- 1Shaanxi Joint Laboratory of Artificial Intelligence, Shaanxi University of Science and Technology, Xi’an, China
- 2The School of Electronic Information and Artificial Intelligence, Shaanxi University of Science and Technology, Xi’an, China
- 3The School of Electronic and Information Engineering, Lanzhou Jiaotong University, Lanzhou, China
Medical image segmentation plays an important role in clinical applications, such as disease diagnosis and treatment planning. On the premise of ensuring segmentation accuracy, segmentation speed is also an important factor to improve diagnosis efficiency. Many medical image segmentation models based on deep learning can improve the segmentation accuracy, but ignore the model complexity and inference speed resulting in the failure of meeting the high real-time requirements of clinical applications. To address this problem, an asymmetric lightweight medical image segmentation network, namely AL-Net for short, is proposed in this paper. Firstly, AL-Net employs the pre-training RepVGG-A1 to extract rich semantic features, and reduces the channel processing to ensure the lower model complexity. Secondly, AL-Net introduces the lightweight atrous spatial pyramid pooling module as the context extractor, and combines the attention mechanism to capture the context information. Thirdly, a novel asymmetric decoder is proposed and introduced into AL-Net, which not only effectively eliminates redundant features, but also makes use of low-level features of images to improve the performance of AL-Net. Finally, the reparameterization technology is utilized in the inference stage, which effectively reduces the parameters of AL-Net and improves the inference speed of AL-Net without reducing the segmentation accuracy. The experimental results on retinal vessel, cell contour, and skin lesions segmentation datasets show that AL-Net is superior to the state-of-the-art models in terms of accuracy, parameters and inference speed.
1 Introduction
Medical image segmentation refers to the process of dividing medical images into several non-overlapping regions according to some similarity characteristics of medical images. Medical image segmentation is of great significance for understanding the content of medical images and discovering lesion objects. It is not only the basis of biomedical image analysis, such as medical image registration and 3D reconstruction, but also plays an extremely important role in clinical diagnosis and treatment.
In recent years, with the development of deep learning, medical image segmentation based on deep learning has made remarkable progress and become a hot topic in the field of medical image analysis. Many classical semantic segmentation models (Liu et al., 2015; Ronneberger et al., 2015; Shelhamer et al., 2015; Chen et al., 2017; Chen et al., 2018; Gu et al., 2019; Zhou et al., 2019; Zhou et al., 2020; Chen et al., 2021) usually adopt the idea of extracting pixel-level features, such as the end-to-end fully convolutional network (FCN) (Shelhamer et al., 2015) and U-shape Net (U-Net) (Ronneberger et al., 2015). The above two types of segmentation models are mainly composed of encoder and decoder. Meanwhile, more and more contextual feature extraction modules are also employed in medical image segmentation. Firstly, Medical image segmentation models usually employ the most popular feature extractors (Shelhamer et al., 2015); (Gu et al., 2019); (Simonyan and Zisserman, 2014); (He et al., 2016); (Valanarasu et al., 2021) as encoders, such as VGG and ResNet, but the improvement of segmentation accuracy usually leads to the increase of model complexity. Secondly, context information is indispensable for image feature extraction. At present, most prominent semantic feature extraction modules are implemented based on dilated convolution (Chen et al., 2018); (Gu et al., 2019) and multi-scale pooling (Liu et al., 2015); (Gu et al., 2019); (Jie et al., 2018). In order to effectively focus on semantic features, attention mechanism is widely used to extract semantic information (Li et al., 2019); (Ni et al., 2019); (Le et al., 2020). Thirdly, medical image segmentation models are mostly improved on the basis of U-Net (Ronneberger et al., 2015); (Zhou et al., 2020). U-Net uses skip connection to effectively supplement low-level features, but it leads to information redundancy. In addition, on the basis of ensuring the segmentation accuracy, the segmentation speed is an important factor in applying the medical image segmentation model to clinical treatment. However, these models ignore the inference speed and model complexity to pursue the segmentation accuracy, they are not suitable for some clinical applications, such as image-guided surgery, online adaptive radiotherapy and real-time disease monitoring, which have high real-time requirements for image segmentation task.
To solve the above problems, we propose a lightweight asymmetric medical image segmentation network, namely AL-Net for short. Our main contributions are summarized as follows.
1) We introduce RepVGG-A1 as the encoder of AL-Net to extract powerful semantic features, and select Lite R-ASPP as the context information extraction module to ensure that the model can effectively capture the context features and has smaller parameters and lower model complexity.
2) We design an asymmetric decoder using skip connection and convolution operation for medical image segmentation. This decoder not only fully integrate the low-level features of images, but also eliminate the feature redundancy to further improve the segmentation accuracy.
3) We integrate the re-parameterization technology in the inference stage of AL-Net. Therefore, the inference model of AL-Net has only 3.45 M parameters, and achieves the best balance between speed and accuracy on the dataset of retinal vessel, cell contour and skin image, respectively, which is better than the existing models.
The structure of the remainder of this paper is organized as follows. Section 2 introduces the related work of this paper. Section 3 mainly describes the AL-Net in detail. Section 4 demonstrates the performance of AL-Net. Finally, Section 5 makes the conclusion for this paper.
2 Related Work
In recent years, medical image segmentation based on deep learning has made great progress. In this section, we mainly introduce three general components of medical image segmentation network and the popular lightweight architecture.
2.1 General Components of Network
Medical image segmentation network usually includes encoder, decoder and context extraction module. In this section, we discuss these modules in detail.
Encoder: The semantic segmentation model based on deep learning (Szegedy et al., 2016a; Le et al., 2019; Jns et al., 2020) uses the encoder to extract high-level semantic information. U-Net selects the most powerful convolutional neural network VGG (Simonyan and Zisserman, 2014) as the encoder to capture high-level semantic information, but VGG limits the richness of image features due to the simple structure (Ronneberger et al., 2015). Because more and more powerful convolutional neural networks are proposed, the medical image segmentation network can choose a more advanced convolutional neural network as the backbone to extract more abundant image features (Gu et al., 2019); (Zhou et al., 2020); (Chen et al., 2021). For example, Context Encoder Network (CE-Net) (Gu et al., 2019) selects ResNet-34 (He et al., 2016) as the encoder, because the parameters of ResNet-34 are moderate and gradient dispersion can be avoided through residual connection. The backbone of U-Net++ (Zhou et al., 2020) is ResNet-101 with deeper network layers. TransUNet (Chen et al., 2021) added transformers to the encoder to extract more advanced features. However, these networks have huge structures and many parameters, which makes the model training and inference process consume a long time and computational resources. In order to further reduce the training and inference time of the network within limited computing resources, a series of lightweight backbones have emerged, such as Inception (Szegedy et al., 2016a); (Szegedy et al., 2015); (Szegedy et al., 2016b); (Sergey IoffeSzegedy, 2015), DenseNet (Huang et al., 2017) and RefineNet (Nekrasov et al., 2018); (Lin et al., 2017). Although the lightweight structure of the network is realized using these lightweight backbones, the segmentation accuracy has made an unexpected sacrifice. Re-parameterization technology can effectively avoid the contradiction between model lightweight and segmentation accuracy. Recently, Ding et al. (Ding et al., 2021) use re-parameterization technology to realize multi-branch training and single branch inference, which opens up another way for the selection of encoder.
Decoder: The decoder is used to recover the spatial information of images step by step, but the earliest decoder only performs up-sampling, which will lead to the inability to recover the spatial information of images. Then, U-Net (Ronneberger et al., 2015) proposes a U-shaped decoder, which is composed of up-sampling and skip connection to supplement the detailed information lost in the encoder stage. However, the simple connection is easy to cause the loss of important semantic information in the process of high-level and low-level semantic information fusion. To solve this problem, scholars have proposed a variety of decoders to improve feature fusion (Ibtehaz and Rahman, 2020); (Alom et al., 2019); (Zheng et al., 2020). Nabil et al. (Ibtehaz and Rahman, 2020) used the residual path to replace the skip connection of the U-shaped decoder in the decoder of multiResUnet, so as to eliminate the semantic difference caused by the fusion of the low-level features of the encoder and the high-level features of the decoder. Zhou et al. (2020) improved the decoding ways of U-Net and proposed U-Net++ with nested dense skip connection path with deep monitoring. Alom et al. (2019) added a dual attention mechanism composed of spatial and channel attention in the last two layers of the decoder. Zheng et al. (2020) applied transformer to image segmentation and designed three different decoders based on the output serialization characteristics of transformer. However, these works only focus on improving the segmentation accuracy, ignoring the issue that many branch structures lead to a significantly slow inference speed.
Context extraction module: To maintain the semantic information extracted in the encoding stage, many modules for extracting image context information are proposed. ParseNet fuses global context information from image level to solve the problem of insufficient actual receptive field (Liu et al., 2015). DeepLabv2 proposes the atrous spatial pyramid pooling (ASPP) module to effectively capture contextual features by expanding receptive fields (Chen et al., 2018). DeepLabv3 combines image level information and employs parallel atrous convolution layers with different dilated rates to capture multi-scale information (Chen et al., 2017). Nekrasov et al. (Nekrasov et al., 2018) designed the chained residual pooling (CRP) module and used it to capture context features from high-resolution images and improve the performance of semantic segmentation. To solve the problem of object size change in image segmentation, Gu et al. (Gu et al., 2019) proposed dense atrous convolution (DAC) module and residual multi-kernel pooling (RMP) module, which rely on the effective receptive fields to detect objects with different sizes. However, most of these modules only retain context information. For medical images with complex background, it is of great significance to focus the objects with sufficient context information. Hu et al. (Jie et al., 2018) proposed the squeeze and excitation (SE) module, which can automatically improve the useful features according to the importance and suppress the features that contribute less to the current task, so as to enhance the features and improve the segmentation performance.
2.2 Lightweight Segmentation
In recent years, the lightweight design of semantic segmentation network has gradually become a hot topic of image segmentation task, which has attracted the attention of many scholars. SegNAS3D (Wong and Moradi, 2019) uses network architecture search to solve the problem of network structure optimization in 3D image segmentation, which greatly reduces the complexity of model. In order to pursue the real-time performance of the model, Nekrasov et al. (2018) employed the lightweight RefineNet (Lin et al., 2017) as the backbone network. ICNet (Zhao et al., 2018) uses image cascade and branch training to accelerate the convergence of model. BiSeNet (Yu et al., 2018); (Yu et al., 2021) realized a lightweight model based on double branch structure, which uses different paths to extract spatial and semantic information. In addition, other models use common components to reduce the amount of computation. For example, DMFNet (Chen et al., 2019) divides the channels into multiple groups, and introduces weighted three-dimensional extended convolution to reduce parameters and improve the inference efficiency of model. Xception (Chollet, 2017) and MobileNets (Howard et al., 2017); (Sandler et al., 2018) employed deep separable convolution to effectively improve the inference speed. Dense-Inception U-Net (Zhang et al., 2020) combines the lightweight backbone Inception and dense module to extract high-level semantic information with lightweight encoder. ShuffleNets (Ma et al., 2018); (Zhang et al., 2018) proposed group convolution and channel shuffling, which greatly reduced the computational cost compared with the advanced models. However, lightweight segmentation networks for relatively complex medical images are less than them for natural images.
Some scholars have also designed lightweight segmentation networks for medical images. However, on the premise of ensuring accuracy, there are few medical image segmentation networks that achieve both low complexity and high inference speed. nnU-Net (Isensee et al., 2020) improves the adaptability of the network by preprocessing the data and post-processing the segmentation results, but comes at cost of increasing model parameters. U-Net++ (Zhou et al., 2020) uses small parameters to achieve good segmentation accuracy, but ignores the inference time of the model. And lightweight V-Net (Lei et al., 2020) guarantees segmentation accuracy and fewer parameters by employing depth-wise convolution and point-wise convolution, but does not improve the inference time of the model. In addition, Tarasiewicz et al. (2021) trained multiple skinny networks over all image planes and proposed Lightweight U-Nets, which obtains accurate brain tumor delineation from multi-modal MRIs. PyConvU-Net (Li et al., 2021) replaces all conventional convolution layers in the U-Net with the pyramidal convolution, which makes the segmentation accuracy better and the parameters less. However, the inference speed of PyConvU-Net still needs to be improved.
3 Asymmetric Lightweight Network
To ensure the segmentation accuracy and improve the segmentation speed, we proposed an asymmetric lightweight network for medical image segmentation. Figure 1 shows the network structure of the proposed AL-Net. In Figure 1, AL-Net consists of three important components, which are encoder, semantic extraction module and decoder. Compared to other classical models, the encoder of AL-Net does not involve residual connection, which makes AL-Net occupy less memory in the training stage. Meanwhile, AL-Net improves the ability of feature representation and generalization by designing multi-branch parallel structure in each convolution layer. The semantic extraction module is usually a pyramid structure, and the semantic extraction module employed by AL-Net are LR-ASPP (Howard et al., 2019). Compared with other pyramid modules, it can not only capture different-sized objects by the multi-core effective receptive field, but also combine the attention mechanism to more effectively deal with the low-contrast medical image segmentation task. In addition, LR-ASPP also uses a large pooling kernel with a large stride and only one 1 × 1 convolution, which save some computation and make AL-Net more lightweight. In order to effectively map the low-resolution features of the encoding stage to the pixel-level classification features with the original resolution in the decoding stage, we are inspired by the DeepLabV3+ and U-Net, and design a decoder with asymmetric structure and apply it to AL-Net, which is more suitable for medical image segmentation. This decoder uses 3 × 3 convolutions on the basis of U-shaped decoder instead of using skip connections directly, which can not only fully integrate the low-level semantic information, but also effectively eliminate the redundant features of images. It is more conducive to extract accurately object contours of medical images. In addition, in the inference stage of AL-Net, we employ the re-parameterization technology and optimize the multi-branch structure of encoder to single-branch structure, which improves the inference speed of AL-Net.
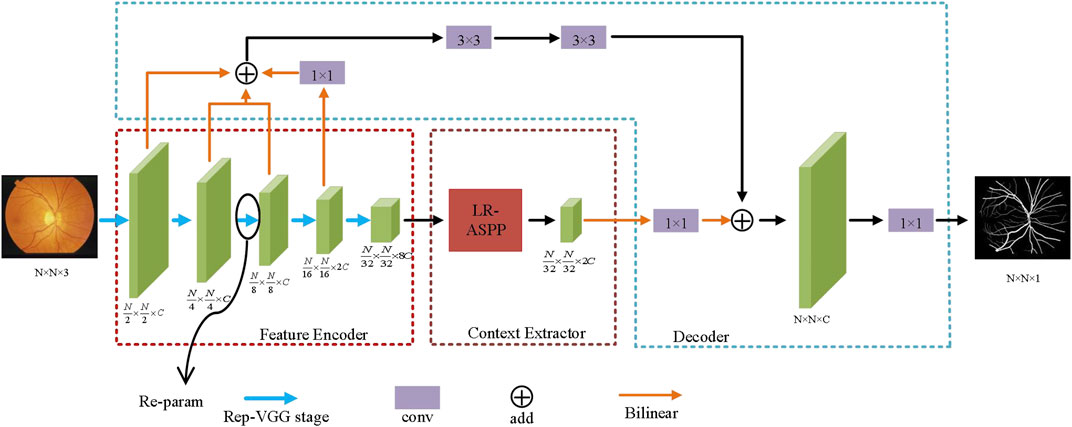
FIGURE 1. The structure of AL-Net. After the training of AL-Net, each RepVGG block in the encoder is processed by reparameterization technology, so as to improve the inference efficiency of AL-Net.
3.1 Encoder Module
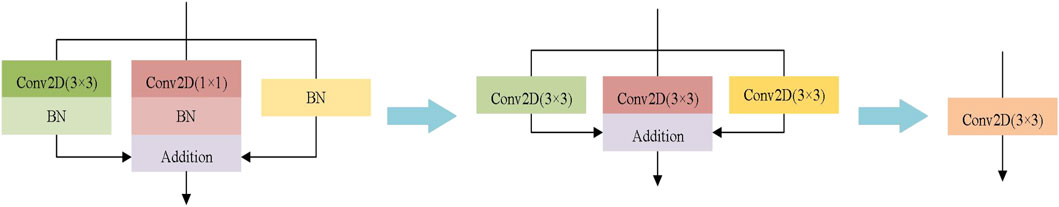
The encoder plays an important role in image segmentation and feature extraction. The early medical image segmentation network, such as U-Net, chose VGG as encoder, which is always composed of convolution, ReLU and pooling. With the development of deep learning technology, the encoders of medical image segmentation network usually choose better modules, such as Inception, ResNet and DenseNet, which make the medical image segmentation model more and more complex. Although complex models may have higher accuracy than simple models, the multi-branch structure of complex models makes the model difficult to implement, and increases the inference time and memory utilization. In order to guarantee the segmentation accuracy and reduce the model complexity, the encoder of AL-Net employs the RepVGG-A1, which has the same ability of feature representation as Res-Net34 but has fewer parameters. RepVGG block is shown in Figure 2. The RepVGG-A1 is designed for the image classification task of ImageNet, and there are few classes for medical image segmentation task. Therefore, there is channel redundancy when RepVGG-A1 is utilized as the backbone of AL-Net. We decrease the channels of the backbone of AL-Net without significantly reducing performance. Specially, the stride convolution is used by the RepVGG to replace the pooling operation, which avoids the possibility of losing the spatial information of images. In addition, the RepVGG can employ re-parameterization technology to transform multi-branch structure into single-branch structure to improve the inference speed effectively.
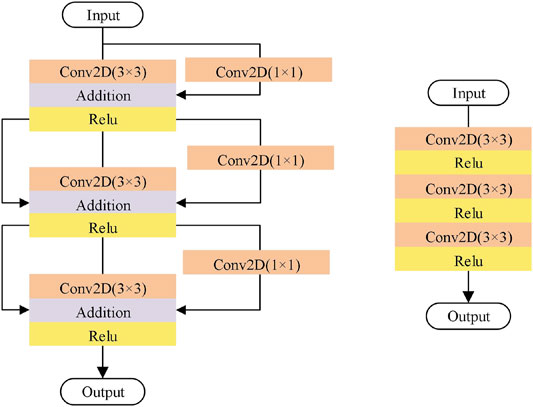
FIGURE 2. RepVGG block. RepVGG-A1 is divided into five stages, and the number of the layers of each stage are 1, 2, 4, 14 and 1, respectively. Training stage (Left): in the first layer of each stage, down sampling is carried out through convolution with step size of 2, and there is no identity branch. Inference stage (Right): AL-Net becomes a single branch after reparameterization. Only the three layers of one stage are shown here.
3.2 Context Extractor Module
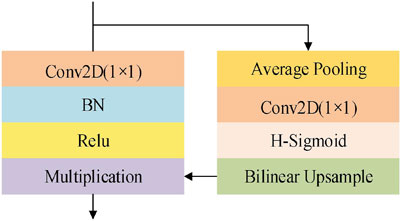
The context extraction module is used to extract contextual semantic information and generate more high-level feature maps. Currently popular context extraction modules, such as ASPP, can enrich spatial information, but do not have a specific direction of feature response. Medical images have the characteristics of high complexity, lack of simple linear features, and the gray of the background and objects are similar. Therefore, compared with natural images, it is more necessary for medical images to optimize the channel dimension. In this paper, the Lite R-ASPP firstly discards the atrous convolution that spends a lot of computational cost. Then, Lite R-ASPP can realize the information integration between channels by simplifying the four branches into two branches. Lite R-ASPP employs the global average pooling to prevent over fitting by regularizing the structure of the whole network. The global context information and semantic features are extracted by global average pooling to realize the global distribution on the feature channel. The feature is compressed into an attention vector. Finally, hard sigmoid with faster computational speed is employed as the activation function to realize the weight normalization, so as to recalibrate the semantic dependencies of the original features in the channel dimension, and highlight the key features and filter the background information. In addition, Lite R-ASPP uses only 1 × 1 convolution, which effectively reduces the parameters. Lite R-ASPP is shown in Figure 3.
3.3 Decoder Module
In the popular architecture of image segmentation networks, the decoder only has a simple upsampling process, which may lead to the loss of spatial information. To address this issue, U-Net uses skip connection to fuse the feature maps in the encoding and decoding stage to obtain richer spatial information. However, this connection mode produces many redundant low-level features. To solve this problem, we design an asymmetric decoder, namely A-Decoder. Firstly, A-Decoder still fully integrates the low-level features in the encoding stage to supplement the high-resolution information and recover more edge information of objects in medical images. However, instead of using skip connection for symmetrical structure directly, 3 × 3 convolution is used after fusing low-level features, which effectively reduces redundant features. Then, it is fused with the high-level features from the context extraction module to refine the contour information of objects. Finally, A-Decoder apply a 1 × 1 convolution to reduce the number of channels. A-Decoder is shown in Figure 1. In addition, A-Decoder can effectively reduce the fusion times of skip connections and supplement the same amount of spatial information with less computation. A-Decoder can be expressed as:
Where
3.4 Loss Function
We utilize loss function to supervise the training process of AL-Net. Binary cross entropy is defined as a measure of the difference between two probability distributions for a given random variable or set of events. It is widely used in classification and segmentation tasks, and segmentation is a kind of pixel-level classification. Therefore, binary cross entropy loss function works well in the segmentation tasks. In addition, Dice coefficient is an ensemble similarity measure, which usually used to calculate the similarity of two samples. Dice coefficient can maximize the segmentation objects, thus preventing the learning process from falling into the local minimum. To effectively segment objects in medical images, AL-Net employs a composite loss function that combines Dice coefficient and binary cross entropy. The loss function employed by AL-Net is shown in Equation (4):
Where,
Where,
3.5 Re-Parameterization
AL-Net is a convolutional neural network with multi-branch structure. This structure is beneficial to model training and improve the segmentation accuracy, but it will lead to a long inference time. In the inference stage, reparameterization technology can couple multiple branches into a single branch, which can speed up the inference procedure of AL-Net without sacrificing accuracy. Generally, the encoder has the greatest impact on the performance of image segmentation model. The encoder of AL-Net consists of three branches: identity branch, 1 × 1 convolution and 3 × 3 convolution. In the procedure of reparameterization, the identity branch can be regarded as a degenerative 1 × 1 convolution and 1 × 1 convolution can be regarded as a degenerative 3 × 3 convolution. Thus, 3 × 3 convolution, 1 × 1 convolution and the batch standardization layer in the training model can be reconstructed into a 3 × 3 convolution in the inference model. After reparameterization, the encoder of AL-Net becomes a single branch structure with only 3 × 3 convolution layer. The procedure of reparameterization for each block is shown in Figure 4. The reparameterized AL-Net can effectively reduce the amount of parameters and shorten the segmentation time.
4 Experiments
In this section, we first introduce the datasets, experimental setup and evaluation criteria. Then, the ablation studies for AL-Net are carried out on the cell contour segmentation datasets. Finally, AL-Net is compared with other state-of-the-art segmentation models in terms of parameters, segmentation speed and accuracy, and the results on three datasets of retinal vessels, cell contour and skin lesions are shown and discussed.
4.1 Datasets
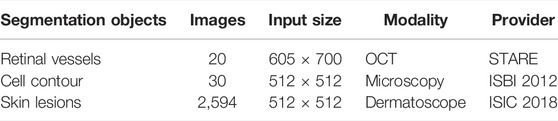
In order to evaluate the performance of AL-Net, we conducted segmentation experiments on three medical image datasets, which are shown in Table 1. These datasets are derived from the most common medical imaging modalities, including microscopy, dermatoscope and optical coherence tomography.
1) Retinal vessels. This dataset is a color fundus image dataset for retinal vessel segmentation (Hoover and Goldbaum, 2003), which includes ten lesion images and ten healthy images. The size of each image is 605 × 700.
2) Cell contour. This dataset is obtained by transmission electron microscopy from the serial segment of the ventral nerve zone of Drosophila melanogaster, with a total of 30 images. Each image has complete cell and membrane labels, and the size of each image is 512 × 512 (Cardona et al., 2010).
3) Skin lesions. This dataset is provided by ISIC 2018 (Tschandl et al., 2018); (Allan, 2019) for melanoma detection, including 2,594 images of skin lesion. The size of each image is 2,166 × 3,188, which is resampled to 512 × 512 in this experiment.
Two steps of data augmentation are carried out for these datasets to avoid the risk of over fitting caused by little data. Firstly, each image is expanded to eight times of the original image by turning horizontally, vertically and diagonally, respectively. Then, each image is translated up, down, left and right, respectively. The above data augmentation methods cannot change the data distribution, meanwhile can avoid over-fitting in the training process and effectively improve the generalization ability of the model.
The division of training dataset and test dataset of cell contour and skin lesions segmentation dataset is consistent with the official description. Retinal vessel segmentation dataset is randomly divided into the training dataset and test dataset according to 7:3. The training dataset and test dataset are separately expanded using the data augmentation methods described above. Then, the validation dataset is divided from the training dataset, and the proportion of training dataset, validation dataset and test dataset is 5:2:3. In the experiment, the training dataset is used for model training, the test dataset is used to evaluate the model, and the validation dataset is used to evaluate the model performance in the process of model training to obtain the best model.
4.2 Experimental Setup
All models involved in the experiment are implemented on the cloud service platform, which is equipped with NVIDIA Tesla P100 GPU with 16 GB memory. PyTorch is chosen as the framework of deep learning. In AL-Net, Adam is used as the optimizer. The initial learning rate is set to 5e-3 and the batch size is set to 4. In addition, we use the automatic attenuation strategy of the learning rate, where the step size is 1 and the attenuation factor
4.3 Evaluation Measures
We evaluate the performance of AL-Net from three aspects: model complexity, inference speed and segmentation accuracy. The model complexity is measured by the parameters of the model, and the inference speed is evaluated by the inference time of the model for a single image. For the sake of fairness, the inference time is the average time of performing ten segmentation processes for each sample after hardware preheating. We choose the accuracy (Acc) and intersection union ratio (IoU) to evaluate the segmentation accuracy of all models.
Accuracy refers to the ratio of object results in all prediction results, which is shown in Eq. 7:
IoU represents the similarity or overlap between the predicted object and the ground truth, which is computed as follows:
Where
4.4 Ablation Study
In this paper, we design a lightweight segmentation network for medical images on the premise of ensuring the segmentation accuracy, which mainly includes three contributions. First, RepVGG-A1, which realizes lightweight design using the re-parameterization technology, is selected as the backbone of AL-Net. Secondly, we integrate LR-ASPP which is a lightweight context information extraction module into AL-Net. Thirdly, we design a decoder with asymmetric structure in term of the characters of medical images. To validate the efficiency of three contributions, we conduct the ablation study on the test dataset of cell contour images.
4.4.1 Ablation Study for RepVGG-A1
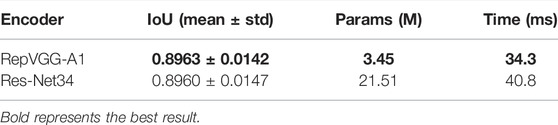
Encoder plays an important role in extracting features for the image segmentation model. High performance encoder is of great significance to image segmentation model. Since we choose RepVGG-A1 as the encoder of AL-Net, we compare it with Res-Net34 to validate the performance of RepVGG-A1. RepVGG-A1 and Res-Net34 are respectively used as encoders of AL-Net. The results of segmentation accuracy and speed are shown in Table 2.
In Table 2, the IoU value of these models is similar, but the parameters of AL-Net using RepVGG-A1 in the inference stage is 3.45 M, which is only 1/6 of AL-Net using Res-Net34. Moreover, the inference time of AL-Net using RepVGG-A1 is 34.3 ms, which is faster than that of AL-Net using Res-Net34. To sum up, compared with Res-Net34, our encoder has higher accuracy, faster speed and smaller parameters.
4.4.2 Ablation Study for LR-ASPP Block
The context extraction module is an important component to enhance the ability of the feature representation of the model. We employed LR-ASPP as the semantic feature extractor of AL-Net and compared it with the classical context extraction module, such as ASPP. The results are shown in Table 3.
In Table 3, the parameters of AL-Net using LR-ASPP is only 3.45 M, which is nearly 1/3 less than that of AL-Net using ASPP. Compared to AL-Net using ASPP, the inference time for each image is also shortened from 42.4 to 34.3 ms, and the segmentation accuracy is effectively improved. The main reason is that LR-ASPP has fewer branches and combines the attention mechanism, which is more suitable for medical images than ASPP.
4.4.3 Ablation Study for A-Decoder
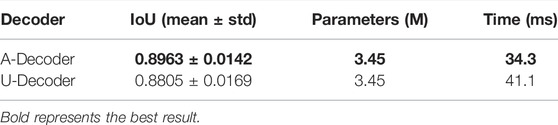
In order to improve the performance of AL-Net, we also designed an asymmetric decoder A-Decoder, which is compared with the popular symmetric U-Decoder. The results are shown in Table 4.
It can be seen from Table 4 that the inference speed of the AL-Net with A-Decoder is 34.3 ms, which is 16.5% faster than that of the model with U-Decoder, and the IoU is increased from 0.8805 to 0.8963. This is due to the fact that A-Decoder not only retains sufficient low-level semantic information with less feature fusion, but also skillfully employs 3 × 3 convolution and refines low-level features to improve performance of the model.
4.5 Comparison With Other Methods
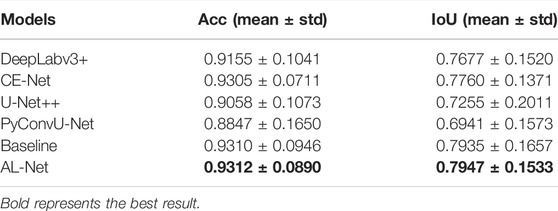
In this section, we compare AL-Net with three semantic segmentation models with excellent performance in recent years (DeepLabv3+, CE-Net, PyConvU-Net and U-Net++) and its baseline (replacing the encoder of U-Net with RepVGG-A1) in terms of speed and accuracy. We analyze the parameters and speed of these models, and report the experimental results in terms of their accuracy.
4.5.1 Inference Speed
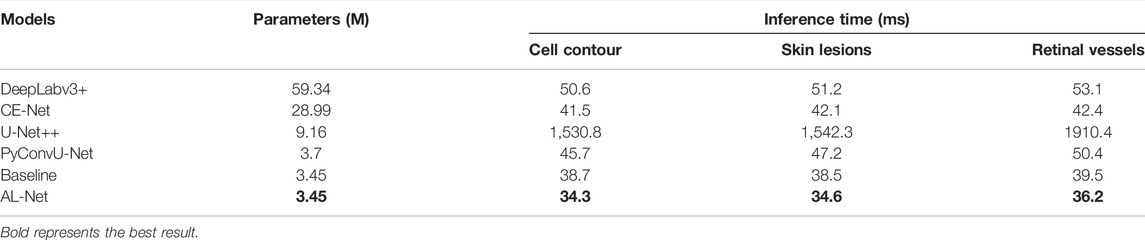
Inference speed is an important basis for evaluating the performance of medical image segmentation model, especially in practical application. We use the above six models to test the inference speed on the datasets of retinal vessel, cell contour and skin lesions segmentation, respectively. For the sake of fairness, all experiments are conducted in the same environment, and we record the average inference time of ten executions. The experiment results are shown in Table 5.
In Table 5, the parameters of AL-Net is only 3.45 M, which is equivalent to baseline and much smaller than other models. The parameters of AL-Net is only 1/25 of that of DeepLabv3+, which is 5.71 M less than that of U-Net++. In addition, the inference time of AL-Net for input images with different sizes is significantly shorter than that of other models. For the input image with size 512 × 512 on the cell contour dataset, the inference time of AL-Net is 34.3 ms, which is 44.6 times shorter than U-Net++. For the skin lesions dataset, the inference time of AL-Net is 34.6 ms, which is 1.5 times faster than DeepLabv3+. For each image with size 700 × 605 on the retinal vessel dataset, the inference time of AL-Net is 36.2 ms, which is shorter than other models. Because U-Net++ has only 9.16 M parameters but many branches, which reduces the parallelism of the model, the inference time of U-Net++ is much longer than other models. However, AL-Net achieves the high inference speed due to the single branch structure in the inference stage. AL-Net is also faster than DeepLabv3+ and CE-Net. There are two main reasons. One is that the decoder of AL-Net has a simple single branch structure in the inference stage, and the other is that the encoder of AL-Net has fewer fusion operations, which saves a lot of time.
4.5.2 Accuracy Analysis
In this section, we will show some visualization examples and experimental results of segmentation accuracy on retinal vessels, cell contours and skin injury segmentation datasets.
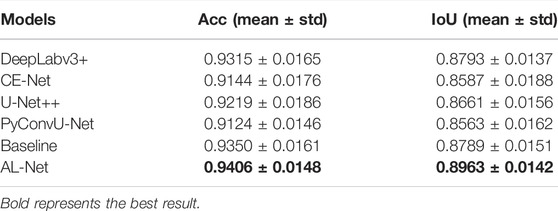
Retinal vessels: the segmentation results on the retinal vessel dataset are shown in Figure 5. In Figure 5, the segmentation results of AL-Net are closest to the ground truth, and there are inaccurate segmentation boundaries in the segmentation results of other models. For example, the segmentation results of DeepLabv3+ are the coarsest and cannot interpret the details of retinal vessels. U-Net++ cannot completely segment the ends of blood vessels. Baseline and CE-Net lead to over segmentation and incorrectly segment objects from background. The accuracy evaluation results of the above six models on the retinal vessels dataset are shown in Table 6. The Acc and IoU of AL-Net is 0.9728 and 0.6851, respectively, which is better than other models. In conclusion, the performance of AL-Net for retinal vessels segmentation is significantly better than other models.
In the retinal vessels dataset, AL-Net first benefits from the semantic extraction module, which combines the channel attention module, so that AL-Net can not only capture the high-level context information, but also optimize the channel dimension. Secondly, compared with other models, the advantage of AL-Net comes from the decoder. There is only one layer of low-level information combined with DeepLabv3+, which is far from enough for medical images. Baseline, U-Net++, PyConvU-Net and CE-Net simply transmit low-level features to the decoder through skip connection. However, the decoder of AL-Net integrates low-level features and applies 3 × 3 convolution to refine the features, which makes AL-Net more suitable for segmenting small objects and plays a gain effect on segmenting retinal vessels.
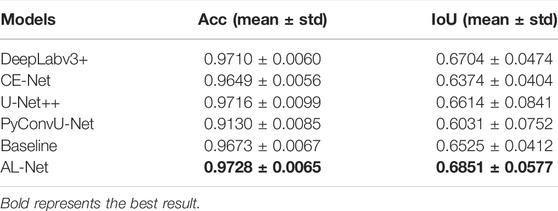
Cell contour: Figure 6 shows the segmentation results of six models on the cell contour segmentation dataset. In Figure 6, the segmentation results of AL-Net are more consistent with the ground truth, and the segmentation results of other models are discontinuous at the foreground edge. In addition, the segmentation results of U-Net++ are also disturbed by complex noise. The accuracy evaluation of these models is shown in Table 7. In Table 7, the value of
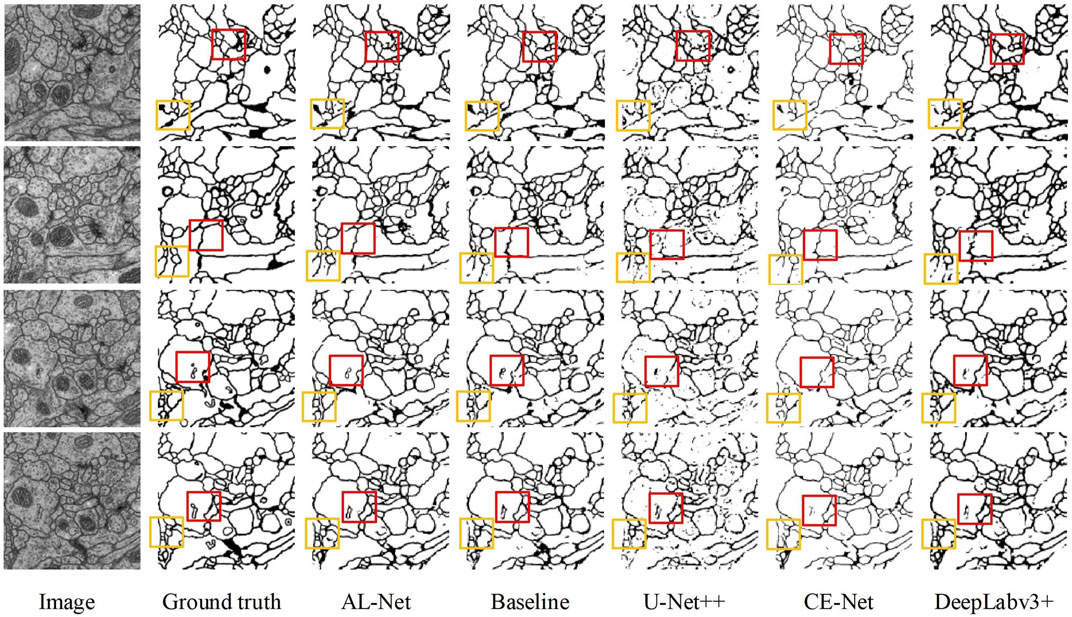
FIGURE 6. The segmentation results of five models for cell contour images. The red and yellow boxes respectively represent the segmentation differences of five models in the same position.
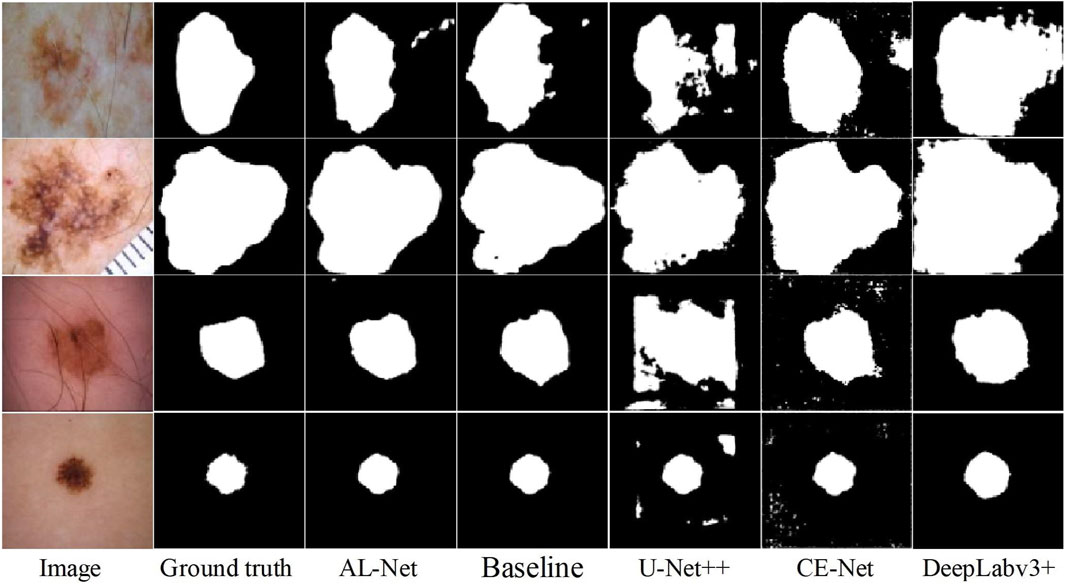
Skin lesions: The visualization and accuracy evaluation of segmentation results on the skin lesions dataset using six models are shown in Figure 7 and Table 8, respectively. In Figure 7, compared with other models, the segmentation results of AL-Net are obviously closer to the ground truth. In Table 8, the
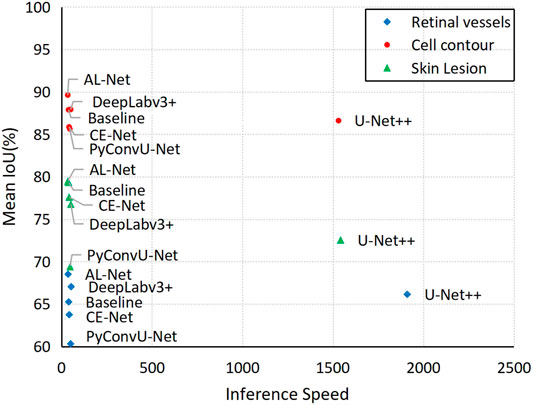
Figure 8 shows the evaluation of the inference speed and accuracy of the six models for three different datasets. As can be seen from Figure 8, AL-Net has the faster inference speed and the higher performance than the other models for the three datasets, which more intuitively proves the efficiency and effectiveness of AL-Net.
5 Conclusion
Aiming at the problems of large parameters and slow inference speed of medical image segmentation model, an asymmetric lightweight semantic segmentation network AL-Net is proposed in this paper. The encoder of AL-Net is trained through multi-branch structure to extract powerful medical image features. The context extraction module of AL-Net captures the context features and recalibrates the feature response in the channel direction by explicitly modeling the interdependence between channels, which is more suitable for segmenting medical images. The decoder of AL-Net not only makes full use of the low-level semantic information, but also combines 3 × 3 convolution to effectively eliminate redundant features. Finally, the reparameterization technology simplifies the inference procedure of AL-Net and improves the inference speed of AL-Net. The total parameters of AL-Net are only 3.45 M. Meanwhile, compared with the state-of-the-art models, AL-Net has achieved the best accuracy and the fastest speed on three datasets of retinal vessel, cell contour and skin lesions.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: STARE: http://cecas.clemson.edu/∼ahoover/stare/. ISIB2012: http://brainiac2.mit.edu/isbi_challenge/. ISIC2018: https://challenge.isic-archive.com/data/.
Author Contributions
YN and XD put forward the innovative ideas of the article, FW and TL designed and completed some experiments, YN and XD wrote the article, SW and XZ made important revisions to the article.
Funding
This work is partly supported by National Natural Science Foundation of China under Grant Nos. 61861024, 61871259, and 61762058, Natural Science Foundation of Gansu Province of China under Grant No.20JR5RA404, Natural Science Basic Research Program of Shaanxi(Program No.2021JC-47), Key Research and Development Program of Shaanxi (Program No. 2021ZDLGY08-07), and Shaanxi Joint Laboratory of Artificial Intelligence (Program No. 2020SS-03).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Allan, C., Halpern et al. “Skin lesion Analysis Toward Melanoma Detection 2018: A Challenge Hosted by the International Skin Imaging Collaboration (ISIC),” arXiv:1902.03368, 2019.
Alom, M. Z., Yakopcic, C., Hasan, M., Taha, T. M., and Asari, V. K. (2019). Recurrent Residual U-Net for Medical Image Segmentation. J. Med. Imaging (Bellingham) 6 (1), 014006. doi:10.1117/1.JMI.6.1.014006
Cardona, A., Saalfeld, S., Preibisch, S., Schmid, B., Cheng, A., Pulokas, J., et al. (2010). An Integrated Micro- and Macroarchitectural Analysis of the Drosophila Brain by Computer-Assisted Serial Section Electron Microscopy. Plos Biol. 8 (10). doi:10.1371/journal.pbio.1000502
Chen, C., Liu, X., Ding, M., Zheng, J., and Li, J. (2019). “3D Dilated Multi-Fiber Network for Real-Time Brain Tumor Segmentation in MRI,” in 22nd International Conference on Medical Image Computing and Computer-Assisted Intervention, 184–192. doi:10.1007/978-3-030-32248-9_21
Chen, L.-C., Papandreou, G., Kokkinos, I., Murphy, K., and Yuille, A. L. (2018). DeepLab: Semantic Image Segmentation with Deep Convolutional Nets, Atrous Convolution, and Fully Connected CRFs. IEEE Trans. Pattern Anal. Mach. Intell. 40 (4), 834–848. doi:10.1109/tpami.2017.2699184
Chen, L. C., Papandreou, G., Schroff, F., and Adam, H. (2017). “Rethinking Atrous Convolution for Semantic Image Segmentation,” in IEEE Conference on Computer Vision and Pattern Recognition. arXiv:1706.05587.
Chollet, F. (2017). “Xception: Deep Learning with Depthwise Separable Convolutions,” in IEEE Conference on Computer Vision and Pattern Recognition, 1251–1258. doi:10.1109/cvpr.2017.195
Chen, J., Lu, Y., Yu, Q., Luo, X., and Zhou, Y., “TransUNet: Transformers Make Strong Encoders for Medical Image Segmentation,” arXiv:2102.04306, 2021.
Ding, X., Zhang, X., Ma, N., Han, J., Ding, G., and Sun, J. (2021). “RepVGG: Making VGG-Style ConvNets Great Again,” in IEEE Conference on Computer Vision and Pattern Recognition, 13733–13742. doi:10.1109/cvpr46437.2021.01352
Gu, Z., Cheng, J., Fu, H., Zhou, K., Hao, H., Zhao, Y., et al. (2019). CE-net: Context Encoder Network for 2D Medical Image Segmentation. IEEE Trans. Med. Imaging 38, 2281–2292. doi:10.1109/TMI.2019.2903562
He, K., Zhang, X., Ren, S., and Sun, J. (2016). “Deep Residual Learning for Image Recognition,” in IEEE Conference on Computer Vision and Pattern Recognition, 770–778.
Hoover, A., and Goldbaum, M. (2003). Locating the Optic Nerve in a Retinal Image Using the Fuzzy Convergence of the Blood Vessels. IEEE Trans. Med. Imaging 22 (8), 951–958. doi:10.1109/tmi.2003.815900
Howard, A. G., Zhu, M., Chen, B., Kalenichenko, D., Wang, W., Weyand, T., et al. (2017). “MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications,” in IEEE Conference on Computer Vision and Pattern Recognition. arXiv:1704.04861.
Howard, A., Sandler, M., Chu, G., Chen, L. C., Chen, B., Tan, M., et al. (2019). “Searching for MobileNetV3,” in IEEE International Conference on Computer Vision, 1314–1324.
Huang, G., Liu, Z., Laurens, V., and Weinberger, K. Q. (2017). “Densely Connected Convolutional Networks,” in IEEE Conference on Computer Vision and Pattern Recognition, 2261–2269.
Ibtehaz, N., and Rahman, M. S. (2020). MultiResUNet : Rethinking the U-Net Architecture for Multimodal Biomedical Image Segmentation. Neural Networks 121, 74–87. doi:10.1016/j.neunet.2019.08.025
Isensee, F., Jaeger, P. F., Kohl, S. A. A., Petersen, J., and Maier-Hein, K. H. (2020). nnU-Net: a Self-Configuring Method for Deep Learning-Based Biomedical Image Segmentation. Nat. Methods 18, 203–211. doi:10.1038/s41592-020-01008-z
Jie, H., Li, S., Gang, S., and Albanie, S. (2018). “Squeeze-and-Excitation Networks,” in IEEE Conference on Computer Vision and Pattern Recognition, 7132–7141.
Jns, A., Si, Y., Mhy, A., Halim Yulius, M., Su, X., Kien Yee, Y. E., et al. (2020). Incorporating Convolutional Neural Networks and Sequence Graph Transform for Identifying Multilabel Protein Lysine Ptm Sites. Chemometrics Intell. Lab. Syst. 206, 104171. doi:10.1016/j.chemolab.2020.104171
Le, N. Q. K., Ho, Q.-T., Yapp, E. K. Y., Ou, Y.-Y., and Yeh, H.-Y. (2020). DeepETC: A Deep Convolutional Neural Network Architecture for Investigating and Classifying Electron Transport Chain's Complexes. Neurocomputing 375, 71–79. doi:10.1016/j.neucom.2019.09.070
Le, N. Q. K., Yapp, E. K. Y., Nagasundaram, N., and Yeh, H.-Y. (2019). Classifying Promoters by Interpreting the Hidden Information of DNA Sequences via Deep Learning and Combination of Continuous FastText N-Grams. Front. Bioeng. Biotechnol. 7, 305. doi:10.3389/fbioe.2019.00305
Lei, T., Zhou, W., Zhang, Y., Wang, R., Meng, H., and Nandi, A. K. (2020). “Lightweight V-Net for Liver Segmentation,” in ICASSP 2020 - 2020 IEEE International Conference on Acoustics, Speech and Signal Processing (Virtual Barcelona: ICASSP), 1379–1383. doi:10.1109/ICASSP40776.2020.9053454
Li, C., Fan, Y., and Cai, X. (2021). PyConvU-Net: PyConvU-Net: a Lightweight and Multiscale Network for Biomedical Image Segmentation. BMC Bioinformatics 22 (14). doi:10.1186/s12859-020-03943-2
Li, R., Li, M., Li, J., and Zhou, Y. (2019). “Connection Sensitive Attention U-NET for Accurate Retinal Vessel Segmentation,” in IEEE Conference on Computer Vision and Pattern Recognition. arXiv:1903.05558.
Lin, G., Milan, A., Shen, C., and Reid, I. (2017). “RefineNet: Multi-Path Refinement Networks for High-Resolution Semantic Segmentation,” in IEEE Conference on Computer Vision and Pattern Recognition, 1925–1934. doi:10.1109/cvpr.2017.549
Liu, W., Rabinovich, A., and Berg, A. C. (2015). “ParseNet: Looking Wider to See Better,” in IEEE Conference on Computer Vision and Pattern Recognition. arXiv:1506.04579.
Ma, N., Zhang, X., Zheng, H. T., and Sun, J. (2018). “ShuffleNet V2: Practical Guidelines for Efficient CNN Architecture Design,” in Proceedings of the European Conference on Computer Vision, 116–131. doi:10.1007/978-3-030-01264-9_8
Nekrasov, V., Shen, C., and Reid, I. (2018). “Light-Weight RefineNet for Real-Time Semantic Segmentation,” in IEEE Conference on Computer Vision and Pattern Recognition. arXiv:1810.03272.
Ni, Z.-L., Bian, G.-B., Zhou, X.-H., Hou, Z.-G., Xie, X.-L., Wang, C., et al. (2019). “RAUNet: Residual Attention U-Net for Semantic Segmentation of Cataract Surgical Instruments,” in 26th International Conference on Neural Information Processing, 139–149. doi:10.1007/978-3-030-36711-4_13
Ronneberger, O., Fischer, P., and Brox, T. (2015). “U-net: Convolutional Networks for Biomedical Image Segmentation,” in International Conference on Medical Image Computing and Computer-Assisted Intervention, 234–241. doi:10.1007/978-3-319-24574-4_28
Sandler, M., Howard, A., Zhu, M., Zhmoginov, A., and Chen, L. C. (2018). “MobileNetV2: Inverted Residuals and Linear Bottlenecks,” in IEEE Conference on Computer Vision and Pattern Recognition, 4510–4520.
Sergey Ioffe , , and Szegedy, Christian. (2015). “Batch normalization:Accelerating Deep Network Training by Reducing Internal Covariate Shift,” in International Conference on Machine Learning, 448–456.
Shelhamer, E., Long, J., and Darrell, T. (2015). Fully Convolutional Networks for Semantic Segmentation. IEEE Trans. Pattern Anal. Mach Intell. 39 (4), 640–651. doi:10.1109/TPAMI.2016.2572683
Simonyan, K., and Zisserman, A. (2014). “Very Deep Convolutional Networks for Large-Scale Image Recognition,” in IEEE Conference on Computer Vision and Pattern Recognition. arXiv:1409.1556.
Szegedy, C., Ioffe, S., Vanhoucke, V., and Alemi, A. (2016). “Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning,” in 31st AAAI Conference on Artificial Intelligence, 4278–4284.
Szegedy, C., Liu, W., Jia, Y., Sermanet, P., and Rabinovich, A. (2015). “Going Deeper with Convolutions,” in IEEE Conference on Computer Vision and Pattern Recognition, 1–9.
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., and Wojna, Z. (2016). “Rethinking the Inception Architecture for Computer Vision,” in IEEE Conference on Computer Vision and Pattern Recognition, 2818–2826. doi:10.1109/cvpr.2016.308
Tarasiewicz, T., Kawulok, M., and Nalepa, J. (2021). “Lightweight U-Nets for Brain Tumor Segmentation,” in Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes. Lecture Notes in Computer Science. Editors A. Crimi,, and S. Bakas (Cham: Springer), Vol. 12659, 3–14. doi:10.1007/978-3-030-72087-2_1
Tschandl, P., Rosendahl, C., and Kittler, H. (2018). The HAM10000 Dataset, a Large Collection of Multi-Source Dermatoscopic Images of Common Pigmented Skin Lesions. Sci. Data 5 (1), 180161–180169. doi:10.1038/sdata.2018.161
Valanarasu, J., Oza, P., Hacihaliloglu, I., and Patel, V., “Medical Transformer: Gated Axial-Attention for Medical Image Segmentation,” arXiv:2102.10662, 2021.
Wong, K. C. L., and Moradi, M. (2019). “SegNAS3D: Network Architecture Search with Derivative-free Global Optimization for 3D Image Segmentation,” in 22nd International Conference on Medical Image Computing and Computer-Assisted Intervention, 393–401. doi:10.1007/978-3-030-32248-9_44
Yu, C., Gao, C., Wang, J., Yu, G., Shen, C., and Sang, N. (2021). BiSeNet V2: Bilateral Network with Guided Aggregation for Real-Time Semantic Segmentation. Int. J. Comp. Vis., 1–18. doi:10.1007/s11263-021-01515-2
Yu, C., Wang, J., Peng, C., Gao, C., Yu, G., and Sang, N. (2018). “BiSeNet: Bilateral Segmentation Network for Real-Time Semantic Segmentation,” in European Conference on Computer Vision, 334–349. doi:10.1007/978-3-030-01261-8_20
Zhang, X., Zhou, X., Lin, M., and Sun, J. (2018). “ShuffleNet: An Extremely Efficient Convolutional Neural Network for Mobile Devices,” in Proceedings of the IEEE conference on Computer Vision and Pattern Recognition, 6848–6856. doi:10.1109/cvpr.2018.00716
Zhang, Z., Wu, C., Coleman, S., and Kerr, D. (2020). DENSE-INception U-Net for Medical Image Segmentation. Comp. Methods Programs Biomed. 192, 105395. doi:10.1016/j.cmpb.2020.105395
Zhao, H., Qi, X., Shen, X., Shi, J., and Jia, J. (2018). “ICNet for Real-Time Semantic Segmentation on High-Resolution Images,” in European Conference on Computer Vision, 418–434. doi:10.1007/978-3-030-01219-9_25
Zheng, S., Lu, J., Zhao, H., Zhu, X., and Zhang, L. (2020). Rethinking Semantic Segmentation from a Sequence-To-Sequence Perspective with Transformers. IEEE Conf. Comp. Vis. Pattern Recognition, 6881–6890.
Zhou, S., Nie, D., Adeli, E., Yin, J., Lian, J., and Shen, D. (2019). High-Resolution Encoder-Decoder Networks for Low-Contrast Medical Image Segmentation. IEEE Trans. Image Process. 29 (99), 461–475. doi:10.1109/TIP.2019.2919937
Keywords: deep learning, convolutional neural network, medical image segmentation, lightweight model, contextual encoder, asymmetric decoder
Citation: Du X, Nie Y, Wang F, Lei T, Wang S and Zhang X (2022) AL-Net: Asymmetric Lightweight Network for Medical Image Segmentation. Front. Sig. Proc. 2:842925. doi: 10.3389/frsip.2022.842925
Received: 24 December 2021; Accepted: 21 March 2022;
Published: 02 May 2022.
Edited by:
Jong-Seok Lee, Yonsei University, South KoreaReviewed by:
Jakub Nalepa, Silesian University of Technology, PolandYeejin Lee, Seoul National University of Science and Technology, South Korea
Copyright © 2022 Du, Nie, Wang, Lei, Wang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tao Lei, bGVpdGFvQHN1c3QuZWR1LmNu
 Xiaogang Du
Xiaogang Du Yinyin Nie
Yinyin Nie Fuhai Wang1,2
Fuhai Wang1,2 Tao Lei
Tao Lei Song Wang
Song Wang