
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. RNA Res. , 03 August 2023
Sec. Non-coding RNA
Volume 1 - 2023 | https://doi.org/10.3389/frnar.2023.1240954
During interphase, the nucleus contains a multitude of transcripts that influence the function of chromatin and global structure of the nucleus. Nuclear transcripts include nascent mRNAs in the process of transcription and mRNA processing, spliceosomal RNAs which catalyze mRNA processing, rRNAs that are being transcribed and processed to assemble functional ribosomes, and sno- and scaRNAs that participate in rRNA processing and modification. In addition, there are long noncoding RNAs (lncRNA) that associate with chromatin to control gene expression, or can even influence locus function in the case of centromeres and telomeres. Most of our knowledge of the functions of nuclear RNAs come from studies of interphase cells when the nuclear envelope separates nuclear and cytoplasmic contents. However, during mitosis the nuclear envelope breaks down, resulting in the mixing of nuclear and cytoplasmic components. Much less is known about the regulation and function of nuclear RNAs during mitosis. In this review, we discuss the cell cycle-dependent localization of different categories of RNAs, how the trans-acting factors SAF-A and Ki-67 regulate mitotic RNA localization, and describe how select categories of RNAs are inherited from the previous cell cycle in G1.
In interphase the genome is organized into a series of chromatin loops through the loop extrusion activity of the cohesin complex and the insulation factor CTCF (Dixon et al., 2012; Rao et al., 2014; Nora et al., 2017; Rao et al., 2017). Upon mitotic entry, the nucleus undergoes rapid and extensive remodeling to prepare the chromosomes for segregation. At the beginning of prophase, the kinases Plk1, AurKB, and Cdk1 phosphorylate cohesin (Waizenegger et al., 2000; Losada et al., 2002) and associated factors to remove all cohesin loops and the majority of cohesive cohesin from chromosome arms (Naumova et al., 2013; Gibcus et al., 2018). Concomitant with the loss of cohesin from chromosomes, the condensin I and II complexes are loaded onto chromatin to create a series of nested chromatin loops that dramatically compact chromatin (Gibcus et al., 2018).
At approximately the same time that chromatin begins to change shape and condense in prophase, many changes occur that affect the function of nuclear RNAs. Transcription by all three nuclear RNA polymerases is sharply attenuated (Gottesfeld and Forbes, 1997) and transcription associated factors largely dissociate from chromosomes (Festuccia et al., 2017; Gonzalez et al., 2021). In addition, the nucleolus dissolves and nucleolar components redistribute to the cytoplasm and chromosome surface (Van Hooser et al., 2005; Boisvert et al., 2007; Hernandez-Verdun, 2011). Finally, all known chromatin-associated RNAs are released from chromatin and relocalized to the cytoplasm (Sharp et al., 2020). Collectively, the macromolecular and structural changes associated with mitosis impact the localization and function of all known nuclear RNAs. This review will discuss the regulatory mechanisms and the importance of redistributing nuclear RNAs during mitosis.
Genome-wide RNA profiling in human cells demonstrated that a large proportion of the genome is transcribed, with around 75% being involved in the production of diverse RNA molecules (Djebali et al., 2012). This includes both protein-coding messenger RNAs (mRNAs) and non-coding RNAs (ncRNAs), such as transfer RNA (tRNA), ribosomal RNA (rRNA), small nucleolar RNA (snoRNA), small nuclear RNA (snRNA), microRNA (miRNA), enhancer-associated RNA (eRNA), and long non-coding RNA (lncRNA) (Jurica and Moore, 2003; Licatalosi and Darnell, 2010; Phizicky and Hopper, 2010; Lam et al., 2014; Dupuis-Sandoval et al., 2015; Quinn and Chang, 2016; Sloan et al., 2017; Statello et al., 2021). Chromatin-associated RNAs have a wide variety of functions during interphase that have been described in several recent reviews, highlighting a variety of techniques available for studying the functions and interactions of nuclear lncRNAs (Kato and Carninci, 2020; Mattick et al., 2023). Our review will focus specifically on the regulation and function of chromatin-RNA interactions during mitosis. RNA molecules can interact with chromatin using several different mechanisms. Newly transcribed RNAs (called nascent RNAs) remain at their site of synthesis, creating interactions between the RNA and nearby chromatin. Secondly, RNAs may be released from their site of transcription and interact with genomic loci at a distance through the binding of specific adapter proteins (Li and Fu, 2019; Xiao et al., 2019). A third mechanism of chromatin-RNA interaction is the formation of R-loops, in which RNA and dsDNA form a triple helix termed a R-loop. The formation, regulation and functions of R-loops are described in a recent review (Niehrs and Luke, 2020).
During mitosis most transcription ceases, resulting in the removal of nascent RNAs from chromatin. However, centromeres are transcriptionally active during mitosis, and RNAPII and centromere RNA (cenRNA) persist at centromere chromatin (Ferri et al., 2009; Du et al., 2010; Chan et al., 2012; Quénet and Dalal, 2014; Rošić et al., 2014; Liu et al., 2015; Perea-Resa et al., 2020; Chen et al., 2021). The centromere is typically composed entirely of simple repetitive sequences, with chromatin containing a specific histone, CENP-A, that replaces the canonical H3 histone in the nucleosome core particle (Earnshaw et al., 1986; Palmer et al., 1987; Palmer et al., 1991; Sullivan et al., 1994; Sullivan et al., 2001; McKinley and Cheeseman, 2016). Specialized centromeric chromatin has been implicated in the persistence of active transcription during mitosis.
Recent genome-wide studies conducted on mouse and human cells have revealed that many nucleolar ncRNAs, including snRNA, snoRNA and rRNA, are present in mitotic chromosome preparations (Mondal et al., 2010; Werner and Ruthenburg, 2015; Meng et al., 2016; Sridhar et al., 2017; Werner et al., 2017; Bell et al., 2018; Bonetti et al., 2020; Shen et al., 2022). These results are consistent with previous reports showing that some of these RNAs are not displaced from chromatin during mitosis but become part of the perichromosomal layer made up of RNAs and nucleolar proteins (Boisvert et al., 2007). At present, the mitotic functions of perichromosomal layer RNAs are not well understood.
Another example of chromatin-associated RNA during mitosis is Telomeric Repeat-containing RNA (TERRA). That TERRA is associated with transcriptionally silent metaphase chromosome ends suggests that after its synthesis, at least a portion of TERRA remains bound to telomeres. This association implies that TERRA may have a role in maintaining the integrity and function of telomeres, even during the condensed state of chromosomes in metaphase (Azzalin et al., 2007).
While cenRNA and TERRA RNA mark important structural features of metaphase chromosomes, recent papers have demonstrated that silencing of nascent RNA transcription and eviction of most lncRNA or mRNA from chromatin is required for accurate chromosome segregation (Perea-Resa et al., 2020; Sharp et al., 2020). Perichromosomal layer RNAs may act in concert with perichromosomal proteins to contribute to the overall function of the layer, which is to promote chromosome individualization and reformation of the nucleus at the end of mitosis (Cuylen et al., 2016; Cuylen-Haering et al., 2020; Fujimura et al., 2020; Ma et al., 2022). A recent study of tracking chromatin-associated RNA during chromatin maturation across the cell cycle showed that GA-rich-, alpha-, and TERRA are still associated with replicated DNA in G1 after mitosis (Gylling et al., 2020). Collectively these results show that multiple pathways exist to regulate the association or dissociation of transcripts with mitotic chromatin. We note that the net effect of mitotic RNA localization is to create condensed chromosomes with RNAs present at centromeres, telomeres, and in the perichromosomal layer. We speculate these changes are important for creating important spatial cues for regulatory proteins, or by changing biochemical conditions present on the surface of mitotic chromosomes due to the presence or absence of highly charged RNA molecules.
The centromere is the specific chromosomal region that controls the proper segregation of chromosomes during cell division. It acts as the assembly site for the kinetochore that binds to spindle microtubules, which play a crucial role in segregating replicated sister chromosomes during mitosis and meiosis. Most eukaryotic centromeres are structured based on short tandem repeats that form higher-order repeats (Pardue and Gall, 1970; Manuelidis and Wu, 1978; Altemose et al., 2022). However, the specific organization of the centromere varies significantly among species and exhibits substantial variation between individuals within the same species (Wade et al., 2009; Shang et al., 2010; Melters et al., 2013). This diversity in centromere organization might reflect the evolutionary plasticity of centromeric DNA sequences and their ability to adapt to species-specific or individual-specific requirements for proper chromosome segregation (Henikoff et al., 2001).
Importantly, although there is a lack of sequence conservation between organisms, recent studies have demonstrated that transcription plays a conserved role at centromeres (Corless et al., 2020). The centromere exhibits a high enrichment of specific histone modifications, including histone H3 mono-methylation on lysine 4 (H3K4me1), as well as H3K4me2, H3K36me2, and H3K36me3, H4K20me1, and H3K9ac (Sullivan and Karpen, 2004; Vakoc et al., 2006; Muramoto et al., 2010; Ribeiro et al., 2010; Bergmann et al., 2011; Bergmann et al., 2012; Hori et al., 2014). These histone modifications contribute to the establishment of an open chromatin structure, facilitating the activity of RNA polymerase II (RNAPII). Interestingly, histone marks associated with transcriptional repression, H3K9me2/3, are also detected on centromeres (Fischle et al., 2005; Bergmann et al., 2012). Recent studies demonstrated that centromeres are stably transcribed during cell cycle (Ferri et al., 2009; Du et al., 2010; Chan et al., 2012; Quénet and Dalal, 2014; Rošić et al., 2014; Liu et al., 2015; Perea-Resa et al., 2020; Chen et al., 2021), suggesting that centromere-specific chromatin may promote transcription.
Two elements of mitotic centromere transcription have been proposed to impact centromere structure and function. In the first scenario, the transcription process itself leads to chromatin remodeling, which may be an important step for the loading of CENP-A into centromeric nucleosomes to maintain centromeric chromatin (Bobkov et al., 2018; Talbert and Henikoff, 2018; Ghosh and Lehner, 2022). Recent studies showed that centromere transcription is regulated by CENP-B, CENP-I, CENP-C, and interactions with nucleoli (Bury et al., 2020; Chen et al., 2021; Hirai et al., 2022). In addition, one group suggested that centromere transcription maintains centromere cohesion (Chen et al., 2021). Thus, one function for mitotic centromere transcription may be to remodel the structure or composition of centromeric chromatin.
Second, the specific RNAs produced by centromere transcription may also play an active role in accurate kinetochore assembly (Hall et al., 2014). For example, CENP-C associates with cen-RNA, and its interaction is required for CENP-C localization (Wong et al., 2007; Du et al., 2010; Rošić et al., 2014; Grenfell et al., 2016b; McNulty et al., 2017). Targeted knockdown of cen-RNA has been shown to decrease the deposition of CENP-A, and cen-RNA derived from one centromere can partially compensate for the depletion of cen-RNA transcripts on another centromere (Liu et al., 2015; Ling and Yuen, 2019). Cen-RNA also interacts directly with the Chromosome Passenger Complex (CPC), including Aurora-B kinase, which controls accurate attachment between kinetochores and spindle microtubules (Carmena et al., 2012). Recent studies revealed that Aurora-B kinase localization and function at centromeres are controlled by its RNA binding at centromeres during mitosis (Ideue et al., 2014; Jambhekar et al., 2014; Grenfell et al., 2016a; Blower, 2016; Kabeche et al., 2018). Despite these findings, which domain of Aurora-B interacts with RNA and how RNA may affect its kinase activity are still unknown. In addition to cenRNA, one recent study showed that a chromosome 17-derived lncRNA, CCTT, specifically localizes on all centromeres and is required for proper CENP-C localization (Zhang et al., 2022). Therefore, centromere and kinetochore assembly are regulated by not only cen-RNAs but also lncRNAs from chromosome arm regions.
During mitosis, transcription of most genes is dramatically reduced and components of the RNAPII transcription machinery are displaced from chromatin, with the exception of the centromere region (Cho et al., 1998; Gibcus et al., 2018; Gottesfeld and Forbes, 1997; Liang et al., 2015; Naumova et al., 2013; Palozola et al., 2017; Parsons and Spencer, 1997; Prescott and Bender, 1962; Shermoen and O'Farrell, 1991). Mitotic RNAPII removal is regulated by phosphorylation of components of the transcriptional initiation complex (Segil et al., 1996; Bellier et al., 1997; Akoulitchev and Reinberg, 1998) and activation of positive transcription elongation factor β (P-TEFb (Liang et al., 2015), which promotes transcriptional elongation and RNAPII runoff. However, these global mechanisms did not explain the persistence of active RNAPII at centromeres. During mitosis, cohesin complexes are removed from chromosomes in a spatial and temporal pattern that correlates with transcriptional silencing. Cohesin is released from chromosome arms by the cohesin release factor WAPL but is protected at centromeres by Shugoshin proteins (Losada et al., 2002; Gandhi et al., 2006; Kueng et al., 2006; Nasmyth and Haering, 2009). During interphase, cohesin influences gene transcription through its interaction with Mediator, formation of topologically associated domains (TADs) and recruitment of RNAPII (Wendt et al., 2008; Remeseiro et al., 2012; Yan et al., 2013; Izumi et al., 2015; Merkenschlager and Nora, 2016; Rao et al., 2017; Mach et al., 2022). A recent study demonstrated that retention of the cohesin complex resulted in RNAPII remaining on mitotic chromosomes, resulting in nascent transcription from arm regions and chromosome missegregation (Perea-Resa et al., 2020). Thus, cohesin controls the chromosome-wide pattern of transcriptional silencing during mitosis (Figure 1).
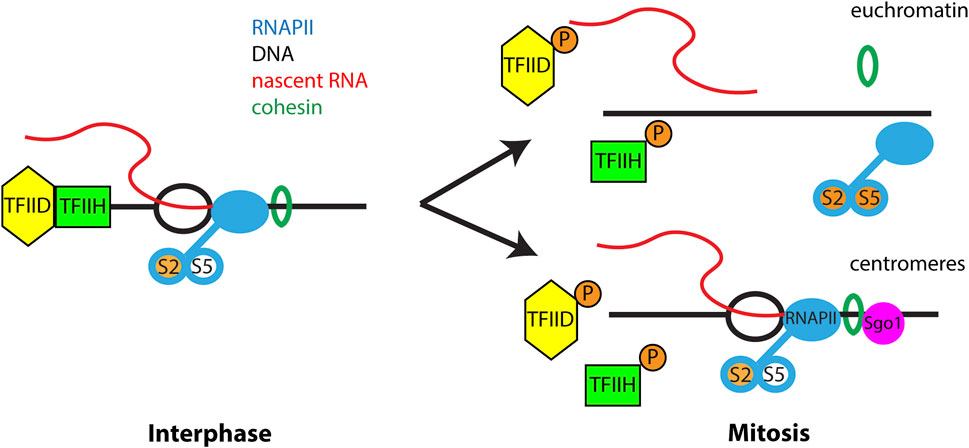
FIGURE 1. Transcriptional regulation during mitosis. During interphase RNAPII actively transcribes chromatin and is phosphorylated at Ser 2 of the C-terminal domain during transcriptional elongation. Serine-5 of the CTD is phosphorylated during transcriptional pausing and is absent from elongating RNAPII. TFIID and TFIIH present at promoter regions are important for transcriptional initiation. Cohesin regulates RNAPII transcription through an unknown mechanism. During mitosis phosphorylation of TFIIH, TFIID and hyperphosphorylation of RNAPII block new transcription initiation. In addition, phosphorylation of multiple proteins trigger release of cohesin from euchromatin which is important for removal of elongating RNAPII from chromosome arms. However, at centromeres Sgo1 protects cohesin from removal from chromatin, leading to retention of RNAPII at the mitotic centromere. Importantly, only elongating RNAPII is retained at mitotic centromeres and all factors and RNAPII modifications associated with transcriptional initiation and pausing are absent from the centromere.
The prototypical lncRNA is the XIST RNA that functions in X chromosome inactivation. The XIST RNA is transcribed from one of the two X chromosomes in female cells and initiates an epigenetic cascade to silence one of the two X chromosomes, resulting in equal gene dosage between male and female cells. XIST RNA localizes in cis to the inactive X (Xi) chromosome in interphase cells, and is required for gene dosage compensation in female (XX) cells (Lyon, 1961; Brown et al., 1991; Clemson et al., 1996). Interphase XIST RNA appears to be punctate, as determined by fluorescent in situ hybridization (FISH), revealing around 50 molecules of RNA acting on the entire inactive X chromosome (Cerase et al., 2014; Sunwoo et al., 2015; Markaki et al., 2021; Rodermund et al., 2021). Interestingly, as the cell enters mitosis, XIST particles begin to separate from chromatin in prophase (Clemson et al., 1996), eventually releasing completely from mitotic chromatin in prometaphase. This overall distribution of signal suggests XIST RNA does not associate with Xi chromatin during mitosis (Clemson et al., 1996).
Additional evidence shows other chromatin-bound lncRNAs dissociate from their respective targets during mitosis. A broad screen of predicted lncRNAs using single-molecule RNA-FISH revealed a wide distribution of interphase nuclear lncRNA localization. However, similar to XIST RNA, the nuclear FISH foci of all tested lncRNAs were displaced from mitotic chromatin (Cabili et al., 2015). The same cell cycle-dependent RNA localization pattern is observed for MALAT and NEAT1 (Clemson et al., 2009; Tripathi et al., 2010). In addition, interphase chromosomes are coated in cis by repetitive RNAs recognized by Cot-1 RNA FISH probes. Cot-1 RNAs (primarily composed of LINE and SINE elements) also dissociate from mitotic chromosomes and fail to re-enter the nucleus in G1 (Hall et al., 2014). Collectively these results demonstrate that all tested chromatin-associated lncRNAs are released into the cytoplasm during mitosis, primarily through a mechanism involving phosphorylation of SAF-A by the Aurora-B kinase (discussed in detail below).
Telomeres share characteristics with heterochromatin, as they possess HP1 (heterochromatin protein 1) proteins and exhibit heterochromatic histone marks, such as trimethylation of lysine 9 on histone 3 (H3K9me3), lysine 27 on histone 3 (H3K27me3) and trimethylation of lysine 20 on histone 4 (H4K20me3) (Ng et al., 2003; Perrini et al., 2004; Canudas et al., 2011; Montero et al., 2018; Jezek and Green, 2019). Similar to centromeres, human telomeres are also labeled with histone modifications associated with transcriptional activation, H4K20me1 and H3K27ac, as well as the histone acetyltransferase p300 (Cubiles et al., 2018). Moreover, the telomeric TTAGGG repeats lack the CpG sequence which is susceptible to methylation by DNA methyltransferases (DNMTs) in vertebrates (Li et al., 1992; Okano et al., 1998; Okano et al., 1999). However, the subtelomeres adjacent to the telomeres contain a high density of CpG sequences, which are methylated in somatic cells of the human genome (de Lange et al., 1990; Brock et al., 1999; Steinert et al., 2004). As a result, genes that are positioned close to telomeres are subjected to variegated transcriptional silencing (Baur et al., 2001). However, telomeres themselves are transcribed into a lncRNA known as TERRA. Telomere transcription is a highly conserved characteristic across eukaryotes, as it has been observed in various phyla, and it is important to facilitate heterochromatinization at telomeres (Azzalin et al., 2007; Schoeftner and Blasco, 2008; Deng et al., 2009).
TERRA levels display dynamic changes throughout the cell cycle. In human cells, the levels of TERRA are at their highest during G1/S phase and gradually decrease as the cell progresses toward the late S/G2 phase (Porro et al., 2010; Flynn et al., 2011; Arnoult et al., 2012). This cell cycle-dependent fluctuation in TERRA levels reflects the regulation and modulation of telomere function during different cell cycle stages. The expression of TERRA is controlled by the chromatin organizing factor CCCTC-binding factor (CTCF) and the cohesin protein Rad21. In human cells, the presence of CTCF and Rad21 promotes the recruitment of RNAPII to the TERRA promoter region. Depletion of Rad21 or CTCF using short hairpin RNA (shRNA) leads to a loss of RNAPII binding to TERRA promoters, resulting in a subsequent decrease in TERRA levels (Deng et al., 2012; Porro et al., 2014). The transcription of TERRA is also regulated by CpG methylation controlled by DNMT activities. The loss of DNMT activities leads to reduction of CpG methylation in the TERRA promoter at subtelomeres and overexpression of the TERRA, resulting in telomere shortening (Yehezkel et al., 2008; Nergadze et al., 2009). A recent study showed that deletion of CTCF-sites from subtelomeric regions leads to a notable reduction in TERRA expression from the modified telomere in human cells (Beishline et al., 2017). Furthermore, there is a significant decrease in the recruitment of histone H3K4me3 to the same chromosome ends (Beishline et al., 2017). This suggests that subtelomeric CTCF-binding sites play a crucial role in promoting TERRA expression and facilitating the recruitment of histone modifications associated with transcription at the telomeric region.
During mitosis, cohesin and RNAPII are removed from telomere regions, however, TERRA is detected in telomeres during mitosis, suggesting that TERRA is stably bound to telomeres and not associated via nascent transcription. Presumably, the association of TERRA with mitotic chromosomes is achieved through continued association with one or more of the known interacting proteins. For example, TERRA transcripts in mammalian cells associate with various heterochromatic marks, including H3K9me3 and HP1 proteins. They also interact with several chromatin remodeling complexes, such as Suv39h1 (a histone methyltransferase), nucleolar remodeling complex (NoRC), histone acetyltransferase complex (NuA), and BAF-type SWI/SNF nucleosome remodeling complex (Porro et al., 2010; Postepska-Igielska et al., 2013; Scheibe et al., 2013). Furthermore, recent studies have revealed that TERRA transcripts associate with the histone methyltransferase complex known as Polycomb Repressive Complex 2 (PRC2). This association occurs through a direct interaction TERRA with EZH2 and SUZ12 which are components of PRC2 (Chu et al., 2017; Wang et al., 2017; Montero et al., 2018). TERRA interacts with the shelterin components Telomeric repeat-binding factor1 (TRF1) and TRF2, which contribute to TERRA localization at telomeres (Deng et al., 2009; Lee et al., 2018). Additionally, recent work examining RNAs associated with recently replicated chromatin identified a family of GA-rich repeat RNAs homologous to TERRA and alpha satellite that are associated with replicating chromatin and chromatin in G1, suggesting that these transcripts may be a stable component of some chromatin loci (Gylling et al., 2020). However, whether there is continued physical associations between TERRA and these interactors during mitosis is still unclear.
As chromosomes condense in prophase, Aurora-B orchestrates the bulk removal of RNA from mitotic chromosomes (Sharp et al., 2020). This is most clearly visualized by labeling cells with 5-ethynyluridine (EU), a RNA analog that is readily incorporated into RNA as it is transcribed (Jao and Salic, 2008). In intact cells undergoing mitosis, chromosome arms display marked exclusion of RNA, in a pattern that is maintained through the end stages of mitosis. In contrast, cells treated with the Aurora-B inhibitor barasertib have chromosomes enriched with RNA, in a perichromosomal layer-like conformation suggestive of interaction primarily on the chromosome surface.
Additional evidence for the role for Aurora-B in the regulation of mitotic RNA localization is derived from FISH experiments using specific probes for individual RNAs. Notably, the XIST and MALAT-1/NEAT2 lncRNAs are cytoplasmic during mitosis (Clemson et al., 1996; Tripathi et al., 2010), yet show ectopic localization on chromatin after Aurora-B inhibition (Hall et al., 2009; Sharp et al., 2020). The same general trend is observed for the O-linked N-Acetylglucosamine transferase mRNA that undergoes nuclear retention (Sharp et al., 2020). Together, these specific examples corroborate the localization pattern reported for EU-RNA, arguing that Aurora-B has a central role organizing mitotic localization of chromatin-associated and/or nuclear-retained RNA.
The principal target of Aurora-B in this process is the SAF-A protein (Sharp et al., 2020). Originally identified as a heterogeneous nuclear RNA-binding protein (hnrnp U (Dreyfuss et al., 1984; Kiledjian and Dreyfuss, 1992; Romig et al., 1992)), SAF-A is markedly abundant and represents a major RNA binding activity in the interphase nucleus (Cho et al., 1998; Caudron-Herger et al., 2019). SAF-A has multiple modular domains: the DNA-binding SAP domain (Aravind and Koonin, 2000; Kipp et al., 2000); a highly charged acidic domain enriched with aspartic acid and glutamic acid residues; a SPRY domain of unknown function; an AAA + -type ATPase domain controlling its oligomeric state (Nozawa et al., 2017); a RGG-type RNA binding domain with multiple repeats of arginine and glycine (Kiledjian and Dreyfuss, 1992); and a low-complexity domain termed an intrinsically disordered region (IDR) (Sakaguchi et al., 2016) (Figure 2A). The multiple modalities of SAF-A domain structure are likely coordinated to impact its various roles in lncRNA localization, splicing, and euchromatin decondensation (Hasegawa et al., 2010; Huelga et al., 2012; Nozawa et al., 2017; Fan et al., 2018; Xiao et al., 2019).
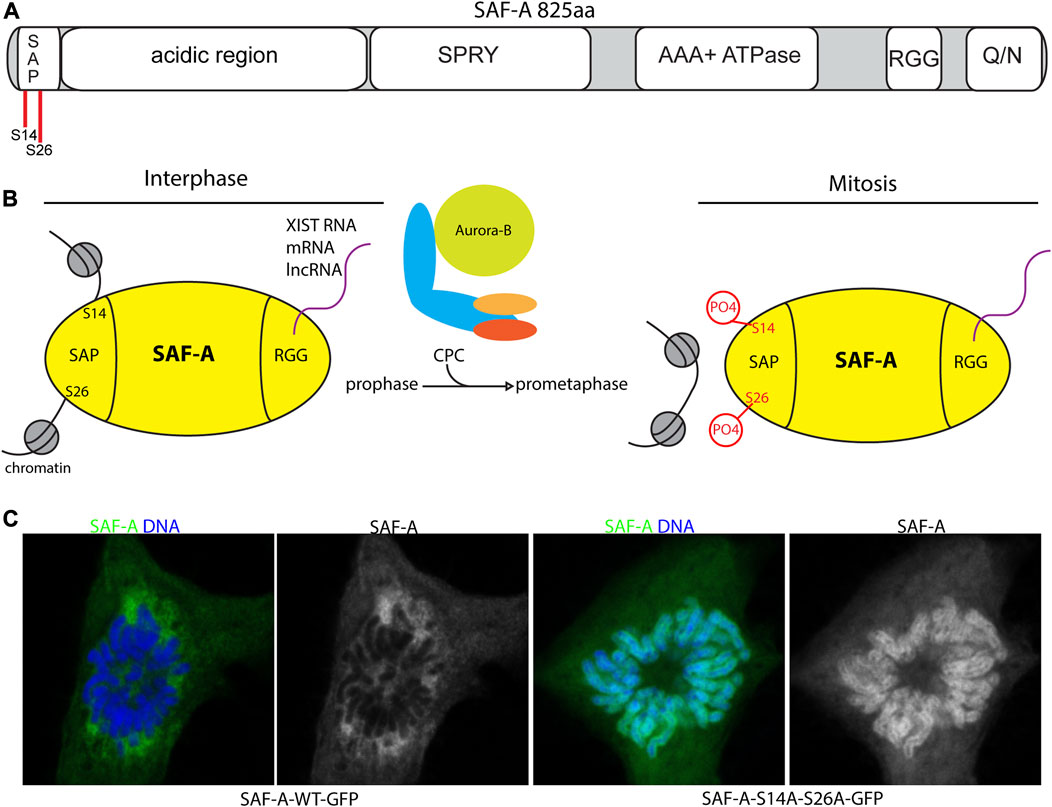
FIGURE 2. SAF-A regulates chromatin associated RNAs during mitosis. (A). Domain structure of SAF-A depicting the major domains associated with various SAF-A functions: A N-terminal SAP-type DNA binding domain, a large acidic region of unknown function, a SPRY domain of unknown function, a AAA + ATPase domain important for protein oligomerization, a RGG-type RNA binding domain, and a C-terminal Q/N-rich domain of unknown function. (B). Model of SAF-A chromatin binding during interphase and mitosis. During interphase SAF-A binds to chromatin through the N-terminal SAP-domain and RNA through the C-terminal RGG domain. During mitosis the CPC phosphorylates two serines in the SAP domain to trigger release of SAF-A:RNA complexes from chromatin. (C). Images of WT and SAP-domain mutant SAF-A during mitosis. SAF-A-AA is retained on the surface of mitotic chromosomes (Sharp et al., 2020; Blower et al., 2023).
The two nucleic acid binding activities of SAF-A are distinctly regulated during cell division. SAF-A binds to a subset of mRNAs and lncRNAs, representing approximately one-tenth of all expressed RNAs throughout the cell cycle (Sharp et al., 2020), and hydrodynamic analysis indicates all of the SAF-A is engaged with RNA cargo (Caudron-Herger et al., 2019). In contrast, the activation of Aurora-B in early mitosis controls SAF-A localization through negative regulation of DNA/chromatin binding, resulting in the eviction of SAF-A:RNA complexes from chromosomes (Sharp et al., 2020). This pathway therefore defines a two-component phosphoregulatory switch that functions to reduce the overall RNA content on mitotic chromatin (Figure 2B).
Although SAF-A phosphopeptides with the Aurora-B consensus sequence are interspersed throughout the N-terminal domains, multiple lines of evidence point to two key serines in the SAP domain as the critical target of Aurora-B in controlling SAF-A RNP localization. Specifically, when the SAP domain residues S14 and S26 are mutated to alanine, mimicking the unphosphorylated state, SAF-A has a longer residence time on chromatin, resulting in mitotic chromosomes covered in a sheath of SAF-A:RNA complexes in the perichromosomal layer (Sharp et al., 2020; Blower et al., 2023) (Figures 2B, C). In short, the SAF-A phosphomutant phenocopies Aurora-B inhibition. Conversely, structural and biochemical data show a SAF-A phosphomimetic allele profoundly interrupts the DNA binding properties of SAF-A due a gain in negative surface charge. When expressed in vivo, a SAF-A phosphomimetic allele renders SAF-A chemically resistant to the effects of Aurora-B inhibition. Together these data unambiguously point to SAF-A SAP domain as the key target for Aurora-B in mitotic RNA localization.
When the Aurora-B and SAF-A phosphoregulatory switch is disrupted, the fidelity of chromosome transmission to daughter cells is acutely compromised (Sharp et al., 2020; Teves, 2020). Cells expressing the SAF-A phosphomutant allele have a delay in anaphase onset, misaligned chromosomes, defects in chromosome and spindle morphology, and an overall increase in the rate of chromosome missegregation. These effects are likely due to reduced function of proteins involved in chromosome segregation: for example, CENP-E localization to the kinetochore is reduced in the SAF-A phosphomutant, and morphological phenotypes point to reduced Kif22 kinesin function. In sum, the Aurora-B control of mitotic SAF-A-RNA localization is an important pathway to ensure proper genome stability during cell division.
We note although epistasis analysis points to SAF-A being the primary RNP target of Aurora-B, it is possible that eviction of SAF-A from chromatin impacts secondary RNPs as well. For example, like SAF-A, mutation of the Ciz1 RNA-binding protein causes defective localization of the XIST RNA in interphase cells (Ridings-Figueroa et al., 2017; Sunwoo et al., 2017). Genetic analysis in mice suggests SAF-A is upstream of Ciz1; when SAF-A function is disrupted, Ciz1 is still bound to Xist RNA particles even as they become disengaged from contact with the Xi (Sunwoo et al., 2017). It is possible that an analogous scenario occurs during early mitosis, with the dissociation of SAF-A-RNA complexes from chromatin triggering a network of other RNPs disengaging from chromatin. In the case of the Xi, this scenario could impact not just Ciz1, but possibly other Xist RNA localization factors, such as hnrnp K or Polycomb group proteins (Colognori et al., 2019).
Among the proteins that control RNA localization to mitotic chromosomes one of the most well studied is Ki-67. Ki-67 is widely used as a marker for cell proliferation in cancer cells (Sun and Kaufman, 2018). During interphase, Ki-67 localizes to the nucleolus where it may play a role in nucleolar assembly and organization of heterochromatin. Ki-67 is a very large protein comprised of several different modular domains: a N-terminal FHA domain, a PP1-binding domain, an unstructured domain containing 16 repeats of the Ki-67 domain, and a C-terminal LR chromatin-binding domain (Figure 3A). The FHA and PP1-binding domains recognize phosphorylated proteins and recruit PP1γ to mitotic chromosomes, respectively. The LR domain aids in chromatin binding through its leucine and arginine-rich properties. However, function of the tandem repeats domain is not well understood (Sun and Kaufman, 2018). During mitosis, Ki-67 is released from the nucleolus and relocalizes to the perichromosomal layer (Figure 3B). Interestingly, the perichromosomal layer constitutes approximately a third of the total chromosomal volume (Booth et al., 2016; Booth and Earnshaw, 2017).
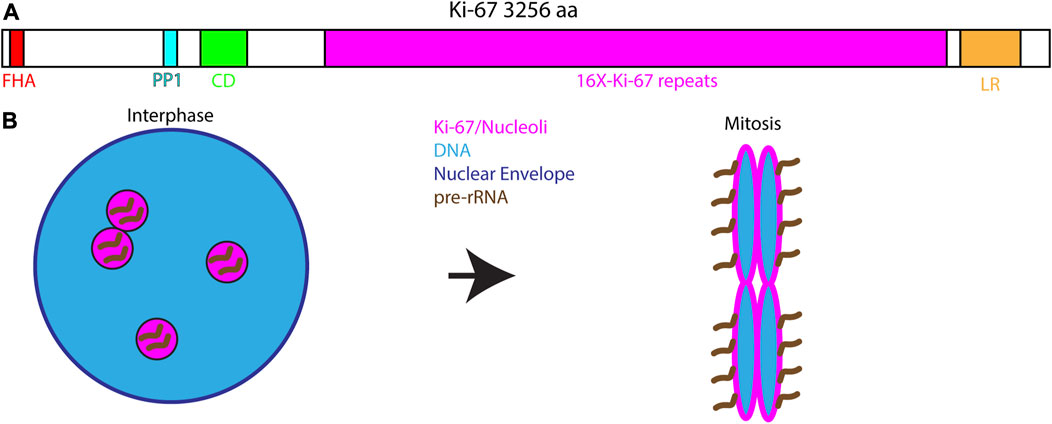
FIGURE 3. Ki-67 and the perichromosomal layer. (A). Domain structure of Ki-67 depicting the major conserved domains: A N-terminal Forkhead-associated domain (FHA) that recognizes phosphorylated proteins, a Protein Phosphatase 1 interacting motif, a Conserved Domain of unknown function, 16 repeats of a simple Ki67-specific repeat that is heavily phosphorylated during mitosis and a C-terminal Leucine/Arginine-rich domain important for interacting with DNA and chromatin. (B). During interphase Ki-67 localizes to the nucleolus where it is associated with pre-rRNA. At the onset of mitosis Ki-67 redistributes to the chromosomal surface where it serves and the assembly point for the perichromosomal layer. Ki-67 is necessary for targeting the entire perichromosomal layer to the chromosome surface during mitosis including pre-rRNA.
Ki-67 is but one of many proteins and RNAs to relocalize from the nucleolus to the perichromosomal layer in mitosis (Van Hooser et al., 2005), however its incorporation into the layer is critical for perichromosomal layer function (Booth et al., 2014). Loss of Ki-67 is associated with a loss of chromosome individualization, such that mitotic chromosomes clump into a single mass of chromatin (Cuylen et al., 2016; Takagi et al., 2016). Based on these finding it has been proposed that Ki-67 forms an extended structure during mitosis that serves as a surfactant to prevent chromosomes from sticking to one another (Cuylen et al., 2016).
Interestingly, chromatin organization by Ki-67 appears to be substantially different than that induced by the condensin complexes. Double mutants of Ki-67 and condensin result in a dramatic chromosome condensation defect much more severe than either single mutant alone, suggesting two parallel pathways of chromatin organization during mitosis (Takagi et al., 2018). Knockout of Ki-67 results in the loss of almost all proteins associated with the perichromosmal layer by both proteomics and Correlated Light Electron Microscopy (CLEM) (Booth et al., 2016). This suggests that Ki-67 serves as a hub for the assembly of the perichromosomal layer and that loss of the perichromomsal layer results in chromosome clustering during mitosis.
Recent work has validated the observation that unprocessed pre-rRNA associates with mitotic chromosomes. This study also found that Ki-67 is required for tethering pre-rRNA to mitotic chromosomes and that Ki-67 uses multiple different domains for RNA binding (Ma et al., 2022). Collectively these studies show that disassembly of the nucleolus during mitosis results in chromosomal coating by the nucleolar remnants in the form of the perichromosomal layer. Ki-67 is the foundation of the perichromosomal layer which may assemble through tethering pre-rRNA to mitotic chromosomes. Confoundingly, the mouse knockout of Ki-67 is viable and fertile and it is possible to establish constitutive Ki-67 knockout cell lines, suggesting that redundant mechanisms exist to compensate for the dramatic chromosome morphology changes observed after loss of Ki-67 (Cidado et al., 2016; Sobecki et al., 2016).
As outlined in the previous section, most chromatin-associated transcripts are released during mitosis. At the completion of mitosis, segregated chromosomes coalesce and a nuclear envelope rapidly separates the chromatin from the cytoplasm. However, very little is known about the fate of chromatin-associated transcripts at the beginning of the next cell cycle. In theory, nuclear RNAs could either re-associate with chromatin at the end of mitosis and be reused in the next cell cycle or could remain in the cytoplasm where they would presumably be degraded.
The most well-studied case of reuse of nuclear RNAs comes from work on the nucleolus. During interphase, the nucleolus is the site of rRNA transcription, processing, modification and the assembly site for mature ribosomes. During mitosis, rRNA transcription ceases and the nucleolus dissociates. Dissociated nucleolar components assume different localization patterns during mitosis: residency in the perichromosomal layer (Figure 3B) dispersed in the cytoplasm, or present in cytoplasmic foci termed Nucleolus-Derived Foci (NDF) during anaphase and telophase (Figure 4). Interestingly, partially-processed rRNA and U3 and U8 snoRNAs localize to both the perichromosomal layer and NDF (Dundr et al., 1997; Dundr and Olson, 1998; Dundr et al., 2000). The perichromosomal layer associates with the surface of all mitotic chromosomes from prometaphase until late anaphase. The contents of the perichromosomal layer are important for the individualization of chromosomes during mitosis and for the exclusion of cytoplasmic material from the nucleus at the end of mitosis (Cuylen-Haering et al., 2020). Contents of the perichromosomal layer are reincorporated into the nucleus in early G1. NDF particles first appear in late anaphase and disappear by early G1. Protein components of the NDF exchange rapidly, suggesting that the NDF contents may also participate in reestablishment of the nucleolus in G1 (Dundr et al., 2000). Partially processed pre-RNA has been hypothesized to play an important role in reestablishing the nucleolus in G1. As a result of association with the perichromosomal layer, some unprocessed pre-rRNA and some snoRNAs are inherited by the nucleus in G1 where they likely contribute to the rapid resumption of ribosome assembly.
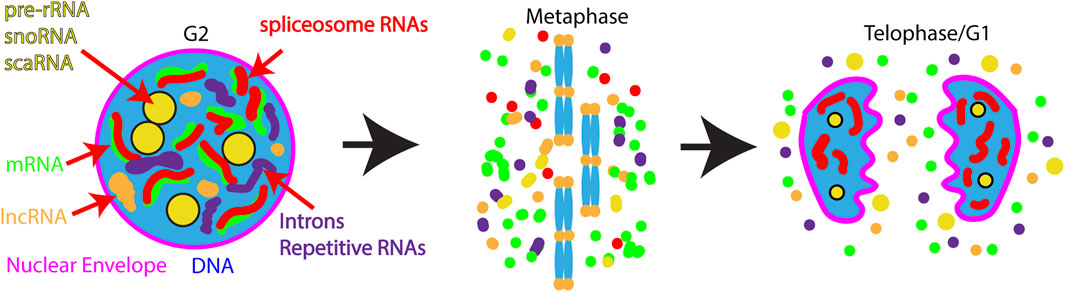
FIGURE 4. Model for nuclear RNA regulation during and after mitosis. During interphase many transcripts are enriched in the nucleus including: pre-rRNA, lncRNAs, sno-, sca- RNAs, nascent mRNAs, introns, and spliceosomal RNAs. During mitosis the vast majority of these transcripts are released into the cytoplasm. Exceptions to RNA release from chromatin are the centromeres and telomeres where cenRNA and TERRA transcripts are retained. At the end of mitosis, some nucleolar RNAs re-enter the nucleus along with splicosomal RNAs, while a fraction of nucleolar RNAs are retained in foci in the cytoplasm termed NDF (Nucleolus Derived Foci). In contrast lncRNAs, mRNAs, and introns remain in the cytoplasm where they may be subject to programmed degradation.
In contrast to nucleolar-associated transcripts, much less is known about the fate of other nuclear RNAs following mitosis. Early work on the XIST lncRNA showed that this transcript dissociates from chromatin during mitosis and remains in the cytoplasm during G1 following transcriptional inhibition. However, this study used Actinomycin D to inhibit transcription in G1 which been shown to have several detrimental effects on cells including: changes in protein binding to DNA, loss of transcription, loss of nuclear import, and changes in RNA half-lives (Sawicki and Godman, 1971; Clemson et al., 1996), so it is not clear if XIST RNA can re-enter the nucleus in G1. More recent work studying repetitive RNAs associated with transcriptionally active portions of the genome also found that these transcripts dissociate from mitotic chromatin and remain in the cytoplasm in G1 cells in response to the inclusion of several different transcription inhibitors (Hall et al., 2014). Additionally, our recent work found that the lncRNAs MALAT1 and NEAT1 dissociate from chromatin during mitosis and remain in the cytoplasm during G1 in response to transcriptional inhibition (Blower et al., 2023). This is consistent with previous work showing that reassembly of paraspeckles, which require NEAT1 lncRNA for assembly, requires de novo transcription in G1 (Mao et al., 2011). Collectively, these studies indicate that most studied lncRNAs are not inherited by G1 nuclei but remain in the cytoplasm in G1 cells where they may undergo programmed degradation. Interestingly, this result suggests that chromatin:lncRNA interactions are reset every time a cell passes through mitosis. This may provide the cell with an opportunity to change chromatin:RNA interactions which could be important for changing transcriptional programs during development.
In a recent study, we examined inheritance of nuclear RNAs in a transcript-independent manner using metabolic labeling of RNA. We found that approximately half of all nuclear RNAs are inherited by nuclei in G1 including prominent foci associated with the G1 nucleolus. In contrast to lncRNAs, we found that the spliceosomal RNA U2 is rapidly inherited by daughter nuclei in a nuclear import-dependent manner (Blower et al., 2023). This is likely a result of the fact that spliceosomal RNAs normally exit the nucleus following transcription where they undergo maturation in the cytoplasm before they are reimported into the nucleus (Fischer et al., 2011). This result suggests that the majority of spliceosomal RNAs will be inherited by G1 nuclei. The differences in behavior between snRNAs and lncRNAs may be a result of the fact that snRNAs are normally imported into the nucleus as part of their maturation cycle, while lncRNAs do not exit the nucleus during interphase and may not have a nuclear import pathway. To fully understand the implications of differential nuclear RNA inheritance following mitosis it will be necessary to perform a genome-wide analysis of inheritance.
The current evidence suggests that all lncRNAs follow the same progression as XIST/NEAT1/MALAT1 and do not reenter the cell after completion of mitosis, but rather accumulate in the nucleus through transcriptional restart early G1 (Clemson et al., 1996). Interestingly, mRNA is degraded in two waves during the mitosis-to-G1 transition, as observed by single-cell sequencing and pulse-labeling. This temporal regulation of RNA decay contributes to the controlled gene expression necessary for proper cell cycle progression. The cytoplasmic deadenylase CNOT1 was identified as a main contributor of RNA degradation from the previous cell cycle (Krenning et al., 2022). The pre-existing nuclear lncRNAs that reside in the cytoplasm following the reformation of the nuclear envelope in G1 could be degraded by similar mechanisms. However, many lncRNAs are not polyadenylated (Sun et al., 2018), making degradation by CNOT1 in the mitosis-to-G1 transition unlikely. Interestingly, these typically-nuclear lncRNAs are exposed to a new environment in this cell cycle transition, raising the possibility that residency in the cytoplasm alters lncRNP composition.
Mitosis exposes nuclear lncRNAs to cytoplasmic ribosomes which could lead to lncRNA translation and the production of short peptides. Although lncRNAs contain much smaller open reading frames (ORFs) compared to mRNAs, ribosomal footprints were identified on lncRNAs across six eukaryotic species, including human, mouse, and zebrafish (Ruiz-Orera et al., 2014), confirming that some lncRNAs have translational potential. Studies show an increased instance of ribosomal binding to XIST and other lncRNAs during mitosis (Tanenbaum et al., 2015). Additionally, ribosomal footprints revealed preferential binding to the lncRNA primary ORF (Ruiz-Orera et al., 2014). Therefore, we speculate that lncRNAs could undergo nonsense mediated decay (NMD) during the mitosis to G1 transition. Nonsense mediated decay tags aberrant transcripts for decay, as denoted by the presence of a premature stop codon. This process requires ribosomal binding, which will detect an exon junction complex downstream of a stop codon, signaling for NMD (Nickless et al., 2017). Since lncRNAs do not typically contain long, translatable ORFs, reside in the cytoplasm during mitosis, and do not appear to reenter the nucleus in G1, we speculate that they may interact with ribosomes to trigger NMD, resulting in a general turnover of lncRNAs in early G1. Further work is necessary to evaluate lncRNA kinetics during the mitosis to G1 transition.
Mitosis is a time of vast changes to the nuclear structure and function of cells. A variety of recent studies have provided insight into the regulation and function of nuclear RNAs during and after mitosis. While recent work has provided a variety of insights into the mechanisms that control nuclear RNA localization and transcriptional regulation there are many open questions related to the functions of changing nuclear RNA regulation. Important mechanistic questions include: how are cohesin dynamics linked to RNAPII activity during and after mitosis? How does mitotic transcriptional silencing affect transcription in the next cell cycle? What is the full spectrum of nuclear RNAs that are re-imported into the nucleus in G1? How are cytoplasmic lncRNAs degraded in G1 cells? What are the functional consequences of a failure to degrade cytoplasmic lncRNAs during G1? We expect that future work will advance our understanding of nuclear RNA regulation during this dramatic stage of the cell cycle.
ES, JS, RW, and MB wrote and edited the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by funding from the NIH to MB. (GM122893, GM144352).
We would like to thank the members of the MB Lab for helpful discussions.
The author MB declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Akoulitchev, S., and Reinberg, D. (1998). The molecular mechanism of mitotic inhibition of TFIIH is mediated by phosphorylation of CDK7. Genes. & Dev. 12, 3541–3550. doi:10.1101/gad.12.22.3541
Altemose, N., Logsdon, G. A., Bzikadze, A. V., Sidhwani, P., Langley, S. A., Caldas, G. V., et al. (2022). Complete genomic and epigenetic maps of human centromeres. Science 376, eabl4178. doi:10.1126/science.abl4178
Aravind, L., and Koonin, E. V. (2000). Sap - a putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 25, 112–114. doi:10.1016/s0968-0004(99)01537-6
Arnoult, N., Van Beneden, A., and Decottignies, A. (2012). Telomere length regulates TERRA levels through increased trimethylation of telomeric H3K9 and HP1α. Nat. Struct. Mol. Biol. 19, 948–956. doi:10.1038/nsmb.2364
Azzalin, C. M., Reichenbach, P., Khoriauli, L., Giulotto, E., and Lingner, J. (2007). Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318, 798–801. doi:10.1126/science.1147182
Baur, J. A., Zou, Y., Shay, J. W., and Wright, W. E. (2001). Telomere position effect in human cells. Science 292 (292), 2075–2077. doi:10.1126/science.1062329
Beishline, K., Vladimirova, O., Tutton, S., Wang, Z., Deng, Z., and Lieberman, P. M. (2017). CTCF driven TERRA transcription facilitates completion of telomere DNA replication. Nat. Commun. 8 (8), 2114. doi:10.1038/s41467-017-02212-w
Bell, J. C., Jukam, D., Teran, N. A., Risca, V. I., Smith, O. K., Johnson, W. L., et al. (2018). Chromatin-associated RNA sequencing (ChAR-seq) maps genome-wide RNA-to-DNA contacts. eLife 7, e27024. doi:10.7554/elife.27024
Bellier, S., Chastant, S., Adenot, P., Vincent, M., Renard, J. P., and Bensaude, O. (1997). Nuclear translocation and carboxyl-terminal domain phosphorylation of RNA polymerase II delineate the two phases of zygotic gene activation in mammalian embryos. EMBO J. 16, 6250–6262. doi:10.1093/emboj/16.20.6250
Bergmann, J. H., Jakubsche, J. N., Martins, N. M., Kagansky, A., Nakano, M., Kimura, H., et al. (2012). Epigenetic engineering: Histone H3K9 acetylation is compatible with kinetochore structure and function. J. Cell. Sci. 125, 411–421. doi:10.1242/jcs.090639
Bergmann, J. H., Rodríguez, M. G., Martins, N. M., Kimura, H., Kelly, D. A., Masumoto, H., et al. (2011). Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 30, 328–340. doi:10.1038/emboj.2010.329
Blower, M. D. (2016). Centromeric transcription regulates Aurora-B localization and activation. Cell. Rep. 15, 1624–1633. doi:10.1016/j.celrep.2016.04.054
Blower, M. D., Wang, W., and Sharp, J. A. (2023). Differential nuclear import regulates nuclear RNA inheritance following mitosis. Mol. Biol. Cell. 34, ar32. doi:10.1091/mbc.e23-01-0004
Bobkov, G. O. M., Gilbert, N., and Heun, P. (2018). Centromere transcription allows CENP-A to transit from chromatin association to stable incorporation. J. Cell. Biol. 217, 1957–1972. doi:10.1083/jcb.201611087
Boisvert, F. M., van Koningsbruggen, S., Navascués, J., and Lamond, A. I. (2007). The multifunctional nucleolus. Nat. Rev. Mol. Cell. Biol. 8, 574–585. doi:10.1038/nrm2184
Bonetti, A., Agostini, F., Suzuki, A. M., Hashimoto, K., Pascarella, G., Gimenez, J., et al. (2020). RADICL-seq identifies general and cell type-specific principles of genome-wide RNA-chromatin interactions. Nat. Commun. 11 (11), 1018. doi:10.1038/s41467-020-14337-6
Booth, D. G., Beckett, A. J., Molina, O., Samejima, I., Masumoto, H., Kouprina, N., et al. (2016). 3D-CLEM reveals that a major portion of mitotic chromosomes is not chromatin. Mol. Cell. 64 (64), 790–802. doi:10.1016/j.molcel.2016.10.009
Booth, D. G., and Earnshaw, W. C. (2017). Ki-67 and the chromosome periphery compartment in mitosis. Trends Cell. Biol. 27, 906–916. doi:10.1016/j.tcb.2017.08.001
Booth, D. G., Takagi, M., Sanchez-Pulido, L., Petfalski, E., Vargiu, G., Samejima, K., et al. (2014). Ki-67 is a PP1-interacting protein that organises the mitotic chromosome periphery. Elife 3, e01641. doi:10.7554/elife.01641
Brock, G. J., Charlton, J., and Bird, A. (1999). Densely methylated sequences that are preferentially localized at telomere-proximal regions of human chromosomes. Gene 240, 269–277. doi:10.1016/s0378-1119(99)00442-4
Brown, C. J., Ballabio, A., Rupert, J. L., Lafreniere, R. G., Grompe, M., Tonlorenzi, R., et al. (1991). A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349, 38–44. doi:10.1038/349038a0
Bury, L., Moodie, B., Ly, J., McKay, L. S., Miga, K. H., and Cheeseman, I. M. (2020). Alpha-satellite RNA transcripts are repressed by centromere-nucleolus associations. eLife 9, e59770. doi:10.7554/elife.59770
Cabili, M. N., Dunagin, M. C., McClanahan, P. D., Biaesch, A., Padovan-Merhar, O., Regev, A., et al. (2015). Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 16, 20. doi:10.1186/s13059-015-0586-4
Canudas, S., Houghtaling, B. R., Bhanot, M., Sasa, G., Savage, S. A., Bertuch, A. A., et al. (2011). A role for heterochromatin protein 1γ at human telomeres. Genes. & Dev. 25 (25), 1807–1819. doi:10.1101/gad.17325211
Carmena, M., Wheelock, M., Funabiki, H., and Earnshaw, W. C. (2012). The chromosomal passenger complex (CPC): From easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell. Biol. 13, 789–803. doi:10.1038/nrm3474
Caudron-Herger, M., Rusin, S. F., Adamo, M. E., Seiler, J., Schmid, V. K., Barreau, E., et al. (2019). R-DeeP: Proteome-wide and quantitative identification of RNA-dependent proteins by density gradient ultracentrifugation. Mol. Cell. 75, 184–199. doi:10.1016/j.molcel.2019.04.018
Cerase, A., Smeets, D., Tang, Y. A., Gdula, M., Kraus, F., Spivakov, M., et al. (2014). Spatial separation of Xist RNA and polycomb proteins revealed by superresolution microscopy. Proc. Natl. Acad. Sci. 111 (111), 2235–2240. doi:10.1073/pnas.1312951111
Chan, F. L., Marshall, O. J., Saffery, R., Won Kim, B., Earle, E., Choo, K. H., et al. (2012). Active transcription and essential role of RNA polymerase II at the centromere during mitosis. Proc. Natl. Acad. Sci. 109, 1979–1984. doi:10.1073/pnas.1108705109
Chen, Y., Zhang, Q., Teng, Z., and Liu, H. (2021). Centromeric transcription maintains centromeric cohesion in human cells. J. Cell. Biol. 220, e202008146. doi:10.1083/jcb.202008146
Cho, R. J., Campbell, M. J., Winzeler, E. A., Steinmetz, L., Conway, A., Wodicka, L., et al. (1998). A genome-wide transcriptional analysis of the mitotic cell cycle. Mol. Cell. 2, 65–73. doi:10.1016/s1097-2765(00)80114-8
Chu, H. P., Cifuentes-Rojas, C., Kesner, B., Aeby, E., Lee, H. G., Wei, C., et al. (2017). TERRA RNA antagonizes ATRX and protects telomeres. Cell. 170, 86–101.e16. doi:10.1016/j.cell.2017.06.017
Cidado, J., Wong, H. Y., Rosen, D. M., Cimino-Mathews, A., Garay, J. P., Fessler, A. G., et al. (2016). Ki-67 is required for maintenance of cancer stem cells but not cell proliferation. Oncotarget 7 (7), 6281–6293. doi:10.18632/oncotarget.7057
Clemson, C. M., Hutchinson, J. N., Sara, S. A., Ensminger, A. W., Fox, A. H., Chess, A., et al. (2009). An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell. 33 (33), 717–726. doi:10.1016/j.molcel.2009.01.026
Clemson, C. M., McNeil, J. A., Willard, H. F., and Lawrence, J. B. (1996). XIST RNA paints the inactive X chromosome at interphase: Evidence for a novel RNA involved in nuclear/chromosome structure. J. Cell. Biol. 132, 259–275. doi:10.1083/jcb.132.3.259
Colognori, D., Sunwoo, H., Kriz, A. J., Wang, C. Y., and Lee, J. T. (2019). Xist deletional analysis reveals an interdependency between xist RNA and polycomb complexes for spreading along the inactive X. Mol. Cell. 74 (74), 101–117.e10. doi:10.1016/j.molcel.2019.01.015
Corless, S., Hocker, S., and Erhardt, S. (2020). Centromeric RNA and its function at and beyond centromeric chromatin. J. Mol. Biol. 432 (432), 4257–4269. doi:10.1016/j.jmb.2020.03.027
Cubiles, M. D., Barroso, S., Vaquero-Sedas, M. I., Enguix, A., Aguilera, A., and Vega-Palas, M. A. (2018). Epigenetic features of human telomeres. Nucleic Acids Res. 46 (46), 2347–2355. doi:10.1093/nar/gky006
Cuylen, S., Blaukopf, C., Politi, A. Z., Muller-Reichert, T., Neumann, B., Poser, I., et al. (2016). Ki-67 acts as a biological surfactant to disperse mitotic chromosomes. Nature 535 (535), 308–312. doi:10.1038/nature18610
Cuylen-Haering, S., Petrovic, M., Hernandez-Armendariz, A., Schneider, M. W. G., Samwer, M., Blaukopf, C., et al. (2020). Chromosome clustering by Ki-67 excludes cytoplasm during nuclear assembly. Nature 587, 285–290. doi:10.1038/s41586-020-2672-3
de Lange, T., Shiue, L., Myers, R. M., Cox, D. R., Naylor, S. L., Killery, A. M., et al. (1990). Structure and variability of human chromosome ends. Mol. Cell. Biol. 10, 518–527. doi:10.1128/mcb.10.2.518
Deng, Z., Norseen, J., Wiedmer, A., Riethman, H., and Lieberman, P. M. (2009). TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell. 35 (35), 403–413. doi:10.1016/j.molcel.2009.06.025
Deng, Z., Wang, Z., Stong, N., Plasschaert, R., Moczan, A., Chen, H. S., et al. (2012). A role for CTCF and cohesin in subtelomere chromatin organization, TERRA transcription, and telomere end protection. EMBO J. 31 (31), 4165–4178. doi:10.1038/emboj.2012.266
Dixon, J. R., Selvaraj, S., Yue, F., Kim, A., Li, Y., Shen, Y., et al. (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485 (485), 376–380. doi:10.1038/nature11082
Djebali, S., Davis, C. A., Merkel, A., Dobin, A., Lassmann, T., Mortazavi, A., et al. (2012). Landscape of transcription in human cells. Nature 489, 101–108. doi:10.1038/nature11233
Dreyfuss, G., Choi, Y. D., and Adam, S. A. (1984). Characterization of heterogeneous nuclear RNA-protein complexes in vivo with monoclonal antibodies. Mol. Cell. Biol. 4, 1104–1114. doi:10.1128/mcb.4.6.1104
Du, Y., Topp, C. N., and Dawe, R. K. (2010). DNA binding of centromere protein C (CENPC) is stabilized by single-stranded RNA. PLoS Genet. 6 (6), e1000835. doi:10.1371/journal.pgen.1000835
Dundr, M., Meier, U. T., Lewis, N., Rekosh, D., Hammarskjold, M. L., and Olson, M. O. (1997). A class of nonribosomal nucleolar components is located in chromosome periphery and in nucleolus-derived foci during anaphase and telophase. Chromosoma 105, 407–417. doi:10.1007/bf02510477
Dundr, M., Misteli, T., and Olson, M. O. (2000). The dynamics of postmitotic reassembly of the nucleolus. J. Cell. Biol. 150 (150), 433–446. doi:10.1083/jcb.150.3.433
Dundr, M., and Olson, M. O. (1998). Partially processed pre-rRNA is preserved in association with processing components in nucleolus-derived foci during mitosis. Mol. Biol. Cell. 9, 2407–2422. doi:10.1091/mbc.9.9.2407
Dupuis-Sandoval, F., Poirier, M., and Scott, M. S. (2015). The emerging landscape of small nucleolar RNAs in cell biology. Wiley Interdiscip. Rev. RNA 6, 381–397. doi:10.1002/wrna.1284
Earnshaw, W., Bordwell, B., Marino, C., and Rothfield, N. (1986). Three human chromosomal autoantigens are recognized by sera from patients with anti-centromere antibodies. J. Clin. Investigation 77, 426–430. doi:10.1172/jci112320
Fan, H., Lv, P., Huo, X., Wu, J., Wang, Q., Cheng, L., et al. (2018). The nuclear matrix protein HNRNPU maintains 3D genome architecture globally in mouse hepatocytes. Genome Res. 28, 192–202. doi:10.1101/gr.224576.117
Ferri, F., Bouzinba-Segard, H., Velasco, G., Hubé, F., and Francastel, C. (2009). Non-coding murine centromeric transcripts associate with and potentiate Aurora B kinase. Nucleic Acids Res. 37, 5071–5080. doi:10.1093/nar/gkp529
Festuccia, N., Gonzalez, I., Owens, N., and Navarro, P. (2017). Mitotic bookmarking in development and stem cells. Development 144, 3633–3645. doi:10.1242/dev.146522
Fischer, U., Englbrecht, C., and Chari, A. (2011). Biogenesis of spliceosomal small nuclear ribonucleoproteins. WIREs RNA 2, 718–731. doi:10.1002/wrna.87
Fischle, W., Tseng, B. S., Dormann, H. L., Ueberheide, B. M., Garcia, B. A., Shabanowitz, J., et al. (2005). Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438, 1116–1122. doi:10.1038/nature04219
Flynn, R. L., Centore, R. C., O'Sullivan, R. J., Rai, R., Tse, A., Songyang, Z., et al. (2011). TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature 471 (471), 532–536. doi:10.1038/nature09772
Fujimura, A., Hayashi, Y., Kato, K., Kogure, Y., Kameyama, M., Shimamoto, H., et al. (2020). Identification of a novel nucleolar protein complex required for mitotic chromosome segregation through centromeric accumulation of Aurora B. Nucleic Acids Res. 48 (48), 6583–6596. doi:10.1093/nar/gkaa449
Gandhi, R., Gillespie, P. J., and Hirano, T. (2006). Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr. Biol. 16 (16), 2406–2417. doi:10.1016/j.cub.2006.10.061
Ghosh, S., and Lehner, C. F. (2022). Incorporation of CENP-A/CID into centromeres during early Drosophila embryogenesis does not require RNA polymerase II-mediated transcription. Chromosoma 131, 1–17. doi:10.1007/s00412-022-00767-2
Gibcus, J. H., Samejima, K., Goloborodko, A., Samejima, I., Naumova, N., Nuebler, J., et al. (2018). A pathway for mitotic chromosome formation. Science 359, eaao6135. doi:10.1126/science.aao6135
Gonzalez, I., Molliex, A., and Navarro, P. (2021). Mitotic memories of gene activity. Curr. Opin. Cell. Biol. 69, 41–47. doi:10.1016/j.ceb.2020.12.009
Gottesfeld, J. M., and Forbes, D. J. (1997). Mitotic repression of the transcriptional machinery. Trends Biochem. Sci. 22, 197–202. doi:10.1016/s0968-0004(97)01045-1
Grenfell, A. W., Heald, R., and Strzelecka, M. (2016a). Correction: Mitotic noncoding RNA processing promotes kinetochore and spindle assembly in Xenopus. J. Cell. Biol. 214, 783. doi:10.1083/jcb.20160402908222016c
Grenfell, A. W., Heald, R., and Strzelecka, M. (2016b). Mitotic noncoding RNA processing promotes kinetochore and spindle assembly in Xenopus. J. Cell. Biol. 214 (214), 133–141. doi:10.1083/jcb.201604029
Gylling, H. M., Gonzalez-Aguilera, C., Smith, M. A., Kaczorowski, D. C., Groth, A., and Lund, A. H. (2020). Repeat RNAs associate with replication forks and post-replicative DNA. RNA. Sep. 26 (9), 1104–1117. doi:10.1261/rna.074757.120
Hall, L. L., Byron, M., Pageau, G., and Lawrence, J. B. (2009). AURKB-mediated effects on chromatin regulate binding versus release of XIST RNA to the inactive chromosome. J. Cell. Biol. 186, 491–507. doi:10.1083/jcb.200811143
Hall, L. L., Carone, D. M., Gomez, A. V., Kolpa, H. J., Byron, M., Mehta, N., et al. (2014). Stable C0T-1 repeat RNA is abundant and is associated with euchromatic interphase chromosomes. Cell. 156 (156), 907–919. doi:10.1016/j.cell.2014.01.042
Hasegawa, Y., Brockdorff, N., Kawano, S., Tsutui, K., Tsutui, K., and Nakagawa, S. (2010). The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev. Cell. 19, 469–476. doi:10.1016/j.devcel.2010.08.006
Henikoff, S., Ahmad, K., and Malik, H. S. (2001). The centromere paradox: Stable inheritance with rapidly evolving DNA. Science 293, 1098–1102. doi:10.1126/science.1062939
Hernandez-Verdun, D. (2011). Assembly and disassembly of the nucleolus during the cell cycle. Nucleus 2, 189–194. doi:10.4161/nucl.2.3.16246
Hirai, H., Shogaki, Y., and Sato, M. (2022). The Mis6 inner kinetochore subcomplex maintains CENP-A nucleosomes against centromeric non-coding transcription during mitosis. Commun. Biol. 5 (5), 818. doi:10.1038/s42003-022-03786-y
Hori, T., Shang, W. H., Toyoda, A., Misu, S., Monma, N., Ikeo, K., et al. (2014). Histone H4 Lys 20 monomethylation of the CENP-A nucleosome is essential for kinetochore assembly. Dev. Cell. 29 (29), 740–749. doi:10.1016/j.devcel.2014.05.001
Huelga, S. C., Vu, A. Q., Arnold, J. D., Liang, T. Y., Liu, P. P., Yan, B. Y., et al. (2012). Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep. 1 (2), 167–178. doi:10.1016/j.celrep.2012.02.001
Ideue, T., Cho, Y., Nishimura, K., and Tani, T. (2014). Involvement of satellite I noncoding RNA in regulation of chromosome segregation. Genes. Cells 19, 528–538. doi:10.1111/gtc.12149
Izumi, K., Nakato, R., Zhang, Z., Edmondson, A. C., Noon, S., Dulik, M. C., et al. (2015). Germline gain-of-function mutations in AFF4 cause a developmental syndrome functionally linking the super elongation complex and cohesin. Nat. Genet. 47, 338–344. doi:10.1038/ng.3229
Jambhekar, A., Emerman, A. B., Schweidenback, C. T., and Blower, M. D. (2014). RNA stimulates Aurora B kinase activity during mitosis. PLoS One 9, e100748. doi:10.1371/journal.pone.0100748
Jao, C. Y., and Salic, A. (2008). Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl. Acad. Sci. U. S. A. 105, 15779–15784. doi:10.1073/pnas.0808480105
Jezek, M., and Green, E. M. (2019). Histone modifications and the maintenance of telomere integrity. Cells 8, 199. doi:10.3390/cells8020199
Jurica, M. S., and Moore, M. J. (2003). Pre-mRNA splicing: Awash in a sea of proteins. Mol. Cell. 12, 5–14. doi:10.1016/s1097-2765(03)00270-3
Kabeche, L., Nguyen, H. D., Buisson, R., and Zou, L. (2018). A mitosis-specific and R loop-driven ATR pathway promotes faithful chromosome segregation. Science 359, 108–114. doi:10.1126/science.aan6490
Kato, M., and Carninci, P. (2020). Genome-wide technologies to study RNA-chromatin interactions. Noncoding RNA 6, 20. doi:10.3390/ncrna6020020
Kiledjian, M., and Dreyfuss, G. (1992). Primary structure and binding activity of the hnRNP U protein: Binding RNA through RGG box. EMBO J. 11, 2655–2664. doi:10.1002/j.1460-2075.1992.tb05331.x
Kipp, M., Gohring, F., Ostendorp, T., van Drunen, C. M., van Driel, R., Przybylski, M., et al. (2000). SAF-Box, a conserved protein domain that specifically recognizes scaffold attachment region DNA. Mol. Cell. Biol. 20, 7480–7489. doi:10.1128/mcb.20.20.7480-7489.2000
Krenning, L., Sonneveld, S., and Tanenbaum, M. E. (2022). Time-resolved single-cell sequencing identifies multiple waves of mRNA decay during the mitosis-to-G1 phase transition. eLife 11, e71356. doi:10.7554/elife.71356
Kueng, S., Hegemann, B., Peters, B. H., Lipp, J. J., Schleiffer, A., Mechtler, K., et al. (2006). Wapl controls the dynamic association of cohesin with chromatin. Cell. 127, 955–967. doi:10.1016/j.cell.2006.09.040
Lam, M. T., Li, W., Rosenfeld, M. G., and Glass, C. K. (2014). Enhancer RNAs and regulated transcriptional programs. Trends Biochem. Sci. 39, 170–182. doi:10.1016/j.tibs.2014.02.007
Lee, Y. W., Arora, R., Wischnewski, H., and Azzalin, C. M. (2018). TRF1 participates in chromosome end protection by averting TRF2-dependent telomeric R loops. Nat. Struct. Mol. Biol. 25, 147–153. doi:10.1038/s41594-017-0021-5
Li, E., Bestor, T. H., and Jaenisch, R. (1992). Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 69, 915–926. doi:10.1016/0092-8674(92)90611-f
Li, X., and Fu, X. D. (2019). Chromatin-associated RNAs as facilitators of functional genomic interactions. Nat. Rev. Genet. 20, 503–519. doi:10.1038/s41576-019-0135-1
Liang, K., Woodfin, A. R., Slaughter, B. D., Unruh, J. R., Box, A. C., Rickels, R. A., et al. (2015). Mitotic transcriptional activation: Clearance of actively engaged pol II via transcriptional elongation control in mitosis. Mol. Cell. 60, 435–445. doi:10.1016/j.molcel.2015.09.021
Licatalosi, D. D., and Darnell, R. B. (2010). RNA processing and its regulation: Global insights into biological networks. Nat. Rev. Genet. 11, 75–87. doi:10.1038/nrg2673
Ling, Y. H., and Yuen, K. W. Y. (2019). Point centromere activity requires an optimal level of centromeric noncoding RNA. Proc. Natl. Acad. Sci. 116 (116), 6270–6279. doi:10.1073/pnas.1821384116
Liu, H., Qu, Q., Warrington, R., Rice, A., Cheng, N., and Yu, H. (2015). Mitotic transcription installs Sgo1 at centromeres to coordinate chromosome segregation. Mol. Cell. 59 (59), 426–436. doi:10.1016/j.molcel.2015.06.018
Losada, A., Hirano, M., and Hirano, T. (2002). Cohesin release is required for sister chromatid resolution, but not for condensin-mediated compaction, at the onset of mitosis. Genes. Dev. 16 (16), 3004–3016. doi:10.1101/gad.249202
Lyon, M. F. (1961). Gene action in the X-chromosome of the mouse (Mus musculus L). Nature 190, 372–373. doi:10.1038/190372a0
Ma, K., Luo, M., Xie, G., Wang, X., Li, Q., Gao, L., et al. (2022). Ribosomal RNA regulates chromosome clustering during mitosis. Cell. Discov. 8 (8), 51. doi:10.1038/s41421-022-00400-7
Mach, P., Kos, P. I., Zhan, Y., Cramard, J., Gaudin, S., Tünnermann, J., et al. (2022). Cohesin and CTCF control the dynamics of chromosome folding. Nat. Genet. 54, 1907–1918. doi:10.1038/s41588-022-01232-7
Manuelidis, L., and Wu, J. C. (1978). Homology between human and simian repeated DNA. Nature 276 (276), 92–94. doi:10.1038/276092a0
Mao, Y. S., Sunwoo, H., Zhang, B., and Spector, D. L. (2011). Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat. Cell. Biol. 13, 95–101. doi:10.1038/ncb2140
Markaki, Y., Chong, J. G., Wang, Y., Jacobson, E. C., Luong, C., Tan, S. Y. X., et al. (2021). Xist nucleates local protein gradients to propagate silencing across the X chromosome. Cell. 184, 6212–6192. doi:10.1016/j.cell.2021.11.028
Mattick, J. S., Amaral, P. P., Carninci, P., Carpenter, S., Chang, H. Y., Chen, L. L., et al. (2023). Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell. Biol. 24, 430–447. doi:10.1038/s41580-022-00566-8
McKinley, K. L., and Cheeseman, I. M. (2016). The molecular basis for centromere identity and function. Nat. Rev. Mol. Cell. Biol. 17, 16–29. doi:10.1038/nrm.2015.5
McNulty, S. M., Sullivan, L. L., and Sullivan, B. A. (2017). Human centromeres produce chromosome-specific and array-specific alpha satellite transcripts that are complexed with CENP-A and CENP-C. Dev. Cell. 42, 226–240.e6. doi:10.1016/j.devcel.2017.07.001
Melters, D. P., Bradnam, K. R., Young, H. A., Telis, N., May, M. R., Ruby, J. G., et al. (2013). Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol. 14, R10. doi:10.1186/gb-2013-14-1-r10
Meng, Y., Yi, X., Li, X., Hu, C., Wang, J., Bai, L., et al. (2016). The non-coding RNA composition of the mitotic chromosome by 5'-tag sequencing. Nucleic Acids Res. 44 (44), 4934–4946. doi:10.1093/nar/gkw195
Merkenschlager, M., and Nora, E. P. (2016). CTCF and cohesin in genome folding and transcriptional gene regulation. Annu. Rev. Genomics Hum. Genet. 17 (17), 17–43. doi:10.1146/annurev-genom-083115-022339
Mondal, T., Rasmussen, M., Pandey, G. K., Isaksson, A., and Kanduri, C. (2010). Characterization of the RNA content of chromatin. Genome Res. 20, 899–907. doi:10.1101/gr.103473.109
Montero, J. J., López-Silanes, I., Megías, D., F Fraga, M., Castells-García, Á., and Blasco, M. A. (2018). TERRA recruitment of polycomb to telomeres is essential for histone trymethylation marks at telomeric heterochromatin. Nat. Commun. 9 (9), 1548. doi:10.1038/s41467-018-03916-3
Muramoto, T., Müller, I., Thomas, G., Melvin, A., and Chubb, J. R. (2010). Methylation of H3K4 Is required for inheritance of active transcriptional states. Curr. Biol. 20 (20), 397–406. doi:10.1016/j.cub.2010.01.017
Nasmyth, K., and Haering, C. H. (2009). Cohesin: Its roles and mechanisms. Annu. Rev. Genet. 43, 525–558. doi:10.1146/annurev-genet-102108-134233
Naumova, N., Imakaev, M., Fudenberg, G., Zhan, Y., Lajoie, B. R., Mirny, L. A., et al. (2013). Organization of the mitotic chromosome. Science 342 (342), 948–953. doi:10.1126/science.1236083
Nergadze, S. G., Farnung, B. O., Wischnewski, H., Khoriauli, L., Vitelli, V., Chawla, R., et al. (2009). CpG-island promoters drive transcription of human telomeres. RNA. Dec 15, 2186–2194. doi:10.1261/rna.1748309
Ng, H. H., Ciccone, D. N., Morshead, K. B., Oettinger, M. A., and Struhl, K. (2003). Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: A potential mechanism for position-effect variegation. Proc. Natl. Acad. Sci. 100, 1820–1825. doi:10.1073/pnas.0437846100
Nickless, A., Bailis, J. M., and You, Z. (2017). Control of gene expression through the nonsense-mediated RNA decay pathway. Cell. Biosci. 7, 26. doi:10.1186/s13578-017-0153-7
Niehrs, C., and Luke, B. (2020). Regulatory R-loops as facilitators of gene expression and genome stability. Nat. Rev. Mol. Cell. Biol. 21, 167–178. doi:10.1038/s41580-019-0206-3
Nora, E. P., Goloborodko, A., Valton, A. L., Gibcus, J. H., Uebersohn, A., Abdennur, N., et al. (2017). Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell. 169, 930–944.e22. doi:10.1016/j.cell.2017.05.004
Nozawa, R. S., Boteva, L., Soares, D. C., Naughton, C., Dun, A. R., Buckle, A., et al. (2017). SAF-A regulates interphase chromosome structure through oligomerization with chromatin-associated RNAs. Cell. 169, 1214–1227.e18. doi:10.1016/j.cell.2017.05.029
Okano, M., Bell, D. W., Haber, D. A., and Li, E. (1999). DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 99 (99), 247–257. doi:10.1016/s0092-8674(00)81656-6
Okano, M., Xie, S., and Li, E. (1998). Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 19, 219–220. doi:10.1038/890
Palmer, D. K., O'Day, K., Trong, H. L., Charbonneau, H., and Margolis, R. L. (1991). Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc. Natl. Acad. Sci. U. S. A. 88, 3734–3738. doi:10.1073/pnas.88.9.3734
Palmer, D. K., O'Day, K., Wener, M. H., Andrews, B. S., and Margolis, R. L. (1987). A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J. Cell. Biol. 104, 805–815. doi:10.1083/jcb.104.4.805
Palozola, K. C., Donahue, G., Liu, H., Grant, G. R., Becker, J. S., Cote, A., et al. (2017). Mitotic transcription and waves of gene reactivation during mitotic exit. Science 358, 119–122. doi:10.1126/science.aal4671
Pardue, M. L., and Gall, J. G. (1970). Chromosomal localization of mouse satellite DNA. Science 168 (168), 1356–1358. doi:10.1126/science.168.3937.1356
Parsons, G. G., and Spencer, C. A. (1997). Mitotic repression of RNA polymerase II transcription is accompanied by release of transcription elongation complexes. Mol. Cell. Biol. 17, 5791–5802. doi:10.1128/mcb.17.10.5791
Perea-Resa, C., Bury, L., Cheeseman, I. M., and Blower, M. D. (2020). Cohesin removal reprograms gene expression upon mitotic entry. Mol. Cell. 78, 127–140.e7. doi:10.1016/j.molcel.2020.01.023
Perrini, B., Piacentini, L., Fanti, L., Altieri, F., Chichiarelli, S., Berloco, M., et al. (2004). HP1 controls telomere capping, telomere elongation, and telomere silencing by two different mechanisms in Drosophila. Mol. Cell. 15 (15), 467–476. doi:10.1016/j.molcel.2004.06.036
Phizicky, E. M., and Hopper, A. K. (2010). tRNA biology charges to the front. Genes. & Dev. 24 (24), 1832–1860. doi:10.1101/gad.1956510
Porro, A., Feuerhahn, S., Delafontaine, J., Riethman, H., Rougemont, J., and Lingner, J. (2014). Functional characterization of the TERRA transcriptome at damaged telomeres. Nat. Commun. 5 (5), 5379. doi:10.1038/ncomms6379
Porro, A., Feuerhahn, S., Reichenbach, P., and Lingner, J. (2010). Molecular dissection of telomeric repeat-containing RNA biogenesis unveils the presence of distinct and multiple regulatory pathways. Mol. Cell. Biol. 30, 4808–4817. doi:10.1128/mcb.00460-10
Postepska-Igielska, A., Krunic, D., Schmitt, N., Greulich-Bode, K. M., Boukamp, P., and Grummt, I. (2013). The chromatin remodelling complex NoRC safeguards genome stability by heterochromatin formation at telomeres and centromeres. EMBO Rep. 14, 704–710. doi:10.1038/embor.2013.87
Prescott, D. M., and Bender, M. A. (1962). Synthesis of RNA and protein during mitosis in mammalian tissue culture cells. Exp. Cell. Res. 26, 260–268. doi:10.1016/0014-4827(62)90176-3
Quénet, D., and Dalal, Y. (2014). A long non-coding RNA is required for targeting centromeric protein A to the human centromere. Elife 3 (3), e03254. doi:10.7554/eLife.03254
Quinn, J. J., and Chang, H. Y. (2016). Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 17, 47–62. doi:10.1038/nrg.2015.10
Rao, S. S., Huntley, M. H., Durand, N. C., Stamenova, E. K., Bochkov, I. D., Robinson, J. T., et al. (2014). A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 159, 1665–1680. doi:10.1016/j.cell.2014.11.021
Rao, S. S. P., Huang, S. C., Glenn St Hilaire, B., Engreitz, J. M., Perez, E. M., Kieffer-Kwon, K. R., et al. (2017). Cohesin loss eliminates all loop domains. Cell. 171, 305–320. doi:10.1016/j.cell.2017.09.026
Remeseiro, S., Cuadrado, A., Gómez-López, G., Pisano, D. G., and Losada, A. (2012). A unique role of cohesin-SA1 in gene regulation and development. EMBO J. 31, 2090–2102. doi:10.1038/emboj.2012.60
Ribeiro, S. A., Vagnarelli, P., Dong, Y., Hori, T., McEwen, B. F., Fukagawa, T., et al. (2010). A super-resolution map of the vertebrate kinetochore. Proc. Natl. Acad. Sci. U. S. A. 107 (107), 10484–10489. doi:10.1073/pnas.1002325107
Ridings-Figueroa, R., Stewart, E. R., Nesterova, T. B., Coker, H., Pintacuda, G., Godwin, J., et al. (2017). The nuclear matrix protein CIZ1 facilitates localization of Xist RNA to the inactive X-chromosome territory. Genes. & Dev. 31, 876–888. doi:10.1101/gad.295907.117
Rodermund, L., Coker, H., Oldenkamp, R., Wei, G., Bowness, J., Rajkumar, B., et al. (2021). Time-resolved structured illumination microscopy reveals key principles of Xist RNA spreading. Science 372, eabe7500. doi:10.1126/science.abe7500
Romig, H., Fackelmayer, F. O., Renz, A., Ramsperger, U., and Richter, A. (1992). Characterization of SAF-A, a novel nuclear DNA binding protein from HeLa cells with high affinity for nuclear matrix/scaffold attachment DNA elements. EMBO J. 11, 3431–3440. doi:10.1002/j.1460-2075.1992.tb05422.x
Rošić, S., Köhler, F., and Erhardt, S. (2014). Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division. J. Cell. Biol. 207 (207), 673–349. doi:10.1083/jcb.20140409711122014c
Ruiz-Orera, J., Messeguer, X., Subirana, J. A., and Alba, M. M. (2014). Long non-coding RNAs as a source of new peptides. Elife 3 (3), e03523. doi:10.7554/eLife.03523
Sakaguchi, T., Hasegawa, Y., Brockdorff, N., Tsutsui, K., Tsutsui, K. M., Sado, T., et al. (2016). Control of chromosomal localization of xist by hnRNP U family molecules. Dev. Cell. 39, 11–12. doi:10.1016/j.devcel.2016.09.022
Sawicki, S. G., and Godman, G. C. (1971). On the differential cytotoxicity of actinomycin D. J. Cell. Biol. 50, 746–761. doi:10.1083/jcb.50.3.746
Scheibe, M., Arnoult, N., Kappei, D., Buchholz, F., Decottignies, A., Butter, F., et al. (2013). Quantitative interaction screen of telomeric repeat-containing RNA reveals novel TERRA regulators. Genome Res. 23, 2149–2157. doi:10.1101/gr.151878.112
Schoeftner, S., and Blasco, M. A. (2008). Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell. Biol. 10, 228–236. doi:10.1038/ncb1685
Segil, N., Guermah, M., Hoffmann, A., Roeder, R. G., and Heintz, N. (1996). Mitotic regulation of TFIID: Inhibition of activator-dependent transcription and changes in subcellular localization. Genes. Dev. 10 (10), 2389–2400. doi:10.1101/gad.10.19.2389
Shang, W. H., Hori, T., Toyoda, A., Kato, J., Popendorf, K., Sakakibara, Y., et al. (2010). Chickens possess centromeres with both extended tandem repeats and short non-tandem-repetitive sequences. Genome Res. 20, 1219–1228. doi:10.1101/gr.106245.110
Sharp, J. A., Perea-Resa, C., Wang, W., and Blower, M. D. (2020). Cell division requires RNA eviction from condensing chromosomes. J. Cell. Biol. 219, e201910148. doi:10.1083/jcb.201910148
Shen, W., Zhang, Y., Shi, M., Ye, B., Yin, M., Li, P., et al. (2022). Profiling and characterization of constitutive chromatin-enriched RNAs. iScience 25 (25), 105349. doi:10.1016/j.isci.2022.105349
Shermoen, A. W., and O'Farrell, P. H. (1991). Progression of the cell cycle through mitosis leads to abortion of nascent transcripts. Cell. 67, 303–310. doi:10.1016/0092-8674(91)90182-x
Sloan, K. E., Warda, A. S., Sharma, S., Entian, K. D., Lafontaine, D. L. J., and Bohnsack, M. T. (2017). Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 14 (14), 1138–1152. doi:10.1080/15476286.2016.1259781
Sobecki, M., Mrouj, K., Camasses, A., Parisis, N., Nicolas, E., Lleres, D., et al. (2016). The cell proliferation antigen Ki-67 organises heterochromatin. eLife 5 (5), e13722. doi:10.7554/elife.13722
Sridhar, B., Rivas-Astroza, M., Nguyen, T. C., Chen, W., Yan, Z., Cao, X., et al. (2017). Systematic mapping of RNA-chromatin interactions in vivo. Curr. Biol. 27 (27), 610–612. doi:10.1016/j.cub.2017.01.068
Statello, L., Guo, C. J., Chen, L. L., and Huarte, M. (2021). Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell. Biol. 22, 96–118. doi:10.1038/s41580-020-00315-9
Steinert, S., Shay, J. W., and Wright, W. E. (2004). Modification of subtelomeric DNA. Mol. Cell. Biol. 24, 4571–4580. doi:10.1128/mcb.24.10.4571-4580.2004
Sullivan, B. A., Blower, M. D., and Karpen, G. H. (2001). Determining centromere identity: Cyclical stories and forking paths. Nat. Rev. Genet. 2, 584–596. doi:10.1038/35084512
Sullivan, B. A., and Karpen, G. H. (2004). Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat. Struct. Mol. Biol. 11, 1076–1083. doi:10.1038/nsmb845
Sullivan, K. F., Hechenberger, M., and Masri, K. (1994). Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J. Cell. Biol. 127, 581–592. doi:10.1083/jcb.127.3.581
Sun, Q., Hao, Q., and Prasanth, K. V. (2018). Nuclear long noncoding RNAs: Key regulators of gene expression. Trends Genet. 34, 142–157. doi:10.1016/j.tig.2017.11.005
Sun, X., and Kaufman, P. D. (2018). Ki-67: More than a proliferation marker. Chromosoma 127, 175–186. doi:10.1007/s00412-018-0659-8
Sunwoo, H., Colognori, D., Froberg, J. E., Jeon, Y., and Lee, J. T. (2017). Repeat E anchors Xist RNA to the inactive X chromosomal compartment through CDKN1A-interacting protein (CIZ1). Proc. Natl. Acad. Sci. U. S. A. 114, 10654–10659. doi:10.1073/pnas.1711206114
Sunwoo, H., Wu, J. Y., and Lee, J. T. (2015). The Xist RNA-PRC2 complex at 20-nm resolution reveals a low Xist stoichiometry and suggests a hit-and-run mechanism in mouse cells. Proc. Natl. Acad. Sci. U. S. A. 112 (112), E4216–E4225. doi:10.1073/pnas.1503690112
Takagi, M., Natsume, T., Kanemaki, M. T., and Imamoto, N. (2016). Perichromosomal protein Ki67 supports mitotic chromosome architecture. Genes. cells. 21, 1113–1124. doi:10.1111/gtc.12420
Takagi, M., Ono, T., Natsume, T., Sakamoto, C., Nakao, M., Saitoh, N., et al. (2018). Ki-67 and condensins support the integrity of mitotic chromosomes through distinct mechanisms. J. Cell. Sci. 131, jcs212092. doi:10.1242/jcs.212092
Talbert, P. B., and Henikoff, S. (2018). Transcribing centromeres: Noncoding RNAs and kinetochore assembly. Trends Genet. 34, 587–599. doi:10.1016/j.tig.2018.05.001
Tanenbaum, M. E., Stern-Ginossar, N., Weissman, J. S., and Vale, R. D. (2015). Regulation of mRNA translation during mitosis. eLife 4, e07957. doi:10.7554/elife.07957
Teves, S. S. (2020). Function through absence: Active RNA exclusion from chromosomes leads to proper cell division. J. Cell. Biol. 219, e202009193. doi:10.1083/jcb.202009193
Tripathi, V., Ellis, J. D., Shen, Z., Song, D. Y., Pan, Q., Watt, A. T., et al. (2010). The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 39 (39), 925–938. doi:10.1016/j.molcel.2010.08.011
Vakoc, C. R., Sachdeva, M. M., Wang, H., and Blobel, G. A. (2006). Profile of histone lysine methylation across transcribed mammalian chromatin. Mol. Cell. Biol. 26, 9185–9195. doi:10.1128/mcb.01529-06
Van Hooser, A. A., Yuh, P., and Heald, R. (2005). The perichromosomal layer. Chromosoma 114, 377–388. doi:10.1007/s00412-005-0021-9
Wade, C. M., Giulotto, E., Sigurdsson, S., Zoli, M., Gnerre, S., Imsland, F., et al. (2009). Genome sequence, comparative analysis, and population genetics of the domestic horse. Science 326 (326), 865–867. doi:10.1126/science.1178158
Waizenegger, I. C., Hauf, S., Meinke, A., and Peters, J. M. (2000). Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 103 (103), 399–410. doi:10.1016/s0092-8674(00)00132-x
Wang, X., Goodrich, K. J., Gooding, A. R., Naeem, H., Archer, S., Paucek, R. D., et al. (2017). Targeting of polycomb repressive complex 2 to RNA by short repeats of consecutive guanines. Mol. Cell. 65 (65), 1056–1067. doi:10.1016/j.molcel.2017.02.003
Wendt, K. S., Yoshida, K., Itoh, T., Bando, M., Koch, B., Schirghuber, E., et al. (2008). Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451, 796–801. doi:10.1038/nature06634
Werner, M. S., and Ruthenburg, A. J. (2015). Nuclear fractionation reveals thousands of chromatin-tethered noncoding RNAs adjacent to active genes. Cell. Rep. 12 (12), 1089–1098. doi:10.1016/j.celrep.2015.07.033
Werner, M. S., Sullivan, M. A., Shah, R. N., Nadadur, R. D., Grzybowski, A. T., Galat, V., et al. (2017). Chromatin-enriched lncRNAs can act as cell-type specific activators of proximal gene transcription. Nat. Struct. Mol. Biol. 24, 596–603. doi:10.1038/nsmb.3424
Wong, L. H., Brettingham-Moore, K. H., Chan, L., Quach, J. M., Anderson, M. A., Northrop, E. L., et al. (2007). Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res. 17, 1146–1160. doi:10.1101/gr.6022807
Xiao, R., Chen, J. Y., Liang, Z., Luo, D., Chen, G., Lu, Z. J., et al. (2019). Pervasive chromatin-RNA binding protein interactions enable RNA-based regulation of transcription. Cell. 178 (178), 107–121.e18. doi:10.1016/j.cell.2019.06.001
Yan, J., Enge, M., Whitington, T., Dave, K., Liu, J., Sur, I., et al. (2013). Transcription factor binding in human cells occurs in dense clusters formed around cohesin anchor sites. Cell. 154 (154), 801–813. doi:10.1016/j.cell.2013.07.034
Yehezkel, S., Segev, Y., Viegas-Pequignot, E., Skorecki, K., and Selig, S. (2008). Hypomethylation of subtelomeric regions in ICF syndrome is associated with abnormally short telomeres and enhanced transcription from telomeric regions. Hum. Mol. Genet. 17 (17), 2776–2789. doi:10.1093/hmg/ddn177
Keywords: RNA, lncRNA, Mitosis, hnRNP-U/SAF-A, XIST, centromere/cenRNA
Citation: Sparago E, Watanabe R, Sharp JA and Blower MD (2023) Dynamic redistribution and inheritance of chromatin:RNA interactions during cell division. Front. RNA Res. 1:1240954. doi: 10.3389/frnar.2023.1240954
Received: 15 June 2023; Accepted: 21 July 2023;
Published: 03 August 2023.
Edited by:
Greg L. Stewart, University of British Columbia, CanadaReviewed by:
Eugene V. Sheval, Lomonosov Moscow State University, RussiaCopyright © 2023 Sparago, Watanabe, Sharp and Blower. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael D. Blower, bWJsb3dlckBidS5lZHU=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.