
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. RNA Res. , 31 May 2024
Sec. Non-coding RNA
Volume 2 - 2024 | https://doi.org/10.3389/frnar.2024.1334464
This article is part of the Research Topic Non-coding RNAs in disease development and progression View all 4 articles
 Sangeeta Dutta*
Sangeeta Dutta* Vijay Suresh Akhade*
Vijay Suresh Akhade* Subhendu Roy Choudhury†
Subhendu Roy Choudhury† Manchanahalli Rangaswamy Satyanarayana Rao‡
Manchanahalli Rangaswamy Satyanarayana Rao‡Advancements and innovations in transcriptomics and computational biology have revealed long non-coding RNAs (lncRNAs) as some of the major regulators of essential biological processes. Their restricted spatial and temporal expressions as well as ability to interact with nucleic acids (DNA and RNA) and proteins make them key players in chromosome integrity, genomic architecture, and transcriptional and post-transcriptional regulation. Their dysregulation has been associated with numerous diseases and pathological conditions, including cancers. Myeloid leukemia is a malignancy of the hematopoietic system, and its pathobiology has been found to have increasing number of lncRNAs with functional significance. This comprehensive review summarizes a majority of the reported lncRNAs in acute myeloid leukemia (AML) and chronic myeloid leukemia (CML), focusing on the regulatory mechanisms by which they modulate the disease progression and pathogenesis, their potential as diagnostics and prognostic markers, and their feasibility as novel therapeutic targets. We also highlight our recent work on the significance of the lncRNA Hmrhl in CML, which has been found to regulate gene transcription at the chromatin level.
Advances and applications of high-throughput sequencing technologies along with whole-genome sequencing have increased the focus of the scientific community on the uncharted territory of non-coding RNAs (Frese et al., 2013; Sun and Kraus, 2015). Although the regulatory roles of most small non-coding RNAs are well-characterized, most of the long non-coding RNAs (lncRNAs), which have lengths exceeding 200 nucleotides, remain relatively unexplored. Scientific research efforts from the last decade have confirmed the associations of thousands of lncRNAs with literally every known biological process (Sun and Kraus, 2015; Schmitz et al., 2016). Their functionality is supported by their dynamic expression patterns during development and differentiation as well as their highly specific spatial localization at the tissue/cellular/subcellular levels (Iyer et al., 2015a; Kopp and Mendell, 2018). From the documented functional roles of this enormous repertoire of lncRNAs, it can be concluded that they regulate gene expression and the epigenetic environment to guide highly precise and complex biological processes; their importance is so far-reaching that it has been considerably underestimated. Regarding the importance of lncRNAs in various biological functions, it is not surprising that they are expressed differentially under various diseases and pathological conditions, such as diabetes, coronary artery disease, neuropsychiatric and neurodegenerative diseases (like schizophrenia and Alzheimer’s), and most notably cancers (Wapinski and Chang, 2011; Mitra et al., 2012; Iyer et al., 2015b; Fatima et al., 2015; Lorenzen and Thum, 2016; Schmitz et al., 2016; Lan et al., 2022; Sivagurunathan et al., 2022). The newly released updated Lnc2Cancer3.0 database contains more data on the associations of lncRNAs with cancers (Gao et al., 2021b); there are 9,254 lncRNA–cancer associations, with 2,659 lncRNAs and their associations with 216 human cancer subtypes. In addition to such associations, the Lnc2Cancer3.0 database includes the experimentally supported regulatory mechanisms and biological functions for cancer-related lncRNAs (http://bio-bigdata.hrbmu.edu.cn/lnc2cancer). Although most of these are not fully characterized in terms of their functional relevance and regulatory mechanisms, their potential clinical applications as prognostic markers in the early detection of cancer and as probable drug targets are being increasingly realized (Rittenhouse et al., 2013; Sahu et al., 2015; Bartonicek et al., 2016; Lorenzi et al., 2019).
Myeloid or myelogenous leukemia is a malignancy of the hematopoietic system arising from various acquired and spontaneous genetic mutations that confer the potential of unchecked proliferation without differentiation on the myeloid progenitor cells (Weiskopf et al., 2017). The two subtypes of myeloid leukemia, namely acute and chronic, are characterized by the time period of disease progression. The more fatal acute myeloid leukemia (AML) progresses rapidly, resulting in the accumulation of immature and non-functional blood cells in the bone marrow. Conversely, chronic myeloid leukemia (CML) progresses more slowly and results in the accumulation of relatively mature but still abnormal blood cells (Kelly and Gilliland, 2002). Given the developments in therapeutics, CML patients often have better prognosis than AML patients. Studies have shown that about 90% of individuals with CML survive for 5 years or more after diagnosis, as compared to 30% in the case of AML (Dong Y. et al., 2020; Pulte et al., 2020).
The implications of lncRNAs in myeloid leukemia have only been highlighted in recent years but are increasing steadily. Herein, we review the current status of lncRNAs in myeloid leukemia and attempt to understand/link their dysregulation, functional, and mechanistic aspects with the pathobiology of this disease along with emphasis on their putative therapeutic potential. We also highlight our recent work on the role of the human meiotic recombination hot spot locus (Hmrhl) lncRNA in the pathobiology of CML in K562 as the cellular model (Choudhury et al., 2021).
The lncRNAs owe their name to their lack of an open reading frame and an arbitrary length classification of more than 200 nucleotides that allows them to be called “long” (Mattick et al., 2023). The best method of classifying this vast cluster of heterogenous lncRNAs is still under debate; however, the prevalent trend for categorizing seems to be dependent on their genomic context with respect to a well-established protein-coding gene (PcG) (Figure 1). They can be intergenic, acting as standalone units between two coding regions without any overlaps, or they can be intragenic, overlapping with PcG annotations. Intragenic lncRNAs can be further classified into sense and antisense based on the overlap of their coding or non-coding sequences with the parent gene; they may also be bidirectional if the transcription of the lncRNA is initiated in close proximity (<1 kb) and opposite orientation to a PcG. The lncRNAs are considered intronic when derived entirely from an intronic region of a PcG (Kung et al., 2013; Jarroux et al., 2017) (Figure 1).
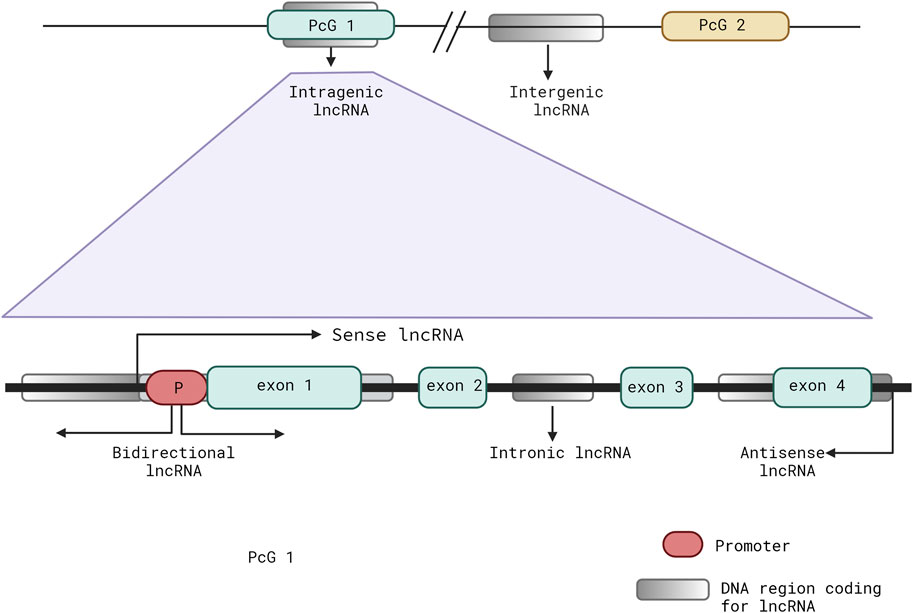
Figure 1. Schematic representation showing the common classifications of long non-coding RNAs (lncRNAs) depending on their locations and biogenesis with respect to a well-established protein-coding gene (PcG).
Although the expressions of lncRNAs are reported in most taxa, from unicellular eukaryotic organisms to primates, they are not conserved well across different or related species (Ulitsky, 2016; Niederer et al., 2017). Even within a species, their expressions are highly specific to the tissue or developmental stages. Their specificity extends to the subcellular level and is restricted to the nucleus, cytoplasm, or both or in other cellular organelles (like the mitochondria, endoplasmic reticulum, nucleoli, and paraspeckles) (Mas-Ponte et al., 2017; Darbellay and Necsulea, 2020; Bridges et al., 2021). The stabilities of the lncRNAs, like their expressions, are also found to be associated with their physiological roles, with regulatory being more stable than housekeeping RNAs, spliced being more stable than unspliced single-exonic RNAs, and cytoplasmic being more stable than nuclear RNAs (Clark et al., 2012; Ayupe and Reis, 2017).
Numerous studies have shown that lncRNAs add yet another layer to the regulatory circuitry controlling gene transcription. Highly specific spatial and temporal expressions of lncRNAs have been directly linked to their regulatory functions in a context-dependent manner (Sun and Kraus, 2015; Kopp and Mendell, 2018). Similar to their expressions, lncRNAs have widely diverse mechanisms of action that usually affect regulation of the PcGs (Yao et al., 2019; Zhao et al., 2020). These properties allow the lncRNAs to act as key players in gene regulation during physiological and developmental processes, including dosage compensation, genomic imprinting, epigenetic regulation, pluripotency, post-transcriptional regulation of mRNAs, and stability/translation modulation of mRNAs (Penny et al., 1996; Sleutels et al., 2002; Ilik et al., 2013; Mercer and Mattick, 2013; Yoon et al., 2013; Yang et al., 2014; Rosa and Ballarino, 2016; Akhade et al., 2017; Yao et al., 2019; Statello et al., 2021).
A significant number of studies have supported the notion that the functional aspects of lncRNAs are solely dependent on their inherent properties, such as the folding patterns that allow binding and interactions with other nucleic acids and proteins to allow modulation of local (in-cis) as well as distal (in-trans) gene regulations (Guo et al., 2016; Zampetaki et al., 2018). LncRNAs can also form DNA–RNA duplex/triplex structures that anchor the associated effectors to active chromatin sites, such as promoters or enhancers (Schmitz et al., 2010; Mondal et al., 2015; Li et al., 2016). Numerous lncRNAs act as chromatin regulators by interacting with the chromatin-modifying complexes and causing selective activation or repression of genes depending on the chromatin complexes (Kopp and Mendell, 2018; Mishra and Kanduri, 2019; Senmatsu and Hirota, 2020). LncRNAs can bind to chromatin complexes and guide them to their specific target loci; for example, the lncRNA ANRIL binds to the Polycomb group of proteins and recruits them to the target gene loci (Yap et al., 2010; Holdt et al., 2013); furthermore, lncRNAs can act as scaffolds/bridges to bring together complexes for their suppressive or activation functions (Jeon and Lee, 2011) (Figure 2). For example, the lncRNA ANRIL bridges the PRC2 and YY1 proteins that are required in the formation of the silencing complex.
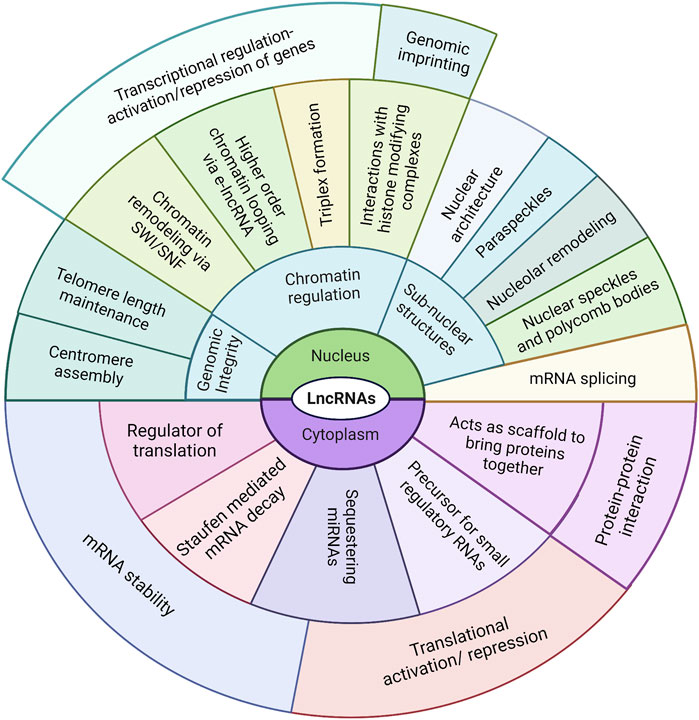
Figure 2. Compilation of known mechanisms employed by lncRNAs to regulate their targets. The regulatory techniques and targets (DNA, RNA, and proteins) vary widely depending on their subcellular localization.
Enhancer-derived lncRNAs (eRNAs) can also promote higher-order chromatin organization, like chromatin looping, to allow interactions between long-distance regulatory elements (like the enhancers and promoters) (Chen H. et al., 2017; Fanucchi and Mhlanga, 2017; Wang Y. et al., 2020). A well-established role of the lncRNAs is in genomic imprinting, a phenomenon in which one of the alleles from the inherited parental pair is inactivated. This is usually achieved by histone/DNA modification, a process in which lncRNAs (such as Xist, Air, and H19) have been known to play major roles (Penny et al., 1996; Sleutels et al., 2002; Ilik et al., 2013). Another interesting mechanism of action of lncRNAs is as sponges, sequestering miRNA with complementary sequences and thereby disrupting their binding with associated targets; lncRNAs can also act as decoys binding to the target itself and leading to stabilization or destabilization of the target mRNA (Yoon et al., 2014; Bayoumi et al., 2016; Sun et al., 2019). LncRNAs also play crucial roles in the assembly, maintenance, and regulation of complex nuclear architectures comprising many subcompartments and domains with nuclear bodies and chromatin, which are the centers of various biological processes (Engreitz et al., 2016; Pisignano et al., 2019; Song Z. et al., 2021) (Figure 2).
Leukemia is a cancer of the early blood-forming cells. Although many leukemias are sporadic, most of them are acquired and typified by recurring chromosomal translocations and point mutations in the genes (Gilliland et al., 2004). Characterization of the leukemogenic genes has led to a two-hit model of pathogenesis. Most leukemias appear to be the consequence of collaborations between one class of mutations or gene rearrangements that confer proliferative and/or survival advantages to the hematopoietic progenitors and a second class of mutations that primarily impair the hematopoietic differentiation and subsequent apoptosis of cells (Kelly and Gilliland, 2002; Gilliland et al., 2004).
The AML phenotype characterized by proliferation and impaired differentiation is usually associated with chromosomal translocations resulting in loss of function in the transcription factors (TFs) along with mutations in the hematopoietic tyrosine kinases (like FLT3 and c-KIT, and in N-RAS and K-RAS) that confer proliferative advantages. This results in rapid accumulation of immature, non-functional blood cells in the bone marrow that later spills into the blood and other organs (Kantarjian et al., 2021; Padmakumar et al., 2021). CMLs are caused by constitutively activated tyrosine kinases, such as BCR/ABL, that confer proliferative and survival advantages to the hematopoietic progenitors but do not affect differentiation, thus resulting in the accumulation of relatively mature but still abnormal blood cells (Perrotti et al., 2010; Ochi et al., 2021).
The continuous generation of specialized blood cells over the lifetime of an organism requires dynamic yet precise gene programming with tight coordination between the cell-lineage specifications, proliferation, and differentiation. Recent comprehensive genome-wide studies on hematopoietic stem cells (HSCs) and lineage-primed multipotent progenitors have revealed that hundreds of lncRNAs are expressed together with lineage-specific TFs that are required for hematopoietic differentiation and cell-fate decisions (Dahariya et al., 2019; Ghafouri-Fard et al., 2021). Luo et al. (2015) performed deep RNA sequencing of HSCs and identified low-expressing lncRNAs in rare HSC populations; they identified and annotated 159 HSC-specific lncRNAs (some involved in HSC self-renewal and differentiation by binding to hematopoietic TF binding sites) with high confidence scores. LncRNAs are perfect regulators for driving specific biological programs like hematopoiesis; this is attributable to the features of lncRNAs, such as their versatility of interactions with both nucleic acids and proteins, along with their highly specific temporal and spatial expressions. This topic has been explored in detail in reviews by Qiu et al. (2021) and Alvarez-Dominguez and Lodish (2017).
Over the past decade, increasing evidence has shown a close relationship between various lncRNAs and the pathophysiology of leukemia. The roles of lncRNAs in leukemia progression, both positive and negative, are attributed to their activities in terms of their specific roles in the differentiation, energy metabolism, malignant proliferation, apoptosis, and drug resistance of the leukemia cells (Gao J. et al., 2020). In a systemic review of 86 articles, Dieter et al. (2020) documented that 3,927 lncRNAs are differentially expressed in various leukemias; further analysis revealed that 12 lncRNAs were consistently dysregulated between leukemic cases and controls (CCAT1, CCDC26, CRNDE, HOTAIR, KCNQ5IT1, LINC00265, MALAT1, PVT1, SNHG5, TUG1, MEG3, and NEAT1).
Findings focusing on the detailed functions and mechanisms of the lncRNAs involved in leukemia pathogenesis are underway, and these specific lncRNAs are expected to serve as diagnostic biomarkers, novel therapeutic targets, and predictors in clinical outcomes. In this review, we summarize the updates on lncRNAs reported for myeloid leukemias (AML and CML), their mode of influence in leukemia progression via various mechanisms, and the methods by which scientists can exploit them as potential drug targets.
Although AML is fairly rare, accounting for only about 1% of all cancers, it is the most prevalent type of leukemia in adults and has an incidence rate of 4.1 per 100,000 individuals every year. The incidence of AML increases with age and constitutes 80% of all adult leukemias. AML is an extremely aggressive malignancy of the hematopoietic system, with a mortality rate of 2.7 per 100,000 individuals per annum (Deschler and Lübbert, 2006; Yi et al., 2020; Vakiti et al., 2021). The malignant transformations in AML can be attributed to chromosomal abnormalities for one group of patients, whereas recurrent somatic mutations in several oncogenes have been reported for the cytogenetically normal other groups (Meyer and Levine, 2014; Abelson et al., 2018; Kishtagari and Levine, 2021).
Numerous studies have characterized lncRNAs and demonstrated their roles in the pathogenesis of AML through the various mechanisms by which they exert oncogenic or tumor-suppressive effects. This review summarizes a few of the well-characterized lncRNAs in terms of functions and mechanisms with respect to AML pathogenesis (Table 1). Based on their mechanism of action in AML, the lncRNAs can be loosely categorized into two groups as those that are epigenetic regulators or/and chromatin modifiers and those that are associated with microRNA sponging to indirectly regulate downstream targets. The lncRNAs may also exert both mechanisms or/and completely different ones. Irrespective of the mechanism, lncRNAs have been found to impact one or multiple cancer phenotypes of AML cells, such as cell survival, proliferation, and differentiation.
Epigenetic regulation, chromatin modifications, interactions, and conformations, are the most well-understood functions of lncRNAs. For example, lncRNAs have been transcribed from the HOX cluster, and HOX gene dysregulations have been widely documented in AML. Many lncRNAs like HOTAIR, HOTAIRM1, HOXB-AS3, HOXA-AS2, and HOXBLINC originating from the HOX cluster are involved in transcription regulation via the epigenetic phenomenon. Silencing and overexpression studies by Feng et al. (2020b) have validated the role of the lncRNA HOXA-AS2 in supporting the cellular proliferation and restraining the differentiation of AML cells. HOXA-AS2 was also shown to epigenetically regulate the expression of its downstream target LATS2 via EZH2 binding (Feng et al., 2020b). LATS2 is a component of the Hippo signaling pathway that regulates cell-cycle progression and apoptosis to inhibit the growth and development of tumors. HOXA-AS2 acts as a modular scaffold for histone-modifying complexes by directly binding with EZH2 and triggering H3K27me3 trimethylation at the promoter of LATS2, thereby suppressing its expression (Feng et al., 2020b).
The HOXA-AS2 lncRNA is also shown to directly regulate SOX4 expression and modulate its downstream target PI3K/AKT pathway (Qu et al., 2020). Given its restricted expression in myeloid cells, the lncRNA HOTAIRM1 transcribed from the intergenic and antisense region of the HOXA gene cluster is one of the most studied lncRNAs in relation to myeloid leukemias (Zhang et al., 2014; Wang and Dostie, 2016); it was found to act as an activator of the proximal HOXA genes in the NB4 cell line and as a repressor of the more distant HOXA 4/5/6 genes in NT2-D1 cells. HOTAIRM1 was found to accomplish these through three-dimensional chromatin organizational changes for interactions with either UTX/MLL or PRC2 complexes; it was shown to delay recruitment of the histone demethylase UTX and transcription of the central HOXA genes by participating in the physical dissociation of the chromatin loops at the proximal end of the cluster (Wang and Dostie, 2016).
Overexpression of HOXBLINC, a HOX-B locus-associated lncRNA, was shown to enhance HSC self-renewal and expand myelopoiesis, leading to the development of AML in mice. Mechanistically, HOXBLINC establishes aberrant expression signatures of the genes found in NPM1 mutants through various mechanisms like MLL1 recruitment, chromatin domain, and cis and trans promoter accessibility; it activates the anterior HOX-B genes by recruiting the MLL1/Setd1a complex at its promoter and maintaining a 3D interactome between the enhancer and promoter (Zhu et al., 2021). The posterior HOX-A locus-associated lncRNA HOTTIP was found to activate the posterior HOXA genes, in addition to the canonical Wnt/β-catenin pathway and various key hematopoietic regulators by coordinating CTCF-defined hematopoietic-associated TADs leading to leukemic transformation (Luo H. et al., 2018, 2019). The HOTAIR lncRNA expressed from the HOX-C locus is recognized as a trans-acting epigenetic repressor of the HOX-D locus genes (Gupta et al., 2010; Bhan and Mandal, 2015). Epigenetic regulation seems to be the primary mechanism of HOTAIR, for which multiple targets have been identified in several studies; it represses p15 expression in trans to maintain leukemogenesis via H3K27 trimethylation of its promoter mediated by PRC2 (Gao et al., 2018). HOTAIR also recruits DNMT3B to increase HOXA5 promoter methylation, causing its suppression as well as leading to increased proliferation and reduced apoptosis of the AML cells (Wang S. L. et al., 2019). The lncRNA HOXB-AS3 transcribed from the HOXB cluster was found to be overexpressed in AML (Papaioannou et al., 2019b); by dissecting the functional aspects of HOXB-AS3, Papaioannou et al. (2019b) showed its interactions with the ErbB3-binding protein 1 (EBP1) and guiding of EBP1 to the ribosomal DNA locus to regulate rRNA expression, facilitating adequate protein production in rapidly proliferating leukemic blasts cells.
In addition to the lncRNAs arising from the HOX cluster, several other lncRNAs have been reported to adopt similar mechanisms of action. For example, the lncRNA USP30-AS1 was found to promote AML pathogenesis by epigenetically regulating USP30 and ANKRD13A in cis, which are known to aid cell proliferation and translocation of the HLA-1 protein from the cell membrane to cytoplasm, leading to tumor immune escape (Zhou et al., 2022). The lncRNA NEAT1 acts as a tumor suppressor and is downregulated in AML. Recently, Miliara et al. (2022) investigated the genome-wide RNA and DNA interactions using RADICL sequencing and showed that NEAT1 binds to the genomic loci of key hematopoietic regulators like RUNX2, SOX6, and FOSL2 while interacting with chromatin regulators KMT2A, KMT5B, and CHD7 to influence AML cell differentiation.
RUNXOR is an unspliced RUNX1-intragenic lncRNA transcribed from an upstream overlapping promoter; translocation of RUNX1 is associated with almost 30–40% of the AML cases. Wang et al. (2015) confirmed that RUNXOR epigenetically regulates RUNX1 in AML via recruitment of EZH2 and H3K27 methylation; they additionally revealed that RUNXOR utilizes its 3′-region to directly interact with the RUNX1 promoters and enhancers, facilitating long-range interchromosomal interactions with the chromatin regions involved in multiple RUNX1 translocations (Wang et al., 2014). Another lncRNA involved in chromatin dynamics to regulate chromatin architecture and status is IRAIN, whose low expression in AML was first identified by Sun et al. (2014) using the RNA-guided chromatin conformation capture (R3C) method and described as a paternally imprinted lncRNA regulating the expression of its parent gene IGF1R in AML in cis. IGF1R is a receptor tyrosine kinase that is abundantly activated in leukemic cells, giving them proliferative and treatment resistance capacities through IGF1R receptor-mediated activation of the PI3K/Akt signaling pathway. IRAIN was found to promote IGF1R expression by allowing long-range intrachromosomal interactions between the IGF1R promoter and a distant intragenic enhancer (Sun et al., 2014). However, the detailed mechanisms and effects of this chromosomal looping remain unexplored.
The number of lncRNAs listed in the microRNA sponging group has increased in recent times, as sponging is one of the common mechanisms of gene regulation (Sun et al., 2022). For example, the lncRNA H19 that is otherwise known for its epigenetic regulation acts as a sponge for miR-326, miR-19a-3p, and miR-29a-3p in the hematopoiesis and AML context (Zhao et al., 2017; Zhao and Liu, 2019; Mofidi et al., 2021). Overexpression of H19 in AML is negatively correlated with miR-326, causing increased transcription of the antiapoptotic gene BCL-2 (Mofidi et al., 2021). H19 was reported to sustain leukemic cell proliferation and limit apoptosis by regulating the expressions of IDH2 and Wnt/β cat effectors by limiting the availabilities of miR-19a-3p and miR-29a-3p, respectively (Zhao et al., 2017; Zhao and Liu, 2019). Recently, HOTAIR was found to regulate AML differentiation via the CEBPBβ/HOTAIR/miR-17-5p/p21 pathway (Hu L. et al., 2021). Utilizing the mechanism of sponging microRNA, HOTAIR was shown to titrate miR-193a, which modulates the expression of its target c-KIT, thus affecting the clonogenic growth of AML cells (Xing et al., 2015).
Several reports have experimentally confirmed the sponging activity of CCAT1 in AML, where it is overexpressed (Chen et al., 2016; Izadifard et al., 2018; Wang C. et al., 2020); it reduces the availability of miR-155 and miR-490-3p, eventually resulting in the upregulation of c-Myc (Chen et al., 2016; Wang C. et al., 2020). CCAT1 represses monocytic differentiation and promotes cell growth via miR-155 while markedly increasing the viability and metastasis of AML cells via the CCAT1/miR-490-3p/MAPK1/c-Myc positive feedback loop (Chen et al., 2016; Wang C. et al., 2020). Very recently, elevated levels of the lncRNA MAFG-AS1 were shown to induce cell growth and epithelial–mesenchymal transitions (EMTs) in AML cells via sponging miR-147b and promoting the expression of HOXA9 indirectly (Yao et al., 2022). Given its repressed expression in AML, the lncRNA TP73-AS1 was revealed to sponge miR-21 and hence upregulate its downstream target PTEN to affect cellular proliferation in AML (Yuan et al., 2021). High expression of the lncRNA DUBR was linked with AML pathogenesis in a recent report by Yin et al. (2021); this study revealed that DUBR sequesters miRNA-142-3P and interacts with FUS protein, increasing their expressions and contributing to cell survival, proliferation, and apoptosis inhibition in AML cells. However, further details regarding these regulations by DUBR remain unexplored (Yin et al., 2021).
Studies have demonstrated that the lncRNA NEAT1 affects the cellular behaviors of AML cells by directly repressing the sponging targets miR-338-3p and miR-23a-3p, consequently modulating the levels of their downstream targets CREBRF and SMC1A, respectively (Zhao et al., 2019; Feng S. et al., 2020). Cancer-associated ANRIL is an antisense lncRNA in the INK4 locus that is transcribed from the short arm of chromosome 9 on p21.3 (Pasmant et al., 2011). In a thorough analysis using experiments on loss and gain of functions Wang C. H. et al. (2020) showed that ANRIL accelerates AML pathogenesis by negatively regulating miR-34a and causing HDAC1 overexpression that in turn inhibits E2F1 recruitment to suppress ASPP2; inhibition of ASPP2 expression restrains apoptosis, promoting aberrant proliferation of the AML cells (Wang C. H. et al., 2020). Upregulation of the lncRNA SNHG1 is correlated with poor prognosis of AML and is associated with the mechanism of sponging microRNAs in AML cells; it has been shown to promote the development of AML through the miR-488-5p/NUP205 axis (Bao et al., 2019). The role of SNHG1 as a competing endogenous lncRNA in the inhibition of the antitumor miR-101 has also been demonstrated (Tian et al., 2019). Downregulating the lncRNA HOTAIRM1 was shown to inhibit proliferation and induce apoptosis in AML cells by negatively regulating miR-148b (Hu et al., 2019). The lncRNA SNHG16 has been reported to have an oncogenic role in AML; it sequesters miR183-5p and causes upregulation of its target gene FOXO1, which is a known promoter of cell proliferation and apoptosis inhibition (Yang R. et al., 2020).
Owing to their ability to bind with both proteins and nucleic acids (DNA and RNA), lncRNAs can coordinate gene regulation at multiple levels through various mechanisms of action, such as regulating the stability of protein/RNA, glucose metabolism, autophagy, signaling pathways, and angiogenesis.
LncRNAs can affect the stability of mRNA or proteins through direct interactions with them; for example, the lncRNA SNHG16 that is overexpressed in AML binds directly to the CELF2 mRNA, resulting in its instability (Shi et al., 2021). CELF2 enhances PTEN activity under normal physiological conditions, which in turn affects PI3K/AKT signaling; under the influence of SNHG16, this pathway is disrupted, endowing proliferative and migration capacities to the AML cells (Shi et al., 2021). Yuan et al. (2019) found that the lncRNA PCAT-1 interacts directly with the FZD6 protein, regulating its stability. FZD6 belongs to a family of G-protein-coupled receptors that are essential components of the Wnt signaling pathway, and its high expression in AML has been identified as a biomarker (Yang et al., 2022). PCAT1 thus enhances cell proliferation and inhibits apoptosis by activating Wnt signaling via FZD6, thus contributing to the pathogenesis of AML (Yuan et al., 2019). In a recent study, the lncRNA UCA1 was shown to indirectly regulate the post-translational m6A methylation of mRNA, in turn upregulating the expressions of CXCR4 and CYP1B1 by affecting the stability of METTL14 in AML (Li J. et al., 2022); it was also found to be involved at the translational level through titration of the hnRNP1 protein, which facilitates translation of p27kip1 (Hughes et al., 2015). Both mechanisms promote migration, invasion, and cell proliferation as well as reduce apoptosis in AML.
Another mechanism by which ANRIL promotes malignant cell survival is by regulating the expression of the key glucose metabolism regulator, i.e., adiponectin receptor (AdipoR1) (Sun L. Y. et al., 2018). Additionally, silencing ANRIL and the adiponectin receptors inhibits the phosphorylation of AMP-activated protein kinase/sirtuin 1 to substantially decrease glycolysis and proliferation of the AML cells (Sun L. Y. et al., 2018). HOTAIRM1 was reported to cause degradation of the chimeric oncoprotein PML-RARA found in AML via autophagy; this process is induced when HOTAIRM1 sponges miR-20a, miR-106a, and miR-125b, affecting their downstream targets ULK1, E2F1, and DRAM2, respectively (Chen Z.-H. et al., 2017).
Recently, the lncRNA SNHG1 was reported to enhance AML pathogenesis via activation of Wnt/β-catenin signaling; SNHG1 sequesters miR-489-3p, resulting in overexpression of SOX12 to stimulate the Wnt signaling pathway and cause AML cell growth (Li C. et al., 2021). The lncRNA DUXAP8, which is downregulated in both AML bone-marrow tissues and cell lines, has been shown to stimulate the expressions of the Wnt/β-catenin pathway proteins, namely, Wnt5a, β-catenin, c-Myc, and cyclin-D1, to inhibit glycolysis and induce apoptosis in AML (Zhai et al., 2021). The antisense lncRNA CD27-AS1 was found to have positive effects on MAPK signaling, leading to cell growth and malignancy in AML; mechanistically, it was found to sponge miR-224-5p, thus increasing the PBX3 levels that are responsible for regulating MAPK signaling (Tao et al., 2021).
The lncRNA SNHG5 was found to be upregulated by YY1 in AML; downstream, SNHG5 was shown to regulate AML angiogenesis by activating the connective tissue growth factor (CTGF)/vascular endothelial growth factor A (VEGFA) by directly targeting miR-26b (Li Z.-J. et al., 2021). Another lncRNA that regulates angiogenesis in AML is H22954, which was found to target the 3’ untranslated region (UTR) of PDGFRA and reduce its half-life, thus inhibiting angiogenesis in AML (Li X. et al., 2022). Another interesting mechanism by which lncRNAs regulate gene expression is at the translational level, such as in the antisense lncRNA PU.1-AS that originates from an intronic promoter in PU.1. As an essential requirement for normal hematopoiesis, PU.1 encodes a key TF and suppresses myeloid leukemia (Cook et al., 2004). PU.1-AS interferes with the translation of PU.1 by competitively binding to the translation initiating factor eIF4A; it was also found to interrupt translational elongation, although the exact mechanism remains unknown (Ebralidze et al., 2008).
We have attempted to include the majority of documented and experimentally validated lncRNAs in the context of AML. Some of the remaining lncRNAs are mentioned in Table 1, along with their functional targets, mechanisms, and references for the convenience of readers (Wang J. et al., 2019; Liu et al., 2019; Dong X. et al., 2020; Zhang F. et al., 2020; Feng et al., 2020c; Xue and Che, 2020; Lu et al., 2021; Zhou et al., 2021).
Although AML occurs in all age groups, children constitute a very small percentage of AML patients. Most of the known lncRNAs in AML were first reported in adults, but there are several recent studies that describe functional lncRNAs in pediatric AML. Schwarzer et al. (2017) reported subtype-specific lncRNA signatures for six major cytogenetic subgroups of pediatric AML: Down syndrome (DS), non-DS acute megakaryoblastic leukemia (AMKL), inv [16], t [8; 21], and AML with KMT2A rearrangements (t [9; 11] and t [10; 11]); furthermore, they defined the core lncRNA stem-cell signature in normal HSCs and pediatric AML blasts, which were significantly correlated with poor survival in an independent cohort of AML patients (Schwarzer et al., 2017). In another study by Porcù et al. (2021), CDK6-AS1 was found to be overexpressed in pediatric AML, leading to an immature phenotype as well as activation of mitochondrial biogenesis in healthy HSCs and primary AML blasts; this study also uncovered the potential role of the CDK6-AS1/CDK6 axis in phenotype differentiation through downregulation of RUNX1 signaling. The UCA1 lncRNA has been studied in pediatric and adult AML cell lines and shown to be oncogenic in function, wherein UCA1 knockdown affects the viability, migration, and invasion of leukemic cells through titrating miRNAs like miR-126, miR-204, miR96-5p, and miR296-3p (Sun M. D. et al., 2018; Li J. J. et al., 2020; Li et al., 2020 J.; Liang et al., 2020). In a more clinically relevant scenario, UCA1 knockdown was shown to suppress the chemoresistance of pediatric AML by inhibiting glycolysis through direct binding with miR-125a (Zhang Y. et al., 2018). There are several examples of lncRNAs that have been characterized in pediatric AML that show correlations to patient prognosis and survival; such examples were summarized in a recent review by Neyazi et al. (2022). In a recent comprehensive study, Vanhooren et al. (2022) performed a miRNA-lncRNA network analysis in leukemic stem cells (LSCs) and leukemic blasts (L-blasts) from pediatric AML patients; this study identified several novel lncRNAs and miRNAs in pediatric AML that could become new biomarkers for risk stratification and targeted therapy in the future. Using RNA-seq data from normal bone marrow and de novo AML pediatric patient samples, followed by regularized Cox proportional hazards modeling of the event-free survival (EFS), Farrar et al. (2022) calculated a 37-gene lncScore that showed a significant correlation with patient survival. Similarly, Guo et al. (2020) proposed a lncRNA risk scoring system based on the expressions of 14 lncRNAs for effectively predicting the prognosis of pediatric AML patients. Tao et al. (2022) constructed a ferroptosis-related lncRNA-mRNA coexpression network to investigate the prognostic roles of ferroptosis-related lncRNAs in pediatric AML patients. Accordingly, a model of 22 ferroptosis-related signatures (lncRNAs and mRNAs) was proposed as an independent prognostic factor of pediatric AML. It would be of interest in the future to look for common lncRNAs in these studies to correlate lncRNA expression signatures to pediatric AML patient survival and prognosis.
In 2018, the global incidence of CML was approximately 1 in 100,000 (Höglund et al., 2015; Hu Y. et al., 2021), accounting for about 15% of newly diagnosed cases of leukemia. Considering that CML is a late-developing disease, the average age range at diagnosis is 57–60 years. Clinically, CML is divided into three phases: an initial chronic phase (CP), an accelerated phase (AP), and a blast phase (BP). CML is usually diagnosed in the CP, which quickly progresses to a blast crisis without effective medical intervention and can lead to death (Sandberg et al., 1971; Zalcberg et al., 1986).
CML is a myeloproliferative disorder originating in the HSC compartment and is predominantly caused by the formation of the chimeric oncogene BCR-ABL1, which is also known as the Philadelphia (Ph) chromosome. The formation of the Ph chromosome is a result of reciprocal translocation between the long arms of chromosomes 9 and 22 t (9; 22) (q34; q11). ABL1 encodes a ubiquitously expressed non-receptor tyrosine kinase responsible for regulating cell-cycle progression, proliferation, DNA repair, and differentiation, among others. Fusion with BCR endows ABL with the ability to become constitutively active, which is a sufficient cause for the development of CML (Sandberg et al., 1971; Chen et al., 2010).
Since the introduction of tyrosine kinase inhibitor (TKI) therapy with imatinib (IM) in 2001 (Goldman, 2000; Kantarjian, 2001) and second-generation TKI in 2007 (Swords et al., 2007), CML has transformed from a life-threatening disease to a manageable chronic condition (Bower et al., 2016). Nevertheless, there are constant search efforts for new treatment strategies and therapeutic targets, especially for the BP and drug resistance as well as BCR-ABL1-independent CML. LncRNAs are emerging as promising candidates in this regard and are expected to serve as diagnostic biomarkers, predictors of clinical outcomes, and therapeutic targets.
Note to readers: Compared to AML, the number of lncRNAs reported and experimentally validated for CML is far fewer. Categorizing them on the basis of mechanism was not suitable; hence, we broadly categorized the studies on lncRNAs in CML into two groups, with the first group focusing on the implications of lncRNAs for drug-resistant CML and tackling the problem of mutations acquired during the treatment course of patients that allow them to escape TKI therapy. The second group focuses on BCR-ABL1-independent CML and/or the general biology behind the pathogenesis of CML (Table 2).
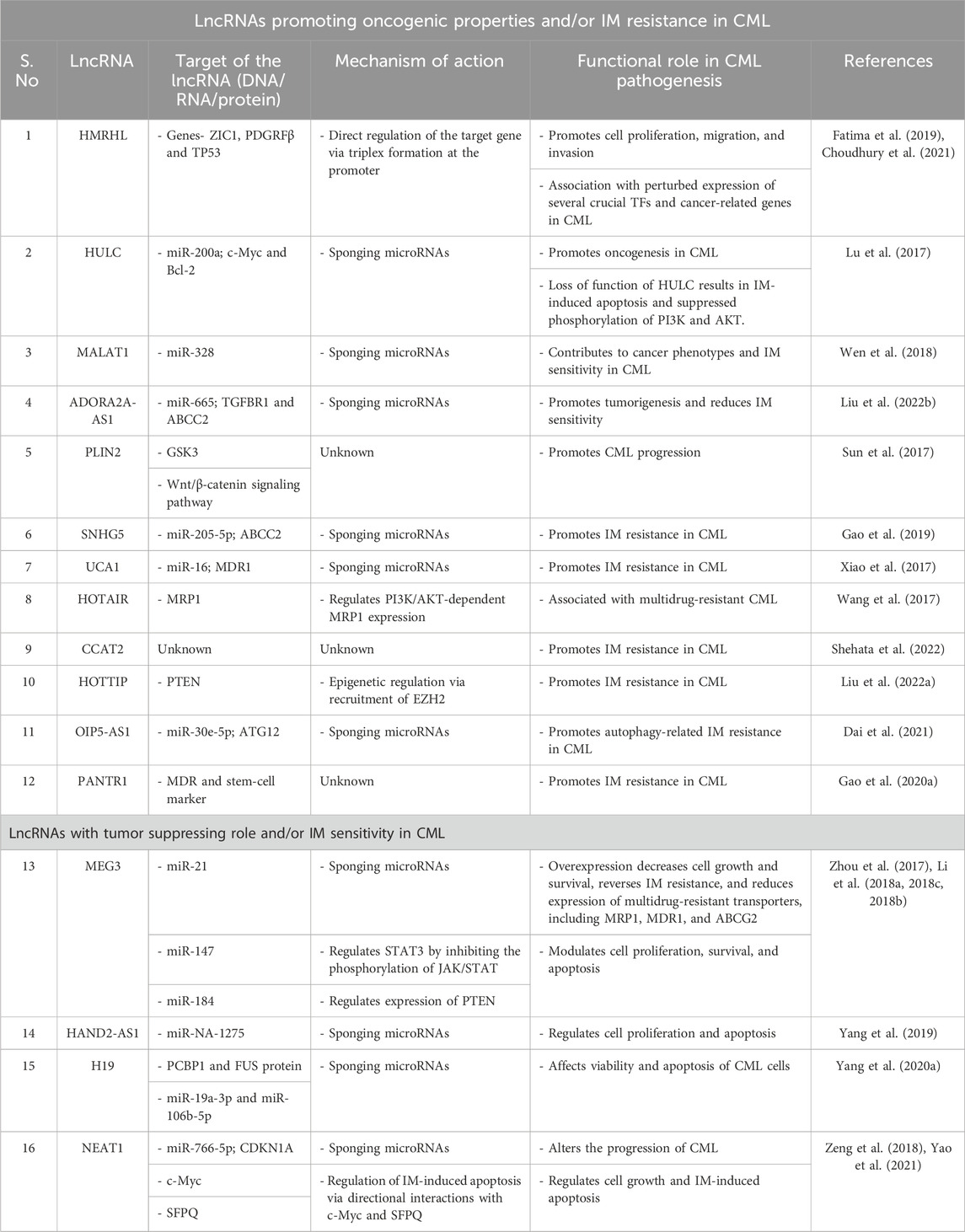
Table 2. Functional roles and action mechanisms of lncRNAs in the pathogenesis and/or IM resistance of CML.
The first line of treatment for CML with the identified BCR-ABL1 oncogene is a competitive TKI like IM that binds to the BCR-ABL1 protein and restrains downstream signal transduction. This drug has greatly improved the 5-year survival rates of CML patients from 34.2% to 80–90% (Kantarjian et al., 2012). However, there are challenges owing to the development of IM resistance, which can be attributed to several mechanisms such as the high copy number of mutant BCL-ABL1 genes, acquired mutations, aberrant expressions of drug transporters, and/or epigenetic alterations. However, the underlying mechanism is still largely unknown. Given the evidence of the involvement of lncRNAs in key biological processes, scientists are now trying to explore the roles of lncRNAs in this regard. For example, the lncRNA SNHG5 was found to be overexpressed in CML patients and IM-resistant cell lines; the study further demonstrated that SNHG5 acts as a competitive endogenous RNA (ceRNA) to sponge away miR-205-5p, upregulating its downstream target ABCC2 and promoting IM resistance in CML (Gao et al., 2019).
Another lncRNA that employs the same mechanism is UCA1, which competitively binds with miR-16, repressing the expression of MDR1 and contributing to IM resistance in CML (Xiao et al., 2017). The lncRNA MEG3 was also found to contribute to IM resistance through possible regulation of miR-21 (Zhou et al., 2017); it was found to be downregulated in CML, and miR-21 expression was observed to have an inverse correlation with MEG3 expression. The study further showed that ectopic expression of MEG3 decreases cell growth and survival, reverses IM resistance, and reduces the expressions of multidrug-resistant transporters, such as MRP1, MDR1, and ABCG2. However, the underlying regulatory mechanisms were unexplored in these works (Zhou et al., 2017; Li et al., 2018b). The role of the overexpressed lncRNA HOTAIR in multidrug-resistant CML was also explored and found to regulate MRP1 expression in a PI3K/AKT-dependent manner (Wang et al., 2017). Recently, the overexpression of another lncRNA, HULC, has been linked with an increase in IM resistance, while the opposite effect was observed for HULC depletion. Mechanistically, HULC was found to regulate the PI3K/AKT pathway by depleting miR-150-5p and thereby modulating MCL1 expression (Han and Ma, 2021). The lncRNA CCAT2 was found to be highly expressed in CML patients and was linked to IM resistance, suggesting that it is a reliable molecular marker for predicting IM responses in CML patients in the CP (Shehata et al., 2022).
Liu J. et al. (2022) have shown that the lncRNA HOTTIP, which is highly expressed in IM-resistant patients and cell lines, recruits EZH2 to suppress the expression of the PTEN gene contributing to IM resistance. A recent report highlighted that autophagy is associated with drug resistance in CML cells; the study revealed that the lncRNA OIP5-AS1 promotes autophagy-related IM resistance in CML by sponging miR-30e-5p and modulating ATG12 levels (Dai et al., 2021). The lncRNA PANTR1 was found to mediate IM resistance by promoting the expressions of MDR and stem-cell markers in CML cell lines (Gao J. J. et al., 2020).
In addition to their association with IM resistance, the roles of lncRNAs in CML pathobiology have been explored widely. This gives us insights into the functional mechanisms involved and potential drug targets, especially in Ph-chromosome-independent CML. For example, several studies have verified the downregulation of the lncRNA MEG3 in CML patient samples as well as cell lines, promoting it as a possible prognostic marker (Zhou et al., 2017; Li et al., 2018c; 2018b); MEG3 was found to modulate cell proliferation, survival, and apoptosis in CML cells. Suppressed expressions of MEG3 by histone deacetylase (HDAC1) and DNA methyltransferases (DNMT1, DNMT3A, DNMT3B) have been reported, indicating the potential clinical applications of demethylation drugs and HDAC inhibitors in the treatment of CML (Li et al., 2018b, 2018c). The expression patterns of numerous microRNAs were found to be correlated with MEG3 expression in CML; for example, MEG3 and miR-147 were observed to be directly correlated, while the expressions of miR-184 and miR21 were inversely correlated (Zhou et al., 2017; Li et al., 2018b, 2018c; Li J. et al., 2018). By deciphering the mechanisms of MEG3 in CML, Li et al. (2018c) showed that MEG3 could regulate STAT3 at least partly by inhibiting the phosphorylation of JAK/STAT through a possible negative feedback loop between MEG3 and STAT3. Direct correlation between the expression patterns of MEG3 and PTEN in CML are also suspected to be involved in the pathogenesis (Li et al., 2018b). Recently, HOTAIR was reported to accelerate CML progression by regulating PTEN; the study confirmed high expression of HOTAIR in the bone-marrow samples from CML patients and showed DNMT1 recruitment to regulate methylation of the PTEN promoter (Song H. et al., 2021).
The expression of the lncRNA HAND2-AS1 was reported to be low in CML patients, which was interestingly also found to decrease further with disease progression from AP to CP. Mechanistically, HAND2-AS1 was found to regulate cell proliferation and apoptosis by sequestering miRNA-1275 (Yang et al., 2019). Increased expression of the lncRNA HULC was also found to be positively correlated with the clinical stages of CML; results from the corresponding study showed that HULC promotes oncogenesis in CML by modulating the expressions of c-Myc and BCL-2 through sponging of miR-200a. The loss of function of HULC resulted in IM-induced apoptosis and suppressed phosphorylation of PI3K and AKT (Lu et al., 2017).
Although scarcely expressed in CML, the lncRNA H19 was found to be a tumor suppressor that affects the viability and apoptosis of CML cells. Using computational and experimental techniques, Yang J. et al. (2020) identified the proteins PCBP1 and FUS as well as microRNAs miR-19a-3p and miR-106b-5p as the targets of H19 in CML. METTL3-mediated m6A modification was found to be responsible for the low expression of the lncRNA NEAT1 in CML; NEAT1 was also found to alter the CML progression through downstream regulation of the miR-766-5p/CDKN1A axis (Yao et al., 2021). However, in another study, NEAT1 was shown to be regulated by c-Myc, and its role in IM-induced apoptosis through interactions with SFPQ (which regulates cell growth and death pathway related genes) was confirmed (Zeng et al., 2018). Overexpression of the protooncogene lncRNA MALAT1 was reported to contribute to cancer phenotypes and IM sensitivity of CML cells via miR-328 targeting (Wen et al., 2018). The lncRNA ADORA2A-AS1 was also found to be overexpressed in CML; using the loss of function study, this lncRNA was shown to exert tumor-promoting activities and reduce IM sensitivity via sponging miR-665 and thereby regulating TGFBR1 and ABCC2 (Liu Y. et al., 2022). The lncRNA PLIN2 was found to promote CML progression via regulation of the GSK3 and Wnt/β-catenin signaling pathways both in vitro and in vivo; high levels of PLIN2 were shown to be the result of regulation by CEBPA, which is also upregulated in CML (Sun et al., 2017).
Lastly, a recent finding by our research group shows high expression of the lncRNA Hmrhl in the CML cell line K562 (Fatima et al., 2019; Choudhury et al., 2021). Hmrhl was discovered as a human homolog of the mouse lncRNA meiotic recombination hot spot locus (mrhl), which was also first reported by our group and has been studied extensively thereafter (Nishant et al., 2004; Ganesan and Rao, 2008; Arun et al., 2012; Akhade et al., 2014; Kataruka et al., 2017; Pal et al., 2021; 2022; Kayyar et al., 2022). With restricted expressions in the testes, liver, kidneys, and spleen, mrhl was found to be a negative regulator of Wnt signaling and a regulator of SOX8 at the chromatin level in mouse spermatogonial cells (Arun et al., 2012; Akhade et al., 2014). The role of mrhl as a chromatin regulator of cellular differentiation and development genes along with its probable importance in the maintenance of the stemness in mouse embryonic stem cells was also established (Pal et al., 2021); the key role of mrhl in neuronal differentiation has also been reported recently (Pal et al., 2022). The lncRNA Hmrhl shares 65% homology with its mouse counterpart mrhl and an identical syntenic locus within the PHKB gene (Figure 3A). Unlike the restricted expressions of mrhl in a few organs, Hmrhl was shown to be ubiquitously expressed in all the organs studied. With a transcript length of 5.5 kb, Hmrhl was found to be larger than mrhl (2.4 kb), which was achieved by acquiring seven different repeat elements (L2b, L2c, MIR, Charlie 15a, AluY, L1PA3, and AluSx) that flank the highly conserved central region. The expression profile of Hmrhl confirmed its deregulation in several cancers. In the CML cell line, Hmrhl was shown to act as an enhancer RNA for its host gene PHKB (Fatima et al., 2019).
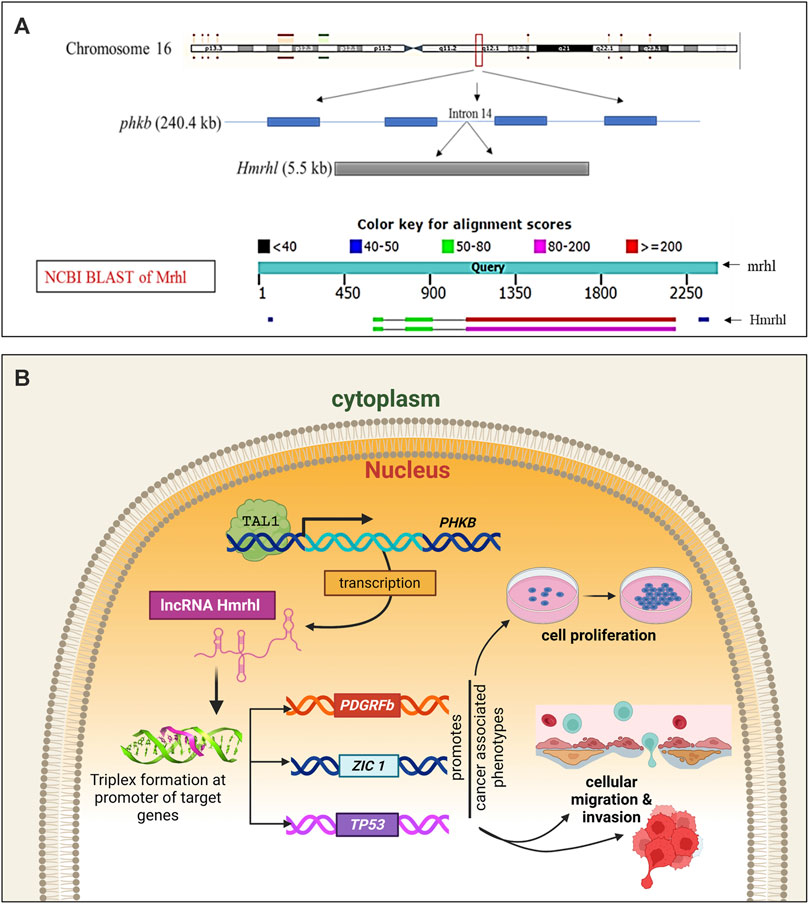
Figure 3. Human meiotic recombination hot spot locus (Hmrhl) lncRNA. (A) Genomic context with respect to its mouse homolog “mrhl” with which it shares 65% sequence homology and syntenic location within the intron of the PHKB gene. (B) Graphical model representing its regulatory mechanism and targets for promoting leukemogenesis of CML in K562 cells. The nuclear-restricted Hmrhl is the only reported lncRNA that acts at the chromatin level in CML by directly interacting with the target genes via triplex formation at their promoter and thereby regulating expression.
Our recently published study (Choudhury et al., 2021) verified the enrichment of Hmrhl within the nucleus and its association with chromatin; this study shows the influence of Hmrhl in promoting cancer-related phenotypes, such as proliferation, migration, and invasion in the CML cell line K562, using gene silencing techniques. By adopting transcriptome-based methods, this report further revealed the association between Hmrhl and the perturbed expressions of several crucial TFs as well as cancer-related genes, highlighting its significance in CML pathobiology. Additionally, the genome-wide occupancy study of Hmrhl indicated its association with several loci throughout the genome, particularly at the intergenic and repetitive element sites along with the other regions. The study further intersected data from RNA-seq and ChIRP-seq, resulting in the identification and selection of TP53, PDGFRβ, and ZIC1 as the possible targets of Hmrhl. Triplex formation at the promoter sites of the target genes was postulated to be the probable regulatory mechanism of Hmrhl (Choudhury et al., 2021) (Figure 3B). Furthermore, the study showed significant rescue effects on cancer-associated cellular phenotypes by overexpression of one of the target genes PDGFRβ in Hmrhl-silenced K562 cells. It was also verified that Hmrhl is regulated by TAL1, a key TF involved in hematopoiesis, in CML (Figure 3B).
An extensive literature search shows that most of the lncRNAs associated with CML exert their functions via microRNA sponging. To the best of our knowledge, Hmrhl is the only lncRNA reported so far that acts via direct interactions with chromatin to regulate its target genes, contributing to the pathobiology of CML.
In addition to studies on the functional and biological significances of lncRNAs in the pathology of myeloid leukemia, several works have focused on translational research to explore the significance of lncRNAs as prognostic biomarkers or drug targets in patients (Figure 4). Many lncRNAs with altered expressions in AML and CML patients have been suggested as prognostic markers for early diagnosis (Table 3) (Wang et al., 2014; Wang X. et al., 2018; Díaz-Beyá et al., 2015; Hao and Shao, 2015; Wu et al., 2015; Fernando et al., 2017; Yao et al., 2017; Ma et al., 2020, 2018; Zhang T. et al., 2018; Zhang W. et al., 2020; Zhang X. et al., 2020; Zhang F. et al., 2020; Luo W. et al., 2018; Izadifard et al., 2018; Jia et al., 2018; Pashaiefar et al., 2018; Qin et al., 2018, 2022; Yang et al., 2018; Papaioannou et al., 2019b; Cai et al., 2019; Chen et al., 2019; El-Khazragy et al., 2019; Huang et al., 2019; Lei et al., 2019; Li et al., 2019; Sellers et al., 2019; Zhang and Tao, 2019; Feng et al., 2020b; Chen et al., 2020; Ketab et al., 2020; Peng et al., 2020; Qu et al., 2020; Tan et al., 2020; Xu et al., 2020; Gao, 2021a; Saad et al., 2021; Gamaleldin et al., 2021; Masoud Eslami et al., 2021; Pavlovic et al., 2021; Salah et al., 2021; Zhu et al., 2021; Pei et al., 2022; Hussein et al., 2023).
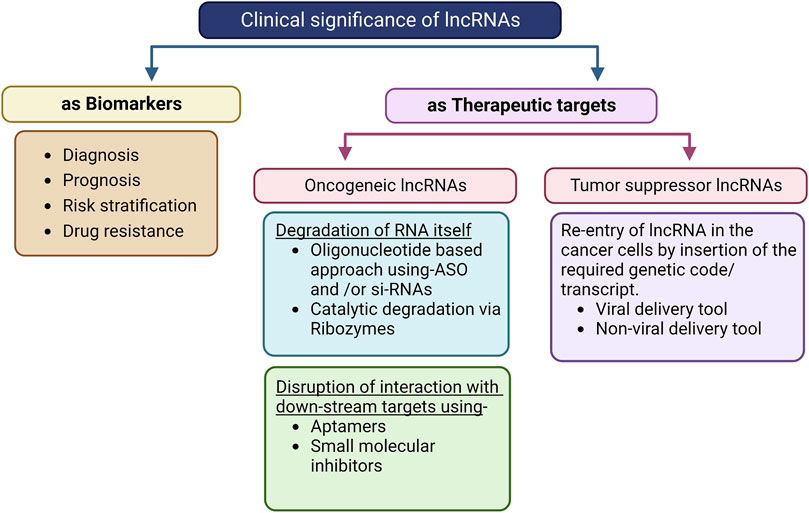
Figure 4. Flowchart depicting the clinical significance of lncRNAs and their incorporation into therapeutics. The presence of some lncRNAs in body fluids like serum, blood, plasma, and urine along with their specific expressions makes them suitable for use as biomarkers using non-invasive detection methods. For example, high levels of the lncRNAs LINC00899, FBXL19-AS1, LINC00460, and LINC00265 in the serum have been suggested as early diagnostic and prognostic biomarkers for AML patients. This method is limited by the stability of the RNA as well as lack of easy, robust, and economical detection methods for clinical use. Oligonucleotide- and catalytic-based approaches are promising for targeting and degrading oncogenic lncRNAs. siRNAs and ASO are already under phase I/II/III clinical trials for some lncRNAs in other diseases. The same approach can be applied in case of AML with targets like HOTAIR, DANCER, and UCA1, and is limited by the stability, delivery method, interferon-induced effects, and binding efficiency of the ribozyme. Aptamer and small-molecular inhibitors can also be used to inhibit the oncogenic effects of lncRNAs by disrupting their interactions with downstream targets. Tumor-suppressive lncRNAs like HOTAIRM1 can be repressed in AML cells to reverse carcinogenic effects using both viral and non-viral delivery channels.
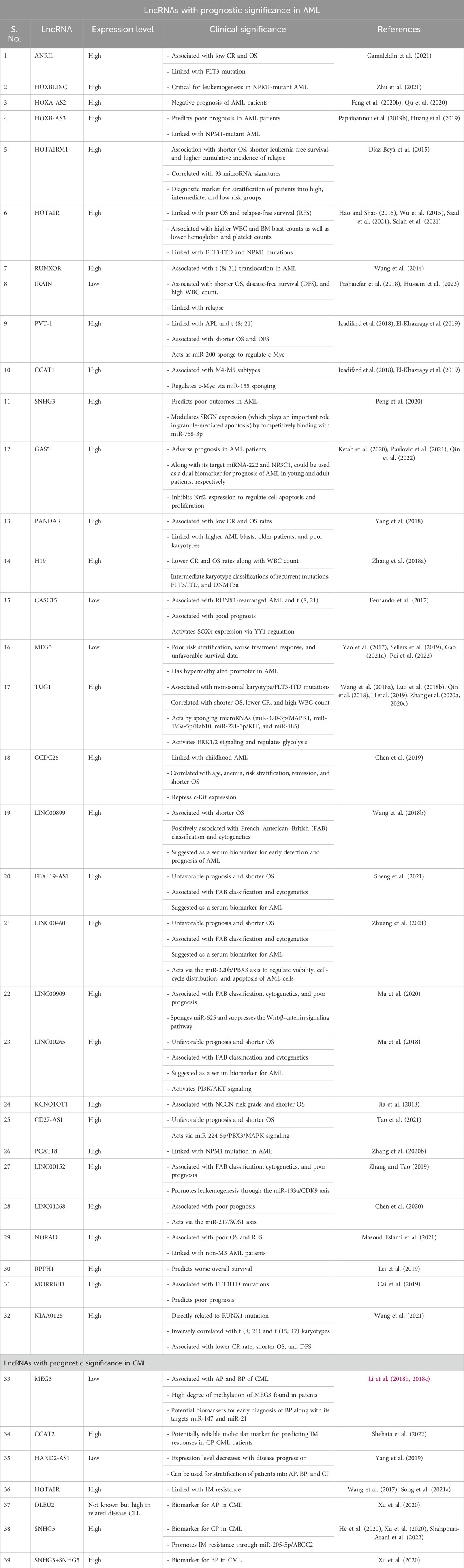
Table 3. Prognostic and clinical significance of documented lncRNAs in myeloid leukemia (AML and CML).
For example, an expression study of HOTAIRM1 in 241 AML patients revealed its association with shorter overall survival, shorter leukemia-free survival, and higher cumulative incidence of relapse (Díaz-Beyá et al., 2015); its expression was also correlated with 33 microRNA signatures, which can be combined and used as a diagnostic marker for stratifying patients into high, intermediate, and low risk groups (Díaz-Beyá et al., 2015). Two separate studies analyzing bone-marrow samples from 178 and 100 AML patients found ANRIL overexpression compared to healthy donors (Tan et al., 2020; Gamaleldin et al., 2021). ANRIL is also associated with low rates of complete remission (CR) and overall survival (OS) along with FLT3 mutation, implying that it could be a valuable prognostic marker for AML (Tan et al., 2020; Gamaleldin et al., 2021). In a study on 119 AML patients, higher PANDAR expression was associated with poor clinical outcomes with low CR and OS rates (Yang et al., 2018). The prognostic value of H19 expression was confirmed in AML patient samples and was correlated with lower CR and OS rates. High levels of H19 are also associated with WBC count and recurrent mutations, FLT3/ITD and DNMT3a in AML. These results were further validated by data analyses on TCGA and GEO (Zhang T. et al., 2018). Low levels of IRAIN are associated with high-risk AML patients, with an adverse prognosis of higher WBC and blast counts, shorter OS, and relapse-free survival (RFS). Resistance to chemotherapy with subsequent relapse was also observed in patients with low IRAIN expressions (Pashaiefar et al., 2018; Hussein et al., 2023). Recently, lower GAS5 expressions during diagnosis have been related to adverse prognosis in AML patients (Ketab et al., 2020; Pavlovic et al., 2021; Qin et al., 2022). Separate studies have associated the expression pattern of GAS5 with the expression profiles of its targets, miRNA-222 and NR3C1, as dual biomarkers for prognosis in young and adult AML patients, respectively (Ketab et al., 2020; Pavlovic et al., 2021). At the molecular level, GAS5 was found to inhibit Nrf2 expression, thereby regulating cell apoptosis and proliferation while further inhibiting the progression of AML (Qin et al., 2022). The tumor suppressor MEG3 is widely reported to have a hypermethylated promoter in AML, and its low expression is correlated with poor risk stratification, worse treatment responses, and unfavorable survival data (Yao et al., 2017; Sellers et al., 2019; He et al., 2020; Gao, 2021a).
Low expression of the lncRNA MEG3 is also linked with the prognosis of CML patients in the AP and BP (Li et al., 2018b, 2018c). These patients also showed higher degrees of methylation of the MEG3 promoter (Li et al., 2018c). The expression patterns of MEG3 and its targets miR-147 and miR-21 could thus be used as potential biomarkers for early diagnosis of CML blast crisis (Li et al., 2018c, 2018b). A study on peripheral blood mononuclear cells from 43 newly diagnosed CML patients showed that enhanced expression of CCAT2 was associated with IM resistance (Shehata et al., 2022); the authors concluded that CCAT2 can therefore be used as a reliable molecular marker for predicting IM responses in CP CML patients. Expression of HAND2-AS1 in the bone-marrow samples of 30 CML patients showed a gradual decline in its level with disease progression from AP to BP to CP; an inverse correlation between HAND2-AS1 and miR-1275 was also shown in the study (Yang et al., 2019). High levels of HOTAIR have been reported in CML patients and are linked with IM resistance (Wang et al., 2017; Song H. et al., 2021); however, the role of HOTAIR as a biomarker has not been suggested yet. Using dynamic network biomarkers (DNBs) and KEGG enrichment analysis, Xu et al. (2020) identified three lncRNAs functioning as ceRNA as potential biomarkers for CML; the authors suggested DLEU2, SNHG3+SNHG5, and SNHG5 as effective biomarkers for AP, BP, and CP owing to their key roles in the pathogenesis of CML (Xu et al., 2020).
Note to readers: For better navigation and searchability, a consolidated supplementary excel sheet (Supplementary File S1) is provided and contains the list of all lncRNAs grouped under various topics based on their roles in AML and CML.
The inherent properties of cell/tissue/disease-specific expressions of lncRNAs make them ideal candidates for diagnosis and prognostic stratification of patients depending on disease progression as well as possible responses to drug resistance. Some of the lncRNAs are reported to be present in bodily fluids (like the blood, plasma, serum, urine, and saliva). This allows non-invasive collection and easy detection of lncRNAs for screening as biomarkers (Badowski et al., 2022; Beylerli et al., 2022; Khawar et al., 2022; Aprile et al., 2023; Li et al., 2023). For example, high levels of the lncRNAs LINC00899, LINC00460, and FBXL19-AS1 in the serum have been suggested as biomarkers for the early clinical detection and prognosis of AML (Wang Y. et al., 2018; Sheng et al., 2021; Zhuang et al., 2021). Developments in transcriptomics technologies offer many techniques like qRT-PCR, RNA sequencing, and microarrays that can be used to detect lncRNAs (Wang et al., 2022). However, there is a need for developing robust and economical assays that are sensitive enough to detect lncRNAs readily and accurately for clinical applications.
For well-characterized lncRNAs, several strategies can be used for targeted treatment. Advanced techniques like the CRISPR/Cas9 for knock-in/-out of specific lncRNAs are under investigation (Sakuma and Yamamoto, 2018). si-RNAs as antisense oligonucleotides (ASOs) can be used in oligonucleotide-based techniques to target overexpressed oncogenic lncRNAs, where they can bind specifically with lncRNAs, initiating their degradation via RNA-induced silencing complex (RISC) or RNase H (Kole et al., 2012; Raguraman et al., 2021; Scharner and Aznarez, 2021; Zhang and Zhang, 2023). Investigations on improved delivery methods, stability of the oligonucleotide, and their long-lasting effects on patients are underway (Glazier et al., 2020; Shadid et al., 2021; Zhu et al., 2023). Another therapeutic strategy to inhibit lncRNA is catalytic degradation using ribozymes. However, their efficiency and specificity to the target are under investigation (Kruger et al., 1982; Pavco et al., 2000; Fedor and Williamson, 2005). Another method of tackling oncogenic lncRNAs is to disrupt their interactions with the targets using aptamers and small molecular inhibitors (Pedram Fatemi et al., 2015; Vitiello et al., 2015; Yang et al., 2017). The use of viral or non-viral delivery tools has also been proposed for tumor suppressor lncRNAs. Whole specific transcripts can also be delivered and re-expressed with functional rescue effects (Nayerossadat et al., 2012; Ibraheem et al., 2014).
Although none of the abovementioned therapeutic strategies were intended for use in myeloid leukemia, many of these are under clinical trials for other cancers, and some are already approved by the USFDA (Pavco et al., 2000; Parker et al., 2009; Smaldone and Davies, 2010; Nguyen et al., 2012; Coelho et al., 2013; Mansoori et al., 2014; Fatima et al., 2015; Liu et al., 2016; De Clara et al., 2017; Titze-de-Almeida et al., 2017; Papaioannou et al., 2019a). Studies on lncRNAs in myeloid leukemia are still in their early stages, but more ongoing research on the functional mechanisms as well as detailed characterizations along with data on the patients, disease progression, and chemoresistance are expected to enable application of these therapeutic strategies to myeloid leukemia.
It can be easily inferred from this review that lncRNAs play crucial roles in the occurrence and progression of myeloid leukemias, AML, and CML. Studies reported thus far have provided valuable insights into the regulatory mechanisms by which lncRNAs control the differentiation patterns, proliferative capacities, and apoptosis abilities of cells in both AML and CML. However, further in-depth studies on the functional mechanisms and regulatory targets of lncRNAs are needed in the context of myeloid leukemias to fully understand the complex pathobiology of the disease and identify promising therapeutic targets. Moreover, more numbers of studies on patient samples with large cohorts are essential to establish the clinical significance of lncRNAs and use them as potential biomarkers for diagnosis, risk stratification, and prognosis. Translation of the present knowledge from bench to bedside still presents a tremendous challenge; however, with the fast pace of ongoing research on lncRNAs and advancements in detection techniques, there is great scope for lncRNAs to provide solutions to the current limitations, which is crucial for precision medicine in myeloid leukemia.
SD: conceptualization, investigation, supervision, validation, visualization, writing–original draft, and writing–review and editing. VA: writing–review and editing. SC: writing–review and editing. MR: conceptualization, funding acquisition, project administration, supervision, validation, writing–original draft, and writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by the Department of Biotechnology of the Government of India (Grant Nos: BT/01/COE/07/09 and DBT/INF/22/SP27679/2018).
SD acknowledges the Department of Biotechnology, Government of India for her postdoctoral fellowship. VA acknowledges funding from the Jawaharlal Nehru Centre for Advanced Scientific Research (JNCASR). SC acknowledges SERB for his national postdoctoral fellowship.
This work was conceived by the late Prof. Manchanahalli Rangaswamy Satyanarayana Rao, a SERB Distinguished Fellow and SERB-YOS Professor. The remaining authors would like to dedicate this article to the memory of Prof. Rao, who was not only an exemplary scientist and mentor but also an exceptional human being.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frnar.2024.1334464/full#supplementary-material
Abelson, S., Collord, G., Ng, S. W. K., Weissbrod, O., Mendelson Cohen, N., Niemeyer, E., et al. (2018). Prediction of acute myeloid leukaemia risk in healthy individuals. Nature 559, 400–404. doi:10.1038/s41586-018-0317-6
Akhade, V. S., Arun, G., Donakonda, S., and Satyanarayana Rao, M. R. (2014). Genome wide chromatin occupancy of mrhl RNA and its role in gene regulation in mouse spermatogonial cells. RNA Biol. 11, 1262–1279. doi:10.1080/15476286.2014.996070
Akhade, V. S., Pal, D., and Kanduri, C. (2017). “Long noncoding RNA: genome organization and mechanism of action,” in Long noncoding RNA biology. Editor M. R. S. Rao (Berlin, Germany: Springer), 47–74. doi:10.1007/978-981-10-5203-3_2
Alvarez-Dominguez, J. R., and Lodish, H. F. (2017). Emerging mechanisms of long noncoding RNA function during normal and malignant hematopoiesis. Blood 130, 1965–1975. doi:10.1182/blood-2017-06-788695
Aprile, M., Costa, V., Cimmino, A., and Calin, G. A. (2023). Emerging role of oncogenic long noncoding RNA as cancer biomarkers. Int. J. Cancer 152, 822–834. doi:10.1002/ijc.34282
Arun, G., Akhade, V. S., Donakonda, S., and Rao, M. R. S. (2012). Mrhl RNA, a long noncoding RNA, negatively regulates Wnt signaling through its protein partner ddx5/p68 in mouse spermatogonial cells. Mol. Cell. Biol. 32, 3140–3152. doi:10.1128/MCB.00006-12
Ayupe, A. C., and Reis, E. M. (2017). “Evaluating the stability of mRNAs and noncoding RNAs,” in Enhancer RNAs, 139–153. doi:10.1007/978-1-4939-4035-6_11
Badowski, C., He, B., and Garmire, L. X. (2022). Blood-derived lncRNAs as biomarkers for cancer diagnosis: the Good, the Bad and the Beauty. npj Precis. Oncol. 6, 40. doi:10.1038/s41698-022-00283-7
Bao, X. L., Zhang, L., and Song, W. P. (2019). LncRNA SNHG1 overexpression regulates the proliferation of acute myeloid leukemia cells through miR-488-5p/NUP205 axis. Eur. Rev. Med. Pharmacol. Sci. 23, 5896–5903. doi:10.26355/eurrev_201907_18334
Bartonicek, N., Maag, J. L. v., and Dinger, M. E. (2016). Long noncoding RNAs in cancer: mechanisms of action and technological advancements. Mol. Cancer 15, 43. doi:10.1186/s12943-016-0530-6
Bayoumi, A., Sayed, A., Broskova, Z., Teoh, J.-P., Wilson, J., Su, H., et al. (2016). Crosstalk between long noncoding RNAs and MicroRNAs in health and disease. Int. J. Mol. Sci. 17, 356. doi:10.3390/ijms17030356
Beylerli, O., Gareev, I., Sufianov, A., Ilyasova, T., and Guang, Y. (2022). Long noncoding RNAs as promising biomarkers in cancer. Non-coding RNA Res. 7, 66–70. doi:10.1016/j.ncrna.2022.02.004
Bhan, A., and Mandal, S. S. (2015). LncRNA HOTAIR: a master regulator of chromatin dynamics and cancer. Biochimica Biophysica Acta (BBA) - Rev. Cancer 1856, 151–164. doi:10.1016/j.bbcan.2015.07.001
Bower, H., Björkholm, M., Dickman, P. W., Höglund, M., Lambert, P. C., and Andersson, T. M.-L. (2016). Life expectancy of patients with chronic myeloid leukemia approaches the life expectancy of the general population. J. Clin. Oncol. 34, 2851–2857. doi:10.1200/JCO.2015.66.2866
Bridges, M. C., Daulagala, A. C., and Kourtidis, A. (2021). LNCcation: lncRNA localization and function. J. Cell Biol. 220 (2), e202009045. doi:10.1083/jcb.202009045
Cai, Z., Ramdas, B., Kotzin, J., Carroll, M. P., Wertheim, G., Williams, A., et al. (2019). Lncrna morrbid regulates myeloid cell survival of AML driven by a combination of FLT3ITD and epigenetic mutations. Blood 134, 915. doi:10.1182/blood-2019-130653
Chen, B., Li, Y., Nie, Y., Tang, A., and Zhou, Q. (2020). Long non-coding RNA LINC01268 promotes cell growth and inhibits cell apoptosis by modulating miR-217/SOS1 axis in acute myeloid leukemia. Braz. J. Med. Biol. Res. 53, e9299. doi:10.1590/1414-431x20209299
Chen, C., Wang, P., Mo, W., Zhang, Y., Zhou, W., Deng, T., et al. (2019). lncRNA-CCDC26, as a novel biomarker, predicts prognosis in acute myeloid leukemia. Oncol. Lett. 18 (3), 2203–2211. doi:10.3892/ol.2019.10591
Chen, H., Du, G., Song, X., and Li, L. (2017a). Non-coding transcripts from enhancers: new insights into enhancer activity and gene expression regulation. Genomics Proteomics Bioinforma. 15, 201–207. doi:10.1016/j.gpb.2017.02.003
Chen, L., Wang, W., Cao, L., Li, Z., and Wang, X. (2016). Long non-coding RNA CCAT1 acts as a competing endogenous RNA to regulate cell growth and differentiation in acute myeloid leukemia. Mol. Cells 39, 330–336. doi:10.14348/molcells.2016.2308
Chen, Y., Peng, C., Li, D., and Li, S. (2010). Molecular and cellular bases of chronic myeloid leukemia. Protein Cell 1, 124–132. doi:10.1007/s13238-010-0016-z
Chen, Z.-H., Wang, W.-T., Huang, W., Fang, K., Sun, Y.-M., Liu, S.-R., et al. (2017b). The lncRNA HOTAIRM1 regulates the degradation of PML-RARA oncoprotein and myeloid cell differentiation by enhancing the autophagy pathway. Cell Death Differ. 24, 212–224. doi:10.1038/cdd.2016.111
Choudhury, S. R., Dutta, S., Bhaduri, U., and Rao, M. R. S. (2021). LncRNA Hmrhl regulates expression of cancer related genes in chronic myelogenous leukemia through chromatin association. Nar. Cancer 3, zcab042. doi:10.1093/narcan/zcab042
Clark, M. B., Johnston, R. L., Inostroza-Ponta, M., Fox, A. H., Fortini, E., Moscato, P., et al. (2012). Genome-wide analysis of long noncoding RNA stability. Genome Res. 22, 885–898. doi:10.1101/gr.131037.111
Coelho, T., Adams, D., Silva, A., Lozeron, P., Hawkins, P. N., Mant, T., et al. (2013). Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N. Engl. J. Med. 369, 819–829. doi:10.1056/NEJMoa1208760
Cook, W. D., McCaw, B. J., Herring, C., John, D. L., Foote, S. J., Nutt, S. L., et al. (2004). PU.1 is a suppressor of myeloid leukemia, inactivated in mice by gene deletion and mutation of its DNA binding domain. Blood 104, 3437–3444. doi:10.1182/blood-2004-06-2234
Dahariya, S., Paddibhatla, I., Kumar, S., Raghuwanshi, S., Pallepati, A., and Gutti, R. K. (2019). Long non-coding RNA: classification, biogenesis and functions in blood cells. Mol. Immunol. 112, 82–92. doi:10.1016/j.molimm.2019.04.011
Dai, H., Wang, J., Huang, Z., Zhang, H., Wang, X., Li, Q., et al. (2021). LncRNA OIP5-AS1 promotes the autophagy-related imatinib resistance in chronic myeloid leukemia cells by regulating miR-30e-5p/ATG12 Axis. Technol. Cancer Res. Treat. 20, 153303382110521. doi:10.1177/15330338211052150
Darbellay, F., and Necsulea, A. (2020). Comparative transcriptomics analyses across species, organs, and developmental stages reveal functionally constrained lncRNAs. Mol. Biol. Evol. 37, 240–259. doi:10.1093/molbev/msz212
De Clara, E., Gourvest, M., Ma, H., Vergez, F., Tosolini, M., Dejean, S., et al. (2017). Long non-coding RNA expression profile in cytogenetically normal acute myeloid leukemia identifies a distinct signature and a new biomarker in NPM1-mutated patients. Haematologica 102, 1718–1726. doi:10.3324/haematol.2017.171645
Deschler, B., and Lübbert, M. (2006). Acute myeloid leukemia: epidemiology and etiology. Cancer 107, 2099–2107. doi:10.1002/cncr.22233
Díaz-Beyá, M., Brunet, S., Nomdedéu, J., Pratcorona, M., Cordeiro, A., Gallardo, D., et al. (2015). The lincRNA HOTAIRM1 located in the HOXA genomic region, is expressed in acute myeloid leukemia, impacts prognosis in patients in the intermediate-risk cytogenetic category, and is associated with a distinctive microRNA signature. Oncotarget 6, 31613–31627. doi:10.18632/oncotarget.5148
Dieter, C., Lourenco, E. D., and Lemos, N. E. (2020). Association of long non-coding RNA and leukemia: a systematic review. Gene 735, 144405. doi:10.1016/j.gene.2020.144405
Dong, X., Xu, X., and Guan, Y. (2020a). LncRNA LINC00899 promotes progression of acute myeloid leukaemia by modulating miR-744-3p/YY1 signalling. Cell Biochem. Funct. 38, 955–964. doi:10.1002/cbf.3521
Dong, Y., Shi, O., Zeng, Q., Lu, X., Wang, W., Li, Y., et al. (2020b). Leukemia incidence trends at the global, regional, and national level between 1990 and 2017. Exp. Hematol. Oncol. 9 (1), 14. doi:10.1186/s40164-020-00170-6
Ebralidze, A. K., Guibal, F. C., Steidl, U., Zhang, P., Lee, S., Bartholdy, B., et al. (2008). PU.1 expression is modulated by the balance of functional sense and antisense RNAs regulated by a shared cis -regulatory element. Genes Dev. 22, 2085–2092. doi:10.1101/gad.1654808
El-Khazragy, N., Elayat, W., Matbouly, S., Seliman, S., Sami, A., Safwat, G., et al. (2019). The prognostic significance of the long non-coding RNAs “CCAT1, PVT1” in t (8; 21) associated acute myeloid leukemia. Gene 707, 172–177. doi:10.1016/j.gene.2019.03.055
Engreitz, J. M., Ollikainen, N., and Guttman, M. (2016). Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat. Rev. Mol. Cell Biol. 17, 756–770. doi:10.1038/nrm.2016.126
Fanucchi, S., and Mhlanga, M. M. (2017). Enhancer-derived lncRNAs regulate genome architecture: fact or fiction? Trends Genet. 33 (6), 375–377. doi:10.1016/j.tig.2017.03.004
Farrar, J. E., Smith, J. L., Othus, M., Huang, B. J., Wang, Y.-C., Ries, R. E., et al. (2022). A robust long non-coding RNA expression classifier for risk stratification in pediatric AML. Blood 140, 9173–9174. doi:10.1182/blood-2022-168296
Fatima, R., Akhade, V. S., Pal, D., and Rao, S. M. (2015). Long noncoding RNAs in development and cancer: potential biomarkers and therapeutic targets. Mol. Cell. Ther. 3, 5. doi:10.1186/s40591-015-0042-6
Fatima, R., Choudhury, S. R., Tr, D., Bhaduri, U., and Rao, M. R. S. (2019). A novel enhancer RNA, Hmrhl, positively regulates its host gene, phkb, in chronic myelogenous leukemia. Non-coding RNA Res. 4, 96–108. doi:10.1016/j.ncrna.2019.08.001
Fedor, M. J., and Williamson, J. R. (2005). The catalytic diversity of RNAs. Nat. Rev. Mol. Cell Biol. 6, 399–412. doi:10.1038/nrm1647
Feng, S., Liu, N., Chen, X., Liu, Y., and An, J. (2020a). Long non-coding RNA NEAT1/miR-338-3p axis impedes the progression of acute myeloid leukemia via regulating CREBRF. Cancer Cell Int. 20, 112. doi:10.1186/s12935-020-01182-2
Feng, Y., Hu, S., Li, L., Peng, X., and Chen, F. (2020b). Long noncoding RNA HOXA-AS2 functions as an oncogene by binding to EZH2 and suppressing LATS2 in acute myeloid leukemia (AML). Cell Death Dis. 11, 1025. doi:10.1038/s41419-020-03193-3
Feng, Y., Hu, S., Li, L., Zhang, S., Liu, J., Xu, X., et al. (2020c). LncRNA NR-104098 inhibits AML proliferation and induces differentiation through repressing EZH2 transcription by interacting with E2F1. Front. Cell Dev. Biol. 8, 142. doi:10.3389/fcell.2020.00142
Fernando, T. R., Contreras, J. R., Zampini, M., Rodriguez-Malave, N. I., Alberti, M. O., Anguiano, J., et al. (2017). The lncRNA CASC15 regulates SOX4 expression in RUNX1-rearranged acute leukemia. Mol. Cancer 16, 126. doi:10.1186/s12943-017-0692-x
Frese, K. S., Katus, H. A., and Meder, B. (2013). Next-generation sequencing: from understanding biology to personalized medicine. Biol. (Basel) 2, 378–398. doi:10.3390/biology2010378
Gamaleldin, M. A., Ghallab, O., and Abo Elwafa, R. (2021). Prognostic significance of long non coding RNA ANRIL and SNHG14 in acute myeloid leukemia. Asian Pac. J. Cancer Prev. 22 (12), 3763–3771. doi:10.31557/apjcp.2021.22.12.3763
Ganesan, G., and Rao, S. M. R. (2008). A novel noncoding RNA processed by Drosha is restricted to nucleus in mouse. RNA 14, 1399–1410. doi:10.1261/rna.838308
Gao, B., Li, S., and Li, G. (2019). Long noncoding RNA (lncRNA) small nucleolar RNA host gene 5 (SNHG5) regulates proliferation, differentiation, and apoptosis of K562 cells in chronic myeliod leukemia. Med. Sci. Monit. 25, 6812–6819. doi:10.12659/MSM.916661
Gao, J., Wang, F., Wu, P., Chen, Y., and Jia, Y. (2020b). Aberrant LncRNA expression in leukemia. J. Cancer 11, 4284–4296. doi:10.7150/jca.42093
Gao, J. J., Zhu, X. P., Wang, M. Q., and Chen, W. Y. (2020a). Relationship between PANTR1 and imatinib resistance of chronic myeloid leukemia cell line K562 and its related mechanisms. Zhongguo Shi Yan Xue Ye Xue Za Zhi 28, 430–435. doi:10.19746/j.cnki.issn.1009-2137.2020.02.012
Gao, S., Zhou, B., Li, H., Huang, X., Wu, Y., Xing, C., et al. (2018). Long noncoding RNA HOTAIR promotes the self-renewal of leukemia stem cells through epigenetic silencing of p15. Exp. Hematol. 67, 32–40.e3. doi:10.1016/j.exphem.2018.08.005
Gao, W. (2021a). Long non-coding RNA MEG3 as a candidate prognostic factor for induction therapy response and survival profile in childhood acute lymphoblastic leukemia patients. Scand. J. Clin. Laboratory Investigation 81, 194–200. doi:10.1080/00365513.2021.1881998
Gao, Y., Shang, S., Guo, S., Li, X., Zhou, H., Liu, H., et al. (2021b). Lnc2Cancer 3.0: an updated resource for experimentally supported lncRNA/circRNA cancer associations and web tools based on RNA-seq and scRNA-seq data. Nucleic Acids Res. 49, D1251–D1258. doi:10.1093/nar/gkaa1006
Ghafouri-Fard, S., Niazi, V., and Taheri, M. (2021). Role of miRNAs and lncRNAs in hematopoietic stem cell differentiation. Non-coding RNA Res. 6, 8–14. doi:10.1016/j.ncrna.2020.12.002
Gilliland, D. G., Jordan, C. T., and Felix, C. A. (2004). The molecular basis of leukemia. Hematology 2004, 80–97. doi:10.1182/asheducation-2004.1.80
Glazier, D. A., Liao, J., Roberts, B. L., Li, X., Yang, K., Stevens, C. M., et al. (2020). Chemical synthesis and biological application of modified oligonucleotides. Bioconjugate Chem. 31, 1213–1233. doi:10.1021/acs.bioconjchem.0c00060
Goldman, J. M. (2000). Tyrosine-kinase inhibition in treatment of chronic myeloid leukaemia. Lancet 355, 1031–1032. doi:10.1016/S0140-6736(00)02029-8
Guo, S., Li, B., Xu, X., Wang, W., Wang, S., Lv, T., et al. (2020). Construction of a 14 lncRNA risk score system predicting survival of children with acute myelocytic leukemia. Exp. Ther. Med. 20, 1521–1531. doi:10.3892/etm.2020.8846
Guo, X., Gao, L., Wang, Y., Chiu, D. K. Y., Wang, T., and Deng, Y. (2016). Advances in long noncoding RNAs: identification, structure prediction and function annotation. Briefings Funct. Genomics 15, 38–46. doi:10.1093/bfgp/elv022
Gupta, R. A., Shah, N., Wang, K. C., Kim, J., Horlings, H. M., Wong, D. J., et al. (2010). Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–1076. doi:10.1038/nature08975
Han, Y., and Ma, Z. (2021). LncRNA highly upregulated in liver cancer regulates imatinib resistance in chronic myeloid leukemia via the miR-150-5p/MCL1 axis. Anticancer Drugs 32, 427–436. doi:10.1097/CAD.0000000000001019
Hao, S., and Shao, Z. (2015). HOTAIR is upregulated in acute myeloid leukemia and that indicates a poor prognosis. Int. J. Clin. Exp. Pathol. 8 (6), 7223.
He, C., Wang, X., Luo, J., Ma, Y., and Yang, Z. (2020). Long noncoding RNA maternally expressed gene 3 is downregulated, and its insufficiency correlates with poor-risk stratification, worse treatment response, as well as unfavorable survival data in patients with acute myeloid leukemia. Technol. Cancer Res. Treat. 19, 153303382094581. doi:10.1177/1533033820945815
Höglund, M., Sandin, F., and Simonsson, B. (2015). Epidemiology of chronic myeloid leukaemia: an update. Ann. Hematol. 94, 241–247. doi:10.1007/s00277-015-2314-2
Holdt, L. M., Hoffmann, S., Sass, K., Langenberger, D., Scholz, M., Krohn, K., et al. (2013). Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks. PLoS Genet. 9, e1003588. doi:10.1371/journal.pgen.1003588
Hu, L., Liu, J., Meng, Y., Zheng, H., Ding, C., Wang, H., et al. (2021a). Long non-coding RNA HOTAIR regulates myeloid differentiation through the upregulation of p21 via miR-17-5p in acute myeloid leukaemia. RNA Biol. 18, 1434–1444. doi:10.1080/15476286.2020.1854520
Hu, N., Chen, L., Li, Q., and Zhao, H. (2019). LncRNA HOTAIRM1 is involved in the progression of acute myeloid leukemia through targeting miR-148b. RSC Adv. 9, 10352–10359. doi:10.1039/C9RA00142E
Hu, Y., Li, Q., Hou, M., Peng, J., Yang, X., and Xu, S. (2021b). Magnitude and temporal trend of the chronic myeloid leukemia: on the basis of the global burden of disease study 2019. JCO Glob. Oncol. 7, 1429–1441. doi:10.1200/GO.21.00194
Huang, H. H., Chen, F. Y., Chou, W. C., Hou, H. A., Ko, B. S., Lin, C. T., et al. (2019). Long non-coding RNA HOXB-AS3 promotes myeloid cell proliferation and its higher expression is an adverse prognostic marker in patients with acute myeloid leukemia and myelodysplastic syndrome. BMC cancer 19, 617–714. doi:10.1186/s12885-019-5822-y
Hughes, J. M., Legnini, I., Salvatori, B., Masciarelli, S., Marchioni, M., Fazi, F., et al. (2015). C/EBPα-p30 protein induces expression of the oncogenic long non-coding RNA UCA1 in acute myeloid leukemia. Oncotarget 6, 18534–18544. doi:10.18632/oncotarget.4069
Hussein, M. A. R., Ahmed, A. E., ElNahass, Y., El-Dahshan, D., and Ali, M. A. M. (2023). Downregulation of IRAIN long non-coding RNA predicts unfavourable clinical outcome in acute myeloid leukaemia patients. Biomarkers 28, 323–340. doi:10.1080/1354750X.2023.2171128
Ibraheem, D., Elaissari, A., and Fessi, H. (2014). Gene therapy and DNA delivery systems. Int. J. Pharm. 459, 70–83. doi:10.1016/j.ijpharm.2013.11.041
Ilik, I. A., Quinn, J. J., Georgiev, P., Tavares-Cadete, F., Maticzka, D., Toscano, S., et al. (2013). Tandem stem-loops in roX RNAs act together to mediate X chromosome dosage compensation in Drosophila. Mol. Cell 51, 156–173. doi:10.1016/j.molcel.2013.07.001
Iyer, M. K., Niknafs, Y. S., Malik, R., Singhal, U., Sahu, A., Hosono, Y., et al. (2015a). The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 47, 199–208. doi:10.1038/ng.3192
Iyer, M. K., Niknafs, Y. S., Malik, R., Singhal, U., Sahu, A., Hosono, Y., et al. (2015b). The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 47, 199–208. doi:10.1038/ng.3192
Izadifard, M., Pashaiefar, H., Yaghmaie, M., Montazeri, M., Sadraie, M., Momeny, M., et al. (2018). Expression analysis of PVT1, CCDC26, and CCAT1 long noncoding RNAs in acute myeloid leukemia patients. Genet. Test. Mol. Biomarkers 22, 593–598. doi:10.1089/gtmb.2018.0143
Jarroux, J., Morillon, A., and Pinskaya, M. (2017). “History, discovery, and classification of lncRNAs,” in, ed. M. R. S. Rao (Springer, Berlin, Germany), 1–46.
Jeon, Y., and Lee, J. T. (2011). YY1 tethers xist RNA to the inactive X nucleation center. Cell 146, 119–133. doi:10.1016/j.cell.2011.06.026
Jia, Z. W., Li, Y., Cui, G. R., Zhao, H. B., Li, P. Y., and Luo, J. M. (2018). Expression and clinical significance of LncRNA KCNQ1OT1 in patients with acute myeloid leukemia. Zhongguo Shi Yan Xue Ye Xue Za Zhi 26 (3), 653–657. doi:10.7534/j.issn.1009-2137.2018.03.004
Kantarjian, H., Kadia, T., DiNardo, C., Daver, N., Borthakur, G., Jabbour, E., et al. (2021). Acute myeloid leukemia: current progress and future directions. Blood Cancer J. 11, 41. doi:10.1038/s41408-021-00425-3
Kantarjian, H., O’Brien, S., Jabbour, E., Garcia-Manero, G., Quintas-Cardama, A., Shan, J., et al. (2012). Improved survival in chronic myeloid leukemia since the introduction of imatinib therapy: a single-institution historical experience. Blood 119, 1981–1987. doi:10.1182/blood-2011-08-358135
Kantarjian, H. (2001). Imatinib mesylate: clinical results in Philadelphia chromosome-positive leukemias. Seminars Oncol. 28, 9–18. doi:10.1016/S0093-7754(01)90098-3
Kataruka, S., Akhade, V. S., Kayyar, B., and Rao, M. R. S. (2017). Mrhl long noncoding RNA mediates meiotic commitment of mouse spermatogonial cells by regulating Sox8 expression. Mol. Cell. Biol. 37, e00632. doi:10.1128/MCB.00632-16
Kayyar, B., Ravikkumar, A. C., Bhaduri, U., and Rao, M. R. S. (2022). Regulation of Sox8 through lncRNA mrhl-mediated chromatin looping in mouse spermatogonia. Mol. Cell. Biol. 42, e0047521. doi:10.1128/mcb.00475-21
Kelly, L. M., and Gilliland, D. G. (2002). GENETICS OF MYELOID LEUKEMIAS. Annu. Rev. Genomics Hum. Genet. 3, 179–198. doi:10.1146/annurev.genom.3.032802.115046
Ketab, F. N. G., Gharesouran, J., Ghafouri-Fard, S., Dastar, S., Mazraeh, S. A., Hosseinzadeh, H., et al. (2020). Dual biomarkers long non-coding RNA GAS5 and its target, NR3C1, contribute to acute myeloid leukemia. Exp. Mol. Pathology 114, 104399. doi:10.1016/j.yexmp.2020.104399
Khawar, M. B., Hamid, S. E., Jan, T., Abbasi, M. H., Idnan, M., and Sheikh, N. (2022). Diagnostic, prognostic and therapeutic potential of long noncoding RNAs in cancer. Mol. Biol. Rep. 49, 2311–2319. doi:10.1007/s11033-022-07180-z
Kishtagari, A., and Levine, R. L. (2021). The role of somatic mutations in acute myeloid leukemia pathogenesis. Cold Spring Harb. Perspect. Med. 11, a034975. doi:10.1101/cshperspect.a034975
Kole, R., Krainer, A. R., and Altman, S. (2012). RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat. Rev. Drug Discov. 11, 125–140. doi:10.1038/nrd3625
Kopp, F., and Mendell, J. T. (2018). Functional classification and experimental dissection of long non coding RNAs. Cell 172, 393–407. doi:10.1016/j.cell.2018.01.011
Kruger, K., Grabowski, P. J., Zaug, A. J., Sands, J., Gottschling, D. E., and Cech, T. R. (1982). Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of tetrahymena. Cell 31, 147–157. doi:10.1016/0092-8674(82)90414-7
Kung, J. T. Y., Colognori, D., and Lee, J. T. (2013). Long noncoding RNAs: past, present, and future. Genetics 193, 651–669. doi:10.1534/genetics.112.146704
Lan, Z., Chen, Y., Jin, J., Xu, Y., and Zhu, X. (2021). Long non-coding RNA: insight into mechanisms of alzheimer’s disease. Front. Mol. Neurosci. 14, 821002. doi:10.3389/fnmol.2021.821002
Lei, B., He, A., Chen, Y., Cao, X., Zhang, P., Liu, J., et al. (2019). Long non-coding RNA RPPH1 promotes the proliferation, invasion and migration of human acute myeloid leukemia cells through down-regulating miR-330-5p expression. EXCLI J. 18, 824–837. doi:10.17179/excli2019-1686
Li, C., Gao, Q., Wang, M., and Xin, H. (2021a). LncRNA SNHG1 contributes to the regulation of acute myeloid leukemia cell growth by modulating miR-489-3p/SOX12/Wnt/β-catenin signaling. J. Cell. Physiology 236, 653–663. doi:10.1002/jcp.29892
Li, G., Zheng, P., Wang, H., Ai, Y., and Mao, X. (2019). Long non-coding RNA TUG1 modulates proliferation, migration, and invasion of acute myeloid leukemia cells via regulating miR-370-3p/MAPK1/ERK. OncoTargets Ther. Vol. 12, 10375–10388. doi:10.2147/ott.s217795
Li, J., Li, Z., Bai, X., Chen, X., Wang, M., Wu, Y., et al. (2022a). LncRNA UCA1 promotes the progression of AML by upregulating the expression of CXCR4 and CYP1B1 by affecting the stability of METTL14. J. Oncol. 2022, 1–13. doi:10.1155/2022/2756986
Li, J., Wang, M., and Chen, X. (2020b). Long non-coding RNA UCA1 modulates cell proliferation and apoptosis by regulating miR-296-3p/Myc axis in acute myeloid leukemia. Cell Cycle 19, 1454–1465. doi:10.1080/15384101.2020.1750814
Li, J., Zi, Y., Wang, W., and Li, Y. (2018a). Long noncoding RNA MEG3 inhibits cell proliferation and metastasis in chronic myeloid leukemia via targeting miR-184. Oncol. Res. 26, 297–305. doi:10.3727/096504017X14980882803151
Li, J. J., Chen, X. F., Wang, M., Zhang, P. P., Zhang, F., and Zhang, J. J. (2020a). Long non-coding RNA UCA1 promotes autophagy by targeting miR-96-5p in acute myeloid leukaemia. Clin. Exp. Pharmacol. Physiology 47, 877–885. doi:10.1111/1440-1681.13259
Li, M., Zhao, Y., Li, H., Deng, X., and Sheng, M. (2023). Application value of circulating LncRNA in diagnosis, treatment, and prognosis of breast cancer. Funct. Integr. Genomics 23, 61. doi:10.1007/s10142-023-00983-8
Li, X., Rong, J., Li, T., Zhou, Y., and Qi, X. (2022b). LncRNA H22954 inhibits angiogenesis in acute myeloid leukemia through a PDGFA-dependent mechanism. Recent Pat. Anti-Cancer Drug Discov. 17 (4), 427–434. doi:10.2174/1871526522666220321154949
Li, Y., Syed, J., and Sugiyama, H. (2016). RNA-DNA triplex formation by long noncoding RNAs. Cell Chem. Biol. 23, 1325–1333. doi:10.1016/j.chembiol.2016.09.011
Li, Z., Yang, L., Liu, X., Nie, Z., and Luo, J. (2018b). Long noncoding RNA MEG3 inhibits proliferation of chronic myeloid leukemia cells by sponging microRNA21. Biomed. Pharmacother. 104, 181–192. doi:10.1016/j.biopha.2018.05.047
Li, Z., Yang, L., Liu, X., Wang, X., Pan, Y., and Luo, J. (2018c). The long noncoding RNA MEG3 and its target miR-147 regulate JAK/STAT pathway in advanced chronic myeloid leukemia. EBioMedicine 34, 61–75. doi:10.1016/j.ebiom.2018.07.013
Li, Z.-J., Cheng, J., Song, Y., Li, H.-H., and Zheng, J.-F. (2021b). LncRNA SNHG5 upregulation induced by YY1 contributes to angiogenesis via miR-26b/CTGF/VEGFA axis in acute myelogenous leukemia. Lab. Investig. 101, 341–352. doi:10.1038/s41374-020-00519-9
Liang, Y., Li, E., Zhang, H., Zhang, L., Tang, Y., and Wanyan, Y. (2020). Silencing of lncRNA UCA1 curbs proliferation and accelerates apoptosis by repressing SIRT1 signals by targeting miR-204 in pediatric AML. J. Biochem. Mol. Toxicol. 34, e22435. doi:10.1002/jbt.22435
Liu, J., Yang, L., Liu, X., Liu, L., Liu, M., Feng, X., et al. (2022a). lncRNA HOTTIP recruits EZH2 to inhibit PTEN expression and participates in IM resistance in chronic myeloid leukemia. Stem Cells Int. 2022, 1–20. doi:10.1155/2022/9993393
Liu, M. C., Pitcher, B. N., Mardis, E. R., Davies, S. R., Friedman, P. N., Snider, J. E., et al. (2016). PAM50 gene signatures and breast cancer prognosis with adjuvant anthracycline- and taxane-based chemotherapy: correlative analysis of C9741 (Alliance). NPJ Breast Cancer 2, 15023. doi:10.1038/npjbcancer.2015.23
Liu, Y., Gao, X., and Tian, X. (2019). High expression of long intergenic non-coding RNA LINC00662 contributes to malignant growth of acute myeloid leukemia cells by upregulating ROCK1 via sponging microRNA-340-5p. Eur. J. Pharmacol. 859, 172535. doi:10.1016/j.ejphar.2019.172535
Liu, Y., Li, H., Zhao, Y., Li, D., Zhang, Q., Fu, J., et al. (2022b). Knockdown of ADORA2A antisense RNA 1 inhibits cell proliferation and enhances imatinib sensitivity in chronic myeloid leukemia. Bioengineered 13, 2296–2307. doi:10.1080/21655979.2021.2024389
Lorenzen, J. M., and Thum, T. (2016). Long noncoding RNAs in kidney and cardiovascular diseases. Nat. Rev. Nephrol. 12, 360–373. doi:10.1038/nrneph.2016.51
Lorenzi, L., Avila Cobos, F., Decock, A., Everaert, C., Helsmoortel, H., Lefever, S., et al. (2019). Long noncoding RNA expression profiling in cancer: challenges and opportunities. Genes Chromosom. Cancer 58, 191–199. doi:10.1002/gcc.22709
Lu, J., Wu, X., Wang, L., Li, T., and Sun, L. (2021). Long noncoding RNA LINC00467 facilitates the progression of acute myeloid leukemia by targeting the miR-339/SKI pathway. Leukemia Lymphoma 62, 428–437. doi:10.1080/10428194.2020.1832667
Lu, Y., Li, Y., Chai, X., Kang, Q., Zhao, P., Xiong, J., et al. (2017). Long noncoding RNA HULC promotes cell proliferation by regulating PI3K/AKT signaling pathway in chronic myeloid leukemia. Gene 607, 41–46. doi:10.1016/j.gene.2017.01.004
Luo, H., Wang, F., Zha, J., Li, H., Yan, B., Du, Q., et al. (2018a). CTCF boundary remodels chromatin domain and drives aberrant HOX gene transcription in acute myeloid leukemia. Blood 132, 837–848. doi:10.1182/blood-2017-11-814319
Luo, H., Zhu, G., Xu, J., Lai, Q., Yan, B., Guo, Y., et al. (2019). HOTTIP lncRNA promotes hematopoietic stem cell self-renewal leading to AML-like disease in mice. Cancer Cell 36, 645–659.e8. doi:10.1016/j.ccell.2019.10.011
Luo, M., Jeong, M., Sun, D., Park, H. J., Rodriguez, B. A. T., Xia, Z., et al. (2015). Long non-coding RNAs control hematopoietic stem cell function. Cell Stem Cell 16, 426–438. doi:10.1016/j.stem.2015.02.002
Luo, W., Yu, H., Zou, X., Ni, X., and Wei, J. (2018b). Long non-coding RNA taurine-upregulated gene 1 correlates with unfavorable prognosis in patients with refractory or relapsed acute myeloid leukemia treated by purine analogue based chemotherapy regimens. Cancer Biomarkers 23, 485–494. doi:10.3233/CBM-181405
Ma, L., Kuai, W. X., Sun, X. Z., Lu, X. C., and Yuan, Y. F. (2018). Long noncoding RNA LINC00265 predicts the prognosis of acute myeloid leukemia patients and functions as a promoter by activating PI3K-AKT pathway. Eur. Rev. Med. Pharmacol. Sci. 22 (22), 7867–7876. doi:10.26355/eurrev_201811_16412
Ma, L., Wang, Y. Y., and Jiang, P. (2020). LncRNA LINC00909 promotes cell proliferation and metastasis in pediatric acute myeloid leukemia via miR-625-mediated modulation of Wnt/β-catenin signaling. Biochem. Biophysical Res. Commun. 527 (3), 654–661. doi:10.1016/j.bbrc.2020.05.001
Mansoori, B., Sandoghchian Shotorbani, S., and Baradaran, B. (2014). RNA interference and its role in cancer therapy. Adv. Pharm. Bull. 4, 313–321. doi:10.5681/apb.2014.046
Masoud Eslami, M., Soufizomorrod, M., and Ahmadvand, M. (2021). High expression of long noncoding RNA NORAD is associated with poor clinical outcomes in non-M3 acute myeloid leukemia patients. Hematology/Oncology Stem Cell Ther. doi:10.1016/j.hemonc.2021.08.001
Mas-Ponte, D., Carlevaro-Fita, J., Palumbo, E., Hermoso Pulido, T., Guigo, R., and Johnson, R. (2017). LncATLAS database for subcellular localization of long noncoding RNAs. RNA 23, 1080–1087. doi:10.1261/rna.060814.117
Mattick, J. S., Amaral, P. P., Carninci, P., Carpenter, S., Chang, H. Y., Chen, L.-L., et al. (2023). Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 24, 430–447. doi:10.1038/s41580-022-00566-8
Mercer, T. R., and Mattick, J. S. (2013). Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 20, 300–307. doi:10.1038/nsmb.2480
Meyer, S. C., and Levine, R. L. (2014). Translational implications of somatic genomics in acute myeloid leukaemia. Lancet Oncol. 15, e382–e394. doi:10.1016/S1470-2045(14)70008-7
Miliara, S., Neddermeyer, A., Bonetti, A., Shin, J. W., Mujahed, H., Carninci, P., et al. (2022). The biological and prognostic role of long non-coding RNA NEAT1 in acute myeloid leukemia. Res. square (pre-print). doi:10.21203/rs.3.rs-2271358/v1
Mishra, K., and Kanduri, C. (2019). Understanding long noncoding RNA and chromatin interactions: what we know so far. Non-coding RNA 5 (4), 54. doi:10.3390/ncrna5040054
Mitra, S. A., Mitra, A. P., and Triche, T. J. (2012). A central role for long non-coding RNA in cancer. Front. Genet. 3, 17. doi:10.3389/fgene.2012.00017
Mofidi, M., Rahgozar, S., and Pouyanrad, S. (2021). Increased level of long non coding RNA H19 is correlated with the downregulation of miR-326 and BCL-2 genes in pediatric acute lymphoblastic leukemia, a possible hallmark for leukemogenesis. Mol. Biol. Rep. 48, 1531–1538. doi:10.1007/s11033-021-06161-y
Mondal, T., Subhash, S., Vaid, R., Enroth, S., Uday, S., Reinius, B., et al. (2015). MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA–DNA triplex structures. Nat. Commun. 6, 7743. doi:10.1038/ncomms8743
Nayerossadat, N., Maedeh, T., and Ali, P. (2012). Viral and nonviral delivery systems for gene delivery. Adv. Biomed. Res. 1, 27. doi:10.4103/2277-9175.98152
Neyazi, S., Ng, M., Heckl, D., and Klusmann, J.-H. (2022). Long noncoding RNAs as regulators of pediatric acute myeloid leukemia. Mol. Cell Pediatr. 9, 10. doi:10.1186/s40348-022-00142-2
Nguyen, Q. D., Schachar, R. A., Nduaka, C. I., Sperling, M., Basile, A. S., Klamerus, K. J., et al. (2012). Phase 1 dose-escalation study of a siRNA targeting the RTP801 gene in age-related macular degeneration patients. Eye 26, 1099–1105. doi:10.1038/eye.2012.106
Niederer, R. O., Hass, E. P., and Zappulla, D. C. (2017) Long noncoding RNAs in the yeast S. cerevisiae. Berlin, Germany: Springer, 119–132. doi:10.1007/978-981-10-5203-3_4
Nishant, K. T., Ravishankar, H., and Rao, M. R. S. (2004). Characterization of a mouse recombination hot spot locus encoding a Novel non-Protein-Coding RNA. Mol. Cell. Biol. 24, 5620–5634. doi:10.1128/MCB.24.12.5620-5634.2004
Ochi, Y., Yoshida, K., Huang, Y. J., Kuo, M. C., Nannya, Y., Sasaki, K., et al. (2021). Clonal evolution and clinical implications of genetic abnormalities in blastic transformation of chronic myeloid leukaemia. Nat. Commun. 12 (1), 2833. doi:10.1038/s41467-021-23097-w
Padmakumar, D., Chandraprabha, V. R., Gopinath, P., Vimala Devi, A. R. T., Anitha, G. R. J., Sreelatha, M. M., et al. (2021). A concise review on the molecular genetics of acute myeloid leukemia. Leukemia Res. 111, 106727. doi:10.1016/j.leukres.2021.106727
Pal, D., Dutta, S., Iyer, D. P., Shriram, D., Bhaduri, U., and Rao, M. R. S. (2022). Identification of PAX6 and NFAT4 as the transcriptional regulators of the long noncoding RNA mrhl in neuronal progenitors. Mol. Cell. Biol. 42, e0003622. doi:10.1128/mcb.00036-22
Pal, D., Neha, C. V., Bhaduri, U., Zenia, Z., Dutta, S., Chidambaram, S., et al. (2021). LncRNA Mrhl orchestrates differentiation programs in mouse embryonic stem cells through chromatin mediated regulation. Stem Cell Res. 53, 102250. doi:10.1016/j.scr.2021.102250
Papaioannou, D., Nicolet, D., Ozer, H. G., Mrózek, K., Volinia, S., Fadda, P., et al. (2019a). Prognostic and biologic relevance of clinically applicable long noncoding RNA profiling in older patients with cytogenetically normal acute myeloid leukemia. Mol. Cancer Ther. 18, 1451–1459. doi:10.1158/1535-7163.MCT-18-1125
Papaioannou, D., Petri, A., Dovey, O. M., Terreri, S., Wang, E., Collins, F. A., et al. (2019b). The long non-coding RNA HOXB-AS3 regulates ribosomal RNA transcription in NPM1-mutated acute myeloid leukemia. Nat. Commun. 10, 5351. doi:10.1038/s41467-019-13259-2
Parker, J. S., Mullins, M., Cheang, M. C. U., Leung, S., Voduc, D., Vickery, T., et al. (2009). Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 27, 1160–1167. doi:10.1200/JCO.2008.18.1370
Pashaiefar, H., Izadifard, M., Yaghmaie, M., Montazeri, M., Gheisari, E., Ahmadvand, M., et al. (2018). Low expression of long noncoding RNA IRAIN is associated with poor prognosis in non-M3 acute myeloid leukemia patients. Genet. Test. Mol. Biomarkers 22, 288–294. doi:10.1089/gtmb.2017.0281
Pasmant, E., Sabbagh, A., Vidaud, M., and Bièche, I. (2011). ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J. 25, 444–448. doi:10.1096/fj.10-172452
Pavco, P. A., Bouhana, K. S., Gallegos, A. M., Agrawal, A., Blanchard, K. S., Grimm, S. L., et al. (2000). Antitumor and antimetastatic activity of Ribozymes targeting the messenger RNA of vascular endothelial growth factor receptors. Clin. cancer Res. official J. Am. Assoc. Cancer Res. 6, 2094–2103.
Pavlovic, D., Tosic, N., Zukic, B., Pravdic, Z., Vukovic, N. S., Pavlovic, S., et al. (2021). Expression profiles of long non-coding RNA GAS5 and MicroRNA-222 in younger AML patients. Diagnostics 12, 86. doi:10.3390/diagnostics12010086
Pedram Fatemi, R., Salah-Uddin, S., Modarresi, F., Khoury, N., Wahlestedt, C., and Faghihi, M. A. (2015). Screening for small-molecule modulators of long noncoding RNA-protein interactions using AlphaScreen. SLAS Discov. 20, 1132–1141. doi:10.1177/1087057115594187
Peng, L., Zhang, Y., and Xin, H. (2020). lncRNA SNHG3 facilitates acute myeloid leukemia cell growth via the regulation of miR-758-3p/SRGN axis. J. Cell. Biochem. 121 (2), 1023–1031. doi:10.1002/jcb.29336
Penny, G. D., Kay, G. F., Sheardown, S. A., Rastan, S., and Brockdorff, N. (1996). Requirement for Xist in X chromosome inactivation. Nature 379, 131–137. doi:10.1038/379131a0
Perrotti, D., Jamieson, C., Goldman, J., and Skorski, T. (2010). Chronic myeloid leukemia: mechanisms of blastic transformation. J. Clin. Investigation 120, 2254–2264. doi:10.1172/JCI41246
Pisignano, G., Pavlaki, I., and Murrell, A. (2019). Being in a loop: how long non-coding RNAs organise genome architecture. Essays Biochem. 63, 177–186. doi:10.1042/EBC20180057
Porcù, E., Benetton, M., Bisio, V., Da Ros, A., Tregnago, C., Borella, G., et al. (2021). The long non-coding RNA CDK6-AS1 overexpression impacts on acute myeloid leukemia differentiation and mitochondrial dynamics. iScience 24, 103350. doi:10.1016/j.isci.2021.103350
Pulte, D., Jansen, L., and Brenner, H. (2020). Changes in long term survival after diagnosis with common hematologic malignancies in the early 21st century. Blood Cancer J. 10 (5), 56. doi:10.1038/s41408-020-0323-4
Qi, P., Zhou, X., and Du, X. (2016). Circulating long non-coding RNAs in cancer: current status and future perspectives. Mol. Cancer 15, 39. doi:10.1186/s12943-016-0524-4
Qin, J., Bao, H., and Li, H. (2018). Correlation of long non-coding RNA taurine-upregulated gene 1 with disease conditions and prognosis, as well as its effect on cell activities in acute myeloid leukemia. Cancer Biomarkers 23, 569–577. doi:10.3233/CBM-181834
Qin, X., Jiang, Y., Zhang, X., and Li, D. (2022). lncRNA GAS5 induces cell apoptosis in acute myeloid leukemia by targeting Nrf2. Dis. Markers 2022, 1–8. doi:10.1155/2022/5178122
Qiu, Y., Xu, M., and Huang, S. (2021). Long noncoding RNAs: emerging regulators of normal and malignant hematopoiesis. Blood 138, 2327–2336. doi:10.1182/blood.2021011992
Qu, Y., Wang, Y., Wang, P., Lin, N., Yan, X., and Li, Y. (2020). Overexpression of long noncoding RNA HOXA-AS2 predicts an adverse prognosis and promotes tumorigenesis via SOX4/PI3K/AKT pathway in acute myeloid leukemia. Cell Biol. Int. 44, 1745–1759. doi:10.1002/cbin.11370
Raguraman, P., Balachandran, A. A., Chen, S., Diermeier, S. D., and Veedu, R. N. (2021). Antisense oligonucleotide-mediated splice switching: potential therapeutic approach for cancer mitigation. Cancers (Basel) 13, 5555. doi:10.3390/cancers13215555
Rittenhouse, H., Blase, A., Shamel, B., Schalken, J., and Groskopf, J. (2013). The long and winding road to FDA approval of a novel prostate cancer test: our story. Clin. Chem. 59, 32–34. doi:10.1373/clinchem.2012.198739
Rosa, A., and Ballarino, M. (2016). Long noncoding RNA regulation of pluripotency. Stem Cells Int. 2016, 1–9. doi:10.1155/2016/1797692
Saad, R. A. A. A. M., Osman, A. A., Hassan, M. F. A. F., Ibrahim, S. A. M., and El-Sakhawy, Y. N. (2021). HOTAIR expression and prognostic impact in acute myeloid leukemia patients. Egypt. J. Med. Hum. Genet. 22, 1–11. doi:10.1186/s43042-021-00147-y
Sahu, A., Singhal, U., and Chinnaiyan, A. M. (2015). Long noncoding RNAs in cancer: from function to translation. Trends Cancer 1, 93–109. doi:10.1016/j.trecan.2015.08.010
Sakuma, T., and Yamamoto, T. (2018). Acceleration of cancer science with genome editing and related technologies. Cancer Sci. 109, 3679–3685. doi:10.1111/cas.13832
Salah, M., Zawam, H., Fouad, N. B., Soliman, N., and Maksoud, F. A. W. A. (2021). Study of HOTAIR LncRNA in AML patients in context to FLT3-ITD and NPM1 mutations status. Egypt. J. Med. Hum. Genet. 22, 61–69. doi:10.1186/s43042-021-00180-x
Sandberg, A. A., Hossfeld, D. K., Ezdinli, E. Z., and Crosswhite, L. H. (1971). Chromosomes and causation of human cancer and leukemia.VI. Blastic phase, cellular origin, and the Ph1 in CML. Cancer 27, 176–185. doi:10.1002/1097-0142(197101)27:1<176::AID-CNCR2820270125>3.0.CO;2-B
Scharner, J., and Aznarez, I. (2021). Clinical applications of single-stranded oligonucleotides: current landscape of approved and in-development therapeutics. Mol. Ther. 29, 540–554. doi:10.1016/j.ymthe.2020.12.022
Schmitz, K.-M., Mayer, C., Postepska, A., and Grummt, I. (2010). Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 24, 2264–2269. doi:10.1101/gad.590910
Schmitz, S. U., Grote, P., and Herrmann, B. G. (2016). Mechanisms of long noncoding RNA function in development and disease. Cell. Mol. Life Sci. 73, 2491–2509. doi:10.1007/s00018-016-2174-5
Schwarzer, A., Emmrich, S., Schmidt, F., Beck, D., Ng, M., Reimer, C., et al. (2017). The non-coding RNA landscape of human hematopoiesis and leukemia. Nat. Commun. 8, 218. doi:10.1038/s41467-017-00212-4
Sellers, Z. P., Bolkun, L., Kloczko, J., Wojtaszewska, M. L., Lewandowski, K., Moniuszko, M., et al. (2019). Increased methylation upstream of the MEG3 promotor is observed in acute myeloid leukemia patients with better overall survival. Clin. Epigenetics 11, 50. doi:10.1186/s13148-019-0643-z
Senmatsu, S., and Hirota, K. (2020). Roles of lncRNA transcription as a novel regulator of chromosomal function. Genes Genet. Syst. 95, 213–223. doi:10.1266/ggs.20-00024
Shadid, M., Badawi, M., and Abulrob, A. (2021). Antisense oligonucleotides: absorption, distribution, metabolism, and excretion. Expert Opin. Drug Metabolism Toxicol. 17, 1281–1292. doi:10.1080/17425255.2021.1992382
Shehata, A. M. F., Gohar, S. F., Muharram, N. M., and Eldin, S. M. K. (2022). LncRNA CCAT2 expression at diagnosis predicts imatinib response in chronic phase chronic myeloid leukemia patients. Leukemia Res. 116, 106838. doi:10.1016/j.leukres.2022.106838
Sheng, H., Zhang, J., Ma, Y., Zhang, Y., Dai, Y., and Jiang, R. (2021). lncRNA FBXL19-AS1 is a diagnosis biomarker for paediatric patients with acute myeloid leukemia. J. Gene Med. 23, e3317. doi:10.1002/jgm.3317
Shi, M., Yang, R., Lin, J., Wei, Q., Chen, L., Gong, W., et al. (2021). LncRNA-SNHG16 promotes proliferation and migration of acute myeloid leukemia cells via PTEN/PI3K/AKT axis through suppressing CELF2 protein. J. Biosci. 46, 4. doi:10.1007/s12038-020-00127-1
Sivagurunathan, N., Ambatt, A. T. S., and Calivarathan, L. (2022). Role of long non-coding RNAs in the pathogenesis of alzheimer’s and Parkinson’s diseases. Curr. Aging Sci. 15, 84–96. doi:10.2174/1874609815666220126095847
Sleutels, F., Zwart, R., and Barlow, D. P. (2002). The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415, 810–813. doi:10.1038/415810a
Smaldone, M. C., and Davies, B. J. (2010). BC-819, a plasmid comprising the H19 gene regulatory sequences and diphtheria toxin A, for the potential targeted therapy of cancers. Curr. Opin. Mol. Ther. 12, 607–616.
Song, H., Chen, L., Liu, W., Xu, X., Zhou, Y., Zhu, J., et al. (2021a). Depleting long noncoding RNA HOTAIR attenuates chronic myelocytic leukemia progression by binding to DNA methyltransferase 1 and inhibiting PTEN gene promoter methylation. Cell Death Dis. 12, 440. doi:10.1038/s41419-021-03637-4
Song, Z., Lin, J., Li, Z., and Huang, C. (2021b). The nuclear functions of long noncoding RNAs come into focus. Non-coding RNA Res. 6, 70–79. doi:10.1016/j.ncrna.2021.03.002
Statello, L., Guo, C.-J., Chen, L.-L., and Huarte, M. (2021). Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 22, 96–118. doi:10.1038/s41580-020-00315-9
Sun, B., Liu, C., Li, H., Zhang, L., Luo, G., Liang, S., et al. (2020). Research progress on the interactions between long non-coding RNAs and microRNAs in human cancer. Oncol. Lett. 19, 595–605. doi:10.3892/ol.2019.11182
Sun, C., Luan, S., Zhang, G., Wang, N., Shao, H., and Luan, C. (2017). CEBPA-mediated upregulation of the lncRNA PLIN2 promotes the development of chronic myelogenous leukemia via the GSK3 and Wnt/β-catenin signaling pathways. Am. J. Cancer Res. 7, 1054–1067.
Sun, J., Li, W., Sun, Y., Yu, D., Wen, X., Wang, H., et al. (2014). A novel antisense long noncoding RNA within the IGF1Rgene locus is imprinted in hematopoietic malignancies. Nucleic Acids Res. 42, 9588–9601. doi:10.1093/nar/gku549
Sun, L. Y., Li, X. J., Sun, Y. M., Huang, W., Fang, K., Han, C., et al. (2018a). LncRNA ANRIL regulates AML development through modulating the glucose metabolism pathway of AdipoR1/AMPK/SIRT1. Mol. Cancer 17, 127. doi:10.1186/s12943-018-0879-9
Sun, M., and Kraus, W. L. (2015). From discovery to function: the expanding roles of long non coding RNAs in physiology and disease. Endocr. Rev. 36, 25–64. doi:10.1210/er.2014-1034
Sun, M. D., Zheng, Y. Q., Wang, L. P., Zhao, H. T., and Yang, S. (2018b). Long noncoding RNA UCA1 promotes cell proliferation, migration and invasion of human leukemia cells via sponging miR-126. Eur. Rev. Med. Pharmacol. Sci. 22, 2233–2245. doi:10.26355/eurrev_201804_14809
Sun, T., Dong, L., Guo, Y., Zhao, H., and Wang, M. (2022). Revealing key lncRNAs in cytogenetically normal acute myeloid leukemia by reconstruction of the lncRNA–miRNA–mRNA network. Sci. Rep. 12, 4973. doi:10.1038/s41598-022-08930-6
Swords, R., Alvarado, Y., Cortes, J., and Giles, F. J. (2007). Second-generation tyrosine kinase inhibitors as therapy for chronic myeloid leukemia. Curr. Hematol. Malig. Rep. 2, 83–88. doi:10.1007/s11899-007-0012-4
Tao, Y., Wei, L., and You, H. (2022). Ferroptosis-related gene signature predicts the clinical outcome in pediatric acute myeloid leukemia patients and refines the 2017 ELN classification system. Front. Mol. Biosci. 9, 954524. doi:10.3389/fmolb.2022.954524
Tao, Y., Zhang, J., Chen, L., Liu, X., Yao, M., and Zhang, H. (2021). LncRNA CD27-AS1 promotes acute myeloid leukemia progression through the miR-224-5p/PBX3 signaling circuit. Cell Death Dis. 12, 510. doi:10.1038/s41419-021-03767-9
Tian, M., Gong, W., and Guo, J. (2019). Long non-coding RNA SNHG1 indicates poor prognosis and facilitates disease progression in acute myeloid leukemia. Biol. Open 8, bio046417. doi:10.1242/bio.046417
Titze-de-Almeida, R., David, C., and Titze-de-Almeida, S. S. (2017). The race of 10 synthetic RNAi-based drugs to the pharmaceutical market. Pharm. Res. 34, 1339–1363. doi:10.1007/s11095-017-2134-2
Ulitsky, I. (2016). Evolution to the rescue: using comparative genomics to understand long non-coding RNAs. Nat. Rev. Genet. 17, 601–614. doi:10.1038/nrg.2016.85
Vakiti, A., Mewawalla, P., and Wood, S. K. (2021) Acute myeloid leukemia (nursing). Petersburg, Florida, United States: StatPearls Publishing.
Vanhooren, J., Van Camp, L., Depreter, B., de Jong, M., Uyttebroeck, A., Van Damme, A., et al. (2022). Deciphering the non-coding RNA landscape of pediatric acute myeloid leukemia. Cancers (Basel) 14, 2098. doi:10.3390/cancers14092098
Vitiello, M., Tuccoli, A., and Poliseno, L. (2015). Long non-coding RNAs in cancer: implications for personalized therapy. Cell. Oncol. 38, 17–28. doi:10.1007/s13402-014-0180-x
Wang, C., Chen, F., Fan, Z., Yao, C., and Xiao, L. (2020a). lncRNA CCAT1/miR-490-3p/MAPK1/c-Myc positive feedback loop drives progression of acute myeloid leukaemia. J. Biochem. 167, 379–388. doi:10.1093/jb/mvz107
Wang, C. H., Li, Q. Y., Nie, L., Ma, J., Yao, C. J., and Chen, F. P. (2020b). LncRNA ANRIL promotes cell proliferation, migration and invasion during acute myeloid leukemia pathogenesis via negatively regulating miR-34a. Int. J. Biochem. Cell Biol. 119, 105666. doi:10.1016/j.biocel.2019.105666
Wang, H., Guo, R., Hoffman, A. R., Hu, J., and Li, W. (2015). Effect of long noncoding RNA RUNXOR on the epigenetic regulation of RUNX1 in acute myelocytic leukemia. J. Clin. Oncol. 33, 7018. doi:10.1200/jco.2015.33.15_suppl.7018
Wang, H., Li, Q., Tang, S., Li, M., Feng, A., Qin, L., et al. (2017). The role of long noncoding RNA HOTAIR in the acquired multidrug resistance to imatinib in chronic myeloid leukemia cells. Hematology 22, 208–216. doi:10.1080/10245332.2016.1258152
Wang, H., Li, W., Guo, R., Sun, J., Cui, J., Wang, G., et al. (2014). An intragenic long noncoding RNA interacts epigenetically with the RUNX1 promoter and enhancer chromatin DNA in hematopoietic malignancies. Int. J. Cancer 135, 2783–2794. doi:10.1002/ijc.28922
Wang, H., Meng, Q., Qian, J., Li, M., Gu, C., and Yang, Y. (2022). Review: RNA-based diagnostic markers discovery and therapeutic targets development in cancer. Pharmacol. Ther. 234, 108123. doi:10.1016/j.pharmthera.2022.108123
Wang, J., Liu, Z. H., and Yu, L. J. (2019a). Long non-coding RNA LINC00641 promotes cell growth and migration through modulating miR-378a/ZBTB20 axis in acute myeloid leukemia. Eur. Rev. Med. Pharmacol. Sci. 23, 7498–7509. doi:10.26355/eurrev_201909_18864
Wang, S. L., Huang, Y., Su, R., and Yu, Y. Y. (2019b). Silencing long non-coding RNA HOTAIR exerts anti-oncogenic effect on human acute myeloid leukemia via demethylation of HOXA5 by inhibiting Dnmt3b. Cancer Cell Int. 19, 114. doi:10.1186/s12935-019-0808-z
Wang, X., Zhang, L., Zhao, F., Xu, R., Jiang, J., Zhang, C., et al. (2018a). Long non-coding RNA taurine-upregulated gene 1 correlates with poor prognosis, induces cell proliferation, and represses cell apoptosis via targeting aurora kinase A in adult acute myeloid leukemia. Ann. Hematol. 97, 1375–1389. doi:10.1007/s00277-018-3315-8
Wang, X. Q. D., and Dostie, J. (2017). Reciprocal regulation of chromatin state and architecture by HOTAIRM1 contributes to temporal collinear HOXA gene activation. Nucleic Acids Res. 45, 1091–1104. doi:10.1093/nar/gkw966
Wang, Y., Li, Y., Song, H. Q., and Sun, G. W. (2018b). Long non-coding RNA LINC00899 as a novel serum biomarker for diagnosis and prognosis prediction of acute myeloid leukemia. Eur. Rev. Med. Pharmacol. Sci. 22, 7364–7370. doi:10.26355/eurrev_201811_16274
Wang, Y., Nie, H., He, X., Liao, Z., Zhou, Y., Zhou, J., et al. (2020c). The emerging role of super enhancer-derived noncoding RNAs in human cancer. Theranostics 10, 11049–11062. doi:10.7150/thno.49168
Wang, Y. H., Lin, C. C., Hsu, C. L., Hung, S. Y., Yao, C. Y., Lee, S. H., et al. (2021). Distinct clinical and biological characteristics of acute myeloid leukemia with higher expression of long noncoding RNA KIAA0125. Ann. Hematol. 100, 487–498. doi:10.1007/s00277-020-04358-y
Wapinski, O., and Chang, H. Y. (2011). Long non coding RNAs and human disease. Trends Cell Biol. 21, 354–361. doi:10.1016/j.tcb.2011.04.001
Weiskopf, K., Schnorr, P. J., Pang, W. W., Chao, M. P., Chhabra, A., Seita, J., et al. (2017). “Myeloid cell origins, differentiation, and clinical implications,” in Myeloid cells in health and disease (Washington, DC, USA: ASM Press), 857–875. doi:10.1128/9781555819194.ch50
Wen, F., Cao, Y.-X., Luo, Z.-Y., Liao, P., and Lu, Z.-W. (2018). LncRNA MALAT1 promotes cell proliferation and imatinib resistance by sponging miR-328 in chronic myelogenous leukemia. Biochem. Biophysical Res. Commun. 507, 1–8. doi:10.1016/j.bbrc.2018.09.034
Wu, S., Zheng, C., Chen, S., Cai, X., Shi, Y., Lin, B., et al. (2015). Overexpression of long non-coding RNA HOTAIR predicts a poor prognosis in patients with acute myeloid leukemia. Oncol. Lett. 10, 2410–2414. doi:10.3892/ol.2015.3552
Xiao, Y., Jiao, C., Lin, Y., Chen, M., Zhang, J., Wang, J., et al. (2017). lncRNA UCA1 contributes to imatinib resistance by acting as a ceRNA against miR-16 in chronic myeloid leukemia cells. DNA Cell Biol. 36, 18–25. doi:10.1089/dna.2016.3533
Xing, C., Hu, X., Xie, F., Yu, Z., Li, H., Zhou, B., et al. (2015). Long non-coding RNA HOTAIR modulates c-KIT expression through sponging miR-193a in acute myeloid leukemia. FEBS Lett. 589, 1981–1987. doi:10.1016/j.febslet.2015.04.061
Xu, J., Wu, M., Sun, Y., Zhao, H., Wang, Y., and Gao, J. (2020). Identifying dysregulated lncRNA-associated ceRNA network biomarkers in CML based on dynamical network biomarkers. BioMed Res. Int. 2020, 1–8. doi:10.1155/2020/5189549
Xue, F., and Che, H. (2020). The long non-coding RNA LOC285758 promotes invasion of acute myeloid leukemia cells by down-regulating miR-204-5p. FEBS Open Bio 10, 734–743. doi:10.1002/2211-5463.12814
Yang, F., Xue, X., Zheng, L., Bi, J., Zhou, Y., Zhi, K., et al. (2014). Long non-coding RNA GHET1 promotes gastric carcinoma cell proliferation by increasing c-Myc mRNA stability. FEBS J. 281, 802–813. doi:10.1111/febs.12625
Yang, H., Shang, D., Xu, Y., Zhang, C., Feng, L., Sun, Z., et al. (2017). The LncRNA connectivity map: using LncRNA signatures to connect small molecules, LncRNAs, and diseases. Sci. Rep. 7, 6655. doi:10.1038/s41598-017-06897-3
Yang, J., Yin, Z., Li, Y., Liu, Y., Huang, G., Gu, C., et al. (2020a). The identification of long non-coding RNA H19 target and its function in chronic myeloid leukemia. Mol. Ther. - Nucleic Acids 19, 1368–1378. doi:10.1016/j.omtn.2020.01.021
Yang, J. R., Shi, M. X., and Zeng, Y. (2019). LncRNA HAND2-AS1 inhibits proliferation and promotes apoptosis of chronic myeloid leukemia cells by sponging with micRNA-1275. Eur. Rev. Med. Pharmacol. Sci. 23, 2103–2111. doi:10.26355/eurrev_201903_17254
Yang, L., Ma, D., Tang, S., Jiang, T., Yu, J., Wang, L., et al. (2022). Comprehensive genomic analysis for identifying FZD6 as a novel diagnostic biomarker for acute myeloid leukemia. Comput. Math. Methods Med. 2022, 1–14. doi:10.1155/2022/9130958
Yang, L., Zhou, J.-D., Zhang, T.-J., Ma, J.-C., Xiao, G.-F., Chen, Q., et al. (2018). Overexpression of lncRNA PANDAR predicts adverse prognosis in acute myeloid leukemia. Cancer Manag. Res. 10, 4999–5007. doi:10.2147/CMAR.S180150
Yang, R., Ma, D., Wu, Y., Zhang, Y., and Zhang, L. (2020b). <p>LncRNA <em>SNHG16</em> Regulates the Progress of Acute Myeloid Leukemia through miR183-5p–FOXO1 Axis</p>. OncoTargets Ther. 13, 12943–12954. doi:10.2147/OTT.S258684
Yao, F.-Y., Zhao, C., Zhong, F.-M., Qin, T.-Y., Wen, F., Li, M.-Y., et al. (2021). m(6)A modification of lncRNA NEAT1 regulates chronic myelocytic leukemia progression via miR-766-5p/cdkn1a Axis. Front. Oncol. 11, 679634. doi:10.3389/fonc.2021.679634
Yao, H., Duan, M., Lin, L., Wu, C., Fu, X., Wang, H., et al. (2017). TET2 and MEG3 promoter methylation is associated with acute myeloid leukemia in a Hainan population. Oncotarget 8, 18337–18347. doi:10.18632/oncotarget.15440
Yao, Q., Zhang, L., Liu, Z., Yu, L., Wang, Y., Liu, J., et al. (2022). LncRNA MAFG-AS1-induced acute myeloid leukemia development via modulating miR-147b/HOXA9. Environ. Sci. Pollut. Res. 30, 19250–19258. doi:10.1007/s11356-022-23537-0
Yao, R.-W., Wang, Y., and Chen, L.-L. (2019). Cellular functions of long noncoding RNAs. Nat. Cell Biol. 21, 542–551. doi:10.1038/s41556-019-0311-8
Yap, K. L., Li, S., Muñoz-Cabello, A. M., Raguz, S., Zeng, L., Mujtaba, S., et al. (2010). Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by Polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell 38, 662–674. doi:10.1016/j.molcel.2010.03.021
Yi, M., Li, A., Zhou, L., Chu, Q., Song, Y., and Wu, K. (2020). The global burden and attributable risk factor analysis of acute myeloid leukemia in 195 countries and territories from 1990 to 2017: estimates based on the global burden of disease study 2017. J. Hematol. Oncol. 13, 72. doi:10.1186/s13045-020-00908-z
Yin, Z., Shen, H., Gu, C. ming, Zhang, M. qi, Liu, Z., Huang, J., et al. (2021b). MiRNA-142-3P and FUS can be sponged by long noncoding RNA DUBR to promote cell proliferation in acute myeloid leukemia. Front. Mol. Biosci. 8, 754936. doi:10.3389/fmolb.2021.754936
Yoon, J.-H., Abdelmohsen, K., and Gorospe, M. (2013). Posttranscriptional gene regulation by long noncoding RNA. J. Mol. Biol. 425, 3723–3730. doi:10.1016/j.jmb.2012.11.024
Yoon, J.-H., Abdelmohsen, K., and Gorospe, M. (2014). Functional interactions among microRNAs and long noncoding RNAs. Seminars Cell and Dev. Biol. 34, 9–14. doi:10.1016/j.semcdb.2014.05.015
Yuan, Y., Wang, Q., Ma, S. L., Xu, L. Q., Liu, M. Y., Han, B., et al. (2019). lncRNA PCAT-1 interacting with FZD6 contributes to the malignancy of acute myeloid leukemia cells through activating Wnt/β-catenin signaling pathway. Am. J. Transl. Res. 11, 7104–7114.
Yuan, Z., Li, L., Zheng, M., Xu, J., and Wang, W. (2021). lncRNA TP73-AS1 regulates miR-21/PTEN Axis to affect cell proliferation in acute myeloid leukemia. Cancer Biotherapy Radiopharm. 36, 268–272. doi:10.1089/cbr.2019.3142
Zalcberg, J. R., Friedlander, M. L., and Minden, M. D. (1986). Molecular evidence for the clonal origin of blast crisis in chronic myeloid leukaemia. Br. J. Cancer 53, 459–464. doi:10.1038/bjc.1986.73
Zampetaki, A., Albrecht, A., and Steinhofel, K. (2018). Long non-coding RNA structure and function: is there a link? Front. Physiol. 9, 1201. doi:10.3389/fphys.2018.01201
Zeng, C., Liu, S., Lu, S., Yu, X., Lai, J., Wu, Y., et al. (2018). The c-Myc-regulated lncRNA NEAT1 and paraspeckles modulate imatinib-induced apoptosis in CML cells. Mol. Cancer 17, 130. doi:10.1186/s12943-018-0884-z
Zhai, H., Zhao, J., Pu, J., Zhao, P., and Wei, J. (2021). LncRNA-DUXAP8 regulation of the wnt/β-catenin signaling pathway to inhibit glycolysis and induced apoptosis in acute myeloid leukemia. Turkish J. Hematol. 38, 264–272. doi:10.4274/tjh.galenos.2021.2020.0769
Zhang, C., and Zhang, B. (2023). RNA therapeutics: updates and future potential. Sci. China Life Sci. 66, 12–30. doi:10.1007/s11427-022-2171-2
Zhang, F., Li, Q., Zhu, K., Zhu, J., Li, J., Yuan, Y., et al. (2020a). LncRNA LINC00265/miR-485-5p/IRF2-mediated autophagy suppresses apoptosis in acute myeloid leukemia cells. Am. J. Transl. Res. 12, 2451–2462.
Zhang, T., Zhou, J., Zhang, W., Lin, J., Ma, J., Wen, X., et al. (2018a). H19 overexpression promotes leukemogenesis and predicts unfavorable prognosis in acute myeloid leukemia. Clin. Epigenetics 10, 47. doi:10.1186/s13148-018-0486-z
Zhang, W., Liu, Y., Zhang, J., and Zheng, N. (2020b). Long non-coding RNA taurine upregulated gene 1 targets miR-185 to regulate cell proliferation and glycolysis in acute myeloid leukemia cells in vitro. OncoTargets Ther. 13, 7887–7896. doi:10.2147/OTT.S238189
Zhang, X., and Tao, W. (2019). Long noncoding RNA LINC00152 facilitates the leukemogenesis of acute myeloid leukemia by promoting CDK9 through miR-193a. DNA Cell Biol. 38 (3), 236–242. doi:10.1089/dna.2018.4482
Zhang, X., Weissman, S. M., and Newburger, P. E. (2014). Long intergenic non-coding RNA HOTAIRM1 regulates cell cycle progression during myeloid maturation in NB4 human promyelocytic leukemia cells. RNA Biol. 11 (6), 777–787. doi:10.4161/rna.28828
Zhang, X., Yang, L., and Xu, G. (2020c). Silencing of long noncoding RNA TUG1 inhibits viability and promotes apoptosis of acute myeloid leukemia cells by targeting microRNA-221-3p/KIT axis. Clin. Hemorheol. Microcirc. 76, 425–437. doi:10.3233/CH-200906
Zhang, Y., Liu, Y., and Xu, X. (2018b). Knockdown of LncRNA-UCA1 suppresses chemoresistance of pediatric AML by inhibiting glycolysis through the microRNA-125a/hexokinase 2 pathway. J. Cell. Biochem. 119, 6296–6308. doi:10.1002/jcb.26899
Zhao, C., Wang, S., Zhao, Y., Du, F., Wang, W., Lv, P., et al. (2019). Long noncoding RNA NEAT1 modulates cell proliferation and apoptosis by regulating miR-23a-3p/SMC1A in acute myeloid leukemia. J. Cell. Physiology 234, 6161–6172. doi:10.1002/jcp.27393
Zhao, T. F., Jia, H. Z., Zhang, Z. Z., Zhao, X. S., Zou, Y. F., Zhang, W., et al. (2017). LncRNA H19 regulates ID2 expression through competitive binding to hsa-miR-19a/b in acute myelocytic leukemia. Mol. Med. Rep. 16, 3687–3693. doi:10.3892/mmr.2017.7029
Zhao, T.-T., and Liu, X. (2019). LncRNA-H19 inhibits apoptosis of acute myeloid leukemia cells via targeting miR-29a-3p. Eur. Rev. Med. Pharmacol. Sci. 23, 224–231. doi:10.26355/eurrev_201908_18651
Zhao, Y., Teng, H., Yao, F., Yap, S., Sun, Y., and Ma, L. (2020). Challenges and strategies in ascribing functions to long noncoding RNAs. Cancers (Basel) 12, 1458. doi:10.3390/cancers12061458
Zhou, H., Shi, P., Jia, X., and Xue, Q. (2021). Long non coding RNA LINC01018 inhibits the progression of acute myeloid leukemia by targeting miR 499a 5p to regulate PDCD4. Oncol. Lett. 22, 541. doi:10.3892/ol.2021.12802
Zhou, W., Xu, S., Deng, T., Zhou, R., and Wang, C. (2022). LncRNA USP30-AS1 promotes the survival of acute myeloid leukemia cells by cis-regulating USP30 and ANKRD13A. Hum. Cell 35, 360–378. doi:10.1007/s13577-021-00636-7
Zhou, X., Yuan, P., Liu, Q., and Liu, Z. (2017). LncRNA MEG3 regulates imatinib resistance in chronic myeloid leukemia via suppressing MicroRNA-21. Biomol. Ther. 25, 490–496. doi:10.4062/biomolther.2016.162
Zhu, G., Luo, H., Feng, Y., Guryanova, O. A., Xu, J., Chen, S., et al. (2021). HOXBLINC long non-coding RNA activation promotes leukemogenesis in NPM1-mutant acute myeloid leukemia. Nat. Commun. 12, 1956. doi:10.1038/s41467-021-22095-2
Zhu, L., Luo, J., and Ren, K. (2023). Nucleic acid-based artificial nanocarriers for gene therapy. J. Mater Chem. B 11, 261–279. doi:10.1039/D2TB01179D
Keywords: long non-coding RNA, myeloid leukemia, acute myeloid leukemia, chronic myeloid leukemia, prognostic markers, human meiotic recombination hot spot locus
Citation: Dutta S, Akhade VS, Choudhury SR and Rao MRS (2024) Long non-coding RNAs: emerging functional players in the pathobiology and progression of myeloid leukemia. Front. RNA Res. 2:1334464. doi: 10.3389/frnar.2024.1334464
Received: 07 November 2023; Accepted: 24 April 2024;
Published: 31 May 2024.
Edited by:
Michael D. Blower, Boston University, United StatesReviewed by:
Chinedu Anthony Anene, Leeds Beckett University, United KingdomCopyright © 2024 Dutta, Akhade, Choudhury and Rao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sangeeta Dutta, c2FuZ2VldGFkdXR0YTkyQGdtYWlsLmNvbQ==; Vijay Suresh Akhade, dmlqYXlha2hhZGVAZ21haWwuY29t
†Present address: Subhendu Roy Choudhury, Institute for Genome Sciences, University of Maryland, School of Medicine, Baltimore, MD, United States
‡Deceased
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.