- 1Department of Brain and Cognitive Sciences, MIT, Cambridge, MA, United States
- 2School of Life Sciences, Westlake University, Hangzhou, China
- 3Westlake Laboratory of Life Sciences and Biomedicine, Westlake University, Hangzhou, China
- 4Institute of Basic Medical Sciences, Westlake Institute for Advanced Study, Hangzhou, China
Mapping and determining the molecular identity of individual synapses is a crucial step towards the comprehensive reconstruction of neuronal circuits. Throughout the history of neuroscience, microscopy has been a key technology for mapping brain circuits. However, subdiffraction size and high density of synapses in brain tissue make this process extremely challenging. Electron microscopy (EM), with its nanoscale resolution, offers one approach to this challenge yet comes with many practical limitations, and to date has only been used in very small samples such as C. elegans, tadpole larvae, fruit fly brain, or very small pieces of mammalian brain tissue. Moreover, EM datasets require tedious data tracing. Light microscopy in combination with tissue expansion via physical magnification—known as expansion microscopy (ExM)—offers an alternative approach to this problem. ExM enables nanoscale imaging of large biological samples, which in combination with multicolor neuronal and synaptic labeling offers the unprecedented capability to trace and map entire neuronal circuits in fully automated mode. Recent advances in new methods for synaptic staining as well as new types of optical molecular probes with superior stability, specificity, and brightness provide new modalities for studying brain circuits. Here we review advanced methods and molecular probes for fluorescence staining of the synapses in the brain that are compatible with currently available expansion microscopy techniques. In particular, we will describe genetically encoded probes for synaptic labeling in mice, zebrafish, Drosophila fruit flies, and C. elegans, which enable the visualization of post-synaptic scaffolds and receptors, presynaptic terminals and vesicles, and even a snapshot of the synaptic activity itself. We will address current methods for applying these probes in ExM experiments, as well as appropriate vectors for the delivery of these molecular constructs. In addition, we offer experimental considerations and limitations for using each of these tools as well as our perspective on emerging tools.
Introduction
Historical Perspective of Connectomics
Embedded in the concept of the “neuron doctrine” is the principle that neurons communicate through synapses, a striking assumption first made by Ramon y Cajal over a century ago (Ramón y Cajal, 1909). Cajal, in his now-famous illustrations of the silver-stained neurons, was the first person to predict the unique way that neurons are both spatially separated yet connected via the synapse (Llinás, 2003; Dhawale and Bhalla, 2008). Cajal was the first to dream of the form of the synapse, but he and his contemporaries were hindered from directly visualizing them by the limitations of the light microscope. The invention of the electron microscope provided researchers with the first toolkit to truly peer at the synapse (Palay, 1956; Wells, 2005). Quickly after its invention, electron microscopy (EM) produced the first high-resolution images of synaptic vesicles, providing key structural evidence for Cajal’s vision of the way in which neurons connect (Robertson, 1953; De Robertis and Bennett, 1955; Palay and Palade, 1955). Sanford Palay, one of the early pioneers of using EM to study the brain, defined the form of the synapse by two common factors: close proximity of the postsynaptic and presynaptic cells divided by a gap of around 200 Å (20 nm), and the presence of mitochondria and vesicles at the presynaptic terminal (Palay, 1956). As microscopy technology advanced, so did the understanding of the structure and form of the synapse.
The modern neuroscientist has the privilege of access to a great deal more knowledge about the structure and function of the synapse than Cajal and his contemporaries would have had. Synapses can be broadly categorized by which of the two distinct mechanisms of synaptic transmission they use—chemical or electrical (Pereda, 2014). Chemical synapses are those which were first visualized through EM and are the more well-studied of the two varieties (Palay, 1956). In chemical synapses, vesicles from the presynaptic neuron release neurotransmitters into the synaptic cleft, which are then recognized by the postsynaptic cell, and thus specific signal defined by a particular neurotransmitter is transmitted. Electrical synapses, on the other hand, transmit information through a fundamentally different means. At an electrical synapse, the communicating cells are physically connected via gap junctions, allowing ions, and thus voltage, to be transmitted in most cases bidirectionally between neurons (Pereda, 2014). Electrical synapses were discovered through electrophysiological experiments several years after the first confirmation of the existence of chemical synapses through EM (Watanabe, 1958; Furshpan and Potter, 1959), and their role in the central nervous system (CNS) has only relatively recently been of widespread interest (Gibson et al., 1999; Galarreta and Hestrin, 2001; Hormuzdi et al., 2004). The focus of this review will primarily be on chemical synapses, particularly due to their relative abundance compared to electrical synapses in existing connectomes, although the importance of electrical synapses for brain function should not be underestimated.
Broadly, chemical synapses exist as one of two types—inhibitory or excitatory—based on whether they promote or impede an action potential in the postsynaptic neuron, respectively. In the mammalian CNS, the postsynaptic component of most excitatory synapses and of some inhibitory synapses is located on small protrusions known as dendritic spines (Gray, 1959; Chen et al., 2012; Berry and Nedivi, 2017). Synapses can be further characterized by what neurotransmitter the presynaptic neuron releases, as well as what receptors and scaffold proteins exist in the postsynaptic density (PSD) of spines. For example, the postsynaptic scaffold protein PSD-95, which is expressed only at glutamatergic synapses, is strongly associated with excitatory synapses, and the postsynaptic scaffold protein gephyrin, which interacts with GABA and glycine receptors, is strongly associated with inhibitory synapses (El-Husseini et al., 2000; Prange et al., 2004; Sheng and Kim, 2011). Recent studies have shown that mammalian neurons frequently remodel their spine architecture, assembling and removing excitatory and inhibitory postsynaptic sites in a coordinated manner in response to experience (Chen et al., 2012; Villa et al., 2016). Some individual spines are highly dynamic, appearing and disappearing in a manner of days, while others are more persistent (Berry and Nedivi, 2017).
As the wealth of knowledge surrounding the synapse expands further, there is a need for new technologies that can visualize synapses at high resolution and at high-throughputs. One particularly promising area of study that exemplifies this pressing need is connectomics, the study of wiring of neurons at the resolution of the single synapse. Mapping connectomes in model organisms such as C. elegans, Drosophila melanogaster, zebrafish, and mice is an immensely difficult and time-consuming endeavor, historically relying on EM. The colossal density of synapses—as many as 1 trillion synapses per cm3 of cortex in human brains—combined with the extremely precise resolution needed to visualize single synapses makes the mapping of a connectome a herculean endeavor (Tang et al., 2001; Drachman, 2005). The first whole-organism connectome ever produced was of C. elegans hermaphrodite’s 302 neurons and several thousand synapses, which was the result of many years of work from multiple labs and was expanded over time (Albertson et al., 1976; White et al., 1986; Hall and Russell, 1991; Jarrell et al., 2012; Cook S. J. et al., 2019). Recently, the whole-animal synaptic connectome of Platynereis dumerilii larva (Verasztó et al., 2020), the partial adult and larvae Drosophila connectomes (Ohyama et al., 2015; Scheffer et al., 2020; Hulse et al., 2021), the sea squirt Ciona intestinalis connectome (Ryan et al., 2016), a 0.13 mm3 volume of the somatosensory cortex of a young adult mouse (Kasthuri et al., 2015), around 1 mm3 of mouse visual cortex connectome (MICrONS Consortium et al., 2021), and 1 mm3 of the human cerebral cortex (Shapson-Coe et al., 2021) have also been painstakingly reconstructed with EM. Though improvements in EM, such as serial block-face scanning EM, focused ion beam scanning EM, high-throughput serial section scanning EM, and transmission EM, complemented by advanced methods for connectome reconstruction, have facilitated and hastened this process, the imaging of even a partial connectome remains prohibitively demanding of time and resources for most researchers to perform (Xu et al., 2017; Motta et al., 2019; Hubbard et al., 2020; Witvliet et al., 2020). Several ambitious connectomics projects are currently underway, such as the IARPA MICrONS program and the FlyEM project, a multi-lab, multi-year effort which has produced one of the largest and most complete connectomes to date (Dorkenwald et al., 2019; Scheffer et al., 2020; Schneider-Mizell et al., 2020). However, due to the incredible challenge of producing a high-fidelity connectome, only limited volumes of the brain have been mapped so far. This demonstrates the ongoing challenge of imaging synapses at the nanoscale resolution and the need for vast improvements in imaging techniques and technology before connectome reconstruction reaches its full potential.
The speed and resource limitations of traditional methods of connectome reconstruction have significant drawbacks for the usefulness of the connectomes generated. The brain is a highly dynamic structure, and although some synapses and spines are relatively stable, others frequently reassemble, sometimes on the scale of hours (Berry and Nedivi, 2017). A connectome merely represents a snapshot of a brain in a moment in time, and to truly understand the connectivity of an organism, a single connectome will not suffice. Moreover, the connectomes of individual organisms may differ greatly and sometimes unexpectedly (Bergmann et al., 2020; Witvliet et al., 2020). Furthermore, merely knowing the number of synapses that connect two neurons does not provide all of the necessary information for understanding the function of the synapse. The molecular identity of the synapse is incredibly diverse and reveals essential information such as synaptic type and strength, without which a full vision of connectivity cannot be developed, and unfortunately, EM preparation techniques are largely incapable of preserving molecular identity (O’Rourke et al., 2012). On the other hand, optical microscopy is well suited for imaging large samples at high-throughput and compatible with multiplexed imaging required for revealing the molecular identity of synapses. Indeed, high-throughput optical imaging approaches, such as FAST, MOST, and tiling light sheet microscopy, have been already used for whole-brain imaging (Gong et al., 2016; Seiriki et al., 2017; Motta et al., 2019; Winnubst et al., 2019; Chen et al., 2020; Zhong et al., 2021). However, in this case, the resolution is limited by the diffraction of light and thus not sufficient for mapping synaptic connections. Super-resolution microscopy can break the diffraction limit of light but at the cost of greatly reduced throughput and the need for thin sample slicing to maintain point spread function (Sahl et al., 2017; Schermelleh et al., 2019). Expansion Microscopy (ExM), a recently developed tissue processing technique, allows for the imaging of biological specimens at the voxel rates of a diffraction-limited microscope, but with the voxel sizes of a super-resolution microscope (Chen F. et al., 2015; Tillberg et al., 2016). This makes ExM a form of super-resolution microscopy, which relies on swellable polymers to physically expand tissues before imaging (Chen F. et al., 2015). Physical magnification of the specimen occurs at the nanoscale by separating biomolecules, thus enabling subdiffraction limit resolution under a conventional microscope (Tillberg et al., 2016). Here, the authors would like to note that the first method that resulted in brain tissue expansion was reported by Miyawaki and colleagues in 2011 (Hama et al., 2011). However, it was not realized as a way to improve the spatial resolution of imaging until 2015 when Boyden and colleagues introduced the concept of ExM.
ExM has several crucial advantages over EM that make it particularly well suited for visualizing the synapse, particularly for large-scale projects like connectome mapping. For example, the time, labor, equipment, and skill demands of an ExM experiment are substantially less than that of an EM experiment (Wassie et al., 2019). ExM also is compatible with conventional molecular labeling tools and maintains optical microscopy’s ability to image in color, allowing for the use of several fluorescent probes at once or sequentially, thus enabling multiplexing as well as revealing the molecular identity of the synapse in situ (Chen F. et al., 2015; Ku et al., 2016; Wassie et al., 2019; Alon et al., 2021; Payne et al., 2021). ExM is compatible with a wide variety of tissue types and has been used to image brain tissue in many of neuroscience’s most widely used model organisms (Freifeld et al., 2017; Gao R. et al., 2019; Yu et al., 2020) and in monkey specimens (Zhao et al., 2017). ExM has already been successfully utilized to image neural connectivity at the resolution of the single synapse. For example, Gao R. et al. (2019) used ExM in tandem with lattice light-sheet microscopy to visualize synaptic proteins and neuronal morphology at nanoscale resolution in the mouse cortex and Drosophila brain. Shen et al. (2020) also recently used ExM combined with fluorescent labeling and antibody staining to trace likely synaptic connections in neurons while preserving cell-type specific molecular information. There is tremendous potential for ExM to revolutionize the way synapses are imaged and studied. The technology has produced visually stunning results of brain tissue in a variety of model organisms, and most ExM protocols are substantially more compatible with the high-throughput approach needed to tackle the problems of connectomics and beyond in the future.
We start by reviewing the ExM methods that have been already applied for synaptic mapping and imaging using immunostaining and fluorescent protein-based neuronal labeling and tracing, which facilitates assigning synapses to their parent neurons. We also discuss major challenges and limitations of the currently available ExM methods regarding the comprehensive optical connectome. We then summarize some major molecular strategies for visualizing the synapse at high resolution that can be used in combination with ExM for optical connectome. The first strategy involves fusing synaptic scaffold proteins, such as PSD-95, gephyrin, and synaptophysin, with fluorescent markers. Many tools are variations of this general technique and are widely used both for live imaging and for fixed sample preps, and we feature the most commonly used and the most promising for ExM below. The second strategy involves the use of intrabody-based probes known as FingRs, which bind to synaptic scaffold proteins. Although FingRs are a much newer and less established technology than tagged scaffold proteins, they have several key features that make them more suitable for certain applications. We finalize the review by providing experimental considerations and perspectives on ExM technology.
State-of-the-Art ExM Methods for the Optical Connectome
All ExM methods are based on physical magnification of biological sample via hydrogel embedding followed by mechanical homogenization to disrupt intermolecular complexes (so as to remove mechanical resistance to expanding) and subsequent hydrogel swelling usually in water or low salt buffers. To retain proteins in the expanded state, the sample is treated with a reagent to modify amino acid side chains (usually lysine) with a chemical anchor that participates in radical polymerization to covalently bound proteins into polymer mesh. For a more detailed overview of ExM protocols, we refer readers to recent reviews covering the basic principles of hydrogel-based tissue transformation (Wassie et al., 2019; Choi et al., 2021). As ExM employs optical microscopy for imaging it is heavily reliant on fluorescent probes for targeted biomolecule labeling. Immunohistochemistry (IHC), a fixed tissue staining procedure based on antibody labeling, is a widely used technique within neuroscience (Magaki et al., 2019) and at present, it is the most well-validated approach for synaptic protein visualization using ExM. Since the introduction of the ExM concept in 2014, a large diversity of tissue expansion protocols and methods have been developed for various applications and biological samples. The optimization of ExM was focused on three major aspects: (i) improving fluorescent labeling; (ii) increasing spatial resolution; and (iii) diversifying samples (i.e., whole organs and organisms). We briefly review these aspects in the context of the optical connectome.
Depending on the ExM method, antibody staining can be performed either before tissue expansion or after hydrogel embedding and homogenization (Figure 1). In the earlier versions of ExM, fixed tissue is stained with antibodies before it is gelled and expanded, and several particularly useful antibodies for synaptic visualization were demonstrated to be compatible with subsequent tissue expansion (Chen F. et al., 2015; Chozinski et al., 2016). Among the most commonly used antibodies with ExM of brain tissue are Homer and Bassoon (Chen F. et al., 2015; Chozinski et al., 2016; Chang et al., 2017). Staining with antibodies against Homer1 allows for visualization of the post-synaptic components of excitatory synapses in fixed mammalian brain tissue (Gutierrez-Mecinas et al., 2016; Gao R. et al., 2019; Park et al., 2020). Antibodies against Bassoon stain the pre-synaptic components of synapses, and thus are useful to combine with post-synaptic markers like Homer1 (Micheva et al., 2010; Bürgers et al., 2019; Gao R. et al., 2019). Several other important synaptic proteins targeted by immunolabeling with ExM includes glutamate receptor 1, gephyrin, gamma-aminobutyric acid receptor Aα1/Aα2, vesicular glutamate transporter 1, vesicular GABA transporter (vGAT), Rab3A-binding protein (RIM), Shank2, and PSD95 (Chang et al., 2017; Truckenbrodt et al., 2018; Bürgers et al., 2019; Hafner et al., 2019). However, immunostaining of synaptic complexes usually has low efficiency due to the limited accessibility of antibodies to densely packed synapses, where inter-protein distances are smaller than the size of conventional antibodies (Sarkar et al., 2020).
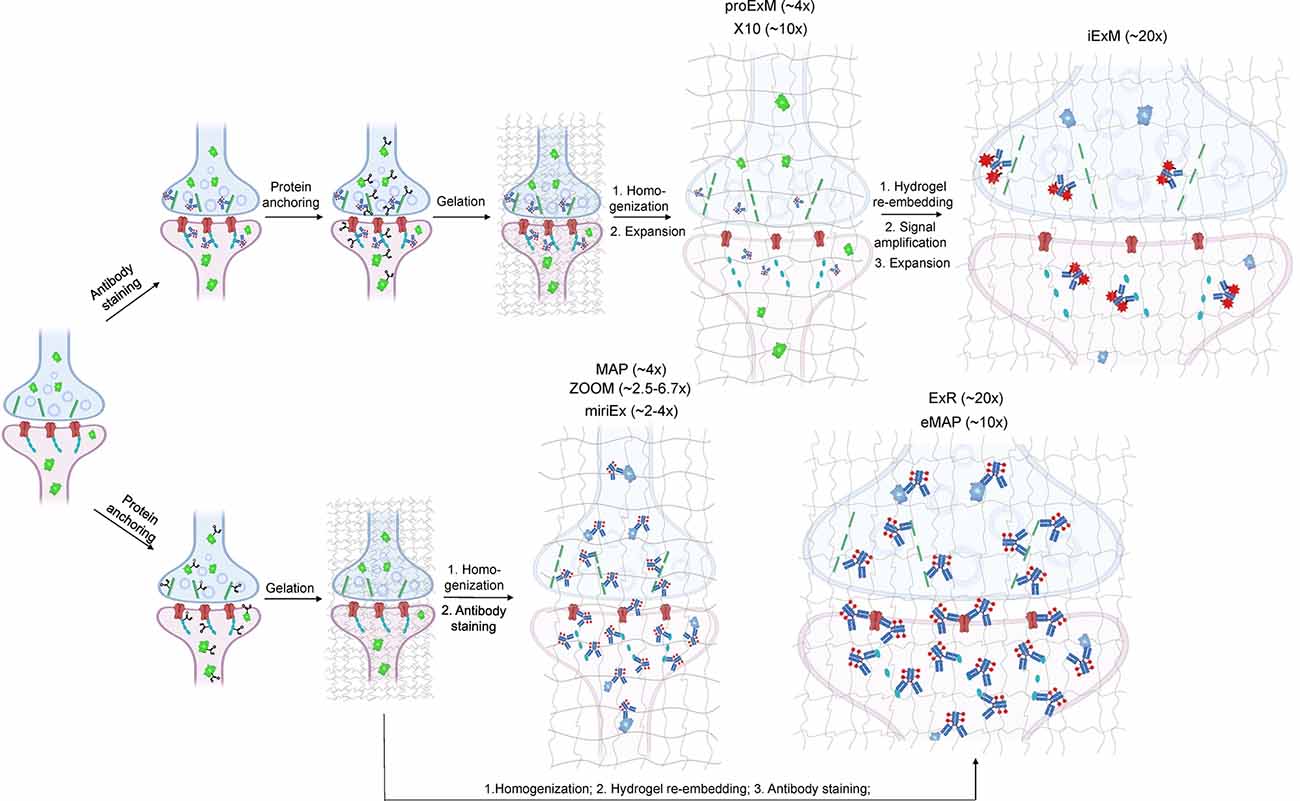
Figure 1. Schematic representations of the ExM workflows with protein retention. Created with Biorender.com.
To overcome this limitation, much effort has been focused on the development of ExM methods that allow antibody staining to be performed after sample homogenization (Figure 1). By utilizing mild chemical treatment with the detergent-containing buffer it was possible to preserve antigens for immunostaining in the expanded samples (Ku et al., 2016; Park et al., 2019). In addition, it was demonstrated that tissue expansion provides better access for antibodies to epitopes of the synaptic proteins, which otherwise might be masked in the dense synaptic protein complexes (Sarkar et al., 2020). Molecular de-crowding via sample expansion significantly increased the efficiency of immunolabeling, which in turn not only improved visualization of synaptic connections but also expanded the list of commercially available antibodies for synaptic proteins compatible with ExM workflow. For example, the recently developed Expansion Revealing (ExR) method was successfully applied to image 7 synaptic proteins important for neural architecture and transmission including the presynaptic proteins Bassoon, RIM1/2, and the P/Q-type Calcium channel Cav2.1 alpha 1A subunit, and the postsynaptic proteins Homer1, Shank3, SynGAP, and PSD95 (Sarkar et al., 2020). This study also allowed us to gain new insights into the nanoscale alignment of presynaptic calcium channels with postsynaptic machinery in intact brain circuits (Sarkar et al., 2020). Earlier Ku et al. (2016) performed systematic screening of commercially available antibodies for synaptic proteins using ExM modification, called magnified analysis of the proteome (MAP). The screening has further extended the list of synaptic proteins present in different types of synapses (e.g., VGluT1 vs. VGluT2, GABABR1, mGluR5, or different receptors) that can allow detection of synapses coming from different neuron types or brain regions with ExM. The very recent modification of MAP, denoted epitope-preserving MAP or eMAP, was optimized to achieve maximal preservation of antigenicity in mouse and marmoset brain tissue, thus increasing success rates of staining with synaptic antibodies to more than 94% (Park et al., 2021).
Another advantage of post-expansion immunostaining is the ability to carry out multiple rounds of labeling and imaging, providing an unprecedent degree of multiplexed super-resolution synaptic proteomic profiling. When expanded tissue samples are imaged in between rounds of immunostaining and antibody stripping, a variety of antibody signals can be converged into a single composite image, allowing for several synaptic proteins to be stained for and imaged at once. For example, the MAP and eMAP protocols were demonstrated to be suitable for highly multiplexed super-resolution imaging via repeated staining and destaining using antibodies for synaptic proteins including Homer1, Bassoon, PSD95, vGluT1, vGluT2, and GABABR1 (Ku et al., 2016). A similar approach was implemented in multi-round immunostaining Expansion Microscopy (miriEX) that involves tissue expansion followed by several iterations of antibody staining and stripping (Shen et al., 2020). The miriEX method was originally applied for iterative immunostaining endogenous synaptic proteins, such as Cannabinoid type 1 receptor, calbindin, and serotonin transporter, in mouse neurons that expressed Brainbow constructs (Shen et al., 2020). While iterative immunolabeling is proven to be a valid approach for multiplexed ExM imaging, it might not be a very practical one due to the need for extremely precise image co-registration between staining steps. Furthermore, this procedure involves buffer exchange and thus it can slightly alter the expansion factor due to osmolarity mismatch. Co-registration of images with varying expansion factors significantly complicates image processing and analysis steps (Alon et al., 2021). Alternatively, multiplexing can be achieved via blind unmixing without reference spectral measurements, allowing up to 15 color imaging (Seo et al., 2021). Seo at el. utilized spectral unmixing on expanded brain samples although only combing four labels for NeuN, GFAP, calretinin, NF-H. The multiplexing approach can significantly increase the utility of ExM for molecular profiling of synapses, however, it still needs to be carefully validated for a large variety of synaptic proteins.
In addition to 3D mapping and molecular profiling of synaptic connections, ExM methods with increased expansion factors can be used to visualize the nanoarchitecture of synapses (Figure 2). The resolution power of ExM is defined by the expansion factor. The majority of the ExM methods enable about four-fold sample expansion in linear dimension, which results in ~70–80 nm of lateral resolution under conventional diffraction-limited microscopes (Tillberg et al., 2016). One strategy to increase the expansion factor is based on the iterative expansion through gel re-embedding, i.e., synthesis of new hydrogel network within an already expanded sample. This strategy was first employed in iterative ExM, or iExM, to achieve ~16–22-fold linear expansion (Chang et al., 2017). The iExM method was carefully validated and characterized by imaging tubulin structures in cultured cells demonstrating isotropic expansion and about 25 nm of effective resolution. The achieved resolution was sufficient to clearly separate post- and pre-synaptic density proteins, such as Homer1, Bassoon, and Gephyrin, from neurotransmitter receptors GluR1 and GABAARα1/α2 in excitatory and inhibitory synapses, respectively (Chang et al., 2017). However, in iExM the proteins are not retained in the expanded state and the staining relies on custom oligonucleotide conjugated antibodies in combination with signal amplification via locked nucleic acid probes. Requirements for customized reagents prevented wide adaptation of iExM. Protein retention with the iterative expansion concept was realized in ExR (Sarkar et al., 2020), tetra-gel-based ExM (Gao et al., 2021), and eMAP (Park et al., 2021). For example, ExR, exhibiting effective resolution comparable to that of iExM, is compatible with commercially available antibodies applied to the expanded samples. Visualizing Cav2.1, PSD95, and RIM1/2 in mouse brain tissue revealed how calcium channel distributions participate in transsynaptic nanoarchitecture. Gel re-embedding complicates and extends sample preparation and treatment steps.
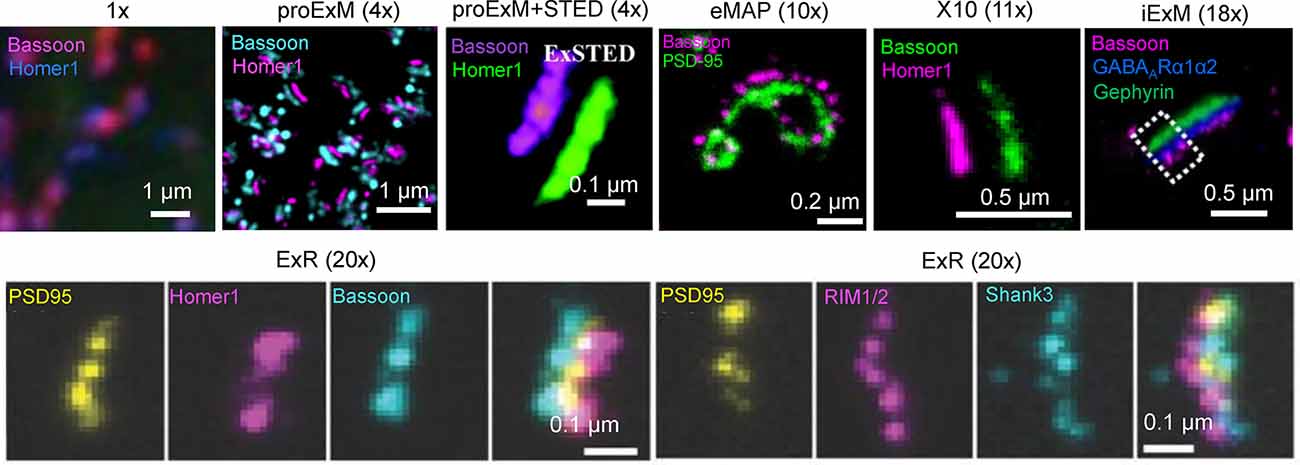
Figure 2. Visualization of synapses in mouse brain tissue using different ExM protocol modifications. The values in parenthesis represent the linear expansion factor for the corresponding image. Adapted from Chen F. et al. (2015), Chang et al. (2017), Li et al. (2018), Truckenbrodt et al. (2018), Gao R. et al. (2019), Sarkar et al. (2020), and Park et al. (2021).
Alternatively, the higher expansion factors (>4-fold) can be achieved by modifying hydrogel chemical composition. For example, by utilizing new hydrogel monomers, Truckenbrodt et al. (2018) developed the X10 ExM method characterized by up to ~11-fold expansion in a single expansion step and compatibility with conventional antibodies. The X10 procedure was used for multi-color imaging of the pre-synaptic active zones and the post-synaptic densities via Homer1, Bassoon, PSD95, and synaptophysin staining with an estimated effective resolution of about 25 nm. While X10 provides the convenience of single step expansion, the hydrogel it uses has poor mechanical properties in the expanded state and its polymerization requires special conditions. The expansion factor of the common ExM hydrogel can be also enhanced by adjusting the cross-linker molar ratio and monomer concentration (Park et al., 2019; Hu et al., 2020; Damstra et al., 2021). For example, the ZOOM and TREx protocols, characterized by the expansion factor of six-10-fold, were used for synaptic visualization using Homer and Bassoon antibodies (Park et al., 2019; Damstra et al., 2021), although the staining of other synaptic proteins still remains to be tested with these methods. It should be also noted that enhancement of resolution can be achieved by combining sample expansion with traditional super-resolution imaging techniques. For example, ExM was combined with stimulated emission depletion microscopy (STED), stochastic optical reconstruction microscopy (STORM), super-resolution optical fluctuation imaging (SOFI) to gain additional improvement in resolution (Gao et al., 2018; Li et al., 2018; Xu et al., 2019; Zwettler et al., 2020; Shi et al., 2021). However, to date, only ExSTED, a combination of ExM with STED, has been applied for synaptic imaging in mouse brain tissue (Li et al., 2018). The proof-of-principle applications of the ExM protocols with increased expansion factor clearly demonstrated the advantages of enhanced resolving power for synaptic nanoarchitecture imaging. However, further validation and characterization are required for establishing the new protocols for routine use in neuroscience research. In particular, it is important to confirm the distortion-free expansion of synaptic complexes, which are known to be very densely packed with proteins and tight proteins complexes.
The ExM methods reviewed above were mostly optimized for cultured cells or thin brain tissue sections. However, neural processes project to local and distal areas throughout the brain, reaching more than 78 cm in total length in the mouse brain (Winnubst et al., 2019). Therefore, imaging of small brain tissue volumes would have limited utility for complete connectome imaging and reconstructions, as registration of neuronal processes from independently expanded and imaged tissue sections from the same brain may not be feasible. It would be ideal if one could expand and image the intact brain with appropriate synaptic and neuron labeling as well as resolution to form a comprehensive connectome. However, this task meets several technical challenges on multiple fronts—expansion procedure, labeling, and imaging itself, which still remain to be fully addressed in practice. For example, four-fold expanded mouse brain with dimensions of 4 × 3 × 6 cm is impossible to image with nanoscale resolution in intact form due to the limited working distances of available objectives. One way to deal with this is to slice the expanded brain upon imaging, a solution already implemented in fMOST technology (Zhong et al., 2021) and in the MouseLight project (Winnubst et al., 2019).
Reducing the expansion factor of the original ExM protocol can facilitate entire brain imaging. For example, two-fold expanded mouse brain using the CUBIC-X protocol can be imaged under a custom light-sheet microscope, albeit with the cost of greatly reduced spatial resolution (Murakami et al., 2018). On the other hand, expansion of small model organisms such as C. elegans, Drosophila, and zebrafish, could be more feasible at present, as expanded samples can be imaged in an intact state using conventional imaging setups. ExM methods were already adopted for super-resolution imaging of Drosophila brain explant (Mosca et al., 2017; Jiang et al., 2018), enabling the mapping of presynaptic sites in the entire brain with lattice light sheet microscopy (Gao R. et al., 2019; Lillvis et al., 2021). It was also demonstrated that zebrafish brain explants (Freifeld et al., 2017) and even whole zebrafish larvae (up to 6 days post-fertilization; Sim et al., 2021) can be expanded and imaged using conventional imaging setups. Similarly to whole zebrafish expansion (Sim et al., 2021), the ExCel method can expand an entire C. elegans specimen using an extensive chemical treatment to ensure isotropic expansion (Yu et al., 2020). However, this treatment reduces the fluorescence of fluorescent proteins and antibodies, requiring a larger amount of antibodies and extended staining time (Yu et al., 2020). To facilitate antibody penetration into expanded samples, stochastic electrotransport (Kim et al., 2015) was shown to speed up antibody diffusion into thick (>5 mm) expanded mouse brain samples (Ku et al., 2016). Immunostaining of magnified samples extends the timeline and increases the cost of sample preparation.
In addition to identifying the synapse itself, an important part of connectomics is tracing synaptic connections to the originating neuron. From this perspective, the expression of fluorescent proteins might be a good alternative to antibodies, as the visualization of genetically encoded fluorescent probes does not require an additional staining step and they can be evenly expressed throughout the plasma membrane and/or cytoplasm. Protein-retention ExM (proExM) was demonstrated to retain native fluorescence of multiple fluorescent proteins in expanded samples, including mouse and monkey brain tissue (Tillberg et al., 2016). Owing to its high brightness and chemical stability, yellow fluorescent protein (YFP) has high performance in ExM and was used for neuronal tracing in mouse tissue (Gao R. et al., 2019). Indeed, cytoplasmic or membranal expression of fluorescent proteins is perhaps one of the most widely used approaches for neuronal tracing using optical microscopy. Fluorescent protein-based technology for neuronal tracing, such as Brainbow, has been already used in combination with ExM (Tillberg et al., 2016; Chang et al., 2017). Brainbow is a transgenic strategy for distinguishing individual cells from their neighbors, due to stochastic fluorescent protein expression that provides individual cells a unique fluorescent signature (Livet et al., 2007; Cai et al., 2013; Weissman and Pan, 2015; Shen et al., 2020). Brainbow allowed for individual neurons to be distinguished from the neighbors in mouse brain slices (Tillberg et al., 2016), while antibodies against gephyrin, Homer1, and Bassoon allowed those synapses to be characterized as excitatory or inhibitory (Shen et al., 2020). Improved versions of Brainbow, such as Tetbow (Sakaguchi et al., 2018), and Bitbow (Li et al., 2021), have been demonstrated to enable highly efficient neuronal morphology reconstruction. Applying proExM to the Bitbow-expressing Drosophila brain made it possible to reconstruct all 21 ventral nerve cord serotonergic neurons out of 26 estimated total. However, the fluorescent signal was amplified using immunostaining (Li et al., 2021). In addition, the expression of fluorescent proteins using the rabies virus is a powerful transneuronal tracing technology (Ugolini, 2011; Kim et al., 2016).
Fluorescent Synaptic Scaffold Proteins
Among the most established strategies we discuss is the fusion of synaptic scaffold proteins with fluorescent markers, which has launched many variations upon the theme of fusing a prominent biologically relevant synaptic protein with a fluorescent protein such as eGFP. The most widely used post-synaptic scaffold proteins involved are PSD-95 (Gray et al., 2006; Cane et al., 2014; Isshiki et al., 2014; Villa et al., 2016) and gephyrin (Craig et al., 1996; Villa et al., 2016), and the most widely used pre-synaptic scaffold protein is synaptophysin (Antonova et al., 2009; Li et al., 2010), and variants of these proteins have been imaged in many of the relevant model organisms used by neuroscientists.
Many of the following tools were developed with in vivo imaging in mind, but they are likely also compatible post-vivo with ExM in fixed tissue. It is our belief that complementing in vivo functional imaging of the synapse with ExM of the fixed tissue presents a powerful opportunity to study the structure and function of the synapse in tandem. Structural and connectomic data are significantly more useful when complemented by functional data: for example, a connectome alone would not reveal the strength of an individual synapse, but a connectome supplemented with functional data would present a much clearer picture of the synapse in question (Turner et al., 2020). ExM is uniquely situated to synergize structural and functional imaging and produce connectomes that are supplemented with custom functional data.
Probes to Visualize Post-synaptic Connections
We begin by describing tools to visualize the postsynaptic scaffold proteins. These approaches enable investigations of structural dynamics in live brains and provide fluorescent markers of synapses for post-fixed tissue expansion. In many researched vertebrate systems, commonly used proteins for fluorescence tagging are postsynaptic density protein 95 (PSD-95), also known as synapse-associated protein 90 (SAP-90), and gephyrin. These particular proteins are found at the postsynaptic density, a dense protein complex found in both excitatory and inhibitory synapses (Sheng and Kim, 2011; Dosemeci et al., 2016). The postsynaptic density of an excitatory neuron can contain several hundred PSD-95 molecules, and the postsynaptic density of an inhibitory synapse can contain tens or even hundreds of gephyrin molecules (Chen X. et al., 2005; Chen et al., 2011; Choii and Ko, 2015). While PSD-95 and gephyrin are among the most common postsynaptic density proteins studied in vertebrates, the postsynaptic density is full of many other proteins which could alternatively be labeled to provide insight into the structure of synapses (Helm et al., 2021).
PSD-95 plays a variety of roles in the postsynaptic density, most notably binding to key excitatory glutamate receptors and promoting the maturation and strengthening of dendritic spines and excitatory synapses (Chen et al., 2011; Cane et al., 2014; Taft and Turrigiano, 2014; Chen X. et al., 2015). This makes PSD-95 a faithful structural surrogate for excitatory synapses in vertebrate synapses. Genetically tagging PSD-95 with exogenous fluorescent proteins permits in vivo tracking of excitatory synapse structural dynamics (Gray et al., 2006; Cane et al., 2014; Isshiki et al., 2014; Villa et al., 2016; Subramanian et al., 2019; Figure 3A). Some of the first experiments to fluorescently label the postsynaptic density in mammalian and zebrafish neurons used a PSD-95-GFP fusion (Craven et al., 1999; Niell et al., 2004). More recently, PSD-95 has been fused to mCherry for imaging of synaptic spines in live mice. For example, using a CPG15/Netrin knock-out mouse and PSD95-mCherry labeling in vivo, Subramanian et al. recently found that GPI-anchored CPG15 interacts with AMPA receptors and recruits PSD-95 to transient, unstable spines, leading to stable long-term synapses (Subramanian et al., 2019). This discovery would have been impossible without a structural marker of PSD-95, underscoring the importance of tools to visualize sub-synaptic structures in vivo. PSD-95 has been imaged in expanded mouse brain slices using antibodies (Sarkar et al., 2020); applying a similar technique to PSD-95-mCherry may even increase brightness in the expanded slice. Importantly, the coupling of live PSD-95 fusion imaging with later tissue expansion creates new possibilities for interrogating function and connectivity side-by-side. PSD-95 fusions such as PSD-95-mCherry allow for the structural dynamics and function of synapses to be interrogated in live brains and are then compatible with tissue expansion and microscopy to resolve the fine details of synaptic connections in fixed brains.
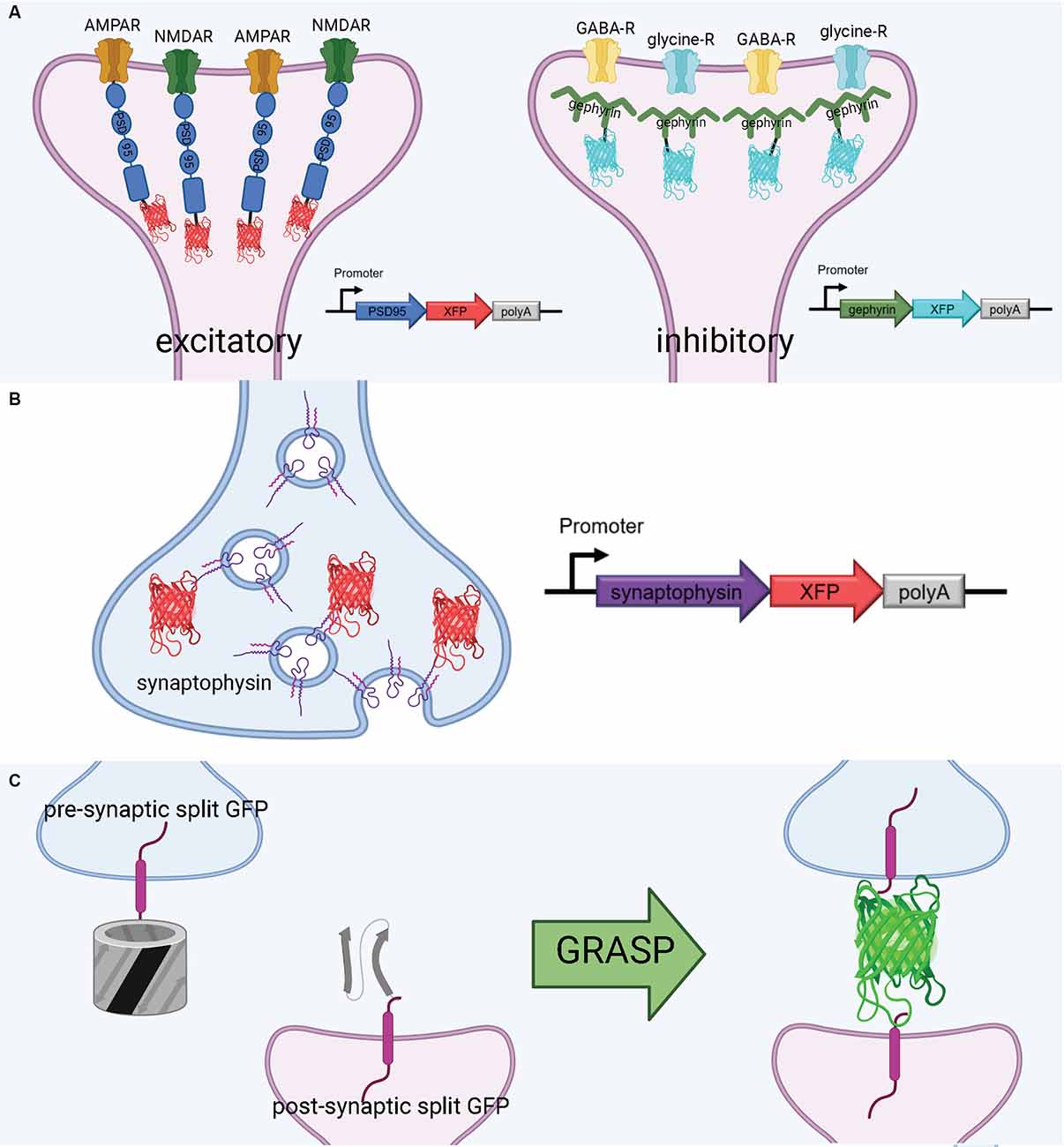
Figure 3. Tools to visualize post-synaptic and pre-synaptic connections. (A) Schematic diagram of post-synaptic labeling of excitatory synapses with exogenous PSD-95-fluorescent protein hybrids and inhibitory synapses with exogenous gephyrin-fluorescent protein hybrids. (B) Schematic diagram of pre-synaptic labeling off synapses with exogenous synaptophysin-fluorescent protein hybrids. (C) Proximate synapse labeling with GFP Recombination Across Synaptic Partners (GRASP). Created with Biorender.com.
Gephyrin is a central postsynaptic scaffold found in the vertebrate CNS exclusively at glycinergic and GABAergic synapses (Craig et al., 1996; Tyagarajan and Fritschy, 2014). Gephyrin tagging with fluorescence proteins has been shown to be a reliable method to mark inhibitory synapses in vitro (Meier and Grantyn, 2004; Vlachos et al., 2013; Dejanovic et al., 2014) and in vivo (Oh et al., 2016; Villa et al., 2016; Figure 3A). The development of a teal-gephyrin as a morphological surrogate for inhibitory synapses revealed that one-third of inhibitory synapses reside on dendritic spines and that inhibitory synapses are clustered and persistently disassemble and reassemble at persistent sites in vivo (Villa et al., 2016). It should be noted that the inhibitory synapses on spines appear to be the most dynamic and plastic inhibitory synapses. Three-color imaging using PSD-95, Gephyrin, and a cell-filling fluorophore resolves three spine types: spines without PSD-95, spines with only PSD-95, and spines with both PSD-95 and gephyrin (Villa et al., 2016; Subramanian et al., 2019). Hence, gephyrin is an effective strategy for labeling inhibitory synapses and can be combined with other fluorescent markers.
In smaller brains, such as that of Drosophila, anterograde synaptic tracing is another option for mapping neural connections. For example, trans-Tango has been used to trace synaptic connections in neurons in several Drosophila brain regions, such as the olfactory and gustatory systems (Talay et al., 2017) and mushroom body (Scaplen et al., 2021). Trans-Tango relies on the Tango assay (Barnea et al., 2008), which is activated when a pan-neuronally expressed presynaptic fusion protein meets a postsynaptic fusion protein (Talay et al., 2017). Trans-Tango exhibits a high signal-to-noise ratio and can be applied to any genetically defined subset of neurons (Talay et al., 2017).
The expression of exogenous synaptic proteins poses the risk of interfering with the normal function of synapses. For example, overexpression of PSD-95 significantly enhances the amplitude of the excitatory postsynaptic current and increases the number of the synapse (El-Husseini et al., 2000; Béïque and Andrade, 2003). To minimize overexpression, the fusion proteins are usually expressed under weak promoters, e.g., the promoter of the polyubiquitin C gene (UBC; Subramanian et al., 2019). An even safer way to label synaptic proteins can be appending the gene of endogenous protein of interest in the genome with the gene of a tag. One approach for labeling synapses without the consequences of relying on exogenous protein expression is Synaptic Tagging with Recombination (STaR; Chen et al., 2014). STaR is a genetic approach which utilizes recombination in Bacterial Artificial Chromosomes to induce the expression of presynaptic and postsynaptic markers. Importantly, these markers can be targeted to specific neurons and are under the control of their endogenous regulatory mechanisms. In Drosophila, Chen et al. (2014) were able to concurrently label both presynaptic and postsynaptic markers by taking advantage of different recombination systems, allowing for visualization of synaptic pairs. The authors used this method to successfully visualize synapses in the Drosophila visual system, although there is potential for the method to be used in other model organisms and their appropriate recombination systems (such as Cre-Lox in mice; Chen et al., 2014). STaR is compatible with fixed brain preparations and thus holds potential for higher resolution usage in combination with ExM.
Just as STaR allows for synaptic protein tagging in Drosophila, there are in vivo genome editing strategies based on the CRISPR-Cas9 technology for mice that have been used to tag cellular proteins with epitopes without interfering with endogenous protein expression (Mikuni et al., 2016; Nishiyama et al., 2017). One such high-throughput option for monitoring endogenous synaptic proteins in live mice at single-cell resolution is SLENDR (Mikuni et al., 2016; Nishiyama et al., 2017). This method involves the delivery of the gene-editing machinery of CRISPR-Cas9 to neural progenitors in developing mouse embryos, traditionally through in utero electroporation. The CRISPR-Cas9 technology allows the user to label proteins of interest with small epitope tags, such as human influenza hemagglutinin (HA), Myc, and V5, as well as large payloads like monomeric EGFP and spaghetti monster fluorescent proteins (smFP; Viswanathan et al., 2015). Alternatively, Suzuki et al. developed a homology-independent targeted integration (HITI) strategy based on CRISPR-Cas9, which enables robust DNA knock-in in neurons of postnatal mammals in vivo (Suzuki et al., 2016). The HITI approach can be implemented using AAV-mediated gene delivery, reaching about 11% of infected neurons in adult mice. More recently, Gao Y. et al. (2019) introduced the homology-independent universal genome engineering (HiUGE) method, characterized by higher efficiency (>20%) and throughput than HITI. The HiUGE method was successfully utilized in neurons in mice to visualize inhibitory postsynaptic density (iPSD) proteome including inhibitory synaptic protein 1 (Insyn1), inhibitory synaptic protein 2 (Insyn2), and Rho GTPase activating protein 32 (Arhgap32; Uezu et al., 2016). Furthermore, by combining HiUGE with retrograde-transported AAV2-retro serotype (Tervo et al., 2016), in the same study Gao et al. performed neural circuit-selective protein manipulations in the well-defined cortico-striatal circuit and the thalamocortical circuit. Alternative CRISPR-Cas9 methods for targeted genomic integration of epitope tags, such as ORANGE (Open Resource for the Application of Neuronal Genome Editing) and TKIT (Targeted Knock-In with Two), were used for targeting PSD95, GluA1, and GluA2 with short epitope tags in vivo in mice with an efficiency of 10–16% (Willems et al., 2020; Fang et al., 2021). The ORANGE system was also validated in vitro for a variety of synaptic proteins, including voltage-dependent Ca2+-channels, Rab3-interacting molecules, Bassoon, Glutamate receptor NMDA 1, etc. Another option for labeling endogenous synaptic proteins is endogenous labeling via exon duplication (ENABLED), which has been utilized specifically to label endogenous PSD-95 in mice. This genetic strategy is particularly notable for connectomics because it minimizes protein overexpression and can label a large subset of neurons (Fortin et al., 2014). These techniques and other similar strategies have the potential to be used to label synaptic proteins, image their dynamics in live mice, and then image synaptic connectivity in expanded brain slices.
Probes to Visualize Pre-synaptic Connections
Much as with postsynaptic proteins, pre-synaptic proteins can be fluorescently labeled to map synaptic connections. A well-established presynaptic protein for labeling synapses, particularly in vertebrates, is synaptophysin, a synaptic vesicle glycoprotein that interacts with the essential synaptic vesicle protein synaptobrevin and is thought to participate in synaptic vesicle release (Wiedenmann and Franke, 1985; Becher et al., 1999). Synaptophysin fused to a fluorescent protein has been used in many circumstances, such as in mice, rat hippocampal neurons, and zebrafish, as a faithful indicator of presynaptic machinery (Meyer and Smith, 2006; Antonova et al., 2009; Li et al., 2010; Figure 3B). Synaptophysin can also be used in tandem with postsynaptic markers, such as PSD-95, to label synaptic connections. Using this approach, Subramanian et al. (2019) could selectively visualize the dynamics of postsynaptic dendritic spines receiving contact from Synaptophysin-labeled presynaptic terminals. In a similar vein, it is also possible to label a postsynaptic marker such as PSD-95 in vivo, and later stain the tissue for a presynaptic marker such as synaptophysin to visualize synaptic connections (Broadhead et al., 2016).
A flexible approach to unambiguously map synaptic connectivity is based on GFP Reconstitution Across Synaptic Partners (GRASP; Feinberg et al., 2008; Figure 3C). Here, two nonfluorescent split-GFP fragments (GFP1–10 corresponding to the first 10 β-strands and GFP11 corresponding to the 11th β-strand of the GFP β-barrel) are tethered to pre- and post- synaptic membranes. Fluorescent GFP is reconstituted only when two neurons, each expressing one of the fragments, are tightly opposed through a synaptic cleft. GRASP, though originally developed in C. elegans, has utility in many model organisms (Feinberg et al., 2008). GRASP first was used in Drosophila in 2009 and has since been widely used and iterated upon (Gordon and Scott, 2009). For example, one of the first enhanced GRASP variants in Drosophila was specifically targeted to synapses by fusion of the presynaptic GFP fragment with Neurexin, a presynaptic cell adhesion protein (Fan et al., 2013). A later variant combines synaptobrevin and GRASP. The syb:GRASP fly chimera allows for in vivo activity-dependent circuit mapping, in contrast with the activity-independent neurexin variant (Macpherson et al., 2015). The same team behind the syb:GRASP fly also developed yellow and cyan GRASP variants for Drosophila, offering the advantages of multicolor labeling. A further enhancement of GRASP in Drosophila is t-GRASP, an activity-independent label which boasts greater signal specificity to the synapse (Shearin et al., 2018).
A mammalian version of GRASP (mGRASP) has had similar success in mapping synaptic connections in mouse brains (Kim et al., 2012; Song et al., 2018). Several variants and enhancements exist, such as CRASP, a cyan fluorescent protein-based variant (Li et al., 2016). Choi et al. (2018) have developed an enhanced GRASP construct, known as eGRASP, which has greater signal intensity than its predecessor. Furthermore, eGRASP expresses either a yellow or cyan fluorescent protein in presynaptic neurons, which allows for visualization of two distinct presynaptic populations which converge on the same postsynaptic neuron.
Neurexin, the presynaptic protein which was targeted in some GRASP variants (Fan et al., 2013), is another synaptic marker of interest, particularly in invertebrate systems (Kim and Emmons, 2017). For example, in C. elegans, neurexin located in the presynapse was shown to have an essential role in the development of the morphology of postsynaptic GABAergic neurons (Philbrook et al., 2018). In particular, neurexin is critical for the development of the spine-like protrusions that appear in C. elegans neurons (Philbrook et al., 2018). Presynaptic neurexin, in conjunction with the postsynaptic marker neuroligin, can be labeled to define synapses in a directional manner, as was demonstrated in live worms in the iBLINC system (Desbois et al., 2015). iBLINC fuses a recombinant biotin ligase with presynaptic neurexin and a small acceptor peptide with postsynaptic neuroligin. In an iBLINC synapse, biotin is transferred from the presynaptic biotin ligase to the postsynaptic acceptor peptide, and when streptavidin fused with fluorescent protein enters the space surrounding the synapse, it fluorescently labels the postsynaptic biotin, and thus the directional connection between the synapses can be observed (Desbois et al., 2015). Although this system was designed with in vivo use in mind, it represents a potential strategy to map synaptic connections in expanded animals, as protein retention expansion microscopy is compatible with streptavidin detection (Tillberg et al., 2016).
Another presynaptic marker used specifically in C. elegans is SYD-2, which has been shown to localize exclusively at the presynaptic active zone of worm neurons (Yeh et al., 2005). A fusion protein of SYD-2 and GFP allowed presynaptic puncta to be labeled in live C. elegans (Yeh et al., 2005). Furthermore, synaptic protein-protein interactions can now be probed in worms with Turbo ID, an enzyme-based proximity labeling strategy (Branon et al., 2018; Artan et al., 2021). TurboID was used to identify protein-protein interactions that a presynaptic protein, ELKS-1, was involved in Artan et al. (2021), representing a blueprint for a potential strategy for synaptic proximity labeling in C. elegans.
An alternative method of visualizing synaptic connections involves genetically-encoded fluorescence-based synapse labeling reagents (Kuljis et al., 2019). Kuljis et al. (2019) recently developed a system which utilizes neuroligin-1, a postsynaptic tag, to target fluorogen-activating protein (FAP) to postsynaptic sites (FAPpost). FAPpost emits a far-red signal upon binding of a small molecule fluorogen, which is applied to brain slices to induce fluorescence. The authors were able to identify synapses belonging to identifiable cell types in the mouse somatosensory cortex. Notably, unlike other constructs for visualizing synapses, FAPpost did not alter synaptic density or neuron firing properties. However, the sparse labeling shown with this technique may not be ideal for assembling complete connectomes.
FingRs
An alternative approach for visualizing synaptic proteins is the use of Fibronectin intrabodies generated with mRNA display (FingRs). These are intrabodies fused to fluorescent proteins which bind to the target endogenous protein and allow for precise visualization of the target’s localization and density (Gross et al., 2013; Figure 4A). FingRs are a newer and less established technology than hybrid fluorescent proteins, and they hold great potential for imaging the synapse despite their recency and lack of widespread use. Unlike many other tools, there has been evidence to show that FingRs do not significantly affect endogenous protein expression, the number and strength of synapses, and synaptic transmission in brain slice (Gross et al., 2013; Bensussen et al., 2020). FingRs are expressed in behaving animals and then quantified using post-mortem histology, and must be expressed in living animals for weeks before they are visualized (Gross et al., 2013; Bensussen et al., 2020). This approach can easily be synthesized with existing ExM protocols. However, unlike standard fluorescent protein techniques, FingRs are not compatible with live imaging.
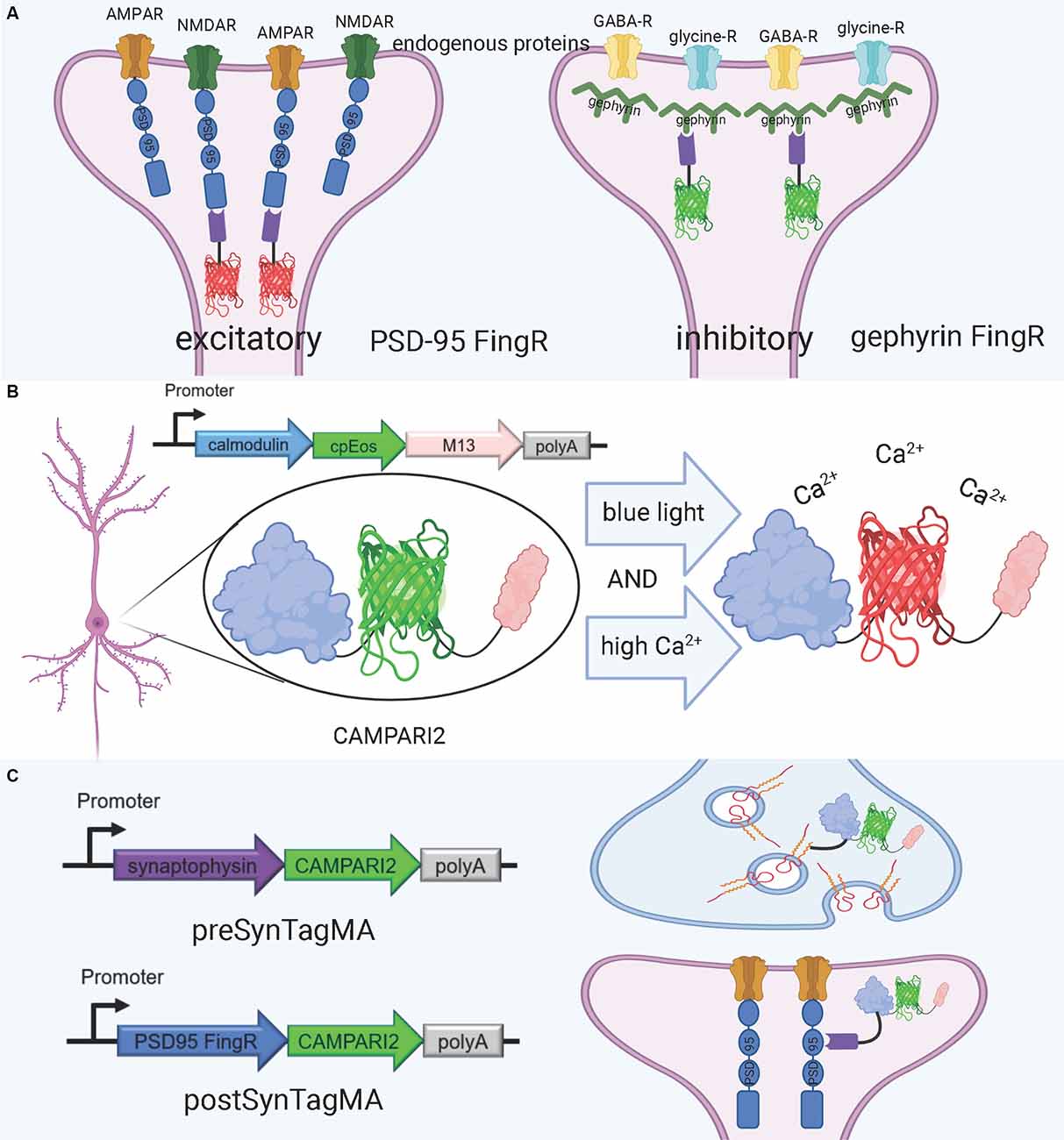
Figure 4. FingRs and related tools. (A) Schematic diagram of FingRs for post-synaptic labeling of excitatory and inhibitory synapses, using endogenous PSD-95 and gephyrin, respectively. (B) Schematic diagram of CAMPARI2, a genetically encoded calcium indicator, which changes fluorescent wavelength in response to combined blue light and high calcium ion levels. CAMPARI2 forms the fluorescent component of SynTagMA. (C) Schematic diagram of pre-SynTagMA, which marks synaptophysin, and post-SynTagMA, which marks endogenous PSD-95 with the aid of a PSD-95-specific FingR. Created with Biorender.com.
FingRs for PSD-95 and gephyrin were the first-ever developed (Gross et al., 2013), and have since proven useful for visualizing changes in synaptic strength in model organisms. Son et al. (2016) used FingRs in live zebrafish and found that FingR expression did not hinder protein expression at the synapse, number of synapses, and synapse function. Additionally, they used FingRs for PSD-95 and gephyrin to show that chronic hypoxia decreases the number of dopaminergic synapses (Son et al., 2016). Cook S. G. et al. (2019) used FingRs to simultaneously image PSD-95, gephyrin, and CaMKII in the hippocampal neural culture and found that amyloid beta (Aβ) interferes with CaMKII activation in stimulus-induced LTP, but not LTD. Xue Han’s group has recently developed further FingR variants, including a red variant which can be used in conjunction with green variants (Bensussen et al., 2020).
A promising tool for understanding the relationship between structure and function in neural circuits is the recently-developed SynTagMA, a genetically encoded sensor for synaptic activity. SynTagMA is a fusion of a modified version of CAMPARI2, an established indicator of active neurons, with FingRs for either PSD-95 or synaptophysin (Moeyaert et al., 2018; Perez-Alvarez et al., 2020; Figures 4B,C). The PSD-95 SynTagMA construct targets post-synaptic terminals, and the synaptophysin SynTagMA construct targets pre-synaptic terminals (Perez-Alvarez et al., 2020). SynTagMA can detect synaptic activity by irreversibly changing from green fluorescence to red in the presence of calcium upon photoactivation by 395–405 nm illumination, generating a snapshot of synaptic activity at a user-defined time (Perez-Alvarez et al., 2020). SynTagMA can simultaneously tag thousands of active synapses. PSD-95-fused SynTagMA has successfully been used in awake head-fixed mice to visualize active synapses, and future experiments using this synaptically localized, photoconvertible calcium sensor will enable further study on the synaptic basis of complex brain function in health and disease. SynTagMA, if used in conjunction with other tools that map connectivity more directly, could be a crucial tool for understanding functional connectivity.
Experimental Considerations
The tools described in this article are all hypothetically compatible with standard ExM protocols and represent a mere subset of the possible ways to image the synapse at nanoscale resolution. Any imaging or staining done pre-expansion, including live imaging, will proceed normally (Figure 5). Afterward, the tissue intended for expansion can be chemically treated according to your ExM protocol of choice (Karagiannis and Boyden, 2018; Wassie et al., 2019). Though some troubleshooting may be involved to receive optimal results, combining previously established synaptic markers with ExM represents exciting possibilities for synaptic mapping in a variety of model organisms. There are, however, a few caveats to be noted about ExM. For example, while the isotropy of the tissue is generally very well preserved in various tissue types, it is important for new ExM users to validate that their tissues expanded in an isotropic manner (Wassie et al., 2019). However, if the protocol is executed correctly, there is no significant rearrangement of the synapse shown when expanded: the relative position of synaptic proteins stays the same after expansion (Zhao et al., 2017; Sarkar et al., 2020). Although the ExM process can degrade some proteins, fortunately, fluorescent proteins are relatively resistant to degradation during the protocol and maintain their brightness (Wassie et al., 2019). Brightness can be improved by processing the tissue with antibodies prior to expansion and choosing ExM protocols that are best equipped to preserve protein integrity suited for individual project needs (Parra-Damas and Saura, 2020). Another important consideration, specifically when the nanoarchitecture of synapses is studied, is the possible distortion upon expansion that may happen at the nanoscale, which is to be validated for every newly developed ExM method or protocol. The gold standard in the field for synapse identification is EM, but ExM views of synapses cannot be correlated with the ultrastructure as seen in EM, as ExM techniques are not compatible with the staining and sample processing done for EM.
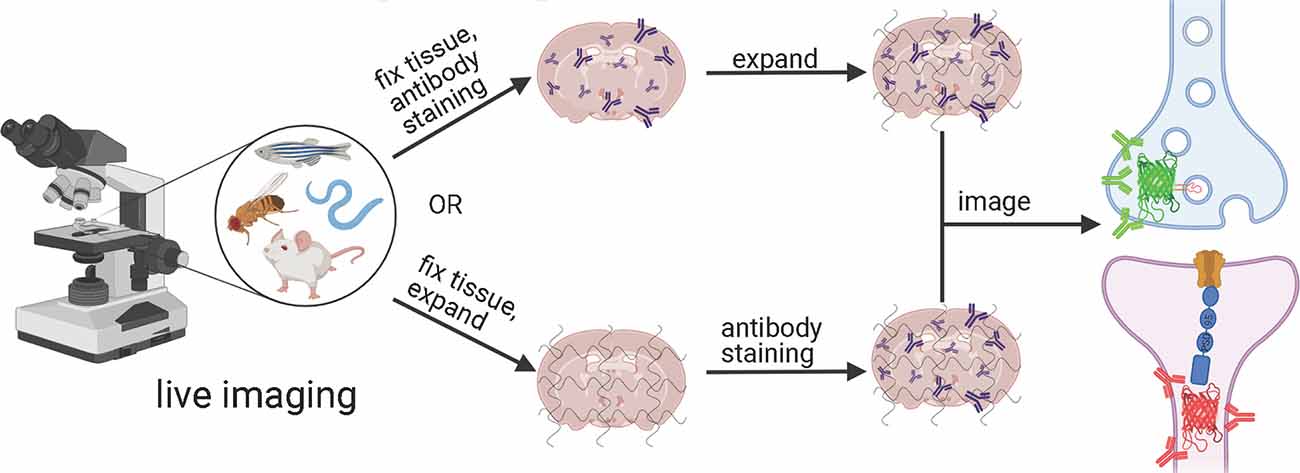
Figure 5. Generalized workflow for combining tools to exogenously label post-synaptic and pre-synaptic neurons in a variety of model organisms with expansion microscopy. Created with Biorender.com.
The choice of gene delivery vector is an important consideration for mapping synaptic connections. One important limitation of many of the tools above, especially several of the sensors designed for use in mammalian brains such as mGRASP and SynTagMA, is that they are typically delivered to live mouse brains via AAV injection. Although AAVs are the favored gene delivery vector for many experiments, this strategy does not guarantee that every single neuron in the target area is labeled (Chan et al., 2017; Wang et al., 2018). For example, when tracing synaptic connectivity in a small volume of the mouse brain, AAVs delivering Brainbow transgenes were not able to label every neuron for connectivity tracing, and the percent of neurons labeled varied widely based on cell type (Shen et al., 2020). For most applications, it is usually not necessary to label every single neuron, but when assembling complete connectomes, it is essential that as many neurons as possible be labeled. Thus, gene delivery strategies must be applied carefully and cautiously to avoid leaving out unlabelled neurons, particularly in mammalian systems. Alternate gene delivery methods for mice, such as the generation of transgenic mouse lines or herpes simplex virus vectors, which have genomic integration rates much closer to 100%, should be considered for connectome-specific applications.
Conclusion/Perspective
There is a great need for the generation of connectomes. The first-ever complete connectome of an organism, the C. elegans connectome, has been indispensable for studying the worm brain. For example, the worm connectome has been combined with ablation experiments to generate circuit maps and has provided the basis for a number of computational models. Taking inspiration from connectomes has also led to biologically-informed innovations in machine learning (Hasani et al., 2020). However, much of the potential of the connectome has remained locked away, particularly because a single connectome cannot possibly represent the full range of connectivities in even a simple nervous system. Connectomes will be most useful when there are multiple, even hundreds, for a single species, let alone for different sexes, developmental stages, and mutants. Furthermore, many of the most commonly used model organisms, particularly zebrafish, mice, and rats, lack anything resembling a complete connectome, and current endeavors to generate these connectomes, though heroic, are incredibly expensive and time-consuming.
ExM has the potential to represent a paradigm shift in connectomics so that any lab with standard microscopy equipment can contribute to the endeavor to map synaptic connections. Rapid advances in synaptic imaging tools and in ExM protocols have paved the way for a powerful synergy: now all that is left is to turn the hypothetical into reality.
Author Contributions
KP: conceptualization, supervision, and funding acquisition. MS: writing—original draft preparation. KP and MS: writing—review and editing, visualization. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by start-up funding from the Foundation of Westlake University, Foundation for Innovative Research Groups of the National Natural Science Foundation of China grant 32050410298 and 32171093, 2020 BBRF Young Investigator Grant number 28961, and MRIC Funding 103536022023 to KP.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Ed Boyden, Mitch Murdock, Yangning Lu, and Orhan Celiker for their helpful comments. We especially thank Mitch Murdock for his support at the beginning of the writing process and Ed Boyden for his support throughout this process. Figures were created with Biorender.com.
References
Albertson, D. G., Thompson, J. N., and Brenner, S. (1976). The pharynx of Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 275, 299–325. doi: 10.1098/rstb.1976.0085
Alon, S., Goodwin, D. R., Sinha, A., Wassie, A. T., Chen, F., Daugharthy, E. R., et al. (2021). Expansion sequencing: spatially precise in situ transcriptomics in intact biological systems. Science 371:eaax2656. doi: 10.1126/science.aax2656
Antonova, I., Lu, F.-M., Zablow, L., Udo, H., and Hawkins, R. D. (2009). Rapid and long-lasting increase in sites for synapse assembly during late-phase potentiation in rat hippocampal neurons. PLoS One 4:e7690. doi: 10.1371/journal.pone.0007690
Artan, M., Barratt, S., Flynn, S. M., Begum, F., Skehel, M., Nicolas, A., et al. (2021). Interactome analysis of C. elegans synapses by TurboID-based proximity labeling. bioRxiv [Preprint]. doi: 10.1016/j.jbc.2021.101094
Barnea, G., Strapps, W., Herrada, G., Berman, Y., Ong, J., Kloss, B., et al. (2008). The genetic design of signaling cascades to record receptor activation. Proc. Natl. Acad. Sci. U S A 105, 64–69. doi: 10.1073/pnas.0710487105
Becher, A., Drenckhahn, A., Pahner, I., Margittai, M., Jahn, R., and Ahnert-Hilger, G. (1999). The synaptophysin-synaptobrevin complex: a hallmark of synaptic vesicle maturation. J. Neurosci. 19, 1922–1931. doi: 10.1523/JNEUROSCI.19-06-01922.1999
Béïque, J.-C., and Andrade, R. (2003). PSD-95 regulates synaptic transmission and plasticity in rat cerebral cortex. J. Physiol. 546, 859–867. doi: 10.1113/jphysiol.2002.031369
Bensussen, S., Shankar, S., Ching, K. H., Zemel, D., Ta, T. L., Mount, R. A., et al. (2020). A viral toolbox of genetically encoded fluorescent synaptic tags. iScience 23:101330. doi: 10.1016/j.isci.2020.101330
Bergmann, E., Gofman, X., Kavushansky, A., and Kahn, I. (2020). Individual variability in functional connectivity architecture of the mouse brain. Commun. Biol. 3:738. doi: 10.1038/s42003-020-01472-5
Berry, K. P., and Nedivi, E. (2017). Spine dynamics: are they all the same. Neuron 96, 43–55. doi: 10.1016/j.neuron.2017.08.008
Branon, T. C., Bosch, J. A., Sanchez, A. D., Udeshi, N. D., Svinkina, T., Carr, S. A., et al. (2018). Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 36, 880–887. doi: 10.1038/nbt.4201
Broadhead, M. J., Horrocks, M. H., Zhu, F., Muresan, L., Benavides-Piccione, R., DeFelipe, J., et al. (2016). PSD95 nanoclusters are postsynaptic building blocks in hippocampus circuits. Sci. Rep. 6:24626. doi: 10.1038/srep24626
Bürgers, J., Pavlova, I., Rodriguez-Gatica, J. E., Henneberger, C., Oeller, M., Ruland, J. A., et al. (2019). Light-sheet fluorescence expansion microscopy: fast mapping of neural circuits at super resolution. Neurophotonics 6:015005. doi: 10.1117/1.NPh.6.1.015005
Cai, D., Cohen, K. B., Luo, T., Lichtman, J. W., and Sanes, J. R. (2013). Improved tools for the brainbow toolbox. Nat. Methods 10, 540–547. doi: 10.1038/nmeth.2450
Cane, M., Maco, B., Knott, G., and Holtmaat, A. (2014). The relationship between PSD-95 clustering and spine stability in vivo. J. Neurosci. 34, 2075–2086. doi: 10.1523/JNEUROSCI.3353-13.2014
Chan, K. Y., Jang, M. J., Yoo, B. B., Greenbaum, A., Ravi, N., Wu, W.-L., et al. (2017). Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 20, 1172–1179. doi: 10.1038/nn.4593
Chang, J.-B., Chen, F., Yoon, Y.-G., Jung, E. E., Babcock, H., Kang, J. S., et al. (2017). Iterative expansion microscopy. Nat. Methods 14, 593–599. doi: 10.1038/nmeth.4261
Chen, Y., Akin, O., Nern, A., Tsui, C. Y. K., Pecot, M. Y., and Zipursky, S. L. (2014). Cell-type specific labeling of synapses in vivo through synaptic tagging with recombination (STaR). Neuron 81, 280–293. doi: 10.1016/j.neuron.2013.12.021
Chen, X., Levy, J. M., Hou, A., Winters, C., Azzam, R., Sousa, A. A., et al. (2015). PSD-95 family MAGUKs are essential for anchoring AMPA and NMDA receptor complexes at the postsynaptic density. Proc. Natl. Acad. Sci. U S A 112, E6983–E6992. doi: 10.1073/pnas.1517045112
Chen, Y., Li, X., Zhang, D., Wang, C., Feng, R., Li, X., et al. (2020). A versatile tiling light sheet microscope for imaging of cleared tissues. Cell Rep. 33:108349. doi: 10.1016/j.celrep.2020.108349
Chen, X., Nelson, C. D., Li, X., Winters, C. A., Azzam, R., Sousa, A. A., et al. (2011). PSD-95 is required to sustain the molecular organization of the postsynaptic density. J. Neurosci. 31, 6329–6338. doi: 10.1523/JNEUROSCI.5968-10.2011
Chen, F., Tillberg, P. W., and Boyden, E. S. (2015). Expansion microscopy. Science 347, 543–548. doi: 10.1126/science.1260088
Chen, J. L., Villa, K. L., Cha, J. W., So, P. T. C., Kubota, Y., and Nedivi, E. (2012). Clustered dynamics of inhibitory synapses and dendritic spines in the adult neocortex. Neuron 74, 361–373. doi: 10.1016/j.neuron.2012.02.030
Chen, X., Vinade, L., Leapman, R. D., Petersen, J. D., Nakagawa, T., Phillips, T. M., et al. (2005). Mass of the postsynaptic density and enumeration of three key molecules. Proc. Natl. Acad. Sci. U S A 102, 11551–11556. doi: 10.1073/pnas.0505359102
Choi, S. W., Guan, W., and Chung, K. (2021). Basic principles of hydrogel-based tissue transformation technologies and their applications. Cell 184, 4115–4136. doi: 10.1016/j.cell.2021.07.009
Choi, J.-H., Sim, S.-E., Kim, J., Choi, D. I., Oh, J., Ye, S., et al. (2018). Interregional synaptic maps among engram cells underlie memory formation. Science 360, 430–435. doi: 10.1126/science.aas9204
Choii, G., and Ko, J. (2015). Gephyrin: a central GABAergic synapse organizer. Exp. Mol. Med. 47:e158. doi: 10.1038/emm.2015.5
Chozinski, T. J., Halpern, A. R., Okawa, H., Kim, H.-J., Tremel, G. J., Wong, R. O. L., et al. (2016). Expansion microscopy with conventional antibodies and fluorescent proteins. Nat. Methods 13, 485–488. doi: 10.1038/nmeth.3833
Cook, S. G., Goodell, D. J., Restrepo, S., Arnold, D. B., and Bayer, K. U. (2019). Simultaneous live imaging of multiple endogenous proteins reveals a mechanism for Alzheimer’s-related plasticity impairment. Cell Rep. 27, 658–665.e4. doi: 10.1016/j.celrep.2019.03.041
Cook, S. J., Jarrell, T. A., Brittin, C. A., Wang, Y., Bloniarz, A. E., Yakovlev, M. A., et al. (2019). Whole-animal connectomes of both Caenorhabditis elegans sexes. Nature 571, 63–71. doi: 10.1038/s41586-019-1352-7
Craig, A. M., Banker, G., Chang, W., McGrath, M. E., and Serpinskaya, A. S. (1996). Clustering of gephyrin at GABAergic but not glutamatergic synapses in cultured rat hippocampal neurons. J. Neurosci. 16, 3166–3177. doi: 10.1523/JNEUROSCI.16-10-03166.1996
Craven, S. E., El-Husseini, A. E., and Bredt, D. S. (1999). Synaptic targeting of the postsynaptic density protein PSD-95 mediated by lipid and protein motifs. Neuron 22, 497–509. doi: 10.1016/s0896-6273(00)80705-9
Damstra, H. G. J., Mohar, B., Eddison, M., Akhmanova, A., Kapitein, L. C., and Tillberg, P. W. (2021). Visualizing cellular and tissue ultrastructure using ten-fold robust expansion microscopy (TREx). bioRxiv [Preprint]. doi: 10.1101/2021.02.03.428837
De Robertis, E. D. P., and Bennett, H. S. (1955). Some features of the submicroscopic morphology of synapses in frog and earthworm. J. Biophys. Biochem. Cytol. 1, 47–58. doi: 10.1083/jcb.1.1.47
Dejanovic, B., Semtner, M., Ebert, S., Lamkemeyer, T., Neuser, F., Lüscher, B., et al. (2014). Palmitoylation of gephyrin controls receptor clustering and plasticity of GABAergic synapses. PLoS Biol. 12:e1001908. doi: 10.1371/journal.pbio.1001908
Desbois, M., Cook, S. J., Emmons, S. W., and Bülow, H. E. (2015). Directional trans-synaptic labeling of specific neuronal connections in live animals. Genetics 200, 697–705. doi: 10.1534/genetics.115.177006
Dhawale, A., and Bhalla, U. S. (2008). The network and the synapse: 100 years after cajal. HFSP J. 2, 12–16. doi: 10.2976/1.2835214
Dorkenwald, S., Turner, N. L., Macrina, T., Lee, K., Lu, R., Wu, J., et al. (2019). Binary and analog variation of synapses between cortical pyramidal neurons. bioRxiv [Preprint]. doi: 10.1101/2019.12.29.890319
Dosemeci, A., Weinberg, R. J., Reese, T. S., and Tao-Cheng, J.-H. (2016). The postsynaptic density: there is more than meets the eye. Front. Synaptic Neurosci. 8:23. doi: 10.3389/fnsyn.2016.00023
Drachman, D. A. (2005). Do we have brain to spare. Neurology 64, 2004–2005. doi: 10.1212/01.WNL.0000166914.38327.BB
El-Husseini, A. E.-D., Schnell, E., Chetkovich, D. M., Nicoll, R. A., and Bredt, D. S. (2000). PSD-95 involvement in maturation of excitatory synapses. Science 290, 1364–1368. doi: 10.1126/science.290.5495.1364
Fan, P., Manoli, D. S., Ahmed, O. M., Chen, Y., Agarwal, N., Kwong, S., et al. (2013). Genetic and neural mechanisms that inhibit Drosophila from mating with other species. Cell 154, 89–102. doi: 10.1016/j.cell.2013.06.008
Fang, H., Bygrave, A. M., Roth, R. H., Johnson, R. C., and Huganir, R. L. (2021). An optimized CRISPR/Cas9 approach for precise genome editing in neurons. eLife 10:e65202. doi: 10.7554/eLife.65202
Feinberg, E. H., VanHoven, M. K., Bendesky, A., Wang, G., Fetter, R. D., Shen, K., et al. (2008). GFP reconstitution across synaptic partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron 57, 353–363. doi: 10.1016/j.neuron.2007.11.030
Fortin, D. A., Tillo, S. E., Yang, G., Rah, J.-C., Melander, J. B., Bai, S., et al. (2014). live imaging of endogenous PSD-95 using ENABLED: a conditional strategy to fluorescently label endogenous proteins. J. Neurosci. 34, 16698–16712. doi: 10.1523/JNEUROSCI.3888-14.2014
Freifeld, L., Odstrcil, I., Förster, D., Ramirez, A., Gagnon, J. A., Randlett, O., et al. (2017). Expansion microscopy of zebrafish for neuroscience and developmental biology studies. Proc. Natl. Acad. Sci. U S A 114, E10799–E10808. doi: 10.1073/pnas.1706281114
Furshpan, E. J., and Potter, D. D. (1959). Transmission at the giant motor synapses of the crayfish. J. Physiol. 145, 289–325. doi: 10.1113/jphysiol.1959.sp006143
Galarreta, M., and Hestrin, S. (2001). Electrical synapses between Gaba-Releasing interneurons. Nat. Rev. Neurosci. 2, 425–433. doi: 10.1038/35077566
Gao, R., Asano, S. M., Upadhyayula, S., Pisarev, I., Milkie, D. E., Liu, T.-L., et al. (2019). Cortical column and whole-brain imaging with molecular contrast and nanoscale resolution. Science 363:eaau8302. doi: 10.1126/science.aau8302
Gao, Y., Hisey, E., Bradshaw, T. W. A., Erata, E., Brown, W. E., Courtland, J. L., et al. (2019). Plug-and-play protein modification using homology-independent universal genome engineering. Neuron 103, 583–597.e8. doi: 10.1016/j.neuron.2019.05.047
Gao, M., Maraspini, R., Beutel, O., Zehtabian, A., Eickholt, B., Honigmann, A., et al. (2018). Expansion stimulated emission depletion microscopy (ExSTED). ACS Nano 12, 4178–4185. doi: 10.1021/acsnano.8b00776
Gao, R., Yu, C.-C., Gao, L., Piatkevich, K. D., Neve, R. L., Munro, J. B., et al. (2021). A highly homogeneous polymer composed of tetrahedron-like monomers for high-isotropy expansion microscopy. Nat. Nanotechnol. 16, 698–707. doi: 10.1038/s41565-021-00875-7
Gibson, J. R., Beierlein, M., and Connors, B. W. (1999). Two networks of electrically coupled inhibitory neurons in neocortex. Nature 402, 75–79. doi: 10.1038/47035
Gong, H., Xu, D., Yuan, J., Li, X., Guo, C., Peng, J., et al. (2016). High-throughput dual-colour precision imaging for brain-wide connectome with cytoarchitectonic landmarks at the cellular level. Nat. Commun. 7:12142. doi: 10.1038/ncomms12142
Gordon, M. D., and Scott, K. (2009). Motor control in a Drosophila taste circuit. Neuron 61, 373–384. doi: 10.1016/j.neuron.2008.12.033
Gray, E. G. (1959). Electron microscopy of synaptic contacts on dendrite spines of the cerebral cortex. Nature 183, 1592–1593. doi: 10.1038/1831592a0
Gray, N. W., Weimer, R. M., Bureau, I., and Svoboda, K. (2006). Rapid redistribution of synaptic PSD-95 in the neocortex in vivo. PLoS Biol. 4:e370. doi: 10.1371/journal.pbio.0040370
Gross, G. G., Junge, J. A., Mora, R. J., Kwon, H.-B., Olson, C. A., Takahashi, T. T., et al. (2013). Recombinant probes for visualizing endogenous synaptic proteins in living neurons. Neuron 78, 971–985. doi: 10.1016/j.neuron.2013.04.017
Gutierrez-Mecinas, M., Kuehn, E. D., Abraira, V. E., Polgár, E., Watanabe, M., and Todd, A. J. (2016). Immunostaining for Homer reveals the majority of excitatory synapses in laminae I-III of the mouse spinal dorsal horn. Neuroscience 329, 171–181. doi: 10.1016/j.neuroscience.2016.05.009
Hafner, A.-S., Donlin-Asp, P. G., Leitch, B., Herzog, E., and Schuman, E. M. (2019). Local protein synthesis is a ubiquitous feature of neuronal pre- and postsynaptic compartments. Science 364:eaau3644. doi: 10.1126/science.aau3644
Hall, D. H., and Russell, R. L. (1991). The posterior nervous system of the nematode Caenorhabditis elegans: serial reconstruction of identified neurons and complete pattern of synaptic interactions. J. Neurosci. 11, 1–22. doi: 10.1523/JNEUROSCI.11-01-00001.1991
Hama, H., Kurokawa, H., Kawano, H., Ando, R., Shimogori, T., Noda, H., et al. (2011). Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat. Neurosci. 14, 1481–1488. doi: 10.1038/nn.2928
Hasani, R., Lechner, M., Amini, A., Rus, D., and Grosu, R. (2020). Liquid time-constant networks. arXiv [Preprint]. Available online at: http://arxiv.org/abs/2006.04439. Accessed March 13, 2021.
Helm, M. S., Dankovich, T. M., Mandad, S., Rammner, B., Jähne, S., Salimi, V., et al. (2021). A large-scale nanoscopy and biochemistry analysis of postsynaptic dendritic spines. Nat. Neurosci. 24, 1151–1162. doi: 10.1038/s41593-021-00874-w
Hormuzdi, S. G., Filippov, M. A., Mitropoulou, G., Monyer, H., and Bruzzone, R. (2004). Electrical synapses: a dynamic signaling system that shapes the activity of neuronal networks. Biochim. Biophys. Acta 1662, 113–137. doi: 10.1016/j.bbamem.2003.10.023
Hu, Y., Chu, X., Chen, T., Pan, Q., Liu, C., Yi, J., et al. (2020). Improving resolving ability of expansion microscopy by varying crosslinker concentration. Chem. Commun. 56, 4176–4179. doi: 10.1039/d0cc00052c
Hubbard, P. M., Berg, S., Zhao, T., Olbris, D. J., Umayam, L., Maitin-Shepard, J., et al. (2020). Accelerated EM connectome reconstruction using 3D visualization and segmentation graphs. bioRxiv [Preprint]. doi: 10.1101/2020.01.17.909572
Hulse, B. K., Haberkern, H., Franconville, R., Turner-Evans, D. B., Takemura, S., Wolff, T., et al. (2021). A connectome of the Drosophila central complex reveals network motifs suitable for flexible navigation and context-dependent action selection. eLife 10:e66039. doi: 10.7554/eLife.66039
Isshiki, M., Tanaka, S., Kuriu, T., Tabuchi, K., Takumi, T., and Okabe, S. (2014). Enhanced synapse remodelling as a common phenotype in mouse models of autism. Nat. Commun. 5:4742. doi: 10.1038/ncomms5742
Jarrell, T. A., Wang, Y., Bloniarz, A. E., Brittin, C. A., Xu, M., Thomson, J. N., et al. (2012). The connectome of a decision-making neural network. Science 337, 437–444. doi: 10.1126/science.1221762
Jiang, N., Kim, H.-J., Chozinski, T. J., Azpurua, J. E., Eaton, B. A., Vaughan, J. C., et al. (2018). Superresolution imaging of Drosophila tissues using expansion microscopy. Mol. Biol. Cell 29, 1413–1421. doi: 10.1091/mbc.E17-10-0583
Karagiannis, E. D., and Boyden, E. S. (2018). Expansion microscopy: development and neuroscience applications. Curr. Opin. Neurobiol. 50, 56–63. doi: 10.1016/j.conb.2017.12.012
Kasthuri, N., Hayworth, K. J., Berger, D. R., Schalek, R. L., Conchello, J. A., Knowles-Barley, S., et al. (2015). Saturated reconstruction of a volume of neocortex. Cell 162, 648–661. doi: 10.1016/j.cell.2015.06.054
Kim, S.-Y., Cho, J. H., Murray, E., Bakh, N., Choi, H., Ohn, K., et al. (2015). Stochastic electrotransport selectively enhances the transport of highly electromobile molecules. Proc. Natl. Acad. Sci. U S A 112, E6274–E6283. doi: 10.1073/pnas.1510133112
Kim, B., and Emmons, S. W. (2017). Multiple conserved cell adhesion protein interactions mediate neural wiring of a sensory circuit in C. elegans. eLife 6:e29257. doi: 10.7554/eLife.29257
Kim, E. J., Jacobs, M. W., Ito-Cole, T., and Callaway, E. M. (2016). Improved monosynaptic neural circuit tracing using engineered rabies virus glycoproteins. Cell Rep. 15, 692–699. doi: 10.1016/j.celrep.2016.03.067
Kim, J., Zhao, T., Petralia, R. S., Yu, Y., Peng, H., Myers, E., et al. (2012). mGRASP enables mapping mammalian synaptic connectivity with light microscopy. Nat. Methods 9, 96–102. doi: 10.1038/nmeth.1784
Ku, T., Swaney, J., Park, J.-Y., Albanese, A., Murray, E., Cho, J. H., et al. (2016). Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nat. Biotechnol. 34, 973–981. doi: 10.1038/nbt.3641
Kuljis, D. A., Park, E., Telmer, C. A., Lee, J., Ackerman, D. S., Bruchez, M. P., et al. (2019). Fluorescence-based quantitative synapse analysis for cell type-specific connectomics. eNeuro 6:ENEURO.0193-19.2019. doi: 10.1523/ENEURO.0193-19.2019
Li, R., Chen, X., Lin, Z., Wang, Y., and Sun, Y. (2018). Expansion enhanced nanoscopy. Nanoscale 10, 17552–17556. doi: 10.1039/c8nr04267e
Li, Y., Guo, A., and Li, H. (2016). CRASP: CFP reconstitution across synaptic partners. Biochem. Biophys. Res. Commun. 469, 352–356. doi: 10.1016/j.bbrc.2015.12.011
Li, L., Tasic, B., Micheva, K. D., Ivanov, V. M., Spletter, M. L., Smith, S. J., et al. (2010). Visualizing the distribution of synapses from individual neurons in the mouse brain. PLoS One 5:e11503. doi: 10.1371/journal.pone.0011503
Li, Y., Walker, L. A., Zhao, Y., Edwards, E. M., Michki, N. S., Cheng, H. P. J., et al. (2021). Bitbow enables highly efficient neuronal lineage tracing and morphology reconstruction in single Drosophila brains. Front. Neural Circuits 15:105. doi: 10.3389/fncir.2021.732183
Lillvis, J. L., Otsuna, H., Ding, X., Pisarev, I., Kawase, T., Colonell, J., et al. (2021). Rapid reconstruction of neural circuits using tissue expansion and lattice light sheet microscopy. bioRxiv [Preprint]. doi: 10.1101/2021.11.14.468535
Livet, J., Weissman, T. A., Kang, H., Draft, R. W., Lu, J., Bennis, R. A., et al. (2007). Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450, 56–62. doi: 10.1038/nature06293
Llinás, R. R. (2003). The contribution of santiago ramon y cajal to functional neuroscience. Nat. Rev. Neurosci. 4, 77–80. doi: 10.1038/nrn1011
Macpherson, L. J., Zaharieva, E. E., Kearney, P. J., Alpert, M. H., Lin, T.-Y., Turan, Z., et al. (2015). Dynamic labelling of neural connections in multiple colours by trans-synaptic fluorescence complementation. Nat. Commun. 6:10024. doi: 10.1038/ncomms10024
Magaki, S., Hojat, S. A., Wei, B., So, A., and Yong, W. H. (2019). “An introduction to the performance of immunohistochemistry,” in Biobanking: Methods and Protocols Methods in Molecular Biology, eds W. H. Yong (New York, NY: Springer), 289–298.
Meier, J., and Grantyn, R. (2004). A gephyrin-related mechanism restraining glycine receptor anchoring at GABAergic synapses. J. Neurosci. 24, 1398–1405. doi: 10.1523/JNEUROSCI.4260-03.2004
Meyer, M. P., and Smith, S. J. (2006). Evidence from in vivo imaging that synaptogenesis guides the growth and branching of axonal arbors by two distinct mechanisms. J. Neurosci. 26, 3604–3614. doi: 10.1523/JNEUROSCI.0223-06.2006
Micheva, K. D., Busse, B., Weiler, N. C., O’Rourke, N., and Smith, S. J. (2010). Single-synapse analysis of a diverse synapse population: proteomic imaging methods and markers. Neuron 68, 639–653. doi: 10.1016/j.neuron.2010.09.024
MICrONS Consortium, M., Bae, J. A., Baptiste, M., Bodor, A. L., Brittain, D., Buchanan, J., et al. (2021). Functional connectomics spanning multiple areas of mouse visual cortex. bioRxiv [Preprint]. doi: 10.1101/2021.07.28.454025
Mikuni, T., Nishiyama, J., Sun, Y., Kamasawa, N., and Yasuda, R. (2016). High-throughput, high-resolution mapping of protein localization in mammalian brain by in vivo genome editing. Cell 165, 1803–1817. doi: 10.1016/j.cell.2016.04.044
Moeyaert, B., Holt, G., Madangopal, R., Perez-Alvarez, A., Fearey, B. C., Trojanowski, N. F., et al. (2018). Improved methods for marking active neuron populations. Nat. Commun. 9:4440. doi: 10.1038/s41467-018-06935-2
Mosca, T. J., Luginbuhl, D. J., Wang, I. E., and Luo, L. (2017). Presynaptic LRP4 promotes synapse number and function of excitatory CNS neurons. eLife 6:e27347. doi: 10.7554/eLife.27347
Motta, A., Berning, M., Boergens, K. M., Staffler, B., Beining, M., Loomba, S., et al. (2019). Dense connectomic reconstruction in layer 4 of the somatosensory cortex. Science 366:eaay3134. doi: 10.1126/science.aay3134
Murakami, T. C., Mano, T., Saikawa, S., Horiguchi, S. A., Shigeta, D., Baba, K., et al. (2018). A three-dimensional single-cell-resolution whole-brain atlas using CUBIC-X expansion microscopy and tissue clearing. Nat. Neurosci. 21, 625–637. doi: 10.1038/s41593-018-0109-1
Niell, C. M., Meyer, M. P., and Smith, S. J. (2004). in vivo imaging of synapse formation on a growing dendritic arbor. Nat. Neurosci. 7, 254–260. doi: 10.1038/nn1191
Nishiyama, J., Mikuni, T., and Yasuda, R. (2017). Virus-mediated genome editing via homology-directed repair in mitotic and postmitotic cells in mammalian brain. Neuron 96, 755–768.e5. doi: 10.1016/j.neuron.2017.10.004
Oh, W. C., Lutzu, S., Castillo, P. E., and Kwon, H.-B. (2016). De novo synaptogenesis induced by GABA in the developing mouse cortex. Science 353, 1037–1040. doi: 10.1126/science.aaf5206
Ohyama, T., Schneider-Mizell, C. M., Fetter, R. D., Aleman, J. V., Franconville, R., Rivera-Alba, M., et al. (2015). A multilevel multimodal circuit enhances action selection in Drosophila. Nature 520, 633–639. doi: 10.1038/nature14297
O’Rourke, N. A., Weiler, N. C., Micheva, K. D., and Smith, S. J. (2012). Deep molecular diversity of mammalian synapses: why it matters and how to measure it. Nat. Rev. Neurosci. 13, 365–379. doi: 10.1038/nrn3170
Palay, S. L. (1956). Synapses in the central nervous system. J. Biophys. Biochem. Cytol. 2, 193–202. doi: 10.1083/jcb.2.4.193
Palay, S. L., and Palade, G. E. (1955). The fine structure of neurons. J. Biophys. Biochem. Cytol. 1, 69–88. doi: 10.1083/jcb.1.1.69
Park, C. E., Cho, Y., Cho, I., Jung, H., Kim, B., Shin, J. H., et al. (2020). Super-resolution three-dimensional imaging of actin filaments in cultured cells and the brain via expansion microscopy. ACS Nano 14, 14999–15010. doi: 10.1021/acsnano.0c04915
Park, H.-E., Choi, D., Park, J. S., Sim, C., Park, S., Kang, S., et al. (2019). Scalable and isotropic expansion of tissues with simply tunable expansion ratio. Adv. Sci. (Weinh) 6:1901673. doi: 10.1002/advs.201901673
Park, J., Khan, S., Yun, D. H., Ku, T., Villa, K. L., Lee, J. E., et al (2021). Epitope-preserving magnified analysis of proteome (eMAP). Sci. Adv. 7: eabf6589. doi: 10.1126/sciadv.abf6589
Parra-Damas, A., and Saura, C. A. (2020). Tissue clearing and expansion methods for imaging brain pathology in neurodegeneration: from circuits to synapses and beyond. Front. Neurosci. 14:914. doi: 10.3389/fnins.2020.00914
Payne, A. C., Chiang, Z. D., Reginato, P. L., Mangiameli, S. M., Murray, E. M., Yao, C.-C., et al. (2021). in situ genome sequencing resolves DNA sequence and structure in intact biological samples. Science 371:eaay3446. doi: 10.1126/science.aay3446
Pereda, A. E. (2014). Electrical synapses and their functional interactions with chemical synapses. Nat. Rev. Neurosci. 15, 250–263. doi: 10.1038/nrn3708
Perez-Alvarez, A., Fearey, B. C., O’Toole, R. J., Yang, W., Arganda-Carreras, I., Lamothe-Molina, P. J., et al. (2020). Freeze-frame imaging of synaptic activity using SynTagMA. Nat. Commun. 11:2464. doi: 10.1038/s41467-020-16315-4
Philbrook, A., Ramachandran, S., Lambert, C. M., Oliver, D., Florman, J., Alkema, M. J., et al. (2018). Neurexin directs partner-specific synaptic connectivity in C. elegans. eLife 7:e35692. doi: 10.7554/eLife.35692
Prange, O., Wong, T. P., Gerrow, K., Wang, Y. T., and El-Husseini, A. (2004). A balance between excitatory and inhibitory synapses is controlled by PSD-95 and neuroligin. Proc. Natl. Acad. Sci. U S A 101, 13915–13920. doi: 10.1073/pnas.0405939101
Ramón y Cajal, S. (1852–1934) A. du texte (1909). Histologie du système nerveux de l’homme et des vertébrés. Cervelet, cerveau moyen, rétine, couche optique, corps strié, écorce cérébrale générale et régionale, grand sympathique / par S. Ramon Cajal. Available online at: https://gallica.bnf.fr/ark:/12148/bpt6k6213192g. Accessed February 25, 2021.
Robertson, J. D. (1953). Ultrastructure of two invertebrate synapses. Proc. Soc. Exp. Biol. Med. 82, 219–223. doi: 10.3181/00379727-82-20071
Ryan, K., Lu, Z., and Meinertzhagen, I. A. (2016). The CNS connectome of a tadpole larva of Ciona intestinalis (L.) highlights sidedness in the brain of a chordate sibling. eLife 5:e16962. doi: 10.7554/eLife.16962
Sahl, S. J., Hell, S. W., and Jakobs, S. (2017). Fluorescence nanoscopy in cell biology. Nat. Rev. Mol. Cell Biol. 18, 685–701. doi: 10.1038/nrm.2017.71
Sakaguchi, R., Leiwe, M. N., and Imai, T. (2018). Bright multicolor labeling of neuronal circuits with fluorescent proteins and chemical tags. eLife 7:e40350. doi: 10.7554/eLife.40350
Sarkar, D., Kang, J., Wassie, A. T., Schroeder, M. E., Peng, Z., Tarr, T. B., et al. (2020). Expansion revealing: decrowding proteins to unmask invisible brain nanostructures. bioRxiv [Preprint]. 10.1101/2020.08.29.273540.
Scaplen, K. M., Talay, M., Fisher, J. D., Cohn, R., Sorkaç, A., Aso, Y., et al. (2021). Transsynaptic mapping of Drosophila mushroom body output neurons. eLife 10:e63379. doi: 10.7554/eLife.63379
Scheffer, L. K., Xu, C. S., Januszewski, M., Lu, Z., Takemura, S., Hayworth, K. J., et al. (2020). A connectome and analysis of the adult Drosophila central brain. eLife 9:e57443. doi: 10.7554/eLife.57443
Schermelleh, L., Ferrand, A., Huser, T., Eggeling, C., Sauer, M., Biehlmaier, O., et al. (2019). Super-resolution microscopy demystified. Nat. Cell Biol. 21, 72–84. doi: 10.1038/s41556-018-0251-8
Schneider-Mizell, C. M., Bodor, A. L., Collman, F., Brittain, D., Bleckert, A. A., Dorkenwald, S., et al. (2020). Chandelier cell anatomy and function reveal a variably distributed but common signal. bioRxiv [Preprint]. doi: 10.1101/2020.03.31.018952
Seiriki, K., Kasai, A., Hashimoto, T., Schulze, W., Niu, M., Yamaguchi, S., et al. (2017). High-speed and scalable whole-brain imaging in rodents and primates. Neuron 94, 1085–1100.e6. doi: 10.1016/j.neuron.2017.05.017
Seo, J., Sim, Y., Kim, J., Kim, H., Cho, I., Yoon, Y.-G., et al. (2021). PICASSO: ultra-multiplexed fluorescence imaging of biomolecules through single-round imaging and blind source unmixing. bioRxiv [Preprint]. doi: 10.1101/2021.01.27.428247
Shapson-Coe, A., Januszewski, M., Berger, D. R., Pope, A., Wu, Y., Blakely, T., et al. (2021). A connectomic study of a petascale fragment of human cerebral cortex. bioRxiv [Preprint]. doi: 10.1101/2021.05.29.446289
Shearin, H. K., Quinn, C. D., Mackin, R. D., Macdonald, I. S., and Stowers, R. S. (2018). t-GRASP, a targeted GRASP for assessing neuronal connectivity. J. Neurosci. Methods 306, 94–102. doi: 10.1016/j.jneumeth.2018.05.014
Shen, F. Y., Harrington, M. M., Walker, L. A., Cheng, H. P. J., Boyden, E. S., and Cai, D. (2020). Light microscopy based approach for mapping connectivity with molecular specificity. Nat. Commun. 11:4632. doi: 10.1038/s41467-020-18422-8
Sheng, M., and Kim, E. (2011). The postsynaptic organization of synapses. Cold Spring Harb. Perspect. Biol. 3:a005678. doi: 10.1101/cshperspect.a005678
Shi, X., Li, Q., Dai, Z., Tran, A. A., Feng, S., Ramirez, A. D., et al. (2021). Label-retention expansion microscopy. J. Cell Biol. 220:e202105067. doi: 10.1083/jcb.202105067
Sim, J., Park, C. E., Cho, I., Min, K., Lee, J.-S., Chong, Y., et al. (2021). Whole-ExM: expansion microscopy imaging of all anatomical structures of whole larval zebrafish. bioRxiv [Preprint]. doi: 10.1101/2021.05.18.443629
Son, J.-H., Keefe, M. D., Stevenson, T. J., Barrios, J. P., Anjewierden, S., Newton, J. B., et al. (2016). Transgenic FingRs for live mapping of synaptic dynamics in genetically-defined neurons. Sci. Rep. 6:18734. doi: 10.1038/srep18734
Song, J. H., Lucaci, D., Calangiu, I., Brown, M. T. C., Park, J. S., Kim, J., et al. (2018). Combining mGRASP and optogenetics enables high-resolution functional mapping of descending cortical projections. Cell Rep. 24, 1071–1080. doi: 10.1016/j.celrep.2018.06.076
Subramanian, J., Michel, K., Benoit, M., and Nedivi, E. (2019). CPG15/neuritin mimics experience in selecting excitatory synapses for stabilization by facilitating PSD95 recruitment. Cell Rep. 28, 1584–1595.e5. doi: 10.1016/j.celrep.2019.07.012
Suzuki, K., Tsunekawa, Y., Hernandez-Benitez, R., Wu, J., Zhu, J., Kim, E. J., et al. (2016). In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 540, 144–149. doi: 10.1038/nature20565
Taft, C. E., and Turrigiano, G. G. (2014). PSD-95 promotes the stabilization of young synaptic contacts. Philos. Trans. R. Soc. B Biol. Sci. 369:20130134. doi: 10.1098/rstb.2013.0134
Talay, M., Richman, E. B., Snell, N. J., Hartmann, G. G., Fisher, J. D., Sorkaç, A., et al. (2017). Transsynaptic mapping of second-order taste neurons in flies by trans-tango. Neuron 96, 783–795.e4. doi: 10.1016/j.neuron.2017.10.011
Tang, Y., Nyengaard, J. R., De Groot, D. M., and Gundersen, H. J. (2001). Total regional and global number of synapses in the human brain neocortex. Synapse 41, 258–273. doi: 10.1002/syn.1083
Tervo, D. G. R., Hwang, B.-Y., Viswanathan, S., Gaj, T., Lavzin, M., Ritola, K. D., et al. (2016). A designer AAV variant permits efficient retrograde access to projection neurons. Neuron 92, 372–382. doi: 10.1016/j.neuron.2016.09.021
Tillberg, P. W., Chen, F., Piatkevich, K. D., Zhao, Y., Yu, C.-C. J., English, B. P., et al (2016). Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nat. Biotechnol. 34, 987–992. doi: 10.1038/nbt.3625
Truckenbrodt, S., Maidorn, M., Crzan, D., Wildhagen, H., Kabatas, S., and Rizzoli, S. O. (2018). X10 expansion microscopy enables 25-nm resolution on conventional microscopes. EMBO Rep. 19:e45836. doi: 10.15252/embr.201845836
Turner, N. L., Macrina, T., Bae, J. A., Yang, R., Wilson, A. M., Schneider-Mizell, C., et al. (2020). Multiscale and multimodal reconstruction of cortical structure and function. bioRxiv [Preprint]. doi: 10.1101/2020.10.14.338681
Tyagarajan, S. K., and Fritschy, J.-M. (2014). Gephyrin: a master regulator of neuronal function. Nat. Rev. Neurosci. 15, 141–156. doi: 10.1038/nrn3670
Uezu, A., Kanak, D. J., Bradshaw, T. W. A., Soderblom, E. J., Catavero, C. M., Burette, A. C., et al. (2016). Identification of an elaborate complex mediating postsynaptic inhibition. Science 353, 1123–1129. doi: 10.1126/science.aag0821
Ugolini, G. (2011). “Chapter 10 - Rabies Virus as a Transneuronal Tracer of Neuronal Connections,” in Advances in Virus Research Research Advances in Rabies, ed A. C. Jackson (New York: Academic Press), 165–202.
Verasztó, C., Jasek, S., Gühmann, M., Shahidi, R., Ueda, N., Beard, J. D., et al. (2020). Whole-animal connectome and cell-type complement of the three-segmented Platynereis dumerilii larva. bioRxiv [Preprint]. doi: 10.1101/2020.08.21.260984
Villa, K. L., Berry, K. P., Subramanian, J., Cha, J. W., Oh, W. C., Kwon, H.-B., et al. (2016). Inhibitory synapses are repeatedly assembled and removed at persistent sites in vivo. Neuron 89, 756–769. doi: 10.1016/j.neuron.2016.01.010
Viswanathan, S., Williams, M. E., Bloss, E. B., Stasevich, T. J., Speer, C. M., Nern, A., et al. (2015). High-performance probes for light and electron microscopy. Nat. Methods 12, 568–576. doi: 10.1038/nmeth.3365
Vlachos, A., Reddy-Alla, S., Papadopoulos, T., Deller, T., and Betz, H. (2013). Homeostatic regulation of gephyrin scaffolds and synaptic strength at mature hippocampal gabaergic postsynapses†. Cereb. Cortex 23, 2700–2711. doi: 10.1093/cercor/bhs260
Wang, Y., Hu, Z., Ju, P., Yin, S., Wang, F., Pan, O., et al. (2018). Viral vectors as a novel tool for clinical and neuropsychiatric research applications. Gen. Psychiatry 31:e000015. doi: 10.1136/gpsych-2018-000015
Wassie, A. T., Zhao, Y., and Boyden, E. S. (2019). Expansion microscopy: principles and uses in biological research. Nat. Methods 16, 33–41. doi: 10.1038/s41592-018-0219-4
Watanabe, A. (1958). The interaction of electrical activity among neurons of lobster cardiac ganglion. Jpn. J. Physiol. 8, 305–318. doi: 10.2170/jjphysiol.8.305
Weissman, T. A., and Pan, Y. A. (2015). Brainbow: new resources and emerging biological applications for multicolor genetic labeling and analysis. Genetics 199, 293–306. doi: 10.1534/genetics.114.172510
Wells, W. A. (2005). The discovery of synaptic vesicles. J. Cell Biol. 168, 12–13. doi: 10.1083/jcb1681fta2
White, J. G., Southgate, E., Thomson, J. N., and Brenner, S. (1986). The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 314, 1–340. doi: 10.1098/rstb.1986.0056
Wiedenmann, B., and Franke, W. W. (1985). Identification and localization of synaptophysin, an integral membrane glycoprotein of Mr 38,000 characteristic of presynaptic vesicles. Cell 41, 1017–1028. doi: 10.1016/s0092-8674(85)80082-9
Willems, J., Jong, A. P. H. D., Scheefhals, N., Mertens, E., Catsburg, L. A. E., Poorthuis, R. B., et al. (2020). ORANGE: a CRISPR/Cas9-based genome editing toolbox for epitope tagging of endogenous proteins in neurons. PLoS Biol. 18:e3000665. doi: 10.1371/journal.pbio.3000665
Winnubst, J., Bas, E., Ferreira, T. A., Wu, Z., Economo, M. N., Edson, P., et al. (2019). Reconstruction of 1,000 projection neurons reveals new cell types and organization of long-range connectivity in the mouse brain. Cell 179, 268–281.e13. doi: 10.1016/j.cell.2019.07.042
Witvliet, D., Mulcahy, B., Mitchell, J. K., Meirovitch, Y., Berger, D. R., Wu, Y., et al. (2020). Connectomes across development reveal principles of brain maturation in C. elegans. Nature 596, 257–261. doi: 10.1038/s41586-021-03778-8
Xu, C. S., Hayworth, K. J., Lu, Z., Grob, P., Hassan, A. M., García-Cerdán, J. G., et al. (2017). Enhanced FIB-SEM systems for large-volume 3D imaging. eLife 6:e25916. doi: 10.7554/eLife.25916
Xu, H., Tong, Z., Ye, Q., Sun, T., Hong, Z., Zhang, L., et al. (2019). Molecular organization of mammalian meiotic chromosome axis revealed by expansion STORM microscopy. Proc. Natl. Acad. Sci. U S A 116, 18423–18428. doi: 10.1073/pnas.1902440116
Yeh, E., Kawano, T., Weimer, R. M., Bessereau, J.-L., and Zhen, M. (2005). Identification of genes involved in synaptogenesis using a fluorescent active zone marker in Caenorhabditis elegans. J. Neurosci. 25, 3833–3841. doi: 10.1523/JNEUROSCI.4978-04.2005
Yu, C.-C. J., Barry, N. C., Wassie, A. T., Sinha, A., Bhattacharya, A., Asano, S., et al. (2020). Expansion microscopy of C. elegans. eLife 9:e46249. doi: 10.7554/eLife.46249
Zhao, Y., Bucur, O., Irshad, H., Chen, F., Weins, A., Stancu, A. L., et al. (2017). Nanoscale imaging of clinical specimens using pathology-optimized expansion microscopy. Nat. Biotechnol. 35, 757–764. doi: 10.1038/nbt.3892
Zhong, Q., Li, A., Jin, R., Zhang, D., Li, X., Jia, X., et al. (2021). High-definition imaging using line-illumination modulation microscopy. Nat. Methods 18, 309–315. doi: 10.1038/s41592-021-01074-x
Keywords: expansion microscopy, optical connectome, super-resolution, electron miscroscopy, synaptic labeling
Citation: Sneve MA and Piatkevich KD (2022) Towards a Comprehensive Optical Connectome at Single Synapse Resolution via Expansion Microscopy. Front. Synaptic Neurosci. 13:754814. doi: 10.3389/fnsyn.2021.754814
Received: 07 August 2021; Accepted: 17 December 2021;
Published: 18 January 2022.
Edited by:
Thomas A. Blanpied, University of Maryland, United StatesReviewed by:
Kristina D. Micheva, Stanford University, United StatesJoachim H. R. Lübke, Helmholtz Association of German Research Centres (HZ), Germany
Copyright © 2022 Sneve and Piatkevich. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kiryl D. Piatkevich, a2lyeWwucGlhdGtldmljaEB3ZXN0bGFrZS5lZHUuY24=
 Madison A. Sneve
Madison A. Sneve Kiryl D. Piatkevich
Kiryl D. Piatkevich