
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. RNA Res., 10 August 2023
Sec. RNP Biogenesis and Function
Volume 1 - 2023 | https://doi.org/10.3389/frnar.2023.1226610
This article is part of the Research TopicRibonucleoprotein formation and regulationView all 5 articles
 Pauline Adjibade1,2,3
Pauline Adjibade1,2,3 Rachid Mazroui1,2,3*
Rachid Mazroui1,2,3*Stress granules (SG) are macro-complexes that assemble as phase-separated and dynamic RNA biocondensates in the cytoplasm of the eukaryotic cell when the initiation step of the general translation of mRNAs is stalled. This occurs mainly as an adaptive cell response to either environmental (i.e., radiation, exposure to chemical drugs), pathological (i.e., viral treatment), physiological (i.e., oxygen-, amino acids-, and glucose-deprivation), or therapeutic (i.e., treatment with anti-cancer drugs) translational stress. SG also formed when translation initiation is blocked through stress-independent events including alteration of the activities of specific translation initiation factors and RNA-binding proteins. Both stress-dependent and–independent inhibition of translation initiation results in the accumulation of untranslated mRNAs, considered as integral components of SG. Consistently, in vivo assays of SG assembly combined with in vitro-based assembly of SG-like biocondensates studies support a fundamental role of the accumulation of untranslated mRNA in initiating the formation of SG, which then further promote their repression, potentially in a feed-back regulatory mechanism. The potential role of SG in actively repressing translation of associated mRNAs has been supported by a number of functional studies, establishing SG as critical regulatory sites of RNA homeostasis, in particular during stress. The view that the SG environment restricts translation of associated mRNAs was however challenged in studies showing that stress-induced translation repression can occur similarly in absence and presence of SG, leading to the emerging concept that formation of SG and translation repression are uncoupled processes. While it still a debate if mRNA recruitment to SG contributes to their translation repression, recent finding reported translation of reporter mRNAs in SG, suggesting rather an active translational role of SG. In this review, we describe the main translational signaling pathways that regulate the biology of SG, summarize current data supporting RNA as an integral functional component of SG, and then discuss evidence supporting or not the role of SG in regulating translation either negatively or positively during stress.
SG are described as a stress-induced membraneless phase-dense cytoplasmic bio-condensates of RNA, translation initiation factors (eIFs), and RNA-binding proteins (RBPs) that assemble when translation initiation is blocked (Riggs et al., 2020). SG are observed in most tested organisms ranging from yeast to mammals, establishing their formation as an evolutionary conserved stress-induced eukaryotic response. In mammals, SG form in virtually any analysed type of cells including neurons (Aramburu-Nunez et al., 2022), muscle cells (Kubinski and Claus, 2022), and blood cells (Ghisolfi et al., 2012), or tissues such as brain (DeGracia and Hu, 2007; DeGracia et al., 2008; Degracia and Hu, 2013) and muscles (Kubinski and Claus, 2022), either promoting cell adaptation to stress required for maintaining the normal cell physiology, or contributing to the development of stress-associated diseases including neurodegenerative pathologies (Hu et al., 2022) and cancer (Aulas et al., 2020; Lee and Namkoong, 2022). Stress-inducing SG includes environmental stress such as radiation (Moeller et al., 2004; Moutaoufik et al., 2014), exposure to chemical drugs such as arsenite (Kedersha et al., 2005), pathological stress such as viral infection (Mazroui et al., 2006), or physiological stress due to oxygen-(Moeller et al., 2004; Fahling, 2009), amino acids (Damgaard and Lykke-Andersen, 2011)-, or glucose-deprivation (Buchan et al., 2011; Wang et al., 2022), and chemotherapeutic stress generated during therapy (Fournier et al., 2010; Adjibade et al., 2015; Szaflarski et al., 2016; Adjibade et al., 2020). Stress-inducing SG generally triggers a global reprograming of gene expression characterized by a general inhibition of translation initiation, while translation of mRNAs encoding stress adaptation functions is prioritized (Yamasaki and Anderson, 2008). This stress-induced translational reprograming of gene expression generally involves the activation of a translational initiation regulatory signaling pathway characterized by the induction of phosphorylation of the translation initiation factor eIF2α (Holcik and Sonenberg, 2005; Wek et al., 2006; Koromilas, 2014), a master inducer of SG during stress (Advani and Ivanov, 2019). In this review, the term of translational stress refers to any type of stress that induces SG concomitantly with the inhibition of translation initiation, which generally involves the induction of phosphorylation of eIF2α.
Translation of the majority of cellular mRNAs, known as cap-dependent translation, initiates via a mechanism that involves the early recognition of their 5′end m7GTP cap structure by the eIF4F complex consisting of the cap-binding protein eIF4E, the RNA helicase eIF4A known to unwind secondary structures in the 5′ untranslated region (UTR) of mRNAs, and the scaffolding eIF4GI (Gingras et al., 1999; Sonenberg and Dever, 2003; Sonenberg, 2008; Jackson et al., 2010). eIF4F recruits eIF3 bound to the 43S preinitiation complex (43S PIC) consisting of the 40S ribosomal subunit, the eIF2.GTP.Met-tRNAiMet ternary complex (TC), and additional eIFs such as eIF1 and eIF1A. The assembled 43S PIC scans the 5′UTR of eIF4F-bound mRNA in the 5′ to 3′ direction for the first translation initiation codon. The recognition of the start codon induces conformational changes of 43S PIC resulting in the release of eIFs such as eIF1, to form 48S complex. Factors such as eIF5 and 5B are then recruited to 48S complex, which is also joined by the 60S ribosomal subunit forming an 80S complex competent for translation (Jackson et al., 2010). However, in response to translational stress, the rate of this cap-dependent translation initiation is globally reduced, which is due mainly to the induction of the phosphorylation of eIF2α (Holcik and Sonenberg, 2005; Wek et al., 2006; Koromilas, 2014).
eIF2α is phosphorylated on serine 51 by one of the four kinases (Holcik and Sonenberg, 2005; Donnelly et al., 2013), that are activated in a stress-type dependant manner: protein kinase R (PKR), PERK (PKR-like endoplasmic reticulum kinase), GCN2 (general control non-derepressible 2), and HRI (heme-regulated inhibitor). Phosphorylation of eIF2α by either kinase inhibits translation initiation by triggering the formation of a non-functional TC (Gebauer and Hentze, 2004). The incorporation of this aberrant TC into the 43S is believed to induce formation of 48S complexes that cannot be converted into functional 80S, which then stall and accumulate in an inactive form (Kedersha et al., 2002), though the formation of such stalled inactive 48S complexes in vivo has never been demonstrated. Phosphorylated eIF2α-induced inhibition of translation initiation also allows the elongating translating ribosomes (polysomes) run-off mRNAs, increasing the pool of free mRNPs consisting of untranslated mRNAs bound to specific RBPs. Accumulation of such mRNPs has a tendency to condense in a spatially organized network of protein-protein (Kedersha et al., 2016; Panas et al., 2016), protein-RNA, and RNA-RNA (Jain et al., 2016; Van Treeck et al., 2018; Van Treeck and Parker, 2018; Van Treeck and Parker, 2019) multivalent interactions, that can assemble in a phase-separation (Kroschwald et al., 2015; Molliex et al., 2015; Van Treeck et al., 2018) and density transitions processes leading to hierarchical assembly of RNA granules that are reminiscent to SG (Deniz, 2020). In this model, the excess of the free pool of untranslated RNPs and condensation of its RNA component may be central in initiating the formation of SG. This RNA condensation-based mode of SG assembly has been widely recapitulated in in vitro experiments of the formation of biocondensates using RNA and recombinant RBPs, validating the central role of untranslated mRNPs in triggering the formation of SG-like condensates (Deniz, 2020), either downstream or concomitant with the inhibition of translation initiation.
The requirement of untranslated mRNPs in nucleating SG is also consistent with previous in vivo data reporting the assembly of SG in a stress-independent manner. We (Mazroui et al., 2006) and others (Dang et al., 2006) have initially showed that targeting eIF4A, the RNA helicase required for stabilizing the interaction between the cap-binding complex and RNA, either genetically using specific siRNAs, or pharmacologically using hippuristanol and pateamine, is sufficient to induce SG. This formation of SG does not require the activation of a stress response that normally triggers phosphorylation of eIF2α. Recent mechanistic in vivo and in vitro studies showed that eIF4A, through its helicase activity, antagonises SG formation, in part by reducing RNA condensation (Tauber et al., 2020), further supporting the possibility that condensation of mRNPs such as those released from dissociating polysomes upon the inhibition of translation initiation, contributes to the formation of SG. Collectively, current data strongly support the integral role of untranslated mRNPs in nucleating SG, which is consistent with proteomic, transcriptomic, and epitranscriptomic data revealing mRNPs as main components of SG (Jain et al., 2016; Protter and Parker, 2016; Anders et al., 2018). Whether the formation of SG, driven by mRNP condensates, acts as a feed-back translational regulatory mechanism that forces SG-associated mRNPs to maintain an untranslated conformation, is still a matter of a debate. In this review, we highlight data investigating i) the interplay between the formation of SG and translational signaling pathways, ii) the role of RNA in regulating SG, and iii) discuss current data implicating SG in translation regulation.
Formation of SG is driven by various pathways and factors (summarised in Table 1; see Figures 1, 2) that regulate translation (Panas et al., 2016). The main translational regulatory pathway shown to trigger the assembly of SG is the phosphorylation of eIF2α (Kedersha and Anderson, 2002; Kedersha et al., 2013), a well-established modified translational factor acting as both a general inhibitor of translation initiation (Holcik and Sonenberg, 2005) and activator of specific translation (Koromilas, 2014), during translational stress. eIF2α phosphorylation-dependent formation of SG occurs during various types of translational stress including oxidative (i.e., arsenite treatment), reticulum, nutritional and proteotoxic stress, This establishes SG a novel and reliable marker of translational stress, downstream of eIF2α phosphorylation, potentially serving as a refuge of untranslated mRNAs that have been either translationally stalled with ribosomes at the initiation step due to eIF2α phosphorylation, or released from collapsing polysomes that cannot resume additional rounds of translation (Kedersha and Anderson, 2009). While this view supports a potential functional link between the induction of SG and the phosphorylation of eIF2α in reprograming translation during stress, the two processes can be however uncoupled. This is well-illustrated in studies showing that SG can also form independently of eIF2α phosphorylation, upon either inhibiting the formation of TC or affecting the activity of specific eIFs (Table 1; see also Figure 2).

FIGURE 1. Non-canonical and canonical SG. (A) Non-canonical SG assemble as a result of either eIF4F inactivation that is mediated by hypophosphorylated 4EBP1 induced during either hypoxia, or treatment with chemotherapeutic agents such as vinorelbine, or upon eIF4GI cleavage triggered by poliovirus infection. (B) Formation of canonical SG is initiated when a stress activates one of the four stress eIF2α kinases leading to the phosphorylation of eIF2α and downstream inhibition of translation initiation. Additional translation pathways involved in the formation of canonical SG include the activation of mTORC1 and inactivation of eIF4A.
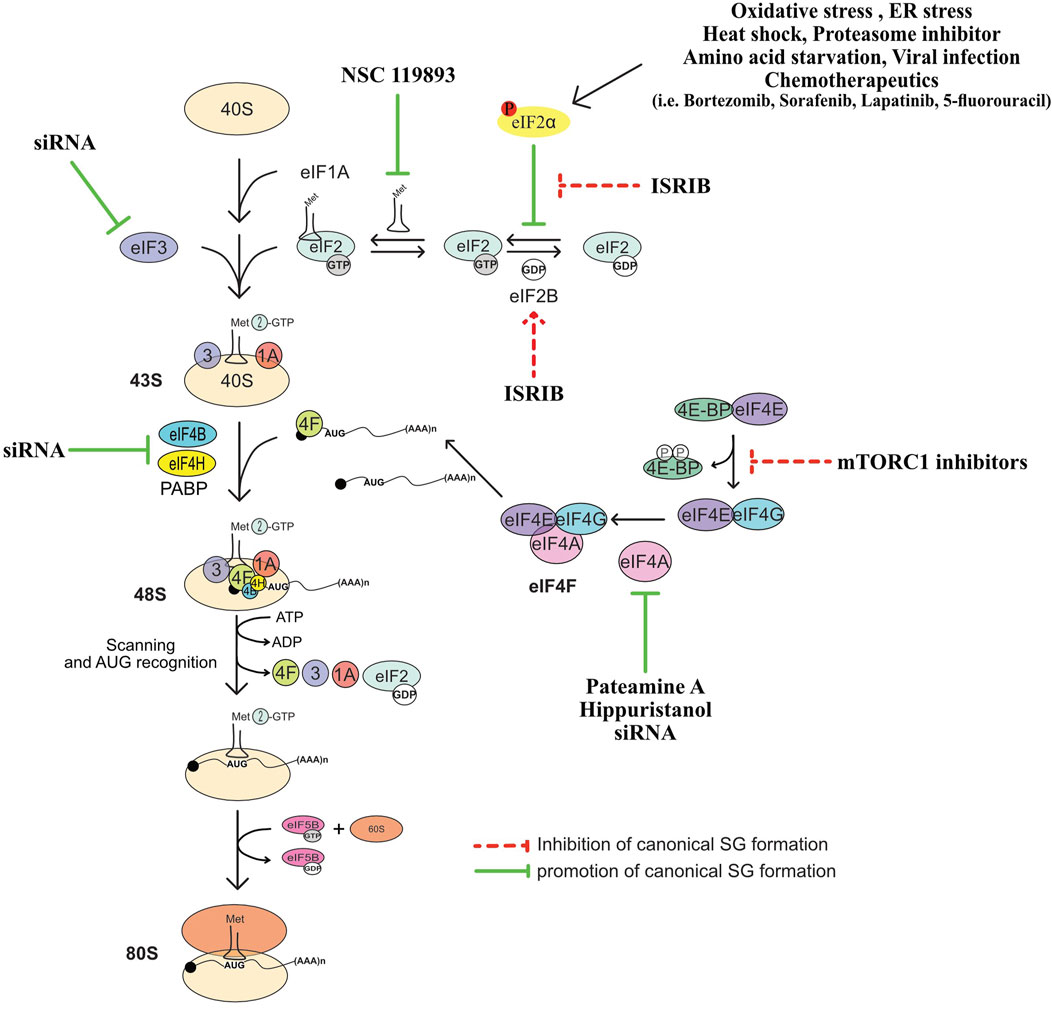
FIGURE 2. Summary of translation initiation steps and factors whose regulation affect the formation of SG. See text for more details.
NSC 119893 is a chemical compound that inhibits translation initiation by specifically preventing the association of Met-tRNAi with eIF2 and limiting the availability of TC for translation (Robert et al., 2006). Because NSC 119893 targets TC, it should not allow the formation of stalled 48S complexes. Our group showed however that treatment of human cells with NSC 119893 induces SG, which as expected occurs without induction of eIF2α phosphorylation and do not trigger the formation of translationally stalled 48S complexes (Mokas et al., 2009). Whether NSC-induced SG is driven by the accumulation of untranslated mRNPs caused by the collapse of polysomes, is a possibility that remain to be tested. Nevertheless, these data show that targeting TC is sufficient to drive SG, providing initial evidence that the formation of SG is a translational response mechanism that is activated when the rate of translation initiation is reduced.
In initial studies testing the contribution of eIFs in SG formation, we and others have found that targeting eIF4A with two translational initiation inhibitors, namely, hippuristanol that binds selectively to the eIF4A carboxy-terminal domain and inhibits RNA interaction by stabilizing eIF4A in a closed conformation (Bordeleau et al., 2006a; Lindqvist et al., 2008), and pateamine A that stabilizes RNA-bound eIF4A and stimulates its helicase activity while inducing its depletion from the eIF4F complex (Bordeleau et al., 2005; Low et al., 2005; Bordeleau et al., 2006a; Bordeleau et al., 2006b; Bordeleau et al., 2008; Cencic et al., 2009), is sufficient to drive the formation of SG. Consistently, depletion of eIF4A with specific siRNAs induces SG. Depleting eIF4H and eIF4B, the two auxiliary eIF4A factors, is also sufficient to induce SG (Mokas et al., 2009), though less efficiently than pharmacological inhibition of translation initiation. Whether the three proteins (eIF4A, eIF4H and 4B) act in concert to dampen SG is currently unknown. Nevertheless, these studies raised the possibility that specific eIFs may play additional and protective roles by actively antagonising fortuitous or constitutive formation of SG that would impact either translation or SG-associated signaling pathways under physiological growth conditions. In this case, the activity of such SG-inhibitory factors should be antagonised during stress to facilitate the formation of SG.
Among eIFs, eIF4A is the most abundant translation initiation factor (Duncan et al., 1987) making it a strong candidate for antagonising SG. Accordingly, overexpression of eIF4A in U2OS prevents the formation of SG upon arsenite treatment (Tauber et al., 2020). This effect is mediated by its ATP-dependent RNA binding that stimulates its RNA helicase activity. The authors further assessed this assumption using an SG-deficient U2OS model (Tauber et al., 2020). These cells are unable to form SG upon arsenite treatment as they lack G3BPs, the main RBP known to nucleate SG (Tourriere et al., 2001), potentially by inducing RNA condensation. Using this model, the authors showed that either depleting eIF4A, or inactivating its helicase activity using hippuristanol, rescue the formation of SG upon arsenite treatment (Tauber et al., 2020). On contrary, pateamine A which inhibits eIF4A’s function in translation while stimulating its RNA binding and helicase activity (Bordeleau et al., 2005; Bordeleau et al., 2006a) fails to restore that formation of SG (Tauber et al., 2020). Thus, targeting the helicase activity of eIF4A is sufficient to rescue the formation of SG in absence of the G3BP-inducing RNA condensation, indicating that eIF4A antagonises SG, possibly by preventing the formation of RNA condensates through its helicase activity. This is consistent with in vitro data using eIF4A mutants showing that the ATP-dependent RNA binding of eIF4A, which stimulates its helicase activity, is required for antagonising the assembly of RNA granules (Tauber et al., 2020). Thus, besides the well-characterised role of the helicase activity of eIF4A in driving translation, this activity is also necessary to prevent condensation of untranslated RNAs and associated formation of SG. However, the role played by eIF4A in limiting SG by preventing condensation of RNA is independent from its canonical translational role played as a component of eIF4F, which by unwinding the 5′UTR of target mRNAs, it stabilises their interaction with 40S during translation initiation. On contrary, the possibility that eIF4A drives the formation of SG by acting as a translational repressor was raised. Rocaglamide (Roc A), an inhibitor of translation initiation, converts eIF4A to a translational repressor by clamping it onto mRNA, thus preventing the scanning of the 5′UTR of mRNA by the 40S and subsequent formation of the 48S ribosome (Iwasaki et al., 2016), inducing RNA condensation. Consistently, by altering the function of eIF4A in antagonising RNA condensation, Roc A induces SG in various human cell lines such as U2OS or HAP1 leukemia cells (Kedersha et al., 2016; Aulas et al., 2017a; Aulas et al., 2017b), further supporting the assumption that eIF4A acts as a chaperone that prevents RNA condensation and associated formation of SG.
The finding that eIF4A prevents the formation of SG provides opportunities to test the possibility that this formation constitutes a general downstream target of specific eIFs. However, besides eIF4A, no additional eIFs known to activate translation has been shown to antagonise SG, potentially reflecting a unique function of eIF4A as an RNA helicase required for the prevention of RNA condensation-mediated SG formation.
The role of the eIF4F complex in regulating SG has been investigated under various translational stresses. As an initial finding, the formation of SG was reported to occur in response to poliovirus infection, which inhibits host protein synthesis in part via the cleavage of eIF4G (Mazroui et al., 2006). However, because poliovirus infection induces eIF2α phosphorylation, albeit at a minimal level (Mazroui et al., 2006), it is difficult to conclude that poliovirus induces SG by disrupting eIF4F as a result of eIF4G cleavage. Follow-up studies (Piotrowska et al., 2010; Yang et al., 2018) showed that SG induced by either poliovirus or related virus such as picornavirus EV71, and coxsackievirus infections, are atypical as they lack G3BP1 and a series of eIFs and thus are compositionally distinguished from canonical SG (Figure 1). The formation of these virus-induced SG requires the cleavage of eIF4GI, are independent of eIF2α phosphorylation, and they can specifically sequester cellular mRNAs but not viral mRNAs. How the cleavage of eIF4GI triggers the formation of such SG remained unknown, though it may involves the disruption of eIF4F (Yang et al., 2018).
Atypical SG (Figure 1) lacking classical SG components such as eIF3 and eIF5A also form in mammalian cells upon nutritional (Fujimura et al., 2012), hypoxia (Emara et al., 2012), or chemotherapeutic (Szaflarski et al., 2016) stress, respectively. The assembly of these non-conventional SG involves both a partial inactivation of eIF4F and the phosphorylation of eIF2α. Stress-inducing inactivation of the eIF4F is generally mediated by hypophosphorylated form of 4EBP1, a specific competitor of eIF4GI for eIF4E binding (Haghighat et al., 1995; Haghighat et al., 1996; Haghighat and Sonenberg, 1997). Hypophosphorylation of 4EBP1 (Haghighat et al., 1995; Haghighat et al., 1996; Haghighat and Sonenberg, 1997; Gingras et al., 2001) occurs when its upstream mTORC1 kinase is inactivated (Hara et al., 2002; Kim et al., 2002), resulting in the loss of the active eIF4F complex (Richter and Sonenberg, 2005; Thoreen et al., 2009; Topisirovic and Sonenberg, 2011; Thoreen et al., 2012). Both hypophosphorylation of 4EBP1 and the loss of eIF4F has been observed to occur concomitantly with the formation of the non-conventional SG (Emara et al., 2012; Fujimura et al., 2012; Szaflarski et al., 2016), suggesting that 4EBP1-mediated disruption of eIF4F drives the formation of such SG. Consistently, preventing the loss of the eIF4F by depleting 4EBP1 (Emara et al., 2012; Fujimura et al., 2012; Szaflarski et al., 2016) downregulates that formation of SG. These data are a validation that 4EBP1 promotes the formation of a specific class of SG. The underlying mechanism of 4EBP1-induced SG remains unknown, though it may involve accumulation of free untranslated mRNAs resulting from the loss of eIF4F. It is however intriguing that in contrast to SG-nucleating proteins, 4EBP1 do not localise at any type of SG, making unlikely the possibility that 4EBP1 directly nucleates SG. Also, none of the above studies used inhibitors or knockdowns testing whether this 4EBP1-mediated SG formation depends on its upstream regulatory mTORC1. The assembly of similar 4EBP1-dependent SG has also been documented to occur in mammalian cells such as HeLa, and human retinal pigment epithelial (RPE1) cells, as a consequence of the ATP depletion due to the blockade of glycolysis (Wang et al., 2022). However, these 4EBP1-dependent SG do not involve the phosphorylation of eIF2α, raising the possibility that their assembly requires additional pathways not previously reported, and which are potentially energy-dependent. The main energy-dependent pathway known to regulate SG involves the eIF4A helicase activity that antagonises RNA condensation (Tauber et al., 2020). In this model, ATP hydrolysis is required for the binding of eIF4A to untranslated RNA, preventing its condensation and SG formation. The finding that glycolysis blockade reduces the association of eIF4A with RNA targets (Wang et al., 2022) is conform with the assumption that reducing the intracellular ATP level dampens the binding capacity of eIF4A to RNA, resulting in its condensation. It remains however unclear if this decreased binding of eIF4A to RNA contributes to the formation of 4EBP1-dependent SG, and whether eIF4A antagonises that formation. As an additional emerging question is whether 4EBP1 drive the formation of SG by inhibiting the activity of eIF4A, or by inducing its displacement from eIF4E promoting the condensation of eIF4E-bound RNA.
However, depletion of 4EBP1 do not affect the assembly of conventional SG (Figure 1) (Fournier et al., 2013). These results ruling out an essential role for 4EBP1 in promoting the formation of classical SG is also consistent with the finding that 4EBP1 is excluded from SG (Sukarieh et al., 2009). Significantly, expressing hypophosphorylated 4EBP1, but not the 4EBP1 control mutant that cannot alter eIF4F, inhibited the formation of SG (Fournier et al., 2013). These data established 4EBP1 as a novel factor that antagonises the assembly of SG, potentially by disrupting the eIF4F complex. As a support, we and others have found that disrupting eIF4F by inducing 4EBP1 hypophosphorylation upon inhibition of mTORC1 either pharmacological using pp242 and torin1 inhibitors or genetically by downregulating mTOR, prevents the assembly of conventional SG in mammalian cells (Fournier et al., 2013; Mazan-Mamczarz et al., 2015; Sfakianos et al., 2018; Heberle et al., 2019), while TSC2 deficiency, known to hyper-activate mTORC1, increases their number (Kosmas et al., 2021). Similarly, both mTORC1-activating kinases PI3Ks and MAPK14 promote SG formation (Brown et al., 2011; Heberle et al., 2019), further supporting that a stress-activated signaling network converging on mTORC1-mediated phosphorylation of 4EBP1 promotes SG formation, potentially involving the activity of the downstream eIF4F complex.
Validation of the SG-promoting role of eIF4F is brought in depletion and pharmacological targeting experiments showing that eIF4E interaction with eIF4G1 drives the formation of conventional SG, which is also consistent with data showing that poliovirus-mediated cleavage of eIF4GI inhibits the formation of SG (Yang et al., 2018). The finding that canonical translation initiation factors promote the formation of SG is intriguing, thought predicting that they may play specific functions during translational stress by driving the formation of RNA granules. Based on both yeast (Hilliker et al., 2011) and mammalian (Fournier et al., 2013; Mazan-Mamczarz et al., 2015; Sfakianos et al., 2018; Heberle et al., 2019) studies, the SG-promoting functions of the eIF4F complex involves an eIF4G-mediated recruitment of specific translational repressors that nucleate SG, though other possibilities may exist. The requirement of eIF4F for the assembly of SG may involve its role in forming the 48S complex whose accumulation as stalled complexes during stress induces SG. This assumption is consistent with the finding that eIF3, whose interaction with eIF4F is required for the formation of 48S, acts also as a SG-assembling factor (Ohn et al., 2008). Alternatively, eIF4F may drive SG formation by acting as a complex that sequesters its eIF4A component preventing its function in antagonising RNA condensation. Although evidence supporting this assumption is still lacking, it is consistent with the finding that the three components of the eIF4F complex are also core components of SG. In any case, the opposite role of the eIF4E and eIF4GI relative to eIF4A in regulating SG during stress is surprising as the three proteins function as part of the same eIF4F complex to activate translation.
eIF5A was initially described as a translation initiation factor involved in the first peptide bond formation (Kemper et al., 1976; Safer et al., 1976). Since then, depletion experiments revealed that eIF5A promotes peptide transfer during translation elongation (Schuller et al., 2017). An RNAi screen conducted in human RDG3 and U2OS cells identified eIF5A as a factor required for the assembly of SG during oxidative stress (Ohn et al., 2008). Interestingly, eIF5A promotes that formation of SG by enhancing the rate of translation elongation that allows ribosome run-off mRNAs and their subsequent condensation, when translation initiation is inhibited (Li et al., 2010). The possibility that the rate of translation elongation affects the assembly of SG is consistent with earliest studies (Kedersha et al., 2000; Kedersha et al., 2002) using translation elongation inhibitors. In these studies, the formation of SG was efficiently prevented upon treatment with either cycloheximide or emetine, which both neutralise the elongation step by stalling polysomes, thereby reducing the pool of free mRNPs available for initiating SG. On the contrary, treatment with puromycin, a tRNA analog that induces premature termination by destabilizing polysomes, enhances SG formation upon stress (Kedersha et al., 2000). The inhibition of translation elongation may also allow translation initiation complexes whose activity is reduced by stress to complete their round of initiation and then be trapped in polysomes, further decreasing the pool of translation initiation complexes available for SG. Similarly, elongation slowdown that naturally occurs during mitosis induces stalling of polysomes and render mitotic cells refractory to SG formation even if translation is inhibited at the same time (Sivan et al., 2007). Thus, the initiation of SG requires an ongoing active translation elongation. In this case, factors such as eEFs and RBPs involved in translation elongation should also contribute to the formation of SG, when translation initiation is blocked, a possibility that remains to be tested.
RBPs affect the rate of translation either at the initiation step by regulating the formation of translation initiation complexes, or at the elongation step by regulating the polysomes runoff mRNAs. Among RBPs required for the formation of SG, many regulate translation initiation (Ohn et al., 2008), while none has been characterised as translation elongation regulatory factors (Table 2; see also Figure 3). G3BP1 is a selected SG-associated RBP whose role in SG formation has been extensively studied (Alam and Kennedy, 2019). Like many other SG components known to form condensates, G3BP1 contains intrinsically disordered regions (IDRs). IDR-IDR interactions are necessary for the in vitro phase separation of a number of SG factors including hnRNPA1, FUS, and TDP43 (Molliex et al., 2015; Patel et al., 2015; Wang et al., 2018; Conicella et al., 2020a; Conicella et al., 2020b), enables G3BP1 ability to form condensates (Yang et al., 2020) and to act as a scaffold factor for nucleating SG (Guillen-Boixet et al., 2020). G3BP1 drives SG formation also by inducing the condensation of untranslated RNPs that are released from dissociating polysomes during the inhibition of translation initiation (Guillen-Boixet et al., 2020), either directly or indirectly by antagonising the activity of RNA helicases such as eIF4A (Tauber et al., 2020; Yang et al., 2020). The latter possibility is supported by the finding that depleting eIF4A or inactivating its helicase activity rescue stress-induced SG in cells lacking G3BP1 (Tauber et al., 2020). However, G3BP1 may also alter SG upstream of RNA condensation, by regulating translation elongation. Consistently, G3BP1 was reported to regulate translation either positively or negatively, involving interactions with either the 40S ribosomal subunits (Kedersha et al., 2016; Gotte et al., 2019), with translation elongation regulators such as poly (ADP ribose) polymers (Mateyak and Kinzy, 2013), or with the RBP named Caprin 1 (Kedersha et al., 2016). While these G3BP’s interactions have been shown to promote SG formation under stress, it remains unknown if the underlying mechanisms involve translation elongation regulation.
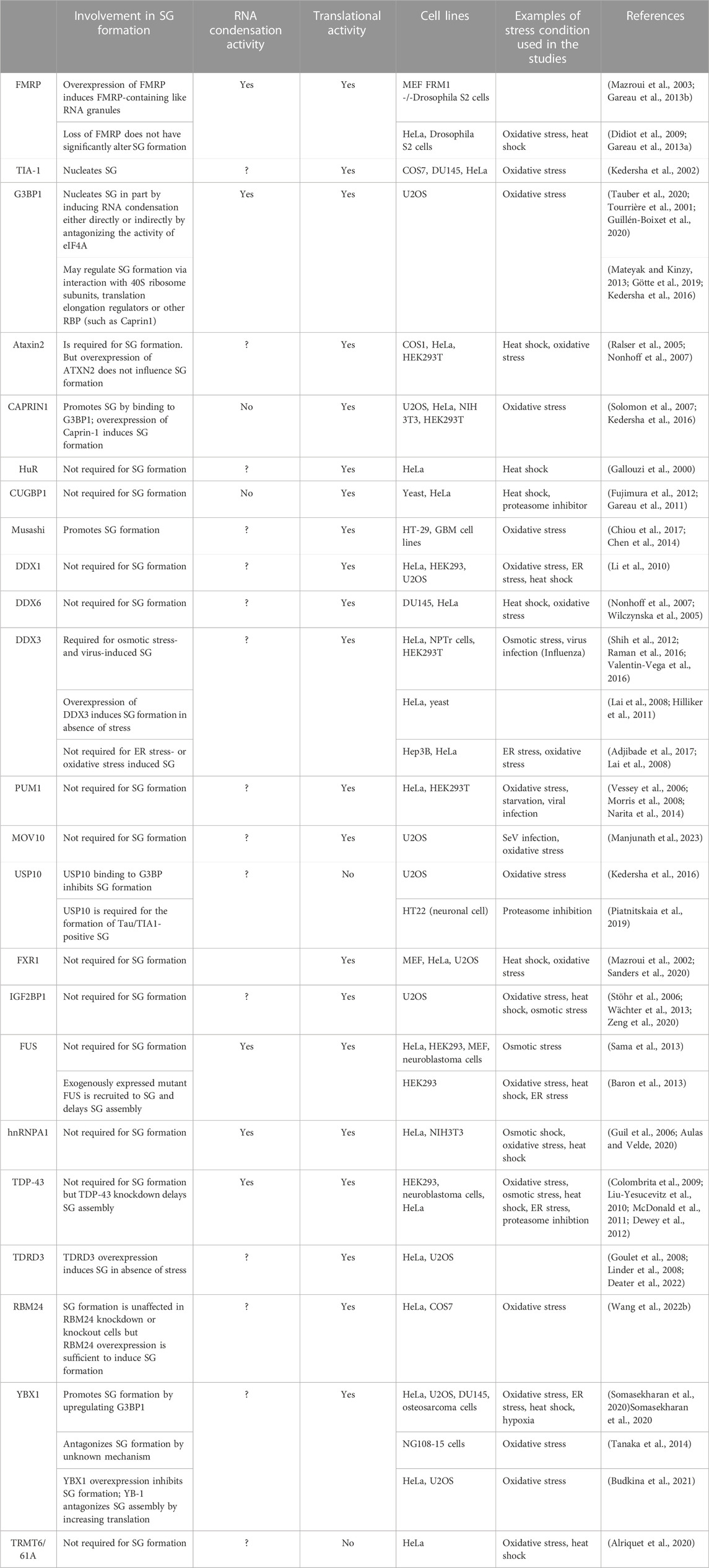
TABLE 2. Role in SG formation of a list of SG-associated RBPs with RNA condensation or translation activity.

FIGURE 3. List of RBPs involved in the formation of SG (see Table 2 for details).
Additional RBPs with both translational and RNA condensation activities include FMRP, a core SG component (Mazroui et al., 2002; Mazroui et al., 2003). Initially, FMRP was shown to repress translation initiation by inactivating eIF4F (Napoli et al., 2008). More recent studies implicated FMRP in repressing translation elongation by stalling polysomes (Darnell et al., 2011), while mechanistic studies showed that the drosophila homologue of FMRP inhibits translation elongation by blocking the binding of tRNA and translation elongation factors on the ribosome (Chen et al., 2014). Validation of the translational role of FMRP was provided in experiments showing that it represses translation as a phospho-protein, a process prevented by its methylation (Tsang et al., 2019). Methylation of FMRP also antagonises its ability to phase separate with RNA in vitro, while FMRP-RNA phase separation is enhanced by its phosphorylation (Tsang et al., 2019). The possibility that FMRP induces RNA condensation by phase separation is consistent with its ability to nucleate RNA granules in in vitro experiments (Tsang et al., 2019). Similarly, we have shown that overexpressing FMRP in either mammalian or drosophila cells induces formation of FMRP-containing cytoplasmic RNA granules. These FMRP-granules resemble SG as they contain SG components including mRNAs, eIF4E and PABP (Mazroui et al., 2003; Gareau et al., 2013b), and are dynamic foci, as demonstrated by FRAP experiments (Gareau et al., 2013b). However, on contrary to SG, the formation of FMRP-granules occurs without inhibition of the translation initiation, raising the possibility that formation of such RNA granules is potentially driven by RNA condensation, independently of the inhibition of translation initiation. Whether FMRP induces RNA condensation by regulating translation elongation is not known. Intriguingly, while FMRP-induced RNA condensation nucleates RNA granules, is it not sufficient to induce SG, because the loss of FMRP do not significantly alters SG (Didiot et al., 2009; Gareau et al., 2013a). It is however possible that the loss of the activity of FMRP in driving SG is compensated by similar activities of FXR1 and FXR2, the two highly translational regulators and functional homologues of FMRP. Additional RBPs that regulate translation and induce RNA condensates through their phase separation properties include FUS, hnRNPA1 and TDP-43 (Portz et al., 2021). However, none of these proteins has been reported as essential for the formation of mammalian SG upon stress. This suggests that the RNA condensation mediated by individual RBP by itself is not sufficient to drive SG formation. On the other hand, other RBPs with translational activity such as Caprin 1 promote SG formation in vivo, yet it does not form RNA condensates in vitro (Kedersha et al., 2016). This finding indicates that RBPs may also promote SG formation independently of RNA condensation. Clearly, while they are accumulating evidence predicting that RNA condensation mediated by RBPs contribute to the formation SG (Table 2), it remains unclear if this RNA condensation is a general mechanism associated with their translational activity.
RNA is an integral component of SG. It is estimated that 78–90% of mammalian SG are composed of RNA, with a major fraction of the SG-transcriptome (75%) is composed of mRNA (Khong et al., 2017; Curdy et al., 2023). However, the role of mRNA in regulating the biology of SG remains not fully understood (Table 3). In vitro studies showed that RNA molecules can form homo- and heterotypic interactions with other RNA molecules, leading to RNA condensation and phase separation with RBPs, nucleating RNA granules (Campos-Melo et al., 2021). While there is no direct experimental evidence that demonstrate the essentiality of RNA condensation in SG assembly, it is accepted that this assembly involves the accumulation of free mRNAs that are released from dissociating polysomes when the initiation step of translation is blocked (Kedersha et al., 2000). Accordingly, cell delivery of nucleic acids that cannot associate with polysomes is sufficient to trigger SG (Bounedjah et al., 2014) further indicating that increasing the pool of free RNA contributes to the assembly of SG, potentially through a phase separation and condensation processes. This also raised the possibility that SG originate as a consequence of an altered balance between polysomal- and free-RNA, involving the action of specific RBPs. In normal growth conditions, the low level of free RNA may preclude intensive RNA-RNA interaction and association with aggregating RBPs. Upon stress, the accumulation of free RNA causes an extensive RNA-RNA interactions that trap specific aggregating RBPs, which further promote RNA condensation, inducing SG. Clearly, better understanding of the role of RNA in regulating SG requires further studies investigating the underlying mechanisms regulating the sorting of mRNAs in SG inducing their nucleation.
To better understand the role of RNA in assembling SG, a number of transcriptomic studies have been developed (Table 4; see also Figure 4), providing catalogues of mRNAs that are enriched in SG, and which may be critical for their assembly. As an initial study, Khong et al interrogated the transcriptome of SG that are induced by arsenite treatment of U2OS-stably expressing G3BP1, and isolated by G3BP1 immunoprecipitation as core SG (Khong et al., 2017). Thousands of different RNA species have been found in those SG, and proportionally, there is more RNA than protein in SG as compared to the cytoplasm, further supporting a predominant role of RNA in regulating SG. Quantification of RNAseq data obtained from purified SG-G3BP1 relative to total RNA show that only ∼10% of bulk mRNAs is enriched in SG, though most mRNA species are detected in SG. The minimal fraction of mRNAs accumulating in SG also parallels the fraction of polyA + mRNA detected in SG by oligo (dT) FISH (Khong et al., 2017). The finding that only ∼10% of total cellular mRNA accumulates in SG also led to the conclusion that SG may not impact global translation, though it may affect translation of specific mRNAs that are highly accumulating in SG. However, the translatome itself is minimal when compared to the total transcriptome, indicating that a significant fraction of mRNAs in the cell is kept untranslated. At this stage, there is no direct estimation of the relative fraction of mRNAs that accumulate in SG as compared to the fraction of mRNAs that are actively translated, i.e., associated with translating polysomes. Such estimations are important to define the percentage of the translatome that is trapped in SG, better predicting the impact of SG has on translation. Bioinformatics analysis of the identified transcriptome provided evidence that mRNAs highly accumulating in the core SG-G3BP1 tend to have longer length. These mRNAs were inferred to have poor translation efficiency (Khong et al., 2017), as they have been previously shown to be less loaded by ribosomes (Sidrauski et al., 2015a; Sidrauski et al., 2015b). This finding was validated in a Padron et al. 2019 study using the RNA proximity labeling technique, APEX-seq. APEX is a proximity labeling approach that was originally developed to identify in vivo protein-protein interactions. It employs an engineered ascorbate peroxidase enzyme (APEX2) to convert a cell-permeable biotin-tyramide substrate into a highly reactive free radical that labels aromatic amino acids in proteins within few nanometers (Rhee et al., 2013) that can be isolated by streptavidin pull-down and identified by sequencing. Padron et al adapted APEX to identify SG-associated RNAs. They fused APEX2 to the core SG component eIF4A1, which is then expressed in HEK293T cells, validating efficient in vivo biotinylation of total RNA through APEX2 reaction. Similarly to endogenous eIF4A1, APEX-eIF4A1 localizes in SG upon heat shock. RNA in close proximity with APEX-eIF4A1 in stressed cells are then biotinylated. Modified RNA are pulled-down and identified by APEX-seq. Data showed a significant overlap between RNA identified by APEX-seq, potentially representing heat shock-induced SG transcriptome, with those identified in Khong et al. 2017 study. Overexpression of eIF4A1 was however previously shown to impact arsenite-induced SG (Tauber et al., 2020). Expressing APEX-eIF4A1 may thus also impact heat shock-SG, which in this case will preclude the identification of a complete fraction of SG-mRNAs by APEX. Moreover, little correlation was observed between SG-mRNAs identified by APEX-seq upon treatment with the eIF4A1 inhibitor hippuristanol (Padron et al., 2019) with those identified associating with arsenite-induced SG (Khong et al., 2017), possibly reflecting differences between the mechanisms underlying the recruitment of mRNAs into arsenite-SG with those into hippuristanol-SG. Nevertheless, mRNAs identified by APEX-seq have also long length with lower translational efficiency, similar to those shown to accumulate in SG-G3BP1induced by arsenite (Khong et al., 2017). mRNAs with high translation rate and shorter length seem to be either excluded from SG or associated transiently and less stably with SG. The observation that mRNAs with high translation efficiency are less associating with SG is intriguing considering that the assembly of SG requires the accumulation and condensation of free mRNAs that are released from translating polysomes. Nevertheless, the observation that longer and poorly translated mRNAs are highly enriched in SG is consistent with the in vivo single molecule mRNA translation reporter assays showing that longer and repressed reporter mRNAs interact stably with SG (Moon et al., 2019). The preferential enrichment of mRNAs that have poor translation efficiency and longer length in SG may reflect a high tendency to condensate by engaging in extensive RNA-RNA interaction and by associating with multiple aggregating RBPs, relative to mRNAs with higher rate of translation and shorter length. In any case, the cis- (i.e., RNA elements) and trans- (i.e., RBPs interactors) mechanisms underlying the enrichment of longer and low translated mRNAs in SG remained to be defined. SG-associated mRNAs tend also to have shorter half live, which may indicate that their selective association with SG protects their rapid decay, though this assumption was challenged in previous yeast studies showing that genetic alteration of the formation of SG do not affect the decay of tested mRNAs (Buchan et al., 2008), and in recent reporter assays showing that the disassembly of SG do not accelerate the decay of associated reporter mRNAs (Wilbertz et al., 2019). The SG-transcriptome was also captured in HEK cells expressing the SG nucleating TIA protein using a modified PAR-CLIP (Anders et al., 2018). This approach is based on photoactivatable ribonucleoside cross-linking reaction of RNAs to TIA that stabilizes SG, allowing their isolation by immunoprecipitation using anti-TIA antibodies, followed by RNAseq. This approach permitted the capture of additional mRNAs as compared to the list of RNAs associated with SG-G3BP1 (Khong et al., 2017). Comparing the SG-transcriptome obtained by PAR-CLIP with Ribo-seq data which classifies mRNAs as high or low translated RNA, also support previous finding that mRNAs accumulating in SG have low translational level.
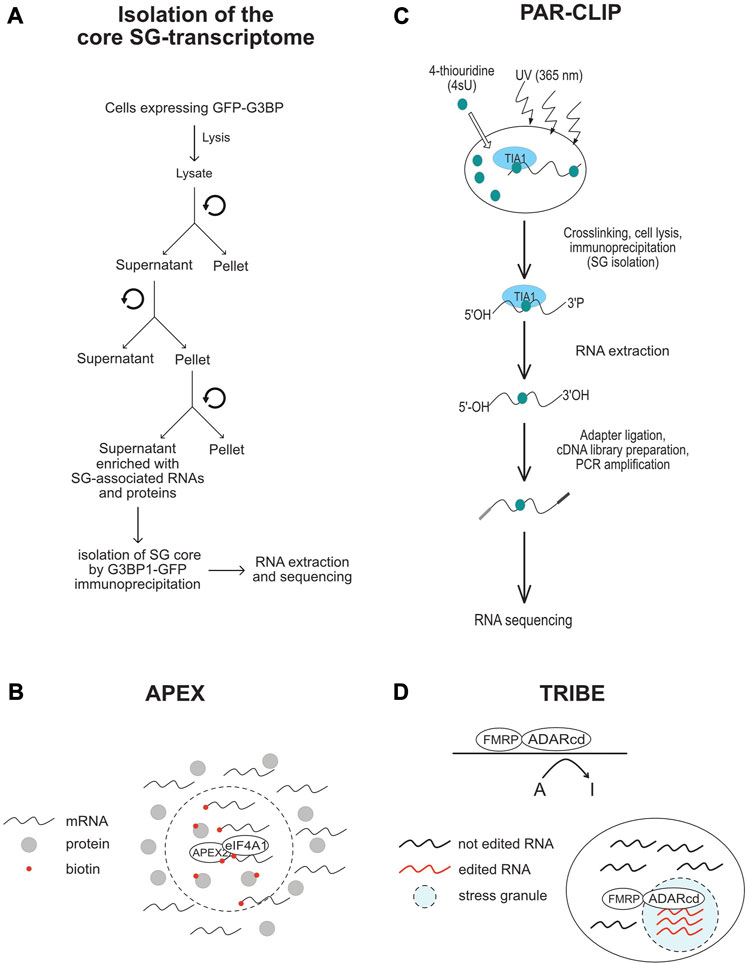
FIGURE 4. Schematic representation of the methods that are used in SG-transcriptomic studies described in the text. (A) Isolation of core SG-transcriptome centrifugation (B) APEX2-seq. (C) Workflow of a PAR-CLIP experiment. (D) TRIBE-seq.
To further define the nature of mRNAs accumulating in SG, Van Leeuwen et al used the TRIBE (target of RNA-binding proteins identified by editing) technique to analyse the SG-transcriptome of drosophila embryonic and neuronal cells (van Leeuwen et al., 2022). In TRIBE, the SG component dFMRP (Gareau et al., 2013a) is fused to the catalytic domain of the DNA/RNA-editing enzyme ADAR (ADARcd) that deaminates adenosine-to-inosine on RNA molecules, an editing event that can be quantified upon sequencing. Upon arsenite stress, dFMRP-ADARcd-V5 is recruited to SG, potentially with associated-edited RNA to SG. dFMRP-ADARcd-V5 may also edit target RNAs in SG. By analysing the editing frequency of isolated RNA, the authors found a number of RNA to be specifically edited upon arsenite treatment, potentially corresponding to the SG-transcriptome, though a subset may represent edited mRNAs that are not associated with SG. In particular, expressing dFMRP by itself induces RNA granules that resemble SG (Gareau et al., 2013b), and thus it may be difficult to discriminate RNA edited in FMRP-granules and those in SG. dFMRP mutants that do not associate with SG, while still bind RNA may be used in TRIBE as control eliminating RNA that are edited independently of their association with SG. It remained also unclear whether the mRNAs identified by TRIBE are selectively enriched in SG, and whether they have reduced rate of translation, similar to SG-transcriptomes identified by other studies. Using ADARcd-V5 fused to other RBPs that associate (such as ELAV or Rasputin) or not (DCP1) with SG should also help define the SG transcriptome. Despite these gaps, TRIBE offers an opportunity to analyse SG-associated RNA in single cells, which may reveal specificities in the RNA composition of SG and probably also its role played at individual cells. Collectively, these transcriptomic studies are highly informative on the identity of mRNAs that associate with SG, providing evidence that the sorting of mRNAs in SG depends on their translational rate. However, while these data further support the assumption that mRNAs are not equally sorted during stress in SG, whether this sorting involves specific cis- and trans- RNA based mechanisms, remained an open question.
Growing evidence indicate that important aspects of mRNA metabolism including splicing, stability, localization and translation are affected not only by cis-acting mRNA sequences such as the binding motifs of RBPs, but also by specific secondary structures adopted by mRNAs upon folding, and by chemical modifications occurring at their bases (Lewis et al., 2017). While it is unclear if the localisation of mRNAs in SG relies on specific cis-acting sequences and secondary structures, recent data implicated specific chemical modifications of mRNA in their sorting during stress, potentially promoting their accumulation as untranslated mRNPs in SG through interaction with specific RBPs (Table 5).
Well-studied RNA internal chemical modifications include N6-methyladenosine (m6A), N6, 2′-O-dimethyladenosine (m6A), deamination of adenine to inosine (I), 5-methylcytidine (m5C) and 5- hydroxylmethylcytidine (hm5C), N1-methyladenosine (m1A) and pseudouridine Ψ) (Roden and Gladfelter, 2021). Among the most abundant internal mRNA modification is m6A that regulates RNA stability, translation, and localization (Dominissini et al., 2012; Meyer et al., 2012; Wang X. et al., 2014; Wang Y. et al., 2014; Wang and He, 2014; Huang and Yin, 2018). This reversible modification occurs through a highly regulated process that involves the action of writer methyltransferases that transfer the methyl group to the mRNA, reader enzymes that recognise the modified mRNA, and erasers that remove the modification (He and He, 2021). The original first observations that support the possibility that m6A regulates mRNA sorting to SG and/or the dynamics of SG are based on data showing that m6A disrupts RNA binding to SG components including G3BPs, USP10, Caprin 1, and RBM24 (Arguello et al., 2017; Edupuganti et al., 2017). We and others have used anti-m6A antibodies in immunofluorescence experiments detecting m6A signals in HeLa- and U2OS-SG that are induced by a variety of stresses including arsenite (Anders et al., 2018; Adjibade et al., 2020; Fu and Zhuang, 2020), supporting the possibility that SG contain m6A-(modified) RNAs. However, data documenting if the m6A methyltransferases associate or not with SG, which would support a possibility that m6A may also occur in SG, are still lacking. Nevertheless, co-depleting the core methyltransferase components of the “writer” m6A complex, namely, METTL3, METTL14, and WTAP significantly abrogates the m6A signal within arsenite-induced SG that form in U2OS-expressing G3BP1 (Anders et al., 2018). While these depletion data validate the association of m6A-RNAs with SG, it indicates that interfering with this modification does not alter SG (Anders et al., 2018), which is also consistent with the study of Ries et al. showing that SG are normally formed in m6A-deficient (ΔMETTL3) mouse embryonic cells (mES) cells (Ries et al., 2019). However, 50% of the SG-mRNA transcriptome isolated by PAR-CLIP are m6A-modified (Anders et al., 2018) suggesting that modified RNA are selectively sorted to SG. The possibility that m6A promotes mRNAs sorting to SG is further supported by quantitative studies showing that the level of m6A in SG-associated mRNAs, is elevated during arsenite treatment (Anders et al., 2018). More correlative analyses between the m6A ratios of mRNAs established by m6A-seq (Anders et al., 2018) with RNA-seq data defining mRNAs enriched in the core SG (Khong et al., 2017), showed that mRNAs harboring high level of m6A are significantly enriched in SG, while those lacking m6A are largely depleted from SG (Anders et al., 2018). These data are consistent with immunofluorescence quantification showing an enrichment of m6A signal in SG induced in arsenite-treated U2OS (Fu and Zhuang, 2020), further supporting the selective sorting of m6A-modified RNAs to SG. The lack of m6A methyltransfreases from SG also precludes the possibility that the accumulation of m6A-modified RNAs in SG reflects their local methylation, while it supports their sorting to SG as methylated mRNAs.
m6A-RNAs are selectively recognised by YTHDFs(1–3), a family of readers that bind the m6A moiety with their YTH domain (Dominissini et al., 2012) and regulate various aspects of RNA homeostasis. Interestingly, Anders et al. (Anders et al., 2018) showed that interfering with the activity of YTHDF3 reduced the level of m6A-mRNAs in SG that form in U2OS-stably expressing G3BP1 and treated with arsenite, thought it does not affect the formation of SG. This result combined with those localizing YTHDFs in SG (Wang et al., 2015; Fu and Zhuang, 2020), supports a possible role of YTHDFs in sorting methylated mRNAs clients to SG under oxidative stress. How YTHDFs relocate m6A-mRNAs in SG is still unknown, though it may involve a phase separation process. Accordingly, YTHDFs contain low-complexity sequences that promote their phase separation observed in vitro and in (mES) cells (Ries et al., 2019). This phase separation is markedly enhanced by mRNAs that contain multiple, but not single, m6A residues (Ries et al., 2019), providing a possible mechanism underlying the selective recruitment of m6A-mRNAs in SG. Alternatively, YTHDFs-mediated m6A-mRNAs localization in SG may involve association between YTHDFs and its partners known as SG components including eIF3 and specific RBPs with known roles in translational control such as YBX1, IGF2BP1, and G3BP1. These interactions are either RNA-dependent as reported between YTHDF1 and G3BP1, or–independent as shown between YTHDF1 and eIF3, potentially underlying the role of YTHDFs in translation of methylated mRNAs (Wang et al., 2015). PAR-CLIP combined with Ribo-seq experiments in WT and m6A methyltransferase (METTL3)-deficient HeLa cells showed that YTHDFs promote ribosome occupancy of its target mRNAs, indicating that YTHDFs promote translation efficiency in an m6A-dependent manner, potentially at the initiation step (Wang et al., 2015). Whether this role of YTHDF in translation accounts for the sorting of m6A-mRNAs in SG remained however unknown.
There are also conflicting data supporting or not the role of the association of YTHDFs with SG. in regulating their dynamics. Fu et al. reported an essential role played by YTHDF1 and 3 in SG induction upon arsenite treatment of U2OS (Fu and Zhuang, 2020), contradicting initial data showing a dispensable role of YTHDFs in assembling SG in U2OS-G3BP1 (Anders et al., 2018; Ries et al., 2019). The formation of SG in U2OS-G3BP1 lacking YTHDFs is intriguing. This may be due however to the overexpression of G3BP1 impacting the effect of YTHDFs depletion in SG formation. The role of YTHDFs in the dynamics of SG was also assessed in mutagenesis studies. Overexpression of a dominant negative mutant of YTHDF1 that inhibits the binding of endogenous YTHDF to m6A-RNAs resulted in an impaired formation of SG in arsenite-treated U2OS (Fu and Zhuang, 2020), supporting the assumption that binding of YTHDFs with m6A-RNAs facilitates the formation of SG. However, both the selective association of m6A-mRNAs with SG and the role of YTHDFs binding with m6A-RNAs in facilitating the formation of SG was challenged in a more recent study (Khong et al., 2022). The dispensable role of YTHDFs in SG formation was concluded based on single-molecule FISH experiments performed in mES cells showing that selected m6A-mRNAs partition similarly into SG in both wild-type and m6A-deficient (ΔMETTL3) mES cells (Khong et al., 2022), also precluding a major role of the modification in the association of mRNAs with SG. Furthermore, tethering YTHDFs proteins to reporter mRNAs does not significantly induced their enrichment in SG (Khong et al., 2022), questioning the role of the YTHDFs binding with m6A-RNAs in their sorting and accumulation into SG. While YTHDFs are considered as the main canonical m6A readers, specific core SG components such as FMRP and its homologs FXR1 and FXR2, and G3BPs (Edupuganti et al., 2017) were recently identified as RNA sequence context-dependent m6A readers or regulators.
Through RNA pull-downs in HeLa cell lysates followed by quantitative mass-spectrometry-based proteomics, FMRP and FXR1/2 were identified as new binders of m6A-mRNA sequences, while G3BPs and its partners USP10 and Caprin1 preferentially bind the unmodified versions (Edupuganti et al., 2017). The preferential binding of FMRP to mRNA targets on m6A sites was validated by comparing FMRP PAR-CLIP data (Ascano et al., 2012) with those of m6A-seq (Meyer et al., 2012). A significant fraction of FMRP-targets was also identified in Wang et al. study as YTHDF1-binding mRNAs in their YTHDF1 PAR-CLIP experiments (Wang et al., 2015). However, on contrary to YTHDF1 that binds mainly to m6A-RNAs, FMRP binds m6A-containing mRNAs, but it is not strictly dependent on m6A for mRNA binding, raising the possibility that FMRP and YTHDF1 may compete for binding to m6A sites on mRNA, accounting for their opposite role in translation repression and activation of target mRNAs, respectively (Edupuganti et al., 2017). Consistently, preventing YTHDFs binding to target mRNAs by reducing the level of m6A through METTL3 depletion increases the inhibitory effects of FMRP on translation. This is because many putative FMRP-target sites corresponding to m6A become available for FMRP binding due to the loss of YTHDF (Edupuganti et al., 2017). A direct binding of FMRP to m6A on mRNA rather than indirectly to unmethylated regions of m6A-containing mRNAs was confirmed by combining PAR-CLIP, m6A-seq, and methylated RNA immunoprecipitation (MeRIP) in HEK293T cells (Hsu et al., 2019). This is consistent with in vitro data showing that FMRP phase-separates with m6A-modified binding RNA probe but not with the unmodified probe, forming RNA granules (Zhang et al., 2022). While these accumulating data support a role played by the SG-associating FMRP as an m6A reader, it remains unknown if FMRP mediates the localization of selected m6A-modified targets in SG. Because FMRP binds also to unmodified targets, it may compensate for the loss of the activity of YTHDFs in permitting the association of unmodified target mRNAs with SG, also accounting for the observation that the localization of YTHDFs-unmodified targets in SG is not affected by METTL3 depletion. On contrary, the finding that the association of m6A-mRNAs with SG is reduced in YTHDFs-depleted cells (Anders et al., 2018), precludes the possibility that FMRP may significantly compensate the loss of YTHDFs in aggregating YTHDF-modified targets into SG, and thus may not be sufficient for their association with SG. In addition to FMRP, G3BP1 and its partners Caprin1 and USP10 were also identified as SG components that bind YTHDFs-target mRNAs. On contrary to FMRP whose binding to mRNAs is not significantly affected by m6A, the binding of G3BP1 to mRNAs is negatively affected by the modification since the protein binds preferentially to unmodified mRNAs (Edupuganti et al., 2017). Reducing m6A levels on YTHDFs-mRNA targets in METTL3-depleted cells may thus activate the binding of G3BP1 to those mRNAs, inducing their localization to SG, despite the loss of YTHDFs binding. Clearly, all these are speculative possibilities that may be verified after knocking down candidate RBPs in METTL-depleted cells under conditions that induce SG formation.
Although less abundant than m6A, m1A modification increases during stress (Dominissini et al., 2016) and m1A-modified RNAs also localize in mammalian SG induced by either arsenite or heat shock. On contrary to m6A-methylating enzymes which do not associate with SG, the m1A-generating methyltransferase enzyme TRMT6/61A (Xiong et al., 2018a; Xiong et al., 2018b) do associate with SG (Alriquet et al., 2021). This finding is intriguing since the sequestration of TRMT6/61A in SG may a priori impact m1A in the cytoplasm of stressed cells, preventing the accumulation of m1A-modified mRNAs. However, there is possibility that TRMT6/61A may modify its target mRNAs once they are transferred to SG, resulting in their accumulation. Alternatively, a residual fraction TRMT6/61A that do not associate with SG may be sufficient to modify its target in the cytoplasm during stress, which then aggregate into SG through the action of specific readers. At this stage, however, no m1A reader has been described, raising the question of how m1A-modified mRNAs selectively aggregate into SG. Although additional studies are needed to better define the role played by RNA modifications in the association of target mRNA in SG, it become clear that such modifications constitute novel underlying mechanisms involved in the accumulation of mRNAs in SG.
The role of SG in translation is still a matter of debate (Table 6). Originally, SG were described as sites where untranslated mRNAs are trapped, potentially contributing to their repression (Kedersha and Anderson, 2009; Protter and Parker, 2016; Youn et al., 2018; Ivanov et al., 2019; Youn et al., 2019; Riggs et al., 2020). This assumption was based on several observations: i) SG form specifically during cell growth conditions that inhibit translation initiation, resulting in the accumulation of untranslated mRNAs, ii) SG disassemble when translation is recovered, reducing the pool of untranslated mRNAs, iii) mRNAs, such as Hsp70 and 90 mRNAs, whose expression is significantly increased during stress, are excluded from SG, iv) transcriptomic studies establishing that mRNAs preferentially accumulating in SG are generally low-expressed species due to their poor translation rate, and v) proteomic studies identifying RBPs involved in translation repression as key SG components, while excluding the association of the 60S ribosomal subunits with SG. On contrary, the premise that SG do not significantly contribute in maintaining the global inhibition of translation during stress was first provided by yeast studies showing that altering genetically SG has no effect on general translation (Buchan et al., 2008). The assumption that mammalian SG do not contribute to translation reprograming is largely based on observations that SG-deficient cells such as those lacking G3BPs show global inhibition of translation (Kedersha et al., 2016). However, deep analysis of translation using translatomic studies showed significant changes in translation rates between SG-forming and -deficient cells. Whether the observed differential translation is SG-dependent remains unknown. A dispensable role of SG in translation repression was also provided in reporter assays, showing that translation of specific reporter mRNAs (such as Renilla luciferase mRNAs) is inhibited throughout the cytosol, regardless of granule localization, during the stress response (Halstead et al., 2015; Horvathova et al., 2017). However, targeted approaches supported the above assumption that SG may regulate translation of specific associated mRNAs. These approaches combined FISH experiments localizing target mRNAs in SG, polysomes profiling assessing their distribution on translating polysomes, and depletion experiments testing the effects of abrogating SG on the expression of target mRNAs. Among tested mRNAs, p21 mRNA was shown to accumulate in HeLa-SG upon proteasome inhibition, potentially through binding to the SG component CUGBP1, while its expression remained low (Gareau et al., 2011). p21 mRNA became highly expressed during prolonged proteasome inhibitor treatment once it is released from dissociating SG. These correlative results suggest that p21 mRNA associated with SG is kept untranslated, though direct evidence remained to be established. A similar potential SG-mediated temporal translation has been described, either during IL1b-stimulated human osteoarthritis chondrocytes for the regulation of cyclooxygenase 2 (Cox-2) translation (Ansari and Haqqi, 2016), or during T-cell activation for the expression of the inhibitory immune checkpoints mRNAs including PDCD1, CTLA4, TIM3, and LAG3 mRNAs (Franchini et al., 2019). Ansari et al. study showed that stimulation of human osteoarthritis chondrocytes with IL1b induces an endoplasmic reticulum (ER) stress response characterized by phosphorylation of eIF2α that triggers the formation of SG. FISH experiments revealed a quantitative accumulation of Cox-2 mRNA in SG while no cox-2 protein was produced. Prolonged treatment with IL induces dephosphorylation of eIF2α and SG disassembly that correlates with Cox-2 expression. Maintaining Cox-2 mRNA in IL1b-induced SG by inhibiting SG clearance prevents Cox-2 expression, while preventing their formation accelerates Cox-2 expression. Interfering with the association of Cox-2 mRNA with SG by targeting its SG partner HuR, also accelerates Cox-2 expression, further supporting a SG-mediated temporal regulation of Cox-2 mRNA translation. Additional studies are required to determine whether this SG-mediated temporal regulation is general during cytokine-stimulated chondrocytes. Similarly, Franchini et al. reports that T-cell activation causes a stress response that induces inhibitory immune checkpoints mRNAs to form RNP-G3BP1 complexes, which aggregate into SG as untranslated form (Franchini et al., 2019). These mRNPs are then translated following maturation and disassembly of SG, consistent with a SG-mediated temporal regulation of the expression of inhibitory immune checkpoints mRNAs in activated T-cells.
Similar to T-cell, B-cell activation induces a stress response that triggers the assembly of functional SG. In this study (Diaz-Munoz et al., 2017), the impact of SG on translation of mRNAs-encoding DNA damage response factors was then tested upon treatment of activated B-cells with the DNA damaging etoposide drug, identifying p53 mRNA as one mRNA target. In activated B-cells, p53 mRNA was quantitatively present in SG but no corresponding protein was produced, indicating its translation repression. Treatment of activated B-cells with etoposide had no effect on the abundance of p53 mRNA, neither affect SG number and/or size. Similar treatment decreases however the level of p53 mRNA in SG as determined by FISH, while it induces its association with translating polysomes (Diaz-Munoz et al., 2017). It was thus concluded that such etoposide treatment induces the release of p53 mRNA from assembled SG, which then associates with polysomes, resulting in its expression. The loss of p53 mRNA from SG parallels with its dissociation from TIA, a well-known SG component, suggesting that in absence of etoposide treatment, p53 mRNA is trapped in SG as untranslated form via TIA (Diaz-Munoz et al., 2017). While the Diaz-Munoz et al. study supports a role of SG in the control of p53 mRNA temporal translation, validations are required by combining strategies that target SG, with those that allow accurate cellular quantification of p53 mRNA such as smiFISH. Including TIA mutants that associate with SG but cannot bind p53 mRNA would also be helpful to confirm the functional role of TIA in targeting p53 mRNA into SG. On contrary, experiments using p53 reporter mRNA lacking the TIA binding sites would help validate the association and dissociation of p53 mRNA from SG via TIA.
SG were also implicated in the selective translation repression of a class of mRNAs that contain hallmark 5′-terminal oligopyrimidine tracts (5′TOP) proximal to the cap 5′end, known as 5′TOP mRNAs (Damgaard and Lykke-Andersen, 2011). These TOP mRNAs encode protein biosynthesis factors whose expression is selectively regulated in response to growth conditions of mammalian cells, mainly via the conserved 5′TOP element (Avni et al., 1994; Avni et al., 1997). In Damgaard et al. study (Damgaard and Lykke-Andersen, 2011), it was found that in response to amino acid starvation, TOP mRNAs accumulate in SG as untranslated mRNAs. The translational regulation of the TOP mRNAs involves binding of TIA to their 5′TOP element, blocking their translation initiation and causing dissociation from polysomes. However, in mRNA reporter studies, TIA is able to dissociate the β-globin reporter mRNA containing the 5′end the 5′TOP element from polysomes independently of its SG association. This suggests that the association of TIA with SG is dispensable for its ability to repress translation of TOP mRNAs, contradicting the possibility that SG cause translation repression of TOP mRNAs, though direct evidence is still lacking. Nevertheless, this study further supports SG as storage sites of untranslated mRNAs. It is intriguing however that even mRNAs that are transcriptionally induced during stress can be stored into SG as untranslated mRNAs. Such mRNAs include those encoded by stress-responsive genes such as glucose-repressed genes and cytokines that are specifically transcribed under glucose starvation conditions (Zid and O'Shea, 2014), and T-cell activation (Scheu et al., 2006), respectively. Whether the sequestration of these mRNAs in SG contributes to their translation repression, regulating specific pathways such as glucose metabolism and immunity, remained to be tested.
By trapping mRNAs in untranslated form, SG may also provide an environment that affects the rate of translation of specific SG-excluded mRNAs, which is necessary for the cell to cope with stress, a possibility that has not been addressed experimentally. This assumption is consistent with the finding that mRNAs that do not associate with SG such Hsp70 and 90 are highly translated under various SG-inducing stresses including heat shock and arsenite treatment (Kedersha et al., 2002; Stohr et al., 2006) or glucose starvation (Zid and O'Shea, 2014). These mRNAs belong to a class of specific mRNAs-encoding stress response functions that are transcriptionally induced and preferentially translated during various types of stress, while translation of bulk mRNAs is repressed. Hsp mRNAs contain long structured 5′UTR that allows their preferential translation via specialised mechanisms (Holcik et al., 2000; Holcik and Sonenberg, 2005; Wek et al., 2006). Whether such 5′UTR-mediated mechanisms of translation prevent the aggregation of mRNAs in SG is still not known.
Omic studies including polysome-seq, RNAseq and LC-MS/MS further validated Hsp mRNAs among a specific set of mRNAs whose enrichment in polysomes is induced by arsenite treatment, correlating with the increase in their corresponding proteins (Somasekharan et al., 2020). Hsp mRNAs were also scored in APEX-seq as associating with APEX-G3BP1 in arsenite-treated cells. As expected, FISH confirmed that those G3BP1-target mRNAs do not associate with SG, excluding their aggregation with G3BP1 in SG. Intriguingly, depletion of G3BP1 enhanced translation of its Hsp mRNA targets, resulting in their hyper-expression (Somasekharan et al., 2020). This finding suggests that besides its role in aggregating untranslated mRNAs in SG, G3BP1 may also act as a translational repressor of Hsp mRNAs independently of SG association, preventing their hyper-expression. However, depletion of G3BP1 also prevents the formation of arsenite-induced SG, which may indirectly result in the observed enhanced translation of Hsp mRNAs. In this case, SG would prevent hyper-expression of Hsp mRNAs, potentially by trapping specific RBPs, preventing their effect in driving translation of Hsps mRNAs, and thus buffering their expression. Using complementary strategies that deplete both SG and specific SG components involved in Hsp mRNAs translation, would be helpful to resolve the issue. By comparing APEX-G3BP1-seq with polysome-seq data, additional categories of mRNAs that are associated with G3BP1 and are either enriched- or depleted from polysomes during arsenite treatment were identified (Somasekharan et al., 2020). As a paradigm of the class of polysomes-depleted mRNAs, BAX mRNA was selected validating its association with SG, its exclusion from polysomes, and its reduced translation. The translation of Bax mRNA was restored in SG-lacking stressed cells owing to G3BP1 depletion. This finding raised the possibility that arsenite-induced downregulation of the expression of Bax mRNA involves its aggregation into SG through G3BP1 binding. An additional class of mRNAs that have been identified in the above study (Somasekharan et al., 2020) contains those whose enrichment into polysomes in arsenite-treated cells requires dissociation from G3BP1. As a representative, HIF-1 mRNA was selected for functional studies, excluding its association with SG, while validating its enhanced translation during arsenite treatment. Although the association of HIF-1 mRNA with G3BP1 is lost during arsenite treatment, depletion of G3BP1 enhanced HIF-1 mRNA translation. This finding suggests that during arsenite treatment, the aggregation of G3BP1 with SG results in its dissociation from HIF-1 mRNA inducing its loading into polysomes and translation. Collectively, these data indicate that reprograming translation of mRNAs during translational stress involves multiple direct and indirect mechanisms involving SG assembly.
One class of mRNAs known to be translationally activated specifically during translation stress is the class of short upstream open reading frames (uORFs) mRNAs (Holcik and Sonenberg, 2005; Wek et al., 2006), which encode stress response functions. Stress-induced translation of such uORFs mRNAs occurs in a phospho-eIF2α-dependent manner, though the molecular mechanisms that drive this phospho-eIF2α-mediated uORF mRNAs translation remained obscure. The main PeIF2α-uORFs mRNA paradigm encodes for the Activating Transcription Factor 4 (ATF4), a master ER stress-induced protein required for reprogramming gene expression towards survival functions. While ER stress had no effect on the abundance of ATF4 mRNA, it significantly induces its loading into translating polysomes, resulting in its preferential translation. Two mechanisms that promote phosphor-eIF2α-mediated uORFs mRNA translation have been reported. The first underlying mechanism reported in HeLa involves the recognition of the 5′end cap of ATF4 mRNA by the DDX3 RNA helicase-containing eIF4F, upon ER stress (Adjibade et al., 2017). The second mechanism was shown to occur in starved-mouse embryonic mouse cells, involving demethylation of a specific and conserved m6A located at the 5′UTR of ATF4 mRNA (Zhou et al., 2018). FISH experiments showed however that a sub-fraction of ATF4 mRNA associates with ER stress-induced SG (Adjibade et al., 2017). This finding raises the possibility that aggregation of a sub-fraction of ATF4 mRNA into SG, potentially in an untranslated form, prevents the lethal overproduction of ATF4. The mechanisms underlying the localization of ATF4 mRNA in SG remained unknown. As a possibility, we speculate that the fraction of ATF4 mRNAs that associate with SG may represent m6A–modified ATF4 mRNAs that escaped demethylation. Investigating the m6A status of the ATF4 mRNAs that are associating with SG may reveal an epitranscriptomic mechanism of methylation/demethylation of ATF4 mRNA at the m6A target site, inducing either its translation in the cytoplasm, or its aggregation in SG as untranslated form. A similar scenario has been described as a mechanism regulating translation of androgen receptor (AR) mRNA during stress (Somasekharan et al., 2022). While in unstressed prostate cancer cells, m6A-modified AR mRNA is preferentially translated, unmodified AR mRNA is kept untranslated. Treatment of prostate cancer cells with AR pathway inhibition stress (ARPI) induces the release of m6A-modified AR mRNA from actively translating polysomes, leading to a significant reduction of AR mRNA translation (Somasekharan et al., 2022). FISH experiments show that upon ARPI stress, AR mRNA associates with SG that are positive for G3BP1 and YTHDF3 (Somasekharan et al., 2022), the two AR mRNA interactors. However, on contrary to YTHDF3 that recognizes m6A-modified AR mRNA, G3BP1 binds preferentially to unmodified AR mRNA, validating previous data (Edupuganti et al., 2017). This m6A-dependent differential association of AR mRNA with YTHDF3 and G3BP1 is also recapitulated in in vitro SG-resembling droplet assays, showing that m6A-modified AR mRNAs phase-separate with the YTHDF3, but not with G3BP1, into droplets, while unmodified-AR mRNAs phase separate exclusively with G3BP1. This raised the possibility that AR mRNAs consist of two pools of m6A-modified and -unmodified forms that may co-exist as two separate YTHDF3 and G3BP1 clusters, respectively within SG. This is supported by STED microscopy images showing separate clusters of YTHDF3 and G3BP1 in SG formed in ARPI stressed cells, while proximity labeling assays with antibodies against m6A and YTHDF3 or G3BP1 showed the association of YTHDF3, but not G3BP1, with m6A-modified mRNAs in ARPI-stressed cells (Somasekharan et al., 2022). Together, these data suggest that m6A-modified AR mRNAs partition into specific SG clusters that kept the message untranslated. Investigating this possibility may also reveal if m6A constitutes a general mechanism that mediates partitioning of target mRNAs into specific translation repression clusters within SG.
The assumption that SG consists of separate clusters also predicts the possibility that such SG clusters, representing different RNPs complexes, may regulate specific aspects of RNA metabolism including translation, stability, and modifications. SG may also affect differentially RNA metabolism during their maturation. mRNAs in SG may thus be spatially organized in active translation and repressed translation condensates. With the development of approaches that allows imaging of translation in vivo, it become possible to monitor translation of reporter mRNAs in SG.
The sunTag in vivo single molecule translation reporter assay (Pichon et al., 2016; Wang et al., 2016; Wu et al., 2016; Yan et al., 2016) allows imaging of target reporter mRNAs via its fused MS2 binding site, simultaneously of SunTag visualisation of the nascent peptide chains. The SunTag consists of an array of antibody epitopes that are recognized by genetically encoded single-chains antibodies (scFv). To establish this Nascent Chain Tracking system, Moon et al. (Moon et al., 2019) used U2OS expressing both a fluorescent MS2 coat protein (Grimm et al., 2015) to label the co-expressed reporter mRNA containing the MS2 binding site, and a fluorescent antibody fragments (Fab) to label epitopes at the N-terminus of the nascent peptide encoded by the reporter mRNA. Using this system, individually translated mRNAs are labelled by both MCP and Fab, while non-translated mRNAs are labelled by only MCP. This Nascent Chain Tracking system was expressed in U2OS-expressing GFP-G3BP1 (Kedersha et al., 2008), examining translation status of single reporter KDM5B mRNA molecules during arsenite stress and recovery in and out of SG-G3BP1. Quantification studies show that 98% of the reporter mRNAs associated with SG is translationally repressed as they lack the SunTag nascent chains signal, consistent with the current model of the association of untranslated mRNAs with SG. The observation that 1–2% of the SG-associated reporter mRNAs retained nascent chains is conform with the possibility that a subset of mRNAs associates with SG, while they are still bound to translating polysomes, though it may also indicate that some residual translation occurs in SG. The analysis of translation of the reporter mRNAs during the recovery phase of arsenite treatment showed that such translation did not resume until SG had dissolved completely, implying that SG disassembly is a prerequisite for translation resumption, which may also support the possibility that SG dampens translation of associated mRNAs. This assumption is however challenged in the Wilbertz et al. study (Wilbertz et al., 2019). Using a similar SunTag translational assays, they show that the localization of a 5′TOP-Renilla luciferase mRNA into SG during arsenite stress does not alter its capacity to resume translation compared to a transcript that remained in the cytosol, when arsenite stress has been relieved (Wilbertz et al., 2019). This conclusion is based on the finding that the fraction of the reporter mRNA that resumes translation during the recovery phase is significantly higher that the fraction of the corresponding mRNA that was associated with SG. In any case, because both KDM5B and TOP reporter mRNAs are translationally repressed during arsenite treatment, it was not possible to assess directly the contribution of SG in repressing their translation.
To address this question, Mateju et al. (Mateju et al., 2020) assessed translation of an ATF4 mRNA reporter whose corresponding endogenous mRNA is actively translated during stress. To establish the SunTag system, Mateju et al. (Mateju et al., 2020) used HeLa cells that are stably co-expressing ScFv-GFP and Halo-tagged MCP (Voigt et al., 2017). At its 5′end, the reporter mRNA encodes the SunTag array in frame with Renilla luciferase, enabling the measurement of protein synthesis by two complementary approaches. The reporter mRNA also contains a destabilised FKBP domain to enhance the degradation of the mature protein thus facilitating the detection of the nascent peptide during translation (Banaszynski et al., 2006). The reporter mRNA contains the 5′UTR of ATF4 mRNA that allows translation during stress. ATF4-SunTag was efficiently translated during arsenite treatment that also induced robust SG formation (Mateju et al., 2020). Translation of the ATF4-SunTag mRNA was then monitored in SG that are depicted as SNAP-tag-labelled G3BP1. Both non-translating- (no SunTag signal) and -translating ATF4 SunTag mRNAs were present in SG, which is consistent with our previous data showing the association of ATF4 mRNA with SG induced in human Hep3B cells treated with ER stress (Adjibade et al., 2015). A fraction of the SG-associated reporter mRNAs showed a SunTag signal, which was blocked by puromycin treatment, indicating active translation. The fraction of ATF4 reporter mRNAs that is associating with SG is however minimal as the major fraction of the ATF4 reporter mRNAs is found in the cytoplasm, and which may be responsible for the bulk translation of the reporter mRNA during stress. Similarly, the SunTag was used detecting translation of the 5′TOP-suntag reporter mRNA (Mateju et al., 2020). This translation was equally low in both the cytoplasm and in SG during stress. The limited translation of the TOP reporter mRNA is consistent with the repression of endogenous TOP mRNAs during stress, while it indicates that even low expressed mRNAs can be translated in SG, though minimally. This study pioneering reporter translation in SG constitutes one of the main important advancements in the field of SG, though the impact of such translation in the post-transcriptional regulation that occurs during stress remains to be demonstrated. This will require the development of translation approaches that provide experimental evidence that endogenous RNAs are translated in SG. The establishment of an in vitro translation assay using purified SG would also help to understand translation and its regulation in SG. Nevertheless, the finding that SG environment is somehow permissive to translation constitutes an additional example of the growing evidence (Parker et al., 2022) demonstrating translation in RNA biocondensates.
PA helped collecting pertinent literature, critically reading the review and making the associated table RM wrote the first draft of the review. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adjibade, P., Grenier St-Sauveur, V., Bergeman, J., Huot, M. E., Khandjian, E. W., and Mazroui, R. (2017). DDX3 regulates endoplasmic reticulum stress-induced ATF4 expression. Sci. Rep. 7, 13832. doi:10.1038/s41598-017-14262-7
Adjibade, P., Simoneau, B., Ledoux, N., Gauthier, W. N., Nkurunziza, M., Khandjian, E. W., et al. (2020). Treatment of cancer cells with Lapatinib negatively regulates general translation and induces stress granules formation. PLoS One 15, e0231894. doi:10.1371/journal.pone.0231894
Adjibade, P., St-Sauveur, V. G., Quevillon Huberdeau, M., Fournier, M. J., Savard, A., Coudert, L., et al. (2015). Sorafenib, a multikinase inhibitor, induces formation of stress granules in hepatocarcinoma cells. Oncotarget 6, 43927–43943. doi:10.18632/oncotarget.5980
Advani, V. M., and Ivanov, P. (2019). Translational control under stress: reshaping the translatome. Bioessays 41, e1900009. doi:10.1002/bies.201900009
Alam, U., and Kennedy, D. (2019). Rasputin a decade on and more promiscuous than ever? A review of G3BPs. Biochimica Biophysica Acta (BBA) - Mol. Cell Res. 1866, 360–370. doi:10.1016/j.bbamcr.2018.09.001
Alriquet, M., Calloni, G., Martinez-Limon, A., Delli Ponti, R., Hanspach, G., Hengesbach, M., et al. (2021). The protective role of m1A during stress-induced granulation. J. Mol. Cell Biol. 12, 870–880. doi:10.1093/jmcb/mjaa023
Anders, M., Chelysheva, I., Goebel, I., Trenkner, T., Zhou, J., Mao, Y., et al. (2018). Dynamic m(6)A methylation facilitates mRNA triaging to stress granules. Life Sci. Alliance 1, e201800113. doi:10.26508/lsa.201800113
Ansari, M. Y., and Haqqi, T. M. (2016). Interleukin-1β induced stress granules sequester COX-2 mRNA and regulates its stability and translation in human OA chondrocytes. Sci. Rep. 6, 27611. doi:10.1038/srep27611
Aramburu-Nunez, M., Custodia, A., Perez-Mato, M., Iglesias-Rey, R., Campos, F., Castillo, J., et al. (2022). Stress granules and acute ischemic stroke: beyond mRNA translation. Int. J. Mol. Sci. 23, 3747. doi:10.3390/ijms23073747
Arguello, A. E., DeLiberto, A. N., and Kleiner, R. E. (2017). RNA chemical proteomics reveals the N(6)-methyladenosine (m(6)A)-Regulated protein-RNA interactome. J. Am. Chem. Soc. 139, 17249–17252. doi:10.1021/jacs.7b09213
Ascano, M., Mukherjee, N., Bandaru, P., Miller, J. B., Nusbaum, J. D., Corcoran, D. L., et al. (2012). FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature 492, 382–386. doi:10.1038/nature11737
Aulas, A., Fay, M. M., Lyons, S. M., Achorn, C. A., Kedersha, N., Anderson, P., et al. (2017a). Stress-specific differences in assembly and composition of stress granules and related foci. J. Cell Sci. 130, 927–937. doi:10.1242/jcs.199240
Aulas, A., Fay, M. M., Szaflarski, W., Kedersha, N., Anderson, P., and Ivanov, P. (2017b). Methods to classify cytoplasmic foci as mammalian stress granules. J. Vis. Exp. JoVE, 55656. doi:10.3791/55656
Aulas, A., Finetti, P., Lyons, S. M., Bertucci, F., Birnbaum, D., Acquaviva, C., et al. (2020). Revisiting the concept of stress in the prognosis of solid tumors: a role for stress granules proteins? Cancers (Basel) 12, 2470. doi:10.3390/cancers12092470
Avni, D., Biberman, Y., and Meyuhas, O. (1997). The 5' terminal oligopyrimidine tract confers translational control on TOP mRNAs in a cell type- and sequence context-dependent manner. Nucleic Acids Res. 25, 995–1001. doi:10.1093/nar/25.5.995
Avni, D., Shama, S., Loreni, F., and Meyuhas, O. (1994). Vertebrate mRNAs with a 5'-terminal pyrimidine tract are candidates for translational repression in quiescent cells: characterization of the translational cis-regulatory element. Mol. Cell Biol. 14, 3822–3833. doi:10.1128/mcb.14.6.3822
Banaszynski, L. A., Chen, L. C., Maynard-Smith, L. A., Ooi, A. G., and Wandless, T. J. (2006). A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell 126, 995–1004. doi:10.1016/j.cell.2006.07.025
Bordeleau, M. E., Cencic, R., Lindqvist, L., Oberer, M., Northcote, P., Wagner, G., et al. (2006a). RNA-mediated sequestration of the RNA helicase eIF4A by Pateamine A inhibits translation initiation. Chem. Biol. 13, 1287–1295. doi:10.1016/j.chembiol.2006.10.005
Bordeleau, M. E., Matthews, J., Wojnar, J. M., Lindqvist, L., Novac, O., Jankowsky, E., et al. (2005). Stimulation of mammalian translation initiation factor eIF4A activity by a small molecule inhibitor of eukaryotic translation. Proc. Natl. Acad. Sci. U. S. A. 102, 10460–10465. doi:10.1073/pnas.0504249102
Bordeleau, M. E., Mori, A., Oberer, M., Lindqvist, L., Chard, L. S., Higa, T., et al. (2006b). Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat. Chem. Biol. 2, 213–220. doi:10.1038/nchembio776
Bordeleau, M. E., Robert, F., Gerard, B., Lindqvist, L., Chen, S. M., Wendel, H. G., et al. (2008). Therapeutic suppression of translation initiation modulates chemosensitivity in a mouse lymphoma model. J. Clin. Invest. 118, 2651–2660. doi:10.1172/jci34753
Bounedjah, O., Desforges, B., Wu, T. D., Pioche-Durieu, C., Marco, S., Hamon, L., et al. (2014). Free mRNA in excess upon polysome dissociation is a scaffold for protein multimerization to form stress granules. Nucleic Acids Res. 42, 8678–8691. doi:10.1093/nar/gku582
Brown, J. A., Roberts, T. L., Richards, R., Woods, R., Birrell, G., Lim, Y. C., et al. (2011). A novel role for hSMG-1 in stress granule formation. Mol. Cell. Biol. 31, 4417–4429. doi:10.1128/mcb.05987-11
Buchan, J. R., Muhlrad, D., and Parker, R. (2008). P bodies promote stress granule assembly in Saccharomyces cerevisiae. J. Cell Biol. 183, 441–455. doi:10.1083/jcb.200807043
Buchan, J. R., Yoon, J. H., and Parker, R. (2011). Stress-specific composition, assembly and kinetics of stress granules in Saccharomyces cerevisiae. J. Cell Sci. 124, 228–239. doi:10.1242/jcs.078444
Campos-Melo, D., Hawley, Z. C. E., Droppelmann, C. A., and Strong, M. J. (2021). The integral role of RNA in stress granule formation and function. Front. Cell Dev. Biol. 9, 621779. doi:10.3389/fcell.2021.621779
Cencic, R., Carrier, M., Galicia-Vazquez, G., Bordeleau, M. E., Sukarieh, R., Bourdeau, A., et al. (2009). Antitumor activity and mechanism of action of the cyclopenta[b]benzofuran, silvestrol. PLoS One 4, e5223. doi:10.1371/journal.pone.0005223
Chen, E., Sharma, M. R., Shi, X., Agrawal, R. K., and Joseph, S. (2014). Fragile X mental retardation protein regulates translation by binding directly to the ribosome. Mol. Cell 54, 407–417. doi:10.1016/j.molcel.2014.03.023
Conicella, A. E., Dignon, G. L., Zerze, G. H., Schmidt, H. B., D'Ordine, A. M., Kim, Y. C., et al. (2020a). TDP-43 alpha-helical structure tunes liquid-liquid phase separation and function. Proc. Natl. Acad. Sci. U. S. A. 117, 5883–5894. doi:10.1073/pnas.1912055117
Conicella, A. E., Huang, R., Ripstein, Z. A., Nguyen, A., Wang, E., Lohr, T., et al. (2020b). An intrinsically disordered motif regulates the interaction between the p47 adaptor and the p97 AAA+ ATPase. Proc. Natl. Acad. Sci. U. S. A. 117, 26226–26236. doi:10.1073/pnas.2013920117
Curdy, N., Lanvin, O., Cerapio, J. P., Pont, F., Tosolini, M., Sarot, E., et al. (2023). The proteome and transcriptome of stress granules and P bodies during human T lymphocyte activation. Cell Rep. 42, 112211. doi:10.1016/j.celrep.2023.112211
Damgaard, C. K., and Lykke-Andersen, J. (2011). Translational coregulation of 5'TOP mRNAs by TIA-1 and TIAR. Genes Dev. 25, 2057–2068. doi:10.1101/gad.17355911
Dang, Y., Kedersha, N., Low, W. K., Romo, D., Gorospe, M., Kaufman, R., et al. (2006). Eukaryotic initiation factor 2α-independent pathway of stress granule induction by the natural product pateamine A. J. Biol. Chem. 281, 32870–32878. doi:10.1074/jbc.m606149200
Darnell, J. C., Van Driesche, S., Zhang, C., Hung, K. Y., Mele, A., Fraser, C. E., et al. (2011). FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–261. doi:10.1016/j.cell.2011.06.013
Degracia, D., and Hu, B. (2013). The editorial for this special issue. Transl. Stroke Res. 4, 579–580. doi:10.1007/s12975-013-0302-1
DeGracia, D. J., and Hu, B. R. (2007). Irreversible translation arrest in the reperfused brain. J. Cereb. Blood Flow. Metab. 27, 875–893. doi:10.1038/sj.jcbfm.9600388
DeGracia, D. J., Jamison, J. T., Szymanski, J. J., and Lewis, M. K. (2008). Translation arrest and ribonomics in post-ischemic brain: layers and layers of players. J. Neurochem. 106, 2288–2301. doi:10.1111/j.1471-4159.2008.05561.x
Deniz, A. A. (2020). Networking and dynamic switches in biological condensates. Cell 181, 228–230. doi:10.1016/j.cell.2020.03.056
Diaz-Munoz, M. D., Kiselev, V. Y., Le Novere, N., Curk, T., Ule, J., and Turner, M. (2017). Tia1 dependent regulation of mRNA subcellular location and translation controls p53 expression in B cells. Nat. Commun. 8, 530. doi:10.1038/s41467-017-00454-2
Didiot, M. C., Subramanian, M., Flatter, E., Mandel, J. L., and Moine, H. (2009). Cells lacking the fragile X mental retardation protein (FMRP) have normal RISC activity but exhibit altered stress granule assembly. Mol. Biol. Cell 20, 428–437. doi:10.1091/mbc.e08-07-0737
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206. doi:10.1038/nature11112
Dominissini, D., Nachtergaele, S., Moshitch-Moshkovitz, S., Peer, E., Kol, N., Ben-Haim, M. S., et al. (2016). The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature 530, 441–446. doi:10.1038/nature16998
Donnelly, N., Gorman, A. M., Gupta, S., and Samali, A. (2013). The eIF2α kinases: their structures and functions. Cell Mol. Life Sci. 70, 3493–3511. doi:10.1007/s00018-012-1252-6
Duncan, R., Milburn, S. C., and Hershey, J. W. (1987). Regulated phosphorylation and low abundance of HeLa cell initiation factor eIF-4F suggest a role in translational control. Heat shock effects on eIF-4F. J. Biol. Chem. 262, 380–388. doi:10.1016/s0021-9258(19)75938-9
Edupuganti, R. R., Geiger, S., Lindeboom, R. G. H., Shi, H., Hsu, P. J., Lu, Z., et al. (2017). N(6)-methyladenosine (m(6)A) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol. 24, 870–878. doi:10.1038/nsmb.3462
Emara, M. M., Fujimura, K., Sciaranghella, D., Ivanova, V., Ivanov, P., and Anderson, P. (2012). Hydrogen peroxide induces stress granule formation independent of eIF2α phosphorylation. Biochem. Biophys. Res. Commun. 423, 763–769. doi:10.1016/j.bbrc.2012.06.033
Fahling, M. (2009). Cellular oxygen sensing, signalling and how to survive translational arrest in hypoxia. Acta Physiol. 195, 205–230. doi:10.1111/j.1748-1716.2008.01894.x
Fournier, M. J., Coudert, L., Mellaoui, S., Adjibade, P., Gareau, C., Cote, M. F., et al. (2013). Inactivation of the mTORC1-eukaryotic translation initiation factor 4E pathway alters stress granule formation. Mol. Cell. Biol. 33, 2285–2301. doi:10.1128/mcb.01517-12
Fournier, M. J., Gareau, C., and Mazroui, R. (2010). The chemotherapeutic agent bortezomib induces the formation of stress granules. Cancer Cell Int. 10, 12. doi:10.1186/1475-2867-10-12
Franchini, D. M., Lanvin, O., Tosolini, M., Patras de Campaigno, E., Cammas, A., Pericart, S., et al. (2019). Microtubule-driven stress granule dynamics regulate inhibitory immune checkpoint expression in T cells. Cell Rep. 26, 94–107.e7. doi:10.1016/j.celrep.2018.12.014
Fu, Y., and Zhuang, X. (2020). m(6)A-binding YTHDF proteins promote stress granule formation. Nat. Chem. Biol. 16, 955–963. doi:10.1038/s41589-020-0524-y
Fujimura, K., Sasaki, A. T., and Anderson, P. (2012). Selenite targets eIF4E-binding protein-1 to inhibit translation initiation and induce the assembly of non-canonical stress granules. Nucleic Acids Res. 40, 8099–8110. doi:10.1093/nar/gks566
Gareau, C., Fournier, M. J., Filion, C., Coudert, L., Martel, D., Labelle, Y., et al. (2011). p21(WAF1/CIP1) upregulation through the stress granule-associated protein CUGBP1 confers resistance to bortezomib-mediated apoptosis. PLoS One 6, e20254. doi:10.1371/journal.pone.0020254
Gareau, C., Houssin, E., Martel, D., Coudert, L., Mellaoui, S., Huot, M. E., et al. (2013a). Characterization of fragile X mental retardation protein recruitment and dynamics in Drosophila stress granules. PLoS One 8, e55342. doi:10.1371/journal.pone.0055342
Gareau, C., Martel, D., Coudert, L., Mellaoui, S., and Mazroui, R. (2013b). Characterization of fragile X mental retardation protein granules formation and dynamics in Drosophila. Biol. Open 2, 68–81. doi:10.1242/bio.20123012
Gebauer, F., and Hentze, M. W. (2004). Molecular mechanisms of translational control. Nat. Rev. Mol. Cell Biol. 5, 827–835. doi:10.1038/nrm1488
Ghisolfi, L., Dutt, S., McConkey, M. E., Ebert, B. L., and Anderson, P. (2012). Stress granules contribute to alpha-globin homeostasis in differentiating erythroid cells. Biochem. Biophysical Res. Commun. 420, 768–774. doi:10.1016/j.bbrc.2012.03.070
Gingras, A. C., Raught, B., Gygi, S. P., Niedzwiecka, A., Miron, M., Burley, S. K., et al. (2001). Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 15, 2852–2864. doi:10.1101/gad.912401
Gingras, A. C., Raught, B., and Sonenberg, N. (1999). eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68, 913–963. doi:10.1146/annurev.biochem.68.1.913
Gotte, B., Panas, M. D., Hellstrom, K., Liu, L., Samreen, B., Larsson, O., et al. (2019). Separate domains of G3BP promote efficient clustering of alphavirus replication complexes and recruitment of the translation initiation machinery. PLoS Pathog. 15, e1007842. doi:10.1371/journal.ppat.1007842
Grimm, J. B., English, B. P., Chen, J., Slaughter, J. P., Zhang, Z., Revyakin, A., et al. (2015). A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods 12, 244–250. doi:10.1038/nmeth.3256
Guillen-Boixet, J., Kopach, A., Holehouse, A. S., Wittmann, S., Jahnel, M., Schlüßler, R., et al. (2020). RNA-induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell 181, 346–361.e17. doi:10.1016/j.cell.2020.03.049
Haghighat, A., Mader, S., Pause, A., and Sonenberg, N. (1995). Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 14, 5701–5709. doi:10.1002/j.1460-2075.1995.tb00257.x
Haghighat, A., and Sonenberg, N. (1997). eIF4G dramatically enhances the binding of eIF4E to the mRNA 5'-cap structure. J. Biol. Chem. 272, 29398–21680. doi:10.1016/s0021-9258(18)50898-x
Haghighat, A., Svitkin, Y., Novoa, I., Kuechler, E., Skern, T., and Sonenberg, N. (1996). The eIF4G-eIF4E complex is the target for direct cleavage by the rhinovirus 2A proteinase. J. Virol. 70, 8444–8450. doi:10.1128/jvi.70.12.8444-8450.1996
Halstead, J. M., Lionnet, T., Wilbertz, J. H., Wippich, F., Ephrussi, A., Singer, R. H., et al. (2015). An RNA biosensor for imaging the first round of translation from single cells to living animals. Science 347, 1367–1671. doi:10.1126/science.aaa3380
Hara, K., Maruki, Y., Long, X., Yoshino, K., Oshiro, N., Hidayat, S., et al. (2002). Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110, 177–189. doi:10.1016/s0092-8674(02)00833-4
He, P. C., and He, C. (2021). m(6) A RNA methylation: from mechanisms to therapeutic potential. EMBO J. 40, e105977. doi:10.15252/embj.2020105977
Heberle, A. M., Razquin Navas, P., Langelaar-Makkinje, M., Kasack, K., Sadik, A., Faessler, E., et al. (2019). The PI3K and MAPK/p38 pathways control stress granule assembly in a hierarchical manner. Life Sci. Alliance 2, e201800257. doi:10.26508/lsa.201800257
Hilliker, A., Gao, Z., Jankowsky, E., and Parker, R. (2011). The DEAD-box protein Ded1 modulates translation by the formation and resolution of an eIF4F-mRNA complex. Mol. Cell 43, 962–972. doi:10.1016/j.molcel.2011.08.008
Holcik, M., Sonenberg, N., and Korneluk, R. G. (2000). Internal ribosome initiation of translation and the control of cell death. Trends Genet. 16, 469–473. doi:10.1016/s0168-9525(00)02106-5
Holcik, M., and Sonenberg, N. (2005). Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 6, 318–327. doi:10.1038/nrm1618
Horvathova, I., Voigt, F., Kotrys, A. V., Zhan, Y., Artus-Revel, C. G., Eglinger, J., et al. (2017). The dynamics of mRNA turnover revealed by single-molecule imaging in single cells. Mol. Cell 68, 615–625.e9. doi:10.1016/j.molcel.2017.09.030
Hsu, P. J., Shi, H., Zhu, A. C., Lu, Z., Miller, N., Edens, B. M., et al. (2019). The RNA-binding protein FMRP facilitates the nuclear export of N (6)-methyladenosine-containing mRNAs. J. Biol. Chem. 294, 19889–19895. doi:10.1074/jbc.ac119.010078
Hu, L., Mao, S., Lin, L., Bai, G., Liu, B., and Mao, J. (2022). Stress granules in the spinal muscular atrophy and amyotrophic lateral sclerosis: the correlation and promising therapy. Neurobiol. Dis. 170, 105749. doi:10.1016/j.nbd.2022.105749
Huang, J., and Yin, P. (2018). Structural insights into N(6)-methyladenosine (m(6)A) modification in the transcriptome. Genomics, Proteomics Bioinforma. 16, 85–98. doi:10.1016/j.gpb.2018.03.001
Ivanov, P., Kedersha, N., Anderson, P., and Anderson, P. (2019). Stress granules and processing bodies in translational control. Cold Spring Harb. Perspect. Biol. 11, a032813. doi:10.1101/cshperspect.a032813
Iwasaki, S., Floor, S. N., and Ingolia, N. T. (2016). Rocaglates convert DEAD-box protein eIF4A into a sequence-selective translational repressor. Nature 534, 558–561. doi:10.1038/nature17978
Jackson, R. J., Hellen, C. U., and Pestova, T. V. (2010). The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127. doi:10.1038/nrm2838
Jain, S., Wheeler, J. R., Walters, R. W., Agrawal, A., Barsic, A., and Parker, R. (2016). ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164, 487–498. doi:10.1016/j.cell.2015.12.038
Kedersha, N., and Anderson, P. (2009). Regulation of translation by stress granules and processing bodies. Prog. Mol. Biol. Transl. Sci. 90, 155–185. doi:10.1016/S1877-1173(09)90004-7
Kedersha, N., and Anderson, P. (2002). Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30, A117–A969. doi:10.1042/bst030a117
Kedersha, N., Chen, S., Gilks, N., Li, W., Miller, I. J., Stahl, J., et al. (2002). Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell 13, 195–210. doi:10.1091/mbc.01-05-0221
Kedersha, N., Cho, M. R., Li, W., Yacono, P. W., Chen, S., Gilks, N., et al. (2000). Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 151, 1257–1268. doi:10.1083/jcb.151.6.1257
Kedersha, N., Ivanov, P., and Anderson, P. (2013). Stress granules and cell signaling: more than just a passing phase? Trends Biochem. Sci. 38, 494–506. doi:10.1016/j.tibs.2013.07.004
Kedersha, N., Panas, M. D., Achorn, C. A., Lyons, S., Tisdale, S., Hickman, T., et al. (2016). G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J. Cell Biol. 212, 845–860. doi:10.1083/jcb.201508028
Kedersha, N., Stoecklin, G., Ayodele, M., Yacono, P., Lykke-Andersen, J., Fritzler, M. J., et al. (2005). Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169, 871–884. doi:10.1083/jcb.200502088
Kedersha, N., Tisdale, S., Hickman, T., and Anderson, P. (2008). Real-time and quantitative imaging of mammalian stress granules and processing bodies. Methods Enzymol. 448, 521–552. doi:10.1016/S0076-6879(08)02626-8
Kemper, W. M., Berry, K. W., and Merrick, W. C. (1976). Purification and properties of rabbit reticulocyte protein synthesis initiation factors M2Balpha and M2Bbeta. J. Biol. Chem. 251, 5551–5557. doi:10.1016/s0021-9258(17)33095-8
Khong, A., Matheny, T., Huynh, T. N., Babl, V., and Parker, R. (2022). Limited effects of m(6)A modification on mRNA partitioning into stress granules. Nat. Commun. 13, 3735. doi:10.1038/s41467-022-31358-5
Khong, A., Matheny, T., Jain, S., Mitchell, S. F., Wheeler, J. R., and Parker, R. (2017). The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol. Cell 68, 808–820.e5. doi:10.1016/j.molcel.2017.10.015
Kim, D. H., Sarbassov, D. D., Ali, S. M., King, J. E., Latek, R. R., Erdjument-Bromage, H., et al. (2002). mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110, 163–175. doi:10.1016/s0092-8674(02)00808-5
Koromilas, A. E. (2015). Roles of the translation initiation factor eIF2α serine 51 phosphorylation in cancer formation and treatment. Biochim. Biophys. Acta 1849, 871–880. doi:10.1016/j.bbagrm.2014.12.007
Kosmas, K., Filippakis, H., Khabibullin, D., Turkiewicz, M., Lam, H. C., Yu, J., et al. (2021). TSC2 interacts with HDLBP/vigilin and regulates stress granule formation. Mol. Cancer Res. 19, 1389–1397. doi:10.1158/1541-7786.mcr-20-1046
Kroschwald, S., Maharana, S., Mateju, D., Malinovska, L., Nuske, E., Poser, I., et al. (2015). Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. Elife 4, e06807. doi:10.7554/elife.06807
Kubinski, S., and Claus, P. (2022). Protein network analysis reveals a functional connectivity of dysregulated processes in ALS and SMA. Neurosci. Insights 17, 263310552210877. doi:10.1177/26331055221087740
Lee, J. I., and Namkoong, S. (2022). Stress granules dynamics: benefits in cancer. BMB Rep. 55, 577–586. doi:10.5483/bmbrep.2022.55.12.141
Lewis, C. J., Pan, T., and Kalsotra, A. (2017). RNA modifications and structures cooperate to guide RNA-protein interactions. Nat. Rev. Mol. Cell Biol. 18, 202–210. doi:10.1038/nrm.2016.163
Li, C. H., Ohn, T., Ivanov, P., Tisdale, S., and Anderson, P. (2010). eIF5A promotes translation elongation, polysome disassembly and stress granule assembly. PLoS One 5, e9942. doi:10.1371/journal.pone.0009942
Lindqvist, L., Imataka, H., and Pelletier, J. (2008). Cap-dependent eukaryotic initiation factor-mRNA interactions probed by cross-linking. Rna 14, 960–969. doi:10.1261/rna.971208
Low, W. K., Dang, Y., Schneider-Poetsch, T., Shi, Z., Choi, N. S., Merrick, W. C., et al. (2005). Inhibition of eukaryotic translation initiation by the marine natural product pateamine A. Mol. Cell 20, 709–722. doi:10.1016/j.molcel.2005.10.008
Mateju, D., Eichenberger, B., Voigt, F., Eglinger, J., Roth, G., and Chao, J. A. (2020). Single-molecule imaging reveals translation of mRNAs localized to stress granules. Cell 183, 1801–1812.e13. doi:10.1016/j.cell.2020.11.010
Mateyak, M. K., and Kinzy, T. G. (2013). ADP-ribosylation of translation elongation factor 2 by diphtheria toxin in yeast inhibits translation and cell separation. J. Biol. Chem. 288, 24647–24655. doi:10.1074/jbc.m113.488783
Mazan-Mamczarz, K., Peroutka, R. J., Steinhardt, J. J., Gidoni, M., Zhang, Y., Lehrmann, E., et al. (2015). Distinct inhibitory effects on mTOR signaling by ethanol and INK128 in diffuse large B-cell lymphoma. Cell Commun. Signal 13, 15. doi:10.1186/s12964-015-0091-0
Mazroui, R., Huot, M. E., Tremblay, S., Boilard, N., Labelle, Y., and Khandjian, E. W. (2003). Fragile X Mental Retardation protein determinants required for its association with polyribosomal mRNPs. Hum. Mol. Genet. 12, 3087–3096. doi:10.1093/hmg/ddg335
Mazroui, R., Huot, M. E., Tremblay, S., Filion, C., Labelle, Y., and Khandjian, E. W. (2002). Trapping of messenger RNA by Fragile X Mental Retardation protein into cytoplasmic granules induces translation repression. Hum. Mol. Genet. 11, 3007–3017. doi:10.1093/hmg/11.24.3007
Mazroui, R., Sukarieh, R., Bordeleau, M. E., Kaufman, R. J., Northcote, P., Tanaka, J., et al. (2006). Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2α phosphorylation. Mol. Biol. Cell 17, 4212–4219. doi:10.1091/mbc.e06-04-0318
Meyer, K. D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C. E., and Jaffrey, S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell 149, 1635–1646. doi:10.1016/j.cell.2012.05.003
Moeller, B. J., Cao, Y., Li, C. Y., and Dewhirst, M. W. (2004). Radiation activates HIF-1 to regulate vascular radiosensitivity in tumors: role of reoxygenation, free radicals, and stress granules. Cancer Cell 5, 429–441. doi:10.1016/s1535-6108(04)00115-1
Mokas, S., Mills, J. R., Garreau, C., Fournier, M. J., Robert, F., Arya, P., et al. (2009). Uncoupling stress granule assembly and translation initiation inhibition. Mol. Biol. Cell 20, 2673–2683. doi:10.1091/mbc.e08-10-1061
Molliex, A., Temirov, J., Lee, J., Coughlin, M., Kanagaraj, A. P., Kim, H. J., et al. (2015). Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133. doi:10.1016/j.cell.2015.09.015
Moon, S. L., Morisaki, T., Khong, A., Lyon, K., Parker, R., and Stasevich, T. J. (2019). Multicolour single-molecule tracking of mRNA interactions with RNP granules. Nat. Cell Biol. 21, 162–168. doi:10.1038/s41556-018-0263-4
Moutaoufik, M. T., El Fatimy, R., Nassour, H., Gareau, C., Lang, J., Tanguay, R. M., et al. (2014). UVC-induced stress granules in mammalian cells. PLoS One 9, e112742. doi:10.1371/journal.pone.0112742
Napoli, I., Mercaldo, V., Boyl, P. P., Eleuteri, B., Zalfa, F., De Rubeis, S., et al. (2008). The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell 134, 1042–1054. doi:10.1016/j.cell.2008.07.031
Ohn, T., Kedersha, N., Hickman, T., Tisdale, S., and Anderson, P. (2008). A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat. Cell Biol. 10, 1224–1231. doi:10.1038/ncb1783
Padron, A., Iwasaki, S., and Ingolia, N. T. (2019). Proximity RNA labeling by APEX-seq reveals the organization of translation initiation complexes and repressive RNA granules. Mol. Cell 75, 875–887.e5. doi:10.1016/j.molcel.2019.07.030
Panas, M. D., Ivanov, P., and Anderson, P. (2016). Mechanistic insights into mammalian stress granule dynamics. J. Cell Biol. 215, 313–323. doi:10.1083/jcb.201609081
Parker, D. M., Winkenbach, L. P., and Osborne Nishimura, E. (2022). It's just a phase: exploring the relationship between mRNA, biomolecular condensates, and translational control. Front. Genet. 13, 931220. doi:10.3389/fgene.2022.931220
Patel, A., Lee, H. O., Jawerth, L., Maharana, S., Jahnel, M., Hein, M. Y., et al. (2015). A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077. doi:10.1016/j.cell.2015.07.047
Pichon, X., Bastide, A., Safieddine, A., Chouaib, R., Samacoits, A., Basyuk, E., et al. (2016). Visualization of single endogenous polysomes reveals the dynamics of translation in live human cells. J. Cell Biol. 214, 769–781. doi:10.1083/jcb.201605024
Piotrowska, J., Hansen, S. J., Park, N., Jamka, K., Sarnow, P., and Gustin, K. E. (2010). Stable formation of compositionally unique stress granules in virus-infected cells. J. Virol. 84, 3654–3665. doi:10.1128/jvi.01320-09
Portz, B., Lee, B. L., and Shorter, J. (2021). FUS and TDP-43 phases in health and disease. Trends Biochem. Sci. 46, 550–563. doi:10.1016/j.tibs.2020.12.005
Protter, D. S., and Parker, R. (2016). Principles and properties of stress granules. Trends Cell Biol. 26, 668–679. doi:10.1016/j.tcb.2016.05.004
Rhee, H. W., Zou, P., Udeshi, N. D., Martell, J. D., Mootha, V. K., Carr, S. A., et al. (2013). Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339, 1328–1331. doi:10.1126/science.1230593
Richter, J. D., and Sonenberg, N. (2005). Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433, 477–480. doi:10.1038/nature03205
Ries, R. J., Zaccara, S., Klein, P., Olarerin-George, A., Namkoong, S., Pickering, B. F., et al. (2019). m(6)A enhances the phase separation potential of mRNA. Nature 571, 424–428. doi:10.1038/s41586-019-1374-1
Riggs, C. L., Kedersha, N., Ivanov, P., and Anderson, P. (2020). Mammalian stress granules and P bodies at a glance. J. Cell Sci. 133, jcs242487. doi:10.1242/jcs.242487
Robert, F., Kapp, L. D., Khan, S. N., Acker, M. G., Kolitz, S., Kazemi, S., et al. (2006). Initiation of protein synthesis by hepatitis C virus is refractory to reduced eIF2.GTP.Met-tRNA(i)(Met) ternary complex availability. Mol. Biol. Cell 17, 4632–4644. doi:10.1091/mbc.e06-06-0478
Roden, C., and Gladfelter, A. S. (2021). RNA contributions to the form and function of biomolecular condensates. Nat. Rev. Mol. Cell Biol. 22, 183–195. doi:10.1038/s41580-020-0264-6
Safer, B., Adams, S. L., Kemper, W. M., Berry, K. W., Lloyd, M., and Merrick, W. C. (1976). Purification and characterization of two initiation factors required for maximal activity of a highly fractionated globin mRNA translation system. Proc. Natl. Acad. Sci. U. S. A. 73, 2584–2588. doi:10.1073/pnas.73.8.2584
Scheu, S., Stetson, D. B., Reinhardt, R. L., Leber, J. H., Mohrs, M., and Locksley, R. M. (2006). Activation of the integrated stress response during T helper cell differentiation. Nat. Immunol. 7, 644–651. doi:10.1038/ni1338
Schuller, A. P., Wu, C. C., Dever, T. E., Buskirk, A. R., and Green, R. (2017). eIF5A functions globally in translation elongation and termination. Mol. Cell 66, 194–205.e5. doi:10.1016/j.molcel.2017.03.003
Sfakianos, A. P., Mellor, L. E., Pang, Y. F., Kritsiligkou, P., Needs, H., Abou-Hamdan, H., et al. (2018). The mTOR-S6 kinase pathway promotes stress granule assembly. Cell Death Differ. 25, 1766–1780. doi:10.1038/s41418-018-0076-9
Sidrauski, C., McGeachy, A. M., Ingolia, N. T., and Walter, P. (2015a). The small molecule ISRIB reverses the effects of eIF2α phosphorylation on translation and stress granule assembly. Elife 4, e05033. doi:10.7554/elife.05033
Sidrauski, C., Tsai, J. C., Kampmann, M., Hearn, B. R., Vedantham, P., Jaishankar, P., et al. (2015b). Pharmacological dimerization and activation of the exchange factor eIF2B antagonizes the integrated stress response. Elife 4, e07314. doi:10.7554/elife.07314
Sivan, G., Kedersha, N., and Elroy-Stein, O. (2007). Ribosomal slowdown mediates translational arrest during cellular division. Mol. Cell. Biol. 27, 6639–6646. doi:10.1128/mcb.00798-07
Somasekharan, S. P., Saxena, N., Zhang, F., Beraldi, E., Huang, J. N., Gentle, C., et al. (2022). Regulation of AR mRNA translation in response to acute AR pathway inhibition. Nucleic Acids Res. 50, 1069–1091. doi:10.1093/nar/gkab1247
Somasekharan, S. P., Zhang, F., Saxena, N., Huang, J. N., Kuo, I. C., Low, C., et al. (2020). G3BP1-linked mRNA partitioning supports selective protein synthesis in response to oxidative stress. Nucleic Acids Res. 48, 6855–6873. doi:10.1093/nar/gkaa376
Sonenberg, N., and Dever, T. E. (2003). Eukaryotic translation initiation factors and regulators. Curr. Opin. Struct. Biol. 13, 56–63. doi:10.1016/s0959-440x(03)00009-5
Sonenberg, N. (2008). eIF4E, the mRNA cap-binding protein: from basic discovery to translational researchThis paper is one of a selection of papers published in this Special Issue, entitled CSBMCB — systems and Chemical Biology, and has undergone the Journal's usual peer review process. Biochem. Cell Biol. 86, 178–183. doi:10.1139/o08-034
Stohr, N., Lederer, M., Reinke, C., Meyer, S., Hatzfeld, M., Singer, R. H., et al. (2006). ZBP1 regulates mRNA stability during cellular stress. J. Cell Biol. 175, 527–534. doi:10.1083/jcb.200608071
Sukarieh, R., Sonenberg, N., and Pelletier, J. (2009). The eIF4E-binding proteins are modifiers of cytoplasmic eIF4E relocalization during the heat shock response. Am. J. Physiology-Cell Physiology 296, C1207–C1217. doi:10.1152/ajpcell.00511.2008
Szaflarski, W., Fay, M. M., Kedersha, N., Zabel, M., Anderson, P., and Ivanov, P. (2016). Vinca alkaloid drugs promote stress-induced translational repression and stress granule formation. Oncotarget 7, 30307–30322. doi:10.18632/oncotarget.8728
Tauber, D., Tauber, G., Khong, A., Van Treeck, B., Pelletier, J., and Parker, R. (2020). Modulation of RNA condensation by the DEAD-box protein eIF4A. Cell 180, 411–426.e16. doi:10.1016/j.cell.2019.12.031
Thoreen, C. C., Chantranupong, L., Keys, H. R., Wang, T., Gray, N. S., and Sabatini, D. M. (2012). A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485, 109–113. doi:10.1038/nature11083
Thoreen, C. C., Kang, S. A., Chang, J. W., Liu, Q., Zhang, J., Gao, Y., et al. (2009). An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 284, 8023–8032. doi:10.1074/jbc.m900301200
Topisirovic, I., and Sonenberg, N. (2011). mRNA translation and energy metabolism in cancer: the role of the MAPK and mTORC1 pathways. Cold Spring Harb. Symposia Quantitative Biol. 76, 355–367. doi:10.1101/sqb.2011.76.010785
Tourriere, H., Gallouzi, I. E., Chebli, K., Capony, J. P., Mouaikel, J., van der Geer, P., et al. (2001). RasGAP-associated endoribonuclease G3Bp: selective RNA degradation and phosphorylation-dependent localization. Mol. Cell. Biol. 21, 7747–7760. doi:10.1128/mcb.21.22.7747-7760.2001
Tsang, B., Arsenault, J., Vernon, R. M., Lin, H., Sonenberg, N., Wang, L. Y., et al. (2019). Phosphoregulated FMRP phase separation models activity-dependent translation through bidirectional control of mRNA granule formation. Proc. Natl. Acad. Sci. U. S. A. 116, 4218–4227. doi:10.1073/pnas.1814385116
van Leeuwen, W., VanInsberghe, M., Battich, N., Salmen, F., van Oudenaarden, A., and Rabouille, C. (2022). Identification of the stress granule transcriptome via RNA-editing in single cells and in vivo. Cell Rep. Methods 2, 100235. doi:10.1016/j.crmeth.2022.100235
Van Treeck, B., and Parker, R. (2018). Emerging roles for intermolecular RNA-RNA interactions in RNP assemblies. Cell 174, 791–802. doi:10.1016/j.cell.2018.07.023
Van Treeck, B., and Parker, R. (2019). Principles of stress granules revealed by imaging approaches. Cold Spring Harb. Perspect. Biol. 11, a033068. doi:10.1101/cshperspect.a033068
Van Treeck, B., Protter, D. S. W., Matheny, T., Khong, A., Link, C. D., and Parker, R. (2018). RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc. Natl. Acad. Sci. U. S. A. 115, 2734–2739. doi:10.1073/pnas.1800038115
Voigt, F., Zhang, H., Cui, X. A., Triebold, D., Liu, A. X., Eglinger, J., et al. (2017). Single-molecule quantification of translation-dependent association of mRNAs with the endoplasmic reticulum. Cell Rep. 21, 3740–3753. doi:10.1016/j.celrep.2017.12.008
Wang, C., Han, B., Zhou, R., and Zhuang, X. (2016). Real-time imaging of translation on single mRNA transcripts in live cells. Cell 165, 990–1001. doi:10.1016/j.cell.2016.04.040
Wang, J., Choi, J. M., Holehouse, A. S., Lee, H. O., Zhang, X., Jahnel, M., et al. (2018). A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell 174, 688–699.e16. doi:10.1016/j.cell.2018.06.006
Wang, T., Tian, X., Kim, H. B., Jang, Y., Huang, Z., Na, C. H., et al. (2022). Intracellular energy controls dynamics of stress-induced ribonucleoprotein granules. Nat. Commun. 13, 5584. doi:10.1038/s41467-022-33079-1
Wang, X., and He, C. (2014). Reading RNA methylation codes through methyl-specific binding proteins. RNA Biol. 11, 669–672. doi:10.4161/rna.28829
Wang, X., Lu, Z., Gomez, A., Hon, G. C., Yue, Y., Han, D., et al. (2014a). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120. doi:10.1038/nature12730
Wang, X., Zhao, B. S., Roundtree, I. A., Lu, Z., Han, D., Ma, H., et al. (2015). N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell 161, 1388–1399. doi:10.1016/j.cell.2015.05.014
Wang, Y., Li, Y., Toth, J. I., Petroski, M. D., Zhang, Z., and Zhao, J. C. (2014b). N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 16, 191–198. doi:10.1038/ncb2902
Wek, R. C., Jiang, H. Y., and Anthony, T. G. (2006). Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 34, 7–11. doi:10.1042/bst0340007
Wilbertz, J. H., Voigt, F., Horvathova, I., Roth, G., Zhan, Y., and Chao, J. A. (2019). Single-molecule imaging of mRNA localization and regulation during the integrated stress response. Mol. Cell 73, 946–958.e7. doi:10.1016/j.molcel.2018.12.006
Wu, B., Eliscovich, C., Yoon, Y. J., and Singer, R. H. (2016). Translation dynamics of single mRNAs in live cells and neurons. Science 352, 1430–1435. doi:10.1126/science.aaf1084
Xiong, X., Li, X., Wang, K., and Yi, C. (2018a). Perspectives on topology of the human m(1)A methylome at single nucleotide resolution. Rna 24, 1437–1442. doi:10.1261/rna.067694.118
Xiong, X., Li, X., and Yi, C. (2018b). N(1)-methyladenosine methylome in messenger RNA and non-coding RNA. Curr. Opin. Chem. Biol. 45, 179–186. doi:10.1016/j.cbpa.2018.06.017
Yamasaki, S., and Anderson, P. (2008). Reprogramming mRNA translation during stress. Curr. Opin. Cell Biol. 20, 222–226. doi:10.1016/j.ceb.2008.01.013
Yan, X., Hoek, T. A., Vale, R. D., and Tanenbaum, M. E. (2016). Dynamics of translation of single mRNA molecules in vivo. Cell 165, 976–989. doi:10.1016/j.cell.2016.04.034
Yang, P., Mathieu, C., Kolaitis, R. M., Zhang, P., Messing, J., Yurtsever, U., et al. (2020). G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell 181, 325–345.e28. doi:10.1016/j.cell.2020.03.046
Yang, X., Hu, Z., Fan, S., Zhang, Q., Zhong, Y., Guo, D., et al. (2018). Picornavirus 2A protease regulates stress granule formation to facilitate viral translation. PLoS Pathog. 14, e1006901. doi:10.1371/journal.ppat.1006901
Youn, J. Y., Dunham, W. H., Hong, S. J., Knight, J. D. R., Bashkurov, M., Chen, G. I., et al. (2018). High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol. Cell 69, 517–532.e11. doi:10.1016/j.molcel.2017.12.020
Youn, J. Y., Dyakov, B. J. A., Zhang, J., Knight, J. D. R., Vernon, R. M., Forman-Kay, J. D., et al. (2019). Properties of stress granule and P-body proteomes. Mol. Cell 76, 286–294. doi:10.1016/j.molcel.2019.09.014
Zhang, G., Xu, Y., Wang, X., Zhu, Y., Wang, L., Zhang, W., et al. (2022). Dynamic FMR1 granule phase switch instructed by m6A modification contributes to maternal RNA decay. Nat. Commun. 13, 859. doi:10.1038/s41467-022-28547-7
Zhou, J., Wan, J., Shu, X. E., Mao, Y., Liu, X. M., Yuan, X., et al. (2018). N(6)-Methyladenosine guides mRNA alternative translation during integrated stress response. Mol. Cell 69, 636–647.e7. doi:10.1016/j.molcel.2018.01.019
Keywords: stress granules, translation factors, RNA-binding proteins, RNA modification, RNA condensation
Citation: Adjibade P and Mazroui R (2023) Stress granules: stress-induced cytoplasmic mRNPs compartments linked to mRNA translational regulatory pathways. Front. RNA Res. 1:1226610. doi: 10.3389/frnar.2023.1226610
Received: 21 May 2023; Accepted: 21 July 2023;
Published: 10 August 2023.
Edited by:
A. Gregory Matera, University of North Carolina at Chapel Hill, United StatesReviewed by:
Imed Gallouzi, McGill University, CanadaCopyright © 2023 Adjibade and Mazroui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rachid Mazroui, cmFjaGlkLm1henJvdWlAY3JjaHVkZXF1ZWJlYy51bGF2YWwuY2E=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.