
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. RNA Res., 05 June 2023
Sec. RNP Biogenesis and Function
Volume 1 - 2023 | https://doi.org/10.3389/frnar.2023.1197990
This article is part of the Research TopicRibonucleoprotein formation and regulationView all 5 articles
Cajal bodies (CBs) are subnuclear domains that contribute to the biogenesis of several different classes of ribonucleoproteins (RNPs), including small nuclear RNPs. Only some cell types contain abundant CBs, such as neuronal cells and skeletal muscle, but CBs are invariant features of transformed cells. In contrast, coilin, the CB marker protein, is a ubiquitously expressed nuclear protein, but the function of coilin in cell types that lack CBs is not well understood. We have previously shown that coilin promotes microRNA biogenesis by promoting phosphorylation of DGCR8, a component of the microprocessor. Here, we identify seven additional residues of DGCR8 with decreased phosphorylation upon coilin knockdown. In addition to phosphorylation, the addition of a small ubiquitin-like modifier (SUMO) to DGCR8 also increases its stability. Because of coilin’s role in the promotion of DGCR8 phosphorylation, we investigated whether coilin is involved in DGCR8 SUMOylation. We show that coilin knockdown results in global decrease of protein SUMOylation, including decreased DGCR8 and Sp100 (a PML body client protein) SUMOylation and decreased SMN expression. Alternatively, we found that coilin expression rescued Sp100 SUMOylation and increased DGCR8 and SMN levels in a coilin knockout cell line. Furthermore, we found that coilin facilitates RanGAP1 SUMOylation, interacts directly with components of the SUMOylation machinery (Ubc9 and SUMO2), and, itself, is SUMOylated in vitro and in vivo. In summary, we have identified coilin as a regulator of DGCR8 phosphorylation and a promotor of protein SUMOylation with SUMO E3 ligase-like activity.
Cajal bodies (CBs) are subnuclear domains that function in the biogenesis of ribonucleoproteins (RNPs) including telomerase (Kiss, 2004). Coilin is required for CB formation and maintenance (Tucker et al., 2001; Collier et al., 2006; Liu et al., 2009). CBs are found in some cell types (such as neuronal cells) but not in others (such as fibroblasts) (Pena et al., 2001; Young et al., 2001; Hearst et al., 2009) (Spector et al., 1992). However, CBs are invariant features of transformed cells (Spector et al., 1992; Hearst et al., 2009). CBs contain additional proteins including the survival of motor neuron (SMN) protein and WRAP53 (TCAB1/WDR79) (Liu and Dreyfuss, 1996; Matera and Frey, 1998; Carvalho et al., 1999; Young et al., 2000; Mahmoudi et al., 2010). We have previously shown a functional relationship between coilin, the CB marker protein, and microRNA (miRNA) biogenesis. Specifically, we observed that coilin knockdown (KD) decreases mature miRNA and DGCR8 (Logan et al., 2020; Lett et al., 2021). DGCR8 and Drosha are major components of the microprocessor, which crops primary-miRNA generating precursor-miRNA during miRNA biogenesis (Han et al., 2004; Landthaler et al., 2004; Tomari and Zamore, 2005; Yeom et al., 2006). Importantly, we observed decreased miRNA biogenesis and decreased DGCR8 protein upon coilin KD in cell lines with CBs (HeLa) or lacking this structure (WI-38). Upon further investigation, we found that coilin not only interacts with DGCR8 but also promotes DGCR8 stability by promoting phosphorylation at S377 (Lett et al., 2021).
Protein SUMOylation regulates multiple biological processes, including cell division, DNA repair, and cellular metabolism (Eifler and Vertegaal, 2015; Sarangi and Zhao, 2015; Kamynina and Stover, 2017; Zhu et al., 2022). SUMO is conjugated to lysine residues in target proteins catalyzed by SUMO-specific activating (E1), conjugating (E2, Ubc9), and ligating (E3) enzymes (Wang and Dasso, 2009). There are three major SUMO proteins: SUMO1, SUMO2, and SUMO3 (Matunis et al., 1996; Mahajan et al., 1997). The SUMO groups linked to target proteins can interact with other proteins through SUMO-interaction motifs (SIMs). Thus, SUMO modification provides a platform to enhance protein–protein interaction (Minty et al., 2000). SUMOylation can influence protein localization, activity, and stability. Dysregulation of protein SUMOylation has been implicated in a wide variety of diseases (Li et al., 2003; Bawa-Khalfe and Yeh, 2010; Kho et al., 2011; Kessler et al., 2012; O’Rourke et al., 2013; Weetman et al., 2013; Luo et al., 2014; Ferdaoussi et al., 2015; Heo et al., 2015; Yu et al., 2015; Hasegawa et al., 2016).
One mechanism by which SUMOylation leads to disease is by influencing miRNA biogenesis (Herbert et al., 2013; Zhu et al., 2015; Zhu et al., 2016). MiRNAs negatively regulate gene expression by influencing the stability or the translational efficiency of target mRNAs (Pratt and MacRae, 2009). The microprocessor must be heavily regulated as disruption of miRNA function is associated with a variety of diseases (Lekka and Hall, 2018). DGCR8 is regulated by Drosha at the mRNA level and by post-translational modifications, including phosphorylation, SUMOylation, and acetylation (Han et al., 2004; Kadener et al., 2009; Triboulet et al., 2009; Wada et al., 2012; Herbert et al., 2013; Zhu et al., 2015; Zhu et al., 2016). Phosphorylation of DGCR8 has been shown to increase protein stability, and at least 23 phosphorylation sites have been identified (Herbert et al., 2013). The phosphorylation of DGCR8 promotes its SUMOylation, which acts synergistically to further increase protein stability. SUMOylation also affects the affinity of the Microprocessor for certain pri-miRNAs and the cellular localization of DGCR8 (Zhu et al., 2015; Zhu et al., 2016).
The CB-enriched protein SMN is also a substrate for SUMOylation. SMN functions in many aspects of RNP biogenesis, such as small nuclear RNP (snRNP) biogenesis (Liu and Dreyfuss, 1996; Liu et al., 1997; Pellizzoni et al., 1998; Wan et al., 2005; Workman et al., 2012) and mRNA axonal transport (Fallini et al., 2012). A deficiency of SMN causes the neurodegenerative disease spinal muscular atrophy (Lefebvre et al., 1995). SMN has a SIM-like motif in the Tudor domain (Tapia et al., 2014; Riboldi et al., 2021). Interestingly, loss of the SIM motif in SMN disrupts the formation of both the SMN complex and CBs, severely impairing snRNP biogenesis (Tapia et al., 2014; Riboldi et al., 2021). Disruption of SMN SUMOylation results in aberrant accumulation of SMN in the cytoplasm (Riboldi et al., 2021). In contrast, hyper-SUMOylation of SMN marks the protein for degradation via the proteasome (Zhang et al., 2021). In mice, a deficiency of SUMO/sentrin-specific protease 2 (SENP2), which deconjugates SUMO1 from SMN, results in hyper-SUMOylated SMN, increased degradation of SMN, and eventually an SMN deficiency (Zhang et al., 2021). SENP2-deficient mice also develop an SMA-like phenotype including flaccid paralysis, decreased motor neuron number, decreased muscle fiber number and size, and inhibition of CB formation (Zhang et al., 2021).
CBs are known to associate with the PML body (Sun et al., 2005). PML bodies are dynamic nuclear protein aggregates that function in a variety of cell processes, including DNA repair, apoptosis, and carcinogenesis (Salomoni and Pandolfi, 2002; Bernardi and Pandolfi, 2007; Bernardi et al., 2008). The PML protein is the scaffold of PML bodies (Maul et al., 2000). SUMOylation of the PML protein regulates the number and size of PML bodies (Hirano and Udagawa, 2022). Mutant PML that can no longer be SUMOylated is unable to recruit Sp100 and other PML body client proteins (Zhong et al., 2000). Sp100 is also SUMOylated by SUMO1, 2, and 3, which increases its stability (Sternsdorf et al., 1999). The interaction between SIM motifs and SUMOs on PML body proteins contributes to the liquid–liquid phase separation of these domains (Van Damme et al., 2010).
Here, we further investigate coilin’s role in the post-translational modification of DGCR8. Tandem MS confirmed our previous finding of decreased phosphorylation at S377 and identified seven additional sites with decreased phosphorylation after coilin KD. Because of the closely intertwined relationship of phosphorylation and SUMOylation of DGCR8, we examined changes in protein SUMOylation in primary and transformed cells upon coilin KD and found a global reduction in protein SUMO1 and SUMO2/3 modification. We also observed decreased SUMOylation of DGCR8 and Sp100 as well as decreased SMN expression. Moreover, we found that expression of coilin in coilin knockout (KO) mouse embryonic fibroblasts (MEF) resulted in increased Sp100 SUMOylation and increased expression of DGCR8 and SMN. We then conducted in vitro SUMOylation assays to investigate how coilin promotes SUMOylation and found that coilin promotes RanGAP1 SUMOylation similar to SUMO E3 ligases. Deletion of residues 121–291, which are in the central, intrinsically disordered region of coilin (Makarov et al., 2013), did not promote SUMOylation of RanGAP1 to the same extent as wildtype (WT) coilin. Furthermore, we found that coilin interacts directly with components of the SUMOylation machinery and can be auto-SUMOylated in vitro. We also show that coilin is SUMOylated in vivo. Given the known connections and interactions between coilin with CBs, PML bodies, DGCR8, SMN, and the biogenesis of snRNPs, small nucleolar RNPs, small Cajal body-specific RNPs, telomerase, and miRNAs (Grande et al., 1996; Hebert et al., 2001; Sun et al., 2005; Strzelecka et al., 2010; Broome and Hebert, 2013; Enwerem et al., 2014; Logan et al., 2020; Lett et al., 2021; Logan et al., 2021; Logan et al., 2022), the identification of coilin as a promoter of SUMOylation provides important mechanistic insights into the function of coilin in cell types with or without CBs.
To expand on our previous findings that coilin functions in miRNA biogenesis by promoting the phosphorylation of DGCR8 at S377 (Lett et al., 2021), we examined the phosphorylation status of DGCR8 after 72 h control or coilin KD. HeLa cells were transfected with FLAG-DGCR8 DNA 48 h after siRNA treatment. The lysate was generated and subjected to immunoprecipitation (IP) with anti-FLAG beads 24 h later. The immunoprecipitates were run on SDS-PAGE gel and Coomassie-stained, and the DGCR8 band was excised and subjected to phospho-proteomic analysis. As shown in Figure 1A and Supplementary Table S1, this analysis confirmed that S377 phosphorylation is decreased upon coilin KD and identified seven additional residues that are hypophosphorylated in coilin KD compared to control KD, including one phosphorylation site that had not yet been identified, S314. We did not detect any residues that were hyperphosphorylated with coilin KD compared to the control. All the mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD036370.

FIGURE 1. Coilin positively regulates DGCR8 phosphorylation and global SUMOylation. (A) DGCR8 phosphopeptide analysis. HeLa cells were transfected with the negative control or coilin siRNA for 48 h and then transfected with FLAG-DGCR8 for 24 h, followed by IP. IP complexes were isolated and subjected to MS/MS analysis. The abundance ratios for residues shown to be hypophosphorylated upon coilin KD compared to control KD are shown. (B) Analysis of global protein SUMOylation after coilin and Ubc9 reduction. HeLa cells were transfected with control, coilin, and Ubc9 siRNA for 72 h. Cells were harvested, and lysates were subjected to Western blotting. Anti-SUMO2/3 antibody was used to assess global protein SUMOylation. Anti-coilin and anti-Ubc9 antibodies were used to confirm siRNA knockdown.
Phosphorylation and SUMOylation of DGCR8 have been shown to act synergistically to increase protein stability, and DGCR8 phosphorylation induces SUMOylation (Zhu et al., 2015). Because we found that coilin KD reduced DGCR8 phosphorylation and protein levels (Lett et al., 2021), we hypothesized that coilin KD would also affect protein SUMOylation. To test this, we transfected HeLa cells with control, coilin, or Ubc9 siRNA for 72 h (Figure 1B). We examined total SUMO2/3 protein SUMOylation via Western blotting and saw a decrease in SUMO2/3 signal in cells with coilin KD compared to the control (top panel, lane 2 compared to lane 1). Because the mature forms of SUMO2 and 3 are approximately 97% identical, there is no available antibody that can differentiate between the two. As expected, KD of Ubc9, the only SUMO E2 enzyme in humans, greatly decreased the number of proteins modified by SUMO2/3 (top panel, lane 3 compared to lane 1). Ubc9 and coilin KD were verified by probing with the appropriate antibodies.
While the data in Figure 1 suggest a role for coilin in protein SUMOylation, we examined the effects of the reduction of coilin and other CB proteins in cells with and without CBs (Figure 2) to provide further support for our hypothesis. To explore whether SUMOylation is a function of nucleoplasmic coilin or CBs, we utilized cells expressing abundant (HeLa) or few (WI-38) CBs (Spector et al., 1992). Endogenous SUMO1 and SUMO2/3 levels were examined by Western blotting from cells in which coilin, SMN, and WRAP53 were reduced. HeLa lysate showed decreased SUMO1 (Figure 2A) and SUMO2/3 (Figure 2B) with coilin KD (lane 2) compared to the control (lane 1). However, we did not see a comparable decrease in SUMO1 or SUMO2/3 signals after SMN or WRAP53 KD (lanes 3 and 4, respectively). Quantification of the SUMO signals normalized to actin confirmed a statistically significant decrease in SUMO1 (Figure 2C) and SUMO2/3 (Figure 2D) after coilin KD in HeLa cells. For SUMO2/3, there was no statistical difference with SMN and WRAP53 KD. However, while there was not a significant change in SUMO1 levels with WRAP53 KD, we did see a statistically significant decrease in SUMO1 with SMN KD. Additionally, we saw a decrease in SUMO2/3 signal using three different siRNAs to KD coilin (Supplementary Figure S1A).
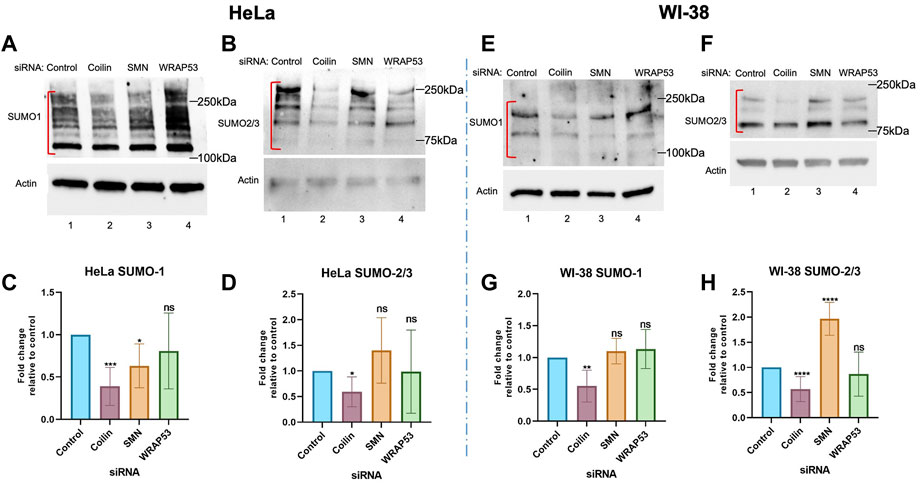
FIGURE 2. Coilin knockdown in cells with and without CBs reduces protein SUMOylation. HeLa (A,B) and WI-38 (E,F) cells were transfected with indicated siRNA for 72 h, followed by Western blotting to assess endogenous SUMO1 [(A) HeLa; (E) WI-38] and SUMO2/3 [(B) HeLa; (F) WI-38] levels. Actin was detected to assess protein loading. Quantifications of HeLa SUMO1 (C) and SUMO2/3 (D) and WI-38 SUMO1 (G) and SUMO2/3 (H) normalized to actin with the control ratio set to 1 are shown. N > 4, including three biological replicates. Error bars represent SD. *p < 0.0332, **p < 0.0021, ***p < 0.0002, ****p < 0.0001. ns, not significant. Brackets indicate the SUMO1 or 2/3 signal used for the quantification.
Our results from a primary cell line that has few CBs, WI-38 (Figures 2E–H), are similar to those obtained with HeLa. Specifically, SUMO1 (Figure 2E) and SUMO2/3 (Figure 2F) signals were decreased after coilin reduction compared to the control (lane 2 compared to lane 1), but we did not see a reduction in SUMO1 or SUMO2/3 signal with SMN (lane 3) or WRAP53 (lane 4) KDs. Quantification of SUMO1 (Figure 2G) and SUMO2/3 (Figure 2H) revealed a significant reduction in protein SUMOylation upon coilin KD comparable to that seen in HeLa cells. SUMO1 levels with SMN and WRAP KDs were not significantly changed compared to the control, and neither were SUMO2/3 levels with WRAP53 KD. However, SMN KD in WI-38 cells caused a significant increase in SUMO2/3 signal compared to the control. The increase of SUMO2/3 SUMOylation upon 72 h SMN KD in WI-38 is surprising but may reflect the deleterious consequences of SMN KD in this primary cell line that result in reduced cell numbers. All siRNA KDs were confirmed via Western blotting (Supplementary Figure S1). Overall, these data indicate that nucleoplasmic coilin, not strictly CBs, is involved in protein SUMOylation.
To provide further evidence of coilin’s role in protein SUMOylation, we examined endogenous SUMO1 and SUMO2/3 protein modifications in the wild-type MEF (MEF26) and coilin knockout MEF (MEF42) cell lines. The MEF42 cell line was generated by deleting 85% of the coilin gene’s coding sequence, a 10 kb deletion encompassing exon 2 through exon 7 (Tucker et al., 2001). Coilin KO was confirmed by Western blotting (Figure 3A). As shown in Figure 3, protein SUMOylation in WT MEF26 and coilin KO MEF42 cells was assessed via Western blotting. Coilin KO resulted in decreased protein modification with both SUMO1 (Figure 3B, lane 2 vs. lane 1) and SUMO2/3 (Figure 3D, lane 2 vs. lane 1). Similar to the results observed after coilin KD in HeLa and WI-38 cells, quantification of SUMO1 (Figure 3C) and SUMO2/3 (Figure 3E) revealed a significant (∼50%) decrease in MEF42 compared to MEF26 cells.
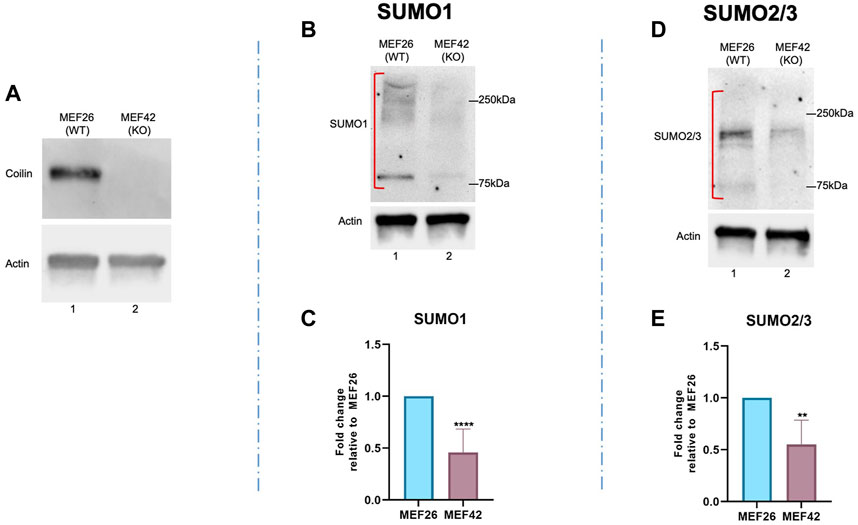
FIGURE 3. Coilin KO results in a global decrease in SUMO1 and SUMO2/3 protein modification. (A) Coilin KO was confirmed via Western blotting of MEF26 (WT) and MEF42 (KO) lysates by probing with anti-coilin antibodies. Anti-actin antibodies were used to assess protein loading. Protein was isolated from wild-type MEF cells (MEF26) and coilin KO cells (MEF42) and subjected to SDS PAGE, followed by Western blotting. Proteins modified by SUMO1 (B) and SUMO2/3 (D) were detected with appropriate antibodies. Blots were also probed with actin to assess loading. Quantifications of SUMO1 (C) and SUMO2/3 (E) signals normalized to actin with the MEF26 ratio set to 1. Error bars represent SD. **p < 0.0021, ****p < 0.0001. Red brackets indicate the SUMO1 or 2/3 signal used for quantification. N > 5, including two biological replicates.
Our data thus far demonstrate a role for coilin in protein SUMOylation. Next, we aimed to identify specific proteins whose SUMOylation is disrupted by coilin reduction. Our previously published results showing a relationship between coilin and miRNA biogenesis, DGCR8 protein levels, and DGCR8 post-translational modification (Logan et al., 2020; Lett et al., 2021; Logan et al., 2021; 2022) inspired us to first examine DGCR8 SUMOylation. As shown in Figure 4A, HeLa cells were transfected with control or coilin siRNA, followed by transfection with FLAG-DGCR8 and His-SUMO1. His-SUMO1-conjugated proteins were pulled down with nickel-nitrilotriacetic acid (Ni-NTA) beads, and Western blotting was used to visualize total FLAG-DGCR8 and SUMOylated FLAG-DGCR8 (top panel, lane 3). Compared to the control (lane 3), the amount of SUMOylated FLAG-DGCR8 (bracket) was decreased upon coilin KD (lane 4). We also observed decreased total SUMO1 signals in the coilin KD input and pulldown lanes compared to the control (bottom panel; compare the SUMO1 signals in lanes 2 and 4 to those in lanes 1 and 3). These data further support the role of coilin in the regulation of the microprocessor and miRNA biogenesis.
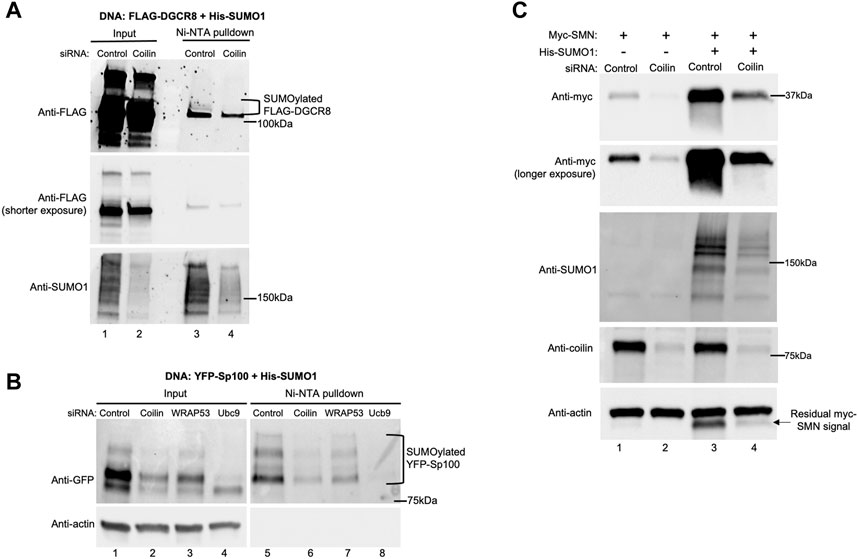
FIGURE 4. Coilin KD decreases SUMOylation of DGCR8 and Sp100 and SMN expression. HeLa cells were transfected with indicated siRNA for 48 h, followed by DNA transfection with His-SUMO1 and FLAG-DGCR8 (A) or YFP-Sp100 (B) for 24 h. Ni-NTA beads were used to pull down proteins modified with His-tagged SUMO1. Lysates were incubated with Ni-NTA beads for 1 h, followed by washing, SDS-PAGE, and Western blotting. YFP-Sp100 was detected with anti-GFP antibodies, and FLAG-DGCR8 was detected with anti-FLAG antibodies. Input lanes represent 4% of lysate used for pulldown. (C) HeLa cells were transfected with the control or coilin siRNA for 48 h, followed by transfection with myc-SMN alone or myc-SMN and His-SUMO1 for 24 h. Western blotting was used to analyze myc-SMN expression, and the blot was probed with indicated antibodies.
Another process that is heavily dependent upon protein SUMOylation is PML body formation. Pathological conditions, such as viral infections, disrupt PML bodies by inhibition of PML protein SUMOylation and abolishment of SUMOylated Sp100 (Kim et al., 2011; Schilling et al., 2017). For our studies, we examined Sp100 SUMOylation (Figure 4B). HeLa cells were transfected with siRNA, followed by DNA transfection with YFP-Sp100 and His-SUMO1. Ni-NTA beads were used to pull down proteins modified by His-SUMO1, followed by Western blotting and detection of total (top, left panel) and SUMOylated Sp100 (top, right panel). When probing for YFP-Sp100 (top panel), we found a decrease in SUMOylated YFP-Sp100 in the coilin KD compared to the control and WRAP53 KDs (lane 6 compared to lanes 5 and 7, respectively; SUMOylated YFP-Sp100 bands are denoted with a bracket). Additionally, we found less total YFP-Sp100 in coilin KD compared to the control and WRAP53 KDs (lane 2 compared to lanes 1 and 3, respectively). However, because we also see less total YFP-Sp100 in the Ubc9 KD (lane 4 compared to lane 1), and the actin probing (bottom panel) indicates equal protein loading, we attribute the decrease in total YFP-Sp100 to the protein being less stable in the coilin KD background due to decreased SUMOylation.
The final SUMOylated protein that we investigated is SMN (Figure 4C). SMN is a known CB protein that interacts directly with coilin (Hebert et al., 2001; Hebert et al., 2002; Toyota et al., 2010; Tapia et al., 2014). SMN also has a SIM motif in its Tudor domain, indicating SMN interacts with other SUMOylated proteins (Tapia et al., 2014; Riboldi et al., 2021). SUMOylation of SMN modulates its stability, localization, and degradation (Tapia et al., 2014; Riboldi et al., 2021; Zhang et al., 2021). After control and coilin KDs, HeLa cells were transfected with myc-SMN alone or in combination with His-SUMO1. When probing for myc-SMN, we observed an increased myc-SMN expression when co-transfected with His-SUMO1 for both the control (lane 3 compared to lane 1) and the coilin KD (lane 4 compared to lane 2). However, myc-SMN expression was decreased in the coilin KD compared to the control with (lane 4 compared to lane 3) and without His-SUMO1 (lane 2 compared to lane 1). Coilin KD was confirmed (lanes 2 and 4 compared to lanes 1 and 3), and the blot was probed with actin to assess protein loading (bottom panel). We also conducted Ni-NTA pulldowns to evaluate the level of His-SUMO1 conjugated SMN, with and without coilin KD, but were unable to detect SUMOylated SMN (our unpublished observations). We hypothesize that the lack of detected SUMOylated SMN in this assay is a consequence of there being fewer SUMOylated residues in SMN compared to those found in Sp100.
After finding that coilin KD decreases DGCR8 and Sp100 SUMOylation and SMN expression, we investigated whether the opposite would also be true—that coilin expression would increase SUMOylation and/or expression. To explore what region of coilin may be responsible for this activity, in addition to full-length coilin, we also utilized a coilin construct with a deletion within coilin’s intrinsically disordered region (Δ121–291: deletion of aa 121–291). This region of coilin is predicted to be highly SUMOylated and contains one of coilin’s three predicted SIMs. Therefore, we hypothesize that deletion of this region would hinder coilin’s ability to promote protein SUMOylation. Coilin KO MEF42 cells were transfected with FLAG-DGCR8 alone (Figure 5A, lane 1), FLAG-DGCR8 and His-SUMO1 (lane 2), FLAG-DGCR8, His-SUMO1, and WT myc-coilin (lane 3), or FLAG-DGCR8, His-SUMO1, and myc-coilin Δ121–291 (lane 4). Western blot of the lysates showed the highest FLAG-DGCR8 expression when co-transfected with His-SUMO1 and WT myc-coilin (top panel, lane 3 compared to lanes 1, 2, and 4), followed by co-transfection with His-SUMO1 and myc-coilin Δ121–291 (top panel, lane 4 compared to lanes 1 and 2). Because FLAG-DGCR8 expression increased with the addition of His-SUMO1 (lane 2 compared to lane 1) and further increased with the addition of WT myc-coilin (lane 3 compared to lane 2), we believe this increased expression is due to greater protein stability via promotion of SUMOylation by coilin. Furthermore, MEF42 cells transfected with His-SUMO1 and Δ121–291 displayed greater expression of FLAG-DGCR8 than those transfected with His-SUMO1 (top panel, lane 4 compared to lane 2) but less expression than those transfected with His-SUMO1 and WT myc-coilin (top panel, lane 4 compared to lane 3), indicating that this region contributes to the promotion of DGCR8 expression. However, it is also possible that deletion of a large region of the intrinsically disordered region in coilin, such as that found in the Δ121–291 mutant, disrupts the N- and C-terminal structured domains of coilin, thereby altering coilin function. Transfections were confirmed with anti-coilin (second panel) and anti-SUMO1 (third panel) antibodies, and actin levels were used to assess total protein loading (bottom panel).
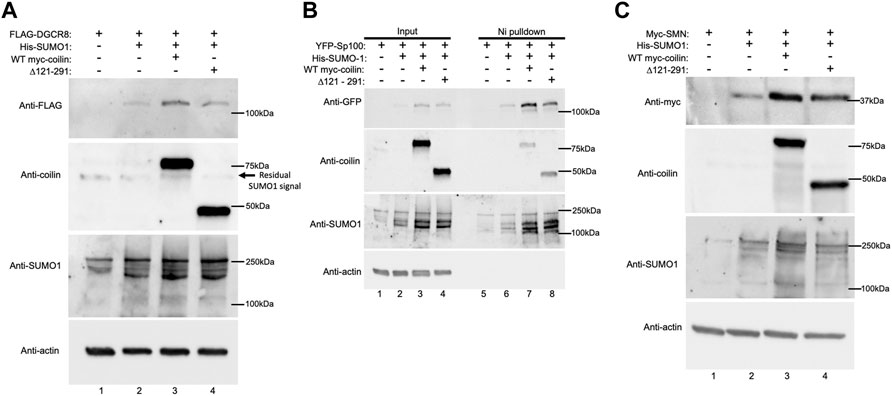
FIGURE 5. Exogenous coilin expression increases Sp100 SUMOylation and expression of DGCR8 and SMN. (A) MEF42 cells were transfected with FLAG-DGCR8 alone (lane 1), FLAG-DGCR8 and His-SUMO1 (lane 2), FLAG-DGCR8, His-SUMO1, and WT myc-coilin (lane 3), and FLAG-DGCR8, His-SUMO1, and myc-coilin ∆121–291 (lane 4) for 24 h. Cells were lysed, and lysates were subjected to SDS-PAGE, followed by Western blotting. (B) MEF42 cells were transfected with YFP-Sp100 alone (lanes 1 and 5), YFP-Sp100 and His-SUMO1 (lanes 2 and 6), YFP-Sp100, His-SUMO1, and WT myc-coilin (lanes 3 and 7), and YFP-Sp100, His-SUMO1, and myc-coilin ∆121–291 (lanes 4 and 8). Lysates from cells transfected with YFP-Sp100 were incubated with Ni beads for 1 h to pull down His-SUMO1-modified proteins. Western blotting was used to analyze the input lysate (lanes 1–4) and pulldown proteins (lanes 5–8). The blot was probed with indicated antibodies. Input represents 4% of lysate used for pulldown. (C) MEF42 cells were transfected with myc-SMN alone (lane 1), myc-SMN and His-SUMO1 (lane 2), myc-SMN, His-SUMO1, and WT myc-coilin (lane 3) and myc-SMN, His-SUMO1, and myc-coilin ∆121–291 (lane 4) for 24 h. Western blotting was used to analyze myc-SMN and FLAG-DGCR8 expression and was probed with indicated antibodies.
In contrast to DGCR8, Sp100 is a highly SUMOylated protein. Therefore, changes in Sp100 SUMOylation are more easily detected compared to DGCR8. To explore the effect of coilin expression on Sp100 SUMOylation, coilin KO MEF42 cells were transfected with YFP-Sp100 alone (Figure 5B, lanes 1 and 5), YFP-Sp100 and His-SUMO1 (lanes 2 and 6), YFP-Sp100, His-SUMO1, and WT myc-coilin (lanes 3 and 7), or YFP-Sp100, His-SUMO1, and myc-coilin Δ121–291 (lanes 4 and 8). Ni-NTA beads were used to pull down proteins modified with His-SUMO1. Input lysates and pulldowns were subjected to Western blotting. When transfecting with YFP-Sp100 alone, we did not detect any SUMOylated YFP-SP100 after Ni-NTA pulldown (top panel, lane 5) but we did when YFP-Sp100 was co-transfected with His-SUMO1 (top panel, lane 6). Interestingly, we see an increase in SUMOylated YFP-Sp100 when myc-coilin Δ121–291 was expressed (top panel, lane 8 compared to lane 6) and a further increase with the addition of WT myc-coilin (top panel, lane 7 compared to lane 8). Like DGCR8, we see a similar pattern in total YFP-SP100 expression with the highest expression occurring when WT myc-coilin was co-transfected (top panel, lane 3), followed by myc-coilin Δ121–291 (top panel, lane 4). Transfections were confirmed with anti-coilin (second panel) and anti-SUMO1 (third panel) antibodies, and actin expression was used to assess protein loading (bottom panel).
Finally, we investigated SMN expression in coilin KO MEFs (Figure 5C). MEF42 cells were transfected with myc-SMN alone (lane 1), myc-SMN and His-SUMO1 (lane 2), myc-SMN, His-SUMO1, and WT myc-coilin (lane 3), or myc-SMN, His-SUMO1, and myc-coilin Δ121–291 (lane 4). Western blotting was used to visualize differences in myc-SMN expression. Similar to the results observed with FLAG-DGCR8 and YFP-Sp100, the highest expression of myc-SMN occurred when myc-SMN was co-transfected with His-SUMO1 and WT myc-coilin (top panel, lane 3). Again, co-transfection of myc-SMN with His-SUMO1 and myc-coilin Δ121–291 yielded greater expression than myc-SMN alone and myc-SMN co-transfected with His-SUMO1, but less than WT myc-coilin (top panel, lane 4 compared to lanes 1, 2, and 3, respectively). Transfections were confirmed (second and third panels), and actin levels were used to assess total protein loading (bottom panel). In summary, all three proteins, SMN, DGCR8, and Sp100, displayed the greatest increase of expression when WT myc-coilin was expressed, while Sp100 also showed increased SUMOylation. In all coilin expression experiments in MEF42, the total SUMO1 signal was greatest in reactions containing WT myc-coilin. We also conducted Ni-NTA pulldowns in the MEF42 background with DGCR8 and SMN but were unable to detect SUMOylated DGCR8 or SMN by this assay (unpublished observations). Both DGCR8 and SMN have fewer SUMOylated residues compared to Sp100, and we speculate that this accounts for our inability to detect SUMOylated DGCR8 and SMN in the coilin KO background.
One way in which coilin could be promoting protein SUMOylation is by altering the levels or activity of the SUMO conjugation (Ubc9) or deconjugation (SENPs) enzymes. However, we did not see any change in Ubc9 or SENP1 levels after coilin KD in primary (WI-38) or transformed (HeLa) cells (Supplementary Figures S2A–D). This suggests coilin might be enhancing SUMOylation via E3 ligase-like activity. To test this idea, we conducted in vitro SUMOylation assays to investigate whether coilin possesses E3 ligase-like activity. These reactions were conducted using a decreased amount of E1 and E2 enzymes in order to evaluate E3 ligase activity, as previously described (Yang et al., 2018b). His-T7-tagged coilin or WRAP53 partially purified from E. coli were added to SUMOylation reactions containing diluted E1 and E2 enzymes, SUMO1, Mg-ATP, and a RanGAP1 fragment as the target protein. Reactions were subjected to SDS-PAGE, Western blotting, and probing with anti-SUMO1 antibodies (Figure 6A). Reactions using the standard amount of E1 and E2 enzymes were also conducted, without and with Mg-ATP, to verify that SUMO1 was enzymatically added to RanGAP1 (lanes 4 and 5). The RanGAP1-SUMO1 signal when coilin was added to the reaction (top panel, lane 2) is greater than that observed for elution buffer control (lane 1) or WRAP53 (lane 3), indicating that coilin may possess E3 ligase-like activity. The presence of partially purified coilin and WRAP53 proteins was confirmed by probing for coilin (middle panel) and T7 (bottom panel), respectively. Quantification of SUMOylated RanGAP1 showed a two-fold increase when coilin was included in the reaction compared to the elution buffer control or WRAP53 (Figure 6B).
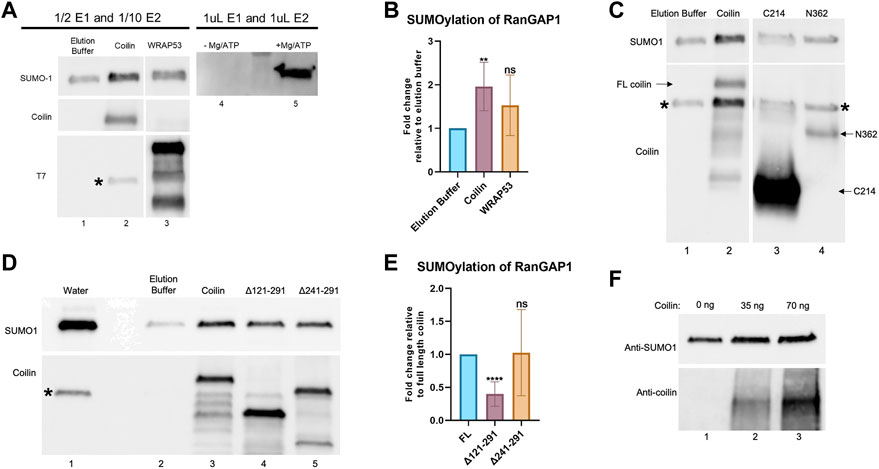
FIGURE 6. Coilin promotes SUMOylation of RanGAP1 in vitro. (A) In vitro SUMOylation assay, containing a RanGAP1 fragment as the target, decreased amounts of E1 and E2 enzymes, SUMO1, Mg-ATP, and elution buffer (lane 1), His-T7-coilin (lane 2), or His-T7-WRAP53 (lane 3). Reactions with undiluted amounts of E1 and E2 with (lane 5) and without (lane 4) Mg/ATP are shown for comparison. Reactions were subjected to SDS-PAGE, Western blotting, and probing with indicated antibodies. * denotes residual SUMO1 signal. (B) Quantification of in vitro SUMOylation assays with elution buffer set to 1 (N = 6; **p < 0.0021; ns, not significant). (C) In vitro SUMOylation of RanGAP1 was conducted as described in (A) with elution buffer (lane 1), His-T7-coilin (lane 2), His-T7-coilin C-terminal fragment (C214, lane 3), and His-T7-coilin N-terminal fragment (N362, lane 4). The blot was probed with anti-SUMO1 (top) and anti-coilin (bottom) antibodies. Arrows denote the location of full-length (FL) coilin, C214, and N362. * denotes a residual SUMO1 signal. Note: the data shown in lanes 1 and 2 are obtained from the same experiment shown in panel A, lanes 1 and 2. (D) In vitro SUMOylation of RanGAP1 was conducted as described in (A) with water (lane 1), elution buffer (lane 2), His-T7-coilin (lane 3), His-T7-coilin Δ121–291 (lane 4), and His-T7-coilin Δ241–291 (lane 5). Western blot of the assay was probed with anti-SUMO1 (top) and anti-coilin (bottom) antibodies. * denotes a residual SUMO1 signal. (E) Quantification of in vitro SUMOylation assays normalized to full-length coilin (N = 8; ****p < 0.0001; ns, not significant). (F) In vitro SUMOylation assay including RanGAP1 fragment substrate, E1 and E2 enzymes, SUMO1, Mg/ATP, and varying amounts of highly purified coilin (0 ng–lane 1, 35 ng–lane 2, and 70 ng–lane 3). Western blot of the assay was probed with anti-SUMO1 (top) and anti-coilin (bottom) antibodies.
Known SUMO E3 ligases can be divided into two groups: those that contain SIMs and those with SP-RING domains. Coilin has three predicted SIMs (aa 29–32, 145–148, and 512–515). To begin an analysis of the region in coilin responsible for this putative SUMO E3 ligase-like activity, we conducted additional in vitro SUMOylation assays (using the same conditions in 6A) with an N-terminal (aa 1–362, N362) or C-terminal (aa 362–576, C214) coilin fragment (Figure 6C). N362 is missing one SIM (512–515), while C214 is missing two SIMs (29–32 and 145–148). Neither of these fragments promoted the SUMOylation of RanGAP1 to the level observed with full-length coilin (top panel, lane 2 compared to lanes 3 and 4), indicating that all three SIMs or the central, intrinsically disordered region of coilin spanning these two fragments may be essential for this activity. Full-length coilin, C214, and N362 fragments were detected with anti-coilin antibodies (bottom panel, lanes 2, 3, and 4, respectively; labeled with arrows). We next examined coilin constructs with deletions in coilin’s intrinsically disordered region (Δ121–291: deletion of aa 121–291 and Δ241–291: deletion of aa 241–291). The Δ121–291 coilin construct is also missing one SIM (145–148). In vitro SUMOylation assays (using the same conditions as 6A) revealed that Δ121–291 was not capable of promoting the SUMOylation of RanGAP1 to the same extent as full-length coilin (Figure 6D, upper panel, lane 4 compared to lane 3). Anti-coilin antibodies were used to detect full-length coilin, Δ121–291, and Δ241–291 (bottom panel, lanes 3, 4, and 5, respectively). Quantification of SUMOylated RanGAP1, when corrected for the amount of coilin construct present and normalized to that obtained for reactions using full-length coilin, revealed that reactions containing Δ121–291 had less SUMOylated RanGAP1. No significant difference in the ability of Δ241–291 and full-length coilin to promote RanGAP1 SUMOylation (Figure 6E) was observed. The data indicate that amino acids 121–241 are required to achieve coilin’s full ability to promote SUMOylation. However, this section of coilin is not solely responsible for the activity as deletion of these residues reduced but did not eliminate the SUMOylation ability of coilin as evidenced by the enrichment of the SUMO1 signal with the addition of Δ121–291 compared to an elution buffer (Figure 6D, compare lane 2 to lane 4).
Even though bacteria are not capable of SUMOylating proteins, and no SUMOylation enzymes have been identified in bacteria, we next conducted experiments with highly purified bacterially expressed coilin (Supplementary Figure S2E) to confirm that coilin is, in fact, responsible for the promotion of RanGAP1 SUMOylation. We have previously published the coilin purification protocol, which involves bacterial expression of GST-coilin, GST-tag removal, RNase/DNase treatment, SDS-PAGE, electroelution of the excised SDS-PAGE coilin band, SDS removal, and dialysis against high salt PBS (Broome and Hebert, 2013). An amount (35 or 70 ng) of highly purified coilin in high-salt PBS was added to SUMOylation assays containing decreased amounts of E1 and E2, RanGAP1, SUMO1, and Mg/ATP. A reaction containing decreased amounts of E1 and E2, RanGAP1, SUMO1, Mg/ATP, and high-salt PBS was used as the control (Figure 6F). Western blot of these assays revealed that the addition of 35 ng of highly purified coilin resulted in increased SUMOylation of RanGAP1 compared to no coilin (top panel, lane 2 compared to lane 1), and the addition of 75 ng of coilin further increased RanGAP1 SUMOylation (top panel, lane 3 compared to lane 2). The addition of coilin to the assays was confirmed by probing with anti-coilin antibodies (bottom panel).
Our observation that coilin is involved in protein SUMOylation in vitro and in vivo led us to investigate whether coilin can directly interact with the SUMOylation machinery. To test this, we performed Ni-NTA pulldown assays using pure Ubc9 and SUMO2 and partially purified His-T7 tagged coilin and WRAP53 (Figure 7). Ubc9 was incubated with Ni beads alone (Figure 7A, lane 2), Ni beads and His-T7-coilin (lane 3), or Ni beads and His-T7-WRAP53 (lane 4) for 1 h at 4°C. The beads were then extensively washed and subjected to Western blotting and probing for Ubc9. As shown in Figure 7A, reactions containing coilin recovered the most Ubc9 compared to the amount recovered by WRAP53 or beads alone. The presence of His-T7-tagged coilin and WRAP53 was confirmed using the appropriate antibodies (bottom panel). Ni-NTA pulldowns utilizing SUMO2 were performed in the same manner (Figure 7B), and we observed similar results. Specifically, the most SUMO2 was recovered in reactions containing His-T7-coilin (top panel, lane 3). The presence of His-T7-tagged coilin and WRAP53 was confirmed with appropriate antibodies (bottom panel). We also found a direct interaction between SUMO1 and coilin (Supplementary Figure S2F).
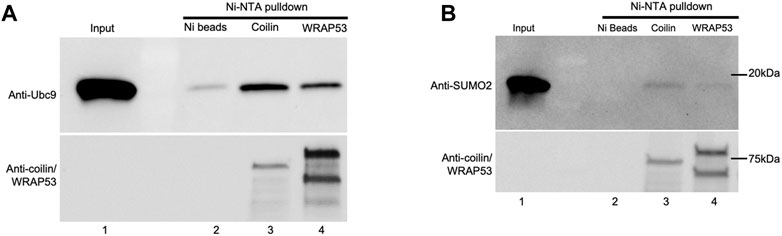
FIGURE 7. Coilin interacts directly with Ubc9 and SUMO2. His-T7-tagged coilin and WRAP53 were partially purified from bacteria and incubated with 1 ul Ubc9 (A) and SUMO2 (B) for 1 h at 4°C along with Ni-NTA beads. Beads were then washed and subjected to SDS-PAGE, Western blotting, and protein detection. Input [lane 1, (A,B)] represents 100% (1 ul) of protein used for pulldown. Ubc9 and SUMO2 were incubated with Ni-NTA beads alone as a negative control [(A), lane 2 and (B), lane 2, respectively]. Western blot of input and the Ni pulldowns were probed with indicated antibodies.
Auto-SUMOylation, the ability to self-SUMOylate in the absence of an E3 ligase, is one of the hallmarks of bona fide SUMO E3 ligases (Pichler et al., 2017). Therefore, we next examined if coilin is SUMOylated in cells and is capable of auto-SUMOylation in vitro. Using highly purified coilin (Supplementary Figure S2E), we performed in vitro auto-SUMOylation assays with (Figure 8A, lane 2) and without Mg/ATP (lane 1). Western blot of the assays was probed with anti-SUMO1 antibodies that detected SUMOylated coilin (top panel, lane 2 compared to lane 1). In addition to bands corresponding in size to SUMOylated coilin, we detected a slight decrease in unconjugated SUMO1 when Mg/ATP was present (top panel, lane 2 compared to lane 1, denoted with arrow). To evaluate if coilin is SUMOylated in vivo and begin an examination of the coilin regions that are SUMOylated, HeLa cells were transfected with WT myc-coilin or myc-coilin Δ121–291 with and without His-SUMO1 for 24 h, followed by Ni-NTA pulldown of His-SUMO1 (Figure 8B). Western blot of input lysates (lanes 2–5) and pulldowns (lanes 6–7) were probed with anti-coilin antibodies. Bands corresponding in size to SUMOylated WT myc-coilin in both the input and the Ni-NTA pulldown lanes that were co-transfected with His-SUMO1 were detected (top panel, lane 3 compared to lane 2 and lane 7 compared to 6, respectively). However, we did not detect any SUMOylated myc-coilin Δ121–291 when co-transfected with His-SUMO1 in the input or pulldown lanes (top panel, lane 5 compared to lane 4 and lane 9 compared to lane 8, respectively), indicating that amino acids 121–291 contain SUMOylated lysine residues. However, it is also possible that this deletion alters the coilin structure and inhibits SUMOylation. The blot was also probed with anti-SUMO1 antibodies to confirm transfections (bottom panel).
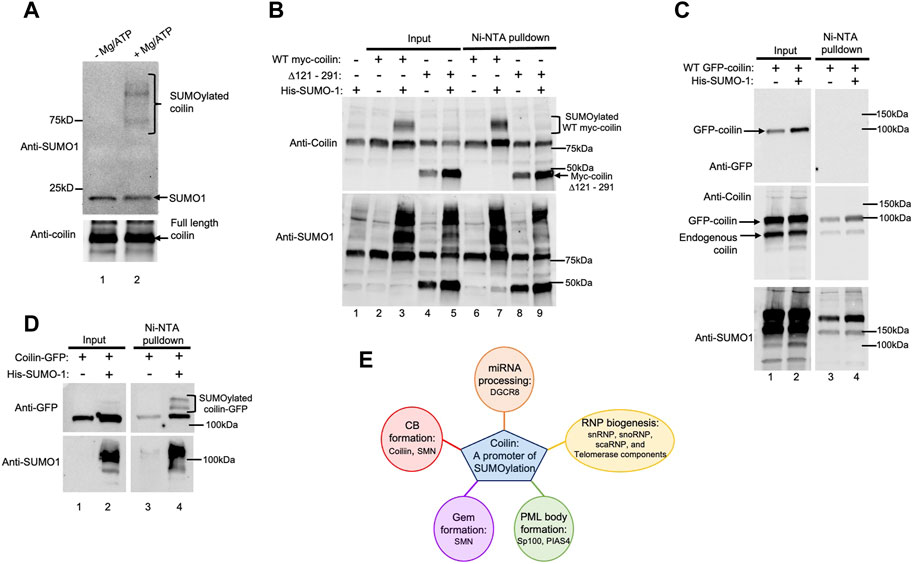
FIGURE 8. Coilin is auto-SUMOylated in vitro and SUMOylated in vivo. (A) In vitro SUMOylation assays containing 70 ng of highly purified coilin as the substrate, E1 and E2 enzymes, and SUMO1 with (lane 2) and without Mg/ATP (lane 1). SUMOylated coilin and unconjugated SUMO1 are denoted. (B) HeLa cells were transfected with WT myc-coilin or myc-coilin ∆121–291 with and without His-SUMO1 (WT myc-coilin alone: lanes 2 and 6; WT myc-coilin + His-SUMO1: lanes 3 and 7; myc-coilin ∆121–291 alone: lanes 4 and 8; myc-coilin ∆121–291 + His–SUMO1: lanes 5 and 9). (C) HeLa cells were transfected with GFP-coilin with (lanes 2 and 4) and without His-SUMO1 (lanes 1 and 3). (D) HeLa cells were transfected with coilin-GFP with (lanes 2 and 4) and without His-SUMO1 (lanes 1 and 3). All Ni-NTA pulldowns (B–D) were incubated on Ni beads for 1 h, followed by SDS-PAGE and Western blotting. Inputs represent 4% of lysate used for pulldowns. Blots were probed with anti-coilin and anti-SUMO1 antibodies. Brackets indicate SUMOylated coilin. (E) Schematic of possible target proteins of coilin-promoted SUMOylation and impacted cell processes.
Lastly, we examined the SUMOylation of coilin with a GFP tag located on the N-terminus (GFP-coilin; Figure 8C) or the C-terminus (coilin-GFP; Figure 8D). These two constructs exhibit different localization patterns when expressed in cells. Specifically, GFP-coilin has clear enrichment in the nucleoplasm, as does endogenous coilin, in addition to enrichment in CBs. In contrast, coilin-GFP has relatively less nucleoplasmic signal and can generate numerous and large CBs when overexpressed (Shpargel et al., 2003). We were curious whether this localization difference could be due to changes in SUMOylation that are influenced by the location of the GFP tag on the N-terminus or C-terminus of coilin. To test this concept, HeLa cells were transfected with GFP-coilin with and without His-SUMO1, followed by Ni-NTA pulldown of His-SUMO1 (Figure 8C). Western blot of input lysates (lanes 1 and 2) and pulldowns (lanes 3 and 4) was probed with anti-GFP (top panel), anti-coilin (middle panel), and anti-SUMO1 (bottom panel) antibodies. However, bands corresponding in size to SUMOylated GFP-coilin were not detected in pulldown lanes co-transfected with His-SUMO1 (lane 4 compared to lane 3). Interestingly, we were able to detect SUMOylated coilin-GFP. HeLa cells were transfected with coilin-GFP with and without His-SUMO1, followed by Ni-NTA pulldown of His-SUMO1 modified proteins (Figure 8D). Input lysates and pulldowns were subjected to SDS-PAGE, followed by Western blotting. Anti-GFP antibodies detected bands corresponding in size to SUMOylated coilin-GFP in the pulldown lane that was co-transfected with His-SUMO1 (top right panel, lane 4 compared to lane 3; indicated by bracket). These bands were also slightly detected in the input lane co-transfected with His-SUMO1 (top left panel, lane 2 compared to lane 1). The signal of these bands was enriched in the Ni-NTA pulldown lane, indicating the specificity of the pulldown. His-SUMO1 transfections were confirmed by probing with anti-SUMO1 antibodies (bottom panels).
Here, we have further explored coilin’s role in the regulation of miRNA biogenesis and identified a new role of coilin in protein modification. We have shown by Western blot utilizing siRNA KD and coilin KO cell lines, Ni-NTA pulldown assays, and in vitro SUMOylation experiments that coilin plays a role in protein SUMOylation. The expression of DGCR8 is tightly regulated because it is required for normal miRNA biogenesis and normal cell functions. Dysregulation of DGCR8 and associated aberrant expression of miRNAs is detected in many diseases such as schizophrenia and several kinds of cancers (Horikawa et al., 2008; Brzustowicz and Bassett, 2012; Earls et al., 2012; Kitagawa et al., 2013; Kim et al., 2014). Post-translational modifications are one of the many mechanisms utilized to regulate DGCR8 protein levels. Phosphorylation has been shown to produce a graded increase in DGCR8 stability and is conducted at least in part by extracellular signal-regulated kinases (ERKs) linking the microprocessor activity to extracellular signals. Furthermore, HeLa cells transfected with a phospho-mimic DGCR8 exhibit increased cell proliferation and migration (Herbert et al., 2013). Therefore, phosphorylation of DGCR8 not only increased microprocessor levels but also altered the miRNA profile (Herbert et al., 2013). We have previously reported coilin as a regulator of miRNA biogenesis at least in part by promoting the phosphorylation of DGCR8 at S377 and increasing protein stability (Lett et al., 2021). Here, we identified seven additional residues of DGCR8 whose phosphorylation is promoted by coilin. Our previous data in combination with our current finding that coilin KD causes global reduction in protein SUMOylation have led us to believe that coilin is specifically involved in DGCR8 SUMOylation either directly or indirectly by promoting its phosphorylation. DGCR8 has at least four SUMOylation sites (Supplementary Figure S3A). SUMOylation of K707 acts synergistically with phosphorylation to increase DGCR8 stability and also affects the direct functions of pri-miRNAs, while K259 SUMOylation is responsible for nuclear localization (Zhu et al., 2015; Zhu et al., 2016). Our data demonstrate that coilin possesses SUMO E3 ligase-like activity and promotes the SUMOylation of DGCR8, indicating yet another role for coilin in the regulation of the microprocessor. However, coilin’s role in DGCR8 phosphorylation and how coilin-promoted phosphorylation contributes to DGCR8 stability compared to SUMOylation is not yet clear. Future studies will aim to elucidate the mechanism by which coilin promotes DGCR8 phosphorylation. We also plan to identify and mutate lysine residues in DGCR8 that are SUMOylated by coilin and examine whether coilin-mediated SUMOylation, phosphorylation, or both contribute to DGCR8 protein stability.
We also investigated coilin’s role in SUMOylation of the PML body client protein, Sp100, and CB protein, SMN. The protein–protein interactions afforded by SUMO proteins and SIMs are required for PML body condensation, as unmodified PML protein is unable to recruit client proteins (Hirano and Udagawa, 2022). Our results showed decreased SUMOylation of Sp100 with coilin reduction and increased Sp100 SUMOylation with coilin expression. Similarly, we have shown that coilin reduction decreases SMN expression, while coilin expression enhances SMN expression. Our data also demonstrate that WT coilin enhances Sp100 SUMOylation and SMN expression more so than coilin Δ121–291, indicating that this region is necessary to achieve coilin’s full potential in these processes. CBs are known to associate with PML bodies, possibly via coilin’s interaction with PIAS4, a known SUMO E3 ligase (Sun et al., 2005). Future studies will investigate whether coilin contributes to PIAS4 SUMOylation or SUMOylation of other PML body client proteins, as well as PML body formation.
In contrast to ubiquitin E2 conjugation enzymes, SUMO E2s do not require an E3 ligase for conjugation. E3 protein ligases often facilitate this process through two mechanisms: recruiting the E2∼SUMO and substrate into a complex to promote specificity or stimulating the ability of the E2 enzyme to discharge SUMO to substrates. SUMO E3 ligases typically fall into one of two categories: those with a SIM motif or an SP-RING domain (Pichler et al., 2017). Coilin has three predicted SIM motifs (Beauclair et al., 2015) (Supplementary Figure S3B). Moreover, SUMO E3 ligases without a SIM motif or SP-RING domain have been reported, but how they function is not clear, and their classification as a SUMO E3 ligase is controversial (Hochstrasser, 2001; Pichler et al., 2017). Pichler et al. (2017) have identified hallmarks of bona fide SUMO E3 ligases and steps for identifying bona fide SUMO E3 ligases. All known SUMO E3 ligases are heavily automodified and function at substoichiometric concentrations relative to substrates. Detailed structural analyses are required to investigate how the E3 ligases discharge activated SUMO from the E2, as all known E3s interact with Ubc9 and SUMO to bind activated SUMOs in nearly identical conformations (Pichler et al., 2017). Our in vitro SUMOylation assays demonstrated coilin’s ability to promote RanGAP1 SUMOylation in a concentration-dependent manner and indicated that amino acids 121–291 are essential for attaining coilin’s full activity. This region of coilin is within its intrinsically disordered region and contains one of the three predicted SIM motifs (aa 145–148) (Beauclair et al., 2015). However, deletion of this segment only reduced, but did not abolish, coilin’s ability to promote RanGAP1 SUMOylation. The N362 coilin fragment, which contains the amino acids in question but is missing the C-terminal SIM (aa 512–414), did not enhance the SUMOylation of RanGAP1. Therefore, all three SIMs may be required to achieve coilin’s full SUMO E3 ligase-like activity. Further support for the hypothesis that coilin may be functioning as an E3 ligase came from our Ni-NTA pulldown assays demonstrating that coilin interacts directly with Ubc9 and SUMO2, one of the hallmarks of bona fide E3s. Additionally, automodification of known SUMO E3 ligases has been shown to modulate SUMO E3 ligase activity (Ihara et al., 2005; Jansen and Vertegaal, 2021). Therefore, we explored the SUMOylation status of coilin in vitro and in vivo. We found that highly purified coilin is capable of auto-SUMOylation in vitro, and WT myc-coilin and coilin-GFP are SUMOylated in vivo, while myc-coilin Δ121–291 is not. These data suggest that most SUMOylated lysine residues exist in the region of amino acids 121–291. Large-scale proteomic analyses have demonstrated that coilin has 11 identified SUMO2 sites (K127, K151, K160, K204, K209, K274, K281, K293, K297, K444, and K496) (Xiao et al., 2015; Hendriks et al., 2017), and seven of these are located between amino acids 121 and 291. Interestingly, we also found that GFP-coilin, in contrast to coilin-GFP, is not highly SUMOylated. This could be due to alternative protein folding depending on where the GFP tag is located, making the protein more or less accessible to the SUMOylation machinery. Our current studies are investigating how SUMOylation regulates coilin localization in the nucleoplasm and CB. Because SUMOylation plays a role in PML body formation, it is possible that increased coilin-GFP SUMOylation contributes to the tendency of this fusion protein to form many and large CBs when overexpressed, with correspondingly little nucleoplasmic enrichment (Shpargel et al., 2003).
Together, our data provide evidence indicating a clear role for coilin in protein SUMOylation and serve as the foundation for further studies aiming to identify coilin as a SUMO E3 ligase. Detailed structural analyses of how coilin interacts with Ubc9 and activated SUMO are needed in order to be able to classify coilin as an E3 ligase. However, because coilin has a large region of intrinsic disorder, structural analyses may be difficult to achieve. Our future studies will focus on further investigation of coilin domains responsible for its E3 ligase-like activity. Specifically, we will create a variety of coilin constructs with one or more SIM deletions and investigate the ability of these constructs to promote SUMOylation. We will also aim to identify other coilin target proteins and characterize cell processes dependent upon coilin-promoted SUMOylation. Although the RanGAP1 fragment can serve as a SUMO substrate for coilin-promoted in vitro SUMOylation, it is possible that RanGAP1 is not a target protein for coilin in vivo. The most likely targets for coilin’s SUMO E3 ligase-like activity in vivo would be proteins that coilin normally interacts with, such as components of snRNPs, snoRNPs, scaRNPs, telomerase, and miRNA biogenesis. Many snRNP, snoRNP, and telomerase components are SUMOylated, such as SmE, SmB/B′, Prp3 (snRNP components), Mop58, Nhp2, Nopp140 (snoRNP components), and dyskerin (telomerase components) (Westman and Lamond, 2011; Miyagawa et al., 2014; Pozzi et al., 2017; Yalcin et al., 2017; Zalzman et al., 2020). Many of these proteins are enriched in CBs and are known to interact with coilin (Isaac et al., 1998; Xu et al., 2005; Poole and Hebert, 2016). Taken together, our data show a role for coilin in the promotion of SUMOylation. Although far from proven, this coilin SUMO E3 ligase-like activity likely contributes to the biogenesis of several different classes of RNPs in addition to influencing the formation of nuclear bodies, including CBs (Figure 8E).
Very interestingly, although CBs are not found in all cell types, CBs are invariant features of transformed cells (Spector et al., 1992), (Hearst et al., 2009). Many studies have shown that expression of the SUMO E1, E2, and E3 ligases appears to be enhanced in numerous cancers (Coppola et al., 2009; Bellail et al., 2014; Liu et al., 2015; Seeler and Dejean, 2017). Specifically, in cancers with activating mutations of the Ras oncogene (∼25% of human cancers), inhibition of the SUMO E1 and E2 enzymes is lethal (Yu et al., 2015). Furthermore, myc-driven tumors with lower SUMO E1 expression are correlated with longer metastasis-free survival (Kessler et al., 2012). In patients with acute lymphoblastic leukemia, higher coilin expression has been identified as a negative prognostic indicator (Yue et al., 2018). Identification of coilin as an E3 ligase in conjunction with further investigation to identify more target proteins will provide insights into the transformation process. Given how many proteins are known to be SUMOylated, it does not appear that there are enough identified SUMO E3 ligases. Therefore, there must be other proteins with SUMO E3 ligase activity that have yet to be identified (Pichler et al., 2017). Our data clearly demonstrate that coilin promotes protein SUMOylation, is capable of auto-SUMOylation, and interacts directly with Ubc9 and SUMO2, identifying coilin as a potential SUMO E3 ligase pending further investigation.
HeLa and WI-38 cells were obtained from the American Type Culture Collection (ATCC). Cells were cultured as previously described (Enwerem et al., 2014). MEF26 and MEF42 cell lines were previously described (Tucker et al., 2001). All siRNAs were obtained from Integrated DNA Technologies (Coralville, IA) and utilized with RNAiMax (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s protocol. The negative control, coilin 2, coilin A, coilin 3′UTR, SMN, and WRAP53 siRNAs were previously described (Poole et al., 2016; Logan et al., 2018). The sequences of the Ubc9 siRNA used are forward (5′-rGrUrUrUrGrGrCrArArGrArArCrUrUrGrUrUrUrArCrArACA-3′) and reverse (5′-rUrGrUrUrGrUrArArArCrArArGrUrUrCrUrUrGrCrCrArArArCrCrA-3′). All siRNA transfections were conducted for 72 h. In experiments with both siRNA and DNA transfections, HeLa cells were treated with siRNA for 48 h, followed by DNA transfection for 24 h resulting in 72 h knockdown and 24 h plasmid expression. DNA transfections in HeLa cells were conducted using FuGene HD (Promega, Madison, WI); those conducted in WI-38 and MEF42 cells utilized Lipofectamine 2000 (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s protocols. His-SUMO1, YFP-Sp100, and FLAG-DGCR8 plasmids were obtained from Addgene (Watertown, MA, United States). GFP-coilin (Hebert and Matera, 2000) and coilin-GFP plasmids (Shpargel et al., 2003) previously contained a point mutation (K496E) shown to impact CB formation (Basello et al., 2022). For the work shown here, we used constructs containing the WT sequence. Myc-tagged SMN was a kind gift of G. Dreyfuss (HHMI, University of Pennsylvania, Philadelphia). WT myc-coilin was obtained from the Lamond lab (University of Dundee) and was mutated to form the deletion construct myc-coilin Δ121–291.
Cells were lysed in RIPA as previously described (Poole et al., 2016). Lysate (15 μL) was run on a precast 10% Mini-PROTEAN Gel (Bio-Rad Laboratories, Hercules, California, United States). For Ni-NTA pulldown experiments, 20 uL of 2x SDS loading buffer was added to the beads. Pulldowns and input lysates were run on a precast 10% Mini-Protean Gel (Bio-Rad Laboratories, Hercules, CA, United States). In vitro SUMOylation assays were run on gradient 4%–20% precast Mini-Protean Gels (Bio-Rad Laboratories, Hercules, CA, United States). Western blotting was conducted using a Trans-Blot Turbo Transfer System according to the manufacturer’s protocol (Bio-Rad, Hercules, CA, United States). The primary antibodies used were: anti-coilin rabbit polyclonal antibody (sc-32860, Santa Cruz Biotechnology Inc., Dallas, TX, United States); anti-SUMO-1 rabbit polyclonal antibody (4930, Cell Signaling Technology, Danvers, MA, United States); anti-SUMO-2/3 rabbit monoclonal antibody (4971, Cell Signaling Technology, Danvers, MA, United States); anti-Ubc9 rabbit monoclonal antibody (4786, Cell Signaling Technology, Danvers, MA, United States); anti-SENP1 rabbit polyclonal antibody (25349-1-AP, Proteintech, Rosemont, IL, United States); anti-GFP mouse monoclonal antibody (11814460001, Sigma Aldrich, St. Louis, MO, United States); anti-β-actin mouse monoclonal antibody (3700, Cell Signaling Technology, Danvers, MA, United States); anti-SMN mouse monoclonal antibody (610646, BD Transduction Laboratories, San Jose, CA, United States); anti-WRAP53 rabbit polyclonal antibody (A301-442A, Bethyl Laboratories Inc., Montgomery, TX, United States); and anti-FLAG mouse monoclonal antibody (F3165, Sigma-Aldrich, St. Louis, MO, United States). Secondary antibodies used were goat anti-mouse HRP (31440, Thermo Fisher Scientific, Waltham, MA, United States) and goat anti-rabbit HRP (31460, Thermo Fisher Scientific, Waltham, MA, United States). The secondary antibodies were detected with SuperSignal West Pico Chemiluminescent Substrate (34580, Thermo Fisher Scientific, Waltham, MA, United States) following the manufacturer’s suggested protocol. Imaging was performed on a ChemiDoc (Bio-Rad, Hercules, CA, United States). Adjustments to images were made using the transformation settings on Image Lab software and applied across the entire image.
His-T7-tagged coilin and WRAP53, after transformation into E. coli BL21(DE3)pLysS cells, were induced and isolated using Ni-NTA beads (30210, Qiagen, Germantown, MD, United States) as previously described (Hebert et al., 2001). SUMO1 and Ubc9 proteins used in Ni-NTA pulldowns were from the in vitro SUMOylation kit (ab139470, Abcam, Cambridge, MA, United States). SUMO2 and Ubc9 were added to tubes containing PBS, 50 uL 50% Ni-NTA beads, and His-T7-coilin or His-T7-WRAP53. For Ni-NTA pulldowns using cell lysate, the cells were lysed in modified RIPA buffer (50 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% NP-40, 0.25% Na-Deoxycholate, no EDTA, no SDS). Prior to lysis, 20 uL of 1 M N-ethylmaleimide per mL mRIPA was added to inhibit SENPs. Proteins and cell lysates were incubated with the Ni-NTA beads for 1 h at 4°C. The beads were then washed three times with PBS for protein pulldowns and with modified RIPA for lysate pulldowns, followed by the addition of 20 uL of 2x loading buffer.
Preparation of the excised SDS-PAGE bands, LC/MS/MS, and data analysis were performed by the Cornell Institute of Biotechnology Proteomics and Metabolomics Facility. The protein band from an SDS-PAGE gel was sliced into ∼1 mm cubes and subjected to in-gel digestion, followed by extraction of the tryptic peptide, as reported previously (Yang et al., 2007). The excised gel pieces were washed consecutively with 200 μL distilled/deionized water, followed by 50 mM ammonium bicarbonate, 50% acetonitrile, and, finally, 100% acetonitrile. The dehydrated gel pieces were reduced with 50 μL of 10 mM DTT in 100 mM ammonium bicarbonate for 1 h at 56°C and alkylated with 50 μL of 55 mM iodoacetamide in 100 mM ammonium bicarbonate at room temperature in the dark for 45 min. The wash steps described previously were repeated. The gel was then dried and rehydrated with trypsin (Promega) at an estimated 1:10 w/w ratio in 50 mM ammonium bicarbonate, 10% ACN (100 ul used to overlay gel @ 10 ng/µl trypsin, total enzyme used 1 µg). The gel was incubated at 37°C for 18 h. The digested peptides were extracted twice with 200 μL of 50% acetonitrile and 5% formic acid and once with 200 μL of 75% acetonitrile and 5% formic acid. Extractions from each sample were pooled and filtered with a 0.22 um spin filter (Costar Spin-X from Corning) and dried in the speed vacuum. Each sample was reconstituted in 0.5% formic acid prior to LC-MS/MS analysis.
The in-gel tryptic digests were reconstituted in 20 μL of 0.5% FA for nanoLC-ESI-MS/MS analysis, which was carried out using an Orbitrap Fusion™ Tribrid™ (Thermo Fisher Scientific, San Jose, CA, United States) mass spectrometer equipped with a nanospray Flex Ion Source and coupled with a Dionex UltiMate 3000 RSLCnano system (Thermo, Sunnyvale, CA, United States) (Thomas et al., 2017; Yang et al., 2018a). The gel-extracted peptide samples (2.5 μL) were injected onto a PepMap C-18 RP nano trapping column (5 μm, 100 µm i.d. × 20 mm) at 20 μL/min flow rate for rapid sample loading and then separated on a PepMap C-18 RP nano column (2 μm, 75 μm × 25 cm) at 35°C. The tryptic peptides were eluted in a 90-min gradient of 5%–35% acetonitrile (ACN) in 0.1% formic acid at 300 nL/min, followed by an 8-min ramping to 90% can in 0.1% FA and an 8 min hold at 90% ACN in 0.1% FA. The column was re-equilibrated with 0.1% FA for 25 min prior to the next run. The Orbitrap Fusion was operated in positive ion mode with the spray voltage set at 1.9 kV and source temperature at 275°C with advanced peak determination. External calibration for FT, IT, and quadrupole mass analyzers was performed. In data-dependent acquisition (DDA) analysis, the instrument was operated using an FT mass analyzer in an MS scan to select precursor ions, followed by three “Top Speed” data-dependent CID ion trap MS/MS scans at 1.6 m/z quadrupole isolation for precursor peptides with multiple charged ions above a threshold ion count of 10,000 and normalized collision energy of 30%. MS survey scans at a resolving power of 120,000 (fwhm at m/z 200) for the mass range of m/z 375–1,600. Dynamic exclusion parameters were set at 35 s of exclusion duration with ±10 ppm exclusion mass width. All data were acquired using Xcalibur 4.3 operation software (Thermo Fisher Scientific).
The DDA raw files for CID MS/MS were subjected to database searches using Proteome Discoverer (PD) 2.4 software (Thermo Fisher Scientific, Bremen, Germany) with the SEQUEST HT algorithm. The processing workflow used precursor-based quantification. The PD 2.4 processing workflow containing an additional node of Minora Feature Detector for precursor ion-based quantification was used for protein identification. The database search was conducted against the Homo sapiens UniProt database, which has 20,315 sequences. Two missed trypsin cleavage sites were allowed. The peptide precursor tolerance was set to 10 ppm, and fragment ion tolerance was set to 0.6 Da. Variable modification of methionine oxidation, deamidation of asparagine/glutamine, phosphorylation of serine/threonine/tyrosine, and fixed modification of cysteine carbamidomethylation were set for the database search. To confidently localize phosphorylation sites, the phosphoRS 3.0 node integrated into the PD 2.4 workflow was also used. The algorithm of phosphoRS 3.0 software enables automated and confident localization of phosphosites by determining individual probability values for each putatively phosphorylated site within the validated peptide sequences (Taus et al., 2011). Only high-confidence peptides defined by Sequest HT with a 1% FDR by percolator were considered for the peptide identification. All phosphorylated peptides were validated by manual inspection of the relevant MS/MS spectra. The occupancy rates of all phosphorylated peptides were calculated based on the peak area of XICs versus their counterpart native peptides in different charge states using Xcalibur software under the assumption that ionization efficiency is the same or similar between the native peptide and its phosphorylated form. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD036370.
A SUMOylation kit was utilized for the in vitro SUMOylation assays (ab139470, Abcam, Cambridge, MA, United States). The amounts of buffer, Mg-ATP, RanGAP1, and SUMO1 were added according to the manufacturer’s suggested protocol. However, where noted, we used decreased amounts of E1 activation and E2 conjugation enzymes (1/2 and 1/10 the suggested amounts, respectively), as it has been shown that using smaller amounts of E1 and E2 allows better visualization of changes caused by the addition of an E3 ligase (Yang et al., 2018b). His-tagged full-length coilin, WRAP53, and coilin fragments C214, N362, Δ121–291, and Δ241–291 were induced and isolated as previously described after transformation into E. coli BL21 (DE3) pLysS cells (Hebert et al., 2001). Elution buffer (50 mM NaH2PO4, 300 mM NaCl, and 250 mM imidazole, pH 8) used to elute the His-tagged proteins was used as a control, and ∼50 ng of partially purified proteins was added to the reactions. For coilin auto-SUMOylation assays and RanGAP1 SUMOylation assays, GST-coilin was expressed by E. coli BL21 (DE3) pLysS, followed by removal of the GST tag and purification as previously described (Broome and Hebert, 2012; Broome et al., 2013). Highly purified coilin (35 and 70 ng) were added to the SUMOylation assay of RanGAP1 in Figure 6F, and 70 ng of purified coilin was used in the coilin autoSUMOylation assay. Coilin auto-SUMOylation assays were performed using 1 uL E1 and E2, Mg/ATP, and SUMO1. The reactions were run for 1 h at 37°C, followed by SDS-PAGE, Western blotting, and probing with the indicated antibodies.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: http://www.proteomexchange.org/, PXD036370.
KL and MH designed and performed experimental work and analyzed the data. KL drafted the manuscript. DM and ST also performed experimental work. All authors contributed to the article and approved the submitted version.
This work was supported by funds from the Department of Cell and Molecular Biology at the University of Mississippi Medical Center. The Proteomics Facility of Cornell University acknowledges NIH SIG grant 1S10 OD017992-01 grant for support of the Orbitrap Fusion mass spectrometer.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author MH declared that they were an editorial board member of Frontiers at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frnar.2023.1197990/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | siRNA knockdown confirmation in HeLa and WI-38 cells. HeLa (A–C) and WI-38 (D,E) cells were transfected with indicated siRNAs. Knockdowns were confirmed by probing with anti-coilin [(A,B) left panel, and (C)], anti-WRAP53 [(B) right panel, (D)], and anti-SMN (B,C) antibodies. Additionally, global SUMOylation was detected with anti-SUMO2/3 antibodies after coilin KD using various siRNAs (A). Blots were also probed with actin to assess protein loading.
SUPPLEMENTARY FIGURE S2 | Coilin KD does not alter the protein levels of Ubc9 or SENP1. Ubc9 (A,B) and SENP1 (C,D) protein levels in HeLa (A,C) and WI-38 (B,D) cells after 72 h KD with negative control, coilin, SMN, and WRAP53 siRNA. (E) Highly purified coilin and BSA were subjected to SDS-PAGE, followed by Coomassie staining. Full-length coilin and coilin degradation fragments are denoted with arrows. (F) Ni-NTA pulldown demonstrating coilin’s interaction with SUMO1 in vitro. Partially purified His-tagged coilin was incubated with SUMO1 and Ni-NTA beads (lane 3) for 1 h at 4°C. SUMO1 was also incubated with Ni beads alone as a negative control (lane 2). Input (lane 1) represents 100% (1 uL) of protein used for pulldowns. Western blots of input and the Ni pulldowns were probed with the indicated antibodies.
SUPPLEMENTARY FIGURE S3 | Schematic of DGCR8 (A) and coilin (B) proteins. Structured regions of the proteins are labeled. Green diamonds denote phosphosites of DGCR8 (A), and S, T, and Y reflect phosphorylated serine, threonine, and tyrosine residues, respectively. Red balloons mark sites of SUMOylation. Predicted SIMs are shown in green boxes.
SUPPLEMENTARY TABLE S1 | Quantification of DGCR8 phosphorylated peptides between the negative control and coilin knockdown. All proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD036370.
Basello, D. A., Matera, A. G., and Staněk, D. (2022). A point mutation in human coilin prevents Cajal body formation. J. Cell Sci. 135, jcs259587. doi:10.1242/jcs.259587
Bawa-Khalfe, T., and Yeh, E. T. (2010). SUMO losing balance: SUMO proteases disrupt SUMO homeostasis to facilitate cancer development and progression. Genes & Cancer 1, 748–752. doi:10.1177/1947601910382555
Beauclair, G., Bridier-Nahmias, A., Zagury, J. F., Saib, A., and Zamborlini, A. (2015). JASSA: A comprehensive tool for prediction of SUMOylation sites and SIMs. Bioinformatics 31, 3483–3491. doi:10.1093/bioinformatics/btv403
Bellail, A. C., Olson, J. J., and Hao, C. (2014). SUMO1 modification stabilizes CDK6 protein and drives the cell cycle and glioblastoma progression. Nat. Commun. 5, 4234. doi:10.1038/ncomms5234
Bernardi, R., and Pandolfi, P. P. (2007). Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell Biol. 8, 1006–1016. doi:10.1038/nrm2277
Bernardi, R., Papa, A., and Pandolfi, P. P. (2008). Regulation of apoptosis by PML and the PML-NBs. Oncogene 27, 6299–6312. doi:10.1038/onc.2008.305
Broome, H. J., Carrero, Z. I., Douglas, H. E., and Hebert, M. D. (2013). Phosphorylation regulates coilin activity and RNA association. Biol. Open 2, 407–415. doi:10.1242/bio.20133863
Broome, H. J., and Hebert, M. D. (2013). Coilin displays differential affinity for specific RNAs in vivo and is linked to telomerase RNA biogenesis. J. Mol. Biol. 425, 713–724. doi:10.1016/j.jmb.2012.12.014
Broome, H. J., and Hebert, M. D. (2012). In vitro RNase and nucleic acid binding activities implicate coilin in U snRNA processing. PLoS One 7, e36300. doi:10.1371/journal.pone.0036300
Brzustowicz, L. M., and Bassett, A. S. (2012). miRNA-mediated risk for schizophrenia in 22q11.2 deletion syndrome. Front. Genet. 3, 291. doi:10.3389/fgene.2012.00291
Carvalho, T., Almeida, F., Calapez, A., Lafarga, M., Berciano, M. T., and Carmo-Fonseca, M. (1999). The spinal muscular atrophy disease gene product, SMN: A link between snRNP biogenesis and the cajal (coiled) body. J. Cell Biol. 147, 715–728. doi:10.1083/jcb.147.4.715
Collier, S., Pendle, A., Boudonck, K., van Rij, T., Dolan, L., and Shaw, P. (2006). A distant coilin homologue is required for the formation of cajal bodies in Arabidopsis. Mol. Biol. Cell 17, 2942–2951. doi:10.1091/mbc.e05-12-1157
Coppola, D., Parikh, V., Boulware, D., and Blanck, G. (2009). Substantially reduced expression of PIAS1 is associated with colon cancer development. J. Cancer Res. Clin. Oncol. 135, 1287–1291. doi:10.1007/s00432-009-0570-z
Earls, L. R., Fricke, R. G., Yu, J., Berry, R. B., Baldwin, L. T., and Zakharenko, S. S. (2012). Age-dependent microRNA control of synaptic plasticity in 22q11 deletion syndrome and schizophrenia. J. Neurosci. 32, 14132–14144. doi:10.1523/jneurosci.1312-12.2012
Eifler, K., and Vertegaal, A. C. O. (2015). SUMOylation-mediated regulation of cell cycle progression and cancer. Trends Biochem. Sci. 40, 779–793. doi:10.1016/j.tibs.2015.09.006
Enwerem, I. I., Velma, V., Broome, H. J., Kuna, M., Begum, R. A., and Hebert, M. D. (2014). Coilin association with Box C/D scaRNA suggests a direct role for the Cajal body marker protein in scaRNP biogenesis. Biol. Open 3, 240–249. doi:10.1242/bio.20147443
Fallini, C., Bassell, G. J., and Rossoll, W. (2012). Spinal muscular atrophy: The role of SMN in axonal mRNA regulation. Brain Res. 1462, 81–92. doi:10.1016/j.brainres.2012.01.044
Ferdaoussi, M., Dai, X., Jensen, M. V., Wang, R., Peterson, B. S., Huang, C., et al. (2015). Isocitrate-to-SENP1 signaling amplifies insulin secretion and rescues dysfunctional beta cells. J. Clin. Investigation 125, 3847–3860. doi:10.1172/jci82498
Grande, M. A., van der Kraan, I., van Steensel, B., Schul, W., de The, H., van der Voort, H. T., et al. (1996). PML-Containing nuclear bodies: Their spatial distribution in relation to other nuclear components. J. Cell Biochem. 63, 280–291. doi:10.1002/(sici)1097-4644(19961201)63:3<280::aid-jcb3>3.0.co;2-t
Han, J., Lee, Y., Yeom, K. H., Kim, Y. K., Jin, H., and Kim, V. N. (2004). The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 18, 3016–3027. doi:10.1101/gad.1262504
Hasegawa, M., Nonaka, T., and Masuda-Suzukake, M. (2016). α-Synuclein: Experimental pathology. Cold Spring Harb. Perspect. Med. 6, a024273. doi:10.1101/cshperspect.a024273
Hearst, S. M., Gilder, A. S., Negi, S. S., Davis, M. D., George, E. M., Whittom, A. A., et al. (2009). Cajal-body formation correlates with differential coilin phosphorylation in primary and transformed cell lines. J. Cell Sci. 122, 1872–1881. doi:10.1242/jcs.044040
Hebert, M. D., and Matera, A. G. (2000). Self-association of coilin reveals a common theme in nuclear body localization. Mol. Biol. Cell 11, 4159–4171. doi:10.1091/mbc.11.12.4159
Hebert, M. D., Shpargel, K. B., Ospina, J. K., Tucker, K. E., and Matera, A. G. (2002). Coilin methylation regulates nuclear body formation. Dev. Cell 3, 329–337. doi:10.1016/s1534-5807(02)00222-8
Hebert, M. D., Szymczyk, P. W., Shpargel, K. B., and Matera, A. G. (2001). Coilin forms the bridge between Cajal bodies and SMN, the spinal muscular atrophy protein. Genes Dev. 15, 2720–2729. doi:10.1101/gad.908401
Hendriks, I. A., Lyon, D., Young, C., Jensen, L. J., Vertegaal, A. C., and Nielsen, M. L. (2017). Site-specific mapping of the human SUMO proteome reveals co-modification with phosphorylation. Nat. Struct. Mol. Biol. 24, 325–336. doi:10.1038/nsmb.3366
Heo, K. S., Le, N. T., Cushman, H. J., Giancursio, C. J., Chang, E., Woo, C. H., et al. (2015). Disturbed flow-activated p90RSK kinase accelerates atherosclerosis by inhibiting SENP2 function. J. Clin. Invest. 125, 1299–1310. doi:10.1172/jci76453
Herbert, K. M., Pimienta, G., DeGregorio, S. J., Alexandrov, A., and Steitz, J. A. (2013). Phosphorylation of DGCR8 increases its intracellular stability and induces a progrowth miRNA profile. Cell Rep. 5, 1070–1081. doi:10.1016/j.celrep.2013.10.017
Hirano, S., and Udagawa, O. (2022). SUMOylation regulates the number and size of promyelocytic leukemia-nuclear bodies (PML-NBs) and arsenic perturbs SUMO dynamics on PML by insolubilizing PML in THP-1 cells. Arch. Toxicol. 96, 545–558. doi:10.1007/s00204-021-03195-w
Hochstrasser, M. (2001). SP-RING for SUMO: New functions bloom for a ubiquitin-like protein. Cell 107, 5–8. doi:10.1016/s0092-8674(01)00519-0
Horikawa, Y., Wood, C. G., Yang, H., Zhao, H., Ye, Y., Gu, J., et al. (2008). Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma. Clin. Cancer Res. 14, 7956–7962. doi:10.1158/1078-0432.ccr-08-1199
Ihara, M., Yamamoto, H., and Kikuchi, A. (2005). SUMO-1 modification of PIASy, an E3 ligase, is necessary for PIASy-dependent activation of Tcf-4. Mol. Cell. Biol. 25, 3506–3518. doi:10.1128/mcb.25.9.3506-3518.2005
Isaac, C., Yang, Y., and Meier, U. T. (1998). Nopp140 functions as a molecular link between the nucleolus and the coiled bodies. J. Cell Biol. 142, 319–329. doi:10.1083/jcb.142.2.319
Jansen, N. S., and Vertegaal, A. C. O. (2021). A chain of events: Regulating target proteins by SUMO polymers. Trends Biochem. Sci. 46, 113–123. doi:10.1016/j.tibs.2020.09.002
Kadener, S., Rodriguez, J., Abruzzi, K. C., Khodor, Y. L., Sugino, K., Marr, M. T., et al. (2009). Genome-wide identification of targets of the drosha-pasha/DGCR8 complex. RNA 15, 537–545. doi:10.1261/rna.1319309
Kamynina, E., and Stover, P. J. (2017). The roles of SUMO in metabolic regulation. Adv. Exp. Med. Biol. 963, 143–168. doi:10.1007/978-3-319-50044-7_9
Kessler, J. D., Kahle, K. T., Sun, T., Meerbrey, K. L., Schlabach, M. R., Schmitt, E. M., et al. (2012). A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science 335, 348–353. doi:10.1126/science.1212728
Kho, C., Lee, A., Jeong, D., Oh, J. G., Chaanine, A. H., Kizana, E., et al. (2011). SUMO1-dependent modulation of SERCA2a in heart failure. Nature 477, 601–605. doi:10.1038/nature10407
Kim, B., Lee, J. H., Park, J. W., Kwon, T. K., Baek, S. K., Hwang, I., et al. (2014). An essential microRNA maturing microprocessor complex component DGCR8 is up-regulated in colorectal carcinomas. Clin. Exp. Med. 14, 331–336. doi:10.1007/s10238-013-0243-8
Kim, Y. E., Lee, J. H., Kim, E. T., Shin, H. J., Gu, S. Y., Seol, H. S., et al. (2011). Human cytomegalovirus infection causes degradation of Sp100 proteins that suppress viral gene expression. J. Virol. 85, 11928–11937. doi:10.1128/jvi.00758-11
Kiss, T. (2004). Biogenesis of small nuclear RNPs. J. Cell Sci. 117, 5949–5951. doi:10.1242/jcs.01487
Kitagawa, N., Ojima, H., Shirakihara, T., Shimizu, H., Kokubu, A., Urushidate, T., et al. (2013). Downregulation of the microRNA biogenesis components and its association with poor prognosis in hepatocellular carcinoma. Cancer Sci. 104, 543–551. doi:10.1111/cas.12126
Landthaler, M., Yalcin, A., and Tuschl, T. (2004). The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 14, 2162–2167. doi:10.1016/j.cub.2004.11.001
Lefebvre, S., Burglen, L., Reboullet, S., Clermont, O., Burlet, P., Viollet, L., et al. (1995). Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80, 155–165. doi:10.1016/0092-8674(95)90460-3
Lekka, E., and Hall, J. (2018). Noncoding RNAs in disease. FEBS Lett. 592, 2884–2900. doi:10.1002/1873-3468.13182
Lett, K. E., Logan, M. K., McLaurin, D. M., and Hebert, M. D. (2021). Coilin enhances phosphorylation and stability of DGCR8 and promotes miRNA biogenesis. Mol. Biol. Cell 32, br4. doi:10.1091/mbc.e21-05-0225
Li, Y., Wang, H., Wang, S., Quon, D., Liu, Y. W., and Cordell, B. (2003). Positive and negative regulation of APP amyloidogenesis by sumoylation. Proc. Natl. Acad. Sci. U. S. A. 100, 259–264. doi:10.1073/pnas.0235361100
Liu, J. L., Wu, Z., Nizami, Z., Deryusheva, S., Rajendra, T. K., Beumer, K. J., et al. (2009). Coilin is essential for Cajal body organization in Drosophila melanogaster. Mol. Biol. Cell 20, 1661–1670. doi:10.1091/mbc.e08-05-0525
Liu, Q., and Dreyfuss, G. (1996). A novel nuclear structure containing the survival of motor neurons protein. EMBO J. 15, 3555–3565. doi:10.1002/j.1460-2075.1996.tb00725.x
Liu, Q., Fischer, U., Wang, F., and Dreyfuss, G. (1997). The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell 90, 1013–1021. doi:10.1016/s0092-8674(00)80367-0
Liu, X., Xu, Y., Pang, Z., Guo, F., Qin, Q., Yin, T., et al. (2015). Knockdown of SUMO-activating enzyme subunit 2 (SAE2) suppresses cancer malignancy and enhances chemotherapy sensitivity in small cell lung cancer. J. Hematol. Oncol. 8, 67. doi:10.1186/s13045-015-0164-y
Logan, M. K., Burke, M. F., and Hebert, M. D. (2018). Altered dynamics of scaRNA2 and scaRNA9 in response to stress correlates with disrupted nuclear organization. Biol. Open 7, bio037101. doi:10.1242/bio.037101
Logan, M. K., Lett, K. E., and Hebert, M. D. (2021). The Cajal body protein coilin is a regulator of the miR-210 hypoxamiR and influences MIR210HG alternative splicing. J. Cell Sci. 134, jcs258575. doi:10.1242/jcs.258575
Logan, M. K., Lett, K. E., McLaurin, D. M., and Hebert, M. D. (2022). Coilin as a regulator of NF-kB mediated inflammation in preeclampsia. Biol. Open 11, bio059326. doi:10.1242/bio.059326
Logan, M. K., McLaurin, D. M., and Hebert, M. D. (2020). Synergistic interactions between Cajal bodies and the miRNA processing machinery. Mol. Biol. Cell 31, 1561–1569. doi:10.1091/mbc.e20-02-0144
Luo, H. B., Xia, Y. Y., Shu, X. J., Liu, Z. C., Feng, Y., Liu, X. H., et al. (2014). SUMOylation at K340 inhibits tau degradation through deregulating its phosphorylation and ubiquitination. Proc. Natl. Acad. Sci. U. S. A. 111, 16586–16591. doi:10.1073/pnas.1417548111
Mahajan, R., Delphin, C., Guan, T., Gerace, L., and Melchior, F. (1997). A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88, 97–107. doi:10.1016/s0092-8674(00)81862-0
Mahmoudi, S., Henriksson, S., Weibrecht, I., Smith, S., Soderberg, O., Stromblad, S., et al. (2010). WRAP53 is essential for Cajal body formation and for targeting the survival of motor neuron complex to Cajal bodies. PLoS Biol. 8, e1000521. doi:10.1371/journal.pbio.1000521
Makarov, V., Rakitina, D., Protopopova, A., Yaminsky, I., Arutiunian, A., Love, A. J., et al. (2013). Plant coilin: Structural characteristics and RNA-binding properties. PLoS One 8, e53571. doi:10.1371/journal.pone.0053571
Matera, A. G., and Frey, M. R. (1998). Coiled bodies and gems: Janus or gemini? Am. J. Hum. Genet. 63, 317–321. doi:10.1086/301992
Matunis, M. J., Coutavas, E., and Blobel, G. (1996). A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 135, 1457–1470. doi:10.1083/jcb.135.6.1457
Maul, G. G., Negorev, D., Bell, P., and Ishov, A. M. (2000). Review: Properties and assembly mechanisms of ND10, PML bodies, or PODs. J. Struct. Biol. 129, 278–287. doi:10.1006/jsbi.2000.4239
Minty, A., Dumont, X., Kaghad, M., and Caput, D. (2000). Covalent modification of p73α by SUMO-1. J. Biol. Chem. 275, 36316–36323. doi:10.1074/jbc.m004293200
Miyagawa, K., Low, R. S., Santosa, V., Tsuji, H., Moser, B. A., Fujisawa, S., et al. (2014). SUMOylation regulates telomere length by targeting the shelterin subunit Tpz1(Tpp1) to modulate shelterin-Stn1 interaction in fission yeast. Proc. Natl. Acad. Sci. U. S. A. 111, 5950–5955. doi:10.1073/pnas.1401359111
O'Rourke, J. G., Gareau, J. R., Ochaba, J., Song, W., Rasko, T., Reverter, D., et al. (2013). SUMO-2 and PIAS1 modulate insoluble mutant huntingtin protein accumulation. Cell Rep. 4, 362–375. doi:10.1016/j.celrep.2013.06.034
Pellizzoni, L., Kataoka, N., Charroux, B., and Dreyfuss, G. (1998). A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell 95, 615–624. doi:10.1016/s0092-8674(00)81632-3
Pena, E., Berciano, M. T., Fernandez, R., Ojeda, J. L., and Lafarga, M. (2001). Neuronal body size correlates with the number of nucleoli and Cajal bodies, and with the organization of the splicing machinery in rat trigeminal ganglion neurons. J. Comp. Neurol. 430, 250–263. doi:10.1002/1096-9861(20010205)430:2<250::aid-cne1029>3.0.co;2-l
Pichler, A., Fatouros, C., Lee, H., and Eisenhardt, N. (2017). SUMO conjugation - a mechanistic view. Biomol. Concepts 8, 13–36. doi:10.1515/bmc-2016-0030
Poole, A. R., Enwerem, I. I., Vicino, I. A., Coole, J. B., Smith, S. V., and Hebert, M. D. (2016). Identification of processing elements and interactors implicate SMN, coilin and the pseudogene-encoded coilp1 in telomerase and box C/D scaRNP biogenesis. RNA Biol. 13, 955–972. doi:10.1080/15476286.2016.1211224
Poole, A. R., and Hebert, M. D. (2016). SMN and coilin negatively regulate dyskerin association with telomerase RNA. Biol. Open 5, 726–735. doi:10.1242/bio.018804
Pozzi, B., Bragado, L., Will, C. L., Mammi, P., Risso, G., Urlaub, H., et al. (2017). SUMO conjugation to spliceosomal proteins is required for efficient pre-mRNA splicing. Nucleic Acids Res. 45, 6729–6745. doi:10.1093/nar/gkx213
Pratt, A. J., and MacRae, I. J. (2009). The RNA-induced silencing complex: A versatile gene-silencing machine. J. Biol. Chem. 284, 17897–17901. doi:10.1074/jbc.r900012200
Riboldi, G. M., Faravelli, I., Kuwajima, T., Delestree, N., Dermentzaki, G., De Planell-Saguer, M., et al. (2021). Sumoylation regulates the assembly and activity of the SMN complex. Nat. Commun. 12, 5040. doi:10.1038/s41467-021-25272-5
Salomoni, P., and Pandolfi, P. P. (2002). The role of PML in tumor suppression. Cell 108, 165–170. doi:10.1016/s0092-8674(02)00626-8
Sarangi, P., and Zhao, X. (2015). SUMO-mediated regulation of DNA damage repair and responses. Trends Biochem. Sci. 40, 233–242. doi:10.1016/j.tibs.2015.02.006
Schilling, E. M., Scherer, M., Reuter, N., Schweininger, J., Muller, Y. A., and Stamminger, T. (2017). The human cytomegalovirus IE1 protein antagonizes PML nuclear body-mediated intrinsic immunity via the inhibition of PML de novo SUMOylation. J. Virol. 91, 020499–e2116. doi:10.1128/jvi.02049-16
Seeler, J. S., and Dejean, A. (2017). SUMO and the robustness of cancer. Nat. Rev. Cancer 17, 184–197. doi:10.1038/nrc.2016.143
Shpargel, K. B., Ospina, J. K., Tucker, K. E., Matera, A. G., and Hebert, M. D. (2003). Control of Cajal body number is mediated by the coilin C-terminus. J. Cell Sci. 116, 303–312. doi:10.1242/jcs.00211
Spector, D. L., Lark, G., and Huang, S. (1992). Differences in snRNP localization between transformed and nontransformed cells. Mol. Biol. Cell 3, 555–569. doi:10.1091/mbc.3.5.555
Sternsdorf, T., Jensen, K., Reich, B., and Will, H. (1999). The nuclear dot protein sp100, characterization of domains necessary for dimerization, subcellular localization, and modification by small ubiquitin-like modifiers. J. Biol. Chem. 274, 12555–12566. doi:10.1074/jbc.274.18.12555
Strzelecka, M., Trowitzsch, S., Weber, G., Luhrmann, R., Oates, A. C., and Neugebauer, K. M. (2010). Coilin-dependent snRNP assembly is essential for zebrafish embryogenesis. Nat. Struct. Mol. Biol. 17, 403–409. doi:10.1038/nsmb.1783
Sun, J., Xu, H., Subramony, S. H., and Hebert, M. D. (2005). Interactions between Coilin and PIASy partially link Cajal bodies to PML bodies. J. Cell Sci. 118, 4995–5003. doi:10.1242/jcs.02613
Tapia, O., Lafarga, V., Bengoechea, R., Palanca, A., Lafarga, M., and Berciano, M. T. (2014). The SMN Tudor SIM-like domain is key to SmD1 and coilin interactions and to Cajal body biogenesis. J. Cell Sci. 127, 939–946. doi:10.1242/jcs.138537
Taus, T., Kocher, T., Pichler, P., Paschke, C., Schmidt, A., Henrich, C., et al. (2011). Universal and confident phosphorylation site localization using phosphoRS. J. Proteome Res. 10, 5354–5362. doi:10.1021/pr200611n
Thomas, C. J., Cleland, T. P., Zhang, S., Gundberg, C. M., and Vashishth, D. (2017). Identification and characterization of glycation adducts on osteocalcin. Anal. Biochem. 525, 46–53. doi:10.1016/j.ab.2017.02.011
Tomari, Y., and Zamore, P. D. (2005). MicroRNA biogenesis: Drosha can't cut it without a partner. Curr. Biol. 15, R61–R64. doi:10.1016/j.cub.2004.12.057
Toyota, C. G., Davis, M. D., Cosman, A. M., and Hebert, M. D. (2010). Coilin phosphorylation mediates interaction with SMN and SmB. Chromosoma 119, 205–215. doi:10.1007/s00412-009-0249-x
Triboulet, R., Chang, H. M., Lapierre, R. J., and Gregory, R. I. (2009). Post-transcriptional control of DGCR8 expression by the Microprocessor. RNA 15, 1005–1011. doi:10.1261/rna.1591709
Tucker, K. E., Berciano, M. T., Jacobs, E. Y., LePage, D. F., Shpargel, K. B., Rossire, J. J., et al. (2001). Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J. Cell Biol. 154, 293–308. doi:10.1083/jcb.200104083
Van Damme, E., Laukens, K., Dang, T. H., and Van Ostade, X. (2010). A manually curated network of the PML nuclear body interactome reveals an important role for PML-NBs in SUMOylation dynamics. Int. J. Biol. Sci. 6, 51–67. doi:10.7150/ijbs.6.51
Wada, T., Kikuchi, J., and Furukawa, Y. (2012). Histone deacetylase 1 enhances microRNA processing via deacetylation of DGCR8. EMBO Rep. 13, 142–149. doi:10.1038/embor.2011.247
Wan, L., Battle, D. J., Yong, J., Gubitz, A. K., Kolb, S. J., Wang, J., et al. (2005). The survival of motor neurons protein determines the capacity for snRNP assembly: Biochemical deficiency in spinal muscular atrophy. Mol. Cell. Biol. 25, 5543–5551. doi:10.1128/mcb.25.13.5543-5551.2005
Wang, Y., and Dasso, M. (2009). SUMOylation and deSUMOylation at a glance. J. Cell Sci. 122, 4249–4252. doi:10.1242/jcs.050542
Weetman, J., Wong, M. B., Sharry, S., Rcom-H'cheo-Gauthier, A., Gai, W. P., Meedeniya, A., et al. (2013). Increased SUMO-1 expression in the unilateral rotenone-lesioned mouse model of Parkinson's disease. Neurosci. Lett. 544, 119–124. doi:10.1016/j.neulet.2013.03.057
Westman, B. J., and Lamond, A. I. (2011). A role for SUMOylation in snoRNP biogenesis revealed by quantitative proteomics. Nucleus 2, 30–37. doi:10.4161/nucl.2.1.14437
Workman, E., Kolb, S. J., and Battle, D. J. (2012). Spliceosomal small nuclear ribonucleoprotein biogenesis defects and motor neuron selectivity in spinal muscular atrophy. Brain Res. 1462, 93–99. doi:10.1016/j.brainres.2012.02.051
Xiao, Z., Chang, J. G., Hendriks, I. A., Sigurethsson, J. O., Olsen, J. V., and Vertegaal, A. C. (2015). System-wide analysis of SUMOylation dynamics in response to replication stress reveals novel small ubiquitin-like modified target proteins and acceptor lysines relevant for genome stability. Mol. Cell. Proteomics 14, 1419–1434. doi:10.1074/mcp.o114.044792
Xu, H., Pillai, R. S., Azzouz, T. N., Shpargel, K. B., Kambach, C., Hebert, M. D., et al. (2005). The C-terminal domain of coilin interacts with Sm proteins and U snRNPs. Chromosoma 114, 155–166. doi:10.1007/s00412-005-0003-y
Yalcin, Z., Selenz, C., and Jacobs, J. J. L. (2017). Ubiquitination and SUMOylation in telomere maintenance and dysfunction. Front. Genet. 8, 67. doi:10.3389/fgene.2017.00067
Yang, S., Li, X., Liu, X., Ding, X., Xin, X., Jin, C., et al. (2018a). Parallel comparative proteomics and phosphoproteomics reveal that cattle myostatin regulates phosphorylation of key enzymes in glycogen metabolism and glycolysis pathway. Oncotarget 9, 11352–11370. doi:10.18632/oncotarget.24250
Yang, W. S., Campbell, M., Kung, H. J., and Chang, P. C. (2018b). In vitro SUMOylation assay to study SUMO E3 ligase activity. J. Vis. Exp., 56629. doi:10.3791/56629
Yang, Y., Thannhauser, T. W., Li, L., and Zhang, S. (2007). Development of an integrated approach for evaluation of 2-D gel image analysis: Impact of multiple proteins in single spots on comparative proteomics in conventional 2-D gel/MALDI workflow. Electrophoresis 28, 2080–2094. doi:10.1002/elps.200600524
Yeom, K. H., Lee, Y., Han, J., Suh, M. R., and Kim, V. N. (2006). Characterization of DGCR8/Pasha, the essential cofactor for Drosha in primary miRNA processing. Nucleic Acids Res. 34, 4622–4629. doi:10.1093/nar/gkl458
Young, P. J., Le, T. T., Dunckley, M., thi Man, N., Burghes, A. H., and Morris, G. E. (2001). Nuclear gems and cajal (coiled) bodies in fetal tissues: Nucleolar distribution of the spinal muscular atrophy protein, SMN. Exp. Cell Res. 265, 252–261. doi:10.1006/excr.2001.5186
Young, P. J., Le, T. T., thi Man, N., Burghes, A. H., and Morris, G. E. (2000). The relationship between SMN, the spinal muscular atrophy protein, and nuclear coiled bodies in differentiated tissues and cultured cells. Exp. Cell Res. 256, 365–374. doi:10.1006/excr.2000.4858
Yu, B., Swatkoski, S., Holly, A., Lee, L. C., Giroux, V., Lee, C. S., et al. (2015). Oncogenesis driven by the Ras/Raf pathway requires the SUMO E2 ligase Ubc9. Proc. Natl. Acad. Sci. U. S. A. 112, E1724–E1733. doi:10.1073/pnas.1415569112
Yue, Z. X., Gao, R. Q., Gao, C., Liu, S. G., Zhao, X. X., Xing, T. Y., et al. (2018). The prognostic potential of coilin in association with p27 expression in pediatric acute lymphoblastic leukemia for disease relapse. Cancer Cell Int. 18, 106. doi:10.1186/s12935-018-0600-5
Zalzman, M., Meltzer, W. A., Portney, B. A., Brown, R. A., and Gupta, A. (2020). The role of ubiquitination and SUMOylation in telomere Biology. Curr. Issues Mol. Biol. 35, 85–98. doi:10.21775/cimb.035.085
Zhang, Y., Chen, X., Wang, Q., Du, C., Lu, W., Yuan, H., et al. (2021). Hyper-SUMOylation of SMN induced by SENP2 deficiency decreases its stability and leads to spinal muscular atrophy-like pathology. J. Mol. Med. Berl. 99, 1797–1813. doi:10.1007/s00109-021-02130-x
Zhong, S., Muller, S., Ronchetti, S., Freemont, P. S., Dejean, A., and Pandolfi, P. P. (2000). Role of SUMO-1-modified PML in nuclear body formation. Blood 95, 2748–2752. doi:10.1182/blood.v95.9.2748.009k31a_2748_2752
Zhu, C., Chen, C., Chen, R., Deng, R., Zhao, X., Zhang, H., et al. (2016). K259-SUMOylation of DGCR8 promoted by p14ARF exerts a tumor-suppressive function. J. Mol. Cell Biol. 8, 456–458. doi:10.1093/jmcb/mjw030
Zhu, C., Chen, C., Huang, J., Zhang, H., Zhao, X., Deng, R., et al. (2015). SUMOylation at K707 of DGCR8 controls direct function of primary microRNA. Nucleic Acids Res. 43, 7945–7960. doi:10.1093/nar/gkv741
Keywords: Cajal body, coilin, SUMOlyation, E3 ligase, phosphorylation, RNP, DGCR8
Citation: Lett KE, McLaurin DM, Tucker SK and Hebert MD (2023) The Cajal body marker protein coilin is SUMOylated and possesses SUMO E3 ligase-like activity. Front. RNA Res. 1:1197990. doi: 10.3389/frnar.2023.1197990
Received: 31 March 2023; Accepted: 18 May 2023;
Published: 05 June 2023.
Edited by:
Miroslav Dundr, Rosalind Franklin University of Medicine and Science, United StatesReviewed by:
Yuichiro Takagi, Indiana University School of Medicine, United StatesCopyright © 2023 Lett, McLaurin, Tucker and Hebert. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael D. Hebert, bWhlYmVydEB1bWMuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.