
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. RNA Res. , 25 September 2024
Sec. RNA Processing and Regulation
Volume 2 - 2024 | https://doi.org/10.3389/frnar.2024.1460913
This article is part of the Research Topic RNA Methylation: Detection, Deposition, and Functions View all 6 articles
Decoration of cellular RNAs with modified RNA nucleosides is an important layer of gene expression regulation. Throughout the transcriptome, RNA modifications influence the folding, stability and function of RNAs as well as their interactions with RNA-binding proteins. Although first detected more than 50 years ago, the modified nucleosides N2-methylguanosine (m2G) and N2,N2-dimethylguanosine (m22G) have recently come to the fore through the identification and characterization of the human methyltransferases (MTases) responsible for their installation. In tRNAs, m2G and m22G are present at the junctions between the acceptor stem and the D-arm, and the D-arm and the anticodon stem loop. Here, we review the current knowledge on the effects of mono- and di-methylation of N2 of guanosine on base-pairing and provide an overview of m2(2)G sites in cytosolic and mitochondrial tRNAs. We highlight key features of m2G and m22G MTases, and describe how these enzymes specifically recognize their RNA substrates and target nucleosides. We also discuss the impact of these modifications on tRNA functions, their dynamic regulation and their implications in disease.
Ribonucleic acids (RNAs) are multifunctional cellular molecules that serve as carriers of genetic information as well as components of ribonucleoprotein complexes and catalysts of diverse chemical reactions. To fulfill such varied functionalities, RNA molecules require greater chemical diversity than that provided by the four standard ribonucleosides (adenosine (A), cytidine (C), guanosine (G) and uridine (U)). This extended functional capability is achieved by the modification of RNA nucleosides (Shi et al., 2020). Modified nucleosides in RNAs can arise through the incoroporation of non-canonical nucleosides, such as queosine, the isomerization of canonical nucleoside, e.g. the conversion of uridine to pseudouridine (Ψ), or by the addition of chemical groups to pre-exisiting nucleosides, e.g. methylation, acetylations, etc (Ontiveros et al., 2019). Across the three domains of life, to date, around 170 different types of modified RNA nucleosides have been identified (Cappannini et al., 2023). These modified nucleosides influence the biogenesis, localization and functionality of various classes of RNAs (Shi et al., 2020).
Transfer RNAs (tRNA) are the most extensively and diversely modified class of RNAs, with approximately half of the known naturally occurring modified nucleosides being present within them (Cui et al., 2023; Phizicky and Hopper, 2023; Suzuki, 2021). Eukaryotic nuclear-encoded tRNAs contain, on average, 13 modified nucleosides (Pan, 2018). By contrast, five and eight RNA modifications are typically present in mitochondrial and bacterial tRNAs, respectively (Suzuki et al., 2020; Zhang et al., 2022a). This abundance of modified nucleosides in tRNAs reflects their important functions in fine-tuning tRNA structures and maintaining tRNA functionality during protein synthesis.
To fulfil their functions as adaptor molecules during translation, each tRNA is specifically charged with a cognate amino acid (Giegé and Eriani, 2023). The aminoacyl-tRNA then delivers the amino acid to the ribosome where, upon recognition of a messenger RNA (mRNA) codon complementary to the tRNA anticodon, the amino acid is added to the nascent polypeptide chain. In this context, modified nucleosides in the anticodon loop play important roles in ensuring the specificity and fidelity of anticodon–codon base-pairing and that the anticodon loop is in an optimal conformation for efficient decoding (Smith et al., 2024; Zhou et al., 2021). By contrast, modifications in other regions of tRNAs (the acceptor stem, the D- and T-arms and the connecting loops) typically contribute to tRNA folding and structure maintenance, thus regulating tRNA stability, aminoacylation and interactions with ribosomes (Yared et al., 2024; Zhang et al., 2022a). Several tRNA modifications are also implicated in regulating the cleavage of tRNAs into tRNA-derived fragments (tRFs), which fulfil cellular functions beyond those of canonical tRNAs (Kuhle et al., 2023; Muthukumar et al., 2024).
While Ψ is the most common modified nucleoside found in tRNAs (Fernandez-Vizarra et al., 2006), methylation is the most prominent type of tRNA modification (Höbartner et al., 2024). Nucleosides in tRNAs can carry methyl groups on the 2′-hydroxyls of ribose moieties or on any type of nucleobase. The most common methylated nucleoside in tRNAs is N1-methyladenosine (m1A), which is present at various positions of many tRNAs from different organisms (Oerum et al., 2017). 5-methyluridine (m5U) in the T-loop is one of the most conserved tRNA modifications found across all three domains of life (Huang et al., 2021), whereas 3-methylcytidine (m3C) is a eukaryotic-specific modification present at particular positions of a subset of tRNAs (Bohnsack et al., 2022). N2-methylguanosine (m2G) and N2,N2-dimethylguanosine (m22G) are present in eukaryotic, bacterial, archaeal, mitochondrial, and chloroplast-derived tRNAs (Canaday et al., 1980; Hirata et al., 2016; Purushothaman et al., 2005; Stark et al., 2002; Suzuki et al., 2020), indicating a universally conserved functional relevance of this type of modifications.
Guanosine can be either mono- or di-methylated on the exocyclic nitrogen attached to carbon 2, forming m2G or m22G (Figure 1). Both types of methylation lead to non-standard conformations across the carbon 4–carbon 5 bond and affect nucleoside stacking interactions (Ginell and Parthasarathy, 1978). However, the presence of one versus two methyl groups differentially influences the base-pairing properties of the nucleoside. Mono-methylation of G to m2G does not affect canonical Watson-Crick base-pairing (m2G–C) and m2G is also able to form non-canonical base pairings (m2G·A and m2G·U; Figures 1A, B; Rife et al., 1998). The methyl group of m2G can adopt either s-cis or s-trans energetically stable rotamers. In canonical m2G–C base pairing, the methyl moiety is in the s-trans orientation, whereas it is in the s-cis orientation when m2G base-pairs with A. As a result of the free rotation around the carbon 2–nitrogen bond, both the s-trans and s-cis orientations are possible when m2G is base-paired with U (Figure 1B; Rife et al., 1998). In contrast to the mono-methylation, di-methylation of nitrogen 2 of G to m22G disturbs Watson-Crick base-pairing with C (Pallan et al., 2008). However, N2 of G is not involved in the formation of non-canonical base pairing, so m22G is still able to base-pair with A and U (Figures 1A, C; Steinberg and Cedergren, 1995). However, di-methylation changes the base-pairing mode of G with A from a sheared base-pair to a more stable imino-hydrogen bonded form (Pallan et al., 2008).
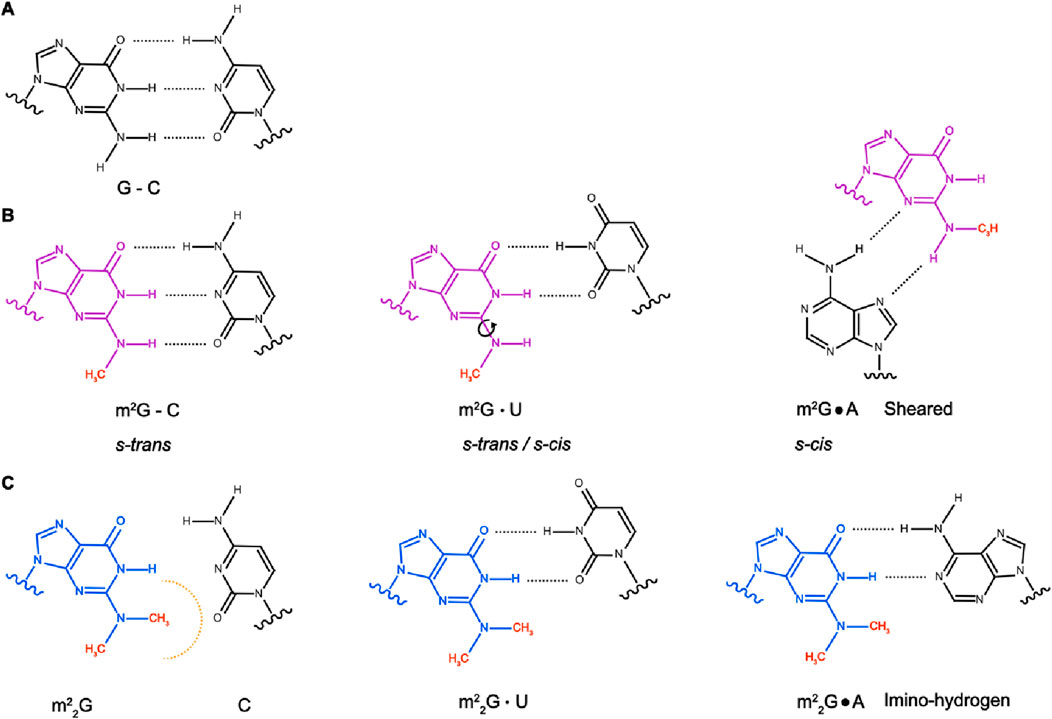
Figure 1. Base-pairing properties of G, m2G and m22G. (A) Canonical base-pairing between guanosine (G) and cytidine (C). (B) N2-methylguanosine (m2G) forms canonical base-pairing with C, with the methyl group in s-trans orientation. m2G also forms non-canonical base-pairing with U, where the methyl group can adopt either s-trans or s-cis orientation, and sheared base-pairing with A, with the methyl group in s-cis orientation. (C) Conversion of G to N2,N2-dimethylguanosine (m22G) hinders the formation of G-C canonical base-pairing but does not affect base-pairing with U. Compared to G, m22G alters the non-canonical base-pairing with A from the sheared to the imino-hydrogen form.
Quantitative detection methods with high sensitivity are essential for analyses of RNA modifications (Helm and Motorin, 2017). Several computational tools are available to predict m2G sites in RNAs based on nucleoside chemical properties, sequence-derived features and position-specific scoring matrices (Ao et al., 2022; Chen et al., 2019; Ryvkin et al., 2013). Experimentally, similar to other modified nucleosides, m2G and m22G can be detected by high-performance liquid chromatography (HPLC) coupled with mass spectrometry (MS) (Wang et al., 2023; Zhang et al., 2022b). Total RNA, enriched RNA populations or specific target RNAs can be digested into nucleosides, separated by reverse-phase chromatography, then detected by MS. The retention time and mass-to-charge ratio acquired by MS determines the presence of m2G and m22G and allows its quantification in the analyte (Yoluç et al., 2021). Partial digestion of tRNAs followed by MS can identify not only the presence of these modified nucleosides but also their position within the tRNA sequence (Jones et al., 2023).
The inability of m22G to form canonical base-pairing causes reverse transcriptase termination at the modification site and truncated cDNAs can be detected through the use of radioactively or fluorescently labelled primers when prior knowledge of the modification site is available (Funk et al., 2020). Alternatively, truncated cDNAs could be ligated to sequencing adaptors allowing mapping of m22G modifications in a transcriptome-wide manner. By contrast, the ability of m2G to base-pair as unmodified G makes its detection by approaches other than MS challenging. However, the high oxidizability of G has been exploited for the development of the m2G detection approach PhOxi-Seq (Chung Kim Chung et al., 2022; Klimontova et al., 2024); oxidation of m2G in the presence of a photoactivated catalyst leads to its conversion to 2,5-diamino-4H-imidazole-4-one, which base-pairs with G. This altered base-pairing induces clear mutational signatures during sequencing, enabling site-specific detection of m2G either within specific RNAs or within the transcriptome. A similar mutational signature arises from the presence m1G necessitating selective demethylation of m1G prior to photo-oxidation for the unambiguous detection of m2G. Importantly, m2G is a precursor of m22G (Edqvist et al., 1992), meaning that detected m2G sites have the potential to represent intermediates during m22G formation, and therefore, parallel detection of both modification types is desirable. Interestingly, antibodies against m22G are commercially available, potentially offering an alternative approach for detection of the modified nucleoside in cellular RNAs.
Bacterial and archaeal ribosomal RNAs (rRNAs) contain m2G, m22G has been detected in mRNAs from the yeast Saccharomyces cerevisiae (Sc) and the U6 small nuclear RNA (snRNA) of higher eukaryotes contains an m2G, but m2G and m22G are predominantly found in tRNAs (De Crécy-Lagard et al., 2019; Jones et al., 2023; Noon et al., 1998; Sergiev et al., 2007; Wang et al., 2023; Yang et al., 2024).
m2G is present at position six of cytoplasmic tRNAs from different species including the eubacterium Thermus thermophilus (Tt), the thermophilic methanogenic archaeon Methanocaldococcus jannaschii (Mj), Bos taurus (Bt) and humans (Figure 2; Purushothaman et al., 2005; Wang et al., 2023; Yang et al., 2021). Intriguingly, in specific tRNA isoacceptors from some species (for example, Loligo bleekeri and Thermococcus kodakarensis (Tko)), G6 is unmethylated, but m2G is present on the opposite strand of the acceptor stem at position 67, which base-pairs with position 6 (Figure 2A; Hirata et al., 2019; Matsuo et al., 1995). Despite the high conservation of m2G6 across mammalian tRNAs, bovine mitochondrial tRNA (mt-tRNA)Asp contains m2G6 (Suzuki and Suzuki, 2014) while this modification is lacking in the human counterpart (Suzuki et al., 2020). Furthermore, in human, bovine and chicken tRNATrp(CCA), m2G is present at position seven rather than position 6 (Yang et al., 2021).
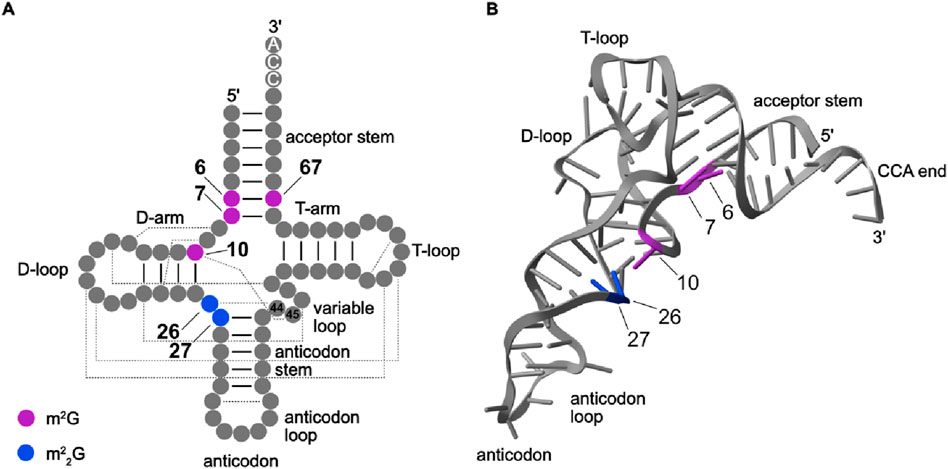
Figure 2. Secondary and tertiary structures indicating positions m2G and m22G in tRNAs. (A) The secondary structure of a canonical human tRNA with the positions of m2G (purple) and m22G (blue) modifications indicated. tRNA secondary structure and positions according to Sprinzl and Vassilenko, 2005. Base-pairings within stems are shown by black lines and base-pairings leading to tertiary structure formation are indicated by dashed lines. (B) The tertiary structure of tRNAAsp (PDB: 8OMR) is shown as a representative human tRNA. The nucleobases are shown as sticks, and m2G and m22G modifications present in human tRNAs are colored as in (A).
Many archaeal and eukaryotic tRNAs also, or instead, contain N2-methylated guanosines at position 10 (Figure 2; Bourgeois et al., 2017b; Hirata et al., 2019; Kuchino et al., 1982; Nishida et al., 2022; Purushothaman et al., 2005; RajBhandary et al., 1968; Suzuki et al., 2020; Wang et al., 2020). While structural studies have suggested the presence of the m2G10 at bacterial tRNAs (Stark et al., 2002), it remains unclear if this modification is indeed present. Another example of species-specific guanosine methylation patterns in tRNAs is that while eukaryotic tRNAs typically contain m2G10, in many archaeal tRNAs, G10 is di-methylated to form m22G10 (Edqvist et al., 1995).
m2G is also installed at position 26 of many tRNAs (Figure 2; Edqvist et al., 1992). In most cytosolic tRNAs, m2G26 is a precursor to the m22G modifications present at this position (Cappannini et al., 2023; Holley et al., 1965). Only in very few cases, such as tRNAiMet, is the m2G26 modification (partially) retained (Kawamura et al., 2014; Walker, 1983; Xiong et al., 2023). By contrast, only one of the four human mt-tRNAs in which m2G26 is installed is further methylated to form m22G (Suzuki et al., 2020). The tandem occurrence of adjacent m22Gs at positions 26 and 27 has been reported in tRNAs from the eubacterium Aquifex aeolicus (tRNACys; Awai et al., 2009), and bovine and human cytosolic tRNATyr (Johnson et al., 1985; Edqvist et al., 1995; Hwang et al., 2024; Zhang et al., 2024a). Interestingly, in the archaeon Sulfolobus acidocalclarius, tRNAMet contains N2, N2, 2′-O-trimethylguanosine (m22Gm26) where the ribose methylation is mediated by an RNA-guided 2′-O-methyltransferase (Kuchino et al., 1982).
Despite organism-specific variations in the extent of methylation (mono- versus di-methylation), guanosine N2 modifications are present within specific tRNA regions. m2Gs at positions 6/7 base-pair with nucleosides 67/66 forming the base of the acceptor stem, while m2Gs at position 10 base-pair with nucleosides at position 25 terminating the D-arm as well as forming tertiary interactions with nucleosides at position 45 (Figure 2A). By contrast, position 26 of eukaryotic tRNAs is in the hinge region between the D-arm and the anticodon stem (Figure 2B). The presence of m2G at positions 6, 7 and 67 within the acceptor stem is consistent with retention of the Watson-Crick base-pairing capacity of the mono-methylated form of G. The long-range tertiary interactions formed by m2(2)G10 and m2(2)G26 within tRNAs rationalize the presence of either N2 mono- and di-methylated G at this position (see Section 2.5.1). Although m22G prevents Watson-Crick base-pairing, in tRNATyr, m22G27 associates with U/A44 via non-canonical base-pairing, thus avoiding disruption of the anticodon stem (Johnson et al., 1985; van Tol et al., 1987).
The deposition of m2G and m22G modifications is catalyzed by Rossmann fold methyltransferases (MTases) that use S-adenosyl-L-methionine (SAM) as a methyl group donor. Similar to the methylation of amino groups on DNA and proteins, the methylation reactions catalyzed by m2G and m22G RNA MTases follow an SN2-like substitution mechanism (Figure 3A; Abdelraheem et al., 2022; Awai et al., 2011; Hirata et al., 2016; Wang et al., 2020). The nitrogen atom of the exocyclic amino group of the guanine base initiates a nucleophilic attack onto the methyl group of SAM and when the nitrogen 2–carbon bond is formed, the carbon–sulfur bond within SAM is cleaved. The target base is thereby methylated and S-adenosyl-L-homocysteine (SAH), the derivate of SAM lacking the transferred methyl group, is released. This leaves the methylated nucleoside positively charged and a basic amino acid side chain within the MTase active site abstracts a proton from the former amino group so that the methylated nucleoside becomes uncharged.
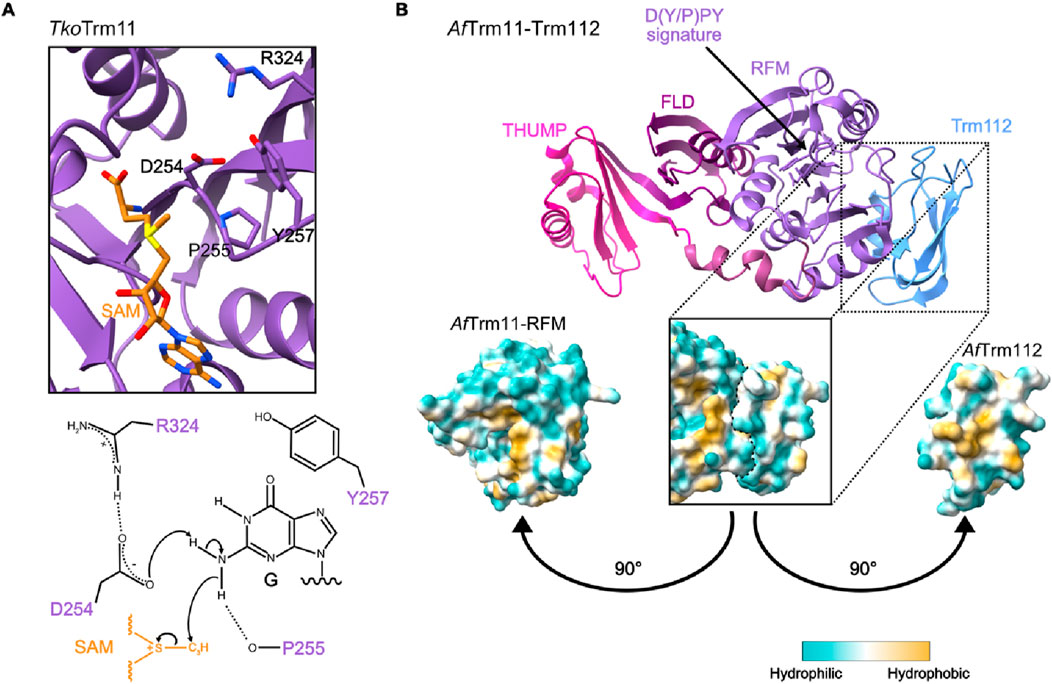
Figure 3. Catalytic mechanism and structure of m2G10 MTase Trm11. (A) The catalytic center of the tRNA m2(2)G10 methyltransferase Trm11 from Thermococcus kodakarensis (Tko) (PDB: 5E72; top) and the proposed mechanism of the SN2-like substitution reaction (bottom). R324 promotes the base-like activity of aspartic acid 254 (D254) to deprotonate the exocyclic amino group of guanosine (G), making it a nucleophile. This intermediate is stabilized by stacking interaction with tyrosine 257 (Y257) and able to attack the methyl group of S-adenosylmethionine (SAM). (B) Representative structure of a Trm11-Trm112 complex from Archaeoglobus fulgidus (Af; PDB: 6ZXW) with different domains of Trm11 and Trm112 highlighted (THUMP - THioUridine synthases, RNA Methylases and Pseudouridine synthases; FLD–ferredoxin-like domain; RFM–Rossmann-fold MTase. The D(Y/P)PY signature is indicated with an arrow. The lipophilicity potential map (bottom) shows the hydrophobic patches on the interacting surfaces of the Trm11 RFM domain and Trm112.
MjTrm14 was the first m2G6 MTase identified when it was discovered to target tRNACys (Menezes et al., 2011). Subsequently, m2G6 in TttRNAPhe (the only bacterial tRNA that contains this modification) was shown to be introduced by TtTrmN, a eubacterial ortholog of MjTrm14 (Roovers et al., 2012). Despite the low sequence similarity between MjTrm14 and TtTrmN, crystallographic analyses revealed similar tertiary structures of these two MTases (Fislage et al., 2012). In human cells, the writer enzyme responsible for the m2G6/7 modifications in cytosolic tRNAs is THUMPD3 (Yang et al., 2021). THUMPD3 has a domain architecture similar to MjTrm14/TtTrmN, suggesting evolutionary conservation of the characteristic features of these MTases. This fundamental domain architecture of m2G6 MTases extends to Trm11/TRMT11, an enzyme conserved from archaea through eukaryotes that methylates N2 of guanosines at position 10 of tRNAs. Trm11 from archaea such as Pyrococcus abyssi (Pa) and Tko not only catalyzes the installation of m2G10 but can also further methylate it to m22G10 (Armengaud et al., 2004; Hirata et al., 2016). However, ScTrm11 and human TRMT11 exclusively install m2G10 (Purushothaman et al., 2005; Wang et al., 2023).
The catalytic domains of these m2G MTases are characterized by the sequence motif [D/N/S]-P-[P/I]-[F/Y/W/H] and this highly conserved signature specifies them as m2G methyltransferases (Figure 3B; Bujnicki, 2000; Hirata et al., 2016; Wang et al., 2020). To initiate methyl group transfer from SAM to the substrate, the [D/N/S]-P residues interact with the target exocyclic amino group of the nucleobase via hydrogen bonds. From its planar position, the lone pair of electrons of the amino group is orientated towards the methyl group of the bound SAM to elicit the nucleophilic attack. The [F/Y/W/H] residue forms π–π stacking interactions with the nucleic acid that are essential to stabilize the reaction intermediate. As the m2G MTases target base-paired purines, it is assumed that the base is flipped out for methylation.
Another striking feature of these m2G MTases is that the catalytic domain is typically accompanied by an N-terminal ferredoxin-like domain (FLD) and an associated thiouridine synthases, RNA methylases and pseudouridine synthases (THUMP) domain (Figure 3B). The THUMP fold is an ancient RNA-binding domain that has been shown to assist diverse RNA modification enzymes in binding their targets (Aravind and Koonin, 2001). In the context of different tRNA modification enzymes, the THUMP domains bind the 3’ -CCA tails that are added to tRNAs post-transcriptionally and ultimately act as docking sites for conjugated amino acids (Fislage et al., 2012; Hirata et al., 2016; Hori, 2023; McCleverty et al., 2007; Neumann et al., 2014; Nishida et al., 2022; Randau et al., 2009; Waterman et al., 2006). Consistent with the importance of such interactions for substrate binding, the presence of a 3’ -CCA end is strictly required for the association of THUMPD3 with substrate tRNAs (Yang et al., 2021). Likewise, structural analysis revealed that the THUMP domain of Trm11 from the Archaeoglobus fulgidus (Af) archaeon recognizes the 3’ -CCA end of tRNAs (Bourgeois et al., 2017b). Lack of the 3’ -CCA and/or destabilization of the acceptor stem significantly reduces substrate methylation by AfTrm11 and human THUMPD3 (Bourgeois et al., 2017b; Yang et al., 2021), indicating coordinated action of the THUMP and MTase domains. It is proposed that the FLD and following linker region adjust the distance between the THUMP fold and the catalytic domain, enabling capture of the tRNA’s 3′-CCA end by the THUMPD fold to be coordinated with correct positioning of particular nucleosides in the enzyme’s active site for modification (see, for example, Bourgeois et al., 2017b; Hirata et al., 2016; Neumann et al., 2014). Thus, the N-terminal non-catalytic regions of m2G MTases fulfil dual functions in i) recognizing particular features of target RNAs to ensure recruitment to appropriate substrates and ii) acting as molecular rulers to direct methylation of the specific target nucleosides. This mode of structure-based target recognition contrasts with the sequence motif-based recognition of some modification enzymes (see, for example, Aoyama et al., 2020; Kleiber et al., 2022; Linder et al., 2015). However, it is ideally suited for MTases, such as Trm11/TRMT11 and THUMPD3, that methylate a broad spectrum of similarly folded RNAs; the diverse sequences contexts of m2G6 and m2(2)G10 in tRNAs (Sajek et al., 2019) are only compatible with structure-based target recognition by the cognate MTases. Beyond the acceptor stem and 3’ -CCA, tertiary interactions between the D- and T-arms are also important recognition elements for these m2G MTases; nucleoside substitutions in the T- and variable loops that disrupt these connections perturb methylation of G10 by ScTrm11 (Purushothaman et al., 2005). Furthermore, it has been shown that ScTrm11-mediated methylation of G10 is sensitive to both the length of the variable loop and nucleoside substitutions at position 38 within the anticodon loop (Nishida et al., 2022), suggesting that diverse tRNA features influence substrate methylation by m2G MTases.
The prokaryotic m2G6 MTases (TtTrmN and MjTrm14) and the m2G10 MTases of some archaea (e.g., Pa/TkoTrm11) function independently to catalyze their target modifications (Menezes et al., 2011; Roovers et al., 2012). By contrast, the human m2G6 and m2G10 tRNA MTases (THUMPD3 and TRMT11, respectively), ScTrm11 and the archaeal m2G10 MTases (e.g., AfTrm11 and Halloferax volcanii (Hv) Trm11) associate with Trm112/TRMT112 to form MTase complexes (Figure 3B; Wang et al., 2023; Yang et al., 2021). Trm112/TRMT112 is a hub MTase cofactor protein composed of a zinc-binding domain and a four-stranded β-sheet (Heurgué-Hamard et al., 2006). Human TRMT112 interacts with at least seven MTases, increasing their stabilities and substrate affinities as well as modulating their catalytic activities (Bourgeois et al., 2017a; 2017b; Brūmele et al., 2021; Gao et al., 2020; Lacoux et al., 2020; Leetsi et al., 2019; Õunap et al., 2015; Van Tran et al., 2019; Wang et al., 2023; Yang et al., 2021). Interactions of THUMPD3, human TRMT11 and Trm11 from some archaea with Trm112/TRMT112 have been demonstrated by proximity labelling and co-immunoprecipitation experiments, and in vitro co-expression approaches have confirmed direct cofactor–MTase interactions (Brūmele et al., 2021; Van Tran et al., 2018; Wang et al., 2023; Yang et al., 2021) Structural analysis of AfTrm11 revealed that, similar to other Trm112–MTase complexes (Létoquart et al., 2014; Van Tran et al., 2019), Trm112 binds to the catalytic domain on the face opposite to the SAM binding pocket (Wang et al., 2020). While experimentally derived structures of the human TRMT11-TRMT112 and THUMPD3-TRMT112 complexes are lacking, modelling of the THUMPD3-TRMT112 complex strongly supports analogous interactions (Yang et al., 2021). Stabilization of the binding interface relies on assembly of a β-zipper formed by hydrogen bonds between specific β-strands in the MTase and cofactor. There can be several consequences of Trm112/TRMT112 binding, which include the shielding of hydrophobic patches on the surface of cognate MTases (Figure 3B; Hirata et al., 2016; Wang et al., 2020). For example, the anticipated TRMT112 binding site of THUMPD3 is strongly hydrophobic and recombinant THUMPD3 alone forms aggregates in vitro, while THUMPD3 co-purified with TRMT112 is soluble (Yang et al., 2021). AfTrm112 prevents the thermal denaturation of AfTrm11 in vitro, suggesting the importance of this cofactor in an archaeon with an optimal growth temperature of 83°C (Wang et al., 2020). Lack of TRMT112 has been shown to reduce TRMT11 and THUMPD3 levels in human cells (Brūmele et al., 2021), highlighting the role that cofactor association plays in maintaining the solubility and stability of non-bacterial m2G MTases. Beyond regulating MTase stability, interaction with Trm112/TRMT112 can influence the catalytic activity of m2G MTases. Although AfTrm11 is an active MTase alone, in the presence of Trm112, its catalytic activity is strongly enhanced (Wang et al., 2020). By contrast, ScTrm11 and human TRMT11 only display methylation activity in the presence of ScTrm112/TRMT112. Interestingly, the mechanistic basis of stimulation of Trm11/TRMT11 by Trm112/TRMT112 differs between species; while SAM binding by ScTrm11 is enhanced by ScTrm112, AfTrm112 does not influence the affinity of AfTrm11 for SAM but rather accelerates the catalytic step of the methylation reaction (Bourgeois et al., 2017b; Wang et al., 2020). Similar to eukaryotic Trm11/TRMT11, THUMPD3 alone lacks in vitro methylation activity as it fails to bind either substrate tRNAs or SAM, properties that are restored upon formation of the THUMPD3–TRMT112 complex (Brūmele et al., 2021; Wang et al., 2023; Yang et al., 2021).
Distinct from the evolutionarily-conserved and architecturally-related m2G MTases targeting the acceptor stem and D-arm, the writer enzymes responsible for m2G26, m22G26 and m22G27 modifications in tRNAs are Trm1/TRMT1 and TRMT1L (Figure 4A; Dewe et al., 2017; Ellis et al., 1986; Hwang et al., 2024; Zhang et al., 2024a; Constantinesco et al., 1999; 1998). On the structural level, these enzymes both contain Rossmann-fold MTase domains and zinc fingers (ZnF) of the CCCH type (Figure 4B; Awai et al., 2011; Bujnicki et al., 2002; D’Oliviera et al., 2022; Ihsanawati et al., 2008). Interestingly, despite TRMT1 and TRMT1L sharing the same two domains, they are arranged differently with the ZnF of TRMT1 C-terminal of the MTase domain and the MTase domain of TRMT1L being preceded by the ZnF. CCCH-type ZnF domains are associated with RNA binding (Fu and Blackshear, 2017), suggesting that this region of TRMT1 and TRMT1L may contribute to substrate interactions. Consistent with this, removal of the TRMT1 ZnF ablates substrate methylation (Dewe et al., 2017; Zhang et al., 2024a). It is interesting to note that the Trm112/TRMT112 cofactor of the m2(2)G6/7/10 MTases, which do not themselves contain a ZnF, is also a ZnF-containing protein.
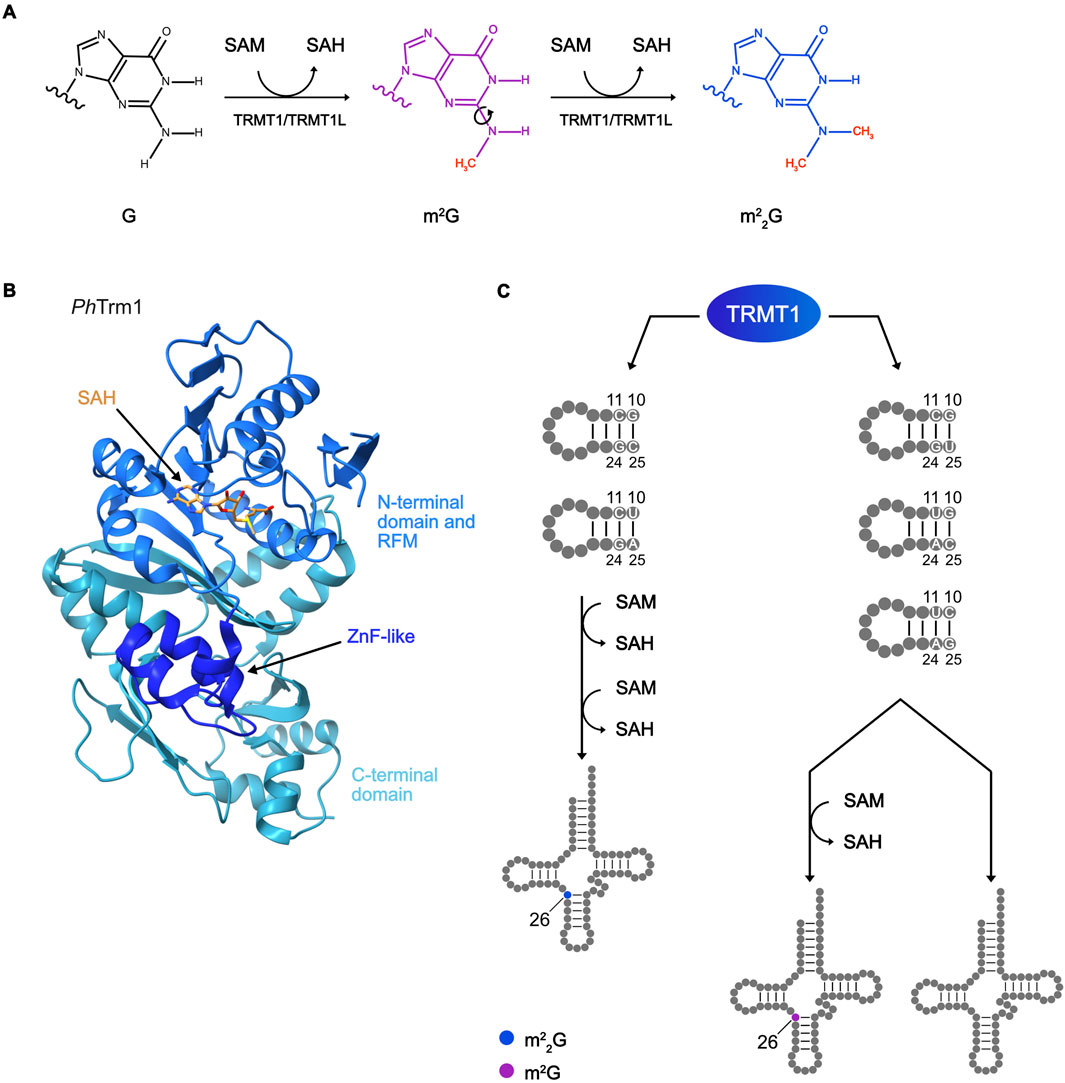
Figure 4. Methylation scheme of m22G installation, structure of an m22G MTase, and its substrate specificity. (A) Reaction scheme of m22G deposition, showing the two subsequent methylation reactions using S-adenosylmethionine (SAM) as a methyl group donor and releasing S-adenosylhomocysteine (SAH). (B) Representative structure of Trm1 m22G MTase from Pyrococcus horikoshii (Ph; PDB: 2YTZ) with N-terminal, Rossmann-fold MTase (RFM), and C-terminal domains highlighted. The zinc finger (ZNF)-like motif and bound SAH are indicated with arrows. (C) Diagram showing the mechanism of TRMT1 substrate specificity for m2G or m22G deposition depending on the sequence of the target tRNA D-arm.
A distinguishing architectural feature of Trm1/TRMT1 and TRMT1L is the presence of a mitochondrial targeting sequence (MTS) at the N-terminal end of Trm1/TRMT1 that is lacking from TRMT1L. Cellular fractionation and immunofluorescence microscopy demonstrate the presence of Trm1/TRMT1 in the cytosol/mitochondria and nucleus, while TRMT1L is enriched in nuclei/nucleoli (Dewe et al., 2017; Hwang et al., 2024; Li et al., 1989). These sub-cellular localizations are in line with the dual functions of Trm1/TRMT1 in methylating G26 in cytosolic and mitochondrial tRNAs and the identification of the nuclear-encoded tRNATyr as the sole target of TRMT1L (Hwang et al., 2024; Xiong et al., 2023). Although TRMT1 is detected in the cytosol (Dewe et al., 2017), it is likely that m22G26 modifications of cytosolic tRNAs take place prior to their nuclear export. This notion is supported by the finding that ScTrm1 is enriched at the inner nuclear membrane, to which it is targeted by a specific sequence motif (Diaz-Muñoz et al., 2014; Lai et al., 2009). As the human m2G MTases THUMPD3 and TRMT11 lack nuclear localization signals and are present exclusively in the cytosol (Brūmele et al., 2021), this suggests that m22G26/27 tRNA modifications are installed prior to m2G6/7/10 modifications. Lack of the MTS of TRMT1 obliterates methylation of mt-tRNAs while installation of m2G26/m22G26 modifications in cytosolic tRNAs is unaffected (Dewe et al., 2017). The expressed Trm1/TRMT1 protein contains an MTS and it is unclear how sufficient levels of cytosolic Trm1/TRMT1 are maintained to enable efficient tRNA methylation in this compartment.
While the methylation reactions catalyzed by Trm1/TRMT1 and TRMT1L follow the SN2-like mechanism described for the m2G MTases, details of how the sequential methylation reactions take place remain unclear. To accomplish di-methylation, following the first methylation reaction, release of SAH is required and the MTase must re-bind SAM to enable the second methylation reaction to occur (Figure 4A). Formation of m22G is a thus two-step reaction, but it remains unknown if the MTases dissociates from the substrate after the first methylation step and re-binds, or whether SAH release and recharging with SAM take place on target-bound MTases. While ScTrm1 catalyzes m22G26 formation with high efficiency on some tRNA substrates, the mono-methylated m2G intermediate is detected in in vitro methylation reactions taking place on mutated or non-endogenous tRNAs (Edqvist et al., 1994). This suggests that specific features of the tRNA substrates determine whether they are mono- or di-methylated. Strikingly, while cytosolic TRMT1 predominantly introduces m22G26, within mitochondria, only mt-tRNAIle contains m22G26 and all other TRMT1 targets contain m2G26. It has been shown that ScTrm1 recognizes the G10:C25 and C11:G24 base-pairs in the D-stem along with nucleosides in the variable loop to specifically deposit m22G26 modification (Edqvist et al., 1995; 1992). The human ortholog TRMT1 is able to catalyze m22G26 formation in mt-tRNAIle, which contains a U10:A25 base pair and a deeper investigation of human TRMT1 recognition elements revealed that it requires C11:G24 and G10:C25 or U10:A25 base pairs for m22G26 modification (Figure 4C; Xiong et al., 2023). tRNAs not satisfying these criteria (i.e., cytosolic tRNAiMet and the mt-tRNAs with modified G26, except mt-tRNAIle) are only mono-methylated by TRMT1 to carry m2G26. It is possible that tRNAs not satisfying the criteria for efficient m22G26 installation have a lower affinity for Trm1/TRMT1 and dissociate from the enzyme after the first methylation step, but such mechanistic details remain to be clarified. Notably, both TRMT1 and archaeal Trm11 display dual activities as m2G and m22G MTases whereas eukaryotic TRMT11 and the G6/7 MTases MjTrm14/TtTrmN/THUMPD3 only install m2G (Armengaud et al., 2004; Hirata et al., 2016; Menezes et al., 2011; Roovers et al., 2012; Wang et al., 2023; Xiong et al., 2023; Yang et al., 2021). The identification of tRNA elements influencing mono- versus di-methylation by TRMT1 (Xiong et al., 2023) raises the intriguing question whether features of the substrate tRNAs of MjTrm14/TtTrmN/THUMPD3 and eukaryotic TRMT11 dictate their action as m2G, rather than m22G, MTases or whether the nature of the methylation reaction catalyzed is inherent to the MTases themselves.
Modified nucleosides can influence RNAs directly by modulating RNA folding and/or RNA–RNA interactions. Alternatively, recognition of specific modified RNA nucleosides by proteins termed “readers” can have functional consequences, such as destabilization or altered translation (Lewis et al., 2017; Roundtree et al., 2017). To date, no proteins specifically recognizing m2G or m22G have been identified, implying that these modifications predominantly exert direct effects on the RNAs that contain them. At the level of base-pairing, chemical and molecular dynamics studies show that m2G:C and G:C base-pairs in RNA duplexes and GNRA tetraloops are isoenergetic, indicating that the m2G modification does not directly contribute to RNA structural stability (Bavi et al., 2013; Rife et al., 1998). The presence of a methyl group may weakly stabilize RNA structures due to improved hydrophobic interactions, but this effect is canceled out by mild destabilization due to the loss of rotational entropy of the methyl group (Rife et al., 1998).
Modifications within the core of tRNAs are typically thought to have stabilizing effects, and although the precise molecular function(s) of the m2(2)G modifications remain to be clarified, these methylations likely contribute to tRNA structure by regulating the formation of tertiary interactions within the tRNA core. Interactions between the G10-C25 base-pair and nucleoside 45 in the variable loop as well as between m22G26 and nucleoside 44 are important for maintaining the L-shaped tRNA architecture (Figure 5A; Shi and Moore, 2000). In human cells, when m2G10 is lacking, the global structures of tRNAs determined by native gel electrophoresis are not noticeably affected (Wang et al., 2023), suggesting that the impact of this modification on tRNA architecture is subtle. This notion is supported by the observation that m2G10 alone does not influence the folding of mt-tRNALys, but that it augments the stabilizing effect of m1A9 modifications (Kobitski et al., 2011).
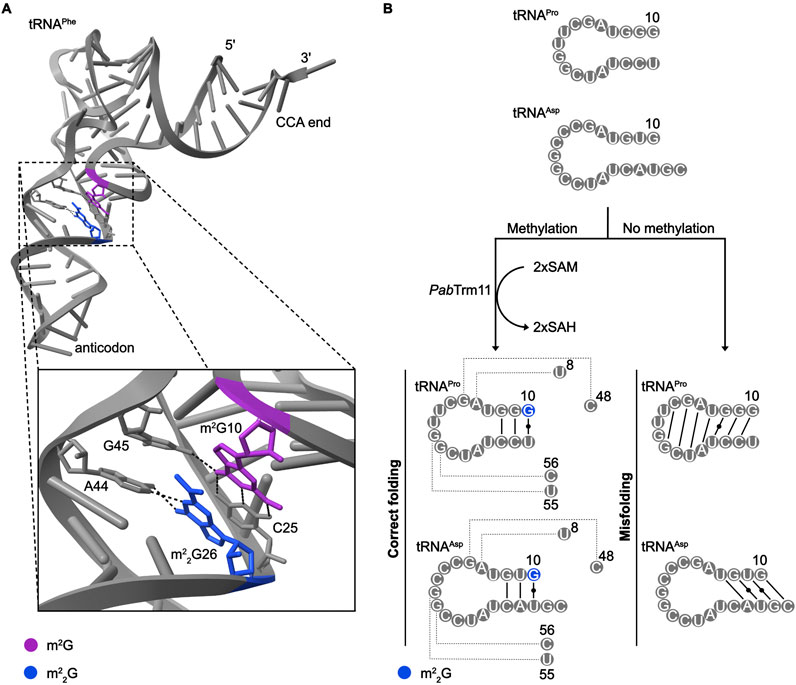
Figure 5. Influence of m2(2)G modifications on tRNA folding and structure. (A) Interactions of the m2G10-C25 base-pair with G45 and m22G26 with A44 in S. cerevisiae tRNAPhe (PBD: 1EHZ). (B) Alternative base-pairing interactions within Pyrococcus abyssi (Pab) tRNAPro and tRNAAsp depending on the presence of m22G10.
When m22G is present in eukaryotic tRNAs, it is primarily found linking the D-arm and anticodon stem (Edqvist et al., 1992) where it has been suggested to act as a molecular hinge (Holbrook et al., 1978). As di-methylation of G strongly impacts its base-pairing properties, it is likely that the presence of m22G26 limits the formation of alternative, sub-optimal tRNAs conformations that then represent an energetic hurdle requiring to be overcome to allow correct folding (Steinberg and Cedergren, 1995; Urbonavičius et al., 2006). m22G26 is much more prominent within cytosolic tRNAs than mitochondrial tRNAs, which may reflect the ability of many mt-tRNAs to adopt non-canonical architectures. Consistent with the notion that m22G26 promotes correct tRNA folding, the presence of m22G generally shift the equilibrium of duplex-hairpin structures toward the hairpin form (Pallan et al., 2008). The importance of m22G in ensuring proper tRNA folding is emphasized by the presence of this modification at position 10 in some hyperthermophilic archaea; in this context, the di-methylation of G10, rather than the mono-methylation typically observed in eukaryotes, may play an important role in ensuring the establishment of correctly folded tRNAs at high temperatures (Hirata et al., 2019). For example, in the archaeon P. abyssi, N2 dimethylation of G10 has been proposed to impede the formation of aberrant base-pairings in tRNAPro and tRNAAsp, thus shifting the equilibrium towards correctly folded tRNAs (Figure 5B; Urbonavičius et al., 2006). Potentially linked to defective folding, lack of m22G26 modifications is associated with decreased tRNA stability (Porat et al., 2023; Zhang et al., 2024a). Interestingly, the levels of acp3U and dihydrouridine modifications in several tRNAs depends on the m22G27 MTase TRMT1L (Hwang et al., 2024; Zhang et al., 2024b); within this modification circuit, it remains to be determined if m22G27 is a pre-requisite for the other modifications because of changes in tRNA architecture that it induces or whether its presence is directly sensed by the corresponding modification enzymes.
Beyond the direct effects of m22G26 modifications on tRNA structure, a non-catalytic role for the Schizosaccharomyces pombe (Sp) m22G26 MTase Trm1 in chaperoning pre-tRNA folding has also been proposed (Porat et al., 2023; Vakiloroayaei et al., 2017). In vitro, both wild-type and catalytically inactive Trm1 display RNA strand annealing and dissociation activities that are characteristic of RNA chaperones (Porat et al., 2023). Supporting a functional relevance of this additional activity of SpTrm1, the comined lack of SpTrm1 and the well-established RNA chaperone La is synthetically lethal (Vakiloroayaei et al., 2017).
Beyond the molecular level, cellular phenotypes associated with m22G, and especially m2G, tRNA modifications have proved challenging to identify. While the absence of modified nucleosides within the anticodon loop typically leads to demonstrable defects on translation, the effects of individual RNA modifications in other regions of tRNAs are often more subtle. This likely reflects the fact that the dense network of modified nucleosides present in most tRNAs functions co-operatively to fine-tune tRNA structure and function. As has been observed for other types of modifications, such as ribosomal RNA 2′-O-methylations and pseudouridylations (King et al., 2003; Liang et al., 2009), the absence of an individual m2G/m22G modifications often does not induce an obvious phenotype, but the combined loss of two or more modifications can lead to significant phenotypic effects. For example, in S. cerevisiae, the simultaneous deletion of the trm11 and trm1 genes (i.e., preventing the formation of m2G10 and m22G26) increases doubling time and affects cell morphology (Purushothaman et al., 2005). Re-expression of either Trm11 or Trm1 leads to restoration of the wild-type phenotype, indicating that both m2G10 and m22G26 modifications are physiologically relevant. The mutual connection between these two modifications is rationalized by the tertiary structure of yeast tRNAPhe, which reveals that m2G10 and m22G26 are stacked on each other with methyl groups pointed to the same face of the tRNA (Purushothaman et al., 2005). Thus, a single hydrophobic group on the surface of the tRNA provided by either of these methylation marks may be enough to ensure normal tRNA folding and function. Notably, TRMT1-deficient human cells exhibit a decreased proliferation rate (Dewe et al., 2017), perhaps suggesting that m22G26 is more functionally relevant in human cells than in yeast. Similarly, while knockout of the m2G6/7 writer THUMPD3 in HEK293T cells mildly reduces growth rate (Yang et al., 2021), knockout of THUMPD3 or the m2G10 MTase TRMT11 in HCT116 cells does not significantly perturb cell growth (Wang et al., 2023). However, when both THUMPD3 and TRMT11, and their cognate m2G6/7 and m2G10 tRNA modifications, are lacking, cellular proliferation is significantly impaired (Wang et al., 2023).
Given the function of tRNAs as adaptor molecules during translation, defects in protein synthesis likely underpin the perturbed growth of cells lacking m2G and/or m22G in tRNAs. Generally, these modifications could modulate different aspects of translation by impacting tRNA charging or molecular interactions between tRNAs and ribosomes. Recent data demonstrate that lack of THUMPD3 and/or TRMT11 does not affect the aminoacylation status of m2G6-, m2G10-or m2G6+10-containing tRNAs (Wang et al., 2023), indicating that in contrast to other tRNA modifications (Clifton et al., 2021; Gieg and Eriani, 2023), these methylation marks are not recognized by the tRNA aminoacyl synthetases. However, nascent protein synthesis is globally affected by lack of THUMPD3 in HEK293T cells (Yang et al., 2021), and the absence of both THUMPD3 and TRMT11 from HCT116 cells causes reduced monosome/polysome levels and impairs global translation in human HCT116 cells (Wang et al., 2023). As tRNA structure and charging are not substantially affected by lack of m2G6/7/10, the molecular basis of these translation defects remains to be determined.
Absence of the m22G26 tRNA modifications in the tRNA hinge region also affects translation. In S. pombe, the m22G modification has been suggested to enhance the activity of a nonsense suppressor tRNA in decoding the UAA stop codon, thus contributing to the regulation of translation termination (Niederberger et al., 1999). The nascent synthesis of proteins is globally down-regulated in cells lacking TRMT1, suggesting a role for m22G26 modifications in cytosolic tRNAs in maintaining efficient basal translation (Dewe et al., 2017). The effects of lack of TRMT1 on mitochondrial translation are, however, more nuanced. While the levels of CYB and ATP6 are unaffected by lack of m2G/m22G26 in mt-tRNAs, increased expression of the Complex I and IV subunits ND5/CO-I, ND4, ND2, ND1, and CO-2/CO-3 is observed upon TRMT1 knockout (Dewe et al., 2017). While the molecular basis of this phenotype is unclear, it is striking as sub-stoichiometric mt-tRNA modifications are typically associated with impaired, rather than enhanced, mitochondrial translation (Bohnsack and Sloan, 2018). Interestingly, due to sequence specificities, cytoplasmic tRNAiMet and tRNAeMet contain m2G26 and m22G26, respectively (Cappannini et al., 2023). It has, therefore, been suggested that the presence of the mono- and di-methylated forms fulfill distinct roles in translation initiation and translation elongation (Xiong et al., 2023). However, precisely how the differently methylated forms of G26 might optimize the alternative tRNAMet forms for these different functions is still unknown.
In mitochondria, the presence of m22G26 in mt-tRNAIle is not only necessary for efficient translation, but has also been shown to impact tRNA processing (Zhang et al., 2021). Transcription of the mitochondrial genome, composed of a light and heavy DNA strand, synthesizes polycistronic RNA precursors in which the mt-rRNAs and mt-mRNAs are typically separated by mt-tRNAs that are excised to release the individual RNAs (Jedynak-Slyvka et al., 2021; Ojala et al., 1981). m22G26 in mt-tRNAIle is detected within nascent, unprocessed mt-RNA, where it is implicated in regulating mt-tRNA processing (Zhang et al., 2021). The presence of m22G26 affects folding of mt-tRNAIle, thus altering interactions between the heavy and light strand transcripts such that the ds mt-RNA is cleaved more efficiently when m22G26 in mt-tRNAIle is present than when it is lacking (Zhang et al., 2021).
m2G and m22G modifications are exclusively present in the 5′-half of eukaryotic tRNAs. When tRNAs are cleaved to produce tRFs, those arising from the 5′ half are typically the most stable and can have tRNA-independent functions in gene expression regulation (Tosar et al., 2018). The functional implications of m2G and m22G in tRNA-derived fragments are currently poorly understood, but a possible epigenetic function has been suggested (Chen et al., 2016). RNA-mediated transgenerational epigenetic inheritance may rationalize the observed connection between paternal diet and metabolic disorders in offspring (Ng et al., 2010). The high levels of m2G in sperm tRFs of mice fed a high-fat diet therefore raise the possibility of a role for this modification related to epigenetic inheritance (Chen et al., 2016). Alongside potential roles in fine-tuning tRNA structure, such a function could increase the selective pressure to maintain m2G within the 5′ half of tRNAs.
The RNA modification landscape is dynamic as modifications can be differentially introduced in specific conditions and some methylation marks can be actively removed by demethylases (Roundtree et al., 2017). Nucleosides in tRNAs are typically constitutively modified, and consistent with this, MS analyses suggest high stoichiometries of m2G and m22G within tRNAs (Suzuki and Suzuki, 2014; Wang et al., 2023; Xiong et al., 2023; Yang et al., 2021). A potential exception is m2G10 of sctRNAVal(ACC), which has recently been shown to be sub-stoichiometrically methylated (Nishida et al., 2022). Interestingly, an association between the activity of RNA polymerase III, which synthesizes nuclear-encoded tRNAs, and the efficiency of m22G26 modification has been suggested in S. cerevisiae and human cells (Arimbasseri et al., 2015). When tRNA synthesis is reduced, m22G is more abundant, and when the RNA polymerase III repressor Maf1 is lacking and tRNAs levels are elevated, the incorporation of m22G26 is decreased. A model is proposed in which Trm1/TRMT1 becomes limiting when tRNAs are overexpressed, leading to sub-stoichiometric m22G26.
As well as differential installation depending on tRNA abundance, m22G26 can be actively erased. The mitochondria-localized α-ketogluterate- and Fe2+-dependent AlkB family member, ALKBH7, has been shown to demethylate m22G26 in mt-tRNAIle (Zhang et al., 2021). In vitro, ALKBH7 efficiently demethylates m22G to yield G, without substantial accumulation of the m2G intermediate. As described above, the presence of m22G26 in mt-tRNAIle influences processing of the polycistronic mt-RNA and demethylation of this position by ALKBH7 slows processing of the transcript, perhaps helping ensure accurate cleavage (Zhang et al., 2021). An atomic resolution structure of ALKBH7 revealed a solvent-exposed active site and negatively charged surface that was initially anticipated to repel RNAs due to their negatively charged backbone (Wang et al., 2014). However, ALKBH7 contains nucleic acid-binding loops, also present in other AlkB family members that target RNAs, and this region likely mediates interactions with the substrate RNA (Zhang et al., 2021). The ability of ALKBH7 to efficiently demethylate m22G26 and the identification of key amino acids required for this activity draws parallels with the development of mutant forms of Escherichia coli AlkB that can be leveraged to demethylated m22G sites in vitro. As m22G blocks reverse transcriptase progression, its removal is a pre-requisite for tRNA sequencing approaches that rely on reverse transcription read-through, such as hydro-tRNAseq and ARM-seq (Padhiar et al., 2024). Wild-type AlkB efficiently demethylates m1A and m3C (Zheng et al., 2014) and an aspartic acid 135 to serine (D135S) substitution allows demethylation of N1-methylgaunosine to G. The removal of m22G is particularly challenging due to the deeply buried position of m22G26 into the tRNA structure, however, D135S and leucine 118 to valine (L118V) substitutions that mimic key residues of ALKBH7 enable selective and efficient conversion of m22G to m2G (Dai et al., 2017). This activity differs from that of ALKBH7, which fully demethylates m22G to G, (Zhang et al., 2021). However, as m2G maintains its canonical base-pairing ability, the removal of one methyl group from m22G is sufficient to allow reverse transcriptase progression and subsequent tRNA sequencing.
Alterations in m2G and m22G stoichiometries have been observed in S. cerevisiae tRNAs upon exposure to cellular stress (Chan et al., 2010), suggesting that regulation of these modifications could contribute to the dynamic adaptation of gene expression. More specifically, exposure to hydrogen peroxide leads to an increase in m22G, while the levels of m2G are decreased in a dose-dependent manner. The assessment of changes in the levels of these modified nucleosides in a non-site-specific manner limits the mechanistic understanding of the observed dynamics. These alterations in m2G and m22G levels in tRNAs may arise due to differential expression of the MTases responsible for installing these modifications in oxidizing conditions, but it is also possible that the catalytic activity of these enzymes is affected by the stress condition. To date, no m2(2)G demethylase has been identified in yeast, but in case stress-induced changes in the m2(2)G landscape also occur in human cells, it is possible that ALKBH7 is dynamically regulated to modulate the erasing of specific methylation marks. Post-translational modifications are an efficient way to modulate protein functions, and high-throughput phosphoproteomics identify numerous sites within functionally important regions of the m2G and m22G MTases (Hornbeck et al., 2015). This raises the possibility that stress-induced post-translational modification dynamics may contribute to the regulation of m2G and m22G modifications in tRNAs. Lack of TRMT1 or TRMT1L leads to hypersensitivity to oxidative stress, suggesting an important role for m22G26/27 in cell survival in this condition (Dewe et al., 2017; Hwang et al., 2024). Elevated levels of reactive oxygen species (ROS) are observed in the absence of TRMT1 (Dewe et al., 2017), but it remains unclear how these enzymes and/or their cognate modifications contribute to maintaining redox homeostasis in cells.
Increasing evidence suggests co-ordination of the metabolic statuses of cells with their epitranscriptomes (Liu et al., 2024). Such connections likely arise as a result of the use of various metabolic intermediates as modification enzyme cofactors. For example, α-ketogluterate is essential for oxidative demethylation reactions taking place on RNAs (Fedeles et al., 2015), such as the demethylation of m22G26 in mt-tRNAIle by ALKBH7, and the methyl group donor in most cellular methylation reactions, including that generate m2G and m22G, is SAM (Sun et al., 2021). SAM is synthesized from the sulfur-containing amino acid methionine, and it has been speculated that in the archaeon M. jannaschii, Trm14-dependent methylation of m2G6 of tRNACys, which is charged with another sulfur-containing amino acids may play a role in the regulation of protein synthesis depending on sulfur availability (Menezes et al., 2011).
Given the importance of the epitranscriptome for gene expression regulation and the critical roles of modifications in tRNAs for their functionality in protein synthesis, unsurprisingly, dysfunction or abnormal expression of the >50 tRNA modifying enzymes and their cofactor proteins are associated with human diseases (Chujo and Tomizawa, 2021).
Alternations in the expression levels of the m2G MTase TRMT11, its TRMT112 cofactor and the m22G demethylase ALKBH7 are observed in various cancers. For example, overexpression of TRMT11 in high-grade gliomas is associated with increased cellular proliferation, invasion, migration and glial tumor growth (Di et al., 2013). Over-expression of TRMT112 is observed in most cancer types (Xu et al., 2022), highlighting its potential as a prognostic predictor. In ovarian serous carcinoma, ALKBH7 expression correlates positively with the pathological stage of the disease (Cai et al., 2021) and higher expression of ALKBH7 is observed in lung adenocarcinoma where it is associated with shorter overall survival time in patients (Wu et al., 2021). A single-nucleotide polymorphism in ALKBH7 that leads to an arginine 191 to glutamine (R191Q) substitution is correlated with prostate cancer, and perturbed co-substrate binding by this variant suggests that defective m22G demethylation underlies this association (Walker et al., 2017). These and other studies indicate the potential of ALKBH7 demethylase as a target for anticancer therapy (Chen et al., 2022).
Aberrant tRNA modifications are generally linked to metabolic diseases and neurological disorders, while defects in mt-tRNA modifications often leads to mitochondrial encephalomyopathies (Bednářová et al., 2017; Bohnsack and Sloan, 2018; Boughanem et al., 2023; Liu et al., 2024). The high metabolic demands and complex development of the brain rationalize the profound association of defective protein synthesis with these conditions. For example, consistent with its mitochondrial localization, ALKBH7 is involved in programmed cell necrosis and fatty acid metabolism (Fu et al., 2013; Solberg et al., 2013), and mice lacking ALKBH7 display an obesity phenotype (Solberg et al., 2013). Moreover, the m22G26 and m2G26 modifications deposited by TRMT1 in cytoplasmic and mitochondrial tRNAs are associated with microcephaly, intellectual disabilities, and epilepsy (Blaesius et al., 2018; Davarniya et al., 2015; Najmabadi et al., 2011). The autosomal recessive mutations of TRMT1 associated with these diseases often affect the enzyme’s catalytic activity, cause expression of truncated proteins that cannot associate with target RNAs or lead to nonsense-mediated mRNA decay (Blaesius et al., 2018; Zhang et al., 2020). TRMT1 variants associated with intellectual disabilities, which lack m22G deposition activity, cannot rescue normal cellular translation or oxidative stress sensitivity in TRMT1-deficient human cells (Dewe et al., 2017). However, as TRMT1 targets both cytoplasmic and mitochondrial tRNAs, further investigation is needed to clarify how aberrantly modified tRNAs in these two translation systems contribute to the pathogenesis. Interestingly, it was recently shown that upon neuronal activation, TRMT1 relocalizes from mitochondrial into distinct nuclear foci, suggesting that sequesteration may contribute to the dynamic regulation of m22G26 modifications in the context of neuronal function (Jonkhout et al., 2021).
Interestingly, infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) reduces the levels of m22G modifications and it has recently emerged that TRMT1 is a target of the SARS-CoV-2 protease Nsp5 (Lu and Zhou, 2023; Zhang et al., 2024b). Nsp5 cleaves TRMT1 within the 530QANFT534 sequence that matches the consensus motif of SARS-CoV-2 polyprotein cleavage sites (D’Oliviera et al., 2022). Nsp5-mediated processing leads to TRMT1 cleavage fragments that contain either the catalytic MTase domain or the ZNF that is required for RNA substrate interaction, and uncoupling of these two features renders TRMT1 functionally inactive. Lack of TRMT1 reduces SARS-CoV-2 RNA replication and particle infectivity, leading to the hypothesis that the virus might self-limit its proliferation by altering the host translation machinery through TRMT1 degradation and reduced levels of m22G-modified tRNAs (Zhang et al., 2024b). A role for m2G7 in tRNATrp in regulating the replication of the Avian myeloblastosis virus (AMV) has also been proposed (Keith and Heyman, 1990). During budding, retroviruses package host tRNAs, predominantly tRNATrp, to prime viral reverse RNA-DNA synthesis (Waters and Mullin, 1977). Unmethylated tRNATrp can fulfil this function but tRNATrp containing m2G7 is not packaged into viral particles (Keith and Heyman, 1990). This suggests that the methylation mark may act as a selectivity determinant for packaging, potentially through altering interactions with retroviral proteins.
While m2G and m22G modifications have long been recognized as key features of many cytosolic and mitochondrial tRNAs, recent years have seen rapid advances in the understanding of how these methylation marks are installed and how they contribute to tRNA function. The identification of MTases responsible for installing m2G and m22G tRNA modifications has revealed, on the one hand, many common features shared between these enzymes and, on the other hand, shown how each protein is specifically adapted to appropriately methylate particular tRNA nucleosides. A growing body of evidence suggests dynamic regulation of m2G and m22G in (mt-)tRNAs, linking these methylations to the adaptive response to cellular stress. The development of novel detection methods for m2G would expedite quantitative analyses of the stoichiometries of this type of modification in different cell types and tissues, various growth and stress conditions, and during development in the future. Deeper understanding of the molecular basis of pathogenic conditions associated with defects in m2G and m22G will be driven by molecular and biochemical studies that dissect the precise roles of these modifications in regulating tRNA function.
JP: Visualization, Writing–original draft, Writing–review and editing. KB: Conceptualization, Funding acquisition, Writing–original draft, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Deutsche Forschungsgemeinschaft (DFG) via SFB1565 (project number 469281184; P12 to KB).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdelraheem, E., Thair, B., Varela, R. F., Jockmann, E., Popadić, D., Hailes, H. C., et al. (2022). Methyltransferases: functions and applications. ChemBioChem 23, e202200212. doi:10.1002/cbic.202200212
Ao, C., Zou, Q., and Yu, L. (2022). RFhy-m2G: identification of RNA N2-methylguanosine modification sites based on random forest and hybrid features. Methods 203, 32–39. doi:10.1016/j.ymeth.2021.05.016
Aoyama, T., Yamashita, S., and Tomita, K. (2020). Mechanistic insights into m6A modification of U6 snRNA by human METTL16. Nucleic Acids Res. 48, 5157–5168. doi:10.1093/nar/gkaa227
Aravind, L., and Koonin, E. V. (2001). THUMP - a predicted RNA-binding domain shared by 4-thiouridine, pseudouridine synthases and RNA methylases. Trends Biochem. Sci. 26, 215–217. doi:10.1016/S0968-0004(01)01826-6
Arimbasseri, A. G., Blewett, N. H., Iben, J. R., Lamichhane, T. N., Cherkasova, V., Hafner, M., et al. (2015). RNA polymerase III output is functionally linked to tRNA dimethyl-G26 modification. PLoS Genet. 11, e1005671. doi:10.1371/journal.pgen.1005671
Armengaud, J., Urbonavičius, J., Fernandez, B., Chaussinand, G., Bujnicki, J. M., and Grosjean, H. (2004). N2-methylation of guanosine at position 10 in tRNA is catalyzed by a THUMP domain-containing, S-adenosylmethionine-dependent methyltransferase, conserved in archaea and eukaryota. J. Biol. Chem. 279, 37142–37152. doi:10.1074/jbc.M403845200
Awai, T., Kimura, S., Tomikawa, C., Ochi, A., Ihsanawati, , Bessho, Y., et al. (2009). Aquifex aeolicus tRNA (N2,N2-Guanine)-dimethyltransferase (Trm1) catalyzes transfer of methyl groups not only to guanine 26 but also to guanine 27 in tRNA. J. Biol. Chem. 284, 20467–20478. doi:10.1074/jbc.M109.020024
Awai, T., Ochi, A., Ihsanawati, , Sengoku, T., Hirata, A., Bessho, Y., et al. (2011). Substrate tRNA recognition mechanism of a multisite-specific tRNA methyltransferase, Aquifex aeolicus Trm1, based on the X-ray crystal structure. J. Biol. Chem. 286, 35236–35246. doi:10.1074/jbc.M111.253641
Bavi, R. S., Sambhare, S. B., and Sonawane, K. D. (2013). MD simulation studies to investigate iso-energetic conformational behaviour of modified nucleosides m2G and m2 2G present in tRNA. Comput. Struct. Biotechnol. J. 5, e201302015. doi:10.5936/csbj.201302015
Bednářová, A., Hanna, M., Durham, I., Vancleave, T., England, A., Chaudhuri, A., et al. (2017). Lost in translation: defects in transfer RNA modifications and neurological disorders. Front. Mol. Neurosci. 10, 135. doi:10.3389/fnmol.2017.00135
Blaesius, K., Abbasi, A. A., Tahir, T. H., Tietze, A., Picker-Minh, S., Ali, G., et al. (2018). Mutations in the tRNA methyltransferase 1 gene TRMT1 cause congenital microcephaly, isolated inferior vermian hypoplasia and cystic leukomalacia in addition to intellectual disability. Am. J. Med. Genet. Part A 176, 2517–2521. doi:10.1002/ajmg.a.38631
Bohnsack, K. E., Kleiber, N., Lemus-Diaz, N., and Bohnsack, M. T. (2022). Roles and dynamics of 3-methylcytidine in cellular RNAs. Trends Biochem. Sci. 47, 596–608. doi:10.1016/j.tibs.2022.03.004
Bohnsack, M. T., and Sloan, K. E. (2018). The mitochondrial epitranscriptome: the roles of RNA modifications in mitochondrial translation and human disease. Cell. Mol. Life Sci. 75, 241–260. doi:10.1007/s00018-017-2598-6
Boughanem, H., Böttcher, Y., Tomé-Carneiro, J., López de las Hazas, M. C., Dávalos, A., Cayir, A., et al. (2023). The emergent role of mitochondrial RNA modifications in metabolic alterations. Wiley Interdiscip. Rev. RNA 14, e1753. doi:10.1002/wrna.1753
Bourgeois, G., Létoquart, J., van Tran, N., and Graille, M. (2017a). Trm112, a protein activator of methyltransferases modifying actors of the eukaryotic translational apparatus. Biomolecules 7, 7. doi:10.3390/biom7010007
Bourgeois, G., Marcoux, J., Saliou, J. M., Cianférani, S., and Graille, M. (2017b). Activation mode of the eukaryotic m2G10 tRNA methyltransferase Trm11 by its partner protein Trm112. Nucleic Acids Res. 45, 1971–1982. doi:10.1093/nar/gkw1271
Brūmele, B., Mutso, M., Telanne, L., Õunap, K., Spunde, K., Abroi, A., et al. (2021). Human trmt112-methyltransferase network consists of seven partners interacting with a common co-factor. Int. J. Mol. Sci. 22, 13593. doi:10.3390/ijms222413593
Bujnicki, J. M. (2000). Phylogenomic analysis of 16S rRNA:(guanine-N2) methyltransferases suggests new family members and reveals highly conserved motifs and a domain structure similar to other nucleic acid amino-methyltransferases. FASEB J. 14, 2365–2368. doi:10.1096/fj.00-0076com
Bujnicki, J. M., Leach, R. A., Debski, J., and Rychlewski, L. (2002). Bioinformatic analyses of the tRNA: (guanine 26, N2,N2)-dimethyltransferase (Trm1) family. J. Mol. Microbiol. Biotechnol. 4, 405–415.
Cai, Y., Wu, G., Peng, B., Li, J., Zeng, S., Yan, Y., et al. (2021). Expression and molecular profiles of the AlkB family in ovarian serous carcinoma. Aging 13, 9679–9692. doi:10.18632/aging.202716
Canaday, J., Guillemaut, P., and Weil, J. H. (1980). The nucleotide sequences of the initiator transfer RNAs from bean cytoplasm and chloroplasts. Nucleic Acids Res. 8, 999–1008. doi:10.1093/nar/8.5.999
Cappannini, A., Ray, A., Purta, E., Mukherjee, S., Boccaletto, P., Moafinejad, S. N., et al. (2023). MODOMICS: a database of RNA modifications and related information. 2023 update. Nucleic Acids Res. 52, D239–D244. doi:10.1093/nar/gkad1083
Chan, C. T. Y., Dyavaiah, M., DeMott, M. S., Taghizadeh, K., Dedon, P. C., and Begley, T. J. (2010). A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet. 6, e1001247. doi:10.1371/journal.pgen.1001247
Chen, K., Shen, D., Tan, L., Lai, D., Han, Y., Gu, Y., et al. (2022). A pan-cancer analysis reveals the prognostic and immunotherapeutic value of ALKBH7. Front. Genet. 13, 822261. doi:10.3389/fgene.2022.822261
Chen, Q., Yan, M., Cao, Z., Li, X., Zhang, Y., Shi, J., et al. (2016). Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 351, 397–400. doi:10.1126/science.aad7977
Chen, W., Song, X., Lv, H., and Lin, H. (2019). iRNA-m2G: identifying N2-methylguanosine sites based on sequence-derived information. Mol. Ther. - Nucleic Acids 18, 253–258. doi:10.1016/j.omtn.2019.08.023
Chujo, T., and Tomizawa, K. (2021). Human transfer RNA modopathies: diseases caused by aberrations in transfer RNA modifications. FEBS J. 288, 7096–7122. doi:10.1111/febs.15736
Chung Kim Chung, K., Mahdavi-Amiri, Y., Korfmann, C., and Hili, R. (2022). PhOxi-seq: single-nucleotide resolution sequencing of N2-methylation at guanosine in RNA by photoredox catalysis. J. Am. Chem. Soc. 144, 5723–5727. doi:10.1021/jacs.2c00670
Clifton, B. E., Fariz, M. A., Uechi, G. I., and Laurino, P. (2021). Evolutionary repair reveals an unexpected role of the tRNA modification m1G37 in aminoacylation. Nucleic Acids Res. 49, 12467–12485. doi:10.1093/nar/gkab1067
Constantinesco, F., Benachenhou, N., Motorin, Y., and Grosjean, H. (1998). The tRNA(guanine-26,N2-N2) methyltransferase (Trm1) from the hyperthermophilic archaeon Pyrococcus furiosus: cloning, sequencing of the gene and its expression in Escherichia coli. Nucleic Acids Res. 26, 3753–3761. doi:10.1093/nar/26.16.3753
Constantinesco, F., Motorin, Y., and Grosjean, H. (1999). Characterisation and enzymatic properties of tRNA(guanine 26, N2,N2)-dimethyltransferase (Trm1p) from Pyrococcus furiosus. J. Mol. Biol. 291, 375–392. doi:10.1006/jmbi.1999.2976
Cui, W., Zhao, D., Jiang, J., Tang, F., Zhang, C., and Duan, C. (2023). tRNA modifications and modifying enzymes in disease, the potential therapeutic targets. Int. J. Biol. Sci. 19, 1146–1162. doi:10.7150/ijbs.80233
Dai, Q., Zheng, G., Schwartz, M. H., Clark, W. C., and Pan, T. (2017). Selective enzymatic demethylation of N2,N2-dimethylguanosine in RNA and its application in high-throughput tRNA sequencing. Angew. Chem. - Int. Ed. 56, 5017–5020. doi:10.1002/anie.201700537
Davarniya, B., Hu, H., Kahrizi, K., Musante, L., Fattahi, Z., Hosseini, M., et al. (2015). The role of a novel TRMT1 gene mutation and rare GRM1 gene defect in intellectual disability in two azeri families. PLoS One 10, e0129631. doi:10.1371/journal.pone.0129631
De Crécy-Lagard, V., Boccaletto, P., Mangleburg, C. G., Sharma, P., Lowe, T. M., Leidel, S. A., et al. (2019). Survey and summary: matching tRNA modifications in humans to their known and predicted enzymes. Nucleic Acids Res. doi:10.1093/nar/gkz011
Dewe, J. M., Fuller, B. L., Lentini, J. M., Kellner, S. M., and Fu, D. (2017). TRMT1-Catalyzed tRNA modifications are required for redox homeostasis to ensure proper cellular proliferation and oxidative stress survival. Mol. Cell. Biol. 37 (21), e00214-e00217. doi:10.1128/mcb.00214-17
Di, K., Linskey, M. E., and Bota, D. A. (2013). TRIM11 is overexpressed in high-grade gliomas and promotes proliferation, invasion, migration and glial tumor growth. Oncogene 32, 5038–5047. doi:10.1038/onc.2012.531
Diaz-Muñoz, G., Harchar, T. A., Lai, T. P., Shen, K. F., and Hopper, A. K. (2014). Requirement of the spindle pole body for targeting and/or tethering proteins to the inner nuclear membrane. Nucl. (United States) 5, 352–366. doi:10.4161/nucl.29793
D’Oliviera, A., Geissler, E. P., and Mugridge, J. S. (2022). Recognition and cleavage of human tRNA methyltransferase TRMT1 by the SARS-CoV-2 main protease. FASEB J. 36. doi:10.1096/fasebj.2022.36.s1.r4632
Edqvist, J., Blomqvist, K., and Straaby, K. (1994). Structural elements in yeast tRNAs required for homologous modification of guanosine-26 into dimethylguanosine-26 by the yeast Trm1 tRNA-modifying enzyme. Biochemistry 33, 9546–9551. doi:10.1021/bi00198a021
Edqvist, J., Grosjean, H., and Stråby, K. B. (1992). Identity elements for N2-dimethylation of guanosine-26 in yeast tRNAs. Nucleic Acids Res. 20, 6575–6581. doi:10.1093/nar/20.24.6575
Edqvist, J., Ståby, K. B., and Grosjean, H. (1995). Enzymatic formation of N2,N2-dimethylguanosine in eukaryotic tRNA: importance of the tRNA architecture. Biochimie 77, 54–61. doi:10.1016/0300-9084(96)88104-1
Ellis, S. R., Morales, M. J., Li, J. M., Hopper, A. K., and Martin, N. C. (1986). Isolation and characterization of the TRM1 locus, a gene essential for the N2,N2-dimethylguanosine modification of both mitochondrial and cytoplasmic tRNA in Saccharomyces cerevisiae. J. Biol. Chem. 261, 9703–9709. doi:10.1016/s0021-9258(18)67571-4
Fedeles, B. I., Singh, V., Delaney, J. C., Li, D., and Essigmann, J. M. (2015). The AlkB family of Fe(II)/α-ketoglutarate-dependent dioxygenases: repairing nucleic acid alkylation damage and beyond. J. Biol. Chem. 290, 20734–20742. doi:10.1074/jbc.R115.656462
Fernandez-Vizarra, E., Berardinelli, A., Valente, L., Tiranti, V., and Zeviani, M. (2006). Nonsense mutation in pseudouridylate synthase 1 (PUS1) in two brothers affected by myopathy, lactic acidosis and sideroblastic anaemia (MLASA). J. Med. Genet. 44, 173–180. doi:10.1136/jmg.2006.045252
Fislage, M., Roovers, M., Tuszynska, I., Bujnicki, J. M., Droogmans, L., and Versées, W. (2012). Crystal structures of the tRNA:m2G6 methyltransferase Trm14/TrmN from two domains of life. Nucleic Acids Res. 40, 5149–5161. doi:10.1093/nar/gks163
Fu, D., Jordan, J. J., and Samson, L. D. (2013). Human ALKBH7 is required for alkylation and oxidation-induced programmed necrosis. Genes Dev. 27, 1089–1100. doi:10.1101/gad.215533.113
Fu, M., and Blackshear, P. J. (2017). RNA-binding proteins in immune regulation: a focus on CCCH zinc finger proteins. Nat. Rev. Immunol. 17, 130–143. doi:10.1038/nri.2016.129
Funk, H. M., Zhao, R., Thomas, M., Spigelmyer, S. M., Sebree, N. J., Bales, R. O., et al. (2020). Identification of the enzymes responsible for m2,2G and acp3U formation on cytosolic tRNA from insects and plants. PLoS One 15, e0242737. doi:10.1371/journal.pone.0242737
Gao, J., Wang, B., Yu, H., Wu, G., Wan, C., Liu, W., et al. (2020). Structural insight into HEMK2–TRMT112-mediated glutamine methylation. Biochem. J. 477, 3833–3838. doi:10.1042/BCJ20200594
Giegé, R., and Eriani, G. (2023). The tRNA identity landscape for aminoacylation and beyond. Nucleic Acids Res. 51, 1528–1570. doi:10.1093/nar/gkad007
Ginell, S. L., and Parthasarathy, R. (1978). Conformation of N2-methylguanosine, A modified nucleoside of tRNA. Biochem. Biophysical Res. Commun. 84, 886–894. doi:10.1016/0006-291X(78)91666-2
Helm, M., and Motorin, Y. (2017). Detecting RNA modifications in the epitranscriptome: predict and validate. Nat. Rev. Genet. 18, 275–291. doi:10.1038/nrg.2016.169
Heurgué-Hamard, V., Graille, M., Scrima, N., Ulryck, N., Champ, S., Van Tilbeurgh, H., et al. (2006). The zinc finger protein Ynr046w is plurifunctional and a component of the eRF1 methyltransferase in yeast. J. Biol. Chem. 281, 36140–36148. doi:10.1074/jbc.M608571200
Hirata, A., Nishiyama, S., Tamura, T., Yamauchi, A., and Hori, H. (2016). Structural and functional analyses of the archaeal tRNA m2G/m22G10 methyltransferase aTrm11 provide mechanistic insights into site specificity of a tRNA methyltransferase that contains common RNA-binding modules. Nucleic Acids Res. 44, 6377–6390. doi:10.1093/nar/gkw561
Hirata, A., Suzuki, T., Nagano, T., Fujii, D., Okamoto, M., Sora, M., et al. (2019). Distinct modified nucleosides in tRNAtrp from the hyperthermophilic archaeon Thermococcus kodakarensis and requirement of tRNA m2G10/m2 2G10 methyltransferase (archaeal Trm11) for survival at high temperatures. J. Bacteriol. 201, e00448-19. doi:10.1128/JB.00448-19
Höbartner, C., Bohnsack, K. E., and Bohnsack, M. T. (2024). How natural enzymes and synthetic ribozymes generate methylated nucleotides in RNA. Annu. Rev. Biochem. 93, 109–137. doi:10.1146/annurev-biochem-030222-112310
Holbrook, S. R., Sussman, J. L., Warrant, R. W., and Kim, S. H. (1978). Crystal structure of yeast phenylalanine transfer RNA. J. Mol. Biol. 123, 631–660. doi:10.1016/0022-2836(78)90210-3
Holley, R. W., Apgar, J., Everett, G. A., Madison, J. T., Marquisee, M., Merrill, S. H., et al. (1965). Structure of a ribonucleic acid. Science 147, 1462–1465. doi:10.1126/science.147.3664.1462
Hori, H. (2023). Transfer RNA modification enzymes with a thiouridine synthetase, methyltransferase and pseudouridine synthase (THUMP) domain and the nucleosides they produce in tRNA. Genes (Basel) 14, 382. doi:10.3390/genes14020382
Hornbeck, P. V., Zhang, B., Murray, B., Kornhauser, J. M., Latham, V., and Skrzypek, E. (2015). PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–D520. doi:10.1093/nar/gku1267
Huang, H., Li, H., Pan, R., Wang, S., and Liu, X. (2021). tRNA modifications and their potential roles in pancreatic cancer. Archives Biochem. Biophysics 714, 109083. doi:10.1016/j.abb.2021.109083
Hwang, S.-P., Liao, H., Barondeau, K., Han, X., Herbert, C., Mcconie, H., et al. (2024). TRMT1L-catalyzed m22G27 on tyrosine tRNA is required for efficient mRNA translation and cell survival under oxidative stress. bioRxiv. doi:10.1101/2024.05.02.591343
Ihsanawati, , Nishimoto, M., Higashijima, K., Shirouzu, M., Grosjean, H., Bessho, Y., et al. (2008). Crystal structure of tRNA N2,N2-guanosine dimethyltransferase Trm1 from Pyrococcus horikoshii. J. Mol. Biol. 383, 871–884. doi:10.1016/j.jmb.2008.08.068
Jedynak-Slyvka, M., Jabczynska, A., and Szczesny, R. J. (2021). Human mitochondrial rna processing and modifications: overview. Int. J. Mol. Sci. 22, 7999. doi:10.3390/ijms22157999
Johnson, G. D., Pirtle, I. L., and Pirtle, R. M. (1985). The nucleotide sequence of tyrosine tRNAQ*ΨA from bovine liver. Archives Biochem. Biophysics 236, 448–453. doi:10.1016/0003-9861(85)90647-2
Jones, J. D., Simcox, K. M., Kennedy, R. T., and Koutmou, K. S. (2023). Direct sequencing of total Saccharomyces cerevisiae tRNAs by LC–MS/MS. RNA 29, 1201–1214. doi:10.1261/rna.079656.123
Jonkhout, N., Cruciani, S., Santos Vieira, H. G., Tran, J., Liu, H., Liu, G., et al. (2021). Subcellular relocalization and nuclear redistribution of the RNA methyltransferases TRMT1 and TRMT1L upon neuronal activation. RNA Biol. 18, 1905–1919. doi:10.1080/15476286.2021.1881291
Kawamura, T., Anraku, R., Hasegawa, T., Tomikawa, C., and Hori, H. (2014). Transfer RNA methyltransferases from Thermoplasma acidophilum, a thermoacidophilic archaeon. Int. J. Mol. Sci. 16, 91–113. doi:10.3390/ijms16010091
Keith, G., and Heyman, T. (1990). Heterogeneities in vertebrate tRNAsTrp avian retroviruses package only as a primer the tRNATrp lacking modified m2G in positon 7. Nucleic Acids Res. 18, 703–710. doi:10.1093/nar/18.4.703
King, T. H., Liu, B., McCully, R. R., and Fournier, M. J. (2003). Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol. Cell 11, 425–435. doi:10.1016/S1097-2765(03)00040-6
Kleiber, N., Lemus-Diaz, N., Stiller, C., Heinrichs, M., Mai, M. M. Q., Hackert, P., et al. (2022). The RNA methyltransferase METTL8 installs m3C32 in mitochondrial tRNAsThr/Ser(UCN) to optimise tRNA structure and mitochondrial translation. Nat. Commun. 13, 209. doi:10.1038/s41467-021-27905-1
Klimontova, J. M., Chung Kim Chung, K., Zhang, H., Kouzarides, T., Bannister, A. J., and Hili, R. (2024). PhOxi-seq detects enzyme-dependent m2G in multiple RNA types. bioRxiv. doi:10.1101/2024.08.15.608094
Kobitski, A. Y., Hengesbach, M., Seidu-Larry, S., Dammertz, K., Chow, C. S., van Aerschot, A., et al. (2011). Single-molecule FRET reveals a cooperative effect of two methyl group modifications in the folding of human mitochondrial tRNA Lys. Chem. & Biol. 18, 928–936. doi:10.1016/j.chembiol.2011.03.016
Kuchino, Y., Ihara, M., Yabusaki, Y., and Nishimura, S. (1982). Initiator tRNAs from archaebacteria show common unique sequence characteristics. Nature 298, 684–685. doi:10.1038/298684a0
Kuhle, B., Chen, Q., and Schimmel, P. (2023). tRNA renovatio: rebirth through fragmentation. Mol. Cell 83, 3953–3971. doi:10.1016/j.molcel.2023.09.016
Lacoux, C., Wacheul, L., Saraf, K., Pythoud, N., Huvelle, E., Figaro, S., et al. (2020). The catalytic activity of the translation termination factor methyltransferase Mtq2-Trm112 complex is required for large ribosomal subunit biogenesis. Nucleic Acids Res. 48, 12310–12325. doi:10.1093/nar/gkaa972
Lai, T. P., Stauffer, K. A., Murthi, A., Shaheen, H. H., Peng, G., Martin, N. C., et al. (2009). Mechanism and a peptide motif for targeting peripheral proteins to the yeast inner nuclear membrane. Traffic 10, 1243–1256. doi:10.1111/j.1600-0854.2009.00956.x
Leetsi, L., Õunap, K., Abroi, A., and Kurg, R. (2019). The common partner of several methyltransferases TRMT112 regulates the expression of N6AMT1 isoforms in mammalian cells. Biomolecules 9, 422. doi:10.3390/biom9090422
Létoquart, J., Huvelle, E., Wacheul, L., Bourgeois, G., Zorbas, C., Graille, M., et al. (2014). Structural and functional studies of Bud23-Trm112 reveal 18S rRNA N7-G1575 methylation occurs on late 40S precursor ribosomes. Proc. Natl. Acad. Sci. U. S. A. 111, E5518–E5526. doi:10.1073/pnas.1413089111
Lewis, C. J. T., Pan, T., and Kalsotra, A. (2017). RNA modifications and structures cooperate to guide RNA-protein interactions. Nat. Rev. Mol. Cell Biol. 18, 202–210. doi:10.1038/nrm.2016.163
Li, J. M., Hopper, A. K., and Martin, N. C. (1989). N2,N2-dimethylguanosine-specific tRNA methyltransferase contains both nuclear and mitochondrial targeting signals in Saccharomyces cerevisiae. J. cell Biol. 109, 1411–1419. doi:10.1083/jcb.109.4.1411
Liang, X. H., Liu, Q., and Fournier, M. J. (2009). Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA 15, 1716–1728. doi:10.1261/rna.1724409
Linder, B., Grozhik, A. V., Olarerin-George, A. O., Meydan, C., Mason, C. E., and Jaffrey, S. R. (2015). Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12, 767–772. doi:10.1038/nmeth.3453
Liu, W. W., Zheng, S. Q., Li, T., Fei, Y. F., Wang, C., Zhang, S., et al. (2024). RNA modifications in cellular metabolism: implications for metabolism-targeted therapy and immunotherapy. Signal Transduct. Target. Ther. 9, 70. doi:10.1038/s41392-024-01777-5
Lu, J. L., and Zhou, X. L. (2023). SARS-CoV-2 main protease Nsp5 cleaves and inactivates human tRNA methyltransferase TRMT1. J. Mol. Cell Biol. 15, mjad024. doi:10.1093/jmcb/mjad024
Matsuo, M., Yokogawa, T., Nishikawa, K., Watanabe, K., and Okada, N. (1995). Highly specific and efficient cleavage of squid tRNALys catalyzed by magnesium ions. J. Biol. Chem. 270, 10097–10104. doi:10.1074/jbc.270.17.10097
McCleverty, C. J., Hornsby, M., Spraggon, G., and Kreusch, A. (2007). Crystal structure of human Pus10, A novel pseudouridine synthase. J. Mol. Biol. 373, 1243–1254. doi:10.1016/j.jmb.2007.08.053
Menezes, S., Gaston, K. W., Krivos, K. L., Apolinario, E. E., Reich, N. O., Sowers, K. R., et al. (2011). Formation of m 2G6 in Methanocaldococcus jannaschii tRNA catalyzed by the novel methyltransferase Trm14. Nucleic Acids Res. 39, 7641–7655. doi:10.1093/nar/gkr475
Muthukumar, S., Li, C. T., Liu, R. J., and Bellodi, C. (2024). Roles and regulation of tRNA-derived small RNAs in animals. Nat. Rev. Mol. Cell Biol. 25, 359–378. doi:10.1038/s41580-023-00690-z
Najmabadi, H., Hu, H., Garshasbi, M., Zemojtel, T., Abedini, S. S., Chen, W., et al. (2011). Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature 478, 57–63. doi:10.1038/nature10423
Neumann, P., Lakomek, K., Naumann, P. T., Erwin, W. M., Lauhon, C. T., and Ficner, R. (2014). Crystal structure of a 4-thiouridine synthetase-RNA complex reveals specificity of tRNA U8 modification. Nucleic Acids Res. 42, 6673–6685. doi:10.1093/nar/gku249
Ng, S. F., Lin, R. C. Y., Laybutt, D. R., Barres, R., Owens, J. A., and Morris, M. J. (2010). Chronic high-fat diet in fathers programs β-cell dysfunction in female rat offspring. Nature 467, 963–966. doi:10.1038/nature09491
Niederberger, C., Gräub, R., Costa, A., Desgrès, J., and Schweingruber, M. E. (1999). The tRNA N2,N2-dimethylguanosine-26 methyltransferase encoded by gene trm1 increases efficiency of suppression of an ochre codon in Schizosaccharomyces pombe. FEBS Lett. 464, 67–70. doi:10.1016/S0014-5793(99)01679-8
Nishida, Y., Ohmori, S., Kakizono, R., Kawai, K., Namba, M., Okada, K., et al. (2022). Required elements in tRNA for methylation by the eukaryotic tRNA (guanine-N2-) methyltransferase (Trm11-Trm112 complex). Int. J. Mol. Sci. 23, 4046. doi:10.3390/ijms23074046
Noon, K. R., Bruenger, E., and McCloskey, J. A. (1998). Posttranscriptional modifications in 16S and 23S rRNAs of the archaeal hyperthermophile Sulfolobus solfataricus. J. Bacteriol. 180, 2883–2888. doi:10.1128/jb.180.11.2883-2888.1998
Oerum, S., Dégut, C., Barraud, P., and Tisné, C. (2017). m1A post-transcriptional modification in tRNAs. Biomolecules 7, 20. doi:10.3390/biom7010020
Ojala, D., Montoya, J., and Attardi, G. (1981). TRNA punctuation model of RNA processing in human mitochondria. Nature 290, 470–474. doi:10.1038/290470a0
Ontiveros, R. J., Stoute, J., and Liu, K. F. (2019). The chemical diversity of RNA modifications. Biochem. J. 476, 1227–1245. doi:10.1042/BCJ20180445
Õunap, K., Leetsi, L., Matsoo, M., and Kurg, R. (2015). The stability of ribosome biogenesis factor WBSCR22 is regulated by interaction with TRMT112 via ubiquitin-proteasome pathway. PLoS One 10, e0133841. doi:10.1371/journal.pone.0133841
Padhiar, N. H., Katneni, U., Komar, A. A., Motorin, Y., and Kimchi-Sarfaty, C. (2024). Advances in methods for tRNA sequencing and quantification. Trends Genet. 40, 276–290. doi:10.1016/j.tig.2023.11.001
Pallan, P. S., Kreutz, C., Bosio, S., Micura, R., and Egli, M. (2008). Effects of 2-dimethylguanosine on RNA structure and stability: crystal structure of an RNA duplex with tandem m22G:A pairs. RNA 14, 2125–2135. doi:10.1261/rna.1078508
Pan, T. (2018). Modifications and functional genomics of human transfer RNA. Cell Res. 28, 395–404. doi:10.1038/s41422-018-0013-y
Phizicky, E. M., and Hopper, A. K. (2023). The life and times of a tRNA. RNA 29, 898–957. doi:10.1261/rna.079620.123
Porat, J., Vakiloroayaei, A., Remnant, B. M., Talebi, M., Cargill, T., and Bayfield, M. A. (2023). Crosstalk between the tRNA methyltransferase Trm1 and RNA chaperone La influences eukaryotic tRNA maturation. J. Biol. Chem. 299, 105326. doi:10.1016/j.jbc.2023.105326
Purushothaman, S. K., Bujnicki, J. M., Grosjean, H., and Lapeyre, B. (2005). Trm11p and Trm112p are both required for the formation of 2-methylguanosine at position 10 in yeast tRNA. Mol. Cell Biol. 25, 4359–4370. doi:10.1128/mcb.25.11.4359-4370.2005
RajBhandary, U. L., Stuart, A., and Chang, S. H. (1968). Studies on polynucleotides. LXXXII. Yeast phenylalanine transfer ribonucleic acid: partial digestion with ribonuclease T-1 and derivation of the total primary structure. J. Biol. Chem. 243, 584–591. doi:10.1016/s0021-9258(18)93645-8
Randau, L., Stanley, B. J., Kohlway, A., Mechta, S., Xiong, Y., and Söll, D. (2009). A cytidine deaminase edits c to u in transfer RNAs in archaea. Science 324, 657–659. doi:10.1126/science.1170123
Rife, J. P., Cheng, C. S., Moore, P. B., and Strobel, S. A. (1998). N2-methylguanosine is iso-energetic with guanosine in RNA duplexes and GNRA tetraloops. Nucleic Acids Res. 26, 3640–3644. doi:10.1093/nar/26.16.3640
Roovers, M., Oudjama, Y., Fislage, M., Bujnicki, J. M., Versées, W., and Droogmans, L. (2012). The open reading frame TTC1157 of Thermus thermophilus HB27 encodes the methyltransferase forming N 2-methylguanosine at position 6 in tRNA. RNA 18, 815–824. doi:10.1261/rna.030411.111
Roundtree, I. A., Evans, M. E., Pan, T., and He, C. (2017). Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200. doi:10.1016/j.cell.2017.05.045
Ryvkin, P., Leung, Y. Y., Silverman, I. M., Childress, M., Valladares, O., Dragomir, I., et al. (2013). HAMR: high-throughput annotation of modified ribonucleotides. RNA 19, 1684–1692. doi:10.1261/rna.036806.112
Sajek, M. P., Woźniak, T., Sprinzl, M., Jaruzelska, J., and Barciszewski, J. (2020). T-psi-C: user friendly database of tRNA sequences and structures. Nucleic Acids Res. 48, D256-D260. doi:10.1093/nar/gkz922
Sergiev, P. V., Bogdanov, A. A., and Dontsova, O. A. (2007). Ribosomal RNA guanine-(N2)-methyltransferases and their targets. Nucleic Acids Res. 35, 2295–2301. doi:10.1093/nar/gkm104
Shi, H., Chai, P., Jia, R., and Fan, X. (2020). Novel insight into the regulatory roles of diverse RNA modifications: Re-defining the bridge between transcription and translation. Mol. Cancer 19, 78. doi:10.1186/s12943-020-01194-6
Shi, H., and Moore, P. B. (2000). The crystal structure of yeast phenylalanine tRNA at 1.93 resolution: a classic structure revisited. RNA 6, 1091–1105. doi:10.1017/S1355838200000364
Smith, T. J., Giles, R. N., and Koutmou, K. S. (2024). Anticodon stem-loop tRNA modifications influence codon decoding and frame maintenance during translation. Seminars Cell & Dev. Biol. 154, 105–113. doi:10.1016/j.semcdb.2023.06.003
Solberg, A., Robertson, A. B., Aronsen, J. M., Rognmo, Ø., Sjaastad, I., Wisløff, U., et al. (2013). Deletion of mouse Alkbh7 leads to obesity. J. Mol. Cell Biol. 5, 194–203. doi:10.1093/jmcb/mjt012
Sprinzl, M., and Vassilenko, K. S. (2005). Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 33, D139–D140. doi:10.1093/nar/gki012
Stark, H., Rodnina, M. V., Wieden, H. J., Zemlin, F., Wintermeyer, W., and Van Heel, M. (2002). Ribosome interactions of aminoacyl-tRNA and elongation factor Tu in the codon-recognition complex. Nat. Struct. Biol. 9, 849–854. doi:10.1038/nsb859
Steinberg, S., and Cedergren, R. (1995). A correlation between N2-dimethylguanosine presence and alternate tRNA conformers. RNA 1, 886–891.
Sun, Q., Huang, M., and Wei, Y. (2021). Diversity of the reaction mechanisms of SAM-dependent enzymes. Acta Pharm. Sin. B 11, 632–650. doi:10.1016/j.apsb.2020.08.011
Suzuki, T. (2021). The expanding world of tRNA modifications and their disease relevance. Nat. Rev. Mol. Cell Biol. 22, 375–392. doi:10.1038/s41580-021-00342-0
Suzuki, T., and Suzuki, T. (2014). A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res. 42, 7346–7357. doi:10.1093/nar/gku390
Suzuki, T., Yashiro, Y., Kikuchi, I., Ishigami, Y., Saito, H., Matsuzawa, I., et al. (2020). Complete chemical structures of human mitochondrial tRNAs. Nat. Commun. 11, 4269. doi:10.1038/s41467-020-18068-6
Tosar, J. P., Gámbaro, F., Darré, L., Pantano, S., Westhof, E., and Cayota, A. (2018). Dimerization confers increased stability to nucleases in 5 halves from glycine and glutamic acid tRNAs. Nucleic Acids Res. 46, 9081–9093. doi:10.1093/nar/gky495
Urbonavičius, J., Armengaud, J., and Grosjean, H. (2006). Identity elements required for enzymatic formation of N2,N 2-dimethylguanosine from N2-monomethylated derivative and its possible role in avoiding alternative conformations in archaeal tRNA. J. Mol. Biol. 357, 387–399. doi:10.1016/j.jmb.2005.12.087
Vakiloroayaei, A., Shah, N. S., Oeffinger, M., and Bayfield, M. A. (2017). The RNA chaperone La promotes pre-TRNA maturation via indiscriminate binding of both native and misfolded targets. Nucleic Acids Res. 45, 11341–11355. doi:10.1093/nar/gkx764
van Tol, H., Stange, N., Gross, H. J., and Beier, H. (1987). A human and a plant intron-containing tRNATyr gene are both transcribed in a HeLa cell extract but spliced along different pathways. EMBO J. 6, 35–41. doi:10.1002/j.1460-2075.1987.tb04715.x
van Tran, N., Ernst, F. G. M., Hawley, B. R., Zorbas, C., Ulryck, N., Hackert, P., et al. (2019). The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 47, 7719–7733. doi:10.1093/nar/gkz619
van Tran, N., Muller, L., Ross, R. L., Lestini, R., Létoquart, J., Ulryck, N., et al. (2018). Evolutionary insights into Trm112-methyltransferase holoenzymes involved in translation between archaea and eukaryotes. Nucleic Acids Res. 46, 8483–8499. doi:10.1093/nar/gky638
Walker, A. R., Silvestrov, P., Müller, T. A., Podolsky, R. H., Dyson, G., Hausinger, R. P., et al. (2017). ALKBH7 variant related to prostate cancer exhibits altered substrate binding. PLoS Comput. Biol. 13, e1005345. doi:10.1371/journal.pcbi.1005345
Walker, R. T. (1983). Mycoplasma evolution. A review of the use of ribosomal and transfer RNA nucleotide sequences in the determination of phylogenetic relationships. Yale J. Biol. Med. 56, 367–372.
Wang, C., Ulryck, N., Herzel, L., Pythoud, N., Kleiber, N., Guérineau, V., et al. (2023). N2-methylguanosine modifications on human tRNAs and snRNA U6 are important for cell proliferation, protein translation and pre-mRNA splicing. Nucleic Acids Res. 51, 7496–7519. doi:10.1093/nar/gkad487
Wang, C., van Tran, N., Jactel, V., Guérineau, V., and Graille, M. (2020). Structural and functional insights into Archaeoglobus fulgidus m2G10tRNA methyltransferase Trm11 and its Trm112 activator. Nucleic Acids Res. 48, 11068–11082. doi:10.1093/nar/gkaa830
Wang, G., He, Q., Feng, C., Liu, Y., Deng, Z., Qi, X., et al. (2014). The atomic resolution structure of human alkb homolog 7 (ALKBH7), a key protein for programmed necrosis and fat metabolism. J. Biol. Chem. 289, 27924–27936. doi:10.1074/jbc.M114.590505
Waterman, D. G., Ortiz-Lombardía, M., Fogg, M. J., Koonin, E. V., and Antson, A. A. (2006). Crystal structure of Bacillus anthracis ThiI, a tRNA-modifying enzyme containing the predicted RNA-binding THUMP domain. J. Mol. Biol. 356, 97–110. doi:10.1016/j.jmb.2005.11.013
Waters, L. C., and Mullin, B. C. (1977). Transfer RNA into RNA tumor viruses. Prog. Nucleic Acid Res. Mol. Biol. 20, 131–160. doi:10.1016/S0079-6603(08)60471-7
Wu, G., Yan, Y., Cai, Y., Peng, B., Li, J., Huang, J., et al. (2021). ALKBH1-8 and FTO: potential therapeutic targets and prognostic biomarkers in lung adenocarcinoma pathogenesis. Front. Cell Dev. Biol. 9, 633927. doi:10.3389/fcell.2021.633927
Xiong, Q. P., Li, J., Li, H., Huang, Z. X., Dong, H., Wang, E. D., et al. (2023). Human TRMT1 catalyzes m2G or m22G formation on tRNAs in a substrate-dependent manner. Sci. China Life Sci. 66, 2295–2309. doi:10.1007/s11427-022-2295-0
Xu, H., Jiang, C., Yao, F., Liang, H., Yan, H., Chen, D., et al. (2022). Pan-cancer analysis reveals the relation between TRMT112 and tumor microenvironment. J. Oncol. 2022, 1–11. doi:10.1155/2022/1445932
Yang, W.-Q., Ge, J.-Y., Zhang, X., Zhu, W.-Y., Lin, L., Shi, Y., et al. (2024). THUMPD2 catalyzes the N2 -methylation of U6 snRNA of the spliceosome catalytic center and regulates pre-mRNA splicing and retinal degeneration. Nucleic Acids Res. 52, 3291–3309. doi:10.1093/nar/gkad1243
Yang, W. Q., Xiong, Q. P., Ge, J. Y., Li, H., Zhu, W. Y., Nie, Y., et al. (2021). THUMPD3-TRMT112 is a m2G methyltransferase working on a broad range of tRNA substrates. Nucleic Acids Res. 49, 11900–11919. doi:10.1093/nar/gkab927
Yared, M. J., Marcelot, A., and Barraud, P. (2024). Beyond the anticodon: tRNA core modifications and their impact on structure, translation and stress adaptation. Genes (Basel) 15, 374. doi:10.3390/genes15030374
Yoluç, Y., Ammann, G., Barraud, P., Jora, M., Limbach, P. A., Motorin, Y., et al. (2021). Instrumental analysis of RNA modifications. Crit. Rev. Biochem. Mol. Biol. 56, 178–204. doi:10.1080/10409238.2021.1887807
Zhang, K., Eldin, P., Ciesla, J. H., Briant, L., Lentini, J. M., Ramos, J., et al. (2024a). Proteolytic cleavage and inactivation of the TRMT1 tRNA modification enzyme by SARS-CoV-2 main protease. Elife 12. doi:10.7554/eLife.90316
Zhang, K., Lentini, J. M., Prevost, C. T., Hashem, M. O., Alkuraya, F. S., and Fu, D. (2020). An intellectual disability-associated missense variant in TRMT1 impairs tRNA modification and reconstitution of enzymatic activity. Hum. Mutat. 41, 600–607. doi:10.1002/humu.23976
Zhang, K., Manning, A. C., Lentini, J. M., Howard, J., Dalwigk, F., Maroofian, R., et al. (2024b). Human TRMT1 and TRMT1L paralogs ensure the proper modification state. Stab. Funct. tRNAs. doi:10.1101/2024.05.20.594868
Zhang, L. S., Xiong, Q. P., Peña Perez, S., Liu, C., Wei, J., Le, C., et al. (2021). ALKBH7-mediated demethylation regulates mitochondrial polycistronic RNA processing. Nat. Cell Biol. 23, 684–691. doi:10.1038/s41556-021-00709-7
Zhang, W., Foo, M., Eren, A. M., and Pan, T. (2022a). tRNA modification dynamics from individual organisms to metaepitranscriptomics of microbiomes. Mol. Cell 82, 891–906. doi:10.1016/j.molcel.2021.12.007
Zhang, Y., Lu, L., and Li, X. (2022b). Detection technologies for RNA modifications. Exp. Mol. Med. 54, 1601–1616. doi:10.1038/s12276-022-00821-0
Zheng, G., Fu, Y., and He, C. (2014). Nucleic acid oxidation in DNA damage repair and epigenetics. Chem. Rev. 114, 4602–4620. doi:10.1021/cr400432d
Keywords: transfer RNA, RNA stability, translation, mitochondria, aminoacylation, RNA modification, epitranscriptome, methyltransferase
Citation: Petrosyan J and Bohnsack KE (2024) N2-methylguanosine and N2, N2-dimethylguanosine in cytosolic and mitochondrial tRNAs. Front. RNA Res. 2:1460913. doi: 10.3389/frnar.2024.1460913
Received: 07 July 2024; Accepted: 10 September 2024;
Published: 25 September 2024.
Edited by:
Sunny Sharma, The State University of New Jersey, United StatesCopyright © 2024 Petrosyan and Bohnsack. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Katherine E. Bohnsack, a2F0aGVyaW5lLmJvaG5zYWNrQG1lZC51bmktZ29ldHRpbmdlbi5kZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.