
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Signal Process., 05 April 2022
Sec. Image Processing
Volume 2 - 2022 | https://doi.org/10.3389/frsip.2022.857313
This article is part of the Research TopicAdvances in Biomedical Image Segmentation and Analysis using Deep LearningView all 6 articles
Unet architectures are being investigated for automatic image segmentation of bones in CT scans because of their ability to address size-varying anatomies and pathological deformations. Nonetheless, changes in mineral density, narrowing of joint spaces and formation of largely irregular osteophytes may easily disrupt automatism requiring extensive manual refinement. A novel Unet variant, called CEL-Unet, is presented to boost the segmentation quality of the femur and tibia in the osteoarthritic knee joint. The neural network embeds region-aware and two contour-aware branches in the decoding path. The paper features three main technical novelties: 1) directed connections between contour and region branches progressively at different decoding scales; 2) pyramidal edge extraction in the contour branch to perform multi-resolution edge processing; 3) distance-weighted cross-entropy loss function to increase delineation quality at the sharp edges of the shapes. A set of 700 knee CT scans was used to train the model and test segmentation performance. Qualitatively CEL-Unet correctly segmented cases where the state-of-the-art architectures failed. Quantitatively, the Jaccard indexes of femur and tibia segmentation were 0.98 and 0.97, with median 3D reconstruction errors less than 0.80 and 0.60 mm, overcoming competitive Unet models. The results were evaluated against knee arthroplasty planning based on personalized surgical instruments (PSI). Excellent agreement with reference data was found for femoral (0.11°) and tibial (0.05°) alignments of the distal and proximal cuts computed on the reconstructed surfaces. The bone segmentation was effective for large pathological deformations and osteophytes, making the techniques potentially usable in PSI-based surgical planning, where the reconstruction accuracy of the bony shapes is one of the main critical factors for the success of the operation.
Biomedical image segmentation has lately undergone an unprecedented push forward thanks to the rapid accumulation of evidences about the promising performance of a novel generation of deep convolutional neural networks (Litjens et al., 2017; Isensee et al., 2021). Such networks entail encoder-decoder (E-D) architectures for processing both two-dimensional (2D) images and three-dimensional (3D) volumes, where the function of the decoder path is to cast the low-resolution encoder feature maps to high-resolution feature maps for pixel-wise classification. The Unet (Ronneberger et al., 2015), a particular type of symmetric E-D network with horizontal skip connections between the encoder and decoder paths, proved effective in a wide range of medical domains ranging from orthopaedics (Norman et al., 2018; Noguchi et al., 2020; Marzorati et al., 2020), oncology (Huang et al., 2018; Jin et al., 2020; Li et al., 2020), neurology (Gadosey et al., 2020; McKinley et al., 2021) even up to histology (Falk et al., 2019; Long, 2020; Zhou et al., 2020). In the wake traced by these researches and aiming at addressing general purpose biomedical image segmentation tasks, there has been recently a remarkable attempt to gather and consolidate most of the earlier Unet developments into a unique computational framework called nnUnet (Isensee et al., 2021). Despite such a network outperformed most of the earlier proposals in the literature to a large extent, across many international challenges (D12 PROMISE12, D16 CHAOS, MICCAI 2019, MICCAI 2020, D20-D23 Cell Tracking Challenge, just to cite few), the ambition of the work was partially mitigated, recognizing some fundamental aspects worth of further investigations. Authors argued actually that: 1) very similar architectures may lead to very varying results across datasets; 2) specific tasks may require tailored loss function and highly domain-specific target metrics as well; 3) none of the commonly used architectural modifications, such as for instance batch normalization, residual connections, and attention layers, may ensure a necessary condition for reliable performance. In general, how to exploit task-dependent knowledge for combining network hyper-parameters, training setups, and loss functions remained elusive. All these considerations have definitely wiped out the idea that a single architecture, in a one-fits-all mode, can address all segmentation applications. Concerning the osseous shape segmentation from X-ray and CT images, 2D and 3D Unet models have been investigated for musculoskeletal analysis (e.g., bone age assessment), computer-assisted diagnostics, and therapy purposes (e.g., joint replacement planning and bone tumor resection). Weak bone boundaries, narrowness of joint space, variability in bone density, size and shape were acknowledged to be the main barriers to automatic bone delineation (Ambellan et al., 2019; Chang et al., 2019). In order to account for hand bone scale variations during growth in children, 2D Unet, applied to X-ray images, was tailored by leveraging multi-resolution with different filter sizes (Ding et al., 2019). Mandibular bones in cranio-facial CT were segmented using a 2D Unet processing concurrently three orthogonal planes (Qiu et al., 2019). In order to handle large variations of shape and density, automatic femur segmentation from CT scans was addressed by enhancing the Unet with an in-line task-specific edge detection processor and harnessing fusion of multi-scale features (Chen et al., 2019). The quality of bone segmentation in whole-body CT scans was evaluated by comparing three alternative Unet setups harnessing 2D axial slices only, axial, sagittal and coronal slices in bundle, and an approach where the network was pre-trained using an unsupervised technique (Klein et al., 2019). With a similar purpose, Unet architecture was explored by evaluating different data augmentation strategies to improve bone segmentation on whole-body CT scans (Noguchi et al., 2020). However, such studies did not addressed severe pathological bone deformation induced by osteophytes formation, and disregarded how the effects of local segmentation errors may impact differently across clinical applications. In (Marzorati et al., 2020), our group described one 3D-Unet model tailored to bone segmentation in knee CT images, with a specific clinical aim towards knee replacement applications based on personalized surgical instruments (PSI) (Shih et al., 2020). PSI technique has been acknowledged to be very demanding as submillimetric shape reconstruction is fundamental for the success of the knee surgery, especially at the areas of contact with the PSI of both femur and tibia (Anderl et al., 2016; Cerveri et al., 2017; Ogura et al., 2019). Provided that the coupling of the PSI to the true bone geometry determines the bone resection alignment, uncontrolled segmentation uncertainty may lead to unexpected angular deviations away from the surgical planning [clinical acceptance less than 1° (Gong et al., 2019; Shi et al., 2020)] affecting the mechanical corrections of the knee joint or even leading to the withdrawal of the PSI technique in favour of the traditional invasive surgery based on intra-medullary tools (Ogura et al., 2019). In (Marzorati et al., 2020) our group showcased (over a dataset of 200 retrospective CT scans) that the achieved segmentation quality was substantially good enough to cope with PSI requirements. However, some severe deformations and variability of the osseous density were shown to be still critical affecting the segmentation quality especially at the boundaries of condylar regions of the femur and tibial plateau. In the present work, we evolved this Unet model (Marzorati et al., 2020) into an innovative architecture, based on a two-channel decoding branch, to simultaneously predict region and contour masks, and exploit the contour information to improve the quality of the segmentation at the shape boundaries. In order to achieve this goal, in the decoding path a contour-aware (CA) branch was enabled in parallel to the region-aware (RA) branch. In (Chen et al., 2019), the authors proposed a modified Unet architecture including an edge-detector path. However, the information processed by such a path was not explicitly fed into the region path, but only used to constrain the global loss function during the training. In the present work, uni-directional residual connections from CA to RA branch (Figure 1) were inserted enabling the explicit aggregation of contour features to region masks at increasing scales. According to some recent contributions in the literature (Dangi et al., 2019; Ma et al., 2020), which used the distance maps to constraint the loss function, we extended such paradigm by implementing a modification of the cross-entropy loss function embedding the contribution of the distance-weighted map to increase delineation quality at the sharp edges of the shapes. The boundary reconstruction in the CA branch were boosted by exploiting pyramidal edge extraction. In order to improve the training quality and extend the generalization results, the dataset was increased to 700 knees, featuring many different degrees of bone deformation and osteoarthritis. Specific evaluation metrics of the segmentation quality were proposed to cope with PSI-based surgical planning requirements. In this regard, traditional Dice index and global 3D reconstruction accuracy do not provide enough information to the clinical operator about the effects of the segmentation residuals on the surgical planning. In this paper, errors in salient regions of the shape were specifically evaluated and the derived bone cutting alignment error was quantified.
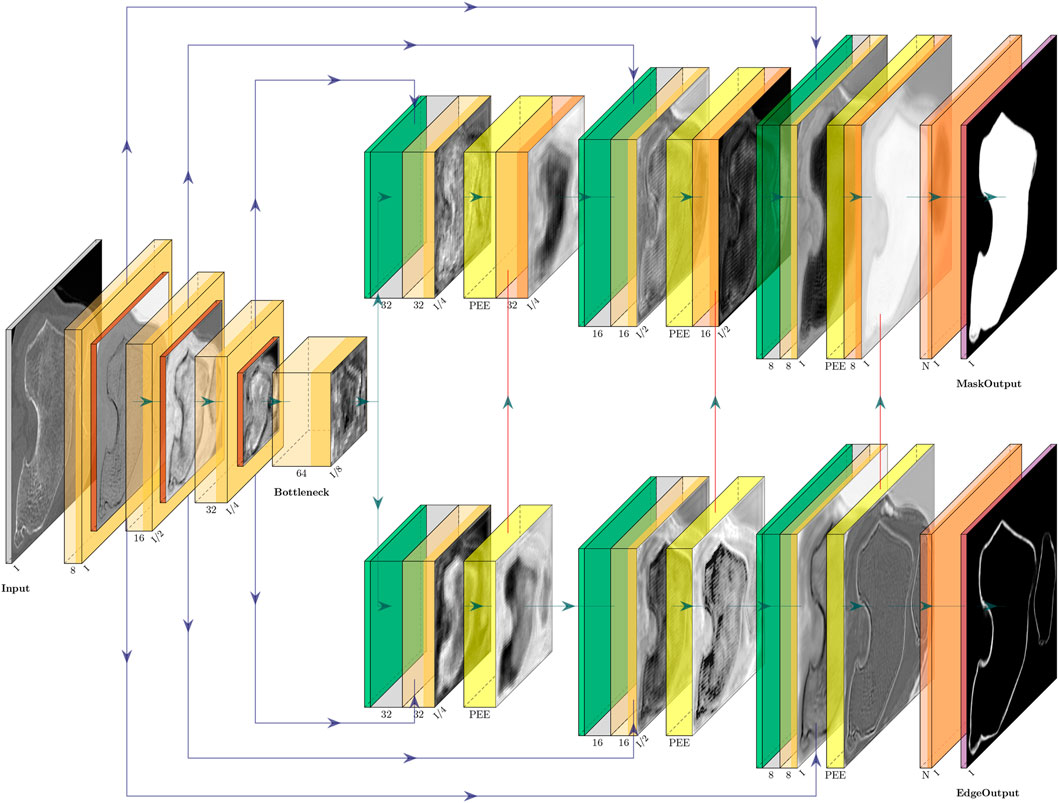
FIGURE 1. CEL-Unet architecture featuring three E–D layers. Left part represents the encoding path ending into the bottleneck level. Skip connections (horizontal blue connections) arise from the encoding path directed towards the two branches of up-sampling decoding path. The upper branch is devoted to the segmentation of bone regions. The lower decoding branch is devoted to edge detection. In order to enhance the region segmentation, the output (vertical red connections) of each decoding layer into the edge detection branch aggregates to the region decoding branch to the corresponding scale. Pyramidal edge extraction (PEE) is implemented in each level of the edge detection branch.
The developed network, called CEL-Unet, processed a CT volume of the knee (size: Nx, Ny, Nz) to produce in output a corresponding labeled volume with the semantically segmented femur and tibia. According to the Unet architecture, the CEL-Unet embedded an encoder path for feature extraction at decreasing spatial resolution, along with a final encoding block, called the bottleneck of the network. In each encoding processing block (ePB), convolutional with linear activation, rectified-linear-unit (Relu) and down-sampling max-pooling layers were embedded. Taking a cue from ablation test results in the earlier work of our group (Marzorati et al., 2020), the number of ePB was set to three, the initial number of feature maps was set to 8 and doubled when moving from one ePB to the next one. Each feature map was characterized by a filter size and stride length of 3 × 3 × 3 and 1 × 1 × 1, respectively. The max-pooling layers featured filter size and stride length of 2 × 2 × 2 and 2 × 2 × 2, respectively. The bottleneck featured convolutional (64 feature maps) and Relu layers only (Figure 1). Unlike the Unet, the decoding path of the CEL-Unet was split into two parallel branches, one devoted to region segmentation (RA, region-aware branch) and the other one addressing edge detection (CA, contour-aware branch). Likewise the encoder path, increasing spatial resolution was enabled through upsampling (transpose deconvolution) in both decoding paths, where the number of convolutional feature maps, starting from 32, were halved when moving from one decoding processing block (dPB) to the next one. Likewise the encoder, convolution filter size was 3 × 3 × 3 with stride length 1 × 1 × 1. Skip connections between the encoder and the two decoder paths were allowed according to the Unet architecture. The last layer of the region-aware decoding path was a 1 × 1 × 1 depth convolution block with three output channels, representing three classes, namely background, femur and tibia, respectively, followed by a Softmax activation featuring a tensor of size Nx × Ny × Nz × 3. Likewise, the output of the CA branch had the same structure of the RA branch output. In order to empower region segmentation, two structural setups were deployed in the contour branch. First, vertical unidirectional skip connections between the output of each processing block and the corresponding one in the region branch were inserted. This enabled the aggregation between edge and region features at progressively increasing spatial scales. Second, a devoted processing module, implemented through the pyramidal edge extraction (PEE) paradigm (Wang et al., 2020), was concatenated to the corresponding processing block of the region branch (Figure 2). Being i and xi the current up-sampling stage and the input to the corresponding PEE module, an initial depth-convolution Fi(x), featuring Pi feature maps with a filter size of 1 × 1 × 1, was computed. The edge features were computed by the difference between Fi(x) and average pooling avg processors at increasing scale s as:
where
where S and Fi were the number of pyramid scales, the convolution parameters and the output feature map of PEE module at current stage, respectively. This allowed to further increase the robustness of thin edge detection exploiting multiple granularity of edge features. Considering the number of feature maps in each PEE module equivalent to that of the corresponding input, the number of trainable parameters of the CEL-Unet was 558298.
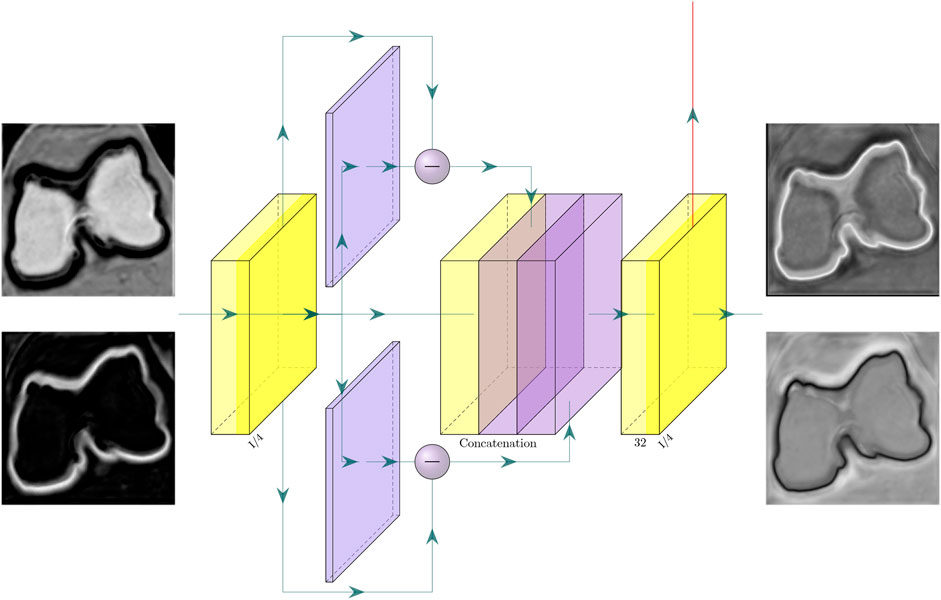
FIGURE 2. Pyramidal edge extraction (PEE) module in one level of the edge decoding branch. For sake of clarity, only two pyramid-edge filters were considered. The input to the PEE module consists of the set of activation maps (for simplicity here only 2 were depicted), being the output of the previous layer in the contour-aware decoding branch. Likewise, the module output consists of the set of activation maps (again for simplicity here only 2 were depicted), which are delivered to the next level of the contour-aware decoding branch (horizontal arrow), and to the same level of the region-aware decoding branch (vertical arrow).
The formulation of the loss function, based on Dice index and cross-entropy, leveraged the information conveyed by both decoding branches thus exploiting both region and contour labels. Based on the region label, Dice index was considered and tailored to the multi-class problem. For each class, a coefficient, proportional to the number of voxels belonging to the specific class was used to weight the corresponding Dice contribution. This way the different frequencies of voxels for each class were compensated and the overall loss function re-balanced. Assuming C classes and N voxels, the Dice formula can be written as:
where yc and
where Pc is the number of voxels belonging to class c, respectively. In order to fully exploit the mini-batch paradigm in the training, the kc factors was computed at run-time for each specific batch. Exploiting contour label, the euclidean distance transform (EDT), which assigns to each voxel the value of its distance from the closest voxel belonging to the boundary of the corresponding target label (Figure 3) was taken into account. In order to correctly scale EDT as a loss function, the distance weight map (DWM) was computed by the negative exponential transformation as:
where γ and σ are constant factors. The cross-entropy loss was therefore spatially weighted with DWM to specifically enhance the importance of contour and near-surface voxels, obtaining the voxel-wise distance cross-entropy

FIGURE 3. From left to right. Original CT slice, binary image, euclidean distance transform and distance weight map (DWM). Lighter voxels in the DWM indicate shorter distances from the bone contour.
Similarly, reversed distance cross-entropy was considered as:
Summarizing, two losses were designed, one for region branch
where
700 axial knee CT scans of patients who underwent PSI-based total knee replacement (TKR) surgery, between 2015 and 2020, were retrospectively available in anonymized form by Medacta International SA (Castel San Pietro, Switzerland). At diagnosis time, the patients reported knee instability and local knee pain. Clinical findings were advanced osteoarthritis with different degrees of cartilage defects, femoral osteophytes and shape abnormalities mainly at the condylar regions of the distal femur and at the tibial plateau. Corresponding deviation of the clinical axis alignment from normality was confirmed by radiological assessment (Table 1). CT scans were acquired with different imaging equipment, mostly at 512 × 512 pixels and 600 slices on average, with variable pixel size, ranging from 0.3 to 0.4 mm, and axial slicing ranging from 0.3 to 1.0 mm. Along with the CT images, the dataset entailed distal femur and proximal tibia segmentation masks. The corresponding reconstructed surfaces were available and stored in STL format. Along with the reconstructed surfaces, the dataset included the corresponding planning surfaces. Surgical indications were available on planning surfaces in terms of PSI mask contact points and contact areas, along with planar sections indicating the planned cuts. Expert radiological operators manually performed the image segmentation of the osseous portion of the bones using Mimics software (Materialize, Belgium). For increasing reliability, each dataset was segmented by one expert radiologist and revised by the surgeon who later performed the TKR. Because of the imaging equipment and acquisition protocol variability, no common segmentation protocol was adopted and no data about segmentation uncertainty was available. As a function of the particular centering of the knee joint in the CT scan, the distal femur was segmented up to 2–4 cm away from the frontal notch of the trochlear region along the femur shaft. Concurrently, the length of the tibia segmented shaft was variable across the patient dataset in a range of about 2–3 cm. For this study, 500 cases, out of the 700, were randomly selected (203 males and 297 females—272 left against 228 right knees) in the overall set to be used for training. The remaining 200 cases were used to independently test the segmentation performance. As originating from different scanning equipment, the CT scans underwent preprocessing to make the pixel intensity distribution consistent and to arrange spatial dimensions to cope with network input. First, pixels belonging to filling background and air were automatically identified in the images, according to information gathered from the DICOM header, and the corresponding intensity values put both to zero. The remaining image pixels underwent intensity normalization taking into account of different intensity scale encoding (e.g., Hounsfield units, 12-bit raw pixels, 16-bit raw pixels). Then, each scan was cropped first in the axial direction to remove the slices where reference segmentation was not available. Then, a further crop was attained by a bounding box with a margin of 2 mm about reference 3D geometries. Finally, all the cropped volumes where re-sampled to a size of 192 × 192 × 192, corresponding to an average voxel resolution of 0.45 × 0.45 × 0.85 mm3. In order to perform analysis of the 3D reconstruction errors, all the surfaces were re-sampled and smoothed to 30,000 faces and 15,000 vertices.

TABLE 1. Patient data: age range, male/female ratio, right/left ratio. Clinical data reported as mean (SD) value: Hip-knee-ankle (HKA, physiological value: 180°) angle, femoral varus/valgus (FVV, physiological value: 0°) angle, tibial varus/valgus (TVV, physiological value: 0°) angle, tibial slope (TS, physiological value: 7° posteriorly) and external rotation (ER, physiological value: 0°).
The training was optimized with ADAM (Adaptive Moment Estimation) with a learning rate of 10−4. Parameters γ and σ in Eq. 5 were set to 1 and 0.5, respectively. Parameter α in Eq. 8 was set to 1 and decreased by a factor of 0.005 each iteration up to α = 0.5. α reached a value of 0.5 at the 100-th iteration, and from that iteration on, it was kept constant. This schedule was conceived to allow dice loss to constrain weight learning at the beginning and progressively introducing the effect of cross-entropy based on distance weighted map reducing progressively the role of the dice loss component. Parameter β in Eq. 8 was set to the ratio between the number of shape boundary voxels and the total number of voxels in the batch to balance the two contributions in
The quality of femoral and tibial segmentation was evaluated in terms of clinical impact on the surgical planning in the total knee replacement based on MyKnee technology by Medacta. The planning feasibility, verified on a subset of 20 samples (randomly selected in the test set), was evaluated in terms of the angular error alignment with respect to the distal cutting plane for the femur and the proximal cutting plane of the tibia. These two cuts represent the main surgical resections which determine the recovery of the physiological mechanical axis alignment of the lower limb. Error alignment was computed in both frontal and sagittal projection planes. As earlier mentioned, the dataset used in this work included the corresponding planning surfaces embedding the planes of resection. In order to compute the corresponding resection planes on the reconstructed surfaces, four landmarks were picked in the reference planning surface, along the resection sulcus, two frontally and two posteriorly (Figure 4). The corresponding landmarks in the reconstructed surface were computed by minimal distance. For each bone, the normal direction of the plane fit to the four points was computed in the planning and reconstructed surfaces and the in-between angular deviation was projected on both frontal and sagittal anatomical planes, obtaining the two clinically relevant measures (Cerveri et al., 2010; Pietsch et al., 2013; Gong et al., 2019).
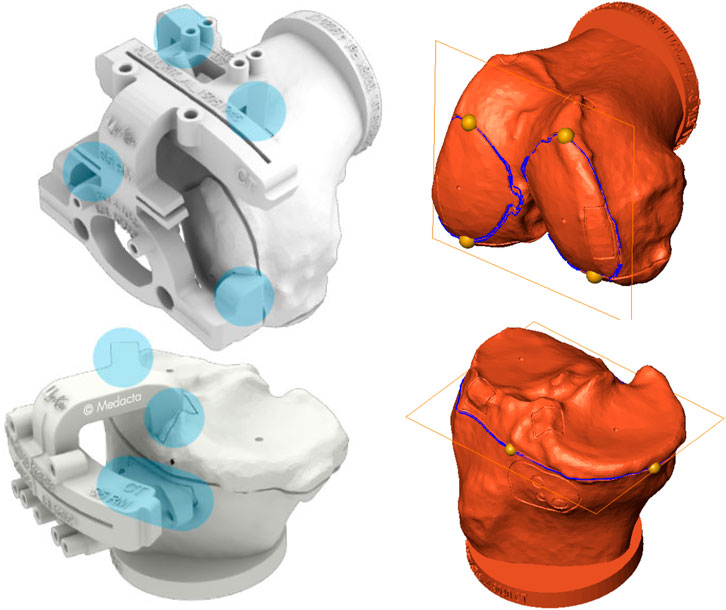
FIGURE 4. Left: Femoral and tibial PSI of MyKnee system (courtesy of Medacta International SA) with the contact areas highlighted in light blue. As it can be appreciated, the main contact femur areas are at distal condylar level. Likewise, tibial contact areas are located at the plateau mainly. Right: Planning surfaces for case #184 with distal femoral and proximal tibial cutting planes. Four yellow landmarks, taken on the cutting profiles (blue curves) of the two planning surfaces, were mapped on the reconstructed surfaces for estimating the corresponding cutting planes and compute angular deviations.
The ablation study was carried out using the CEL-Unet trained using the loss function reported in Eq. 8. Hyper-parameter effects on the segmentation accuracy were evaluated by two independent tests about the PEE role and the number of encoding/decoding layers, respectively. In the first test, the number of pyramid-edge feature maps was varied from zero (no PEE), two, three and four, using (3, 5), (3, 5, 7), and (3, 5, 7, 9) as filter sizes, respectively. Three layers in all encoding and decoding paths were considered. Results (median Jaccard error) proved the beneficial effect of using PEE with two (0.97 against 0.95) and three (0.98 against 0.95) filters (Table 2). Using four filters did not provide improvement in the segmentation accuracy. Minimal additional computational effort was measured during training when using PEE (less than 2 s per iteration). In the second test, the convolutional layers were either inserted or removed symmetrically in the encoding path and in the two decoding paths, setting consequently the bottleneck layer. According to the results of the previous ablation test, three pyramid-edge feature maps (filter sizes: 3, 5, 7) were considered in the PEE modules. Four (8-16-32-64-128-64-32-16-8), three (8-16-32-64-32-16-8) and two (8-16-32-16-8) layers were taken into account. The results, reported in Table 3, showcased that changing the number of convolutional layers produced very small changes in the Jaccard metrics for both femur and tibia, with a full range in the interval of the median value equal to 0.95–0.98, with the first architecture providing slightly poorer results. No statistically significant difference (p
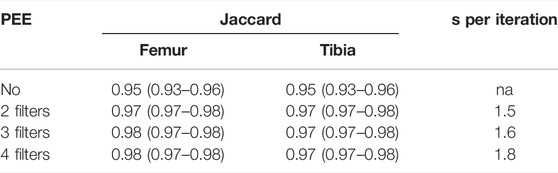
TABLE 2. Results (median and IQR values) of the ablation test performed on the CEL-Unet about the effect of PEE module. The additional computational effort (seconds per iteration), with respect to the removal of the PEE module, were reported.
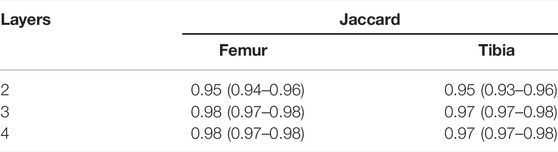
TABLE 3. Results (median and IQR values) of the ablation test performed on the CEL-Unet and a different number of convolutional layers.
The training convergence of the proposed CEL-Unet was achieved after 100 epochs with similar metrics results in both training and validation sets, this supporting the view that the network was appropriately trained avoiding over-fitting. Without loss of generality, the evolution of the Jaccard index was depicted in (Figure 5), computed for both femur and tibia and split on a coronal, sagittal and axial planes for one case in the training set and one case in the validation set. As expected initial accuracy was very poor, while training approached high accuracy after 20 epochs. Despite some rare periods of failure (see the peaks after epoch 50 in Figure 5), the training reached a progressively stable convergence without overfitting, with the metric on validation data consistent with the metric on training data. These results were valid for both the tibia and the femur and for all three orthogonal slices. The test of the segmentation quality was carried out by comparing the proposed model with the state-of-the-art Unet architecture and with the network proposed in (Chen et al., 2019), herewith termed Chen-Unet, (Table 4), which was adapted to process 3D scans in input. For the traditional Unet, hyper-parameters were set according to the extensive results of the ablation tests previously performed by our group and reported in (Marzorati et al., 2020). Namely, three convolutional layers in both encoding and decoding paths were considered. The number of feature maps were 8, 16, 32 (32, 16, 8) for the encoder (decoder) path. The bottle-neck had 64 feature maps. Each processing block embedded a convolution with linear activation, a Relu layer and a max-pooling layer. The filter size was 3 × 3 × 3 in all the convolutional processing. Globally, this model featured 351435 trainable parameters and was trained using three different cost functions, namely Dice (D-Unet), Focal (Shi et al., 2020) (F-Unet) and distance-weighted cross-entropy (DCE-Unet) losses. According to the literature, the Chen-Unet featured two parallel branches in the decoding path, one for region segmentation and the other one for contour delineation, while disregarding vertical skip connections between the two branches, featuring 482894 trainable parameters. Cross-entropy loss was used in the training with the contribution of both region and edge outputs (Chen et al., 2019). The comparative analysis was performed over the 200 CT samples in the test set. As a main result, all the median values of Jaccard, recall, and precision indexes were all larger than 0.95 and 0.92 for femur and tibia, respectively (Figure 6). Specifically, Chen-Unet and CEL-Unet outperformed the traditional Unet, trained with Dice and Focal losses (Table 5), while DCE-Unet attained similar results. F-Unet provided the poorest recall index for the tibia (median value: 0.94, IQR: 0.85–0.97). The statistical comparison supported the superiority of the CEL-Unet with respect to the traditional Unet and the Chen-Unet as well (Table 6). Specifically, CEL-Unet, with respect to the Chen-Unet, showed greater accuracy in the femur featuring statistical difference for Jaccard index (p = 0.001) and recall (p = 0.03), while the precision was very similar (p = 0.80). Despite the difference in the tibial Jaccard index was not significant (p = 0.07), the precision gained by the CEL-Unet was superior to that provided by the Chen-Unet. According to the results, we can summarize that CEL-Unet provided slightly better recall than precision values. However, the difference in between was very small, which allowed to conclude that the proposed model was able to ensure high accuracy reducing under- and over-segmentation altogether. The visual inspection (Figure 7) of some critical cases confirmed lower accuracy of the F-Unet and DCE-Unet testified by mainly under-segmentation (cases #11 and #62) and partial label confusion (cases #11, #62 and #194). D-Unet provided slightly better results, despite affected by over-segmentation (case #176). Chen-Unet showed label confusion in the femoral shaft (cases #176 and #194) and partial under-segmentation of the tibial plateau in the case #11 (axial slice 62) and #194. On the contrary, CEL-Unet was able to correctly separate tibia from the femur with nice agreement with the reference segmentation. The analysis of narrow interface segmentation between femur and patella, and osteophytes, showed the ability of the CEL-Unet to correctly delineate the femoral profile of the trochlear sulcus avoiding both over- and under-segmentation (Figure 8), showing conversely the Chen-Unet under-segmentation, especially in the trochlear anterior boundary for case #15 and in the lateral condyle for case #181.
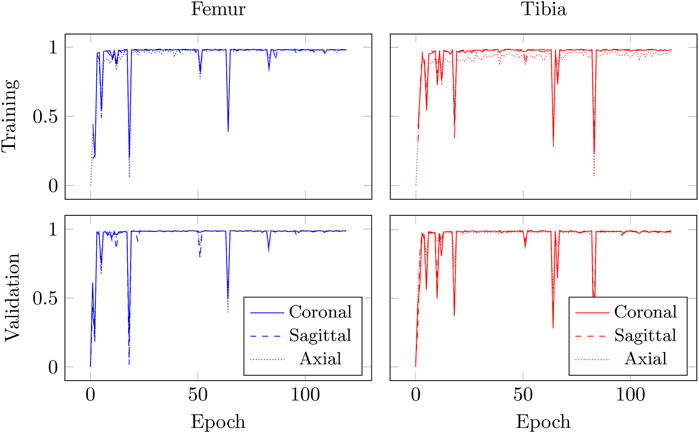
FIGURE 5. Evolution of Jaccard index, measured on coronal, sagittal, and axial slices, for case #2 (training set) and case #199 (validation set) over 120 epochs for CEL-UNet model during training.

TABLE 4. Comparison pf the proposed solution with literature Unet models and different loss functions.
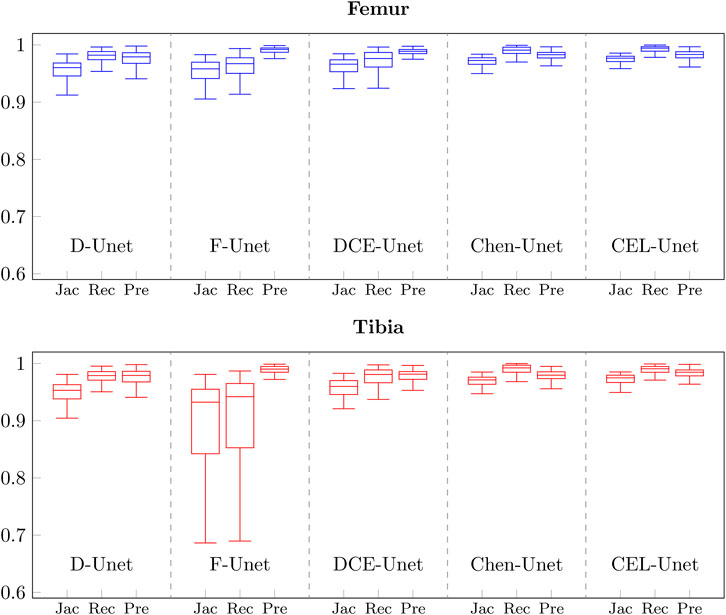
FIGURE 6. Boxplots of Jaccard, Recall, and Precision values obtained on the test set with the five different network architectures described in Table 4 for femur (blue) and tibia (red).
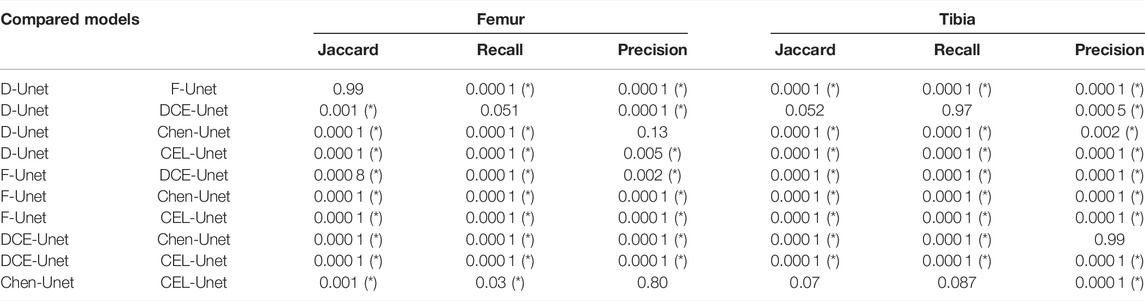
TABLE 6. Statistical results (p values) about comparison of the five segmentation networks. Asterisk means statistical difference at 5%.
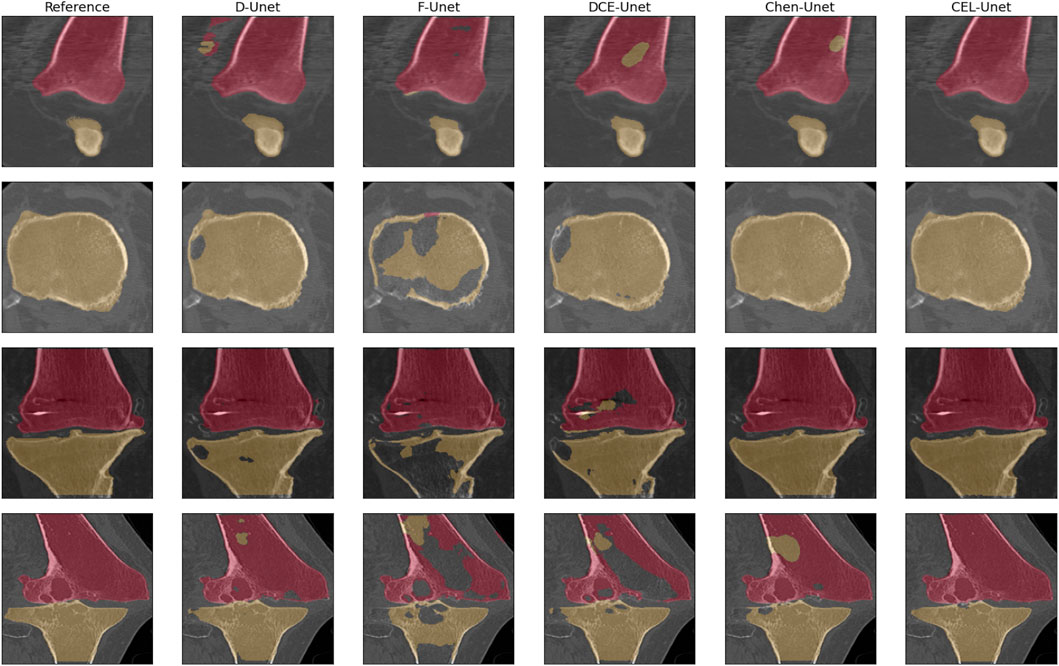
FIGURE 7. Qualitative comparison of the segmentation of femur (purple) and tibia (light brown) in 4 critical cases (case #176 coronal slice: 44; case #11, axial slice: 62; case #11, coronal slice 78; case #194, coronal slice 82).
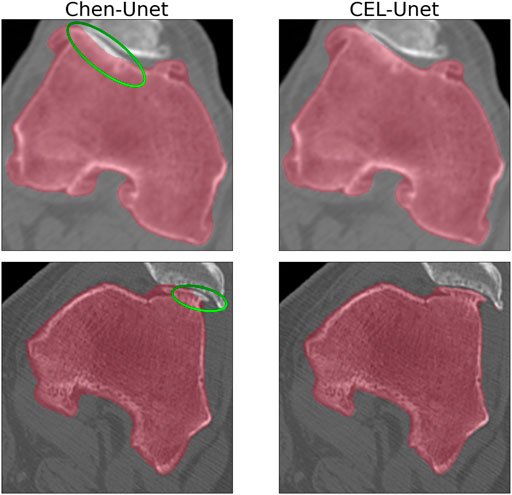
FIGURE 8. Axial slice detail about the segmentation at the femur-patella interface for cases #15 and #181. Comparison between Chen-Unet and CEL-Unet. Under-segmentation (Chen-Unet), highlighted by the green ellipse, at femur-patellar interface can be appreciated.
In order to analyse the global RMS error distributions, each reconstructed surface in the test set was matched to the corresponding reference surface. All the five error distributions were synthesized again in terms of median value and IRQ (Figure 9). The median values were all less than 1 mm, with higher accuracy for the reconstructed tibia shapes. According to the segmentation results, the three traditional Unet models showed poorer median values than both Chen-Unet and CEL-Unet, featuring larger variability as well. Overall, CEL-Unet obtained IRQ ranges less than 0.25 mm for both shapes. The statistical analysis revealed significant difference (p = 0.008) for femur shape in favour of CEL-Unet, whilst for the tibia no difference was detected (p = 0.20). Considering the comparison between femur and tibia, significant differences were found (p = 0.0001) in favor of tibia reconstruction accuracy in all the models except the F-Unet. The 3D Hausdorff distance errors on the femur (median/IRQ) for Chen-Unet and CEL-Unet were 4.79 mm (4.07,5.75) and 4.44 mm (3.88,5.31), respectively, featuring a statistical difference (p = 0.003). As for the tibia, the 3D Hausdorff distance errors were 5.32 mm (4.13,6.50) and 4.34 mm (3.61,5.42), respectively, featuring a statistical difference (p = 0.0001). Visual representation of the reconstructed surfaces (patients #15, #29, #69 and #181), with superimposed distance map with respect to the corresponding reference shapes, supported the above quantitative results (Figure 10). As it can be noticed, especially for patient #69, the proposed model achieved better results in the intercondylar space and on the tibial plateau.
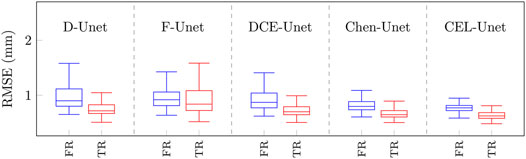
FIGURE 9. Boxplots of global RMS errors (mm) obtained on the test set with the five different network architectures. Boxplots in blue refer to the femur class, while boxplots in red refer to the tibial class. FR: Femur RMSE; TR: Tibia RMSE.
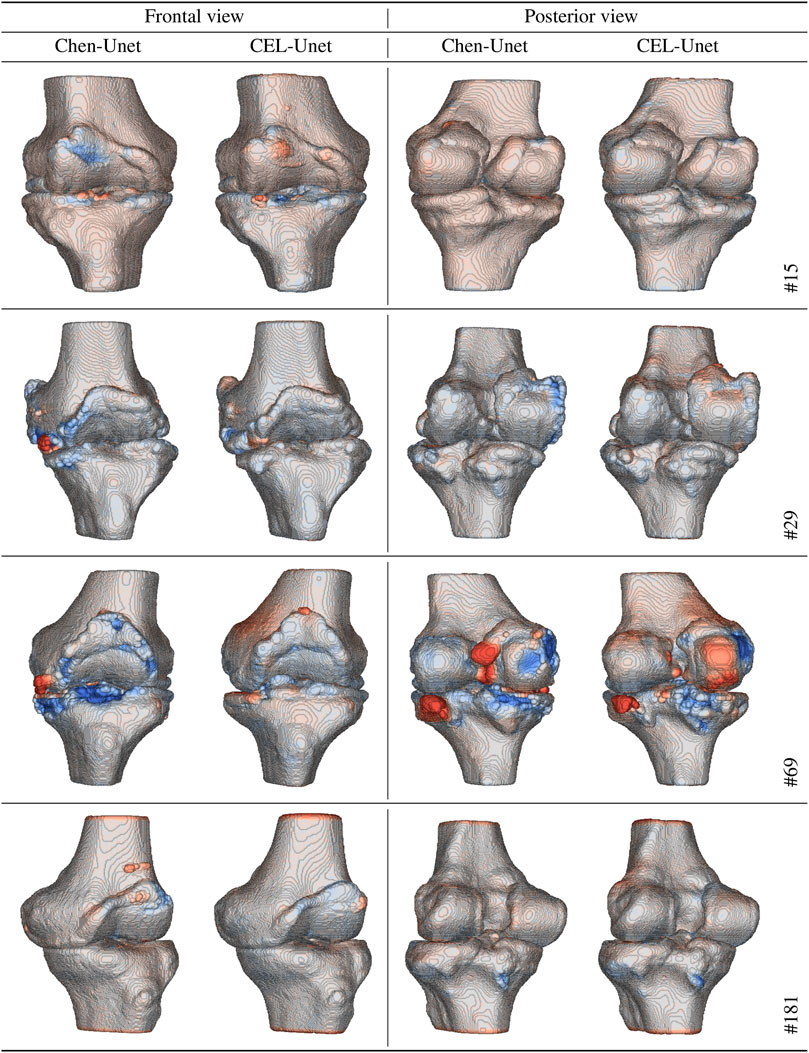
FIGURE 10. Reconstructed shapes (frontal and posterior views) with color distance map with respect to the reference shapes for four patients in the test set (#15, #29, #69, and #181). Red tones mean over-segmentation. Blue tones mean under-segmentation.
In order to study the segmentation accuracy at areas, strategic for PSI-based surgical planning, of the two shapes, a surface processing procedure was designed and developed to automatically split the two femur condylar regions and the tibial plateau in four parts, respectively. For the femur, lateral and medial parts in both posterior and anterior condylar areas were attained. Likewise for the tibia, lateral and medial parts in both posterior and anterior plateau areas were attained. Without lack of generality, in the analysis of local errors, D-Unet and F-Unet models were disregarded. Results showed similar accuracy across the four areas, for both femur (median range: 1 mm) and tibia (median range: 0.5 mm) as reported in Figure 11, being such a difference expected due to much relevant condylar deterioration at femur level. The computed alignment errors showed on average very high accuracy for both Chen-Unet and CEL-Unet (Table 7). All the median values were less than 0.15° for both femoral and tibial alignments. No statistical difference (p > 0.25) was found between the two models, although in few cases (e.g., #69) the frontal alignment errors in the Chen-Unet model for the femur and tibia were greater than 2°.
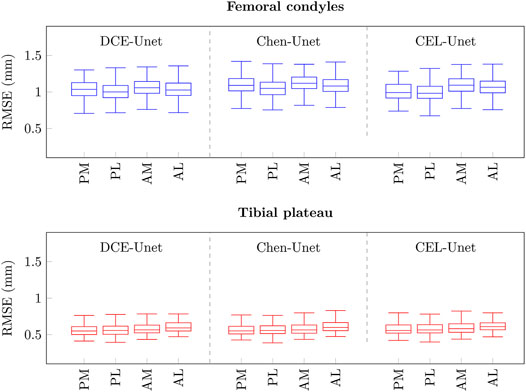
FIGURE 11. Boxplots of local RMS errors (mm) at femoral condyle (blue), and tibial plateau (red) areas. P: posterior, A: anterior, M: medial, L: lateral.

TABLE 7. Alignment errors, namely median (IQR) values, of the distal and proximal cuts, for femur and tibia respectively, obtained using reconstructed surface from Chen-Unet and CEL-Unet segmentations.
Automatic bone segmentation in CT scans is being seemingly acknowledged to pose fewer obstacles than the segmentation of other anatomical regions in different image modalities, whereby tissues feature very similar colorimetric and textural clues. Nevertheless, pathological conditions affecting mineral density, leading to cartilage damages and inducing bone deformations may reduce sensibly the accuracy causing both over- and under-segmentation (Chang et al., 2019; Yun et al., 2020). It is therefore required an extensive effort of manual refinement, performed by expert radiologists. In complicated cases, this activity may easily take more than half an hour (Ambellan et al., 2019; León-Muñoz et al., 2019). Thus, research is still ongoing towards the development of automatic and accurate techniques, especially in the domain of surgical planning. In PSI-based knee replacement surgery, the segmentation of femur and tibia bones is mandatory to obtain the 3D geometries, which are matched in the planning stage to the designed cutting masks. However, two main critical issues are to be acknowledged: 1) it is mandatory to ensure accuracy in specific bony regions in contact with the cutting mask, which usually are affected by the largest deformation and osteophytes; 2) cartilage wearing induces progressing thinning of the articular space between femur and tibia. In principle, addressing both issues may be conflicting. As pointed out, the success of such surgical technique demands for accurate segmentation of femoral condyles especially, coping with deformations and osteophytes. Over-segmentation may ensure to capture such specificity but may increase the confusion at the boundary between femur and tibia. Conversely, under-segmentation leaves the osseous boundaries less susceptible to mislabeling at risk however of reducing the quality of osteophyte segmentation. In this paper, we proposed a novel CNN network, called CEL-Unet, devoted to the segmentation of femur an tibia bones in CT scans, with specific focus on the balance between over- and under-segmentation. Results on an independent set of 200 cases showcased the excellent quality of the segmentation that was superior to state-of-the art Unet models (Chen et al., 2019; Marzorati et al., 2020; Isensee et al., 2021). Along with the traditional region-aware branch in the decoding path, the network exploited a contour-aware branch working in parallel, similar to the Chen-Unet model proposed in (Chen et al., 2019). Differently to such a model however, progressive skip connections between contour- and region-aware branches were introduced. In the Chen model, the information shared by the two tasks was only related to the encoder path. The CEL-Unet conversely shared information also in the decoding path. According to the results in Table 6, the segmentation accuracy of the femur was statistically in favour of the CEL-Unet, for the tibia no statistical difference was found between the two models though. Considering that the femur undergoes greater deformations than the tibia, this generally increases the complexity of its segmentation. The obtained results indirectly supported the view the vertical skip connections in the decoding path are useful for increasing the segmentation accuracy of the CEL-Unet. This effect looks likes more relevant in presence of larger deformation and osteophytes (condylar surfaces in the distal femur). Two additional technical innovations were adopted, namely the use of distance weight map in the loss function and the PEE module in the contour-aware branch, which was then concatenated to corresponding decoding level in the region-aware branch (cfr. Figure 1). The validity of such technical advancements was substantiated by quantitative (Table 2) and qualitative (Figure 7) analysis. It was shown that statistically CEL-Unet overcome all the competitive models. Specifically, CEL-Unet showed the ability to accurately segment critical cases where the traditional models had failed (cfr. Figure 7), unveiling ability to address narrow femur-patella interface and osteophytes with superior ability than the competitive Chen-Unet model (cfr. Table 6 and Figure 10). The clinical impact of the obtained segmentation highlighted the valuable span of the proposed work, proved by few tenths of a degree of error in the calculation of surgical cutting planes (cfr. Table 7). Segmentation analysis at femoral condyles and tibial plateau confirmed 3D errors compatible with accurate matching of the resection mask with the true anatomy.
In the latest few years, deep networks and Unet models for bone segmentation in X-Ray images, CT and MRI scans addressed different anatomical regions such as head bones, spine, pelvic bone, lower limb bones up to hand bones. With the aim of assisting surgical planning, deep CNN were proposed to segment skull surface in 20 CT scans reporting 92% of sensitivity and 3D reconstruction errors in the range of 1.5 mm (Minnema et al., 2018). 2D Unet for processing the three anatomical planes in cranio-facial CT was developed to segment mandibular bones reporting dice index of 93% and surface errors of 1.4 mm (Qiu et al., 2019). Focusing on the vertebral bodies, CT segmentation of 32 scans using deep CNN provided sensitivity of 97% and 3D surface errors of 7.4 mm (Vania et al., 2019). Pelvic bone segmentation in 30 dual energy CT scans was addressed by the traditional Unet achieving Dice index of about 96% (González Sánchez et al., 2020). 2D Unet model was demonstrated to obtain 94% of sensitivity to segment wrist and finger bones (Ding et al., 2019). 53 low-quality low-dose whole-body CT scans were segmented using a traditional Unet model leading to dice index of 95% (Klein et al., 2019). 2D Unet was applied to multi-label segmentation of 12 different structures in knee joint by processing 20 MRI scans achieving a mean Dice index for femur and tibia of about 90% (Zhou et al., 2018). Basically, despite all these results are in agreement with our achievements, we highlight that the large number of the samples available in this work, and the heterogeneity of the spanned pathological severity, makes the validation of the segmentation quality robust to a larger extent. Along with the attained quality, considering that the segmentation of one single volume takes less than 10 s, the computational time was compatible with clinical settings. In addition, in main literature works data were collected using a single CT scan system. In the present work, the CT images featured different pixel encoding and were acquired with four different scanners, namely Philips, Canon Medical Systems, GE Medical Systems and Toshiba. This variability showcased a larger generality of the obtained results.
In the available dataset, knee bones featured deformations and irregularities, particularly close to the intra-articular spaces. Also, conspicuous osteophytes were detected especially surrounding condylar and trochlear femur areas. As we have shown, traditional Dice and cross-entropy loss functions may fail providing poor segmentation. As far as the addressed clinical application is concerned, shape profiles are to be fully considered during preoperative planning and manufacturing of the cutting guides so as to ensure accurate anatomical matching of the PSI in the surgical setup. From the general observation that most segmentation errors are found along the boundaries of the anatomies (Kasten et al., 2020) and in order to cope with the specific clinical requirements, a new loss function was designed that specifically focused on edge voxels by exploiting the distance weighted map. The DWM took its root from the EDT, which assigns to each voxel the value of its distance from the closest voxel belonging to the boundary of the target structure. The basic idea was to use the DWM to constrain the cross-entropy to assign more importance to segmentation errors occurring at the boundaries voxels, while providing lower importance to the ones inside the shape profile. Nonetheless, DCE alone was proved to bias in some cases the segmentation leading to holes corresponding to misclassified voxels (cfr. Figure 7). For this reason, in the CEL-Unet, DCE was combined with the traditional dice coefficient as defined in Eq. 8 and its contribution was progressively elicited during training according to the factor α, which progressively focused the optimizer on shape edges. The heuristic trade-off (0.5) for α ensured the consistent balance between region and contour segmentation. As far as the network architecture is concerned, we developed an contour-aware decoding path, parallel to the region-aware decoding path, enabling directed vertical residual connections towards the region-detector path. This follows the approach pursued in (Zhou et al., 2020) focusing on enhanced skip connections to aggregate features of varying semantic scales at the decoder sub-networks. In this work, we extended such an approach by allowing pyramidal edge extraction (cfr. Figure 2) to enrich the edge details to be used in the corresponding decoding level in the region branch. In this study, we addressed the risk of overfitting two ways: 1) by hyper-parameter ablation (see par. 3.1); 2) by usage of a large and heterogeneous set of CT volumes of the knee. In addition, the training was automatically stopped as soon as the validation loss did no longer decrease with a patience factor equal to 25. The nice balance between recall and precision results (cfr. Figure 6) supported the expectation. Finally, a couple of issues may deserve attention. From a technical point of view, a batch size of two was chosen for the training in order to ensure computational feasibility with the allotted memory resources. Basically, this led to a an input data tensor of size 2 × 192 × 192 × 192×3, considering three classes namely the background, the femur and the tibia. Assuming 8 feature maps in the first convolutional layer, the memory allocation just for the first processing step was about 340 MB. The role of larger batches are planned for future analysis. From a clinical point of view, the work showcased the feasibility of automatic bone segmentation for knee surgical planning based on personalized instruments. A system like that is to be still through of as a support to the radiological analysis assuming that the physicians should provide a final refinement of the results before acceptance. Due to the use of the artificial intelligence paradigm, implementation in the surgical planning arena would require addressing current questions relevant to the reliability, interpretability and explainability of the results. This last point is recognized to be fundamental especially when the result of the segmentation is not satisfactory. Future activities are planned to address such issues with an approach the aims to embed into the network additional automatic tools able to provide information easily readable by the physician about the overall quality assurance of the results.
Knee replacement based on PSI has been very recently reported to improve functional kinematics with respect to traditional surgery, and it is increasingly recognized as a reliable technique to use in advanced osteoarthritis conditions especially in case of bone deformity, which can prevent the intra-medullar alignment. Nonetheless, digital shape of the bones is mandatory in the pre-operative planning phase, requiring CT/MRI scan acquisition and intensive manual delineation of the images. We have shown that the proposed network is effective for bone segmentation in knee CT scans. Overall, it delineates automatically femur and tibia profiles with high accuracy also in case of large pathological deformations and in presence of osteophytes. This makes it potentially usable for surgical planning with particular interest for knee surgery based on personalized surgical instruments where the reconstruction accuracy of the bony shapes is one of the main critical factors for the success of the operation. From the achieved outcomes, we point out that high-quality segmentation and automatism are both ensured which brings the use of image data tools, based on intelligent image processing tools, incrementally closer to clinical translation.
The datasets presented in this article are not readily available because The image dataset used in this work cannot be made publicly available. Requests to access the datasets should be directed to cGlldHJvLmNlcnZlcmlAcG9saW1pLml0.
The studies involving human participants were reviewed and approved by the MEDACTA INTERNATIONAL SPA. The patients/participants provided their written informed consent to participate in this study.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
MR and LM are supported by the PhD Program in Bioengineering at Politecnico di Milano, with funding by National Center of HadronTherapy in Oncology (Pave, Italy) and Medacta International (CH), respectively. PC and LM are supported by Regional Operative Program-European Fund for Regional Development 2014-2020-Lombardia Region, ARIA-2020-403, ID 138.360.852. This study received funding from MEDACTA INTERNATIONAL SPA (CH) and National Center of HadronTherapy in Oncology (IT). The funder MEDACTA had the following involvement with the study: providing the data. All authors declare no other competing interests.
This study received funding from MEDACTA INTERNATIONAL SPA (CH) and National Center of HadronTherapy in Oncology (IT). The funder MEDACTA had the following involvement with the study: providing the data. All authors declare no other competing interests.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors would like to thank Medacta International SA for providing images and clinical data.
Ambellan, F., Tack, A., Ehlke, M., and Zachow, S. (2019). Automated Segmentation of Knee Bone and Cartilage Combining Statistical Shape Knowledge and Convolutional Neural Networks: Data from the Osteoarthritis Initiative. Med. image Anal. 52, 109–118. doi:10.1016/j.media.2018.11.009
Anderl, W., Pauzenberger, L., Kölblinger, R., Kiesselbach, G., Brandl, G., Laky, B., et al. (2016). Patient-Specific Instrumentation Improved Mechanical Alignment, while Early Clinical Outcome Was Comparable to Conventional Instrumentation in Tka. Knee Surg. Sports Traumatol. Arthrosc. 24, 102–111. doi:10.1007/s00167-014-3345-2
Cerveri, P., Marchente, M., Bartels, W., Corten, K., Simon, J.-P., and Manzotti, A. (2010). Towards Automatic Computer-Aided Knee Surgery by Innovative Methods for Processing the Femur Surface Model. Int. J. Med. Robotics Comput. Assist. Surg. 6, 350–361. doi:10.1002/rcs.345
Cerveri, P., Sacco, C., Olgiati, G., Manzotti, A., and Baroni, G. (2017). 2d/3d Reconstruction of the Distal Femur Using Statistical Shape Models Addressing Personalized Surgical Instruments in Knee Arthroplasty: A Feasibility Analysis. Int. J. Med. Robotics Comput. Assist. Sur 13, e1823. doi:10.1002/rcs.1823
Chang, Y., Yuan, Y., Guo, C., Wang, Y., Cheng, Y., and Tamura, S. (2019). Accurate Pelvis and Femur Segmentation in Hip Ct with a Novel Patch-Based Refinement. IEEE J. Biomed. Health Inform. 23, 1192–1204. doi:10.1109/JBHI.2018.2834551
Chen, F., Liu, J., Zhao, Z., Zhu, M., and Liao, H. (2019). Three-Dimensional Feature-Enhanced Network for Automatic Femur Segmentation. IEEE J. Biomed. Health Inform. 23, 243–252. doi:10.1109/JBHI.2017.2785389
Dangi, S., Linte, C. A., and Yaniv, Z. (2019). A Distance Map Regularized Cnn for Cardiac Cine Mr Image Segmentation. Med. Phys. 46, 5637–5651. doi:10.1002/mp.13853
Ding, L., Zhao, K., Zhang, X., Wang, X., and Zhang, J. (2019). A Lightweight U-Net Architecture Multi-Scale Convolutional Network for Pediatric Hand Bone Segmentation in X-ray Image. IEEE Access 7, 68436–68445. doi:10.1109/ACCESS.2019.2918205
Falk, T., Mai, D., Bensch, R., Çiçek, Ö., Abdulkadir, A., Marrakchi, Y., et al. (2019). U-Net: Deep Learning for Cell Counting, Detection, and Morphometry. Nat. Methods 16, 67–70. doi:10.1038/s41592-018-0261-2
Gadosey, P. K., Li, Y., Agyekum, E. A., Zhang, T., Liu, Z., Yamak, P. T., et al. (2020). SD-UNet: Stripping Down U-Net for Segmentation of Biomedical Images on Platforms with Low Computational Budgets. Diagnostics 10, 110. doi:10.3390/diagnostics10020110
Gong, S., Xu, W., Wang, R., Wang, Z., Wang, B., Han, L., et al. (2019). Patient-Specific Instrumentation Improved Axial Alignment of the Femoral Component, Operative Time and Perioperative Blood Loss after Total Knee Arthroplasty. Knee Surg. Sports Traumatol. Arthrosc. 27, 1083–1095. doi:10.1007/s00167-018-5256-0
González Sánchez, J. C., Magnusson, M., Sandborg, M., Carlsson Tedgren, Å., and Malusek, A. (2020). Segmentation of Bones in Medical Dual-Energy Computed Tomography Volumes Using the 3d U-Net. Physica Med. 69, 241–247. doi:10.1016/j.ejmp.2019.12.014
Huang, Y. J., Dou, Q., Wang, Z. X., Liu, L. Z., Jin, Y., Li, C. F., et al. (2018). 3d Roi-Aware U-Net for Accurate and Efficient Colorectal Tumor Segmentation. arXiv preprint arXiv:1806.10342.
Isensee, F., Jaeger, P. F., Kohl, S. A. A., Petersen, J., and Maier-Hein, K. H. (2021). Nnu-Net: A Self-Configuring Method for Deep Learning-Based Biomedical Image Segmentation. Nat. Methods 18, 203–211. doi:10.1038/s41592-020-01008-z
Jin, Q., Meng, Z., Sun, C., Cui, H., and Su, R. (2020). RA-UNet: A Hybrid Deep Attention-Aware Network to Extract Liver and Tumor in CT Scans. Front. Bioeng. Biotechnol. 8, 605132. doi:10.3389/fbioe.2020.605132
Kasten, Y., Doktofsky, D., and Kovler, I. (2020). End-to-end Convolutional Neural Network for 3d Reconstruction of Knee Bones from Bi-planar X-ray Images. Cham: Springer, 123–133. doi:10.1007/978-3-030-61598-7_12
Klein, A., Warszawski, J., Hillengaß, J., and Maier-Hein, K. H. (2019). Automatic Bone Segmentation in Whole-Body Ct Images. Int. J. CARS 14, 21–29. doi:10.1007/s11548-018-1883-7
León-Muñoz, V. J., Martínez-Martínez, F., López-López, M., and Santonja-Medina, F. (2019). Patient-Specific Instrumentation in Total Knee Arthroplasty. Expert Rev. Med. devices 16, 555–567. doi:10.1080/17434440.2019.1627197
Li, W., Qin, S., Li, F., and Wang, L. (2020). MAD‐UNet: A Deep U‐Shaped Network Combined with an Attention Mechanism for Pancreas Segmentation in CT Images. Med. Phys. 48, 329–341. doi:10.1002/mp.14617
Litjens, G., Kooi, T., Bejnordi, B. E., Setio, A. A. A., Ciompi, F., Ghafoorian, M., et al. (2017). A Survey on Deep Learning in Medical Image Analysis. Med. image Anal. 42, 60–88. doi:10.1016/j.media.2017.07.005
Long, F. (2020). Microscopy Cell Nuclei Segmentation with Enhanced U-Net. BMC Bioinformatics 21, 8. doi:10.1186/s12859-019-3332-1
Ma, J., Wei, Z., Zhang, Y., Wang, Y., Lv, R., Zhu, C., et al. (2020). “How Distance Transform Maps Boost Segmentation Cnns: An Empirical Study,” in Proceedings of the Third Conference on Medical Imaging with Deep Learning (PMLR), Montreal, QC, July 6–8, 2020. Editors T. Arbel, I. Ben Ayed, M. de Bruijne, M. Descoteaux, H. Lombaert, and C. Pal, 121, 479–492.
Marzorati, D., Sarti, M., Mainardi, L., Manzotti, A., and Cerveri, P. (2020). Deep 3d Convolutional Networks to Segment Bones Affected by Severe Osteoarthritis in Ct Scans for Psi-Based Knee Surgical Planning. IEEE Access 8, 196394–196407. doi:10.1109/ACCESS.2020.3034418
McKinley, R., Wepfer, R., Aschwanden, F., Grunder, L., Muri, R., Rummel, C., et al. (2021). Simultaneous Lesion and Brain Segmentation in Multiple Sclerosis Using Deep Neural Networks. Sci. Rep. 11, 1087. doi:10.1038/s41598-020-79925-4
Minnema, J., van Eijnatten, M., Kouw, W., Diblen, F., Mendrik, A., and Wolff, J. (2018). Ct Image Segmentation of Bone for Medical Additive Manufacturing Using a Convolutional Neural Network. Comput. Biol. Med. 103, 130–139. doi:10.1016/j.compbiomed.2018.10.012
Noguchi, S., Nishio, M., Yakami, M., Nakagomi, K., and Togashi, K. (2020). Bone Segmentation on Whole-Body Ct Using Convolutional Neural Network with Novel Data Augmentation Techniques. Comput. Biol. Med. 121, 103767. doi:10.1016/j.compbiomed.2020.103767
Norman, B., Pedoia, V., and Majumdar, S. (2018). Use of 2d U-Net Convolutional Neural Networks for Automated Cartilage and Meniscus Segmentation of Knee Mr Imaging Data to Determine Relaxometry and Morphometry. Radiology 288, 177–185. doi:10.1148/radiol.2018172322
Ogura, T., Le, K., Merkely, G., Bryant, T., and Minas, T. (2019). A High Level of Satisfaction after Bicompartmental Individualized Knee Arthroplasty with Patient-Specific Implants and Instruments. Knee Surg. Sports Traumatol. Arthrosc. 27, 1487–1496. doi:10.1007/s00167-018-5155-4
Pietsch, M., Djahani, O., Hochegger, M., Plattner, F., and Hofmann, S. (2013). Patient-Specific Total Knee Arthroplasty: The Importance of Planning by the Surgeon. Knee Surg. Sports Traumatol. Arthrosc. 21, 2220–2226. doi:10.1007/s00167-013-2624-7
Qiu, B., Guo, J., Kraeima, J., Glas, H. H., Borra, R. J. H., Witjes, M. J. H., et al. (2019). Automatic Segmentation of the Mandible from Computed Tomography Scans for 3d Virtual Surgical Planning Using the Convolutional Neural Network. Phys. Med. Biol. 64, 175020. doi:10.1088/1361-6560/ab2c95
Ronneberger, O., Fischer, P., and Brox, T. (2015). “U-net: Convolutional Networks for Biomedical Image Segmentation,” in Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. Editors N Navab, J Hornegger, WM Wells, and AF Frangi (Cham: Springer International Publishing), 234–241. doi:10.1007/978-3-319-24574-4_28
Shi, T., Jiang, H., and Zheng, B. (2020). A Stacked Generalization U-Shape Network Based on Zoom Strategy and its Application in Biomedical Image Segmentation. Comput. Methods Programs Biomed. 197, 105678. doi:10.1016/j.cmpb.2020.105678
Shih, K.-S., Lin, C.-C., Lu, H.-L., Fu, Y.-C., Lin, C.-K., Li, S.-Y., et al. (2020). Patient-Specific Instrumentation Improves Functional Kinematics of Minimally-Invasive Total Knee Replacements as Revealed by Computerized 3d Fluoroscopy. Comput. Methods Programs Biomed. 188, 105250. doi:10.1016/j.cmpb.2019.105250
Vania, M., Mureja, D., and Lee, D. (2019). Automatic Spine Segmentation from Ct Images Using Convolutional Neural Network via Redundant Generation of Class Labels. J. Comput. Des. Eng. 6, 224–232. doi:10.1016/j.jcde.2018.05.002
Wang, R., Chen, S., Ji, C., Fan, J., and Li, Y. (2020). Boundary-aware Context Neural Network for Medical Image Segmentation. arXiv:2005.00966.
Yun, Y.-J., Ahn, B.-C., Kavitha, M. S., and Chien, S.-I. (2020). An Efficient Region Precise Thresholding and Direct Hough Transform in Femur and Femoral Neck Segmentation Using Pelvis Ct. IEEE Access 8, 110048–110058. doi:10.1109/ACCESS.2020.3001578
Zhou, Z., Siddiquee, M. M. R., Tajbakhsh, N., and Liang, J. (2020). Unet++: Redesigning Skip Connections to Exploit Multiscale Features in Image Segmentation. IEEE Trans. Med. Imaging 39, 1856–1867. doi:10.1109/TMI.2019.2959609
Keywords: deep convolutional neural networks, UNET, pyramidal edge extraction, bone segmentation, osteophytes
Citation: Rossi M, Marsilio L, Mainardi L, Manzotti A and Cerveri P (2022) CEL-Unet: Distance Weighted Maps and Multi-Scale Pyramidal Edge Extraction for Accurate Osteoarthritic Bone Segmentation in CT Scans. Front. Sig. Proc. 2:857313. doi: 10.3389/frsip.2022.857313
Received: 18 January 2022; Accepted: 22 February 2022;
Published: 05 April 2022.
Edited by:
Stavros Karkanis, University of Thessaly, GreeceReviewed by:
Michalis Savelonas, University of Thessaly, GreeceCopyright © 2022 Rossi, Marsilio, Mainardi, Manzotti and Cerveri. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pietro Cerveri, cGlldHJvLmNlcnZlcmlAcG9saW1pLml0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.