- 1Soil Resources, Department of Environmental Systems Science, Swiss Federal Institute of Technology (ETH) Zurich, Zurich, Switzerland
- 2Division of Terrestrial Ecosystem Research, Department of Microbiology and Ecosystem Science, Center of Microbiology and Environmental Systems Science, Vienna, Austria
- 3Department of Environment and Biodiversity, Paris Lodron University of Salzburg, Salzburg, Austria
- 4Environmental Genomics and Systems Biology, Lawrence Berkeley National Laboratory, Berkeley, CA, United States
Introduction: Organic phosphorus (Po) compounds constitute an important pool in soil P cycling, but their decomposition dynamics are poorly understood. Further, it has never been directly tested whether low molecular weight Po compounds are taken up by soil microbes in an intact form, which reduces the dependence of their P acquisition on extracellular phosphatases.
Methods: We investigated the short-term fate (24 h) of five 33P-labelled Po compounds (teichoic acids, phospholipids, DNA, RNA and soluble organophosphates) and 33P-labelled inorganic P (Pi) in two soils.
Results: We found indications that soil microbial breakdown of phosphodiesters was limited by the depolymerization step, and that direct microbial uptake of Po occurred to a substantial extent.
Discussion: We postulate a trade-off between direct Po uptake and complete extracellular Po mineralization. These findings have profound consequences for our understanding of microbial P cycling in soils.
1. Introduction
Phosphorus (P) is a key constituent of the cellular machinery across all life forms, and it is increasingly recognized that the P cycle has important implications for global carbon (C) and nitrogen (N) cycling, as the dynamics of these cycles are closely interlinked (1). In contrast to the high demand for P by organisms (2–4), the bioavailability of P in soils - the prime source of P in terrestrial ecosystems – can be low, thereby often creating a situation of P limitation in ecosystems. The amount of bioavailable P in soils thus represents a crucial bottleneck in P cycling, as it sets a frame for how much P can enter the biotic component of the terrestrial P cycle.
Bioavailable P in soils is considered to be mainly supplied in inorganic form (Pi) by desorption/dissolution processes from the solid phase and by decomposition and mineralization of organic P (Po) compounds, with their relative contribution strongly varying between ecosystems (5, 6). Organic P constitutes 20-80% of total soil P (7, 8), and is therefore an essential component in the terrestrial P cycle. The pool of Po consists of a highly heterogeneous group of compounds, such as phosphate monoesters (phosphomonoesters), phosphate diesters (phosphodiesters) and organic phosphonates (9). Moreover, a range of organic phosphorus compounds in soil still remains uncharacterized (10, 11). A considerable fraction of extractable soil Po has been shown to consist of large molecules (10), and to be closely associated with large structures of soil organic matter (11). Different Po compounds have different chemical properties, pathways of decomposition and therefore degrees of bioavailability to soil microbes. Microbial turnover and root cell lysis release especially phosphodiesters into soils, as those are the major P components of all living microbial and plant cells, including nucleic acids (DNA and RNA), phospholipids and teichoic acids (12–14). Different from previous estimates of the predominance of phosphomonoesters (e.g. phytates – inositol polyphosphates) in soil Po, 2D-NMR and back-calculation of the in situ soil Po speciation showed that phosphodiesters like RNA (41%), phospholipids (28%) and DNA (22%) dominate the soil Po pool (15).
Major strides have been recently made towards a better understanding of soil P speciation, but also in soil Pi dynamics (16–19) and in linking microbial community structure with soil P availability (20–22). In contrast to these advances, much less is known about (i) the pathways and controls of soil Po decomposition and Po mineralization and (ii) whether or not microbes can directly utilize low molecular weight Po compounds (23, 24). The Po decomposition process involves cleavage of the compounds by extracellular phosphatases, which are produced by plants and microbes (23). The cleavage of phosphodiesters is mediated by specific phosphodiesterases (25–27), and the final dephosphorylation of the formed phosphomonoesters to free Pi is performed by acid and alkaline phosphomonoesterases (28). If Pi is the main form acquired by all organisms to meet their P demand and given that phosphomonoesterases are abundant and ubiquitous in soils, soil P cycling might be rate-limited by the depolymerization and cleavage of diesters to smaller oligomers and to monomers (phosphomonoesters), while dephosphorylation of phosphomonoesters to Pi is rapid (Figure 1). In this case, the initial breakdown step may determine the rate of Po mineralization. An analogous situation is known from soil N transformation processes, where depolymerization of high molecular weight organic N compounds such as proteins, chitin or peptidoglycan represents the bottleneck of soil N cycling (29–32).
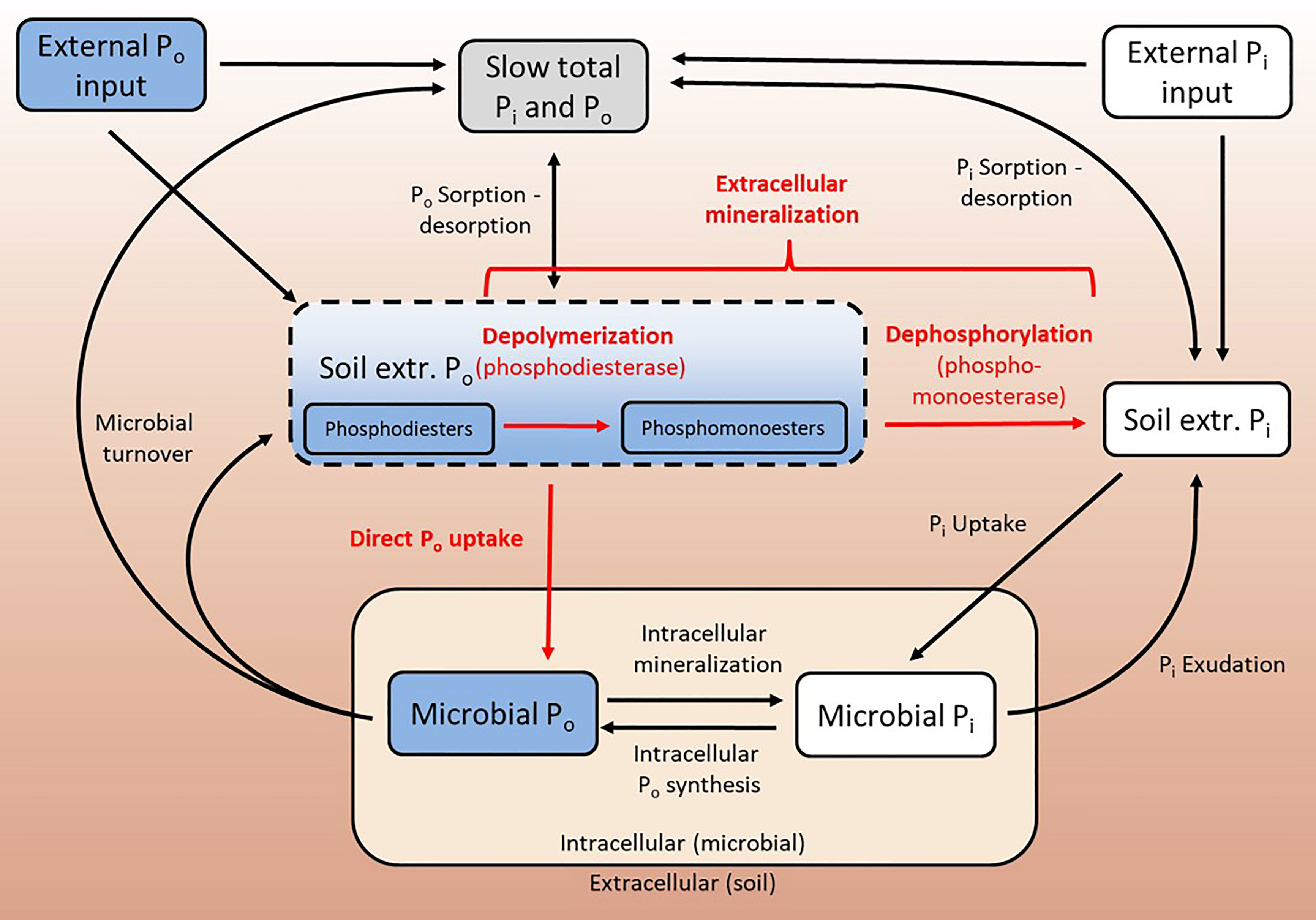
Figure 1 Schematic representation of soil P cycle processes. Bio-available Po is mineralized extracellularly, where phosphodiesters are depolymerized by phosphodiesterases, yielding smaller phosphomonoesters. Phosphomonoesters can be dephosphorylated by phosphomonoesterases, and the resulting bio-available Pi can be taken up by the microbial community. In microbes, Po compounds are synthesized and mineralized. Pi can be exuded by microbes, and Po is mainly released into soil via microbial turnover. Soil Pi (and Po) are subject to sorption and desorption processes. The current paradigm is that bio-available Po cannot be directly taken up by soil microbes. Fluxes in red are in the focus of this study.
Tracing the pathways and measuring the gross rates of Po decomposition requires homogeneously radio-isotopically labelled (33P or 32P) and purified Po compounds, which are rarely (nucleotides, glucose-6-phosphate) or not at all commercially available and very expensive to obtain by custom synthesis. To our knowledge, few studies have therefore directly traced the decomposition and fate of radiotracer-labelled organic P compounds in soils. To date, only radio-P labelled RNA (33), a cytosine dinucleotide (34), phytate (35), glucose-6-P (36, 37) and bacterial or fungal necromass (38, 39) have been studied. Further, except for Harrison (33), studies have been carried out with only one Po compound per study, thus impeding comparative assessments of decomposition dynamics of various Po forms in different soils. Actual decomposition dynamics of most Po compounds in soil and their controls have therefore remained largely elusive.
The current paradigm in soil microbial ecology is that microbes fully meet their P demand by the uptake of Pi. Utilization of Po therefore only occurs after complete extracellular mineralization of Po through phosphatases to Pi (Figure 1) (23, 24, 40). For this purpose, soil microbes produce a wide range of phosphomonoesterases (acid and alkaline phosphatases, phytases, phosphonatases) and phosphodiesterases (nucleases, phospholipases, glycerophosphodiesterases). However, some of the responsible genes are restricted to smaller phylogenetic groups while others are relatively ubiquitous (22, 41). Transporters for Pi-uptake are ubiquitous and well-studied in bacteria (39) and fungi (42). Pi is efficiently taken up against steep concentration gradients by high affinity phosphate:H+ symporters (PHT protein family). In contrast, transporters for Po compounds are less thoroughly studied. While some Po uptake systems were only recently discovered (43), others are well understood in model organisms (44). For example, uptake systems for glycerol-3-phosphate (GlpT), hexose-6-phosphate (UhpT) and phosphoglycerate (PgpT) belonging to the organophosphate:phosphate antiporter (OPA) family are well known (41, 45), and have been characterized in several species of bacteria and eukaryotes (39, 46, 47). Despite awareness of the existence of microbial Po transporters (39, 46, 48), we lack knowledge about the occurrence and potential ecological relevance of direct microbial Po uptake in the soil environment (41). Direct uptake of Po compounds could shortcut the extracellular mineralization step and potentially directly fuel salvage pathways or precede intracellular mineralization. Microbial use of Po would therefore not be exclusively regulated by the production of extracellular enzymes, but rather be subject to metabolic controls that involve the balance between extracellular Po mineralization and direct uptake.
In this study, we therefore assessed the decomposition kinetics of five 33P-labelled Po compounds and addressed the following research questions (1): Is the mineralization rate of phosphodiester compounds (teichoic acids, phospholipids, DNA, RNA) limited by the initial depolymerization and cleavage steps to monoesters, or by dephosphorylation of the intermediate phosphomonoester to free Pi? (2) Do soil microbes directly take up and utilize Po in an intact form or only after complete mineralization to Pi? To tackle these questions, we developed a novel protocol to produce 33P-labelled biochemical fractions that are relevant constituents of the soil Po pool: cell walls (containing wall teichoic acids, TA), lipids including phospholipids (PL), DNA, RNA and soluble organophosphates (SOP). We added these five 33Po-fractions in purified form as well as 33Pi to two soils (temperate cropland and temperate pasture; Table 1) and traced the radioisotope with high temporal resolution over a time span of 24 h into the extractable Po and Pi pools of soils and microbes, utilizing sequential P extraction and isobutanol fractionation of Po and Pi.
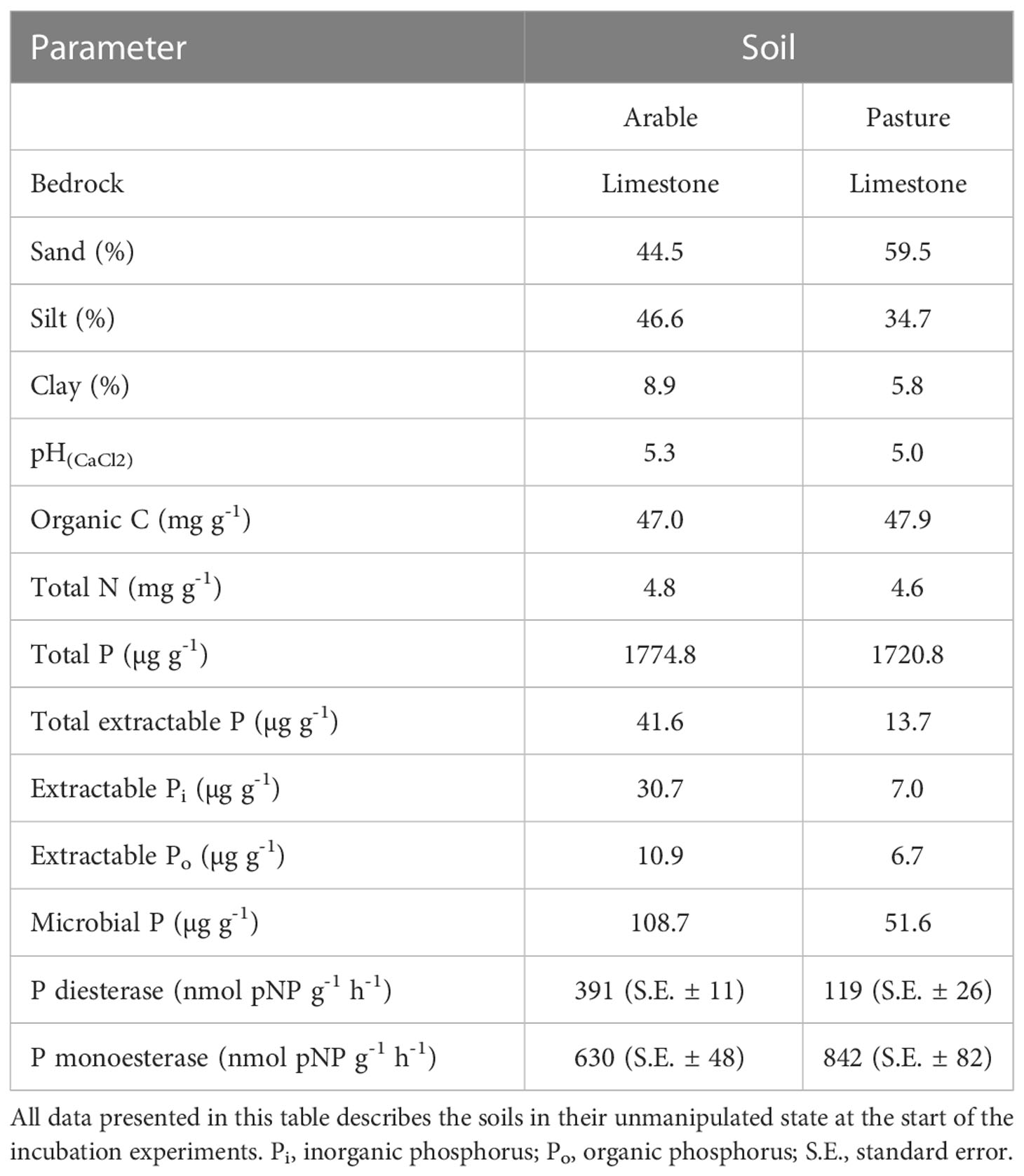
Table 1 Physicochemical properties and potential extracellular enzyme activities of the two soils used in this study.
2. Materials and methods
2.1. Soil characterization and experimental design
Two agricultural topsoils (0-15 cm, Luvisols on limestone) were collected from an arable field and a pasture site grazed by cattle at Moarhof [Trautenfels, Styria, Austria; 47°31’04’’ N, 14°04’31’’ E; 640 m a.s.l.; MAT: 7°C; MAP: 1230 mm; see (49)] in September 2016. We selected the arable and pasture soils under the assumption that contrasting management should lead to major differences in organic vs. inorganic P cycling. Phosphorus cycling in the pasture should be more dominated by organic P, through intense topsoil rooting and high microbial activity, while cycling in the cropland should be dominated by inorganic P through decadal mineral P fertilizer amendment. Soils were sieved to 2 mm and stored air dried, and two weeks prior to the experiments they were rewetted to 60% of their water holding capacity and kept at room temperature (25°C). Sand content was determined by wet sieving to 0.05 mm, after suspending the soil in 5% (w/v) sodium hexametaphosphate, clay content was calculated by using a pipette method (50) and silt content was calculated by difference. Soil pH was measured in 10 mM CaCl2 in a 1:5 (w/v) soil slurry using an ISFET electrode (Sentron, Netherlands). Soil organic C and total soil N were measured by elemental analyser (CE1110, Thermo Fisher), after removal of carbonates by HCl treatment. Potential activities of extracellular phosphomonoesterases and -diesterases were measured one time after two weeks of pre-incubation, directly before the start of the experiment. The measurements were done photometrically in triplicates using p-nitrophenyl (pNP) coupled substrates (51), at native soil pH using a modified universal buffer (MUB, exact description in the reference) (52). Briefly, 1.5 g of soil were dissolved in 40 ml of MUB and dispersed by ultrasonication with an energy input of 8.5 J ml-1. Subsequently, the soil slurry was mixed 1:1 (v/v) with 5 mM substrate solutions (pNP-phosphate for phosphomonoesterase, and bis-pNP-phosphate for phosphodiesterase, in MUB) in Eppendorf tubes, using wide pore pipette tips. After incubation for 1 h at 20°C under continuous shaking, the tubes were centrifuged at 10 000 g, and the supernatant was mixed with 1 M NaOH (10:1 (v/v)) to terminate enzyme activity and start the color reaction. Samples were diluted and transferred to a 96-well microtiter plate, and in addition a standard row was prepared by a 1:2 dilution series of 1 mM p-nitrophenol, as well as substrate controls without soil slurry. Absorbance of all wells was read at 410 nm with a microplate photometer (Tecan Infinite M200, Tecan Trading AG). Values were corrected for substrate controls, and potential extracellular enzyme activities were expressed as nmol pNP g-1 h-1. Soil total P was determined colorimetrically with the malachite-green method (53) after ignition (450°C, 5 h) of 0.5 g dry soil and subsequent extraction with 20 mL 0.5 M H2SO4 for 16 h. Soil Pi was determined on unignited soils using the same protocol, and total Po by difference. In brief, the malachite green method was applied to quantify Pi in soil extracts (after dilution with deionized water) as follows: 200 µL of the respective extracted solution were pipetted into a microtiter plate. First, 40 µL of malachite green reagent A (16.8 ml of 95 – 97% H2SO4 and 1.76 g (NH4)6Mo7O24 4H2O, filled up to 100 ml with H2O) were added, and after 10 min 40 µL of reagent B (0.875 g polyvinyl alcohol (MW = 72 000 g mol-1) with 87 mg malachite green oxalate in 250 ml H2O) were added. After 45 min of incubation, the absorbance was read at 610 nm with a microplate photometer. A 1:2 dilution series of 32 µM KH2PO4 was used as concentration standards. Total dissolved P (TDP) and Pi were measured in 0.5 M NaHCO3 extracts after acidification with 2.75 M H2SO4 (10% (v/v)), with or without acid persulfate digestion with 5% (v/v) acidic persulfate (0.5 M H2SO4 + 0.5 M Na2S2O8, autoclaved at 121°C for 60 min) (54), using the malachite green method. Microbial P was measured using chloroform (CHCl3) fumigation (55). Soil was incubated in CHCl3 saturated atmosphere for 48 h to lyse microbial cells. After fumigation, acid persulfate-digestion as described above was performed on 0.5 M NaHCO3 extracts, yielding TDP of the soil plus the microbial biomass. Pi concentration was measured colorimetrically by the malachite green method. Microbial P was calculated as the difference in TDP between fumigated and unfumigated samples, applying an extraction factor kEP of 0.4 based on an established literature value (55). Soil parameters are given in Table 1.
2.2. Production of 33P-labelled organic compounds
To produce and purify 33P-labelled organic compounds, the gram-positive bacterium Bacillus subtilis was grown in liquid medium amended with 33P-labelled orthophosphoric acid. We then applied optimized biochemical fractionation protocols to fractionate the 33P enriched culture into the five biochemical compound groups for use as substrates in the incubation experiment: cell walls containing wall teichoic acids (TA, glycerophosphate diester polymers), lipids including phospholipids (PL), DNA, RNA and soluble organophosphates (SOP). We did not investigate the fate of inositol phosphates, which are major P storage compounds in higher plants. Inositol phosphates, particularly myo-inositol hexakisphosphate (IP6), often accumulate in soils and constitute a large proportion of the soil phosphomonoester Po pool, due to their resistance to mineralization and strong sorption (56, 57). Though microbes have been reported to produce IP6, the production of 33P-labelled IP6 by plants or microbes remains very complicated, and we therefore did not include this compound class in this study. To keep losses of 33P-substrate by radioactive decay as low as possible, the entire labelling and biochemical fractionation procedure was performed twice, directly before each of the two incubation experiments with either soil. A detailed description of the enrichment and fractionation protocol is provided in the supplementary material (Supplementary S1.1 Production of 33P-labelled organic compounds).
2.3. Incubation experiment
We performed the incubation experiment twice, each time for one soil (i.e., arable and pasture). Each of the five 33P-labelled organic substrates plus inorganic 33P (Pi) were applied individually to the soils. The 33P activity that was applied to each sample ranged between 1.5 and 44 kBq, depending on substrates and soils (Table 1). A schematic overview of the workflow of the incubation experiments is given in Figure S1.
Every soil x substrate combination was incubated in triplicates in 50 mL polypropylene tubes at room temperature, and incubations were stopped at seven time points (0, 0.5, 1, 2, 4, 10, 24 h). All substrates were applied in 200 µL deionized water on 2 g fresh soil equilibrated at 60% WHC, except for PL. Because of its amphiphilic character, the PL fraction was first dissolved in chloroform (CHCl3) and added onto 180 mg acid washed glass beads (≤106 µm) in small aliquots. After the evaporation of CHCl3, the beads were mixed with the soil aliquots and 200 µL deionized water was added. At the end of the incubations, we performed a sequential P extraction with 0.5 M NaHCO3 to extract available soil P (Psoil) and subsequently microbial P (Pmic). The detailed protocol is given in the supplementary material (Supplementary S1.2 Sequential extraction). To separate Pi and Po, isobutanol fractionation was conducted as described previously (58, 59), on aliquots of Psoil and Pmic, in which Pi binds to acidified molybdate which then is partitioned into the isobutanol phase, while Po remains in the aqueous phase. The solution ratios applied were 1.5 mL extract plus 1.5 mL acidified molybdate, 3 mL isobutanol and 3 mL Milli-Q water. In all P fractions (Po and Pi of each, Psoil and Pmic) as well as in unfractionated Psoil and Pmic extracts after acidification by addition of 10% (v/v) 2.75 M H2SO4, 33P was quantified via liquid scintillation counting after addition of Ultima Gold Scintillation cocktail (10:1 and 16:1 liquid scintillation cocktail to sample ratio for isobutanol and aqueous samples, respectively) using a Tri-Carb1600-TR counter (Packard, PerkinElmer). To separate high molecular weight (HMW) and low molecular weight (LMW) fractions in the nucleic acid treatments, 1.5 mL of the Psoil extract was centrifuged through Amicon Ultra - 2 mL Centrifugal Filters (3 kDa molecular weight cut-off) for 90 min (6500 g, 15°C). The ultrafiltrate was then readjusted to 1.5 mL and rejoined the common workflow before isobutanol fractionation, whereas the concentrate was directly measured by liquid scintillation counting. Technical recoveries of added 33P without soil were run in triplicates for all substrates (200 µL organic and inorganic 33P substrates added to 20 mL 0.5 M NaHCO3), in order to assess their behavior during isobutanol fractionation and to measure recoveries by 0.5 M NaHCO3 extraction. To avoid potential contamination of Pi in the isobutanol phase with extracted phospholipids, 2.5 mL of Psoil and Pmic of the PL treatment were briefly mixed with 1 mL of CHCl3 and phases were separated before isobutanol fractionation of the aqueous phase.
Concentrations of Pi were measured colorimetrically in the isobutanol Pi fractions by applying the molybdenum-blue method (58), and in Psoil and Pmic after acidification with 2.75 M H2SO4 (10% (v/v)) by using the malachite-green method (53). Total dissolved P (TDP) was also quantified by the malachite-green method after digestion of Po in Psoil and Pmic with 5% (v/v) acidic persulfate (0.5 M H2SO4 + 0.5 M Na2S2O8, autoclaved at 121°C for 60 min). Po concentrations were then calculated as difference between TDP and Pi.
2.4. Calculations and statistical analysis
All measured 33P activities were corrected for radioactive decay. Technical recoveries showed that large organic molecules do not partition into the aqueous Po fraction during isobutanol fractionation, but are precipitated at the interlayer between the two phases. The aqueous fraction of the isobutanol fractionation therefore does not represent the entire 33Po fraction. For this reason, in the treatments with TA, PL and SOP, we calculated the activity of 33Po in soil as 33Psoil (i.e. total 33P in Psoil - extract before isobutanol fractionation) minus 33Pi, and similarly the activity of 33Po in the microbial pool as 33Pmic (i.e. total 33P in Pmic extract before isobutanol fractionation) minus 33Pi. In the treatment with inorganic P, the 33P activities in the Pi and Po fraction of soil were proportionally fitted to Psoil (i.e. total 33P in Psoil extract before isobutanol fractionation). In the nucleic acid treatments, the 33P activities in the Pi and Po fraction of soil were proportionally fitted to Psoil minus P in the HMW fraction (i.e. total 33P in Psoil - extract before isobutanol fractionation minus 33P in HMW-NA fraction). This is because 33P in the concentrate of the ultrafiltration was quantified directly without undergoing isobutanol fractionation, and therefore was not affected by the bias arising through interlayer precipitation. 33P recoveries in the different organic and inorganic fractions of Psoil and Pmic were calculated relative to the decay-adjusted technical recoveries, and specific activities (SA) of the Po pools were calculated as Bq µg-1 Po-P using the Po concentrations obtained with acidic persulfate digestion and subsequent malachite-green quantification.
We calculated the degree of the mineralization of 33P labelled Po-substrates as the percentage of soil extractable 33P present in inorganic form. This calculation was done for each time point and all five 33P-labelled organic substrates and is subsequently referred to as the degree of substrate mineralization. This measure informs on the form (Po vs. Pi) of available P (derived from the 33P-labelled substrate) in the extractable soil pool.
We estimated the biosynthesis of 33Po from 33Pi taken up for all organic treatments, based on our observations of Po biosynthesis in the Pi treatments. For this, we assumed that the microbial communities of each soil allocate a fixed percentage of the available amendment-derived Pi into intracellular Po biosynthesis. Based on the values from the Pi treatment, we calculated this percentage for each soil and time point as the microbial 33Po activity over the soil 33Pi activity *100. To calculate the activities of 33P invested into biosynthesis for each soil, time point and treatment, we multiplied the observed soil 33Pi activities in the organic treatments with this factor. Finally, microbial 33Po was corrected for biosynthesis by subtraction of this activity, and to obtain 33Po:33Pi ratios of cumulative 33P uptake, the same activity was added to microbial 33Pi. By basing these calculations for each treatment and time point on the actual measured soil 33Pi activities, the estimate of Po biosynthesis from Pi with this method accounts for Pi sorption over time, extracellular substrate mineralization and Pi contaminations of the added substrates.
All statistical analyses were performed with R statistical software (R 3.5.2). Phosphatase activities and 33Pi recoveries (Pi treatment, at 0 h and 10 h autoclaved) were compared between the soils with either Student`s t-Test or Welch`s t-Test after testing homogeneity of variance with the Bartlett test. After testing the assumption of variance homogeneity with Levene’s tests, ANOVA was performed to test whether soil Pi or TDP pools changed after substrate addition. Post-hoc-tests (Fisher’s least significant difference test) were applied to test for pair-wise differences between the treatments and native pool sizes. After testing the assumption of variance homogeneity with Levene’s tests, two-way-ANOVA was applied to test for differences in extraction efficiencies between the two soils and the six substrates. Linear regression was applied to the 33Po:33Pi ratios of microbes versus the degree of substrate mineralization of extractable soil P for all organic substrates in both soils. The assumption of heteroscedasticity of the models was tested with the Breusch-Pagan test, and in case of violation weighted least squares regression was applied, using the inverse of the fitted values as weights. We tested for differences in the dynamics of the labelled substrates between the soils, and for changes of 33P recovery over time. For this, we applied factorial repeated measures ANOVA to the 33P recoveries of all pools, using the R-package “ez”. The assumption of sphericity was tested with the Mauchley’s test, and in case of violation Greenhouse-Geisser (GG) adjustment was applied. Significant outliers from the time series were identified with Grubb`s test.
3. Results
3.1. General observations about the experimental design
The addition of organic 33P substrates only caused negligible changes in extractable soil Pi (-8.8% to + 6.3%) and Po (+ 0.8% to + 77.3%) concentrations relative to the Pi treatment (no ‘spike’, Table S2). This means that soil P concentrations of the treatments were similar to natural in situ conditions. More specifically, after 33Po substrate additions (at time point zero), only the extractable soil Po pool of the pasture soil varied significantly between treatments (ANOVA, p-value < 0.05), increasing by 1%-77% relative to unamended controls, i.e. reaching 101% to 177% of the native Po pool. Addition of 33Po substrate neither affected extractable soil Pi pools in pasture (ANOVA, p-value = 0.34) nor in arable (ANOVA, p-value = 0.58) soils.
Mineralization, uptake and immobilization processes of the applied 33P compounds took place very rapidly, within the first 2 to 4 h of incubation (Figure 2), and markedly slowed down afterwards. This shows that the short observation period of 24 h was suitable to observe trends in the decomposition of the various Po substrates, while minimizing the potential effect of intesa PC10 ndelay po ang response ng pc rfering longer-term processes such as remineralization and microbial turnover.
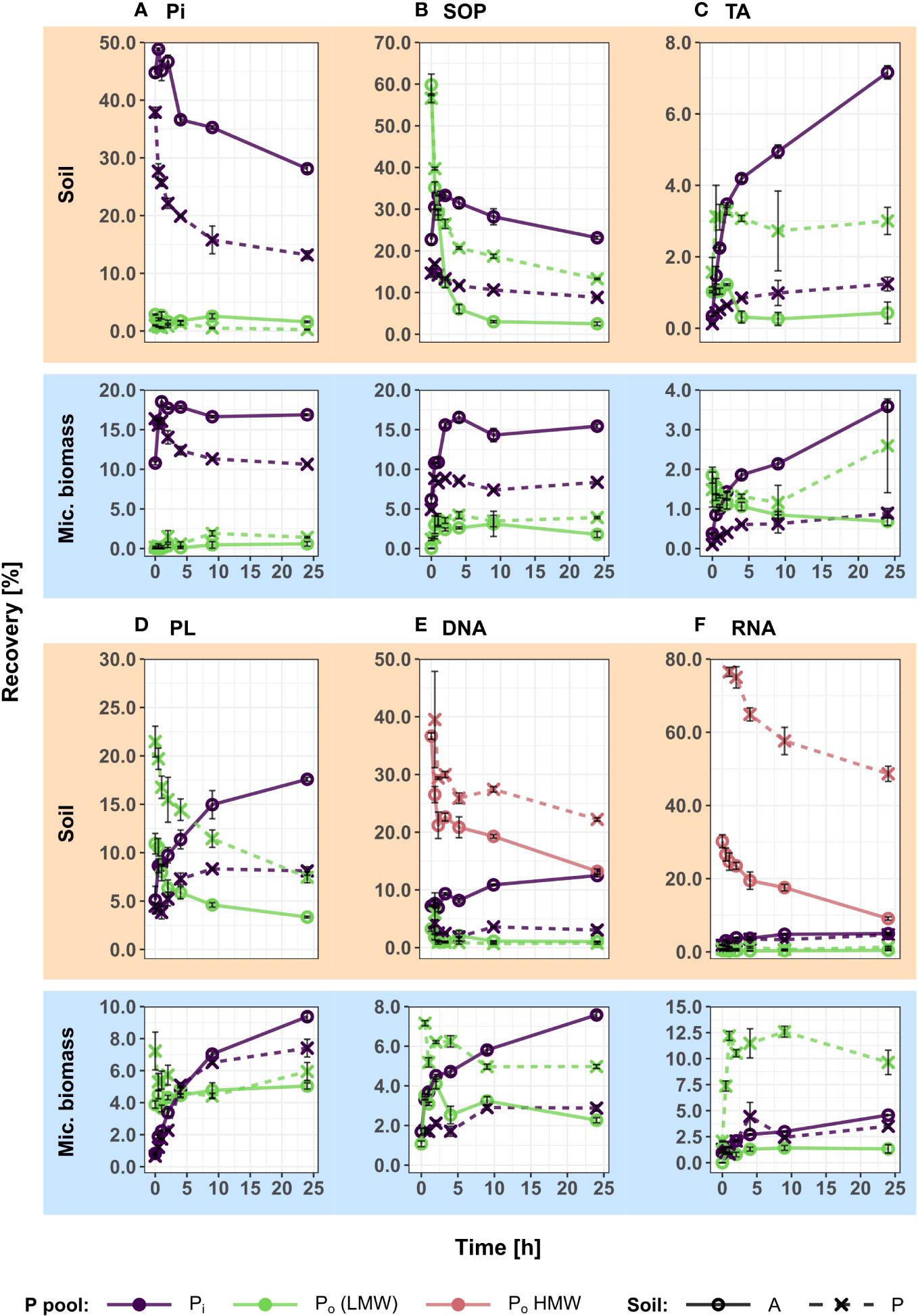
Figure 2 Recoveries of experimentally added 33P in extractable soil and microbial organic (Po) and inorganic (Pi) phosphorus pools over 24 h, showing the integrated temporal dynamics of decomposition processes, sorption and microbial P uptake for six different P compounds. 33P recoveries (mean of triplicates ± S.E.) of all pools and time points are displayed as percentages of the initially added 33P activity. In the nucleic acid treatments (E, F), soil 33Po is fractionated into low molecular weight (LMW) Po (<3kDa) and high molecular weight (HMW) Po (>3kDa) size classes. In all other treatments (A-D), Po represents extracted Po, without size fractionation. In the case of TA (C) and PL (D) additions, extractable Po does not represent the intact added substrates (which are unextractable), but organic break down products. In the case of Po additions (B-F), microbial 33Po recovery was corrected for cell internal biosynthesis of Po, as described in detail in the methods section. Pi, inorganic phosphate; TA, cell wall teichoic acids; PL, phospholipids; SOP, soluble organophosphates.
3.2. Recovery dynamics of 33P in the extractable soil pool
3.2.1. General recovery dynamics of 33P in the extractable soil pool
The 33P recovery kinetics of the amended substrates (Pi, SOP, TA, PL, DNA, RNA) from soil showed differences and commonalities between the different 33Po compounds as well as between the two soils (Figure 2 and Table 2). Soil 33Po recoveries decreased significantly over time in all Po treatments except for TA. Soil 33Po recoveries of the substrates and their breakdown products (except for LMW-DNA, i.e. the low molecular weight DNA fraction) differed significantly between the two soils. Further, the temporal dynamics of SOP and NA treatments were significantly different between the two soils (Table 2). We found higher extractable soil 33Pi recoveries (from 3.4% for TA to 28.9% for SOP averaged across all time points) in the arable soil for all Po treatments. This could be caused by lower 33Pi sorption, and/or potentially higher extracellular mineralization activity in the arable soil as compared to the pasture soil. Pi sorption, assessed as the non-extractable fraction of added 33Pi, was significantly lower in arable soil (mean 51.0% ± 1.9% standard deviation (S.D.)) than in pasture soil (66.9% ± 5.9% S.D.) (Welch`s t-Test, p-value < 0.05). Moreover, phosphodiesterase activity was 3-fold higher in the arable soil (Student`s t-Test, p-value < 0.05) with 391 nmol pNP g-1 h-1, whereas phosphomonoesterase activities did not differ significantly between the soils (Student`s t-Test, p-value = 0.07) with 630 nmol pNP g-1 h-1 for the arable soil and 842 nmol pNP g-1 h-1 for the pasture soil (Table 1).
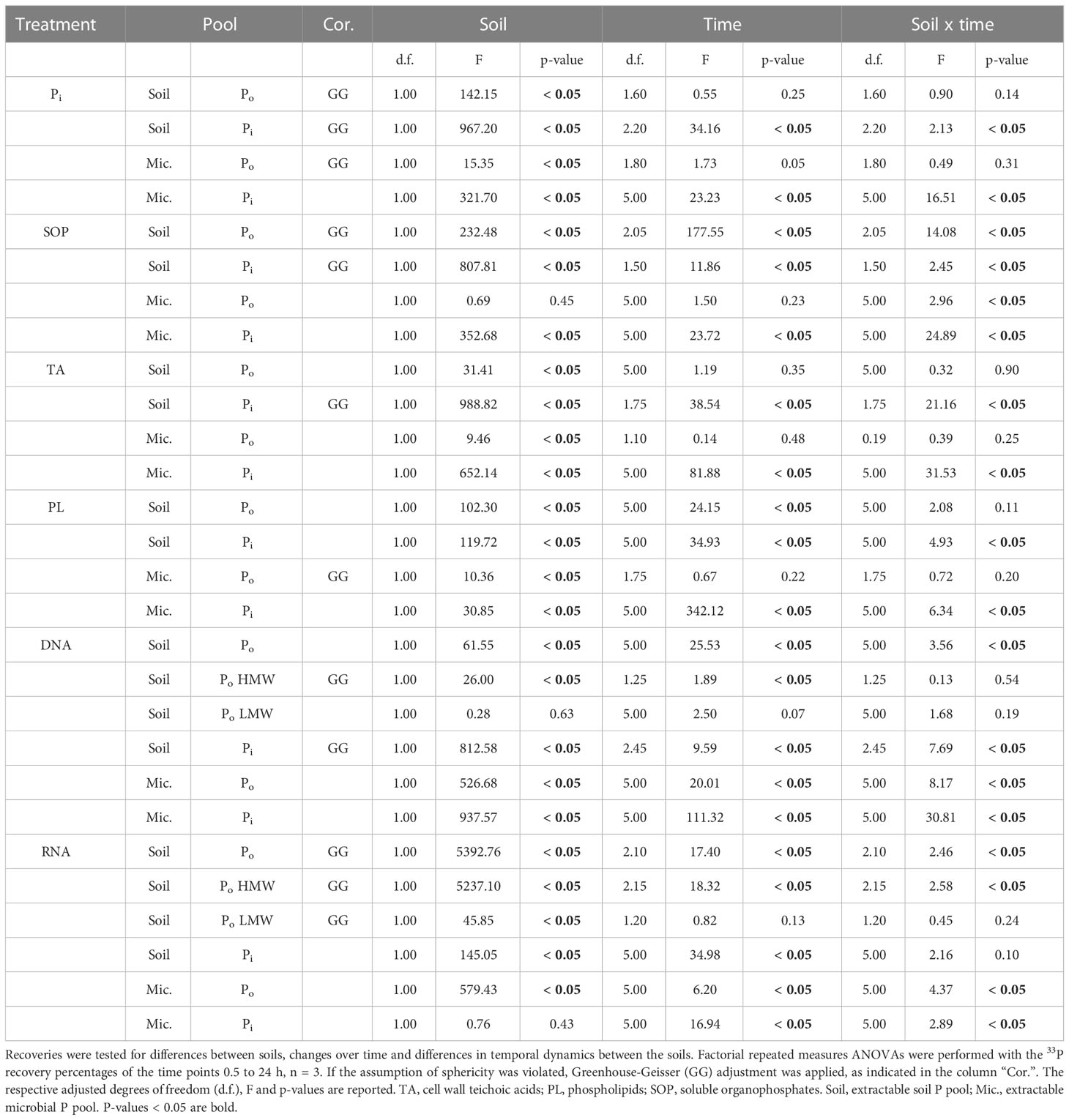
Table 2 Differences of 33P recovery from extractable soil and microbial organic (Po) and inorganic (Pi) phosphorus pools, across time and between soils.
3.2.2. Recovery dynamics of soluble organophosphates in the extractable soil pool
In the SOP treatments, the soil 33Po pool declined rapidly, leading to almost complete mineralization in the arable soil after 10 h. In the pasture soil, mineralization slowed down after 4 h as indicated by the time kinetics of 33Pi formation from 33Po, reaching Pi recovery values of 8.8% (± 0.3% S.D.) (Figure 2B).
3.2.3. Recovery dynamics of teichoic acids and phospholipids in the extractable soil pool
In the case of TA and PL additions, extractable soil 33Po continued to decline over the course of 24 h, with a slowdown after 2 to 4 h (Figures 2C, D). In both treatments, only a small fraction of the applied 33P was recovered in soil 33Po (1.3% for TA and 16.2% for PL, averaged across both soils at 0 h), and this pool did not change significantly over time when TA was added, while it significantly declined in the PL treatments (Table 2). TA and PL are phosphodiesters which contain subunits of glycerophosphate. Both phosphodiester compounds are not directly extractable with NaHCO3 as such, due to the following: in the case of TA their polymeric form and covalent binding within bacterial cell walls and in the case of PL their hydrophobicity render them unextractable by NaHCO3. The labelled Po in TA and PL therefore only becomes extractable after breakdown of the compounds into smaller and water-soluble units, such as glycerol-3-phosphate. The data therefore can be assumed to show the integral of two consecutive steps in the decomposition processes of 33P labelled PL and TA: breakdown to small water-soluble molecules (extractable soil 33Po), followed by complete mineralization (to soil 33Pi).
3.2.4. Recovery dynamics of nucleic acids in the extractable soil pool
In both nucleic acid (NA) treatments (DNA and RNA), ultrafiltration was performed to separate soil 33Po into low molecular weight-NA (<3 kDa; LMW) and high molecular weight-NA (>3 kDa; HMW) fractions. The LMW-fraction thus contained nucleotide monomers and small oligomers [max. 4 to 9 nucleotides, depending on the degree of hydration (60)], whereas the HMW-fraction contained NA oligomers and polymers larger than that. Again, the data showed two steps in the decomposition process of these 33P labelled compounds: 33Po in the LMW-NA fraction is an indicator for breakdown of HMW compounds into oligonucleotides and mononucleotides, and soil 33Pi is a measure for complete mineralization of the added 33P-NA by phosphomonoesterases. In both soils, recovery of HMW-33Po decreased significantly (from 51.6% at 0 h to 23.7% after 24 h, averaged across both NA treatments and both soils), while recovery of LMW-33Po remained very low at all times without significant changes (between 1.5% at 0 h and 0.9% after 24 h, averaged across both NA treatments and both soils) and recovery of 33Pi increased slowly in the arable soil and stagnated in the pasture soil (from 4.6% to 8.7% and between 2.9% and 3.9%, respectively, averaged across of both NA treatments) (Figures 2E, F; Table 2).
3.3. Microbial uptake of 33Po and 33Pi
3.3.1. Direct uptake of 33Po
With the experimental approach in this study, 33P derived from labelled Po-substrates can be traced into the extractable organic fraction of the microbial biomass pool (Pmic). However, 33Po in Pmic can stem from direct uptake of intact Po forms (pathway 1) and from extracellular 33Po mineralization to 33Pi, followed by cellular 33Pi uptake and subsequent intracellular 33Po biosynthesis (pathway 2). To disentangle the source of measured intracellular 33Po between both pathways, we estimated intracellular 33Po biosynthesis using incubations with 33Pi addition. Only a small amount of 33P was recovered in the microbial Po pools of these control treatments (0 to 2.5%, Figure 2A), indicating that cell internal biosynthesis of extractable Po during the 24 h incubation period was limited. To correct microbial 33Po pools of the Po treatments for intracellular Po biosynthesis, we assumed that microbes allocated a fixed percentage (as observed in the 33Pi additions) of the available substrate-derived Pi into biosynthesis. While strong increases of available Pi could theoretically change microbial P metabolism via induction of Pi assimilation or formation of polyphosphates, this should not have been triggered in our treatments. After substrate addition, soil Pi concentrations in the Po treatments did not differ significantly from the Pi treatments (Table S2), supporting the assumption that microbial P metabolism was comparable between treatments. The observation of microbial 33Po that remains after accounting for Po biosynthesis from Pi in all treatments thus indicates direct uptake of intact 33Po compounds into the microbial biomass.
Biosynthesis-corrected recoveries in the microbial 33Po pool after 24 h are therefore indicative for direct uptake of intact 33Po compounds into the soil microbial biomass. They ranged between 0.7% (± 0.2% S.D.) for TA in the arable soil and 9.6% (± 2.0% S.D.) for RNA in the pasture soil. Biosynthesis-corrected recoveries of 33P in the microbial Po-pool were significantly lower in arable (SOP: 2.4%, TA: 1.2%, PL: 4.4%, DNA: 2.8%, RNA: 1.0%, averaged across all time points) than in pasture soil (SOP: 3.0%, TA: 1.5%, PL: 5.5%, DNA: 5.8%, RNA: 9.4%, averaged across all time points), except for the SOP treatment which did not differ significantly between soils (Table 2).
3.3.2. Temporal dynamics of microbial 33Po and 33Pi uptake
In the SOP and NA treatments, the temporal dynamics of microbial 33Po recoveries varied significantly between the soils. Across soils the temporal changes of microbial 33Po recoveries were not significant in the TA, PL and SOP treatments. In the SOP and NA treatments, extractable microbial 33Po recoveries increased rapidly initially and then remained stable (Figures 2B-F).
The 33Po:33Pi ratio of Pmic is an indicator for the amount of substrate-derived Po that is directly taken up into microbial biomass relative to the amount of substrate-derived Po that is taken up as Pi after external mineralization. The ratio is calculated for successive timesteps (0.5 to 24 h), based on timestep-integrated and biosynthesis-corrected 33Po and 33Pi recoveries. In the arable soil, microbial 33Po:33Pi ratios were lower than in the pasture soil (0.68 compared to 2.86, averaged across all Po treatments and all considered time points). However, three potential methodological problems need to be considered: (i) Intracellular dephosphorylation of 33Po causes an increase in 33Pi and a decrease in this ratio, underestimating intact Po uptake. (ii) Continued extraction of soil Pi in the apparent Pmic pool could lead to an overestimation of 33Pi in Pmic and a subsequent underestimation of the microbial 33Po:33Pi ratio. (iii) On the other hand, sorption of microbial 33Pi to soil during the extraction could lead to an opposite bias. As the pasture soil showed a higher Pi sorption capacity (66.9% compared to 51.0% in arable soil), these unquantified processes obstruct a direct comparison of the microbial 33Po:33Pi ratio between the soils and may skew absolute values of the ratio. However, any such affect can be assumed to be independent of incubation time, thus allowing to consider the relative temporal patterns of treatment-specific microbial 33Po:33Pi ratios. The 33Po:33Pi ratios of Pmic decreased over time for most substrates, and this decrease was significantly related to the decline of available 33Po in the soil (Figure 3; Table S3). For all Po amendments we found that the microbial 33Po:33Pi ratios were well above 0 (in many cases above 1, ranging up to 8), indicating uptake of intact Po compounds. Furthermore, the dynamics of the 33Po:33Pi ratio of Pmic differed between soils (e.g. PL and TA versus NA dynamics) and between different Po treatments. The microbial 33Po:33Pi ratio for RNA after 1 h incubation time in pasture soil was a significant outlier (Grubb`s test, G = 1.661, p-value = 0.06) and was excluded from further analysis.
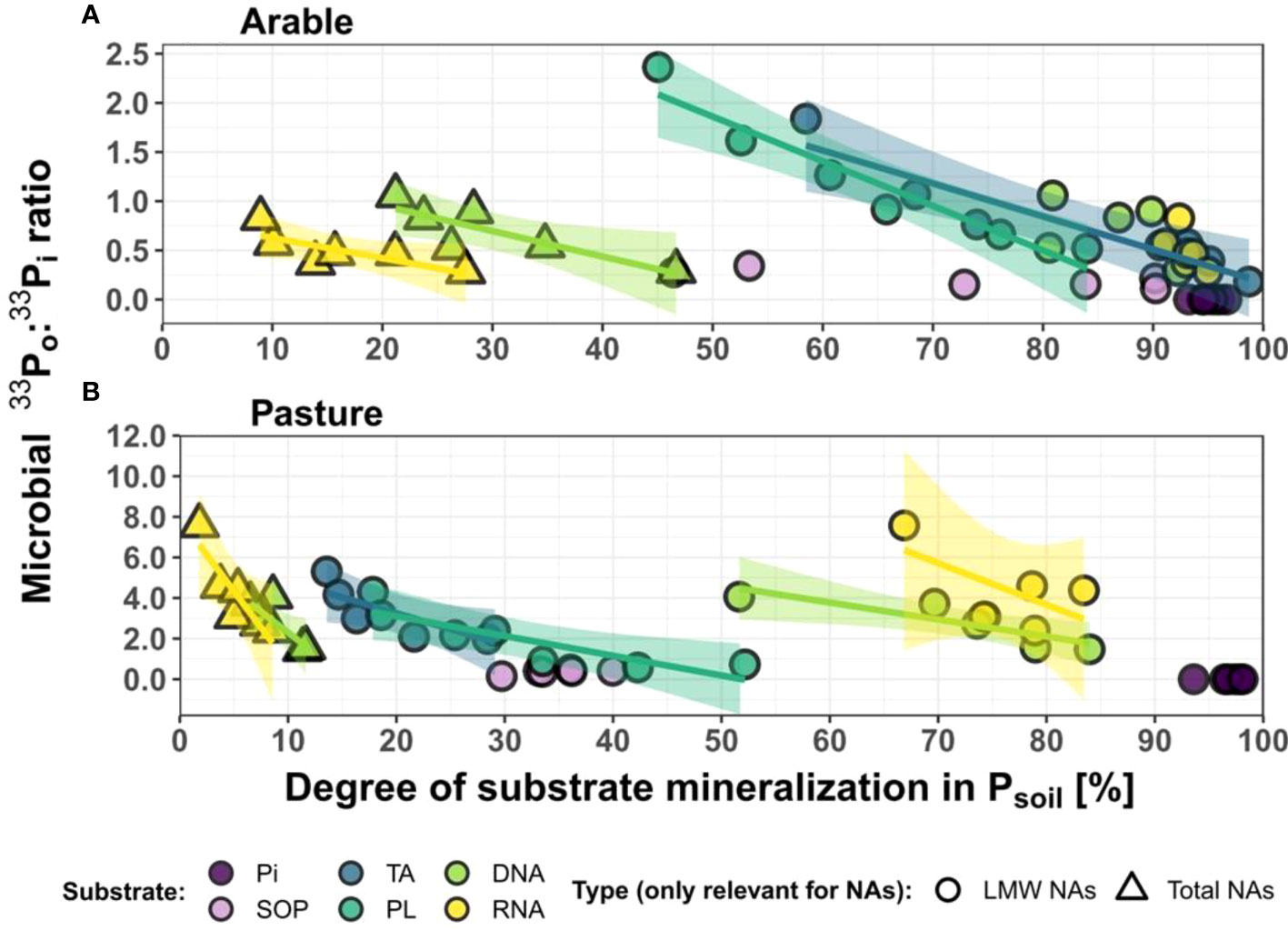
Figure 3 Ratios of cumulative Po:Pi uptake from labelled substrates decline with progressing substrate mineralization in Psoil mineralization in Psoil. , shown for the arable (A) and pasture (B) soil. Data points are the means of triplicates for time points 0.5 to 24 h (the temporal pattern within each treatment proceeds from left to right). For both nucleic acid treatments (DNA, RNA), substrate mineralization is given for for total extractable NAs [high molecular weight (HMW) + low molecular weight (LMW)] and for the LMW fraction only. Fitted significant models are shown with 95% confidence intervals. For model descriptions, see Table S3. Microbial 33Po:33Pi ratios are calculated based on 33P activity [Bq] and were corrected for biosynthesis to obtain the 33Po:33Pi ratio of cumulative 33P uptake as explained in detail in the “methods” section. The degree of substrate mineralization is the percentage of extractable soil 33P in inorganic form, i.e. Pi. Pi = inorganic phosphate; TA, cell wall teichoic acids; PL, phospholipids; SOP, soluble organophosphates.
4. Discussion
4.1. Decomposition dynamics of 33Po compounds
4.1.1. Decomposition dynamics of teichoic acids and phospholipids
We observed comparable trends in the decomposition dynamics of TA and PL, which both contain P in the form of glycerophosphates, either polymeric in wall teichoic acids or linked to fatty acids in phospholipids. We found that the extractable intermediate breakdown products (33Po) of TA and PL were mineralized to a higher degree in the arable than in the pasture soil (72.7% compared to 26.1%, averaged across both treatments and all considered time points; see the degree of substrate mineralization, x-axis in Figure 3). This suggests that depolymerization by phosphodiesterases may be the rate limiting step of TA and PL decomposition in the arable soil (high degree of substrate mineralization). In the pasture soil, either mineralization through phosphomonoesterases seems to (co)limit complete TA and PL decomposition (lower degree of substrate mineralization), or a larger amount of mineralized P may be rendered bio-unavailable through higher Pi sorption as compared to the arable soil. The lack of accumulation of intermediate breakdown products in 33Psoil over time demonstrates that glycerophosphate is promptly consumed, either by the direct use by microbes or by rapid dephosphorylation to 33Pi by phosphomonoesterases, in line with earlier studies [e.g (61, 62)]. The observed decomposition and mineralization of TA is in contrast with the hypothesis that teichoic acids are resistant to decomposition (63). Mechanisms for the breakdown of wall teichoic acids under P limiting conditions are known (64, 65), and it has also been observed that soil bacteria can produce specialized phosphodiesterases (glycerophosphodiesterases) to utilize exogenous teichoic acid as the sole P source (66), thereby releasing glycerol-3-phosphate (27, 67, 68). Interestingly, enzymes from the same highly conserved family (glycerophosphodiesterases) are known to be involved in the breakdown of both TA and PL (69). We also observed rapid initial breakdown of PL, similar to Tollefson and McKercher (70). There are at least three alternative pathways for the first step of soil microbial breakdown of PL, all yielding different breakdown products (71–73). Since we observed direct uptake of 33Po (see discussion below), and transporters are only known for glycerol-3-phosphate, we hypothesize that a substantial part of the PL breakdown occurred via a pathway which yields glycerophosphate, choline and fatty acids as products (71). This pathway likely involves phospholipase D, which cleaves off the headgroup from the phospholipid (e.g. choline, ethanolamine) leaving phosphatidic acid behind, and phospholipase B (or A1 plus A2) that cleaves phosphatidic acid into two fatty acids and one glycerol-3-phosphate (26, 74, 75).
4.1.2. Decomposition dynamics of nucleic acids
The second group of compounds with commonalities in decomposition dynamics are the nucleic acids. While the 33P activity recovered in HMW fractions in Psoil decreased (from 51.6% to 23.7% over 24 h, averaged across both NA treatments and both soils), the recovered activity of 33P in the LMW fractions remained low (between 1.5% and 0.9% over 24 h, averaged across both NA treatments and both soils) and the recovery of 33Pi generally increased over time (from 3.7% to 6.3% over 24 h, averaged across both NA treatments and both soils) (Figures 2E, F; Table 2). Intact nucleic acids (which are recovered in the HMW-NA pool) need to be broken down into nucleotides (which are recovered in the LMW-NA pool) before mineralization. Therefore, the LMW-NA pool reflects the net product of inputs via HMW-NA breakdown and outputs due to mineralization and microbial uptake (and sorption). There is a constant flux of 33P through the LMW-NA pool, as nucleotides are the intermediate product between depolymerization and mineralization. The constant and low recoveries of LMW-33Po (Figures 2E, F) therefore strongly indicate that substrate-derived nucleotides do not accumulate. This means that NA depolymerization to free nucleotides and oligonucleotides is the rate-limiting step in the decomposition of nucleic acids in soil, while dephosphorylation of phosphomonoesters (nucleotides) occurs rapidly and therefore is not rate limiting. Extracellular DNA in soil is thought to be cleaved into duplex oligonucleotides of 400 bp length by restriction endonucleases; studies in pure culture showed cleavage of HMW-DNA to oligonucleotides of ~7 bp likely by endonucleases and further hydrolysis via exonucleases into mononucleotides by DNases, although this has not yet been shown in situ in soils (76). Others observed that fragmentation dynamics during DNA decomposition varied between different soils (77). While it is still not clear which phosphodiesterases of the different nuclease types are involved in soil NA decomposition and who produces those in soils (77–79), our findings are in line with earlier observations. Phosphodiesterase activity was previously found to be rate-limiting in labile Po turnover in pasture soils (80), and other studies reported rapid mineralization of free added nucleotides (61, 81). It has been implied that RNA can be used as a viability marker as it degrades more rapidly in soils in comparison with DNA, although comparable half-lives of 0.5 - 1.5 days of DNA and RNA in soils have also been reported previously (82–84). In this study, we found similar recovery dynamics of HMW-DNA and HMW-RNA within each soil, pointing to similar decomposition rates of environmental DNA and RNA.
4.1.3. Decomposition dynamics of soluble organophosphates
Recovery of intact organophosphates (SOP) from Psoil decreased rapidly (58.2% to 13.4% over the first 4 h, averaged across both soils), initially with concomitant but moderate increases in the soil 33Pi pool (18.7% to 21.6% over the first 4 h, averaged across both soils). The SOP substrate presumably consisted mostly of phosphomonoesters (sugar phosphates, organic acid phosphates, nucleotides, nucleotide sugars, cofactors etc.), which are likely to be rapidly mineralized in soil by the ubiquitous acid and alkaline phosphomonoesterases. However, after one hour 33Pi in Psoil also started to decrease, with continued decreases in 33Po. This indicates that other processes aside from mineralization must have contributed to the 33P removal from Psoil. Patterns of 33Pi recovery resembled those observed in the Pi treatments and were thus likely caused by sorption of 33Pi and by microbial uptake. However, it is unlikely that sorption played a large role in the observed 33Po decreases, since another study (81) found very rapid completion – within 15 to 45 minutes – of sorption processes of small phosphate monoesters in an Eutric Cambisol under meadow. It has been proposed that sorption dynamics of Po compounds depend on the size and structure of their organic moieties, which have lower surface affinities than phosphate groups (85–87). Small molecules as in the SOP treatment may therefore exhibit comparatively rapid initial sorption upon addition to soil because the effect of the organic moieties over the phosphate groups may be less dominant due to their small size. In addition, phosphate monoesters also have a high ability to form insoluble complexes with polyvalent cations (86). The same study which observed rapid completion of initial sorption of phosphate monoesters (81) found rapid initial mineralization and respiratory utilization of 14C-labelled glucose-6-phosphate and adenosine phosphates, gradually slowing down after 6 and 24 h, respectively. Two studies using 14C and 33P labelling (36, 37) also found rapid utilization of glucose-6-phosphate, with no more major changes in 33P recovery after 2 to 3 days. Another possible explanation could be rapid uptake of 33P, and assimilation into non-extractable Pmic. 33P that is used to build up structures that are embedded in macromolecules (e.g. teichoic acids in cell walls) or that are hydrophobic (e.g. phospholipids) may not be extractable anymore with the methods applied in this study. Assimilation of labelled substrate into non-extractable microbial pools has been observed previously elsewhere (88).
4.2. Direct microbial uptake of Po versus uptake of Pi after extracellular mineralization
4.2.1. Direct uptake of 33Po in general
The low microbial Po biosynthesis in soils amended with Pi clearly points to intermittent storage (e.g. polyphosphates) and slow metabolism of Pi in the soil microbial communities, showing a decoupling of uptake and metabolism of Pi. Direct Po uptake contributed to 33P uptake in all Po treatments, and in both soils (Figure 2). The contribution of direct Po uptake to cumulative 33P uptake into Pmic seemed lower in the arable than in the pasture soil (with a 33Po:33Pi ratio of Pmic of 0.68 compared to 2.86, averaged across all Po treatments and all considered time, see also Figure 3). This could eventually be caused by a higher sorptive loss of microbial 33Pi during extraction in the pasture soil. However, the arable soil also had a 4-fold higher native extractable Pi concentration than the pasture soil (30.7 µg g-1 compared to 7.0 µg g-1, see Table 1), and a higher percentage of 33Psoil was present as Pi (72.7% compared to 26.1%, averaged across both treatments and all considered time points, Figure 3). Both these aspects render a real difference between the soils plausible. The 33Po:33Pi ratios of Pmic decreased in both soils, as the progression of extracellular substrate mineralization over 24 h decreased 33Po availability through substrate consumption and concomitantly increased 33Pi (Figure 3). Direct Po uptake therefore decreases with observational time due to the continuous action of the phosphomonoesterases converting 33Po to 33Pi. Hence, these patterns reveal that the Po : Pi ratio of cumulative uptake of Po-derived P is ultimately governed by the form in which it is available. This actually causes a situation in soils where microbial Po uptake transporters compete with extracellular phosphomonoesterases for organic substrates. Direct uptake of organic P may be of greater relative (and absolute) importance under conditions of low Pi availability, regardless of whether low Pi availability is caused by lower extracellular mineralization or higher Pi sorption.
The observed stagnation of 33Po accumulation in Pmic of the TA, PL and SOP treatments (Table 2) is likely to be the net result of several (possible) processes: (i) In situ production and decomposition of unlabelled Po compounds in Psoil, contributing to a decrease in the specific activities of soil Po pools (Figure S2), thus leading to a decline in net 33Po accumulation at constant Po uptake rates, (ii) intracellular Po mineralization decreasing the microbial 33Po recoveries, and (iii) the intracellular production of insoluble (non-extractable) 33Po diesters. For instance, another study found that labelled DNA, which was cleaved and then assimilated by bacterioplankton, became non-extractable with time (88). Overall, the above mentioned processes would contribute to an underestimation of direct Po uptake in this study.
4.2.2. Direct uptake of nucleic acids
It is commonly thought that NA-derived P cannot be taken up in an intact organic-bound form (i.e. nucleotide). Rather it is thought that nucleotides are first dephosphorylated, and that P-free nucleosides and Pi are taken up independently by nucleoside transporters and PHT transporters, respectively (89, 90). Empirical evidence for direct uptake of intact LMW nucleotides in non-parasitic organisms is scarce (91), but recently transporters of the nucleotide transport protein (NTT) family, previously known from intracellular parasites, have been found in free-living prokaryotes and eukaryotes (43). NTTs either exchange nucleotides across membranes (e.g. ADP for ATP, ADP for NAD+) or are H+:nucleotide symporters allowing efficient net uptake of nucleotides. NTTs could thus play an important role in the environment, contributing to the direct uptake of NA-derived P. Alternatively to nucleotide uptake, polymeric NA, such as DNA molecules, might also be actively transported across the cell envelope and into the cytoplasm, by a process called transformation in bacterial cells. Transformation aims at DNA molecules with ranges reported from 300 bp to more than 50 kb length (92, 93), and therefore would clearly relate to DNA-Po from the HMW fraction in this study. This ability is taxonomically widespread but it is unknown whether it is common in soil microbes (76, 93, 94). Furthermore, RNA presumably cannot be taken up in intact form by any process analogous to transformation, although other studies found indications for intact uptake of HMW-RNA in yeast (95) and for intact, energy-dependent uptake of oligonucleotides including RNA by Candida albicans (96). Our data suggest that direct uptake of 33Po derived from DNA and RNA may have taken place via transporters for nucleotides (NTT), after decomposition of LMW-NA to oligo- and mononucleotides by exo- and endonucleases.
4.2.3. Direct uptake of teichoic acid and phospholipid decomposition products
Glycerolphosphodiesterases (GlpQ) hydrolyze wall teichoic acids and phospholipids, yielding glycerol-3-phosphate (G3P) amongst other byproducts. Uptake transporters for G3P (GlpT) are well known and belong to the organophosphate:phosphate antiporter (OPA) family. Often GlpT and GlpQ are co-localized (41). Recently, a novel potential Po transporter was found in bacteria, which is a component of the phosphorus utilization system (PUS) clusters. These contain the genes of the two-component PusCD system alongside with phosphodiesterases, where PusC and D likely represent a G3P transporter and a lipoprotein cap (45). The known diversity of G3P transporters therefore renders direct uptake of G3P likely.
4.2.4. Direct uptake of soluble organophosphates
We also observed direct uptake of Po compounds from the group of soluble organophosphates. However, in this treatment, the mineralization proceeded so rapidly that 33Pi uptake outpaced that of 33Po right from the beginning, without major changes in the 33Po:33Pi ratio of Pmic (Figure 3). The fraction of soluble organophosphates derived from bacterial cells is a complex mixture of compounds (sugar phosphates, nucleotide sugars, nucleotides, phosphoamino acids, organic polyphosphates, phosphonates etc.) and it is possible that only some of them can be taken up directly in an intact form, depending on the presence of suitable uptake transporters. For sugar phosphates, several transporters belonging to the OPA family are known, and these have been found in a variety of bacteria (44, 46, 97). There is also evidence for direct uptake of hexose phosphates in fungi (98). Another uptake system from the same family is known to transport phosphoenolpyruvate and phosphoglyceric acid into bacteria, and further transporters for several phosphonates have been characterized (46, 48).
4.2.5. Cell internal fate of directly taken up 33Po
In our study, the 33Po taken up directly remained at least partially in organic form, showing that complete intracellular mineralization did not take place within 24 h. The P derived from Po compounds could undergo phosphoryl transfer to ATP and enter central metabolic pathways (48). Shortcutting cell internal metabolic pathways would be beneficial by saving costs for associated enzymes and skipping energy requiring steps. Salvage pathways using intact nucleotides have been suggested to be an important use of extracellular DNA (43, 76, 93). All antiporters of the OPA family transport intermediates of the glycolytic pathway (99), e.g. glycerol-3-phosphate, an intermediate of phospholipid synthesis (47, 73), which could directly contribute to the host metabolism. Po uptake via OPA antiporters, however, has been suggested to be inefficient for bacterial P nutrition, because it requires efflux of Pi or Po anions in order to take up Po (46, 48). Similarly, many of the nucleotide transporter proteins (NTT) exchange ADP for ATP, or NAD+ for ADP, serving purposes of energy gain with little net gain in P. Only a few NTTs for unidirectional proton-driven nucleotide import are known (100). However, efficient re-uptake of secreted Pi by phosphate:H+ symporters (PHT family) might allow for continuous Po uptake with net P gain. While Pi uptake clearly serves P nutrition, P containing organic molecules could also be used as sources of energy and C (36, 37) and in some cases N.
4.3. Ecological implications of direct Po uptake by microbes
As direct microbial uptake of Po is not perceived as an integrated component of the soil P cycle, we here consider some aspects that possibly have important ecological implications compared to complete extracellular mineralization. (i) The ability of direct uptake of Po compounds reduces the dependence of P acquisition on extracellular phosphatases, especially phosphomonoesterases, since phosphate diesters still require a certain degree of extracellular enzymatic breakdown. Firstly, the efficacy of extracellular phosphatases in soils can be reduced by suboptimal pH, or by the presence of metal ions, polyvalent anions or chelating agents (56, 101), condensed organic molecules (102), and by sorption to clay minerals (103). Secondly, extracellular enzyme production requires an investment of N (and C), and thus, under N-limiting conditions, excretion of phosphatases may not be an adequate P acquisition strategy. N additions have been shown to enhance extracellular phosphatase activity under a wide range of conditions (104), suggesting that microbes can allocate excess N to phosphatase enzyme production. Under N-limiting conditions direct Po uptake may be beneficial, because of reduced N investment into phosphatase production. (ii) Complete mineralization by extracellular enzymes might promote the risk of losing cleaved Pi to soil Pi sorption/precipitation processes. Sorption of Pi can remove a considerable fraction of free Pi or of mineralized P in soils, particularly in highly weathered soils with high content of Fe/Al oxyhydroxides and aluminosilicates (105). Since Po sorption is weaker and slower than that of Pi (106, 107), Pi becomes more rapidly inaccessible compared to Po. (iii) The general advantage of direct access to organic P substrates could also affect plant-microbe interactions, as plants and microbes compete for available P over short timescales (23, 108). Since there is limited evidence for direct Po uptake by plants (109), P taken up in organic form by microbes might not be directly available to plants at any time, and microbes possessing this capability could therefore have an advantage over plants.
4.4. Methodological limitations
The applied methodology captures net changes in pool sizes (33P recoveries relative to the activity added) over time, and therefore does not allow conclusions on underlying gross process rates. However, extractable soil 33P pools (33Po and 33Pi in Psoil) provide a realistic picture of biologically available P and of the involved ecologically relevant net P processes. Aside from this, it must be considered that 33Pi and 33Po are subject to abiotic immobilization processes such as sorption to soil particles and precipitation with cations (as indicated by soil Po specific activities, Figure S2). Estimates of extracellular mineralization after accounting for Pi sorption indicate that mineralization of 33P labelled organic substrates is likely underestimated by the quantification of 33Pi in the extractable soil P pool (Figure S3). This is caused by continuous sorption of parts of the 33Pi that is produced by mineralization of 33Po compounds. However, we argue that this ongoing sorption of 33Pi does not confound the interpretation of the data when viewing Psoil as the available P pool; it just prohibits conclusions about gross mineralization rates. As for sorption of 33Po, several studies that investigated the time kinetics of sorption of organic P compounds found a rapid slowdown of sorption after the first 60 min (81, 85, 87). According to this, losses of 33Po from the extractable soil pool by sorption should strongly decline after the fast initial sorption of added 33Po substrate is complete. Further, phosphomonoesters are known to have a higher affinity for complexation with cations and get more strongly stabilized by sorption than phosphodiesters (86). To directly quantify sorption of Po compounds in soil remains an open challenge. There is yet no method for isotope pool dilution approaches or isotope exchange kinetic approaches (which are often used to assess Pi sorption and the size of the bioavailable Pi pool) which reliably inhibits microbial activity as well as the activity of extracellular phosphatases without affecting soil structure (6, 18, 110). However, as long as biotic mineralization of added 33Po compounds cannot be reliably inhibited without affecting sorption dynamics, Po sorption dynamics in real soils remain hard to determine, and therefore empirical data on Po sorption will remain constrained to artificial abiotic systems. Because of the above mentioned processes, the observed decreases in soil 33Po and the dynamics of soil 33Pi do not necessarily reflect biological processes alone, and gaps in the balance between apparent Po mineralization, the amount of soil 33Pi recovery and microbial uptake of 33P can arise. Similar restrictions apply to the quantification of Po uptake; several possibilities for hidden gross fluxes of 33P – for instance incorporation into unextractable microbial Po pools or intracellular Po mineralization – might lead to an underestimation of actual Po uptake.
Further uncertainty of the applied methodology comes from the unknown fate of polyphosphates during P extraction and fractionation of Pi and Po from the soil. In the case of high soil Pi supply, bacteria and fungi can take up Pi in excess of their demand and store it in the form of inorganic polyphosphates, serving as intermittent stores for Pi and energy (111). The behavior of such polyphosphates during isobutanol fractionation is unknown, and polyphosphates might theoretically be partitioned into the microbial Po fraction as an artefact. However, we can safely exclude that all observed microbial 33Po is polyphosphate synthesized from 33Pi, as this would require that polyphosphate synthesis had not occurred in the 33Pi treatment, but consistently in all 33Po treatments. Moreover, significant in situ synthesis of polyphosphates by soil microbes is only expected under high Pi availability, while in this study substrate additions did not lead to significant changes in concentrations of available soil Pi relative to the Pi control treatments (Table 1).
4.5. Conclusions
We presented evidence that decomposition of all tested phosphodiester compounds (cell wall teichoic acids, phospholipids, DNA and RNA) was rate-limited by the initial breakdown step carried out by phosphodiesterases, while the resulting simple phosphomonoesters (such as nucleotides and glycerophosphates) were rapidly dephosphorylated by phosphomonoesterases in most cases. This indicates that depolymerization sets the pace for the decomposition of high molecular weight Po compounds, with phosphodiesterase-activities representing the bottleneck in Po decomposition. This study did not allow for direct quantification of the abiotic sorption of intact Po compounds. However, we could show that abiotic sorption processes strongly reduced 33Pi recoveries, and likely also competed with microbial uptake of Pi after extracellular Po mineralization.
Up to date, the possibility of direct microbial uptake of Po in soils has been widely overlooked. Here we presented indications that this process occurred to a substantial extent in two agricultural soils. Thus, we showed that direct microbial uptake of Po constitutes a P-acquisition pathway in the soil P cycle that does not require complete extracellular mineralization and subsequent Pi uptake. We further found that cumulative net microbial Po uptake seemed to be driven by the availability of intact phosphomonoesters. Direct Po uptake could be competing or complementary to extracellular Po mineralization, depending on whether it is subject to different or to interconnected controls. Further research will be necessary to investigate the interplay of these two pathways, to assess the quantitative importance of direct Po uptake in soil systems, and to elucidate the ecological consequences of this process. Systematic studies of the distribution of phosphatases and Po uptake transporters across the soil microbial diversity will be necessary to answer these questions. Organic P plays a central role in the P cycle of most soils, and accounting for direct microbial Po uptake and the potential of phosphodiesterase activities to control overall Po mineralization rates will substantially enhance our understanding of soil P cycling (Figure 1).
Data availability statement
The raw data supporting the conclusions of this article is available in the Supplementary Material as Data Sheet 2.
Author contributions
WW designed the experiment, DW and WW developed the methods with contributions from YH. DW, JP and DZ conducted the laboratory work, DW analyzed the data, and DW and WW interpreted the data and wrote the manuscript with contributions from MM. All authors contributed to the article and approved the submitted version.
Funding
We are grateful to the continuous support by the University of Vienna, in terms of basic funding, instruments, and lab space, without which this study would not have been possible.
Acknowledgments
We would like to thank Johann Püspök and Sabine Maringer for their support in the laboratory, and two reviewers for their helpful comments on the manuscript. D.W. would like to thank Sonja Reiterlehner deeply for all her support and patience.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsoil.2023.1097965/full#supplementary-material
References
1. Wang YP, Law RM, Pak B. A global model of carbon, nitrogen and phosphorus cycles for the terrestrial biosphere. Biogeosciences. (2010) 7(7):2261–82. doi: 10.5194/bg-7-2261-2010
2. Kattge J, Díaz S, Lavorel S, Prentice IC, Leadley P, Bönisch G, et al. TRY - a global database of plant traits: TRY - a GLOBAL DATABASE OF PLANT TRAITS. Glob Change Biol (2011) 17(9):2905–35. doi: 10.1111/j.1365-2486.2011.02451.x
3. Xu X, Thornton PE, Post WM. A global analysis of soil microbial biomass carbon, nitrogen and phosphorus in terrestrial ecosystems: Global soil microbial biomass c, n and p. Glob Ecol Biogeogr (2013) 22(6):737–49. doi: 10.1111/geb.12029
4. Zechmeister-Boltenstern S, Keiblinger KM, Mooshammer M, Peñuelas J, Richter A, Sardans J, et al. The application of ecological stoichiometry to plant–microbial–soil organic matter transformations. Ecol Monogr (2015) 85(2):133–55. doi: 10.1890/14-0777.1
5. Frossard E, Condron LM, Oberson A, Sinaj S, Fardeau JC. Processes governing phosphorus availability in temperate soils. J Environ Qual (2000) 29(1):15–23. doi: 10.2134/jeq2000.00472425002900010003x
6. Bünemann EK. Assessment of gross and net mineralization rates of soil organic phosphorus – a review. Soil Biol Biochem (2015) 89:82–98. doi: 10.1016/j.soilbio.2015.06.026
7. Harrison AF. Soil organic phosphorus: a review of world literature. Wallingford, UK: CABI Publishing (1987).
8. Yang X, Post WM. Phosphorus transformations as a function of pedogenesis: A synthesis of soil phosphorus data using hedley fractionation method. Biogeosciences. (2011) 8(10):2907–16. doi: 10.5194/bg-8-2907-2011
9. Darch T, Blackwell MSA, Hawkins JMB, Haygarth PM, Chadwick D. A meta-analysis of organic and inorganic phosphorus in organic fertilizers, soils, and water: Implications for water quality. Crit Rev Environ Sci Technol (2014) 44(19):2172–202. doi: 10.1080/10643389.2013.790752
10. Reusser JE, Tamburini F, Neal AL, Verel R, Frossard E, McLaren TI. The molecular size continuum of soil organic phosphorus and its chemical associations. Geoderma. (2022) 412:115716. doi: 10.1016/j.geoderma.2022.115716
11. Reusser JE, Piccolo A, Vinci G, Savarese C, Cangemi S, Cozzolino V, et al. Phosphorus species in sequentially extracted soil organic matter fractions. Geoderma. (2023) 429:116227. doi: 10.1016/j.geoderma.2022.116227
12. Magid J, Tiessen H, Condron L. Dynamics of organic phosphorus in soils under natural and agricultural ecosystems. In: Piccolo A, editor. Humic substances in terrestrial ecosystems. Amsterdam, Netherlands: Elsevier Science B. V. (1996). doi: 10.1016/B978-044481516-3/50012-8
13. Oberson A, Joner EJ. Microbial turnover of phosphorus in soil. In: Turner BL, Frossard E, Baldwin D, editors. Organic phosphorus in the environment. Wallingford, UK: CABI Publishing (2005).
14. Bünemann E, Prusisz B, Ehlers K. Characterization of phosphorus forms in soil microorganisms. In: Bünemann E K, Oberson A, Frossard E, editors. Phosphorus in action, soil biology 26. Heidelberg, Germany: Springer (2011).
15. Vincent AG, Schleucher J, Giesler R, Wardle DA. Soil phosphorus forms show only minor changes across a 5000-year-old boreal wildfire chronosequence. Biogeochemistry. (2022) 159(1):15–32. doi: 10.1007/s10533-022-00910-2
16. Achat DL, Augusto L, Gallet-Budynek A, Loustau D. Future challenges in coupled c–N–P cycle models for terrestrial ecosystems under global change: a review. Biogeochemistry. (2016) 131(1–2):173–202. doi: 10.1007/s10533-016-0274-9
17. Helfenstein J, Jegminat J, McLaren TI, Frossard E. Soil solution phosphorus turnover: derivation, interpretation, and insights from a global compilation of isotope exchange kinetic studies. Biogeosciences. (2018) 15(1):105–14. doi: 10.5194/bg-15-105-2018
18. Wanek W, Zezula D, Wasner D, Mooshammer M, Prommer J. A novel isotope pool dilution approach to quantify gross rates of key abiotic and biological processes in the soil phosphorus cycle. Biogeosciences. (2019) 16(15):3047–68. doi: 10.5194/bg-16-3047-2019
19. Wang Y, Huang Y, Augusto L, Goll DS, Helfenstein J, Hou E. Toward a global model for soil inorganic phosphorus dynamics: Dependence of exchange kinetics and soil bioavailability on soil physicochemical properties. Glob Biogeochem Cycles (2022) 36(3). doi: 10.1029/2021GB007061
20. Liu J, Cade-Menun BJ, Yang J, Hu Y, Liu CW, Tremblay J, et al. Long-term land use affects phosphorus speciation and the composition of phosphorus cycling genes in agricultural soils. Front Microbiol (2018) 9:1643. doi: 10.3389/fmicb.2018.01643
21. Lu J, Jia P, Feng S, Wang Y, Zheng J, Ou S, et al. Remarkable effects of microbial factors on soil phosphorus bioavailability: A country-scale study. Glob Change Biol (2022) 3;gcb:16213. doi: 10.1111/gcb.16213
22. Siles JA, Starke R, Martinovic T, Parente Fernandes ML, Orgiazzi A, Bastida F. Distribution of phosphorus cycling genes across land uses and microbial taxonomic groups based on metagenome and genome mining. Soil Biol Biochem (2022) 174:108826. doi: 10.1016/j.soilbio.2022.108826
23. Richardson AE, Simpson RJ. Soil microorganisms mediating phosphorus availability update on microbial phosphorus. Plant Physiol (2011) 156(3):989–96. doi: 10.1104/pp.111.175448
24. George TS, Giles CD, Menezes-Blackburn D, Condron LM, Gama-Rodrigues AC, Jaisi D, et al. Organic phosphorus in the terrestrial environment: a perspective on the state of the art and future priorities. Plant Soil (2018) 427(1–2):191–208. doi: 10.1007/s11104-017-3391-x
25. Yang W. Nucleases: diversity of structure, function and mechanism. Q Rev Biophys (2011) 44(1):1–93. doi: 10.1017/S0033583510000181
26. Lidbury IDEA, Murphy ARJ, Fraser TD, Bending GD, Jones AME, Moore JD, et al. Identification of extracellular glycerophosphodiesterases in pseudomonas and their role in soil organic phosphorus remineralisation. Sci Rep (2017) 7(1):2179. doi: 10.1038/s41598-017-02327-6
27. Walter A, Unsleber S, Rismondo J, Jorge AM, Peschel A, Gründling A, et al. Phosphoglycerol-type wall and lipoteichoic acids are enantiomeric polymers differentiated by the stereospecific glycerophosphodiesterase GlpQ. J Biol Chem (2020) 295(12):4024–34. doi: 10.1074/jbc.RA120.012566
28. Nannipieri P, Giagnoni L, Landi L, Renella G. Role of phosphatase enzymes in soil. In: Bünemann EK, Oberson A, Frossard E, editors. Phosphorus in action, soil biology 26. Heidelberg, Germany: Springer (2011).
29. Schimel JP, Bennett J. NITROGEN MINERALIZATION: CHALLENGES OF a CHANGING PARADIGM. Ecology. (2004) 85(3):591–602. doi: 10.1890/03-8002
30. Jan MT, Roberts P, Tonheim SK, Jones DL. Protein breakdown represents a major bottleneck in nitrogen cycling in grassland soils. Soil Biol Biochem (2009) 41(11):2272–82. doi: 10.1016/j.soilbio.2009.08.013
31. Wanek W, Mooshammer M, Blöchl A, Hanreich A, Richter A. Determination of gross rates of amino acid production and immobilization in decomposing leaf litter by a novel 15N isotope pool dilution technique. Soil Biol Biochem (2010) 42(8):1293–302. doi: 10.1016/j.soilbio.2010.04.001
32. Hu Y, Zheng Q, Noll L, Zhang S, Wanek W. Direct measurement of the in situ decomposition of microbial-derived soil organic matter. Soil Biol Biochem (2020) 141:107660. doi: 10.1016/j.soilbio.2019.107660
33. Harrison AF. Labile organic phosphorus mineralization in relationship to soil properties. Soil Biol Biochem (1982) 14(4):343–51. doi: 10.1016/0038-0717(82)90004-9
34. Jayachandran K, Schwab AP, Hetricic BAD. Mineralization of organic phosphorus by vesicular-arbuscular mycorrhizal fungi. Soil Biol Biochem (1992) 24(9):897–903. doi: 10.1016/0038-0717(92)90012-M
35. Martin JK, Cartwright B. The comparative plant availability of 32 p myo -inositol hexaphospha.te and KH 232 PO 4 added to soils. Commun Soil Sci Plant Anal (1971) 2(5):375–81.
36. Spohn M, Kuzyakov Y. Phosphorus mineralization can be driven by microbial need for carbon. Soil Biol Biochem (2013) 61:69–75. doi: 10.1016/j.soilbio.2013.02.013
37. Heuck C, Weig A, Spohn M. Soil microbial biomass C:N:P stoichiometry and microbial use of organic phosphorus. Soil Biol Biochem (2015) 85:119–29. doi: 10.1016/j.soilbio.2015.02.029
38. Kapoor KK, Haider K. Mineralization and plant availability of phosphorus from biomass of hyaline and melanic fungi. Soil Sci Soc Am J (1982) 46(5):953–7. doi: 10.2136/sssaj1982.03615995004600050014x
39. van Veen HW. Phosphate transport in prokaryotes: molecules, mediators and mechanisms. Antonie Van Leeuwenhoek (1997) 17:299–315.
40. Richardson AE, George TS, Hens M, Simpson RJ. Utilization of soil organic phosphorus by higher plants. In: Turner BL, Frossard E, Baldwin D, editors. Organic phosphorus in the environment. Wallingford, UK: CABI Publishing (2005).
41. Park Y, Solhtalab M, Thongsomboon W, Aristilde L. Strategies of organic phosphorus recycling by soil bacteria: acquisition, metabolism, and regulation. Environ Microbiol Rep (2022) 14(1):3–24. doi: 10.1111/1758-2229.13040
42. Beever RE, Burns DJW. Phosphorus uptake, storage and utilization by fungi. In: Advances in botanical research. London: Academic Press INC). (1981) 127–219. Available at: https://linkinghub.elsevier.com/retrieve/pii/S0065229608600348.
43. Major P, Embley TM, Williams TA. Phylogenetic diversity of NTT nucleotide transport proteins in free-living and parasitic bacteria and eukaryotes. Genome Biol Evol (2017) 9(2):480–7. doi: 10.1093/gbe/evx015
44. Winkler HH. Distribution of an inducible hexose-phosphate transport system among various bacteria. J Bacteriol (1973) 116(2):1079–81. doi: 10.1128/jb.116.2.1079-1081.1973
45. Lidbury IDEA, Borsetto C, Murphy ARJ, Bottrill A, Jones AME, Bending GD, et al. Niche-adaptation in plant-associated bacteroidetes favours specialisation in organic phosphorus mineralisation. ISME J (2021) 15(4):1040–55. doi: 10.1038/s41396-020-00829-2
46. Maloney PC, Ambudkar SV, Sonna LA, Varadhachary’ A. Anion-exchange mechanisms in bacteria. Microbiol Rev (1990) 54:17. doi: 10.1128/mr.54.1.1-17.1990
47. Lemieux MJ, Huang Y, Wang DN. Glycerol-3-phosphate transporter of escherichia coli: Structure, function and regulation. Res Microbiol (2004) 155(8):623–9. doi: 10.1016/j.resmic.2004.05.016
48. Wanner BL. Phosphorus assimilation and control of the phosphate regulon. In: Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, et al, editors. Escherichia coli and salmonella: Cellular and molecular biology. American society for microbiology, ed. 2 . (Washington, D.C.: American Society for Microbiology Press) (1996).
49. Zhang S, Zheng Q, Noll L, Hu Y, Wanek W. Environmental effects on soil microbial nitrogen use efficiency are controlled by allocation of organic nitrogen to microbial growth and regulate gross n mineralization. Soil Biol Biochem (2019) 135:304–15. doi: 10.1016/j.soilbio.2019.05.019
50. Miller WP, Miller DM. A micro-pipette method for soil mechanical analysis. Commun Soil Sci Plant Anal (1987) 18(1):1–15. doi: 10.1080/00103628709367799
51. Sinsabaugh RL, Klug MJ, Collins HP, Yeager PE, Petersen SO. Characterizing soil microbial communities. In: Robertson GP, Coleman DC, Bledsoe CS, Sollins P, editors. Standard soil methods for long-term ecological research. Oxford: Oxford University Press (1999). Available from: https://lter.kbs.msu.edu/pub/2713
52. Tabatabai MA. Soil enzymes. In: Weaver RW, Angle S, Bottomley P, Bezdicek D, Smith S, Tabatabai A, et al, editors. Methods of soil analysis part 2 microbiological and biochemical properties. Madison, USA: Soil Science Society of America (1994).
53. D’Angelo E, Crutchfield J, Vandiviere M. Rapid, sensitive, microscale determination of phosphate in water and soil. J Environ Qual (2001) 30(6):2206–9. doi: 10.2134/jeq2001.2206
54. Rowland AP, Haygarth PM. Determination of total dissolved phosphorus in soil solutions. J Environ Qual (1997) 26(2):410–5. doi: 10.2134/jeq1997.00472425002600020011x
55. Brookes PC, Powlson DS, Jenkinson DS. Measurement of microbial biomass phosphorus in soil. Soil Biol Biochem (1982) 14(4):319–29. doi: 10.1016/0038-0717(82)90001-3
56. Quiquampoix H, Mousain D. Enzymatic hydrolysis of organic phosphorus. In: Turner BL, Frossard E, Baldwin D, editors. Organic phosphorus in the environment. Wallingford, UK: CABI Publishing (2005).
57. Amadou I, Faucon MP, Houben D. New insights into sorption and desorption of organic phosphorus on goethite, gibbsite, kaolinite and montmorillonite. Appl Geochem (2022) 143:105378. doi: 10.1016/j.apgeochem.2022.105378
58. Jayachandran K, Schwab AP. Hetrick BAD. partitioning dissolved inorganic and organic phosphorus using acidified molybdate and isobutanol. Soil Sci Soc Am J (1992) 56(3):762–5. doi: 10.1016/0038-0717(92)90012-M
59. Mooshammer M, Wanek W, Schnecker J, Wild B, Leitner S, Hofhansl F, et al. Stoichiometric controls of nitrogen and phosphorus cycling in decomposing beech leaf litter. Ecology. (2012) 93(4):770–82. doi: 10.1890/11-0721.1
60. Corbridge DEC. Phosphorus: Chemistry, biochemistry and technology, sixth edition. Boca Raton, USA: CRC Press (2013).
61. Bowman RA, Cole CV. Transformations of organic phosphorus substrates in soils evaluated by NaHCO3 extraction. Soil Sci (1978) 125(1):49–54. doi: 10.1097/00010694-197801000-00008
62. Drobníková V. Factors influencing the determination of phosphatases in soil. Folia Microbiol (Praha) (1961) 6(4):260–7. doi: 10.1007/BF02872531
63. Mclaughlin M, Alston A, Martin J. Phosphorus cycling in wheat-pasture rotations. III. organic phosphorus turnover and phosphorus cycling. Soil Res (1988) 26(2):343. doi: 10.1071/SR9880343
64. Fischer W. Physiology of lipoteichoic acids in bacteria. In: Advances in microbial physiology. Elsevier (1988). p. 233–302. Available at: https://linkinghub.elsevier.com/retrieve/pii/S0065291108603495.
65. Myers CL, Li FKK, Koo BM, El-Halfawy OM, French S, Gross CA, et al. Identification of two phosphate starvation-induced wall teichoic acid hydrolases provides first insights into the degradative pathway of a key bacterial cell wall component. J Biol Chem (2016) 291(50):26066–82. doi: 10.1074/jbc.M116.760447
66. Wise EM, Glickman RS, Teimer E. Teichoic acid hydrolase activity in soil bacteria. Proc Natl Acad Sci (1972) 69(1):233–7. doi: 10.1073/pnas.69.1.233
67. Kusser W, Fiedler F. A novel glycerophosphodiesterase from Bacillus pumilus. FEBS Lett (1984) 30;166(2):301–6. doi: 10.1016/0014-5793(84)80100-3
68. Jorge AM, Schneider J, Unsleber S, Xia G, Mayer C, Peschel A. Staphylococcus aureus counters phosphate limitation by scavenging wall teichoic acids from other staphylococci via the teichoicase GlpQ. J Biol Chem (2018) 293(38):14916–24. doi: 10.1074/jbc.RA118.004584
69. Corda D, Mosca MG, Ohshima N, Grauso L, Yanaka N, Mariggiò S. The emerging physiological roles of the glycerophosphodiesterase family. FEBS J (2014) 281(4):998–1016. doi: 10.1111/febs.12699
70. Tollefson TS, McKercher RB. The degradation of 14C-labelled phosphatidyl choline in soil. Soil Biol Biochem (1983) 15(2):145–8. doi: 10.1016/0038-0717(83)90095-0
71. Hayaishi O, Kornberg A. Metabolism of phospholipides by bacterial enzymes. J Biol Chem (1954) 206(2):647–63. doi: 10.1016/S0021-9258(19)50833-X
72. Ölinger R, Margesin R, Kandeler E. Enzymes involved in phosphorus metabolism. In: Schinner F, Öhliner R, Kandeler E, Margesin R, editors. Methods in soil biology. (Berlin: Springer) (1996). doi: 10.1007/978-3-642-60966-4
73. Larson TJ, Ehrmann M, Boos W. Periplasmic glycerophosphodiester phosphodiesterase of escherichia coli, a new enzyme of the glp regulon. J Biol Chem (1983) 258(9):5428–32. doi: 10.1016/S0021-9258(20)81908-5
74. Jorge AM, Schneider J, Unsleber S, Göhring N, Mayer C, Peschel A. Utilization of glycerophosphodiesters by Staphylococcus aureus: Staphylococcus aureus uses GPDs. Mol Microbiol (2017) 103(2):229–41. doi: 10.1111/mmi.13552
75. Barman A, Gohain D, Bora U, Tamuli R. Phospholipases play multiple cellular roles including growth, stress tolerance, sexual development, and virulence in fungi. Microbiol Res (2018) 209:55–69. doi: 10.1016/j.micres.2017.12.012
76. Levy-Booth DJ, Campbell RG, Gulden RH, Hart MM, Powell JR, Klironomos JN, et al. Cycling of extracellular DNA in the soil environment. Soil Biol Biochem (2007) 39(12):2977–91. doi: 10.1016/j.soilbio.2007.06.020
77. Blum SAE, Lorenz MG, Wackernagel W. Mechanism of retarded DNA degradation and prokaryotic origin of DNases in nonsterile soils. Syst Appl Microbiol (1997) 20(4):513–21. doi: 10.1016/S0723-2020(97)80021-5
78. Torti A, Lever MA, Jørgensen BB. Origin, dynamics, and implications of extracellular DNA pools in marine sediments. Mar Genomics (2015) 24:185–96. doi: 10.1016/j.margen.2015.08.007
79. Wasmund K, Pelikan C, Schintlmeister A, Wagner M, Watzka M, Richter A, et al. Genomic insights into diverse bacterial taxa that degrade extracellular DNA in marine sediments. Nat Microbiol (2021) 6(7):885–98. doi: 10.1038/s41564-021-00917-9
80. Turner BL, Haygarth PM. Phosphatase activity in temperate pasture soils: Potential regulation of labile organic phosphorus turnover by phosphodiesterase activity. Sci Total Environ (2005) 344(1–3):27–36. doi: 10.1016/j.scitotenv.2005.02.003
81. Fransson AM, Jones DL. Phosphatase activity does not limit the microbial use of low molecular weight organic-p substrates in soil. Soil Biol Biochem (2007) 39(5):1213–7. doi: 10.1016/j.soilbio.2006.11.014
82. Pietramellara G, Ascher J, Borgogni F, Ceccherini MT, Guerri G, Nannipieri P. Extracellular DNA in soil and sediment: fate and ecological relevance. Biol Fertil Soils (2009) 45(3):219–35. doi: 10.1007/s00374-008-0345-8
83. Dubelman S, Fischer J, Zapata F, Huizinga K, Jiang C, Uffman J, et al. Environmental fate of double-stranded RNA in agricultural soils. Smagghe G editor PloS One (2014) 9(3):e93155. doi: 10.1371/journal.pone.0093155
84. Kunadiya MB, Burgess TI, Dunstan W, White D, StJ. Hardy GE. Persistence and degradation of Phytophthora cinnamomi DNA and RNA in different soil types. Environ DNA (2021) 3(1):92–104. doi: 10.1002/edn3.127
85. Shang C, Huang PM, Stewart JWB. Kinetics of adsorption of organic and inorganic phosphates by short-range ordered precipitate of alumnium. Can J Soil Sci (1990) 70(3):461–70. doi: 10.4141/cjss90-045
86. Celi L, Barberis E. Abiotic stabilization of organic phosphorus in the environment. In: Turner BL, Frossard E, Baldwin D, editors. Organic phosphorus in the environment. (Wallingford, UK: CABI Publishing) (2005).
87. Yan YP, Liu F, Li W, Liu F, Feng XH, Sparks DL. Sorption and desorption characteristics of organic phosphates of different structures on aluminium (oxyhydr)oxides: Sorption-desorption of organic p on Al oxyhydroxides. Eur J Soil Sci (2014) 65(2):308–17. doi: 10.1111/ejss.12119
88. Jørgensen N, Jacobsen C. Bacterial uptake and utilization of dissolved DNA. Aquat Microb Ecol (1996) 11:263–70. doi: 10.3354/ame011263
89. Acimovic Y, Coe IR. Molecular evolution of the equilibrative nucleoside transporter family: Identification of novel family members in prokaryotes and eukaryotes. Mol Biol Evol (2002) 19(12):2199–210. doi: 10.1093/oxfordjournals.molbev.a004044
90. Cabrita MA, Baldwin SA, Young JD, Cass CE. Molecular biology and regulation of nucleoside and nucleobase transporter proteins in eukaryotes and prokaryotes. Biochem Cell Biol (2002) 80(5):623–38. doi: 10.1139/o02-153
91. Wickner RB. Mutants of Saccharomyces cerevisiae that incorporate deoxythymidine-5′-Monophosphate into deoxyribonucleic acid in vivo. J Bacteriol (1974) 117(1):252–60. doi: 10.1128/jb.117.1.252-260.1974
92. Finkel SE, Kolter R. DNA As a nutrient: Novel role for bacterial competence gene homologs. J Bacteriol (2001) 183(21):6288–93. doi: 10.1128/JB.183.21.6288-6293.2001
93. Mell JC, Redfield RJ. Natural competence and the evolution of DNA uptake specificity. J Bacteriol (2014) 196(8):1471–83. doi: 10.1128/JB.01293-13
94. Chen I, Dubnau D. DNA Uptake during bacterial transformation. Nat Rev Microbiol (2004) 2(3):241–9. doi: 10.1038/nrmicro844
95. Bloemers HPJ, Koningsberger VV. Uptake of large molecular weights radioactive RNA by protoplasts of saccharomyces carlsbergensis. Nature (1967) 214:487–8. doi: 10.1038/214487a0
96. Disney MD, Haidaris CG, Turner DH. Uptake and antifungal activity of oligonucleotides in Candida albicans. Proc Natl Acad Sci (2003) 18;100(4):1530–4. doi: 10.1073/pnas.0337462100
97. Dietz GW. The hexose phosphate transport system of escherichia coli. In: Meister A, editor. Advances in enzymology and related areas of molecular biology, volume 44. Hoboken, USA: John Wiley & Sons, Inc. (1976).
98. Lyon AJE, Lucas RL. The phosphorus metabolism of rhizopus stolonifer and chaetomium sp. with respect to phosphorus translocation. New Phytol (1969) 68(4):971–6. doi: 10.1111/j.1469-8137.1969.tb06497.x
99. Albermann C, Weiner M, Tröndle J, Weuster-Botz D, Sprenger GA. Utilization of organophosphate:phosphate antiporter for isotope-labeling experiments in e. coli FEMS Microbiol Lett (2014) 361(1):52–61. doi: 10.1111/1574-6968.12612
100. Haferkamp I, Schmitz-Esser S, Wagner M, Neigel N, Horn M, Neuhaus HE. Tapping the nucleotide pool of the host: novel nucleotide carrier proteins of protochlamydia amoebophila. Mol Microbiol (2006) 60(6):1534–45. doi: 10.1111/j.1365-2958.2006.05193.x
101. Nash DM, Haygarth PM, Turner BL, Condron LM, McDowell RW, Richardson AE, et al. Using organic phosphorus to sustain pasture productivity: A perspective. Geoderma. (2014) 221–222:11–9. doi: 10.1016/j.geoderma.2013.12.004
102. Joanisse GD, Bradley RL, Preston CM, Munson AD. Soil enzyme inhibition by condensed litter tannins may drive ecosystem structure and processes: the case of kalmia angustifolia. New Phytol (2007) 175(3):535–46. doi: 10.1111/j.1469-8137.2007.02113.x
103. Tietjen T G, Wetzel R. Extracellular enzyme-clay mineral complexes: Enzyme adsorption, alteration of enzyme activity, and protection from photodegradation. Aquat Ecol (2003) 37(4):331–9. doi: 10.1023/B:AECO.0000007044.52801.6b
104. Marklein AR, Houlton BZ. Nitrogen inputs accelerate phosphorus cycling rates across a wide variety of terrestrial ecosystems. New Phytol (2012) 193(3):696–704. doi: 10.1111/j.1469-8137.2011.03967.x
105. Barrow NJ. Soil phosphate chemistry and the p-sparing effect of previous phosphate applications. Plant Soil (2015) 397(1–2):401–9. doi: 10.1007/s11104-015-2514-5
106. Lilienfein J, Qualls RG, Uselman SM, Bridgham SD. Adsorption of dissolved organic and inorganic phosphorus in soils of a weathering chronosequence. Soil Sci Soc Am J (2004) 68(2):620–8. doi: 10.2136/sssaj2004.6200
107. Condron LM, Turner BL, Cade-Menun BJ. Chemistry and dynamics of soil organic phosphorus. In: Sims JT, Sharpley AN, editors. Phosphorus: Agriculture and the environment agronomy 46. Madison, USA: Soil Science Society of America, Inc. (2005).
108. Vandecar KL, Lawrence D, Wood T, Oberbauer SF, Das R, Tully K, et al. Biotic and abiotic controls on diurnal fluctuations in labile soil phosphorus of a wet tropical forest. Ecology. (2009) 90(9):2547–55. doi: 10.1890/08-1516.1
109. Richardson AE, Lynch JP, Ryan PR, Delhaize E, Smith FA, Smith SE, et al. Plant and microbial strategies to improve the phosphorus efficiency of agriculture. Plant Soil (2011) 349(1–2):121–56. doi: 10.1007/s11104-011-0950-4
110. Frossard E, Sinaj S. The isotope exchange kinetic technique: A method to describe the availability of inorganic nutrients. applications to K, p, s and zn. Isotopes Environ Health Stud (1997) 33(1–2):61–77. doi: 10.1080/10256019808036360
Keywords: organic phosphorus, soil microbes, phosphorus uptake, decomposition, depolymerization, microbial activity, phosphatase, soil phosphorus cycle
Citation: Wasner D, Prommer J, Zezula D, Mooshammer M, Hu Y and Wanek W (2023) Tracing 33P-labelled organic phosphorus compounds in two soils: New insights into decomposition dynamics and direct use by microbes. Front. Soil Sci. 3:1097965. doi: 10.3389/fsoil.2023.1097965
Received: 14 November 2022; Accepted: 18 January 2023;
Published: 23 February 2023.
Edited by:
Federica Tamburini, ETH Zürich, SwitzerlandReviewed by:
Sebastian Loeppmann, University of Kiel, GermanySteffen A. Schweizer, Technical University of Munich, Germany
Copyright © 2023 Wasner, Prommer, Zezula, Mooshammer, Hu and Wanek. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniel Wasner, ZGFuaWVsLndhc25lckB1c3lzLmV0aHouY2g=; Wolfgang Wanek, d29sZmdhbmcud2FuZWtAdW5pdmllLmFjLmF0
 Daniel Wasner
Daniel Wasner Judith Prommer2
Judith Prommer2 David Zezula
David Zezula Maria Mooshammer
Maria Mooshammer Yuntao Hu
Yuntao Hu Wolfgang Wanek
Wolfgang Wanek