
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Synaptic Neurosci. , 13 September 2022
Volume 14 - 2022 | https://doi.org/10.3389/fnsyn.2022.939793
This article is part of the Research Topic Synaptogenesis in Neuropathology View all 4 articles
Synapses are the basic units for information processing and storage in the nervous system. It is only when the synaptic connection is established, that it becomes meaningful to discuss the structure and function of a circuit. In humans, our unparalleled cognitive abilities are correlated with an increase in the number of synapses. Additionally, genes involved in synaptogenesis are also frequently associated with neurological or psychiatric disorders, suggesting a relationship between synaptogenesis and brain physiology and pathology. Thus, understanding the molecular mechanisms of synaptogenesis is the key to the mystery of circuit assembly and neural computation. Furthermore, it would provide therapeutic insights for the treatment of neurological and psychiatric disorders. Multiple molecular events must be precisely coordinated to generate a synapse. To understand the molecular mechanisms underlying synaptogenesis, we need to know the molecular components of synapses, how these molecular components are held together, and how the molecular networks are refined in response to neural activity to generate new synapses. Thanks to the intensive investigations in this field, our understanding of the process of synaptogenesis has progressed significantly. Here, we will review the molecular mechanisms of synaptogenesis by going over the studies on the identification of molecular components in synapses and their functions in synaptogenesis, how cell adhesion molecules connect these synaptic molecules together, and how neural activity mobilizes these molecules to generate new synapses. Finally, we will summarize the human-specific regulatory mechanisms in synaptogenesis and results from human genetics studies on synaptogenesis and brain disorders.
The word “synapse,” coined by Charles Sherrington, is used to describe the place at which communication occurs between neurons. The existence of the synapse was consolidated by electron microscopy after years of debate (De Robertis and Franchi, 1956; Sotelo, 2020). The study of synapses started with histology and was later joined by biophysics, molecular biology, and biochemistry. It is now well-recognized that there are two kinds of synapses in the nervous system: electrical and chemical synapses. The electrical synapses allow an ensemble of cells to be activated simultaneously, as compared to chemical synapses where there is a delay in the transmission of signals. There has been accumulating evidence supporting the importance of electrical synapses in physiological processes (Pereda, 2014), but here we will primarily review the molecular mechanisms of the synaptogenesis of chemical synapses. Synapses are composed of three parts: the pre-synaptic terminal, synaptic cleft, and post-synaptic compartment. Synapses transform the electrical signals into chemical signals and then into electric signals again. They help living animals survive their environments, are the targets of predators, and are often affected under diseases. Consequently, understanding the molecular genetic basis of synaptogenesis may help us understand how genes instruct the wirings of the nervous system and direct the flow of information (Colon-Ramos, 2009), how the environment shapes behaviors, and how diseases arise, especially in humans.
Synapse formation involves the interactions between neurons. As a result, neuron differentiation, migration, axon guidance, and dendrite morphogenesis can all influence the outcomes of synaptogenesis (Batool et al., 2019). Nevertheless, even if all these processes are accomplished, forming synapses still requires precise regulations (Chia et al., 2013). To gain insights into the underlying mechanisms required to generate a synapse, one needs to first know the components that constitute a synapse and how they are assembled together. Both pre- and post-synaptic compartments contain electron-rich structures which are named the “cytomatrix” and “post-synaptic density,” respectively. These structures consist of mostly scaffold proteins that are connected intercellularly by cell adhesion molecules and are anchored by the cytoskeleton intracellularly. Accordingly, forming synapses usually requires a few prerequisites. First, synaptic molecules must be assembled at pre- and post-synaptic sites instead of randomly in the cytosol. Second, pre- and post-synaptic compartments where these synaptic molecules dwell must be aligned and connected accurately by cell adhesion molecules. Third, the cytoskeleton must be organized and coordinated so as to support the synaptic structure. In addition, synapses are dynamic and can be influenced by neural activities. Thus, by manipulating neural activities, we can examine the molecular events that impact the destinies of synapses. Last but not least, synaptogenesis, though largely conserved, has been constantly reshaped during evolution (Schmidt and Polleux, 2021), which endows humans with unprecedented cognitive capabilities and provides avenues to develop treatments and therapies for neurological disorders and diseases.
To better parse the process of synaptogenesis, we have organized this review into four parts: (I) the assembly of intracellular synaptic proteins, most of which are scaffold proteins (Jin and Garner, 2008; Sheng and Kim, 2011); (II) cell adhesion molecules that bridge these proteins intercellularly (Sudhof, 2021); (III) the cytoskeleton system that supports the structure of the synapse; and (IV) neural activity that drives the formation of new synapse (Zito and Svoboda, 2002; Pan and Monje, 2020). These aspects coordinated with each other, were renovated during evolution, and, when gone awry, are frequently associated with impaired brain formation or cognitive abilities. We will review each of these aspects using some well-studied examples and summarize their relationships to the development of diseases.
To study the molecular mechanisms of synapse formation, the first step is to identify the molecules that constitute the synapses and examine their ways of assembly through protein interactions during synaptogenesis, because most of them are used to define synapses. Studies on synaptic transmission led to the observations that the pre-synaptic terminals contain ready-to-be-released vesicles docking on the “cytomatrix” and that post-synaptic compartment named post-synaptic density (PSD) contain “patches” that are electron-enriched (Phillips et al., 2001). Endeavors to purify synaptic proteins and examine their effects on synaptic transmission, led by Sudhof et al. (1993), discovered synaptic vesicle proteins such as synaptophysin, synapsin, synaptobrevin/VAMP 1/2, syntaxin, and SNAP-25 as well as their functions in synaptic transmission. Although genetic evidence from knockout mice suggests that most of them have no effects on the structure of synapses (Rosahl et al., 1995; Ahnert-Hilger et al., 1996; Fujiwara et al., 2006; Kwon and Chapman, 2011), they have laid the foundation for further probing the building blocks of synapses that enable the vesicles to dock. Advances in biochemistry, molecular biology, immunology, and other fields such as imaging (Fortin et al., 2014) have provided versatile approaches to dissect the building blocks of synaptic components and describe the protein meshwork for synaptic transmission, transforming the search into a more feasible and fruitful field of study (Sudhof, 1995; De Camilli and Takei, 1996).
One approach is genetic screening. For example, using Synaptobrevin/VAMP fused with GFP (Hallam and Jin, 1998; Nonet, 1999), Zhen and Jin (1999) performed screenings on C. elegans regarding the localization of synaptic vesicles and discovered Liprin-α which was also named syd-2 (short for synaptic defective). Liprin-α encodes a synaptic scaffold protein and was first identified through the yeast-two-hybridization as a binding protein for the leukocyte common antigen-related protein (LAR) transmembrane protein (Serra-Pages et al., 1995, 1998). There are four members of LIPRIN-α in humans with distinct expression patterns: LIPRIN-α1 and LIPRIN-α4 are widely expressed across the body, whereas LIPRIN-α2 and LIPRIN-α3 are enriched in the brain (Serra-Pages et al., 1998). LIPRIN-α contains multiple domains including two coiled-coil (CC) domains, a single alpha helix (SAH) domain, and three sterile-α-motif (SAM) domains (Figure 1). The N-terminal coiled-coil domains are sufficient to mediate the homo- and heterodimerization between liprins, and C-terminal regions interact with LARs (Serra-Pages et al., 1995, 1998). When co-expressed together with LARs in cell lines, Liprin-α was sufficient to promote the clustering of LARs (Serra-Pages et al., 1995, 1998), suggesting that through oligomerization, LIPRIN-αs provided a platform for the assembly of other synaptic proteins. They interact with multiple synaptic proteins that are essential for the development of both pre- and post-synaptic compartments (Spangler and Hoogenraad, 2007; Xie et al., 2021). At the pre-synaptic site, SYD-2/Liprin-α recruits the synaptic protein ELKS/CAST to promote the assembly of the active zone (Dai et al., 2006). ELKS is a protein around 120 KD and contains four coiled-coil domains indicating its role in associating with other protein components. CAST is also a member of this protein family that is structurally homologous but with a differential expression pattern as compared to ELKS in that CAST is highly expressed in the brain whereas ELKS is broadly expressed (Hida and Ohtsuka, 2010). ELKS interacts with Liprin-α through the coiled-coil domain and mediates the function of Liprin-α in recruiting synaptic vesicles to the active zone in worms (Dai et al., 2006). Indeed, ELKS/CAST can bind with RIM1 (Rab interacting molecules) and regulates synaptic transmission, consistent with its role in the generation of functional synapses (Takao-Rikitsu et al., 2004). ELKS also interacts directly with Bassoon and Piccolo through the coiled-coil domain (Takao-Rikitsu et al., 2004), suggesting that it plays a pivotal role in the assembly of active zone proteins. Piccolo and Bassoon were identified from a study by Craig Garner in 1996 in which a cDNA library from the rat brain was screened with antisera against the synaptic fraction and the positive clones were selected as candidates for synaptic proteins (Garner et al., 1988; Langnaese et al., 1996). They are structurally related proteins both containing zinc-finger regions and coiled-coil domains (Takao-Rikitsu et al., 2004). Through the same approach, a few other synaptic proteins were discovered in addition to Piccolo (Cases-Langhoff et al., 1996; Fenster et al., 2000) and Bassoon (Dieck et al., 1998), such as SAP90 (also known as PSD95) (Kistner et al., 1993), SAP97 (Muller et al., 1995), and SAP102 (Muller et al., 1996).
Another approach is homologous cloning of genes identified from other model organisms or tissues such as C. elegans or fruit flies. For example, the mammalian homolog of unc-13, Munc-13, was found in this way (Brose et al., 1995); identified and cloned by Brenner (1974) and Maruyama and Brenner (1991), unc-13 causes abnormal neurotransmitter release but normal anatomical structure when mutated in worms (Richmond et al., 1999). In mammals, there are three isoforms of Munc-13 that are enriched in the brain. However, neither Munc-13-1 nor Munc-13-2 has been found to be essential for synaptic morphogenesis in mice (Rosenmund et al., 2002). Using known synaptic proteins as baits to probe unknown candidates is also an effective strategy. For example, by fusing the cytoplasmic domain of SYNTAXIN A with glutathione-s-transferase (GST) and performing pull-down assays followed by amino acid sequencing, Munc-18/STXBP1 was discovered (Hata et al., 1993b); through yeast two-hybrid screen using the C-terminal of NEUREXIN, Cask was cloned (Hata et al., 1996); and using the active mutant of Rab3C or its isolated domains, the active zone protein RIM (Rab3 Interacting Molecules) and its interaction partners such as RIM-BP, ELKS, and CAST were identified (Wang et al., 1997, 2000, 2002) [sub-cellular fractionation of mammals’ synaptic fraction also independently discovered CAST (Ohtsuka et al., 2002)]. Among these molecules, mice deficient in Munc18-1 die immediately after birth but the differentiation of synapses appears normal (Verhage et al., 2000). Cultured neurons from Cask knockout mice developed a similar number of synapses when compared with wild-type counterparts, indicating that Cask is not essential for the formation of structurally sound synapses (Atasoy et al., 2007). Studies on the morphology from cultured neurons where RIM-BPs were removed showed that there are no significant differences between knockout animals and wild-type controls (Acuna et al., 2015). These studies suggested that synaptic proteins coordinate to regulate the formation and function of synapses.
Through ion channels such as NR2B (Grin2b) that are located in the post-synaptic region, Sheng and colleagues performed yeast hybridization and GST-pull-down assays to uncover the interactomes of post-synaptic proteins. Using the cytoplasmic domain of the NR2B subunit of NMDA receptors, Sheng and colleagues discovered that PSD95 is the scaffold protein for NR2B (Cho et al., 1992). PSD95 contains three PDZ (PSD95, Dlg4, and ZO-1) domains, one SH3 domain, and one GK domain (Figure 1), indicating that it organizes the synaptic proteins at the post-synaptic site. Indeed, further studies have demonstrated that it interacts with GKAP through its GK domain (Kim et al., 1997), while GKAP interacts with SHANK proteins in the PDZ domains (Naisbitt et al., 1999). SHANK proteins are also synaptic scaffold proteins that contain PDZ domains and other domains (Figure 1). They interact with Homer (Brakeman et al., 1997), which on the other hand interacts with mGluR and IR3 receptors. The PDZ domains of PSD95 also interact with the protein TARP, which is an auxiliary subunit of AMPAR receptors. In such a way, PSD95 acts as a core that enables the coupling between different kinds of receptors and controls the excitability of synapses. In order to do so, PSD95 clusters together through the N-terminal cysteine formed a di-sulfate bond, which is regulated by the phosphorylation of CDK5 (Morabito et al., 2004). PSD95 may also form clusters with other members in the same family and thus increase the diversity of molecules that can be anchored within synapses (Kim et al., 1996). Additionally, PSD95 interacts with SPAR (Spine-associated RapGAP) via its GK domain (Pak et al., 2001). Overexpression of SPAR in cultured hippocampal neurons increases both the density and diameter of dendritic spines, likely through its effect on the re-organization of the actin cytoskeleton. In contrast, the PDZ domains of PSD95 interact with SynGAP1 which is a RasGAP to restrict spine maturation (Chen et al., 1998; Kim et al., 1998; Clement et al., 2012).
Synaptic proteins need to be transported to the synaptic terminals to execute their synaptic functions (Ahmari et al., 2000; Zhai et al., 2001). In neurons, the anterograde trafficking of cargos through microtubules is mainly achieved through the Kinesin family motor proteins (Hirokawa and Takemura, 2005). Mutations in the kinesin family gene have been shown to cause deficits in synaptic vesicle trafficking (Hall and Hedgecock, 1991; Yonekawa et al., 1998; Pack-Chung et al., 2007). Liprin-α interacts directly with KIF1A through its coiled-coil domain (Shin et al., 2003; Miller et al., 2005), and mutations in Liprin-α showed decreased anterograde movement of synaptic vesicles (Miller et al., 2005). Interestingly, in unc-104/KIF1A mutated worms, the localization of syd-2 is unchanged (Zhen and Jin, 1999), suggesting other distinct mechanisms underlying its trafficking. Further studies have shown that to ensure the synaptic localization of syd-2, its retrograde trafficking by dynein was inhibited by cyclin-dependent kinases pct-1 and cdk-5 (Ou et al., 2010). In addition, the cyclin-dependent kinase 5 pathway promotes synaptogenesis by facilitating the trafficking of synaptic vesicles to the newly formed synapses through KIF1A (Park et al., 2011). Transportation of Piccolo–Bassoon vesicles is mediated by KIF5B which also mediates the activity-dependent pre-synaptic assembly (Cai et al., 2007). The interactions between Bassoon and dynein light chains mediate the retrograde trafficking of vesicles (Fejtova et al., 2009). When fused with fluorescent proteins or through antibody labeling, these synaptic proteins provide important insights into how pre-synaptic and post-synaptic assembly happen, both spatially and temporally. For example, Bassoon clustering occurs in parallel with both inhibitory and excitatory synaptogenesis (Zhai et al., 2000). Piccolo and Bassoon are transported in a protein complex associated with vesicles, which may be the basis for the formation of the active zone (Zhai et al., 2001). In contrast, PSD95 accumulates rapidly after the establishment of new contacts (Bresler et al., 2001), and the post-synaptic scaffold proteins are recruited to the synaptic sites gradually (Bresler et al., 2004).
These complex synaptic machinery must deposit at specific locations during synaptogenesis. The theory of liquid–liquid phase separation provides insights into how this phenomenon may occur. Recently, it has been shown that both SYD-2 and ELKS can undergo liquid–liquid phase separation before condensation and that such a step is essential for recruiting other active zone proteins (McDonald et al., 2020). The mechanisms underlying the liquid–liquid phase separation seem to be initialized by the oligomerization of Liprin-α followed by the phase separation of ELKS and the recruitment of RIM-α to the condensates (Liang et al., 2021). At the post-synaptic compartment, PSD95 and SynGAP1 interactions have also been reported to be essential for the formation of liquid condensates (Zeng et al., 2016). In addition, both the combinations of PSD95-GKAP and SHANK-Homer are able to form condensed droplets and recruit their interacting proteins, such as NR2B and SynGAP1 (Chen et al., 2020). Given that dendritic spines and axonal buttons are highly specialized compartments, and that the density of these molecules is extremely high, it is reasonable to propose that liquid–liquid phase separation plays an important role in promoting the specification of synapses.
Synaptic proteins assemble at synapses through protein–protein interactions and govern synaptic signaling and plasticity (Sheng and Kim, 2002). With accumulated information on these molecules, we are now able to describe a sketch of the synaptic network and “zoom in” to examine their role in synaptogenesis (Figure 2). Since most of the proteins mentioned above are scaffold proteins that provide platforms for the assembly of the synaptic machinery, overexpressing or knocking them down usually affects synaptic strength or morphogenesis (Hung et al., 2008; Wang et al., 2014; Yoon et al., 2021). Mutations in these genes, for example, SHANK1/2/3 and SYNGAP1 to name a few, are frequently associated with abnormal synaptogenesis and neurological or psychiatric disorders such as intellectual disability and autism (Table 1).
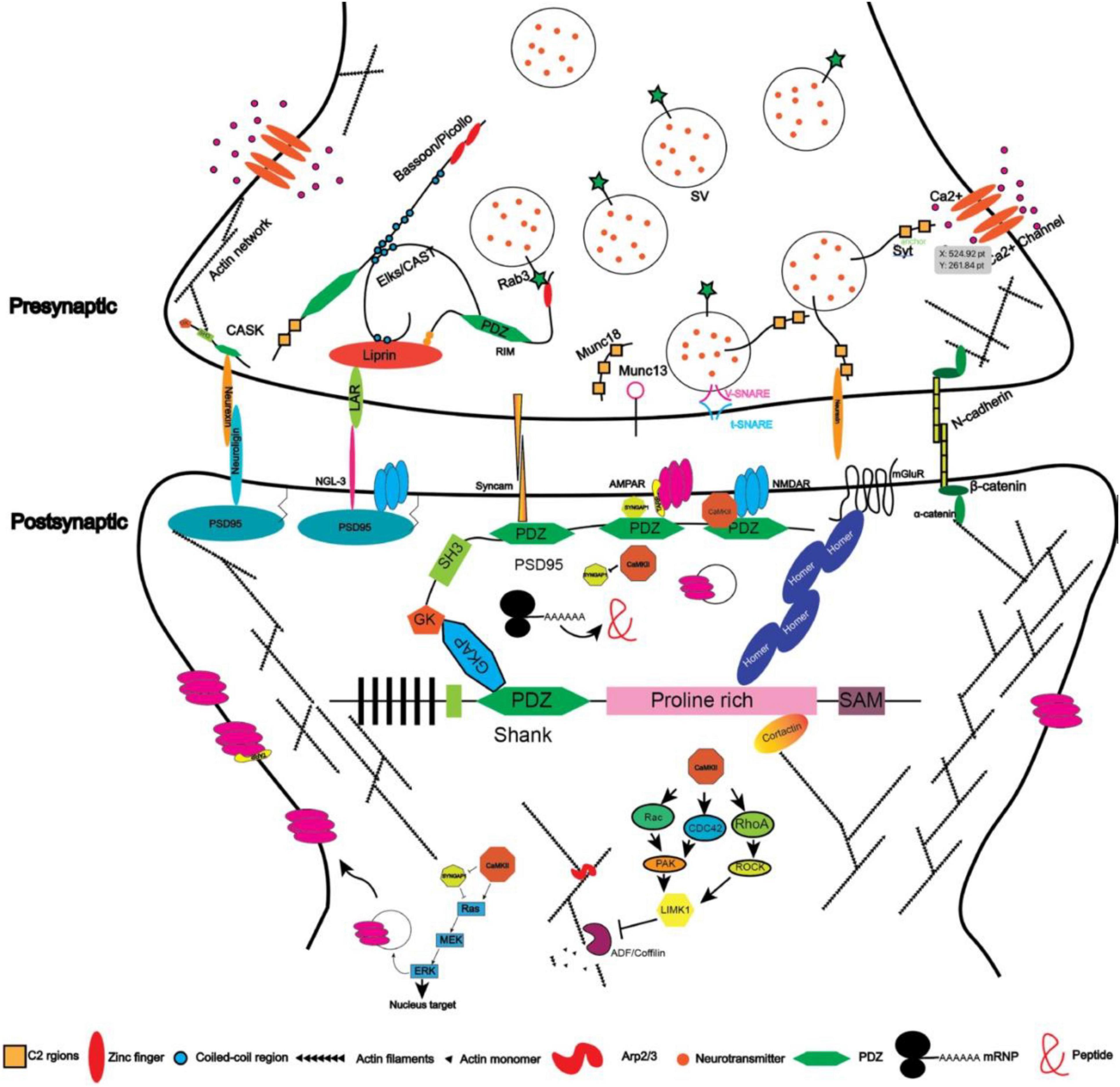
Figure 2. Illustration depicting the protein network in excitatory synapses (protein size not scaled).
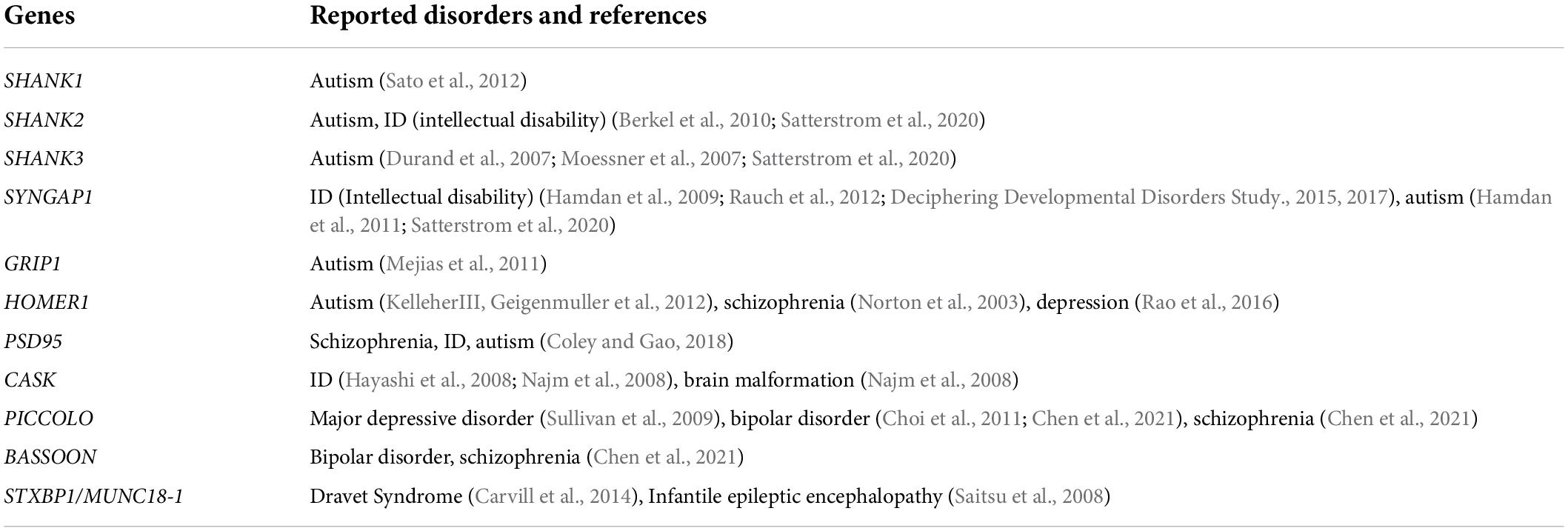
Table 1. Scaffold proteins that have been reported to be associated with neurological or psychiatric disorders from human genetics studies.
Scaffold proteins beneath the synaptic membrane act as the core for the assembly of various molecular modules and thus provide the platform for synaptogenesis. Nevertheless, these platforms must be connected across the synaptic cleft. Cell adhesion molecules are the best candidates to bridge pre- and post-synaptic components. Consistently, it was discovered that cell adhesion molecules are transported together with pre-synaptic components-formed particles, and that cell adhesion molecule also interact with scaffold proteins, suggesting that cell adhesion molecule may mediate the transcellular connections between pre- and post-synaptic terminals (Phillips et al., 2001). Moreover, in artificial synapse-inducing assays, cell adhesion molecules such as Syncam1 (Biederer et al., 2002), Neurexin (Graf et al., 2004), Neuroligin (Scheiffele et al., 2000), and NGL-3 (Woo et al., 2009) have been shown to be sufficient to induce the formation of synapses (Sudhof, 2018), highlighting the importance of cell adhesion molecules. Similarly, evidence from RNAi/shRNA-mediated knockdown and knockout animals also underlines the essential roles of cell adhesion molecules in synaptogenesis. Though these assays have their own caveats, they have provided unique perspectives for our understanding of the processes of synapse formation.
The leukocyte common antigen-related family receptors protein-tyrosine phosphatases (LAR-RPTPs) are composed of extracellular Ig domains and fibronectin domains at the extracellular side, a single transmembrane domain, and a cytoplasmic region that includes two phosphatase domains. The phosphatase domain proximal to the membrane is catalytic active, whereas the distal one is catalytic inactive (Um and Ko, 2013). In vertebrates, there are three genes encoding the proteins of this family: LAR, also called RPTPF; RPTδ, also called PTPRD; and RPTσ, also called RPTPS. They distribute both pre- and post-synaptically. Loss-of-function studies on LAR-RPTPs by RNAi-mediated knocking down resulted in a dramatic reduction in synapse numbers (Dunah et al., 2005). Intracellularly, LAR-RPTPs bind to the scaffold protein Liprin-α and transcellularly, they bind to various cell adhesion molecules including NGL-3, synaptic adhesion-like molecules (SALM), TrkC, and slit and trk like(Slitrks) (Um and Ko, 2013). Their associations with specific cell adhesion molecules are controlled by alternative splicing, indicating functional diversity and specificity (Han et al., 2018). Among them, NGL-3 belongs to the netrin G ligand family and is a single transmembrane protein anchored at the post-synaptic sites through interactions with PSD95. The interaction between LAR and NGL-3 induces the formation of excitatory synapses in cultured neurons (Woo et al., 2009). The phenotypes of mice lacking NGL-3 depends on the genetic background: mice with the C57BL/6J background show reduced growth and abnormal brain anatomy, while mice with a hybrid genetic background exhibit normal gross brain development but impaired synaptic transmission (Lee et al., 2019). SALMs are a family of cell adhesion molecules that also bind to the PDZ domain of PSD95. They have eight leucine-rich repeats, one Ig domain, and one fibronectin domain. Mice lacking SALMs show varying extents of abnormality in synapse development depending on which specific member of the family was knocked out (Lie et al., 2018). SALMs regulate synapse development by inducing the clustering of PSD95 and recruiting NR1 (Lie et al., 2018). Overexpressing and knocking down Slitrks can promote and reduce excitatory and inhibitory synapses, respectively, in cultured neurons by interacting with PTPδ and PTPσ (Yim et al., 2013). The detailed interactions among these cell adhesion molecules and their function in synapse development need to be further examined to explain the diverse phenotypes from knockout mice. The complicated interactions will likely compensate for the loss of others. Consistently, mice carrying the mutated LAR without the cytoplasmic phosphatase domains exhibited impaired spatial learning and hyperactive behaviors (Kolkman et al., 2004), whereas mice lacking LAR showed disrupted glucose homeostasis (Ren et al., 1998). Mice lacking RPTσ, on the contrary, showed increased synapse density (Horn et al., 2012). Studies on triple knockout mice either in vivo using virus carrying Cre recombinase or in cultured neurons showed that PTPδ, PTPσ, and LAR triple knockout do not affect the development and function of synapses (Sclip and Sudhof, 2020; Emperador-Melero et al., 2021).
Neurexins were discovered through the chromatography purification of the targets of α-latrotoxin (Ushkaryov et al., 1992). Through the studies of neurexins, Ca2+ sensor synaptotagmin was found to interact with their cytoplasmic domains (Brose et al., 1992); the ligands of neurexins, neuroligins, were identified as well (Ichtchenko et al., 1995). In total, there are three neurexin genes, each encoding α- and β- isoforms driven by distinct promoters, and four neuroligin genes with two splice sites differentially distributed in these four genes (Craig and Kang, 2007; Sudhof, 2017). Pre-synaptic neurexins are anchored at the synapses by binding with the scaffold protein CASK, while post-synaptic neuroligins are anchored by binding with PSD95 and gephyrin (Craig and Kang, 2007). Neuroligin 1 is mainly expressed at excitatory synapses, while neuroligin 2 is enriched at inhibitory synapses (Song et al., 1999). When expressed in non-neural cells, neurexins and neuroligins can, respectively, induce the differentiation of post-synaptic and pre-synaptic parts, whereas neuroligins containing autism-associated mutations fail to do so (Scheiffele et al., 2000; Graf et al., 2004; Chubykin et al., 2005; Poulopoulos et al., 2009), suggesting their important roles in the initiation of synaptic terminal specification and synapse formation (Hata et al., 1993a). Consistent with this idea, both loss- and gain-of-function studies have validated the importance of neurexins on synapse formation in cultured neurons (Prange et al., 2004; Chih et al., 2005) and in vivo (Chen et al., 2017), though the function of neuroligins on synapse formation in vivo was inconsistent with that of in vitro studies (Varoqueaux et al., 2006; Shaw and Salmon, 2017). One of the possible reasons for the differential effects of neuroligin 1/2 is that the overexpression effects in vitro resulted from the use-dependent stabilization of synapses instead of newly generated ones (Chubykin et al., 2007). These results also suggested that neurexins have other receptors or ligands that are used for synapse formation. Indeed, several lines of evidence demonstrated that neurexins can bind to Cbln1 (Uemura et al., 2010), C1ql2/3 (Matsuda et al., 2016), LRRTM2 (de Wit et al., 2009; Ko et al., 2009; Siddiqui et al., 2010), GABA(A) receptors (Zhang et al., 2010), and latrophilin (Boucard et al., 2012). The interactions between neurexins and these molecules are isoform-specific, and their functions are synaptic type specific (Chih et al., 2006; Sudhof, 2017). For example, neuroligin 1 splice site b determines if it can bind to α-neurexin but does not affect its binding with β-neurexin. In addition, the neuroligin that binds with β-neurexin only promotes synaptogenesis, while the isoform binding to both neurexin isoforms stimulates the enlargement of synapses. LRRTM2 binds to neurexins lacking splice site #4 to induce the formation of excitatory synapses. In line with the diverse interactions of neurexins and neuroligins, various mutations in neuroligins have been reported to be associated with autism disorders; and some syndromic phenotypes can be recapitulated in mice (Sudhof, 2008; Reichelt and Dachtler, 2015).
N-cadherins are single transmembrane proteins that localize both pre- and post-synaptically. They consist of extracellular ectodomains that mediate homophilic interactions, a transmembrane domain, and a cytoplasmic domain that connects it to the cytoskeleton system through β- and α-catenins (Bruses, 2006; Figure 2). Evidence supporting N-cadherins’ functions in synaptogenesis include (1) its colocalization with the synaptic marker synaptophysin and PSD95 at the stage of synapse formation (Benson and Tanaka, 1998); (2) its interacting protein β-catenin begins to localize at the dendritic protrusions at the beginning of synaptogenesis in cultured hippocampal neurons, and cultured neurons from β-catenin knockout mice showed thinner dendritic protrusions; and (3) overexpressing the ectodomain deleted N-cadherins in cultured neurons causes the same phenotype as that of the loss of β-catenin in cultured neurons, indicating an essential role of N-cadherins and β-catenin in synapse formation (Togashi et al., 2002; Okuda et al., 2007). However, another study showed that overexpressing only the cytoplasmic domain of N-cadherin in cultured neurons has no effects on either the level of dendritic and synaptic PSD95 and Bassoon but reduced the amplitude of mEPSCs (Peng et al., 2009), suggesting a function of N-cadherin in synaptic transmission. The discrepancies between the two studies might be due to the development stages the experiments were performed with the former at around div (days in vitro) 21, while the latter at div12, suggesting that the functions of N-cadherin vary at different developmental stages. In line with this, knocking down N-cadherin in cultured neurons reduces spine density only at the early stage, suggesting that N-cadherin is required for the initiation but not the maintenance of synapses (Saglietti et al., 2007). Although in vivo knockout of N-cadherin in the neocortex and hippocampus disrupted tissue structures such as the cortical lamination and radial glia fiber orientation, there were no obvious effects on spine morphology in cultured hippocampal neurons after Cre mediated knockout in culture (Kadowaki et al., 2007). These results appear to indicate that N-cadherin maybe not essential in the formation of synapses in vivo or that there could be other adhesion molecules that can replace N-cadherin to associate with catenins and regulate synapse formation (Yamagata et al., 2018). Nevertheless, it should be noted that acute knockout may take time to remove the protein, and thus when the protein is completely gone, the time window for synaptogenesis may have passed. It is also possible that N-cadherin is important for neural activity-dependent synaptogenesis or pruning. The evidence supporting this hypothesis comes from experiments in which neuronal depolarization, induced by extracellular K+, leads to an increased membrane level of N-cadherin. The mechanisms behind this have been reported to be protein kinase D mediated phosphorylation and the ensuing enhanced interaction with β-cateinin (Tanaka et al., 2000; Tan et al., 2010; Bian et al., 2015; Cen et al., 2018). In addition, mutations in β-catenin have been reported to be associated with autism (O’Roak et al., 2012a,b; Sanders et al., 2012; Krumm et al., 2014) and a dominant mutation in humans that causes intellectual disability has been shown to reduce the affinity between β-catenin and N-cadherin (Sanders et al., 2012). These discoveries and the fact that constitutive N-cadherin knockout is embryonic lethal underline the importance of N-cadherin in neural development and suggest that further examination of conditional knockout animals of N-cadherin is necessary for analyzing synaptogenesis.
After connections mediated by cell adhesion molecules are established between pre-synaptic terminals and post-synaptic compartments, the highly differentiated sub-cellular regions need to be anchored. The cellular cytoskeleton system provides support to these molecules and thus the building of synapses. We will summarize the function of regulations of cytoskeleton organization.
The actin cytoskeleton is the main cytoskeleton system that supports the structure of the dendritic spines as well as the growth cone of axons (Landis and Reese, 1983). The synaptic scaffold proteins are anchored at the actin framework (Kuriu et al., 2006). However, the actin system is quite dynamic, and the kinetics of actin re-organization underlies the structural changes in dendritic spines (Okabe, 2020). In addition, neural activity also remodels synaptic structure by acting on the actin system (Hotulainen and Hoogenraad, 2010). Actin can be classified as G-actin and F-actin: G-actin is a monomer, while F-actin is a polymer. The balance between G- and F-actin is controlled by actin-associated proteins such as profilin and capping proteins (Amann and Pollard, 2000). The branching of actin filaments is mainly regulated by the Arp2/3 complex while severing filaments and dissociating actin are achieved through the function of cofilin (Amann and Pollard, 2000; Goley and Welch, 2006). The branching dynamics of actin affect the curvature of the plasma membrane, so it determines the fate and shape of the synapses. For example, the head of the dendritic spines usually contains more highly branched actin than the base (Korobova and Svitkina, 2010; Basu and Lamprecht, 2018). Imaging studies also revealed rapid actin dynamics along with the formation of dendritic spines (Fischer et al., 1998). Moreover, when actin undergoes remodeling that is mediated by synaptic activity, it allows the entry of the microtubules into the dendritic spines (Schätzle et al., 2018), possibly facilitating the trafficking of synaptic proteins. Mutations in the actin-regulating molecules impair synaptic plasticity and are associated with neurodevelopmental deficits and cognitive disorders (Bednarek and Caroni, 2011; Lian and Sheen, 2015; Yan et al., 2016; Hlushchenko et al., 2018; Cousin et al., 2021; Qi et al., 2021).
In the nervous system, synapses are formed enthusiastically at the beginning which usually culminates with more synapses than required (Figure 3). It is possible that such consequences are supposed to provide enough neural connections for organisms to learn and memorize their encounters. When the organisms gradually adapt to the environments, the synapse network will be reshaped and stabilized by neural activity to operate efficiently. The openings of various channels are the basis for neural activity and will be followed by the spreading of signals from activated synapses to surrounding regions and nuclei. The chemical reactions induced by ion influx or efflux reorganize the structure of synapses (Ebert and Greenberg, 2013).
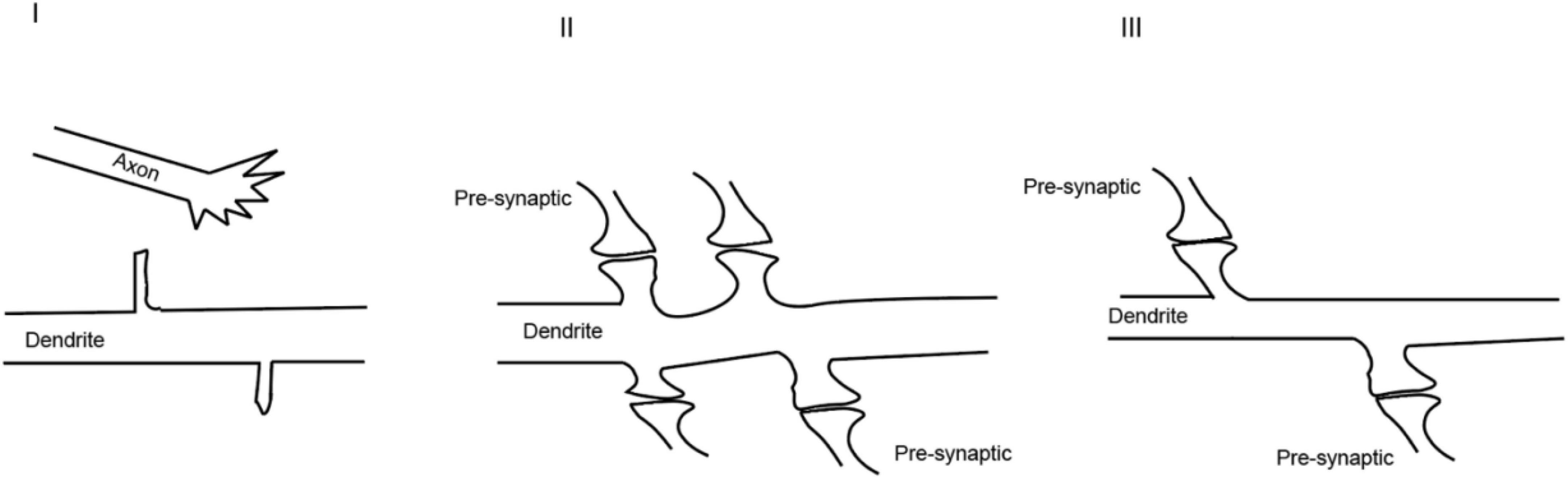
Figure 3. Dynamic process of synaptogenesis: (I) axonal and dendritic growth; (II) excessive synaptogenesis; (III) pruning of the synapses.
Studies from the visual system provided a good amount of insights into how neural activities instruct the formation of neural circuits. Reshaping of the neural circuits mediated by neural activity has been demonstrated systematically by studies in the visual system by Hubel and Wiesel (2012) in the 1970s. Parallelly, studies from neuromuscular junctions suggested the essential roles of neural activity in the elimination of muscle innervating axonal terminals of motor neurons (Colman and Lichtman, 1993). Moreover, ocular dominance can form in animals that have undergone binocular deprivation, underscoring the importance of spontaneous activity on synaptogenesis (Shatz, 1990). In a series of pioneering studies led by Katz and Shatz (1996), retinal waves in the early developmental stages were demonstrated to be crucial for the connectivity of retinal ganglion cells to the thalamus. Cholinergic transmission has been shown to be required for the generation of spontaneous waves (Feller et al., 1996). With the advances in imaging techniques and available genetic tools, it has been demonstrated that retinal waves instruct the activity of the midbrain superior colliculus and primary visual cortex (Ackman et al., 2012), and that disrupting the naïve spontaneous activity by deleting the β2-acetylcholine receptor or using optogenetics interferes with the normal retinotopic map refinement (Xu et al., 2011; Zhang et al., 2011; Burbridge et al., 2014).
These studies indicated that the formation of functional synapses occurs at the cost of removing other synapses. However, one of the ensuing questions is whether the reshaping of the neural connectivity happens at the synaptic level and, if yes (Zuo et al., 2005), what are the molecules that are acted on by the neural activity. In addition, evidence on whether there would be de novo synaptogenesis after neural activity was still limited at the time. Disrupting neural activity by genetically ablating neurotransmitter release caused a reduction in synapse number, which provided indirect but important evidence supporting the role of neural activity in synaptogenesis (Andreae and Burrone, 2014). Nevertheless, there have been several lines of evidence supporting that there is a cause-and-effect relationship between neural activity and synaptogenesis. First, it has been reported that neural activity can lead to the switch between different types of neurotransmitters (Spitzer, 2017), suggesting that there may be newly generated synaptic types. Second, the treatment of cultured neurons with potassium chloride (KCl) or bicuculline which induces depolarization increased the spine density (Papa and Segal, 1996). This observation was later validated by time-lapse imaging on the same segment of denrites (Hamilton et al., 2012). Third, Maletic-Savatic et al. (1999) observed the rapid formation of dendritic protrusions after tetanic stimulation, certifying that neural activity induces the formation of potential dendritic spines. In addition, Engert and Bonhoeffer performed experiments where they induced neural activity locally and imaged the dynamics of dendritic spines on slices. They found that, accompanied by long-term potentiation induction, there are newly generated spines (Engert and Bonhoeffer, 1999). This study provided evidence that neural activity promotes synaptogenesis within an intact circuit. Recently, it has been corroborated elegantly by the discoveries that the local application of glutamate or GABA with high frequency uncaging at places that originally have no spines will be sufficient to induce the de novo formation of dendritic spines in the cortex of developing mice (Kwon and Sabatini, 2011; Oh et al., 2016). Finally, in vivo studies suggested strong associations between physiological level neural activity and synaptogenesis. Using two-photon microscopy, it has been shown that there is rapid synapse formation during motor activity in the cortex of developing mice (Xu et al., 2009). At the adult stage, although the turnover speed of dendritic spines becomes lower (Grutzendler et al., 2002), it has also been demonstrated that after stimulation, there exists rapid formation of both excitatory and inhibitory synapses in freely moving animals (Knott et al., 2002). These studies provided compelling evidence that neural activity reshapes the circuits through synaptogenesis. Moreover, the dendritic spines are quite dynamic and can be generated independently or derived from the existing ones by splitting (Zito and Svoboda, 2002). Neural activity can also change the motility of pre-synaptic terminals in a retrograde manner (Gan, 2003; Tashiro et al., 2003). There are a few other models to mimic and study the effects of neural activity on synapse formation, such as environmental enrichment (Kondo et al., 2012) and fear conditioning (Lai et al., 2012) to name a few.
As for the underlying molecular mechanisms for activity-dependent synaptogenesis, multiple aspects are believed to be involved. For example, accumulated evidence shows that neural activity re-organizes the cytoskeleton system and regulates the production of neurotrophic factors from various levels and thus affects synaptogenesis (Ebert and Greenberg, 2013). Most of these mechanistic studies are conducted in vitro due to technical obstacles, but there is some in vivo evidence as well. We will summarize these investigations here.
Under most circumstances, neural activity utilizes Ca2+ to activate kinase CaMKII or phosphatases such as calcineurin to affect synaptogenesis (Figure 2). Imaging studies suggested that the activation of CaMKII is restricted to the dendritic spine (Nishiyama and Yasuda, 2015). One of the pathways activated by CaMKII-mediated phosphorylation is the Ras signaling pathway, whose activation results from the inactivation of the RasGAP, Syngap1 (Gamache et al., 2020). On the one hand, the activated Ras can promote the synaptic trafficking of AMPA receptors and enhance transmission efficiency (Zhu et al., 2002). The GluR2 subunit of AMPAR has been reported to interact with the extracellular domain of N-cadherin and promote the morphogenesis of dendritic spines (Passafaro et al., 2003), suggesting function and structure coupling. On the other hand, the activated Ras can relay the signal through PI3K-Akt-mTOR to promote the formation of mushroom-shaped spines (Kumar et al., 2005). The Ras signaling pathway can also send signals to the nucleus by activating ERK, which will phosphorylate CREB (Tang and Yasuda, 2017). Other small GTPases such as Rho/Rac/CDC42 have also been shown to be involved in regulating spine morphology through the cytoskeleton system (Lawler, 1999; Tada and Sheng, 2006; Figure 2). Activated Ras and RhoA signaling can spread to nearby spines whereas the CDC42 signaling is restricted in the activated spines (Nishiyama and Yasuda, 2015). With different temporal kinetics of these three signaling pathways, they coordinated to mediate input-specific structural plasticity of dendritic spines: usually, the signal pathways that will be relayed to nearby synapses or the nucleus have a longer activation time window and vice versa (Hedrick et al., 2016). Of the molecules involved, the loss-of-function mutation of SYNGAP1 has been reported to be a cause of intellectual disability (Jeyabalan and Clement, 2016). The loss of one copy of Syngap1 in mice will result in premature development of synapses and impaired synaptic plasticity after stimulation (Clement et al., 2012).
In 1986, (Greenberg et al., 1986) performed experiments by stimulating the differentiated PC12 cells with a cholinergic agonist and discovered that this treatment induced rapid transcription, which was demonstrated later both in cultured neurons and in vivo (Greenberg et al., 1986; Sheng and Greenberg, 1990). These discoveries opened the direction that neural activity might also affect neural connectivity at the level of transcription. Indeed, the signaling from synapses can be transduced to the nucleus to initiate transcription. After the initiation of transcription, these products will be transported to synapses both non-selectively and selectively and act locally at the activated synapses. Multiple mechanisms can contribute to this process such as the cis-elements in the mRNAs (Steward and Worley, 2001), trans-factors such as RNA-binding proteins (Fujii et al., 2005; Doyle and Kiebler, 2011), remodeling of the cytoskeletons system (Schätzle et al., 2018), and local translation and degradation (Tai and Schuman, 2008; Bingol and Sheng, 2011; Glock et al., 2017). In such a way, transcription and local translation supply the materials to support the activity-induced structure changes (Sutton and Schuman, 2006; Ebert and Greenberg, 2013). The effectors that relay information from synapses to the nucleus are usually the calcium-activated CamKII–CamKIV signaling pathways. In this process, a series of transcription factors act coordinately to control the expression of downstream genes to adapt to the external changes as well as to maintain internal homeostasis. Under some circumstances, some membrane-associated proteins will undergo cleavage and translocate into the nucleus to function as transcription factors. For example, the intracellular C-terminal domain of neuregulin 1 has been reported to enhance the transcription of PSD95 in response to elevated neural activity (Bao et al., 2004).
The neural activity also regulates the synapse by regulating neurotrophic factors (Vicario-Abejon et al., 2002). There are four main kinds of neurotrophic factors: NGF, BDNF, NT3, and NT4. The receptors for these neurotrophic factors include TrkA, TrkB, TrkC, and p75NTR. All neurotrophins bind to p75NTR but with lower affinity than to the Trk receptor family. Trk receptors belong to the receptor tyrosine kinase family and, when binding with their ligands these receptors undergo endocytosis and retrogradely relay the neurotrophic factors to the nucleus (Sharma et al., 2010). TrkA is the main receptor for NGF, TrkB for BDNF and NT4, and TrkC for NT3 which can also bind to TrkA and TrkB but with lower efficiency. Neurotrophic factors were recognized as important players in the formation of synapses first at neural muscular junctions and later in the central nervous system as well. Mice with TrkB and TrkC knockout showed a reduced number of excitatory synapses at the stage of synaptogenesis (Martinez et al., 1998), and mice with conditional deletion of TrkB showed reduced synapse density at the CA1 regions (Luikart et al., 2005). Among the neurotrophic factors, BDNF is the most prominent, and accumulating evidence supports its role in the development of synapses. The mRNA of BDNF can be targeted to dendrites by the alternative 3′UTR (An et al., 2008) and BDNF can mediate neural activity-regulated synapse formation in various ways. First, it can undergo proteolytic maturation after synaptic stimulation (Nagappan et al., 2009; Yang et al., 2009); second, the BDNF receptor TrkB can undergo activity-dependent exocytosis (Lu, 2003); and third, BDNF and its signaling can activate the local translation, thus augmenting the contrast between activated versus non-activated synapses (Kang and Schuman, 1996; Baj et al., 2016). Moreover, the BDNF-TrkB signaling mediated structural change in dendritic spines is NMDAR-CaMKII dependent (Harward et al., 2016).
Under the influence of neural activity, synaptogenesis not only enables neurons to adapt to new conditions but also acts as a feedback mechanism to maintain neural homeostasis. Activity-dependent neurotransmitter switching is one way to achieve such a goal (Spitzer, 2017). Recently, it has been reported that neural activity can promote the differentiation of parvalbumin interneurons (Donato et al., 2013) and NMDAR-mediated inhibitory potentiation (Chiu et al., 2018). In cultured neurons, elevating neural activity by tetanic stimulation or blocking GABA transmission promotes gephyrin clustering mediated by CaMKII phosphorylation (Flores et al., 2015). At the transcriptional level, transcription factor NPAS4 can maintain the overall activity at a proper level by promoting the transcription of Bdnf, thus leading to the formation of inhibitory synapses at somatic regions in excitatory neurons, and excitatory synapses in inhibitory neurons (Lin et al., 2008; Bloodgood et al., 2013; Spiegel et al., 2014). The transcription factor MEF2 can also mediate the activity-induced homeostasis via transcriptional activation of arc and synGAP to put a restriction on synaptogenesis (Flavell et al., 2006).
Glia is another major cell type in the nervous system other than neurons. Astrocyte is one of the most abundant glia types in the brain. In the central nervous system, the astrocytes are generated after neurogenesis. In studies by Barres and colleagues, it was found that when co-cultured with neurons, astrocytes promoted a higher number of synapses on retinal ganglion neurons (Ullian et al., 2001; Freeman, 2005), and this phenomenon was observed in other types of neurons such as motor neurons (Ullian et al., 2004). Astrocytes regulate synapse formation mainly in a non–cell-autonomous manner, by para-secretion (Christopherson et al., 2005; Eroglu et al., 2009) or through cell adhesion molecules-mediated cell interactions (Stogsdill et al., 2017). In a forward genetic screen in C. elegans, it was identified that the glia cells secreted UNC-6 (netrin), which instructed the synaptogenesis between AIY and RIA neurons through regulating pre-synaptic assembly and the axon guidance via its receptor UNC-40 (DCC) (Colon-Ramos et al., 2007). For secreted netrin to act properly, the position of the astrocyte is required to be retained through its interaction with epithelial cells (Shao et al., 2013). The Wnt signaling pathway has also been reported to regulate synaptogenesis via both pre- and post-synaptic sites (Park and Shen, 2012; Shi et al., 2018). These reports highlighted the conserved function of astrocytes on synaptogenesis. In addition, astrocytes can promote microglia-mediated synaptic elimination (Allen and Eroglu, 2017). Though it has been shown that astrocytes are quite heterogeneous across brain regions, if and how astrocytes selectively influence synaptogenesis is still an open question. Moreover, astrocytes can influence synaptic connectivity by modulating the interaction between microglia and neurons. Neurons express the complement system molecules C1q after they were co-cultured with astrocytes, although the mechanisms for astrocytes to stimulate the upregulated C1q remain unknown (Stevens et al., 2007; Stephan et al., 2012). The “tagged” spines will undergo phagocytosis and thus the synaptic elimination by microglia. How the microglia only engulf the synapse and leave the remaining part of the neuron intact is unknown. When this process goes awry, it is usually associated with psychiatric diseases. In a genetic screen for genes associated with the susceptibility to schizophrenia, variants in the complement component C4 were found to be associated with dysfunction in synapse elimination (Sekar et al., 2016). Overexpression of C4A/B causes excess pruning of synapses (Yilmaz et al., 2021) through the phagocytosis function of microglia (Schafer et al., 2012), suggesting their contributions to the pathogenesis of Alzheimer’s disease. Microglia can influence specific types of synapses via both active and feedback mechanisms by responding to changes from the active neurons. For example, it has been shown that neural activity enhances the expression of Fn14 in thalamic relaying neurons to promote synapse formation; however, at the same time, the expression of Fn14’s ligand TWEAK increases in microglia, which will restrict the number of spines in relay neurons at places where microglia interact with (Cheadle et al., 2020). It has also been found that microglia can respond to the released GABA neurotransmitter and sculpt the number of synapses (Favuzzi et al., 2021). Neurons express CD47 to exhibit a “don’t eat me” signal and are prevented from being engulfed by microglia (Lehrman et al., 2018). Thus, the homeostatic interactions between astrocytes/microglia and neurons are pivotal for the proper wiring and connectivity of the nervous system (Schafer and Stevens, 2013; Hammond et al., 2018).
Given its importance for information processing, the development of the synapse provides an avenue for the understanding of the unrivaled cognitive abilities of humans. It has been observed that accompanied by the increase in the size of the human brain, the spine density is higher in human cortical neurons compared to other primates or rodents (Defelipe, 2011; Schmidt and Polleux, 2021). The most prominent feature of synaptogenesis in humans is the protracted developmental window (Liu et al., 2012). One molecule that has been reported to prevent the premature assembly of a functional synapse is SYNGAP1. SYNGAP1 encodes a RasGAP that negatively regulates the Ras-GTPase activity and thus prevents the formation of actin filaments and the insertion of AMPAR into the synapse. Haploinsufficiency of SYNGAP1 leads to autism-like spectrum disorders, highlighting the importance of SYNGAP1 dosage in synaptogenesis (Clement et al., 2012). Neurons derived from human iPSCs where the level of SYNGAP1 was reduced showed premature neural activity (LIamosas et al., 2020), further highlighting the temporal importance of synaptogenesis.
Another gene that has been reported to regulate human-specific synaptogenesis is SRGAP2. In mice, there is only one copy of Srgap2. It contains mainly three functional domains: the F-BAR domains, RhoGAP domain, and SH3 domains. Loss of Srgap2 in juvenile mice in vivo resulted in increased spine density accompanied by decreased spine head width, which commonly reflects the maturation of the synapse. The increase in spine density will persist to adulthood (Charrier et al., 2012), indicating that in mice, Srgap2 restricts the generation but promotes the maturation of synapses. Srgap2 is required for the maturation of both excitatory and inhibitory synapses at the expense of spine density, and the spine density regulation depends on both the F-bar domain and RhoGAP domain, whereas the regulation of inhibitory synapses density on the dendritic shaft depends on RhoGAP only (Fossati et al., 2016). During evolution, SRGAP2 genes were duplicated partially three times only in humans which are named SRGAP2B/C/D, with SRGAP2C expressed at the highest level (Dennis et al., 2012). The product of SRGAP2C only contains the F-BAR domain and when overexpressed in mice, it phenocopies the effects of loss of function of Srgap2A, suggesting its inhibitory effects on Srgap2A. These results suggested that the human-specific duplication generated product prolonged the synaptogenesis of humans. Bioinformatic analysis suggested that chromosome aberrations potentially disrupt the SRGAP2A genes that are associated with patients with brain malformations and psychiatric diseases, such as epilepsy, while the diploid copy number of the duplicated gene SRGAP2C is highly stable (Dennis et al., 2012).
Recently, it has been reported that in the human prefrontal cortex (PFC), which is an evolutionarily expanded region during evolution and has been reported to have a higher density of synapses, CBLN2 (cerebellin 2 precursor) is differentially expressed across mice, rhesus macaques, and human (Johnson et al., 2009; Shibata et al., 2021b). Overexpression of CBLN2 in mice is sufficient to increase spinogenesis (Shibata et al., 2021b). CBLN2 belongs to the cerebellin gene family and is a secreted glycoprotein. It bridges the interaction between neurexins and glutamate receptor σ2, and such a function has been demonstrated in the cerebellum of mice to promote synaptogenesis (Tao et al., 2018). Further examination revealed that the differential expression pattern of CBLN2 across species is due to the variations in its enhancers that can be regulated by Sox5 and retinal acid signaling pathways (Tao et al., 2018; Shibata et al., 2021a). Noteworthy, retinal acid signaling pathways have been reported to be associated with psychiatric disorders such as autism spectrum disorders and schizophrenia (Wan et al., 2009; Moreno-Ramos et al., 2015; Huang et al., 2020; Reay and Cairns, 2020). In summary, this evidence not only provides novel insights into the molecular mechanisms of synaptogenesis but also strengthens the associations between abnormal synaptogenesis and psychiatric disorders.
Synapse formation is fundamental for the precise wiring of the nervous system. It is a coordinated process that mobilizes various cellular and molecular events. Nevertheless, there are still many unanswered questions in the field. First, it remains unclear how post-synaptic sites are precisely aligned to the pre-synaptic releasing site. Although cell adhesion molecules have provided insightful clues to this question, the in vivo functions of these molecules are not always consistent with in vitro studies and thus need to be clarified in greater detail. Currently, the widely accepted criteria to define the importance of molecules in synapse development are through genetically modified model organisms or human genetic studies. Second, the temporal sequence of the synapse assembly should be elucidated in more detail. For example, does the synapse formation start from the contact between pre- and post-synaptic sites, or is the cytoskeleton system below the synaptic membrane organized first to prepare a platform for the cell–cell contact? Technical advances for multiplexing labeling and live cell imaging are likely to provide more clues. Third, it is now agreed that synapse formation is not random but rather stereotyped at least in the developmental stage. How the specificity of synaptic contacts is achieved remains largely an open question, especially in mammals. It has been demonstrated in invertebrates that alternative splicing of cell adhesion molecules governs the contact-dependent synaptic specificity, and recently there has been evidence that mice cell adhesion molecules, indeed, govern the wiring specificity in the retina (Sanes and Zipursky, 2020) and hippocampus (Berns et al., 2018; Li et al., 2020). Nevertheless, because of the complexity of the central nervous system, especially the brain, further investigations are still needed to get a comprehensive picture of this landscape. Importantly, it has been shown that most neurological and psychiatric disorders can be attributed to neural developmental disorders, particularly anomalies in synapse development such as synaptic transmission and dendritic spine morphogenesis (Penzes et al., 2011; Zoghbi and Bear, 2012). Studies on how genetic variants contribute to abnormal synapse formation still have a long way to go. With the reduced cost of genomic sequencing and advances in gene therapies, we will be able to unravel the molecular genetics of synapse formation and provide accessible and reliable ways to treat and cure neurological and neuropsychiatric diseases (Sztainberg et al., 2015; Benger et al., 2018; Hill and Meisler, 2021).
CQ designed the manuscript. CQ and L-DL wrote the manuscript. IF and SM helped to revise the manuscript. All authors contributed to the article and approved the submitted version.
The authors appreciate the comments of Yishi Jin on the manuscript and Rothem Kovner on the editing of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ackman, J. B., Burbridge, T. J., and Crair, M. C. (2012). Retinal waves coordinate patterned activity throughout the developing visual system. Nature 490, 219–225. doi: 10.1038/nature11529
Acuna, C., Liu, X., Gonzalez, A., Sudhof, T. C., and Ps Mediate, R. I. M.-B. (2015). Tight Coupling of Action Potentials to Ca(2+)-Triggered Neurotransmitter Release. Neuron 87, 1234–1247. doi: 10.1016/j.neuron.2015.08.027
Ahmari, S. E., Buchanan, J., and Smith, S. J. (2000). Assembly of presynaptic active zones from cytoplasmic transport packets. Nat. Neurosci. 3, 445–451. doi: 10.1038/74814
Ahnert-Hilger, G., Kutay, U., Chahoud, I., Rapoport, T., and Wiedenmann, B. (1996). Synaptobrevin is essential for secretion but not for the development of synaptic processes. Eur. J. Cell Biol. 70, 1–11.
Allen, N. J., and Eroglu, C. (2017). Cell Biology of Astrocyte-Synapse Interactions. Neuron 96, 697–708. doi: 10.1016/j.neuron.2017.09.056
Amann, K. J., and Pollard, T. D. (2000). Cellular regulation of actin network assembly. Curr. Biol. 10:R728–R730. doi: 10.1016/S0960-9822(00)00751-X
An, J. J., Gharami, K., Liao, G. Y., Woo, N. H., Lau, A. G., Vanevski, F., et al. (2008). Distinct role of long 3’ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell 134, 175–187. doi: 10.1016/j.cell.2008.05.045
Andreae, L. C., and Burrone, J. (2014). The role of neuronal activity and transmitter release on synapse formation. Curr. Opin. Neurobiol. 27, 47–52. doi: 10.1016/j.conb.2014.02.008
Atasoy, D., Schoch, S., Ho, A., Nadasy, K. A., Liu, X., Zhang, W., et al. (2007). Deletion of CASK in mice is lethal and impairs synaptic function. Proc. Natl. Acad. Sci. U.S.A. 104, 2525–2530. doi: 10.1073/pnas.0611003104
Baj, G., Pinhero, V., Vaghi, V., and Tongiorgi, E. (2016). Signaling pathways controlling activity-dependent local translation of BDNF and their localization in dendritic arbors. J. Cell Sci. 129, 2852–2864. doi: 10.1242/jcs.177626
Bao, J., Lin, H., Ouyang, Y., Lei, D., Osman, A., Kim, T. W., et al. (2004). Activity-dependent transcription regulation of PSD-95 by neuregulin-1 and Eos. Nat. Neurosci. 7, 1250–1258. doi: 10.1038/nn1342
Basu, S., and Lamprecht, R. (2018). The Role of Actin Cytoskeleton in Dendritic Spines in the Maintenance of Long-Term Memory. Front. Mol. Neurosci. 11:143. doi: 10.3389/fnmol.2018.00143
Batool, S., Raza, H., Zaidi, J., Riaz, S., Hasan, S., and Syed, N. I. (2019). Synapse formation: From cellular and molecular mechanisms to neurodevelopmental and neurodegenerative disorders. J. Neurophysiol. 121, 1381–1397. doi: 10.1152/jn.00833.2018
Bednarek, E., and Caroni, P. (2011). beta-Adducin is required for stable assembly of new synapses and improved memory upon environmental enrichment. Neuron 69, 1132–1146. doi: 10.1016/j.neuron.2011.02.034
Benger, M., Kinali, M., and Mazarakis, N. D. (2018). Autism spectrum disorder: Prospects for treatment using gene therapy. Mol. Autism 9:39. doi: 10.1186/s13229-018-0222-8
Benson, D. L., and Tanaka, H. (1998). N-cadherin redistribution during synaptogenesis in hippocampal neurons. J. Neurosci. 18, 6892–6904. doi: 10.1523/JNEUROSCI.18-17-06892.1998
Berkel, S., Marshall, C. R., Weiss, B., Howe, J., Roeth, R., Moog, U., et al. (2010). Mutations in the SHANK2 synaptic scaffolding gene in autism spectrum disorder and mental retardation. Nat. Genet. 42, 489–491. doi: 10.1038/ng.589
Berns, D. S., DeNardo, L. A., Pederick, D. T., and Luo, L. (2018). Teneurin-3 controls topographic circuit assembly in the hippocampus. Nature 554, 328–333. doi: 10.1038/nature25463
Bian, W. J., Miao, W. Y., He, S. J., Qiu, Z., and Yu, X. (2015). Coordinated Spine Pruning and Maturation Mediated by Inter-Spine Competition for Cadherin/Catenin Complexes. Cell 162, 808–822. doi: 10.1016/j.cell.2015.07.018
Biederer, T., Sara, Y., Mozhayeva, M., Atasoy, D., Liu, X., Kavalali, E. T., et al. (2002). SynCAM, a synaptic adhesion molecule that drives synapse assembly. Science 297, 1525–1531. doi: 10.1126/science.1072356
Bingol, B., and Sheng, M. (2011). Deconstruction for reconstruction: The role of proteolysis in neural plasticity and disease. Neuron 69, 22–32. doi: 10.1016/j.neuron.2010.11.006
Bloodgood, B. L., Sharma, N., Browne, H. A., Trepman, A. Z., and Greenberg, M. E. (2013). The activity-dependent transcription factor NPAS4 regulates domain-specific inhibition. Nature 503, 121–125. doi: 10.1038/nature12743
Boucard, A. A., Ko, J., and Sudhof, T. C. (2012). High affinity neurexin binding to cell adhesion G-protein-coupled receptor CIRL1/latrophilin-1 produces an intercellular adhesion complex. J. Biol. Chem. 287, 9399–9413. doi: 10.1074/jbc.M111.318659
Brakeman, P. R., Lanahan, A. A., O’Brien, R., Roche, K., Barnes, C. A., Huganir, R. L., et al. (1997). Homer: A protein that selectively binds metabotropic glutamate receptors. Nature 386, 284–288. doi: 10.1038/386284a0
Brenner, S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. doi: 10.1093/genetics/77.1.71
Bresler, T., Ramati, Y., Zamorano, P. L., Zhai, R., Garner, C. C., and Ziv, N. E. (2001). The dynamics of SAP90/PSD-95 recruitment to new synaptic junctions. Mol. Cell Neurosci. 18, 149–167. doi: 10.1006/mcne.2001.1012
Bresler, T., Shapira, M., Boeckers, T., Dresbach, T., Futter, M., Garner, C. C., et al. (2004). Postsynaptic density assembly is fundamentally different from presynaptic active zone assembly. J. Neurosci. 24, 1507–1520. doi: 10.1523/JNEUROSCI.3819-03.2004
Brose, N., Hofmann, K., Hata, Y., and Sudhof, T. C. (1995). Mammalian homologues of Caenorhabditis elegans unc-13 gene define novel family of C2-domain proteins. J. Biol. Chem. 270, 25273–25280. doi: 10.1074/jbc.270.42.25273
Brose, N., Petrenko, A. G., Sudhof, T. C., and Jahn, R. (1992). Synaptotagmin: A calcium sensor on the synaptic vesicle surface. Science 256, 1021–1025. doi: 10.1126/science.1589771
Bruses, J. L. (2006). N-cadherin signaling in synapse formation and neuronal physiology. Mol. Neurobiol. 33, 237–252. doi: 10.1385/MN:33:3:237
Burbridge, T. J., Xu, H. P., Ackman, J. B., Ge, X., Zhang, Y., Ye, M. J., et al. (2014). Visual circuit development requires patterned activity mediated by retinal acetylcholine receptors. Neuron 84, 1049–1064. doi: 10.1016/j.neuron.2014.10.051
Cai, Q., Pan, P. Y., and Sheng, Z. H. (2007). Syntabulin-kinesin-1 family member 5B-mediated axonal transport contributes to activity-dependent presynaptic assembly. J. Neurosci. 27, 7284–7296. doi: 10.1523/JNEUROSCI.0731-07.2007
Carvill, G. L., Weckhuysen, S., McMahon, J. M., Hartmann, C., Moller, R. S., Hjalgrim, H., et al. (2014). GABRA1 and STXBP1: Novel genetic causes of Dravet syndrome. Neurology 82, 1245–1253. doi: 10.1212/WNL.0000000000000291
Cases-Langhoff, C., Voss, B., Garner, A. M., Appeltauer, U., Takei, K., Kindler, S., et al. (1996). Piccolo, a novel 420 kDa protein associated with the presynaptic cytomatrix. Eur. J. Cell Biol. 69, 214–223.
Cen, C., Luo, L. D., Li, W. Q., Li, G., Tian, N. X., Zheng, G., et al. (2018). PKD1 Promotes Functional Synapse Formation Coordinated with N-Cadherin in Hippocampus. J. Neurosci. 38, 183–199. doi: 10.1523/JNEUROSCI.1640-17.2017
Charrier, C., Joshi, K., Coutinho-Budd, J., Kim, J. E., Lambert, N., de Marchena, J., et al. (2012). Inhibition of SRGAP2 function by its human-specific paralogs induces neoteny during spine maturation. Cell 149, 923–935. doi: 10.1016/j.cell.2012.03.034
Cheadle, L., Rivera, S. A., Phelps, J. S., Ennis, K. A., Stevens, B., Burkly, L. C., et al. (2020). Sensory Experience Engages Microglia to Shape Neural Connectivity through a Non-Phagocytic Mechanism. Neuron 108, 451–468.e9. doi: 10.1016/j.neuron.2020.08.002
Chen, C. H., Huang, Y. S., Liao, D. L., Huang, C. Y., Lin, C. H., and Fang, T. H. (2021). Identification of Rare Mutations of Two Presynaptic Cytomatrix Genes BSN and PCLO in Schizophrenia and Bipolar Disorder. J. Pers. Med. 11:1057. doi: 10.3390/jpm11111057
Chen, H. J., Rojas-Soto, M., Oguni, A., and Kennedy, M. B. (1998). A synaptic Ras-GTPase activating protein (p135 SynGAP) inhibited by CaM kinase II. Neuron 20, 895–904. doi: 10.1016/S0896-6273(00)80471-7
Chen, L. Y., Jiang, M., Zhang, B., Gokce, O., and Sudhof, T. C. (2017). Conditional Deletion of All Neurexins Defines Diversity of Essential Synaptic Organizer Functions for Neurexins. Neuron 94, 611–625.e4. doi: 10.1016/j.neuron.2017.04.011
Chen, X., Wu, X., Wu, H., and Zhang, M. (2020). Phase separation at the synapse. Nat. Neurosci. 23, 301–310. doi: 10.1038/s41593-019-0579-9
Chia, P. H., Li, P., and Shen, K. (2013). Cell biology in neuroscience: Cellular and molecular mechanisms underlying presynapse formation. J. Cell Biol. 203, 11–22. doi: 10.1083/jcb.201307020
Chih, B., Engelman, H., and Scheiffele, P. (2005). Control of excitatory and inhibitory synapse formation by neuroligins. Science 307, 1324–1328. doi: 10.1126/science.1107470
Chih, B., Gollan, L., and Scheiffele, P. (2006). Alternative splicing controls selective trans-synaptic interactions of the neuroligin-neurexin complex. Neuron 51, 171–178. doi: 10.1016/j.neuron.2006.06.005
Chiu, C. Q., Martenson, J. S., Yamazaki, M., Natsume, R., Sakimura, K., Tomita, S., et al. (2018). Input-Specific NMDAR-Dependent Potentiation of Dendritic GABAergic Inhibition. Neuron 97, 368–377.e3. doi: 10.1016/j.neuron.2017.12.032
Cho, K. O., Hunt, C. A., and Kennedy, M. B. (1992). The rat brain postsynaptic density fraction contains a homolog of the Drosophila discs-large tumor suppressor protein. Neuron 9, 929–942. doi: 10.1016/0896-6273(92)90245-9
Choi, K. H., Higgs, B. W., Wendland, J. R., Song, J., McMahon, F. J., and Webster, M. J. (2011). Gene expression and genetic variation data implicate PCLO in bipolar disorder. Biol. Psychiatry 69, 353–359. doi: 10.1016/j.biopsych.2010.09.042
Christopherson, K. S., Ullian, E. M., Stokes, C. C., Mullowney, C. E., Hell, J. W., Agah, A., et al. (2005). Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell 120, 421–433. doi: 10.1016/j.cell.2004.12.020
Chubykin, A. A., Atasoy, D., Etherton, M. R., Brose, N., Kavalali, E. T., Gibson, J. R., et al. (2007). Activity-dependent validation of excitatory versus inhibitory synapses by neuroligin-1 versus neuroligin-2. Neuron 54, 919–931. doi: 10.1016/j.neuron.2007.05.029
Chubykin, A. A., Liu, X., Comoletti, D., Tsigelny, I., Taylor, P., and Sudhof, T. C. (2005). Dissection of synapse induction by neuroligins: Effect of a neuroligin mutation associated with autism. J. Biol. Chem. 280, 22365–22374. doi: 10.1074/jbc.M410723200
Clement, J. P., Aceti, M., Creson, T. K., Ozkan, E. D., Shi, Y., Reish, N. J., et al. (2012). Pathogenic SYNGAP1 mutations impair cognitive development by disrupting maturation of dendritic spine synapses. Cell 151, 709–723. doi: 10.1016/j.cell.2012.08.045
Coley, A. A., and Gao, W. J. (2018). PSD95: A synaptic protein implicated in schizophrenia or autism? Prog. Neuropsychopharmacol. Biol. Psychiatry 82, 187–194. doi: 10.1016/j.pnpbp.2017.11.016
Colman, H., and Lichtman, J. W. (1993). Interactions between nerve and muscle: Synapse elimination at the developing neuromuscular junction. Dev. Biol. 156, 1–10. doi: 10.1006/dbio.1993.1054
Colon-Ramos, D. A. (2009). Synapse formation in developing neural circuits. Curr. Top Dev. Biol. 87, 53–79. doi: 10.1016/S0070-2153(09)01202-2
Colon-Ramos, D. A., Margeta, M. A., and Shen, K. (2007). Glia promote local synaptogenesis through UNC-6 (netrin) signaling in C. elegans. Science 318, 103–106. doi: 10.1126/science.1143762
Cousin, M. A., Creighton, B. A., Breau, K. A., Spillmann, R. C., Torti, E., Dontu, S., et al. (2021). Pathogenic SPTBN1 variants cause an autosomal dominant neurodevelopmental syndrome. Nat. Genet. 53, 1006–1021. doi: 10.1038/s41588-021-00886-z
Craig, A. M., and Kang, Y. (2007). Neurexin-neuroligin signaling in synapse development. Curr. Opin. Neurobiol. 17, 43–52. doi: 10.1016/j.conb.2007.01.011
Dai, Y., Taru, H., Deken, S. L., Grill, B., Ackley, B., Nonet, M. L., et al. (2006). SYD-2 Liprin-alpha organizes presynaptic active zone formation through ELKS. Nat. Neurosci. 9, 1479–1487. doi: 10.1038/nn1808
De Camilli, P., and Takei, K. (1996). Molecular mechanisms in synaptic vesicle endocytosis and recycling. Neuron 16, 481–486. doi: 10.1016/S0896-6273(00)80068-9
De Robertis, E., and Franchi, C. M. (1956). Electron microscope observations on synaptic vesicles in synapses of the retinal rods and cones. J. Biophys. Biochem. Cytol. 2, 307–318. doi: 10.1083/jcb.2.3.307
de Wit, J., Sylwestrak, E., O’Sullivan, M. L., Otto, S., Tiglio, K., Savas, J. N., et al. (2009). LRRTM2 interacts with Neurexin1 and regulates excitatory synapse formation. Neuron 64, 799–806. doi: 10.1016/j.neuron.2009.12.019
Deciphering Developmental Disorders Study. (2015). Large-scale discovery of novel genetic causes of developmental disorders. Nature 519, 223–228. doi: 10.1038/nature14135
Deciphering Developmental Disorders Study. (2017). Prevalence and architecture of de novo mutations in developmental disorders. Nature 542, 433–438. doi: 10.1038/nature21062
Defelipe, J. (2011). The evolution of the brain, the human nature of cortical circuits, and intellectual creativity. Front. Neuroanat. 5:29. doi: 10.3389/fnana.2011.00029
Dennis, M. Y., Nuttle, X., Sudmant, P. H., Antonacci, F., Graves, T. A., Nefedov, M., et al. (2012). Evolution of human-specific neural SRGAP2 genes by incomplete segmental duplication. Cell 149, 912–922. doi: 10.1016/j.cell.2012.03.033
Dieck, S. Tom, Sanmarti-Vila, L., Langnaese, K., Richter, K., Kindler, S., Soyke, A., et al. (1998). Bassoon, a novel zinc-finger CAG/glutamine-repeat protein selectively localized at the active zone of presynaptic nerve terminals. J. Cell. Biol. 142, 499–509. doi: 10.1083/jcb.142.2.499
Donato, F., Rompani, S. B., and Caroni, P. (2013). Parvalbumin-expressing basket-cell network plasticity induced by experience regulates adult learning. Nature 504, 272–276. doi: 10.1038/nature12866
Doyle, M., and Kiebler, M. A. (2011). Mechanisms of dendritic mRNA transport and its role in synaptic tagging. EMBO J. 30, 3540–3552. doi: 10.1038/emboj.2011.278
Dunah, A. W., Hueske, E., Wyszynski, M., Hoogenraad, C. C., Jaworski, J., Pak, D. T., et al. (2005). LAR receptor protein tyrosine phosphatases in the development and maintenance of excitatory synapses. Nat. Neurosci. 8, 458–467. doi: 10.1038/nn1416
Durand, C. M., Betancur, C., Boeckers, T. M., Bockmann, J., Chaste, P., Fauchereau, F., et al. (2007). Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat. Genet. 39, 25–27. doi: 10.1038/ng1933
Ebert, D. H., and Greenberg, M. E. (2013). Activity-dependent neuronal signalling and autism spectrum disorder. Nature 493, 327–337. doi: 10.1038/nature11860
Emperador-Melero, J., de Nola, G., and Kaeser, P. S. (2021). Intact synapse structure and function after combined knockout of PTPdelta, PTPsigma, and LAR. eLife 10:e66638. doi: 10.7554/eLife.66638
Engert, F., and Bonhoeffer, T. (1999). Dendritic spine changes associated with hippocampal long-term synaptic plasticity. Nature 399, 66–70. doi: 10.1038/19978
Eroglu, C., Allen, N. J., Susman, M. W., O’Rourke, N. A., Park, C. Y., Ozkan, E., et al. (2009). Gabapentin receptor alpha2delta-1 is a neuronal thrombospondin receptor responsible for excitatory CNS synaptogenesis. Cell 139, 380–392. doi: 10.1016/j.cell.2009.09.025
Favuzzi, E., Huang, S., Saldi, G. A., Binan, L., Ibrahim, L. A., Fernandez-Otero, M., et al. (2021). GABA-receptive microglia selectively sculpt developing inhibitory circuits. Cell 184:5686. doi: 10.1016/j.cell.2021.10.009
Fejtova, A., Davydova, D., Bischof, F., Lazarevic, V., Altrock, W. D., Romorini, S., et al. (2009). Dynein light chain regulates axonal trafficking and synaptic levels of Bassoon. J. Cell Biol. 185, 341–355. doi: 10.1083/jcb.200807155
Feller, M. B., Wellis, D. P., Stellwagen, D., Werblin, F. S., and Shatz, C. J. (1996). Requirement for cholinergic synaptic transmission in the propagation of spontaneous retinal waves. Science 272, 1182–1187. doi: 10.1126/science.272.5265.1182
Fenster, S. D., Chung, W. J., Zhai, R., Cases-Langhoff, C., Voss, B., Garner, A. M., et al. (2000). Piccolo, a presynaptic zinc finger protein structurally related to bassoon. Neuron 25, 203–214. doi: 10.1016/S0896-6273(00)80883-1
Fischer, M., Kaech, S., Knutti, D., and Matus, A. (1998). Rapid actin-based plasticity in dendritic spines. Neuron 20, 847–854. doi: 10.1016/S0896-6273(00)80467-5
Flavell, S. W., Cowan, C. W., Kim, T. K., Greer, P. L., Lin, Y., Paradis, S., et al. (2006). Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science 311, 1008–1012. doi: 10.1126/science.1122511
Flores, C. E., Nikonenko, I., Mendez, P., Fritschy, J. M., Tyagarajan, S. K., and Muller, D. (2015). Activity-dependent inhibitory synapse remodeling through gephyrin phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 112:E65–E72. doi: 10.1073/pnas.1411170112
Fortin, D. A., Tillo, S. E., Yang, G., Rah, J. C., Melander, J. B., Bai, S., et al. (2014). imaging of endogenous PSD-95 using ENABLED: A conditional strategy to fluorescently label endogenous proteins. J. Neurosci. 34, 16698–16712. doi: 10.1523/JNEUROSCI.3888-14.2014
Fossati, M., Pizzarelli, R., Schmidt, E. R., Kupferman, J. V., Stroebel, D., Polleux, F., et al. (2016). SRGAP2 and Its Human-Specific Paralog Co-Regulate the Development of Excitatory and Inhibitory Synapses. Neuron 91, 356–369. doi: 10.1016/j.neuron.2016.06.013
Freeman, M. R. (2005). Glial control of synaptogenesis. Cell 120, 292–293. doi: 10.1016/j.cell.2005.01.021
Fujii, R., Okabe, S., Urushido, T., Inoue, K., Yoshimura, A., Tachibana, T., et al. (2005). The RNA binding protein TLS is translocated to dendritic spines by mGluR5 activation and regulates spine morphology. Curr. Biol. 15, 587–593. doi: 10.1016/j.cub.2005.01.058
Fujiwara, T., Mishima, T., Kofuji, T., Chiba, T., Tanaka, K., Yamamoto, A., et al. (2006). Analysis of knock-out mice to determine the role of HPC-1/syntaxin 1A in expressing synaptic plasticity. J. Neurosci. 26, 5767–5776. doi: 10.1523/JNEUROSCI.0289-06.2006
Gamache, T. R., Araki, Y., and Huganir, R. L. (2020). Twenty Years of SynGAP Research: From Synapses to Cognition. J. Neurosci. 40, 1596–1605. doi: 10.1523/JNEUROSCI.0420-19.2020
Gan, W. B. (2003). Glutamate-dependent stabilization of presynaptic terminals. Neuron 38, 677–678. doi: 10.1016/S0896-6273(03)00333-7
Garner, C. C., Farmer, L., and Matus, A. (1988). Single-shot cloning of multiple cDNAs coding for a set of related microtubule-associated proteins. Gene 71, 483–490. doi: 10.1016/0378-1119(88)90065-0
Glock, C., Heumuller, M., and Schuman, E. M. (2017). mRNA transport & local translation in neurons. Curr. Opin. Neurobiol. 45, 169–177. doi: 10.1016/j.conb.2017.05.005
Goley, E. D., and Welch, M. D. (2006). The ARP2/3 complex: An actin nucleator comes of age. Nat. Rev. Mol. Cell Biol. 7, 713–726. doi: 10.1038/nrm2026
Graf, E. R., Zhang, X., Jin, S. X., Linhoff, M. W., and Craig, A. M. (2004). Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell 119, 1013–1026. doi: 10.1016/j.cell.2004.11.035
Greenberg, M. E., Ziff, E. B., and Greene, L. A. (1986). Stimulation of neuronal acetylcholine receptors induces rapid gene transcription. Science 234, 80–83. doi: 10.1126/science.3749894
Grutzendler, K., Kasthuri, N., and Gan, W. B. (2002). Long-term dendritic spine stability in the adult cortex. Nature 420, 812–816. doi: 10.1038/nature01276
Hall, D. H., and Hedgecock, E. M. (1991). Kinesin-related gene unc-104 is required for axonal transport of synaptic vesicles in C. elegans. Cell 65, 837–847. doi: 10.1016/0092-8674(91)90391-B
Hallam, S. J., and Jin, Y. (1998). lin-14 regulates the timing of synaptic remodelling in Caenorhabditis elegans. Nature 395, 78–82. doi: 10.1038/25757
Hamdan, F. F., Daoud, H., Piton, A., Gauthier, J., Dobrzeniecka, S., Krebs, M. O., et al. (2011). De novo SYNGAP1 mutations in nonsyndromic intellectual disability and autism. Biol. Psychiatry 69, 898–901. doi: 10.1016/j.biopsych.2010.11.015
Hamdan, F. F., Gauthier, J., Spiegelman, D., Noreau, A., Yang, Y., Pellerin, S., et al. (2009). Mutations in SYNGAP1 in autosomal nonsyndromic mental retardation. N. Engl. J. Med. 360, 599–605. doi: 10.1056/NEJMoa0805392
Hamilton, A. M., Oh, W. C., Vega-Ramirez, H., Stein, I. S., Hell, J. W., Patrick, G. N., et al. (2012). Activity-dependent growth of new dendritic spines is regulated by the proteasome. Neuron 74, 1023–1030. doi: 10.1016/j.neuron.2012.04.031
Hammond, T. R., Robinton, D., and Stevens, B. (2018). Microglia and the Brain: Complementary Partners in Development and Disease. Annu. Rev. Cell Dev. Biol. 34, 523–544. doi: 10.1146/annurev-cellbio-100616-060509
Han, K. A., Ko, J. S., Pramanik, G., Kim, J. Y., Tabuchi, K., Um, J. W., et al. (2018). PTPsigma Drives Excitatory Presynaptic Assembly via Various Extracellular and Intracellular Mechanisms. J. Neurosci. 38, 6700–6721. doi: 10.1523/JNEUROSCI.0672-18.2018
Harward, S. C., Hedrick, N. G., Hall, C. E., Parra-Bueno, P., Milner, T. A., Pan, E., et al. (2016). Autocrine BDNF-TrkB signalling within a single dendritic spine. Nature 538, 99–103. doi: 10.1038/nature19766
Hata, Y., Butz, S., and Sudhof, T. C. (1996). CASK: A novel dlg/PSD95 homolog with an N-terminal calmodulin-dependent protein kinase domain identified by interaction with neurexins. J. Neurosci. 16, 2488–2494. doi: 10.1523/JNEUROSCI.16-08-02488.1996
Hata, Y., Davletov, B., Petrenko, A. G., Jahn, R., and Sudhof, T. C. (1993a). Interaction of synaptotagmin with the cytoplasmic domains of neurexins. Neuron 10, 307–315. doi: 10.1016/0896-6273(93)90320-Q
Hata, Y., Slaughter, C. A., and Sudhof, T. C. (1993b). Synaptic vesicle fusion complex contains unc-18 homologue bound to syntaxin. Nature 366, 347–351. doi: 10.1038/366347a0
Hayashi, S., Mizuno, S., Migita, O., Okuyama, T., Makita, Y., Hata, A., et al. (2008). The CASK gene harbored in a deletion detected by array-CGH as a potential candidate for a gene causative of X-linked dominant mental retardation. Am. J. Med. Genet A 146A, 2145–2151. doi: 10.1002/ajmg.a.32433
Hedrick, N. G., Harward, S. C., Hall, C. E., Murakoshi, H., McNamara, J. O., and Yasuda, R. (2016). Rho GTPase complementation underlies BDNF-dependent homo- and heterosynaptic plasticity. Nature 538, 104–108. doi: 10.1038/nature19784
Hida, Y., and Ohtsuka, T. (2010). CAST and ELKS proteins: Structural and functional determinants of the presynaptic active zone. J. Biochem. 148, 131–137. doi: 10.1093/jb/mvq065
Hill, S. F., and Meisler, M. H. (2021). Antisense Oligonucleotide Therapy for Neurodevelopmental Disorders. Dev. Neurosci. 43, 247–252. doi: 10.1159/000517686
Hirokawa, N., and Takemura, R. (2005). Molecular motors and mechanisms of directional transport in neurons. Nat. Rev. Neurosci. 6, 201–214. doi: 10.1038/nrn1624
Hlushchenko, I., Khanal, P., Abouelezz, A., Paavilainen, V. O., and Hotulainen, P. (2018). ASD-Associated De Novo Mutations in Five Actin Regulators Show Both Shared and Distinct Defects in Dendritic Spines and Inhibitory Synapses in Cultured Hippocampal Neurons. Front. Cell Neurosci. 12:217. doi: 10.3389/fncel.2018.00217
Horn, K. E., Xu, B., Gobert, D., Hamam, B. N., Thompson, K. M., Wu, C. L., et al. (2012). Receptor protein tyrosine phosphatase sigma regulates synapse structure, function and plasticity. J. Neurochem. 122, 147–161. doi: 10.1111/j.1471-4159.2012.07762.x
Hotulainen, P., and Hoogenraad, C. C. (2010). Actin in dendritic spines: Connecting dynamics to function. J. Cell Biol. 189, 619–629. doi: 10.1083/jcb.201003008
Huang, G., Osorio, D., Guan, J., Ji, G., and Cai, J. J. (2020). Overdispersed gene expression in schizophrenia. NPJ Schizophr. 6:9. doi: 10.1038/s41537-020-0097-5
Hubel, D., and Wiesel, T. (2012). David Hubel and Torsten Wiesel. Neuron 75, 182–184. doi: 10.1016/j.neuron.2012.07.002
Hung, A. Y., Futai, K., Sala, C., Valtschanoff, J. G., Ryu, J., Woodworth, M. A., et al. (2008). Smaller dendritic spines, weaker synaptic transmission, but enhanced spatial learning in mice lacking Shank1. J. Neurosci. 28, 1697–1708. doi: 10.1523/JNEUROSCI.3032-07.2008
Ichtchenko, K., Hata, Y., Nguyen, T., Ullrich, B., Missler, M., Moomaw, C., et al. (1995). Neuroligin 1: A splice site-specific ligand for beta-neurexins. Cell 81, 435–443. doi: 10.1016/0092-8674(95)90396-8
Jeyabalan, J., and Clement, J. P. (2016). SYNGAP1: Mind the Gap. Front. Cell Neurosci. 10:32. doi: 10.3389/fncel.2016.00032
Jin, Y., and Garner, C. C. (2008). Molecular mechanisms of presynaptic differentiation. Annu. Rev. Cell Dev. Biol. 24, 237–262. doi: 10.1146/annurev.cellbio.23.090506.123417
Johnson, M. B., Kawasawa, Y. I., Mason, C. E., Krsnik, Z., Coppola, G., Bogdanovic, D., et al. (2009). Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron 62, 494–509. doi: 10.1016/j.neuron.2009.03.027
Kadowaki, M., Nakamura, S., Machon, O., Krauss, S., Radice, G. L., and Takeichi, M. (2007). N-cadherin mediates cortical organization in the mouse brain. Dev. Biol. 304, 22–33. doi: 10.1016/j.ydbio.2006.12.014
Kang, H., and Schuman, E. M. (1996). A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science 273, 1402–1406. doi: 10.1126/science.273.5280.1402
Katz, L. C., and Shatz, C. J. (1996). Synaptic activity and the construction of cortical circuits. Science 274, 1133–1138. doi: 10.1126/science.274.5290.1133
Kelleher, R. J. III, Geigenmuller, U., Hovhannisyan, H., Trautman, E., Pinard, R., Rathmell, B., et al. (2012). High-throughput sequencing of mGluR signaling pathway genes reveals enrichment of rare variants in autism. PLoS One 7:e35003. doi: 10.1371/journal.pone.0035003
Kim, E., Cho, K. O., Rothschild, A., and Sheng, M. (1996). Heteromultimerization and NMDA receptor-clustering activity of Chapsyn-110, a member of the PSD-95 family of proteins. Neuron 17, 103–113. doi: 10.1016/S0896-6273(00)80284-6
Kim, E., Naisbitt, S., Hsueh, Y. P., Rao, A., Rothschild, A., Craig, A. M., et al. (1997). GKAP, a novel synaptic protein that interacts with the guanylate kinase-like domain of the PSD-95/SAP90 family of channel clustering molecules. J. Cell Biol. 136, 669–678. doi: 10.1083/jcb.136.3.669
Kim, J. H., Liao, D., Lau, L. F., and Huganir, R. L. (1998). SynGAP: A synaptic RasGAP that associates with the PSD-95/SAP90 protein family. Neuron 20, 683–691. doi: 10.1016/S0896-6273(00)81008-9
Kistner, U., Wenzel, B. M., Veh, R. W., Cases-Langhoff, C., Garner, A. M., Appeltauer, U., et al. (1993). SAP90, a rat presynaptic protein related to the product of the Drosophila tumor suppressor gene dlg-A. J. Biol. Chem. 268, 4580–4583. doi: 10.1016/S0021-9258(18)53433-5
Knott, G. W., Quairiaux, C., Genoud, C., and Welker, E. (2002). Formation of dendritic spines with GABAergic synapses induced by whisker stimulation in adult mice. Neuron 34, 265–273. doi: 10.1016/S0896-6273(02)00663-3
Ko, J., Fuccillo, M. V., Malenka, R. C., and Sudhof, T. C. (2009). LRRTM2 functions as a neurexin ligand in promoting excitatory synapse formation. Neuron 64, 791–798. doi: 10.1016/j.neuron.2009.12.012
Kolkman, M. J., Streijger, F., Linkels, M., Bloemen, M., Heeren, D. J., Hendriks, W. J., et al. (2004). Mice lacking leukocyte common antigen-related (LAR) protein tyrosine phosphatase domains demonstrate spatial learning impairment in the two-trial water maze and hyperactivity in multiple behavioural tests. Behav. Brain Res. 154, 171–182. doi: 10.1016/j.bbr.2004.02.006
Kondo, L., Takei, Y., and Hirokawa, N. (2012). Motor protein KIF1A is essential for hippocampal synaptogenesis and learning enhancement in an enriched environment. Neuron 73, 743–757. doi: 10.1016/j.neuron.2011.12.020
Korobova, F., and Svitkina, T. (2010). Molecular architecture of synaptic actin cytoskeleton in hippocampal neurons reveals a mechanism of dendritic spine morphogenesis. Mol. Biol. Cell 21, 165–176. doi: 10.1091/mbc.e09-07-0596
Krumm, N., O’Roak, B. J., Shendure, J., Eichler, E. E., and de, A. (2014). novo convergence of autism genetics and molecular neuroscience. Trends Neurosci. 37, 95–105. doi: 10.1016/j.tins.2013.11.005
Kumar, V., Zhang, M. X., Swank, M. W., Kunz, J., and Wu, G. Y. (2005). Regulation of dendritic morphogenesis by Ras-PI3K-Akt-mTOR and Ras-MAPK signaling pathways. J. Neurosci. 25, 11288–11299. doi: 10.1523/JNEUROSCI.2284-05.2005
Kuriu, T., Inoue, A., Bito, H., Sobue, K., and Okabe, S. (2006). Differential control of postsynaptic density scaffolds via actin-dependent and -independent mechanisms. J. Neurosci. 26, 7693–7706. doi: 10.1523/JNEUROSCI.0522-06.2006
Kwon, H. B., and Sabatini, B. L. (2011). Glutamate induces de novo growth of functional spines in developing cortex. Nature 474, 100–104. doi: 10.1038/nature09986
Kwon, S. E., and Chapman, E. R. (2011). Synaptophysin regulates the kinetics of synaptic vesicle endocytosis in central neurons. Neuron 70, 847–854. doi: 10.1016/j.neuron.2011.04.001
Lai, C. S., Franke, T. F., and Gan, W. B. (2012). Opposite effects of fear conditioning and extinction on dendritic spine remodelling. Nature 483, 87–91. doi: 10.1038/nature10792
Landis, D. M., and Reese, T. S. (1983). Cytoplasmic organization in cerebellar dendritic spines. J. Cell Biol. 97, 1169–1178. doi: 10.1083/jcb.97.4.1169
Langnaese, K., Seidenbecher, C., Wex, H., Seidel, B., Hartung, K., Appeltauer, U., et al. (1996). Protein components of a rat brain synaptic junctional protein preparation. Brain Res. Mol. Brain Res. 42, 118–122. doi: 10.1016/S0169-328X(96)00147-7
Lawler, S. (1999). Regulation of actin dynamics: The LIM kinase connection. Curr. Biol. 9:R800–R802. doi: 10.1016/S0960-9822(99)80493-X
Lee, H., Shin, W., Kim, K., Lee, S., Lee, E. J., Kim, J., et al. (2019). NGL-3 in the regulation of brain development, Akt/GSK3b signaling, long-term depression, and locomotive and cognitive behaviors. PLoS Biol. 17:e2005326. doi: 10.1371/journal.pbio.2005326
Lehrman, E. K., Wilton, D. K., Litvina, E. Y., Welsh, C. A., Chang, S. T., Frouin, A., et al. (2018). CD47 Protects Synapses from Excess Microglia-Mediated Pruning during Development. Neuron 100, 120–134.e6. doi: 10.1016/j.neuron.2018.09.017
Li, J., Xie, Y., Cornelius, S., Jiang, X., Sando, R., Kordon, S. P., et al. (2020). Alternative splicing controls teneurin-latrophilin interaction and synapse specificity by a shape-shifting mechanism. Nat. Commun. 11:2140. doi: 10.1038/s41467-020-16029-7
LIamosas, A. V., Vij, R., Kilinc, M., Bijoch, L., Rojas, C., Reich, A., et al. (2020). Human SYNGAP1 Regulates the Development of Neuronal Activity by Controlling Dendritic and Synaptic Maturation. Biorxiv [Preprint]. doi: 10.1101/2020.06.01.127613
Lian, G., and Sheen, V. L. (2015). Cytoskeletal proteins in cortical development and disease: Actin associated proteins in periventricular heterotopia. Front. Cell Neurosci. 9:99. doi: 10.3389/fncel.2015.00099
Liang, M., Jin, G., Xie, X., Zhang, W., Li, K., Niu, F., et al. (2021). Oligomerized liprin-alpha promotes phase separation of ELKS for compartmentalization of presynaptic active zone proteins. Cell Rep. 34:108901. doi: 10.1016/j.celrep.2021.108901
Lie, E., Li, Y., Kim, R., and Kim, E. (2018). SALM/Lrfn Family Synaptic Adhesion Molecules. Front. Mol. Neurosci. 11:105. doi: 10.3389/fnmol.2018.00105
Lin, Y., Bloodgood, B. L., Hauser, J. L., Lapan, A. D., Koon, A. C., Kim, T. K., et al. (2008). Activity-dependent regulation of inhibitory synapse development by Npas4. Nature 455, 1198–1204. doi: 10.1038/nature07319
Liu, X., Somel, M., Tang, L., Yan, Z., Jiang, X., Guo, S., et al. (2012). Extension of cortical synaptic development distinguishes humans from chimpanzees and macaques. Genome Res. 22, 611–622. doi: 10.1101/gr.127324.111
Lu, B. (2003). BDNF and activity-dependent synaptic modulation. Learn. Mem. 10, 86–98. doi: 10.1101/lm.54603
Luikart, B. W., Nef, S., Virmani, T., Lush, M. E., Liu, Y., Kavalali, E. T., et al. (2005). TrkB has a cell-autonomous role in the establishment of hippocampal Schaffer collateral synapses. J. Neurosci. 25, 3774–3786. doi: 10.1523/JNEUROSCI.0041-05.2005
Maletic-Savatic, M., Malinow, R., and Svoboda, K. (1999). Rapid dendritic morphogenesis in CA1 hippocampal dendrites induced by synaptic activity. Science 283, 1923–1927. doi: 10.1126/science.283.5409.1923
Martinez, A., Alcantara, S., Borrell, V., Del Rio, J. A., Blasi, J., Otal, R., et al. (1998). Silos-Santiago, and E. Soriano, TrkB and TrkC signaling are required for maturation and synaptogenesis of hippocampal connections. J. Neurosci. 18, 7336–7350. doi: 10.1523/JNEUROSCI.18-18-07336.1998
Maruyama, I. N., and Brenner, S. (1991). A phorbol ester/diacylglycerol-binding protein encoded by the unc-13 gene of Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 88, 5729–5733. doi: 10.1073/pnas.88.13.5729
Matsuda, K., Budisantoso, T., Mitakidis, N., Sugaya, Y., Miura, E., Kakegawa, W., et al. (2016). Transsynaptic Modulation of Kainate Receptor Functions by C1q-like Proteins. Neuron 90, 752–767. doi: 10.1016/j.neuron.2016.04.001
McDonald, N. A., Fetter, R. D., and Shen, K. (2020). Assembly of synaptic active zones requires phase separation of scaffold molecules. Nature 588, 454–458. doi: 10.1038/s41586-020-2942-0
Mejias, R., Adamczyk, A., Anggono, V., Niranjan, T., Thomas, G. M., Sharma, K., et al. (2011). Gain-of-function glutamate receptor interacting protein 1 variants alter GluA2 recycling and surface distribution in patients with autism. Proc. Natl. Acad. Sci. U.S.A. 108, 4920–4925. doi: 10.1073/pnas.1102233108
Miller, K. E., DeProto, J., Kaufmann, N., Patel, B. N., Duckworth, A., and Van Vactor, D. (2005). Direct observation demonstrates that Liprin-alpha is required for trafficking of synaptic vesicles. Curr. Biol. 15, 684–689. doi: 10.1016/j.cub.2005.02.061
Moessner, R., Marshall, C. R., Sutcliffe, J. S., Skaug, J., Pinto, D., Vincent, J., et al. (2007). Contribution of SHANK3 mutations to autism spectrum disorder. Am. J. Hum. Genet. 81, 1289–1297. doi: 10.1086/522590
Morabito, M. A., Sheng, M., and Tsai, L. H. (2004). Cyclin-dependent kinase 5 phosphorylates the N-terminal domain of the postsynaptic density protein PSD-95 in neurons. J. Neurosci. 24, 865–876. doi: 10.1523/JNEUROSCI.4582-03.2004
Moreno-Ramos, O. A., Olivares, A. M., Haider, N. B., de Autismo, L. C., and Lattig, M. C. (2015). Whole-Exome Sequencing in a South American Cohort Links ALDH1A3, FOXN1 and Retinoic Acid Regulation Pathways to Autism Spectrum Disorders. PLoS One 10:e0135927. doi: 10.1371/journal.pone.0135927
Muller, B. M., Kistner, U., Kindler, S., Chung, W. J., Kuhlendahl, S., Fenster, S. D., et al. (1996). SAP102, a novel postsynaptic protein that interacts with NMDA receptor complexes in vivo. Neuron 17, 255–265. doi: 10.1016/S0896-6273(00)80157-9
Muller, B. M., Kistner, U., Veh, R. W., Cases-Langhoff, C., Becker, B., Gundelfinger, E. D., et al. (1995). Molecular characterization and spatial distribution of SAP97, a novel presynaptic protein homologous to SAP90 and the Drosophila discs-large tumor suppressor protein. J. Neurosci. 15, 2354–2366. doi: 10.1523/JNEUROSCI.15-03-02354.1995
Nagappan, G., Zaitsev, E., Senatorov, V. V. Jr., Yang, J., Hempstead, B. L., and Lu, B. (2009). Control of extracellular cleavage of ProBDNF by high frequency neuronal activity. Proc. Natl. Acad. Sci. U.S.A. 106, 1267–1272. doi: 10.1073/pnas.0807322106
Naisbitt, S., Kim, E., Tu, J. C., Xiao, B., Sala, C., Valtschanoff, J., et al. (1999). Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron 23, 569–582. doi: 10.1016/S0896-6273(00)80809-0
Najm, J., Horn, D., Wimplinger, I., Golden, J. A., Chizhikov, V. V., Sudi, J., et al. (2008). Mutations of CASK cause an X-linked brain malformation phenotype with microcephaly and hypoplasia of the brainstem and cerebellum. Nat. Genet 40, 1065–1067. doi: 10.1038/ng.194
Nishiyama, J., and Yasuda, R. (2015). Biochemical Computation for Spine Structural Plasticity. Neuron 87, 63–75. doi: 10.1016/j.neuron.2015.05.043
Nonet, M. L. (1999). Visualization of synaptic specializations in live C. elegans with synaptic vesicle protein-GFP fusions. J. Neurosci. Methods 89, 33–40. doi: 10.1016/S0165-0270(99)00031-X
Norton, N., Williams, H. J., Williams, N. M., Spurlock, G., Zammit, S., Jones, G., et al. (2003). Mutation screening of the Homer gene family and association analysis in schizophrenia. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 120B, 18–21. doi: 10.1002/ajmg.b.20032
Oh, W. C., Lutzu, S., Castillo, P. E., and Kwon, H. B. (2016). De novo synaptogenesis induced by GABA in the developing mouse cortex. Science 353, 1037–1040. doi: 10.1126/science.aaf5206
Ohtsuka, T., Takao-Rikitsu, E., Inoue, E., Inoue, M., Takeuchi, M., Matsubara, K., et al. (2002). Cast: A novel protein of the cytomatrix at the active zone of synapses that forms a ternary complex with RIM1 and munc13-1. J. Cell Biol. 158, 577–590. doi: 10.1083/jcb.200202083
Okabe, S. (2020). Regulation of actin dynamics in dendritic spines: Nanostructure, molecular mobility, and signaling mechanisms. Mol. Cell Neurosci. 109:103564. doi: 10.1016/j.mcn.2020.103564
Okuda, T., Yu, L. M., Cingolani, L. A., Kemler, R., and Goda, Y. (2007). beta-Catenin regulates excitatory postsynaptic strength at hippocampal synapses. Proc. Natl. Acad. Sci. U.S.A. 104, 13479–13484. doi: 10.1073/pnas.0702334104
O’Roak, B. J., Vives, L., Fu, W., Egertson, J. D., Stanaway, I. B., Phelps, I. G., et al. (2012a). Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 338, 1619–1622. doi: 10.1126/science.1227764
O’Roak, B. J., Vives, L., Girirajan, S., Karakoc, E., Krumm, N., Coe, B. P., et al. (2012b). Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485, 246–250. doi: 10.1038/nature10989
Ou, C. Y., Poon, V. Y., Maeder, C. I., Watanabe, S., Lehrman, E. K., Fu, A. K., et al. (2010). Two cyclin-dependent kinase pathways are essential for polarized trafficking of presynaptic components. Cell 141, 846–858. doi: 10.1016/j.cell.2010.04.011
Pack-Chung, E., Kurshan, P. T., Dickman, D. K., Schwarz, T. L., and Drosophila, A. (2007). kinesin required for synaptic bouton formation and synaptic vesicle transport. Nat. Neurosci. 10, 980–989. doi: 10.1038/nn1936
Pak, D. T., Yang, S., Rudolph-Correia, S., Kim, E., and Sheng, M. (2001). Regulation of dendritic spine morphology by SPAR, a PSD-95-associated RapGAP. Neuron 31, 289–303. doi: 10.1016/S0896-6273(01)00355-5
Pan, Y., and Monje, M. (2020). Activity Shapes Neural Circuit Form and Function: A Historical Perspective. J. Neurosci. 40, 944–954. doi: 10.1523/JNEUROSCI.0740-19.2019
Papa, M., and Segal, M. (1996). Morphological plasticity in dendritic spines of cultured hippocampal neurons. Neuroscience 71, 1005–1011. doi: 10.1016/0306-4522(95)00490-4
Park, M., and Shen, K. (2012). WNTs in synapse formation and neuronal circuitry. EMBO J. 31, 2697–2704. doi: 10.1038/emboj.2012.145
Park, M., Watanabe, S., Poon, V. Y., Ou, C. Y., Jorgensen, E. M., and Shen, K. (2011). CYY-1/cyclin Y and CDK-5 differentially regulate synapse elimination and formation for rewiring neural circuits. Neuron 70, 742–757. doi: 10.1016/j.neuron.2011.04.002
Passafaro, I., Nakagawa, T., Sala, C., and Sheng, M. (2003). Induction of dendritic spines by an extracellular domain of AMPA receptor subunit GluR2. Nature 424, 677–681. doi: 10.1038/nature01781
Peng, Y. R., He, S., Marie, H., Zeng, S. Y., Ma, J., Tan, Z. J., et al. (2009). Coordinated changes in dendritic arborization and synaptic strength during neural circuit development. Neuron 61, 71–84. doi: 10.1016/j.neuron.2008.11.015
Penzes, L., Cahill, M. E., Jones, K. A., VanLeeuwen, J. E., and Woolfrey, K. M. (2011). Dendritic spine pathology in neuropsychiatric disorders. Nat. Neurosci. 14, 285–293. doi: 10.1038/nn.2741
Pereda, A. E. (2014). Electrical synapses and their functional interactions with chemical synapses. Nat. Rev. Neurosci. 15, 250–263. doi: 10.1038/nrn3708
Phillips, G. R., Huang, J. K., Wang, Y., Tanaka, H., Shapiro, L., and Zhang, W. (2001). The presynaptic particle web: Ultrastructure, composition, dissolution, and reconstitution. Neuron 32, 63–77. doi: 10.1016/S0896-6273(01)00450-0
Poulopoulos, A., Aramuni, G., Meyer, G., Soykan, T., Hoon, M., and Papadopoulos, T. (2009). Neuroligin 2 drives postsynaptic assembly at perisomatic inhibitory synapses through gephyrin and collybistin. Neuron 63, 628–642. doi: 10.1016/j.neuron.2009.08.023
Prange, O., Wong, T. P., Gerrow, K., Wang, Y. T., and El-Husseini, A. (2004). A balance between excitatory and inhibitory synapses is controlled by PSD-95 and neuroligin. Proc. Natl. Acad. Sci. U.S.A. 101, 13915–13920. doi: 10.1073/pnas.0405939101
Qi, C., Feng, I., Costa, A. R., Pinto-Costa, R., Neil, J. E., Caluseriu, O., et al. (2021). Variants in ADD1 cause intellectual disability, corpus callosum dysgenesis, and ventriculomegaly in humans. Genet. Med. 24, 319–331. doi: 10.1016/j.gim.2021.09.014
Rao, S., Lam, M. H., Yeung, V. S., Wing, Y. K., and Waye, M. M. (2016). Association of HOMER1 rs2290639 with suicide attempts in Hong Kong Chinese and the potentially functional role of this polymorphism. Springerplus 5:767. doi: 10.1186/s40064-016-2404-1
Rauch, A., Wieczorek, D., Graf, E., Wieland, T., Endele, S., Schwarzmayr, T., et al. (2012). Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: An exome sequencing study. Lancet 380, 1674–1682. doi: 10.1016/S0140-6736(12)61480-9
Reay, W. R., and Cairns, M. J. (2020). The role of the retinoids in schizophrenia: Genomic and clinical perspectives. Mol. Psychiatry 25, 706–718. doi: 10.1038/s41380-019-0566-2
Reichelt, A. C., and Dachtler, J. (2015). “The Role of Neurexins and Neuroligins in Autism,” in The Molecular Basis of Autism, ed. S. H. Fatemi (New York, NY: Springer), 361–381. doi: 10.1007/978-1-4939-2190-4_17
Ren, J. M., Li, P. M., Zhang, W. R., Sweet, L. J., Cline, G., Shulman, G. I., et al. (1998). Transgenic mice deficient in the LAR protein-tyrosine phosphatase exhibit profound defects in glucose homeostasis. Diabetes 47, 493–497. doi: 10.2337/diabetes.47.3.493
Richmond, J. E., Davis, W. S., and Jorgensen, E. M. (1999). UNC-13 is required for synaptic vesicle fusion in C. elegans. Nat. Neurosci. 2, 959–964. doi: 10.1038/14755
Rosahl, T. W., Spillane, D., Missler, M., Herz, J., Selig, D. K., Wolff, J. R., et al. (1995). Essential functions of synapsins I and II in synaptic vesicle regulation. Nature 375, 488–493. doi: 10.1038/375488a0
Rosenmund, C., Sigler, A., Augustin, I., Reim, K., Brose, N., and Rhee, J. S. (2002). Differential control of vesicle priming and short-term plasticity by Munc13 isoforms. Neuron 33, 411–424. doi: 10.1016/S0896-6273(02)00568-8
Saglietti, L., Dequidt, C., Kamieniarz, K., Rousset, M. C., Valnegri, P., Thoumine, O., et al. (2007). Extracellular interactions between GluR2 and N-cadherin in spine regulation. Neuron 54, 461–477. doi: 10.1016/j.neuron.2007.04.012
Saitsu, H., Kato, M., Mizuguchi, T., Hamada, K., Osaka, H., Tohyama, J., et al. (2008). De novo mutations in the gene encoding STXBP1 (MUNC18-1) cause early infantile epileptic encephalopathy. Nat. Genet. 40, 782–788. doi: 10.1038/ng.150
Sanders, S. J., Murtha, M. T., Gupta, A. R., Murdoch, J. D., Raubeson, M. J., Willsey, A. J., et al. (2012). De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 485, 237–241. doi: 10.1038/nature10945
Sanes, J. R., and Zipursky, S. L. (2020). Synaptic Specificity, Recognition Molecules, and Assembly of Neural Circuits. Cell 181, 1434–1435. doi: 10.1016/j.cell.2020.05.046
Sato, D., Lionel, A. C., Leblond, C. S., Prasad, A., Pinto, D., and Walker, S. (2012). SHANK1 Deletions in Males with Autism Spectrum Disorder. Am. J. Hum. Genet. 90, 879–887. doi: 10.1016/j.ajhg.2012.03.017
Satterstrom, F. K., Kosmicki, J. A., Wang, J., Breen, M. S., De Rubeis, S., An, J. Y., et al. (2020). Buxbaum, Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell 180, 568–584.e23. doi: 10.1016/j.cell.2019.12.036
Schafer, D. P., and Stevens, B. (2013). Phagocytic glial cells: Sculpting synaptic circuits in the developing nervous system. Curr. Opin. Neurobiol. 23, 1034–1040. doi: 10.1016/j.conb.2013.09.012
Schafer, D. P., Lehrman, E. K., Kautzman, A. G., Koyama, R., Mardinly, A. R., Yamasaki, R., et al. (2012). Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 74, 691–705. doi: 10.1016/j.neuron.2012.03.026
Schätzle, P., Esteves da Silva, M., Tas, R. P., Katrukha, E. A., Hu, H. Y., Wierenga, C. J., et al. (2018). Activity-Dependent Actin Remodeling at the Base of Dendritic Spines Promotes Microtubule Entry. Curr. Biol. 28, 2081–2093.e6. doi: 10.1016/j.cub.2018.05.004
Scheiffele, P., Fan, J., Choih, J., Fetter, R., and Serafini, T. (2000). Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell 101, 657–669. doi: 10.1016/S0092-8674(00)80877-6
Schmidt, E. R. E., and Polleux, F. (2021). Genetic Mechanisms Underlying the Evolution of Connectivity in the Human Cortex. Fron.t Neural Circuits 15:787164. doi: 10.3389/fncir.2021.787164
Sclip, A., and Sudhof, T. C. (2020). LAR receptor phospho-tyrosine phosphatases regulate NMDA-receptor responses. eLife 9:e53406. doi: 10.7554/eLife.53406
Sekar, A., Bialas, A. R., de Rivera, H., Davis, A., Hammond, T. R., Kamitaki, N., et al. (2016). Schizophrenia risk from complex variation of complement component 4. Nature 530, 177–183. doi: 10.1038/nature16549
Serra-Pages, C., Kedersha, N. L., Fazikas, L., Medley, Q., Debant, A., and Streuli, M. (1995). The LAR transmembrane protein tyrosine phosphatase and a coiled-coil LAR-interacting protein co-localize at focal adhesions. EMBO J. 14, 2827–2838. doi: 10.1002/j.1460-2075.1995.tb07282.x
Serra-Pages, C., Medley, Q. G., Tang, M., Hart, A., and Streuli, M. (1998). Liprins, a family of LAR transmembrane protein-tyrosine phosphatase-interacting proteins. J. Biol. Chem. 273, 15611–15620. doi: 10.1074/jbc.273.25.15611
Shao, Z., Watanabe, S., Christensen, R., Jorgensen, E. M., and Colon-Ramos, D. A. (2013). Synapse location during growth depends on glia location. Cell 154, 337–350. doi: 10.1016/j.cell.2013.06.028
Sharma, N., Deppmann, C. D., Harrington, A. W., St Hillaire, C., Chen, Z. Y., Lee, F. S., et al. (2010). Long-distance control of synapse assembly by target-derived NGF. Neuron 67, 422–434. doi: 10.1016/j.neuron.2010.07.018
Shatz, C. J. (1990). Impulse activity and the patterning of connections during CNS development. Neuron 5, 745–756. doi: 10.1016/0896-6273(90)90333-B
Shaw, H. S., and Salmon, C. K. (2017). Refining the Roles of Neuroligins in Synapse Development and Function: A Reductionist Conditional Knock-out Approach. J. Neurosci. 37, 11769–11771. doi: 10.1523/JNEUROSCI.2492-17.2017
Sheng, M., and Greenberg, M. E. (1990). The regulation and function of c-fos and other immediate early genes in the nervous system. Neuron 4, 477–485. doi: 10.1016/0896-6273(90)90106-P
Sheng, M., and Kim, E. (2011). The postsynaptic organization of synapses. Cold Spring Harb. Perspect. Biol. 3:a005678. doi: 10.1101/cshperspect.a005678
Sheng, M., and Kim, M. J. (2002). Postsynaptic signaling and plasticity mechanisms. Science 298, 776–780. doi: 10.1126/science.1075333
Shi, Y., Li, Q., and Shao, Z. (2018). Wnts Promote Synaptic Assembly Through T-Cell Specific Transcription Factors in Caenorhabditis elegans. Front. Mol. Neurosci. 11:194. doi: 10.3389/fnmol.2018.00194
Shibata, M., Pattabiraman, K., Lorente-Galdos, B., Andrijevic, D., Kim, S. K., Kaur, N., et al. (2021a). Regulation of prefrontal patterning and connectivity by retinoic acid. Nature 598, 483–488. doi: 10.1038/s41586-021-03953-x
Shibata, M., Pattabiraman, K., Muchnik, S. K., Kaur, N., Morozov, Y. M., Cheng, X., et al. (2021b). Hominini-specific regulation of CBLN2 increases prefrontal spinogenesis. Nature 598, 489–494. doi: 10.1038/s41586-021-03952-y
Shin, H., Wyszynski, M., Huh, K. H., Valtschanoff, J. G., Lee, J. R., Ko, J., et al. (2003). Association of the kinesin motor KIF1A with the multimodular protein liprin-alpha. J. Biol. Chem. 278, 11393–11401. doi: 10.1074/jbc.M211874200
Siddiqui, T. J., Pancaroglu, R., Kang, Y., Rooyakkers, A., and Craig, A. M. (2010). LRRTMs and neuroligins bind neurexins with a differential code to cooperate in glutamate synapse development. J. Neurosci. 30, 7495–7506. doi: 10.1523/JNEUROSCI.0470-10.2010
Song, J. Y., Ichtchenko, K., Sudhof, T. C., and Brose, N. (1999). Neuroligin 1 is a postsynaptic cell-adhesion molecule of excitatory synapses. Proc. Natl. Acad. Sci. U.S.A. 96, 1100–1105. doi: 10.1073/pnas.96.3.1100
Spangler, S. A., and Hoogenraad, C. C. (2007). Liprin-alpha proteins: Scaffold molecules for synapse maturation. Biochem. Soc. Trans. 35, 1278–1282. doi: 10.1042/BST0351278
Spiegel, I., Mardinly, A. R., Gabel, H. W., Bazinet, J. E., Couch, C. H., Tzeng, C. P., et al. (2014). Npas4 regulates excitatory-inhibitory balance within neural circuits through cell-type-specific gene programs. Cell 157, 1216–1229. doi: 10.1016/j.cell.2014.03.058
Spitzer, N. C. (2017). Neurotransmitter Switching in the Developing and Adult Brain. Annu. Rev. Neurosci. 40, 1–19. doi: 10.1146/annurev-neuro-072116-031204
Stephan, A. H., Barres, B. A., and Stevens, B. (2012). The complement system: An unexpected role in synaptic pruning during development and disease. Annu. Rev. Neurosci. 35, 369–389. doi: 10.1146/annurev-neuro-061010-113810
Stevens, B., Allen, N. J., Vazquez, L. E., Howell, G. R., Christopherson, K. S., Nouri, N., et al. (2007). The classical complement cascade mediates CNS synapse elimination. Cell 131, 1164–1178. doi: 10.1016/j.cell.2007.10.036
Steward, K., and Worley, P. F. (2001). A cellular mechanism for targeting newly synthesized mRNAs to synaptic sites on dendrites. Proc. Natl. Acad. Sci. U.S.A. 98, 7062–7068. doi: 10.1073/pnas.131146398
Stogsdill, J. A., Ramirez, J., Liu, D., Kim, Y. H., Baldwin, K. T., Enustun, E., et al. (2017). Astrocytic neuroligins control astrocyte morphogenesis and synaptogenesis. Nature 551, 192–197. doi: 10.1038/nature24638
Sudhof, T. C. (1995). The synaptic vesicle cycle: a cascade of protein-protein interactions. Nature 375, 645–653. doi: 10.1038/375645a0
Sudhof, T. C. (2008). Neuroligins and neurexins link synaptic function to cognitive disease. Nature 455, 903–911. doi: 10.1038/nature07456
Sudhof, T. C. (2017). Synaptic Neurexin Complexes: A Molecular Code for the Logic of Neural Circuits. Cell 171, 745–769. doi: 10.1016/j.cell.2017.10.024
Sudhof, T. C. (2018). Towards an Understanding of Synapse Formation. Neuron 100, 276–293. doi: 10.1016/j.neuron.2018.09.040
Sudhof, T. C. (2021). The cell biology of synapse formation. J. Cell Biol. 220:e202103052. doi: 10.1083/jcb.202103052
Sudhof, T. C., De Camilli, P., Niemann, H., and Jahn, R. (1993). Membrane fusion machinery: Insights from synaptic proteins. Cell 75, 1–4. doi: 10.1016/S0092-8674(05)80077-7
Sullivan, P. F., de Geus, E. J., Willemsen, G., James, M. R., Smit, J. H., Zandbelt, T., et al. (2009). Genome-wide association for major depressive disorder: A possible role for the presynaptic protein piccolo. Mol Psychiatry 14, 359–375. doi: 10.1038/mp.2008.125
Sutton, M. A., and Schuman, E. M. (2006). Dendritic protein synthesis, synaptic plasticity, and memory. Cell 127, 49–58. doi: 10.1016/j.cell.2006.09.014
Sztainberg, Y., Chen, H. M., Swann, J. W., Hao, S., Tang, B., Wu, Z., et al. (2015). Reversal of phenotypes in MECP2 duplication mice using genetic rescue or antisense oligonucleotides. Nature 528, 123–126. doi: 10.1038/nature16159
Tada, T., and Sheng, M. (2006). Molecular mechanisms of dendritic spine morphogenesis. Curr. Opin. Neurobiol. 16, 95–101. doi: 10.1016/j.conb.2005.12.001
Tai, H. C., and Schuman, E. M. (2008). Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nat. Rev. Neurosci. 9, 826–838. doi: 10.1038/nrn2499
Takao-Rikitsu, E., Mochida, S., Inoue, E., Deguchi-Tawarada, M., Inoue, M., Ohtsuka, T., et al. (2004). Physical and functional interaction of the active zone proteins, CAST, RIM1, and Bassoon, in neurotransmitter release. J. Cell Biol. 164, 301–311. doi: 10.1083/jcb.200307101
Tan, Z. J., Peng, Y., Song, H. L., Zheng, J. J., and Yu, X. (2010). N-cadherin-dependent neuron-neuron interaction is required for the maintenance of activity-induced dendrite growth. Proc. Natl. Acad. Sci. U.S.A. 107, 9873–9878. doi: 10.1073/pnas.1003480107
Tanaka, H., Shan, W., Phillips, G. R., Arndt, K., Bozdagi, O., Shapiro, L., et al. (2000). Molecular modification of N-cadherin in response to synaptic activity. Neuron 25, 93–107. doi: 10.1016/S0896-6273(00)80874-0
Tang, S., and Yasuda, R. (2017). Imaging ERK and PKA Activation in Single Dendritic Spines during Structural Plasticity. Neuron 93, 1315–1324.e3. doi: 10.1016/j.neuron.2017.02.032
Tao, W., Diaz-Alonso, J., Sheng, N., and Nicoll, R. A. (2018). Postsynaptic delta1 glutamate receptor assembles and maintains hippocampal synapses via Cbln2 and neurexin. Proc. Natl. Acad. Sci. U.S.A. 115:E5373–E5381. doi: 10.1073/pnas.1802737115
Tashiro, A., Dunaevsky, A., Blazeski, R., Mason, C. A., and Yuste, R. (2003). Bidirectional regulation of hippocampal mossy fiber filopodial motility by kainate receptors: A two-step model of synaptogenesis. Neuron 38, 773–784. doi: 10.1016/S0896-6273(03)00299-X
Togashi, H., Abe, K., Mizoguchi, A., Takaoka, K., Chisaka, O., and Takeichi, M. (2002). Cadherin regulates dendritic spine morphogenesis. Neuron 35, 77–89. doi: 10.1016/S0896-6273(02)00748-1
Uemura, T., Lee, S. J., Yasumura, M., Takeuchi, T., Yoshida, T., Ra, M., et al. (2010). Trans-synaptic interaction of GluRdelta2 and Neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell 141, 1068–1079. doi: 10.1016/j.cell.2010.04.035
Ullian, E. M., Harris, B. T., Wu, A., Chan, J. R., and Barres, B. A. (2004). Schwann cells and astrocytes induce synapse formation by spinal motor neurons in culture. Mol. Cell Neurosci. 25, 241–251. doi: 10.1016/j.mcn.2003.10.011
Ullian, E. M., Sapperstein, S. K., Christopherson, K. S., and Barres, B. A. (2001). Control of synapse number by glia. Science 291, 657–661. doi: 10.1126/science.291.5504.657
Um, J. W., and Ko, J. (2013). LAR-RPTPs: Synaptic adhesion molecules that shape synapse development. Trends Cell Biol. 23, 465–475. doi: 10.1016/j.tcb.2013.07.004
Ushkaryov, Y. A., Petrenko, A. G., Geppert, M., and Sudhof, T. C. (1992). Neurexins: Synaptic cell surface proteins related to the alpha-latrotoxin receptor and laminin. Science 257, 50–56. doi: 10.1126/science.1621094
Varoqueaux, F., Aramuni, G., Rawson, R. L., Mohrmann, R., Missler, M., Gottmann, K., et al. (2006). Neuroligins determine synapse maturation and function. Neuron 51, 741–754. doi: 10.1016/j.neuron.2006.09.003
Verhage, M., Maia, A. S., Plomp, J. J., Brussaard, A. B., Heeroma, J. H., Vermeer, H., et al. (2000). Synaptic assembly of the brain in the absence of neurotransmitter secretion. Science 287, 864–869. doi: 10.1126/science.287.5454.864
Vicario-Abejon, C., Owens, D., McKay, R., and Segal, M. (2002). Role of neurotrophins in central synapse formation and stabilization. Nat. Rev. Neurosci. 3, 965–974. doi: 10.1038/nrn988
Wan, C., Shi, Y., Zhao, X., Tang, W., Zhang, M., Ji, B., et al. (2009). Positive association between ALDH1A2 and schizophrenia in the Chinese population. Prog. Neuropsychopharmacol. Biol. Psychiatry 33, 1491–1495. doi: 10.1016/j.pnpbp.2009.08.008
Wang, S. H., Celic, I., Choi, S. Y., Riccomagno, M., Wang, Q., Sun, L. O., et al. (2014). Dlg5 regulates dendritic spine formation and synaptogenesis by controlling subcellular N-cadherin localization. J. Neurosci. 34, 12745–12761. doi: 10.1523/JNEUROSCI.1280-14.2014
Wang, Y., Liu, X., Biederer, T., and Sudhof, T. C. (2002). A family of RIM-binding proteins regulated by alternative splicing: Implications for the genesis of synaptic active zones. Proc. Natl. Acad. Sci. U.S.A. 99, 14464–14469. doi: 10.1073/pnas.182532999
Wang, Y., Okamoto, M., Schmitz, F., Hofmann, K., and Sudhof, T. C. (1997). Rim is a putative Rab3 effector in regulating synaptic-vesicle fusion. Nature 388, 593–598. doi: 10.1038/41580
Wang, Y., Sugita, S., and Sudhof, T. C. (2000). The RIM/NIM family of neuronal C2 domain proteins. Interactions with Rab3 and a new class of Src homology 3 domain proteins. J. Biol. Chem. 275, 20033–20044. doi: 10.1074/jbc.M909008199
Woo, J., Kwon, S. K., Choi, S., Kim, S., Lee, J. R., Dunah, A. W., et al. (2009). Trans-synaptic adhesion between NGL-3 and LAR regulates the formation of excitatory synapses. Nat. Neurosci. 12, 428–437. doi: 10.1038/nn.2279
Xie, X., Liang, M., Yu, C., and Wei, Z. (2021). Liprin-alpha-Mediated Assemblies and Their Roles in Synapse Formation. Front. Cell Dev. Biol. 9:653381. doi: 10.3389/fcell.2021.653381
Xu, H. P., Furman, M., Mineur, Y. S., Chen, H., King, S. L., Zenisek, D., et al. (2011). An instructive role for patterned spontaneous retinal activity in mouse visual map development. Neuron 70, 1115–1127. doi: 10.1016/j.neuron.2011.04.028
Xu, T., Yu, X., Perlik, A. J., Tobin, W. F., Zweig, J. A., Tennant, K., et al. (2009). Rapid formation and selective stabilization of synapses for enduring motor memories. Nature 462, 915–919. doi: 10.1038/nature08389
Yamagata, M., Duan, X., and Sanes, J. R. (2018). Cadherins Interact With Synaptic Organizers to Promote Synaptic Differentiation. Front. Mol. Neurosci. 11:142. doi: 10.3389/fnmol.2018.00142
Yan, Z., Kim, E., Datta, D., Lewis, D. A., and Soderling, S. H. (2016). Synaptic Actin Dysregulation, a Convergent Mechanism of Mental Disorders? J. Neurosci. 36, 11411–11417. doi: 10.1523/JNEUROSCI.2360-16.2016
Yang, J., Siao, C. J., Nagappan, G., Marinic, T., Jing, D., McGrath, K., et al. (2009). Neuronal release of proBDNF. Nat. Neurosci. 12, 113–115. doi: 10.1038/nn.2244
Yilmaz, M., Yalcin, E., Presumey, J., Aw, E., Ma, M., Whelan, C. W., et al. (2021). Overexpression of schizophrenia susceptibility factor human complement C4A promotes excessive synaptic loss and behavioral changes in mice. Nat. Neurosci. 24, 214–224. doi: 10.1038/s41593-020-00763-8
Yim, Y. S., Kwon, Y., Nam, J., Yoon, H. I., Lee, K., Kim, D. G., et al. (2013). Slitrks control excitatory and inhibitory synapse formation with LAR receptor protein tyrosine phosphatases. Proc. Natl. Acad. Sci. U.S.A. 110, 4057–4062. doi: 10.1073/pnas.1209881110
Yonekawa, Y., Harada, A., Okada, Y., Funakoshi, T., Kanai, Y., Takei, Y., et al. (1998). Defect in synaptic vesicle precursor transport and neuronal cell death in KIF1A motor protein-deficient mice. J. Cell Biol. 141, 431–441. doi: 10.1083/jcb.141.2.431
Yoon, S., Piguel, N. H., Khalatyan, N., Dionisio, L. E., Savas, J. N., and Penzes, P. (2021). Homer1 promotes dendritic spine growth through ankyrin-G and its loss reshapes the synaptic proteome. Mol. Psychiatry 26, 1775–1789. doi: 10.1038/s41380-020-00991-1
Zeng, M., Shang, Y., Araki, Y., Guo, T., Huganir, R. L., and Zhang, M. (2016). Phase Transition in Postsynaptic Densities Underlies Formation of Synaptic Complexes and Synaptic Plasticity. Cell 166, 1163–1175.e12. doi: 10.1016/j.cell.2016.07.008
Zhai, R. G., Vardinon-Friedman, H., Cases-Langhoff, C., Becker, B., Gundelfinger, E. D., Ziv, N. E., et al. (2001). Assembling the presynaptic active zone: A characterization of an active one precursor vesicle. Neuron 29, 131–143. doi: 10.1016/S0896-6273(01)00185-4
Zhai, R., Olias, G., Chung, W. J., Lester, R. A., Dieck, S. Tom, Langnaese, K., et al. (2000). Temporal appearance of the presynaptic cytomatrix protein bassoon during synaptogenesis. Mol. Cell Neurosci. 15, 417–428. doi: 10.1006/mcne.2000.0839
Zhang, C., Atasoy, D., Arac, D., Yang, X., Fucillo, M. V., Robison, A. J., et al. (2010). Neurexins physically and functionally interact with GABA(A) receptors. Neuron 66, 403–416. doi: 10.1016/j.neuron.2010.04.008
Zhang, J., Ackman, J. B., Xu, H. P., and Crair, M. C. (2011). Visual map development depends on the temporal pattern of binocular activity in mice. Nat. Neurosci. 15, 298–307. doi: 10.1038/nn.3007
Zhen, M., and Jin, Y. (1999). The liprin protein SYD-2 regulates the differentiation of presynaptic termini in C. elegans. Nature 401, 371–375. doi: 10.1038/43886
Zhu, J. J., Qin, Y., Zhao, M., Van Aelst, L., and Malinow, R. (2002). Ras and Rap control AMPA receptor trafficking during synaptic plasticity. Cell 110, 443–455. doi: 10.1016/S0092-8674(02)00897-8
Zito, K., and Svoboda, K. (2002). Activity-dependent synaptogenesis in the adult Mammalian cortex. Neuron 35, 1015–1017. doi: 10.1016/S0896-6273(02)00903-0
Zoghbi, H. Y., and Bear, M. F. (2012). Synaptic dysfunction in neurodevelopmental disorders associated with autism and intellectual disabilities. Cold Spring Harb. Perspect. Biol. 4:a009886. doi: 10.1101/cshperspect.a009886
Keywords: scaffold protein, cell adhesion molecule (CAM), cytoskeleton, neural activity, evolution, disease
Citation: Qi C, Luo L-D, Feng I and Ma S (2022) Molecular mechanisms of synaptogenesis. Front. Synaptic Neurosci. 14:939793. doi: 10.3389/fnsyn.2022.939793
Received: 09 May 2022; Accepted: 27 July 2022;
Published: 13 September 2022.
Edited by:
Yiran Gu, East China Normal University, ChinaReviewed by:
Zhiyong Shao, Fudan University, ChinaCopyright © 2022 Qi, Luo, Feng and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cai Qi, Y2FpLnFpQHlhbGUuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.