
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Sens. , 28 October 2022
Sec. Lab-on-a-Chip Devices
Volume 3 - 2022 | https://doi.org/10.3389/fsens.2022.1012775
This article is part of the Research Topic Editors’ Showcase: Lab-on-a-chip Devices View all 8 articles
Extensive research is focused on the development of highly sensitive, rapid on-site diagnostic devices. The lateral flow strip (LFS) is a paper-based point-of-care diagnostic device, which is highly promising because of its ease of use and low cost. Despite these advantages, LFS device is still less popular than other methods such as enzyme-linked immunosorbent assay (ELISA) or real-time polymerase chain reaction (qPCR) due to its low sensitivity. Here, we have developed a fluorescence-based lateral flow strip (f-LFS) device for DNA detection using a molecular beacon (MB), a short hairpin-forming DNA strand tagged with a fluorophore-quencher pair. Each paper and membrane component of f-LFS device was carefully selected based on their physicochemical properties including porosity, surface functionality, and autofluorescence. The limit of detection (LOD) of this device was substantially improved to 2.1 fg/mL by adding MgCl2 to the reaction buffer and narrowing the test membrane dimension. Also, a portable fluorescence detection system for f-LFS was developed using a multi-pixel photon counter (MPPC), a sensitive detector detecting the signal on site. We anticipate that this highly sensitive paper-based diagnostic device can be utilized for on-site diagnosis of various diseases.
Various methods are available for biomolecular diagnoses, such as real-time polymerase chain reaction (qPCR), high-performance liquid chromatography (HPLC), enzyme-linked immunosorbent assay (ELISA), etc (Matuszewski et al., 2003; Postollec et al., 2011; Aydin, 2015; Choi et al., 2019). Although these techniques enable precise and accurate detection, they require expertise and involve complicated procedures. To overcome these drawbacks, researchers have developed various microfluidic devices, e.g. digital microfluidics, modular microfluidics, and paper-based microdevices which utilize loop-mediated isothermal amplification (LAMP) of DNA or RNA, surface-enhanced Raman scattering (SERS) detection, or qPCR (Wang et al., 2009; Lo et al., 2013; Choi et al., 2015; Song et al., 2019). Amongst these recently developed methods, paper-based diagnostic devices (i.e., lateral flow strip, LFS) are some of the most promising technologies (Jafry et al., 2017; Cho et al., 2019; Jafry et al., 2019). They are capable of fast and on-site detection of various conditions or diseases; e.g., infectious organisms, drug addiction, and disease monitoring crucial to human physiology, along with animal disease and environmental testing (Anfossi et al., 2018; Land et al., 2019; Lim et al., 2019; Di Nardo et al., 2021).
The commercially available LFS device consists of cellulose-based sample pad and absorbent pad, a glass fiber-based conjugate pad, and a nitrocellulose-based test membrane (St John and Price, 2014; Sajid et al., 2015). The test line in the middle of the test membrane contains antibody-conjugated gold nanoparticles, which give rise to a color change when bound with antigen molecules. There are two distinct detection methods for LFS; sandwich and competitive immunoassays. The sandwich immunoassay is a method using two antibodies, which bind to different sites on the antigen (Cox et al., 2019). A labeled detector antibody moves through the test membrane with the antigens of interest. The antigen-antibody complex is captured by an immobilized, antigen-specific antibody, resulting in a colored line (Ching, 2015). The competitive immunoassay is based on antibody-antigen interactions in which the number of antigen-binding sites on the antibody is limited (Clarke, 2004). In this assay, the antigen of interest is immobilized on the test membrane. If antigen is present in the sample, it will bind with the detector antibody, and the complex will flow past the immobilized antigen, resulting in no colored line (Ching, 2015). This LFS device has been used for detecting not only antigen but also nucleic acids (Chen and Yang, 2015; Yao et al., 2019). In these applications, nucleic acid probes conjugated to the nanoparticles bind with their complementary target strands to cause nanoparticle aggregation and coloring of the test line. However, these conventional LFS devices suffer from low throughput (single target detection per loading) and low sensitivity.
To address these drawbacks, numerous devices have been developed since LFS device was invented and patented in the 1970s (Deutsch and Mead, 1978). To detect multiple targets for LFS, some research groups have reported the use of multiple test lines (Song et al., 2014; Peng et al., 2016; Wang et al., 2018). However, such LFS devices need a larger footprint and sample volume, and the nonlinear velocity reduction along a longer test line produces an inconsistent reaction time. Therefore, other types of LFS were developed to detect multiple antigens in one test line using nanoparticles of multiple shapes and colors. For example, several groups fabricated such LFS devices using red nanoparticles and blue nanoparticles (Kim et al., 2019; Rodriguez-Quijada et al., 2020). They conjugated different antibodies to the red nanoparticles and blue nanoparticles to detect multiple targets on one test line. When one antigen is detected, the test line turns blue or red, and when two antigens are detected simultaneously, the test line turns purple.
Additionally, the general limit of detection (LOD) of LFS is as low as 0.1–10 ng/mL when it is detected by color appearance or change (Nguyen et al., 2020). Several studies have been conducted to modify the materials or the detection method to improve LOD. Parolo et al. improved LOD to ∼ 200 pg/mL by functionalizing the surface of gold nanoparticles with an enzyme called horseradish peroxidase and adding a metal enhancer (Parolo et al., 2013). The enzyme produces insoluble and non-flowing chromogens, which are then captured by the enhancers on the gold nanoparticles, leading to a 5-fold increase in the color intensity of the test line. The color intensity of LFS was also enhanced by a double-targeted detection method, where antibodies of a target antigen and proteins are conjugated to small-sized gold nanoparticles, and antibodies of the protein to large-sized gold nanoparticles (Choi et al., 2010). The smaller gold nanoparticles were caught by antigens at the test line, and sequentially the larger nanoparticles are caught by the smaller gold nanoparticles. This double-targeted detection method increased the sensitivity ∼ 10- to 100-fold (LOD ∼ 10–100 pg/mL). Takalkar et al. enhanced the LOD of LFS ∼15-fold up to ∼ 67 pg/mL by combining large silica nanorods with a large number of gold nanoparticles (Takalkar et al., 2016).
Recently, researchers employed fluorescence to improve sensitivity using the portable fluorescence reader or fluorescence scanner, etc (Han et al., 2020; Lee et al., 2021). Berlina et al. and Qu et al. used quantum dots instead of gold nanoparticles for LFS (Berlina et al., 2013; Qu et al., 2016). They could quantitatively determine the antigen level from the signal-to-noise ratio (SNR) of the fluorescence signal. Lou et al. attached a fluorophore to a specific part of the antibody and use this to functionalize polystyrene microspheres (Lou et al., 2018). This functionalization greatly increased the fluorescence intensity of the polystyrene microspheres, resulting in ∼31-fold increase in sensitivity (LOD ∼ 32 pg/mL). Wang et al. fabricated a magnetic particle coated with three layers of quantum dots; the Fe3O4 triple QD-shell nanocomposite (named as MagTQD) (Wang et al., 2021). This nanocomposite particle had numerous quantum dots on the surface, which could improve the detection limit by about a factor of 2000 (LOD ∼ 0.5 pg/mL). Ao et al. fabricated the LFS device using the near infrared-II (NIR-II, 1,000–1700 nm) emitting lead sulfide quantum dots (Ao et al., 2022). They concentrated the quantum dots in porous silica particles to amplify the fluorescence intensity and improved the sensitivity of the LFS by about a factor of 9 (LOD ∼ 110 pg/mL).
In this study, we report a sensitive and rapid point-of-care fluorescence-based lateral flow strip device (f-LFS device). We used molecular beacons (MB), hairpin-shaped molecules composed of a single-stranded oligonucleotide probe labeled with a fluorophore and a quencher, to detect human immunodeficiency virus type 1 (HIV-1) as the target. To improve the detection sensitivity, the materials for the LFS device including paper and membrane were selected based on measurements of porosity, pore size, total intrusion volume, and autofluorescence. And, a portable fluorescence detection system was developed using multi-pixel photon counter (MPPC). The reactivity between the designed MB and the target nucleic acid in solution was enhanced by adding MgCl2 which serves as a charge screening agent. Furthermore, we improved the sensitivity by reducing the width of the test membrane, thus increasing the target concentration during flow. We expect the f-LFS device to be widely adopted in the diagnostic field due to its rapid processing, high sensitivity, low cost, and ease of fabrication.
Multiple Grade 1 chromatography papers, CF3, CF5, standard 17, glass fiber filter, and FF170HP were purchased from Whatman, United Kingdom, and AP1004700 and GFCP203000 were purchased from Millipore, United States. Deoxyribonucleic acid (DNA) oligos and Molecular Beacon (MB) conjugated with fluorophore (6-carboxyfluorescein; 6-FAM)-quencher (black hole quencher-1; BHQ-1) pair were purchased from Integrated DNA Technologies (IDT), United States. Sodium chloroacetate, N-(3-Dimethylaminopropyl)-N′-ethyl carbodiimide hydrochloride (EDC), N-hydroxysuccinimide (NHS), cystamine dihydrochloride, iodoacetic anhydride, 1,2-dichloroethane and tris(2-carboxyethyl)phosphine hydrochloride (TCEP) were purchased from Sigma-Aldrich, United States. Sodium hydroxide and acetone were purchased from Samchun Chemicals, Rep. of Korea. Polyethylene glycol spacer (maleimide-polyethylene glycol (PEG)-NH2, MW 2000) was purchased from Nanocs, United States. N, N-Dimethylformamide (99.8% purity) was purchased from Alfa Aesar, United States. The nucleic acid purifying kit (Oligo clean and concentrator) was purchased from Zymo Research, United States.
The paper and membrane characteristics of porosity, pore size, and total intrusion volume were measured using the automatic porosimeter (AutoPore IV, Micromeritics, United States) at Gacheon University, Rep. of Korea. A paper was placed inside a glass penetrometer bulb, which was then sealed and loaded into the automatic porosimeter. Afterwards, pressure was applied to permeate mercury through the paper, and the porosity, pore size, and total intrusion volume of the paper were calculated. The fiber width was calculated by averaging the fiber radii observed by the scanning electron microscope (SEM, JSM7000F, JEOL Ltd., Japan) at Sungkyunkwan University, Rep. of Korea.
For imaging, an inverted microscope (Ti-U, Nikon, Japan) with a 2X magnification objective lens was used. For fluorescence imaging of 6-carboxyfluorescein (6-FAM), we used a mercury lamp as the excitation source with an excitation bandpass filter (465–495 nm) and an emission filter (513–557 nm), the center wavelengths of which overlap with the excitation and emission wavelengths of 6-carboxyfluorescein (6-FAM), respectively. Images were taken using a CMOS camera (UCMOS03100KPA, Hangzhou ToupTek Photonics Co., Ltd., China).
For MB to be immobilized on glass fiber, N, N-Bis(α-iodoacetyl)-2, 2′-dithiobis (ethylamine) (BIDBE) was synthesized to create the thiol group (-SH) on MB, following the literature procedure (Ludueña et al., 1981). First, 18 mM of cystamine dihydrochloride solution (1 mL) in 0.1 M of NaOH was mixed with 220 mM of iodoacetic anhydride (0.25 mL) in 1, 2-dichloroethane solution, and the mixture was vortexed for 1 min. The white precipitate in this solution was collected via centrifugation (Centrifuge 5430R, Eppendorf, Germany) for 15 min (12,000 rcf and 20°C) and dried in a vacuum oven. After drying, the precipitate was dissolved in acetone and centrifuged (12,000 rcf and 15°C) for 15 min to remove any remaining iodoacetate. The supernatant was collected from the centrifuged solution and dried to obtain white BIDBE powder. For linking BIDBE to phosphorothioate on MB and reducing the disulfide bond in BIDBE, we followed a previously published protocol (Lee et al., 2007; Lee et al., 2010). To make BIDBE conjugated MB, 1 mM of MB containing phosphorothioate modification (10 μL) was mixed with 100 mM of BIDBE solution (20 μL, dissolved in DMF) and 10 mM of phosphate buffer (36 μL, pH 7.0) followed by heating at 50°C for 5–6 h. After heating, BIDBE conjugated MB (BIDBE-MB) was washed and collected via centrifugation using the Oligo clean and concentrator. To cleave the disulfide bond from BIDBE, 12 μM of BIDBE-MB (100 μL), 50 mM of acetate buffer (1 μL, pH 5.2) and 1 mM of TCEP (5 μL) were mixed and incubated at room temperature for 1 h, followed by washing and collecting via centrifugation using the Oligo clean and concentrator kit again.
To attach thiol-modified MB to glass fibers in the test membrane, we terminated the glass surface with maleimide groups in a two-step process. The hydroxyl group (-OH) of glass fiber was substituted with a carboxyl group (-COOH) in a reaction with a solution of 3 M of sodium chloroacetate and 10 wt% of sodium hydroxides at 90°C for 4 h, followed by washing with DI water for 10 min twice. The surface-modified glass fiber was immersed in 10 mM of EDC solution followed by 10 mM of NHS solution at room temperature for 3 h. EDC and NHS solutions were used at 1 mL per 1 cm2 of glass fiber. Afterwards, the substrate was washed thoroughly with DI water and immersed in 10 nM of maleimide-PEG-NH2 solution at room temperature for 12 h to functionalize the glass fiber surface. After maleimide incorporation, the surface was washed using DI water. Finally, MB-thiol was applied to the surface for conjugation to the PEG polymer on glass fiber.
A CO2 laser cutter (VLS 6.60, Universal Laser Systems, United States) and a blade cutter (silhouette cameo 2, Silhouette America, United States) were used to cut the selected papers and membranes after characterization. To construct the f-LFS device, the sample pad, test membrane and absorbent pad were fixed to a lateral flow backing card (Adhesive polyester film, Fancylobby, Rep. of Korea) with an overlap of 1 mm between them. The test line was formed by drawing MB solution onto the membrane using an in-house linear motor control system connected with the reagent dispenser (Claremont Biosolutions LLC, United States) and a syringe pump (PHD Ultra Advanced Syringe Pump, Harvard Apparatus, United States). After several trials, we found the optimal flow rate and concentration of MB solution (20 μL/min at 0.5 μM), linear motor speed (150 mm/min), and number of drawings (one time) needed to form the test line. The inverted microscope and a multi-pixel photon counter (MPPC) (C11208 MPPC module, Hamamatsu Photonics, Japan) were used to detect the fluorescence intensity from the f-LFS device and MB before and after binding with target DNA. To check the effect of the MgCl2 on the interaction between the MB and target, a multimode microplate reader (Varioskan LUX, ThermoFisher, United States) was used to measure the fluorescence intensity in DI water. To evaluate the selectivity and sensitivity of f-LFS, 150 μL of the target and non-target DNA molecules were placed on the sample pad at concentrations ranging from 0.21 to 210 fg/mL. The fluorescence intensities with and without DNA at the test line were measured using the fluorescence measurement system consists of MPPC, and the signal-to-noise ratio (SNR) was calculated.
For the conventional LFS device, the conjugate pad holds nanoparticles functionalized with antibodies or nucleic acid probes. In comparison, our f-LFS device does not have a conjugate pad, and MBs were directly attached to the fibers of the test membrane. Our f-LFS device (0.5 cm × 9.8 cm; 1-mm overlap between paper and membrane) consists of a sample pad (0.5 cm × 2 cm), MB-functionalized test membrane (0.5 cm × 4 cm), and an absorbent pad (0.5 cm × 4 cm), which are all placed on an adhesive backing card as illustrated in Figure 1A. Activating the paper surface with carboxyl groups and subsequent reactions with EDC/NHS and maleimide-PEG-NH2 expose maleimide groups on the surface. These maleimides react with the thiol groups on MB, leading to MB-anchored paper shown in Figure 1B.
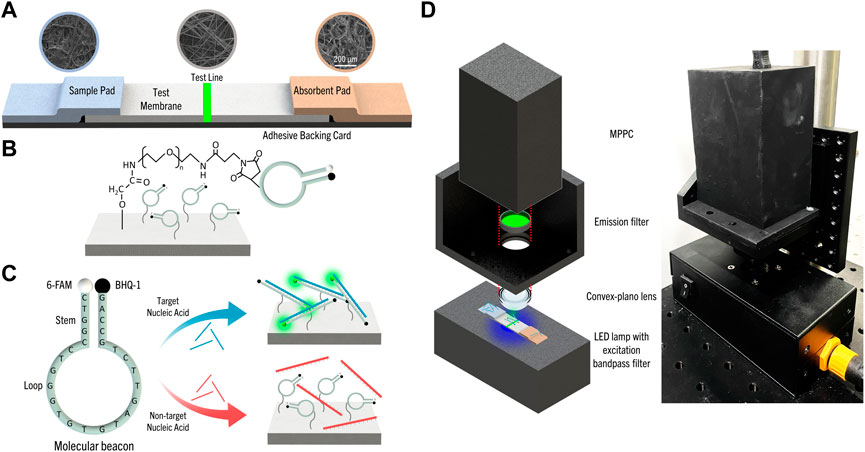
FIGURE 1. (A) Schematic of f-LFS device and SEM images of each part of f-LFS device; sample pad: CF3, test membrane: GFCP203000, and absorbent pad: AP1004700. (B) Schematic of MB attachment to the paper made of glass fiber using maleimide-PEG-NH2. (C) The MB-based detection mechanism of the target nucleic acid in f-LFS device. (D) The portable MPPC fluorescence detection system.
The schematic shown in Figure 1C represents the mechanism for detecting the target nucleic acid using MB. HIV-1, one of the viruses that require continuous monitoring of infection and on-site diagnosis has various mutations, such as U3, R, US and the U3 region has low variability among the gene regions of HIV-1 (Tatt et al., 2001; Mbondji-Wonje et al., 2018). Therefore, the U3 region of HIV-1 was selected as the target for detecting HIV-1 and our MB is based on the U3 region of HIV-1 (5′-CTGGCCCTGGTGTGTAGTTCTGCC-3′) with the addition of 5′-AG-3′ to the 3′-end for detecting short nucleic acid target molecules, such as microRNA, which are biomarkers for disease detection (Lee et al., 2022). The complementary sequences 5′-CTGGC-3′ and 5′-GCCAG-3′ at the ends form the stem, and the rest of the sequence forms the loop. The fluorophore, 6-FAM, and the quencher, BHQ-1, were incorporated at 5′- and 3′-end respectively. Using the OligoAnalyzerTM Tool (IDT, United States), the Gibbs free energy for binding of the MB to its complementary target (5′-GGCAGAACTACACACCAGGGCCAGGGATCA-3′) is calculated to be -47.64 kcal/mol. We also designed a non-target DNA with the sequence 5′-GCTTTCCTTACGTGACGTCGATCGGAGTCT-3′, whose Gibbs free energy for binding to MB is -4.64 kcal/mol, much weaker than the target as expected. The free energy between MBs of the same sequence is also much lower at -9.28 kcal/mol. Hence, fluorescence emission from MB predominantly arises from binding with target DNA.
The portable fluorescence detection system was developed, as shown in Figure 1D, to measure the selectivity and sensitivity of fluorescence, quantitatively. The light source of a LED light module (29 mm-3030–7 LED-CL, WELED, Rep. of Korea) was used as the excitation source with the excitation band-pass filter (465–495 nm) and the emission band-pass filter (513–557 nm) for detecting 6-FAM functionalized in MB. Also, the system had a convex plano lens to focus and collect the emission from the test line only. And, a very sensitive MPPC was used for fluorescence measurement unlike most strip tests which are detected with naked eyes and have sensitivity issues, so that the control line was not introduced in the developed LFS (Kang and Kim, 2020; Jung et al., 2022).
To fabricate the f-LFS device, we first selected suitable paper and membrane materials. The selected materials and their characteristics such as the porosity, pore diameter, and total intrusion volume are listed in Table 1. The list includes cellulose (chromatography paper, CF3, CF5, and AP1004700), glass fiber (GFCP203000, standard 17, and glass fiber filter), and nitrocellulose (FF170HP). Here, the porosity is the ratio of the void to the total volume of paper, and the total intrusion volume is the absorbed volume of liquid for unit mass of paper.
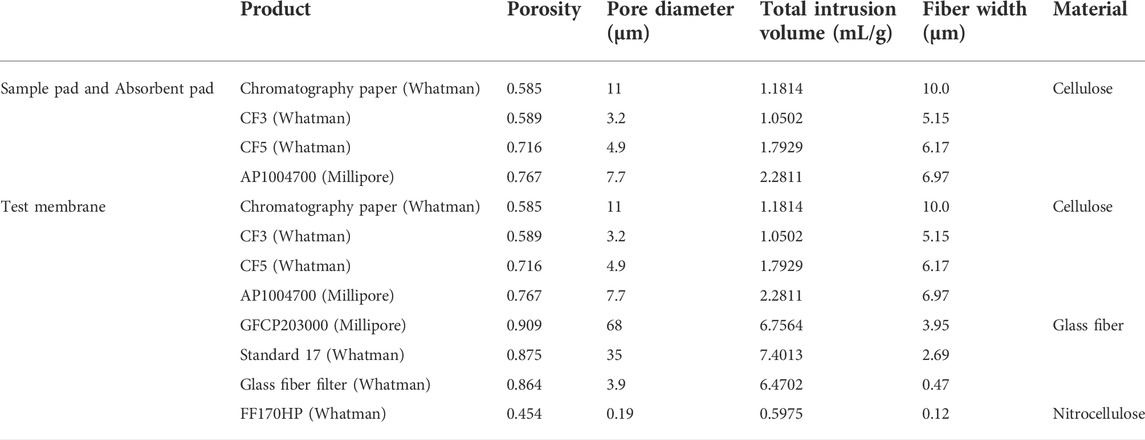
TABLE 1. Physical characteristics of papers for choosing sample pad, absorbent pad, and test membrane.
The total intrusion volume is the primary factor for selecting the sample pad and absorbent pad. Papers with a large total intrusion volume retain fluid better than those with a small total intrusion volume. Therefore, small intrusion volume is suitable for the sample pad, but large intrusion volume for the absorbent pad. Among the products, CF3 was selected as the sample pad because of its smallest total intrusion volume, and AP1004700 from Millipore was selected as the absorbent pad for having the largest total intrusion volume.
The absence of autofluorescence of the material is essential for selecting the test membrane. We measured the fluorescence intensities of the test membrane candidates: (i) FF170HP for nitrocellulose, (ii) chromatography paper for cellulose, and (iii) GFCP203000 for glass fiber (Figure 2A). Because GFCP203000 showed the lowest level of autofluorescence, we concluded that glass fiber was the most suitable material for the test membrane. Next, we took images of different glass fibers functionalized with MB (Figure 2B) and calculated their standard deviations of pixel intensity to quantify the uniformity of MB functionalization; (i) glass fiber filter, (ii) standard 17, and (iii) GFCP203000. The standard deviation was calculated as
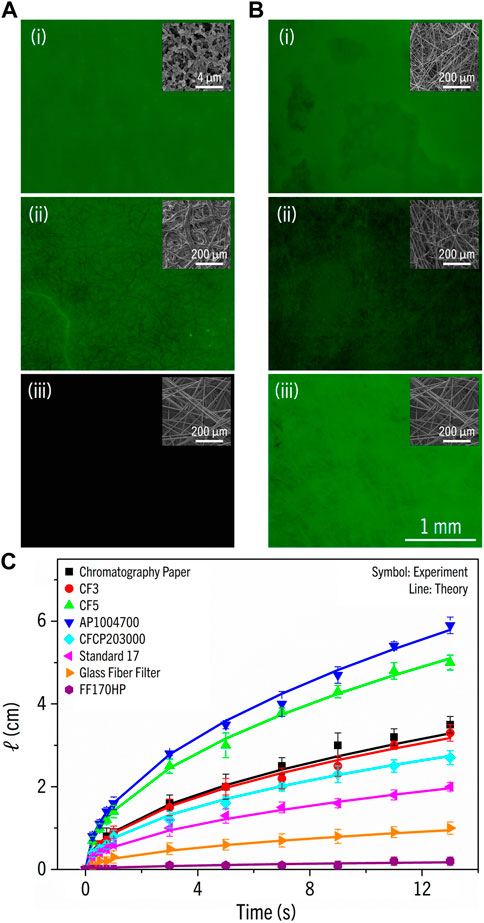
FIGURE 2. (A) The fluorescence images of papers: (i) FF170HP, (ii) chromatography paper, and (iii) GFCP203000. Images were taken with the excitation wavelength λ = 465–495 nm and the emission wavelength λ = 513–557 nm. The insets are SEM images. (B) The fluorescence images of glass fibers functionalized with molecular beacon; (i) glass fiber filter, (ii) standard 17, and (iii) GFCP203000. The insets are SEM images. (C) The flow penetration length (cm) vs. time (s) for eight test membranes.
In lateral flow strip, the reaction time must be optimized for the signal to reflect the true reactivity between MB and the target nucleic acid; if the reaction time is too long, nonspecific reactions may produce a false negative. The fluid flow through a porous medium such as paper where flow penetration is induced by surface tension can be described by the Lucas-Washburn equation. The penetration length is expressed as (Mendez et al., 2010; Jafry et al., 2016):
where
where
The reactivity between MB and the target DNA in a buffer without salt was extremely low, as indicated by the measured fluorescence close to the base line level (blue line, Figure 3A). This low reactivity is expected without salt because of the repulsion between the negatively charged phosphate backbones of MB and target. Hence, we added MgCl2 to the buffer, which is known to increase the hybridization rate of DNA due to effective charge screening of DNA by the divalent ions (Misra and Draper, 1999). Consistent with this prediction, the fluorescence intensity of MB increased as Mg2+ concentration increased (Figure 3A). In a control experiment, we also showed that MgCl2 alone is not responsible for this fluorescence increase (Figure 3B).
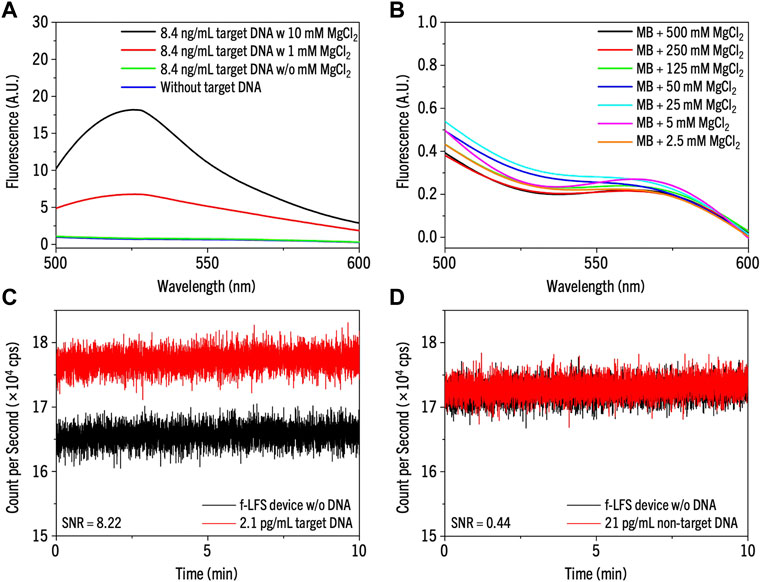
FIGURE 3. (A,B) The reactivity test in the solution of (A) only MB without target DNA (blue line) and MB with 8.4 ng/mL target DNA with MgCl2 of 10 mM (black), 1 mM (red), and without MgCl2 (green), and (B) MB without target DNA varying the concentration of MgCl2 from 2.5 mM to 500 mM. (C,D) The selectivity test of the MB on f-LFS device with 500 mM of MgCl2 for (C) 2.1 pg/mL of target DNA and (D) 21 pg/mL of non-target DNA.
After we confirmed the specificity of hybridization between MB and target DNA in solution, we tested the same reaction on the f-LFS platform. We spotted 2.1 pg/mL of the target DNA and 21 pg/mL of the non-target DNA on the sample pad of the f-LFS device and waited until the DNA sample reached the MB-coated test line on the test membrane. The fluorescence signal was then measured using MPPC. We chose SNR larger than 3 as the positive signal for target DNA detection (Sharma et al., 2014). Figures 3C,D show the results with the target and the non-target DNA solutions, respectively. Although we used a 10-fold lower target DNA concentration (2.1 pg/mL) compared to the non-target DNA (21 pg/mL), the SNR value from target DNA (8.22) was much higher than that from non-target DNA (0.44), suggesting high specificity of our MB-based f-LFS device.
We also tried different methods to increase the sensitivity of the device. We reasoned that increasing the effective concentrations of MB and target DNA at the point of detection would increase the sensitivity, and confining the reactants to a narrower area could bring about such an effect (Eriksson et al., 2019). Hence, we reduced the width of the test membrane to 0.3 cm and measured the limit of detection (LOD) by applying 150 μL of target DNA solution to the MB-coated test line while varying DNA concentrations from 210 to 0.21 fg/mL. As shown in Figure 4, SNR of the original f-LFS device dropped below 3 at 2.1 fg/mL of target DNA, and therefore, the LOD was determined to be 21 fg/mL. In comparison, the modified f-LFS device with the narrower test membrane yielded SNR = 3.23 at 2.1 fg/mL of target DNA, resulting in a higher LOD at 2.1 fg/mL. Our f-LFS device thus represents a significant improvement over LFS device of previous studies (Mao et al., 2009; Liu et al., 2019).
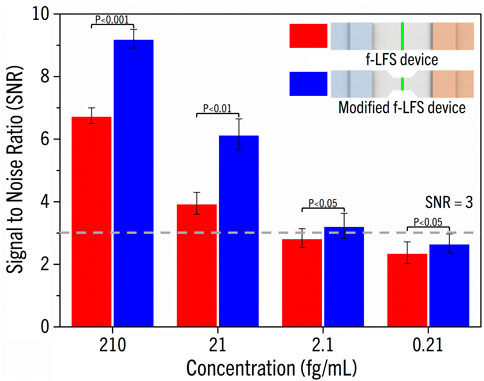
FIGURE 4. SNR showing for both f-LFS and modified f-LFS devices. The LOD of f-LFS is 21 fg/mL, and 2.1 fg/mL for modified f-LFS.
A novel f-LFS device was fabricated using a sample pad, an absorbent pad, and a test membrane. Each component was selected based on its measured mechanical properties (pore size, porosity and total intrusion volume) and autofluorescence. We selected CF3 with a small total intrusion volume and AP1004700 with a large intrusion volume as the sample pad and the absorbent pad, respectively. To maximize the SNR and avoid false positives, we selected GFCP203000 with the minimum autofluorescence and large void size as the test membrane. To test f-LFS performance, we selected the U3 region with low variability among the genetic regions from HIV-1 and designed a MB labeled with a dye-quencher pair. The MB was loaded on the test membrane using an in-house test line drawer. We showed that MgCl2 is critical for enhancing the binding reaction between MB and target DNA, probably because of the screening capability of Mg2+. The portable and sensitive detection system was developed using MPPC. We tested f-LFS devices with different widths and showed that the narrower one performed better than the wider one, with a 10-fold improvement in LOD from 21 fg/mL to 2.1 fg/mL. In conclusion, we demonstrated the excellent performance of a novel MB-based f-LFS device for DNA detection. We believe that this portable f-LFS device can find many applications for point-of-care disease diagnosis.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
This work was supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (IPET) through the Animal Disease Management Technology Development Program, funded by the Ministry of Agriculture, Food and Rural Affairs (MAFRA) (No. 118094-03), by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF 2019R1A6A1A03033215), and the Ministry of Science, ICT & Future Planning (NRF 2020R1A2C3010568), and by Brain Pool program funded by the Ministry of Science and ICT through the National Research Foundation of Korea (NRF 2021H1D3A2A01099478).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Anfossi, L., Di Nardo, F., Cavalera, S., Giovannoli, C., and Baggiani, C. (2018). Multiplex lateral flow immunoassay: An overview of strategies towards high-throughput point-of-need testing. Biosensors 9 (1), 2. doi:10.3390/bios9010002
Ao, L., Liao, T., Huang, L., Lin, S., Xu, K., Ma, J., et al. (2022). Sensitive and simultaneous detection of multi-index lung cancer biomarkers by an NIR- fluorescence lateral-flow immunoassay platform. Chem. Eng. J. 436, 135204. doi:10.1016/j.cej.2022.135204
Aydin, S. (2015). A short history, principles, and types of ELISA, and our laboratory experience with peptide/protein analyses using ELISA. Peptides 72, 4–15. doi:10.1016/j.peptides.2015.04.012
Berlina, A. N., Taranova, N. A., Zherdev, A. V., Vengerov, Y. Y., and Dzantiev, B. B. (2013). Quantum dot-based lateral flow immunoassay for detection of chloramphenicol in milk. Anal. Bioanal. Chem. 405 (14), 4997–5000. doi:10.1007/s00216-013-6876-3
Chen, A., and Yang, S. (2015). Replacing antibodies with aptamers in lateral flow immunoassay. Biosens. Bioelectron. 71, 230–242. doi:10.1016/j.bios.2015.04.041
Cho, H. H., Kim, S. J., Jafry, A. T., Lee, B., Heo, J. H., Yoon, S., et al. (2019). A paper‐based platform for long‐term deposition of nanoparticles with exceptional redispersibility, stability, and functionality. Part. Part. Syst. Charact. 36 (6), 1800483. doi:10.1002/ppsc.201800483
Choi, D. H., Lee, S. K., Oh, Y. K., Bae, B. W., Lee, S. D., Kim, S., et al. (2010). A dual gold nanoparticle conjugate-based lateral flow assay (LFA) method for the analysis of troponin I. Biosens. Bioelectron. 25 (8), 1999–2002. doi:10.1016/j.bios.2010.01.019
Choi, J. R., Tang, R., Wang, S., Abas, W. A. B. W., Pingguan-Murphy, B., and Xu, F. (2015). Paper-based sample-to-answer molecular diagnostic platform for point-of-care diagnostics. Biosens. Bioelectron. 74, 427–439. doi:10.1016/j.bios.2015.06.065
Choi, J. R., Yong, K. W., Choi, J. Y., and Cowie, A. C. (2019). Emerging point-of-care technologies for food safety analysis. Sensors 19 (4), 817. doi:10.3390/s19040817
Clarke, W. (2004). Immunoassays for therapeutic drug monitoring and clinical toxicology. Drug Monit. Clin. Chem. 5, 95–112. doi:10.1016/S1567-7192(04)80005-6
Cox, K. L., Devanarayan, V., Kriauciunas, A., Manetta, J., Montrose, C., and Sittampalam, S. (2019). Immunoassay methods. Assay guidance manual. Bethesda, MD: Eli Lilly & Company and the National Center for Advancing Translational Sciences.
Deutsch, M. E., and Mead, L. W. (1978). Test device. U.S. Patent No. 4,094,647. Washington, DC: U.S. Patent and Trademark Office.
Di Nardo, F., Chiarello, M., Cavalera, S., Baggiani, C., and Anfossi, L. (2021). Ten years of lateral flow immunoassay technique applications: Trends, challenges and future perspectives. Sensors 21 (15), 5185. doi:10.3390/s21155185
Eriksson, E., Lysell, J., Larsson, H., Cheung, K. Y., Filippini, D., and Mak, W. C. (2019). Geometric flow control lateral flow immunoassay devices (GFC-LFIDs): A new dimension to enhance analytical performance. Research 2019, 8079561–8079568. doi:10.34133/2019/8079561
Gebart, B. R. (1992). Permeability of unidirectional reinforcements for RTM. J. Compos. Mater. 26 (8), 1100–1133. doi:10.1177/002199839202600802
Han, Y. D., Chun, H. J., and Yoon, H. C. (2020). Low-cost point-of-care biosensors using common electronic components as transducers. BioChip J. 14 (1), 32–47. doi:10.1007/s13206-020-4104-8
Jafry, A. T., Lee, H., Tenggara, A. P., Lim, H., Moon, Y., Kim, S.-H., et al. (2019). Double-sided electrohydrodynamic jet printing of two-dimensional electrode array in paper-based digital microfluidics. Sensors Actuators B Chem. 282, 831–837. doi:10.1016/j.snb.2018.11.135
Jafry, A. T., Lim, H., Kang, S. I., Suk, J. W., and Lee, J. (2016). A comparative study of paper-based microfluidic devices with respect to channel geometry. Colloids Surfaces A Physicochem. Eng. Aspects 492, 190–198. doi:10.1016/j.colsurfa.2015.12.033
Jafry, A. T., Lim, H., Sung, W.-K., and Lee, J. (2017). Flexible time–temperature indicator: A versatile platform for laminated paper-based analytical devices. Microfluid. Nanofluidics 21 (3), 57–13. doi:10.1007/s10404-017-1883-x
Jung, D. H., Kim, Y., Cho, H. H., Lee, B., Suh, S.-J., Heo, J. H., et al. (2022). Automatic quantification of living cells via a non-invasive achromatic colorimetric sensor through machine learning-assisted image analysis using a smartphone. Chem. Eng. J. 450, 138281. doi:10.1016/j.cej.2022.138281
Kang, J., and Kim, M.-G. (2020). Advancements in DNA-assisted immunosensors. BioChip J. 14 (1), 18–31. doi:10.1007/s13206-020-4103-9
Kim, J., Cao, X. E., Finkelstein, J. L., Cárdenas, W. B., Erickson, D., and Mehta, S. (2019). A two-colour multiplexed lateral flow immunoassay system to differentially detect human malaria species on a single test line. Malar. J. 18 (1), 313–410. doi:10.1186/s12936-019-2957-x
Land, K. J., Boeras, D. I., Chen, X.-S., Ramsay, A. R., and Peeling, R. W. (2019). REASSURED diagnostics to inform disease control strategies, strengthen health systems and improve patient outcomes. Nat. Microbiol. 4 (1), 46–54. doi:10.1038/s41564-018-0295-3
Lee, J. H., Wernette, D. P., Yigit, M. V., Liu, J., Wang, Z., and Lu, Y. (2007). Site-specific control of distances between gold nanoparticles using phosphorothioate anchors on DNA and a short bifunctional molecular fastener. Angew. Chem. Int. Ed. Engl. 119 (47), 9006–9010. doi:10.1002/anie.200702569
Lee, J. H., Wong, N. Y., Tan, L. H., Wang, Z., and Lu, Y. (2010). Controlled alignment of multiple proteins and nanoparticles with nanometer resolution via backbone-modified phosphorothioate DNA and bifunctional linkers. J. Am. Chem. Soc. 132 (26), 8906–8908. doi:10.1021/ja103739f
Lee, J. S., Ahn, J. J., Kim, S. J., Yu, S. Y., Koh, E. J., Kim, S. H., et al. (2021). POCT detection of 14 respiratory viruses using multiplex RT-PCR. BioChip J. 15 (4), 371–380. doi:10.1007/s13206-021-00037-w
Lee, W.-J., Kim, K.-J., Hossain, M., Cho, H.-Y., and Choi, J.-W. (2022). DNA–gold nanoparticle conjugates for intracellular MiRNA detection using surface-enhanced Raman spectroscopy. BioChip J. 16 (1), 33–40. doi:10.1007/s13206-021-00042-z
Lim, H., Jafry, A. T., and Lee, J. (2019). Fabrication, flow control, and applications of microfluidic paper-based analytical devices. Molecules 24 (16), 2869. doi:10.3390/molecules24162869
Liu, Q., Lin, Y., Xiong, J., Wu, L., Hou, X., Xu, K., et al. (2019). Disposable paper-based analytical device for visual speciation analysis of Ag (I) and silver nanoparticles (AgNPs). Anal. Chem. 91 (5), 3359–3366. doi:10.1021/acs.analchem.8b04609
Liukkonen, A. (1997). Contact angle of water on paper components: Sessile drops versus environmental scanning electron microscope measurements. Scanning 19 (6), 411–415. doi:10.1002/sca.4950190604
Lo, S.-J., Yang, S.-C., Yao, D.-J., Chen, J.-H., Tu, W.-C., and Cheng, C.-M. (2013). Molecular-level dengue fever diagnostic devices made out of paper. Lab. Chip 13 (14), 2686–2692. doi:10.1039/c3lc50135c
Lou, D., Fan, L., Cui, Y., Zhu, Y., Gu, N., and Zhang, Y. (2018). Fluorescent nanoprobes with oriented modified antibodies to improve lateral flow immunoassay of cardiac troponin I. Anal. Chem. 90 (11), 6502–6508. doi:10.1021/acs.analchem.7b05410
Ludueña, R. F., Roach, M. C., Trcka, P. P., and Weintraub, S. (1981). N, N-Bis (α-iodoacetyl)-2, 2′-dithiobis (ethylamine), a reversible crosslinking reagent for protein sulfhydryl groups. Anal. Biochem. 117 (1), 76–80. doi:10.1016/0003-2697(81)90694-1
Mao, X., Ma, Y., Zhang, A., Zhang, L., Zeng, L., and Liu, G. (2009). Disposable nucleic acid biosensors based on gold nanoparticle probes and lateral flow strip. Anal. Chem. 81 (4), 1660–1668. doi:10.1021/ac8024653
Matuszewski, B. K., Constanzer, M., and Chavez-Eng, C. (2003). Strategies for the assessment of matrix effect in quantitative bioanalytical methods based on HPLC− MS/MS. Anal. Chem. 75 (13), 3019–3030. doi:10.1021/ac020361s
Mbondji-Wonje, C., Dong, M., Wang, X., Zhao, J., Ragupathy, V., Sanchez, A. M., et al. (2018). Distinctive variation in the U3R region of the 5'Long Terminal Repeat from diverse HIV-1 strains. PloS one 13 (4), e0195661. doi:10.1371/journal.pone.0195661
Mendez, S., Fenton, E. M., Gallegos, G. R., Petsev, D. N., Sibbett, S. S., Stone, H. A., et al. (2010). Imbibition in porous membranes of complex shape: Quasi-stationary flow in thin rectangular segments. Langmuir 26 (2), 1380–1385. doi:10.1021/la902470b
Misra, V. K., and Draper, D. E. (1999). The interpretation of Mg2+ binding isotherms for nucleic acids using Poisson-Boltzmann theory. J. Mol. Biol. 294 (5), 1135–1147. doi:10.1006/jmbi.1999.3334
Nabovati, A., Llewellin, E. W., and Sousa, A. C. (2009). A general model for the permeability of fibrous porous media based on fluid flow simulations using the lattice Boltzmann method. Compos. Part A Appl. Sci. Manuf. 40 (6-7), 860–869. doi:10.1016/j.compositesa.2009.04.009
Nguyen, V.-T., Song, S., Park, S., and Joo, C. (2020). Recent advances in high-sensitivity detection methods for paper-based lateral-flow assay. Biosens. Bioelectron. 152, 112015. doi:10.1016/j.bios.2020.112015
Parolo, C., de la Escosura-Muñiz, A., and Merkoçi, A. (2013). Enhanced lateral flow immunoassay using gold nanoparticles loaded with enzymes. Biosens. Bioelectron. 40 (1), 412–416. doi:10.1016/j.bios.2012.06.049
Peng, J., Wang, Y., Liu, L., Kuang, H., Li, A., and Xu, C. (2016). Multiplex lateral flow immunoassay for five antibiotics detection based on gold nanoparticle aggregations. RSC Adv. 6 (10), 7798–7805. doi:10.1039/c5ra22583c
Postollec, F., Falentin, H., Pavan, S., Combrisson, J., and Sohier, D. (2011). Recent advances in quantitative PCR (qPCR) applications in food microbiology. Food Microbiol. 28 (5), 848–861. doi:10.1016/j.fm.2011.02.008
Qu, H., Zhang, Y., Qu, B., Kong, H., Qin, G., Liu, S., et al. (2016). Rapid lateral-flow immunoassay for the quantum dot-based detection of puerarin. Biosens. Bioelectron. 81, 358–362. doi:10.1016/j.bios.2016.03.008
Rodriguez-Quijada, C., Gomez-Marquez, J., and Hamad-Schifferli, K. (2020). Repurposing old antibodies for new diseases by exploiting cross-reactivity and multicolored nanoparticles. ACS Nano 14 (6), 6626–6635. doi:10.1021/acsnano.9b09049
Sajid, M., Kawde, A.-N., and Daud, M. (2015). Designs, formats and applications of lateral flow assay: A literature review. J. Saudi Chem. Soc. 19 (6), 689–705. doi:10.1016/j.jscs.2014.09.001
Sharma, H., Wood, J. B., Lin, S., Corn, R. M., and Khine, M. (2014). Shrink-induced silica multiscale structures for enhanced fluorescence from DNA microarrays. Langmuir 30 (37), 10979–10983. doi:10.1021/la501123b
Song, R., Abbasi, M. S., and Lee, J. (2019). Fabrication of 3D printed modular microfluidic system for generating and manipulating complex emulsion droplets. Microfluid. Nanofluidics 23 (7), 92–11. doi:10.1007/s10404-019-2258-2
Song, S., Liu, N., Zhao, Z., Njumbe Ediage, E., Wu, S., Sun, C., et al. (2014). Multiplex lateral flow immunoassay for mycotoxin determination. Anal. Chem. 86 (10), 4995–5001. doi:10.1021/ac500540z
St John, A., and Price, C. P. (2014). Existing and emerging technologies for point-of-care testing. Clin. Biochem. Rev. 35 (3), 155–167.
Takalkar, S., Xu, H., Chen, J., Baryeh, K., Qiu, W., Zhao, J. X., et al. (2016). Gold nanoparticle coated silica nanorods for sensitive visual detection of microRNA on a lateral flow strip biosensor. Anal. Sci. 32 (6), 617–622. doi:10.2116/analsci.32.617
Tatt, I. D., Barlow, K. L., Nicoll, A., and Clewley, J. P. (2001). The public health significance of HIV-1 subtypes. Aids 15, S59–S71. doi:10.1097/00002030-200100005-00009
Wang, C., Cheng, X., Liu, L., Zhang, X., Yang, X., Zheng, S., et al. (2021). Ultrasensitive and simultaneous detection of two specific SARS-CoV-2 antigens in human specimens using direct/enrichment dual-mode fluorescence lateral flow immunoassay. ACS Appl. Mat. Interfaces 13 (34), 40342–40353. doi:10.1021/acsami.1c11461
Wang, G., Lim, C., Chen, L., Chon, H., Choo, J., Hong, J., et al. (2009). Surface-enhanced Raman scattering in nanoliter droplets: Towards high-sensitivity detection of mercury (II) ions. Anal. Bioanal. Chem. 394 (7), 1827–1832. doi:10.1007/s00216-009-2832-7
Wang, Q., Liu, Y., Wang, M., Chen, Y., and Jiang, W. (2018). A multiplex immunochromatographic test using gold nanoparticles for the rapid and simultaneous detection of four nitrofuran metabolites in fish samples. Anal. Bioanal. Chem. 410 (1), 223–233. doi:10.1007/s00216-017-0714-y
Keywords: paper-based diagnostic device, molecular beacon (MB), nucleic acid detection, point-of-care (POC), lateral flow strip (LFS)
Citation: Moon Y, Moon H, Chang J, Kim HD, Lee JH and Lee J (2022) Development of a highly sensitive lateral flow strip device for nucleic acid detection using molecular beacons. Front. Sens. 3:1012775. doi: 10.3389/fsens.2022.1012775
Received: 05 August 2022; Accepted: 13 October 2022;
Published: 28 October 2022.
Edited by:
Chang-Soo Lee, Chungnam National University, South KoreaReviewed by:
Nandi Zhou, Jiangnan University, ChinaCopyright © 2022 Moon, Moon, Chang, Kim, Lee and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jung Heon Lee, amhsZWU3QHNra3UuZWR1; Jinkee Lee, bGVlLmppbmtlZUBza2t1LmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.