- 1Air Force Research Laboratory, 711th Human Performance Wing, Airman Bioengineering Division, Dayton, OH, United States
- 2The Henry M. Jackson Foundation, Bethesda, MD, United States
- 3National Research Council, National Academy of Sciences, Engineering and Medicine, Washington, DC, United States
Manned spaceflight exposes humans to extreme environmental conditions, including microgravity exposures. The effects of microgravity during spaceflight could lead to changes in brain structure, gene expression, and vascular physiology. Given the known physiological effects, it is highly likely that there are microgravity-initiated proteomic differentials in the brain, possibly domain specific. MALDI-TOF (matrix-assisted laser desorption/ionization time of flight) Imaging Mass Spectrometry allows the visualization of the spatial distribution of highly abundant intact proteins in tissue specimens. This study utilized this technique to visualize global proteomic changes induced by microgravity exposure in brain tissue received from the Rodent Research-1 Center for the Advancement of Science in Space (CASIS)/National Aeronautics and Space Administration (NASA). Proteome profiles were obtained from isolated whole brain tissue from microgravity exposed, Habitat control, and baseline. While a total of 135 mass peaks equating to individual proteins were identified, statistical analysis determined that there were no significant differences in the spectra profiles from the three test groups utilizing this methodology, possibly due to sample collection logistics rather than lack of cellular response.
1 Introduction
The age of space travel is in an exciting new era where the commercialization of space is fast becoming a reality. For decades, humans have been launching into space while subjected to extreme environmental conditions such as microgravity and radiation. Comprehensive studies to determine the impacts of spaceflight are being performed to understand the effects on the entirety of the human system. (Demontis et al., 2017; Van Ombergen et al., 2017) NASA’s Twin study was pivotal in developing an integrated systems description of the effects of 1 year of spaceflight on the human body. (Garrett-Bakelman et al., 2019) This study collected physiological, telomeric, transcriptomic, epigenetic, proteomic, metabolomic, immune, microbiome, cardiovascular, vision-related, and cognitive data to investigate long term molecular pathway alterations due to lengthy exposures. (Demontis et al., 2017) Despite the tremendous data output from this collaborative research, there were limitations in examining potentially subtle molecular changes induced in the brain by microgravity.
The brain is arguably the most complex organ in the human body. The enormous network of cells, containing billions of neurons and interconnections is responsible for receiving, processing, and executing cognition and perception, controlling motor activities and the storage of memories. (Kandel, 2001) The absence of gravity can affect the brain, possibly inhibiting cognitive function. (Grabherr and Mast, 2010; Mammarella, 2020) Neuroasthenia, also known as “space fog”, has been reported by astronauts for some time and prompted extensive cognitive testing. (Kanas et al., 2001; Basner et al., 2015) However, these cognitive evaluation data have not shown statistical linkage to self-reported cognitive decrement, possibly due to individual responsivity as well as certain stressors not influenced by microgravity (Vaughan et al., 2013; Strangman et al., 2014).
Despite discrepancies in cognitive evaluation, it has long been observed that long term spaceflight can change brain physiology and affect the sensory-motor system. (Blaber et al., 2010; Roberts et al., 2017) Visualizing the changes in brain physiology on human astronauts and pilots has been performed via diffusion magnetic resonance imaging (dMRI). This instrumentation allows for the observation of Free Water shifts, grey matter, and white matter distribution. Disrupted white matter structural connectivity in various brain regions have been linked with several previous spaceflight missions, mission duration and preflight to postflight balance declines. (Basner et al., 2015) Equivalent changes in brain physiology have also been observed within U2 pilots in the United States Air Force (Nelson et al., 2013). Invasive analytical imaging cannot be performed to examine the human brain proteome, therefore MALDI TOF imaging mass spectrometry (IMS) instrumentation was utilized to search for proteomic shifts due to the effects of spaceflight in a well-characterized mouse model. MALDI-TOF-IMS is a label-free technique that can be used for global screening of proteomic, metabolomic or lipidomic changes on the surface of tissue samples, providing a bridge between histology and deep molecular mass spectrometric analysis in tissue-based research (Aichler and Walch, 2015).
Gaining insight on protein-associated changes in rodent brains following microgravity exposure provides insight on which regions may be impacted. Mice are an excellent candidate to study any potential effects because of the shared genetic homology (Uhl and Warner, 2015). In this study, nine mice brains were evaluated using MALDI-TOF-IMS. There were three separate groupings: Baseline, Habitat Control and Flight. The highest abundance protein ions were visualized and spatially resolved by brain region. The hippocampus and cerebellum are regions of interest because it is well known that activity between these regions contribute to human spatial navigation and motor control functions that are critical functions for astronauts (Iglói et al., 2015).
This study used MALDI-TOF-IMS to characterize proteomic shifts within different regions. Insight into any shifting behavior can provide a target for in depth proteomic analysis to fully characterize the observed effect. Understanding how the body is affected by space exploration will help shape space travel development and influence the proper measures that must be taken to safeguard against the effects of microgravity.
2 Materials and methods
2.1 Animal study
This study was conducted under an IACUC approved protocol in a facility accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC), International, in accordance with the Guide for the Care and Use of Laboratory Animals (NRC, 2011) and approved by the NASA Institutional Animal Care and Use Committee.
The animal transport was conducted on the as part of the resupply launch mission CRS-16. Mature (30–52 weeks old) female BALB/cAnNTac mice (Taconic Biosciences, Rensselaer, NY) were used in this study. The Habitat Control (HC) group was kept at standard Earth gravity in the specialized animal containment (called the Rodent Habitat), a part of the ISS Rodent Habitat Hardware System. The Baseline group (BL) was prepared just prior to launch of the microgravity-exposed group and consists of animals at Earth gravity in normal animal cages. The microgravity-exposed group (FL) were placed in the Rodent Transporter containment cages, launched aboard a Falcon 9 rocket, docked at the International Space Station (ISS), then transferred to a Rodent Habitat using the Animal Access Unit. On the ISS, the animals were fed ad libitum using NASA Type 12 Nutrient-upgraded Rodent Food Bars diet plus unrestricted water. The animals were on board the ISS at microgravity a total of 36 days. At Day 37, the animals were placed in a Rodent Transporter cage for return transport to Earth. Within 3 days of FL mice returning to standard Earth gravity (9.80665 m/s2), all group animals were euthanized via ketamine/xylazine overdose, the whole brain removed and quickly frozen in liquid nitrogen. The tissue samples were kept at −80°C prior to MALDI analyses. Dissection and initial tissue collection was handled by CASIS and shipped to the Air Force Research Laboratory (AFRL) overnight on dry ice. Upon receipt, the samples were weighed, measured, and prepared for analysis.
2.2 Sample preparation for MALDI-TOF analysis
Frozen whole brain tissues were cyrosectioned along the sagittal axis in 12 μm sections (landmark p56, Allen Brain Atlas) using an Advantic Cryostat in preparation for MALDI-TOF-IMS. Sagittal orientation was selected in order to capture maximal brain region coverage in a 2D image (Fernandes et al., 2016). Brain slices were mounted on Indium-Tin-Oxide (ITO) coated slides using a gelatin/CMC solution (Gemperline et al., 2014) and adhered under vacuum. Once mounted, the slides were stored in a −80°C freezer until ready for imaging. All brain tissues were randomized for processing as well as for imaging.
2.3 MALDI-TOF IMS slide preparation for protein analysis
ITO slides with mounted tissue sections were acclimated to room temperature in a vacuum desiccator for 1 h and washed using the Carnoy method to target proteins. (Puchtler et al., 1970) Prior to MALDI-TOF imaging, an optical image of the tissue ITO slide was generated using a high resolution scanner at a dpi 4,800. This image was utilized to program the MALDI-TOF instrument. A total of 10 mg/ml Sinapinic Acid matrix was applied to the ITO slide using a Bruker ImagePrep sprayer and imaged using a Bruker Ultraflextreme MALDI-TOF (Bruker Corp.). All samples were imaged in linear positive mode, utilizing a 1.8 0 in the 29 kDa mass range with continuous 50 µm raster.
2.4 Sample analysis
Mass spectra and images were obtained from intact protein ions using one brain slice section from each animal from the BL, HC and FL groups (3 animals per group). Individual spectra were obtained for each sample (Figure 1). To compare the microgravity proteome to both the HC and BL groups, the spectra were averaged for each exposure group to take in account biological variability. The developed averaged spectra for each group was then utilized to generate ROC curves to evaluate if there were significant changes between exposure groups. Receiver operating characteristic (ROC) curves were generated using SCiLs Lab software (Bruker Corp.) to compare the averaged spectra between the BL, HC, and FL samples. A two-way analysis of variance (ANOVA) comparison was performed utilizing GraphPad Prism V7.0 to confirm the data.
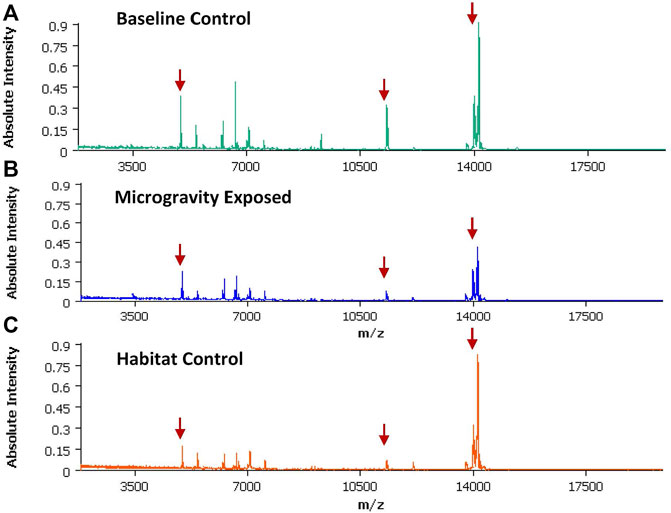
FIGURE 1. An averaged spectra generated SCiLs Lab software utilizing spectra data generated from a total of three brain sections (3 different animals) within the same exposure group. Red arrows indicate the m/z used to create the spectral images discussed below (A) Averaged spectra from Baseline (BL) group; (B) Averaged spectra from the microgravity group (FL); (C) Average spectra from the Habitat (HC) group.
3 Results
MALDI-TOF imaging captures the highest abundance proteins on the surface of the tissue sample. A total of 135 intact protein ions were acquired via mass spectrum across tissue slices generated from each of the 9 brains (3 per exposure group) analyzed. All images were normalized by Total Ion Current (TIC). An average mass spectra (Table 1) was generated for each group as described above to account for biological variability between animals (Figure 1). For each m/z peak on the spectra acquired, an image was generated with the corresponding ion. Select sample images reflecting m/z 6,652 (Figure 3), m/z 11,308 (Figure 4), and m/z 14,144 (Figure 5) from each exposure group indicate some areas of high level proteomic alterations (approximate hypothalamic region, Figure 5C, FL), but no uniformity within groups or across groups were noted. Landmark notation of specific brain regions was difficult due to brain tissue distortion after collection during freezing. The cerebellum was noted (Figure 2) but other more subtle domains could not be discerned.
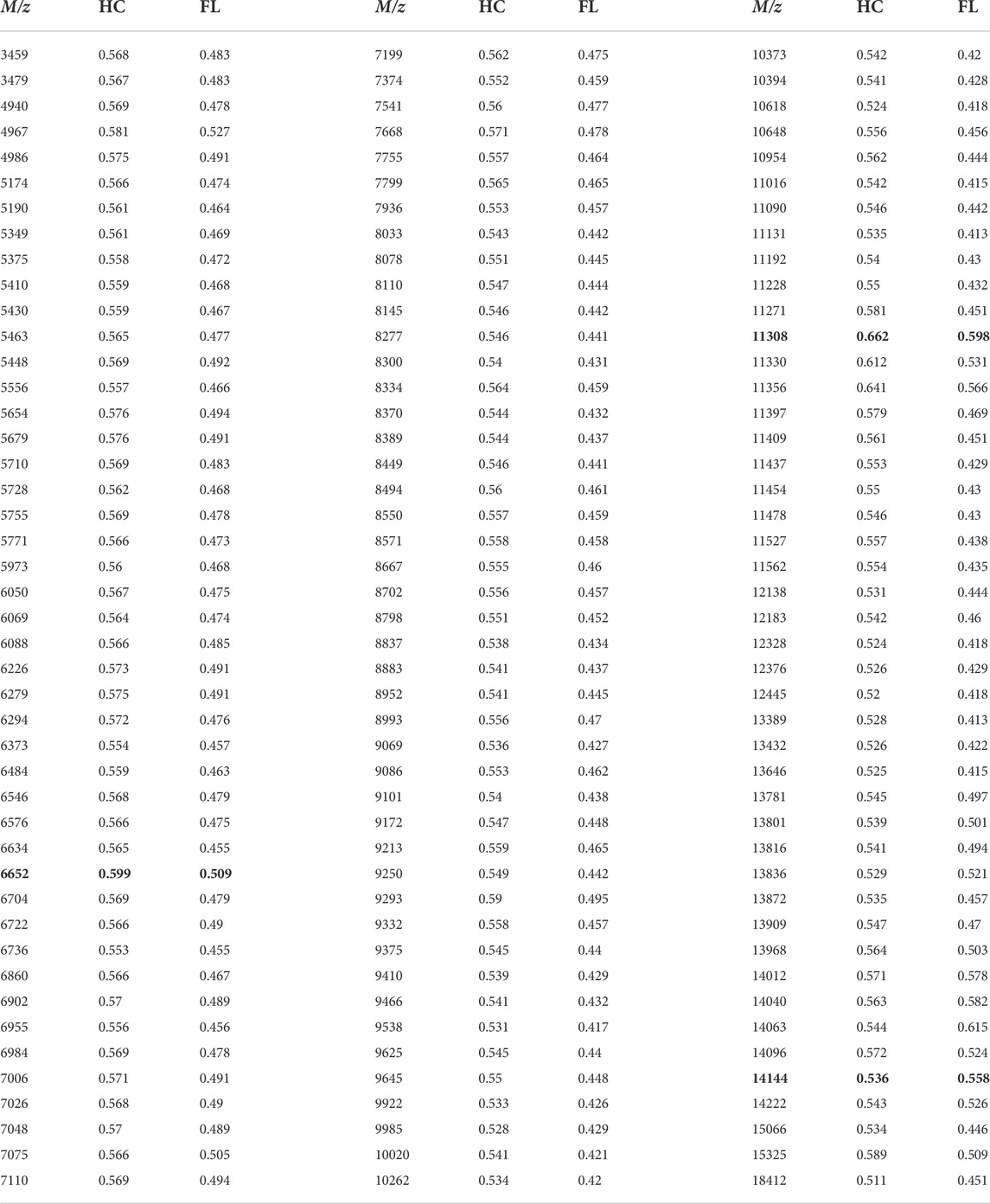
TABLE 1. AUC values generated from ROC curves for each m/z. Data from Figures 3–5 are highlighted in yellow. BL = Baseline; HC = Habitat; FL = Microgravity exposed.
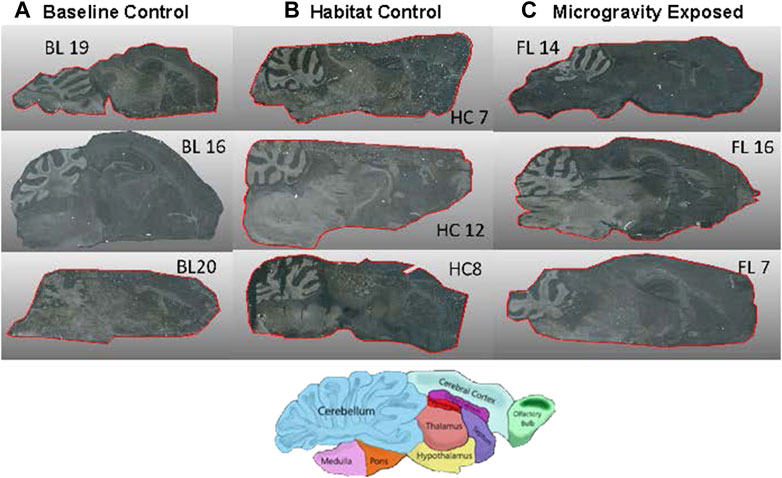
FIGURE 2. High Resolution images of 12 µm tissue samples from three animals per exposure group. Diagram indicates brain domains in orientation as spectral images. (A) BL = Baseline control, (B) HC = Habitat Control, (C) FL = Microgravity exposed.
Receiver operating characteristic (ROC) curves were generated in SCiLs Lab software to determine significant change that occurred during exposure to the microgravity environment found onboard the ISS. Area under the curve (AUC) values between 0.3 and 0.7 convey that there were no significant shifts seen between FL and HC compared against the Baseline. To confirm this data, Prism was utilized to perform statistical analysis (Table 2). A 2-way ANOVA comparison was utilized to verify that there were no significant changes determined between the protein ions detected. There were no adjusted p-values that indicated statistically significant changes in the microgravity brain proteome compared to baseline or habitat control values. There are several potential study design variables, discussed below, which may have impacted our ability to parse out the proteomic signature differences between the exposure groups.
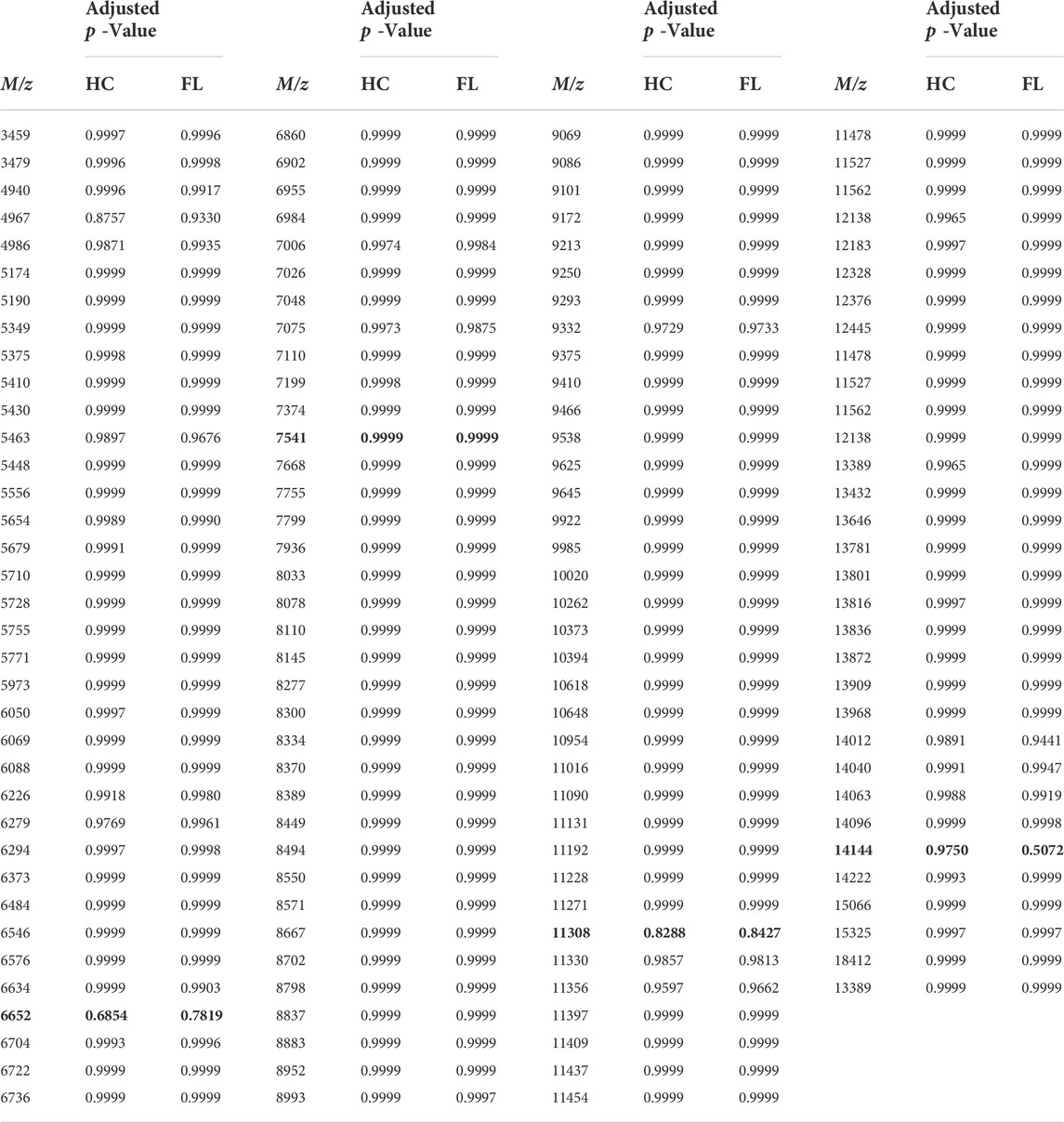
TABLE 2. p values resulting from a two-way ANOVA Statistical Analysis of spectra values, as compared to Baseline. HC = Habitat; FL = Microgravity exposed. Specific m/z comparisons from spectral images (Figures 3–5) are highlighted in yellow.
4 Discussion
MALDI-TOF-IMS can be utilized to include differential expression of lipids and metabolites, in addition to proteins, to create a time-specific map of spatially-connected molecular alterations. (Murphy et al., 2009; Aichler and Walch, 2015) Such multi-omic molecular maps are especially important in the multi-domain brain, where targeted Omic analyses are problematic if one does not have a priori knowledge on which or now many brain regions are modulated in response to a given external exposure or condition. Identification of the specific locales of differential expression allow for a secondary targeted Omics approach, more likely to produce statistically stronger data as the tissue analyzed has first been identified as relevant to the response rather than generated via a “tissue-averaged” signal. Precise isolation of very small tissue regions can then be accomplished using targeted laser dissection for in-depth discovery of mechanistic pathways. Thus, imaging is the critical initial approach to use an integrated top down and bottom up Omics approach for the identification of molecules of interest in a targeted brain region. (Ntai et al., 2016)
Technology and software have made it possible to construct three dimensional images of the organ being analyzed using MALDI-IMS capabilities. (Seeley and Caprioli, 2012; Vos et al., 2020) This would provide an ultimate mapping of spatial shifts in protein, lipids and metabolites. (Seeley and Caprioli, 2011) The employment of the full Omic suite could be employed on multiple organs in vivo studies allowing for an understanding of how all the biological functions are impacted by spaceflight. Bridging the medicinal and biochemical studies together can lead to steps to develop countermeasures to combat detrimental effects caused by microgravity.
This study aimed to examine proteome shifts initiated by microgravity by spatially resolving surface protein ions on brain tissue samples in order to visualize proteomic shifts between different exposure groups. The results also served to observe biological variation between each animal in different brain regions. After 36 days in a microgravity environment, no significant changes in protein expression were observed using MALDI-TOF-IMS within the highest abundant intact protein ions that were detected. Other research has indicated that microgravity does have an effect on protein expression, although these studies only utilized simulated microgravity. (Sarkar et al., 2006; Ishikawa et al., 2017) These published experiments had the distinct advantage of performing dissection and tissue preparation quickly after exposure. In this study, the logistics of retrieval of the animals from ISS and tissue dissection post-spaceflight posed an unavoidable delay from exposure time to tissue collection, a delay of over 3 days. Therefore, it is likely that the failure to identify statistically significant proteome alterations was due to this time lapse, allowing the mice in this study reacclimatized in the period before dissection, and that any unique proteome signatures induced by the microgravity exposure were lost. A previous proteomic study by Mao et al. utilizing LC-MS analysis confirmed that there were 26 significant protein changes in the mouse brain after spaceflight. (Mao et al., 2018) The samples were processed for analysis within 3–5 h after Space Shuttle Atlantis returned to Earth after a 13 days space mission. However, the study did not confirm whether the proteome alterations were induced by the microgravity or due to the stress induced from landing. To parse out stress vs. microgravity-induced alternations, future studies should include a secondary control group that could mimic the launch exposure without gravitational changes, possibly by use of NASA astronaut training equipment.
In addition, the ability to perform dissection and sample collection on board the ISS would clarify the lack of proteome responsivity to microgravity between the microgravity exposed (FL) animals and the control sets as analyzed by MALDI-TOF-IMS. MALDI-TOF-IMS is a complex technique and sample preparation is a critical issue. In this study, despite care, the brain morphology were not uniform across the specimens (Figure 2). As such, cryosectioning of the brain tissue using landmarks for spatial sectioning provided its own unique challenges in order to acquire comparable sagittal sections for imaging. However, certain brain regions are clearly observable (note cerebellum in Figure 2 and corresponding proteome activity in Figures 3–5).
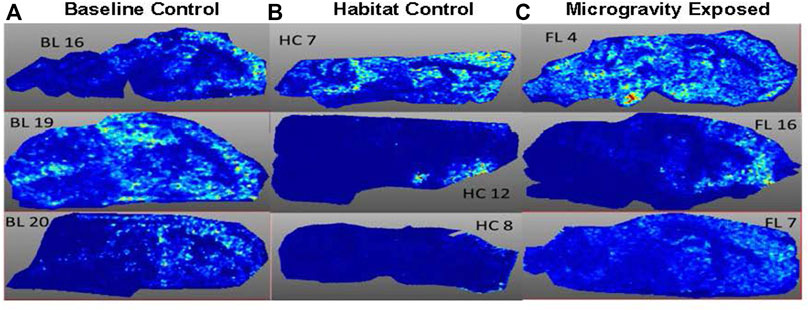
FIGURE 3. Spatial Image obtained at m/z 6,652 from three animals per exposure group. (A) BL = Baseline group; (B) HC = Habitat control group; (C) FL = Microgravity exposed group.
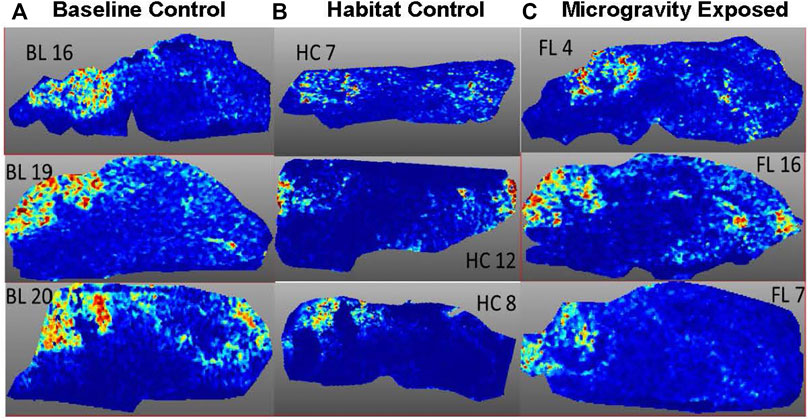
FIGURE 4. Spatial Image obtained at m/z 11,308 from three animals per exposure group. (A) BL = Baseline control, (B) HC = Habitat control, (C) FL = Microgravity exposed.
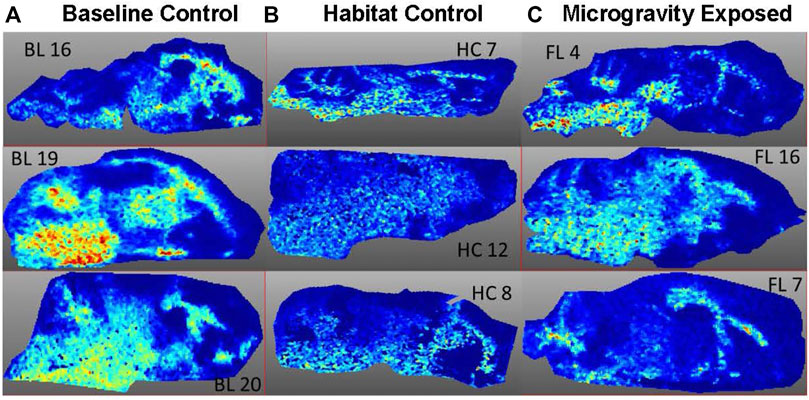
FIGURE 5. Image of m/z 14,144 from three animals per exposure group. (A) BL = Baseline, (B) HC = Habitat, (C) FL = Microgravity exposed.
Data quality with respect to signal intensity and reproducibility is also impacted by sample handling. (Watrous and Dorrestein, 2013) Matrix application is critical to protonating the proteins on the surface tissue and influences sensitivity. Matrix crystal formation is critical in this application. Improving deposition or sublimation in the matrix application process can possibly lead to an increase in reproducibility of application and better sensitivity of analytes (Hankin et al., 2007; Gemperline et al., 2014).
With future improvements in sample collection as discussed above, MALDI-TOF-IMS analysis of a second set of microgravity-exposed brain tissue could reveal not only the global proteome alterations but brain domain and spatial locale of those shifts. With further analyses to identify the proteins within these proteome alteration ‘hot spots’, an in-depth understanding of pathway alterations underpinning the subtle cognitive neuro and/or physiological effects of microgravity could be determined. Such effects have been seen using MRI analysis of post-flight astronauts and include increases in brain ventricular volume and spaceflight-associated neuro-ocular syndrome (Roberts et al., 2019).
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was reviewed and approved by This study was conducted under an IACUC approved protocol in a facility accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC), International, in accordance with the Guide for the Care and Use of Laboratory Animals (NRC, 2011) and approved by the NASA Institutional Animal Care and Use Committee.
Author contributions
CV, HC, and CM wrote original CASIS proposal. CV, HC, and AD developed methodologies, conducted sample analyses, and completed final statistical evaluations. JS conducted sample preparation and analyses. CM provided project guidance and management. All authors contributed to the development of the manuscript, and CM provided final edits and submissions.
Funding
MALDI effort was funded by the Air Force Research Laboratory, 711 Human Performance Wing, Airman Systems Directorate. Research reported in this paper was supported by the International Space Station U.S. National Laboratory under UA-2019-019 number AR-007262. The views expressed in this study are those of the authors and do not reflect the official policy or position of the United States Air Force, Department of Defense, NASA, or the United States Government. This material is declared a work of the United States Government and is not subject to copyright protection in the United States of America. This article was designated Distribution A, approved for public release, distribution unlimited by MSC/PA-2021-0046; AFRL-2021-0495 on 24 March 2021.
Acknowledgments
The authors would like to thank the CASIS organization for providing the specimens and Derek Smith, Case Western University, for discussions on ISS research opportunities.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aichler, M., and Walch, A. (2015). MALDI Imaging mass spectrometry: Current frontiers and perspectives in pathology research and practice. Lab. Invest. 95, 422–431. doi:10.1038/labinvest.2014.156
Basner, M., Savitt, A., Moore, T. M., Port, A. M., McGuire, S., Ecker, A. J., et al. (2015). Development and validation of the <I>Cognition</I> test battery for spaceflight. Aerosp. Med. Hum. Perform. 86, 942–952. doi:10.3357/amhp.4343.2015
Blaber, E., Marçal, H., and BurnsBioastronautics, B. P. (2010). Bioastronautics: The influence of microgravity on astronaut health. Astrobiology 10, 463–473. doi:10.1089/ast.2009.0415
Demontis, G. C., Germani, M. M., Caiani, E. G., Barravecchia, I., Passino, C., and Angeloni, D. (2017). Human pathophysiological adaptations to the space environment. Front. Physiol. 8, 547. doi:10.3389/fphys.2017.00547
Fernandes, A. M., Vendramini, P. H., Galaverna, R., Schwab, N. V., Alberici, L. C., Augusti, R., et al. (2016). Direct visualization of neurotransmitters in rat brain slices by desorption electrospray ionization mass spectrometry imaging (DESI - ms). J. Am. Soc. Mass Spectrom. 27 (12), 1944–1951. doi:10.1007/s13361-016-1475-0
Garrett-Bakelman, F. E., Darshi, M., Green, S. J., Gur, R. C., Lin, L., Macias, B. R., et al. (2019). The NASA twins study: A multidimensional analysis of a year-long human spaceflight. Science 364, eaau8650. doi:10.1126/science.aau8650
Gemperline, E., Rawson, S., and Li, L. (2014). Optimization and comparison of multiple MALDI matrix application methods for small molecule mass spectrometric imaging. Anal. Chem. 86, 10030–10035. doi:10.1021/ac5028534
Grabherr, L., and Mast, F. W. (2010). Effects of microgravity on cognition: The case of mental imagery. J. Vestib. Res. 20 (1), 53–60. doi:10.3233/ves-2010-0364
Hankin, J. A., Barkley, R. M., and Murphy, R. C. (2007). Sublimation as a method of matrix application for mass spectrometric imaging. J. Am. Soc. Mass Spectrom. 18, 1646–1652. doi:10.1016/j.jasms.2007.06.010
Iglói, K., Doeller, C. F., Paradis, A-L., Benchenane, K., Berthoz, A., Burgess, N., et al. (2015). Interaction between hippocampus and cerebellum Crus I in Sequence-based but not Place based navigation. Cereb. Cortex 25, 4146–4154. doi:10.1093/cercor/bhu132
Ishikawa, C., Li, H., Ogura, R., Yoshimura, Y., Kudo, T., Shirakawa, M., et al. (2017). Effects of gravity changes on gene expression of BDNF and serotonin receptors in the mouse brain. PLoS One 12, e0177833. doi:10.1371/journal.pone.0177833
Kanas, N., Salnitskiy, V., Gushin, V., Weiss, D. S., Grund, E. M., Flynn, C., et al. (2001). Asthenia - does it exist in space? Psychosom. Med. 63, 874–880. doi:10.1097/00006842-200111000-00004
Kandel, E. R. (2001). The molecular biology of memory storage: A dialogue between genes and synapses. Science 294, 1030–1038. doi:10.1126/science.1067020
Mammarella, N. (2020). The effect of microgravity-like conditions on high-level cognition: A review. Front. Astron. Space Sci. 7, 25. doi:10.3389/fspas.2020.00006
Mao, X. W., Sandberg, L. B., Gridley, D. S., Herrmann, E. C., Zhang, G., Raghavan, R., et al. (2018). Proteomic analysis of mouse brain subjected to spaceflight. Int. J. Mol. Sci. 20 (1), 7. doi:10.3390/ijms20010007
Murphy, R. C., Hankin, J. A., and Barkley, R. M. (2009). Imaging of lipid species by MALDI mass spectrometry. J. Lipid Res. 50, 317–322. doi:10.1194/jlr.r800051-jlr200
Nelson, K. A., Daniels, G. J., Fournie, J. W., and Hemmer, M. J. (2013). Optimization of whole-body zebrafish sectioning methods for mass spectrometry imaging. J. Biomol. Tech. 24, 119–127. doi:10.7171/jbt.13-2403-002
Ntai, I., LeDuc, R. D., Fellers, R. T., Erdmann-Gilmore, P., Davies, S. R., Rumsey, J., et al. (2016). Integrated bottom-up and top-down proteomics of patient-derived breast tumor xenografts. Mol. Cell. Proteomics 15, 45–56. doi:10.1074/mcp.m114.047480
Puchtler, H., Waldrop, F. S., Meloan, S. N., Terry, M. S., and Conner, H. M. (1970). Methacarn (methanol-Carnoy) fixation. Histochemie 21, 97–116. doi:10.1007/bf00306176
Roberts, D. R., Albrecht, M. H., Collins, H. R., Asemani, D., Chatterjee, A. R., Spampinato, M. V., et al. (2017). Effects of spaceflight on astronaut brain structure as indicated on MRI. N. Engl. J. Med. 377, 1746–1753. doi:10.1056/nejmoa1705129
Roberts, D. R., Asemani, D., Nietert, P. J., Eckert, M. A., Inglesby, D. C., Bloomberg, J. J., et al. (2019). Prolonged microgravity affects human brain structure and function. AJNR. Am. J. Neuroradiol. 40, 1878–1885. doi:10.3174/ajnr.a6249
Sarkar, P., Sarkar, S., Ramesh, V., Hayes, B. E., Thomas, R. L., Wilson, B. L., et al. (2006). Proteomic analysis of mice hippocampus in simulated microgravity environment. J. Proteome Res. 5, 548–553. doi:10.1021/pr050274r
Seeley, E. H., and Caprioli, R. M. (2012). 3D imaging by mass spectrometry: A new frontier. Anal. Chem. 84, 2105–2110. doi:10.1021/ac2032707
Seeley, E. H., and Caprioli, R. M. (2011). MALDI imaging mass spectrometry of human tissue: Method challenges and clinical perspectives. Trends Biotechnol. 29 (3), 136–143. doi:10.1016/j.tibtech.2010.12.002
Strangman, G. E., Sipes, W., and Beven, G. (2014). Human cognitive performance in spaceflight and analogue environments. Aviat. space Environ. Med. 85 (10), 1033–1048. doi:10.3357/asem.3961.2014
Uhl, E. W., and Warner, N. J. (2015). Mouse models as predictors of human responses: Evolutionary medicine. Curr. Pathobiol. Rep. 3, 219–223. doi:10.1007/s40139-015-0086-y
Van Ombergen, A., Demertzi, A., Tomilovskaya, E., Jeurissen, B., Wuyts, F. L., Kozlovskaya, I. B., et al. (2017). The effect of spaceflight and microgravity on the human brain. J. Neurol. 264, 18–22. doi:10.1007/s00415-017-8427-x
Vaughan, L., Leng, I., Dagenbach, D., Resnick, S. M., Rapp, S. R., Jennings, J. M., et al. (2013). Intraindividual variability in domain-specific cognition and risk of mild cognitive impairment and dementia. Curr. Gerontol. Geriatr. Res. 2013, 495793. doi:10.1155/2013/495793
Vos, D. R. N., Ellis, S. R., Balluff, B., and Herren, R. M. A. (2020). Experimental and data analysis considerations for three-dimensional mass spectrometry imaging in biomedical research. Mol. Imaging Biol. 23, 149–159. doi:10.1007/s11307-020-01541-5
Keywords: microgravity, MALDI, brain, proteomics, International Space Station, in vivo, mice, proteome
Citation: Vigil C, Daubenspeck A, Coia H, Smith J and Mauzy C (2022) Matrix-assisted laser desorption/ionization analysis of the brain proteome of microgravity-exposed mice from the International Space Station. Front. Space Technol. 3:971229. doi: 10.3389/frspt.2022.971229
Received: 16 June 2022; Accepted: 26 October 2022;
Published: 16 November 2022.
Edited by:
Jack J. W. A. van Loon, VU Amsterdam, NetherlandsReviewed by:
Khaled Y. Kamal, Texas A&M University, United StatesHolly Hyde Birdsall, Baylor College of Medicine, United States
Copyright © 2022 Vigil, Daubenspeck, Coia, Smith and Mauzy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Camilla Mauzy, Y2FtaWxsYS5tYXV6eUB1cy5hZi5taWw=
 Correy Vigil1
Correy Vigil1 Camilla Mauzy
Camilla Mauzy