- Head of Functional Genomics, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, United States
The biological importance of RNA has expanded as our appreciation of the complexity of its multiple types, structures, chemical compositions and biological roles. Research in RNA has been instrumental in revealing insights into fundamental biological processes including: the organization of information within genomes, the mechanisms of control of gene expression at the transcriptional (providing scaffolds for transcription factors and chromatin-modifying proteins) and post-transcriptional (RNA editing and modifications, translation, sponging) levels, spatiotemporal localization of elements involved in developmental and cell biology, and the evolution of first RNA genomes. Most recently, studies of RNA have expanded their clinical roles as diagnostics to the realm of therapeutic treatment for detected diseases. Finally, advances in RNA studies have been prompted by and contributed to the development of many novel methodological and computational approaches. The future of RNA research will add even more to our understanding of the origins of endophenotypes and these findings will be the focus of the Frontiers in RNA Research.
Introduction
Our understanding of the versatility and importance of RNA has expanded dramatically since its role was initially characterized as nucleic acid found associated with ribosomes and in protein synthesis. (Siekevitz and Zamecni, 1981). Long considered an enabling intermediary between the cell’s genetic information storehouse (DNA) and its workhorse element (protein), the discovery of the many types of RNAs found both in intra- and intercellular locations, their functional roles and their mechanisms of action have provided new insights in biology. While this growth of knowledge has been impressive, there continues to be an evolution in our understanding and appreciation of RNA’s remarkable capabilities. This growth has opened for us a landscape of new fundamental and applied questions in many academic and commercial disciplines.
Annotation of genomes
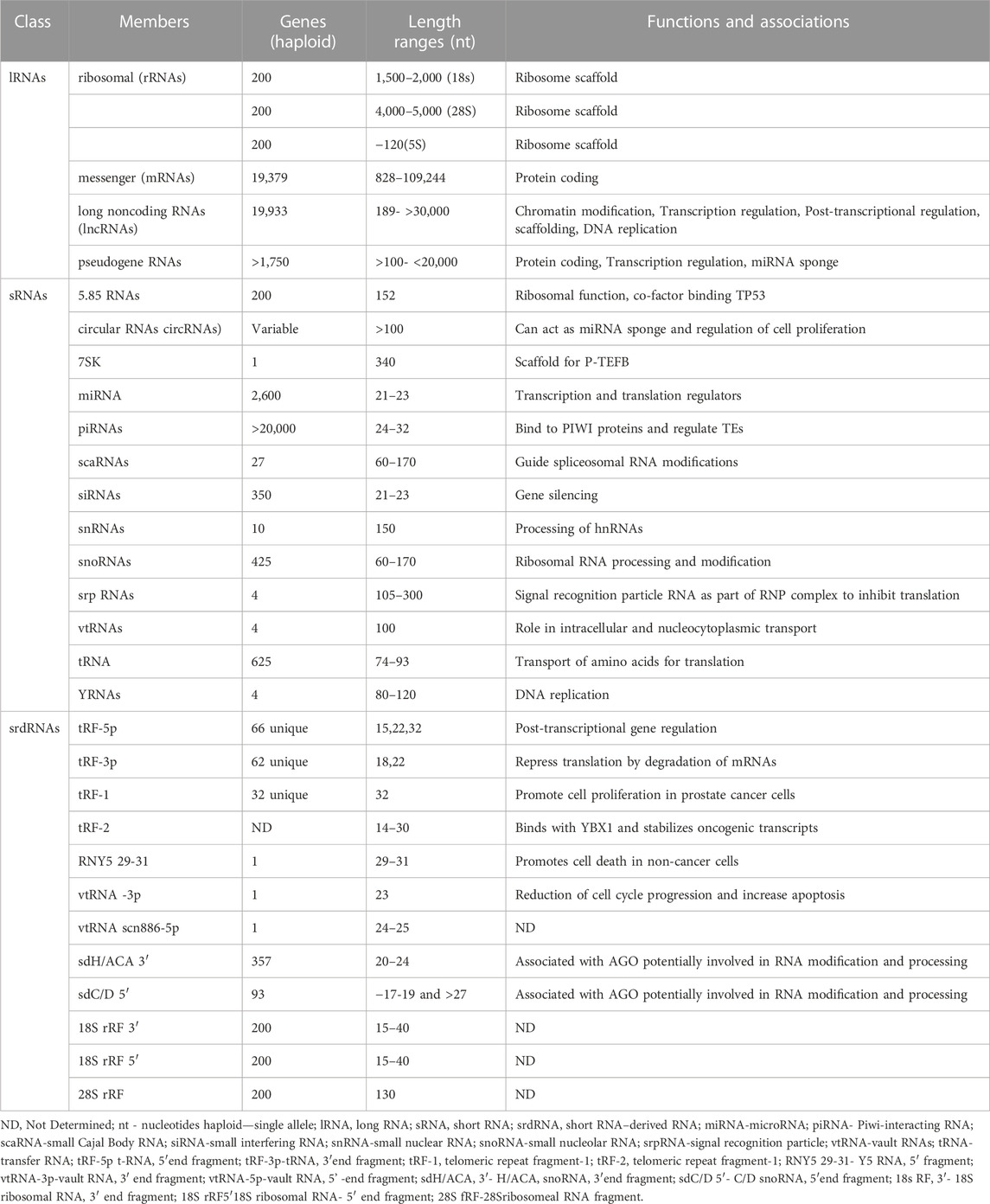
A consensus statement concerning a current and coherent picture of the roles of long noncoding RNAs (lncRNAs) recently has been published containing a suggested classification system for all coding and non-coding RNAs (Mattick et al., 2023). This classification system is straightforward and is based simply on the lengths of RNAs. Table 1 provides an additional view of RNA classification that is divided into three broad RNA classes. Each class is characterized by its RNA membership and the molecular origin of each class member. Information concerning the number of genes encoding each member, their approximate nucleotide lengths and, if known, at least one representative function and/or its molecular association is provided. The first class is composed of long (L) RNAs (lRNAs) (>200 nucleotides [nt]) lengths. The 4 members of the lRNAs class range from the well-studied messenger (mRNAs), ribosomal (rRNAs), and pseudogene RNAs to the emerging long non-coding RNAs (lncRNAs). Currently, there are also 13 members of the short RNA class (<200 nt) RNAs (sRNAs) ranging from transfer RNAs (tRNAs) to more recently identified functional sRNAs. There is also a third class of sRNA-derived RNAs (srdRNAs), which comprises 12 members, each of which are processed fragments of sRNAs. Overall, each class comprises multiple members, each of which are products processed from longer precursors. Based on the continuing growth in the membership of the sRNA and srdRNA classes, the catalog of RNAs is likely incomplete due to their low expression levels and their expression in only specific cell types (discussed below).
There is a sense that the annotation of animal genomes is relatively stable and this is commonly based on the relatively stable status of the human protein coding gene annotations (https://www.gencodegenes.org/human/stats.html) This sense comes from the observation the number of protein coding have been decreasing in number (2.5%) over the last decade. However, this intuition is misleading for lncRNA and sRNA genes. An increase of 4,058 (20%) lncRNA genes and a decrease of 1,970 (21%) sRNA genes has occurred over the same time period. In addition, many srdRNA products have only recently been identified and associated with specific biological functions (Table 1). However, the identification and mapping of srdRNAs have significantly lagged behind the protein-coding class and other ncRNAs primarily due to the uncertainty of their biological functions and their mapping positions in the genome. This is especially the case for some srdRNAs, which consist of complementary sequences to repeat elements, allowing them, for example, to control retrotransposon replication (Schorn et al., 2017).
The ongoing development and application of genome editing tools (e.g., CRISPR-Cas systems, prime editing, and base editing methodologies) strongly depend upon the availability of accurate and well-annotated genomes that contain a complete complement of coding and non-coding genes and transcripts. Until recently, with the completion of the Telomere-to-Telomere (T2T) Consortium’s T2T-CHM13 assembly, a contiguous sequence of the human genome was absent (Nurk et al., 2022). The results of this project have provided an additional 1956 gene predictions, 99 of which are predicted to be new annotated protein coding genes. Hence, the exploration and characterization of the human genome and its RNA products continues.
RNA binding partners and RNA functions
Outside of membrane-enclosed compartments, RNAs exist in inter-and intra-cellular domains protected by cofactors (proteins, carbohydrates, lipids) that are specifically or non-specifically interacting with them. RNA-binding proteins (RBPs) are the most well-studied of these cofactors. The importance of RBPs is underscored by the size of this family of genes. Depending on the evidence used to identify RBP genes, it is estimated that the size of the RBP family varies from approximately 1550 (7.9%) (Van Nostrand et al., 2020) to 4,000 (21%) (Gebauer et al., 2021) of 19,393 human protein-coding genes. Since RNAs are composed of highly structured and/or intrinsically disordered regions, the diversity of RBP genes is consistent with these structural properties. Since mRNAs and lncRNAs may have both of these structural characteristics, and since binding to an RNA may involve more than one RBP, a great deal of uncertainty in identifying a complete catalog of RBPs for each RNA persists. Additionally, the identity of RBPs interacting with each RNA and the binding location on the RNA remain active areas of study.
Ribonucleoprotein (RNP) complexes composed of RBPs and their target RNAs form an integral part of almost all biological operations. However, a key question that has been the focus of many studies is what are the functional roles of noncoding RNAs that are part of RBP complexes? Of the 19,928 human annotated lncRNA genes (https://www.gencodegenes.org/human/stats.html), fewer than approximately 150 of these have a validated biological function and for only a fraction of these is there a mechanistic understanding of this function. To address this issue, many multi-omic studies performed on single cells, cell lines, and tissues have sought to identify the subcellular locations at which specific RNP complexes operate. In a subset of these experiments, the identities of the protein and RNA binding components are identified. Most helpful is when spatial proteomic and transcriptomics can be performed on the same cells of origin, helping to provide subcellular locations and the identity of the RNAs in RNP complexes (Vandereyken et al., 2023). In addition, cross-linking immunoprecipitation sequencing (CLIP-seq) and selective 2′-hydroxyl acylation analysis by primer extension followed by mutational profiling (SHAPE-MaP) have been instrumental in determining secondary structures to annotate RBP binding sites for coding and lncRNAs in a transcriptome-wide fashion (Smola et al., 2015; Schmitz et al., 2016). The combination of having the binding protein identity and specific subcellular location provides insight into the role of the RNA. However, there is a need for the development of 3D structural modeling tools, akin to the protein structure modeler AlphaFold (Jumper et al., 2021) that can utilize these data.
Chemical modification of RNAs
The function of mRNA as an essential element in the translation process leading to protein synthesis has been understood since 1961, as is their subcellular locations in the nucleus and in the cytosolic ribosomes. However, there are numerous steps involving mRNAs and associated noncoding RNAs in the pre-transcription and post-transcription processes that remain unknown, such as the control of mRNA expression levels, the factors leading to predominance among all of a gene’s isoforms, the role of enhancer RNAs, the mechanisms of allele-specific expression, the influence of epigenetic modifications on transcription rates and splicing, and the role and regulation of RNA editing. More recently, discoveries in mRNAs have triggered new avenues of investigation. One of these involves the chemical modification of RNA ribonucleotides, which have been shown to regulate mRNA stability and to affect diverse biological processes. These modifications include: N6-methyladenosine (m6A), N6,2′-O-dimethyladenosine (m6Am), 8-oxo-7,8-dihydroguanosine (8-oxoG), pseudouridine (Ψ), 5-methylcytidine (m5C), and N4-acetylcytidine (ac4C). Beyond mRNAs, these modifications have been observed as part of a constellation of signals on many lncRNA and sRNAs, including snRNA in the spliceosome, and rRNAs affecting RNA expression and processing. There are still many questions surrounding the functions of these chemical modifications as well as the function of the modifications and the regulation of their location and temporal appearance.
A relatively recent discovery of a new type of RNA modification involving glycan-associated RNAs has been described (Flynn et al., 2021). These glycoRNAs occupy the luminal face of several types of cell membranes but most abundantly decorate the surface membranes of human peripheral blood monocyte cells (PBMCs). These RNAs with sialic acid–containing glycans modify a group of well-characterized sRNAs, including tRNAs, snRNAs, snoRNAs, and Y RNAs. Blocking the expression of specific glycoRNAs leads to the inhibition of monocyte and endothelial cell interactions. The study of these modified RNAs is ongoing and focus on how the RNAs reach the external surface of the cell, what type of binding exists between the glycan and RNA, what proteins compose the RNP complexes, and the how the glycoRNAs are protected when exposed to the external cellular environment remains unexplained.
Intercellular RNA communication
In addition to RNAs resident on the external surfaces of cells, many different types of RNAs have been detected outside of their cell of origin, housed in vesicles that are produced and released via different pathways and as free-floating RNPs. Again, it is important to note that in both conditions the RNAs are associated with other types of molecules (lipids, carbohydrates, and proteins). This property of exosomes or RNPs presents the opportunity for the packaging of multiple functional co-factors with RNA. Of note is the observation that the majority of the packaged and protected RNAs are sRNA or srdRNA and that the landscape of RNAs found outside of a cell is not necessarily a reflection of the profile found within the cell of origin. Rather, the most abundant RNA of a particular type detected outside of the cell often is a low copy number of the same sRNA type within the cell or vice versa, suggesting that there may be enrichment mechanisms for specific RNAs to be selected for packaging and extracellular release (Nechooshtan et al., 2020). It is also the case that the processing of tRNAs into tRF-5p and tRF-3p occurs outside of the cell by RNase I, which is inactive inside the cell. The mechanism by which sRNA or srdRNA enrichment is conducted is unknown but is of interest.
Both in the case of RNAs that are vesicle-bound or part of a RNP complex, there is abundant cytological and RNA-seq evidence that RNAs are transported into neighboring and distal cells (Chakrabortty et al., 2015). Interestingly, this capability has also been the focus of long-term and ongoing applied research efforts. RNA was conceived as a therapeutic in 1989 (Malone et al., 1989) and has followed a long and tortuous path leading to its current use as an immune-inducing agent against SARS-CoV-2 in vaccines to prevent Coronavirus Disease 2019 (COVID-19) (Dolgin, 2021). The use of mRNA and other sRNAs as therapeutics and in genetic engineering continue to be part of many ongoing pharmaceutical, academic, and commercial efforts. However, the technical issues facing the use of RNAs in these applied arenas are numerous and, thus, will be focus of research efforts for the foreseeable future.
Clinical challenges
For decades, both coding and ncRNAs have been useful in providing diagnostic and prognostic information as biomarkers for hundreds of clinical conditions. This is underscored by the 22,712 clinical studies that have evaluated, or are evaluating, both types of RNAs as markers (see ClinicalTrials.gov database). Many of these studies have examined autopsy organs and tissues affected by the disease state. These markers may not be useful if the expression or generation of a processed product of an RNA biomarker is tissue-specific, as is the case with many ncRNAs. Thus, an ongoing challenge is to determine if such RNA biomarkers can be detected in accessible samples or represented in the genome sequence or epigenome of living affected individuals.
The relatively recent emergence of FDA-approved RNA-based therapeutics has been enabled by the increased capabilities in the synthesis, production, and novel delivery of RNA (Damase et al., 2021). RNA approved therapeutics consist of many categories of RNA such as mRNAs, siRNAs, miRNAs, tRNAs and aptamers using lipid, polymer, silica, gold, and carbon nanoparticles as delivery and stability modalities. The continued success of the use of RNA as a therapeutic approach partially rests on insuring the stability of RNA. In addition to using the carrier modalities mentioned above, employing a variety of base modifications in the synthesis of the RNA therapeutic is also being investigated. Finally, with the recent success of mRNA-based vaccines, the checkered history of RNAi- and oligonucleotide-based drugs (Levin, 2019) could be understandably forgotten. In large measure, the inconsistency of these forms of RNA therapeutics stemmed from the challenges of off-target effects. Devising ways of avoiding these issues remains a focus of many clinical efforts.
Conclusion
There have been many fundamental and important scientific and medical contributions made by those in the RNA field. The challenges mentioned in this perspective represent frontiers that demark landscapes of opportunity. These and many other areas f RNA studies highlight the focus of Frontiers in RNA Research. This journal will seek to communicate noteworthy findings and address the outstanding challenges confronting the RNA field and to provide a resource for the scientific community.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Chakrabortty, S. K., Prakash, A., Nechooshtan, G., Hearn, S., and Gingeras, T. R. (2015). Extracellular vesicle-mediated transfer of processed and functional RNY5 RNA. RNA 21, 1966–1979. doi:10.1261/rna.053629.115
Damase, T. R., Sukhovershin, R., Boada, C., Taraballi, F., Pettigrew, R. I., and Cooke, J. P. (2021). The limitless future of RNA therapeutics. Front. Bioeng. Biotechnol. 9, 628137. doi:10.3389/fbioe.2021.628137
Dolgin, E. (2021). The tangled history of mRNA vaccines. Nature 597, 318–324. doi:10.1038/d41586-021-02483-w
Flynn, R. A., Pedram, K., Malaker, S. A., Batista, P. J., Smith, B. A., Johnson, A. G., et al. (2021). Small RNAs are modified with N-glycans and displayed on the surface of living cells. Cell 184, 3109–3124 e22. doi:10.1016/j.cell.2021.04.023
Gebauer, F., Schwarzl, T., Valcárcel, J., and Hentze, M. W. (2021). RNA-binding proteins in human genetic disease. Nat. Rev. Genet. 22, 185–198. doi:10.1038/s41576-020-00302-y
Jumper, J. E., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi:10.1038/s41586-021-03819-2
Levin, A. A. (2019). Treating disease at the RNA level with oligonucleotides. N. Engl. J. Med. 380, 57–70. doi:10.1056/NEJMra1705346
Malone, R. W., Felgner, P. L., and Verma, I. M. (1989). Cationic-liposome mediated RNA transfection. Proc. Natl. Acad. Sci. U. S. A. 86, 6077–6081. doi:10.1073/pnas.86.16.6077
Mattick, J. S., Amaral, P. P., Carninci, P., Carpenter, S., Chang, H. Y., Chen, L. L., et al. (2023). Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Bio. [Epub ahead of print]. doi:10.1038/s41580-022-00566-8
Nechooshtan, G., Yunusov, D., Chang, K., and Gingeras, T. R. (2020). Processing by RNAse 1 forms t—RNA halves and distinct Y-RNA fragments in the extracellular environment. Nucleic Acids Res. 48 (14), 8035–8049. doi:10.1093/nar/gkaa526
Nurk, S., Koren, S., Rhie, A., Rautiainen, M., Bzikadze, A. V., Mikheenko, A., et al. (2022). The complete sequence of a human genome. Science 376, 44–53. doi:10.1126/science.abj6987
Schmitz, S. U., Grote, P., and Herrmann, B. G. (2016). Mechanisms of long noncoding RNA function in development and disease. Cell Mol. Life Sci. 73, 2491–2509. doi:10.1007/s00018-016-2174-5
Schorn, A. J., Gutbrod, M. J., LeBlanc, C., and Martienssen, R. (2017). LTR-retrotransposon control by tRNA-derived small RNAs. Cell 170 (1), 61–71.e11. doi:10.1016/j.cell.2017.06.013
Siekevitz, P., and Zamecnik, P. C. (1981). Ribosomes and protein synthesis. J. Cell Biol. 91, 53s–65s. doi:10.1083/jcb.91.3.53s
Smola, M., Calabrese, J. M., and Weeks, K. M. (2015). Detection of RNA-Protein interactions in living cells with SHAPE. Biochemistry 54, 6867–6875. doi:10.1021/acs.biochem.5b00977
Van Nostrand, E. L., Freese, P., Pratt, G. A., Wang, X., Wei, X., Xiao, R., et al. (2020). A large-scale binding and functional map of human RNA-binding proteins. Nature 583, 711–719. doi:10.1038/s41586-020-2077-3
Keywords: RNA, long non coding RNA, RNA binding protein, genome annotation, RNA function, mRNA, rRNA (ribosomal RNA), RNA biomarkers
Citation: Gingeras TR (2023) Current frontiers in RNA research. Front. RNA Res. 1:1152146. doi: 10.3389/frnar.2023.1152146
Received: 27 January 2023; Accepted: 21 April 2023;
Published: 22 May 2023.
Edited by:
Chandrasekhar Kanduri, University of Gothenburg, SwedenReviewed by:
Peng Yao, University of Rochester, United StatesPieter Mestdagh, Ghent University, Belgium
John Stanley Mattick, University of New South Wales, Australia
Copyright © 2023 Gingeras. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thomas R. Gingeras, Z2luZ2VyYXNAY3NobC5lZHU=
 Thomas R. Gingeras
Thomas R. Gingeras