
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Space Technol., 29 March 2022
Sec. Microgravity
Volume 3 - 2022 | https://doi.org/10.3389/frspt.2022.853980
This article is part of the Research TopicTechnologies for handling, preparation, and liquid-based analysis of fluidic samples in spaceView all 6 articles
For the last 15 years, small satellites known as CubeSats have been used to investigate the effects of the space environment on biological organisms. All biological CubeSat missions flown to date have performed studies in low Earth orbit (LEO), each one improving its biological support sub-systems from the last. An upcoming NASA biological CubeSat mission, BioSentinel, will launch as a secondary payload on Artemis 1 and eventually reach a heliocentric orbit beyond LEO, and the protection of Earth’s magnetosphere. The main objectives of BioSentinel are 1) to investigate the biological effects of the deep space radiation environment and 2) to develop our technological capacity to support biological research in deep space. The instruments and subsystems within BioSentinel have heritage from previous CubeSat missions (e.g., fluidics, optics, thermal control), but are extended on many levels. BioSentinel improves upon the materials and design (e.g., decreased card vapor permeability to maintain low humidity; the addition of a fluidic manifold with internal check-valves, desiccant chambers, and bubble traps for each individual fluidic card) and adds new tools for discovery (e.g., onboard LET spectrometer). The main objective of this Perspective is to emphasize the evolution of the fluidic systems used in past and ongoing NASA biological CubeSat missions and highlight aspects of these systems that can be optimized for future experimentation beyond LEO.
Human spaceflight has not ventured past Low Earth Orbit (LEO) since Apollo 17 in 1972, leaving space biology experiments to be mainly conducted on the International Space Station (ISS). The ISS sits below the Van Allen radiation belts, thus protecting astronauts (and the biological experiments onboard) from the harmful effects of deep space radiation. Spacecraft sent beyond LEO have been limited and mostly non-biological, focusing on reconnaissance missions to planetary bodies in our solar system such as the Moon and Mars. To support future human exploration beyond LEO, significant technological and biomedical countermeasures are necessary. NASA’s Artemis Program aims in the short term to land astronauts back on the Moon by 2024, and in the long term to eventually inhabit it for long durations as well as build a cislunar space station or Lunar Gateway. The Moon sits 1,000x further from Earth than the ISS, and beyond the protection of the Van Allen radiation belts. Thus, such missions would require astronauts to spend long durations exposed to the harmful effects of deep space, particularly galactic cosmic radiation (GCR) and reduced gravity.
To understand how to best mitigate these threats, more biological experimentation in such environments is necessary. While many biological studies in space have occurred in LEO on the ISS (Kanapskyte et al., 2021), this limits the extent of knowledge on how space affects biology to a single space environment. In addition, at least one astronaut has often been required to participate in performing experiments, limiting the quantity of experiments that can be conducted at a time. An alternative approach is to perform autonomous experiments using biological CubeSats – small, relatively inexpensive satellites that can conduct a variety of biological experiments in free space (Massaro Tieze et al., 2020; Kanapskyte et al., 2021). CubeSats are built to a standard originally developed in 1999 to support academic spaceflight access; each CubeSat is a combination of one or more cubical “unit” volumes (1U = 10 × 10 × 10 cm), allowing compatibility with a set of common interfaces and support subsystem technologies (Twiggs and Puig-Suari, 2020).
The core of biological CubeSat missions to date are their intricate fluidic systems, which deliver and manipulate fluids such as culture media, antibiotics, and colorimetric or fluorescent dyes to their biological subjects (e.g., bacteria or yeast, and eventually human cells). BioSentinel, NASA’s first biological CubeSat designed for deep space, possesses the most complex and versatile fluidic system to date – a fluidic manifold with internal check valves, desiccant chambers, and bubble traps for efficient reagent delivery to each individual fluidic card (Ricco et al., 2020; Padgen et al., 2021). Additional insights, like improved fluidic card and manifold materials, and the physical separation of the fluidic manifolds from reagent reservoirs, keep humidity at a minimum since the biological samples inside the fluidic cards are integrated and flown in desiccated state. Given that fluidic systems are the lifeforce of a successful biological mission, further innovation and advancement is necessary in preparation for future long-duration missions beyond LEO.
Our desire to venture to deep space highlights knowledge gaps in biological research, particularly on the biological effects of the deep space radiation environment. Biological CubeSats could help fill these gaps by performing biological experiments in deep space in an autonomous manner. In recent years, NASA has successfully operated five biological CubeSats, namely GeneSat-1, PharmaSat, O/OREOS, SporeSat, and EcAMSat, the majority of which achieved full mission success (Massaro Tieze et al., 2020; Kanapskyte et al., 2021). Each CubeSat mission optimized and added to the fluidic systems and technologies of its predecessors, extending technological innovation with every mission. A summary of the main features and differences between their fluidic systems (including BioSentinel), and how fluidic systems have evolved over time, are shown in Table 1 and Figure 1, respectively.
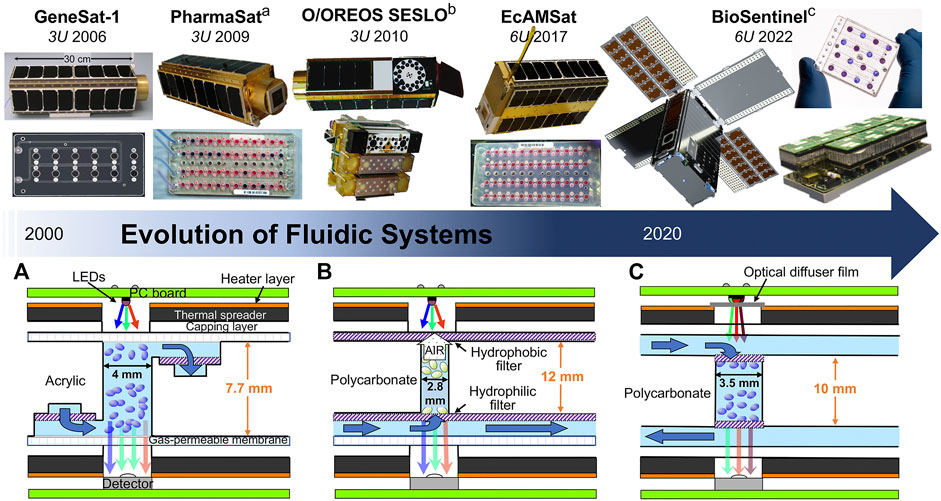
FIGURE 1. Evolution of fluidic systems in NASA’s biological CubeSats. Timeline of NASA’s biological CubeSats, unit size, and launch date (top). Cross-sections of PharmaSat (A), O/OREOS SESLO (B), and BioSentinel (C) fluidic cards (bottom). Layers include PC boards (green), heater layer (orange), thermal spreaders with temperature sensors (dark grey), capping layer (white), gas-permeable membrane (hatched white), filters (hatched purple), fluidic wells containing biology (shaded blue, ovals represent biological cells), and the optical system (LEDs above, detector chip below). Growth media are supplied through fluidic channels (also shaded blue). Dark blue arrows indicate direction of flow. Dimensions listed in millimeters (not to scale).
The first biological payload to fly into space on a CubeSat platform was NASA’s GeneSat-1 mission in 2006 (Ricco et al., 2007). The goal of the 3U CubeSat was to observe the effects of microgravity on gene expression in two strains of Escherichia coli (E. coli) modified to produce a green fluorescent protein (GFP) constitutively. The payload included an acrylic fluidic card, liquid growth medium storage, an optical sensing system, thermal control, and connection and control support hardware (e.g., data logging).
The fluidic card was fabricated from multiple laser-cut acrylic layers laminated together with pressure-sensitive-adhesive interlayers (ALine, Inc.) and contained 12 wells (110 μl each), including two solid-state reference wells and one well without biology (Figure 1). The fluidic card also served as a manifold that allowed delivery of fluids to the wells. Furthermore, every well possessed 0.5-μm porosity nylon membrane filters at both ends to ensure that E. coli cells stayed within the wells during nutrient flow-through. The bottom surface of the fluidic card was a 0.5-mm-thick layer of optical quality acrylic, to ensure high quality optical measurements with minimal background fluorescence. A medical-grade polyethylene vinyl acetate (PEVA) bag filled with nutrient medium was connected to the fluidic card via an inlet valve. A second PEVA bag connected to the fluidic card was used to collect waste. To prevent alignment issues due to moving parts during experiment operation, each of the 12 wells had a dedicated optical subassembly consisting of a fluorescence excitation blue LED light (470 nm), excitation and emission filters, and an intensity-to-frequency photodetector. In addition to the fluorescent GFP signaling for gene expression detection, a green LED light (Osram) was used to obtain absorbance (optical density via light scattering) measurements (Ricco et al., 2007). The thermal system consisted of a pair of custom Kapton (Minco) film heaters mated with 3-mm-thick, black-anodized aluminum plates (to minimize light reflections) as thermal spreaders that sandwiched the card. Temperature stability (±0.5°C) and monitoring was provided by a system microcontroller and six temperature sensors (Analog Devices) distributed over the thermal spreaders to monitor fluidic card temperature. A micromachined pressure transducer (Motorola) and a thin-film capacitive humidity sensor (Sensirion) were also present.
Nine card microwells were inoculated with E. coli prior to flight (as well as the fluid filling of all tubing and channels). The only in-space fluidic operation was the replacement of stasis medium with nutrient medium for cell growth. To initiate the experiment during spaceflight, the fluidic card was activated by remote command (49 h after launch) to bring the card temperature from ambient ∼10°C to the operating temperature of 34°C. After ∼1 h for temperature stabilization, the solenoid valve was opened automatically to deliver growth medium plus antibiotic (Luria broth +2.4 mg/ml carbenicillin) into the wells to displace the stasis buffer and start cell growth. Both fluorescence (corresponding to GFP production) and cell growth (corresponding to absorbance) were monitored via the optical sensor measurements (Ricco et al., 2007). Cell growth (from light scattering measurements) was first observed ∼2.8 h after valve opening, and by 10 h all nine microwells containing cells showed measurable growth. Importantly, most of the wells (6 out of 9) exhibited green fluorescence by 20 h after growth medium addition; eventually, the remaining wells also produced GFP signals (Ricco et al., 2007). Overall, data were obtained over ∼100 h following valve activation. With a design and development process of under 3 years, GeneSat-1 proved that CubeSats can perform innovative biological experiments in space and served as the first steppingstone for all future biological CubeSats.
The goal of the PharmaSat 3U CubeSat mission, launched in 2009, was to measure the response to an antifungal drug in Saccharomyces cerevisiae (budding yeast) cells. As with GeneSat-1, PharmaSat used optical measurements to detect cell population and metabolic activity changes over time (Ricco et al., 2011). While inheriting many basic design components of GeneSat-1, such as a multiwell fluidic card, LED lights, and the 1U bus, PharmaSat also expanded and innovated two main features of its fluidic system. The bus is the section of a satellite or spacecraft that contains the primary components required to power the onboard instruments, navigate in space, and communicate to the ground.
First, PharmaSat successfully expanded the fluidic card (Micronics, Inc.; laser-cut poly(methylmethacrylate) layers laminated with pressure-sensitive acrylic adhesive) from 12 wells to a larger layout containing 48 100-μl wells and 11 solid-state reference wells containing absorbance standards (Figure 1). The wells were divided into four 12-well banks served by four independent sets of fluidic inlets and outlets supplying nutrients and antifungal agent. Yeast cells were confined within the microwells by 1.2-μm pore-size nylon membrane filters (Sterlitech). A 51-μm-thick layer of optical-quality polystyrene covered the tops and bottoms of the fluidic cards, providing gas permeability for CO2 and O2 exchange and optical transparency for the 3-color absorbance measurements. The fluid delivery system consisted of 14 solenoid-operated valves, two pumps, a bubble trap, and nine medical-grade fluoropolymer bags for growth medium, dye, antifungal agent, and waste. Compared to on Earth, where fluid density differences can cause gravity-driven fluid movement and mixing of reagents, in space, medium exchange within fluidic cards can only occur by diffusion. For this reason, the fluidic cards were designed to have the entrance and exit of fluids diagonally opposite one another, to minimize the distance the fluid needs to travel in order to mix.
Second, instead of utilizing GFP expression as a reporter, PharmaSat used the oxidation-reduction potential (redox) indicator dye alamarBlue (Invitrogen) to detect changes in metabolism. Each of the wells had a dedicated set of three LEDs (470, 525, and 615 nm), with an intensity-to-frequency photodetector. These LED lights allowed for the detection of changes in both cell growth and in metabolic activity: alamarBlue transitions from an oxidized blue form to a reduced pink form when cellular metabolic processes are active (Ricco et al., 2011).
Similar to GeneSat-1, PharmaSat used a pair of custom Kapton-film, patterned heaters adhered to 3-mm-thick, black-anodized aluminum plates that served as thermal spreaders and sandwiched the fluidic card to provide thermal uniformity throughout the mission. Temperature stability (±0.3°C) was ensured using a system microcontroller and six temperature sensors distributed over the thermal spreaders. Similarly, a micromachined pressure transducer (Motorola) and a thin-film capacitive humidity sensor (Sensirion) were onboard the CubeSat. As with GeneSat-1, the biology experiment was activated remotely ∼47 h after orbital deployment, by heating cards from the ambient ∼6–16°C to a constant operating temperature of 27°C. Once the temperature stabilized (after ∼3.6 h), yeast growth was activated in four sequential sets of 12 wells each. With each set, the fluidic system supplied growth medium and alamarBlue to initiate cell growth (i.e., 12-h recovery period), followed by mixing and delivery of the antifungal agent at four different concentrations (0 “control”, 0.13, 0.50, and 2.0 μg/ml). Optical measurements were performed every 15 min and the fluidic card was maintained at 27°C. The experiment was terminated ∼48 h after addition of the antifungal agent (Ricco et al., 2011).
The O/OREOS (Organism/Organic Exposure to Orbital Stresses) CubeSat, launched in 2010, was the first demonstration of two distinct experiments on a single autonomous satellite. The first, SESLO (Space Environment Survivability of Living Organisms), examined the effects of microgravity and LEO space radiation in spores of the bacterium Bacillus subtilis. The second, SEVO (Space Environment Viability of Organics), investigated the photodegradation of biomarkers and bio-building blocks via UV-visible spectroscopy (Nicholson et al., 2011). Each occupied a 1U volume of the 3U CubeSat. Although SEVO was a biochemical spaceflight experiment, SESLO’s use of living cells makes it a direct successor to GeneSat-1 and PharmaSat.
SESLO returned to the 12-well card format of GeneSat-1, though this time three cards were present instead of one. For the first time, the biological organism was desiccated prior to payload assembly, to keep the bacterial spores in stasis, then rehydrated to re-activate cell growth in space. This allowed for multi-time-point activation of the experiment – at three different time points (14, 97, and ∼180 days). The SESLO fluidic system was composed of three “bioblock” modules (Figure 1), each with 12 wells (75 μl each) interconnected by microfluidic channels to a fluidic reservoir filled with spore germination medium containing the alamarBlue dye, previously used in PharmaSat. After delivery of growth medium to the fluidic wells was initiated via solenoid valves, a similar 3-color LED illumination system (470, 525, and 615 nm) was utilized to monitor spore germination, cell growth, and metabolic activity (Nicholson et al., 2011). Germination, growth, and metabolism were monitored for 48 h at the different time points. Its higher-inclination orbit within LEO (∼650 km above Earth; 72°) compared to GeneSat-1 and PharmaSat (∼450 km; 40°) positioned the CubeSat so that ∼30% of its orbits passed through the South Atlantic Anomaly, exposing it to ∼15x more ionizing radiation. Exposure of the biological samples was measured in situ using two pairs of radiation-sensitive field-effect transistors (radFETs).
Overall, O/OREOS expanded upon prior technologies through the introduction of three fluidically and optically independent “bioblocks”, enabling activation and delivery of fluids at three different time points throughout the 6-month mission.
Tasked with investigating the effect of microgravity on spores of the fern Ceratopteris richardii, SporeSat, a 3U CubeSat, launched in 2014 (∼330 km above Earth; 51.6°). This mission did not involve active fluidic components, which is why it is minimally described here. SporeSat, however, was the first CubeSat to use lab-on-a-chip devices (Park et al., 2017). These lab-on-a-chip devices or biology compact discs (bioCDs) utilized ion-selective electrodes to investigate the effects of microgravity on calcium signaling. SporeSat carried three 50-mm bioCD lab-on-a-chip devices for real-time measurements, two of which rotated onboard using minicentrifuges to generate artificial gravity while the third one remained stationary as a microgravity control. Even though the mission successfully demonstrated the use of ion-selective electrodes and artificial gravity technologies in LEO, the red LED lights necessary for spore germination failed on the ground and during spaceflight.
EcAMSat launched in 2017 (∼400 km above Earth) and was the first 6U biological CubeSat and the first 6U satellite deployed from the ISS. Its main objective was to investigate the effects of microgravity on dose-dependent antibiotic response and resistance in uropathogenic E. coli (Padgen et al., 2020).
EcAMSat borrowed heavily from PharmaSat’s fluidic and optical systems (∼90% commonality), repurposing the 48-well fluidic card and 3-color LED optical system to measure cell growth and metabolic activity (Figure 1). Even though the experimental payload occupied 3U of the total volume, a 6U format provided 50% more solar panel power to keep bacterial experiments at the required 37°C for extended periods of time (for comparison, PharmaSat’s yeast required 27°C). Swapping of the biological organism also required adaptation of the fluidic card. Instead of the 1.2-μm porous nylon filters of PharmaSat, EcAMSat required 0.2-μm polytetrafluoroethylene laminated membrane filters (Sterlitech). During the spaceflight mission, bacteria were first grown to stationary phase and then exposed to various doses of the antibiotic gentamicin for ∼46 h. A metering pump was used to generate different doses of gentamicin by extracting precise amounts from the antibiotic bag. Following the incubation with antibiotic, metabolic activity was measured using the redox dye alamarBlue for an additional 48-h period (Padgen et al., 2020). The entire biology experiment lasted 156.5 h.
NASA plans to launch 10 secondary payloads onboard Artemis 1, all 6U CubeSats. Among them is BioSentinel, NASA’s first interplanetary biological satellite and the first deep space bioscience mission past LEO since 1972 (Massaro Tieze et al., 2020; Ricco et al., 2020). BioSentinel’s primary science objective is to assess the effect of deep space ionizing radiation on DNA and cell damage response, using S. cerevisiae (budding yeast) as a model organism. The response to space radiation will be monitored via cell growth and metabolic activity using alamarBlue and a 3-color LED light detection system, building on the heritage of PharmaSat, O/OREOS, and EcAMSat (Ricco et al., 2020; Padgen et al., 2021). BioSentinel innovates on the fluidic systems from prior CubeSat missions at multiple levels.
The 4U BioSensor instrument, the core of the BioSentinel CubeSat, contains 18 microfluidic cards, each one with 16 100-μl wells, for a total of 288 wells (Figure 1) (Padgen et al., 2021). This dramatically increases the experimental size and capability of the BioSentinel CubeSat in comparison to PharmaSat and EcAMSat, which used a single 48-well fluidic card. In addition, the fluidic cards were designed with improved biocompatibility – constructed from thermally robust polymers and adhesives and low-permeability, high optical quality Zeonor cover layers to better protect dried cells from humidity. Compared to previous missions, BioSentinel cards have polycarbonate filters positioned within the optical path of each well (i.e., GeneSat-1, PharmaSat, and EcAMSat had filters outside of the optical path as to not perturb the optical signals). BioSentinel minimizes disturbance in the optical signal by using optically clear materials. The 18 cards are divided into two independent banks of 9 cards, mounted on two independent fluidic manifolds (Figure 1). One important improvement compared to previous missions is the use of fused polycarbonate to manufacture the cards and manifolds, which allows for high-temperature sterilization via autoclaving. Previous missions using acrylic relied primarily on ethylene oxide sterilization, which can lead to the release of toxic volatiles from plastic materials over time. Proper delivery of the combined growth medium and redox dye fluids is controlled by a series of individual 3-way valves and bubble traps integrated into the manifolds upstream of each card (Figure 2). Further upstream, mixing of the growth medium and the redox dye during spaceflight is performed by a system of timed peristaltic pumps and valves, which are contained within two smaller manifolds placed in between the 9-card manifolds and the reagent bags. Toggling between streams of alamarBlue and growth medium allows controlled dilution of the reagents into each well. Importantly, the alamarBlue dye and growth medium are stored in separate bags, to prevent the dye from being prematurely reduced by the medium itself (without cells).
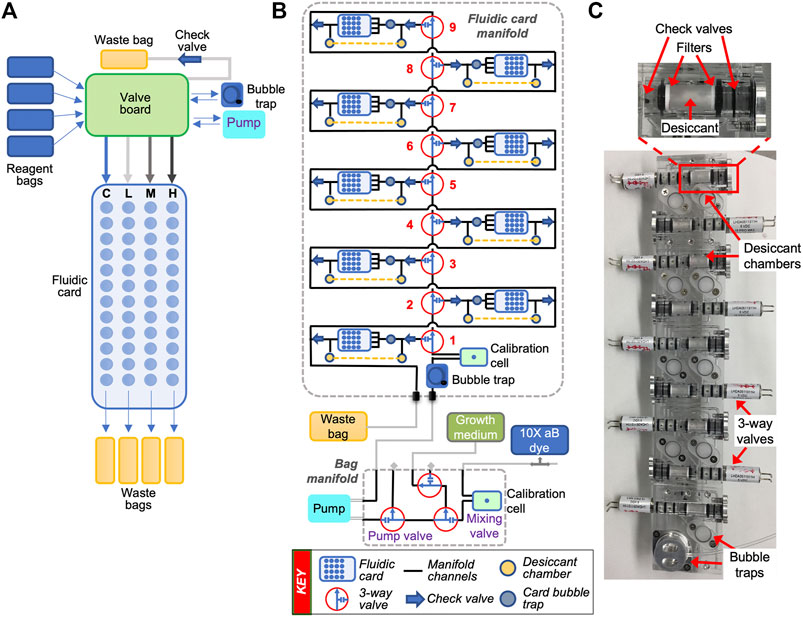
FIGURE 2. Fluidic schematics of a single-card system and a multi-card manifold-based delivery system. (A) Simplified schematic of EcAMSat’s fluidic system showing flow directions through a valve board and a single fluidic card. Each bank of 12 wells is supplied by a common inlet ending in four independent waste bags. C, L, M, H = control, low, medium, and high banks of the fluidic card, referring to the different doses of antibiotic delivered to each bank. A more detailed schematic has been published previously (Padgen et al., 2020). (B) Fluidic schematic of the BioSensor system in BioSentinel. Shown is one set of the fluidic card and bag manifolds, including fluidic cards, valves, bubble traps, optical calibration cells, desiccant chambers, reagent bags, pump, and waste bag. (C) Photo of BioSentinel’s monolithic card manifold showing the valves, bubble traps, and the desiccant assemblies for each card position. The inset on top shows the components of the desiccant assembly in more detail.
Similar to O/OREOS SESLO, the biological samples are desiccated inside the microfluidic wells, which allows for stasis for >20 months prior to activation in space (Santa Maria et al., 2020). A new feature in BioSentinel is the addition of desiccant chambers adjacent to each card within the fluidic manifolds (Figure 2). These chambers contain approximately 80 mg of Drierite desiccant separated from the fluidic path by hydrophobic filters to ensure that the desiccant is in vapor contact with the biological samples within the cards before fluid delivery. Two check valves were added on both sides of the desiccant chamber to provide additional humidity barriers. The inlet check valve prevents humidity from entering the card from the main fluidic line while the outlet valve prevents backflow into the card. This maintains the desiccated yeast cells at a low relative humidity prior to card activation. Importantly, and to prevent bubble formation when the fluid passes through the desiccant chamber, a small bubble trap was integrated between the desiccant chamber and each fluidic card (see fluidic schematic in Figure 2).
Delivery of fluids and rehydration of the yeast cells will be performed at different time points throughout the mission. Cell growth and metabolic activity will be monitored optically using the 3-color LED detection system (Padgen et al., 2021). Two LED lights aid in the detection of the oxidized (blue, 630 nm) and reduced (pink, 570 nm) forms of the alamarBlue redox dye. A third LED light (850 nm) will measure light scattering produced by cell growth (turbidity) at a wavelength which is unaffected by the dye.
To maintain thermal control, Kapton-film-based heaters are adhered to aluminum plates attached to the top and bottom of each fluidic card in addition to a temperature detector embedded in the middle of the card. This dedicated thermal control system allows for active cards to be kept at the experimental growth temperature (23°C) while keeping the remaining cards and fluidic components above freezing (∼5°C). Improved thermal control of each individual card enables activation of two cards at a time, without affecting the temperature profile of the surrounding cards. This also allows experimental activation, two cards at a time, at increasing distances from Earth, up to nine times throughout the entire mission duration. Based on preliminary data from our team, we expect each two-card experiment to last approximately 7 days given the age of the yeast cells at the time of launch and activation in space (Santa Maria et al., unpublished observations). An onboard miniature linear energy transfer (LET) spectrometer will be used to correlate biological response to cumulative ionizing radiation dose and to characterize the space radiation environment (Ricco et al., 2020).
NASA is planning the next autonomous experiments in preparation for future crewed Artemis missions. Still in its formulation and design stages, the Lunar Exploration Instrument for space biology Applications (LEIA) program will use the BioSensor instrument originally designed for the BioSentinel mission. Instead of interplanetary space, however, LEIA is intended to be a secondary payload on a Commercial Lunar Payload Services (CLPS) lander. To ensure optimal compatibility and performance on the surface of the Moon, most of the spacecraft components will be removed, leaving the payload fluidic, optical, and thermal systems in place, controlled by an updated electrical power system (EPS) that also manages communications with the lander. Minor modifications permit potential upgrades to the fluidic system, including the possibility of running multiple experiments on a single payload (one on each of two fluidic manifolds), or of running fluid exchange of different reagent types (antibiotics or nutrient drop-out media) to run different experiments at once. Radiation sensors better suited for the lunar surface environment (i.e., galactic cosmic rays and secondary neutrons) may also be included as part of the LEIA instrument.
GeneSat-1, PharmaSat, and EcAMSat shared many similarities in their fluidic systems (Figure 1). First, they had a similar flow path – fluids flowed from the bottom of one side of the well, to the top on the other side of the well. Second, fluids flowed through small hose-barbs built onto the fluidic cards. These were attached, via additional tubing, to a network of pumps, valves, and reagent and waste bags. Additionally, fluidic cards for GeneSat-1, PharmaSat, and EcAMSat had filters that were positioned outside of the optical path, as to not perturb the signal for growth (and metabolism). By contrast, despite being within the optical path, BioSentinel’s filters minimize disturbance to the optical signal because they are made of optically clear materials; polycarbonate track etched (PCTE) filters appear transparent, due to the size and spacing of the pores.
The fluidic systems of O/OREOS SESLO and BioSentinel are similar in their integration of many fluidic components into a solid-state polymer manifold. Importantly, shifting fluidic systems from a single, large-card format (PharmaSat and EcAMSat) to a parent manifold, with simpler smaller cards attached, greatly improves the capabilities of CubeSat fluidic systems. For O/OREOS SESLO, the card was its own manifold - including a fluid reservoir, air pump (that pushed air onto the reservoir to move fluid into wells), a valve, and the experimental wells. By incorporating all the parts into a single assembly, O/OREOS SESLO was a step in the direction of the monolithic manifolds being used in BioSentinel and future life detection missions. By contrast, the pumps, valves, and other fluidic components are external to the fluidic cards for BioSentinel, on their own separate manifold (Figure 2).
There are a few other differences between the fluidic systems. Although all the individual fluidic card wells housing biological samples held ∼100 ul volume, GeneSat-1 wells were 6.5 mm in diameter, whereas PharmaSat and EcAMSat were 4 mm (BioSentinel is 3.5 mm). GeneSat-1 had an air permeable membrane on one side, whereas PharmaSat and EcAMSat had permeable membranes on both sides. For O/OREOS SESLO, each well dead-ended into a hydrophobic filter, to encourage air to be pushed out of the filter while the well was being filled. Because there was only a single channel inlet, there was no possibility of fluid exchange for O/OREOS SESLO. There was no waste line and no destination for the fluid to flow once the well was filled. Conversely, BioSentinel was specifically designed for fluid exchange. Fluid flows into the wells to revive the yeast (allowing growth and metabolic measurements to be taken), then flows out the waste line to a designated waste bag (Figure 2).
One of the main advantages of using small satellites like CubeSats for biological research is that they can perform autonomous experiments in harsh environments without human presence. Throughout the progression of biological CubeSats, the basic technologies behind fluidic-based instruments – components and materials for moving fluid, performing optical and electrochemical measurements, and controlling temperature – have become more capable and efficient. The challenge in adapting these advancements for biological CubeSats is miniaturizing already-existing instruments and procedures used on Earth to fit into small spaces and automating them to account for limited and delayed communications. More specifically to space biology applications, there is a need to improve current detection instrumentation, fluid sample processing and manipulation, and long-term preservation capability of the biological samples.
The past and present missions discussed above have included small, dedicated sensors for each replicate (or fluidic well). Except for SporeSat, all CubeSats discussed here used a copy of the same LED lights and photodetectors for every well to detect fluorescence, reduction of alamarBlue, or optical density, and in SporeSat, each spore had its own ion-selective electrode. Because of the high variability naturally found in biological systems, as well as the often-subtle effects of the spaceflight environment under study, future experiments are likely to continue to need large numbers of replicates; however, under this paradigm, the requirement to have one sensor per sample greatly limits the variety of sensors that could feasibly be used. More complex instruments, like brightfield or fluorescence microscopes, flow cytometers, mass spectrometers, or microchip capillary electrophoresis chambers, might be able to fit into a CubeSat, but including one for every well is not realistic. Thus, complex detection instruments would likely exist in a format as a single, centralized instrument with an intricate fluidic system for pushing samples to and from for secondary processing (i.e., sample de-salting, concentration or dilution, calibration, or fluoro-labeling). Such a system comes with its own set of challenges, introducing operational complexity (i.e., more fluidic channels = more opportunity for bubble formation) and cross-contamination risks. However, passing a sample from one sensor to the next would also greatly increase the number of measurements that could be conducted on a single sample. SPLIce (Sample Processor for Life on Icy Worlds), ELSAH (Enceladus Life Signatures and Habitability), and MICA (Microfluidic Icy-world Chemistry Analyzer) are a few integrated life detection payloads designed and developed at NASA (in collaboration with Johns Hopkins University, Tufts University, University of Alberta, and Honeybee Robotics) that take advantage of this format (Chinn et al., 2017; Noell et al., 2019).
Thus far, biological CubeSats have been designed to control sub-milliliter-range fluid volumes. This allows use of mostly off-the-shelf components, including standards fluidic parts (valves, tubing, fittings, and bubble traps), optical sensors (surface-mount LEDs and photodiodes), and manufacturing and assembly methods (e.g., milling and layering of thermoplastics). There is significant potential in smaller-scale microfluidic technology. These include “3D-printed” approaches, such as the use of stereophotolithography with materials like the silicon-based organic polymer polydimethylsiloxane (PDMS), and emerging techniques such as laser-fused glass micromachining (Wlodarczyk et al., 2019). These advanced microfluidic manufacturing techniques offer more than just the possibility of improved miniaturization (or greater functionality within the same footprint). For example, the concept of “reactionware”, in which reagents are embedded directly into the fluidic surface, could allow new types of assays to be implemented autonomously (Symes et al., 2012). The intrinsic flexibility of PDMS can be used to create membrane valves that can separate microchannels to specifically block or allow channel flow within a single channel (Abate and Weitz, 2008). Similarly, cells can be preferentially isolated based on their size in microfluidic channels which expand and contract in width (Dhar et al., 2018). Many of these microfluidic techniques are still not fully developed or commercialized on Earth; however, if successfully developed for space biological applications, they could reduce payload volume and mass, allow more replicates to be included, enable automation of more complex and multi-step experimental procedures, reduce cross-contamination risks, and allow a wider array of sensors to be integrated on small platforms like CubeSats.
Another technology that could improve mission flexibility and robustness is single-cell microencapsulation. One of the biggest restrictions on biology for missions beyond LEO, is their ability to survive the limitations of long-duration spaceflight (i.e., extended prelaunch periods and long flight durations, where loaded biology could sit for many months without human interaction). Our preliminary studies suggest that by encapsulating single biological cells inside semi-permeable hydrogel microcapsules could improve the long-term storage capability of biological cells for long duration spaceflight (Ng et al., unpublished data). Furthermore, microencapsulation of cells could allow for better performance and miniaturization of certain assays. The encapsulated cells are suspended in the inner liquid core of the capsule, providing an unconstrained growing environment (van Zee et al., 2022). Meanwhile, the hydrogel shell allows diffusion of media, waste, and small assay reagents into and out of the capsule while trapping the cell, large molecules like genomic DNA, and cell secretions with binding antibodies embedded within the hydrogel. These microcapsules enable replacement of buffers and addition of assay reagents, while the cell is kept within the capsule (Leonaviciene et al., 2020). This enables assay readout on the single cell level, which could help to identify rare cells and cell subpopulations in the space environment. Similar encapsulations without the hydrogel capsule have been used for a wide range of single cell omics investigations (Matula et al., 2020).
New and improved sensor technologies, beyond the relatively simple LED/photodiode absorbance and fluorescence assays implemented so far, also hold significant promise. Absorption spectrometry using tunable laser diodes (allowing measurements across multiple wavelengths) and specialized high-gain optical cells (reducing the needed unit path length, and thus required fluid volumes) have been used for some years in commercial applications, and similar implementations have been designed for fluorescence and Raman spectroscopy (Rushworth et al., 2012). Beyond optical sensors, miniaturized electrochemical sensors – such as ion-selective electrodes, which are widely used to measure pH, and can be customized to assay specific solutes such as dissolved oxygen or the oxidized/reduced states of a chemical species – have a long history for medical applications (Bakker et al., 2008; Ozbek et al., 2020). More recently, dielectric spectroscopy has emerged as a low-cost, low-footprint method of assessing microbiological health and function (Russel et al., 2018; Flores-Cosio et al., 2020).
An additional challenge associated with more complex detectors, like microscopes, is the transmission of large raw data files back to Earth, along with the required metadata – including dimensions, image type, bit-depth, pixel size, and microscope settings. A common discussion among astrophysicists and astrobiologists surrounding the topic of microscopy feasibility for a CubeSat is file size and image resolution. This could be mitigated by adding onboard data processing capabilities (i.e., an onboard computer), and potentially leverage artificial intelligence, to pre-analyze data and transmit smaller, processed data files. The Korea Aerospace Research Institute (KARI) has taken a first step to address this with their High-Resolution Image and Video CubeSat (HiREV) platform carrying an onboard 866-MHz processor computer, with a commercial off-the-shelf (COTS) camera payload that can capture 5-m high-resolution color image and video (Cho et al., 2019).
These sensor, replicate, and data return considerations are also affected by the duration of the experiment. The missions described here have used each well for a single microbial growth cycle, from rehydration of desiccated cells in stasis to exhaustion of nutrients in the liquid growth medium within the well volume. Design modifications to the flow paths and controls would allow the same basic approach – fluidics parts, materials, sensors, and manufacturing techniques – to be extended to continuous or iterative growth cycles, as in a bioreactor. However, the small size of CubeSats creates severe limitations on the volume and mass of consumables (e.g., additional consumables required to support longer-term growth) that can be carried. Additionally, the longer the experimental payload remains active, the more significant the cost of mission operations, including communications and data return, will become. These factors will require careful tradeoff in experiment design to take best advantage of the CubeSat format.
Nations worldwide are more determined than ever to send humans beyond LEO, but there are significant gaps in our knowledge on the associated biological risks. Biological CubeSats have demonstrated that they can be valuable assets to aid fill these gaps beyond the limitations of the ISS, allowing for data on the biological response of key model organisms to microgravity, space radiation, and other factors generated in and beyond LEO. The data gathered from BioSentinel will be the first indication of how biology responds to the deep space environment and will help define mitigations for future crewed missions beyond LEO. Importantly, it will also give an initial demonstration for how the CubeSat systems (i.e., fluidic, electrical, command and data handling, communications, etc.) fare against the austere conditions in deep space. Accordingly, we expect biological CubeSats to further distinguish themselves to the scientific community and to become excellent educational and scientific tools worldwide.
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.
BH, SN, and SSM conceptualized this work. BH, SN, LL, DG, and SSM wrote the paper. All authors have read and agreed to the published version of the manuscript.
The BioSentinel project is supported by the Advanced Exploration Systems Program Office at NASA Headquarters.
LCL is employed by Logyx, LLC.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank past and present members of the NASA BioSentinel project and the Space Life Sciences Training Program (SLSTP).
Abate, A. R., and Weitz, D. A. (2008). Single-layer Membrane Valves for Elastomeric Microfluidic Devices. Appl. Phys. Lett. 92, 243509. doi:10.1063/1.2945797
Bakker, E., Bhakthavatsalam, V., and Gemene, K. L. (2008). Beyond Potentiometry: Robust Electrochemical Ion Sensor Concepts in View of Remote Chemical Sensing. Talanta 75, 629–635. doi:10.1016/j.talanta.2007.10.021
Chinn, T. N., Lee, A. K., Boone, T. D., Tan, M. X., Chin, M. M., Mccutcheon, G. C., et al. (2017). “Sample Processor for Life on Icy Worlds (SPLIce): Design and Test Results,” in Proceedings of the International Conference on Miniaturized Systems for Chemistry and Life Sciences (MicroTAS 2017), Savannah, GA, USA, October 22–26, 2017.
Cho, D.-H., Choi, W.-S., Kim, M.-K., Kim, J.-H., Sim, E., and Kim, H.-D. (2019). High-Resolution Image and Video CubeSat (HiREV): Development of Space Technology Test Platform Using a Low-Cost CubeSat Platform. Int. J. Aerospace Eng. 2019, 1–17. doi:10.1155/2019/8916416
Dhar, M., Lam, J. N., Walser, T., Dubinett, S. M., Rettig, M. B., and Di Carlo, D. (2018). Functional Profiling of Circulating Tumor Cells with an Integrated Vortex Capture and Single-Cell Protease Activity Assay. Proc. Natl. Acad. Sci. U.S.A. 115, 9986–9991. doi:10.1073/pnas.1803884115
Flores-Cosío, G., Herrera-López, E. J., Arellano-Plaza, M., Gschaedler-Mathis, A., Kirchmayr, M., and Amaya-Delgado, L. (2020). Application of Dielectric Spectroscopy to Unravel the Physiological State of Microorganisms: Current State, Prospects and Limits. Appl. Microbiol. Biotechnol. 104, 6101–6113. doi:10.1007/s00253-020-10677-x
Kanapskyte, A., Hawkins, E. M., Liddell, L. C., Bhardwaj, S. R., Gentry, D., and Santa Maria, S. R. (2021). Space Biology Research and Biosensor Technologies: Past, Present, and Future. Biosensors 11, 38. doi:10.3390/bios11020038
Leonaviciene, G., Leonavicius, K., Meskys, R., and Mazutis, L. (2020). Multi-step Processing of Single Cells Using Semi-permeable Capsules. Lab. Chip 20, 4052–4062. doi:10.1039/d0lc00660b
Massaro Tieze, S., Liddell, L. C., Santa Maria, S. R., and Bhattacharya, S. (2020). BioSentinel: A Biological CubeSat for Deep Space Exploration. Astrobiology 20. doi:10.1089/ast.2019.2068
Matuła, K., Rivello, F., and Huck, W. T. S. (2020). Single‐Cell Analysis Using Droplet Microfluidics. Adv. Biosys. 4, 1900188. doi:10.1002/adbi.201900188
Nicholson, W. L., Ricco, A. J., Agasid, E., Beasley, C., Diaz-Aguado, M., Ehrenfreund, P., et al. (2011). The O/OREOS mission: First Science Data from the Space Environment Survivability of Living Organisms (SESLO) Payload. Astrobiology 11, 951–958. doi:10.1089/ast.2011.0714
Noell, A. C., Jaramilllo, E. A., Kounaves, S. P., Hecht, M. H., Harrison, D. J., Quinn, R. C., et al. (2019). “MICA: Microfluidic Icy-World Chemistry Analyzer,” in Proceedings of the 2019 Astrobiology Science Conference (AbSciCon), Bellevue, WA, USA, June 24–28, 2019.
Özbek, O., Berkel, C., and Isildak, Ö. (2020). Applications of Potentiometric Sensors for the Determination of Drug Molecules in Biological Samples. Crit. Rev. Anal. Chem., 1–12. doi:10.1080/10408347.2020.1825065
Padgen, M. R., Chinn, T. N., Friedericks, C. R., Lera, M. P., Chin, M., Parra, M. P., et al. (2020). The EcAMSat Fluidic System to Study Antibiotic Resistance in Low Earth Orbit: Development and Lessons Learned from Space Flight. Acta Astronautica 173, 449–459. doi:10.1016/j.actaastro.2020.02.031
Padgen, M. R., Liddell, L. C., Bhardwaj, S. R., Gentry, D., Marina, D., Parra, M., et al. (2021). BioSentinel: A Biofluidic Nanosatellite Monitoring Microbial Growth and Activity in Deep Space. Astrobiology 21. doi:10.1089/ast.2020.2305
Park, J., Salmi, M. L., Wan Salim, W. W. A., Rademacher, A., Wickizer, B., Schooley, A., et al. (2017). An Autonomous Lab on a Chip for Space Flight Calibration of Gravity-Induced Transcellular Calcium Polarization in Single-Cell Fern Spores. Lab. Chip 17, 1095–1103. doi:10.1039/c6lc01370h
Ricco, A. J., Beasley, C., Giovangrandi, L., Henschke, M., Kitts, C., Levine, L., et al. (2007). “Autonomous Genetic Analysis System to Study Space Effects on Microorganisms: Results from Orbit,” in Proceedings of the TRANSDUCERS 2007–2007 International Solid-State Sensors, Actuators and Microsystems Conference, Lyon, France, June 10–14, 2007. doi:10.1109/SENSOR.2007.4300065
Ricco, A. J., Maria, S. R. S., Hanel, R. P., and Bhattacharya, S. (2020). BioSentinel: A 6U Nanosatellite for Deep-Space Biological Science. IEEE Aerosp. Electron. Syst. Mag. 35, 6–18. doi:10.1109/MAES.2019.2953760
Ricco, A. J., Parra, M., Niesel, D., Piccini, M., Ly, D., McGinnis, M., et al. (2011). “PharmaSat: Drug Dose Response in Microgravity from a Free-Flying Integrated Biofluidic/optical Culture-And-Analysis Satellite,” in Proceedings of the SPIE 7929, Microfluidics, BioMEMS, and Medical Microsystems IX, San Francisco, CA, USA, January 22–27, 2011. doi:10.1117/12.881082
Rushworth, C. M., Davies, J., Cabral, J. T., Dolan, P. R., Smith, J. M., and Vallance, C. (2012). Cavity-enhanced Optical Methods for Online Microfluidic Analysis. Chem. Phys. Lett. 554, 1–14. doi:10.1016/j.cplett.2012.10.009
Russel, M., Sophocleous, M., JiaJia, S., Xu, W., Xiao, L., Maskow, T., et al. (2018). High-frequency, Dielectric Spectroscopy for the Detection of Electrophysiological/biophysical Differences in Different Bacteria Types and Concentrations. Analytica Chim. Acta 1028, 86–95. doi:10.1016/j.aca.2018.04.045
Santa Maria, S. R., Marina, D. B., Massaro Tieze, S., Liddell, L. C., and Bhattacharya, S. (2020). BioSentinel: Long-TermSaccharomyces cerevisiaePreservation for a Deep Space Biosensor Mission. Astrobiology 20. doi:10.1089/ast.2019.2073
Symes, M. D., Kitson, P. J., Yan, J., Richmond, C. J., Cooper, G. J. T., Bowman, R. W., et al. (2012). Integrated 3D-Printed Reactionware for Chemical Synthesis and Analysis. Nat. Chem 4, 349–354. doi:10.1038/nchem.1313
Twiggs, R. J., and Puig-Suari, J. (2020). “Introduction: The History of the CubeSat,” in CubeSat Handbook: From Mission Design to Operations. Editors C. Cappelletti, S. Battistini, and B.K. Malphrus (Cambridge, MA: Academic Press), xxi–xxviii.
van Zee, M., de Rutte, J., Rumyan, R., Williamson, C., Burnes, T., Radakovits, R., et al. (2022). High-throughput Selection of Cells Based on Accumulated Growth and Division Using PicoShell Particles. Proc. Natl. Acad. Sci. U.S.A. 119 (4), e2109430119. doi:10.1073/PNAS.2109430119
Keywords: space biology, CubeSat, fluidics, deep space, space radiation
Citation: Harandi B, Ng S, Liddell LC, Gentry DM and Santa Maria SR (2022) Fluidic-Based Instruments for Space Biology Research in CubeSats. Front. Space Technol. 3:853980. doi: 10.3389/frspt.2022.853980
Received: 13 January 2022; Accepted: 09 March 2022;
Published: 29 March 2022.
Edited by:
Timothy Granata, Lucerne University of Applied Sciences and Arts, SwitzerlandReviewed by:
Aaron Noell, NASA Jet Propulsion Laboratory (JPL), United StatesCopyright © 2022 Harandi, Ng, Liddell, Gentry and Santa Maria. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sergio R. Santa Maria, c2VyZ2lvLnNhbnRhbWFyaWFAbmFzYS5nb3Y=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.