- 1School of Life Sciences, University of Nottingham, Nottingham, United Kingdom
- 2School of Biosciences, University of Nottingham, Loughborough, United Kingdom
- 3School of Veterinary Sciences, University of Nottingham, Loughborough, United Kingdom
Eukaryotic mRNA cap structures directly influence mRNA stability, translation, and immune recognition. While the significance of the mRNA cap itself has been well-established, recent research has revealed the intricate modifications to the nucleotides immediately adjacent to the cap. These include 2′-O-methylation (Cap1, Cap2) and N6-methyladenosine (m6Am), which recent work suggests may be dynamically regulated, interdependent, and vital for gene-regulation. This perspective explores the expanding role of cap-adjacent modifications. Cap1 is found on metazoan mRNA, viruses which infect metazoans, and the COVID-19 mRNA vaccines. We combined basic local alignment and 2D-TLC to track cap-adjacent modifying proteins and the modifications themselves to profile their prevalence throughout eukaryotes. We confirm that Cap1 is broadly metazoan specific, but surprisingly also in a brown algae. Additionally, we find genomic presence of the eraser, FTO more predictive of the occurrence of m6Am than the m6Am-writer, PCIF1. We then outline future directions to understand and exploit these enigmatic modifications.
Introduction
Eukaryotic cells finely control protein production, partly by modulating mRNA production, transport, translation and decay. A component of this is the inverted guanosine “cap” connected to the first nucleotide transcribed from DNA. The cap is added co-transcriptionally to RNAP-II transcribed RNAs (e.g., mRNA, lncRNA, eRNAs) and is involved throughout the mRNA lifecycle (Galloway and Cowling, 2019).
Over 150 RNA modifications have been identified (Cappannini et al., 2024), leading to the establishment of the “epitranscriptomics” field. Following early descriptions of essential roles in organism development (Zhong et al., 2008), m6A has become the prototypical epitranscriptomic modification, found mostly around the stop codon of eukaryotic mRNA (Meyer et al., 2012; Dominissini et al., 2012). Epitranscriptomics is characterised through “writer,” “reader,” and “eraser” proteins which add, bind, or erase modifications, respectively (Figure 1A).
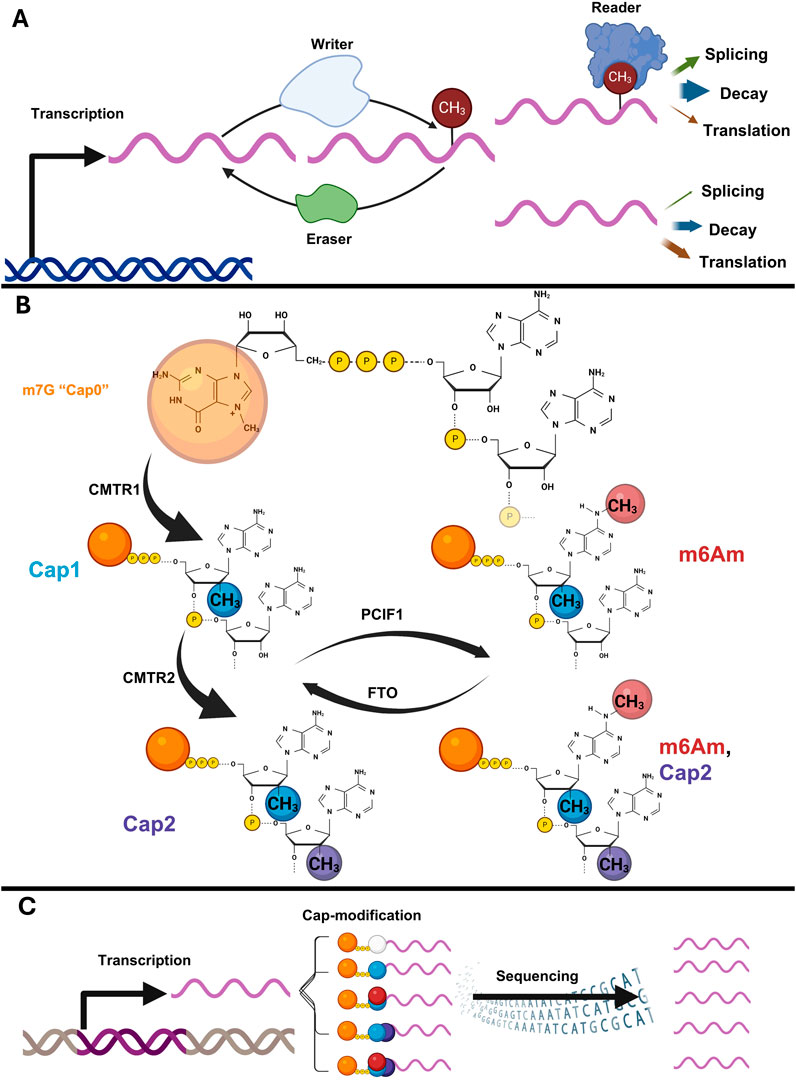
Figure 1. (A) Epitranscriptomics seeks to understand writers, erasers, readers, and downstream consequences of modifications on RNA metabolism. (B) Cap-adjacent modification by CMTR1, CMTR2, and PCIF1, the latter of which can be reversed by FTO. (C) Cap-modifications expand the potential molecular diversity of mRNA transcribed from single genes, but such information is lost during NGS.
In animals, cap-adjacent nucleotides undergo complex modifications (Figure 1B). CMTR1 and CMTR2 methylate the 2′-O ribose position of first (Cap1) and second (Cap2) nucleosides respectively (Bélanger et al., 2010; Werner et al., 2011). When Cap1 nucleoside is adenosine, PCIF1 can methylate to create m6Am (Akichika et al., 2019; Boulias et al., 2019; Sendinc et al., 2019; Sun et al., 2019). RNA-sequencing library preparation loses modification information, making modifications undetectable by standard next-generation sequencing (NGS) (Figure 1C).
This perspective reviews cap-adjacent modifications and their assay. We profile cap-adjacent modifications through selected eukaryotes, confirming broad specificity to metazoans. However, we surprisingly find writers and Cap1 in brown algae. We consider the next steps towards understanding and exploiting cap-adjacent modifications.
Determinants of cap-adjacent modifications
The timing, interplay, and cellular locations of cap-adjacent modifications is poorly understood. A key step in describing the mRNA cap is in understanding the regulation of the writer enzymes and their substrates.
The additions of the Cap1 and m6Am modifications likely occur co-transcriptionally. Both CMTR1 and PCIF1 contain WW-domains, enabling their recruitment to pre-mRNA via the carboxy terminal domain (CTD) of the transcribing RNAP-II (Fan et al., 2003; Haline-Vaz et al., 2008). The CTD of RNAP-II undergoes extensive phosphorylation during the transcription cycle, and this facilities both CMTR1 (Inesta-Vaquera et al., 2018) and PCIF1 binding (Garg et al., 2023; Fan et al., 2003; Li et al., 2024). Cap-adjacent modification is not always dependent upon N7 methylation of the cap (m7G). In vitro, the Vaccinia Cap1 writer is active on unmodified caps but more efficient on m7G-caps (Werner et al., 2011), however human CMTR1 does not (Bélanger et al., 2010). Structural studies have suggested that sequence specificity is unlikely (Smietanski et al., 2014), and none has been reported.
CMTR2 and its methylating activity is found in the cytoplasm (Langberg and Moss, 1981; Werner et al., 2011; Despic and Jaffrey, 2023). Human CMTR2 can methylate regardless of the m7G or Cap1 (Werner et al., 2011), suggesting Cap2 may exist without Cap1. Such “Cap2-1” mRNAs have distinct properties when transfected into cells (Drazkowska et al., 2022). Drosophila melanogaster and Caenorhabditis elegans CMTR1 and CMTR2 act redundantly (Dix et al., 2022; Haussmann et al., 2022). However, in human and mouse, they do not (Dohnalkova et al., 2023).
PCIF1 requires m7G and Cap1-adenosine to create m6Am structures (Sendinc et al., 2019; Boulias et al., 2019). Early work found cytoplasmic m6Am-modifying activity (Keith et al., 1978) and this is corroborated by descriptions of m6Am on the mRNA of viruses which replicate in the cytoplasm (Tartell et al., 2021). m6Am is reversible; the obesity related protein, FTO, erases m6A (Jia et al., 2011), including the m6A within Cap-adjacent m6Am, which may actually be FTO’s preferred substrate (reverting m6Am to Cap1-Am) (Mauer and Jaffrey, 2018; Mauer et al., 2017). PCIF1 has functions beyond catalysing m6Am. For example, it is catalytically inactive in drosophila, but essential for fertility (Franco et al., 2023). Perturbing mouse PCIF1 catalytic activity does not affect fertility, but causes reduced body weights (Pandey et al., 2020).
With gene-regulatory roles, but no identified sequence preference, CMTR1 and CMTR2 are likely regulated by proteins beyond RNAP-II. The first identified was DHX15, an RNA helicase that reduces CMTR1 activity (Inesta-Vaquera et al., 2018), but can unwind complex 5′ untranslated regions to facilitate CMTR1 activity on structured mRNAs (Toczydlowska-Socha et al., 2018). Lukoszek et al. (2024) identified a p-patch domain that, when phosphorylated by CK2, enhances CMTR1 activity.
Roles of cap-adjacent modifications
Early work identified gene-regulatory roles for cap-adjacent modifications. In sea urchin, histone mRNAs were found to gain cap-adjacent modifications following fertilisation, increasing their polysome loading (Caldwell and Emerson, 1985). Similarly, Cap1 facilitates mRNA translation during oocyte maturation (Kuge et al., 1998; Kuge and Richter, 1995). Recent findings corroborate this; CMTR1 was found to be important for normal histone gene expression in embryonic stem cells (Liang et al., 2022); and mouse CMTR1 knockout impairs fertility (Dohnalkova et al., 2023). A regulatory role is consistent with direct measurements of Cap-adjacent modifications by Kruse et al. (2011) which described varying levels of Cap1 and m6Am across mouse tissues, even among mRNAs derived from the same gene.
Cap-adjacent modifications mark mRNA as “self”, preventing sequestration by IFIT-1 (Daffis et al., 2010) and de-capping by DXO (Picard-Jean et al., 2018). The foreign RNA sensor, RIG-I, can be triggered by double-stranded Cap0-RNA (Devarkar et al., 2016). However, removing Cap1 from viral mRNA lowers infectivity without increased RIG-I binding or interferon signalling (Daffis et al., 2010). This aligns with work in which the productivity of transfected mRNA is enhanced by Cap1 when crude RNA is transfected into cells, likely due to contamination with immune-stimulatory byproducts such as uncapped RNA (Drazkowska et al., 2022; Sikorski et al., 2020). Evading the host cell’s own response is likely why Cap1 is essential for translation of a subset of mRNAs during the type I interferon response (Williams et al., 2020), and CMTR1 activity required for both the immune response and, interestingly, influenza propagation (Lukoszek et al., 2024).
Whole-organism Drosophila Cap1 knockout is viable, but with reduced memory and learning performance (Haussmann et al., 2022). Mechanistically, this is due to altered binding of the cap-binding complex to mRNA, perturbing the spatio-temporal control over translation required in neurons. Although recent work has shown that CMTR1 is required for neuronal development (Lee et al., 2020), and differentiation (Liang et al., 2023) the drosophila phenotype could be rescued by expression in the developed brain (Haussmann et al., 2022) – demonstrating a role distinct from differentiation. In mouse, CMTR1 knockout is embryo-lethal (Dohnalkova et al., 2023). However, Dohnalkova et al. (2023) developed a conditional, tissue-specific knockout via floxing which indicated a role for CMTR1 in mammalian fertility. These authors also found that CMTR1 knockout in liver caused chronic activation of the interferon pathway, suggesting that the immune-stimulatory potential of Cap0 mRNA is tissue-specific (Dohnalkova et al., 2023).
In recent years, the role of m6Am in cap-dependent translation has been reported, though with opposing impacts described (Mauer et al., 2017; Sendinc et al., 2019). It seems likely that this variation is due to varied sequence features, variations in cell contexts, or other modifications in this region.
Where are cap-modifications (and how do we know)?
Technological advancement drives epitranscriptomics. The earliest methods relied on feeding cell cultures radioisotope and separating purified, labelled nucleotides by charge and hydrophobicity. This was used to detect a “fifth ribonucleotide” in hitherto discarded fractions of yeast RNA (Davis and Allen, 1957). Methylated bases in tRNA and rRNA are far more common than in mRNA, and were thus identified first (Littlefield and Dunn, 1958). The purification of mRNA from other species (Perry et al., 1972; Grady et al., 1978), enabled rejection of earlier claims that only rRNA and tRNA contained modifications (Borek and Srinivasan, 1966) and described reported modified nucleotides in mRNA (Perry and Kelley, 1974). Similar dependence on technologies is necessarily true for the study of cap-adjacent modifications.
Methods deployed to resolve cap-adjacent nucleotides included thin layer, column, and liquid chromatography, and thin layer electrophoresis (Heckle et al., 1977; Adams and Cory, 1975; Wei C. et al., 1975; Wei et al., 1976; Wei C. M. et al., 1975; Kuo et al., 1986). Similar principles apply to RNA from tissues and cell lines in techniques used now, including:
Thin layer chromatography (TLC)
De-capping and de-phosphorylating purified mRNA exposes a 5′ end which can be radiolabelled with 32P and resolved by 2-Dimensional TLC (Kruse et al., 2011). Labelled RNA can be hybridised to DNA probes to isolate specific mRNAs and measure Cap-adjacent modifications. This cannot resolve Cap2 or guanine-cap modifications, and shows a relatively high abundance of Cap0 compared to other methods. However, the low quantities of input RNA required make it valuable in the analysis of RNA extracted from rare samples, and as a result, it or similar methods are commonly used (Dix et al., 2022; Haussmann et al., 2022; Mauer et al., 2017).
Mass-spectrometry (MS)
Nuclease P1 digestion degrades mRNA to single nucleotides and cap-dinucleotides. These can be subjected to liquid chromatography-tandem MS (LC/MS-MS) to detect global levels of cap-adjacent methylation on dinucleotide fragments (Galloway et al., 2020), high-performance liquid-chromatography can be used to enrich dinucleotides before LC/MS-MS (Wang et al., 2019). Detecting dinucleotides enables discrimination of the methylation status of the cap itself as well as the identity of nucleotides in Cap1. Mass-spectrometry require greater input quantities of RNA (approx.2 µg) compared to radioisotope incorporation methods (approx.50 ng), and cannot detect Cap2.
Recapping
Treatment with yeast de-capping enzyme leaves mRNA with diphosphate 5′ termini to which vaccinia capping enzyme can add 32P-radiolabelled guanosine. Labelled RNA is digested with RNAse I, which cannot cleave 2′-O-ribose methylated sites and PAGE used to resolve the resultant cap-fragments. This method can detect Cap0 through to the ribose modifications found on the hypermethylated Cap IV structure of trypanosome brucei, but not nucleotide identity, G-cap-methylation status, or m6Am (Dix et al., 2022; Haussmann et al., 2022).
Sequencing
The existence of m6A immunoprecipitation methods (Bodi et al., 2010; Meyer et al., 2012; Dominissini et al., 2012) made early work on m6Am possible. As these methods rely on fragmentation and peak calling, deconvoluting 5′ UTR m6A from cap-adjacent m6Am is challenging (Meyer and Jaffrey, 2017). To solve this, m6Am-exo-seq treats fragmented RNA with an exonuclease to degrade uncapped fragments prior to immunoprecipitation and sequencing. This dramatically improves the specificity of m6Am mapping (Sendinc et al., 2019). An intriguing method, CAPturAM uses a modified, recombinant PCIF1 to propargylate Cap1 adenosines for selective biotinylation and isolation with streptavidin (Muthmann et al., 2023). This has the advantage of being independent of antibodies, but is arguably akin to protein-crosslinking experiments in that it determines potential targets, rather than the current methylation status of sequenced mRNA.
Despic and Jaffrey (2023) developed the first transcriptome-wide sequencing method for 2′-O-ribose methylation, CLAM-cap-seq. This reverse transcribes de-capped mRNA, before using circligase to join the mRNA 5′ end to the 3′ end of the cDNA. RNAseT2 is then used to degrade the RNA. RNAseT2 cannot cleave 2′-ribose modified RNA, leaving a “tag” on the 3′ end of the cDNA detectable via bioinformatics. This is a complex method in terms of reagent availability, library preparation, and downstream bioinformatics. For example, analysis must account for a number (typically 0–4) of random nucleotides introduced during library preparation, and then detect a palindrome by combining the 3′ cDNA nucleotides with their reverse complement derived from the RNA. The palindrome length describes the cap structure, four for Cap1 (two cDNA plus two RNA nucleotides), and six for Cap2. This method discards Cap0, and cannot capture m6Am – potentially due to inefficiencies in reverse transcription (Archer et al., 2016).
Cap-adjacent prevalence across organisms
Most methods describe ubiquitous Cap1, approximately 30% of Cap1-adenosines are further modified to m6Am, and approximately 50% of transcripts are Cap2 (Heckle et al., 1977; Adams and Cory, 1975; Wei C. et al., 1975; Wei et al., 1976; Wei C. M. et al., 1975). Cap1 is found in animals (Furuichi, 2015) while m6Am in only found in vertebrates (Dix et al., 2022; Furuichi, 2015; Sendinc et al., 2019). Despite early reports to the contrary (Furuichi and Shatkin, 2000) there is no cap-adjacent methylation, or the requisite writer enzymes, found in plants. We sought to expand this understanding, by combining BLAST analysis and TLC to track the cap-adjacent writers through animals (Figure 2). We selected representative species across the evolutionary tree; mammals, birds, fish, amphibians, insects and cephalopods. We focussed on brain tissue, as the nervous system has the most clearly described role for cap-adjacent modifications beyond innate immunity. We added a brown algae to this analysis as, interestingly, an FTO orthologue has been reported in Ectocarpus siliculosus (Robbens et al., 2008).
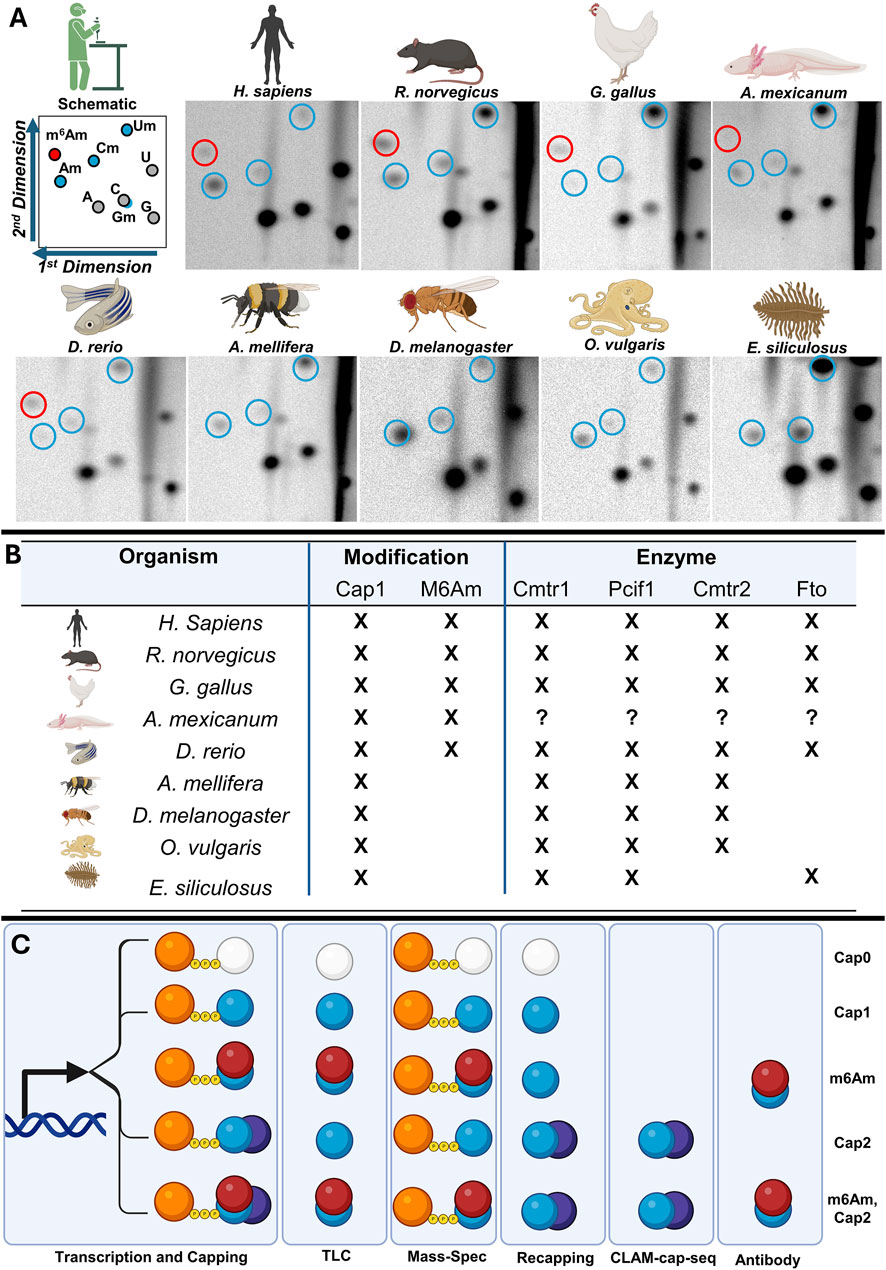
Figure 2. Cap-adjacent modifications are found throughout the animal kingdom. We used thin layer chromatography (TLC) to measure global cap-adjacent modification in each of the organisms listed (A). We annotated each of these with their enzyme in the genome and presence (X) or absence (blank) by TLC (B). The methods used to map or measure cap-adjacent modifications can only detect sub populations of the modification (C). For TLCs (A) N = 3, representative TLCs shown, replicates are found in Supplementary Data 1. BLAST data are found in Supplementary Data 2. Circles denote modifications, with colour denoting Cap1 (blue) or m6Am (red).
As expected, our 2D-TLC found Cap1 in all animals. Curiously, our orthologue searches found that a species of algae, Ectocarpus siliculosus has orthologues for CMTR1 (39.67% identity, 39.67% coverage) and PCIF1 (36.67% identity, 35% coverage) in addition to FTO (27.97% identity, 85% coverage) (Figure 2B, Supplementary Data 2; Robbens et al., 2008). TLC on E. siliculosus confirmed presence of Cap1, but not m6Am in this algae. Beyond this, our work corroborated the canon, finding m6Am specific to vertebrates, but at greater levels in H. sapiens, R. norvegicus, D. rerio and G. gallus and lower levels in A. mexicanum. Our orthologue search found PCIF1 orthologues in more species than we detect m6Am. Notably, FTO – an annotated m6Am eraser - correlated better with m6Am.
Kruse et al., posed that the variations they detected in cap-adjacent modifications may reflect differences in start site selection. This seems plausible - in vertebrates, approximately 70% of the genes have dispersed promoters (Sandelin et al., 2007; Juven-Gershon and Kadonaga, 2010), with mammals exhibiting a further increase in dispersed transcription initiation (Carninci et al., 2006). This would fit a model in which cap-adjacent modifications facilitates gene regulation by increasing the potential molecular diversity of mRNAs derived from a single gene.
Discussion; what next?
Though enigmatic, cap-modifications clearly act as a layer of selective gene-regulation. A dynamic mRNA cap is a fascinating possibility, whether by demethylation via erasers or by new deposition in the cytoplasm by PCIF1 and CMTR2; a feature which appears unique to the cap region.
Elucidating the roles of cap-adjacent modifications requires developing methods and understanding of the full complex cap. This includes co-occurring modifications within this small region of RNA and the identity of the nucleotide itself. There has been progress here, deploying co-transcriptional capping to generate mRNAs with complex caps such as Cap2 with m6Am in the first position (m6Am,Cap2), or without Cap1 (Cap2-1). However, mapping these structures to native mRNA has not been achieved even in the latest methods. The 2′-O-ribose modifications can stall reverse transcriptase under limiting dNTP conditions. This has been used to map internal 2′-O-ribose modifications (Incarnato et al., 2017). However, since Cap-adjacent modification occurs at the terminal end of the mRNA, such a method is not easily translated to these modifications – it would also not be possible for such a method to discern Cap1 from Cap2 – or detect m6Am. Direct-RNA sequencing (DRS) can capture multiple modifications on the same mRNA (Acera Mateos et al., 2024) and thus holds promise. Such Oxford Nanopore Technologies (ONT) methods are steadily improving (Liu-Wei et al., 2024). However, being kmer-based, Nanopore-sequencing loses accuracy as the terminal end of the RNA passes through the pore.
Cap-adjacent modifications proved essential in the development of the COVID-19 mRNA vaccines. Beyond vaccines, there are a swathe of potential uses for targetted expression of proteins in vivo - not least CRISPR/Cas editing (for review see Paunovska et al., 2022; Qin et al., 2022). Future RNA therapeutic and vaccine molecules are likely to seek to target specific cell contexts, tissues, or expression characteristics. Under hypoxic conditions, for example, efficacies of current mRNA molecules vary by cell type (Ma and Fenton, 2023). Indeed, recent studies using in vitro transcribed mRNA with defined cap structures found cell-type specific effects (Drazkowska et al., 2022; Sikorski et al., 2020), suggesting that Cap-adjacent modifications offer a useful design modality. Beyond this, understanding the in vivo function and interactors of cap-modifications promises new therapeutics targetting the mRNA cap of viruses (for review see Ferron et al., 2012) or to modulate host gene expression in disease.
Epitranscriptomics examines writers, readers, and erasers, and the effects of nucleotide modifications on the RNA itself. A similarly holistic approach to the “Epicapome” is developing. Compared to typical individual epitranscriptomic modifications, the Epicapome is complicated by the varied nucleotides affected by the modification, the partially interdependent nature of the modifications, and non-catalytic functions of the writer proteins. While there is no clear sequence motif, it is certainly true that we know where on the mRNA to start!
Methods
Portions of Figures 1, 2 and Supplementary Data 1 were created with BioRender.com.
RNA extractions
RNA from frozen Human brain tissue was extracted using AllPrep DNA/RNA kit (Qiagen, Germany). Animal tissues were either collected as frozen samples or stored in RNA later and processed using the mirVana miRNA kit (ThermoFisher Scientific, Uited States), with phenol. Ectocarpus siliculosus (CCAP1310/4) RNA was extracted according to Greco et al. (2014).
Thin layer chromatography
TLCs were performed as described in Kruse et al. (2011). Following extraction and poly(A) isolation, RppH(New England Biolabs (NEB), United States) was used to remove the 5′Cap of the mRNA in Thermopol buffer (NEB). This leaves a single phosphate group which we removed with CIP (NEB) in the same reaction. PNK (Thermofisher, United Kingdom) was then used to add a radioactive 5′ phosphate (ATP, [γ-32P]- 3000Ci/mmol 10mCi/mL EasyTide, Revvity, Wales) to the exposed nucleotide. After clean-up to remove residual [γ-32P]-ATP, mRNA was digested to individual nucleotides with P1 nuclease (Sigma-Aldrich, United States). 1 μL of the labelled nucleotides were added to cellulose TLC (Merck, Germany) plates and run in two dimensions before visualisation via phosphor storage screens and imaging. All nucleotides are separated on this method, but only those that were exposed prior to radiolabelling are visible via the phosphor imaging.
BLAST
To identify orthologues of the cap-methyltransferases CMTR1, CMTR2, PCIF1 and the m6Am demethylase FTO the human proteins were blasted for somewhat similar sequences (blastp) against the proteomes of the organisms studied by TLC.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving human tissue were approved by Wales Research Ethics Committee. The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired from Human brain tissue, acquired from the Parkinson’s UK Brain Bank, Imperial College. Its use is covered by the ethics approval that the Parkinson’s UK Brain Bank at Imperial College London has as a Research Tissue Bank by the Wales Research Ethics Committee (Ref. No. 08/MRE09/31+5). The material transfer agreement was between the Imperial Brain Bank and HMK, and used under the HTA licence 12265. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements. Animal tissues were gifts from other groups, from past studies, originally collected in accordance with their own local ethical rules, legislation, and institutional requirements.
Author contributions
EB: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Software, Validation, Visualization, Writing–original draft, Writing–review and editing. RF: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Supervision, Validation, Writing–review and editing. HK: Conceptualization, Data curation, Funding acquisition, Project administration, Resources, Supervision, Validation, Visualization, Writing–review and editing. NA: Conceptualization, Data curation, Formal Analysis, Software, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing.
Funding
The authors declare that financial support was received for the research, authorship, and/or publication of this article. EB received funding via BBSRC-DTP studentship (BB/M008770/1).
Acknowledgments
We thank the following for providing animal tissue; Michael Garle, Andrew Johnson, Martin Gering, Reinhard Stoger, Marios Georgiou, and Catrin Rutland (University of Nottingham); Alix Harvey, Marine Biological Association; Marie-Mathilde Perrineau, Scottish Association for Marine Science.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frnar.2024.1485307/full#supplementary-material
SUPPLEMENTARY DATA 1 | Replicate TLCs of species from Figure 2.
SUPPLEMENTARY DATA 2 | Tables showing query coverage and percentage identity for genes examined across species compared to human (CMTR1, CMTR2, PCIF1, FTO).
References
Acera Mateos, P. J., J Sethi, A., Ravindran, A., Srivastava, A., Woodward, K., Mahmud, S., et al. (2024). Prediction of m6A and m5C at single-molecule resolution reveals a transcriptome-wide co-occurrence of RNA modifications. Nat. Commun. 15 (1), 3899. doi:10.1038/s41467-024-47953-7
Adams, J. M., and Cory, S. (1975). Modified nucleosides and bizarre 5'-termini in mouse myeloma mRNA. Nature 255, 28–33. doi:10.1038/255028a0
Akichika, S., Hirano, S., Shichino, Y., Suzuki, T., Nishimasu, H., Ishitani, R., et al. (2019). Cap-specific terminal N (6)-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science 363, eaav0080. doi:10.1126/science.aav0080
Archer, N., Walsh, M. D., Shahrezaei, V., and Hebenstreit, D. (2016). Modeling enzyme processivity reveals that RNA-seq libraries are biased in characteristic and correctable ways. Cell Syst. 3, 467–479.e12. doi:10.1016/j.cels.2016.10.012
Bélanger, F., Stepinski, J., Darzynkiewicz, E., and Pelletier, J. (2010). Characterization of hMTr1, a human Cap1 2'-O-ribose methyltransferase. J. Biol. Chem. 285, 33037–33044. doi:10.1074/jbc.m110.155283
Bodi, Z., Button, J. D., Grierson, D., and Fray, R. G. (2010). Yeast targets for mRNA methylation. Nucleic Acids Res. 38, 5327–5335. doi:10.1093/nar/gkq266
Borek, E., and Srinivasan, P. R. (1966). The methylation of nucleic acids. Annu. Rev. Biochem. 35, 275–298. doi:10.1146/annurev.bi.35.070166.001423
Boulias, K., Toczydłowska-Socha, D., Hawley, B. R., Liberman, N., Takashima, K., Zaccara, S., et al. (2019). Identification of the m(6)Am methyltransferase PCIF1 reveals the location and functions of m(6)Am in the transcriptome. Mol. Cell 75, 631–643.e8. doi:10.1016/j.molcel.2019.06.006
Caldwell, D. C., and Emerson, C. P. (1985). The role of cap methylation in the translational activation of stored maternal histone mRNA in sea urchin embryos. Cell 42, 691–700. doi:10.1016/0092-8674(85)90126-6
Cappannini, A., Ray, A., Purta, E., Mukherjee, S., Boccaletto, P., Moafinejad, S. N., et al. (2024). MODOMICS: a database of RNA modifications and related information. 2023 update. Nucleic Acids Res. 52, D239–d244. doi:10.1093/nar/gkad1083
Carninci, P., Sandelin, A., Lenhard, B., Katayama, S., Shimokawa, K., Ponjavic, J., et al. (2006). Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 38, 626–635. doi:10.1038/ng1789
Daffis, S., Szretter, K. J., Schriewer, J., Li, J., Youn, S., Errett, J., et al. (2010). 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468, 452–456. doi:10.1038/nature09489
Davis, F. F., and Allen, F. W. (1957). Ribonucleic acids from yeast which contain a fifth nucleotide. J. Biol. Chem. 227, 907–915. doi:10.1016/s0021-9258(18)70770-9
Despic, V., and Jaffrey, S. R. (2023). mRNA ageing shapes the Cap2 methylome in mammalian mRNA. Nature 614, 358–366. doi:10.1038/s41586-022-05668-z
Devarkar, S. C., Wang, C., Miller, M. T., Ramanathan, A., Jiang, F., Khan, A. G., et al. (2016). Structural basis for m7G recognition and 2'-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I. Proc. Natl. Acad. Sci. U. S. A. 113, 596–601. doi:10.1073/pnas.1515152113
Dix, T. C., Haussmann, I. U., Brivio, S., Nallasivan, M. P., Hadzhiev, Y., Müller, F., et al. (2022). CMTr mediated 2'-O-ribose methylation status of cap-adjacent nucleotides across animals. Rna 28, 1377–1390. doi:10.1261/rna.079317.122
Dohnalkova, M., Krasnykov, K., Mendel, M., Li, L., Panasenko, O., Fleury-Olela, F., et al. (2023). Essential roles of RNA cap-proximal ribose methylation in mammalian embryonic development and fertility. Cell Rep. 42, 112786. doi:10.1016/j.celrep.2023.112786
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206. doi:10.1038/nature11112
Drazkowska, K., Tomecki, R., Warminski, M., Baran, N., Cysewski, D., Depaix, A., et al. (2022). 2′-O-Methylation of the second transcribed nucleotide within the mRNA 5′ cap impacts the protein production level in a cell-specific manner and contributes to RNA immune evasion. Nucleic Acids Res. 50, 9051–9071. doi:10.1093/nar/gkac722
Fan, H., Sakuraba, K., Komuro, A., Kato, S., Harada, F., Hirose, Y., et al. (2003). PCIF1, a novel human WW domain-containing protein, interacts with the phosphorylated RNA polymerase II. Biochem. Biophysical Res. Commun. 301, 378–385. doi:10.1016/s0006-291x(02)03015-2
Ferron, F., Decroly, E., Selisko, B., and Canard, B. (2012). The viral RNA capping machinery as a target for antiviral drugs. Antivir. Res. 96 (1), 21–31. doi:10.1016/j.antiviral.2012.07.007
Franco, G., Taillebourg, E., Delfino, E., Homolka, D., Gueguen, N., Brasset, E., et al. (2023). The catalytic-dead Pcif1 regulates gene expression and fertility in Drosophila. Rna 29, 609–619. doi:10.1261/rna.079192.122
Furuichi, Y. (2015). Discovery of m(7)G-cap in eukaryotic mRNAs. Proc. Jpn. Acad. Ser. B 91, 394–409. doi:10.2183/pjab.91.394
Furuichi, Y., and Shatkin, A. J. (2000). Viral and cellular mRNA capping: past and prospects. Adv. Virus Res. 55, 135–184. doi:10.1016/s0065-3527(00)55003-9
Galloway, A., Atrih, A., Grzela, R., Darzynkiewicz, E., Ferguson, M. A. J., and Cowling, V. H. (2020). CAP-MAP: cap analysis protocol with minimal analyte processing, a rapid and sensitive approach to analysing mRNA cap structures. Open Biol. 10, 190306. doi:10.1098/rsob.190306
Galloway, A., and Cowling, V. H. (2019). mRNA cap regulation in mammalian cell function and fate. Biochimica Biophysica Acta (BBA) - Gene Regul. Mech. 1862, 270–279. doi:10.1016/j.bbagrm.2018.09.011
Garg, G., Dienemann, C., Farnung, L., Schwarz, J., Linden, A., Urlaub, H., et al. (2023). Structural insights into human co-transcriptional capping. Mol. Cell 83, 2464–2477.e5. doi:10.1016/j.molcel.2023.06.002
Grady, L. J., North, A. B., and Campbell, W. P. (1978). Complexity of poly(A+) and poly(A-) polysomal RNA in mouse liver and cultured mouse fibroblasts. Nucleic Acids Res. 5, 697–712. doi:10.1093/nar/5.3.697
Greco, M., Sáez, C. A., Brown, M. T., and Bitonti, M. B. (2014). A simple and effective method for high quality Co-extraction of genomic DNA and total RNA from low biomass Ectocarpus siliculosus, the model Brown alga. PLOS ONE 9, e96470. doi:10.1371/journal.pone.0096470
Haline-Vaz, T., Silva, T. C., and Zanchin, N. I. (2008). The human interferon-regulated ISG95 protein interacts with RNA polymerase II and shows methyltransferase activity. Biochem. Biophys. Res. Commun. 372, 719–724. doi:10.1016/j.bbrc.2008.05.137
Haussmann, I. U., Wu, Y., Nallasivan, M. P., Archer, N., Bodi, Z., Hebenstreit, D., et al. (2022). CMTr cap-adjacent 2′-O-ribose mRNA methyltransferases are required for reward learning and mRNA localization to synapses. Nat. Commun. 13, 1209. doi:10.1038/s41467-022-28549-5
Heckle, W. L., Fenton, R. G., Wood, T. G., Merkel, C. G., and Lingrel, J. B. (1977). Methylated nucleosides in globin mRNA from mouse nucleated erythroid cells. J. Biol. Chem. 252, 1764–1770. doi:10.1016/s0021-9258(17)40616-8
Incarnato, D., Anselmi, F., Morandi, E., Neri, F., Maldotti, M., Rapelli, S., et al. (2017). High-throughput single-base resolution mapping of RNA 2′-O-methylated residues. Nucleic Acids Res. 45 (3), 1433–1441. doi:10.1093/nar/gkw810
Inesta-Vaquera, F., Chaugule, V. K., Galloway, A., Chandler, L., Rojas-Fernandez, A., Weidlich, S., et al. (2018). DHX15 regulates CMTR1-dependent gene expression and cell proliferation. Life Sci. Alliance 1, e201800092. doi:10.26508/lsa.201800092
Jia, G., Fu, Y., Zhao, X., Dai, Q., Zheng, G., Yang, Y., et al. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887. doi:10.1038/nchembio.687
Juven-Gershon, T., and Kadonaga, J. T. (2010). Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev. Biol. 339, 225–229. doi:10.1016/j.ydbio.2009.08.009
Keith, J. M., Ensinger, M. J., and Moss, B. (1978). HeLa cell RNA (2′-O-methyladenosine-N6-)-methyltransferase specific for the capped 5′-end of messenger RNA. J. Biol. Chem. 253, 5033–5039. doi:10.1016/s0021-9258(17)34652-5
Kruse, S., Zhong, S., Bodi, Z., Button, J., Alcocer, M. J., Hayes, C. J., et al. (2011). A novel synthesis and detection method for cap-associated adenosine modifications in mouse mRNA. Sci. Rep. 1, 126. doi:10.1038/srep00126
Kuge, H., Brownlee, G. G., Gershon, P. D., and Richter, J. D. (1998). Cap ribose methylation of c-mos mRNA stimulates translation and oocyte maturation in Xenopus laevis. Nucleic Acids Res. 26, 3208–3214. doi:10.1093/nar/26.13.3208
Kuge, H., and Richter, J. D. (1995). Cytoplasmic 3′ poly(A) addition induces 5′ cap ribose methylation: implications for translational control of maternal mRNA. EMBO J. 14, 6301–6310. doi:10.1002/j.1460-2075.1995.tb00320.x
Kuo, K. C., Smith, C. E., Shi, Z., Agris, P. F., and Gehrke, C. W. (1986). Quantitative measurement of mRNA cap 0 and cap 1 structures by high-performance liquid chromatography. J. Chromatogr. B Biomed. Sci. Appl. 378, 361–374. doi:10.1016/s0378-4347(00)80732-x
Langberg, S. R., and Moss, B. (1981). Post-transcriptional modifications of mRNA. Purification and characterization of cap I and cap II RNA (nucleoside-2′-)-methyltransferases from HeLa cells. J. Biol. Chem. 256, 10054–10060. doi:10.1016/s0021-9258(19)68740-5
Lee, Y. L., Kung, F. C., Lin, C. H., and Huang, Y. S. (2020). CMTR1-Catalyzed 2'-O-ribose methylation controls neuronal development by regulating Camk2α expression independent of RIG-I signaling. Cell Rep. 33, 108269. doi:10.1016/j.celrep.2020.108269
Liang, S., Almohammed, R., and Cowling, V. H. (2023). The RNA cap methyltransferases RNMT and CMTR1 co-ordinate gene expression during neural differentiation. Biochem. Soc. Trans. 51, 1131–1141. doi:10.1042/bst20221154
Liang, S., Silva, J. C., Suska, O., Lukoszek, R., Almohammed, R., and Cowling, V. H. (2022). CMTR1 is recruited to transcription start sites and promotes ribosomal protein and histone gene expression in embryonic stem cells. Nucleic Acids Res. 50, 2905–2922. doi:10.1093/nar/gkac122
Littlefield, J. W., and Dunn, D. B. (1958). The occurrence and distribution of thymine and three methylated-adenine bases in ribonucleic acids from several sources. Biochem. J. 70, 642–651. doi:10.1042/bj0700642
Liu-Wei, W., van der Toorn, W., Bohn, P., Hölzer, M., Smyth, R. P., and Von Kleist, M. (2024). Sequencing accuracy and systematic errors of nanopore direct RNA sequencing. BMC Genomics 25, 528. doi:10.1186/s12864-024-10440-w
Li, Y., Wang, Q., Xu, Y., and Li, Z. (2024). Structures of co-transcriptional RNA capping enzymes on paused transcription complex. Nat. Commun. 15, 4622. doi:10.1038/s41467-024-48963-1
Lukoszek, R., Inesta-Vaquera, F., Brett, N. J. M., Liang, S., Hepburn, L. A., Hughes, D. J., et al. (2024). CK2 phosphorylation of CMTR1 promotes RNA cap formation and influenza virus infection. Cell Rep. 43, 114405. doi:10.1016/j.celrep.2024.114405
Mauer, J., and Jaffrey, S. R. (2018). FTO, m(6) A(m), and the hypothesis of reversible epitranscriptomic mRNA modifications. FEBS Lett. 592, 2012–2022. doi:10.1002/1873-3468.13092
Mauer, J., Luo, X., Blanjoie, A., Jiao, X., Grozhik, A. V., Patil, D. P., et al. (2017). Reversible methylation of m(6)A(m) in the 5′ cap controls mRNA stability. Nature 541, 371–375. doi:10.1038/nature21022
Ma, Y., and Fenton, O. S. (2023). An efficacy and mechanism driven study on the impact of hypoxia on lipid nanoparticle mediated mRNA delivery. J. Am. Chem. Soc. 145, 11375–11386. doi:10.1021/jacs.3c02584
Meyer, K. D., and Jaffrey, S. R. (2017). Rethinking m(6)A readers, writers, and erasers. Annu. Rev. Cell Dev. Biol. 33, 319–342. doi:10.1146/annurev-cellbio-100616-060758
Meyer, K. D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C. E., and Jaffrey, S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646. doi:10.1016/j.cell.2012.05.003
Muthmann, N., Albers, M., and Rentmeister, A. (2023). CAPturAM, a chemo-enzymatic strategy for selective enrichment and detection of physiological CAPAM-targets. Angewandte Chemie Int. ed. Engl. 62, e202211957. doi:10.1002/anie.202211957
Pandey, R. R., Delfino, E., Homolka, D., Roithova, A., Chen, K. M., Li, L., et al. (2020). The mammalian cap-specific m(6)Am RNA methyltransferase PCIF1 regulates transcript levels in mouse tissues. Cell Rep. 32, 108038. doi:10.1016/j.celrep.2020.108038
Paunovska, K., Loughrey, D., and Dahlman, J. E. (2022). Drug delivery systems for RNA therapeutics. Nat. Rev. Genet. 23 (5), 265–280. doi:10.1038/s41576-021-00439-4
Perry, R. P., and Kelley, D. E. (1974). Existence of methylated messenger RNA in mouse L cells. Cell 1, 37–42. doi:10.1016/0092-8674(74)90153-6
Perry, R. P., La Torre, J., Kelley, D. E., and Greenberg, J. R. (1972). On the lability of poly(A) sequences during extraction of messenger RNA from polyribosomes. Biochimica Biophysica Acta (BBA) - Nucleic Acids Protein Synthesis 262, 220–226. doi:10.1016/0005-2787(72)90236-5
Picard-Jean, F., Brand, C., Tremblay-Létourneau, M., Allaire, A., Beaudoin, M. C., Boudreault, S., et al. (2018). 2'-O-methylation of the mRNA cap protects RNAs from decapping and degradation by DXO. PLoS One 13, e0193804. doi:10.1371/journal.pone.0193804
Qin, S., Tang, X., Chen, Y., Chen, K., Fan, N., Xiao, W., et al. (2022). mRNA-based therapeutics: powerful and versatile tools to combat diseases. Sig Transduct. Target Ther. 7 (1), 166. doi:10.1038/s41392-022-01007-w
Robbens, S., Rouzé, P., Cock, J. M., Spring, J., Worden, A. Z., and Van de Peer, Y. (2008). The FTO gene, implicated in human obesity, is found only in vertebrates and marine algae. J. Mol. Evol. 66, 80–84. doi:10.1007/s00239-007-9059-z
Sandelin, A., Carninci, P., Lenhard, B., Ponjavic, J., Hayashizaki, Y., and Hume, D. A. (2007). Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat. Rev. Genet. 8, 424–436. doi:10.1038/nrg2026
Sendinc, E., Valle-Garcia, D., Dhall, A., Chen, H., Henriques, T., Navarrete-Perea, J., et al. (2019). PCIF1 catalyzes m6Am mRNA methylation to regulate gene expression. Mol. Cell 75, 620–630.e9. doi:10.1016/j.molcel.2019.05.030
Sikorski, P. J., Warminski, M., Kubacka, D., Ratajczak, T., Nowis, D., Kowalska, J., et al. (2020). The identity and methylation status of the first transcribed nucleotide in eukaryotic mRNA 5′ cap modulates protein expression in living cells. Nucleic Acids Res. 48, 1607–1626. doi:10.1093/nar/gkaa032
Smietanski, M., Werner, M., Purta, E., Kaminska, K. H., Stepinski, J., Darzynkiewicz, E., et al. (2014). Structural analysis of human 2'-O-ribose methyltransferases involved in mRNA cap structure formation. Nat. Commun. 5, 3004. doi:10.1038/ncomms4004
Sun, H., Zhang, M., Li, K., Bai, D., and Yi, C. (2019). Cap-specific, terminal N6-methylation by a mammalian m6Am methyltransferase. Cell Res. 29, 80–82. doi:10.1038/s41422-018-0117-4
Tartell, M. A., Boulias, K., Hoffmann, G. B., Bloyet, L. M., Greer, E. L., and Whelan, S. P. J. (2021). Methylation of viral mRNA cap structures by PCIF1 attenuates the antiviral activity of interferon-β. Proc. Natl. Acad. Sci. U. S. A. 118, e2025769118. doi:10.1073/pnas.2025769118
Toczydlowska-Socha, D., Zielinska, M. M., Kurkowska, M., Almeida, C. F., Asthac, F., Stefaniak, F., et al. (2018). Human RNA cap1 methyltransferase CMTr1 cooperates with RNA helicase DHX15 to modify RNAs with highly structured 5′ termini. Philosophical Trans. R. Soc. B Biol. Sci. 373, 20180161. doi:10.1098/rstb.2018.0161
Wang, J., Alvin Chew, B. L., Lai, Y., Dong, H., Xu, L., Balamkundu, S., et al. (2019). Quantifying the RNA cap epitranscriptome reveals novel caps in cellular and viral RNA. Nucleic Acids Res. 47, e130. doi:10.1093/nar/gkz751
Wei, C. M., Gershowitz, A., and Moss, B. (1975). Methylated nucleotides block 5' terminus of HeLa cell messenger RNA. Cell 4, 379–386. doi:10.1016/0092-8674(75)90158-0
Wei, C. M., Gershowitz, A., and Moss, B. (1976). 5'-Terminal and internal methylated nucleotide sequences in HeLa cell mRNA. Biochemistry 15, 397–401. doi:10.1021/bi00647a024
Wei, C., Gershowitz, A., and Moss, B. (1975). N6, O2′-dimethyladenosine a novel methylated ribonucleoside next to the 5' terminal of animal cell and virus mRNAs. Nature 257, 251–253. doi:10.1038/257251a0
Werner, M., Purta, E., Kaminska, K. H., Cymerman, I. A., Campbell, D. A., Mittra, B., et al. (2011). 2′-O-ribose methylation of cap2 in human: function and evolution in a horizontally mobile family. Nucleic Acids Res. 39, 4756–4768. doi:10.1093/nar/gkr038
Williams, G. D., Gokhale, N. S., Snider, D. L., and Horner, S. M. (2020). The mRNA cap 2'-O-methyltransferase CMTR1 regulates the expression of certain interferon-stimulated genes. mSphere 5, e00202–e00220. doi:10.1128/msphere.00202-20
Keywords: mRNA, epitranscriptome, mRNA cap, gene expression, translation, epicapome
Citation: Bellows E, Fray RG, Knight HM and Archer N (2024) The expanding role of cap-adjacent modifications in animals. Front. RNA Res. 2:1485307. doi: 10.3389/frnar.2024.1485307
Received: 23 August 2024; Accepted: 18 September 2024;
Published: 01 October 2024.
Edited by:
Sunny Sharma, Rutgers, The State University of New Jersey, United StatesReviewed by:
Luis E. Contreras-Llano, University of California, Davis, United StatesCopyright © 2024 Bellows, Fray, Knight and Archer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nathan Archer, bmF0aGFuLmFyY2hlckBub3R0aW5naGFtLmFjLnVr
†ORCID: Eleanor Bellows: orcid.org/0000-0003-0055-8330; Rupert G. Fray: orcid.org/0000-0002-3935-8945; Helen M. Knight: orcid.org/0000-0003-3267-9591; Nathan Archer: orcid.org/0000-0002-3356-6161
 Eleanor Bellows
Eleanor Bellows Rupert G. Fray
Rupert G. Fray Helen M. Knight1†
Helen M. Knight1† Nathan Archer
Nathan Archer