
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Signal Process., 10 November 2022
Sec. Biomedical Signal Processing
Volume 2 - 2022 | https://doi.org/10.3389/frsip.2022.984901
This article is part of the Research TopicTime-Frequency and Machine Learning Applications for Biomedical SignalsView all 4 articles
Recently, the development of learning-based algorithms has shown a crucial role to extract features of vital importance from multi-spectral photoacoustic imaging. In particular, advances in spectral photoacoustic unmixing algorithms can identify tissue biomarkers without a priori information. This has the potential to enhance the diagnosis and treatment of a large number of diseases. Here, we investigated the latest progress within spectral photoacoustic unmixing approaches. We evaluated the sensitivity of different unsupervised Blind Source Separation (BSS) techniques such as Principal Component Analysis (PCA), Independent Component Analysis (ICA), and Non-negative Matrix Factorization (NNMF) to distinguish absorbers from spectral photoacoustic imaging. Besides, the performance of a recently developed superpixel photoacoustic unmixing (SPAX) framework has been also examined in detail. Near-infrared spectroscopy (NIRS) has been used to validate the performance of the different unmixing algorithms. Although the NNMF has shown superior unmixing performance than PCA and ICA in terms of correlation and processing time, this is still prone to unmixing misinterpretation due to spectral coloring artifact. Thus, the SPAX framework, which also compensates for the spectral coloring effect, has shown improved sensitivity and specificity of the unmixed components. In addition, the SPAX also reveals the most and less prominent tissue components from sPAI at a volumetric scale in a data-driven way. Phantom experimental measurements and in vivo studies have been conducted to benchmark the performance of the BSS algorithms and the SPAX framework.
In recent years, there has been special attention on developing new learning-based signal processing approaches as it plays a crucial role in medical image analysis and its interpretation. In addition to the enhanced visualization of the images, one of the main aims of these advanced signal-processing approaches is to identify specific signal markers that can distinguish physiological and pathological conditions. Commonly, expert radiologists are required for analyzing and interpreting biomedical images. Although this is a standard operating procedure, in most cases the approach is tedious, user-dependent, and can be biased. To this end, the development and implementation of an automated feature extraction algorithm will have an added value as this can be used also as a surrogate marker to improve the diagnosis and interpretation (Ali et al., 2008; Chowdhary and Acharjya, 2020).
Photoacoustic is an emerging biomedical imaging modality that has the potential to noninvasively extract molecular tissue composition at a volumetric scale (Cassidy and Radda, 2005). The approach is real-time and over the last decade, it has been used in many preclinical applications such as tissue inflammatory monitoring (Jo et al., 2017; Jo et al., 2018; Xie et al., 2020), vascular remodeling (Hu and Wang, 2010; Arabul et al., 2019; Sangha and Goergen, 2020; Iskander-Rizk et al., 2021), cancer oxygen saturation changes (Li et al., 2008; Stoffels et al., 2015; Bayer et al., 2017; Li et al., 2018; Lawrence et al., 2019), and biodistribution of exogenous contrast agents (Yang et al., 2019; Lavaud et al., 2020; Liu et al., 2020). Although the photoacoustic imaging technology is well established in preclinical research, the clinical aspect of the technology is comparatively limited as it has regulatory concerns and requires local ethical approvals to conduct clinical research studies. This scenario could change soon as most of these technology providers are looking into the translational aspect of this technology and applying for Food and Drug Administration (FDA) approval. Recently, FDA has granted premarket approval to one of the industrial providers of breast cancer photoacoustic imaging technology, which helps physicians to differentiate between benign and malignant lesions (Kuniyil Ajith Singh et al., 2020). This clearly demonstrates the expanding potential of photoacoustic technology for clinical applications. However, to facilitate this translation, a significant improvement to the hardware and image analysis algorithms is necessary. Specifically, multi-spectral photoacoustic imaging (sPAI) mode enables the detection of molecular components of vital importance. In sPAI the tissues are illuminated with pulsed laser light at different wavelengths within the visible or near-infrared (NIR) spectral range (Kim et al., 2015; Allen and Beard, 2016; Das et al., 2021; Park et al., 2022). The light absorbed by the tissue chromophores induces a thermoelastic expansion which consequently generates pressure waves that are recorded just as conventional ultrasound signals. Thus, the contrast of PA imaging is due to light-absorbing molecules that can be either endogenous to tissue, (e.g., hemoglobin, melanin, water, and lipid) or administered exogenous agents. Therefore, the photoacoustic image represents the spatial distribution of the initial pressure generated by the optical contrast of a variety of tissue chromophores (Wang, 2017). The light absorption is dependent on the wavelength, consequently, different wavelengths of optical excitation are absorbed differently by the distinct tissue components. Thus, the generated sPAI data include per pixel the spectral fingerprints of multiple absorbers present within the tissue (Deán-Ben and Razansky, 2014).
Despite the high spatial resolution of sPAI, a pixel usually corresponds to more than one chromophore present in the tissue. Thus, the spectrum per pixel is a linear combination of the spectral responses of all the absorbers. However, to characterize the tissue composition from sPAI, the spectral profile of each individual chromophore must be efficiently distinguished. Thus, advanced spectral unmixing methods are required to solve this problem (Manwar et al., 2020).
To date, the most commonly used spectral unmixing methods are based on differential or linear fitting algorithms (Keshava, 2003). But generally, these algorithms require user interaction to provide the expected source spectral curves as an input to unmix the signals. For translational research with patients, these types of supervised spectral unmixing can be challenging, as the spectral signature of the tissues differs with respect to the disease conditions. Thus, the reliability of these spectral fitting methods depends on the a priori knowledge of the spectral information including all potential components and the quality of the data.
The automatic spectral unmixing algorithms that do not require a priori spectral information and can effectively unmix the data without user interaction are an ideal choice to overcome all these limitations. Recently there have been a lot of efforts in the implementation of deep learning (DL) architectures to solve the unmixing problem in sPAI (Lee et al., 2017; Cai et al., 2018; Gröhl et al., 2021; Yang et al., 2021). Although DL approaches can lead to ultra-high accurate results, the data set constructed to train the network determines the generalization ability and robustness of the learning model. As a result, since sPAI is an innovative technology, there are currently insufficient data sets for deep neural network training. Consequently, common remedies to this problem include using simulated data for proof-of-concept verification (Allman et al., 2018; Gröhl et al., 2019), conducting transfer learning with “real” experimental data (Jnawali et al., 2019), or techniques of data augmentation (Rodrigues et al., 2021). To generate these PA in silico data, numerical simulations of the forward light diffusion (“light-IN”) and backward US propagation (“sound-OUT”) processes are required. Hence, the current limitation of these approaches is that they are application-specific and challenging to generalize for “unseen” cases.
On the other hand, unsupervised feature learning methods have resulted effective in learning representations without training data sets. However, many existing feature-learning algorithms are hard to use and require extensive hyperparameter tuning (Durairaj et al., 2020). Blind source separation (BSS) algorithms are a class of more sophisticated approaches, requiring only one hyperparameter, as the number of features to learn (Jutten and Karhunen, 2004; Benyamin et al., 2020). These are based on modeling the data, making assumptions about their distribution, and solving an optimization problem. Although the BSS methods perform well on image object recognition, and video/audio classification (Smita et al., 2014), these are not optimized for spectral photoacoustic unmixing. Specifically, the sPAI unmixing is an ill-posed inverse problem where the a priori information related to the spectral fingerprints of tissue components and their spatial distribution are both unknown. Glatz et al. 2011 have shown that BSS techniques such as Principal Component Analysis (PCA) and Independent Component Analysis (ICA) have great potential to solve the sPAI unmixing problem efficiently, without prior knowledge of the constituent spectra (Buehler et al., 2010).
Although these BSS approaches are promising to detect the spectral components, the light fluence variation along the depth could be a major challenge, because the absorption spectra of the tissue components along the depth can be perceived as altered and thus creating misinterpretations in the unmixing process. Specifically, in sPAI the detected spectra are not unique per each molecular component. This is because the acquired absorption spectrum is influenced by the position of the moiety within the tissue. Thus, the actual spectrum is altered due to the light fluence
In one of our previous works (Grasso et al., 2020), we optimized a specialized class of BSS algorithm, the Non-negative Matrix Factorization (NNMF) for the spectral unmixing of sPAI. This method has great potential to accurately extract the prominent tissue chromophores from sPAI. However, the problem of automatically distinguishing weak absorbers, such as lipids, from prominent tissue chromophores like hemoglobin makes the unmixing procedure more complex by leading to incomplete identification of tissue constituents. Thus, recently we have implemented a superpixel photoacoustic unmixing (SPAX) framework that is capable to overcome the limitations of the classical BSS approaches (Grasso et al., 2022). The SPAX algorithm is a unique data processing procedure, that has the potential to reveal weak and strong chromophores and it is also extended to compensate for the spectral coloring artifact.
In this study, we have investigated the performance of different blind source separation (BSS) algorithms, typically used for automated unmixing in sPAI. Specifically, a comparative analysis of Principal Component Analysis (PCA), Independent Component Analysis (ICA), and Non-negative Matrix Factorization (NNMF) has been conducted through an experimental phantom study at different signal-to-noise ratio (SNR) conditions. These are promising methods to automatically distinguish prominent absorbers. On the other end, their sensitivity for distinguishing less prominent absorbers and components with similar spectra is still limited. Thus the unmixing performance of the innovative SPAX framework has been investigated in detail. The SPAX framework that allows automated unmixing at a volumetric scale has been also evaluated by performing an in vivo case study. The near-infrared spectroscopy (NIRS) has been used to measure the pure absorption spectra of the chromophores involved in the study. Finally, a cross-correlation analysis has been performed to benchmark the unmixing performance of the different unmixing algorithms with NIRS.
Spectral photoacoustic imaging (sPAI) can be modeled as a set of mixed observations
The matrices
The following subsections initially provide the theoretical background of the main BSS methods. Moreover, a summary of the features of the different BSS approaches is reported in Table 1. Finally, we discussed the characteristics of a newly developed approach named superpixel photoacoustic unmixing (SPAX) framework.
The singular value decomposition (SVD) and principal component analysis (PCA) are related methods that rely on the hypothesis that the source components
where
Principal component analysis (PCA) identifies orthogonal vectors for dimension reduction by performing an eigen-decomposition of the covariance matrix of the observed data
The first PC (which corresponds to the eigenvectors) represents the subspace in which the variance of the data is the largest. Therefore, the variance (which corresponds to the eigenvalues) is related to the respective prominence of the principal component. Often the SVD/PCA method is used as a priori step of other blind approaches, which consequently use the achieved components as a starting point of the optimization process.
Independent component analysis (ICA) is another BSS method that assumes that the observed data are a superimposition of a number of stochastically independent components (Hyvärinen, 2013). This is based on the hypothesis that the source components are maximally independent and non-Gaussian. The non-Gaussianity of the source data is the crucial assumption that makes this method more effective than PCA (uncorrelated and Gaussian). Following the Central Limit Theorem, the sum of independent variables tends toward a normal distribution (Kwak and Kim, 2017). Therefore, with the non-Gaussianity assumption, the independence hypothesis of the unmixed components is stricter and consequently more robust than uncorrelatedness. Usually, the ICA algorithm requires a pre-processing step of mixed data centering and whitening to reduce the correlation of the observed data. During the centering step the mean is subtracted from all the mixed signals. Then the whitening includes two main steps: a decorrelation (covariance is zero) and scaling (unit variance) of the centered mixed signals. After the centering and whitening, the mixed data
Here, the reconstruction independent component analysis (RICA) method has been used to minimize the cost function and estimate the source components
Like ICA, non-negative matrix factorization (NNMF) is a BSS method that performs a factorization of mixed observations
Lee and Seung (2001) have proposed the Multiplicative Update Rules to solve this optimization problem. Since no elements are negative, the LMM can be intuitively interpreted as a process of generating the original data by linear combinations (parts-based) of the prominent components (subtraction operations are not involved). This is known as a parts-based linear representation for non-negative data. Besides, literature studies have shown that imposing the positivity condition enhances the convergence of the optimization algorithm used for the unmixing problem (Lipovetsky, 2009; Hervé, 2010).
In addition, the NNMF approach is considered a special version of PCA including the non-negativity constraint and without orthogonality of factors (Sotiras et al., 2015). Although both approaches are similar, there is a fundamental difference between them. While the goal of PCA is to reduce the correlated observed variables, factor analysis aims to explain the correlation between the variables underlining the latent factors causing the observations. Furthermore, considering another comparison, NNMF and ICA are two closely related unmixing approaches. The main difference is that NNMF over ICA can recover the sources without requiring knowledge about the mean and variance of the sources.
Table 1 shows a comparison of the different BSS algorithms including the details of the mixture model and assumptions as well as the cost function model and optimization iterative algorithms adopted by the BSS approaches to unmix the source spectra
The BSS methods mainly enable the detection of prominent molecular targets, which may not lead to complete tissue characterization. In addition, blind approaches incorporate uncertainty in their performance, which depends on the choice of the hyperparameter (number of endmembers to separate). Moreover, these are also affected by spectral coloring.
Figure 1 schematically summarizes the main procedures implemented within the Superpixel Photoacoustic Unmixing (SPAX) framework. Specifically, the SPAX framework uses multi-spectral photoacoustic images co-registered with high-resolution ultrasound images (US-sPAI) as input to automatically identify the tissue spectral components obtained as output. First, the SPAX framework models the light fluence distribution to compensate for the spectral coloring, thus preventing spectral unmixing misinterpretations. The compensation approach utilizes Ultrasound image segmentation and spectral Monte Carlo simulations based on a predefined library of optical properties. Within the SPAX framework, this spectral decoloring approach is crucial to mitigate the inhomogeneities caused by the non-uniform light fluence distribution along the depth. Specifically, the approach implements an automated segmentation of the ultrasound image to identify the skinline, which is used as a watershed to distinguish the tissue structures and the background. Besides, for complex tissue structures, the user can refine the US segmentation by selecting some regions of interest for active contours, thus resulting in a semi-automated approach. Then, each segmented cluster is labeled with spectral optical properties gathered from literature in a predefined library. Therefore, this represents the numerical domain used for spectral light fluence simulations implemented in MCXLAB (Yan and Fang, 2020). Finally, the obtained fluence maps at multiple wavelengths are used to correct the acquired sPAI (Grasso et al., 2022).
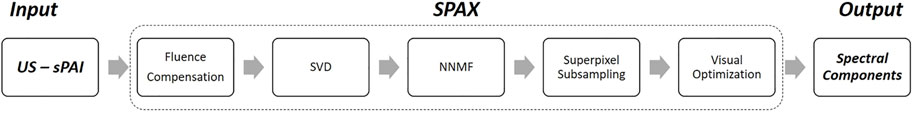
FIGURE 1. Schematic flowchart of the SPAX framework including the main procedures from input to output.
After the sPAI correction, the rank hyper-parameter can be automatically tuned by performing the SVD pre-analysis of the mixture. The core of the SPAX framework is based on non-negative superpixel unmixing, which enables the automated detection of tissue chromophores with improved sensitivity and specificity. To solve the ill-conditioned problem, positivity constraints and multiplicative update rule have been used to improve the convergence of the blind iterative method. Besides, a novel superpixel subsampling approach has been implemented to differentiate the less prominent molecular components, from the prominent absorbers. Finally, imaging optimization methods are applied to improve the visualization of the distribution maps of the detected spectral components. Therefore, the SPAX offers several advantages over the simple blind unmixing approaches. Details of the algorithm and the validation studies are described elsewhere (Grasso et al., 2022).
High-resolution Ultrasound (US) and spectral Photoacoustic imaging (sPAI) have been acquired by using the platform Vevo LAZR-X (FUJIFILM VisualSonics, Inc., Toronto, ON, Canada). The imaging setup includes a high-frequency US system, an optical parametric oscillator (OPO) integrated Nd:YAG nanosecond pulsed laser, and the animal imaging platform. The system is equipped with a linear US transducer array (MX250) that consists of
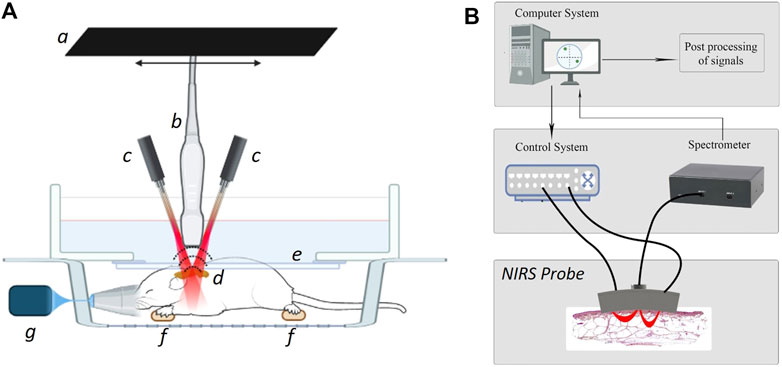
FIGURE 2. Schematic of the ultrasound-photoacoustic (US-PA) whole-body acquisition setup (A) that includes: a linear stepper motor (a), the ultrasound transducer (b) with optical fibers on the sides (c) to illuminate the tissue chromophores (d), optical and acoustic compatible membrane (e) in a water bath to maintain the sound coupling, ECG electrodes to monitor the physiological parameters in vivo (f), and the anesthesia system (g). Schematic of the near-infrared spectroscopy (NIRS) optical probe setup (B) that also includes: a control system for the optical source, a spectrometer for the detection, and a computer for the signals post-processing.
Figure 2B depicts the schematic of a portable near-infrared spectroscopic (NIRS) probe that has been used to validate the spectral absorption curves of the chromophores involved in the study. To this end, a customized NIRS probe was designed at the Optical lab in OsloMet (Oslo Metropolitan University, Norway) (Hassan et al., 2021a; Hassan et al., 2021b). Unlike commercially available solutions, this NIRS probe is designed to target two regions of interest at two different depths. Specifically, the OsloMet’s probe uses a broadband LED light source (650–1,050 nm) and works with two depth ranges (2–2.25 mm and 3–3.5 mm respectively). In addition, the optical probe’s light sources ensure the safety of the operations, in compliance with the NIR safety guidelines (IEC, 2015). This NIRS probe uses a source-detector pair that generates a banana-shaped optical path that narrows at the source and detector’s locations and broadens as it penetrates deeper into the tissue (Feng et al., 1995). The depth of a photon before detection is proportional to the square root of the source-detector separation (Weiss et al., 1989). Hence, optical depth is generally 1/2 to 1/3 of the source-detector distance (Papazoglou et al., 2006). The spectra from the optical probe are then collected by a spectrometer (Avaspec 2048x14, Avantes BV, Netherlands). An integration time of 4 s, was selected to make sure that enough photons are collected by the detector. In this experiment, a distance of 6 mm between the source and detector was used to get a focal depth of 2–2.5 mm and the wavelength range has been fixed to 680–970 nm as the one used for sPAI. Finally, the obtained spectra from NIRS have been used as a reference to be compared with the automatically unmixed signatures from sPAI.
To obtain the spectral curves for the correlation analysis, the NIRS measurements have been performed on the dyes used for the vessel-mimicking phantoms. Blue, green oil colors (Winsor and Newton, London, United Kingdom), and India Ink (Higgins Ink, Leeds, MA, United States) were used, and the colors have been overspread on white paper. These have been placed at the optimal distance from the NIRS probe during the measurements using a transparent gel pad as a spacer. This guarantees the optimal position of the sample at the focal zone of the NIRS probe design. Figure 3 depicts the absorption spectra of the dyes measured via the OsloMet NIRS probe. While the Blue Oil Color absorption spectrum shows the highest absorption at 680 nm and has a quasi-exponential decrease afterward, the Green Oil Color shows wider absorption until around 800–830 nm before decreasing at higher wavelengths. Finally, India Ink shows a broad absorption spectrum within the 680–900 nm region and a gradual absorption decrease toward 900 nm.
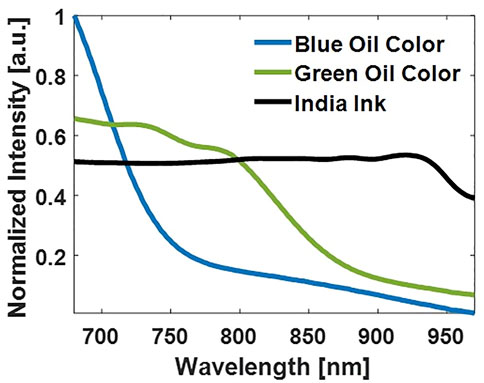
FIGURE 3. Absorption spectra of the dyes used within the vessel-mimicking phantoms measured by the NIRS probe.
Phantom measurements were first performed to investigate the sensitivity of the BSS unmixing algorithms at different signal-to-noise ratio (SNR) conditions. Three vessel-mimicking phantoms have been fabricated with different optical properties. A vessel-mimicking shape has been painted with green and blue oil colors (Winsor and Newton, London, United Kingdom) on white paper (see Figure 4A). After the paint has dried, this has been embedded in each turbid agarose solution. The paper is carefully retrieved from the agarose to sequentially reuse it in all of the phantoms. All the phantoms have been designed to have a reduced scattering coefficient of ∼5 cm−1. Thus, the phantoms were made with 1.5% (w/v) agar (Alfa Aesar, Heysham, Lancaster) and 2% (w/v) Intralipid (20%, Sigma-Aldrich, Canada). To challenge the unmixing algorithms, in two of the phantoms, black India Ink (Higgins Ink, Leeds, MA, United States) was added to the agar-Intralipid mixture before solidification, to simulate background absorption. Specifically, these resulted in a background absorption of 0.05% v/v and 0.1% v/v respectively. Hence, this leads to three vessel-mimicking phantoms that have no background absorption, medium background absorption, and high background absorption respectively.
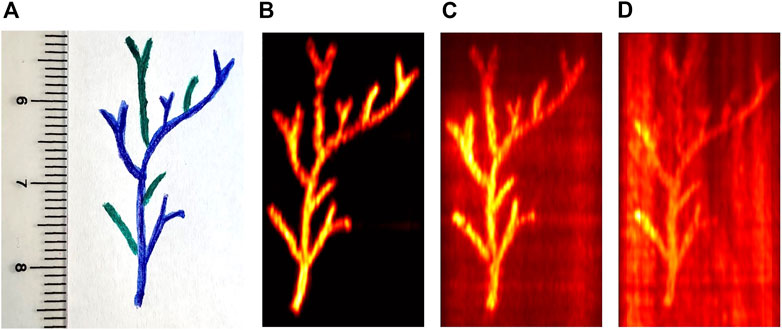
FIGURE 4. Vessel-mimicking phantom shape (A) and the single wavelength photoacoustic images acquired at 700 nm for the three phantoms with different levels of absorbing background. Specifically, without India Ink (B), with 0.05% v/v India Ink (C), and with 0.1% v/v India Ink (D) as background absorption.
Figures 4B–D show the PA image at a single wavelength (
The in vivo experiments were conducted at FUJIFILM Sonosite/VisualSonics facility in Amsterdam, Netherlands. All the experiments involving animals were in full compliance with the protocol (AV D2450020173644) evaluated and approved by the Animal Use and Ethics Committee (CEUA) of Netherlands. These were in accordance with FELASA guidelines and the National Law for Laboratory Animal Experimentation (Law No.18.611). Three female CD-1 mice models (Envigo, Horst, the Netherlands) 9 weeks old, were used to image the cervical-thoracic region. During imaging acquisition, the mice were anesthetized with isoflurane (2–3% by volume with 0.8l/min gas flow). The US and sPAI acquisition was performed with the animal in its prone position, with the transducer aligned perpendicularly to image the region of interest. After in vivo acquisition, the adipose tissue from the cervical-thoracic area was dissected post-mortem and used for spectral measurements conducted via a portable NIR spectrophotometer (see Section 2.3). We focused on this anatomical area where the interscapular adipose tissue is present.
The vessel-mimicking phantoms have been used to evaluate the unmixing performances of PCA, ICA, and NNMF, which have been applied to unmix the absorbers included within the phantoms. Overall, the analysis has been performed on the three phantoms, that have no background absorption, 0.05% v/v, and 0.1% v/v India Ink as background absorption. Thus, the phantoms without background absorption and medium/high background absorption result in sPAI with SNR values of 30dB, 15dB, and 8dB, respectively. The only hyperparameter of these BSS methods was the number of endmembers. This has been manually selected to 2 in the absence of background absorption and 3 in the other cases to unmix blue/green oil colors and India ink. Figure 5 presents the source spectra obtained as a result of the different BSS unmixing methods. The detection sensitivity of the BSS methods depends not only on the ability to unmix the molecular targets of interest from the absorbing background but is also hampered by the SNR, which indicates the quality of the images. Specifically, the presence of bulk absorption at different concentrations resembles possible real scenarios where it is more complex to reveal the absorber targets of interest. The unmixing results show that all three BSS methods managed to isolate the target components (Blue and Green Oil Colors) and the background (India Ink), when present. However, PCA and ICA algorithms exhibited unwanted negative values in the spectra, which are attributed to the non-constrained optimization process. Besides, the BSS methods show unique solutions only up to sign ambiguities. In particular, PCA and ICA are more prone to error when the sources of interest have low variance. Thus, often the iterative optimization procedure converges into inverted unmixed components as compared to the expected spectra.
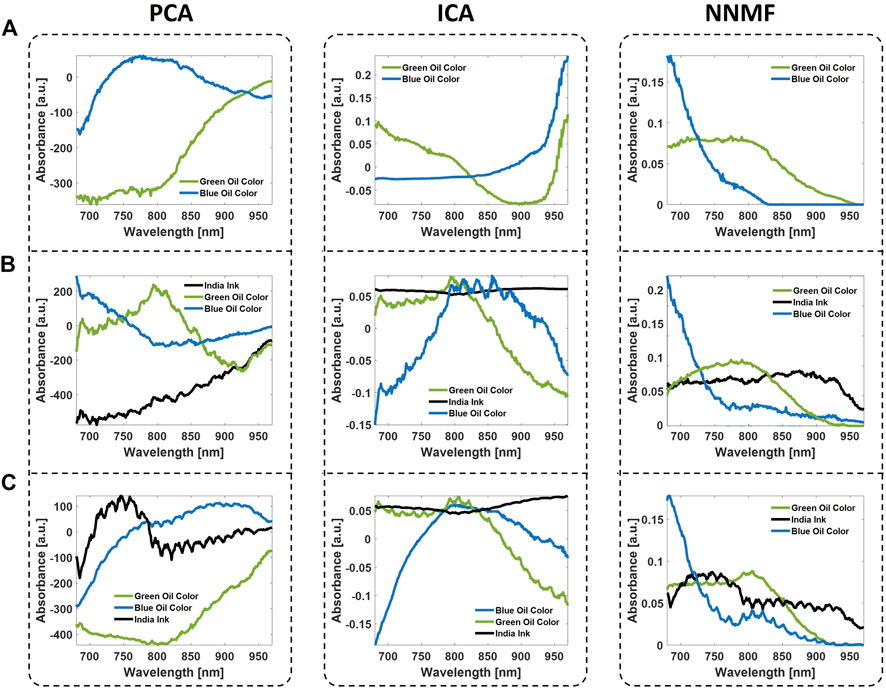
FIGURE 5. Unmixed spectra obtained from the vessel-mimicking phantoms with no background absorption (row (A)), medium background absorption (row (B)), and high background absorption (row (C)). The unmixing has been performed by means of the BSS algorithms PCA, ICA, and NNMF.
On the other hand, the phantom studies showed that NNMF could detect the vessel-tree branches colored in green and blue with higher accuracy than PCA and ICA. High performance of the NNMF method was obtained even in the case where the background was highly absorbing (0.1% v/v India Ink) and the SNR value was almost three times lower (8 dB). The NNMF accurately unmixed the components and the detected spectra are very close to the expected spectra measured by the NIR spectrophotometer (see Figure 3). From the unmixing results, all the phantoms qualitatively demonstrate the superiority of the NNMF as a detection method in resolving the different absorbers from sPAI. Furthermore, the non-negativity constraint was found to maintain the most robust and accurate unmixing performance in all experiments.
Finally, the unmixed source components from sPAI, by applying the different BSS methods, have been compared using the correlation with the curves measured via NIRS. Table 2 reports the spectral correlation values obtained from the different phantoms. Although in some instances PCA and ICA show higher absolute correlation values than NNMF, often this results in negative correlation values. The latter leads to the identification of the components but with opposite signs. On the contrary, the NNMF performance is superior and more robust in all the vessel-mimicking phantoms conditions.

TABLE 2. Correlation values between reference spectra obtained via NIRS probe and the unmixed spectra via BSS algorithms and SPAX framework for the vessel-mimicking phantoms with different background absorption.
Besides, Table 3 reports the processing time of the BSS algorithms used in the experimental phantoms. Here we have performed the processing on a portable laptop with Intel(R) Core(TM) i7-9750H CPU 2.60GHz and 16.0 GB of RAM. Although the processing time depends on the dimensionality reduction chosen by the user (number of endmembers), PCA has shown the advantage of being faster than ICA when background absorption is present. On the other hand NNMF, despite the non-negativity constraint, seems computationally faster to implement and leads to converging to more interpretable data.
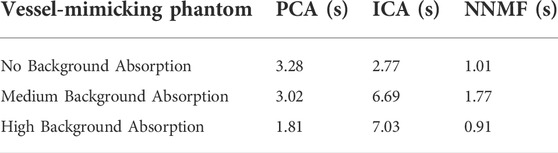
TABLE 3. Unmixing processing time of the BSS algorithms (PCA, ICA, and NNMF) to distinguish the source components from the vessel-mimicking phantoms with different background absorption.
Since the main focus of the experimental vessel-mimicking phantoms was to test the BSS unmixing algorithms’ performance, especially their accuracy in detecting the spectral signatures of the components, in this study, the local fluence was assumed to be constant. This is a reasonable assumption for the design of the vessel-mimicking phantoms since the absorbers are all placed at the same distance from the excitation light source.
Figure 6 shows the unmixing results obtained on the vessel-mimicking phantoms with the recently developed method named Superpixel spectral Photoacoustic Unmixing (SPAX) framework. The SPAX is more robust to errors thanks to the eigenvalues-based choice of the rank unmixing dimension (number of endmembers). This SVD pre-analysis within the SPAX ensures that significant information is not lost during the dimensionality reduction. Thus, as shown in Figure 6 from the SVD analysis, the SPAX has automatically evaluated the optimal number of endmembers in the phantoms. Depending on the absence or presence of the bulk absorber, the number of endmembers has been automatically chosen as 2 or 3, respectively. Besides, Figure 6 shows the unmixed source spectra and respective abundance maps obtained as the final output of the SPAX from the different phantoms. These outputs are obtained following all the procedures within SPAX: NNMF, Superpixel Subsampling, and visual optimization (as shown in the flowchart of the SPAX in Figure 1). Even for the phantom with a highly absorbing background (0.1% v/v India Ink), the SPAX has shown high unmixing performance by extracting the source components and abundance maps with high accuracy.
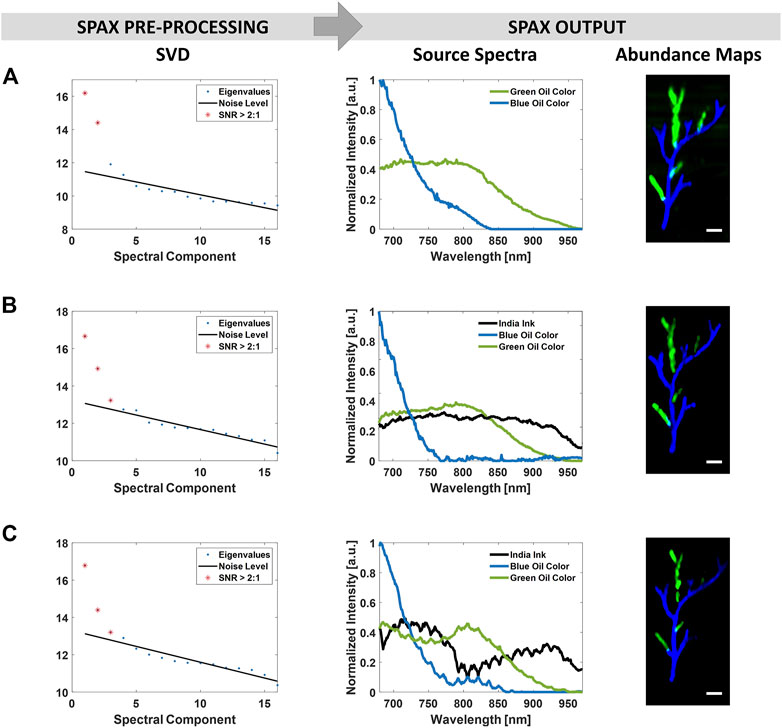
FIGURE 6. SPAX framework applied on the vessel-mimicking phantoms without background absorption (row (A)), with 0.05% v/v India Ink (row (B)), and 0.1% v/v India Ink (row (C)) as background absorption. The SVD analysis, implemented within the SPAX, automatically determines the optimal number of source components that is significantly above the noise floor. This has resulted as 2 in (A) and 3 in (B–C) where in addition to blue/green oil colors, the India ink is present as a background absorber. The unmixed spectra and abundance maps obtained as the SPAX outputs show the extracted dyes from the vessel-mimicking phantoms. The scale bar size is 2 mm.
The correspondence between the unmixed abundance maps by the SPAX framework and the known phantom design was determined by visual inspection. To validate the accuracy of the obtained unmixing results the spectral correlation between the reference spectra measured by the NIR spectrophotometer and those unmixed by SPAX is performed. Table 2 includes the obtained correlation values. Pearson’s correlation coefficients have been estimated in the three phantoms, and the values have been reported for the India Ink within the background, and blue/green targets. The correlation values obtained by SPAX showed its superior unmixing performance as compared to the other BSS algorithms. Only in the phantom with high background absorption, the performance is slightly lower than pure NNMF, this might be caused by the re-iterations included within the SPAX that automatically lead the optimization process to the convergence. These re-iterations might cause some numerical errors in case of convergence to a local minimum.
Although the phantom experiments have enabled the evaluation of the unmixing performance of the different algorithms, the spectral changes along the depth were not considered. Compared to the conventional BSS approaches the SPAX framework, also includes a fluence correction procedure. Figure 7A shows the source components unmixed via SPAX algorithm on the interscapular region of the mouse in vivo. Although the sPAX automatically extracted seven components, here we reported five identified components of interest: oxygenated hemoglobin, deoxygenated hemoglobin, skinline, white adipose tissue (WAT), and water. These have a high correlation with the theoretical absorption spectra, reported in the literature, of oxygenated hemoglobin, deoxygenated hemoglobin, melanin, fat, and water respectively (Jacques, 2013).
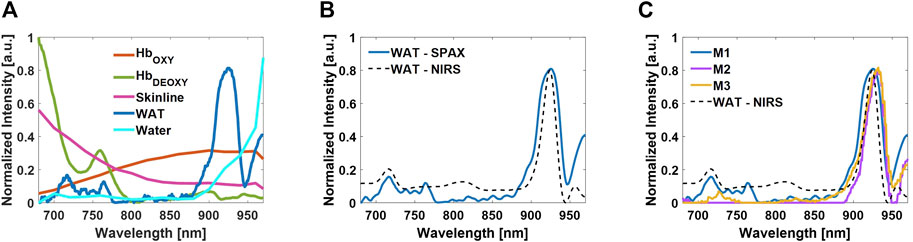
FIGURE 7. Tissue component spectra unmixed by the SPAX from the cervical-thoracic area of the mouse in vivo. Oxygenated hemoglobin, deoxygenated hemoglobin, skinline, WAT, and water are the identified components from the seven source components (A). Comparison between the spectrum of WAT extracted by SPAX and the WAT absorption spectrum measured via NIRS (B). Comparison between the WAT spectrum extracted by SPAX from three different animals in vivo and the WAT absorption spectrum measured via NIRS (C).
For a qualitative validation of the spectra, spectral measures of endogenous chromophore via NIRS have been conducted. The interscapular fat pad has been dissected from the cervical-thoracic area of the mouse post-mortem and the sample has been prepared to contain pure fat. Thus, to avoid confounding factors and ensure the measure of the absorption spectrum from the fat only. NIRS measurements have been performed and used as a reference to validate the unmixed spectra via SPAX. Finally, the obtained NIR spectrum has been compared with the WAT spectrum automatically unmixed in vivo from sPAI. Figure 7B shows the overlay of the measured spectrum of WAT via NIR spectrophotometer and the spectrum automatically unmixed via SPAX from the sPAI data. Spectrally, the WAT absorbs primarily in the 900–950 nm region, as shown by the measured signature of the dissected interscapular fat pad acquired by the NIR spectrophotometer. The obtained WAT spectrum, unmixed via SPAX, shows a characteristic peak at around 930 nm, which is representative of absorption by lipids. Besides, the correlation values, between the unmixed spectrum of WAT and the NIRS reference, show that the SPAX approach could reveal the WAT presence even with skin intact during in vivo experiments with
In addition to the unmixed spectra, Figure 8 shows the abundance maps of the components of interest obtained from SPAX during in vivo acquisitions. The whole-body US image is obtained simultaneously during the sPAI acquisition, thus enabling the recognition of anatomical structures such as the cervical-thoracic area with high accuracy (see Figure 8A). Figure 8B, depicts the oxy/deoxy hemoglobin distribution map in red/green respectively. Figure 8C, shows the distribution map of WAT in the blue colormap that matches the WAT distribution within the interscapular region as confirmed by the schematic reported in Figure 8D. High fidelity and quality of the tissue component unmixing maps were estimated with in situ visualization performed after dissection. In particular, as observed in Figure 8D from the schematic of the interscapular adipose tissue of a mouse, the WAT has an arrangement between the shoulders adjacent to the interscapular brown adipose tissue (iBAT) which has a bilateral symmetrical shape. The SPAX framework has distinguished WAT in vivo, non-invasively, and label-free. Therefore, it has been assessed that the adipose tissue can be separated from other tissue chromophores in vivo, within the NIR range of 680–970 nm. This has also been confirmed from ex vivo evaluations where it is possible to verify that the WAT is localized within the cervical-thoracic region and shows the same arrangement between the shoulders as automatically obtained in vivo via SPAX (Reber et al., 2018; Bagchi and Macdougald, 2019).
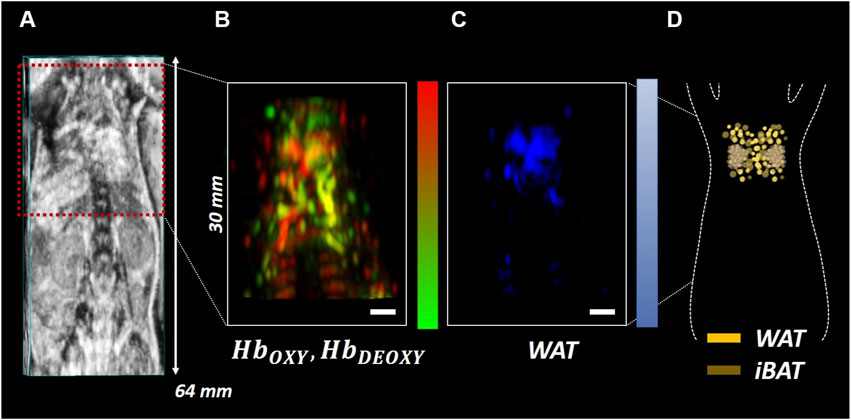
FIGURE 8. Whole-body US image of the mouse in a prone position with the cervical-thoracic area highlighted by the red ROI (A); Oxy/deoxy hemoglobin (B) and WAT (C) abundance maps obtained via SPAX depicted in red/green and blue respectively. Schematic of a mouse interscapular adipose tissue including WAT and iBAT (D). The scale bar size is 2 mm.
The interscapular fat pad represents the largest brown adipose tissue (BAT) depot, located on the dorsal side of the animal, immediately inferior to the shoulders as shown from the rodents’ atlas of adipose tissue by Zhang et al. (2018). The interscapular BAT (iBAT) is covered with white adipose tissue (WAT), which is a highly efficient energy storage. Conventionally, the WAT is a weaker absorber that shows an absorption intensity of around two orders lower than BAT, thus it is difficult to be automatically differentiated (Reber et al., 2018). In this study, the fat spectrum identified by SPAX (see Figure 7), from healthy animals, shows the characteristic WAT peak at 930 nm, also confirmed by NIRS measurements. According to the adipose tissue atlas (Zhang et al., 2018), the distribution map of fat obtained via SPAX is also matching the WAT distribution depicted in the schematic (see Figure 8C,D). On contrary, the iBAT significantly contributes to the nonshivering thermogenic response and regulates the body temperature homeostasis. Thus iBAT can be activated via cold exposure. Once activated, the iBAT becomes highly vascularized which may contribute to greater absorption than WAT. Beyond WAT, as demonstrated elsewhere (Reber et al., 2018), the activation of BAT can be also indirectly monitored by tracking hemodynamic changes as a marker of BAT’s metabolic state. Thus the activation of BAT can be also monitored via sPAI. However, in this study, we focused on the label-free detection of less prominent tissue components such as WAT, via SPAX framework.
Besides, although the axial depth of the in vivo images is 30 mm, the fat pad deposited within the interscapular region is a superficial target. Therefore, in this case, the additional benefits of SPAX in terms of spectral fluence compensation at depth is not so evident. However, our previous study (Grasso et al., 2022) shows in detail the improvements and high robustness of the SPAX for unmixing absorbers positioned at deeper imaging depths.
Spectral photoacoustic imaging (sPAI) has shown high potential to determine detailed information on molecular tissue composition. Conventionally, linear unmixing (LU) and blind source separation (BSS) approaches are implemented to differentiate the tissue components from sPAI. Although the linear spectral unmixing method (Erhayiem et al., 2011; Reber et al., 2018) is widely used, this has some drawbacks as it requires user interaction to provide the expected spectral signatures of the tissue components. Due to unpredictable spectral changes in disease conditions or in clinical scenarios, linear unmixing is challenging and can lead to suboptimal results. Thus, unsupervised approaches that can automatically reveal any spectral change or fingerprints that might be related to pathological conditions are beneficial. Over the years, blind source separation (BSS) algorithms have shown promising automated unmixing performances in sPAI. Thus the BSS methods present a possible solution to this ill-posed unmixing problem and open up more photoacoustic applications both in preclinical and translational research.
Here, we evaluated the unmixing performance of various BSS algorithms to solve the sPAI unmixing problem in an unsupervised way. The performance of previously reported blind unmixing algorithms as principal component analysis (PCA), independent component analysis (ICA), and non-negative matrix factorization (NNMF) have been investigated in detail on experimental vessel-mimicking phantoms. Although these different methods are all part of the BSS category of algorithms, they implement different hypotheses, constraints, and optimization procedures, thus producing different results. Besides, near-infrared spectroscopy (NIRS) technology has been used as a reference to validate the unmixed spectra obtained by the different unmixing algorithms.
The phantom experiment has shown that all the studied BSS algorithms achieved the separation of the absorbers and PCA has shown the advantage of being faster than ICA when background absorption is present. On the other hand NNMF, despite the non-negativity constraint, computationally appears to be the fastest method to implement and converges to more interpretable data. Furthermore, the cross-correlation values measured between the reference NIR spectra and the unmixed spectra by the NNMF range between 77.9–97.7%. Thus, this has confirmed that the NNMF enabled more accurate unmixing performance for sPAI, as compared to PCA and ICA. Specifically, the NNMF has shown promising unmixing performance even in more complex scenarios, where the phantom has a highly absorbing background that confounds the included absorbers of interest and thus drastically reduces the quality/SNR of the sPAI. This confirms that the imposed positivity constraints are crucial to unmix tissue chromophores from sPAI. While NNMF has shown superior unmixing performance compared to the other BSS approaches, this is sensitive to uncertainties and misinterpretation, especially of less prominent chromophores at depth. In particular, NNMF assumes that the most prominent absorbers such as hemoglobin, are the sole tissue absorbers, not accounting for the presence of other absorbing molecules (e.g., extrinsic molecules, melanin, lipids, etc.) that are weaker absorbers. Therefore the less prominent absorbers are discarded within the NNMF unmixing, thus leading to incomplete tissue composition. Besides, the unmixing result is hampered by the choice of the hyperparameter and is jeopardized by the spectral coloring effect. Specifically, the excitation light fluence used in sPAI generally decreases with depth, and thus degrades the image uniformity, causing spatial fluence variations within the tissue. Therefore the NNMF could lead to significant errors in unmixing the tissue spectra, especially in deeper areas where spectral coloring is dominant.
Therefore, in addition to the classical BSS algorithms, a recently developed unmixing framework named superpixel photoacoustic unmixing (SPAX), which also considers these aspects, has been explored (Grasso et al., 2022). Specifically, the SPAX framework includes multiple procedures to accurately identify the tissue composition by minimizing the user interaction and prior knowledge about the tissue components. Specifically, within the SPAX an initial fluence compensation step based on US image segmentation and spectral Monte Carlo simulation enables to mitigate unmixing misinterpretations caused by the spectral coloring artifact at depth. The singular value decomposition (SVD) analysis, included within the SPAX, allows the automated choice of the hyperparameter. Therefore, the SPAX framework has shown higher sensitivity than the classic BSS methods for sPAI. Specifically, the SPAX approach is more robust against nonlinearities caused by inaccurate fluence estimation, converging to source components strengthened by a meaningful positivity constraint. Besides, when the mixing matrix is more ill-conditioned, as in the case of low SNR, SPAX provides significantly more accurate results as it is based on a parts-based decomposition, including the innovative superpixel subsampling approach.
We have validated the SPAX on preclinical in vivo studies focusing on the automated detection of less and most prominent endogenous chromophores within the cervical-thoracic area. Specifically, the SPAX has shown the ability to resolve not only hemoglobin, considered the main source of endogenous contrast in sPAI, but also less prominent tissue features such as white adipose tissue (WAT) which is an important biomarker in several diseases (Erhayiem et al., 2011). Finally, the results obtained from the cervical-thoracic area via SPAX have been compared to the NIR spectroscopic measures. A Pearson’s correlation value of 96% has been obtained for the WAT, thus showing the high sensitivity of SPAX to unmix WAT from highly absorbing components. Therefore, it has been assessed that by using SPAX the WAT can be separated from other tissue chromophores in vivo, non-invasively, and label-free within the NIR range of 680–970 nm.
Although the SPAX framework has demonstrated exemplary performance as a data-driven unmixing approach for sPAI, additional aspects can still compromise the performance of spectral unmixing. For instance, the accuracy of the spectral photoacoustic imaging reconstruction could also impact the unmixing. Specifically, the recorded photoacoustic spectral signals can be corrupted by many noise sources. To overcome this limitation, advanced image reconstruction techniques, which lead to a reduced alteration caused by artifacts and noise (Han et al., 2015), have been developed to guarantee higher quality sPAI and, consequently, more robust unmixing results. Moreover, spectral unmixing still has absolute quantification limits caused by additional uncertainties within the spectral coloring compensation. Specifically, improved measurements of the tissue properties, system response, and Gruneisen parameters are still required. To this end, Brochu et al. (2017) have demonstrated to retrieve the tissue optical properties iteratively using finite element-based simulations, thus no predefined library of properties is needed. Besides, deep learning approaches based on non-explicit light fluence estimation have been proposed (Li et al., 2022) for application-specific tasks. Thus, in the future, developing deep learning approaches that can be data-driven based would be beneficial.
Furthermore, blind approaches like SPAX produce a number of components that need to be visually inspected by an expert to select the most relevant one based on experience and prior knowledge or determined using e.g., a spectrophotometer. This may introduce uncertainties, especially, in pathological conditions where there is no theoretical comparison with the spectral components automatically unmixed. Thus, future implementations of prediction models based on simulations could help to recognize meaningful features and their relation with specific disease conditions. Finally, to promote the clinical translation of the sPAI technology real-time unmixing processing would be beneficial. Typically, spectral unmixing techniques are not feasible for real-time applications due to the long acquisition time associated with transmitting multiple laser wavelengths in sPAI. Thus, further efforts would be required to adjust the tradeoff between acquisition time and spectral resolution. Currently, the SPAX is optimized for spectral PAI including a spectral decoloring approach based on high-resolution US images and multi-wavelength Monte-Carlo light fluence simulations. This approach is ideal for accurate and highly sensitive offline unmixing analysis. However, opportunely choosing a minimum number of excitation wavelengths during PAI acquisition, which maintains the accuracy and sensitivity of the spectral unmixing approach, would reduce the acquisition and processing times. As already demonstrated by Luke et al. (2013), the implementation of advanced algorithms for the selection of significant wavelengths could facilitate unsupervised unmixing using an optimum number of wavelengths, thus minimizing the acquisition time and speeding up the analysis.
In addition to the number of wavelengths, the linear motor step size and consequently the number of slices of 3D multi-wavelength data also limits the real-time acquisition. Thus, the motor step size can be selected to minimize imaging artifacts by maintaining a good compromise between speed of acquisition, resolution, and continuity of volumetric imaging. Besides, the optimization of spectral Monte Carlo simulations for fluence compensation by using GPU-based implementations would also speed up the computational processing time, encouraging the use of SPAX in further applications. By optimizing these parameters, acquisition and processing time can be improved, thus facilitating the translation of the approach into clinical practice.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The animal study was reviewed and approved by Animal Use and Ethics Committee (CEUA) of Netherlands (Protocol: AV D2450020173644).
VG performed experiments, designed data analysis, and wrote the first draft of the manuscript. JJ conceptualized the idea and design of the study, edited the article draft, and contributed on supervision, project administration, and funding acquisition. HWH performed validation experiments and wrote sections of the manuscript. PM edited the article draft and contributed to study administration. RWR contributed to co-supervision and project administration. All authors contributed to manuscript revision, read, and approved the submitted version.
This publication is part of a project that has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement No 811226.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ahn, J. H., Oh, J. H., and Choi, S. (2007). Learning principal directions: Integrated-squared-error minimization. Neurocomputing 70 (7–9), 1372–1381. doi:10.1016/j.neucom.2006.06.004
Ali, M., Magee, D., and Dasgupta, U. (2008). Signal processing overview of ultrasound systems for medical imaging. Texas: SPRAB12.
Allen, T. J., and Beard, P. C. (2016). High power visible light emitting diodes as pulsed excitation sources for biomedical photoacoustics. Biomed. Opt. Express 7 (4), 1260. doi:10.1364/boe.7.001260
Allman, D., Reiter, A., and Bell, M. A. L. (2018). Photoacoustic source detection and reflection artifact removal enabled by deep learning. IEEE Trans. Med. Imaging 37 (6), 1464–1477. doi:10.1109/tmi.2018.2829662
Arabul, M. U., Rutten, M. C. M., Bruneval, P., van Sambeek, M. R. H. M., van de Vosse, F. N., and Lopata, R. G. P. (2019). Unmixing multi-spectral photoacoustic sources in human carotid plaques using non-negative independent component analysis. Photoacoustics 15, 100140. doi:10.1016/j.pacs.2019.100140
Bagchi, D. P., and Macdougald, O. A. (2019). Identification and dissection of diverse mouse adipose depots. J. Vis. Exp. 2019 (149). doi:10.3791/59499
Bayer, C. L., Wlodarczyk, B. J., Finnell, R. H., and Emelianov, S. Y. (2017). Ultrasound-guided spectral photoacoustic imaging of hemoglobin oxygenation during development. Biomed. Opt. Express 8 (2), 757. doi:10.1364/boe.8.000757
Bell, A. J., and Sejnowski, T. J. (1995). An information-maximization approach to blind separation and blind deconvolution. Neural Comput. 7 (6), 1129–1159. doi:10.1162/neco.1995.7.6.1129
Benyamin, M., Genish, H., Califa, R., Wolbromsky, L., Ganani, M., Wang, Z., et al. (2020). Autoencoder based blind source separation for photoacoustic resolution enhancement. Sci. Rep. 10 (1), 21414. doi:10.1038/s41598-020-78310-5
Berry, M. W., Browne, M., Langville, A. N., Pauca, V. P., and Plemmons, R. J. (2007). Algorithms and applications for approximate nonnegative matrix factorization. Comput. Stat. Data Anal. 52 (1), 155–173. doi:10.1016/j.csda.2006.11.006
Brochu, F. M., Brunker, J., Joseph, J., Tomaszewski, M. R., Morscher, S., and Bohndiek, S. E. (2017). Towards quantitative evaluation of tissue absorption coefficients using light fluence correction in optoacoustic tomography. IEEE Trans. Med. Imaging 36, 322–331. doi:10.1109/tmi.2016.2607199
Buehler, A., Herzog, E., Razansky, D., and Ntziachristos, V. (2010). Video rate optoacoustic tomography of mouse kidney perfusion. Opt. Lett. 35, 2475. doi:10.1364/ol.35.002475
Bulsink, R., Singh, M. K. A., Xavierselvan, M., Mallidi, S., Steenbergen, W., and Francis, K. J. (2021). Oxygen saturation imaging using LED-based photoacoustic system. Sensors Switz. 21 (1), 283. doi:10.3390/s21010283
Cai, C., Deng, K., Ma, C., and Luo, J. (2018). End-to-end deep neural network for optical inversion in quantitative photoacoustic imaging. Opt. Lett. 43 (12), 2752. doi:10.1364/ol.43.002752
Cassidy, P. J., and Radda, G. K. (2005). Molecular imaging perspectives. J. R. Soc. Interface 2, 133–144. doi:10.1098/rsif.2005.0040
Chowdhary, C. L., and Acharjya, D. P. (2020). Segmentation and feature extraction in medical imaging: A systematic review. Procedia Comput. Sci. 167, 26–36. doi:10.1016/j.procs.2020.03.179
Cox, B. T., Arridge, S. R., and Beard, P. C. (2009). Estimating chromophore distributions from multiwavelength photoacoustic images. J. Opt. Soc. Am. A 26 (2), 443–455. doi:10.1364/josaa.26.000443
Cox, B. T., Arridge, S. R., Köstli, K. P., and Beard, P. C. (2006). Two-dimensional quantitative photoacoustic image reconstruction of absorption distributions in scattering media by use of a simple iterative method. Appl. Opt. 45 (8), 1866. doi:10.1364/ao.45.001866
Das, D., Sharma, A., Rajendran, P., and Pramanik, M. (2021). Another decade of photoacoustic imaging. Phys. Med. Biol. 66, 05TR01. doi:10.1088/1361-6560/abd669
Deán-Ben, X. L., and Razansky, D. (2014). Adding fifth dimension to optoacoustic imaging: Volumetric time-resolved spectrally enriched tomography. Light. Sci. Appl. 3, e137. doi:10.1038/lsa.2014.18
Durairaj, D. A., Agrawal, S., Johnstonbaugh, K., Chen, H., Karri, S. P. K., and Kothapalli, S-R. (2020). “Unsupervised deep learning approach for photoacoustic spectral unmixing,” in Photons plus ultrasound: Imaging and sensing (SPIE), 173–181. doi:10.1117/12.2546964
Erhayiem, B., Dhingsa, R., Hawkey, C. J., and Subramanian, V. (2011). Ratio of visceral to subcutaneous fat area is a biomarker of complicated crohn’s disease. Clin. Gastroenterol. Hepatol. 9 (8), 684–687. doi:10.1016/j.cgh.2011.05.005
Feng, S., Zeng, F-A., and Chance, B. (1995). Photon migration in the presence of a single defect: A perturbation analysis. Appl. Opt. 34 (19), 3826. doi:10.1364/ao.34.003C826
Glatz, J., Deliolanis, N. C., Buehler, A., Razansky, D., and Ntziachristos, V. (2011). Blind source unmixing in multi-spectral optoacoustic tomography. Opt. Express 19 (4), 3175. doi:10.1364/oe.19.003175
Grasso, V., Holthof, J., and Jose, J. (2020). An automatic unmixing approach to detect tissue chromophores from multispectral photoacoustic imaging. Sensors Switz. 20, 3235. doi:10.3390/s20113235
Grasso, V., Willumeit-Rӧmer, R., and Jose, J. (2022). Superpixel spectral unmixing framework for the volumetric assessment of tissue chromophores: A photoacoustic data-driven approach. Photoacoustics 26, 100367. doi:10.1016/j.pacs.2022.100367
Gröhl, J., Kirchner, T., Adler, T., and Maier-Hein, L. (2019). Estimation of blood oxygenation with learned spectral decoloring for quantitative photoacoustic imaging (LSD-qPAI). Available from http://arxiv.org/abs/1902.05839.
Gröhl, J., Schellenberg, M., Dreher, K., and Maier-Hein, L. (2021). Deep learning for biomedical photoacoustic imaging: A review. Photoacoustics 22, 100241. doi:10.1016/j.pacs.2021.100241
Han, Y., Tzoumas, S., Nunes, A., Ntziachristos, V., and Rosenthal, A. (2015). Sparsity-based acoustic inversion in cross-sectional multiscale optoacoustic imaging. Med. Phys. 42 (9), 5444–5452. doi:10.1118/1.4928596
Hassan, H. W., Grasso, V., Korostynska, O., Khan, H., Jose, J., and Mirtaheri, P. (2021). An overview of assessment tools for determination of biological Magnesium implant degradation. Med. Eng. Phys. 93, 49–58. doi:10.1016/j.medengphy.2021.05.016
Hassan, H. W., Mathew, A., Khan, H., Korostynska, O., and Mirtaheri, P. (2021). “Feasibility study of multi-wavelength optical probe to analyze magnesium implant degradation effects,” in Proceedings of IEEE sensors (IEEE).
Hervé, L. (2010). Nonnegative matrix factorization: A blind spectra separation method for in vivo fluorescent optical imaging. J. Biomed. Opt. 15 (5), 056009. doi:10.1117/1.3491796
Hu, S., and Wang, L. V. (2010). Photoacoustic imaging and characterization of the microvasculature. J. Biomed. Opt. 15 (1), 011101. doi:10.1117/1.3281673
Hyvärinen, A. (1999). Fast and robust fixed-point algorithms for independent component analysis. IEEE Trans. Neural Netw. 10 (3), 626–634. doi:10.1109/72.761722
Hyvärinen, A. (2013). Independent component analysis: Recent advances. Phil. Trans. R. Soc. A 371, 20110534. doi:10.1098/rsta.2011.0534
IEC (2015). Iec 80601-2-71: 2015 medical electrical equipment – Part 2-71: Particular requirements for the basic safety and essential performance of functional near-infrared spectroscopy (NIRS) equipment. Int. Stand 2015 (2-71), EN80601.
Iskander-Rizk, S., Visscher, M., Moerman, A. M., Korteland, S. A., Van der Heiden, K., Van der Steen, A. F. W., et al. (2021). Micro Spectroscopic Photoacoustic (μsPA) imaging of advanced carotid atherosclerosis. Photoacoustics 22, 100261. doi:10.1016/j.pacs.2021.100261
Jacques, S. L. (2013). Optical properties of biological tissues: A review. Phys. Med. Biol. 58, R37–R61. doi:10.1088/0031-9155/58/11/r37
Jnawali, K., Chinni, B., Dogra, V., and Rao, N. (2019). “Transfer learning for automatic cancer tissue detection using multispectral photoacoustic imaging,” in Proceedings volume 10950, medical imaging 2019: Computer-aided diagnosis (San Diego, California, United States: SPIE Medical Imaging), 109503W. doi:10.1117/12.2506950
Jo, J., Tian, C., Xu, G., Sarazin, J., Schiopu, E., Gandikota, G., et al. (2018). Photoacoustic tomography for human musculoskeletal imaging and inflammatory arthritis detection. Photoacoustics 12, 82–89. doi:10.1016/j.pacs.2018.07.004
Jo, J., Xu, G., Cao, M., Marquardt, A., Francis, S., Gandikota, G., et al. (2017). A functional study of human inflammatory arthritis using photoacoustic imaging. Sci. Rep. 7 (1), 15026. doi:10.1038/s41598-017-15147-5
Jutten, C., and Karhunen, J. (2004). Advances in blind source separation (BSS) and independent component analysis (ICA) for nonlinear mixtures. Int. J. Neural Syst. 14 (5), 267–292. doi:10.1142/s012906570400208x
Kim, J., Lee, D., Jung, U., and Kim, C. (2015). Photoacoustic imaging platforms for multimodal imaging. Ultrasonography 34 (2), 88–97. doi:10.14366/usg.14062
Kuniyil Ajith Singh, M., Sato, N., Ichihashi, F., and Sankai, Y. (2020). Clinical translation of photoacoustic imaging—opportunities and challenges from an industry perspective. Springer.
Kwak, S. G., and Kim, J. H. (2017). Central limit theorem: The cornerstone of modern statistics. Korean J. Anesthesiol. 70 (2), 144. doi:10.4097/kjae.2017.70.2.144
Lavaud, J., Henry, M., Gayet, P., Fertin, A., Vollaire, J., Usson, Y., et al. (2020). Noninvasive monitoring of liver metastasis development via combined multispectral photoacoustic imaging and fluorescence diffuse optical tomography. Int. J. Biol. Sci. 16 (9), 1616–1628. doi:10.7150/ijbs.40896
Lawrence, D. J., Escott, M. E., Myers, L., Intapad, S., Lindsey, S. H., and Bayer, C. L. (2019). Spectral photoacoustic imaging to estimate in vivo placental oxygenation during preeclampsia. Sci. Rep. 9 (1), 558. doi:10.1038/s41598-018-37310-2
Le, Q. V., Karpenko, A., Ngiam, J., and Ng, A. Y. (2011). ICA with reconstruction cost for efficient overcomplete feature learning. Proc. Adv. Neural Inform. Process. Syst., 1017–1025.
Lee, D. D., and Seung, H. S. (2001). Algorithms for non-negative matrix factorization. Adv. Neural Inf. Process. Syst. 13, 556–562.
Lee, J. G., Jun, S., Cho, Y. W., Lee, H., Kim, G. B., Seo, J. B., et al. (2017). Deep learning in medical imaging: General overview. Korean J. Radiology 18, 570–584. doi:10.3348/kjr.2017.18.4.570
Li, J., Wang, C., Chen, T., Lu, T., Li, S., Sun, B., et al. (2022). Deep learning-based quantitative optoacoustic tomography of deep tissues in the absence of labeled experimental data. Optica 9 (1), 32. doi:10.1364/optica.438502
Li, M., Tang, Y., and Yao, J. (2018). Photoacoustic tomography of blood oxygenation: A mini review. Photoacoustics 10, 65–73. doi:10.1016/j.pacs.2018.05.001
Li, M. L., Oh, J. T., Xie, X., Ku, G., Wang, W., Li, C., et al. (2008). Simultaneous molecular and hypoxia imaging of brain tumors in vivo using spectroscopic photoacoustic tomography. Proc. IEEE 96 (3), 481–489. doi:10.1109/jproc.2007.913515
Lipovetsky, S. (2009). PCA and SVD with nonnegative loadings. Pattern Recognit. DAGM. 42 (1), 68–76. doi:10.1016/j.patcog.2008.06.025
Liu, Y., Wu, P. C., Guo, S., Chou, P. T., Deng, C., Chou, S. W., et al. (2020). Low-toxicity FePt nanoparticles for the targeted and enhanced diagnosis of breast tumors using few centimeters deep whole-body photoacoustic imaging. Photoacoustics 19, 100179. doi:10.1016/j.pacs.2020.100179
Luke, G. P., Nam, S. Y., and Emelianov, S. Y. (2013). Optical wavelength selection for improved spectroscopic photoacoustic imaging. Photoacoustics 1 (2), 36–42. doi:10.1016/j.pacs.2013.08.001
Manwar, R., Zafar, M., and Xu, Q. (2020). Signal and image processing in biomedical photoacoustic imaging: A review. Optics 2 (1), 1–24. doi:10.3390/opt2010001
Matrone, G., Savoia, A. S., Caliano, G., and Magenes, G. (2015). The delay multiply and sum beamforming algorithm in ultrasound B-mode medical imaging. IEEE Trans. Med. Imaging 34, 940–949. doi:10.1109/tmi.2014.2371235
Oblefias, W. R., Soriano, M. N., and Saloma, C. A. (2004). SVD vs PCA : Comparison of performance in an imaging spectrometer. Science 16, 1.
Papazoglou, E. S., Weingarten, M. S., Zubkov, L., Zhu, L., Tyagi, S., and Pourrezaei, K. (2006). Optical properties of wounds: Diabetic versus healthy tissue. IEEE Trans. Biomed. Eng. 53 (6), 1047–1055. doi:10.1109/tbme.2006.873541
Park, E. Y., Lee, H., Han, S., Kim, C., and Kim, J. (2022). Photoacoustic imaging systems based on clinical ultrasound platform. Exp. Biol. Med. 247, 551–560. doi:10.1177/15353702211073684
Reber, J., Willershäuser, M., Karlas, A., Paul-Yuan, K., Diot, G., Franz, D., et al. (2018). Non-invasive measurement of Brown fat metabolism based on optoacoustic imaging of hemoglobin gradients. Cell Metab. 27 (3), 689–701. doi:10.1016/j.cmet.2018.02.002
Rodrigues, J., Amin, A., Raghushaker, C. R., Chandra, S., Joshi, M. B., Prasad, K., et al. (2021). Exploring photoacoustic spectroscopy-based machine learning together with metabolomics to assess breast tumor progression in a xenograft model ex vivo. Lab. Invest. 101 (7), 952–965. doi:10.1038/s41374-021-00597-3
Rutledge, D. N., and Jouan-Rimbaud Bouveresse, D. (2013). Independent components analysis with the JADE algorithm. TrAC Trends Anal. Chem. 50, 22–32. doi:10.1016/j.trac.2013.03.013
Sangha, G. S., and Goergen, C. J. (2020). Label-free photoacoustic and ultrasound imaging for murine atherosclerosis characterization. Apl. Bioeng. 4 (2), 026102. doi:10.1063/1.5142728
Santosh Kumar, S., Bharathi, S. H., and Archana, M. (2016). “Non-negative matrix based optimization scheme for blind source separation in automatic speech recognition system,” in Proceedings of the international conference on communication and electronics systems (ICCES). doi:10.1109/CESYS.2016.7889860
Smita, S., Biswas, S., and Solanki, S. S. (2014). Audio signal separation and classification: A review paper. Int. J. Innov. Res. Comput. Commun. Eng. 2 (11).
Sotiras, A., Resnick, S. M., and Davatzikos, C. (2015). Finding imaging patterns of structural covariance via Non-Negative Matrix Factorization. Neuroimage 108, 1–16. doi:10.1016/j.neuroimage.2014.11.045
Stoffels, I., Morscher, S., Helfrich, I., Hillen, U., Leyh, J., Lehy, J., et al. (2015). Metastatic status of sentinel lymph nodes in melanoma determined noninvasively with multispectral optoacoustic imaging. Sci. Transl. Med. 7 (317), 317ra199. doi:10.1126/scitranslmed.aad1278
Tipping, M. E., and Bishop, C. M. (1999). Probabilistic principal component analysis. J. R. Stat. Soc. B 61 (3), 611–622. doi:10.1111/1467-9868.00196
Tzoumas, S., Nunes, A., Olefir, I., Stangl, S., Symvoulidis, P., Glasl, S., et al. (2016). Eigenspectra optoacoustic tomography achieves quantitative blood oxygenation imaging deep in tissues. Nat. Commun. 7, 12121. doi:10.1038/ncomms12121
Weiss, G. H., Nossal, R., and Bonner, R. F. (1989). Statistics of penetration depth of photons re-emitted from irradiated tissue. J. Mod. Opt. 36 (3), 349–359. doi:10.1080/09500348914550381
Xie, Z., Yang, Y., He, Y., Shu, C., Chen, D., Zhang, J., et al. (2020). In vivo assessment of inflammation in carotid atherosclerosis by noninvasive photoacoustic imaging. Theranostics 10 (10), 4694–4704. doi:10.7150/thno.41211
Yan, S., and Fang, Q. (2020). Hybrid mesh and voxel based Monte Carlo algorithm for accurate and efficient photon transport modeling in complex bio-tissues. Biomed. Opt. Express 11 (11), 6262. doi:10.1364/boe.409468
Yang, C., Lan, H., Gao, F., and Gao, F. (2021). Review of deep learning for photoacoustic imaging. Photoacoustics 21, 100215. doi:10.1016/j.pacs.2020.100215
Yang, Q., Jin, H., Gao, Y., Lin, J., Yang, H., and Yang, S. (2019). Photostable iridium(III)-Cyanine complex nanoparticles for photoacoustic imaging guided near-infrared photodynamic therapy in vivo. ACS Appl. Mat. Interfaces 11 (17), 15417–15425. doi:10.1021/acsami.9b04098
Yuan, Z., Wang, Q., and Jiang, H. (2007). Reconstruction of optical absorption coefficient maps of heterogeneous media by photoacoustic tomography coupled with diffusion equation based regularized Newton method. Opt. Express 15 (26), 18076. doi:10.1364/oe.15.018076
Zhang, F., Hao, G., Shao, M., Nham, K., An, Y., Wang, Q., et al. (2018). An adipose tissue atlas: An image-guided identification of human-like BAT and beige depots in rodents. Cell Metab. 27 (1), 252–262. doi:10.1016/j.cmet.2017.12.004
Keywords: blind source separation, photoacoustic, spectral coloring, spectral imaging, ultrasound, unsupervised machine learning
Citation: Grasso V, Hassan HW, Mirtaheri P, Willumeit-Rӧmer R and Jose J (2022) Recent advances in photoacoustic blind source spectral unmixing approaches and the enhanced detection of endogenous tissue chromophores. Front. Sig. Proc. 2:984901. doi: 10.3389/frsip.2022.984901
Received: 02 July 2022; Accepted: 31 October 2022;
Published: 10 November 2022.
Edited by:
James Joseph, University of Dundee, United KingdomReviewed by:
Mithun Kuniyil Ajith Singh, Cyberdyne,Inc., NetherlandsCopyright © 2022 Grasso, Hassan, Mirtaheri, Willumeit-Rӧmer and Jose. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jithin Jose, aml0aGluLmpvc2VAZnVqaWZpbG0uY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.