- 1NanoScience Technology Center, University of Central Florida, Orlando, FL, United States
- 2Department of Physics, College of Sciences, University of Central Florida, Orlando, FL, United States
- 3Burnett School of Biomedical Sciences, College of Medicine, University of Central Florda, Orlando, FL, United States
- 4Departmnet of Material Science and Engineering, College of Engineering and Computer Science University of Central Florida, Orlando, FL, United States
Gelsolin is a calcium (Ca2+) dependent, pH sensitive actin-binding protein that regulates actin filament dynamics to remodel the actin cytoskeleton. It is known that gelsolin binding induces conformational changes of actin filaments, leading to filament severing. However, the influence of physiological conditions, such as pH variations, on gelsolin-mediated filament severing activities, mechanics and conformations remains unclear despite their role in actin-actin interactions. Using Total Internal Reflection Fluorescence (TIRF) microscopy imaging and pyrene fluorescence assays, we demonstrate that filament severing efficiencies by gelsolin are enhanced in acidic conditions. In addition, analysis of filament thermal fluctuations using TIRF reveals that gelsolin binding stiffens actin filaments. Furthermore, we show that gelsolin binding induces conformational changes in filaments by measuring the filament half-pitch using high resolution Atomic Force Microscopy imaging. Together, our results suggest that pH modulation plays a key role in gelsolin-mediated filament severing activities, bending mechanics, and conformational changes, which have implications in many cellular processes including cell motility and morphogenesis.
1 Introduction
The actin cytoskeleton forms, within the cell, dynamic architectures regulating the mechanical and structural properties necessary for motility, morphogenesis, force generation and membrane trafficking (Blanchoin et al., 2014; Pollard and Cooper, 2009; Pollard and Borisy, 2003). Numerous actin binding proteins (ABPs) contribute to remodeling the actin cytoskeleton by modulating its assembly/disassembly dynamics and mechanics (Merino et al., 2020). Gelsolin is a calcium (Ca2+) dependent, pH-sensitive ABP that nucleates, severs, and caps actin filaments (Kinosian et al., 1998; Nag et al., 2013; Sun et al., 1999), serving as a multifunctional regulator of cellular processes and metabolism. Actin filament length regulation by gelsolin is crucial in controlling cell structure and movement of the filopodia and lamellipodia (Silacci et al., 2004). Gelsolin has also been linked to many diseases such as amyloidosis, cancer, and cardiovascular diseases (Spinardi and Witke, 2007).
Intracellular pH plays an important role in regulating the actin cytoskeleton dynamics and many ABPs including gelsolin. Acidic pH enhances actin filament elongation rates and decreases critical concentrations of actin (Crevenna et al., 2013; Zimmerle and Frieden, 1988). Moreover, previous studies have shown that low pH (<6) increases gelsolin-actin complex formation and induces gelsolin-mediated severing by lowering the transient Ca2+ requirement for gelsolin activation (Lamb et al., 1993; Lagarrigue et al., 2003; Garg et al., 2011; Fan et al., 2017). While cytoplasmic pH is maintained within the neutral pH (range of 7.0–7.5), cellular acidification in response to biological processes such as apoptosis (Kothakota et al., 1997; Matsuyama et al., 2000), stress responses (Jin et al., 2022) and pathological conditions (Decker et al., 2021; Salameh et al., 2014) can influence intracellular pH processes. When an influx of hydrogen (H+) and Ca2+ ions occurs near the plasma membrane, large pH fluctuations may impact the reorganization of the actin cytoskeleton as well as gelsolin activation (Ritter et al., 1998). Although the effects of Ca2+ concentrations on gelsolin-mediated filament severing have been extensively studied (Kinosian et al., 1998; Selden et al., 1998), the effects of varying pH on filament severing activity of gelsolin are not well understood.
The effects of gelsolin on actin filament mechanics and conformations are also important for understanding filament severing, as gelsolin-induced structural changes and mechanics precedes severing. A previous time resolved phosphorescence (TPA) and absorption anisotropy (TAA) spectroscopy study demonstrated that gelsolin binding at filament barbed ends propagates conformational change that alters filament dynamics and reduces torsional rigidity (Prochniewicz et al., 1996). Furthermore, small angle x-ray scattering (SAXS) experiments conducted at low pH determined that severing occurs because of gelsolin-induced structural changes in filaments (Garg et al., 2011). This supports the hypothesis that changes in actin filament mechanics and conformations may be linked to gelsolin-induced severing. However, how gelsolin affects filament bending mechanics and conformational changes at varying pH has not been known.
Here, we investigated the effects of intracellular pH and gelsolin binding on actin cytoskeleton dynamics by studying severing activity, mechanics, and conformations of actin filaments in neutral and acidic conditions. We evaluated gelsolin-mediated filament severing and bending mechanics using Total Internal Fluorescence (TIRF) microscopy and pyrene fluorescence assays, and measured filament height and half-pitch changes with gelsolin binding using Atomic Force Microscopy (AFM) imaging. This study demonstrates how acidic pH enhances gelsolin-mediated filament severing activities and how gelsolin binding regulates the mechanics and conformations of actin filaments in physiological conditions.
2 Materials and methods
2.1 Actin purification and sample preparation
Rhodamine-labeled actin from rabbit skeletal muscle and human recombinant plasma gelsolin were purchased from Cytoskeleton Inc (Denver, CO). Unlabeled actin was purified from rabbit skeletal muscle acetone powder (Pel-Freeze Biologicals Inc., Rogers, AR), then gel filtered over a Sephacryl S-300 size exclusion column equilibrated in buffer A [2 mM Tris-HCl pH 8, 0.2 mM CaCl2, 1 mM NaN3, 0.2 mM, adenosine 5-triphosphate (ATP), and 0.5 mM dithiothreitol (DTT)] as previously described (Pardee and Spudich, 1982). Purified actin was fluorescently labeled with Alexa-488 succimidyl ester dye as previously described (Kang et al., 2012; Park et al., 2022), yielding a labeling efficiency of ∼0.18–0.5 fluorophores per actin monomer. Ca2+-G-actin (actin monomer) underwent cation exchange to form Mg2+-G-actin by the addition of MgCl2 at a concentration equal to the initial G-actin concentration plus 10 μM and 0.2 mM ethylene glycolbis (β-aminoethyl ether)-N,N,N’N-tetraacetic acid (EGTA).
2.2 Total internal reflection fluorescence (TIRF) microscopy imaging and analysis
Actin polymerization was initiated at 1-2 µM actin (50% Rhodamine labeled or 20% Alexa 488 labeled) using 1/10th total volume of 10X KMCI polymerization buffer (100 mM imidazole, 500 mM KCl, 20 mM MgCl2, 3 mM CaCl2, 10 mM ATP, and 10 mM DTT, pH 7.5 or 6.0) at room temperature for 1-2 h. Actin was diluted to 0.74–1 μM, following the addition of 5.5–10 nM gelsolin to obtain a gelsolin to actin molar ratio of 1:134 or 1:100. Samples were diluted using optical imaging buffer containing 0.2 mg/mL glucose oxidase, 1 mg/mL catalase and 15 mM glucose to minimize photobleaching then fixed to 0.1% v/v poly-L-lysine treated coverslips. Time-dependent TIRF imaging was conducted over various time (0, 1, 5, 10, 20, and 30 min) intervals. Images were acquired using a Nikon Eclipse Ti TIRF microscope equipped with a Hamamatsu Image EM X2 CCD Camera, a ×100 oil immersion objective, Nikon LU-N4 laser and Nikon Imaging Software (NIS) Elements (version 4.50) (pixel size = 0.16 µm) Average filament lengths were measured using ImageJ (NIH) (Schindelin et al., 2012) and the Persistence Software (Graham et al., 2014). Statistical analysis was performed using ANOVA one-way and post hoc Tukey tests. Length distributions were plotted using the Gaussian (Equation 1) and log-normal (Equation 2) functions to fit the data:
where
where
2.3 Flow cell preparation and analysis
Functionalized flow cells were modified from previously described protocols (Heidings et al., 2020; Winterhoff et al., 2016). Coverslips were cleaned by sonicating at 60°C in 1M KOH, 1M HCl, and 70% ethanol for 45 min, then thoroughly rinsed with warm ddH2O. 200 nM of N-ethylmalemide (NEM)-inactivated myosin and 1 mg/mL bovine serum albumin (BSA) solutions were separately loaded into a flow cell and left to incubate for 5 min before loading filaments into the flow cell. Images were taken every 1 s to visualize filament severing before and after the addition of gelsolin. To quantify the change in average filament length over time, the data points were fitted with exponential decay functions. The fit yielded the decay time constant (t1) which is inversely proportional to the rate of gelsolin severing (Heidings et al., 2020). Negative control experiments were done with actin filaments and gelsolin in the absence of Ca2+ using 1X KMI polymerization buffer (10 mM imidazole, 50 mM KCl, 2 mM MgCl2, 1 mM ATP, and 1 mM DTT, pH 7.5 or 6.0).
2.4 Pyrene assays
Bulk actin depolymerization was measured using pyrene-labeled G-actin (1–2 μM, 10% pyrene labeled) as previously described (Vemula et al., 2021; Zhang et al., 2006). F-actin samples were diluted below the actin critical concentration (∼100 nM) in 1X KMCI buffer without or with gelsolin [(gelsolin]: [actin]),1:125]. The time course of pyrene fluorescence intensities was monitored using the SpectraMax Gemini XPS microplate reader equipped with a Molecular Devices SoftMax Pro software (Version 7.0) (λex = 365 nm and λem = 407 nm) for 5 min (Δt = 1 s) at room temperature. Negative control experiments were done with actin filaments and gelsolin in the absence of Ca2+ using 1X KMI polymerization buffer (10 mM imidazole, 50 mM KCl, 2 mM MgCl2, 1 mM ATP, and 1 mM DTT, pH 7.5 or 6.0).
2.5 Filament bending mode analysis
Bending persistence lengths of actin filaments with or without gelsolin were measured by analyzing thermal bending modes (Castaneda et al., 2019; Demosthene et al., 2024). A molar ratio of 1:185 [(gelsolin): (actin)] was used to prevent tumbling of short filaments. Coverslips were incubated in a buffer solution containing 1 mg/mL BSA, 50 mM Tris-HCl (pH 7.5), 1 mM NaN3, and 150 mM NaCl for 1 h to prevent filaments from adhering to the glass. Actin samples were dispensed onto BSA-treated coverslips, then sequential time-lapse images of thermally fluctuating filaments in the absence or presence of gelsolin at pH 7.5 or 6.0 were recorded using a Hamamatsu ORCA-Flash 4.0 V3 Digital CMOS camera (pixel size = 0.07 µm). Time-lapse videos were recorded for 1 min at 7–10 frames/s using the Nikon Imaging Software (NIS) Elements (version 4.50).
Fourier decomposition of filament shape with respect to the tangent angle [θ(s)] along the chosen segment length was processed as indicated by Equation 4 (Castaneda et al., 2019; Gittes et al., 1993):
Amplitude of the cosine is denoted by an and is nonzero when filaments are in a relaxed state, n is the mode number and L is the segment length of the filament. The amplitude variance
Flexural rigidity of actin filaments can also be defined using Equation 6:
where κ is the flexural rigidity of the filament, Lp is the persistence length, and kBT is the thermal energy (Gittes et al., 1993).
2.6 Molecular visualization
PyMOL software was used to visualize interactions of gelsolin domains 1, 2 and 3 (G1-G3) [PDB ID: 1RGI (Burtnick et al., 2004)] and gelsolin domains 4, 5 and 6 (G4-G6) [PBD ID: 1H1V (Choe et al., 2002)] with actin monomers. Ribbon diagram of actin is shown in hot pink and pink and gelsolin is shown in green, lime green and cyan. G1-G3 amino acid residues, Ala-116, Gly-114, and Asp-109 and actin residue, Glu-167 were represented as ball and stick structures. G4-G6 residues, Pro-494, Gly-492, Asp-487 and actin residue, Glu167՛ (adjacent actin monomer) were also represented as ball and stick. Actin and gelsolin complexes were shown to form salt bridges through Ca2+.
2.7 AFM imaging and analysis
A gelsolin to actin molar ratio of 1:100 or 1:134 [(actin) = 1 µM] was used for AFM experiments. These ratios are consistent with those chosen for steady-state TIRF imaging and yielded the best AFM images. Freshly cleaved mica substrates were coated 0.1% v/v (3-Aminopropyl)triethoxysilane (APTES) before putting actin. To determine the height of filaments with or without gelsolin in air, the AFM (Park NX10, Parks Systems Corp., South Korea) was operated in non-contact mode (NCM) at a scanning rate of 0.8–1 Hz, 512 × 512 or 1,024 × 1,024 points and scan size range of 0.5–1.0 µm. PPP NCHR (Nanosensors, Switzerland) AFM probe was used with a nominal spring constant of 42 N/m and nominal resonance frequency of 300–330 kHz. Height images were corrected using minimal data processing. The minimum height of the images was fixed to zero and height profiles were extracted using the Gwyddion Software (Nečas and Klapetek, 2012). Helical half-pitch conformational changes were determined by measuring the periodic variations corresponding to G-actin subunits along the filament longitudinal profile section (Ikawa et al., 2007; Gilmore et al., 2013). Statistical analysis was performed using ANOVA one-way and post hoc Tukey tests.
3 Results and discussion
3.1 Filament severing efficiency and disassembly by gelsolin increases at lower pH
To evaluate the effect of pH on gelsolin-mediated actin filament severing, we calculated the steady-state average filament length (Lavg) using TIRF microscopy imaging (Figure 1A). The Lavg (Figure 1B) and length distributions (Supplementary Figures S1, S2) were used to determine filament severing efficiency in the presence of gelsolin and varying pH. 1 min after the addition of gelsolin to actin filaments (1 μM, 20% Alexa 488-labeled) at pH 7.5 (at a molar ratio of 1:100), the average filament lengths decreased from Lavg, pH7.5 = 5.2 ± 0.2 µm to Lavg, pH 7.5, gel = 3.2 ± 0.1 µm (± standard error of mean, S.E.M) (Figure 1B). At pH 6.0, the control average filament length significantly decreased compared to pH 7.5 (Lavg, pH6.0 = 4.5 ± 0.1 µm) in the absence of gelsolin. Upon addition of gelsolin at pH 6.0, Lavg, pH6.0, gel decreased further to 2.4 µm. At 5 min, Lavg, pH 7.5, gel=3.2 ± 0.1 µm and Lavg, pH 6.0, gel = 2.3 µm were recorded, and at 10 min, Lavg, pH 7.5, gel=2.9 ± 0.1 µm and Lavg, pH 6.0, gel = 2.3 ± 0.1 µm were recorded. Overall, the actin filaments at pH 6.0 are significantly shorter than those at pH 7.5. 15 min after gelsolin addition, average filament lengths were similar at pH 6.0 (Lavg, pH 6.0, gel = 2.3 ± 0.1 µm) and pH 7.5 (Lavg, pH 7.5, gel=2.4 ± 0.1 µm). We also assessed the effect of pH on steady-state filament length distribution in the absence and presence of gelsolin. This data was fitted using the Gaussian and Log-normal functions (Supplementary Figures S1, S2) and indicates that the length distribution in the presence of gelsolin at pH 6.0 narrows down earlier than at pH 7.5.
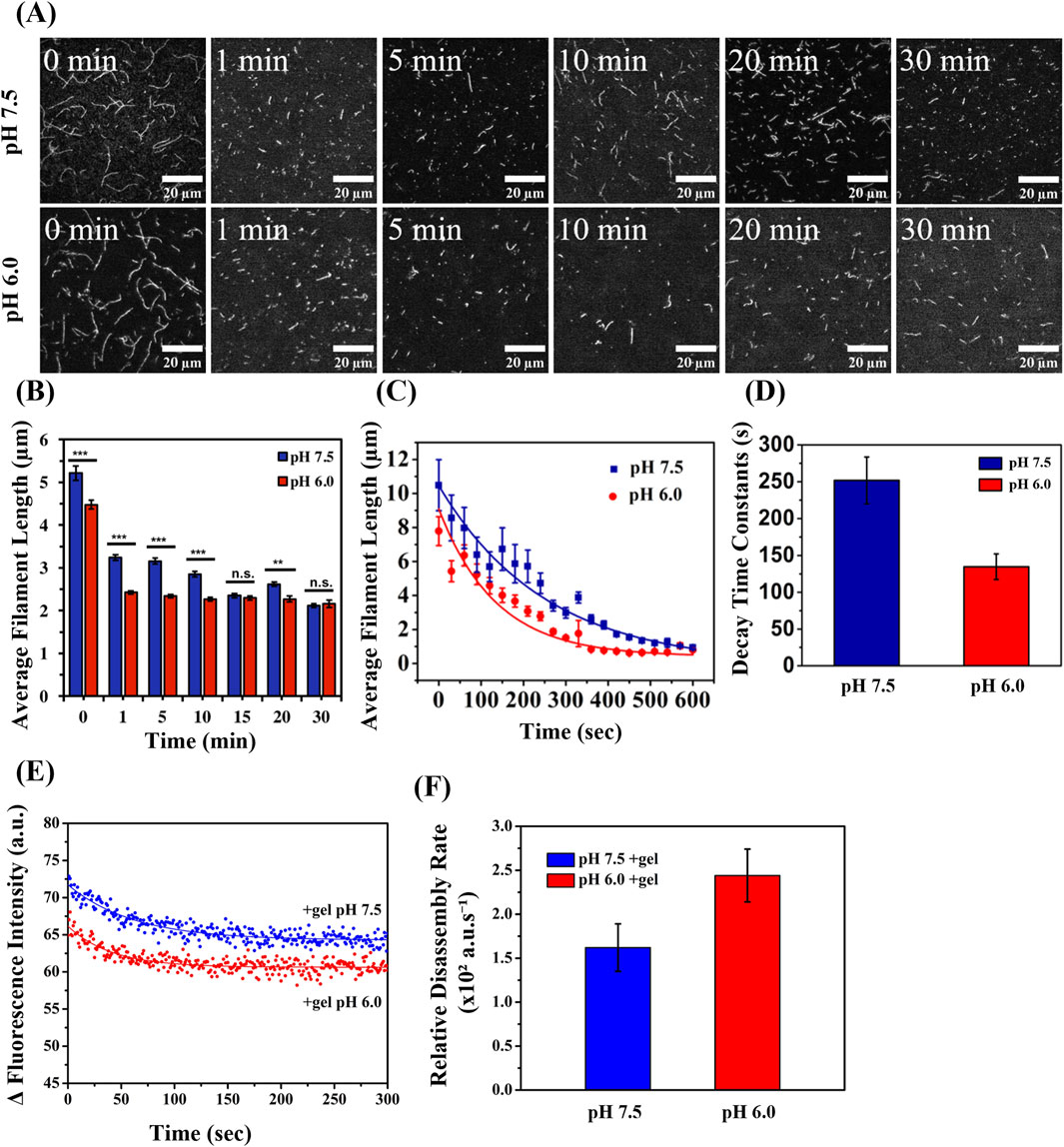
Figure 1. Filament Severing Efficiency and Disassembly by Gelsolin at pH 6.0 and 7.5. (A) Representative TIRF microscopy images of steady-state actin filaments (1 μM, 20% Alexa 488-labeled) with gelsolin (10 nM) at pH 7.5 and 6.0 at the indicated time. (Scale bars, 20 µm) (B) Steady-state actin filament lengths at pH 7.5 and 6.0 in the presence and absence of gelsolin. Statistical analysis was performed using the Turkey test (N = 34, *p < 0.05, **p < 0.01, ***p < 0.001) (N = 519–4,542). (C) Real-time actin filament lengths in varied pH (7.5 and 6.0) fit with an exponential decay function Equation 3. (D) Decay time constantpH7.5 = 251.82 ± 31.7 s, decay time constantpH6.0 = 134.71 ± 17.37 s. Buffer 1X KMCI: 10 mM imidazole, pH 7.5 or 6.0, 50 mM KCl, 2 mM MgCl2, 0.3 mM CaCl2, 1 mM ATP, 1 mM DTT (N = 34–182). Uncertainty bars represent standard error of the mean (S.E.M.). (E) Pyrene-labeled actin (10% labeled) was polymerized and diluted to 100 nM in 1X KMCI: 10 mM imidazole, pH 7.5 or 6.0, 50 mM KCl, 2 mM MgCl2, 0.3 mM CaCl2, 1 mM ATP, 1 mM DTT supplemented with 0.8 nM gelsolin at a gelsolin to actin molar ratio of 1:125 (N = 7). (F) Relative disassembly rates were derived from the inverse of the decay constants (disassembly ratepH7.5 = 1.61 ± 0.27 × 102 a.u./s, disassembly ratepH6.0 = 2.43 ± 0.30 × 102 a.u./s). Uncertainty bars represent the inverse of the standard error of the mean (S.E.M.).
Next, we directly visualized real-time filament severing activities utilizing a functionalized flow cell (Supplementary Videos S1, S2). Isolated filaments in their native shapes were tracked after the addition of gelsolin to monitor the evolution of Lavg over time. To determine the relative rate of gelsolin severing, the data was fitted using Equation 3 (Heidings et al., 2020) (Figure 1C). The decay time constant t1 at pH 7.5 was 251 ± 32 s and decreased to 135 ± 18 s at pH 6.0 (Figure 1D). A lower decay time constant indicates that gelsolin severed filaments more efficiently at pH 6.0 than at pH 7.5. Overall, we observed shorter Lavg at pH 6.0 compared to pH 7.5 in the absence of gelsolin. This agrees with a previous study that showed pH influences actin polymerization, resulting in filaments with shorter length at pH 5.8 than at pH 7.8 (Crevenna et al., 2013). This effect was attributed to modulation of the nuclei formation from G-actin by pH (Crevenna et al., 2013). Earlier studies suggested that average filament lengths or shortening were independent of pH (Wang et al., 1989), but recent studies and our data suggests this is not the case. When the effects of both pH and gelsolin were combined, a rapid decrease in average filament length and increase in severing rates were revealed using the real-time severing assays.
To understand the effect of pH on gelsolin-mediated filament disassembly, we used bulk pyrene assays to monitor variations in fluorescence intensities of pyrene-labeled actin over time (Lamb et al., 1993; Fan et al., 2017). The time-dependent fluorescence intensity (Figure 1E) was fitted using an exponential decay function which yielded decay constants (t1) for pH 7.5 (61.75 ± 3.70 s) and 6.0 (41.02 ± 3.33 s). We then derived the relative disassembly rates of actin filaments in the presence of gelsolin from the inverse of t1, which provided a rate of 1.61 ± 0.27 × 102 a.u./s at pH 7.5 and 2.43 ± 0.30 × 102 a.u./s at pH 6.0) (Figure 1F). This corresponds to a relative disassembly rate that was 1.5-fold faster at pH 6.0 in the presence of gelsolin. In the absence of gelsolin, no distinct change in fluorescence intensity was noted at pH 7.5 and 6.0 (Supplementary Figure S3). Both of our real-time severing activity data and bulk pyrene fluorescence data are consistent with previous studies that showed enhanced severing activity and actin depolymerization in the presence of full-length gelsolin or gelsolin fragment [residues 28–161 which consists of linker; (G1+)] at pH 5.0–6.0 (Lamb et al., 1993; Fan et al., 2017).
To test whether pH alone affected gelsolin-mediated actin severing, we performed negative controls in the absence of 0.3 mM CaCl2 using real-time severing assays. At pH 7.5 we observed no significant change in average filament length after the addition of gelsolin for negative controls (Supplementary Video S3; Supplementary Figure S4). In contrast, severing for negative control at pH 6.0 in the presence of gelsolin had a decrease in average filament length from 8.6 µm to 0.9 µm and decay time constantpH6.0 = 164.32 s (Supplementary Video S4). Gelsolin-mediated filament severing efficiency for negative controls at pH 6.0 was reduced compared to similar conditions supplemented with Ca2+. Using real-time severing assays, we showed that gelsolin-induced severing is inefficient at neutral pH in the absence of Ca2+. To confirm this data, we performed bulk pyrene fluorescence assays for both pH conditions. In the absence of Ca2+ we determined severing rates decreased by more than half at pH 6.0 (1.64 × 102 a.u./s) and an even greater decrease was quantified at pH 7.5 (0.12 × 102 a.u./s) in the presence of gelsolin (Supplementary Figures S5A,B). While Ca2+ plays an important role in inducing gelsolin-mediated severing, pH is also an important factor in modulating gelsolin for filament severing. Previous studies have shown that acidic pH (pH 5) lowers the requirement for Ca2+ due to bypassing gelsolin’s Ca2+ sensitive C-tail latch, while neutral pH conditions (pH 8) remain Ca2+ dependent (Lamb et al., 1993; Lagarrigue et al., 2003; Garg et al., 2011). This suggests that acidic pH dominates in regulating gelsolin severing while in the absence or presence of Ca2+.
3.2 Actin and gelsolin-bound filament bending stiffness increases in the presence of gelsolin at neutral and acidic pH
We further evaluated the effect of pH on filament bending mechanics in the absence or presence of gelsolin by using TIRF microscopy imaging. Thermal bending modes of freely fluctuating filaments were analyzed and filament bending persistence lengths were calculated (Supplementary Videos S5–S8; Figure 2). The mode amplitude variance [var (an)] calculated from the bending mode analysis (Castaneda et al., 2019; Gittes et al., 1993) (see Method section for details) indicates that thermal bending is affected by varying pH and gelsolin. In the absence of gelsolin, thermal bending was lower at pH 7.5 (Figure 2A) than at pH 6.0 (Figure 2B), as evidenced by reduced var(an) (Supplementary Videos S5, S6). “In the case of gelsolin-bound filaments, we observed reduced thermal bending and var(an) for both pH conditions compared to the control (Supplementary Videos S7, S8; Figures 2C, 2D). Reduced thermal bending and variance correlates to higher average bending Lp of filaments at pH 7.5 (Lp, pH7.5 = 17.3 ± 1.8 μm) in comparison to bending Lp of filaments at pH 6.0 (Lp, pH6.0 = 14.8 ± 1.3 μm) (Figure 2E) in the absence of gelsolin. Using the Lp values, we calculated the average flexural rigidity κ (Equation 6) to be 7.12 × 10−26 N m2 and 6.10 × 10−26 N m2 at pH 7.5 and 6.0, respectively, suggesting that actin filaments are slightly less rigid in acidic conditions. In addition, bending Lp at pH 7.5 (Lp, +gel pH 7.5 = 20.6 ± 1.2 μm) was 19% higher than the control Lp at pH 7.5, while Lp at pH 6.0 (Lp, +gel pH 6.0 = 18.6 ± 1.0 μm) was 26% higher than the control Lp at pH 6.0 (Figure 2F). Respective κ values were 8.55 × 10−26 N m2 and 7.67 × 10−26 N m2. Our results reveal lower Lp and κ values in acidic conditions for control and gelsolin-bound filaments at pH 6.0, with bending rigidity for control filaments at pH 6.0 being significantly lower than gelsolin-bound filaments at pH 7.5. While our study focuses on a more rigid actin species, Mg2+-F-actin, the trend is consistent with a previous study reporting that persistence lengths of Ca2+-F-actin decreases with decreasing pH (Arii and Hatori, 2008). Lowered flexural rigidity at pH 6 may be a result of the rapid depolymerization and instabilities at filament ends (Fan et al., 2017; Wioland et al., 2018), that may also impact the structure and helical pitch.
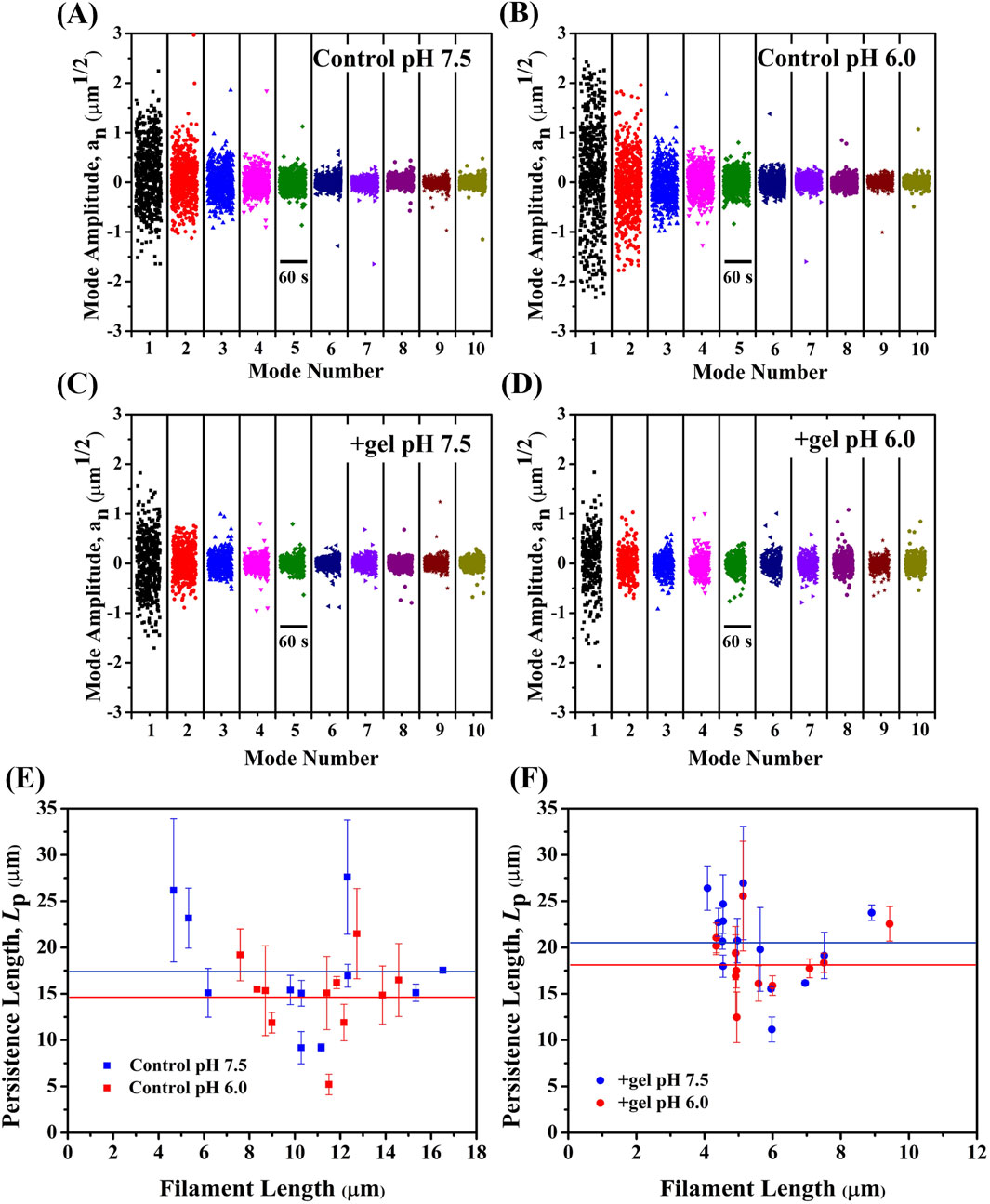
Figure 2. Gelsolin binding increases filament bending stiffness at pH 7.5 and 6.0. Amplitudes of 10 bending modes were plotted as a function of time for subpanels (t = 60 s) (A) Control pH 7.5 (B) Control pH 6.0 (C) with gelsolin pH 7.5 (D) with gelsolin pH 6.0. (E) Representative bending Lp versus filament length graph for control at pH 7.5 and 6.0. Blue solid line represents average Lp for control pH 7.5 (17.3 ± 6.1 µm), red solid line is control pH 6.0 (14.8 ± 4.2 µm). (F) +gel pH 7.5 and +gel pH 6.0 bending Lp versus filament length was plotted. Blue solid line represents + gel pH 7.5 (20.6 ± 4.4 µm) and red solid line is +gel pH 6.0 (18.6 ± 3.4 µm). N = 11–14 filaments per condition and uncertainty bars representing standard error of the mean (S.E.M.).
Comparing bending stiffness between control and gelsolin-bound filaments demonstrates that gelsolin binding renders filaments stiffer, indicating gelsolin’s ability to modulate filament bending mechanics. This agrees with a previous study showing that gelsolin binding regulates actin mechanics, where torsional rigidity had a three-fold decrease due to conformational changes (Prochniewicz et al., 1996). Yet, the mechanism contributing to gelsolin’s modulation of bending stiffness of actin filaments remains unclear. Another essential ABP, cofilin, was found to reduce both torsional (Prochniewicz et al., 2005) and flexural rigidity (McCullough et al., 2008) when binding to actin. However, we found that gelsolin binding increases actin filament flexural rigidity rather than decreasing it as seen with cofilin. We speculate different severing mechanisms are at play with cofilin and gelsolin. Cofilin was found to affect filament twist (Huehn et al., 2018), making filaments more flexible in twisting and bending (Prochniewicz et al., 2005; McCullough et al., 2008; Kang et al., 2014). Cofilin severing occurs at or near boundaries between bare actin and cofilin-bound segments, at sites of local mechanical and structural discontinuities (Elam et al., 2013; Tanaka et al., 2018). In contrast, gelsolin initiates severing by first attaching gelsolin domain 2 (G2) at the side of actin subdomain (SD) 1 and 2 of longitudinally adjacent monomers. Simultaneous repositioning of the gelsolin domain 1 and 2 (G1-G2) and gelsolin domain 3 and 4 (G3-G4) linkers allow gelsolin domain 1 (G1) and gelsolin domain 4 (G4) to bind between actin SD 1 and 3 of laterally adjacent monomers to induce steric clashing of inter subunit bonds (Nag et al., 2013).
G1 and G4 compete with actin monomers for binding, which induces severing, and also becomes stabilized by gelsolin type-1 Ca2+ binding sites formed between actin residue Glu 167 and gelsolin residues at the gelsolin/actin interfaces (Figure 3) (Nag et al., 2013; Burtnick et al., 2004). Glu167 has been shown to form ‘stiffness cation binding sites’ with adjacent monomers in SD 2 to regulate filament stiffness (Kang et al., 2012). Hence, gelsolin binding allows actin Glu167 to form a bond with residues at gelsolin Ca2+ binding sites. Potassium (K+) and magnesium ions (Mg2+) get swapped with Ca2+ which binds between actin residue Glu167 and G1 residues (Asp109, Gly114, and Ala116) and adjacent actin residue Glu167՛ and G4 residues (Asp487, Gly492, and Pro494) (Figures 3A, B) (Nag et al., 2013; Burtnick et al., 2004; Choe et al., 2002; Barrie et al., 2024). This may modulate the bending and torsional rigidity of gelsolin-bound filaments. Furthermore, Fan et al. (2017) recently revealed that protonation of gelsolin residues (His29, Asp109, and His151) at acidic pH (<6.0) in Ca2+ free conditions induces local structural changes of G1+ and G1+/actin interfaces to enhance binding and pH-dependent severing. We can speculate that structural rearrangement on gelsolin/actin interfaces from Ca2+ and pH will also impact filament mechanics as it did for severing dynamics.
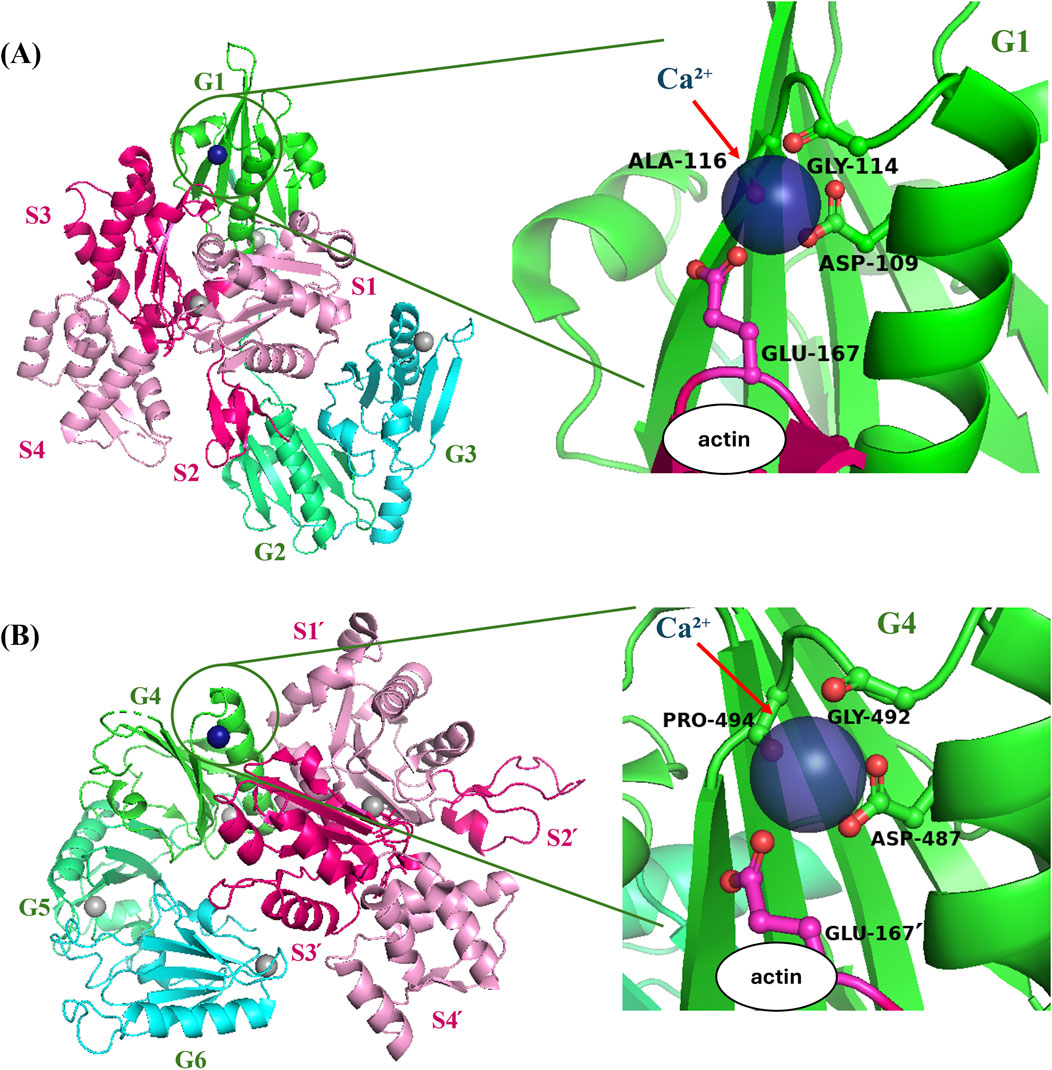
Figure 3. Gelsolin residues interact with actin residue Glu167 at the gelsolin/actin interface (A) Ribbon diagram of actin is shown in hot pink (SD2 and SD3) and pink (SD 1 and SD4), and gelsolin domains are shown in green (G1), lime green (G2) and cyan (G3), visualized using PyMOL. Gray spheres represent Ca2+ bound to either actin or gelsolin. The green circles show the zoomed in area of protein complexes in the right panel. G1-G3: actin complex [PDB ID:1RGI (36)] is formed through Ca2+ (navy blue sphere) salt bridge of actin and gelsolin amino acid residues. (B) Gelsolin domains are shown in green (G4), lime green (G5) and cyan (G6). Actin domains and residues are labeled with (՛), signifying adjacent actin monomer and residues. G4–G6:actin structure [PDB ID:1H1V (Choe et al., 2002)] also forms salt bridge between actin and gelsolin residues.
Although it is not well understood how the effects of gelsolin binding on single filament mechanics are linked to the mechanical properties of cells, we anticipate its role in regulating cell stress and elastic modulus by referring to other severing ABPs. Actin Depolymerization Factor (ADF) and cofilin causes fluidization of the actin cytoskeleton and cell softening by remodeling contracted, solid filaments into relaxed, fluid-like filaments (Vakhrusheva et al., 2022). Increased bending stiffness of single filaments by gelsolin may modulate the viscoelasticity of cytoskeletal networks, thereby regulating overall cell mechanics.
3.3 Filament conformational changes are regulated by gelsolin in neutral and acidic pH conditions
We investigated conformational changes of F-actin associated with gelsolin binding at varied pH using AFM imaging. Changes in filament height was evaluated from topography images (Figure 4A). Selected lateral line profiles along filaments were used to extract their height values (Figure 4B) (Nečas and Klapetek, 2012; Ikawa et al., 2007). In the absence of gelsolin, we observed average filament height values of 5 nm at pH 7.5 and pH 6.0 (Figure 4C). The addition of gelsolin at pH 7.5 resulted in a decrease of the average height value (4 nm), which was slightly lower than the control. In contrast, the average filament height at pH 6.0 increased to 6 nm. When comparing gelsolin-bound filaments at both pH conditions, we observed significantly higher filament heights in acidic pH. We also noted that our height values were lower for actin filaments than previous values reported in High Speed (HS) AFM studies (∼8–10 nm) (Matusovsky et al., 2019; Ngo et al., 2015). This may be attributed to their use of mica supported bilayers in solution.
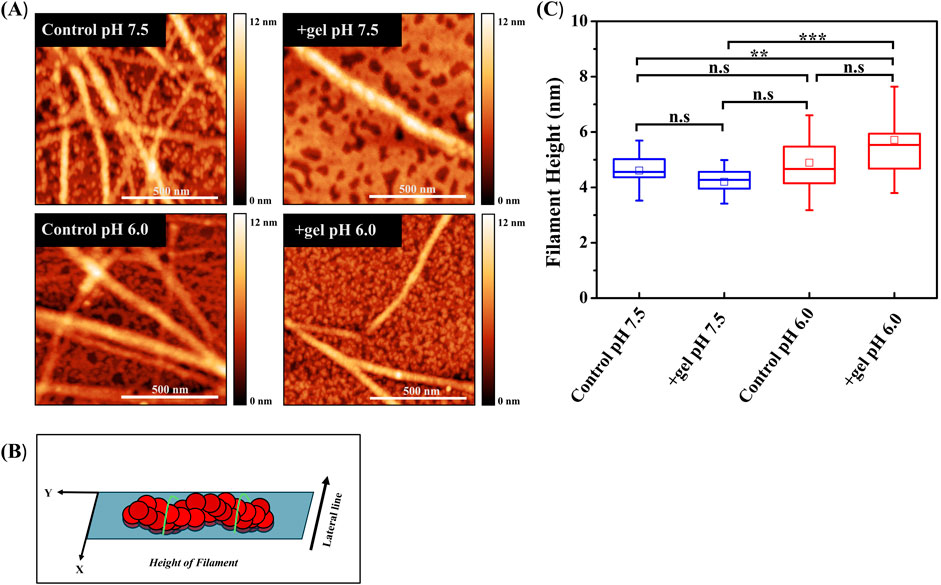
Figure 4. Gelsolin binding induces conformational changes in actin filament. (A) Representative AFM height images of control actin filaments and gelsolin-bound filaments at pH 7.5 and 6.0 (scale bar = 500 nm). (B) The schematic represents F-actin orientation with respect to analysis, where lateral green lines signify the area along which filament height was analyzed. (C) Box plot of actin filament heights in the absence (control) or presence of gelsolin at pH 7.5 and pH 6.0. Box signifies 25%–75% of data, whiskers represent SD, and smaller squares are the mean. Buffer 1X KMCI: 10 mM imidazole, pH 7.5 or 6.0, 50 mM KCl, 2 mM MgCl2, 0.3 mM CaCl2, 1 mM ATP, 1 mM DTT with gelsolin to actin molar ratio of 1:100 or 1:134 [(actin) = 1 µM]. Statistical analysis was performed using Tukey test; N = 13–30, [*p < 0.05, **p < 0.01, ***p < 0.001, n.s (not significant)].
To further understand the effects of pH on gelsolin-mediated conformational changes, we examined actin filament helical structure from topography images (Figure 5A). Longitudinal line profiles along filaments were utilized to extract half-pitch values (Figure 5B). We reported the half-pitch of actin filaments at neutral pH to be 39 ± 2 nm (Figure 5C), with gelsolin binding significantly lowering half-pitch to 32 ± 1 nm. Similarly, at pH 6.0 we also observed a significant decrease in actin half-pitch in the presence of gelsolin (39 ± 1 nm to 32 ± 1 nm). Control actin filaments exhibited a large variance and distribution in half-pitch values at both pH conditions. Our half-pitch values were higher than previously reported for actin (35–38 nm) and cofilin-bound filaments (26.9 nm) that underwent shortening and/or overtwisting (Matusovsky et al., 2019; Ngo et al., 2015; Shao et al., 2000; Jegou and Romet-Lemonne, 2020). Recent AFM studies observed a wide distribution for half-pitch and height values, indicative of structural polymorphism (Matusovsky et al., 2019; Ngo et al., 2015). This can be supported by a Molecular Dynamics (MD) simulation study demonstrating how gelsolin segement-1 modulated the orientation, and filament twist of different actin variants (Lee and Kang, 2020). Our data suggest that overtwisting of filaments may occur in the presence of gelsolin at pH 7.5 and 6.0.
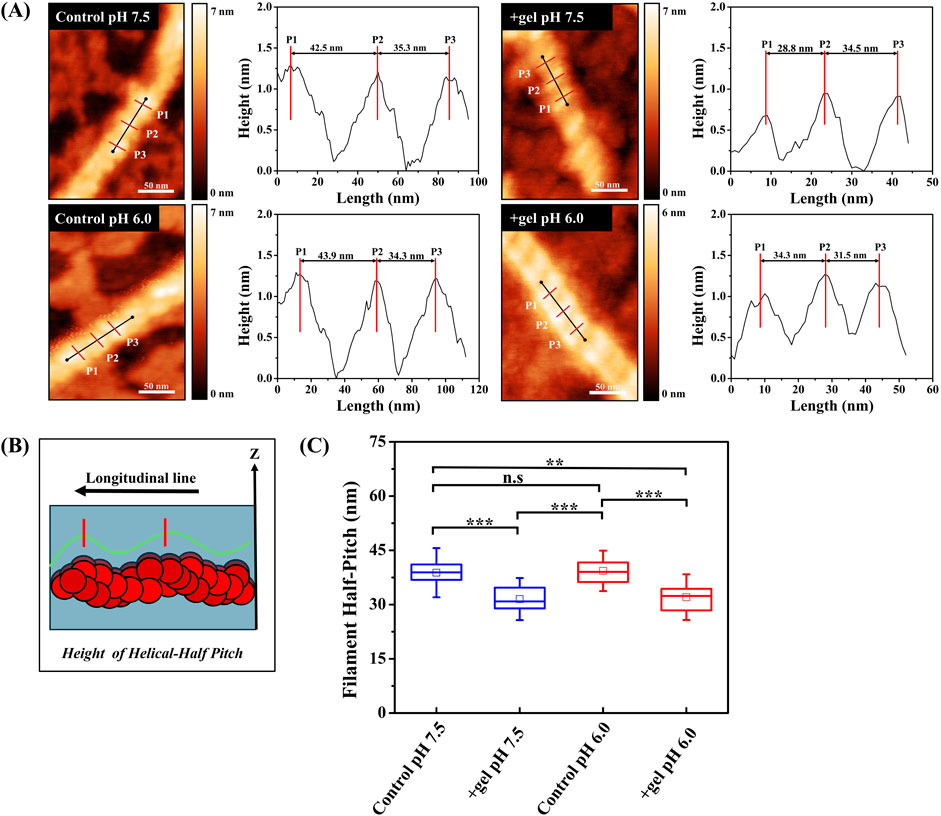
Figure 5. Gelsolin binding induces shortening of filament helical half-pitch. (A) Representative AFM height images of control actin filaments and gelsolin-bound filaments at pH 7.5 and 6.0 with helical half-pitch lengths profiles. Vertical red lines signify the peak of cross-over points or helical half-pitches. Scale bar = 50 nm. (B) The schematic represents F-actin orientation with respect to analysis, where the longitudinal green line signifies area along which filament half-pitch lengths were analyzed, and the red vertical lines indicates the peak of filament half-pitch. (C) Box plot of half-pitches for filaments in the absence or presence of gelsolin at pH 7.5 and 6.0. Box signifies 25%–75% of data, whiskers represent SD, and smaller squares are the mean. Buffer 1X KMCI: 10 mM imidazole, pH 7.5 or 6.0, 50 mM KCl, 2 mM MgCl2, 0.3 mM CaCl2, 1 mM ATP, 1 mM DTT with gelsolin to actin molar ratio of 1:100 or 1:134. Statistical analysis was performed using Tukey test; N = 9–14, [*p < 0.05, **p < 0.01, **p < 0.001, n.s (not significant)].
The helical structures of actin filaments have implications in mechanical properties including filament torsional and bending rigidity (Jegou and Romet-Lemonne, 2020). Applied mechanical torque or twisting by ABPs modifies helical pitch uniformly or non-uniformly along actin filaments. While the previous study by Prochniewicz et al. (Prochniewicz et al., 1996) demonstrates gelsolin’s effects on filament torsional rigidity, further mechanical experiments utilizing magnetic tweezers or optical tweezers can determine the point where torsional rigidity and helical pitch are no longer uniform. Moreover, we speculate that gelsolin causes overtwisting of filament half-pitch as this is the most probable consequence of disrupting longitudinal contacts postulated by models (Schramm et al., 2017).
4 Conclusion
In summary, we have investigated how gelsolin-mediated actin filament severing, mechanics, and conformations are modulated at neutral and acidic pH. We proposed a potential mechanism which links gelsolin-induced helical changes to single filament bending mechanics and severing efficiencies (Figure 6). Gelsolin binding rearranges intersubunit contacts as it competes for actin monomers and shortens filament helical twist. Capping of filament barbed ends by gelsolin increases bending stiffness at neutral and acidic pH, with acidic pH conditions undergoing increased thermal fluctuations. Another consequence of these conformational changes is actin severing. Enhanced severing activities occur in acidic pH due to overriding the requirements for Ca2+ in gelsolin activation and severing.
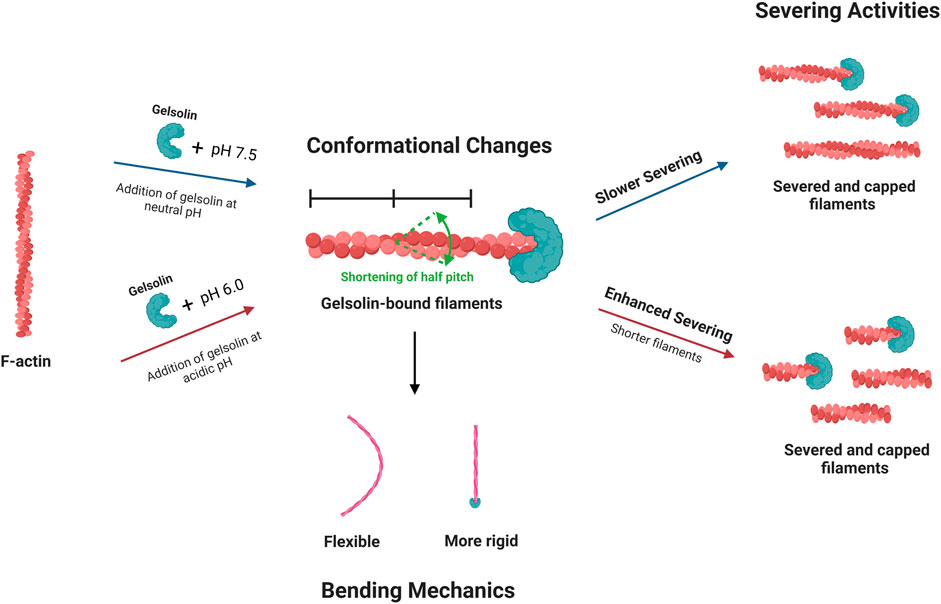
Figure 6. A schematic suggests potential mechanism for the effects of pH on gelsolin-induced filament severing, conformational changes and bending mechanics. Gelsolin binding may regulate actin filament helical structure and bending mechanics. pH conditions can affect gelsolin-mediated severing efficiencies and disassembly dynamics, influencing filament lengths and cytoskeleton remodeling.
We show that gelsolin-mediated filament severing efficiencies and disassembly dynamics increase at low pH. Furthermore, our results indicate that gelsolin binding to actin filaments at intracellular pH conditions regulates the bending mechanics and helical pitch of filaments. Gelsolin’s effects on filament severing activities, bending stiffness, and structural changes at varied pH have implications in cell mechanics. This study shows how gelsolin and pH play critical roles in actin cytoskeleton regulation and rapid remodeling.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
TD: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Writing–original draft, Writing–review and editing. CT: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Writing–original draft, Writing–review and editing. SP: Data curation, Formal Analysis, Investigation, Writing–review and editing. NC: Writing–original draft, Writing–review and editing. LT: Investigation, Writing–original draft, Writing–review and editing. EK: Conceptualization, Funding acquisition, Investigation, Project administration, Supervision, Validation, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The research reported in this publication is supported by the National Science Foundation under the CAREER Award 1943266 (to EK).
Acknowledgments
This content is solely the responsibility of the authors and does not represent the National Science Foundation. We acknowledge the University of Central Florida ORCGS Doctoral Fellowship (to TD). We thank the Kang group members as well as the UCF Nanoscience Technology Center for their support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be considered as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frsfm.2025.1530439/full#supplementary-material
References
Arii, Y., and Hatori, K. (2008). Relationship between the flexibility and the motility of actin filaments: effects of pH. Biochem. biophysical Res. Commun. 371 (4), 772–776. doi:10.1016/j.bbrc.2008.04.135
Barrie, K. R., Rebowski, G., and Dominguez, R. (2024). Mechanism of actin filament severing and capping by gelsolin. bioRxiv. 32, 237–242. doi:10.1038/s41594-024-01412-5
Blanchoin, L., Boujemaa-Paterski, R., Sykes, C., and Plastino, J. (2014). Actin dynamics, architecture, and mechanics in cell motility. Physiol. Rev. 94 (1), 235–263. doi:10.1152/physrev.00018.2013
Burtnick, L. D., Urosev, D., Irobi, E., Narayan, K., and Robinson, R. C. (2004). Structure of the N-terminal half of gelsolin bound to actin: roles in severing, apoptosis and FAF. EMBO J. 23 (14), 2713–2722. doi:10.1038/sj.emboj.7600280
Castaneda, N., Lee, M., Rivera-Jacquez, H. J., Marracino, R. R., Merlino, T. R., and Kang, H. (2019). Actin filament mechanics and structure in crowded environments. J. Phys. Chem. B 123 (13), 2770–2779. doi:10.1021/acs.jpcb.8b12320
Choe, H., Burtnick, L. D., Mejillano, M., Yin, H. L., Robinson, R. C., and Choe, S. (2002). The calcium activation of gelsolin: insights from the 3 Å structure of the G4–G6/actin complex. J. Mol. Biol. 324 (4), 691–702. doi:10.1016/s0022-2836(02)01131-2
Crevenna, A. H., Naredi-Rainer, N., Schonichen, A., Dzubiella, J., Barber, D. L., Lamb, D. C., et al. (2013). Electrostatics control actin filament nucleation and elongation kinetics. J. Biol. Chem. 288 (17), 12102–12113. doi:10.1074/jbc.m113.456327
Decker, Y., Németh, E., Schomburg, R., Chemla, A., Fülöp, L., Menger, M. D., et al. (2021). Decreased pH in the aging brain and Alzheimer's disease. Neurobiol. aging 101, 40–49. doi:10.1016/j.neurobiolaging.2020.12.007
Demosthene, B., Kravchuk, P., Harmon, C. L., Kalae, A., and Kang, E. H. (2024). Small organic osmolytes accelerate actin filament assembly and stiffen filaments. Cytoskeleton. doi:10.1002/cm.21927
Elam, W. A., Kang, H., and Enrique, M. (2013). Biophysics of actin filament severing by cofilin. FEBS Lett. 587 (8), 1215–1219. doi:10.1016/j.febslet.2013.01.062
Fan, J.-S., Goh, H., Ding, K., Xue, B., Robinson, R. C., and Yang, D. (2017). Structural basis for pH-mediated regulation of F-actin severing by gelsolin domain 1. Sci. Rep. 7 (1), 45230. doi:10.1038/srep45230
Garg, R., Peddada, N., Sagar, A., Nihalani, D., and Ashish, (2011). Visual insight into how low pH alone can induce actin-severing ability in gelsolin under calcium-free conditions. J. Biol. Chem. 286 (23), 20387–20397. doi:10.1074/jbc.m111.236943
Gilmore, J. L., Kumeta, M., and Takeyasu, K. (2013). AFM investigation of the organization of actin bundles formed by actin-binding proteins. J. Surf. Eng. Mater. Adv. Technol. 3 (04), 13–19. doi:10.4236/jsemat.2013.34a1002
Gittes, F., Mickey, B., Nettleton, J., and Howard, J. (1993). Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J. Cell Biol. 120 (4), 923–934. doi:10.1083/jcb.120.4.923
Graham, J. S., McCullough, B. R., Kang, H., Elam, W. A., Cao, W., and De La Cruz, E. M. (2014). Multi-platform compatible software for analysis of polymer bending mechanics. PloS one 9 (4), e94766. doi:10.1371/journal.pone.0094766
Heidings, J. B., Demosthene, B., Merlino, T. R., Castaneda, N., and Kang, E. H. (2020). Gelsolin-mediated actin filament severing in crowded environments. Biochem. Biophysical Res. Commun. 532 (4), 548–554. doi:10.1016/j.bbrc.2020.08.041
Huehn, A., Cao, W., Elam, W. A., Liu, X., Enrique, M., and Sindelar, C. V. (2018). The actin filament twist changes abruptly at boundaries between bare and cofilin-decorated segments. J. Biol. Chem. 293 (15), 5377–5383. doi:10.1074/jbc.ac118.001843
Ikawa, T., Hoshino, F., Watanabe, O., Li, Y., Pincus, P., and Safinya, C. R. (2007). Molecular scale imaging of F-actin assemblies immobilized on a photopolymer surface. Phys. Rev. Lett. 98 (1), 018101. doi:10.1103/physrevlett.98.018101
A. Jegou, and G. Romet-Lemonne (2020). “The many implications of actin filament helicity,” Seminars in cell & developmental biology (Elsevier).
Jin, X., Zhou, M., Chen, S., Li, D., Cao, X., and Liu, B. (2022). Effects of pH alterations on stress-and aging-induced protein phase separation. Cell. Mol. Life Sci. 79 (7), 380. doi:10.1007/s00018-022-04393-0
Kang, H., Bradley, M. J., Cao, W., Zhou, K., Grintsevich, E. E., Michelot, A., et al. (2014). Site-specific cation release drives actin filament severing by vertebrate cofilin. Proc. Natl. Acad. Sci. 111 (50), 17821–17826. doi:10.1073/pnas.1413397111
Kang, H., Bradley, M. J., McCullough, B. R., Pierre, A., Grintsevich, E. E., Reisler, E., et al. (2012). Identification of cation-binding sites on actin that drive polymerization and modulate bending stiffness. Proc. Natl. Acad. Sci. U. S. A. 109 (42), 16923–16927. doi:10.1073/pnas.1211078109
Kinosian, H. J., Newman, J., Lincoln, B., Selden, L. A., Gershman, L. C., and Estes, J. E. (1998). Ca2+ regulation of gelsolin activity: binding and severing of F-actin. Biophysical J. 75 (6), 3101–3109. doi:10.1016/s0006-3495(98)77751-3
Kothakota, S., Azuma, T., Reinhard, C., Klippel, A., Tang, J., Chu, K., et al. (1997). Caspase-3-generated fragment of gelsolin: effector of morphological change in apoptosis. Science 278 (5336), 294–298. doi:10.1126/science.278.5336.294
Lagarrigue, E., Ternent, D., Maciver, S. K., Fattoum, A., Benyamin, Y., and Roustan, C. (2003). The activation of gelsolin by low pH: the calcium latch is sensitive to calcium but not pH. Eur. J. Biochem. 270 (20), 4105–4112. doi:10.1046/j.1432-1033.2003.03803.x
Lamb, J., Allen, P., Tuan, B., and Janmey, P. (1993). Modulation of gelsolin function. Activation at low pH overrides Ca2+ requirement. J. Biol. Chem. 268 (12), 8999–9004. doi:10.1016/s0021-9258(18)52970-7
Lee, M., and Kang, E. H. (2020). Molecular dynamics study of interactions between polymorphic actin filaments and gelsolin segment-1. Proteins: structure. Funct. Bioinforma. 88 (2), 385–392. doi:10.1002/prot.25813
Matsuyama, S., Llopis, J., Deveraux, Q. L., Tsien, R. Y., and Reed, J. C. (2000). Changes in intramitochondrial and cytosolic pH: early events that modulate caspase activation during apoptosis. Nat. cell Biol. 2 (6), 318–325. doi:10.1038/35014006
Matusovsky, O. S., Mansson, A., Persson, M., Cheng, Y.-S., and Rassier, D. E. (2019). High-speed AFM reveals subsecond dynamics of cardiac thin filaments upon Ca2+ activation and heavy meromyosin binding. Proc. Natl. Acad. Sci. 116 (33), 16384–16393. doi:10.1073/pnas.1903228116
McCullough, B. R., Blanchoin, L., Martiel, J. L., and De la Cruz, E. M. (2008). Cofilin increases the bending flexibility of actin filaments: implications for severing and cell mechanics. J. Mol. Biol. 381 (3), 550–558. doi:10.1016/j.jmb.2008.05.055
Merino, F., Pospich, S., and Raunser, S. (2020). “Towards a structural understanding of the remodeling of the actin cytoskeleton,” in Seminars in cell & developmental biology (Elsevier).
Nag, S., Larsson, M., Robinson, R. C., and Burtnick, L. D. (2013). Gelsolin: the tail of a molecular gymnast. Cytoskeleton 70 (7), 360–384. doi:10.1002/cm.21117
Nečas, D., and Klapetek, P. (2012). Gwyddion: an open-source software for SPM data analysis. Open Phys. 10 (1), 181–188. doi:10.2478/s11534-011-0096-2
Ngo, K. X., Kodera, N., Katayama, E., Ando, T., and Uyeda, T. Q. (2015). Cofilin-induced unidirectional cooperative conformational changes in actin filaments revealed by high-speed atomic force microscopy. elife 4, e04806. doi:10.7554/elife.04806
Pardee, J. D., and Spudich, J. A. (1982). Purification of muscle actin. Methods Enzymol. 85 (Pt B), 164–181. doi:10.1016/0076-6879(82)85020-9
Park, J., Kravchuk, P., Krishnaprasad, A., Roy, T., and Kang, E. H. (2022). Graphene enhances actin filament assembly kinetics and modulates NIH-3T3 fibroblast cell spreading. Int. J. Mol. Sci. 23 (1), 509. doi:10.3390/ijms23010509
Pollard, T. D., and Borisy, G. G. (2003). Cellular motility driven by assembly and disassembly of actin filaments. Cell 112 (4), 453–465. doi:10.1016/s0092-8674(03)00120-x
Pollard, T. D., and Cooper, J. A. (2009). Actin, a central player in cell shape and movement. science 326 (5957), 1208–1212. doi:10.1126/science.1175862
Prochniewicz, E., Janson, N., Thomas, D. D., and Enrique, M. (2005). Cofilin increases the torsional flexibility and dynamics of actin filaments. J. Mol. Biol. 353 (5), 990–1000. doi:10.1016/j.jmb.2005.09.021
Prochniewicz, E., Zhang, Q., Janmey, P. A., and Thomas, D. D. (1996). Cooperativity in F-actin: binding of gelsolin at the barbed end affects structure and dynamics of the whole filament. J. Mol. Biol. 260 (5), 756–766. doi:10.1006/jmbi.1996.0435
Ritter, M., Schratzberger, P., Rossmann, H., Wöll, E., Seiler, K., Seidler, U., et al. (1998). Effect of inhibitors of Na+/H+-exchange and gastric H+/K+ ATPase on cell volume, intracellular pH and migration of human polymorphonuclear leucocytes. Br. J. Pharmacol. 124(4), 627–638. doi:10.1038/sj.bjp.0701864
Salameh, A. I., Ruffin, V. A., and Boron, W. F. (2014). Effects of metabolic acidosis on intracellular pH responses in multiple cell types. Am. J. Physiology-Regulatory, Integr. Comp. Physiology 307 (12), R1413–R1427. doi:10.1152/ajpregu.00154.2014
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. methods 9 (7), 676–682. doi:10.1038/nmeth.2019
Schramm, A. C., Hocky, G. M., Voth, G. A., Blanchoin, L., Martiel, J.-L., and Enrique, M. (2017). Actin filament strain promotes severing and cofilin dissociation. Biophysical J. 112 (12), 2624–2633. doi:10.1016/j.bpj.2017.05.016
Selden, L. A., Kinosian, H. J., Newman, J., Lincoln, B., Hurwitz, C., Gershman, L. C., et al. (1998). Severing of F-actin by the amino-terminal half of gelsolin suggests internal cooperativity in gelsolin. Biophysical J. 75 (6), 3092–3100. doi:10.1016/s0006-3495(98)77750-1
Shao, Z., Shi, D., and Somlyo, A. V. (2000). Cryoatomic force microscopy of filamentous actin. Biophysical J. 78 (2), 950–958. doi:10.1016/s0006-3495(00)76652-5
Silacci, P., Mazzolai, L., Gauci, C., Stergiopulos, N., Yin, H., and Hayoz, D. (2004). Gelsolin superfamily proteins: key regulators of cellular functions. Cell. Mol. Life Sci. CMLS 61, 2614–2623. doi:10.1007/s00018-004-4225-6
Spinardi, L., and Witke, W. (2007). “Gelsolin and diseases,” in Calcium signalling and disease: molecular pathology of calcium, 55–69.
Sun, H. Q., Yamamoto, M., Mejillano, M., and Yin, H. L. (1999). Gelsolin, a multifunctional actin regulatory protein. J. Biol. Chem. 274 (47), 33179–33182. doi:10.1074/jbc.274.47.33179
Tanaka, K., Takeda, S., Mitsuoka, K., Oda, T., Kimura-Sakiyama, C., Maéda, Y., et al. (2018). Structural basis for cofilin binding and actin filament disassembly. Nat. Commun. 9 (1), 1860. doi:10.1038/s41467-018-04290-w
Vakhrusheva, A., Murashko, A., Trifonova, E., Efremov, Y. M., Timashev, P., and Sokolova, O. (2022). Role of actin-binding proteins in the regulation of cellular mechanics. Eur. J. Cell Biol. 101 (3), 151241. doi:10.1016/j.ejcb.2022.151241
Vemula, V., Huber, T., Usaj, M., Bugyi, B., and Mansson, A. (2021). Myosin and gelsolin cooperate in actin filament severing and actomyosin motor activity. J. Biol. Chem. 296, 100181. doi:10.1074/jbc.ra120.015863
Wang, F., Sampogna, R. V., and Ware, B. R. (1989). pH dependence of actin self-assembly. Biophysical J. 55 (2), 293–298. doi:10.1016/s0006-3495(89)82804-8
Winterhoff, M., Bruhmann, S., Franke, C., Breitsprecher, D., and Faix, J. (2016). Visualization of actin assembly and filament turnover by in vitro multicolor TIRF microscopy. Methods Mol. Biol. 1407, 287–306. doi:10.1007/978-1-4939-3480-5_21
Wioland, H., Jegou, A., and Romet-Lemonne, G. (2018). Quantitative variations with pH of actin depolymerizing factor/Cofilin’s multiple actions on actin filaments. Biochemistry 58 (1), 40–47. doi:10.1021/acs.biochem.8b01001
Zhang, Y., Vorobiev, S. M., Gibson, B. G., Hao, B., Sidhu, G. S., Mishra, V. S., et al. (2006). A CapG gain-of-function mutant reveals critical structural and functional determinants for actin filament severing. EMBO J. 25 (19), 4458–4467. doi:10.1038/sj.emboj.7601323
Keywords: actin cytoskeleton, gelsolin, severing, bending mechanics, filament conformation, intracellular pH
Citation: Douglas TV, Toland CA, Paulin SA, Castaneda N, Tetard L and Kang EH (2025) Gelsolin-mediated actin filament severing, mechanics, and conformational changes at neutral and acidic pH. Front. Soft Matter 5:1530439. doi: 10.3389/frsfm.2025.1530439
Received: 18 November 2024; Accepted: 27 February 2025;
Published: 24 March 2025.
Edited by:
Thomas A. Vilgis, Max Planck Institute for Polymer Research, GermanyCopyright © 2025 Douglas, Toland, Paulin, Castaneda, Tetard and Kang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ellen H. Kang, ZWxsZW4ua2FuZ0B1Y2YuZWR1
 Taylor V. Douglas
Taylor V. Douglas Claire A. Toland1
Claire A. Toland1 Nicholas Castaneda
Nicholas Castaneda Laurene Tetard
Laurene Tetard Ellen H. Kang
Ellen H. Kang