- 1Human Factors and Behavioral Neurobiology Department, Embry-Riddle Aeronautical University, Daytona Beach, FL, United States
- 2Mechanical Engineering Department, Embry-Riddle Aeronautical University, Daytona Beach, FL, United States
- 3Division of Sciences and Technology, Universidad Ana G. Mendez Cupey Campus, San Juan, PR, United States
Over the past few decades there has been a steady increase in interest in the study of the role of space environment in the genetic and phenotypical changes of microorganisms. More specifically, there are concerns with astronaut health being compromised during missions to the Moon and beyond from changes in many conditions. These include changes in the physiology of bacteria leading to alterations directly related to human health such as virulence and antibiotic resistance or to the functioning of life support systems such as the increase in biofilm formation in the water supply or treatment components. The effects of space conditions on microorganisms have been studied for more than a decade; however, there is still a need to determine the impact of the physiological effect of microgravity not only of bacterial growth, but also on the different virulence-related phenotypes that might contribute to phenotypic plasticity and microbial adaptation. This study focuses on deciphering the phenotypical changes of the commensal bacterium E. coli K12 after growth under simulated microgravity conditions using a 2D microgravity analog. Using a 2D clinostat, Escherichia coli was grown up to 22 days and used to measure changes in phenotypes commonly related to virulence. The phenotypes measured included cell population growth, biofilm development and the response to acidic pH and oxidative stress. Results from our studies showed the tendency to enhanced biofilm formation and a decreased resistance to oxidative stress and to grow under acidic conditions. These results suggest that microgravity regulates the adaptation and phenotypic plasticity of E. coli that could lead to changes in virulence.
Introduction
Environmental conditions in space, such as microgravity, radiation, and extreme temperature fluctuations, present challenges to the survival and growth of organicms, including bacteria (Milojevic and Weckwerth, 2020). Microgravity, in particular, affects bacterial behavior and physiology such as the regulation of growth rates, gene expression, and resistance to antibiotics, among others (Nickerson, et al., 2000; Klaus and Howard, 2006; Castro et al., 2011). Given the constraints to experiment under real microgravity conditions, the use of analogs allows us to simulate some of the effects of microgravity on cells such as reduced sedimentation due to a constant state of free falling and the reduction of shear forces (Klaus, 2001). These analogs significantly increase our capacity to formulate better hypotheses and to predict biological changes at different levels. For example, experiments using E. coli have reported changes in growth rate and biofilm development, reduction in the resistance to acidic pH, alterations in the transport and utilization of metal ions, increase in antibiotic resistance, increase in membrane fluidity and the regulation of gene expression (Lynch et al., 2006; Kim and Rhee, 2016; Yim et al., 2020; Allen et al., 2022; Kim et al., 2022). Another important observation has been the expression of virulence factors such as the LT-1 toxin in entero-toxigenic Escherichia coli (Chopra et al., 2006) and the increase in adhesiveness of adherent-invasive E. coli to mammalian gastrointestinal cells (Allen et al., 2008). These responses were observed in cultures under aerated conditions typical for E. coli and after the exposure to simulated microgravity that lasted between 12 and 48 h, suggesting E. coli cells sensed and responded to a change in their physical environment within only a small number of generations. Although there are multiple studies that emphasize the effect of microgravity in bacterial growth, only a few addresses the effect of prolonged exposure to microgravity in bacterial growth dynamics and other phenotypes. An important consideration is that in a real-life scenario E. coli cells in the human microbiome are subjected to microgravity conditions that last from several days to weeks during stays at the International Space Station, creating the possibility of responses or adaptations not previously observed in short-term experiments. To address this issue, two studies that grew E. coli for 1,000 generations under simulated microgravity reported mutations in genes coding for pili biogenesis (surA), adhesion (fimH) and osmotic stress (betA) (Tirumalai et al., 2017), as well as the increase in resistance to the antibiotics chloramphenicol and cefalotin that persisted for over 100 generations after the exposure to the treatment (Tirumalai et al., 2019), suggesting that prolonged exposure to microgravity selects for phenotypes better adapted to this unusual condition. In this study, we exposed E. coli to simulated microgravity conditions for 22 days, equivalent to approximately 128 generations, in rich media and under non-aerated conditions to induce the microaerophilic growth common to the human gut. Escherichia coli is one of the most well studied bacterial species and it is widely reported to be an important member of the healthy human gut microbiome (Martinson and Walk, 2020); it is also a commonly used model system to study alterations in both its genotype and phenotype when exposed to environmental stressors (Jang et al., 2017). Furthermore, the wide variety and consistency of the altered stress responses reported in this model system have been useful for understanding the overall adaptive mechanisms that occur under the effects of short exposure to microgravity (Aunins et al., 2018). Our objective was to measure the regulation of three phenotypes (biofilm development, resistance to oxidative stress and acid tolerance stress) commonly associated with virulence in our model species, when E. coli strain K12 was exposed to 22 days under clinorotation using the recently developed microgravity analog called the EagleStat (Topolski et al., 2022). This system is a device that is specifically designed to be ergonomically efficient and to allow manipulation of multiple microbiological samples under fractional gravitational loads or microgravity. Prolonged exposure to simulated microgravity has been reported before in Stenotrophomonas maltophilia cultures using a high-aspect rotating vessel for a total of 14 days (Su et al., 2021). Our experiment reports a similar treatment using E. coli K12 exposed 22 days of simulated microgravity that describes phenotypical changes known to contribute to virulence.
Materials and methods
Prolonged exposure of bacteria to simulated microgravity
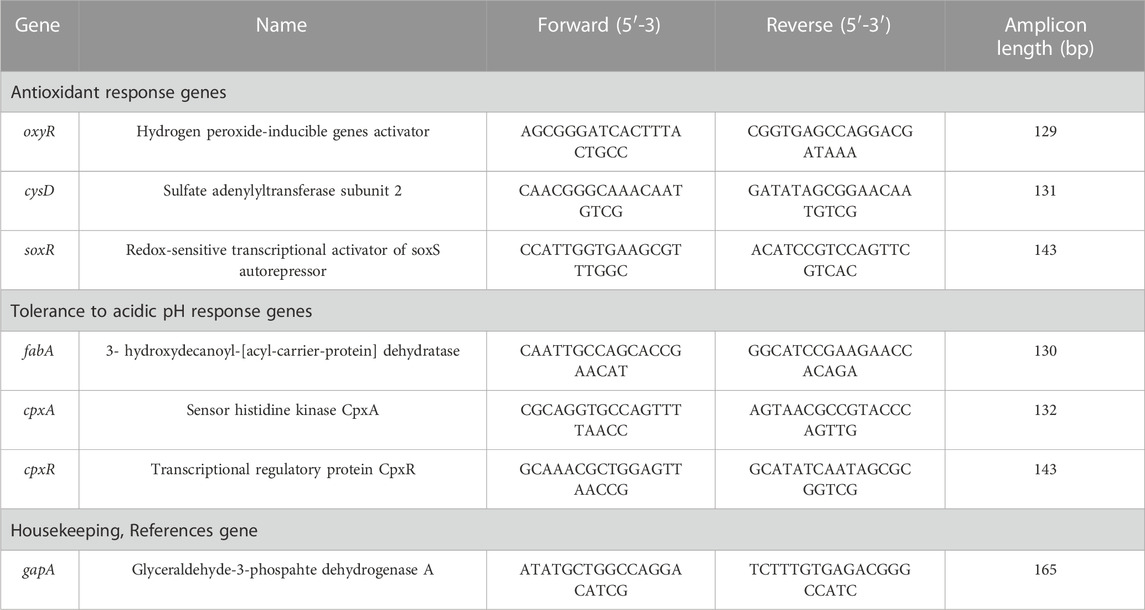
Three overnight cultures of E. coli K12 MG1655 were grown in nutrient broth (0.3% beef extract, 0.5% peptone, 1% sodium chloride) at 30°C in a microbiological incubator under constant agitation (200 rpm) for 15 h. The prolonged exposure to simulated microgravity was performed using a 2D clinostat designed for microbial studies as previously described in Topolski et al. (2022). In brief, 2 mL aliquots of overnight cultures diluted 1:100 in nutrient broth (NB) were transferred to 2 mL screwcap microtubes while avoiding the presence of visible air bubbles to ensure low-shear force conditions required for microgravity simulation. The tubes were mounted on the clinostat on their respective simulated microgravity (SM) and gravity (G) positions (Figure 1) inside a microbiological incubator at 30°C and constant rotation at 8 rpm. For the prolonged exposure to SM and G conditions, 20 μL of each tube were transferred to new tubes containing 1980 μL of nutrient both for a 1:100 dilution, daily, for 22 days. On each transfer, aliquots of 100 μL of culture were mixed with an equal volume of 50% glycerol and frozen at −80°C.
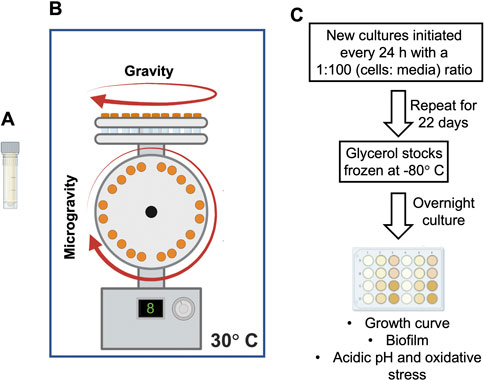
FIGURE 1. Overview of the simulated microgravity experiment on Escherichia coli cultures. (A) Two mL screw-cap tube used as growth vessel. (B) The 2D clinostat used in this study accommodates both simulated microgravity and gravity treatments simultaneously (Topolski et al., 2022). (C) Flow diagram summarizing the cultures’ treatment, storage, re-growth, and the assays performed in this study.
Bacterial growth dynamics
In preparation for all the assays here reported, overnight cultures (OC) from days 4 and 22 samples were prepared by inoculating 2 mL of nutrient broth with 10 μL of each glycerol stock and grown at 30°C under constant agitation at 200 rpm for 15 h. The growth of the cultures was measured from 100 μL aliquots transferred to a 96 well plate to read optical density at 630 nm using a microplate reader (Biotek, Winoosky, VT) and their density adjusted to 0.5 units of absorbance to proceed with the experiment. Normalized OC were diluted 1:100 in nutrient broth and 1 mL aliquots were transferred into multiple wells of a 24-well microtiter plate, using separate plates for days 4 and 22. The plates were incubated at 30°C and their absorbance was measure every hour for 15 h as previously described. Cell count was estimated from serial 10-fold dilutions and plating 10 µL drops, in triplicate, on NB agar plates.
Biofilm formation
Biofilm biomass was measured using the tetrazolium XTT and crystal violet (CV) assays (Kuhn et al., 2003; O’Toole, 2011). For this purpose, 1 mL aliquots of 1:100 dilutions of overnight cultures were prepared in nutrient broth and transferred to individual wells on a flat-bottom, polystyrene 24-well microtiter plate (Corning, Sigma-Aldrich CLS3628, St. Louis, MO) and incubated for 24 h under static conditions at 30°C. After incubation, planktonic cells (those not forming biofilms) were removed by shaking the plate and attached cells (biofilm formers) were washed three times with sterile saline solution (0.85% sodium chloride). Three wells on the plate were reserved for sterile negative controls (only sterile media added). For the CV assay, 1 mL of crystal violet (1% aqueous solution) was added to each well and incubated at room temperature for 20 min. After incubation, the CV was transferred to a waste container and the plate was rinsed five times with sterile saline solution to remove the unbound dye. After the rinse, the wells were treated with 1 mL of 95% isopropanol to solubilize the CV and optical density at 550 nm was measured from three 100 μL aliquots on a 96-well microplate.
For the XTT assay, plates rinsed as previously described for the CV assay were treated with 1 mL of a solution containing 0.010 mol/L menadione (Sigma-Aldrich, St. Louis, MO) stock solution (diluted in acetone) mixed with XTT/Saline solution (0.5 g of XTT -2,3-bis-2-methoxy-4-nitro-5-sulfophenyl-5-phenylaminocarbonyl-2H-tetrazolium hydroxide-from Sigma diluted in Saline Solution) at a final concentration of 1 μmol/L. The plates were then covered in aluminum foil and incubated for 2 h at 37°C degrees. The XTT was actively and effectively reduced by metabolically active cells (biofilm formers) and the original clear solution was transformed into an orange solution that can be measured at an absorbance of 490 nm. Three technical replicates (from each biological replicate) were transferred to a 96-well microplate and measured at the respective optical density. For both CV and XTT assays, negative controls were treated and measured for experiment quality.
Oxidative stress resistance assay
To measure resistance to oxidative stress, planktonic cells were diluted 1:100 in nutrient broth containing 3 mM hydrogen peroxide (Millipore Sigma). Media without hydrogen peroxide was used as control. One mL aliquots were transferred to a 24 well microplate (in triplicate with unamended and cell-free media as controls) and incubated for 1 h at 37°C and constant agitation at 150 rpm. After incubation, the cultures were centrifuged at 10,000 rpm for 5 min, the supernatant was removed, and the cell pellet was rinsed twice with saline solution before starting a 10-fold serial dilution for drop plating to evaluate colony forming units.
Acidic pH tolerance assay
For growth in acidic media, cells were diluted 1:100 in nutrient broth buffered to pH 4.5 using HCl. One mL aliquots were transferred to a 24 well microplate (in triplicate with media pH 7.5 as control) and incubated for 1 h at 37°C with constant agitation at 150 rpm. After incubation, the cultures were serially diluted, 10-fold, in saline solution and plated using the drop plate technique to estimate CFU/mL.
Differential gene expression
The differential expression of genes coding for enzymes involved in the antioxidant response (oxyR, soxR, cysD) and tolerance to acidic pH (fabA, cpxR, cpxA) was analyzed using RT-qPCR (Table 1). For this purpose, cDNA was synthesized from total RNA with the PR1MA qMax first strand cDNA synthesis flex kit (Midsci, Valley Park, MO), using a program of priming at 25°C for 5 min, reverse transcription at 46°C for 20 min and RT inactivation at 95°C for 1 min. Quantitative PCR reactions, in 10 μL volumes, were ran in triplicate using the Applied Biosystems PowerUp SYBR green master mix (Applied Biosystems), 0.5 μM of each primer and 1 ng of cDNA as template. The relative gene expression of the target genes (Table 1) was normalized against the expression of the gapA gene using the efficiency-corrected model by Pfaffl et al., 2002. Each comparison was calculated from 7 to 9 Ct values from three biological replicates.
Results
Growth and biofilm formation, after exposure to simulated microgravity
We present the results of E. coli cultures grown under SM for 4 and 22 days, corresponding to approximately 53 and 128 generations, respectively. The long generation times observed (1.8–4 h) were most likely due to the microaerophilic conditions expected to develop on the micro tubes used as growth vessels and the absence of headspace required to attain the low shear conditions needed for microgravity simulation. Our experiments show that SM did not regulate the number of cells on cultures from either exposure time (Figures 2A, B). However, the biofilm biomass (CV) and metabolic activity (XTT) assays showed a significant increase in cultures grown under simulated microgravity, compared to those from the gravity control (Figures 2C, D), similar to a previous observation in 24 h cultures (Topolski et al., 2022).
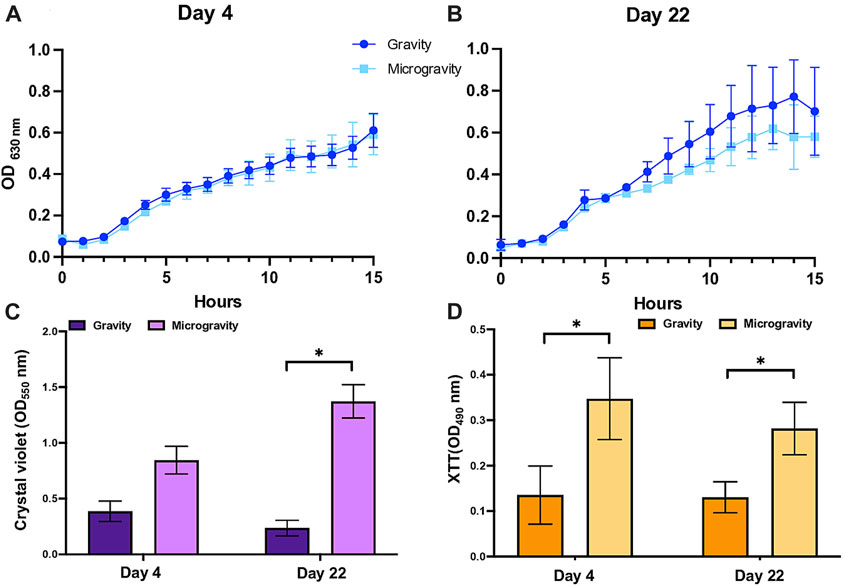
FIGURE 2. Growth and biofilm development of Escherichia coli upon exposure to simulated microgravity. No significant difference in growth dynamics observed between simulated microgravity and gravity cultures after (A) 4 and (B) 22 days of treatment. (C) Biofilm biomass measured with crystal violet. (D) Biofilm metabolic activity measured with XTT. The optical density, crystal violet and XTT values are the mean of three independent biological replicates (n = 3) and the bars indicate standard deviations. Asterisks indicate a p-value <0.05 from a t-test.
Acidic pH and oxidative stress tolerance assay
We tested the effect on microgravity on the regulation of tolerance to media acidified to pH 4.5 and amended with 3 mM hydrogen peroxide, using media pH 7.5 and unamended media as controls, respectively. Our data shows a marked increase in the culture’s sensitivity to acidic pH when cultures were grown under simulated microgravity conditions that did not change, significantly, with the number of generations (Figure 3A). Similarly, the effect of oxidative stress induced by the addition of hydrogen peroxide caused a decrease in cell viability in cultures that experienced reduced gravity, but only after cells grew under this condition for 22 days (Figure 3B).
FIGURE 3. Regulation of the acid tolerance (A) and oxidative stress (C) in Escherichia coli by simulated microgravity and the differential expression of genes coding for proteins related to growth under acidic pH (B) and to the response to oxidative stress (D). Bar graphs represent the mean of triplicated experiments (n = 3) with standard deviation as error bars. The asterisks represent significant differences between simulated microgravity and gravity cultures (∗ < 0.05).
Differential expression of genes involved in the response to oxidative stress and tolerance to acid pH
We measured the differential expression of genes involved in the tolerance of E. coli to acidic pH (fabA, cpxR and cpxA) and the response to oxidative stress (oxyR, soxR, cysD). Our results show that expression of these genes initially not regulated in cells growing under SM conditions for 4 days, decreased upon the prolonged exposure to the treatment for 22 days (Figures 3B, D).
Discussion
Human presence in space becomes more frequent, longer and it will expand beyond the limits of the International Space Station. In their quest to reach other planets, humans will rely not only on traditional life support systems to provide sustenance and protection, but also on their relationship with other organisms such as bacteria that significantly influence their interaction with the immediate physical environment. For this reason, it is imperative that we understand better the potential for the metabolic regulation of microbial activity in response to components of the space environment such as microgravity. The primary objective of this investigation was to examine the influence of the continuous exposure to simulated microgravity, from several days to weeks, under oxygen-limited conditions. The use of clinostats and other microgravity analogs present limitations in their results as they only partially mimic the effects of real microgravity conditions, a condition previously discussed (Ferranti et al., 2021; Allen et al., 2022). However, their use allows us to explore the biological effects of near weightlessness in bacterial cultures in a systematic way. Our results showed no significant difference in the growth kinetics between SM and G cultures on either exposure time (Figure 1), similar to the response in a previous experiment using a different microgravity analog with comparable growth conditions (Allen et al., 2022) and the device here discussed in 24 h old cultures (Topolski et al., 2022). This could be explained as the result of the cellular homeostatic control present in E. coli in the event that the constant state of free-falling imposed by clinorotation was in fact sensed as a stress. However, when we analyze the results of our biofilm assays, we observed that SM conditions selected for cells capable of forming increasing amounts of biofilm biomass, as well as a higher metabolic activity within biofilms, suggesting that our treatment led to stable phenotype changes in our model. It is known that biofilm development provides bacteria with advantages such as a higher resistance to unfavorable environments and antimicrobial agents by enhancing their ability to colonize solid surfaces or to alter the expression of genes, which translates into the potential increase in virulence and/or antibiotic resistance (Katongole et al., 2020; Behzadi et al., 2022). In contrast, the reduction in the viability of cells under acidic (days 4 and 22) and oxidative stress (day 22), supported by the downregulation of several genes involved in these responses, suggest that either SM favors the growth of cells with an impaired ability to survive the passage through the stomach, colonize the gut surfaces and to express virulence genes or it regulates the expression of the genes coding for enzymes involved in these processes. Although perceived as desirable traits, it is important to consider the majority of E. coli strains are not pathogenic, but form commensalistic relationships with humans (Martinson and Walk, 2020), hence the changes in the growth and survival of this organism could alter its dynamics in the gut of an astronaut during long-term space residence. Interestingly, our results are in accordance with a previous report for E. coli in spaceflight for 12 days that found a comparable reduction in viability after exposure to an acidic environment (Liu et al., 2023). Two other experiments showed responses comparable to ours, one with the downregulation of the soxR gene during a 49-h experiment exploring the effect of antibiotics on E. coli at the ISS (Aunins et al., 2018) and a second with the decreased survival of Salmonella enterica that grew the cells in a microgravity analog for 10 h (Wilson et al., 2002). The interesting finding of decreased viability under oxidative stress, induced by hydrogen peroxide, in prolonged exposure to microgravity reaffirms a complicated network of regulation to stress that may be attributed to interfering gene expression profiles caused by microgravity; however, our study shows that the soxR global regulator pathway is affected by oxidative stress after 22 days of exposure to microgravity. One interesting finding also reported by Aunins et al., 2018 is that downregulation of soxR was parallel to an upregulation of genes responsible of antibiotic resistance (particularly those that target the 30 S ribosomal subunit). Future studies in our system will include a comprehensive profile of antibiotic resistance patterns under prolonged exposure to microgravity. Our findings include a higher susceptibility to acidic conditions when cells were grown under microgravity. This finding can be explained by observing the higher quantity of biofilms formed under microgravity. Biofilms are known to have high tolerance to low pH. However, the Extra Polymeric Substances (EPS) that are part of the biofilm matrix may create zones of variable pH that could expose individual cells (without the EPS matrix) to low pH and affect dramatically their viability. Future studies will include exposure of cells grown under microgravity to high pH environments and the formation of alkaliphilic biofilms that have been reported to confer a greater level of control and survival in adverse environmental conditions (Charles et al., 2022). The composition of the EPS matrix will also be studied since it has been reported that alkaliphilic biofilms contain a higher amount of DNA when compared to protein, cellulose or lipid content (Charles et al., 2022). This data suggest that the direct effect of an acidic environment and oxidative stress conveyed by the condition of microgravity can increase biofilm formation along with resistance to antibiotics, a phenotype that will be addressed in the future. Overall, our experiment adds to the previously reported need to develop more realistic simulations of how microorganisms grow in space by exploring the interaction of the prolonged exposure to microgravity with microaerophilic conditions on the selection for phenotypes directly related to cell survival and the expression of virulence in the human gut.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
AC: Conceptualization, Investigation, Methodology, Writing–review and editing. CT: Conceptualization, Data curation, Methodology, Writing–review and editing. JH: Methodology, Writing–review and editing. MV: Formal Analysis, Methodology, Visualization, Writing–review and editing. NB: Methodology, Visualization, Writing–review and editing. MB: Methodology, Writing–review and editing. HC: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded, in part, by the ERAU Office of Undergraduate Research through the Spark Grants program. The participation of Natalie Baez was possible through the REU NSF program (grant number 2050887). The opinions, findings, and conclusions, or recommendations expressed are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Allen, C. A., Niesel, D. W., and Torres, A. G. (2008). The effects of low-shear stress on Adherent-invasive Escherichia coli. Environ. Microbiol. 10 (6), 1512–1525. doi:10.1111/j.1462-2920.2008.01567.x
Allen, L. A., Kalani, A. H., Estante, F., Rosengren, A. J., Stodieck, L., Klaus, D., et al. (2022). Simulated micro-lunar, and Martian gravities on earth—effects on Escherichia coli growth, phenotype, and sensitivity to antibiotics. Life 12 (9), 1399. doi:10.3390/life12091399
Aunins, T. R., Erickson, K. E., Prasad, N., Levy, S. E., Jones, A., Shrestha, S., et al. (2018). Spaceflight modifies escherichia coli gene expression in response to antibiotic exposure and reveals role of oxidative stress response. Front. Microbiol. 9, 310. doi:10.3389/fmicb.2018.00310
Behzadi, P., Gajdács, M., Pallós, P., Ónodi, B., Stájer, A., Matusovits, D., et al. (2022). Relationship between biofilm-formation, phenotypic virulence factors and antibiotic resistance in environmental Pseudomonas aeruginosa. Pathogens 11 (9), 1015. doi:10.3390/pathogens11091015
Castro, S. L., Nelman-Gonzalez, M., Nickerson, C. A., and Ott, C. M. (2011). Induction of attachment-independent biofilm formation and repression of Hfq expression by low-fluid-shear culture of Staphylococcus aureus. Appl. Environ. Microbiol. 77 (18), 6368–6378. doi:10.1128/aem.00175-11
Charles, C. J., Rout, S. P., Jackson, B. R., Boxall, S. A., Akbar, S., and Humphreys, P. N. (2022). The evolution of alkaliphilic biofilm communities in response to extreme alkaline pH values. Microbiol. Open 11 (4), e1309. doi:10.1002/mbo3.1309
Chopra, V., Fadl, A. A., Sha, J., Chopra, S., Galindo, C. L., and Chopra, A. K. (2006). Alterations in the virulence potential of enteric pathogens and bacterial-host cell interactions under simulated microgravity conditions. J. Toxicol. Environ. Health A 69 (14), 1345–1370. doi:10.1080/15287390500361792
Ferranti, F., Del Bianco, M., and Pacelli, C. (2021). Advantages and limitations of current microgravity platforms for space biology research. Appl. Sci. 11, 68. doi:10.3390/app11010068
Jang, J., Hur, H. G., Sadowsky, M. J., Byappanahalli, M. N., Yan, T., and Ishii, S. (2017). Environmental Escherichia coli: ecology and public health implications—a review. J. Appl. Microbiol. 123 (3), 570–581. doi:10.1111/jam.13468
Katongole, P., Nalubega, F., Florence, N. C., Asiimwe, B., and Andia, I. (2020). Biofilm formation, antimicrobial susceptibility and virulence genes of Uropathogenic Escherichia coli isolated from clinical isolates in Uganda. BMC Infect. Dis. 20 (1), 453–456. doi:10.1186/s12879-020-05186-1
Kim, H. W., Park, B. H., Park, H., Baek, B. C., Choi, I. G., and Rhee, M. S. (2022). Low-shear modeled microgravity affects metabolic networks of Escherichia coli O157: H7 EDL933: further insights into space-microbiology consequences. Food Res. Int. 154, 111013. doi:10.1016/j.foodres.2022.111013
Kim, H. W., and Rhee, M. S. (2016). Influence of low-shear modeled microgravity on heat resistance, membrane fatty acid composition, and heat stress-related gene expression in Escherichia coli O157: H7 ATCC 35150, ATCC 43889, ATCC 43890, and ATCC 43895. Appl. Environ. Microbiol. 82 (10), 2893–2901. doi:10.1128/AEM.00050-16
Klaus, D. M., and Howard, H. N. (2006). Antibiotic efficacy and microbial virulence during space flight. Trends Biotechnol. 24 (3), 131–136. doi:10.1016/j.tibtech.2006.01.008
Kuhn, D. M., Balkis, M., Chandra, J., Mukherjee, P. K., and Ghannoum, M. A. (2003). Uses and limitations of the XTT assay in studies of Candida growth and metabolism. J. Clin. Microbiol. 41 (1), 506–508. doi:10.1128/jcm.41.1.506-508.2003
Liu, Y., Xu, C., Zhao, G., Wang, Y., Zhu, Y., Yin, Y., et al. (2023). Effect of spaceflight on the phenotype and proteome of Escherichia coli. Open Life Sci. 18 (1), 20220576.
Lynch, S. V., Mukundakrishnan, K., Benoit, M. R., Ayyaswamy, P. S., and Matin, A. (2006). Escherichia coli biofilms formed under low-shear modeled microgravity in a ground-based system. Appl. Environ. Microbiol. 72 (12), 7701–7710. doi:10.1128/AEM.01294-06
Martinson, J. N., and Walk, S. T. (2020). Escherichia coli residency in the gut of healthy human adults. EcoSal Plus 9 (1), 10–1128. doi:10.1128/ecosalplus.esp-0003-2020
Milojevic, T., and Weckwerth, W. (2020). Molecular mechanisms of microbial survivability in outer space: a systems biology approach. Front. Microbiol. 11, 923. doi:10.3389/fmicb.2020.00923
Nickerson, C. A., Ott, C. M., Mister, S. J., Morrow, B. J., Burns-Keliher, L., and Pierson, D. L. (2000). Microgravity as a novel environmental signal affecting Salmonella enterica serovar Typhimurium virulence. Infect. Immun. 68 (6), 3147–3152. doi:10.1128/iai.68.6.3147-3152.2000
O’Toole, G. A. (2011). Microtiter dish biofilm formation assay. J. Vis. Exp. 47, 2437. doi:10.3791/2437
Pfaffl, M. W., Horgan, G. W., and Dempfle, L. (2002). Relative expression software tool REST for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30, e36–e10. doi:10.1093/nar/30.9.e36
Su, X., Guo, Y., Fang, T., Jiang, X., Wang, D., Li, D., et al. (2021). Effects of simulated microgravity on the physiology of Stenotrophomonas maltophilia and multiomic analysis. Front. Microbiol. 12, 701265. doi:10.3389/fmicb.2021.701265
Tirumalai, M. R., Karouia, F., Tran, Q., Stepanov, V. G., Bruce, R. J., Ott, C. M., et al. (2017). The adaptation of Escherichia coli cells grown in simulated microgravity for an extended period is both phenotypic and genomic. npj Microgravity 3 (1), 15. doi:10.1038/s41526-017-0020-1
Tirumalai, M. R., Karouia, F., Tran, Q., Stepanov, V. G., Bruce, R. J., Ott, C. M., et al. (2019). Evaluation of acquired antibiotic resistance in Escherichia coli exposed to long-term low-shear modeled microgravity and background antibiotic exposure. Mbio 10 (1), 026377–e11128. doi:10.1128/mBio.02637-18
Topolski, C., Divo, E., Li, X., Hicks, J., Chavez, A., and Castillo, H. (2022). Phenotypic and transcriptional changes in Escherichia coli K12 in response to simulated microgravity on the EagleStat, a new 2D microgravity analog for bacterial studies. Life Sci. Space Res. 34, 1–8. doi:10.1016/j.lssr.2022.04.003
Wilson, J. W., Ott, C. M., Ramamurthy, R., Porwollik, S., McClelland, M., Pierson, D. L., et al. (2002). Low-Shear modeled microgravity alters the Salmonella enterica serovar typhimurium stress response in an RpoS-independent manner. Appl. Environ. Microbiol. 68 (11), 5408–5416. doi:10.1128/AEM.68.11.5408-5416.2002
Keywords: Escherichia coli, microgravity, biofilm, oxidative stress, antibiotic resistance
Citation: Chavez A, Topolski C, Hicks J, Villafania M, Baez N, Burke M and Castillo H (2024) Phenotypical changes in Escherichia coli K12 after prolonged exposure to simulated microgravity. Front. Space Technol. 4:1282850. doi: 10.3389/frspt.2023.1282850
Received: 24 August 2023; Accepted: 30 November 2023;
Published: 10 January 2024.
Edited by:
Michael Lebert, University of Erlangen Nuremberg, GermanyReviewed by:
Sebastian Strauch, University of the Region of Joinville, BrazilPeter Rolf Richter, Friedrich-Alexander-Universität Erlangen-Nürnberg, Germany
Madhan Tirumalai, University of Houston, United States
Copyright © 2024 Chavez, Topolski, Hicks, Villafania, Baez, Burke and Castillo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hugo Castillo, Y2FzdGlsaDJAZXJhdS5lZHU=
 Alba Chavez
Alba Chavez Collin Topolski2
Collin Topolski2 Mitchell Villafania
Mitchell Villafania Hugo Castillo
Hugo Castillo