
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Surg. , 16 July 2024
Sec. Genitourinary Surgery and Interventions
Volume 11 - 2024 | https://doi.org/10.3389/fsurg.2024.1429831
Clinical decisions based on the test results for prostate-specific antigen often result in overdiagnosis and overtreatment. Multiparametric magnetic resonance imaging (mpMRI) can be used to identify high-grade prostate cancer (HGPCa; Gleason score ≥3 + 4); however, certain limitations remain such as inter-reader variability and false negatives. The combination of mpMRI and prostate cancer (PCa) biomarkers (prostate-specific antigen density, Proclarix, TMPRSS2:ERG gene fusion, Michigan prostate score, ExoDX prostate intelliscore, four kallikrein score, select molecular diagnosis, prostate health index, and prostate health index density) demonstrates high accuracy in the diagnosis of HGPCa, ensuring that patients avoid unnecessary prostate biopsies with a low leakage rate. This manuscript describes the characteristics and diagnostic performance of each biomarker alone and in combination with mpMRI, with the intension to provide a basis for decision-making in the diagnosis and treatment of HGPCa. Additionally, we explored the applicability of the combination protocol to the Asian population.
Prostate cancer (PCa) is the most common cancer and second leading cause of cancer-related death in males, with a serious effect on their health (1). Prostate-specific antigen (PSA) is the most commonly used screening tool for PCa in the clinic; however, not all patients with PCa have elevated PSA levels, and it is challenging to distinguish PCa from benign diseases, such as prostatitis and benign prostatic hyperplasia (BPH), by relying on PSA ≥ 4 ng/ml alone. Patients with PSA level of 4–10 ng/ml are usually at low-risk for PCa (Gleason score = 6), and the preferred treatment option for patients with low-risk PCa is active surveillance (AS) (2–4). Therefore, clinical decisions based exclusively on PSA are likely to subject patients to unnecessary prostate biopsies and biopsy-induced complications, such as rectal bleeding, infection, urinary retention, and erectile dysfunction, severely impacting their quality of life (5).
Multiparametric magnetic resonance imaging (mpMRI) includes three sequences of T2-weighted imaging (T2WI), diffusion-weighted imaging (DWI), and dynamic contrast enhancement (DCE). A score in the range of 1–5 is obtained based on the prostate imaging reporting and data system (PI-RADS), with the risk of developing high-grade prostate cancer (HGPCa; Gleason score ≥3 + 4) increasing with the score. To accurately diagnose HGPCa and screen candidates for surgery and radiotherapy, the National Comprehensive Cancer Network (NCCN) guidelines recommend that patients with elevated PSA undergo mpMRI before biopsy (3). Clinicians recommend prostate biopsy for patients with PI-RADS ≥ 4 lesions; patients with PI-RADS ≤ 2 lesions do not need to be biopsied. In contrast, PI-RADS 3 lesions lies in the gray area with mpMRI examination; how to guide the biopsy decision in this group of patients has become a significant challenge in the clinic (6). The inter-reader variability is a limitation of mpMRI, reaching 22.0% false negatives in patients with PI-RADS ≤ 3 lesions (7, 8). Therefore, it is necessary to explore new detection methods or feasible combined programs to compensate for the shortcomings of PSA and mpMRI, thus improving the diagnostic accuracy. Biomarkers with a high sensitivity and specificity, convenient sampling, and easy detection for combined testing is the preferred examination method.
This article reviews the research progress of biomarkers of HGPCa and their combined application with mpMRI, aiming to provide a reference for the accurate diagnosis of HGPCa. Although Asians have a lower incidence of PCa than African Americans and Caucasians, approximately 50.0% of Asian patients with PCa have HGPCa as the pathological outcome (9, 10). Notedly, approximately 80.0%–90.0% of clinical trials on mpMRI combined with biomarkers are conducted in Caucasians (11–13), while fewer are conducted in Asian populations. Therefore, we also explored the effectiveness of the combined programs in the Asian population.
PSAD is a PSA derivative, which is calculated with the following formula: PSAD = serum PSA/prostate volume (PV). The diagnosis of HGPCa using the PSAD is susceptible to the influence of the PV. In the PV intervals defined by <50 cm3, 50 ≤ PV ≤ 75 cm3, and >75 cm3, PSAD is a predictor of HGPCa in the range of small [odds ratio (OR): 2.13, P = 0.030] to intermediate (OR: 2.80, P = 0.010) PVs at a threshold value of 0.15 [ng/ml]/cm3. However, when PV is >75 cm3, PSAD is not associated with HGPCa (OR: 0.28, P = 0.370) (14). Therefore, the ability of PSAD to recognize HGPCa in patients with a very large PV is limited.
Thrombospondin-1 (THBS1) and cathepsin D (CTSD) are two glycoproteins identified to be associated with prostate carcinogenesis by mass spectrometry-based proteomics (15). Klocker et al. (16) combined THBS1, CTSD, total PSA (tPSA), free PSA (fPSA), and age to develop Proclarix, an in vitro diagnostic test for predicting HGPCa. The sensitivity, specificity, and negative predictive value (NPV) of the test for diagnosing HGPCa in patients with a PSA of 4–10 ng/ml and PV of 35–250 cm3 were 90.0%, 43.0%, and 95.0%, respectively (16). The diagnostic performance of Proclarix is not affected by the PV. Furthermore, it appears to be a good tool for differentiating between HGPCa and BPH. Clinicians using Proclarix to guide biopsy decisions can avoid unnecessary prostate biopsies in 18.2% of patients and reduce mpMRI exams by 25.4%, with a resulting HGPCa misdiagnosis rate of 2.6% (17).
Androgen-regulated transmembrane serine protease 2 (TMPRSS2) is genetically fused with erythrocyte transformation-specific related genes (ERGs), leading to the overexpression of ERG proteins, which promote the occurrence and progression of PCa. Tumor cells that detach into the urine after a digital rectal examination (DRE) can be detected by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and fluorescence in situ hybridization (18). Considering its incidence in Asian patients with PCa is only 27.0% (19), this tool is not suitable for the diagnosis of PCa in Asia.
Tomlins et al. constructed a logistic regression model by combining the RNA copy numbers of blood PSA, post-DRE urinary prostate cancer antigen 3 (PCA3), and T2-E fusion genes. This MiPS model produces a range of scores from 0 to 100, reflecting the likelihood of detecting HGPCa on prostate biopsy. Its ability to diagnose HGPCa was superior to a single metric or a combination of the two metrics in this model (20). Although MiPS demonstrated the ability to monitor disease progression in the clinical trial by Eyrich et al. (21), the results of this single-center, small-sample (n = 52) study cannot be generalized to the entire patient population under AS.
The EPI measures the mRNA copy number of three genes, ERG, PCA3, and SPDEF, in the exosomes of prostate tumor cells released into the urine without DRE. RT-qPCR results in a score range of 0–100 that can be used to infer the likelihood of patients with PSA in the gray region to develop HGPCa (22). EPI had an area under the curve (AUC) of 0.700 and an NPV of 90.1% for diagnosing HGPCa when using 15.6 as a threshold to guide biopsy decisions, thus avoiding unnecessary biopsies in 26.0% of patients (23–25). Based on its NPV and AUC, EPI is more suitable as an exclusionary indicator. The idea that EPI was suitable for excluding patients with biopsy-negative, low-risk PCa was confirmed in a follow-up study by Tutrone's team (26). Over 92.0% of the patients with an EPI < 15.6 were low-risk patients who were much less likely to progress to HGPCa at 2.5 years after the EPI examination compared to patients with an EPI > 15.6 (7.9% vs. 26.8%, P < 0.001). The EPI reflects disease progression over long time periods and is, therefore, suitable before biopsy and may be applicable to patients under AS.
The 4Kscore is a predictive model of HGPCa risk that combines four different kinin-releasing enzymes in the blood: tPSA, fPSA, intact PSA (iPSA), and human kallikrein-related peptidase 2 (hk2), as in addition to three clinical variables (age, DRE, and previous biopsy results) to construct a risk prediction model for HGPCa (27). The 4Kscore showed high accuracy in diagnosing HGPCa in patients before biopsy and those under AS, with AUCs of 0.779 and 0.780, respectively (27, 28).
SelectMDx is a logistic regression model constructed by detecting mRNA levels of HOXC6 and DLX1 gene transcripts in urine samples after DRE, combined with the following clinical factors: age, DRE, PSA, PSAD, and family history. It has an AUC for the diagnosis of HGPCa before the initial biopsy of 0.684 (29), and a concordance index of only 0.670 for predicting progression in patients under AS (30). Therefore, SelectMDx should be explored in combined protocols to accurately diagnose HGPCa and monitor patients under AS.
The PHI measures blood levels of fPSA, tPSA, and [−2] PSA prosoma ([−2] proPSA). The HGPCa prediction PHI score is calculated by the following formula: PHI =[([−2] proPSA/fPSA) × tPSA1/2]. The Food and Drug Administration has approved PHI for patients aged >50 years with a PSA level 4–10 ng/ml and negative DRE (31). However, thus, far, there is no consensus on the PHI threshold; the current PHI thresholds used in the literature to diagnose HGPCa range from 27 to 67, corresponding to different sensitivities and specificities (32). In areas with a high incidence of PCa, in high-risk populations, or when mpMRI is not available, it is recommended to use a threshold of 62, which has 89.0% specificity, and to combine it with PSA to avoid unnecessary biopsies or MRI. In areas of lower incidence or in non-high-risk populations, it is recommended to use a threshold of 27, which has 100.0% sensitivity and could help avoid missed diagnoses. Multiple PHI examinations are an effective way to monitor disease progression in patients under AS. The risk of progression to HGPCa in patients with PHI ≥ 36 is 2.12 times higher than in patients with PHI < 27 (hazard ratio (HR) = 2.12, 95% confidence interval (CI): 1.00–4.50, P = 0.002) (33).
Similar to PSAD, the diagnostic performance of PHID is limited by the PV, which is indicated for patients with a PV ≤ 50 cm3 (34). In 306 patients with a median PV of 37.9 cm3 and PSA 4–10 ng/ml, PHID diagnosed HGPCa with an AUC of up to 0.826, which is superior to PSA, PSAD, and PHI (35). In the group of patients with a PV > 50 cm3, PHID demonstrated a diagnostic power similar to PSA (AUC: 0.686 vs. 0.700, respectively) (34).
Clinical practitioners have further experimented with PSAD thresholds of <0.15 [ng/ml]/cm3 to improve its ability to diagnose HGPCa for PI-RADS ≤ 3 lesions, which is based on testing the effectiveness of PSAD in conjunction with mpMRI. Among them, 0.10 [ng/ml]/cm3 is one of the most studied thresholds (36, 37). In the group of patients with PI-RADS 2 lesions with pre-biopsy and AS, PSAD with 0.10 [ng/ml]/cm3 as a threshold was advantageous in terms of the NPV (96.2% vs. 89.7%) and leakage rate (3.8% vs. 10.3%) when compared with 0.15 [ng/ml]/cm3; however, this advantage was not significant in patients with PI-RADS 3 lesions (38, 39). Therefore, patients with PIRADS ≤ 2 and PSAD < 0.10 [ng/ml]/cm3 do not need to be biopsied, patients with PIRADS 3 and PSAD > 0.15 [ng/ml]/cm3 should be biopsied, and patients with HGPCa risk between these should undergo biopsy according to their wishes. This is consistent with risk reporting in the European Association of Urology Guidelines (40). Because PSAD is affected by PV (14), clinicians should calculate the PV after mpMRI and determine whether patients with PI-RADS ≤ 3 lesions can be further diagnosed definitively by PSAD. Adjusting for conditions can potentially improve the accuracy of PSAD in diagnosing HGPCa in patients with PI-RADS ≤ 3 lesions.
The combination of Proclarix and mpMRI significantly improved the accuracy of diagnosing HGPCa compared to Proclarix alone and mpMRI alone (41) (Table 1). In a cohort of patients with PI-RADS ≤ 2 lesions, Proclarix enabled 30.0% of patients to avoid biopsy while accurately detecting all patients with HGPCa missed by mpMRI. In a cohort of patients with PI-RADS 3 lesions, Proclarix enabled 21.3% of patients to avoid biopsy and detected all patients with HGPCa (43). Therefore, for patients with PI-RADS ≤ 3 lesions, the highly sensitive Proclarix is an excellent complementary test.
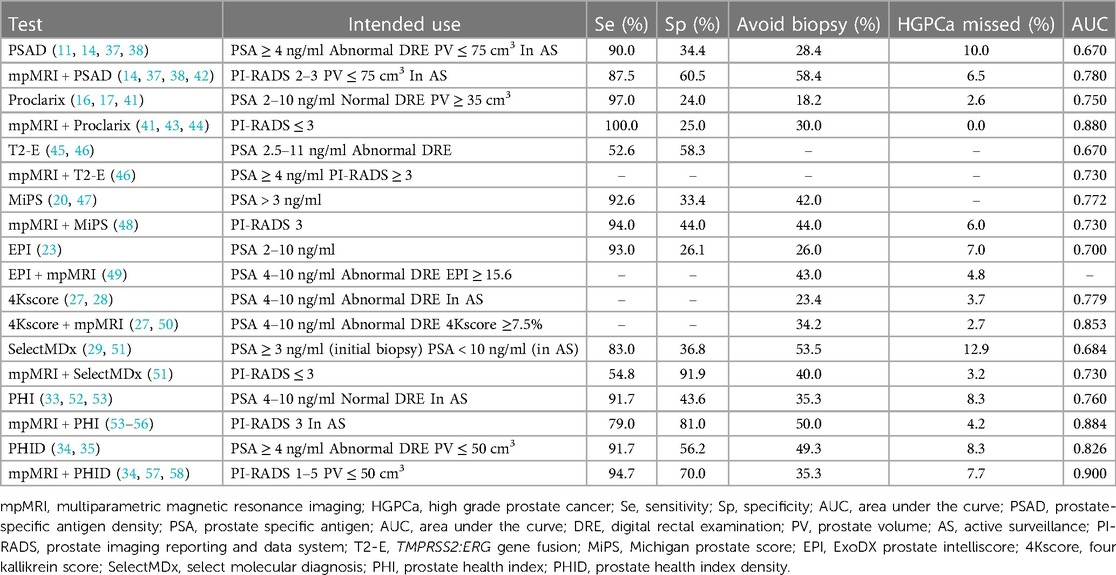
Table 1 Diagnostic performance of mpMRI combined biomarkers vs. biomarkers alone in patients with HGPCa.
In a prospective study that included 158 patients, the combination of T2-E and mpMRI did not show an advantage in the diagnostic accuracy of HGPCa compared to a baseline model consisting of age, PSA, previous biopsy history, and family history (AUC: 0.730 vs. 0.740) (46). The high economic cost of this combined protocol, the lack of improvement in diagnostic accuracy, and the low incidence of T2-E gene fusion in Asian patients make the combination unsuitable for HGPCa diagnosis in Asians (19, 46).
In patients with PI-RADS 3 lesions, using MiPS as the basis for biopsy decisions would allow 44.0% of patients to avoid biopsy and miss only 6.0% of cases of HGPCa (48). However, the accuracy of MiPS combined with mpMRI for diagnosing HGPCa was lower than that of MiPS alone (AUC: 0.730 vs. 0.772), which is possibly due to differences in disease severity between the patients in the two studies (20, 48). It should be noted that in the validation cohort in which Tomlins et al. developed the MiPS model, the prevalence of HGPCa was 68.0%, and the median MiPS was 50, whereas in the study evaluating mpMRI in combination with MiPS, the percentage of patients with HGPCa was 49.0% and the median MiPS was 35.4 (20, 48). When comparing the ability to diagnose HGPCa, a high prevalence of HGPCa in a patient cohort can mask MiPS misdiagnosis.
The EPI-mpMRI regimen with anterior biomarkers ensured that more unnecessary biopsies were avoided than the mpMRI-EPI regimen (43.0% vs. 19.3%) with a low missed diagnosis rate (4.8%) while reducing the use of mpMRI by 39.9% (49); therefore, it greatly reduces the medical burden on the patients. According to the recommendation of de la Calle et al. (49), patients with suspected PCa should first undergo an EPI examination, and if the result is <15.6, a PSA follow-up examination could be performed. For patients with 15.6 ≤ EPI < 19, the decision for a subsequent biopsy or PSA follow-up examination should be made according to the mpMRI result. If the EPI ≥ 19, then prostate biopsy should be performed after mpMRI examination.
The 4Kscore-mpMRI protocol resulted in 39.4% of patients avoiding biopsy, a reduction in the use of mpMRI by 29.5%, and a missed diagnosis rate of only 5.6% (49, 59). According to this examination protocol, patients can first undergo a 4Kscore examination, and if the result is <7.5%, they could choose PSA follow-up examination. If 7.5% ≤4Kscore <20.0%, the decision for a subsequent biopsy or PSA follow-up examination is made based on the results of the mpMRI. Furthermore, if the result of 4Kscore is ≥20.0%, prostate biopsy is performed after the mpMRI examination. Given the ability of the 4Kscore combined with mpMRI to diagnose HGPCa and the ability of the 4Kscore to monitor disease progression (27, 28), 4Kscore can be added to the monitoring methods for patients under AS in conjunction with mpMRI for the early detection of disease progression, which is in line with the current NCCN guidelines, encouraging the use of mpMRI to monitor the condition of patients under AS (3).
A meta-analysis (29) including data from 1,328 patients showed that SelectMDx alone and mpMRI alone had a similar accuracy in the diagnosis of HGPCa. In contrast, the combination of the two ensured a higher number of true-positive cases (527/1,000) and fewer false-negative cases (13/1,000). In recent years, researchers have devoted themselves to studying the combined strategy of SelectMDx and mpMRI (51, 60, 61). Maggi et al. (51) designed seven examination strategies for SelectMDx and mpMRI to diagnose HGPCa and found that the mpMRI-SelectMDx protocol was the optimal choice by comparison. Patients with PSA > 3 ng/ml and/or abnormal DRE were first examined by mpMRI, and prostate biopsy was performed if mpMRI was positive (PI-RADS > 3). If PI-RADS was ≤3, SelectMDx was performed, and a biopsy was performed only if SelectMDx results were abnormal. Clinicians following this protocol to guide biopsy decisions can avoid unnecessary biopsies in 40.0% of patients with a missed diagnosis rate of only 3.2% for HGPCa.
The ability of PHI to predict HGPCa in specimens from biopsy or radical prostatectomy is further enhanced when coupled with mpMRI (55, 62, 63). Diagnostic data on PHI-mpMRI (64) and mpMRI-PHI protocols (53, 54, 65) suggest that the mpMRI-PHI protocol is the better choice. When a PHI threshold of 27 was used, mpMRI-PHI had a sensitivity of 100.0% and an AUC of 0.884 for diagnosing HGPCa in a PI-RADS 3 population (32, 56). Thus, PHI could be used as a complementary screening tool after mpMRI to guide biopsy decisions in patients with PI-RADS 3 lesions. If mpMRI is used with PHI to rule out low-risk patients under AS, the NPV can be as high as 98.0% while enabling 20.0% of patients to avoid an unnecessary prostate biopsy (54).
The PHI demonstrated a similar diagnostic ability to PHID in patients with PSA 4–10 ng/ml and negative DRE (AUC: 0.760 vs. 0.770). In contrast, PHID, as a derivative of PHI, broadened the scope of use and further improved the diagnostic accuracy. In patients with PSA > 10 ng/ml, PHID was a superior diagnostic tool for HGPCa to PHI (AUC: 0.840 vs. 0.790) (52). Given that PHID is affected by the PV (34), the decision of whether to perform PHID in patients with PSA ≥ 4 ng/ml and/or negative or suspicious DRE could be based on the PV on the mpMRI report. In a group of patients with a median PV ≤ 50 cm3 and PSA ≥ 4 ng/ml, the AUC for PHID combined with mpMRI to diagnose HGPCa was as high as 0.900 (58).
Among the abovementioned combined protocols, mpMRI with Proclarix, EPI, 4Kscore, PHI, and PHID performed exceptionally well in diagnosing HGPCa, with all demonstrating a high accuracy (AUC > 0.850). Combination regimens that use anterior biomarkers, such as EPI-mpMRI and 4Kscore-mpMRI, can help to avoid unnecessary prostate biopsies and reduce the use of mpMRI, conducive to reducing the healthcare burden on the patients. If the mpMRI results are negative or suspicious, elevated PSA persists, and PCa is highly suspected, clinicians could choose to assess the Proclarix, PHI, or PHID for further clarifications and avoid prostate biopsy if subsequent biomarker tests are negative. Considering the ability to diagnose HGPCa and the frequency of T2-E fusion genes, mpMRI combined with T2-E was found to not be suitable for the diagnosis of HGPCa in Asians.
Germline mutations, DNA methylation, molecular alterations of key PCa genes, genome-wide expression profiles, and epidemiologic characteristics differ significantly among patients with PCa of different races due to genetic factors, living environment, and medical care (66, 67). The incidence of PCa in European Americans is 68 cases per 100,000 men, with a 10-year survival rate of 86.0%, while the incidence of PCa in Asians ranges from 2 to 10 cases per 100,000 men, with a 10-year survival rate of 36.2%. Genetically, the three most common mutated genes and mutation frequencies are BRCA2 (2.6%), BRCA1 (1.3%), and HOXB13 (1.3%) in African Americans, and BRCA2 (4.1%), BRCA1 (2.7%), and ATM (2.7%) in Asians, with mutations in BRCA2 associated with a poor prognosis (67–70). The incidence of PCa in Asia is lower than in Europe and North America; however, the prognosis is poor. This inspires us not to neglect the early and accurate diagnosis of Asian patients with PCa, and there is a need to explore the applicability of diagnostic protocols in Asian populations. Although fewer clinical trials on mpMRI combined with biomarkers have been conducted in Asian populations, limited data suggest that such combination protocols have good diagnostic accuracy in Asian populations (Table 2).
Theoretically, biomarkers that can recognize HGPCa can also be used to monitor disease progress in patients under AS. In the future, the accuracy of combined mpMRI and biomarkers in the diagnosis of HGPCa could be evaluated in prospective, multicenter cohorts of patients under AS. The combined protocol could be extended from pre-biopsy patients to patients under AS to provide them with additional monitoring options. The accuracy of mpMRI combined with biomarkers for the diagnosis of HGPCa in Asians needs to be further evaluated in large-sample, prospective clinical trials.
MpMRI-Proclarix, mpMRI-PHI, mpMRI-PHID, 4Kscore-mpMRI, and EPI-mpMRI were shown to improve the diagnostic accuracy of HGPCa, enabling patients to avoid unnecessary biopsy or mpMRI. Moreover, mpMRI combined with biomarkers to diagnose HGPCa is feasible in Asians.
S-lL: Data curation, Investigation, Writing – original draft, Writing – review & editing. M-yZ: Supervision, Writing – review & editing. QW: Conceptualization, Methodology, Writing – review & editing. YT: Conceptualization, Data curation, Project administration, Resources, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article.
This study was supported by the National Natural Science Foundation of China (82360587), the Guangxi Medical & Health Appropriate Technology Development and Promotion Application Project (S2019052), the “139” Plan for Cultivating High-level and Key Talents in Guangxi Medicine of China (G201903036), and the Guangxi Natural Science Foundation Project (2018GXNSFAA138061, 2024GXNSFDA010022).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. (2023) 73(1):17–48. doi: 10.3322/caac.21763
2. Albertsen PC. Prostate cancer screening and treatment: where have we come from and where are we going? BJU Int. (2020) 126(2):218–24. doi: 10.1111/bju.15153
3. Mason BR, Eastham JA, Davis BJ, Mynderse LA, Pugh TJ, Lee RJ, et al. Current Status of MRI and PET in the NCCN guidelines for prostate cancer. J Natl Compr Canc Netw. (2019) 17(5):506–13. doi: 10.6004/jnccn.2019.7306
4. Merriel SWD, Funston G, Hamilton W. Prostate cancer in primary care. Adv Ther. (2018) 35(9):1285–94. doi: 10.1007/s12325-018-0766-1
5. Borghesi M, Ahmed H, Nam R, Schaeffer E, Schiavina R, Taneja S, et al. Complications after systematic, random, and image-guided prostate biopsy. Eur Urol. (2017) 71(3):353–65. doi: 10.1016/j.eururo.2016.08.004
6. Guerra-Lacambra M, Yanez-Castillo Y, Folgueral-Corral M, Melgarejo-Segura MT, Del Carmen Cano-Garcia M, Sanchez-Tamayo FJ, et al. Results of fusion prostate biopsy comparing with cognitive and systematic biopsy. J Cancer Res Clin Oncol. (2023) 149(16):15085–90. doi: 10.1007/s00432-023-05293-x
7. Park KJ, Choi SH, Lee JS, Kim JK, Kim MH, Jeong IG. Risk stratification of prostate cancer according to PI-RADS(R) version 2 categories: meta-analysis for prospective studies. J Urol. (2020) 204(6):1141–9. doi: 10.1097/JU.0000000000001306
8. Hsi RA, Dinh TK, Greer M, Bensen C, Mitchell MA, Li AY, et al. Performance of multiparametric prostate magnetic resonance imaging validated by targeted and systematic transperineal biopsies. BJUI Compass. (2023) 4(1):96–103. doi: 10.1002/bco2.184
9. McKay RR, Gold T, Zarif JC, Chowdhury-Paulino IM, Friedant A, Gerke T, et al. Tackling diversity in prostate cancer clinical trials: a report from the diversity working group of the IRONMAN registry. JCO Glob Oncol. (2021) 7:495–505. doi: 10.1200/go.20.00571
10. Zlotta AR, Egawa S, Pushkar D, Govorov A, Kimura T, Kido M, et al. Prevalence of prostate cancer on autopsy: cross-sectional study on unscreened Caucasian and Asian men. J Natl Cancer Inst. (2013) 105(14):1050–8. doi: 10.1093/jnci/djt151
11. Cussenot O, Renard-Penna R, Montagne S, Ondet V, Pilon A, Guechot J, et al. Clinical performance of magnetic resonance imaging and biomarkers for prostate cancer diagnosis in men at high genetic risk. BJU Int. (2023) 131(6):745–54. doi: 10.1111/bju.15968
12. Drevik J, Dalimov Z, Uzzo R, Danella J, Guzzo T, Belkoff L, et al. Utility of PSA density in patients with PI-RADS 3 lesions across a large multi-institutional collaborative. Urol Oncol. (2022) 40(11):490.e1–490.e6. doi: 10.1016/j.urolonc.2022.08.003
13. Wagaskar VG, Levy M, Ratnani P, Sullimada S, Gerenia M, Schlussel K, et al. A SelectMDx/magnetic resonance imaging-based nomogram to diagnose prostate cancer. Cancer Rep (Hoboken). (2023) 6(1):e1668. doi: 10.1002/cnr2.1668
14. Omri N, Kamil M, Alexander K, Alexander K, Edmond S, Ariel Z, et al. Association between PSA density and pathologically significant prostate cancer: the impact of prostate volume. Prostate. (2020) 80(16):1444–9. doi: 10.1002/pros.24078
15. Cima I, Schiess R, Wild P, Kaelin M, Schuffler P, Lange V, et al. Cancer genetics-guided discovery of serum biomarker signatures for diagnosis and prognosis of prostate cancer. Proc Natl Acad Sci U S A. (2011) 108(8):3342–7. doi: 10.1073/pnas.1013699108
16. Klocker H, Golding B, Weber S, Steiner E, Tennstedt P, Keller T, et al. Development and validation of a novel multivariate risk score to guide biopsy decision for the diagnosis of clinically significant prostate cancer. BJUI Compass. (2020) 1(1):15–20. doi: 10.1002/bco2.8
17. Morote J, Campistol M, Celma A, Regis L, de Torres I, Semidey ME, et al. The efficacy of proclarix to select appropriate candidates for magnetic resonance imaging and derived prostate biopsies in men with suspected prostate cancer. World J Mens Health. (2022) 40(2):270–9. doi: 10.5534/wjmh.210117
18. Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. (2005) 310(5748):644–8. doi: 10.1126/science.1117679
19. Kong DP, Chen R, Zhang CL, Zhang W, Xiao GA, Wang FB, et al. Prevalence and clinical application of TMPRSS2-ERG fusion in Asian prostate cancer patients: a large-sample study in Chinese people and a systematic review. Asian J Androl. (2020) 22(2):200–7. doi: 10.4103/aja.aja_45_19
20. Tomlins SA, Day JR, Lonigro RJ, Hovelson DH, Siddiqui J, Kunju LP, et al. Urine TMPRSS2:eRG plus PCA3 for individualized prostate cancer risk assessment. Eur Urol. (2016) 70(1):45–53. doi: 10.1016/j.eururo.2015.04.039
21. Eyrich NW, Wei JT, Niknafs YS, Siddiqui J, Ellimoottil C, Salami SS, et al. Association of MyProstateScore (MPS) with prostate cancer grade in the radical prostatectomy specimen. Urol Oncol. (2022) 40(1):4.e1–7. doi: 10.1016/j.urolonc.2021.09.007
22. Kretschmer A, Tutrone R, Alter J, Berg E, Fischer C, Kumar S, et al. Pre-diagnosis urine exosomal RNA (ExoDx EPI score) is associated with post-prostatectomy pathology outcome. World J Urol. (2022) 40(4):983–9. doi: 10.1007/s00345-022-03937-0
23. McKiernan J, Donovan M J, Margolis E, Partin A, Carter B, Brown G, et al. A prospective adaptive utility trial to validate performance of a novel urine exosome gene expression assay to predict high-grade prostate cancer in patients with prostate-specific antigen 2–10ng/ml at initial biopsy. Eur Urol. (2018) 74(6):731–8. doi: 10.1016/j.eururo.2018.08.019
24. Tutrone R, Donovan MJ, Torkler P, Tadigotla V, McLain T, Noerholm M, et al. Clinical utility of the exosome based ExoDx prostate(IntelliScore) EPI test in men presenting for initial biopsy with a PSA 2–10ng/ml. Prostate Cancer Prostatic Dis. (2020) 23(4):607–14. doi: 10.1038/s41391-020-0237-z
25. Margolis E, Brown G, Partin A, Carter B, McKiernan J, Tutrone R, et al. Predicting high-grade prostate cancer at initial biopsy: clinical performance of the ExoDx (EPI) prostate intelliscore test in three independent prospective studies. Prostate Cancer Prostatic Dis. (2022) 25(2):296–301. doi: 10.1038/s41391-021-00456-8
26. Tutrone R, Lowentritt B, Neuman B, Donovan MJ, Hallmark E, Cole TJ, et al. Exodx prostate test as a predictor of outcomes of high-grade prostate cancer—an interim analysis. Prostate Cancer Prostatic Dis. (2023) 26(3):596–601. doi: 10.1038/s41391-023-00675-1
27. Thomas J, Atluri S, Zucker I, Reis I, Kwon D, Kim E, et al. A multi-institutional study of 1,111 men with 4K score, multiparametric magnetic resonance imaging, and prostate biopsy. Urol Oncol. (2023) 41(10):430.e9–430.e16. doi: 10.1016/j.urolonc.2023.07.001
28. Lin DW, Newcomb LF, Brown MD, Sjoberg DD, Dong Y, Brooks JD, et al. Evaluating the four kallikrein panel of the 4Kscore for prediction of high-grade prostate cancer in men in the canary prostate active surveillance study. Eur Urol. (2017) 72(3):448–54. doi: 10.1016/j.eururo.2016.11.017
29. Sari Motlagh R, Yanagisawa T, Kawada T, Laukhtina E, Rajwa P, Aydh A, et al. Accuracy of SelectMDx compared to mpMRI in the diagnosis of prostate cancer: a systematic review and diagnostic meta-analysis. Prostate Cancer Prostatic Dis. (2022) 25(2):187–98. doi: 10.1038/s41391-022-00538-1
30. Fiorella D, Marenco JL, Mascaros JM, Borque-Fernando A, Esteban LM, Calatrava A, et al. Role of PCA3 and SelectMDx in the optimization of active surveillance in prostate cancer. Actas Urol Esp (Engl Ed). (2021) 45(6):439–46. doi: 10.1016/j.acuroe.2020.10.013
31. Ferro M, De Cobelli O, Lucarelli G, Porreca A, Busetto GM, Cantiello F, et al. Beyond PSA: the role of prostate health index (phi). Int J Mol Sci. (2020) 21(4):1184. doi: 10.3390/ijms21041184
32. Agnello L, Vidali M, Giglio RV, Gambino CM, Ciaccio AM, Lo Sasso B, et al. Prostate health index (PHI) as a reliable biomarker for prostate cancer: a systematic review and meta-analysis. Clin Chem Lab Med. (2022) 60(8):1261–77. doi: 10.1515/cclm-2022-0354
33. de la Calle CM, Jing Y, Mamawala MM, Landis P, Macura KJ, Trock BJ, et al. Baseline prostate health index risk category and risk category changes during active surveillance predict grade reclassification. Urol Oncol. (2023) 41(11):455.e1–455.e6. doi: 10.1016/j.urolonc.2023.08.011
34. Filella X, Foj L, Wijngaard R, Luque P. Value of PHI and PHID in the detection of intermediate- and high-risk prostate cancer. Clin Chim Acta. (2022) 531:277–82. doi: 10.1016/j.cca.2022.04.992
35. Boo Y, Chung JH, Kang M, Sung HH, Jeon HG, Jeong BC, et al. Comparison of prostate-specific antigen and its density and prostate health Index and its density for detection of prostate cancer. Biomedicines. (2023) 11(7):1912. doi: 10.3390/biomedicines11071912
36. Pellegrino F, Stabile A, Sorce G, Quarta L, Robesti D, Cannoletta D, et al. Added value of prostate-specific antigen density in selecting prostate biopsy candidates among men with elevated prostate-specific antigen and PI-RADS ≥3 lesions on multiparametric magnetic resonance imaging of the prostate: a systematic assessment by PI-RADS score. Eur Urol Focus. (2023) S2405-4569(23):00223–7. doi: 10.1016/j.euf.2023.10.006
37. Frisbie JW, Van Besien AJ, Lee A, Xu L, Wang S, Choksi A, et al. PSA density is complementary to prostate MP-MRI PI-RADS scoring system for risk stratification of clinically significant prostate cancer. Prostate Cancer Prostatic Dis. (2023) 26(2):347–52. doi: 10.1038/s41391-022-00549-y
38. Lokeshwar SD, Nguyen J, Rahman SN, Khajir G, Ho R, Ghabili K, et al. Clinical utility of MR/ultrasound fusion-guided biopsy in patients with lower suspicion lesions on active surveillance for low-risk prostate cancer. Urol Oncol. (2022) 40(9):407.e21–407.e27. doi: 10.1016/j.urolonc.2022.06.005
39. Nguyen T-A, Fourcade A, Zambon A, Saout K, Deruelle C, Joulin V, et al. Optimal PSA density threshold and predictive factors for the detection of clinically significant prostate cancer in patient with a PI-RADS 3 lesion on MRI. Urol Oncol Semin Orig Invest. (2023) 41(8):354.e11–354.e18. doi: 10.1016/j.urolonc.2023.05.005
40. Schoots IG, Padhani AR. Risk-adapted biopsy decision based on prostate magnetic resonance imaging and prostate-specific antigen density for enhanced biopsy avoidance in first prostate cancer diagnostic evaluation. BJU Int. (2021) 127(2):175–8. doi: 10.1111/bju.15277
41. Morote J, Pye H, Campistol M, Celma A, Regis L, Semidey M, et al. Accurate diagnosis of prostate cancer by combining Proclarix with magnetic resonance imaging. BJU Int. (2023) 132(2):188–95. doi: 10.1111/bju.15998
42. Sigle A, Borkowetz A, von Hardenberg J, Drerup M, Kornienko K, Kwe J, et al. Prediction of significant prostate cancer in equivocal magnetic resonance imaging lesions: a high-volume international multicenter study. Eur Urol Focus. (2023) 9(4):606–13. doi: 10.1016/j.euf.2023.01.020
43. Morote J, Campistol M, Regis L, Celma A, de Torres I, Semidey ME, et al. Who with suspected prostate cancer can benefit from Proclarix after multiparametric magnetic resonance imaging? Int J Biol Markers. (2022) 37(2):218–23. doi: 10.1177/03936155221081537
44. Morote J, Campistol M, Triquell M, Celma A, Regis L, de Torres I, et al. Improving the early detection of clinically significant prostate cancer in men in the challenging prostate imaging-reporting and data system 3 category. Eur Urol Open Sci. (2022) 37:38–44. doi: 10.1016/j.euros.2021.12.009
45. Montoya Perez I, Jambor I, Pahikkala T, Airola A, Merisaari H, Saunavaara J, et al. Prostate cancer risk stratification in men with a clinical suspicion of prostate cancer using a unique biparametric MRI and expression of 11 genes in apparently benign tissue: evaluation using machine-learning techniques. J Magn Reson Imaging. (2020) 51(5):1540–53. doi: 10.1002/jmri.26945
46. Lazzeri M, Fasulo V, Lughezzani G, Benetti A, Soldà G, Asselta R, et al. Prospective evaluation of the role of imaging techniques and TMPRSS2:ERG mutation for the diagnosis of clinically significant prostate cancer. Front Oncol. (2022) 12:968384. doi: 10.3389/fonc.2022.968384
47. Sanda MG, Feng Z, Howard DH, Tomlins SA, Sokoll LJ, Chan DW, et al. Association between combined TMPRSS2:ERG and PCA3 RNA urinary testing and detection of aggressive prostate cancer. JAMA Oncol. (2017) 3(8):1085–93. doi: 10.1001/jamaoncol.2017.0177
48. Tosoian JJ, Singhal U, Davenport MS, Wei JT, Montgomery JS, George AK, et al. Urinary MyProstateScore (MPS) to rule out clinically-significant cancer in men with equivocal (PI-RADS 3) multiparametric MRI: addressing an unmet clinical need. Urology. (2022) 164:184–90. doi: 10.1016/j.urology.2021.11.033
49. de la Calle CM, Fasulo V, Cowan JE, Lonergan PE, Maggi M, Gadzinski AJ, et al. Clinical utility of 4Kscore((R)), ExosomeDx and magnetic resonance imaging for the early detection of high grade prostate cancer. J Urol. (2021) 205(2):452–60. doi: 10.1097/JU.0000000000001361
50. Falagario UG, Martini A, Wajswol E, Treacy PJ, Ratnani P, Jambor I, et al. Avoiding unnecessary magnetic resonance imaging (MRI) and biopsies: negative and positive predictive value of MRI according to prostate-specific antigen density, 4Kscore and risk calculators. Eur Urol Oncol. (2020) 3(5):700–4. doi: 10.1016/j.euo.2019.08.015
51. Maggi M, Del Giudice F, Falagario UG, Cocci A, Russo GI, Di Mauro M, et al. SelectMDx and multiparametric magnetic resonance imaging of the prostate for men undergoing primary prostate biopsy: a prospective assessment in a multi-institutional study. Cancers (Basel). (2021) 13(9):2047. doi: 10.3390/cancers13092047
52. Rius Bilbao L, Valladares Gomez C, Aguirre Larracoechea U, Pereira Arias JG, Arredondo Calvo P, Urdaneta Salegui LF, et al. Do PHI and PHI density improve detection of clinically significant prostate cancer only in the PSA gray zone? Clin Chim Acta. (2023) 542:117270. doi: 10.1016/j.cca.2023.117270
53. Hsieh PF, Li WJ, Lin WC, Chang H, Chang CH, Huang CP, et al. Combining prostate health index and multiparametric magnetic resonance imaging in the diagnosis of clinically significant prostate cancer in an Asian population. World J Urol. (2020) 38(5):1207–14. doi: 10.1007/s00345-019-02889-2
54. Schwen ZR, Mamawala M, Tosoian JJ, Druskin SC, Ross AE, Sokoll LJ, et al. Prostate health Index and multiparametric magnetic resonance imaging to predict prostate cancer grade reclassification in active surveillance. BJU Int. (2020) 126(3):373–8. doi: 10.1111/bju.15101
55. Mo LC, Zhang XJ, Zheng HH, Huang XP, Zheng L, Zhou ZR, et al. Development of a novel nomogram for predicting clinically significant prostate cancer with the prostate health index and multiparametric MRI. Front Oncol. (2022) 12:1068893. doi: 10.3389/fonc.2022.1068893
56. Fan YH, Pan PH, Cheng WM, Wang HK, Shen SH, Liu HT, et al. The prostate health index aids multi-parametric MRI in diagnosing significant prostate cancer. Sci Rep. (2021) 11(1):1286. doi: 10.1038/s41598-020-78428-6
57. Zhou Y, Qi W, Cui J, Zhong M, Lv G, Qu S, et al. Construction and comparison of different models in detecting prostate cancer and clinically significant prostate cancer. Front Oncol. (2022) 12:911725. doi: 10.3389/fonc.2022.911725
58. Druskin SC, Tosoian JJ, Young A, Collica S, Srivastava A, Ghabili K, et al. Combining prostate health index density, magnetic resonance imaging and prior negative biopsy status to improve the detection of clinically significant prostate cancer. BJU Int. (2018) 121(4):619–26. doi: 10.1111/bju.14098
59. Marzouk K, Ehdaie B, Vertosick E, Zappala S, Vickers A. Developing an effective strategy to improve the detection of significant prostate cancer by combining the 4Kscore and multiparametric MRI. Urol Oncol. (2019) 37(10):672–7. doi: 10.1016/j.urolonc.2019.07.010
60. Hendriks RJ, van der Leest MMG, Israël B, Hannink G, YantiSetiasti A, Cornel EB, et al. Clinical use of the SelectMDx urinary-biomarker test with or without mpMRI in prostate cancer diagnosis: a prospective, multicenter study in biopsy-naïve men. Prostate Cancer Prostatic Dis. (2021) 24(4):1110–9. doi: 10.1038/s41391-021-00367-8
61. Busetto GM, Del Giudice F, Maggi M, De Marco F, Porreca A, Sperduti I, et al. Prospective assessment of two-gene urinary test with multiparametric magnetic resonance imaging of the prostate for men undergoing primary prostate biopsy. World J Urol. (2021) 39(6):1869–77. doi: 10.1007/s00345-020-03359-w
62. Gentile F, La Civita E, Della Ventura B, Ferro M, Cennamo M, Bruzzese D, et al. A combinatorial neural network analysis reveals a synergistic behaviour of multiparametric magnetic resonance and prostate health Index in the identification of clinically significant prostate cancer. Clin Genitourin Cancer. (2022) 20(5):e406–10. doi: 10.1016/j.clgc.2022.04.013
63. Ferro M, Crocetto F, Bruzzese D, Imbriaco M, Fusco F, Longo N, et al. Prostate health index and multiparametric MRI: partners in crime fighting overdiagnosis and overtreatment in prostate cancer. Cancers (Basel). (2021) 13(18):4723. doi: 10.3390/cancers13184723
64. Kim L, Boxall N, George A, Burling K, Acher P, Aning J, et al. Clinical utility and cost modelling of the phi test to triage referrals into image-based diagnostic services for suspected prostate cancer: the PRIM (phi to RefIne mri) study. BMC Med. (2020) 18(1):95. doi: 10.1186/s12916-020-01548-3
65. Carbunaru S, Stinson J, Babajide R, Hollowell CMP, Yang X, Sekosan M, et al. Performance of prostate health index and PSA density in a diverse biopsy-naïve cohort with mpMRI for detecting significant prostate cancer. BJUI Compass. (2021) 2(6):370–6. doi: 10.1002/bco2.91
66. Boehm BE, York ME, Petrovics G, Kohaar I, Chesnut GT. Biomarkers of aggressive prostate cancer at diagnosis. Int J Mol Sci. (2023) 24(3):2185. doi: 10.3390/ijms24032185
67. Hinata N, Fujisawa M. Racial differences in prostate cancer characteristics and cancer-specific mortality: an overview. World J Mens Health. (2022) 40(2):217–27. doi: 10.5534/wjmh.210070
68. Hassanipour S, Delam H, Arab-Zozani M, Abdzadeh E, Hosseini SA, Nikbakht HA, et al. Survival rate of prostate cancer in Asian countries: a systematic review and meta-analysis. Ann Glob Health. (2020) 86(1):2. doi: 10.5334/aogh.2607
69. Pettaway CA. African American and Asian males: what do we know about germline predisposition to prostate cancer. Can J Urol. (2019) 26(5 Suppl 2):27–8.31629423
70. Vernooij RWM, van Oort I, de Reijke TM, Aben KKH. Nationwide treatment patterns and survival of older patients with prostate cancer. J Geriatr Oncol. (2019) 10(2):252–8. doi: 10.1016/j.jgo.2018.06.010
71. Zhang CC, Tu X, Lin TH, Cai DM, Yang L, Nie L, et al. The role of prostate-specific antigen density and negative multiparametric magnetic resonance imaging in excluding prostate cancer for biopsy-naïve men: clinical outcomes from a high-volume center in China. Asian J Androl. (2022) 24(6):615–9. doi: 10.4103/aja202220
72. Ye C, Ho JN, Kim DH, Song SH, Kim H, Lee H, et al. The prostate health Index and multi-parametric MRI improve diagnostic accuracy of detecting prostate cancer in Asian populations. Investig Clin Urol. (2022) 63(6):631–8. doi: 10.4111/icu.20220056
73. Kim JY, Jeon SS, Chung JH, Lee SS, Park SW. How to avoid prostate biopsy in men with prostate image-reporting and data system 3 lesion? Development and external validation of new biopsy indication using prostate health index density. Prostate Int. (2023) 11(3):167–72. doi: 10.1016/j.prnil.2023.07.001
74. Chen Y, Xu D, Ruan M, Li H, Lin G, Song G. A prospective study of the prostate health index density and multiparametric magnetic resonance imaging in diagnosing clinically significant prostate cancer. Investig Clin Urol. (2023) 64(4):363–72. doi: 10.4111/icu.20230060
75. Chiu PK, Leow JJ, Chiang CH, Mok A, Zhang K, Hsieh PF, et al. Prostate health index density outperforms prostate-specific antigen density in the diagnosis of clinically significant prostate cancer in equivocal magnetic resonance imaging of the prostate: a multicenter evaluation. J Urol. (2023) 210(1):88–98. doi: 10.1097/ju.0000000000003450
Keywords: high-grade prostate cancer, multiparametric magnetic resonance imaging, biomarker, pre-diagnosis, Asian
Citation: Li S-l, Zha M-y, Wang Q and Tang Y (2024) Advances in multiparametric magnetic resonance imaging combined with biomarkers for the diagnosis of high-grade prostate cancer. Front. Surg. 11:1429831. doi: 10.3389/fsurg.2024.1429831
Received: 8 May 2024; Accepted: 27 June 2024;
Published: 16 July 2024.
Edited by:
Guiming Liu, Northeast Ohio Medical University, United StatesReviewed by:
Po-Fan Hsieh, China Medical University Hospital, Taiwan© 2024 Li, Zha, Wang and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qi Wang, wangqi@stu.gxmu.edu.cn; Yong Tang, yong_tang_md@hotmail.com
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.