
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Space Technol. , 03 January 2022
Sec. Microgravity
Volume 2 - 2021 | https://doi.org/10.3389/frspt.2021.773244
 Katharina Siems1†
Katharina Siems1† Daniel W. Müller2†
Daniel W. Müller2† Laurens Maertens3,4
Laurens Maertens3,4 Aisha Ahmed2
Aisha Ahmed2 Rob Van Houdt3
Rob Van Houdt3 Rocco L. Mancinelli5
Rocco L. Mancinelli5 Sandra Baur6
Sandra Baur6 Kristina Brix6
Kristina Brix6 Ralf Kautenburger6
Ralf Kautenburger6 Nicol Caplin7
Nicol Caplin7 Jutta Krause7
Jutta Krause7 Rene Demets7
Rene Demets7 Marco Vukich7
Marco Vukich7 Alessandra Tortora8
Alessandra Tortora8 Christian Roesch9
Christian Roesch9 Gudrun Holland10
Gudrun Holland10 Michael Laue10
Michael Laue10 Frank Mücklich2
Frank Mücklich2 Ralf Moeller1*
Ralf Moeller1*Maintaining crew health and safety are essential goals for long-term human missions to space. Attaining these goals requires the development of methods and materials for sustaining the crew’s health and safety. Paramount is microbiological monitoring and contamination reduction. Microbial biofilms are of special concern, because they can cause damage to spaceflight equipment and are difficult to eliminate due to their increased resistance to antibiotics and disinfectants. The introduction of antimicrobial surfaces for medical, pharmaceutical and industrial purposes has shown a unique potential for reducing and preventing biofilm formation. This article describes the development process of ESA’s BIOFILMS experiment, that will evaluate biofilm formation on various antimicrobial surfaces under spaceflight conditions. These surfaces will be composed of different metals with and without specified surface texture modifications. Staphylococcus capitis subsp. capitis, Cupriavidus metallidurans and Acinetobacter radioresistens are biofilm forming organisms that have been chosen as model organisms. The BIOFILMS experiment will study the biofilm formation potential of these organisms in microgravity on the International Space Station on inert surfaces (stainless steel AISI 304) as well as antimicrobial active copper (Cu) based metals that have undergone specific surface modification by Ultrashort Pulsed Direct Laser Interference Patterning (USP-DLIP). Data collected in 1 x g has shown that these surface modifications enhance the antimicrobial activity of Cu based metals. In the scope of this, the interaction between the surfaces and bacteria, which is highly determined by topography and surface chemistry, will be investigated. The data generated will be indispensable for the future selection of antimicrobial materials in support of human- and robotic-associated activities in space exploration.
An enduring human presence in space is required to achieve the primary goals of space programs worldwide. As longer human missions, (e.g., to Moon and Mars) are being planned for the foreseeable future, the optimization of isolated self-sustaining habitats is of prime importance. One key aspect in this regard is extensive microbial control. The ubiquity of microorganisms in crewed spacecrafts such as the International Space Station (ISS) poses several critical risks to astronauts as well as to structural spacecraft materials. Decreasing unwanted microbial growth and biofilm formation within a space craft will also aid in inhibiting forward and backward contamination of extraterrestrial systems as for improved adherence to planetary protection protocols. Nevertheless, microorganisms have shown unparalleled potential for regenerative life support systems, such as MELiSSA, which has been designed to enable long-term manned missions in space (Lasseur et al., 2010).
Microbiological studies have shown the pervasive presence of microorganisms aboard the ISS associated with dust particles (Checinska et al., 2015; Mora et al., 2016), filter debris (Venkateswaran et al., 2014; Be et al., 2017), air vents (Lang et al., 2017), walls and panels (Singh and Wade, 2014), as well as touch surfaces such as laptop keyboards and handrails (Lang et al., 2017). The microbial population described in these studies is mostly composed of human-associated strains, but environmental organisms are also present. Several bacterial and fungal strains have been characterized in detail, often with specific attention towards antimicrobial resistance (AMR) and stress responses (Mijnendonckx et al., 2013; Romsdahl et al., 2018; Seuylemezian et al., 2018). The prevalence of AMR genes in the ISS microbiome was examined in a longitudinal study, indicating multiple multidrug-resistant organisms (Urbaniak et al., 2018). A similar study showed the persistence of several opportunistic pathogens, including Klebsiella pneumoniae and Staphylococcus aureus (Singh et al., 2018) that showed an increase in AMR genes and virulence observed over the duration of the study. These results are worrying in light of a decreased immune function of astronauts during spaceflight (Sonnenfeld and Shearer, 2002; Sonnenfeld et al., 2003; Aponte et al., 2006). In contrast, it has been proposed that these health concerns are unjustified, and that the risks accompanying unchecked microbial contamination are associated with degradation of equipment integrity rather than human health-related (Mora et al., 2019). This hypothesis is validated by the low number of observed health issues and infections of crew members (Crucian et al., 2016).
Monitoring and controlling microbial contamination on the ISS has been reviewed by Van Houdt and Leys (2020). While air and surface contamination levels were generally within acceptable levels, onboard potable water sources were found to be unsuited for human consumption in 60% of cases (Bruce et al., 2005), leading to increased costs and loss of crew time. Despite repeated decontamination attempts, (e.g., with silver), microbial counts rapidly increased above acceptable levels. These results could be caused by the possible prevalence of silver resistance mechanisms in contaminating cells (Mijnendonckx et al., 2013), as well as adaptation to the oligotrophic environment by entering a viable but non-culturable (VBNC) state (Davey, 2011; Schottroff et al., 2018) that could be more resistant to extant biocides. Lastly, the formation of biofilms on equipment surfaces could impair the functioning of biocidal applications.
Biofilms have been defined as aggregated cell communities enveloped in self-produced extracellular polymeric substances (EPS) often associated with surfaces (Flemming et al., 2016). Microorganisms in biofilms are often more resistant to antimicrobial agents because of a decreased permeation of these agents into the biofilm (Koo et al., 2017), and a high cellular heterogeneity including dormant cells with increased stress resistance (Singh et al., 2017). Biofilm formation on spacecraft materials has been observed in the ISS (Novikova et al., 2006; Perrin et al., 2018), as well as in the Mir space station (Klintworth et al., 1999; Ilyin, 2005). The presence of biofilms is known to cause damage to structural equipment via polymer deterioration, metal corrosion and biofouling. Microbial cells in space, in the planktonic phase as well as in biofilms, are not subjected to the same net gravitational field as on Earth due to a lack of convection, decreasing mixing in the system. Consequently, mass transfer between the cell and its immediate environment is limited to diffusion (Horneck et al., 2010). This leads to depletion of nutrients and accumulation of metabolites in the immediate extracellular environment that elicits a biological response in the cell. To study the effects of spaceflight on biofilm formation several experiments have been conducted (reviewed by Zea et al. (2018). For example, Pseudomonas aeruginosa is able to form biofilms in microgravity (McLean et al., 2001) and shows an increased propensity for biofilm formation dependent on nutrient availability and flagella-driven cell motility (Kim et al., 2013). This effect of motility on mixing rates in the fluid surrounding the cell has been hypothesized to partially negate the effects of microgravity and decreased convective phenomena (Horneck et al., 2010). In microgravity, Escherichia coli clustered together in liquid medium rather than being homogeneously distributed in the medium, indicating enhanced biofilm formation in space (Zea et al., 2017). Lastly, it must be noted that the function of quorum sensing mechanisms that depend on small signaling molecules can be impacted in microgravity. Cell-cell communication via quorum sensing plays a key role in biofilm formation and adaptation to a changing environment, but also bacterial virulence (Li and Tian, 2012). An increased concentration of N-acyl-homoserine lactones, that function as signaling compounds, was shown in Rhodospirillum rubrum in simulated microgravity while cell density was unaltered (Mastroleo et al., 2013).
The prevalence of multispecies biofilms and their possible effects on human health and equipment integrity, in combination with the frequent problems with bacterial contamination of drinking water, illustrates the need for advanced, integrated decontamination systems (Van Houdt et al., 2012). One promising alternative for existing antimicrobial strategies is the use of copper (Cu) surfaces. Metallic Cu is extremely efficient in killing microorganisms in atmospherically dry conditions, with near-complete inactivation in minutes to hours (Vincent et al., 2018). Among the modes of action of copper toxicity direct cell-to-metal contact is important in the inactivation kinetics and has led to the term “contact killing” (Mathews et al., 2013). Additional killing modes or parameters include ambient temperature and Cu concentration in any alloys used (Wilks et al., 2005; Noyce et al., 2006). Interestingly, in wet applications the antimicrobial effects of Cu surfaces are diminished, having inactivation times of several hours (Dwidjosiswojo et al., 2011; Maertens et al., 2020). The antimicrobial potential of Cu surfaces is currently being studied in drinking water production, air conditioning and in hospital settings (Vincent et al., 2016).
Copper, while an indispensable micronutrient, rapidly induces toxic effects in bacteria upon exposure to excess concentrations. A key aspect of Cu toxicity is the catalysis of intracellular reactive oxygen species (ROS) production via a Fenton-like reaction (Lemire et al., 2013). In addition, membrane and DNA damage, as well as inactivation of Fe-S clusters have been reported (Vincent et al., 2018). Amino acid and lipid metabolism in space-grown bacteria are often pre-adapted to increased oxidative stress, that has been linked to an elevated dose of ionizing radiation in space (Crabbe et al., 2011; Li J. et al., 2014). Interestingly, copper toxicity has been shown to induce the VBNC state in bacterial cells suspended in oligotrophic media, which is sometimes associated with subsequent recovery and virulence (Dwidjosiswojo et al., 2011; Li L. et al., 2014; Dopp et al., 2017; Maertens et al., 2021). In addition, bacteria have evolved a multitude of mechanisms to maintain Cu homeostasis. While import mechanisms have been relatively rarely studied (Lutkenhaus, 1977; Balasubramanian et al., 2011; Ekici et al., 2012), many Cu export systems are known (Arguello et al., 2013). These exporters are supplemented by cytoplasmic and periplasmic Cu chelators and chaperones, and the integration of such systems into complex regulatory networks can lead to high levels of Cu resistance (Monsieurs et al., 2011; Monsieurs et al., 2015).
The antibacterial efficiency of Cu surfaces has been proven to be directly linked to the quantitative release of Cu ions, independent of the composition of the surface (purely metallic or embedded in matrix material) or Cu particle size in the case of nanoparticles (Hans et al., 2013; Hans et al., 2014; Sun et al., 2018; Bastos et al., 2020). This mechanism is also responsible for the decreasing antibacterial properties with depleting Cu content, and is why additional surface functionalization focusing on increased antibacterial efficiency is key to further improvement of Cu-based antimicrobial surfaces. Surface functionalization involving surface chemistry (Tripathy et al., 2018; Maikranz et al., 2020) and topography (Helbig et al., 2016) can alter bacterial adhesion by repelling or additional killing mechanisms, similar to the mechanical destruction of the cell wall on Cicada Wings (Tripathy et al., 2017). However, inherent antimicrobial capacities might as well be decreased alongside surface functionalization due to degraded Cu ion release (Boinovich et al., 2019) emphasizing the need for a considered approach to surface modification. Ion release itself was shown to be mainly driven by corrosive interaction with the contacting medium, where Cu scavenging of bacteria alongside nutrient ingestion additionally increases ion release rates by preventing the formation of a passivating Cu-oxide layer (Luo et al., 2019). In this context, bacterial mortality on Cu surfaces has recently been shown to be at least as dependent on metabolic activity as on sufficiently high Cu ion release rates. Here, topographical and chemical surface functionalization by Ultrashort Pulsed Direct Laser Interference Patterning (USP-DLIP) at the scale of single bacterial cells (Pattern periodicity P = Bac) targeting increased bacterial adhesion induced an amplification of Cu sensitivity of E. coli (Müller et al., 2021).
Copper containing antimicrobial surfaces have been successfully applied before in clinical environments. The replacement of frequent-touch items from standard materials to copper alloys reduced the overall microbial burden significantly. It also led to a decline in contamination with antibiotic resistant microorganisms such as vancomycin resistant enterococci and methicillin resistant S. aureus (Karpanen et al., 2012). Regarding the astronauts’ health and the integrity of the spacecraft, the application of copper containing antimicrobial surfaces and coatings can be beneficial also for spaceflight purposes (Hahn et al., 2017; Zea et al., 2020). However, antimicrobial copper surfaces especially those with surface functionalization have not been tested for their potential against biofilm formation under spaceflight conditions yet and it is not known to what extent the antimicrobial activity might change in microgravity.
The BIOFILMS (Biofilm Inhibition on Flight equipment and on board the ISS using microbiologically Lethal Metal Surfaces) experiment proposal was submitted and accepted by ESA within an open call for experiments in 2014 (ILSRA-2014-054). The goal of the experiment is to compare how biofilms are formed in microgravity in a liquid environment on inhibiting and non-inhibiting metal surfaces to investigate the antibacterial efficacy of several copper-based antimicrobial surface pattern designs under different space-relevant gravity conditions. Within a single experiment different microbial species will be tested and the different metal surfaces will be cross-compared from different gravity regimes. To accomplish this, three specifically selected bacterial species will be exposed to topographically functionalized metallic surfaces with and without inherent antimicrobial material properties under microgravity (µg) (ISS and space travel conditions), 0.4 x g (approximated gravity level on Mars) and 1 x g (terrestrial gravity control). Post-flight, the metal surfaces alongside the adhering biofilms will be subjected to in-depth microbiological and molecular biological analyses as well as high-resolution spectroscopic and metallographic characterization of the metal surfaces to determine altered surface properties.
The BIOFILMS project will evaluate the applicability of topographically functionalized antimicrobial contact surfaces for contamination control in extra-terrestrial environments, including a focussed investigation on the role of the actual bacteria/substrate contact. A recent study following a similar approach of functionalized Cu contact surfaces has shown enhanced antibacterial efficiency in close relation to altered bacteria/surface interaction on Earth in 1 x g (Müller et al., 2021). Expectedly, the data generated will be indispensable for the future selection of antimicrobial materials in support of astronaut-/robotic-associated activities in space exploration. An overview of the whole timeline of the BIOFILMS experiment can be found in the Supplementary Material (Supplementary Figure S1).
The bacterial cells of the three species included in BIOFILMS need to be stored in the flight hardware as pre-test inoculation cultures until the experiment is activated aboard the ISS. For this, we considered liquid buffers that fulfilled specific requirements. Namely, the storage buffer needs to limit bacterial activity during the storage phase, but keep the cells in an intact and viable condition allowing growth when the experiment is activated. Furthermore, in the selection process for the liquid growth medium that will be present during the incubation and biofilm formation phase of the experiment various factors were considered. Since different bacterial species need different amounts of nutrient for growth and biofilm formation, the growth medium will need to support growth and biofilm formation of all three selected strains. Furthermore, a growth medium that only includes low concentrations of available nutrients was sought to more closely resemble realistic environmental conditions. Another important point in the selection process was the comparability to other spaceflight experiments, that can be limited in case different types of growth media are used. After the cultivation phase the experiment will be fixed to prevent further bacterial activity during onboard storage, download from the station and transport and storage post flight. The selection criteria for a chemical fixative were the compatibility with the chosen bacterial strains, the metal sample plates and the experimental hardware. Additionally, the chemical hazard of the fixative had to be minimized.
The bacterial species and strains for BIOFILMS were selected for 1) their ability to form biofilms, 2) their relevance for spaceflight and 3) their potential risk to the crew. Spaceflight relevance was defined here as either relevant for the astronaut’s health or the integrity of spaceflight equipment. Moreover, species and strains were considered that had been isolated from spacecrafts or the International Space Station itself. We only considered bacteria that are classified in the lowest risk group to eliminate possible health risks in the unlikely event of exposure. The selected strains were tested for their ability to grow in the chosen growth medium, their biofilm formation on inert surfaces and their compatibility with the experimental setup and chosen fixative. Furthermore, an appropriate protocol for preparing the pre-test inoculation cultures for flight was developed.
The concept of antimicrobial efficacy involves both topographical influence on bacterial adhesion (Helbig et al., 2016) as well as inherent bactericidal capacities of the materials. Since, the goal of the BIOFILMS experiment is to evaluate topographically functionalized antimicrobial surfaces under space conditions, the selection of the materials itself and the method for generating the different topographies was an integral part of the experiment. As inert non-antibacterial reference surface stainless steel (AISI 304) was selected for evaluating bacterial biofilm formation. Furthermore, copper (OF-Cu) and brass as a copper alloy (CuZn37) were selected as appropriate metal surfaces with antimicrobial properties. In this selection process for the antimicrobial surfaces it was important that the metal itself had been well studied for its antimicrobial properties and that it was suitable for topographic functionalization. In the experiment, each material will be tested with three topography modifications: smooth, patterned at or very near the bacterial scale (P ≈ Bac), patterned smaller than the bacterial scale (P < Bac). With this we will include the effect of topography on bacteria-surface interactions. The proposed effective interplay of mechanisms is listed in Table 1 for each of the surface modifications.
Because the setup for the BIOFILMS experiment will be based on a single species biofilm formation microbial contamination on the metal sample plates will be prevented in advance of the experiment. Copper containing surfaces may form additional oxides during passivation in contact with water, therefore a specific sterilization treatment with minimal contact with water was developed, because the standard sterilization methods such as autoclaving and treatment with 70% ethanol are not suitable. We tested different combinations of ultrasound, ethanol and UV-C treatment. To test the sterilization efficacy of each, smooth steel surfaces were intentionally contaminated with bacteria (Staphylococcus capitis subsp. capitis strain DSM 111179) by placing the surfaces into petri dishes with 10 ml of liquid growth medium (R2A) inoculated with 107 CFU/ml and incubated overnight at 37°C. Next, the plates were treated either with different concentrations of ethanol in tubes in the ultrasonic bath at 50% for 30 min (Sonorex Digital 10P, Bandelin, Berlin, Germany) followed by a UV-C treatment (total dose 840 J/m2), autoclaved (positive control) or not treated (negative control). To verify effective sterilization the sample plates were placed in Erlenmeyer flasks containing 20 ml of liquid growth medium (R2A) and incubated overnight at 37°C in a shaking incubator at 100 rpm. The sterilization procedure was scored effective in the absence of visual growth.
For the hardware design of the BIOFILMS experiment several scientific requirements were stated. The basic principle for the hardware design had to allow bacterial biofilm formation on different metal sample plates in a controlled oxygenated environment that will permit distinct activation and termination of the experiment with little crew time involved. The specific scientific requirements are listed below:
(1) The experiment hardware needs to be equipped with culture chambers, each one hosting five metal sample plates.
(2) Each chamber should offer sufficient room to accommodate all five metal sample plates without touching or damaging their surfaces during their placement into or removal from the chamber.
(3) The bacteria need to be uploaded as “ready-to-start” pre-test cultures. The volume of a pre-test culture should be 0.25 ml for each culture chamber. The experiment hardware needs to be capable of activating the pre-test cultures by the addition of liquid growth medium.
(4) The experiment hardware must be capable of transferring the activated culture into the culture chamber containing the metal sample plates. The transfer should occur directly after activation.
(5) The experiment hardware must be capable of filling the complete culture chamber, covering all five metal sample plates with growth medium and activated bacterial cultures. The chamber must remain free of liquid until the cultures are introduced to prevent premature chemical alterations of the metal sample plates.
(6) The growth medium must not be stirred after the chamber is filled.
(7) The introduction of air bubbles must be prevented, since these may obstruct the biofilm formation in an unpredictable and non-verifiable way.
(8) The experiment hardware must be capable of providing the bacterial cultures with oxygen throughout incubation.
(9) The experiment hardware must be capable of replacing the liquid growth medium with a fixing solution.
(10) The culture chambers must be sterilizable (preferably via autoclaving).
Moreover, a safety requirement is to maintain a level of containment that will prevent any leakage from the experiment hardware container once it is aboard the ISS. Furthermore, aboard the ISS, the experiment must be integrated into Kubik; a temperature-controlled cube shaped facility with a centrifuge in its center to simulate gravity. Therefore, the outer measures and electronic connections need to be compatible between the experiment hardware and Kubik.
The experiment hardware was designed and produced by Kayser Italia, SRL, Livorno LI, Italy. During the design process, two prototypes, the reduced model and the scientific model, were provided to the science team for initial tests before the actual flight models were produced. Pre-flight hardware tests included a biocompatibility test and a science verification test (SVT). Additionally, an experiment sequence test was performed to validate the overall set-up including the respective timeframes of the experiment.
To confirm that all materials used in the BIOFILMS hardware are biocompatible, thereby ensuring inhibition of bacterial growth would not occur, a biocompatibility test was performed. All materials (Table 2) were autoclaved in advance, put into test tubes with plastic lids, and covered with 5 ml of liquid growth medium (double concentrated R2A). An additional tube was inoculated without including any material (positive control) and a set of blank samples (materials without bacteria) was included. The optical density at 600 nm (OD600nm) of an overnight culture in double concentrated R2A of the bacterial strains was measured with an absorbance reader (ELx808, BioTek). An appropriate amount of overnight culture was used to inoculate the medium in the test tubes so that final OD600nm was at 0.1. For the positive control bacteria were cultivated in medium only. After 5 h and 15 days of incubation at room temperature (20–22°C), tubes were vortexed and OD600nm was determined.
For the science verification test the science success criteria were defined to determine if the hardware design allows for the successful implementation of the experiment and at the same time ensures that the scientific questions are answered. For BIOFILMS the following science success criteria were defined:
(1) Successful integration of samples (5 metal plates per hardware unit) and fluids into the experiment units (0.25 ml of bacteria suspended in buffer in the bacterial reservoir, 2.1 ml growth medium in the medium reservoir and 2.1 ml chemical fixative in the fixative reservoir)
(2) Successful filling of the culture chambers with growth medium (activation of experiment)
(3) Successful termination of experiments with injection of the fixative into the respective culture chambers (termination of experiment)
(4) Successful growth and even distribution of biofilm in the chambers including steel sample plates
(5) Detection of traces of bacteria in the chambers containing the antimicrobial sample plates
(6) Successful completion of the experiment without contamination by other bacteria
The timeline for the test itself consisted of 2 days for assembly and filling the experiment hardware, 7 days of storage at 4°C followed by culturing for 14 days at 20°C. After this, the experiment hardware was shipped at 20°C to the science team for hardware disassembly o and sample retrieval from the experiment units. Samples were analyzed via microscopy.
The BIOFILMS experiment will investigate biofilm formation of three different bacterial species on three types of metal sample plates with three different surface topographies in different gravitational conditions. As a control, blank samples will be included as well and the ground experiments will be performed in the same setup. In the experiment gravity parameters important for spaceflight were selected to study the corresponding biofilm morphology and antimicrobial activity. These gravity parameters are microgravity (µg), Mars gravity (0.38 x g) and Earth gravity (1 x g) as control. For the simulation of the gravity parameters on the ISS, two Kubik facilities aboard the ISS are going to be used. To accommodate all sample types and combinations throughout three flights with limited space available in the two Kubik facilities, a specific flight plan was developed that allows evaluation of the sample types such that a successful comparison among the sample types and references can be obtained throughout the different gravitational parameters.
The experiment sequence must be identical for all three BIOFILMS flights. Certain factors such as temperature and duration can vary within the experiment sequence itself due to their difficulty to control. Therefore, certain temperature ranges and time periods were established that define the minimum and maximum temperature fluctuations and duration of incubation and storage time. The experimental design for the ground-based experiments was developed that provides for a detailed comparison between space and ground conditions. For testing purposes, the whole experiment sequence was simulated during the experiment sequence test over a 2-month time period.
For the onboard storage of the bacteria inside the BIOFILMS hardware phosphate buffered saline (PBS) was selected to ensure there is an osmotic balance between the bacterial cells and the ambient medium. Also, storage in PBS keeps the metabolic activity of the bacteria minimal compared to storage in a nutrient rich medium. The make-up of the PBS to be used for BIOFILMS contains 7.0 g/L Na2HPO4, 3.0 g/L KH2PO4, 4.0 g/L NaCl in ddH2O with pH 7.5 and is sterilized by autoclaving. The selected liquid growth medium that provides nutrients for the bacteria during the incubation phase of experiment is double concentrated R2A medium (Teknova R0005: 0.5 g/L yeast extract, 0.5 g/L peptone, 0.5 g/L casamino acids, 0.5 g/L glucose, 0.5 g/L soluble starch, 0.3 g/L sodium pyruvate, 0.3 g/L KH2HPO4, 0.05 g/L MgSO4∙7H2O in ddH20; pH 7.2). This medium was selected because: (I) It supports growth of the selected bacterial species, (II) it is a growth medium that contains only small amounts of nutrients resembling a more realistic biofilm formation scenario, and (III) it was already used in previous spaceflight experiments such as BioRock (Loudon et al., 2018; Cockell et al., 2020) and therefore allows comparability with the published results.
The chosen fixative for BIOFILMS is a mixture of 1% paraformaldehyde (PFA; Merck 104005) and 2.5% glutaraldehyde (GA; Roth 3778.1) in 0.05 M Hepes buffer (pH 7.2–7.4). This fixative is a standard for ultrastructural research, especially for clinical specimens. It is a variant of the Karnovsky´s fixative (4% PFA, 5% GA) (Karnovsky, 1965) which uses higher concentrations and is suitable for a wide range of biological samples (Bozzola and Russell, 1999). The reason for using 1% of PFA instead of 4%, as used in Karnovsky´s fixative, is that at this concentration a substantial fraction of the PFA is persistently present as formaldehyde in the mixture providing a stable fixative over time.
Three bacterial species with different spaceflight implications were selected for the BIOFILMS experiment: Acinetobacter radioresistens, Staphylococcus capitis and Cupriavidus metallidurans. In the following sections, the rationale for specific bacterial strain selection is presented.
Members of the genus Acinetobacter are non-motile, Gram-negative and strictly aerobic bacteria that can form biofilms (Martí et al., 2011; Towner et al., 2013). The cells are rod shaped, but become more coccoid as they approach and enter stationary phase (Henriksen, 1973). Most clinically relevant is Acinetobacter baumannii, a global pathogen that causes serious infections particularly in intensive care units and is known for its antibiotic resistance (Peleg et al., 2008). For the BIOFILMS experiment A. radioresistens strain 50v1 isolated from the surface of the Mars Odyssey spacecraft prior to flight (La Duc et al., 2003) (Figure 1A) was selected. Studies show that strain 50v1 is remarkably resistant to extreme conditions including UV-radiation, hydrogen peroxide and desiccation (La Duc et al., 2003; McCoy et al., 2012). Furthermore, 50v1 is not only highly resistant to cleaning reagents such as Kleenol 30, but can biodegrade them (Mogul et al., 2018).

FIGURE 1. Selected bacterial species for BIOFILMS. Scanning Electron Microscopy images of Acinetobacter radioresistens strain 50v1 (A), Staphylococcus capitis subsp. capitis strain DSM 111179 (B) and Cupriavidus metallidurans strain CH34 (C) after 18 h incubation in R2A liquid growth medium at 37°C (S. capitis subsp. capitis) or 30°C (A. radioresistens and C. metallidurans) at constant agitation of 150 rpm. Cells were fixed with 1% PFA, 2.5% GA in 0.05 M Hepes buffer for 2 h at room temperature and stored at 4°C afterwards. Scale bars represent 500 nm.
S. capitis is a Gram-positive skin-associated bacterium isolated predominantly from the human head (Kloos and Schleifer, 1975; Byrd et al., 2018). S. capitis has the ability to form biofilms and is a causative agent of infections such as endocarditis, urinary tract infections and catheter induced bacteremia (Cui et al., 2013). In contrast to most other staphylococcal species, S. capitis is classified as a biosafety level 1 according to the U.S. Public Health Service Guidelines. In BIOFILMS we use the strain S. capitis subsp. capitis DSM 111179 (Figure 1B) isolated from an exposure experiment aboard the ISS, where different materials, including antimicrobial materials containing silver and ruthenium, were exposed to the ISS indoor environment (Sobisch et al., 2019). The materials were in the pathway of crew members and therefore were exposed to microorganisms originating from the astronauts’ skin, hair and clothing. DSM 111179 was isolated after 19 months from V2A steel (Sobisch et al., 2019). The strain was selected for BIOFILMS because of its strong biofilm formation, its phylogenetic proximity to other clinically relevant staphylococcal species and its origin from the space station indoor environment.
C. metallidurans is a non-pathogenic β-proteobacterial species. The cells are Gram-negative peritrichously flagellated rods that are resistant to high concentrations of metal ions (Vandamme and Coenye, 2004). The species has often been isolated from harsh, oligotrophic, artificial environments, including the ISS (Mijnendonckx et al., 2013). For BIOFILMS, C. metallidurans type strain CH34 will be used (Figure 1C), which was first isolated from a zinc decantation basin in a non-ferrous metallurgical plant near Engis, Belgium (Mergeay et al., 1978). Strain CH34 has since become a model organism for bacterial metal resistance, owing to its tolerance to a surprisingly wide range of metal ions. This tolerance is mediated by a large number of gene clusters associated with metal resistance as well as its adaptation potential during metal exposure (Janssen et al., 2010; Monsieurs et al., 2015; Ali et al., 2019; Mijnendonckx et al., 2019; Van Houdt et al., 2021). These properties have led to the selection of CH34 for several other space experiments (Leys et al., 2009; Loudon et al., 2018). For BIOFILMS, this strain was selected for its Cu tolerance, its ability to survive in harsh environments and its previous space experiments.
During the selection process, the strains were tested for several parameters that are important for the space experimental setup. These parameters include, but are not limited to: growth in R2A liquid medium, biofilm formation on reference surfaces (smooth stainless steel), inactivation by the selected fixative. All the parameters were fulfilled by the selected strains (Table 3).
The bacterial culture preparation for flight will occur prior to the assembly of the hardware to assure that any contamination can be detected before the cultures are loaded into the hardware. For backup, three individual cultures will be prepared and cultivated on double concentrated R2A agar plates for 48 h. DSM 111179 is cultivated at 37°C and the other two strains are cultivated at 30°C. After cultivation (colonies formed) the plates are stored at room temperature. For preparation of the flight bacterial samples, 50 ml of a double concentrated R2A liquid culture will be inoculated in 200 ml Erlenmeyer flasks containing the growth medium, and incubated as described above for 16–18 h in two shaking incubators at 200 rpm. Cultures will be transferred to 50 ml tubes and centrifuged for 10 min at 4000 x g. Cultures are washed three times with 30 ml phosphate buffered saline and the final cell pellet is resuspended in 10 ml phosphate buffered saline. One mL of each culture is aliquoted into individual 1.5 ml tubes and stored at 4°C until they are inoculated into the experimental hardware. The final concentration is approximately 109 colony forming units per ml (CFU/ml) for all three bacterial species. This high concentration is used to assure sufficient cell survival due to the possible 1-month time lag for storage and uploading before the initiation of the experiment. When the experiment is initiated the bacterial sample is diluted by the addition of growth medium (dilution factor: 8.4).
The three different metals addressed in Materials and Methods will be evaluated for their ability to prevent or affect microbial growth and thus biofilm formation in the BIOFILMS hardware. To fit into the experimental hardware designed for the flight experiments, 1 mm sheets of of-Cu (Cu >99.95%), CuZn37 (Cu 63%, Zn 37%) (Wieland, Ulm, Germany) and AISI 304 Stainless Steel (Brio, Lüdenscheid, Germany) are cut into plates of 10 × 25 mm2 dimension. The Cu-based metals are mirror-polished on an automated TegraPol-21 system (Struers, Willich, Germany). The steel sample plates, however, are provided with polished surfaces. The polished plates of each material are used as reference samples of smooth surface topography and as starting material for topographic surface functionalization.
Topographic surface functionalization is done via USP-DLIP, whereby both micro- and nanometer pattern scales are achieved in high pattern quality (Müller et al., 2020a) under low chemical surface modification (Müller et al., 2020b). This technique enables both matching the required size relation to single bacterial cells while retaining the inherent antibacterial capacity of the Cu-based substrates (Müller et al., 2021). The USP-DLIP setup consists of a Ti:Sapphire Spitfire laser system (Spectra Physics, Irvine, US) that emits ultrashort pulses of tp = 100 fs pulse duration (Full Width at Half Maximum, FWHM) at a centered wavelength λ of 800 nm at 1 kHz pulse frequency. The optical setup induces two beam interference by dividing the seed beam in a diffractive optical element (DOE) and overlapping the individual beams on the sample surface using a lens system. By varying the incident angle
To achieve topographies in the size range of single bacterial cells (P ≈ Bac) the periodicity is set to 3 µm. Surface patterning is conducted in scanning mode at a fluence of 3 J/cm2 and 90% pulse overlap, that induces homogeneous surface properties on the three different metallic materials as described in Müller et al. (2020a). For pattern scales below a single bacterial cell size (P < Bac) the periodicity is set to 800 nm, while patterning is conducted modifying the lasing parameters according to Müller et al. (2020a) to achieve comparable pattern morphology on each material. The laser processed samples are subsequently selected to undergo immersion etching in 3% citric acid in an ultrasonic bath to remove process-induced surface oxide and sub-structures (Müller et al., 2020b). SEM images of the USP-DLIP patterned surface topographies after post-processing are displayed in Figure 2.
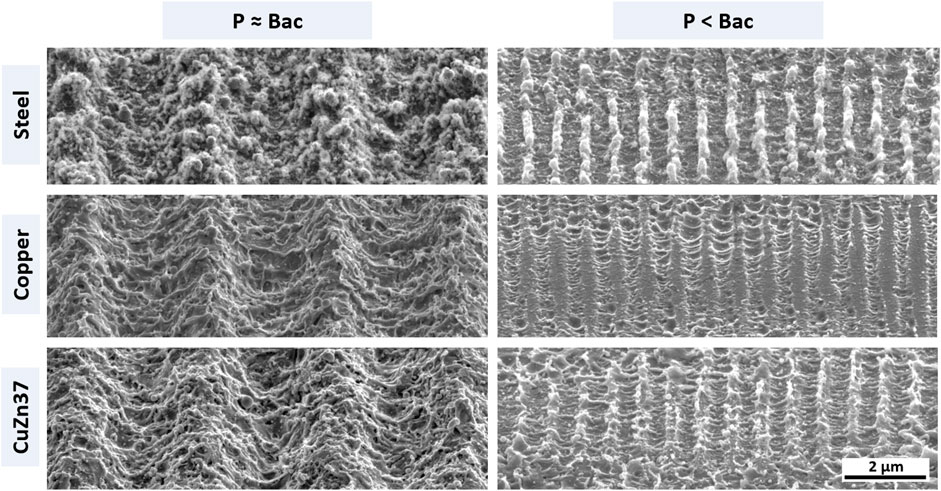
FIGURE 2. SEM images of topographically functionalized surfaces of the three different metallic materials used in the experiments. The left column lists patterns targeting retaining bacterial adhesion due to increased contact area. The right column presents sub-µm scaled patterns aspiring a repelling effect on bacterial cells by decreased contact area. The single scale bar applies for all sub-images.
Figure 3 shows the efficacy of the different tested sterilization methods. Visual bacterial growth was only observed in the untreated control (Figure 3A). All other sterilization methods did not show visual growth, thus effectively inactivating and removing all contaminants (Figures 3B–D). The bactericidal activity of ethanol is normally achieved at a concentration of 70%. However, ≥99.9% ethanol in combination with ultrasound and UV-C treatment was shown to be equally effective. The latter approach prevents the metal sample plates from coming in contact with water prior to the start of the experiment, which could induce chemical alterations of the surfaces. Therefore, this combination was selected for the BIOFILMS experiment with the exact protocol being:
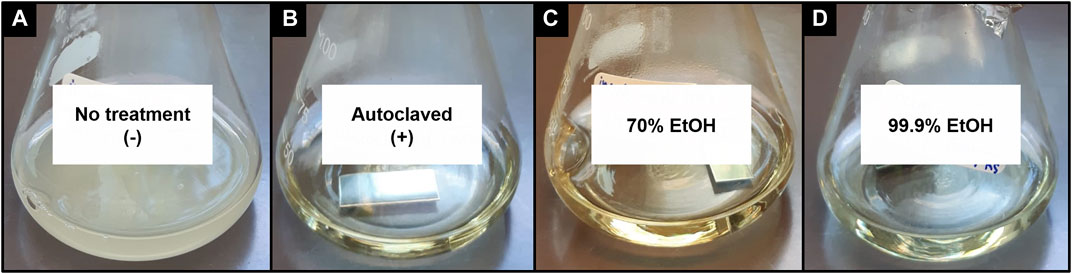
FIGURE 3. Smooth steel sample plates after incubation in growth medium overnight at 37°C in a shaking incubator at 100 rpm after deliberate contamination and different sterilization methods: (A) no treatment as negative control, (B) autoclaved as positive control, (C) incubated in the ultrasonic bath with 70% ethanol + UV-C treatment, (D) incubated with 99.9% ethanol + UV-C treatment.
Metal sample plates are spray rinsed with ethanol (≥99.9%). The plates are then placed into 5 ml tubes and submerged in 4 ml ethanol (≥99.9%). The tubes are placed in an ultrasonic bath and sonicated for 30 min. The ultrasonic treatment removes all unwanted particles on the surface such as dust and other particles. After that, the metal sample plates are placed into sterile petri dishes with sterile tweezers and air dried in a tilted state by leaning them against the wall of the petri dish. Once the samples are dried completely, the samples are UV-C irradiated with a dose of 420 J/m2 on each side. Sample plates are stored in sterile petri dishes until they are integrated into the BIOFILMS hardware.
The BIOFILMS hardware was manufactured by Kayser Italia. Before the actual flight model was produced, two prototypes were built. The first prototype, the Reduced Model (RM), only fits one metal sample plate and served as a first testbed. Here, the bacteria in the growth medium were injected directly into the hardware, where they were incubated in direct contact with the metal sample plate. Figure 4 shows the principle of experiment activation and gas exchange during the experiment. For oxygen exchange, a gas permeable membrane is placed on top of the sample plate to allow gas exchange with the air pocket that is located above (Figure 5G). During assembly, the membrane first is in direct contact with the metal sample plate, without any space in between. When the medium with the bacteria is injected in between the metal plate and the sample plate, the gas permeable membrane bulges upwards.
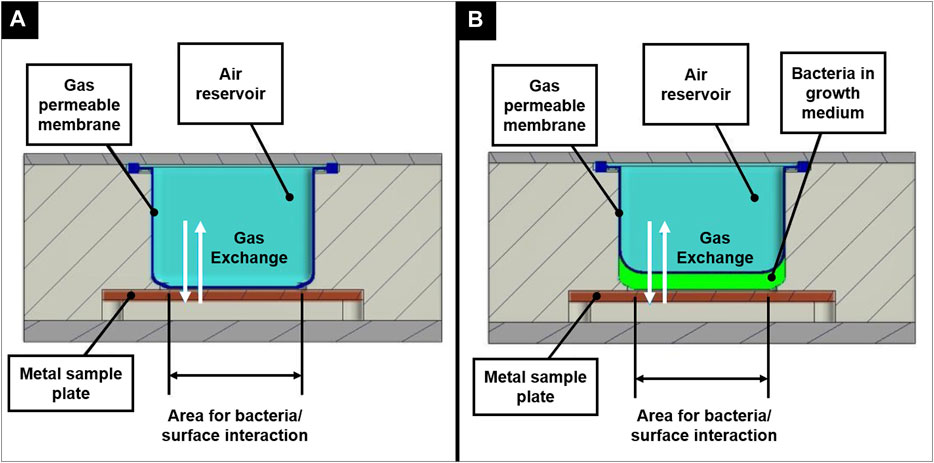
FIGURE 4. Principle of experiment activation shown for the Reduced Model in an inactivated state (A) and after activation (B), illustrating the concept of oxygen exchange during the incubation period.
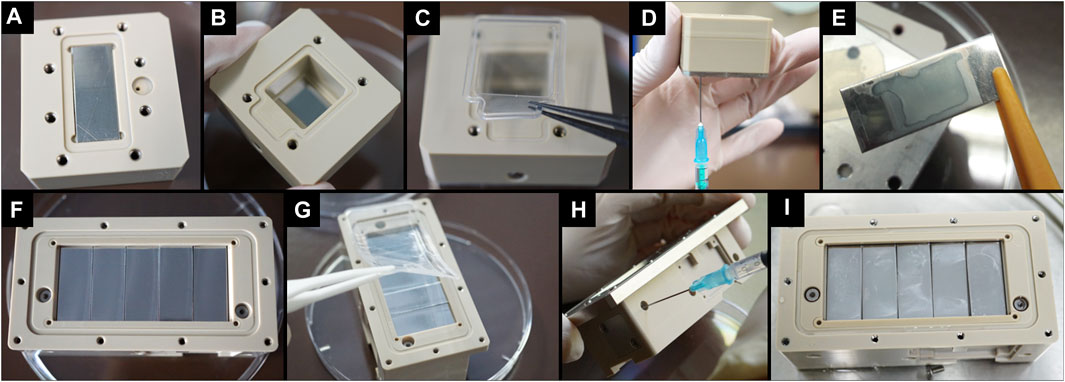
FIGURE 5. Overview of the assembly process and filling process of the BIOFILMS experiment unit prototypes. (A–D) Assembly and filling of Reduced Model (RM) as prototype for first tests on basic experimental concept. (E) Image of a smooth stainless steel sample plate that was incubated inside the RM with S. capitis subsp. capitis DSM111179 for 24 h at 37°C. (F–H) Assembly and filling of Scientific Model (SM) as prototype for further testing of the actual setup of the flight models. (I) Top view of the SM after incubation of smooth stainless steel plates with S. capitis subsp. capitis DSM111179 for 10 days at RT.
After the successful first tests with the RM, the Scientific Model (SM) was developed, in the same way as the experiment units of the flight models would later be designed, and was used for testing whether this design fulfils the scientific requirements of the experiment. The assembly and filling process of the RM and SM are depicted in Figure 5. In addition, the figure shows how the metal plates (in this case smooth stainless steel) appear after incubation in the experiment units with bacteria in growth medium (Figures 5E,I). The tests with the SM showed that the hardware design is suitable for the BIOFILMS experiment since all requirements were met (Scientific requirements of the experimental hardware) and therefore the same design was incorporated into the experiment units of the flight models. Since the flight hardware requires a double level of containment to prevent leakage of contents in any case. The experiment unit is integrated into an outer container, the experiment container, that is inserted into Kubik during the experiment sequence. Inside each experiment container is one experiment unit that contains the sample plates, bacteria, growth medium, fixative and electronic equipment (Figure 5). The whole BIOFILMS experimental hardware system is the assembled experiment unit inside the experiment hardware. For BIOFILMS, the experiment units are specifically designed to allow biofilm formation in a controlled environment on the antimicrobial and inert sample plates. Five sample plates can be accommodated on the bottom of the culture chamber inside the experiment unit. The gas permeable membrane is placed on top of the sample plates to allow gas exchange with the air pocket located above (Figure 5G). The experiment unit has three reservoirs that are separated from each other and from the culture chamber by flat head valves. One reservoir has a capacity of 0.25 ml and serves as a means of storage for the bacterial inoculation suspensions. The other two reservoirs have a capacity of 2.1 ml and are filled with sterile syringes and needles during assembly with the growth medium and fixative, respectively (Figure 5H). The BIOFILMS experiment is activated by a release system with springs and pistons that are controlled by internal electronics. The initiation of the experiment is done by a ground controller providing Kubik power to the experiment containers. This enables the experiment to be conducted inside Kubik without extensive crew interaction. When the experiment is initiated, a spring is released that pushes a piston through the medium reservoir. By doing so, the medium is flushed through the bacterial reservoir into the culture chamber, delivering the bacterial cells. After activation, the experiment is incubated for 14 days at 20°C. During the incubation period, the bacteria are incubated in the culture chamber in spatial proximity to the sample plates. The contact area for bacteria/surface interaction is 2.1 cm2 (10 mm × 21 mm) per sample plate, totaling to 10.5 cm2. The experiment is terminated by activating the release system of the fixative reservoir forcing the fixative directly into the culture chamber. The fixative mixes with the liquid inside the chamber and chemically fixes the experiment to prevent further microbial activity.
The biocompatibility test with all materials that were incorporated in the BIOFILMS hardware revealed that none of the materials were inhibitory to microbial growth (Figure 6). For A. radioresistens and C. metallidurans, a strong increase in OD600nm was observed between 5 h and 15 days of incubation in all test tubes, including the test tube without any material. The OD600nm for S. capitis subsp. capitis remained the same (3, 4B, 4C, 4D, 5, 6, Blank) or increased only slightly between the two measuring points (1, 2A, 2B, 2C, 2D, 4A, 7). However, this seemed to be due to insufficient mixing of the vial before taking an aliquot for determining the OD600nm rather than actual inhibition of growth induced by the material. Microbial growth was clearly visible as turbidity around the materials on the bottom in the test tubes as shown in Figure 6 with a white arrow indicating strong cell agglomeration.
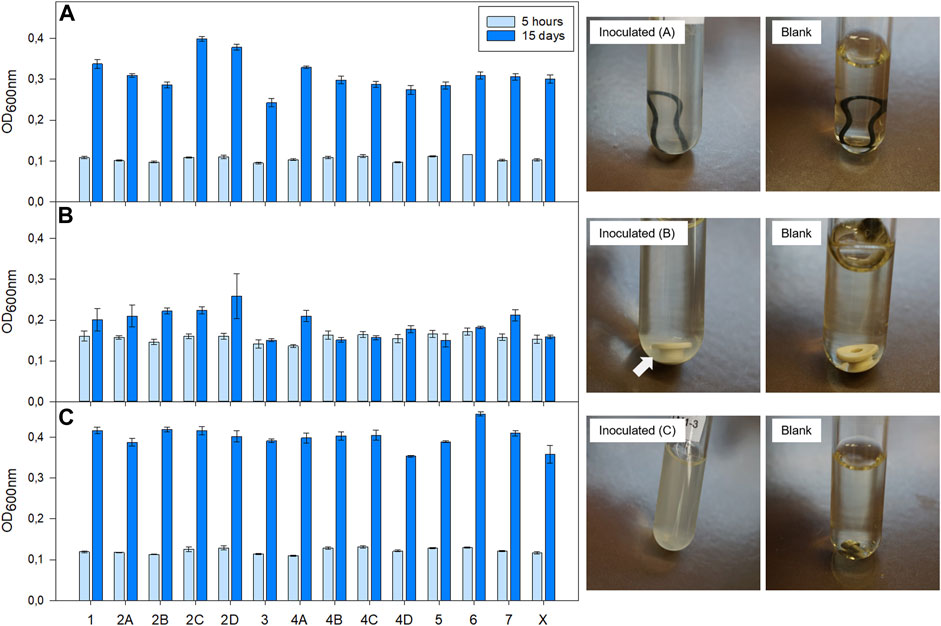
FIGURE 6. Results of biocompatibility test with bacteria selected for BIOFILMS: (A) A. radioresistens 50v1, (B) S. capitis subsp. capitis DSM 111179, (C) C. metallidurans CH34. Bacteria were incubated at room temperature (20–22°C) in a test tube with double concentrated R2A growth medium in presence of different hardware related materials: (1) AISI 316 stainless steel, (2A) Silicone a, (2B) Silicone b, (2C) Silicone a with Vaseline, (2D) Silicone b with Vaseline, (3) Silicone for LSR injection molding, (4A) Ethylene propylene diene monomer a, (4B) Ethylene propylene diene monomer b, (4C) Ethylene propylene diene monomer a with silicone grease (4D) Ethylene propylene diene monomer b with silicone grease, (5) Silicone mL, (6) Polyetheretherketone, (7) AISI 304 stainless steel (X) positive control without material. Optical density was measured at in a 96-well-plate a wavelength of 600 nm (OD600 nm) after 5 h and 15 days post inoculation with an absorbance reader. Pictures on the right are exemplary for bacterial growth in the presence of material 4B (A), material 6 (B) and material 7 (C). The respective blank samples are shown as well.
During the science verification test it was shown that the design of the BIOFILMS hardware is able to meet the defined science success criteria. The hardware was filled according to the protocol and after incubation, an evenly distributed biofilm formation was observed on the reference surfaces. Detailed results of the science verification test can be found in Supplementary Table S1. During the test, one of the experiment units became contaminated. Nevertheless, all other units including the blank units had no problems with contamination, which is why the test was considered as an overall success. However, the protocol for sterilization of the experiment units and the sample plates was revised accordingly.
Within the BIOFILMS experiment, biofilm formation will be investigated under three different gravitational levels aboard the ISS. Gravity levels tested will be microgravity (µg), an approximation of Martian gravity (0.4 x g) and Earth gravity (1 x g) as a control. Additionally, ground experiments on Earth will be performed. The two Kubiks that are currently located in the Columbus module aboard the station will be employed. An astronaut will integrate the experiment hardware into the two Kubiks, which are controlled by a ground controller. Once the Kubik power buses deliver power to the experiment containers, the programmed experiment timeline is executed automatically, meaning that the experiment is activated and terminated automatically once the experiment container has been integrated. This way the required crew time can be kept to a minimum. Kubik can be set to specific temperatures, for BIOFILMS this is 20°C. Furthermore, each Kubik has eight slots for experiment containers, where gravitational forces can be created via centrifugation (Figure 7A). Here, the gravitational vector is directed towards the sample plates within the BIOFILMS experiment. One Kubik will be set to a centrifugal setting resembling 1 x g and the other one to 0.4 x g. Moreover, in each Kubik, four slots in the corners outside of the centrifugal rotor are used to subject the experiment containers to µg.
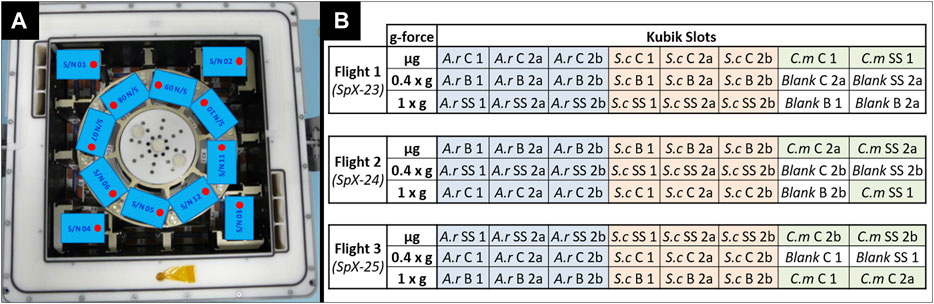
FIGURE 7. (A) Exemplary accommodation of BIOFILMS experiment containers inside one Kubik aboard the ISS. 8 slots are located inside the centrifugal rotor where gravitational forces (1 x g/0.4 x g) are resembled via centrifugation. 4 slots are located outside the rotor where µg is acting on the containers. (B) Flight plan for the three flights of the BIOFILMS experiment with A.r.: A. radioresistens, S.c.: S. capitis subsp. capitis, C.m.: C.metallidurans for the three selected bacterial species and C: Copper, B: Brass and SS: Stainless steel for the types of metals. The numbers account for the different types of metal topography with 1: smooth, 2a: USP-DLIP pattern at P ≈ Bac and 2b: USP-DLIP pattern at P < Bac.
Because there are only two Kubiks available at the station and each has a limited number of slots, a specific flight plan had to be developed to accommodate all sample types that require testing during the BIOFILMS experiment. In total the BIOFILMS experiment requires 72 slots. Biofilm formation of A. radioresistens and S. capitis subsp. capitis will be tested on steel, copper and brass with the three metal topographies (Smooth, P ≈ Bac, P < Bac). Due to limited space for sample accommodation, C. metallidurans will be tested only on steel and copper surfaces with the three types of metal topographies. The innovative aspect of the BIOFILMS experiment is the use of different metal surface topographies for increasing the antimicrobial efficacy. Therefore, the direct comparison of the different topographies had the highest priority. Importantly, each flight set had to include at least one sample set with stainless steel, because it is the reference surface without antimicrobial efficacy and at least one blank sample, to exclude the occurrence of outside contamination. The flight plan is depicted in (Figure 7B). With this flight plan, the direct effect of the metal topographies on the biofilm formation of one bacterial species can be compared on one type of metal in gravitational condition each. The comparison between the different flights and therefore, the comparison between the different types of metals and gravity conditions can still be made by evaluating all influencing factors such as temperature variances, differences in storage time pre and post incubation, incubation time and period as well as possible disturbances during upload and download to the station.
For each flight of the BIOFILMS experiment, the experimental sequence will be identical. However, due to unpredictable events that can occur during launch or integration on board, shifts in time and temperature range can occur and the conditions might differ slightly between flights. Sample preparation and analysis is carried out with sufficient lead time ahead of the actual flight experiment. For each flight, two sets of ground experiments will be performed. One will be performed in parallel to the flight with the same bacterial cultures. The other ground experiment will be performed at a later stage when all data from the flight are retrieved and the exact condition from flight, such as storage duration and temperature, can be recreated on Earth. The experiment sequence for BIOFILMS is listed in following:
I. Pre-flight preparations in science lab
(1) USP-DLIP sample production and pre-flight analysis
(2) Preparation of growth medium and fixative
(3) Preparation of pre-test inoculation cultures
(4) Filling and assembly of the experiment hardware
II. Mission Operations
(1) Transport of experiment hardware to the launch site
(2) Upload in phase change material
(3) On-board cold storage at +4°C
(4) Integration into Kubik
(5) Automated supply of medium
(6) Incubation period: culturing at µg, 0.4 x g, 1 x g for 14 days at 20°C
(7) Automated supply of fixative
(8) On board storage
(9) Download, transport and hand-over to science team
III. Post-flight sample laboratory analysis
(1) Analysis of biofilms, metal surfaces and growth medium
For each step of the experimental sequence specific time frames and temperature ranges are required (Figure 8). All steps have an optimal temperature profile and time frame, but certain deviations from the optimum can be tolerated within the experimental setup. The first steps are the pre-flight preparation of the metal sample plates and the pre-test of bacterial cultures as well as the preparation of the growth medium and fixative. Followed by the assembly and filling of the experiment hardware. The experiment is packed and transported to the launch site for upload to the ISS in its stored state. The time between filling the hardware and activation of the experiment aboard the ISS should be as short as possible and must not exceed 1 month. The temperature for this time period should optimally be 4°C but can range between 1 and 8°C. The incubation period begins when the experiment is integrated into Kubik and the medium is supplied to the culture chamber. The optimal duration for incubation is 14 days but can be shortened to a minimum of 10 ays and lengthened to 20 days maximum. The optimal temperature during incubation is 20°C but can range between 18 and 22°C. The incubation period is terminated by the addition of fixative into the culture chamber. After fixation the experiment hardware will be stored until download at ambient temperature. Here the tolerable temperature is between 15 and 30°C. The preferred timeframe for storage, download, transport and handover to the science team should be as short as possible but can be extended up to 90 days maximum. Within the experiment sequence test it was shown that this anticipated experiment sequence is suitable for the successful implementation of the experiment.
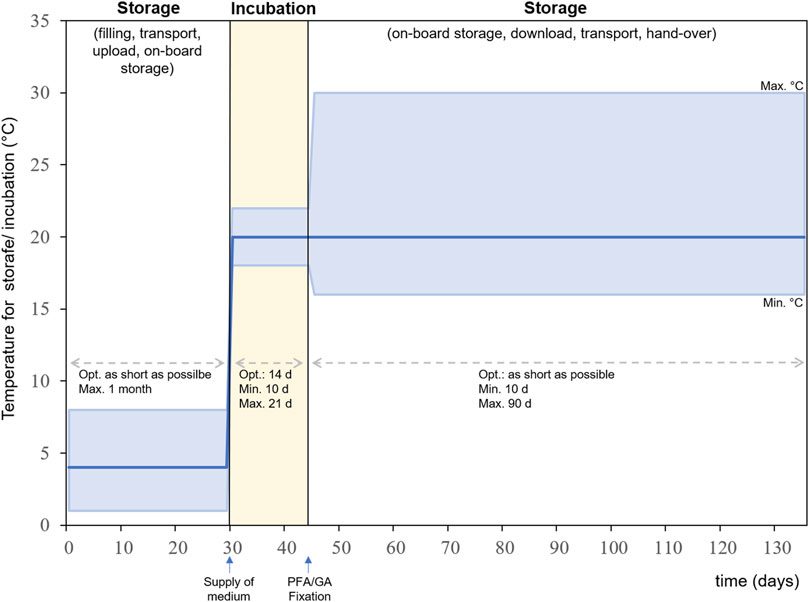
FIGURE 8. Experiment sequence for BIOFILMS showing the timeline for storage and incubation and the according temperature ranges. Dark blue line: optimal temperature, upper light blue line: maximum temperature, lower light blue line: minimum temperature.
Post flight microbiological analysis will include scanning electron microscopy (SEM) and fluorescence microscopy of the bacterial cultures and biofilms. The SEM analysis will allow detailed insights into biofilm morphology, matrix structure and overall integrity of the bacteria. Via Focused Ion Beam cross sectioning (FIB/SEM), biofilm thickness and structure in direct relation to the available surface topography can be assessed by high resolution imaging. For the fluorescence microscopic analysis, different fluorescent dyes will be used such as DAPI for straining nucleic acids and biofilm matrix straining dyes such as FITC-marked wheat germ agglutinin (WGA). A typical image of a Staphylococcus capitis (DSM 111179) biofilm on the smooth stainless-steel reference surface from the BIOFILMS science verification test is shown in Figure 9.
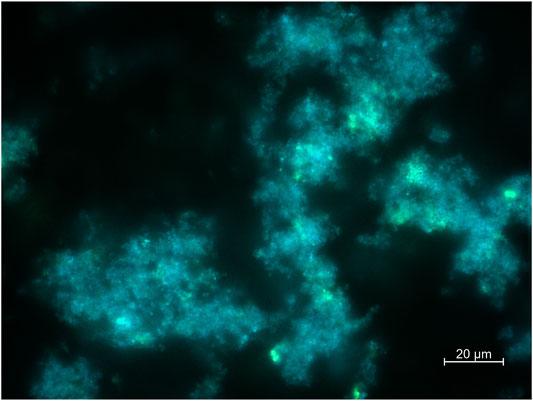
FIGURE 9. Exemplary fluorescence microscopic image of Staphylococcus capitis subsp. capitis biofilm on the smooth stainless steel reference sample plate from the BIOFILMS science verification test. DSM 111179 was incubated for 14 days at 20°C within the BIOFILMS flight hardware with an initial cold storage time of 7 days at 4°C and a post-incubation storage time of 2 days at 20°C in a fixed state. Staining was performed with DAPI (5 µg/ml) and FITC conjugated WGA (10 µg/ml). Imaging was performed by using an Axio Imager M2 with AxioCamMR3. Scale bar represents 20 µm.
By comparing the data from the pre-flight experiments, the topographical and chemical influence of the bacteria (e.g. through corrosive material removal) on the substrate is determined. This allows both the ability to determine the damaging influence of bacterial biofilms on the surface, and local damage providing an indication of the presence and activity of bacteria during the experiment (an example of localized corrosion sites is illustrated in Figure 10).
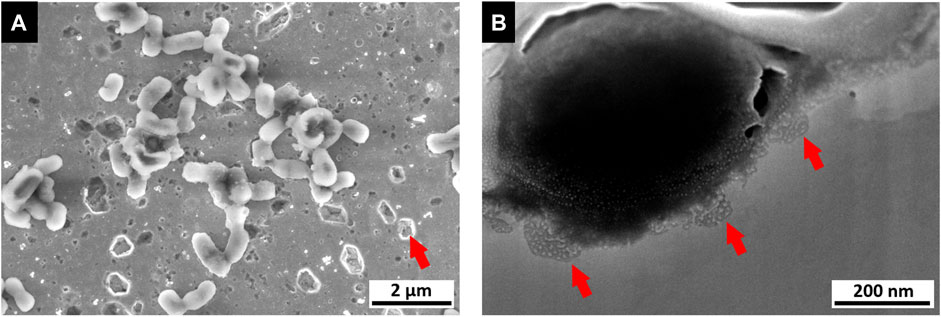
FIGURE 10. Image of a microbial corrosion analysis (data obtained from contact killing experiment). (A) SEM image of a polished copper surface exhibiting sites of pitting corrosion, especially in close vicinity to bacteria (E. coli as tested reference organism). (B) FIB/SEM cross section of single bacteria cell on copper surface exhibiting smaller sites of pitting corrosion below.
This provides data to determine any differences in bacterial behavior due to changes in gravity, as well as the weighting of short-scale interactions such as Van der Waals forces on colonization behavior through close comparison with the proteomic analyses. Localized surface disruption, oxide growth and chemical surface modification are investigated localized via FIB/SEM and energy dispersive X-ray spectroscopy (EDS), while phase analysis of the substrate and oxide films is conducted via grazing incidence X-ray diffraction (GI-XRD). Additionally, inductively coupled plasma mass spectrometry (ICP-MS) is used for the analysis of metal ions diffused into the medium. The method allows the determination of metal ions in a wide dynamic concentration range from parts-per-trillion (ppt) to parts-per-million (ppm).
The BIOFILMS experiment successfully passed all preflight tests including biocompatibility, science verification test (SVT) and experiment sequence test (EST), indicating that the hardware is biocompatible and meets the scientific requirements. The hardware itself, including the two prototypes, allows for the cultivation of bacteria in direct spatial proximity to the different metal sample plates. As such, biofilm formation on the inert reference (stainless steel) and antimicrobial surfaces can be compared, thereby exposing inhibitory effects. Although the extended results of the SVT and EST are out of the scope of this article, both proved that the hardware designed by Kayser Italia fulfilled the scientific requirements of the experiment, allowing biofilm formation on the inert reference surfaces including both smooth and P ≈ Bac patterned surfaces. P < Bac patterned surfaces were not tested within the scope of the pre-flight tests. However, the antimicrobial surfaces, both brass and copper showed an inhibiting effect on bacterial biofilm formation with a stronger effect caused by the P ≈ Bac patterned copper surfaces, which is in good correspondence to preliminary results (Müller et al., 2021). The data that will be generated within the BIOFILMS project will be valuable for the future selection of antimicrobial materials in support of human- and robotic-associated activities in space exploration.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
KS, DM, LM, and AA performed the pre-flight experiments, USP-DLIP sample preparation and characterization, analyzed the data, and wrote the manuscript. RM, RH, RLM, and FM were responsible for the conception and design of the experiment and manuscript preparation. DM, GH, and ML are responsible for the electron microscopic images of the bacterial strains while the latter two also contributed in the manuscript preparation. SB, KZ, and RK carried out the element trace analysis and evaluations of the metal release from the different surfaces. NC, JK, MV, AT, and CR contributed to the realization of the experiment, the hardware development, the pre-flight tests and the manuscript evaluation.
KS and RM were supported by the DLR grant FuE-Projekt “ISS LIFE” (Programm RF-FuW, Teilprogramm 475). DM and AA were supported by German Aerospace Center—Space Administration (DLR) within the project “Investigation of antimicrobial metal surfaces under space conditions—An effective strategy to prevent microbial biofilm formation” (project number 50WB1930). LM and RH were supported by the European Space Agency (ESA-PRODEX) and the Belgian Science Policy (Belspo) through the BIOFILMS project (C4000129318). RLM was supported by NASA Space Biology grant 80NSSC18K0751 (BIOFILMS) to the Bay Area Environmental Research Institute.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank the team of the Aerospace Microbiology Group at the German Aerospace Center (DLR) in Cologne (Köln) namely, Andrea Schröder, Stella Koch, Erika Muratov, Marta Cortesao, Yen Ly and Katharina Runzheimer for their help in the preparation and performance of the pre-flight tests such as SVT and EST. We express our gratitude to Elisabeth Grohmann for cordially providing the Staphylococcus capitis subsp. capitis strain. This research is part of the PhD theses of KS, DM, and LM.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frspt.2021.773244/full#supplementary-material
Ali, M. M., Provoost, A., Maertens, L., Leys, N., Monsieurs, P., Charlier, D., et al. (2019). Genomic and Transcriptomic Changes that Mediate Increased Platinum Resistance in Cupriavidus metallidurans. Genes 10 (1), 63. doi:10.3390/genes10010063
Aponte, V. M., Finch, D. S., and Klaus, D. M. (2006). Considerations for Non-invasive In-Flight Monitoring of Astronaut Immune Status with Potential Use of MEMS and NEMS Devices. Life Sci. 79 (14), 1317–1333. doi:10.1016/j.lfs.2006.04.007
Argüello, J. M., Raimunda, D., and Padilla-Benavides, T. (2013). Mechanisms of Copper Homeostasis in Bacteria. Front. Cel. Infect. Microbiol. 3, 73. doi:10.3389/fcimb.2013.00073
Balasubramanian, R., Kenney, G. E., and Rosenzweig, A. C. (2011). Dual Pathways for Copper Uptake by Methanotrophic Bacteria. J. Biol. Chem. 286 (43), 37313–37319. doi:10.1074/jbc.M111.284984
Bastos, C. A. P., Faria, N., Wills, J., Malmberg, P., Scheers, N., Rees, P., et al. (2020). Copper Nanoparticles Have Negligible Direct Antibacterial Impact. NanoImpact 17, 100192. doi:10.1016/j.impact.2019.100192
Be, N. A., Avila-Herrera, A., Allen, J. E., Singh, N., Checinska Sielaff, A., Jaing, C., et al. (2017). Whole Metagenome Profiles of Particulates Collected from the International Space Station. Microbiome 5 (1), 81. doi:10.1186/s40168-017-0292-4
Boinovich, L. B., Kaminsky, V. V., Domantovsky, A. G., Emelyanenko, K. A., Aleshkin, A. V., Zulkarneev, E. R., et al. (2019). Bactericidal Activity of Superhydrophobic and Superhydrophilic Copper in Bacterial Dispersions. Langmuir 35 (7), 2832–2841. doi:10.1021/acs.langmuir.8b03817
Bozzola, J. J., and Russell, L. D. (1999). Electron Microscopy: Principles and Techniques for Biologists. Burlington: Jones & Bartlett Learning.
Bruce, R. J., Ott, C. M., Skuratov, V. M., and Pierson, D. L. (2005). Microbial Surveillance of Potable Water Sources of the International Space Station. SAE Tech. Paper 114, 283–292. doi:10.4271/2005-01-2886
Byrd, A. L., Belkaid, Y., and Segre, J. A. (2018). The Human Skin Microbiome. Nat. Rev. Microbiol. 16 (3), 143–155. doi:10.1038/nrmicro.2017.157
Checinska, A., Probst, A. J., Vaishampayan, P., White, J. R., Kumar, D., Stepanov, V. G., et al. (2015). Microbiomes of the Dust Particles Collected from the International Space Station and Spacecraft Assembly Facilities. Microbiome 3, 50. doi:10.1186/s40168-015-0116-3
Cockell, C. S., Santomartino, R., Finster, K., Waajen, A. C., Eades, L. J., Moeller, R., et al. (2020). Space Station Biomining experiment Demonstrates Rare Earth Element Extraction in Microgravity and Mars Gravity. Nat. Commun. 11 (1), 5523. doi:10.1038/s41467-020-19276-w
Crabbé, A., Schurr, M. J., Monsieurs, P., Morici, L., Schurr, J., Wilson, J. W., et al. (2011). Transcriptional and Proteomic Responses of Pseudomonas aeruginosa PAO1 to Spaceflight Conditions Involve Hfq Regulation and Reveal a Role for Oxygen. Appl. Environ. Microbiol. 77 (4), 1221–1230. doi:10.1128/AEM.01582-10
Crucian, B., Babiak-Vazquez, A., Johnston, S., Pierson, D., Ott, C. M., and Sams, C. (2016). Incidence of Clinical Symptoms during Long-Duration Orbital Spaceflight. Int. J. Gen. Med. 9, 383–391. doi:10.2147/ijgm.S114188
Cui, B., Smooker, P. M., Rouch, D. A., Daley, A. J., and Deighton, M. A. (2013). Differences between Two Clinical Staphylococcus Capitis Subspecies as Revealed by Biofilm, Antibiotic Resistance, and Pulsed-Field Gel Electrophoresis Profiling. J. Clin. Microbiol. 51 (1), 9–14. doi:10.1128/JCM.05124-11
Davey, H. M. (2011). Life, Death, and In-Between: Meanings and Methods in Microbiology. Appl. Environ. Microbiol. 77 (16), 5571–5576. doi:10.1128/AEM.00744-11
Dopp, E., Richard, J., Dwidjosiswojo, Z., Simon, A., and Wingender, J. (2017). Influence of the Copper-Induced Viable but Non-culturable State on the Toxicity of Pseudomonas aeruginosa towards Human Bronchial Epithelial Cells In Vitro. Int. J. Hyg. Environ. Health 220 (8), 1363–1369. doi:10.1016/j.ijheh.2017.09.007
Dwidjosiswojo, Z., Richard, J., Moritz, M. M., Dopp, E., Flemming, H.-C., and Wingender, J. (2011). Influence of Copper Ions on the Viability and Cytotoxicity of Pseudomonas aeruginosa under Conditions Relevant to Drinking Water Environments. Int. J. Hyg. Environ. Health 214, 485–492. doi:10.1016/j.ijheh.2011.06.004
Ekici, S., Yang, H., Koch, H.-G., and Daldal, F. (2012). Novel Transporter Required for Biogenesis of Cbb 3 -Type Cytochrome C Oxidase in Rhodobacter Capsulatus. mBio 3 (1), 1–11. doi:10.1128/mBio.00293-11
Flemming, H.-C., Wingender, J., Szewzyk, U., Steinberg, P., Rice, S. A., and Kjelleberg, S. (2016). Biofilms: an Emergent Form of Bacterial Life. Nat. Rev. Microbiol. 14 (9), 563–575. doi:10.1038/nrmicro.2016.94
Hahn, C., Hans, M., Hein, C., Mancinelli, R. L., Mücklich, F., Wirth, R., et al. (2017). Pure and Oxidized Copper Materials as Potential Antimicrobial Surfaces for Spaceflight Activities. Astrobiology 17 (12), 1183–1191. doi:10.1089/ast.2016.1620
Hans, M., Erbe, A., Mathews, S., Chen, Y., Solioz, M., and Mücklich, F. (2013). Role of Copper Oxides in Contact Killing of Bacteria. Langmuir 29 (52), 16160–16166. doi:10.1021/la404091z
Hans, M., Támara, J. C., Mathews, S., Bax, B., Hegetschweiler, A., Kautenburger, R., et al. (2014). Laser Cladding of Stainless Steel with a Copper-Silver alloy to Generate Surfaces of High Antimicrobial Activity. Appl. Surf. Sci. 320, 195–199. doi:10.1016/j.apsusc.2014.09.069
Helbig, R., Günther, D., Friedrichs, J., Rößler, F., Lasagni, A., and Werner, C. (2016). The Impact of Structure Dimensions on Initial Bacterial Adhesion. Biomater. Sci. 4 (7), 1074–1078. doi:10.1039/C6BM00078A
Henriksen, S. D. (1973). Moraxella, Acinetobacter, and the Mimeae. Bacteriol. Rev. 37 (4), 522–561. doi:10.1128/br.37.4.522-561.1973
Horneck, G., Klaus, D. M., and Mancinelli, R. L. (2010). Space Microbiology. Microbiol. Mol. Biol. Rev. 74 (1), 121–156. doi:10.1128/MMBR.00016-09
Ilyin, V. K. (2005). Microbiological Status of Cosmonauts during Orbital Spaceflights on Salyut and Mir Orbital Stations. Acta Astronautica 56 (9-12), 839–850. doi:10.1016/j.actaastro.2005.01.009
Janssen, P. J., Van Houdt, R., Moors, H., Monsieurs, P., Morin, N., Michaux, A., et al. (2010). The Complete Genome Sequence of Cupriavidus metallidurans Strain CH34, a Master Survivalist in Harsh and Anthropogenic Environments. PLoS One 5 (5), e10433. doi:10.1371/journal.pone.0010433
Karnovsky, M. J. (1965). A Formaldehyde Glutaraldehyde Fixative of High Osmolality for Use in Electron Microscopy. J. Cel Biol 27 (2), 1A–149A. Abstracts of Papers Presented at the Fifth Annual Meeting: The American Society for Cell Biology.
Karpanen, T. J., Casey, A. L., Lambert, P. A., Cookson, B. D., Nightingale, P., Miruszenko, L., et al. (2012). The Antimicrobial Efficacy of Copper Alloy Furnishing in the Clinical Environment: A Crossover Study. Infect. Control. Hosp. Epidemiol. 33 (1), 3–9. doi:10.1086/663644
Kim, W., Tengra, F. K., Young, Z., Shong, J., Marchand, N., Chan, H. K., et al. (2013). Spaceflight Promotes Biofilm Formation by Pseudomonas aeruginosa. PLoS One 8 (4), e62437. doi:10.1371/journal.pone.0062437
Klintworth, R., Reher, H. J., Viktorov, A. N., and Bohle, D. (1999). Biological Induced Corrosion of Materials II: New Test Methods and Experiences from MIR Station. Acta Astronaut 44 (7-12), 569–578. doi:10.1016/s0094-5765(99)00069-7
Kloos, W. E., and Schleifer, K. H. (1975). Isolation and Characterization of Staphylococci from Human Skin II. Descriptions of Four New Species: Staphylococcus warneri, Staphylococcus capitis, Staphylococcus hominis, and Staphylococcus simulans. Int. J. Syst. Bacteriol. 25(1), 62–79. doi:10.1099/00207713-25-1-62
Koo, H., Allan, R. N., Howlin, R. P., Stoodley, P., and Hall-Stoodley, L. (2017). Targeting Microbial Biofilms: Current and Prospective Therapeutic Strategies. Nat. Rev. Microbiol. 15 (12), 740–755. doi:10.1038/nrmicro.2017.99
La Duc, M. T., Nicholson, W., Kern, R., and Venkateswaran, K. (2003). Microbial Characterization of the Mars Odyssey Spacecraft and its Encapsulation Facility. Environ. Microbiol. 5 (10), 977–985. doi:10.1046/j.1462-2920.2003.00496.x
Lang, J. M., Coil, D. A., Neches, R. Y., Brown, W. E., Cavalier, D., Severance, M., et al. (2017). A Microbial Survey of the International Space Station (ISS). PeerJ 5, e4029. doi:10.7717/peerj.4029
Lasseur, C., Brunet, J., de Weever, H., Dixon, M., Dussap, G., Godia, F., et al. (2010). MELiSSA: The European Project of Closed Life Support System. Grav Space Biol. 23 (2), 3–12.
Lemire, J. A., Harrison, J. J., and Turner, R. J. (2013). Antimicrobial Activity of Metals: Mechanisms, Molecular Targets and Applications. Nat. Rev. Microbiol. 11 (6), 371–384. doi:10.1038/nrmicro3028
Leys, N., Baatout, S., Rosier, C., Dams, A., s’Heeren, C., Wattiez, R., et al. (2009). The Response of Cupriavidus metallidurans CH34 to Spaceflight in the International Space Station. Antonie Van Leeuwenhoek 96 (2), 227–245. doi:10.1007/s10482-009-9360-5
Li, J., Liu, F., Wang, Q., Ge, P., Woo, P. C. Y., Yan, J., et al. (2014a). Genomic and Transcriptomic Analysis of NDM-1 Klebsiella pneumoniae in Spaceflight Reveal Mechanisms Underlying Environmental Adaptability. Sci. Rep. 4, 6216. doi:10.1038/srep06216
Li, L., Mendis, N., Trigui, H., Oliver, J. D., and Faucher, S. P. (2014b). The Importance of the Viable but Non-culturable State in Human Bacterial Pathogens. Front. Microbiol. 5, 258. doi:10.3389/fmicb.2014.00258
Li, Y.-H., and Tian, X. (2012). Quorum sensing and Bacterial Social Interactions in Biofilms. Sensors 12 (3), 2519–2538. doi:10.3390/s120302519
Loudon, C.-M., Nicholson, N., Finster, K., Leys, N., Byloos, B., Van Houdt, R., et al. (2018). BioRock: New Experiments and Hardware to Investigate Microbe-mineral Interactions in Space. Int. J. Astrobiology 17 (4), 303–313. doi:10.1017/S1473550417000234
Luo, J., Hein, C., Ghanbaja, J., Pierson, J.-F., and Mücklich, F. (2019). Bacteria Accumulate Copper Ions and Inhibit Oxide Formation on Copper Surface during Antibacterial Efficiency Test. Micron 127, 102759. doi:10.1016/j.micron.2019.102759
Lutkenhaus, J. F. (1977). Role of a Major Outer Membrane Protein in Escherichia coli. J. Bacteriol. 131 (2), 631–637. doi:10.1128/jb.131.2.631-637.1977
Maertens, L., Coninx, I., Claesen, J., Leys, N., Matroule, J.-Y., and Van Houdt, R. (2020). Copper Resistance Mediates Long-Term Survival of Cupriavidus Metallidurans in Wet Contact with Metallic Copper. Front. Microbiol. 11, 1208. doi:10.3389/fmicb.2020.01208
Maertens, L., Matroule, J.-Y., and Van Houdt, R. (2021). Characteristics of the Copper‐induced Viable‐but‐non‐culturable State in Bacteria. World J. Microbiol. Biotechnol. 37 (3), 1–9. doi:10.1007/s11274-021-03006-5
Maikranz, E., Spengler, C., Thewes, N., Thewes, A., Nolle, F., Jung, P., et al. (2020). Different Binding Mechanisms of Staphylococcus aureus to Hydrophobic and Hydrophilic Surfaces. Nanoscale 12 (37), 19267–19275. doi:10.1039/d0nr03134h
Martí, S., Rodríguez-Baño, J., Catel-Ferreira, M., Jouenne, T., Vila, J., Seifert, H., et al. (2011). Biofilm Formation at the Solid-Liquid and Air-Liquid Interfaces by Acinetobacter Species. BMC Res. Notes 4 (1), 5. doi:10.1186/1756-0500-4-5
Mastroleo, F., Van Houdt, R., Atkinson, S., Mergeay, M., Hendrickx, L., Wattiez, R., et al. (2013). Modelled Microgravity Cultivation Modulates N-Acylhomoserine Lactone Production in Rhodospirillum rubrum S1H Independently of Cell Density. Microbiology 159 (Pt 12), 2456–2466. doi:10.1099/mic.0.066415-0
Mathews, S., Hans, M., Mücklich, F., and Solioz, M. (2013). Contact Killing of Bacteria on Copper Is Suppressed if Bacterial-Metal Contact Is Prevented and Is Induced on Iron by Copper Ions. Appl. Environ. Microbiol. 79 (8), 2605–2611. doi:10.1128/aem.03608-12
McCoy, K. B., Derecho, I., Wong, T., Tran, H. M., Huynh, T. D., La Duc, M. T., et al. (2012). Insights into the Extremotolerance of Acinetobacter radioresistens 50v1, a Gram-Negative Bacterium Isolated from the Mars Odyssey Spacecraft. Astrobiology 12 (9), 854–862. doi:10.1089/ast.2012.0835
McLean, R. J. C., Cassanto, J. M., Barnes, M. B., and Koo, J. H. (2001). Bacterial Biofilm Formation under Microgravity Conditions. FEMS Microbiol. Lett. 195 (2), 115–119. doi:10.1111/j.1574-6968.2001.tb10507.x
Mergeay, M., Houba, C., and Gerits, J. (1978). Extrachromosomal Inheritance Controlling Resistance to Cadmium, Cobalt, Copper and Zinc Ions: Evidence from Curing in a Pseudomonas [proceedings]. Arch. Int. Physiol. Biochim. 86 (2), 440–442. PMID: 81018.
Mijnendonckx, K., Ali, M. M., Provoost, A., Janssen, P., Mergeay, M., Leys, N., et al. (2019). Spontaneous Mutation in the AgrRS Two-Component Regulatory System of Cupriavidus metallidurans results in Enhanced Silver Resistance. Metallomics 11 (11), 1912–1924. doi:10.1039/c9mt00123a
Mijnendonckx, K., Provoost, A., Ott, C. M., Venkateswaran, K., Mahillon, J., Leys, N., et al. (2013). Characterization of the Survival Ability of Cupriavidus metallidurans and Ralstonia rickettii from Space-Related Environments. Microb. Ecol. 65 (2), 347–360. doi:10.1007/s00248-012-0139-2
Mogul, R., Barding, G. A., Lalla, S., Lee, S., Madrid, S., Baki, R., et al. (2018). Metabolism and Biodegradation of Spacecraft Cleaning Reagents by Strains of Spacecraft-Associated Acinetobacter. Astrobiology 18 (12), 1517–1527. doi:10.1089/ast.2017.1814
Monsieurs, P., Hobman, J., Vandenbussche, G., Mergeay, M., and Van Houdt, R. (2015). “Response of Cupriavidus metallidurans CH34 to Metals,” in Metal Response in Cupriavidus metallidurans. Editors M. Mergeay,, and R. Van Houdt (Switzerland: Springer International Publishing), 45–89. doi:10.1007/978-3-319-20594-6_3
Monsieurs, P., Moors, H., Van Houdt, R., Janssen, P. J., Janssen, A., Coninx, I., et al. (2011). Heavy Metal Resistance in Cupriavidus metallidurans CH34 Is Governed by an Intricate Transcriptional Network. Biometals 24 (6), 1133–1151. doi:10.1007/s10534-011-9473-y
Mora, M., Perras, A., Alekhova, T. A., Wink, L., Krause, R., Aleksandrova, A., et al. (2016). Resilient Microorganisms in Dust Samples of the International Space Station-Survival of the Adaptation Specialists. Microbiome 4 (65), 1–21. doi:10.1186/s40168-016-0217-7
Mora, M., Wink, L., Kögler, I., Mahnert, A., Rettberg, P., Schwendner, P., et al. (2019). Space Station Conditions Are Selective but Do Not Alter Microbial Characteristics Relevant to Human Health. Nat. Commun. 10 (1), 3990. doi:10.1038/s41467-019-11682-z
Müller, D. W., Fox, T., Grützmacher, P. G., Suarez, S., and Mücklich, F. (2020a). Applying Ultrashort Pulsed Direct Laser Interference Patterning for Functional Surfaces. Sci. Rep. 10 (1), 3647. doi:10.1038/s41598-020-60592-4
Müller, D. W., Holtsch, A., Lößlein, S., Pauly, C., Spengler, C., Grandthyll, S., et al. (2020b). In-Depth Investigation of Copper Surface Chemistry Modification by Ultrashort Pulsed Direct Laser Interference Patterning. Langmuir 36 (45), 13415–13425. doi:10.1021/acs.langmuir.0c01625
Müller, D. W., Lößlein, S., Terriac, E., Brix, K., Siems, K., Moeller, R., et al. (2021). Increasing Antibacterial Efficiency of Cu Surfaces by Targeted Surface Functionalization via Ultrashort Pulsed Direct Laser Interference Patterning. Adv. Mater. Inter. 8 (5), 2001656. doi:10.1002/admi.202001656
Novikova, N., De Boever, P., Poddubko, S., Deshevaya, E., Polikarpov, N., Rakova, N., et al. (2006). Survey of Environmental Biocontamination on Board the International Space Station. Res. Microbiol. 157 (1), 5–12. doi:10.1016/j.resmic.2005.07.010
Noyce, J. O., Michels, H., and Keevil, C. W. (2006). Use of Copper Cast Alloys to Control Escherichia coli O157 Cross-Contamination during Food Processing. Appl. Environ. Microbiol. 72 (6), 4239–4244. doi:10.1128/AEM.02532-05
Peleg, A. Y., Seifert, H., and Paterson, D. L. (2008). Acinetobacter Baumannii : Emergence of a Successful Pathogen. Clin. Microbiol. Rev. 21 (3), 538–582. doi:10.1128/cmr.00058-07
Perrin, E., Bacci, G., Garrelly, L., Canganella, F., Bianconi, G., Fani, R., et al. (2018). Furnishing Spaceship Environment: Evaluation of Bacterial Biofilms on Different Materials Used inside International Space Station. Res. Microbiol. 169 (6), 289–295. doi:10.1016/j.resmic.2018.04.001
Romsdahl, J., Blachowicz, A., Chiang, A. J., Singh, N., Stajich, J. E., Kalkum, M., et al. (2018). Characterization of Aspergillus niger Isolated from the International Space Station. mSystems 3 (5), e00112–18. doi:10.1128/mSystems.00112-18
Schottroff, F., Fröhling, A., Zunabovic-Pichler, M., Krottenthaler, A., Schlüter, O., and Jäger, H. (2018). Sublethal Injury and Viable but Non-culturable (VBNC) State in Microorganisms during Preservation of Food and Biological Materials by Non-thermal Processes. Front. Microbiol. 9, 2773. doi:10.3389/fmicb.2018.02773
Seuylemezian, A., Vaishampayan, P., Cooper, K., and Venkateswaran, K. (2018). Draft Genome Sequences of Acinetobacter and Bacillus Strains Isolated from Spacecraft-Associated Surfaces. Genome Announc 6 (6), e01554–17. doi:10.1128/genomeA.01554-17
Singh, N. K., Wood, J. M., Karouia, F., and Venkateswaran, K. (2018). Succession and Persistence of Microbial Communities and Antimicrobial Resistance Genes Associated with International Space Station Environmental Surfaces. Microbiome 6 (1), 204. doi:10.1186/s40168-018-0585-2
Singh, N., and Wade, J. T. (2014). Identification of Regulatory RNA in Bacterial Genomes by Genome-Scale Mapping of Transcription Start Sites. Methods Mol. Biol. 1103, 1–10. doi:10.1007/978-1-62703-730-3_1
Singh, S., Singh, S. K., Chowdhury, I., and Singh, R. (2017). Understanding the Mechanism of Bacterial Biofilms Resistance to Antimicrobial Agents. Tomicroj 11, 53–62. doi:10.2174/1874285801711010053
Sobisch, L.-Y., Rogowski, K. M., Fuchs, J., Schmieder, W., Vaishampayan, A., Oles, P., et al. (2019). Biofilm Forming Antibiotic Resistant Gram-Positive Pathogens Isolated from Surfaces on the International Space Station. Front. Microbiol. 10, 543. doi:10.3389/fmicb.2019.00543
Sonnenfeld, G., Butel, J. S., and Shearer, W. T. (2003). Effects of the Space Flight Environment on the Immune System. Rev. Environ. Health 18 (1), 1–18. doi:10.1515/reveh.2003.18.1.1
Sonnenfeld, G., and Shearer, W. T. (2002). Immune Function during Space Flight. Nutrition 18 (10), 899–903. doi:10.1016/s0899-9007(02)00903-6
Sun, C., Li, Y., Li, Z., Su, Q., Wang, Y., and Liu, X. (20182018). Durable and Washable Antibacterial Copper Nanoparticles Bridged by Surface Grafting Polymer Brushes on Cotton and Polymeric Materials. J. Nanomater. 2018, 1–7. doi:10.1155/2018/6546193
Towner, K. J., Bergogne-Bérézin, E., and Fewson, C. (2013). The Biology of Acinetobacter: Taxonomy, Clinical Importance, Molecular Biology, Physiology, Industrial Relevance. Luxemburg: Springer Science & Business Media.
Tripathy, A., Kumar, A., Sreedharan, S., Muralidharan, G., Pramanik, A., Nandi, D., et al. (2018). Fabrication of Low-Cost Flexible Superhydrophobic Antibacterial Surface with Dual-Scale Roughness. ACS Biomater. Sci. Eng. 4 (6), 2213–2223. doi:10.1021/acsbiomaterials.8b00209
Tripathy, A., Sen, P., Su, B., and Briscoe, W. H. (2017). Natural and Bioinspired Nanostructured Bactericidal Surfaces. Adv. Colloid Interf. Sci. 248, 85–104. doi:10.1016/j.cis.2017.07.030
Urbaniak, C., Sielaff, A. C., Frey, K. G., Allen, J. E., Singh, N., Jaing, C., et al. (2018). Detection of Antimicrobial Resistance Genes Associated with the International Space Station Environmental Surfaces. Sci. Rep. 8 (1), 814. doi:10.1038/s41598-017-18506-4
Van Houdt, R., and Leys, N. (2020). “Monitoring the Microbial Burden in Manned Space Stations,” in Stress Challenges and Immunity in Space. Editor A. Chouker (Springer), 463–475. doi:10.1007/978-3-030-16996-1_25
Van Houdt, R., Mijnendonckx, K., and Leys, N. (2012). Microbial Contamination Monitoring and Control during Human Space Missions. Planet. Space Sci. 60 (1), 115–120. doi:10.1016/j.pss.2011.09.001
Van Houdt, R., Vandecraen, J., Leys, N., Monsieurs, P., and Aertsen, A. (2021). Adaptation of Cupriavidus metallidurans CH34 to Toxic Zinc Concentrations Involves an Uncharacterized ABC-type Transporter. Microorganisms 9 (2), 309. doi:10.3390/microorganisms9020309
Vandamme, P., and Coenye, T. (2004). Taxonomy of the Genus Cupriavidus: a Tale of Lost and Found. Int. J. Syst. Evol. Microbiol. 54 (6), 2285–2289. doi:10.1099/ijs.0.63247-0
Venkateswaran, K., Vaishampayan, P., Cisneros, J., Pierson, D. L., Rogers, S. O., and Perry, J. (2014). International Space Station Environmental Microbiome - Microbial Inventories of ISS Filter Debris. Appl. Microbiol. Biotechnol. 98 (14), 6453–6466. doi:10.1007/s00253-014-5650-6
Vincent, M., Duval, R. E., Hartemann, P., and Engels‐Deutsch, M. (2018). Contact Killing and Antimicrobial Properties of Copper. J. Appl. Microbiol. 124 (5), 1032–1046. doi:10.1111/jam.13681
Vincent, M., Hartemann, P., and Engels-Deutsch, M. (2016). Antimicrobial Applications of Copper. Int. J. Hyg. Environ. Health 219 (7 Pt A), 585–591. doi:10.1016/j.ijheh.2016.06.003
Wilks, S. A., Michels, H., and Keevil, C. W. (2005). The Survival of Escherichia coli O157 on a Range of Metal Surfaces. Int. J. Food Microbiol. 105 (3), 445–454. doi:10.1016/j.ijfoodmicro.2005.04.021
Zea, L., Larsen, M., Estante, F., Qvortrup, K., Moeller, R., Dias de Oliveira, S., et al. (2017). Phenotypic Changes Exhibited by E. coli Cultured in Space. Front. Microbiol. 8, 1598. doi:10.3389/fmicb.2017.01598
Zea, L., McLean, R. J. C., Rook, T. A., Angle, G., Carter, D. L., Delegard, A., et al. (2020). Potential Biofilm Control Strategies for Extended Spaceflight Missions. Biofilm 2, 100026. doi:10.1016/j.bioflm.2020.100026
Keywords: biofilms, direct laser interference patterning technique (DLIP), contamination (equipment), antimicobial, surfaces
Citation: Siems K, Müller DW, Maertens L, Ahmed A, Van Houdt R, Mancinelli RL, Baur S, Brix K, Kautenburger R, Caplin N, Krause J, Demets R, Vukich M, Tortora A, Roesch C, Holland G, Laue M, Mücklich F and Moeller R (2022) Testing Laser-Structured Antimicrobial Surfaces Under Space Conditions: The Design of the ISS Experiment BIOFILMS. Front. Space Technol. 2:773244. doi: 10.3389/frspt.2021.773244
Received: 09 September 2021; Accepted: 29 November 2021;
Published: 03 January 2022.
Edited by:
Cosimo Buffone, Tianjin University of Commerce, ChinaReviewed by:
Po Bian, Hefei Institutes of Physical Science (CAS), ChinaCopyright © 2022 Siems, Müller, Maertens, Ahmed, Van Houdt, Mancinelli, Baur, Brix, Kautenburger, Caplin, Krause, Demets, Vukich, Tortora, Roesch, Holland, Laue, Mücklich and Moeller. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ralf Moeller, cmFsZi5tb2VsbGVyQGRsci5kZQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.