- 1Division for Therapies against Intractable Diseases, Institute for Comprehensive Medical Science, Fujita Health University, Toyoake, Japan
- 2Genome and Transcriptome Analysis Center, Fujita Health University, Toyoake, Japan
- 3Department of Biochemistry, Faculty of Medicine, Kindai University, Osaka-Sayama, Japan
- 4Department of Bioscience and Genetics, National Cerebral and Cardiovascular Center Research Institute, Suita, Japan
Introduction
Adult rodent skeletal muscles are composed of four types of myofibers (Schiaffino and Reggiani, 1996, 2011; Bassel-Duby and Olson, 2006), which are present in different proportions in different muscles. Slow-twitch (type I) fibers have fatigue resistance properties by virtue of large numbers of mitochondria. The remaining three types are fast-twitch (type II) fibers, subclassified as types IIA, IIX, and IIB. Of these, type IIA fibers exhibit the slowest shortening velocity and display fatigue resistance due to a high density of mitochondria. Conversely, type IIB fibers have the fastest shortening velocity and exhibit exercise intolerance due to a low density of mitochondria, while type IIX fibers are intermediate. Although the predominant fiber types in each muscle are determined during embryonic and fetal myogenesis (Lu et al., 1999; Agbulut et al., 2003), functional adaptations can lead to the alteration of these proportions through changes in gene expression (Swoap et al., 2000; Oh et al., 2005).
Vestigial is a nuclear protein that is essential for Drosophila wing formation and muscle differentiation (Paumard-Rigal et al., 1998; Simon et al., 2016). Four mammalian vestigial-like proteins (Vgll1, Vgll2, Vgll3, and Vgll4) have been identified, which support the function of TEA domain (TEAD) transcription factors in a tissue-specific manner (Maeda et al., 2002; Mielcarek et al., 2002; Pobbati and Hong, 2013). One of these proteins, Vgll2/VITO-1, is expressed in adult skeletal muscles (Maeda et al., 2002; Mielcarek et al., 2002) and plays a role in in vitro muscle differentiation (Maeda et al., 2002; Günther et al., 2004). Our previous study using Vgll2 knockout (KO) mice revealed that Vgll2 regulates skeletal muscle fiber composition by repressing the neonatal expression of sex-determining region Y (SRY)-box 6 (Sox6), trans-acting transcription factor 3 (Sp3), and purine-rich element-binding protein B (Purβ), which are transcriptional repressors of slow-twitch fiber-related genes (Honda et al., 2017).
In humans, long-term resistance training not only induces muscle hypertrophy but also increases the proportion of slower muscle fibers by increasing and decreasing the number of type IIA and type IIX fibers, respectively (Hather et al., 1991; Adams et al., 1993; Williamson et al., 2001; Bickel et al., 2011). Like resistance training, mechanical overload (MOV, also called chronic overload) by synergistic ablation of the soleus and gastrocnemius muscles induces hypertrophy in the plantaris muscles. MOV also modifies the fiber type proportions in the plantaris muscles toward slower fibers by shifting type IIX and IIB fibers to type I and IIA fibers, through the activation of multiple genes related to muscle contractility in mice (Karasseva et al., 2003; Ji et al., 2007; McGee et al., 2008; Pérez-Schindler et al., 2013). We previously showed that MOV increases Vgll2 protein expression and activity, enhancing the functional adaptation of skeletal muscle (Honda et al., 2019). In addition, mice lacking Vgll2 exhibited limited fiber type transition after MOV due to the repression of slow muscle genes, which are increased by MOV in wild-type mice. Vgll2 KO mice also displayed downregulated expression of several genes involved in oxidative metabolism. Thus, Vgll2 is essential for the fast-to-slow shift of muscle fibers after MOV and acts by regulating the expression of downstream genes at the transcriptional level (Honda et al., 2019). However, gene expression changes downstream of Vgll2 in skeletal muscle subjected to MOV have not been systematically identified. To examine molecular network changes in skeletal muscle after MOV, we conducted a comparative gene expression analysis in wild-type and Vgll2 KO mice under both sedentary and MOV conditions. Raw data files have been deposited to the Sequence Read Archive of the DNA Data Bank of Japan (DDBJ) under Accession No. DRA008472.
Methods
Animals
Vgll2 KO mice and wild-type littermates on a C57BL/6J background were generated as previously described (Honda et al., 2017) and housed in cages at 24°C with a 12:12-h light–dark cycle. Animal experiments were approved by the Animal Care and Use Committee of the National Cerebral and Cardiovascular Center in Japan and conducted under institutional and national guidelines.
Synergistic Ablation
MOV was performed as previously described (McGee et al., 2008; Honda et al., 2019). In brief, for synergistic ablation of the plantaris, the soleus and gastrocnemius were surgically removed from 10-weeks-old male wild-type littermates (Wild_MOV) and Vgll2 KO mice (KO_MOV) under anesthesia (2.5% isoflurane). Mice that underwent sham surgeries were used as controls (Wild_sham, KO_sham). To explore the results of long-term effects of MOV, 6 weeks after surgery, the mice were sacrificed, and the plantaris muscles were collected and submerged in RNAlater (Thermo Fisher Scientific, Waltham, MA, USA) for RNA isolation.
RNA Isolation and Library Preparation
Total RNA was extracted from the plantaris muscles using the miRNeasy Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions and quantified using a NanoDrop spectrophotometer (Thermo Fisher Scientific). Poly(A)+ RNA was purified from 1 μg of total RNA using the NEBNext Poly(A) mRNA Magnetic Isolation Module (New England Biolabs, Ipswich, MA, USA) and used for RNA sequencing (RNA-Seq) library preparation with the NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs), according to the manufacturer's protocol. The quantity of the libraries was assessed using a Library Quantification Kit (Takara, Shiga, Japan) and a Thermal Cycler Dice Real Time System TP800 (Takara); their quality was assessed with a DNA 1000 Kit and a Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA).
Transcriptome Analysis
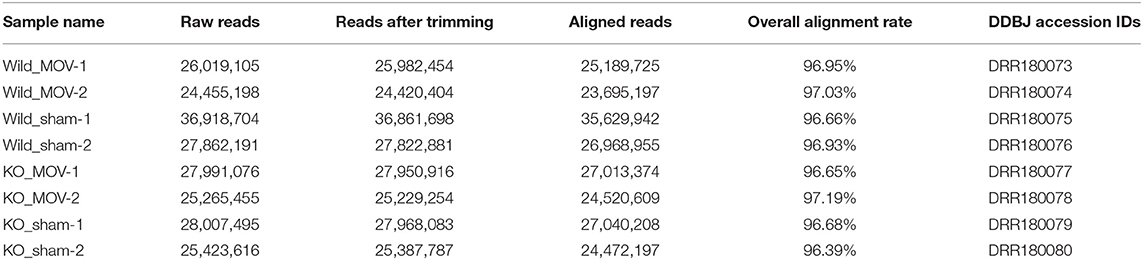
RNA-Seq libraries were sequenced with 124-bp single-end reads using a HiSeq 1500 system (Illumina, San Diego, CA, USA) at Fujita Health University. Two biological replicates were performed per sample. Base calling was conducted using bcl2fastq ver. 1.8.4 software. The RNA-Seq raw data have been deposited in the DDBJ Sequence Read Archive under Accession No. DRA008472. DDBJ accession IDs for each sample are listed in Table 1. Raw sequence data were cleaned using FastQC ver. 0.11.3 software (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and the command “–Q 33 –t 20 –l 30”. Trimmed reads were aligned to the mouse reference genome (mm10) using HISAT2 ver. 2.0.5 (Pertea et al., 2016) using default parameters. SAMtools ver. 1.3.1 software (Li et al., 2009) was used to convert SAM files, which contain aligned reads, into BAM files. A summary of sample names, raw reads, and aligned reads is provided in Table 1.
Statistical Analysis of Differentially Expressed Genes
Statistical analysis of RNA-Seq data was performed as previously described (Hitachi et al., 2019). In brief, to count the aligned reads, HTSeq ver. 0.6.0 software (Anders et al., 2015) was used against the Mus_musculus_UCSC_mm10.gtf file with the optional command “–stranded=no –format=bam”. We compared gene expression levels in Wild_sham mice with those in Wild_MOV, KO_sham, and KO_MOV mice using DESeq2 software ver. 1.12.4 (Love et al., 2014) using the Wald test. Differentially expressed genes with a false discovery rate (adjusted P-value, padj) <0.001 and log2-fold change >1 or <-1 were considered significant. The expression levels of all genes in each experimental group are listed in Table S1.
Using these parameters, we identified 454 differentially expressed genes (398 upregulated and 56 downregulated) after MOV in wild-type mice. Although in the absence of MOV, statistically significant expression changes were observed in only 18 genes (11 upregulated and 7 downregulated) in Vgll2 KO mice compared to wild-type mice; in the presence of MOV, the expression levels of 1,889 genes (1,336 upregulated and 553 downregulated), including a majority of genes whose expression was altered by MOV in wild-type mice, were significantly altered in Vgll2 KO mice compared with wild-type mice subjected to the sham operation (Figure 1A). Intriguingly, although Vgll2 is required to activate the expression of genes involved in the shift from fast to slow muscle fibers by MOV (Honda et al., 2019), Vgll2 deficiency eventually enhanced the effects of MOV on gene expression (Figure 1B). Therefore, these results suggest that Vgll2 plays a role in the functional adaptation of skeletal muscle by MOV beyond the fast-to-slow shifting of muscle fibers.
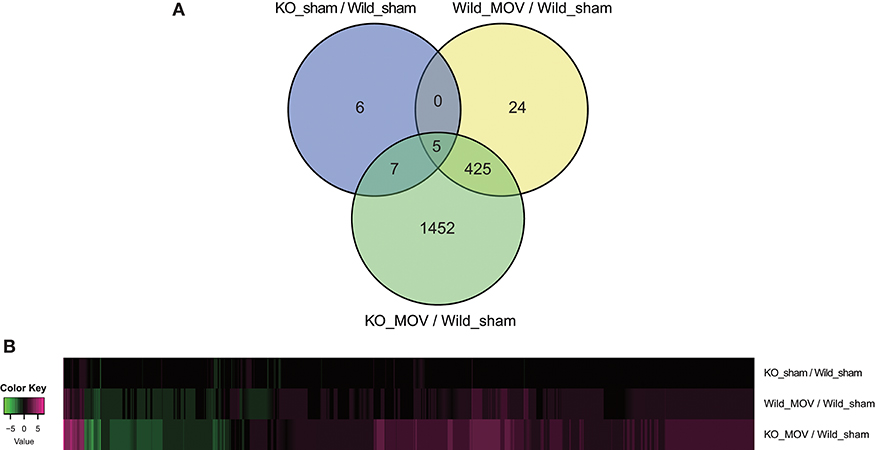
Figure 1. Summary of differentially expressed genes in the skeletal muscle of Vgll2 knockout (KO) mice subjected to mechanical overload (MOV). (A) Venn diagram of differentially expressed genes between sets of experimental groups. (B) Heat map displaying differentially expressed genes in each experimental group (log2 ratio scale).
These data will be helpful in elucidating the skeletal muscle molecular networks that respond to MOV and allow in-depth analysis of expression changes in genes regulated by Vgll2 under both sedentary and MOV conditions. Interestingly, fusion genes involving Vgll2 have been identified in the spindle cell variant of rhabdomyosarcoma (Alaggio et al., 2016). Thus, our data identifying genes downstream of Vgll2 could also help reveal the molecular mechanisms underlying not only MOV but also human diseases caused by Vgll2 fusion genes.
Data Availability Statement
The RNA-Seq raw data have been deposited in the DDBJ Sequence Read Archive under Accession No. DRA008472 (https://ddbj.nig.ac.jp/DRASearch/study?acc=DRP005135).
Ethics Statement
The animal study was reviewed and approved by the Animal Care and Use Committee of the National Cerebral and Cardiovascular Center in Japan.
Author Contributions
KH, HI, HK, HO, and KT conducted the experiments. KH and MH designed the experiments and prepared the manuscript.
Funding
This work was supported by Grants-in-Aid for Scientific Research (KAKENHI; 15K16501, 17K08646, and 19K11486) from the Japan Society for the Promotion of Science.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Editage (www.editage.jp) for English language editing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fspor.2019.00041/full#supplementary-material
Table S1. Plantaris RNA-Seq results.
References
Adams, G. R., Hather, B. M., Baldwin, K. M., and Dudley, G. A. (1993). Skeletal muscle myosin heavy chain composition and resistance training. J. Appl. Physiol. 74, 911–915. doi: 10.1152/jappl.1993.74.2.911
Agbulut, O., Noirez, P., Beaumont, F., and Butler-Browne, G. (2003). Myosin heavy chain isoforms in postnatal muscle development of mice. Biol. Cell. 95, 399–406. doi: 10.1016/S0248-4900(03)00087-X
Alaggio, R., Zhang, L., Sung, Y. S., Huang, S. C., Chen, C. L., Bisogno, G., et al. (2016). A molecular study of pediatric spindle and sclerosing rhabdomyosarcoma: identification of novel and recurrent VGLL2-related fusions in infantile cases. Am. J. Surg. Pathol. 40, 224–235. doi: 10.1097/PAS.0000000000000538
Anders, S., Pyl, P. T., and Huber, W. (2015). HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. doi: 10.1093/bioinformatics/btu638
Bassel-Duby, R., and Olson, E. N. (2006). Signaling pathways in skeletal muscle remodeling. Annu. Rev. Biochem. 75, 19–37. doi: 10.1146/annurev.biochem.75.103004.142622
Bickel, C. S., Cross, J. M., and Bamman, M. M. (2011). Exercise dosing to retain resistance training adaptations in young and older adults. Med. Sci. Sports. Exerc. 43, 1177–1187. doi: 10.1249/MSS.0b013e318207c15d
Günther, S., Mielcarek, M., Krüger, M., and Braun, T. (2004). VITO-1 is an essential cofactor of TEF1-dependent muscle-specific gene regulation. Nucleic Acids Res. 32, 791–802. doi: 10.1093/nar/gkh248
Hather, B. M., Tesch, P. A., Buchanan, P., and Dudley, G. A. (1991). Influence of eccentric actions on skeletal muscle adaptations to resistance training. Acta Physiol. Scand. 143, 177–185. doi: 10.1111/j.1748-1716.1991.tb09219.x
Hitachi, K., Nakatani, M., Takasaki, A., Ouchi, Y., Uezumi, A., Ageta, H., et al. (2019). Myogenin promoter-associated lncRNA Myoparr is essential for myogenic differentiation. EMBO Rep. 20:e47468. doi: 10.15252/embr.201847468
Honda, M., Hidaka, K., Fukada, S.-I., Sugawa, R., Shirai, M., Ikawa, M., et al. (2017). Vestigial-like 2 contributes to normal muscle fiber type distribution in mice. Sci. Rep. 7:7168. doi: 10.1038/s41598-017-07149-0
Honda, M., Tsuchimochi, H., Hitachi, K., and Ohno, S. (2019). Transcriptional cofactor Vgll2 is required for functional adaptations of skeletal muscle induced by chronic overload. J. Cell Physiol. 234, 15809–15824. doi: 10.1002/jcp.28239
Ji, J., Tsika, G. L., Rindt, H., Schreiber, K. L., McCarthy, J. J., Kelm, R. J., et al. (2007). Purα and Purβ collaborate with Sp3 to negatively regulate β-myosin heavy chain gene expression during skeletal muscle inactivity. Mol. Cell Biol. 27, 1531–1543. doi: 10.1128/MCB.00629-06
Karasseva, N., Tsika, G., Ji, J., Zhang, A., Mao, X., and Tsika, R. (2003). Transcription enhancer factor 1 binds multiple muscle MEF2 and A/T-rich elements during fast-to-slow skeletal muscle fiber type transitions. Mol. Cell Biol. 23, 5143–5164. doi: 10.1128/mcb.23.15.5143-5164.2003
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Lu, B. D., Allen, D. L., Leinwand, L. A., and Lyons, G. E. (1999). Spatial and temporal changes in myosin heavy chain gene expression in skeletal muscle development. Dev. Biol. 216, 312–326. doi: 10.1006/dbio.1999.9488
Maeda, T., Chapman, D. L., and Stewart, A. F. R. (2002). Mammalian vestigial-like 2, a cofactor of TEF-1 and MEF2 transcription factors that promotes skeletal muscle differentiation. J. Biol. Chem. 277, 48889–48898. doi: 10.1074/jbc.M206858200
McGee, S. L., Mustard, K. J., Hardie, D. G., and Baar, K. (2008). Normal hypertrophy accompanied by phosphorylation and activation of AMP-activated protein kinase alpha1 following overload in LKB1 knockout mice. J. Physiol. (Lond). 586, 1731–1741. doi: 10.1113/jphysiol.2007.143685
Mielcarek, M., Günther, S., Krüger, M., and Braun, T. (2002). VITO-1, a novel vestigial related protein is predominantly expressed in the skeletal muscle lineage. Mech. Dev. 119, S269–S274. doi: 10.1016/S0925-4773(03)00127-8
Oh, M., Rybkin, I. I., Copeland, V., Czubryt, M. P., Shelton, J. M., van Rooij, E., et al. (2005). Calcineurin is necessary for the maintenance but not embryonic development of slow muscle fibers. Mol. Cell Biol. 25, 6629–6638. doi: 10.1128/MCB.25.15.6629-6638.2005
Paumard-Rigal, S., Zider, A., Vaudin, P., and Silber, J. (1998). Specific interactions between vestigial and scalloped are required to promote wing tissue proliferation in Drosophila melanogaster. Dev. Genes Evol. 208, 440–446. doi: 10.1007/s004270050201
Pérez-Schindler, J., Summermatter, S., Santos, G., Zorzato, F., and Handschin, C. (2013). The transcriptional coactivator PGC-1α is dispensable for chronic overload-induced skeletal muscle hypertrophy and metabolic remodeling. Proc. Natl. Acad. Sci. U.S.A. 110, 20314–20319. doi: 10.1073/pnas.1312039110
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., and Salzberg, S. L. (2016). Transcript-level expression analysis of RNA-Seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667. doi: 10.1038/nprot.2016.095
Pobbati, A. V., and Hong, W. (2013). Emerging roles of TEAD transcription factors and its coactivators in cancers. Cancer Biol. Ther. 14, 390–398. doi: 10.4161/cbt.23788
Schiaffino, S., and Reggiani, C. (1996). Molecular diversity of myofibrillar proteins: gene regulation and functional significance. Physiol. Rev. 76, 371–423. doi: 10.1152/physrev.1996.76.2.371
Schiaffino, S., and Reggiani, C. (2011). Fiber types in mammalian skeletal muscles. Physiol. Rev. 91, 1447–1531. doi: 10.1152/physrev.00031.2010
Simon, E., Faucheux, C., Zider, A., Thézé, N., and Thiébaud, P. (2016). From vestigial to vestigial-like: the Drosophila gene that has taken wing. Dev. Genes Evol. 226, 297–315. doi: 10.1007/s00427-016-0546-3
Swoap, S. J., Hunter, R. B., Stevenson, E. J., Felton, H. M., Kansagra, N. V., Lang, J. M., et al. (2000). The calcineurin–NFAT pathway and muscle fiber-type gene expression. Am. J. Physiol. Cell Physiol. 279, C915–C924. doi: 10.1152/ajpcell.2000.279.4.C915
Keywords: skeletal muscle adaptation, resistance training, synergistic ablation, RNA sequencing, vestigial-like 2
Citation: Hitachi K, Inagaki H, Kurahashi H, Okada H, Tsuchida K and Honda M (2019) Deficiency of Vgll2 Gene Alters the Gene Expression Profiling of Skeletal Muscle Subjected to Mechanical Overload. Front. Sports Act. Living 1:41. doi: 10.3389/fspor.2019.00041
Received: 17 July 2019; Accepted: 17 September 2019;
Published: 09 October 2019.
Edited by:
John Joseph McCarthy, University of Kentucky, United StatesReviewed by:
Ferdinand Von Walden, Karolinska Institute (KI), SwedenVandre Casagrande Figueiredo, University of Kentucky, United States
Copyright © 2019 Hitachi, Inagaki, Kurahashi, Okada, Tsuchida and Honda. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Masahiko Honda, bWhvbmRhQG1lZC5raW5kYWkuYWMuanA=
 Keisuke Hitachi
Keisuke Hitachi Hidehito Inagaki2
Hidehito Inagaki2 Hiroki Kurahashi
Hiroki Kurahashi Kunihiro Tsuchida
Kunihiro Tsuchida Masahiko Honda
Masahiko Honda