- 1Genetics Department, Faculty of Agriculture, Zagazig University, Zagazig, Egypt
- 2Department of Botany, Hansraj College, University of Delhi, Delhi, India
- 3ICAR-National Institute of Biotic Stress Management, Raipur, India
- 4Animal Production Department, Faculty of Agriculture, Zagazig University, Zagazig, Egypt
- 5Biochemistry Department, Faculty of Agriculture, Zagazig University, Zagazig, Egypt
- 6Horticulture Department, Faculty of Agriculture, Zagazig University, Zagazig, Egypt
- 7Agricultural Research Center, ARC, Giza, Egypt
- 8Poultry Department, Faculty of Agriculture, Zagazig University, Zagazig, Egypt
- 9College of Liberal Arts and Sciences, University of Florida, Gainesville, FL, United States
Food security is the situation in which all people around the globe, in all circumstances must maintain constant physical and financial access to adequate nutritious, clean, and safe food. This is why food security is regarded as a major global issue. Domestically and internationally, many factors affect food production and quality; these include environmental climate change conditions, disasters, emerging new diseases, political issues, and shifts in the global economy. Additionally, rapid population growth necessitates adjustments to food and crop production. The current agricultural production strategies negatively affect soil fertility and cause many defects such as desertification, deforestation, and increased soil salinity. Additionally, the high use of chemical fertilizers and pesticides affects human health and food production. The production of numerous strategic crops, including rice, maize, soybeans, and wheat, was reduced due to the aforementioned factors. The global hunger index increased, and millions of people were placed in the food insecurity red zone. Governments and societies attempted to address the problem by creating a new one: a higher yield of low-quality main food crops, which can give rise to hidden hunger, a serious issue that violates people’s rights to a healthy and nutritious diet and inevitably causes food insecurity. In an attempt to end hunger, the Green Revolution produced high-yielding varieties of crops; however, this came at the expense of massive amounts of chemical pesticides, fertilizers, and other agricultural activities and practices that ignored the negative effects on the environment and nutritional value. Yet, billions of people worldwide suffer from iron, zinc, iodine, and vitamin A deficiency, making hidden hunger, malnutrition, and micronutrient deficiencies a severe and widespread nutritional issue in developing nations. The most common nutritional condition affecting poor and rural populations, especially those in low-income developing countries, is iron and zinc deficiency. To decrease and close the gap between crop production and food consumption, there is a need to increase agricultural productivity. Here we discussed the current status of food insecurity and malnutrition status where many research efforts and developments to enhance food quality and production for food crops with the help of genome editing tools and applications for sustainable food production.
1 Introduction
The world economy continues to suffer from the COVID-19 pandemic and the most recent conflict in Ukraine (2022), which has raised the cost of fuel such as Gasoline followed by an increase in food prices (Figure 1). Greater than the level observed before the COVID-19 pandemic (Figure 2). With real issues like climate change, problems with food and chain supplements, and many other serious issues. The situation is utterly unstable and puts global food security at risk. Hidden hunger or malnutrition is a serious problem that violates people’s right to a healthy and nutritious diet and exacerbates food insecurity. United Nations defined food security as “a lack of consistent access to food which diminishes dietary quality, disrupts normal eating patterns and can have negative consequences for nutrition, health and wellbeing.” According to the global report on food crises (GRFC), more than 193 million people worldwide will be faced with food insecurity (Report on food crises, 2022). The United Nations World Food Program (UNWFP) predicts that the percentage of the population who face hunger will rise by 17% shortly and the percentage is likely to be increased so further. Long- and short-term insecurity implications can affect life and threaten the food security of millions of people worldwide. Low and no economic resources in developing countries which considered as another issue especially since the resources get exhausted during the COVID-19 pandemic time (Figure 3). Conflicts extreme weather change conditions and disease pandemics around the world force more than 139 million people to deal with food insecurity and nutrition crisis problems. Osendarp et al. (2021) stated that the number of children who will be wasted is around 9.3 million, the death will be raised to 168 thousand, and almost 2.6 million children will be stunted by 2022 (Figures 4, 5).
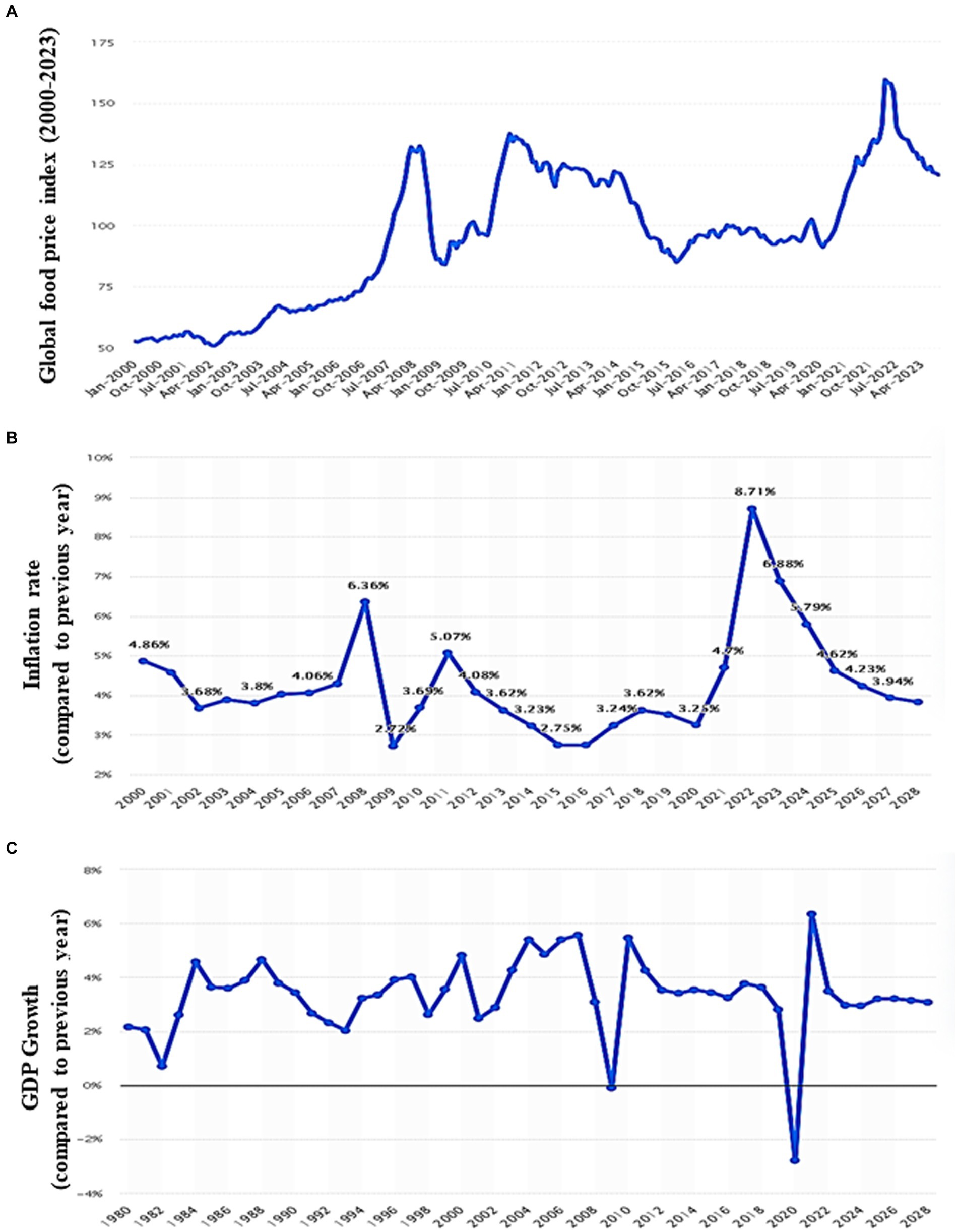
Figure 1. Global food price, inflation rate, and gross domestic product charts. (A) Monthly food price index worldwide from 2000 to 2023 (including meat, dairy, cereals, vegetable oils, and sugar). (B) Global inflation rate from 2000 to 2022, with forecasts until 2028. (C) Growth of the global gross domestic product (GDP) from 1980 to 2022, with forecasts until 2028 (compared to the previous year; Source: FAO; Statistica, 2024).
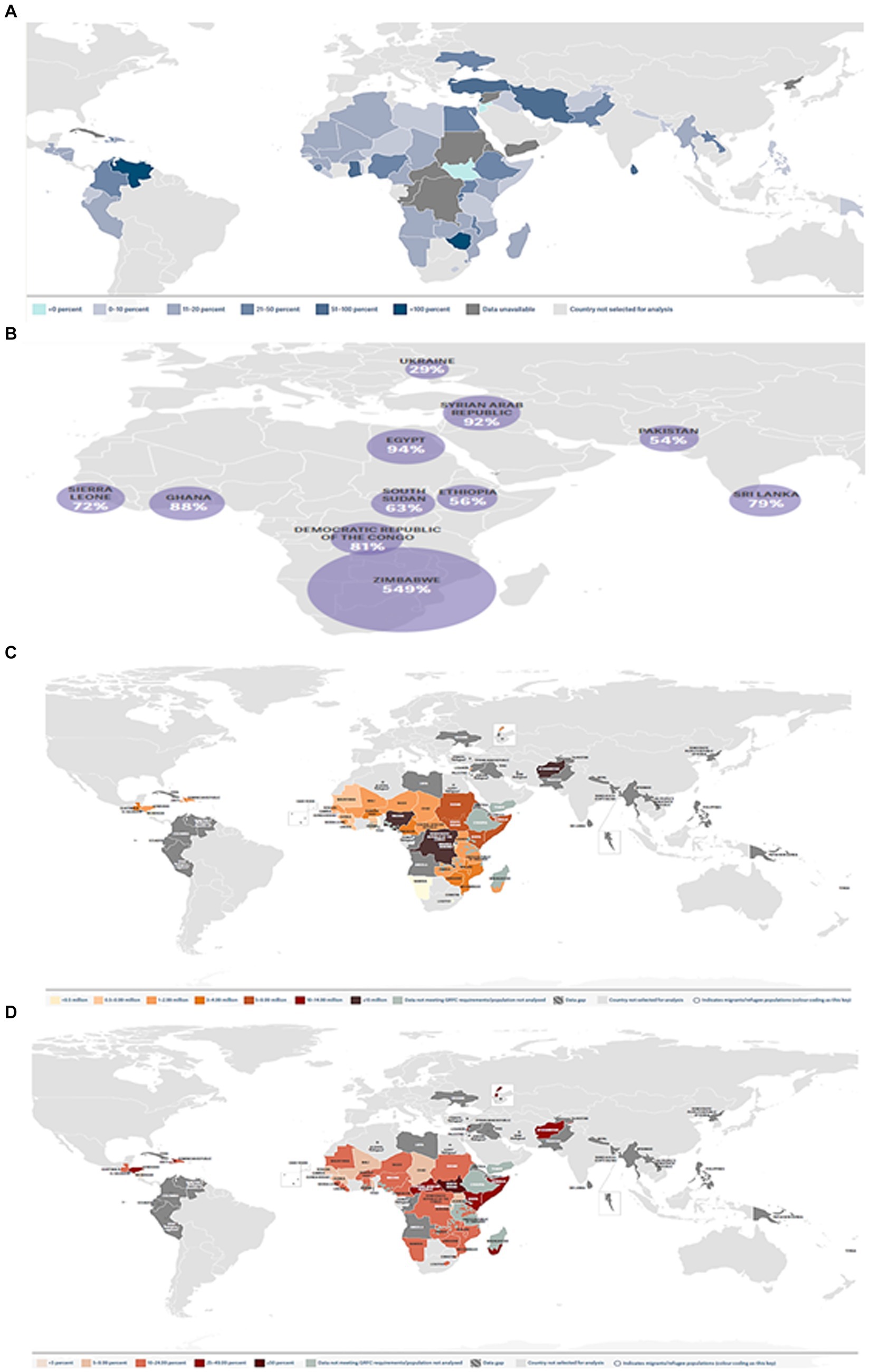
Figure 2. The effect of the COVID-19 pandemic and Ukraine’s war on the global economy. (A) % increase in the price of staple foods in countries/territories (Source: WFP Dataviz, 2023). (B) Increase of food prices had increased in countries/territories, with food inflation being over 10 percent in 47 out of the 73 countries/territories (WFP, 2023, February). (C) Numbers of people projected to be in 38 countries/territories in 2023. (D) Share of people projected to be in Crisis in 38 countries/territories in 2023.
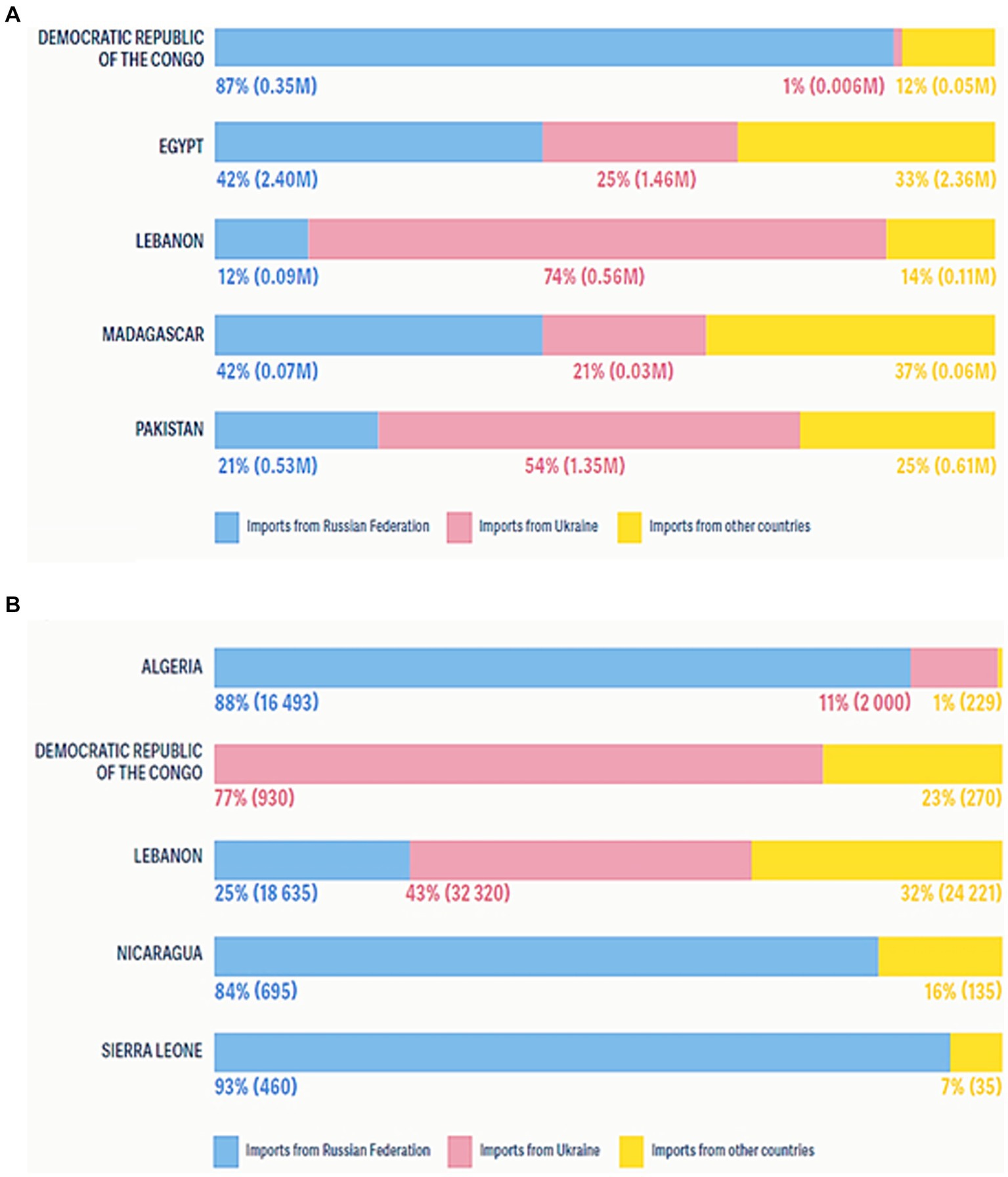
Figure 3. Countries identified with major food crises were net food importers and price fluctuations due to the COVID-19 pandemic and Ukraine’s war on the global economy. (A) Shares of wheat import volumes from the Russian Federation and Ukraine in 2021, in tonnes. (B) Shares of sunflower oil import volumes from the Russian Federation and Ukraine in 2021, in tonnes.
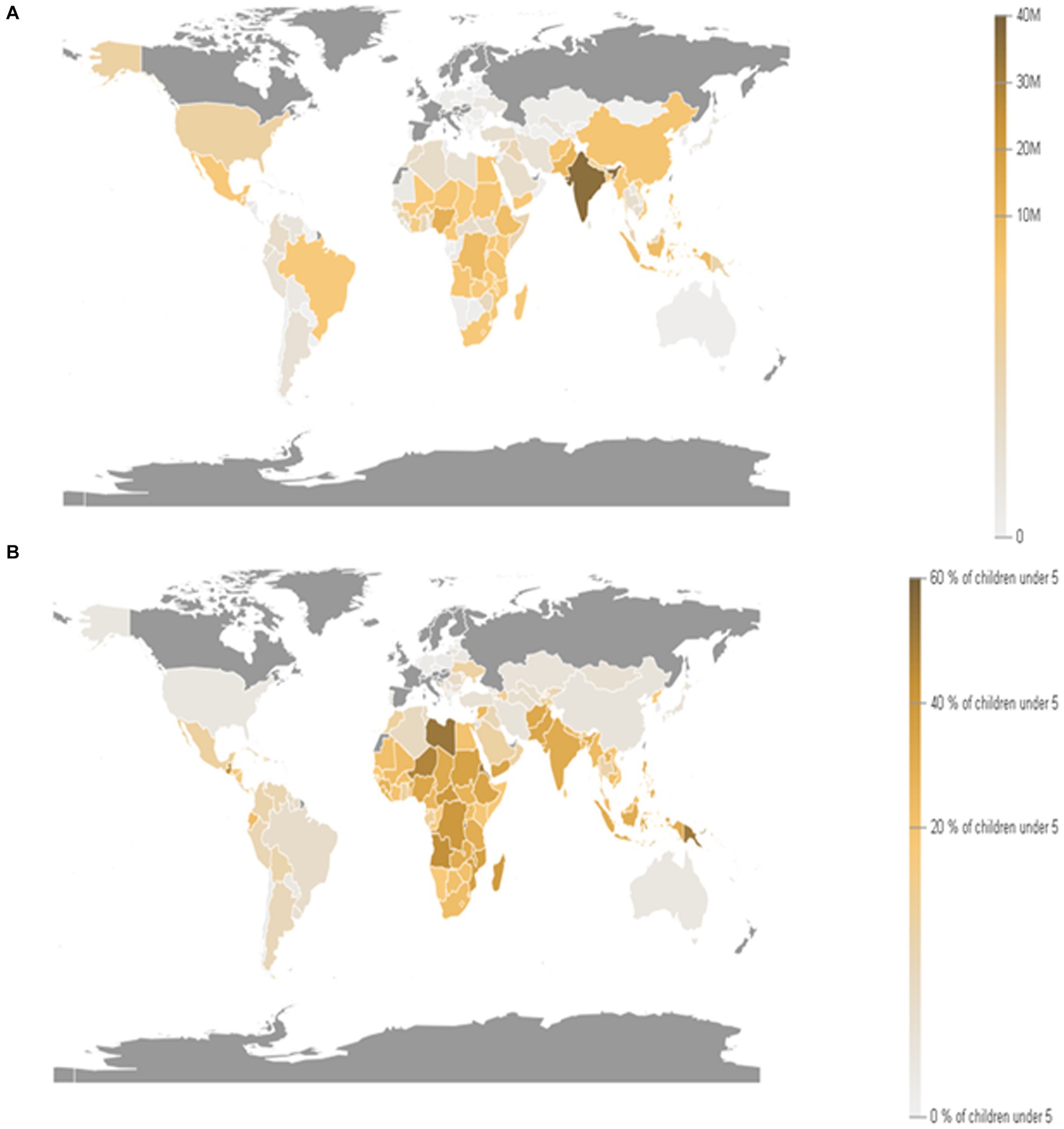
Figure 4. Prevalence of stunting among children under 5 years of age. (A) children moderately or severely stunted (under 5 years old). (B) Proportion of children moderately or severely stunted (under 5 years old; Source: Department of Economic and Social Affairs statistics, United Nations, 2024).
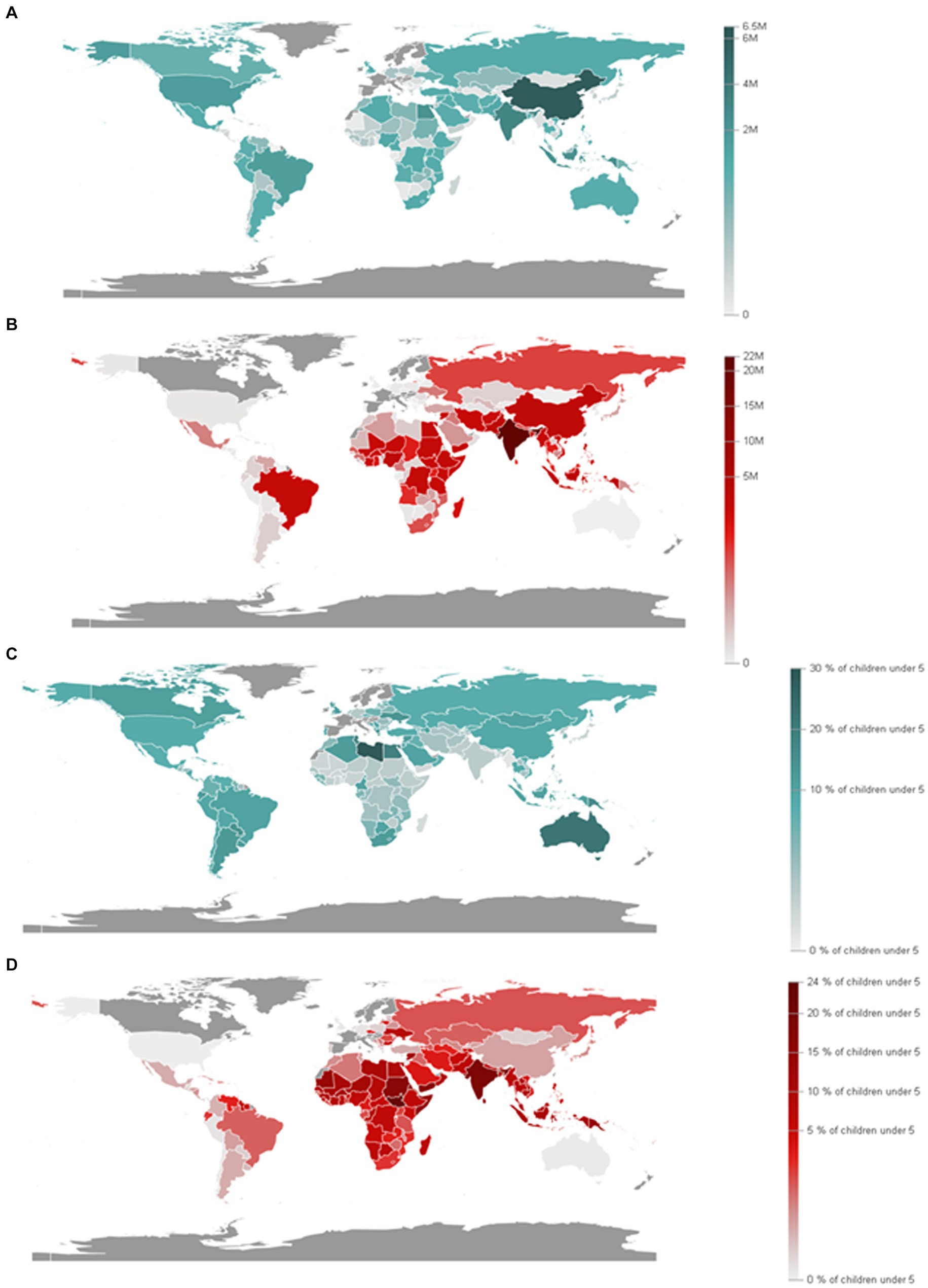
Figure 5. Prevalence of malnutrition among children under 5 years of age, by type wasting and overweight. (A) Children moderately or severely overweight (under 5 years old). (B) Children moderately or severely wasted (under 5 years old). (C) Proportion of children moderately or severely overweight (under 5 years old). (D) Children moderately or severely wasted (under 5 years old). (Source: Department of Economic and Social Affairs Statistics, United Nations, 2024).
According to the World Food Programme (2020), there are an estimated more than 97 million people who experience chronic food insecurity and this number is going to be increased in the coming years. This can have a negative impact on the Sustainable Development Goal (SDG2) which aims to end hunger (Zero hunger), as well as the global nutrition targets for 2025 (FAO, 2020; The Bill and Melinda Gates Foundation, 2020; World Food Programme, 2020). Proximately 2 million women will be added to the list of those who suffer from anemia in newborns with malnutrition and low body mass index (Figure 6) according to the United Nations (2021a,b).
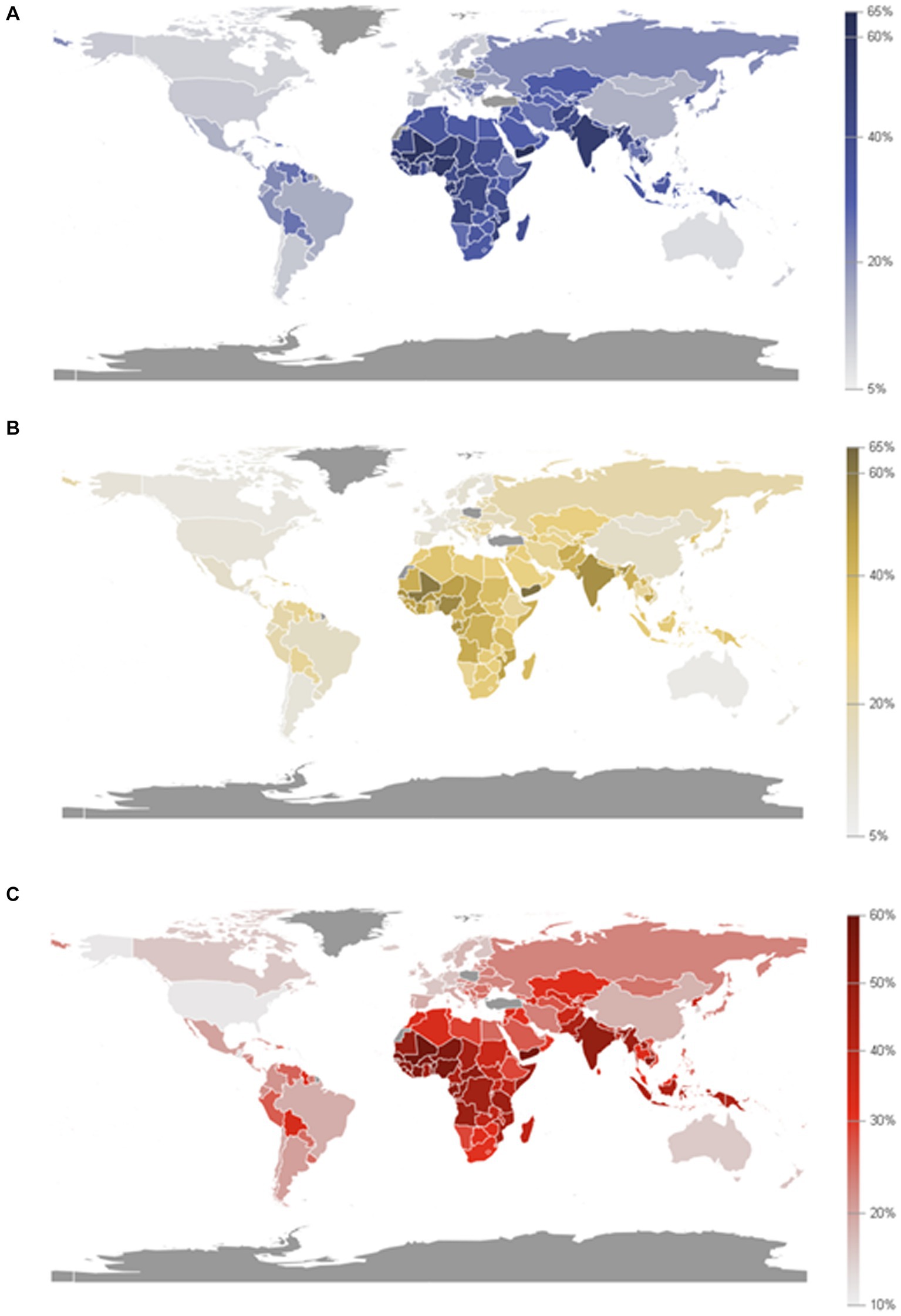
Figure 6. Prevalence of anemia in women aged 15 to 49 years old, by pregnancy status (%). (A) Proportion of women aged 15 to 49 years with anemia (15 to 49 years old, female). (B) Proportion of women aged 15 to 49 years with anemia, non-pregnant (15 to 49 years old, female). (C) Proportion of women aged 15 to 49 years with anemia, pregnant (15 to 49 years old, female). (Source: Department of Economic and Social Affairs Statistics, United Nations, 2024).
In order to meet the expected increase in population by 2050, it is necessary to ensure that more than 70% of the world’s food production will be sufficient to meet the growing demand for food as the world’s population grows faster than it has before in previous decades. The population’s enormous and quick growth will make it challenging to produce high-yield, quality, and nutrient-rich crops (Naidoo and Fisher, 2020; Chiwona et al., 2021).
In an attempt to end hunger, the Green Revolution (GR) produced high-yielding varieties; however, this came with the extensive use of high amounts of harmful chemicals and fertilizers without regard for the dietary value or the environmental and ecological consequences. The Food and Agriculture Organization (FAO) of the United Nations estimates that 820 million people worldwide suffer from chronic hunger (FAOSTAT, 2018). According to the data from the Food and Agriculture Organization (FAO) of the United Nations (FAOSTAT, 2018), 820 million people worldwide suffer from hunger. One of the most highly reported countries; is India which has 194.4 million undernourished people according to “The State of Food Security and Nutrition in the Word” FAO report, 2022. Since hunger is the greatest threat to human health, more than 2 billion people are estimated to lack regular access to nutritious food, including 8 % of people in Europe and North America countries. This number includes both those who are suffering from food insecurity and hunger worldwide.1 Belarus ranked as 1st out of the top 20 countries with a Global Hunger Index (GHI) less than 5, India is ranked 111th out of 125 countries in the 2023 GHI while China ranked as 4th out of the top 20 countries in the GHI based on many indicators such as children under the age of 5 years, the rate of child mortality and percentage of undernourished people (GHI, 2023). In developing countries, where half of the population suffers from micronutrient deficiencies in one or more essential mineral elements have emerged as the primary threat to over two-thirds of the global population (Stein, 2010). People who are malnourished are more prone to infections and experience delays in their physical and mental development (WHO, 2022). The lack of microelements affects cognitive functions such as memory and the ability to learn (Gordon, 2003). Here, we discuss the current status of micronutrient availability and malnutrition in addition to the role of crop improvement efforts to enhance and maintain food security insurance in the world.
2 Overview of plant genome editing technologies
Many genome editing technologies have been developed such as meganucleases, Zinc Finger nuclease (ZFN), Transcription activator-like effector nucleases (TALEN), CRISPR/Cas, Base editors, CRISPRi-CRISPRa editors and Prime editors.
2.1 Meganucleases
Meganucleases are endo deoxyribonucleases with recognition sites of 12 to 40 bp double-stranded DNA. They are the most specific natural restriction enzymes, with two main families of enzymes intron and intein. Meganucleases contain five major families based on cleavage site and recognition site, including LAGLIDADG, His-Cysbox, PD-(D/E) XK, GIY-YIG, and HNH (Silva et al., 2011). The most characterized endonucleases are I-DmoI, I-SceI, and I-CreI. I-SecI is the most precision meganuclease used in editing in many eukaryotic organisms, and it has been used to increase HDR frequency in Nicotiana tabacum. Two approaches to creating tailor-made meganucleases include creating small changes and variations to amino acid sequences followed by functional protein selection with variation in the recognition site and developing chimeric meganucleases with a new recognition site. Directed Nuclease Editor (DNE) is used for targeting gene sequences in cotton plants precisely to a predetermined site. Meganucleases have limitations due to the recognition sequence motif involved in each target site and the construction of sequence-specific enzymes for each sequence (Boissel et al., 2014; Daboussi et al., 2014).
2.2 Zinc finger nucleases
Zinc Finger nucleases (ZFNs) are chimeric restriction enzymes that create double-strand breaks into targeted gene sequences. They bind to the target gene’s DNA, generate an active nuclease complex, and repair the DNA through NHEJ or HR pathways. ZFNs have been used in gene editing in Arabidopsis, soybean genes, and other plants. They have been used to produce efficient variation in the Leafy cotyledon1-Like 4 gene. However, the complex construct of using two separate motifs targeted with two proximal sites makes the method more complex (Maeder et al., 2008; Weeks et al., 2016).
2.3 Transcription activator-like effector nucleases
Plant pathogens, particularly Xanthomonas species, produce effectors that produce a DNA binding domain associated with the endonuclease catalytic domain FokI. These effectors nucleases, introduced into plant host cells through the type III secretion system, produce double-strand breaks in DNA sequences, allowing targeted genome/chromosomal modifications like insertion, deletion, and replacements. TAL effectors have a similar structure with two specific DNA binding proteins and FokI endonucleases, which form dimers due to cuts in the DNA target sequence.
2.4 Clustered regularly interspaced short palindromic repeats/Cas system
CRISPR/Cas9 technology offers gene multiplexing, targeting multiple genes with a single molecular structure (Forner et al., 2015; Svitashev et al., 2015; Sharma et al., 2017). Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein (Cas) have evolved into a versatile genome editing tool with numerous applications. The CRISPR/Cas9 system, first identified in Escherichia Coli in 1987, is an RNA-guided genome editing tool that detects, identifies, and cleaves a specified target sequence of desired genes using the Protospacer adjacent motif (PAM) sequence. This mechanism leads to double-strand breaks, enhances cell repair mechanisms, and enhances mutation development (Mojica et al., 2005; Jinek et al., 2012).
The CRISPR/Cas system has been used for targeted genome modification in bacteria, yeast, plants, monkeys, and humans. The system has been used in gene-editing therapy, detection and quantification of SARS-CoV-2 RNA, and gene therapy in Alzheimer’s disease. The CRISPR/Cas13-based approach has been developed for SARS-CoV-2 detection and quantification (Kumar et al., 2020). New protein-Cpf1 or modified CRISPR/Cas9 techniques have been developed, including prime editing, base editing, and RNA editing. Two new genome editing technologies for the activation and interference of genes are CRISPRa (for activation) and CRISPRi (for interference). The CRISPR/Cas9 technique has been awarded the Nobel Prize in Chemistry for its efficiency in genome editing (Koonin et al., 2017).
CRISPR/Cas systems are divided into two classes, six types, and 33 subtypes. Class I systems are less commonly used in genome editing experiments, while Class II systems are commonly used due to their multifunctional Cas enzymes. Class I systems contain multiple Cas enzymes targeting DNA and can be subclassified into three types (I, III, and IV) and 16 subtypes (Makarova et al., 2020). Each type has its own Cas protein, such as type I for Cas3, type II for Cas9, and type III for Cas10. Class II systems are most commonly used in genome editing due to their small size and high efficiency. Type II systems are associated with two Cas genes, Cas1 and Cas2, which play a crucial role in acquiring new spacers from invading viruses. Some Cas9 orthologues have distinct recognition and targeting requirements, such as Cas12a variants for engineered genome editing. Cas13, dominant in Type VI, contains RuvC or HNH nuclease domains and targets RNA sequences directly, making it suitable for epigenetic editing (Makarova et al., 2020; Xie et al., 2020).
2.4.1 Mode of action
Trans-activating crRNAs (tracrRNAs) in cells form a stem-loop structure that allows Cas enzymes to bind. A single guide RNA (sgRNA) directs the Cas9 nuclease, which is derived from Streptococcus pyogenes (SpCas9). The Cas9 requires a protospacer adjacent motif (PAM) sequence of “NGG” in the target DNA sequence of the gene of interest. After an invasion of a bacterial cell by injecting pathogen genetic material, the phage genome gets cleaved into small pieces using a Cas protein in the bacterial cell. The cleaved sequences are incorporated into the CRISPR protein, providing immunity against the same phage by degrading its DNA with crRNA molecules (Jinek et al., 2012). CRISPR arrays are transcribed into a single long RNA stretch and processed into CRISPR RNA (crRNAs) short RNA fragment, which directs the Cas enzyme to cleave target nucleic acids. CRISPR/Cas systems provide self-defense mechanisms for archaea and bacteria, protecting viral DNA and RNA. Cas9 contains two nuclease domains: the RuvC-like domain that cleaves PAM with a noncomplementary DNA strand and the HNH domain that cleaves the guide-RNA bound complementary target DNA strand (Mohanraju et al., 2016). A large number of Cas9 variants have been identified for genome editing applications, with the type II CRISPR/Cas9 system being the most simple, used, and efficient for genome editing in crop plants (Veillet et al., 2020; Zhang et al., 2021).
The CRISPR system consists of three main steps: Adaptation (spacer acquisition), Expression (crRNA processing and target binding), and Interference (target DNA cleavage). Adaptation involves incorporating foreign plasmid spacers into the host genome, using Cas1 and Cas2 as integrases. Expression involves the biogenesis of crRNA, processing, and target binding. Targeting complexes are produced by fastening gRNAs with Cas enzymes. Interference involves cleaving the target, with Cas nuclease searching for complementary sequences. System types include Cas3 in type I, Cas9 in type II, and Cas7 and Cas10 in type III. SpCas9, a bacterium used in genome editing, has a conserved core with two major nucleic acid binding recognition lobes: REC (recognition) and NUC (nuclease). Mutations in the REC lobe reduce DNA cleavage activity. The small NUC nuclease lobe is highly conserved and can be converted into nickases for single-strand breaks. Cas9 nickases can be used for Homology Directed Repair (HDR) mechanisms. The Cas9-sgRNA complex binds to DNA, scans the complementary target site sequence, and cleaves the target sequence, resulting in double-strand breaks (DSB). The DNA is repaired using NHEJ or HDR, with the ability to generate monoallelic, biallelic, or homozygous mutations. HDR-mediated DNA repair requires a homologous template (Jinek et al., 2014; Nishimasu et al., 2014; Song et al., 2016).
2.4.1.1 Non-homologous end joining
Non-homologous end joining (NHEJ) is a DNA ligase that links broken DNA ends to repair the sequence, leading to mutations and frameshifts in the amino acid sequence. This process is the primary DSB fixing pathway in plant cells, resulting in gene knockout (KO). CRISPR/Cas9-based knockout has been used in gene function studies and has modified traits for stress tolerance, disease resistance, yield, nutritional improvement, and male sterility. In wheat, gene knockouts are difficult due to their functional redundancy. However, knockouts have been successfully used in polyploidy species to develop fungal-resistant wheat, increase vitamin E levels in barley, and enhance monounsaturated fatty acid content in seeds. The CRISPR/Cas system is also used for breeding herbicide-resistant crops by altering PDS, ALS, and EPSPS genes in tomatoes. CRISPR/Cas9-based knockout approaches have also been used in cotton, tobacco, Arabidopsis, and rice. For example, the OsERF922 gene in rice has been knocked out to enhance blast-resistant lines, while the SBEIIb gene has been knocked out to enhance amylose content (Pannunzio et al., 2017; Khaled et al., 2022, 2023; Yang et al., 2022).
2.4.1.2 Homology directed repair
HDR-mediated Gene Replacement is an error-free repair pathway that inserts a gene copy or homologous gene to fill the DSB as a homologous DNA template. This technique allows for targeted gene insertion at precise DNA locations and overexpression studies. Both NHEJ and HDR can be used to repair DSBs generated by CRISPR/Cas-based genome editing. The genome editing frequency using HDR is lower in plants compared to NHEJ, but it can be enhanced by using vectors for donor template delivery. HDR efficiency depends on factors such as CRISPR/Cas system, delivery system cell type, cell cycle stage, donor DNA template concentration, and length of the homologous arms (Sander and Joung, 2014; Feng et al., 2021). Donor template preference, such as PCR templates with preferred mutations, synthetic oligonucleotides, and linear and double-stranded plasmid DNA, can affect HDR efficiency. Single-stranded DNA (ssDNA) templates are usually more efficient than plasmid donors for small sequence changes. Modified Cas9 can increase HDR efficiency. HDR-mediated genome editing in animal cells is easier than using HDR in plant cells, with only up to 10% success. In animals, more than 30% efficiency has been achieved with some modifications using single-stranded oligodeoxynucleotides (ssODNs) instead of a plasmid donor. Two tandem repeat-HDR (TR-HDR) approaches have been developed for delivering a large copy of the donor template into the rice and maize genome. HDR-mediated knockin(s) in rice and potatoes resulted in tolerance to herbicides and improved self-life in tomatoes (Lin et al., 2014; Wang M. et al., 2017; Liu et al., 2019).
2.4.1.3 Base editing
Base editors (BEs) are a CRISPR/Cas tool for specific single base changes and modifications. They are generated through the fusion of a dCas9 nuclease with an engineered base converter enzyme, without inducing DSB. Three types of base editors have been generated: cytosine base editors (CBEs), adenine base editors (ABEs), and glycosylase base editors (GBEs). CBEs catalyze the conversion of a single C to a T, initiating a change from a C/G pair to a T/A pair. Adenine base editors (ABEs) consist of an evolved transfer RNA adenosine deaminase (TadA*) fused with dCas9. GBEs convert C-to-A and C-to-G, respectively. CRISPR-BE helps in optimizing and developing D10A nCas9 fused with cytidine deaminase enzymes. ABE base editors have high catalytic activity for point mutations. However, base editing approaches have issues such as range of editing, bystander, and editing off-target. RNA base editing in plants includes REPAIR for A-to-I (G) replacement and RESCUE for specific C-to-U exchange (Komor et al., 2016; Zong et al., 2018; Lin et al., 2020; Richter et al., 2020; Zhao et al., 2021).
CRISPRi and CRISPRa are extensions of CRISPR/Cas technology, which can repress or activate multiple target genes without detectable off-target effects. CRISPRi targets Cas enzymes to transcription factors and associated proteins, allowing gene expression to be repressed or enhanced. CRISPRa contains dCas9 fused with a transcriptional activator, allowing targeted enhancement of gene expression. CRISPRa can be used for identifying long noncoding RNA loci and epigenetic studies. CRISPRi efficiency can reach up to 86% repression, depending on sgRNA quality and target site. CRISPRi can be enhanced by fusing dCas9 with gene expression repressors, such as the Krüppel associated box (KRAB) domain of Kox1, the WPRW domain of Hes1, and the chromoshadow domain of HP1α (Cheng et al., 2013; Konermann et al., 2013; Qi et al., 2013; Konermann et al., 2015).
2.4.1.4 Prime editing
Prime editing is a new genome editing technology that uses a search-and-replace strategy to produce desired edits without generating double-strand breaks. It involves 12 base conversions, insertions, and deletions without generating double-strand breaks. The prime editing guide RNA (pegRNA) plays a role in producing sgRNAs and RT templates. The prime editing system uses a 3″ flap primer to transcribe the desired sequence, while the 5″ flap is cleaved via FEN1. The edited strand is ligated after 5″ flap digestion, forming a heteroduplex of edited and unedited strands. Prime editing efficiency depends on an appropriate pegRNA with low off-target events. Prime editing systems have been used in humans and plants, such as rice and wheat. The PPE-R system has increased the efficiency of the prime editing system in plants, but efficiency is lower in plants (Anzalone et al., 2019; Kantor et al., 2020; Scholefield and Harrison, 2021).
3 The current status of micronutrients and malnutrition
In developing nations, however, millions of people suffer from iron, vitamin A, Zinc, and iodine deficiency, which is called malnutrition, hidden hunger, or micronutrient deficiencies. These deficiencies are severe to public health and have widespread nutritional value. The most common dietary deficiencies found to be among rural people (WHO, 2022). In order to ensure environmental sustainability and the development of global partnerships, estimated to be achieved by 2035,2 micronutrient malnutrition is highly associated with health risks that become a serious obstacle in achieving the Millennium Development Goals (MDG), which include eliminating extreme poverty and hunger, improving education systems, empowering women, reducing child mortality significantly, promoting gender equality, improving maternal health, and combating diseases (Cakmak, 2008; Wessells and Brown, 2012). The most important and frequently deficient minerals in human diets are iron and zinc (Stein, 2010). On the other hand, Iodine is necessary for the prevention of brain damage and mental capacity, and vitamin A; is necessary to prevent infections, blindness, visual impairment, and infections3 as well as some populations may also have dietary deficiencies in other important and essential minerals such as copper, magnesium and calcium (Genc et al., 2005).
To decrease the gap between crop production and food consumption by 2030, sustainable development goals should be applied, so agriculture productivity must be increased (Wood et al., 2021). Numerous techniques and approaches were used to increase crop productivity and quality improvement. Stable and strategically important crops such as wheat, maize, and rice must be produced at higher levels in order to solve and decrease the gap. Since rice is a stable food for over 90% of Asians, who are thought to make up half of the world’s population, it may be the answer to the problem of meeting high-calorie food demands. Lists China and India as the world’s top rice producers and consumers. In the last several years Global rice consumers increased in the 2022 crop year. In the 2009 crop year, 437 million metric tons were consumed, while in the 2022/2023 crop year, about 517.184 million metric tons of rice were consumed worldwide (Statistica, 2024). During previous decades, genetic crop improvement techniques have played a significant role in preventing millions of people from starvation and food crises worldwide. Nowadays, with limited time for improving crops, new and efficient methods are needed to achieve sustainable agricultural development through a combination of advanced technologies such as genome editing, genetic engineering, and synthetic biology to modify and improve the genetic background of strategic crops.
The World Health Organization states that there are three types of malnutrition; Undernutrition, micronutrient malnutrition, Diet-related noncommunicable diseases (NCDs), and overweight. According to WHO data, almost 462 million children were sufferers, 39 million were obese children and almost 2 billion were overweight. Micronutrient malnutrition is caused by a deficiency of minerals and vitamins which is necessary for body development and growth. Undernutrition, which includes overweight with abnormal body mass weight, stunting, and wasting.4
Since undernutrition accounts for over 45 percent of deaths in children under 5 years old, particularly in low-income nations. The United Nations has been actively working to find solutions and end malnutrition worldwide by enacting laws and policies that support sustainable agriculture and meet the nutrition targets of the sustainable development agenda by 2030.
Most biological processes, plants, animals, and humans require enough supplements of micronutrients such as iron and zinc. Both humans and animals cannot provide their own needs from micronutrients and rely on plants to consume as a part of their diet, which is a vital source of nutrients but not a balanced diet that provides an adequate supplement of required minerals (Table 1), 1.2 billion people worldwide suffer from iron deficiency with half of those cases being found in China and India (Majumder et al., 2019). The majority of people eat an unbalanced diet which low in nutrients, surviving mostly on cereals which are a stable source of carbohydrates but deficient in micronutrients which are needed on a daily basis. The main cause of malnutrition is diets based on cereals in developing countries. Cereal grains have low levels of micronutrients such as Vitamin A, Zinc, and Iron. According to World Health Organization estimates more than 17 % of the population is at risk of numerous infectious diseases. Three main strategies are available to overcome and address the Zinc/Iron nutrition issues; food biofortification, a balanced diet, and medical supplements with needed micronutrients. The majority of developing nations cannot implement these strategies because they are expensive, and require a lot of money and infrastructure. Enhancing and improving the levels of micronutrient elements in cereal crops through advanced genetic engineering and biotechnological procedures might enhance and could be considered as a potential strategy to overcome the limitations that face poor countries.
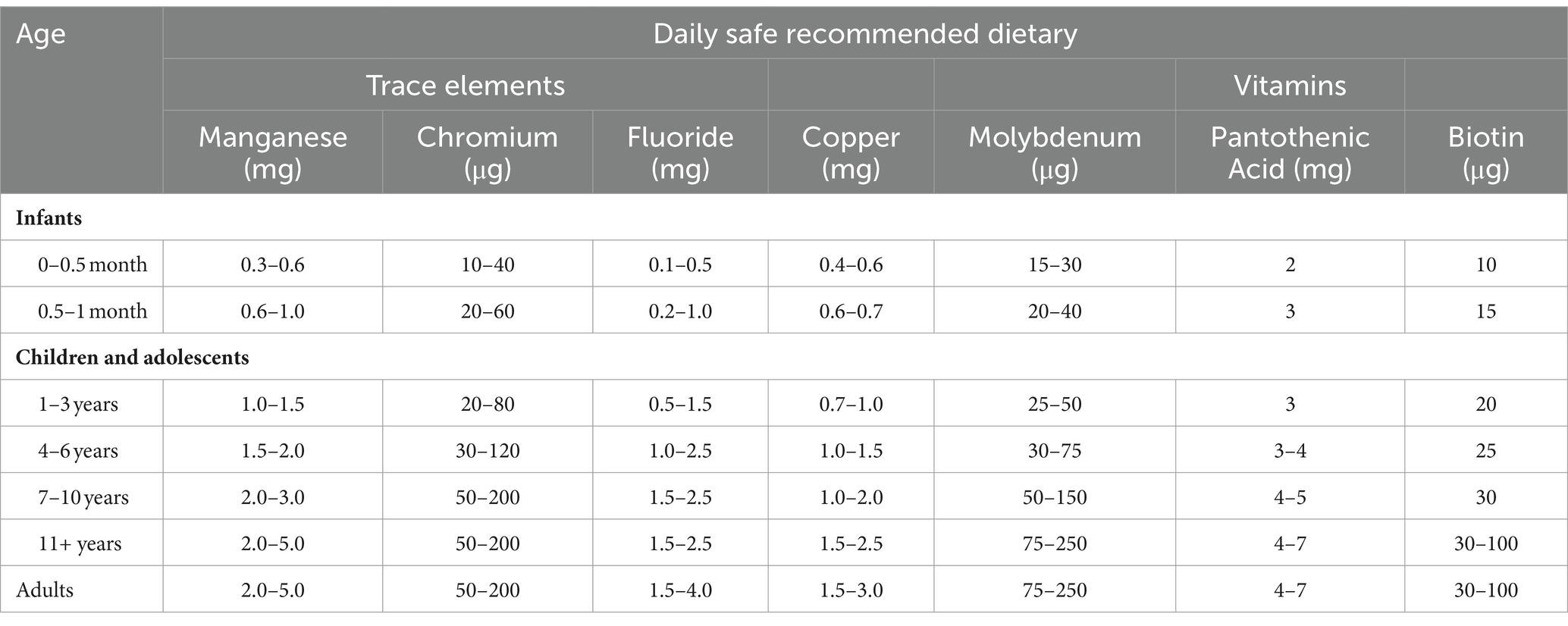
Table 1. Daily safe recommended dietary for vitamins and trace elements [National Research Council (US), 1989].
There is a need to develop highly biofortified cereal crops such as wheat, rice, and maize as a long-term solution to micronutrient malnutrition and health risks. The human body needs certain essential nutrients in small amounts for proper metabolism regulation and physical development. For the body to stay healthy, it needs both macronutrients and micronutrients (Table 2). Certain nutrients are necessary for a daily diet. Micronutrient element nutrients are vital for all living organisms and are involved in a wide range of biological, metabolic, enzymatic, and biochemical processes as well as regulatory functions. Neurological disorders, heart diseases, anemia, immune disorders, gastrointestinal issues, and central nervous problems can all result from Zinc deficiency (Pan et al., 2017; Ajeesh et al., 2020). The primary component of oxidoreductase, myoglobin, and hemoglobin—all of which are necessary for the transportation of oxygen—is iron. Hair loss, anemia, pagophagia, restless legs syndrome, pallor, exhaustion, and morbidity can all result from low iron levels (Jimenez et al., 2015). Iron deficiency in plants also lowers chlorophyll concentration, which negatively impacts plant growth and development as well as crop productivity (Malik and Maqbool, 2020).
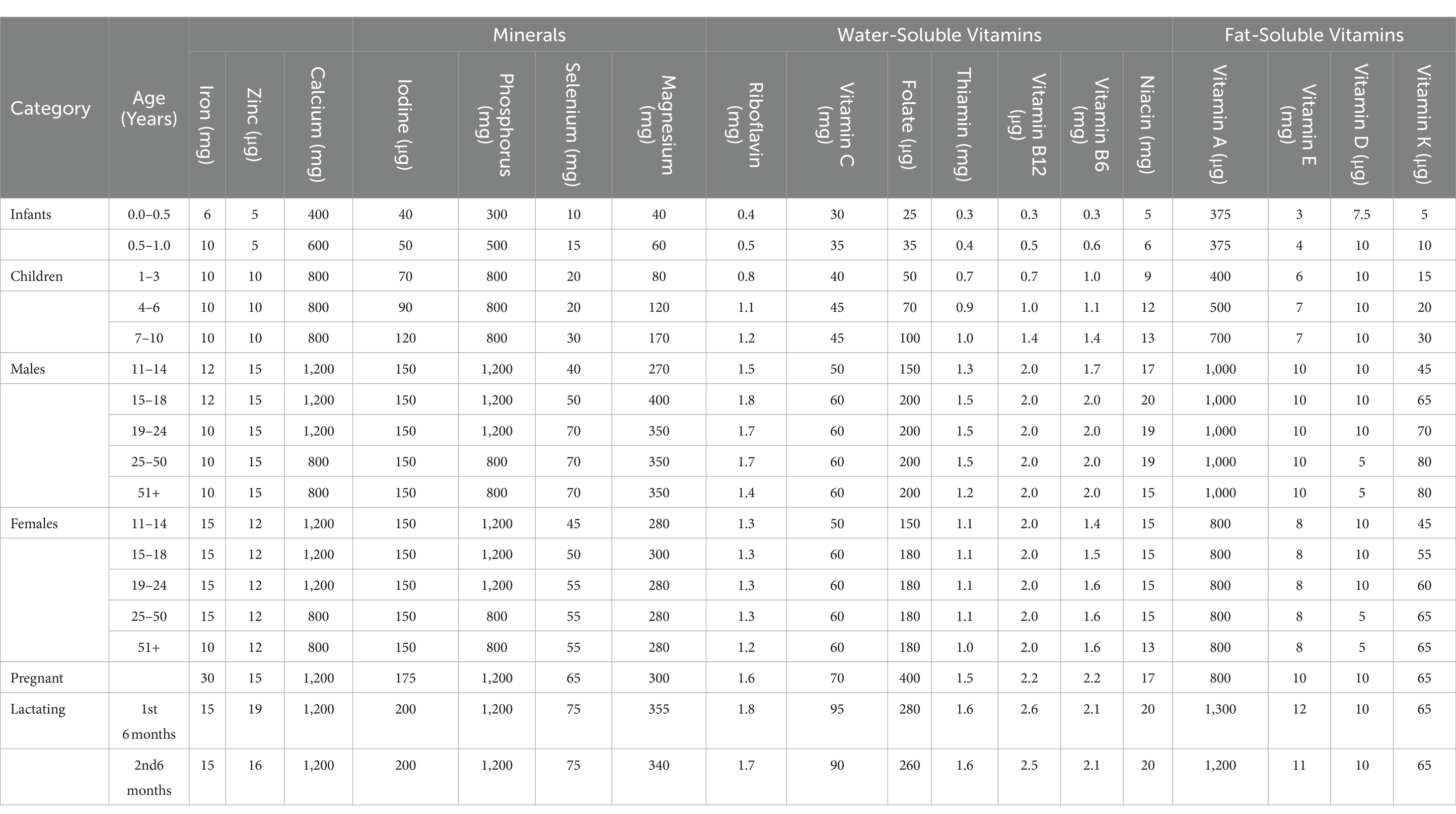
Table 2. Daily safe recommended dietary for vitamins and trace elements [National Research Council (US), 1989] for the maintenance of good nutrition in United States of America.
A zinc deficiency affects billions of people worldwide as a result of inadequate dietary intake. Zinc must be given to the body on a daily basis because it cannot be stored in the body for an extended period of time. Zinc is essential for the immune system’s operation and plays a key role in the growth and division of cells. Children’s impaired neurodevelopment and goiter, or enlargement of the thyroid tissues, are brought on by iodine deficiency, as iodine is necessary for the thyroid hormones’ biosynthesis. The formation of neurotransmitters, various metabolic processes, and a healthy immune system all depend on vitamin B and its eight forms; deficiencies in this nutrient cause inflammation, lethargy, and depression (Bryan et al., 2002; Kumssa et al., 2015; Zimmermann and Boelaert, 2015).
In addition to its significant role as an antioxidant, vitamin E plays a preventive role in the regulation of liquid packaging and the prevention of neurological, ophthalmological, cardiovascular, and cancer diseases as well as the prevention of plate late aggregation. Due to its antioxidant qualities, vitamin C is necessary for the synthesis of collagen, amino acids, energy metabolism, and immunity. Joint and bone issues can result from a vitamin C deficiency. The primary functions of vitamin A are in the regulation of genes, enhancement of the visual system, cell growth and function, immune system maintenance, and reproduction. A deficiency in vitamin A can result in nyctalopia, maternal mortality, and problems during pregnancy and lactation (Simpson et al., 2011; Rizvi et al., 2014; Maggini et al., 2017).
4 Soil is an essential play factor for micro and macronutrient availability
Human iron and zinc deficiency (hidden hunger) are highly related and associated with iron and zinc deficiency in the soil. For the majority of the population on the planet cereal crops serve as their primary source of food and micronutrient supplements for humans. Reduced Zinc availability in the soil is the cause of low zinc concentration in the seeds (Cakmak, 2008; Zhang et al., 2010; Horuz et al., 2019). The main and primary source of numerous macro and microelements necessary for plants, humans, and animal growth and development is soil. Plants are considered a major source of energy and food for most organisms such as animals and humans. Many and numerous factors influence the availability of phytonutrients such as soil texture, moisture, pH, farming practices, and many other factors. These factors also influence how the plant roots can absorb essential minerals and interaction activity with other soil elements. The scarcity of micronutrients and the soil chemical structure may affect plant root structure and lead to changes in root structure in an attempt to increase nutrient uptake by excreting H+ ions which aid in root elongation and growth (Alloway, 2009; Welch et al., 2013).
A shortage and deficiency of micronutrients in the soil have a negative impact on crop quality and yield. More than 5 % of Europeans and 19 % of Asians prefer plant-based foods and products over animal products. Plant products are high in phytate and lower the body’s ability to absorb and bioavailability absorbance of the micronutrients. The number of vegetarians worldwide has increased recently which may affect their health condition. Frequent alcohol use reduces the body’s ability to absorb essential vital nutrients. The maximum percentage of iron and zinc deficiency in global soil was approximately 3 and 49%, respectively (Sillanpää, 1990; Hillesund et al., 2021). To determine and quantify the phytoavailable concentration of micronutrients in agricultural soil that could enhance crop productivity, nitrogen use efficiency, and nutritional quality of crops, a soil map for nutrition deficiency is necessary. Crops with enhanced content of micronutrients through breeding programs showed normal yield and growth with no effect due to biofortification improvement (Graham et al., 2001). Haas et al. (2005) reported that a fed experiment was conducted using biofortified rice, the rice was fed to the Philippine women for 9 months as a trial experiment. The results indicated a 17 percent increase in ferritin in the blood serum of the women, which was attributed to the consumption of this biofortified rice line (IR68144-3B-2-2-3).
Agroecology takes an integrated approach to agriculture, including local and Native knowledge and participatory procedures. It aims to encourage ecosystem services and biodiversity like the pollination process and control of pests while reducing the need for additional inputs (Nogué et al., 2024). Several and numerous techniques and methods including improved agronomic techniques, development of genetic variations, genetic engineering, transgenic techniques, and biofortification through genetic improvement and breeding were used to achieve and provide sustainable biofortified crop production.
5 Strategic crops for maintaining food security stability
Since the world’s population is expanding much faster than it was in previous decades and food consumption is rising daily, it will be necessary to guarantee global food security. Rice considered one of the most extensively growing and consumed crops also crucial for preventing hunger and helping in the stability of food security. Most of the population relies on rice and its products as a stable food and primary source of energy and nutrients.
Numerous variables control rice nutrient content determined by several factors including the type of rice variety, how it is processed (Polished or not), soil type, and nutrition quality. Polished Rice is found to be high in fiber and low in essential nutrients, it is not a balanced diet that is suitable for long-term use as it puts the human body at risk of multiple nutritional deficiencies (Bouis and Saltzman, 2017). Many rice lines have been developed as a good source for enhanced vitamins, and micro and macro elements. World Health Organization aimed to address malnutrition on a global scale by implementing various strategies and also providing guidelines to produce and improve biofortified rice and other crops with quality maintenance after cooking (IFPRI, 2013; IRRI Rice Knowledge Bank, 2013).
Climate change poses enormous difficulties to smallholder agricultural systems, prompting the development of innovative technologies to increase food security and environmental health. Genome editing is a powerful technology that can play an important part in this process. However, the problem is to create an enabling atmosphere that promotes research, innovation, and openness. Policy ideas include boosting public investment in crops, prioritizing genetics and trait development, rewarding start-up businesses, and producing environmentally friendly goods. A clear, open, and predictable regulatory structure is required. Research shows that even a small temperature increase of 1°C reduces crop production. The African continent is projected to face greater temperature rises compared to the majority of the globe (Pickson and Boateng, 2022; Wachira, 2022).
Numerous problems, including abiotic and abiotic stresses, impact rice production, these stresses affect rice grain quality which might negatively affect the malnutrition status of the population worldwide who depends on rice as a diet on a daily basis.
6 Crop improvement and food security insurance using genome editing technology
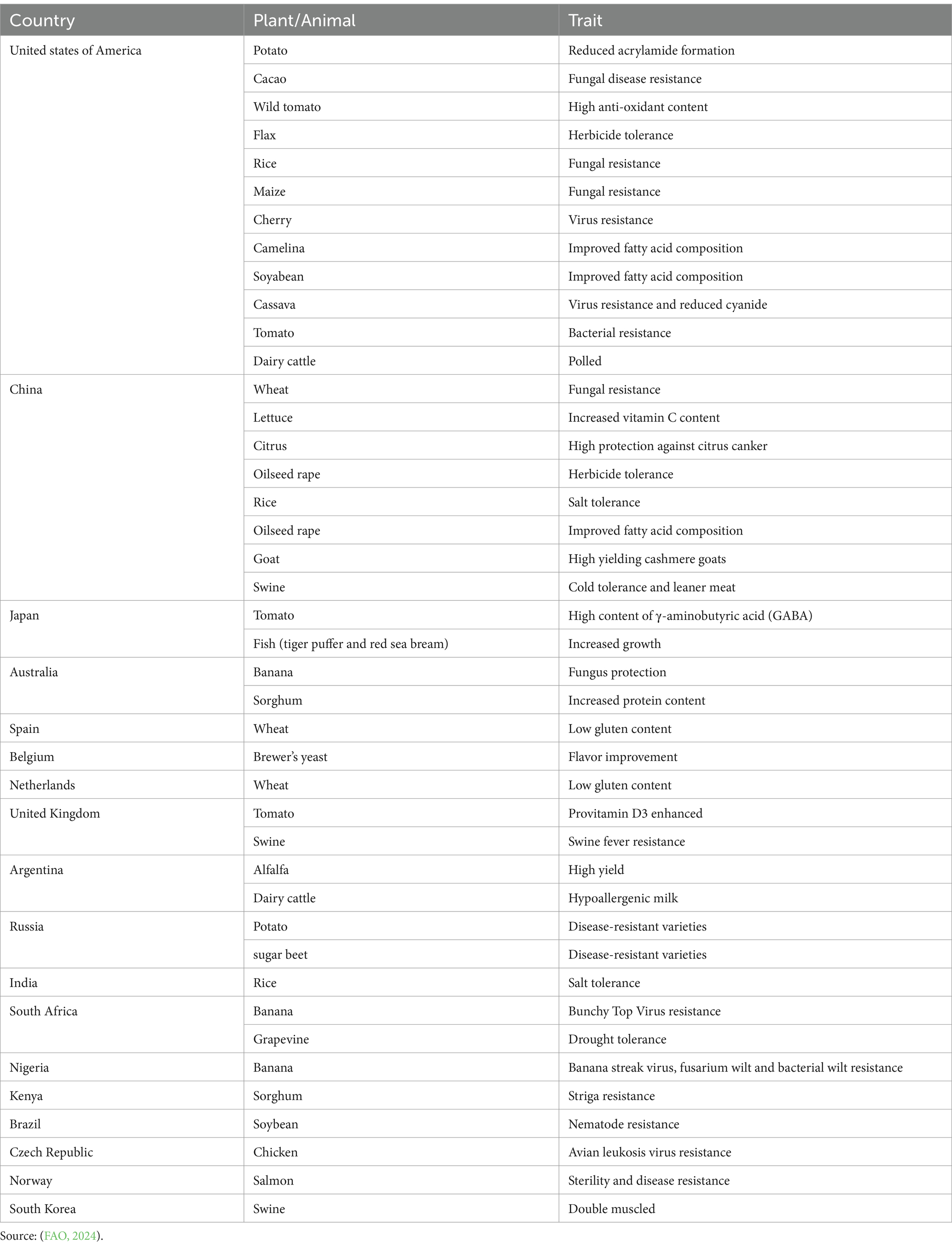
Obtaining a suitable genetically stable variety with a high degree of homozygosity through conventional breeding programs takes time. This method is less successful and may transfer unwanted characteristics. Nevertheless, CRISPR/Cas9-based genome editing offers a less successful method of producing healthy plants in less than a year. Along with the technique’s adaptability, effectiveness, and specificity, a variety of applications can be made using it, including single knockin or knockout, prime editing, multiplex gene editing, CRISPRa, CRISPRi, base editing, and more (Table 3). In 2024, the European Parliament voted to support a proposal to make it easier to authorize plants generated that cannot be distinguished from traditionally developed plants (European Parliament, 2024; Green Transition of Farmers, 2024). Liu et al. (2021) reported that in addition to high yield and nutritional quality, CRISPR/Cas genome editing technology enhanced crop resistance to biotic and abiotic stresses and produced plants with medicinal qualities, such as anti-oxidation, anti-inflammatory, and anti-cancerous molecules.
Editing the genome to produce desired genes for crop biofortification will improve macro- and micronutrients and improve human health and food security. The majority of model and non-model plants as well as significant and strategic crops, including rice, wheat, maize, tomato, potato, soybean, cotton, and many others, were grown using this technology. This decrease in agricultural output has been observed to cause ripple impacts, resulting in a mean decline of 1.3 percent in economic development for every degree of temperature. In addition, new data indicates a growing worldwide issue that includes a rise in malnutrition and starvation, which is made worse by a growing number of harsh weather occurrences.
Biotic and abiotic stresses have a significant impact on crop yield and nutritional quality. Plant resistance to biotic and abiotic stresses has increased thanks to genome editing techniques based on CRISPR/Cas. Numerous genes exhibit stress sensitivity and function as negative regulators in the plant’s reaction to both biotic and abiotic stressors (Filiz and Vatansever, 2018). The loss of function of the (MLO) MILDEW-RESISTANCE LOCUS O genes led to increased resistance to powdery mildew disease in a number of crops. Susceptible gene manipulation can improve crop tolerance to a variety of diseases and pests. Applications for CRISPR/Cas genome editing have altered many plants to make them resistant to viral, fungus, and bacterial diseases.
These are just a few examples of the many plant genes that can be knocked out using CRISPR/Cas9-based knockouts for enhanced resistance such as crt1a gene knockout in oilseed rape and Arabidopsis thaliana resulted in Verticillium longisporum resistance (Pröbsting et al., 2020), Verticillium dahlia resistance was enhanced in cotton after 14–3-3 gene was knocked out (Zhang et al., 2018), for Magnaporthe oryzae and Xanthomonas oryzae resistance in rice, OsERF922 and Os8N3 genes was knocked out using CRISPR/Cas9 (Wang et al., 2016; Kim et al., 2019), Knockout of pmr4 gene, gave powdery mildew resistance in tomato (Santillán Martínez et al., 2020). In cucumber, knock out of the eif4e gene through CRISPR/Cas9-based knockout resulted in resistance to Cucumber vein yellowing virus (CVYV), Papaya ring spot mosaic virus-W (PRSMV-W) and Zucchini yellow mosaic virus (Chandrasekaran et al., 2016). milo gene knockout using CRISPR/Cas9, which gave resistance to powdery mildew (PM) in many plants such as wheat, grapevine, and tomato. Many genes in different plants, CRISPR/Cas9 technology were used for enhancing and give resistance to many plant infections and diseases such as Clpsk1 for Fusarium oxysporum resistance in watermelon CsWRKY22 for Xanthomonas citri resistance in orange, CsLOB1 for Xanthomonas citri resistance in citrus, LIPOXYGENASE 3 (lox3) for Ustilagomaydis resistance in maize, SlJAZ2 for Pseudomonas syringae resistance in tomato (Peng et al., 2017; Ortigosa et al., 2019; Wang et al., 2019; Pathi et al., 2020; Zhao et al., 2020).
There are not many genes known to act as negative regulators in plants in response to abiotic stresses. Plant root development and abiotic stress response are regulated by numerous genes. Tomatoes that had their arf4 transcription factor gene knocked out via CRISPR/Cas9 exhibited increased salt tolerance (Bouzroud et al., 2020). The arg gene was edited using CRISPR/Cas9 to increase a plant’s resistance to drought and nitrogen shortage (Peng et al., 2021). Cotton showed increased root number and differentiation upon arg gene knockout (Wang Y. L. et al., 2017). Cui et al. (2020) reported that Gs3 and dep1 gene knockouts in rice increased the plant tolerance to many abiotic stresses such as chilling, salinity, and drought. CRISPR/Cas systems for enhancing crop yield and nutritional quality.
Several genes regulate the crop yield trait. Crop yield is negatively impacted by certain genes that function as negative regulators. Grain size and number per panicle in rice are negatively regulated by genes like Dep1, Gs3, and Gn1a. Grain size and number were increased by gs3, gn1a, and dep1 gene knockout (Li et al., 2016; Xu et al., 2016). A frameshift mutation in rice cultivars through knocking out resulted in yield improvement when the early heading date 1 gene was knocked out (Wu et al., 2020). Grain weight was improved and increased by knocking out thousand-grain weight 2, 5, and 6 genes (Zuo and Li, 2014; Xu et al., 2016). Rice grain weight and number were increased by OsPAO5 gene knockout using CRISPR/Cas9 (Yusong et al., 2021). The number of plant tillers in switchgrass was increased when the (TB1) teosinte branched 1 gene was knocked out using CRISPR/Cas9 (Liu et al., 2020). The durum wheat a-amylase/trypsin inhibitor genes were edited (Camerlengo et al., 2020).
Editing the maize SWEET13 gene family, which functions as a sucrose using CRISPR/Cas9 (Bezrutczyk et al., 2018). In another case, using CRISPR/Cas9 to produce waxy and sweet maize seeds by editing WX and SH2 genes (Qi et al., 2013; Dong et al., 2019; Gao et al., 2020). The role of HvPTST1 in starch accumulation and its interaction with (GBSSI) granule-bound starch Synthase I in barley grain have been characterized by means of Protein Targeting to Starch 1 through CRISPR/Cas9-based editing (Zhong et al., 2019). In rapeseed, knockout BnaMAX1 alleles enhanced yield with many numbers of branches (Zeng et al., 2020). Multiple amino acids and fatty acids accumulated after the SBEIIb gene was knocked out (Baysal et al., 2020). Gamma-aminobutyric acid (GABA) fortified rice was produced by CRISPR/Cas9 knockout of the GAD3 gene (Akama et al., 2020). The TaGW2 gene has three homologs in hexaploid wheat: TaGW2-A1, TaGW2-B1, and TaGW2-D1. TaGW2 knockouts resulted in increased grain yield and weight with a high grain protein content (Zhang et al., 2018).
Additionally, the knockout of the sucrose transporter SWEET11 gene resulted in a decrease in sucrose concentration levels in the embryo sacs, which affected the mature caryopses starch content and led to defective grain filling (Ma et al., 2017). When the FAD2-1 gene, which codes for the enzyme (FAD2) fatty acid desaturase 2, was mutated using CRISPR/Cas9, the amount of oleic acid was doubly produced and linoleic acid was absent (Abe et al., 2018). Endo et al. (2016) reported that the OsOr rice gene, which is the ortholog of the Orange (Or) gene, was edited using CRISPR-Cas9 to increase the accumulation of b-carotene in the rice callus.
The sugar metabolism vacuolar invertase genes, INV2 and NV3, were double knocked out, resulting in lower amylose content, lower weight, smaller seeds, and higher sucrose levels (Deng et al., 2020). The phospholipase D gene (OsPLDa1) was knocked out using CRISPR/Cas9, which decreased the amount of phytic acid (PA) which acts as anti-nutritional agents that decrease the availability of phosphorus, iron, and zinc in rice grains (Khan et al., 2019). OsSPS1 and OsSPS11 knockout decreased total SPS activity and caused starch to accumulate; plant growth was unaffected (Hashida et al., 2016). ITP5/6 K mutants in rice were created using CRISPR/Cas9 knockout and generated low rice grains phosphatidic acid content (Jiang et al., 2019). Anthocyanin content was decreased after using CRISPR/Cas9 to edit OsF30H, OsDFR, and OsLDOX genes (Jung et al., 2019). Reduced cellulose content and decreased expression of MYB58/63, MYB61L, and MYB86L genes, which are linked to secondary cell walls, were observed upon knockout of transcription factor SND2 (Ye et al., 2018).
By utilizing CRISPR/Cas9 technology to knock out two rice genes; Nramp5 and LCT1, low cadmium (Cd) rice was produced, resulting in a decrease in cadmium levels within the grains (Tang L. et al., 2017; Songmei et al., 2019; Wang et al., 2019). Fragrant rice was produced through the editing of Betaine aldehyde dehydrogenase (Badh2)'s first exon with (T) thymine as an extra base (Shao et al., 2017). Takeda et al. (2019) reported that more guaiacyl was accumulated as a result of the knockout of the Coniferaldehyde 5-hydroxylase gene, which increased the amount of galactan, Xylan, and arabinan in the culm and leaf.
Zhu et al. (2019) used CRISPR-Cas9, to create a frame-shift deletion in the Rc and Rd genes was introduced, producing red color grains with high anthocyanidin and proanthocyanidin content without affecting any agronomic traits. Roca Paixão et al. (2019) reported that drought stress in Arabidopsis was exacerbated by activating the endogenous promoter of AREB1 using CRISPRa for the ABA-responsive element-binding protein 1 (AREB1) and ABRE-bindingfactor2 (ABF2). Gene-editing approaches have been employed in the actual breeding of plants and animals to improve the efficiency and precision of certain essential processes. Only a few have been commercialized yet (Table 4). CAST (2024) reviewed the Applications, Benefits, and Challenges of Genome Edited Crops, for more than a decade, genome editing methods have been utilized to enhance crops as well as human and animal health. They are currently being used for a variety of crops and trait combinations, benefiting both producers and consumers. Regulatory frameworks are being revised to guarantee successful implementation. Genome editing will be critical for establishing a competitive economy and resolving issues.
Many crops have benefited from multi-trait improvement through the application of multiplexed genome editing. Numerous techniques and approaches have been used to improve crop yields, tolerance to extreme biotic and abiotic stresses, and improved nutritional quality and value.
7 Food insecurity and malnutrition rates in African developing countries
The United Nations Sustainable Development Goals aim toward the eradication of all kinds of malnutrition and hunger worldwide by the year 2030 (Mohajan, 2022). Wars and conflicts have contributed to the prevalence of food insecurity in many African nations (Otekunrin et al., 2020). Nutrition and food security are important human rights, however, in the African continent, certain individuals lack these necessities, which makes it one of Africa’s most significant concerns. The COVID-19 epidemic has made the situation worse (Atukunda et al., 2021). The population of Africa reached roughly 1.46 billion, which is predicted to reach 1.71 billion by 2030 which is an additional stress to what is already excised such as extreme weather, war and conflict over politics, ineffective leadership and government instability, and climate change leading to malnutrition and food insecurity in most African nations (Worldometer, 2023). Africa boasts a notably youthful population, with 41 % under 15 years old and around nineteenth percent aged from 15 to 24. As a consequence, food production must expand to meet the feed and nutritional demands (Giller, 2020; Mohajan, 2022). By 2022, the rate of undernourishment in Africa was 19.7%, having roughly 278 million individuals undernourished, and this number is anticipated to rise to 25 % by the year 2030 (Giller, 2020; Mohajan, 2022; Maniragaba et al., 2023).
Farming is critical to the economics of many African countries, especially those with high poverty rates. Yet, it is commonly acknowledged that climate change presents among the most significant risks to agricultural output and food security (Mendelsohn, 2008). Abigarl et al. (2024) reviewed the application and importance of CRISPR-mediated genome editing for climate change and sustainable technologies that can help smallholder farmers in low-middle-income countries of Africa to adapt to climate change without productivity loss. Unfortunately, certain African nations are anticipated to incur severe consequences, including some possibly facing a shocking 50 % reduction in agricultural yield by 2030 with an incredible 90 % decrease in crop revenue by 2,100, mainly owing to significant global warming and its accompanying unpredictability (Serdeczny et al., 2017). African countries must continue to implement national and international efforts to alleviate malnutrition and poverty, among which is the CRISPR-mediated production of climate change-tolerant crops.
8 Combining conventional agricultural techniques with CRISPR-based genome editing in addition to current rules and regulations in developing countries
In Africa, combining traditional agricultural techniques alongside CRISPR-based plant breeding provides a chance to transform the agricultural industry by tackling significant issues confronting farmers. This combination enables the production of crops with higher resistance to biotic stresses and increased tolerance to abiotic challenges with enhanced nutritional value (Tripathi et al., 2022). Through leveraging the enormous variety of indigenous crop types, CRISPR-based crop improvement may preserve cultural value simultaneously improving agricultural performance as well as adaptation to changes in the climate. Agro-ecological ideas typically underpin African traditional methods of agriculture, such as intercropping, rotation of crops, and the application of organic fertilizers (Wang et al., 2020).
This CRISPR-Cas technique enhances organically and farming operations by minimizing the demand for chemical fertilizers and encouraging environmentally friendly farming (Ogaugwu et al., 2019). Furthermore, the use of CRISPR may assist combat hunger by increasing the nutritional value of basic crops. To successfully implement CRISPR-based reproduction, scientists, as well as must work together to exchange knowledge (Wang et al., 2020). Protecting farmers’ rights to conserve and share seeds encourages seed independence and equal advantages for communities nearby (Zaidi et al., 2020; Tripathi et al., 2022). Building capacities and transfer of technology enable indigenous researchers to modify and use innovations in their environments (Montenegro, 2020). In general, this combination supports small-scale farmers as well as helps to preserve African conventional habits by agreeing with current scientific breakthroughs. (Boluwade, 2020; Buchholzer and Frommer, 2023).
The African Union’s Agenda 2063 contains ambitions to use genome editing to increase agricultural output. Nigeria and Kenya have been paving the way in developing legislation for case-by-case approvals of genome-edited crops. The Kenyan government, the National Biosafety Authority of Kenya published recommendations describing a method for excluding genome-edited organisms and their products from the provisions of the Biosafety Act, which classifies them as conventional varieties (Guarango, 2022; Masehela and Barros, 2023; Ongu et al., 2023). Similarly, Nigeria’s National Biosafety Management Agency approved rules on genome editing, enabling modified crops to be categorized as traditional cultivars. Researchers and scientists in developing countries may struggle to gain the necessary funds to effectively utilize genome editing technology. Efforts concentrating on transferring technology as well as capacity development have become essential to equip local scientists with the ability to employ this technology in agriculture (Ahmad et al., 2021). Small-scale farmers serve a vital part in Africa’s economic industry, therefore protecting their legal entitlements to utilize agricultural materials as well as assuring the availability of non-genetically modified seeds (Ogaugwu et al., 2019). Masehela and Barros (2023) initiated that CRISPR-based genome editing for agricultural improvement provides significant benefits to the production of food and agriculture, but it is essential to guarantee that these advantages are distributed fairly among farmers.
9 Food security requires roles and regulations such as social issues, ethical, as well as health concerns for better use of genome editing
Global policy roles and regulations about genome editing CRISPR/Cas9 technology have revolutionized and improved breeding technologies for target gene precision. It is necessary to alter and regulate the current regulatory framework. Products created by traditional breeding methods can be replaced by those created through gene editing technologies. With particular delivery systems, transgene-free editing technologies have produced inheritable alterations and modifications. The concept concentrates on crops developed through controlled mutagenesis, including methods such as cisgenesis and intragenesis, that alter the genetic makeup without adding foreign DNA (Voigt, 2023).
In breeding programs, gene-edited varieties serve as genetic resources for the introduction of beneficial traits aimed at improving crops. It includes these crops and associated nutritional derivatives, however excludes those transformed and that include gene material of another organism, and continue to be subjected to existing genetically modified organism’s regulations (Dima et al., 2023).
CRISPR-based product regulation across African countries differs per each country due to many variables that include national, and international policies, current biotechnology rules, and regulations (Martínez-Fortún et al., 2022; Tripathi et al., 2022).
CRISPR-Cas9-based crops can be culturally acceptable by merging traditional agricultural practices with CRISPR-edited crop breeding projects in Africa. This technique can adhere to agroecological concepts, engage local communities, and prioritize small-scale producers. Nevertheless, the democratization of the CRISPR-Cas9 approach is critical for its adoption in the African continent, as cultural acceptability levels range among areas.
Engagement with the community is critical to the development and adoption of Genome edited crops. To guarantee equal advantages for everyone involved, ethical concerns about the safety, regulation, and benefit-sharing of gene-edited goods must be considered.
Knowing how to generate transgene-free plants can help modify some countries’ strict laws governing the editing of crop plants’ genomes. Transgene-free plants without genetic disorders can be produced in a variety of ways and techniques. Non-genetically modified organisms (GMOs) include plants altered by DNA-free editing techniques. Three classes of genome editing techniques exist: SDN-1, SDN-2, and SDN-3. SDN-1 employs nonhomologous end joining (NHEJ) and microhomology-mediated end-joining (MMEJ) for insertions/deletions (indels) without the need for a donor DNA template, depending on the kind of mutation being introduced to the plant or organism. A small homologous DNA fragment that differs from the targeted DNA template by a few base pairs is involved in SDN-2 (Wolter and Puchta, 2017).
Creation of exact and minute genetic alterations at the target site, from small indels to point mutations. Double-strand cuts are identified by the host repair system and are concurrently repaired by introducing predefined mutations and using a donor DNA fragment (a DNA template for homology-directed repair; HDR). Using a large donor DNA template of the desired gene, SDN-3 entails inserting entire DNA cassettes into a target site. If the donor comes from an unrelated species, insertion by HDR or NHEJ and a transgenic plant result. The classification of genome-edited crops as recombinant (GM) or nonrecombinant (non-GM) is a contentious issue. The safety of CRISPR-based genome-edited crops for consumption becomes crucial. To guarantee that these crops do not include unanticipated allergies or poisons, rigorous testing and procedures for risk evaluation should be implemented. Involving members of the public and interested parties in the decisions being made for genome-edited crops promotes openness and assists solve problems and aspirations. African countries investigating the utilization of genome-edited crops have to carefully evaluate whether this corresponds with trade pacts and access to market standards. Certain areas impose prohibitions on genetically modified products and crops (Wadvalla, 2022).
Schmidt et al. (2020) stated that the final product would be subject to the same legal framework as traditionally produced plant species because SDN-1 products are nearly universally recognized as non-GMO. To foster sustainable implementation, developing countries require strong and open laws and regulations for genome-edited crop use and acceptance. Responsible leadership is required to sustain the safety requirements and ethical values throughout the entire procedure. Different national policymakers have different views on the regulation and acceptance of edited crops, and the release of modified crops is governed by the policies formulated for Genetically Modified Organisms (GMOs) organizations. Concerns about ethics to motivate the deployment of genome editing technology in particular in modifying genes have possible adverse health consequences, which include allergies and toxicity (Ayanoğlu et al., 2020).
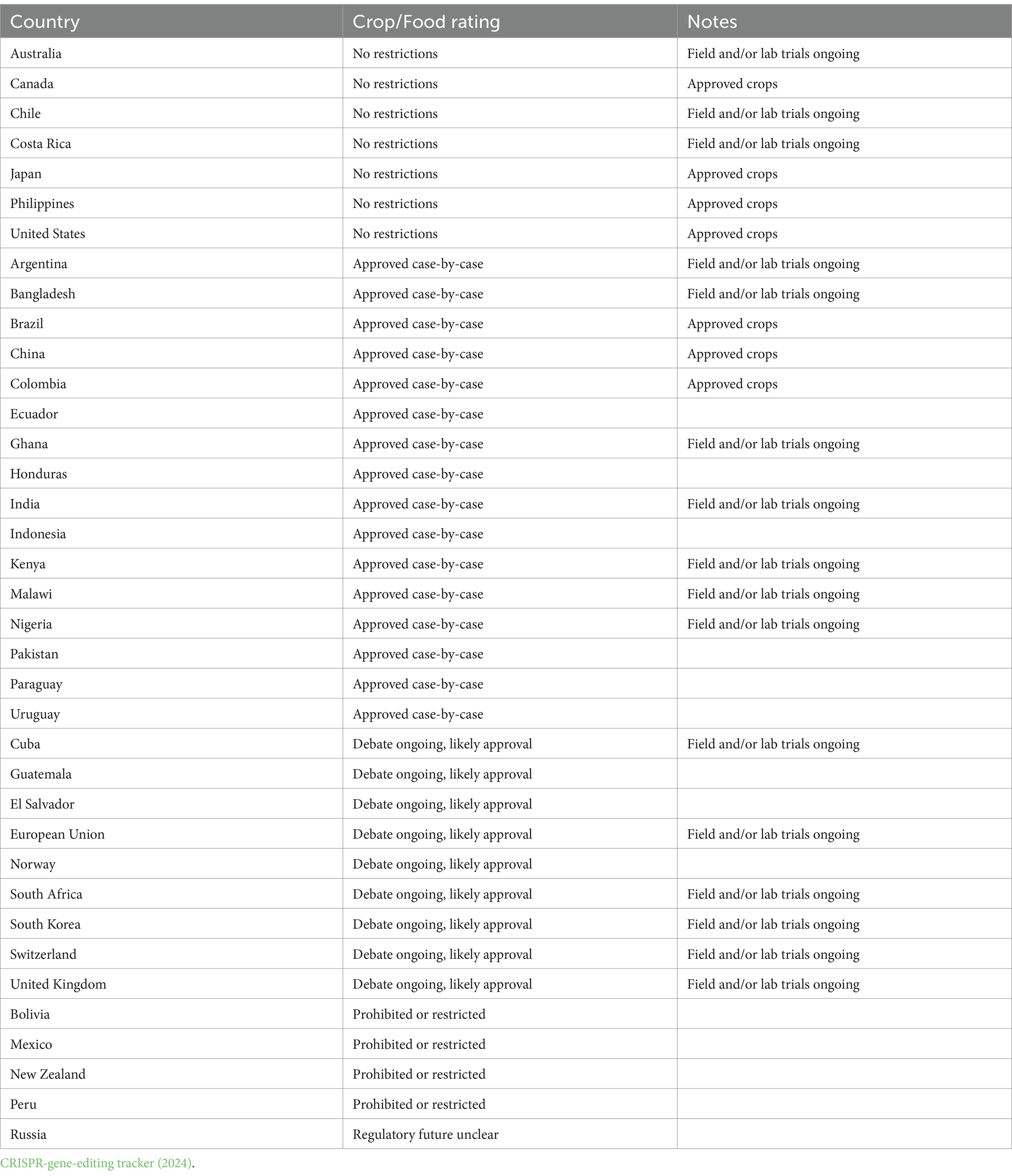
The adoption of such standards by Nigeria and Kenya supports the African Union’s Pan-African goals, and additional African nations might think about enacting similar laws to capitalize on the promise of genome modification in the agricultural sector. Voigt (2023) and Sprink and Wilhelm (2024) reported that The European Commission has suggested new restrictions for plants generated using novel genomic techniques such as genome editing. New genomic techniques offer sophisticated genetically modified approaches, such as CRISPR technology, that allow potentially greater depth and quick changes to plant genomes than conventional breeding methods. Gene editing index rating and regulatory status of some countries or regions for what crops are approved or in development, and reactions (Table 5). The widespread use of genome editing technologies in African countries raises a variety of social, and ethical issues that need to be tackled effectively to enable sustainable and long-term deployment. The continent’s different cultures have different opinions regarding editing the genome, and some people might be concerned about how genome-edited crops might impact conventional agricultural techniques. To tackle such problems and foster community acceptability, awareness-raising actions along with open discussions are essential (Tripathi et al., 2022). Resolving such social, and ethical challenges requires cooperation from governments, universities, farmers, and foreign collaborators. Developing nations can maximize the promise of genome editing technology to improve food availability and economic sustainability.
10 Conclusion and future perspectives
Ensuring food security is imperative, and increasing global food production is necessary to meet the projected rise in population by 2050. A lot of work and techniques have been put into improving crop production and improvement with high grain yield, tolerance, and resistance to extreme biotic and abiotic stresses with enhanced nutritional quality and value. However, the large and rapid increase in population will make it difficult for crop production with high quality and nutrients. Stable and strategically important crops like rice, wheat, and maize must be produced at higher levels in order to close this gap. Climate change poses a challenge to smallholder farming practices, and developing climate-resilient crops is critical for their adaptation. Genome editing of crops can be culturally acceptable by combining conventional methods with genome editing tools in Africa and developing countries. Concentrate on ecological principles, and regional expertise, especially small-scale producers. CRISPR-Cas9 technology must be democratized beforehand to enable it to be adopted, as cultural acceptability levels vary among locations. Collaboration with community members is critical to the creation and adoption of genome-edited crops. Ethical issues surrounding the safety, regulation, and gain-sharing of gene-edited products must be addressed to ensure that everyone involved benefits effectively.
To enhance and ensure the maximum use of genome editing, some recommendations have to be considered and kept in mind firstly, increase governmental funding for main crops to find genetic diversity for high quality and yield crops that can be produced and served for commercialization by the private sector. Secondly, increase funding and open more opportunities for genomic studies, and gene and trait discovery for the benefit of society, in addition to Creating incentives for start-up enterprises to use innovative breeding and genome editing methods.
Ethics statement
The studies involving human participants were reviewed and approved by Wolaiata Sodo University (WSU) ethics committee. Written informed consent to participate in this study was provided by the participants
Author contributions
FK: Conceptualization, Formal analysis, Writing – original draft, Writing – review & editing. SM: Conceptualization, Writing – original draft, Writing – review & editing. PA: Writing – original draft. UA-M: Writing – original draft. HE-A: Writing – original draft. AG: Writing – original draft. SA-M: Writing – original draft. RH: Writing – original draft. HW: Writing – original draft. RF: Writing – original draft. ON: Conceptualization, Resources, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
1. ^https://www.un.org/en/sections/issues-depth/food/index.html
2. ^http://sustainabledevelopment.org
4. ^https://who.int./news-room/fact-sheets/detail/malnutrition
References
Abe, K., Araki, E., Suzuki, Y., Toki, S., and Saika, H. (2018). Production of high oleic/low linoleic rice by genome editing. Plant Physiol. Biochem. 131, 58–62. doi: 10.1016/j.plaphy.2018.04.033
Abigarl, N., Angela, S. M., Sizo, M., and Enetia, D. B. (2024). CRISPR-Cas9 genome editing in crop breeding for climate change resilience: implications for smallholder farmers in Africa. J. Agri. Food R. 16:101132. doi: 10.1016/j.jafr.2024.101132
Ahmad, S., Tang, L., Shahzad, R., Mawia, A. M., Rao, G. S., Jamil, S., et al. (2021). CRISPR based crop improvements: a way forward to achieve zero hunger. J. Agric. Food Chem. 69, 8307–8323. doi: 10.1021/acs.jafc.1c02653
Ajeesh, K. T. P., Maharajan, T., Victor, R. G., Ignacimuthu, S., and Antony, C. S. (2020). Structure, function, regulation and phylogenetic relationship of ZIP family transporters of plants. Front. Plant Sci. 11:662. doi: 10.3389/fpls.2020.00662
Akama, K., Akter, N., Endo, H., Kanesaki, M., Endo, M., and Toki, S. (2020). An in vivo targeted deletion of the calmodulin-binding domain from rice glutamate decarboxylase 3 (OsGAD3) increases γ-aminobutyric acid content in grains. Rice 13:20. doi: 10.1186/s12284-020-00380-w
Alloway, B. (2009). Soil factors associated with zinc deficiency in crops and humans. Environ. Geochem. Health 31, 537–548. doi: 10.1007/s10653-009-9255-4
Anzalone, A. V., Randolph, P. B., Davis, J. R., Sousa, A. A., Koblan, L. W., Levy, J. M., et al. (2019). Search and- replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157. doi: 10.1038/s41586-019-1711-4
Atukunda, P., Eide, W. B., Kardel, K. R., and Westerberg, A. C. (2021). Unlocking the potential for achievement of the UN sustainable development goal 2 ‘zero hunger’–in Africa: targets, strategies, synergies and challenges. Food Nutr. Res. 65:7686. doi: 10.29219/fnr.v65.7686
Ayanoğlu, F. B., Elçin, A. E., and Elçin, Y. M. (2020). Bioethical issues in genome editing by CRISPR-Cas9 technology. Turk. J. Biol. 44, 110–120. doi: 10.3906/biy-1912-52
Baysal, C., He, W., Drapal, M., Villorbina, G., Medina, V., Capell, T., et al. (2020). Inactivation of rice starch branching enzyme IIb triggers broad and unexpected changes in metabolism by transcriptional reprogramming. Proc. Natl. Acad. Sci. USA 117, 26503–26512. doi: 10.1073/pnas.2014860117
Ben Shlush, I., Samach, A., Melamed-Bessudo, C., Ben-Tov, D., Dahan-Meir, T., Filler-Hayut, S., et al. (2021). CRISPR/Cas9 induced somatic recombination at the CRTISO locus in tomato. Genes 12:59. doi: 10.3390/genes12010059
Bezrutczyk, M., Hartwig, T., Horschman, M., Char, S. N., Yang, J., Yang, B., et al. (2018). Impaired phloem loading in zmsweet13a,b,c sucrose transporter triple knock-out mutants in Zea mays. New Phytol. 218, 594–603. doi: 10.1111/nph.15021
Boissel, S., Jarjour, J., Astrakhan, A., Adey, A., Gouble, A., Duchateau, P., et al. (2014). megaTALs: a rare-cleaving nuclease architecture for therapeutic genome engineering. Nucleic Acids Res. 42:1224. doi: 10.1093/nar/gkt1224
Boluwade, E. (2020). Government of Nigeria Approved National Biosafety Guideline on Gene Editing, 2021. December 2020, Available at: https://apps.fas.usda.gov/newgainapi/api/Report/DownloadReportByFileName?fileName=Government%20of%20Nigeria%20approved%20National%20Biosafety%20Guideline%20on%20Gene%20 Editing_Lagos_Nigeria_02-08-2021 (Accessed July 1, 2024).
Bouis, H. E., and Saltzman, A. (2017). Improving nutrition through biofortification: A review of evidence from HarvestPlus, 2003 through 2016. Glob. Food Sec. 12, 49–58. doi: 10.1016/j.gfs.2017.01.009
Bouzroud, S., Gasparini, K., Hu, G. J., MAM, B., Rosa, B. L., Fahr, M., et al. (2020). Down regulation and loss of auxin response factor 4 function using CRISPR/Cas9 alters plant growth, stomatal function and improves tomato tolerance to salinity and osmotic stress. Genes 11:272. doi: 10.3390/genes11030272
Bryan, J., Calvaresi, E., and Hughes, D. (2002). Short-term folate, vitamin B- 12 or vitamin B-6 supplementation slightly affects memory performance but not mood in women of various ages. J. Nutr. 132, 1345–1356. doi: 10.1093/jn/132.6.1345
Buchholzer, M., and Frommer, W. B. (2023). An increasing number of countries regulate genome editing in crops. New Phytol. 237, 12–15. doi: 10.1111/nph.18333
Cakmak (2008). Enrichment of cereal grains with zinc: agronomic or genetic biofortification? Plant Soil 302, 1–17. doi: 10.1007/s11104-007-9466-3
Camerlengo, F., Frittelli, A., Sparks, C., Doherty, A., Martignago, D., Larré, C., et al. (2020). CRISPR-Cas9 multiplex editing of the α-amylase/trypsin inhibitor genes to reduce allergen proteins in durum wheat. Front. Sustain. Food Syst. 4, 2571–2581. doi: 10.3389/fsufs.2020.00104
CAST (2024). Council for Agricultural Science and Technology (CAST). Applications, benefits, and challenges of genome edited crops. Issue Paper 74. Ames, Iowa: CAST.
Cermák, T., Baltes, N. J., Cegan, R., Zhang, Y., and Voytas, D. F. (2015). High-frequency, precise modification of the tomato genome. Genome Biol. 16, 1–15. doi: 10.1186/s13059-015-0796-9
Chandrasekaran, J., Brumin, M., Wolf, D., Leibman, D., Klap, C., Pearlsman, M., et al. (2016). Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 17, 1140–1153. doi: 10.1111/mpp.12375
Chao, S., Cai, Y., Feng, B., Jiao, G., Sheng, Z., Luo, J., et al. (2019). Editing of rice Isoamylase gene ISA1 provides insights into its function in starch formation. Rice Sci. 26, 77–87. doi: 10.1016/j.rsci.2018.07.001
Che, J., Yamaji, N., and Ma, J. F. (2021). Role of a vacuolar iron transporter OsVIT2 in the distribution of iron to rice grains. New Phytol. 230, 1049–1062. doi: 10.1111/nph.17219
Cheng, A. W., Wang, H. Y., Yang, H., Shi, L., Katz, Y., Theunissen, T. W., et al. (2013). Multiplexed activation of endogenous genes by CRISPR-on, an RNA guided transcriptional activator system. Cell Res. 23, 1163–1171. doi: 10.1038/cr.2013.122
Chiwona, K. L., Amuakwa-M, F., Wamala, L. C., Amuakwa- Mensah, S., Hatab, A. A., Made, N., et al. (2021). COVID-19: from health crises to food security anxiety and policy implications. Ambio 50, 794–811. doi: 10.1007/s13280-020-01481-y
Connorton, J. M., Jones, E. R., Rodríguez-Ramiro, I., Fairweather-Tait, S., Uauy, C., and Balk, J. (2017). Wheat vacuolar iron transporter TaVIT2 transports Fe and Mn and is effective for biofortification. Plant Physiol. 174, 2434–2444. doi: 10.1104/pp.17.00672
CRISPR-gene-editing tracker (2024). Available at: www.crispr-gene-editing-regs-tracker.geneticliteracyproject.org/ (Accessed July 3, 2024).
Cui, Y., Jiang, N., and Xu, Z. J. (2020). Heterotrimeric G protein are involved in the regulation of multiple agronomic traits and stress tolerance in rice. BMC Plant Biol. 20:90. doi: 10.1186/s12870-020-2289-6
D’Ambrosio, C., Stigliani, A. L., and Giorio, G. (2018). CRISPR/Cas9 editing of carotenoid genes in tomato. Transgenic Res. 27, 367–378. doi: 10.1007/s11248-018-0079-9
Daboussi, F., Leduc, S., Maréchal, A., Dubois, G., Guyot, V., Perez-Michaut, C., et al. (2014). Genome engineering empowers the diatom Phaeodactylum tricornutum for biotechnology. Nat. Commun. 5:3831. doi: 10.1038/ncomms4831
Deng, H., Tan, S. W., Gao, X. Q., Zou, C., Xu, C., Tu, K., et al. (2020). Cdk5 knocking out mediated by CRISPR-Cas9 genome editing for PD-L1 attenuation and enhanced antitumor immunity. Acta Pharm. Sin. B 10, 358–373. doi: 10.1016/j.apsb.2019.07.004
Deng, L., Wang, H., Sun, C., Li, Q., Jiang, H., Du, M., et al. (2018). Efficient generation of pink-fruited tomatoes usingCRISPR/Cas9 system. J. Genet. Genom. 45, 51–54. doi: 10.1016/j.jgg.2017.10.002
Dima, O., Custers, R., Veirman, D., and Inz’e D, L. (2023). EU legal proposal for genome-edited crops hints at a science-based approach. Trends Plant Sci. 28, 1350–1353. doi: 10.1016/j.tplants.2023.09.014
Dong, L., Qi, X., Zhu, J., Liu, C., Zhang, X., Cheng, B., et al. (2019). Super sweet and waxy: meeting the diverse demands for specialty maize by genome editing. Plant Biotechnol. J. 17, 1853–1855. doi: 10.1111/pbi.13144
Dong, O. X., Yu, S., Jain, R., Zhang, N., Duong, P. Q., Butler, C., et al. (2020). Marker–free carotenoid–enriched rice generated through targeted gene insertion using CRISPR–Cas9. Nat. Commun. 11:1178. doi: 10.1038/s41467-020-14981-y
Endo, A., Masafumi, M., Kaya, H., and Toki, S. (2016). Efficient targeted mutagenesis of rice and tobacco genomes using Cpf1 from Francisella novicida. Sci. Rep. 6:38169. doi: 10.1038/srep38169
European Parliament . (2024). New genomic techniques: MEPs back rules to support. European Parliament.
FAO . (2020). The state of food security and nutrition in the world 2020. Transforming food systems for affordable healthy diets. Rome, Italy. Available at: https://www.fao.org/publications/sofia/2020/en/ (Accessed June 8, 2024).
FAO report . (2022). The State of Food Security and Nutrition in the World 2022. Transforming food systems for affordable healthy diets. Rome, Italy. Available at: https://www.fao.org/3/cb7099en/cb7099en.pdf (Accessed June 8, 2024).
Feng, Y. L., Liu, S. C., Chen, R. D., and Xie, A. (2021). Target binding and residence: a new determinant of DNA double-strand break repair pathway choice in CRISPR/Cas9 genome editing. J. Zhejiang Univ. Sci. B Biomed. Biotechnol. 22, 73–86. doi: 10.1631/jzus.B2000282
Filiz, E., and Vatansever, R. (2018). Genome-wide identification of mildew resistance locus O (MLO) genes in tree model poplar (Populus trichocarpa): powdery mildew management in woody plants. Eur. J. Plant Pathol. 152, 95–109. doi: 10.1007/s10658-018-1454-3
Forner, J., Pfeiffer, A., Langenecker, T., Manavella, P. A., and Lohmann, J. U. (2015). Correction: germline-transmitted genome editing in Arabidopsis thaliana using TAL-effector-nucleases. PLoS One 10:0133945. doi: 10.1371/journal.pone.0133945
Gao, H., Gadlage, M. J., Lafitte, H. R., Lenderts, B., Yang, M., Schroder, M., et al. (2020). Superior field performance of waxy corn engineered using CRISPR–Cas9. Nat. Biotechnol. 38, 579–581. doi: 10.1038/s41587-020-0444-0
Genc, Y., Humphries, J. M., Lyons, G. H., and Graham, R. D. (2005). Exploriting genotypic variation in plant nutrient accumulation to alternative micronutrient deficiency in populations. J. Trace Elem. Med. Biol. 18, 319–324. doi: 10.1016/j.jtemb.2005.02.005
GHI . (2023). Available at: https://www.globalhungerindex.org/ (Accessed July 7, 2024).
Giller, K. E. (2020). The food security conundrum of sub-Saharan Africa. Glob. Food Sec. 26:100431. doi: 10.1016/j.gfs.2020.100431
Global Report on Food Crises . (2022). Available at: www.reliefweb.int/ (Accessed July 3, 2024).
Gordon, S. (2003). Alternative activation of macrophages. Nat. Rev. Immunol. 3, 23–35. doi: 10.1038/nri978
Graham, R. D., Welch, R. M., and Bouis, H. E. (2001). Addressing micronutrient malnutrition through enhancing the nutritional quality of staple foods: principles, perspectives and knowledge gaps. Adv. Agron. 70, 77–142. doi: 10.1016/S0065-2113(01)70004-1(01)70004-1
Green Transition of Farmers . (2024). European Parliament. Available at: https://go.nature.com/3VUn5Ia. (Accessed February 7, 2024).
Guarango, P. M. (2022). Guidelines for determining the regulatory process of genome edited organisms and products in Kenya. Kenya: National Biosafety Authority of Kenya.
Haas, J. D., Beard, J. L., Murray-Kolb, A. M., Del, M., Felix, A., and Gregorio, G. (2005). Iron biofortified rice improves the iron stores of non-anaemic Filipino women. J. Nutr. 135, 2823–2830. doi: 10.1093/jn/135.12.2823
Hashida, Y., Hirose, T., Okamura, M., Hibara, K., Ohsugi, R., and Aoki, N. (2016). A reduction of sucrose phosphate synthase (SPS) activity affects sucrose/starch ratio in leaves but does not inhibit normal plant growth in rice. Plant Sci. 253, 40–49. doi: 10.1016/j.plantsci.2016.08.017
Hillesund, E. R., Øverby, N. C., Valen, E. L., and Engeset, D. (2021). Alcohol consumption among students and its relationship with nutritional intake: a cross-sectional study. Public Health Nutr. 24, 2877–2888. doi: 10.1017/S1368980020004450
Horuz, A., Turan, M., Akinoglu, G., Ozcan, C., Gunes, A., Korkmaz, A., et al. (2019). Effects of phosphogypsum waste application on corn (Zea mays L.) yield and nutrient contents. Fresenius Environ. Bull. 28, 8814–8822.
Hua, K., Tao, X., Liang, W., Zhang, Z., Gou, R., and Zhu, J. K. (2020). Simplified adenine base editors improve adenine base editing efficiency in rice. Plant Biotech. J. 18, 770–778. doi: 10.1111/pbi.13244
Ibrahim, S., Saleem, B., Rehman, N., Zafar, S. A., Naeem, M. K., and Khan, M. R. (2021). CRISPR/Cas9 mediated disruption of inositol Pentakisphosphate 2-kinase 1(TaIPK1) reduces phytic acid and improves iron and zinc accumulation in wheat grains. J adv Vet 37, 33–41. doi: 10.1016/j.jare.2021.07.006
IFPRI . (2013). Available at: https://www.ifpri.org/sites/default/files/ghi/2014/feature_1818.html (Accessed July 3 2024).
IRRI Rice Knowledge Bank . (2013). Available at: knowledgebank.irri.org. (Accessed April 20, 2013).
Jiang, M., Liu, Y., Liu, Y., Tan, Y., Huang, J., and Shu, Q. (2019). Mutation of inositol 1,3,4-trisphosphate 5/6-kinase6 impairs plant growth and Phytic acid synthesis in Rice. Plan. Theory 8:114. doi: 10.3390/plants8050114
Jimenez, K., Kulnigg-Dabsch, S., and Gasche, C. (2015). Management of iron deficiency anemia. Gastroenterol. Hepatol. 11, 241–250.
Jin, S., Zong, Y., Gao, Q., Zhu, Z., Wang, Y., Qin, P., et al. (2019). Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science 364, 292–295. doi: 10.1126/science.aaw7166
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Jinek, M., Jiang, F., Taylor, D. W., Sternberg, S. H., Kaya, E., Ma, E., et al. (2014). Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 343:1247997. doi: 10.1126/science.1247997
Jung, Y. J., Lee, H. J., Kim, J. H., Kim, D. H., Kim, H. K., Cho, Y. G., et al. (2019). CRISPR/Cas9-targeted mutagenesis of F3′H, DFR and LDOX, genes related to anthocyanin biosynthesis in black rice (Oryza sativa L.). Plant. Biotechnol. Rep. 13, 521–531. doi: 10.1007/s11816-019-00579-4
Kantor, A., McClements, M. E., and MacLaren, R. E. (2020). CRISPR-Cas9 DNA Base-editing and prime-editing. Int. J. Mol. Sci. 21:6240. doi: 10.3390/ijms21176240
Khaled, F. A. M., Kaul, T., Agrawal, P. K., Sopory, S. K., and Nanda, R. (2023). Improving rice quality by enhancing Fe-Zn content in grains using multiplex CRISPR/Cas9-based genome editing. PhD thesis. New Delhi: International Centre for Genetic Engineering and Biotechnology.
Khaled, F. A. M., Tanushri, K., Pawan, K. A., Arulprakash, T., Rashmi, K., and Sudhir, K. S. (2022). Function identification and characterization of Oryza sativa ZRT and IRT-like proteins computationally for nutrition and biofortification in rice. J. Biomol. Struct. Dyn. 41, 7490–7510. doi: 10.1080/07391102.2022.2118169
Khan, M. S. S., Basnet, R., Islam, S. A., and Shu, Q. (2019). Mutational analysis of OsPLD1 reveals its involvement in phytic acid biosynthesis rice grains. J. Agric. Food Chem. 67, 11436–11443. doi: 10.1021/acs.jafc.9b05052
Kim, Y. A., Moon, H., and Park, C. J. (2019). CRISPR/Cas9-targeted mutagenesis of Os8N3 in rice to confer resistance to Xanthomonas oryzae pv. Oryzae. Rice 12:67. doi: 10.1186/s12284-019-0325-7
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A., and Liu, D. R. (2016). Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424. doi: 10.1038/nature17946
Konermann, S., Brigham, M. D., and Trevino, A. E. (2013). Optical control of mammalian endogenous transcription and epigenetic states. Nature 500, 472–476. doi: 10.1038/nature12466
Konermann, S., Brigham, M. D., and Trevino, A. E. (2015). Genome scale transcriptional activation by an engineered CRISPRCas9 complex. Nature 517, 583–588. doi: 10.1038/nature14136
Koonin, E. V., Makarova, K. S., and Zhang, F. (2017). Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 37, 67–78. doi: 10.1016/j.mib.2017.05.008
Kumar, P., Malik, Y. S., Ganesh, B., Rahangdale, S., Saurabh, S., Natesan, S., et al. (2020). CRISPR-Cas system: an approach with potentials for COVID-19 diagnosis and therapeutics. Front. Cell. Infect. Microbiol. 10:576875. doi: 10.3389/fcimb.2020.576875
Kumssa, D. B., Joy, E. J., Ander, E. L., Watts, M. J., Young, S. D., Walker, S., et al. (2015). Dietary calcium and zinc deficiency risks are decreasing but remain prevalent. Sci. Rep. 5:10974. doi: 10.1038/srep10974
Lee, K., Zhang, Y., Kleinstiver, B. P., Guo, J. A., Aryee, M. J., Miller, J., et al. (2019). Activities and specificities of CRISPR/Cas9 and Cas12a nucleases for targeted mutagenesis in maize. Plant Biotechnol. J. 17, 362–372. doi: 10.1111/pbi.12982
Li, J., Meng, X., Zong, Y., Chen, K., Zhang, H., Liu, J., et al. (2016). Gene replacements and insertions in rice by intron targeting using CRISPR–Cas9. Nat. Plants 2:16139. doi: 10.1038/nplants.2016.139
Li, M., Li, X., Zhou, Z., Wu, P., Fang, M., Pan, X., et al. (2016). Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in Rice using a CRISPR/Cas9 system. Front. Plant Sci. 7:377. doi: 10.3389/fpls.2016.00377
Li, H., Pinot, F., Sauveplane, V., Werck-Reichhart, D., Diehl, P., Schreiber, L., et al. (2010). Cytochrome P450 family member CYP704B2 catalyzes the hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. Plant Cell 22, 173–190. doi: 10.1105/tpc.109.070326
Li, J., Sun, Y., Du, J., Zhao, Y., and Xia, L. (2017). Generation of targeted point mutations in Rice by a modified CRISPR/Cas9 system. Mol. Plant 10, 526–529. doi: 10.1016/j.molp.2016.12.001
Li, C., Zong, Y., Wang, Y., Jin, S., Zhang, D., Song, Q., et al. (2018). Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol. 19, 59. doi: 10.1186/s13059-018-1443-z
Li, X., Wang, Y., Chen, S., Tian, H., Fu, D., Zhu, B., et al. (2018). Lycopene is enriched in tomato fruit by CRISPR/Cas9-mediated multiplex genome editing. Front. Plant Sci. 9:559. doi: 10.3389/fpls.2018.00559
Li, C., Zhang, R., Meng, X. B., Chen, S., Zong, Y., Lu, C., et al. (2020). Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors. Nat. Biotechnol. 38, 875–882. doi: 10.1038/s41587-019-0393-7
Li, X., Zhou, W., Ren, Y., Tian, X., Lv, T., Wang, Z., et al. (2017). High-efficiency breeding of early-maturing rice cultivars via CRISPR/Cas9-mediated genome editing. J. Genet. Genom. 44, 175–178. doi: 10.1016/j.jgg.2017.02.001
Liang, Z., Chen, K., Zhang, Y., Liu, J., Yin, K., Qiu, J. L., et al. (2018). Genome editing of bread wheat using biolistic delivery of CRISPR/Cas9 in vitro transcripts or ribonucleoproteins. Nat. Protoc. 13, 413–430. doi: 10.1038/nprot.2017.145
Liang, Z., Zhang, K., Chen, K., and Gao, C. (2014). Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J. Genet. Genomics 41, 63–68. doi: 10.1016/j.jgg.2013.12.001
Lin, S., Staahl, B. T., Alla, R. K., and Doudna, J. A. (2014). Enhanced homology directed human genome engineering by controlled timing of CRISPR/ Cas9 delivery. eLife 3:304766. doi: 10.7554/eLife.04766
Lin, Q. P., Zong, Y., Xue, C. X., Wang, S., Jin, S., Zhu, Z., et al. (2020). Prime genome editing in rice and wheat. Nat. Biotechnol. 38, 582–585. doi: 10.1038/s41587-020-0455-x
Liu, L., Gallagher, J., Arevalo, E. D., Chen, R., Skopelitis, T., Wu, Q., et al. (2021). Enhancing grain-yield-related traits by CRISPR-Cas9 promoter editing of maize CLE genes. Nat. Plants 7, 287–294. doi: 10.1038/s41477-021-00858-5
Liu, J., Orlova, N., Oakes, B. L., Ma, E., Spinner, H. B., Baney, K. L. M., et al. (2019). CasX enzymes comprise a distinct family of RNA-guided genome editors. Nature 566, 218–223. doi: 10.1038/s41586-019-0908-x
Liu, Y., Wang, W. L., Yang, B., Currey, C., and Fei, S. Z. (2020). Functional analysis of the teosinte branched 1 gene in the tetraploid switchgrass (Panicum virgatum L.) by CRISPR/Cas9-directed mutagenesis. Front. Plant Sci. 11:572193. doi: 10.3389/fpls.2020.572193
Ma, L., Zhang, D., Miao, Q., Yang, J., Xuan, Y., and Hu, Y. (2017). Essential role of sugar transporter OsSWEET11 during the early stage of rice grain filling. Plant Cell Physiol. 58, 863–873. doi: 10.1093/pcp/pcx040
Macovei, A., Sevilla, N. R., Cantos, C., Jonson, G. B., Slamet-Loedin, I., Cermák, T., et al. (2018). Novel alleles of rice eIF4G generated by CRISPR/Cas9-targeted mutagenesis confer resistance to Rice tungro spherical virus. Plant Biotechnol. J. 16, 1918–1927. doi: 10.1111/pbi.12927
Maeder, M. L., Thibodeau-Beganny, S., Osiak, A., Wright, D. A., Anthony, R. M., Eichtinger, M., et al. (2008). Rapid "open-source" engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol. Cell 31, 294–301. doi: 10.1016/j.molcel.2008.06.016
Maggini, S., Maldonado, P., Cardim, P., Fernandez Newball, C., and Sota Latino, E. (2017). Vitamins C, D and zinc: synergistic roles in immune function and infections. Vitamin Miner. 6:167. doi: 10.4172/2376-1318.1000167
Majumder, S., Datta, K., and Datta, S. K. (2019). Rice biofortification: high Iron, zinc, and vitamin-A to fight against —hidden hunger. Agronomy 9, –803. doi: 10.3390/agronomy9120803
Makarova, K. S., Wolf, Y. I., Iranzo, J., Shmakov, S. A., Alkhnbashi, O. S., Brouns, S. J. J., et al. (2020). Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 18, 67–83. doi: 10.1038/s41579-019-0299-x
Malik, K. A., and Maqbool, A. (2020). Transgenic crops for biofortification. Front. Sustain. Food Syst. 4:571402. doi: 10.3389/fsufs.2020.571402
Malzahn, A., Lowder, L., and Qi, Y. (2017). Plant genome editing with TALEN and CRISPR. Cell Biosci. 7:21. doi: 10.1186/s13578-017-0148-4
Maniragaba, V. N., Atuhaire, L. K., and Rutayisire, P. C. (2023). Undernutrition among the children below five years of age in Uganda: a spatial analysis approach. BMC Public Health 23:390. doi: 10.1186/s12889-023-15214-9
Mao, X., Zheng, Y., Xiao, K., Wei, Y., Zhu, Y., Cai, Q., et al. (2018). OsPRX2 contributes to stomatal closure and improves potassium deficiency tolerance in rice. Biochem. Biophys. Res. Commun. 495, 461–467. doi: 10.1016/j.bbrc.2017.11.045
Martínez-Fortún, J., Phillips, D. W., and Jones, H. D. (2022). Natural and artificial sources of genetic variation used in crop breeding: a baseline comparator for genome editing. Front. Genome Ed. 4:937853. doi: 10.3389/fgeed.2022.937853
Masehela, T. S., and Barros, E. (2023). The African continent should consider a harmonized consultative and collaborative effort towards coordinated policy and regulatory guidelines across the fields of biotechnology. Front. Bioeng. Biotechnol. 11:1211789. doi: 10.3389/fbioe.2023.1211789
Mendelsohn, R. (2008). The impact of climate change on agriculture in developing countries. J. Nat. Resour. Pol. Res. 1, 5–19. doi: 10.1080/19390450802495882
Miao, C., Xiao, L., Hua, K., Zou, C., Zhao, Y., Bressan, R. A., et al. (2018). Mutations in a subfamily of abscisic acid receptor genes promote rice growth and productivity. Proc. Natl. Acad. Sci. USA 115, 6058–6063. doi: 10.1073/pnas.1804774115
Mohajan, H. K. (2022). Food insecurity and malnutrition of Africa: a combined attempt can reduce them. J. Econ. Dev. Environ. People 11, 24–34. doi: 10.26458/jedep.v1i1.716
Mohanraju, P., Makarova, K. S., Zetsche, B., Zhang, F., Koonin, E. V., and van der Oost, J. (2016). Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science 353:5147. doi: 10.1126/science.aad5147
Mojica, F. J. M., Díez-Villaseñor, C., García-Martínez, J., and Soria, E. (2005). Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 60, 174–182. doi: 10.1007/s00239-004-0046-3
Montenegro, M. (2020). Democratizing CRISPR? Stories, practices, and politics of science and governance on the agricultural gene editing frontier. Elem Sci Anth 8:405. doi: 10.1525/elementa.405
Naidoo, R., and Fisher, B. (2020). Reset sustainable development goals for a pandemic world. Nature 583, 198–201. doi: 10.1038/d41586-020-01999-x
National Research Council (US) (1989). Recommended dietary allowances. 10th Edn. Washington, DC: National Academies Press (US).
Nieves-Cordones, M., Mohamed, S., Tanoi, K., Kobayashi, N. I., Takagi, K., Vernet, A., et al. (2017). Production of low-Cs + rice plants by inactivation of the K+ transporter OsHAK1 with the CRISPR -Cas system. Plant J. 92, 43–56. doi: 10.1111/tpj.13632
Nishimasu, H., Ran, F. A., Hsu, P. D., Konermann, S., Shehata, S. I., Dohmae, N., et al. (2014). Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156, 935–949. doi: 10.1016/j.cell.2014.02.001
Nogué, F., Causse, M., Debaeke, P., Déjardin, A., Lemarié, S., Richard, G., et al. (2024). Can genome editing help transitioning to agroecology? iScience 27:109159. doi: 10.1016/j.isci.2024.109159
Nonaka, S., Arai, C., Takayama, M., Matsukura, C., and Ezura, H. (2017). Efficient increase of ɣ aminobutyric acid (GABA) content in tomato fruits by targeted mutagenesis. Sci. Rep. 7:705. doi: 10.1038/s41598-017-06400-y
Ogaugwu, C. E., Agbo, S. O., and Adekoya, M. A. (2019). CRISPR in sub-Saharan Africa: applications and education. Trends Biotechnol. 37, 234–237. doi: 10.1016/j.tibtech.2018.07.012
Ongu, I., Olayide, P., Alexandersson, E., Mugwanya Zawedde, B., and Eriksson, D. (2023). Biosafety regulatory frameworks in Kenya, Nigeria, Uganda and Sweden and their potential impact on international R&D collaborations. GM Crops Food 14, 1–17. doi: 10.1080/21645698.2023.2194221
Ortigosa, A., Gimenez-Ibanez, S., Leonhardt, N., and Solano, R. (2019). Design of a bacterial speck resistant tomato by CRISPR/Cas9-mediated editing of SlJAZ2. Plant Biotechnol. J. 17, 665–673. doi: 10.1111/pbi.13006
Osendarp, S., Akuaoku, J. K., Black, R., Headey, D., Ruel, M., Scott, N., et al. (2021). The COVID-19 crisis will exacerbate maternal and child undernutrition and child mortality in low- and middle-income countries. Nat. Food 2, 476–484. doi: 10.1038/s43016-021-00319-4
Otekunrin, O., Sawicka, B., and Ayinde, I. (2020). Three decades of fighting against hunger in Africa: Progress, challenges and opportunities. World Nutr. 11, 86–111. doi: 10.26596/wn.202011386-111
Pan, Z., Choi, S., Ouadid-Ahidouch, H., Yang, J. M., Beattie, J. H., and Korichneva, I. (2017). Zinc transporters and dysregulated channels in cancers. Front. Biosci. (Landmark Ed) 22, 623–643. doi: 10.2741/4507
Pannunzio, N. R., Watanabe, G., and Lieber, M. R. (2017). Nonhomologous DNA end joining for repair of DNA double-Strand breaks. J. Biol. Chem. 293, 10512–10523. doi: 10.1074/jbc.TM117.000374
Pathi, K. M., Rink, P., Budhagatapalli, N., Betz, R., Saado, I., Hiekel, S., et al. (2020). Engineering smut resistance in maize by site-directed mutagenesis of LIPOXYGENASE 3. Front. Plant Sci. 11:543895. doi: 10.3389/fpls.2020.543895
Peng, A. H., Chen, S. C., Lei, T. G., Xu, L., He, Y., Wu, L., et al. (2017). Engineering canker resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol. J. 15, 1509–1519. doi: 10.1111/pbi.12733
Peng, R. H., Jones, D. C., Liu, F., and Zhang, B. (2021). From sequencing to genome editing for cotton improvement. Trends Biotechnol. 39, 221–224. doi: 10.1016/j.tibtech.2020.09.001
Pickson, R. B., and Boateng, E. (2022). Climate change: a friend or foe to food security in Africa? Environ. Dev. Sustain. 24, 4387–4412. doi: 10.1007/s10668-021-01621-8
Pröbsting, M., Schenke, D., Hossain, R., Häder, C., Thurau, T., Wighardt, L., et al. (2020). Loss of function of CRT1A (calreticulin) reduces plant susceptibility to Verticillium longisporum in both Arabidopsis thaliana and oilseed rape (Brassica napus). Plant Biotechnol. J. 18, 2328–2344. doi: 10.1111/pbi.13394
Qi, L. S., Larson, M. H., Gilbert, L. A., Doudna, J. A., Weissman, J. S., Arkin, A. P., et al. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183. doi: 10.1016/j.cell.2013.02.022
Richter, M. F., Zhao, K. T., Eton, E., Lapinaite, A., Newby, G. A., Thuronyi, B. W., et al. (2020). Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat. Biotechnol. 38, 883–891. doi: 10.1038/s41587-020-0453-z
Rizvi, S., Raza, S. T., Ahmed, F., Ahmad, A., Abbas, S., and Mahdi, F. (2014). The role of vitamin E in human health and some diseases. Sultan Qaboos Univ. Med. J. 14, e157–e165
Roca Paixão, J. F., Gillet, F. X., Ribeiro, T. P., Bournaud, C., Lourenço-Tessutti, I. T., Noriega, D. D., et al. (2019). Improved drought stress tolerance in Arabidopsis by CRISPR/dCas9 fusion with a histone acetyltransferase. Sci. Rep. 9:8080. doi: 10.1038/s41598-019-44571-y
Rodríguez-Leal, D., Lemmon, Z. H., Man, J., Bartlett, M. E., and Lippman, Z. B. (2017). Engineering quantitative trait variation for crop improvement by genome editing. Cell 171, 470–480.e8. doi: 10.1016/j.cell.2017.08.030
Sanchez-Leon, S., Gil-Humanes, J., Ozuna, C. V., Gimenez, M. J., Sousa, C., Voytas, D. F., et al. (2018). Low-gluten, non transgenic wheat engineered with CRISPR/Cas9. Plant Biotechnol. J. 16, 902–910. doi: 10.1111/pbi.12837
Sander, J. D., and Joung, J. K. (2014). CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 32, 347–355. doi: 10.1038/nbt.2842
Santillán Martínez, M. I., Bracuto, V., Koseoglou, E., Appiano, M., Jacobsen, E., Visser, R. G. F., et al. (2020). CRISPR/Cas9-targeted mutagenesis of the tomato susceptibility gene PMR4 for resistance against powdery mildew. BMC Plant Biol. 20:284. doi: 10.1186/s12870-020-02497-y
Schmidt, S. M., Belisle, M., and Frommer, W. B. (2020). The evolving landscape around genome editing in agriculture: many countries have exempted or move to exempt forms of genome editing from GMO regulation of crop plants. EMBO Rep. 21:e50680. doi: 10.15252/embr.202050680
Scholefield, J., and Harrison, P. T. (2021). Prime editing – an update on the field. Gene Ther. 28, 396–401. doi: 10.1038/s41434-021-00263-9
Serdeczny, O., Adams, S., Baarsch, F., Coumou, D., Robinson, A., Hare, W., et al. (2017). Climate change impacts in sub-Saharan Africa: from physical changes to their social repercussions. Reg. Environ. Chang. 17, 1585–1600. doi: 10.1007/s10113-015-0910-2
Shan, Q., Wang, Y., Li, J., Zhang, Y., Chen, K., Liang, Z., et al. (2013). Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 31, 686–688. doi: 10.1038/nbt.2650
Shao, G., Xie, L., Jiao, G., Wei, X., Sheng, Z., Tang, S., et al. (2017). CRISPR/CAS9-mediated editing of the fragrant gene Badh2 in rice. Chin. J. Rice Sci. 31, 216–222. doi: 10.1111/ppl.13871
Sharma, S., Kaur, R., and Singh, A. (2017). Recent advances in CRISPR/Cas mediated genome editing for crop improvement. Plant Biotechnol. Rep. 11, 193–207. doi: 10.1007/s11816-017-0446-7
Shi, J., Gao, H., Wang, H., Lafitte, H. R., Archibald, R. L., Yang, M., et al. (2017). ARGOS 8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 15, 207–216. doi: 10.1111/pbi.12603
Sillanpää, M. (1990). “Micronutrient assessment at country level: an international study” in Soils Bulletin No. 63. ed. M. Sillanpää (Rome, Italy: FAO), 208.
Silva, G., Poirot, L., Galetto, R., Smith, J., Montoya, G., Duchateau, P., et al. (2011). Meganucleases and other tools for targeted genome engineering: perspectives and challenges for gene therapy. Curr. Gene Ther. 11, 11–27. doi: 10.2174/156652311794520111
Simpson, J. L., Bailey, L. B., Pietrzik, K., Shane, B., and Holzgreve, W. (2011). Micronutrients and women of reproductive potential: required dietary intake and consequences of dietary deficiency or excess. J. Matern. Neonatal Med. 24, 1–24. doi: 10.3109/14767051003678226
Song, G., Jia, M., Chen, K., Kong, X., Khattak, B., Xie, C., et al. (2016). CRISPR/Cas9: A powerful tool for crop genome editing. Crop J. 4, 75–82. doi: 10.1016/j.cj.2015.12.002
Songmei, L., Jiang, J., Liu, Y., Meng, J., Xu, S., Tan, Y., et al. (2019). Characterization and evaluation of OsLCT1 and OsNramp5 mutants generated through CRISPR/Cas9-mediated mutagenesis for breeding low cd Rice. Rice Sci. 26, 88–97. doi: 10.1016/j.rsci.2019.01.002
Sprink, T., and Wilhelm, R. (2024). “Genome editing in biotech regulations worldwide” in A Roadmap for Plant Genome Editing. eds. A. Ricroch, D. Eriksson, D. Miladinovic, J. Sweet, K. Van Laere, and E. W. Gientka (Cham: Springer).
Statistica . (2024). Available at: https://www.statista.com/statistics/ (Accessed January 8, 2024).
Stein, A. J. (2010). Global impacts of human mineral malnutrition. Plant Soil 335, 133–154. doi: 10.1007/s11104-009-0228-2
Sun, Y., Jiao, G., Liu, Z., Zhang, X., Li, J., Guo, X., et al. (2017). Generation of high-amylose rice through CRISPR/Cas9-mediated targeted mutagenesis of starch branching enzymes. Front. Plant Sci. 8:298. doi: 10.3389/fpls.2017.00298
Sun, S. K., Xu, X. J., Tang, Z., Tang, Z., Huang, X. Y., Wirtz, M., et al. (2021). A molecular switch in sulfur metabolism to reduce arsenic and enrich selenium in rice grain. Nat. Commun. 12:1392. doi: 10.1038/s41467-021-21282-5
Sun, Y., Zhang, X., Wu, C., He, Y., Ma, Y., Hou, H., et al. (2016). Engineering herbicide-resistant rice plants through CRISPR/Cas9-mediated homologous recombination of acetolactate synthase. Mol. Plant 9, 628–631. doi: 10.1016/j.molp.2016.01.001
Svitashev, S., Young, J. K., Schwartz, C., Gao, H., Falco, S. C., and Cigan, A. M. (2015). Targeted mutagenesis, precise gene editing and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 169, 931–945. doi: 10.1104/pp.15.00793
Takeda, Y., Suzuki, S., Tobimatsu, Y., Osakabe, K., Osakabe, Y., Ragamustari, S. K., et al. (2019). Lignin characterization of rice CONIFERALDEHYDE 5-HYDROXYLASE loss-of-function mutants generated with the CRISPR/Cas9 system. Plant J. 97, 543–554. doi: 10.1111/tpj.14141
Tang, X., Lowder, L. G., Zhang, T., Malzahn, A. A., Zheng, X., Voytas, D. F., et al. (2017). A CRISPR–Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat. Plants 3, 1–5. doi: 10.1038/nplants.2017.18
Tang, L., Mao, B., Li, Y., Lv, Q., Zhang, L., Chen, C., et al. (2017). Knockout of OsNramp5 using the CRISPR/Cas9 system produces low cd-accumulating indica rice without compromising yield. Sci. Rep. 7:14438. doi: 10.1038/s41598-017-14832-9
The Bill and Melinda Gates Foundation (2020). COVID-19: A global Perspectives-2020 goal keepers report. Washington, DC: The Bill and Melinda Gates Foundation.
Tripathi, L., Dhugga, K. S., Ntui, V. O., Runo, S., Syombua, E. D., Muiruri, S., et al. (2022). Genome editing for sustainable agriculture in Africa. Front. Genome Ed. 4:697. doi: 10.3389/fgeed.2022.876697
United Nations . (2021a). Secretary-General’s chair summary and statement of action on the UN food systems summit. Food Systems Summit, September 23. https://www.un.org/en/food-systems-summit/news/making-food-systems-work-people-planet-and-prosperity (Accessed July 3, 2024).
United Nations . (2021b). Nearly 300 commitments highlights Summit’s inclusive process to accelerate action. September 23. https://www.un.org/en/food-syste ms-summit/news/nearly-300-commitmentshighlights-summit%E280%99s-inclusive-process (Accessed July 3, 2024).
United Nations . (2024a). Available at: http://sustainabledevelopment.un.org (Accessed July 3, 2024).
United Nations . (2024b). Available at: https://www.un.org/en/sections/issues-depth/food/index.html (Accessed July 3, 2024).
Veillet, F., Kermarrec, M.-P., Chauvin, L., Chauvin, J. E., and Nogué, F. (2020). CRISPR-induced indels and base editing using the Staphylococcus aureus Cas9 in potato. PLoS One 15:e0235942. doi: 10.1371/journal.pone.0235942
Voigt, B. (2023). EU regulation of gene-edited plants—a reform proposal. Front. Genome Ed. 5. doi: 10.3389/fgeed.2023.1119442
Vu, T. V., Sivankalyani, V., Kim, E.-J., Doan, D. T. H., Tran, M. T., Kim, J., et al. (2020). Highly efficient homology-directed repair using CRISPR/Cpf1-geminiviral replicon in tomato. Plant Biotechnol. J. 18, 2133–2143. doi: 10.1111/pbi.13373
Wachira, K. (2022). University world news Africa edition: Countries spend less than 1% of GDP on research, in: In proceedings of the IAU 2022, 16th general conference. Dublin, Ireland: University World News Africa Edition. University World News, London.
Wadvalla, B. (2022). Advancing biotechnology to solve Africa’s food challenges. Natl. Biotechnol. 40, 904–907. doi: 10.1038/d44148-022-00106-8
Wang, D., Samsulrizal, N. H., Yan, C., Allcock, N. S., Craigon, J., Blanco-Ulate, B., et al. (2019). Characterization of CRISPR mutants targeting genes modulating pectin degradation in ripening tomato. Plant Physiol. 179, 544–557. doi: 10.1104/pp.18.01187
Wang, L. J., Chen, S. C., Peng, A. H., Xie, Z., He, Y., Zou, X., et al. (2019). CRISPR/Cas9-mediated editing of CsWRKY22 reduces susceptibility to Xanthomonas citri subsp. citri in Wanjincheng orange (Citrus sinensis (L.) Osbeck). Plant Biotechnol. Rep. 13, 501–510. doi: 10.1007/s11816-019-00556-x
Wang, T., Li, Y., Fu, Y., Xie, H., Song, S., Qiu, M., et al. (2019). Mutation at different sites of metal transporter gene OsNramp5 affects cd accumulation and related agronomic traits in Rice (Oryza sativa L.). Front. Plant Sci. 11:1081. doi: 10.3389/fpls.2019.01081
Wang, Y., Cheng, X., Shan, Q., Zhang, Y., Liu, J., Gao, C., et al. (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. doi: 10.1038/nbt.2969
Wang, J., Kuang, H., Zhang, Z., Yang, Y., Yan, L., Zhang, M., et al. (2020). Generation of seed lipoxygenase-free soybean using CRISPR-Cas9. Crop J. 8, 432–439. doi: 10.1016/j.cj.2019.08.008
Wang, M., Mao, Y., Lu, Y., Tao, X., and Zhu, J. K. (2017). Multiplex gene editing in Rice using the CRISPR-Cpf1 system. Mol. Plant 10, 1011–1013. doi: 10.1016/j.molp.2017.03.001
Wang, Y. L., Meng, Z. G., Liang, C. Z., Meng, Z., Wang, Y., Sun, G., et al. (2017). Increased lateral root formation by CRISPR/Cas9-mediated editing of arginase genes in cotton. Sci. China Life Sci. 60, 524–527. doi: 10.1007/s11427-017-9031-y
Wang, F., Wang, C., Liu, P., Lei, C., Hao, W., Gao, Y., et al. (2016). Enhanced Rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS One 11:e0154027. doi: 10.1371/journal.pone.0154027
Weeks, D. P., Spalding, M. H., and Yang, B. (2016). Use of designer nucleases for targeted gene and genome editing in plants. Plant Biotechnol. 14, 483–495. doi: 10.1111/pbi.12448
Welch, R. M., Graham, R. D., and Cakmak, I. (2013). Linking agricultural production practices to improving human nutrition and health, expert paper written for ICN2. In: Second International Conference on Nutrition Preparatory Technical Meeting Rome, November, 2013, pp. 13–15.
Wessells, K. R., and Brown, K. H. (2012). Estimating the global prevalence of zinc deficiency: results based on zinc availability in national food supplies and the prevalence of stunting. PLoS One 7:E50568. doi: 10.1371/journal.pone.0050568
Wolter, F., and Puchta, H. (2017). Knocking out consumer concerns and regulator’s rules: efficient use of CRISPR/Cas ribonucleoprotein complexes for genome editing in cereals. Genome Biol. 18:43. doi: 10.1186/s13059-017-1179-1
Wood, J., Wong, C., and Paturi, S. (2021). Addressing food security in constrained urban environments. Trop. Const. Environ. Sustain. Adapt. 1, 169–186. doi: 10.1007/978-981-33-4631-4_10
World Food Programme (2020). WFP global update on COVID-19: November 2020. Rome, Italy: World Food Programme.
Worldometer . (2023). Africa Population (Live). Available at: https://www.worldometers.info/world-population/africa-population/ (Accessed July 1 2024).
Wu, M. J., Liu, H. Q., Lin, Y., Chen, J., Fu, Y., Luo, J., et al. (2020). In-frame and frame-shift editing of the Ehd1 gene to develop japonica rice with prolonged basic vegetative growth periods. Front. Plant Sci. 11:307. doi: 10.3389/fpls.2020.00307
Xie, H. H., Ge, X. L., Yang, F. Y., Wang, B., Li, S., Duan, J., et al. (2020). High-fidelity SaCas9 identified by directional screening in human cells. PLoS Biol. 18:e3000747. doi: 10.1371/journal.pbio.3000747
Xie, K., and Yang, Y. (2013). RNA-guided genome editing in plants using a CRISPR–Cas system. Mol. Plant 6, 1975–1983. doi: 10.1093/mp/sst119
Xu, R., Qin, R., Li, H., Li, J., Yang, J., and Wei, P. (2019). Enhanced genome editing in rice using single transcript unit CRISPR-LbCpf1 systems. Plant Biotechnol. J. 17, 553–555. doi: 10.1111/pbi.13028
Xu, R., Yang, Y., Qin, R., Li, H., Qiu, C., Li, L., et al. (2016). Rapid improvement of grain weight via highly efficient CRISPR/Cas9-mediated multiplex genome editing in rice. J. Genet. Genom. 43, 529–532. doi: 10.1016/j.jgg.2016.07.003
Yang, S. H., Kim, E., and Park, H. (2022). Selection of the high efficient sgRNA for CRISPR-Cas9 to edit herbicide related genes, PDS, ALS, and EPSPS in tomato. Appl. Biol. Chem. 65:13. doi: 10.1186/s13765-022-00679-w
Ye, Y., Wu, K., Chen, J., Liu, Q., Wu, Y., Liu, B., et al. (2018). OsSND2, a NAC family transcription factor, is involved in secondary cell wall biosynthesis through regulating MYBs expression in rice. Rice (NY) 11:36. doi: 10.1186/s12284-018-0228-z
Yin, X., Biswal, A. K., Dionora, J., Perdigon, K. M., Balahadia, C. P., Mazumdar, S., et al. (2017). CRISPR-Cas9 and CRISPR-Cpf1 mediated targeting of a stomatal developmental gene EPFL9 in rice. Plant Cell Rep. 36, 745–757. doi: 10.1007/s00299-017-2118-z
Yu, Q.-H., Wang, B., Li, N., Tang, Y., Yang, S., Yang, T., et al. (2017). CRISPR/Cas9-induced targeted mutagenesis and gene replacement to generate long-shelf life tomato lines. Sci. Rep. 7, 1–9. doi: 10.1038/s41598-017-12262-1
Yusong, L., Gaoneng, S., Guiai, J., Zhonghua, S., Lihong, X., Shikai, H., et al. (2021). Targeted mutagenesis of POLYAMINE OXIDASE 5 that negatively regulates mesocotyl elongation enables the generation of direct-seeding rice with improved grain yield. Mol. Plant 14, 344–351. doi: 10.1016/j.molp.2020.11.007
Yuste-Lisbona, F. J., Fernández-Lozano, A., Pineda, B., Bretones, S., Ortíz-Atienza, A., García-Sogo, B., et al. (2020). ENO regulates tomato fruit size through the floral meristem development network. Proc. Natl. Acad. Sci. USA 117, 8187–8195. doi: 10.1073/pnas.1913688117
Zaidi, S. S., Mahas, A., Vanderschuren, H., and Mahfouz, M. M. (2020). Engineering crops of the future: CRISPR approaches to develop climate-resilient and disease-resistant plants. Genome Biol. 21:289. doi: 10.1186/s13059-020-02204-y
Zeng, D., Li, X., Huang, J., Li, Y., Cai, S., Yu, W., et al. (2020). Engineered Cas9 variant tools expand targeting scope of genome and base editing in rice. Plant Biotechnol. J. 18, 1348–1350. doi: 10.1111/pbi.13293
Zhang, Y., Bai, Y., Wu, G., Zou, S., Chen, Y., Gao, C., et al. (2017). Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat. Plant J. 91, 714–724. doi: 10.1111/tpj.13599
Zhang, Z. N., Ge, X. Y., Luo, X. Y., Wang, P., Fan, Q., Hu, G., et al. (2018). Simultaneous editing of two copies of Gh14-3-3d confers enhanced transgene-clean plant defense against Verticillium dahliae in allotetraploid upland cotton. Front. Plant Sci. 9:842. doi: 10.3389/fpls.2018.00842
Zhang, Y., Li, D., Zhang, D. B., Zhao, X., Cao, X., Dong, L., et al. (2018). Analysis of the functions of TaGW2 homoeologs in wheat grain weight and protein content traits. Plant J. 94, 857–866. doi: 10.1111/tpj.13903
Zhang, Y., Liang, Z., Zong, Y., Wang, Y., Liu, J., Chen, K., et al. (2016). Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 7:12617. doi: 10.1038/ncomms12617
Zhang, Y., Malzahn, A. A., Sretenovic, S., and Qi, Y. (2019). The emerging and uncultivated potential of CRISPR technology in plant science. Nat. Plants 5, 778–794. doi: 10.1038/s41477-019-0461-5
Zhang, Y., Ren, Q., Tang, X., Liu, S., Malzahn, A. A., Zhou, J., et al. (2021). Expanding the scope of plant genome engineering with Cas12a orthologs and highly multiplexable editing systems. Nat. Commun. 12:1944. doi: 10.1038/s41467-021-22330-w
Zhang, F., Shen, J., Zhang, J., Zuo, Y., Li, L., and Chen, X. (2010). Rhizosphere processes and management for improving nutrient use efficiency and crop productivity: implications for China. Advn. Agron. 107, 1–32. doi: 10.1016/S0065-2113(10)07001-X
Zhang, X., Wang, J., Huang, J., Lan, H., Wang, C., Yin, C., et al. (2012). Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc. Natl. Acad. Sci. U. S. A. 109, 21534–21539. doi: 10.1073/pnas.1219776110
Zhang, J., Zhang, H., Botella, J. R., and Zhu, J. K. (2018). Generation of new glutinous rice by CRISPR/Cas9-targeted mutagenesis of the waxy gene in elite rice varieties. J. Integr. Plant Biol. 60, 369–375. doi: 10.1111/jipb.12620
Zhao, D., Li, J., Li, S., Xin, X., Hu, M., Price, M. A., et al. (2020). CRISPR/Cas9- mediated mutagenesis of Clpsk1 in watermelon to confer resistance to fusarium oxysporum f. sp. niveum. Plant Cell Rep. 39, 589–595. doi: 10.1007/s00299-020-02516-0
Zhao, D. D., Li, J., Li, S. W., Xin, X., Hu, M., Price, M. A., et al. (2021). Glycosylase base editors enable C-to-A and C-to-G base changes. Nat. Biotechnol. 39, 35–40. doi: 10.1038/s41587-020-0592-2
Zhong, Y., Blennow, A., Kofoed-Enevoldsen, O., Jiang, D., and Hebelstrup, K. H. (2019). Protein targeting to starch 1 is essential for starchy endosperm development in barley. J. Exp. Bot. 70, 485–496. doi: 10.1093/jxb/ery398
Zhu, Y., Lin, Y., Chen, S., Liu, H., Chen, Z., Fan, M., et al. (2019). CRISPR/Cas9-mediated functional recovery of the recessive rc allele to develop red rice. Plant Biotechnol. J. 17, 2096–2105. doi: 10.1111/pbi.13125
Zhu, J., Song, N., Sun, S., Yang, W., Zhao, H., Song, W., et al. (2016). Efficiency and inheritance of targeted mutagenesis in maize using CRISPR-Cas9. J. Genet. Genom. 43, 25–36. doi: 10.1016/j.jgg.2015.10.006
Zimmermann, M. B., and Boelaert, K. (2015). Iodine deficiency and thyroid disorders. Lancet Diabetes Endocrinol. 3, 286–295. doi: 10.1016/S2213-8587(14)70225-6
Zong, Y., Song, Q. N., Li, C., Jin, S., Zhang, D., Wang, Y., et al. (2018). Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat. Biotechnol. 36, 950–953. doi: 10.1038/nbt.4261
Zong, Y., Wang, Y., Li, C., Zhang, R., Chen, K., Ran, Y., et al. (2017). Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat. Biotechnol. 35, 438–440. doi: 10.1038/nbt.3811
Zsögön, A., Cermák, T., Naves, E. R., Notini, M. M., Edel, K. H., Weinl, S., et al. (2018). De novo domestication of wild tomato using genome editing. Nat. Biotechnol. 36, 1211–1216. doi: 10.1038/nbt.4272
Keywords: malnutrition deficiency, climate change, genome editing, sustainable development, hidden hunger, micronutrients
Citation: Khaled F, Mehta S, Agrawal PK, Abdel-Monem UM, El-Akkad H, Gendy AS, Al-Maracy SHA, Heakel RMY, Wafa HA, Fathy R and Narayan OP (2024) The era of genome editing: current updates and the status of food and nutrition security. Front. Sustain. 5:1378712. doi: 10.3389/frsus.2024.1378712
Edited by:
Ahmed Ibrahim Osman, Queen’s University Belfast, United KingdomReviewed by:
Janpriy Sharma, University of Trento, ItalyA. Amarender Reddy, National Institute of Agricultural Extension Management, India
Copyright © 2024 Khaled, Mehta, Agrawal, Abdel-Monem, El-Akkad, Gendy, Al-Maracy, Heakel, Wafa, Fathy and Narayan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Om Prakash Narayan, b20udHVmdHNAZ21haWwuY29t; Fathy Khaled, a2hhbGVkZ2VuZUB6dS5lZHUuZWc=
 Fathy Khaled
Fathy Khaled Sahil Mehta
Sahil Mehta Pawan Kumar Agrawal
Pawan Kumar Agrawal Usama M. Abdel-Monem4
Usama M. Abdel-Monem4 Hend El-Akkad
Hend El-Akkad Om Prakash Narayan
Om Prakash Narayan