
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Synaptic Neurosci. , 28 July 2022
Volume 14 - 2022 | https://doi.org/10.3389/fnsyn.2022.926570
This article is part of the Research Topic Role of Protein Palmitoylation in Synaptic Plasticity and Neuronal Differentiation, Volume II View all 6 articles
 Barbara Barylko1
Barbara Barylko1 Per Niklas Hedde2,3
Per Niklas Hedde2,3 Clinton A. Taylor1
Clinton A. Taylor1 Derk D. Binns1
Derk D. Binns1 Yu-Kai Huang3
Yu-Kai Huang3 Gemma Molinaro4
Gemma Molinaro4 Kimberly M. Huber4
Kimberly M. Huber4 David M. Jameson2
David M. Jameson2 Joseph P. Albanesi1*
Joseph P. Albanesi1*Calmodulin kinase-like vesicle-associated (CaMKv), a pseudokinase belonging to the Ca2+/calmodulin-dependent kinase family, is expressed predominantly in brain and neural tissue. It may function in synaptic strengthening during spatial learning by promoting the stabilization and enrichment of dendritic spines. At present, almost nothing is known regarding CaMKv structure and regulation. In this study we confirm prior proteomic analyses demonstrating that CaMKv is palmitoylated on Cys5. Wild-type CaMKv is enriched on the plasma membrane, but this enrichment is lost upon mutation of Cys5 to Ser. We further show that CaMKv interacts with another regulator of synaptic plasticity, Arc/Arg3.1, and that the interaction between these two proteins is weakened by mutation of the palmitoylated cysteine in CamKv.
Calmodulin kinase-like vesicle-associated (“CaMKv”), a member of the Ca2+/calmodulin-dependent protein kinase family, is almost exclusively expressed in brain and endocrine tissues (Human Protein Atlas). Because it lacks key residues required in other kinases for ATP binding, as well as the consensus autophosphorylation motif (RXXS/T) required for maintenance of CaMKII activity, CaMKv is believed to be a pseudokinase. Consistent with this view, kinase activity was not detected in in vitro assays (Godbout et al., 1994) and mutation of a key ATP-binding residue did not interfere with its ability to rescue activities that were lost in CaMKv knockdown neurons (Liang et al., 2016).
In a report describing CaMKv function in the nervous system, Liang et al. (2016) showed that CaMKv is critical for the maintenance of dendritic spines. They further showed that CaMKv expression is induced by sensory experience in vivo and by synaptic activity in cultured neurons and that overexpression of CaMKv increases spine density, whereas its depletion reduces spine density and results in impaired spatial memory. Although mechanisms underlying these synaptic functions remain to be fully elucidated, they are likely to involve regulation of the actin cytoskeleton in dendritic spines, as CaMKv was shown to interact with and inhibit the Rho guanine nucleotide exchange factor (GEF), GEF-H1, thereby suppressing RhoA activation and RhoA-dependent reduction in spine density (Liang et al., 2016).
Despite the evident importance of CaMKv in neuronal function, almost nothing is known about the protein itself. Indeed, since its discovery in 1994 by Sutcliffe’s group (Godbout et al., 1994; who named it “1G5”), only three publications focusing on CaMKv have been published (Won et al., 2006; Liang et al., 2016; Sussman et al., 2020). CaMKv was first identified as a protein associated with neuronal vesicles (Godbout et al., 1994) and was more recently found to partially localize to the plasma membrane (Liang et al., 2016; Sussman et al., 2020). However, our analysis of secondary structure prediction programs, e.g., TMHMM-2.0 (Krogh et al., 2001) and CCTOP (Dobson et al., 2015), failed to reveal a transmembrane domain. Thus, the mechanism that underlies CaMKv binding to membranes is not understood. A relatively large percentage of peripheral membrane proteins in dendrites undergo palmitoylation, the covalent modification of cysteines with 16-carbon palmitoyl chains (Zarêba-Kozioł et al., 2018). CaMKv was detected in three large-scale screens aimed at identifying palmitoylated proteins (Kang et al., 2008; Wan et al., 2013; Collins et al., 2017). In the present study, we confirm that endogenous CaMKv undergoes palmitoylation in neurons and that recombinant CaMKv undergoes palmitoylation in heterologous cells. We further show that mutation of the palmitoylation site, Cys5, results in the displacement of CaMKv from the plasma membrane. Finally, we report for the first time that CaMKv interacts directly with Arc (Activity-regulated cytoskeleton-associated protein) (Lyford et al., 1995), also known as Arg 3.1 (Activity-regulated gene 3.1) (Link et al., 1995), an activity-dependent immediate-early gene product that regulates synaptic plasticity and is required for the formation of long-term memories (Epstein and Finkbeiner, 2018; Zhang and Bramham, 2021). Our data indicate that the CaMKv-Arc interaction is influenced by CaMKv palmitoylation.
Mouse CaMKv cDNA (Myc-DDK-tagged at the C-terminus) and mouse monoclonal anti-DDK antibody (TA50011) were from Origene. Fluorescently labeled secondary antibodies for Infrared Imaging System were from LI-COR. Cloning reagents were from Thermo Scientific. Primers and Lipofectamines were from Invitrogen. Reagents for electrophoresis and immunoblotting were from Bio-Rad. Phosphatase inhibitors cocktail (PhosSTOP) was from Roche. Thiopropyl Sepharose, reagents for analysis of palmitoylation, tissue culture, and other reagents were from Sigma.
Arc-mCherry construct was generated as described in Hedde et al. (2022). Fluorescently tagged CaMKv was generated by subcloning into pEGFP-C1vector (Contech) using CaMKv-Myc-DDK as a template. This CaMKv-pEGFP construct was then used as a template to generate the C5S point mutant by site-directed mutagenesis. All DNA constructs were verified by sequencing.
Bacterially-expressed glutathione-S-transferase-labeled Arc (GST-Arc) was purified on Glutathione Sepharose 4B resin in buffer contained 20 mM HEPES (pH 8.0), 100 mM NaCl, 1 mM tris(2-carboxyethyl)phosphine (TCEP), 0.2 mM PMSF, and protease inhibitor cocktail (10 μg/ml each of N-p-tosyl-L-lysine chloromethyl ester, N-p-tosyl-L-arginine methyl ester, N-p-tosyl-L-lysine chloromethyl ketone, leupeptin, and pepstatin), as described (Byers et al., 2015). GST-Arc was released from the resin using 50 mM glutathione and dialyzed against 20 mM HEPES, pH 7.5, 0.1 M NaCl, 0.2 mM PMSF and 1 mM TCEP. To remove potential aggregates, Samples of Arc were centrifuged at 214,000 × g for 15 min at 4°C prior to all assays.
Palmitoylation was detected using the Acyl-Resin Assisted Capture (Acyl-RAC) method (Forrester et al., 2011), as described in detail in Barylko et al. (2018). Briefly, cells or whole brain were solubilized with 2.5% SDS in 100 mM HEPES (pH 7.5), 1 mM EDTA, 0.2 mM PMSF, protease inhibitor cocktail, and 50 mM dithiothreitol (DTT). Cells were then incubated at 40°C for 0.5 h (to reduce potential S–S bonds), then for an additional 4 h with methyl methanethiosulfonate (MMTS) to block free thiols. Excess MMTS was removed by protein precipitation and washing with acetone. Dried pellets were re-solubilized in 1% SDS and mixed with thiopropyl-Sepharose resin. Half of the sample was incubated with hydroxylamine (NH2OH) to cleave thioester bonds, the other half was incubated with 2 M NaCl (to control for false positives). Proteins with free thiols (i.e., from cysteines that were originally palmitoylated before NH2OH treatment) are captured on the resin. After extensive washing, proteins released from the resin were analyzed by SDS-PAGE and identified by immunoblotting.
Calmodulin kinase-like vesicle-associated-Myc-DDK was expressed in HeLa cells and cell lysates were incubated overnight with GST-Arc or GST alone (control) bound to glutathione resin. After low-speed centrifugation (1 min at 500 × g), samples were washed, proteins were eluted with glutathione, and electrophoresed. CaMKv and Arc were recognized by immunoblotting with anti-DDK and anti-GST antibodies, respectively.
MCF-10A cells plated on imaging dishes coated with fibronectin were transfected with wild-type and mutant CaMKv-EGFP and Arc-mCherry using Lipofectamine 3000. Cells were imaged at room temperature 20–24 h after transfection for a maximum duration of 90 min with a Zeiss LSM880 laser scanning microscope set up for FLIM. EGFP fluorescence was excited at 880 nm (two-photon excitation, 80 MHz) and detected at 510–560 nm with a 40×, NA 1.2 water immersion lens in non-descanned mode using a hybrid photomultiplier detector (HPM-100, Becker & Hickl, Germany) coupled to a FLIMBox (ISS, Champaign, IL, United States). Per data set, 35 frames of 256 × 256 pixels were acquired with a pixel dwell time of 16 μs. Data were analyzed in SimFCS. Before FLIM, the presence of both donor (EGFP) and acceptor (mCherry) were verified with 488 and 594-nm excitation.
HEK293, HeLa, and MCF-10A cells (ATCC) were cultured in DMEM supplemented with 10% fetal bovine serum and antibiotics. They were transfected with either Lipofectamine 2000 (HEK293, HeLa) or Lipofectamine 3000 (MCF-10A) according to the manufacturer’s instructions and were used 20–24 h after transfection. Protein concentration was determined using the modified Lowry method according to Peterson (1977) with BSA as a standard. SDS-PAGE was carried out according to Laemmli (1970). For immunoblotting, proteins were transferred to nitrocellulose, and immunoblotted with the indicated antibodies. Bound primary antibodies were detected and quantified using fluorescently labeled secondary antibody in the LI-COR Odyssey system.
Calmodulin kinase-like vesicle-associated contains an N-terminal kinase domain with 39% identity and 59% similarity to the kinase domain of CaMKIIα (CaMK2a), a central calmodulin-binding domain, and an extended C-terminal intrinsically disordered domain (∼ residues 330–501) (Figure 1A). Figure 1B shows a structural alignment of CaMK2a (obtained by X-ray crystallography) with residues 1–329 of CaMKv (predicted by AlphaFold; Jumper et al., 2021; Varadi et al., 2022). Residues T7-K300 of CaMK2a align well both structurally and by sequence with residues S18-K314 of CaMKv. However, the two proteins diverge in sequence and structure in the remaining C-terminal regions. The expanded view of the putative CaMKv active site (Figure 1C) shows that His168 replaces the glycine of the Asp-Phe-Gly (DFG) motif, which is highly conserved among kinases and which plays a critical role in catalysis. In addition, Asn145 in the catalytic loop is an aspartate in active kinases. We note that mutation of this Asp to Asn is commonly used to abolish kinase activity. Thus, it is unlikely that CaMKv expresses kinase activity.
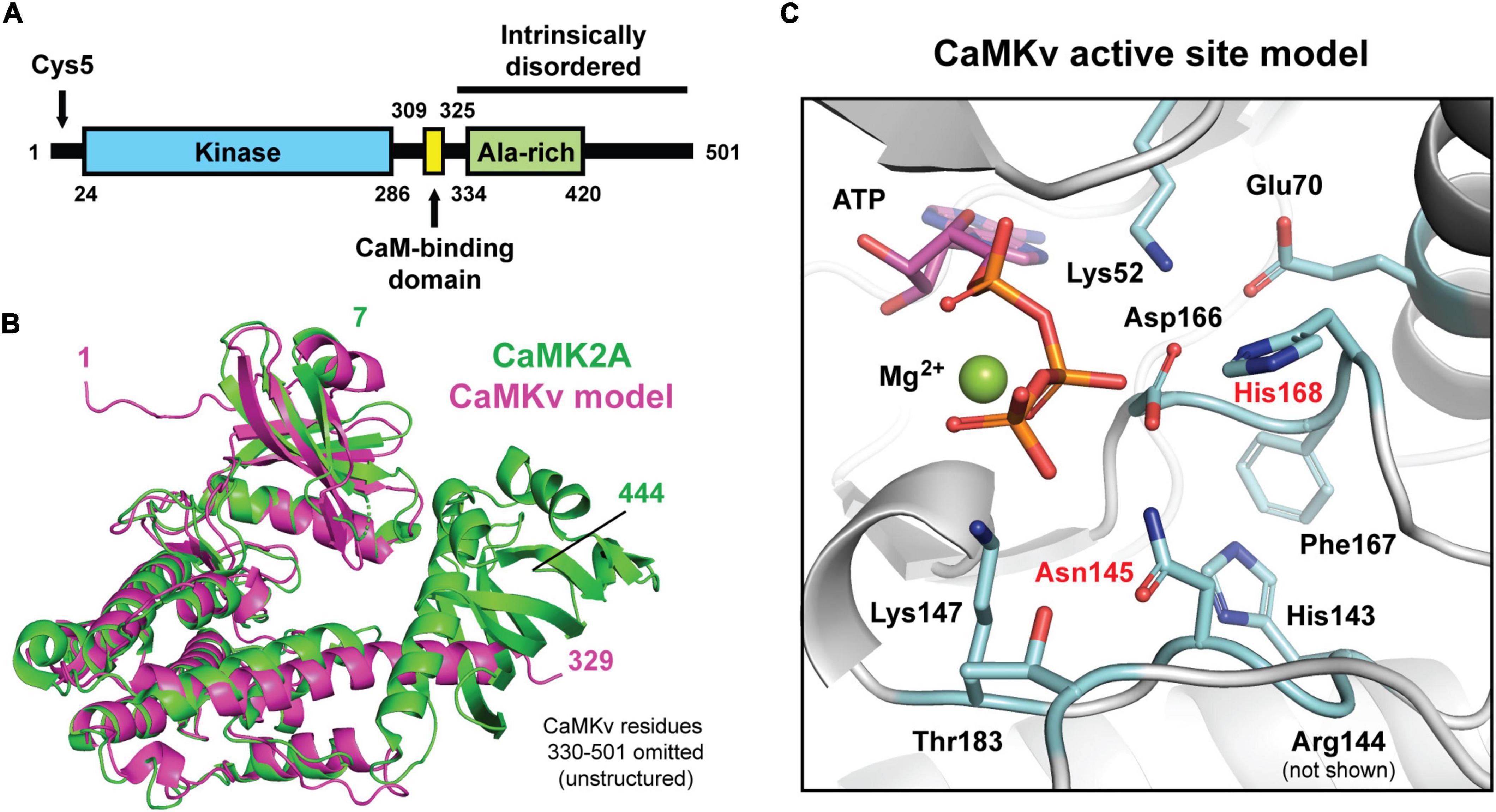
Figure 1. Predicted secondary and tertiary structure of calmodulin kinase-like vesicle-associated (CaMKv). (A) Domains of CaMKv. The intrinsically disordered region was predicted using the PONDR (Predictor of Natural Disorder Region) program. The entire sequence from residues 325–351 had PONDR scores ranging from 0.8 to 1.0. (B) Structural alignment (Cα r.m.s.d = 1.21 Å) of the full-length crystal structure of human CAMK2A (green; PDB: 3SOA) and a predicted model of residues 1–329 of human CaMKv derived from the AlphaFold Protein Structure Database (magenta; entry Q8NCB2). Unstructured residues 330–501 of CaMKv were omitted. (C) Close-up view of the predicted active site of CaMKv with modeled placement of ATP and Mg2+ based on an alignment to a crystal structure of CAMK2A bound to ATP (PDB: 6XBX). Residues located at positions that are important for catalysis in protein kinases are colored blue. Residues labeled in red differ from those typically found in active kinases.
Using the Acyl-RAC approach we confirmed that endogenous CaMKv is palmitoylated in mouse brain (Figure 2A) and recombinant CaMKv is palmitoylated in HeLa cells (Figure 2B). As expected, endogenous palmitoylated CaMKv was detected in mouse brain membranes, but not in cytosolic fractions (Supplementary Figure 2A). However, only 5–10% of membrane-associated CaMKv was palmitoylated, indicating that this modification is not essential for membrane binding, but may instead contribute to specific subcellular targeting. When synaptosomal membranes were centrifuged on a sucrose step gradient, a portion of CaMKv distributed to low buoyant density fractions, commonly termed “lipid rafts,” which are enriched in palmitoylated proteins (Levental et al., 2010; Supplementary Figure 2B). To determine if palmitoylation influences the subcellular distribution of CaMKv, we localized wild-type CaMKv-EGFP and a palmitoylation-deficient CaMKv-EGFP mutant in heterologous (MCF-10) cells (Figure 3). Cysteine 5 (1MPFGCVTLGD10) was identified as the palmitoylation site of mouse CaMKv in a thioacylation screen (Collins et al., 2017). As shown in Figures 3A,B, wild-type CaMKv-EGFP undergoes palmitoylation in HeLa cells, whereas C5S-CaMKv-EGFP does not. In MCF-10 cells, wild-type CaMKv-EGFP shows a diffuse cytoplasmic distribution, but with a pronounced enrichment on the plasma membrane (Figure 3C, top four panels). This plasma membrane enrichment is essentially abrogated in cells expressing C5S-CaMKv-EGFP (Figure 3C, bottom four panels). During our studies, we found that the position of the EGFP tag influenced the distribution of wild-type CaMKv. Whereas CaMKv-EGFP displays pronounced plasma membrane localization in HEK-293 cells, as it does in MCF-10 cells, EGFP-CaMKv is almost entirely cytoplasmic (Supplementary Figure 1), suggesting that an N-terminal tag may suppress palmitoylation of Cys5.
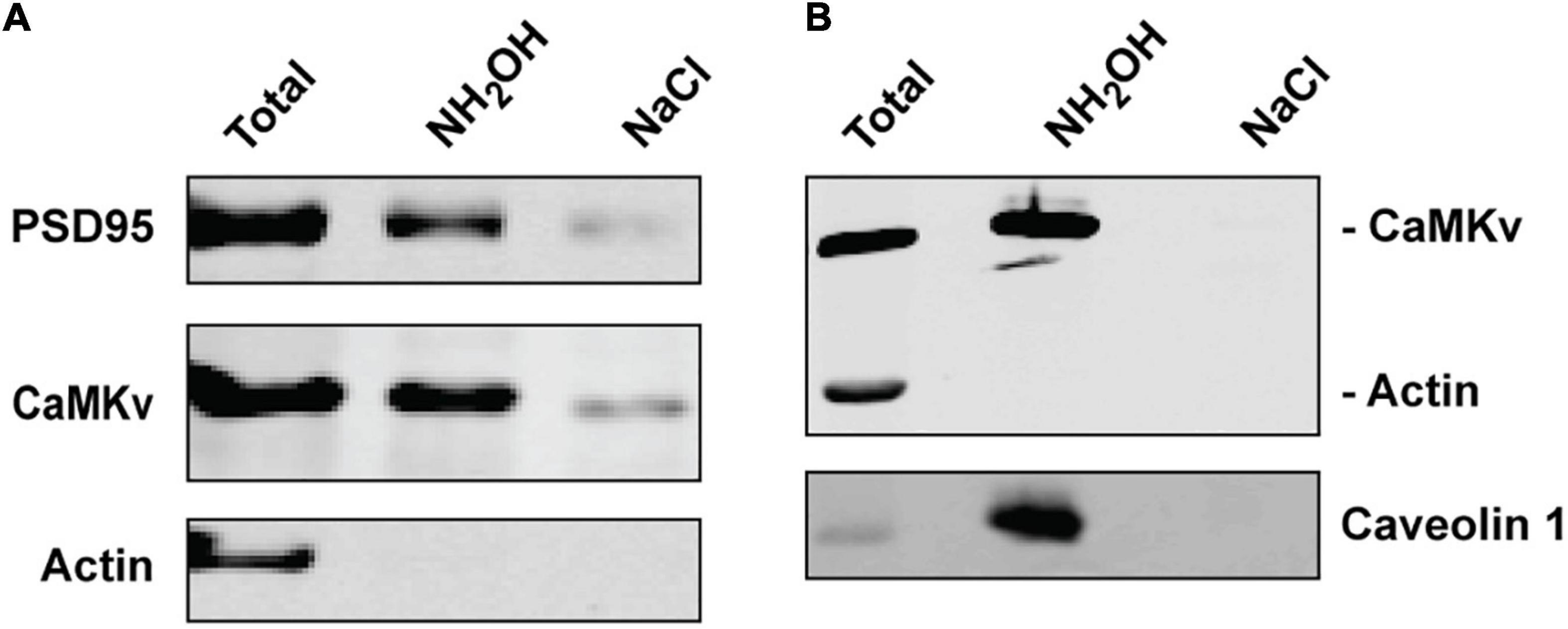
Figure 2. Palmitoylation of calmodulin kinase-like vesicle-associated (CaMKv). (A) Palmitoylation of endogenous CaMKv in mouse brain homogenate detected by Acyl-RAC (see “Materials and methods” section). PSD95 and actin are shown as positive and negative controls, respectively. Approximately 5% of input was palmitoylated in six separate experiments. (B) Palmitoylation of CaMKv expressed in HeLa cells. Caveolin and actin are positive and negative controls, respectively. Approximately 7% of input was palmitoylated in two separate experiments.
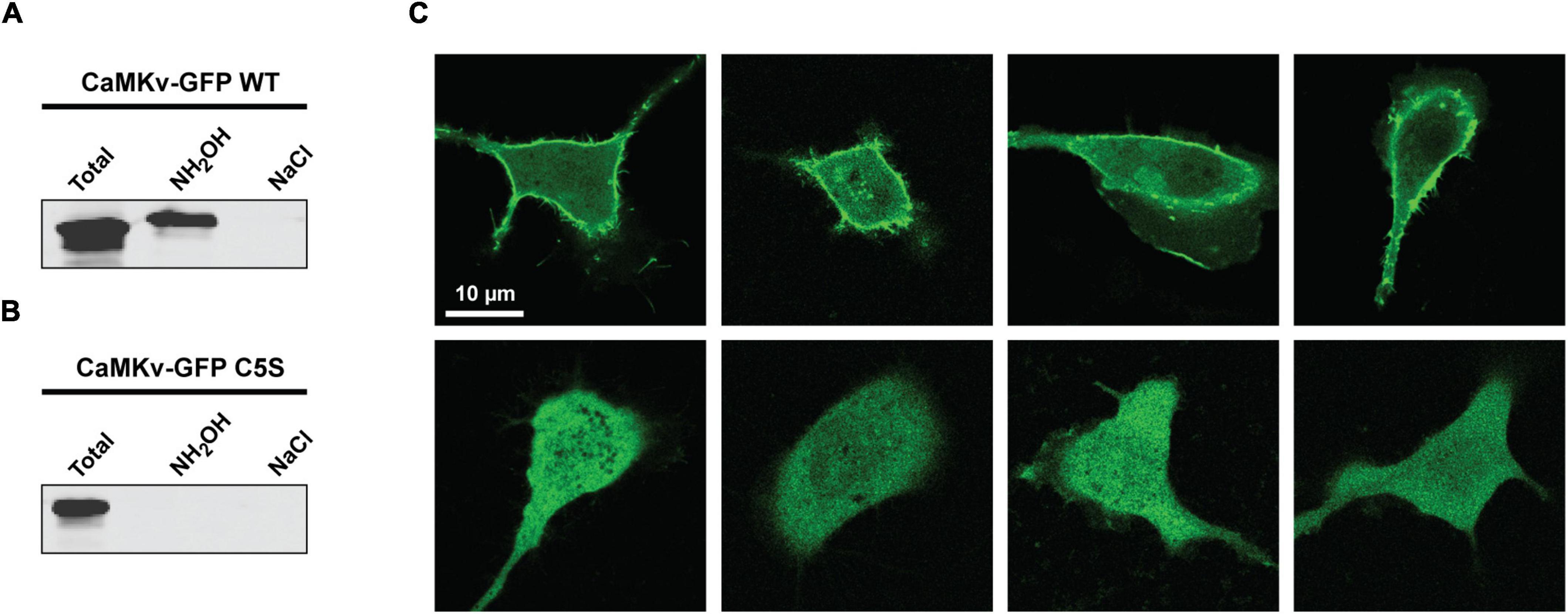
Figure 3. Effect of suppressing palmitoylation on the subcellular distribution of calmodulin kinase-like vesicle-associated (CaMKv). (A,B) Acyl-RAC analysis showing palmitoylation of wild-type CaMKv-EGFP (A) but not of the C5S mutant of CaMKv-EGFP in HEK-293 cells (B). Approximately 4% of CaMKv-EGFP was palmitoylated in three separate experiments. (C) Localization of wild-type CaMKv-EGFP (top four panels) and (C5S)-CaMKv-EGFP (bottom four panels) in MCF-10A cells.
The finding that Arc binds directly to CaMKII (Donai et al., 2003) prompted us to examine whether it may also binds to CaMKv. Indeed, we found that CaMKv expressed in HeLa cells is pulled down by purified GST-Arc but not by GST alone (Figure 4A). To test whether CaMKv and Arc are likely to interact directly, we turned to measurements of Förster resonance energy transfer (FRET), which occurs if an acceptor fluorophore comes within a few nm of a donor. Energy transfer causes a reduction in both the intensity and lifetime of the donor. However, lifetime measurements are generally a more robust reporter of FRET in living cells (Padilla-Parra and Tramier, 2012). We expressed wild-type- and C5S-CaMKv-EGFP (the donor), either alone or together with Arc-mCherry (the acceptor) in MCF-10A cells and acquired lifetime images by pulsed 880 nm two-photon excitation. The fluorescence lifetime of each pixel was calculated and displayed using phasor plots (Malacrida et al., 2021; Figures 4B–E), which are 2D histograms of the pixels of cell images. In cells expressing only CaMKv-EGFP (Figures 4C,E) the center of mass of the pixel distribution falls on the universal semicircle, indicating a single exponential lifetime decay as expected for unquenched EGFP. Instead, for cells that were co-transfected with Arc-mCherry (Figures 4B,D), a comet tail extending from this region toward the zero- lifetime point (S = 0, G = 1) was observed, indicating reduced donor lifetime and, hence, the presence of FRET. Comparing the number of pixels falling outside the universal circle when Arc-mCherry is co-expressed with wild-type-CaMKv (Figure 4B) vs. the palmitoylation-deficient C5S mutant (Figure 4D), it is evident that mutation of the palmitoylation site reduces the extent of FRET between Arc and CaMKv. Quantification is provided in Figure 4F. Although palmitoylation of CaMKv apparently enhances its localization to the plasma membrane (Figure 3), we note that Arc is predominantly cytoplasmic when expressed in MCF-10A cells and that FRET between Arc-mCherry and CaMKv-EGFP was most evident in the cytoplasm (Supplementary Figure 3).
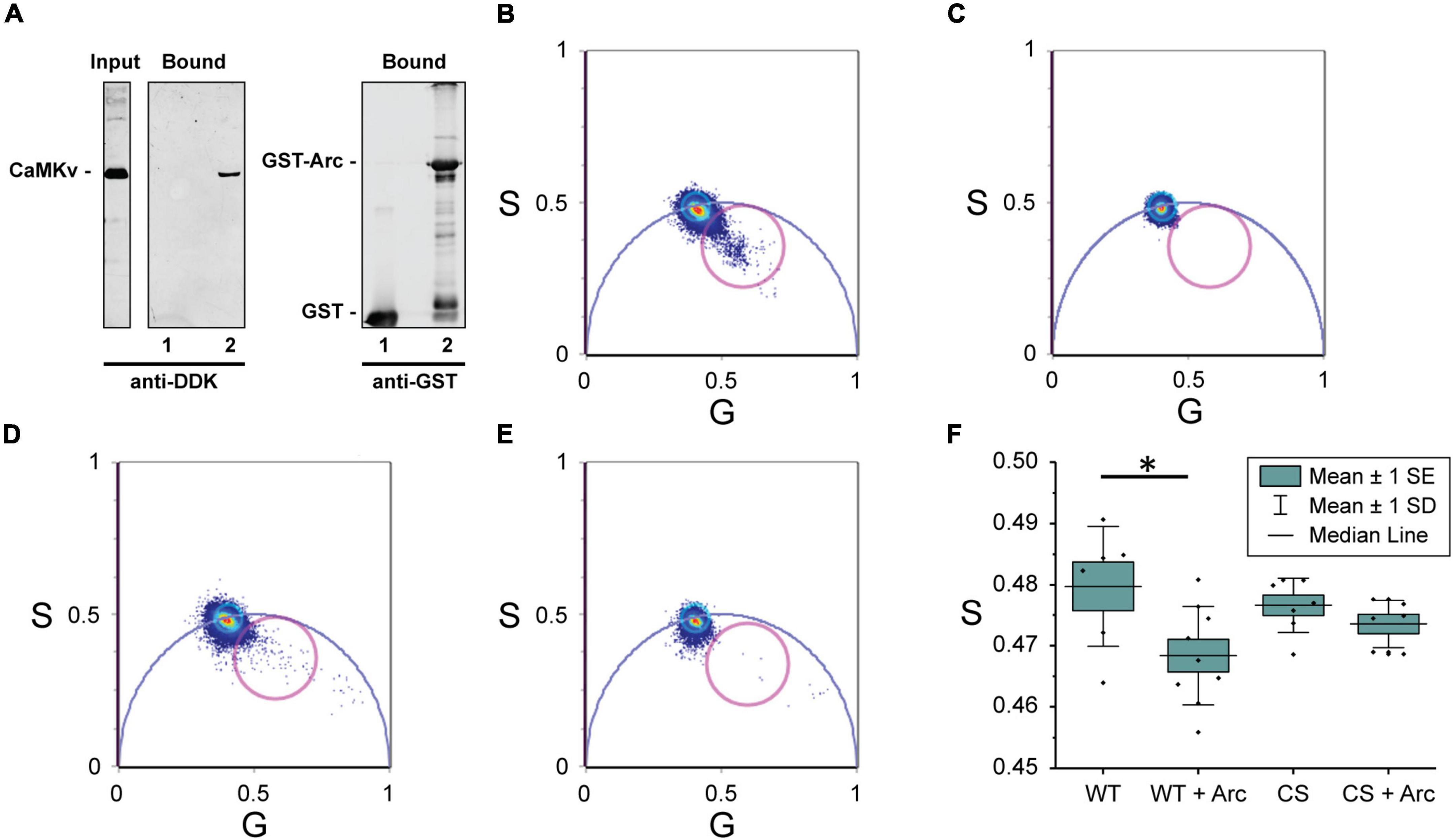
Figure 4. Interaction of calmodulin kinase-like vesicle-associated (CaMKv) with Arc (A) CaMKv binding to GST-Arc (see “Materials and methods” section). HeLa cell extract expressing CaMKv-myc-DDK was incubated with purified GST (control) or GST-Arc (10 μM) with glutathione beads. Samples were centrifuged and pelleted proteins were electrophoresed and blotted with anti-DDK or anti-GST. Lane 1: GST alone (control); Lane 2: GST-Arc. Input represents 20% of CaMKv in the incubation mixture. Similar results were obtained in three separate experiments. (B–F) FRET/FLIM analysis. Phasor plot pixel lifetime distribution of cells expressing: (B) wild-type-CaMKv-EGFP and Arc-mCherry, (C) wild-type CaMKv-EGFP (donor alone), (D) C5S-CaMKv-EGFP and Arc-mCherry, and (E) C5S-CaMKv-EGFP (donor alone). Points within magenta circles represent pixels in which reduction of donor lifetime (i.e., FRET) is observed. (F) Quantification of data in Figures 4B–E. Asterisk indicates significant difference (p < 0.05) using Mann-Whitney nonparametric test.
Here we confirm results from proteomic screens that CaMKv undergoes palmitoylation in cells and show that this modification is important for its targeting to the plasma membrane. Unlike most other forms of lipidation, palmitoylation is reversible and, hence, may be responsive to changes in neuronal conditions. Indeed, CaMKv was identified as one of 121 proteins that were differentially palmitoylated in the mouse hippocampus in response to context-dependent fear conditioning (Nasseri et al., 2021; preprint). In that study, an increase in palmitoylation of CaMKv was observed, suggesting that it may translocate to the plasma membrane upon neuronal activation. Although only about 5% of CaMKv was palmitoylated in unstimulated brain tissue, the specific pool of acylated CaMKv may play an important role in synaptic function, as was already shown for the relatively small pools of palmitoylated Arc (∼5–10%) (Barylko et al., 2018) and PICK1 (∼1%) (Thomas et al., 2013).
We also found that CaMKv interacts with Arc in a palmitoylation-sensitive manner. Arc is a positive regulator of AMPA receptor (AMPAR) endocytosis and, hence, plays a critical role in LTD. However, Arc is also required for late-stage LTP and its deletion interferes with the formation of spatial, taste, and fear memories. Like CaMKv, Arc’s promotion of late-stage LTP has been ascribed to its ability to regulate the actin cytoskeleton within dendritic spines (Bramham, 2008). There are several striking similarities between Arc and CaMKv (Liang et al., 2016). First, translation of both CaMKv and Arc is induced in dendrites in response to synaptic activity. Second, both proteins are required for late-stage LTP and spatial learning. Third, they both localize to the plasma membrane and post-synaptic density. Fourth, they both have been implicated in regulation of the actin cytoskeleton. The functional significance of the CaMKv-Arc interaction remains to be determined. We suggest that palmitoylation of Arc and CaMKv induces their colocalization to the same membrane subdomains, perhaps lipid rafts, where their coordinated activities function to regulate the postsynaptic actin cytoskeleton.
A recent report identified CaMKv as a potential immunotherapeutic target in MYCN-amplified neuroblastoma, due to its inordinately high expression in these tumors compared to normal human tissues (Sussman et al., 2020). CaMKv was found in both membrane and soluble fractions of neuroblastoma cell lines, and plasma membrane (as well as cytoplasmic) staining was detected. Notably, the study suggested that CaMKv, as a potential transmembrane protein, may be susceptible to therapeutic targeting by anti-CaMKv antibodies. However, in light of the presence of CaMKv in both membrane-bound and soluble pools, the absence of a predicted membrane-spanning motif, and our finding that palmitoylation is a likely plasma membrane targeting mechanism, it will be important to experimentally test whether or not CaMKv is indeed a transmembrane protein.
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
JA, BB, DJ, and KH planned and designed the study. BB, PH, CT, DB, Y-KH, and GM performed the experiments. All authors participated in writing and editing the manuscript and approved the submitted version.
This work was funded by NIH grants R21MH119516 (DJ and JA), RO1GM121536 (JA), and RO1HD052731 (KH).
Alexander Pope for technical assistance. FLIM experiments were performed at the Laboratory for Fluorescence Dynamics (LFD) at the University of California, Irvine (UCI). The LFD was supported jointly by the National Institute of General Medical Sciences of the National Institutes of Health (P41GM103540) and UCI.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnsyn.2022.926570/full#supplementary-material
Barylko, B., Wilkerson, J. R., Cavalier, S. H., Binns, D. D., James, N. G., Jameson, D. M., et al. (2018). Palmitoylation and membrane binding of Arc/Arg3.1: a potential role in synaptic depression. Biochemistry 57, 520–524. doi: 10.1021/acs.biochem.7b00959
Bramham, C. R. (2008) Local protein synthesis, actin dynamics, and LTP consolidation. Curr. Opin. Neurobiol. 18, 524–531. doi: 10.1016/j.conb.2008.09.013
Byers, C. E., Barylko, B., Ross, J. A., Southworth, D. R., James, N. G., and Taylor, C. A. IV, et al. (2015). Enhancement of dynamin polymerization and GTPase activity by Arc/Arg3.1. Biochim. Biophys. Acta 1850, 1310–1318. doi: 10.1016/j.bbagen.2015.03.002
Collins, M. O., Woodley, K. T., and Choudhary, J. S. (2017). Global, site-specific analysis of neuronal protein S-acylation. Sci. Rep. 7:4683. doi: 10.1038/s41598-017-04580-1
Dobson, L., Reményi, I., and Tusnády, G. E. (2015). CCTOP: a consensus constrained topoly prediction web server. Nucleic Acids Res. 43:W408–W112. doi: 10.1093/nar/gkv451
Donai, H., Sugiura, H., Ara, D., Yoshimura, Y., Yamagata, K., and Yamauchi, T. (2003). Interaction of Arc with CaM kinase II and stimulation of neurite extension by Arc in neuroblastoma cells expressing CaM kinase II. Neurosci. Res. 47, 399–408. doi: 10.1016/j.neures.2003.08.004
Epstein, I., and Finkbeiner, S. (2018). The Arc of Cognition: Signaling Cascades Regulating Arc and Implications for Cognitive Function and Disease. Semin. Cell Dev. Biol. 77, 63–72. doi: 10.1016/j.semcdb.2017.09.023
Forrester, M. T., Hess, D. T., Thompson, J. W., Hultman, R., Moseley, M. A., Stamler, J. S., et al. (2011). Sitespecific analysis of protein S-acylation by resin-assisted capture. J. Lipid. Res. 52, 393–398. doi: 10.1194/jlr.D011106
Godbout, M., Erlander, M. G., Hasel, K. W., Danielson, P. E., Wong, K. K., Battenberg, E. L., et al. (1994). 1G5: a calmodulin-binding, vesicle-associated, protein kinase-like protein enriched in forebrain neurites. J. Neurosci. 14, 1–13. doi: 10.1523/JNEUROSCI.14-01-00001.1994
Hedde, P. N., Barylko, B., Binns, D. D., Jameson, D. M., and Albanesi, J. P. (2022). Differential mobility and self-association of Arc/Arg3.1 in the cytoplasm and nucleus of living cells. ACS Chem. Neurosci. 13, 876–882. doi: 10.1021/acschemneuro.1c00744
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi: 10.1038/s41586-021-03819-2
Kang, R., Wan, J., Arstikaitis, P., Takahashi, H., Huang, K., Bailey, A. O., et al. (2008). Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature 456, 904–909. doi: 10.1038/nature07605
Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. doi: 10.1038/227680a0
Levental, I., Grzybek, M., and Simons, K. (2010). Greasing their way: lipid modifications determine protein association with membrane rafts. Biochemistry 49, 6305–6316. doi: 10.1021/bi100882y
Liang, Z., Zhan, Y., Shen, Y., Wong, C. C., Yates, J. R. III, Plattner, F., et al. (2016). The pseudokinase CaMKv is required for the activity-dependent maintenance of dendritic spines. Nat. Commun. 7:13282. doi: 10.1038/ncomms13282
Link, W., Konietzko, U., Kauselmann, G., Krug, M., Schwanke, B., Frey, U., et al. (1995). Somatodendritic Expression of an Immediate Early Gene Is Regulated by Synaptic Activity. Proc. Natl. Acad. Sci. U.S.A. 92, 5734–5738. doi: 10.1073/pnas.92.12.5734
Lyford, G. L., Yamagata, K., Kaufmann, W. E., Barnes, C. A., Sanders, L. K., Copeland, N. G., et al. (1995). Arc, a Growth Factor and Activity-Regulated Gene, Encodes a Novel Cytoskeleton-Associated Protein That Is Enriched in Neuronal Dendrites. Neuron 14, 433–445. doi: 10.1016/0896-6273(95)90299-6
Malacrida, L., Ranjit, S., Jameson, D. M., and Gratton, E. (2021). The phasor plot: a universal circle to advance fluorescence lifetime analysis and interpretation. Annu. Rev. Biophys. 50, 575–593. doi: 10.1146/annurev-biophys-062920-063631
Nasseri, G., Matin, N., Tosefsky, K., Stacey, G., Flibotte, S., Hollman, R., et al. (2021). Synaptic Activity-Dependent Changes in the Hippocampal Palmitoylome. bioRxiv [Preprint]. doi: 10.1101/2021.11.26.470153
Padilla-Parra, S., and Tramier, M. (2012). FRET microscopy in the living cell: different approaches, strengths and weaknesses. Bioessays 34, 369–376. doi: 10.1002/bies.201100086
Peterson, G. L. (1977). A simplification of the protein assay method of Lowry et al. which is more generally applicable Anal. Biochemistry 83, 346–356. doi: 10.1016/0003-2697(77)90043-4
Sussman, R. T., Rokita, J. L., Huang, K., Raman, P., Rathi, K. S., Martinez, D., et al. (2020). Front. Oncology 10:302. doi: 10.3389/fonc.2020.00302
Thomas, G. M., Hayashi, T., Huganir, R. L., and Linden, D. J. (2013). DHHC8-dependent PICK1 palmitoylation is required for induction of cerebellar long-term synaptic depression. J. Neurosci. 33, 15401–15407. doi: 10.1523/JNEUROSCI.1283-13.2013
Varadi, M., Anyango, S., Deshpande, M., Nair, S., Natassia, C., Yordanova, G., et al. (2022). AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high accuracy models. Nucleic Acids Res. 50:D439–D444. doi: 10.1093/nar/gkab1061
Wan, J., Savas, J. N., Roth, A. F., Sanders, S. S., Singaraja, R. R., Hayden, M. R., et al. (2013). Tracking brain palmitoylation change: predominance of glial change in a mouse model of Huntington’s disease. Chem. Biol. 20, 1421–1434. doi: 10.1016/j.chembiol.2013.09.018
Won, M., Ro, H., Park, H. C., Kim, K. E., Huh, T. L., Kim, C. H., et al. (2006). Dynamic expression patterns of zebrafish 1G5 (1G5z), a calmodulin kinase-like gene in the developing nervous system. Dev. Dyn. 235, 835–842. doi: 10.1002/dvdy.20693
Zarêba-Kozioł, M., Figiel, I., Bartkowiak-Kaczmarek, A., and Włodarczyk, J. (2018). Insights into protein Spalmitoylation in synaptic plasticity and neurological disorders: potential and limitations of methods for detection and analysis. Front. Mol. Neurosci. 11:175. doi: 10.3389/fnmol.2018.00175
Keywords: CaMKv, palmitoylation, pseudokinase, plasma membrane, Arc/Arg3.1, CaMKv-EGFP, Arc-mCherry
Citation: Barylko B, Hedde PN, Taylor CA, Binns DD, Huang Y-K, Molinaro G, Huber KM, Jameson DM and Albanesi JP (2022) Palmitoylation-regulated interactions of the pseudokinase calmodulin kinase-like vesicle-associated with membranes and Arc/Arg3.1. Front. Synaptic Neurosci. 14:926570. doi: 10.3389/fnsyn.2022.926570
Received: 22 April 2022; Accepted: 21 June 2022;
Published: 28 July 2022.
Edited by:
Akira Yoshii, University of Illinois at Chicago, United StatesReviewed by:
Oksana Sorokina, University of Edinburgh, United KingdomCopyright © 2022 Barylko, Hedde, Taylor, Binns, Huang, Molinaro, Huber, Jameson and Albanesi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joseph P. Albanesi, Sm9zZXBoLkFsYmFuZXNpQFVUU291dGh3ZXN0ZXJuLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.