- 1Nephrology Section, Medicine Service Line, Durham VA Health Care System, Durham, NC, United States
- 2Nephrology Division, Department of Internal Medicine, Duke University School of Medicine, Durham, NC, United States
- 3Space Policy Institute, Elliott School of International Affairs, George Washington University, Washington, DC, United States
- 4Nephrology Section, Department of Medicine, Tulane University School of Medicine, New Orleans, LA, United States
- 5Research Service Line, Durham VA Health Care System, Durham, NC, United States
- 6The Bionetics Corporation, Mail Code: Bio-2, Kennedy Space Center Florida, Brevard County, FL, United States
- 7Faculty of Pharmaceutical Sciences, The University of British Columbia, Vancouver, BC, Canada
- 8Departments of Otorhinolaryngology, Immunology, and Psychiatry, Baylor College of Medicine, Houston, TX, United States
- 9Otolaryngology Section, Surgery Service Line, Durham VA Health Care System, Durham, NC, United States
Artemis is a NASA initiative to return US astronauts to the moon and beyond. Artemis includes a robust science program. A key to space-based biological science is the ability to construct high-quality, efficacious scientific hardware cheaply, and in a short time frame. We report the design and fabrication of affordable new flight hardware to support the growth of Chlamydomonas reinhardtii, a single-cell green alga on the Artemis-1 mission. C. reinhardtii produces two products of great interest to space travel—lipids as a source of bioproducts and hydrogen a source of fuel. We will use this hardware to grow a library of mutant strains to identify C. reinhardtii genes that optimize survival and fuel production during exposure to the combined impacts of space radiation and microgravity in the cosmic space environment beyond the Van Allen belts. Using fitness as a readout, a library of mapped insertion mutant strains will be grown competitively during exposure to space radiation and microgravity. This report describes the hardware and the science verification test that validates its utility for missions ahead. This hardware can be easily adapted to a broad range of uses and microbes, and its low cost will make space science affordable and practical.
Introduction
Artemis-1 is the first in a series of NASA flights designed to return astronauts to the moon and beyond. The Artemis spacecraft is an Orion capsule sitting atop a space launch system rocket (https://www.nasa.gov/artemisprogram). The first flight is an unmanned translunar demonstration sortie with capsule recovery after a water landing.
The Space Biology Program, based out of Kennedy Space Center in Florida, was invited to include four science payloads in the Artemis-1 flight. The payloads had to be autonomous for any activation or deactivation steps and provide their own power and temperature control, as necessary, throughout ~2 weeks on the launch pad and during a mission of 60+ days. These requirements place significant constraints on the selection of the biological system and the experimental design.
Chlamydomonas reinhardtii is a single-cell green alga that meets the Artemis-1 payload design criteria and is a leading candidate organism for bioproduction of fuel: biodiesel, oxygen, and hydrogen for space travel (Scaife et al., 2015; Batyrova and Hallenbeck, 2017). This project will identify strains of Chlamydomonas with optimized fitness during exposure to the combined impacts of space radiation and microgravity in the cosmic space environment beyond the Van Allen belts. Two factors drove our choice to study C. reinhardtii. First, among the model microalgae, C. reinhardtii is a known efficient biological producer of hydrogen (Gaffron and Rubin, 1942; Vijayaraghavan et al., 2009; Dasgupta et al., 2010; Scoma et al., 2012; Batyrova and Hallenbeck, 2017) and is already being exploited as an industrial biotechnology platform or “cell factory” (Grobbelaar, 2010; Scaife et al., 2015). Second, Chlamydomonas is well-suited for genetic characterization, including a library of mapped insertion mutant strains that allow for an unbiased identification of the genes mediating the best long-term survival advantage during exposure to space radiation and microgravity (Alfred et al., 2012; Blaby et al., 2014; Gallaher et al., 2015; Li et al., 2016). For example, those strain(s) that show the best survival advantage outside the Van Allen belt can serve as parent strains for further genetic engineering to enhance hydrogen production, as well as diverse other initiatives using biomass for spaceflight purposes (Scaife et al., 2015).
Chlamydomonas has a track record of robust viability in space, and its low resource requirements and its ability to maintain viability for weeks prior to light-induced growth activation make it ideally suited spaceflight applications (Mergenhagen and Mergenhagen, 1987, 1989).
We worked with the Bionetics Corporation to design and fabricate a low-cost hardware, with integral power, optimized to support the growth of C. reinhardtii within the constraints of the Artemis-1 mission profile. The hardware has been named “Moonshot” by the engineers who did the design and fabrication work. The system provides blue and red light to each of three Petri dishes for continuous 6 h alternating with 18 h of darkness and can do so for at least 60 days. The hardware monitors and records the temperature of the experiment, battery power levels, and acceleration levels in three perpendicular axes.
Materials and Methods
Chemicals and reagents were purchased from Sigma Inc. (St. Louis, MO) unless otherwise noted. A stab of C. reinhardtii (CC-125 wild-type mt+ [137c]) and Hutner's trace elements were purchased from the Chlamydomonas Resource Center collection at the University of Minnesota (Minneapolis, MN). Annexin-binding buffer “5X concentrate” and annexin V Alexa Fluor 488 conjugate were purchased from Life Technologies (Carlsbad, CA). Spectrophotometry was performed on a Molecular Devices M5e spectrophotometer (San Jose, CA). Flow cytometry was performed on a Becton–Dickinson Accuri C+ flow cytometer (Franklin Lakes, NJ).
Hardware Specifications
NASA's requirements for experimental hardware on the Artemis mission included the following:
• not require external power
• not require uplink or downlink
• monitor and record the temperature of the experiment
• monitor and record acceleration levels
• run continuously for a total of 48 days (the initial estimated duration of the mission was 21–40 days from launch to splashdown)
• withstand temperatures of 54–107°F
• weigh less than 8 lb
• fit within 5.75 × 11.68 × 5 inches, including relief for Nomex bag
The experimental requirements for Chlamydomonas growth were that three 10-cm-diameter 15-mm-tall circular Petri dishes be illuminated with blue (~450 nm) and red (~660 nm) lights for continuous 6 h out of every 24 h for 60 days.
Moonshot Hardware
Bionetics custom “Moonshot” hardware is an ultralow-power growth system for use on Artemis-1. Each of the three growth modules holds one standard 100-mM-diameter circular Petri dish. The plates are illuminated with blue (~450 nm) and red (~660 nm) light-emitting diode (LED) lights continuously for 6 h in every 24 h. The combination of red and blue light is optimum for Chlamydomonas (Oldenhof et al., 2006; Li et al., 2016). The hardware monitors and records the temperature of the experiment, the power level, and draw-on in the batteries (as an indication that the lights were turned on and off), as well as the acceleration levels in three axes.
The hardware was designed using the computer-aided design and drafting program SolidWorks. It is largely composed of milled aluminum parts. The electrical boards were robotically fabricated using standard FR-4 fiberglass.
Hardware Validation
The functional and biological support properties of the Moonshot hardware were validated during a science verification test (SVT) for the Artemis-1 mission. The engineering testing was performed in the actual flight hardware November 26, 2019, to through January 31, 2020; a 60-days period was chosen to reflect the best guess at the time for the duration of the Artemis-1 mission. The biological validation was performed on prototype hardware.
Protocols
A loop inoculum of C. reinhardtii was placed into liquid Tris–acetate–phosphate (TAP) media [1 M Tris base 20 mL, phosphate K2HPO4/KH2PO4 buffer 1 mL, Hutner's trace metals 1 mL, 10 mL of solution A (NH4Cl, MgSP4·7H2O, CaCl2·2H2O), glacial acetic acid 1 mL to adjust to pH 7] and made up to 1 L with distilled water. After a lag of ~21 days, which is typical for previously refrigerated samples, the Chlamydomonas grew robustly.
TAP media plus 12 g agar per liter was autoclaved, and 10 mL was poured into 100-mm-diameter Petri dishes, and 20 mL into single-cell rectangular Omniplates (Nunc). Dark green, robustly growing Chlamydomonas were inoculated, 5 μL per spot, onto each plate in a 96-well matrix of colonies for the Omniplate and 52 spot matrix for the round Petri dishes. The plates were sealed with Parafilm, and the Chlamydomonas spots allowed to adhere to the agar for 2 days at room temperature in a box containing red and blue light coming on contemporaneously for 6 h out of a 24-h cycle. The round Petri dishes were then moved to the flight hardware prototype and the rectangular Omniplates to the spectrophotometer to initiate the SVT.
At the end of the 60-days SVT, six colonies of Chlamydomonas in round plates from the flight hardware prototype were harvested for flow cytometric assays of viability and lipid content. Samples in the rectangular Omniplates were used to measure Chlamydomonas protein accumulation throughout the SVT. The Omniplates, which have the dimensions of an Society for Laboratory Automation and Screening standard microplate, fit the sample drawer of the spectrophotometer and allowed for precise alignment of the colonies in the detector for repetitive daily reads over the 60-days SVT. The lighting and temperature of the round Petri plates and rectangular Omniplates were matched throughout the SVT.
Measurement of Chlamydomonas Proteins
Chlamydomonas colonies were assayed by spectrophotometry on a daily basis throughout the 60-days SVT to define protein content over time. To match the illumination provided by the Moonshot flight hardware, a light box was placed over the spectrophotometer so as to illuminate the Chlamydomonas when the plate drawer was in the open position. A loop program directed the spectrophotometer to retract the plate into the machine, take the absorbance readings at six wavelengths, and then return the drawer to the open configuration so that the illumination of the plate could resume. The six wavelengths were chosen to measure chlorophyll A (436 nm), chlorophyll B (472 nm), photosystem I and II (700 nm), cytochrome f (554 nm), and total protein (800 nm). The detector read the absorbances in the same eight colonies each time. The light box over the spectrophotometer cycled in the same 6-h-on/18-h-off pattern as the flight hardware prototype.
Viability and Lipid Analysis
At the end of the SVT, six colonies were harvested from the round Petri dishes in the Moonshot hardware and assayed for lipid content and viability by flow cytometry. Colonies containing ~107 cells were resuspended in annexin-binding buffer, vortexed vigorously to dissociate the Chlamydomonas, and filtered through 70-μm nylon to remove aggregates. For viability assays, 100-μL aliquots of each sample were stained with 5 μL of Alexa Fluor 488 annexin and 1 μL of 1 mg/mL propidium iodide (PI) and incubated at room temperature for 15 min. Live Chlamydomonas populations are both annexin-negative and PI-negative; apoptotic Chlamydomonas take up annexin, but not PI; and necrotic Chlamydomonas take up PI alone. To measure lipid content, 100-μL aliquots were permeabilized with 10 μL of 0.2% Triton X and stained with 5 μL of Nile red (100 μg/mL in methanol) (Halim and Webley, 2015; Terashima et al., 2015). Viability was also verified by observing whether the Chlamydomonas resume growth when inoculated into fresh media.
Flow Cytometry
In flow cytometry analyses, we counted 5,000 Chlamydomonas and recorded forward and side scatter in addition to fluorescent parameters using log amplified photomultipliers. With every run, quality control was performed on the instrument with three color fluorescent beads, followed by assay of five control tubes: no dyes, annexin alone (488-nm excitation 533/30-nm emission), PI alone (488-nm excitation 585/40-nm emission), annexin plus PI, and Nile red alone (488-nm excitation 585/40-nm emission). All flow cytometry values are in arbitrary fluorescence units. While PI and Nile red have brighter emission at 675/25 nm in many cells types, chlorophyll A and B absorption precludes using these wavelengths in Chlamydomonas (Halim and Webley, 2015; Terashima et al., 2015).
Statistics
Statistical analyses are presented as mean ± standard error of n = 5 or 6, as indicated. Significance was determined using Student t-test. All comparisons shown are also significant using Kolmogorov–Smirnov summation statistics for large data sets.
Results
Hardware Design
The Bionetics “Moonshot” custom hardware consist of three module types: two battery modules, three LED lit growth modules, and a control board. The total hardware mass with margin is 5.44 lb, and it measures 7.05 ± 0.02 × 5.67 ± 0.02 × 4.45 ± 0.02 inches (Figure 1A).
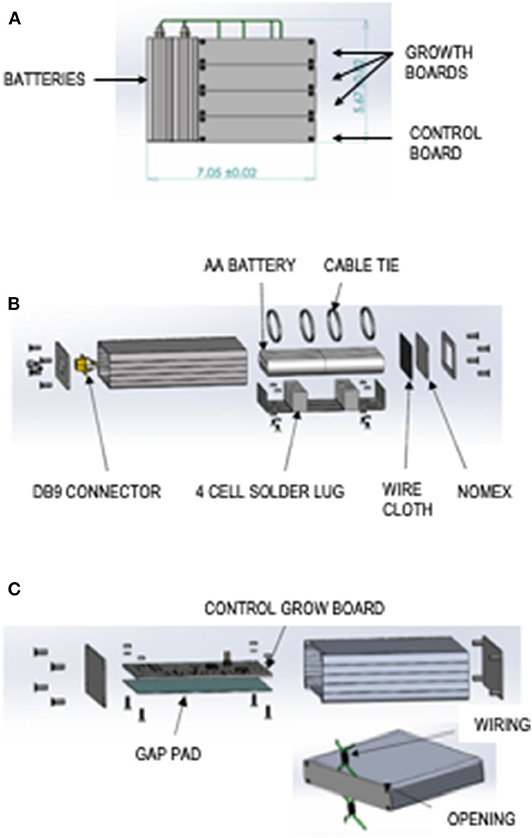
Figure 1. Moonshot hardware design. (A) Flight configuration of the assembled “Moonshot” hardware. (B) Battery Module exploded view. (C) Control Board Module exploded view.
The features of the battery module can be seen in an exploded view schematic (Figure 1B). Components of the battery module include a silver extruded aluminum enclosure with metal end panels, eight AA Energizer batteries, a PPTC resettable fuse, four keystone electronics, battery holder AA cell solder lug cable ties, Nomex fabric, small-mesh particle-filtering wire cloth, and 4 × 4-inch gap pad.
The features of the control growth module can be seen in an exploded view schematic (Figure 1C). Components of the control growth module include a silver extruded aluminum enclosure with metal end panels, a gap pad, and a control board. Each module has an opening in the lid and base for wiring. The control board electrical design overview includes an ultralow-power PIC18 microcontroller unit with internal real-time clock, 16 MB of flash for data storage, temperature sensors, accelerometers, and universal asynchronous receiver/transmitter to universal serial bus (USB). The control board can be powered by USB or 9- to 14-V battery input.
The features of the LED growth module can be seen in an exploded view schematic (Figure 2A). Components of the LED growth module include a silver extruded aluminum enclosure with metal end panels, a 110-mm-diameter and 15-mm-high cylindrical polystyrene Petri dish mount, a Petri dish holder, a gap pad, a thermistor temperature sensor, and an LED illumination board. The Petri dish holders are polycarbonate, and the lid lined with reflective polytetrafluoroethylene. Figures 2B–D shows the LED array in the base of the growth module. Figure 2E shows a plate of Chlamydomonas incubated in the growth module for 60 days and verifies that the colonies grow robustly and evenly across the plate.
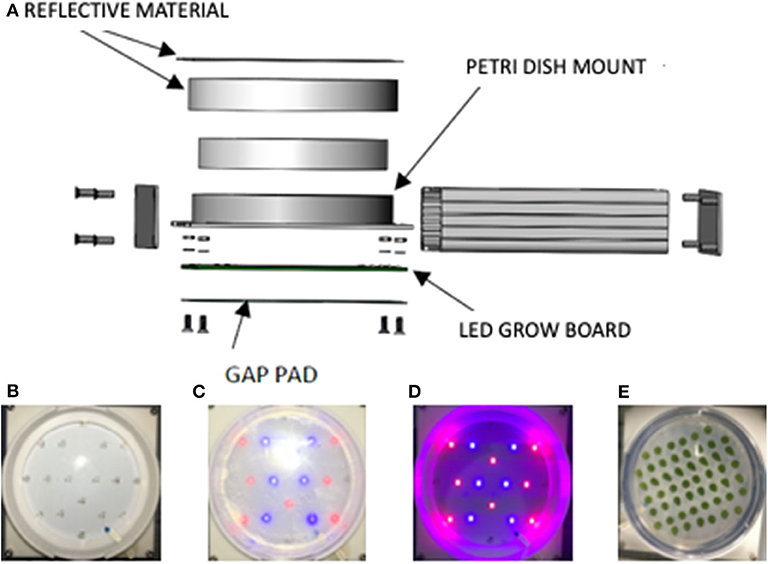
Figure 2. Moonshot hardware growth module design. (A) Growth Module exploded view. (B) LED grow board “off.” (C) LED grow board “on.” (D) LED grow board “on” in dark environment to demonstrate red and blue LED lamps. (E) Petri plate spotted with Chlamydomonas and incubated 60 days in the flight hardware with cycling lights. The spots are dark green, consistent with robust chlorophyll production, and of uniform intensity indicative of uniform lighting across the plate.
Hardware Performance
The hardware performed nominally throughout the verification test. The maximum temperature increase was 5°F, well within the envelope of tolerable experimental conditions (Figure 3A). At the end of the 60-days test, the batteries still had more than 20% of power remaining (Figure 3B). The power profile validated that the lights came on for 6 out of 24 h daily, as planned (Figure 3C). Acceleration recordings show serial measurements of gravity in the z axis, but no acceleration in other directions as expected (Figure 3D).
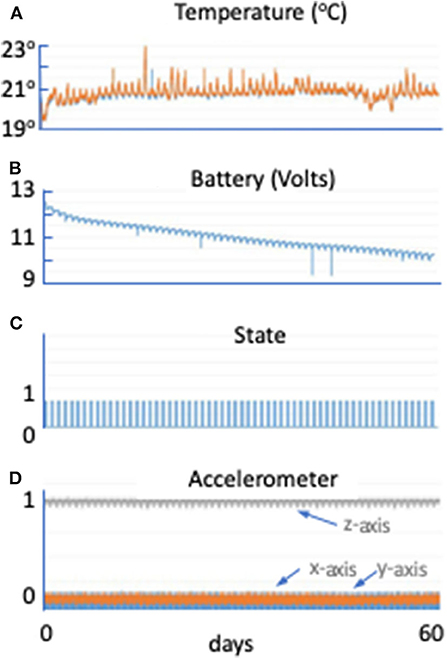
Figure 3. Moonshot hardware performance. (A) Temperature (°C) in the hardware (Petri dish in blue and control board in orange) across the 60-days SVT (minutes). The Petri dish remained between 19.5 and 23°C, which will support Chlamydomonas growth. (B) Battery voltage (in volts) in the hardware across the 60-days SVT (in minutes). The scale on the y axis is from 9 to 13 V. At least 10 V remain in the battery at the end of the 60-days period. (C) State is a unitless parameter. If it is higher than 1, it means the unit just started; if it is 1, it means the LEDs are on; if it is zero, it means the LEDs are off, allowing up to document hardware functioning during the flight. (D) Accelerometer data in g in the hardware across the 60-days SVT (in minutes). Gray line shows the z direction (Earth gravity). Orange and blue are x and y directions.
The hardware has been flight-certified by NASA and passed all materials compatibility and other safety reviews. Electromagnetic interference testing was not performed as NASA safety ruled that the low voltages, amperage, and fluxes in the hardware were too trivial to interfere with the Orion capsule integrated circuit technology. Certainly, electromagnetic interference testing can similarly be completed simply and affordably, if necessary. Vibration testing is pending if NASA later mandates, but all costs are already built into the costs stated.
Protein Measurement
The kinetics of the accumulated chlorophyll A, chlorophyll B, photosystem I and II, cytochrome f, and total protein are shown in Figure 4. We observed that the levels of these specific proteins and protein levels in general increased as the Chlamydomonas grew for the first few days and reached equilibrium for the rest of the expected 60-days Artemis-1 mission profile.
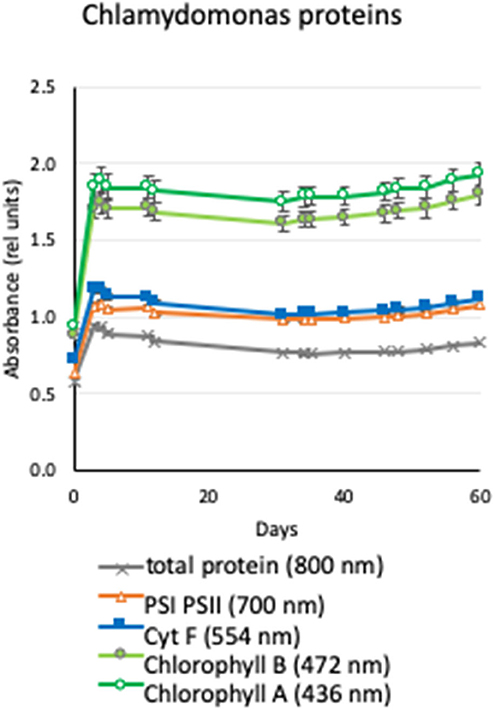
Figure 4. Production of proteins by Chlamydomonas under conditions of the Moonshot hardware. Agar plates were spotted with Chlamydomonas and incubated in a spectrophotometer adapted to provide cycling illumination that matched the conditions of the Moonshot hardware. Serial absorbance measurements were collected daily for 60 days to quantify chlorophyll A (436 nm, open green circles), chlorophyll B (472-nm, filled green circles), cytochrome f (554 nm, filled blue squares), photosystem I and II (700 nm; open orange triangles), and total protein (800 nm; gray X). Values shown are the net absorbance above background (plain agar) in relative units, and error bars show the ±standard error of the mean of eight replicate colonies.
Viability
The fresh 7-days Chlamydomonas liquid culture used to inoculate the agar plates contained 88 ± 1% live cells, 12 ± 1% apoptotic (i.e., dying) cells, and 0.2 ± 0.1% dead cells (Figure 5). At the end of the 60-days SVT, 13 ± 1% of the Chlamydomonas were viable, 13 ± 1% were apoptotic, and 74 ± 1% were dead. The quantity of nonviable cells was an expected finding given the fact that cell growth ceases after 7 days, and viability drops shortly thereafter (Nakamura et al., 1986). Importantly, a live population remains, as Chlamydomonas maintained for 60 days in the Moonshot hardware grew robustly when inoculated into fresh TAP media (data not shown).
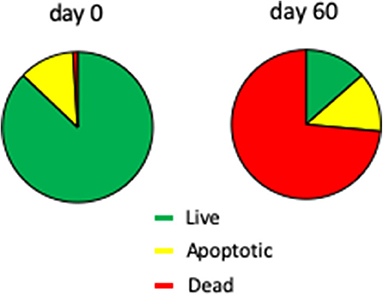
Figure 5. Viability of Chlamydomonas incubated in Moonshot hardware. Agar plates were spotted with Chlamydomonas and incubated in the flight hardware with cycling illumination for 60 days. Colonies were harvested at 0 and 60 days and assayed for viability and apoptosis by flow cytometry. Pie charts show the % of the total Chlamydomonas population that is viable (green; annexin-negative and PI-negative), apoptotic (yellow; annexin-positive but PI-negative), or dead (red; PI-positive).
The amount of total lipid present in the Chlamydomonas, as measured by Nile red fluorescence, increased from 14,888 ± 146 in fresh zero-day cultures to 37,281 ± 244 arbitrary fluorescence units in the 60-days SVT cultures, mean ± standard error, n = 5, p < 0.05 by Student t-test (Figure 6). These observations suggest that lipid production is robust on the Moonshot platform and demonstrate that Nile red staining will be a useful metric to gauge overall lipid production (Halim and Webley, 2015; Terashima et al., 2015).
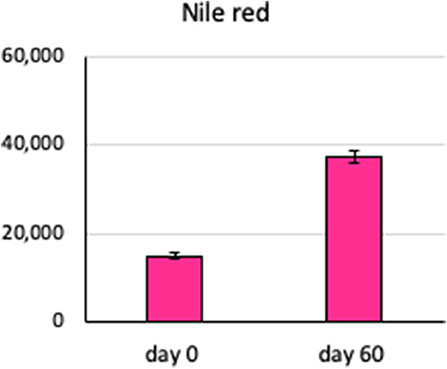
Figure 6. Lipid content of Chlamydomonas incubated in Moonshot hardware. Agar plates were spotted with Chlamydomonas and incubated in the flight hardware with cycling illumination for 60 days. Colonies were harvested at 60 days and assayed for lipid content by uptake of Nile red and flow cytometry and compared to fresh Chlamydomonas cultures. Bars show the lipid content, measured as uptake of Nile red dye, in arbitrary fluorescence units, at 0 and 60 days. Error bars are ±SEM of six replicates.
Discussion
The space sector is one of the economy's most rapidly evolving elements. Launch costs are plummeting dramatically led by cost efficiencies in vehicle design and manufacture, including reusable rocket elements pioneered by Space-X. If we are to use the environment of space for basic science research and development of biotechnology products, we must develop a similar transformation. The common quote of “5 years and 5 million dollars” to fly a payload (personal communications) has scared off numerous cell biology, pharmaceutical, business, and other investors. We need more efficient and less expensive ways to produce flight hardware.
Advances in materials, microelectronics, and molecular and cellular biology technologies provide a firm basis to produce cost-effective customized spaceflight hardware. Following this paradigm, we report the design, construction, and validation of a simple cost-effective flight hardware for growth of the green algae C. reinhardtii. The new hardware, termed “Moonshot,” is scheduled for use in the Orion capsule of the first Artemis mission, a circumnavigation of the moon.
New materials, new technologies, and new understanding of cell biology are allowing a revolution in space biology hardware design and fabrication. For instance, we now only need picograms of RNA or DNA to sequence a genome or transcriptome. Strain banks with individual deletion of every gene in the genome and insertion of both gene-specific and universal primers afford the opportunity to pursue sophisticated science and determine whether microgravity has a role in defining drug targets. These types of modern tools need to be integrated into the policies and procedures of spaceflight providers. The incorporation of the space biology policies and procedures into spaceflight providers' standard operating procedures will plummet cost and exponentially facilitate access to flight options.
We show here that you can build space flight biology hardware for an order of magnitude less than the going rate. The hardware met all the validation criteria for power use, power cycling, temperature control, g-force recording in three axes, data capture, and downloading for analysis, materials, and biological compatibility. This type of hardware will be absolutely key in the new space business environment.
In the past, flight certification of new hardware involved acoustic testing at Marshall Space Flight Center in Huntsville, Alabama; bench review at Johnson Space Center in Houston, Texas; electromagnetic interference testing and safety committee reviews at Kennedy Space Center; and vibration testing at a variety of sites. The geographic distribution of flight certification activities added time, effort, and substantial cost. Flight certification functionalities are now centralized at Kennedy Space Center and/or the adjacent Bionetics facility. This makes flight certification facile and affordable, and these expenses are included in the costs quoted.
Payload cost efficiency is achievable. Our laboratory flew renal cells on STS-90 and performed the very first gene arrays on a cell biology sample from space (Hammond et al., 1999). We flew a repeat, which cost $100,000, all up, for hardware, science, and salaries on STS-106 (Hammond et al., 2000). A large determinant of the controlled cost was the hardware, the BioServe GAP, which was simple and very well-validated in space and on the ground. Prior flight experience grandfathered it through many of the flight safety testing requirements (Hoehn et al., 2004). This proved that space science can be done on a modest budget. There are other examples. We have provided plates of yeast spotted on plates to NASA for no cost to validate the Spectrum hardware just installed on the International Space Station. The cost to us was minimal. NASA Ames Research Center has been building and flying 10 × 10 × 10-cm CubeSats at modest cost since 2006 (Hunter and Korsmeyer, 2015).
It should not be forgotten, however, that NASA has consistently and reliably provided the huge launch costs to move the hardware into space—in the order of a few tens of thousands of dollars per pound. New launch systems partially mitigate these costs by reuse of many rocket elements. To this end, we use light materials wherever possible, facilitating the two-pronged approach of lower launch cost and lighter payloads.
Moonshot was designed to support the growth of light-dependent algae. It is scalable equipment that can serve as a blueprint for other incubators, such as those requiring control of CO2, temperature, and humidity and holding more plates at the same time. The hardware is light enough to be easily returned to Earth for recovery of performance metrics and samples. Alternatively, the SIM card and biological samples alone can be harvested and returned to Earth for analysis. In this case, the hardware can be held on a space vehicle for repeated use or jettisoned to burn up on re-entry.
NASA's stated purpose is to hand off spaceflight vehicles and scientific exploitation of the International Space Station to commercial groups. To embed biotechnology as a commercially viable entity, while enhancing utilization of the spaceflight revolution, we need to be cost-effective, while understanding the biotechnology and hardware development tradeoffs such as materials and electronic compatibility with space flight systems.
The study of algae in space is driven by the industrial-scale applications relevant to spaceflight and its ground-based spinoffs, most commonly embodied in the form of seaweed. Rockweed, a brown macroalgae, is harvested from the sea and processed into liquid fertilizer for crops and dry ingredients to feed farm animals (Hafting et al., 2015). We have all eaten carrageenan extracted from the red seaweed known as Irish moss, a phycocolloid that is the basis for junket, pie fillings, cake mixes, Twinkies, ice cream and processed meats (McKim et al., 2019). Another red seaweed product, agar, is ubiquitous in microbiology laboratories worldwide (Lee et al., 2017). The options are endless: burnt algae can replace wood (sparing forests) to provide potash for glass manufacture, and algae can be incorporated into products from plastics to biofuels (Khan et al., 2018). Algae products may have the potential to be a critical tool in maintaining long-term manned spaceflight. Algae products simply need water, heat, sunlight, and a simple carbon source (Matula and Nabity, 2019). In return, algae can provide biologicals and nutritional supplements, as well as fuel—all of which are critical spaceflight provisions. Powerful genetic tools are available to understand the survival of C. reinhardtii mutants during photosynthesis and many other biological processes (Li et al., 2016). Specifically, a large pool of mapped deletion strains allows sophisticated genome-wide analysis of the pathways of algae survival (Li et al., 2016). Insight into the genetic traits that confer increased survival in the environment of microgravity and deep space radiation can guide the development of “space-hardy” strains of Chlamydomonas to accompany us in our exploration of space (Figure 7). Liquid cultures of these “space-hardy” strains of Chlamydomonas could be propagated for production purposes using NASA growth hardware, such as VEGGIE (https://www.nasa.gov/content/veggie-plant-growth-system-activated-on-international-space-station) being developed for plants (Zhang et al., 2020).
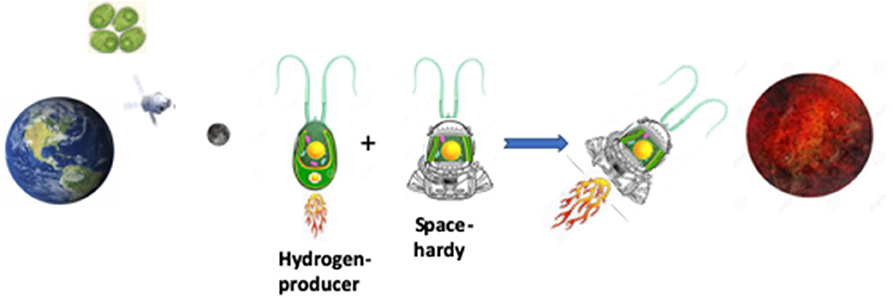
Figure 7. The goal of the Moonshot payload experiments of Artemis-1 is to identify space-hardy strains of Chlamydomonas that can be used to generate space-hardy hydrogen-producing strains of Chlamydomonas for use as fuel for space travel.
The benefits of the use of space radiation environment and research in the field of astrobiology conducted in space, experimental space facilities, and hardware have been reviewed, presented, and discussed (Cottin et al., 2017). The Moonshot hardware is designed to reduce the cost and complexity of space flight experiments. As it is robust and self-supporting, not requiring external power or other resources, the Moonshot hardware can also be applied for ground-based studies. Moonshot is aligned for use in the study of astrobiology. To quote Martins et al., “the ways the Earth is used by astrobiologists: (i) by conducting planetary field analog studies…, (ii) by conducting planetary field analog studies…, and (iii) by exposing terrestrial samples to simulated space or planetary environments and producing a sample analog to investigate changes in minerals, biosignatures, and microorganisms” (Martins et al., 2017).
Conclusions
The current report shows that one can build, validate, and utilize space flight hardware at a very reasonable cost. You just need to understand materials, cell biology, and the policies and procedures of your integration and launch partners.
Although the results of the Artemis-1 experiment are to be established, the closest guide is that the techniques to be used in this proposal have already been used to identify genes required by yeast to survive in space using genome-wide yeast deletion collections cultured during spaceflight for 10 days on the International Space Station (Nislow et al., 2015). Even though those studies were carried out within the protection of the Van Allen belt, it was apparent that DNA and RNA damage repair was critical for yeast survival.
We conclude that a multipurpose, flight-certified hardware can be designed, manufactured, and validated with unprecedented timing and cost efficiency.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
TH managed the project, collaborated on study design, and performed all the flow cytometry. PA managed the Chlamydomonas cultures and performed the spectrophotometry. HW and JR designed and fabricated the flight hardware and prototypes. CN and GG optimized the Chlamydomonas culture conditions and provided critical commentary on the manuscript. HB collaborated on study design, acted as the data warden, storing and verifying all data, and performed all the statistics in a blinded manner. All authors agree to be accountable for the content of the work and all contributed prose and/or figures to the manuscript.
Funding
NASA Grant 80NSSC19K0706 under solicitation NNH18ZTT001N-EM1 to the Institute for Medical Research supported these studies.
Conflict of Interest
HW and JR are employed by the company Bionetics Corporation.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer DF declared a shared affiliation with one of the authors, TH, to the handling editor at time of review.
Acknowledgments
This material is the result of work supported with resources and the use of facilities at the Durham Veterans Affairs Health Care System and Duke University School of Medicine. Contents do not represent the views of the Department of Veterans Affairs or the United States of America. CN and GG are both supported as Tier 1 Canadian Research Chairs. We want to thank Dr. Jens Hauslage at the Institut für Luf- und Raumfahrtmedizin for hardware design conceptualization.
References
Alfred, S. E., Surendra, A., Le, C., Lin, K., Mok, A., Wallace, I. M., et al. (2012). A phenotypic screening platform to identify small molecule modulators of Chlamydomonas reinhardtii growth, motility and photosynthesis. Genome Biol. 13:R105. doi: 10.1186/gb-2012-13-11-r105
Batyrova, K., and Hallenbeck, P. C. (2017). Hydrogen production by a Chlamydomonas reinhardtii strain with inducible expression of photosystem II. Int. J. Mol. Sci. 18:647. doi: 10.3390/ijms18030647
Blaby, I. K., Blaby-Haas, C. E., Tourasse, N., Hom, E. F., Lopez, D., Aksoy, M., et al. (2014). The Chlamydomonas genome project: a decade on. Trends Plant Sci. 19, 672–680. doi: 10.1016/j.tplants.2014.05.008
Cottin, H., Kotler, J. M., Billi, D., Cockell, C., Demets, R., Ehrenfreund, P., et al. (2017). Space as a tool for astrobiology: review and recommendations for experimentations in earth orbit and beyond. Space Sci. Rev. 209, 83–181. doi: 10.1007/s11214-017-0365-5
Dasgupta, C. N., Gilbert, J. J., Lindblad, P., Heidorn, T., Borgvang, S. A., Skjanes, K., et al. (2010). Recent trends on the development of photobiological processes and photobioreactors for the improvement of hydrogen production. Int. J. Hydrogen Energy 35, 10218–10238. doi: 10.1016/j.ijhydene.2010.06.029
Gaffron, H., and Rubin, J. (1942). Fermentative and photochemical production of hydrogen in algae. J. Gen. Physiol. 26, 219–240. doi: 10.1085/jgp.26.2.219
Gallaher, S. D., Fitz-Gibbon, S. T., Glaesener, A. G., Pellegrini, M., and Merchant, S. S. (2015). Chlamydomonas genome resource for laboratory strains reveals a mosaic of sequence variation, identifies true strain histories, and enables strain-specific studies. Plant Cell 27, 2335–2352. doi: 10.1105/tpc.15.00508
Grobbelaar, J. U. (2010). Microalgal biomass production: challenges and realities. Photosyn. Res. 106, 135–144. doi: 10.1007/s11120-010-9573-5
Hafting, J. T., Craigie, J. S., Stengel, D. B., Loureiro, R. R., Buschmann, A. H., Yarish, C., et al. (2015). Prospects and challenges for industrial production of seaweed bioactives. J. Phycol. 51, 821–837. doi: 10.1111/jpy.12326
Halim, R., and Webley, P. A. (2015). Nile red staining for oil determination in microalgal cells: a new insight through statistical modelling. Int. J. Chem. Eng. 2015:695061. doi: 10.1155/2015/695061
Hammond, T. G., Benes, E., O'Reilly, K. C., Wolf, D. A., Linnehan, R. M., Taher, A., et al. (2000). Mechanical culture conditions effect gene expression: gravity-induced changes on the space shuttle. Physiol. Genomics 3, 163–173. doi: 10.1152/physiolgenomics.2000.3.3.163
Hammond, T. G., Lewis, F. C., Goodwin, T. J., Linnehan, R. M., Wolf, D. A., Hire, K. P., et al. (1999). Gene expression in space. Nat. Med. 5:359. doi: 10.1038/7331
Hoehn, A., Klaus, D. M., and Stodieck, L. S. (2004). A modular suite of hardware enabling spaceflight cell culture research. J. Gravit. Physiol. 11, 39–49.
Hunter, R., and Korsmeyer, D. J. (2015). A Review of NASA Ames CubeSat Program. NTRS - NASA Technical Reports Server. Available online at: https://ntrs.nasa.gov/citations/20160007401 (accessed September 6, 2016).
Khan, M. I., Shin, J. H., and Kim, J. D. (2018). The promising future of microalgae: current status, challenges, and optimization of a sustainable and renewable industry for biofuels, feed, and other products. Microb. Cell Fact. 17:36. doi: 10.1186/s12934-018-0879-x
Lee, W. K., Lim, Y. Y., Leow, A. T., Namasivayam, P., Ong Abdullah, J., and Ho, C. L. (2017). Biosynthesis of agar in red seaweeds: a review. Carbohydr. Polym. 164, 23–30. doi: 10.1016/j.carbpol.2017.01.078
Li, X., Zhang, R., Patena, W., Gang, S. S., Blum, S. R., Ivanova, N., et al. (2016). An indexed, mapped mutant library enables reverse genetics studies of biological processes in Chlamydomonas reinhardtii. Plant Cell 28, 367–387. doi: 10.1105/tpc.15.00465
Martins, Z., Cottin, H., Kotler, J. M., Carrasco, N., Cockell, C. S., de la Torre Noetzel, R., et al. (2017). Earth as a tool for astrobiology—a European Perspective. Space Sci. Rev. 209, 43–81. doi: 10.1007/s11214-017-0369-1
Matula, E. E., and Nabity, J. A. (2019). Failure modes, causes, and effects of algal photobioreactors used to control a spacecraft environment. Life Sci. Space Res. 20, 35–52. doi: 10.1016/j.lssr.2018.12.001
McKim, J. M., Willoughby, J. A. Sr, Blakemore, W. R., and Weiner, M. L. (2019). Clarifying the confusion between poligeenan, degraded carrageenan, and carrageenan: a review of the chemistry, nomenclature, and in vivo toxicology by the oral route. Crit. Rev. Food Sci. Nutr. 59, 3054–3073. doi: 10.1080/10408398.2018.1481822
Mergenhagen, D., and Mergenhagen, E. (1987). The biological clock of Chlamydomonas reinhardii in space. Eur. J. Cell Biol. 43, 203–207.
Mergenhagen, D., and Mergenhagen, E. (1989). The expression of a circadian rhythm in two strains of Chlamydomonas reinhardii in space. Adv. Space Res. 9, 261–270. doi: 10.1016/0273-1177(89)90082-3
Nakamura, S., Itoh, S., and Kuroiwa, T. (1986). Behavior of chloroplast nucleus during chloroplast development and degeneration in Chlamydomonas reinhardii. Plant Cell Physiol. 27, 775–784. doi: 10.1093/oxfordjournals.pcp.a077163
Nislow, C., Lee, A. Y., Allen, P. L., Giaever, G., Smith, A., Gebbia, M., et al. (2015). Genes required for survival in microgravity revealed by genome-wide yeast deletion collections cultured during spaceflight. Biomed. Res. Int. 2015:976458. doi: 10.1155/2015/976458
Oldenhof, H., Zachleder, V., and Van Den Ende, H. (2006). Blue- and red-light regulation of the cell cycle in Chlamydomonas reinhardtii (Chlorophyta). J. Phycol. 41, 313–320. doi: 10.1080/09670260600699920
Scaife, M. A., Nguyen, G. T., Rico, J., Lambert, D., Helliwell, K. E., and Smith, A. G. (2015). Establishing Chlamydomonas reinhardtii as an industrial biotechnology host. Plant J. 82, 532–546. doi: 10.1111/tpj.12781
Scoma, A., Krawietz, D., Faraloni, C., Giannelli, L., Happe, T., and Torzillo, G. (2012). Sustained H(2) production in a Chlamydomonas reinhardtii D1 protein mutant. J. Biotechnol. 157, 613–619. doi: 10.1016/j.jbiotec.2011.06.019
Terashima, M., Freeman, E. S., Jinkerson, R. E., and Jonikas, M. C. (2015). A fluorescence-activated cell sorting-based strategy for rapid isolation of high-lipid Chlamydomonas mutants. Plant J. 81, 147–159. doi: 10.1111/tpj.12682
Vijayaraghavan, K., Karthik, R., and Nalini, S. P. K. (2009). Hydrogen production by Chlamydomonas reinhardtii under light driven sulfur deprived condition. Int. J. Hydrogen Energy 34, 7964–7970. doi: 10.1016/j.ijhydene.2009.08.010
Keywords: Artemis-1, flight hardware, Chlamydomonas, microgravity, cosmic radiation, biofuels
Citation: Hammond TG, Allen PL, Wells HW, Russick JM, Nislow C, Giaever G and Birdsall HH (2020) Moonshot: Affordable, Simple, Flight Hardware for the Artemis-1 Mission and Beyond. Front. Space Technol. 1:593523. doi: 10.3389/frspt.2020.593523
Received: 10 August 2020; Accepted: 12 October 2020;
Published: 11 November 2020.
Edited by:
Cosimo Buffone, Tianjin University of Commerce, ChinaReviewed by:
Dahlene N. Fusco, Tulane University, United StatesFrancesco Romanò, Arts et Métiers ParisTech, France
Anselmo Cecere, University of Naples Federico II, Italy
Copyright © 2020 Hammond, Allen, Wells, Russick, Nislow, Giaever and Birdsall. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Timothy G. Hammond, aGFtbW9uZG91dG9mb2ZmaWNlQHlhaG9vLmNvbQ==
 Timothy G. Hammond
Timothy G. Hammond Patricia L. Allen5
Patricia L. Allen5 Corey Nislow
Corey Nislow Holly H. Birdsall
Holly H. Birdsall