experiments (Blount et al., 2018) with the same setup starting from passages 72 when growth was still acceptable to investigate if the sudden drop in the cell density is reproducible. All such attempts ended with drastic reductions in cell population.
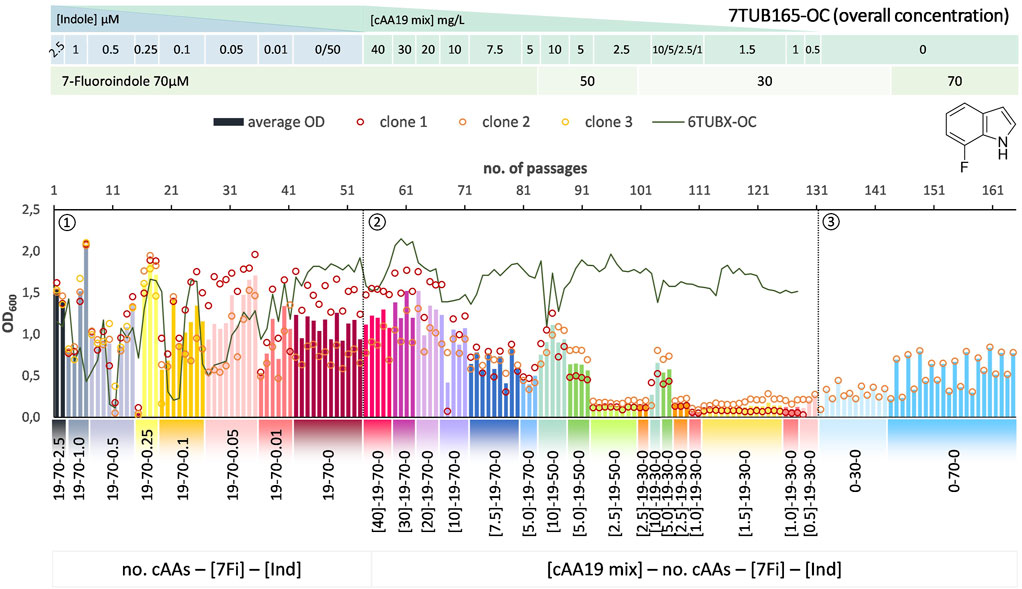
FIGURE 6. Cultivation scheme of the 7TUB165-OC lineage. Optical density (OD600) is plotted against the number of passages (reinoculation steps); average OD600 is shown as bars and individual clones are shown as circles. The corresponding 6TUBX-OC counterpart is depicted as green line. ALE phases ①, ②, ③ and medium composition are illustrated; in the OC approach the concentration (mg/L) of the cAA19 mix is indicated in squared brackets [cAA conc.]-a-b-c. Note the relaxed growth conditions by means of increased amino acid supply and reduced 7Fi concentration, depicted in the legends.
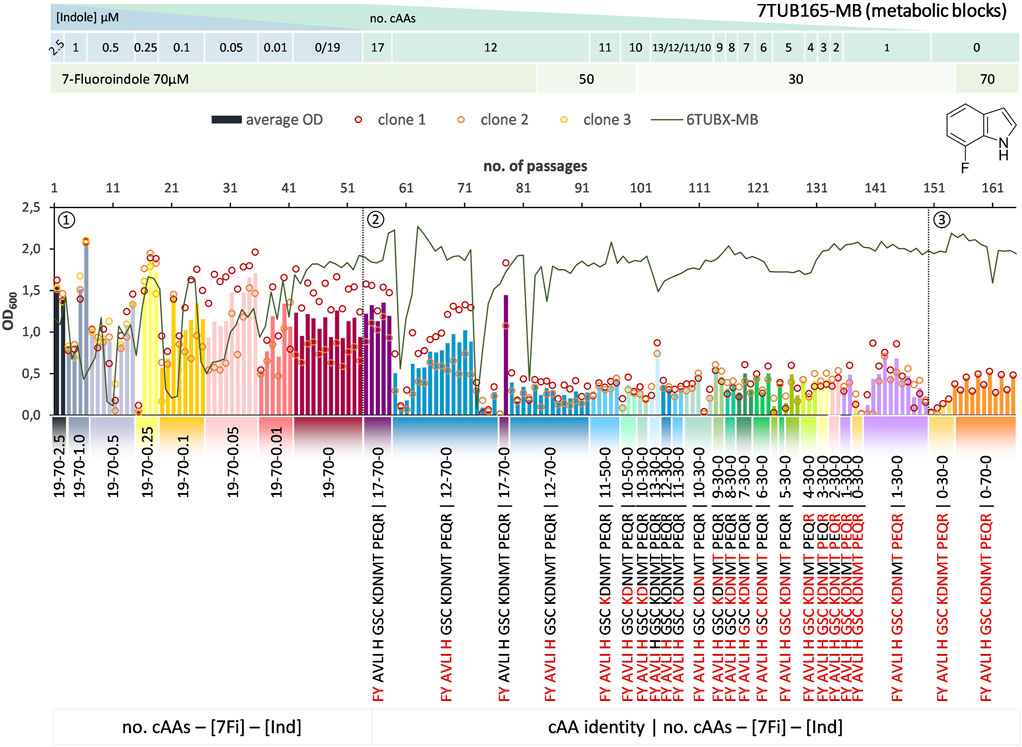
FIGURE 7. Cultivation scheme of the 7TUB165-MB lineage. Optical density (OD600) is plotted against the number of passages (reinoculation steps); average OD600 is shown as bars and individual clones are shown as circles. The corresponding 6TUBX-MB counterpart (mean of MB4 and MB3) is depicted as green line. ALE phases ①, ②, ③ and medium composition are illustrated; in the MB approach the amino acid composition is shown where red letters representing removed cAAs. Note the relaxed growth conditions by means of increased amino acid supply and reduced 7Fi concentration, depicted in the legends.
In order to rescue the cells, we reduced the concentration of 7Fi while temporarily increasing the cAA supply both starting with passage 84 (displayed in the legends of Figures 6, 7). Finally, the cells slowly recovered upon the relaxed selection pressure and the concentration of 7Fi could again be raised to 70 µM. Still, this process demonstrated that cAAs had to be removed extremely conservatively (compared to 6Fi): in smaller steps in the OC approach, and not at all as metabolic blocks in the MB approach, but rather removed on an individual basis. Even then not all clones survived the treatment, and the population sizes were considerably lower compared to the 6Fi adaptations. Taken together, this underlines the great challenge for E. coli to accept and adapt to 7Fi. Based on the described hurdles, a noteworthy difference between the OC and the MB approach is difficult to be determined. The OC and MB approaches both produced functionally adapted cells, however, from an experimental point of view the OC approach was easier to implement.
3.4 Growth characteristics of evolved strains
We conducted extensive growth experiments of all strains in different media that reflect the conditions along the adaptation trajectory (Figure 8B, upper panel). Overall, the impression about the adaptation success gained from the cultivation schemes discussed above (Figures 3–7) is reflected in the growth curves (Figure 8A). 6Fi adapted cells show better growth performance than 7Fi adapted cells. The final isolates of the 6Fi adapted cells exhibit similar growth behaviors to one another in their final growth medium (NMM0-70-0), although 6TUB165-MB4 appears to be the best adapted lineage. In contrast, the 7Fi cells exhibit a significant increased lag time and decreased growth rate, which is more pronounced in the 7TUB165-MB strain, which also shows a lower maximum OD600 value.
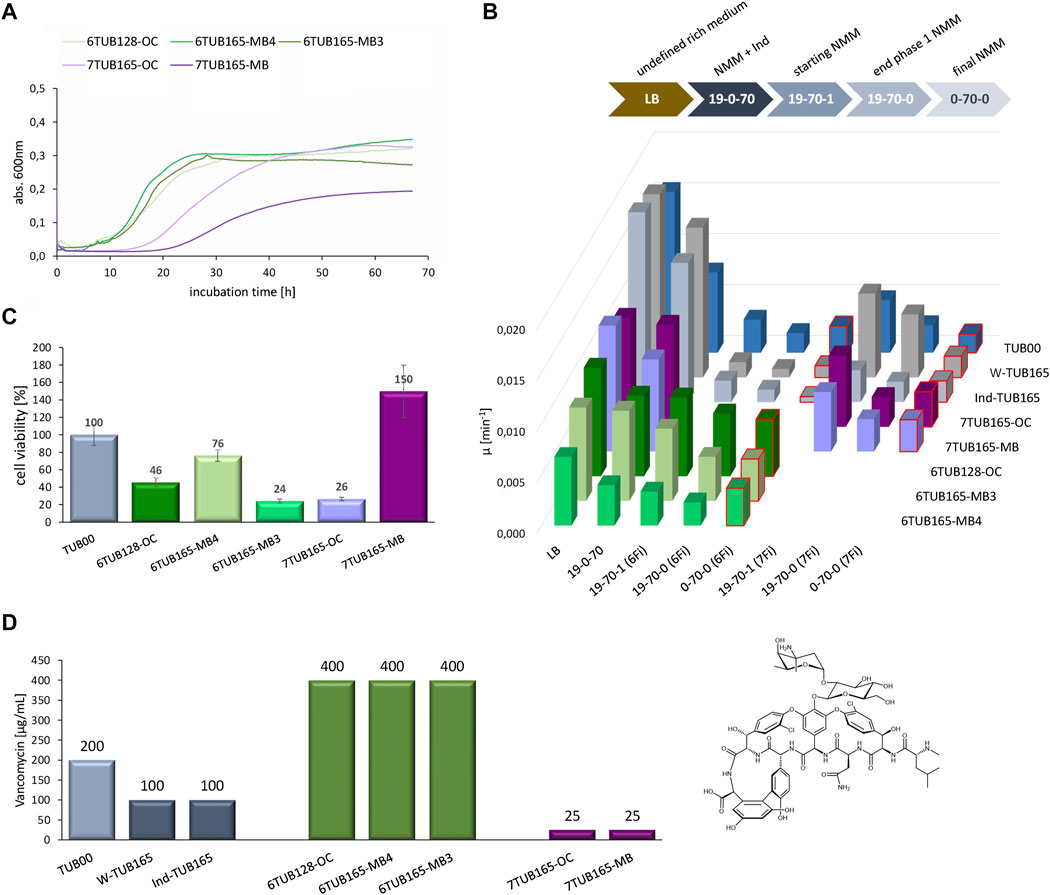
FIGURE 8. Growth characteristics of adapted strains (6TUB128-OC, 6TUB165-MB4, 6TUB165-MB3, 7TUB165-OC, 7TUB165-MB and W-TUB165, Ind-TUB165) in comparison to the ancestral TUB00. (A) Entire growth curves of final isolates of 6Fi adapted (shades of green) and 7Fi adapted (shades of purple) strains measured by growth in their final medium composition NMM0-70-0 (70 µM 6Fi or 7Fi, respectively). (B) The specific growth rate (µ) rate was calculated from the slope of exponential phase using the program GrowthRates (Hall et al., 2013); values obtained in final medium composition are red framed and characteristics of the different growth media are depicted above. (C) Cell viability was determined using CCK-8 assay (Cell Counting Kit-8); values of F-adapted strains were normalized to TUB00 (set 100%). (D) Investigation of cell membrane integrity by antibiotic susceptibility testing. The minimal inhibitory concentrations (MIC) of vancomycin (right structure) are shown for the ancestral TUB00 (light grey), two positive controls W-TUB165 and Ind-TUB165 (grey), 6Fi adapted strains 6TUB128-OC, 6TUB165-MB4 and 6TUB165-MB3 (green) as well as 7Fi adapted strains 7TUB165-OC and 7TUB165-MB (purple). Cells were treated with a dilution series of vancomycin (0–400 μg/mL) and growth after 24 h incubation at 30°C, 200 rpm is shown.
Furthermore, Figure 8B provides an overview of the growth of all investigated strains along the adaptation course given by calculation of the specific growth rate (Hall et al., 2013) (for comprehensive view see detailed growth curves in Supplementary Figures S11A–H). Generally, the growth correlates with the concentration of provided nutrients. Interestingly, the ancestral TUB00 tolerates 7Fi better than 6Fi, although based on the cultivation schemes (Figures 6, 7) we had considered 7Fi to be more toxic. The growth performance of the positive controls in NMM19-0-70 approaches that in LB (Supplementary Figures S11B, C), which is comprehensible because these strains were adapted to grow on minimal medium.
Among the 6Fi adapted cells, the OC lineage exhibits the highest specific growth rate in all tested media. However, as also seen in the MB3 lineage, there is no further growth improvement observable after the transition from NMM19-70-0 to NMM0-70-0. Remarkably, the MB4 lineage displays such enhanced growth behavior. Although its growth rates are lower compared to both other lineages, in 6Fi supplemented NMM, the highest growth rate is observed in the final medium NMM0-70-0, which is only surpassed by growth in the ancestral medium. This finding is even more evident in the entire growth curves (Supplementary Figures S11D–F). This observation strongly indicates a fluoroindole preferring phenotype, which is also supported by the subpopulation screening results.
Both 7Fi-adapted strains show decreased growth rates along with reducing nutrient supply (cAAs, Ind). Indole supplemented media enables enhanced growth in the absence of 7Fi (NMM19-0-70 > NMM19-70-1), which is a clear signal for an incomplete adaptation, as these cells still strongly prefer indole over 7Fi. However, the growth performance in 7Fi supplemented medium with and without cAAs addition (NMM19-70-0, NMM0-70-0) is equal, suggesting that removal of the canonical amino acids is well accepted.
We completed the picture of bacterial fitness by determination of the cell viability using a CCK-8 assay (Figure 8C), which measures the bio-reduction of tetrazolium salts (WST-8) (Yang et al., 2021). The viability of all 6Fi adapted lineages correlates very well with the growth parameters, whereas among the 7Fi adapted strains there is a discrepancy found for 7TUB165-MB. This strain shows a cell viability of 150% compared to TUB00, although this seems inconsistent with growth curves and parameters.
Additionally, we investigated the membrane integrity of the adapted cells through determination of the susceptibility towards the antibiotic vancomycin, which is inactive against Gram-negative E. coli because the intrusion through the (intact) outer membrane is blocked (Figure 8D, Supplementary Figure S12). Compared to TUB00 (MIC 200 μg/mL) the minimal inhibitory concentration of the positive controls (MIC 100 μg/mL) shows increased vancomycin sensitivity. This may be due to temperature: on the one hand, membrane permeability in general is influenced by temperature, and on the other hand, cold stress is known to increase E. coli’s susceptibility to vancomycin (Stokes et al., 2016). The 7Fi adapted cells show a strongly increased vancomycin sensitivity (MIC 25 μg/mL), which could be attributed to a membrane rearrangement (Agostini et al., 2021). Interestingly, the strains adapted to 6Fi exhibit the opposite effect being highly resistant to vancomycin treatment (MIC 400 μg/mL). A potential explanation could be the accumulation of phosphatidic acid, a standard membrane component. However, it is assumed that an increased phosphatidic acid level hinders the penetration of vancomycin and thus causes resistance (Sutterlin et al., 2014). Certainly, additional research is needed to explore this hypothesis in the context of the adapted strains.
3.5 Proteome-wide replacement of Trp by 6FTrp or 7FTrp
Global substitution of Trp for FTrp was shown by protein nano-electrospray-ionization-mass-spectrometry coupled with liquid chromatography (nLC-ESI-MS/MS) within the limits of the method. Trp-containing peptides were exclusively detected in TUB00 and both positive controls, while FTrp-containing peptides were only present in the isolates of adapted strains (Supplementary Figure S9). Despite of our careful experimental setup (strict prevention of Trp import in the growth medium and Fi substrates), the following background Trp incorporation was detected. Occurrence of FTrp-containing peptides in positive controls: TUB00 (0.0225% ± 0.0318), Ind-TUB165 (0.0878% ± 0.0098), W-TUB165 (0.0485% ± 0.0485), and occurrence of Trp-containing peptides in adapted strains: 6TUB128-OC (0.2309% ± 0.1315), 6TUB165-MB4 (0.1702% ± 0.0555), 6TUB165-MB3 (0.1269% ± 0.1269), 7TUB165-OC (0.3984% ± 0.0822) and 7TUB165-MB (0.5154% ± 0.1173); the mean of three biological replicates is reported, respectively. These values are within the false discovery rate threshold of 1% for peptide identification of the method.
Furthermore, the 6Fi and 7Fi adapted strains were tested for their robustness in driving recombinant protein expression (Kuthning et al., 2016). Two different variants of the green fluorescent protein (GFP) model protein were used for heterologous expression: 1) the enhanced green fluorescent protein EGFP, which harbors one single Trp residue at position 57 in its primary sequence, and 2) the enhanced cyan fluorescent protein ECFP, which additionally contains a Trp in the chromophore at position 66 (cartoons in Figure 9).
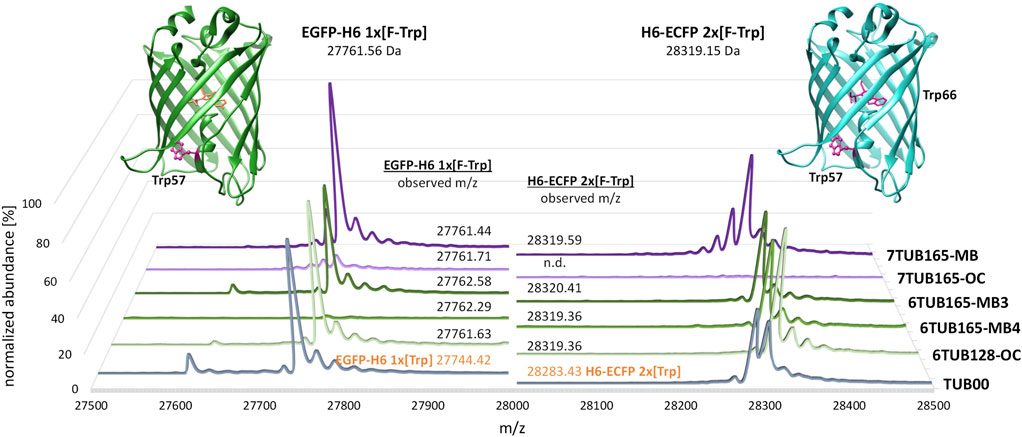
FIGURE 9. Deconvoluted mass spectra of C-terminal His-tagged enhanced green fluorescent protein (EGFP-H6, left) and N-terminal His-tagged enhanced cyan fluorescent protein (H6-ECFP, right). MS spectra of expression products of 6Fi adapted strains (6TUB128-OC, 6TUB165-MB4 and 6TUB165-MB3) are colored in green and respective spectra of proteins expressed with 7Fi adapted strains (7TUB165-OC and 7TUB165-MB) are colored in purple. EGFP-H6 and H6-ECFP expressed in TUB00 are colored in grey and have calculated masses of 27,743.79 Da and 28,283.38 Da, respectively. In addition to the dominant mass of the full-length protein, the spectra of the EGFPs show a second signal assigned to a truncated product after processing of the initial methionine (masses: 27,630 Da for FTrp-EGFP and 27,613 Da for Trp-EGFP, Δm/z −131 Da). The structures of EGFP (PDB ID 2Y0G) (Royant and Noirclerc-Savoye, 2011) and ECFP (PDB ID 2WSN) (Lelimousin et al., 2009) from Aequorea victoria were drawn using UCSF Chimera (Pettersen et al., 2004); Trp-positions are highlighted.
Cells of the final isolates of all adapted strains (6TUB128-OC, 6TUB165-MB4, 6TUB165-MB3 and 7TUB165-OC, 7TUB165-MB) and TUB00 were transformed with a plasmid bearing the sequence of either His-tagged EGFP (C-terminal EGFP-H6) or ECFP (N-terminal H6-ECFP). The recombinant protein expression was performed under the same conditions as used for adaptation, meaning cultivation in NMM0-70-0 containing either 6Fi or 7Fi at 30 °C; followed by isolation and purification using Ni-NTA affinity chromatography and analysis via LC-ESI-MS (Supplementary Figure S13 and Supplementary Table S6).
Almost all adapted strains support recombinant expression of GFP variants in which all Trp residues are quantitatively replaced by fluorinated counterparts. Mass spectrometric analyses of 6FTrp- and 7FTrp-substituted EGFP and ECFP show homogeneous mass profiles associated with an expected mass shift of 18 Da (which correlates with a H-to-F exchange on the aromatic ring of the indole). The expected theoretical values for translated EGFP and ECFP after chromophore maturation (Lundqvist et al., 2019) are consistent with the observed masses (Figure 9). However, the amount of 6FTrp-tagged EGFP expressed with 6TUB165-MB4 was very low; and the 7TUB165-OC strain was either able to produce only small amounts of 7FTrp-EGFP or was even unable to form 7FTrp-ECFP at all. The successful expression confirmed that in the adapted strains protein synthesis was also reprogrammed as Trp was completely replaced by 6FTrp or 7FTrp, respectively.
4 Discussion
We conducted ALE experiments encompassing the adaptation of an auxotrophic metabolic prototype in the presence of the antimetabolites 6Fi and 7Fi. This resulted in five differently adapted independent strains that grew stably for over 400 days, 165 passages and about 1,000 generations (Supplementary Table S5). These relinquished their dependence on canonical Trp and instead acquired the ability to use the fluorinated counterparts for all cellular processes. This convincingly demonstrates once again the feasibility of assimilation of fluoroindoles and trophic reassignment of the UGG codon by fluorinated tryptophan, the reproducibility of our previous study on the adaptation towards 4Fi and 5Fi (Agostini et al., 2021) as well as of the ALE process itself.
The ALE process was expanded to include, for the first time, two positive controls long-term cultivated on either Trp or indole, as well as a new approach for amino acid removal. The positive controls will allow us to distinguish between general consequences of long-term propagation in minimal medium and those that are due to the presence of fluorine. They can also be consulted for our other Trp-based ALEs (Hoesl et al., 2015; Agostini et al., 2021) since we conserved the same adaptation setup. Our novel OC approach provides an excellent alternative for adaptation of challenging substrates and might be more suitable to apply for already stressed cells, as shown in case of 7-fluoroindole. In our opinion, this straightforward technique is more comfortable in laboratory handling but in addition we believe that the metabolic burden to the cells of biosynthesizing missing amino acids is reduced with this method (compared to the MB approach). However, we refrain from favoring one approach over the other, as both have yielded the desired adaptation, despite variations in cellular outcomes. For instance, while the lineage 7TUB165-OC exhibits notably enhanced growth compared to 7TUB165-MB, an opposite trend is observed concerning recombinant protein expression capacity.
Based on the adaptation process itself and subsequent characterization experiments we observed considerable differences on the acceptance for 6Fi and 7Fi. Whereas 6Fi is unambiguously better tolerated by E. coli, 7Fi presents a considerable challenge evident through the intricate nature of the adaptation process and the compromised growth performance (Supplementary Figures S7, S11 and Supplementary Table S5). We attribute these differences to the physicochemical properties of the constitutional isomers, such as polarity and lipophilicity (Agostini et al., 2021).
Contrary to 4-, 5- and 6-fluoroindole/Trp, the Ind/Trp isomer fluorinated at position 7 is so far rather underrepresented in studies on biosynthetically incorporation and only a few have investigated the differences of all four isomers in specific proteins in direct comparison (Budisa et al., 2004; Kenward et al., 2018; Tobola et al., 2018; Zlatopolskiy et al., 2018). Why these isomers have not made much of an appearance remains speculative, however, the chemical synthesis and use of both 7Fi (Allen et al., 1955) and 7FTrp (Nevinsky et al., 1974; Lee and Phillips, 1991) was already described many decades ago. However, the proteome-wide insertion of 7-fluorotryptophan (by cellular conversion of 7-fluoroindole) described here represents the first study of its kind involving this ncAA.
Phenotypic analysis showed that the 6Fi adapted strains emerged a subpopulation that had started to become nearly independent of indole and even prefer 6Fi, approaching full xenobiotic “addiction”. Equally astonishing was the finding that all 6Fi adapted lineages showed an increased resistance to vancomycin, which is unique among all other 4-, 5-, (Agostini et al., 2021) and 7Fi adapted strains.
We demonstrated proteome-wide substitution of Trp by FTrp for all lineages, and that our fluorine adapted strains can drive overexpression of fully FTrp substituted proteins as we showed for two types of enhanced GFPs. This opens up opportunities to overcome limitations of automated solid-phase peptide synthesis (SPPS) by harnessing the ribosomal synthesis of adapted cells (Völler et al., 2017). However, the application of classical molecular biology methods such as amber stop codon suppression (SCS) and selective pressure incorporation (SPI) (Hoesl and Budisa, 2012) also benefit from the abolished dependence on canonical amino acids. However, although E. coli had acquired the competence to use the fluorinated substrates as integrated metabolic intermediates, the adapted strains remained (still) facultative fluorotryptophan/tryptophan users.
Although largely ignored by nature, a life based on fluorine is both an interesting concept and an absolutely conceivable scenario (Budisa et al., 2014). Fluorine-containing building blocks have been extensively used to investigate and modify proteins and their interactions (Marsh and Suzuki, 2014; Berger et al., 2017; Monkovic et al., 2022). However, the adaptation of living organisms represents a step forward in exploring and understanding the consequences of global fluorination. At this point, we would like to highlight the outstanding achievements of the lineages we report here to cope with the myriad consequences associated with the exchange of a hydrogen against a fluorine atom in an essential building block (note the existence of more than 20,000 Trp positions only in the proteome) (Tolle et al., 2023). Ultimately, this affects the complex interplay of cellular processes that include not only protein interactions through incorporated FTrp such as π-π stacking, hydrogen bonding, and cation-π interactions (Barik, 2020), but also all metabolic processes to which Ind as signaling molecule (Zarkan et al., 2020) and Trp or downstream metabolites contribute to.
Even a cursory comparison of the cultivation schemes of all four substrates shows parallels in the adaptation behavior of 4Fi and 6Fi, which are well tolerated, while 5Fi and 7Fi present adaptation challenges. Since with this study, the set of E. coli bacteria adapted to all mono-fluorinated indoles (4-, 5-, 6- and 7Fi) is complete we can now investigate the influence of the position of the F-substituent on cellular adaptability, metabolic pathways, regulatory networks, and protein folding quality. Ongoing experiments in our labs focus on the different biological information levels (DNA, RNA, proteins and metabolites) by means of multi-OMICs approaches that could unravel a rationale behind adaptation towards xenobiotics and fluorine in particular. Enlightening the responsible mechanisms of how a former antimetabolite was incorporated into biomass will push advancements in scientific fields such as synthetic biology and biotechnology, but also across the board of environmental concerns such as bioremediation and biocontainment (Atashgahi et al., 2018).
Data availability statement
Proteomics data have been deposited on ProteomeXchange, https://proteomecentral.proteomexchange.org/. Project Name: Adaptive Laboratory Evolution of E. coli towards usage of fluoroindole. Accession number: PXD048225.
Author contributions
CT-K: Conceptualization, Data curation, Investigation, Methodology, Visualization, Writing–original draft. AB: Supervision, Writing–review and editing. LA: Data curation, Methodology, Writing–review and editing. NB: Conceptualization, Project administration, Supervision, Writing–review and editing. BK: Conceptualization, Project administration, Supervision, Writing–review and editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
A preprint of the article has been published online on BioRxiv and can be found at doi: https://doi.org/10.1101/2023.09.25.559291
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsybi.2023.1345634/full#supplementary-material
References
Agostini, F., Sinn, L., Petras, D., Schipp, C. J., Kubyshkin, V., Berger, A. A., et al. (2021). Multiomics analysis provides insight into the laboratory evolution of Escherichia coli toward the metabolic usage of fluorinated indoles. ACS Cent. Sci. 7, 81–92. doi:10.1021/acscentsci.0c00679
Akashi, H., and Gojobori, T. (2002). Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 99, 3695–3700. doi:10.1073/pnas.062526999
Allen, F. L., Brunton, J. C., and Suschitzky, H. (1955). Heterocyclic fluorine compounds. Part II. Bz-Monofluoroindoles. J. Chem. Soc., 1283–1286. doi:10.1039/jr9550001283
Atashgahi, S., Sánchez-Andrea, I., Heipieper, H. J., van der Meer, J. R., Stams, A. J. M., and Smidt, H. (2018). Prospects for harnessing biocide resistance for bioremediation and detoxification. Science 360, 743–746. doi:10.1126/science.aar3778
Azim, M. K., and Budisa, N. (2008). Docking of tryptophan analogs to trytophanyl-tRNA synthetase: implications for non-canonical amino acid incorporations. Biol. Chem. 389, 1173–1182. doi:10.1515/BC.2008.133
Bacher, J. M., and Ellington, A. D. (2001). Selection and characterization of Escherichia coli variants capable of growth on an otherwise toxic tryptophan analogue. J. Bacteriol. 183, 5414–5425. doi:10.1128/jb.183.18.5414-5425.2001
Barik, S. (2020). The uniqueness of tryptophan in biology: properties, metabolism, interactions and localization in proteins. Int. J. Mol. Sci. 21, 8776. doi:10.3390/ijms21228776
Berger, A. A., Völler, J. S., Budisa, N., and Koksch, B. (2017). Deciphering the fluorine code - the many hats fluorine wears in a protein environment. Acc. Chem. Res. 50, 2093–2103. doi:10.1021/acs.accounts.7b00226
Blount, Z. D., Lenski, R. E., and Losos, J. B. (2018). Contingency and determinism in evolution: replaying life’s tape. Sci. 362, eaam5979. doi:10.1126/science.aam5979
Budisa, N., Kubyshkin, V., and Schulze-Makuch, D. (2014). Fluorine-rich planetary environments as possible habitats for life. Life 4, 374–385. doi:10.3390/life4030374
Budisa, N., Pal, P. P., Alefelder, S., Birle, P., Krywcun, T., Rubini, M., et al. (2004). Probing the role of tryptophans in Aequorea victoria green fluorescent proteins with an expanded genetic code. Biol. Chem. 385, 191–202. doi:10.1515/BC.2004.038
Budisa, N., Steipe, B., Demange, P., Eckerskorn, C., Kellermann, J., and Huber, R. (1995). High-level biosynthetic substitution of methionine in proteins by its analogs 2-aminohexanoic acid, selenomethionine, telluromethionine and ethionine in Escherichia coli. Eur. J. Biochem. 230, 788–796. doi:10.1111/j.1432-1033.1995.tb20622.x
Calero, P., Volke, D. C., Lowe, P. T., Gotfredsen, C. H., O’Hagan, D., and Nikel, P. I. (2020). A fluoride-responsive genetic circuit enables in vivo biofluorination in engineered Pseudomonas putida. Nat. Commun. 11, 5045. doi:10.1038/s41467-020-18813-x
Carvalho, M. F., and Oliveira, R. S. (2017). Natural production of fluorinated compounds and biotechnological prospects of the fluorinase enzyme. Crit. Rev. Biotechnol. 37, 880–897. doi:10.1080/07388551.2016.1267109
Connolly, J. P. R., Roe, A. J., and O’Boyle, N. (2021). Prokaryotic life finds a way: insights from evolutionary experimentation in bacteria. Crit. Rev. Microbiol. 47, 126–140. doi:10.1080/1040841X.2020.1854172
Dall’Angelo, S., Bandaranayaka, N., Windhorst, A. D., Vugts, D. J., van der Born, D., Onega, M., et al. (2013). Tumour imaging by Positron Emission Tomography using fluorinase generated 5-[¹⁸F]fluoro-5-deoxyribose as a novel tracer. Nucl. Med. Biol. 40, 464–470. doi:10.1016/j.nucmedbio.2013.02.006
Deng, H., O’Hagan, D., and Schaffrath, C. (2004). Fluorometabolite biosynthesis and the fluorinase from Streptomyces cattleya. Nat. Prod. Rep. 21, 773–784. doi:10.1039/b415087m
Deutsch, E. W., Bandeira, N., Perez-Riverol, Y., Sharma, V., Carver, J., Mendoza, L., et al. (2023). The ProteomeXchange Consortium at 10 years: 2023 update. Nucleic Acids Res. 51 (D1), D1539–D1548.
Dragosits, M., and Mattanovich, D. (2013). Adaptive laboratory evolution - principles and applications in industrial biotechnology. Microb. Cell Fact. 12, 64. doi:10.1186/1475-2859-12-64
Eustáquio, A. S., O’Hagan, D., and Moore, B. S. (2010). Engineering fluorometabolite production: fluorinase expression in Salinispora tropica yields fluorosalinosporamide. J. Nat. Prod. 73, 378–382. doi:10.1021/np900719u
Goss, R. J. M., and Newill, P. L. A. (2006). A convenient enzymatic synthesis of L-halotryptophans. Chem. Commun., 4924–4925. doi:10.1039/b611929h
Gresham, D., and Dunham, M. J. (2014). The enduring utility of continuous culturing in experimental evolution. Genomics 104, 399–405. doi:10.1016/j.ygeno.2014.09.015
Gribble, G. W. (2003). The diversity of naturally produced organohalogens. Chemosphere 52, 289–297. doi:10.1016/S0045-6535(03)00207-8
Gribble, G. W. A. (2015). A recent survey of naturally occurring organohalogen compounds. Environ. Chem. 12, 396–405. doi:10.1071/en15002
Gupta, G. N., Srivastava, S., Khare, S. K., and Prakash, V. (2014). Extremophiles: an overview of microorganism from extreme environment. Int. J. Agric. Enviroment Biotechnol. 7, 371–379. doi:10.5958/2230-732x.2014.00258.7
Hall, B. G., Acar, H., Nandipati, A., and Barlow, M. (2013). Growth rates made easy. Mol. Biol. Evol. 31, 232–238. doi:10.1093/molbev/mst187
Harper, D. B., and OHagan, D. (1994). The fluorinated natural products. Nat. Prod. Rep. 11, 123–133. doi:10.1039/np9941100123
Hoesl, M. G., and Budisa, N. (2012). Recent advances in genetic code engineering in Escherichia coli. Curr. Opin. Biotechnol. 23, 751–757. doi:10.1016/j.copbio.2011.12.027
Hoesl, M. G., Oehm, S., Durkin, P., Darmon, E., Peil, L., Aerni, H. R., et al. (2015). Chemical evolution of a bacterial proteome. Angew. Chem. - Int. Ed. 54, 10030–10034. doi:10.1002/anie.201502868
Kenward, C., Shin, K., and Rainey, J. K. (2018). Mixed fluorotryptophan substitutions at the same residue expand the versatility of 19F protein NMR spectroscopy. Chem. Eur. J. 24, 3391–3396. doi:10.1002/chem.201705638
Kublik, A., Deobald, D., Hartwig, S., Schiffmann, C. L., Andrades, A., von Bergen, M., et al. (2016). Identification of a multi-protein reductive dehalogenase complex in Dehalococcoides mccartyi strain CBDB1 suggests a protein-dependent respiratory electron transport chain obviating quinone involvement. Environ. Microbiol. 18, 3044–3056. doi:10.1111/1462-2920.13200
Kuthning, A., Durkin, P., Oehm, S., Hoesl, M. G., Budisa, N., and Süssmuth, R. D. (2016). Towards biocontained cell factories: an evolutionarily adapted Escherichia coli strain produces a new-to-nature bioactive lantibiotic containing thienopyrrole-alanine. Sci. Rep. 6, 33447. doi:10.1038/srep33447
LaCroix, R. A., Palsson, B. O., and Feist, A. M. (2017). A model for designing adaptive laboratory evolution experiments. Appl. Environ. Microbiol. 83, e03115–e03116. doi:10.1128/AEM.03115-16
Lee, M., and Phillips, R. S. (1991). Synthesis and resolution of 7-fluorotryptophans. Bioorg. Med. Chem. Lett. 1, 477–480. doi:10.1016/s0960-894x(01)81109-4
Lelimousin, M., Noirclerc-Savoye, M., Lazareno-Saez, C., Paetzold, B., Le Vot, S., Chazal, R., et al. (2009). Intrinsic dynamics in ECFP and cerulean control fluorescence quantum yield. Biochemistry 48, 10038–10046. doi:10.1021/bi901093w
Lundqvist, M., Thalén, N., Volk, A.-L., Hansen, H. G., von Otter, E., Nygren, P.-Å., et al. (2019). Chromophore pre-maturation for improved speed and sensitivity of split-GFP monitoring of protein secretion. Sci. Rep. 9, 310. doi:10.1038/s41598-018-36559-x
Markakis, K., Lowe, P. T., Davison-Gates, L., O’Hagan, D., Rosser, S. J., and Elfick, A. (2020). An engineered E. coli strain for direct in vivo fluorination. ChemBioChem 21, 1856–1860. doi:10.1002/cbic.202000051
Marsh, E. N. G., and Suzuki, Y. (2014). Using 19F NMR to probe biological interactions of proteins and peptides. ACS Chem. Biol. 9, 1242–1250. doi:10.1021/cb500111u
Mat, W. K., Xue, H., and Wong, J. T. F. (2010). Genetic code mutations: the breaking of a three billion year invariance. PLoS One 5, e12206. doi:10.1371/journal.pone.0012206
Mavrommati, M., Daskalaki, A., Papanikolaou, S., and Aggelis, G. (2022). Adaptive laboratory evolution principles and applications in industrial biotechnology. Biotechnol. Adv. 54, 107795. doi:10.1016/j.biotechadv.2021.107795
Merino, N., Aronson, H. S., Bojanova, D. P., Feyhl-Buska, J., Wong, M. L., Zhang, S., et al. (2019). Living at the extremes: extremophiles and the limits of life in a planetary context. Front. Microbiol. 10, 780. doi:10.3389/fmicb.2019.00780
Merkel, L., and Budisa, N. (2012). Organic fluorine as a polypeptide building element: in vivo expression of fluorinated peptides, proteins and proteomes. Org. Biomol. Chem. 10, 7241–7261. doi:10.1039/c2ob06922a
Monkovic, J. M., Gibson, H., Sun, J. W., and Montclare, J. K. (2022). Fluorinated protein and peptide materials for biomedical applications. Pharmaceuticals 15, 1201. doi:10.3390/ph15101201
Nevinsky, G. A., Favorova, O. O., Lavrik, O. I., Petrova, T. D., Kochkina, L. L., and Savchenko, T. I. (1974). Fluorinated tryptophans as substrates and inhibitors of the ATP-(32P)PPi exchange reaction catalysed by tryptophanyl tRNA synthetase. FEBS Lett. 43, 135–138. doi:10.1016/0014-5793(74)80985-3
O’Hagan, D., and Deng, H. (2015). Enzymatic fluorination and biotechnological developments of the fluorinase. Chem. Rev. 115, 634–649. doi:10.1021/cr500209t
O’Hagan, D., Schaffrath, C., Cobb, S. L., Hamilton, J. T. G., and Murphy, C. D. (2002). Biosynthesis of an organofluorine molecule. Nature 416, 279. doi:10.1038/416279a
Perez-Riverol, Y., Bai, J., Bandla, C., Hewapathirana, S., García-Seisdedos, D., Kamatchinathan, S., et al. (2022). The PRIDE database resources in 2022: a Hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50 (D1), D543–D552.
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., et al. (2004). UCSF Chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. doi:10.1002/jcc.20084
Royant, A., and Noirclerc-Savoye, M. (2011). Stabilizing role of glutamic acid 222 in the structure of enhanced green fluorescent protein. J. Struct. Biol. 174, 385–390. doi:10.1016/j.jsb.2011.02.004
Sanada, M., Miyano, T., Iwadare, S., Williamson, J. M., Arison, B. H., Smith, J. L., et al. (1986). Biosynthesis of fluorothreonine and fluoroacetic acid by the thienamycin producer, Streptomyces cattleya. J. Antibiot. 39, 259–265. doi:10.7164/antibiotics.39.259
Sandberg, T. E., Salazar, M. J., Weng, L. L., Palsson, B. O., and Feist, A. M. (2019). The emergence of adaptive laboratory evolution as an efficient tool for biological discovery and industrial biotechnology. Metab. Eng. 56, 1–16. doi:10.1016/j.ymben.2019.08.004
Sangwan, N., Verma, H., Kumar, R., Negi, V., Lax, S., Khurana, P., et al. (2014). Reconstructing an ancestral genotype of two hexachlorocyclohexane-degrading Sphingobium species using metagenomic sequence data. ISME J. 8, 398–408. doi:10.1038/ismej.2013.153
Seidel, K., Kühnert, J., and Adrian, L. (2018). The complexome of Dehalococcoides mccartyi reveals its organohalide respiration-complex is modular. Front. Microbiol. 9, 1130. doi:10.3389/fmicb.2018.01130
Stokes, J. M., French, S., Ovchinnikova, O. G., Bouwman, C., Whitfield, C., and Brown, E. D. (2016). Cold stress makes Escherichia coli susceptible to glycopeptide antibiotics by altering outer membrane integrity. Cell Chem. Biol. 23, 267–277. doi:10.1016/j.chembiol.2015.12.011
Sutterlin, H. A., Zhang, S., and Silhavya, T. J. (2014). Accumulation of phosphatidic acid increases vancomycin resistance in Escherichia coli. J. Bacteriol. 196, 3214–3220. doi:10.1128/JB.01876-14
Tack, D. S., Ellefson, J. W., Thyer, R., Wang, B., Gollihar, J., Forster, M. T., et al. (2016). Addicting diverse bacteria to a noncanonical amino acid. Nat. Chem. Biol. 12, 138–140. doi:10.1038/nchembio.2002
Tobola, F., Lelimousin, M., Varrot, A., Gillon, E., Darnhofer, B., Blixt, O., et al. (2018). Effect of noncanonical amino acids on protein-carbohydrate interactions: structure, dynamics, and carbohydrate affinity of a lectin engineered with fluorinated tryptophan analogs. ACS Chem. Biol. 13, 2211–2219. doi:10.1021/acschembio.8b00377
Tolle, I., Oehm, S., Hoesl, M. G., Treiber-Kleinke, C., Peil, L., Bozukova, M., et al. (2023). Evolving a mitigation of the stress response pathway to change the basic chemistry of life. Front. Synth. Biol. 1, 1248065. doi:10.3389/fsybi.2023.1248065
Vasi, F., Travisano, M., and Lenski, R. E. (1994). Long-term experimental evolution in Escherichia coli. II. Changes in life-history traits during adaptation to a seasonal environment. Am. Nat. 144, 432–456. doi:10.1086/285685
Völler, J. S., Dulic, M., Gerling-Driessen, U. I. M., Biava, H., Baumann, T., Budisa, N., et al. (2017). Discovery and investigation of natural editing function against artificial amino acids in protein translation. ACS Cent. Sci. 3, 73–80. doi:10.1021/acscentsci.6b00339
Wang, Z., DeWitt, J. C., Higgins, C. P., and Cousins, I. T. (2017). A never-ending story of per- and polyfluoroalkyl substances (PFASs)? Environ. Sci. Technol. 51, 2508–2518. doi:10.1021/acs.est.6b04806
Watkins-Dulaney, E., Straathof, S., and Arnold, F. (2021). Tryptophan synthase: biocatalyst extraordinaire. ChemBioChem 22, 5–16. doi:10.1002/cbic.202000379
Wilcox, M. (1974). The enzymatic synthesis of L-tryptophan analogues. Anal. Biochem. 59, 436–440. doi:10.1016/0003-2697(74)90296-6
Wong, H. E., Huang, C.-J., and Zhang, Z. (2018). Amino acid misincorporation propensities revealed through systematic amino acid starvation. Biochemistry 57, 6767–6779. doi:10.1021/acs.biochem.8b00976
Wong, J. T. F. (1983). Membership mutation of the genetic code: loss of fitness by tryptophan. Proc. Natl. Acad. Sci. U. S. A. 80, 6303–6306. doi:10.1073/pnas.80.20.6303
Yang, X., Zhong, Y., Wang, D., and Lu, Z. (2021). A simple colorimetric method for viable bacteria detection based on cell counting Kit-8. Anal. Methods 13, 5211–5215. doi:10.1039/d1ay01624e
Yoshida, S., Hiraga, K., Takehana, T., Taniguchi, I., Yamaji, H., Maeda, Y., et al. (2016). A bacterium that degrades and assimilates poly(ethylene terephthalate). Sci. 351, 1196–1199. doi:10.1126/science.aad6359
Yu, A. C. S., Yim, A. K. Y., Mat, W. K., Tong, A. H. Y., Lok, S., Xue, H., et al. (2014). Mutations enabling displacement of tryptophan by 4-fluorotryptophan as a canonical amino acid of the genetic code. Genome Biol. Evol. 6, 629–641. doi:10.1093/gbe/evu044
Zarkan, A., Liu, J., Matuszewska, M., Gaimster, H., and Summers, D. K. (2020). Local and universal action: the paradoxes of indole signalling in bacteria. Trends Microbiol. 28, 566–577. doi:10.1016/j.tim.2020.02.007
Zlatopolskiy, B. D., Zischler, J., Schäfer, D., Urusova, E. A., Guliyev, M., Bannykh, O., et al. (2018). Discovery of 7-[18F]fluorotryptophan as a novel positron emission tomography (PET) probe for the visualization of tryptophan metabolism in vivo. J. Med. Chem. 61, 189–206. doi:10.1021/acs.jmedchem.7b01245
Keywords: adaptive laboratory evolution (ALE), fluoroindole, fluorotryptophan, xenonutrient, proteome-wide Trp fluorination, codon reassignment, synthetic life
Citation: Treiber-Kleinke C, Berger AA, Adrian L, Budisa N and Koksch B (2024) Escherichia coli adapts metabolically to 6- and 7-fluoroindole, enabling proteome-wide fluorotryptophan substitution. Front. Synth. Biol. 1:1345634. doi: 10.3389/fsybi.2023.1345634
Received: 28 November 2023; Accepted: 27 December 2023;
Published: 16 January 2024.
Edited by:
Chenguang Fan, University of Arkansas, United StatesReviewed by:
Noah Reynolds, University of Illinois at Springfield, United StatesJohn D. (Nick) Fisk, University of Colorado Denver, United States
Copyright © 2024 Treiber-Kleinke, Berger, Adrian, Budisa and Koksch. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Beate Koksch, YmVhdGUua29rc2NoQGZ1LWJlcmxpbi5kZQ==; Nediljko Budisa, bmVkaWxqa28uYnVkaXNhQHR1LWJlcmxpbi5kZQ==, bmVkaWxqa28uYnVkaXNhQHVtYW5pdG9iYS5jYQ==
 Christin Treiber-Kleinke
Christin Treiber-Kleinke Allison Ann Berger1
Allison Ann Berger1 Lorenz Adrian
Lorenz Adrian Nediljko Budisa
Nediljko Budisa Beate Koksch
Beate Koksch