
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Public Health, 19 March 2025
Sec. Infectious Diseases: Epidemiology and Prevention
Volume 13 - 2025 | https://doi.org/10.3389/fpubh.2025.1568194
This article is part of the Research TopicOne Health Approach to Mycobacterial Infections in Veterinary ScienceView all 8 articles
Tuberculosis (TB), caused by members of the Mycobacterium tuberculosis complex (MTBC), is a significant global health threat, with millions of cases diagnosed annually and an annual death toll exceeding 1.6 million. Zoonotic TB (zTB), transmitted between animals and humans, remains poorly understood and difficult to control. This narrative review examined current evidence of the emergence and transmission pathways of Mycobacterium orygis, a recently defined member of MTBC. The structured searches for published literature and genome sequence with relevant metadata were conducted using NCBI PubMed and GenBank, respectively. Population diversity was investigated using phylogenomic analysis. Despite significant gaps in current laboratory diagnostic capacity for TB, M. orygis has been documented in 14 countries from 5 continents across 17 host species. Many cases (≈40%) were diagnosed around The Subcontinent and associated with a diverse range of mammalian hosts. In India, zTB due to M. orygis appeared to be more prevalent than disease associated with M. bovis (another zoonotic member of the MTBC). The whole genome sequencing of M. orygis isolates highlighted high diversity associated with different ecological niches. The increasing world-wide prevalence of M. orygis, especially in Asia, highlighted its emergence as a significant pathogen with zoophilic and anthropophilic potential. The reviewed evidence suggested multiple transmission pathways between humans and domesticated and wild mammalian hosts. Enhanced TB laboratory diagnostics and surveillance are imperative for mitigating the spread of zTB including one caused by M. orygis in areas of established and currently unrecognized endemicity.
Tuberculosis (TB) is an ancient disease that kills more people every year than any other infectious disease (1, 2). There are approximately 11 million cases globally, resulting in up to 1.6 million deaths annually (3). In 2024 the World Health Organization (WHO) reported a global reduction in the number of people diagnosed with TB and receiving treatment in the year prior. However, this is only a slight reversal in the treatment disruptions caused by the COVID-19 pandemic (4). TB disproportionally affects people in low-income and middle-income countries (LMICs). South Asia (SA), which includes India, Nepal, Pakistan, Bangladesh, and Sri Lanka, accounts for almost 40% of global cases (5). TB thrives in impoverished regions where access to healthcare services is limited and social determinants of poor outcomes such as overcrowding and poor ventilation are common (6). There is a direct association between poverty and higher rates of TB, especially in ethnic minorities (7).
Tuberculosis is caused by genetically related members of the Mycobacterium tuberculosis complex (MTBC), which includes both animal- and human-adapted (M. tuberculosis sensu stricto) lineages. The MTBC consists of over 10 species of mycobacteria with highly conserved genomes (8, 9). In addition, each of these species occupy a distinct ecological niche and infect a range of host species (10). Globally, of the eight members of the MTBC that cause TB in humans, the primary causative agent is M. tuberculosis. However, seven other species responsible for both human and zoonotic infections have been identified, including M. orygis, M. bovis, M. caprae, M. canettii, M. africanum, M. pinnipedii, and M. microti (11). Zoonotic tuberculosis (zTB) has been previously defined as a human infection caused by M. bovis; however, it is now understood zTB can be a manifestation of TB in humans, caused by an animal adapted lineage and acquired from infected animals (12). Urbanization and the destruction of natural wildlife habitats have increased the risk of zTB and approximately one-third of all zoonotic cases originate from Asia (13). The genetic similarity between the MTBC species demands high-resolution diagnostic methods to accurately identify the causative agent, as well as to decipher transmission dynamics and drug resistance in implicated pathogens. However, timely recognition of zTB remains challenging and zTB has become a significant public health concern globally with the potential to disrupt production and trade of animal products for food and other applications. This review described the emerging evidence of the global spread and diversity in M. orygis genomes, affected hosts and pathways of transmission.
Previously known as Oryx bacillus or the antelope clade, M. orygis has been infrequently reported in association with zTB over the past 30 years, although the number of case reports has recently increased due to advances in molecular diagnostics (Figure 1). In 1976, Lomme et al. observed granulomatous lesions in oryx’s that was suspected to be infected with M. tuberculosis; however, the oryx failed skin antigen tests and TB was confirmed by microscopic and postmortem examination (14). It has been subsequently reported that it was possible to have a negative tuberculous skin test (TST) in M. orygis positive cases (15). The first confirmed case of Oryx bacillus was diagnosed in 1987 by Van Soolingen et al. but was not reported until 1994 (16). Molecular typing using DNA fingerprinting revealed IS6110 differences between M. bovis isolates and two Oryx leucoryx (Arabian oryx) isolates collected from the wildlife parks in the Netherlands and one isolate from a previous study (16). It should also be noted that methods for differentiation between members of the MTBC was in its infancy during that time. Formal nomenclature of M. orygis was proposed in 2012 following both molecular and genomic analyses demonstrating that Oryx bacillus, while genetically related to other members of MTBC, also possessed distinct genomic signatures which satisfied the definition for a new subspecies (17). Since then, reported case numbers have gradually increased, particularly over the past 10 years (Figure 1) (18).
As of March 2024, M. orygis was reported in 14 countries with 127 human and 123 animal infections across 17 unique host species (Table 1 and Figure 2). The only continents where the organism has yet to be reported are South America and Antarctica. A significant proportion of cases (84/250 or 33.6%) have been reported from India, Pakistan, Nepal, and Bangladesh. Importantly, these cases are associated with a diverse host range, spanning multiple mammalian species (19, 20). In the Subcontinent, the identification and reporting of M. orygis cases was primarily done through prospective studies (21). At the same time, in non-endemic countries, retrospective analysis of archived samples accounted for a more significant proportion of reported cases (Table 1) Identifying the MTBC subspecies is often not a priority or is unavailable, leading to gaps in epidemiological data and unrecognized transmission pathways (22).

Figure 2. Global heat map of human and animal M. orygis infections reported between 1987 and 2024 using the data consolidated in Table 1. Cases are scaled according to the gradient bar and regions in gray indicate countries that have not yet reported any M. orygis infections.
Approximately 66% of reported M. orygis infections, whether in animals or humans, were diagnosed outside the TB endemic areas of South Asia. Of the 250 reported cases, 150 were reported in High-Income Countries (HIC) that are described traditionally as low-burden TB countries [<10/100,000 cases of TB] (23). Among the low-burden countries where M. orygis infections were reported, Canada, New Zealand, and the United Kingdom stood out, and interestingly, they all have endemic reservoirs of bovine tuberculosis. Concerningly, Canada and the USA, two of the most affected high-income countries, have recognized wild host species for M. orygis, including members of the Bovidae family (such as buffalo) and deer (24). Although low numbers of bovine TB have been reported in the USA and the Netherlands (<0.5% of total TB cases) (24, 25), and complete eradication of this disease has been achieved in Australia and Norway (24), the potential threat M. orygis poses to cattle populations within these countries cannot be dismissed. Furthermore, these unexpected cases have drawn significant attention from clinicians and epidemiologists. For instance, in 2012, a dairy worker from India apparently transmitted M. orygis to a cow in New Zealand, a country with a low incidence of TB (26). Similarly, in New York, USA, a female patient initially from South Asia developed lymph node TB secondary to an M. orygis infection (15). Such cases underscore the complex dynamics of M. orygis transmission, highlighting the need to track the pathogen’s ability to cross geographic and species boundaries.
Although global context on the prevalence of M. orygis remained understudied, certain geographical regions exhibit notably higher case densities, particularly in South Asia (Figures 2, 3) (27). For example, Duffy et al. showed that of 940 mycobacterial cultures collected between 2018 and 2019 in India the prevalence of M. orygis was 0.7%, surpassing M. bovis at 0.5%. Cultures identified as MTBC members other than M. tuberculosis, or those with inconclusive PCR results, were then further analyzed using whole-genome sequencing (WGS) to ensure accurate identification (28). This study provided a pivotal argument that zTB should be redefined to include other MTBC members and not just exclusively limited to M. bovis.
This regional concentration of M. orygis infections in Asia contrasts sharply with data from countries like the United States and Canada, where reported cases of M. orygis remain low. In the United States, approximately 9,000 cases of MTBC infection are reported annually, with the vast majority attributed to M. tuberculosis. However, between 2005 and 2017, data from New York State indicated a minimal incidence of M. orygis, though this likely does not provide a comprehensive representation of the national prevalence (29). The true prevalence is unknown, as these cases were sourced exclusively from New York State’s Health Department, potentially overlooking cases from the animal or agriculture sector.
Importation of infected animals into low TB incidence countries has been also a potential route for geographical spread highlighting the importance of quarantine measures to protect humans from potential exposure of zTB (29). Crossing of geographical borders, be it human movement via migration or import of animals, from countries with a high incidence of TB could be a plausible explanation for the of global spread of zTB caused by M. orygis. In Canada, a significant proportion of M. orygis cases (>70%) have been linked to individuals with South Asian origins. One study reported that all detected infections (21/21) were associated with this region (30). This trend aligns with the country’s diverse population and underscores the importance of understanding geographic and demographic patterns in M. orygis transmission, with approximately 57 cases reported in the last decade (8). Concerns also arise regarding zoonotic transmission, especially given the endemicity of known host species of M. orygis within Canada. In the USA, all reported macaque infections were linked to South Asian imports, and the nine human cases primarily originated in South Asia. This influx highlights the need for improved surveillance and detection measures. Canada’s experience with M. orygis disease has evolved notably since the first reported case in 1997. While only four cases were documented between 2007 and 2010, there has been a substantial rise, with 57 cases reported over the past decade (8). Similarly, the United Kingdom (UK), where all MTBC isolates responsible for human are routinely sequenced, has reported only 25 clinical isolates to date, suggesting a low prevalence or challenges with culture-confirmation of zTB in humans (31).
Despite significant advancements in identifying and reporting M. orygis cases, the true burden of this pathogen remains underrepresented due to diagnostic challenges and underreporting. The trend may suggest both progress and ongoing challenges in global TB control efforts, exacerbated by the aftermath of the SARS-CoV-2 pandemic. The spread of M. orygis highlights the critical need for continuous vigilance and the refinement of surveillance strategies to accurately assess the burden of this pathogen and mitigate its spread effectively.
From a total of 250 cases reported to date, 67 were diagnosed in cattle, 56 in ‘other mammalian hosts’ and 127 in humans, with an apparent disparity in host infections (Figures 3, 4). Instances of human infection occurred the most compared to any other host species, with humans having the highest relative frequency, accounting for half (51.2%) of reported cases. While the frequency is high for humans, it is difficult to rule out a sampling bias due to more active investigation and reporting of zTB in humans as opposed to TB in animals. Nonetheless, M. orygis has been isolated from the following animal hosts: Arabian Oryx, African Buffalo, Antelope, Bison, Blackbuck, Blue Bull, Buffalo, Camel, Cattle, Cynomolgus Macaque, Deer, East African Oryx, Gazelle, Greater One Horned Rhinoceros, Rhesus Monkey, Sambar, Spotted Deer, and Waterbuck (Table 1). Notably, all infections reported to date from the African continent (including animals or specimens imported into Europe that were identified subsequently) appeared to be in wild/ captured animals, specifically in the Bovidae family (17, 32, 33). Additional research is warranted to elucidate the transmission dynamics of M. orygis, from and within Africa, as current data remains limited.
Current evidence suggests that M. orygis infections cause typical TB-like symptoms with both latent and active TB forms documented (15, 25, 26). Of all reported cases in animals and humans with known manifestations (Table 1; 250 cases), 59% exhibited only pulmonary disease (PTB), 41% had extrapulmonary disease (EPTB), and 32% presented with both PTB and EPTB. In humans the distribution was relatively uniformed, with 52% presenting with PTB, 48% with EPTB, and 30% with both PTB and EPTB. The observation of a more significant proportion of PTB than EPTB in M. orygis infections is consistent with the literature concerning M. tuberculosis and M. africanum (34) but not consistent with M. bovis (26, 35). A potential risk for the development EPTB from M. orygis has been observed via the oral consumption route of M. orygis bacilli (36, 37).
Susceptibility to M. orygis infections of different gender remains poorly understood. Riopel et al. showed a higher frequency of female than male patients in human cases infected with M. orygis (28, 30). Of the cases to date where the gender of the host species was reported, 26.5% (27/102) were male, and 73.5% (75/102) were female The ratio becomes even more skewed upon examining only human cases, with 25.3% male (20/79) and 74.7% (59/79) female. This trend is not consistent with typical MTBC epidemiological data. A study conducted in Victoria, Australia, concluded from less than 5,000 cases that the ratio of male to female TB cases was [1.2:1] and that males generally seek diagnosis and treatment sooner (38). In 2016, a German study analyzed 5,915 cases of TB, indicating an incidence rate nationally of 7.2:100,000, but with males having an incidence more than double the female incidence [9.9:100,000 versus 4.6:100,000] (39). The reasons for the observed disparity in M. orygis infections remained unclear. While occupational and environmental exposures could play a role, particularly in regions where women have an increased contact with cattle, definitive conclusions cannot be drawn (40, 41). Further research is needed to determine whether the pattern reflected true biological susceptibility, differences in exposure or biases in infection reporting (41, 42).
Animal host-to-host transmission has been documented in the wild. Rahim et al. identified isolates of M. orygis from two captured rhesus monkeys and 18 dairy cows and showed the same ‘Mycobacterial interspersed repetitive unit-variable number tandem repeat’ [MIRU-VNTR] (43) pattern in both monkeys and 15 cattle, suggesting a possible spread of this strain within the area (19). Similarly, Lipworth et al. identified 24 isolates within the United Kingdom, identifying two clusters of probable human-to-human transmission, with one genomic cluster having zero single nucleotide polymorphism (SNP) difference amongst the genomes, and the other cluster having members with up to 6 SNP difference (31). One of the most notable transmission cases was the reverse zoonotic case in New Zealand observed by Dawson et al. (26). Even though this is a rare occurrence, it is not impossible, and clinically consistent with disease caused by M. tuberculosis that has been observed to occur as a reverse zoonotic transmission (44). The occurrence of reverse zoonosis (zooanthroponosis) (26) (Figure 4), challenges current definition of zTB. While the definition of zTB has been proposed to include all members of the MTBC, the bidirectional flow of mycobacterial infections between humans and animals emphasizes the need for a more inclusive and nuanced definition of zTB.
There is limited evidence on drug resistance in M. orygis in the literature. Sumanth et al. identified resistance-associated genes in M. orygis isolates in 3 out of 8 patients in southern India. One patient was infected by a M. orygis strain which had a frameshift in gid gene, suggesting resistance to streptomycin while possible hetero-resistance was detected in the remaining two (i.e., proportion of sequencing reads with resistance conferring mutations present in rrs, rplC and embB in one of the cases, gyrB gene in the other) (45). All isolates, however, appeared phenotypically susceptible to first line anti-tuberculous agents.
Treatment of M. orygis follows a standard MTBC treatment regime; first line anti-tuberculous drugs (rifampicin, isoniazid, pyrazinamide and ethambutol) are employed for disease caused by wild-type susceptible strains. Treatment could be extended for the disseminated disease when necessary, or second line anti-tuberculous agents used for more resistant strains (17, 45).
Differentiating strains within the MTBC, including M. orygis, has historically been a challenge, necessitating the development of specialized molecular diagnostic assays as phenotypic presentation and morphology of M. orygis is typical of MTBC members. An early differentiation within MTBC was performed using restriction fragment length polymorphism (RFLP) typing within IS6110 (16). However, this method has limited resolution and require a large amount of target DNA (46). MIRU-VNTR typing has also been critical in distinguishing M. orygis within the MTBC by identifying unique tandem repeat loci, such as the undetectable 2163b loci, which are characteristic of M. orygis strains (30). This method has effectively identified strain clusters in wildlife and livestock, highlighting distinct transmission dynamics and host adaptations (21, 47).
While members of MTBC are highly genetically related, each species carry insertions/deletions known as region of differences (RD) (17). RD analysis was long considered the ‘gold standard’ for MTBC identification and is still used in many centers. Similar to other MTBC species, such as M. caprae and M. bovis, M. orygis lacks specific regions, RD7, RD8, RD9, RD10, and unique regions RDcap, RDbovis and RDoryx1. Moreover, M. orygis has preserved regions of difference, including RD1, RD2, RD4, RD5a, RD6, RD13, and N-RD25bovis/caprae. The presence of RD12 in ‘oryx bacillus’ was reported in one earlier paper, although Van Ingen et al. and Rahim et al. found a deletion (17, 19). Clarification via NCBI and the European Nucleotide Archive confirmed the deletion of RD12 but affirmed the presence of RD1, RD2, RD4, RD5a, RD6, and RD13 in M. orygis. All major MTBC RD differences have been laid out (Table 2). Importantly, there are more specific regions within M. orygis genome which can be targeted for further speciation such as RD2seal, RD12oryx and RDcap_Spain1 (9), among others. RD analysis can also be used in conjunction with SNPs to clarify speciation (9, 48, 49).
Whole Genome Sequencing (WGS) provides the ultimate high-resolution identification of MTBC species and insights into the phylogeny and the evolution within the MTBC. As of Nov 2024, there were 153 M. orygis genomic sequences uploaded onto public repositories. Inferences into the phylogeny of M. orygis revealed genomic diversity with pairwise SNP differences ranging from 0 to 523 core SNPS. Interestingly, genomes that clustered together (0 SNPs) did not always show stratification by host, suggesting a complex transmission dynamic that warrants further investigation (Figure 5). The analysis of virulence genes has pinpointed crucial loci within the ESX-1 secretion system, including the ESAT-6 and CFP-10 genes, which contribute to pathogenicity in M. orygis (50), namely, disruption of host cell membranes and regulation of host immune response (51, 52).
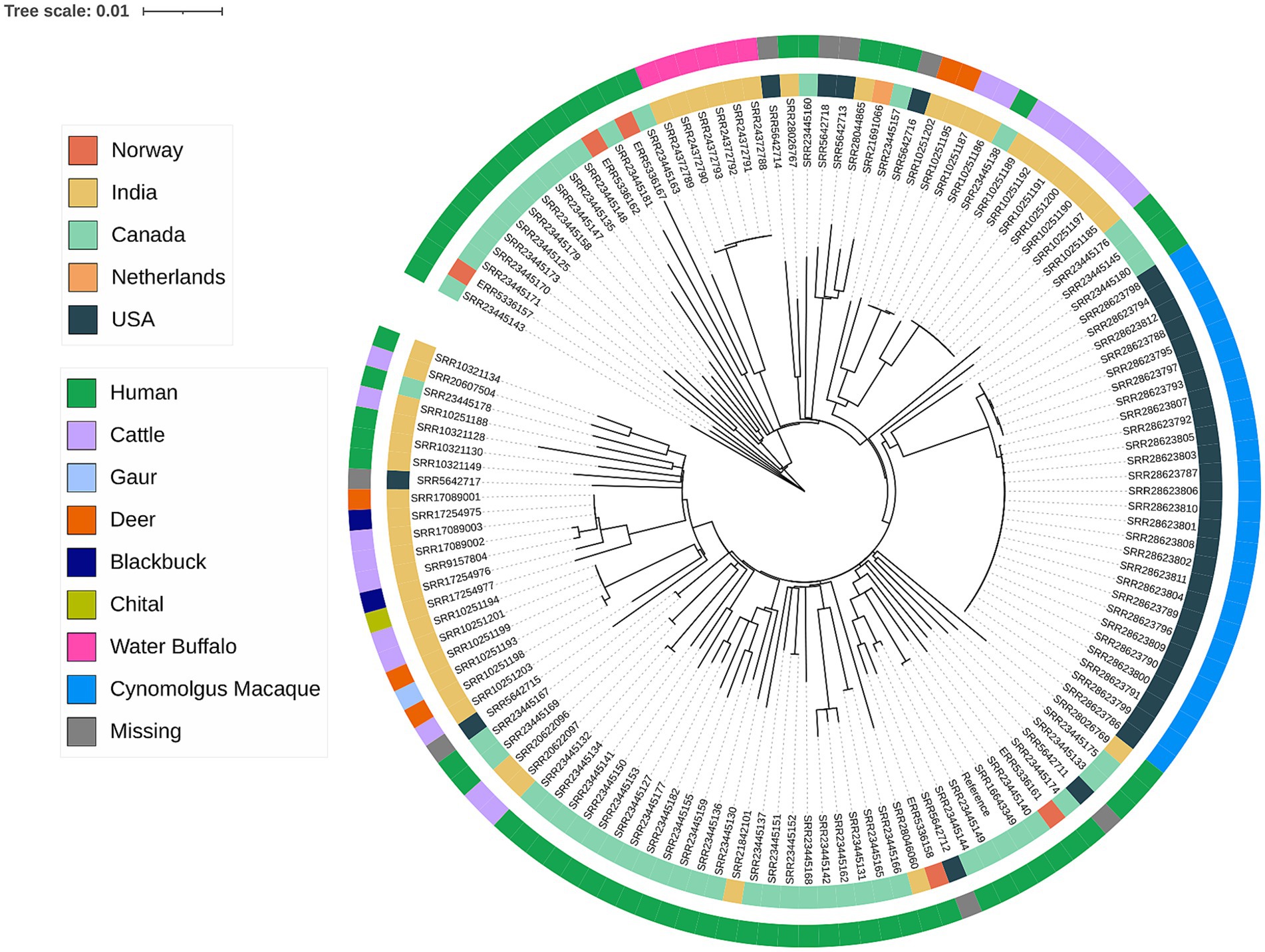
Figure 5. Phylogenetic tree of available M. orygis genomes. Sequencing reads or contigs were aligned to M. orygis strain 51,145 (GenBank accession CP063804) as the reference. Genomes that aligned to less than 90% of the reference were omitted. A maximum-likelihood tree was inferred from the whole genome SNP alignment using IQ-TREE version 2.3.6 using the GTR + F + R2 model with 1,000 replicates for ultrafast bootstrap. Phylogenetic tree was generated using I-TOL (https://itol.embl.d).
The continuing advancement of sequencing technologies and accessibility of WGS have aided our understanding of zTB, paving the way for faster diagnostics and genomic surveillance. Targeted metagenomics was recently endorsed by the WHO as a method for rapid and accurate TB diagnosis (53). This style of sequencing focuses on specific genomic regions, such as 16S rRNA or pathogen-specific SNPs, to provide precise insights into microbial diversity and adaptations in less time than conventional culture-based methods. M. orygis-specific SNPs incorporated into targeted metagenomics offers a high-resolution diagnostic tool for its identification and characterization (54). Additionally, adapting RD-based SNPs into a PCR panel could further improve targeted diagnostics in the short term, enabling accurate detection and analysis of genetic variations.
In conclusion, there is increasing evidence of M. orygis emergence and spread as a human and zoonotic pathogen, particularly in South Asia. The understanding of the organism’s transmission and magnitude of its emergence is complicated by the complexities of tuberculosis epidemiology and the challenges in accurate diagnosis and reporting. The higher frequency of female patients in M. orygis infections, in contrast to M. tuberculosis, suggests possible distinct risk factors, highlighting the need for further research into the epidemiology of the disease. Furthermore, there are concerns that M. orygis may be circulating in wild animal and livestock populations with significant implications for the transmission and spread of the disease within domesticated and food producing animals. To address these challenges, it is essential to prioritize investment into enhance laboratory diagnosis, surveillance strategies for zTB and epidemiological studies. Strengthening these efforts will be key in controlling the spread of zTB and alleviating its impact on both human and animal populations globally.
BH: Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft. ES: Conceptualization, Investigation, Methodology, Resources, Supervision, Validation, Visualization, Writing – review & editing. TC: Data curation, Methodology, Project administration, Resources, Validation, Writing – review & editing. VS: Conceptualization, Funding acquisition, Methodology, Supervision, Writing – review & editing, Data curation, Formal analysis, Investigation, Project administration, Resources, Software, Validation, Visualization, Writing – original draft.
The author(s) declare that no financial support was received for the research and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declare that no Gen AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Bhargava, A, and Bhargava, M. Tuberculosis deaths are predictable and preventable: comprehensive assessment and clinical care is the key. J Clin Tuberc Other Mycobact Dis. (2020) 19:100155. doi: 10.1016/j.jctube.2020.100155
2. Barberis, I, Bragazzi, NL, Galluzzo, L, and Martini, M. The history of tuberculosis: from the first historical records to the isolation of Koch's bacillus. J Prev Med Hyg. (2017) 58:E9–E12. doi: 10.15167/2421-4248/jpmh2017.58.1.728
3. Centers for Disease Control and Prevention. Tuberculosis (TB). Centers for Disease Control and Prevention. (2023). Available online at: https://www.cdc.gov/tb/default.htm (Accessed January 14, 2025).
4. World Health Organization. Global tuberculosis report 2024. World Health Organization. (2024). Available online at: https://www.who.int/teams/global-tuberculosis-programme/tb-reports/global-tuberculosis-report-2024 (Accessed January 14, 2025).
5. Basnyat, B, Caws, M, and Udwadia, Z. Tuberculosis in South Asia: a tide in the affairs of men. Multidiscip Respir Med. (2018) 13:1–8. doi: 10.1186/s40248-018-0122-y
6. Lönnroth, K, Jaramillo, E, Williams, BG, Dye, C, and Raviglione, M. Drivers of tuberculosis epidemics: the role of risk factors and social determinants. Soc Sci Med. (2009) 68:2240–6. doi: 10.1016/j.socscimed.2009.03.041
7. Spence, DP, Hotchkiss, J, Williams, CS, and Davies, PD. Tuberculosis and poverty. Br Med J. (1993) 307:759–61. doi: 10.1136/bmj.307.6907.759
8. Islam, MR, Sharma, MK, Khun Khun, R, Shandro, C, Sekirov, I, Tyrrell, GJ, et al. Whole genome sequencing-based identification of human tuberculosis caused by animal-lineage Mycobacterium orygis. J Clin Microbiol. (2023) 61:e00260–23. doi: 10.1128/jcm.00260-23
9. Bespiatykh, D, Bespyatykh, J, Mokrousov, I, and Shitikov, E. A comprehensive map of mycobacterium tuberculosis complex regions of difference. Msphere. (2021) 6:10–128. doi: 10.1128/mSphere.00535-21
10. Coscolla, M. Biological and epidemiological consequences of MTBC diversity. In: Gagneux S, (editor). Strain variation in the Mycobacterium tuberculosis complex: its role in biology, epidemiology and control Vol. 1019. Cham: Advances in Experimental Medicine and Biology, Springer (2017) p. 95–116.
11. Kock, R, Michel, AL, Yeboah-Manu, D, Azhar, EI, Torrelles, JB, Cadmus, SI, et al. Zoonotic tuberculosis–the changing landscape. Int J Infect Dis. (2021) 113:S68–72. doi: 10.1016/j.ijid.2021.02.091
12. Duffy, SC, Marais, B, Kapur, V, and Behr, MA. Zoonotic tuberculosis in the 21st century. Lancet Infect Dis. (2024) 24:339–41. doi: 10.1016/S1473-3099(24)00059-8
13. World Health Organization. Roadmap for zoonotic tuberculosis. (2017). Available online at: https://www.who.int/publications/i/item/9789241513043 (Accessed January 14, 2025).
14. Lomme, JR, Thoen, CO, Himes, EM, Vinson, JW, and King, RE. Mycobacterium tuberculosis infection in two east African oryxes. J Am Vet Med Assoc. (1976) 169:912–4.
15. Marcos, LA, Spitzer, ED, Mahapatra, R, Ma, Y, Halse, TA, Shea, J, et al. Mycobacterium orygis lymphadenitis in New York, USA. Emerg Infect Dis. (2017) 23:1749–51. doi: 10.3201/eid2310.170490
16. Van Soolingen, D, De Haas, PE, Haagsma, J, Eger, T, Hermans, PW, Ritacco, V, et al. Use of various genetic markers in differentiation of Mycobacterium bovis strains from animals and humans and for studying epidemiology of bovine tuberculosis. J Clin Microbiol. (1994) 32:2425–33. doi: 10.1128/jcm.32.10.2425-2433.1994
17. van Ingen, J, Rahim, Z, Mulder, A, Boeree, MJ, Simeone, R, Brosch, R, et al. Characterization of Mycobacterium orygis as M. tuberculosis complex subspecies. Emerg Infect Dis. (2012) 18:653–5. doi: 10.3201/eid1804.110888
18. Thapa, J, Gordon, SV, Nakajima, C, and Suzuki, Y. Threat from Mycobacterium orygis-associated tuberculosis in South Asia. Lancet Microbe. (2022) 3:e641–2. doi: 10.1016/S2666-5247(22)00149-5
19. Rahim, Z, Thapa, J, Fukushima, Y, Van Der Zanden, AG, Gordon, SV, Suzuki, Y, et al. Tuberculosis caused by Mycobacterium orygis in dairy cattle and captured monkeys in Bangladesh: a new scenario of tuberculosis in South Asia. Transbound Emerg Dis. (2017) 64:1965–9. doi: 10.1111/tbed.12596
20. Refaya, AK, Ramanujam, H, Ramalingam, M, Rao, GV, Ravikumar, D, Sangamithrai, D, et al. Tuberculosis caused by Mycobacterium orygis in wild ungulates in Chennai, South India. Transbound Emerg Dis. (2022) 69:e3327–33. doi: 10.1111/tbed.14613
21. Thapa, J, Paudel, S, Sadaula, A, Shah, Y, Maharjan, B, Kaufman, GE, et al. Mycobacterium orygis–associated tuberculosis in free-ranging rhinoceros, Nepal, 2015. Emerg Infect Dis. (2016) 22:570–2. doi: 10.3201/eid2203.151929
22. Figueredo, LJ, Miranda, SS, Santos, LB, Manso, CG, Soares, VM, Alves, S, et al. Cost analysis of smear microscopy and the Xpert assay for tuberculosis diagnosis: average turnaround time. Rev Soc Bras Med Trop. (2020) 53:e20200314. doi: 10.1590/0037-8682-0314-2020
23. World Health Organization. TB incidence. World Health Organization. Available online at: https://www.who.int/teams/global-tuberculosis-programme/tb-reports/global-tuberculosis-report-2023/tb-disease-burden/1-1-tb-incidence (Accessed January 14, 2025).
24. De la Rua-Domenech, R. Human Mycobacterium bovis infection in the United Kingdom: incidence, risks, control measures and review of the zoonotic aspects of bovine tuberculosis. Tuberculosis. (2006) 86:77–109. doi: 10.1016/j.tube.2005.05.002
25. Lombard, JE, Patton, EA, Gibbons-Burgener, SN, Klos, RF, Tans-Kersten, JL, Carlson, BW, et al. Human-to-cattle Mycobacterium tuberculosis complex transmission in the United States. Front Vet Sci. (2021) 8:691192. doi: 10.3389/fvets.2021.691192
26. Dawson, KL, Bell, A, Kawakami, RP, Coley, K, Yates, G, and Collins, DM. Transmission of Mycobacterium orygis (M. tuberculosis complex species) from a tuberculosis patient to a dairy cow in New Zealand. J Clin Microbiol. (2012) 50:3136–8. doi: 10.1128/JCM.01652-12
27. Rufai, SB, McIntosh, F, Poojary, I, Chothe, S, Sebastian, A, Albert, I, et al. Complete genome sequence of Mycobacterium orygis strain 51145. Microbiol Resource Announc. (2021) 10:10–128. doi: 10.1128/MRA.01279-20
28. Duffy, SC, Srinivasan, S, Schilling, MA, Stuber, T, Danchuk, SN, Michael, JS, et al. Zoonotic tuberculosis in India: looking beyond Mycobacterium bovis. bioRxiv [Preprint]. (2019). doi: 10.1101/847715
29. Swisher, SD. Outbreak of Mycobacterium orygis in a shipment of Cynomolgus macaques imported from Southeast Asia—United States, February–May 2023. MMWR Morb Mortal Wkly Rep. (2024) 73:145–8. doi: 10.15585/mmwr.mm7307a2
30. Riopel, ND, Long, R, Heffernan, C, Tyrrell, GJ, Shandro, C, Li, V, et al. Characterization of Mycobacterium orygis, Mycobacterium bovis, and Mycobacterium caprae infections in humans in Western Canada. J Infect Dis. (2024) 230:e789–97. doi: 10.1093/infdis/jiae124
31. Lipworth, S, Jajou, R, De Neeling, A, Bradley, P, Van Der Hoek, W, Maphalala, G, et al. SNP-IT tool for identifying subspecies and associated lineages of Mycobacterium tuberculosis complex. Emerg Infect Dis. (2019) 25:482–8. doi: 10.3201/eid2503.180894
32. Pittius, NC, Perrett, KD, Michel, AL, Keet, DF, Hlokwe, T, Streicher, EM, et al. Infection of African buffalo (Syncerus caffer) by oryx bacillus, a rare member of the antelope clade of the Mycobacterium tuberculosis complex. J Wildl Dis. (2012) 48:849–57. doi: 10.7589/2010-07-178
33. Parsons, SD, Miller, MA, and van Helden, PD. The Mycobacterium tuberculosis complex in Africa In: Dibaba AB, Kriek NPJ, Thoen CO, (editors). Tuberculosis in animals: an African perspective. Cham: Springer (2019). 73–86.
34. Arnedo-Pena, A, Romeu-Garcia, MA, Meseguer-Ferrer, N, Vivas-Fornas, I, Vizcaino-Batllés, A, Safont-Adsuara, L, et al. Pulmonary versus extrapulmonary tuberculosis associated factors: a case-case study. Microbiol Insights. (2019) 12:1178636119840362. doi: 10.1177/1178636119840362
35. Majoor, CJ, Magis-Escurra, C, van Ingen, J, Boeree, MJ, and van Soolingen, D. Epidemiology of Mycobacterium bovis disease in humans, the Netherlands, 1993–2007. Emerg Infect Dis. (2011) 17:457–63. doi: 10.3201/eid1703.101111
36. Behr, M, Danchuk, S, Duffy, S, Sullivan, J, Rufai, SB, McIntosh, F, et al. M. orygis hypervirulence provides new insights into tuberculosis pathogenesis. Res Square [Preprint] (2024).
37. Duffy, SC. Epidemiology and pathogenesis of bovine mycobacteria [dissertation]. Montreal: McGill University Montreal (2023).
38. Dale, K, Tay, E, Trauer, JM, Trevan, P, and Denholm, J. Gender differences in tuberculosis diagnosis, treatment and outcomes in Victoria, Australia, 2002–2015. Int J Tuberc Lung Dis. (2017) 21:1264–71. doi: 10.5588/ijtld.17.0338
39. Brodhun, B, Altmann, D, Hauer, B, and Haas, W. Bericht zur Epidemiologie der Tuberkulose in Deutschland für. Robert Koch Institute (2020).
40. Ishaq, W, and Memon, SQ. Roles of women in agriculture: a case study of rural Lahore, Pakistan. JSSH. (2016) 1:1–11.
41. Hatherall, B, Newell, JN, Emmel, N, Baral, SC, and Khan, MA. “Who will marry a diseased girl?” marriage, gender, and tuberculosis stigma in Asia. Qual Health Res. (2019) 29:1109–19. doi: 10.1177/1049732318812427
42. Srinivasan, S, Easterling, L, Rimal, B, Niu, XM, Conlan, AJ, Dudas, P, et al. Prevalence of bovine tuberculosis in India: a systematic review and meta-analysis. Transbound Emerg Dis. (2018) 65:1627–40. doi: 10.1111/tbed.12915
43. De Beer, JL, Kremer, K, Ködmön, C, Supply, P, and Van Soolingen, D. Global network for the molecular surveillance of tuberculosis 2009. First worldwide proficiency study on variable-number tandem-repeat typing of Mycobacterium tuberculosis complex strains. J Clin Microbiol. (2012) 50:662–9. doi: 10.1128/JCM.00607-11
44. Stephens, N, Vogelnest, L, Lowbridge, C, Christensen, A, Marks, GB, Sintchenko, V, et al. Transmission of Mycobacterium tuberculosis from an Asian elephant (Elephas maximus) to a chimpanzee (Pan troglodytes) and humans in an Australian zoo. Epidemiol Infection. (2013) 141:1488–97. doi: 10.1017/S095026881300068X
45. Sumanth, LJ, Suresh, CR, Venkatesan, M, Manesh, A, Behr, MA, Kapur, V, et al. Clinical features of human tuberculosis due to Mycobacterium orygis in southern India. J Clin Tuberc Other Mycobact Dis. (2023) 32:100372. doi: 10.1016/j.jctube.2023.100372
46. Gori, A, Bandera, A, Marchetti, G, Degli Esposti, A, Catozzi, L, Nardi, GP, et al. Spoligotyping and Mycobacterium tuberculosis. Emerg Infect Dis. (2005) 11:1242–8. doi: 10.3201/eid1108.040982
47. Guimaraes, AM, and Zimpel, CK. Mycobacterium bovis: from genotyping to genome sequencing. Microorganisms. (2020) 8:667. doi: 10.3390/microorganisms8050667
48. Smith, NH, Kremer, K, Inwald, J, Dale, J, Driscoll, JR, Gordon, SV, et al. Ecotypes of the Mycobacterium tuberculosis complex. J Theor Biol. (2006) 239:220–5. doi: 10.1016/j.jtbi.2005.08.036
49. Napier, G, Campino, S, Merid, Y, Abebe, M, Woldeamanuel, Y, Aseffa, A, et al. Robust barcoding and identification of Mycobacterium tuberculosis lineages for epidemiological and clinical studies. Genome Med. (2020) 12:1. doi: 10.1186/s13073-020-00817-3
50. Mukherjee, A, Dandapat, P, Haque, MZ, Mandal, S, Jana, PS, Samanta, S, et al. Computational analysis of hypothetical proteins from Mycobacterium orygis identifies proteins with therapeutic and diagnostic potentials. Anim Gene. (2023) 29:200154. doi: 10.1016/j.angen.2023.200154
51. Anes, E, Pires, D, Mandal, M, and Azevedo-Pereira, JM. ESAT-6 a major virulence factor of Mycobacterium tuberculosis. Biomol Ther. (2023) 13:968. doi: 10.3390/biom13060968
52. Kanabalan, RD, Lee, LJ, Lee, TY, Chong, PP, Hassan, L, Ismail, R, et al. Human tuberculosis and Mycobacterium tuberculosis complex: a review on genetic diversity, pathogenesis and omics approaches in host biomarkers discovery. Microbiol Res. (2021) 246:126674. doi: 10.1016/j.micres.2020.126674
53. World Health Organization. WHO operational handbook on tuberculosis. Module 3: diagnosis-rapid diagnostics for tuberculosis detection World Health Organization (2024).
54. Cole, S, Brosch, R, Parkhill, J, Garnier, T, Churcher, C, Harris, D, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. (1998) 396:190. doi: 10.1038/24206
55. Lavender, CJ, Globan, M, Kelly, H, Brown, LK, Sievers, A, Fyfe, JA, et al. Epidemiology and control of tuberculosis in Victoria, a low-burden state in South-Eastern Australia, 2005–2010. Int J Tuberc Lung Dis. (2013) 17:752–8. doi: 10.5588/ijtld.12.0791
56. Refaya, AK, Kumar, N, Raj, D, Veerasamy, M, Balaji, S, Shanmugam, S, et al. Whole-genome sequencing of a Mycobacterium orygis strain isolated from cattle in Chennai, India. Microbiol Resource Announc. (2019) 8:10–128. doi: 10.1128/MRA.01080-19
57. Karthik, K, Subramanian, S, Vinoli Priyadharshini, M, Jawahar, A, Anbazhagan, S, Kathiravan, RS, et al. Whole genome sequencing and comparative genomics of Mycobacterium orygis isolated from different animal hosts to identify specific diagnostic markers. Front Cell Infect Microbiol. (2023) 13:1302393. doi: 10.3389/fcimb.2023.1302393
58. Prajapati, SK. Detection of tuberculous mycobacteria (M. orygis) and non-tuberculous Mycobacteria (M. smegmatis & M. kansasii) in cattle and buffaloes. [Doctoral dissertation]. Ludhiana: Guru Angad Dev Veterinary and Animal Sciences University (2021).
59. Sharma, M, Mathesh, K, Dandapat, P, Mariappan, AK, Kumar, R, Kumari, S, et al. Emergence of Mycobacterium orygis–associated tuberculosis in wild ruminants, India. Emerg Infect Dis. (2023) 29:661–3. doi: 10.3201/eid2903.221228
60. Jawahar, A, Raj, GD, Pazhanivel, N, and Karthik, K. Gross and histopathological features of tuberculosis in cattle, buffalo and spotted deer (Axis axis) caused by Mycobacterium orygis. J Comp Pathol. (2024) 208:15–9. doi: 10.1016/j.jcpa.2023.10.010
61. Thapa, J, Nakajima, C, Maharjan, B, Poudel, A, and Suzuki, Y. Molecular characterization of Mycobacterium orygis isolates from wild animals of Nepal. Jpn J Vet Res. (2015) 63:151–8. doi: 10.14943/jjvr.63.3.151
62. Huard, RC, Fabre, M, de Haas, P, Claudio Oliveira Lazzarini, L, van Soolingen, D, Cousins, D, et al. Novel genetic polymorphisms that further delineate the phylogeny of the Mycobacterium tuberculosis complex. J Bacteriol. (2006) 188:4271–87. doi: 10.1128/JB.01783-05
63. Brosch, R, Gordon, SV, Marmiesse, M, Brodin, P, Buchrieser, C, Eiglmeier, K, et al. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc Natl Acad Sci. (2002) 99:3684–9. doi: 10.1073/pnas.052548299
64. Eldholm, V, Rønning, JO, Mengshoel, AT, and Arnesen, T. Import and transmission of Mycobacterium orygis and Mycobacterium africanum, Norway. BMC Infect Dis. (2021) 21:562. doi: 10.1186/s12879-021-06269-3
65. Maqsood, R, Duffy, SC, Bin Rashid, H, Gill, SS, Jabeen, C, Arshad, N, et al. Molecular detection and characterization of the Mycobacterium tuberculosis complex subspecies responsible for bovine tuberculosis in Punjab, Pakistan. Microbiol Spectr. (2024) 12:e02692-23. doi: 10.1128/spectrum.02692-23
66. Hlokwe, TM, and Mogano, RM. Utility of xpert® MTB/RIF ultra assay in the rapid diagnosis of bovine tuberculosis in wildlife and livestock animals from South Africa. Preventive Vet Med. (2020) 177:104980. doi: 10.1016/j.prevetmed.2020.104980
67. Mostowy, S, Inwald, J, Gordon, S, Martin, C, Warren, R, Kremer, K, et al. Revisiting the evolution of Mycobacterium bovis. J Bacteriol. (2005) 187:6386–95. doi: 10.1128/JB.187.18.6386-6395.2005
68. Kinne, J, Johnson, B, Jahans, KL, Smith, NH, Ul-Haq, A, and Wernery, U. Camel tuberculosis–a case report. Trop Anim Health Prod. (2006) 38:207–13. doi: 10.1007/s11250-006-4366-8
69. Wernery, U, Kinne, J, Jahans, KL, Vordermeier, HM, Esfandiari, J, Greenwald, R, et al. Tuberculosis outbreak in a dromedary racing herd and rapid serological detection of infected camels. Vet Microbiol. (2007) 122:108–15. doi: 10.1016/j.vetmic.2007.01.012
70. Love, DM, Garner, MM, Lyashchenko, KP, Sikar-Gang, A, Bradway, DS, Robbe-Austerman, S, et al. Tuberculosis caused by Mycobacterium orygis in a greater one-horned rhinoceros (Rhinoceros unicornis): first report in the western hemisphere. J Zoo Wildl Med. (2020) 50:1000–4. doi: 10.1638/2018-0084
Keywords: Mycobacterium orygis , tuberculosis, zoonotic infections, genomics, epidemiology
Citation: Hugh BT, Sim EM, Crighton T and Sintchenko V (2025) Emergence of Mycobacterium orygis: novel insights into zoonotic reservoirs and genomic epidemiology. Front. Public Health. 13:1568194. doi: 10.3389/fpubh.2025.1568194
Received: 29 January 2025; Accepted: 05 March 2025;
Published: 19 March 2025.
Edited by:
Maria Emilia Eirin, CONICET Institute of Agrobiotechnology and Molecular Biology (IABIMO), ArgentinaReviewed by:
Ariel H. Magallon-Tejada, Gorgas Memorial Institute of Health Studies, PanamaCopyright © 2025 Hugh, Sim, Crighton and Sintchenko. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Benedict T. Hugh, QmVuZWRpY3QuSHVnaEBoZWFsdGgubnN3Lmdvdi5hdQ==; Vitali Sintchenko, dml0YWxpLnNpbnRjaGVua29AaGVhbHRoLm5zdy5nb3YuYXU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.