
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Surg., 28 April 2022
Sec. Neurosurgery
Volume 9 - 2022 | https://doi.org/10.3389/fsurg.2022.880048
This article is part of the Research TopicOptic Pathway Glioma: A multidisciplinary entity, posing dilemmas in diagnosis and managementView all 8 articles
 Orit Barinfeld1,2†
Orit Barinfeld1,2† Alon Zahavi1,2,3†
Alon Zahavi1,2,3† Shirel Weiss1,2
Shirel Weiss1,2 Helen Toledano1,4
Helen Toledano1,4 Shalom Michowiz1,5
Shalom Michowiz1,5 Nitza Goldenberg-Cohen2,6*
Nitza Goldenberg-Cohen2,6*
Objective: To investigate pediatric low-grade gliomas for alterations in IDH1, IDH2, CDKN2A, MYB, and MYBL1.
Materials and Methods: DNA and RNA were extracted from 62 pediatric gliomas. Molecular methods included PCR, RT-PCR, and RNA sequencing; Sanger sequencing was used for validation.
Results: Analysis for hotspot genetic alterations in IDH1 R132 and IDH2 R172 (45 and 33 samples) was negative in all cases. CDKN2A deletions were detected in exons 1 and 2 in 1 (pleomorphic xanthoastrocytoma) sample of 9 samples analyzed. Of 10 samples analyzed for MYB translocation, 4 each were positive for translocations with exon 2 and exon 3 of PCDHGA1. Six samples showed MYBL rearrangement. The lack of IDH1/2 genetic alterations is in accordance with the literature in pediatric tumors. Alterations in MYB, MYBL were recently reported to characterize diffuse grade II, but not grade I, gliomas.
Conclusion: We optimized methods for analyzing gene variations and correlated the findings to pathological grade. The high incidence of MYB and MYBL need further evaluation. We also compared DNA, RNA, and RNA sequencing results for fusion, translocation, and genetic alterations. More accurate identification of the underlying biology of pediatric gliomas has implications for the development of targeted treatment.
Brain tumors are the most common solid tumors in children, with a prevalence of 2.4–4 per 100,000. More than 40% are low-grade gliomas (LGGs) originating from glial cells, usually astrocytes (1–5). LGGs comprise a heterogeneous group of tumors. The most common is pilocytic astrocytoma which accounts for 30% of all central nervous system tumors in children (6).
LGGs (grade I-II) have less aggressive biological behavior than higher-grade gliomas (grade III-IV), with better response to treatment and better prognosis. The first line of treatment is surgical resection unless the tumor is located in less accessible areas such as the diencephalon, brain stem, or optic pathways (2, 6) when chemotherapy or radiation may be necessary. All these modalities especially radiation have significant side effects, such as cognitive impairment, vascular injury, and secondary malignancies. Pilocytic astrocytoma is associated with a 10-year overall survival rate of more than 85% (3), but the rate is much lower for grade II tumors (4, 6).
Until fairly recently, relatively little was known about the molecular changes that promote the formation or growth of LGGs in children. There is now clear evidence that dysregulated signaling of the Ras/Raf/MEK/ERK mitogen-activated protein kinase (MAPK) pathway plays an important role in glioma biology (5). Serine/threonine-protein kinase B-raf is a member of the Raf family of kinases and serves as a downstream effector in the MAPK pathway. Molecular studies have reported ERK/MAPK activation via focal gains at band 7q34 resulting in a fusion gene, BRAF-KIAA1549, in up to 80% of pilocytic astrocytomas of the cerebellum (7, 8). Activation of the Ras/Raf pathway can also result from oncogenic mutations of the BRAF gene itself, usually substitution of glutamic acid for valine at amino acid 600 (V600E), although they are less commonly identified in pediatric LGG (3%–10%) (6, 7, 9). These findings highlight the importance of the ERK/MAPK pathway both in the development of pediatric LGG and as a possible therapeutic target. Studies in various phases of development are already in progress assessing both BRAF and downstream MEK inhibitors for the treatment of gliomas in children (6, 10–12). Gene fusions may also provide a means of molecular classification of pilocytic astrocytomas in the future.
The commonest alterations in LGG in children are in BRAF. Either a fusion or a genetic alteration. In our previous study of BRAF genetic alterations in 45 samples of pediatric LGGs (13), we found an increased BRAF copy number in 18 of the 47 primary samples tested; 15 of them (83.3%) were pilocytic astrocytomas. A BRAF genetic alteration was found in 3 of 48 primary tumors, all with a normal BRAF copy number and no GNAQ genetic alteration. One sample had a GNAQ209 genetic alteration (Q209P626) with a normal BRAF gene. None of the tumors had a GNA11Q209 genetic alteration. Recurrent or progressive tumors, analyzed in 4 patients, had the same molecular genotype as their primary. The upregulation of the RAS/MAPK pathway is a major factor in the molecular pathways of pediatric LGGs and data are accumulating from large studies (6).
Aberrations in PTEN (14) and IDH1/2 (15) genes have previously been detected in adult high-grade astrocytomas (15, 16) but not in any pediatric astrocytoma samples examined. This supports previous findings that some molecular changes in astrocytomas may be age-dependent. CDKN2A genetic alterations are found in low grade gliomas often with BRAF genetic alterations and correlate with later clinical progression (6, 15).
Recently, attention has been addressed to three new genes that may be involved in the genetic profile of grade II diffuse gliomas: MYB, MYBL1, and FGFR1. MYB, an oncogene not previously implicated in gliomagenesis, was found to be activated in a diverse subset of pediatric LGGs (17). Novel rearrangements were identified, usually translocation or fusion with exon 2 or exon 3 of PCDHGA1. Rearrangements were also identified in the MYBL1 oncogene in addition to duplication and fusion of the tyrosine kinase domain in FGFR1 (6). These events may be exclusive to grade II gliomas and are likely to have an important role in tumorigenesis (18). The findings have implications for improving the identification and treatment of grade II gliomas, especially those involving the thalamus that are not amenable to complete surgical resection.
The aim of the present study was to screen pediatric grade I and II LGGs for genetic alterations in candidate genes IDH1/2, CDKN2A, MYB and MYBL1 in 62 pLGG in order to investigate their potential involvement in pediatric gliomagenesis. Molecular methods included PCR, RT-PCR, and RNA sequencing; Sanger sequencing was used for validation. Nowadays, next generation sequencing (NGS) panel for wide screening may be recommended as an alternative.
Paraffin-embedded or fresh-frozen samples were collected from 62 pediatric patients with various grade I (n = 45) or grade II (n = 17 LGGs, as follows: pilocytic astrocytoma (n = 34), ganglioglioma (n = 8), diffuse astrocytoma (n = 6), pleomorphic xanthoastrocytoma (n = 6), pilomyxoid astrocytoma (n = 3), subependymal giant cell astrocytoma (n = 2), astroblastoma (n = 1), gangliocytoma (n = 1), oligodendroglioma (n = 1). Data on patient demographics, tumor location, treatment, response to treatment, and survival are shown in Table 1.
Polymerase chain reaction (PCR) DNA analysis for IDH1 and IDH2 was performed in 45 and 34 samples, respectively. All findings were negative. Of the 62 samples, 14 did not show a sequence for either gene (N/A) and 7 were evaluated for only one gene (N/P) (Table 1).
Real-time PCR (RT-PCR) was performed on 9 grade II pediatric glioma samples. A deletion was detected in exon 1 and exon 2 of CDKN2A in one sample (no. 54) of pleomorphic xanthoastrocytoma. The cycle threshold (Ct) values are shown in Figure 1.
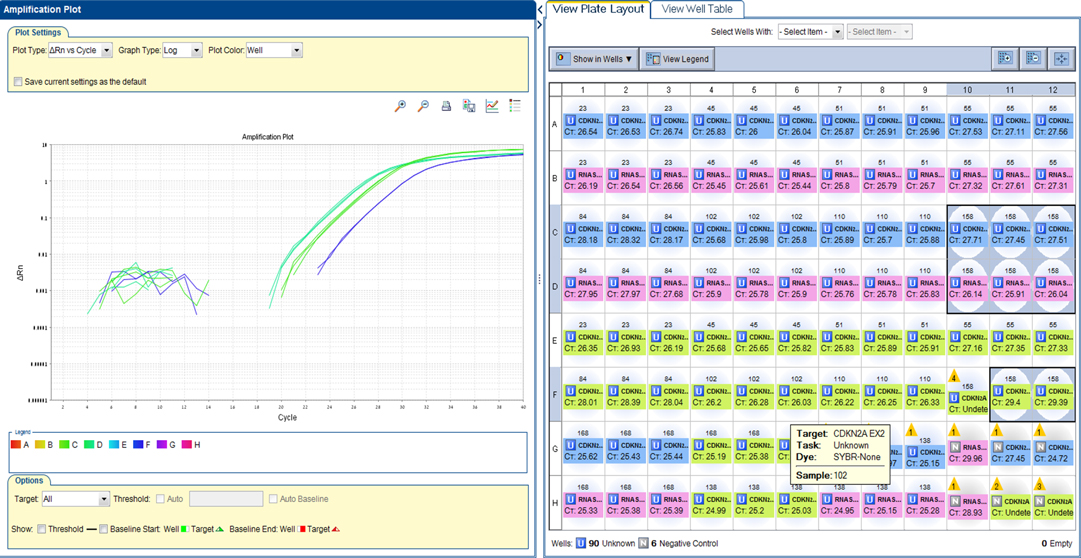
Figure 1. CDKN2A plot. A deletion was detected in exon 1 and exon 2 of CDKN2A in one sample (no. 54) of pleomorphic xanthoastrocytoma. RTPCR plot, showing Reference gene RNaseP - 26 Ct. Please note CDKN2A Exon 1–27 Ct (middle green) and CDKN2A Exon 2–29 Ct (right blue). A ratio of <0.5 was considered a deletion.
Two samples of grade II LGG were initially analyzed for MYB translocation using rapid amplification of cDNA ends (RACE): one diffuse astrocytoma (no. 51) and one pilomyxoid astrocytoma (no. 55) (Figure 2A–D). When primers 1 + 3 were used, the RACE gel showed bands at 700, 600, and 450 k in both samples. With the primer combination 2 + 3, two bands were detected, at 200 and 380 bp.
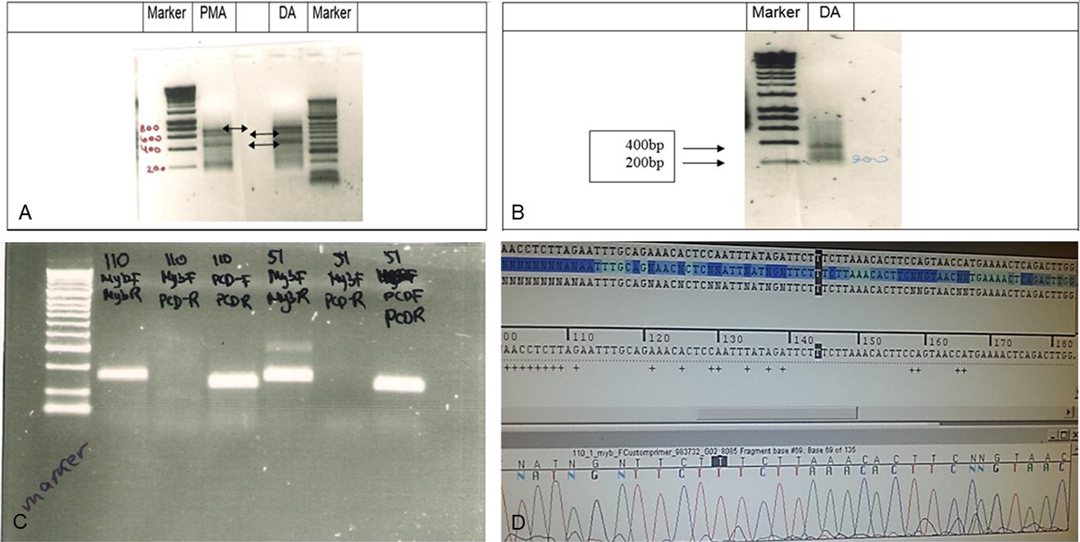
Figure 2. RACE analysis for MYB translocation. (A) RACE analysis of Grade II glioma samples 55 (PMA,102) and 51 (DA). RACE gel results showing the bands (amplicons by primers combination 1 + 3). Note the bands at 700k, 600k and 450k (arrows) in both samples If translocation occurred, then the PCR reaction will give a shorter amplicon. (B) RACE results primers 2 + 3 in sample 51 show 2 bands, 200 and 380 bp. (C) PCR products in gel. Left to right: Marker, Sample 53 (110) primer combination of MYB F + R, MYB F+ PCDHGA1 R, PCDHGA1 F + R, sample 51 combination of primers of MYB F + R, MYB F+ PCDHGA1 R, PCDHGA1 F + R. Only 53(110) showed light band in MYB F+ PCDHGA1 R combination, suggesting translocation of the MYB. (D) Sequence of the normal MYB in sample 55. Validation PCR only showed the normal MYB sequence.
Although we detected RACE products of translocation in the gel, direct sequencing showed only the normal MYB sequence, probably because the bands were weak (Figure 2D). The PCR results with a combination of 3 primers included MYB (forward + reverse)[(primers (1) + (2)], MYB (forward) + PCDHGA1(reverse)[primers(1) + (4)], PCDHGA1(forward + reverse) [primers (3) + (4)] in samples 53 and 51. A light band was detected in sample 53 using the 1 + 4 [MYB (forward) + PCDHGA1(reverse)] primer combination.
RT-PCR with a different combination of primers was used as an alternative method to detect MYB translocation with PCDHGA1 in 10 samples (Table 2). In samples without MYB translocation, the reactions with primers 1 + 2 or 3 + 4 led to a product, and in samples with MYB translocation with exon 2 of PCDHGA1, the reactions with primers 1 + 4 led to a product. Product was detected in sample 53 with primers 1 + 2 and 3 + 4 and also in sample 51 (Figure 3A–C). Five samples were positive for translocation of exon 9 of MYB with exon 2 of PCDHGA1: nos. 53, 55 (pilomyxoid astrocytoma), no. 62 (oligodendroglioma), and no. 60 (pleomorphic xanthoastrocytoma), and 4 were positive for translocation of exon 9 of MYB with exon 3 of PCDHGA1 (primer combination 5 + 6): nos. 51 (diffuse astrocytoma), 50 (ganglioglioma), 62 (oligodendroglioma), and 60 (pleomorphic xanthoastrocytoma).
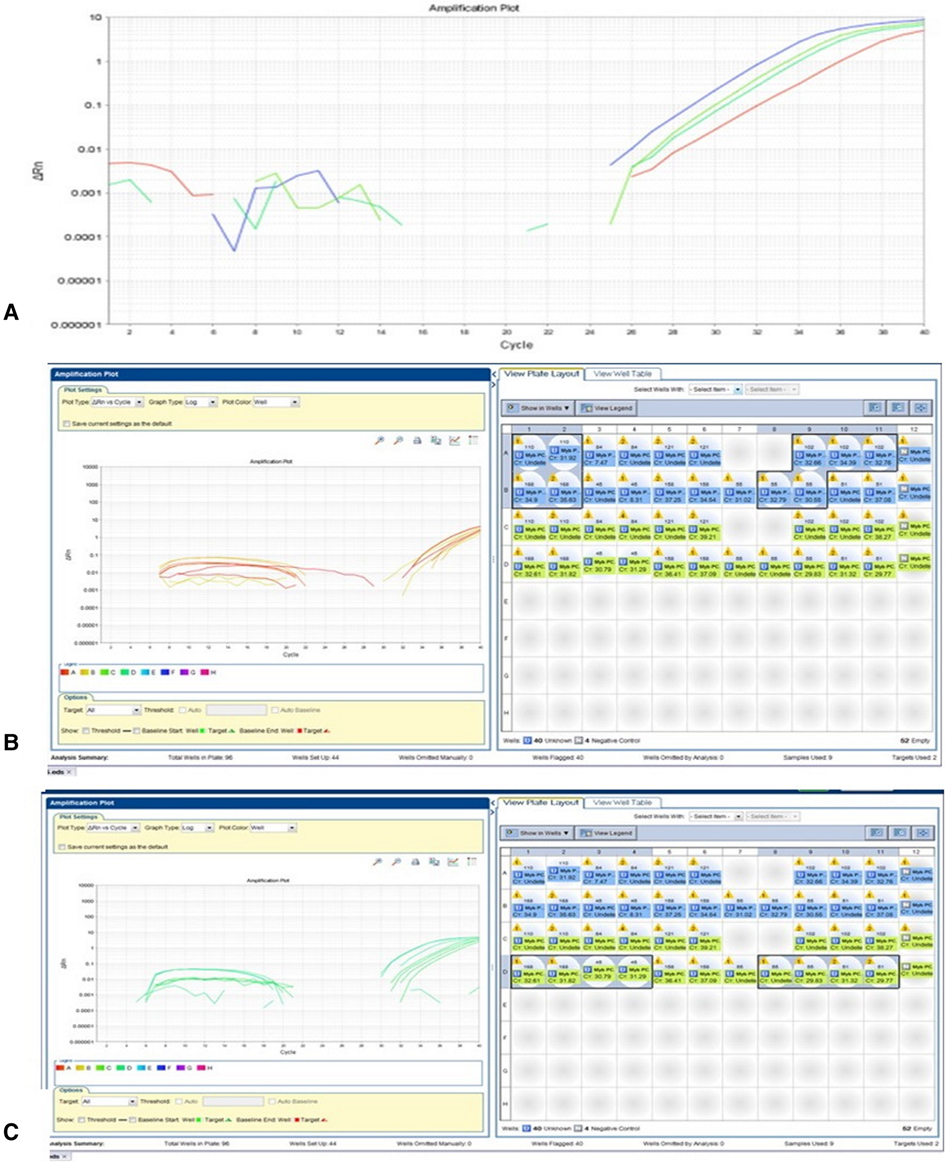
Figure 3. MYB translocation with exon 2 or 3 of PCDHGA1 By RT-PCR. (A) As predicted, the product was detected in sample 53 (110) with primers MYB F + R (blue) and PCDHGA1 F + R (upper green); sample 51 showed product in primers MYB F + R combination (lower green) and PCDHGA1 F + R (red). (B) RT-PCR results of MYB exon 9 and exon 2 of PCDHGA1. Note 4 positive samples for this translocation 53(110), 55(102) – PMA; 168- oligodendroglioma; 60(55)-PXA. (C) RT-PCR results of MYB exon 9 and exon 3 of PCDHGA1. MYB forward & PCDHGA1 reveres show product only in translocation samples. Note 4 positive samples for this translocation (51- DA; 50(45)- ganglioglioma;168 - oligodendroglioma; 60(55) - PXA).
Of the 10 samples, 5 were positive for translocation of MYB with exon 2 of PCDHGA1, and 4 were positive for MYB translocation with exon 3 of PCDHGA1. All samples positive for a translocation were negative for BRAF except sample 51.
To determine whether MYBL1 duplication-truncation was present in grade II pediatric LGGs, PCR for genomic DNA and RT-PCR were used. Ten samples of grade II gliomas were analyzed at the DNA level (Tables 3 and 4, Figure 3). A band at 350 bp on the gel represents a PCR product of the reaction. Eight samples were positive, as follows: no. 50 (ganglioglioma), 52, 51 and 49 (diffuse astrocytoma), 55 and 53 (pilomyxoid astrocytoma), 54 (pleomorphic xanthoastrocytoma), and 62 (oligodendroglioma). The remaining sample, no. 60 (pleomorphic xanthastrocytoma), was negative.
When PCR products of genomic DNA were run on gel, we detected a band at 350 bp in 6 samples: nos. 49, 51 and 52 (diffuse astrocytoma), 50 (ganglioglioma, and 55 and 53 (pilomyxoid astrocytoma). In one sample (no. 60, pleomorphic xanthoastrocytoma), the band was undetected, and in one (no. 54, pleomorphic xanthoastrocytoma), the results were equivocal. Sample 62 (oligodendroglioma) was negative in the gel but positive on RT-PCR. The data are summarized in Table 4.
It was clear that there were discordant results for MYBL1, so we appllied several different methods in an attempt to reach conclusive results.
Analysis of mRNA (converted to cDNA) was performed. The presence of amplification suggested positive MYBL1-trunc2 (Figures 4A,B). Four samples were positive for MYBL1-trunc2: nos. 50 (ganglioglioma), 58 (pilocytic astrocytoma), 60 (pleomorphic xanthoastrocytoma), and 52 (diffuse astrocytoma) (Table 4). Four samples were negative for MYBL1-trunc2: nos. 54 (pleomorphic xanthoastrocytoma), 62 (oligodendroglioma), 53 (pilomyxoid astrocytoma), and 61 (pleomorphic xanthoastrocytoma).

Figure 4. MYBL-1 duplication–truncation by RT-PCR. Genomic DNA of Glioma grade II: Negative (n = 1): 60(55)-PXA. Positive (n = 8): 49–55 and 168. Key of gel order: 50(45) GG, 52(84) FA, 51 FA, 49(23) FA, 55(102) PMA, 53(110) PMA, 54(158) PXA, 168 ODG. Of note, sample 168 was negative on gel, although positive in RTPCR.
RNA sequencing results for BRAF showed no genetic alteration in 6 samples examined, probably because of low coverage in the area. On Sanger sequencing, 2 patients had the BRAF V600E genetic alteration.
RNA sequencing analysis of CDKN2A revealed one genetic alteration with high coverage quality, rs11515, in 5 of 6 patients. RT-PCR was performed on 9 grade II LGG samples. Deletions were detected in one of them (no. 54) in both exon 1 and exon 2 of CDKN2A (Figure 5).
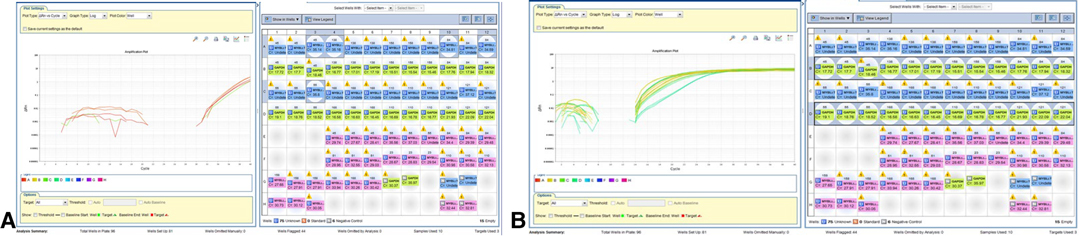
Figure 5. Positive MYBL1-trunc2. (A) Detection of MYBL-1 trunc 2 positive samples are shown:50(45), 58(138), 60(55), 52(84). Negative 4 samples: 54(158) PXA, 168 ODG, 53 (110) PMA, and 121 PXA. (B) Expression of normalized GAPDH gene in grade II gliomas samples.
RNA sequencing of IDH1 and IDH2 revealed no genetic alteration in IDH1 and 13 potential single nucleotide polymorphisms (SNPs) in IDH2 (n = 6). Eight of the SNPs were known intron variants with no pathogenic effect, 3 were unknown intron variants and were found to be heterozygous, and 2 were homozygous unknown intron variants and are under investigation.
In the present study, we used multiple newly implemented techniques to investigate some of the genetic alterations in IDH1, IDH2, CDKN2A, MYB and MYBL1 in pediatric low-grade gliomas. All these genes are relevant to the pathogenesis of LGGs, particularly the less studied and less common grade II gliomas. Nowadays, with the accumulation of data, the preferred method should be NGS (11, 19, 20).
In the present study, we showed that none of the LGGs had a genetic alteration in IDH1/2. This finding is in line with reports in the literature that IDH1/2 genetic alterations appear almost exclusively in adult gliomas (10, 21). In adults with grade II-III gliomas, lack of an IDH genetic alteration indicates higher risk of transformation to a higher-grade tumor and poorer prognosis (22). Additionally, the grade II gliomas most often associated with BRAF genetic alterations were pleomorphic xanthoastrocytomas, similar to the findings of Myung et al. (23) Most samples with MYB- PCDHGA1 translocation and MYBL1 genetic rearrangement did not have a BRAF genetic alteration (9). The rate of BRAF genetic alteration in the grade II samples (3/9, 33%) was significantly higher than the reported rate for grade I pilocytic astrocytoma (13). We found MYB- translocations with exon 2 or 3 of PCDHGA1 5/10 and 4/10 grade II gliomas respectively, and MYBL1 rearrangements in 6/9 of the samples examined. In grade II gliomas, the MYB translocations (with exon 2 of PCDHGA1 in 5 samples and exon 3 of PCDHGA1 in 4 samples) and the MYBL1 rearrangement (in 6 samples) were technically difficult to prove and validate, which could serve as an indication for differentiating grade I from grade II-III gliomas, which have a potentially more aggressive outcome. These data are higher than reported elsewhere, and further studies and analytical methods are required.
Our detection of different combinations of BRAF genetic alterations, MYB- PCDHGA1 translocations, and MYBL1 rearrangement in the same samples contradicts the hypothesis of Zhang et al. (21) that BRAF and MYB I genetic alterations are mutually exclusive. The limited amount of sample material that was available for the study prevented us from confirming or repeating the results. Nevertheless, given that we used several different methods (PCR, RT-PCR, RACE, RNA sequencing), we assume the findings are correct.
Although Ramiksson et al. (9) showed the same results when analyzing DNA and cDNA (mRNA), we were unable to find a complete match. The DNA results are most likely more reliable, as mRNA is sensitive and minimal material was available. Samples 45 (ganglioglioma) and 84 (diffuse astrocytoma) were positive by all methods, but samples 158 (pleomorphic xanthoastrocytoma), 168 (oligodendroglioma), and 110 (pilomyxoid astrocytoma) were positive only on DNA analysis. (Some findings were confirmed in gel.) We cannot explain the positive cDNA and negative DNA results of sample 55.
In this study, we were pioneers in the implementation of new molecular methods to the analysis of data of our cohort. Further efforts must be taken to keep involved in the state-of-the art molecular methods, understand and use novel methods to detect translocations, genetic rearrangements, and abnormal levels of expression of several candidate genes in pediatric LGGs (23, 24). Our results are in line with the recent leading publications (6, 9, 10, 24) although we need to further explain the relative high occurrence of genetic alterations detected here in MYBL and MYB. At present, chemotherapy and radiotherapy are the mainstay of treatment for unresectable or progressive disease. It is clear that genetic abnormalities and activating genetic alterations are significant events in tumorigenesis in pediatric LGG and further exploration of the molecular pathways underlying their development will pave the way for the formulation of targeted treatments. BRAF inhibitors are already in use in the treatment of BRAF mutated high and low grade gliomas LGGs (25, 26). cMYC inhibitor may hold promise for the management of tumors caused by MYB and MYBL1 translocations, Downstream MEK inhibitors are already in phase 2 in clinical trials for children with BRAF fused gliomas and phase 1 studies with FGFR1 inhibitors have started recently.
Archival samples of pediatric LGGs (WHO grades I–II) were obtained from the Department of Pathology, Rabin Medical Center. Fresh tissues were also collected and analyzed after informed consent was obtained from the parents. Inclusion criteria were patient age <18 years and sufficient paraffin-embedded or frozen tissue for analysis. Tumor classification and cellularity were reviewed by a neuropathologist. Demographic and clinical data were collected from the medical files. The study was approved by the national and institutional review boards.
Paraffin-embedded tissues or frozen tissues were used for DNA isolation. In brief, 10 µm sections were stained with hematoxylin-eosin, and the slides were reviewed by a pathologist. The areas with an estimated content of more than 75% tumor cells were then separated by microdissection (no. 11 surgical blade) from 5 consecutive 10-µm unstained paraffin-embedded sections of each block. The tissues were deparaffinized and incubated overnight in 1% sodium dodecyl sulfate and proteinase K 0.5 mg/mL. DNA was purified by phenol chloroform extraction and ethanol precipitation and dissolved in 50 µL of distilled water, as described previously (13). All fresh tissues removed at surgery were taken from areas immediately adjacent to the tumor that was submitted to pathology.
Tumor tissue was snap-frozen in liquid nitrogen. Total RNA was isolated using TRIzol reagent (Invitrogen, Life Technologies, Carlsbad, CA) according to the manufacturer’s protocol and then reverse transcribed into complementary DNA (cDNA) using random hexamers (Amersham Biosciences, Buckinghamshire, United Kingdom) and Moloney murine leukemia virus reverse transcriptase (Promega, Madison, WI).
IDH1 and IDH2 analysis using specific designed primers for Sanger analysis.
RT-PCR was used to check deletions in CDKN2A. The ratio between the CDKN2A gene Ct and the reference gene RNase Ct was calculated. A ratio of <0.5 was considered a deletion. The primers for exons 1 and 2 of CDKN2A and the reference gene RNase are shown in Table 5 (24).
Oncogenic genetic alterations in MYB and MYBL1 were analyzed with rapid amplification of cDNA ends (RACE) using specific primers designed for the RACE reaction. The method entails initiation of first strand cDNA synthesis at the poly(A) tail of mRNA using the adapter primer and then destroying the original mRNA template with RNase. Specific cDNA is amplified by PCR using a gene-specific primer, which anneals in a region of known exon sequences, and an adapter primer that targets the 5´ terminus. The primers are listed in Table 6.
To identify MYB translocation and MYBL1 rearrangement, RNA was extracted from the tissue and converted to cDNA, as described (13). Triplicates of cDNA from LGG samples were analyzed. The mastermix contained 1 µg (1 µL), forward primer 0.5 µL (10 µM), reverse primers 0.5 µL (10 µM), Syber Fast 5 µL and H2O 3 µL. The total reaction volume (10 mL) was loaded into a 96-well plate and run on StepOne System software, v 2.2 (Applied Biosystems, Foster City, CA, USA) in real time. The primers are listed in Table 2. The Ct levels of product expression were calculated.
To investigate whether MYB and BRAF genetic alterations are mutually exclusive, we analyzed MYB and BRAF rearrangements in the same samples.
After DNA extraction, each PCR amplification was performed in a 50 µL reaction volume containing 150 ng of sample DNA as a template using specific primers for the relevant genes. PCR parameters were as follows: denaturation for 5 min at 95°C; 35 cycles of 1 min at 95°C; and annealing for 1 min at 56–60°C and for 1 min at 72°C with Taq polymerase. The PCR product was amplified on 2% agarose gel and visualized with ethidium bromide staining.
Direct sequencing of the PCR products was performed with reagents and an analyzer (Big Dye Terminator Cycle Sequencing and ABI PRISM 3700 DNA Analyzer; Applied Biosystems).
For RNA sequencing, we used 5 samples of grade II LGGs (2 pilomyxoid astrocytoma, 2 diffuse astrocytoma, 1 pleomorphic xanthoastrocytoma) and one sample of grade I LGG (pilocytic astrocytoma).
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
The studies involving human participants were reviewed and approved by Bnai Zion Medical Center Local Ethics Committee. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
OB, HT and SM collected the data, OB and SW performed molecular analysis, HT, AZ and NG-C analyzed the results, HT, AZ and NG-C wrote the main manuscript, all authors reviewed the manuscript. All authors contributed to the article and approved the submitted version.
This work was part of the master thesis of Orit Barinfeld, Tel Aviv University. Supported in part by the Isabelle and Zangvyl Krieger Fund, Baltimore, MD, USA; Israel Cancer Association; Novartis Pharma Ltd Israel. Claire and Amedee Maratier Institute for the Study of Blindness and Visual Disorders, Sackler Faculty of Medicine, Tel Aviv University, Tel Aviv, Israel.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Tatevossian RG, Lawson ARJ, Forshew T, Hindley GFL, Ellison DW, Sheer D. MAPK pathway activation and the origins of pediatric low-grade astrocytomas. J Cell Physiol. (2010) 222:509–14. doi: 10.1002/jcp.21978
2. Gnekow AK, Kortmann RD, Pietsch T, Emser A. Low grade chiasmatic-hypothalamic glioma - carboplatin and vincristin chemotherapy effectively defers radiotherapy within a comprehensive treatment strategy. Klin Pädiatr. (2004) 216:331–42. doi: 10.1055/s-2004-832355
3. Ohgaki H, Kleihues P. Population-based studies on incidence, survival rates, and genetic alterations in astrocytic and oligodendroglial gliomas. J Neuropathol Exp Neurol. (2005) 64:479–89. doi: 10.1093/jnen/64.6.479
4. Fisher PG, Tihan T, Goldthwaite PT, Wharam MD, Carson BS, Weingart JD, et al. Outcome analysis of childhood low-grade astrocytomas. Pediatr Blood Cancer. (2008) 51:245–50. doi: 10.1002/pbc.21563
5. Lyustikman Y, Momota H, Pao W, Holland EC. Constitutive activation of Raf-1 induces glioma formation in mice. Neoplasia. (2008) 10:501–10. doi: 10.1593/neo.08206
6. Ryall S, Zapotocky M, Fukuoka K, Nobre L, Guerreiro Stucklin A, Bennett J, et al. Integrated molecular and clinical analysis of 1,000 pediatric low-grade gliomas. Cancer Cell. (2020) 37:569–83.e5. doi: 10.1016/j.ccell.2020.03.011
7. Pfister S, Janzarik WG, Remke M, Ernst A, Werft W, Becker N, et al. BRAF gene duplication constitutes a mechanism of MAPK pathway activation in low-grade astrocytomas. J Clin Invest. (2008) 118:1739–49. doi: 10.1172/JCI33656
8. Eisenhardt AE, Olbrich H, Röring M, Janzarik W, Anh TN, Cin H, et al. Functional characterization of a BRAF insertion mutant associated with pilocytic astrocytoma. Int J Cancer. (2011) 129:2297–303. doi: 10.1002/ijc.25893
9. Ramkissoon LA, Horowitz PM, Craig JM, Ramkissoon SH, Rich BE, Schumacher SE, et al. Genomic analysis of diffuse pediatric low-grade gliomas identifies recurrent oncogenic truncating rearrangements in the transcription factor MYBL1. Proc Natl Acad Sci USA. (2013) 110:8188–93. doi: 10.1073/pnas.1300252110
10. Ryall S, Tabori U, Hawkins C. Pediatric low-grade glioma in the era of molecular diagnostics. Acta Neuropathol Comm. (2020) 8. doi: 10.1186/s40478-020-00902-z
11. Jones DTW, Kieran MW, Bouffet E, Alexandrescu S, Bandopadhayay P, Bornhorst M, et al. Pediatric low-grade gliomas: next biologically driven steps. Neuro-Oncology. (2018) 20:160–73. doi: 10.1093/neuonc/nox141
12. Miklja Z, Pasternak A, Stallard S, Nicolaides T, Kline-Nunnally C, Cole B, et al. Molecular profiling and targeted therapy in pediatric gliomas: review and consensus recommendations. Neuro-Oncology. (2019) 21:968–80. doi: 10.1093/neuonc/noz022
13. Laviv Y, Toledano H, Michowiz S, Dratviman-Storobinsky O, Turm Y, Fichman-Horn S, et al. BRAF, GNAQ, and GNA11 mutations and copy number in pediatric low-grade glioma. FEBS Open Bio. (2012) 2:129–34. doi: 10.1016/j.fob.2012.05.004
14. Broniscer A, Baker SJ, West AN, Fraser MM, Proko E, Kocak M, et al. Clinical and molecular characteristics of malignant transformation of low-grade glioma in children. J Clin Oncol. (2007) 25:682–9. doi: 10.1200/JCO.2006.06.8213
15. Dougherty MJ, Santi M, Brose MS, Ma C, Resnick AC, Sievert AJ, et al. Activating mutations in BRAF characterize a spectrum of pediatric low-grade gliomas. Neuro-Oncology. (2010) 12:621–30. doi: 10.1093/neuonc/noq007
16. Pollack IF, Hamilton RL, James CD, Finkelstein SD, Burnham J, Yates AJ, et al. Rarity of PTEN deletions and EGFR amplification in malignant gliomas of childhood: results from the Children’s Cancer Group 945 cohort. J Neurosurg. (2006) 105(5 Suppl):418–24. doi: 10.3171/ped.2006.105.5.418
17. Tatevossian RG, Tang B, Dalton J, Forshew T, Lawson AR, Ma J, et al. MYB upregulation and genetic aberrations in a subset of pediatric low-grade gliomas. Acta Neuropathol. (2010) 120:731–43. doi: 10.1007/s00401-010-0763-1
18. Von Deimling A, Korshunov A, Hartmann C. The next generation of glioma biomarkers: MGMT methylation, BRAF fusions and IDH1 mutations. Brain Pathol. (2011) 21:74–87. doi: 10.1111/j.1750-3639.2010.00454.x
19. Rubin JB, Finlay JL. Pediatric low-grade gliomas: a brave new world. Neuro-Oncology. (2018) 20:149–50. doi: 10.1093/neuonc/nox221
20. Packer RJ, Pfster S, Bouffet E, Avery R, Bandopadhayay P, Bornhorst M, et al. Pediatric low-grade gliomas: implications of the biologic era. Neuro-Oncology. (2017) 19:750–61. doi: 10.1093/neuonc/now209
21. Zhang J, Wu G, Miller CP, Tatevossian RG, Dalton JD, Tang B, et al. Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet. (2013) 45:602–12. doi: 10.1038/ng.2611
22. Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. (2009) 360:765–73. doi: 10.1056/NEJMoa0808710
23. Myung JK, Cho H, Park CK, Kim SK, Lee SH, Park SH. Analysis of the BRAF(V600E) mutation in central nervous system tumors. Translat Oncol. (2012) 5:430–6. doi: 10.1593/tlo.12328
24. Bieńkowski M, Piaskowski S, Stoczyńska-Fidelus E, Szybka M, Banaszczyk M, Witusik-Perkowska M, et al. Screening for EGFR amplifications with a novel method and their significance for the outcome of glioblastoma patients. PLoS One. (2013) 8:e65444. doi: 10.1371/journal.pone.0065444
25. Raabe E, Kieran MW, Cohen KJ. New strategies in pediatric gliomas: molecular advances in pediatric low-grade gliomas as a model. Clin Cancer Res. (2013) 19:4553–8. doi: 10.1158/1078-0432.CCR-13-0662
Keywords: glioma, pediatric, molecular, IDH1, MYB, MYBL1, CDKN2A
Citation: Barinfeld O, Zahavi A, Weiss S, Toledano H, Michowiz S and Goldenberg-Cohen N (2022) Genetic Alteration Analysis of IDH1, IDH2, CDKN2A, MYB and MYBL1 in Pediatric Low-Grade Gliomas. Front. Surg. 9:880048. doi: 10.3389/fsurg.2022.880048
Received: 20 February 2022; Accepted: 14 April 2022;
Published: 28 April 2022.
Edited by:
Zohreh Habibi, Tehran University of Medical Sciences, IranReviewed by:
Naureen Mushtaq, Aga Khan University, PakistanCopyright © 2022 Barinfeld, Zahavi, Weiss, Toledano, Michowiz and Goldenberg-Cohen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nitza Goldenberg-Cohen bmNvaGVuMUBnbWFpbC5jb20=
†These authors have contributed equally to this work
Speciality section: This article was submitted to Neurosurgery, a section of the journal Frontiers in Surgery
Abbreviations: IDH1, isocitrate dehydrogenase 1; IDH2, isocitrate dehydrogenase 2; CDKN2A, Cyclin Dependent Kinase Inhibitor 2A; MYB, myeloblastosis; MYBL1, MYB Proto-Oncogene Like 1; PCDHGA1, Protocadherin Gamma Subfamily A, 1; LGG, Low grade glioma; Ras, Rat sarcoma virus; Raf, Rapidly Accelerated Fibrosarcoma; ERK, extracellular signal-regulated kinases; MAPK (MEK), mitogen-activated protein kinase; GNAQ, Guanine Nucleotide binding protein, Alpha Stimulating activity polypeptide.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.