- 1Department of Electrical Engineering, Eindhoven University of Technology, Eindhoven, Netherlands
- 2Department of Engineering, King’s College London, London, United Kingdom
Simulation is an indispensable tool in both engineering and the sciences. In simulation-based modeling, a parametric simulator is adopted as a mechanistic model of a physical system. The problem of designing algorithms that optimize the simulator parameters is the focus of the emerging field of simulation-based inference (SBI), which is often formulated in a Bayesian setting with the goal of quantifying epistemic uncertainty. This work studies Bayesian SBI that leverages a parameterized quantum circuit (PQC) as the underlying simulator. The proposed solution follows the well-established principle that quantum computers are best suited for the simulation of certain physical phenomena. It contributes to the field of quantum machine learning by moving beyond the likelihood-based methods investigated in prior work and accounting for the likelihood-free nature of PQC training. Experimental results indicate that well-motivated quantum circuits that account for the structure of the underlying physical system are capable of simulating data from two distinct tasks.
1 Introduction
1.1 Context and motivation
Simulation has been an indispensable tool for the understanding and discovery of complex and open-ended phenomena in situ via the study of dynamic systems and processes in silico (Lavin et al., 2021). The studied phenomena run the gamut of scale and domain, from biology (Dada and Mendes, 2011) and climatology (Vautard et al., 2013) to economics and the social sciences (Elshafei et al., 2016). In simulation-based modeling, a parametric simulator is adopted as a mechanistic model of a physical system. Given specific parameter values, the simulator produces synthetic data. The general modeling principle is that parameters that lead to synthetic data close to the actual observations from the physical system are considered the most plausible ones to explain the measurements.
However, there are important challenges that have limited the adoption of simulators in many settings of scientific and engineering relevance. On the one hand, computational costs may motivate the imposition of simplifying assumptions, which may render the results unusable for reliable hypothesis testing. On the other hand, at a methodological level, simulators are poorly suited for statistical inference as they inherently provide only implicit access to the likelihood of an observation. In fact, simulators can sample from a distribution, but they cannot, typically, quantify the probability of a simulation output (Song et al., 2020; Simeone, 2022). These problems are currently being tackled using novel machine learning tools and probabilistic programming in the emerging field of simulation-based inference (SBI) (Cranmer et al., 2020).
In a frequentist setting, SBI produces point estimates for the simulator parameters, failing to capture epistemic uncertainty arising from the access to limited data from the physical system. Alternatively, adopting a Bayesian formulation, a distribution on the model parameter space can be optimized in order to reflect a probabilistic notion of uncertainty (Cranmer et al., 2020).
1.2 Quantum SBI
As illustrated in Figure 1, in this work, we study Bayesian SBI that leverages a parameterized quantum circuit (PQC) as the underlying simulator. PQCs are the subject of the field of quantum machine learning (Schuld and Petruccione, 2021). They consist of quantum circuits with a fixed ansatz whose parameters, typically rotation angles for some of the gates, are optimized using a classical computer. PQCs can be readily implemented on existing noisy intermediate scale quantum (NISQ) hardware, and are viewed as a potential means to demonstrate practical use cases for quantum computing.
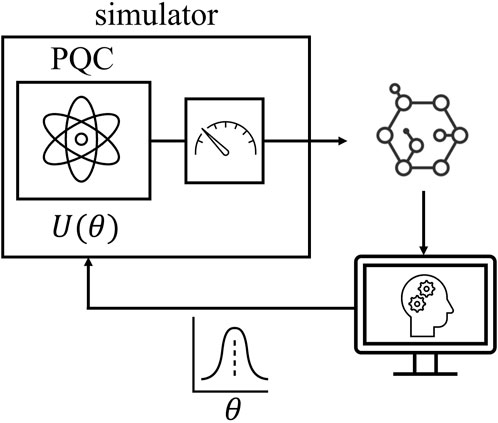
Figure 1. Quantum simulation-based Bayesian inference: A simulator based on a parameterized quantum circuit (PQC) is trained via a likelihood-free Bayesian inference algorithm to serve as a simulator for a physical process of interest.
The motivation for the proposed solution, integrating PQCs with SBI, is twofold. First, from the perspective of SBI, by leveraging quantum circuits as simulators, we follow the well-established principle that quantum computers are best suited for the simulation of certain physical phenomena, especially at the microscopic scale (Georgescu et al., 2014). Second, from the viewpoint of quantum machine learning, Bayesian learning methods have been argued to be potentially beneficial as they can be better account for uncertainty in the model space, enhancing test-time performance (Duffield et al., 2022). Our work thus contributes to the literature on quantum machine learning by moving beyond the likelihood-based method investigated in (Duffield et al., 2022) by leveraging state-of-the-art likelihood-free SBI methods.
1.3 Main contributions
This work explores the application of Bayesian SBI for the training of simulators implemented as PQCs. The main contributions are as follows.
2 Bayesian simulation-based inference
Let us assume the availability of a data set
To this end, we fix a class of parameterized simulators
We focus on the problem of inferring the parameter vector
where
is the likelihood evaluated on the data set
of data point
Since the likelihood
The rest of the paper is organized as follows. Section 2 presents the problem of Bayesian SBI. Section 3, 4 review Bayesian SBI methods based on sampling and surrogate functions, respectively. Section 5 presents the proposed approach based on quantum simulators. Section 6 presents experimental results, and Section 7 concludes the paper.
3 Bayesian SBI via sampling
In this section, we describe sampling-based Bayesian SBI.
3.1 Model
As shown in Figure 2, sampling-based Bayesian SBI, also known as ABC, models the data-generating mechanism via a hierarchical probability distribution. In it, the simulator
1. A model parameter is drawn from the prior
2. The simulator outputs conditionally independent and identically distributed latent variables
3. And the data set
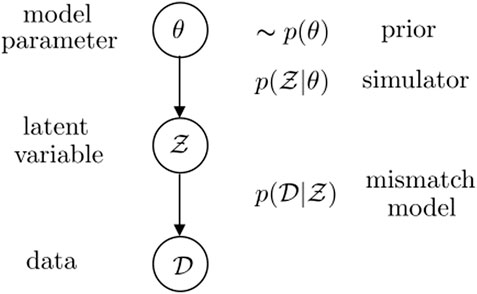
Figure 2. Probabilistic graphical model adopted by sampling-based Bayesian SBI. We follow here the definition of mismatch model given in (Schmon et al., 2020).
The distribution
if no such mismatch is expected, one can set
where
More generally, the choice of the mismatch model must account for requirements of accuracy and efficiency, and it is typically specified as (Wilkinson, 2013; Schmon et al., 2020)
where
By (Equation 7), the data
where
where
By the model in Figure 2, the posterior distribution over model parameters and samples produced by the simulator given the data set
by marginalizing out the simulator’s outputs
in the special case in which no mismatch is accounted for in the model, i.e., when
which corresponds to the conventional posterior distribution (Equation 1) in the ideal case of a well-specified model.
The goal of ABC is to produce samples
3.2 Rejection-sampling ABC
Rejection-sampling ABC (RS-ABC) iteratively draws candidate samples
To this end, for each candidate model parameter sample
The sample
The distribution of an accepted sample can be computed as
Therefore, in order for the distribution (Equation 14) to match the desired posterior (Equation 10), one can set
In particular, if
3.3 Metropolis-Hastings ABC
RS-ABC typically produces a low rate of acceptance of the generated samples, particularly when data are sufficiently high dimensional. To see this, consider the common case in which the prior
To overcome this drawback, reference (Marjoram et al., 2003) proposed Metropolis-Hastings ABC (MH-ABC). MH-ABC proceeds to sample from Equation 10) in a sequential manner. To elaborate, let us denote as
accordingly, a new parameter
Let
this equality can be ensured by setting
using Equations 10, 17, we finally get the acceptance probability adopted by MH-ABC as
4 Bayesian SBI via surrogates
Unlike sampling-based methods, surrogate-based methods use the simulator, along with the data set
4.1 Ratio estimation
Ratio estimation (RE) applies contrastive learning to estimate the ratio between the likelihood
given an estimate
In practice, the unnormalized posterior in (Equation 22) can be used, without the need for an explicit normalization, to obtain samples
RE methods train a binary classifier to distinguish between data sets generated according to the distributions at the numerator and denominator of the ratio (Equation 21). To this end, for any fixed value
To generate data sets in the first class, one directly runs the simulator with the given value
The binary classifier takes as input a data set
By the construction of the data set, a data set
furthermore, the posterior distribution is
writing
and
Making the approximation
4.2 Amortized ratio estimation
To improve the performance of RE, amortization techniques can be used, whereby the classifier is amortized using the parameters from the simulator. To explain, note that the true ratio can be equivalently expressed as
This modification suggests a way to train the binary classifier to distinguish between dependent sample-parameter pairs
In cases when the divergence between the densities is large, the classifier can obtain almost perfect accuracy with a relatively poor estimate of the density ratio. This failure mode is known as the density-chasm problem, and can be overcome by transporting samples from one distribution to the other, creating a chain of intermediate data sets. The density-ratio between consecutive datasets along this chain can be then accurately estimated via classification. The chained ratios are then combined via a telescoping product to obtain an estimate of the original density-ratio. This method is referred to as telescopic amortized RE (Montel et al., 2023).
Finally, as practical note, we emphasize that, both in the amortized and non-amortized settings, to avoid numerical errors one can extract the logit,
5 Quantum bayesian simulation-based inference
In the previous sections, we have reviewed sampling-based and surrogate-based Bayesian SBI techniques. In the proposed quantum Bayesian SBI system, illustrated in Figure 1, both classes of methods are applicable. The key new element is the introduction of a PQC as the simulator
5.1 Parameterized quantum circuits as simulators
The proposed quantum SBI solution aims at developing simulators for the generation of a quantity of interest
As reviewed in (Schuld and Petruccione, 2021; Simeone et al., 2022), PQCs implement a parameterized unitary transformation
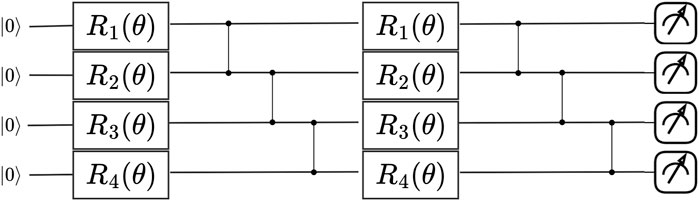
Figure 3. An example of a PQC which serves as a simulator of a physical process. In this work, we propose to treat the PQC as a likelihood-free model that can be trained via Bayesian simulation-based inference.
Initializing the register of
Furthermore, by Born’s rule, the probability distribution
where
5.2 Choosing the ansatz
In general, choosing a good ansatz for the quantum simulator entails a difficult trade-off between adherence to the physics of the problem and complexity of implementation. In particular, if prior knowledge about the structure of the data are available, this may be encoded as an inductive bias into the choice of the quantum circuit architecture, assuming that the complexity of the implementation allows it.
The typical way to encode structure into the ansatz is to leverage symmetries in the data. Symmetries refer to transformations of the data that leave it invariant or change it in a predictable, equivariant manner. For example, the binding energy of a molecule does not change by permuting the order of the atoms, and a picture of a cat still depicts a cat regardless of the position of the cat within the image. This prior knowledge can be encoded into the simulator ansatz as a geometric prior. Notable examples include quantum graph neural networks (QGNNs) (Verdon et al., 2019; Mernyei et al., 2022) and quantum convolutional neural network (QCNNs) (Cong et al., 2019), which preserve equivariance to permutations and shifts, respectively. Other examples include quantum recurrent neural networks for time series processing (Nikoloska et al., 2023).
In the absence of prior knowledge, or when the practitioner is concerned with efficient hardware implementation, they may choose to use a hardware-efficient architecture (HEA). Such architectures use only single qubit and two qubit gates, placed along the existing connectivity of the quantum computer, which are easily implemented on both gate-based or pulse-based NISQ machines (Zulehner et al., 2018; Gyongyosi and Imre, 2021).
6 Results
In this section, we provide experimental results to validate the proposed concept of quantum Bayesian SBI.
6.1 Tasks
6.1.1 Generating bars-and-stripes images
We first consider the classical small-scale benchmark problem of generating
6.1.2 Simulating molecular topologies
In this second task, which is closer to a real-life application of the proposed method, the task of the simulator is to generate valid molecular structures, i.e., valid primary topologies, for 4-atom molecules comprised of carbon (C), hydrogen (H), boron (B), oxygen (O), or nitrogen (N) atoms. Knowing a valid molecular topology, specifying which atom is covalently bonded to which other atom, is crucial for determining classical potentials for biomolecules. Each sample consists of a
6.2 Simulator ansatz and hyperparameters
We consider four circuit architectures. All of the considered architectures are comprised of four qubits and two layers.
6.2.1 QCNN
For BAS, an image dataset, we employ a QCNN. QCNN is a translation-equivariant model that uses convolution layers and applies a single quasi-local unitary (Cong et al., 2019). Each pixel is represented by a qubit. We do not employ pooling, and the quasi-local unitary is applied on pairs of qubits. To determine the
6.2.2 QGNN
For molecular topologies, we employ a QGNN. as molecules can be well represented as graphs. A QGNN is an permutation-equivariant ansatz (Verdon et al., 2019). Each atom is represented by a qubit. To determine whether a covalent bond is present between each atom pair
6.2.3 HEA
As a basic benchmark, for both tasks, we also implement an HEA, which consists of general single qubit gates, i.e., rotations described by three angles, and by CNOT gates applied in a cyclical manner across all pairs successive qubits. The same observables described above are considered to extract information from the output states for the two tasks.
6.2.4 Separable circuits
Finally, to gauge the potential benefits of entanglement, we adopt a mean-field, or separable, ansatz that consists solely of general single-qubit gates. The resulting circuits can be efficiently simulated on classical computers for any number of qubits, with no need for quantum hardware. Therefore, this setup essentially represents a classical benchmark. The same observables are again applied for the two tasks.
6.3 SBI algorithms
As a representative of sampling-based schemes, we implement RS-SBI with the classical kernel (Equation 8) with summary statistics given by the histogram of the generated samples
6.4 Evaluation and performance metrics
We are interested in evaluating the adherence of the distribution of the samples produced by the simulator to the ground-truth data-generating distribution. To this end, for any fixed simulator parameters
In Bayesian SBI, the model parameter
Each draw of the model parameter vector
As a benchmark learning algorithm, we also show the performance of a scheme that produces a point estimate for the parameters
6.5 Results
The distributions of the TVD for both sampling- and surrogate-based Bayesian SBI schemes are shown in Figure 4 for the two tasks under study. For this figure, we adopt the best-performing ansatz for each task, namely, the QCNN and QGNN, respectively. It is observed that, by accounting for the uncertainty on the likelihood, Bayesian SBI schemes can outperform conventional frequentist techniques. In fact, samples produced from the posterior distribution can yield significantly lower TVD values, which indicate a closer match of the ground-truth distribution. Furthermore, the spread of the distribution produced by Bayesian SBI strategies is task-dependent. Similarly, the choice between sampling-based and surrogate-based schemes is also seen to depend on the task, with the latter having a clear advantage in the BAS task.
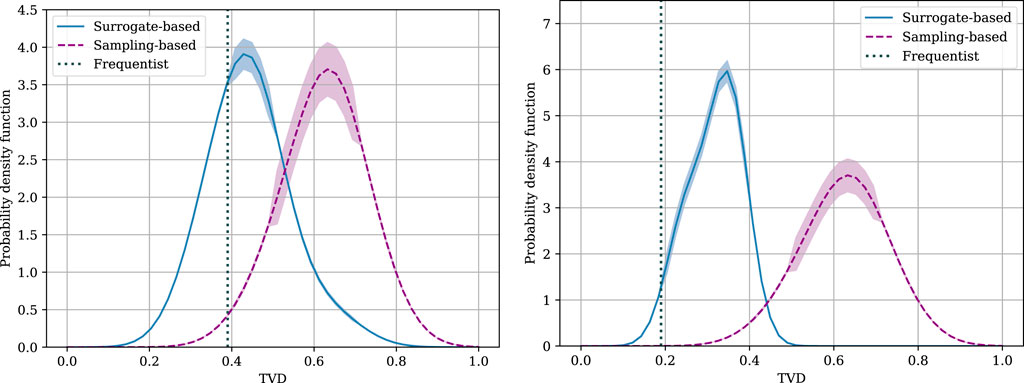
Figure 4. Distribution of the TVD between distribution of the samples produced by the simulator and ground-truth distribution for the BAS data set (left), and for the primary molecular structure task (right).
We now analyze the impact of different ansatzes by showing in Figure 5 distributions of the TVD for various architectures of the quantum simulator. Whilst we do not claim that quantum circuits are provably better than classical counterparts for the problem at hand, the separable circuits is observed to result in a very large TVD. In contrast, for both tasks, the symmetry-preserving simulators–QCNN for the BAS task and QGNN for the molecular topology task–result in the smallest TVD between the generated samples and the true distribution, suggesting that encoding inductive-biases in the simulator is indeed helpful for SBI.
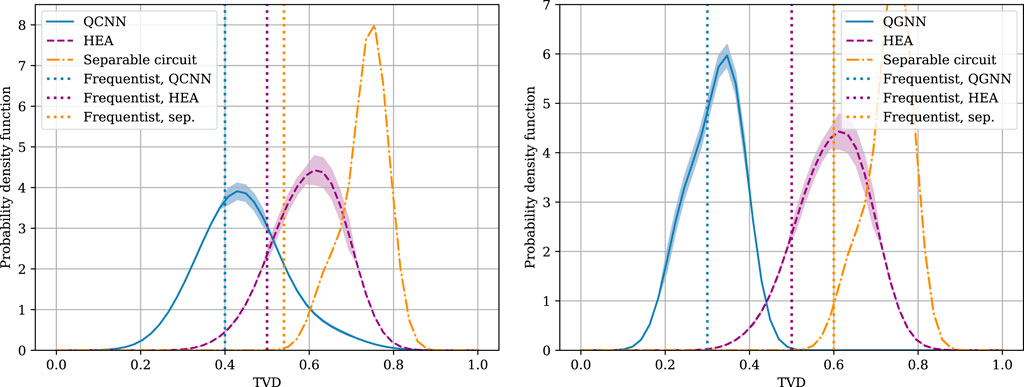
Figure 5. Distribution of the TVD between distribution of the samples produced by the simulator and ground-truth distribution for the BAS data set (left), and for the primary molecular structure task (right).
7 Concluding remarks
Simulation intelligence is an emerging multi-disciplinary topic that views simulation as a central tool for design and discovery (Lavin et al., 2021). The scope and reach of the field are only expected to grow in importance with the fast development of generative artificial intelligence tools and with the spread of digital twinning as a framework for engineering complex systems (Ruah et al., 2023). Quantum circuits are known to be efficient solutions to implementing samplers from complex distributions in discrete spaces. This property makes quantum circuit appealing as co-processors for the controlled generation of latent random variables (Nikoloska and Simeone, 2022). In this context, this work has taken a few steps towards the idea of integrating quantum circuits as simulators in a simulation-based process.
The main aim of this article is to provide readers with a background in quantum machine learning with an introduction to Bayesian SBI tools. Many problems are left open to future investigations, including the investigation of larger-scale use cases, the implementation on NISQ computers, and the analysis of the impact of quantum noise.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
IN: Data curation, Investigation, Software, Validation, Visualization, Writing–original draft, Writing–review and editing. OS: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The work of OS was partially supported by the European Union’s Horizon Europe project CENTRIC (101096379), by the Open Fellowships of the EPSRC (EP/W024101/1), by the EPSRC project (EP/X011852/1), and by the United Kingdom Government under Project REASON.
Acknowledgments
The authors acknowledge the contribution of Hari Hara Suthan Chittoor in the early stages of this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Beaumont, M. A. (2019). Approximate Bayesian computation. Annu. Rev. Statistics Its Appl. 6, 379–403. doi:10.1146/annurev-statistics-030718-105212
Beaumont, M. A., Zhang, W., and Balding, D. J. (2002). Approximate Bayesian computation in population genetics. Genetics 162, 2025–2035. doi:10.1093/genetics/162.4.2025
Bergholm, V., Izaac, J., Schuld, M., Gogolin, C., Ahmed, S., Ajith, V., et al. (2018). Pennylane: automatic differentiation of hybrid quantum-classical computations. arXiv Prepr. arXiv:1811.04968.
Brooks, S., Gelman, A., Jones, G., and Meng, X. L. (2011). Handbook of Markov chain Monte Carlo. United Kingdom: CRC Press.
Cong, I., Choi, S., and Lukin, M. D. (2019). Quantum convolutional neural networks. Nat. Phys. 15, 1273–1278. doi:10.1038/s41567-019-0648-8
Cranmer, K., Brehmer, J., and Louppe, G. (2020). The frontier of simulation-based inference. Proc. Natl. Acad. Sci. 117, 30055–30062. doi:10.1073/pnas.1912789117
Dada, J. O., and Mendes, P. (2011). Multi-scale modelling and simulation in systems biology. Integr. Biol. 3, 86–96. doi:10.1039/c0ib00075b
Duffield, S., Benedetti, M., and Rosenkranz, M. (2022). Bayesian learning of parameterised quantum circuits. arXiv Prepr. arXiv:2206.07559.
Elshafei, Y., Tonts, M., Sivapalan, M., and Hipsey, M. (2016). Sensitivity of emergent sociohydrologic dynamics to internal system properties and external sociopolitical factors: implications for water management. Water Resour. Res. 52, 4944–4966. doi:10.1002/2015wr017944
Georgescu, I. M., Ashhab, S., and Nori, F. (2014). Quantum simulation. Rev. Mod. Phys. 86, 153–185. doi:10.1103/revmodphys.86.153
Gyongyosi, L., and Imre, S. (2021). Scalable distributed gate-model quantum computers. Sci. Rep. 11, 5172. doi:10.1038/s41598-020-76728-5
Hastings, W. K. (1970). Monte Carlo sampling methods using Markov chains and their applications. Biometrika 57, 97–109. doi:10.1093/biomet/57.1.97
Hermans, J., Begy, V., and Louppe, G. (2020). “Likelihood-free mcmc with amortized approximate ratio estimators,” in International conference on machine learning. United Kingdom: PMLR, 4239–4248.
Lavin, A., Krakauer, D., Zenil, H., Gottschlich, J., Mattson, T., Brehmer, J., et al. (2021). Simulation intelligence: towards a new generation of scientific methods. arXiv Prepr. arXiv:2112.03235.
MacKay, D. J., and Mac Kay, D. J. (2003). Information theory, inference and learning algorithms. United Kingdom: Cambridge University Press.
Marjoram, P., Molitor, J., Plagnol, V., and Tavaré, S. (2003). Markov chain Monte Carlo without likelihoods. Proc. Natl. Acad. Sci. 100, 15324–15328. doi:10.1073/pnas.0306899100
Mernyei, P., Meichanetzidis, K., and Ceylan, I. I. (2022). “Equivariant quantum graph circuits,” in International conference on machine learning. United Kingdom: PMLR, 15401–15420.
Montel, N. A., Alvey, J., and Weniger, C. (2023). Scalable inference with autoregressive neural ratio estimation. arXiv Prepr. arXiv:2308.08597.
Nikoloska, I., and Simeone, O. (2022). “Quantum-aided meta-learning for Bayesian binary neural networks via born machines,” in 2022 IEEE 32nd international Workshop on machine Learning for signal processing (MLSP) (IEEE), 1–6.
Nikoloska, I., Simeone, O., Banchi, L., and Veličković, P. (2023). Time-warping invariant quantum recurrent neural networks via quantum-classical adaptive gating. Mach. Learn. Sci. Technol. 4, 045038. doi:10.1088/2632-2153/acff39
Papamakarios, G., Sterratt, D., and Murray, I. (2019). “Sequential neural likelihood: fast likelihood-free inference with autoregressive flows,” in The 22nd international Conference on artificial Intelligence and statistics (PMLR), 837–848.
Price, L. F., Drovandi, C. C., Lee, A., and Nott, D. J. (2018). Bayesian synthetic likelihood. J. Comput. Graph. Statistics 27, 1–11. doi:10.1080/10618600.2017.1302882
Qin, C., Wen, Z., Lu, X., and Van Roy, B. (2022). An analysis of ensemble sampling. arXiv Prepr. arXiv:2203.01303.
Ragone, M., Braccia, P., Nguyen, Q. T., Schatzki, L., Coles, P. J., Sauvage, F., et al. (2022). Representation theory for geometric quantum machine learning. arXiv Prepr. arXiv:2210.07980.
Ruah, C., Simeone, O., and Al-Hashimi, B. (2023). A Bayesian framework for digital twin-based control, monitoring, and data collection in wireless systems. IEEE J. Sel. Areas Commun. 41, 3146–3160. doi:10.1109/jsac.2023.3310093
Schmon, S. M., Cannon, P. W., and Knoblauch, J. (2020). Generalized posteriors in approximate Bayesian computation. arXiv preprint arXiv:2011.08644.
Simeone, O. (2022). Machine learning for engineers. Cambridge University Press. doi:10.1017/9781009072205
Simeone, O., et al. (2022). An introduction to quantum machine learning for engineers. Found. Trends® Signal Process. 16, 1–223. doi:10.1561/2000000118
Sisson, S. A., and Fan, Y. (2010). “Likelihood-free Markov chain Monte Carlo,” in arXiv preprint arXiv:1001.2058.
Song, J., Meng, C., and Ermon, S. (2020). Denoising diffusion implicit models. arXiv preprint arXiv:2010.02502.
Sunnåker, M., Busetto, A. G., Numminen, E., Corander, J., Foll, M., and Dessimoz, C. (2013). Approximate Bayesian computation. PLoS Comput. Biol. 9, e1002803. doi:10.1371/journal.pcbi.1002803
Thomas, O., Dutta, R., Corander, J., Kaski, S., and Gutmann, M. U. (2022). Likelihood-free inference by ratio estimation. Bayesian Anal. 17, 1–31. doi:10.1214/20-ba1238
Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones, L., Gomez, A. N., et al. (2017). Attention is all you need. Adv. neural Inf. Process. Syst. 30.
Vautard, R., Gobiet, A., Jacob, D., Belda, M., Colette, A., Déqué, M., et al. (2013). The simulation of european heat waves from an ensemble of regional climate models within the euro-cordex project. Clim. Dyn. 41, 2555–2575. doi:10.1007/s00382-013-1714-z
Verdon, G., McCourt, T., Luzhnica, E., Singh, V., Leichenauer, S., and Hidary, J. (2019). “Quantum graph neural networks,” in arXiv preprint arXiv:1909, 12264.
Wilkinson, R. D. Approximate Bayesian computation (ABC) gives exact results under the assumption of model error. Stat. Appl. Genet. Mol. Biol. 12 (2013) 129–141. doi:10.1515/sagmb-2013-0010
Keywords: simulation-based inference, quantum computing, Bayesian methods, quantum machine learning, Bayesian inference
Citation: Nikoloska I and Simeone O (2024) An introduction to Bayesian simulation-based inference for quantum machine learning with examples. Front. Quantum Sci. Technol. 3:1394533. doi: 10.3389/frqst.2024.1394533
Received: 01 March 2024; Accepted: 05 August 2024;
Published: 29 August 2024.
Edited by:
Julio De Vicente, Universidad Carlos III de Madrid, SpainReviewed by:
Gabriel Nathan Perdue, Fermilab Accelerator Complex, Fermi National Accelerator Laboratory (DOE), United StatesLaszlo Gyongyosi, Budapest University of Technology and Economics, Hungary
Copyright © 2024 Nikoloska and Simeone. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ivana Nikoloska, aS5uaWtvbG9za2FAdHVlLm5s
 Ivana Nikoloska
Ivana Nikoloska Osvaldo Simeone
Osvaldo Simeone