- 1Shaanxi Joint Laboratory of Artificial Intelligence, Shaanxi University of Science and Technology, Xi’an, China
- 2The School of Electronic Information and Artificial Intelligence, Shaanxi University of Science and Technology, Xi’an, China
- 3Unmanned Intelligent Control Division, China Electronics Technology Group Corporation Northwest Group Corporation, Xi’an, China
- 4Electronic and Electrical Engineering, Brunel University London, London, United Kingdom
- 5School of Mechanical Engineering, Xi’an Jiaotong University, Xi’an, China
Deep convolutional neural networks (DCNNs) have been widely used in medical image segmentation due to their excellent feature learning ability. In these DCNNs, the pooling operation is usually used for image down-sampling, which can gradually reduce the image resolution and thus expands the receptive field of convolution kernel. Although the pooling operation has the above advantages, it inevitably causes information loss during the down-sampling of the pooling process. This paper proposes an effective weighted pooling operation to address the problem of information loss. First, we set up a pooling window with learnable parameters, and then update these parameters during the training process. Secondly, we use weighted pooling to improve the full-scale skip connection and enhance the multi-scale feature fusion. We evaluated weighted pooling on two public benchmark datasets, the LiTS2017 and the CHAOS. The experimental results show that the proposed weighted pooling operation effectively improve network performance and improve the accuracy of liver and liver-tumor segmentation.
Introduction
Accurate segmentation of livers and liver tumors can assist doctors in better diagnosis and help doctors make better medical plans. Therefore, liver and liver tumor segmentations have always been one of the research hotspots in the field of medical image analysis. However, because the liver has a similar density with other nearby organs, it is difficult to find the liver boundary accurately in a CT image of the abdomen for non-professionals (Li et al., 2015). Relying on manual labeling of liver regions is not only time-consuming and labor-intensive, tedious and inefficient, but also requires high-level professional technical expertise for delineating labelers. Therefore, automatic or semi-automatic liver segmentation algorithms have become a research goal in the field of medical image analysis (Furukawa et al., 2017).
Before the advent of deep learning (LeCun et al., 2015), three popular image segmentation algorithms were often used for liver segmentation: algorithms based on gray values (Adams and Bischof, 1994; ChenyangXu and Prince, 1998; Lei et al., 2018), algorithms based on statistical shape models (Heimann et al., 2006; Zhang et al., 2010; Tomoshige et al., 2014), and algorithms based on texture features (Gambino et al., 2010; Ji et al., 2013). These traditional image segmentation algorithms often employ only image low-level features such as edge, shape, texture, etc., but do not employ the image semantic information with strong representation ability. Thus, they only provide low image segmentation accuracy and show poor generalization. In recent years, with the rapid development of deep learning (Hinton and Salakhutdinov, 2006; Tu et al., 2017; Tu et al., 2018; Yang et al., 2021) in the field of computer vision, especially after the emergence of fully connected convolutional neural networks (Shelhamer et al., 2017), researchers have begun to use deep learning methods for image segmentation. The emergence of the U-Net (Ronneberger et al., 2015) network model has greatly promoted the development of medical image segmentation (Lei et al., 2020a). Since then, this end-to-end segmentation network (Nie et al., 2016) has become the benchmark for medical image segmentations. U-Net is a completely symmetrical encoder-decoder structure. In the encoder part, the network gradually extracts the deep semantic information of images, and then in the decoder part, feature maps are gradually restored into a segmentation map. The skip connection enables the network to fuse all levels of feature information from the encoder during the decoding process, which enables the network to obtain more refined segmentation results. Due to the great success of U-Net, various improved U-Nets have been proposed (Guo et al., 2019; De Sio et al., 2021). These improved networks can be roughly grouped into two categories. The first category of methods often employs a new network backbone instead of the convolution in the original network encoder-decoder part, such as VGG (Simonyan and Zisserman, 2014), ResNet (He et al., 2016), DenseNet (Huang et al., 2017), GhostNet (Han et al., 2020), etc. The second category of method often adds some new function modules to U-Net to enhance network performance, such as attention U-Net (Oktay et al., 2018), CE-Net (Gu et al., 2019), QAU-Net (Hong et al., 2021), and RA-UNet (Jin et al., 2018). In addition, R2-UNet (Alom et al., 2018) adopts circular convolution, that can use the same feature map to extract information multiple times, and make full use of the potentially useful information in the feature map. UNet++ (Zhou et al., 2020) explores the impact of different depths of U-Net on network performance and adopts a new skip connection to gather features of different semantic scales. UNet3+ (Huang et al., 2020) further proposes a full-scale skip connection to fuse the low-level information and high-level semantics of feature maps of different sizes. LV-Net (Lei et al., 2020b) uses a lightweight network to segment the liver. Furthermore, there are some improved networks such as DefU-Net (Lei et al., 2021), CE-Net (Gu et al., 2019) and MSB-Net (Shao et al., 2019) that use multi-scale feature fusion to enhance the feature representation of the network.
These networks perform pooling operations to achieve down-sampling multiple times in the encoder part. The purpose is to gradually pass down the feature information of images, and in this process, the feature information of the image space and channel is continuously integrated, and finally extracted deep semantic information. Due to the characteristics of pooling, both the average pooling (Wang et al., 2021) or the maximum pooling (Nagi et al., 2011; Giusti et al., 2013; Graham, 2014; Bulo et al., 2017) will inevitably lead to the loss of some image feature information. Skip connection is a common operation in medical image segmentation networks. In order to achieve various skip connections, researchers usually use pooling operation to process feature maps to the same size, which will also cause the loss of feature information. This is especially so, when we need to change the feature map ruler on a large scale to realize skip connection (Huang et al., 2020), the information loss caused by the pooling operation will be more.
In order to solve the problem of image information loss caused by pooling operation, this paper proposes a weighted pooling (Golan et al., 2012; Zhu et al., 2019) operation. The operation is to allow images to be trained with parameters during the process of down-sampling. Using weighted pooling operation, we can change the size of the feature map while reducing information loss, which provides conditions for achieving more types of skip connection, especially for the situation where a large range of feature map size needs to be changed. In summary, we have made the following contributions:
1) We propose a weighted pooling operation that can reduce the loss of feature information of images while performing the reduction of the image resolution.
2) We demonstrate that the weighted pooling operation is helpful to the realization and improvement of various skip connections.
3) Based on the weighted pooling operation and the full-scale skip connection, we design a novel U-shaped network for liver and liver-tumor segmentation, and the proposed network achieves better performance than U-Net.
The rest of the paper is organized as follows. In Methods section, we introduce the principle and implementation method of weighted pooling in detail, and then explain the improvement method of weighted pooling on skip connection, and finally introduce the network we propose. In Experiments section, we use experiments to demonstrate the effectiveness of weighted pooling; Finally, in Conclusion section we present conclusions and plan our future work.
Methods
In this section, we will sequentially introduce the weighted pooling operation and the improved skip connection based on the weighted pooling operation. Then we design a new U-shaped network for liver and liver tumor segmentation.
Weighted Pooling Operation
Pooling operation is very common in convolutional neural networks (Seo et al., 2020), and its purpose is to perform down-sampling on an image so that convolution kernels can gradually obtain a larger receptive field and fuse more image context information. The traditional pooling operation uses the maximum pooling or the average pooling, that is, taking the maximum value of pixels or the average value of pixels in a window, and traversing the entire feature map to achieve the purpose of down-sampling after determining the step size. Taking the maximum pooling as an example, the pooling operation can be expressed as
The weighted pooling operation is to give each pixel in the window a learnable parameter, and then let the window traverse the entire feature map, which can achieve the purpose of down-sampling and reduce the loss of image information. Specifically, similar to the process of pooling operation, we can set a matrix window with parameters for the feature map of each channel. The size of the window can be determined by itself according to different tasks or the degree of down-sampling. After determining the step size, let the window slide on the feature map of each channel to traverse the entire feature map. For example, we perform pooling operation using a 2 × 2 window with a step size of 2, the pooling process is shown in Figure 1.
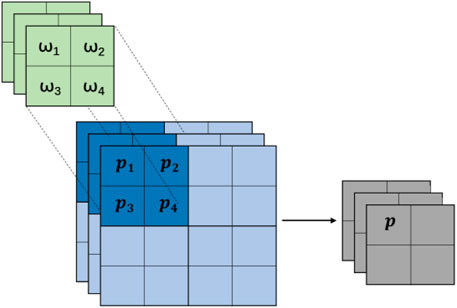
FIGURE 1.
According to Figure 1, we understand the basic principle of the weighted pooling, and the final pixel value can be expressed as:
where
According to (1), we can design the corresponding parameter matrix window, but the existing modules allow us to implement weighted pooling very conveniently. We can perform the convolution operation on the feature map of each channel separately, because the convolution operation is the process of sliding the matrix window with parameters on the feature map, which is the same as the idea of the weighted pooling. The window size in the pooling process is generally even, which is more conducive to our down-sampling, because odd-numbered windows will have more stringent requirements on the resolution of the feature map, so the weighted pooling also chooses even-numbered convolution kernels. To perform the operation, and then set different step lengths to achieve different down-sampling requirements. In different tasks, we can freely choose even-numbered convolution kernels of different sizes to perform weighted pooling operation.
The Full-Scale Skip Connection
A key factor for the success of U-Net is the use of skip connection (Milletari et al., 2016; Jin et al., 2017; Guo et al., 2021), which enables the network to fuse low-level and high-level feature information during the decoding process, and finally obtains a more accurate segmentation result. It is worth noting that there is a premise for using skip connection. The premise is that the feature maps must have the same size. Due to the limitation of the size of the feature map, skip connection is not arbitrary.
For vanilla skip connection, to obtain the feature map of the same size, researchers often use the pooling operation to change the size of the feature map. According to the previous analysis, the use of pooling to change the size of the feature map will inevitably lead to the loss of image information, especially when the image size needs to be changed in a large range, just like the full-scale skip connection in U-Net3+. The proposed weighted pooling operation can well solve the problem, which provides the possibility for the proposal of more effective skip connections, which is also another important contribution of this work. The improvement of weighted pooling to skip connection is shown in Figure 2. This paper mainly uses weighted pooling to improve full-scale skip connection.
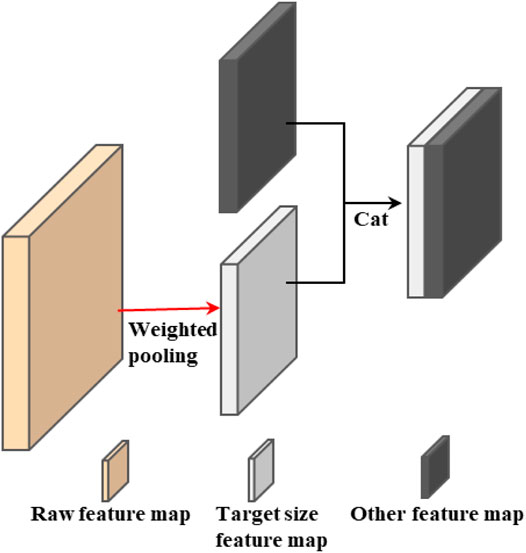
FIGURE 2. The use of weighted pooling reduces the loss of information while changing the size of the feature map.
It can be seen from Figure 2 that we can use weighted pooling to change the size of the feature map to the target size, which will facilitate our subsequent skip connection and provide more possibilities for the design of the network model.
The Proposed Network
This paper uses the weight pooling module and the improved full-scale skip connection to design a network for liver and liver-tumor segmentation. The network framework is shown in Figure 3.
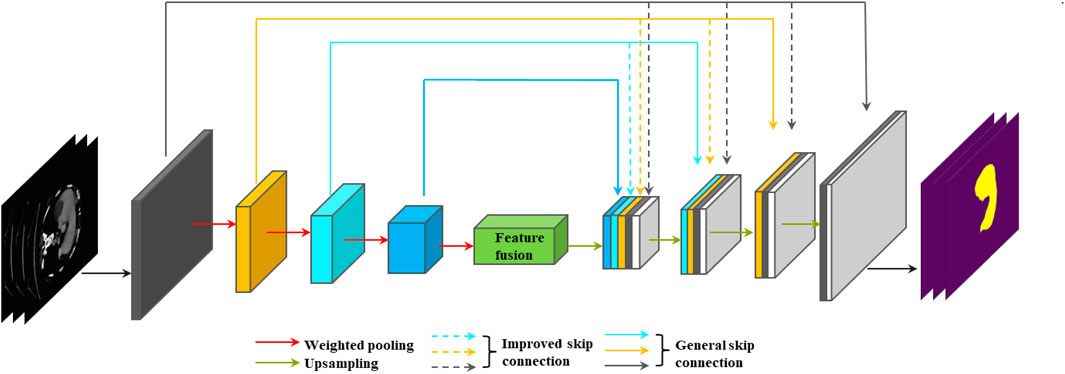
FIGURE 3. Liver and liver-tumor segmentation network designed using weighted pooling and improved full-scale skip connection.
In Figure 3, we can see that the network is an enhanced version of U-Net. The entire network can still be divided into two parts: the encoder and the decoder. The convolution modules of the encoder and the decoder use deep separable convolution (Chollet, 2017), which can greatly reduce the number of parameters of the network while maintaining segmentation performance of the network. In the encoder part, all down-sampling operations use the weighted pooling module to replace the original maximum pooling module, which can reduce the loss of image information. For the first and second down-sampling, we used a 2 × 2 window for weighted pooling, which is conducive to the extraction of image edge information and detail information. Small windows are often adopted since they can do this well. For the third and fourth down-sampling, we used 4 × 4 and 8 × 8 windows respectively for weighted pooling. We hope that the deeper layer of the encoder can fuse the image information of the larger area of the feature map, which is conducive to the decoding process and finally obtains more accurate image segmentation results. A large window is often adopted since it can do this well. For the feature map containing deep semantic information finally obtained by the encoder, we use the SE module (Jie et al., 2017) to perform feature fusion. In the decoder part, for upsampling, we use the combination of bilinear interpolation (Accadia et al., 2003; Kirkland, 2010) and 1 × 1 convolution (Szegedy et al., 2015) to replace the original deconvolution operation (Noh et al., 2016), which can reduce the parameters at the same time, and avoid the checkerboard-like phenomenon in the feature map.
Full-scale skip connection can combine low-level appearance information and high-level semantic information from feature maps of different sizes to clarify better the location and boundary of the liver. As shown in Figure 3, it is clear that each layer of the convolution module of the decoder combines the feature maps of all layers in the encoder. Compared with the original skip connection of U-Net, the full-scale skip connection is integrated from the network as a whole with sufficient information, and these features at different scales can obtain fine-grained details and coarse-grained semantics. As shown in Figure 3, every time the encoder part is down-sampled, the resolution of the feature map will become half of the original feature map. To achieve a full-scale skip connection, we must ensure that the size of the feature map is consistent. Then we must use pooling to make the feature map resolution consistent. But we can see in Figure 3 that the largest difference in size between the two feature maps is 8 times, which means that we need to use pooling to change the feature map to 1/8 of the original. The information loss in the middle will be huge. When we use weighted pooling to change the size of the feature map, we can reduce the loss of information, because we will learn with parameters when we change the resolution of the feature map. In this paper, we use an operation with a window size of 2 × 2 and a step size of 2 to change the resolution of the feature map to 1/2, and use an operation with a window size of 4 × 4 and a step size of 4 to change the resolution of the feature map to 1/4, and use an operation with a window size of 8 × 8 and a step size of 8 to change the resolution of the feature map to 1/8.
Experiments
Dataset and Pre-Processing
In order to evaluate the effects of the weighted pooling module on improving the performance of the liver and liver tumor segmentation network, we used the LiTS2017 (Liver Tumor Segmentation Challenge) dataset and CHAOS (Combined Healthy Abdominal Organ Segmentation) dataset as experimental data.
The LiTS2017 dataset contains 131 cases of labeled abdominal 3D CT scan images, in which the in-plane resolution ranges from 0.55 to 1 mm, the slice pitch ranges from 0.45 to 6 mm, and each image size is 512 × 512. The CHAOS dataset is a small dataset that contains 20 3D data, where the image size is 512 × 512. The entire experiments use the axial 2D slice images of the LiTS2017 and CHAOS dataset. We constructed the training set and validation set using 90 patients and 10 patients. Then the other 30 patients are considered as the test set. For the CHAOS, it was split into 16 patients for training and 4 patients for test.
Medical CT axial slices are different from normal axial slices. The former can obtain values ranging from −1,000 to 3,000, while the latter can only obtain values ranging from 0 to 255. In order to eliminate interference and enhance the liver area, we selected the [−200, 250] HU range and performed a normalization process.
Experimental Setup and Evaluation Metrics
All the algorithms in this experiment are implemented on the NVIDIA GeForce RTX 3090 Ti server. The neural network chooses pytorch 1.7.0 as the framework for training and execution. In the training process, the learning rate is set to 0.001, and this experiment did not use the dynamic learning rate. The number of training rounds in this experiment is set to 100.
The most commonly used and effective index for medical image segmentation is Dice score. The value of Dice ranges from 0 to 1. The larger the value, the higher the accuracy of segmentation. The value of Dice for perfect segmentation is 1. This experiment uses the average Dice of all slices in the test dataset as the evaluation metrics.
Ablation Study
This paper is mainly to study the effect of the weighted pooling operation on the liver and liver-tumor segmentation. The paper mainly highlights two contributions. The first is that the weighted pooling operation can be used to replace the traditional pooling module to reduce the loss of image feature information, and the second is that the weighted pooling operation can improve various skip connections. This paper improves the full-scale skip connection. In order to explore the validity of these contributions, we conducted two sets of ablation experiments on the LiTS2017 dataset and CHAOS dataset.
The Effectiveness of the Weighted Pooling Operation
We let the U-Net, CE-Net, and U-Net++ networks to be trained on the liver and liver-tumor training dataset respectively, and then get the segmentation accuracy of the liver and liver-tumor on the test dataset. Then, we use the weighted pooling operation instead of the maximum pooling in the U-Net, CE-Net, U-Net++. Then the new network is trained on the liver and liver-tumor training dataset respectively, and the segmentation accuracy of the liver and liver-tumor on the test dataset is obtained.
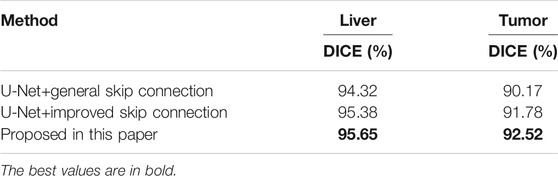
In Table 1, when we use the weighted pooling operation to replace the maximum pooling module, the segmentation accuracy of liver and liver-tumor are improved, which verifies the first contribution of the paper. Especially for the CHAOS dataset with a small amount of data, reducing the information loss in the pooling process can more effectively improve the network performance. In Figure 4, we can see the difference of the segmentation results more clearly.
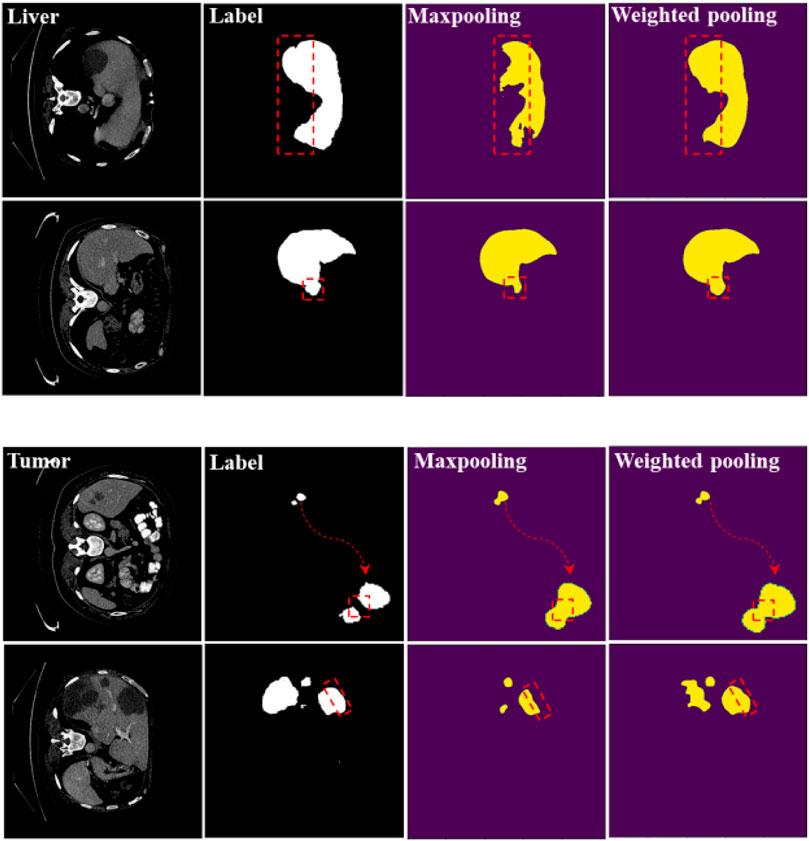
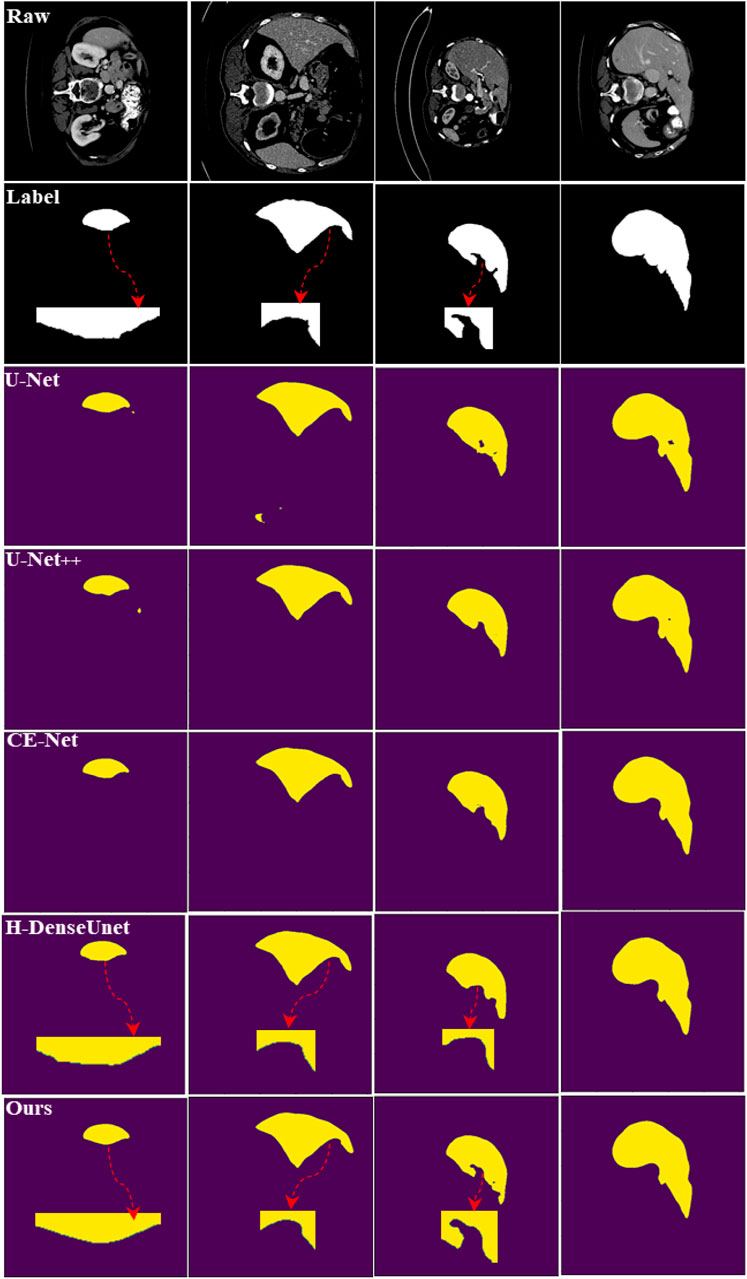
FIGURE 4. Segmentation results of maximum pooling and weighted pooling. The experimental network is U-Net.
We have marked the main differences in the segmentation results with red dashed lines. We can see from the segmentation results that weighted pooling is more powerful. The first set of liver segmentation results show that weighted pooling can reduce the loss of information. The second set of liver segmentation results show that weighted pooling has better feature learning capabilities. The segmentation results of the two groups of liver-tumor show that weighted pooling is more conducive to the segmentation of small target objects, and weighted pooling has a better ability to learn detailed information.
The Improvement Effect of Full-Scale Skip Connection
The experiment process is similar to the first set of experiments. We trained U-Net with a general full-scale skip connection and U-Net with an improved full-scale skip connection, and then obtained the accuracy on the liver and liver-tumor test datasets.
In Table 2, when we use the improved full-scale skip connection, the segmentation accuracy of the liver and liver-tumor are improved, which demonstrates the improvement effect of full-scale skip connection based on the weighted pooling. In Figure 5, we can also see the difference in segmentation results. We have marked the main differences in the segmentation results with red dashed lines. We can see from the segmentation results that weighted pooling enhances the information transmission of skip connection, and weighted pooling reduces the information loss in skip connection.
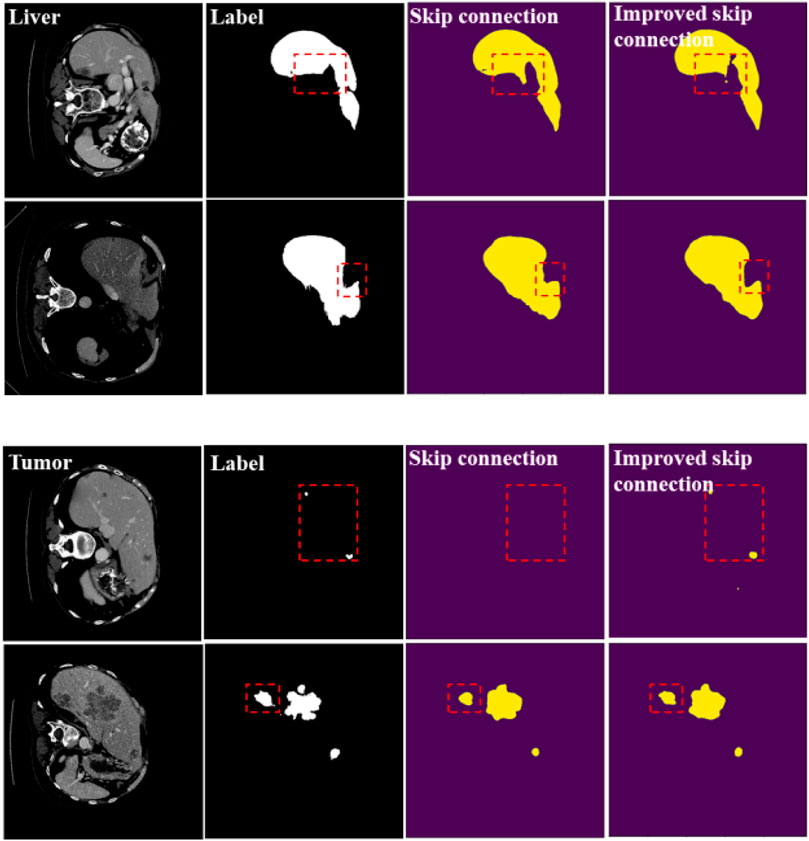
FIGURE 5. Segmentation results of general skip connection and improved skip connection. The experimental network is U-Net.
Comparative Experiment
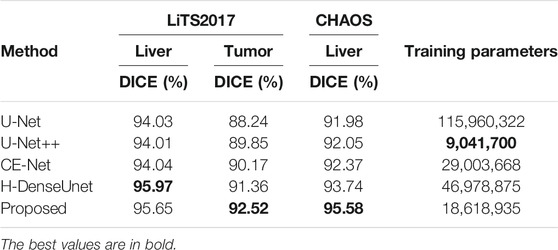
In order to test the performance of the proposed network, we conducted a set of comparative experiments on LiTS2017 dataset and CHAOS dataset. We trained several popular medical image segmentation networks, including U-Net, U-Net++, CE-Net, H-DenseUnet (Li et al., 2017), etc., and then obtained the segmentation accuracy of liver and liver-tumor of these networks on the test dataset. In Table 3 for the LiTS2017 dataset, we can see that the segmentation accuracy of the network proposed in the paper is higher than that of some popular medical image segmentation networks, except that H-DenseUnet has a slightly higher liver segmentation accuracy than our network. Although the liver segmentation accuracy of H-DenseUnet is slightly higher than our method, its network parameters are higher than our network. Fewer parameters means that our network takes up less memory and consumes less training time. At the same time, our network is easier to migrate to other devices. Our method achieves a good balance between computational complexity and performance. For the CHAOS dataset, our network achieved the best performance, which shows that our network has better learning ability in the case of less data.
In Figure 6, we can see the liver segmentation results of different methods. Our method performs well on detailed information and edge information, which shows that our method reduces the loss of information and better combines global feature information. The red dashed line marks some typical differences between our method and H-DenseUnet.
Conclusion
In this work, we have rethought pooling operation in DCNNs for liver and liver-tumor segmentations. We have found that the vanilla pooling operation suffers from a problem of information loss leading to performance degradation of networks for image segmentations. To overcome this, we have proposed a weighted pooling operation to solve the problem. The weighted pooling operation allows an image to be learned with parameters during the down-sampling process, which reduces the image resolution while ensuring the integrity of the image information. Using the weighted pooling operation can easily make the feature map size consistent, which is helpful for the realization and improvement of various skip connections. The final experiments demonstrate the effectiveness of the weighted pooling operation for liver and liver-tumor segmentation. In the future, we will explore the availability of weighted pooling for segmentation of other types of images.
Data Availability Statement
Publicly available datasets have been analyzed in this study. This data can be found here: https://academictorrents.com/details/27772adef6f563a1ecc0ae19a528b956e6c803ce.
Author Contributions
JL and TL proposed the innovative ideas of the paper. WZ and XD designed and completed some experiments. JL and MX wrote the paper together. TL and AN made important revisions to the paper.
Funding
This work was supported in part by Natural Science Basic Research Program of Shaanxi (Program No. 2021JC-47), in part by the National Natural Science Foundation of China under Grant 61871259, Grant 61861024, in part by Key Research and Development Program of Shaanxi (Program No. 2021ZDLGY08-07), in part by Serving Local Special Program of Education Department of Shaanxi Province (21JC002), and in part by Xi’an Science and Technology program (21XJZZ0006).
Conflict of Interest
Author WZ was employed by the company China Electronics Technology Group Corporation Northwest Group Corporation.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Lei would like to thank Brunel University London for a visiting position in 2020 to collaborate with Nandi.
References
Accadia, C., Mariani, S., Casaioli, M., Lavagnini, A., and Speranza, A. (2003). Sensitivity of Precipitation Forecast Skill Scores to Bilinear Interpolation and a Simple Nearest-Neighbor Average Method on High-Resolution Verification Grids. Auk 133.2, 129–130. doi:10.1175/1520-0434(2003)018<0918:sopfss>2.0.co;2
Adams, R., and Bischof, L. (1994). Seeded Region Growing. IEEE Trans. Pattern Anal. Machine Intell. 16 (6), 641–647. doi:10.1109/34.295913
Alom, M. Z., Hasan, M., Yakopcic, C., Taha, T. M., and Asari, V. K. (2018). Recurrent Residual Convolutional Neural Network Based on U-Net (R2U-Net) for Medical Imagesegmentation. arXiv:1802.06955.
Bulo, S. R., Neuhold, G., and Kontschieder, P. (2017). “Loss Max-Pooling for Semantic Image Segmentation,” in IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, July 21-26, 2017, 7082–7091. doi:10.1109/cvpr.2017.749
Chenyang Xu, C., and Prince, J. L. (1998). Snakes, Shapes, and Gradient Vector Flow. IEEE Trans. Image Process. 7 (3), 359–369. doi:10.1109/83.661186
Chollet, F. (2017). “Xception: Deep Learning with Depthwise Separable Convolutions,” in 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, July 21-26, 2017 (IEEE).
De Sio, C., Velthuis, J. J., Beck, L., Pritchard, J. L., and Hugtenburg, R. P. (2021). r-UNet: Leaf Position Reconstruction in Upstream Radiotherapy Verification. IEEE Trans. Radiat. Plasma Med. Sci. 5, 272–279. doi:10.1109/TRPMS.2020.2994648
Furukawa, D., Shimizu, A., and Kobatake, H. (2017). “Automatic Liver Segmentation Method Based on Maximum a Posterior Probability Estimation and Level Set Method,” in Proc. Int. Conf. Med. Image Comput. Comput.Assist. Intervent. (MICCAI), Quebec City, Canada, September 10–14, 2017, 117–124.
Gambino, O., Vitabile, S., Re, G. L., Tona, G. L., Librizzi, S., Pirrone, R., et al. (2010). “Automatic Volumetric Liver Segmentation Using Texturebased Region Growing,” in 2010 International Conference on Complex, Intelligent and Software Intensive Systems, Krakow, Poland, February 15-18, 2010, 146–152. doi:10.1109/cisis.2010.118
Giusti, A., Cirean, D. C., Masci, J., Gambardella, L. M., and Schmidhuber, J. (2013). “Fast Image Scanning with Deep Max-Pooling Convolutional Neural Networks,” in 2013 IEEE International Conference on Image Processing, Melbourne, VIC, Australia, September 15-18, 2013.
Golan, D., Erlich, Y., and Rosset, S. (2012). Weighted Pooling-Ppractical and Cost-Effective Techniques for Pooled High-Throughput Sequencing. Bioinformatics 28 (12), i197–i206. doi:10.1093/bioinformatics/bts208
Gu, Z., Cheng, J., Fu, H., Zhou, K., Hao, H., Zhao, Y., et al. (2019). CE-net: Context Encoder Network for 2D Medical Image Segmentation. IEEE Trans. Med. Imaging 38 (10), 2281–2292. doi:10.1109/tmi.2019.2903562
Guo, C., Szemenyei, M., Yi, Y., Wang, W., Chen, B., and Fan, C. (2021). “SA-UNet: Spatial Attention U-Net for Retinal Vessel Segmentation,” in 2020 25th International Conference on Pattern Recognition (ICPR), Milan, Italy, January 10-15, 2021.
Guo, Z., Li, X., Huang, H., Guo, N., and Li, Q. (2019). Deep Learning-Based Image Segmentation on Multimodal Medical Imaging. IEEE Trans. Radiat. Plasma Med. Sci. 3 (2), 162–169. doi:10.1109/trpms.2018.2890359
Han, K., Wang, Y., Tian, Q., Guo, J., Xu, C., and Xu, C. (2020). “GhostNet:More Features from Cheap Operations,” in Proc. IEEE Conf. Comput.Vis. Pattern Recogn., Seattle, WA, USA, June 13-19, 2020, 1580–1589. doi:10.1109/cvpr42600.2020.00165
He, K., Zhang, X., Ren, S., and Sun, J. (2016). “Deep Residual Learning for Image Recognition,” in Proc. IEEE Conf. Comput. Vis. Pattern Recogn., Las Vegas, NV, USA, June 27-30, 2016, 770–778. doi:10.1109/cvpr.2016.90
Heimann, T., Wolf, I., and Meinzer, H.-P. (2006). “Active Shape Models for a Fully Automated 3D Segmentation of the Liver - an Evaluation on Clinical Data,” in International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Copenhagen, Denmark, October 1-6, 2006, 41–48. doi:10.1007/11866763_6
Hinton, G. E., and Salakhutdinov, R. R. (2006). Reducing the Dimensionality of Data with Neural Networks. Science 313 (5786), 504–507. doi:10.1126/science.1127647
Hong, L., Wang, R., Lei, T., Du, X., and Wan, Y. (2021). “QAU-NET: Quartet Attention U-Net for Liver and Liver-Tumor Segmentation,” in 2021 IEEE International Conference on Multimedia and Expo (ICME), Shenzhen, China, July 5-9, 2021 (IEEE).
Huang, G., Liu, Z., an Der Maaten, L. V., and Weinberger, K. Q. (2017). “Densely Connected Convolutional Networks,” in Proc. IEEE Conf. Comput. Vis.Pattern Recogn., Honolulu, HI, USA, July 21-26, 2017, 2261–2269. doi:10.1109/cvpr.2017.243
Huang, H., Lin, L., Tong, R., Hu, H., and Wu, J. (2020). “UNet 3+: A Full-Scale Connected UNet for Medical Image Segmentation,” in ICASSP 2020 - 2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, May 4-8, 2020 (IEEE).
Ji, H., He, J., Yang, X., Deklerck, R., and Cornelis, J. (2013). ACM-based Automatic Liver Segmentation from 3-D CT Images by Combining Multiple Atlases and Improved Mean-Shift Techniques. IEEE J. Biomed. Health Inform. 17 (3), 690–698. doi:10.1109/jbhi.2013.2242480
Jie, H., Li, S., Gang, S., and Albanie, S. (2017). Squeeze-and-Excitation Networks. IEEE Trans. Pattern Anal. Machine Intelligence 42, 2011–2023. doi:10.1109/TPAMI.2019.2913372
Jin, Q., Meng, Z., Sun, C., Wei, L., and Su, R. (2018). RA-UNet: A Hybrid Deep Attention-Aware Network to Extract Liver and Tumor in CT Scans. arXiv:1811.01328.
Jin, Y., Kuwashima, S., and Kurita, T. (2017). “Fast and Accurate Image Super Resolution by Deep CNN with Skip Connection and Network in Network,” in International Conference on Neural Information Processing, Kyoto, Japan, October 16-21, 2017.
LeCun, Y., Bengio, Y., and Hinton, G. (2015). Deep Learning. Nature 521, 436–444. doi:10.1038/nature14539
Lei, T., Jia, X., Zhang, Y., He, L., Meng, H., and Nandi, A. K. (2018). Significantly Fast and Robust Fuzzy C-Means Clustering Algorithm Based on Morphological Reconstruction and Membership Filtering. IEEE Trans. Fuzzy Syst. 26 (5), 3027–3041. doi:10.1109/tfuzz.2018.2796074
Lei, T., Wang, R., Wan, Y., Zhang, B., Meng, H., and Nandi, A. K. (2020). Medical Image Segmentation Using Deep Learning: A Survey. arXiv:2009.13120.
Lei, T., Wang, R., Zhang, Y., Liu, Y. C., and Nandi, A. K. (2021). DefED-Net: Deformable Encoder-Decoder Network for Liver and Liver Tumor Segmentation. IEEE Trans. Radiat. Plasma Med. Sci.99, 1. doi:10.1109/TRPMS.2021.3059780
Lei, T., Zhou, W., Zhang, Y., Wang, R., and Nandi, A. K. (2020). “Lightweight V-Net for Liver Segmentation,” in ICASSP 2020 - 2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, May 4-8, 2020 (IEEE).
Li, G., Chen, X., Shi, F., Zhu, W., Tian, J., and Xiang, D. (2015). Automatic Liver Segmentation Based on Shape Constraints and Deformable Graph Cut in CT Images. IEEE Trans. Image Process. 24 (12), 5315–5329. doi:10.1109/tip.2015.2481326
Li, X., Chen, H., Qi, X., Dou, Q., Fu, C.-W., and Heng, P.-A. (2017). H-DenseUNet: Hybrid Densely Connected UNet for Liver and Liver Tumor Segmentation from CT Volumes. IEEE Trans. Med. Imaging 37, 2663–2674. doi:10.1109/tmi.2018.2845918
Milletari, F., Navab, N., and Ahmadi, S. A. (2016). “V-net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation,” in 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, October 25-28, 2016 (IEEE).
Nagi, J., Ducatelle, F., Di Caro, G. A., Ciresan, D., Meier, U., Giusti, A., et al. (2011). “Max-pooling Convolutional Neural Networks for Vision-Based Hand Gesture Recognition,” in IEEE International Conference on Signal and Image Processing Applications, Kuala Lumpur, Malaysia, November 16-18, 2011 (IEEE).
Nie, D., Wang, L., Gao, Y., and Shen, D. (2016). “Fully Convolutional Networks for Multi-Modality Isointense Infant Brain Image Segmentation,” in Proc. IEEE Conf. Int. Symp. Biomed. Imag., Prague, Czech Republic, April 13-16, 2016, 1342–1345. doi:10.1109/isbi.2016.7493515
Noh, H., Hong, S., and Han, B. (2016). “Learning Deconvolution Network for Semantic Segmentation,” in 2015 IEEE International Conference on Computer Vision (ICCV), Santiago, Chile, December 7-13, 2015.
Oktay, O., Schlemper, J., Folgoc, L. L., Lee, M., Heinrich, M., Misawa, K., et al. (2018). Attention U-Net: Learning where to Look for the Pancreas. arXiv:1804.03999.
Ronneberger, O., Fischer, P., and Brox, T. (2015). “U-net: Convolutional Networks for Biomedical Image Segmentation,” in Proc. Int. Conf. Med. Image Comput. Comput. Assist. Intervent. (MICCAI), Munich, Germany, October 5-9, 2015, 234–241. doi:10.1007/978-3-319-24574-4_28
Seo, H., Huang, C., Bassenne, M., Xiao, R., and Xing, L. (2020). Modified U-Net (mU-Net) with Incorporation of Object-dependent High Level Features for Improved Liver and Liver-Tumor Segmentation in CT Images. IEEE Trans. Med. Imaging 39, 1316–1325. doi:10.1109/TMI.2019.2948320
Shao, Q., Gong, L., Ma, K., Liu, H., and Zheng, Y. (2019). “Attentive CT Lesion Detection Using Deep Pyramid Inference with Multi-Scale Booster,” in Proc. Int. Conf. Med. Image Comput. Comput. Assist. Intervent. (MICCAI), Shenzhen, China, October 13–17, 2019, 301–309. doi:10.1007/978-3-030-32226-7_34
Shelhamer, E., Long, J., and Darrell, T. (2017). Fully Convolutional Networks for Semantic Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 39 (4), 640–651. doi:10.1109/tpami.2016.2572683
Simonyan, K., and Zisserman, A. (2014). Very Deep Convolutional Networks for Large-Scale Image Recognition, . arXiv:1409.1556.
Szegedy, C., Wei, L., Jia, Y., Sermanet, P., and Rabinovich, A. (2015). “Going Deeper with Convolutions,” in 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, June 7-12, 2015 (IEEE).
Tomoshige, S., Oost, E., Shimizu, A., Watanabe, H., and Nawano, S. (2014). A Conditional Statistical Shape Model with Integrated Error Estimation of the Conditions; Application to Liver Segmentation in Non-contrast CT Images. Med. Image Anal. 18 (1), 130–143. doi:10.1016/j.media.2013.10.003
Tu, Z., Guo, Z., Xie, W., Yan, M., Veltkamp, R. C., Li, B., et al. (2017). Fusing Disparate Object Signatures for Salient Object Detection in Video. Pattern Recognition 72, 285–299. doi:10.1016/j.patcog.2017.07.028
Tu, Z., Xie, W., Qin, Q., Poppe, R., Veltkamp, R. C., Li, B., et al. (2018). Multi-Stream CNN: Learning Representations Based on Human-Related Regions for Action Recognition. Pattern Recognition 79, 32–43. doi:10.1016/j.patcog.2018.01.020
Wang, S. H., Govindaraj, V., Gorriz, J. M., Zhang, X., and Zhang, Y. D. (2021). Explainable Diagnosis of Secondary Pulmonary Tuberculosis by Graph Rank-Based Average Pooling Neural Network. J. Ambient Intelligence Humanized Comput. 13, 1–14. doi:10.1007/s12652-021-02998-0
Yang, S., Wang, J., Zhang, N., Deng, B., Pang, Y., and Azghadi, M. R. (2021). CerebelluMorphic: Large-Scale Neuromorphic Model and Architecture for Supervised Motor Learning. IEEE Trans. Neural Netw. Learn. Syst. (99), 1–15. doi:10.1109/tnnls.2021.3057070
Zhang, X., Tian, J., Deng, K., Wu, Y., and Li, X. (2010). Automatic Liver Segmentation Using a Statistical Shape Model with Optimal Surface Detection. IEEE Trans. Biomed. Eng. 57 (10), 2622–2626. doi:10.1109/tbme.2010.2056369
Zhou, Z., Siddiquee, M. M. R., Tajbakhsh, N., and Liang, J. (2020). UNet++: Redesigning Skip Connections to Exploit Multiscale Features in Image Segmentation. IEEE Trans. Med. Imaging 39, 1856–1867. doi:10.1109/TMI.2019.2959609
Keywords: image segmentation, deep learning, weighted pooling, U-net, skip connection
Citation: Lei J, Lei T, Zhao W, Xue M, Du X and Nandi AK (2022) Rethinking Pooling Operation for Liver and Liver-Tumor Segmentations. Front. Sig. Proc. 1:808050. doi: 10.3389/frsip.2021.808050
Received: 02 November 2021; Accepted: 20 December 2021;
Published: 10 January 2022.
Edited by:
Yuming Fang, Jiangxi University of Finance and Economics, ChinaCopyright © 2022 Lei, Lei, Zhao, Xue, Du and Nandi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Asoke K. Nandi, YXNva2UubmFuZGlAYnJ1bmVsLmFjLnVr; Tao Lei, bGVpdGFvQHN1c3QuZWR1LmNu
 Junchao Lei1,2
Junchao Lei1,2 Tao Lei
Tao Lei Asoke K. Nandi
Asoke K. Nandi